Abstract
Approximately 1% of those infected with HIV-1 develop broad and potent serum cross-neutralizing antibody activities. It is unknown whether or not the development of such immune responses affects the replication of the contemporaneous autologous virus. Here, we defined a pathway of autologous viral escape from contemporaneous potent and broad serum neutralizing antibodies developed by an elite HIV-1-positive (HIV-1+) neutralizer. These antibodies potently neutralize diverse isolates from different clades and target primarily the CD4-binding site (CD4-BS) of the viral envelope glycoprotein. Viral escape required mutations in the viral envelope glycoprotein which limited the accessibility of the CD4-binding site to the autologous broadly neutralizing anti-CD4-BS antibodies but which allowed the virus to infect cells by utilizing CD4 receptors on their surface. The acquisition of neutralization resistance, however, resulted in reduced cell entry potential and slower viral replication kinetics. Our results indicate that in vivo escape from autologous broadly neutralizing antibodies exacts fitness costs to HIV-1.
INTRODUCTION
In the months following infection with HIV-1, the majority of infected subjects mount a neutralizing antibody (NAb) response against the autologous virus but not against heterologous viruses (29, 30). HIV-1, however, is capable of continuously evolving to escape this autologous NAb response (24, 30). Later during the course of infection, approximately 15% of HIV-1-infected subjects develop neutralizing antibody responses against diverse heterologous HIV-1 viruses, and approximately 1% of HIV-positive (HIV+) subjects develop exquisitely potent and broad neutralizing antibody responses (4, 8, 13, 19, 21, 23, 32). In several well-characterized cases of such “elite neutralizers,” the cross-neutralizing activities of sera were determined to be due to NAbs that target a small number of structurally conserved regions of env (4, 6, 13, 17, 19, 21, 32), including the CD4-binding site (CD4-BS) (1, 7, 35, 44, 46), and conserved elements of the variable loops V2 and V3, with the later specificities being glycan dependent (27, 39).
The breadth of serum cross-neutralizing antibody activities in HIV-1+ subjects positively correlates with plasma viral load (8, 23, 31, 32), and recently a number of broadly neutralizing monoclonal antibodies (MAbs) have been isolated from HIV+ patients, including elite neutralizers (27, 35, 39, 44, 47). Several studies have reported on viral escape from the autologous neutralizing antibody responses that do not display broad neutralization (29, 30, 41), and different in vitro methodologies have been used to define the pathways of viral escape from such responses. Alanine-scanning mutagenesis of the envelope glycoprotein has been used to identify those amino acids that are critical for the recognition by broadly neutralizing antibodies (bNAbs); however, these studies do not report on whether the virus actually modifies those amino acids in order to escape in vivo. Also, in vitro escape neutralization experiments with known bNAbs are beset with limitations, since the MAb/virus pairs examined have been isolated from different HIV+ subjects at different times of the HIV/AIDS epidemic (28, 45). Thus, the mechanisms by which the contemporaneous virus escapes the action of the autologous broadly neutralizing antibodies in vivo remain largely unknown. It is also unknown whether viral escape from the autologous broadly neutralizing antibodies leads to the emergence and predominance of viral variants with reduced cell entry and replication potential.
We previously reported that the plasma of HIV-1+ subject VC10042 (infected with a clade B virus) exhibits extraordinary broad and potent neutralizing activity that targets primarily the CD4-BS (32) (see additional results in Fig. S1 in the supplemental material). Although plasma and purified plasma IgG from VC10042 potently neutralizes a variety of different viruses from diverse genetic clades (Fig. 1A; see Fig. S1 in the supplemental material), this subject maintained plasma viral loads of greater than 104 viral RNA copies per ml during the period of observation, as well as stable CD4+ and CD8+ T cell numbers (over 2 decades of infection in the absence of antiretroviral treatment) (see Fig. S2 in the supplemental material). Thus, the study of subjects such as VC10042 not only provides information on the pathways of viral escape from potent and broadly neutralizing antibodies during natural HIV-1infection but also informs on any adverse effects that such an escape may have on HIV-1 in vivo.
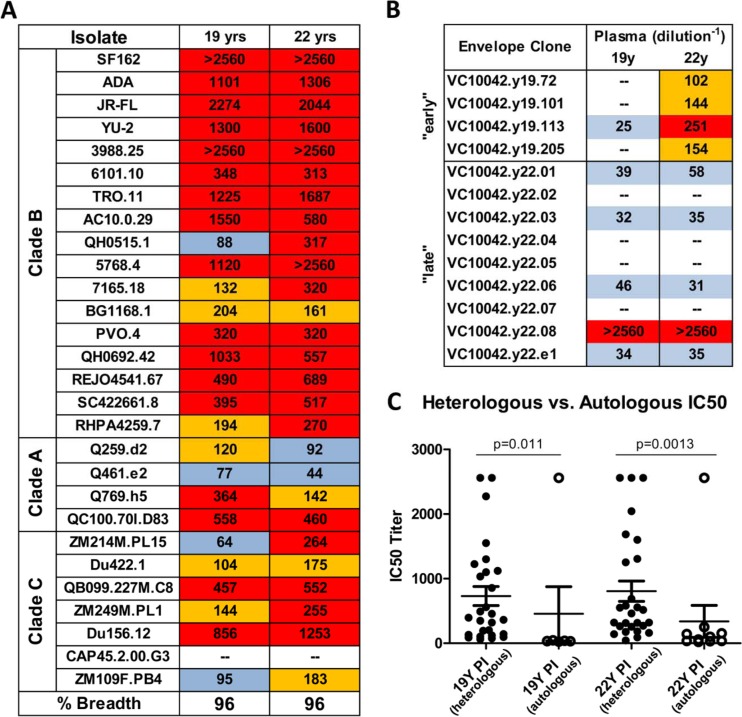
Fig 1.
Neutralization breadth and potency of plasma from subject VC10042. (A) Heterologous neutralization profile of plasma from VC10042 against isolates from clades A, B, and C. (B) Neutralization of autologous plasma viral clones isolated at 19 and 22 years postinfection. The numbers reported are the IC50, the plasma dilution at which viral cell entry was inhibited by 50% in the TZM-bl pseudovirus assay. IC50 values for the clade B and two clade A (Q259.d2 and Q461.e2) viruses were reported previously (32). (C) Statistical comparison of the plasma IC50 values against the heterologous and autologous viruses. The IC50 values against the heterologous and autologous isolates for plasma collected at 19 and 22 years of infection were compared by a Mann-Whitney U test.
In the present study, we defined the neutralization phenotype of contemporaneous circulating viral env clones to autologous NAbs and known bNAbs and the receptor usage of these clones and defined the molecular basis for viral escape. Our study reveals that in the presence of potent and broad anti-CD4-BS antibody responses, viral escape variants emerge, which retain their abilities to utilize the CD4 and CCR5 receptors. However, this adaptation is associated with defects in virus cell entry potential and leads to slower viral replication kinetics.
MATERIALS AND METHODS
Subject.
Subject VC10042 is an HIV+ African American male who acquired HIV-1 clade B in 1984 and is part of the Vanderbilt cohort, as previously reported (32). Plasma samples were obtained for multiple time points beginning 19 years postseroconversion and as late as 22.5 years postseroconversion. At the time of sample collection, VC10042 was coinfected with both hepatitis B and hepatitis C viruses, both of which were acquired at an unknown time. During the period of observation, subject VC10042 maintained steady CD4+ T cell counts, was without antiretroviral therapy, and had no AIDS-defining illness. Plasma samples were heat inactivated at 54°C for 1 h and centrifuged at 17,000 × g for 10 min prior to use in any assay.
Envelope amplifications.
Viral RNA was isolated from 250 μl of plasma using the QIAamp Ultrasense virus kit (Invitrogen, Carlsbad, CA). Viral RNA was reverse transcribed with Superscript III reverse transcriptase (Invitrogen) using oligo(dT) primers. Viral envelope sequences were amplified from cDNA by PCR as previously described (18). Briefly, cDNA template was amplified by nested PCR using the following primer pairs, as described by Li et al. (18): outer sense primer 5′-TAGAGCCCTGGAAGCATCCAGGAAG-3′, nucleotides (nt) 5852 to 5876; outer antisense primer 5′-TTGCTACTTGTGATTGCTCCATGT-3′, nt 8912 to 8935; inner sense primer 5′-GATCAAGCTTTAGGCATCTCCTATGGCAGGAAGAAG-3′, nt 5957 to 5982; and inner antisense primer 5′-AGCTGGATCCGTCTCGAGATACTGCTCCCACCC-3′, nt 8881 to 8903. High-fidelity Pfx DNA polymerase (Invitrogen) was used in all PCRs. The entire env-rev cassette was cloned into the pcDNA3.1-TOPO (Invitrogen) directional cloning vector for expression. Single clones were verified by BigDye cycle sequencing.
Pseudovirus production and virus coreceptor usage assays.
Pseudoviruses were produced by cotransfecting 293T cells plated at a density of 5 × 105 cells/ml with 8 μg of the pNL4-3.Rev−.Env−.Luc+ HIV backbone and 4 μg of env plasmid DNA, using Genejuice (Novagen) as a transfection reagent. After incubation at 37°C for 3 days, the clarified cell supernatant was tested for p24 content and for functional entry in TZM-bl cells. Virus coreceptor usage studies were performed in U87 cells bearing the CD4 receptor with and without coreceptors, as well as coreceptors alone without the CD4 receptor. Briefly, cell-free virus (including positive-control virus) was titrated, added to U87 cells, and left for 3 days. The medium was then removed, the cells were lysed in SteadyLite Plus luciferase reagent (Perkin-Elmer), and the cell-associated luciferase was read on a Fluoroskan luminometer. Viral entry is reported as the relative light units (RLU) associated with the cell lysates at a single viral input (10 ng p24).
Neutralization assays.
Neutralization assays were performed using VC10042 envelope clone variants in the TZM-bl cell-based pseudovirus assay, as previously described (32). Briefly, plasma samples were titrated 2-fold from 1:20 to 1:2,560 and were incubated for 90 min at 37°C in the presence of single-round competent virions. The virus-plasma mixture was added to TZM-bl cells that were plated at a density of 3 × 103 cells per well in a 96-well plate 24 h prior to inoculation; 72 h later, the cell supernatants were removed and 100 μl of SteadyLite Plus (Perkin-Elmer) was added to the cells of each well. The cell-associated luciferase activity (luminescence) for each well was determined on a Fluoroskan luminometer (Thermo Scientific). The neutralization values reported here are the 50% inhibitory concentrations (IC50s), i.e., the plasma dilution at which viral entry was inhibited by 50% compared to that in the absence of plasma. When MAbs were used instead of plasma, the IC50 values reported are the antibody concentrations (μg/ml) that inhibit infection by 50%. MAb VRC01 was kindly provided by J. Mascola (Vaccine Research Center [VRC], NIH), and MAbs 4E10, 2F5, 2G12, and b12 were purchased from Polymun Scientific (Vienna, Austria).
Generation of reversion mutations in VC10042 env clones.
Reversion mutations were introduced into the autologous env gp160 by mutagenesis. Mutagenesis reactions were carried out using the QuikChange mutagenesis kit (Stratagene). High-pressure liquid chromatography (HPLC)-purified complementary primer pairs coding for the desired point mutations were added to buffer, deoxynucleoside triphosphates (dNTPs), and DNA polymerase and denatured at 94°C for 10 min, followed by 19 cycles with the following conditions: 94°C for 30 s, 58°C for 30 s, and 72°C for 4 min. The reaction products were digested with DpnI for 1 h at 37°C to remove parental plasmids and were transformed into DH5α cells (Invitrogen). Single plasmid DNA clones were verified by DNA sequencing and then used to produce gp160 variants as described below. For testing as a pseudovirus in neutralization and entry assays, three amino acid changes were introduced into clone VC10042.y22.05: H364S, R373M, and N386D. The new plasmids bearing the reversion mutations were used to produce pseudovirus for the entry and neutralization assays as described above. R373M was also introduced into seven VC10042 env clones for use in replication competence assays (described below): VC10042.y19.72, VC10042.y19.205, VC10042.y22.01, VC10042.y22.03, VC10042.y22.04, VC10042.y22.05, and VC10042.y22.07.
Replication competence viral kinetic assay.
Replication-competent HIV-1 vectors were constructed by inserting the entire gp160 sequence from the autologous env clones amplified from the plasma of VC10042 into the TN6-green fluorescent protein (GFP) replication-competent backbone (26). Each DNA construct was transfected into 293T cells in the presence of Opti-MEM (Invitrogen, Carlsbad, CA) and Genejuice (EMD, Gibbstown, NJ) for 4 h, followed by a complete change of medium to complete Dulbecco modified Eagle medium (DMEM) (Cellgro, Manassas, VA) supplemented with 10% fetal bovine serum (FBS). Cell supernatants were collected after 3 days, centrifuged at 16,000 × g for 10 min, and stored at −80C. Cell-free supernatants containing virus were assayed for p24 content by p24 capture enzyme-linked immunosorbent assay (ELISA) according to the manufacturer's instructions (Zeptometrix, Buffalo, NY). Naïve human peripheral blood mononuclear cells (PBMCs) were isolated from fresh whole blood by centrifugation through a Ficoll gradient and resuspension in complete RPMI medium (Cellgro, Manassas, VA) supplemented with 10% fetal bovine serum (Invitrogen, Carlsbad CA), 10 ng/ml interleukin-2 (IL-2) (Sigma-Aldrich, St. Louis, MO), and 3 μg/ml phytohemagglutinin (PHA) for 3 days. A total of 15 × 106 activated PBMCs were then inoculated with replication-competent viruses for 3 h at 37°C. At that time, the cells were resuspended in fresh complete RPMI at 3 × 106 cells/ml. Supernatant samples were collected at 3- to 4-day intervals, with 50% of the growth medium being collected and replaced with fresh complete RPMI. At several sampling points, the viability of the culture was assessed by trypan blue dye exclusion staining to ensure that the cultures remained healthy. All samples were assayed for p24 content by p24 capture ELISA (Zeptometrix).
RESULTS
env sequence analysis of the circulating virus.
To better define the replication potential and neutralization phenotype of the virus circulating in VC10042 in the presence of potent and broad anti-CD4-BS NAbs, we amplified viral env sequences from plasma samples isolated from two time points that plasma samples were available from this subject: approximately 19 years postinfection (ypi) and approximately 22 ypi. From 19 ypi, more than 200 clones were amplified and sequenced, while from 22 ypi, more than 70 clones were amplified and sequenced. Surprisingly, the amino acid sequences of the gp120 envelope subunit from 19 ypi were highly homogeneous, varying by only 0.2% (see Fig. S3 in the supplemental material), while higher heterogeneity (5%) was evident for the gp41 envelope subunit. Five unique clonotypes were identified by phylogenetic analysis, with a diversity of 2.14%. In contrast, a high degree of variation in amino acid sequences (27.4% in gp120 and 11.3% in gp41) and lengths of the variable loops V1, V2, V4, and V5, as well as in the number of N-linked glycosylation sites (between 23 and 35), was evident in the sequences from 22 ypi (see Fig. S4 in the supplemental material). Nine unique clonotypes were identified, with a diversity of 21.93%.
These observations indicate that the contemporaneous virus was continuously evolving in the presence of broadly neutralizing antibodies and were suggestive of viral efforts to escape the action of the autologous neutralizing antibody response. Phylogenetic analysis of the env clones circulating in VC10042 confirmed that the virus circulating in that subject was clade B, separated by a node with bootstrap support of 100 (see Fig. S5 in the supplemental material), although the VC10042 env clones form distinct clusters separated from heterologous clade B sequences by a node with a bootstrap value of 65. Similarly, the 19-ypi clones cluster at a node with bootstrap support of 92, while the majority of the 22-ypi clones cluster separately from the 19-ypi clones (bootstrap support of 90).
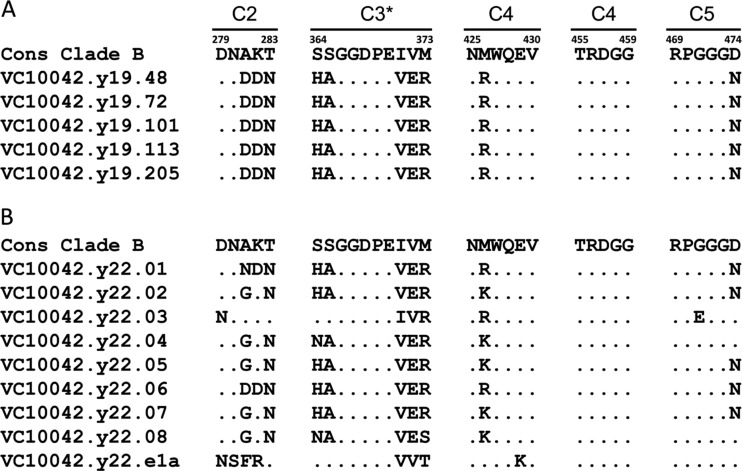
Significant variation in the 19- and 22-ypi sequences was evident among residues that are known to contact the CD4 receptor, compared to the consensus clade B env sequence (Fig. 2). This variability was located predominantly at both “ends” of the CD4 binding loop (denoted by an asterisk in Fig. 2), as well the portion of the C2 region of gp120 that partially constitutes the CD4-BS (Fig. 2). In contrast, very little variability was observed within the center of the CD4 binding loop (residues 366 to 370), in agreement with recently reported heterogeneity analysis within the CD4 binding loop of clade B isolates (45). The regions of the CD4-BS that are formed by the three-dimensional association of elements of the C4 and C5 regions of gp120 were highly conserved among both the 19- and 22-ypi sequences. It should be noted, however, that many of the predominant amino acid changes within the CD4 binding loop are present in both the 19- and 22-ypi env sequences (Fig. 2), indicating that those changes likely occurred sometime before 19 years postinfection.
Fig 2.
Sequence variation of the VC10042 env clones within the five discontinuous peptide regions that constitute the CD4-BS. (A) Representative sequences isolated from plasma collected at 19 years postinfection. (B) Representative sequences isolated from plasma collected at 22 years postinfection. Sequence identities are denoted by periods, while variable residues are listed by amino acid symbol. The C3 region constitutes the CD4 binding loop, residues 364 to 373, and is denoted by an asterisk. The amino acid residue position, in HXB2 numbering, is listed at the beginning and end of each peptide sequence.
The virus requires CD4 and CCR5 for cell entry.
Recently it was reported that HIV-1 has the ability to escape from serum broadly neutralizing anti-CD4-BS antibodies without altering its ability to utilize the CD4 receptor for entry (43). Because of the extensive sequence changes within the CD4-BS of the VC10042 viral env (Fig. 2) and because the broad neutralizing activity of this subject's plasma was due almost exclusively to anti-CD4-BS antibodies (32) (see Fig. S1 in the supplemental material), we tested whether viruses isolated from VC10042 had a reduced or altered ability to interact with CD4 molecules on the surface of target cells. To define the receptor/coreceptor requirement of the virus circulating in VC10042, we generated single-round competent virions expressing the 19- and 22-ypi env clones and examined their relative abilities to enter U87 cells expressing various combinations of CD4, CCR2, CCR5, and CXCR4. We found that the virions were unable to enter cells that expressed either CCR5 or CXCR4 but not CD4, CD4 alone, CD4 and CXCR4, or CD4 and CCR2 (Table 1). However, because we isolated viral clones exclusively from peripheral blood samples, we cannot rule out the presence of viruses in other compartments that utilize CD4 and CXCR4. In contrast, all clones were competent to enter cells expressing CD4 and CCR5, indicating that the circulating clones utilize CD4 and CCR5 for entry into target cells. Thus, despite the above-mentioned changes within the CD4-BS, none of these clones acquired the ability to efficiently mediate entry through CD4-independent means.
Table 1.
Entry of individual plasma viral clones into U87 cells
| ypia | Envelope clone | Viral entry (RLU) into U87 cells at 3 ng p24 with the following receptor/coreceptor combinationb: |
||||
|---|---|---|---|---|---|---|
| CD4 | CD4/CCR5 | CD4/CXCR4 | CCR5 | CXCR4 | ||
| 19 | VC10042.y19.48 | — | 79,530 | — | — | — |
| VC10042.y19.72 | — | 4,834,620 | — | — | — | |
| VC10042.y19.101 | — | 3,544,620 | — | — | — | |
| VC10042.y19.113 | — | 746,125 | — | — | — | |
| VC10042.y19.205 | — | 5,224,620 | — | — | — | |
| 22 | VC10042.y22.01 | — | 1,428,341 | — | — | — |
| VC10042.y22.02 | — | 2,052,091 | — | — | — | |
| VC10042.y22.03 | — | 1,888,341 | — | — | — | |
| VC10042.y22.05 | — | 2,192,091 | — | — | — | |
| VC10042.y22.06 | — | 1,062,591 | — | — | — | |
| VC10042.y22.07 | — | 3,079,591 | — | — | — | |
| VC10042.y22.08 | — | 155,461 | — | — | — | |
| VC10042.y22.e1 | — | 190,561 | — | — | — | |
Clones were isolated from plasma collected at the indicated year postinfection.
Each clone was tested for its ability to enter into U87 cell lines bearing the indicated combinations of CD4 receptors and the CCR5 and CXCR4 coreceptors. Values reported are the relative light units (RLU) obtained by viral entry into the target cell. —, no entry was observed above background, relative to negative controls (RLU recorded with viral particles not expressing HIV env).
Neutralization phenotype of the circulating virus.
We next determined the neutralization phenotype of the viral clones circulating in VC10042 using autologous plasma collected at 19 and 22 years postinfection. The majority of clones were either completely resistant or only moderately susceptible to neutralization by the contemporaneous plasma (Fig. 1B). In fact, plasma from VC10042 was significantly more effective (Fig. 1C) in neutralizing a wide range of heterologous viruses from multiple clades than the autologous virus (Fig. 1A and B). One clone circulating at 22 years postinfection, VC10042.y22.08, was highly susceptible to neutralization by the autologous plasma (Fig. 1B), more so than most heterologous isolates evaluated. Interestingly, the heterologous isolates that the plasma neutralized as potently as VC10042.y22.08 also are known to be very sensitive to anti-CD4-BS NAbs (18, 44). It is unclear how this env clone persisted at 22 ypi in the presence of potent autologous neutralizing antibodies. It is possible that such clones persist by utilizing cell-to-cell transmission to evade potent anti-CD4-BS bNAbs, as has been recently described (2). Transmission from cell to cell would allow such clones to persist and bud virus into the plasma, regardless of their sensitivity to the autologous plasma antibody response.
Since the viral variants circulating in VC10042 were significantly less susceptible to the autologous NAbs than heterologous viruses, we examined whether the VC10042 viral clones are more generally resistant to antibody-mediated neutralization. To this end, we examined the neutralization susceptibilities of the above-described viral clones to anti-HIV antibodies that neutralize diverse tier 1, 2, and 3 viruses from several HIV-1 clades (Tables 2 and 3)(16, 44, 47). Consistent with the requirement for CD4-mediated entry, all clones were sensitive to neutralization by CD4-IgG(2) (Table 2). Nine bNAbs to the CD4-BS were tested, including several recently described anti-CD4-BS NAbs termed highly active agonistic CD4-BS antibodies (HAADs) (35). With a few exceptions (discussed below) the viral clones were resistant to these antibodies. Clones VC10042.y22.08 and VC10042.y22.e1 were susceptible to b12 and VRC01, and clone VC10042.y22.03 was susceptible to MAbs 12A12 and 3BNC117. We also examined MAb NIH45-46G54W for its ability to neutralize these viral variants. MAb NIH45-46G54W is derived from MAb NIH45-46 (isolated from the same subject as MAb VRC01) by inserting a tryptophan at position 54 of its CDR2 region (7). The G54W modification in the CDRH2 allows NIH45-46G54W to penetrate into the Phe43 cavity of the CD4-BS on gp120, far deeper than NIH45-46 or VRC01, and engage not only the bridging sheet and the outer domain of gp120 but also the inner domain of gp120. MAb NIH45-46G54W displayed a different neutralization profile against the VC10042 viral clones than the other anti-CD4-BS MAbs tested. Interestingly, NIH45-46G54W neutralized the same VC10042 viral clones as the autologous plasma (all the 19-ypi clones and 22-ypi clones VC10042.y22.01, -03, -06, -08, and -e1), while it failed to neutralize those clones that the autologous plasma failed to neutralize (VC10042.y22.02, -04, -05, and -0.7).
Table 2.
Neutralization phenotype of VC10042 pseudotyped env clones with autologous plasma and with monoclonal antibodies that target the CD4-BS [as well as IgG(2)-CD4]
| ypia | Envelope clone | TZM-bl neutralization assay IC50b with the following: |
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Plasma (dilution−1) |
MAb (μg/ml) |
|||||||||||||||||
| NIH45-46G54W | b12 | VRC01 | HAAD antibodies |
IgG(2)-CD4 | Anticore antibodies |
|||||||||||||
| 19y | 22y | 12A12 | 12A21 | 3BNC55 | 3BNC117 | 1NC9 | 1B2530 | 1-74 | 4-277 | 4-252 | 4-77 | 1-752 | ||||||
| 19 | VC10042.y19.72 | — | 102 | 7.19 | — | — | — | — | — | — | — | — | 1.35 | — | — | — | — | — |
| VC10042.y19.101 | — | 144 | 7.9 | — | — | — | — | — | — | — | — | 1.5 | — | — | — | — | — | |
| VC10042.y19.113 | 25 | 251 | 5.22 | — | — | — | — | — | — | — | — | >0.03 | — | — | — | — | — | |
| VC10042.y19.205 | — | 154 | 5.62 | — | — | — | — | — | — | — | — | 1.09 | — | — | — | — | — | |
| 22 | VC10042.y22.01 | 39 | 58 | 3.55 | — | — | — | — | — | — | — | — | 0.15 | — | — | — | — | — |
| VC10042.y22.02 | — | — | — | — | — | — | — | — | — | — | — | 0.22 | — | — | — | — | — | |
| VC10042.y22.03 | 32 | 35 | 3.02 | — | — | 15.6 | — | — | 2.82 | — | — | 3.52 | — | — | — | — | — | |
| VC10042.y22.04 | — | — | — | — | — | — | — | — | — | — | — | 0.28 | — | — | — | — | — | |
| VC10042.y22.05 | — | 20 | — | — | — | — | — | — | — | — | — | 0.18 | — | — | — | — | — | |
| VC10042.y22.06 | 46 | 31 | 12.2 | — | — | — | — | — | — | — | — | 1.59 | — | — | — | — | — | |
| VC10042.y22.07 | — | — | — | — | — | — | — | — | — | — | — | 0.31 | — | — | — | — | — | |
| VC10042.y22.08 | >2,560 | >2,560 | 3.21 | 0.11 | 0.018 | — | — | — | — | — | — | <0.03 | — | — | — | — | — | |
| VC10042.y22.e1 | 34 | 35 | 4.64 | 0.32 | 0.026 | — | — | — | — | — | — | 0.33 | — | <0.2 | <0.2 | <0.2 | <0.2 | |
Clones were isolated from plasma collected at the indicated year postinfection.
The values reported are the IC50, i.e., the antibody concentration (μg/ml) or plasma dilution at which viral entry is inhibited by 50%. HAAD, highly active agonistic anti-CD4-BS antibody and dependent on the D368R mutation. Anticore, recognizing elements of the CD4-BS but not affected by the D368R mutations. —, no neutralizing activity was observed.
Table 3.
Neutralization phenotype of VC10042 pseudotyped env clones with known monoclonal antibodies that target a wide array of different epitopes on the HIV-1 envelope
| ypia | Envelope clone | TZM-bl neutralization assay IC50b(μg/ml) with MAb: |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| PG9 | PG16 | 2G12 | 447-52D | 2F5 | 4E10 | CD4 induced |
|||||
| 3BC176 | 3BC315 | 17b | X5 | ||||||||
| 19 | VC10042.y19.72 | — | — | — | — | — | 6.06 | 0.33 | 1.11 | — | — |
| VC10042.y19.101 | — | — | — | — | — | 9.69 | 0.51 | 2.15 | — | — | |
| VC10042.y19.113 | — | — | — | — | — | 7.22 | <0.195 | 1.9 | — | — | |
| VC10042.y19.205 | — | — | — | — | — | 9.19 | <0.195 | 1.47 | — | — | |
| 22 | VC10042.y22.01 | — | — | — | — | — | — | — | — | — | — |
| VC10042.y22.02 | — | — | — | — | — | 15.9 | — | — | — | — | |
| VC10042.y22.03 | — | — | — | — | — | — | 1.9 | — | — | — | |
| VC10042.y22.04 | — | — | — | — | — | — | — | — | — | — | |
| VC10042.y22.05 | — | — | — | — | — | 13.6 | 3.08 | — | — | — | |
| VC10042.y22.06 | — | — | — | — | — | — | — | — | — | — | |
| VC10042.y22.07 | — | — | — | — | — | — | — | — | — | — | |
| VC10042.y22.08 | — | — | — | — | — | 0.06 | — | — | — | — | |
| VC10042.y22.e1 | — | — | — | — | — | 0.65 | — | — | — | — | |
Clones were isolated from plasma collected at the indicated year postinfection.
The values reported are the IC50, i.e., the antibody concentration (μg/ml) at which viral entry is inhibited by 50%. CD4 induced, antibodies that target CD4-induced epitopes. —, no neutralizing activity was observed.
We also tested several anticore NAbs, antibodies that target an epitope that overlaps the CD4-BS but are not affected by the D368R mutation (34) (Table 2). With one exception (clone VC10042.y22.e1) all clones were resistant to anti-core NAbs. Clone VC10042.y22.e1 was susceptible to four of five anticore NAbs tested. Thus, although clones VC10042.y22.08 and VC10042.y22.e1 were the only clones susceptible to MAbs b12 and VRC01, only one of them (VC10042.y22.e1) was also susceptible to anticore NAbs.
All VC10042 clones were resistant to the anti-V3 MAb 447-52D, as well as MAb 2G12, which recognizes an epitope formed exclusively by mannose residues present on specific positions within gp120 (33) (Table 3). In addition, all clones were resistant to MAbs PG9 and PG16, which recognize a complex glycopeptide epitope at the base of the V2 loop (22), and to the anti-MPER MAb 2F5 (Table 3), although the canonical 2F5 epitope is present in all clones examined (see Fig. S2 and S3 in the supplemental material). In contrast, various degrees of sensitivity to a second anti-MPER MAb, 4E10, were observed even though all clones express the canonical 4E10 epitope (see Fig. S2 and S3 in the supplemental material). Interestingly, while all of the 19-ypi clones were sensitive to MAb 4E10, only four of nine of the 22-ypi clones were susceptible to that MAb. The 17b and X5 MAbs, which recognize CD4-induced epitopes (1, 25, 36), did not neutralize any of the clones tested (Table 3). However, two recently identified broadly neutralizing MAbs, 3BC176 and 3BC315, whose epitopes are also CD4 induced but which are critically dependent on the virion-associated trimeric form of env (15), displayed a very different neutralizing activity than 17b and X5. All of the 19-ypi env clones were sensitive to 3BC176 and 3BC315 (Table 3). Two 22-ypi clones, VC10042.y22.03 and VC10042.y22.06, were sensitive to 3BC176 but not to 3BC315. The remaining 22-ypi clones were resistant to both 3BC176 and 3BC315 (Table 3). Interestingly, these two MAbs utilize the germ line gene VH1-2*02, a germ line gene reported to be associated with a higher likelihood of conferring neutralizing activity and which is commonly utilized among broadly neutralizing antibodies (35, 39, 42, 44), perhaps explaining their capacity to neutralize the virus while MAbs 17b and X5 do not.
Collectively, these findings indicate that the viruses circulating in VC10042 evolved in a way that allows escape from autologous potent bNAbs and from potent heterologous anti-CD4-BS bNAbs while maintaining its requirement for cellular CD4 for entry. Therefore, viral env clones circulating in VC10042 had adopted conformations allowing them to engage CD4 molecules on target cell surfaces, while simultaneously limiting the accessibility of the CD4-BS to anti-CD4-BS NAbs.
Mechanism of escape from the autologous anti-CD4-BS bNAbs.
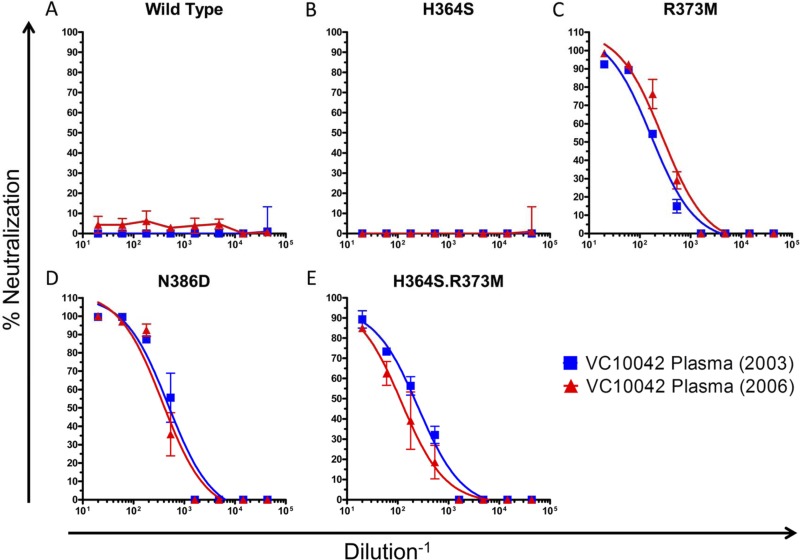
In order to define potential pathways of escape from the autologous NAbs, we focused on the CD4-BS of the viral envelope. We focused on positions within the CD4-BS that have been reported to influence the accessibility of that site to anti-CD4-BS antibodies, specifically, positions 364, 373, and 386 (10, 45). We introduced single and combination reversion mutations at these positions on the backbone of clone VC10042.y22.05, which was resistant to the autologous plasma and to the heterologous anti-CD4-BS NAbs tested (Tables 2 and 3). Position 364 is located within the CD4 binding loop, and the consensus amino acid at that position is a Ser (S). Ten of 14 VC10042 clones had a His (H) at that position (Fig. 2), and a His at that position is known to mediate escape from MAbs b12 and VRC01 (20, 45). However, the H364S reversion mutant viral clone (VC10042.y22.05.H364S) was as resistant to the autologous plasma as the wild-type (wt) clone (Fig. 3B). This suggested to us that position 364 most likely was not involved in offering escape from autologous NAbs. Another possible mechanism of escape involves positions 373 and 386. When present together, R373 and N386 are thought to interact on the “back side” of the CD4-BS (see Fig. S6 [inset] in the supplemental material) and indirectly occlude the Phe43 cavity (10), the 153-Å3 cavity at the interface between gp120 and CD4 (16), thus preventing some anti-CD4-BS antibodies from inserting into the Phe43 cavity on gp120 (10, 45). The consensus amino acid at position 373 is a Met (M); however, an Arg (R) was present in all neutralization-resistant VC10042 clones examined (Fig. 2). Interestingly, the one neutralization-sensitive clone, VC10042.y22.08, contained neither the H364 mutation nor the R373/N386 amino acid pair. Reversion of the R to M led to a complete change in neutralization phenotype and rendered the virus highly susceptible to autologous plasma (Fig. 3C). As expected, when the R373M mutation was combined with the H364S mutation, the virus was also highly susceptible to neutralization by the autologous plasma (Fig. 3E). All resistant clones in VC10042 had the R373/N386 combination (see Fig. S3 and S4 in the supplemental material), which is extremely rare in circulating HIV-1 (see below). Replacing the Asn at 386 by an Asp (N386D) alone also rendered the virus susceptible to the autologous plasma (Fig. 3D). Combined, these results suggest that the steric occlusion of the Phe43 cavity on gp120 was necessary to render the circulating virus resistant to the autologous NAbs.
Fig 3.
Neutralization sensitivity of clone VC10042.y22.05, bearing reversion mutations to autologous plasma from two time points during observation. (A) Unaltered clone; (B to D) three introduced reversion mutations, H364S (B), R373M (C), and N386D (D); (E) generated H364S.R373M double mutant (E).
The R373M/H364S combination mutation also rendered the virus susceptible to heterologous anti-CD4-BS MAbs, such as VRC01 and NIH45-46G54W (see Fig. S7 in the supplemental material). However, some differences were observed between the autologous anti-CD4-BS Abs and the heterologous anti-CD4-BS MAbs tested in their abilities to neutralize the remaining revertant viruses. This is expected since these heterologous MAbs were isolated from patients infected by viruses different from the one replicating in VC10042, and even though these patients, like VC10042, generated potent and broad anti-CD4-BS antibodies, the pathways of viral escape in these patients share common features but also some differences. However, we cannot at this point exclude the possibility that any differences in the neutralizing susceptibility of the reversion mutant viruses to the autologous plasma and the heterologous anti-CD4-BS MAbs are due to the presence of additional neutralizing specificities in the plasma.
Distinct entry and replication potentials of neutralization-resistant and neutralization-sensitive viruses.
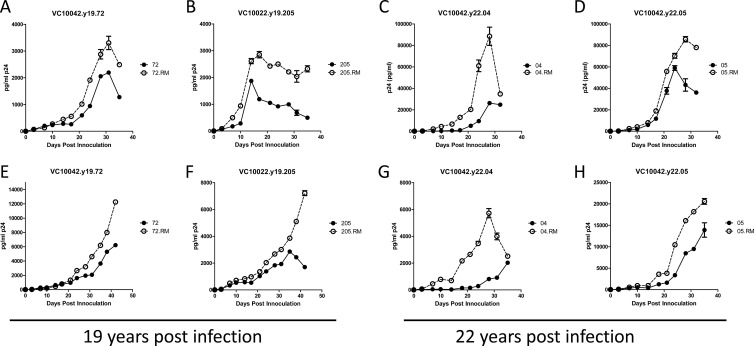
The R373/N386 amino acid combination is extraordinarily rare in the Los Alamos National Laboratory HIV-1 sequence database and is present in only 12 of 3,034 unique subjects (0.39%) in the database (www.hiv.lanl.gov/content/sequence/HIV/mainpage.html). The rarity of this amino acid combination among circulating isolates suggests that this amino acid combination may be associated with reduced viral fitness (10). To address this possibility, we introduced the entire env genes from seven resistant clones from both the 19-ypi and 22-ypi time points (VC10042.y19.72, VC10042.y19.205, VC10042.y22.01, VC10042.y22.03, VC10042.y22.04, VC10042.y22.05, and VC10042.y22.07) into the backbone of the replication competent TN6-GFP virus (26), and we also constructed the corresponding R373M reversion mutants. The replication kinetics of the seven viral pairs were then compared in PBMCs (Fig. 4). In three cases (VC10042.y22.01, VC10042.y22.03, and VC10042.y22.07), the R373M reversion mutation resulted in poorly replication-competent viruses (data not shown). In the other four cases, VC10042.y19.72, VC10042.y19.205, VC10042.y22.04, and VC10042.y22.05, the R373M reversion neutralization-sensitive variants replicated faster and to higher titers than the corresponding wild-type, neutralization-resistant viral clones (Fig. 4A-H).
Fig 4.
Entry and replication potentials of neutralization-resistant and neutralization-sensitive, revertant viral clones. Replication kinetics in human PBMCs of VC10042 wild-type viral clones VC10042.Y19.72, VC10042.Y19.205, VC10042.y22.04, and VC10042.y22.05 and of their corresponding R373M reversion variants are shown. Closed circles and solid lines, wild type; open circles and dotted lines, paired R373M reversion mutant. Results from two independent replication trials performed in PBMCs from two different donors (A to D and E to H) are shown. The average p24 concentrations and the standard deviations from replicate wells in duplicate experiments are shown.
It is notable that some of the viral variants into which we introduced the reversion mutation resulted in viruses that did not replicate well. Potentially, these viruses adopted this escape mechanism during the course of infection but subsequently adopted compensatory mutations that restored replicative capacity. Thus, the introduction of R373M may have disrupted the CD4-BS in these viruses and, rather than restoring a more efficient CD4 receptor interaction, resulted in a loss of replication.
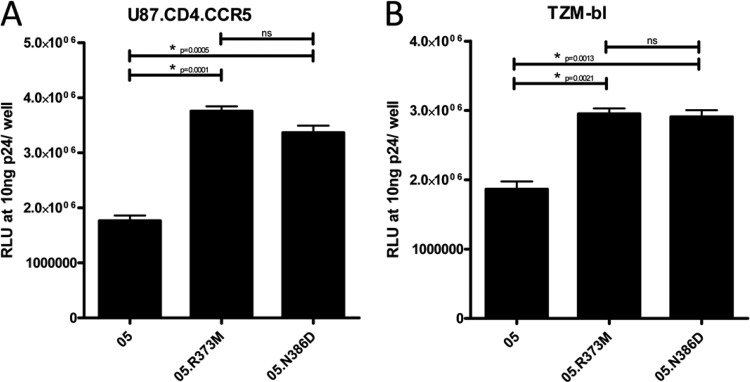
We hypothesized that the reduction in replication kinetics of the resistant clones was due to differences in the virus's ability to enter target cells, as the R/N amino acid combination is thought to alter the interaction between gp120 and the CD4 receptor (10). We assessed the effect of the R373M reversion on viral entry by generating pseudovirus from clones VC10042.y22.05 and VC10042.y22.05.R373M and compared their abilities to enter into U87.CD4.CCR5 and TZM-bl cells (Fig. 5A and B, respectively). Virions expressing the R373M reversion mutant env entered target cells with a significantly higher ability than the wild-type clones. These findings imply that the presence of the R373/N386 residues may alter the viral envelope's affinity for the CD4 receptor, and thus, the reduction in viral replication in viruses containing these residues described above is likely due to an impaired ability to utilize the CD4 receptor for viral entry. However, further studies will be required to determine whether the differences we record in entry potential between the wt virus and the reversion mutants are relevant in vivo.
Fig 5.
Pseudotyped viral entry of the wild type, neutralization-resistant VC10042.y22.05 clone and of two reversion mutants, R373M and N386D, measured in U87.CD4.CCR5 (A) and TZM-bl (B) cell lines. Viral entry in RLU at 10 ng p24/well is reported, with statistically significant differences in entry denoted by an asterisk. Entry among the isolates were compared using a two-tailed unpaired t test, with a significance cutoff of P = 0.05. Where appropriate, significant P values are listed. NS, P values are nonsignificant.
Collectively, the above neutralization, replication, and entry results indicate that the development of potent and broad NAbs in subject VC10042 exerted selection pressure on the virus to alter its env conformation in a specific way. While these changes allowed it to retain its ability to engage cellular CD4 and CCR5 molecules on target cells, they also limited the accessibility of its Phe43 cavity within the CD4-BS to autologous and heterologous potent and broadly neutralizing anti-CD4-BS antibodies. A consequence of this viral adaptation is less efficient entry and slower replication kinetics into target cells.
DISCUSSION
Approximately 10 to 20% of those infected by HIV-1 develop cross-neutralizing antibody responses at approximately 2.5 to 3 years after infection (4, 6, 8, 19, 21, 23, 32). Recently, several potent and broadly neutralizing MAbs have been isolated from such individuals (1, 5, 34, 35, 39, 40, 44, 46). Of particular interest for vaccine development are those broadly neutralizing MAbs that target the CD4-BS of the HIV envelope glycoprotein in very similar manner as the CD4 receptor (35, 39, 46). Understanding whether the development of potent neutralizing anti-CD4-BS antibodies by HIV+ subjects inflicts replication costs to the contemporaneous, autologous virus will provide missing information that is relevant to vaccine development and therapeutic approaches against HIV.
HIV+ patient VC10042 developed potent and broad plasma neutralizing antibody responses which are due primarily to IgG antibodies that target primarily the CD4-BS of the viral envelope glycoprotein (32). The neutralizing activity of the VC10042 anti-CD4-BS antibodies against the contemporaneous virus is similar to that of MAb NIH45-46G54W but very different from those of all other anti-CD4-BS MAbs tested. Collectively our results indicate that the anti-CD4-BS bNAbs developed by VC10042 penetrate deep into the Phe43 cavity of the CD4-BS, as does MAb NIH45-46G54W. We note, however, that the presence of a Trp at position 54 is not by itself sufficient to confer potent and broad anti-HIV neutralizing activities to anti-CD4-BS antibodies. For example, VRC03 has a longer CDR3 than VRC01 and a Trp at position W54 and yet is less potent and less broad than VRC01 (or NIH45-46G54W) (46). Therefore, additional interactions between the anti-CD4-BS bNAbs developed by VC10042 and the HIV envelope gp120 may be required for these antibodies to potently neutralize diverse isolates.
While the viral variants circulating in VC10042 required cellular CD4 receptors to enter target cells and were susceptible to neutralization by CD4-IgG(2), they were resistant to neutralization by the autologous and most heterologous anti-CD4-BS NAbs tested. The ability to utilize cellular CD4 and escape from some of the most potent and broadly neutralizing antibodies known was due to the presence of an arginine at position 373 and an asparagine at position 386 (R373/N386) of gp120. The R373/N368 combination is extremely rare in the known circulating HIVs (10). It is possible that the rarity of this escape mutation reflects the rate at which potent anti-CD4-BS bNAb responses occur during natural infection. However, our results suggest that one reason is that the R/N-based escape mechanism imparts a fitness cost to the virus.
There are some caveats to this study that warrant discussion here. Because samples were unavailable before 19 years postinfection, it is not possible to evaluate the precise pathways of evolution over time that the virus followed in response to presence of the autologous anti-CD4-BS NAbs. Indeed, the escape residues R/N were present already in the sequences at 19 ypi, indicating that escape from the autologous anti-CD4-BS bNAbs had occurred sometime earlier during infection. Thus, we are not able to determine how the autologous antibody response drives viral evolution or to evaluate viral diversity at 20 years postinfection. However, our primary goal here was to understand the potential mechanisms that the virus is able to utilize in order to escape from anti-CD4-BS bNAbs and to understand the potential phenotypic consequences of such escape.
Further, the apparent lack of diversity in the gp120 env sequences isolated from 19 ypi is surprising, but without samples prior to 19 ypi we can only speculate about the basis for these observations. However, we note that at the time the 19-ypi sample was collected, there was a marked downward trend in serum viral load from 19 to 20 ypi (see Fig. S2 in the supplemental material). Viral loads dropped from 27,401 copies per ml to 10,043, before rebounding to more than 20,000 copies per ml by 22 ypi (at the time when we observed more sequence diversity). Further, at 19 ypi, the VC10042 env clones were sensitive to neutralization by the CD4-induced bNAbs 3BC176 and 3BC315, but they had escaped by 22 ypi (Tables 2 and 3). Thus, the observed homogeneity in gp120 at 19 ypi may be a result of selection pressure from similar types of bNAbs, followed by subsequent escape and sequence diversification during this time period. We note that there was much higher diversity in the gp41 subunit (5%), which supports the hypothesis that the minimal diversity on gp120 was due to selection pressure exerted by the anti-gp120 antibodies generated by VC10042.
It has been reported that HIV-1 is able to escape the action of autologous NAbs with narrow breadth of neutralizing activities without any reduction in fitness (11, 38). However, recent reports suggest that in some cases, escape from the autologous neutralizing antibody responses can result in a circulating virus that exhibits a reduced ability to replicate (3, 24). It is worth noting that in these cases, the observed reduction in viral fitness was not absolute and resulted in a decreased ability to replicate rather than a complete loss of fitness (similar to our findings here). However, in some of these studies the epitope specificities of the neutralizing activities were not known, while we focus here directly on the impact of escape from autologous bNAbs that target the CD4-BS. Escape through the R373/N386 mutation combination was not inconsequential for the virus circulating in VC10042, because it reduced its cell entry and replicative potential. Our findings suggest that the epitope specificity of the autologous response may determine whether escape will impart a fitness cost to the virus or whether the virus is able to escape without penalty. In cases where autologous NAbs target conserved regions of env that are vital for viral function, such as the CD4-BS, escape mutations may be more likely to exact a fitness cost.
At the time of collection of the plasma samples examined here, patient VC10042 was infected for 2 decades with HIV-1 without any signs of disease and had not received antiretroviral treatment, with a set point viral load (average of 4.23 log10 RNA copies per ml) that is below the average viral load set point for the majority of HIV infections (4.46 to 5.05 log10 RNA copies per ml) (9, 12, 14, 37). The development, therefore, of broadly neutralizing anti-CD4-BS antibodies in VC10042 may have contributed in some measure to the control of viral replication. Of course, additional antibody functions and T cell-mediated antiviral immune responses could participate in controlling viral replication in VC10042. Further work will be necessary to determine whether the fitness differences we observed are sufficient to impact viral load and influence the course of disease. Since we had access to samples from VC10042 at only approximately 2 decades following infection and not from earlier time points, we do not know at which point during infection these broad anti-CD4-BS NAbs emerged in this subject, nor do we know the precise sequence of mutations that allowed the autologous virus to escape such antibodies. In sum, our study indicates that the development during HIV-1 infection of broad and potent neutralizing antibodies that target the CD4-BS does not abrogate the ability of the autologous virus to engage the CD4 and CCR5 receptors. As a result, the infected individual remains viremic. However, the pathway that leads to viral neutralization resistance may also result in reductions in the viruses' ability to enter CD4+ cells and replicate in vivo.
Supplementary Material
ACKNOWLEDGMENTS
This study was funded by NIH/NIAID grants R33 AI405089 to D.N.S. and R01 AI081625 to L.S.
We thank G. Sellhorn and Z. Caldwell for providing us with the purified gp120 envelope proteins utilized in this study and J. R. Mascola, D. Dimitrov, and J. Robinson for providing MAbs. We also acknowledge J. R. Mascola and M. C. Nussenzweig for many discussions on the manuscript and the interpretation of the results. We acknowledge S. Kalams for providing samples from VC10042. We thank P. Bjorkman and A. West for providing MAbs and for helpful comments and suggestions on the manuscript. We also thank C. Derdeyn for providing the TN6-GFP system used in the viral replication studies.
Footnotes
Published ahead of print 12 September 2012
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Abdelwahab SF, et al. 2003. HIV-1-suppressive factors are secreted by CD4+ T cells during primary immune responses. Proc. Natl. Acad. Sci. U. S. A. 100:15006–15010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Abela IA, et al. 2012. Cell-cell transmission enables HIV-1 to evade inhibition by potent CD4bs directed antibodies. PLoS Pathog. 8:e1002634 doi:10.1371/journal.ppat.1002634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bar KJ, et al. 2012. Early low-titer neutralizing antibodies impede HIV-1 replication and select for virus escape. PLoS Pathog. 8:e1002721 doi:10.1371/journal.ppat.1002721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Binley JM, et al. 2008. Profiling the specificity of neutralizing antibodies in a large panel of plasmas from patients chronically infected with human immunodeficiency virus type 1 subtypes B and C. J. Virol. 82:11651–11668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bonsignori M, et al. 2011. Analysis of a clonal lineage of HIV-1 envelope V2/V3 conformational epitope-specific broadly neutralizing antibodies and their inferred unmutated common ancestors. J. Virol. 85:9998–10009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dhillon AK, et al. 2007. Dissecting the neutralizing antibody specificities of broadly neutralizing sera from human immunodeficiency virus type 1-infected donors. J. Virol. 81:6548–6562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Diskin R, et al. 2011. Increasing the potency and breadth of an HIV antibody by using structure-based rational design. Science 334:1289–1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Doria-Rose NA, et al. 2010. Breadth of HIV-specific neutralizing activity in sera: clustering analysis and association with clinical variables. J. Virol. 84:1631–1636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dorrucci M, et al. 2007. Temporal trends in postseroconversion CD4 cell count and HIV load: the Concerted Action on Seroconversion to AIDS and Death in Europe Collaboration, 1985-2002. J. Infect. Dis. 195:525–534 [DOI] [PubMed] [Google Scholar]
- 10. Duenas-Decamp MJ, Peters P, Burton D, Clapham PR. 2008. Natural resistance of human immunodeficiency virus type 1 to the CD4bs antibody b12 conferred by a glycan and an arginine residue close to the CD4 binding loop. J. Virol. 82:5807–5814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Frost SD, et al. 2005. Characterization of human immunodeficiency virus type 1 (HIV-1) envelope variation and neutralizing antibody responses during transmission of HIV-1 subtype B. J. Virol. 79:6523–6527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gras L, et al. 2009. Viral load levels measured at set-point have risen over the last decade of the HIV epidemic in the Netherlands. PLoS One 4:e7365 doi:10.1371/journal.pone.0007365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gray ES, et al. 2009. Antibody specificities associated with neutralization breadth in plasma from human immunodeficiency virus type 1 subtype C-infected blood donors. J. Virol. 83:8925–8937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Herbeck JT, et al. 2008. Lack of evidence for changing virulence of HIV-1 in North America. PLoS One 3:e1525 doi:10.1371/journal.pone.0001525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Klein F, et al. 2012. Broad neutralization by a combination of antibodies recognizing the CD4 binding site and a new conformational epitope on the HIV-1 envelope protein. J. Exp. Med. 209:1469–1479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kwong PD, et al. 1998. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393:648–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lavine CL, et al. 2012. High-mannose glycan-dependent epitopes are frequently targeted in broad neutralizing antibody responses during infection of human immunodeficiency virus type 1. J. Virol. 86:2153–2164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li M, et al. 2005. Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J. Virol. 79:10108–10125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Li Y, et al. 2007. Broad HIV-1 neutralization mediated by CD4-binding site antibodies. Nat. Med. 13:1032–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li Y, et al. 2011. Mechanism of neutralization by the broadly neutralizing HIV-1 monoclonal antibody VRC01. J. Virol. 85:8954–8967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li Y, et al. 2009. Analysis of neutralization specificities in polyclonal sera derived from human immunodeficiency virus type 1-infected individuals. J. Virol. 83:1045–1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. McLellan JS, et al. 2011. Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9. Nature 480:336–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mikell I, et al. 2011. Characteristics of the earliest cross-neutralizing antibody response to HIV-1. PLoS Pathog. 7:e1001251 doi:10.1371/journal.ppat.1001251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Moore PL, et al. 2009. Limited neutralizing antibody specificities drive neutralization escape in early HIV-1 subtype C infection. PLoS Pathog. 5:e1000598 doi:10.1371/journal.ppat.1000598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Moulard M, et al. 2002. Broadly cross-reactive HIV-1-neutralizing human monoclonal Fab selected for binding to gp120-CD4-CCR5 complexes. Proc. Natl. Acad. Sci. U. S. A. 99:6913–6918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Neumann T, et al. 2005. T20-insensitive HIV-1 from naive patients exhibits high viral fitness in a novel dual-color competition assay on primary cells. Virology 333:251–262 [DOI] [PubMed] [Google Scholar]
- 27. Pejchal R, et al. 2011. A potent and broad neutralizing antibody recognizes and penetrates the HIV glycan shield. Science 334:1097–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pietzsch J, et al. 2010. Human anti-HIV-neutralizing antibodies frequently target a conserved epitope essential for viral fitness. J. Exp. Med. 207:1995–2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Richman DD, Wrin T, Little SJ, Petropoulos CJ. 2003. Rapid evolution of the neutralizing antibody response to HIV type 1 infection. Proc. Natl. Acad. Sci. U. S. A. 100:4144–4149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rong R, et al. 2009. Escape from autologous neutralizing antibodies in acute/early subtype C HIV-1 infection requires multiple pathways. PLoS Pathog. 5:e1000594 doi:10.1371/journal.ppat.1000594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sajadi MM, et al. 2011. Correlation between circulating HIV-1 RNA and broad HIV-1 neutralizing antibody activity. J. Acquir. Immune Defic. Syndr. 57:9–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sather DN, et al. 2009. Factors associated with the development of cross-reactive neutralizing antibodies during human immunodeficiency virus type 1 infection. J. Virol. 83:757–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Scanlan CN, et al. 2002. The broadly neutralizing anti-human immunodeficiency virus type 1 antibody 2G12 recognizes a cluster of alpha1→2 mannose residues on the outer face of gp120. J. Virol. 76:7306–7321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Scheid JF, et al. 2009. Broad diversity of neutralizing antibodies isolated from memory B cells in HIV-infected individuals. Nature 458:636–640 [DOI] [PubMed] [Google Scholar]
- 35. Scheid JF, et al. 2011. Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science 333:1633–1637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Thali M, et al. 1993. Characterization of conserved human immunodeficiency virus type 1 gp120 neutralization epitopes exposed upon gp120-CD4 binding. J. Virol. 67:3978–3988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Troude P, et al. 2009. No evidence of a change in HIV-1 virulence since 1996 in France. AIDS 23:1261–1267 [DOI] [PubMed] [Google Scholar]
- 38. van Gils MJ, et al. 2010. Rapid escape from preserved cross-reactive neutralizing humoral immunity without loss of viral fitness in HIV-1-infected progressors and long-term nonprogressors. J. Virol. 84:3576–3585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Walker LM, et al. 2011. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature 477:466–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Walker LM, et al. 2009. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science 326:285–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wei X, et al. 2003. Antibody neutralization and escape by HIV-1. Nature 422:307–312 [DOI] [PubMed] [Google Scholar]
- 42. West AP, Jr, Diskin R, Nussenzweig MC, Bjorkman PJ. 2012. Structural basis for germ-line gene usage of a potent class of antibodies targeting the CD4-binding site of HIV-1 gp120. Proc. Natl. Acad. Sci. U. S. A. 109:E2083–E2090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wu X, et al. 2012. Selection pressure on HIV-1 envelope by broadly neutralizing antibodies to the conserved CD4-binding site. J. Virol. 86:5844–5856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wu X, et al. 2010. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science 329:856–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wu X, et al. 2009. Mechanism of human immunodeficiency virus type 1 resistance to monoclonal antibody B12 that effectively targets the site of CD4 attachment. J. Virol. 83:10892–10907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wu X, et al. 2011. Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing. Science 333:1593–1602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zhou T, et al. 2010. Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science 329:811–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.