Abstract
CrkRS (Cdc2-related kinase, Arg/Ser), or cyclin-dependent kinase 12 (CKD12), is a serine/threonine kinase believed to coordinate transcription and RNA splicing. While CDK12/CrkRS complexes were known to phosphorylate the C-terminal domain (CTD) of RNA polymerase II (RNA Pol II), the cyclin regulating this activity was not known. Using immunoprecipitation and mass spectrometry, we identified a 65-kDa isoform of cyclin K (cyclin K1) in endogenous CDK12/CrkRS protein complexes. We show that cyclin K1 complexes isolated from mammalian cells contain CDK12/CrkRS but do not contain CDK9, a presumed partner of cyclin K. Analysis of extensive RNA-Seq data shows that the 65-kDa cyclin K1 isoform is the predominantly expressed form across numerous tissue types. We also demonstrate that CDK12/CrkRS is dependent on cyclin K1 for its kinase activity and that small interfering RNA (siRNA) knockdown of CDK12/CrkRS or cyclin K1 has similar effects on the expression of a luciferase reporter gene. Our data suggest that cyclin K1 is the primary cyclin partner for CDK12/CrkRS and that cyclin K1 is required to activate CDK12/CrkRS to phosphorylate the CTD of RNA Pol II. These properties are consistent with a role of CDK12/CrkRS in regulating gene expression through phosphorylation of RNA Pol II.
INTRODUCTION
Cyclin-dependent kinases (CDKs) regulate a variety of cellular processes, including cell cycle progression, transcription, and RNA processing (52). Their activity requires heterodimeric interactions with specific cyclins where the CDK is the catalytic subunit and the cyclin is the regulatory subunit (52). CDKs/cyclins typically are the nexus of signal transduction pathways for the regulation of central cellular processes.
Many of the ∼20 human CDKs, such as CDK1, -2, -3, -4, -5, -6, -10, and -11p58, which partner with members of the cyclin A, B, C, D, and E families, are involved in cell cycle progression (3, 43, 52). Cyclins A, B, D, and E, which interact with CDK1, -2, -3, and -6, show fluctuating expression levels during different phases of the cell cycle (25). CDK7, -8, -9, and -11 are involved in transcriptional regulation (20). CDK9, CDK11p110, CDK12/CrkRS (Cdc2-related kinase, Arg/Ser), and CDK13/CDC2L5 have been reported to have roles in both transcription and pre-mRNA processing (4, 6, 9, 10, 19, 26, 37, 38). The multiple functions of CDKs are due, in part, to their interactions with different cyclin partners which modulate their localization and activity (15, 42, 67).
Previous studies have demonstrated a link between transcription and pre-mRNA processing (29, 45, 47). The multisubunit RNA polymerase II (RNA Pol II) links transcription and cotranscriptional maturation of pre-mRNA through differential recruitment of transcription and RNA processing factors to the C-terminal domain (CTD) of its largest subunit, Rbp1 (24, 37). The integration of these two processes is tightly regulated by posttranslational modification of the CTD. CDK7, -8, and -9 differentially phosphorylate the amino acid residues in the heptad repeat of the CTD to regulate the activation of elongation during transcription. The CTD, which in humans consists of 52 tandem heptaresidue repeats with a consensus sequence of Tyr1Ser2Pro3Thr4Ser5Pro6Ser7, serves as the recognition platform for interaction with a variety of transcription factors, cofactors, and pre-mRNA processing factors (18, 37).
In mammals, positive transcription elongation factor b (P-TEFb) complexes are composed of CDK9 as the catalytic subunit and one of the cyclins T1, T2a, T2b, or K (48). The ratio of two CDK9 isoforms, CDK9p42 and CDK9p55, generated through alternative transcription start sites (56, 57), is different in various tissues and changes when cells are challenged by different stimuli (40, 56). Both CDK9 isoforms interact with cyclins T1, T2a/b, and K; these different CDK9/cyclin heterodimers add cellular functional complexity to P-TEFb complexes (15, 23, 53, 58, 65, 67). For example, interaction of CDK9p42/cyclin T1 with the viral Tat protein facilitates the transactivation of the long terminal repeat of human immunodeficiency virus type 1 (HIV-1) and HIV-2 and is essential for productive viral replication (22). Recently, cyclin K was found to inhibit HIV-1 gene expression and replication through its interaction with HIV-1 Nef (32). CDK9/cyclin T2 complexes interact with PKNα and Rho-activated Ser/Thr kinase and are involved in regulating terminal differentiation in muscle cells through activation of Myo-D (12, 58). CDK9/cyclin K complexes have also been proposed to function in replication stress response to maintain genome integrity (65). Thus, the interaction of CDK9 with specific cyclins modulates CDK9 kinase activity to effect different cellular outcomes (15, 65, 67).
Cyclin K was originally identified as a 357-residue protein that restores cell cycle progression and rescues the lethality of deletions of the G1 cyclins CLN1, CLN2, and CLN3 in Saccharomyces cerevisiae (17). Cyclin K was independently identified in a yeast two-hybrid screen for interactors of human CDK9 (23). CDK9/cyclin K complexes can phosphorylate the CTD of RNA Pol II in vitro and can functionally substitute for CDK9/cyclin T1 in in vitro transcription assays (17, 23, 39). More recent studies have suggested that a larger isoform of cyclin K interacts with CDK12 and CDK13 (6, 13). Although many aspects of cyclin K function remain to be elucidated, it appears that its gene can be regulated by p53 and that it can function in DNA damage response and endoderm development (6, 46, 65, 67).
CTD kinases are evolutionarily conserved in eukaryotes. In S. cerevisiae, there are two CDK homologs with high similarity to CDK9; the Ctk1/Ctk2 (CTDK-1) complex and the Bur1/Bur2 complex (7, 49, 64). It was assumed that CDK9 performs the functions of CTDK-1 and Bur1 in higher eukaryotes (63). However, phylogenetic studies suggest that Bur1 is homologous with CDK9, while Ctk1 is homologous with CDK12/CrkRS and CDK13/CDC2L5 (28, 41). Supporting this observation, it was recently shown that Drosophila melanogaster CDK12/cyclin K appears to be a transcription elongation phase CTD kinase (4). Although the kinase domains of CDK12/CrkRS and CDK13/CDC2L5 are very similar to that of Ctk-1, Drosophila CDK12 (dCDK12) and human CDK12/CrkRS and CDK13/CDC2L5 are much larger proteins with additional structural features that suggest functions in signal transduction and pre-mRNA processing. Both CDK12/CrkRS and CDK13/CDC2L5 have N-terminal arginine/serine (RS) domains which are frequently found in the SR family of RNA processing factors (35, 55). In addition, they contain proline-rich regions and several potential SRC homology 3 (SH3) domains, both of which could recruit protein interaction partners (35). SH3 domains are well-characterized protein interaction modules frequently found in proteins that are involved in signaling pathways regulating a wide variety of biological functions (44). Previous studies have shown that dCDK12 and human CDK12/CrkRS and CDK13/CDC2L5 can phosphorylate the CTD of RNA Pol II and that CDK12/CrkRS colocalizes with a hyperphosphorylated form of RNA Pol II (4, 35). Coexpression experiments suggested that CDK12/CrkRS and CDK13/CDC2L5 can interact with cyclin L, an RS domain-containing cyclin, to facilitate alternative splicing of a model substrate (9, 10). CDK13/CDC2L5 has been shown to interact with ASF/SF2 to affect pre-RNA splicing in vivo (19). These observations make CrkRS/CDK12 and CDK13/CDC2L5 well suited for the regulation and coordination of transcription and pre-mRNA processing.
We employed immunoprecipitation-mass spectrometry (IP-MS) in an unbiased approach to study CDK/cyclin interactions of CDK12/CrkRS, CDK9, and CDK11p110. We provide transcriptome and protein evidence by mass spectrometry for the expression of a 65-kDa form of cyclin K in human-derived cell lines. We also employ data from enzymatic, small interfering RNA (siRNA), and promoter-reporter assays to establish that CDK12/CrkRS functionally requires cyclin K1 to activate its CDK kinase functions.
MATERIALS AND METHODS
Plasmids and RT-PCR.
CDK12 isoform 1 (GenBank accession number NM_016507.2), CDK9 (GenBank accession number NM_001261.3), cyclin L1 (GenBank accession number NM_020307.2), and cyclin K2 (GenBank accession number NM_003858.3) were obtained from the Mammalian Gene Collection through Open Biosystems and cloned into the Creator System (11). cDNAs were then transferred by Cre recombination into mammalian expression vectors with 3×FLAG or double Myc (dMyc) epitope tags. Cyclin T1 cDNA was generated by reverse transcription (RT)-PCR (RT-PCR kit, catalog number 12574-030; Invitrogen) from HeLa total RNA (Ambion; catalog number AM7852) using cyclin T1 primers 5′-CTGCCTTCTGGTTGAAGCAC-3′ and 5′-CTTAGGAAGGGGTGGAAGTGGTG-3′. The cyclin T1 sequence we obtained matched the cyclin T1 reference sequence (GenBank sequence accession number NM_001240.2). The cyclin K1 cDNA was also generated by RT-PCR from HeLa mRNA using primers designed to the 5′ ATG start site (primer 3, 5′-ATGAAGGAGAATAAAGAAAATTC-3′) and the 3′ untranslated region (UTR) (primer 2, 5′-TACCCTGCTGCTTACCCACT-3′). To distinguish between cyclin K1 and cyclin K2 isoform cDNAs, RT-PCR primer 1 (5′-CATCTGCCACCAAATCCTG-3′) was used. The cyclin T1 and K1 cDNAs were cloned into the Creator System as described above and expressed in mammalian cells with either a 3×FLAG or dMyc epitope tag. RNA Pol II CTD cDNA was generated by RT-PCR using HeLa total RNA (Ambion; catalog number AM7852) with primers 5′-GCCATGTCTCCCAGCTACTCG-3′ and 5′-TCAGTTCTCCTCGTCACTGTCATC-3′ and cloned into a glutathione S-transferase (GST) expression vector (Creator System plasmid V1544) for bacterial expression (GST-CTD).
Mammalian cell culture.
HEK293A and HeLa cells were cultured in Dulbecco's modified Eagle's medium (DMEM) with 10% fetal bovine serum (FBS). K562 cells (a gift from X. Jiang) were cultured in RPMI, 10% FBS, 10−4 M β-mercaptoethanol. PTX0002 and V180 cell lines were generated by stable transfection of plasmids expressing 3×FLAG-CDK12 isoform 1 or 3×FLAG alone, respectively, into HEK293A cells.
Bacterial culture and GST-CTD purification.
The GST-CTD plasmid was transformed into BL21(DE3)pLysS Escherichia coli cells (Invitrogen) and cultured in LB medium at 37°C until mid-log phase. Cells were induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 3 h at 37°C and harvested. Cells were lysed as previously described (35), and GST-CTD was purified using a 1-ml GSTrap 4B column (GE Healthcare).
Antibodies.
CDK12 antibodies were obtained from J. Pines (35) and Novus Biologicals (NB100-07001, H00051755-MO2A). Anti-FLAG antibody was purchased from Sigma (F1804). Anti-Myc antibodies were purchased from Roche (11667149001). CDK9 (sc-7331), cyclin T1 (sc-8127), and CDK11 (sc-928) antibodies were purchased from Santa Cruz Biotechnology. RNA Pol II CTD antibodies were purchased from Covance (anti-Ser2P H5, anti-Ser5P H14, and anti-CTD 8WG16).
Immunoprecipitation and mass spectrometry.
Plasmids expressing 3×FLAG-tagged CDKs or cyclins were transfected using calcium phosphate into HEK293A cells. At 60 h posttransfection, 108 cells were isolated and lysed in MS lysis buffer (Tris-buffered saline [TBS], pH 7.5, 1 mM EDTA, 0.1% NP-40, 0.05% [wt/vol] deoxycholate, 10 mM β-glycerophosphate, 2 mM NaVO4, and Roche Complete EDTA-free protease inhibitors). Protein lysates were clarified by centrifugation at 13,000 rpm for 30 min at 4°C, precleared with 50 μl packed Sepharose 4B beads, and mixed overnight with 20 μl packed M2-FLAG-agarose (Sigma) at 4°C. Bound protein complexes were washed 5 times with MS lysis buffer and once with 50 mM ammonium bicarbonate. Anti-FLAG antibody-immunoprecipitated complexes were eluted with 3×FLAG peptide (400 ng/ml) at room temperature for 30 min or by boiling in 2× SDS-PAGE Laemmli sample buffer. Protein complexes eluted with 3×FLAG peptide were lyophilized and resuspended in SDS-PAGE sample buffer. Immunoprecipitation of endogenous proteins was performed essentially as described above, using 10 to 20 μg of antibody protein G-agarose (GE Healthcare), and eluted by boiling in 2× SDS-PAGE Laemmli sample buffer. The protein complexes were then resolved by SDS-PAGE (4 to 12% bis-Tris; Invitrogen) and stained with colloidal Coomassie stain (8% [wt/vol] ammonium sulfate, 1.6% [vol/vol] phosphoric acid, 0.08% [wt/vol] Coomassie G-250, 20% [vol/vol] methanol). Each lane was then cut into 16 gel slices, with each slice corresponding to a known molecular weight range, and subjected to in-gel trypsin digestion (34).
High-performance liquid chromatography–electrospray ionization–mass spectrometry (HPLC-ESI-MS) was performed on a 4000 QTrap mass spectrometer (Applied Biosystems/Sciex) coupled to an Agilent 1100 Nano-HPLC using a nano-ESI interface. Samples were desalted using an on-line trap column (Zorbax 300SB-C18, 5 μm, 5 by 0.3 mm; Agilent), and chromatography coupled to electrospray ionization was performed on a 75-μm by 150-mm reverse-phase column (Reprosil-Pur C18; Dr.Maisch GmbH) using buffer A (5% HPLC grade acetonitrile, 0.1% formic acid) and buffer B (90% acetonitrile, 0.1% formic acid) in a linear gradient of 0 to 20% B for 23 min, 20 to 39% B for 9 min at a flow rate of 300 nl/min. Tandem MS (MS-MS) spectrum assignment was performed using X!Tandem (http://www.thegpm.org/TANDEM/) and Mascot (Matrix Science) software against the IPI human protein database (http://www.uniprot.org). HEK293A cells transfected with empty vector constructs or prebleed IgG were used as negative-control samples.
RNA-Seq analysis of cyclin K gene exon and exon-exon junction expression.
RNA-Seq data of mRNA libraries available at the Michael Smith Genome Sciences Centre production-scale sequencing facility (British Columbia Cancer Research Centre, Vancouver British Columbia, Canada; http://www.bcgsc.ca/platform/sequencing/) were analyzed for cyclin K mRNA structure using two methods (see below). The libraries were prepared using standardized protocols similar to those described in Griffith et al. (27) and varied for read length and lanes of Illumina GAII sequencing obtained.
ALEXA-Seq method.
Using the ALEXA-Seq analysis algorithm (27) (http://www.alexaplatform.org/), 291 lanes of paired-end reads of Illumina GAII RNA-Seq data from 38 human tissues and cell lines were mapped to a database of all possible theoretically valid exons and exon-exon junctions of known Ensembl (21) (http://www.ensembl.org/) (version 49) genes. Specifically, the donor sites of each known exon in a gene were connected to all known downstream acceptor sites of the gene. Across the 38 libraries, a total of 5.06 billion reads were mapped, of which 321,890 reads mapped to cyclin K gene exons and exon-exon junctions. Differential expression values for the gene features were determined as the log2 difference in average coverage values and normalized for library read depth as described previously (27). The type and distribution of the 38 human RNA-Seq libraries were as follows: A, tumors, 24 (7 neuroblastoma, 6 oligodendroglioma, 5 lymphoma, 4 multiple myeloma, and 2 colorectal); and B, normal tissues, 14 (6 breast, 5 B cell, 2 brain, and 1 human embryonic stem cell [hESC]).
HMMSplicer method.
A total of 1,774 lanes of paired-end Illumina GAII RNA-Seq data from 599 human tissues and cell lines were processed using the splice-aware aligner HMMSplicer (16) to identify expressed canonical (GU-AG, GC-AG, and AU-CA) and noncanonical exon-exon junctions and the read count and confidence score associated with each. HMMSplicer does not rely on existing annotation and is capable of finding novel splicing events. A total of 4.32 billion reads were found to map to 693,238 exon-exon junctions (at least 10 reads across all libraries). Each library yielded, on average, 129,644 distinct exon-exon junctions (with >1 observation). Of the 314,935 known junctions in our database, 223,714 (71%) were detected in at least one library (and with ≥10 reads across all libraries); exon junctions with <10 reads across all libraries were removed. Exon junction reads that mapped to the cyclin K gene were identified; libraries with <20 total cyclin K gene junction reads were removed. Across the remaining 570 libraries, there were 552,300 total cyclin K gene exon-exon junction reads (mean = 969, median = 593, and range = 20 to 20,425); 98.3% were canonical GU-AG junctions. There were 34 intragenic junctions (average HMMSplicer score = 1,123) of which 10 mapped to the cyclin K1 mRNA structure (average score = 1,373). The final types and distribution of the 570 human RNA-Seq libraries used were as follows: A, tumors, 206 (88 ovarian, 89 breast, 8 gastrointestinal, 6 brain, 4 colon, 4 kidney, 4 lung, 2 melanoma, and 1 urinary); B, leukemias, 189 (170 acute myeloid leukemia [AML], 13 acute lymphoblastic leukemia [ALL], and 6 undetermined); C, lymphomas, 116 (93 diffuse large B-cell lymphoma [DLBCL], 18 undetermined, and 5 follicular lymphoma [FL]); D, normal tissues, 37 (9 tonsil, 6 breast, 2 blood, and 1 each adipose, adrenal, bone marrow, brain, colon, heart, hESC, kidney, liver, lung, lymph, muscle, ovary, prostate, testes, thyroid, and white blood cell); E, other cancers, 13; F, other, 7; and G, cancer cell lines, 2.
Immunoprecipitation and Western blotting.
Plasmids expressing 3×FLAG-CDK12 or 3×FLAG-CDK9 were cotransfected using calcium phosphate in combination with plasmids expressing dMyc-cyclin K1, K2, T1, and L1 into HEK293A cells. Protein complexes were prepared as described above and immunoprecipitated with anti-FLAG or anti-Myc antibodies, and Western blots were probed with anti-Myc or anti-FLAG antibodies, respectively. MultiGauge (FujiFilm) was used to quantify bands.
siRNA knockdown and CTD phosphorylation assay.
An amount of 6 × 106 PTX0002 cells (stably expressing 3×FLAG-CDK12) was transfected with 1.2 μmol of an equimolar mixture of two siRNA sequences designed against two different regions of the 3′ UTR of the cyclin K gene (CCNK) (see Table S1 in the supplemental material) or a negative-control siRNA (catalog number 12935200; Invitrogen) using XtremeGene siRNA transfection reagent (Roche). Twenty-four hours later, the cells were transfected with 10 μg of plasmids expressing dMyc-cyclin K1 or an empty plasmid using Lipofectamine 2000 (Invitrogen). The medium was changed 6 h posttransfection, and cells were harvested at 72 h post-siRNA transfection. Twenty-five percent of the cells were used to prepare total RNA for quantitative (qPCR) (see below), and 75% of the cells were used to prepare protein lysates. Protein lysates were clarified and precleared with Sepharose 4B (Sigma) as described above and incubated with 5 μg of anti-FLAG antibody overnight at 4°C with constant rotation. Anti-FLAG-bound 3×FLAG-CDK12 protein complexes were precipitated with 20 μl packed protein G-agarose (GE Healthcare) for 30 min at 4°C and washed 5 times with MS lysis buffer, followed by 2 washes with kinase assay buffer (50 mM Tris, pH 7.5, 100 mM NaCl, 10 mM MgCl2, 0.1% [vol/vol] NP-40, 1 mM dithiothreitol [DTT], 20 μM β-glycerophosphate, Roche Complete EDTA-free protease inhibitors). After the last wash, protein-bound beads were resuspended in 30 μl kinase buffer supplemented with 50 μM ATP, 1 μg of purified GST-CTD, and 10 μCi of [γ-32P]ATP and incubated at 30°C for 1 h, and then the kinase reaction was stopped with the addition of 6× SDS-PAGE loading dye. Samples were heated at 85°C for 5 min, resolved using 4 to 12% SDS-PAGE, and imaged with a FujiFilm FLA-7000 scanner. Phosphor bands were quantified using FujiFilm MultiGauge software.
Luciferase assays.
To examine the effect of siRNA dosage on transcription, 1.2 × 105 HeLa cells were reverse transfected with various amounts of siRNA designed against CDK12 or CDK9 using Lipofectamine 2000 (Invitrogen). The negative-control siRNA used in these experiments was either CDK12 negative-control siRNA (cytomegalovirus [CMV] transcription experiment using pRL-CMV plasmid [Promega]) or CDK9 negative-control siRNA (simian virus 40 [SV40] transcription experiment using pGL3-Promoter plasmid [Promega]). Forty-eight hours after siRNA transfection, 100 ng of reporter plasmid (pRL-CMV or pGL3-Promoter) expressing luciferase was transfected, and at 72 h after siRNA transfection, cells were lysed with luciferase lysis buffer (Promega) and the assay was performed according to the manufacturer's instructions. The total protein concentration was determined by Bradford assay. To determine the dependence of transcription on CDK12 and cyclin K, 10 pmol CDK12 or cyclin K siRNA designed against the 3′ UTR of each transcript was cotransfected with plasmids expressing ectopic 3×FLAG-CDK12 or 3×FLAG-cyclin K1, respectively, or an empty vector control. Forty-eight hours posttransfection, 100 ng of reporter plasmid (pRL-CMV or pGL3-Promoter) was transfected, and cells were harvested at 72 h posttransfection and assayed for transcription as described above.
RESULTS
CDK12/CrkRS interacts with a 65-kDa isoform of cyclin K.
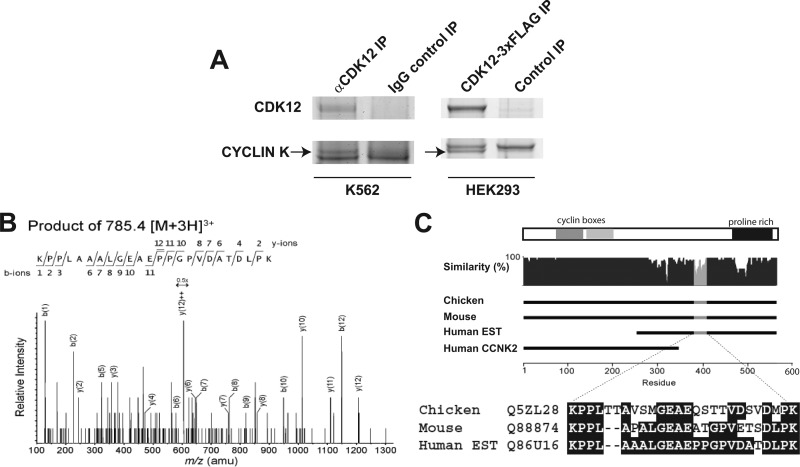
We employed IP-MS as an unbiased approach to identify possible cyclin partners from endogenous CDK12/CrkRS complexes isolated from HeLa, K562, and HEK293 cells. Mass spectra that matched cyclin K peptides were identified in SDS-polyacrylamide gel slices corresponding to a molecular-mass range between 60 and 80 kDa (Fig. 1A and Table 1). Hereinafter, CDK12/CrkRS will be called CDK12 for its association with cyclin K.
Fig 1.
Identification of a novel isoform of cyclin K in CDK12/CrkRS protein complexes. (A) Colloidal Coomassie stain of immunoprecipitated endogenous CDK12/CrkRS complexes from K562 cells and 3×FLAG-CDK12/CrkRS complexes from HEK293A cells. Cyclin K peptides were found in a 65- to 80-kDa gel slice. (B) MS-MS spectra of a cyclin K1 peptide. (C) Sequence of a human C-terminal cyclin K1 peptide found by MS-MS aligned to the corresponding chicken and mouse cyclin K peptides. The predicted cyclin boxes and a proline-rich domain of the cyclin K1 isoform are shown in the bar above the alignment.
Table 1.
CDK12/cyclin K interactions identified by IP-MSa
| Sample | Cell type | No. of unique peptides/total no. of peptides (X!Tandem score) |
Cyclin K1-specific peptide observedb |
|||||
|---|---|---|---|---|---|---|---|---|
| CDK12 | Cyclin K | Amino acid position |
Sequence | X!Tandem score | Unique | |||
| Start | End | |||||||
| CDK12 endogenous IP | HeLaS3 | 8/9 (−65.0) | 2/2 (−12.2) | — | ||||
| CDK12 endogenous IP | HEK293 | 20/28 (−169.4) | 4/7 (−29.9) | 306 | 321 | DPQQPAQQQQPAQQPK | −1.9 | Y |
| 375 | 398 | KPPLAAALGEAEPPGPVDATDLPK | −3.6 | Y | ||||
| CDK12 endogenous IP | K562 | 16/26 (−143.5) | 7/8 (−55.5) | 306 | 321 | DPQQPAQQQQPAQQPK | −3.1 | Y |
| 336 | 346 | AVVVSPKEENK | −1.3 | Y | ||||
| 375 | 398 | KPPLAAALGEAEPPGPVDATDLPK | −4.9 | Y | ||||
| CDK12-3×FLAG anti-FLAG IP | HEK293A | 18/28 (−158.6) | 2/2 (−10.6) | — | ||||
| HEK293A | 10/15 (−84.3) | 1/2 (−8.5) | — | |||||
| 3×FLAG-CDK12 anti-FLAG IP | HEK293A | 24/47 (−224.2) | 3/4 (−18.9) | 375 | 398 | KPPLAAALGEAEPPGPVDATDLPK | −2.7 | Y |
| 450 | 459 | MMKTEGPSYG | −1.2 | Y | ||||
CDK12/CrkRS complexes were immunoprecipitated, and cyclin K peptides were identified by MS. Cyclin K1-specific peptides were observed in CDK12 immunoprecipitates from various cell lines. CDK12-3×FLAG, C-terminally FLAG-tagged CDK12; 3×FLAG-CDK12, N-terminally FLAG-tagged CDK12.
Individual X!Tandem expected value scores for cyclin K1-specific peptides are shown. Y, yes; —, no cyclin K1-specific peptide detected.
To more comprehensively identify cyclin partners of CDK12, we expressed N-terminally and C-terminally tagged 3×FLAG-CDK12 proteins in HEK293A cells. IP-MS analysis of anti-FLAG-immunoprecipitated 3×FLAG-CDK12 complexes again identified mass spectra that matched cyclin K peptides in the 60- to 80-kDa gel slices (Fig. 1A and Table 1). No other cyclins were present in any of our CDK12 immunoprecipitates.
We were intrigued by the reproducibility of our IP-MS experiments in identifying cyclin K peptides within the 60- to 80-kDa range. Previous studies on cyclin K showed that human cyclin K is a 43-kDa protein that interacts with CDK9 and is involved in transcription (17, 23, 39). However, we identified a number of peptides which suggested the existence of a larger isoform of cyclin K when we searched the MS data against databases that include translated expressed sequence tags (ESTs) (Fig. 1B and C). Genomic arrangement of the cyclin K gene (CCNK) and EST data suggested an alternative isoform of cyclin K similar to mouse and chicken cyclin K with a predicted molecular mass of 64 kDa (Fig. 1C). The deduced reference sequence for human cyclin K isoform 1 (cyclin K1; GenBank accession number NM_001099402.1) encodes a 580-amino-acid protein with a molecular mass of 64 kDa that was predicted based on available EST data in NCBI. Recently, a nominal 70-kDa form of cyclin K was identified by Western blotting using cyclin K antibodies and analysis of available EST data (6). Cyclin K2 is defined as the original 357-amino-acid, 43-kDa isoform of cyclin K (GenBank accession number NM_003858.3). Our IP-MS spectra have identified novel cyclin K peptides that match the predicted amino acid sequence of cyclin K1 (Table 1). These peptides were observed on multiple occasions in CDK12 immunoprecipitates from both endogenous CDK12 and ectopic 3×FLAG-CDK12 complexes. This is the first protein sequence evidence to support the expression of human cyclin K1 protein.
Cyclin K1 is the predominant isoform of cyclin K.
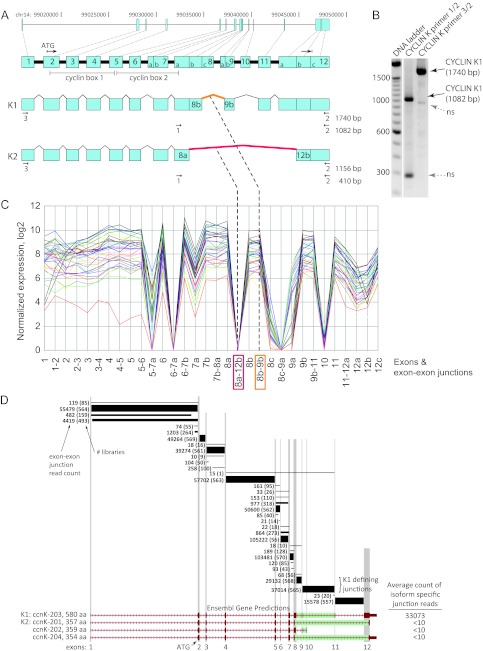
To address the question of whether each cyclin K isoform is expressed at the mRNA level, we performed RT-PCR using primers which are common to the cyclin K1 and K2 open reading frame (ORF) but differ in the size of the amplification products due to alternative splicing (Fig. 2A and B). For cyclin K1 and K2, the first 7 exons are shared but different splice donors in exon 8 and different splice acceptors in exons 9a (cyclin K1) and 12b (cyclin K2) are used. For both isoforms, the last 150 bp of exon 12 are shared but, due to a frameshift, different amino acids are encoded. Sequencing of these RT-PCR products generated from HeLa mRNA showed that 49/49 clones isolated conformed to the cyclin K1 isoform, suggesting that cyclin K1 was the predominant isoform in HeLa cells.
Fig 2.
Cyclin K1 is the predominant cyclin K mRNA isoform. (A) Cyclin K gene exon arrangement and splicing. The exon arrangement of the CCNK locus on chromosome 14 is depicted. Splicing pathways that produce a 43-kDa isoform (cyclin K2) and a 64-kDa isoform (cyclin K1) are shown; subdivisions in exons 7, 8, 9, and 12 represent different known splice acceptors and donors observed in the mRNA and EST data. The numbered arrows depict primers and RT-PCR product sizes that were used to discriminate between the cyclin K1 and K2 isoforms. The 8b-9b (orange) and 8a-12b (red) exon-exon junctions differentiate the expression of the cyclin K1 and K2 isoforms, respectively. (B) Cyclin K1 mRNA is present in HeLa cells. RT-PCR of HeLa poly(A)-enriched RNA was performed with primer pairs 1/2 and 3/2 that were designed to differentiate between cDNAs for cyclin K1 and K2 (see panel A). RT-PCR products corresponding to the predicted size for cyclin K1 are indicated. Sequencing of the ∼300-bp and ∼950-bp bands showed that they are not cyclin K related, and they are denoted as nonspecific (ns). (C) RNA-Seq analysis of known cyclin K exons and exon junctions. The normalized expression of observed combinations of known splice donors and acceptor sites (Fig. 2A) for the CCNK locus for 38 human cancer and cell line mRNA libraries is shown. (D) Detection of cyclin K exon-exon junctions by HMMSplicer analysis of RNA-Seq data. Exon-exon junction RNA-Seq read counts (552,300 total) from 570 human cancer and cell mRNA libraries that mapped to the CCNK locus using HMMSplicer are shown. The junctions putatively connect the exons (vertical gray bars) for the Ensembl-predicted gene transcripts (red, bottom). Total read count among the 570 libraries for each junction read is shown; in parentheses is the number of libraries where the junction read was observed. The thickness of each line approximates the relative abundance of the junctions. In the exon 8 region, three different splice donor sites (Fig. 2A) are used between four Ensembl transcripts. The green bars represent the junctions or junction combinations that would uniquely define each of the Ensembl-predicted isoforms. The number of counts observed for each of the isoform-defining junctions is shown in the right-hand column. For the K1 isoform (CCNK-203), the isoform-specific read count, 33,073, is the mean of the counts of the two K1-defining junctions (29,132 and 37,014).
To address the generality of expression of the cyclin K1 and K2 isoforms, we examined massively parallel short-read RNA-Seq data from mRNA libraries of various human tumors and cell lines available at our research center. RNA-Seq data from 38 libraries across 7 human tissue types were mapped to all possible combinations of known CCNK gene exons and splice sites using the ALEXA-Seq gene model-based alternative splicing algorithm (27). There were zero reads for the 8a-12b exon-exon junction specific for expressing cyclin K2, while the 8b-9a exon-exon junction used in expressing cyclin K1 exhibited high relative expression (Fig. 2C). In addition, we used the splice-aware de novo aligner HMMSplicer (16) to identify any canonical and noncanonical exon-exon junctions involving annotated or novel unannotated CCNK gene exons in RNA-Seq data from 570 libraries across more than 22 human tissue types. CCNK gene exons 1 to 7 are common to the four Ensembl-annotated transcripts (Fig. 2D) (http://www.ensembl.org/), while the splicing of exon 8 with downstream exons differentiates the expression of the isoforms; three different splice donor sites in exon 8 are used in expressing the four isoforms. In total, 552,300 exon-exon junction reads were mapped to the CCNK locus. Most of the reads (542,746, 98.3%) mapped to the canonical junctions defined by the expression of the cyclin K1 isoform (CCNK-203) (Fig. 2D). The combination of the exon 8-9 junction with 29,132 reads and the exon 9-11 junction with 37,014 reads is uniquely specific for the cyclin K1 isoform. The junctions that uniquely specify the other three Ensembl gene predictions each had less than 10 total reads between all 570 libraries. For the CCNK-202 isoform, predicted to encode cyclin K2, and the CCNK-204 isoform, the exon/intron junctions are unusual and noncanonical and may represent artifacts. In addition, each of the cyclin K1-associated junctions was present in 557 to 570 of the 570 total libraries. We conclude that the cyclin K1 isoform is the highly predominant cyclin K isoform present in human tissues.
There were a total of 3,232 non-cyclin K1-expressing junction reads (0.6%) in the coding regions of the CCNK locus (exons 2 to 12) which would be predicted to alter the cyclin K reading frame. All of these junctions are canonical GU-AG intron junctions except for one GC-AG intron (8). Many of these junction reads were observed with just 1 or 2 reads in the libraries in which they were observed (data not shown) and may represent sequencing or mapping artifacts. However, 1,841 of these reads (57%) were present in just two junctions; the junctions with 977 reads and 864 reads are present in 318 and 273 different libraries, respectively (Fig. 2D). These putative alternative splicing events affect the structure of the second cyclin box domain (Fig. 2A). These data indicate that in some human cancers and tissues, significant levels of alternative splicing of cyclin K transcripts may occur.
Cyclin K1 interacts poorly with CDK9 in endogenous complexes.
Previously, it has been shown that cyclin K interacts with CDK9 and stimulates transcription in a nuclear runoff assay (23, 39). To test this protein-protein interaction, we performed IP-MS analysis on endogenous CDK9 complexes isolated by immunoprecipitation of CDK9 from HEK293 and HeLa cells. We observed the presence of cyclin T1 peptides but not cyclin K peptides (Table 2). Next, we ectopically expressed 3×FLAG-tagged CDK9 in HEK293A cells and isolated recombinant CDK9 complexes to analyze the protein constituents by IP-MS. Both N- and C-terminal 3×FLAG-CDK9 complexes contained multiple cyclin T1 peptides. In one sample, we were able to detect one unique peptide each for cyclin T2 and cyclin K. This suggests that cyclin T2 and K may interact with only a small proportion of CDK9 molecules within these cells (Table 2).
Table 2.
Cyclin interactions of CDK9 identified by IP-MSa
| Bait protein | Cell line | No. of unique peptides/total no. of peptides (X!Tandem score) |
|||
|---|---|---|---|---|---|
| CDK9 | Cyclin: |
||||
| K1/K2 | T1 | T2 | |||
| Endogenous CDK9 | HEK293 | 13/18 (−108.5) | 0 | 18/34 (−169.3) | 0 |
| HeLaS3 | 15/17 (−118.9) | 0 | 4/4 (−27.2) | 0 | |
| 3×FLAG-CDK9 | HEK293A | 13/27 (−116.2) | 0 | 3/3 (−18.4) | 0 |
| CDK9-3×FLAG | HEK293A | 15/31 (−145.4) | 0 | 11/13 (−98.8) | 0 |
| HEK293A | 23/96 (−227) | 1/1 (−1.4) | 12/12 (−98.7) | 1/3 (−18.2) | |
Endogenous CDK9 and N-terminally (3×FLAG-CDK9) and C-terminally tagged (CDK9-3×FLAG) proteins were immunoprecipitated, and the complex was analyzed for cyclin proteins.
Cyclin K interacts primarily with CDK12 in endogenous complexes.
Previous studies have reported interactions between rat Cdk12 and cyclins L1 and L2 that modulate alternative splicing in vitro (9). Cyclin L1 was also shown to interact with CDK13/CDC2L5 and facilitate alternative splicing (10). Contrary to these reports, we were unable to detect cyclin L1 or L2 in any of our endogenous and ectopically expressed CDK12 immunoprecipitates by MS. To better understand which CDKs associate with a particular cyclin, we ectopically expressed epitope-tagged cyclin K1, K2, T1, and L1 and performed IP-MS on the isolated cyclin complexes. IP-MS analysis of 3×FLAG-cyclin L1 complexes showed the presence of CDK11 peptides, confirming previous reports that CDK11 interacts with cyclin L1 (30, 42, 60). However, we failed to detect any CDK13/CDC2L5 or CDK12 peptides in two replicates of 3×FLAG-cyclin L1 immunoprecipitates (Table 3). Analysis of 3×FLAG-cyclin T1 complexes revealed the presence of CDK9 peptides exclusively. Lastly, we found that 3×FLAG-tagged cyclin K1 and K2 both interact predominantly with CDK12, with only a few unique peptides detected that correspond to CDK13/CDC2L5 and CDK9 (Table 3). The lower numbers of peptides observed for CDK13/CDC2L5 and CDK9 in these ectopically expressed cyclin K1 and K2 immunoprecipitates suggested that the number of cyclin K molecules associated with CDK13/CDC2L5 and CDK9 is less than the number of cyclin K molecules associated with CDK12.
Table 3.
CDK interactions of cyclin K1, K2, T1, and L1 identified by IP-MSa
| Bait protein | Cell line | No. of unique peptides/total no. of peptides (X!Tandem score) |
||||
|---|---|---|---|---|---|---|
| Cyclin | CDK: |
|||||
| 12 | 13 (Cdc2L5) | 9 | 11 | |||
| Cyclin L1-3×FLAG | HEK293A | 4/4 (−24.8) | 0 | 0 | 0 | 1/1 (−1.2) |
| HEK293A | 3/3 (−18.7) | 0 | 0 | 0 | 9/14 (−69.4) | |
| 3×FLAG-cyclin T1 | HEK293A | 5/5 (−36.7) | 0 | 0 | 5/6 (−33.9) | 0 |
| Endogenous cyclin T1 | HEK293 | 1/1 (−1.6) | 0 | 0 | 14/18 (−114.3) | 0 |
| HeLaS3 | 11/32 (−98.9) | 0 | 0 | 16/25 (−136.3) | 0 | |
| Cyclin K2-3×FLAG | HEK293A | 12/17 (−102.9) | 14/16 (−112.8) | 8/11 (−83.8) | 0 | 0 |
| HEK293A | 4/4 (−29.5) | 8/9 (−66.6) | 1/2 (−10.5) | 1/2 (−7.9) | ||
| 3×FLAG-cyclin K1 | HEK293A | 16/60 (−157.5) | 8/14 (−66.3) | 3/5 (−23.1) | 0 | 0 |
| HEK293A | 17/92 (−187.9) | 6/12 (−44.8) | 1/6 (−17.2) | 2/6 (−17.1) | 0 | |
Endogenous and overexpressed epitope-tagged cyclin complexes were isolated by immunoprecipitation and analyzed by MS for CDKs. Cyclin L1- or K2-3×FLAG, N-terminally FLAG-tagged cyclin L1 or K2; 3×FLAG-cyclin T1 or K1, C-terminally FLAG-tagged cyclin T1 or K1.
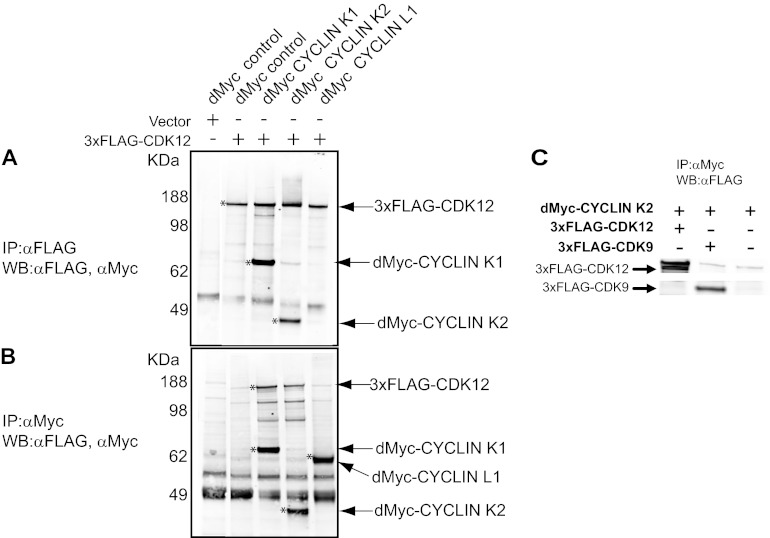
To validate our IP-MS results, we performed IP-Western blot experiments using ectopic expression of CDKs and cyclins. Ectopic expression of 3×FLAG-CDK9 and dMyc-cyclin K2 showed an interaction, as previously reported (Fig. 3C) (23, 39). Expression of 3×FLAG-CDK12 in combination with dMyc-cyclin L1 did not show an interaction even though cyclin K1, K2, and L1 were expressed at similar levels (Fig. 3A and B). However, expression of 3×FLAG-CDK12 in combination with dMyc-cyclin K1 or dMyc-cyclin K2 showed an interaction between CDK12 and cyclin K1 and between CDK12 and cyclin K2 (Fig. 3A). Together, our IP-MS and Western blot results strongly suggest that CDK12 predominantly interacts with cyclin K in vivo.
Fig 3.
Immunoprecipitation-Western blotting validation of CDK12-cyclin K interaction. (A and B) Western blot analyses of anti-FLAG-immunoprecipitated complexes probed with anti-FLAG and anti-Myc antibodies (A) and of anti-Myc-immunoprecipitated complexes probed with anti-FLAG and anti-Myc antibodies (B). An asterisk denotes the protein indicated by the arrow on the right. dMyc, 2×Myc. (C) dMyc-cyclin K2 was immunoprecipitated with anti-Myc, and the blots probed with anti-FLAG antibody. Both 3×FLAG-CDK12 and 3×FLAG-CDK9 interact with dMyc-cyclin K2.
To estimate the relative proportions of the cyclin partners for a given CDK or the relative proportions of the CDK partners for a given cyclin, we employed an MS technique called multiple reaction monitoring (MRM) to analyze immunoprecipitates for multiple specific cyclin or CDK partners. MRM measures the ion intensity of specific fragments from selected peptides (parent ions), producing ion chromatograms that are proportional to the abundance of the parent peptide; many peptides can be measured simultaneously, allowing for multiplex analysis of the relative proportions of different CDK or cyclin populations. MRM analysis of CDK complexes showed that endogenous or ectopic CDK12 interacts with cyclin K1 and not cyclin K2 or cyclin L1 and that endogenous or ectopic CDK9 interacts predominantly with cyclin T1 in HEK293A cells. MRM analysis of ectopic cyclin complexes in HEK293A cells showed reciprocal results: cyclin K1 interacts predominantly with CDK12 and CDK13, cyclin T1 interacts with CDK9 exclusively, and cyclin L1 interacts predominantly with CDK11. A detailed description of the MRM study can be found in Figures S1 and S2 in the supplemental material.
The CDK12/cyclin K complex is required to phosphorylate the C-terminal domain of RNA Pol II in vitro.
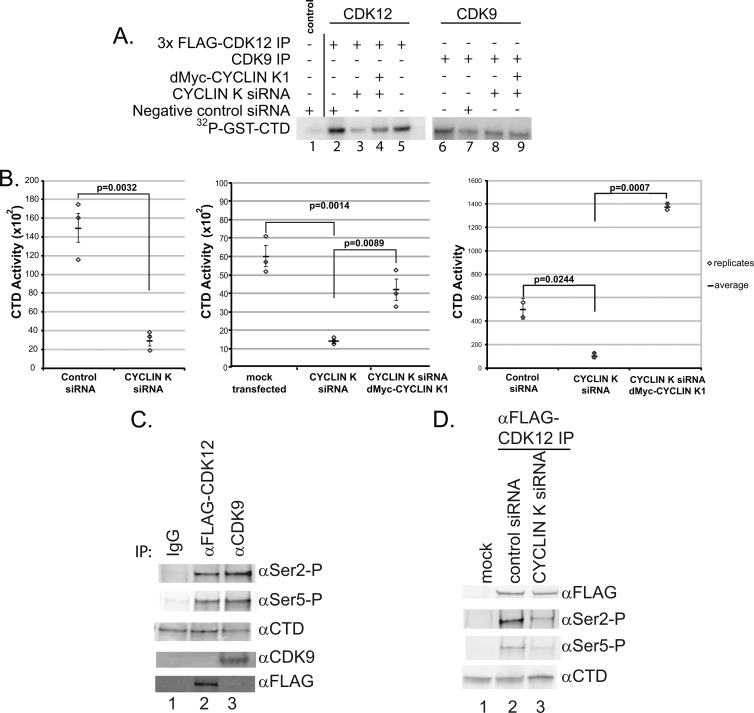
To investigate the dependence of CDK12 on cyclin K for phosphorylation of the RNA Pol II CTD, we performed in vitro CTD phosphorylation assays using CDK12 protein complexes isolated from cells stably expressing 3×FLAG-CDK12 where we modulated the expression of cyclin K using siRNA and ectopic expression. In this cell line, we determined the mRNA levels of CDK12 and cyclin K by qRT-PCR (see Fig. S3 in the supplemental material). CDK12 expression was normalized against the control cell line which expresses only endogenous CDK12. The HEK293A/3×FLAG-CDK12 stable cell line typically expresses CDK12 mRNA at levels 9- to 11-fold higher than the level in the control cell line (see Fig. S3B). We designed siRNAs against a 3′ untranslated region (UTR) shared by both cyclin K1 and K2. The 3′ UTR cyclin K siRNAs proved to be effective in knocking down cyclin K mRNA levels (>60%) in HEK293A cells as determined by qRT-PCR (see Fig. S3A). We next proceeded to isolate 3×FLAG-CDK12 complexes by immunoprecipitation using an anti-FLAG antibody and assaying the CDK12 protein complex for CTD phosphorylation activity in vitro. In cells treated with cyclin K siRNAs, we noted a decrease in CTD phosphorylation that was consistent and significant in 3 replicate experiments (Fig. 4A, lanes 2 and 3, and B). CTD phosphorylation was partially restored or increased with the expression of ectopic dMyc-cyclin K1 (Fig. 4A, lanes 3 and 4, and B). Based on our results, we conclude that CDK12 can be activated by cyclin K for phosphorylation of the CTD of RNA Pol II.
Fig 4.
CDK12 is dependent on cyclin K to phosphorylate the C-terminal domain of RNA Pol II. (A) Cyclin K expression modulates CTD phosphorylation by CDK12. 3×FLAG-CDK12 complexes isolated from cells stably expressing 3×FLAG-CDK12 that were modulated with cyclin K siRNA designed against the 3′ UTR of the CCNK gene showed a significant decrease in RNA Pol II CTD phosphorylation compared to the results for the negative control (lane 3 versus lane 2). CTD phosphorylation was restored with the expression of dMyc-cyclin K from an exogenous plasmid (lane 4). Lane 1, a negative-control anti-FLAG IP from a control cell line not expressing 3×FLAG-CDK12; lanes 2 to 9, anti-FLAG-CDK12 or endogenous CDK9 complexes isolated from a cell line stably expressing 3×FLAG-CDK12; lane 5, cells were mock transfected with an empty dMyc vector and without siRNA. (B) Graphical representation of the results of 3 replicate experiments showing CDK12 dependence on cyclin K to phosphorylate RNA Pol II CTD. The vertical range and horizontal mark are the standard deviation and the mean, respectively. (C) Immunoprecipitated CDK12 and CDK9 phosphorylate Ser2 and Ser5 of the CTD heptad in vitro. “IP” denotes the Western analysis of the immunoprecipitated complexes used in the CTD assays. (D) siRNA knockdown of cyclin K in CD K12 immunoprecipitates showed a decrease in Ser2 and Ser5 phosphorylation relative to the results of a control siRNA knockdown.
To test whether the phosphorylation of the CTD by CDK9 is dependent on cyclin K, we isolated CDK9 complexes from the same cells treated with cyclin K siRNAs. Cyclin K siRNA and cyclin K1 ectopic expression had little to no effect on CDK9 phosphorylation of the CTD (Fig. 4A, lanes 7, 8, and 9). This agrees with our previous observations that CDK9 interacts predominantly with cyclin T1 and that CDK9/cyclin K complexes may constitute a small subset of P-TEFb (positive transcription elongation factor b) complexes within a cell.
We next investigated which serine residues of the CTD heptad repeat are phosphorylated by CDK12 and whether phosphorylation of specific serine residues in the CTD by CDK12 is activated by cyclin K. CDK9 has been shown to phosphorylate Ser2 and Ser5 of the CTD heptad repeat (1, 50, 51, 66). We performed an in vitro CTD phosphorylation reaction using immunoprecipitated CDK12 complexes isolated from cells treated with negative-control siRNA or cyclin K siRNA to determine whether the presence of cyclin K would have a differential effect on phosphorylation of Ser2 and Ser5 of the CTD. Like CDK9, CDK12 can phosphorylate Ser 2 and Ser5 of the CTD (Fig. 4C). We also found that the phosphorylation of Ser2 and Ser5 was potentiated in CDK12 complexes isolated from cells treated with cyclin K siRNA, suggesting that the phosphorylation of Ser2 and Ser5 by CDK12 is dependent on the presence of cyclin K in vitro (Fig. 4D).
CDK12 affects gene expression in vivo.
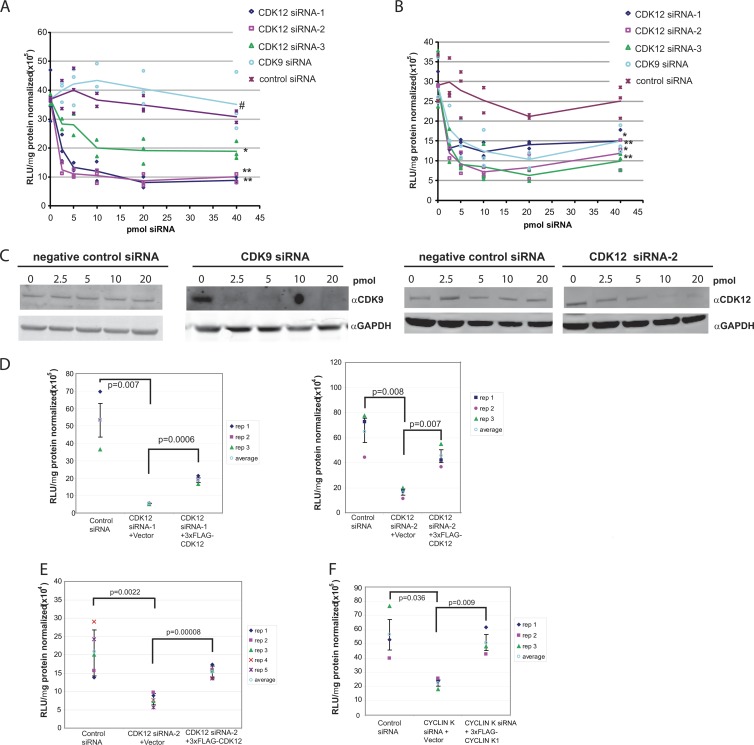
Phosphorylation of RNA Pol II CTD can modulate the transcription of genes and promoter/reporter constructs (29). To determine whether CDK12 can affect the expression of a reporter gene, we transfected HeLa cells with various doses of siRNA against CDK12 and CDK9. Total cellular lysates were assayed for the ability to affect the expression of firefly luciferase (hereinafter Fluc) or Renilla reniformis luciferase (hereinafter Rluc) from two different promoters. We found that Rluc expression from a CMV-driven reporter plasmid significantly decreased in a CDK12 siRNA dose-dependent manner, whereas the negative-control siRNA and CDK9 siRNA did not show a significant siRNA dose-dependent effect (Fig. 5A). Three different CDK12 siRNA molecules, two designed against the 3′ UTR of the CDK12 gene and one for the ORF, were assayed and showed similar dose-dependent effects on Rluc gene expression (Fig. 5A). This suggests that CDK12 protein expression can affect protein expression from a CMV promoter.
Fig 5.
CDK12 modulates CMV and SV40 promoter-driven transcription. (A and B) SiRNA dose-dependent knockdown by 3 different CDK12 siRNAs and a CDK9 siRNA of transcription driven by CMV (A) or SV40 (B). Transcription is measured by Rluc (A) or Flu (B) activity. Lines represent the averages of 3 replicates. Significance was calculated for the siRNA treatments versus the negative control using the 5 to 40 pmol data with the Mann-Whitney U test [two-sided; P(2)], as follows: in panel A, * indicates P(2) < 0.0002 (CDK12 siRNA-3), ** indicates P(2) < 0.0001 (CDK12 siRNA-1 and siRNA-2), and # indicates P(2) = 0.1135; in panel B, * indicates P(2) < 0.00011 (CDK12 siRNA-1 and siRNA-3 and ** indicates P(2) <0.0001 (CDK12 siRNA-2 and CDK9 siRNA). (C) Western blots showing examples of CDK9 siRNA and CDK12 siRNA-2 knockdown of CDK9 and CDK12 protein expression, respectively. (D) CDK12 siRNAs reduce SV40-mediated transcription. CDK12 siRNA knockdown of SV40-driven transcriptional activities was rescued by ectopic expression of CDK12. (E) CDK12 siRNA-2 reduces CMV-mediated transcription. CDK12 siRNA knockdown of CMV transcriptional activities was also rescued by ectopic expression of CDK12. P values indicated are significant (P < 0.05) and reproducible across different CDK12 siRNA molecules. (F) Transcription from the CMV promoter was sensitive to cyclin K expression, demonstrating that CDK12 was dependent on cyclin K for transcriptional activity (P < 0.004). (D to F) The vertical range is the standard deviation.
To show that the CDK12 transcriptional effect was not restricted to the CMV promoter, we tested an SV40 promoter driving the Fluc gene. All three CDK12 siRNA molecules decreased the expression of luciferase from an SV40-driven promoter in a dose-dependent manner similar to that observed with the CMV-driven reporter (Fig. 5B). We noted that CDK9 siRNA also decreased the expression of luciferase in a dose-dependent manner. Western blot analysis of CDK12 and CDK9 expression after treatment with siRNA showed that CDK9 and CDK12 protein levels were decreased (Fig. 5C). Thus, the siRNA dose-dependent decrease in SV40-mediated gene expression correlates with decreases in CDK12 and CDK9 protein expression.
To further test the dependence of CMV-Rluc and SV40-Fluc expression on CDK12 expression, we sought to restore reporter expression in CDK12 siRNA-treated cells with the expression of ectopic CDK12. HeLa cells were independently transfected with two different CDK12 3′-UTR siRNAs in combination with a plasmid expressing ectopic 3×FLAG-CDK12 or a control plasmid. For both promoters, cells cotransfected with CDK12 3′-UTR siRNAs and a control plasmid showed a significant decrease in reporter protein expression compared to the expression level in cells cotransfected with a negative-control siRNA and control plasmid (Fig. 5D and E). Cells cotransfected with CDK12 3′-UTR siRNAs and the 3×FLAG-CDK12 plasmid showed significant restoration of reporter protein activity from both promoters (Fig. 5D and E). These results support our earlier finding that CDK12 specifically affects protein expression from both CMV and SV40 promoters by demonstrating that the reduction of reporter protein activity due to CDK12 siRNA treatment can be relieved by the expression of ectopic CDK12 protein.
We next sought to link CDK12 dependence on cyclin K to its effect on reporter protein expression. We found that cyclin K 3′-UTR siRNA treatment significantly decreased CMV-driven reporter expression and that reporter expression could be restored with the addition of ectopically expressed cyclin K1 (Fig. 5F).
Our data convincingly show that cyclin K and CDK12 physically interact, that both CDK12 and cyclin K can modulate reporter gene expression, and that the phosphorylation of RNA Pol II by CDK12 is modulated by cyclin K, suggesting that the CDK12/cyclin K complex directly affects gene expression, possibly through modulation of RNA Pol II activity by CTD phosphorylation.
DISCUSSION
We have identified a 65-kDa isoform of cyclin K (cyclin K1) as the predominant cyclin partner for human CDK12, previously named CrkRS. Cyclin K1 is required to activate CDK12 to phosphorylate Ser2 and Ser5 of RNA Pol II CTD in vitro, and CDK12/cyclin K affects the expression of two reporter genes under the control of different promoters.
Truncated cyclin K cDNAs were first identified through yeast 2-hybrid experiments as a cyclin partner for human CDK9 (17, 23). The cyclin K primary structure was subsequently defined as a 357-amino-acid (43-kDa) protein according to a single cDNA clone (GenBank accession number BC015935) from human uterine tissue made in a large-scale human cDNA cloning project (59). This cDNA is currently referred to as cyclin K2. Our analysis of >500,000 RNA-Seq exon-exon junction read sequences mapped to the CCNK locus from 570 mRNA libraries and more than 22 human tissue types and tumor samples indicated that >99% of the junction reads are consistent with the expression of the cyclin K1 isoform primary structure. For the other three isoforms of cyclin K, none had Ensembl-annotated junction reads supporting their presence above background levels. RT-PCR analysis of HeLa cyclin K gene transcripts also exclusively supported the cyclin K1 isoform. Recently, the reference sequence of cyclin K2 (GenBank accession number NM_003858.3) was suppressed by GenBank because of poor support and use of nonconsensus splice sites. Indeed, our inspection of the cyclin K2 entries in EST and cDNA databases appears to indicate that they were all derived from the BC015935 clone. We therefore conclude that the 65-kDa cyclin K1 is the predominant cyclin K protein in humans. However, the RNA-Seq data also indicate that novel alternative splice versions of cyclin K mRNAs may exist at low frequencies. Most of the novel exon junction reads in the ORF alter the primary sequence of the cyclin K gene in the exon 6 and 7 region, which encodes the second cyclin box-like domain that forms part of the CDK binding surface in the crystal structure of cyclin K (2). The truncated protein products predicted from these transcripts may have altered CDK12 binding properties or function. In addition, there is RNA-Seq evidence for novel alternative splice events (∼10%) that affect the 5′ UTR of cyclin K but not the ORF. The existence and biological significance of these putative novel cyclin K mRNAs remain to be investigated. Our analysis shows that querying large numbers of existing RNA-Seq libraries is a rapid means to comprehensively define the structural variants of human mRNAs.
Our MS results are the first observation of cyclin K1 at the protein sequence level and show no evidence for the existence of the cyclin K2 protein. Structurally, the 580-amino-acid sequence of cyclin K1 encodes an ∼200-amino-acid proline-rich C-terminal region conserved in metazoans that is not present in cyclin K2 (Fig. 1C). Proline-rich protein domains are often sites for the binding of regulatory proteins (61, 62). We propose that this region may facilitate cyclin K protein-protein interactions that regulate CDK12/cyclin K1 function.
Cyclin K has been identified as a CDK9 cyclin partner in several studies. Ectopic coexpression of CDK9 and cyclin K was shown to phosphorylate the CTD of RNA Pol II or to modulate transcription and gene expression (17, 23, 39). Fusion of cyclin K to Rev modulates a Rev-responsive promoter/reporter gene (39). These results led to the inclusion of cyclin K as a cyclin partner for CDK9 in P-TEFb complexes. Several biological functions have also been attributed to cyclin K and CDK9/cyclin K. Overexpression or siRNA-mediated knockdown of cyclin K showed that cyclin K could regulate mesoderm induction in Xenopus laevis and modulate replication stress response and genome integrity in human cells (65, 67). These results were interpreted as cyclin K regulating CDK9 function based on the presumed association of cyclin K and CDK9. Using an unbiased approach employing IP-MS of endogenous and ectopic CDK12, CDK9, cyclin K1, and cyclin T1 complexes in three human cell lines (HEK293, K562, and HeLa), we showed that the primary cyclin partner for CDK12 is cyclin K1, that cyclin K1 predominantly interacts with CDK12 and CDK13, and that cyclin K1 is not a common cyclin partner for CDK9. While we do not observe detectable levels of endogenous CDK9/cyclin K complexes, we cannot rule out this complex at low levels or in other tissues or biological states. In our experiments with ectopic expression of CDK9 and cyclin K1, we can force low levels of CDK9/cyclin K1 interaction, but these are much lower than the levels of CDK9/cyclin T1 or CDK12/cyclin K1 complexes. Thus, the roles of cyclin K in mesoderm induction, replication stress response, and genomic stability could be interpreted as CDK12/cyclin K functions. This is further corroborated by a recent study showing that CDK12/cyclin K complexes helped maintain genomic stability through regulation of DNA damage response genes, such as BRCA1, ATR, and FANCI (6). Therefore, previous functions attributed to cyclin K through a CDK9 interaction should be interpreted with caution.
Further evidence that CDK9/cyclin K interactions may lack structural and biological significance is shown by the crystal structure of the N-terminal region of cyclin K that encompasses the cyclin box domains (residues 11 to 267) (2). It was noted that the cyclin K surface on the CDK9 binding interface differed significantly from the same interface on cyclin T1. It was hypothesized that cyclin T1 and K may have different affinities for CDK9. Similarly, analysis of the CDK9/cyclin T1 co-crystal structure reveals that some of the critical cyclin T1 residues involved in binding CDK9 are different in cyclin K (5). Our data indicate that the different chemical nature of the interface relates primarily to the preferential binding of cyclin K to CDK12 or CDK13. This is important since inhibitors that interfere with CDK9/cyclin T1 or CDK9/cyclin K binding or P-TEFb kinase activity are being pursued for therapeutic applications in AIDS, heart hypertrophy, and cancer (2, 52). Our data suggest that CDK9-, cyclin K-, and P-TEFb-based inhibitors may also modulate functions associated with CDK12 or CDK13.
Rat Cdk12 and mouse Cdk13 have previously been shown to interact with cyclin L1, an RS domain-containing cyclin known to be involved in splicing (9, 10, 14, 42). These studies were performed using ectopic coexpression of rat Cdk12 or mouse Cdk13 with rat cyclin L. It was shown that rat cyclin L1 enhanced the alternative splicing of a model pre-mRNA splicing substrate (9, 10). We did not detect cyclin L1 in any of our CDK12 immunoprecipitates by IP-Western blotting, MS-MS, or MRM. However, we did detect cyclin L1 in endogenous CDK11 immunoprecipitates and CDK11 in ectopic 3×FLAG-cyclin L1 immunoprecipitates using both MS-MS and MRM. These data indicate that CDK11 and cyclin L1 interact primarily with each other. CDK11 has been shown to interact with cyclin L1 and L2 to differentially affect splicing in vivo (42). We cannot rule out that CDK12 and CDK13 interact with cyclin L1 in vivo at low levels or in other tissues or biological states; however, this interaction was below the limits of our detection methods. Our results argue that cyclin K1 and not cyclin L1 is the primary cyclin partner of CDK12 and CDK13. This observation was further corroborated by a recent publication (6). We propose that, because both CDK12 and CDK13 contain their own RS domain, they do not need the RS domain in cyclin L1 to function in the spliceosome and that cyclin K1 may be required to activate CDK12 and CDK13 for splicing functions in addition to their kinase functions.
Recently, it was reported that a 70-kDa cyclin K interacts with CDK12, that CDK12/cyclin K affects the expression of DNA damage response genes, and that CDK12 affects Ser2 and Ser5 phosphorylation of RNA Pol II CTD in vivo (6). However, the report did not show a functional dependence of CDK12 on cyclin K for these activities. In addition to clarifying cyclin K1's structure and its CDK interactions, we have demonstrated that cyclin K1 can activate CDK12 CTD kinase activity and that immunoprecipitated complexes of CDK12 were able to phosphorylate Ser2 and Ser5 of RNA Pol II CTD in vitro in a cyclin K1-dependent manner. Whether CDK12 phosphorylates Ser5 in vivo remains to be examined (although knockdown of Drosophila Cdk12 in vivo did not result in lower levels of phosphorylated Ser5) (4). We cannot rule out the possibility that human CDK12 can phosphorylate other residues, such as Ser7, or only specific heptads in the 52-heptad repeat of the human RNA Pol II CTD.
Using CMV and SV40 promoter reporter assays, we showed that CDK12 modulated the expression of genes driven by two different promoters and that cyclin K1 also modulated CMV-driven reporter gene expression. Combined, our data show that the CDK12/cyclin K1 complex can phosphorylate Ser2 and Ser5 of RNA Pol II CTD, that CDK12/cyclin K can affect gene expression from different promoters, and that both these activities are dependent on cyclin K expression. Interestingly, we observed that CDK9 only modulated reporter gene expression from the SV40 promoter, whereas CDK12 modulated reporter gene expression from both SV40 and CMV promoters. Previous studies show that CDK9 and CDK12 have distinct differences in localization within the cell nucleus, suggesting different activities (4). These observations are consistent with the functional differences between the yeast Bur1 and Ctk-1 complexes, the homologues of CDK9 and CDK12, respectively (28, 41). The Bur1 complex has been shown to phosphorylate Rad6 to regulate histone H2B monoubiquitination, which is associated with transcriptionally active chromatin (31). Likewise, CDK9 has been shown to interact with and phosphorylate UBE2A to regulate H2B monoubiquitination (54). In yeast, the Ctk-1 complex is the major RNA Pol II CTD serine 2 kinase and is involved in the recruitment of histone methyltransferase Set2 and other transcription factors during the mid- and late elongation phases of gene transcription (33, 36). Similarly, Cdk12 has been shown to phosphorylate serine 2 both in vitro and in vivo and appears to be associated with transcription elongation (4). Thus, CDK12/cyclin K and CDK9/cyclin T1 complexes may have different roles in regulating RNA Pol II transcription. Further exploration of the functions of CDK12 and CDK13 are needed to identify CDK12-/CDK13-associated functions, as well as some previously attributed to CDK9, which could have broad relevance in the control of many cellular processes and diseases.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the British Columbia Cancer Agency and the National Science and Engineering Council of Canada (G.B.M).
We thank Samantha H. B. Morin for statistical analysis.
Footnotes
Published ahead of print 17 September 2012
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1. Ahn SH, Kim M, Buratowski S. 2004. Phosphorylation of serine 2 within the RNA polymerase II C-terminal domain couples transcription and 3′ end processing. Mol. Cell 13:67–76 [DOI] [PubMed] [Google Scholar]
- 2. Baek K, Brown RS, Birrane G, Ladias JA. 2007. Crystal structure of human cyclin K, a positive regulator of cyclin-dependent kinase 9. J. Mol. Biol. 366:563–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Balk SP, Knudsen KE. 2008. AR, the cell cycle, and prostate cancer. Nucl. Recept. Signal. 6:e001 doi:10.1621/nrs.06001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bartkowiak B, et al. 2010. CDK12 is a transcription elongation-associated CTD kinase, the metazoan ortholog of yeast Ctk1. Genes Dev. 24:2303–2316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Baumli S, et al. 2008. The structure of P-TEFb (CDK9/cyclin T1), its complex with flavopiridol and regulation by phosphorylation. EMBO J. 27:1907–1918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Blazek D, et al. 2011. The cyclin K/Cdk12 complex maintains genomic stability via regulation of expression of DNA damage response genes. Genes Dev. 25:2158–2172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Buratowski S. 2009. Progression through the RNA polymerase II CTD cycle. Mol. Cell 36:541–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Burset M, Seledtsov IA, Solovyev VV. 2001. SpliceDB: database of canonical and non-canonical mammalian splice sites. Nucleic Acids Res. 29:255–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chen HH, Wang YC, Fann MJ. 2006. Identification and characterization of the CDK12/cyclin L1 complex involved in alternative splicing regulation. Mol. Cell. Biol. 26:2736–2745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chen HH, Wong YH, Geneviere AM, Fann MJ. 2007. CDK13/CDC2L5 interacts with L-type cyclins and regulates alternative splicing. Biochem. Biophys. Res. Commun. 354:735–740 [DOI] [PubMed] [Google Scholar]
- 11. Colwill K, et al. 2006. Modification of the Creator recombination system for proteomics applications—improved expression by addition of splice sites. BMC Biotechnol. 6:13 doi:10.1186/1472-6750-6-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cottone G, et al. 2006. Pkn is a novel partner of cyclin T2a in muscle differentiation. J. Cell. Physiol. 207:232–237 [DOI] [PubMed] [Google Scholar]
- 13. Dai Q, et al. 2012. Cyclin K-containing kinase complexes maintain self-renewal in murine embryonic stem cells. J. Biol. Chem. 287:25344–25352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. de Graaf K, et al. 2004. Characterization of cyclin L2, a novel cyclin with an arginine/serine-rich domain: phosphorylation by DYRK1A and colocalization with splicing factors. J. Biol. Chem. 279:4612–4624 [DOI] [PubMed] [Google Scholar]
- 15. De Luca A, De Falco M, Baldi A, Paggi MG. 2003. Cyclin T: three forms for different roles in physiological and pathological functions. J. Cell. Physiol. 194:101–107 [DOI] [PubMed] [Google Scholar]
- 16. Dimon MT, Sorber K, DeRisi JL. 2010. HMMSplicer: a tool for efficient and sensitive discovery of known and novel splice junctions in RNA-Seq data. PLoS One 5:e13875 doi:10.1371/journal.pone.0013875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Edwards MC, Wong C, Elledge SJ. 1998. Human cyclin K, a novel RNA polymerase II-associated cyclin possessing both carboxy-terminal domain kinase and Cdk-activating kinase activity. Mol. Cell. Biol. 18:4291–4300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Egloff S, Murphy S. 2008. Cracking the RNA polymerase II CTD code. Trends Genet. 24:280–288 [DOI] [PubMed] [Google Scholar]
- 19. Even Y, et al. 2006. CDC2L5, a Cdk-like kinase with RS domain, interacts with the ASF/SF2-associated protein p32 and affects splicing in vivo. J. Cell. Biochem. 99:890–904 [DOI] [PubMed] [Google Scholar]
- 20. Fisher RP. 2005. Secrets of a double agent: CDK7 in cell-cycle control and transcription. J. Cell Sci. 118:5171–5180 [DOI] [PubMed] [Google Scholar]
- 21. Flicek P, et al. 2011. Ensembl. Nucleic Acids Res. 39:D800–D806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fraldi A, Licciardo P, Majello B, Giordano A, Lania L. 2001. Distinct regions of cyclinT1 are required for binding to CDK9 and for recruitment to the HIV-1 Tat/TAR complex. J. Cell Biochem. Suppl. Suppl. 36:247–253 [DOI] [PubMed] [Google Scholar]
- 23. Fu TJ, Peng J, Lee G, Price DH, Flores O. 1999. Cyclin K functions as a CDK9 regulatory subunit and participates in RNA polymerase II transcription. J. Biol. Chem. 274:34527–34530 [DOI] [PubMed] [Google Scholar]
- 24. Fuda NJ, Ardehali MB, Lis JT. 2009. Defining mechanisms that regulate RNA polymerase II transcription in vivo. Nature 461:186–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Giacinti C, Giordano A. 2006. RB and cell cycle progression. Oncogene 25:5220–5227 [DOI] [PubMed] [Google Scholar]
- 26. Glover-Cutter K, et al. 2009. TFIIH-associated Cdk7 kinase functions in phosphorylation of C-terminal domain Ser7 residues, promoter-proximal pausing, and termination by RNA polymerase II. Mol. Cell. Biol. 29:5455–5464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Griffith M, et al. 2010. Alternative expression analysis by RNA sequencing. Nat. Methods 7:843–847 [DOI] [PubMed] [Google Scholar]
- 28. Guo Z, Stiller JW. 2004. Comparative genomics of cyclin-dependent kinases suggest co-evolution of the RNAP II C-terminal domain and CTD-directed CDKs. BMC Genomics 5:69 doi:10.1186/1471-2164-5-69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hirose Y, Ohkuma Y. 2007. Phosphorylation of the C-terminal domain of RNA polymerase II plays central roles in the integrated events of eucaryotic gene expression. J. Biochem. 141:601–608 [DOI] [PubMed] [Google Scholar]
- 30. Hu D, Mayeda A, Trembley JH, Lahti JM, Kidd VJ. 2003. CDK11 complexes promote pre-mRNA splicing. J. Biol. Chem. 278:8623–8629 [DOI] [PubMed] [Google Scholar]
- 31. Keogh MC, Podolny V, Buratowski S. 2003. Bur1 kinase is required for efficient transcription elongation by RNA polymerase II. Mol. Cell. Biol. 23:7005–7018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Khan SZ, Mitra D. 2011. Cyclin K inhibits HIV-1 gene expression and replication by interfering with CDK9-cyclin T1 interaction in Nef dependent manner. J. Biol. Chem. 286:22943–22954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kim M, Ahn SH, Krogan NJ, Greenblatt JF, Buratowski S. 2004. Transitions in RNA polymerase II elongation complexes at the 3′ ends of genes. EMBO J. 23:354–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kinter M, Sherman NE. 2000. Protein sequencing and identification using tandem mass spectrometry. Wiley-Interscience, New York, NY [Google Scholar]
- 35. Ko TK, Kelly E, Pines J. 2001. CrkRS: a novel conserved Cdc2-related protein kinase that colocalises with SC35 speckles. J. Cell Sci. 114:2591–2603 [DOI] [PubMed] [Google Scholar]
- 36. Krogan NJ, et al. 2003. Methylation of histone H3 by Set2 in Saccharomyces cerevisiae is linked to transcriptional elongation by RNA polymerase II. Mol. Cell. Biol. 23:4207–4218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lenasi T, Barboric M. 2010. P-TEFb stimulates transcription elongation and pre-mRNA splicing through multilateral mechanisms. RNA Biol. 7:145–150 [DOI] [PubMed] [Google Scholar]
- 38. Li T, Inoue A, Lahti JM, Kidd VJ. 2004. Failure to proliferate and mitotic arrest of CDK11(p110/p58)-null mutant mice at the blastocyst stage of embryonic cell development. Mol. Cell. Biol. 24:3188–3197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lin X, Taube R, Fujinaga K, Peterlin BM. 2002. P-TEFb containing cyclin K and Cdk9 can activate transcription via RNA. J. Biol. Chem. 277:16873–16878 [DOI] [PubMed] [Google Scholar]
- 40. Liu H, Herrmann CH. 2005. Differential localization and expression of the Cdk9 42k and 55k isoforms. J. Cell. Physiol. 203:251–260 [DOI] [PubMed] [Google Scholar]
- 41. Liu J, Kipreos ET. 2000. Evolution of cyclin-dependent kinases (CDKs) and CDK-activating kinases (CAKs): differential conservation of CAKs in yeast and metazoa. Mol. Biol. Evol. 17:1061–1074 [DOI] [PubMed] [Google Scholar]
- 42. Loyer P, et al. 2008. Characterization of cyclin L1 and L2 interactions with CDK11 and splicing factors: influence of cyclin L isoforms on splice site selection. J. Biol. Chem. 283:7721–7732 [DOI] [PubMed] [Google Scholar]
- 43. Malumbres M. 2007. Cyclins and related kinases in cancer cells. J. BUON 12(Suppl 1):S45–S52 [PubMed] [Google Scholar]
- 44. Mayer BJ. 2001. SH3 domains: complexity in moderation. J. Cell Sci. 114:1253–1263 [DOI] [PubMed] [Google Scholar]
- 45. Moore MJ, Proudfoot NJ. 2009. Pre-mRNA processing reaches back to transcription and ahead to translation. Cell 136:688–700 [DOI] [PubMed] [Google Scholar]
- 46. Mori T, et al. 2002. Cyclin K as a direct transcriptional target of the p53 tumor suppressor. Neoplasia 4:268–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Perales R, Bentley D. 2009. “Cotranscriptionality”: the transcription elongation complex as a nexus for nuclear transactions. Mol. Cell 36:178–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Peterlin BM, Price DH. 2006. Controlling the elongation phase of transcription with P-TEFb. Mol. Cell 23:297–305 [DOI] [PubMed] [Google Scholar]
- 49. Qiu H, Hu C, Hinnebusch AG. 2009. Phosphorylation of the Pol II CTD by KIN28 enhances BUR1/BUR2 recruitment and Ser2 CTD phosphorylation near promoters. Mol. Cell 33:752–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ramanathan Y, et al. 2001. Three RNA polymerase II carboxyl-terminal domain kinases display distinct substrate preferences. J. Biol. Chem. 276:10913–10920 [DOI] [PubMed] [Google Scholar]
- 51. Ramanathan Y, Reza SM, Young TM, Mathews MB, Pe'ery T. 1999. Human and rodent transcription elongation factor P-TEFb: interactions with human immunodeficiency virus type 1 tat and carboxy-terminal domain substrate. J. Virol. 73:5448–5458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Romano G, Giordano A. 2008. Role of the cyclin-dependent kinase 9-related pathway in mammalian gene expression and human diseases. Cell Cycle 7:3664–3668 [DOI] [PubMed] [Google Scholar]
- 53. Romano G, et al. 1999. Regulatory functions of Cdk9 and of cyclin T1 in HIV tat transactivation pathway gene expression. J. Cell. Biochem. 75:357–368 [PubMed] [Google Scholar]
- 54. Shchebet A, Karpiuk O, Kremmer E, Eick D, Johnsen SA. 2012. Phosphorylation by cyclin-dependent kinase-9 controls ubiquitin-conjugating enzyme-2A function. Cell Cycle 11:2122–2127 [DOI] [PubMed] [Google Scholar]
- 55. Shepard PJ, Hertel KJ. 2009. The SR protein family. Genome Biol. 10:242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Shore SM, Byers SA, Dent P, Price DH. 2005. Characterization of Cdk9(55) and differential regulation of two Cdk9 isoforms. Gene 350:51–58 [DOI] [PubMed] [Google Scholar]
- 57. Shore SM, Byers SA, Maury W, Price DH. 2003. Identification of a novel isoform of Cdk9. Gene 307:175–182 [DOI] [PubMed] [Google Scholar]
- 58. Simone C, et al. 2002. Activation of MyoD-dependent transcription by cdk9/cyclin T2. Oncogene 21:4137–4148 [DOI] [PubMed] [Google Scholar]
- 59. Strausberg RL, et al. 2002. Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences. Proc. Natl. Acad. Sci. U. S. A. 99:16899–16903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Trembley JH, Hu D, Slaughter CA, Lahti JM, Kidd VJ. 2003. Casein kinase 2 interacts with cyclin-dependent kinase 11 (CDK11) in vivo and phosphorylates both the RNA polymerase II carboxyl-terminal domain and CDK11 in vitro. J. Biol. Chem. 278:2265–2270 [DOI] [PubMed] [Google Scholar]
- 61. Walker KK, Levine AJ. 1996. Identification of a novel p53 functional domain that is necessary for efficient growth suppression. Proc. Natl. Acad. Sci. U. S. A. 93:15335–15340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wang X, et al. 2006. The proline-rich domain and the microtubule binding domain of protein tau acting as RNA binding domains. Protein Pept. Lett. 13:679–685 [DOI] [PubMed] [Google Scholar]
- 63. Wood A, Shilatifard A. 2006. Bur1/Bur2 and the Ctk complex in yeast: the split personality of mammalian P-TEFb. Cell Cycle 5:1066–1068 [DOI] [PubMed] [Google Scholar]
- 64. Yao S, Neiman A, Prelich G. 2000. BUR1 and BUR2 encode a divergent cyclin-dependent kinase-cyclin complex important for transcription in vivo. Mol. Cell Biol. 20:7080–7087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Yu DS, et al. 2010. Cyclin-dependent kinase 9-cyclin K functions in the replication stress response. EMBO Rep. 11:876–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Zhou M, et al. 2000. Tat modifies the activity of CDK9 to phosphorylate serine 5 of the RNA polymerase II carboxyl-terminal domain during human immunodeficiency virus type 1 transcription. Mol. Cell. Biol. 20:5077–5086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Zhu H, Doherty JR, Kuliyev E, Mead PE. 2009. CDK9/cyclin complexes modulate endoderm induction by direct interaction with Mix.3/mixer. Dev. Dyn. 238:1346–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.