Abstract
The dynamics of protein conformational changes, from protein folding to smaller changes, such as those involved in ligand binding, are governed by the properties of the conformational energy landscape. Different techniques have been used to follow the motion of a protein over this landscape and thus quantify its properties. However, these techniques often are limited to short timescales and low-energy conformations. Here, we describe a general approach that overcomes these limitations. Starting from a nonnative conformation held by an aromatic disulfide bond, we use time-resolved spectroscopy to observe nonequilibrium backbone dynamics over nine orders of magnitude in time, from picoseconds to milliseconds, after photolysis of the disulfide bond. We find that the reencounter probability of residues that initially are in close contact decreases with time following an unusual power law that persists over the full time range and is independent of the primary sequence. Model simulations show that this power law arises from subdiffusional motion, indicating a wide distribution of trapping times in local minima of the energy landscape, and enable us to quantify the roughness of the energy landscape (4–5 kBT). Surprisingly, even under denaturing conditions, the energy landscape remains highly rugged with deep traps (>20 kBT) that result from multiple nonnative interactions and are sufficient for trapping on the millisecond timescale. Finally, we suggest that the subdiffusional motion of the protein backbone found here may promote rapid folding of proteins with low contact order by enhancing contact formation between nearby residues.
Keywords: photochemical trigger, subdiffusion
Major advances have been made in recent years in understanding dynamic aspects of protein conformational changes, particularly protein folding; however, many issues remain to be solved (1). Among these are the properties of the unfolded protein ensemble and the role of residual structure of denatured proteins in promoting folding (2), the heterogeneity of microscopic folding pathways (3), and the existence of multiple distinct, but only transiently populated, intermediates (4). Particularly for fast-folding proteins, the idea of downhill folding, i.e., the absence of a significant barrier, has been suggested as an alternative mechanism (5, 6), but it is not clear to what extent fast-folding proteins make use of this mechanism. On the other hand, technical progress has made it possible to observe multiple folding and unfolding events in millisecond all-atom molecular dynamics simulations. Such simulations have shown that some proteins always follow the same folding pathway, whereas others have several different pathways (7). Moreover, individual folding events occur with submicrosecond transit times through a distinct transition state but are separated by long waiting times, which yield the experimentally observed folding times (8).
The idea of motion on a rugged energy landscape (9–11) has been used widely to describe conformational changes in proteins, including protein folding. As a result of the multitude and varying strength of local and nonlocal interactions of the polypeptide backbone, side chains, and solvent molecules, the multidimensional conformational energy landscape is thought to consist of a hierarchy of local minima of varying depths (Fig. 1). The properties of this energy landscape govern the motion of the polypeptide backbone in real space; thus, a full understanding of processes such as protein folding or ligand binding will require a better characterization of this landscape. In practice, motion on the multidimensional energy landscape must be approximated by diffusion on an idealized low-dimensional surface, the speed of which is limited by solvent viscosity and internal friction arising from the roughness of the full energy landscape (12). It is noteworthy that energy landscape roughness has been implicated directly in causing nonexponential folding kinetics (5, 6). Similarly, the experimentally observed increase of the transition state transit time at higher temperatures was explained by stronger internal friction due to increased energy landscape roughness (13, 14).
Fig. 1.
Rugged energy landscape (schematic). (A) One-dimensional representation of the multidimensional potential energy landscape in conformational space, encompassing two large-scale minima (basins) corresponding to significantly different conformations and multiple local minima of varying depths, including conformations with short (solid arrows) and long (dotted arrows) trapping times. The Inset indicates the hierarchy of local minima with different depths on different length scales. It should be noted that only minima with a depth >kBT will significantly affect motion over the landscape. Also indicated is the jump activation barrier, E, for one particular minimum. (B) The exponential distribution, g(E), of E with a width of 3.5 kBT, which is indicated by our experimental results (solid line), compared with a distribution with a width of kBT (dotted line), which would yield normal diffusion of the polypeptide backbone, both normalized to 1 at E = 0; the hatched area highlights the thermal energy kBT. Also indicated are the trapping times associated with E, with the double-sided arrows corresponding to a range of the preexponential factor of 1011 to 1013 s−1.
Some progress has been made in experimentally observing motion over the energy landscape on short timescales. In particular, intrapeptide contact formation has been investigated extensively using fluorescence and triplet quenching (15–19). These experiments observe intrachain loop formation from an equilibrium distribution of initial conformations and provide important information on polypeptide backbone diffusion, including the effects of excluded volume and chain stiffness. Nonexponential dynamics were observed on the picosecond timescale and were assigned to motion of the polypeptide backbone within a local basin of the energy landscape (17). Diffusion over larger distances in conformational space involves transitions over higher energy barriers separating the local basins and has been observed on the nano- to microsecond timescale. Structural changes corresponding to transitions over even larger barriers, e.g., those due to formation of secondary and tertiary structure, should occur on longer timescales but are not accessible with methods whose timescale is limited by the intrinsic lifetime of the reporter state (18). More recently, single-molecule fluorescence correlation spectroscopy was used, which in principle extends the time window to milliseconds but is affected by microsecond triplet blinking (20). Similarly, ligand-binding experiments extend the time window to milliseconds but are limited to specific proteins (21).
An alternative approach for the observation of backbone dynamics is described here; it uses geminate recombination of a photocleaved aromatic disulfide bond that initially holds the protein in a nonnative conformation (Fig. 2) (22, 23). This method observes the opposite of loop formation, namely the separation of two residues that initially are in close contact. As the residues separate as the result of backbone motion, their encounter probability decreases, which leads to a decreasing rate of geminate recombination. Thus, the decay of the transient thiyl radical concentration, which is measured readily using their strong absorbance near 500 nm (22), may be used to monitor the dynamics of the polypeptide backbone. Whereas quenching experiments observe loop formation in equilibrium, which excludes all conformations that lie higher than a few kBT on the potential energy landscape (where kB is the Boltzmann constant and T the temperature), disulfide recombination monitors processes far from equilibrium, providing complementary information, importantly with no intrinsic time limit.
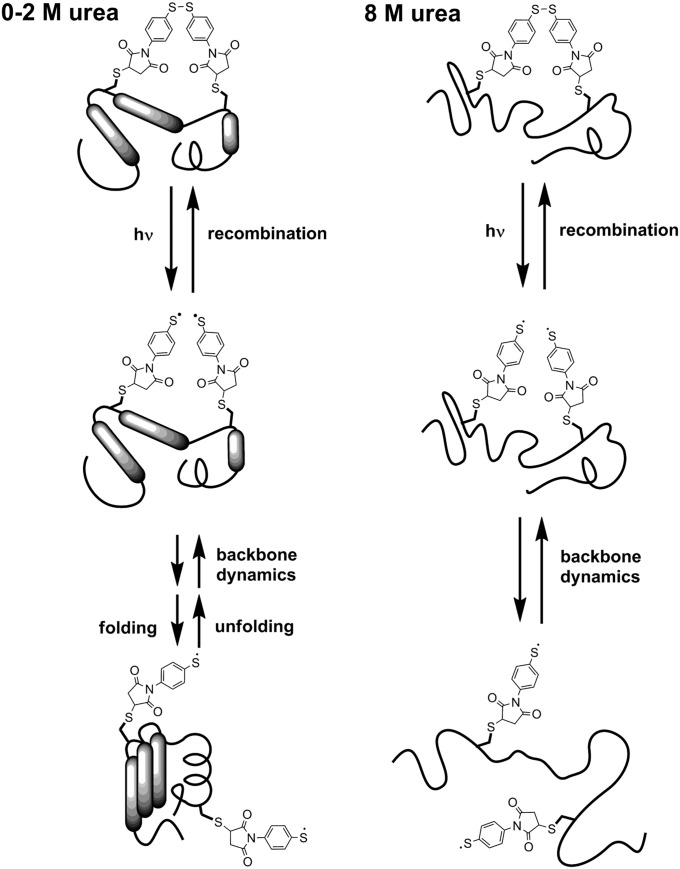
Fig. 2.
Schematic representation of the processes following disulfide bond photolysis of cross-linked proteins. Following cross-link photolysis, recombination to the original disulfide bond competes with backbone motions that separate the thiyl radicals and lead to folding of the protein (0–2 M urea) or to full equilibration between all unfolded conformations (8 M urea).
The disulfide recombination method requires an aromatic disulfide bond because nonaromatic thiyl radicals do not recombine fast enough to report on backbone dynamics (24). Consequently, this approach has been used only for short peptides (22, 23) that can be synthesized using solid-phase techniques. Although advances in nonnatural amino acid mutagenesis and chemical ligation methods allow the functionalization of proteins with photosensitive moieties, examples of intramolecular photosensitive cross-linkers still are rare. We recently introduced an aromatic disulfide cross-link into a 174-amino acid protein (the N-terminal domain of phosphoglycerate kinase from Geobacillus steareothermophylus, N-PGK) (25). At low denaturant concentrations (0–2 M urea), the cross-linked protein has a molten-globule–like conformation with reduced tertiary structure and reduced stability (SI Appendix, Appendix II). When the disulfide cross-link is cleaved by addition of a reducing agent, the protein folds into the native structure (25). Thus, photolysis of the disulfide bond at low urea concentrations is expected to trigger structural changes of the protein from the partially unfolded toward the folded structure (Fig. 2). At 8 M urea, both the cross-linked and the open protein are denatured, but photolysis of the disulfide bond will alter the range of conformations accessible at equilibrium (25). Here, we report the recombination dynamics of the thiyl radicals after photolysis of cross-linked N-PGK, for both low and high concentrations of urea, and show that their reencounter probability decreases with time following a universal power law over nine orders of magnitude in time, from picoseconds to milliseconds, regardless of primary sequence or the presence of secondary or tertiary structure. Model simulations show that this power law arises from subdiffusional motion of the residues, indicating a wide distribution of trapping times in local minima of the energy landscape and enabling us to quantify the energy landscape roughness. Furthermore, we suggest that the subdiffusional motion of the protein backbone is a likely candidate for promoting rapid folding, particularly of proteins with low contact order, by enhancing contact formation between nearby residues.
Results
Thiyl Radical Recombination Dynamics.
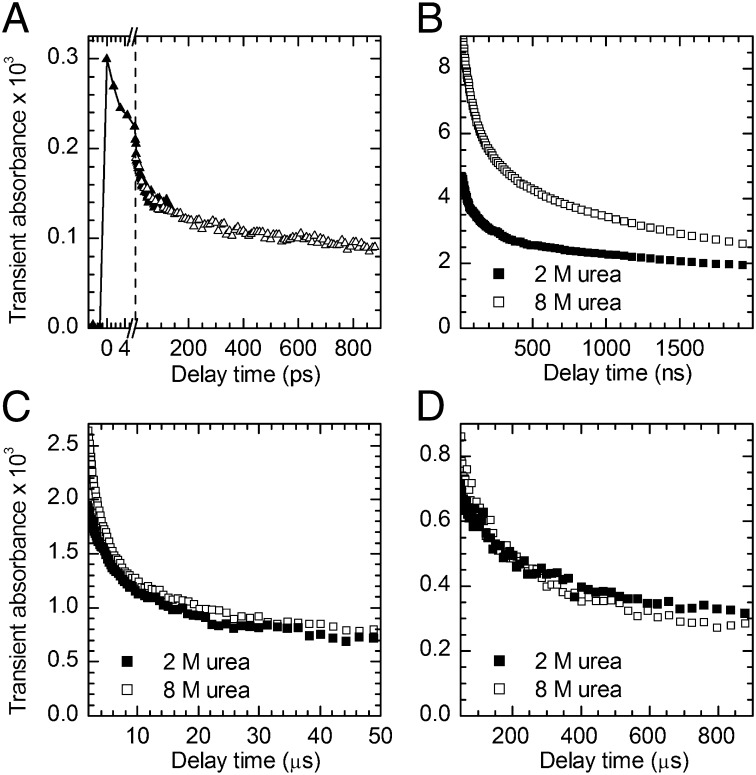
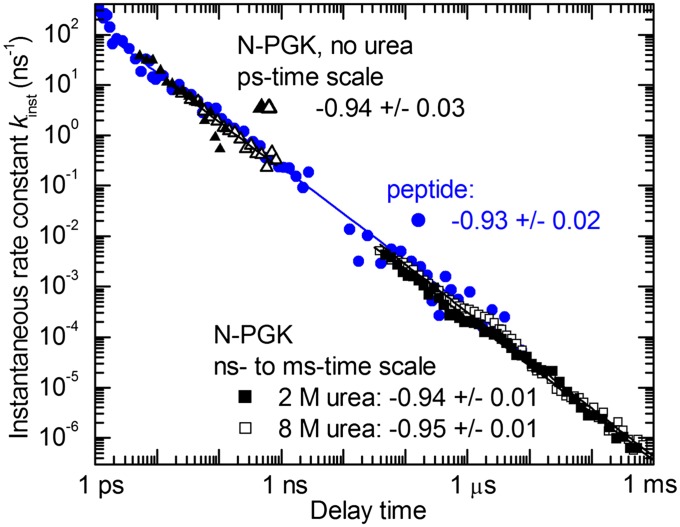
The transient absorbance near 500 nm, which is a direct measure of thiyl radical concentration (22), was recorded from 1 ps to 1 ms after UV photolysis of cross-linked N-PGK (Fig. 3). The fast appearance of the absorbance, Fig. 3A, confirms aromatic disulfide bond splitting within 1.5 ps after absorption of a UV photon, as found in model aromatic disulfides (22, 26). The decay of this transient absorbance shows that, initially, geminate recombination is very fast because of the radicals being generated in close proximity. As the protein relaxes toward its new equilibrium conformation, the radicals are separated to increasingly larger average distances. On longer timescales and for low concentrations of urea, the radicals may be held apart further by the formation of secondary and tertiary structure. Both effects lead to a decrease of the probability of a reencounter, and hence to a slowing of the recombination of the surviving thiyl radicals. As a consequence, thiyl radical recombination spans many orders of magnitude in time. Fig. 4 shows that the instantaneous rate constant for recombination, kinst(t) = −[dc(t)/dt]/c(t), where t is the time after photolysis and c(t) is the surviving radical concentration, follows a highly unusual power law, kinst(t) ∼ t−0.94(±0.03), over the full experimental time window. Most intriguingly, the same power law is found in the absence of significant secondary or tertiary structure (8 M urea) as under conditions of persistent native-like secondary structure (0–2 M urea).† Although the timescales that were investigated are shorter (picoseconds to a few microseconds), the same power law also had been found for synthetic α-helical peptides (Fig. 4) (22). Our results on N-PGK now show that the kinst(t) ∼ t−0.94 power law is an intrinsic property of the polypeptide backbone and does not depend on details of sequence or secondary and tertiary structure.
Fig. 3.
Transient thiyl radical absorbance decay after UV photolysis of cross-linked N-PGK. (A) Absorbance changes at 560 nm on the picosecond timescale, in the absence of urea; shown are two separate measurements, indicated by △ and ▲ symbols, scaled to account for different experimental conditions (note the scale break indicated by the dashed line). (B–D) Absorbance changes at 488 nm on the nanosecond–millisecond timescale in the presence of 2 M and 8 M urea, respectively.
Fig. 4.
Time dependence of the instantaneous rate constant kinst(t) for thiyl radical recombination after photolysis of an aromatic disulfide cross-linker. Shown are the results for N-PGK (black), two separate measurements on the picosecond timescale in the absence of urea and measurements on the nano- to millisecond timescale in the presence of 2 M and 8 M urea, respectively, and α-helical model peptides (22) (blue). Solid lines are the results of power law fits; also given are the powers obtained from these fits.
Simulation of Residue Encounter Probability.
To identify the origin of this dynamic behavior, we undertook a series of numerical simulations, incorporating various aspects of polypeptide dynamics. These simulations calculate the time-dependent survival probability, P(t), of two radicals that initially are in close contact and undergo diffusion-controlled geminate recombination. From P(t), the instantaneous rate constant kinst(t) = −[dP(t)/dt]/P(t) can be calculated and compared with the experimental results. Full details of the simulations and their results are given in SI Appendix, Appendix III.
For normal diffusion, the mean square displacement of a residue undergoing random walk increases linearly with time, <r2(t)> ∼ t. For untethered radical pairs undergoing normal diffusion, the solution for the time-dependent survival probability during geminate recombination can be expressed in analytical closed form (29), which agrees well with experimental results for aromatic thiyl radicals (30). The time dependence of kinst(t) is determined by the diffusion-controlled probability of a reencounter between the two radicals at the radical pair contact distance at which recombination takes place. Initially, only diffusion in one dimension (away from each other) is of significance, yielding a power law kinst(t) ∼ t−0.5; this turns into kinst(t) ∼ t−1.5 once diffusion has taken place over a length scale comparable to the radical pair contact distance, when diffusion in all three dimensions becomes relevant. With typical values for backbone diffusion coefficients from quenching experiments (15–18) and aromatic thiyl contact distances from experiments on model compounds (30), this transition is estimated to occur on the nanosecond timescale (SI Appendix, Fig. S3). However, this neglects the tethering effect of the polypeptide chain that links the two radicals, which can be approximated by assuming the radicals diffuse under the constraints of a harmonic potential. Numerical simulations of the survival probability of geminately recombining radicals under this constraint show the same initial behavior as for freely diffusing radicals, with a transition from kinst(t) ∼ t−0.5 to kinst(t) ∼ t−1.5 on the nanosecond timescale. This is followed by conformational equilibration in the potential within ∼1 μs, when kinst(t) becomes time independent (SI Appendix, Fig. S5).
Other important aspects of the dynamic behavior of the polypeptide backbone are chain stiffness and excluded volume effects. These effects tend to disfavor conformations with the radicals in close contact, and thus might be expected to lead to faster initial separation of the radicals. We accounted for these effects by numerically simulating radical diffusion and recombination in various modified potentials derived from theoretical models, simulations, or experimental results for the equilibrium end-to-end distance distribution of short peptide sections. Full details and mathematical expressions of these potentials, as well as the results of our simulations, are given in SI Appendix, Appendix III. All these simulations yielded only minor modifications of the general recombination behavior in a harmonic potential (SI Appendix, Figs. S6–S10) and thus failed to reproduce the experimental behavior of kinst(t). Therefore, the observation that kinst(t) ∼ t−0.94 over the full time range from picoseconds to milliseconds is not consistent with normal diffusion of the peptide sections to which the radicals are bound, even when accounting for tethering, chain stiffness, and excluded volume effects.
On the other hand, simulations in which the peptide residues are assumed to follow “subdiffusional” behavior successfully account for the unusual power law, kinst(t) ∼ t−0.94. Subdiffusion means that the mean square displacement of an unconstrained particle is nonlinear in time, <r2(t)> ∝ tα with α < 1, and implies quasi-random motion with a wide distribution of trapping times (31). Our subdiffusion simulations use the results by Seki et al. (32) for the survival probability, P(t), of a pair of particles undergoing subdiffusive motion that are subject to geminate recombination. Subdiffusive motion is simulated using a random walk model with a fixed jump size and a broad waiting time distribution, which results from an activated jump rate, γ(E) = γr exp(−E/kBT), with an exponential distribution of the jump activation energy, E, g(E) ∼ exp(−αE/kBT) (Fig. 1B). Here, γr is the maximum (nonactivated) jump rate and kBT/α characterizes the width of the jump activation energy distribution g(E), which has been shown to result in subdiffusive behavior, i.e., the mean square displacement is nonlinear in time: <r2(t)> ∝ tα. Further mathematical details are given in SI Appendix, Appendix III.
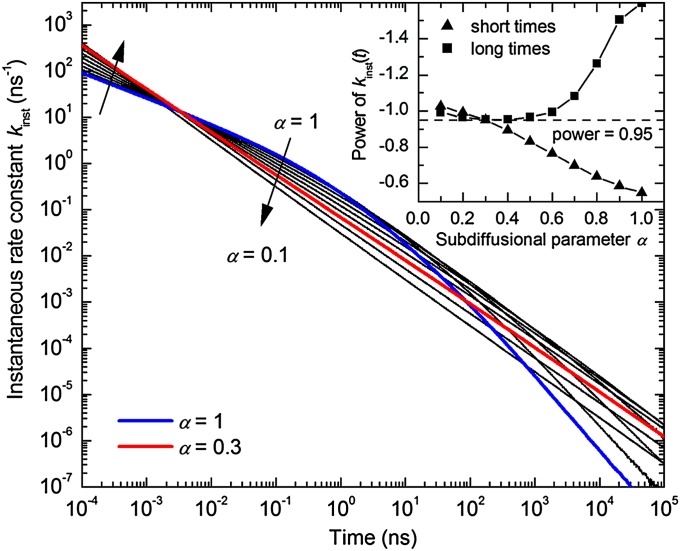
Fig. 5 shows typical results for the instantaneous rate constant, kinst(t), obtained from these simulations for different values of the subdiffusional parameter α. As expected, for normal diffusion (α = 1), this model yields the same result as described above, i.e., an initial power law kinst(t) ∼ t−0.5 that turns into kinst(t) ∼ t−1.5 at later times, with the transition time determined by the radical pair contact distance. Upon decreasing α, i.e., for increasingly subdiffusional behavior, the power of kinst(t) for the initial phase increases, whereas that for the later phase decreases. When α is around 0.3, the two phases of kinst(t) merge into a single power law, kinst(t) ∝ t−0.95. This time dependence holds over at least nine orders of magnitude in time in our simulations and closely resembles the experimental observation (Fig. 4); the results do not significantly depend on the other model parameters used in the simulations (SI Appendix, Fig. S11). We conclude that the experimentally observed kinst(t) ∼ t−0.94 power law is the result of subdiffusional motion of the residues holding the thiyl radicals after disulfide bond photolysis.
Fig. 5.
Simulated time dependence of the instantaneous rate constant, kinst(t), for geminate recombination of thiyl radicals undergoing subdiffusive motion, for values of the subdiffusion parameter α ranging from 1 (normal diffusion) to 0.1 in steps of 0.1. Model parameters: initial pair separation r0 = 7.2 Å, contact distance σ = 7.2 Å, and diffusion constant D = 4 Å2/ns (SI Appendix, Appendix III). Highlighted are the curves for α = 1 (blue) and α = 0.3 (red). (Inset) Power of the kinst(t) time dependence obtained from power law fits at short and long times, respectively, for different values of the subdiffusion parameter α.
These simulations ignore the tethering effect of the polypeptide backbone. As in the case of normal diffusion, it is expected that the radicals eventually reach an equilibrium distribution and that the instantaneous rate constant for recombination, kinst(t), then does not change any further with time. In principle, this might be simulated by assuming subdiffusive motion in a harmonic potential, which may be described by the fractional Fokker–Planck equation (33), although this is beyond the scope of the current paper. However, it has been shown that the approach to equilibrium for this situation is significantly slower than in the case of normal diffusion (33); it is reasonable to assume that this slower equilibration in the case of subdiffusion contributes to the fact that no leveling off of kinst(t) is observed in our experimental data, even at the longest timescale (1 ms).
Discussion
Roughness of the Protein Energy Landscape.
Our results show that the polypeptide backbone in proteins is not subject to normal diffusional behavior. Instead, a subdiffusional model is required to account for the experimental observations. Subdiffusional behavior of certain aspects of polypeptide backbone dynamics has been reported on the pico- to nanosecond timescale from neutron-scattering experiments (34) and molecular dynamics simulations (35), and on the millisecond to second timescale from single-molecule electron transfer measurements (36). However, these results refer to small-scale equilibrium fluctuations, and it is not clear to what extent their conclusions can be extrapolated to large-scale nonequilibrium conformational changes. Our experimental results were obtained under nonequilibrium conditions and thus show that intraprotein subdiffusion not only occurs during small-scale equilibrium fluctuations, but also governs large-scale motions, such as those taking place during protein folding.
In general, subdiffusional behavior arises from the random motion of particles that encounter local traps with a wide distribution of trapping times (31). In the context of intraprotein residue subdiffusion, the distribution of trapping times is the result of a wide range of barrier heights (or depths of local minima) on the conformational energy landscape (37) (Fig. 1); as the polypeptide backbone relaxes toward the new equilibrium distribution following photolysis of the disulfide bond, some proteins become trapped in local minima that hold the radicals apart, preventing their rapid recombination. Thus, our results yield experimental confirmation of the existence and relevance of a wide range of barrier heights, due to a rugged potential energy landscape, even during large-scale protein conformational changes. Furthermore, the observation that good agreement between the simulated time dependence of kinst(t) and the experimentally observed power law, kinst(t) ∝ t−0.94, is found for a value of α near ∼0.3 (Fig. 5) allows us to estimate the width of the barrier height distribution and thus the roughness of the potential energy landscape. Assuming an exponential distribution of the jump activation barriers, as used in the model by Seki et al. (32), a value of α near ∼0.3 corresponds to an average barrier height <E> ∼3–4 kBT, and a root-mean-squared roughness, ε = <E2>1/2, which is the parameter normally used to quantify roughness, of ∼4–5 kBT.
Potential transient interactions that cause local minima of the energy landscape are hydrogen bonding, electrostatic interactions between ionized or polar residues, and hydrophobic interactions between nonpolar residues, including van der Waals forces and π–π aromatic interactions. Protein engineering experiments indicate that a single hydrophobic side chain contact, such as a Phe–Phe aromatic interaction, contributes about 5 kJ/mol (2 kBT) to the stability of a folded protein (38); a single hydrogen bond contributes about 2–7 kJ/mol (∼1–3 kBT) (39) and a surface-exposed salt bridge contributes about 2 kJ/mol (∼ kBT) (40). These values are of the same order of magnitude as the landscape roughness determined here, which suggests that many local minima may arise from a single interaction. However, because a broad distribution of trapping energies is required to account for subdiffusional behavior, a significant fraction of the minima is deeper than 2 kBT, and thus must be a result of the combined effect of two or more such interactions.
The roughness of the folding energy landscape found here is somewhat smaller than the roughness of protein–ligand interaction landscapes (5–8 kBT) of bimolecular protein complexes (41) and similar to the roughness of bacteriorhodopsin (4–6 kBT) (42) suggested by the temperature dependence of single-molecule dynamic force spectroscopy data. Similar results on filamin might be interpreted as arising from a landscape roughness of 4 kBT, although a completely different interpretation of the experimental observations is preferred by the authors (43), raising some uncertainty over the interpretation of such force-stretching experiments. Roughness values of 1.3–2.6 kBT were suggested for the folding energy landscape from slow backbone diffusion observed in unfolded proteins (27, 28), but these values are based on problematic estimates of the speed of diffusion of the backbone in the absence of roughness. From similar observations, a roughness of 1.7 kBT was suggested for unstructured oligopeptides (15), which may reflect weak residue–residue interactions in such peptides. It must be noted that all these estimates of energy landscape roughness are based on the theory of diffusion in a one-dimensional rough potential (44), which normally is assumed to be the projection of the multidimensional potential onto the reaction coordinate. However, motion of a protein does take place on a multidimensional landscape, which modifies the diffusion behavior; for example, high barriers can be circumvented, which is not possible in a one-dimensional potential, and it is not clear to what extent the use of the one-dimensional diffusion theoretical treatment may distort these results. In contrast, the roughness parameter determined here directly refers to the local variation of the potential energy of the multidimensional energy landscape; in any case, this may not be the same as the roughness of the idealized reaction potential, which effectively averages over many conformations and microscopic reaction pathways.
Deep Traps on the Protein Energy Landscape.
Subdiffusion arising from a wide range of trapping times should persist only up to timescales corresponding to the longest trapping times (31), and normal diffusional behavior should be found at longer times. Our experiments show that in N-PGK, these trapping times extend up to milliseconds. Such trapping times suggest a barrier of the order of 50 kJ/mol (20 kBT), assuming a typical value for the preexponential factor in the range 1011 to 1013 s−1. As detailed above, individual residue–residue interaction energies have values not greater than ∼5 kJ/mol. This indicates that although the average barrier height is of the order 3–4 kBT, proteins temporarily adopt conformations that are stabilized by multiple interactions, leading to deep minima on the energy landscape. The range of long trapping times observed in these experiments therefore implies that the protein samples a diverse ensemble of states that contain multiple native or nonnative interactions.
Conformational dynamics on the millisecond timescale also are accessible from NMR measurements. Under denaturant conditions comparable to our 2 M urea experiments, a collapsed, compact state of N-PGK with some nonnative secondary structure and many nonnative tertiary contacts rapidly forms and is substantially populated before folding to the native state (45). Population of this molten globule state leads to NMR line broadening, which indicates that the component species have trapping times on the millisecond timescale (46). Indeed, the NMR line broadening (47) and complex interconversion behavior over a wide range of timescales (48) that are typical features of molten globule states are readily accounted for by subdiffusional behavior on a rugged energy landscape, as proposed here.
Significantly, we also observe subdiffusional behavior with trapping times up to milliseconds in N-PGK at denaturant concentrations for which the protein does not fold and population of the collapsed state no longer is detectable by NMR (8 M urea). This indicates that even under denaturing conditions, the energy landscape of real proteins remains rugged, to the extent of providing energy minima having a depth of ∼20 kBT, despite its large-scale features being completely different from those found under folding conditions (a wide basin encompassing the multitude of unfolded conformations vs. a deep minimum at the native structure).
Our observations substantially extend the timescale of the relevance of hierarchical energy landscapes in unfolded proteins. Previously, these had been observed only for short oligopeptides (17), in which they are limited to the sub-nanosecond timescale and full conformational equilibration is achieved within 1 μs (15–17) because the peptide design excluded strong residue–residue interactions. Close inspection of triplet-quenching experiments on denatured proteins shows slight indications of nonexponential dynamics on the 10–100-μs timescale that might indicate deeper traps (18). The existence of significant interactions in unfolded proteins had been suggested previously on the basis of photochemically induced dynamic nuclear polarization NOE (49) and paramagnetic relaxation enhancement (50) NMR experiments and from the low value and unusual denaturant dependence of their intrapolymer diffusion constant (18). Similarly, the radius of gyration of the unfolded state of staphylococcal nuclease indicated the formation of transient hydrophobic clusters (51). Our results now confirm that even under conditions in which they are assumed to be fully unfolded, proteins may become trapped in a wide range of states, some of which are long-lived and stabilized by multiple (nonnative) interactions.
Relevance for Protein Folding.
In general, the roughness of the energy landscape determines the speed at which the protein moves over this landscape and therefore is relevant for the dynamics of large-scale structural changes, such as those occurring during protein folding, as well as the rate of small-scale conversion between different conformations at or near equilibrium, such as those required for ligand binding. Quantitative information on this parameter so far has been rare. Here, we report experimental results indicating significant overall roughness (∼4–5 kBT) as well as the existence of deep traps (>20 kBT), even in denatured proteins, most likely from multiple nonnative interactions; this energy roughness results in strongly subdiffusional behavior of the polypeptide backbone, i.e., the mean square displacement of a polypeptide segment is nonlinear in time, <r2(t)> ∝ tα with α << 1. Any theoretical treatment of internal polypeptide motion must account for this behavior. In particular, this may have important consequences for the theoretical treatment of one-dimensional diffusion along the (folding) reaction coordinate, e.g., for estimating reaction rates using Kramers’ theory (12) or simulating diffusion in idealized potentials (8, 13). Currently, such simulations include the roughness of the multidimensional energy surface by assuming an effective friction coefficient; an investigation of the extent to which subdiffusional behavior (in 3D real space) does affect the validity of this approach is beyond the scope of the current paper.
The role of transient nonnative interactions in unfolded proteins for the speed of folding was discussed previously. Such nonnative contacts may bring closer together backbone sections that are far from each other in the primary sequence but close in the native structure, thus accelerating their folding. Such interactions are thought to be responsible for “abnormal” Φ-values (52). More specifically, lattice model simulations directly suggested that they could significantly enhance the rate of folding (52). Folding kinetics experiments using mutations of surface hydrophobic residues are in agreement with this proposal (53). Our observation of the existence of such interactions even in denatured proteins yields further support for this suggestion.
It is even more intriguing to speculate about the role intraprotein subdiffusion may play in promoting fast folding of proteins with low contact order, i.e., those in which many native interactions are formed by residues that are close in the primary sequence. Compared with normal diffusion, subdiffusion enhances the probability of encountering a nearby interaction partner within a given time (54). Thus, protein structures with low contact order are even more likely to form their native interactions rapidly if the residues undergo subdiffusional motion than if they followed normal diffusion. Subdiffusion of the backbone as a result of the highly rugged landscape, therefore, may be an important contributing factor for the observed correlation between the folding times of small proteins and their contact order (55).
Concluding Remarks.
We have presented a unique approach for characterizing the roughness of a protein’s potential energy landscape. Geminate recombination following photolysis of an aromatic disulfide bond provides evidence for subdiffusional behavior of the protein backbone, with the mean square displacement of residues showing sublinear time dependence, <r2(t)> ∼ tα, with α ∼0.3, over nine orders of magnitude in time, from picoseconds to milliseconds. This observation shows the existence of a wide range of trapping times and provides a measure of the roughness of the potential energy landscape, which was found to be of the order 4–5 kBT. However, there are potential energy minima in proteins, stabilized by multiple interactions, which are far deeper than this and extend at least up to 20 kBT. Most intriguingly, both these results also were observed under denaturing conditions.
Materials and Methods
Disulfide cross-linked N-PGK in 50 mM sodium phosphate buffer, pD 6.6–7, 100 mM NaCl was prepared as described in ref. 25 and urea added as indicated. The addition of 2 M urea has no effect on the secondary structural content of the open form of N-PGK, and only a small effect on that of the cross-linked form (SI Appendix, Fig. S1). For measurements on the picosecond timescale, disulfide photolysis was achieved with 100-fs laser pulses at 275 nm and absorbance changes were probed using optically delayed 100-fs laser pulses at 560 nm. Measurements on slower timescales used 5-ns laser pulses at 266 nm for disulfide photolysis; continuous light and a fast detector were used for probing absorbance changes. Sample concentration and excitation energy were lower for measurements on the picosecond timescale than for those on the nanosecond–millisecond timescale. For all measurements, samples were flowed or rotated continuously to avoid photodamage; no signal degradation was observed during the course of the measurements. Dynamics of the transient absorbance changes within the first few picoseconds may be affected by dielectric and vibrational relaxation (22, 26) and were excluded from further analysis. See SI Appendix, Appendix I for full experimental details.
Supplementary Material
Acknowledgments
The authors thank Dr. Julia Weinstein (University of Sheffield) for the loan of an Ar-ion laser. Financial support from the Biotechnology and Biological Sciences Research Council is gratefully acknowledged.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
†kinst values for N-PGK in the absence and presence of denaturant differ by up to a factor of 2, which is difficult to see in Fig. 4, because the y-scale extends over nine orders of magnitude. This may be a result of the effect of denaturant on the intramolecular diffusion coefficient D (27, 28), although a change in D will have only weak effects on kinst(t), because separation of radicals, which competes with their recombination, and the reverse process, which is required for geminate recombination, are both diffusion controlled and thus slow down or accelerate in parallel.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1211764109/-/DCSupplemental.
References
- 1.Sosnick TR, Barrick D. The folding of single domain proteins—have we reached a consensus? Curr Opin Struct Biol. 2011;21(1):12–24. doi: 10.1016/j.sbi.2010.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McCarney ER, Kohn JE, Plaxco KW. Is there or isn’t there? The case for (and against) residual structure in chemically denatured proteins. Crit Rev Biochem Mol Biol. 2005;40(4):181–189. doi: 10.1080/10409230591008143. [DOI] [PubMed] [Google Scholar]
- 3.Krishna MMG, Englander SW. A unified mechanism for protein folding: Predetermined pathways with optional errors. Protein Sci. 2007;16(3):449–464. doi: 10.1110/ps.062655907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brockwell DJ, Radford SE. Intermediates: Ubiquitous species on folding energy landscapes? Curr Opin Struct Biol. 2007;17(1):30–37. doi: 10.1016/j.sbi.2007.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sabelko J, Ervin J, Gruebele M. Observation of strange kinetics in protein folding. Proc Natl Acad Sci USA. 1999;96(11):6031–6036. doi: 10.1073/pnas.96.11.6031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eaton WA. Searching for “downhill scenarios” in protein folding. Proc Natl Acad Sci USA. 1999;96(11):5897–5899. doi: 10.1073/pnas.96.11.5897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lindorff-Larsen K, Piana S, Dror RO, Shaw DE. How fast-folding proteins fold. Science. 2011;334(6055):517–520. doi: 10.1126/science.1208351. [DOI] [PubMed] [Google Scholar]
- 8.Shaw DE, et al. Atomic-level characterization of the structural dynamics of proteins. Science. 2010;330(6002):341–346. doi: 10.1126/science.1187409. [DOI] [PubMed] [Google Scholar]
- 9.Ansari A, et al. Protein states and proteinquakes. Proc Natl Acad Sci USA. 1985;82(15):5000–5004. doi: 10.1073/pnas.82.15.5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frauenfelder H, Sligar SG, Wolynes PG. The energy landscapes and motions of proteins. Science. 1991;254(5038):1598–1603. doi: 10.1126/science.1749933. [DOI] [PubMed] [Google Scholar]
- 11.Onuchic JN, Wolynes PG. Theory of protein folding. Curr Opin Struct Biol. 2004;14(1):70–75. doi: 10.1016/j.sbi.2004.01.009. [DOI] [PubMed] [Google Scholar]
- 12.Hagen SJ. Solvent viscosity and friction in protein folding dynamics. Curr Protein Pept Sci. 2010;11(5):385–395. doi: 10.2174/138920310791330596. [DOI] [PubMed] [Google Scholar]
- 13.Liu F, Nakaema M, Gruebele M. The transition state transit time of WW domain folding is controlled by energy landscape roughness. J Chem Phys. 2009;131(19):195101. doi: 10.1063/1.3262489. [DOI] [PubMed] [Google Scholar]
- 14.Cellmer T, Henry ER, Hofrichter J, Eaton WA. Measuring internal friction of an ultrafast-folding protein. Proc Natl Acad Sci USA. 2008;105(47):18320–18325. doi: 10.1073/pnas.0806154105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lapidus LJ, Eaton WA, Hofrichter J. Measuring the rate of intramolecular contact formation in polypeptides. Proc Natl Acad Sci USA. 2000;97(13):7220–7225. doi: 10.1073/pnas.97.13.7220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Möglich A, Krieger F, Kiefhaber T. Molecular basis for the effect of urea and guanidinium chloride on the dynamics of unfolded polypeptide chains. J Mol Biol. 2005;345(1):153–162. doi: 10.1016/j.jmb.2004.10.036. [DOI] [PubMed] [Google Scholar]
- 17.Fierz B, et al. Loop formation in unfolded polypeptide chains on the picoseconds to microseconds time scale. Proc Natl Acad Sci USA. 2007;104(7):2163–2168. doi: 10.1073/pnas.0611087104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Singh VR, Kopka M, Chen Y, Wedemeyer WJ, Lapidus LJ. Dynamic similarity of the unfolded states of proteins L and G. Biochemistry. 2007;46(35):10046–10054. doi: 10.1021/bi700270j. [DOI] [PubMed] [Google Scholar]
- 19.Soranno A, Longhi R, Bellini T, Buscaglia M. Kinetics of contact formation and end-to-end distance distributions of swollen disordered peptides. Biophys J. 2009;96(4):1515–1528. doi: 10.1016/j.bpj.2008.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nettels D, Hoffmann A, Schuler B. Unfolded protein and peptide dynamics investigated with single-molecule FRET and correlation spectroscopy from picoseconds to seconds. J Phys Chem B. 2008;112(19):6137–6146. doi: 10.1021/jp076971j. [DOI] [PubMed] [Google Scholar]
- 21.Hagen SJ, Carswell CW, Sjolander EM. Rate of intrachain contact formation in an unfolded protein: temperature and denaturant effects. J Mol Biol. 2001;305(5):1161–1171. doi: 10.1006/jmbi.2000.4366. [DOI] [PubMed] [Google Scholar]
- 22.Volk M, et al. Peptide conformational dynamics and vibrational Stark effects following photoinitiated disulfide cleavage. J Phys Chem B. 1997;101(42):8607–8616. [Google Scholar]
- 23.Volk M. Fast initiation of peptide and protein folding processes. Eur J Org Chem. 2001;2001(13):2605–2621. [Google Scholar]
- 24.Kolano C, Helbing J, Bucher G, Sander W, Hamm P. Intramolecular disulfide bridges as a phototrigger to monitor the dynamics of small cyclic peptides. J Phys Chem B. 2007;111(38):11297–11302. doi: 10.1021/jp074184g. [DOI] [PubMed] [Google Scholar]
- 25.Milanesi L, et al. A method for the reversible trapping of proteins in non-native conformations. Biochemistry. 2008;47(51):13620–13634. doi: 10.1021/bi801362f. [DOI] [PubMed] [Google Scholar]
- 26.Ernsting NP. Solvation of photolytically generated p-aminophenylthiyl radicals studied by sub-picosecond transient absorption. Chem Phys Lett. 1990;166(3):221–226. [Google Scholar]
- 27.Nettels D, Gopich IV, Hoffmann A, Schuler B. Ultrafast dynamics of protein collapse from single-molecule photon statistics. Proc Natl Acad Sci USA. 2007;104(8):2655–2660. doi: 10.1073/pnas.0611093104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Waldauer SA, Bakajin O, Lapidus LJ. Extremely slow intramolecular diffusion in unfolded protein L. Proc Natl Acad Sci USA. 2010;107(31):13713–13717. doi: 10.1073/pnas.1005415107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shin KJ, Kapral R. Kinetic theory of reactive pair dynamics in liquids. J Chem Phys. 1978;69(8):3685–3696. [Google Scholar]
- 30.Scott TW, Liu SN. Picosecond geminate recombination of phenylthiyl free-radical pairs. J Phys Chem. 1989;93(4):1393–1396. [Google Scholar]
- 31.Metzler R, Klafter J. The restaurant at the end of the random walk: Recent developments in the description of anomalous transport by fractional dynamics. J Phys Math Gen. 2004;37(31):R161–R208. [Google Scholar]
- 32.Seki K, Wojcik M, Tachiya M. Recombination kinetics in subdiffusive media. J Chem Phys. 2003;119(14):7525–7533. [Google Scholar]
- 33.Metzler R, Klafter J. The random walk’s guide to anomalous diffusion: A fractional dynamics approach. Phys Rep. 2000;339(1):1–77. [Google Scholar]
- 34.Paciaroni A, et al. Coupled relaxations at the protein-water interface in the picosecond time scale. J R Soc Interface. 2009;6(Suppl 5):S635–S640. doi: 10.1098/rsif.2009.0182.focus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Senet P, Maisuradze GG, Foulie C, Delarue P, Scheraga HA. How main-chains of proteins explore the free-energy landscape in native states. Proc Natl Acad Sci USA. 2008;105(50):19708–19713. doi: 10.1073/pnas.0810679105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang H, et al. Protein conformational dynamics probed by single-molecule electron transfer. Science. 2003;302(5643):262–266. doi: 10.1126/science.1086911. [DOI] [PubMed] [Google Scholar]
- 37.Luo G, Andricioaei I, Xie XS, Karplus M. Dynamic distance disorder in proteins is caused by trapping. J Phys Chem B. 2006;110(19):9363–9367. doi: 10.1021/jp057497p. [DOI] [PubMed] [Google Scholar]
- 38.Serrano L, Bycroft M, Fersht AR. Aromatic-aromatic interactions and protein stability. Investigation by double-mutant cycles. J Mol Biol. 1991;218(2):465–475. doi: 10.1016/0022-2836(91)90725-l. [DOI] [PubMed] [Google Scholar]
- 39.Fersht AR. The hydrogen bond in molecular recognition. Trends Biochem Sci. 1987;12(1):301–304. [Google Scholar]
- 40.Horovitz A, Serrano L, Avron B, Bycroft M, Fersht AR. Strength and co-operativity of contributions of surface salt bridges to protein stability. J Mol Biol. 1990;216(4):1031–1044. doi: 10.1016/S0022-2836(99)80018-7. [DOI] [PubMed] [Google Scholar]
- 41.Rico F, Moy VT. Energy landscape roughness of the streptavidin-biotin interaction. J Mol Recognit. 2007;20(6):495–501. doi: 10.1002/jmr.841. [DOI] [PubMed] [Google Scholar]
- 42.Janovjak H, Knaus H, Muller DJ. Transmembrane helices have rough energy surfaces. J Am Chem Soc. 2007;129(2):246–247. doi: 10.1021/ja065684a. [DOI] [PubMed] [Google Scholar]
- 43.Schlierf M, Rief M. Temperature softening of a protein in single-molecule experiments. J Mol Biol. 2005;354(2):497–503. doi: 10.1016/j.jmb.2005.09.070. [DOI] [PubMed] [Google Scholar]
- 44.Zwanzig R. Diffusion in a rough potential. Proc Natl Acad Sci USA. 1988;85(7):2029–2030. doi: 10.1073/pnas.85.7.2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reed MAC, et al. The denatured state under native conditions: A non-native-like collapsed state of N-PGK. J Mol Biol. 2006;357(2):365–372. doi: 10.1016/j.jmb.2005.12.080. [DOI] [PubMed] [Google Scholar]
- 46.Cliff MJ, et al. The denatured state of N-PGK is compact and predominantly disordered. J Mol Biol. 2009;385(1):266–277. doi: 10.1016/j.jmb.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 47.Redfield C. Using nuclear magnetic resonance spectroscopy to study molten globule states of proteins. Methods. 2004;34(1):121–132. doi: 10.1016/j.ymeth.2004.03.009. [DOI] [PubMed] [Google Scholar]
- 48.Mok KH, Nagashima T, Day IJ, Hore PJ, Dobson CM. Multiple subsets of side-chain packing in partially folded states of alpha-lactalbumins. Proc Natl Acad Sci USA. 2005;102(25):8899–8904. doi: 10.1073/pnas.0500661102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mok KH, et al. A pre-existing hydrophobic collapse in the unfolded state of an ultrafast folding protein. Nature. 2007;447(7140):106–109. doi: 10.1038/nature05728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Felitsky DJ, Lietzow MA, Dyson HJ, Wright PE. Modeling transient collapsed states of an unfolded protein to provide insights into early folding events. Proc Natl Acad Sci USA. 2008;105(17):6278–6283. doi: 10.1073/pnas.0710641105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schroer MA, et al. High-pressure SAXS study of folded and unfolded ensembles of proteins. Biophys J. 2010;99(10):3430–3437. doi: 10.1016/j.bpj.2010.09.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li L, Mirny LA, Shakhnovich EI. Kinetics, thermodynamics and evolution of non-native interactions in a protein folding nucleus. Nat Struct Biol. 2000;7(4):336–342. doi: 10.1038/74111. [DOI] [PubMed] [Google Scholar]
- 53.Viguera AR, Vega C, Serrano L. Unspecific hydrophobic stabilization of folding transition states. Proc Natl Acad Sci USA. 2002;99(8):5349–5354. doi: 10.1073/pnas.072387799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Guigas G, Weiss M. Sampling the cell with anomalous diffusion—the discovery of slowness. Biophys J. 2008;94(1):90–94. doi: 10.1529/biophysj.107.117044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Baker D. A surprising simplicity to protein folding. Nature. 2000;405(6782):39–42. doi: 10.1038/35011000. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.