ABSTRACT
Staphylococcus aureus is a human commensal that at times turns into a serious bacterial pathogen causing life-threatening infections. For the delicate control of virulence, S. aureus employs the agr quorum-sensing system that, via the intracellular effector molecule RNAIII, regulates virulence gene expression. We demonstrate that the presence of the agr locus imposes a fitness cost on S. aureus that is mediated by the expression of RNAIII. Further, we show that exposure to sublethal levels of the antibiotics ciprofloxacin, mupirocin, and rifampin, each targeting separate cellular functions, markedly increases the agr-mediated fitness cost by inducing the expression of RNAIII. Thus, the extensive use of antibiotics in hospitals may explain why agr-negative variants are frequently isolated from hospital-acquired S. aureus infections but rarely found among community-acquired S. aureus strains. Importantly, agr deficiency correlates with increased duration of and mortality due to bacteremia during antibiotic treatment and with a higher frequency of glycopeptide resistance than in agr-carrying strains. Our results provide an explanation for the frequent isolation of agr-defective strains from hospital-acquired S. aureus infections and suggest that the adaptability of S. aureus to antibiotics involves the agr locus.
IMPORTANCE
Staphylococcus aureus is the most frequently isolated pathogen in intensive care units and a common cause of nosocomial infections, resulting in a high degree of morbidity and mortality. Surprisingly, a large fraction (15 to 60%) of hospital-isolated S. aureus strains are agr defective and lack the main quorum-sensing-controlled virulence regulatory system. This is a problem, as agr-defective strains are associated with a mortality level in bacteremic infections and a probability of glycopeptide resistance greater than those of other strains. We show here that agr-negative strains have a fitness advantage over agr-positive strains in the presence of sublethal concentrations of some antibiotics and that the fitness defect of agr-positive cells is caused by antibiotic-mediated expression of the agr effector molecule RNAIII. These results offer an explanation of the frequent isolation of agr-defective S. aureus strains in hospitals and will influence how we treat S. aureus infections.
Introduction
In growing bacterial populations, even small changes in fitness are rapidly manifested in subpopulations with different growth rates (1). A classic example is resistance to streptomycin. In the presence of the antibiotic, resistant cells have an enormous selective advantage, whereas in its absence, the resistance imposes a fitness cost that results in a large reduction in the growth rate compared to that of sensitive cells (2, 3). Similarly for bacterial pathogens, virulence factor expression may be disadvantageous outside a host but needed for infection, as in the case of the Salmonella enterica serovar Typhimurium type III secretion system (4). In Pseudomonas aeruginosa, it is less obvious what confers the fitness difference than for the LasR quorum-sensing (QS) system, where virulence factor expression is selected against both in vitro and in vivo (5). Thus, what confers maximized fitness under one set of conditions may be counterselected under different environmental conditions (6, 7, 4) and the exact components providing the selective pressure are often not known.
QS allows for a coordinated response to cell density and environmental changes and is commonly employed by bacteria to control virulence gene expression (8, 9). A particularly well-studied QS system is encoded by the agr (accessory gene regulator) locus in the human pathogen Staphylococcus aureus (10). The signal molecule of agr is a posttranslationally modified peptide termed the autoinducing peptide (AIP) that is formed and excreted by the combined activity of AgrB and ArgD. At high concentrations, the signal is perceived by a classical two-component signal transduction system composed of the membrane-bound histidine kinase AgrC and the response regulator AgrA, both of which are encoded by the agr locus. Upon the binding of AIPs, AgrC activates AgrA by His-dependent phosphorylation. AgrA, in turn, induces the expression of a stable RNA, RNAIII, as well as that of the RNAII transcript containing agrA, agrB, agrC, and agrD, resulting in a feedback loop (11, 12). RNAIII is the key intracellular effector molecule of agr, and as its concentration increases with cell density, it induces the expression of extracellular virulence factors while repressing the expression of cell wall-associated proteins. Independently of RNAIII, AgrA directly controls the expression of α and β phenol-soluble modulins and, via an unknown mechanism, participates in the downregulation of genes involved in carbohydrate and amino acid metabolism (13). Together, AgrA and RNAIII interconnect metabolism and virulence gene expression in response to cell density (14, 13).
The central role of agr in S. aureus virulence has been verified in a large number of in vivo models, including septic arthritis (15), skin abscesses (16, 17), osteomyelitis (18), and endocarditis (19), in which agr-defective strains display less virulence than wild-type (WT) strains. The agr locus is functional in essentially all community-acquired S. aureus strains, and the locus is considered important for the high virulence of these strains (20), as well as for their transmission between hosts (21). Also, subinhibitory concentrations of antibiotics are known to modulate virulence gene expression in S. aureus in a process likely involving agr (22). In contrast, agr-negative isolates frequently arise in hospital infections (23). Here, it has been estimated that 15 to 60% of S. aureus-associated infections display agr dysfunctions, whereas carriage of agr-negative strains by healthy individuals is unusual outside hospital settings (prevalence, ~4%) and is associated with previous hospital exposure (21, 24–28). Even though virulence gene expression is compromised in agr-deficient isolates, they still give rise to concern. Clinical studies indicate that agr-defective variants have reduced susceptibility to thrombin-induced platelet microbicidal proteins and are linked with an increased duration of and mortality due to S. aureus bacteremia (24, 29–31). In terms of resistance to antimicrobials, agr-negative strains are known to display intermediate resistance or heteroresistance to glycopeptides such as vancomycin (glycopeptide intermediate-level resistant S. aureus [GISA] and hetero-GISA) (32) and a laboratory-generated agr-negative strain demonstrated a small but reproducible increase in vancomycin heteroresistance (32).
The observation that agr-deficient strains frequently arise in the hospital environment, where the antibiotic pressure is expected to be high, led us to investigate (i) if there is a fitness cost associated with agr and (ii) if this effect is enhanced during growth in the presence of antibiotics. Growth competition experiments demonstrated that agr-negative strains displayed greater fitness than the isogenic WT strain in the presence of the antibiotics ciprofloxacin, mupirocin, and rifampin but not vancomycin. The fitness cost of carrying an intact agr operon in the presence of antibiotics was correlated with the ability to induce RNAIII expression. The study described here possibly explains the frequent isolation of agr-defective strains in clinical settings and identifies antibiotics as a factor that modulates the competition between QS-positive and -negative cells.
RESULTS
Fitness cost of agr expression.
To assess if there is an impact on fitness associated with agr, we compared the growth of WT S. aureus Newman to that of a Newman ΔagrA mutant strain that does not produce any detectable amounts of RNAIII, as determined by quantitative PCR (qPCR). Fitness was assessed by using three growth parameters, namely, the exponential growth rate, the CFU count at stationary phase, and the outcome of competition between the two strains when inoculated at a 1:1 ratio and grown for ~8 cell divisions. The competition assay showed that the ΔagrA mutant strain exhibited a fitness advantage over the WT strain, with a relative competitive fitness of 1.07 determined as previously described (33, 34) (Fig. 1, TSB [tryptic soy broth]). However, when cultured individually, the WT and ΔagrA mutant strains multiplied with identical growth rates in exponential phase (OD [optical density], 0.02 to 0.08) and reached the same final cell density, as measured by CFU counting at stationary phase (see Fig. S1 in the supplemental material). The difference in fitness between the two strains when grown in competition was observed in late exponential phase/early stationary phase and continued until stationary phase (Fig. 2A). Thus, the reduced fitness of the WT compared with that of the ΔagrA mutant may be related to the induction of agr at this growth stage (Fig. 2B).
FIG 1 .
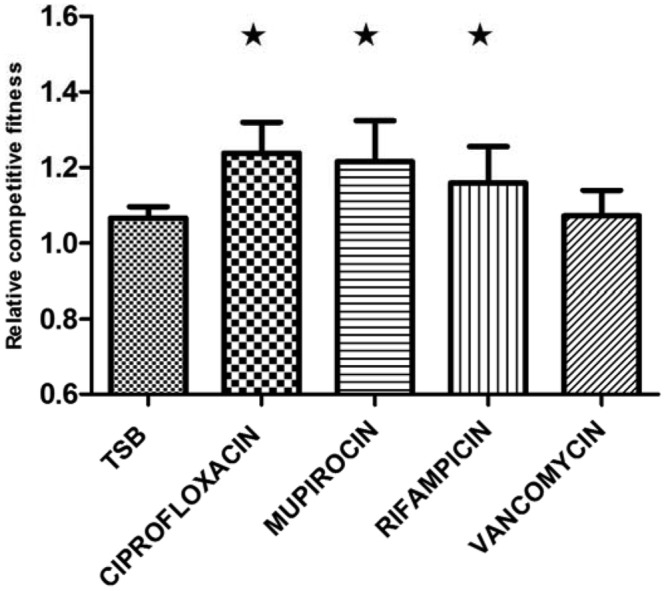
agr and antibiotics influence fitness. Relative competitive fitness of the ΔagrA mutant compared to that of the WT grown in the absence or presence of antibiotics after 48 h. The relative competitive fitness of the WT is 1. A star indicates a significant difference (P < 0.05) from the TSB control. Means with error bars indicating 95% confidence intervals are presented.
FIG 2 .
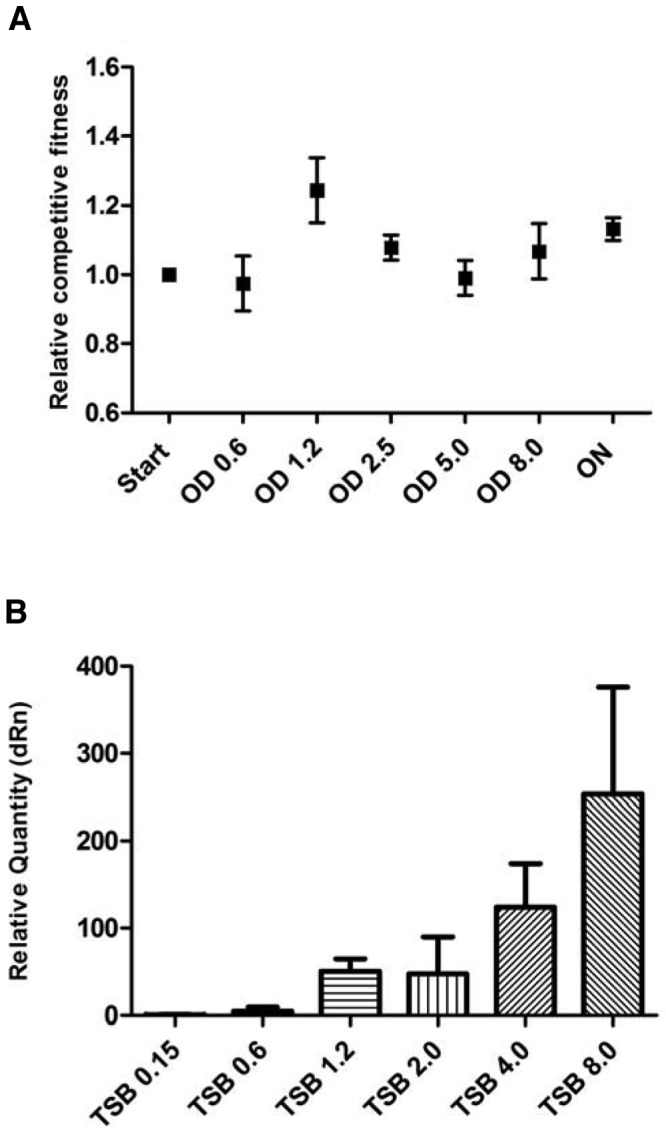
Effect of agr status on the competition ratio at various cell ODs. (A) Competitive fitness of the ΔagrA mutant relative to that of the WT grown in TSB as a function of OD with the last sampling point after 24 h of incubation. (B) RNAIII expression levels of WT cells in TSB normalized to the expression level of RNAIII at an OD of 0.15. Means with error bars indicating 95% confidence intervals are presented. ON, 24 hours; dRn, baseline subtracted fluorescent reading normalized to the reference dye.
agr imposes a fitness cost during antibiotic treatment.
To determine if the presence of antibiotics affects the cost of carrying an intact agr operon, we conducted competition assays (~8 cell divisions) of the WT and ΔagrA mutant strains in TSB growth medium supplemented with 1.0 µg/ml ciprofloxacin, 0.2 µg/ml mupirocin, 0.02 µg/ml rifampin, or 1.0 µg/ml vancomycin. These antibiotics were chosen to be clinically relevant and to target various cellular functions and were supplied at concentrations near their MICs in order to mimic a failed, sublethal antibiotic treatment (MICs are shown in Table S1 in the supplemental material).
In the growth medium supplemented with ciprofloxacin, mupirocin, or rifampin, the ΔagrA mutant strain exhibited a significantly higher relative competitive fitness than the WT strain, with values of 1.24 (P value of 0.0129) in ciprofloxacin, 1.21 (P value of 0.0018) in mupirocin, and 1.16 (P value of 0.05) in rifampin (Fig. 1). In the presence of vancomycin, however, no significant fitness increase was observed in the ΔagrA mutant strain, with a relative competitive fitness of 1.07 (P value of 0.65) (Fig. 1). Again, the higher relative competitive fitness of the ΔagrA mutant could not be attributed to differences in the exponential growth rates or final yields (endpoint CFU counts) of the individual strains (see Fig. S1 in the supplemental material). Thus, the observed fitness advantage of the ΔagrA mutant strain in competition with the WT is substantially enhanced by the presence of certain antibiotics.
RNAIII is responsible for the fitness cost imposed by agr.
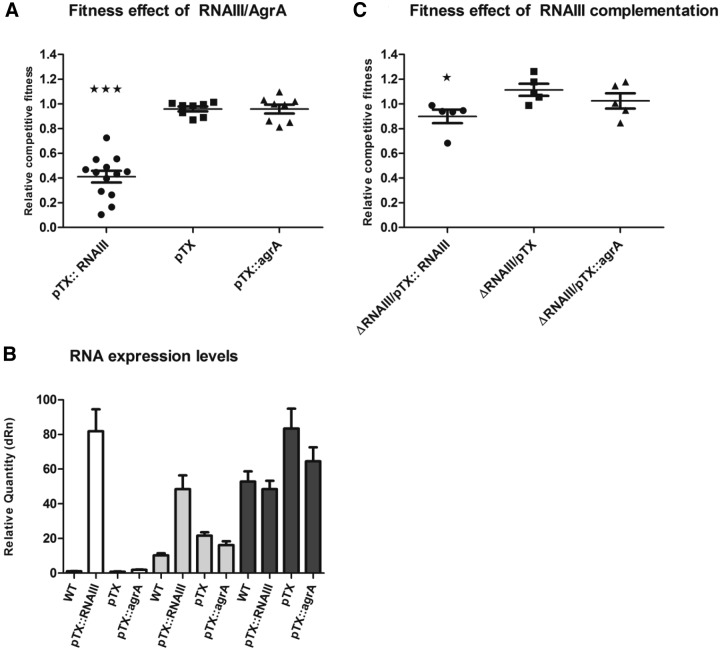
The agr QS regulon is composed of (i) the RNAIII-controlled virulence factors and (ii) the AgrA-regulated genes (13). In order to determine which of these regulatory pathways is responsible for the agr-mediated decrease in fitness, we assayed the relative fitness in TSB without antibiotics of WT cells and a ΔRNAIII mutant constitutively overexpressing either the AgrA response regulator or RNAIII. The fitness assay was designed with pairwise competitions between the plasmid-lacking WT and strains constitutively overexpressing either agrA (pTX::agrA) or RNAIII (pTX::RNAIII) or carrying an empty expression vector as a control. The competition assay showed that overexpression of agrA did not alter fitness (0.96; P value of 0.98) relative to that of the control strain with the empty pTX vector. In contrast, a large fitness cost was observed when RNAIII was overexpressed, decreasing the relative competitive fitness to 0.40 (P value of <0.0001), in contrast to the 0.96 relative competitive fitness of the strain carrying the pTX plasmid (Fig. 3A).
FIG 3 .
Overexpression of RNAIII reduces fitness. (A) Relative competitive fitness in TSB of the WT strain carrying the pTX vector expressing RNAIII or agrA or without an insert. A relative competitive fitness level of <1 is a result of decreased fitness of pTX-related gene expression. Three stars indicate a significant difference (P < 0.001) from the WT strain carrying the pTX vector without an insert. Means with error bars indicating 95% confidence intervals are shown. (B) RNAIII expression levels in TSB of WT cells carrying the pTX vector, pTX::RNAIII, or pTX::agrA and WT cells not carrying the pTX vector quantified at ODs of 0.15 (white), 0.6 (gray), and 1.2 (black). The relative expression level of RNAIII is set to the WT strain’s RNAIII expression levels at an OD of 0.15. Means with standard deviations are shown. (C) Relative competitive fitness in TSB of ΔRNAIII mutant strains carrying the pTX vector expressing RNAIII or agrA or without an insert. Means with error bars indicating 95% confidence intervals are shown. A star indicates a significant difference (P < 0.05) from the ΔRNAIII mutant strain carrying the pTX vector without an insert. dRn, baseline subtracted fluorescent reading normalized to the reference dye.
In order to confirm that RNAIII expression is increased in cells carrying pTX::RNAIII but not in cells carrying pTX::agrA, we determined the RNAIII levels by reverse transcription (RT)-qPCR at three separate ODs, 0.15, 0.6, and 1.2. At two out of three OD sample points (0.15 and 0.6), we observed an increase in the RNAIII levels of the pTX::RNAIII-carrying strain over that of cells carrying the pTX::agrA or pTX plasmid (Fig. 3B). In order to corroborate the observed fitness cost due to increased RNAIII levels in the WT strain, we conducted competitions between the WT lacking the pTX plasmid and the ΔRNAIII mutant strain (no RNAIII expression) constitutively overexpressing agrA (pTX::agrA) or RNAIII (pTX::RNAIII) or carrying the vector (pTX) (Fig. 3C). Overexpression of RNAIII in the ΔRNAIII background caused a large decrease in fitness (0.89; P value of <0.018) relative to that of the ΔRNAIII mutant strain carrying the empty pTX vector (Fig. 3C). Compared to the WT strain carrying the pTX vector (Fig. 3A) the ΔRNAIII mutant (pTX::RNAIII) strain exhibited a significant increase in fitness (1.11; P value of <0.005) (Fig. 3C). Overexpression of agrA in the ΔRNAIII background did not significantly alter fitness (1.02, P value of 0.34) relative to that seen with the overexpression of agrA in the WT background (fitness, 0.96) (Fig. 3A). On the basis of these observations, we conclude that RNAIII expression is responsible for the fitness cost to agr-positive S. aureus cells.
RNAIII expression levels in the presence of antibiotics.
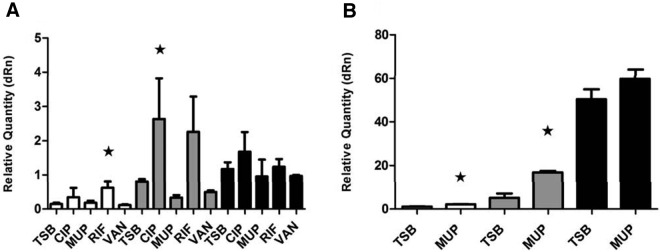
Previous studies have shown that translational inhibitors at sublethal concentrations affect virulence gene expression in S. aureus (22, 35) and that mupirocin induces RNAIII expression in strain Newman (36). Therefore, we examined the hypothesis that the fitness decrease associated with some antibiotics may be mediated via an antibiotic-dependent increase in RNAIII expression. For this purpose, RNA was isolated from WT cells at various ODs in the absence or presence of antibiotics supplemented at the concentrations used in the competition assays. RNAIII transcript levels in antibiotic-treated cultures relative to those in nontreated TSB cultures were assessed by RT-qPCR. The results show that, relative to untreated cultures, RNAIII expression was increased by mupirocin at ODs (600 nm) of 0.15 and 0.6 and by rifampin and ciprofloxacin at ODs of 2.0 and 4.0, whereas vancomycin did not affect RNAIII levels (Fig. 4). In conclusion, the antibiotics that decrease the fitness of strains carrying agr relative to that of the ΔagrA mutant strain also increase RNAIII expression, particularly at high cell densities and in the presence of vancomycin, which did not enhance the agr-mediated fitness cost or stimulate RNAIII expression (Fig. 4).
FIG 4 .
Induction of RNAIII expression by antibiotics. (A and B) RNAIII expression levels of WT cells in TSB in the absence or presence of the antibiotics ciprofloxacin (CIP), mupirocin (MUP), rifampin (RIF), and vancomycin (VAN). A star indicates a significant difference in the relative RNAIII expression level from that of the TSB control at the same OD. The ODs tested were 2.0 (white), 4.0 (gray), and 8.0 (black) in panel A and 0.15 (white), 0.6 (gray), and 1.2 (black) in panel B. The relative expression levels are normalized to the TSB control RNAIII expression levels of one sample at an OD of 8.0 in panel A and an OD of 0.15 in panel B. Means with standard deviations are shown. dRn, baseline subtracted fluorescent reading normalized to the reference dye.
DISCUSSION
S. aureus strains defective in the agr QS system are known to arise spontaneously under laboratory conditions (37–39), and among clinical isolates, agr-negative variants are surprisingly common (23, 24). The data presented here show that the expression of RNAIII, the effector molecule of the agr QS system, is responsible for the fitness cost of agr and, importantly, that the fitness cost is enhanced by the presence of antibiotics that induce the expression of RNAIII. The inverse correlation between fitness and RNAIII expression is corroborated by the findings that (i) ectopic expression of RNAIII is sufficient to reduce fitness (Fig. 3); (ii) the fitness cost is displayed in the transition to and during the stationary growth phase, where RNAIII is maximally expressed (Fig. 2); and (iii) conditions that do not affect RNAIII expression, such as vancomycin exposure, do not influence fitness (Fig. 1 and 4).
Fitness costs associated with QS are not restricted to S. aureus. In the opportunistic human pathogen P. aeruginosa, the LasR-LasI QS system controls the expression of a number of genes needed for infection (40). Mutants lacking LasR arise spontaneously and show a selective advantage in competition with WT bacteria (41). Importantly, they are also isolated from a variety of infections, where they can develop from QS-positive cells within a matter of days (42–46). In a social context, lasR and agr mutants may be considered “cheaters” that fail to contribute to the production of “public goods,” such as the catabolic enzymes needed during infection (41). However, mixed infections carrying both WT and QS-deficient cells occur, suggesting that there could be an interplay between cheaters and providers and that labor division is a deliberate strategy during the infection process (47, 48).
We show that in S. aureus, the expression of RNAIII is instrumental in the agr-associated fitness cost and that antibiotics that induce RNAIII expression also have a negative impact on fitness (Fig. 3). Beyond QS-controlled gene expression, little is known of the elements that impose the fitness burden. It has been speculated that the expression of a large number of gene products in response to a quorum signal creates a metabolic burden in WT cells and selects for QS-deficient variants (42). However, when the strains are cultivated individually, there are only minor differences in growth, although in some cases, QS mutants grow to a final cell density greater than that of the WT (41, 49, 50). In our studies, both the WT Newman and ΔagrA mutant strains multiplied with identical growth rates and reached the same final cell density; therefore, if fitness costs are to be explained by growth differences, they must be subtle and apparent only in limited parts of the growth cycle. Another explanation for the fitness differences seen may be the quorum-controlled production of autolysins that occurs in both S. aureus and P. aeruginosa in the stationary growth phase (50, 51). At high pHs, lasR mutant cells reach up to 10-fold greater cell numbers than WT cells because of pH-mediated autolysis (50). If QS-dependent lysis is prominent in stationary phase, the QS-negative cells that fail to undergo lysis may have an advantage observed as increased fitness.
The results reported here have important clinical implications. It has long been recognized that agr-deficient mutants are often isolated in the hospital setting but rarely in the community (25). Given the central role of agr in virulence, these observations are puzzling. Our findings suggest that the fitness cost of carrying agr is enhanced by the presence of some antibiotics and that treatment with those antibiotics will select for agr-deficient mutants. This notion is supported by a study reporting that bacteremic patients who had received fluoroquinolone or beta-lactam antibiotic treatment prior to hospitalization displayed an approximately 2-fold greater probability of harboring agr-dysfunctional strains than those who had not received any treatment prior to admission (26). In another study, isogenic S. aureus isolates were periodically recovered from the bloodstream of a patient undergoing chemotherapy. Among the 31 loci affected in the last multidrug-resistant isolate was agrC, resulting in the inactivation of agr (52). In this case, the ability of agr-negative strains to develop a GISA or hetero-GISA phenotype (32) may have been instrumental in the final development of vancomycin resistance and in the fatal outcome of the infection. Our results show that some antibiotics select for agr-negative variants of S. aureus, and this phenomenon should be taken into consideration when designing antimicrobial chemotherapy.
MATERIALS AND METHODS
Strains, media, and MIC determination.
The strains used in this study are derivatives of S. aureus Newman if not otherwise specified (see Table S2 in the supplemental material). All strains were grown at 37°C in TSB with or without ciprofloxacin (Bayer Schering Pharma), mupirocin (GlaxoSmithKline), rifampin (Sigma-Aldrich), and vancomycin (Sigma-Aldrich). MICs were determined by (i) using E-test strips (Biodisk) according to the manufacturer’s instructions and (ii) broth dilution assay according to EUCAST instructions (E.Dis 5) including the reference strain S. aureus ATCC 25923. Construction of ΔagrA and ΔRNAIII mutants was performed by transduction with phage φ80α (53).
Quantitation of RNAIII expression by qPCR.
RNA was isolated by using the SV RNeasy Mini Kit (Qiagen). RNA was converted to cDNA by using the high-capacity cDNA RT kit (Applied Biosystems) with an RNase inhibitor. The cDNA was used as the template for real-time qPCRs with the primers listed in Table S3 in the supplemental material and the Maxima SYBR green/ROX qPCR Master Mix. PCR products were detected by using the MX3000P qPCR system (Stratagene Products/Agilent Technologies), and the results were analyzed with the MxPro software (version 4.10; Stratagene).
Fitness measurements.
Three different assays were used to estimate fitness. (i) Growth rates were determined by growing bacteria in TSB medium with or without antibiotics at 37°C and measuring OD (600 nm) over time with a Bioscreen C reader (Labsystems) by using a 100-well honeycomb plate filled at 300 µl/well with an overnight culture diluted to 106 bacteria/ml in TSB growth medium. The relative fitness of the strains was calculated as the ratio of their doubling times (tDs) as follows: tD(WT)/tD(mutant). (ii) CFU counts at stationary phase were determined after 24 and 48 h for cells grown in 30 ml of TSB medium in a 300-ml narrow-neck Erlenmeyer flask with or without antibiotics shaken at 200 rpm with a starting inoculum of ~107 bacteria/ml. (iii) Competition experiments to assay relative competitive fitness after 24 and 48 h, W, of the tetracycline-resistant ΔagrA mutant or a plasmid (pTX, pTX::RNAIII, or pTX::agrA)-carrying strain compared to that of the tetracycline-susceptible WT strain was calculated by using the following formula (33): W = ln(RF/RI)/ln(SF/SI), where RI and SI refer to the CFU counts of resistant and susceptible cells at the start of the competition assay, respectively, and RF and SF refer to the numbers of resistant and susceptible cells at the endpoint of the competition assay. The experimental conditions of the competition assay were as follows. Overnight cultures of mutant and WT cells were diluted to ~107 bacteria/ml, mixed at a 1:1 ratio, and allowed to compete for 7 to 8 generations, reaching stationary phase (under the same culturing conditions as those used to determine CFU counts). The ratio of the endpoint CFU counts of the mutant and WT cells (competition ratio) was determined by spreading suitable dilutions on TSB agar plates with and without tetracycline at 2 µg/ml. The number of biological replicates used in the competition assay was between 5 and 12 for each condition assayed.
SUPPLEMENTAL MATERIAL
Effect of agr status on exponential growth and CFU counts in stationary growth phase. (A) Relative fitness measured in the exponential growth phase of the WT (white bars) and the ΔagrA mutant (gray bars) grown in the absence or presence of antibiotics. The generation time of the WT strain in TSB was 26 min. (B) CFU counts of the WT (white bars) and the ΔagrA mutant (gray bars) after 24 and 48 h in TSB. (C) CFU count of the WT (white bars) and the ΔagrA mutant (gray bars) grown in the presence of antibiotics for 48 h. Means with error bars indicating 95% confidence intervals are presented. Download Figure S1, PDF file, 0.3 MB.
MICs (E-test results) of the antibiotics used in this study.
Strains used in the study.
Primers used in the study.
ACKNOWLEDGMENTS
We thank Dan Andersson, Sophie Maisnier-Patin, and Anders Folkesson for constructive comments on the manuscript; Vi Phuong Thi Nguyen for technical help; and Michael Otto, Christiane Wolz, and Eva Morfeldt for kindly providing us with plasmids and strains. Bayer Schering Pharma is acknowledged for the generous gift of ciprofloxacin.
J.H. and H.I. are supported by grants from the Danish Research Council of Independent Research (274-08-0531) and Lundbeck (R31-A2472), and W.P. is supported by grants from the Danish Research Council of Independent Research (09-069656) and Ung Elitforskarpris (09-076146). The funders had no role in study design, data collection and analysis, the decision to publish, or preparation of the manuscript.
Footnotes
Citation Paulander W, et al. 2012. Antibiotic-mediated selection of quorum-sensing-negative Staphylococcus aureus. mBio 3(6):e00459-12. doi:10.1128/mBio.00459-12
REFERENCES
- 1. Gullberg E, et al. 2011. Selection of resistant bacteria at very low antibiotic concentrations. PLoS Pathog. 7:e1002158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Maisnier-Patin S, Berg OG, Liljas L, Andersson DI. 2002. Compensatory adaptation to the deleterious effect of antibiotic resistance in Salmonella typhimurium. Mol. Microbiol. 46:355–366 [DOI] [PubMed] [Google Scholar]
- 3. Ruusala T, Andersson D, Ehrenberg M, Kurland CG. 1984. Hyper-accurate ribosomes inhibit growth. EMBO J. 3:2575–2580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sturm A, et al. 2011. The cost of virulence: retarded growth of Salmonella typhimurium cells expressing type III secretion system 1. PLoS Pathog. 7:e1002143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. D’Argenio DA, et al. 2007. Growth phenotypes of Pseudomonas aeruginosa lasR mutants adapted to the airways of cystic fibrosis patients. Mol. Microbiol. 64:512–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Paulander W, Maisnier-Patin S, Andersson DI. 2009. The fitness cost of streptomycin resistance depends on rpsL mutation, carbon source and RpoS (sigmaS). Genetics 183:539–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rudkin JK, et al. 2012. Methicillin resistance reduces the virulence of healthcare-associated methicillin-resistant Staphylococcus aureus by interfering with the agr quorum sensing system. J. Infect. Dis. 205:798–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ji G, Beavis RC, Novick RP. 1995. Cell density control of staphylococcal virulence mediated by an octapeptide pheromone. Proc. Natl. Acad. Sci. U. S. A. 92:12055–12059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Antunes LC, Ferreira RB, Buckner MM, Finlay BB. 2010. Quorum sensing in bacterial virulence. Microbiology 156:2271–2282 [DOI] [PubMed] [Google Scholar]
- 10. Recsei P, et al. 1986. Regulation of exoprotein gene expression in Staphylococcus aureus by agar. Mol. Gen. Genet. 202:58–61 [DOI] [PubMed] [Google Scholar]
- 11. Novick RP, Geisinger E. 2008. Quorum sensing in staphylococci. Annu. Rev. Genet. 42:541–564 [DOI] [PubMed] [Google Scholar]
- 12. Novick RP. 2003. Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol. Microbiol. 48:1429–1449 [DOI] [PubMed] [Google Scholar]
- 13. Queck SY, et al. 2008. RNAIII-independent target gene control by the agr quorum-sensing system: insight into the evolution of virulence regulation in Staphylococcus aureus. Mol. Cell 32:150–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chevalier C, et al. 2010. Staphylococcus aureus RNAIII binds to two distant regions of coa mRNA to arrest translation and promote mRNA degradation. PLoS Pathog. 6:e1000809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Abdelnour A, Arvidson S, Bremell T, Rydén C, Tarkowski A. 1993. The accessory gene regulator (agr) controls Staphylococcus aureus virulence in a murine arthritis model. Infect. Immun. 61:3879–3885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mayville P, et al. 1999. Structure-activity analysis of synthetic autoinducing thiolactone peptides from Staphylococcus aureus responsible for virulence. Proc. Natl. Acad. Sci. U. S. A. 96:1218–1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wright JS, III, Jin R, Novick RP. 2005. Transient interference with staphylococcal quorum sensing blocks abscess formation. Proc. Natl. Acad. Sci. U. S. A. 102:1691–1696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gillaspy AF, et al. 1995. Role of the accessory gene regulator (agr) in pathogenesis of staphylococcal osteomyelitis. Infect. Immun. 63:3373–3380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cheung AL, et al. 1994. Diminished virulence of a sar-/agr- mutant of Staphylococcus aureus in the rabbit model of endocarditis. J. Clin. Invest. 94:1815–1822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cheung GY, Wang R, Khan BA, Sturdevant DE, Otto M. 2011. Role of the accessory gene regulator agr in community-associated methicillin-resistant Staphylococcus aureus pathogenesis. Infect. Immun. 79:1927–1935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Shopsin B, et al. 2010. Mutations in agr do not persist in natural populations of methicillin-resistant Staphylococcus aureus. J. Infect. Dis. 202:1593–1599 [DOI] [PubMed] [Google Scholar]
- 22. Joo HS, Chan JL, Cheung GY, Otto M. 2010. Subinhibitory concentrations of protein synthesis-inhibiting antibiotics promote increased expression of the agr virulence regulator and production of phenol-soluble modulin cytolysins in community-associated methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 54:4942–4944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Traber KE, et al. 2008. agr function in clinical Staphylococcus aureus isolates. Microbiology 154:2265–2274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fowler VG, Jr, et al. 2004. Persistent bacteremia due to methicillin-resistant Staphylococcus aureus infection is associated with agr dysfunction and low-level in vitro resistance to thrombin-induced platelet microbicidal protein. J. Infect. Dis. 190:1140–1149 [DOI] [PubMed] [Google Scholar]
- 25. Shopsin B, et al. 2008. Prevalence of agr dysfunction among colonizing Staphylococcus aureus strains. J. Infect. Dis. 198:1171–1174 [DOI] [PubMed] [Google Scholar]
- 26. Butterfield JM, et al. 2011. Predictors of agr dysfunction in methicillin-resistant Staphylococcus aureus (MRSA) isolates among patients with MRSA bloodstream infections. Antimicrob. Agents Chemother. 55:5433–5437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tsuji BT, Rybak MJ, Cheung CM, Amjad M, Kaatz GW. 2007. Community- and health care-associated methicillin-resistant Staphylococcus aureus: a comparison of molecular epidemiology and antimicrobial activities of various agents. Diagn. Microbiol. Infect. Dis. 58:41–47 [DOI] [PubMed] [Google Scholar]
- 28. Tsuji BT, MacLean RD, Dresser LD, McGavin MJ, Simor AE. 2011. Impact of accessory gene regulator (agr) dysfunction on vancomycin pharmacodynamics among Canadian community and health-care associated methicillin-resistant Staphylococcus aureus. Ann. Clin. Microbiol. Antimicrob. 10:20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Schweizer ML, et al. 2011. Increased mortality with accessory gene regulator (agr) dysfunction in Staphylococcus aureus among bacteremic patients. Antimicrob. Agents Chemother. 55:1082–1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Moise PA, et al. 2010. Factors influencing time to vancomycin-induced clearance of nonendocarditis methicillin-resistant Staphylococcus aureus bacteremia: role of platelet microbicidal protein killing and agr genotypes. J. Infect. Dis. 201:233–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sakoulas G, et al. 2005. Reduced susceptibility of Staphylococcus aureus to vancomycin and platelet microbicidal protein correlates with defective autolysis and loss of accessory gene regulator (agr) function. Antimicrob. Agents Chemother. 49:2687–2692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sakoulas G, et al. 2002. Accessory gene regulator (agr) locus in geographically diverse Staphylococcus aureus isolates with reduced susceptibility to vancomycin. Antimicrob. Agents Chemother. 46:1492–1502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gagneux S, et al. 2006. The competitive cost of antibiotic resistance in Mycobacterium tuberculosis. Science 312:1944–1946 [DOI] [PubMed] [Google Scholar]
- 34. Lenski RE, Rose MR, Simpson SC, Tadler SC. 1991. Long-term experimental evolution in Escherichia coli. I. Adaptation and divergence during 2,000 generations. Am. Nat. 138:1315–1341 [Google Scholar]
- 35. Subrt N, Mesak LR, Davies J. 2011. Modulation of virulence gene expression by cell wall active antibiotics in Staphylococcus aureus. J. Antimicrob. Chemother. 66:979–984 [DOI] [PubMed] [Google Scholar]
- 36. Geiger T, et al. 2010. Role of the (p)ppGpp synthase RSH, a RelA/SpoT homolog, in stringent response and virulence of Staphylococcus aureus. Infect. Immun. 78:1873–1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Somerville GA, et al. 2002. In vitro serial passage of Staphylococcus aureus: changes in physiology, virulence factor production, and agr nucleotide sequence. J. Bacteriol. 184:1430–1437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Adhikari RP, Arvidson S, Novick RP. 2007. A nonsense mutation in agrA accounts for the defect in agr expression and the avirulence of Staphylococcus aureus 8325-4 traP::kan. Infect. Immun. 75:4534–4540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chen J, Novick RP. 2007. svrA, a multi-drug exporter, does not control agr. Microbiology 153:1604–1608 [DOI] [PubMed] [Google Scholar]
- 40. Van Delden C, Iglewski BH. 1998. Cell-to-cell signaling and Pseudomonas aeruginosa infections. Emerg. Infect. Dis. 4:551–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Diggle SP, Griffin AS, Campbell GS, West SA. 2007. Cooperation and conflict in quorum-sensing bacterial populations. Nature 450:411–414 [DOI] [PubMed] [Google Scholar]
- 42. Sandoz KM, Mitzimberg SM, Schuster M. 2007. Social cheating in Pseudomonas aeruginosa quorum sensing. Proc. Natl. Acad. Sci. U. S. A. 104:15876–15881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Köhler T, Buckling A, van Delden C. 2009. Cooperation and virulence of clinical Pseudomonas aeruginosa populations. Proc. Natl. Acad. Sci. U. S. A. 106:6339–6344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Cabrol S, Olliver A, Pier GB, Andremont A, Ruimy R. 2003. Transcription of quorum-sensing system genes in clinical and environmental isolates of Pseudomonas aeruginosa. J. Bacteriol. 185:7222–7230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Heurlier K, Dénervaud V, Haas D. 2006. Impact of quorum sensing on fitness of Pseudomonas aeruginosa. Int. J. Med. Microbiol. 296:93–102 [DOI] [PubMed] [Google Scholar]
- 46. Bjarnsholt T, et al. 2010. Quorum sensing and virulence of Pseudomonas aeruginosa during lung infection of cystic fibrosis patients. PLoS One 5:e10115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Czárán T, Hoekstra RF. 2009. Microbial communication, cooperation and cheating: quorum sensing drives the evolution of cooperation in bacteria. PLoS One 4:e6655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Morris JJ, Lenski RE, Zinser ER. 2012. The black queen hypothesis: evolution of dependencies through adaptive gene loss. mBio 3(2):e00036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. An D, Danhorn T, Fuqua C, Parsek MR. 2006. Quorum sensing and motility mediate interactions between Pseudomonas aeruginosa and Agrobacterium tumefaciens in biofilm cocultures. Proc. Natl. Acad. Sci. U. S. A. 103:3828–3833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Heurlier K, et al. 2005. Quorum-sensing-negative (lasR) mutants of Pseudomonas aeruginosa avoid cell lysis and death. J. Bacteriol. 187:4875–4883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Memmi G, Nair DR, Cheung A. 2012. Role of ArlRS in autolysis in methicillin-sensitive and methicillin-resistant Staphylococcus aureus strains. J. Bacteriol. 194:759–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Mwangi MM, et al. 2007. Tracking the in vivo evolution of multidrug resistance in Staphylococcus aureus by whole-genome sequencing. Proc. Natl. Acad. Sci. U. S. A. 104:9451–9456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Novick RP. 1991. Genetic systems in staphylococci. Methods Enzymol. 204:587–636 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Effect of agr status on exponential growth and CFU counts in stationary growth phase. (A) Relative fitness measured in the exponential growth phase of the WT (white bars) and the ΔagrA mutant (gray bars) grown in the absence or presence of antibiotics. The generation time of the WT strain in TSB was 26 min. (B) CFU counts of the WT (white bars) and the ΔagrA mutant (gray bars) after 24 and 48 h in TSB. (C) CFU count of the WT (white bars) and the ΔagrA mutant (gray bars) grown in the presence of antibiotics for 48 h. Means with error bars indicating 95% confidence intervals are presented. Download Figure S1, PDF file, 0.3 MB.
MICs (E-test results) of the antibiotics used in this study.
Strains used in the study.
Primers used in the study.