Abstract
Respiration, photosynthesis and metabolism require the transfer of electrons through and between proteins over relatively long distances. It is critical that this electron transfer (ET) occur with specificity to avoid cellular damage, and at a rate which is sufficient to support the biological activity. A multi-step hole hopping mechanism could, in principle, enhance the efficiency of long range ET through proteins as it does in organic semiconductors. To explore this possibility, two different ET reactions that occur over the same distance within the protein complex of the diheme enzyme MauG and different forms of methylamine dehydrogenase (MADH) were subjected to kinetic and thermodynamic analysis. An ET mechanism of single-step direct electron tunneling from diferrous MauG to the quinone form of MADH is consistent with the data. In contrast, the biosynthetic ET from preMADH, which contains incompletely synthesized tryptophan tryptophylquinone, to the bis-Fe(IV) form of MauG is best described by a two-step hole hopping mechanism. Experimentally-determined values of ET distance matched the distances determined from the crystal structure that would be expected for single-step tunneling and multi-step hopping, respectively. Experimentally-determined relative values of electronic coupling (HAB) for the two reactions correlated well with the relative HAB values predicted from computational analysis of the structure. The rate of the hopping-mediated ET reaction is also ten-fold greater than that of the single-step tunneling reaction despite having a smaller overall driving force for the reaction. These data provide insight into how the intervening protein matrix and redox potentials of the electron donor and acceptor determine whether the ET reaction proceeds via single-step tunneling or multi-step hopping.
Long range electron transfer (ET) is a fundamental process to the fields of chemistry, biology and physics. Two different mechanisms for long range ET have been considered. One is direct electron tunneling which occurs in a single step reaction from electron donor to acceptor (1–5), and the other one is multi-step hopping (6). To fully describe the relevance of these processes to biology, two key questions remain to be answered. How does ET occur over large distances through and between proteins with speed and specificity? How does the protein matrix and nature of the redox centers determine contributions of electron tunneling and hole hopping to the overall mechanism of ET? This study determines, analyzes and compares the temperature dependencies of the rates of two different ET reactions that occur between the diheme enzyme MauG (7) and different forms of methylamine dehydrogenase (MADH) (8), which bears the tryptophan tryptophylquinone (TTQ) redox cofactor. One reaction is ET from diferrous MauG to the quinone form of MADH. The other is ET from a biosynthetic precursor of MADH (preMADH) (9) to a high valent bis-Fe(IV) form (10) of MauG. The results demonstrate that the former reaction occurs via single-step electron tunneling while the latter reaction occurs via multi-step hole hopping.
During long range electron tunneling through proteins, the amino acid residues that mediate ET do not undergo a change in redox state. They are simply a conductive matrix. In contrast, during hopping certain intervening amino acid residues are reversibly oxidized and reduced and serve as intermediate staging points or stepping stones for the hops. Two alternative hopping mechanisms are possible. If the electron donor reduces the intermediate which in turn reduces the electron acceptor, then this would be electron hopping. If the electron acceptor oxidizes the intermediate which in turn oxidizes the electron donor, then this would be hole hopping. Hopping has been described in non-biological systems such as organic semiconductors which have been described as hole charge transporters (11) and through molecular wires (12). In biological systems, hole hopping through DNA has been demonstrated (13–17). The purine base guanine is the preferred hopping relay as it the easiest base to oxidize. The efficiency of DNA as a charge carrier depends in large part on the spacing between guanine bases. While it is not likely relevant to metabolism, hopping does allow rapid charge transfer through DNA. Documentation of hopping through proteins has been difficult to achieve and evidence for this mechanism has only been obtained for a few systems, ribonucleotide reductase (18, 19), DNA photolyase (20), an engineered ruthenium labeled azurin (21) and cytochrome c peroxidase (22). A recent sitedirected mutagenesis study of tryptophan 199 of MauG suggested that this residue mediates hole hopping during the long range ET that is required for MauG-dependent TTQ biosynthesis (23). Thus, the MauG system provides a rare opportunity to characterize a natural physiologically relevant hopping mechanism in a protein system.
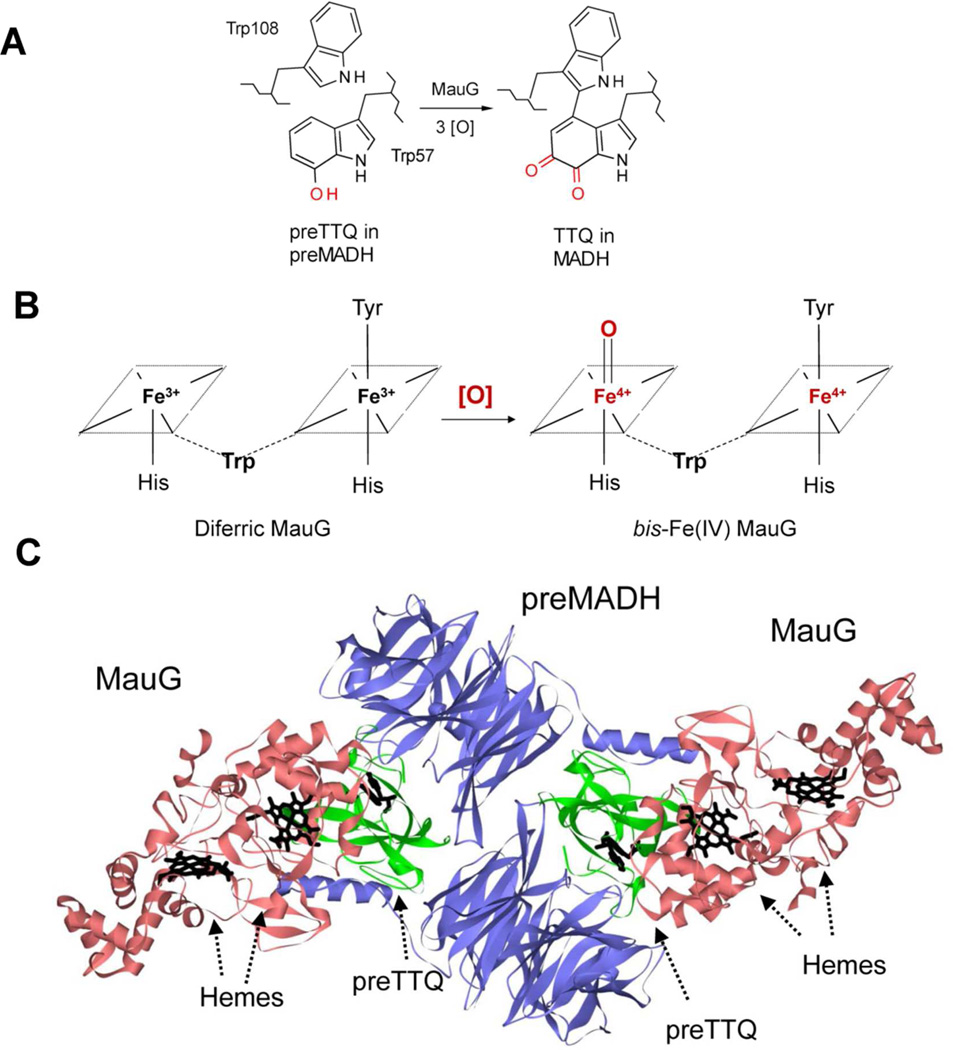
MauG is a diheme enzyme that catalyzes the final steps in the biosynthesis of the protein-derived cofactor, TTQ in MADH (24–26). The biosynthetic process is a six-electron oxidation of the precursor protein, preMADH, which possesses monohydroxylated residue βTrp57 (Figure 1A). The completion of TTQ biosynthesis requires that βTrp57 is covalently crosslinked to βTrp108, a second oxygen atom is inserted into the indole ring of βTrp57, and the quinol species is oxidized to the quinone (9). These reactions require the formation of a high valent bis-Fe(IV) intermediate of MauG in which one heme is present as Fe(IV)=O with an axial His ligand and the other is Fe(IV) with no exogenous ligand (10) and axial ligands provided by His and Tyr residues (24) (Figure 1B). Significantly, the crystal structure of the MauG-preMADH complex revealed that βTrp57 and βTrp108 do not make direct contact with either heme of MauG (24) (Figure 1C). In fact, the distance between the side-chain of βTrp108 of preMADH and the iron of the oxygen-binding five-coordinate heme is 40.1 Å, and the closest distance from the residues of preMADH that are modified to the iron of the six-coordinate heme is 19.4 Å. Nonetheless, addition of H2O2 to MauG-preMADH crystals causes synthesis of the mature TTQ cofactor in crystallo demonstrating that the orientation of the proteins in the crystallized complex is catalytically active (24). Thus, these catalytic reactions absolutely require long range ET. This study applies Marcus theory (3) to analyze the temperature dependencies of the rates of two different ET reactions that occur between these proteins (Figure 2). One is a non-biosynthetic ET reaction from diferrous MauG to the quinone form of MADH (27). The other is a biosynthetic ET reaction from preMADH to the bis-Fe(IV) form of MauG (28). The results are consistent with the former reaction occurring via single-step electron tunneling while the latter reaction occurring via multi-step hole hopping.
Figure 1.
A. MauG catalyzes the conversion of monohydroxylated βTrp57 and βTrp108 of preMADH to TTQ. [O] is oxidation equivalents. B. The bis-Fe(IV) state of MauG is formed by reaction of [O] with diferric MauG. The residues which form the axial ligands of the hemes and an intervening Trp residue which is believed to facilitate communication between the hemes are indicated. C. The crystal structure of the MauG-preMADH complex (PDB ID: 3L4M) (24) is shown with MauGs colored red; preMADH α subunits colored blue, and preMADH β subunits colored green. The hemes of MauG and βTrp108 and mono-hydroxylated βTrp57 of preMADH are drawn in a stick representation and colored black.
Figure 2.
Schematic depiction of the electron transfer reactions (A) from diferrous MauG to quinone MADH and (B) from preMADH to the nearest heme of bis-Fe(IV) MauG.
Experimental Methods
Protein expression and purification
Recombinant MauG (7) and native MADH (29) were purified from P. denitrificans as described previously. PreMADH was expressed in Rhodobacter sphaeroides and purified as described previously (9).
Electron Transfer from Diferrous MauG to Quinone MADH
The single-turnover kinetics of this reaction were studied as described previously (27) using a Shimadzu Multispec- 1501 spectrophotometer. Reactions were performed under anaerobic conditions to ensure that the MauG remained reduced prior to and during reaction with MADH. Each reaction was performed in 0.01 M potassium phosphate buffer, pH 7.5, at the indicated temperature. The reaction mixture contained 1.5 µM of the limiting reactant, diferrous MauG, and a saturating concentration of quinone MADH (35 µM, Kd=10.1 µM). The reaction was monitored by the decrease in absorbance at 550 nm which corresponds to the conversion of diferrous MauG to diferric MauG (7).
Electron Transfer from preMADH to bis-Fe(IV) MauG
The single-turnover kinetics of this reaction were studied as described previously (28) using an Online Instrument Systems (OLIS, Bogart, GA) RSM stopped-flow spectrophotometer. Kinetic data collected in the rapidscanning mode were reduced by factor analysis using the singular-value decomposition algorithm and then globally fit using the fitting routines of OLIS Global Fit. Each reaction was performed in 0.01 M potassium phosphate buffer, pH 7.5, at the indicated temperature. The reaction mixture contained 2.0 IM of the limiting reactant, bis-Fe(IV) MauG, and a saturating concentration of preMADH (8 µM, Kd=1.5 µM). Reactions were monitored from 3662–446 nm to observe the conversion of bis-Fe(IV) MauG to diferric MauG.
Analysis of kinetic data
Kinetic data were analyzed using the kinetic model described in eq 1 where E and S are either diferrous MauG and quinone MADH (Figure 2A) or bis-Fe(IV) MauG and preMADH (Figure 2B). The conversions of ES to E’P are shown in Figure 2. In each of the single-turnover kinetic studies, the observed rate constant (kobs) was best fit to a single exponential relaxation. The limiting first-order rate constant for each reaction (kET) was determined from the concentration dependence of the observed rate using eq 2. In each case, fits of the data to eq 2 yielded r2 values of 0.99, and use of more complex kinetic models did not improve the fit of the data.
| (1) |
| (2) |
Calculation of predicted values of electronic coupling
Calculations of relative values of electronic coupling (HAB) were performed using the HARLEM computer program (30) to analyze the crystal structure of the MauG-preMADH complex (PDB code 3L4M (24)) using the direct distance approach of Dutton and coworkers (4).
Results and Discussion
Electron transfer from diferrous MauG to oxidized MADH
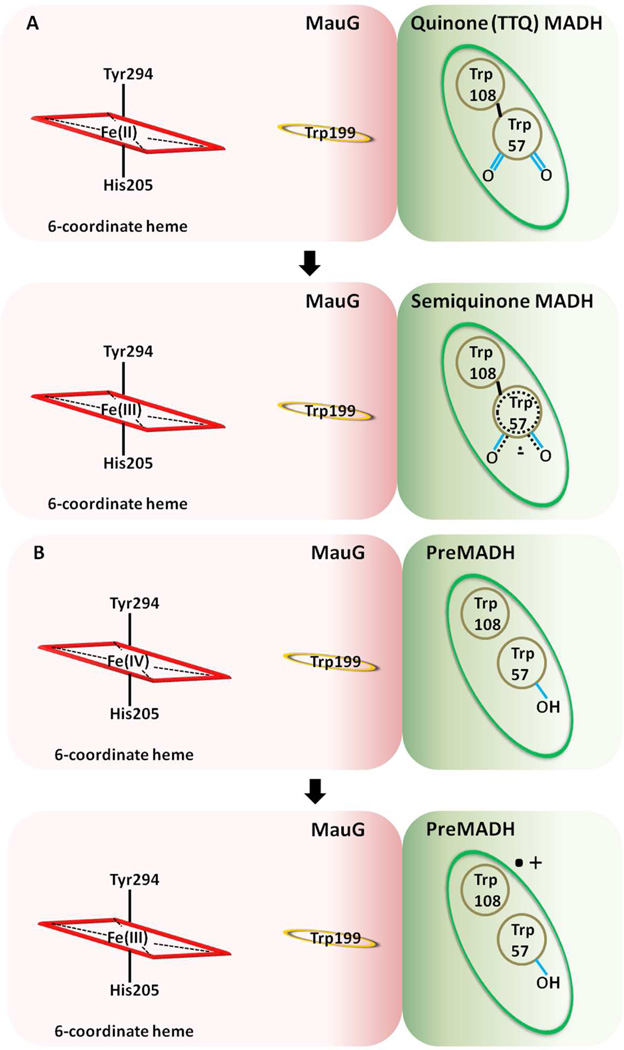
The ET reaction from diferrous MauG to the oxidized TTQ of MADH is not involved in TTQ biosynthesis, but is thermodynamically favorable (27). The ability to monitor this reaction has proven to be useful as a tool to determine whether mutations of MauG specifically affect the ability of MauG to stabilize the bis-Fe(IV) state or otherwise disrupt ET (23, 31). This reaction does not involve the formation of the high valent bis-Fe(IV) species. As the Em values of the donor and acceptor redox couples in this reaction are not sufficiently positive or negative to oxidize or reduce amino acid residues, ET is not expected to occur via a hopping mechanism.
For a single-step electron tunneling reaction the rate is described by eqs 3 and 4 (3). The first-order rate constant for ET from a donor and an acceptor (kET) varies predictably with temperature (T), ΔG°, the reorganization energy (λ) comprising inner-sphere (ligand) and outer-sphere (solvent) nuclear rearrangement, and electron coupling (HAB) which describes the extent of overlap between the localized donor and acceptor wave functions. HAB exhibits an exponential dependence on the distance between the donor and acceptor. The parameter β is used to quantitate the nature of the intervening medium with respect to its efficiency to mediate ET. The donor to acceptor distance is r, and r0 is the close contact distance (3Å). k0 is the characteristic frequency of nuclei (k0=1013 s−1) which is the maximum ET rate when donor and acceptor are in van der Waals’ contact and λ=−ΔG°. Planck’s constant (h) and the gas constant (R) are also included.
| (3) |
| (4) |
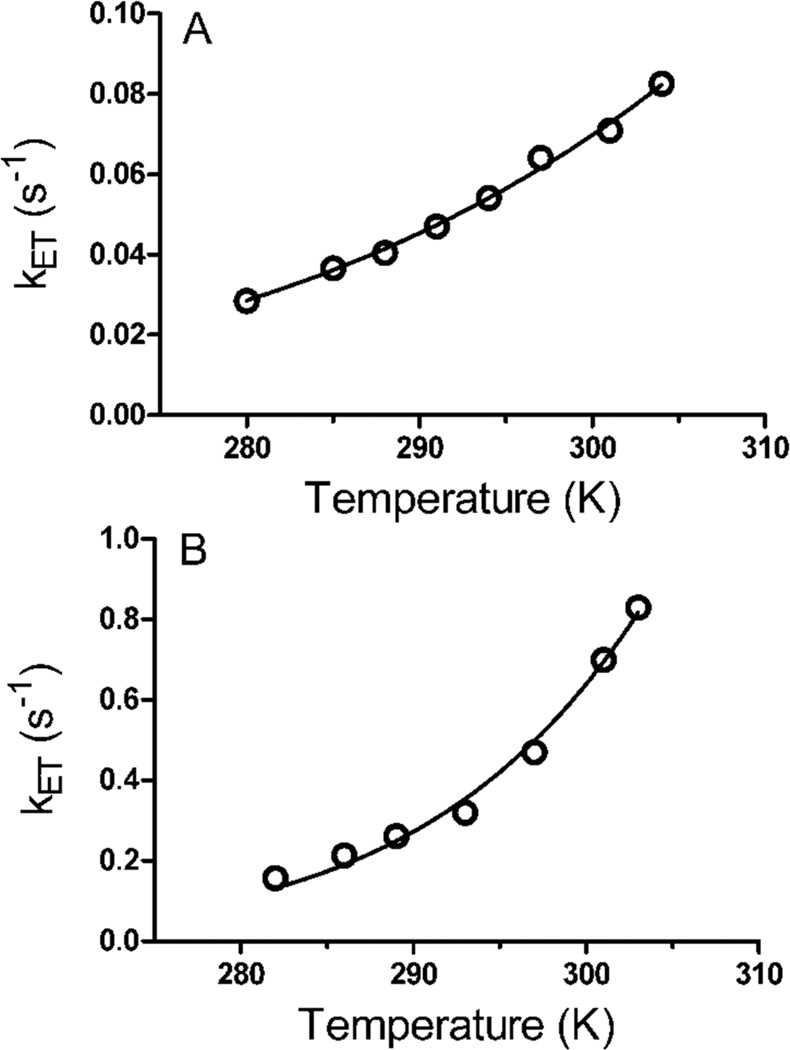
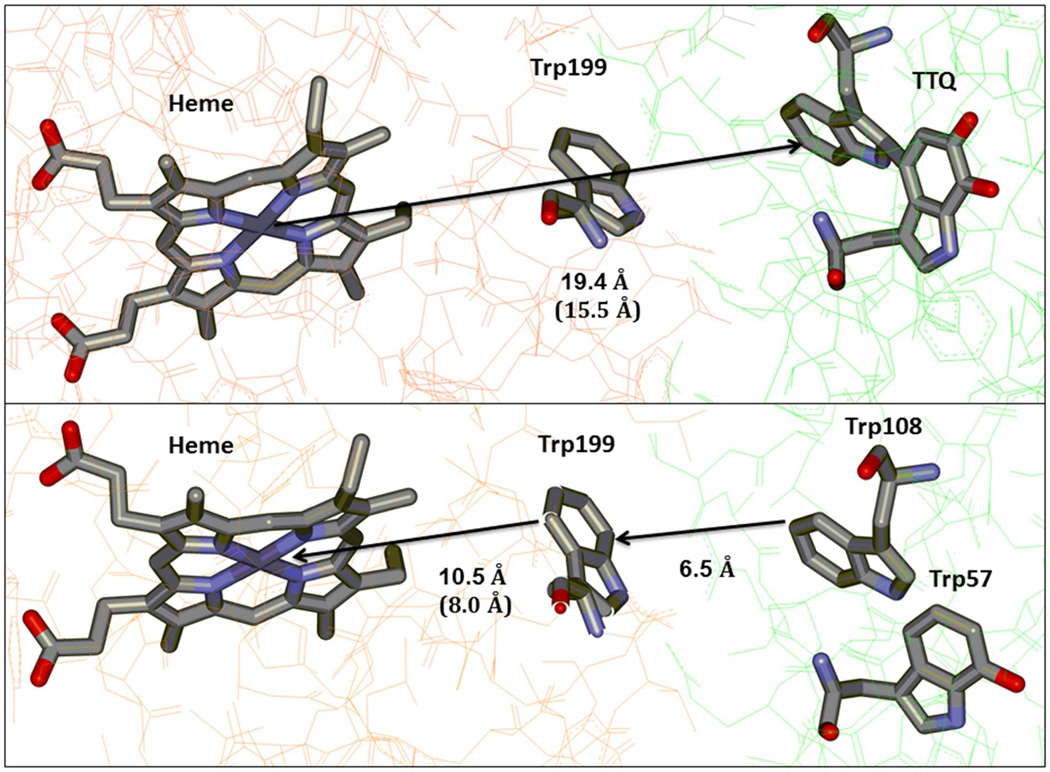
The temperature dependence of the ET reaction from diferrous MauG to the oxidized TTQ of MADH was studied under single turnover conditions over a temperature range from 7 °C to 31 °C (Figure 3a). Unfortunately MauG was not stable above the higher temperature and so the range could not be extended. Over this range the kET varied from 0.028 to 0.083 s−1. In each case the reaction kinetics fit well to a single exponential relaxation. The reaction under study is a sequential two-electron transfer process. The Em values for the bis-Fe(III)/bis-Fe(II) redox couple and the quinone/quinol MADH redox couple are known. Each is a two-electron oxidation-reduction. For MauG, the Em values for the first and second electron reductions are −159 mV and −244 mV, respectively (32). For MADH, the Em values for the quinone/semiquinone and semiquinone/quinol couples are +14mV and +190 mV, respectively (33). Thus, the ΔEm values for the first and second ET are +258 and +349 mV, respectively, and indicate that the rate of the first ET will be much slower than the second. The observed single phase kinetics is consistent with a relatively slow ET followed by a faster ET with the observed rate being that of the first ET step. Fit of the data by eq 3 using ΔG° = −2.49 kJ•mol−1, which corresponds to the first ET, yields HAB= 0.013 ± 0.002 cm−1 and λ = 1.8 ± 0.1 eV. While the λ value may seem large for an ET reaction it is consistent with results of previous studies of the well-characterized ET reaction from the reduced TTQ quinol form of MADH to its natural redox partner, amicyanin, which yielded a λ of 2.3 eV (33, 34). This λ value was obtained both from analysis of the temperature dependence of the rate of that reaction and from the ΔG° dependence of the reaction. The latter was determined by measuring the forward and reverse ET rates between different redox forms of MADH (i.e., quinone, semiquinone, quinol) and amicyanin, each of which exhibit a different ΔG° (33). The large magnitude of λ for ET reactions involving MADH was attributed to the TTQ cofactor, a large organic cofactor with charge delocalized over a large area and partial exposure to solvent (35). Analysis of the data by eq 4 allows estimation of the ET distance given a particular β value. Analysis of the data using a range of β values from 1.1–1.4 Å−1, which is typically assumed for protein ET reactions (4, 36, 37), yielded an ET distance of 17.4–21.3 Å (Table 1). Previous site-directed mutagenesis studies indicated that the nearer six-coordinate heme rather than the five-coordinate heme is the entry and exit point for electrons into and from the diheme system of MauG during these interprotein ET reactions (23). The distance in the crystal structure from βTrp108 of TTQ to the six-coordinate heme iron is 19.4 Å, or 15.5 Å to the porphyrin ring, which is in good agreement with the experimentally determined value. These results are consistent with ET from diferrous MauG to the oxidized TTQ of MADH proceeding via single-step electron tunneling (Figure 4a).
Figure 3.
The dependence on temperature of the rate of electron transfer from (A) diferrous MauG to quinone MADH and from (B) preMADH to bis-Fe(IV) MauG. The lines are fits of the data by equations 3 and 4. The two sets of fits are superimposable.
Table 1.
Comparative analysis of electron transfer reactions
| Parameters | Diferrous MauG to quinone MADH |
PreMADH to bisFe(IV) MauG |
|---|---|---|
| ΔG°(kJ mol−1) | −24.9 | (−28.9) – (+28.9)a |
| HAB (cm−1) | 0.013 ± 0.003 | 17.9 — 20.2 ± 12.8a |
| r (Å, β=1.1 Å−1) | 21.3 ± 0.4 | 8.1 ± 1.2 |
| r (Å, β=1.4 Å−1) | 17.4 ± 0.3 | 7.0 ± 0.9 |
| λ(eV) | 1.80 ± 0.04 | 1.97 — 3.18 ± 0.14a |
As discussed in the text, a range of ΔG°values corresponding toΔEm values from −300 mV to +300 mV was used when analyzing this reaction. The corresponding range of HAB and λ values are listed for this reaction. The r values which were obtained did not vary with ΔG° over this range.
Figure 4.
Schematic representation of (Top panel) electron tunneling from the nearest heme of diferrous MauG to quinone MADH and (Bottom panel) hole hopping via Trp199 from preMADH to the nearest heme of bis-Fe(IV) MauG. Distances to and from the heme are listed both as to and from the heme Fe and the porphyrin ring (in parentheses). The figure was drawn using the coordinates from PDB ID: 3L4O (top) and 3L4M (bottom).
Electron transfer from preMADH to bis-Fe(IV) MauG
The initial ET reaction in the six-electron oxidation of preMADH is the reduction of the bis-Fe(IV) species to diferric by preMADH (28). The exact nature of the product of this reaction that is derived from preMADH is still under investigation, but it is known that a protein-based radical species forms on preMADH concomitant with the reduction of the bis-Fe(IV) species (10). This reaction was monitored under single-turnover conditions over a temperature range from 9 °C to 30 °C (Figure 2b). As stated earlier, MauG was not stable above the higher temperature and so the range could not be extended. Over this range the kET varied from 0.16 to 0.83 s−1, rates which are approximately ten-fold greater than kET for the reaction from diferrous MauG to quinone MADH. It should be noted that the kcat for the overall steady-state reaction of MauG-dependent TTQ biosynthesis from preMADH is 0.2 s−1 at 30 °C (38, 39). Thus, the rates determined here are kinetically competent and indicate that some other reaction step is rate-limiting for the overall biosynthetic reaction in the steady-state.
Analysis of this reaction by eqs 3 and 4 is complicated by the fact that the redox potentials of the electron donor and acceptor are not known. However, it is possible to estimate the ΔEm for the reaction from values in the literature of similar redox centers. The Em value for the bis-Fe(IV)/diferric MauG redox couple is unknown, but Em values for Fe(IV)/Fe(III) couples in many heme-dependent peroxidases have been determined and these values range from 724–1160 mV (40). The Em value associated with the reduction of preMADH is also unknown, however, the residues of preMADH which are modified are tryptophans and it is likely that the product is a tryptophan radical (10). Em values for the Trp radical/Trp redox couple have been determined to be in the range of 890–1080 mV (41, 42), which is similar to that for Fe(IV)/Fe(III) couples. For the purpose of this study, it is the experimentally-determined values of HAB and r that are key in distinguishing whether an ET reaction occurs via single-step tunneling or multi-step hopping. When fitting the data with eq 4, uncertainty of the value of ΔG° will affect the accuracy of the value obtained for λ but will have essentially no effect on the value obtained for r, since λ and r are present in different exponential terms. When fitting the data with eq 3, again uncertainty of the value of ΔG° will affect the value obtained for λ. It will also affect the value obtained for HAB but the effect will be minimal. This can be seen in Table 1 which shows the values that were obtained when fitting the data using a range of values of ΔG°, which correspond to ΔEm values ranging from −300 to +300 mV. This range was chosen given the similar Em values reported for Fe(IV)/Fe(III) and Trp radical/Trp redox couples (discussed above).
In multi-step hopping, the rate of each hopping step follows eqs 3 and 4. Each hopping step will exhibit a distinct set of values of ET distance, HAB, ΔG° and λ. In the general case where the slowest hopping step becomes rate determining, the analysis of the temperature dependence of the observed rate of the reaction will yield parameters that describe the electron tunneling associated with the rate-determining hopping step. In this case, the hundred-fold greater HAB value for the reaction of preMADH with bis-Fe(IV) MauG compared to that of the reaction of diferrous MauG with quinone MADH is consistent with the rate-limiting electron tunneling segment in the hopping mechanism being much shorter than the total distance. Analysis of these data with β ranging from 1.1–1.4 Å−1 yields an ET distance of 7.0–8.1 Å. This distance is not consistent with direct electron tunneling from preMADH to the nearest heme. It was previously shown that mutation of Trp199 of MauG, which lies midway between βTrp108 and the nearest heme, abolished TTQ biosynthesis activity, although it did not disrupt the ET reaction from diferrous MauG to quinone MADH (23). Inspection of the crystal structure of the MauG-preMADH complex indicates that the closest distance between an atom of Trp199 and the six-coordinate heme iron is 10.5 Å, or 8.0 Å to the porphyrin ring, and the closest distance between an atom of Trp199 and an atom of Trp108 of preMADH is 6.5 Å (Figure 4b). Either of these distances for the putative rate-determining hopping step would be consistent with the experimentally determined distance and evidence that this ET reaction proceeds by a hole hopping mechanism. As discussed above, while reliable values for HAB and r were obtained in this analysis, values obtained for λ span a wide range due to the uncertainty in ΔG°. The lowest value of λ of 1.9 eV which was obtained when assuming a ΔEm of −300 mV is comparable to the value of λ obtained for the ET from diferrous MauG to quinone MADH. This does not prove, but suggests that the rate limiting hopping segment for ET from preMADH to bis-Fe(IV) MauG may be endergonic by ~200–300 mV.
It should be noted that if the observed rate of the ET reaction from preMADH to bis-Fe(IV) MauG is that of a hop involving Trp199, the Trp199 radical/Trp199 redox couple will be relevant to the determination of ΔG° for the hopping segment. As stated earlier, it is expected that this value will be similar to those for the Fe(IV)/Fe(III) couple, and that which describes the oxidation of the Trp residues that form TTQ. This being the case, one would expect a substantial fraction of Trp199 might be oxidized prior to preMADH binding which would lead to a kinetic phase with spectral characteristics different from that of heme reduction. This was not observed and this is explained by inspection of the crystal structure of the MauG-preMADH complex (24). Trp199 is hydrogen bonded across the MauG-preMADH interface with Glu101 of preMADH. This structural feature allows the proton to remain close to the transiently oxidized Trp199. For this reason Trp199 probably is not oxidized until after it binds preMADH.
Applicability of electron transfer theory
While each of the two reactions under study are two-electron oxidation-reduction reactions, in each case the reaction kinetics fit well to a single exponential relaxation. This means that in each case the kinetic mechanism for the reaction is a relatively slow one-electron transfer followed by a faster one-electron transfer. As such, no intermediate state accumulates and is not observed. The observed rate in this case is that of the slower initial ET event. This is important as the analysis of these reaction rates by eqs 3 and 4 is only valid for a single-electron transfer process. Given the complexity of the interprotein ET reactions that are under study it is also important to consider the possibility that the observed reaction may be gated by a non-ET process such as a conformational change. It was previously shown that the kinetics of each of these ET reactions exhibit saturation behavior, a hyperbolic dependence of rate on concentration (27, 28). Thus, the observed rate constant is independent of the protein-protein binding step and the rate of binding is not rate limiting for the observed reaction rate. This does not rule out the possibility of a rate limiting intracomplex conformational change. However, it should be noted that MauG-dependent TTQ biosynthesis has been shown to occur in crystallo in the MauG-preMADH complex (24). This does rule out a requirement for a large conformational change for the reaction to occur. The possibility of a more subtle conformational change that may contribute to the observed ET rate cannot be excluded; not necessarily a very slow process which gates the ET reaction but perhaps one which results in kinetically coupled ET (5). This would be true if the relatively slow ET is preceded by a rapid but unfavorable (i.e., Keq <1) adiabatic reaction step. In this case, analysis of such reactions by eqs 3 and 4 may yield erroneous values for λ, HABor r (43). However, the strong correlation of the experimentally-determined values of r with the distances seen in the crystal structure of the complex provides additional support for the validity of the experimentally determined parameters in Table 1.
HAB is related to the ET distance and nature of the intervening medium between ET donor and acceptor. It has been shown for several systems that analysis of the temperature dependence of the rate of a gated ET reaction will yield unreasonably large values of HAB and correspondingly unreasonably small values of r (37). That was not the case in this study. The experimentally-determined HAB values are well below the non-adiabatic limit, i.e. ~80 cm−1 (44), also strongly suggest that these data describe a true ET reaction (5). To further examine the relevance of these HAB values, the crystal structure of the MauG-preMADH complex was analyzed with the HARLEM computer program (30). Relative values of HAB were calculated for the single-step electron tunneling from residue βTrp108 of preMADH to the 6-coordinate heme of MauG, and the electron tunneling segments in the Trp199-mediated hole hopping mechanism (Table 2). Comparison of these predicted values with the experimentally-determined values shows a strong correlation, consistent with the interpretation that the rate of ET in the reaction from preMADH to bis-Fe(IV) MauG is enhanced by the hole hopping mechanism relative to single-step electron tunneling.
Table 2.
Comparison of predicted values of electronic coupling with experimentally-determined values.
| Predicted values determined from the structurea | ||
|---|---|---|
| Donor | Acceptor | Relative HAB |
| Direct single-step tunneling | ||
| preMADH βTrp108 | MauG 6-coordinate heme |
1.0 |
| Tunneling segments in Trp199-mediated hopping | ||
| preMADH βTrp108 | MauG Trp199 | 1837 |
| MauG residue 199 | MauG 6-coordinate heme |
1317 |
| Experimentally determined valueb | 1400–1600 | |
The HARLEM computer program (30) was used to calculate relative values of HAB from the crystal structures of the MauG-preMADH complex using the direct distance approach Dutton and coworkers (4). These HAB values are dimensionless and therefore cannot be directly compared to the experimentally determined values. However, relative values for putative hopping segments and the single-step direct tunneling path can be compared with the experimental values. For ease of comparison all values were normalized to that for the HAB value for the overall ET reaction; the shortest distance between an atom of residue βTrp108 of preMADH and the 6-coordinate heme of MauG.
Calculated from the values presented in Table 1.
Summary
Electron tunneling can be an efficient mechanism for ET. However, as indicated in eq 4kET decreases exponentially with distance. This precludes efficient long range ET in the absence of a very large driving force (i.e., - ΔG°) for the reaction. This presents a challenge in nature because in order to support biological activity it is often necessary to transfer electrons over large distances, through and between proteins with a relatively low driving force. Hopping provides an alternative mechanism to overcome the distance dependence of ET. In molecular electronics there is a competition between tunneling and hopping mechanisms. Shorter distances (bridges) between electron donor and acceptor allow larger overlap between donor and acceptor wavefunctions which favor a tunneling (superexchange) mechanism. However, at longer distances hopping will dominate. This is also observed for ET through DNA where tunneling is observed at shorter distances and hopping dominates at longer distances. Thus, in these systems one observes an apparent contradiction, a strong distance dependence of kET at shorter distances and a weak or lack of distance dependence of kET at longer distances.
For the first time, a comparative Marcus analysis of tunneling and hopping mechanisms of ET through a natural protein system is presented in this study and the requirements for hopping during ET through proteins are illustrated. In proteins, hopping relays must be amino acid residues that can be reversibly oxidized and reduced. In nature, protein ET reactions are not initiated by an applied voltage as in non-biological systems. The driving force for biological protein ET reactions depends on the ΔEm of the donor and acceptor redox centers. To initiate hopping, a redox center with a sufficiently high potential to oxidize an intervening amino acid residue is required. In MauG, the ET reaction is initiated by the formation of a high valent bis-Fe(IV) species which then oxidizes residue Trp199 at the protein surface via electron tunneling over a relatively short distance. The preMADH substrate bound to the MauG protein surface is in turn oxidized via electron tunneling over another relatively short distance by the Trp199 radical species to complete the reaction. In the hopping mechanism, the electron which leaves the electron donor is not the same as the electron which enters the electron acceptor. Rather it is an electron which is displaced from the intermediate. This allows the overall ET reaction to occur more rapidly than via single-step long range electron tunneling between donor and acceptor. Furthermore, while a high potential oxidizing species in needed to initiate the reaction, the overall ΔG° for the reaction is near zero. This study demonstrates that if these requirements for hopping are met, ET and even catalysis can occur over very long distances with little if any driving force via hole hopping through and between proteins.
ACKNOWLEDGEMENTS
We thank Yu Tang for technical assistance.
This was supported by NIH grant GM-41574 (VLD).
Abbreviations
- ET
electron transfer
- MADH
methylamine dehydrogenase
- TTQ
tryptophan tryptophylquinone
- preMADH
the biosynthetic precursor protein of MADH with incompletely synthesized TTQ
- bis-Fe(IV) MauG
redox state of MauG with one heme as Fe(IV)=O and the other as Fe(IV)
- Em
oxidation-reduction midpoint potential
- λ
reorganization energy
- HAB
electron coupling
- r
distance between electron donor and acceptor
REFERENCES
- 1.Beratan DN, Onuchic JN, Hopfield JJ. Electron tunneling through covalent and noncovalent pathways in proteins. J. Chem. Phys. 1987;86:4488–4498. [Google Scholar]
- 2.Gray HB, Winkler JR. Electron tunneling through proteins. Q Rev Biophys. 2003;36:341–372. doi: 10.1017/s0033583503003913. [DOI] [PubMed] [Google Scholar]
- 3.Marcus RA, Sutin N. Electron transfers in chemistry and biology. Biochim. Biophys. Acta. 1985;811:265–322. [Google Scholar]
- 4.Page CC, Moser CC, Chen X, Dutton PL. Natural engineering principles of electron tunnelling in biological oxidation-reduction. Nature. 1999;402:47–52. doi: 10.1038/46972. [DOI] [PubMed] [Google Scholar]
- 5.Davidson VL. Protein control of true, gated and coupled electron transfer reactions. Acc. Chem. Res. 2008;41:730–738. doi: 10.1021/ar700252c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Giese B, Graber M, Cordes M. Electron transfer in peptides and proteins. Curr. Opin. Chem. Biol. 2008;12:755–759. doi: 10.1016/j.cbpa.2008.08.026. [DOI] [PubMed] [Google Scholar]
- 7.Wang Y, Graichen ME, Liu A, Pearson AR, Wilmot CM, Davidson VL. MauG, a novel diheme protein required for tryptophan tryptophylquinone biogenesis. Biochemistry. 2003;42:7318–7325. doi: 10.1021/bi034243q. [DOI] [PubMed] [Google Scholar]
- 8.Davidson VL. Pyrroloquinoline quinone (PQQ) from methanol dehydrogenase and tryptophan tryptophylquinone (TTQ) from methylamine dehydrogenase. Adv. Protein Chem. 2001;58:95–140. doi: 10.1016/s0065-3233(01)58003-1. [DOI] [PubMed] [Google Scholar]
- 9.Pearson AR, De La Mora-Rey T, Graichen ME, Wang Y, Jones LH, Marimanikkupam S, Agger SA, Grimsrud PA, Davidson VL, Wilmot CM. Further insights into quinone cofactor biogenesis: probing the role of mauG in methylamine dehydrogenase tryptophan tryptophylquinone formation. Biochemistry. 2004;43:5494–5502. doi: 10.1021/bi049863l. [DOI] [PubMed] [Google Scholar]
- 10.Li X, Fu R, Lee S, Krebs C, Davidson VL, Liu A. A catalytic di-heme bis-Fe(IV) intermediate, alternative to an Fe(IV)=O porphyrin radical. Proc. Natl. Acad. Sci. USA. 2008;105:8597–8600. doi: 10.1073/pnas.0801643105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Afzali A, Dimitrakopoulos CD, Breen TL. High-performance, solution-processed organic thin film transistors from a novel pentacene precursor. J Am Chem Soc. 2002;124:8812–8813. doi: 10.1021/ja0266621. [DOI] [PubMed] [Google Scholar]
- 12.Berlin YA, Hutchison GR, Rempala P, Ratner MA, Michl J. Charge Hopping in Molecular Wires as a Sequence of Electron-Transfer Reactions. J. Phys. Chem. A. 2003;107:3970–3980. [Google Scholar]
- 13.Genereux JC, Barton JK. Mechanisms for DNA charge transport. Chem Rev. 2010;110:1642–1662. doi: 10.1021/cr900228f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Giese B. Electron transfer in DNA. Curr Opin Chem Biol. 2002;6:612–618. doi: 10.1016/s1367-5931(02)00364-2. [DOI] [PubMed] [Google Scholar]
- 15.Lewis FD, Letsinger RL, Wasielewski MR. Dynamics of photoinduced charge transfer and hole transport in synthetic DNA hairpins. Acc Chem Res. 2001;34:159–170. doi: 10.1021/ar0000197. [DOI] [PubMed] [Google Scholar]
- 16.Bixon M, Giese B, Wessely S, Langenbacher T, Michel-Beyerle ME, Jortner J. Long-range charge hopping in DNA. Proc Natl Acad Sci U S A. 1999;96:11713–11716. doi: 10.1073/pnas.96.21.11713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Giese B, Amaudrut J, Kohler AK, Spormann M, Wessely S. Direct observation of hole transfer through DNA by hopping between adenine bases and by tunnelling. Nature. 2001;412:318–320. doi: 10.1038/35085542. [DOI] [PubMed] [Google Scholar]
- 18.Stubbe J, Nocera DG, Yee CS, Chang MC. Radical initiation in the class I ribonucleotide reductase: long-range proton-coupled electron transfer? Chem. Rev. 2003;103:2167–2201. doi: 10.1021/cr020421u. [DOI] [PubMed] [Google Scholar]
- 19.Baldwin J, Krebs C, Ley BA, Edmondson DE, Huynh BH, Bollinger JH. Mechanism of rapid electron transfer during oxygen activation in the R2 subunit of Escherichia coli ribonucleotide reductase. 1. Evidence for a transient tryptophan radical. J Am Chem Soc. 2000;122:12195–12206. [Google Scholar]
- 20.Lukacs A, Eker AP, Byrdin M, Brettel K, Vos MH. Electron hopping through the 15 A triple tryptophan molecular wire in DNA photolyase occurs within 30 ps. J Am Chem Soc. 2008;130:14394–14395. doi: 10.1021/ja805261m. [DOI] [PubMed] [Google Scholar]
- 21.Shih C, Museth AK, Abrahamsson M, Blanco-Rodriguez AM, Di Bilio AJ, Sudhamsu J, Crane BR, Ronayne KL, Towrie M, Vlcek A, Jr, Richards JH, Winkler JR, Gray HB. Tryptophan-accelerated electron flow through proteins. Science. 2008;320:1760–1762. doi: 10.1126/science.1158241. [DOI] [PubMed] [Google Scholar]
- 22.Seifert JL, Pfister TD, Nocek JM, Lu Y, Hoffman BM. Hopping in the electron-transfer photocycle of the 1:1 complex of Zn-cytochrome c peroxidase with cytochrome c. J Am Chem Soc. 2005;127:5750–5751. doi: 10.1021/ja042459p. [DOI] [PubMed] [Google Scholar]
- 23.Abu Tarboush N, Jensen LMR, Yukl ET, Geng J, Liu A, Wilmot CM, Davidson VL. Mutagenesis of tryptophan199 suggets that hopping is required for MauG-dependent tryptophan tryptophylquinone biosynthesis. Proc Natl Acad Sci U S A. 2011;108:16956–16961. doi: 10.1073/pnas.1109423108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jensen LM, Sanishvili R, Davidson VL, Wilmot CM. In crystallo posttranslational modification within a MauG/pre-methylamine dehydrogenase complex. Science. 2010;327:1392–1394. doi: 10.1126/science.1182492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li X, Jones LH, Pearson AR, Wilmot CM, Davidson VL. Mechanistic possibilities in MauG-dependent tryptophan tryptophylquinone biosynthesis. Biochemistry. 2006;45:13276–13283. doi: 10.1021/bi061497d. [DOI] [PubMed] [Google Scholar]
- 26.Wang Y, Li X, Jones LH, Pearson AR, Wilmot CM, Davidson VL. MauG-dependent in vitro biosynthesis of tryptophan tryptophylquinone in methylamine dehydrogenase. J Am Chem Soc. 2005;127:8258–8259. doi: 10.1021/ja051734k. [DOI] [PubMed] [Google Scholar]
- 27.Shin S, Abu Tarboush N, Davidson VL. Long-range electron transfer reactions between hemes of MauG and different forms of tryptophan tryptophylquinone of methylamine dehydrogenase. Biochemistry. 2010;49:5810–5816. doi: 10.1021/bi1004969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee S, Shin S, Li X, Davidson V. Kinetic mechanism for the initial steps in MauG-dependent tryptophan tryptophylquinone biosynthesis. Biochemistry. 2009;48:2442–2447. doi: 10.1021/bi802166c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Davidson VL. Methylamine dehydrogenases from methylotrophic bacteria. Methods Enzymol. 1990;188:241–246. doi: 10.1016/0076-6879(90)88040-h. [DOI] [PubMed] [Google Scholar]
- 30.Kurnikov IV. HARLEM Computer Program. University of Pittsburgh; 2000. http://harlem.chem.cmu.edu/ [Google Scholar]
- 31.Abu Tarboush N, Jensen LM, Feng M, Tachikawa H, Wilmot CM, Davidson VL. Functional importance of tyrosine 294 and the catalytic selectivity for the bis-Fe(IV) state of MauG revealed by replacement of this axial heme ligand with histidine. Biochemistry. 2010;49:9783–9791. doi: 10.1021/bi101254p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li X, Feng M, Wang Y, Tachikawa H, Davidson VL. Evidence for redox cooperativity between c-type hemes of MauG which is likely coupled to oxygen activation during tryptophan tryptophylquinone biosynthesis. Biochemistry. 2006;45:821–828. doi: 10.1021/bi052000n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brooks HB, Davidson VL. Free energy dependence of the electron transfer reaction between methylamine dehydrogenase and amicyanin. J. Am. Chem. Soc. 1994;116:11201–11202. [Google Scholar]
- 34.Brooks HB, Davidson VL. Kinetic and thermodynamic analysis of a physiologic intermolecular electron-transfer reaction between methylamine dehydrogenase and amicyanin. Biochemistry. 1994;33:5696–5701. doi: 10.1021/bi00185a005. [DOI] [PubMed] [Google Scholar]
- 35.Sun D, Chen ZW, Mathews FS, Davidson VL. Mutation of αPhe55 of methylamine dehydrogenase alters the reorganization energy and electronic coupling for its electron transfer reaction with amicyanin. Biochemistry. 2002;41:13926–13933. doi: 10.1021/bi026654x. [DOI] [PubMed] [Google Scholar]
- 36.Onuchic JN, Beratan DN, Winkler JR, Gray HB. Pathway analysis of protein electron-transfer reactions. Ann. Rev. Biophys. Biomol. Struct. 1992;21:349–377. doi: 10.1146/annurev.bb.21.060192.002025. [DOI] [PubMed] [Google Scholar]
- 37.Davidson VL. What controls the rates of interprotein electron-transfer reactions. Acc Chem Res. 2000;33:87–93. doi: 10.1021/ar9900616. [DOI] [PubMed] [Google Scholar]
- 38.Li X, Fu R, Liu A, Davidson VL. Kinetic and physical evidence that the diheme enzyme MauG tightly binds to a biosynthetic precursor of methylamine dehydrogenase with incompletely formed tryptophan tryptophylquinone. Biochemistry. 2008;47:2908–2912. doi: 10.1021/bi702259w. [DOI] [PubMed] [Google Scholar]
- 39.Feng M, Jensen LM, Yukl ET, Wei X, Liu A, Wilmot CM, Davidson VL. Proline 107 is a major determinant in maintaining the structure of the distal pocket and reactivity of the high-spin heme of MauG. Biochemistry. 2012;51:1598–1606. doi: 10.1021/bi201882e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Battistuzzi G, Bellei M, Bortolotti CA, Sola M. Redox properties of heme peroxidases. Arch Biochem Biophys. 2010;500:21–36. doi: 10.1016/j.abb.2010.03.002. [DOI] [PubMed] [Google Scholar]
- 41.DeFelippis MR, Murthy CP, Faraggi M, Klapper MH. Pulse radiolytic measurement of redox potentials: the tyrosine and tryptophan radicals. Biochemistry. 1989;28:4847–4853. doi: 10.1021/bi00437a049. [DOI] [PubMed] [Google Scholar]
- 42.Tommos C, Skalicky JJ, Pilloud DL, Wand AJ, Dutton PL. De novo proteins as models of radical enzymes. Biochemistry. 1999;38:9495–9507. doi: 10.1021/bi990609g. [DOI] [PubMed] [Google Scholar]
- 43.Davidson VL. Effects of kinetic coupling on experimentally determined electron transfer parameters. Biochemistry. 2000;39:4924–4928. doi: 10.1021/bi992671j. [DOI] [PubMed] [Google Scholar]
- 44.Winkler JR, Gray HB. Electron transfer in ruthenium-modified proteins. Chem. Rev. 1992;92:369–379. doi: 10.1007/BF02110099. [DOI] [PubMed] [Google Scholar]