Abstract
Neurodegenerative diseases affect the lives of millions of patients and their families. Due to the complexity of these diseases and our limited understanding of their pathogenesis, the design of therapeutic agents that can effectively treat these diseases has been challenging. Huntington disease (HD) is one of several neurological disorders with few therapeutic options. HD, like numerous other neurodegenerative diseases, involves extensive neuronal cell loss. One potential strategy to combat HD and other neurodegenerative disorders is to intervene in the execution of neuronal cell death. Inhibiting neuronal cell death pathways may slow the development of neurodegeneration. However, discovering small molecule inhibitors of neuronal cell death remains a significant challenge. Here, we review candidate therapeutic targets controlling cell death mechanisms that have been the focus of research in HD, as well as an emerging strategy that has been applied to developing small molecule inhibitors—fragment-based drug discovery (FBDD). FBDD has been successfully used in both industry and academia to identify selective and potent small molecule inhibitors, with a focus on challenging proteins that are not amenable to traditional high-throughput screening approaches. FBDD has been used to generate potent leads, pre-clinical candidates, and has led to the development of an FDA approved drug. This approach can be valuable for identifying modulators of cell-death-regulating proteins; such compounds may prove to be the key to halting the progression of HD and other neurodegenerative disorders.
Keywords: Huntington disease, cell death, fragment-based drug discovery, neurodegenerative diseases1
1. Introduction
Neurodegenerative diseases encompass a large class of disorders that affect millions of individuals around the world. Broadly, such diseases can be defined as those that selectively and progressively induce neuronal death or dysfunction, especially in midlife. Currently, there are few effective therapies for such disorders. The number of patients affected by neurodegenerative diseases, such as Alzheimer’s disease (AD), Parkinson’s disease (PD), multiple sclerosis (MS), amyotrophic lateral sclerosis (ALS), and HD, is estimated to increase over time due, to the growing size of the elderly population (Hebert et al., 2001; Thrall, 2005). The cost of care in the US for AD patients alone surpasses $100 billion (Alzheimer’s Association, 2010). Delaying the onset or slowing the development of neurodegeneration would have a significant benefit: it would decrease the economic burden and improve the quality of life of affected individuals and their families (Alzheimer’s Association, 2010; Thrall, 2005).
Here, we focus on one neurological disorder, HD, and emerging drug discovery approaches involving fragment-based and computational drug design that can be applied to developing small molecule inhibitors of proteins that induce cell-death. We begin by reviewing what is known about HD (section 2) and then examine the pro-cell-death targets that may be of therapeutic benefit to HD (section 3). In the final section, we describe FBDD and how it may be used in drug discovery efforts for HD and other neurodegenerative diseases (section 4).
2. Huntington disease
There have been several extensive reviews published on HD (Imarisio et al., 2008; Ross and Tabrizi, 2011; Walker, 2007; Zuccato et al., 2010). Here, we provide only a brief overview of the disease.
2.1 Brief history
The first patients exhibiting the characteristic dance-like movements, chorea (χορεία), can be traced back to 1237 in Erfurt, Germany (Park and Park, 1990). Large epidemics were later documented in 1374 and 1518 across Europe (Waller, 2009). These cases were termed chorea by Paracelsus, 16th century German-Swiss physician, but today they are referred to as dancing mania (Jummani and Okun, 2001; Osler, 1894; Park and Park, 1990) and may have been due to mass hysteria during the Black Death pandemic in Europe (Krack, 1999). Subsequently, many acquired forms of chorea, such as a complication from rheumatic fever or from drug use, in addition to genetic causes, have been identified (Wild and Tabrizi, 2007); the most prevalent of the genetic chorea disorders is HD. The features and symptoms of this hereditary chorea were first described in detail by a New York physician, George Huntington, in an 1872 paper titled “On Chorea” (Huntington, 1872; Huntington, 2003), which described the disease as it was manifest in East Hampton families. Because of his description, this genetic chorea became known as Huntington’s disease. Since then, understanding of the pathology and molecular mechanism of HD has advanced dramatically, but few therapeutics have been developed.
2.2 Pathology and characteristics
HD is a fatal, neurodegenerative trinucleotide-repeat disorder. Pathologically, HD is characterized by expansion of a cytosine-adenine-guanine (CAG) repeat in the coding region of the huntingtin gene (HTT), located on chromosome 4p16.3 (The Huntington’s Disease Collaborative Research Group, 1993). The CAG repeats in HTT are translated into a polyglutamine (polyQ) sequence in the N-terminal region of the huntingtin (Htt) protein. HD is one of nine polyQ disorders, which include the spinocerebellar ataxias (SCA1, 2, 3, 6, 7, and 17), spinal bulbar muscular atrophy, and dentatorubral-pallidoluysian atrophy; HD is the most prevalent member of this group. (For a thorough characterization and epidemiology of spinocerebellar ataxias and dentatorubral-pallidoluysian diseases, see the review by Schols et al. (Schols et al., 2004)).
HD typically occurs in midlife, but extensive CAG expansion leads to a juvenile onset form of the disease. In unaffected individuals, there is an average of 19 CAG repeats in the huntingtin gene, but HD patients acquire 36 to 121 CAG repeats (Kremer et al., 1994). Patients with 36 to 39 repeats show reduced penetrance for the disease and can be asymptomatic for many years (Quarrell et al., 2007; Rubinsztein et al., 1996).
HD is inherited in an autosomal dominant manner, though “sporadic HD” has been shown to occur in patients with an asymptomatic father with an intermediate allele containing 30-35 CAG repeats (De Rooij et al., 1993; Goldberg et al., 1993; Hendricks et al., 2009). The length of the expanded CAG correlates with an earlier age at which the symptoms of the disease manifest and with a more severe form of HD. Thus, in juvenile cases of HD, patients have over 63 CAG repeats and the disease progresses more rapidly than in patients with fewer repeats (Telenius et al., 1993), leading to mortality within 11 years of onset (compared to 15-20 years typical for adult onset HD) (Foroud et al., 1999).
Long tracks of CAG repeats are prone to replication errors in meiosis (Kremer et al., 1995), leading to expansion or contraction of the CAG repeats. Expansions of more than seven repeats are passed down from the paternal line to the offspring 96% of the time (Kremer et al., 1995), due to greater CAG instability in spermatogenesis than in oogenesis (Zuhlke et al., 1993). Meanwhile, a reduction in the size of the CAG track is caused predominantly by maternal inheritance of the HTT gene (Kremer et al., 1995; Zuhlke et al., 1993).
Typically, HD is associated with loss of motor control, resulting in uninhibited movements of muscles in the face, body, legs, and arms. This is a consequence of gradual and continuous degeneration of neurons in the caudate and putamen. Especially affected are medium spiny neurons in the striatum that project to the globus pallidus externa, part of the indirect basal ganglia pathway (Mink, 1996). The death of these medium spiny neurons leads to a loss of inhibition of the thalamus, hyper activation of the direct basal ganglia circuit (Lobo, 2009), and an increase in undesired motor signals to the body and eyes. As HD progresses, there is an increasing rate of cell death in other areas of the brain, including the basal ganglia, thalamus, and cerebral cortex (Vonsattel and DiFiglia, 1998). As a result, HD patients experience emotional, cognitive, and motivational difficulties, in addition to motor complications, changes in behavior, mood swings, weight loss, irritability, anxiety, and depression (Foroud et al., 1999). Cognitively, there is a strain on short-term memory, error correction, long term planning, organization, and concentration (Walker, 2007). As the disease develops, HD patients develop difficulties in coordination, including speaking, swallowing and walking (Foroud et al., 1999). Mortality generally arises from complications, such as pneumonia and heart disease (Sorensen and Fenger, 1992; Zuccato et al., 2010).
2.3 Epidemiology
Worldwide, HD has the highest prevalence among Caucasians of European descent, estimated to be 5 to 7.5 out of 100,000 individuals (Warby et al., 2011). The greatest concentration of HD patients is seen in the Lake Maracaibo region of Venezuela at 700 cases per 100,000 (Warby et al., 2011), while Asian populations have the lowest prevalence of HD; in Japan, the prevalence is 0.65 per 100,000 (Nakashima et al., 1996). Data on African populations are lacking, but the disease is generally of low occurrence in such populations (Harper, 1992; Scrimgeour, 1981). Higher prevalence is seen in South Africa (Hayden and Beighton, 1982), United Republic of Tanzania (Scrimgeour, 1981), and Egypt (Kandil et al., 1994).
Warby and colleagues proposed an explanation for HD prevalence differences: they were able to identify, in Western European populations, two groups of single nucleotide polymorphisms located near HTT that are inherited together with the long CAG repeats of the gene (Warby et al., 2009). These two groups of single nucleotide polymorphisms make up two haplotypes that are highly predisposed to CAG instability (Warby et al., 2009). In other areas of the world, such as China and Japan, these haplotypes are not abundant in the population, and thus the mutation rates of CAG-track expansion are low (Warby et al., 2011).
2.4 Huntingtin gene
It has been almost 20 years since the discovery of the genetic cause of HD, the huntingtin gene, and yet the molecular pathways that lead to neurodegeneration in HD remain poorly defined. The huntingtin gene, identified in 1993 by a collective effort (The Huntington’s Disease Collaborative Research Group, 1993), is a large 180 kb DNA sequence that consists of 67 exons (Ambrose et al., 1994) and differs in sequence from any previously identified gene (The Huntington’s Disease Collaborative Research Group, 1993). The pathogenic CAG expansion is found in the first exon of the gene. Messenger RNA generated from HTT can be one of two lengths, 13.7 kb or 10.3 kb (Lin et al., 1993); these vary only at their 3’ untranslated polyadenylated tail. Neurons preferentially express the longer mRNA transcript (Lin et al., 1993), but the significance of this is not fully understood.
2.5 Huntingtin protein
Wild-type HTT mRNA is translated into a 348 kDa protein (The Huntington’s Disease Collaborative Research Group, 1993) that is ubiquitously expressed. Purifying recombinant Htt protein has been challenging due to the protein’s large size, low solubility, and poor bacterial expression (Li et al., 2006), making it difficult to obtain a structure of the full length protein. Most of what is known about the Htt protein structure comes from molecular modeling and biophysical studies on either polyglutamine peptides or the N-terminal portion of the protein (Darnell et al., 2007). To date, the only success at generating a recombinant full-length wild-type Htt protein, although at low yields, was seen by James A. Huntington’s group using insect cells (Li et al., 2006).
2.5.1 Structure
In 2009, Kim and colleagues generated seven crystals containing exon 1 of wild-type Htt with 17 glutamines in the polyQ region (PDB entries: 3IO4, 3IO6, 3IOR, 3IOT, 3IOU, 3IOV, 3IOW) (Kim et al., 2009). Their studies confirmed the predicted secondary structure for the N-terminus of the Htt protein. The CAG repeats in HTT translate into a polyQ track that starts at amino acid 18. The 17 amino acids preceding the polyQ sequence adopt an alpha helix (Kim et al., 2009). Directly following the polyQ region is a sequence of 11 consecutive proline residues, followed by several mixed proline/glutamine residues, and then another proline-enriched tract (Kim et al., 2009). These polyproline and mixed P/Q regions form a polyproline type II helix (Darnell et al., 2007; Kim et al., 2009) that is proposed to be involved in interactions with multiple binding partners (Qin et al., 2004). Furthermore, Kim et al. found that the wild-type polyQ track of 17 Q is able to adopt multiple conformations in the crystals: an alpha helix, an extended loop, and a random coil (Kim et al., 2009). They speculated that this floppy polyQ stretch can be stabilized in the full length Htt protein by the proline-rich regions that follow it (Kim et al., 2009). Several studies postulated that the expansion to 36 or more glutamines can change the conformation of the polyQ region into a beta sheet or other conformation (Darnell et al., 2007).
Additional structural features of the Htt protein were identified by analyzing the protein sequence. Andrade and Bork originally identified the presence of 10 HEAT tandem arrays, named after the initial four proteins found to contain these repeats (huntingtin, elongation factor 3, subunit A of protein phosphatase 2A, and TOR1) present within the first 1,710 N-terminal amino acids of the protein (Andrade and Bork, 1995). A later analysis with an alternative algorithm revealed the presence of 36 HEAT-like sequences throughout the Htt protein (Takano and Gusella, 2002).
These HEAT domains consist of 37-45 amino acids that form a helix-turn-helix secondary structure and are repeated multiple times in proteins. The alpha helices in each repeated unit are anti-parallel, but stack in tandem with neighboring HEAT repeats in a parallel fashion, so that each alpha helix in a HEAT unit is parallel to its counterpart in the next HEAT unit (Groves et al., 1999). Takano et al. proposed that having these alpha helical coils throughout Htt makes the protein adopt a solenoid tertiary structure that serves as a potential protein-protein interaction platform (Takano and Gusella, 2002). Preliminary biophysical characterizations by Li et al. on the full-length wild-type Htt (Li et al., 2006), as well as crystallization studies on other HEAT repeat proteins (Cingolani et al., 1999; Groves et al., 1999), support the notion of this superhelical solenoid platform. Furthermore, the fact that other HEAT-containing proteins are found in multi-protein complexes and recruit other proteins, may explain why the Htt has been reported to interact with numerous binding partners (Takano and Gusella, 2002).
2.5.2 Function
Wild-type Htt is an essential protein that is necessary for embryogenesis and development. Expression of truncated murine Htt protein generated by inactivation of either exon 5 (Nasir et al., 1995), the promoter and exon 1 (Zeitlin et al., 1995), or deletion of exon 4 and exon 5 (Duyao et al., 1995) results in early embryonic lethality in mice. Mice heterozygous for disrupted exon 5 of the murine huntingtin gene showed significant loss of neurons in the basal ganglia and resulting behavioral abnormalities (Nasir et al., 1995; O’Kusky et al., 1999). These findings suggest that wild-type huntingtin is important in maintaining basal ganglia neurons during adulthood (O’Kusky et al., 1999). Additional studies using transgenic mice carrying the human mutant HTT on a yeast artificial chromosome (YAC) showed that increasing the expression of endogenous murine wild-type huntingtin can protect against mutant huntingtin-induced apoptosis in vivo (Leavitt et al., 2001).
The precise role of wild-type huntingtin is unknown, but it has been implicated in numerous cellular pathways and shown to interact with a variety of proteins that are involved in clathrin-mediated endocytosis (Metzler et al., 2001; Singaraja et al., 2002), intracellular trafficking (Gauthier et al., 2004), transcriptional regulation (Dunah et al., 2002; Zhai et al., 2005; Zuccato et al., 2003), and cell survival (Leavitt et al., 2006; Zhang et al., 2006).
Mutant huntingtin has aberrant interactions with its binding partners, and loss of some or all of these interactions may contribute to HD pathogenesis. Furthermore, homozygous inactivation of murine huntingtin supports the idea that HD arises due to a gain-of-function caused by the expanded polyQ track (Duyao et al., 1995; Nasir et al., 1995; Zeitlin et al., 1995), which may be involved in aberrant interactions (Cui et al., 2006; Morfini et al., 2009). Expansion of the polyQ track in Htt leads to formation of neuronal nuclear aggregates and induces the pathology associated with HD (Bates, 2003).
2.6 Pathogenic mechanisms in HD
The presence of mutant huntingtin in the cell results in many cellular problems. It causes activation of pro-apoptotic proteins, mitochondria dysfunction, impaired axonal transport, transcriptional dysregulation, excitotoxicity, altered metabolism and energy levels, and aberrant calcium regulation. These cellular dysfunctions occur in early stages of HD, prior to the demise of the striatal cells. When left unchecked for a prolonged period, they ultimately lead to neuronal death. The exact mechanism of how mutant huntingtin causes such cellular dysfunction is not clear. In the following section, we briefly review proposed pathogenic cellular mechanisms.
2.6.1. Excitotoxicity
Excitotoxicity in HD involves overstimulation of neuronal glutamate receptors, especially ionotropic N-methyl-D-aspartic acid receptors (NMDARs), that leads to excessive influx of calcium ions and eventually results in activation of cell death. Several factors have been proposed to be responsible for induced NMDAR overactivation in HD (Raymond, 2003).
First, impaired glutamate reuptake by astrocytes has been proposed, which results in substantial glutamate accumulation in the synapse and overactivation of NMDARs. Hassel and colleagues reported a 43% reduction of glutamate uptake and defective excitatory amino-acid transporter 2 (EAAT2) (also known as glutamate transporter 1, GLT1) function in postmortem brains of HD patients (Hassel et al., 2008). These researchers also observed that the rate of glutamate uptake was inversely related to CAG repeat length in portmortem brains of HD patients (Hassel et al., 2008); the rate was lower in brain tissues with a larger number of CAG repeats in mutant HTT (Hassel et al., 2008). Multiple studies in mice also found decreased EAAT2/GLT1 expression and function in the presence of mutant huntingtin, well before neuronal cell death occurred (Behrens et al., 2002; Estrada-Sanchez et al., 2009; Lievens et al., 2001). R6/2 HD mice treated with ceftriaxone, a β-lactam antibiotic that can activate EAAT2/GLT1 promoter (Rothstein et al., 2005), had elevated EAAT2/GLT1 expression in the striatum and cortex in early and late stages of HD (Miller et al., 2008; Sari et al., 2010). Furthermore, increasing EAAT2/GLT1 levels with ceftriaxone was able to ameliorate the HD phenotype in R6/2 mice (Miller et al., 2008) and was neuroprotective in an ALS mouse model (Rothstein et al., 2005). Huang and colleagues found that palmitoylation, which is the covalent attachment of palmitic acid, to cys-38 of EAAT2/GLT1 was important for EAAT2/GLT1 reuptake of glutamate by glial cells (Huang et al., 2010). Significant loss of palmitoylation at this site was observed in YAC128 HD mice (Huang et al., 2010).
Another factor that can lead to overactivation of NMDARs is altered energy metabolism, due to dysfunctioning mitochondria. NMDAR and other ionotropic neuronal receptors require a resting membrane potential between −70 to −80 mV. This resting potential is sustained by Na+/K+ ATPase, which pumps Na+ ions out and K+ ions into cells in an ATP-dependent manner. When mitochondria are damaged and there is less ATP production to support the proper Na+/K+ ion gradients, this leads to cell depolarization (Hara and Snyder, 2007). Under normal resting potential, there is a Mg+2ion in the NMDAR that blocks entry of calcium into neurons, even if the channel is open. However, when the membrane is depolarized the Mg+2 is lost from the NMDAR and the channel allows free entry of Ca+2, once glutamate and glycine bind to the NMDAR (Hara and Snyder, 2007).
2.6.2 Mitochondrial dysfunction
Mitochondria are one of the most important organelles for neuronal survival. They are the main providers of energy to neurons, and in addition to supporting common cellular functions are needed to maintain resting membrane potential and neurotransmitter uptake and release. Additionally, mitochondria are involved in activation of apoptosis, calcium buffering, and produce reactive oxygen species. Thus, damage to mitochondria can have serious repercussions. Impaired mitochondrial function has been observed in multiple models of HD.
Evidence exists for mitochondrial metabolic dysfunction in HD. Weight loss is a pronounced factor in HD patients, and has been observed in HD mouse models. HD patients experience weight loss in the early (Aziz et al., 2008; Djousse et al., 2002) and late stages of the disease (Kirkwood et al., 2001). Severe chorea, increased energy expenditure, and difficulties swallowing may account for weight loss in patients with moderate HD (Pratley et al., 2000). In early stages of the disease, when HD patients have few involuntary movements (Djousse et al., 2002) weight loss is indicative of mitochondria dysfunction. Aziz et al. found a direct correlation between the number of CAG repeats and the rate of weight loss (Aziz et al., 2008). A larger number of CAG repeats in HD patients in the early stages of the disease and in R6/2 mice resulted in greater weight loss and increased caloric consumption in mice (Aziz et al., 2008). These authors showed that these results were not dependent on chorea or other motor activities, but rather might be due to a hypermetabolic cellular state, due to impaired mitochondria (Aziz et al., 2008).
Some researchers propose that weight loss in HD is indicative of impaired glycolysis. Monitoring 18F-fluorodeoxyglucose with positron emission tomography showed decreased glucose metabolism in the striatum of symptomatic and presymptomatic HD patients (Kuhl et al., 1982; Martin et al., 1992; Mazziotta et al., 1987). In addition to these same findings, Powers W.J. et al. also observed no change in the metabolic rate of oxygen and an increase in the ratio of oxygen to glucose metabolism in the cortex of HD patients (Powers et al., 2007). The authors conclude that no change in oxygen metabolism, but a decrease in glucose metabolism might be due to impaired glycolysis in astrocytes (Powers et al., 2007). Shifting the metabolism from breaking down glucose to ketone bodies by keeping mice on a diet high in fat and low in carbohydrates, decreased weight loss in R6/2 1J mice (Ruskin et al., 2011).
Applying 31P magnetic resonance spectroscopy, several groups were able to monitor high-energy metabolites, such as phosphocreatine, to study oxidative metabolism in HD patients during exercise. In cells needing energy sources that can be mobilized rapidly, such as in neurons and muscle, the energy reserves are stored as phosphocreatine. When energy is needed, such as during exercise, phosphate is transferred from phosphocreatine to ADP to generate ATP and creatine. When ATP is abundant, such as during a resting state, creatine can be converted to phosphocreatine in mitochondria to replenish this high-energy metabolite. In symptomatic HD patients and asymptomatic carriers with mutated HTT, there was a decreased rate of ATP generation after exercise, as monitored by phosphocreatine regeneration (Saft et al., 2005) and a reduced ratio of phosphocreatine to inorganic phosphate (Lodi et al., 2000). This is indicative of inadequate energy generation and dysfunctional oxidative metabolism in the mitochondria.
Impaired mitochondrial complex II, III, IV functions in the electron transport chain have been shown in mitochondria isolated from postmortem brain tissues of HD patients (Benchoua et al., 2006; Browne et al., 1997; Gu et al., 1996; Mann et al., 1990). Primates treated with complex II inhibitor, 3-nitropropionic acid, produced selective neuronal death in the striatum similar to what is observed in HD phenotype (Brouillet et al., 1995). Additionally, overexpressing the subunits of complex II was neuroprotective in striatal neurons transfected with mutant Htt (Benchoua et al., 2006).
There is reduced number, size, and altered morphology of mitochondria in rat cortical neurons transfected with mutant HTT (Song et al., 2011) and in HD patients (Kim et al., 2010). Overexpressing mutant Htt increased mitochondria fragmentation, reduced ATP levels, and induced caspase-3 activation in HeLa cells (Wang et al., 2009). A study by Song and collaborators showed that in fibroblasts from HD patients and in HD mice, mutant Htt induced mitochondrial fragmentation and decreases mitochondrial transport, which led to neuronal cell death (Song et al., 2011). This mitochondria fragmentation occurred before the onset of cell death (Song et al., 2011). Furthermore, mutant Htt, but not wild-type Htt, colocalized and bound to DRP1 (dynamin-related protein-1) in YAC128 mice and in brain tissues from HD patients, and activated DRP1 GTPase activity (Song et al., 2011). DRP1 K38A, which is a dominant-negative, enzymatically inactive mutant, was able to rescue neurons from mutant-Htt-induced cell death, reduce mitochondrial fragmentation and improve mitochondrial transport (Song et al., 2011) Expressing proteins that promote mitochondria fusion, such as Mfn2, was also able to prevent mitochondrial fragmentation, cell death and restore ATP levels in HeLa cells expressing mutant Htt (Wang et al., 2009). RNAi knockdown of DRP1 was able to reduce motility defects in C. elegans expressing mutant Htt (Wang et al., 2009). Targeting DRP1 protein with small-molecules might turn out to be a beneficial therapy for preventing cell death due to mitochondria fragmentation in HD.
Another consequence of malfunctioning mitochondria in HD is seen in disturbed calcium homeostasis. One of the normal functions of the mitochondria is to serve as a buffer for calcium ions. Calcium is taken up by the mitochondrial calcium uniporter (MCU), in the inner mitochondrial membrane, and is stored in the mitochondrial matrix until it is needed for release (Collins and Meyer, 2010; Viola and Hool, 2010). In HD, Panov et al. found that mitochondria from HD patients and HD transgenic mice had lower capacity to retain calcium, had lower membrane potential than controls, and the rate of calcium uptake was slower in mitochondria from HD YAC72 mice than YAC18 mice (Panov et al., 2002). Additionally, the researchers showed that mutant Htt was localized in the mitochondrial outer membrane in mitochondria isolated from brains of YAC72 mice (Panov et al., 2002) and from STHdhQ111/HdhQ111 cultured striatal neurons from knock-in embryonic mice (Choo et al., 2004). This disburbance of calcium ion homeostasis point to mitochondria impairement in HD.
There are also numerous studies that show reduced expression of PGC-1α (peroxisome proliferator-activated receptor-γ coactivator 1α), a transcriptional regulator of mitochondrial genes and antioxidant enzymes, in HD models. Mitochondrial transcription requires three proteins for activation: mitochondrial RNA polymerase (POLRMT), Tfam (mitochondrial transcription factor A), and one or both of TFB1M or TFB2M isoforms (mitochondrial transcription factor B1 and B2) (Scarpulla, 2008). Tfam, TFB1M, TFB2M, as well as cytochrome c and some subunits of electron transfer chain are encoded by nuclear genes, which are regulated by Nrf1 and Nrf2 (nuclear respiratory factors 1 and 2) transcription factors (Gleyzer et al., 2005). PGC-1α binds to Nrf1 and Nrf2 to trans-activate transcription of these genes (Gleyzer et al., 2005). In HD, there was an observed reduction in mRNA levels of PGC-1α in the medium spiny neurons of mutant Htt knockin mice, in cultured STHdhQ111 striatal cells, and reduced PGC-1α gene transcription in brains of R6/2 mice (Cui et al., 2006). Furthermore, postmortem caudate of presymptomatic HD patients also had lower PGC-1α mRNA levels, as well as decreased levels of PGC-1α-targeted mitochondrial genes (Cui et al., 2006). Overexpression of PGC-1α was able to rescue from mutant-Htt-induced toxicity in striatal cells (Cui et al., 2006). Mice that had PGC-1α knocked out and mutant Htt knocked in (HD(CAG)140+/− /PGC-1α+/−), showed worse motor performance and earlier degeneration in the striatum than mutant Htt knockin mice (Cui et al., 2006). This decrease in PGC-1α transcription in HD, which leads to earlier degeneration of striatum, was due to mutant Htt interference with CREB-mediated gene transcription (Cui et al., 2006). The PGC-1α gene promoter contains cAMP response elements needed for CREB binding and activation of transcription. As described in section 2.6.3, mutant Htt can interact with TAFII130/TAF4 (subunit of TFIID) and CREB, sequestering them from forming functional transcription complex (Cui et al., 2006).
2.6.3 Transcriptional dysregulation
Interference with the normal process of transcription has been proposed as another pathogenic mechanism in HD. One piece of evidence to support this hypothesis is a set of observations that wild-type Htt is predominately a cytosolic protein. However, when mutant Htt is present, the protein is cleaved and the N-terminal fragments translocate into the nucleus, forming intranuclear inclusion bodies, which were observed in the cortex and striatum in postmortem brains of HD patients (DiFiglia et al., 1997).
In addition, some transcription factors contain glutamine-rich sequences that allow them to recruit other proteins to the site of DNA interaction. One such transcription factor is specificity protein 1 (Sp1). Sp1 binds to the GC-box in promoters and recruits transcription factor II D (TFIID) to DNA, through its glutamine-rich sequence interaction with one of the TFIID subunits, TAFII130 (also known as TAF4) (Dunah et al., 2002). This causes binding of the rest of the transcription factors (TFIIA, TFIIB, TFIIF, TFIIE, and TFIIH), recruitment of RNA polymerase II, and initiation of transcription. When mutant Htt is present in the nucleus, its polyQ track binds to the glutamine-rich region of Sp1 and impedes Sp1 binding to specific DNA promoters and to TAFII130/TAF4 (Dunah et al., 2002; Li et al., 2002). This inhibits expression of selective Sp1-dependent genes that are downregulated in HD, such as dopamine D2 receptor and nerve growth factor receptor (Chen-Plotkin et al., 2006; Dunah et al., 2002; Li et al., 2002).
Furthermore, mutant Htt can directly interact with the transcription machinery. Soluble, not aggregated, mutant Htt directly binds and sequesters TAFII130/TAF4 (subunit of TFIID) and RAP30 (subunit of TFIIF) (Dunah et al., 2002; Zhai et al., 2005). The two subunits of TFIIF, RAP30 and RAP74, are needed together to recruit RNA polymerase II to the site of transcription. Mutant Htt may compete with RAP74 for binding to the N-terminal region of RAP30, thus preventing the formation of full TFIIF complex and inhibiting transcription (Zhai et al., 2005).
Several other studies showed that expression of genes that rely on the transcription factor CREB (cAMP response element binding) were also downregulated in HD (see also section 2.6.2 on transcription of mitochondrial gene regulator PGC-1α). CREB-mediated transcription relies on the coactivator CBP (CREB binding protein) for initiation of transcription (Kwok et al., 1994). CBP is a 265 kDa protein with multiple domains that aid its binding to the DNA and basal transcription factors, a KIX domain for interaction with phosphorylated CREB, a HAT domain for its histone acetyltransferase activity (McManus and Hendzel, 2001), and a stretch of 18 polyglutamines in the C-terminal portion of the protein (Nucifora et al., 2001). Nucifora Jr. and colleagues showed that mutant Htt interacts with the polyQ region in CBP in mouse model of HD and in postmortem brains of HD patients (Nucifora et al., 2001). They reported that CBP is sequestered by mutant Htt into intranuclear inclusions, preventing CBP-mediated transcription and reducing CBP histone acetyltransferase activity (Nucifora et al., 2001). Overexpressing CBP rescued cells from mutant-Htt-induced cell death (Nucifora et al., 2001).
2.7 Treatment
2.7.1 Medication
Currently, there is no treatment that can stop, postpone, or reverse the progression of HD. There are however, several drugs that can ameliorate the symptoms of HD. In 2008, tetrabenazine was approved by the U.S. FDA to control the involuntary sporadic movements of the extremities and face associated with chorea (Hayden et al., 2009; Wang et al., 2010). It is the only medication in United States solely prescribed for controlling muscular movement disorders. Tetrabenazine decreases the uptake of dopamine by inhibiting vesicular monoamine transporter 2 (VMAT2) (Hayden et al., 2009).
Antipsychotic medications, such as haloperidol, clozapine, chlorpromazine, and olanzapine, which function by decreasing dopamine levels, are also sometimes prescribed to reduce chorea (Jankovic, 2009; Phillips et al., 2008). A benefit of these agents is that they also help control the psychotic features that may be associated with HD.
Anticonvulsant medications, such as clonazepam, valproic acid, divalproex, and lamotrigine, may also decrease chorea (Adam and Jankovic, 2008). An antiparkinsonian medication, amantadine, has also shown success in controlling chorea (Bonelli and Wenning, 2006; Lucetti et al., 2003). To cope with depression, suicidal thoughts and compulsiveness, anti-depressants such as fluoxetine, escitalopram, sertraline, and nortriptyline have been prescribed to HD patients (Phillips et al., 2008). Furthermore, anti-anxiety medications, such as diazepam, benzodiazepines, paroxetine, and venlafaxin are used to calm patients, and to mitigate anxiety associated with the disease. Drugs such as lithium, valproate, and carbamazepine are used to ameliorate mania and mood swings in HD patients (Bonelli and Wenning, 2006). Unfortunately, many of these medications have adverse side effects that can worsen HD symptoms.
2.7.2 Non-medication therapies
Physical therapy and occupational therapy in the earlier stages of the disease can help improve motor coordination; exercises can strengthen muscles and improve balance and posture (Bilney et al., 2003). Psychiatric therapy can help reduce anxiety, depression and stress, as well as help cope with the changes associated with the disease. Additionally, decreasing stress and anxiety can help manage the chorea. Speech language pathologists can work with HD patients to help them with communication and swallowing problems, and determine if augmentative or alternative communication devices would be helpful (Bilney et al., 2003).
2.7.3 Drugs in development
Table 1 provides a list of small molecules, RNAi, gene therapy, and biological agents in preclinical or clinical stages of testing. Below we provide details of some of these experimental therapeutic agents.
Table 1.
Drugs currently in development for treatment of HD.
| Company | Compound | Type | Stage | Mechanism | Application | References |
|---|---|---|---|---|---|---|
| Accera, Inc. | AC-0523 | Small Molecule |
Preclinical | Restores impaired mitochondrial function |
HD | (Accera, Inc., 2012) |
| Alnylam Pharmaceuticals, Inc. Medtronic, Inc. CHDI Foundation, Inc. |
ALN-HTT | siRNA | Preclinical | Silences huntingtin mRNA |
HD | (DiFiglia et al., 2007; Stiles et al., 2011) |
| Avicena Group, Inc. |
HD-02 (Creatine monohydrate) |
Small Molecule |
Phase III | Improves energy metabolism |
HD | (Dedeoglu et al., 2003; Ferrante et al., 2000; Hersch et al., 2006) |
| Ceregene, Inc. | CERE-120 (AAV- neurturin) |
Gene Therapy |
Preclinical (HD) Phase II (PD) |
Promotes striatal survival through activation of RET tyrosine kinase pathway |
HD, PD | (Alberch et al., 2002; Perez-Navarro et al., 2000; Ramaswamy et al., 2007; Ramaswamy et al., 2009) |
| Chaperone Therapeutics, Inc. |
Lead compounds |
Small Molecule |
Preclinical | Activates HSF1 that leads to upregulation of heat-shock proteins |
AD, ALS, CJD, HD, PD |
(Neef et al., 2010) |
| Charite University Berlin, Germany |
EGCG | Small Molecule |
Phase II | Reduces mutant huntingtin aggregates |
HD | (Ehrnhoefer et al., 2006) |
| Cortex Pharmaceuticals, Inc. |
CX929 (ampakines) |
Small Molecule |
Preclinical | Allosterically activates AMPA receptor resulting in the production of BDNF |
HD | (Simmons et al., 2009; Simmons et al., 2011) |
| Edison Pharmaceuticals, Inc. |
EPI-743 | Small molecule |
Preclinical (HD) Phase II (IMDs) |
Potent Coenzyme Q10 analog that targets NQO1 |
IMDs, HD | (Edison Pharmaceuticals, 2011; Enns et al., 2012) |
| Genervon Biopharmaceuticals, LLC |
GM6 (MNTF analog) |
Protein biologic |
Preclinical (HD) Phase II (stroke, PD) |
Activates neurogenesis and anti-apoptosis pathways |
HD, AD, PD, ALS, MS, stroke, SCI |
(Genervon Biopharmaceuticals, 2012; Yu et al., 2008) |
| The Huntington Study Group NINDS University of Rochester Massachusetts General Hospital |
CoQ10 | Small molecule |
Phase III | Anti-oxidant and restores impaired mitochondrial function |
HD | (Koroshetz et al., 1997) |
| Indus Biotech | INDUS815C | Small molecule |
Preclinical | SIRT2 inhibitor | HD | (Luthi-Carter et al., 2010) |
| Isis Pharmaceuticals, Inc. CHDI Foundation, Inc. |
Modified antisense oligo- nucleotides |
siRNA | Preclinical | Selectively inhibit mutant huntingtin translation |
HD | (Isis Pharmaceuticals, Inc., 2011; Carroll et al., 2011; Gagnon et al., 2010) |
| Link Medicine Corp. |
LNK-754 | Small molecule |
Phase I (AD) Preclinical (HD) |
Inhibits farnesyl- transferase and stimulates autophagy |
AD, PD, HD | (Sommer and Stacy, 2008) |
| Neuralstem, Inc. | Region specific CNS stem cells |
Stem cell | Preclinical (HD) Phase I (ALS) |
Transplanted region specific stem cells will generate new neural circuits and provide neurotrophic pro- survival factors for existing neurons |
HD, ALS, SCI, stroke |
(Neuralstem, Inc., 2011) |
| Neurologix, Inc. Aegera Therapeutics, Inc. |
XIAP | Gene Therapy |
Preclinical | XIAP inhibits caspases and leads to degradation of Smac/Diablo |
HD | (Neurologix, Inc., 2011; Goffredo et al., 2005; Nakamura et al., 2010) |
| NeuroSearch A/S | Huntexil® (pridopidine; ACR16) |
Small molecule |
Phase III | Dopaminergic stabilizer |
HD | (de Yebenes et al., 2011; Lundin et al., 2010) |
| Novartis Pharmaceuticals |
AFQ056 | Small molecule |
Phase II (HD) Phase II (PD) Phase III (FXS) |
Reducing chorea | HD, PD, FXS |
(Levenga et al., 2011) |
| NsGene A/S | NsG33 (Meteorin) |
Protein biologic |
Preclinical | Novel neurotrophic factor that promotes survival of striatal neurons |
HD, NP | (NsGene A/S, 2012; Jorgensen et al., 2011) |
| Oryzon | LSD-1/ MAO-B |
Small molecule |
Preclinical | Inhibits both the lysine-specific demethylase-1 and monoamine oxidase B enzyme |
HD, PD, AD | (Oryzon, 2012) |
| Prana Biotechnology, Ltd. |
PBT2 (8-hydroxy- quinoline analog) |
Small Molecule |
Phase II | Prevents the formation of metal-mediated mutant protein aggregates and restores depleted metal ion levels in neurons |
AD, HD | (Adlard et al., 2008; Crouch et al., 2011; Nguyen et al., 2005; Tardiff et al., 2012) |
| Prosensa | PRO289 (antisense oligo- nucleotide) |
siRNA | Preclinical | Prevents the production of mutant huntingtin mRNA and protein |
HD | (Prosensa, 2012) |
| Raptor Pharmaceutical, Corp. |
RP103 (DR Cysteamine) |
Small molecule |
Phase II (HD) Phase III (NC) |
Delayed release of cysteamine, which can increase levels of BDNF in neurons |
HD, NC | (Borrell-Pages et al., 2006; Gibrat and Cicchetti, 2011) |
| ReNeuron | ReN005 | ReN005 Stem cell | Preclinical (HD) Phase I (stroke) |
Implanted into the brain to replace neuronal loss and improve motor functions |
HD, stroke | (ReNeuron, 2011) |
| Repligen Corp. | Lead Compounds |
Small molecule |
Preclinical | HDAC3 inhibitors | HD | (Jia et al., 2012) |
| Siena Biotech S.p.A. Elixir Pharmaceuticals, Inc. |
Selisistat (SEN196/ SEN0014196/ EX-527) |
Small molecule |
Phase II | SIRT1, class III histone deacetylase, inhibitor |
HD | (Gray, 2010) |
| University of Iowa NINDS CHDI Foundation, Inc. |
Citalopram (Celexa®) |
Small molecule |
Phase II | Selective serotonin reuptake inhibitor |
HD | (ClinicalTrials.gov, 2012) |
| Varinel |
VAR10300 (M30 and analogs) |
Small molecule |
Preclinical | Iron chelator and an irreversible inhibitor of brain- selective MAO-A and MAO-B |
AD, ALS, HD, PD, FA |
(Gal et al., 2005; Zheng et al., 2005) |
| VistaGen Therapeutics , Inc. |
AV-101 (L-4-chloro- kynurenine) |
Small molecule |
Phase I (NP) Preclinical (HD) |
Antagonist of the glycine-B site on NMDA receptor |
HD, PD, NP epilepsy |
(Campesan et al., 2011; Wu et al., 2000) |
| Wellstat Therapeutics, Corp. |
PN401 (Uridine Triacetate) |
Small molecule |
Preclinical | Delivers high concentration of uridine that is neuroprotective |
AD, HD, PD | (Saydoff et al., 2003; Saydoff et al., 2006) |
Alzheimer’s disease (AD); Amyotrophic lateral sclerosis/Lou Gehrig’s disease (ALS); Creutzfeldt–Jakob disease (CJD); Fragile X syndrome (FXS); Friedreich’s ataxia (FA); Huntington disease (HD); Inherited mitochondrial diseases (IMDs); Nephropathic Cystinosis (NC); Neuropathic pain (NP); Parkinson’s disease (PD); Spinal cord injury (SCI)
An RNAi therapeutic, ALN-HTT, is being developed in collaboration with Alnylam Pharmaceuticals, Inc., Medtronic, Inc., and CHDI Foundation, Inc. to silence the huntingtin mRNA and decrease mutant Htt protein production (Alnylam Pharmaceuticals, 2011). Alnylam Pharmaceuticals has proposed to file for Investigational New Drug (IND) application with the FDA in 2012 (Alnylam Pharmaceuticals, 2011) and shortly thereafter begin Phase I clinical trials. Experiments in mice showed that direct RNAi delivery into the brain provided protection against cell death for striatal neurons, decreased aggregation of mutant Htt protein, and improved motor functions (DiFiglia et al., 2007; Harper et al., 2005; Machida et al., 2006).
Link Medicine Corp. is developing the small molecule LNK-754 for the treatment of multiple neurodegenerative diseases, including AD, PD, ALS, and HD (Link Medicine, 2012). LNK-754 is an inhibitor of farnesyltransferase (Sommer and Stacy, 2008) that can induce the degradation of misfolded and aggregated proteins by stimulating autophagy, without inhibiting mTOR. In PD, studies have shown that inhibition of farnesyltransferases decreased alpha synuclein protein levels in cell culture and was neuroprotective (Liu et al., 2009). Furthermore, there have been two completed Phase I clinical studies with LNK-754 in healthy individuals and individuals with mild AD with promising outcomes. Phase II studies for AD have not commenced at this time. Trials for this therapeutic agent in HD are currently in development.
The gene therapy drug CERE-120 (AAV-NTN), developed by Ceregene, Inc., has shown promise in PD. CERE-120 is a viral vector containing the gene for neurturin, a pro-survival neurotrophic factor. It has shown neuroprotection in murine models of PD (Quarrell et al., 2007), has been well tolerated in Phase I human clinical trials, and is currently being evaluated in Phase II trials for PD. Furthermore, in preclinical trials CERE-120 decreased striatal cell loss in HD animal models (Perez-Navarro et al., 2000). It is now being evaluated in preclinical phase for advancement of CERE-120 into a Phase I trial for HD.
The small molecule pridopidine (Huntexil® or ACR16) is being evaluated in advanced clinical trials by NeuroSearch A/S for treatment of symptoms of HD. This compound has successfully completed a Phase III clinical study in Europe (MermaiHD study) and Phase II in US (HART study) (de Yebenes et al., 2011; Lundin et al., 2010). Patients receiving pridopidine have shown improvements in voluntary movements, had no serious adverse effects, and were able to tolerate the drug at high doses (de Yebenes et al., 2011). This drug is a dopamine stabilizing agent that binds to a D2 dopamine receptor (de Yebenes et al., 2011; Lundin et al., 2010). What makes pridopidine and this class of dopamine stabilizers unique is that depending on the levels of dopamine in the cells, it will either have stimulatory or inhibitory effects (Lundin et al., 2010).
2.8 Summary
There have been advances in understanding the causes of HD. This knowledge has translated into multiple therapeutic strategies, many of which are in development. Most of the investigational interventions for HD have focused on controlling dopamine levels, altering the mutant huntingtin mRNA and protein directly, or providing cell death protection to neurons in the striatum. These strategies have resulted in several therapeutic agents in preclinical and clinical trials. However, additional targets involved in cell death pathways might provide more substantial protection to neurons in the presence of mutant huntingtin protein. In the next section, we review cell death mechanisms in HD and some of the potential drug targets in these pathways.
3. Regulated cell death pathways and HD targets
Classical programmed cell death was initially used to describe apoptosis, a tightly regulated, active, self-terminating process that Kerr morphologically characterized in 1972 to contrast with accidental cell death (Kerr et al., 1972). Today, there are numerous cell death pathways that have been identified. In addition to the well-established regulated cell death pathway of apoptosis, novel pathways and processes involving cell death are being identified, such as ferroptosis (Dixon et al., 2012), entosis (Overholtzer et al., 2007), paraptosis (Sperandio et al., 2000), Netosis (Wartha and Henriques-Normark, 2008), and necroptosis (Degterev et al., 2005; Vandenabeele et al., 2010), to name a few. Most of the literature indicates that in striatal neurons of HD patients and in HD animal models, there are markers for apoptotic cell death (discussed more in section 3.1). We begin by discussing the cell death components implicated in HD, and then describe autophagy, the pro-survival pathway that is impaired in HD and their relevance to HD pathology.
3.1 Apoptosis pathway
Morphologically and biochemically, apoptosis is a distinct form of cell death. A cell that has been instructed to undergo apoptosis begins to contract, reducing the size of its cytoplasm, condensing its chromatin, and digesting the nucleus into fragments (Kerr et al., 1972). This is accompanied by the formation of apoptotic bodies — unequal sized, cell membrane-bound blebs that surround organelles, condensed chromatin, and cytoplasmic components — that pinch-off and get consumed by phagocytes for further degradation (Kerr et al., 1972; Kroemer et al., 2009).
The biochemical pathways of apoptosis are divided into two distinct routes: the extrinsic pathway and the intrinsic pathway. Both of these pathways activate a set of specific cysteine endoproteases, known as caspases (Alnemri et al., 1996), that carry out the execution order of the cell by cleaving protein substrates, and result in the characteristic morphology of apoptosis (Kroemer et al., 2009).
In the extrinsic pathway, the death stimulus activates a cell surface TNF family death receptor, causing oligomerization of its intracellular domain and binding of the adaptor protein, FADD (Boldin et al., 1996; Muzio et al., 1996). FADD contains a death effector domain (DED) that can bind to the DED of pro-caspase-8. Binding of FADD to pro-caspase-8 brings into close proximity other pro-caspase-8 molecules, causing pro-caspase-8 to dimerize and self-activate. Activated caspase-8 can then cleave and activate effector caspases, such as capsase-3, caspase-6, and caspase-7, which in turn execute the apoptosis program.
In the intristic pathway, the death stimulus results in the activation of pro-apoptotic proteins of the Bcl-2 family, Bax and Bak, that penetrate the mitochondrial outer membrane, causing the release of cytochrome c. Once released, cytochrome c binds to APAF-1, causing the heptamerization of APAF-1, recruitment of pro-caspase-9, formation of the apoptosome, and activation of caspase-9. Once activated, the apoptosome-caspase-9 complex cleaves effector caspases, capsase-3, caspase-6, and caspase-7, to launch the apoptosis program.
3.1.1. Apoptosis in HD
In HD, there are many markers of activated apoptotic machinery but at present there is no consensus on the cell death pathway that leads to the demise of the cell (reviewed in (Hickey and Chesselet, 2003)). In an inducible PC12 cell model of HD upon expression of mutant Htt there is release of cytochrome C from the mitochondria and activation of caspase-3, 6, and 7, which leads to cell death. This mutant Htt induced death can be rescued with Bcl-2 protein overexpression or with general caspase inhibitor, Boc-D-FMK (Hoffstrom et al., 2010)(Aiken et al., 2004). In cultures of rat striatal neurons expressing mutant Htt there is evidence of chromatin condensation (Rigamonti et al., 2000; Saudou et al., 1998), DNA laddering (Rigamonti et al., 2000), and activation of caspase-3 (Rigamonti et al., 2000). The cell death in primary striatal neurons could be rescued with co-expression of anti-apoptotic protein BclXL or caspase-3 inhibitor (Saudou et al., 1998). Additionally, wild-type Htt has been shown to protect from several apoptotic stimuli such as caspase-3, caspase-9, and BAK activation in immortalized striatal cells (Rigamonti et al., 2000). Medium spiny neurons from YAC128 transgenic mice also showed intrinsic caspase-dependent apoptotic death due to glutamate-induced Ca2+ signaling (Tang et al., 2005). All these in HD models point to the intrinsic caspase-dependent apoptosis mechanism.
Studies of proteins that can interact with Htt demonstrate that HIP1 can activate the intrinsic apoptosis cascade, implying that the loss of Htt-HIP1 interaction leads to this apoptotic death in HD (Choi et al., 2006). However, HIP1 interaction with Hippi can also activate caspase-8 dependent extrinsic cell death (Gervais et al., 2002). In this study, the cell death induced by expression of HIP1 and Hippi could not be rescued with co-expression of Bcl-2 protein, but could be ameliorated with inhibitors of the extrinsic apoptosis pathway (Gervais et al., 2002).
Furthermore, studies show involvement of caspase-1 in HD (Wang et al., 2005) (Ona et al., 1999). Caspase-1 activation is involved in a form of cell death, termed pyroptosis, which has different biochemical features than apoptosis altogether (Galluzzi et al., 2012).
In the striatum from postmortem brains of HD patients Kiechle and colleagues show that there was activated caspase-9, released cytochrome c into the cytoplasm, and activated caspase-3 only in the severe neuropathological grades samples (Grade 3 and Grade 4) (Kiechle et al., 2002). They observed that in R6/2 mice caspase-9 and cytochrome c release was also present only at the endstage of the disease (Kiechle et al., 2002). Even though their findings indicate intrinsic caspase-dependent apoptosis, this pathway does not initiate until later in the course of the disease; thus something else is responsible for the demise of neurons in the early neuropathological grades (Kiechle et al., 2002).
Even though there is no consensus right now on the cell death pathway that leads to the demise of the cell, pro-apoptotic machinery and cell death inducing proteins are activated in HD, and inhibiting them may serve as beneficial therapeutic targets (Hickey and Chesselet, 2003).
3.2 Pro-cell-death targets
There have been numerous reports of the involvement of pro-apoptotic and death-inducing proteins in HD that are activated as a result of mutant huntingtin protein expression. Inhibiting these proteins can delay the toxic effects of mutant huntingtin by postponing cell death and the onset of the disease. Here, we review these studies.
3.2.1 Caspase-1
Caspase-1 is not a typical pro-apoptotic protease in that it is not involved in the apoptotic cascade, however, it is capable of activating a form of cell death, termed pyroptosis (Galluzzi et al., 2012). Originally discovered as interleukin-1β converting enzyme (ICE), caspase-1 functions to cleave interleukin substrates, interleukin (IL)-1β and IL-18, in the inflammatory response (Thornberry et al., 1992). A recent study also demonstrated that caspase-1 can activate caspase-7, but not caspase-3, and thus linking inflammation to cell death response (Galluzzi et al., 2012; Lamkanfi et al., 2008). The optimal sequence in the substrate for caspase-1 cleavage is WEHD (Thornberry et al., 1997).
Mutating the active site cysteine in caspase-1 delayed the progression of symptoms in the R6/2 mouse model of HD by 20% (Ona et al., 1999). Additionally, increased levels of interleukin-1β were detected in the cortex of patients with moderate to severe cases of HD (Ona et al., 1999). These findings indicate that inflammation may trigger cell death in these patients.
Minocycline, a tetracycline-derivative antibiotic, which can bind to the 30S ribosome in prokaryotes and inhibit translation, has been also shown to inhibit caspase-1 activation in R6/2 mice, while tetracycline has no effect (Chen et al., 2000). Minocycline was tested in Phase II clinical trials, but failed to produce significant effects compared to placebo. Although minocycline was not successfully developed, caspase-1 should not be eliminated as a possible target. Minocycline may bind multiple targets, aside from caspase-1 (Wang et al., 2003), and thus lack the potency and specificity needed for an effective drug. A selective caspase-1 inhibitor, VX-765, is in development by Vertex Pharmaceuticals for the treatment of epilepsy (Vertex Pharmaceuticals, 2010), and is currently in Phase II trials. Despite the failure of minocycline to ameliorate HD, identification of a specific caspase-1 inhibitor, such as VX-765, or an inhibitor of its downstream targets in the inflammatory pathway may still prove to be useful in delaying HD.
3.2.2 Caspase-3
Caspase-3, formerly known as apopain and CPP32, is a key executioner of apoptosis. Caspase-3 has over 100 specific substrates that it cleaves to carry out apoptosis, including cytoskeletal proteins, ribonuclear proteins, proteins involved in DNA repair, cell cycle, and chromatin structure, protein kinases, and other caspase zymogens (Earnshaw et al., 1999). The optimal consensus sequence of its substrate is DEVD (Thornberry et al., 1997). As an effector caspase, its structure consists of a 30 amino acid pro-domain at the N-terminus, followed by a long and a short domain. To be activated, the short domain is separated from the long domain by cleavage, and then a second cut is made between the pro-domain and the long domain. These domains then tetramerize, forming a catalytically active caspase-3 enzyme, consisting of 2 long and 2 short domains.
Caspase-3 has been shown to have structural and biochemical similarities to another effector, caspase-7. Genetic deletion of caspase-3 in some mouse strains, such as C57BL/6J mice, can be compensated for by the presence of other caspases, while in other strains, such as 129X1/SvJ, caspase-3 deletion results in embryonic lethality (Leonard et al., 2002). Caspase-7 null C57BL/6J mice underwent normal development (Lakhani et al., 2006). Cells from mice with the long pro-domain deleted from either caspase-3 or caspase-7 undergo normal apoptosis when treated with apoptotic stimuli. Deletion of both caspase-3 and caspase-7 caused mice to die shortly after birth (Lakhani et al., 2006). Furthermore, having at least one allele of caspase-3 is necessary to have nuclear fragmentation, chromatin condensation, and the morphology of apoptosis (Lakhani et al., 2006).
In addition to its vital function in apoptosis, caspase-3 has been shown to cleave the huntingtin protein at amino acids 513 and 552 (Goldberg et al., 1996). Generation of these fragments is believed to be one of the toxic events contributing to HD. Irreversible small molecule inhibitors of pan-caspases have been shown to prevent caspase-3 cleavage of the huntingtin protein and protect cortical neurons from undergoing cell death (Leyva et al., 2010). The reversible, caspase-3-specific, small molecule M826 was also neuroprotective in a rat model of HD (Toulmond et al., 2004), indicating that one or more functions of these caspase inhibitors could be beneficial for the treatment of HD.
3.2.3 Caspase-6
Caspase-6 is the third effector caspase (in addition to caspase-3 and 7). In its inactive zymogen form, it contains a short prodomain that is cleaved during activation (Crawford and Wells, 2010). Activation of caspase-6 can be accomplished via initiator caspase-8 or 10, and in some cases, by caspase-3. Once activated, caspase-6 can cleave numerous substrates. Most of the substrates of caspase-6 can also be cleaved by caspase-3, except for the nuclear lamins A and C (Ruchaud et al., 2002; Takahashi et al., 1996). Caspase-6 cleaves substrate peptides with the optimal sequence VEHD (Thornberry et al., 1997). However, cleavage at other sites has been observed as well (Klaiman et al., 2008), such as cleavage of huntingtin at an IVLD sequence (Wellington et al., 2000). Overexpression of caspase-6 activates apoptosis, while genetic deletion of caspase-6 prevents chromatin condensation during apoptosis (Ruchaud et al., 2002), demonstrating its central role in this cell death pathway.
Besides its role in apoptosis, caspase-6 has been shown to cleave the mutant huntingtin protein at amino acid 586 (Wellington et al., 2000), which generates a toxic huntingtin fragment that translocates to the nucleus and induces neurodegeneration. The presence of caspase-6 has been seen in post-mortem brain tissues of HD patients and mice. Additionally, active caspase-6 has been detected in murine and human brain tissues, prior to the onset of HD or the activation of the apoptotic cascade (Graham et al., 2010), indicating it my be an early event in HD pathology. In 2006, Graham and colleagues showed that mutating the caspase-6 cleavage site in the human huntingtin protein rescued mice from HD (Graham et al., 2006). Since then, several small molecule inhibitors have been developed to inhibit caspase-6 (Chu et al., 2009; Leyva et al., 2010). Optimization of these compounds for clinical trials in still in process.
3.2.4 Caspase-8
Caspase-8 is one of three initiator caspases, along with caspase-9 and 10, which begin the activation of the apoptotic caspase cascade. Like all initiator caspases, caspase-8 in its inactive, zymogen form, contains a pro-domain at its N-terminus followed by a long domain and a short domain. The pro-domain contains two death effector domains (DED) with which pro-caspase-8 can interact with the adaptor protein FADD. The close proximity of two molecules of pro-caspase-8 bound to FADD causes pro-caspase-8 to dimerize, self-cleave, and form the active structure containing two long domains and two short domains (Earnshaw et al., 1999). The preferred sequence of substrates for caspase-8 is LETD (Thornberry et al., 1997).
Sanchez et al. found increased caspase-8 activation in striatal neurons and in the brain tissue of HD patients as a result of an expanded poly-Q track (Sanchez et al., 1999). Gervais and colleagues proposed a link connecting mutant huntingtin protein and caspase-8 activation (Gervais et al., 2002). According to their model, wild-type huntingtin interacts with protein HIP1 (huntingtin interacting protein) via its polyglutamine region. When this polyQ tract gets expanded, HIP1 loses its affinity for mutant huntingtin and instead is free to interact with newly discovered Hippi protein (Hip-1 protein interactor) (Gervais et al., 2002). Both Hippi and HIP1 contain pseudo-DED domains. Formation of the Hippi-HIP1 complex recruits the binding of two pro-caspase-8 proteins. The “proximity-induced autocatalytic maturation” results in an active caspase-8 enzyme, ready to initiate the apoptosis cascade (Gervais et al., 2002).
3.2.5 Caspases, all
The broad spectrum caspase inhibitor, ZVAD-fmk, has been shown to induce beneficial outcomes in the R6/2 mouse model of HD by prolonging survival and improving motor functions (Ona et al., 1999). Furthermore, multiple caspases have been found to cleave huntingtin into toxic N-terminal fragments: caspase-2 at amino acid 552, caspase-3 at residues 513 and 552, caspase-6 at amino acid 586, and caspase-7 at amino acid 513 and 552. Thus, it might be beneficial to develop a pan-caspase inhibitor that prevents all forms of huntingtin protein cleavage and also inhibits the apoptotic cascade.
3.2.6 HIP1
HIP1 has been identified in a yeast two-hybrid screen as a pro-apoptotic protein that is expressed at high levels in the brain and can interact with wild-type huntingtin in neuronal cells (Hackam et al., 2000; Kalchman et al., 1997; Metzler et al., 2003; Wanker et al., 1997). Structurally there have been three domains identified in HIP1—an AP180 N-terminal homology (ANTH) domain (Ford et al., 2001; Ford et al., 2002; Legendre-Guillemin et al., 2004), a coiled-coiled domain that contains the pseudo DED region, and a talin Hip1/R/Sla2 actin-tethering C-terminal homology (THATCH) domain (Brett et al., 2006). The ANTH domain can mediate binding to the 3-phosphoinositides, such as phosphatidylinositol-3,4-bisphosphate and phosphatidylinositol-3,5-bisphosphate, thus allowing the protein to dock to the plasma membrane (Hyun et al., 2004). The coiled-coiled domain spans the mid-portion of the HIP1 protein and is the site where the majority of protein partners interact with HIP1; some of these are clathrin light and heavy chains (Legendre-Guillemin et al., 2005; Waelter et al., 2001), polyglutamine-track-containing androgen receptor (Mills et al., 2005), HIP1 protein interactor (HIPPI) (Gervais et al., 2002; Niu and Ybe, 2008), NMDA receptor (Metzler et al., 2007), and of course huntingtin protein (Hackam et al., 2000(Kalchman et al., 1997)). The THATCH domain can interact and bind to filamentous actin (F-actin) in the cytoskeleton (Wilbur et al., 2008).
The precise function of HIP1 has not been defined. Its most studied involvement is in clathrin-mediated endocytosis, where HIP1 enables the formation of clathrin-coated vesicles and clathrin-coated pits (Legendre-Guillemin et al., 2005; Metzler et al., 2001; Waelter et al., 2001). In neurons, clathrin-mediated endocytosis is essential for recycling of neurotransmitter vesicles from presynaptic membranes and regulating the internalization of cell surface receptors. (For a thorough review of clathrin-mediated endocytosis, see the paper by McMahon, H.T. and Boucrot, E. (McMahon and Boucrot, 2011)). Parker and colleagues, studying the C. elegans homolog of HIP1, hipr-1, found that hipr-1 can localize to the presynaptic nerve terminals and there it serves a neuroprotective role against mutant-huntingtin-induced toxicity (Parker et al., 2007). A study by Metzler and colleagues showed that HIP1 is enriched in postsynaptic compartments in primary neurons (Metzler et al., 2003). They also observed that in HIP1−/− primary neurons there is a decline in internalization of GluR1-containing AMPA (α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid) receptors upon AMPA or glutamate stimulation (Metzler et al., 2003). Furthermore, the same group later also showed that HIP1 can modulate the downstream effects of NMDA receptor activation (Metzler et al., 2007). They reported that HIP1−/− striatal and cortical neurons had lower NMDA-induced excitotoxicity, less TUNEL-stained apoptotic cell death, less caspase-3 activation, and attenuated NMDA-induced AMPA receptor internalization than wild type neurons (Metzler et al., 2007). Additionally, HIP1 was shown to perturb huntingtin phosphorylation at serine 421, which is involved in neuronal survival, during NMDA-induced excitotoxicity (Metzler et al., 2007). This evidence suggests that HIP1 enhances NMDA-mediated cell death when it is located in the postsynaptic compartments, but may be neuroprotective when it’s in the presynaptic nerve terminals.
As mentioned (section 3.2.4), HIP1 interacts via the polyglutamine region of huntingtin, but has decreased affinity for huntingtin protein when the polyQ track becomes expanded (Kalchman et al., 1997; Wanker et al., 1997). A number of lines of evidence support the notion that it is this unbound-to-huntingtin HIP1 that is toxic to the neuronal survival, because that pool of HIP1 is free to interact with apoptotic partners. First, studies that overexpressed HIP1 found induced apoptosis in cells and neurons (Choi et al., 2006; Hackam et al., 2000). Second, co-expression of wild type huntingtin and HIP1 decreased apoptosis, but co-expression of mutant huntingtin and HIP1 did not have the same effect (Hackam et al., 2000). Third, overexpression of just the pseudo-DED region of HIP1 resulted in the same apoptotic phenotype in cells, which was prevented by mutating a conserved DED residue, Phe-398, in this pseudo DED region of HIP1 (Hackam et al., 2000). Second, free HIP1 has been found to interact with the Hippi protein (Hip-1 protein interactor) (Gervais et al., 2002). Both Hippi and HIP1 contain pseudo-DED regions that favor interaction. Formation of the Hippi-HIP1 complex recruits the binding of two pro-caspase-8 molecules. Then, “proximity-induced autocatalytic maturation” results in an active caspase-8 enzyme, which is able to initiate the apoptosis cascade (Gervais et al., 2002). Lastly, overexpressed HIP1 has also been shown to interact with pro-caspase-9 and Apaf-1 (Choi et al., 2006).
3.2.7 Protein disulfide isomerase
Protein disulfide isomerase (PDI) (EC 5.3.4.1) is a molecular chaperone protein and part of a large superfamily of disulfide oxidoreductases (Wilkinson and Gilbert, 2004). PDI is predominantly located in the endoplasmic reticulum and is responsible for catalyzing the proper formation (oxidation), incorrect bond elimination (reduction), and rearrangement (isomerization) of native disulfide bonds (Ellgaard and Ruddock, 2005; Hatahet and Ruddock, 2009). In 2010, a novel function of PDI was discovered that linked PDI to induction of cell death upon accumulation of misfolded protein, as is seen in HD and other neurodegenerative diseases (Hoffstrom et al., 2010). Hoffstrom et al. showed that in the presence of mutant huntingtin aggregates, PDI accumulates in the membrane spaces connecting the endoplasmic reticulum and mitochondria, known as mitochondrial-associated membranes (MAMs), and catalyzes formation of disulfide-linked Bak oligomers (Hoffstrom et al., 2010). This causes Bak to permeabilize the mitochondrial outer-membrane, initiating the apoptotic cascade. Inhibiting the catalytic activity of PDI with small molecules or reducing PDI levels with RNAi was neuroprotective.
3.2.8. NMDA receptor
As mentioned (section 2.6.1), in HD, there is over activation of NMDAR that leads to excessive influx of calcium and eventually apoptosis. Data have emerged suggesting that there are two relevant types of NMDA receptors. Synaptic NMDARs, which are present in the postsynaptic membrane and are composed of NR1/NR2A subunits (Benarroch, 2011), are important for neuronal survival; extrasynaptic NMDARs, which are distal from the synapse at the perisynaptic membrane sites and are composed of NR1/NR2B subunits (Benarroch, 2011), are neurotoxic. A more thorough description of NMDAR structures and functions is reviewed in (Benarroch, 2011). Studies with mice expressing both the mutant huntingtin gene and overexpressing NMDA NR2B subunit showed increased degeneration of striatal neurons (Heng et al., 2009). Furthermore there is an increase in the expression, activity, and current of extrasynaptic NMDAR in the striatum of HD mice, and even in pre-symptomatic HD mice (Milnerwood et al., 2010). Selectively inhibiting extra-synaptic NR2B subunits with low concentrations of memantine ameliorates the HD phenotype (Okamoto et al., 2009).
3.2.9. DRP1
As described in more detail in section 2.6.2, DRP1 is a promising new target in treatment of HD. Mutant huntingtin when bound to DRP1, activated DRP1 GTPase activity (Song et al., 2011) inducing mitochondria fission. Targeting DRP1 protein with small-molecules can be a beneficial therapy for preventing cell death due to mitochondria fragmentation in HD.
3.3 Autophagy
Autophagy is a cellular process normally initiated to protect the cell during the times of starvation, stress, bacterial infection, and low nutrient conditions (Nixon, 2006; Yorimitsu and Klionsky, 2005). Under normal conditions autophagy is used in development, to fight off bacterial and viral infection, and to recycle damaged or degraded organelles (Nixon, 2006). It is also used as a mechanism to deplete the cell of misfolded protein aggregates that are present in many neurodegenerative diseases (Ravikumar et al., 2002).
There are three defined types of autophagy processes: microautophagy, chaperone-mediated autophagy, and macroautophagy. Microautophagy is the least understood process of the three and involves the engulfment of cytoplasmic components to be degraded directly by the lysosome (Yorimitsu and Klionsky, 2005). Chaperone-mediated autophagy is a process that degrades only a selective population of proteins, with the aid of the heat shock cognate protein 70 kDa (hsc70). Hsc70 recognizes and binds to a KFERQ-like motif (Dice, 1990) in the cytosolic protein substrate and targets it to the lysosome, where this Hsp70-substrate complex is recognized by the lysosome associated membrane protein type 2A (LAMP-2A) (Cuervo and Dice, 1996). A second chaperone protein, lysosomal Hsc73, assists in translocating the substrate across the lysosomal membrane for degradation (Cuervo and Dice, 1996). The last process, macroautophagy, has a distinct morphological characteristic that involves the presence of cytoplasmic double-membrane vesicles. In macroautophagy, the intracellular components to be dismantled are enclosed by a double-layer membrane called the autophagosome (Yorimitsu and Klionsky, 2005), which fuses with a lysosome to cause degradation of the cargo by lysosomal hydrolases.
The macroautophagy pathway, which is vital for cell survival, is impaired in HD and in other protein misfolding neurodegenerative diseases (Iwata et al., 2005; Martinez-Vicente et al., 2010; Nixon et al., 2005; Ravikumar et al., 2002). Inhibiting macroautophagy in mice by selectively deleting the gene required for proper formation of the autophagosomes, Atg5, causes neurodegeneration and formation of cytoplasmic aggregates in neurons (Hara et al., 2006). In HD, the proper recognition of damaged organelles and protein aggregates is a defective step in autophagy, resulting in the formation of almost empty autophagosomes (Martinez-Vicente et al., 2010). Accumulation of mutant huntingtin protein aggregates in cells results in cell death. Small molecule inhibitors that promote autophagy have been shown to be neuroprotective: inhibiting mTOR, a key negative regulator of autophagy, with rapamycin in a fly model of HD decreased neurodegeneration of photoreceptor cells (Ravikumar et al., 2004). Furthermore, mice treated with the rapamycin ester analog, CCI-779, had decreased mTOR signaling in the striatum and cortex, fewer mutant huntingtin aggregates, and performed better in motor trials (Ravikumar et al., 2004). Thus, developing therapeutic compounds that can reactivate autophagy in HD may be beneficial.
4. Drug design
Identifying potential therapeutic targets in HD is only the first stage of creating new therapies. The next challenge is designing inhibitors for those targets. Since the first introduction of Pfizer’s “high-throughput screen” in 1948 (Janzen, 2002), the high-throughput screening (HTS) approach for lead elicitation has become ubiquitous within the pharmaceutical industry (Fischer and Hubbard, 2009; Janzen, 2002) and academia. However, limitations of this method have started to emerge and a growing need for additional methodologies to identify and develop potent lead compounds led to the rise of fragment-based drug design.
4.1 High-throughput screening
The initial step of HTS generally constitutes screening a library of approximately 106 large molecular weight compounds (Carr et al., 2005) and identifying compounds that give a phenotype of interest or modulate a specific protein target. The activity of each hit compound is then validated in a secondary screen and their chemical structures verified by mass spectrometry and nuclear magnetic resonance (NMR). Once the activity of a hit can be reproduced and its identity is confirmed, closely related structural analogs are synthesized and tested to assess which chemical moieties are responsible for the biological effects. This structure-activity relationship (SAR) profile provides the information necessary for chemically optimizing the hit into a lead compound, i.e. optimizing the hit to contain more drug-like properties, favorable pharmacokinetics, and improved potency and selectivity. The lead compound is then tested in vivo and once again optimized to decrease toxicity and improve ADME (absorption, distribution, metabolism, and excretion) properties. The optimized lead compound is thus developed into a candidate for clinical trials.
HTS has dramatically changed the industry and has seen many hits developed into therapeutic candidates. Although frequently successful, the method suffers from low hit rates, time consuming hit-to-lead optimization steps, and a lack of chemical diversity in the libraries (Carr et al., 2005; Langdon et al., 2011). In fact, several studies have now indicated that many small-molecule libraries are represented by only a few number of scaffolds and tend to encompass the same structural moieties (Krier et al., 2006; Langdon et al., 2011; Lipkus et al., 2008). This lack of chemical diversity can be problematic for identifying new compound leads. Furthermore, HTS’s low success rate with challenging drug targets has required researchers to look for progressive approaches in order to identify promising lead candidates for their targets of interest.
4.2 Fragment-based drug design
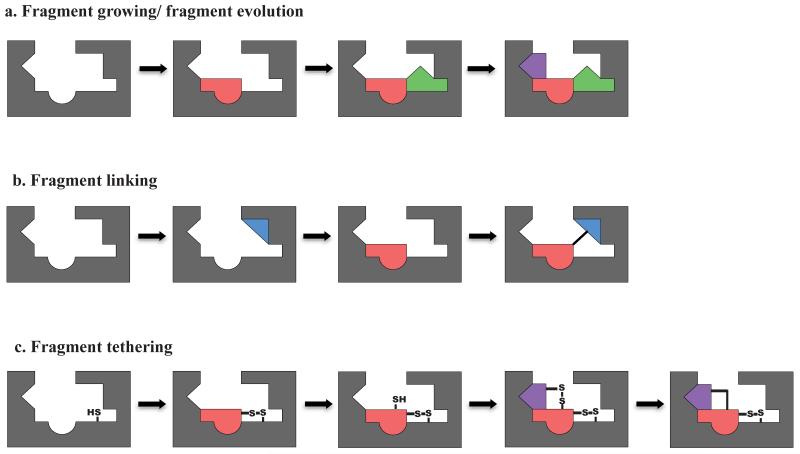
Over the past 16 years, fragment-based drug discovery (FBDD) has been gaining momentum as a parallel methodology to identify potent drug compounds (Alex and Flocco, 2007). The principle idea behind FBDD involves using less chemically complex and smaller compounds than are used for traditional HTS, to obtain information about the binding site of a protein target. Fragments that are active against a given target are optimized into typical “drug-size” compounds by either linking together active fragments that are binding in the adjacent pockets (“linking” method) (Shuker et al., 1996), chemically expanding the active fragment to cover a larger portion of the binding site (“growing” method), or assembling the active fragments on the protein through formation of a covalent bond, such as a disulfide bond, between the protein and the bound fragments (“tethering” method) (Erlanson et al., 2000). Figure 1 illustrates the three fragment optimization strategies.
Figure 1.
Strategies for optimizing fragments into “drug-size” compounds. Fragment growing/evolution strategy (a) is most similar to the traditional hit-to-lead optimization process. The active fragment (red) is chemically modified until a new chemical moiety (green) improves the binding affinity for the target by acquiring additional contacts with the target. The process can be further iterated to incorporate more favorable functional groups and interactions (purple). Structural binding information is not necessary for this method, but it does minimize the time for chemical optimization. Computation modeling can also facilitate in this process. A fragment linking strategy (b) requires knowing structurally where the fragments interact with the target protein. Active fragments in the neighboring binding pocket (red and blue) can be linked together to optimize potency for the protein. Care must be taken however, in designing the linker not to alter the original binding pose of the active fragments. The fragment tethering approach (c) makes use of a modified fragment library, designed to contain a single disulfide bond on each fragment, and an endogenous or engineered cysteine near the binding pocket of the protein. Under mild reducing conditions the fragment that has an affinity for the protein will be able to form a stable disulfide bond with the target (red). This active fragment can then be modified to include a thiol group that will be able to react with another disulfide fragment (purple). Lastly, the disulfide linker between the two active fragments is replaced by another more rigid linker. Figure adapted from (Erlanson, 2011).
The fragments used for FBDD typically contain 8 to 15 atoms (not including hydrogens) and have molecular weights between 100 and 250 Daltons. Because of their small size, such fragments can interact with proteins only weakly (dissociation constant, Kd, in the high micromolar to low millimolar range). As a result of their weak interactions, sensitive methods are needed to detect the binding of fragments. In 1996, Shuker SB et al. successfully constructed a nanomolar inhibitor to FK506 binding protein (FKBP) using what they called “SAR by NMR”, a fragment “linking” method, and have shown that this method can be beneficial in reducing the time and money spent in the drug development process (Shuker et al., 1996). Their proof of concept, as well as advances in biochemical techniques such as NMR spectroscopy, X-ray crystallography, isothermal titration calorimetry, surface plasmon resonance, mass spectrometry, and improvements in computational methods for molecular docking and the development of high concentration biological assays for screening fragments, played a major role in the recent successes of FBDD (Alex and Flocco, 2007).
FBDD offers a number of advantages over other screening techniques, such as HTS, that have attracted many companies to this method. First, there are a fewer compounds in fragment libraries than there are in HTS libraries; in general, only a few thousand fragments are needed compared to the nearly one million compounds used for HTS (Rees et al., 2004). This reduces the cost and screening time in the lead development process. A second advantage is that fragment libraries are more diverse and thus sample more of the chemical space than their larger molecular weight counterparts (Hann et al., 2001; Leach and Hann, 2011). This is because for each added atom in a molecule, there is an additional point of chemical variation that leads to an exponentially increasing total number of compounds (Blum and Reymond, 2009). Fragments, having fewer heavy atoms than drug-size compounds (approximately 36 atoms for a molecule with a molecular weight of 500 Daltons) thus make up a more encompassing set that can be tested experimentally or computationally than their drug-size counterparts (Erlanson, 2006). A third advantage is that fragments are more efficient at binding to their target than larger, more complex compounds (Hann et al., 2001). As compounds increase in size, the probability of complementing the target’s binding site decreases exponentially (Hann et al., 2001; Leach and Hann, 2011). These two points, a more chemically diverse library and a better match with the binding site, leads to a fourth benefit of FBDD— higher hit rates (Schuffenhauer et al., 2005). A fifth advantage is that FBDD generates multiple starting points for lead series. If more than one chemically diverse fragment binds to the same protein pocket, each of these hits can be optimized independently into a potent lead, giving multiple opportunities for success in the event that some of the lead compounds fail during development for compound-specific reasons (Murray and Rees, 2009).
FBDD can also reduce the time to bring a compound to market. It took less than 6 years from the initial discovery of a weak binding fragment to receive FDA approval of the optimized drug, Zelboraf (vemurafenib, PLX4032), for the treatment of melanomas with B-Raf V600E mutation (Bollag et al., 2010; Plexxikon, 2011; Tsai et al., 2008). This is a short timeline for a drug development process.
There have been several excellent reviews published that detail successful development of screening fragments into potent leads and pre-clinical compound candidates (Alex and Flocco, 2007; Erlanson, 2011; Hajduk and Greer, 2007). Of these compounds, it is important to mention those that have promise in neurodegenerative diseases, especially in HD. Starting from a weakly interacting fragment, two groups from Sunesis Pharmaceuticals, Inc. developed a potent small molecule inhibitors of caspase-3 (Allen et al., 2003; Choong et al., 2002) and a reversible caspase-1 inhibitor (O’Brien et al., 2005) using the fragment tethering method. A group from Astex Therapeutics, Ltd. (Congreve et al., 2007; Murray et al., 2007) and a group from AstraZeneca Pharmaceuticals (Edwards et al., 2007; Geschwindner et al., 2007) implemented the fragment growing approach to find inhibitors of β-secretase, BACE-1, an enzyme that cleaves amyloid precursor protein (APP) into a pathogenic β-amyloid peptide.
FBDD offers numerous benefits, especially to challenging and novel proteins that are hard to target with traditional methods, such as HTS. Implementing FBDD can be a valuable tool for finding inhibitors of pro-apoptotic proteins that can be utilized to slow the progression of HD.
5. Conclusions
Developing pharmaceutical agents to counteract neurodegenerative diseases has always posed a grand challenge. Primarily, our understanding of the molecular pathways involved in these diseases is still limited. Furthermore, the proteins that are known to be involved in neurodegenerative disorders are not easy targets to modulate with traditional approaches. To overcome the first problem, research has focused on the most downstream effectors of neurodegeneration—cell death pathways. There is an increasing amount of literature that points to the involvement of apoptosis in HD pathology and the down-regulation of the pro-survival autophagic response. Thus, inhibiting pro-apoptotic proteins or stimulating the activation of autophagy with small molecules may lead to neuroprotection. As a solution to the second problem, one can apply the FBDD methodology to target proteins, which have not been amenable to classical methods of drug discovery. Since its implementation, FBDD has proven to be an approach competitive with traditional screening methods and an efficient way to identify potent drug compounds. In HD specifically, much still needs to be learned about wild-type and mutant Htt function, but the FBDD approach has the potential to provide therapeutic compounds to the HD community.
Highlights.
Huntington disease is one of numerous neurodegenerative disorders
Inhibiting neuronal cell death is one strategy to combat Huntington disease
Inhibiting cell-death proteins or activating autophagy leads to neuroprotection
Numerous proteins are not amenable to classical methods of drug discovery
Fragment-based drug discovery is an alternative method to identify potent drugs
Acknowledgements
We would like to thank R.R. Letso and B.E. Kaplan for their thorough review of the manuscript. We apologize to investigators whose work we did not include in this review due to time and space restrictions. BRS is an Early Career Scientist of the Howard Hughes Medical Institute, and is supported by additional funding from the Arnold and Mabel Beckman Foundation, NYSTAR and the National Institutes of Health (R01CA097061, R01GM085081 and R01CA161061). AK is supported by the Training Program in Molecular Biophysics T32GM008281.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Abbreviations: HD, Huntington disease; AD, Alzheimer’s disease; PD, Parkinson’s disease; MS, multiple sclerosis; ALS, amyotrophic lateral sclerosis; FBDD, fragment-based drug discovery; HTT, huntingtin gene; Htt, huntingtin protein; CAG, cytosine-adenine-guanine; Q, glutamine; DED, death effector domain; HIP1, huntingtin interacting protein 1; NMDAR, N-methyl-D-aspartic acid receptor; EAAT2, excitatory amino-acid transporter 2; GLT1, glutamate transporter 1; DRP1, dynamin-related protein-1; PGC-1α, peroxisome proliferator-activated receptor-γ coactivator 1α; CREB, cAMP response element binding;
References
- Accera, Inc. [Retrieved February 5, 2012];Product Pipeline. 2012 from www.accerapharma.com/products.html.
- Adam OR, Jankovic J. Symptomatic treatment of Huntington disease. Neurotherapeutics. 2008;5:181–197. doi: 10.1016/j.nurt.2008.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adlard PA, Cherny RA, Finkelstein DI, Gautier E, Robb E, Cortes M, Volitakis I, Liu X, Smith JP, Perez K, Laughton K, Li QX, Charman SA, Nicolazzo JA, Wilkins S, Deleva K, Lynch T, Kok G, Ritchie CW, Tanzi RE, Cappai R, Masters CL, Barnham KJ, Bush AI. Rapid restoration of cognition in Alzheimer’s transgenic mice with 8-hydroxy quinoline analogs is associated with decreased interstitial Abeta. Neuron. 2008;59:43–55. doi: 10.1016/j.neuron.2008.06.018. [DOI] [PubMed] [Google Scholar]
- Alberch J, Perez-Navarro E, Canals JM. Neuroprotection by neurotrophins and GDNF family members in the excitotoxic model of Huntington’s disease. Brain Res Bull. 2002;57:817–822. doi: 10.1016/s0361-9230(01)00775-4. [DOI] [PubMed] [Google Scholar]
- Aiken CT, Tobin AJ, Schweitzer ES. A cell-based screen for drugs to treat Huntington’s disease. Neurobiol Dis. 2004;16:546–555. doi: 10.1016/j.nbd.2004.04.001. [DOI] [PubMed] [Google Scholar]
- Alex AA, Flocco MM. Fragment-based drug discovery: what has it achieved so far? Curr Top Med Chem. 2007;7:1544–1567. doi: 10.2174/156802607782341082. [DOI] [PubMed] [Google Scholar]
- Allen DA, Pham P, Choong IC, Fahr B, Burdett MT, Lew W, DeLano WL, Gordon EM, Lam JW, O’Brien T, Lee D. Identification of potent and novel small-molecule inhibitors of caspase-3. Bioorg Med Chem Lett. 2003;13:3651–3655. doi: 10.1016/j.bmcl.2003.08.024. [DOI] [PubMed] [Google Scholar]
- Alnemri ES, Livingston DJ, Nicholson DW, Salvesen G, Thornberry NA, Wong WW, Yuan J. Human ICE/CED-3 protease nomenclature. Cell. 1996;87:171. doi: 10.1016/s0092-8674(00)81334-3. [DOI] [PubMed] [Google Scholar]
- Alnylam Pharmaceuticals, Inc. [Retrieved May 3, 2011];News Release: Alnylam Launches “Alnylam 5×15” Product Strategy and Provides Guidance and Goals for 2011. 2011 from http://phx.corporate-ir.net/phoenix.zhtml?c=148005&p=irol-newsArticle&ID=1513530&highlight=
- Alzheimer’s Association [Retrieved April 20, 2011];Changing the Trajectory of Alzheimer’s Disease: A National Imperative. 2010 from www.alz.org/trajectory.
- Ambrose CM, Duyao MP, Barnes G, Bates GP, Lin CS, Srinidhi J, Baxendale S, Hummerich H, Lehrach H, Altherr M, et al. Structure and expression of the Huntington’s disease gene: evidence against simple inactivation due to an expanded CAG repeat. Somat Cell Mol Genet. 1994;20:27–38. doi: 10.1007/BF02257483. [DOI] [PubMed] [Google Scholar]
- Andrade MA, Bork P. HEAT repeats in the Huntington’s disease protein. Nat Genet. 1995;11:115–116. doi: 10.1038/ng1095-115. [DOI] [PubMed] [Google Scholar]
- Aziz NA, van der Burg JM, Landwehrmeyer GB, Brundin P, Stijnen T, Roos RA. Weight loss in Huntington disease increases with higher CAG repeat number. Neurology. 2008;71:1506–1513. doi: 10.1212/01.wnl.0000334276.09729.0e. [DOI] [PubMed] [Google Scholar]
- Bates G. Huntingtin aggregation and toxicity in Huntington’s disease. Lancet. 2003;361:1642–1644. doi: 10.1016/S0140-6736(03)13304-1. [DOI] [PubMed] [Google Scholar]
- Behrens PF, Franz P, Woodman B, Lindenberg KS, Landwehrmeyer GB. Impaired glutamate transport and glutamate-glutamine cycling: downstream effects of the Huntington mutation. Brain. 2002;125:1908–1922. doi: 10.1093/brain/awf180. [DOI] [PubMed] [Google Scholar]
- Benarroch EE. NMDA receptors: recent insights and clinical correlations. Neurology. 2011;76:1750–1757. doi: 10.1212/WNL.0b013e31821b7cc9. [DOI] [PubMed] [Google Scholar]
- Benchoua A, Trioulier Y, Zala D, Gaillard MC, Lefort N, Dufour N, Saudou F, Elalouf JM, Hirsch E, Hantraye P, Deglon N, Brouillet E. Involvement of mitochondrial complex II defects in neuronal death produced by N-terminus fragment of mutated huntingtin. Mol Biol Cell. 2006;17:1652–1663. doi: 10.1091/mbc.E05-07-0607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilney B, Morris ME, Perry A. Effectiveness of physiotherapy, occupational therapy, and speech pathology for people with Huntington’s disease: a systematic review. Neurorehabil Neural Repair. 2003;17:12–24. doi: 10.1177/0888439002250448. [DOI] [PubMed] [Google Scholar]
- Blum LC, Reymond JL. 970 million druglike small molecules for virtual screening in the chemical universe database GDB-13. J Am Chem Soc. 2009;131:8732–8733. doi: 10.1021/ja902302h. [DOI] [PubMed] [Google Scholar]
- Boldin MP, Goncharov TM, Goltsev YV, Wallach D. Involvement of MACH, a novel MORT1/FADD-interacting protease, in Fas/APO-1- and TNF receptor-induced cell death. Cell. 1996;85:803–815. doi: 10.1016/s0092-8674(00)81265-9. [DOI] [PubMed] [Google Scholar]
- Bollag G, Hirth P, Tsai J, Zhang J, Ibrahim PN, Cho H, Spevak W, Zhang C, Zhang Y, Habets G, Burton EA, Wong B, Tsang G, West BL, Powell B, Shellooe R, Marimuthu A, Nguyen H, Zhang KY, Artis DR, Schlessinger J, Su F, Higgins B, Iyer R, D’Andrea K, Koehler A, Stumm M, Lin PS, Lee RJ, Grippo J, Puzanov I, Kim KB, Ribas A, McArthur GA, Sosman JA, Chapman PB, Flaherty KT, Xu X, Nathanson KL, Nolop K. Clinical efficacy of a RAF inhibitor needs broad target blockade in BRAF-mutant melanoma. Nature. 2010;467:596–599. doi: 10.1038/nature09454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonelli RM, Wenning GK. Pharmacological management of Huntington’s disease: an evidence-based review. Curr Pharm Des. 2006;12:2701–2720. doi: 10.2174/138161206777698693. [DOI] [PubMed] [Google Scholar]
- Borrell-Pages M, Canals JM, Cordelieres FP, Parker JA, Pineda JR, Grange G, Bryson EA, Guillermier M, Hirsch E, Hantraye P, Cheetham ME, Neri C, Alberch J, Brouillet E, Saudou F, Humbert S. Cystamine and cysteamine increase brain levels of BDNF in Huntington disease via HSJ1b and transglutaminase. J Clin Invest. 2006;116:1410–1424. doi: 10.1172/JCI27607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brett TJ, Legendre-Guillemin V, McPherson PS, Fremont DH. Structural definition of the F-actin-binding THATCH domain from HIP1R. Nat Struct Mol Biol. 2006;13:121–130. doi: 10.1038/nsmb1043. [DOI] [PubMed] [Google Scholar]
- Brouillet E, Hantraye P, Ferrante RJ, Dolan R, Leroy-Willig A, Kowall NW, Beal MF. Chronic mitochondrial energy impairment produces selective striatal degeneration and abnormal choreiform movements in primates. Proc Natl Acad Sci U S A. 1995;92:7105–7109. doi: 10.1073/pnas.92.15.7105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browne SE, Bowling AC, MacGarvey U, Baik MJ, Berger SC, Muqit MM, Bird ED, Beal MF. Oxidative damage and metabolic dysfunction in Huntington’s disease: selective vulnerability of the basal ganglia. Ann Neurol. 1997;41:646–653. doi: 10.1002/ana.410410514. [DOI] [PubMed] [Google Scholar]
- Campesan S, Green EW, Breda C, Sathyasaikumar KV, Muchowski PJ, Schwarcz R, Kyriacou CP, Giorgini F. The kynurenine pathway modulates neurodegeneration in a Drosophila model of Huntington’s disease. Curr Biol. 2011;21:961–966. doi: 10.1016/j.cub.2011.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr RA, Congreve M, Murray CW, Rees DC. Fragment-based lead discovery: leads by design. Drug Discov Today. 2005;10:987–992. doi: 10.1016/S1359-6446(05)03511-7. [DOI] [PubMed] [Google Scholar]
- Carroll JB, Warby SC, Southwell AL, Doty CN, Greenlee S, Skotte N, Hung G, Bennett CF, Freier SM, Hayden MR. Potent and selective antisense oligonucleotides targeting single-nucleotide polymorphisms in the Huntington disease gene / allele-specific silencing of mutant huntingtin. Mol Ther. 2011;19:2178–2185. doi: 10.1038/mt.2011.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Ona VO, Li M, Ferrante RJ, Fink KB, Zhu S, Bian J, Guo L, Farrell LA, Hersch SM, Hobbs W, Vonsattel JP, Cha JH, Friedlander RM. Minocycline inhibits caspase-1 and caspase-3 expression and delays mortality in a transgenic mouse model of Huntington disease. Nat Med. 2000;6:797–801. doi: 10.1038/77528. [DOI] [PubMed] [Google Scholar]
- Chen-Plotkin AS, Sadri-Vakili G, Yohrling GJ, Braveman MW, Benn CL, Glajch KE, DiRocco DP, Farrell LA, Krainc D, Gines S, MacDonald ME, Cha JH. Decreased association of the transcription factor Sp1 with genes downregulated in Huntington’s disease. Neurobiol Dis. 2006;22:233–241. doi: 10.1016/j.nbd.2005.11.001. [DOI] [PubMed] [Google Scholar]
- Choi SA, Kim SJ, Chung KC. Huntingtin-interacting protein 1-mediated neuronal cell death occurs through intrinsic apoptotic pathways and mitochondrial alterations. FEBS Lett. 2006;580:5275–5282. doi: 10.1016/j.febslet.2006.08.076. [DOI] [PubMed] [Google Scholar]
- Choo YS, Johnson GV, MacDonald M, Detloff PJ, Lesort M. Mutant huntingtin directly increases susceptibility of mitochondria to the calcium-induced permeability transition and cytochrome c release. Hum Mol Genet. 2004;13:1407–1420. doi: 10.1093/hmg/ddh162. [DOI] [PubMed] [Google Scholar]
- Choong IC, Lew W, Lee D, Pham P, Burdett MT, Lam JW, Wiesmann C, Luong TN, Fahr B, DeLano WL, McDowell RS, Allen DA, Erlanson DA, Gordon EM, O’Brien T. Identification of potent and selective small-molecule inhibitors of caspase-3 through the use of extended tethering and structure-based drug design. J Med Chem. 2002;45:5005–5022. doi: 10.1021/jm020230j. [DOI] [PubMed] [Google Scholar]
- Chu W, Rothfuss J, Chu Y, Zhou D, Mach RH. Synthesis and in vitro evaluation of sulfonamide isatin Michael acceptors as small molecule inhibitors of caspase-6. J Med Chem. 2009;52:2188–2191. doi: 10.1021/jm900135r. [DOI] [PubMed] [Google Scholar]
- Cingolani G, Petosa C, Weis K, Muller CW. Structure of importin-beta bound to the IBB domain of importin-alpha. Nature. 1999;399:221–229. doi: 10.1038/20367. [DOI] [PubMed] [Google Scholar]
- ClinicalTrials.gov [Retrieved February 15, 2012];CIT-HD: Study in Huntington’s Disease. 2012 from http://clinicaltrials.gov/ct2/show/study/NCT00271596?cond=Huntington&intr=citalopra m&rank=1.
- Collins S, Meyer T. Cell biology: A sensor for calcium uptake. Nature. 2010;467:283. doi: 10.1038/467283a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Congreve M, Aharony D, Albert J, Callaghan O, Campbell J, Carr RA, Chessari G, Cowan S, Edwards PD, Frederickson M, McMenamin R, Murray CW, Patel S, Wallis N. Application of fragment screening by X-ray crystallography to the discovery of aminopyridines as inhibitors of beta-secretase. J Med Chem. 2007;50:1124–1132. doi: 10.1021/jm061197u. [DOI] [PubMed] [Google Scholar]
- Crawford ED, Wells JA. Caspase Substrates and Cellular Remodeling. Annu Rev Biochem. 2010 doi: 10.1146/annurev-biochem-061809-121639. [DOI] [PubMed] [Google Scholar]
- Crouch PJ, Savva MS, Hung LW, Donnelly PS, Mot AI, Parker SJ, Greenough MA, Volitakis I, Adlard PA, Cherny RA, Masters CL, Bush AI, Barnham KJ, White AR. The Alzheimer’s therapeutic PBT2 promotes amyloid-beta degradation and GSK3 phosphorylation via a metal chaperone activity. J Neurochem. 2011;119:220–230. doi: 10.1111/j.1471-4159.2011.07402.x. [DOI] [PubMed] [Google Scholar]
- Cuervo AM, Dice JF. A receptor for the selective uptake and degradation of proteins by lysosomes. Science. 1996;273:501–503. doi: 10.1126/science.273.5274.501. [DOI] [PubMed] [Google Scholar]
- Cui L, Jeong H, Borovecki F, Parkhurst CN, Tanese N, Krainc D. Transcriptional repression of PGC-1alpha by mutant huntingtin leads to mitochondrial dysfunction and neurodegeneration. Cell. 2006;127:59–69. doi: 10.1016/j.cell.2006.09.015. [DOI] [PubMed] [Google Scholar]
- Darnell G, Orgel JP, Pahl R, Meredith SC. Flanking polyproline sequences inhibit beta-sheet structure in polyglutamine segments by inducing PPII-like helix structure. J Mol Biol. 2007;374:688–704. doi: 10.1016/j.jmb.2007.09.023. [DOI] [PubMed] [Google Scholar]
- De Rooij KE, De Koning Gans PA, Skraastad MI, Belfroid RD, Vegter-Van Der Vlis M, Roos RA, Bakker E, Van Ommen GJ, Den Dunnen JT, Losekoot M. Dynamic mutation in Dutch Huntington’s disease patients: increased paternal repeat instability extending to within the normal size range. J Med Genet. 1993;30:996–1002. doi: 10.1136/jmg.30.12.996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Yebenes JG, Landwehrmeyer B, Squitieri F, Reilmann R, Rosser A, Barker RA, Saft C, Magnet MK, Sword A, Rembratt A, Tedroff J. Pridopidine for the treatment of motor function in patients with Huntington’s disease (MermaiHD): a phase 3, randomised, double-blind, placebo-controlled trial. Lancet Neurol. 2011;10:1049–1057. doi: 10.1016/S1474-4422(11)70233-2. [DOI] [PubMed] [Google Scholar]
- Dedeoglu A, Kubilus JK, Yang L, Ferrante KL, Hersch SM, Beal MF, Ferrante RJ. Creatine therapy provides neuroprotection after onset of clinical symptoms in Huntington’s disease transgenic mice. J Neurochem. 2003;85:1359–1367. doi: 10.1046/j.1471-4159.2003.01706.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degterev A, Huang Z, Boyce M, Li Y, Jagtap P, Mizushima N, Cuny GD, Mitchison TJ, Moskowitz MA, Yuan J. Chemical inhibitor of nonapoptotic cell death with therapeutic potential for ischemic brain injury. Nat Chem Biol. 2005;1:112–119. doi: 10.1038/nchembio711. [DOI] [PubMed] [Google Scholar]
- Dice JF. Peptide sequences that target cytosolic proteins for lysosomal proteolysis. Trends Biochem Sci. 1990;15:305–309. doi: 10.1016/0968-0004(90)90019-8. [DOI] [PubMed] [Google Scholar]
- DiFiglia M, Sapp E, Chase KO, Davies SW, Bates GP, Vonsattel JP, Aronin N. Aggregation of huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain. Science. 1997;277:1990–1993. doi: 10.1126/science.277.5334.1990. [DOI] [PubMed] [Google Scholar]
- DiFiglia M, Sena-Esteves M, Chase K, Sapp E, Pfister E, Sass M, Yoder J, Reeves P, Pandey RK, Rajeev KG, Manoharan M, Sah DW, Zamore PD, Aronin N. Therapeutic silencing of mutant huntingtin with siRNA attenuates striatal and cortical neuropathology and behavioral deficits. Proc Natl Acad Sci U S A. 2007;104:17204–17209. doi: 10.1073/pnas.0708285104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon SJ, Lemberg KM, Lamprecht MR, Skouta R, Zaitsev EM, Gleason CE, Patel DN, Bauer AJ, Cantley AM, Yang WS, Morrison B, III, Stockwell BR. Ferroptosis: an iron-dependent form of non-apoptotic cell death. Cell. 2012;149:1060–1072. doi: 10.1016/j.cell.2012.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djousse L, Knowlton B, Cupples LA, Marder K, Shoulson I, Myers RH. Weight loss in early stage of Huntington’s disease. Neurology. 2002;59:1325–1330. doi: 10.1212/01.wnl.0000031791.10922.cf. [DOI] [PubMed] [Google Scholar]
- Dunah AW, Jeong H, Griffin A, Kim YM, Standaert DG, Hersch SM, Mouradian MM, Young AB, Tanese N, Krainc D. Sp1 and TAFII130 transcriptional activity disrupted in early Huntington’s disease. Science. 2002;296:2238–2243. doi: 10.1126/science.1072613. [DOI] [PubMed] [Google Scholar]
- Duyao MP, Auerbach AB, Ryan A, Persichetti F, Barnes GT, McNeil SM, Ge P, Vonsattel JP, Gusella JF, Joyner AL, et al. Inactivation of the mouse Huntington’s disease gene homolog Hdh. Science. 1995;269:407–410. doi: 10.1126/science.7618107. [DOI] [PubMed] [Google Scholar]
- Earnshaw WC, Martins LM, Kaufmann SH. Mammalian caspases: structure, activation, substrates, and functions during apoptosis. Annu Rev Biochem. 1999;68:383–424. doi: 10.1146/annurev.biochem.68.1.383. [DOI] [PubMed] [Google Scholar]
- Edison Pharmaceuticals Inc [Retrieved April 20, 2011];Technology. 2011 from edisonpharma.com/RnD.aspx.
- Edwards PD, Albert JS, Sylvester M, Aharony D, Andisik D, Callaghan O, Campbell JB, Carr RA, Chessari G, Congreve M, Frederickson M, Folmer RH, Geschwindner S, Koether G, Kolmodin K, Krumrine J, Mauger RC, Murray CW, Olsson LL, Patel S, Spear N, Tian G. Application of fragment-based lead generation to the discovery of novel, cyclic amidine beta-secretase inhibitors with nanomolar potency, cellular activity, and high ligand efficiency. J Med Chem. 2007;50:5912–5925. doi: 10.1021/jm070829p. [DOI] [PubMed] [Google Scholar]
- Ehrnhoefer DE, Duennwald M, Markovic P, Wacker JL, Engemann S, Roark M, Legleiter J, Marsh JL, Thompson LM, Lindquist S, Muchowski PJ, Wanker EE. Green tea (-)-epigallocatechin-gallate modulates early events in huntingtin misfolding and reduces toxicity in Huntington’s disease models. Hum Mol Genet. 2006;15:2743–2751. doi: 10.1093/hmg/ddl210. [DOI] [PubMed] [Google Scholar]
- Ellgaard L, Ruddock LW. The human protein disulphide isomerase family: substrate interactions and functional properties. EMBO Rep. 2005;6:28–32. doi: 10.1038/sj.embor.7400311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enns GM, Kinsman SL, Perlman SL, Spicer KM, Abdenur JE, Cohen BH, Amagata A, Barnes A, Kheifets V, Shrader WD, Thoolen M, Blankenberg F, Miller G. Initial experience in the treatment of inherited mitochondrial disease with EPI-743. Molecular Genetics and Metabolism. 2012;105:91–102. doi: 10.1016/j.ymgme.2011.10.009. [DOI] [PubMed] [Google Scholar]
- Erlanson DA, Braisted AC, Raphael DR, Randal M, Stroud RM, Gordon EM, Wells JA. Site-directed ligand discovery. Proc Natl Acad Sci U S A. 2000;97:9367–9372. doi: 10.1073/pnas.97.17.9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erlanson DA. Fragment-based lead discovery: a chemical update. Curr Opin Biotechnol. 2006;17:643–652. doi: 10.1016/j.copbio.2006.10.007. [DOI] [PubMed] [Google Scholar]
- Erlanson DA. Introduction to Fragment-Based Drug Discovery. Top Curr Chem. 2011 doi: 10.1007/128_2011_180. [DOI] [PubMed] [Google Scholar]
- Estrada-Sanchez AM, Montiel T, Segovia J, Massieu L. Glutamate toxicity in the striatum of the R6/2 Huntington’s disease transgenic mice is age-dependent and correlates with decreased levels of glutamate transporters. Neurobiol Dis. 2009;34:78–86. doi: 10.1016/j.nbd.2008.12.017. [DOI] [PubMed] [Google Scholar]
- Ferrante RJ, Andreassen OA, Jenkins BG, Dedeoglu A, Kuemmerle S, Kubilus JK, Kaddurah-Daouk R, Hersch SM, Beal MF. Neuroprotective effects of creatine in a transgenic mouse model of Huntington’s disease. J Neurosci. 2000;20:4389–4397. doi: 10.1523/JNEUROSCI.20-12-04389.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer M, Hubbard RE. Fragment-based ligand discovery. Mol Interv. 2009;9:22–30. doi: 10.1124/mi.9.1.7. [DOI] [PubMed] [Google Scholar]
- Ford MG, Pearse BM, Higgins MK, Vallis Y, Owen DJ, Gibson A, Hopkins CR, Evans PR, McMahon HT. Simultaneous binding of PtdIns(4,5)P2 and clathrin by AP180 in the nucleation of clathrin lattices on membranes. Science. 2001;291:1051–1055. doi: 10.1126/science.291.5506.1051. [DOI] [PubMed] [Google Scholar]
- Ford MG, Mills IG, Peter BJ, Vallis Y, Praefcke GJ, Evans PR, McMahon HT. Curvature of clathrin-coated pits driven by epsin. Nature. 2002;419:361–366. doi: 10.1038/nature01020. [DOI] [PubMed] [Google Scholar]
- Foroud T, Gray J, Ivashina J, Conneally PM. Differences in duration of Huntington’s disease based on age at onset. J Neurol Neurosurg Psychiatry. 1999;66:52–56. doi: 10.1136/jnnp.66.1.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagnon KT, Pendergraff HM, Deleavey GF, Swayze EE, Potier P, Randolph J, Roesch EB, Chattopadhyaya J, Damha MJ, Bennett CF, Montaillier C, Lemaitre M, Corey DR. Allele-selective inhibition of mutant huntingtin expression with antisense oligonucleotides targeting the expanded CAG repeat. Biochemistry. 2010;49:10166–10178. doi: 10.1021/bi101208k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gal S, Zheng H, Fridkin M, Youdim MB. Novel multifunctional neuroprotective iron chelator-monoamine oxidase inhibitor drugs for neurodegenerative diseases. In vivo selective brain monoamine oxidase inhibition and prevention of MPTP-induced striatal dopamine depletion. J Neurochem. 2005;95:79–88. doi: 10.1111/j.1471-4159.2005.03341.x. [DOI] [PubMed] [Google Scholar]
- Galluzzi L, Vitale I, Abrams JM, Alnemri ES, Baehrecke EH, Blagosklonny MV, Dawson TM, Dawson VL, El-Deiry WS, Fulda S, Gottlieb E, Green DR, Hengartner MO, Kepp O, Knight RA, Kumar S, Lipton SA, Lu X, Madeo F, Malorni W, Mehlen P, Nunez G, Peter ME, Piacentini M, Rubinsztein DC, Shi Y, Simon HU, Vandenabeele P, White E, Yuan J, Zhivotovsky B, Melino G, Kroemer G. Molecular definitions of cell death subroutines: recommendations of the Nomenclature Committee on Cell Death 2012. Cell Death Differ. 2012;19:107–120. doi: 10.1038/cdd.2011.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauthier LR, Charrin BC, Borrell-Pages M, Dompierre JP, Rangone H, Cordelieres FP, De Mey J, MacDonald ME, Lessmann V, Humbert S, Saudou F. Huntingtin controls neurotrophic support and survival of neurons by enhancing BDNF vesicular transport along microtubules. Cell. 2004;118:127–138. doi: 10.1016/j.cell.2004.06.018. [DOI] [PubMed] [Google Scholar]
- Genervon Biopharmaceuticals, LLC [Retrieved February 5, 2012];Pipeline. 2012 from www.genervon.com/genervon/medicines_pipeline.php.
- Gervais FG, Singaraja R, Xanthoudakis S, Gutekunst CA, Leavitt BR, Metzler M, Hackam AS, Tam J, Vaillancourt JP, Houtzager V, Rasper DM, Roy S, Hayden MR, Nicholson DW. Recruitment and activation of caspase-8 by the Huntingtin-interacting protein Hip-1 and a novel partner Hippi. Nat Cell Biol. 2002;4:95–105. doi: 10.1038/ncb735. [DOI] [PubMed] [Google Scholar]
- Geschwindner S, Olsson LL, Albert JS, Deinum J, Edwards PD, de Beer T, Folmer RH. Discovery of a novel warhead against beta-secretase through fragment-based lead generation. J Med Chem. 2007;50:5903–5911. doi: 10.1021/jm070825k. [DOI] [PubMed] [Google Scholar]
- Gibrat C, Cicchetti F. Potential of cystamine and cysteamine in the treatment of neurodegenerative diseases. Prog Neuropsychopharmacol Biol Psychiatry. 2011;35:380–389. doi: 10.1016/j.pnpbp.2010.11.023. [DOI] [PubMed] [Google Scholar]
- Gleyzer N, Vercauteren K, Scarpulla RC. Control of mitochondrial transcription specificity factors (TFB1M and TFB2M) by nuclear respiratory factors (NRF-1 and NRF-2) and PGC-1 family coactivators. Mol Cell Biol. 2005;25:1354–1366. doi: 10.1128/MCB.25.4.1354-1366.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goffredo D, Rigamonti D, Zuccato C, Tartari M, Valenza M, Cattaneo E. Prevention of cytosolic IAPs degradation: a potential pharmacological target in Huntington’s Disease. Pharmacol Res. 2005;52:140–150. doi: 10.1016/j.phrs.2005.01.006. [DOI] [PubMed] [Google Scholar]
- Goldberg YP, Andrew SE, Theilmann J, Kremer B, Squitieri F, Telenius H, Brown JD, Hayden MR. Familial predisposition to recurrent mutations causing Huntington’s disease: genetic risk to sibs of sporadic cases. J Med Genet. 1993;30:987–990. doi: 10.1136/jmg.30.12.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg YP, Nicholson DW, Rasper DM, Kalchman MA, Koide HB, Graham RK, Bromm M, Kazemi-Esfarjani P, Thornberry NA, Vaillancourt JP, Hayden MR. Cleavage of huntingtin by apopain, a proapoptotic cysteine protease, is modulated by the polyglutamine tract. Nat Genet. 1996;13:442–449. doi: 10.1038/ng0896-442. [DOI] [PubMed] [Google Scholar]
- Graham RK, Deng Y, Slow EJ, Haigh B, Bissada N, Lu G, Pearson J, Shehadeh J, Bertram L, Murphy Z, Warby SC, Doty CN, Roy S, Wellington CL, Leavitt BR, Raymond LA, Nicholson DW, Hayden MR. Cleavage at the caspase-6 site is required for neuronal dysfunction and degeneration due to mutant huntingtin. Cell. 2006;125:1179–1191. doi: 10.1016/j.cell.2006.04.026. [DOI] [PubMed] [Google Scholar]
- Graham RK, Deng Y, Carroll J, Vaid K, Cowan C, Pouladi MA, Metzler M, Bissada N, Wang L, Faull RL, Gray M, Yang XW, Raymond LA, Hayden MR. Cleavage at the 586 amino acid caspase-6 site in mutant huntingtin influences caspase-6 activation in vivo. J Neurosci. 2010;30:15019–15029. doi: 10.1523/JNEUROSCI.2071-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray SG. Targeting histone deacetylases for the treatment of Huntington’s disease. CNS Neurosci Ther. 2010;16:348–361. doi: 10.1111/j.1755-5949.2010.00184.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groves MR, Hanlon N, Turowski P, Hemmings BA, Barford D. The structure of the protein phosphatase 2A PR65/A subunit reveals the conformation of its 15 tandemly repeated HEAT motifs. Cell. 1999;96:99–110. doi: 10.1016/s0092-8674(00)80963-0. [DOI] [PubMed] [Google Scholar]
- Gu M, Gash MT, Mann VM, Javoy-Agid F, Cooper JM, Schapira AH. Mitochondrial defect in Huntington’s disease caudate nucleus. Ann Neurol. 1996;39:385–389. doi: 10.1002/ana.410390317. [DOI] [PubMed] [Google Scholar]
- Hackam AS, Yassa AS, Singaraja R, Metzler M, Gutekunst CA, Gan L, Warby S, Wellington CL, Vaillancourt J, Chen N, Gervais FG, Raymond L, Nicholson DW, Hayden MR. Huntingtin interacting protein 1 induces apoptosis via a novel caspase-dependent death effector domain. J Biol Chem. 2000;275:41299–41308. doi: 10.1074/jbc.M008408200. [DOI] [PubMed] [Google Scholar]
- Hajduk PJ, Greer J. A decade of fragment-based drug design: strategic advances and lessons learned. Nat Rev Drug Discov. 2007;6:211–219. doi: 10.1038/nrd2220. [DOI] [PubMed] [Google Scholar]
- Hann MM, Leach AR, Harper G. Molecular complexity and its impact on the probability of finding leads for drug discovery. J Chem Inf Comput Sci. 2001;41:856–864. doi: 10.1021/ci000403i. [DOI] [PubMed] [Google Scholar]
- Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, Yokoyama M, Mishima K, Saito I, Okano H, Mizushima N. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- Hara MR, Snyder SH. Cell signaling and neuronal death. Annu Rev Pharmacol Toxicol. 2007;47:117–141. doi: 10.1146/annurev.pharmtox.47.120505.105311. [DOI] [PubMed] [Google Scholar]
- Harper PS. The epidemiology of Huntington’s disease. Hum Genet. 1992;89:365–376. doi: 10.1007/BF00194305. [DOI] [PubMed] [Google Scholar]
- Harper SQ, Staber PD, He X, Eliason SL, Martins IH, Mao Q, Yang L, Kotin RM, Paulson HL, Davidson BL. RNA interference improves motor and neuropathological abnormalities in a Huntington’s disease mouse model. Proc Natl Acad Sci U S A. 2005;102:5820–5825. doi: 10.1073/pnas.0501507102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassel B, Tessler S, Faull RL, Emson PC. Glutamate uptake is reduced in prefrontal cortex in Huntington’s disease. Neurochem Res. 2008;33:232–237. doi: 10.1007/s11064-007-9463-1. [DOI] [PubMed] [Google Scholar]
- Hatahet F, Ruddock LW. Protein disulfide isomerase: a critical evaluation of its function in disulfide bond formation. Antioxid Redox Signal. 2009;11:2807–2850. doi: 10.1089/ars.2009.2466. [DOI] [PubMed] [Google Scholar]
- Hayden MR, Beighton P. Genetic aspects of Huntington’s chorea: results of a national survey. Am J Med Genet. 1982;11:135–141. doi: 10.1002/ajmg.1320110203. [DOI] [PubMed] [Google Scholar]
- Hayden MR, Leavitt BR, Yasothan U, Kirkpatrick P. Tetrabenazine. Nat Rev Drug Discov. 2009;8:17–18. doi: 10.1038/nrd2784. [DOI] [PubMed] [Google Scholar]
- Hebert LE, Beckett LA, Scherr PA, Evans DA. Annual incidence of Alzheimer disease in the United States projected to the years 2000 through 2050. Alzheimer Disease & Associated Disorders. 2001;15:169. doi: 10.1097/00002093-200110000-00002. [DOI] [PubMed] [Google Scholar]
- Hendricks AE, Latourelle JC, Lunetta KL, Cupples LA, Wheeler V, MacDonald ME, Gusella JF, Myers RH. Estimating the probability of de novo HD cases from transmissions of expanded penetrant CAG alleles in the Huntington disease gene from male carriers of high normal alleles (27-35 CAG) Am J Med Genet A. 2009;149A:1375–1381. doi: 10.1002/ajmg.a.32901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heng MY, Detloff PJ, Wang PL, Tsien JZ, Albin RL. In vivo evidence for NMDA receptor-mediated excitotoxicity in a murine genetic model of Huntington disease. J Neurosci. 2009;29:3200–3205. doi: 10.1523/JNEUROSCI.5599-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hersch SM, Gevorkian S, Marder K, Moskowitz C, Feigin A, Cox M, Como P, Zimmerman C, Lin M, Zhang L, Ulug AM, Beal MF, Matson W, Bogdanov M, Ebbel E, Zaleta A, Kaneko Y, Jenkins B, Hevelone N, Zhang H, Yu H, Schoenfeld D, Ferrante R, Rosas HD. Creatine in Huntington disease is safe, tolerable, bioavailable in brain and reduces serum 8OH2′dG. Neurology. 2006;66:250–252. doi: 10.1212/01.wnl.0000194318.74946.b6. [DOI] [PubMed] [Google Scholar]
- Hickey MA, Chesselet MF. Apoptosis in Huntington’s disease. Prog Neuropsychopharmacol Biol Psychiatry. 2003;27:255–265. doi: 10.1016/S0278-5846(03)00021-6. [DOI] [PubMed] [Google Scholar]
- Hoffstrom BG, Kaplan A, Letso R, Schmid RS, Turmel GJ, Lo DC, Stockwell BR. Inhibitors of protein disulfide isomerase suppress apoptosis induced by misfolded proteins. Nat Chem Biol. 2010;6:900–906. doi: 10.1038/nchembio.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang K, Kang MH, Askew C, Kang R, Sanders SS, Wan J, Davis NG, Hayden MR. Palmitoylation and function of glial glutamate transporter-1 is reduced in the YAC128 mouse model of Huntington disease. Neurobiol Dis. 2010;40:207–215. doi: 10.1016/j.nbd.2010.05.027. [DOI] [PubMed] [Google Scholar]
- Huntington G. On Chorea. The Medical and Surgical Reporter: A Weekly Journal. 1872;26:317–321. [Google Scholar]
- Huntington G. On chorea. George Huntington, M.D. J Neuropsychiatry Clin Neurosci. 2003;15:109–112. doi: 10.1176/jnp.15.1.109. [DOI] [PubMed] [Google Scholar]
- Huntington Study Group Pre2CARE Investigators Safety and tolerability of high-dosage coenzyme Q10 in Huntington’s disease and healthy subjects. Movement Disorders. 2010;25:1924–1928. doi: 10.1002/mds.22408. [DOI] [PubMed] [Google Scholar]
- Hyun TS, Rao DS, Saint-Dic D, Michael LE, Kumar PD, Bradley SV, Mizukami IF, Oravecz-Wilson KI, Ross TS. HIP1 and HIP1r stabilize receptor tyrosine kinases and bind 3-phosphoinositides via epsin N-terminal homology domains. J Biol Chem. 2004;279:14294–14306. doi: 10.1074/jbc.M312645200. [DOI] [PubMed] [Google Scholar]
- Imarisio S, Carmichael J, Korolchuk V, Chen CW, Saiki S, Rose C, Krishna G, Davies JE, Ttofi E, Underwood BR, Rubinsztein DC. Huntington’s disease: from pathology and genetics to potential therapies. Biochem J. 2008;412:191–209. doi: 10.1042/BJ20071619. [DOI] [PubMed] [Google Scholar]
- Isis Pharmaceuticals, Inc. [Retrieved February 15, 2012];Press Release. Isis and CHDI Foundation Renew Drug Development Collaboration for Huntington’s Disease. 2011 from http://ir.isispharm.com/phoenix.zhtml?c=222170&p=irol-newsArticle&ID=1595491&highlight=
- Iwata A, Christianson JC, Bucci M, Ellerby LM, Nukina N, Forno LS, Kopito RR. Increased susceptibility of cytoplasmic over nuclear polyglutamine aggregates to autophagic degradation. Proc Natl Acad Sci U S A. 2005;102:13135–13140. doi: 10.1073/pnas.0505801102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankovic J. Treatment of hyperkinetic movement disorders. Lancet Neurol. 2009;8:844–856. doi: 10.1016/S1474-4422(09)70183-8. [DOI] [PubMed] [Google Scholar]
- Janzen WP, editor. High Throughput Screening: Methods and Protocols. first ed Humana Press; Totowa, NJ: 2002. [Google Scholar]
- Jia H, Pallos J, Jacques V, Lau A, Tang B, Cooper A, Syed A, Purcell J, Chen Y, Sharma S, Sangrey GR, Darnell SB, Plasterer H, Sadri-Vakili G, Gottesfeld JM, Thompson LM, Rusche JR, Marsh JL, Thomas EA. Histone deacetylase (HDAC) inhibitors targeting HDAC3 and HDAC1 ameliorate polyglutamine-elicited phenotypes in model systems of Huntington’s disease. Neurobiology of Disease. 2012 doi: 10.1016/j.nbd.2012.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen JR, Emerich DF, Thanos C, Thompson LH, Torp M, Bintz B, Fjord-Larsen L, Johansen TE, Wahlberg LU. Lentiviral delivery of meteorin protects striatal neurons against excitotoxicity and reverses motor deficits in the quinolinic acid rat model. Neurobiol Dis. 2011;41:160–168. doi: 10.1016/j.nbd.2010.09.003. [DOI] [PubMed] [Google Scholar]
- Jummani R, Okun M. Sydenham chorea. Arch Neurol. 2001;58:311–313. doi: 10.1001/archneur.58.2.311. [DOI] [PubMed] [Google Scholar]
- Kalchman MA, Koide HB, McCutcheon K, Graham RK, Nichol K, Nishiyama K, Kazemi-Esfarjani P, Lynn FC, Wellington C, Metzler M, Goldberg YP, Kanazawa I, Gietz RD, Hayden MR. HIP1, a human homologue of S. cerevisiae Sla2p, interacts with membrane-associated huntingtin in the brain. Nat Genet. 1997;16:44–53. doi: 10.1038/ng0597-44. [DOI] [PubMed] [Google Scholar]
- Kandil MR, Tohamy SA, Fattah MA, Ahmed HN, Farwiez HM. Prevalence of chorea, dystonia and athetosis in Assiut, Egypt: a clinical and epidemiological study. Neuroepidemiology. 1994;13:202–210. doi: 10.1159/000110380. [DOI] [PubMed] [Google Scholar]
- Kerr JF, Wyllie AH, Currie AR. Apoptosis: a basic biological phenomenon with wide-ranging implications in tissue kinetics. Br J Cancer. 1972;26:239–257. doi: 10.1038/bjc.1972.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiechle T, Dedeoglu A, Kubilus J, Kowall NW, Beal MF, Friedlander RM, Hersch SM, Ferrante RJ. Cytochrome C and caspase-9 expression in Huntington’s disease. Neuromolecular Med. 2002;1:183–195. doi: 10.1385/NMM:1:3:183. [DOI] [PubMed] [Google Scholar]
- Kim J, Moody JP, Edgerly CK, Bordiuk OL, Cormier K, Smith K, Beal MF, Ferrante RJ. Mitochondrial loss, dysfunction and altered dynamics in Huntington’s disease. Hum Mol Genet. 2010;19:3919–3935. doi: 10.1093/hmg/ddq306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MW, Chelliah Y, Kim SW, Otwinowski Z, Bezprozvanny I. Secondary structure of Huntingtin amino-terminal region. Structure. 2009;17:1205–1212. doi: 10.1016/j.str.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkwood SC, Su JL, Conneally P, Foroud T. Progression of symptoms in the early and middle stages of Huntington disease. Arch Neurol. 2001;58:273–278. doi: 10.1001/archneur.58.2.273. [DOI] [PubMed] [Google Scholar]
- Klaiman G, Petzke TL, Hammond J, Leblanc AC. Targets of caspase-6 activity in human neurons and Alzheimer disease. Mol Cell Proteomics. 2008;7:1541–1555. doi: 10.1074/mcp.M800007-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koroshetz WJ, Jenkins BG, Rosen BR, Beal MF. Energy metabolism defects in Huntington’s disease and effects of coenzyme Q10. Annals of Neurology. 1997;41:160–165. doi: 10.1002/ana.410410206. [DOI] [PubMed] [Google Scholar]
- Krack P. Relicts of dancing mania: the dancing procession of Echternach. Neurology. 1999;53:2169–2172. doi: 10.1212/wnl.53.9.2169. [DOI] [PubMed] [Google Scholar]
- Kremer B, Goldberg P, Andrew SE, Theilmann J, Telenius H, Zeisler J, Squitieri F, Lin B, Bassett A, Almqvist E, et al. A worldwide study of the Huntington’s disease mutation. The sensitivity and specificity of measuring CAG repeats. N Engl J Med. 1994;330:1401–1406. doi: 10.1056/NEJM199405193302001. [DOI] [PubMed] [Google Scholar]
- Kremer B, Almqvist E, Theilmann J, Spence N, Telenius H, Goldberg YP, Hayden MR. Sex-dependent mechanisms for expansions and contractions of the CAG repeat on affected Huntington disease chromosomes. Am J Hum Genet. 1995;57:343–350. [PMC free article] [PubMed] [Google Scholar]
- Krier M, Bret G, Rognan D. Assessing the scaffold diversity of screening libraries. J Chem Inf Model. 2006;46:512–524. doi: 10.1021/ci050352v. [DOI] [PubMed] [Google Scholar]
- Kroemer G, Galluzzi L, Vandenabeele P, Abrams J, Alnemri ES, Baehrecke EH, Blagosklonny MV, El-Deiry WS, Golstein P, Green DR, Hengartner M, Knight RA, Kumar S, Lipton SA, Malorni W, Nunez G, Peter ME, Tschopp J, Yuan J, Piacentini M, Zhivotovsky B, Melino G. Classification of cell death: recommendations of the Nomenclature Committee on Cell Death 2009. Cell Death Differ. 2009;16:3–11. doi: 10.1038/cdd.2008.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhl DE, Phelps ME, Markham CH, Metter EJ, Riege WH, Winter J. Cerebral metabolism and atrophy in Huntington’s disease determined by 18FDG and computed tomographic scan. Ann Neurol. 1982;12:425–434. doi: 10.1002/ana.410120504. [DOI] [PubMed] [Google Scholar]
- Kwok RP, Lundblad JR, Chrivia JC, Richards JP, Bachinger HP, Brennan RG, Roberts SG, Green MR, Goodman RH. Nuclear protein CBP is a coactivator for the transcription factor CREB. Nature. 1994;370:223–226. doi: 10.1038/370223a0. [DOI] [PubMed] [Google Scholar]
- Lakhani SA, Masud A, Kuida K, Porter GA, Jr., Booth CJ, Mehal WZ, Inayat I, Flavell RA. Caspases 3 and 7: key mediators of mitochondrial events of apoptosis. Science. 2006;311:847–851. doi: 10.1126/science.1115035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamkanfi M, Kanneganti TD, Van Damme P, Vanden Berghe T, Vanoverberghe I, Vandekerckhove J, Vandenabeele P, Gevaert K, Nunez G. Targeted peptidecentric proteomics reveals caspase-7 as a substrate of the caspase-1 inflammasomes. Mol Cell Proteomics. 2008;7:2350–2363. doi: 10.1074/mcp.M800132-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langdon SR, Brown N, Blagg J. Scaffold diversity of exemplified medicinal chemistry space. J Chem Inf Model. 2011;51:2174–2185. doi: 10.1021/ci2001428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leach AR, Hann MM. Molecular complexity and fragment-based drug discovery: ten years on. Curr Opin Chem Biol. 2011;15:489–496. doi: 10.1016/j.cbpa.2011.05.008. [DOI] [PubMed] [Google Scholar]
- Leavitt BR, Guttman JA, Hodgson JG, Kimel GH, Singaraja R, Vogl AW, Hayden MR. Wild-type huntingtin reduces the cellular toxicity of mutant huntingtin in vivo. Am J Hum Genet. 2001;68:313–324. doi: 10.1086/318207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leavitt BR, van Raamsdonk JM, Shehadeh J, Fernandes H, Murphy Z, Graham RK, Wellington CL, Raymond LA, Hayden MR. Wild-type huntingtin protects neurons from excitotoxicity. J Neurochem. 2006;96:1121–1129. doi: 10.1111/j.1471-4159.2005.03605.x. [DOI] [PubMed] [Google Scholar]
- Legendre-Guillemin V, Wasiak S, Hussain NK, Angers A, McPherson PS. ENTH/ANTH proteins and clathrin-mediated membrane budding. J Cell Sci. 2004;117:9–18. doi: 10.1242/jcs.00928. [DOI] [PubMed] [Google Scholar]
- Legendre-Guillemin V, Metzler M, Lemaire JF, Philie J, Gan L, Hayden MR, McPherson PS. Huntingtin interacting protein 1 (HIP1) regulates clathrin assembly through direct binding to the regulatory region of the clathrin light chain. J Biol Chem. 2005;280:6101–6108. doi: 10.1074/jbc.M408430200. [DOI] [PubMed] [Google Scholar]
- Leonard JR, Klocke BJ, D’Sa C, Flavell RA, Roth KA. Strain-dependent neurodevelopmental abnormalities in caspase-3-deficient mice. J Neuropathol Exp Neurol. 2002;61:673–677. doi: 10.1093/jnen/61.8.673. [DOI] [PubMed] [Google Scholar]
- Levenga J, Hayashi S, de Vrij FM, Koekkoek SK, van der Linde HC, Nieuwenhuizen I, Song C, Buijsen RA, Pop AS, Gomezmancilla B, Nelson DL, Willemsen R, Gasparini F, Oostra BA. AFQ056, a new mGluR5 antagonist for treatment of fragile X syndrome. Neurobiol Dis. 2011;42:311–317. doi: 10.1016/j.nbd.2011.01.022. [DOI] [PubMed] [Google Scholar]
- Leyva MJ, Degiacomo F, Kaltenbach LS, Holcomb J, Zhang N, Gafni J, Park H, Lo DC, Salvesen GS, Ellerby LM, Ellman JA. Identification and evaluation of small molecule pan-caspase inhibitors in Huntington’s disease models. Chem Biol. 2010;17:1189–1200. doi: 10.1016/j.chembiol.2010.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li SH, Cheng AL, Zhou H, Lam S, Rao M, Li H, Li XJ. Interaction of Huntington disease protein with transcriptional activator Sp1. Mol Cell Biol. 2002;22:1277–1287. doi: 10.1128/mcb.22.5.1277-1287.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Serpell LC, Carter WJ, Rubinsztein DC, Huntington JA. Expression and characterization of full-length human huntingtin, an elongated HEAT repeat protein. J Biol Chem. 2006;281:15916–15922. doi: 10.1074/jbc.M511007200. [DOI] [PubMed] [Google Scholar]
- Lievens JC, Woodman B, Mahal A, Spasic-Boscovic O, Samuel D, Kerkerian-Le Goff L, Bates GP. Impaired glutamate uptake in the R6 Huntington’s disease transgenic mice. Neurobiol Dis. 2001;8:807–821. doi: 10.1006/nbdi.2001.0430. [DOI] [PubMed] [Google Scholar]
- Lin B, Rommens JM, Graham RK, Kalchman M, MacDonald H, Nasir J, Delaney A, Goldberg YP, Hayden MR. Differential 3′ polyadenylation of the Huntington disease gene results in two mRNA species with variable tissue expression. Hum Mol Genet. 1993;2:1541–1545. doi: 10.1093/hmg/2.10.1541. [DOI] [PubMed] [Google Scholar]
- Link Medicine [Retrieved February 7, 2012];Our Approach. 2012 from http://www.linkmedicine.com/science/approach.asp.
- Lipkus AH, Yuan Q, Lucas KA, Funk SA, Bartelt WF, 3rd, Schenck RJ, Trippe AJ. Structural diversity of organic chemistry. A scaffold analysis of the CAS Registry. J Org Chem. 2008;73:4443–4451. doi: 10.1021/jo8001276. [DOI] [PubMed] [Google Scholar]
- Liu Z, Meray RK, Grammatopoulos TN, Fredenburg RA, Cookson MR, Liu Y, Logan T, Lansbury PT., Jr. Membrane-associated farnesylated UCH-L1 promotes alpha-synuclein neurotoxicity and is a therapeutic target for Parkinson’s disease. Proc Natl Acad Sci U S A. 2009;106:4635–4640. doi: 10.1073/pnas.0806474106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobo MK. Molecular profiling of striatonigral and striatopallidal medium spiny neurons past, present, and future. Int Rev Neurobiol. 2009;89:1–35. doi: 10.1016/S0074-7742(09)89001-6. [DOI] [PubMed] [Google Scholar]
- Lodi R, Schapira AH, Manners D, Styles P, Wood NW, Taylor DJ, Warner TT. Abnormal in vivo skeletal muscle energy metabolism in Huntington’s disease and dentatorubropallidoluysian atrophy. Ann Neurol. 2000;48:72–76. [PubMed] [Google Scholar]
- Lucetti C, Del Dotto P, Gambaccini G, Dell’ Agnello G, Bernardini S, Rossi G, Murri L, Bonuccelli U. IV amantadine improves chorea in Huntington’s disease: an acute randomized, controlled study. Neurology. 2003;60:1995–1997. doi: 10.1212/01.wnl.0000068165.07883.64. [DOI] [PubMed] [Google Scholar]
- Lundin A, Dietrichs E, Haghighi S, Goller ML, Heiberg A, Loutfi G, Widner H, Wiktorin K, Wiklund L, Svenningsson A, Sonesson C, Waters N, Waters S, Tedroff J. Efficacy and safety of the dopaminergic stabilizer Pridopidine (ACR16) in patients with Huntington’s disease. Clin Neuropharmacol. 2010;33:260–264. doi: 10.1097/WNF.0b013e3181ebb285. [DOI] [PubMed] [Google Scholar]
- Luthi-Carter R, Taylor DM, Pallos J, Lambert E, Amore A, Parker A, Moffitt H, Smith DL, Runne H, Gokce O, Kuhn A, Xiang Z, Maxwell MM, Reeves SA, Bates GP, Neri C, Thompson LM, Marsh JL, Kazantsev AG. SIRT2 inhibition achieves neuroprotection by decreasing sterol biosynthesis. Proceedings of the National Academy of Sciences. 2010;107:7927–7932. doi: 10.1073/pnas.1002924107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machida Y, Okada T, Kurosawa M, Oyama F, Ozawa K, Nukina N. rAAV-mediated shRNA ameliorated neuropathology in Huntington disease model mouse. Biochem Biophys Res Commun. 2006;343:190–197. doi: 10.1016/j.bbrc.2006.02.141. [DOI] [PubMed] [Google Scholar]
- Mann VM, Cooper JM, Javoy-Agid F, Agid Y, Jenner P, Schapira AH. Mitochondrial function and parental sex effect in Huntington’s disease. Lancet. 1990;336:749. doi: 10.1016/0140-6736(90)92242-a. [DOI] [PubMed] [Google Scholar]
- Martin WR, Clark C, Ammann W, Stoessl AJ, Shtybel W, Hayden MR. Cortical glucose metabolism in Huntington’s disease. Neurology. 1992;42:223–229. doi: 10.1212/wnl.42.1.223. [DOI] [PubMed] [Google Scholar]
- Martinez-Vicente M, Talloczy Z, Wong E, Tang G, Koga H, Kaushik S, de Vries R, Arias E, Harris S, Sulzer D, Cuervo AM. Cargo recognition failure is responsible for inefficient autophagy in Huntington’s disease. Nat Neurosci. 2010;13:567–576. doi: 10.1038/nn.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazziotta JC, Phelps ME, Pahl JJ, Huang SC, Baxter LR, Riege WH, Hoffman JM, Kuhl DE, Lanto AB, Wapenski JA, et al. Reduced cerebral glucose metabolism in asymptomatic subjects at risk for Huntington’s disease. N Engl J Med. 1987;316:357–362. doi: 10.1056/NEJM198702123160701. [DOI] [PubMed] [Google Scholar]
- McMahon HT, Boucrot E. Molecular mechanism and physiological functions of clathrin-mediated endocytosis. Nat Rev Mol Cell Biol. 2011;12:517–533. doi: 10.1038/nrm3151. [DOI] [PubMed] [Google Scholar]
- McManus KJ, Hendzel MJ. CBP, a transcriptional coactivator and acetyltransferase. Biochem Cell Biol. 2001;79:253–266. [PubMed] [Google Scholar]
- Metzler M, Legendre-Guillemin V, Gan L, Chopra V, Kwok A, McPherson PS, Hayden MR. HIP1 functions in clathrin-mediated endocytosis through binding to clathrin and adaptor protein 2. J Biol Chem. 2001;276:39271–39276. doi: 10.1074/jbc.C100401200. [DOI] [PubMed] [Google Scholar]
- Metzler M, Li B, Gan L, Georgiou J, Gutekunst CA, Wang Y, Torre E, Devon RS, Oh R, Legendre-Guillemin V, Rich M, Alvarez C, Gertsenstein M, McPherson PS, Nagy A, Wang YT, Roder JC, Raymond LA, Hayden MR. Disruption of the endocytic protein HIP1 results in neurological deficits and decreased AMPA receptor trafficking. EMBO J. 2003;22:3254–3266. doi: 10.1093/emboj/cdg334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzler M, Gan L, Wong TP, Liu L, Helm J, Georgiou J, Wang Y, Bissada N, Cheng K, Roder JC, Wang YT, Hayden MR. NMDA receptor function and NMDA receptor-dependent phosphorylation of huntingtin is altered by the endocytic protein HIP1. J Neurosci. 2007;27:2298–2308. doi: 10.1523/JNEUROSCI.5175-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller BR, Dorner JL, Shou M, Sari Y, Barton SJ, Sengelaub DR, Kennedy RT, Rebec GV. Up-regulation of GLT1 expression increases glutamate uptake and attenuates the Huntington’s disease phenotype in the R6/2 mouse. Neuroscience. 2008;153:329–337. doi: 10.1016/j.neuroscience.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills IG, Gaughan L, Robson C, Ross T, McCracken S, Kelly J, Neal DE. Huntingtin interacting protein 1 modulates the transcriptional activity of nuclear hormone receptors. J Cell Biol. 2005;170:191–200. doi: 10.1083/jcb.200503106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milnerwood AJ, Gladding CM, Pouladi MA, Kaufman AM, Hines RM, Boyd JD, Ko RW, Vasuta OC, Graham RK, Hayden MR, Murphy TH, Raymond LA. Early increase in extrasynaptic NMDA receptor signaling and expression contributes to phenotype onset in Huntington’s disease mice. Neuron. 2010;65:178–190. doi: 10.1016/j.neuron.2010.01.008. [DOI] [PubMed] [Google Scholar]
- Mink JW. The basal ganglia: focused selection and inhibition of competing motor programs. Prog Neurobiol. 1996;50:381–425. doi: 10.1016/s0301-0082(96)00042-1. [DOI] [PubMed] [Google Scholar]
- Morfini GA, You YM, Pollema SL, Kaminska A, Liu K, Yoshioka K, Bjorkblom B, Coffey ET, Bagnato C, Han D, Huang CF, Banker G, Pigino G, Brady ST. Pathogenic huntingtin inhibits fast axonal transport by activating JNK3 and phosphorylating kinesin. Nat Neurosci. 2009;12:864–871. doi: 10.1038/nn.2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray CW, Callaghan O, Chessari G, Cleasby A, Congreve M, Frederickson M, Hartshorn MJ, McMenamin R, Patel S, Wallis N. Application of fragment screening by X-ray crystallography to beta-secretase. J Med Chem. 2007;50:1116–1123. doi: 10.1021/jm0611962. [DOI] [PubMed] [Google Scholar]
- Murray CW, Rees DC. The rise of fragment-based drug discovery. Nat Chem. 2009;1:187–192. doi: 10.1038/nchem.217. [DOI] [PubMed] [Google Scholar]
- Muzio M, Chinnaiyan AM, Kischkel FC, O’Rourke K, Shevchenko A, Ni J, Scaffidi C, Bretz JD, Zhang M, Gentz R, Mann M, Krammer PH, Peter ME, Dixit VM. FLICE, a novel FADD-homologous ICE/CED-3-like protease, is recruited to the CD95 (Fas/APO-1) death--inducing signaling complex. Cell. 1996;85:817–827. doi: 10.1016/s0092-8674(00)81266-0. [DOI] [PubMed] [Google Scholar]
- Nakamura T, Wang L, Wong CC, Scott FL, Eckelman BP, Han X, Tzitzilonis C, Meng F, Gu Z, Holland EA, Clemente AT, Okamoto S, Salvesen GS, Riek R, Yates JR, 3rd, Lipton SA. Transnitrosylation of XIAP regulates caspase-dependent neuronal cell death. Mol Cell. 2010;39:184–195. doi: 10.1016/j.molcel.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima K, Watanabe Y, Kusumi M, Nanba E, Maeoka Y, Nakagawa M, Igo M, Irie H, Ishino H, Fujimoto A, Goto J, Takahashi K. Epidemiological and Genetic Studies of Huntington’s Disease in the San-in Area of Japan. Neuroepidemiology. 1996;15:126–131. doi: 10.1159/000109899. [DOI] [PubMed] [Google Scholar]
- Nasir J, Floresco SB, O’Kusky JR, Diewert VM, Richman JM, Zeisler J, Borowski A, Marth JD, Phillips AG, Hayden MR. Targeted disruption of the Huntington’s disease gene results in embryonic lethality and behavioral and morphological changes in heterozygotes. Cell. 1995;81:811–823. doi: 10.1016/0092-8674(95)90542-1. [DOI] [PubMed] [Google Scholar]
- Neef DW, Turski ML, Thiele DJ. Modulation of heat shock transcription factor 1 as a therapeutic target for small molecule intervention in neurodegenerative disease. PLoS Biol. 2010;8:e1000291. doi: 10.1371/journal.pbio.1000291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuralstem, Inc. [Retrieved February 15, 2012];Neuralstem Cell Therapy: Repairing and Replacing Damaged Cells. 2011 from http://www.neuralstem.com/cell-therapy.
- Neurologix, Inc. [Retrieved February 5, 2012];Product Pipeline. Huntington’s Disease. 2011 from http://www.neurologix.net/products/huntingtons.html.
- Nguyen T, Hamby A, Massa SM. Clioquinol down-regulates mutant huntingtin expression in vitro and mitigates pathology in a Huntington’s disease mouse model. Proc Natl Acad Sci U S A. 2005;102:11840–11845. doi: 10.1073/pnas.0502177102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu Q, Ybe JA. Crystal structure at 2.8 A of Huntingtin-interacting protein 1 (HIP1) coiled-coil domain reveals a charged surface suitable for HIP1 protein interactor (HIPPI) J Mol Biol. 2008;375:1197–1205. doi: 10.1016/j.jmb.2007.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nixon RA, Wegiel J, Kumar A, Yu WH, Peterhoff C, Cataldo A, Cuervo AM. Extensive involvement of autophagy in Alzheimer disease: an immuno-electron microscopy study. J Neuropathol Exp Neurol. 2005;64:113–122. doi: 10.1093/jnen/64.2.113. [DOI] [PubMed] [Google Scholar]
- Nixon RA. Autophagy in neurodegenerative disease: friend, foe or turncoat? Trends Neurosci. 2006;29:528–535. doi: 10.1016/j.tins.2006.07.003. [DOI] [PubMed] [Google Scholar]
- NsGene A/S [Retrieved February 15, 2012];NsG33 - Meteorin. 2012 from http://nsgene.dk/NsGene-413.aspx.
- Nucifora FC, Jr., Sasaki M, Peters MF, Huang H, Cooper JK, Yamada M, Takahashi H, Tsuji S, Troncoso J, Dawson VL, Dawson TM, Ross CA. Interference by huntingtin and atrophin-1 with cbp-mediated transcription leading to cellular toxicity. Science. 2001;291:2423–2428. doi: 10.1126/science.1056784. [DOI] [PubMed] [Google Scholar]
- O’Brien T, Fahr BT, Sopko MM, Lam JW, Waal ND, Raimundo BC, Purkey HE, Pham P, Romanowski MJ. Structural analysis of caspase-1 inhibitors derived from Tethering. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2005;61:451–458. doi: 10.1107/S1744309105010109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto S, Pouladi MA, Talantova M, Yao D, Xia P, Ehrnhoefer DE, Zaidi R, Clemente A, Kaul M, Graham RK, Zhang D, Vincent Chen HS, Tong G, Hayden MR, Lipton SA. Balance between synaptic versus extrasynaptic NMDA receptor activity influences inclusions and neurotoxicity of mutant huntingtin. Nat Med. 2009;15:1407–1413. doi: 10.1038/nm.2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Kusky JR, Nasir J, Cicchetti F, Parent A, Hayden MR. Neuronal degeneration in the basal ganglia and loss of pallido-subthalamic synapses in mice with targeted disruption of the Huntington’s disease gene. Brain Res. 1999;818:468–479. doi: 10.1016/s0006-8993(98)01312-2. [DOI] [PubMed] [Google Scholar]
- Ona VO, Li M, Vonsattel JP, Andrews LJ, Khan SQ, Chung WM, Frey AS, Menon AS, Li XJ, Stieg PE, Yuan J, Penney JB, Young AB, Cha JH, Friedlander RM. Inhibition of caspase-1 slows disease progression in a mouse model of Huntington’s disease. Nature. 1999;399:263–267. doi: 10.1038/20446. [DOI] [PubMed] [Google Scholar]
- Oryzon [Retrieved February 15, 2012];Press Release 2012. Oryzon nominates bispecific LSD1/MAOB inhibitor as drug candidate to enter preclinical development in Huntington’s Disease. 2012 from http://www.oryzon.com/files/PRESS_RELEASE_2002-2012.pdf.
- Osler SW. On Chorea and Choreiform Affections. H.K. Lewis; London: 1894. Historical note. Etiology; pp. 3–19. [Google Scholar]
- Overholtzer M, Mailleux AA, Mouneimne G, Normand G, Schnitt SJ, King RW, Cibas ES, Brugge JS. A nonapoptotic cell death process, entosis, that occurs by cell-in-cell invasion. Cell. 2007;131:966–979. doi: 10.1016/j.cell.2007.10.040. [DOI] [PubMed] [Google Scholar]
- Panov AV, Gutekunst CA, Leavitt BR, Hayden MR, Burke JR, Strittmatter WJ, Greenamyre JT. Early mitochondrial calcium defects in Huntington’s disease are a direct effect of polyglutamines. Nat Neurosci. 2002;5:731–736. doi: 10.1038/nn884. [DOI] [PubMed] [Google Scholar]
- Park RH, Park MP. Saint Vitus’ dance: vital misconceptions by Sydenham and Bruegel. J R Soc Med. 1990;83:512–515. doi: 10.1177/014107689008300814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker JA, Metzler M, Georgiou J, Mage M, Roder JC, Rose AM, Hayden MR, Neri C. Huntingtin-interacting protein 1 influences worm and mouse presynaptic function and protects Caenorhabditis elegans neurons against mutant polyglutamine toxicity. J Neurosci. 2007;27:11056–11064. doi: 10.1523/JNEUROSCI.1941-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Navarro E, Akerud P, Marco S, Canals JM, Tolosa E, Arenas E, Alberch J. Neurturin protects striatal projection neurons but not interneurons in a rat model of Huntington’s disease. Neuroscience. 2000;98:89–96. doi: 10.1016/s0306-4522(00)00074-9. [DOI] [PubMed] [Google Scholar]
- Phillips W, Shannon KM, Barker RA. The current clinical management of Huntington’s disease. Mov Disord. 2008;23:1491–1504. doi: 10.1002/mds.21971. [DOI] [PubMed] [Google Scholar]
- Plexxikon, Inc. [Retrieved August 30, 2011];Oncology. Melanoma & BRAF Mutant Cancer Program. 2011 from www.plexxikon.com/file.cfm/13/docs/Plexxikon_Melanoma_v15%203%20final_080211.pdf.
- Powers WJ, Videen TO, Markham J, McGee-Minnich L, Antenor-Dorsey JV, Hershey T, Perlmutter JS. Selective defect of in vivo glycolysis in early Huntington’s disease striatum. Proc Natl Acad Sci U S A. 2007;104:2945–2949. doi: 10.1073/pnas.0609833104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratley RE, Salbe AD, Ravussin E, Caviness JN. Higher sedentary energy expenditure in patients with Huntington’s disease. Ann Neurol. 2000;47:64–70. [PubMed] [Google Scholar]
- Prosensa [Retrieved February 16, 2012];HD-Huntington Disease. 2012 from http://prosensa.eu/patients-and-family/huntington-disease.
- Qin ZH, Wang Y, Sapp E, Cuiffo B, Wanker E, Hayden MR, Kegel KB, Aronin N, DiFiglia M. Huntingtin bodies sequester vesicle-associated proteins by a polyproline-dependent interaction. J Neurosci. 2004;24:269–281. doi: 10.1523/JNEUROSCI.1409-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quarrell OW, Rigby AS, Barron L, Crow Y, Dalton A, Dennis N, Fryer AE, Heydon F, Kinning E, Lashwood A, Losekoot M, Margerison L, McDonnell S, Morrison PJ, Norman A, Peterson M, Raymond FL, Simpson S, Thompson E, Warner J. Reduced penetrance alleles for Huntington’s disease: a multi-centre direct observational study. J Med Genet. 2007;44:e68. doi: 10.1136/jmg.2006.045120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaswamy S, McBride JL, Herzog CD, Brandon E, Gasmi M, Bartus RT, Kordower JH. Neurturin gene therapy improves motor function and prevents death of striatal neurons in a 3-nitropropionic acid rat model of Huntington’s disease. Neurobiol Dis. 2007;26:375–384. doi: 10.1016/j.nbd.2007.01.003. [DOI] [PubMed] [Google Scholar]
- Ramaswamy S, McBride JL, Han I, Berry-Kravis EM, Zhou L, Herzog CD, Gasmi M, Bartus RT, Kordower JH. Intrastriatal CERE-120 (AAV-Neurturin) protects striatal and cortical neurons and delays motor deficits in a transgenic mouse model of Huntington’s disease. Neurobiol Dis. 2009;34:40–50. doi: 10.1016/j.nbd.2008.12.005. [DOI] [PubMed] [Google Scholar]
- Ravikumar B, Duden R, Rubinsztein DC. Aggregate-prone proteins with polyglutamine and polyalanine expansions are degraded by autophagy. Hum Mol Genet. 2002;11:1107–1117. doi: 10.1093/hmg/11.9.1107. [DOI] [PubMed] [Google Scholar]
- Ravikumar B, Vacher C, Berger Z, Davies JE, Luo S, Oroz LG, Scaravilli F, Easton DF, Duden R, O’Kane CJ, Rubinsztein DC. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat Genet. 2004;36:585–595. doi: 10.1038/ng1362. [DOI] [PubMed] [Google Scholar]
- Raymond LA. Excitotoxicity in Huntington disease. Clinical Neuroscience Research. 2003;3:121–128. [Google Scholar]
- Rees DC, Congreve M, Murray CW, Carr R. Fragment-based lead discovery. Nat Rev Drug Discov. 2004;3:660–672. doi: 10.1038/nrd1467. [DOI] [PubMed] [Google Scholar]
- ReNeuron ReNeuron presents further pre-clinical stem cell data at US conference; 27/10/04; [Retrieved February 5, 2012]. 2011. from http://www.reneuron.co.uk/monthly-archive/91-reneuron-presents-further-pre-clinical-stem-cell-data-at-us-conference-271004. [Google Scholar]
- Rigamonti D, Bauer JH, De-Fraja C, Conti L, Sipione S, Sciorati C, Clementi E, Hackam A, Hayden MR, Li Y, Cooper JK, Ross CA, Govoni S, Vincenz C, Cattaneo E. Wild-type huntingtin protects from apoptosis upstream of caspase-3. J Neurosci. 2000;20:3705–3713. doi: 10.1523/JNEUROSCI.20-10-03705.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross CA, Tabrizi SJ. Huntington’s disease: from molecular pathogenesis to clinical treatment. Lancet Neurol. 2011;10:83–98. doi: 10.1016/S1474-4422(10)70245-3. [DOI] [PubMed] [Google Scholar]
- Rothstein JD, Patel S, Regan MR, Haenggeli C, Huang YH, Bergles DE, Jin L, Dykes Hoberg M, Vidensky S, Chung DS, Toan SV, Bruijn LI, Su ZZ, Gupta P, Fisher PB. Beta-lactam antibiotics offer neuroprotection by increasing glutamate transporter expression. Nature. 2005;433:73–77. doi: 10.1038/nature03180. [DOI] [PubMed] [Google Scholar]
- Rubinsztein DC, Leggo J, Coles R, Almqvist E, Biancalana V, Cassiman JJ, Chotai K, Connarty M, Crauford D, Curtis A, Curtis D, Davidson MJ, Differ AM, Dode C, Dodge A, Frontali M, Ranen NG, Stine OC, Sherr M, Abbott MH, Franz ML, Graham CA, Harper PS, Hedreen JC, Hayden MR, et al. Phenotypic characterization of individuals with 30-40 CAG repeats in the Huntington disease (HD) gene reveals HD cases with 36 repeats and apparently normal elderly individuals with 36-39 repeats. Am J Hum Genet. 1996;59:16–22. [PMC free article] [PubMed] [Google Scholar]
- Ruchaud S, Korfali N, Villa P, Kottke TJ, Dingwall C, Kaufmann SH, Earnshaw WC. Caspase-6 gene disruption reveals a requirement for lamin A cleavage in apoptotic chromatin condensation. EMBO J. 2002;21:1967–1977. doi: 10.1093/emboj/21.8.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruskin DN, Ross JL, Kawamura M, Jr., Ruiz TL, Geiger JD, Masino SA. A ketogenic diet delays weight loss and does not impair working memory or motor function in the R6/2 1J mouse model of Huntington’s disease. Physiol Behav. 2011;103:501–507. doi: 10.1016/j.physbeh.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saft C, Zange J, Andrich J, Muller K, Lindenberg K, Landwehrmeyer B, Vorgerd M, Kraus PH, Przuntek H, Schols L. Mitochondrial impairment in patients and asymptomatic mutation carriers of Huntington’s disease. Mov Disord. 2005;20:674–679. doi: 10.1002/mds.20373. [DOI] [PubMed] [Google Scholar]
- Sanchez I, Xu CJ, Juo P, Kakizaka A, Blenis J, Yuan J. Caspase-8 is required for cell death induced by expanded polyglutamine repeats. Neuron. 1999;22:623–633. doi: 10.1016/s0896-6273(00)80716-3. [DOI] [PubMed] [Google Scholar]
- Sari Y, Prieto AL, Barton SJ, Miller BR, Rebec GV. Ceftriaxone-induced up-regulation of cortical and striatal GLT1 in the R6/2 model of Huntington’s disease. J Biomed Sci. 2010;17:62. doi: 10.1186/1423-0127-17-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saudou F, Finkbeiner S, Devys D, Greenberg ME. Huntingtin acts in the nucleus to induce apoptosis but death does not correlate with the formation of intranuclear inclusions. Cell. 1998;95:55–66. doi: 10.1016/s0092-8674(00)81782-1. [DOI] [PubMed] [Google Scholar]
- Saydoff JA, Liu LS, Garcia RA, Hu Z, Li D, von Borstel RW. Oral uridine pro-drug PN401 decreases neurodegeneration, behavioral impairment, weight loss and mortality in the 3-nitropropionic acid mitochondrial toxin model of Huntington’s disease. Brain Res. 2003;994:44–54. doi: 10.1016/j.brainres.2003.09.049. [DOI] [PubMed] [Google Scholar]
- Saydoff JA, Garcia RA, Browne SE, Liu L, Sheng J, Brenneman D, Hu Z, Cardin S, Gonzalez A, von Borstel RW, Gregorio J, Burr H, Beal MF. Oral uridine pro-drug PN401 is neuroprotective in the R6/2 and N171-82Q mouse models of Huntington’s disease. Neurobiol Dis. 2006;24:455–465. doi: 10.1016/j.nbd.2006.08.011. [DOI] [PubMed] [Google Scholar]
- Scarpulla RC. Transcriptional paradigms in mammalian mitochondrial biogenesis and function. Physiol Rev. 2008;88:611–638. doi: 10.1152/physrev.00025.2007. [DOI] [PubMed] [Google Scholar]
- Schols L, Bauer P, Schmidt T, Schulte T, Riess O. Autosomal dominant cerebellar ataxias: clinical features, genetics, and pathogenesis. Lancet Neurol. 2004;3:291–304. doi: 10.1016/S1474-4422(04)00737-9. [DOI] [PubMed] [Google Scholar]
- Schuffenhauer A, Ruedisser S, Marzinzik AL, Jahnke W, Blommers M, Selzer P, Jacoby E. Library design for fragment based screening. Curr Top Med Chem. 2005;5:751–762. doi: 10.2174/1568026054637700. [DOI] [PubMed] [Google Scholar]
- Scrimgeour EM. Huntington’s disease in Tanzania. J Med Genet. 1981;18:200–203. doi: 10.1136/jmg.18.3.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuker SB, Hajduk PJ, Meadows RP, Fesik SW. Discovering high-affinity ligands for proteins: SAR by NMR. Science. 1996;274:1531–1534. doi: 10.1126/science.274.5292.1531. [DOI] [PubMed] [Google Scholar]
- Simmons DA, Rex CS, Palmer L, Pandyarajan V, Fedulov V, Gall CM, Lynch G. Up-regulating BDNF with an ampakine rescues synaptic plasticity and memory in Huntington’s disease knockin mice. Proc Natl Acad Sci U S A. 2009;106:4906–4911. doi: 10.1073/pnas.0811228106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons DA, Mehta RA, Lauterborn JC, Gall CM, Lynch G. Brief ampakine treatments slow the progression of Huntington’s disease phenotypes in R6/2 mice. Neurobiology of Disease. 2011;41:436–444. doi: 10.1016/j.nbd.2010.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singaraja RR, Hadano S, Metzler M, Givan S, Wellington CL, Warby S, Yanai A, Gutekunst CA, Leavitt BR, Yi H, Fichter K, Gan L, McCutcheon K, Chopra V, Michel J, Hersch SM, Ikeda JE, Hayden MR. HIP14, a novel ankyrin domain-containing protein, links huntingtin to intracellular trafficking and endocytosis. Hum Mol Genet. 2002;11:2815–2828. doi: 10.1093/hmg/11.23.2815. [DOI] [PubMed] [Google Scholar]
- Sommer DB, Stacy MA. What’s in the pipeline for the treatment of Parkinson’s disease? Expert Rev Neurother. 2008;8:1829–1839. doi: 10.1586/14737175.8.12.1829. [DOI] [PubMed] [Google Scholar]
- Song W, Chen J, Petrilli A, Liot G, Klinglmayr E, Zhou Y, Poquiz P, Tjong J, Pouladi MA, Hayden MR, Masliah E, Ellisman M, Rouiller I, Schwarzenbacher R, Bossy B, Perkins G, Bossy-Wetzel E. Mutant huntingtin binds the mitochondrial fission GTPase dynamin-related protein-1 and increases its enzymatic activity. Nat Med. 2011;17:377–382. doi: 10.1038/nm.2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorensen SA, Fenger K. Causes of death in patients with Huntington’s disease and in unaffected first degree relatives. J Med Genet. 1992;29:911–914. doi: 10.1136/jmg.29.12.911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperandio S, de Belle I, Bredesen DE. An alternative, nonapoptotic form of programmed cell death. Proc Natl Acad Sci U S A. 2000;97:14376–14381. doi: 10.1073/pnas.97.26.14376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiles DK, Zhang Z, Ge P, Nelson B, Grondin R, Ai Y, Hardy P, Nelson PT, Guzaev AP, Butt MT, Charisse K, Kosovrasti V, Tchangov L, Meys M, Maier M, Nechev L, Manoharan M, Kaemmerer WF, Gwost D, Stewart GR, Gash DM, Sah DWY. Widespread suppression of huntingtin with convection-enhanced delivery of siRNA. Experimental Neurology. 2011;233:463–471. doi: 10.1016/j.expneurol.2011.11.020. [DOI] [PubMed] [Google Scholar]
- Takahashi A, Alnemri ES, Lazebnik YA, Fernandes-Alnemri T, Litwack G, Moir RD, Goldman RD, Poirier GG, Kaufmann SH, Earnshaw WC. Cleavage of lamin A by Mch2 alpha but not CPP32: multiple interleukin 1 beta-converting enzyme-related proteases with distinct substrate recognition properties are active in apoptosis. Proc Natl Acad Sci U S A. 1996;93:8395–8400. doi: 10.1073/pnas.93.16.8395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano H, Gusella JF. The predominantly HEAT-like motif structure of huntingtin and its association and coincident nuclear entry with dorsal, an NF-kB/Rel/dorsal family transcription factor. BMC Neurosci. 2002;3:15. doi: 10.1186/1471-2202-3-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang TS, Slow E, Lupu V, Stavrovskaya IG, Sugimori M, Llinas R, Kristal BS, Hayden MR, Bezprozvanny I. Disturbed Ca2+ signaling and apoptosis of medium spiny neurons in Huntington’s disease. Proc Natl Acad Sci U S A. 2005;102:2602–2607. doi: 10.1073/pnas.0409402102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tardiff DF, Tucci ML, Caldwell KA, Caldwell GA, Lindquist S. Different 8-Hydroxyquinolines Protect Models of TDP-43 Protein, alpha-Synuclein, and Polyglutamine Proteotoxicity through Distinct Mechanisms. J Biol Chem. 2012;287:4107–4120. doi: 10.1074/jbc.M111.308668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telenius H, Kremer H, Thellmann J, Andrew S, Almqvist E, Anvret M, Greenberg C, Greenberg J, Lucotte G, Squltierl F. Molecular analysis of juvenile Huntington disease: the major influence on (CAG) n repeat length is the sex of the affected parent. Human molecular genetics. 1993;2:1535–1540. doi: 10.1093/hmg/2.10.1535. [DOI] [PubMed] [Google Scholar]
- The Huntington’s Disease Collaborative Research Group A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- Thornberry NA, Bull HG, Calaycay JR, Chapman KT, Howard AD, Kostura MJ, Miller DK, Molineaux SM, Weidner JR, Aunins J, et al. A novel heterodimeric cysteine protease is required for interleukin-1 beta processing in monocytes. Nature. 1992;356:768–774. doi: 10.1038/356768a0. [DOI] [PubMed] [Google Scholar]
- Thornberry NA, Rano TA, Peterson EP, Rasper DM, Timkey T, Garcia-Calvo M, Houtzager VM, Nordstrom PA, Roy S, Vaillancourt JP, Chapman KT, Nicholson DW. A combinatorial approach defines specificities of members of the caspase family and granzyme B. Functional relationships established for key mediators of apoptosis. J Biol Chem. 1997;272:17907–17911. doi: 10.1074/jbc.272.29.17907. [DOI] [PubMed] [Google Scholar]
- Thrall JH. Prevalence and costs of chronic disease in a health care system structured for treatment of acute illness. Radiology. 2005;235:9–12. doi: 10.1148/radiol.2351041768. [DOI] [PubMed] [Google Scholar]
- Toulmond S, Tang K, Bureau Y, Ashdown H, Degen S, O’Donnell R, Tam J, Han Y, Colucci J, Giroux A, Zhu Y, Boucher M, Pikounis B, Xanthoudakis S, Roy S, Rigby M, Zamboni R, Robertson GS, Ng GY, Nicholson DW, Fluckiger JP. Neuroprotective effects of M826, a reversible caspase-3 inhibitor, in the rat malonate model of Huntington’s disease. Br J Pharmacol. 2004;141:689–697. doi: 10.1038/sj.bjp.0705662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai J, Lee JT, Wang W, Zhang J, Cho H, Mamo S, Bremer R, Gillette S, Kong J, Haass NK, Sproesser K, Li L, Smalley KS, Fong D, Zhu YL, Marimuthu A, Nguyen H, Lam B, Liu J, Cheung I, Rice J, Suzuki Y, Luu C, Settachatgul C, Shellooe R, Cantwell J, Kim SH, Schlessinger J, Zhang KY, West BL, Powell B, Habets G, Zhang C, Ibrahim PN, Hirth P, Artis DR, Herlyn M, Bollag G. Discovery of a selective inhibitor of oncogenic B-Raf kinase with potent antimelanoma activity. Proc Natl Acad Sci U S A. 2008;105:3041–3046. doi: 10.1073/pnas.0711741105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenabeele P, Galluzzi L, Vanden Berghe T, Kroemer G. Molecular mechanisms of necroptosis: an ordered cellular explosion. Nat Rev Mol Cell Biol. 2010;11:700–714. doi: 10.1038/nrm2970. [DOI] [PubMed] [Google Scholar]
- Vertex Pharmaceuticals, Inc. [Retrieved June 3, 2011];VX-765 (Epilepsy) 2010 from www.vrtx.com/current-projects/drug-candidates/pipeline.html.
- Viola HM, Hool LC. Cross-talk between L-type Ca2+ channels and mitochondria. Clin Exp Pharmacol Physiol. 2010;37:229–235. doi: 10.1111/j.1440-1681.2009.05277.x. [DOI] [PubMed] [Google Scholar]
- Vonsattel JP, DiFiglia M. Huntington disease. J Neuropathol Exp Neurol. 1998;57:369–384. doi: 10.1097/00005072-199805000-00001. [DOI] [PubMed] [Google Scholar]
- Waelter S, Scherzinger E, Hasenbank R, Nordhoff E, Lurz R, Goehler H, Gauss C, Sathasivam K, Bates GP, Lehrach H, Wanker EE. The huntingtin interacting protein HIP1 is a clathrin and alpha-adaptin-binding protein involved in receptor-mediated endocytosis. Hum Mol Genet. 2001;10:1807–1817. doi: 10.1093/hmg/10.17.1807. [DOI] [PubMed] [Google Scholar]
- Walker FO. Huntington’s disease. Lancet. 2007;369:218–228. doi: 10.1016/S0140-6736(07)60111-1. [DOI] [PubMed] [Google Scholar]
- Waller J. A forgotten plague: making sense of dancing mania. Lancet. 2009;373:624–625. doi: 10.1016/s0140-6736(09)60386-x. [DOI] [PubMed] [Google Scholar]
- Wang H, Lim PJ, Karbowski M, Monteiro MJ. Effects of overexpression of huntingtin proteins on mitochondrial integrity. Hum Mol Genet. 2009;18:737–752. doi: 10.1093/hmg/ddn404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Chen X, Li Y, Tang TS, Bezprozvanny I. Tetrabenazine is neuroprotective in Huntington’s disease mice. Mol Neurodegener. 2010;5:18. doi: 10.1186/1750-1326-5-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Zhu S, Drozda M, Zhang W, Stavrovskaya IG, Cattaneo E, Ferrante RJ, Kristal BS, Friedlander RM. Minocycline inhibits caspase-independent and - dependent mitochondrial cell death pathways in models of Huntington’s disease. Proc Natl Acad Sci U S A. 2003;100:10483–10487. doi: 10.1073/pnas.1832501100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Wang H, Figueroa BE, Zhang WH, Huo C, Guan Y, Zhang Y, Bruey JM, Reed JC, Friedlander RM. Dysregulation of receptor interacting protein-2 and caspase recruitment domain only protein mediates aberrant caspase-1 activation in Huntington’s disease. J Neurosci. 2005;25:11645–11654. doi: 10.1523/JNEUROSCI.4181-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanker EE, Rovira C, Scherzinger E, Hasenbank R, Walter S, Tait D, Colicelli J, Lehrach H. HIP-I: a huntingtin interacting protein isolated by the yeast two-hybrid system. Hum Mol Genet. 1997;6:487–495. doi: 10.1093/hmg/6.3.487. [DOI] [PubMed] [Google Scholar]
- Warby SC, Montpetit A, Hayden AR, Carroll JB, Butland SL, Visscher H, Collins JA, Semaka A, Hudson TJ, Hayden MR. CAG expansion in the Huntington disease gene is associated with a specific and targetable predisposing haplogroup. Am J Hum Genet. 2009;84:351–366. doi: 10.1016/j.ajhg.2009.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warby SC, Visscher H, Collins JA, Doty CN, Carter C, Butland SL, Hayden AR, Kanazawa I, Ross CJ, Hayden MR. HTT haplotypes contribute to differences in Huntington disease prevalence between Europe and East Asia. Eur J Hum Genet. 2011;19:561–566. doi: 10.1038/ejhg.2010.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wartha F, Henriques-Normark B. ETosis: a novel cell death pathway. Sci Signal. 2008;1:pe25. doi: 10.1126/stke.121pe25. [DOI] [PubMed] [Google Scholar]
- Wellington CL, Singaraja R, Ellerby L, Savill J, Roy S, Leavitt B, Cattaneo E, Hackam A, Sharp A, Thornberry N, Nicholson DW, Bredesen DE, Hayden MR. Inhibiting caspase cleavage of huntingtin reduces toxicity and aggregate formation in neuronal and nonneuronal cells. J Biol Chem. 2000;275:19831–19838. doi: 10.1074/jbc.M001475200. [DOI] [PubMed] [Google Scholar]
- Wilbur JD, Chen CY, Manalo V, Hwang PK, Fletterick RJ, Brodsky FM. Actin binding by Hip1 (huntingtin-interacting protein 1) and Hip1R (Hip1-related protein) is regulated by clathrin light chain. J Biol Chem. 2008;283:32870–32879. doi: 10.1074/jbc.M802863200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wild EJ, Tabrizi SJ. The differential diagnosis of chorea. Practical Neurology. 2007;7:360–373. doi: 10.1136/pn.2007.134585. [DOI] [PubMed] [Google Scholar]
- Wilkinson B, Gilbert HF. Protein disulfide isomerase. Biochim Biophys Acta. 2004;1699:35–44. doi: 10.1016/j.bbapap.2004.02.017. [DOI] [PubMed] [Google Scholar]
- Wu HQ, Lee SC, Schwarcz R. Systemic administration of 4-chlorokynurenine prevents quinolinate neurotoxicity in the rat hippocampus. Eur J Pharmacol. 2000;390:267–274. doi: 10.1016/s0014-2999(00)00024-8. [DOI] [PubMed] [Google Scholar]
- Yorimitsu T, Klionsky DJ. Autophagy: molecular machinery for self-eating. Cell Death Differ. 2005;12(Suppl 2):1542–1552. doi: 10.1038/sj.cdd.4401765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Zhu H, Ko D, Kindy MS. Motoneuronotrophic factor analog GM6 reduces infarct volume and behavioral deficits following transient ischemia in the mouse. Brain Research. 2008;1238:143–153. doi: 10.1016/j.brainres.2008.08.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeitlin S, Liu JP, Chapman DL, Papaioannou VE, Efstratiadis A. Increased apoptosis and early embryonic lethality in mice nullizygous for the Huntington’s disease gene homologue. Nat Genet. 1995;11:155–163. doi: 10.1038/ng1095-155. [DOI] [PubMed] [Google Scholar]
- Zhai W, Jeong H, Cui L, Krainc D, Tjian R. In vitro analysis of huntingtin-mediated transcriptional repression reveals multiple transcription factor targets. Cell. 2005;123:1241–1253. doi: 10.1016/j.cell.2005.10.030. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Leavitt BR, van Raamsdonk JM, Dragatsis I, Goldowitz D, MacDonald ME, Hayden MR, Friedlander RM. Huntingtin inhibits caspase-3 activation. EMBO J. 2006;25:5896–5906. doi: 10.1038/sj.emboj.7601445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H, Gal S, Weiner LM, Bar-Am O, Warshawsky A, Fridkin M, Youdim MB. Novel multifunctional neuroprotective iron chelator-monoamine oxidase inhibitor drugs for neurodegenerative diseases: in vitro studies on antioxidant activity, prevention of lipid peroxide formation and monoamine oxidase inhibition. J Neurochem. 2005;95:68–78. doi: 10.1111/j.1471-4159.2005.03340.x. [DOI] [PubMed] [Google Scholar]
- Zuccato C, Tartari M, Crotti A, Goffredo D, Valenza M, Conti L, Cataudella T, Leavitt BR, Hayden MR, Timmusk T, Rigamonti D, Cattaneo E. Huntingtin interacts with REST/NRSF to modulate the transcription of NRSE-controlled neuronal genes. Nat Genet. 2003;35:76–83. doi: 10.1038/ng1219. [DOI] [PubMed] [Google Scholar]
- Zuccato C, Valenza M, Cattaneo E. Molecular mechanisms and potential therapeutical targets in Huntington’s disease. Physiol Rev. 2010;90:905–981. doi: 10.1152/physrev.00041.2009. [DOI] [PubMed] [Google Scholar]
- Zuhlke C, Riess O, Bockel B, Lange H, Thies U. Mitotic stability and meiotic variability of the (CAG)n repeat in the Huntington disease gene. Hum Mol Genet. 1993;2:2063–2067. doi: 10.1093/hmg/2.12.2063. [DOI] [PubMed] [Google Scholar]