Bazooka/PAR-3 apicolateral polarity landmarks are established by the combination of two basolateral displacement activities in the Drosophila embryo. Basolateral PAR-1 activity acts redundantly with a basal-to-apical transport mechanism. With disruption of either mechanism alone Bazooka can polarize, but disruption of both blocks polarization.
Abstract
Polarity landmarks guide epithelial development. In the early Drosophila ectoderm, the scaffold protein Bazooka (Drosophila PAR-3) forms apicolateral landmarks to direct adherens junction assembly. However, it is unclear how Bazooka becomes polarized. We report two mechanisms acting in concert to displace Bazooka from the basolateral membrane. As cells form during cellularization, basally localized Bazooka undergoes basal-to-apical transport. Bazooka requires its three postsynaptic density 95, discs large, zonula occludens-1 (PDZ) domains to engage the transport mechanism, but with the PDZ domains deleted, basolateral displacement still occurs by gastrulation. Basolateral PAR-1 activity appears to act redundantly with the transport mechanism. Knockdown of PAR-1 sporadically destabilizes cellularization furrows, but basolateral displacement of Bazooka still occurs by gastrulation. In contrast, basolateral Bazooka displacement is blocked with disruption of both the transport mechanism and phosphorylation by PAR-1. Thus Bazooka is polarized through a combination of transport and PAR-1–induced dispersion from basolateral membranes. Our work complements recent findings in Caenorhabditis elegans and thus suggests the coupling of transport and dispersion is a common protein polarization strategy.
INTRODUCTION
Epithelial cell polarity underlies tissue structure and function. Cells polarize by recruiting or repelling molecules from primary polarity landmarks (Nelson, 2003; Goldstein and Macara, 2007; St Johnston and Ahringer, 2010; Laprise and Tepass, 2011). The scaffold protein Bazooka (Baz/PAR-3) forms polarity landmarks for spot adherens junction (AJ) assembly in Drosophila (Harris and Peifer, 2004). In Drosophila embryos, the first epithelium forms by cellularization—the simultaneous compartmentalization of ∼6000 nuclei by invaginating plasma membranes (Mazumdar and Mazumdar, 2002; Lecuit, 2004; Harris et al., 2009). As lateral membranes form, Baz can localize to apicolateral spot AJ assembly sites independently of AJ proteins (Harris and Peifer, 2004) and normally recruits and aggregates separately formed cadherin–catenin clusters (McGill et al., 2009). With loss of Baz, AJs mislocalize, and epithelial disruption occurs by gastrulation (Muller and Wieschaus, 1996; Harris and Peifer, 2004). During cellularization, three mechanisms normally position Baz: 1) an apicolateral actin-based scaffold, 2) basal-to-apical dynein-based transport, and 3) an undefined mechanism evident from analysis of dynein mutants in which basally mislocalized Baz redistributes apically by gastrulation (Harris and Peifer, 2005).
PAR-1 is a serine/threonine kinase that localizes basolaterally in mammalian, Drosophila follicular, and Drosophila embryonic epithelia (Bohm et al., 1997; Benton and St Johnston, 2003b; Bayraktar et al., 2006). In the Drosophila follicular epithelium, PAR-1 phosphorylation displaces basolateral Baz apparently by inhibiting Baz oligomerization and PAR complex assembly (Benton and St Johnston, 2003b). More recent work in mammalian cell culture has shown that the scaffolding adaptor protein GAB1 recruits PAR-1 and PAR-3 into a transient complex promoting inhibitory phosphorylation of PAR-3 important for proper apical–basal polarity (Yang et al., 2012). In contrast to these substantial effects, knockdown of PAR-1 only leads to slight apical-to-basal shifts of AJs and Baz in the Drosophila embryonic ectoderm (Bayraktar et al., 2006; Wang et al., 2012). Of interest, a normal local down-regulation of PAR-1 protein levels leads to a slight apical-to-basal shift of Baz and AJs important for epithelial folding events in the early embryo (Wang et al., 2012). It has been unclear why normal or experimentally induced reductions of PAR-1 lead to only modest changes in Baz localization in the early Drosophila embryo. We find that these subtle changes are due to buffering in the system. Specifically, PAR-1–induced dispersion functions redundantly with the basal-to-apical transport mechanism and the apical scaffold to polarize Baz.
RESULTS AND DISCUSSION
Basal-to-apical transport of Baz::GFP requires its PDZ domains
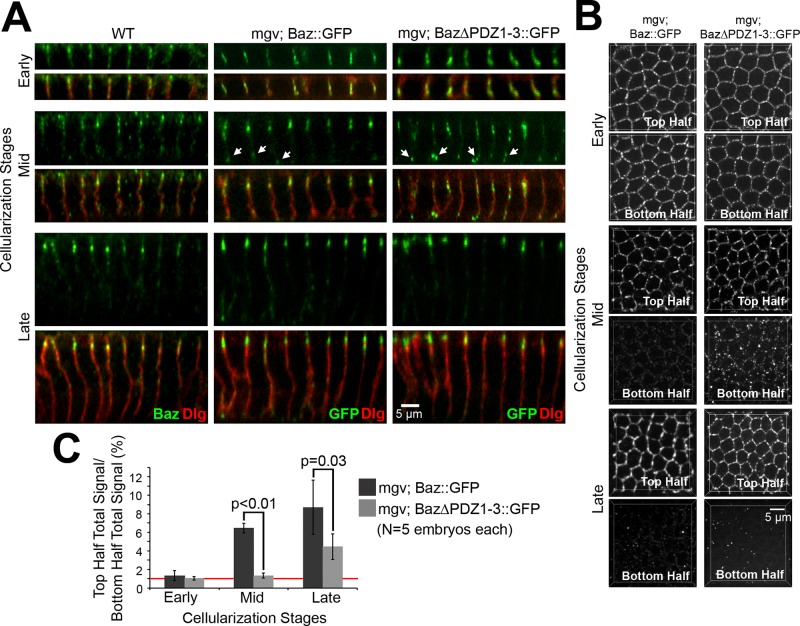
Dynein has been shown to function in the apical positioning of Baz at cellularization, and basal-to-apical Baz transport has been observed in Baz::green fluorescent protein (GFP) overexpression experiments in which Baz::GFP puncta initially mislocalized basally but then translocated apically during mid-late cellularization (Harris and Peifer, 2005). Baz is composed of multiple molecular interaction domains: an N-terminal oligomerization domain, three postsynaptic density 95, discs large, zonula occludens-1 (PDZ) domains, an aPKC-binding site, and a phosphoinositide-binding site (reviewed in Laprise and Tepass, 2011). PDZ domains are versatile molecular interaction modules (reviewed in Nourry et al., 2003), and PDZ1 of mammalian PAR-3 has been implicated in interactions with dynein light intermediate chain 2 (Schmoranzer et al., 2009). Thus, to further understand how the Baz::GFP translocations occur, we focused on the three PDZ domains of Baz. We first analyzed a construct with all three domains deleted. We compared the distribution of Baz::GFP and Baz∆PDZ1-3::GFP at early, mid, and late cellularization after maternal expression with a maternal-α4-tubulin-GAL4-VP16 (mgv) driver line. In contrast to endogenous Baz, overexpression of Baz::GFP and Baz∆PDZ1-3::GFP led to puncta accumulation along the full lateral membrane at early cellularization (Figure 1A). At midcellularization, both Baz::GFP and Baz∆PDZ1-3::GFP puncta mislocalized basally with enrichment at the base of the furrows, but the mislocalization was greater for Baz∆PDZ1-3::GFP (Figure 1A), despite it having similar if not lower total cortical levels versus Baz::GFP (Supplemental Table S1). Of note, both Baz::GFP and Baz∆PDZ1-3::GFP were mainly apicolateral by late cellularization, similar to endogenous Baz (Figure 1A).
FIGURE 1:
The PDZ domains of Baz promote its apical enrichment during cellularization. (A) Fixed early (5- to 7-μm furrows), mid (12- to 14-μm furrows), and late (19- to 21-μm furrows) cellularization embryos. Endogenous Baz vs. Baz::GFP and Baz∆PDZ1-3::GFP expressed maternally with mgv. Discs large (Dlg) shows furrows. Arrows show basal Baz puncta. (B) Projections of top and bottom halves of lateral membranes. (C) Total fluorescence intensity ratios of top vs. bottom halves of lateral membranes. Red line shows 1:1 top:bottom ratio.
To quantify the distributions, we created three-dimensional data sets for the top and bottom halves of lateral membranes at early, mid, and late cellularization (Figure 1B) and made three-dimensional masks around Baz puncta to quantify their total intensities in each half of the membrane (Figure 1C). At early cellularization, the fluorescence intensities in the top and bottom halves of the short lateral membranes were similar for both Baz::GFP and Baz∆PDZ1-3::GFP (top:bottom ratios of ∼1:1). At midcellularization, the top:bottom ratio of Baz∆PDZ1-3::GFP (∼1:1) was significantly lower than that of Baz::GFP (∼6:1; p < 0.01). By late cellularization, both Baz::GFP and Baz∆PDZ1-3::GFP showed apical enrichment (top:bottom ratios of ∼8:1 and ∼4:1, respectively). Thus apical recruitment of Baz∆PDZ1-3::GFP is delayed versus Baz::GFP.
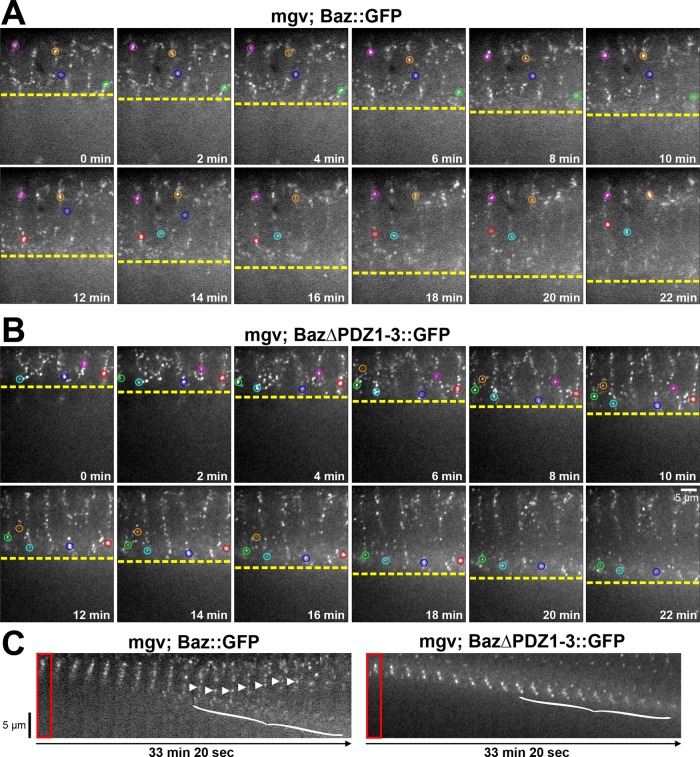
To test whether the delayed Baz∆PDZ1-3::GFP apical localization was due to a defect in basal-to-apical translocation, we performed live-cell imaging of the ectoderm apical–basal axis in embryo cross sections. At the onset of cellularization, Baz::GFP puncta localized along the full lateral membrane (Figure 2A and Supplemental Movie S1). As the furrows elongated, Baz::GFP puncta either remained apical or followed the furrows basally. Then puncta underwent basal-to-apical translocations (Figure 2C, arrowheads, and Supplemental Movie S1; six of six embryos displayed translocations). Translocations occurred until late cellularization, when most Baz::GFP puncta were properly positioned apicolaterally (Figure 2, A and C, and Supplemental Movie S1). Baz::GFP puncta failing to undergo basal-to-apical translocation dispersed locally by the end of cellularization (Figure 2C, bracket). Baz∆PDZ1-3::GFP puncta displayed a similar early cellularization distribution (Figure 2B). As cellularization continued, some Baz∆PDZ1-3::GFP puncta remained apical, but most followed the furrow basally, with enrichment at the furrow base, until late cellularization, when they locally dispersed (Figure 2, B and C, and Supplemental Movie S2). Significantly, the basal Baz∆PDZ1-3::GFP puncta did not undergo basal-to-apical translocations (Supplemental Movie S2; zero of six embryos displayed translocations). Thus the PDZ domains are needed for the basal-to-apical transport of Baz puncta during cellularization.
FIGURE 2:
The PDZ domains of Baz are required for its basal-to-apical transport. (A, B) Cellularizing embryos over 22 min. Single confocal planes of central cross sections. Circles track puncta in plane for extended periods. Constructs expressed maternally with mgv. (A) As furrow canals progress (yellow line), Baz::GFP puncta stay apical (pink, orange, and blue circles), move basally (green circles), or translocate basally to apically (red and cyan circles). (B) As furrow canals progress (yellow line), Baz∆PDZ1-3::GFP puncta move basally with them (cyan, blue, red, green, pink, and orange circles) without apical translocations. (C) Kymographs of cellularizing embryo cross sections. Baz::GFP puncta translocate apically (arrowheads). Basal Baz::GFP and Baz∆PDZ1-3::GFP puncta disperse at late cellularization (brackets; apical puncta lack photobleaching).
The distribution of Baz∆PDZ1-3::GFP is similar to that of endogenous Baz in dynein mutants at each stage of cellularization (Harris and Peifer, 2005). With disruption of either the PDZ domains or dynein, basal-to-apical transport fails at midcellularization, but apical polarization occurs by the end of cellularization (Figure 2; Harris and Peifer, 2005). Thus the basal-to-apical movement may involve direct or indirect interactions between the PDZ domains and dynein, and an additional mechanism removes residual basal Baz at later stages. Of interest, this latter mechanism could be overwhelmed when Baz∆PDZ1-3::GFP is highly overexpressed with a maternal-triple-driver-GAL4 (MTD). In this case, most Baz∆PDZ1-3::GFP puncta remained at the base of lateral membranes into gastrulation (Supplemental Figure S1).
Each single Baz PDZ domain can rescue the transport defect of Baz∆PDZ1-3
To investigate which PDZ domains are responsible for the basal-to-apical transport, we analyzed constructs in which each domain was singly added to Baz∆PDZ1-3::GFP (McKinley et al., 2012). A fragment of mammalian PAR-3 that contains the N-terminal oligomerization domain and PDZ1 can directly bind dynein light intermediate chain 2 (Schmoranzer et al., 2009). We hypothesized that Baz might engage dynein in a similar way. However, addition of any single PDZ domain to Baz∆PDZ1-3::GFP could restore basal-to-apical transport events; Baz∆PDZ2-3::GFP (two of six embryos had translocation events), Baz∆PDZ1+3::GFP (two of six embryos had translocation events), and Baz∆PDZ1-2::GFP (six of six embryos had translocation events; Supplemental Movies S3–S5). It is possible that each PDZ domain interacts with dynein directly or indirectly or that the constructs are influenced by endogenous Baz via homo-oligomerization (Benton and St Johnston, 2003a). However, PDZ3 had the strongest apparent effect, suggesting that it might be most closely linked to the transport machinery.
PAR-1 overlaps with Baz at early cellularization but segregates basally by gastrulation
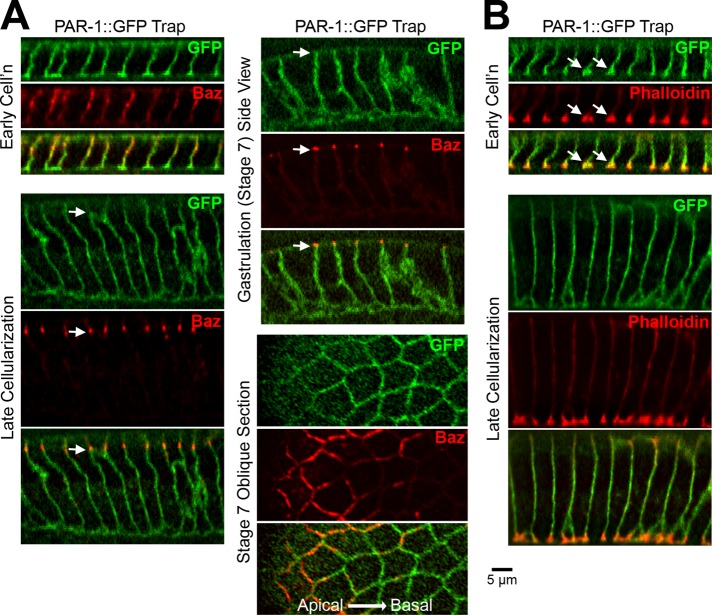
Because PAR-1 phosphorylates and displaces basal Baz in follicle cells (Benton and St Johnston, 2003b), we hypothesized that PAR-1 might be responsible for the additional mechanism that displaces basal Baz in the early ectoderm. We first analyzed the distribution of PAR-1 during cellularization and gastrulation (Figure 3A). Previous PAR-1 antibody staining showed PAR-1 along the full length of cellularization furrows, with enrichment at furrow canals (Bayraktar et al., 2006). After cellularization, the antibody detected PAR-1 on basolateral, apicolateral, and apical membranes, and this distribution was used to argue against a role for PAR-1 in Baz localization during embryonic epithelial polarity establishment (Bayraktar et al., 2006). To confirm that this staining reflects PAR-1 and to compare PAR-1 to Baz, we analyzed a GFP-tagged form of endogenous PAR-1 (a PAR-1 GFP trap line). At early cellularization, PAR-1::GFP localized over the full plasma membrane, overlapping with apicolateral Baz and localizing to the apical surface (Figure 3A), and was strongly enriched at phalloidin-stained furrow canals (Figure 3, A and B, arrows in B). PAR-1 apical localization and furrow canal enrichment decreased through mid and late cellularization (Figure 3, A and B). By late cellularization, segregation of PAR-1 below Baz became evident. This segregation was stronger at gastrulation (stage 7; Figure 3A) and was maintained at later stages (unpublished data). Oblique optical sections of the epithelium confirmed the enrichment of Baz apical to the strongest membrane concentration of PAR-1 (Figure 3A). Of note, the timing of PAR-1 segregation to the basolateral region roughly coincides with the detection of an additional Baz displacement mechanism after disruption of the PDZ domains (Figure 2C) or dynein (Harris and Peifer, 2005).
FIGURE 3:
PAR-1 segregates from apicolateral Baz by gastrulation. (A) Fixed PAR-1::GFP trap embryos. PAR-1 segregates below endogenous Baz beginning at late cellularization (arrows). Oblique stage 7 section shows Baz above PAR-1. (B) PAR-1::GFP trap embryos stained with phalloidin. Note PAR-1 and actin furrow canal coenrichment at early cellularization (arrows).
PAR-1 has major effects on cellularization furrows but subtle effects on Baz localization
To determine how PAR-1 affects Baz localization, we expressed short hairpin RNAs (shRNAs) to knock down PAR-1 gene expression. We used two par-1 shRNA lines in two different expression vectors (Valium20 and Valium22) and targeting two different sequences in par-1. They were expressed maternally with the mgv driver. Both produced similar defects, although the Valium22 line had stronger effects (the Valium22 shRNA also led to undetectable PAR1::GFP with the same microscope settings as PAR1::GFP imaged without RNA interference [RNAi; unpublished data]). The most obvious defect was the sporadic absence of actin-positive cellularization furrows (Figure 4A, arrowheads). Despite furrow absence, microtubule baskets that surround each nucleus appeared to form normally (Figure 4A). Given that cellularization furrows depend on actomyosin accumulations at the furrow canals (Mazumdar and Mazumdar, 2002; Lecuit, 2004; Harris et al., 2009) and PAR-1 is enriched at furrow canals, PAR-1 may normally promote actomyosin activity at these sites, as recently shown in Drosophila border cells (Majumder et al., 2012).
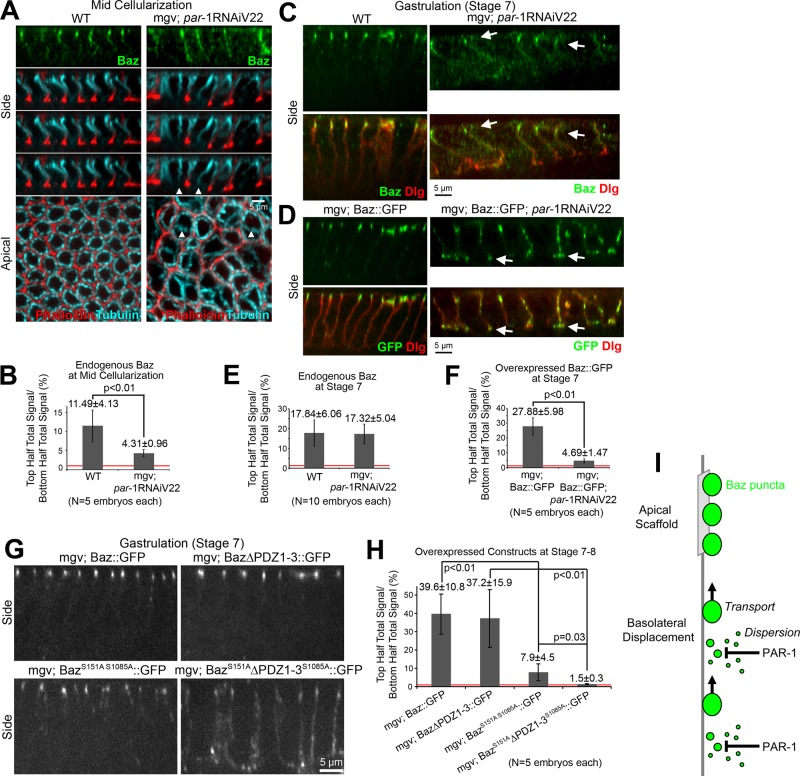
FIGURE 4:
PAR-1 phosphorylation contributes redundantly to Baz polarization. (A) Fixed midcellularization embryos. In WT, Baz localizes apicolaterally, phalloidin stains overall furrows and actin-rich furrow canals, and apical-basal microtubule bundles form baskets around nuclei. With maternal expression of par-1 shRNA (Valium22 line) with mgv, Baz mislocalizes basally and furrows are lost sporadically (arrowheads), but microtubule baskets are present. (B) Total fluorescence intensity ratios of endogenous Baz puncta in the top vs. bottom halves of lateral membranes. (C, D) Fixed stage 7 embryos. (C) With and without PAR-1 RNAi, endogenous Baz is apicolaterally enriched despite PAR-1 RNAi morphological defects. Dlg shows basolateral membranes. (D) Insertion of UAS-Baz::GFP at attp40 (chromosome 2) allowed maternal coexpression with par-1 shRNA (Valium22 line) at attp2 (chromosome 3; other Baz constructs at attp2). With coexpression, Baz::GFP mislocalized basally, accumulating at the base of lateral membranes (arrows). (E, F) Total fluorescence intensity ratios of puncta in the top vs. bottom halves of cells. (G) Fixed stage 7 embryo side views with Baz::GFP, Baz∆PDZ1-3::GFP, BazS151AS1085A::GFP, and BazS151A∆PDZ1-3S1085A::GFP expressed maternally with mgv. (H) Total fluorescence intensity ratios of protein puncta in top vs. bottom halves of cells. Red lines show 1:1 top:bottom ratios. (I) Model of Baz polarization.
Next, we examined how PAR-1 RNAi affected endogenous Baz. At midcellularization, Baz often spread basally with PAR-1 RNAi versus wild type (WT; Figure 4A). Quantification revealed significantly decreased Baz apical enrichment with PAR-1 knockdown versus WT at midcellularization, although Baz was still apically enriched with PAR-1 RNAi (Figure 4B). The overall intensity of Baz with PAR-1 knockdown was similar to WT (Supplemental Table S1). To test whether Baz mislocalization persisted, we probed gastrulating (stage 7) embryos. The embryos were morphologically disrupted, in part due to large multinucleated cells arising from the cellularization defects. Nonetheless, endogenous Baz was apically enriched, similar to WT (Figure 4, C and E). This relatively subtle effect on Baz is consistent with past PAR-1 RNAi experiments, which resulted in subtle basolateral shifts of AJs and Baz at gastrulation (Bayraktar et al., 2006; Wang et al., 2012).
Saturating the Baz apical scaffold reveals a redundant role for PAR-1 in Baz::GFP localization
We hypothesized that the other Baz-positioning mechanisms might compensate for the PAR-1 knockdown. Thus we challenged the apical scaffold for Baz by overexpressing Baz::GFP with and without PAR-1 RNAi. In the WT background, Baz::GFP localized apicolaterally at gastrulation (stage 7; Figure 4D). With PAR-1 RNAi, Baz::GFP mislocalized basally, often concentrating at the base of the lateral membranes at stage 7 (Figure 4D, arrows, F). Thus PAR-1 apparently functions in displacing basolateral Baz, but at normal Baz expression levels, the apical Baz scaffold can largely compensate for PAR-1 loss.
Disabling the Baz transport mechanism reveals a redundant role for PAR-1 in Baz::GFP localization
To test for redundancies between the basal-to-apical transport mechanism and PAR-1 activity, we investigated whether PAR-1 phosphorylation was responsible for the dissipation of basally mislocalized Baz∆PDZ1-3::GFP puncta by gastrulation. To focus on the effect of PAR-1 on Baz and to avoid potential effects of PAR-1 loss on microtubules (Shulman et al., 2000; Doerflinger et al., 2003), we mutated the known PAR-1 phosphorylation sites in Baz (Benton and St Johnston, 2003b) to alanine in both full-length Baz::GFP and Baz∆PDZ1-3::GFP. We examined gastrulating embryos (stages 7–8) to assess whether Baz puncta disengaged from the basal-to-apical transport mechanism could be dispersed with the disruption of PAR-1 phosphorylation. Baz::GFP and Baz∆PDZ1-3::GFP were both strongly apically enriched at this stage (Figure 4G). BazS151AS1085A::GFP displayed slightly more basal localization but still had substantial apical enrichment (Figure 4G) and appeared to undergo basal-to-apical translocations during cellularization (Supplemental Movie S6). In contrast, BazS151A∆PDZ1-3S1085A::GFP showed strong basal mislocalization along the full lateral membrane, with only slight enrichment at AJs (Figure 4G). These relationships were confirmed by quantifications of embryos with indistinguishable total cortical amounts of each construct (Figure 4H and Supplemental Table S1). Thus PAR-1 is responsible for the third Baz polarization mechanism operating during early epithelial development in Drosophila.
The coupling of transport and diffusion as a general polarization strategy
Our overexpression, structure–function, and loss-of-function analyses indicate that Baz polarization in the Drosophila embryonic ectoderm involves the coordinated activities of an apical scaffold, basal-to-apical transport, and basal PAR-1-induced dispersion (Figure 4I). It is important to note that our overall conclusions are largely based on targeted mutations of Baz constructs that were subsequently overexpressed in a wild-type background. This strategy avoided the confounding multiple effects of perturbing overall dynein or PAR-1 activity, but basal mislocalization and the subsequent apical relocalization mechanisms could only be assayed when the apical scaffold was saturated by overexpression of the constructs. In addition, overexpression in a wild-type background could allow endogenous Baz to have some effects on the constructs through oligomerization (Benton and St Johnston, 2003a), a concern that may be particularly relevant to our inability to map a specific PDZ domain essential for transport.
With these caveats in mind, the coupling of polarized transport and PAR-1–induced dispersion to displace basolateral Baz is nonetheless quite intriguing. Recently, polarized transport has been shown to function with PAR-1–induced dispersion to polarize PAR-3 in the Caenorhabditis elegans one-cell embryo. Here the sperm centrosome elicits a posterior-to-anterior actomyosin flow that transports PAR-3 to the anterior (Goldstein and Hird, 1996; Munro et al., 2004; Jenkins et al., 2006). However, with perturbation of this transport, PAR-3 can still polarize anteriorly. This polarization arises from the sperm centrosome also stabilizing a posterior domain of PAR-1, which phosphorylates and displaces PAR-3 (Motegi et al., 2011). Although the transport mechanisms differ, being microtubule based in Drosophila and actomyosin based in C. elegans, their coupling with PAR-1-induced dispersion reveals a common strategy for cell polarization. How exactly transport and dispersion are coupled is unclear in either case. Possibly PAR-1 phosphorylation breaks Baz/PAR-3 complexes into smaller units for easier transport. Alternatively, PAR-1–induced dispersion may simply clear residual Baz/PAR-3 that fails to be transported. In the latter case, it is also unclear what would happen to Baz/PAR-3 after its dispersal from the plasma membrane. It could possibly be destroyed or dephosphorylated and redeployed.
MATERIALS AND METHODS
Fly stocks and crosses
UASp Baz constructs inserted at attp2 on chromosome 3 (Baz::GFP, Baz∆PDZ2-3::GFP, Baz∆PDZ1+3::GFP, Baz∆PDZ1-2::GFP, and Baz∆PDZ1-3::GFP) were generated previously (McKinley et al., 2012). par-1 shRNA lines in the pValium22 and pValium20 vectors inserted at attp2 were from the Bloomington Drosophila Stock Center (BDSC; Indiana University, Bloomington, IN; BDSC #32410 and #35342). All UAS constructs were expressed maternally using either mgv (a gift from Mark Peifer, University of North Carolina at Chapel Hill, Chapel Hill, NC) or a MTD from the BDSC (BDSC #31777: P{otu-GAL4::VP16.R}1, w*; P{GAL4-nos.NGT}40; P{GAL4::VP16-nos.UTR}CG6325MVD1; BDSC#31777). The PAR-1::GFP gene trap line was from the Flytrap collection (line CC01981). WT was yellow white.
To coexpress Baz::GFP with par-1 shRNA, we targeted the vector containing Baz::GFP (McKinley et al., 2012) to the attp40 recombination site on chromosome 2 (BestGene, Chino Hills, CA). The transgenic flies were crossed to the par-1 shRNA lines in the pValium22 vectors inserted at attp2 on chromosome 3 to synthesize a double-homozygous line for coexpression of both UAS constructs.
To mutate the known PAR-1 phosphorylation sites in Baz (Benton and St Johnston, 2003b), we used PCR to mutate S151 to alanine (forward primer, GTCGCAGCGCTGATCCCAATC; reverse complement, GATTGGGATCAGCGCTGCGAC) and S1085 to alanine (forward primer, GAAGAAGTCCTCGGCGTTGGAGTCG; reverse complement, CGACTCCAACGCCGAGGACTTCTTC) in Baz::GFP and Baz∆PDZ1-3::GFP using an established approach (McKinley et al., 2012). The constructs were confirmed by sequencing and inserted at attp2 on chromosome 3 (BestGene).
Embryo staining
For tubulin and actin staining, embryos were washed with 0.1% Triton X-100, dechorionated with 50% bleach, washed with 0.1% Triton X-100, fixed for 10 min in 1:1 10% formaldehyde in phosphate-buffered saline (PBS):heptane, devitellinized by hand peeling, and stained. For other stainings, embryos were fixed for 20 min in 1:1 3.7% formaldehyde in PBS:heptane and then devitellinized in methanol. Blocking and staining were in PBS containing 1% goat serum, 0.1% Triton X-100. and 1% sodium azide. Antibodies used were as follows: mouse anti-Dlg (1:100; Developmental Studies Hybridoma Bank, University of Iowa, Iowa City, IA); mouse anti-tubulin (1:100; Developmental Studies Hybridoma Bank); rabbit anti-Baz (1:3500; raised in our lab against GST-Baz1-311); and rabbit anti-GFP (1:2000; Abcam, Cambridge, MA). F-actin was stained with Alexa Fluor 568–conjugated phalloidin (1:200; Invitrogen, Carlsbad, CA). Secondary antibodies were conjugated to Alexa Fluor 488, Alexa Fluor 546, and Alexa Fluor 647 (Invitrogen).
Imaging
Dechorionated live embryos were mounted in halocarbon oil (series 700; Halocarbon Products, Beech Island, SC) on a gas-permeable membrane (petriPERM; Sigma-Aldrich, St. Louis, MO). Fixed embryos were mounted in Aqua Polymount (Polysciences, Warrington, PA). Images were collected by a spinning disk confocal system (Quorum Technologies, Guelph, ON, Canada) at room temperature using a 63× Plan Apochromat, numerical aperture 1.4, objective (Carl Zeiss, Jena, Germany) with a piezo top plate and an electron-multiplying charge-coupled device camera (Hamamatsu Photonics, Hamamatsu, Japan). Z-stacks had 300-nm step sizes. Images were analyzed with Volocity software (PerkinElmer, Waltham, MA).
Postacquisition image analysis and manipulation
For top:bottom total intensity ratios, Imaris software, version 6.2 (Bitplane, Zurich, Switzerland), was used to analyze three-dimensional data sets collected with identical settings. Z-stacks taken at each stage were split in half along the z-axis to obtain top and bottom halves. The depth of the furrow was used to determine the bottom of the Z-stack. Puncta selection was standardized for each experiment. The same data were used for calculations of total puncta intensities by adding the top- and bottom-half quantifications and then dividing by the number of cells.
Photoshop (Adobe, San Jose, CA) was used for figure preparation. Input levels were adjusted so that the main signal range spanned the entire output grayscale. Images were resized by bicubic interpolation without noticeable changes at normal viewing magnifications.
Statistics
Comparisons were done using Student's t-tests. Means are shown with SD.
Supplementary Material
Acknowledgments
We thank Cao Guo Yu and Pil Gyu Park for technical assistance. This work was supported by a Canadian Institutes of Health Research Operating Grant. R.F.A.M was supported by an Ontario Graduate Scholarship. T.J.C.H. holds a Tier 2 Canada Research Chair.
Abbreviations used:
- AJ
adherens junction
- Baz
Bazooka
- BDSC
Bloomington Drosophila Stock Center
- mgv
maternal-α4-tubulin-GAL4-VP16
- MTD
maternal-triple-driver-GAL4
- PDZ
postsynaptic density 95, discs large, zonula occludens-1
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E12-09-0655) on September 26, 2012.
REFERENCES
- Bayraktar J, Zygmunt D, Carthew RW. Par-1 kinase establishes cell polarity and functions in Notch signaling in the Drosophila embryo. J Cell Sci. 2006;119:711–721. doi: 10.1242/jcs.02789. [DOI] [PubMed] [Google Scholar]
- Benton R, St Johnston D. A conserved oligomerization domain in Drosophila Bazooka/PAR-3 is important for apical localization and epithelial polarity. Curr Biol. 2003a;13:1330–1334. doi: 10.1016/s0960-9822(03)00508-6. [DOI] [PubMed] [Google Scholar]
- Benton R, St Johnston D. Drosophila PAR-1 and 14-3-3 inhibit Bazooka/PAR-3 to establish complementary cortical domains in polarized cells. Cell. 2003b;115:691–704. doi: 10.1016/s0092-8674(03)00938-3. [DOI] [PubMed] [Google Scholar]
- Bohm H, Brinkmann V, Drab M, Henske A, Kurzchalia TV. Mammalian homologues of C. elegans PAR-1 are asymmetrically localized in epithelial cells and may influence their polarity. Curr Biol. 1997;7:603–606. doi: 10.1016/s0960-9822(06)00260-0. [DOI] [PubMed] [Google Scholar]
- Doerflinger H, Benton R, Shulman JM, St Johnston D. The role of PAR-1 in regulating the polarised microtubule cytoskeleton in the Drosophila follicular epithelium. Development. 2003;130:3965–3975. doi: 10.1242/dev.00616. [DOI] [PubMed] [Google Scholar]
- Goldstein B, Hird SN. Specification of the anteroposterior axis in Caenorhabditis elegans. Development. 1996;122:1467–1474. doi: 10.1242/dev.122.5.1467. [DOI] [PubMed] [Google Scholar]
- Goldstein B, Macara IG. The PAR proteins: fundamental players in animal cell polarization. Dev Cell. 2007;13:609–622. doi: 10.1016/j.devcel.2007.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris TJ, Peifer M. Adherens junction-dependent and -independent steps in the establishment of epithelial cell polarity in Drosophila. J Cell Biol. 2004;167:135–147. doi: 10.1083/jcb.200406024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris TJ, Peifer M. The positioning and segregation of apical cues during epithelial polarity establishment in Drosophila. J Cell Biol. 2005;170:813–823. doi: 10.1083/jcb.200505127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris TJ, Sawyer JK, Peifer M. How the cytoskeleton helps build the embryonic body plan: models of morphogenesis from Drosophila. Curr Top Dev Biol. 2009;89:55–85. doi: 10.1016/S0070-2153(09)89003-0. [DOI] [PubMed] [Google Scholar]
- Jenkins N, Saam JR, Mango SE. CYK-4/GAP provides a localized cue to initiate anteroposterior polarity upon fertilization. Science. 2006;313:1298–1301. doi: 10.1126/science.1130291. [DOI] [PubMed] [Google Scholar]
- Laprise P, Tepass U. Novel insights into epithelial polarity proteins in Drosophila. Trends Cell Biol. 2011;21:401–408. doi: 10.1016/j.tcb.2011.03.005. [DOI] [PubMed] [Google Scholar]
- Lecuit T. Junctions and vesicular trafficking during Drosophila cellularization. J Cell Sci. 2004;117:3427–3433. doi: 10.1242/jcs.01312. [DOI] [PubMed] [Google Scholar]
- Majumder P, Aranjuez G, Amick J, McDonald JA. Par-1 controls myosin-II activity through myosin phosphatase to regulate border cell migration. Curr Biol. 2012;22:363–372. doi: 10.1016/j.cub.2012.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazumdar A, Mazumdar M. How one becomes many: blastoderm cellularization in Drosophila melanogaster. BioEssays. 2002;24:1012–1022. doi: 10.1002/bies.10184. [DOI] [PubMed] [Google Scholar]
- McGill MA, McKinley RF, Harris TJ. Independent cadherin-catenin and Bazooka clusters interact to assemble adherens junctions. J Cell Biol. 2009;185:787–796. doi: 10.1083/jcb.200812146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinley RF, Yu CG, Harris TJ. Assembly of Bazooka polarity landmarks through a multifaceted membrane-association mechanism. J Cell Sci. 2012;125:1177–1190. doi: 10.1242/jcs.091884. [DOI] [PubMed] [Google Scholar]
- Motegi F, Zonies S, Hao Y, Cuenca AA, Griffin E, Seydoux G. Microtubules induce self-organization of polarized PAR domains in Caenorhabditis elegans zygotes. Nat Cell Biol. 2011;13:1361–1367. doi: 10.1038/ncb2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller HA, Wieschaus E. armadillo, bazooka, and stardust are critical for early stages in formation of the zonula adherens and maintenance of the polarized blastoderm epithelium in Drosophila. J Cell Biol. 1996;134:149–163. doi: 10.1083/jcb.134.1.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro E, Nance J, Priess JR. Cortical flows powered by asymmetrical contraction transport PAR proteins to establish and maintain anterior-posterior polarity in the early C. elegans embryo. Dev Cell. 2004;7:413–424. doi: 10.1016/j.devcel.2004.08.001. [DOI] [PubMed] [Google Scholar]
- Nelson WJ. Adaptation of core mechanisms to generate cell polarity. Nature. 2003;422:766–774. doi: 10.1038/nature01602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nourry C, Grant SG, Borg JP. PDZ domain proteins: plug and play! Sci STKE. 2003;2003:RE7. doi: 10.1126/stke.2003.179.re7. [DOI] [PubMed] [Google Scholar]
- Schmoranzer J, Fawcett JP, Segura M, Tan S, Vallee RB, Pawson T, Gundersen GG. Par3 and dynein associate to regulate local microtubule dynamics and centrosome orientation during migration. Curr Biol. 2009;19:1065–1074. doi: 10.1016/j.cub.2009.05.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulman JM, Benton R, St Johnston D. The Drosophila homolog of C. elegans PAR-1 organizes the oocyte cytoskeleton and directs oskar mRNA localization to the posterior pole. Cell. 2000;101:377–388. doi: 10.1016/s0092-8674(00)80848-x. [DOI] [PubMed] [Google Scholar]
- St Johnston D, Ahringer J. Cell polarity in eggs and epithelia: parallels and diversity. Cell. 2010;141:757–774. doi: 10.1016/j.cell.2010.05.011. [DOI] [PubMed] [Google Scholar]
- Wang YC, Khan Z, Kaschube M, Wieschaus EF. Differential positioning of adherens junctions is associated with initiation of epithelial folding. Nature. 2012;484:390–393. doi: 10.1038/nature10938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Xue B, Umitsu M, Ikura M, Muthuswamy SK, Neel BG. The signaling adaptor gab1 regulates cell polarity by acting as a par protein scaffold. Mol Cell. 2012;47:469–483. doi: 10.1016/j.molcel.2012.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.