Abstract
The “Hippo” signaling pathway has emerged as a major regulator of cell proliferation and survival in metazoans. The pathway, as delineated by genetic and biochemical studies in Drosophila, consists of a kinase cascade regulated by cell-cell contact and cell polarity that inhibits the transcriptional coactivator Yorkie and its proliferative, anti-differentiation, antiapoptotic transcriptional program. The core pathway components are the GC kinase Hippo, which phosphorylates the noncatalytic polypeptide Mats/Mob1 and, with the assistance of the scaffold protein Salvador, phosphorylates the ndr-family kinase Lats. In turn phospho-Lats, after binding to phospho-Mats, autoactivates and phosphorylates Yorkie, resulting in its nuclear exit. Hippo also uses the scaffold protein Furry and a different Mob protein to control another ndr-like kinase, the morphogenetic regulator Tricornered. Architecturally homologous kinase cascades consisting of a GC kinase, a Mob protein, a scaffolding polypeptide and an ndr-like kinase are well described in yeast; in S. cerevisiae e.g., the MEN pathway promotes mitotic exit whereas the RAM network, using a different GC kinase, Mob protein, scaffold and ndr-like kinase, regulates cell polarity and morphogenesis. In mammals, the Hippo orthologues Mst1 and Mst2 utilize the Salvador ortholog WW45/Sav1 and other scaffolds to regulate the kinases Lats1/Lats2 and ndr1/ndr2. As in Drosophila, murine Mst1/Mst2, in a redundant manner, negatively regulate the Yorkie ortholog YAP in the epithelial cells of the liver and gut; loss of both Mst1 and Mst2 results in hyperproliferation and tumorigenesis that can be largely negated by reduction or elimination of YAP. Despite this conservation, considerable diversification in pathway composition and regulation is already evident; in skin e.g., YAP phosphorylation is independent of Mst1Mst2 and Lats1Lats2. Moreover, in lymphoid cells, Mst1/Mst2, under the control of the Rap1 GTPase and independent of YAP, promotes integrin clustering, actin remodeling and motility while restraining the proliferation of naïve T cells. This review will summarize current knowledge of the structure and regulation of the kinases Hippo/Mst1&2, their noncatalytic binding partners, Salvador and the Rassf polypeptides, and their major substrates Warts/Lats1&2, Trc/ndr1&2, Mats/Mob1 and FOXO.
Keywords: Protein kinase, Hippo, Mst1/2, Mob1, Lats1/2, ndr1/2
1. Introduction and overview
The Hippo tumor suppressor pathway was defined in Drosophila, primarily through screens for genes whose loss of function results in tissue overgrowth. A considerable number of such genes had been identified by analysis of focal overgrowths in mutants undergoing late larval lethality, however no coherent pathways had emerged[1]. A more comprehensive approach to the identification of genes encoding candidate tumor suppressors (TS) was achieved using the Flp/Frt recombination system to enable expression of homozygous lethal mutations as mosaics in viable flies[2]. One of the first and most robust overgrowth phenotypes identified in such screens resulted from mutations in the Warts[3] or Lats[4] gene, which encodes a protein kinase within the AGC family. Two mammalian homologs, Lats1[5] and Lats2[6]/KPM[7] were identified and biallelic deletion of mouse Lats1 resulted in soft-tissue sarcomas and ovarian stromal cell tumours[8], validating its tumor suppressor function. Mutations of Salvador[9] (also called Shar-pei[10]) gave a similar but milder overgrowth phenotype as Warts/Lats, characterized by overproliferation, inhibition of differentiation and a loss of developmental apoptosis. Salvador encodes a non-catalytic protein that binds through its WW domains directly to a PPXY motif on the aminoterminal, noncatalytic segment of Warts/Lats[9]. The following year, five groups independently described the Hippo kinase[11–15], whose mutations result in a phenotype corresponding closely to that of Warts/Lats. Hippo binds and phosphorylates Salvador; moreover Hippo and Salvador individually and synergistically promote Lats phosphorylation in S2 cells[11]. The Hippo kinase domain is homologous to the Ste20 family of protein kinases, and the aminoterminal location of the catalytic domain places it among the Germinal Center (GC) kinases, most closely related to Mst1/stk4 and Mst2/stk3[16]; transgenic expression of human Mst2/stk3 in the fly greatly ameliorates the Hippo phenotype[11]. This finding defined a conserved kinase cascade wherein Salvador binds Lats and Hippo, enabling Hippo to efficiently phosphorylate and activate Lats. An additional component of the core cascade was identified as the noncatalytic protein Mats, mutations in whose gene also phenocopies the Hippo phenotype[17]; Mats binds directly to Warts/Lats. The mammalian Mats orthologs Mob1A and Mob1B were independently identified as preferred substrates of Mst1 and Mst2; Mst1/2-catalyzed phosphorylation of Mob1A/B enhances Mob1A/B’s ability to bind and activate the Lats1 (and ndr1) kinase[18].
The function of the Drosophila Hippo pathway is the inhibition of the transcriptional coactivator Yorkie, inasmuch as deletion of yorkie reverses the LOF phenotypes of Hippo, Warts/Lats, Salvador and Mats[19]. Yorkie inhibition results from its phosphorylation by Warts/Lats at a set of HXRXXS motifs, resulting in the binding of 14-3-3, which mediates Yorkie nuclear exit. The upstream regulation of the Hippo pathway is a much more complex matter; genetic evidence indicates that regulatory inputs are provided in a combinatorial fashion by multiple cell surface transmembrane complexes that mediate and respond to cell-cell contact and cell polarity, the actin cytoskeleton, the intracellular FERM domain proteins Merlin and Expanded, and the WW domain protein Kibra[20–22]. There is scant biochemical data however describing how these inputs are conveyed to the core kinase cassette.
As indicated above, each of the core components of the Hippo kinase cassette are conserved and duplicated in mammalian genomes, except for Salvador (also called WW45) and yorkie (=YAP), although the transcriptional coactivator and YAP paralogue TAZ is also negatively regulated by this pathway[23]. Inhibition of YAP, the mammalian ortholog of Yorkie, by phosphorylation at Ser127 (in human YAP; Ser112 in mouse YAP and Ser168 in Yorkie), serves to best define the operation of the “Hippo” pathway. Studies in mammalian cell culture[24] as well as inactivation of the mouse genes encoding the structural homologues of the core Drosophila kinase cassette indicates that a Hippo-like TS pathway is operative in MEFs[25], in those epithelia studied thusfar[25–32] and in neurons[33]. Nevertheless, inactivation of YAP in some tissues can occur independently of Mst1/Mst2 (MEFs[25], keratinocytes[31]) and perhaps of Lats1/Lats2 (liver[25]) through elements not as yet identified. In Drosophila, Hippo also phosphorylates and activates the kinase Tricornered[34], a regulator of morphogenesis and the orthologue of the mammalian kinases ndr1/ndr2. Although the mechanism of ndr1/ndr2 regulation by phosphorylation provided the model for Lats1/Lats2 regulation[35], upstream control of the Mst1/Mst2-ndr1/ndr2 pathway[36] and the identification of its critical outputs[37] has, in comparison to Mst1/Mst2-Lats1/Lats2-YAP/TAZ, received relatively little attention. Moreover, inactivating mutations of the human Mst1 gene have been recently described in several families who presented with a primary immunodeficiency syndrome involving both T and B cells[38,39]. Mst1 and, to a lesser extent, Mst2 are expressed at highest abundance in the lymphoid system[40], and Mst1-deficient mice also exhibit a deficiency in the number and functionality of naïve T cells[40–43]; this defect is entirely independent of signaling to Lats1/Lats2 and YAP, but due in part to a lack of phospho-Mob1 regulation of the Dock-C subfamily of Rac1 guanyl nucleotide exchange proteins[44]. This chapter is devoted to a review of the structure and regulation and of the kinases Hippo/Mst1&2, their noncatalytic binding partners, and their known physiologic substrates Warts/Lats1&2, Trc/ndr1&2, Mats/Mob1 and FOXO1/3.
2. The “Hippo” kinases, Hippo, Mst1 and Mst2
Mst1 was identified by Chernoff and colleagues, who sought new kinases related to Ste20 using degenerate PCR[45,46]. Krs1(=Mst2) and Krs2(=Mst1) were identified independently by Erikson and colleagues, who purified a ~60kDa kinase activated by severe stress (e.g., arsenite, 0.25M; heat shock, 55°C), or okadaic acid, as detected by “in-gel” assay of cell extracts[47]; this probably corresponded to a kinase they observed previously in cells containing a temperature-sensitive vSrc, that became activated 24 hours after the cells were shifted to the permissive temperature[48]. The Mst1/Mst2 catalytic domain showed homology with the Germinal Center kinase (GCK) subgroup of the Ste20 kinase family. The GCKs are readily subdivided into eight subfamilies, all containing a Ste20-related catalytic domain beginning near their aminoterminus. Each subfamily is distinguished by the unique sequence and domain structures of their carboxyterminal tail[16]. Importantly, a single representative of each of these eight subfamilies is encoded in the Drosophila genome; the group II GCKs e.g., are comprised of Mst1and Mst2, Hippo/CG11228 and the C. elegans cst-1/cst-2. The similarity of the Mst1/Mst2 domain structure to Hippo and the ability of Mst2 to compensate for Hippo deficiency in the fly eye[11], predicts that there will be considerable similarity in the biochemical properties of Hippo and its mammalian orthologues, and a recent report characterizing Hippo biochemistry supports this conclusion[49,50].
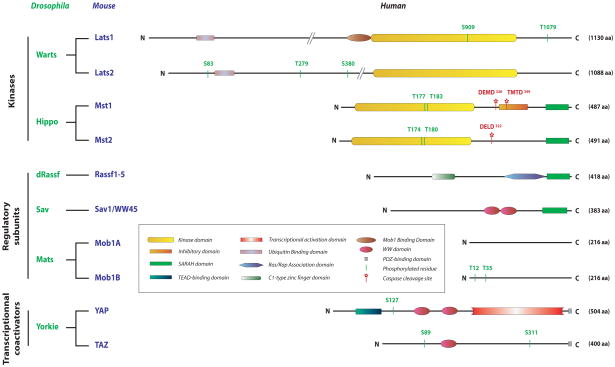
2.1 The SARAH domain
The domain structure of group II GCKs is well maintained in Hippo (669AA) and the shorter Mst1 (487AA) and Mst2 (491AA) polypeptides (~75% identical to each other) and is distinguished by a specialized coiled coil domain near the carboxyterminus (Hippo AA607-655) known as a SARAH domain, an acronym derived from the three gene families that contain homologous domains, i.e., Salvador/Sav1-WW45, Rassf(1–6) and Hippo/Mst1/Mst2[51] (Figure 1). The Mst1 SARAH domain (AA432-480) forms a hairpin of helical segments, with the short N-terminal helical segment (AA433–437) bent back ~45° toward the C-terminal helix (AA441–480); the latter mediates formation of SARAH/SARAH homo/heterodimers through an antiparallel, head-to-tail interaction stabilized by hydrophobic residues[53]. The Rassf polypeptides 1–6 (reviewed in [53]) are noncatalytic polypeptides that all contain near their carboxyterminus a Ras-Rap Association (RA) domain followed immediately by a SARAH domain, through which all six can heterodimerize with Mst1 or Mst2. When transiently expressed, Mst1 or Mst2 is each retrieved as a SARAH domain-mediated homodimer[49,54]. Addition in vitro of an equimolar amount of recombinant Rassf5 (also called Nore1) SARAH domain to the Mst1 SARAH domain displaces the Mst1 SARAH homodimer entirely into an Rassf5-SARAH/Mst1-SARAH heterodimer, utilizing the same Mst1 residues that mediate the Mst1 SARAH homodimer[52]. The Kd of the Mst1/Rassf5 heterodimer is in the nM range, suggesting that this is a constitutive dimer; consistent with this view, immunoprecipitates of Mst1 from mouse lymphoid cells contain Rassf5B[55] and deletion of both Mst1 and Mst2 from the lymphoid compartment results in the complete disappearance of the Rassf5B polypeptide without alteration in Rassf5B mRNA abundance[40,44].
Figure 1.
Domain structure of Mst1/2 and selected substrates and scaffolds.
The structure of the Sav1-WW45 SARAH domain and the Sav1/Mst1 SARAH heterodimer has not been described, but differs from that of the Rassf5/Mst1 heterodimer, inasmuch as the recombinant Sav1/WW45 SARAH domain perturbs a different set of residues on C13/N15-labelled Mst1than does Rassf5[52]. Moreover, addition of the Sav1/WW45 SARAH domain to the Rassf5/Mst1 SARAH heterodimer does not alter the pattern of signals arising from C13/N15-labelled Mst1 detected by NMR, nor is a heterotrimer detectable upon cross-linking or gel filtration[52]. These results suggest that the affinity of the Rassf5 SARAH domain for Mst1 is substantially higher than that of the Sav1/WW45 SARAH domain; moreover, should Sav1/WW45 be present in excess of Rassf5, this behavior predicts that complexes of Mst1 with Nore1 and with Sav1/WW45 will be mutually exclusive. Consistent with this, immunoprecipitates of either Salvador or dRASSF from extracts of Drosophila Kc cells each contain Hippo, but not each other, nor is a trimeric complex detectable when all three recombinant polypeptides are co-overexpressed[56]. However coimmunoprecipitation of endogenous WW45 and Rassf1A from extracts of HeLa [57] and H1792 (a metastatic lung adenocarcinoma; [58]) cells has been reported. Thus, Rassf1A may interact differently with Sav1/Mst1-2 than does Rassf5 (although direct comparisons have not been reported) and further studies will be needed to unravel the interactions between other Rassfs and Sav1 and Mst1-2, to determine whether Mst1 or Mst2 heterotrimers involving Sav1 and Rassf2,3,4 or 6 do exist [e.g., 59] and if so, to define their regulation and output. Evidence from mouse studies so far suggests that Rassf5/Mst1 and Sav1-WW45/Mst1 complexes occur in different tissues and respond to entirely distinct upstream inputs.
2.2 Hippo/Mst1/Mst2 regulation
2.2.1. Autoactivation
When extracted from mammalian cells after transient expression, Mst1 or Mst2 is retrieved predominantly as a dimer of the full length polypeptides, whose activity can be substantially enhanced in vitro by preincubation with Mg++ATP; This in vitro activation results from an auto-transphosphorylation of the Mst1 (at Ser183) or Mst2 (at Ser180) activation loop catalyzed within the homodimer; Mst1[Ser177] is also phosphorylated during activation, however [Ser177Ala] has wildtype activity, whereas [Ser183Ala] is inactive[54]. Coexpression of excess Mst1ATP site mutant [K59R] with wildtype Mst1 results in dimers capable of extensive phosphorylation of the Mst1[K59R] activation loop, that are nevertheless inactive due to failure of transphosphorylation of the Mst1 WT activation loop[54]. Mst1[K59R] (or Mst2[K56R]) thereby serves effectively as a dominant inhibitor of both Mst1 and Mst2[18]. Deletion of the SARAH domain or site-specific mutations (e.g., L444P) that abolish Mst1 homodimerization greatly reduce the ability to autoactivate in vitro and convert the rate of Mst1 phosphorylation from a concentration-independent process to first order kinetics[54]. Internal deletions between the Mst1 catalytic domain and the SARAH domain, e.g., Δ331–394, increase kinase activity about ten-fold during transient expression[49], pointing to the presence in this region of an autoinhibitory domain.
2.2.2. Activation during apoptosis and in normal liver
Another important feature of the segment between the kinase and SARAH domains of mammalian Mst1/Mst2 is the presence of cleavage sites for caspase 3[60–62] located just distal to the catalytic domain (Mst1-DEMD326; Mst2-DELD321; human Mst1, caspase 6/7 TMTD349[63]); such caspase cleave sites are lacking in Hippo. Numerous proapoptotic stimuli result in the generation of ~36kDa Mst1/Mst2 polypeptides that are catalytically active, as shown by their more extensive activation loop phosphorylation as compared to the full length Mst1/Mst2 polypeptides. Moreover, although the spontaneous fractional activation of full length recombinant Mst1 (~4%) or Mst2 (~20%) during transient expression is modest[54], transiently expressed Mst1 (or Mst2) itself is a potent initiator of apoptosis, yielding abundant Mst1(or Mst2) active catalytic fragments[60,64]. Caspase cleavage alone is likely not sufficient for activation; transiently expressed recombinant Mst1[1-330] has low activity and Ser183 phosphorylation. Thus Mst1/Mst2 activation precedes and probably promotes susceptibility to caspase cleavage[54,65]. Recombinant overexpressed Mst1/Mst2 initiate apoptosis through p53- and Jnk-dependent pathways[60,64], and the activated caspase 3 may simply cleave preferentially the already activated Mst1/Mst2 polypeptides. The mechanism(s) by which diverse proapoptotic stimuli activate endogenous Mst1/Mst2 is however not known.
The caspase-cleaved, activated Mst1/2 catalytic fragments differ from full length Mst1/Mst2 in several respects. Full length recombinant Mst1/Mst2 polypeptides are almost entirely cytoplasmic, although they cycle through the nucleus, as shown by their ability to be trapped in the nucleus by treatment of cells with the Crm1 inhibitor Leptomycin, or by mutation of the dual nuclear export signals (Mst1=AA361-370; AA444-451)[64,66]. In contrast, the caspase cleaved 36kDa Mst1/Mst2 polypeptides exhibit unimpeded nuclear access. Moreover, not only is the caspase cleaved Mst1 fragment more active than the full length due to the loss of the autoinhibitory domain, but its substrate selectivity is altered as well; Mst1 cleavage e.g. reduces affinity for FOXO while enhancing affinity for Histone H2B[67].
Immunoblots of extracts from normal mouse spleen (or MEFs) for Mst1 and Mst2 demonstrate that essentially all of the kinase polypeptides are present as 55–60 kDa polypeptides. In contrast, the majority of immunoreactive Mst1 and a significant fraction of the Mst2 polypeptides in liver extracts are visualized at 36kDa[25]. Immmunoblot with phosphospecific antibodies directed against the Mst1/Mst2 activation loop ([Ser183-P]/[Ser180-P] respectively) gives little or no signal with a splenic (or MEF) extract whereas the liver extracts exhibit an abundant 36 kDa phosphoprotein that by immunoprecipitation is predominantly derived from Mst1. Thus normal liver contains a substantial amount of the constitutively active, monomeric (~40 kDa by gel filtration) Mst1 (and some Mst2) catalytic fragment. A plausible hypothesis is that these catalytically active Mst1/Mst2 fragments are generated by caspase 3 cleavage of a preactivated Mst1; nevertheless, other markers of apoptosis and caspase 3 activation, including the 17 kDa activated caspase 3 are not evident (but are readily elicited by FAS activation in vivo). Nonapoptotic functions of caspase 3 are well documented, e.g. in the differentiation of several cellular lineages[68–70], however the mechanism by which Mst1/Mst2 in liver are activated and cleaved is not known nor is direct evidence for the participation of caspase 3 available. It is tempting to speculate that the apparent abundance of constitutively active Mst1/Mst2 helps maintain the normally low rate of hepatocyte proliferation, however 36 kDa activated Mst1/Mst2 fragments are also detectable in extracts of mouse intestinal epithelium, a cellular compartment with one of the highest rates of cellular turnover.
2.2.3. Regulation by Salvador/WW45
Salvador (Sav, 608AA) encodes tandem WW domains (AA425-456/459-491), through which it binds Lats, and a SARAH domain near its carboxyterminus that mediates binding to Hippo (Figure 1). These domains are preserved in mammalian Sav1/WW45 (383AA) and, although the more aminoterminal AA sequences in Salvador and WW45 diverge considerably, WW45 binds Mst1/2 and Guo et.al., identified endogenous Lats1 as well as Mst2 in a WW45 immunoprecipitate from a HeLa cell extract[57]. Coexpression of Hippo with Sav or Lats results in the phosphorylation of both, whereas expression of all three further enhances Lats (but not Sav) phosphorylation, as visualized by electrophoretic slowing; the Hippo and Sav SARAH domains are required for this effect[11]. Similar responses are reported with coexpression of WW45, Lats1 and Mst2 [57,71]. The impact of WW45 on the regulation of Mst1 and Lats1/2 in more physiologic conditions was examined in mouse primary embryonic keratinocytes, which can be induced to differentiate by addition of Ca++ at mM levels. This stimulus is accompanied by increase Mst1/2[Ser183/180] phosphorylation, by the formation of a complex containing Mst1, Lats1/Lats2 and WW45, and by Lats1/Lats2 and YAP phosphorylation. In addition, whereas Lats is constitutively nuclear, a portion of Mst1 relocalizes to the nucleus and YAP relocalizes to the cytoplasm presumably reflecting its phosphorylation and binding of 14-3-3. In embryonic keratinocytes lacking WW45 due to germline deletion, these responses fail to occur but can be restored by expression of wildtype WW45, but not by a mutant WW45 lacking the SARAH domain[26]. These results illustrate several important features of the mammalian Hippo pathway, at least in Ca++ stimulated embryonic keratinocytes; first they show that the formation of the WW45/Mst1-2/Lats1-2 complex is itself a regulated rather than a constitutive process. Moreover, they indicate that WW45 is required for the physiologic activation of Mst1/Mst2. The biochemical processes that underlie these two features remain to be defined. It should be noted however, that embryonic deletion of Mst11 and Mst2 from mouse skin does not result in YAP dephosphorylation and activation[31]; thus YAP phosphorylation during Ca++-induced keratinocyte differentiation in vitro may not recapitulate the mechanism operative in vivo, or perhaps an alternate mechanism of YAP phosphorylation is recruited in the Mst1Mst2 null keratinocyte.
2.2.4. TAO-an upstream activating kinase for Hippo
Treatment of cells with the protein phosphatase (PP)2A/PP1 inhibitor okadaic acid can activate endogenous or recombinant Mst1 and Mst2 completely[47,49,54]. Such activation of the recombinant Mst1/Mst2, which are found as homodimers, can be easily explained by unopposed ongoing trans-molecular autophosphorylation, as occurs in vitro by addition of Mg++ATP to recombinant Mst1/Mst2[54]. Endogenous Mst1/Mst2 polypeptides however are likely bound through their SARAH domain as a monomer to a regulatory protein subunit (WW45 or an Rassf protein) and/or to a scaffold (e.g., c-Raf) and therefore are perhaps unable to readily autophosphorylate; if so, some additional element is needed for Mst1/Mst2 activation, either to promote autophosphorylation or to catalyze the activating phosphorylation. The ability to autoactivate does not preclude a role for an upstream activating kinase, and e.g. Nek9[72,73] and p38α[74,75] utilize both mechanisms for activation in vivo. Screens using shRNA depletion of Drosophila Ste20 kinases[76] or all 250 protein kinases[77] yielded TAO as a negative regulator of Yorkie output. Loss of TAO function increased proliferation in manner dependent on Hippo, Lats and Yorkie, and was necessary for the ability of the upstream regulators Merlin, expanded and Kibra to suppress Yorkie output. Moreover, TAO was shown capable of directly phosphorylating Hippo on the activation loop, and the mammalian TAO kinase was capable of similarly phosphorylating Mst1/Mst2 and activating Mst2.
The ability of one GCK to activate a second GCK mirrors the arrangement in the S. pombe Septation Initiation Network (SIN) pathway, where the GC kinase cdc7 (40% identical to Mst1 catalytic domain), activates sid1[78], another GC kinase (46% and 40% identical to Mst4 and Mst1 catalytic domains, respectively) which phosphorylates and together with Mob1 activates the protein kinase sid2[79,80] (35% and 32% identical to the ndr1/2 and Lats1/2 catalytic domains, respectively). There are three mammalian TAO kinases (TAOKs)[81–84], which constitute the group VIII GCK subfamily. They share long, relatively conserved but rather nondescript carboxyterminal tails. Although GCKs often function as MAP4Ks, TAOK1 can function as a MAP3K, i.e., directly phosphorylate and activate the MAP2Ks 3 and 6 (which activate the p38 MAPKs) and 4 (which activates JNKs)[81,82]. The physiological situations wherein the TAOKs serve as MAP3Ks are however not clear; thus TAOK3 (identified as JIK) inhibits JNK activation and is itself strongly inhibited by EGFR activation[83]. TAOK1 also acts as the immediate upstream kinase for the microtubule affinity regulating kinase MARK, a group II CaMK homologous with the KIN1/PAR-1/Snf1 kinases, that regulates microtubule stability by phosphorylation of a variety of microtubule-associated proteins (MAPs)[85]. This appears to be a physiologic role of the TAOKs, inasmuch as Drosophila TAO regulates microtubular stability[86] and TAOK1 was isolated in a screen for regulators of mitotic progression, and shown as well to regulate the function of the BubR1 (kinase) and MAD2 spindle checkpoint components[87]. Regulation of the TAOKs is as yet poorly defined; regulation by the Merlin/Expanded/Kibra complex may occur although their direct association with TAO is yet to be shown. Signals from the actin or microtubular network, likely upstream inputs to the Hippo pathway[88–90], may communicate with TAOK1, however the molecular elements linking the cytoskeleton to the Hippo pathway are as yet unknown.
2.2.5. Regulation by Rassf polypeptides
The human Rassf polypeptides encompass the protein products of ten genes (Rassf1-10); all ten contain a Ras-Rap association (RA) domain, the basis of their shared name, however only Rassf1-6 contain a SARAH domain and thus the ability to bind Mst1/Mst2[53,91]. The founding member, Nore1[92], renamed Rassf5, was discovered through its ability to bind preferentially and with high affinity to Ras-GTP over Ras-GDP, and subsequently to the GTP liganded form of several other Ras-like small GTPases[93]. Rassf(1–6) all share homologous ~225AA carboxyterminal segments that contain an RA domain followed immediately by a SARAH domain. Rassf1 and Rassf5 are expressed as two predominant splice variants that differ only in their aminoterminal segments upstream of the RA domain. The aminotermini of the longer isoforms (Rassf1A and Rassf5A/Nore1A) contain one or more proline-rich motifs upstream of a C1-type zinc finger; the aminoterminal segments of the shorter variants (Rassf1C and Rassf5B) are nondescript. The Rassf polypeptides gained prominence with the discovery of Rassf1, which is encoded by a gene located on Chr3p21, a region frequently deleted in lung cancer. Importantly, expression of the mRNA encoding the longer Rassf1A transcript is usually silenced in lung cancers and derived cell lines by promoter hypermethylation; reexpression of Rassf1A in these lines strongly suppresses proliferation and induces apoptosis[94]. Rassf1A has emerged as perhaps the most frequently epigenetically silenced human tumor suppressor gene[95].
The relationship of Mst1/Mst2 with the Rassf proteins was first suggested by the retrieval of Mst1 in a two-hybrid screen with Rassf1A. In turn a two-hybrid screen with Mst1 retrieved Rassf(1–6) through their carboxyterminal SARAH domains and Mst1 was shown to occur as a constitutive partner of endogenous Rassf5[96]. Seeking a function for the Rassf5/Mst1 complex as a candidate Ras-GTP effector, it was demonstrated that the apoptosis induced by overexpression of val12-Ki-Ras is blocked by coexpression with the SARAH domain of either Mst1 or Rassf5. The val12-Ha-Ras oncogene, which binds PI-3 kinase much better than does val12-Ki-Ras, does not elicit apoptosis. In contrast, val12/gly37-Ha-Ras, which cannot bind c-Raf or PI-3 kinase but is unimpaired in its ability to bind Rassf5, elicits apoptosis with an efficacy near that of val12-Ki-Ras; this apoptotic response is also inhibited by overexpression of the Mst1 or Rassf5 SARAH domains as well as by recombinant PI-3 kinase[96]. PI-3 kinase is anti-apoptotic through several mechanisms[97], including the ability of Akt to directly phosphorylate and inhibit Mst1/Mst2. Recently it was suggested that val12-Ki-Ras may also activate Akt by a paracrine mechanism involving increased EGF production, which activates the endogenous wildtype Ras and in turn, PI-3 kinase[98].
Whether val12-Ki-Ras-induced apoptosis has a physiologic counterpart is unclear; consequently, the finding that the endogenous Rassf5/Mst1 complex can mediate this action did not clarify the physiologic function(s) of this complex or the identity its physiologic upstream regulator(s). Several years later, Kinashi and colleagues identified Rassf5B (the predominant Rassf5/Nore1 isoform in lymphoid tissues, which they dubbed RAPL) as an intermediate in the pathway by which T cell antigen receptor stimulation, through activation of the Rap1 GTPase results in the “inside-out” activation of the integrin LFA-1[99,100]. Thus in naïve T cells, Rap1-GTP generated in response to T cell receptor stimulation binds to the Rap-Ras association (RA) domain of Rassf5B and the complex of Rap1GTP-Rassf5B–Mst1 is recruited from its basal localization in the Golgi to the immune synapse, concomitant with Mst1 activation. Thus in the T cell, the Rassf5B/Mst1-2 complex is inactive until an activating upstream signal (Rap1-GTP) appears. Studies in mouse T cells lacking expression of Mst1 and Rassf5b strongly support the requirement for this complex in the Rap1-GTP regulation of integrin activation[101]. In contrast, despite evidence from gene deletion that Rassf5A may be a tumor suppressor[102], the physiologic function(s) of the Rassf5A/Mst1 complex remain poorly understood.
The precise mechanism of Mst1 activation in complex with Rassf5 and Rap1-GTP is not known. Coexpression of Mst1 with an excess of any of the recombinant Rassf5 or Rassf1 polypeptides displaces the Mst1 homodimer into an Rassf/Mst1 heterodimer, greatly inhibits the extent of Mst1[Ser183] phosphorylation in vivo and abolishes completely the ability of Mst1 to autoactivate in vitro[54]. This behavior implies that the endogenous Rassf5B/Mst1 heterodimers are inactive awaiting an activating input, analogous e.g., to the heterodimeric, cyclic-AMP-activated protein kinase. Coexpression of Mst1 with excess of Rassf5 and activated forms of Ras does not result in the activation of the bulk of Mst1, however the Mst1 polypeptides retrieved in association with Ras-GTP show a higher extent of Ser183 phosphorylation than does the bulk Mst1[54]. This behavior suggests that a continued association of the Rassf5/Mst1 complex with the activated GTPase is required to maintain kinase activation. This may reflect rapid dephosphorylation once freed from the GTPase, and is possibly a mechanism to restrict Mst1 action, e.g., to limit the nuclear actions of Mst1. Ras/Rap1-induced activation of Mst1 may occur simply because membrane localization increases Mst1 concentration sufficiently to enable transphosphorylation; fusion of the aminoterminal myristoylation motif of cSrc onto Mst1 is sufficient to strongly activate Mst1[54]. Nevertheless, the participation of upstream kinases in the activation of Mst1 and Mst2 is a possibility that has received insufficient attention until the recent description of the ability of TAO to activate Hippo[76,77]. The restricted and transient nature of Ras/Rap1 activation of Mst1/2 differs from that of the Raf kinase, which subsequent to binding Ras-GTP is converted to a stably active form through altered phosphorylation and 14-3-3 binding[103,104].
As compared with Rassf5B, the activating inputs operating upstream of Rassf1A and the regulation of Mst1/2 bound to Rassf1A (or to Rassf2,3,4,6) are poorly understood. Although val12-Ras can associate with Rassf1 when both are overexpressed[105] the affinity of Ras- and Rap1-GTP for the Rassf1 RA domain is more than thirty fold lower than for the Rassf5/nore1B RA domain[106]. Recombinant Rassf1A, through its ability to heterodimerize with Rassf5A, can associate with recombinant activated Ras[93], however recruitment of endogenous Rassf1A by ras-like GTPases has not been observed and probably does not occur. In contrast to the finding that Rassf1A and Rassf1C are only inhibitory when coexpressed in excess of Mst1 (conditions wherein all Mst1 polypeptides are bound by an Rassf polypeptide)[54], several groups have reported that transient expression of Rassf1A, but not the shorter Rassf1C, is accompanied by activation of endogenous or coexpressed Mst1 or Mst2[57,107,108]. The differences in experimental conditions that account for these differing results are not known. Rassf1A and Rassf1C share identical RA and SARAH domains and bind Mst1 comparably, so that the selective ability of Rassf1A to activate Mst1 must depend on its unique aminoterminal segment. Assuming that, as for the Rassf5B/Mst1 complex, upstream inputs exist that can activate Rassf1A-bound Mst1/2, activation of Mst1 selectively by Rassf1A may occur through the binding of the Rassf1A aminoterminal segment to an element that activates the bound Rassf1A/Mst1-2 complex. Alternatively, the activation of Mst1 by Rassf1A might occur indirectly, e.g. through an ability of Rassf1A to trigger pathways that activate Mst1/Mst2, such as those that are recruited, e.g., during apoptosis.
Several reports have implicated Rassf1A in the mechanism by which FAS and TNFα promote the activation of Mst1/Mst2 as well as apoptosis[107–109]. RNAi-induced depletion of Rassf1A reduces the ability of FAS or TNF-R1 activation to promote Mst1/Mst2 activation and also partially inhibits the apoptotic response; depletion of Mst1/Mst2 also reduces the apoptotic response. Foley et.al.[110], have observed the association of Rassf1A with TNF-R1 and DR4 death receptors, mediated by the C1 zinc finger unique to the Rassf1A aminoterminus. Nevertheless, the recruitment of Rassf1A to these receptors is delayed by several hours after receptor activation and is dependent on prior recruitment of the BH3 protein MOAP-1(also called MAP-1). The increase in Mst1/Mst2 kinase activity in response to FAS or TNFα (+cycloheximide) also is not evident however until 3–4 hours after receptor stimulation[109], suggesting that an indirect mechanism of activation is operative. Apoptosis is known to recruit many amplifiers in a feed-forward, self-magnifying cascade and the contribution of apoptosis itself to Mst1/Mst2 activation by death receptors is unclear. Agents such as staurosporine or osmostress, which activate apoptosis through the intrinsic pathway independently of death receptors, can activate Mst1/Mst2 with a lag similar to that seen with stimulation of death receptors[54]. The very delayed response of Mst1/Mst2 activity to death receptor stimulation, the ability of recombinant Rassf1A itself to promote apoptosis (an effect only partially inhibited by deletion of its SARAH domain[110]) and the ability of apoptosis, through undefined mediators, to activate Mst1/Mst2 (prior to their caspase-catalyzed cleavage) leave open the mechanism of Mst1/Mst2 regulation downstream of death receptors and the role of Rassf1A. It is likely that Rassf1A, like Rassf5B, acts as the recipient/mediator of an activating signal for the bound Mst1/Mst2, however the identity of this input remains unclear[111].
2.2.6. Regulation of Mst2 by RAF1
The Raf kinases are MAP3Ks regulated directly by Ras-GTP whose best defined physiologic substrates are MAP2K1/2, the activators of Erk1 and Erk2[103,104]. Inactivation of the murine RAF1 (also called c-Raf or Raf-1) gene results in embryonic lethality due to widespread apoptosis[112,113]. Knockin of a mutant RAF1 gene (YY340/341FF) that lacks the ability to phosphorylate and activate the MAP2K1/2 and that fails to activate Erk1/2 is nevertheless able to rescue fully from lethality[114], prompting a search for kinase-independent functions of RAF1 relevant to the control of apoptosis. Proteomic studies have identified several protein kinases that coprecipitate with RAF1, including Mst2[115] and ASK1[116], both proapoptotic kinases, and Rokα[117,118]. RAF1 antagonizes the activation of these kinases through a mechanism independent of RAF1 catalytic activity. Kolch and coworkers[115] propose that RAF1 binding to Mst2 SARAH domain interdicts Mst2 homodimerization and autoactivation, and recruits protein phosphates 2A thereby promoting Mst2 inactivation. RAF1 catalytic activity is dispensable for these functions, so that RAF1 serves as a scaffold, functioning like its paralogue KSR[119,120]. In cell culture, overexpression of Rassf1A (but not Rassf1C) is reported to disrupt the RAF1/Mst2 complex, which may occur as well in response to FAS. The Rassf1A/Mst2 complex is proposed to phosphorylate YAP, and promote p73 activation which results in apoptosis[108].
Defining the physiologic relevance of biochemical responses uncovered in cell culture can be challenging, and the in vivo context wherein the regulation of Mst2 by the RAF1 polypeptide is biologically significant is not clear. Notably, there is no genetic evidence in support of Mst2 regulation by Raf kinases. Mst2 does not associate with BRaf [115], the closest mammalian homolog of Drosophila Raf, leading Matallanas et.al., to infer that Hippo is not regulated by dRaf [104]. We retrieved no viable Mst2−/−Raf1−/− offspring in multiple litters obtained by intercrossing Mst2−/−Raf-1+/− mice (Zhou and Avruch, unpublished). Nevertheless, tissue-specific gene deletion might be better suited for uncovering a functional relationship between RAF1 and Mst2. By example, cardiac-specific RAF1 inactivation causes heart dysfunction and dilatation as well as cardiac fibrosis; these phenotypes are reversed by concomitant deletion of ASK1[121]. Similar experiments with RAF1 and Mst2 might provide better evidence to support a role for RAF1 as a negative regulator of Mst2 activation in vivo. A different functional relationship between RAF1 and Mst2, i.e., an ability of Mst2 to promote RAF1 signaling, was recently described. RNAi depletion of Mst2 or Lats1/2 in several cell lines was found to impair EGF-induced Erk activation concomitant with diminished levels of the protein phosphatase 2A catalytic subunit and increased levels of the inhibitory RAF1(Ser259) phosphorylation[122]. Further surprises concerning the relationships between the Ras/Raf and Hippo pathways are likely to emerge.
2.2.7. Regulation of Mst1/Mst2 by Akt
Although genetic interactions between dAkt and Hippo have not been described, a series of reports support the likelihood that Akt is a physiologically relevant negative regulator of Mst1 and perhaps Mst2. In the initial description of Mst1, it was observed that EGF treatment of COS cells reduced the kinase activity of transiently expressed Mst1 by ~50%, as measured by an in-gel kinase assay[45]. The decrement was maximal at one minute and returned to baseline by thirty minutes; TPA, LPA, serum were without effect. Despite the ability of okadaic acid treatment to activate endogenous and recombinant Mst1/Mst2 in cells[47,54], treatment in vitro of transiently expressed Mst1 with protein phosphatase 2A caused a 3–4 fold increase in kinase activity, pointing to the presence of an inhibitory phosphorylation[45]. Constitutively active variants of PI-3 kinase[96,123] and Akt[123,124] inhibit apoptosis induced by recombinant Mst1 as well as Mst1 cleavage and nuclear translocation; in addition, increased activation of endogenous Mst1 is evident in Akt1-null as compared with WT MEFs and after RNAi depletion of Akt2[123]. Mst1 and Mst2 contain one canonical consensus sequence for Akt phosphorylation RLRNKT120/117L, which is well conserved in Hippo (RLRKKT132L), and several other related but variant motifs that are not conserved. Mst1[T120] was identified as a site of Akt-catalyzed phosphorylation in response to IGF-1; Mst1[T120D] exhibits greatly reduced Mst1 kinase activity in vitro, and reduced caspase cleavage and nuclear localization in cells. Mst1[T120A] by contrast exhibits higher levels of caspase cleavage and nuclear localization as compared with Mst1 WT[123]. The Mst1 nuclear export signals (AA361–370 and 444–451) are removed by caspase cleavage at D326, resulting in constitutive nuclear localization[64,65]. Nuclear residence of Mst1[1-326] can be greatly reduced by a T120D mutation or by overexpression of activated Akt; the latter does not alter the nuclear localization of Mst1[1-326/T120A][123]. IGF1 and Akt were subsequently reported to also inhibit Mst2 activity, susceptibility to caspase cleavage and apoptotic efficacy, concomitant with phosphorylation of Mst2[T117][125]. These findings may explain an earlier report that serum withdrawal from NIH3T3 cells produced an increase in the activity of a 33kDa kinase evident by in-gel assay reactive that was reactive with an anti-Mst2 antibody. This kinase activity was first evident four hours after withdrawal and maximal by twelve hours; readdition of serum reduced kinase activity over a six hour interval[126].
A second, Mst2-specific mechanism by which Akt-catalyzed phosphorylation regulates Mst2 function was proposed by Romano and coworkers[127]; they report that Mst2[T117] phosphorylation reduces Mst2 binding to Rassf1A while increasing its binding to RAF1, thereby promoting Mst2 inhibition beyond that achieved by the phosphorylation itself. Conversely, the enhanced binding of Mst2 to Rassf1A enabled by [T117] dephosphorylation further enhances Mst2 activity. The relative importance of this mechanism as compared with the direct effects of phosphorylation on Mst2 function is not known.
Ye and colleagues[124] identified Mst1[T387] (KRRDET387M) as an avid site of Akt phosphorylation in vitro; EGF treatment of cells stimulated T387 phosphorylation, a response suppressed by inhibitors of PI-3 kinase but not of PKC. The Mst1[T387E] mutant exhibited reduced cleavage in response to H2O2, reduced ability to induce apoptosis and was selectively impaired in its ability to phosphorylate FOXO3, a modification that enhances FOXO3 nuclear translocation and proapoptotic activity; the [T387A] mutant exhibited opposite effects. A phosphorylation in this region of Mst2 has not been observed. Interestingly, the protein phosphatase M family members PHLPP1/2 (pleckstrin homology (PH) domain leucine-rich repeat protein phosphatase), the enzymes that dephosphorylates Akt[S473][128], has been found in complex with Mst1 in cells and in vitro catalyzes the dephosphorylation of T387. Overexpression of PHLPP is accompanied by a modestly increased Mst1[T183] phosphorylation, as well as activation of Jnk, p38 and induction of apoptosis; PHLPP activation of the stress kinases can be partially suppressed by coexpression of the kinase-inactive Mst1[K59R] mutant[129]. The PHLPP polypeptide contains RA and PH domains and a leucine-rich segment that binds nucleotide-free Ras, however the regulation of PHLPP activity in vivo is obscure. Nevertheless, the possibility of some linked reciprocal regulation of Akt and Mst1 is attractive. Another mechanism for such reciprocal regulation was suggested by the finding of a complex of Akt and Mst1 in the prostate cancer cell line LNCaP[130]. Mst1 and Mst2 were shown to bind and inhibit an activated wildtype Akt; full length Mst1 as well as the Mst1 fragments 1–330 and 331–487 all bind to Akt near its carboxyterminus and all can inhibit Akt activity when overexpressed. Overexpression of these fragments in a zebrafish model suppressed the phenotypes elicited by activated Akt. Whether this inhibition reflects simply the ability of an overexpressed Akt substrate to interfere with the binding of other substrates, or a physiologic regulatory mechanism is not known.
Recently, genetic evidence from Drosophila indicates the occurrence of reciprocal regulation between Ins/IGF1 receptor and the Hippo signaling pathways. Thus the ability of dp110 PI-3 kinase to promote cell proliferation depends strongly on yorkie, and is accompanied by a diminution of Yorkie(Ser168) phosphorylation. Moreover, dp110 causes no additional overgrowth in a hippo-null background. Conversely, inactivation of wts results in activation of dS6K accompanied by downregulated expression of Tsc1, Tsc2 and PTEN, and increased expression of Ins/IGF1R and PDK1 [131]. The extent to which dp110 PI-3 kinase regulation of yorkie signaling is mediated by Akt phosphorylation of Hippo is not clear, inasmuch as Akt depletion in S2R+ cells did not completely abolish the effect of insulin on Yorkie phosphorylation. Nevertheless, Ye et. al. [132] independently observed that loss of Hippo signaling upregulates Akt expression through Yorkie, and depletion of Akt reduced the overgrowth caused by Mats deficiency by ~40%. Thus the occurrence of cross regulation between the Hippo and Ins/IGF1R pathways is firmly established.
3. Hippo/Mst1/Mst2 substrates
The following discussion is restricted to candidate substrates that are directly phosphorylated in vitro by Mst1/Mst2 at sites modified in vivo and for which in vivo evidence, usually genetic, supports a role for Hippo, Mst1 or Mst2 as the operative kinase. Many early studies of recombinant Mst1/Mst2 demonstrated that overexpression resulted in activation of the stress kinases p38 and JNK/SAPK[60,88,133,134], however no evidence supports the activation of these pathways as physiologic outputs of Mst1/Mst2 rather than indirect responses to a stress state engendered by Mst1/Mst2 overexpression. Similarly, the evidence is weak that Mst1[135], as opposed to PKC[136] is the kinase responsible for Histone H2B[Ser14] phoshorylation during apoptosis.
3.1. Warts/Lats and Lats1/Lats2
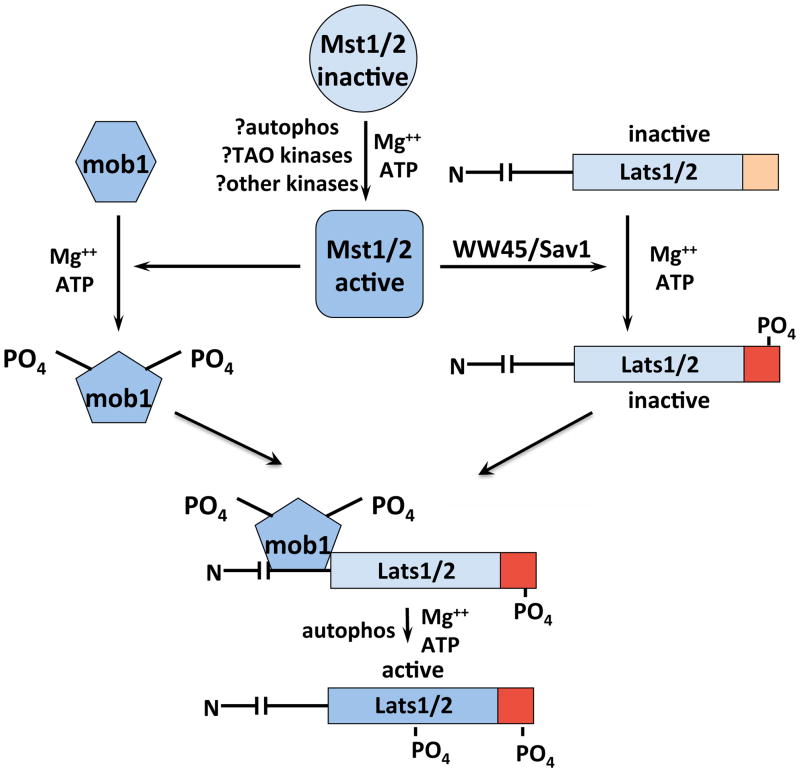
The findings that Hippo loss of function mutations gave a phenotype closely resembling that caused by inactivation of Warts or Sav and strongly suppressed the phenotypes caused by overexpression of those elements, together with the ability of all three polypeptides to physically interact, pointed to the operation of a kinase cascade. Wu et.al.[11], demonstrated that Hippo and Sav individually promoted slowing of Warts electrophoretic mobility and together did so synergistically. A similar slowing of Lats1 was induced by cotranslation in vitro with Mst2. Such slowing usually reflects phosphorylation but not necessarily regulation, however recombinant Mst1 or Mst2 (but not Mst4), in the absence of Sav1/WW45, phosphorylated Lats1 and Lats2 in vitro and increased the ability of Lats1/Lats2 to autphosphorylate, whereas Lats1/Lats2 did not modify Mst1/Mst2[137]. Human Lats1 is an 1130 AA polypeptide containing a kinase domain between AA705-1013 of the AGC subfamily, most closely related to the kinases ndr1/ndr2. Immediately carboxyterminal to the Lats1 catalytic domain is a more hydrophobic segment extending to AA1091 that shares ~ 35–40% identity to the corresponding segments carboxyterminal to the catalytic domains of several other AGC kinases, e.g. Akt, S6K1, PKC, ndr. Deletion of Lats1 AA1-588 did not alter the ability of Lats1 to be activated by Mst2 in vitro or in cells by okadaic acid, but Lats1[662–1130] was unresponsive to those treatments. A set of sites phosphorylated on kinase dead-Lats1[589–1130] in vitro by Mst2 were identified by mass spectroscopy, and the five sites conserved in Lats2 were mutated to Ala; only Lats1[S909A] and [T1079A] were no longer activatable by Mst2. Lats1[T1079E] had low basal activity but was activated by Mst2 whereas [S909D or E] was entirely inactive[138]. Previous work had established that activation of the closely related ndr1/ndr2 kinases occurred through phosphorylation at the homologous sites (S281/282 and T444/442)[35]; S281/282 was modified by autophosphorylation whereas T444/442 phosphorylation required an upstream kinase, first identified as Mst3[138]. The homologues of Lats1 and ndr1 in S. cerevisae are the kinases Dbf2 and Cbk1 respectively[139], which regulate late steps in mitosis (see 3.5). Genetic evidence indicating that the Mob1 and Mob2 polypeptides are required for the action of Dbf2 and Cbk1 respectively led Hemmings and coworkers to examine the effect of the human Mob1 proteins on ndr1/ndr2[140,141] and subsequently on Lats1/Lats2[142]. Mob1 was found to stimulate the autophosphorylation of both ndr and Lats in vitro; in cells, coexpression with Mob1 per se did not promote activation but augmented activation by okadaic acid an additional 2–4 fold. Mob1 binds to a highly conserved, positively charged segment in both the ndr and Lats kinases located immediately aminoterminal to the catalytic domain, and mutation in this segment (or deletion, as in Lats1[662–1130]) strongly inhibits activation by okadaic acid. The ndr and Lats kinases both also contain a unique, basically charged insert in their activation loop not found in other AGC family kinases; mutations in this insert increase kinase activity substantially and reduce the stimulatory effect of Mob1 on autophosphorylation [140,142]. Thus it appears that the binding of Mob1 reduces autoinhibition by this kinase insert, enabling autophosphorylation within the activation loop. In summary, Mst1/Mst2 phosphorylates the Lats1/Lats2 hydrophobic domain [T1079/T1041], a modification necessary but not sufficient for activation. The binding of Mob1 to a basic region aminoterminal to the catalytic domain relieves an autoinhibitory interaction mediated by a basic insert in the activation loop, enabling Lats autophosphorylation [S909/872]; as with a subset of other AGC kinases such as Akt, S6K1 and others, this dual phosphorylation is required for Lats1/Lats2 (and ndr1/ndr2) activation[143] (Figure 2).
Figure 2.
Mechanism of Mst1/2 activation of Lats1/2. See text for details.
3.2. Tricornered and ndr1/ndr2
Tricornered (Trc) is the Drosophila ortholog of ndr1/ndr2[139]. Genetic evidence places Hippo upstream of both Lats and Trc in sensory neurons and studies in S2 cells and biochemical studies demonstrate an ability of Hippo to associate with and phosphorylate Trc at the hydrophobic site S449[22]. Interestingly, dTOR complex 2 is also required for Trc[S449] phosphorylation[144]; whether dTORC2 and Hippo serve as alternative activators or somehow interact to regulate Trc is not known. Trc loss of function results in recessive lethality[145]; the mosaic LOF phenotypes are in epithelia, with split hairs and bristles, and in sensory neurons, which show increased dendritic branching but a loss of tiling, i.e., the ability to cover a sensory field without the crossing of dendrites from other neurons of the same type. Lats mutants tile normally but the dendritic branches are not maintained[22]. Little is known regarding the Trc downstream effectors, save that they involve control of cadherins and the actin cytoskeleton[146]. The function of Trc depends absolutely on the scaffold protein Furry[147–149], a 3479AA polypeptide with homologues in yeast and mammalian cells that are also required for ndr signaling[150]. As with Lats1/Lat2[140], ndr1/ndr2 possess a basic region aminoterminal to the catalytic domain that mediates Mob1 binding, and an autoinhibitory segment inserted into the proximal part of the activation loop[140]. The binding of phospho-Mob1 enables ndr1 disinhibition and stimulates autophosphorylation of the activation loop; full activation results from phosphorylation at the ndr1/ndr2 hydrophobic site by Mst1, Mst2 or Mst3, which is facilitated by ndr binding of phospho-Mob1[109] (not true of Mst1Mst2-catalyzed Lats1/Lats2 phosphorylation). Each of these inputs is linked to a distinct output-in G1, ndr1 phosphorylates p21[S146] (its only known in vivo substrate) promoting p21 degradation; Mst3 appears to be necessary uniquely for ndr1 activation in G1[37,137]. Mst1 regulates the ability of ndr1 to promote centrosome duplication early in S[151–153] and Mst1, in complex with Rassf1A, is reported to activate ndr1 in response to FAS, resulting in apoptosis[109]. In mitosis, Mst2 together with Fry and Mob2 mediate the ability ndr1 to promote chromosome attachment at the metaphase spindle[150]. The likelihood is high that specific scaffolds mediate each of these outputs, however the composition of these complexes and mechanistic insight into their operation remain largely unknown.
3.3. Mats and Mob1A/Mob1B
The Mob genes were discovered in yeast through mutations that interfere with mitotic exit, and the Mob polypeptides were shown to bind and function as obligatory cofactors for the yeast Dbf-related (i.e., ndr-Lats-like) protein kinases[154]. There are four Mob-related gene families in humans[see supplement in ref. 18] and the newly revised and simplified nomenclature will be used here. The Mob polypeptides range from 216AA (Mob1A and Mob1B, 95% identical) to 268AA. Mob1A and Mob1B bind and coactivate ndr1/2 and Lats1/2. Mob2 binds to ndr1/ndr2 competitively with Mob1and is inhibitory but does not bind Lats1/Lats2[155]. The three Mob3 polypeptides and the single Mob4 do not appear to regulate ndr/Lats kinases. The participation of Mob1 in the Hippo pathway, although anticipated from the composition of the yeast MEN/SIN pathways, was established by the description of the drosophila Mob as tumor suppressor (Mats). Mats loss of function phenocopies loss of the other core Hippo elements and is rescued by mouse Mob1A; the Mats polypeptide was shown to bind and activate the Lats kinase[17].
The importance of Mob1 phosphorylation was first suggested by the observation that Mob1 extracted from okadaic acid-treated cells showed much stronger binding to ndr1 in vitro whereas ndr1 extracted from okadaic acid treated cells was unaltered in its ability to bind Mob1[140]. Subsequently, Wei et.al.[156] demonstrated that Mats coprecipitated with Hippo and could be phosphorylated by Hippo in vitro, resulting in enhanced Mats binding to Lats and Lats1 activity towards YAP. Independently, Praskova et.al.[18], using activated Mst1 to phosphorylate fractionated extracts from mouse L1210 B cell leukemia cells, identified a mixture of Mob1A/Mob1B as the dominant Mst1 substrate. It bears emphasis that Mob1A/B are the only Mst1/Mst2 substrates identified thusfar whose phosphorylation does not require, nor is facilitated in vivo or in vitro by a scaffold. Mst1 and Mst2 catalyze a near-stoichiometric phosphorylation of Mob1B at two sites, Thr12 and Thr35, at a rate >100 fold faster than Mst1 phosphorylation of histone H2B. Thr35 phosphorylation facilitates Thr12 phosphorylation. Phosphorylatable residues at these sites are absent in Mob2 and Mob4 but present in the Mob3 polypeptides; nevertheless, Mst1/Mst2 phosphorylate Mob3B (identified as Mobkl2B by the previous nomenclature) at ~5% the rate of Mob1B. Little or no binding of Mob1 to Mst1[K59R]/Mst2[K56R] is evident and Mob1 binding to wildtype Mst1/Mst2 is greatly enhanced if those kinases are first activated. Once Mob1 is phosphorylated, its affinity for Mst1/Mst2 diminishes greatly whereas its affinity for Lats1 is increased (Figure 2).
The phosphorylation of Mob1 at Thr12 and Thr35 depends strongly and perhaps absolutely on Mst1 and Mst2. Mst3 is unable to phosphorylate Mob1 in vitro, and expression of Mst2[K56R] in cell culture suppresses or eliminates Mob1 phosphorylation by okadaic acid or H2O2[18]. More importantly Mob1 phosphorylation in mouse T cells is greatly diminished by inactivation of the Mst1 gene[40], and dual inactivation of Mst1 and Mst2 in a tissue-specific manner eliminates Mob1 phosphorylation entirely from mouse thymocytes[44], liver[25] and intestinal epithelium[32], the compartments in which this has been examined thusfar. Thus Mob1 phosphorylation is a reliable monitor of Mst1/Mst2 activation.
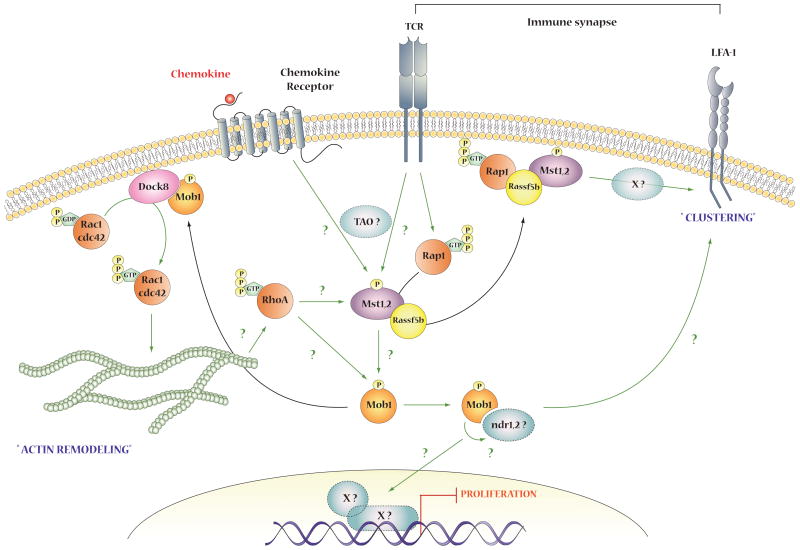
Although the ability of phospho-Mob1A/B to bind and activate Lats1/Lats2 (and ndr1/ndr2) is indisputable, this outcome does not always accompany Mst1/Mst2 activation and Mob1 phosphorylation. Thus deletion of Mst1 and Mst2 from mouse liver results in the immediate onset of hyperproliferation; Mob1 phosphorylation is lost but Lats1/Lats2 phosphorylation is minimally altered. The Mst1/Mst2-deficient livers develop multifocal hepatocellular carcinomas. Cell lines derived from these tumors show continued Lats1/Lats2 phosphorylation; reexpression of Mst1 dramatically suppresses proliferation and induces apoptosis of these cells with little change in Lats1/Lats2 phosphorylation. Doubly surprising is that in spite of the minimal change in Lats1/Lats2 phosphorylation subsequent to elimination of Mst1/Mst2 from mouse liver, there is essentially complete loss of YAP[Ser112] phosphorylation. These findings suggest that a kinase other than Lats1/Lats2 is the Mst1/Mst2-regulated YAP[Ser112] kinase in liver, and preliminary experiments point to the existence of such a kinase[25]. Another example of the variability of Lats1/Lats2 as an effector of Mst1/Mst2 is provided by the responses of naïve T cells to activation of the T cell antigen receptor (TCR)[40]. Anti-CD3 plus anti-CD28 treatment of naïve T cells from wildtype mice produces rapid Mst1 activation and strongly increases Mob1 phosphorylation. Mst1-deficient naïve T cells exhibit a nearly complete loss of TCR-stimulated Mob1 phosphorylation, significantly reduced transwell migration and no clustering of LFA-1. Lats1/Lats2[S909] and [T1079] phosphorylation in wildtype T cells is modestly stimulated by TCR activation, but largely unaffected by the deletion of Mst1, and YAP[S112] phosphorylation is unaltered. Only with the supraphysiologic stimuli of PMA+ionomycin or H2O2 is the increase in Lats1/Lats2 (and ndr1/ndr2) and YAP phosphorylation observed to be diminished in T cells lacking Mst1. These findings indicate that in naïve T cells, Lats1/Lats2 and YAP are not primary effectors of Mst1; Mst1 regulation of e.g., LFA-1 clustering must be mediated by another Mst1/Mst2 substrate (?ndr1/ndr2) or through a Mob1-P target other than Lats1/Lats2.
A novel Mob1-P effector was recently identified in studies of Mst1/Mst2 deficient thymocytes[44]. Mice with double deletion of Mst1/Mst2 from the lymphoid compartment have essentially no T cells in their circulation or secondary lymphoid organs; thymocyte development proceeds normally, but the Mst1/Mst2-deficient thymocytes exhibit a high level of ongoing apoptosis and thymic egress is reduced by >90%. A proximate cause of failed egress of the Mst1/Mst2-deficient mature thymocytes is the inability of chemokines such as CCL19/21 and sphingosine-1 phosphate (the latter being the immediate stimulus to egress) to promote GTP charging of Rac1 and RhoA, actin polarization and migration. We demonstrated that DOCK8, a Rac1 guanyl nucleotide exchange protein (GEF), binds specifically to Mob1-P; cotransfection with Mob1 results in a modest increase in DOCK8 stimulation of Rac1 GTP charging, despite the minimal phosphorylation and colocalization of transiently expressed Mob1 with Dock8. By contrast, fusion of the cSrc myristoylation motif to the Mob1 aminoterminus targets Mob1 to the membrane, markedly increases its phosphorylation and causes extensive colocalization with DOCK8; these responses do not occur with Myr-Mob1[T12/35A]. In wildtype thymocytes, CCL19/21 induces the binding of DOCK8 to Mob1 and this interaction is lost entirely in Mst1/Mst2 deficient thymocytes. There are eleven DOCK polypeptides, subclasses A-D[157]; DOCK8, which is especially abundant in thymocytes is, together with the more widely expressed DOCK6 and 7, a member of the Class C subfamily. We find that DOCK7 also binds specifically to Mob1-P whereas the class B DOCK2 does not. Thus DOCK-C subfamily of guanyl nucleotide exchangers are Mob1-P effectors. It is likely that other Mob1-P interactors remain to be discovered.
3.4. FOXO1/FOXO3
The FOXO family of transcription factors (FOXO1, 3, 4 and 6) regulate expression of genes concerned with inhibition of cell cycle progression, hematopoietic differentiation, stress-resistance, e.g., repair of DNA damage and scavenging of reactive oxygen species, apoptosis and carbohydrate metabolism[158]. Somatic triple deletion of FOXO1,3 and 4 results in thymomas and hemangiomas, establishing a tumor suppressor role for these proteins[159], although the tumor spectrum is surprisingly narrow. FOXO polypeptides are regulated by phosphorylation[160,161]; they are inhibited by the kinases Akt, Sgk, IKKβ, CDK2 and TAK1 which all promote FOXO nuclear exit, usually mediated by binding of 14-3-3 and in some instances followed by ubiquitination and degradation[162]. Conversely, phosphorylation by Jnk, activated by a variety of stressors, and by Mst1, promote FOXO nuclear residence and transcriptional activity.
Bonni and coworkers[163] demonstrated that Mst1 can phosphorylate FOXO3 (and subsequently, FOXO1) principally Ser207 (Ser212 in FOXO1), a conserved site in the forkhead domain. This phosphorylation interdicts 14-3-3 binding, promotes FOXO nuclear residence and transcriptional activity. The site shows increased phosphorylation in H2O2-treated primary neurons that is Mst1 and Mst2 dependent; depletion of FOXO1/3 or Mst1 suppresses H2O2-induced neuronal apoptosis[164].
The C. elegans FOXO orthologue daf-16 is known to control worm lifespan and aging; elimination of daf-16 shortens lifespan and the increased longevity of worms mutant in the insulin receptor/PI-3 kinase pathway is reversed completely by inactivation of daf-16[165]. The C. elegans genome encodes two Mst1 homologues, cst-1 and cst-2; feeding of cst-1 RNAi depletes both and shortens lifespan markedly without affecting development to adulthood. More specifically, transgenic overexpression of cst-1 delays the appearance of aging phenotypes and prolongs lifespan, an effect that was prevented fully by feeding daf-16 RNAi[163]. Thus the Ins/IGF-1R pathway and Mst1 may converge on daf-16 in the regulation of C. elegans aging.
Mst1 null naïve T cells exhibit high levels of reactive oxygen species, increased susceptibility to paraquat- and γ-radiation-induced ROS and lower abundance of SOD2 and catalase, both FOXO transcriptional targets. Choi et.al.[43] found low levels of FOXO1 and 3 polypeptides in the Mst1 null naïve T cells and transgenic expression of Mst1 or FOXO1[S207D] opposed the excessive ROS. Conversely, Zhou et.al. did not find the levels of FOXO polypeptides or S207/212 phosphorylation to be diminished in Mst1 null naïve T cells[40] or in Mst1/Mst2 doubly deficient thymocytes[44]. Thus, although positive regulation of FOXO1/3 by Mst1/Mst2 is likely a physiologic regulatory event, the circumstances in vivo that engender Mst1/Mst2 phosphorylation of FOXO1/3 phosphorylation are not known.
3.5 Mitotic functions of the Mst1/2 and Lats1/2 kinases
Well before the discovery of the Hippo pathway, work in yeast had identified kinase cascades that regulated late events in mitosis whose central elements are one or two kinases with a GCK-like catalytic domain that activates a kinase with Lats/ndr-like catalytic domain, the activity of the latter dependent on the binding of a Mob protein. Specifically, the S. cerevisiae Mitotic Exit Network (MEN) pathway where the GC kinase Cdc15p (41% identical to the Mst1/Mst2 catalytic domain) activates the ndr/Lats-like kinase Dbf2 (38% identical to Lats1/Lats2 catalytic domain), which binds Mob1p; and the S. pombe Septation Initiation Network (SIN) pathway, where, as described above, one GCK, cdc7 activates a second GCK sid1, which in collaboration with Mob1, activates the ndr-family kinase sid2. Several excellent reviews of these pathways, which undoubtedly are the progenitors of the metazoan Hippo pathway, are available [166–169]; in addition, a discussion of these yeast kinase cascades and of the mitotic functions of the homologous mammalian GC- and ndr-family kinases is provided in the companion review in this issue by Hergovich and Hemmings.
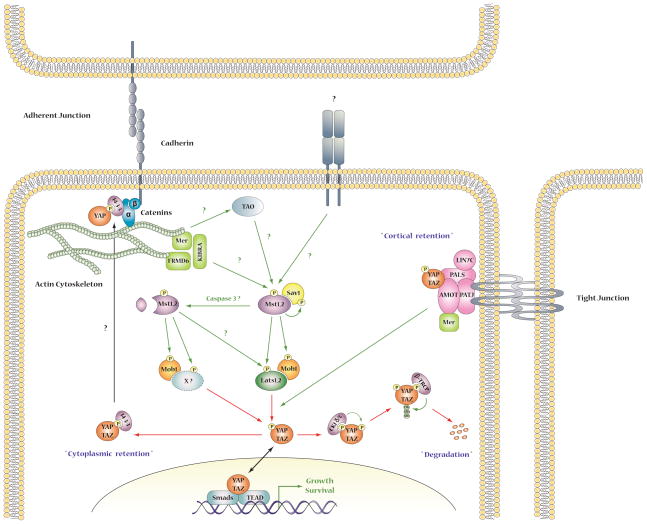
4. Conclusion
The elucidation of the Hippo pathway has uncovered a central mechanism by which cells use information arising from cell-cell contact to regulate cell fate decisions, i.e., proliferation, differentiation or apoptosis. This is a fundamental ability of metazoan cells and one that is violated universally during malignant transformation. The dominant importance of Yorkie and YAP/TAZ as the pathway output is indisputable (Figure 3). The areas of greatest uncertainty how the upstream inputs, e.g. the various cell-cell contact complexes in polarized epithelia and the internal signals arising from the actin and microtubular networks coordinate in the regulation of the Hippo kinase activity, and how other cell fate-determining pathways cross-regulate. In mammalian cells, the biochemical mechanisms underlying the regulation of the Mst1/Mst2 kinases in different tissues appear to be diverse as is best illustrated in T cells, wherein Mst1Mst2 is regulated by the Rap GTPases through Rassf5b, and signal in a Lats/YAP independent manner (Figure 4). Moreover, the regulation of Mst1Mst2 within a single cell may vary depending on the upstream input. Scaffold proteins play a critical role both in the regulation of Mst1/Mst2 activity and in defining its immediate substrates, an area that requires much more investigation. The mechanism and significance of Mst1/Mst2 activation and cleavage in non-apoptotic tissues, e.g., normal liver, requires elucidation. The identity of kinases other than Mst1/Mst2 and Lats1/Lats2 that can regulate YAP/TAZ, and the targets of Mob1-P other than the ndr family kinases and the Dock-C GEFs are questions of immediate interest. Given the overlapping regulation of ndr1/ndr2 with Lats1/Lats2, identification of the targets by which ndr1/ndr2 regulate cell structure and how these morphogenic programs integrate with the proliferative outputs driven by YAP/TAZ are important to define.
Figure 3.
Mst1/2 Regulation and outputs in liver and intestinal epithelia.
Figure 4.
Mst1/2 Regulation and outputs in CD4 or CD8 positive Thymocytes and naïve T cells.
Acknowledgments
The work described herein from the author’s laboratories was supported by NIH grants DK17776 (JA) and CA136567 (to JA and NB), the Sidney Kimmel Foundation for Cancer Research (NB), the Linda J. Verville Cancer Research Foundation (NB), 111 Project of Education of China (NO. B06016, DZ), the Fundamental Research Funds for the Central Universities of China (NO 2010111079, DZ), National Natural Science Foundation of China (NO 81101503, DZ) and Natural Science Foundation of Fujian (NO 2011J05096, DZ).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Watson KL, Justice RW, Bryant PJ. Drosophilia in cancer research: the first fifty tumor suppressor genes. J Cell Sci Suppl. 1994;18:19–33. doi: 10.1242/jcs.1994.supplement_18.4. [DOI] [PubMed] [Google Scholar]
- 2.Xu T, Rubin GM. Analysis of genetic mosaics in developing and adult Drosophila tissues. Development. 1993:1223–37. doi: 10.1242/dev.117.4.1223. [DOI] [PubMed] [Google Scholar]
- 3.Justice RW, Zilian O, Woods DF, Noll M, Bryant PJ. The Drosophila tumor suppressor gene warts encodes a homolog of human myotonic dystrophy kinase and is required for the control of cell shape and proliferation. Genes Dev. 1995;9:534–46. doi: 10.1101/gad.9.5.534. [DOI] [PubMed] [Google Scholar]
- 4.Xu T, Wang W, Zhang S, Stewart RA, Yu W. Identifying tumor suppressors in genetic mosaics: the Drosophila lats gene encodes a putative protein kinase. Development. 1995;121:053–63. doi: 10.1242/dev.121.4.1053. [DOI] [PubMed] [Google Scholar]
- 5.Tao W, Zhang S, Turenchalk GS, Stewart RA, St John MA, Chen W, Xu T. Human homologue of the Drosophila melanogaster lats tumour suppressor modulates CDC2 activity. Nat Genet. 1999;21:177–81. doi: 10.1038/5960. [DOI] [PubMed] [Google Scholar]
- 6.Yabuta N, Fujii T, Copeland NG, Gilbert DJ, Jenkins NA, et al. Structure, expression, and chromosome mapping of LATS2, a mammalian homologue of the Drosophila tumor suppressor gene lats/warts. Genomics. 2000;63:263–70. doi: 10.1006/geno.1999.6065. [DOI] [PubMed] [Google Scholar]
- 7.Hori T, Takaori-Kondo A, Kamikubo Y, Uchiyama T. Molecular cloning of a novel human protein kinase, kpm, that is homologous to warts/lats, a Drosophila tumor suppressor. Oncogene. 2000;19:3101–9. doi: 10.1038/sj.onc.1203659. [DOI] [PubMed] [Google Scholar]
- 8.St John MA, Tao W, Fei X, Fukumoto R, Carcangiu ML, Brownstein DG. Mice deficient of Lats1 develop soft-tissue sarcomas, ovarian tumours and pituitary dysfunction. Nat Genet. 1999;21:182–6. doi: 10.1038/5965. [DOI] [PubMed] [Google Scholar]
- 9.Tapon N, Harvey KF, Bell DW, Wahrer DC, Schiripo TA, Haber DAm, et al. salvador Promotes both cell cycle exit and apoptosis in Drosophila and is mutated in human cancer cell lines. Cell. 2002;110:467–78. doi: 10.1016/s0092-8674(02)00824-3. [DOI] [PubMed] [Google Scholar]
- 10.Kango-Singh M, Nolo R, Tao C, Verstreken P, Hiesinger PR, et al. Shar-pei mediates cell proliferation arrest during imaginal disc growth in Drosophila. Development. 2002;129:5719–30. doi: 10.1242/dev.00168. [DOI] [PubMed] [Google Scholar]
- 11.Wu S, Huang J, Dong J, Pan D. hippo encodes a Ste-20 family protein kinase that restricts cell proliferation and promotes apoptosis in conjunction with salvador and warts. Cell. 2003;22;114(4):445–56. doi: 10.1016/s0092-8674(03)00549-x. [DOI] [PubMed] [Google Scholar]
- 12.Harvey KF, Pfleger CM, Hariharan IK. The Drosophila Mst ortholog, hippo, restricts growth and cell proliferation and promotes apoptosis. Cell. 2003;114:457–67. doi: 10.1016/s0092-8674(03)00557-9. [DOI] [PubMed] [Google Scholar]
- 13.Udan RS, Kango-Singh M, Nolo R, Tao C, Halder G. Hippo promotes proliferation arrest and apoptosis in the Salvador/Warts pathway. Nat Cell Biol. 2003;5:914–20. doi: 10.1038/ncb1050. [DOI] [PubMed] [Google Scholar]
- 14.Pantalacci S, Tapon N, Léopold P. The Salvador partner Hippo promotes apoptosis and cell-cycle exit in Drosophila. Nat Cell Biol. 2003;5:921–7. doi: 10.1038/ncb1051. [DOI] [PubMed] [Google Scholar]
- 15.Jia J, Zhang W, Wang B, Trinko R, Jiang J. The Drosophila Ste20 family kinase dMST functions as a tumor suppressor by restricting cell proliferation and promoting apoptosis. Genes Dev. 2003;17:2514–9. doi: 10.1101/gad.1134003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dan I, Watanabe NM, Kusumi A. The Ste20 group kinases as regulators of MAP kinase cascades. Trends Cell Biol. 2001;11:220–30. doi: 10.1016/s0962-8924(01)01980-8. [DOI] [PubMed] [Google Scholar]
- 17.Lai ZC, Wei X, Shimizu T, Ramos E, Rohrbaugh M, Nikolaidis N, Ho LL, et al. Control of cell proliferation and apoptosis by mob as tumor suppressor, mats. Cell. 2005;120:675–85. doi: 10.1016/j.cell.2004.12.036. [DOI] [PubMed] [Google Scholar]
- 18.Praskova M, Xia F, Avruch J. MOBKL1A/MOBKL1B phosphorylation by MST1 and MST2 inhibits cell proliferation. Curr Biol. 2008;18:311–21. doi: 10.1016/j.cub.2008.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang J, Wu S, Barrera J, Matthews K, Pan D. The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila Homolog of YAP. Cell. 2005;122:421–34. doi: 10.1016/j.cell.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 20.Grusche FA, Richardson HE, Harvey KF. Upstream regulation of the Hippo size control pathway. Curr Biol. 2010;20:R574–82. doi: 10.1016/j.cub.2010.05.023. Review. [DOI] [PubMed] [Google Scholar]
- 21.Yin M, Zhang L. Hippo signaling: a hub of growth control, tumor suppression and pluripotency maintenance. J Genet Genomics. 2011;38:471–81. doi: 10.1016/j.jgg.2011.09.009. [DOI] [PubMed] [Google Scholar]
- 22.Enomoto M, Igaki T. Deciphering tumor-suppressor signaling in flies: genetic link between Scribble/Dlg/Lgl and the Hippo pathways. J Genet Genomics. 2011;38:461–70. doi: 10.1016/j.jgg.2011.09.005. [DOI] [PubMed] [Google Scholar]
- 23.Lei QY, Zhang H, Zhao B, Zha ZY, Bai F, Pei XH, et al. TAZ promotes cell proliferation and epithelial-mesenchymal transition and is inhibited by the Hippo pathway. Mol Cell Biol. 2008;28:2426–36. doi: 10.1128/MCB.01874-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao B, Wei X, Li W, Udan RS, Yang Q, Kim J, et al. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 2007;21:2747–61. doi: 10.1101/gad.1602907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou D, Conrad C, Xia F, Park JS, Payer B, et al. Mst1 and Mst2 maintain hepatocyte quiescence and suppress hepatocellular carcinoma development through inactivation of the Yap1 oncogene. Cancer Cell. 2009;16:425–38. doi: 10.1016/j.ccr.2009.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee JH, Kim TS, Yang TH, Koo BK, Oh SP, et al. A crucial role of WW45 in developing epithelial tissues in the mouse. EMBO J. 2008;27(8):1231–42. doi: 10.1038/emboj.2008.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Song H, Mak KK, Topol L, Yun K, Hu J, et al. Mammalian Mst1 and Mst2 kinases play essential roles in organ size control and tumor suppression. Proc Natl Acad Sci U S A. 2010;107:1431–6. doi: 10.1073/pnas.0911409107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu L, Li Y, Kim SM, Bossuyt W, Liu P, et al. Hippo signaling is a potent in vivo growth and tumor suppressor pathway in the mammalian liver. Proc Natl Acad Sci U S A. 2010 Jan 26;107(4):1437–42. doi: 10.1073/pnas.0911427107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee KP, Lee JH, Kim TS, Kim TH, Park HD, Byun JS, Kim MC, Jeong WI, Calvisi DF, Kim JM, Lim DS. The Hippo-Salvador pathway restrains hepatic oval cell proliferation, liver size, and liver tumorigenesis. Proc Natl Acad Sci U S A. 2010;107:8248–53. doi: 10.1073/pnas.0912203107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cai J, Zhang N, Zheng Y, de Wilde RF, Maitra A, et al. The Hippo signaling pathway restricts the oncogenic potential of an intestinal regeneration program. Genes Dev. 2010 Nov 1;24:2383–8. doi: 10.1101/gad.1978810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schlegelmilch K, Mohseni M, Kirak O, Pruszak J, Rodriguez JR, et al. Yap1 acts downstream of α-catenin to control epidermal proliferation. Cell. 201(144):782–95. doi: 10.1016/j.cell.2011.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou D, Zhang Y, Wu H, Barry E, Yin Y, et al. Mst1 and Mst2 protein kinases restrain intestinal stem cell proliferation and colonic tumorigenesis by inhibition of Yes-associated protein (Yap) overabundance. Proc Natl Acad Sci U S A. 2011;108:E1312–20. doi: 10.1073/pnas.1110428108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cao X, Pfaff SL, Gage FH. YAP regulates neural progenitor cell number via the TEA domain transcription factor. Genes Dev. 2008;22:3320–34. doi: 10.1101/gad.1726608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Emoto K, Parrish JZ, Jan LY, Jan YN. The tumour suppressor Hippo acts with the NDR kinases in dendritic tiling and maintenance. Nature. 2006;443:210–3. doi: 10.1038/nature05090. [DOI] [PubMed] [Google Scholar]
- 35.Millward TA, Hess D, Hemmings BA. Ndr protein kinase is regulated by phosphorylation on two conserved sequence motifs. J Biol Chem. 1999;274:33847–50. doi: 10.1074/jbc.274.48.33847. [DOI] [PubMed] [Google Scholar]
- 36.Hergovich A, Hemmings BA. Mammalian NDR/LATS protein kinases in Hippo tumor suppressor signaling. Biofactors. 2009;35:338–45. doi: 10.1002/biof.47. [DOI] [PubMed] [Google Scholar]
- 37.Cornils H, Kohler RS, Hergovich A, Hemmings BA. Downstream of human NDR kinases: impacting on c-myc and p21 protein stability to control cell cycle progression. Cell Cycle. 2011;10:1897–904. doi: 10.4161/cc.10.12.15826. [DOI] [PubMed] [Google Scholar]
- 38.Abdollahpour H, Appaswamy G, Kotlarz D, Diestelhorst J, Beier R, et al. The phenotype of human STK4 deficiency. Blood. 2012;119:3450–3457. doi: 10.1182/blood-2011-09-378158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nehme NT, Pachlopnik Schmid J, Debeurme F, André-Schmutz I, Lim A, et al. MST1 mutations in autosomal recessive primary immunodeficiency characterized by defective naive T cells survival. Blood. 2012;119:3458–3468. doi: 10.1182/blood-2011-09-378364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou D, Medoff BD, Chen L, Li L, Zhang XF, et al. The Nore1B/Mst1 complex restrains antigen receptor-induced proliferation of naïve T cells. Proc Natl Acad Sci U S A. 2008;105:20321–6. doi: 10.1073/pnas.0810773105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Katagiri K, Katakai T, Ebisuno Y, Ueda Y, Okada T, et al. Mst1 controls lymphocyte trafficking and interstitial motility within lymph nodes. EMBO J. 2009;28:1319–31. doi: 10.1038/emboj.2009.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dong Y, Du X, Ye J, Han M, Xu T, et al. A cell-intrinsic role for Mst1 in regulating thymocyte egress. J Immunol. 2009;183:3865–72. doi: 10.4049/jimmunol.0900678. [DOI] [PubMed] [Google Scholar]
- 43.Choi J, Oh S, Lee D, Oh HJ, Park JY, et al. Mst1-FoxO signaling protects Naïve T lymphocytes from cellular oxidative stress in mice. PLoS One. 2009;4:e8011. doi: 10.1371/journal.pone.0008011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mou F, Praskova M, Xia F, Van Buren D, Hock H, et al. The MST1 and Mst2 kinases control activation of rho family GTPases and thymic egress of mature thymocytes. J Exp Med. 2012;209:741–759. doi: 10.1084/jem.20111692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Creasy CL, Chernoff J. Cloning and characterization of a human protein kinase with homology to Ste20. J Biol Chem. 1995;270:21695–700. doi: 10.1074/jbc.270.37.21695. [DOI] [PubMed] [Google Scholar]
- 46.Creasy CL, Chernoff J. Cloning and characterization of a member of the MST subfamily of Ste20-like kinases. Gene. 1995;167:303–6. doi: 10.1016/0378-1119(95)00653-2. [DOI] [PubMed] [Google Scholar]
- 47.Taylor LK, Wang HC, Erikson RL. Newly identified stress-responsive protein kinases, Krs-1 and Krs-2. Proc Natl Acad Sci U S A. 1996;93:10099–104. doi: 10.1073/pnas.93.19.10099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang HC, Erikson RL. Activation of protein serine/threonine kinases p42, p63, and p87 in Rous sarcoma virus-transformed cells: signal transduction/transformation-dependent MBP kinases. Mol Biol Cell. 1992;3:1329–37. doi: 10.1091/mbc.3.12.1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Creasy CL, Ambrose DM, Chernoff J. The Ste20-like protein kinase, Mst1, dimerizes and contains an inhibitory domain. J Biol Chem. 1996;271:21049–53. doi: 10.1074/jbc.271.35.21049. [DOI] [PubMed] [Google Scholar]
- 50.Jin Y, Dong L, Lu Y, Wu W, Hao Q, et al. Dimerization and cytoplasmic localization regulate Hippo kinase signaling activity in organ size control. J Biol Chem. 2012 doi: 10.1074/jbc.M111.310334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Scheel H, Hofmann K. A novel interaction motif, SARAH, connects three classes of tumor suppressor. Curr Biol. 2003;13:R899–900. doi: 10.1016/j.cub.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 52.Hwang E, Ryu KS, Pääkkönen K, Güntert P, Cheong HK, et al. Structural insight into dimeric interaction of the SARAH domains from Mst1 and RASSF family proteins in the apoptosis pathway. Proc Natl Acad Sci U S A. 2007;104:9236–41. doi: 10.1073/pnas.0610716104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Avruch J, Xavier R, Bardeesy N, Zhang XF, Praskova M, et al. Rassf family of tumor suppressor polypeptides. J Biol Chem. 2009;284:11001–5. doi: 10.1074/jbc.R800073200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Praskova M, Khoklatchev A, Ortiz-Vega S, Avruch J. Regulation of the MST1 kinase by autophosphorylation, by the growth inhibitory proteins, RASSF1 and NORE1, and by Ras. Biochem J. 2004;381(Pt 2):453–62. doi: 10.1042/BJ20040025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Katagiri K, Imamura M, Kinashi T. Spatiotemporal regulation of the kinase Mst1 by binding protein RAPL is critical for lymphocyte polarity and adhesion. Nat Immunol. 2006;7:919–28. doi: 10.1038/ni1374. [DOI] [PubMed] [Google Scholar]
- 56.Polesello C, Huelsmann S, Brown NH, Tapon N. The Drosophila RASSF homolog antagonizes the Hippo pathway. Curr Biol. 2006;16:2459–65. doi: 10.1016/j.cub.2006.10.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Guo C, Tommasi S, Liu L, Yee JK, Dammann R, Pfeifer GP. RASSF1A is part of a complex similar to the Drosophila Hippo/Salvador/Lats tumor-suppressor network. Curr Biol. 2007;17:700–5. doi: 10.1016/j.cub.2007.02.055. [DOI] [PubMed] [Google Scholar]
- 58.Donninger H, Allen N, Henson A, Pogue J, Williams A, Gordon L, et al. Salvador protein is a tumor suppressor effector of RASSF1A with Hippo pathway-independent functions. J Biol Chem. 2011;286:18483–91. doi: 10.1074/jbc.M110.214874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cooper WN, Hesson LB, Matallanas D, Dallol A, von Kriegsheim A, Ward R, et al. RASSF2 associates with and stabilizes the proapoptotic kinase MST2. Oncogene. 2009;28:2988–98. doi: 10.1038/onc.2009.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Graves JD, Gotoh Y, Draves KE, Ambrose D, Han DK, Wright M, et al. Caspase-mediated activation and induction of apoptosis by the mammalian Ste20-like kinase Mst1. EMBO J. 1998;17:2224–34. doi: 10.1093/emboj/17.8.2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lee KK, Ohyama T, Yajima N, Tsubuki S, Yonehara S. MST, a physiological caspase substrate, highly sensitizes apoptosis both upstream and downstream of caspase activation. J Biol Chem. 2001;276:19276–85. doi: 10.1074/jbc.M005109200. [DOI] [PubMed] [Google Scholar]
- 62.Reszka AA, Halasy-Nagy JM, Masarachia PJ, Rodan GA. isphosphonates act directly on the osteoclast to induce caspase cleavage of mst1 kinase during apoptosis. A link between inhibition of the mevalonate pathway and regulation of an apoptosis-promoting kinase. J Biol Chem. 1999;274:34967–73. doi: 10.1074/jbc.274.49.34967. [DOI] [PubMed] [Google Scholar]
- 63.Song JJ, Lee YJ. Differential cleavage of Mst1 by caspase-7/-3 is responsible for TRAIL-induced activation of the MAPK superfamily. Cell Signal. 2008;20:892–906. doi: 10.1016/j.cellsig.2008.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lin Y, Khokhlatchev A, Figeys D, Avruch J. Death-associated protein 4 binds MST1 and augments MST1-induced apoptosis. J Biol Chem. 2002;277:47991–8001. doi: 10.1074/jbc.M202630200. [DOI] [PubMed] [Google Scholar]
- 65.Glantschnig H, Rodan GA, Reszka AA. Mapping of MST1 kinase sites of phosphorylation. Activation and autophosphorylation. J Biol Chem. 2002;277:42987–96. doi: 10.1074/jbc.M208538200. [DOI] [PubMed] [Google Scholar]
- 66.Ura S, Masuyama N, Graves JD, Gotoh Y. Caspase cleavage of MST1 promotes nuclear translocation and chromatin condensation. Proc Natl Acad Sci U S A. 2001;98:10148–53. doi: 10.1073/pnas.181161698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Anand R, Kim AY, Brent M, Marmorstein R. Biochemical analysis of MST1 kinase: elucidation of a C-terminal regulatory region. Biochemistry. 2008;47:6719–26. doi: 10.1021/bi800309m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fernando P, Kelly JF, Balazsi K, Slack RS, Megeney LA. Caspase 3 activity is required for skeletal muscle differentiation. Proc Natl Acad Sci U S A. 2002;99:11025–30. doi: 10.1073/pnas.162172899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Murray TV, McMahon JM, Howley BA, Stanley A, Ritter T, Mohr A, et al. A non-apoptotic role for caspase-9 in muscle differentiation. Cell Sci. 2008;121(Pt 22):3786–93. doi: 10.1242/jcs.024547. [DOI] [PubMed] [Google Scholar]
- 70.Yi CH, Yuan J. The Jekyll and Hyde functions of caspases. Dev Cell. 2009;16:21–34. doi: 10.1016/j.devcel.2008.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Park BH, Lee YH. Phosphorylation of SAV1 by mammalian ste20-like kinase promotes cell death. BMB Rep. 2011;44:584–9. doi: 10.5483/bmbrep.2011.44.9.584. [DOI] [PubMed] [Google Scholar]
- 72.Roig J, Mikhailov A, Belham C, Avruch J. Nercc1, a mammalian NIMA-family kinase, binds the Ran GTPase and regulates mitotic progression. Genes Dev. 2002;16:1640–58. doi: 10.1101/gad.972202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bertran MT, Sdelci S, Regué L, Avruch J, Caelles C, Roig J. Nek9 is a Plk1-activated kinase that controls early centrosome separation through Nek6/7 and Eg5. EMBO J. 2011;30:2634–47. doi: 10.1038/emboj.2011.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dérijard B, Raingeaud J, Barrett T, Wu IH, Han J, Ulevitch RJ, et al. Independent human MAP-kinase signal transduction pathways defined by MEK and MKK isoforms. Science. 1995;267:682–5. doi: 10.1126/science.7839144. [DOI] [PubMed] [Google Scholar]
- 75.Ge B, Gram H, Di Padova F, Huang B, New L, et al. MAPKK-independent activation of p38alpha mediated by TAB1-dependent autophosphorylation of p38alpha. Science. 2002;295:1291–4. doi: 10.1126/science.1067289. [DOI] [PubMed] [Google Scholar]
- 76.Boggiano JC, Vanderzalm PJ, Fehon RG. Tao-1 phosphorylates Hippo/MST kinases to regulate the Hippo-Salvador-Warts tumor suppressor pathway. Dev Cell. 2011;21:888–95. doi: 10.1016/j.devcel.2011.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Poon CL, Lin JI, Zhang X, Harvey KF. The sterile 20-like kinase Tao-1 controls tissue growth by regulating the Salvador-Warts-Hippo pathway. Dev Cell. 2011;21:896–906. doi: 10.1016/j.devcel.2011.09.012. [DOI] [PubMed] [Google Scholar]
- 78.Guertin DA, Chang L, Irshad F, Gould KL, McCollum D. The role of the sid1p kinase and cdc14p in regulating the onset of cytokinesis in fission yeast. EMBO J. 2000;19:1803–15. doi: 10.1093/emboj/19.8.1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hou MC, Salek J, McCollum D. Mob1p interacts with the Sid2p kinase and is required for cytokinesis in fission yeast. Curr Biol. 2000;10:619–22. doi: 10.1016/s0960-9822(00)00492-9. [DOI] [PubMed] [Google Scholar]
- 80.Hou MC, Guertin DA, McCollum D. Initiation of cytokinesis is controlled through multiple modes of regulation of the Sid2p-Mob1p kinase complex. Mol Cell Biol. 2004;24:3262–76. doi: 10.1128/MCB.24.8.3262-3276.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hutchison M, Berman KS, Cobb MH. Isolation of TAO1, a protein kinase that activates MEKs in stress-activated protein kinase cascades. J Biol Chem. 1998;273:28625–32. doi: 10.1074/jbc.273.44.28625. [DOI] [PubMed] [Google Scholar]
- 82.Chen Z, Hutchison M, Cobb MH. Isolation of the protein kinase TAO2 and identification of its mitogen-activated protein kinase/extracellular signal-regulated kinase kinase binding domain. J Biol Chem. 1999;274:28803–7. doi: 10.1074/jbc.274.40.28803. [DOI] [PubMed] [Google Scholar]
- 83.Tassi E, Biesova Z, Di Fiore PP, Gutkind JS, Wong WT. Human JIK, a novel member of the STE20 kinase family that inhibits JNK and is negatively regulated by epidermal growth factor. J Biol Chem. 1999;274:33287–95. doi: 10.1074/jbc.274.47.33287. [DOI] [PubMed] [Google Scholar]
- 84.Moore TM, Garg R, Johnson C, Coptcoat MJ, Ridley AJ, Morris JD. PSK, a novel STE20-like kinase derived from prostatic carcinoma that activates the c-Jun N-terminal kinase mitogen-activated protein kinase pathway and regulates actin cytoskeletal organization. J Biol Chem. 2000;275:4311–22. doi: 10.1074/jbc.275.6.4311. [DOI] [PubMed] [Google Scholar]
- 85.Timm T, Li XY, Biernat J, Jiao J, Mandelkow E, Vandekerckhove J, et al. MARKK, a Ste20-like kinase, activates the polarity-inducing kinase MARK/PAR-1. EMBO J. 2003;22:5090–101. doi: 10.1093/emboj/cdg447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Liu T, Rohn JL, Picone R, Kunda P, Baum B. Tao-1 is a negative regulator of microtubule plus-end growth. J Cell Sci. 2010;123:2708–16. doi: 10.1242/jcs.068726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Draviam VM, Stegmeier F, Nalepa G, Sowa ME, Chen J, Liang A, et al. A functional genomic screen identifies a role for TAO1 kinase in spindle-checkpoint signalling. Nat Cell Biol. 2007;9:556–64. doi: 10.1038/ncb1569. [DOI] [PubMed] [Google Scholar]
- 88.Densham RM, O’Neill E, Munro J, König I, Anderson K, Kolch W, et al. MST kinases monitor actin cytoskeletal integrity and signal via c-Jun N-terminal kinase stress-activated kinase to regulate p21Waf1/Cip1 stability. Mol Cell Biol. 2009;29:6380–90. doi: 10.1128/MCB.00116-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sansores-Garcia L, Bossuyt W, Wada K, Yonemura S, Tao C, Sasaki H, et al. Modulating F-actin organization induces organ growth by affecting the Hippo pathway. EMBO J. 2011;30:2325–35. doi: 10.1038/emboj.2011.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhao B, Li L, Wang L, Wang CY, Yu J, Guan KL. Cell detachment activates the Hippo pathway via cytoskeleton reorganization to induce anoikis. Genes Dev. 2012;26:54–68. doi: 10.1101/gad.173435.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Underhill-Day N, Hill V, Latif F. N-terminal RASSF family: RASSF7-RASSF10. Epigenetics. 2011;6:284–92. doi: 10.4161/epi.6.3.14108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Vavvas D, Li X, Avruch J, Zhang XF. Identification of Nore1 as a potential Ras effector. J Biol Chem. 1998;273:5439–42. doi: 10.1074/jbc.273.10.5439. [DOI] [PubMed] [Google Scholar]
- 93.Ortiz-Vega S, Khokhlatchev A, Nedwidek M, Zhang XF, Dammann R, Pfeifer GP, et al. The putative tumor suppressor RASSF1A homodimerizes and heterodimerizes with the Ras-GTP binding protein Nore1. Oncogene. 2002;21:1381–90. doi: 10.1038/sj.onc.1205192. [DOI] [PubMed] [Google Scholar]
- 94.Dammann R, Li C, Yoon JH, Chin PL, Bates S, Pfeifer GP. Epigenetic inactivation of a RAS association domain family protein from the lung tumour suppressor locus 3p21.3. Nat Genet. 2000;25:315–9. doi: 10.1038/77083. [DOI] [PubMed] [Google Scholar]
- 95.Donninger H, Vos MD, Clark GJ. The RASSF1A tumor suppressor. J Cell Sci. 2007;120:3163–72. doi: 10.1242/jcs.010389. [DOI] [PubMed] [Google Scholar]
- 96.Khokhlatchev A, Rabizadeh S, Xavier R, Nedwidek M, Chen T, Zhang XF, Seed B, Avruch J. Identification of a novel Ras-regulated proapoptotic pathway. Curr Biol. 2002;12:253–65. doi: 10.1016/s0960-9822(02)00683-8. [DOI] [PubMed] [Google Scholar]
- 97.Duronio V. The life of a cell: apoptosis regulation by the PI3K/PKB pathway. Biochem J. 2008;415:333–44. doi: 10.1042/BJ20081056. [DOI] [PubMed] [Google Scholar]
- 98.Matallanas D, Romano D, Al-Mulla F, O’Neill E, Al-Ali W, Crespo P, et al. Mutant K-Ras activation of the proapoptotic MST2 pathway is antagonized by wild-type K-Ras. Mol Cell. 2011;44:893–906. doi: 10.1016/j.molcel.2011.10.016. [DOI] [PubMed] [Google Scholar]
- 99.Katagiri K, Maeda A, Shimonaka M, Kinashi T. RAPL, a Rap1-binding molecule that mediates Rap1-induced adhesion through spatial regulation of LFA-1. Nat Immunol. 2003;4:741–8. doi: 10.1038/ni950. [DOI] [PubMed] [Google Scholar]
- 100.Katagiri K, Ohnishi N, Kabashima K, Iyoda T, Takeda N, Shinkai Y, et al. Crucial functions of the Rap1 effector molecule RAPL in lymphocyte and dendritic cell trafficking. Nat Immunol. 2004;5:1045–51. doi: 10.1038/ni1111. [DOI] [PubMed] [Google Scholar]
- 101.Kinashi T. Integrin regulation of lymphocyte trafficking: lessons from structural and signaling studies. Adv Immunol. 2007;93:185–227. doi: 10.1016/S0065-2776(06)93005-3. [DOI] [PubMed] [Google Scholar]
- 102.Park J, Kang SI, Lee SY, Zhang XF, Kim MS, et al. Tumor suppressor ras association domain family 5 (RASSF5/NORE1) mediates death receptor ligand-induced apoptosis. J Biol Chem. 2010;285:35029–38. doi: 10.1074/jbc.M110.165506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Avruch J, Khokhlatchev A, Kyriakis JM, Luo Z, Tzivion G, Vavvas D, et al. Ras activation of the Raf kinase: tyrosine kinase recruitment of the MAP kinase cascade. Recent Prog Horm Res. 2001;56:127–55. doi: 10.1210/rp.56.1.127. [DOI] [PubMed] [Google Scholar]
- 104.Matallanas D, Birtwistle M, Romano D, Zebisch A, Rauch J, von Kriegsheim A, et al. Raf family kinases: old dogs have learned new tricks. Genes Cancer. 2011;2:232–60. doi: 10.1177/1947601911407323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Vos MD, Ellis CA, Bell A, Birrer MJ, Clark GJ. Ras uses the novel tumor suppressor RASSF1 as an effector to mediate apoptosis. J Biol Chem. 2000;275:35669–72. doi: 10.1074/jbc.C000463200. [DOI] [PubMed] [Google Scholar]
- 106.Wohlgemuth S, Kiel C, Krämer A, Serrano L, Wittinghofer F, Herrmann C. Recognizing and defining true Ras binding domains I: biochemical analysis. J Mol Biol. 2005;348:741–58. doi: 10.1016/j.jmb.2005.02.048. [DOI] [PubMed] [Google Scholar]
- 107.Oh HJ, Lee KK, Song SJ, Jin MS, Song MS, Lee JH, et al. Role of the tumor suppressor RASSF1A in Mst1-mediated apoptosis. Cancer Res. 2006;66:2562–9. doi: 10.1158/0008-5472.CAN-05-2951. [DOI] [PubMed] [Google Scholar]
- 108.Matallanas D, Romano D, Yee K, Meissl K, Kucerova L, Piazzolla D, et al. RASSF1A elicits apoptosis through an MST2 pathway directing proapoptotic transcription by the p73 tumor suppressor protein. Mol Cell. 2007;27:962–75. doi: 10.1016/j.molcel.2007.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Vichalkovski A, Gresko E, Cornils H, Hergovich A, Schmitz D, Hemmings BA. NDR kinase is activated by RASSF1A/MST1 in response to Fas receptor stimulation and promotes apoptosis. Curr Biol. 2008;18:1889–95. doi: 10.1016/j.cub.2008.10.060. [DOI] [PubMed] [Google Scholar]
- 110.Foley CJ, Freedman H, Choo SL, Onyskiw C, Fu NY, Yu VC, et al. Dynamics of RASSF1A/MOAP-1 association with death receptors. Mol Cell Bio. 2008;28:4520–35. doi: 10.1128/MCB.02011-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Gordon M, Baksh S. RASSF1A: Not a prototypical Ras effector. Small Gtpases. 2011;2:148–157. doi: 10.4161/sgtp.2.3.16286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wojnowski L, Stancato LF, Zimmer AM, Hahn H, Beck TW, Larner AC, et al. Craf-1 protein kinase is essential for mouse development. Mech Dev. 1998;76:141–9. doi: 10.1016/s0925-4773(98)00111-7. [DOI] [PubMed] [Google Scholar]
- 113.Mikula M, Schreiber M, Husak Z, Kucerova L, Rüth J, Wieser R, et al. Embryonic lethality and fetal liver apoptosis in mice lacking the c-raf-1 gene. EMBO J. 2001;20:1952–62. doi: 10.1093/emboj/20.8.1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Hüser M, Luckett J, Chiloeches A, Mercer K, Iwobi M, Giblett S, et al. MEK kinase activity is not necessary for Raf-1 function. EMBO J. 2001;20:1940–51. doi: 10.1093/emboj/20.8.1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.O’Neill E, Rushworth L, Baccarini M, Kolch W. Role of the kinase MST2 in suppression of apoptosis by the proto-oncogene product Raf-1. Science. 2004;306:2267–70. doi: 10.1126/science.1103233. [DOI] [PubMed] [Google Scholar]
- 116.Chen J, Fujii K, Zhang L, Roberts T, Fu H. Raf-1 promotes cell survival by antagonizing apoptosis signal-regulating kinase 1 through a MEK-ERK independent mechanism. Proc Natl Acad Sci U S A. 2001;98:7783–8. doi: 10.1073/pnas.141224398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ehrenreiter K, Piazzolla D, Velamoor V, Sobczak I, Small JV, Takeda J, et al. Raf-1 regulates Rho signaling and cell migration. J Cell Biol. 2005;168:955–64. doi: 10.1083/jcb.200409162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Niault T, Sobczak I, Meissl K, Weitsman G, Piazzolla D, Maurer G, et al. From autoinhibition to inhibition in trans: the Raf-1 regulatory domain inhibits Rok-alpha kinase activity. J Cell Biol. 2009;187:335–42. doi: 10.1083/jcb.200906178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Clapéron A, Therrien M. KSR and CNK: two scaffolds regulating RAS-mediated RAF activation. Oncogene. 2007;26:3143–58. doi: 10.1038/sj.onc.1210408. [DOI] [PubMed] [Google Scholar]
- 120.McKay MM, Freeman AK, Morrison DK. Complexity in KSR function revealed by Raf inhibitor and KSR structure studies. Small Gtpases. 2011;2:276–281. doi: 10.4161/sgtp.2.5.17740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Yamaguchi O, Watanabe T, Nishida K, Kashiwase K, Higuchi Y, Takeda T, et al. Cardiac-specific disruption of the c-raf-1 gene induces cardiac dysfunction and apoptosis. J Clin Invest. 2004;114:937–43. doi: 10.1172/JCI20317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kilili GK, Kyriakis JM. Mammalian Ste20-like kinase (Mst2) indirectly supports Raf-1/ERK pathway activity via maintenance of protein phosphatase-2A catalytic subunit levels and consequent suppression of inhibitory Raf-1 phosphorylation. J Biol Chem. 2010;285:15076–87. doi: 10.1074/jbc.M109.078915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Yuan Z, Kim D, Shu S, Wu J, Guo J, Xiao L, et al. Phosphoinositide 3-kinase/Akt inhibits MST1-mediated pro-apoptotic signaling through phosphorylation of threonine 120. J Biol Chem. 2010;285:3815–24. doi: 10.1074/jbc.M109.059675. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 124.Jang SW, Yang SJ, Srinivasan S, Ye K. Akt phosphorylates MstI and prevents its proteolytic activation, blocking FOXO3 phosphorylation and nuclear translocation. J Biol Chem. 2007;282:30836–44. doi: 10.1074/jbc.M704542200. [DOI] [PubMed] [Google Scholar]
- 125.Kim D, Shu S, Coppola MD, Kaneko S, Yuan ZQ, Cheng JQ. Regulation of proapoptotic mammalian ste20-like kinase MST2 by the IGF1-Akt pathway. PLoS One. 2010;5:e9616. doi: 10.1371/journal.pone.0009616. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 126.Wang HC, Fecteau KA. Detection of a novel quiescence-dependent protein kinase. J Biol Chem. 2000;275:25850–7. doi: 10.1074/jbc.M000818200. [DOI] [PubMed] [Google Scholar]
- 127.Romano D, Matallanas D, Weitsman G, Preisinger C, Ng T, Kolch W. Proapoptotic kinase MST2 coordinates signaling crosstalk between RASSF1A, Raf-1, and Akt. Cancer Res. 2010;70:1195–203. doi: 10.1158/0008-5472.CAN-09-3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Warfel NA, Newton AC. Pleckstrin Homology Domain Leucine-rich Repeat Protein Phosphatase (PHLPP): A New Player in Cell Signaling. J Biol Chem. 2012;287:3610–6. doi: 10.1074/jbc.R111.318675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Qiao M, Wang Y, Xu X, Lu J, Dong Y, Tao W, et al. Mst1 is an interacting protein that mediates PHLPPs’ induced apoptosis. Mol Cell. 2010;38:512–23. doi: 10.1016/j.molcel.2010.03.017. [DOI] [PubMed] [Google Scholar]
- 130.Cinar B, Fang PK, Lutchman M, Di Vizio D, Adam RM, Pavlova N, et al. The pro-apoptotic kinase Mst1 and its caspase cleavage products are direct inhibitors of Akt1. EMBO J. 2007;26:4523–34. doi: 10.1038/sj.emboj.7601872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Strassburger K, Tiebe M, Pinna F, Breuhahn K, Teleman AA. Insulin/IGF signaling drives cell proliferation in part via Yorkie/Yap. Dev Biol. 2012;367:187–96. doi: 10.1016/j.ydbio.2012.05.008. [DOI] [PubMed] [Google Scholar]
- 132.Ye X, Deng Y, Lai Z-C. Akt is negatively regulated by Hippo signaling for growth inhibition in Drosophila. Dev Biol. 2012 doi: 10.1016/j.ydbio.2012.06.014. in press. [DOI] [PubMed] [Google Scholar]
- 133.Graves JD, Draves KE, Gotoh Y, Krebs EG, Clark EA. Both phosphorylation and caspase-mediated cleavage contribute to regulation of the Ste20-like protein kinase Mst1 during CD95/Fas-induced apoptosis. J Biol Chem. 2001;276:14909–15. doi: 10.1074/jbc.M010905200. [DOI] [PubMed] [Google Scholar]
- 134.Ura S, Masuyama N, Graves JD, Gotoh Y. MST1-JNK promotes apoptosis via caspase-dependent and independent pathways. Genes Cells. 2001;6:519–30. doi: 10.1046/j.1365-2443.2001.00439.x. [DOI] [PubMed] [Google Scholar]
- 135.Cheung WL, Ajiro K, Samejima K, Kloc M, Cheung P, Mizzen CA, et al. Apoptotic phosphorylation of histone H2B is mediated by mammalian sterile twenty kinase. Cell. 2003;113:507–17. doi: 10.1016/s0092-8674(03)00355-6. [DOI] [PubMed] [Google Scholar]
- 136.Hu Y, Liu Z, Yang SJ, Ye K. Acinus-provoked protein kinase C delta isoform activation is essential for apoptotic chromatin condensation. Cell Death Differ. 2007;14:2035–46. doi: 10.1038/sj.cdd.4402214. [DOI] [PubMed] [Google Scholar]
- 137.Chan EH, Nousiainen M, Chalamalasetty RB, Schäfer A, Nigg EA, Silljé HH. The Ste20-like kinase Mst2 activates the human large tumor suppressor kinase Lats1. Oncogene. 2005;24:2076–86. doi: 10.1038/sj.onc.1208445. [DOI] [PubMed] [Google Scholar]
- 138.Stegert MR, Hergovich A, Tamaskovic R, Bichsel SJ, Hemmings BA. Regulation of NDR protein kinase by hydrophobic motif phosphorylation mediated by the mammalian Ste20-like kinase MST3. Mol Cell Biol. 2005;25:11019–29. doi: 10.1128/MCB.25.24.11019-11029.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Hergovich A, Stegert MR, Schmitz D, Hemmings BA. NDR kinases regulate essential cell processes from yeast to humans. Nat Rev Mol Cell Biol. 2006;7:253–64. doi: 10.1038/nrm1891. [DOI] [PubMed] [Google Scholar]
- 140.Bichsel SJ, Tamaskovic R, Stegert MR, Hemmings BA. Mechanism of activation of NDR (nuclear Dbf2-related) protein kinase by the hMOB1 protein. J Biol Chem. 2004;279:35228–35. doi: 10.1074/jbc.M404542200. [DOI] [PubMed] [Google Scholar]
- 141.Hergovich A, Bichsel SJ, Hemmings BA. Human NDR kinases are rapidly activated by MOB proteins through recruitment to the plasma membrane and phosphorylation. Mol Cell Biol. 2005;25:8259–72. doi: 10.1128/MCB.25.18.8259-8272.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Hergovich A, Schmitz D, Hemmings BA. The human tumour suppressor LATS1 is activated by human MOB1 at the membrane. Biochem Biophys Res Commun. 2006;345:50–8. doi: 10.1016/j.bbrc.2006.03.244. [DOI] [PubMed] [Google Scholar]
- 143.Pearce LR, Komander D, Alessi DR. The nuts and bolts of AGC protein kinases. Nat Rev Mol Cell Biol. 2010;11:9–22. doi: 10.1038/nrm2822. [DOI] [PubMed] [Google Scholar]
- 144.Koike-Kumagai M, Yasunaga K, Morikawa R, Kanamori T, Emoto K. The target of rapamycin complex 2 controls dendritic tiling of Drosophila sensory neurons through the Tricornered kinase signalling pathway. EMBO J. 2009;28:3879–92. doi: 10.1038/emboj.2009.312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Geng W, He B, Wang M, Adler PN. The tricornered gene, which is required for the integrity of epidermal cell extensions, encodes the Drosophila nuclear DBF2-related kinase. Genetics. 2000;156:1817–28. doi: 10.1093/genetics/156.4.1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Fang X, Adler PN. Regulation of cell shape, wing hair initiation and the actin cytoskeleton by Trc/Fry and Wts/Mats complexes. Dev Biol. 2010;341:360–74. doi: 10.1016/j.ydbio.2010.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Cong J, Geng W, He B, Liu J, Charlton J, Adler PN. The furry gene of Drosophila is important for maintaining the integrity of cellular extensions during morphogenesis. Development. 2001;128:2793–802. doi: 10.1242/dev.128.14.2793. [DOI] [PubMed] [Google Scholar]
- 148.He Y, Fang X, Emoto K, Jan YN, Adler PN. The tricornered Ser/Thr protein kinase is regulated by phosphorylation and interacts with furry during Drosophila wing hair development. Mol Biol Cell. 2005;16:689–700. doi: 10.1091/mbc.E04-09-0828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.He Y, Emoto K, Fang X, Ren N, Tian X, Jan YN, et al. Drosophila Mob family proteins interact with the related tricornered (Trc) and warts (Wts) kinases. Mol Biol Cell. 2005;16:4139–52. doi: 10.1091/mbc.E05-01-0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Chiba S, Ikeda M, Katsunuma K, Ohashi K, Mizuno K. MST2- and Furry-mediated activation of NDR1 kinase is critical for precise alignment of mitotic chromosomes. Curr Biol. 2009;19:675–81. doi: 10.1016/j.cub.2009.02.054. [DOI] [PubMed] [Google Scholar]
- 151.Hergovich A, Lamla S, Nigg EA, Hemmings BA. Centrosome-associated NDR kinase regulates centrosome duplication. Mol Cell. 2007;25:625–34. doi: 10.1016/j.molcel.2007.01.020. [DOI] [PubMed] [Google Scholar]
- 152.Hergovich A, Kohler RS, Schmitz D, Vichalkovski A, Cornils H, Hemmings BA. The MST1 and hMOB1 tumor suppressors control human centrosome duplication by regulating NDR kinase phosphorylation. Curr Biol. 2009;19:1692–702. doi: 10.1016/j.cub.2009.09.020. [DOI] [PubMed] [Google Scholar]
- 153.Hergovich A, Cornils H, Hemmings BA. Mammalian NDR protein kinases: from regulation to a role in centrosome duplication. Biochim Biophys Acta. 2008;1784:3–15. doi: 10.1016/j.bbapap.2007.07.017. [DOI] [PubMed] [Google Scholar]
- 154.Hergovich A. MOB control: reviewing a conserved family of kinase regulators. Cell Signal. 2011;23:1433–40. doi: 10.1016/j.cellsig.2011.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Kohler RS, Schmitz D, Cornils H, Hemmings BA, Hergovich A. Differential NDR/LATS interactions with the human MOB family reveal a negative role for human MOB2 in the regulation of human NDR kinases. Mol Cell Biol. 2010;30:4507–20. doi: 10.1128/MCB.00150-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Wei X, Shimizu T, Lai ZC. Mob as tumor suppressor is activated by Hippo kinase for growth inhibition in Drosophila. EMBO J. 2007;26:1772–8. doi: 10.1038/sj.emboj.7601630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Côté JF, Vuori K. GEF what? Dock180 and related proteins help Rac to polarize cells in new ways. Trends Cell Biol. 2007;17:383–93. doi: 10.1016/j.tcb.2007.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Huang H, Tindall DJ. Dynamic FoxO transcription factors. J Cell Sci. 2007;120(Pt 15):2479–87. doi: 10.1242/jcs.001222. [DOI] [PubMed] [Google Scholar]
- 159.Paik JH, Kollipara R, Chu G, Ji H, Xiao Y, Ding Z, et al. FoxOs are lineage-restricted redundant tumor suppressors and regulate endothelial cell homeostasis. Cell. 2007;128:309–23. doi: 10.1016/j.cell.2006.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.van den Berg MC, Burgering BM. Integrating opposing signals toward Forkhead box O. Antioxid Redox Signal. 2011;14:607–21. doi: 10.1089/ars.2010.3415. [DOI] [PubMed] [Google Scholar]
- 161.Boccitto M, Kalb RG. Regulation of Foxo-dependent transcription by post-translational modifications. Curr Drug Targets. 2011;12:1303–10. doi: 10.2174/138945011796150316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Huang H, Tindall DJ. Regulation of FOXO protein stability via ubiquitination and proteasome degradation. Biochim Biophys Acta. 2011;1813:1961–4. doi: 10.1016/j.bbamcr.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Lehtinen MK, Yuan Z, Boag PR, Yang Y, Villén J, Becker EB, et al. A conserved MST-FOXO signaling pathway mediates oxidative-stress responses and extends life span. Cell. 2006;125:987–1001. doi: 10.1016/j.cell.2006.03.046. [DOI] [PubMed] [Google Scholar]
- 164.Yuan Z, Lehtinen MK, Merlo P, Villén J, Gygi S, Bonni A. Regulation of neuronal cell death by MST1-FOXO1 signaling. J Biol Chem. 2009;284:11285–92. doi: 10.1074/jbc.M900461200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Kenyon CJ. The genetics of ageing. Nature. 2010;25;464:504–12. doi: 10.1038/nature08980. [DOI] [PubMed] [Google Scholar]
- 166.Bardin AJ, Amon A. Men and sin: what’s the difference? Nat Rev Mol Cell Biol. 2001;2:815–26. doi: 10.1038/35099020. [DOI] [PubMed] [Google Scholar]
- 167.Krapp A, Gulli MP, Simanis V. SIN and the art of splitting the fission yeast cell. Curr Biol. 2004;14:R722–30. doi: 10.1016/j.cub.2004.08.049. [DOI] [PubMed] [Google Scholar]
- 168.Goyal A, Takaine M, Simanis V, Nakano K. Dividing the spoils of growth and the cell cycle: The fission yeast as a model for the study of cytokinesis. Cytoskeleton (Hoboken) 2011;68:69–88. doi: 10.1002/cm.20500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Rock JM, Amon A. Cdc15 integrates Tem1 GTPase-mediated spatial signals with Polo kinase-mediated temporal cues to activate mitotic exit. Genes Dev. 2011;25:1943–54. doi: 10.1101/gad.17257711. [DOI] [PMC free article] [PubMed] [Google Scholar]