Abstract
In Drosophila, all segments form in the blastoderm where morphogen gradients spanning the entire anterior-posterior axis of the embryo provide positional information. However, in the beetle Tribolium castaneum and most other arthropods, a number of anterior segments form in the blastoderm, and the remaining segments form sequentially from a posterior growth zone during germband elongation. Recently, the cyclic nature of the pair-rule gene Tc-odd-skipped was demonstrated in the growth zone of Tribolium, indicating that a vertebrate-like segmentation clock is employed in the germband stage of its development. This suggests that two mechanisms might function in the same organism: a Drosophila-like mechanism in the blastoderm, and a vertebrate-like mechanism in the germband. Here, we show that segmentation at both blastoderm and germband stages of Tribolium is based on a segmentation clock. Specifically, we show that the Tribolium primary pair-rule gene, Tc-even-skipped (Tc-eve), is expressed in waves propagating from the posterior pole and progressively slowing until they freeze into stripes; such dynamics are a hallmark of clock-based segmentation. Phase shifts between Tc-eve transcripts and protein confirm that these waves are due to expression dynamics. Moreover, by tracking cells in live embryos and by analyzing mitotic profiles, we found that neither cell movement nor oriented cell division could explain the observed wave dynamics of Tc-eve. These results pose intriguing evolutionary questions, as Drosophila and Tribolium segment their blastoderms using the same genes but different mechanisms.
Keywords: Tribolium, Arthropods, Blastoderm, Insects, Segmentation clock, Somitogenesis
INTRODUCTION
Early in embryogenesis, the anterior-posterior (AP) axis of an arthropod embryo is established as a series of body segments. In Drosophila, global, long-range morphogen gradients initiated by maternal factors provide positional information for a downstream cascade of genes that ultimately partitions the AP axis into segments (RiveraPomar and Jackle, 1996). This mechanism is possible in a blastoderm in which morphogens are free to diffuse in a syncytial environment to form gradients that span the entire region to be segmented. However, in most other arthropods, posterior segments are specified sequentially in a cellularized environment out of a posterior pool of cells, referred to as the ‘growth zone’ (Davis and Patel, 2002). For this mode of segmentation, morphogen gradients spanning the entire AP axis have not been discovered.
Vertebrate segmentation (somitogenesis) also occurs in a cellular environment in which segments form sequentially during AP axis elongation. Somitogenesis relies on the ‘clock and wavefront’ mechanism, in which the expression of multiple genes (including genes of the Notch, Wnt and fibroblast growth factor signaling pathways) (Dequeant et al., 2006; Krol et al., 2011) oscillate in the presomatic mesoderm (PSM) posterior to an arrest front [defined by a threshold within the overlapping domain of posterior Wnt and fibroblast growth factor gradients (Aulehla et al., 2003; Dubrulle et al., 2001) and an opposing retinoic acid gradient (Diez del Corral et al., 2003)]. Anterior to the arrest front, oscillation ceases, producing stripes of gene expression. Although the genetic and biochemical details are not yet understood, oscillations seem to be arrested gradually (Giudicelli et al., 2007), resulting in kinematic waves of expression that slow down and shrink as they propagate along the PSM (Palmeirim et al., 1997).
A clock and wavefront model has been proposed to act in arthropods also (Peel et al., 2005), as what appear to be waves of gene expression propagating within the growth zone have been reported in several arthropods (Chipman and Akam, 2008; Chipman et al., 2004; Pueyo et al., 2008; Stollewerk et al., 2003). Recently, it was shown that Tc-odd-skipped (Tc-odd) expression in the growth zone of the Tribolium germband oscillates in a manner that cannot be explained by cell movements (Sarrazin et al., 2012).
Most arthropods form anterior segments in the blastoderm and posterior segments from a posterior growth zone (Davis and Patel, 2002). It is conceivable that these arthropods utilize two different modes of segmentation: a Drosophila-like mechanism in the blastoderm and a clock-based mechanism in the germband. In this Report, we show that the beetle Tribolium castaneum utilizes a clock-based segmentation mechanism in both blastoderm and germband stages of its development. Specifically, we show that the Tribolium primary pair-rule gene Tc-even-skipped (Tc-eve) is expressed in waves propagating in both blastoderm and germband stages. These dynamics are evident from the observed phase shift between Tc-eve transcripts and proteins. By tracking cells in live embryos and by analyzing mitotic profiles, we confirm that the waves of Tc-eve expression in the blastoderm cannot be explained by cell movement or by oriented cell division.
MATERIALS AND METHODS
In situ hybridization and immunocytochemistry
In situ hybridization was performed using digoxigenin (DIG)-labeled RNA probes and an anti-DIG::alkaline phosphatase (AP) antibody (Roche). Signal was developed using NBT/BCIP (BM Purple, Roche), or Fast Red/HNPP (Roche). Immunocytochemistry was performed using anti-EVE (mouse monoclonal antibody 2B8, Hybridoma Bank, University of Iowa) and anti-EN (mouse monoclonal antibody 4D9, Santa Cruz Technology) as primary antibodies, and anti-mouse::POD as secondary antibody (ABC Kit, Vector). Diaminobenzidine (DAB) was used as a substrate to produce a golden brown signal, and AlexaFluor 488-conjugated tyramide (Invitrogen) to give a green fluorescent signal.
Wild-type strains and transgenic lines
All expression analysis was performed using GA-1 strain embryos. Live imaging was carried out using the EFA-nGFP line (Sarrazin et al., 2012).
Live imaging and cell tracking
EFA-nGFP embryos were dechorionated by immersing in 1% bleach for 30 seconds. Embryos were then placed on a microscope glass slide and covered with halocarbon oil 700 (Sigma); no coverslip was used. The time-lapse movie was taken by capturing five focal planes every 5 minutes, over ~11 hours at 26-28°C, on a Leica M205 FA stereoscope at 200× magnification. supplementary material Movie 2 shows a single focal plane at a speed of 6 frames (30 minutes real time) per second. GFP-tagged nuclei were tracked using the ImageJ plugin MTrackJ (Meijering et al., 2012).
Egg collections for developmental time windows
Developmental windows in Fig. 2 were generated by incubating 1-hour egg collections at 23-24°C for the desired length of time. For 3-hour developmental windows (supplementary material Fig. S3), eggs were collected after three hours instead of one.
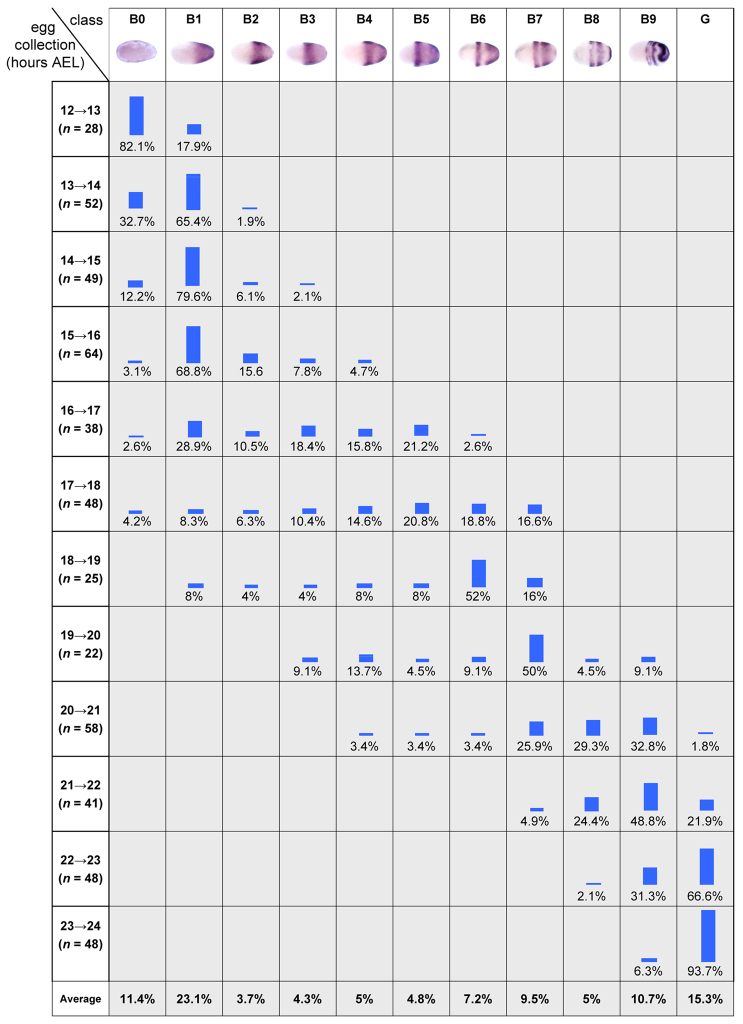
Fig. 2.
Mapping the temporal order of Tc-eve patterns. The proportion of each class of Tc-eve pattern (blastoderm stages B0-B9; all germband stages combined in G) was recorded in egg collections spanning the blastoderm and early germband stages [12-24 hours after egg laying (AEL), at 23-24°C] in 1-hour developmental windows. The last row in the table shows the average percentage of each class over all egg collections, which estimates the proportion of each class in total (spanning the entire 12-24 hour period).
Correlation of time-lapse movie and blastoderm stainings
Based on embryo morphology and nuclear density, blastoderm classes (B0-B9) were correlated with the time-lapse images. The B0 stage is characterized by low nuclear density (up to mitotic cycle 13) and a rounded posterior end; B1-B6 stage embryos have higher nuclear density and the posterior end is still rounded (after mitotic cycle 13); B7 stage is characterized by flattening of the posterior pole; and B8-B9 embryos are identified by primitive pit formation.
Computer simulations
Computer simulations in supplementary material Movies S1 and S3 were generated using Matlab. Source codes are provided in supplementary material Appendix S1.
RESULTS AND DISCUSSION
Waves of Tc-eve gene expression are observed in both blastoderm and germband stages of Tribolium development
The three Tc-eve stripes that form during the blastoderm stage were previously thought to form by subdivision of a broad posterior expression domain (Fig. 1, class B1), through clearing of expression to form the interstripe regions (Fig. 1, classes B6 and B9) (Brown et al., 1997; Patel et al., 1994). By carefully examining a large number of fixed embryos at the blastoderm stage, we found that Tc-eve expression is considerably more dynamic and several intermediate staining patterns are discernible (Fig. 1, classes B0-B9). Each primary Tc-eve stripe first appears as a cap of expression at the posterior pole of the embryo (Fig. 1, classes B1, B4 and B8), which extends anteriorly, clears from the posterior pole, continuously shrinks in width, and eventually freezes into a stable stripe of expression. Later, each primary Tc-eve stripe splits into two secondary (segmental) stripes (Fig. 1, classes B9-G4).
Fig. 1.
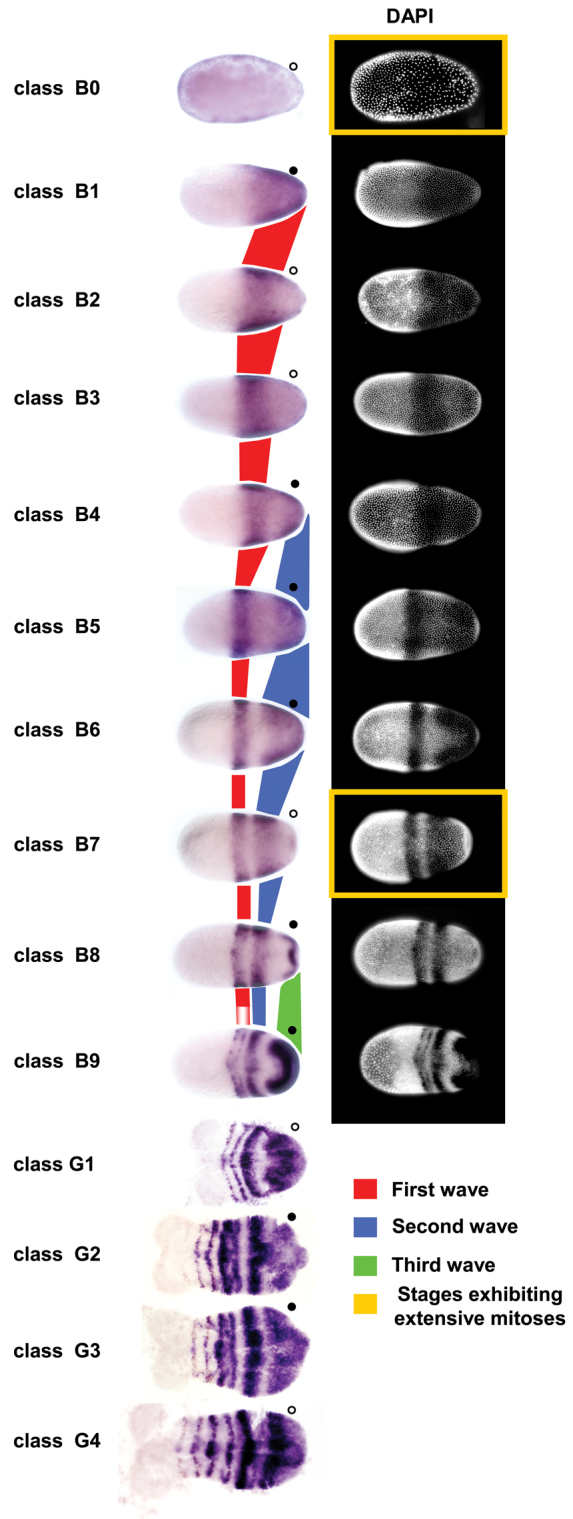
Waves of Tc-eve expression propagate from posterior to anterior in both blastoderm and germband stages of Tribolium development. Progression of Tc-eve expression from early blastoderm to early germband stages is shown. Classes B0-B9 (blastoderm stages) and G1-G4 (germband stages) represent distinct Tc-eve expression patterns, arranged in a putative temporal sequence (see Fig. 2). Propagation of the first three waves of Tc-eve expression is highlighted in red (first stripe), blue (second stripe) and green (third stripe). Tc-eve oscillation in the posterior end of the embryo is marked by circles; high expression levels are marked by filled circles, low levels by open circles. DAPI staining of blastoderm stages is shown on the right. Classes coinciding with extensive mitoses are enclosed in yellow rectangles. Anterior to the left.
The fourth primary Tc-eve stripe starts to form in the germband, with similar dynamics to those of blastodermal stripes. Tc-eve expression emanates from the posterior end of the growth zone (Fig. 1, class G2), expands (Fig. 1, class G3), then shrinks and stabilizes into a stripe at the anterior border of the growth zone (Fig. 1, class G4). The remaining stripes that form during germband elongation show similar dynamics (supplementary material Fig. S1).
To verify that the differences in the blastoderm Tc-eve expression patterns represent a genuine temporal sequence of expression, and are not due to embryo-to-embryo variation, we examined Tc-eve expression in twelve consecutive 1-hour developmental windows, spanning the blastoderm stage (12-24 hours after egg laying at 23-24°C; see Materials and methods). For every time window, we determined the percentage of embryos displaying a given Tc-eve expression pattern (classes B0 to B9 for blastoderm, class G for all germband stages; see Fig. 2). The overall pattern of class distributions supports the notion that the given order of Tc-eve expression classes reflects a genuine temporal sequence. For example, class B3 embryos appear for the first time in the 14-15 hour window, indicating that they are more advanced in age compared with embryos in B1 and B2. Having verified that B0 to B9 represent a genuine temporal sequence of expression, we will henceforth use them as developmental stages.
These observations suggest that Tc-eve dynamics in both the blastoderm and the germband can be described as waves of gene expression that emanate from the posterior end and propagate anteriorly while shrinking in width. These dynamics have the characteristic appearance of kinematic waves generated by an oscillator under the control of a posterior-to-anterior frequency gradient (supplementary material Movie 1) (Palmeirim et al., 1997). The oscillation of Tc-eve expression can be followed in cells at the posterior end of the embryos in Fig. 1 (filled circles represent high expression levels, open circles represent low expression levels).
Tc-eve waves are due to transcription dynamics, rather than cell movement or oriented cell division
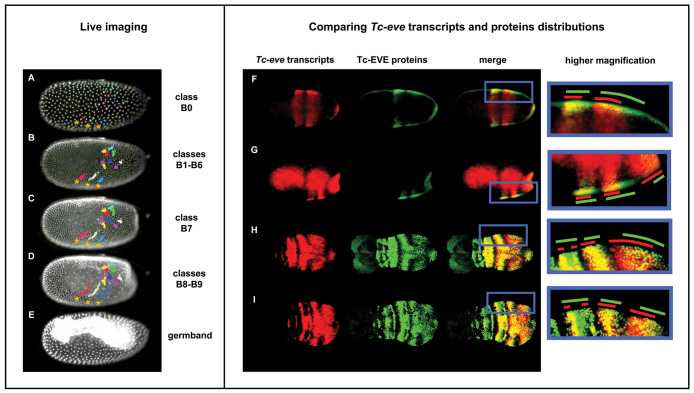
To address the possibility that the observed dynamics of Tc-eve expression are due to cell movement, we used a transgenic line expressing nuclear-localized GFP driven by a Tribolium ubiquitous promoter (EFA-nGFP line) (Sarrazin et al., 2012) to track cell movements in live embryos starting from early blastoderm stage (B0) and continuing through the germband (supplementary material Movie 2; Fig. 3A-E). Blastoderm Tc-eve patterns (B0-B9) were registered with time-lapse movies based on overall morphology and nuclear density in the blastoderm (see Materials and methods). In stages B0 to B7, nuclear movement in most of the blastoderm is very limited and mostly random. We observe some slow movements of the most lateral nuclei in a posterior and ventral direction (Fig. 3A-C, asterisks), which represent the earliest sign of germband condensation, and inward movements of cells at the posterior pole, to produce the characteristic posterior flattening of the late blastoderm. Late in stage B7, a burst of mitoses in the embryonic field differentiates it from the serosa. During stages B8 and B9, the germband anlage rapidly condenses towards the ventral side of the embryo; this involves rapid movements of nuclei from the lateral sides of the blastoderm in a posterior and ventral direction (see three tracks marked by asterisks in Fig. 3D). These movements do not explain the dynamics of Tc-eve expression, which involve much faster changes and shifts of expression from the posterior pole towards the middle of the embryo.
Fig. 3.
Tc-eve waves are due to transcription dynamics, rather than cell movements. (A-E) Stills from live imaging of an EFA-nGFP transgenic Tribolium embryo (supplementary material Movie 2), from early blastoderm (A) to germband (E) stages; the corresponding Tc-eve patterns from Fig. 2 are indicated on the right. Colored circles track the movement of nuclei, showing that cell movement cannot explain Tc-eve dynamics. Three dorsolateral cells are marked with asterisks in A-D. (F-I) Concurrent Tc-eve in situ hybridization (red; first column) and Tc-EVE antibody staining (green; second column) were merged (third column) to reveal the phase shift between mRNA and protein distributions. The fourth column shows higher magnifications of the region of interest; red and green lines mark the extent of Tc-eve transcript and Tc-EVE protein distributions, respectively. The fact that Tc-EVE stripes lag behind those of Tc-eve indicates that Tc-eve stripes propagate as waves from posterior to anterior. The fact that the phase shift decreases as the stripes mature suggests that the waves are slowing down before they eventually freeze and split into secondary stripes. Similar dynamics are seen at different stages. Anterior to the left; ventral is up in A-E.
Oriented cell divisions could, in principle, also contribute to the observed dynamics of gene expression. We therefore examined whether oriented mitoses occur during stages B0-B9. After the synchronous mitoses phase in early blastoderm (class B0), no mitoses were detected in B1- to B5-stage embryos, and only limited random mitoses were detected in stage B6 (n=10 for each class). Stage B7 blastoderms were found to undergo extensive cell division in all cells except those of the serosa. These mitoses are randomly oriented (supplementary material Fig. S2). During stages B8 and B9, mitotic activity decreased and only a very limited number of random mitotic plates were observed. In the germband stage, Sarrazin et al. (Sarrazin et al., 2012) showed that the growth zone extends by convergent extension and experiences low mitotic activity until the formation of the fifth pair-rule stripe (Sarrazin et al., 2012). Hence, neither cell movement nor oriented cell division contributes significantly to Tc-eve dynamics in blastoderm or germband stages.
To confirm directly that Tc-eve waves are due to expression dynamics, we compared the relative distribution of Tc-eve transcripts and Tc-EVE protein within individual embryos (Fig. 3F-I). If Tc-eve is expressed as waves, we expect the short delay between transcription and translation to be reflected in a small shift between the mRNA and protein distributions. When a wave of expression freezes into a stripe, transcripts and proteins should precisely colocalize. Indeed, we observed that the Tc-EVE protein distribution lags behind that of Tc-eve transcripts as each wave propagates from the posterior pole, both in the blastoderm (Fig. 3F,G) and in the germband growth zone (Fig. 3H,I). The two patterns precisely colocalize in the stable/mature primary stripes, which then split into secondary stripes. Interestingly, each primary stripe exhibits smaller phase shift between mRNA and protein as it moves anteriorly (Fig. 3F-I) reflecting a gradual slowing of each stripe as it matures. This outcome is consistent with a model in which the frequency of Tc-eve oscillation changes in a posterior-to-anterior gradient (supplementary material Movie 3).
Determining the frequency of Tc-eve oscillations
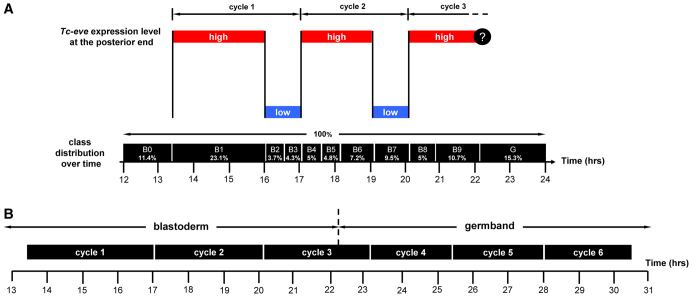
Fixed embryos provide only spatial information about Tc-eve expression (Fig. 1), but by analyzing the distribution of expression patterns over several developmental windows (Fig. 2) we obtained temporal information about Tc-eve dynamics (e.g. its periodicity, or lack thereof). The average percentage of embryos in each class (shown in the last row of Fig. 2) was used to estimate the duration of that class (Fig. 4A, vertical black bars). A waveform of Tc-eve oscillations at the posterior end of the blastoderm was drawn by tracking Tc-eve expression levels (Fig. 1, closed and open circles) during each class (Fig. 4A). From this analysis, the frequency of Tc-eve oscillation at the posterior end of the blastoderm was determined to be approximately one cycle per three hours at 23-24°C, for the first two cycles of Tc-eve.
Fig. 4.
Periodicity of Tc-eve oscillation in the blastoderm and germband. (A) The duration of each class of Tc-eve expression (estimated from Fig. 2, bottom row) is represented by the width of the black boxes. This is equivalent to combining all egg collections into one large collection, spanning 12-24 hours after egg laying, then using the overall percentage of embryos in each class as an estimate of its duration (percentages rather than absolute numbers are used in calculations to correct for differences in the number of eggs in each collection). Tc-eve expression at the posterior end of the blastoderm is shown above the black boxes (red bars, high Tc-eve; blue bars, low Tc-eve; question mark indicates uncertainty due to primitive pit formation). (B) The estimated duration of Tc-eve cycles for blastoderm (taken from A) and germband (determined from supplementary material Fig. S3, for stripes 4, 5 and 6) are represented by the width of the black boxes.
To confirm the periodicity of the first two cycles and to determine whether the third cycle has the same period, we calculated the proportion of embryos undergoing first, second and third cycles of Tc-eve expression in an egg collection spanning the period from 10 to 26 hours after egg laying. We found similar proportions of embryos in each cycle (31.4% in cycle 1, 30.8% in cycle 2 and 37.8% in cycle 3; n=143) indicating that the three cycles are of almost equal duration. A similar analysis of germband stages, using six consecutive 3-hour developmental windows, indicated that the period of Tc-eve cycles in the germband is similar (except for the eighth cycle; supplementary material Fig. S3), but on average slightly shorter than that found in the blastoderm (Fig. 4B). Future experiments with reporter constructs and live imaging will be needed to determine whether this subtle difference is significant. Tc-odd oscillates out of phase with Tc-eve, and its fourth cycle was estimated to be ~95 minutes at 30°C (Sarrazin et al., 2012). This is approximately half the periodicity determined for Tc-eve (3 hours), consistent with a doubling of developmental rates between 24°C and 30°C (Park and Marian Burton, 1948).
Probable molecular basis and implications for segmentation mechanisms
A molecular candidate for the presumed clock is the negative feedback circuit composed of the three Tribolium primary pair-rule genes: Tc-eve, Tc-runt and Tc-odd (Choe et al., 2006). Two of these genes, Tc-eve and Tc-odd, have been found to oscillate in Tribolium (this report) (Sarrazin et al., 2012). The original formulation of the clock and wavefront model postulated an oscillator controlled by a continuously moving gradient (wavefront), within which the oscillation persists and outside of which the oscillation freezes (Cooke and Zeeman, 1976). Interestingly, even a static smooth gradient is capable of generating striped expression (supplementary material Movie 1) (Beck and Varadi, 1972). This could explain the formation of the first three Tc-eve stripes in the blastoderm in the absence of posterior elongation, although the possibility of a continuously retracting wavefront in the blastoderm phase cannot be excluded.
In this report, we showed that the anterior-to-posterior progression of Tc-eve stripe formation reflects an underlying segmentation clock functioning in the Tribolium blastoderm and germband. Sequential segmentation (Liu and Kaufman, 2005; Nakao, 2010) and posterior-to-anterior shifts of pair-rule gene expression (Garcia-Solache et al., 2010) in the blastoderm of other insects have also been reported. Careful examination of stripe-formation dynamics will determine whether these insects utilize clock-based or Drosophila-like segmentation mechanisms. Interestingly, even the expression of posterior pair-rule genes in Drosophila undergo posterior-to-anterior shifts (Keranen et al., 2006). Although limited and not reflecting an overt oscillatory process, as in Tribolium, these shifts might be a vestige of an ancestral clock-based mechanism.
Supplementary Material
Acknowledgements
We are thankful to Andrew Peel for helpful comments on the manuscript; Michelle Gordon and Barb VanSlyke for technical support; and the Bodossaki Foundation for donating the stereoscope used for time-lapse recordings.
Footnotes
Funding
Work in the Brown lab was supported by the National Institutes of Health [grant 5R01HD29594]; and the Kansas IDeA Network of Biomedical Research Excellence (K-INBRE) [grant P20RR016475]. Deposited in PMC for release after 12 months.
Competing interests statement
The authors declare no competing financial interests.
Supplementary material
Supplementary material available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.085126/-/DC1
References
- Aulehla A., Wehrle C., Brand-Saberi B., Kemler R., Gossler A., Kanzler B., Herrmann B. G. (2003). Wnt3a plays a major role in the segmentation clock controlling somitogenesis. Dev. Cell 4, 395-406 [DOI] [PubMed] [Google Scholar]
- Beck M. T., Varadi Z. B. (1972). One, two and three-dimensional spatially periodic chemical reactions. Nature 235, 15-16 [Google Scholar]
- Brown S. J., Parrish J. K., Beeman R. W., Denell R. E. (1997). Molecular characterization and embryonic expression of the even-skipped ortholog of Tribolium castaneum. Mech. Dev. 61, 165-173 [DOI] [PubMed] [Google Scholar]
- Chipman A. D., Akam M. (2008). The segmentation cascade in the centipede Strigamia maritima: involvement of the Notch pathway and pair-rule gene homologues. Dev. Biol. 319, 160-169 [DOI] [PubMed] [Google Scholar]
- Chipman A. D., Arthur W., Akam M. (2004). A double segment periodicity underlies segment generation in centipede development. Curr. Biol. 14, 1250-1255 [DOI] [PubMed] [Google Scholar]
- Choe C. P., Miller S. C., Brown S. J. (2006). A pair-rule gene circuit defines segments sequentially in the short-germ insect Tribolium castaneum. Proc. Natl. Acad. Sci. USA 103, 6560-6564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke J., Zeeman E. C. (1976). A clock and wavefront model for control of the number of repeated structures during animal morphogenesis. J. Theor. Biol. 58, 455-476 [DOI] [PubMed] [Google Scholar]
- Davis G. K., Patel N. H. (2002). Short, long, and beyond: molecular and embryological approaches to insect segmentation. Annu. Rev. Entomol. 47, 669-699 [DOI] [PubMed] [Google Scholar]
- Dequeant M. L., Glynn E., Gaudenz K., Wahl M., Chen J., Mushegian A., Pourquie O. (2006). A complex oscillating network of signaling genes underlies the mouse segmentation clock. Science 314, 1595-1598 [DOI] [PubMed] [Google Scholar]
- Diez del Corral R., Olivera-Martinez I., Goriely A., Gale E., Maden M., Storey K. (2003). Opposing FGF and retinoid pathways control ventral neural pattern, neuronal differentiation, and segmentation during body axis extension. Neuron 40, 65-79 [DOI] [PubMed] [Google Scholar]
- Dubrulle J., McGrew M. J., Pourquie O. (2001). FGF signaling controls somite boundary position and regulates segmentation clock control of spatiotemporal Hox gene activation. Cell 106, 219-232 [DOI] [PubMed] [Google Scholar]
- Garcia-Solache M., Jaeger J., Akam M. (2010). A systematic analysis of the gap gene system in the moth midge Clogmia albipunctata. Dev. Biol. 344, 306-318 [DOI] [PubMed] [Google Scholar]
- Giudicelli F., Ozbudak E. M., Wright G. J., Lewis J. (2007). Setting the tempo in development: an investigation of the zebrafish somite clock mechanism. PLoS Biol. 5, e150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keranen S. V., Fowlkes C. C., Luengo Hendriks C. L., Sudar D., Knowles D. W., Malik J., Biggin M. D. (2006). Three-dimensional morphology and gene expression in the Drosophila blastoderm at cellular resolution II: dynamics. Genome Biol. 7, R124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krol A. J., Roellig D., Dequeant M. L., Tassy O., Glynn E., Hattem G., Mushegian A., Oates A. C., Pourquie O. (2011). Evolutionary plasticity of segmentation clock networks. Development 138, 2783-2792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P. Z., Kaufman T. C. (2005). even-skipped is not a pair-rule gene but has segmental and gap-like functions in Oncopeltus fasciatus, an intermediate germband insect. Development 132, 2081-2092 [DOI] [PubMed] [Google Scholar]
- Meijering E., Dzyubachyk O., Smal I. (2012). Methods for cell and particle tracking. Methods Enzymol. 504, 183-200 [DOI] [PubMed] [Google Scholar]
- Nakao H. (2010). Characterization of Bombyx embryo segmentation process: expression profiles of engrailed, even-skipped, caudal, and wnt1/wingless homologues. J. Exp. Zool. B Mol. Dev. Evol. 314, 224-231 [DOI] [PubMed] [Google Scholar]
- Palmeirim I., Henrique D., Ish-Horowicz D., Pourquie O. (1997). Avian hairy gene expression identifies a molecular clock linked to vertebrate segmentation and somitogenesis. Cell 91, 639-648 [DOI] [PubMed] [Google Scholar]
- Park T., Marian Burton F. (1948). The fecundity and development of the flour beetles, Tribolium Confusum and Tribolium Castaneum, at three constant temperatures. Ecology 29, 368-374 [Google Scholar]
- Patel N. H., Condron B. G., Zinn K. (1994). Pair-rule expression patterns of even-skipped are found in both short- and long-germ beetles. Nature 367, 429-434 [DOI] [PubMed] [Google Scholar]
- Peel A. D., Chipman A. D., Akam M. (2005). Arthropod segmentation: beyond the Drosophila paradigm. Nat. Rev. Genet. 6, 905-916 [DOI] [PubMed] [Google Scholar]
- Pueyo J. I., Lanfear R., Couso J. P. (2008). Ancestral Notch-mediated segmentation revealed in the cockroach Periplaneta americana. Proc. Natl. Acad. Sci. USA 105, 16614-16619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- RiveraPomar R., Jackle H. (1996). From gradients to stripes in Drosophila embryogenesis: Filling in the gaps. Trends Genet. 12, 478-483 [DOI] [PubMed] [Google Scholar]
- Sarrazin A. F., Peel A. D., Averof M. (2012). A segmentation clock with two-segment periodicity in insects. Science 336, 338-341 [DOI] [PubMed] [Google Scholar]
- Stollewerk A., Schoppmeier M., Damen W. G. (2003). Involvement of Notch and Delta genes in spider segmentation. Nature 423, 863-865 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.