Abstract
Francisella tularensis is a Gram-negative bacterium that is highly virulent in humans, causing the disease tularemia. F. novicida is closely related to F. tularensis and exhibits high virulence in mice, but it is avirulent in healthy humans. An F. novicida-specific gene cluster (FTN0451 to FTN0456) encodes two proteins with diguanylate cyclase (DGC) and phosphodiesterase (PDE) domains that modulate the synthesis and degradation of cyclic di-GMP (cdGMP). No DGC- or PDE-encoding protein genes are present in the F. tularensis genome. F. novicida strains lacking either the two DGC/PDE genes (cdgA and cdgB) or the entire gene cluster (strain KKF457) are defective for biofilm formation. In addition, expression of CdgB or a heterologous DGC in strain KKF457 stimulated F. novicida biofilms, even in a strain lacking the biofilm regulator QseB. Genetic evidence suggests that CdgA is predominantly a PDE, while CdgB is predominantly a DGC. The F. novicida qseB strain showed reduced cdgA and cdgB transcript levels, demonstrating an F. novicida biofilm signaling cascade that controls cdGMP levels. Interestingly, KKF457 with elevated cdGMP levels exhibited a decrease in intramacrophage replication and virulence in mice, as well as increased growth yields and biofilm formation in vitro. Microarray analyses revealed that cdGMP stimulated the transcription of a chitinase (ChiB) known to contribute to biofilm formation. Our results indicate that elevated cdGMP in F. novicida stimulates biofilm formation and inhibits virulence. We suggest that differences in human virulence between F. novicida and F. tularensis may be due in part to the absence of cdGMP signaling in F. tularensis.
INTRODUCTION
Tularemia is a serious infectious disease of humans that is caused by the facultative intracellular bacterium Francisella tularensis. Infection with F. tularensis can occur through exposure to infected animals, bites of infected insects or arthropods, or contaminated meat or water or by inhaling airborne bacteria (30, 43). Pneumonic tularemia can lead to high mortality (12, 44). However, the various subspecies of F. tularensis exhibit differing levels of virulence in humans.
F. tularensis subsp. tularensis is highly virulent for humans, with an infectious dose of <10 bacteria (11, 23, 36, 38). F. tularensis subsp. tularensis can be disseminated in an aerosolized form to cause pneumonic tularemia and has been developed into a bioweapon by various countries (9). At the opposite extreme of infectious capability for humans is F. novicida (depending on the classification system, F. novicida is also classified as a subspecies, F. tularensis subsp. novicida) (47). F. novicida is highly attenuated for virulence in humans and is known to cause disease only in immunocompromised individuals (12). However, both F. tularensis subsp. tularensis and F. novicida are highly virulent in mice and utilize a shared set of virulence genes within the Francisella pathogenicity island (FPI) that encode a type VI-like secretion system to escape the phagosome within macrophages and cause disease (2, 29). Thus, the tropisms responsible for the high and low virulence, respectively, of these subspecies in humans are still unclear.
The complete genome sequences of several representative F. tularensis subsp. tularensis and F. novicida strains have revealed a high degree of homology between them, and also highlighted important differences. In comparison to the F. novicida genome, the F. tularensis subsp. tularensis genome is undergoing reduction resulting from rearrangements and the accumulation of pseudogenes (7, 21, 35, 39). Relatively few elements are present in the F. tularensis subsp. tularensis genome that are absent from F. novicida; rather, a number of genes and gene clusters can be found in F. novicida that are absent or disrupted in F. tularensis subsp. tularensis. We have identified a gene cluster present in F. novicida but absent in F. tularensis subsp. tularensis that is involved in the synthesis and degradation of the secondary messenger molecule bis-(3′-5′)-cyclic dimeric GMP (cdGMP).
Elevated cdGMP levels within a large number of bacteria are generally used to control the switch from a free-living planktonic lifestyle to a sedentary biofilm-associated lifestyle (19, 41). Proteins with diguanylate cyclase (DGC) activity, encoded within domains containing a GGDEF motif, synthesize cdGMP, whereas those with phosphodiesterase (PDE) activity, encoded within domains containing an EAL motif, degrade cdGMP; these domains can be found in separate proteins as well as in composite proteins that contain both motifs (GGDEF-EAL) (20). cdGMP signaling generally controls exopolysaccharide expression associated with biofilm development but has also been shown to control a variety of other behaviors in different bacteria, including motility and virulence (17, 41).
F. novicida forms biofilms in vitro, and this behavior is likely linked to its environmental persistence in aquatic habitats as well as possibly within tick and mosquito vectors (10, 26). An orphan response regulator, QseB (FTN1465), is required for normal biofilm development (10), as is the Sec secretion system and several Sec-dependent secreted proteins, some of which are predicted to bind and/or degrade chitin (26). In the current study, we show that high cdGMP levels stimulate F. novicida biofilm formation and inhibit virulence, suggesting that differential cdGMP signaling within Francisella species may contribute in part to their relative virulence in humans.
MATERIALS AND METHODS
Strains, plasmids, and media.
F. novicida strains are isogenic with strain U112 (ATCC 15482) and are listed in Table S1 in the supplemental material. Vibrio cholerae smooth strain A1552 (49) was used for some biofilm studies. F. novicida strains were grown on TSAP broth/agar (24) or Chamberlain's defined medium (CDM) (6), supplemented with antibiotics as appropriate. The concentrations of antibiotics used were as follows: kanamycin, 50 μg ml−1; tetracycline, 10 μg ml−1; ampicillin, 100 μg ml−1; and erythromycin, 150 μg ml−1. F. novicida mutant strain KKF457 [Δ(FTN0451-FTN0456)] was created by splicing by overlap extension (SOE) as described by Liu et al. (24), using the PCR primers listed in Table S2. The cdgA cdgB mutant (KKF539) was constructed by first removing the kanamycin resistance marker from the cdgB transposon mutant tnfn1_pw060323p04q138 (ISFn2/FRT), obtained from a comprehensive F. novicida transposon library (13) (www.beiresources.org/), by FLP-mediated recombination as described previously (2), resulting in strain KKF535. The cdgA transposon mutant tnfn1_pw060328p03q151 (ISFn2), obtained from the comprehensive F. novicida transposon library, was then recombined into the chromosome of this strain via cryotransformation with genomic DNA (22), resulting in strain KKF539. The qseB chiA chiB mutant was constructed in the same manner, utilizing the transposon mutants tnfn1_pw060328p07q138 (qseB), tnfn1_pw060323p01q146 (chiA), and tnfn1_pw060328p03q184 (chiB) from the comprehensive F. novicida transposon library. The transposon insertions in all strains were verified by sequencing. Various experiments with the complementation plasmid pKEK1524 (expresses cdgB) demonstrated a lack of polarity of the cdgB transposon insertion, as shown in Results. To demonstrate a lack of polarity of the cdgA transposon insertion, reverse transcription-PCR (RT-PCR) was performed on the wild-type U112 and KKF538 (cdgA) strains, which showed that transcription of the downstream gene FTN0450 appeared unaffected by the presence of the transposon (see Fig. S1).
Plasmids were constructed using the expression vector pKEK894 as described previously (50) and are listed in Table S1 in the supplemental material. Escherichia coli strain DH5α (15) was used for cloning procedures. Primers used in plasmid construction are listed in Table S2. PCR fragments were digested with NcoI and EcoRI and ligated into similarly digested pKEK894.
Biofilm formation assay.
Overnight cultures of F. novicida strains were used to inoculate 1-ml aliquots of CDM (1:100) in 5-ml borosilicate glass tubes (Falcon), and the cultures were incubated statically at 37°C. At the indicated time points, the liquid medium was removed, and tubes were washed three times with phosphate-buffered saline (PBS) and allowed to air dry. Tubes were then incubated with 1.5 ml 0.4% crystal violet for 30 min, rinsed with distilled water, and air dried. Crystal violet was extracted with 30% acetic acid and quantitated by measuring the optical density at 570 nm (OD570).
RNA isolation and quantitative RT-PCR (qRT-PCR).
Total RNAs were isolated from mid-log-phase (OD600, ∼0.6) F. novicida cultures grown in CDM at 37°C by use of TRIzol reagent (Ambion) and were further purified using an RNeasy miniprep kit (Qiagen). RNA concentrations were determined using a Nano Drop 1000 spectrophotometer (Thermo Scientific). The isolated RNAs were treated with DNase I (Turbo DNA-free kit; Ambion) and reverse transcribed using random primers to produce cDNAs (Invitrogen). cDNAs were amplified with an Applied Biosystems ABI 7500 real-time PCR system, using Sybr green PCR master mix (Applied Biosystems). Gene-specific primers (see Table S2 in the supplemental material) were designed to amplify 120- to 150-bp fragments of each gene. Relative expression values (R) were calculated by using the formula R = 2−(ΔCT target − ΔCT endogenous control), where CT is the fractional threshold cycle (25). The constitutively expressed F. novicida rpoB gene was used as an endogenous control, and a value of 1 was used to standardize the gene expression of the wild-type strain.
Microarray expression analysis.
A whole-genome NimbleGen microarray was developed for multiple Francisella subspecies, using shared probes for common genes as well as unique probes for subspecies-specific genes. Included were probes representing the F. novicida U112 genome, with a maximum of 6 probes/gene. RNA was isolated from bacteria and purified as described above. Double-stranded cDNA was generated and labeled with Cy3 according to the NimbleGen gene expression array protocol, using a SuperScript double-stranded cDNA synthesis kit (Invitrogen) and a DNA labeling kit (NimbleGen). Microarray results were analyzed using ArrayStar software (DNASTAR).
Intramacrophage survival assay.
F. novicida strains were used to infect the J774.1 macrophage cell line (ATCC) at a multiplicity of infection (MOI) of 10:1. The intramacrophage survival assay was performed as previously described (22).
Mouse virulence assays.
Groups of five female 4- to 6-week-old BALB/c mice (Charles River Laboratories) were inoculated intranasally with F. novicida strains in 20 μl PBS. Actual bacterial numbers delivered were determined by plate counts of the inocula, and an approximate 50% lethal dose (LD50) was determined based on the number of surviving mice. Mice were monitored for 30 days after infection. The Institutional Animal Care and Use Committee (IACUC) at UT San Antonio approved all animal procedures.
Microarray data accession number.
Microarray data have been submitted to the NCBI Gene Expression Omnibus (GEO) in accordance with MIAME standards, under accession number GSE39647.
RESULTS
F. novicida contains a cdGMP modulatory gene cluster that is absent in F. tularensis subsp. tularensis.
A gene cluster (FTN0451 to FTN0456 [FTN0451-FTN0456]) was identified in the F. novicida U112 genome that contains six genes that are missing from the F. tularensis subsp. tularensis Schu S4 genome (Fig. 1A and B). The genes are organized into two divergently transcribed operons, FTN0451 and FTN0452 to FTN0456. FTN0451 and FTN0456 encode composite GGDEF-EAL proteins (i.e., containing both DGC and PDE domains) (Fig. 1C), and because evidence below indicates that these proteins modulate cdGMP levels, we have renamed the genes cdgA (cyclic diguanylate modulatory protein A) and cdgB, respectively. CdgA and CdgB also contain PAS domains, which are found in bacterial signaling proteins. CdgA and CdgB are predicted to be localized to the cytoplasm, because they lack obvious signal peptides or transmembrane domains.
Fig 1.
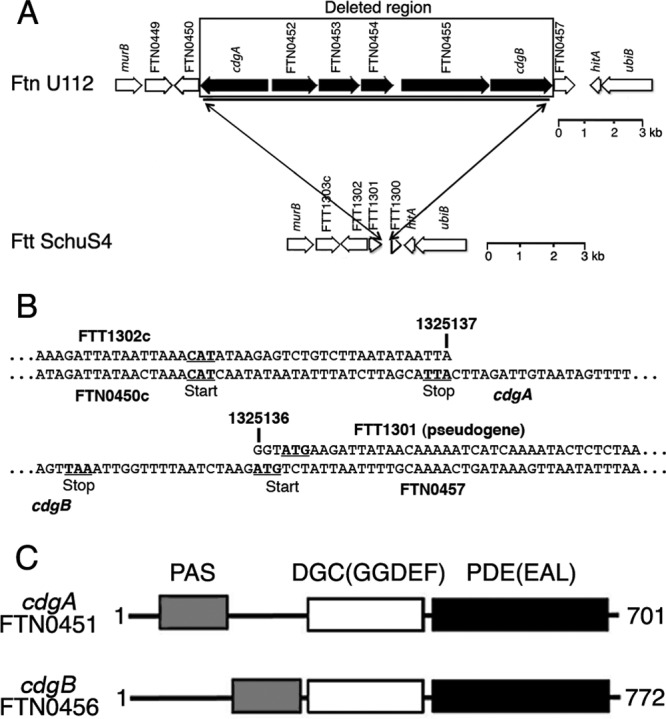
F. novicida FTN0451-FTN0456 gene cluster. (A and B) The 11.3-kbp F. novicida FTN0451-FTN0456 gene cluster is missing in F. tularensis subsp. tularensis. In panel B, the numbers refer to the nucleotides within the Schu S4 genome (GenBank accession no. NC_006570), which are contiguous and missing all coding sequences corresponding to FTN0451 to FTN0456 in F. novicida U112 (GenBank accession no. CP000439). (C) CdgA and CdgB are composite GGDEF-EAL (DGC-PDE) proteins; PAS is a signal transduction domain.
Alignment of the amino acid (aa) sequences of the DGC domains of CdgA and CdgB with those of well-characterized DGCs, i.e., VCA0956 from V. cholerae (45, 46), PleD from Caulobacter crescentus (16, 31), and WspR from Pseudomonas aeruginosa (14, 18, 28), revealed the conservation of critical residues required for nucleotide binding and catalytic activity (see Fig. S2 in the supplemental material). CdgA has a Y substitution (Y331) at a position that normally has a histidine residue important for indirect binding of substrate, as well as substitutions in residues identified as primary and secondary inhibitory sites (48) important for cdGMP inhibition of activity. These changes may correlate with our evidence below suggesting that CdgA is predominantly a PDE rather than a DGC.
Alignment of the aa sequences of the PDE domains of CdgA and CdgB with those of well-characterized PDEs, i.e., VieA from V. cholerae (42, 45, 46), BlrP1 from Klebsiella pneumonia (1), and YkuI from Bacillus subtilis (27), revealed conservation of the critical aspartate and most of the loop 6 residues found in active PDE domains (32), as well as conserved glutamate and lysine residues required for phosphodiesterase activity (33, 37) (see Fig. S3 in the supplemental material). Interestingly, CdgB has an S substitution for T in the highly conserved loop 6 motif DDFGTG (32), as well as a Q substitution for a highly conserved E residue (37), which may correlate with our evidence below suggesting that this protein is primarily a DGC rather than a PDE.
The FTN0452 gene encodes a conserved hypothetical protein with a domain of unknown function (DUF814); however, HHPred analysis (40) identified potential structural homology with DGC domains in the C terminus (aa 314 to 466). Alignment of the FTN0452 protein's aa sequence with those of other DGC domains revealed that many of the critical residues for DGC activity are lacking, including the entire GGDEF motif (see Fig. S4 in the supplemental material), so we suggest that this protein may represent a novel cdGMP binding protein. The FTN0453 gene encodes a protein with homology to the cellulose synthase superfamily (cd06423), likely involved in the synthesis of polysaccharide chains (e.g., cellulose and chitin). The FTN0454 gene encodes a protein with homology to membrane fusion proteins of multidrug resistance pumps (COG1566). The FTN0455 gene encodes a protein with a CheR methyltransferase domain and a CheB methylesterase domain, normally involved in methylation and demethylation, respectively, of chemoreceptors in motile bacteria.
Elevated cdGMP stimulates F. novicida growth yield.
To determine the role of cdGMP signaling in F. novicida, the entire FTN0451-FTN0456 gene cluster was removed and replaced with an erythromycin resistance gene, resulting in strain KKF457 [Δ(FTN0451-FTN0456)]. This strain lacks any DGC- or PDE-encoding genes, similar to F. tularensis subsp. tularensis, and thus can be considered to have no cdGMP. In order to elevate cdGMP levels in this strain, we introduced a well-characterized heterologous DGC from V. cholerae, namely, VCA0956 (45, 46), expressed from an F. novicida promoter. VCA0956 expression is known to elevate intracellular cdGMP levels (4, 45).
The KKF457 strain lacking cdGMP exhibited a growth curve similar to that of the wild-type U112 strain in CDM (Fig. 2A). However, high levels of cdGMP in KKF457 (expressing VCA0956) stimulated a higher OD600 in stationary phase, and this was caused by an ∼100-fold larger number of bacterial cells as determined by plate counts. The growth rate and decline of the strain with high cdGMP appeared similar to those of the strain with low cdGMP, and thus it appears that the stationary-phase yield (cell number) is specifically stimulated by elevated cdGMP. We also introduced a plasmid expressing CdgB into KKF457, and the resulting strain exhibited a growth curve similar to that of KKF457 expressing VCA0956 (Fig. 2A), suggesting that CdgB has predominantly DGC activity (similar to VCA0956) that is able to elevate cdGMP levels in this strain.
Fig 2.
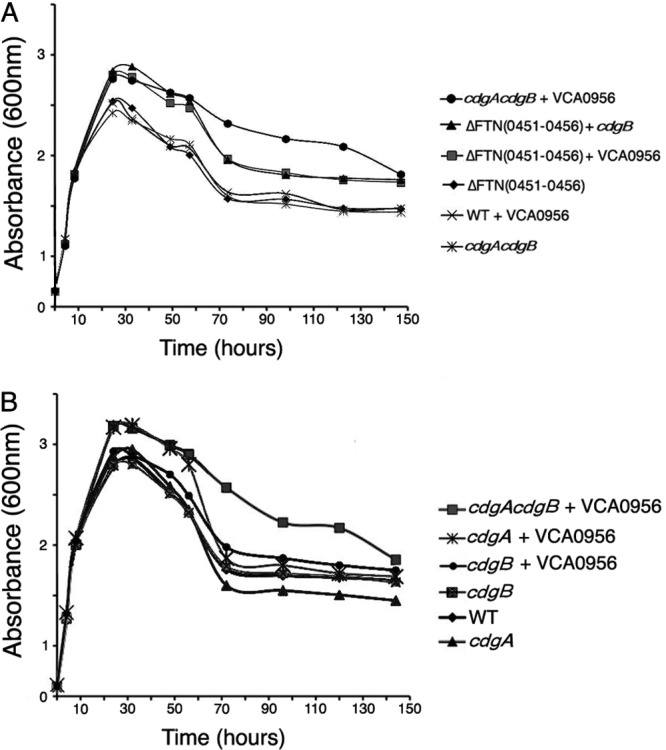
Growth rates and yields of F. novicida with high and low cdGMP levels. (A and B) F. novicida growth curves. F. novicida strains U112 (WT), KKF457 [ΔFTN(0451–0456)], KKF535 (cdgB), KKF538 (cdgA), and KKF539 (cdgA cdgB), carrying plasmid pKEK1524 (+ cdgB) or pKEK1525 (+ VCA0956), were grown at 37°C in CDM.
Interestingly, expression of VCA0956 in the wild-type U112 strain did not stimulate the stationary-phase yield (Fig. 2A), indicating lower cdGMP levels and suggesting that a gene(s) within the FTN0451-FTN0456 cluster has PDE activity capable of reducing cdGMP levels. We created an F. novicida strain lacking only cdgA and cdgB, rather than the entire gene cluster, and this strain grew similarly to the wild-type and KKF457 strains. However, expression of VCA0956 in the cdgA cdgB strain stimulated an increase in growth yield similar to that with expression of VCA0956 in KKF457 (Fig. 2A). These results indicate high levels of cdGMP in this strain, consistent with PDE activity associated with CdgA and/or CdgB.
We further investigated cdGMP stimulation of the growth yield by measuring the growth of F. novicida strains lacking CdgA and/or CdgB (Fig. 2B). As seen previously, the F. novicida cdgA cdgB strain grew similarly to the wild-type U112 strain, but elevating cdGMP in this strain by expression of VCA0956 stimulated the growth yield. However, expression of VCA0956 stimulated the growth yield only in the cdgA mutant, not the cdgB mutant, consistent with CdgA possessing predominantly PDE activity that reduces cdGMP levels. A lack of both CdgA and CdgB was required to see sustained elevated cell counts during the decline phase (>70 h), suggesting that CdgB may possess some PDE activity during this phase. Unfortunately, despite repeated attempts, we were unable to construct a plasmid that expresses CdgA, suggesting that there may be some toxic effect associated with overexpression of this protein. Determination of viable cell counts by plating of samples from several time points during the growth curve analysis revealed that bacterial cell numbers were correlated with optical density. Cumulatively, our results indicate that cdGMP levels are modulated mostly positively by CdgB and mostly negatively by CdgA and that elevated cdGMP levels stimulate the F. novicida growth yield.
Elevated cdGMP stimulates F. novicida biofilm formation.
F. novicida forms biofilms on abiotic and chitinous surfaces (10, 26), and an orphan response regulator, QseB, has been shown to be important for biofilm formation. To investigate the contribution of cdGMP levels to F. novicida biofilm formation, we measured biofilms formed by various F. novicida strains with high or low cdGMP levels (Fig. 3A). As shown previously (10, 26), the wild-type U112 strain formed biofilms on borosilicate glass tubes. A strain lacking the entire FTN0451-FTN0456 cluster (KKF457) or a strain lacking just CdgA and CdgB (cdgA cdgB strain) was defective for biofilm formation, demonstrating that cdGMP signaling contributes to F. novicida biofilm formation. Expression of VCA0956 or CdgB in either KKF457 or the cdgA cdgB strain stimulated more biofilm formation than that of the wild-type strain, demonstrating that high cdGMP levels stimulate F. novicida biofilm formation. These results also further confirm the DGC activity associated with CdgB. Finally, biofilm formation of the individual cdgA and cdgB mutants revealed that a lack of CdgA actually stimulates biofilm formation, whereas a lack of CdgB decreases biofilm formation. These results are consistent with CdgA possessing predominantly PDE activity and CdgB possessing predominantly DGC activity.
Fig 3.
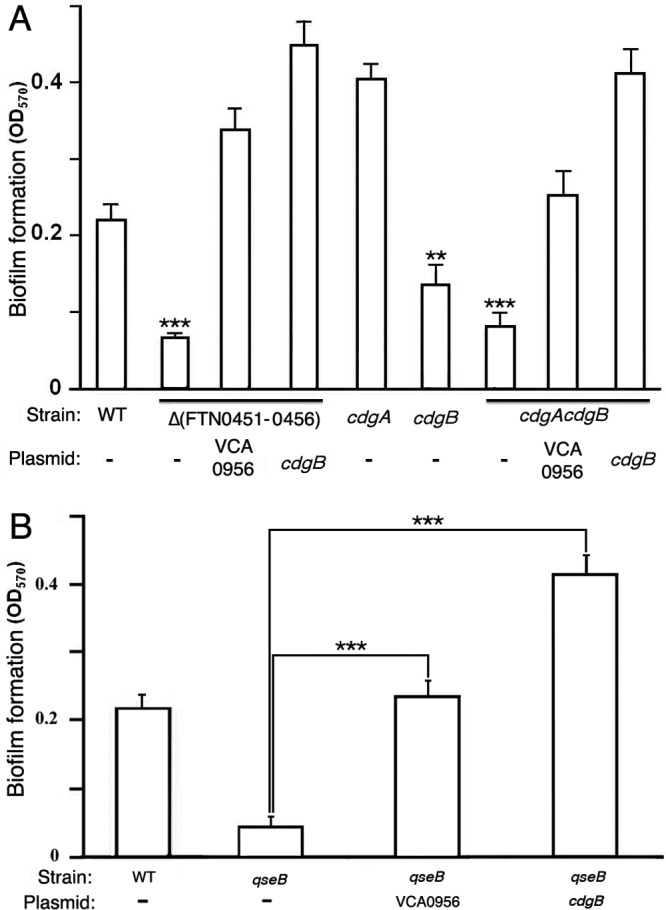
cdGMP stimulates F. novicida biofilm formation. (A) F. novicida strains U112 (WT), KKF457 [Δ(FTN0451-0456)], KKF535 (cdgB), KKF538 (cdgA), and KKF539 (cdgA cdgB), either with no plasmid (−) or carrying plasmid pKEK1524 (cdgB) or pKEK1525 (VCA0956), were assayed for biofilm formation as described in Materials and Methods. **, P < 0.005; ***, P < 0.0005. (B) F. novicida strains U112 (WT) and KKF552 (qseB), either with no plasmid (−) or carrying plasmid pKEK1524 (cdgB) or pKEK1525 (VCA0956), were assayed for biofilm formation. ***, P < 0.0005.
Further confirmation of the DGC activity of CdgB was obtained by transforming V. cholerae strain A1552 with the plasmid expressing CdgB and then measuring biofilm formation (see Fig. S5 in the supplemental material). It has been shown previously that elevated cdGMP levels stimulate V. cholerae biofilm formation (3, 5). As anticipated, A1552 expressing either VCA0956 or CdgB stimulated enhanced biofilm formation, providing additional evidence that CdgB possesses DGC activity.
QseB regulates cdgA and cdgB transcription.
Durham-Colleran et al. (10) previously demonstrated a role for the response regulator QseB in F. novicida biofilm formation. To determine if QseB regulates biofilm formation by regulating cdGMP levels, a qseB mutant was transformed with plasmids expressing either VCA0956 or CdgB, and biofilm formation was measured (Fig. 3B). As shown previously, the F. novicida qseB mutant was defective for biofilm formation compared to the wild-type U112 strain. However, elevation of cdGMP levels in the qseB strain by expression of VCA0956 stimulated biofilm formation, demonstrating that the requirement for QseB can be bypassed by elevated cdGMP levels. This suggests that QseB functions to elevate cdGMP levels in F. novicida and thereby stimulates biofilm formation. Expression of CdgB in the qseB mutant stimulated biofilm formation to a greater extent, further confirming the DGC activity associated with CdgB.
To determine if QseB regulates transcription of cdgA and/or cdgB, qRT-PCR measuring the cdgA and cdgB transcripts was performed with the wild-type and qseB mutant strains (Fig. 4). There was a significant decrease in cdgA and cdgB transcripts in the qseB mutant compared to the wild-type strain, indicating that QseB positively influences cdgA and cdgB transcription.
Fig 4.
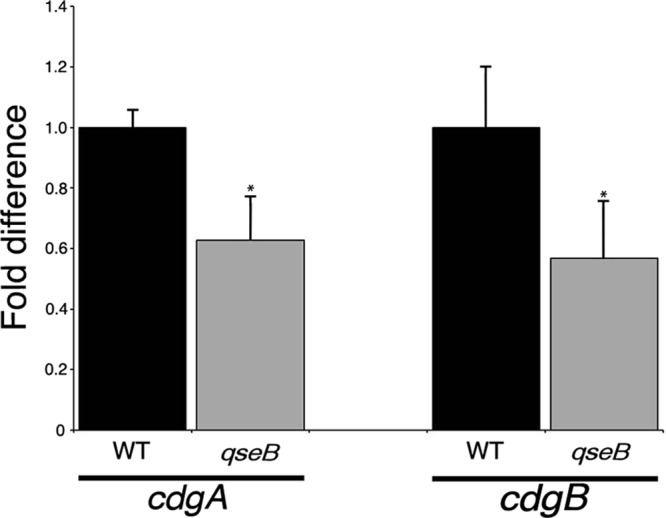
QseB regulates cdgB transcription. F. novicida strains U112 (WT) and KKF552 (qseB) were grown in CDM, and mRNA abundances for cdgA and cdgB were determined by quantitative reverse transcription-PCR (see Materials and Methods for details). Results shown represent three biological replicates consisting of three technical replicates for each strain. *, P < 0.05.
High cdGMP levels decrease F. novicida intramacrophage replication.
Intramacrophage replication is an essential component of Francisella virulence. The ability of F. novicida strains with low and high cdGMP levels to replicate intracellularly was measured in the mouse macrophage-like J774 cell line (Fig. 5). Strains lacking cdGMP [cdgA cdgB or Δ(FTN0451-FTN0456) strain] replicated similarly to the wild-type U112 strain within the J774 cells, indicating that low cdGMP levels do not inhibit intramacrophage replication. In contrast, strains with high cdGMP levels, achieved by expression of VCA0956 or CdgB in the cdgA cdgB or Δ(FTN0451-FTN0456) strain, were defective for intramacrophage replication, exhibiting an approximately 2-log reduction in CFU at 48 h. These results demonstrate that high cdGMP levels inhibit intramacrophage replication.
Fig 5.
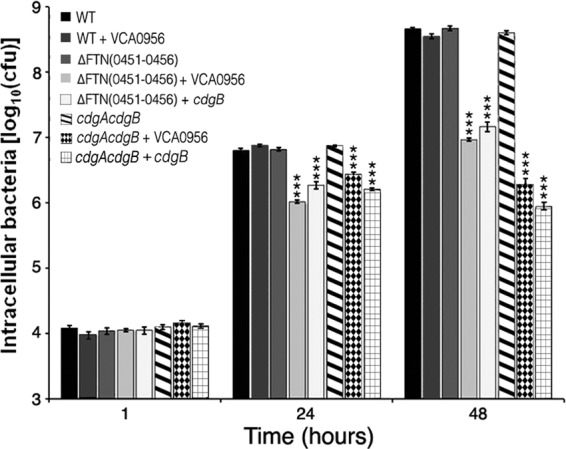
cdGMP inhibits F. novicida intramacrophage replication. F. novicida strains U112 (WT), KKF457 [ΔFTN(0451–0456)], and KKF539 (cdgA cdgB), either without a plasmid or carrying plasmid pKEK1524 (cdgB) or pKEK1525 (VCA0956), were inoculated at an MOI of ∼10:1 into J774 cells, and intracellular bacteria were enumerated at 1, 24, and 48 h (see Materials and Methods for further details). The assay was performed in triplicate. *, P < 0.0005.
Interestingly, expression of VCA0956 in the wild-type U112 strain had no effect on intramacrophage replication. We hypothesized that this was likely because of the PDE activity associated with cdgA reducing intracellular cdGMP levels (see above). To confirm that the PDE activity of CdgA inhibits the elevation of cdGMP levels, we tested the ability of F. novicida cdgA and cdgB strains expressing the DGC VCA0956 to replicate within the J774 cell line (see Fig. S6 in the supplemental material). The cdgA strain expressing VCA0956 was defective for intramacrophage replication at 48 h postinfection, whereas the cdgB strain expressing VCA0956 replicated similarly to the wild-type strain. These results are consistent with CdgA possessing a PDE activity that inhibits elevated cdGMP levels.
High cdGMP levels decrease F. novicida virulence in mice.
To determine the effect of cdGMP levels on F. novicida virulence via the pulmonary route, F. novicida strains with low and high cdGMP levels were inoculated into BALB/c mice intranasally at various inocula, and survival was monitored. Strains lacking either cdgA, cdgB, or both cdgA and cdgB exhibited similarly high levels of virulence as the wild-type strain at relatively low inocula (60 to 115 CFU) (see Fig. S7 in the supplemental material), demonstrating that low levels of cdGMP are not inhibitory to F. novicida virulence. Likewise, the KKF457 strain lacking the entire cdGMP modulatory cluster [Δ(FTN0451-FTN0456)] showed a similarly high level of virulence as the wild-type strain at a relatively low inoculum (110 CFU), with a slightly delayed time to death (Fig. 6), providing further evidence that low levels of cdGMP have little inhibitory effect on F. novicida virulence.
Fig 6.
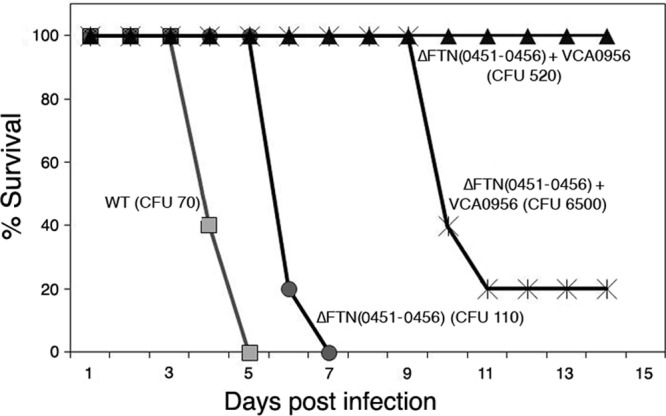
cdGMP inhibits F. novicida virulence in mice. F. novicida strains U112 (WT) and KKF457 [ΔFTN(0451–0456)], either without plasmid or carrying plasmid pKEK1525 (+ VCA0956), were inoculated intranasally into groups of 5 female BALB/c mice at the indicated inocula, and mouse survival was monitored.
In contrast, the high cdGMP levels achieved by expression of VCA0956 in KKF457 attenuated F. novicida virulence (Fig. 6). Strain KKF457 expressing VCA0956 was unable to cause the death of any mice at 520 CFU, whereas 4 of 5 mice succumbed to disease when inoculated with 6,500 CFU of this strain. This result suggested that the apparent LD50 of F. novicida with high cdGMP levels is ∼5,000 CFU. However, because the elevated cdGMP levels were driven by the plasmid-expressed VCA0956 protein in this strain, we tested F. novicida colonies recovered from the spleens of mice that succumbed to infection and discovered that the bacteria recovered from diseased animals had lost the VCA0956 plasmid. This result indicates that the LD50 of F. novicida with high cdGMP levels is likely >6,500 CFU. Since the LD50 of the wild-type F. novicida strain is <10 CFU by the pulmonary route, this demonstrates the strong attenuation of virulence (>650-fold) caused by high cdGMP levels.
cdGMP modulates F. novicida-specific genes.
Our cumulative data indicate that cdGMP positively affects growth yield and biofilm formation and negatively affects intramacrophage replication and virulence in F. novicida. In order to determine which gene(s) modulated by cdGMP might account for these phenotypes, we performed whole-genome microarray transcriptome analysis of F. novicida strains with either a low cdGMP (KKF457) or high (KKF457 expressing VCA0956) level of cdGMP. We utilized a custom-designed NimbleGen array that incorporated probes for F. novicida-specific genes, in addition to the genes shared with other F. tularensis strains, in order to accurately identify transcription changes in these F. novicida strains. The experiment was performed with 2 biological replicates grown in CDM (Table 1).
Table 1.
cdGMP-regulated genes in F. novicida
| Gene | Annotation of gene product | Fold change | Presence in strain Schuh S4 |
|---|---|---|---|
| Upregulated genes | |||
| FTN0010 | Phage terminase | +2.091 | Missing |
| FTN0286 | Transposase | +2.047 | Missing |
| FTN0324 | Anti-sigma factor antagonist | +2.030 | FTT1612 |
| FTN0627 | ChiA (chitinase) | +2.238 | Functional ChiA (44- and 66-aa deletions) |
| FTN0793 | Hypothetical protein | +2.145 | FTT1078; 78-aa C-terminal truncation |
| FTN0879 | Zinc-iron permease | +3.689 | Pseudogene |
| FTN0880 | Cobalamin synthase | +4.379 | CobS |
| FTN1328 | Trehalase | +2.298 | Pseudogene |
| FTN1485 | Chitin-binding protein | +3.438 | Split: FTT1576-FTT1577 |
| FTN1744 | ChiB (chitinase) | +2.983 | ChiB with 124-aa N-terminal truncation |
| FTN1758 | Hypothetical protein | +2.407 | Missing |
| Downregulated genes | |||
| FTN0313 | Acetyltransferase | −2.234 | Pseudogene |
| FTN0714 | Conserved hypothetical protein | −3.299 | Missing |
| FTN0715 | Conserved hypothetical protein | −2.472 | FTT0742; 594-aa C-terminal truncation |
| FTN1021 | Conserved hypothetical protein | −2.423 | FTT0662; 43-aa C-terminal truncation |
| FTN1101 | Lipoprotein, unknown function | −2.721 | Missing |
| FTN1102 | Hypothetical protein | −2.070 | Missing |
| FTN1103 | Conserved hypothetical protein | −2.438 | Missing |
| FTN1104 | Hypothetical protein | −2.293 | FTT1122 (only N-terminal 66 aa) |
Surprisingly few genes showed 2-fold or greater changes in transcript levels between the F. novicida strain with high cdGMP and the strain with low/no cdGMP. This analysis identified 11 genes that were upregulated and 8 genes that were downregulated by high levels of cdGMP (Table 1). Margolis et al. (26) previously demonstrated that the chitinases ChiA and ChiB contribute to F. novicida biofilm formation on chitinous surfaces. Our microarray results indicate that transcription of chiA, chiB, and the gene for a chitin-binding protein (FTN1485) is positively regulated by cdGMP, consistent with the role of these genes in biofilm formation. Other genes positively regulated by cdGMP encode cobalamin synthase, trehalase, zinc-iron permease, and hypothetical proteins.
Interestingly, Margolis et al. (26) identified FTN0714 as a secreted protein that contributes to F. novicida biofilm formation; our microarray analysis identified FTN0714 as being down- rather than upregulated by cdGMP. The microarray analysis provides a snapshot of transcription at a distinct time point, whereas biofilm formation is a dynamic developmental process, so it may be that cdGMP represses some biofilm components that are otherwise upregulated at different time points during this process. Virtually all the other genes downregulated by cdGMP encode proteins of unknown function.
Given that the FTN0451-FTN0456 gene cluster that modulates cdGMP levels is found specifically in F. novicida and absent in F. tularensis subsp. tularensis, it was of interest to determine whether the cdGMP-modulated genes are present in F. tularensis subsp. tularensis. Interestingly, only three of the genes positively regulated by cdGMP in F. novicida (the FTT1612 gene, cobS, and chiA) appear to be fully functional in F. tularensis subsp. tularensis (Table 1); all 8 other genes positively regulated by cdGMP appear to be compromised (pseudogenes, genes with truncations in the coding sequence, or split genes) or completely missing in F. tularensis subsp. tularensis. Likewise, all the genes downregulated by cdGMP in F. novicida are compromised or missing in F. tularensis subsp. tularensis. The ChiA protein from F. tularensis subsp. tularensis Schu S4 contains 44-aa and 66-aa deletions within the C terminus in comparison to the ChiA protein from F. novicida, but Chandler et al. (8) showed that this protein is expressed and functional in Schu S4.
We measured transcript levels of the chiA and chiB genes in the F. novicida strains with high cdGMP [Δ(FTN0451-FTN0456) strain plus VCA0956] or no cdGMP [Δ(FTN0451-FTN0456) strain], as well as the wild-type U112 strain, by qRT-PCR to validate the results of the microarray analysis (Fig. 7). Transcript levels of chiB were significantly lower in the strain with no cdGMP than in the strain with high cdGMP, demonstrating that high cdGMP levels positively regulate chiB transcription. The cdGMP-mediated increase in chiA transcript levels was not statistically significant. Interestingly, the transcript levels for both chiA and chiB were elevated in the KKF457 strain with both high and low cdGMP levels compared to transcript levels in the wild-type strain. We hypothesize that one of the genes in the FTN0451-FTN0456 gene cluster (absent in the KKF457 strain) negatively regulates chiA and chiB transcription.
Fig 7.
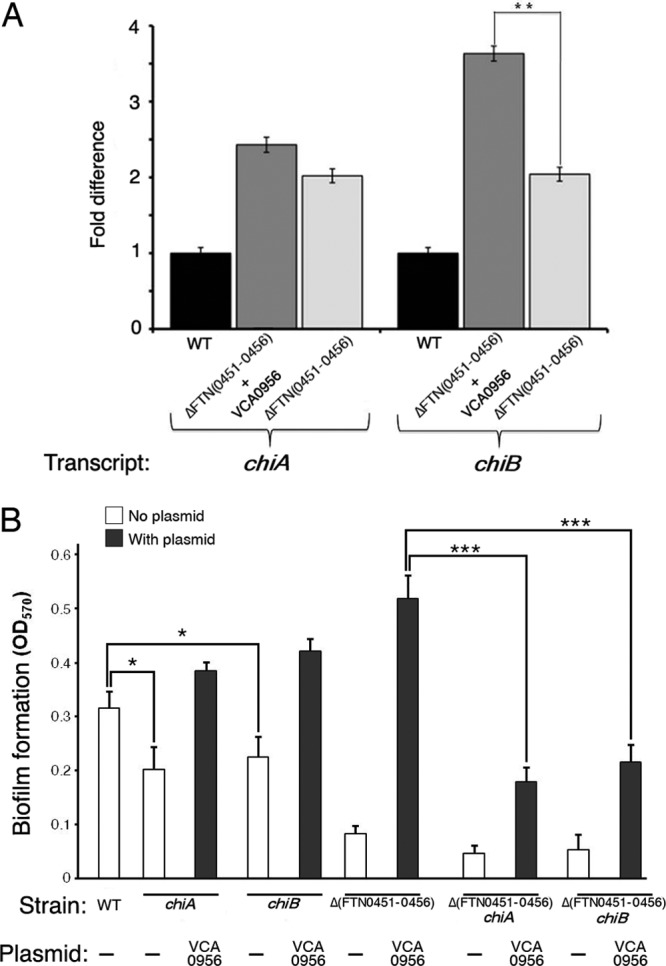
cdGMP controls chiB transcription, which enhances biofilm formation. (A) F. novicida strains U112 (WT) and KKF457 [ΔFTN(0451–0456)], either without plasmid or carrying plasmid pKEK1525 (+ VCA0956), were grown in CDM, and mRNA abundances for chiA and chiB were determined by quantitative reverse transcription-PCR (see Materials and Methods for details). Results shown represent three biological replicates consisting of three technical replicates for each strain. **, P < 0.005. (B) F. novicida strains U112 (WT), KKF555 (chiA), KKF556 (chiB), KKF457 [Δ(FTN0451-0456)], KKF557 [Δ(FTN0451-0456) chiA], and KKF558 [Δ(FTN0451-0456) chiB], either with no plasmid (−) or carrying plasmid pKEK1525 (VCA0956), were assayed for biofilm formation as described in Materials and Methods. *, P < 0.05; ***, P < 0.0005.
To determine if cdGMP-modulated chiA and/or chiB transcription contributes to F. novicida biofilm development on glass surfaces, we first created F. novicida strains containing either chiA or chiB mutations in both the wild-type and Δ(FTN0451-FTN0456) backgrounds. The heterologous DGC VCA0956 was then introduced into these strains, and biofilm formation was assessed (Fig. 7B). Disruption of either chiA or chiB in the wild-type background reduced the ability of F. novicida to form biofilms. As expected, the Δ(FTN0451-FTN0456) strain combined with the chiA or chiB mutations led to significant decreases of biofilm formation. Importantly, raising cdGMP levels in these strains by the introduction of VCA0956 could not fully complement biofilm formation. The Δ(FTN0451-FTN0456) chiA and Δ(FTN0451-FTN0456) chiB strains with VCA0956 showed significant decreases in biofilm formation in comparison to the parental Δ(FTN0451-FTN0456) strain with VCA0956. Thus, cdGMP-dependent regulation of ChiB facilitates biofilm formation on glass surfaces.
DISCUSSION
Although F. novicida and F. tularensis subsp. tularensis are closely related, they exhibit vastly different virulence capabilities in humans, despite both sharing the critical secretion-related virulence genes localized in the FPI. F. tularensis subsp. tularensis is highly virulent in humans, which has resulted in this bacterium being considered a select biothreat agent requiring high-level biocontainment. In contrast, F. novicida is considered avirulent in healthy humans and is exempt from select agent regulations due to its low level of virulence. Comparison of their genomes reveals that in contrast to the F. novicida genome, the F. tularensis subsp. tularensis genome is undergoing decay and is replete with insertion sequence (IS) elements, genomic rearrangements, and pseudogenes (21). This suggests that elements present in the F. novicida genome but absent from the F. tularensis subsp. tularensis genome may have been lost from the F. tularensis subsp. tularensis genome to enhance its virulence (35).
We showed here that the FTN0451-FTN0456 gene cluster, which is found in F. novicida but missing from F. tularensis subsp. tularensis, modulates cdGMP levels. This gene cluster is also missing from the human-pathogenic organism F. tularensis subsp. holarctica. High cdGMP levels positively influence F. novicida biofilm formation and negatively influence virulence, a paradigm seen in other bacterial pathogens that utilize cdGMP signaling (e.g., V. cholerae [45, 46]). cdGMP levels are increased by DGC-containing proteins and decreased by PDE-containing proteins; F. novicida encodes two proteins within the FTN0451-FTN0456 cluster, i.e., CdgA and CdgB, that have both DGC and PDE domains.
Several lines of evidence suggest that CdgB is predominantly a DGC: (i) expression of CdgB stimulates V. cholerae biofilm formation, (ii) expression of CdgB in the absence of CdgA stimulates F. novicida biofilm formation as well as growth yields, and (iii) expression of CdgB in the absence of CdgA inhibits F. novicida intramacrophage growth and virulence in mice. All of these phenotypes could be replicated by replacing CdgB with VCA0956, a well-characterized DGC with no PDE domain. We suggest that the PDE domain of CdgB has very little or no activity and that this may be due to a substitution in the well-conserved loop 6 region; substitutions in this region have been shown to differentiate active from inactive PDE domains (32).
Although we were unable to clone cdgA into an expression vector, for unknown reasons, our evidence still suggests that CdgA is predominantly a PDE. The cdGMP-mediated effects on F. novicida caused by CdgB (increased growth yields and intramacrophage replication) occurred only in the absence of CdgA, indicating that CdgA has an opposing enzymatic activity to that of CdgB. Also, the cdgA mutant exhibited enhanced biofilm formation compared to the wild-type strain, suggesting higher cdGMP levels when CdgA is absent. Finally, elevating cdGMP in F. novicida by the introduction of the heterologous DGC VCA0956 reduced intramacrophage growth only in the absence of CdgA, suggesting that CdgA's PDE activity opposes the activity of VCA0956. The DGC domain of CdgA contains a substitution in a highly conserved histidine residue as well as in inhibitory sites, suggesting that this domain may have decreased or absent enzymatic activity.
Stimulation of biofilm formation is normally associated with persistence within the environment, and the F. novicida genome appears to have a more robust repertoire of genes, which may indicate it is more adept at environmental persistence than F. tularensis subsp. tularensis. Microarray analysis revealed that cdGMP positively regulates genes known to be involved in F. novicida biofilm formation, including the chitinase genes chiA and chiB (26). We confirmed by qRT-PCR that the F. novicida chiB gene is positively regulated by cdGMP. Margolis et al. showed that ChiA and ChiB are important for F. novicida biofilm formation on chitinous surfaces that would be encountered outside the mammalian host. In the current study, we found that ChiA and ChiB also contribute to F. novicida biofilm formation on glass surfaces. In a study by Reif et al., the chiA and/or chiB F. novicida mutant showed no defects in the ability to colonize ticks (34), so perhaps the encoded chitinases facilitate biofilm formation in the environment rather than in insect/arthropod vectors.
ChiA from F. tularensis subsp. tularensis is missing two internal portions, corresponding to aa 663 to 706 and aa 747 to 812 of F. novicida ChiA. Despite this difference, the ChiA proteins from both F. novicida and F. tularensis subsp. tularensis Schu S4 have been shown to possess similar endochitinase activities (8) and to be expressed in both strains. In contrast, F. tularensis subsp. tularensis ChiB is missing the N-terminal 124 aa found in F. novicida ChiB and could not be detected in lysates of F. tularensis subsp. tularensis, unlike F. novicida ChiB. Most of the other cdGMP-regulated genes in F. novicida are either missing or compromised in some manner in F. tularensis subsp. tularensis. This suggests that after the loss of the cdGMP regulatory region, F. tularensis subsp. tularensis is in the process of losing the genes regulated by cdGMP.
The loss of the cdGMP regulon in F. tularensis subsp. tularensis indicates that the absence of cdGMP provides an advantage to this bacterium. We showed that high levels of cdGMP inhibit F. novicida intramacrophage replication and virulence in mice. We suggest that the cdGMP-mediated inhibition of intramacrophage replication and virulence may have driven the loss of the cdGMP regulon in F. tularensis subsp. tularensis. From the microarray analysis, there was no obvious downregulation of known virulence genes in the strain with high cdGMP, and thus the inhibition of intramacrophage replication may be the result of inhibition of a novel virulence gene(s) or, alternately, enhanced expression of a gene(s) that is deleterious to virulence.
Our results have uncovered a regulatory cascade controlling biofilm formation in F. novicida (Fig. 8). The orphan response regulator QseB positively regulates cdgB transcription, presumably upon phosphorylation in response to environmental conditions. As discussed above, CdgB is predominantly a DGC that raises cdGMP levels within the cell. cdGMP, in turn, positively stimulates the transcription of genes that enhance biofilm formation, including chiB. Elevated cdGMP levels also inhibit intramacrophage growth and virulence by an unknown mechanism. QseB is also present in F. tularensis subsp. tularensis Schu S4, but the downstream components are missing or defective. Given that most of the cdGMP-regulated genes in F. novicida are missing or defective in F. tularensis subsp. tularensis, it will be interesting to determine whether induction of cdGMP synthesis in this bacterium will have similar deleterious effects on intramacrophage growth and virulence.
Fig 8.
cdGMP signaling cascade in F. novicida.
Supplementary Material
ACKNOWLEDGMENT
This study was supported by NIH grant PO1 AI57986 to K.E.K.
Footnotes
Published ahead of print 17 September 2012
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1. Barends TR, et al. 2009. Structure and mechanism of a bacterial light-regulated cyclic nucleotide phosphodiesterase. Nature 459:1015–1018 [DOI] [PubMed] [Google Scholar]
- 2. Barker JR, et al. 2009. The Francisella tularensis pathogenicity island encodes a secretion system that is required for phagosome escape and virulence. Mol. Microbiol. 74:1459–1470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Beyhan S, Odell LS, Yildiz FH. 2008. Identification and characterization of cyclic diguanylate signalling systems controlling rugosity in Vibrio cholerae. J. Bacteriol. 190:7392–7405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Beyhan S, Tischler AD, Camilli A, Yildiz FH. 2006. Transcriptome and phenotypic responses of Vibrio cholerae to increased cyclic di-GMP level. J. Bacteriol. 188:3600–3613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Beyhan S, Yildiz FH. 2007. Smooth to rugose phase variation in Vibrio cholerae can be mediated by a single nucleotide change that targets c-di-GMP signalling pathway. Mol. Microbiol. 63:995–1007 [DOI] [PubMed] [Google Scholar]
- 6. Chamberlain RE. 1965. Evaluation of live tularemia vaccine prepared in a chemically defined medium. Appl. Microbiol. 13:232–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Champion MD, et al. 2009. Comparative genomic characterization of Francisella tularensis strains belonging to low and high virulence subspecies. PLoS Pathog. 5:e1000459 doi:10.1371/journal.ppat.1000459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chandler JC, Molins CR, Petersen JM, Belisle JT. 2011. Differential chitinase activity and production within Francisella species, subspecies, and subpopulations. J. Bacteriol. 193:3265–3275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dennis DT, et al. 2001. Tularemia as a biological weapon: medical and public health management. JAMA 285:2763–2773 [DOI] [PubMed] [Google Scholar]
- 10. Durham-Colleran MW, Verhoeven AB, van Hoek ML. 2010. Francisella novicida forms in vitro biofilms mediated by an orphan response regulator. Microb. Ecol. 59:457–465 [DOI] [PubMed] [Google Scholar]
- 11. Eigelsbach HT, Downs CM. 1961. Prophylactic effectiveness of live and killed tularemia vaccines. I. Production of vaccine and evaluation in the white mouse and guinea pig. J. Immunol. 87:415–425 [PubMed] [Google Scholar]
- 12. Ellis J, Oyston PC, Green M, Titball RW. 2002. Tularemia. Clin. Microbiol. Rev. 15:631–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gallagher LA, et al. 2007. A comprehensive transposon mutant library of Francisella novicida, a bioweapon surrogate. Proc. Natl. Acad. Sci. U. S. A. 104:1009–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Guvener ZT, Harwood CS. 2007. Subcellular location characteristics of the Pseudomonas aeruginosa GGDEF protein, WspR, indicate that it produces cyclic-di-GMP in response to growth on surfaces. Mol. Microbiol. 66:1459–1473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hanahan D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:577–580 [DOI] [PubMed] [Google Scholar]
- 16. Hecht GB, Newton A. 1995. Identification of a novel response regulator required for the swarmer-to-stalked-cell transition in Caulobacter crescentus. J. Bacteriol. 177:6223–6229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hengge R. 2009. Principles of c-di-GMP signalling in bacteria. Nat. Rev. Microbiol. 7:263–273 [DOI] [PubMed] [Google Scholar]
- 18. Hickman JW, Tifrea DF, Harwood CS. 2005. A chemosensory system that regulates biofilm formation through modulation of cyclic diguanylate levels. Proc. Natl. Acad. Sci. U. S. A. 102:14422–14427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jenal U. 2004. Cyclic di-guanosine-monophosphate comes of age: a novel secondary messenger involved in modulating cell surface structures in bacteria? Curr. Opin. Microbiol. 7:185–191 [DOI] [PubMed] [Google Scholar]
- 20. Jenal U, Malone J. 2006. Mechanisms of cyclic-di-GMP signaling in bacteria. Annu. Rev. Genet. 40:385–407 [DOI] [PubMed] [Google Scholar]
- 21. Larsson P, et al. 2005. The complete genome sequence of Francisella tularensis, the causative agent of tularemia. Nat. Genet. 37:153–159 [DOI] [PubMed] [Google Scholar]
- 22. Lauriano CM, Barker JR, Nano FE, Arulanandam BP, Klose KE. 2003. Allelic exchange in Francisella tularensis using PCR products. FEMS Microbiol. Lett. 229:195–202 [DOI] [PubMed] [Google Scholar]
- 23. Lauriano CM, et al. 2004. MglA regulates transcription of virulence factors necessary for Francisella tularensis intraamoebae and intramacrophage survival. Proc. Natl. Acad. Sci. U. S. A. 101:4246–4249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Liu J, Zogaj X, Barker JR, Klose KE. 2007. Construction of targeted insertion mutations in Francisella tularensis subsp. novicida. Biotechniques 43:487–490 [DOI] [PubMed] [Google Scholar]
- 25. Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods 25:402–408 [DOI] [PubMed] [Google Scholar]
- 26. Margolis JJ, et al. 2010. Contributions of Francisella tularensis subsp. novicida chitinases and Sec secretion system to biofilm formation on chitin. Appl. Environ. Microbiol. 76:596–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Minasov G, et al. 2009. Crystal structures of YkuI and its complex with second messenger cyclic di-GMP suggest catalytic mechanism of phosphodiester bond cleavage by EAL domains. J. Biol. Chem. 284:13174–13184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Moscoso JA, Mikkelsen H, Heeb S, Williams P, Filloux A. 2011. The Pseudomonas aeruginosa sensor RetS switches type III and type VI secretion via c-di-GMP signalling. Environ. Microbiol. 13:3128–3138 [DOI] [PubMed] [Google Scholar]
- 29. Nano FE, et al. 2004. A Francisella tularensis pathogenicity island required for intramacrophage growth. J. Bacteriol. 186:6430–6436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Oyston PC. 2008. Francisella tularensis: unravelling the secrets of an intracellular pathogen. J. Med. Microbiol. 57:921–930 [DOI] [PubMed] [Google Scholar]
- 31. Paul R, et al. 2004. Cell cycle-dependent dynamic localization of a bacterial response regulator with a novel di-guanylate cyclase output domain. Genes Dev. 18:715–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rao F, et al. 2009. The functional role of a conserved loop in EAL domain-based cyclic di-GMP-specific phosphodiesterase. J. Bacteriol. 191:4722–4731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rao F, Yang Y, Qi Y, Liang ZX. 2008. Catalytic mechanism of cyclic di-GMP-specific phosphodiesterase: a study of the EAL domain-containing RocR from Pseudomonas aeruginosa. J. Bacteriol. 190:3622–3631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Reif KE, et al. 2011. Dermacentor andersoni transmission of Francisella tularensis subsp. novicida reflects bacterial colonization, dissemination, and replication coordinated with tick feeding. Infect. Immun. 79:4941–4946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rohmer L, et al. 2007. Comparison of Francisella tularensis genomes reveals evolutionary events associated with the emergence of human pathogenic strains. Genome Biol. 8:R102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Saslaw S, Carlisle HN. 1961. Studies with tularemia vaccines in volunteers. IV. Brucella aggiutinins in vaccinated and nonvaccinated volunteers challenged with Pasteurella tularensis. Am. J. Med. Sci. 242:166–172 [DOI] [PubMed] [Google Scholar]
- 37. Schmidt AJ, Ryjenkov DA, Gomelsky M. 2005. The ubiquitous protein domain EAL is a cyclic diguanylate-specific phosphodiesterase: enzymatically active and inactive EAL domains. J. Bacteriol. 187:4774–4781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Shen H, Chen W, Conlan JW. 2004. Susceptibility of various mouse strains to systemically- or aerosol-initiated tularemia by virulent type A Francisella tularensis before and after immunization with the attenuated live vaccine strain of the pathogen. Vaccine 22:2116–2121 [DOI] [PubMed] [Google Scholar]
- 39. Siddaramappa S, et al. 2011. Common ancestry and novel genetic traits of Francisella novicida-like isolates from North America and Australia as revealed by comparative genomic analyses. Appl. Environ. Microbiol. 77:5110–5122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Soding J, Biegert A, Lupas AN. 2005. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 33:W244–W248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tamayo R, Pratt JT, Camilli A. 2007. Roles of cyclic diguanylate in the regulation of bacterial pathogenesis. Annu. Rev. Microbiol. 61:131–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Tamayo R, Tischler AD, Camilli A. 2005. The EAL domain protein VieA is a cyclic diguanylate phosphodiesterase. J. Biol. Chem. 280:33324–33330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tarnvik A, Berglund L. 2003. Tularaemia. Eur. Respir. J. 21:361–373 [DOI] [PubMed] [Google Scholar]
- 44. Tarnvik A, Ericsson M, Golovliov I, Sandstrom G, Sjostedt A. 1996. Orchestration of the protective immune response to intracellular bacteria: Francisella tularensis as a model organism. FEMS Immunol. Med. Microbiol. 13:221–225 [DOI] [PubMed] [Google Scholar]
- 45. Tischler AD, Camilli A. 2004. Cyclic diguanylate (c-di-GMP) regulates Vibrio cholerae biofilm formation. Mol. Microbiol. 53:857–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tischler AD, Camilli A. 2005. Cyclic diguanylate regulates Vibrio cholerae virulence gene expression. Infect. Immun. 73:5873–5882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Titball RW, Petrosino JF. 2007. Francisella tularensis genomics and proteomics. Ann. N. Y. Acad. Sci. 1105:98–121 [DOI] [PubMed] [Google Scholar]
- 48. Yang CY, et al. 2011. The structure and inhibition of a GGDEF diguanylate cyclase complexed with (c-di-GMP)(2) at the active site. Acta Crystallogr. D Biol. Crystallogr. 67:997–1008 [DOI] [PubMed] [Google Scholar]
- 49. Yildiz FH, Schoolnik GK. 1999. Vibrio cholerae O1 El Tor: identification of a gene cluster required for the rugose colony type, exopolysaccharide production, chlorine resistance, and biofilm formation. Proc. Natl. Acad. Sci. U. S. A. 96:4028–4033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zogaj X, Chakraborty S, Liu J, Thanassi DG, Klose KE. 2008. Characterization of the Francisella tularensis subsp. novicida type IV pilus. Microbiology 154:2139–2150 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


