Abstract
This article describes an innovative software toolkit that allows the creation of web applications that facilitate the acquisition, integration, and dissemination of multimedia biomedical data over the web, thereby reducing the cost of knowledge sharing. There is a lack of high-level web application development tools suitable for use by researchers, clinicians, and educators who are not skilled programmers. Our Web Interfacing Repository Manager (WIRM) is a software toolkit that reduces the complexity of building custom biomedical web applications. WIRM’s visual modeling tools enable domain experts to describe the structure of their knowledge, from which WIRM automatically generates full-featured, customizable content management systems.
Biomedical research efforts are becoming increasingly reliant on the interoperability of autonomous heterogeneous software applications, involving widespread collaboration by teams of scientists and clinicians across multiple disciplines and institutions. Consequently, there is a need for a new generation of biomedical information systems that facilitate remote collaboration, data sharing, workflow management, and integration of heterogeneous knowledge sources. The diverse nature of experimental data and protocols dictates that each information system be custom-tailored through a domain-specific set of object classes, templates, interfaces, and workflow facilities. This suggests a need for template-based, adaptable frameworks that enable scientists, clinicians, and educators to create their own custom information systems. Such frameworks should provide high-level interfaces that empower domain experts to model the structure of their content and workflow requirements, according to their own domain knowledge.
This article identifies the informatics requirements for such a framework and describes the architecture and implementation of a prototype open source toolkit that begins to meet those requirements: the Web Interfacing Repository Manager (WIRM). WIRM consists of a visual development environment and a high-level programming interface that allows health professionals to rapidly design and implement their own custom web-based interfaces to biomedical content.1 WIRM enables a nonprogrammer to model domain knowledge as object-oriented schemas, using a menu-driven interface. Once schemas are defined, WIRM automatically generates a drill-down web information system for acquiring, querying, navigating, annotating, and editing instances of those schemas.
WIRM has been released as an open source toolkit and has been used to build a wide range of applications for clinicians, researchers, and educators. As estimated by the developers, WIRM reduced implementation time for the applications by 50–75% over more traditional approaches. Planned improvements should increase the usability to point that a growing number of medical professionals will be able to create custom applications that improve the efficiency of their research efforts and expand their capacity to share knowledge.
Background
Need for an Experiment Management System
WIRM was initially developed in response to the complex data management needs of the University of Washington Human Brain Project, an interdisciplinary research project involving data sharing among radiologists, surgeons, neuroscientists, statisticians, computer vision experts, technicians, and students.2,3UW-HBP members must manage a diverse array of biomedical data, including patient records, MRI exams, graphical models, digitized photographs, and tabular experiment results. A system was needed to support data sharing between laboratories and across distributed, heterogeneous platforms. These advanced data management requirements are in fact status quo for current biomedical research projects.4We developed WIRM as a platform for building an experiment management system consisting of interactive, dynamic interfaces that adapt themselves to multiple classes of end-users. For example, the UW-HBP’s experiment management system exports three distinct interfaces for core project members, collaborators, and guests. Each interface allows a different set of operations to be performed, such as viewing patient records, uploading graphical brain models, processing study data, or interfacing with the hospital’s picture archiving system.5
Existing Approaches
A number of vendors supply Laboratory Information Management Systems (LIMS), which are interactive applications for managing biomedical data. However, such applications tend to be boilerplate solutions for commercial industries such as utilities or pharmaceuticals, which deal with high volumes of relatively standardized workflow and data types. As a result, they are not suitable for supporting ad-hoc research that does not conform to a standard industry model.6
Existing clinical information systems, such as Computerized Patient Record Systems (CPR) and Picture Archiving and Communication Systems (PACS), provide clinicians with the ability to store and retrieve records of clinical encounters. However, these systems tend to be inherently rigid: the data models, user interfaces, and functionality are suitable only for basic clinical reporting and do not support the repurposing of clinical content for research and education.
Custom web applications can be built with commercial middleware known as application servers, such as IBM WebSphere (IBM Corp., Armonk, NY) or Cold Fusion (Macromedia, San Fancisco, CA). However, these platforms are designed for corporate business users rather than medical professionals and lack support for biomedical data types, formats, or protocols.8Furthermore, programs built on top of commercial application servers cannot be freely distributed within the research community, because they require the proprietary server software to run. Two open source application servers do exist: Zope (Zope Corp., Fredericksburg, VA) and Enhydra (Lutris, Santa Cruz, CA). However, like their proprietary counterparts, these tools are not designed for managing biomedical content. What is needed is a freely distributable web application development framework tailored specifically for biomedical professionals.
While such a tool does not yet exist, several initiatives, such as OpenEMed (formerly Telemed), define open infrastructures for building large-scale clinical information systems.9OpenEMed is based on the Common Object Request Broker Architecture (CORBA), which allows distributed objects to be assembled to form larger applications.10Although CORBA systems hold promise for enterprise-level application development, they tend to be unwieldy and overly complex from the perspective of smaller projects that do not need to participate in a distributed object network.11
From our experience, independent projects are best built using freestanding components that leverage the power of freely available open source tools. Some examples of open source resources especially relevant to biomedical content management are the MySQL relational database (MySQL AB, Uppsala, Sweden), the ImageMagick graphics software (http://www.imagemagick.org), and the Comprehensive Perl Archive Network (CPAN), a collection of over 900 independent modules dedicated to text parsing, database interfacing, web form processing, and other tasks (http://www.cpan.org). Furthermore, there are numerous open source tools specifically designed to interface with biomedical resources, such as the Central Test Node DICOM toolkit (http://www.erl.wustl.edu/DICOM/ctn.html).
Although a vast wealth of open source software is readily available, much of it remains untapped by the biomedical professionals who need it the most. The tools tend to take the form of low-level building blocks that are designed for expert users, and the details of making them work together can be daunting for scientists and clinicians who are not experienced software developers. Forums such as the Openhealth List (http://www.minoru-development.com/en/healthcare.html) attempt to provide a roadmap to medical open source software. However, to date there is a significant lack of applications that make open source resources accessible to novice users by integrating them and abstracting away their low level details.
Design Objectives
The overall objective of the WIRM project is to build a set of web-based content-management tools that leverage the power of existing open source software in a biomedical context. The tools should preferably give domain experts a large amount of control over the structure of their system and the behavior of the interfaces. Domain experts have the best vantage point from which to understand the requirements of their own applications, and the most laborious stage of implementing a biomedical web application is translating those requirements to a programmer who is unfamiliar with the details of the domain. If the application development tools were suitable for use by the domain experts themselves, systems could be built more quickly and evolve more efficiently.
Given this overall objective, we analyzed target applications in a diverse set of domains, including multimedia databases, teleradiology, clinical structured reporting, medical terminology standards, and experiment management systems. We distilled our findings into the following list of technical requirements that are highly desirable in a biomedical application server. The system that we have implemented to date meets many of these requirements. Others will be met in our planned enhancements, as described in the Discussion section below.
Rapid application development.
The application server should include high-level interfaces for generating web applications quickly and easily by following a stepwise methodology that requires minimal programming. Changes should be easy to test and deploy, without requiring a tedious recompilation procedure. Medical professionals who are not trained programmers require a well-documented, straightforward approach for building complex systems through stepwise refinement.
Visual modeling interfaces.
The system should provide graphical user interfaces that enable domain experts to specify the structure of their knowledge without requiring programming. Through forms and menus, users should be able compose object classes by creating lists of typed attributes and relationships.
Schema evolution.
As users’ understanding of their domain model evolves over time, it should be easy to modify schema definitions to keep up with changes in the domain model.
Automatic form generation.
The system should generate web forms for acquiring and editing instances of data. Form elements should be customized to match the domain schemas that were defined with the modeling interface.
Database connectivity.
The system should be able to interface with all major databases, including Oracle, Sybase, SQL Server, Access, DB2, Informix, MySQL, and MSQL.
Hierarchical navigation.
The system should generate a hypertext navigation interface that provides drill-down access to data stored in databases.
Query support.
Users should be able to pose ad-hoc queries over any combination of attributes.
Document management.
The system should include facilities for uploading and archiving file-based documents and their associated metadata.
Multimedia support.
The system should support management of images and other multimedia, including control over the size and resolution of images and the ability to automatically convert multimedia to formats suitable for viewing over the web.
Context-sensitive, role-based access.
Because medical applications often must support multiple levels of end user, the system should include a high-level programmatic interface for defining context-sensitive interfaces that adapt themselves according to the viewer’s group membership. Views should be tailorable at the granularity of individual object classes.
Robust security.
To protect highly sensitive medical data, the system must use encryption and other security measures.
Scalability.
The system should be able to handle very large data sets and be accessible by a large number of simultaneous users without significant degradation in response time.
Connectivity to biomedical resources.
Biomedical applications often need to communicate with existing multimedia data sources, such as Picture Archiving and Communication Systems (PACS) and medical imaging equipment. The DICOM (Digital Imaging and Communications in Medicine) Standard defines a communication protocol for exchanging medical images and their associated information, allowing diverse groups of medical imaging hardware and software to share data. As most current systems support DICOM, the application server should provide tools for retrieving data from DICOM sources. Similarly, as a growing number of applications use XML to transmit data, the system should provide support for parsing and constructing XML documents.
System Description
Implementation
WIRM is implemented in the Perl programming language. After careful evaluation of candidate languages, including Java, C++, and Python, we chose Perl for its built-in data processing facilities and its unparalleled conciseness: Perl programs require fewer lines of code to perform the same tasks than any of their competitors.12Furthermore, a Perl-based API enables seamless interfacing with CPAN.
WIRM implements web applications using the Common Gateway Interface (CGI) protocol, a standard for interfacing external software with web servers. WIRM works with any web server, preferably the free Apache server (Apache Software Foundation, Forest Hill, MD) running on Linux. In our experience, Linux is the most cost-effective and reliable of all server environments, although WIRM can be easily ported to other Unix platforms or Windows. Regardless of server environment, WIRM clients are platform-independent and compatible with any web browser.
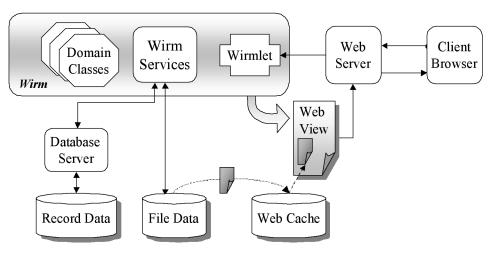
Application behavior is encapsulated in small independent Perl scripts, called wirmlets, that are invoked by the web server. We chose this approach based on years of experience with a wide range of web application development technologies. We have found that breaking an application into manageable chunks greatly reduces the complexity of the development process, allowing construction to proceed in a series of iterative refinements. Figure 1▶ shows how these wirmlets interact with the web server, file repository, and databases to form a multimedia content management system. Each time an end-user clicks on a link or submits a form, the client’s browser sends a request over the Internet to the host’s web server. The web server responds by launching the appropriate wirmlet to handle the request. The wirmlet calls on the services of special APIs built into WIRM to process form variables, query databases, prepare files and images for web transmission, and finally construct a new web page. The page is passed back to the server, which transmits it over the Internet to the user’s waiting browser.
Figure 1 .
Web server invoking a wirmlet.
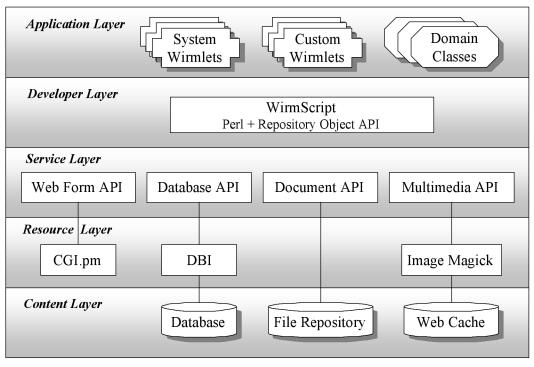
Architecture
WIRM is essentially a framework for integrating existing open source tools, making them easier to use. This is made possible through a five-tiered architecture (Figure 2▶). The layers are as follows:
Figure 2 .
Wirm architecture.
Content layer.
The content layer consists of the repository data and the external systems that contain the data. Records may be stored in any relational database, such as Oracle or MySQL. Files are stored in a protected repository and copied on demand into a web-accessible staging area called the web cache. Files in the web cache are translated into formats suitable for viewing through a web browser. For example, DICOM images are automatically transformed into JPEG images.
Resource layer
The resource layer contains third-party tools that provide much of WIRM’s power. Wherever possible, WIRM leverages existing solutions, and we expect the resource layer to continue to expand as a growing number of tools are integrated into the WIRM framework. All software in the resource layer is open source, which ensures that applications built on WIRM can be freely distributed. Currently, the resource layer includes CGI.pm, a Perl module for handling web forms, DBI.pm, a module for connecting to all major relational databases, and Image Magick, a package for manipulating images.
Service layer
The Perl interfaces exported by the third-party components of the resource layer tend to be overly complex for WIRM’s intended users, who often lack significant programming expertise. The service layer addresses this concern by providing a collection of user-friendly application programmer interfaces (APIs) that abstract away the low-level details of the existing interfaces. The underlying capabilities of the resources remain fully accessible to the system developer, but the service layer APIs make common operations easier to perform by wrapping them in well-documented functions that adhere to a homogenous syntax. There are currently four APIs, one for each tool in the resource layer. The web form API enhances the CGI.pm resource, providing services for creating and parsing interactive form elements, and other shortcuts for generating HTML syntax. The database API encapsulates the DBI module, providing database-independent functions for creating and deleting tables, inserting and removing records, formulating SQL queries, and retrieving query results into tabular data structures. The document API regulates access to the file repository, providing services for copying and retrieving files, managing metadata, assigning unique ID’s, and maintaining version control. The multimedia API provides the media manipulation and transformation operations that are needed to visualize images and other domain data inside web pages. Using the Image Magick utility, the multimedia API works with the document API to copy requested files from the repository into the web cache, where they are transformed into formats suitable for web viewing, and possibly resized and compressed for faster transmission. If this process has already been performed for the desired file, the cached copy is used.
Developer layer.
The developer layer provides a framework for integrating the services of the service layer through a high-level API called WIRMScript. WIRMScript simplifies the process of constructing wirmlets and specifying the behavior of domain objects. WIRMScript exports an object-relational data model to the wirmlet developer. While maintaining the convenience of SQL queries, WIRMScript shields the developer from having to deal explicitly with table structures, allowing data to be treated as objects. In our experience, this abstraction reduces the tedious nature of data manipulation and greatly facilities rapid application development. In addition, WIRMScript provides high-level methods for coordinating the services of the APIs in the third layer.
Application layer.
The Application layer consists of the wirmlets and the domain class definitions. System wirmlets that are packaged with WIRM include the domain modeler, for specifying and evolving class definitions; the content manager, for creating and editing object instances; the object visualizer, for displaying and navigating through repository data; the query composer, for posing queries over repository data; and the admin console, for registering users and controlling authorization. In addition to the system wirmlets, the application layer may also contain custom wirmlets that perform domain-specific operations. For example, the MyPACS application contains a custom wirmlet called AddImages that allows end-users to upload any number of images to their teaching files.
Status Report
We have evaluated WIRM’s feasibility as a tool for supporting each of the following groups of users:
Researchers requiring improved methods for managing data and experiment workflow within and between biomedical laboratories.
Clinicians seeking to organize and disseminate notable cases for diagnostic or teaching purposes.
Educators wanting to build online multimedia archives and knowledge bases.
To demonstrate WIRM’s feasibility, we developed five web-based applications: the Brain Mapper, an experiment management system for a multidisciplinary neuroscience project; MyPACS, a teaching file authoring system for radiologists; Ontolog, a tool for importing and browsing medical vocabularies; the Digital Anatomist Image Collection Manager, an archiving service used by anatomy educators at the University of Washington School of Medicine; and Fathom, a user interface for a natural language processor of medical records. The applications are summarized in Table 1▶. The following sections present overviews of the projects and evaluate WIRM’s effectiveness in the implementation of each application.
Table 1 .
Summary of WIRM-based Applications
| Project | Development Partner | Users | Current Level of Utilization |
|---|---|---|---|
| Brain Mapper Experiment Management System | UW Human Brain Project | Interdisciplinary | Used daily by 14+ project members |
| MyPACS | Seattle Children’s Hospital; Cincinnati Children’s Hospital | Radiologists | Fully operational: over 300 users. |
| Ontolog | None | Clinical system developers | Used by several ongoing terminology projects. |
| Digital Anatomist Image Collection Manager | U. Washington School of Medicine | Professors, Medical students | Used to store several growing image collections for the School of Medicine. |
| Fathom | UCLA Dept. of Radiology | Computer Scientists, clinicians | Supports ongoing NLP research |
Brain Mapper Experiment Management System
The University of Washington Human Brain Project is using WIRM to build the Brain Mapper Experiment Management System (EMS), which manages the workflow of a large group of collaborating scientists. The project’s goal is to develop an information framework for managing cortical stimulation data obtained during neurosurgery. The experiment requires fine-grained collaboration and data sharing among radiologists, neurosurgeons, neuroscientists, statisticians, computer vision experts, database administrators, and a number of technicians, students, and assistants. A wide range of heterogeneous software applications is called upon to interact in a complex workflow process. Patient demographics, MRI exams, surgeries, intra-operative photographs, behavioral experiments, and 3D brain models are all hierarchically modeled as WIRM schemas and views. The EMS provides a repository that acts as a multimedia warehouse consisting of an instantiated view over a wide range of data sources, allowing them to be retrieved and organized at the user’s request, into customized, unified graphical views over the integrated data, facilitating data mining and dissemination.
An important role of the EMS is to assist in the management of the stages of data acquisition and processing. The stages of workflow should be modeled in the EMS, and it should keep track of what has been done on each data object, who did it, what is left to do, and related data. The workflow support is tightly integrated with the Systems Integration facilities, enabling workflow tasks to be automatically recorded and tied to launching the appropriate programs. In the Brain Mapper, the workflow console is patient-centric, that is, all tasks are viewed as parts of the job of acquiring and processing a patient. The main workflow console allows users to register new patients, fetch their exams, update their demographics, upload photographs, and perform other functions. Another important feature is the ability to support an evolving domain model. Users may want to add a new attribute, or rename an existing attribute, or even define a new data type. The system should supply features for these operations. The Brain Mapper EMS provides graphical interfaces for defining new data types and evolving existing types.
The EMS provides a navigational and organizational structure that is consistent and predictable. The system provides a hyperlink-based drill-down interface, allowing users to see an overview of a collection of objects and then retrieve detailed information on a specific object by clicking on it. Because much scientific data are hierarchical in nature, the EMS allows users to traverse data hierarchically. For example, in the Brain Mapper, the users may be presented with a list of patient objects, from which they can click on a single patient to see an overview, from which they can access a detailed view of any part of that patient, such as the MRI exam, which is a hierarchical object in its own right, consisting of multiple series that, in turn, consist of multiple slices. The EMS also provides spatial navigation, in which the user can click on an image-map to retrieve data related to regions on the image. For example, the Brain Mapper allows a user to retrieve information about language sites by clicking on them in the 3D model.
Because current experiments involve multiple classes of users and the data are intended for multiple classes of audience, the EMS should provide multiple, customized interfaces for each class of user. For the Brain Mapper, surgeons, radiologists, and neuroscientists are each interested in a different aspect of the patient record, and should each be provided with a customized view. Furthermore, each refers to a patient by a different preferred label; thus the system should present the preferred identifiers.
The system provides access control over data at an arbitrarily fine granularity. There are two main motivating factors behind this requirement: protecting the privacy of subjects and protecting proprietary data from competitors. Privacy is a central issue in medical informatics, and there is a growing body of research in this area. From the perspective of an EMS, the enforcement of privacy can be considered an aspect of the adaptive user interface. In addition to classifying users by type or interest, the system should classify users according to their privilege level. The Brain Mapper EMS provides three levels of access: privileged, which allows access to everything in the database; collaborator, which allows access to everything except patient identification; and public, which allows access limited to certain parts of a subset of patient records. The distinction between public and collaborator allows us to withhold unpublished data from the general public. The system allows access regulation to be defined at an arbitrarily fine granularity.
The Brain Mapper EMS (Figure 3▶) is being employed on a daily basis by over a dozen researchers at the University of Washington and is continually being adapted as the experiment evolves. The fact that it is used daily by over a dozen researchers across multiple fields of study is a powerful indication of WIRM’s potential. To view a demo of the Brain Mapper, visit <http://wirm.org/brain>.
Figure 3 .
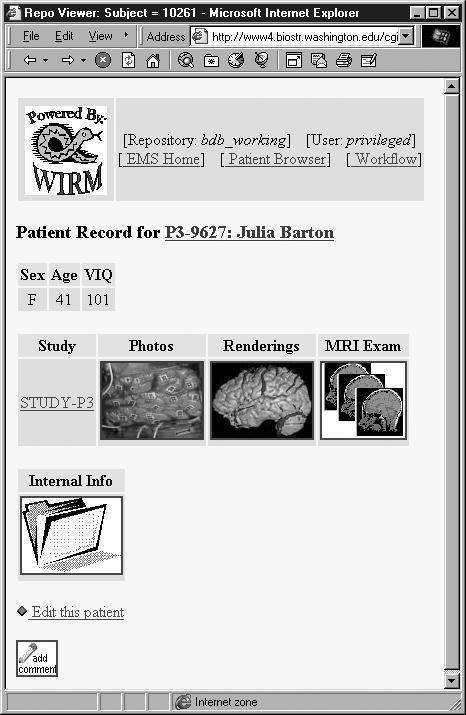
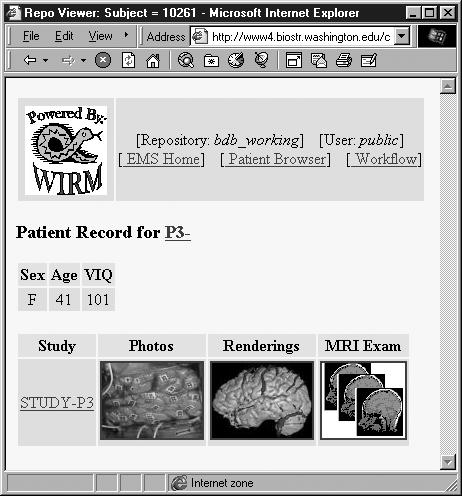
Context-sensitive views in the Brain Mapper EMS.
MyPACS
WIRM was used to build MyPACS (http://mypacs.net), a web-based service that allows radiologists to manage their own online medical image repositories (Figure 4▶).14 Radiologists often keep files of interesting cases for sharing with residents or colleagues or for use in slides or publications. Teaching files have been traditionally stored as hard copies in filing cabinets, but over the past several years a growing number of institutions are publishing their collections online. This has the obvious advantages of widespread dissemination and retrieval by search engines. However, the task of implementing a full-featured online teaching file repository requires significant development effort and web programming expertise. Each hospital that wants an online teaching file repository must create the interfaces for themselves, using ad-hoc methods and tools. Furthermore, they must maintain their own web server, databases, and application software. MyPACS relieves the hospital of these burdens, allowing radiologists to immediately begin authoring cases through their web browser, while maintaining complete control over the content and accessibility of their own collection.
Figure 4 .
MyPACS.
MyPACS allows the case author to upload medical images, enter descriptive information about the case, and input structured data such as patient demographics, image modalities, anatomical structures, and pathological findings. The interface includes a structured reporting tool that guides the user in specifying standardized vocabulary terms. Authors may customize the appearance of their teaching files, including the layout and resolution of images, attributes to be displayed, and other presentation parameters. The author may designate each case as either public or private. For publicly visible cases, sensitive attributes (such as hospital identification numbers) are withheld from nonauthorized users. These features were easy to implement using WIRM’s user management facilities and high-level application programmer’s interface, which facilitates building context-sensitive views over repository data. Teaching files may be retrieved by searching on any parameter, including date, title, pathology, anatomy, or full text searching over the findings in the case. WIRM’s form interface reads the user’s search criteria and translates them into SQL queries, which are handled by WIRM’s database interface. Images can be uploaded in any of over 60 recognizable formats (e.g., JPEG, GIF, BMP, DICOM). For viewing over the web, MyPACS converts images into the browser-friendly JPEG format, but an image can always be retrieved in its original format. Users may specify a preferred image resolution, enabling images to be tailored to screen size and reducing transmission time. The system uses a smooth transform algorithm to preserve image quality during enlargement or reduction. By utilizing the powerful Image Magick programming interface, these features were implemented in under 100 lines of Perl code.
In addition to a teaching file distribution and publishing tool, MyPACS can be used as a convenient system for storing and organizing personal image collections. Authors can upload educational cases as they are encountered, store them in virtual online folders, and later download them for inclusion in publications or PowerPoint presentations. The author of a teaching file retains ownership of the images and case studies that are entered into the system and has complete control over how they are used. Some radiologists are also using MyPACS as a platform for soliciting referrals from remote colleagues.
Ontolog
Medical terminologies such as SNOMED and the UMLS consist of tens of thousands of terms organized into conceptual hierarchies. Although these knowledge resources provide a powerful framework for indexing, integrating, and organizing medical data, there is a lack of manageable interfaces for navigating and visualizing the relationships between the concepts. For example, a part of the Digital Anatomist Foundational Model (the UWDA component of the UMLS)15 can be accessed with a java applet called the Foundational Model Builder,16 but this interface is limited to showing a single hierarchy at a time. Similarly, SNOMED is distributed with a simple text browser, but it is difficult to explore the vast terminology without a more flexible navigation system. This presented an excellent opportunity to test WIRM on interfacing with multiple databases. Thus, we developed Ontolog, a frame-based browser for medical terminologies. The first step consisted of importing the files from their respective data sources into a WIRM repository. Using WIRM’s schema tool, we specified a data model for containing the terms and relationships. Then we used WIRM’s repository object API to build a simple parsing script that connected to the UWDA and SNOMED data sources and import the vocabularies into WIRM’s repository. Once the structures were defined, WIRM automatically generated an interface for navigating the frames.
Ontolog allows the user to view a concept in its full context, not limited to a single attribute dimension. For example, Figure 5▶ depicts the view for the concept superior vena cava, which shows the concept within all its relative hierarchies. To move to a new frame, the user simply clicks on the desired link. Ontolog was used by researchers in the UCLA Telemedicine group towards building a knowledge base that supports clinical record processing.18 Ontolog can be viewed online at http://wirm.org/ontolog.
Figure 5 .
Ontolog frame browser.
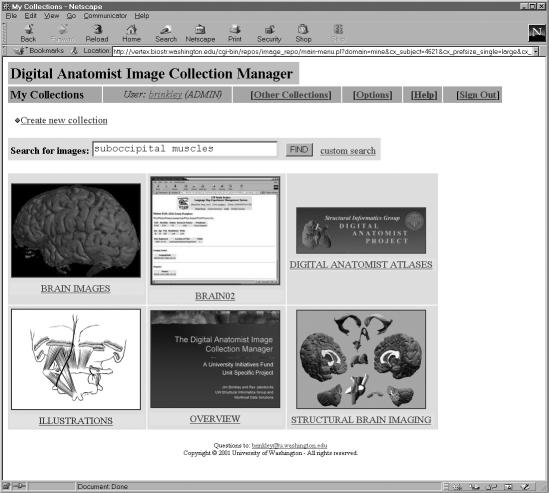
Digital Anatomist Image Collection Manager
The Structural Informatics Group at the University of Washington required a way to organize and disseminate their large image collection, which includes thousands of medical illustrations, radiological images, and computer-generated graphical models from publications, multimedia atlases, and other sources. Existing image archiving solutions, such as CONTENT,19 were considered, but none of them provided the flexibility required by the Digital Anatomist project, which involved a wide range of attributes and classification systems. As a demonstration of WIRM’s effectiveness in building systems for medical educators, we built the Digital Anatomist Image Collection Manager (Figure 6▶), which is available online at <http://www9.biostr.washington.edu/repos/image_repo>.
Figure 6 .
Digital Anatomist Image Collection.
The system allows individual authorized users to upload images from their desktop; to index images by concepts such as source, description and anatomical names from our evolving Foundational Model of Anatomy; and to arrange images in collections and sub collections. Regions of interest on images may be annotated using an image annotation tool we previously developed called AnnotateImage20 and then uploaded along with the images.
Uploaded image collections may be designated readable or writable by the public or by specific user groups. On logging in, a user can search all image collections available to him or her, copy the results of a query to a personal collection, and add new annotations and index terms to the copied images. Collections may be browsed in a drill-down fashion, moving to subcollections and individual images. Images within a collection may be viewed in different sizes, in slide show mode (for online slide shows), and in interactive atlas mode. In the latter mode the user may click on an annotated region in the image, which causes a new image to be displayed showing the outlined region.
The Image Collection Manager is currently being evaluated for its use in managing teaching images. We have also found that it is useful for sharing research images and slide presentations. A goal is to enhance the tool so that anyone with authorization can upload images and dynamically arrange them in slideshows or atlases similar to the images in our existing Digital Anatomist interactive atlases (http://www9.biostr.washington.edu/da.html).
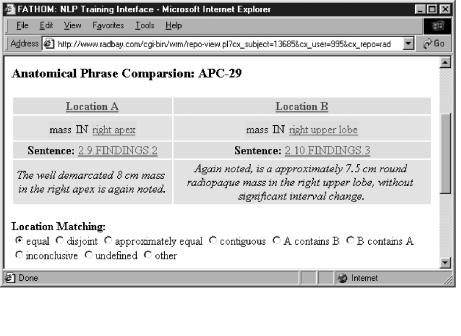
Fathom
Another project using WIRM is Fathom, part of a natural language processing (NLP) system being developed by the UCLA Telemedicine group. The system processes free-text clinical radiology reports and automatically generates structured records about the findings contained therein.21 Structured reports are clinically useful for decision support and outcomes research. The NLP system uses various statistical and machine learning methods to process the records and identify the properties, locations and diagnostic interpretations of each finding mentioned in the text. Fathom (Figure 7▶) is a WIRM-based application that supports the UCLA researchers as they test the effectiveness of various NLP algorithms. In order to evaluate the recall and precision of the NLP engine, the results must be compared to thousands of hand-coded candidates. Fathom reads the candidates from the NLP engine and presents them in web forms to the domain experts, who are able to submit their judgments from any location. In addition, Fathom provides a patient record manager that imports records from the hospital information system, allowing authorized users to retrieve records by patient demographics or by experiment corpus. Fathom allows searching over the raw records or the intermediate data structures used by the various stages of processing. For example, the syntactic parser splits the records into sentences, which are each assigned a sequential identifier for reference by the semantic interpreter. The various processing stages express their output as XML records, which are read by the patient record manager and made available for structured browsing. Perl’s built-in text parsing abilities greatly facilitated the process of importing these records. To view a demo of Fathom, visit <http://wirm.org/fathom>.
Figure 7 .
Fathom training interface.
Discussion
We have used WIRM to create a wide range of information systems for scientists, educators, and clinicians. Our experience building these systems demonstrates the potential impact of WIRM as an enabling technology for biomedical content management. In this section, we evaluate how WIRM meets each of the requirements identified in our analysis.
Rapid application development.
The high-level WIRMScript API makes it possible to create custom web interfaces using brief, high-level commands. Combined with the built-in default interfaces for creating, editing, and viewing object instances, new applications can be created quickly and evolved over time. Our implementation efforts were reduced by an estimated 75% over using the basic building blocks themselves. One key factor in this gain was the ease of testing changes to our system: developers simply reload their browser to see the effects of a code update.
Flexible data model.
WIRM’s object-relational data model was flexible enough to handle all the structures and relationships that we encountered across every project, including complex brain data, hierarchical patient records, and a wide range of multimedia objects. Many of the classes developed for one project were reusable for other projects. For example, the cases used in MyPACS were the basis for the Collections in the Digital Anatomist Image Collection Manager, which shared much of their behavior.
Visual modeling interfaces.
The Domain Modeler Wirmlet serves as a rudimentary interface for declaring new classes. New attributes are assigned names, and the type of each attribute is selected using a drop-down menu. However, the system does not facilitate the specification of ranges of values, which would be a powerful customization feature. Because of the limitations of the Domain Modeler, much of our class definition was done through the programmatic interface rather than the graphical interface. WIRM could benefit from more powerful template-based modeling interfaces.
Schema evolution.
WIRM’s Domain Modeler includes a schema evolver that makes it easy for applications to evolve over time. For example, to add an attribute to a class, the user simply enters the new attribute name and type. In developing MyPACS, we were constantly adding attributes to the structure of a teaching file case, as our users expressed their evolving understanding of how they wanted to organize their data. Without the schema evolver, each change to the underlying class structure would have required a complex series of table definition and copying procedures.
Automatic form generation.
For every class that is defined, the Content Manager wirmlet provides default forms for creating and editing instances of that class. A form element is provided for each attribute, customized for the type of attribute. While the default forms are sufficient for basic object creation and editing, the provided interfaces tend to be less than optimal in terms of esthetics and user-friendliness. For example, if an Image Collection has an attribute named owner of type user, the default form for creating a new collection would have a popup menu showing all possible Users. A more appropriate behavior would be to fill in the owner field the identity of the current logged-in user. Currently, these customizations can only be performed by overriding the default behavior with WIRMScript.
Database connectivity.
Through the DBI interface, WIRM can be used with all major databases. We have tested it with Oracle, MySQL, and MSQL.
Hierarchical navigation.
As the relationships between objects are visualized through hyperlinks, classes composed of sub-parts are naturally rendered as hierarchical web pages. For example, the Brain Mapper Experiment Management System allows a user to start with a group of patients, then select a single patient and drill-down into that patient’s study, exam, series, and finally a single image slice within a series.
Query support.
Through the Query Composer, users are able to pose ad-hoc SQL queries over any combination of attributes. However, the interface is very basic, requiring the user to enter in a Boolean string and does not provide a structured interface for specifying queries over attributes. Consequently, the query system for each application tends to require custom development before it is user-friendly. An improved Query Composer would make this unnecessary.
Document management.
WIRM allows any kind of file to be uploaded, and automatically maintains standard metadata, such as file type, date, owner, etc. Users may effectively specify new metadata fields by encapsulating the built-in File class with custom classes. One aspect lacking in WIRM’s document management facilities is support for versioning. We plan to implement this feature using CVS, an open source version control system, which would allow users to keep track of the changes to files and retrieve earlier versions.
Multimedia support.
WIRM supports management of over seventy types of images and other multimedia. High-level Wirmlet commands allow the developer to convert images across formats and to control their size and resolution. As WIRM applications are browser-based, any multimedia type can be rendered with the appropriate plug-in. For example, users of MyPACS may upload ultrasound files as AVI movies, which can be played using any standard browser-based media player.
Context-sensitive, role-based access.
WIRM excels in its ability to allow the system designer to regulate access control at a fine granularity and to create context-sensitive interfaces that adapt themselves to different classes of end user. This is especially significant for managing medical research data, which require multiple privacy contexts. For example, the UW Human Brain Project’s information system enforces patient privacy by withholding patient identifiers from users who have not signed confidentiality agreements. In addition, each piece of data can be identified as published or unpublished, and only published data are visible by guest users. WIRM makes it easy to support multiple interfaces at the granularity of a data object view, rather than requiring the designer to create a separate site for each user class. In this way, WIRM reduces the task of building an information system into manageable steps.
Robust security.
WIRM uses individual password-authenticated login sessions to enforce access control. Client authentication is maintained through browser-based session identifiers (“cookies”). To protect against unauthorized access to data as it is transmitted over the web, the web server may be configured to encrypt data using secure socket layer protocol.
Scalability.
We have tested WIRM on data sets of hundreds of thousands of records. With proper database indexing, the system can handle millions of records without any noticeable performance degradation. In terms of handling heavy user loads, WIRM has not yet been tested with large numbers of simultaneous accesses. However, all our applications have performed adequately under normal usage, with an average response time of 1–3 seconds per hit. If necessary, WIRM could be made more scalable by using mod_perl, a system for integrating CGI-based applications directly into the web server.
Connectivity to biomedical resources.
WIRM currently is able to display images that are uploaded in the DICOM format, but it does not provide the ability to talk directly with DICOM sources. WIRM users indicated that support for DICOM connectivity should be a high priority. With DICOM connectivity built into the toolkit, WIRM-based applications could be easily integrated with existing medical image systems. For example, MyPACS users must currently rely on external mechanisms for copying images from their DICOM sources to their hard drives, from which they can be uploaded into WIRM as a separate step. If WIRM supported DICOM connectivity, MyPACS could retrieve images directly from hospital PACS systems and/or imaging equipment, and the process of building teaching files would be greatly simplified. In addition, the patient- and image-related metadata stored in DICOM headers could be automatically read into WIRM’s repository. We have identified several open source DICOM toolkits that are compatible with our platform, such as the Central Test Node from the Mallinckrodt Institute (http://www.erl.wustl.edu/DICOM/ctn.html). We intend to integrate one of these toolkits into WIRM, allowing WIRM-based applications to act as DICOM storage class clients and servers.
For XML support, WIRM uses several XML processing modules from CPAN, which enables WIRM-based applications to easily exchange data with the wide range of biomedical applications that are already XML-aware. This effectively makes WIRM an XML server, thereby enabling round-trip interoperability between WIRM and other XML-aware biomedical applications. For example, teaching files can be imported and exported to MyPACS through the Teaching File Modeling Language (http://tfml.org). Additionally, the Digital Anatomist Image Collection Manager uses Image Modeling Language (IML) to import annotated image maps.20
Conclusions
WIRM has been released as open source software and is being adopted by a growing community of users in both academic and industry settings. The WIRM web site (http://WIRM.org) has received hundreds of thousands of hits from tens of thousands of unique visitors, and over 200 developers have signed up to be contributors and/or testers of new features.
WIRM provides an accessible and flexible solution to a critical problem facing the modern biomedical community: how to maximize the benefits afforded by new technology to meet the increasing demands of biomedical content management. Medical professionals should be aided rather than overwhelmed by advancing technology. It is essential that domain experts, who best understand the specific requirements of their own applications, be given the power to design their own individually-tailored solutions. WIRM is poised to fill the technology gap medical professionals face in their clinical, research, and teaching environments, empowering them to effectively manage the acquisition, integration, and dissemination of their knowledge and data.
This work was funded by NIH SBIR grant R44-MH61277-02, NIH Human Brain Project grant MH/DC023210, and National Library of Medicine grant LM06316.
References
- 1.Jakobovits RM, Soderland S, Taira R, Brinkley JF. WIRM: A framework for developing web-based multimedia applications for medical research. Proceedings, 2000 International Conference on Mathematics and Engineering Techniques in Medicine and Biological Sciences, June 2000.
- 2.Hinshaw KP, Poliakov AV, Martin RF, et al. Shape-based cortical surface segmentation for visualization brain mapping. Neuroimage. 2002; 16:295–316. <http://sig.biostr.washington.edu/publications/online/hinshawbrain01.pdf>. [DOI] [PubMed]
- 3.Brinkley JF, Myers LM, Prothero JS, et al. A structural information framework for brain mapping. In: Koslow SH, Huerta MF (eds): Neuroinformatics: An Overview of the Human Brain Project. Mahwah, NJ, Lawrence Erlbaum; 1997, pp 309–34.
- 4.Jakobovits RM, Soderland S, Taira RK, Brinkley JF. Requirements of a web-based experiment management system. In: Proceedings, AMIA Symposium 2000. Los Angeles; 2000, pp 374–8. [PMC free article] [PubMed]
- 5.Jakobovits RM, Modayur B, Brinkley JF. A web-based repository manager for brain mapping data. In Proceedings, AMIA Fall Symposium. Washington, D.C., Oct 28–30, American Medical Informatics Association, 1996, pp 309–13. [PMC free article] [PubMed]
- 6.Gillespie H. Sci. Comp. Automation 1992; 8(12): 48–54. [Google Scholar]
- 7.Ioannidis Y, Livny M, Gupta S, Ponnekanti N. ZOO: A desktop experiment management environment. VLDB, pp 274–85, 1996.
- 8.Brown K. Enterprise Java Programming with IBM Websphere. Boston, Addison Wesley, 2001.
- 9.Siwiki B. National scientists work to create virtual patient record. Health Data Manage. Oct: 96–9, 1997. [PubMed]
- 10.Orfali R, Harkey D. Client/Server Programming with Java and CORBA, 2nd ed. New York, John Wiley & Sons, 1998.
- 11.Vinoski S. CORBA: integrating diverse applications within distributed heterogeneous environments. IEEE Commun Mag. 35(2): 46–55, 1997. [Google Scholar]
- 12.Wall L, Schwartz RL. Programming Perl. Sebastopol, CA, O’Reilly & Associates, Inc., 1991.
- 13.Stein L. Official Guide to Programming with CGI.pm. New York, Wiley & Sons, 1998.
- 14.Weinberger E, Jakobovits RM, Halsted M. MyPACS.net: a web-based teaching file authoring tool. Am J Roentgenol. 2002 [in press]. [DOI] [PubMed]
- 15.Rosse C, Mejino J, Modayur B, et al. Motivation and Organizational Principles for Anatomical Knowledge Representation: the Digital Anatomist Symbolic Knowledge Base. J Am Med Inform Assoc. 1998, 5(1):17–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stalder D, Brinkley JF. The digital anatomist foundational model server. Proceedings of the Perl Conference 3.0, 1999.
- 17.Taira RK, Soderland SG. A statistical natural language processor for medical reports. AMIA Fall Symposium, 1999. [PMC free article] [PubMed]
- 18.Taira R, Soderland S, Jakobovits RM. A Statistical Natural Language Processor for Medical Reports. In Proceedings, 2000 International Conference on Mathematics and Engineering Techniques in Medicine and Biological Sciences, Las Vegas, June 2000.
- 19.Bunker G, Zick G. Collaboration as a key to digital library development. D-LIB Mag. 1999; 5(3).
- 20.Lober WB, Trigg LJ, Bliss D, Brinkley JF. IML: An image markup language. In Proceedings of the AMIA Annual Symposium Washington, DC, pp 403–7, 2001. [PMC free article] [PubMed]