Abstract
In this work we report on how salt concentration and cation species affect DNA translocation in voltage-biased silicon nitride nanopores. The translocation of double-stranded DNA (dsDNA) in linear, circular, and supercoiled forms was measured in salt solutions containing KCl, NaCl, and MgCl2. As the KCl concentrations were decreased from 1M to 0.1M, the time taken by a DNA molecule to pass through a nanopore was shorter and the frequency of the translocation in a folded configuration was reduced, suggesting an increase in DNA electrophoretic mobility and DNA persistence length. When the salt concentration was kept at 1M, but replacing K+ with Na+, longer DNA translocation times (td) were observed. The addition of low concentrations of MgCl2 with 1.6M KCl resulted in longer td and an increased frequency of supercoiled DNA molecules in a branched form. These observations were consistent with the greater counterion charge screening ability of Na+ and Mg2+ as compared to K+. In addition, we demonstrated that dsDNA molecules indeed translocated through a ~10 nm nanopore by PCR amplification and gel electrophoresis. We also compared the dependence of DNA mobility and conformation on KCl concentration and cation species measured at single molecule level by silicon nitride nanopores with existing bulk-based experimental results and theoretical predictions.
Keywords: Counterion charge screening, DNA conformation, DNA mobility, DNA translocation, Silicon nitride nanopore
1 Introduction
The binding of metal ions (cations or counterions) to negatively charged dsDNA molecules play crucial roles in DNA double helix stability and structure [1–4]. In an ionic solution, cations bind to negatively charged DNA molecules due to strong ionic interactions, and the bound counterions can partially neutralize the negative charge of DNA molecules; thereby, reducing their net or effective charge and changing their electrophoretic mobility and shape [2, 3]. The interactions of cations with DNA molecules have been studied experimentally by bulk electrophoresis-based measurements [5–9] and they have also been evaluated theoretically [4, 10–13]. These studies revealed details into how the binding of cations could neutralize the negative charges of DNA molecules, reduce their electrophoretic mobility, and affect their conformation. More recently, the interactions of cations with DNA molecules [14], negatively charged [15] and even neutral [16] polymers have also been studied with nanopores. In this work, we use the chloride salts of three phosphate-binding metal ions K+, Na+, and Mg2+ to study their effects on DNA translocation in silicon nitride nanopores. The goal of this study was to identify the optimum salt conditions to characterize DNA molecules during the development of a solid-state nanopore-based technology as an approach for high throughput DNA sequencing.
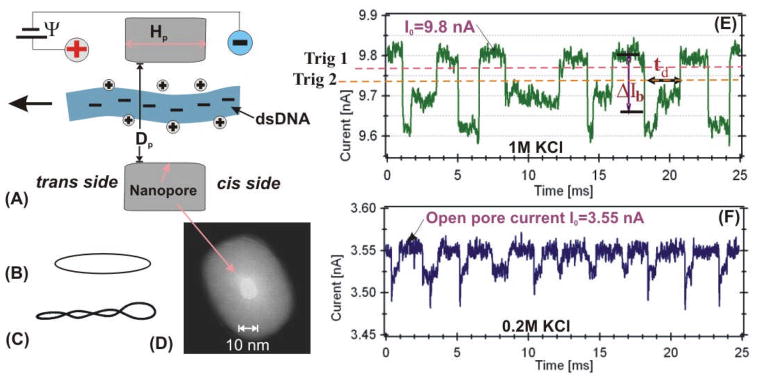
Voltage biased nanometer size pores allow charged particles to be measured one molecule at a time and have been developed for electronic detection and analysis of single biomolecules [17–23]. Motivated by the advancing nanopore-based technology, size adjustable solid-state nanopores have been fabricated with insulating materials such as silicon nitride, silicon dioxide, and aluminum oxides [24–35]. The electric field near a voltage-biased nanopore in an ionic solution can capture a charged DNA molecule (Fig. 1A) and drive it through the pore. A DNA molecule passing through a nanopore induces a measurable transient current drop (Fig. 1E) whose amplitude (ΔIb) and time duration (td) depend on the properties of the pore, solution, DNA, and the applied voltage. Our early studies on dsDNA translocation in solid-state nanopores in a solution containing 1M KCl showed that the time duration, td, was dependent on the pore diameter, Dp [25]. When the diameter was small, Dp<3 nm, very long tds were observed and it was hypothesized that the longer tds were the result of the interactions between a DNA and the nanopore. When the nanopore had Dp ≥ 8 nm, the distributions of the tds were approximately Gaussians and the most probable peak values of td were inversely proportional to the applied voltage Ψ, indicating that those DNA molecules were electrophoretically driven and passed through a nanopore “freely” (24). These observations were consistent with other recent studies that demonstrated the dependence of DNA translocation on nanopore size [36, 37].
Figure 1.
Schematic of a linear DNA molecule translocating through a nanopore (A). Other conformations of DNA measured in this work: circular relaxed (B) and circular supercoiled (C). TEM image of a ~10-nm silicon nitride nanopore (D). Examples of current blockage events for λ DNA measured in 1M KCl (E) and 0.2 M KCl (F) with the ~10 nm pore shown in (D).
DNA molecules are sensitive to the electrolyte environment such as salt concentration and ionic species [38]. In this work, using silicon nitride nanopores with a Dp ≥ 8 nm, we systematically study how the concentration of KCl and cation species affects DNA translocation time td and DNA conformation. To increase the range of td and the amplitude of ΔIb, especially at lower KCl concentrations, DNA molecules of different lengths were used, including: linear DNA of 3 kbp (~1 μm) and 48.5 kbp (~16.5 μm), circular relaxed and circular supercoiled DNA of ~4.4 kbp and 22.5 kbp. Furthermore, we demonstrated that a 1 kbp DNA fragment did indeed translocate through a ~10 nm silicon nitride nanopore using PCR amplification and agarose gel electrophoresis. We also compared our data to with existing experimental results and theoretical predictions.
2. Materials and methods
Solutions
KCl, NaCl, and MgCl2 used in this study were molecular biology or ultra-pure grade. All experiments were performed in a TE buffer (10 mM Tris, 1 mM EDTA, pH=7.5) at room temperature (~22°C). The buffered solutions were filtered through Whatman 0.02 μm nucleopore filters and degased prior to use.
DNA molecules
Three forms of DNA molecules were used in this work: linear 3 kbp plasmid pSP65 [29] and 48.5 kbp λ (Fig. 1A), circular relaxed 5.4 kbp PhiX174 (RFII) (Fig. 1B), circular supercoiled (above 90% supercoiled) ~4.4 kbp pBR322 (Fig. 1C), and a mixture of circular and supercoiled 22.5 kbp pXba (~80%). Gel electrophoresis showed the pXba mixture also contained a small amount (~5%) of linear DNA, data not shown). Except where indicated, the DNA molecules were purchased from New England Biolabs. The final DNA concentration in the cis chamber was ~10 nM for all the measurements.
Nanopores
Nanopores were fabricated in a freestanding ~280 nm thick low stress silicon nitride membrane supported by a 380 μm thick silicone substrate. The size of the freestanding membrane was ~30×30 μm2. The nanopores were made using a combination of focus ion beam milling and feedback controlled ion beam sculpting [24, 39, 40]. The size of nanopores was determined by Transmission Electron Microscope (TEM). Figure 1D shows a TEM image of a ~10 nm pore used for the λ DNA measurement (Fig. 1E and 1F). The thickness of the nanopores (Hp, shown in Fig. 1A) was estimated to be ~15 nm [40]. To minimize pore-to-pore variation in diameter, thickness, and surface charge, a single nanopore or multiple nanopores with approximately the same parameters were used to generate each data set.
Data acquisition
The details of measuring DNA in a solid-state nanopore were described previously [25, 29]. Briefly, a silicon nitride membrane with a nanopore (Fig. 1A) separates two salt solution-filled chambers. A constant bias voltage, Ψ=120 mV is applied to a pair of Ag/AgCl electrodes embedded in the solution. Salt solutions were exchanged by flushing both the cis and trans chambers through fluidic systems. The cis and trans chambers were cast in PDMS [29, 30]. Examples of current drop events produced by λ dsDNA are shown in Fig. 1E and Fig. 1F at 1 M and 0.2 M KCl, respectively. The current blockage events were recorded using an Axopatch 200B (Molecular Devices) in event driven and voltage-clamp mode. The low pass Bessel filter in the Axopatch 200B was set to 10 kHz or 100 kHz. The time response of the Axopatch 200B system at these filter settings was tested and calibrated with synthetic current drops: ideal square pulses of width ranging from 20 to 300 μsec generated from a function generator (Agilent 33250A) (see SI. 1a). When the pulse width was between 25 and 100 μs, the pulse height was attenuated, but the time durations can be measured accurately up to 25 μs under our data analysis routines as described below. The time response of a nanopore membrane was also tested at these filter settings (SI. 1b). Our analysis shows that the time response of a nanopore membrane is less than ~20 μs. Therefore, the bandwidth of the Axopatch and the membrane capacitance would not be a limiting factor for the time durations measured (td ≥50 μs).
Analysis of current blockage events
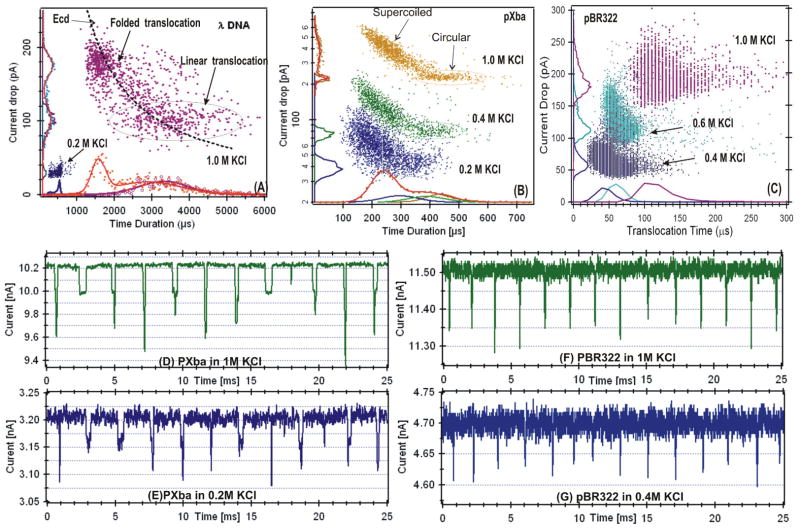
As illustrated in Fig. 1E, current drop events are characterized by its average amplitude ΔIb and time duration td. The integral of an event, referred to as the event charge deficit (ecd), as discussed previously [29], was also calculated. These parameters are extracted using our custom MATLAB routines (see SI. 3 for details), and are presented in the event distribution plots (i.e. Fig. 2A) in which each event, represented by a dot, is defined by its ΔIb and td. In a distribution of events, the presences of different DNA conformations are shown in separable clusters, and the events from the same conformation (cluster) are used for comparison. As an example, the λ DNA event distribution plot shown in Fig. 2A, depicts the cluster of linear and folded translocation events [25]. The td and ΔIb distributions are shown on the axes. The distributions were fitted with Gaussian functions (solid curve). A single Gaussian function was used if the event cluster is well separated or only one form of DNA was used. Two Gaussian functions were used if the events resulting from different DNA conformations were not well resolved. We used the fitted peak values, ΔIbp and tdp, as the most probable current drop and translocation time of the each cluster. The errors to the peak values are the standard deviations of the fittings. A table of the tdp and ΔIbp values for all DNA molecules at all KCl concentrations is included in table 1 for comparison.
Figure 2.
Event distribution plots for: (A) linear λ DNA in 1M and 0.2M KCl, (B) a mixture of circular and supercoiled 22.5 kbp DNA, and (C) supercoiled 4.4 kbp DNA. A separate nanopore was used for the data shown in each panel. Examples of the time histograms of these measurements are shown on the bottom axis. More examples of current blockage events are shown for pXba in 1M KCl (D) and in 0.2M KCl (E), for PBR322 in 1M KCl (F) and in 0.4M KCl (G). For the pBR322 4.4 kbp DNA data recording, the low pass Bessel filter in the Axopatch 200B was set to 100 kHz.
Table 1.
The tdp and ΔIbp values for all DNA molecules measured at all KCl concentrations.
| KCl [M] | λ DNA | pXba1 | pXba2 | pBR322 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Linear | Circular | Super coiled | Circular | Super coiled | ||||||||
| td [μs] | ΔIb[pA]/I0[nA] | td [μs] | ΔIb[pA]/I0[nA] | td [μs] | ΔIb[pA]/I0[nA] | td [μs] | ΔIb[pA]/I0[nA] | td [μs] | ΔIb[pA]/I0[nA] | td [μs] | ΔIb[pA]/I0[nA] | |
| 0.15 | 153 ± 42 | 32/7.0 | 82 ± 27 | 38/7.0 | ||||||||
| 0.2 | 568 ± 37 | 28/3.55 | 294 ± 45 | 42/3.2 | 180 ± 30 | 58/3.2 | 50 ± 16 | 29.6/2.7 | ||||
| 0.35 | 205 ± 52 | 37/14.4 | 108 ± 28 | 47/14.4 | ||||||||
| 0.4 | 1394 ± 270 | 39/4.65 | 386 ± 46 | 79/5.7 | 214 ± 51 | 110/5.7 | 223 ± 30 | 37/16.6 | 139 ± 26 | 47/16.6 | 66 ± 16 | 51.5/4.7 |
| 0.45 | 240 ± 56 | 44/18.3 | 147 ± 32 | 53/18.3 | ||||||||
| 0.6 | 2390 ± 275 | 73/5.4 | 71 ± 10 | 102.1/6.7 | ||||||||
| 0.8 | 2480 ± 310 | 81/6.7 | 83 ± 12 | 111.4/9.2 | ||||||||
| 1.0 | 2944 ± 645 | 104/9.8 | 434 ± 34 | 220/10.3 | 237 ± 31 | 315/10.3 | 495 ± 70 | 79/36.9 | 316 ± 53 | 104/36.9 | 94 ± 8 | 179.6/11.5 |
3. Results and discussion
3.1 ΔIb and td affected by KCl concentration
As the KCl concentration was decreased from 1M to 0.2M, the event distribution plots (Fig. 2), ΔIb versus td, show signature patterns of DNA translocation in solid-state nanopores. For the λ DNA in 1M KCl, the event distribution (Fig. 2A) shows a constant ecd = (ΔIbp·tdp), best fit to ΔIb=ecd/td, consistent with our previously published results for linear dsDNAs [25, 29, 41]. For the linear translocation events, defined as N=1, where N is the number of double-stranded helices in the pore, the most probable values were ΔIbp~104 pA and tdp~2.94±0.32 ms. For the folded or N=2 events, with two helices in the pore, the values were found to be ΔIbp ~180 pA and tdp~1.60±0.26 ms. In contrast, at 0.2M KCl, the event distribution did not show a clear separation between the N=1 and N=2 events as compared to 1M KCl. The most probable values measured were ΔIbp~28 pA and tdp ~ 0.57±0.04 ms.
For the circular form of DNA (22.5 kbp pXba) in 1 M KCl, the event distributions show that the ΔIbp ~220 pA (on the left axis of Fig. 2B), approximately twice the value of ΔIbp for N=1 λ DNA. This was expected since circular DNA must translocate with two helices in the pore (N= 2). For the pXba in a supercoiled form, the ΔIb was larger as the td was shorter as a result of the increased number of superhelical turns [41]. For the 4.4 kbp pBR322 DNA (≥ 90% in circular supercoiled form), the distribution of current blockage events were fit to single Gaussians. The short time durations, td ~430 μs for the pXba (Fig. 2B) and td ~ 100 μs for the pBR322 (Fig. 2C) in 1M KCl, were due to their shorter lengths. The small difference in ΔIb for N=2 could be the result of possible geometric differences along the length of the nanopores used for these two sets of measurements.
The data in Figure 2 demonstrated that as the KCl concentration was lowered, smaller ΔIb were observed. The much smaller ΔIb~20pA for the long (~16.5 μm) linear λ DNA at 0.2M KCl (Fig. 1F) is clearly seen in the recorded data traces compared with the events recorded in 1M KCl (Fig. 1E). These event distributions show that: 1) the ΔIb is approximately proportional to the KCl concentration or solution conductivity and the number of strands (N) in a pore; and 2) td is longer for longer DNA molecules. These observations were consistent with the fact that the DNA molecules were electrophoretically translocated through the nanopores during the measurements.
The previously reported positive spikes seen with SiO2 pores were not observed
The positive spikes at low salt concentrations (<0.4M) reported by other studies [32, 37, 42, 43] using SiO2 pores were not observed in our experiments. This is likely due to the fact that our nanopores were made with silicon nitride (SixN), instead of SiO2. Recent studies using silicon nitride pores similar to ours have shown that SixN nanopores have five times less surface charge density than SiO2 pores [44]. Thus, the expected crossover salt concentration for the positive spikes would be much lower than 120 mM KCl as reported [43].
3.2 Verification of DNA translocation through a nanopore
Shorter DNA translocations through α-hemolysin channel[17] and solid state nanopores[28] from the cis to the trans chamber have been verified by gel electrophoresis previously. To demonstrate that the current blockage events corresponded to the translocation of longer DNA molecules via the nanopore, a segment of 1-kbp of dsDNA was prepared by PCR amplification from the PhiX174 genome. The linear 1-kbp dsDNA was then measured in a translocation experiment using a pore with a Dp ~10 nm pore in 1M KCl. After ~40,000 events, the trans chamber solution was collected and the sample subjected to 30 rounds of PCR amplification using primers specific for the 1-kbp DNA segment. The PCR sample was analyzed by agarose gel electrophoresis and the result (Fig. S3) showed that the sample collected from the trans chamber was indeed the 1 kbp DNA segment initially placed in the cis chamber. Thus, the 1-kbp DNA traversed the nanopore during the translocation experiment (See SI.2).
3.3 Effect of KCl concentration on tdp
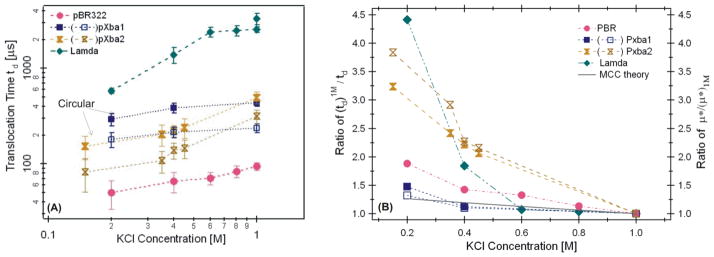
The most probable peak values of tdp as a function of KCl concentration was plotted as shown in Figure 3A. These data demonstrated that the tdp decreased as KCl concentration was lowered for all the DNA molecules measured. The corresponding open pore currents, I0, are approximately linearly proportional to the salt concentration within the range of 0.1 to 1M KCl (Fig. S4 in SI), consistent with our earlier work and that of others [30, 43]. As the KCl concentration was decreased, the experimental results in Figures 2 and 3 support the conclusion that the most probable tdp decreased as KCl concentration was lower, and the decrease in tdp varied with the length and conformation of DNA molecules. For the longest λ DNA molecules examined, tdp decreased by more than a factor of 4 as the KCl concentration was lowered from 1M to 0.2M (Fig. 3B).
Figure 3.
(A) The most probable peak values tdp measured as a function of KCl concentration. The filled square (■) is for the circular and the unfilled square (□) is for the supercoiled pXba measured in a ~10 nm pore. The filled hourglass (⧗) is for the circular and the unfilled hourglass (⧖) is for the supercoiled pXba measured in a ~25 nm pore. (B) The ratio of td1M/td for the DNA molecules measured in (A). The solid line is the theoretically predicted change from MCC theory (see SI.5).
DNA nanopore electrophoretic mobility and effective charge Q*
The interactions between cations and DNA molecules include: 1) specific cation binding or site-bound, 2) territorially binding of condensed counterions, and 3) the Debye-Huckel type interactions that make the DNA electrical potential go to zero exponentially beyond a characteristic Debye screening length [3]. Both specific and territorially binding can reduce a DNA molecule’s net charge Q, resulting in a smaller electrophoretic mobility μ (∝ Q) that can be observed experimentally. In a monovalent salt solution, the average net charge per phosphate on a DNA molecule, Q= (1−θ)q, is lowered from the elementary charge q by a factor of (1− θ), θ is the number of associated bound counterions per phosphate (see SI.6 for details). For example, Q=0.51q in 0.2M KCl and Q =0.38q in 1M NaCl were estimated by gel electrophoresis [5]. However, for a long DNA molecule with only a fraction of the molecule confined in a ~10 nm nanopore with charged walls, the measured DNA electrophoretic mobility as well as its effective charge is more complicated [45]. Below we attempt to use DNA nanopore electrophoretic mobility μ* and the effective charge Q* to describe the change related to td quantitatively. The nanopore electrophoretic mobility μ* is expected to be smaller than its bulk mobility μ by μ* ≈(Hp/Lc)μ due to the fact that only a fraction, Hp/Lc, of a DNA chain is in the pore [46]. Here Lc is the chain length of a DNA molecule.
An increase in μ* or Q* would lead to a shorter td
As KCl concentration was lower, the number of bound K+ ions would be less, leading to a smaller θ and a larger Q or a higher electrophoretic mobility μ. Assuming a DNA molecule moves along the central axis of a nanopore, the electric driving force balances the drag force, FE=−Fdrag [25, 47], and the DNA molecule moves at an average terminal velocity v̄≈Lc/td = μ* E0 (for Lc ≫ Hp). The electric force exerted on the local DNA segment in a pore is FE = SQ* E0, here E0=Ψ/Hp is the electrical field strength and S is the number of phosphates in the pore. The drag force amplitude can be written as Fdrag – fv–ηCfv, with f= ηCf is the friction coefficient, η is the solution viscosity, and Cf is a drag force constant depending on a DNA’s conformation and on the parameters not considered here. The time measured is inversely proportion to a DNA molecule’s mobility μ* and charge Q* as
| (1) |
We use Eq. (1) to emphasize the inverse relationship between td with μ* and Q* under our experimental conditions. The parameters inside the parenthesis are expected to be constants or to change negligibly as KCl concentration varies. Specifically, E0=Ψ/Hp was expected to be a constant, and the number of nucleic acids in a pore S was expected to be the same for the same Hp. The chain length Lc has been shown to have negligible dependence on ionic strength in single molecule experiments [48] and is also supported by theory [11]. Furthermore, an increase in Q* at lower KCl concentrations would increase LDNA due to an increase in Coulomb repulsion between bases, this would increase in td which would be in the opposite direction as what we observed. Thus, LDNA was unlikely to contribute to the decrease in td observed here. We note that the measured td has been found to have a nonlinear relationship with DNA chain length Lc, i.e., td ~ Lcα, where the exponent α was 1.26 [27] and 1.4 [41] for linear DNA molecules, but since Lc remains a constant (the same DNA) for each set of measurements, the rhs of equation (1) holds for any α. The viscosity η in bulk solution had been shown to increase slightly when KCl concentration was lower [49]. Therefore, we do not anticipate that η would contribute to the reduction of td as KCl concentration was decreased.
Electro-osmotic flow effects
The simple model used to derive Eq. (1) has ignored many complex issues involved in DNA translocation such as electro-osmotic flow. For a negatively charged DNA molecule passing through a pore with a negatively charged surface, electro-osmotic flow would act to increase td which has been used to explain that translocation times vary little with salt concentration [43, 47] in SiO2 nanopores. In addition, surface charge effects from a nanopore wall would increase at lower salt due to a longer Debye screening length, 0.3 nm (in 1M KCl) to 1 nm (in 0.1M KCl), which would further increase tdp compared with higher salt concentrations [47].
In summary, we conclude that a shorter td measured at lower KCl concentration suggests an increase in DNA electrophoretic mobility μ* or in DNA net charge Q* or a decrease in Cf. Below we discuss whether the change in Q* could account for all the change in td measured in Figure 3.
The change in Q* could not account for all the change in td1M/td
Comparing the ratio of td1M/td as a function of KCl concentration (Fig. 3B), the plots suggest that the ratio of td1M/td increased as the KCl concentration was lower, but the amount of increase depends on the DNA length and conformation. With the ~10 nm size pores, the ratio of td1M/td0.2M (or μ*0.2M/μ*1M)=1.5, 1.9, 4.5 for the circular ~22.5kbp, supercoiled 4.4 kbp, and the 48.5 kbp λ DNA, respectively. The effective charge Q* per phosphate and viscosity η were expected to vary the same amount as KCl concentration was varied from 1M to 0.2 M, thus the large mobility variations suggest that the drag force constant Cf depended on the conformation and length of DNA molecules. In addition, using the same pXba DNA, the ratio of td1M/td was greater when the measurement was performed with a larger nanopore (~25 nm) (see Fig. 3).
The much larger effect of KCl concentration on λ DNA translocation times must be due to its long length, ~ 16 μm. Considering the length of the local translocating segment of a DNA molecule in a pore is Hp (Fig. 1A), the same regardless its chain length outside the pore, the configuration and motion of the chain outside the pore, such as unwinding upon entering, must be responsible for the greater KCl concentration effect on td of λ DNA. Recent theoretical studies have shown that the transport of DNA through pores is controlled by entropic barriers that accompany their conformational changes [50]. However, the conformation and motion of the DNA chain outside a pore and how it affects td is beyond the scope of this paper.
In conclusion, the variations in the ratio of td1M/td for the DNA molecules shown in Fig. 3 suggest that the effective charge Q* is not the only parameter that affects the td, the other parameters such as DNA chain length, conformation, and nanopore size may all contribute to td.
3.4 KCl concentration on DNA Conformation
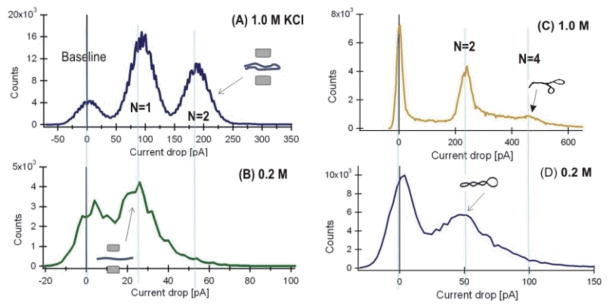
The plots of the instantaneous distribution of ΔIb in 5-μs samples over all events (Fig. 4) show possible DNA configurations measured in nanopores [25, 51]. The quantified blockage currents of N=1 and N=2 peaks at 1.0 M (Fig. 4A) and 0.2 M (Fig. 4B) correspond to one and two strands of λ dsDNA helices in a pore. At 1M KCl, the ratio of DNA at N=2 versus N=1 is 11:16, or 41% DNA molecules in the N=2 conformation (Fig. 4A). In 0.2M KCl, the ratio is 1:10, or only ~9% of the DNA molecules in the N=2 conformation. The much smaller N=2 peak at 0.2 M KCl suggests that the frequency at which a λ DNA molecule was in folded translocation configuration was less favorable.
Figure 4.
The distribution of 5-μs current samples per 2 pA bin of ΔIb at 1 M and 0.2 M KCl. (A) and (B) are for λ DNA data shown in Figure 2A, the insets show the assumed DNA translocation configurations for N=1 and N=2. (C) and (D) are for the supercoiled events of the pXba DNA in Fig. 2B. The baseline peaks at 0 pA corresponds to the open pore level (I0).
For the circular supercoiled pXba DNA in 1.0 M (Fig. 4C) and 0.2 M KCl (Fig. 4D), the quantified blockage currents N=2 and N=4 correspond to two and four strands of dsDNA helices in a pore. The N=4 peak represents DNA molecules in a branched form at 0.2 M KCl that was smaller compared to the same peak in 1M KCl. However, the reduction in the N=4 peak was not as significant as measured for the linear λ DNA.
DNA molecules are less likely to be in folded form at lower KCl concentration
The folding state of a DNA molecule in a nanopore could be caused by either a pre-existing conformation that was captured, or a forced folding or bending by the electric force at the entrance of a nanopore [25]. At low salt concentrations, the reduced probability of detecting DNA in a folded configuration suggested that a DNA molecule had fewer pre-existing folded states or was less bendable by the electric force from the pore as the translocation started. This observation is consistent with other experimental and theoretical studies demonstrating that as the concentration of cations surrounding the DNA decreases, the number of binding counterions to DNA reduces, causing an increase in its effective charge, leading to an increase in its electrophoretic mobility and persistence length [11, 38, 48, 52]. An increase in persistence length, from ~35nm (c=1M) to ~55nm (c=0.1M) [52], would reduce the probability of DNA forming pre-existing folded states and also would be less likely to be bent at the nanopore entrance by an electrical force of the same magnitude (same voltage was applied), which is consistent with our observations (Fig. 4). At higher salt concentrations, due to increased counterion screening, a supercoiled DNA molecule will adopt a highly compact form with more winding and branching than at lower ionic screening [38, 53].
3.5 DNA in more tightly bound Na+ and Mg2+ solution
To further evaluate how cation binding would affect DNA translocation, we performed two more sets of experiments maintaining all of the previous parameters except: 1) K+ was replaced with Na+; and 2) adding of low concentrations of Mg2+ to the KCl solution.
3.5.1 Replacement of K+ with Na+
In a 1M NaCl solution, the open pore current I0 and the current drop ΔIb were both modified by the solution conductivity due to the mobility of Na+ that was reported as 5.19 ×10−4 (cm/s)/(V/cm) in contrast to that of K+ reported as 7.62 ×10−4 (cm/s)/(V/cm) [54]. For a ~12 nm diameter nanopore, the I0 was 9.5 nA for 1M KCl. A 3 kbp linear dsDNA (pSP65) was added to the cis chamber. After ~ 5,000 events were recorded, both cis and trans chambers were extensively flushed with 1M NaCl. The I0 was 7.1 nA for the 1M NaCl solution as expected. The current blockage histograms (Fig. S5A, SI) showed the peak values are ΔIb=105 ± 16 pA for KCl and ΔIb=80 ± 17 pA for NaCl. Histograms of the translocation times (Fig. S5B, SI) showed the peak values are td = 90 ± 17 μs in KCl and td = 135 ± 16 μs in NaCl. That is, tdNa+/tdK+=μ*K+/μ*Na+=135/90=1.5.
In summary, keeping all other parameters the same except for replacing K+ with Na+, a longer td was measured. These experiments demonstrated that the change in td was due to the difference in metal ion binding and suggested that Na+ ions bind to DNA molecules more tightly resulting in a smaller Q* and an increased td, consistent with reports from bulk based measurements. The change in td and in ΔIb observed here are also consistent with recent published nanopore experimental and simulation results [14]. We can estimate the change in Q* using td ~η/Q* (Eq. 1) by assuming the difference in the drag force constant Cf in 1M NaCl versus 1M KCl was negligible. To correct the viscosity difference, the viscosity of these two salt solutions was measured with a GV-2100 (Gilmont Instruments) ball drop viscometer. The measurements showed that the viscosity of the NaCl solution was 10% greater than the KCl solution, in agreement with interpolated published data [55, 56]. That is: A value of is calculated. This relative decrease in Q* for the 3 kpb DNA in NaCl versus KCl is very close to the reported values for DNA in 0.2 M NaCl versus 0.2 M KCl, QNa/Qk = 0.38/0.51=75% was measured by bulk electrophoresis [5].
3.5.2 DNA in more tightly bound Mg2+
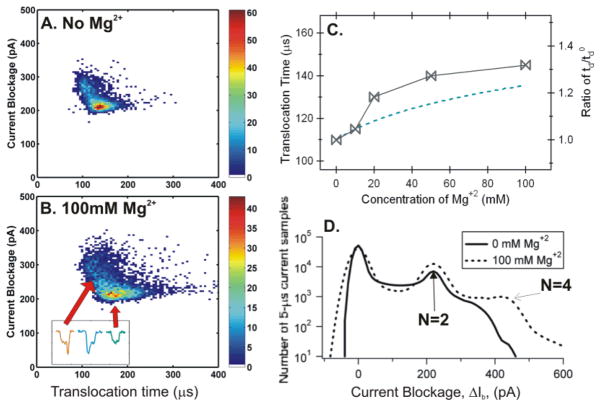
We initially attempted to use the 3 kbp linear DNA to evaluate the effect of MgCl2 addition; however, these studies resulted in blocking of the nanopores at MgCl2 concentration above 10 mM [57]. Thus, a 4.4 kbp circular supercoiled DNA (pBR322) was used for the measurements. The experiment was first performed in 1.6 M KCl solution with 10 mM Tris (pH=7.5), 20% glycerol (its electrical conductivity equivalent to 1M KCl) and 0 mM Mg+2. The same solution with 10, 20, 45, and 100 mM MgCl2 was subsequently exchanged in the chambers and the same experiment was performed. The open pore current I0 and the current blockades ΔIb were approximately the same, regardless of Mg+2 concentration. The peak values in solution from 0 mM Mg+2 (Fig. 5A) to 100 mM Mg+2 (Fig. 5B) were ΔIb=210 ± 11 pA representing nicked circular DNA as reported previously [27]. The translocation time, tdp, was increased from 110 ± 5 μs (at 0 mM Mg+2) to 145 ± 11 μs (at 100 mM Mg+2), a 32% increase in td (Fig. 5c). The increase in td was consistent with early reports that divalent ion Mg2+ binds to DNA more tightly than the monovalent ion K+, and adding divalent salt MgCl2 to a KCl solution would further reduce the net charge Q* and mobility μ of the DNA [7, 11].
Figure 5.
Event number density plots for (A) 0 mM and (B) 100 mM Mg+2 in TE buffer with 20% glycerol. The insert in Fig. 5B shows three events, one corresponding to a circular DNA and the other two corresponded to branched or bent supercoiled DNA. (C) Left, td as a function of the amount of Mg+2 ions added. Right, the change in . The Dashed line is the theoretically predicted values from MCC theory (see SI.4.). (D) Histograms of the number of 5-μs current blockage samples per 2 pA bin for 0 and 100 mM Mg+2. A ~14 nm pore was used with I0 = 11.25 nA.
The increase in td
During the above set of measurements, the only variable was the addition of MgCl2 (≤100 mM) to the KCl solution. The more tightly bound Mg+2 ions would further decrease the effective charge Q* as well as the mobility μ* of the DNA molecules [7, 9, 58], resulting in a longer td as we observed. All parameters, including Ψ, LDNA, N, S, Cf, η, except Q* in Eq. (1) can be considered as constants or would not contribute to the increase in td. The difference in viscosities for a small salt concentration change (≤100 mM) is negligible if interpolated from published data [55]. As the Q* decreases, its length (LDNA) was expected to be shorter due to a decreased Coulomb repulsion, thus, LDNA was unlikely to contribute to the increase in td. Therefore, the change in Q* can be estimated from Eq. (1) by td ~1/Q*. For example, at 100mM Mg+2, the ratio of (Fig. 5C, right). Here is the translocation time at 0 mM Mg+2. Using , the estimated DNA net charge is Q100mM =0.76Q0. In other words, these results show that adding Mg+2 ions can further neutralize a DNA’s charge or decrease its mobility in a KCl solution.
An increase in N=4 conformation
The plots of the distribution of instantaneous ΔIb samples over all events (Fig. 5D) show that the current blockage levels occurred in integer multiples, and the frequency of N=4 configuration increased as more Mg+2 ions were added. At 100 mM Mg+2 (Fig. 5D) the supercoiled DNA had more N=4 configurations inside the nanopore as compared to the absence of Mg+2. These results indicate that the supercoiled DNA molecules were more branched or more bendable as more Mg+2 ions were added. As the Mg+2 concentration increases, the increased counterion screening would further reduce DNA effective charge Q* that could cause a supercoiled DNA molecule to become more tightly wound; eventually winding tightly enough to produce a greater number of branched supercoiled DNA molecules [59–61]. It was also possible that a decrease in the net charge made a DNA molecule be more bendable at the entrance of a nanopore.
Notes: limits of this study
The study of chloride salts on DNA translocation in silicon nitride nanopores described in this work was limited by several factors. First, at salt concentrations lower than 0.1 M, the current drop was too small to be measured accurately. Second, divalent and trivalent counterions bind to DNA molecules more tightly than monovalent ions, having a greater screening of electrostatic charge causing the DNA to precipitate out of solution. The attempts to measure DNA in divalent ion salts alone, Mg+2 and Ca+2, as well as trivalent ions Al+3 and Co(NH3)6+3 were not successful due to DNA precipitation, indicating the limitations to increasing DNA translocation time by reducing DNA net charge or increasing counterion binding. Lastly, the silicon nitride nanopores often became unusable (a change of I0 or too noisy) before a complete set of experiments could be performed.
Concluding Remarks
At single molecule level, we have studied how KCl concentration and metal ion binding affects DNA translocation through solid state nanopores. DNA molecules representing different forms, including linear, circular, and supercoiled, and with wide range of lengths from ~1 μm to ~16.5 μm were measured in this work. As the ionic strength was increased, our nanopore measurements showed that DNA molecules became more folded or branched and their translocation times in a nanopore were longer. By analyzing how the relative DNA translocation times changed as a function of KCl concentration, our results suggest that the change in DNA effective charge due to counterion screening played important roles in DNA translocation dynamics, however, other parameters such as DNA length, conformation, as well as nanopore size also contributed significantly. Moreover, our studies indicate that Na+ is more effective at screening DNA charge than K+, and adding Mg+2 to a KCl solution can further neutralize the charge of DNA molecules. The studies in this work will improve our understanding of nanopore-based DNA sensing and on DNA-counterion binding at single molecule level.
Supplementary Material
Acknowledgments
The authors thank Professor J. Golovchenko for FIB pore preparation, Dr. Slaven Garaj for helpful discussion, John Wang for Matlab programming, and Bradley Ledden for nanopore fabrication. Support of this research has been provided by NIH R21HG003290, NIH R21HG004776, ABI1114, and partially supported by NSF/MRSEC 080054.
A list of abbreviations used in the paper
- SI
- TE buffer
10 mM Tris at pH 7.5 with 1 mM EDTA
References
- 1.Sigel H. Chem Soc Rev. 1993;22:255–267. [Google Scholar]
- 2.McFail-Isom L, Sines CC, Williams LD. Current Opinion in Structural Biology. 1999;9:298–304. doi: 10.1016/S0959-440X(99)80040-2. [DOI] [PubMed] [Google Scholar]
- 3.Bloomfield VA, Crothers DMI, Tinoca J. Nucleic Acids: structures, properties, and functions. University Science Books; 1999. pp. 475–534. [Google Scholar]
- 4.Tan ZJ, Chen SJ. Biophys J. 2006;90:1175–1190. doi: 10.1529/biophysj.105.070904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ross PD, Scruggs RL. Biopolymers. 1964;2:231–236. [Google Scholar]
- 6.Rhee KW, Ware BR. J Chem Phys. 1983;78:3349–3353. [Google Scholar]
- 7.Li AZ, Huang H, Re X, Qi LJ, Marx KA. Biophys J. 1998;74:964–973. doi: 10.1016/S0006-3495(98)74019-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stellwagen E, Stellwagen NC. Biophys J. 2003;84:1855–1866. doi: 10.1016/S0006-3495(03)74993-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ahmad R, Arakawa H, Tajmir-Riahi HA. Biophys J. 2003;84:2460–2466. doi: 10.1016/S0006-3495(03)75050-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feig M, Pettitt BM. Biophys J. 1999;77:1769–1781. doi: 10.1016/S0006-3495(99)77023-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Manning GS. Q Rev Biophys. 1978;2:179–246. doi: 10.1017/s0033583500002031. [DOI] [PubMed] [Google Scholar]
- 12.Manning GS. J Phys Chem. 1981;85:1506–1515. [Google Scholar]
- 13.Luan B, Aksimentiev A. Phys Rev E. 2008:78. [Google Scholar]
- 14.Kowalczyk SW, Wells DB, Aksimentiev A, Dekker C. Nano Lett. 2012;12:1038–1044. doi: 10.1021/nl204273h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oukhaled G, Bacri L, Mathe J, Pelta J, Auvray L. Europhys Lett. 2008:82. doi: 10.1103/PhysRevLett.100.158302. [DOI] [PubMed] [Google Scholar]
- 16.Reiner JE, Kasianowicz JJ, Nablo BJ, Robertson JWF. PNAS. 2010;107:12080–12085. doi: 10.1073/pnas.1002194107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kasianowicz JJ, Brandin E, Branton D, Deamer DW. Proc Natl Acad Sci USA. 1996;93:13770–13773. doi: 10.1073/pnas.93.24.13770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Akeson M, Branton D, Kasianowicz JJ, Brandin E, Deamer DW. Biophys J. 1999;77:3227–3233. doi: 10.1016/S0006-3495(99)77153-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meller A, Nivon L, Brandin E, Golovchenko J, Branton D. Proc Natl Acad Sci USA. 2000;97:1079–1084. doi: 10.1073/pnas.97.3.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gu LQ, Braha O, Conlan S, Cheley S, Bayley H. Nature. 1999;398:686–690. doi: 10.1038/19491. [DOI] [PubMed] [Google Scholar]
- 21.Nakane J, Wiggin M, Marziali A. Biophys J. 2004;87:615–621. doi: 10.1529/biophysj.104.040212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Astier Y, Braha O, Bayley H. J Am Chem Soc. 2006;128:1705–1710. doi: 10.1021/ja057123+. [DOI] [PubMed] [Google Scholar]
- 23.Clarke J, Wu H, Jayasinghe L, Patel A, et al. Proc Natl Acad Sci USA. 2009;106:7702–7707. [Google Scholar]
- 24.Li J, Stein D, McMullan C, Branton D, et al. Nature. 2001;412:166–169. doi: 10.1038/35084037. [DOI] [PubMed] [Google Scholar]
- 25.Li J, Gershow M, Stein D, Brandin E, Golovchenko JA. Nat Mater. 2003;2:611–615. doi: 10.1038/nmat965. [DOI] [PubMed] [Google Scholar]
- 26.Storm AJ, Chen JH, Ling XS, Zandbergen HW, Dekker C. Nat Mater. 2003;2:537–540. doi: 10.1038/nmat941. [DOI] [PubMed] [Google Scholar]
- 27.Storm AJ, Chen JH, Zandbergen HW, Dekker C. Phys Rev E. 2005;71:0519031–05190310. doi: 10.1103/PhysRevE.71.051903. [DOI] [PubMed] [Google Scholar]
- 28.Heng JB, Ho C, Kim T, Timp R, et al. Biophys J. 2004;87:2905–2911. doi: 10.1529/biophysj.104.041814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fologea D, Gershow M, Ledden B, McNabb DS, et al. Nano Lett. 2005;5:1905–1909. doi: 10.1021/nl051199m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fologea D, Uplinger J, Thomas B, McNabb DS, Li J. Nano Lett. 2005;5:1734–1737. doi: 10.1021/nl051063o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gershow M, Golovchenko JA. Nat Nanotechnol. 2007;2:775–779. doi: 10.1038/nnano.2007.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peng H, Ling XS. Nanotechnology. 2009;20 doi: 10.1088/0957-4484/20/18/185101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Venkatesan BM, Shah AB, Zuo JM, Bashir R. Adv Funct Mater. 2010;20:1266–1275. doi: 10.1002/adfm.200902128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Singer A, Wanunu M, Morrison W, Kuhn H, et al. Nano Letters. 2010;10:738–742. doi: 10.1021/nl100058y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wanunu M, Dadosh T, Ray V, Jin J, et al. Nat Nano. 2010;5:807–814. doi: 10.1038/nnano.2010.202. [DOI] [PubMed] [Google Scholar]
- 36.Wanunu M, Sutin J, McNally B, Chow A, Meller A. Biophys J. 2008;95:4716–4725. doi: 10.1529/biophysj.108.140475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dorp Sv, Keyser UF, Dekker NH, Dekker C, Lemay SG. Nat Phys. 2009;5:347–351. [Google Scholar]
- 38.Schlick T, Li B, Olson WK. Biophys J. 1994;67:2146–2166. doi: 10.1016/S0006-3495(94)80732-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stein DM, McMullan CJ, Li J, Golovchenko JA. Rev Sci Instrum. 2004;75:900–905. [Google Scholar]
- 40.Cai Q, Ledden B, Krueger E, Golovchenko JA, Li J. J Appl Phys. 2006:100. doi: 10.1063/1.2216880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fologea D, Brandin E, Uplinger J, Branton D, Li J. Electrophoresis. 2007;28:3168–3192. doi: 10.1002/elps.200700047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chang H, Kosari F, Andreadakis G, Alam MA, et al. Nano Lett. 2004;4:1551–1556. [Google Scholar]
- 43.Smeets RM, Keyser UF, Krapf D, Wu MY, et al. Nano Lett. 2006;6:89–95. doi: 10.1021/nl052107w. [DOI] [PubMed] [Google Scholar]
- 44.Hoogerheide DP, Garaj S, Golovchenko JA. Phys Rev Lett. 2009:102. doi: 10.1103/PhysRevLett.102.256804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Keyser UF, van Dorp S, Lemay SG. Chem Soc Rev. 2009;39:939–947. doi: 10.1039/b902072c. [DOI] [PubMed] [Google Scholar]
- 46.Li J, Talaga DS. J Phys Condens Matter. 2010;22:454129, 454128. doi: 10.1088/0953-8984/22/45/454129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ghosal S. Phys Rev Lett. 2007:98. doi: 10.1103/PhysRevLett.98.238104. [DOI] [PubMed] [Google Scholar]
- 48.Baumann CG, Smith SB, Bloomfield VA, Bustamante C. Proc Natl Acad Sci USA. 1997;94:6185–6190. doi: 10.1073/pnas.94.12.6185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brown WR, Yousef HNS. J Colloid Interface Sci. 2003;264:452–457. doi: 10.1016/S0021-9797(03)00406-5. [DOI] [PubMed] [Google Scholar]
- 50.Muthukumar M. Annu Rev Biophys Biomol Struct. 2007;36:435–450. doi: 10.1146/annurev.biophys.36.040306.132622. [DOI] [PubMed] [Google Scholar]
- 51.Storm AJ, Storm C, Chen J, Zandbergen H, et al. Nano Lett. 2005;5:1193–1197. doi: 10.1021/nl048030d. [DOI] [PubMed] [Google Scholar]
- 52.Manning GS. Biophys J. 2006;91:3607–3616. doi: 10.1529/biophysj.106.089029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Anderson P, Bauer W. Biochemistry. 1978;17:594–601. doi: 10.1021/bi00597a006. [DOI] [PubMed] [Google Scholar]
- 54.Hille B. Ion Channels of Excitable Membranes. Sinauer Associates, Inc; Sunderland, MA: 2001. [Google Scholar]
- 55.Weast RC, editor. CRC Handbook of Chemistry and Physics, 1985–1986. CRC Press, Inc; Boca Raton, FL: 1985. [Google Scholar]
- 56.Nickels L, Allmand AJ. J Phys Chem. 1936;41:861–872. [Google Scholar]
- 57.Bloomfield VA. Curr Opin Struct Biol. 1996;6:334–341. doi: 10.1016/s0959-440x(96)80052-2. [DOI] [PubMed] [Google Scholar]
- 58.Korolev N, Lyubartsev AP, Rupprecht A, Nordenskiold L. Biopolymers. 2001;58:268–278. doi: 10.1002/1097-0282(200103)58:3<268::AID-BIP1004>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 59.Bendar J, Furrer P, Stasiak A, Dubochet J. J Mol Biol. 1994;235:825–847. doi: 10.1006/jmbi.1994.1042. [DOI] [PubMed] [Google Scholar]
- 60.Fogg JM, Kolmakova N, Rees I, Magonov S, et al. J Phys Condens Matter. 2006;18:S145–S159. doi: 10.1088/0953-8984/18/14/S01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xu YC, Bremer H. Nucleic Acids Res. 1997;25:4067–4071. doi: 10.1093/nar/25.20.4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.