Background: A conformational change of the γ subunit of ATP synthase may be critical for enzyme regulation.
Results: A conformational change of γ controls both ADP inhibition and ϵ inhibition.
Conclusion: The γ subunit indirectly regulates the activity by way of ADP inhibition and ϵ inhibition.
Significance: Regulation system of ATP synthase based on the unique molecular structure of the γ subunit was revealed.
Keywords: ATP Synthase, ATPases, Disulfide, Molecular Motors, Redox Regulation, γ Subunit, Central Axis, Conformational Change, Regulation, Slippage
Abstract
The central shaft of the catalytic core of ATP synthase, the γ subunit consists of a coiled-coil structure of N- and C-terminal α-helices, and a globular domain. The γ subunit of cyanobacterial and chloroplast ATP synthase has a unique 30–40-amino acid insertion within the globular domain. We recently prepared the insertion-removed α3β3γ complex of cyanobacterial ATP synthase (Sunamura, E., Konno, H., Imashimizu-Kobayashi, M., and Hisabori, T. (2010) Plant Cell Physiol. 51, 855–865). Although the insertion is thought to be located in the periphery of the complex and far from catalytic sites, the mutant complex shows a remarkable increase in ATP hydrolysis activity due to a reduced tendency to lapse into ADP inhibition. We postulated that removal of the insertion affects the activity via a conformational change of two central α-helices in γ. To examine this hypothesis, we prepared a mutant complex that can lock the relative position of two central α-helices to each other by way of a disulfide bond formation. The mutant obtained showed a significant change in ATP hydrolysis activity caused by this restriction. The highly active locked complex was insensitive to N-dimethyldodecylamine-N-oxide, suggesting that the complex is resistant to ADP inhibition. In addition, the lock affected ϵ inhibition. In contrast, the change in activity caused by removal of the γ insertion was independent from the conformational restriction of the central axis component. These results imply that the global conformational change of the γ subunit indirectly regulates complex activity by changing both ADP inhibition and ϵ inhibition.
Introduction
The F0F1-ATP synthase (F0F1)3 catalyzes synthesis of ATP from ADP and inorganic phosphate using the electrochemical proton gradient formed across chloroplast and cyanobacterial thylakoid membranes, mitochondrial inner membranes, and bacterial plasma membranes by photosynthetic or respiratory electron transfer reaction (1, 2). F0F1 consists of a water-soluble F1 part, which contains catalytic sites for ATP synthesis and hydrolysis, and a membrane-embedded part F0, which is involved in proton translocation. F1 consists of five different subunits with a stoichiometry of α3β3γ1δ1ϵ1 (3) and F0 consists of three different subunits with stoichiometry of a1b2c10–15 (4–6). F1 solely catalyzes the ATP hydrolysis reaction and the minimum catalytic core of F1 is α3β3γ complex (7). In 1994, the first crystal structure of mitochondrial α3β3γ complex was determined (8) revealing an alternating hexagonal arrangement of three α and three β subunits around an α-helical domain containing the N- and C-terminal regions of the γ subunit. Because three β subunits in the structure showed different conformations due to different nucleotide binding situations, the reported structure pointed to the idea of rotation of the γ subunit against the α3β3 ring during catalysis, first proposed by Boyer and co-workers (9). In 1997, rotation of the γ subunit coupled with ATP hydrolysis was directly visualized under an optical microscope by attaching a fluorescent-labeled actin filament to the γ subunit of the α3β3γ complex fixed on a glass surface (10).
When the electrochemical proton gradient across the membrane is insufficient for ATP synthesis, F0F1 can potentially hydrolyze ATP. To prevent this wasteful reverse reaction, the enzyme possesses multiple regulatory mechanisms. The most common regulatory mechanism is ADP inhibition irrespective of the origin of F1-ATPase. Tightly bound MgADP at the catalytic site strongly prevents a ATP hydrolysis reaction but not ATP synthesis (11–15). Recovery from ADP inhibition is accelerated by ATP binding to the noncatalytic sites on the α subunits (16). Single molecule analysis of the catalytic turnover of this enzyme indicates that ADP inhibition can be observed as long pauses during rotation (17). In addition, the ϵ subunit works as an intrinsic inhibitor of ATP hydrolysis in bacterial and chloroplast enzymes. This inhibition is referred to as ϵ inhibition. The C-terminal helix-turn-helix domain of ϵ is important for the inhibition, which adopts a large conformational change from a “retracted” form to an “extended” form (18–21). In the case of chloroplast type F1, the ϵ subunit seems to inhibit ATP hydrolysis activity more strongly than the bacterial ϵ (22–24). Single molecule observations of ϵ inhibition in cyanobacterial and bacterial F1 showed that the ϵ subunit of cyanobacterial F1 completely stops the rotation of the γ subunit (25), whereas that of bacterial F1 decreases the average rotation speed and increases pause duration (26, 27). Again, the ϵ inhibition is thought to be the regulatory mechanism that prevents futile ATP hydrolysis, because this subunit may not inhibit the ATP synthesis reaction (20).
Chloroplast F0F1 possesses another regulatory mechanism: the redox regulation system mediated by thioredoxin (28). This is thanks to the chloroplast F1-γ containing an additional ∼35 amino acid insertion in the middle of the sequence (supplemental Fig. S1) compared with other bacterial and mitochondrial F1-γs (29). Two critical Cys residues are located in this insertion sequence and regulate ATP hydrolysis activity via disulfide bond formation (30).
Compared with the bacterial and mitochondrial F1-γ, the γ subunit of cyanobacterial F0F1 also bears the inserted sequence like spinach CF1-γ, although the sequence lacks nine amino acids including two regulatory Cys residues for redox regulation (31). In our previous studies, the mutant α3β3γ complex was prepared in which the γ insertion sequence of cyanobacterium Thermosynechococcus elongatus BP-1 F1 was deleted (25, 32). The mutant complex obtained showed a large increase in ATP hydrolysis activity as a consequence of the low tendency to lapse into ADP inhibition, and lower sensitivity to ϵ inhibition. Furthermore, the mutant strain of cyanobacterium Synechocystis sp. PCC 6803, whose F1-γ insertion was deleted, showed lower intracellular ATP levels due to insufficient prevention of the ATP hydrolysis activity in the dark (32, 33). These results strongly suggest that the insertion plays an important role in the regulation of ATP hydrolysis activity of cyanobacterial F0F1 in vivo.
However, the location of the insertion seems to be at the bottom of the γ molecule and over 60 Å away from catalytic sites on the α3β3 ring (34, 35). How does the conformational change by removal of the inserted sequence affect the catalytic activity despite the catalytic sites being distant from the inserted sequence in the enzyme complex? Since little contact region is reported in the crystal structure of F1 between the γ subunit and the α3β3 ring (Fig. 1A), we postulated that relative slippage of N- and C-terminal α-helices may connect between the conformational change of the bottom part of γ and a conformational change in the upper part of γ, as suggested by our previous study on the redox regulation of this enzyme complex (36).
FIGURE 1.
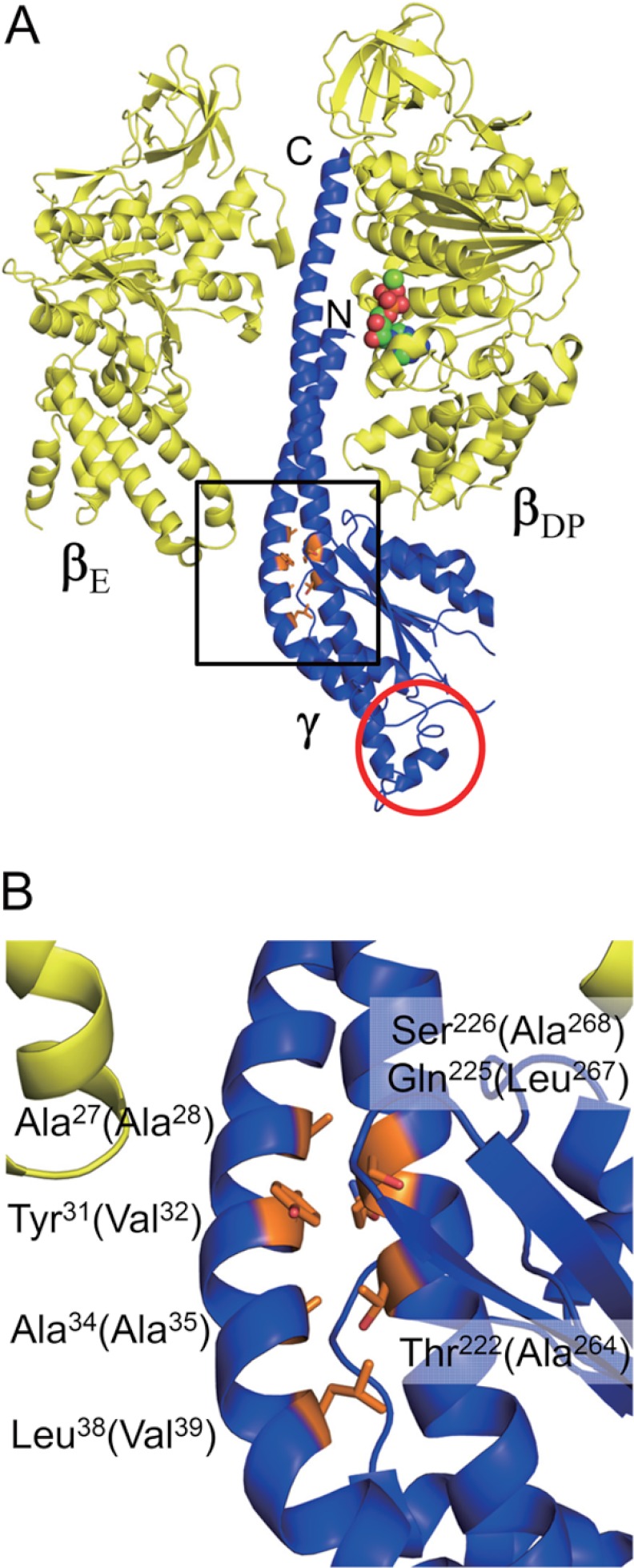
Positions of Cys residues introduced for locking of the central α-helices of the γ subunit. A, structures of βE, βDP (yellow), and γ (blue) subunits in bovine F1 structure (Protein Data Bank code 1E79) are shown. βE and βDP are based on the first crystal structure of F1 (8). N and C termini of α-helices in γ are indicated. Residues of γ, Ala27, Tyr31, Ala34, Leu38, Thr222, Gln225, and Ser226 are shown as a stick model and colored in orange. In βDP, bound MgADP is shown as a sphere model. The apparent position of the inserted sequence of cyanobacterial and chloroplast F1-γ is circled in red. B, magnified structure of the Cys introduced region is marked by a square in A. Residue numbers are those for bovine mitochondrial F1 and the numbers in parentheses correspond to the γ subunit of T. elongatus BP-1. The figures were generated with PyMOL.
To examine our hypothesis, we introduced two Cys residues at an appropriate position in each of the N- and C-terminal α-helices of cyanobacterial F1-γ, allowing us to lock or unlock the relative movement of these two central α-helices. Here we defined the oxidized complex as the locked one and the reduced complex as the unlocked one. The mutant complexes obtained showed significant changes in their ATP hydrolysis activities and sensitivities to ϵ inhibition. In addition, we prepared several lock mutants lacking a γ insertion, and examined the change in ATP hydrolysis activities by locking or unlocking the two central α-helices. Very interestingly, these mutants showed higher ATP hydrolysis activities irrespective of the lock status of the central α-helices. Based on these results, the relationship between the removal of the insertion and the accompanying conformational change of γ is discussed.
EXPERIMENTAL PROCEDURES
Materials
AldrithiolTM-2, ATP, phosphoenolpyruvate, and pyruvate kinase/lactate dehydrogenase were purchased from Sigma. Diamide was obtained from MP Biomedicals (Santa Ana, CA). NADH was purchased from Roche Diagnostics. Other chemicals were of the highest grade commercially available.
Strains
Escherichia coli strains used were DH5α for cloning and BL21(DE3) uncΔ702 (Tcr, ATPase mutant, BL21(DE3) uncΔ702, asnA::Tn10) (37, 38) for expression of α3β3γ complex of T. elongatus BP-1. The latter strain was a kind gift from Dr. C. S. Harwood (University of Iowa).
Construction of Expression Plasmids for Lock Mutants
The expression plasmid for the α3β3γ complex of T. elongatus BP-1 and the expression plasmid of Cys-less α3β3γ complex (all native Cys residues were mutated to Ser) for the single molecule experiment were constructed in the previous study (25). Using the plasmid for the Cys-less complex as template, the expression plasmids for lock mutants were prepared and two Cys residues were introduced into γ. Site-directed mutagenesis was performed by the overlap extension method (39). The primers used for mutagenesis are shown in supplemental Table S1. To prepare the additional lock mutants lacking the γ (Leu198–Val222) insertion, both expression plasmids for the Cys-less complex and the mutant complex lacking the insertion sequence (25) were digested with SacI and NheI. Ligation then yielded the expression plasmid for the Cys-less mutant complex lacking the insertion (hereafter referred as Cys-less/ΔIns). Using the plasmid as a template, Cys residues that allow locking of the central α-helices were introduced as described, and lock mutant plasmids lacking the insertion were obtained.
Expression and Purification of α3β3γ Complex and the ϵ Subunit
Expression and purification of the α3β3γ complex were performed as described (25) with some modifications. After nickel-nitrilotriacetic acid chromatography, proteins were stored at 4 °C in solution containing 20 mm potassium phosphate (pH 8.0), 100 mm KCl, 0.1 mm ADP, 0.1 mm MgCl2, 1 mm DTT, and 55% (w/v) ammonium sulfate before further purification. Then, the ammonium sulfate precipitate was re-dissolved in “HPLC buffer,” which contains 50 mm HEPES/KOH (pH 8.0), 100 mm KCl, 0.1 mm ADP, and 0.1 mm MgCl2, and then purified by gel filtration chromatography on a SuperdexTM 200 column (GE Healthcare) equilibrated with HPLC buffer in advance. The purified complex was stored at −80 °C in 10% glycerol. The ϵ subunit was expressed and purified as described (25). Protein concentrations were measured by the Bradford method with bovine serum albumin as a standard.
Determination of the Optimal Oxidation Conditions
Purified complex (50 μl) was oxidized by varying oxidizing reagents (Aldrithiol-2 and diamide), the concentration of the reagent (50, 100, 500, and 1000 μm), reaction time (30 and 60 min), and protein concentration (0.5 and 1.0 mg/ml). The reaction was stopped by adding 5% (w/v, final concentration) trichloroacetic acid. After centrifugation, the supernatant was removed and the remained oxidant was washed away with 500 μl of acetone. Then, the mixture was centrifuged, and air dried after removing the supernatant. The pellet was dissolved in 50 mm Tris/HCl (pH 7.5) and 1% (w/v) SDS and then electrophoresed on 12% polyacrylamide gel. The optimal oxidation condition was determined by comparing the band intensities of the oxidized γ subunit on the gel.
Purification of Lock Mutant after Oxidation
The ammonium sulfate precipitant as mentioned was re-dissolved in HPLC buffer with a protein concentration of 0.5 mg/ml. The V32C/A268C complex was oxidized by 100 μm Aldrithiol-2 for 60 min at 25 °C, A28C/L267C, V32C/A264C, and A35C/A264C complexes were oxidized by 500 μm Aldrithiol-2 for 60 min at 25 °C, and the V39C/A264C complex was oxidized by 500 μm diamide for 60 min at 25 °C. Oxidized complexes were further purified by gel filtration and stored as described.
Measurement of ATP Hydrolysis Activity
ATP hydrolysis activity was measured by using an ATP regenerating system. The assay mixture contained 50 mm HEPES/KOH (pH 8.0), 100 mm KCl, 2 mm MgCl2, 2 mm ATP, 6.8 to 11 milliunits/ml of pyruvate kinase, 4.5 to 7.5 milliunits/ml of lactate dehydrogenase, 2 mm phosphoenolpyruvate, and 0.2 mm NADH. The assay was carried out at 25 °C. After addition of the ATPase complex, the activity was determined from the steady state slope by monitoring the decrease in NADH absorption at 340 nm with a Jasco spectrophotometer model V650 (Jasco, Tokyo, Japan). To investigate the effect of LDAO on the ATP hydrolysis activity, LDAO (final concentration of 0.1% w/v) was added to the assay mixture before addition of the enzyme complex. In this case, the activity was determined from maximum slope.
Inhibition of ATP Hydrolysis Activity of α3β3γ Complex by the ϵ Subunit
The experimental conditions were the same as described above. The assay was initiated by adding the α3β3γ complex followed by addition of various concentrations of the ϵ subunit. The extent of inhibition of ATP hydrolysis activity was determined by the steady state slope in the presence or absence of the ϵ subunit (25). The titration curve was fitted with the hyperbolic equation, y = A × [ϵ]free/(KD + [ϵ]free), where y represents the percentage of inhibition, A is the maximum inhibition (%), and KD is an apparent equilibrium dissociation constant for the ϵ subunit. To calculate the concentration of the free ϵ subunit, we assumed the complex that bound the ϵ subunit is completely inhibited, thus [ϵ]free = [ϵ]add − [complex] × y/100.
RESULTS
Preparation of Lock Mutant γ to Control the Slippage of α-Helices
In the previous study, we prepared a Cys-less α3β3γ complex of thermophilic cyanobacterial F0F1 for single molecule experiments in which all native Cys at positions 144 and 194 on α, 53 on β, and 90 on γ were substituted with Ser (25). In this study, this complex was referred to as the Cys-less complex and used as a template for preparing lock mutants for two central α-helices in γ. Because no crystal structure information on cyanobacterial or chloroplast F1 α3β3γ complex is available to date, we applied the bovine mitochondrial F1 structure (Protein Data Bank code 1E79) (18) (Fig. 1) instead to design lock mutants. Finally positions Ala28, Val32, Ala35, and Val39 on the N-terminal α-helix, and Ala264, Leu267, and Ala268 on the C-terminal α-helix (residue numbers are those for F1 of T. elongatus BP-1) were selected as candidate residues for Cys substitution. We then prepared nine mutants by a combination of a set of mutations (for example, A28C/L267C, V32C/L267C, and A35C/L267C), which potentially induces N-terminal α-helix slip-up or down by one turn against the C-terminal α-helix after disulfide bond formation. Primers used for the mutation are listed in supplemental Table S1. Site-directed mutagenesis was performed as described under “Experimental Procedures,” and all mutants were successfully expressed in E. coli.
Dynamics of the Mutant γ Subunit in the Complex
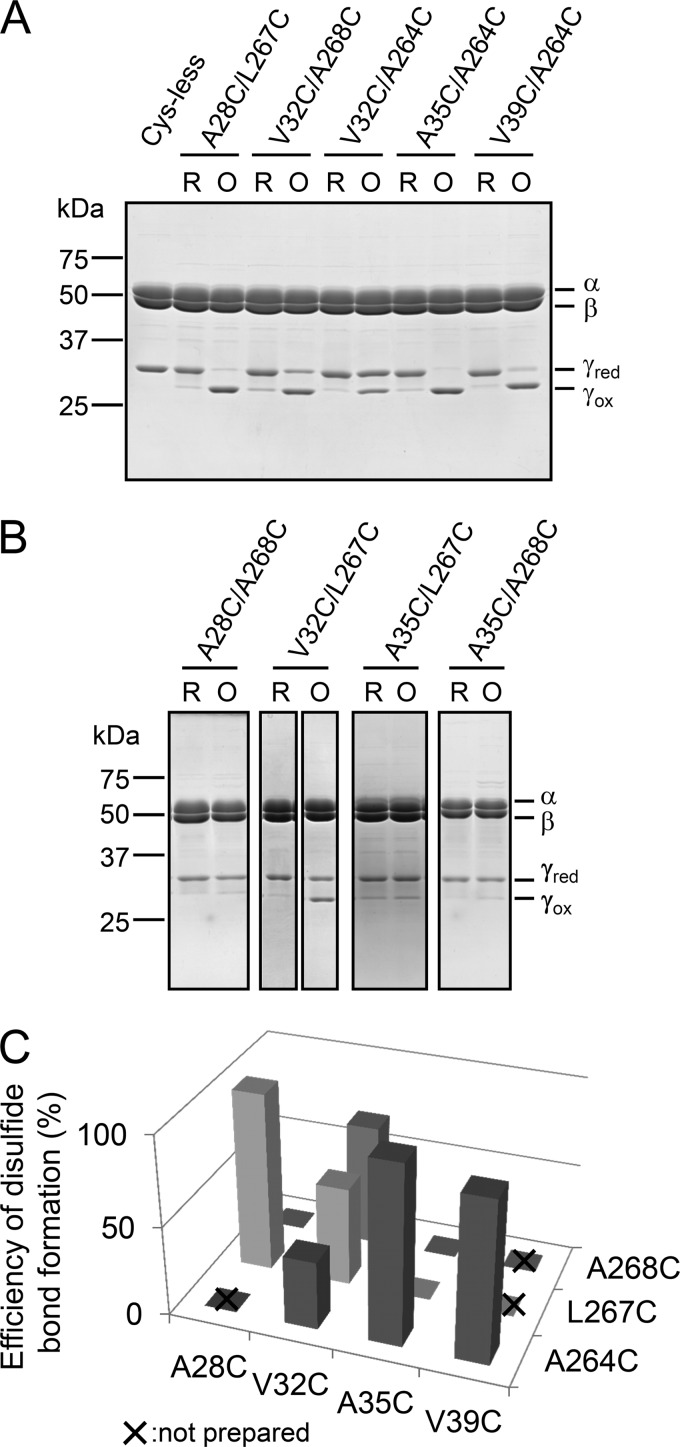
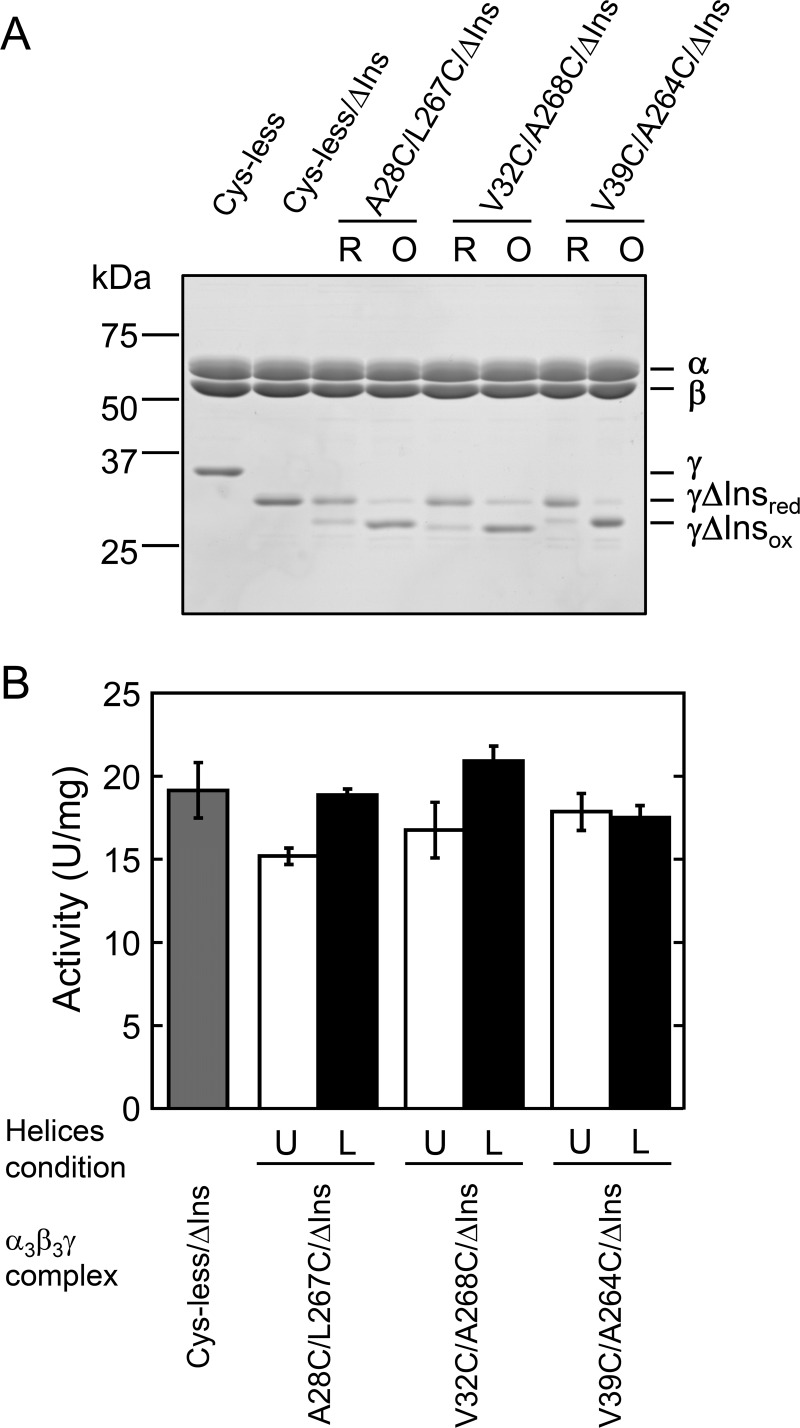
Mutant complexes were basically prepared as the unlocked form after gel filtration chromatography. We then determined disulfide bond formation conditions to lock two central α-helices by changing the concentrations of oxidizing reagents, protein concentrations, and reaction periods (For more detail, see “Experimental Procedures”). Aldrithiol-2 and diamide were used as oxidizing reagents. To examine the efficiency of disulfide bond formation, nonreducing SDS-PAGE analysis, in which 2-mercaptoethanol was omitted from the sample preparation buffer, was performed. When the γ subunit was locked by a disulfide bond, a band shift of γ on the gel was clearly observed as a consequence of conformational constraints. After optimizing oxidation conditions, A28C/L267C, A35C/A264C, and V39C/A264C mutants were mostly oxidized (Fig. 2A), whereas A35C/L267C, A28C/A268C, and A35C/A268C were not (Fig. 2B). The other mutants V32C/L267C, V32C/A268C, and V32C/A264C were partially oxidized by 55, 68, and 38%, respectively, as estimated based on their band intensities (Fig. 2, A and B). Incomplete disulfide bond formation between two Cys residues may be attributed to an excessive distance between thiol groups of the introduced Cys. For example, the distance between side residues of Cys corresponding to V32C and A264C in bovine mitochondrial F1 is 5.0 Å, whereas those of A35C/A264C and V39C/A264C, Cys pairs, are 2.1 Å apart, as measured with PyMOL. In contrast, V32C/L267C and A35C/L267C should have the potential to form a disulfide bond (the distances are 2.3 and 2.7 Å, respectively), although we could not obtain complete disulfide bond formation after oxidation (Fig. 2B). These results imply that the dynamic structure of cyanobacterial F1 in solution might be slightly different from the reported crystal structure. Consequently, the molecular composition of some of the complexes used in this study was heterogeneous due to their incomplete oxidation, somewhat complicating the resulting interpretation. Nevertheless, our key objective was to obtain a series of Cys mutants that could induce slippage of central α-helices after oxidation, and we therefore mainly focused on the series of mutants, V32C/A264C, A35C/A264C, and V39C/A264C, and A28C/L267C and V32C/A268C to assess the effect of slippage of the α-helices.
FIGURE 2.
Nonreducing SDS-PAGE analysis of unlocked and locked mutants. A, purified α3β3γ complexes were electrophoresed on 12% polyacrylamide gel. First lane, Cys-less; second and third lanes, unlocked and locked A28C/L267C; fourth and fifth lanes, unlocked and locked V32C/A268C; sixth and seventh lanes, unlocked and locked V32C/A264C; eighth and ninth, unlocked and locked A35C/A264C; 10th and 11th lanes, unlocked and locked V39C/A264C. B, the oxidation conditions of purified A28C/A268C, V32C/L267C, A35C/L267C, and A35C/A268C complexes were surveyed as described under “Experimental Procedures,” and the most representative results are shown. First and second lanes, unlocked and locked A28C/A268C; third and fourth lanes, unlocked and locked V32C/L267C; fifth and sixth lanes, unlocked and locked A35C/L267C; seventh and eighth lanes, unlocked and locked A35C/A268C. C, efficiency of disulfide bond formation was determined from the band intensities of unlocked and locked γ subunits shown in A and B. The combination which was not prepared in this study is shown by a x.
Change in the ATP Hydrolysis Activity by Unlocking and Locking of Two Central α-Helices
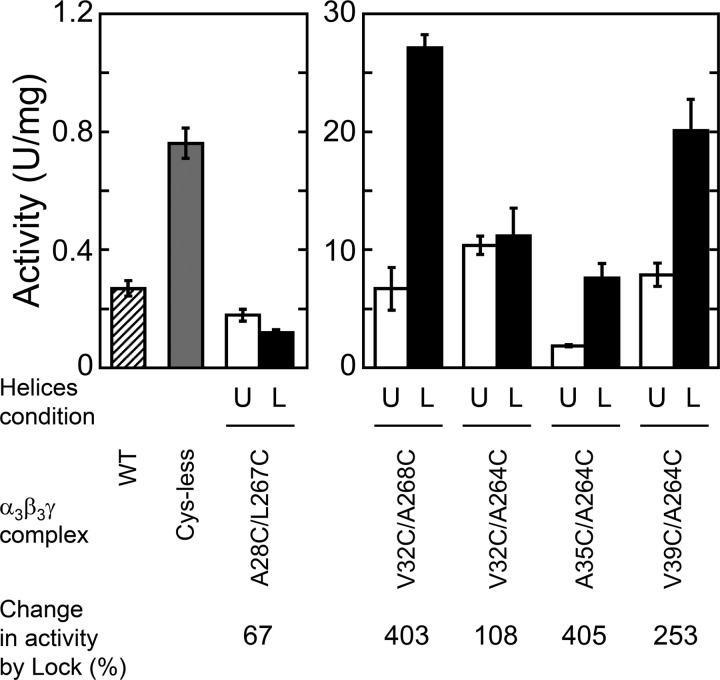
We then measured the change in ATP hydrolysis activities of unlocked and locked form mutants (Fig. 3). Compared with wild type (WT) and Cys-less complexes, unlocked mutants except A28C/L267C showed remarkably higher ATP hydrolysis activities (Fig. 3, open bars), although mutations applied were only Cys substitution of γ, which may not directly affect the catalytic sites. Disulfide bond formation in V32C/A268C and V39C/A264C mutants greatly increased their ATP hydrolysis activities, by about 4- and 2.5-fold, respectively. Although the locked A35C/A264C mutant also showed 4-fold higher ATP hydrolysis activity than the unlocked one, the specific activity obtained for this complex was not much higher than the other two mutants. Irrespective of disulfide bond formation, V32C/A264C showed a stable activity (at around 10 units/mg). The activity of A28C/L267C was much lower than that of WT and the Cys-less complex, and slightly decreased by disulfide bond formation.
FIGURE 3.
ATP hydrolysis activities of unlocked and locked mutants. ATP hydrolysis activities of wild type (WT), Cys-less, and lock mutants were determined at 25 °C in the presence of 2 mm MgATP. The hatched and light gray bars represent the activities of WT and Cys-less, respectively. Open and closed bars represent the activities of unlocked and locked mutants, respectively. The results of three experiments were averaged. Error bar represents S.D. The conditions of α-helices in the mutant complexes were shown as U (unlocked) and L (locked), respectively. In the bottom, the change in the activity by lock is indicated based on the activity of each unlocked mutant as 100%.
Stimulation of ATP Hydrolysis Activity by LDAO
To determine the cause of the strong activation by locking the two central α-helices in the case of V32C/A268C and V39C/A264C, we examined the effect of LDAO on the activity of the mutant complexes (Table 1). As mentioned, F1 has the intrinsic regulation mechanism, ADP inhibition, and the detergent LDAO is known to release this inhibition (40). In the presence of LDAO, ATP hydrolysis activities of all complexes increased significantly, indicating their activities were basically suppressed by ADP inhibition, although only two locked mutants V32C/A268C and V39C/A264C were not significantly affected (Table 1, bold).
TABLE 1.
Effects of LDAO on ATP hydrolysis activities of lock mutants
The activity in the presence of LDAO was measured as described under “Experimental Procedures.” The activity measured in the absence of LDAO was obtained from Fig. 3. The results of three independent experiments were averaged. The results obtained from LDAO less sensitive complexes are in bold.
| Complex | ATP hydrolysis activity |
|||
|---|---|---|---|---|
| Unlocked |
Locked |
|||
| −LDAO | +LDAO | −LDAO | +LDAO | |
| units/mg | units/mg | |||
| WT | 0.27 ± 0.03 | 9.4 ± 0.5 | ||
| Cys-less | 0.76 ± 0.05 | 21.4 ± 0.9 | ||
| A28C/L267C | 0.18 ± 0.02 | 15.4 ± 2.1 | 0.12 ± 0.01 | 11.0 ± 0.3 |
| V32C/A268C | 6.7 ± 1.8 | 30.4 ± 3.7 | 27.1 ± 1.1 | 31.5 ± 2.3 |
| V32C/A264C | 10.4 ± 0.8 | 31.0 ± 1.2 | 11.2 ± 2.4 | 24.7 ± 2.7 |
| A35C/A264C | 1.9 ± 0.1 | 26.7 ± 4.7 | 7.6 ± 1.2 | 35.7 ± 3.3 |
| V39C/A264C | 7.9 ± 1.0 | 35.7 ± 1.5 | 20.1 ± 2.7 | 34.1 ± 5.0 |
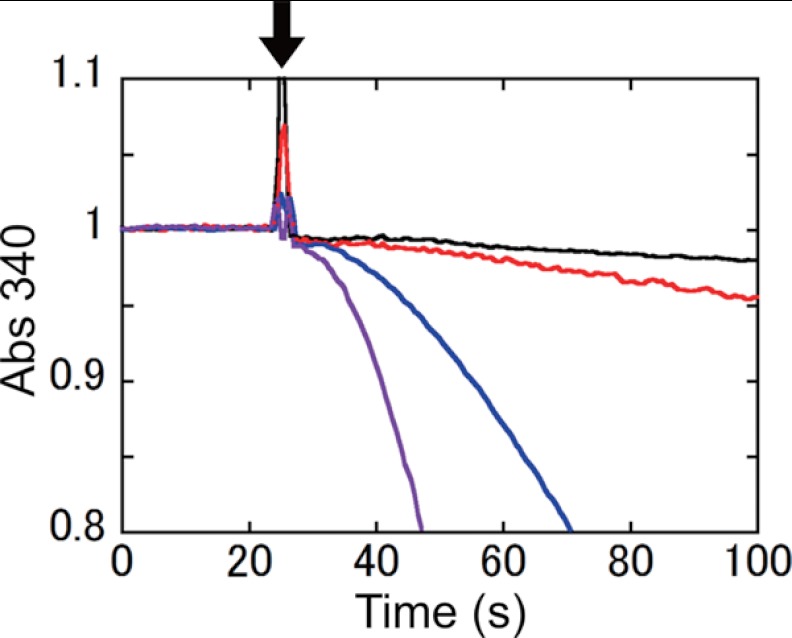
We then compared the initial rates of ATP hydrolysis activities of WT, Cys-less, and unlocked and locked V32C/A268C (Fig. 4). Because the complexes were preincubated in 50 μm MgADP for 10 min, all complexes were expected to lapse into ADP inhibition when the complex was mixed into the reaction mixture solution. Excess ADP carried with the enzyme should be immediately converted to ATP by pyruvate kinase in the solution. Thus, the observed initial rate reflects the release rate of bound ADP from the inhibited complex. The initial rate of locked V32C/A268C recovered much faster than that of WT and Cys-less complexes, indicating that the bound ADP on the locked V32C/A268C complex was released much faster than those on the other complexes.
FIGURE 4.
Time course of ATP hydrolysis by WT, Cys-less, and V32C/A268C lock mutant complexes. The complexes were incubated at 25 °C for 10 min in the presence of 50 μm MgADP in advance. 10 μg of the complexes were added into the assay mixture at 25 s after starting the measurement (down arrow): black line, WT; red line, Cys-less; blue line, unlocked V32C/A268C; purple line, locked V32C/A268C.
Inhibition of ATP Hydrolysis Activity of Unlocked and Locked Mutants by the ϵ Subunit
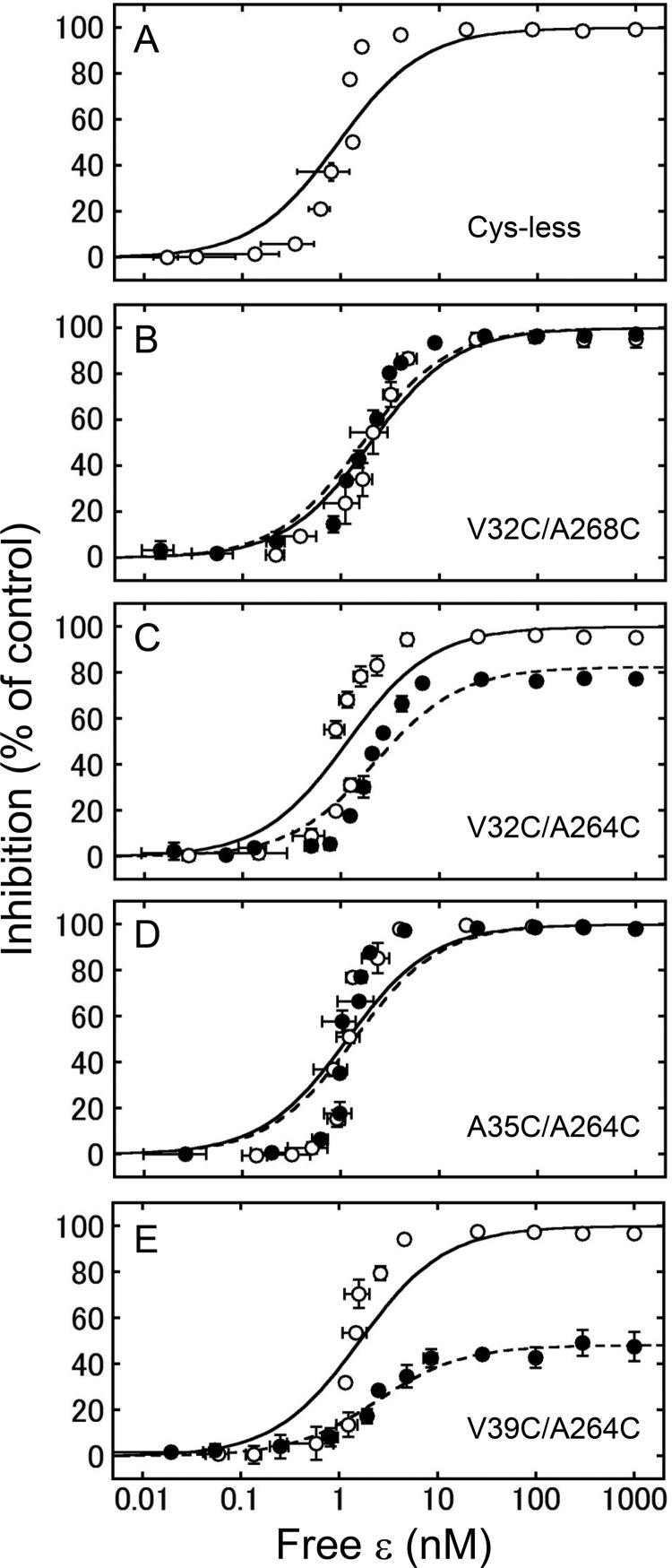
The ϵ subunit of the cyanobacterial F1 inhibits ATP hydrolysis activity by almost 100% (25). We therefore examined whether the conformational change induced by unlocking and locking the central α-helices affected the ϵ inhibition. We thus measured the extent of ϵ inhibition based on the change in ATP hydrolysis activity by addition of various concentrations of the ϵ subunit (Fig. 5). As shown in Fig. 5A, the Cys-less complex was completely inhibited by addition of the ϵ subunit as well as the WT complex (25). All the unlocked complexes and locked V32C/A268C and A35C/A264C complexes were completely inhibited by the ϵ subunit. However, locked V32C/A264C and V39C/A264C complexes were inhibited at around 80 and 50%, respectively. Based on the electrophoresis results, the efficiency of oxidation of the V32C/A264C complex was estimated to be 38% (Fig. 2), indicating that complete oxidation of V32C/A264C may result in about 54% inhibition by the ϵ subunit. We determined the apparent KD value of the ϵ subunit from the observed extent of ϵ inhibition of the activity. Although the theoretical curve did not fit the experimental data very well, the obtained KD values of the Cys-less complex (0.93 ± 0.29 nm, Table 2) were not significantly different compared with the previous study (25, 41). Furthermore, the KD values of the mutants were almost equivalent to that of the Cys-less complex irrespective of their disulfide bond formation (Table 2).
FIGURE 5.
Inhibition of ATP hydrolysis activities of unlocked and locked mutants by the ϵ subunit. The extent of inhibition of the ATP hydrolysis activities of Cys-less (A), V32C/A268C (B), V32C/A264C (C), A35C/A264C (D), and V39C/A264C (E) mutants in unlocked (open circle) and locked (closed circle) forms were measured in the presence of various concentrations of the ϵ subunit. The results of three independent experiments were averaged. The vertical and horizontal error bars indicate the size of S.D. The fitting curves for the data obtained from the Cys-less mutant (solid line in A), and unlocked (solid line) and locked (dotted line) mutants shown in B-E, were calculated from the averaged KD values shown in Table 2.
TABLE 2.
Apparent dissociation constants for the ϵ subunit with lock mutants
Apparent dissociation constants (KD) for the ϵ subunit with unlocked and locked mutants were determined by averaging the results of three independent titration data with the hyperbolic equation as described under “Experimental Procedures.”
| Complex |
KD |
|
|---|---|---|
| Unlocked | Locked | |
| nm | ||
| Cys-less | 0.93 ± 0.29 | |
| V32C/A268C | 2.0 ± 0.6 | 1.6 ± 0.1 |
| V32C/A264C | 1.2 ± 0.2 | 2.2 ± 0.2 |
| A35C/A264C | 1.2 ± 0.2 | 1.4 ± 0.1 |
| V39C/A264C | 1.6 ± 0.5 | 2.4 ± 0.2 |
In contrast, lock of the V39C/A264C complex dramatically changed the extent of ϵ inhibition (Fig. 5E), which might be due to change of the affinity of ϵ to the locked α3β3γ complex. To prove this, the amount of bound ϵ subunit to the complex was determined using gel filtration chromatography (Fig. 6A). The unlocked or locked V39C/A264C mutant complex was first incubated with the ϵ subunit. Then the fraction of the α3β3γϵ complex after gel filtration was collected and electrophoresed on a 15% polyacrylamide gel, and no significant difference in band intensities of ϵ in unlocked and locked complexes was observed (Fig. 6B), implying the complex certainly can bind the ϵ subunit irrespective of the disulfide bond formation on γ.
FIGURE 6.
Binding of the ϵ subunit to V39C/A264C lock mutant. A, purified V39C/A264C complex (1 μm) was mixed with the ϵ subunit (2 μm) for 10 min (245.7 μl). Judging by the KD value, 2 μm ϵ is enough to saturate the binding of ϵ. The excess ϵ subunit (20 μm) was also used to determine the maximum binding. After centrifugation, the mixture of unlocked V39C/A264C complex with 2 μm ϵ (red) and 20 μm ϵ (blue), and the locked V39C/A264C complex with 2 μm ϵ (black) was separated on a SuperdexTM 200 column. B, the peaks corresponding to the α3β3γϵ complex were collected, concentrated by trichloroacetic acid (final 10%), and electrophoresed on 15% polyacrylamide gel. Lane 1, unlocked V39C/A264C mixed with 2 μm ϵ; lane 2, unlocked V39C/A264C mixed with 20 μm ϵ; lane 3, locked V39C/A264C mixed with 2 μm ϵ.
Change in the ATP Hydrolysis Activity by Unlocking and Locking of Insertion Removal Mutants
In the previous study, we found that the insertion region of γ plays a critical role in conferring the ADP inhibition property to the enzyme complex and this is important in prevention of futile ATP hydrolysis activity in vivo (32). To examine the relevance between the removal of the insertion and the relative slippage of two α-helices found in this study, we prepared lock mutants lacking the insertion region in the γ subunit. For this purpose, we deleted the insertion region from three lock mutants, A28C/L267C, V32C/A268C, and V39C/A264C. These mutants showed a significant change in their activities by lock status as shown in Fig. 3. Hereafter, the mutants were referred as A28C/L267C/ΔIns, V32C/A268C/ΔIns, and V39C/A264C/ΔIns, respectively. All mutants were successfully expressed in E. coli, and purified. Using the same oxidation conditions applied for the mutants containing the insertion, all the mutants were oxidized to the same degree (Fig. 7A). We then measured the change in ATP hydrolysis activity of the mutants by locking and unlocking them (Fig. 7B). Although the Cys-less complex and A28C/L267C complex showed very low ATPase activities (Fig. 3), deletion of the insertion significantly accelerated their activities, and Cys-less/ΔIns and unlocked insertion removal mutants showed almost the same activities (15 to 20 units/mg) (Fig. 7B). In contrast, the locked A28C/L267C/ΔIns and V32C/A268C/ΔIns showed a slight increase in their activities (about 1.2-fold) when locked, although Student's t test indicated that the activity of V32C/A268C/ΔIns mutant was not significantly different (p < 0.05). Activity of the V39C/A264C/ΔIns mutant was not affected irrespective of disulfide bond formation.
FIGURE 7.
ATP hydrolysis activities of unlocked and locked mutants without insertion region. A, nonreducing SDS-PAGE analysis of unlocked and locked mutants without the insertion region was performed on 12% polyacrylamide gel. First lane 1, Cys-less; second lane, Cys-less/ΔIns; third and fourth lanes, unlocked and locked A28C/L267C/ΔIns; fifth and sixth lanes, unlocked and locked V32C/A268C/ΔIns; seventh and eighth lanes, unlocked and locked V39C/A264C/ΔIns. B, change in ATP hydrolysis activities of Cys-less/ΔIns and mutants lacking the insertion region by unlocking and locking were measured as described in the legend to Fig. 3. The light gray bar represents the activity of Cys-less/ΔIns. Open and closed bars represent the activities of unlocked and locked mutants, respectively. The results of three independent experiments were averaged. Error bar represents S.D. The conditions of α-helices in the mutant complexes were shown as U (unlocked) and L (locked), respectively.
DISCUSSION
ATP synthase is a very unique enzyme because the reaction catalyzed at the catalytic sites located on the β subunit is coupled to rotation of the central axis γ subunit (10). In addition, this enzyme complex is equipped with multiple systems that work to regulate its activity. Although the major mechanism that acts to regulate its activity is ADP inhibition of the catalytic sites (11, 12, 14, 42), the reason that sites of redox regulation and the ϵ inhibition are structurally distant from the catalytic site is poorly understood. We postulated that the relative slippage of the two central α-helices of the γ subunit, which connect the two active domains for catalysis and regulation in the structure of the complex (8), must be a main cause of the change in the enzyme activity (36). To examine this hypothesis, we prepared several mutant complexes in which the relative slippage of two central α-helices can be locked at a certain position by disulfide bond formation. Accordingly, some of the unlocked and locked mutants prepared in this study showed significant changes in activities caused by disulfide bond formation or reduction. Because the mutated residues for Cys substitution are not supposed to interact with the α3β3 ring directly in the referenced crystal structure of bovine F1 (18), the relative slippage of two central α-helices was expected to be a major cause of any observed change in activity. However, introduction of Cys residues for disulfide formation to the central α-helices also changed the ATP hydrolysis activity compared with the WT and Cys-less complex (Fig. 3), irrespective of the redox conditions of these cysteines. This may imply that the position of these Cys residues introduced into the complex is critical in causing relative slippage of α-helices due to substitution of the side residue moiety. Because the pKa value of the thiol moiety of Cys is around 8.0, the thiol group of the introduced Cys residue is thought to be half-deprotonated, which might affect the slippage of α-helices and alter the activity.
To confirm the cause of the variability of the mutant activities, we examined the effects of LDAO, which can release the enzyme from the ADP inhibition state (Table 1). Consequently, the activities obtained in the presence of LDAO were not significantly different; 22 to 36 units/mg in the case of the unlocked form and 25 to 36 in the case of the locked form. Only the mutant complex A28C/L267C showed very poor activity and the activity did not recover completely even in the presence of LDAO (Table 1). Variability of the mutant activities examined might be attributed to different sensitivities of the complex against ADP inhibition, which is affected by the relative α-helices' position of the γ subunit in the complex.
Where is the significant contact point between γ and the α3β3 ring, which is important in transferring the signal of the conformational change via the slippage of α-helices of γ to the catalytic site(s)? In the reported crystal structure of F1, the contacts mainly consist of a “catch” region and a region near the “DELSEED” sequence (8). Catch is formed by some charged residues in β, especially βE, which does not bind any nucleotides, and in the C-terminal α-helix of γ. He et al. (43) reported that substitution of the highly conserved Arg with Leu in the C-terminal α-helix, which forms a catch, stimulated ATP hydrolysis activity and, substitution of Gln close to this Arg with Ala, decreased the sulfite activation in chloroplast ATP synthase. Sulfite is recognized to release the complex from ADP inhibition like LDAO. Thus, these residues seem to be important in transferring the conformational change of the γ subunit to the catalytic site. In addition, the corresponding mutation analysis in E. coli F1 showed a drastic decrease in activity (44). As the catch region is formed by βE and γ and no nucleotide binds in βE, there are some cooperative interactions among three β subunits as suggested (9). The second contact point between β and γ is the region near the DELSEED sequence on the β subunit. There are a couple of studies that examined the role of this region on catalysis (45–47). Mnatsakanyan et al. (46) and Usukura et al. (47) recently found that the region is important to generate torque and achieve mechanical coupling. Although it is still difficult to assign residues that are responsible to torque generation, this region must also be important for transfer of the signal of the conformational change of the γ subunit to the catalytic site(s).
Richter and co-workers (48) recently prepared several cross-linked mutants of γ to examine the possibility of helical domain movement during rotational catalysis (49). Although they postulated the twisting movement of the α-helices, the position where Cys residues were introduced were very close to those employed in this study. However, all of their mutants showed no significant changes in the activity by disulfide bond formation. In addition, it was difficult to estimate the yield of the oxidized complex by their method, which evaluates accessibility of fluorescent chemicals (see Fig. 3 of Ref. 48). One possible reason for the discrepancy between their study and our study is that the interaction between α3β3 and γ is not exactly the same as that of the native enzyme, in their case because their enzyme is a hybrid complex comprised of α and β subunits from Rhodospirillum rubrum F1 and the γ subunit from spinach chloroplast F1. One of the mutants in their study, the γV31C/A276C complex, did not show any activation by the oxyanion sulfite, implying the enzyme was not recovered from ADP inhibition. Although our complex, V32C/A268C, which corresponds to their γV31C/A276C complex based on the sequence homology, was almost completely released from ADP inhibition and did not show remarkable activation by LDAO when oxidized (Table 1).
When V32C/A268C and A35C/A264C complexes were locked by oxidation, not much constraint was expected to be induced because the expected relative position of α-helices seemed to be similar as those of the unlocked form, which is the position of two α-helices in the crystal structure as mentioned. Their activities were inhibited almost completely by the ϵ subunit irrespective of the locked or unlocked forms of the γ subunit (Fig. 5, B and D). In contrast, the extent of ϵ inhibition was lower when V32C/A264C and V39C/A264C complexes were in the locked states. These results indicate that the possible slippage of α-helix by disulfide bond formation largely affects the extent of ϵ inhibition. Because the ϵ subunit can bind to the locked V39C/A264C complex as well as the unlocked one (Fig. 6), we assumed that the decrease of ϵ inhibition can be attributed to the constraint of the conformational change of the C-terminal domain in ϵ from the extended to retracted form. In the latest E. coli F1 structure, the ϵ subunit is in the extended form, and the C-terminal domain of ϵ can interact with both N- and C-terminal α-helices of γ (21). The relative slippage of two central α-helices may weaken the interaction of γ and ϵ, and consequently the ϵ subunit falls into the retracted form, which cannot inhibit the enzyme (50). Although the inhibitory properties of ϵ inhibition and ADP inhibition appear to be similar, recent single molecule analyses clearly show that these two inhibitions are mechanically different (41, 51).
We finally sought to investigate whether significant activation of the enzyme by removal of the insertion region of cyanobacterial F1-γ could be attributed to the relative slippage of two α-helices found in this study. To address this question, we prepared additional lock mutants lacking the γ insertion region. By deletion of the insertion, basal activities of Cys-less/ΔIns and unlocked deletion mutants were drastically increased (Fig. 7) as observed in the previous studies (25, 32). In contrast, their activities remained relatively unchanged by locking. In the case of A28C/L267C, change in the activity by locking was reversed upon removal of the insertion. Two other mutants, V32C/A268C and V39C/A264C, showed full activities by removal of the insertion and the activities did not change after locking. These results clearly indicate that removal of the insertion region of γ induces another conformational change in γ rather than relative slippage of two α-helices. In the crystal structure of bovine F1 (18), the upper part of the globular domain in γ is in contact with the DELSEED region in βTP and βDP. Therefore, removal of the insertion may potentially result in a conformational change that affects the activity by affecting the structure of the globular domain itself, and this conformational change is transferred to the catalytic site(s) via interaction between the γ and β subunits at the bottom of β. Finally, ADP inhibition must be controlled at the catalytic sites by the induced conformational change of the β subunit.
In the case of the chloroplast ATP synthase, CF1 possesses the additional regulation, redox regulation in the γ subunit. The two functional Cys residues Cys199 and Cys205 (spinach CF1 numbering) involved in this redox regulation are supposed to be located in the bottom of the globular domain of γ close to the c-ring (34). Although we are not currently able to provide a definitive conclusion, redox regulation of chloroplast F1 might be achieved by transferring the conformational change of the disulfide bond formation or reduction to the catalytic site(s) via slippage of the central α-helices or change in the interaction between the upper part of the globular domain of γ and the DELSEED region in βTP and βDP. On this point, further analysis is required to understand how the conformational change caused by redox regulation is transferred to the catalytic site(s) in CF1 enzyme molecule. Previously, we constructed a chimeric cyanobacterial F1 complex in which the 9-amino acid region of spinach CF1-γ including two regulatory Cys was introduced into the cyanobacterial γ subunit (36). Based on the single molecular observation of rotation of this chimeric complex, we concluded that redox regulation is achieved by controlling the probability to lapse into ADP inhibition. In addition, we have reported the reverse of the redox regulation of ATPase caused by deletion of the Glu-Asp-Glu sequence from the insertion region of the γ subunit of spinach CF1 (52, 53). Hence the regulation must certainly be achieved by a slight conformational change of the γ subunit.
In this study, we clearly showed that the conformational change of the γ subunit can regulate ATP hydrolysis activity by controlling other regulatory mechanisms, ADP inhibition, and ϵ inhibition. Further studies, especially analysis of the whole molecular structure of the chloroplast-type F1 including regulatory subunits will lead to a more complete understanding of the complicated regulatory mechanisms of this enzyme complex.
Acknowledgment
We thank S. Hara for helpful suggestions for oxidation of the mutant complex.
This work was supported in part by the Core Research of Evolutional Science and Technology program (CREST) from the Japan Science and Technology Agency (JST) and Grant-in-aid for Scientific Research 22651048 (to T. H.) from the Japan Society for the Promotion of Science.

This article contains supplemental Fig. S1 and Table S1.
- F0F1
- F0F1-ATP synthase
- CF1
- chloroplast F1
- LDAO
- N-dimethyldodecylamine-N-oxide.
REFERENCES
- 1. Boyer P. D. (1997) The ATP synthase. A splendid molecular machine. Annu. Rev. Biochem. 66, 717–749 [DOI] [PubMed] [Google Scholar]
- 2. Yoshida M., Muneyuki E., Hisabori T. (2001) ATP synthase. A marvellous rotary engine of the cell. Nat. Rev. Mol. Cell Biol. 2, 669–677 [DOI] [PubMed] [Google Scholar]
- 3. Yoshida M., Sone N., Hirata H., Kagawa Y., Ui N. (1979) Subunit structure of adenosine triphosphatase. Comparison of the structure in thermophilic bacterium PS3 with those in mitochondria, chloroplasts, and Escherichia coli. J. Biol. Chem. 254, 9525–9533 [PubMed] [Google Scholar]
- 4. Stock D., Leslie A. G., Walker J. E. (1999) Molecular architecture of the rotary motor in ATP synthase. Science 286, 1700–1705 [DOI] [PubMed] [Google Scholar]
- 5. Mitome N., Suzuki T., Hayashi S., Yoshida M. (2004) Thermophilic ATP synthase has a decamer c-ring. Indication of noninteger 10:3 H+/ATP ratio and permissive elastic coupling. Proc. Natl. Acad. Sci. U.S.A. 101, 12159–12164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pogoryelov D., Yu J., Meier T., Vonck J., Dimroth P., Muller D. J. (2005) The c15 ring of the Spirulina platensis F-ATP synthase. F1/F0 symmetry mismatch is not obligatory. EMBO Rep. 6, 1040–1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Matsui T., Yoshida M. (1995) Expression of the wild-type and the Cys-/Trp-less α3β3γ complex of thermophilic F1-ATPase in Escherichia coli. Biochim. Biophys. Acta 1231, 139–146 [DOI] [PubMed] [Google Scholar]
- 8. Abrahams J. P., Leslie A. G., Lutter R., Walker J. E. (1994) Structure at 2.8-Å resolution of F1-ATPase from bovine heart mitochondria. Nature 370, 621–628 [DOI] [PubMed] [Google Scholar]
- 9. Gresser M. J., Myers J. A., Boyer P. D. (1982) Catalytic site cooperativity of beef heart mitochondrial F1 adenosine triphosphatase. Correlations of initial velocity, bound intermediate, and oxygen exchange measurements with an alternating three-site model. J. Biol. Chem. 257, 12030–12038 [PubMed] [Google Scholar]
- 10. Noji H., Yasuda R., Yoshida M., Kinosita K., Jr. (1997) Direct observation of the rotation of F1-ATPase. Nature 386, 299–302 [DOI] [PubMed] [Google Scholar]
- 11. Minkov I. B., Fitin A. F., Vasilyeva E. A., Vinogradov A. D. (1979) Mg2+-induced ADP-dependent inhibition of the ATPase activity of beef heart mitochondrial coupling factor F1. Biochem. Biophys. Res. Commun. 89, 1300–1306 [DOI] [PubMed] [Google Scholar]
- 12. Dunham K. R., Selman B. R. (1981) Regulation of spinach chloroplast coupling factor 1 ATPase activity. J. Biol. Chem. 256, 212–218 [PubMed] [Google Scholar]
- 13. Vasilyeva E. A., Minkov I. B., Fitin A. F., Vinogradov A. D. (1982) Kinetic mechanism of mitochondrial adenosine triphosphatase. ADP-specific inhibition as revealed by the steady-state kinetics. Biochem. J. 202, 9–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yoshida M., Allison W. S. (1983) Modulation by ADP and Mg2+ of the inactivation of the F1-ATPase from the thermophilic bacterium, PS3, with dicyclohexylcarbodiimide. J. Biol. Chem. 258, 14407–14412 [PubMed] [Google Scholar]
- 15. Bald D., Amano T., Muneyuki E., Pitard B., Rigaud J. L., Kruip J., Hisabori T., Yoshida M., Shibata M. (1998) ATP synthesis by F0F1-ATP synthase independent of noncatalytic nucleotide binding sites and insensitive to azide inhibition. J. Biol. Chem. 273, 865–870 [DOI] [PubMed] [Google Scholar]
- 16. Matsui T., Muneyuki E., Honda M., Allison W. S., Dou C., Yoshida M. (1997) Catalytic activity of the α3β3γ complex of F1-ATPase without noncatalytic nucleotide binding site. J. Biol. Chem. 272, 8215–8221 [DOI] [PubMed] [Google Scholar]
- 17. Hirono-Hara Y., Noji H., Nishiura M., Muneyuki E., Hara K. Y., Yasuda R., Kinosita K., Jr., Yoshida M. (2001) Pause and rotation of F1-ATPase during catalysis. Proc. Natl. Acad. Sci. U.S.A. 98, 13649–13654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gibbons C., Montgomery M. G., Leslie A. G., Walker J. E. (2000) The structure of the central stalk in bovine F1-ATPase at 2.4-Å resolution. Nat. Struct. Biol. 7, 1055–1061 [DOI] [PubMed] [Google Scholar]
- 19. Rodgers A. J., Wilce M. C. (2000) Structure of the γ-ϵ complex of ATP synthase. Nat. Struct. Biol. 7, 1051–1054 [DOI] [PubMed] [Google Scholar]
- 20. Tsunoda S. P., Rodgers A. J., Aggeler R., Wilce M. C., Yoshida M., Capaldi R. A. (2001) Large conformational changes of the ϵ subunit in the bacterial F1F0-ATP synthase provide a ratchet action to regulate this rotary motor enzyme. Proc. Natl. Acad. Sci. U.S.A. 98, 6560–6564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cingolani G., Duncan T. M. (2011) Structure of the ATP synthase catalytic complex (F1) from Escherichia coli in an autoinhibited conformation. Nat. Struct. Mol. Biol. 18, 701–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Richter M. L., Patrie W. J., McCarty R. E. (1984) Preparation of the ϵ subunit and ϵ subunit-deficient chloroplast coupling factor 1 in reconstitutively active forms. J. Biol. Chem. 259, 7371–7373 [PubMed] [Google Scholar]
- 23. Weber J., Dunn S. D., Senior A. E. (1999) Effect of the ϵ-subunit on nucleotide binding to Escherichia coli F1-ATPase catalytic sites. J. Biol. Chem. 274, 19124–19128 [DOI] [PubMed] [Google Scholar]
- 24. Keis S., Stocker A., Dimroth P., Cook G. M. (2006) Inhibition of ATP hydrolysis by thermoalkaliphilic F1F0-ATP synthase is controlled by the C terminus of the ϵ subunit. J. Bacteriol. 188, 3796–3804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Konno H., Murakami-Fuse T., Fujii F., Koyama F., Ueoka-Nakanishi H., Pack C. G., Kinjo M., Hisabori T. (2006) The regulator of the F1 motor. Inhibition of rotation of cyanobacterial F1-ATPase by the ϵ subunit. EMBO J. 25, 4596–4604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Haruyama T., Hirono-Hara Y., Kato-Yamada Y. (2010) Inhibition of thermophilic F1-ATPase by the ϵ subunit takes a different path from the ADP-Mg inhibition. Biophysics 6, 59–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sekiya M., Hosokawa H., Nakanishi-Matsui M., Al-Shawi M. K., Nakamoto R. K., Futai M. (2010) Single molecule behavior of inhibited and active states of Escherichia coli ATP synthase F1 rotation. J. Biol. Chem. 285, 42058–42067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mills J. D., Mitchell P., Schürmann P. (1980) Modulation of coupling factor ATPase activity in intact chloroplasts. The role of the thioredoxin system. FEBS Lett. 112, 173–177 [Google Scholar]
- 29. Miki J., Maeda M., Mukohata Y., Futai M. (1988) The γ-subunit of ATP synthase from spinach chloroplasts. Primary structure deduced from the cloned cDNA sequence. FEBS Lett. 232, 221–226 [DOI] [PubMed] [Google Scholar]
- 30. Nalin C. M., McCarty R. E. (1984) Role of a disulfide bond in the γ subunit in activation of the ATPase of chloroplast coupling factor 1. J. Biol. Chem. 259, 7275–7280 [PubMed] [Google Scholar]
- 31. Werner S., Schumann J., Strotmann H. (1990) The primary structure of the γ-subunit of the ATPase from Synechocystis 6803. FEBS Lett. 261, 204–208 [DOI] [PubMed] [Google Scholar]
- 32. Sunamura E., Konno H., Imashimizu-Kobayashi M., Sugano Y., Hisabori T. (2010) Physiological impact of intrinsic ADP inhibition of cyanobacterial F0F1 conferred by the inherent sequence inserted into the γ subunit. Plant Cell Physiol. 51, 855–865 [DOI] [PubMed] [Google Scholar]
- 33. Imashimizu M., Bernát G., Sunamura E., Broekmans M., Konno H., Isato K., Rögner M., Hisabori T. (2011) Regulation of F0F1-ATPase from Synechocystis sp. PCC 6803 by γ and ϵ subunits is significant for light/dark adaptation. J. Biol. Chem. 286, 26595–26602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hisabori T., Ueoka-Nakanishi H., Konno H., Koyama F. (2003) Molecular evolution of the modulator of chloroplast ATP synthase. Origin of the conformational change dependent regulation. FEBS Lett. 545, 71–75 [DOI] [PubMed] [Google Scholar]
- 35. Samra H. S., Gao F., He F., Hoang E., Chen Z., Gegenheimer P. A., Berrie C. L., Richter M. L. (2006) Structural analysis of the regulatory dithiol-containing domain of the chloroplast ATP synthase γ subunit. J. Biol. Chem. 281, 31041–31049 [DOI] [PubMed] [Google Scholar]
- 36. Kim Y., Konno H., Sugano Y., Hisabori T. (2011) Redox regulation of rotation of the cyanobacterial F1-ATPase containing thiol regulation switch. J. Biol. Chem. 286, 9071–9078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Joshi A. K., Ahmed S., Ferro-Luzzi Ames G. (1989) Energy coupling in bacterial periplasmic transport systems. Studies in intact Escherichia coli cells. J. Biol. Chem. 264, 2126–2133 [PubMed] [Google Scholar]
- 38. Nichols N. N., Harwood C. S. (1997) PcaK, a high-affinity permease for the aromatic compounds 4-hydroxybenzoate and protocatechuate from Pseudomonas putida. J. Bacteriol. 179, 5056–5061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ho S. N., Hunt H. D., Horton R. M., Pullen J. K., Pease L. R. (1989) Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77, 51–59 [DOI] [PubMed] [Google Scholar]
- 40. Dunn S. D., Tozer R. G., Zadorozny V. D. (1990) Activation of Escherichia coli F1-ATPase by lauryldimethylamine oxide and ethylene glycol. Relationship of ATPase activity to the interaction of the ϵ and β subunits. Biochemistry 29, 4335–4340 [DOI] [PubMed] [Google Scholar]
- 41. Konno H., Isu A., Kim Y., Murakami-Fuse T., Sugano Y., Hisabori T. (2011) Characterization of the relationship between ADP- and ϵ-induced inhibition in cyanobacterial F1-ATPase. J. Biol. Chem. 286, 13423–13429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Feniouk B. A., Suzuki T., Yoshida M. (2007) Regulatory interplay between proton-motive force, ADP, phosphate, and subunit ϵ in bacterial ATP synthase. J. Biol. Chem. 282, 764–772 [DOI] [PubMed] [Google Scholar]
- 43. He F., Samra H. S., Tucker W. C., Mayans D. R., Hoang E., Gromet-Elhanan Z., Berrie C. L., Richter M. L. (2007) Mutations within the C terminus of the γ subunit of the photosynthetic F1-ATPase activate MgATP hydrolysis and attenuate the stimulatory oxyanion effect. Biochemistry 46, 2411–2418 [DOI] [PubMed] [Google Scholar]
- 44. Greene M. D., Frasch W. D. (2003) Interactions among γR268, γQ269, and the β subunit catch loop of Escherichia coli F1-ATPase are important for catalytic activity. J. Biol. Chem. 278, 51594–51598 [DOI] [PubMed] [Google Scholar]
- 45. Kagawa Y., Hamamoto T. (1996) The energy transmission in ATP synthase. From the γ-c rotor to the α3β3 oligomer fixed by OSCP-b stator via the βDELSEED sequence. J. Bioenerg. Biomembr. 28, 421–431 [DOI] [PubMed] [Google Scholar]
- 46. Mnatsakanyan N., Krishnakumar A. M., Suzuki T., Weber J. (2009) The role of the βDELSEED loop of ATP synthase. J. Biol. Chem. 284, 11336–11345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Usukura E., Suzuki T., Furuike S., Soga N., Saita E., Hisabori T., Kinosita K., Jr., Yoshida M. (2012) Torque generation and utilization in motor enzyme F0F1-ATP synthase. Half-torque F1 with short-sized pushrod helix and reduced ATP Synthesis by half-torque F0F1. J. Biol. Chem. 287, 1884–1891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Samra H. S., He F., Degner N. R., Richter M. L. (2008) The role of specific β-γ subunit interactions in oxyanion stimulation of the MgATP hydrolysis of a hybrid photosynthetic F1-ATPase. J. Bioenerg. Biomembr. 40, 69–76 [DOI] [PubMed] [Google Scholar]
- 49. Richter M. L. (2004) γ-ϵ Interactions regulate the chloroplast ATP synthase. Photosynth. Res. 79, 319–329 [DOI] [PubMed] [Google Scholar]
- 50. Kato-Yamada Y., Yoshida M., Hisabori T. (2000) Movement of the helical domain of the ϵ subunit is required for the activation of thermophilic F1-ATPase. J. Biol. Chem. 275, 35746–35750 [DOI] [PubMed] [Google Scholar]
- 51. Saita E., Iino R., Suzuki T., Feniouk B. A., Kinosita K., Jr., Yoshida M. (2010) Activation and stiffness of the inhibited states of F1-ATPase probed by single-molecule manipulation. J. Biol. Chem. 285, 11411–11417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Konno H., Yodogawa M., Stumpp M. T., Kroth P., Strotmann H., Motohashi K., Amano T., Hisabori T. (2000) Inverse regulation of F1-ATPase activity by a mutation at the regulatory region on the γ subunit of chloroplast ATP synthase. Biochem. J. 352, 783–788 [PMC free article] [PubMed] [Google Scholar]
- 53. Ueoka-Nakanishi H., Nakanishi Y., Konno H., Motohashi K., Bald D., Hisabori T. (2004) Inverse regulation of rotation of F1-ATPase by the mutation at the regulatory region on the γ subunit of chloroplast ATP synthase. J. Biol. Chem. 279, 16272–16277 [DOI] [PubMed] [Google Scholar]