Abstract
Enormous numbers of adult blood cells are constantly regenerated throughout life from hematopoietic stem cells through a series of progenitor stages. Accessibility, robust functional assays, well-established prospective isolation, and successful clinical application made hematopoiesis the classical mammalian stem cell system. Most of the basic concepts of stem cell biology have been defined in this system. At the same time, many long-standing disputes in hematopoiesis research illustrate our still limited understanding. Here we discuss the embryonic development and lifelong maintenance of the hematopoietic system, its cellular components, and some of the hypotheses about the molecular mechanisms involved in controlling hematopoietic cell fates.
Hematopoietic stem cells constantly give rise to more than 10 different types of adult blood cells through a series of progenitor stages. This provides an excellent model for studying the molecular mechanisms of cell-fate control.
1. INTRODUCTION
The blood system contains more than 10 different blood cell types (lineages) with various functions: Leukocytes represent many specialized cell types involved in innate and acquired immunity. Erythrocytes provide O2 and CO2 transport, whereas megakaryocytes generate platelets for blood clotting and wound healing. All blood cell types arise from hematopoietic stem cells (HSCs) that reside mainly in the bone marrow (BM), a major site of adult hematopoiesis. Blood is one of the most regenerative and plastic tissues, and millions of “old” blood cells are replenished with new ones each second during life. In emergency situations such as anemia or infections, blood cell counts rapidly increase. The cell number then declines back to normal after recovery. The lifetimes of various mature blood cell types range from hours to years.
The hematopoietic system is a prime example of successful applied regenerative medicine. For more than 30 years, stem cell transplantation has become a routine treatment for blood disorders and malignant diseases. After the eradication of the patient’s own hematopoietic system, the transplanted donor hematopoietic stem and progenitor cells (HSPCs) provide lifelong reconstitution of the blood system of the patient. The experimental evidence that HSCs naturally migrate back and forth from the BM periodically, as well as the identification of agents that increase HSC mobilization (e.g., granulocyte colony-stimulating factor [G-CSF]), have opened new avenues for stem cell transplantation. However, although stem cell transplants work successfully in the clinic, further improvement of the method is needed to minimize engraftment failure and posttransplant infections. The ex vivo expansion of HSCs would be beneficial for grafts with limiting numbers of HSCs (umbilical cord blood) and for gene therapy approaches for monogenetic inherited blood disorders. However, despite decades of research, the robust expansion (or even maintenance) of HSCs ex vivo is not yet routinely achieved.
HSCs are the most-studied adult stem cells. Five decades ago researchers passed the stage of providing purely descriptive data to start quantitative HSC research (Till and McCulloch 1961; Becker et al. 1963; Siminovitch et al. 1963). Several properties of hematopoietic cells are favorable for stem cell research. First, they are not tightly interconnected in a tissue. Therefore, cells can be physically separated without too much stress. The isolation from peripheral blood is minimally invasive, and millions of cells can easily be harvested. Many blood cell types are naturally capable of extravasating into tightly packed tissues and sustaining enormous shear forces. Therefore, they tolerate isolation by flow cytometry well. This enabled the early correlation of surface marker expression patterns with functional tests examining self-renewal capacity, clonogenicity, and lineage potential, leading to successful prospective enrichment of distinct HSPC populations (Morrison and Weissman 1994; Osawa et al. 1996; Kondo et al. 1997; Akashi et al. 2000; Kiel et al. 2005, Inlay et al. 2009). Last, HSPCs grow into colonies from single cells under appropriate culture conditions, allowing assays at the clonal level.
Only functional tests in vivo can retrospectively determine the true identity of HSCs by demonstrating their unique abilities of long-term or even lifelong self-renewal and multilineage differentiation. Robust HSPC transplants between congenic mouse strains, in which the donor cells differ from the recipient cells by only a single surface marker, allow quantitative functional HSC readouts. Once injected intravenously, even single transplanted HSCs can find their way to the appropriate location in the BM to initiate lifelong blood regeneration (Osawa et al. 1996). Serial transplantation shows that HSC self-renewal can last longer than the normal lifetime of the organism in the mouse model (Morrison and Weissman 1994). The HSC frequency within a mixture of undefined cells can be quantified by transplanting limiting dilutions of cells (Szilvassy et al. 1990).
The murine hematopoietic system is by far the best understood of all species. The generation of genetically modified mouse strains for gain- and loss-of-function studies and for lineage and cell tracing enables the study of hematopoiesis in vivo. Conditional and inducible systems allow the timed and tissue-specific manipulation of gene expression under homeostatic conditions. For this approach, mouse strains expressing Cre recombinase only in specific hematopoietic cell types have proven extremely valuable (Kuhn et al. 1995; de Boer et al. 2003). Murine transplantation models for murine and human HSPCs have been established for decades. The homing and engraftment of murine cells transplanted into congenic mouse strains is very efficient, although not absolute. Sophisticated humanized mouse strains with an incompetent adaptive immune system have been developed to prevent xenograft rejection for reconstitution studies with human BM cells (McDermott et al. 2010). Existing species incompatibilities in receptor–ligand or adhesion molecule binding can be partly compensated for by intrafemoral cell injection (Notta et al. 2011) or by transgenic recipient mice expressing human growth factors (Rongvaux et al. 2011). Less commonly used but extremely valuable model organisms for hematopoiesis research are Danio rerio (zebrafish) (Jing and Zon 2011), and increasingly also Drosophila melanogaster (fruit fly) (Martinez-Agosto et al. 2007). The genetic manipulation of these organisms is faster and cheaper than in the murine model, and genetically modified offspring can be generated within days and weeks. Moreover, their transparent embryos, which develop extremely fast and ex utero, allow relatively easy live imaging.
2. EMBRYONIC HEMATOPOIESIS
The analysis of how blood cells are first produced during embryonic development is not only of great scientific interest, but it can also offer important insights into the cellular and molecular mechanisms that lead to the specification of clinically important HSCs. This could lead to their improved expansion or even de novo generation in vitro from other cells like embryonic stem cells or induced pluripotent stem cells by improved differentiation protocols or from any other cell type by direct reprogramming.
The earliest stages of embryonic development actually happen in the absence of any blood cells. Only after the embryo gets too big to be supplied with oxygen and other essential factors by diffusion are blood cells first generated (Fig. 1). The sites of both de novo HSPC generation and maintenance/expansion are constantly changing in the embryo (Fig. 1). Anatomically, the first hematopoietic cells are generated in the extraembryonic yolk sac before the first heartbeat and later in the allantois and placenta. Finally, HSCs are then generated de novo in the intraembryonic aorta-gonad-mesonephros (AGM) region and the placenta. Interestingly, although all adult and many of the later embryonic blood cells are produced from HSCs (see below), there is a transient population of so-called primitive blood cells in the early embryo. These cells emerge in the extraembryonic yolk sac and placenta and possibly also in intraembryonic sites before the first HSCs. They do not propagate into the adult and are soon replaced by their definitive counterparts in the embryo. They have specific effector functions in the embryo and differ from their adult counterparts. As an example, primitive erythrocytes, in contrast to adult “definitive” erythrocytes, mostly retain their nuclei and express fetal forms of hemoglobin. Other primitive blood cells like macrophages and megakaryocytes exist, but this field of hematopoiesis remains poorly understood and many questions await investigation (Palis et al. 1999; Chen and Zon 2009). In addition, definitive erythroid/myeloid progenitors have been described before HSC formation (Bertrand et al. 2010b). These very recent discoveries of previously unrecognized embryonic cell types and mechanisms of their generation (Chen et al. 2011) well illustrate how little we still know.
Figure 1.
Embryonic hematopoietic development. Anatomical sites and timing of hematopoietic cell generation, maintenance, and expansion during embryonic development. Hematopoietic cells are generated de novo in both extra- (yolk sac, allantois, placenta) and intraembryonic (AGM region) tissues. Cells are migrating to other sites for expansion and ultimately long-term maintenance throughout adult life. dpc, days after conception; wpc, weeks after conception.
The first cells with functional properties of adult HSCs (i.e., regeneration of the hematopoietic system upon transplantation into adult recipient mice; see below) are generated in the intraembryonic AGM region (Muller et al. 1994; Medvinsky and Dzierzak 1996; Ivanovs et al. 2011) and the placenta (Gekas et al. 2005; Ottersbach and Dzierzak 2005). Interestingly, their location quickly changes throughout development. After being generated in the AGM, they soon migrate to the placenta and fetal liver, which are major sites of their expansion. Soon thereafter they migrate to the spleen, and around birth to the BM. BM, and to a reduced extent the spleen, then remain the main sites of HSC-derived hematopoiesis after birth.
The development of the blood system in the embryo is a very complex and dynamic process, and in contrast to most other tissues, different cell types constantly appear in different intra- and extraembryonic tissues (Fig. 1). The transient generation of different embryonic cell types before establishment of HSCs, which then exclusively drive later hematopoiesis, complicates the analysis of how exactly this tissue is generated. This has led to many decades-old disputes about the cellular sources of their de novo generation, differentiation hierarchy, and molecular control (Medvinsky et al. 2011). One of those disputes concerned the cellular source of the first HSCs. Because of the facts that hematopoietic cells (both primitive and definitive) are always found initially in proximity to endothelial cells and that these cell types share many molecular markers, it had been hypothesized for decades that the first hematopoietic cells are actually generated from endothelium (reviewed by Dieterlen-Lievre et al. 2006; Medvinsky et al. 2011). These hemogenic endothelial cells would be integrated into vessel walls but would be able, under the influence of unknown molecular signals, to generate hematopoietic cells. However, proving the existence of hemogenic endothelial cells was surprisingly difficult. For decades, static analyses of cells either before or after the endothelial-to-hematopoietic transition yielded data that could also be explained by the alternative conclusion that hematopoietic cells were generated elsewhere and then migrated toward endothelial cells. Final proof of direct generation of hematopoietic cells from hemogenic endothelium came from continuous single-cell observations of endothelial-to-hematopoietic transitions by time-lapse microscopy (Eilken et al. 2009). Subsequent studies could then also detect this direct transition in vivo in the zebrafish embryo (Bertrand et al. 2010a; Kissa and Herbomel 2010). It is worth mentioning that these studies do not rule out the possible existence of other sources of hemogenic cell types in the embryo. Importantly, comparable timing and location of hemogenic activity in human embryos makes it likely that the same cells, molecules, and mechanisms are also involved in human embryonic hematopoiesis (Ivanovs et al. 2011).
The proof of the existence of hemogenic endothelium now allows the improved analysis of the molecular mechanisms involved in the generation of the first blood cells, including the therapeutically important HSCs. This molecular mechanism is only partially understood. It involves complex signaling from different cell types surrounding developing hemogenic endothelium at different sites and times in the fast-developing and -changing embryo. Mesodermal cells expressing Flk1 and ETV2 were recently identified as the common precursors for endothelial and hematopoietic cells (Lee et al. 2008; Kataoka et al. 2011a). The signals inducing hemogenic fate in these cells and their progeny are the subject of ongoing research. Molecules like membrane-bound Notch ligands or soluble hedgehog, bone morphogenetic protein, or vascular endothelial growth factor, but also shear stress and nitric oxide signaling, have been involved in the induction of the hemogenic program in endothelial cells—possibly even in mesodermal precursors before they have reached endothelial identity (Medvinsky et al. 2011). Within the hemogenic endothelial cell, a number of transcription factors (TFs) seem to be crucial for the induction of the hemogenic program, with Runx1 and CBFβ apparently being critical core factors (Chen et al. 2011).
Importantly, fetal HSCs differ from adult HSCs in their regeneration behavior. In contrast to mostly quiescent adult HSCs, they are actively cycling and regenerate hematopoiesis faster and more robustly upon transplantation. The switch from this fetal to adult HSC behavior occurs in a very short time window a few weeks after birth of a mouse (Bowie et al. 2007). This sharp switch at a defined time is useful for the identification of the involved molecular mechanisms (Kim et al. 2007; He et al. 2011), which could be of great interest for the induction of a clinically preferred fetal phenotype in HSCs.
3. ADULT HEMATOPOIESIS
3.1. The Hematopoietic Differentiation Hierarchy
In adult mammals, HSCs are at the apex of a hierarchy of numerous progenitor cell stages with increasingly restricted lineage potentials that give rise to all blood cell lineages (Fig. 2). Of all blood cells, only HSCs fulfill the criteria for somatic stem cells, namely, long-term (and possibly lifelong) self-renewal and differentiation potential. In the murine system they robustly self-renew (produce more HSCs) while also reconstituting all blood cell lineages for >24 weeks in congenic transplant recipients (Benveniste et al. 2010), and are further able to repopulate secondary recipients. These functional assays have also revealed multipotent progenitor (MPP) populations with intermediate- or short-term repopulating capacity (formerly called IT- and ST-HSCs) with a finite self-renewal potential (Osawa et al. 1996; Benveniste et al. 2010). The stepwise identification of multiple surface markers in the last 25 years enabled the prospective isolation of defined stem and progenitor cell populations by flow cytometry-based cell sorting. Immature multipotent cells express CD117 (c-Kit) and stem cell antigen (Sca)-1 and are low in mature cell marker expression (lineage markers) (Spangrude et al. 1988; Okada et al. 1992; Uchida and Weissman 1992; Morrison and Weissman 1994). About one in 30 cells within this so-called KSL (c-Kit+/Sca-1+/Lineage−) population (0.06% in BM) is an HSC (0.002% in BM), and there is no HSC activity outside the KSL fraction. These findings laid the ground for the identification of additional surface markers for improved prospective HSC identification (Table 1). Currently, the highest purity of at least 50% HSCs in the sorted fraction is obtained with CD150+/CD48−/CD34low KSL cells (Kiel et al. 2005). Moreover, CD150 expression levels might indicate distinct subpopulations of HSCs with predisposed myeloid- or lymphoid-biased reconstitution patterns (see also Sec. 3.2.1) (Morita et al. 2010). CD49b expression could be attributed to intermediate-term repopulating MPPs, a population that shares many functional properties of myeloid-biased HSCs (Benveniste et al. 2010). In addition to surface markers, distinct functional properties of HSCs have been used for prospective isolation, like the increased efflux of the dye Hoechst 33342 (so-called side population, or SP) (Goodell et al. 1996) and low staining for Rhodamine123 dye (Bertoncello et al. 1985). A further discrimination of SPlower and SPhigher HSCs also dissected HSCs with myeloid- and lymphoid-biased repopulation capacity, respectively (Challen et al. 2010). It is important to point out that although transplantation experiments functionally identify HSC properties, these are stress conditions for the HSCs and for the organism. They therefore may not reflect functional properties of homeostatic HSCs. Furthermore, functional HSC reconstitution behavior depends on the chosen transplantation model. This is particularly problematic for xenograft settings with human HSCs repopulating murine recipients. Although this is an extremely important methodology for analyzing human HSCs, it is important to remember that HSC properties and behavior in such a different environment could differ massively from those under conditions in the human niches (McDermott et al. 2010).
Figure 2.
Adult hematopoietic differentiation hierarchy. Long-term self-renewing HSCs are at the apex of a hierarchy of multiple progenitor cell stages giving rise to all blood cell lineages. Distinct HSPC stages have been described by correlating surface marker expression and functional properties for prospective isolation, both in the murine and human systems. Murine hematopoiesis is currently defined in more detail and therefore displayed here. Corresponding human HSPC populations with their markers are indicated. HSCs differentiate into all blood cell lineages via long described (bold arrows) and potentially also or alternatively more recently described differentiation routes (thin arrows). It is important to point out that this model is only a simplified representation of current knowledge and will continue to change. Individual murine HSCs show an intrinsic lineage-biased repopulating ability, generating either more myeloid or lymphoid cells or a balanced mixture. These HSC subgroups can be enriched by detecting various levels of CD150 expression or Hoechst efflux. HSC, hematopoietic stem cell; MPP, multipotent progenitor; LT-, long-term repopulating; IT-, intermediate-term repopulating; ST-, short-term repopulating; LMPP, lymphoid-primed MPP; ELP, early lymphoid progenitor; CLP, common lymphoid progenitor; CMP, common myeloid progenitor; GMP, granulocyte–macrophage progenitor; MEP, megakaryocyte–erythrocyte progenitor; CDP, common dendritic progenitor; MDP, monocyte–dendritic cell progenitor; NK, natural killer cell.
Table 1.
Prospective HSC isolation
| HSPC population | Transplanted cells | Assay/model | HSCs (%) | References |
|---|---|---|---|---|
| Murine | ||||
| Thy-1low/Sca-1+/Lin− | 30 | Competitive | 1.5–3 | Spangrude et al. 1988 |
| KSL | Limiting dilutions | Competitive | 3–7 | Okada et al. 1992; Bryder et al. 2006 |
| Thy-1low KSL | 5–10 | Competitive | 10–20 | Morrison and Weissman 1994 |
| CD34− KSL | 1 | Competitive | 21 | Osawa et al. 1996 |
| SP Rholow/CD45mid/Lin− | 1 | Sublethal W-41 | 42/33 | Uchida et al. 2003; Dykstra et al. 2006 |
| SP CD201+ | 10 | Competitive | >10 | Balazs et al. 2006 |
| CD150+/CD48−/CD41− KSL | 1 | Competitive | 47 | Kiel et al. 2005 |
| CD150+/CD48−/CD49blow KSL | 1 | Sublethal W-41 competitive | 29 | Benveniste et al. 2010 |
| CD150+/CD48−/CD201+/CD45+ | 1 | Sublethal W-41 | 43 | Kent et al. 2009 |
| Human | ||||
| CD34+/CD38−/Lin− | Limiting dilutions | NOD/SCID | <1 | Bhatia et al. 1997 |
| CD34+/CD38−/CD90+/CD45RA−/Lin− | Limiting dilutions | Sublethal NSG | 5 | Majeti et al. 2007; Notta et al. 2011 |
| Rholow/CD49f+/CD34+/CD38−/CD90+/CD45RA−/Lin− | 1 | Sublethal NSG | 15 | Notta et al. 2011 |
HSPC, hematopoietic stem or progenitor cell; KSL, CD117+, Sca-1+, and Lin− cells; SP, side population; Rho, Rhodamine123; W-41, mouse line with homozygous KitW-41J/KitW-41J mutation; NOD/SCID, nonobese diabetic/severe combined immune deficiency mouse line; NSG, NOD/SCID/Il2 γ-chain knockout mouse line; competitive, transplantation in lethally irradiated recipients with “competitor-recipient” HSPCs.
HSCs differentiate into a cascade of progenitor cell stages with declining multilineage potential before unilineage commitment occurs (Kondo et al. 1997; Akashi et al. 2000; Adolfsson et al. 2005; Wilson et al. 2008; Rieger et al. 2009b). Current models of the hematopoietic hierarchy describe a successive loss of individual lineage potentials (Adolfsson et al. 2005; Arinobu et al. 2007; Mansson et al. 2007; Pronk et al. 2007). MPPs first lose their erythroid/megakaryocytic potential and develop into lymphoid-primed MPPs (LMPPs) and early lymphoid progenitors (ELPs) (Adolfsson et al. 2005). Then they lose their myeloid potential and become common lymphoid progenitors (CLPs) (Kondo et al. 1997; Inlay et al. 2009; Schlenner et al. 2010), and finally differentiate into lymphoid cells (Fig. 2). However, the successive restriction to solely lymphoid fate should not imply a picture of default lymphoid cell fate. The opposite might be the case, namely, that active repression of the myeloid cell fate is required to induce lymphoid development. Interestingly, B- and T-cell progenitors retain, at least in vitro, some myeloid potential (Kawamoto et al. 2010). In contrast, lineage-tracing experiments revealed only a minor contribution of myeloid cells from interleukin-7 (IL-7) receptor–positive CLPs in murine homeostatic hematopoiesis (Schlenner et al. 2010). It is important to point out that this model of the hematopoietic differentiation hierarchy clearly only reflects current knowledge and will continue to change over time. In particular, the early progenitor stages are still ill defined, and numerous novel maturation stages and differentiation pathways will certainly be described in coming years.
According to current data, the human hematopoietic differentiation hierarchy closely resembles the murine one (Majeti et al. 2007; Doulatov et al. 2010). However, mostly as a result of more difficult experimentation, and despite their clinical importance, purification methods for human HSPCs are far less advanced than for murine HSPCs (Table 1). Human HSCs express the surface molecule CD34, in contrast to their murine counterparts (Osawa et al. 1996; Dick 2008). However, although many reports describe CD34+/CD38−/Lineage− cells as human HSCs, it is very important to point out that this population comprises <1% functional HSCs (as determined by xenograft transplantation assays). Only recently have improved methods for the prospective enrichment of human HSCs been described that now allow enrichment to purities of ~15% (Notta et al. 2011) (Table 1). Prospective isolation of human hematopoietic progenitors has also recently been improved, allowing the functional and molecular characterization of these important cell types with much improved resolution (Manz et al. 2002; Majeti et al. 2007; Doulatov et al. 2010). However, the percentage of prospectively isolated human HSCs may be underestimated by xenotransplantations because the murine environment is less permissive to human HSCs, and advanced humanized mouse models have improved engraftment efficiency (Rongvaux et al. 2011).
3.2. Molecular Control of Hematopoietic Fate Choices
To continuously regenerate the hematopoietic system, the right number of specific cell types must permanently be generated at the right time and place. To achieve this, correct fate decisions constantly have to be chosen in HSPCs: quiescence versus proliferation, self-renewal versus differentiation, lineage choice, survival versus death, and sessility versus migration (Fig. 3A). Exact timing and sequential order of all choices in each cell underpin normal hematopoiesis, both in steady-state and injury situations. Disruptions of normal cell fate decisions underlie hematological disorders. A comprehensive analysis of their molecular control and the exact interplay of these fate decisions are therefore crucial for a full understanding of normal and malignant hematopoiesis and the development of novel therapies.
Figure 3.
Continuous single-cell observations are required for a comprehensive understanding of dynamic cellular behavior. (A) Individual cell-fate decisions are the reason for the behavior of the complete hematopoietic system in health and disease, and must therefore be quantified. They are influenced by factors like cytokines, extracellular matrix (ECM), or membrane-bound signaling molecules in many different niches. (B) Only continuous single-cell observation allows unambiguous conclusions about cell-fate choices in complex cell systems. Discontinuous input/output analyses—even when done at the single-cell level—cannot distinguish between different combinations of cell-fate decisions leading to the same output of a cellular system, and therefore lead to ambiguous conclusions.
The molecular circuitries underlying HSPC fate choice must enable cells to respond to changing external stimuli but also allow specific cellular states to be stable independent of their environment. As an example, plasticity must enable the choice of HSPCs between multiple lineages but ensure stability of lineage commitment after the choice has been made. The expression, activity, or changing function of fate-control molecules therefore must be well timed, cell type specific, and controllable by other factors. For a comprehensive understanding, these parameters have to be quantified in a cell-stage-specific manner, at the single-cell level, and ideally over time. Experimental analysis of molecular cell-fate control requires demanding technical prerequisites: (1) the prospective isolation of distinct cell stages in sufficient numbers and purity, (2) continuous quantitative measurements of functional molecular activity during differentiation, and (3) the observation of multiple factors in complex genetic programs. Many of these requirements have been achieved individually, but their integrated analysis is still limited by current technologies.
Hematopoiesis is usually analyzed in heterogeneous populations and at individual time points of the process of interest. However, these snapshot analyses only allow limited conclusions about the involved sequence of individual molecular and cellular events (Fig. 3B). Experimental data can then usually be explained by a multitude of hypotheses, leading to ambiguous conclusions. Only when the behavior and fate of each individual cell and its progeny are continuously known can a comprehensive understanding of developmental steps and their molecular control be drawn (Fig. 3B). Ideally, therefore, cellular and molecular HSPC behavior is continuously quantified at the single-cell level and throughout the process of interest (Schroeder 2008). One approach to fulfill these requirements is long-term video microscopy of HSPCs and computer-aided single-cell tracking (Eilken et al. 2009; Rieger et al. 2009a). However, despite its potential, this field is still in its infancy, and because of the required high level of interdisciplinary technical know-how, its use is still limited to few expert laboratories (Schroeder 2011).
3.2.1. A Stem Cell’s Decision: Self-Renewal versus Differentiation
The HSC’s choice between self-renewal and differentiation must be tightly regulated to enable both the generation of differentiated cells and the accurate maintenance of the right HSC number. The BM provides the environment for sustained HSC function, and HSCs rapidly lose their self-renewal capacity once isolated from their in vivo niches. Extrinsic signals from membrane-bound, soluble, or extracellular matrix-associated ligands from the niche are necessary for appropriate HSC behavior. Two major HSC niches are currently proposed to exist in BM: the endosteal osteoblastic niche (Calvi et al. 2003; Zhang et al. 2003; Lo Celso et al. 2009; Xie et al. 2009) and the perivascular endothelial niche (Kiel et al. 2005; Ding et al. 2012). It is unclear if both niches, although spatially separated, fulfill a similar function; if both niches provide distinct properties, for example, for dormant versus cycling HSCs; or if the niche comprises both osteoblasts and endothelial cells working synergistically on HSC function. Also, BM comprises a heterogeneous mixture of various cell types including blood cells, mesenchymal cells, osteoblasts, osteoclasts, endothelial cells, reticular cells, fat cells, and many other less-defined types, which will also influence hematopoietic fates. A multitude of signaling pathways have been shown to be activated in HSCs by the niche (e.g., the cytokine receptors c-Kit and Mpl, Wnt, Notch, Sonic hedgehog, and integrin signaling). However, their precise involvement in HSC maintenance remains surprisingly controversial. For most of them, contradictory conclusions have been drawn in separate studies. Our incomplete understanding of the molecular control of HSC self-renewal is well illustrated by the fact that therapeutically relevant robust maintenance or even expansion of these clinically important cells ex vivo remains elusive without genetic manipulation. Nevertheless, several recent studies with murine and human HSCs describe very promising approaches for the ex vivo expansion of HSCs without genetic manipulation (Zhang et al. 2008; Boitano et al. 2010). A comprehensive discussion of all potential molecular HSC self-renewal modulators goes beyond the scope of this chapter, and can be found elsewhere (Ehninger and Trumpp 2011; Mercier et al. 2011).
At the single-cell level, even HSCs with retrospectively proven functionality (>1% long-term contribution to mature myeloid and lymphoid cells in peripheral blood) are heterogeneous in their reconstitution efficacy and their lineage contribution pattern. They range from 1% to almost 100% total contribution, with varying ratios of myeloid and lymphoid lineages (Muller-Sieburg et al. 2002, 2004; Sieburg et al. 2006; Dykstra et al. 2007; Kent et al. 2009). Importantly, these patterns are conserved through serial transplantations, indicating the existence of stable inheritable stem cell intrinsic programs (Muller-Sieburg et al. 2002, 2004; Sieburg et al. 2006; Dykstra et al. 2007; Kent et al. 2009). Therefore, not only are self-renewal and differentiation controlled by the environment, but intrinsic regulators of HSC functionality exist (Table 2).
Table 2.
Examples of HSC intrinsic molecules implicated in control of self-renewal
| Molecule | Effect on self-renewal (SR) | References |
|---|---|---|
| Transcription factors | ||
| HoxB4 | Expression enhances SR | Antonchuk et al. 2002 |
| Gfi1 | Loss reduces SR upon HSC exhaustion | Hock et al. 2004; Zeng et al. 2004 |
| Evi1 | Loss reduces SR | Kataoka et al. 2011b |
| c-Myc | Loss leads to HSC accumulation with differentiation defect | Laurenti et al. 2008 |
| Signaling modulators | ||
| Pten | Loss reduces SR upon HSC exhaustion | Yilmaz et al. 2006 |
| Lnk | Loss increases SR | Ema et al. 2005; Seita et al. 2007 |
| STAT5A/B | Loss reduces SR | Wang et al. 2009 |
| Epigenetic modifiers | ||
| Dnmt1 | Loss reduces SR by reduced DNA methylation | Broske et al. 2009 |
| Dnmt3a/b | Loss alters SR and/or differentiation | Tadokoro et al. 2007; Challen et al. 2011 |
| Bmi-1 (PRC1) | Loss reduces SR, expression increases SR | Lessard and Sauvageau 2003; Park et al. 2003; Iwama et al. 2004 |
| Suz12 (PRC2) | Loss increases SR | Majewski et al. 2010 |
| Ezh2 (PRC2) | Expression enhances SR, loss reduces SR (mainly in fetal liver) | Herrera-Merchan et al. 2012 |
| Cell cycle regulators | ||
| p57KIP2 | Loss reduces SR upon HSC exhaustion | Matsumoto et al. 2011; Zou et al. 2011 |
| p18Ink4c | Loss increases SR | Yuan et al. 2004 |
| microRNAs | ||
| miR125a | Loss reduces SR and increases apoptosis | Guo et al. 2010 |
| miR125b | Expression enhances SR by blocking apoptosis | O’Connell et al. 2010; Ooi et al. 2010 |
| RNA-binding proteins | ||
| Msi2 | Loss reduces SR | Hope et al. 2010; Ito et al. 2010; Kharas et al. 2010 |
PRC, polycomb repressive complex.
Remarkably, ectopic expression of HoxB4 and in particular of Hox fusion proteins to NUP98 (NUP98–HoxB4 and NUP98–HoxA10 homeodomain) lead to a massive net expansion of HSCs in vitro, while still maintaining their normal multilineage differentiation in vivo (Antonchuk et al. 2002). However, despite years of research, the exact molecular mechanism and the relevant target genes inducing HSC expansion remain poorly defined.
Although all the discussed and numerous additional molecules could be identified to be involved in regulating HSC self-renewal, their exact interplay remains to be unraveled. More sensitive large-scale methods will likely be a key to drawing a more complete picture of the stem cell self-renewal network and to identifying a potential core mechanism of self-renewal control. It is interesting that deletion of many factors that are essential for fetal HSC generation and self-renewal (e.g., Runx1, Tal1, Notch1 and -2, RBP-J, β-catenin, and HoxB4) only have minimal consequences upon deletion in adult HSCs, whereas overexpression often results in enhanced HSC maintenance, self-renewal, and leukemia (Mikkola et al. 2003; Ichikawa et al. 2004).
3.2.2. Hematopoietic Lineage Choice and Stability
Differentiation of multipotent cells into different lineages must be well controlled to enable the timely production of the right number and type of mature cells. Despite intensive research over decades, we are only just beginning to understand how cells manage to establish and maintain a lineage-committed stage at the molecular level. The molecular mechanisms of lineage stability are better defined than those of lineage choice. Here we discuss some exemplary mechanisms of intrinsic and extrinsic control of lineage choice.
The differentiation of a multipotent cell to a specific lineage involves a global change of gene expression. Lineage choice and commitment are accompanied by the induction and maintenance of lineage-affiliated genetic programs. These include not only the expression of lineage-specific genes but also the repression of those specific for other lineages. Stable gene expression requires the presence and activity of a set of distinct TFs, which are integrated in complex networks with other TFs, modulating cofactors, chromatin modifiers, microRNAs, and other regulatory RNAs (Davidson 2010).
Most current knowledge about TF function in hematopoiesis has been gained in static gene-by-gene analyses. Genetically modified mouse models dissect TF function in distinct cell stages during hematopoiesis. These analyses unravel central players of genetic networks but ignore less prominent components that are vital for the orchestration of the whole program and the dynamic regulation of these networks. In the future we will also need to systemically understand the dynamic changes of expression or activity of the whole ensemble of lineage regulators. Deep parallel sequencing methods allowing quantitative whole-genome information about TF binding to DNA, chromatin status, and resulting transcriptional activity will be of great use for this endeavor. Most of these studies have elucidated far more binding sites and DNA motifs than originally predicted, often at sites far from expected promoter regions. These potential cis- or trans-acting sites would have been undetected by conventional methods. However, the technical requirement for high cell numbers interferes with the necessity to analyze distinct primary cell populations at many different time points during differentiation. These usually are very infrequent, and enrichments still yield insufficient numbers and purity. Technical improvement is eagerly awaited to allow these comprehensive analyses from small cell numbers and ideally from single cells (Islam et al. 2011).
3.2.2.1. Stability of Lineage Commitment
In contrast to other tissues, plasticity between lineages, with frequent physiological “transdifferentiation” of cells of one lineage into another, has not been observed in the hematopoietic system. Although maintenance of lineage choice is critical for normal hematopoiesis, its targeted manipulation could also be used to induce dedifferentiation or differentiation in another lineage for therapeutic purposes.
The expulsion of the nucleus during late stages of erythroid maturation is—although implemented for other reasons—probably the most drastic way of preventing the activation of genetic programs of another lineage. In most other cell types, lineage commitment is not entirely an irreversible state. In contrast, it must be actively maintained by sustained commitment factor expression or network stability in committed cells and their progeny. Positive autoregulation of a lineage-specific factor while inhibiting opposing factors leads to stable situations with one factor expressed and the other repressed (Kerenyi and Orkin 2010). One excellent example is the switch from high levels of GATA2 to high levels of GATA1, which precedes erythropoiesis from HSCs and is called the “GATA switch” (Fig. 4A). GATA2 is mainly expressed in early progenitors and induces GATA1 expression, which in turn activates its own expression and represses GATA2. The switch is mediated by the displacement of GATA2 from its own upstream enhancer by increasing levels of the interacting TF pair GATA1 and Friend of GATA1 (FOG1) (Grass et al. 2003). Moreover, the displacement of GATA2 by GATA1–FOG1 at the c-Kit locus results in rearranged chromatin looping and down-regulated c-Kit expression, demonstrating the ability of TFs to directly alter long-range chromatin interactions (Jing et al. 2008). These switches can be implemented rapidly, taking only one specific S phase for their implementation, thus driving cells to the next stage of maturation in very short time (Pop et al. 2010).
Figure 4.
Network motifs for induction and maintenance of lineage commitment. Simplified examples of molecular mechanisms and networks of stable commitment induction and propagation in erythroid (A), B-lymphoid (B), T-lymphoid (C), and myeloid (D) cells. Direct or indirect activation (arrows) and repression (barred lines) of individual factor expression are indicated. Transcription factors are depicted in white, surface receptors in gray. Dashed lines represent indirect interactions.
Lineage commitment in B-cell development is orchestrated in a regulatory network of key TFs with feed-forward regulatory cascades (Fig. 4B). PU.1 and E2A are critical for lymphoid cell-fate determination and induce specific B-lineage commitment factors like early B-cell factor 1 (EBF1) and Pax5. EBF1 is a primary determinant of B-cell fate, and its expression is controlled by high expression of IKAROS and E2A in conjunction with PU.1 and by extrinsic signals of the IL-7 receptor. EBF1-deficient cells arrest in early B-cell development (before the immunoglobulin heavy-chain rearrangement) with a failure to initiate the early B-lineage program of gene expression. E2A might promote the generation of LMPPs by antagonizing megakaryocyte/erythroid-lineage programs and priming the transcription of the B-cell-specific factors EBF1 and Pax5 (Dias et al. 2008). B-cell differentiation in the absence of E2A can be rescued with EBF1 and Pax5 (Seet et al. 2004; Kwon et al. 2008). Progenitors with genetically ablated E2A or EBF1 retain their multilineage potential (with the exception of the erythroid lineage) (Dias et al. 2008; Semerad et al. 2009). EBF1 can antagonize the expression of myeloid determining factors such as PU.1, C/EBPα, and Id2 and induce synergistically with E2A the expression of Pax5 as the main determinant factor of B-cell commitment. The stable B-cell fate is locked in by a feedback loop of Pax5 and IKAROS to sustain EBF1 expression. All factors function in a concerted manner in pro-B cells to ensure the repression of myeloid genes and thus ensure stability of B-cell commitment (Pridans et al. 2008; Decker et al. 2009). Pax5 deletion leads not only to a B-cell differentiation block at the pre-/pro-B-cell stage, but it also enables B cells to dedifferentiate into an LMPP-like stage and to allow entry into normal alternative differentiation paths into various myeloid and lymphoid lineages in vivo and in vitro (Nutt et al. 1999; Rolink et al. 1999; Mikkola et al. 2002; Delogu et al. 2006; Cobaleda et al. 2007). Pax5 is directly involved in epigenetic alterations of B-lineage-specific genes (Schebesta et al. 2007; Gao et al. 2009; McManus et al. 2011). Interestingly, ectopic expression of EBF1 in Pax5-deficient hematopoietic progenitors restricts their alternate lineage potential in vivo (Nutt et al. 1999; Rolink et al. 1999; Mikkola et al. 2002; Delogu et al. 2006; Pongubala et al. 2008). This repression of other lineage-specific molecular programs by Pax5 and partners is an excellent example of how a lineage choice can be kept stable. This mechanism also requires sustained expression and activity of these factors and might therefore be less stable against perturbations than other mechanisms, for example, involving chromatin modifications. Subtle changes and misregulation of individual TFs will directly impact normal differentiation and can lead to leukemia (Rosenbauer et al. 2006).
Epigenetic activation or silencing of lineage-restricting programs by chromatin and DNA modifications may ultimately determine lineage commitment. As one example, the c-fms locus (coding for the macrophage CSF [M-CSF] receptor) becomes epigenetically silenced during B-cell differentiation in a gradual process, inhibiting remaining myeloid potential (Tagoh et al. 2004). However, sustained Pax5 expression is still required for maintaining c-fms locus silencing, because mature B cells rapidly up-regulate M-CSF receptor expression upon Pax5 deletion. These findings well illustrate that TFs function by altering chromatin states and directly recruiting epigenetic modifying complexes (McManus et al. 2011).
In contrast to the intrinsically stabilized B-cell commitment network, T-lineage fate choice remains provisional for an extended period of differentiation with high dependency on extrinsic stimulation from the thymic environment (Fig. 4C). T-cell fate initiation and determination requires persistent extrinsic Notch receptor signaling, which is stimulated by Delta-like 1 and 4 expressing cortical endothelial cells in the thymus (Radtke et al. 1999). Notch signaling turns LMPPs into early T-cell progenitors by inducing a pro-T-cell developmental program with direct and indirect activation of GATA3, Tcf7, and other T-cell-restricting genes, which, once initiated, positively autoregulate their own expression. The activation of a T-lineage program by Notch signaling is, however, not sufficient for T-cell fate determination alone. Additionally, Notch suppresses B-cell-lineage-determining EBF1 and E2A expression and diminishes remaining myeloid potential by down-regulation of PU.1 and C/EBPα (Ordentlich et al. 1998; Smith et al. 2005; Laiosa et al. 2006b). Id2 expression is blocked by Notch signaling to circumvent NK cell development (Ikawa et al. 2001). Conversely, the TF leukemia/lymphoma-related factor (LRF) inhibits Notch signaling in the BM, thus preventing T-cell development. Consequently, the conditional deletion of LRF in HSCs results in the generation of T-lineage progeny in the BM (Maeda et al. 2007).
3.2.2.2. Extrinsic Regulation of Lineage Choice—Instructive versus Selective Function of Cytokines
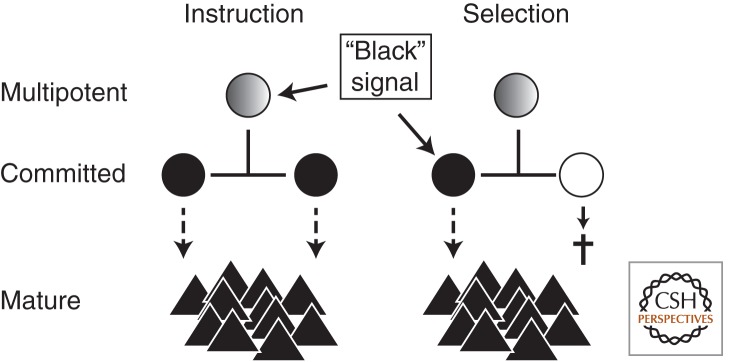
The mechanisms discussed above can explain how a lineage choice, once it is made and implemented, can be maintained. Before this state, molecular changes in uncommitted cells must lead to the actual lineage choice and the induction of these maintenance programs. One way of inducing lineage choice is by signals from outside the cells, for example, by hematopoietic cytokines. Cytokines are very important regulators of hematopoiesis. They affect multiple cell fates of every hematopoietic cell type, including survival, proliferation, maturation, and functional activation (Rieger and Schroeder 2009). In addition to these functions, the instruction of hematopoietic lineage choice by cytokines has been postulated for decades (Metcalf 1998, 2010). However, it proved to be surprisingly difficult to show beyond doubt that cytokines do not merely allow the survival and proliferation of precursors that have already intrinsically—and independently of the presence of a cytokine—committed to one lineage (permissive/selective model) (Fig. 5) (Enver et al. 1998). The pleiotropic, redundant, and cell-type-dependent effects of cytokines are one reason for the difficulty in precisely defining their effects on cell fates (Rieger and Schroeder 2009). Loss-of-function studies have thus provided limited insight. Most studies suggesting lineage instruction by cytokines have therefore relied on ectopic molecular overexpression and/or transformed cell lines. As an example, the ectopic expression of IL-2 receptor converted CLPs from the lymphoid to the myeloid lineage, probably through ectopic up-regulation of granulocyte–macrophage CSF (GM-CSF) receptor upon IL-2 stimulation (Kondo et al. 2000; Iwasaki-Arai et al. 2003). However, the effects of ectopically expressed molecules may not be representing physiologically relevant programs of lineage choice. Only recently has lineage instruction of unmanipulated primary hematopoietic progenitors by cytokines to which they physiologically respond been proven (Rieger et al. 2009a). Culture of granulocyte–macrophage progenitors (GMPs) in only G-CSF or M-CSF will eventually lead to production of only granulocytic or monocytic cells, respectively. In the selective model, GMPs would produce progeny of both lineages even if only one cytokine was present. The absence of one lineage-specific cytokine would then lead to the death of cells that have committed to this “wrong” lineage, leading to a monolineage output (Fig. 5). However, the continuous observation of individual GMPs and all of their progeny throughout the differentiation process demonstrated the lack of cell death events during the cytokine-dependent monolineage differentiation of GMPs (Fig. 5). A purely selective effect could therefore be excluded, demonstrating GMP lineage instruction by these cytokines (Rieger et al. 2009a). However, the involved molecular mechanisms are poorly understood. A signal strength-dependent model of dendritic cell development has recently been proposed, in which dendritic cell development signals are integrated from cytokine receptor expression, downstream signal transduction, and the presence of the cytokines M-CSF, Flt3 ligand, and GM-CSF in specific niches (Schmid et al. 2010). This kind of model could have broader relevance in understanding the role of cytokines in instructing lineage development.
Figure 5.
Possible influence of extracellular signals on production of lineage-committed cells. Extrinsic signals by, for example, hematopoietic cytokines may instruct the lineage choice of multipotent cells. Alternatively, extrinsic signals could merely enable survival, proliferation, and maturation signals of cells that have already committed to one lineage independently of the extrinsic signal. Importantly, static analyses at the start and end of an experiment would lead to identical data in both situations, and continuous single-cell observations are required for definitive conclusions about cytokine function.
Despite this accumulating evidence for lineage instruction by cell-extrinsic signals in some of the more restricted progenitors, their impact on earlier branching points of the hematopoietic hierarchy still remains poorly understood. Cytokines contribute in a major way to the survival and proliferation of different hematopoietic cell types, and not all lineage-affiliated cytokines have a lineage-instructive ability. Even if specific lineages can be instructed by cytokines, this does not preclude the existence of additional cell-intrinsic lineage choice mechanisms that could lead to lineage choice in the absence of cell-extrinsic instructive signals. The capacity of individual extrinsic signals to instruct lineage choice, as well as its integration with other cytokine signals and with other intracellular molecular states like TF expression, must be further analyzed for each individual cytokine, in specific cell types, under chemically defined conditions, and ideally continuously at the single-cell level.
3.2.2.3. Intrinsic Regulation of Lineage Choice by TFs—Decision Makers or Only Executors?
It is clear that individual TFs can instruct lineage choice and are even able to reprogram committed cells, leading to crossing of lineage borders. For example, ectopic expression of GATA1 in multipotent cells or cells committed to other than the megakaryocyte–erythrocyte lineages (e.g., CLPs) enforces the development into erythroid and megakaryocytic cells (Heyworth et al. 2002). C/EBPα and PU.1 are essential for the generation of granulocyte–macrophage progenitors. C/EBPα expression in committed lymphoid cells (B and T cells) instructs the development of macrophages (Xie et al. 2004; Laiosa et al. 2006a,b). Furthermore, committed T cells transdifferentiate into myeloid dendritic cells upon ectopic PU.1 expression (Laiosa et al. 2006b). TFs also actively repress lineage programs. To this end, the myeloid potential as well as PU.1- or C/EBPα-dependent myeloid reprogramming of thymic precursors can be blocked by active Notch signaling (Franco et al. 2006; Laiosa et al. 2006b; Rothenberg 2007), suggesting an instructive extracellular non-cytokine-mediated induction of T-cell identity. Findings like these clearly demonstrate the ability of many lineage-specific TFs to drive cells into one lineage. Upon their up-regulation or activation, they will start the transcription of myriads of direct and indirect target genes and chromatin modifications, which lead to the changing phenotypes of lineage-committing and maturing cells.
However, the ability of TFs to drive a cell into one lineage does not yet explain how the initial decision for this lineage was made—it only explains how this decision is then executed. What led to the up-regulation or activation of, for example, the executing TF remains unclear. One possibility is the instruction of lineage choice by cell-extrinsic signals. In this case, TFs are most likely the final executors, implementing the lineage decision, but are not involved in the actual decision making. Another intriguing possible mechanism is the stochastic output of fluctuating cell-intrinsic networks of TFs. In this case, TFs would not only execute a previously made decision, they would also be the relevant components of the molecular machinery generating the lineage choice. This hypothesis stems from the initially surprising findings that multiple “lineage-specific” TFs are coexpressed in multipotent cells before lineage commitment (“lineage priming”; see, e.g., Miyamoto et al. 2002). These factors are expressed only exclusively in different mature lineages, and they can drive cells into this lineage upon overexpression. Initially, therefore, it was assumed that multipotent cells have the potential to differentiate into multiple lineages because they lack the expression of these lineage-specific TFs. However, with the availability of improved prospective purification of multipotent cells and more sensitive molecular methods for the detection of TF expression, it then became clear that the opposite could be true. Multipotency might in fact be characterized not by the absence of lineage-specific properties but by the presence of properties of multiple lineages, in particular the coexpression of multiple “lineage-specific” TFs. The additional finding that these TFs often inhibit each other by binding mutually or to cofactors of opposing TFs led to the idea that these factors may neutralize each other in uncommitted cells, thus retaining multipotency. Random fluctuations in their expression could then lead to one TF gaining dominance; positive autoregulation and repression of other TFs would lead to a lineage-committed state in which only one lineage-specific TF is expressed (Enver et al. 1998; Cantor and Orkin 2001; Graf and Enver 2009). The production of all lineages would be ensured by the overall wiring of the TF network leading to stable stochastic lineage output. Although not predictable for an individual cell, different cells within a population would fluctuate into decisions for different lineages with specific probabilities. Adaptation of the blood system to stress or injury would then be regulated by the selection of existing lineage-committed progenitors for survival and proliferation (Suda et al. 1984).
Studies on transcriptional pathways that control binary lineage choices in hematopoiesis revealed some similarities in the gene regulatory network circuits of different lineages. Pairs of TFs that mutually antagonize each other’s activity and expression are often involved. One example is the antagonistic interplay between the lineage-determining factors PU.1 and GATA1 as a molecular mechanism of lineage choice between myeloid and megakaryocytic–erythroid fate, respectively (Fig. 4D). PU.1 and GATA1 physically bind each other and cross-antagonize their activity (Zhang et al. 1999, 2000; Stopka et al. 2005; Liew et al. 2006). PU.1 directly inhibits GATA1 DNA-binding capacity, while GATA1 inhibits the transactivation potential of PU.1. Both PU.1 and GATA1 are autoregulatory for their own expression (Nishimura et al. 2000; Okuno et al. 2005; Laiosa et al. 2006a), thereby providing stability to their levels, once expressed. The genetic ablation of these factors underpins their central role in implementing lineage choice. Loss of GATA1 demonstrates its absolute necessity for megakaryocyte/erythrocyte development, whereas PU.1 deficiency leads to a lack of granulocytes, macrophages, and B cells. Furthermore, both factors have instructive lineage commitment ability by implementing lineage-affiliated gene programs (Iwasaki et al. 2003, 2006). However, recent studies suggest a more complicated dynamic implementation. In fish hematopoiesis, the interplay of PU.1 and GATA1 differs in various cell stages during hematopoiesis and is influenced by other factors, such as the transcription intermediate factor 1g (tif1g), a RING domain E3 ubiquitin ligase (Monteiro et al. 2011). Moreover, PU.1 showed positive autoregulation in all analyzed cell stages, but GATA1 only in some of them. C/EBPα and FOG1 have recently been implicated in the lineage choice between myeloid and megakaryocyte/erythrocyte lineages by exhibiting transcriptional cross-regulation, potentially being better candidates for lineage choice making than PU.1 and GATA1 (Mancini et al. 2011).
Again, all of these models suffer from the fact that they are based on expression analyses with very low resolution. Precise timing of changing PU.1 and GATA1 protein levels before and during lineage choice in the individual cell would be critical for this mechanism of lineage choice. However, current expression analyses measure only population averages and/or RNA levels and/or single time points or maturation stages. This static data allows many alternative explanations and is therefore not sufficient to prove the mechanism of lineage choice. Ideally, novel technology will enable the simultaneous quantification of protein levels of multiple TFs, at the single-cell level, continuously with high temporal resolution, and throughout lineage choice of living HSCs.
4. CONCLUSION/OUTLOOK
In conclusion, hematopoiesis is an excellent model for studying the molecular mechanisms of cell-fate control. Being the classical mammalian stem cell model, with high clinical relevance, robust quantitative functional assays, and well-established culture and prospective isolation techniques, it allows us to address the underlying mechanisms of cell-fate control. Most of the basic concepts in stem cell research have been defined in the hematopoietic system, and many novel technical approaches are first developed or applied in hematopoiesis research. Because of the routine use of hematopoietic cells for clinical therapy, the chances for a quick transfer of novel basic insights to patient benefit in the clinic are higher than for most other tissues. Nevertheless, despite decades of successful research, many questions posed very long ago are still awaiting an answer, and long-standing disputes illustrate the need for ever improving technological approaches. These continue to be exciting times in hematopoiesis research.
ACKNOWLEDGMENTS
M.A.R. is thankful for the support of the LOEWE Center for Cell and Gene Therapy Frankfurt (HMWK III L 4- 518/17.004 [2010]) and institutional funds of the Georg-Speyer-Haus. The Georg-Speyer-Haus is funded jointly by the German Federal Ministry of Health (BMG) and the Ministry of Higher Education, Research and the Arts of the state of Hessen (HMWK).
Footnotes
Editors: Patrick P.L. Tam, W. James Nelson, and Janet Rossant
Additional Perspectives on Mammalian Development available at www.cshperspectives.org
REFERENCES
- Adolfsson J, Mansson R, Buza-Vidas N, Hultquist A, Liuba K, Jensen CT, Bryder D, Yang L, Borge OJ, Thoren LA, et al. 2005. Identification of Flt3+ lympho-myeloid stem cells lacking erythro-megakaryocytic potential: A revised road map for adult blood lineage commitment. Cell 121: 295–306 [DOI] [PubMed] [Google Scholar]
- Akashi K, Traver D, Miyamoto T, Weissman IL 2000. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature 404: 193–197 [DOI] [PubMed] [Google Scholar]
- Antonchuk J, Sauvageau G, Humphries RK 2002. HOXB4-induced expansion of adult hematopoietic stem cells ex vivo. Cell 109: 39–45 [DOI] [PubMed] [Google Scholar]
- Arinobu Y, Mizuno S, Chong Y, Shigematsu H, Iino T, Iwasaki H, Graf T, Mayfield R, Chan S, Kastner P, et al. 2007. Reciprocal activation of GATA-1 and PU.1 marks initial specification of hematopoietic stem cells into myeloerythroid and myelolymphoid lineages. Cell Stem Cell 1: 416–427 [DOI] [PubMed] [Google Scholar]
- Balazs AB, Fabian AJ, Esmon CT, Mulligan RC 2006. Endothelial protein C receptor (CD201) explicitly identifies hematopoietic stem cells in murine bone marrow. Blood 107: 2317–2321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker AJ, McCulloch EA, Till JE 1963. Cytological demonstration of the clonal nature of spleen colonies derived from transplanted mouse marrow cells. Nature 197: 452–454 [DOI] [PubMed] [Google Scholar]
- Benveniste P, Frelin C, Janmohamed S, Barbara M, Herrington R, Hyam D, Iscove NN 2010. Intermediate-term hematopoietic stem cells with extended but time-limited reconstitution potential. Cell Stem Cell 6: 48–58 [DOI] [PubMed] [Google Scholar]
- Bertoncello I, Hodgson GS, Bradley TR 1985. Multiparameter analysis of transplantable hemopoietic stem cells: I. The separation and enrichment of stem cells homing to marrow and spleen on the basis of rhodamine-123 fluorescence. Exp Hematol 13: 999–1006 [PubMed] [Google Scholar]
- Bertrand JY, Chi NC, Santoso B, Teng S, Stainier DY, Traver D 2010a. Haematopoietic stem cells derive directly from aortic endothelium during development. Nature 464: 108–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand JY, Cisson JL, Stachura DL, Traver D 2010b. Notch signaling distinguishes 2 waves of definitive hematopoiesis in the zebrafish embryo. Blood 115: 2777–2783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatia M, Wang JC, Kapp U, Bonnet D, Dick JE 1997. Purification of primitive human hematopoietic cells capable of repopulating immune-deficient mice. Proc Natl Acad Sci 94: 5320–5325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boitano AE, Wang J, Romeo R, Bouchez LC, Parker AE, Sutton SE, Walker JR, Flaveny CA, Perdew GH, Denison MS, et al. 2010. Aryl hydrocarbon receptor antagonists promote the expansion of human hematopoietic stem cells. Science 329: 1345–1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowie MB, Kent DG, Dykstra B, McKnight KD, McCaffrey L, Hoodless PA, Eaves CJ 2007. Identification of a new intrinsically timed developmental checkpoint that reprograms key hematopoietic stem cell properties. Proc Natl Acad Sci 104: 5878–5882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broske AM, Vockentanz L, Kharazi S, Huska MR, Mancini E, Scheller M, Kuhl C, Enns A, Prinz M, Jaenisch R, et al. 2009. DNA methylation protects hematopoietic stem cell multipotency from myeloerythroid restriction. Nat Genet 41: 1207–1215 [DOI] [PubMed] [Google Scholar]
- Bryder D, Rossi DJ, Weissman IL 2006. Hematopoietic stem cells: The paradigmatic tissue-specific stem cell. Am J Pathol 169: 338–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvi LM, Adams GB, Weibrecht KW, Weber JM, Olson DP, Knight MC, Martin RP, Schipani E, Divieti P, Bringhurst FR, et al. 2003. Osteoblastic cells regulate the haematopoietic stem cell niche. Nature 425: 841–846 [DOI] [PubMed] [Google Scholar]
- Cantor AB, Orkin SH 2001. Hematopoietic development: A balancing act. Curr Opin Genet Dev 11: 513–519 [DOI] [PubMed] [Google Scholar]
- Challen GA, Boles NC, Chambers SM, Goodell MA 2010. Distinct hematopoietic stem cell subtypes are differentially regulated by TGF-β1. Cell Stem Cell 6: 265–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challen GA, Sun D, Jeong M, Luo M, Jelinek J, Berg JS, Bock C, Vasanthakumar A, Gu H, Xi Y, et al. 2011. Dnmt3a is essential for hematopoietic stem cell differentiation. Nat Genet 44: 23–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang JT, Palanivel VR, Kinjyo I, Schambach F, Intlekofer AM, Banerjee A, Longworth SA, Vinup KE, Mrass P, Oliaro J, et al. 2007. Asymmetric T lymphocyte division in the initiation of adaptive immune responses. Science 315: 1687–1691 [DOI] [PubMed] [Google Scholar]
- Chen AT, Zon LI 2009. Zebrafish blood stem cells. J Cell Biochem 108: 35–42 [DOI] [PubMed] [Google Scholar]
- Chen MJ, Li Y, De Obaldia ME, Yang Q, Yzaguirre AD, Yamada-Inagawa T, Vink CS, Bhandoola A, Dzierzak E, Speck NA 2011. Erythroid/myeloid progenitors and hematopoietic stem cells originate from distinct populations of endothelial cells. Cell Stem Cell 9: 541–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobaleda C, Jochum W, Busslinger M 2007. Conversion of mature B cells into T cells by dedifferentiation to uncommitted progenitors. Nature 449: 473–477 [DOI] [PubMed] [Google Scholar]
- Dahl R, Walsh JC, Lancki D, Laslo P, Iyer SR, Singh H, Simon MC 2003. Regulation of macrophage and neutrophil cell fates by the PU.1:C/EBPα ratio and granulocyte colony-stimulating factor. Nat Immunol 4: 1029–1036 [DOI] [PubMed] [Google Scholar]
- Davidson EH 2010. Emerging properties of animal gene regulatory networks. Nature 468: 911–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer J, Williams A, Skavdis G, Harker N, Coles M, Tolaini M, Norton T, Williams K, Roderick K, Potocnik AJ, et al. 2003. Transgenic mice with hematopoietic and lymphoid specific expression of Cre. Eur J Immunol 33: 314–325 [DOI] [PubMed] [Google Scholar]
- Decker T, Pasca di Magliano M, McManus S, Sun Q, Bonifer C, Tagoh H, Busslinger M 2009. Stepwise activation of enhancer and promoter regions of the B cell commitment gene Pax5 in early lymphopoiesis. Immunity 30: 508–520 [DOI] [PubMed] [Google Scholar]
- Delogu A, Schebesta A, Sun Q, Aschenbrenner K, Perlot T, Busslinger M 2006. Gene repression by Pax5 in B cells is essential for blood cell homeostasis and is reversed in plasma cells. Immunity 24: 269–281 [DOI] [PubMed] [Google Scholar]
- Dias S, Mansson R, Gurbuxani S, Sigvardsson M, Kee BL 2008. E2A proteins promote development of lymphoid-primed multipotent progenitors. Immunity 29: 217–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick JE 2008. Stem cell concepts renew cancer research. Blood 112: 4793–4807 [DOI] [PubMed] [Google Scholar]
- Dieterlen-Lievre F, Pouget C, Bollerot K, Jaffredo T 2006. Are intra-aortic hemopoietic cells derived from endothelial cells during ontogeny? Trends Cardiovasc Med 16: 128–139 [DOI] [PubMed] [Google Scholar]
- Ding L, Saunders TL, Enikolopov G, Morrison SJ 2012. Endothelial and perivascular cells maintain haematopoietic stem cells. Nature 481: 457–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doulatov S, Notta F, Eppert K, Nguyen LT, Ohashi PS, Dick JE 2010. Revised map of the human progenitor hierarchy shows the origin of macrophages and dendritic cells in early lymphoid development. Nat Immunol 11: 585–593 [DOI] [PubMed] [Google Scholar]
- Dykstra B, Ramunas J, Kent D, McCaffrey L, Szumsky E, Kelly L, Farn K, Blaylock A, Eaves C, Jervis E 2006. High-resolution video monitoring of hematopoietic stem cells cultured in single-cell arrays identifies new features of self-renewal. Proc Natl Acad Sci 103: 8185–8190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dykstra B, Kent D, Bowie M, McCaffrey L, Hamilton M, Lyons K, Lee SJ, Brinkman R, Eaves C 2007. Long-term propagation of distinct hematopoietic differentiation programs in vivo. Cell Stem Cell 1: 218–229 [DOI] [PubMed] [Google Scholar]
- Ehninger A, Trumpp A 2011. The bone marrow stem cell niche grows up: Mesenchymal stem cells and macrophages move in. J Exp Med 208: 421–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eilken HM, Nishikawa S, Schroeder T 2009. Continuous single-cell imaging of blood generation from haemogenic endothelium. Nature 457: 896–900 [DOI] [PubMed] [Google Scholar]
- Ema H, Sudo K, Seita J, Matsubara A, Morita Y, Osawa M, Takatsu K, Takaki S, Nakauchi H 2005. Quantification of self-renewal capacity in single hematopoietic stem cells from normal and Lnk-deficient mice. Dev Cell 8: 907–914 [DOI] [PubMed] [Google Scholar]
- Enver T, Heyworth CM, Dexter TM 1998. Do stem cells play dice? Blood 92: 348–351; discussion 352 [PubMed] [Google Scholar]
- Franco CB, Scripture-Adams DD, Proekt I, Taghon T, Weiss AH, Yui MA, Adams SL, Diamond RA, Rothenberg EV 2006. Notch/Delta signaling constrains reengineering of pro-T cells by PU.1. Proc Natl Acad Sci 103: 11993–11998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H, Lukin K, Ramirez J, Fields S, Lopez D, Hagman J 2009. Opposing effects of SWI/SNF and Mi-2/NuRD chromatin remodeling complexes on epigenetic reprogramming by EBF and Pax5. Proc Natl Acad Sci 106: 11258–11263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gekas C, Dieterlen-Lievre F, Orkin SH, Mikkola HK 2005. The placenta is a niche for hematopoietic stem cells. Dev Cell 8: 365–375 [DOI] [PubMed] [Google Scholar]
- Goodell MA, Brose K, Paradis G, Conner AS, Mulligan RC 1996. Isolation and functional properties of murine hematopoietic stem cells that are replicating in vivo. J Exp Med 183: 1797–1806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf T, Enver T 2009. Forcing cells to change lineages. Nature 462: 587–594 [DOI] [PubMed] [Google Scholar]
- Grass JA, Boyer ME, Pal S, Wu J, Weiss MJ, Bresnick EH 2003. GATA-1-dependent transcriptional repression of GATA-2 via disruption of positive autoregulation and domain-wide chromatin remodeling. Proc Natl Acad Sci 100: 8811–8816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S, Lu J, Schlanger R, Zhang H, Wang JY, Fox MC, Purton LE, Fleming HH, Cobb B, Merkenschlager M, et al. 2010. MicroRNA miR-125a controls hematopoietic stem cell number. Proc Natl Acad Sci 107: 14229–14234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He S, Kim I, Lim MS, Morrison SJ 2011. Sox17 expression confers self-renewal potential and fetal stem cell characteristics upon adult hematopoietic progenitors. Genes Dev 25: 1613–1627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera-Merchan A, Arranz L, Ligos JM, de Molina A, Dominguez O, Gonzalez S 2012. Ectopic expression of the histone methyltransferase Ezh2 in haematopoietic stem cells causes myeloproliferative disease. Nat Commun 3: 623. [DOI] [PubMed] [Google Scholar]
- Heyworth C, Pearson S, May G, Enver T 2002. Transcription factor-mediated lineage switching reveals plasticity in primary committed progenitor cells. EMBO J 21: 3770–3781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hock H, Hamblen MJ, Rooke HM, Schindler JW, Saleque S, Fujiwara Y, Orkin SH 2004. Gfi-1 restricts proliferation and preserves functional integrity of haematopoietic stem cells. Nature 431: 1002–1007 [DOI] [PubMed] [Google Scholar]
- Hope KJ, Cellot S, Ting SB, MacRae T, Mayotte N, Iscove NN, Sauvageau G 2010. An RNAi screen identifies Msi2 and Prox1 as having opposite roles in the regulation of hematopoietic stem cell activity. Cell Stem Cell 7: 101–113 [DOI] [PubMed] [Google Scholar]
- Ichikawa M, Asai T, Saito T, Seo S, Yamazaki I, Yamagata T, Mitani K, Chiba S, Ogawa S, Kurokawa M, et al. 2004. AML-1 is required for megakaryocytic maturation and lymphocytic differentiation, but not for maintenance of hematopoietic stem cells in adult hematopoiesis. Nat Med 10: 299–304 [DOI] [PubMed] [Google Scholar]
- Ikawa T, Fujimoto S, Kawamoto H, Katsura Y, Yokota Y 2001. Commitment to natural killer cells requires the helix-loop-helix inhibitor Id2. Proc Natl Acad Sci 98: 5164–5169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inlay MA, Bhattacharya D, Sahoo D, Serwold T, Seita J, Karsunky H, Plevritis SK, Dill DL, Weissman IL 2009. Ly6d marks the earliest stage of B-cell specification and identifies the branchpoint between B-cell and T-cell development. Genes Dev 23: 2376–2381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam S, Kjallquist U, Moliner A, Zajac P, Fan JB, Lonnerberg P, Linnarsson S 2011. Characterization of the single-cell transcriptional landscape by highly multiplex RNA-seq. Genome Res 21: 1160–1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Kwon HY, Zimdahl B, Congdon KL, Blum J, Lento WE, Zhao C, Lagoo A, Gerrard G, Foroni L, et al. 2010. Regulation of myeloid leukaemia by the cell-fate determinant Musashi. Nature 466: 765–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanovs A, Rybtsov S, Welch L, Anderson RA, Turner ML, Medvinsky A 2011. Highly potent human hematopoietic stem cells first emerge in the intraembryonic aorta-gonad-mesonephros region. J Exp Med 208: 2417–2427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwama A, Oguro H, Negishi M, Kato Y, Morita Y, Tsukui H, Ema H, Kamijo T, Katoh-Fukui Y, Koseki H, et al. 2004. Enhanced self-renewal of hematopoietic stem cells mediated by the polycomb gene product Bmi-1. Immunity 21: 843–851 [DOI] [PubMed] [Google Scholar]
- Iwasaki H, Mizuno S, Wells RA, Cantor AB, Watanabe S, Akashi K 2003. GATA-1 converts lymphoid and myelomonocytic progenitors into the megakaryocyte/erythrocyte lineages. Immunity 19: 451–462 [DOI] [PubMed] [Google Scholar]
- Iwasaki H, Mizuno S, Arinobu Y, Ozawa H, Mori Y, Shigematsu H, Takatsu K, Tenen DG, Akashi K 2006. The order of expression of transcription factors directs hierarchical specification of hematopoietic lineages. Genes Dev 20: 3010–3021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki-Arai J, Iwasaki H, Miyamoto T, Watanabe S, Akashi K 2003. Enforced granulocyte/macrophage colony-stimulating factor signals do not support lymphopoiesis, but instruct lymphoid to myelomonocytic lineage conversion. J Exp Med 197: 1311–1322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing L, Zon LI 2011. Zebrafish as a model for normal and malignant hematopoiesis. Dis Model Mech 4: 433–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing H, Vakoc CR, Ying L, Mandat S, Wang H, Zheng X, Blobel GA 2008. Exchange of GATA factors mediates transitions in looped chromatin organization at a developmentally regulated gene locus. Mol Cell 29: 232–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataoka H, Hayashi M, Nakagawa R, Tanaka Y, Izumi N, Nishikawa S, Jakt ML, Tarui H, Nishikawa S 2011a. Etv2/ER71 induces vascular mesoderm from Flk1+PDGFRα+ primitive mesoderm. Blood 118: 6975–6986 [DOI] [PubMed] [Google Scholar]
- Kataoka K, Sato T, Yoshimi A, Goyama S, Tsuruta T, Kobayashi H, Shimabe M, Arai S, Nakagawa M, Imai Y, et al. 2011b. Evi1 is essential for hematopoietic stem cell self-renewal, and its expression marks hematopoietic cells with long-term multilineage repopulating activity. J Exp Med 208: 2403–2416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamoto H, Ikawa T, Masuda K, Wada H, Katsura Y 2010. A map for lineage restriction of progenitors during hematopoiesis: The essence of the myeloid-based model. Immunol Rev 238: 23–36 [DOI] [PubMed] [Google Scholar]
- Kent DG, Copley MR, Benz C, Wohrer S, Dykstra BJ, Ma E, Cheyne J, Zhao Y, Bowie MB, Zhao Y, et al. 2009. Prospective isolation and molecular characterization of hematopoietic stem cells with durable self-renewal potential. Blood 113: 6342–6350 [DOI] [PubMed] [Google Scholar]
- Kerenyi MA, Orkin SH 2010. Networking erythropoiesis. J Exp Med 207: 2537–2541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharas MG, Lengner CJ, Al-Shahrour F, Bullinger L, Ball B, Zaidi S, Morgan K, Tam W, Paktinat M, Okabe R, et al. 2010. Musashi-2 regulates normal hematopoiesis and promotes aggressive myeloid leukemia. Nat Med 16: 903–908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiel MJ, Yilmaz OH, Iwashita T, Yilmaz OH, Terhorst C, Morrison SJ 2005. SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell 121: 1109–1121 [DOI] [PubMed] [Google Scholar]
- Kim I, Saunders TL, Morrison SJ 2007. Sox17 dependence distinguishes the transcriptional regulation of fetal from adult hematopoietic stem cells. Cell 130: 470–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kissa K, Herbomel P 2010. Blood stem cells emerge from aortic endothelium by a novel type of cell transition. Nature 464: 112–115 [DOI] [PubMed] [Google Scholar]
- Kondo M, Weissman IL, Akashi K 1997. Identification of clonogenic common lymphoid progenitors in mouse bone marrow. Cell 91: 661–672 [DOI] [PubMed] [Google Scholar]
- Kondo M, Scherer DC, Miyamoto T, King AG, Akashi K, Sugamura K, Weissman IL 2000. Cell-fate conversion of lymphoid-committed progenitors by instructive actions of cytokines. Nature 407: 383–386 [DOI] [PubMed] [Google Scholar]
- Kuhn R, Schwenk F, Aguet M, Rajewsky K 1995. Inducible gene targeting in mice. Science 269: 1427–1429 [DOI] [PubMed] [Google Scholar]
- Kwon K, Hutter C, Sun Q, Bilic I, Cobaleda C, Malin S, Busslinger M 2008. Instructive role of the transcription factor E2A in early B lymphopoiesis and germinal center B cell development. Immunity 28: 751–762 [DOI] [PubMed] [Google Scholar]
- Laiosa CV, Stadtfeld M, Graf T 2006a. Determinants of lymphoid-myeloid lineage diversification. Annu Rev Immunol 24: 705–738 [DOI] [PubMed] [Google Scholar]
- Laiosa CV, Stadtfeld M, Xie H, de Andres-Aguayo L, Graf T 2006b. Reprogramming of committed T cell progenitors to macrophages and dendritic cells by C/EBPα and PU.1 transcription factors. Immunity 25: 731–744 [DOI] [PubMed] [Google Scholar]
- Laurenti E, Varnum-Finney B, Wilson A, Ferrero I, Blanco-Bose WE, Ehninger A, Knoepfler PS, Cheng PF, MacDonald HR, Eisenman RN, et al. 2008. Hematopoietic stem cell function and survival depend on c-Myc and N-Myc activity. Cell Stem Cell 3: 611–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D, Park C, Lee H, Lugus JJ, Kim SH, Arentson E, Chung YS, Gomez G, Kyba M, Lin S, et al. 2008. ER71 acts downstream of BMP, Notch, and Wnt signaling in blood and vessel progenitor specification. Cell Stem Cell 2: 497–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessard J, Sauvageau G 2003. Bmi-1 determines the proliferative capacity of normal and leukaemic stem cells. Nature 423: 255–260 [DOI] [PubMed] [Google Scholar]
- Liew CW, Rand KD, Simpson RJ, Yung WW, Mansfield RE, Crossley M, Proetorius-Ibba M, Nerlov C, Poulsen FM, Mackay JP 2006. Molecular analysis of the interaction between the hematopoietic master transcription factors GATA-1 and PU.1. J Biol Chem 281: 28296–28306 [DOI] [PubMed] [Google Scholar]
- Lo Celso C, Fleming HE, Wu JW, Zhao CX, Miake-Lye S, Fujisaki J, Cote D, Rowe DW, Lin CP, Scadden DT 2009. Live-animal tracking of individual haematopoietic stem/progenitor cells in their niche. Nature 457: 92–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda T, Merghoub T, Hobbs RM, Dong L, Maeda M, Zakrzewski J, van den Brink MR, Zelent A, Shigematsu H, Akashi K, et al. 2007. Regulation of B versus T lymphoid lineage fate decision by the proto-oncogene LRF. Science 316: 860–866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majeti R, Park CY, Weissman IL 2007. Identification of a hierarchy of multipotent hematopoietic progenitors in human cord blood. Cell Stem Cell 1: 635–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majewski IJ, Ritchie ME, Phipson B, Corbin J, Pakusch M, Ebert A, Busslinger M, Koseki H, Hu Y, Smyth GK, et al. 2010. Opposing roles of polycomb repressive complexes in hematopoietic stem and progenitor cells. Blood 116: 731–739 [DOI] [PubMed] [Google Scholar]
- Mancini E, Sanjuan-Pla A, Luciani L, Moore S, Grover A, Zay A, Rasmussen KD, Luc S, Bilbao D, O’Carroll D, et al. 2011. FOG-1 and GATA-1 act sequentially to specify definitive megakaryocytic and erythroid progenitors. EMBO J 31: 351–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansson R, Hultquist A, Luc S, Yang L, Anderson K, Kharazi S, Al-Hashmi S, Liuba K, Thoren L, Adolfsson J, et al. 2007. Molecular evidence for hierarchical transcriptional lineage priming in fetal and adult stem cells and multipotent progenitors. Immunity 26: 407–419 [DOI] [PubMed] [Google Scholar]
- Manz MG, Miyamoto T, Akashi K, Weissman IL 2002. Prospective isolation of human clonogenic common myeloid progenitors. Proc Natl Acad Sci 99: 11872–11877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Agosto JA, Mikkola HK, Hartenstein V, Banerjee U 2007. The hematopoietic stem cell and its niche: A comparative view. Genes Dev 21: 3044–3060 [DOI] [PubMed] [Google Scholar]
- Matsumoto A, Takeishi S, Kanie T, Susaki E, Onoyama I, Tateishi Y, Nakayama K, Nakayama KI 2011. p57 is required for quiescence and maintenance of adult hematopoietic stem cells. Cell Stem Cell 9: 262–271 [DOI] [PubMed] [Google Scholar]
- McDermott SP, Eppert K, Lechman ER, Doedens M, Dick JE 2010. Comparison of human cord blood engraftment between immunocompromised mouse strains. Blood 116: 193–200 [DOI] [PubMed] [Google Scholar]
- McManus S, Ebert A, Salvagiotto G, Medvedovic J, Sun Q, Tamir I, Jaritz M, Tagoh H, Busslinger M 2011. The transcription factor Pax5 regulates its target genes by recruiting chromatin-modifying proteins in committed B cells. EMBO J 30: 2388–2404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medvinsky A, Dzierzak E 1996. Definitive hematopoiesis is autonomously initiated by the AGM region. Cell 86: 897–906 [DOI] [PubMed] [Google Scholar]
- Medvinsky A, Rybtsov S, Taoudi S 2011. Embryonic origin of the adult hematopoietic system: Advances and questions. Development 138: 1017–1031 [DOI] [PubMed] [Google Scholar]
- Mercier FE, Ragu C, Scadden DT 2011. The bone marrow at the crossroads of blood and immunity. Nat Rev Immunol 12: 49–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metcalf D 1998. Lineage commitment and maturation in hematopoietic cells: The case for extrinsic regulation. Blood 92: 345–347; discussion 352 [PubMed] [Google Scholar]
- Metcalf D 2010. The colony-stimulating factors and cancer. Nat Rev Cancer 10: 425–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkola I, Heavey B, Horcher M, Busslinger M 2002. Reversion of B cell commitment upon loss of Pax5 expression. Science 297: 110–113 [DOI] [PubMed] [Google Scholar]
- Mikkola HK, Klintman J, Yang H, Hock H, Schlaeger TM, Fujiwara Y, Orkin SH 2003. Haematopoietic stem cells retain long-term repopulating activity and multipotency in the absence of stem-cell leukaemia SCL/tal-1 gene. Nature 421: 547–551 [DOI] [PubMed] [Google Scholar]
- Miyamoto T, Iwasaki H, Reizis B, Ye M, Graf T, Weissman IL, Akashi K 2002. Myeloid or lymphoid promiscuity as a critical step in hematopoietic lineage commitment. Dev Cell 3: 137–147 [DOI] [PubMed] [Google Scholar]
- Monteiro R, Pouget C, Patient R 2011. The gata1/pu.1 lineage fate paradigm varies between blood populations and is modulated by tif1γ. EMBO J 30: 1093–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita Y, Ema H, Nakauchi H 2010. Heterogeneity and hierarchy within the most primitive hematopoietic stem cell compartment. J Exp Med 207: 1173–1182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison SJ, Weissman IL 1994. The long-term repopulating subset of hematopoietic stem cells is deterministic and isolatable by phenotype. Immunity 1: 661–673 [DOI] [PubMed] [Google Scholar]
- Muller AM, Medvinsky A, Strouboulis J, Grosveld F, Dzierzak E 1994. Development of hematopoietic stem cell activity in the mouse embryo. Immunity 1: 291–301 [DOI] [PubMed] [Google Scholar]
- Muller-Sieburg CE, Cho RH, Thoman M, Adkins B, Sieburg HB 2002. Deterministic regulation of hematopoietic stem cell self-renewal and differentiation. Blood 100: 1302–1309 [PubMed] [Google Scholar]
- Muller-Sieburg CE, Cho RH, Karlsson L, Huang JF, Sieburg HB 2004. Myeloid-biased hematopoietic stem cells have extensive self-renewal capacity but generate diminished lymphoid progeny with impaired IL-7 responsiveness. Blood 103: 4111–4118 [DOI] [PubMed] [Google Scholar]
- Nishimura S, Takahashi S, Kuroha T, Suwabe N, Nagasawa T, Trainor C, Yamamoto M 2000. A GATA box in the GATA-1 gene hematopoietic enhancer is a critical element in the network of GATA factors and sites that regulate this gene. Mol Cell Biol 20: 713–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notta F, Doulatov S, Laurenti E, Poeppl A, Jurisica I, Dick JE 2011. Isolation of single human hematopoietic stem cells capable of long-term multilineage engraftment. Science 333: 218–221 [DOI] [PubMed] [Google Scholar]
- Nutt SL, Heavey B, Rolink AG, Busslinger M 1999. Commitment to the B-lymphoid lineage depends on the transcription factor Pax5. Nature 401: 556–562 [DOI] [PubMed] [Google Scholar]
- O’Connell RM, Chaudhuri AA, Rao DS, Gibson WS, Balazs AB, Baltimore D 2010. MicroRNAs enriched in hematopoietic stem cells differentially regulate long-term hematopoietic output. Proc Natl Acad Sci 107: 14235–14240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada S, Nakauchi H, Nagayoshi K, Nishikawa S, Miura Y, Suda T 1992. In vivo and in vitro stem cell function of c-kit– and Sca-1–positive murine hematopoietic cells. Blood 80: 3044–3050 [PubMed] [Google Scholar]
- Okuno Y, Huang G, Rosenbauer F, Evans EK, Radomska HS, Iwasaki H, Akashi K, Moreau-Gachelin F, Li Y, Zhang P, et al. 2005. Potential autoregulation of transcription factor PU.1 by an upstream regulatory element. Mol Cell Biol 25: 2832–2845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooi AG, Sahoo D, Adorno M, Wang Y, Weissman IL, Park CY 2010. MicroRNA-125b expands hematopoietic stem cells and enriches for the lymphoid-balanced and lymphoid-biased subsets. Proc Natl Acad Sci 107: 21505–21510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ordentlich P, Lin A, Shen CP, Blaumueller C, Matsuno K, Artavanis-Tsakonas S, Kadesch T 1998. Notch inhibition of E47 supports the existence of a novel signaling pathway. Mol Cell Biol 18: 2230–2239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osawa M, Hanada K, Hamada H, Nakauchi H 1996. Long-term lymphohematopoietic reconstitution by a single CD34-low/negative hematopoietic stem cell. Science 273: 242–245 [DOI] [PubMed] [Google Scholar]
- Ottersbach K, Dzierzak E 2005. The murine placenta contains hematopoietic stem cells within the vascular labyrinth region. Dev Cell 8: 377–387 [DOI] [PubMed] [Google Scholar]
- Palis J, Robertson S, Kennedy M, Wall C, Keller G 1999. Development of erythroid and myeloid progenitors in the yolk sac and embryo proper of the mouse. Development 126: 5073–5084 [DOI] [PubMed] [Google Scholar]
- Park IK, Qian D, Kiel M, Becker MW, Pihalja M, Weissman IL, Morrison SJ, Clarke MF 2003. Bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells. Nature 423: 302–305 [DOI] [PubMed] [Google Scholar]
- Pongubala JM, Northrup DL, Lancki DW, Medina KL, Treiber T, Bertolino E, Thomas M, Grosschedl R, Allman D, Singh H 2008. Transcription factor EBF restricts alternative lineage options and promotes B cell fate commitment independently of Pax5. Nat Immunol 9: 203–215 [DOI] [PubMed] [Google Scholar]
- Pop R, Shearstone JR, Shen Q, Liu Y, Hallstrom K, Koulnis M, Gribnau J, Socolovsky M 2010. A key commitment step in erythropoiesis is synchronized with the cell cycle clock through mutual inhibition between PU.1 and S-phase progression. PLoS Biol 8: e1000484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pridans C, Holmes ML, Polli M, Wettenhall JM, Dakic A, Corcoran LM, Smyth GK, Nutt SL 2008. Identification of Pax5 target genes in early B cell differentiation. J Immunol 180: 1719–1728 [DOI] [PubMed] [Google Scholar]
- Pronk CJ, Rossi DJ, Mansson R, Attema JL, Norddahl GL, Chan CK, Sigvardsson M, Weissman IL, Bryder D 2007. Elucidation of the phenotypic, functional, and molecular topography of a myeloerythroid progenitor cell hierarchy. Cell Stem Cell 1: 428–442 [DOI] [PubMed] [Google Scholar]
- Radtke F, Wilson A, Stark G, Bauer M, van Meerwijk J, MacDonald HR, Aguet M 1999. Deficient T cell fate specification in mice with an induced inactivation of Notch1. Immunity 10: 547–558 [DOI] [PubMed] [Google Scholar]
- Rieger MA, Schroeder T 2009. Analyzing cell fate control by cytokines through continuous single cell biochemistry. J Cell Biochem 108: 343–352 [DOI] [PubMed] [Google Scholar]
- Rieger MA, Hoppe PS, Smejkal BM, Eitelhuber AC, Schroeder T 2009a. Hematopoietic cytokines can instruct lineage choice. Science 325: 217–218 [DOI] [PubMed] [Google Scholar]
- Rieger MA, Smejkal BM, Schroeder T 2009b. Improved prospective identification of megakaryocyte-erythrocyte progenitor cells. Br J Haematol 144: 448–451 [DOI] [PubMed] [Google Scholar]
- Rolink AG, Nutt SL, Melchers F, Busslinger M 1999. Long-term in vivo reconstitution of T-cell development by Pax5-deficient B-cell progenitors. Nature 401: 603–606 [DOI] [PubMed] [Google Scholar]
- Rongvaux A, Willinger T, Takizawa H, Rathinam C, Auerbach W, Murphy AJ, Valenzuela DM, Yancopoulos GD, Eynon EE, Stevens S, et al. 2011. Human thrombopoietin knockin mice efficiently support human hematopoiesis in vivo. Proc Natl Acad Sci 108: 2378–2383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbauer F, Owens BM, Yu L, Tumang JR, Steidl U, Kutok JL, Clayton LK, Wagner K, Scheller M, Iwasaki H, et al. 2006. Lymphoid cell growth and transformation are suppressed by a key regulatory element of the gene encoding PU.1. Nat Genet 38: 27–37 [DOI] [PubMed] [Google Scholar]
- Rothenberg EV 2007. Negotiation of the T lineage fate decision by transcription-factor interplay and microenvironmental signals. Immunity 26: 690–702 [DOI] [PubMed] [Google Scholar]
- Schebesta A, McManus S, Salvagiotto G, Delogu A, Busslinger GA, Busslinger M 2007. Transcription factor Pax5 activates the chromatin of key genes involved in B cell signaling, adhesion, migration, and immune function. Immunity 27: 49–63 [DOI] [PubMed] [Google Scholar]
- Schlenner SM, Madan V, Busch K, Tietz A, Laufle C, Costa C, Blum C, Fehling HJ, Rodewald HR 2010. Fate mapping reveals separate origins of T cells and myeloid lineages in the thymus. Immunity 32: 426–436 [DOI] [PubMed] [Google Scholar]
- Schmid MA, Kingston D, Boddupalli S, Manz MG 2010. Instructive cytokine signals in dendritic cell lineage commitment. Immunol Rev 234: 32–44 [DOI] [PubMed] [Google Scholar]
- Schroeder T 2008. Imaging stem cell driven mammalian regeneration. Nature 453: 345–351 [DOI] [PubMed] [Google Scholar]
- Schroeder T 2011. Long-term single-cell imaging of mammalian stem cells. Nat Methods 8: S30–S35 [DOI] [PubMed] [Google Scholar]
- Seet CS, Brumbaugh RL, Kee BL 2004. Early B cell factor promotes B lymphopoiesis with reduced interleukin 7 responsiveness in the absence of E2A. J Exp Med 199: 1689–1700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seita J, Ema H, Ooehara J, Yamazaki S, Tadokoro Y, Yamasaki A, Eto K, Takaki S, Takatsu K, Nakauchi H 2007. Lnk negatively regulates self-renewal of hematopoietic stem cells by modifying thrombopoietin-mediated signal transduction. Proc Natl Acad Sci 104: 2349–2354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semerad CL, Mercer EM, Inlay MA, Weissman IL, Murre C 2009. E2A proteins maintain the hematopoietic stem cell pool and promote the maturation of myelolymphoid and myeloerythroid progenitors. Proc Natl Acad Sci 106: 1930–1935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieburg HB, Cho RH, Dykstra B, Uchida N, Eaves CJ, Muller-Sieburg CE 2006. The hematopoietic stem compartment consists of a limited number of discrete stem cell subsets. Blood 107: 2311–2316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siminovitch L, McCulloch EA, Till JE 1963. The distribution of colony-forming cells among spleen colonies. J Cell Physiol 62: 327–336 [DOI] [PubMed] [Google Scholar]
- Smith EM, Akerblad P, Kadesch T, Axelson H, Sigvardsson M 2005. Inhibition of EBF function by active Notch signaling reveals a novel regulatory pathway in early B-cell development. Blood 106: 1995–2001 [DOI] [PubMed] [Google Scholar]
- Spangrude GJ, Heimfeld S, Weissman IL 1988. Purification and characterization of mouse hematopoietic stem cells. Science 241: 58–62 [DOI] [PubMed] [Google Scholar]
- Stopka T, Amanatullah DF, Papetti M, Skoultchi AI 2005. PU.1 inhibits the erythroid program by binding to GATA-1 on DNA and creating a repressive chromatin structure. EMBO J 24: 3712–3723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suda T, Suda J, Ogawa M 1984. Disparate differentiation in mouse hemopoietic colonies derived from paired progenitors. Proc Natl Acad Sci 81: 2520–2524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szilvassy SJ, Humphries RK, Lansdorp PM, Eaves AC, Eaves CJ 1990. Quantitative assay for totipotent reconstituting hematopoietic stem cells by a competitive repopulation strategy. Proc Natl Acad Sci 87: 8736–8740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadokoro Y, Ema H, Okano M, Li E, Nakauchi H 2007. De novo DNA methyltransferase is essential for self-renewal, but not for differentiation, in hematopoietic stem cells. J Exp Med 204: 715–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagoh H, Schebesta A, Lefevre P, Wilson N, Hume D, Busslinger M, Bonifer C 2004. Epigenetic silencing of the c-fms locus during B-lymphopoiesis occurs in discrete steps and is reversible. EMBO J 23: 4275–4285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Till JE, McCulloch EA 1961. A direct measurement of the radiation sensitivity of normal mouse bone marrow cells. Radiat Res 14: 213–222 [PubMed] [Google Scholar]
- Uchida N, Weissman IL 1992. Searching for hematopoietic stem cells: Evidence that Thy-1.1lo Lin– Sca-1+ cells are the only stem cells in C57BL/Ka-Thy-1.1 bone marrow. J Exp Med 175: 175–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida N, Dykstra B, Lyons KJ, Leung FY, Eaves CJ 2003. Different in vivo repopulating activities of purified hematopoietic stem cells before and after being stimulated to divide in vitro with the same kinetics. Exp Hematol 31: 1338–1347 [DOI] [PubMed] [Google Scholar]
- Wang Z, Li G, Tse W, Bunting KD 2009. Conditional deletion of STAT5 in adult mouse hematopoietic stem cells causes loss of quiescence and permits efficient nonablative stem cell replacement. Blood 113: 4856–4865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson A, Laurenti E, Oser G, van der Wath RC, Blanco-Bose W, Jaworski M, Offner S, Dunant CF, Eshkind L, Bockamp E, et al. 2008. Hematopoietic stem cells reversibly switch from dormancy to self-renewal during homeostasis and repair. Cell 135: 1118–1129 [DOI] [PubMed] [Google Scholar]
- Xie H, Ye M, Feng R, Graf T 2004. Stepwise reprogramming of B cells into macrophages. Cell 117: 663–676 [DOI] [PubMed] [Google Scholar]
- Xie Y, Yin T, Wiegraebe W, He XC, Miller D, Stark D, Perko K, Alexander R, Schwartz J, Grindley JC, et al. 2009. Detection of functional haematopoietic stem cell niche using real-time imaging. Nature 457: 97–101 [DOI] [PubMed] [Google Scholar]
- Yilmaz OH, Valdez R, Theisen BK, Guo W, Ferguson DO, Wu H, Morrison SJ 2006. Pten dependence distinguishes haematopoietic stem cells from leukaemia-initiating cells. Nature 441: 475–482 [DOI] [PubMed] [Google Scholar]
- Yuan Y, Shen H, Franklin DS, Scadden DT, Cheng T 2004. In vivo self-renewing divisions of haematopoietic stem cells are increased in the absence of the early G1-phase inhibitor, p18INK4C. Nat Cell Biol 6: 436–442 [DOI] [PubMed] [Google Scholar]
- Zeng H, Yucel R, Kosan C, Klein-Hitpass L, Moroy T 2004. Transcription factor Gfi1 regulates self-renewal and engraftment of hematopoietic stem cells. EMBO J 23: 4116–4125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Behre G, Pan J, Iwama A, Wara-Aswapati N, Radomska HS, Auron PE, Tenen DG, Sun Z 1999. Negative cross-talk between hematopoietic regulators: GATA proteins repress PU.1. Proc Natl Acad Sci 96: 8705–8710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Zhang X, Iwama A, Yu C, Smith KA, Mueller BU, Narravula S, Torbett BE, Orkin SH, Tenen DG 2000. PU.1 inhibits GATA-1 function and erythroid differentiation by blocking GATA-1 DNA binding. Blood 96: 2641–2648 [PubMed] [Google Scholar]
- Zhang J, Niu C, Ye L, Huang H, He X, Tong WG, Ross J, Haug J, Johnson T, Feng JQ, et al. 2003. Identification of the haematopoietic stem cell niche and control of the niche size. Nature 425: 836–841 [DOI] [PubMed] [Google Scholar]
- Zhang CC, Kaba M, Iizuka S, Huynh H, Lodish HF 2008. Angiopoietin-like 5 and IGFBP2 stimulate ex vivo expansion of human cord blood hematopoietic stem cells as assayed by NOD/SCID transplantation. Blood 111: 3415–3423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou P, Yoshihara H, Hosokawa K, Tai I, Shinmyozu K, Tsukahara F, Maru Y, Nakayama K, Nakayama KI, Suda T 2011. p57Kip2 and p27Kip1 cooperate to maintain hematopoietic stem cell quiescence through interactions with Hsc70. Cell Stem Cell 9: 247–261 [DOI] [PubMed] [Google Scholar]