In human mitochondria, 1-methyladenosine (m1A) occurs at position 58 of tRNALeu(UUR). In addition, partial m1A58 modifications have been found in human mitochondrial tRNALys and tRNASer(UCN). The authors identified human Trmt61B, which encodes a mitochondria-specific tRNA methyltransferase responsible for m1A58 in these three tRNAs. Trmt61B is dominantly localized to the mitochondria. m1A58 formation in human mitochondrial tRNALeu(UUR) could be reconstituted in vitro using recombinant Trmt61B in the presence of Ado-Met as a methyl donor. Unlike the cytoplasmic tRNA m1A58 methyltransferase that consists of an α2β2 heterotetramer formed by Trmt61A and Trmt6, Trmt61B formed a homo-oligomer (presumably a homotetramer) that resembled the bacterial homotetrameric m1A58 methyltransferase.
Keywords: Trmt61B, 1-methyladenosine, tRNA, RNA modification, mitochondria
Abstract
In human mitochondria, 1-methyladenosine (m1A) occurs at position 58 of tRNALeu(UUR). In addition, partial m1A58 modifications have been found in human mitochondrial tRNALys and tRNASer(UCN). We identified human Trmt61B, which encodes a mitochondria-specific tRNA methyltransferase responsible for m1A58 in these three tRNAs. Trmt61B is dominantly localized to the mitochondria. m1A58 formation in human mitochondrial tRNALeu(UUR) could be reconstituted in vitro using recombinant Trmt61B in the presence of Ado-Met as a methyl donor. Unlike the cytoplasmic tRNA m1A58 methyltransferase that consists of an α2β2 heterotetramer formed by Trmt61A and Trmt6, Trmt61B formed a homo-oligomer (presumably a homotetramer) that resembled the bacterial homotetrameric m1A58 methyltransferase. The bacterial origin of Trmt61B is supported by the results of the phylogenetic analysis.
INTRODUCTION
RNA modifications play pivotal roles in the biogenesis, structural integrity, function, and metabolism of RNA molecules. To date, more than 100 site-specific RNA modifications have been identified (Czerwoniec et al. 2009), the majority of which occur in tRNAs (Juhling et al. 2009).
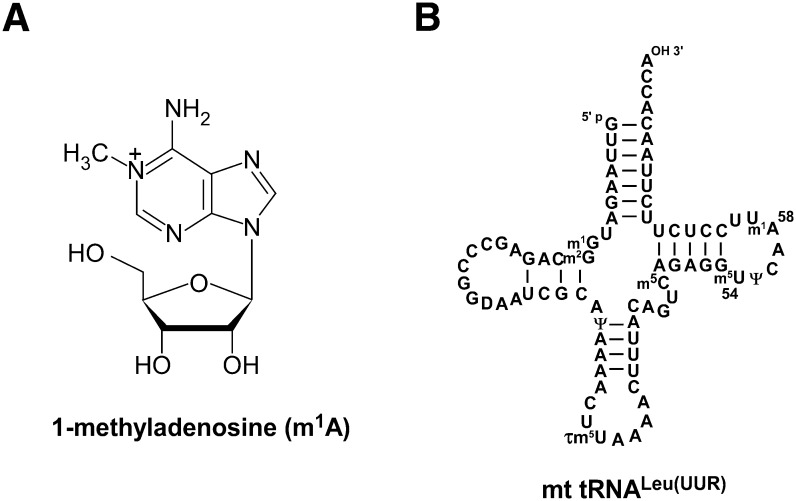
1-Methyladenosine (m1A) (Fig. 1A) has been observed at positions 9, 14, 22, and 58 of tRNAs and is thought to be a primordial RNA modification based on its widespread occurrence in tRNAs in each of the three major phylogenetic domains, as well as in organellar tRNAs (Juhling et al. 2009; Suzuki et al. 2011). m1A at position 58 forms a reverse-Hoogsteen base-pairing with 5-methyluridine (m5U) at position 54 in the T-loop of tRNAs (Robertus et al. 1974). The methylation at the N1 position of A58 confers a positive charge to the elbow region of the tRNA (Agris 1996). This positive charge is believed to stabilize the tertiary structure of tRNAs, especially that of eukaryotic initiator tRNAMet (Basavappa and Sigler 1991). m1A at position 9 in mammalian mitochondrial tRNAs is known to stabilize the canonical cloverleaf structure; when A9 is not methylated, the tRNA folds into a nonfunctional structure (Helm et al. 1998, 1999). In nematode mitochondrial tRNAs lacking the T-arm, m1A9 is an indispensable modification for their aminoacylation and EF-Tu binding (Sakurai et al. 2005). In addition to the role of m1A in tRNA structure and function, m1A is critical for retrovirus replication. All retroviruses use m1A58-containing tRNAs as primers to initiate reverse transcription of the minus strand. During replication of the human immunodeficiency virus-1 (HIV-1), the presence of m1A58 in human tRNALys is required for the accurate termination of plus-strand strong-stop DNA synthesis (Gilboa et al. 1979; Burnett and McHenry 1997).
FIGURE 1.
The chemical structure of 1-methyladenosine and human mitochondrial tRNALeu(UUR). (A) Chemical structure of 1-methyladenosine (m1A). (B) Secondary structure of human mitochondrial tRNALeu(UUR) with modified nucleosides: 1-methylguanosine (m1G), 2-methylguanosine (m2G), dihydrouridine (D), pseudouridine (Ψ), 5-taurinomethyluridine (τm5U), 5-methylcytidine (m5C), and 5-methyluridine (m5U). Nucleoside positions are numbered following conventional guidelines (Steinberg et al. 1993).
In Saccharomyces cerevisiae, the tRNA methyltransferase responsible for m1A58 formation was identified as a two-subunit enzyme encoded by TRM61(GCD14) and TRM6(GCD10) (Anderson et al. 1998, 2000; Ozanick et al. 2007), whose gene products form an α2β2 heterotetramer complex. Trm61p functions as the catalytic subunit for m1A58 formation and is a member of the Rossmann-fold-type tRNA methyltransferase family, which uses S-adenosylmethionine (Ado-Met) as a methyl donor (Anderson et al. 2000; Kozbial and Mushegian 2005). Both Trm61p and Trm6p are required for tRNA binding (Ozanick et al. 2007). Sequence analysis suggests that TRM61 and TRM6 share a common ancestor and arose through gene duplication and divergent evolution (Bujnicki 2001). Both TRM61 and TRM6 have been shown to be essential in yeast. When either gene is inactivated, the initiator tRNAMet is 3′-polyadenylated by the Trf4p/Mtr4p complex and subjected to exosome-mediated degradation (Anderson et al. 1998; Kadaba et al. 2004). Thus, m1A58 is required for the cellular stabilization of initiator tRNAMet in yeast.
The bacterial and archaeal tRNA m1A58 methyltransferase, TrmI, was identified as a homolog of yeast Trm61p (Droogmans et al. 2003; Roovers et al. 2004). Whereas the eukaryotic enzyme forms a heterotetramer, TrmI is a homotetrameric enzyme (Gupta et al. 2001; Roovers et al. 2004; Varshney et al. 2004). In Thermus thermophilus, inactivation of TrmI induces a thermosensitive phenotype (Droogmans et al. 2003). Since TrmI disruption strongly reduced 2-thiolation of 5-methyl-2-thiouridine (s2T) at position 54 (Shigi et al. 2006b), the decreased thermo-tolerance observed in the ΔTrmI strain can be partly explained by the lack of 2-thiolation of s2T54, a key modification required for tRNA thermostability (Yokoyama et al. 1987; Shigi et al. 2006a).
In humans, the majority of cytoplasmic tRNAs bear the m1A58 modification (Juhling et al. 2009). Trmt61A (hTrm61p) and Trmt6 (hTrm6p), the human homologs of yeast Trm61p and Trm6p, respectively, are responsible for m1A58 modification of cytoplasmic tRNAs (Ozanick et al. 2005). In human cells, siRNA-mediated knockdown of either subunit results in a slow-growth phenotype, suggesting that Trmt61A and Trmt6 play a role in cell proliferation (Saikia et al. 2010).
In mammalian mitochondria, all 22 tRNA species are encoded in mitochondrial DNA (Anderson et al. 1981). Among them, three mitochondrial tRNAs have been reported to bear m1A58: human tRNALeu(UUR) (Fig. 1B), bovine tRNASer(UCN), and bovine tRNAGlu (Yokogawa et al. 1991; Yasukawa et al. 2000; Nagao et al. 2009). To date, the functional role and biogenesis of m1A58 in mammalian mitochondrial tRNAs remain to be elucidated (Suzuki et al. 2011). Here, we report on Trmt61B as a mitochondria-specific methyltransferase responsible for m1A58 formation in human mitochondrial tRNAs.
RESULTS AND DISCUSSION
Subcellular localization of Trmt61B in human mitochondria
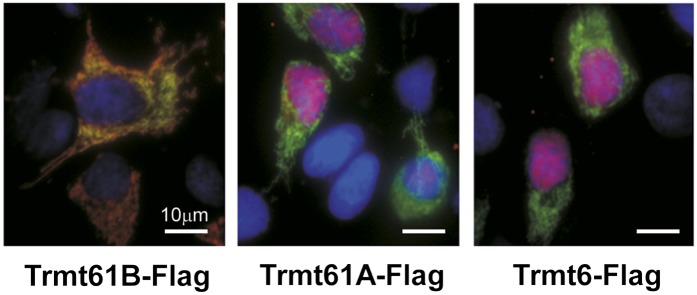
Human Trmt61B is a paralog of Trmt61A with unknown function. To determine whether Trmt61B acts as a methyltransferase for m1A58 in mitochondrial tRNAs, its subcellular localization was examined. Based on an analysis using the WoLF PSORT and Mitoprot programs (Claros and Vincens 1996; Horton et al. 2007), vertebrate Trmt61B proteins were predicted to localize to the mitochondria. Human Trmt61B with a C-terminal Flag tag (Trmt61B-Flag) was transiently expressed in HeLa cells. Immunofluorescence microscopy revealed that Trmt61B-Flag colocalized with GFP-fused MRPL44, a mitochondrial ribosomal protein (Fig. 2; Koc et al. 2001). In addition, we confirmed the mitochondrial localization of Trmt61B, which is fused with C-terminal GFP (Supplemental Fig. 1). For reference, the subcellular localization of transiently expressed Trmt61A-Flag and Trmt6-Flag was also observed in HeLa cells. Both Trmt61A-Flag and Trmt6-Flag localized to the nuclei and not to the mitochondria, in accordance with the nuclear localization of yeast Trm61p and Trm6p (Anderson et al. 1998). These results suggested that Trmt61B might function as a mitochondrial tRNA m1A58 methyltransferase.
FIGURE 2.
Subcellular localization of human Trmt61B and its paralogs. HeLa cells were cotransfected with plasmids expressing MRPL44-GFP (green) and C-terminal Flag-tagged Trmt61B, Trmt61A, or Trmt6. Microscopic images were obtained after immunostaining of Flag-tagged proteins (red) and DAPI staining (blue). (Yellow–orange) An overlap of Flag-tagged proteins and MRPL44-GFP. (Pink) An overlap of Flag-tagged proteins and DAPI-stained nucleus.
Trmt61B is required for m1A58 formation in human mitochondrial tRNAs
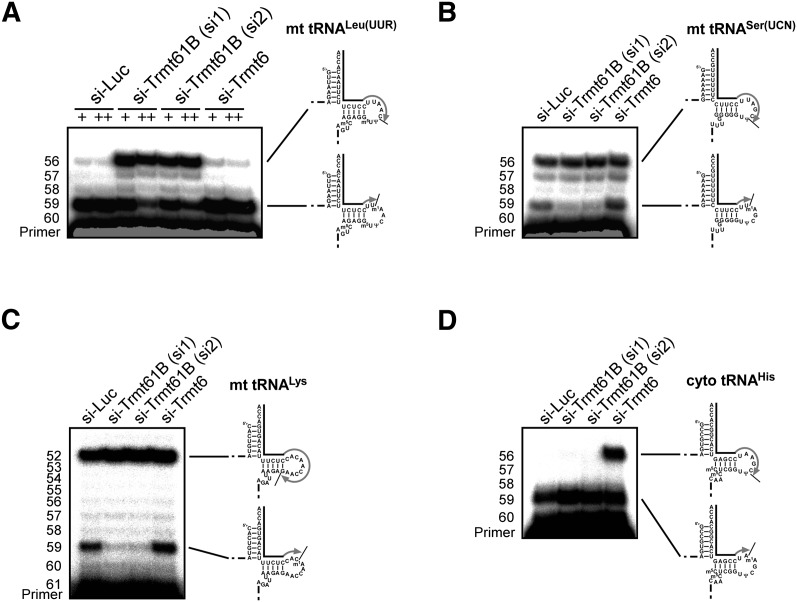
Next, the capacity of Trmt61B to catalyze m1A58 formation in human mitochondrial tRNAs was assessed. Since N1-methylation of m1A inhibits Watson-Crick base-pairing with T, m1A can be detected by primer extension (Burnett and McHenry 1997). To detect m1A58 of human mitochondrial tRNALeu(UUR), a primer complementary to the 3′-terminal region was designed (positions 61–76). Following siRNA-mediated knockdown of Trmt61B, Trmt6, or luciferase mRNA in HeLa cells, total RNA was extracted and used as a template for primer extension. In the luciferase knockdown cells, used as a control, cDNA extension was strongly arrested just before m1A58 of tRNALeu(UUR) and produced a clear band corresponding to position 59 (Fig. 3A). When Trmt61B was knocked down, the cDNA partially extended past position 58 and was stopped at position 56 by inserting dideoxy guanosine (Fig. 3A). Repetitive knockdown of Trmt61B resulted in reduction of the cDNA arrest at position 59. Thus, unmodified adenosine at position 58 becomes evident upon knockdown of Trmt61B. Two different siRNAs (si1 and si2) were used to target Trmt61B mRNA to confirm that the changes in m1A58 level were not due to off-target effects. In contrast, knockdown of Trmt6 did not affect cDNA arrest just before m1A58 of tRNALeu(UUR) and produced no long cDNA. Instead, when cytoplasmic tRNAHis was analyzed to confirm the results of RNAi knockdown, the long cDNA that passed over A58 became evident upon Trmt6 knockdown (Fig. 3D). No involvement of Trmt6 in mitochondrial m1A58 formation was apparent. These results demonstrate that Trmt61B is responsible for m1A58 formation in tRNALeu(UUR).
FIGURE 3.
Detection of hypomodified m1A58 in mitochondrial tRNAs from siRNA-treated cells. After knockdown of luciferase (control), Trmt61B, or Trmt6 mRNAs, primer extension was used to detect methylated or nonmethylated A58 in mitochondrial tRNALeu(UUR) (A), mitochondrial tRNASer(UCN) (B), mitochondrial tRNALys (C), and cytoplasmic tRNAHis (D). Two different siRNAs (si1 and si2) targeting Trmt61B mRNA were used. In A, “+” indicates that the cells were transfected once with siRNA and harvested 4 d after transfection. “++” indicates that the cells were transfected with siRNA a total of three times (every 2 d) and harvested 6 d after the first transfection. In B–D, cells were transfected once with siRNA and harvested 4 d after transfection. The knockdown efficiencies of Trmt61B and Trmt6 mRNAs were quantified by qRT-PCR and normalized to GAPDH mRNA. The steady-state level of Trmt61B mRNA was decreased to 8.0% (si1, +), 8.3% (si1, ++), 18% (si2, +), and 12% (si2, ++), and Trmt6 mRNA was decreased to 17% (si-Trm6, +) and 17% (si-Trm6, ++) compared with si-Luc cells. The primers are shown as solid lines next to the tRNAs, and nascent cDNAs synthesized from the primers are depicted as gray lines.
In human mitochondria, tRNALeu(UUR) is the only tRNA bearing m1A58 reported so far (Suzuki et al. 2011). Because bovine mitochondrial tRNASer(UCN) also contains m1A58 (Yokogawa et al. 1991), human mitochondrial tRNASer(UCN) was examined by primer extension. As expected, an arrest band corresponding to position 59 was observed, as well as a long cDNA band at position 56 of tRNASer(UCN). When Trmt61B was knocked down, the arrest band disappeared, suggesting that tRNASer(UCN) contains partially modified m1A58 (Fig. 3B). Using the same approach, a partial m1A58 modification was also detected in human mitochondrial tRNALys (Fig. 3C). By quantifying the arrest band and long cDNA bands obtained from primer extension (Fig. 3A–C), the frequencies of m1A58 in mitochondrial tRNALeu(UUR), tRNASer(UCN), and tRNALys were estimated to be 97%, 48%, and 27%, respectively. Trmt61B is responsible for m1A58 formation in these three tRNAs.
Because mitochondrial tRNALeu(UUR) contains fully modified m1A58, we speculated that Trmt61B might enhance the processing, stability, or function of tRNALeu(UUR), which is required for translation of the mitochondrial respiratory machinery. To investigate the physiological role of Trmt61B, Trmt61B was knocked down in HeLa cells, and the steady-state levels of mitochondrial tRNALeu(UUR), its processing intermediate (RNA19), and the cellular ATP level were quantified (King et al. 1992). Although a slight increase in RNA19 (∼1.5-fold) was observed, neither a change in the steady-state level of tRNALeu(UUR) nor any measurable loss of ATP level was measured (data not shown). Further studies (e.g., using knockout mice, etc.) will be necessary to identify any physiological roles of m1A58 and Trmt61B.
Trmt61B is a homo-oligomeric tRNA m1A58 methyltransferase
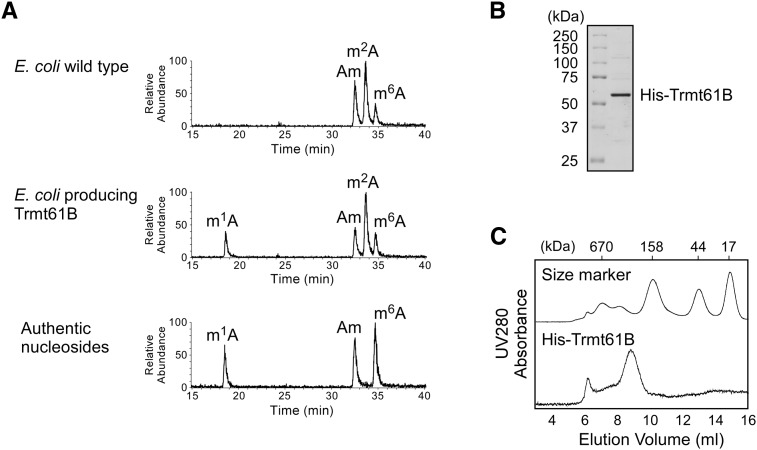
To confirm that Trmt61B is a methyltransferase for m1A58 of mitochondrial tRNAs, human Trmt61B lacking the predicted mitochondrial targeting sequence at the N terminus (amino acid residues 1–34) was recombinantly expressed in Escherichia coli (His-Trmt61B). Total RNAs were extracted, and the total tRNA nucleosides were analyzed by liquid chromatography/mass spectrometry (LC/MS). No m1A was detected in total tRNA from wild-type E. coli cells (Fig. 4A) because E. coli tRNAs do not contain m1A. In contrast, in E. coli overproducing His-Trmt61B, m1A was apparent (Fig. 4A), suggesting that Trmt61B is able to synthesize m1A in E. coli (Fig. 4A).
FIGURE 4.
Trmt61B is responsible for m1A formation in E. coli cells. (A) LC/MS analysis of total tRNA nucleosides from E. coli rosetta cells (top panel) and His-Trmt61B–expressing E. coli rosetta strain (middle panel). Mass chromatograms detecting MH+ (m/z 282) of m1A, Am, m2A, and m6A are shown. Synthetic nucleosides (m1A, Am, and m6A) were also analyzed as a control (bottom panel). (B) Purified His-Trmt61B was resolved by SDS-PAGE and stained with Coomassie brilliant blue. The protein size marker is indicated. (C) Elution profile (UV trace at 280 nm) of His-Trmt61B (bottom) with protein size marker (top) in gel filtration chromatography.
His-Trmt61B (52 kDa) was then purified using a Ni2+-chelating column (Fig. 4B) and analyzed by gel filtration chromatography (Fig. 4C). Based on the analysis of peak elution volumes, His-Trmt61B was observed to be ∼200–300 kDa in size, indicating that Trmt61B forms a homo-oligomeric complex. The crystal structure of Trmt61B (C-terminal 334 amino acids) was solved as a homotetramer (deposited as unknown protein PDB ID: 2B25), similar to that of bacterial TrmI (Gupta et al. 2001; Varshney et al. 2004). Taken together, these data indicate that the His-Trmt61B construct forms a TrmI-like homotetrameric structure.
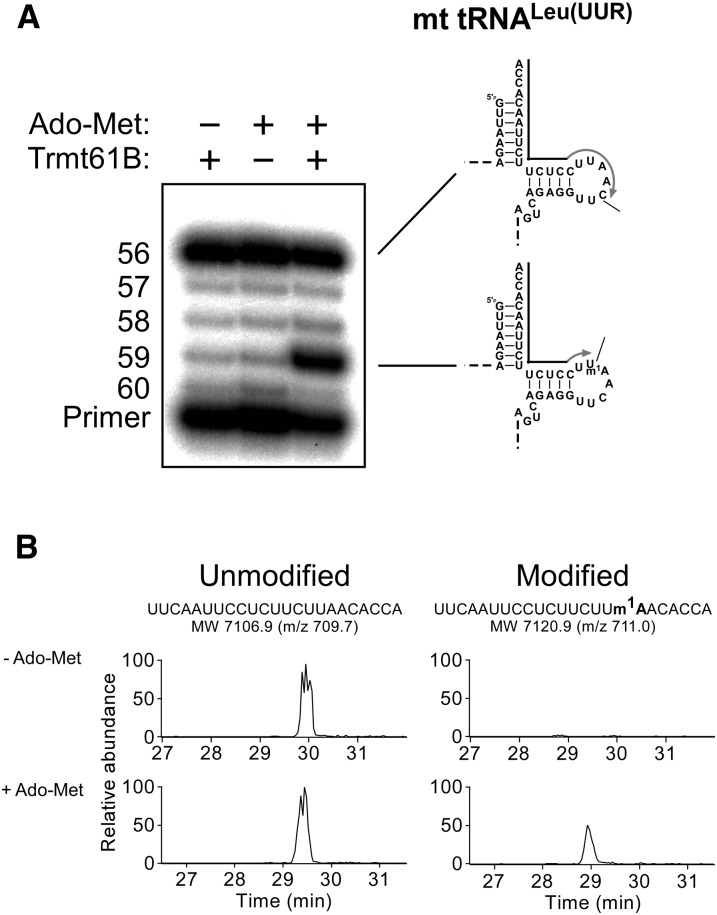
m1A58 formation was then reconstituted in vitro using the recombinant protein. Human mitochondrial tRNALeu(UUR) transcribed in vitro was used as a substrate for the reconstitution. The arrest band at position 59 generated by primer extension was detected only in the presence of both recombinant His-Trmt61B and Ado-Met (Fig. 5A). Next, the product tRNA was digested by RNase T1, and the digest was analyzed by LC/MS. In the absence of Ado-Met, the unmodified 23-mer fragment (positions 54–76) was detected as multiply charged negative ions (m/z 709.7) (Fig. 5B). In addition to the unmodified fragment, the 23-mer fragment containing a single methyl group (m/z 711.0) appeared only in the presence of Ado-Met (Fig. 5B). These results demonstrate that recombinant Trmt61B catalyzed the formation of m1A at position 58 of mitochondrial tRNALeu(UUR) in an Ado-Met-dependent manner. In addition, no additional partner proteins were necessary for Trmt61B to reconstitute m1A58 in vitro.
FIGURE 5.
In vitro reconstitution of m1A58 with recombinant Trmt61B. (A) m1A58 in the tRNALeu(UUR) transcript was detected by primer extension. (B) LC/MS analysis of RNase T1–digested fragments of the tRNALeu(UUR) transcript. Mass chromatograms detecting [M-10H]10− ions for the unmodified 23-mer fragment (positions 54–76, m/z 709.7, and MW 7106.9) and the 23-mer fragment containing a single methyl group (m/z 711.0 and MW 7120.9).
Trmt61B originates from bacterial TrmI
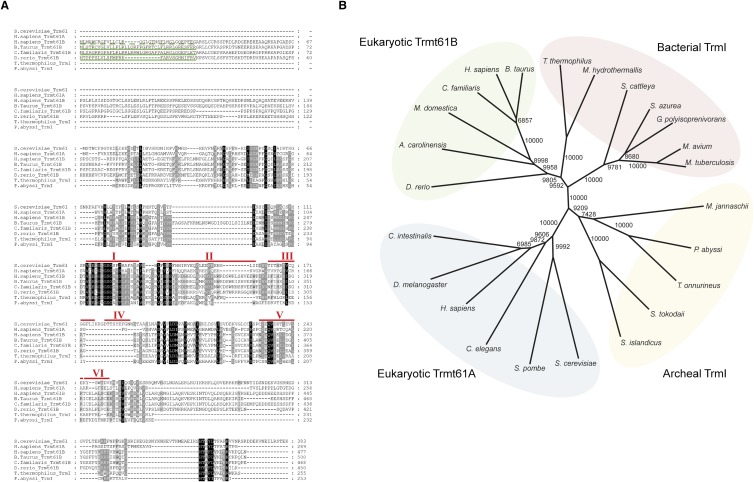
Sequence alignment of tRNA m1A58 methyltransferases from eukaryotes, bacteria, and archaea shows that they possess several conserved motifs (I–VI) specific to Rossmann-fold Ado-Met-dependent methyltransferases (Fig. 6A), including the GxGxG sequence (motif I) for Ado-Met binding, and the conserved carboxylate motif (motif II) for recognition of ribose hydroxyls. Trmt61B homologs were also predicted to contain N-terminal mitochondria-targeting sequences (Fig. 6A). According to the neighbor-joining phylogenetic tree, eukaryotic Trmt61B is closer to bacterial TrmI than to eukaryotic Trmt61A, whereas Trmt61A showed more similarity to the archaeal TrmI (Fig. 6B). The homotetrameric character of functional Trmt61B without additional partner proteins is consistent with the evolutionary assessment of its bacterial origin.
FIGURE 6.
Sequence alignment and phylogenetic analysis of Trmt61B and TrmI family proteins. (A) Sequence alignment of Trmt61B and its homologs from Eukarya, Bacteria, and Archaea. RefSeq or UniProt IDs of these proteins are listed in Supplemental Table S2. The mitochondria-targeting signal (MTS) was predicted using Mitoprot software (Claros and Vincens 1996) and is shown by green underlines. Human MTS is depicted as a dashed line, because the exact end of the MTS could not be predicted. Motifs I–VI of the Rossmann-fold-type methyltransferase in S. cerevisiae Trm61 (Kozbial and Mushegian 2005) are indicated. (B) A nonrooted neighbor-joining tree for Trmt61B and TrmI family proteins was constructed by Clustal_X software (Thompson et al. 1997). The bootstrap value was obtained by 10,000 trials and is indicated at each node. RefSeq or UniProt IDs are listed in Supplemental Table S2.
MATERIALS AND METHODS
Cell culture
HeLa cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum, at 37°C under a humidified atmosphere with 5% CO2.
Fluorescence microscopy
The cDNAs for Trmt61B, Trmt61A, and Trmt6 were amplified by reverse-transcription PCR of total RNA from HeLa cells using the primers listed in Supplemental Table S1 and cloned using the Directional TOPO Cloning Kit (Invitrogen). C-terminal Flag-tagged Trmt61B, Trmt61A, and Trmt6 expression vectors were generated by LR reaction of pDEST 12.2 Flag and directional TOPO clones using LR Clonase (Invitrogen). The C-terminal GFP-fused Trmt61B expression vector was generated by LR reaction of Trmt61B directional TOPO clone and pcDNA-Dest47 Gateway Vector (Invitrogen). The C-terminal GFP-fused MRPL44 expression vector was generated by LR reaction of Human Gateway Entry Clone FLJ12701AAAF (Goshima et al. 2008) and pcDNA-DEST47 Gateway Vector. HeLa cells were grown on uncoated, glass-bottom dishes (Matsunami). Cells were cotransfected with 0.5 μg of Trmt61B-Flag, Trmt61A-Flag, or Trmt6-Flag expression vector and 0.5 μg of MRPL44-GFP expression vector using FuGENE HD (Roche). Cells were cultivated for 48 h, washed with phosphate-buffered saline (PBS), and fixed with 3.7% formaldehyde in PBS for 30 min at room temperature. Cells were then washed three times with PBS, permeabilized with 0.5% Triton in PBS for 10 min at room temperature, and blocked with 2% fetal bovine serum in PBS for 30 min at room temperature. Cells were incubated for 30 min at room temperature with anti-DYKDDDDK antibody (1:500; Wako) diluted in Can Get Signal solution A (Toyobo), followed by 1 h of incubation at room temperature with Cy3-conjugated anti-mouse secondary antibody (1:1000; Millipore) in Can Get Signal solution A. Nuclei were stained with DAPI. For Trmt61B-GFP, 1 μg of expression vector was transfected into HeLa cells grown on an uncoated, glass-bottom dish (Matsunami). Cells were cultivated for 24 h, and a final concentration of 100 nM Mitotracker Red (Molecular Probes) was added to media for 10 min. Fluorescent images were acquired with a Leica DMI6000B microscope equipped with a DFC360FX cooled CCD camera. Images were processed using AF6500 software (Leica).
Quantitative reverse-transcription real-time PCR (qRT-PCR)
qRT-PCR was conducted as was previously described (Chujo et al. 2012). The sequences of the qPCR primers are listed in Supplemental Table S1.
RNAi
siRNAs targeted to Trmt61B, Trmt6, and luciferase mRNAs were designed using the siRNA design algorithm “siExplorer” (http://rna.chem.t.u-tokyo.ac.jp/cgi/siexplorer.htm) (Katoh and Suzuki 2007). The siRNAs used here are listed in Supplemental Table S1. RNAi experiments were conducted with 48 pmol of siRNA (4 nM final concentration), as previously described (Chujo et al. 2012).
Primer extension method to detect m1A58 modification
Primer extension was conducted essentially as described previously (Kirino et al. 2005). The 5′ 32P-labeled primer (0.1 pmol) was incubated with 1 μg of total RNA in a 10-μL solution containing 10 mM Tris-HCl (pH 8.0) and 1 mM EDTA for 2 min at 80°C, and then allowed to cool to 42°C. Alternatively, after in vitro reconstitution (Fig. 5), ∼0.05 pmol of tRNA transcript was mixed with 1 μg of total RNA from E. coli and was treated in the same manner as described above. Subsequently, 1.5 μL of ddH2O, 4 μL of 5× reverse transcription reaction buffer (Invitrogen), 0.5 μL of 1.5 mM d/ddNTP mix, 3 μL of 25 mM MgCl2, and 1 μL of Moloney murine leukemia virus reverse transcriptase (Invitrogen) were added, and the mixture was incubated for 1 h at 42°C. The d/ddNTP mix consisted of dATP, dTTP, and ddGTP for mitochondrial tRNALeu(UUR); dATP, dTTP, dCTP, and ddGTP for mitochondrial tRNASer(UCN); dTTP, dGTP, and ddCTP for mitochondrial tRNALys; and dATP, dTTP, dCTP, and ddGTP for cytoplasmic tRNAHis. Following incubation, 0.1 μg of RNase A (Ambion) was added, and the mixture was incubated again for 10 min at 42°C. The reaction mixture was then subjected to 20% polyacrylamide gel electrophoresis (PAGE) containing 7 M urea. Radiolabeled bands were visualized using a FLA-7000 imager (Fujifilm). The DNA primers are listed in Supplemental Table S1.
Expression and purification of the recombinant Trmt61B protein
The cDNA of Trmt61B lacking the predicted N-terminal mitochondria-targeting signal (amino acid residues 35–477) was amplified by RT-PCR of total RNA from HeLa cells using the primers listed in Supplemental Table S1, which contained HindIII and NheI sites. The PCR product was cloned into the corresponding sites of the pET-28a(+) vector (Novagen) to obtain the expression vector pET-Trmt61B, which produced N-terminal hexahistidine tag-fused Trmt61B (His-Trmt61B). The E. coli rosetta (DE3) strain was transformed with pET-Trmt61B and cultured in LB media containing 50 μg/mL kanamycin and 30 μg/mL chloramphenicol. When the bacteria reached an OD = 0.5, protein expression was induced by the addition of 100 μM IPTG, and the cells were grown for 3 h at 37°C. Cells were harvested and suspended in buffer A (50 mM HEPES-KOH at pH 7.6, 100 mM KCl, 10 mM MgCl2, and 7 mM β-mercaptoethanol), followed by sonication on ice. Cell lysates were cleared by ultracentrifugation at 100,000g for 60 min. The supernatant was loaded onto a nickel-charged HiTrap chelating column (GE Healthcare). After washing off unbound proteins with buffer A, recombinant proteins were eluted with a 50-mL linear gradient from zero to 500 mM imidazole in buffer A. Fractions containing the recombinant proteins were pooled and dialyzed overnight against buffer B (50 mM Tris-HCl at pH 8.0, 100 mM KCl, and 1 mM dithiothreitol). The concentration of purified His-Trmt61B was determined by the BCA protein assay (Thermo Scientific) using bovine serum albumin as a standard.
Preparation of the tRNA fraction from Trmt61B-expressing E. coli cells
The E. coli rosetta (DE3) strain was transformed with pET-Trmt61B and cultured in LB media containing 50 μg/mL kanamycin and 30 μg/mL chloramphenicol. At an OD = 0.5, protein expression was induced by the addition of 100 μM IPTG, and the cells were grown for 3 h at 37°C. Total RNA was extracted from the cells, and the tRNA fraction was purified by denaturing PAGE.
Gel filtration chromatography
Superdex200 (GE Healthcare) was equilibrated with buffer C (50 mM Tris HCl at pH 8.0, 200 mM KCl, and 7 mM 2-mercaptoethanol). Purified His-Trmt61B (25 μg) was injected into the column and eluted with buffer C.
In vitro reconstitution of m1A58 using recombinant Trmt61B
For in vitro reconstitution of m1A58, tRNA was prepared by in vitro transcription with T7 RNA polymerase. Template DNA for mitochondrial tRNALeu(UUR) was generated using the oligo DNAs listed in Supplemental Table S1. tRNA was transcribed by T7 RNA polymerase in vitro (Milligan et al. 1987) and purified by denaturing PAGE. Isolated tRNA was quantified by measuring the optical density at 260 nm. The in vitro reconstitution reaction mixture (50 μL) contained 50 mM NH4OAc, 3 mM MgCl2, 50 mM Tris-HCl (pH 8.0), 1 mM dithiothreitol, 1 mM Ado-Met, 1.6 μM tRNALeu(UUR) transcript, and 1 μM His-Trmt61B. The reaction mixture was incubated for 2 h at 37°C. The reaction was terminated by adding phenol:chloroform isoamylalcohol (Nacalai), and the tRNA was recovered by ethanol precipitation.
RNA mass spectrometry
Nucleoside analysis was performed as previously described (Suzuki et al. 2007). In brief, 10 μg of total tRNA was digested to nucleosides by nuclease P1 (Yamasa) and bacterial alkaline phosphatase A19 (Takara), and then analyzed using a LCQ DUO ion-trap (IT) mass spectrometer (ThermoFinnigan) equipped with an electrospray ionization (ESI) source and HP1100 liquid chromatography system (Agilent Technologies). Specific modified nucleosides were monitored on the mass chromatogram.
After in vitro reconstitution, ∼1 pmol of mitochondrial tRNALeu(UUR) transcript was digested by RNase T1 (Epicentre), and analyzed by capillary liquid chromatography nano-ESI/mass spectrometer (Suzuki et al. 2007; Katoh et al. 2009).
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
We thank Yuriko Sakaguchi for performing RNA mass spectrometry, and Dr. Takeo Suzuki, Dr. Kenjyo Miyauchi, and Dr. Yohei Kirino for their fruitful suggestions. We are also grateful to the other members of the Suzuki laboratory for their valuable advice. This work was supported by Grants-in-Aid for Scientific Research on Priority Areas from the Ministry of Education, Science, Sports, and Culture of Japan (to T.S.).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.035600.112.
REFERENCES
- Agris PF 1996. The importance of being modified: Roles of modified nucleosides and Mg2+ in RNA structure and function. Prog Nucleic Acid Res Mol Biol 53: 79–129 [DOI] [PubMed] [Google Scholar]
- Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, et al. 1981. Sequence and organization of the human mitochondrial genome. Nature 290: 457–465 [DOI] [PubMed] [Google Scholar]
- Anderson J, Phan L, Cuesta R, Carlson BA, Pak M, Asano K, Bjork GR, Tamame M, Hinnebusch AG 1998. The essential Gcd10p–Gcd14p nuclear complex is required for 1-methyladenosine modification and maturation of initiator methionyl-tRNA. Genes Dev 12: 3650–3662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson J, Phan L, Hinnebusch AG 2000. The Gcd10p/Gcd14p complex is the essential two-subunit tRNA(1-methyladenosine) methyltransferase of Saccharomyces cerevisiae. Proc Natl Acad Sci 97: 5173–5178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basavappa R, Sigler PB 1991. The 3 Å crystal structure of yeast initiator tRNA: Functional implications in initiator/elongator discrimination. EMBO J 10: 3105–3111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bujnicki JM 2001. In silico analysis of the tRNA:m1A58 methyltransferase family: Homology-based fold prediction and identification of new members from Eubacteria and Archaea. FEBS Lett 507: 123–127 [DOI] [PubMed] [Google Scholar]
- Burnett BP, McHenry CS 1997. Posttranscriptional modification of retroviral primers is required for late stages of DNA replication. Proc Natl Acad Sci 94: 7210–7215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chujo T, Ohira T, Sakaguchi Y, Goshima N, Nomura N, Nagao A, Suzuki T 2012. LRPPRC/SLIRP suppresses PNPase-mediated mRNA decay and promotes polyadenylation in human mitochondria. Nucleic Acids Res 40: 8033–8047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claros MG, Vincens P 1996. Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur J Biochem 241: 779–786 [DOI] [PubMed] [Google Scholar]
- Czerwoniec A, Dunin-Horkawicz S, Purta E, Kaminska KH, Kasprzak JM, Bujnicki JM, Grosjean H, Rother K 2009. MODOMICS: A database of RNA modification pathways. 2008 update. Nucleic Acids Res 37: D118–D121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Droogmans L, Roovers M, Bujnicki JM, Tricot C, Hartsch T, Stalon V, Grosjean H 2003. Cloning and characterization of tRNA (m1A58) methyltransferase (TrmI) from Thermus thermophilus HB27, a protein required for cell growth at extreme temperatures. Nucleic Acids Res 31: 2148–2156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilboa E, Mitra SW, Goff S, Baltimore D 1979. A detailed model of reverse transcription and tests of crucial aspects. Cell 18: 93–100 [DOI] [PubMed] [Google Scholar]
- Goshima N, Kawamura Y, Fukumoto A, Miura A, Honma R, Satoh R, Wakamatsu A, Yamamoto J, Kimura K, Nishikawa T, et al. 2008. Human protein factory for converting the transcriptome into an in vitro-expressed proteome. Nat Methods 5: 1011–1017 [DOI] [PubMed] [Google Scholar]
- Gupta A, Kumar PH, Dineshkumar TK, Varshney U, Subramanya HS 2001. Crystal structure of Rv2118c: An AdoMet-dependent methyltransferase from Mycobacterium tuberculosis H37Rv. J Mol Biol 312: 381–391 [DOI] [PubMed] [Google Scholar]
- Helm M, Brule H, Degoul F, Cepanec C, Leroux JP, Giege R, Florentz C 1998. The presence of modified nucleotides is required for cloverleaf folding of a human mitochondrial tRNA. Nucleic Acids Res 26: 1636–1643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helm M, Giege R, Florentz C 1999. A Watson-Crick base-pair-disrupting methyl group (m1A9) is sufficient for cloverleaf folding of human mitochondrial tRNALys. Biochemistry 38: 13338–13346 [DOI] [PubMed] [Google Scholar]
- Horton P, Park KJ, Obayashi T, Fujita N, Harada H, Adams-Collier CJ, Nakai K 2007. WoLF PSORT: Protein localization predictor. Nucleic Acids Res 35: W585–W587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juhling F, Morl M, Hartmann RK, Sprinzl M, Stadler PF, Putz J 2009. tRNAdb 2009: Compilation of tRNA sequences and tRNA genes. Nucleic Acids Res 37: D159–D162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadaba S, Krueger A, Trice T, Krecic AM, Hinnebusch AG, Anderson J 2004. Nuclear surveillance and degradation of hypomodified initiator tRNAMet in S. cerevisiae. Genes Dev 18: 1227–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh T, Suzuki T 2007. Specific residues at every third position of siRNA shape its efficient RNAi activity. Nucleic Acids Res 35: e27 doi: 10.1093/nar/gkl1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh T, Sakaguchi Y, Miyauchi K, Suzuki T, Kashiwabara S, Baba T, Suzuki T 2009. Selective stabilization of mammalian microRNAs by 3′ adenylation mediated by the cytoplasmic poly(A) polymerase GLD-2. Genes Dev 23: 433–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King MP, Koga Y, Davidson M, Schon EA 1992. Defects in mitochondrial protein synthesis and respiratory chain activity segregate with the tRNALeu(UUR) mutation associated with mitochondrial myopathy, encephalopathy, lactic acidosis, and strokelike episodes. Mol Cell Biol 12: 480–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirino Y, Goto Y, Campos Y, Arenas J, Suzuki T 2005. Specific correlation between the wobble modification deficiency in mutant tRNAs and the clinical features of a human mitochondrial disease. Proc Natl Acad Sci 102: 7127–7132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koc EC, Burkhart W, Blackburn K, Moyer MB, Schlatzer DM, Moseley A, Spremulli LL 2001. The large subunit of the mammalian mitochondrial ribosome. Analysis of the complement of ribosomal proteins present. J Biol Chem 276: 43958–43969 [DOI] [PubMed] [Google Scholar]
- Kozbial PZ, Mushegian AR 2005. Natural history of S-adenosylmethionine-binding proteins. BMC Struct Biol 5: 19 doi: 10.1186/1472-6807-5-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milligan JF, Groebe DR, Witherell GW, Uhlenbeck OC 1987. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res 15: 8783–8798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagao A, Suzuki T, Katoh T, Sakaguchi Y, Suzuki T 2009. Biogenesis of glutaminyl-mt tRNAGln in human mitochondria. Proc Natl Acad Sci 106: 16209–16214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozanick S, Krecic A, Andersland J, Anderson JT 2005. The bipartite structure of the tRNA m1A58 methyltransferase from S. cerevisiae is conserved in humans. RNA 11: 1281–1290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozanick SG, Bujnicki JM, Sem DS, Anderson JT 2007. Conserved amino acids in each subunit of the heteroligomeric tRNA m1A58 Mtase from Saccharomyces cerevisiae contribute to tRNA binding. Nucleic Acids Res 35: 6808–6819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertus JD, Ladner JE, Finch JT, Rhodes D, Brown RS, Clark BF, Klug A 1974. Structure of yeast phenylalanine tRNA at 3 Å resolution. Nature 250: 546–551 [DOI] [PubMed] [Google Scholar]
- Roovers M, Wouters J, Bujnicki JM, Tricot C, Stalon V, Grosjean H, Droogmans L 2004. A primordial RNA modification enzyme: The case of tRNA (m1A) methyltransferase. Nucleic Acids Res 32: 465–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saikia M, Fu Y, Pavon-Eternod M, He C, Pan T 2010. Genome-wide analysis of N1-methyl-adenosine modification in human tRNAs. RNA 16: 1317–1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai M, Ohtsuki T, Watanabe K 2005. Modification at position 9 with 1-methyladenosine is crucial for structure and function of nematode mitochondrial tRNAs lacking the entire T-arm. Nucleic Acids Res 33: 1653–1661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigi N, Sakaguchi Y, Suzuki T, Watanabe K 2006a. Identification of two tRNA thiolation genes required for cell growth at extremely high temperatures. J Biol Chem 281: 14296–14306 [DOI] [PubMed] [Google Scholar]
- Shigi N, Suzuki T, Terada T, Shirouzu M, Yokoyama S, Watanabe K 2006b. Temperature-dependent biosynthesis of 2-thioribothymidine of Thermus thermophilus tRNA. J Biol Chem 281: 2104–2113 [DOI] [PubMed] [Google Scholar]
- Steinberg S, Misch A, Sprinzl M 1993. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res 21: 3011–3015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Ikeuchi Y, Noma A, Suzuki T, Sakaguchi Y 2007. Mass spectrometric identification and characterization of RNA-modifying enzymes. Methods Enzymol 425: 211–229 [DOI] [PubMed] [Google Scholar]
- Suzuki T, Nagao A, Suzuki T 2011. Human mitochondrial tRNAs: Biogenesis, function, structural aspects, and diseases. Annu Rev Genet 45: 299–329 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG 1997. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25: 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshney U, Ramesh V, Madabushi A, Gaur R, Subramanya HS, RajBhandary UL 2004. Mycobacterium tuberculosis Rv2118c codes for a single-component homotetrameric m1A58 tRNA methyltransferase. Nucleic Acids Res 32: 1018–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasukawa T, Suzuki T, Suzuki T, Ueda T, Ohta S, Watanabe K 2000. Modification defect at anticodon wobble nucleotide of mitochondrial tRNAsLeu(UUR) with pathogenic mutations of mitochondrial myopathy, encephalopathy, lactic acidosis, and stroke-like episodes. J Biol Chem 275: 4251–4257 [DOI] [PubMed] [Google Scholar]
- Yokogawa T, Watanabe Y, Kumazawa Y, Ueda T, Hirao I, Miura K, Watanabe K 1991. A novel cloverleaf structure found in mammalian mitochondrial tRNASer(UCN). Nucleic Acids Res 19: 6101–6105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama S, Watanabe K, Miyazawa T 1987. Dynamic structures and functions of transfer ribonucleic acids from extreme thermophiles. Adv Biophys 23: 115–147 [DOI] [PubMed] [Google Scholar]