Abstract
Male gametogenesis in plants can be impaired by an incompatibility between nuclear and mitochondrial genomes, termed cytoplasmic male sterility (CMS). A sterilizing factor resides in mitochondria, whereas a nuclear factor, Restorer-of-fertility (Rf), restores male fertility. Although a majority of plant Rf genes are thought to encode a family of RNA-binding proteins called pentatrico-peptide repeat (PPR) proteins, we isolated a novel type of Rf from sugar beet. Two BACs and one cosmid clone that constituted a 383-kbp contig covering the sugar beet Rf1 locus were sequenced. Of 41 genes borne by the contig, quadruplicated genes were found to be associated with specific transcripts in Rf1 flower buds. The quadruplicated genes encoded a protein resembling OMA1, a protein known from yeast and mammals to be involved in mitochondrial protein quality control. Construction of transgenic plants revealed that one of the four genes (bvORF20) was capable of restoring partial pollen fertility to CMS sugar beet; the level of restoration was comparable to that evaluated by a crossing experiment. However, the other genes lacked such a capability. A GFP-fusion experiment showed that bvORF20 encoded a mitochondrial protein. The corresponding gene was cloned from rf1rf1 sugar beet and sequenced, and a solitary gene that was similar but not identical to bvORF20 was found. Genetic features exhibited by sugar beet Rf1, such as gene clustering and copy-number variation between Rf1 and rf, were reminiscent of PPR-type Rf, suggesting that a common evolutionary mechanism(s) operates on plant Rfs irrespective of the translation product.
Keywords: cytoplasmic male sterility, Restorer of fertility, sugar beet, nuclear–mitochondrial interaction
AS a phenotypic manifestation of nuclear–mitochondrial incompatibility in plants, cytoplasmic male sterility (CMS) has garnered much interest and has been recorded to occur in >140 plant species (Laser and Lersten 1972). CMS is a maternally inherited trait that inactivates male reproductive function in otherwise normal plants (Schnable and Wise 1998). A genetic model developed to explain CMS suggests that it involves a nuclear–mitochondrial interaction in which a sterility-inducing factor (S) is generated in mitochondria, and one or more nuclear factors, termed restorers of fertility (Rf), capable of inhibiting the action of S (Hanson and Bentolila 2004). According to this model, plants with the S factor and two nonrestoring nuclear alleles, i.e., [S]rfrf, are male sterile (MS), whereas [S]RfRf or [S]Rfrf plants produce functional pollen (Budar et al. 2006; Chase 2007). Plants with N mitochondria lack the S factor and are male fertile irrespective of their nuclear alleles in the Rf locus.
Many S factors have been associated with various unique polypeptides encoded by mitochondrial genomes (Pelletier and Budar 2007). In some cases, the evolutionary origin of the S factor is unclear because the mitochondrial ORF that encodes the unique polypeptide (S-ORF) has no homology within the N mitochondrial genome or with any nucleotide sequences known to date. In other cases, S-ORFs appear to be mosaic of parts of duplicated mitochondrial genes, suggesting that S-ORFs are by-products of mitochondrial genome rearrangement (for reviews, Budar et al. 2004; Kubo and Newton 2008; Kubo et al. 2011).
Nuclear Rfs seem to overcome the action of S factors in different ways, but the mechanisms are obscure. One group of Rfs regulates the expression of S-ORFs at the post-transcriptional level (Fujii and Toriyama 2008). Plants having this type of Rf accumulated fewer S-ORF polypeptides with or without an altered level of S-ORF transcription. Molecular cloning of such Rfs from petunia (Petunia X hybrida hort. ex Vilm), radish (Raphanus sativus L.), and rice (Oryza sativa L.) revealed that these genes encode a class of proteins sharing a common sequence termed a pentatrico-peptide repeat (PPR) (Bentolila et al. 2002; Brown et al. 2003; Desloire et al. 2003; Kazama and Toriyama 2003; Koizuka et al. 2003; Akagi et al. 2004; Komori et al. 2004; Wang et al. 2006; Hu et al. 2012). These proteins constitute a large gene family that is associated with post-transcriptional gene regulation in plant organelles (Schmitz-Linneweber and Small 2008). A genetic association of Rf loci with PPR genes also has been reported from other plants such as CMS-S maize (Zea mays L.), sorghum [Sorghum bicolor (L.) Moench] and Mimulus (Klein et al. 2005; Xu et al. 2009; Barr and Fishman 2010; Jordan et al. 2010).
Rfs distinct from the PPR type, are known, but the current paucity of knowledge precludes further classification. Three non–PPR-type Rfs have been identified to date: maize Rf2a, rice Rf17, and rice Rf2. Maize Rf2a was the first Rf cloned, and encodes a mitochondrial aldehyde dehydrogenase (Cui et al. 1996). However, the functional relationship between URF13-T, a polypeptide encoded by the S-ORF in maize T-type CMS (Dewey et al. 1986), and RF2A proteins is unclear. Rice Rf17 was cloned as an Rf for CW-type CMS (Fujii and Toriyama 2009). The reduced expression of Rf17 in CW mitochondria compromises MS expression, thereby functionally acting as if Rf17 restored male fertility. It remains unknown whether any direct relationships exist between Rf17 and an, as yet, unidentified S-ORF in CW-CMS mitochondria. Genes for glycine-rich proteins have been isolated as rice Rf2 for Lead Rice (LD)-type CMS via map-based cloning (Itabashi et al. 2011). Hu et al. (2012) reported that a PPR-type RF protein, a glycine-rich protein, and a transcript encoding S factor are components of a large mitochondrial complex of 400–500 kDa in Hong-Lian (HL)-type CMS in rice.
Given its importance in hybrid seed production, sugar beet CMS has been extensively studied (Boutry et al. 1984; Lind et al. 1991; Hallden et al. 1992; Ducos et al. 2001). CMS mitochondria of sugar beet are characterized by a unique 39-kDa polypeptide encoded by an N-terminal extension of atp6 (preSatp6) that is missing in N mitochondria (Yamamoto et al. 2005). A precursor polypeptide consisting of preSATP6 and ATP6 is hypothesized to be cleaved into two separate polypeptides, one being the mature ATP6 polypeptide, and the other a preSATP6 polypeptide which subsequently forms a 200-kDa oligomer in the mitochondrial membrane. However, following fertility restoration, the amount of the preSATP6 polypeptide remained unchanged (Yamamoto et al. 2005), an observation that led us to postulate the involvement of a non–PPR-type Rf.
According to a genetic model proposed by Owen (1945), fertility restoration in sugar beet requires two independent genes, X and Z, of which the latter seemed less effective. Genetic mapping of X and Z located these genes on chromosomes III and IV, respectively (Pillen et al. 1993; Schondelmaier and Jung 1997; Hjerdin-Panagopoulos et al. 2002; Bosemark 2006). We previously found that pollen fertility segregated as if it were controlled by a single dominant gene when the sugar beet line NK–198 was used as a pollen parent (Hagihara et al. 2005a), although the level of fertility restoration varied depending on the nuclear genetic background (Hagihara et al. 2005a). The NK–198 Rf was named Rf1 and mapped to a terminal region of chromosome III, suggesting that the Rf1 was an allele of the X locus (Hagihara et al. 2005a). Molecular markers linked to Rf1 were used to isolate BAC clones that covered the Rf1 locus (Hagihara et al. 2005b).
In this study, the nucleotide sequence of a 383-kbp chromosomal region containing the sugar beet Rf1 was determined. From this sequence, we found that an unexpected gene satisfied the following criteria: specific transcription in Rf1 flower buds, partial fertility restoration to transgenic sugar beet (the level of restoration is comparable to that evaluated by a crossing experiment), and mitochondrial localization of the GFP-fused protein. The gene was related to yeast Oma1 known to be involved in quality control of mitochondrial proteins (Kaser et al. 2003). We also found an organizational similarity between sugar beet Rf1 locus and some PPR-type Rf loci in terms of gene clustering and copy-number variation between Rf1 and rf1, suggesting that a common evolutionary mechanism(s) operates on plant Rfs.
Materials and Methods
Plant materials
A restorer line NK–198, three maintainer lines TK–81mm–O, TA–33–O, and NK–219mm–O, and a CMS line NK–219mm–CMS used in this study were developed at the Hokkaido Agricultural Research Center, National Agriculture and Food Research Organization, Japan. Crosses were made by exchanging paper bags over the inflorescences in a greenhouse. Plants were vernalized for 4 months (5°, 24 hr/day) and flowered in the greenhouse. Anther tissues were sampled to examine pollen fertility on the day of anthesis. Pollen fertility was examined by Alexander staining (Alexander 1969).
Isolation of nucleic acids
Total cellular DNA of beet plants was isolated from fresh green leaves by the CTAB-based method described by Doyle and Doyle (1990). DNAs from BAC clones, cosmid clones, and plasmid clones were isolated by an alkaline lysis procedure (Sambrook et al. 1989). Lambda-phage DNA was isolated by a liquid culture method (Sambrook et al. 1989). Isolated DNA was purified by cesium chloride-ethidium bromide (CsCl-EtBr) equilibrium centrifugation when necessary. Total RNA from sugar beets was isolated according to Chomczynski and Sacchi (1987) or by using the RNeasy Plant Mini kit (Qiagen, Hilden, Germany). Residual DNA in the RNA sample was removed by DNase I (Takara Bio, Ohtsu, Japan) digestion in the presence of 8 mM MgCl2.
Subcloning into a cosmid vector
Purified BAC-clone DNA was partially digested with Sau3A I (Takara Bio), then electrophoresed in an agarose gel. DNA fragments of 30–50 kbp were eluted from the gel and partially filled to obtain a 5′-GA-3′ end (0.5 M Tris-HCl pH 7.5, 100 mM MgCl2, 10 mM dithiothreitol, 80 μM dATP, 80 μM dGTP, 2 units Klenow fragments, 30 min at room temperature) to prevent self-ligation. The cosmid vector pWE15 (Stratagene, La Jolla, CA) was completely digested with XhoI and then partially filled to obtain a 5′-TC-3′ end (0.5 M Tris-HCl pH 7.5, 100 mM MgCl2, 10 mM dithiothreitol, 80 μM dCTP, 80 μM dTTP, 2 units Klenow fragments, 30 min at room temperature) to prevent self-ligation. The ligation reaction was carried out using T4 DNA ligase (New England Biolabs, Beverly, MA) in the presence of 10% polyethylene glycol 8000. The ligated DNA sample was precipitated with ethanol and then dissolved in water. Gigapack III Gold (Stratagene) was used for packaging.
Construction of the shotgun library and nucleotide sequencing
Inserts of the lambda-phage clone were amplified with LA-Taq (Takara Bio) according to the instruction manual. Inserts of the cosmid clone were cut out by NotI digestion and recovered from gel slices after electrophoresis. The inserts or whole BAC-clone DNAs were randomly sheared by sonication and then electrophoresed in an agarose gel. DNA fragments of 1.2–1.5 and 2.0–2.5 kbp were eluted from the gel slices. The ends of DNA fragments were blunted by T4 DNA polymerase (Takara Bio) in the presence of dATP, dTTP, dCTP, and dGTP, and then ligated into the HincII site of pUC19. Plasmid DNA was sequenced using a LIC-4200L (Li-COR, Lincoln, NE) or ABI3130 (Applied Biosystems, Foster City, CA) sequencer.
Bioinformatics
Assembly of the nucleotide sequence was done using a Staden package (Staden 1996) and Sequencher 4.0 (Hitachi Software Engineering, Tokyo). Protein-coding regions were predicted by GENESCAN (Burge and Karlin 1997) (http://genes.mit.edu/GENSCAN.html) with an Arabidopsis matrix and the BLASTX program (http://www.ncbi.nlm.nih.gov/). A homology search for putative amino acid sequences was done using BLASTP on the National Center for Biotechnology Information (NCBI) website (http://www.ncbi.nlm.nih.gov/). Intracellular localizations were predicted using TargetP (Emanuelsson et al. 2000) (http://www.cbs.dtu.dk/services/TargetP/) and Predotar (Small et al. 2004) (http://urgi.versailles.inra.fr/predotar/predotar.html). A motif search was undertaken using Pfam (Finn et al. 2006) (http://pfam.sanger.ac.uk/). Repeated sequences were searched with Reputer (Kurtz et al. 2001) (http://bibiserv.techfak.uni-bielefeld.de/reputer/). Multiple sequences were aligned using ClustalW (Chenna et al. 2003) (http://clustalw.ddbj.nig.ac.jp/top-j.html). Nucleotide sequences reported in this study are deposited in the DNA Data Bank of Japan (DDBJ)/GenBank/EMBL under accession nos. AB646133 (4F1), AB646134 (5A3), AB646135 (33E19), and AB646136 (no. 10).
PCR and direct sequencing
Total cellular DNA (5–10 ng) was subjected to PCR amplification using LA-Taq (Takara Bio) or GoTaq Green Master mix (Promega, Madison, WI). Total RNA (2 μg) was reverse transcribed with the SuperScript III First-Strand Synthesis system (Invitrogen, Carlsbad, CA). The resultant cDNA was subjected to PCR amplification. Direct sequencing was achieved using an ABI3130 sequencer (Applied Biosystems).
Hybridization
Colony- and plaque-lift filters were prepared using Hybond N+ membranes (GE Healthcare, Amersham Place, UK) according to the instruction manual. For DNA gel blot analysis, a DNA sample (5 μg) was digested with restriction endonucleases purchased from Takara Bio and electrophoresed in a 1% agarose gel. After denaturation and neutralization, DNA fragments were transferred to Hybond N+ membranes according to the instruction manual. For RNA gel blot analysis, 5 μg RNA was electrophoresed in a 1.5% agarose gel containing 0.66 M formaldehyde and then transferred by capillary action to Hybond N+. The DNA fragment of interest was labeled with 32P using the Megaprime DNA labeling system (GE Healthcare) or with alkaline phosphatase using the AlkPhos Direct DNA labeling system (GE Healthcare). Hybridization was conducted according to the manufacturer’s instructions. Signal bands were detected on X-ray films or with an image analyzer (BAS2000; Fuji Photo Film, Tokyo).
Construction of GFP-fusion genes and transient assays
The pTH2 cloning vector, whose NcoI site includes the initiation codon for GFP, was used (Chiu et al. 1996). Gene segments of interest were PCR amplified with a set of primers, one bearing a SalI and the other an NcoI target sequence (see Supporting Information, Table S1) so that the amplified ORF could fuse in-frame with GFP. The resultant PCR fragments were digested with SalI and NcoI and then ligated into pTH2. A fluorescent signal in mitochondria resulted from the expression of an Arabidopsis F1-ATPase δ-subunit–RFP fusion protein expressed from pMt-R, a derivative plasmid of pWs (Arimura and Tsutsumi 2002). A PCR fragment corresponding to the first 58 amino acids of Arabidopsis RuBisCo activase was amplified and then substituted for the Arabidopsis F1-ATPase δ-subunit region of pMt-R. The resulting plasmid was designated pCp-R (Kitazaki et al. 2011). Plasmid DNA was ethanol precipitated with gold particles of 1 μm diameter (Bio-Rad Laboratories) and then introduced into the epidermal cells of onion bulbs or Welsh onion sheaths using a GIE-III IDERA system (Tanaka, Ishikari, Japan). The fluorescent signal was captured with a BX50 microscope system combined with a digital camera (DP70; Olympus, Tokyo).
Generation of transgenic sugar beets
Genomic DNA fragments containing bvORF19, bvORF20, and bvORF21 were PCR amplified from BAC clone 9C23 (see Table S1 for primer information). Using BP Clonase Enzyme mix (Invitrogen), the genomic DNA fragments were cloned into the donor vector, pDONRzeo, according to the manufacturer’s instruction manual. After verifying the sequence integrity, the inserted DNA fragments were transferred to the binary vector, pMDC123, encoding the bialaphos-resistance gene as a selectable marker (Curtis and Grossniklaus 2003) by using LR Clonase Enzyme mix (Invitrogen). A 5.3-kbp BglII fragment containing bvORF18 was obtained from cosmid clone 4F1 and subcloned into the BamHI site of pBluescript. After verifying the nucleotide sequence, the fragment was excised as a PstI–XbaI fragment and cloned into pMDC123. All the constructs were introduced into Agrobacterium tumefaciens strain LBA4404.
The generation of transgenic sugar beets was accomplished according to an unpublished procedure developed by H. Tamagake (unpublished data). Briefly, leaf explants from aseptic plantlets were laid onto a callus-inducing medium (based on the modified MS medium, where NH4NO3 and 2-(morpholin-4-yl)ethanesulfonic acid (MES) were adjusted 825.0 mg/liter and 250 mg/liter, respectively), containing 0.25 mg/liter 6-benzyladenine (BA) and 2.5 g/liter gellan gum. White, friable calli were cultured in a suspension medium (the modified MS medium containing 0.25 mg/liter BA) for 10 days. After that, calli were co-cultured with Agrobacterium in the suspension medium containing 100 mg/liter acetosyringone for 3–4 days. The calli were washed with the suspension medium containing 100 mg/liter meropenem and 2 mg/liter bialaphos and transferred onto a selection medium (the modified MS medium containing 0.25 mg/liter BA, 8.0 g/liter agar, 50 mg/liter meropenem and 100 mg/liter bialaphos). Calli resistant to bialaphos were regenerated into plantlets on a regeneration medium (the modified MS medium containing 1.0 mg/liter BA, 1.0 mg/liter 2,3,5-triiodobenzonic acid, 1.0 mg/liter abscisic acid, 8.0 g/liter agar, 50 mg/liter meropenem, and 2 mg/liter bialaphos).
Nucleotide sequences of oligonucleotides
Oligonucleotides used in this study are listed in Table S1 and Figure S1.
Results
Nucleotide sequence of the chromosomal region containing sugar beet Rf1
The sugar beet Rf1 had previously been located to a region delimited by two molecular markers, mP-A16 and mCP-L6 (Hagihara et al. 2005b). The region was covered with an array of ordered BAC clones (Hagihara et al. 2005b). To obtain a nucleotide sequence of this region, we selected three of the clones, 5A3, 9C23, and 33E19 (Hagihara et al. 2005b), as sequencing templates. To minimize sequence redundancy, we screened a cosmid clone bridging 5A3 and 33E19, from a sublibrary made from 9C23 by using probes made up of 5A3- and 33E19-BAC ends. As a result, cosmid clone 4F1 was selected for sequencing.
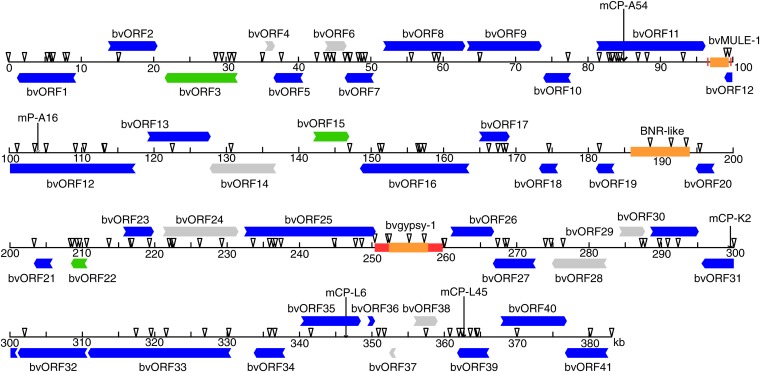
We conducted shotgun sequencing of 5A3, 33E19, and 4F1, yielding 3047, 8058, and 164 independent plasmid sequences, respectively. The plasmid sequences were assembled into three sequences of 156,315; 201,705; and 36,977 bp, respectively. The average coverage was 10.79 for 5A3, 22.64 for 33E19, and 6.2 for 4F1. Overlaps of 4091 bp and 7539 bp occurred between the 4F1 and 5A3 sequences and between the 4F1 and 33E19 sequences, respectively. Therefore, the assembly of 5A3, 33E19, and 4F1 provided a continuous 383,367-bp sequence, with a G + C content of 34.9%. Sequence analysis revealed that target sequences of the five molecular markers (mCP-A54, mP-A16, mCP-K2, mCP-L6, and mCP-L45) that had been mapped to the vicinity of Rf1 (Hagihara et al. 2005b) were included in the assembly in the order predicted by genetic analysis (Figure 1).
Figure 1 .
Organization of a 383-kbp chromosomal region of NK–198 deduced from two BAC clones and a cosmid clone. HindIII restriction sites are shown as triangles. Horizontal arrows indicate predicted genes and their orientation; intronic sequences are omitted. Gray arrows denote the absence of any homologous genes in the database, whereas blue and green arrows indicate the presence of homologous genes in other plants with or without functional assignment, respectively. Orange boxes represent transposable elements, and red boxes show their neighboring repeated sequences. Positions of five molecular markers that were described in Hagihara et al. (2005b) are indicated by vertical arrows.
Potential protein-coding genes in the sequenced region and their transcription
Sequence analysis of the 383,367-bp region identified three potential transposable elements (TEs) (Figure 1). One TE was homologous to the maize mutator element and its related TEs, and was named bvMULE-1 (Beta vulgaris Mutator-like element) (Figure S2). The second TE contained two ORFs (Figure S3). The upstream ORF that encoded 752 amino acid residues (ORF-A in Figure S3) had no homology to any entries in public databases, but a Pfam search identified an RNA recognition motif (RRM). The putative translation product of the second ORF (ORF-B, 1297 amino acid residues) had a high homology to reverse transcriptases of plant long interspersed nuclear elements (LINEs), which include an endonuclease/exonuclease/phosphatase family domain and an RNA-dependent DNA polymerase domain. This structure resembles a group of sugar beet LINEs called BNR (Heitkam and Schmidt 2009). The third TE contained a 4701-bp ORF exhibiting a high homology to Ty3-gypsy–type retroelements, and was named bvgypsy-1 (Figure S4).
Aside from the ORFs encoded by the TEs, 41 genes were predicted. These were named bvORF1–bvORF41 (Figure 1 and Table 1). We surveyed the rest of the sequenced region by BLASTX search to detect any homologous entries in the DDBJ/EMBL/GenBank database but found none. To infer the function of the 41 genes, we conducted a BLASTP search against the DDBJ/EMBL/GenBank database using each of their putative translation products as queries. Although 34 queries matched well with known plant proteins, 7 had no homology to any entries (Table 1). We obtained little information on the possible functions of 3 of the 34 queries with known homologs, as no detailed studies of their homologous entries have been published. The remaining 31 queries retrieved homologous entries whose functions have been fairly well described. Of these entries, Table 1 lists the best matching putative function from the Arabidopsis genome entries and their description from the The Arabidopsis Information Resource (TAIR) database (http://www.arabidopsis.org/).
Table 1 . Characteristics of the genes identified in the 383-kbp region.
| Name of ORFs | Best matched Arabidopsis entries | Transcriptsa | ||||
|---|---|---|---|---|---|---|
| Locus name | Descriptionb | E-value | Anthers | Leaves | Roots | |
| bvORF1 | At2g04940 | Scramblase related | e-80 | NDc | ND | ND |
| bvORF2 | At4g33260 | Putative cdc20 protein | 0 | ND | ND | ND |
| bvORF3 | At5g17210 | Unknown function | 5e-44 | ND | ND | ND |
| bvORF4 | NAd | No hit | NA | ND | ND | ND |
| bvORF5 | At5g57020 | N-myristoyltransferase | 0 | ND | ND | ND |
| bvORF6 | NA | No hit | NA | ND | ND | ND |
| bvORF7 | At5g17170 | Enhancer of sos3-1 (ENH1) | 4e-26 | ND | ND | ND |
| bvORF8 | At4g19490 | Putative homolog of yeast Vps54 | e-139 | ND | ND | ND |
| bvORF9 | At4g19490 | Putative homolog of yeast Vps54 | e-64 | ND | ND | ND |
| bvORF10 | At3g10520 | Class 2 nonsymbiotic hemoglobin | 2e-63 | ND | ND | ND |
| bvORF11 | At2g34780 | MEE22, EMB1611, etc. | 4e-79 | ND | ND | ND |
| bvORF12 | At1g65810 | P loop containing nucleoside triphosphate hydrolases superfamily protein | 0 | + | + | + |
| bvORF13 | At1g65810 | P loop containing nucleoside triphosphate hydrolases superfamily protein | 0 | +e | + | + |
| bvORF14 | NA | No hit | NA | + | + | + |
| bvORF15 | At3g03150 | Unknown function | 3e-15 | + | + | + |
| bvORF16 | At5g42310 | Pentatricopeptide repeat (PPR-like) superfamily protein | 4e-94 | + | + | + |
| bvORF17 | At3g49010 | 60S ribosomal protein L13 | 8e-82 | + | + | + |
| bvORF18 | At5g51740 | Peptidase M48 family protein | 6e-62 | + | + | + |
| bvORF19 | At5g51740 | Peptidase M48 family protein | 4E-52 | + | + | + |
| bvORF20 | At5g51740 | Peptidase M48 family protein | 8E-61 | + | + | + |
| bvORF21 | At5g51740 | Peptidase M48 family protein | 6E-62 | + | + | + |
| bvORF22 | At3g50170 | Unknown function | 2E-71 | + | −f | + |
| bvORF23 | At5g48620 | Disease resistance protein (CC-NBS-LRRg class) family | e-107 | + | + | + |
| bvORF24 | At5g51740 | Peptidase M48 family protein | 8e-06 | + | + | + |
| bvORF25 | At5g35450 | Disease resistance protein (CC-NBS-LRR class) family | e-100 | + | + | + |
| bvORF26 | At1g58390 | Disease resistance protein (CC-NBS-LRR class) family | e-107 | + | + | + |
| bvORF27 | At2g04620 | Cation efflux family protein | e-136 | + | + | + |
| bvORF28 | NA | No hit | NA | – | – | – |
| bvORF29 | NA | No hit | NA | – | – | – |
| bvORF30 | At5g23450 | LCBK1, ATLCBK1, etc. (a sphingosine kinase) | 0 | + | + | + |
| bvORF31 | At4g27870 | Vacuolar iron transporter (VIT) family protein | 2e-31 | + | + | + |
| bvORF32 | At4g27870 | Vacuolar iron transporter (VIT) family protein | 7e-35 | + | + | + |
| bvORF33 | At3g02580 | Brassinosteroid biosynthetic enzyme | + | + | + | |
| bvORF34 | At5g24680 | Peptidase C78, ubiquitin fold modifier-specific peptidase 1/2 | 2e-39 | + | + | – |
| bvORF35 | At3g49590 | Autophagy-related protein 13 | 4e-96 | + | + | + |
| bvORF36 | At5g24660 | RESPONSE TO LOW SULFUR 2 (LSU2) | 2e-13 | ND | ND | ND |
| bvORF37 | NA | No hit | NA | ND | ND | ND |
| bvORF38 | NA | No hit | NA | ND | ND | ND |
| bvORF39 | At5g24650 | Mitochondrial import inner membrane translocase subunit Tim17/Tim22/Tim23 family protein | 5e-65 | ND | ND | ND |
| bvORF40 | At5g24630 | BRASSINOSTEROID-INSENSITIVE4 (a protein that forms part of the topoisomerase VI complex) | 3e-36 | ND | ND | ND |
| bvORF41 | At5g24620 | Pathogenesis-related thaumatin superfamily protein | 2e-76 | ND | ND | ND |
Summary of Figure S1.
Descriptions from TAIR (http://www.arabidopsis.org/).
No data.
Not applicable.
Detected.
Not detected.
N-terminal coiled-coil domain (CC), central nucleotide-binding site domai (NBS) and C-terminal leucine-rich repeat (LRR).
Because Rf1 is a gene for male-fertility restoration, expression patterns of these genes in anthers helps narrow down the coding region of Rf1. RNA samples from NK–198 anthers, leaves, and roots were subjected to reverse transcription (RT)–PCR analysis. Primers for bvORF12–bvORF35, genes located in the region delimited by genetic markers mP-A16 and mCP-L6 (Hagihara et al. 2005b) (see Figure 1), were designed; a single primer set was expected to amplify bvORF18–bvORF21 because these genes were very similar (Figure S5) (quadruplicated genes). Results of the 21 RT–PCR analyses are summarized in Table 1 (see also Figure S1). Transcripts of all genes except bvORF22, bvORF28, bvORF29, and bvORF34 were detected in all organs examined. No amplicon was observed in any organs when the bvORF28- or the bvORF29-specific primer set was used, whereas organ-specific expression was observed in bvORF22 and bvORF34, whose transcript levels were below the detection limit in leaves and roots, respectively.
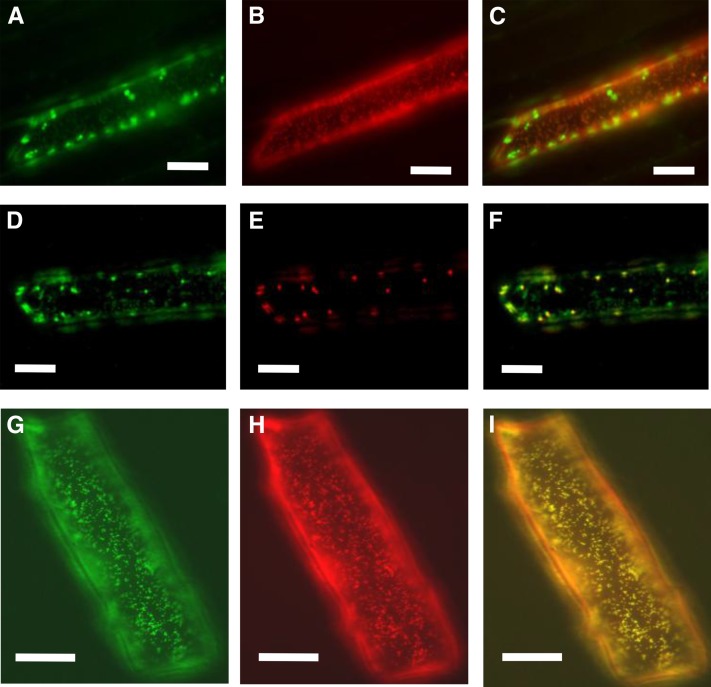
There was a PPR protein gene in the 383-kbp region. Transcripts of this gene, bvORF16, were detected in NK–198 anthers (Table 1). However, because of the amino acid sequence homology between bvORF16 and at5g42310 (Table 1), which presumably is an ortholog of maize crp1, a regulatory gene of plastids (Barkan et al. 1994; Schmitz-Linneweber et al. 2005; Williams-Carrier et al. 2008), it seemed likely that bvORF16 encodes a plastid protein and not a mitochondrial protein. Two programs, TargetP and Predotar, predicted no specific localization for the bvORF16 translation product. We constructed a chimeric GFP gene with 80 N-terminal amino acid residues of bvORF16. The chimeric GFP genes were placed under the control of the 35S promoter of the cauliflower mosaic virus. We bombarded epidermal cells of Welsh onion sheath with plasmids carrying the chimeric GFP gene and observed fluorescent signals. Surprisingly, each of the localized green signals matched with either mitochondria or plastids that were marked by a mitochondrion-targeting RFP or a plastid-targetingRFP (see Materials and Methods), respectively (Figure 2, A–F). Therefore, bvORF16 encodes a dual-targeted PPR protein. As far as we know, no PPR-type Rf reported to date has exhibited this dual-targeting property (Bentolila et al. 2002; Wang et al. 2006). PPR-type Rfs and PPR-type Rf-like (RFL) genes tend to cluster with similar genes on chromosomes (Fujii et al. 2011), unlike bvORF16, a single copy gene in the sugar beet genome (Figure S6). The PPR-type Rfs identified to date belong to a subclass of PPR genes (termed P class) and form a single clade with RFL genes in a phylogenetic tree of P-class PPR genes (Fujii et al. 2011). We examined whether bvORF16, which appears to be a P-class PPR gene, belongs to the clade of RF and RFL by phylogenetic analysis (File S2) and found that bvORF16 clustered together with at5g42310 (labeled as At_CRP1 in File S2) but not with any PPR-type RF or RFL proteins. Therefore, bvORF16 is an atypical Rf candidate.
Figure 2 .
Images of fluorescent signals obtained from transient expression tests. (A–F) Images of epidermal cells of Welsh onion sheath. (G and H) Images of epidermal cells of onion bulb scales. Bars, 50 μm. A and D are green fluorescence images of bvORF16-GFP; B and H are red fluorescence images of mitochondria-targeted RFP; C is a merged image of A and B; E is a red fluorescence image of plastid-targeted RFP; F is a merged image of D and E; G is a green fluorescence image of bvORF20-GFP; and I is a merged image of G and H.
No genes in the 383,367-bp sequence exhibited homology to mitochondrial aldehyde dehydrogenase, glycine-rich protein, or retrograde regulated male sterility protein, which were encoded by maize Rf2a, rice Rf2, or rice Rf17, respectively (Table 1).
The Oma1-Like gene was associated with NK–198-specific transcripts
We previously reported that a 7.0-kbp HindIII fragment that had been subcloned from 37O9 (a BAC clone overlapping with the 5A3, 9C23, and 33E19) detected specific transcripts in flower buds of NK–198 but not of the CMS line, TK–81mm–MS (i.e., [S]rf1rf1) (Hagihara et al. 2005b). During our sequence analysis, we noticed that the 7.0-kbp HindIII fragment included the coding sequence of one of the quadruplicated genes, bvORF19, that resembled yeast Oma1, a peptidase M48 family protein involved in quality control of mitochondrial membrane proteins (Kaser et al. 2003) (Table 1 and see Files S3, S4, and S5). To see whether NK–198-specific transcripts were homologous to bvORF19, RNA gel blot analysis was conducted using the 3′-UTR sequence of bvORF19 as a probe. Because of high sequence homology among bvORF18–bvORF21, the design of specific hybridization probes for bvORF18, bvORF19, bvORF20, and bvORF21 was infeasible. Therefore, our probe simultaneously detected transcripts of the four genes in NK–198 samples. A strong signal appeared in the lane corresponding to NK–198 flower buds, but was hardly seen elsewhere (Figure 3). This result was consistent with our previous results using the 7.0-kbp HindIII fragment of the NK–198 genome (Hagihara et al. 2005b).
Figure 3 .
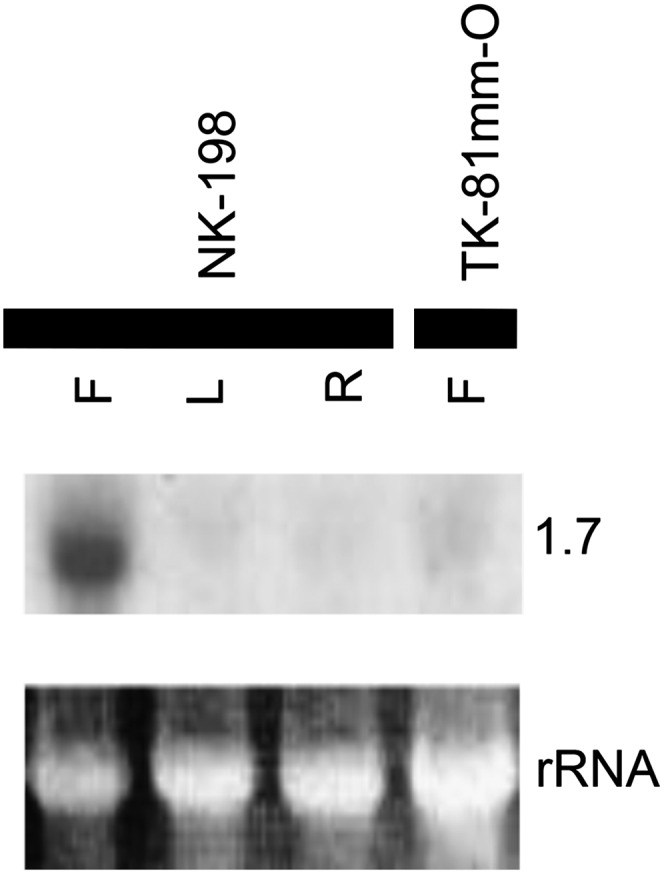
RNA gel blot analysis of the 3′-UTR of bvORF19 hybridized with total RNAs from flower buds (F), leaves (L), and roots (R) of NK–198, and from flower buds of TK–81mm–O. Sizes of signal bands are indicated in kilobases. Images in the bottom row show ethidium-bromide (EtBr-) rRNA after gel electrophoresis.
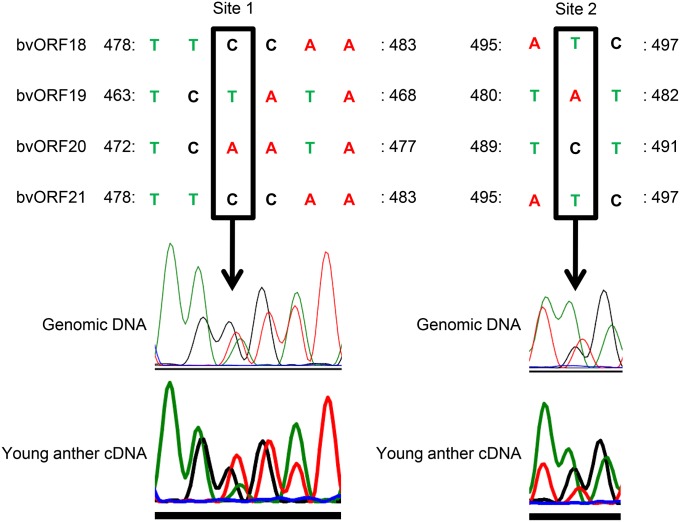
Both RNA gel blot analysis and RT–PCR analysis (see above) revealed that at least one copy of the quadruplicated genes (bvORF18–bvORF21) was expressed in anthers of NK–198, but it remained unclear whether all copies were expressed. Multiple sequence alignment of the bvORF18- to bvORF21-coding regions revealed that bvORF18 and bvORF21 were identical at the nucleotide sequence level, and thus could not be distinguished from each other (Figure S5). On the other hand, the sequences from nucleotide ∼478 to ∼497 provided unique sequence tags for bvORF19 and bvORF20, due to a microsatellite-like polymorphism and nucleotide substitutions (Figure S5). Based on this observation, we set up an assay including direct sequencing of RT–PCR products to detect the sequence tags of the expressed copies. Before we conducted the expression assay, the genomic DNA of NK–198 was subjected to PCR amplification, targeting a region encompassing the polymorphic sites (Figure 4) with primers D-Fw and D-RV to obtain a control template. The sequencing electrophoregram of the control template with the sequencing primer Gre is shown in Figure 4. At polymorphic site 1, a C residue occurs in bvORF18 and bvORF21, whereas T and A are found in bvORF19 and bvORF20, respectively. We next PCR amplified cDNA of NK–198 young anthers (i.e., predehiscence) with the primers D-FW and D-RV. An electrophoregram of the RT–PCR products was obtained using the sequencing primer Gre. The highest peak at site 1 was A, followed by C and T. At polymorphic site 2, the peak of T, indicative of bvORF20, was higher than that of the control (Figure 4), although this may not reflect a significant quantitative difference. These data indicated that all copies of bvORF18–bvORF21 were expressed in anthers.
Figure 4 .
Polymorphic sites in the PCR targets of the quadruplicated genes in NK–198 and electrophoregrams obtained by direct sequencing. The original electrophoregrams were converted to complementary images on the sequencing platform (ABI3130). Red, green, and black lines indicate the signal peaks of adenine, thymine, and cytosine, respectively. Numbers of nucleotides correspond to the sequence alignment shown in Figure S5.
bvORF20 restored partial pollen fertility to CMS sugar beet
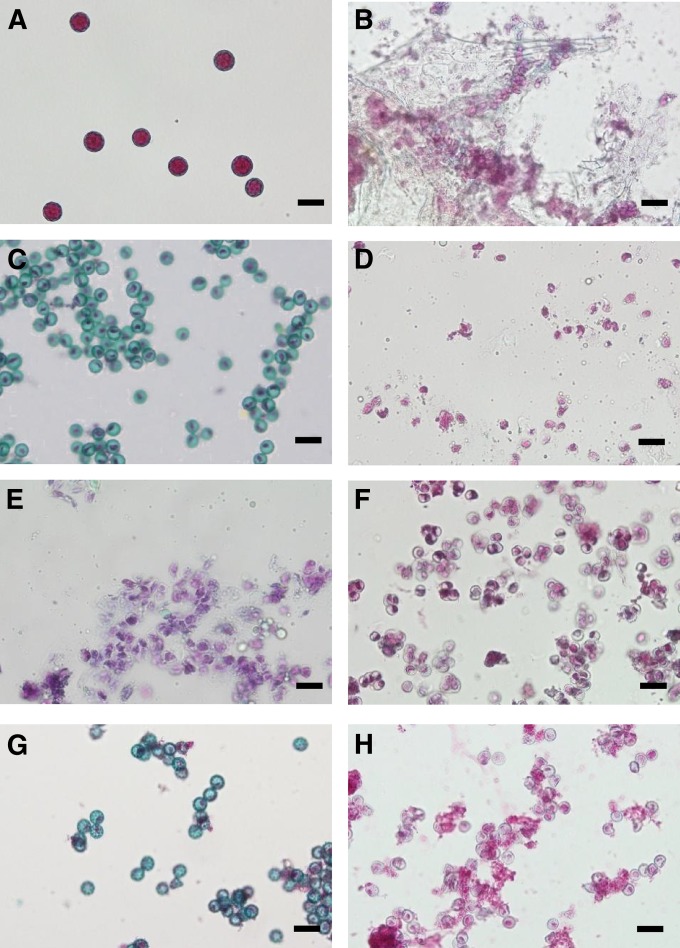
If one of the quadruplicated ORFs is the Rf1 gene, we might expect that the ORF in question could restore pollen fertility when transferred to Owen CMS plants. Sugar beet is known to be quite recalcitrant to regeneration following genetic transformation (Skaracis 2005). One of the present authors also found that regeneration in sugar beet was highly genotype dependent, and a Japanese breeding line, NK–219mm–CMS, had a good shoot regeneration response (H. Tamagake, unpublished data). To examine whether NK–198 actually acted as a restorer of NK–219mm–CMS, we crossed NK–219mm–CMS with NK–198. The F1 progeny (11 plants) were all classified as “partial fertile”; nearly all pollen grains appeared to be well developed morphologically but their cytoplasm was scarcely stained with Alexander’s dye (Figure 5, A–C). Because this phenotype could be clearly distinguished from the completely sterile phenotype of NK–219mm–CMS plants (almost all microspores were aborted at an early stage of microsporogenesis and the exine was poorly developed), we concluded that NK–198 Rf1 restored partial fertility to the NK–219mm–CMS plants, although NK–198 Rf1 restored almost complete fertility to two other sugar beet lines, TK–81mm–CMS and TK–76mm–CMS (Hagihara et al. 2005a). Notably, the effect of NK–198 Rf1 is influenced by the nuclear genetic background (see the result using sugar beet line I-12CMS(R) in Hagihara et al. 2005a).
Figure 5 .
Photographs of anther contents from transgenic and control sugar beets. A–H are images of Alexander’s staining. Bars, 20 µm. (A) Anther contents of a maintainer line, NK–219mm–O. (B) Anther contents of a CMS line, NK–219mm–CMS. (C) Anther contents of an F1 plant (NK–219mm-CMS × NK–198). (D) Anther contents of a transgenic sugar beet transformed with the pMDC123 vector. (E–H) Anther contents of transgenic sugar beets transformed with pBVORF18–pBVORF21, respectively.
To test our hypothesis with transgenic plants, the genomic DNA fragment containing the protein-coding region and its 5′ upstream (2 to 2.5 kbp in length) and 3′ downstream regions (∼500 bp) of bvORF18, bvORF19, bvORF20, or bvORF21 were separately inserted into binary vectors. The resultant constructs were named pBVORF18, pBVORF19, pBVORF20, and pBVORF21, respectively. These constructs were subsequently introduced into NK–219mm–CMS calli by Agrobacterium-mediated transformation. The calli resistant to bialaphos herbicide, a phenotype conferred by the selectable marker on the T-DNA, were transferred to a regeneration medium. The regenerated sugar beet plants contained the bialaphos-resistance gene as shown by PCR analysis using primers BAR5 and BAR6 (data not shown).
We obtained 10 independent transgenic sugar beet plants transformed with pBVORF20, of which 8 exhibited partial fertility (Figure 5G). This partial-fertile phenotype was indistinguishable from that of the F1 progenies of NK–219mm-CMS × NK–198 (Figure 5C). To ascertain the cosegregation of fertility restoration with the transgene, a transgenic plant carrying pBVORF20 was pollinated with the TA–33–O line, which had a maintainer genotype. The 14 F1 plants were either male sterile (8 plants) or partial fertile (6 plants) (Figures S7 and S8). The bialaphos-resistance gene was found to cosegregate with the partial-fertility phenotype (Figure S8).
By contrast, three, nine, and eight transgenic plants were obtained carrying the pBVORF18, pBVORF19, and pBVORF21 constructs, respectively, and they all exhibited complete male sterility, not partial fertility (Figure 5, E, F, and H). These experiments strongly indicated that Rf1 most likely corresponded to bvORF20.
Intracellular localization of bvORF20
The TargetP and Predotar programs predicted that bvORF20 proteins would be localized in mitochondria (scores: TargetP, 0.847; Predotar, 0.85). We constructed chimeric GFP genes with 55 N-terminal amino acid residues of bvORF20 at their 5′ ends. The plasmid carrying the chimeric GFP genes was bombarded into epidermal cells of onion bulbs. The green fluorescent signals matched well with the red signals from the mitochondrial marker construct, pMt-R, which was cobombarded (Figure 2, G–I), confirming that bvORF20 encodes a mitochondrial protein.
Organization of the rf1 allele
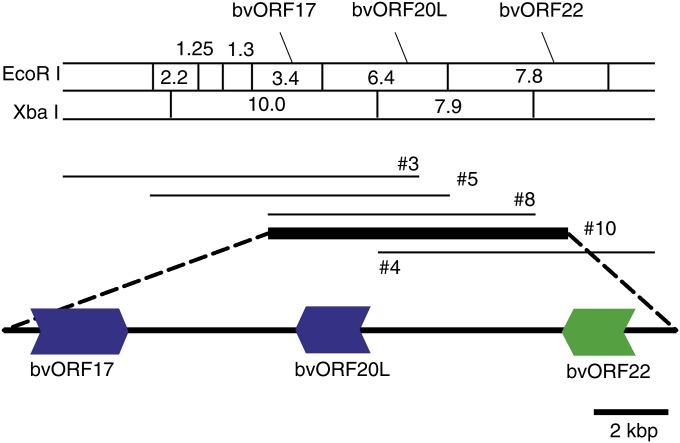
Alteration(s) in nucleotide sequence was expected in the rf1 allele. Using a probe of the 3′-UTR sequence of bvORF19, which is highly conserved among bvORF18–bvORF21, a lambda-phage genomic library of a maintainer line, TK–81mm–O (Matsuhira et al. 2007), was screened, and five recombinant phages were obtained. Restriction mapping of the five clones using EcoRI and XbaI enabled us to assemble these clones into a contig of ∼30 kbp (Figure 6). Gene mapping of bvORF17, bvORF20, and bvORF22 on the physical map was achieved by DNA gel blot analysis, and recombinant phage no. 10 was identified as containing all mapped genes. The insert in recombinant phage no. 10 was subjected to shotgun sequencing. A continuous 16,037-bp region was obtained after assembling 55 independent plasmid clones and subsequent correction of any ambiguities by sequencing PCR fragments encompassing the regions in question. In the 16,037-bp region, we found three homologous genes to bvORF17, bvORF20, and bvORF22, but none of the BNR copies (Figure 6). The order and orientation of the three ORFs was preserved between TK–81mm–O and NK–198, but the bvORF20-like gene was single copy (hereafter named bvORF20L). To examine the copy number of bvORF20L, the conserved 3′-UTR sequence was hybridized to total cellular DNA of TK–81mm–O. The number and size of the signal band was congruent with the sequence data (7.9, 7.0, 5.9, and 1.9 kbp for NK–198 and 5.9 kbp for TK–81mm–O) (Figure 7). A comparison of the amino acid sequences predicted from bvORF20L and its counterparts in NK–198 is shown in Figure S9. bvORF20L is similar to the copies of the quadruplicated genes in NK–198 but not identical to any one of them (see Figure S5 for comparison of nucleotide sequences). Homologies at the amino acid sequence level were 83–85% (vs. bvORF18, bvORF19, bvORF20, and bvORF21). A detailed organizational comparison of this genomic region between TK–81mm–O and NK–198 will be presented elsewhere.
Figure 6 .
Physical map of the chromosomal region containing bvORF17, bvORF20L, and bvORF22 of TK–81mm–O. Sizes of restriction fragments are shown in kilobase pairs. Five recombinant phage clones are indicated. Gene organization deduced from the nucleotide sequence of clone no. 10 is indicated below with a bar. Colors and directions of the horizontal arrows have the same meanings as in Figure 1.
Figure 7 .

DNA gel blot analysis of the 3′-UTR of bvORF19 hybridized with total cellular DNA from NK–198 (lane 1) and TK–81mm–O (lane 2). HindIII restriction endonuclease was used. Size markers are shown on the right (in kilobase pairs).
Discussion
The nucleotide sequence of a 383-kbp chromosomal region containing the Rf1 locus of sugar beet was determined. Forty-one potential genes were found in this region. On this basis the gene density was calculated to be 9.4 kbp/gene, which appeared quite rich, given that the sugar beet’s entire genome is 758 Mbp (Arumuganathan and Earle 1991). This gene density would suggest a total number of sugar beet genes of more than 80,000, an apparent overestimation compared to the total gene numbers of other dicots such as Arabidopsis (25,498), black cottonwood (Populus trichocarpa Torr. & A.Gray; 45,555), and grapevine (Vitis vinifera L.; 30,434) (The Arabidopsis Genome Initiative 2000; Tuskan et al. 2006; Jaillon et al. 2007). Recently, Dohm et al. (2012) reported that a maximum average distance of 30–40 kbp between genes in the sugar beet genome could be assumed according to their physical mapping study. On the other hand, three TEs identified in this study occupied a total of 6% of the sequenced region, which is much less than in other sugar beet chromosomal regions (up to 41.6%) (Schulte et al. 2006).
The 383-kbp region that was sequenced in this study contained neither typical PPR-type Rf gene nor genes related to Rf genes from other plants such as maize Rf2a, rice Rf17, or rice Rf2 (Cui et al. 1996; Fujii and Toriyama 2009; Itabashi et al. 2011). This finding suggests that fertility restoration in sugar beet CMS involves a novel mechanism. This interpretation is consistent with the previous observation that mitochondrial gene expression in sugar beet is apparently unchanged after fertility restoration (Yamamoto et al. 2005).
On the other hand, we found that introduction of bvORF20 as a transgene restored partial fertility to NK–219mm–CMS. A comparable level of fertility restoration was observed in F1 plants of NK–219mm–CMS × NK–198. Although three other ORFs homologous to bvORF20 were encoded in the Rf1 locus, none was capable of restoring male fertility. Therefore, despite their similarity in amino acid sequences, it is unlikely that these three ORFs play a major role in fertility restoration. Compared to bvORF20, the amino acid sequence homology in bvORF18, bvORF19, or bvORF21 is 88–99%. It is possible that one or more of the differences in amino acid sequences is involved in the inability to restore pollen fertility. Additionally, bvORF20L, a bvORF20-related gene found in rf1rf1 sugar beet, encoded an uninterrupted ORF. Homology of the bvORF20L amino acid sequence to bvORF20 was 83%, and the amount of bvORF20L transcripts was greatly reduced compared to Rf1 sugar beet. Either or both of the structural or transcriptional alterations might render bvORF20L an rf1 allele.
As far as we know, bvORF20 homologs (Oma1 group in File S5) are conserved in eukaryotes as single copy genes. For example, the yeast homolog Oma1 is involved in the quality control of mitochondrial membrane proteins with more or less similar activity as that of the matrix AAA protease (Kaser et al. 2003). In mammals, Oma1 functions as a membrane potential-dependent protease, one of whose substrates is OPA1, a GTPase involved in mitochondrial fusion (Ehses et al. 2009; Head et al. 2009). However, bvORF20 appears to lack protease activity because its Zn2+-binding motif in the peptidase M48 domain is His-Gln-Val-Gly-His instead of the conserved His-Glu-x-x-His (Figure S9 and Files S3, S4, and S5). The Glu-to-Gln substitution in this motif was shown to abolish protease activity in yeast Oma1 (Kaser et al. 2003). According to our database search, ORFs homologous to yeast Oma1 preserve the His-Glu-x-x-His motif (File S4). These observations lead us to hypothesize that the function of bvORF20 may not be a protease. On the other hand, if the possible molecular chaperone-like properties of yeast OMA1 (Kaser et al. 2003) are conserved in bvORF20, the bvORF20 protein might interact directly with preSATP6. This protein–protein complex might alter the higher order structure of preSATP6 to make it inactive. Molecular analysis of bvORF20 function is underway.
Concerning the evolution of plant Rf, the tandem gene cluster of bvORF18, bvORF19, bvORF20, and bvORF21 is reminiscent of the organization of the Rf loci of petunia, radish, and rice, whose translation products are PPR proteins (Bentolila et al. 2002; Brown et al. 2003; Desloire et al. 2003; Kazama and Toriyama 2003; Koizuka et al. 2003; Akagi et al. 2004; Komori et al. 2004). The evolutionary significance of such gene clusters may lie in the increased allelic diversity (Touzet and Budar 2004). We should point out an additional similarity that, in both PPR-type Rf loci and the sugar beet Rf1 locus, not all copies but one or several of these are capable of restoring fertility. Therefore, it is possible that a common mechanism has played an important role in the evolution of plant Rfs. We are currently investigating the organizational diversity of Rf1 in B. vulgaris plants to see how these genes have evolved.
Supplementary Material
Acknowledgments
We thank the DNA Sequencing Facility of the Research Faculty of Agriculture, Hokkaido University, for technical assistance. This work was supported in part by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology, Japan; a grant from the Program for Promotion of Basic Research Activities for Innovative Biosciences, Japan; and the Program for Promotion of Basic and Applied Research for Innovations in Bio-Oriented Industry.
Footnotes
Communicating editor: S. Poethig
Literature Cited
- Akagi H., Nakamura A., Yokozeki-Misono Y., Inagaki A., Takahashi H., et al. , 2004. Positional cloning of the rice Rf-1 gene, a restorer of BT-type cytoplasmic male sterility that encodes a mitochondria-targeting PPR protein. Theor. Appl. Genet. 108: 1449–1457 [DOI] [PubMed] [Google Scholar]
- Alexander M. P., 1969. Differential staining of aborted and nonaborted pollen. Stain Technol. 44: 117–122 [DOI] [PubMed] [Google Scholar]
- Arimura S., Tsutsumi N., 2002. A dynamin-like protein (ADL2b), rather than FtsZ, is involved in Arabidopsis mitochondrial division. Proc. Natl. Acad. Sci. USA 99: 5727–5731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative , 2000. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Arumuganathan K., Earle E. D., 1991. Nuclear DNA content of some important plant species. Plant Mol. Biol. Rep. 9: 208–218 [Google Scholar]
- Barkan A., Walker M., Nolasco M., Johnson D., 1994. A nuclear mutation in maize blocks the processing and translation of several chloroplast messenger-RNAs and provides evidence for the differential translation of alternative messenger-RNA forms. EMBO J. 13: 3170–3181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr C. M., Fishman L., 2010. The nuclear component of a cytonuclear hybrid incompatibility in Mimulus maps to a cluster of pentatricopeptide repeat genes. Genetics 184: 455–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentolila S., Alfonso A. A., Hanson M. R., 2002. A pentatricopeptide repeat-containing gene restores fertility to cytoplasmic male-sterile plants. Proc. Natl. Acad. Sci. USA 99: 10887–10892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosemark N. O., 2006. Genetics and breeding, pp. 50–88 in Sugar Beet, edited by Draycott A. P. Blackwell Publishing, Oxford [Google Scholar]
- Boutry M., Faber A. M., Charbonnier M., Briquet M., 1984. Microanalysis of plant mitochondrial protein-synthesis products: detection of variant polypeptides associated with cytoplasmic male-sterility. Plant Mol. Biol. 3: 445–452 [DOI] [PubMed] [Google Scholar]
- Brown G. G., Formanova N., Jin H., Wargachuk R., Dendy C., et al. , 2003. The radish Rfo restorer gene of Ogura cytoplasmic male sterility encodes a protein with multiple pentatricopeptide repeats. Plant J. 35: 262–272 [DOI] [PubMed] [Google Scholar]
- Budar F., Delourme R., Pelletier G., 2004. Male sterility, pp. 43–64 in Biotechnology in Agriculture and Forestry: Brassica, edited by E. C. Pua and C. J. Douglas. Springer-Verlag, Berlin [Google Scholar]
- Budar F., Touzet P., Pelletier G., 2006. Cytoplasmic male sterility, pp. 147–180 in Flowering and Its Manipulation, edited by Ainsworth C. Blackwell Publishing, Oxford [Google Scholar]
- Burge C., Karlin S., 1997. Prediction of complete gene structures in human genomic DNA. J. Mol. Biol. 268: 78–94 [DOI] [PubMed] [Google Scholar]
- Chase C. D., 2007. Cytoplasmic male sterility: a window to the world of plant mitochondrial-nuclear interactions. Trends Genet. 23: 81–90 [DOI] [PubMed] [Google Scholar]
- Chenna R., Sugawara H., Koike T., Lopez R., Gibson T. J., et al. , 2003. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 31: 3497–3500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu W., Niwa Y., Zeng W., Hirano T., Kobayashi H., et al. , 1996. Engineering GFP as a vital reporter in plants. Curr. Biol. 6: 325–330 [DOI] [PubMed] [Google Scholar]
- Chomczynski P., Sacchi N., 1987. Single-step method of RNA isolation by acid guanidium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162: 156–159 [DOI] [PubMed] [Google Scholar]
- Cui X. Q., Wise R. P., Schnable P. S., 1996. The rf2 nuclear restorer gene of male-sterile T-cytoplasm maize. Science 272: 1334–1336 [DOI] [PubMed] [Google Scholar]
- Curtis M. D., Grossniklaus U., 2003. A Gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desloire S., Gherbi H., Laloui W., Marhadour S., Clouet V., et al. , 2003. Identification of the fertility restoration locus, Rfo, in radish, as a member of the pentatricopeptide-repeat protein family. EMBO Rep. 4: 588–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewey R. E., Levings C. S., III, Timothy D. H., 1986. Novel recombination in the maize mitochondrial genome produce a unique transcriptional unit in the Texas male-sterile cytoplasm. Cell 44: 439–449 [DOI] [PubMed] [Google Scholar]
- Dohm J. C., Lange C., Holtgrawe D., Sorensen T. R., Borchardt D., et al. , 2012. Paleohexaploid ancestory for Caryophyllales inferred from extensive gene-based physical and genetic mapping of the sugar beet genome (Beta vulgaris). Plant J. 70: 528–540 [DOI] [PubMed] [Google Scholar]
- Doyle J. J., Doyle J. L., 1990. Isolation of plant DNA from fresh tissue. Focus 12: 13–15 [Google Scholar]
- Ducos E., Touzet P., Saumitou-Laprade P., Vernet P., Cuguen J., 2001. Nuclear effect on mitochondrial protein expression of the CMS Owen cytoplasm in sugar beet. Theor. Appl. Genet. 102: 1299–1304 [Google Scholar]
- Ehses S., Raschke I., Mancuso G., Bernacchia A., Geimer S., et al. , 2009. Regulation of OPA1 processing and mitochondrial fusion by m-AAA protease isoenzymes and OMA1. J. Cell Biol. 187: 1023–1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O., Nielsen H., Brunak S., von Heijne G., 2000. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 300: 1005–1016 [DOI] [PubMed] [Google Scholar]
- Finn R. D., Mistry J., Schuster-Bockler B., Griffiths-Jones S., Hollich V., et al. , 2006. Pfam: clans, web tools and services. Nucleic Acids Res. 34: D247–D251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii S., Toriyama K., 2008. Genome barriers between nuclei and mitochondria exemplified by cytoplasmic male sterility. Plant Cell Physiol. 49: 1484–1494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii S., Toriyama K., 2009. Suppressed expression of RETROGRADE-REGULATED MALE STERILITY restores pollen fertility in cytoplasmic male sterile rice plants. Proc. Natl. Acad. Sci. USA 106: 9513–9518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii S., Bond C. S., Small I. D., 2011. Selection patterns on restorer-like genes reveal a conflict between nuclear and mitochondrial genomes throughout angiosperm evolution. Proc. Natl. Acad. Sci. USA 108: 1723–1728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagihara E., Itchoda N., Habu Y., Iida S., Mikami T., et al. , 2005a Molecular mapping of a fertility restorer gene for Owen cytoplasmic male sterility in sugar beet. Theor. Appl. Genet. 111: 250–255 [DOI] [PubMed] [Google Scholar]
- Hagihara E., Matsuhira H., Ueda M., Mikami T., Kubo T., 2005b Sugar beet BAC library construction and assembly of a contig spanning Rf1, a restorer-of-fertility gene for Owen cytoplasmic male sterility. Mol. Genet. Genomics 274: 316–323 [DOI] [PubMed] [Google Scholar]
- Hallden C., Lind C., Moller I. M., 1992. Variation in mitochondrial translation products in fertile and cytoplasmic male-sterile sugar-beets. Theor. Appl. Genet. 85: 139–145 [DOI] [PubMed] [Google Scholar]
- Hanson M. R., Bentolila S., 2004. Interactions of mitochondrial and nuclear genes that affect male gametophyte development. Plant Cell 16: S154–S169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Head B., Griparic L., Amiri M., Gandre-Babbe S., van der Bliek A. M., 2009. Inducible proteolytic inactivation of OPA1 mediated by the OMA1 protease in mammalian cells. J. Cell Biol. 187: 959–966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitkam T., Schmidt T., 2009. BNR - a LINE family from Beta vulgaris - contains a RRM domain in open reading frame 1 and defines a L1 sub-clade present in diverse plant genomes. Plant J. 59: 872–882 [DOI] [PubMed] [Google Scholar]
- Hjerdin-Panagopoulos A., Kraft T., Rading I. M., Tuvesson S., Nilsson N. O., 2002. Three QTL regions for restoration of Owen CMS in sugar beet. Crop Sci. 42: 540–544 [Google Scholar]
- Hu J., Wang K., Huang W., Liu G., Wang J., et al. , 2012. The rice pentatricopeptide repeat protein RF5 restores fertility in Hong-Lian cytoplasmic male-sterile lines via a complex with the glycine-rich protein GRP162. Plant Cell 24: 109–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itabashi E., Iwata N., Fujii S., Kazama T., Toriyama K., 2011. The fertility restorer gene, Rf2, for Lead Rice-type cytoplasmic male sterility of rice encodes a mitochondrial glycine-rich protein. Plant J. 65: 359–367 [DOI] [PubMed] [Google Scholar]
- Jaillon O., Aury J. M., Noel B., Policriti A., Clepet C., et al. , 2007. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449: 463–467 [DOI] [PubMed] [Google Scholar]
- Jordan D. R., Mace E. S., Henzell R. G., Klein P. E., Klein R. R., 2010. Molecular mapping and candidate gene identification of the Rf2 gene for pollen fertility restoration in sorghum [Sorghum bicolor (L.) Moench.]. Theor. Appl. Genet. 120: 1279–1287 [DOI] [PubMed] [Google Scholar]
- Kaser M., Kambacheld M., Kisters-Woike B., Langer T., 2003. Oma1, a novel membrane-bound metallopeptidase in mitochondria with activities overlapping with the m-AAA protease. J. Biol. Chem. 278: 46414–46423 [DOI] [PubMed] [Google Scholar]
- Kazama T., Toriyama K., 2003. A pentatricopeptide repeat-containing gene that promotes the processing of aberrant atp6 RNA of cytoplasmic male-sterile rice. FEBS Lett. 544: 99–102 [DOI] [PubMed] [Google Scholar]
- Kitazaki K., Kubo T., Kagami H., Matsumoto T., Fujita A., et al. , 2011. A horizontally transferred tRNACys gene in the sugar beet mitochondrial genome: evidence that the gene is present in diverse angiosperms and its transcript is aminoacylated. Plant J. 68: 262–272 [DOI] [PubMed] [Google Scholar]
- Klein R. R., Klein P. E., Mullet J. E., Minx P., Rooney W. L., et al. , 2005. Fertility restorer locus Rf1 of sorghum (Sorghum bicolor L.) encodes a pentatricopeptide repeat protein not present in the colinear region of rice chromosome 12. Theor. Appl. Genet. 111: 994–1012 [DOI] [PubMed] [Google Scholar]
- Koizuka N., Imai R., Fujimoto H., Hayakawa T., Kimura Y., et al. , 2003. Genetic characterization of a pentatricopeptide repeat protein gene, orf687, that restores fertility in the cytoplasmic male-sterile Kosena radish. Plant J. 34: 407–415 [DOI] [PubMed] [Google Scholar]
- Komori T., Ohta S., Murai N., Takakura Y., Kuraya Y., et al. , 2004. Map-based cloning of a fertility restorer gene, Rf-1, in rice (Oryza sativa L.). Plant J. 37: 315–325 [DOI] [PubMed] [Google Scholar]
- Kubo T., Newton K. J., 2008. Angiosperm mitochondrial genomes and mutations. Mitochondrion 8: 5–14 [DOI] [PubMed] [Google Scholar]
- Kubo T., Kitazaki K., Matsunaga M., Kagami H., Mikami T., 2011. Male sterility-inducing mitochondrial genomes: How do they differ? Crit. Rev. Plant Sci. 30: 378–400 [Google Scholar]
- Kurtz S., Choudhuri J. V., Ohlebusch E., Schleiermacher C., Stoye J., et al. , 2001. REPuter: the manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 29: 4633–4642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laser K. D., Lersten N. R., 1972. Anatomy and cytology of microsporogenesis in cytoplasmic male sterile angiosperms. Bot. Rev. 38: 425–454 [Google Scholar]
- Lind C., Hallden C., Moller I. M., 1991. Protein synthesis in mitochondria purified from roots, leaves and flowers of sugar beet. Physiol. Plant. 83: 7–16 [Google Scholar]
- Matsuhira H., Shinada H., Yui-Kurino R., Hamato N., Umeda M., et al. , 2007. An anther-specific lipid transfer protein gene in sugar beet: its expression is strongly reduced in male-sterile plants with Owen cytoplasm. Physiol. Plant. 129: 407–414 [Google Scholar]
- Owen F. V., 1945. Cytoplasmically inherited male-sterility in sugar beets. J. Agric. Res. 71: 423–440 [Google Scholar]
- Pelletier G., Budar F., 2007. The molecular biology of cytoplasmically inherited male sterility and prospects for its engineering. Curr. Opin. Biotechnol. 18: 121–125 [DOI] [PubMed] [Google Scholar]
- Pillen K., Steinrücken G., Hermann R. G., Jung C., 1993. An extended linkage map of sugar beet (Beta vulgaris L.) including nine putative lethal genes and the restorer gene X. Plant Breed. 111: 265–272 [Google Scholar]
- Sambrook J., Fritsch E. F., Maniatis T., 1989. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Schmitz-Linneweber C., Williams-Carrier R., Barkan A., 2005. RNA immunoprecipitation and microarray analysis show a chloroplast pentatricopeptide repeat protein to be associated with the 5′ region of mRNAs whose translation it activates. Plant Cell 17: 2791–2804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz-Linneweber C., Small I., 2008. Pentatricopeptide repeat proteins: a socket set for organelle gene expression. Trends Plant Sci. 13: 663–670 [DOI] [PubMed] [Google Scholar]
- Schnable P. S., Wise R. P., 1998. The molecular basis of cytoplasmic male sterility and fertility restoration. Trends Plant Sci. 3: 175–180 [Google Scholar]
- Schondelmaier J., Jung C., 1997. Chromosomal assignment of the nine linkage groups of sugar beet (Beta vulgaris L.) using primary trisomics. Theor. Appl. Genet. 95: 590–596 [Google Scholar]
- Schulte D., Cai D. G., Kleine M., Fan L. J., Wang S., et al. , 2006. A complete physical map of a wild beet (Beta procumbens) translocation in sugar beet. Mol. Genet. Genomics 275: 504–511 [DOI] [PubMed] [Google Scholar]
- Skaracis G. N., 2005. In vitro culture technique, pp. 247–255 in Genetics and Breeding of Sugar Beet, edited by Biancardi E., Campbell L. G., Skaracis G. N., de Biaggi M. Science Publishers, Plymouth, UK [Google Scholar]
- Small I., Peeters N., Legeai F., Lurin C., 2004. Predotar: a tool for rapidly screening proteomes for N-terminal targeting sequences. Proteomics 4: 1581–1590 [DOI] [PubMed] [Google Scholar]
- Staden R., 1996. The Staden sequence analysis package. Mol. Biotechnol. 5: 233–241 [DOI] [PubMed] [Google Scholar]
- Touzet P., Budar F., 2004. Unveiling the molecular arms race between two conflicting genomes in cytoplasmic male sterility? Trends Plant Sci. 9: 568–570 [DOI] [PubMed] [Google Scholar]
- Tuskan G. A., Difazio S., Jansson S., Bohlmann J., Grigoriev I., et al. , 2006. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313: 1596–1604 [DOI] [PubMed] [Google Scholar]
- Wang Z., Zou Y., Li X., Zhang Q., Chen L., et al. , 2006. Cytoplasmic male sterility of rice with boro II cytoplasm is caused by a cytotoxic peptide and is restored by two related PPR motif genes via distinct modes of mRNA silencing. Plant Cell 18: 676–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams-Carrier R., Kroeger T., Barkan A., 2008. Sequence-specific binding of a chloroplast pentatricopeptide repeat protein to its native group II intron ligand. RNA 14: 1930–1941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X. B., Liu Z. X., Zhang D. F., Liu Y., Song W. B., et al. , 2009. Isolation and analysis of Rice Rf1-orthologus PPR genes co-segregating with Rf3 in maize. Plant Mol. Biol. Rep. 27: 511–517 [Google Scholar]
- Yamamoto M. P., Kubo T., Mikami T., 2005. The 5′-leader sequence of sugar beet mitochondrial atp6 encodes a novel polypeptide that is characteristic of Owen cytoplasmic male sterility. Mol. Genet. Genomics 273: 342–349 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.