Abstract
In plants, basic leucine zipper (bZIP) proteins regulate numerous biological processes such as seed maturation, flower and vascular development, stress signalling and pathogen defence. We have carried out a genome-wide identification and analysis of 125 bZIP genes that exist in the maize genome, encoding 170 distinct bZIP proteins. This family can be divided into 11 groups according to the phylogenetic relationship among the maize bZIP proteins and those in Arabidopsis and rice. Six kinds of intron patterns (a–f) within the basic and hinge regions are defined. The additional conserved motifs have been identified and present the group specificity. Detailed three-dimensional structure analysis has been done to display the sequence conservation and potential distribution of the bZIP domain. Further, we predict the DNA-binding pattern and the dimerization property on the basis of the characteristic features in the basic and hinge regions and the leucine zipper, respectively, which supports our classification greatly and helps to classify 26 distinct subfamilies. The chromosome distribution and the genetic analysis reveal that 58 ZmbZIP genes are located in the segmental duplicate regions in the maize genome, suggesting that the segment chromosomal duplications contribute greatly to the expansion of the maize bZIP family. Across the 60 different developmental stages of 11 organs, three apparent clusters formed represent three kinds of different expression patterns among the ZmbZIP gene family in maize development. A similar but slightly different expression pattern of bZIPs in two inbred lines displays that 22 detected ZmbZIP genes might be involved in drought stress. Thirteen pairs and 143 pairs of ZmbZIP genes show strongly negative and positive correlations in the four distinct fungal infections, respectively, based on the expression profile and Pearson's correlation coefficient analysis.
Keywords: bZIP transcription factor family, maize, phylogenetic analysis, gene expression profile analysis, co-regulatory pathway
1. Introduction
Among several transcription factor families that present exclusively in eukaryotes, the basic leucine zipper (bZIP) transcription factor family is one of the largest and most diverse families. bZIP transcription factors are named according to their common feature, bZIP domain, which is ∼60–80 amino acids in length and consists of a basic region and a Leu zipper.1 The basic region of around 18 amino acid residues with an invariant motif N-x7-R/K-x9 is responsible for nuclear localization and DNA binding, whereas the following Leu zipper motif made up of several heptad repeats of Leu or other bulky hydrophobic amino acids, such as Ile, Val, Phe or Met, is less conserved and mediates the homo- and/or heterodimerization of bZIP proteins. Plant bZIP proteins present a relaxed binding specificity for DNA sequence motifs containing an ACGT core, and preferentially bind to the G-box (CACGTG), C-box (GACGTC) and A-box (TACGTA).2 At the time of DNA binding, the N-terminal half of the basic region inserts into the major groove of double-stranded DNA and the C-terminal half of the Leu zipper mediates dimerization to form a superimposed coiled-coil structure.3,4
Members of the bZIP transcription factor family have been identified or predicted in many eukaryotic genomes. It has been reported that 17 bZIP genes in Saccharomyces cerevisiae, 31 in Caenorhabditis elegans, 27 in Drosophila,5 56 in humans,6 75 in Arabidopsis (Arabidopsis thaliana),7 89 in rice (Oryza sativa),8 92 in Sorghum9 and 131 in soybean (Glycine max).10 In spite of this, only a small part of bZIP transcription factors have been functionally validated in plants. Data show that plant bZIP proteins participate in the differentiation of many organs and tissues, embryogenesis, seed maturation, floral transition and initiation, and vascular development. On the other hand, bZIP proteins are involved in signalling and responses to abiotic/biotic stimuli, including ABA signalling, osmotic, hypoxia, drought, high salinity and cold stresses, and pathogen defence. Available information exhibits that some bZIPs also respond to light irradiation and are involved in photomorphism.
Zea mays (hereafter called maize) is a major cereal crop which is grown for food, feed, fibre and fuel, and also an important model system for basic biological researches.11,12 The maize genome has been completely sequenced in 2009 using the B73 inbred line as a genome donor.13 This provides us a good opportunity to further our research on maize and also enhance our understanding of its grass relatives such as sorghum, wheat and rice. In previous studies, only 12 maize bZIP transcription factors have been isolated and functionally characterized. Among these, the annotation of Opaque-2 (O2) is the most clear; it controls the transcription of α-zein, b-32 and β-prolamin genes and regulates protein accumulation, amino acid and sugar metabolism in maize seeds.14–18 OHP1 and OHP2, mEmBP-1a and mEmBP-1b are also possible zein regulatory proteins that can bind to the O2 box from the 22-kDa zein gene promoter as a homodimer, while OHP1 and OHP2 can also bind as a heterodimeric complex with the O2 target site.19,20 Delayed flowering1 mediates floral transition at the shoot apex serving the same role as liguleless2 (lg2).21–23 Genetic analysis indicates that lg1 and lg2 are uniquely necessary for ligule and auricle development and play different roles in the ligule–auricle induction mechanism.24 Mlip15 is a low temperature-induced gene, its transcript abundance is also strongly increased under salt stress and exogenous abscisic acid treatment, signifying a stress response role of this bZIP member.25 Octopine synthase element-binding factor-1 is expected to have putative roles in one or more aspects of leaf development.26 GBF1 is assumed to be involved in the activation of Adh1, which is a hypoxia-responsive protein.27 Two other ocs-element binding proteins, called OBF3.1 and OBF3.2, sharing 80% amino acid identity with the wheat HBP1b protein, have unknown functions. It is clear that to perform a comprehensive functional exploration for this gene family is a very urgent requirement.28
2. Methods
2.1. Identification of maize bZIP transcription factors
To identify all the putative bZIP proteins in the maize genome, we performed both local BLAST and hidden Markov model profile searches using the maize proteome sequence (ZmB73_5b_FGS_translations.fasta.gz) from the Maize Genome Sequence Project (http://ftp.maizesequence.org/current/filtered-set/) as a database and the known bZIP protein sequences from maize, rice and Arabidopsis as a query. The E-value was set to 1. Then these proteins were subjected to the National Center of Biotechnology Information (NCBI) CD search (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi), SMART (http://smart.embl-heidelberg.de/) and Pfam (http://pfam.sanger.ac.uk/) databases to ensure the presence of the bZIP domain. After removing the repeat and incomplete sequences manually, 170 proteins remained were named ZmbZIP transcription factors. The putative orthologs from Arabidopsis and rice were assigned to corresponding ZmbZIP proteins with the E-value below 1E−10, which were extracted by a BLASTP search from NCBI.
2.2. Phylogenetic reconstruction
All of the phylogenetic trees in this article were reconstructed by the maximum likelihood method using the PhyMl 3.0 software29 with the best model selected by the Akaike information criterion implemented in ProtTest 3.0.30 Bootstrap values from 100 replicates are indicated at each node. The bZIP protein sequences of Arabidopsis (A. thaliana) and rice (O. sativa) were downloaded from the TAIR (http://www.arabidopsis.org/) and TIGR (http://rice.plantbiology.msu.edu/) databases, respectively. Then, the bZIP domain sequences of maize, Arabidopsis and rice, totally 333 sequences were loaded into PhyMl with the model JTT + G. In the same way, the phylogenetic trees generated based on the sequences of the maize bZIP domain, basic and hinge region and leucine zipper defined from the first leucine were best fitted to the model JTT + G + F, LG + G and JTT + I + G + F, respectively. Then the trees were visualized by iTOL.31
2.3. Identification of additional conserved motifs
To identify the additional conserved motifs outside the bZIP domain of ZmbZIP transcription factors, we sent 170 ZmbZIP protein sequences into Multiple Em (Expectation Maximization) for the Motif Elicitation tool (MEME version 4.8.1, http://meme.sdsc.edu/meme/cgi-bin/meme.cgi). The limits of minimum width, maximum width and maximum number of motifs were specified as 10, 50 and 50, respectively, since it excludes the bZIP domain which usually possesses 60–80 amino acids in length. Thirty motifs were finally confirmed because of their low E-value (<E−48). The motifs were numbered according to their order displayed in MEME and were considered as group-specific signatures for their presence of high frequency in the given groups.
2.4. Gene structure and conserved intron splicing site analysis
To define the actual intron–exon arrangements of all the ZmbZIP genes, both gene sequences and corresponding coding sequences were needed and loaded into the Gene Structure Display Server (http://gsds.cbi.pku.edu.cn/). For a better visualization and comparison, the 5′ UTR sequences were removed beforehand. The positions of the bZIP domain were retrieved with PAL2NAL (http://www.bork.embl.de/pal2nal/) by converting a multiple sequence alignment of domain sequences and the corresponding gene sequences into a codon alignment, and then marked in red in the Supplementary Fig. S3.
2.5. Chromosomal localization of genes and detection of duplication events
Each of the ZmbZIP genes was located on maize chromosomes using the GenomePixelizer software (http://www.atgc.org/GenomePixelizer/), which was designed to help visualize the relationships between duplicated genes in chromosomes. We adopted a group-specific colour strategy to mark each gene for better visualization and recognition.
We adopted the definition that two or more ZmbZIP genes intervened by less than eight non-bZIP genes were considered as a gene cluster. The DAGchainer (http://dagchainer.sourceforge.net/) and the GenoPixelizer 2D plotter (http://www.atgc.org/GenoPix_2D_Plotter/) were used to find the duplicated gene pairs which are located in duplicated chromosomal segments, and SyMap (http://www.agcol.arizona.edu/software/symap/), which was developed to compute synteny blocks in a sequenced genome, was applied to discover duplicated regions and analyse larger-scale synteny blocks in maize.
2.6. Construction of a three-dimensional structure
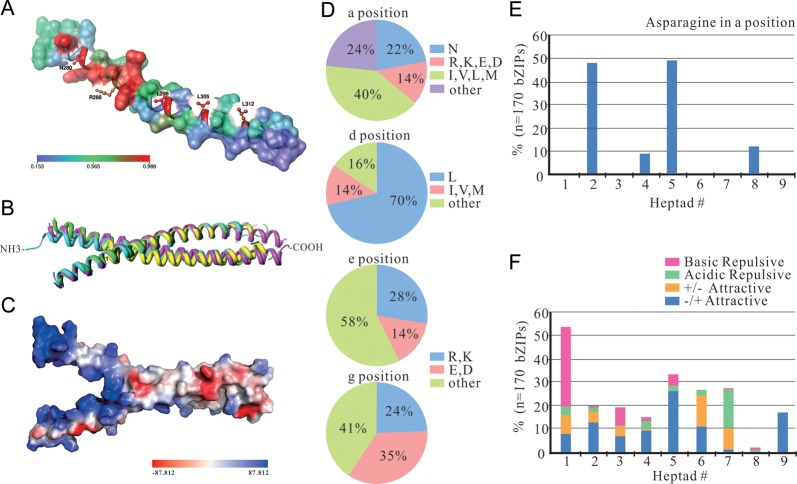
The surface mapping of the evolutionary conservation at each amino acid site of ZmbZIP14 (Fig. 2A) generated by Chimera 1.5.332 was based on the rest of the 169 ZmbZIP protein sequences to show the conserved regions across this transcription factor family. The colour bar shows the gradient of conservation from low to high. The most conserved amino acid residues were labelled. The superposition structure of four crystal structures of bZIP domains (Fig. 2B) were obtained from PDB accession number 1T2K (Homo sapiens, pink), 1DH3 (Mus musculus, green), 1YSA (S. cerevisiae, blue) and 2OQQ (A. thaliana, yellow), which were the most hits of ZmbZIP proteins by homologous modelling. The potential map was generated by Pymol, and the colour bar represents distribution of potential from negative (red) to positive (blue) values.
Figure 2.
The three-dimensional structure analysis and the prediction of dimerization properties of 170 ZmbZIP proteins. (A) Mapping of surface conservation of ZmbZIP14 based on the rest of the 169 ZmbZIP proteins. Conserved sites and regions are labelled and represented in the red sphere, whereas regions lacking conservation are in blue. (B) The superposition structure of bZIP domains of ATF-2 (PDB ID: 1T2K, pink), CREB (PDB ID: 1DH3, green), GCN4 (PDB ID: 1YSA, blue) and HY5 (PDB ID: 2OQQ, yellow). (C) The electrostatic potential map of CREB. The red sphere represents negative potential, whereas the blue sphere represents positive potential. (D) Pie charts depicting the frequency of the amino acids at the a, d, e and g positions of the Leu zippers of all ZmbZIP proteins. (E) Histogram of the frequency of Asn residues present at the a position of the Leu zippers for all ZmbZIP proteins. (F) Histogram of the frequency of attractive or repulsive g↔e’ pairs per heptad for all ZmbZIP proteins.
2.7. Gene expression analysis
To gain an insight into the family-wide expression profile and obtain the genes with highly significant differential expression, microarray-based data analyses of both development and stress responses were performed. To analyse the spatial and temporal expression patterns of ZmbZIP genes during development, transcriptome data of the genome-wide gene expression atlas of maize inbred line B73 made by the NimbleGen microarrary technology was downloaded from Plexdb (ZM37). Gene expression data from the Affymetrix GeneChip array of drought stress as well as the data during four kinds of fungal infection were downloaded from GEO with accession numbers GSE16567, GSE10023, GSE31188, GSE19501 and GSE29747, respectively. All of these microarray data were imported into R and Bioconductor (http://www.bioconductor.org/) for expression analyses. The packages Limma and affy were applied to data processing. Then, the gplots package was used to make the heatmaps.
3. Results and Discussion
3.1. Identification and nomenclature of the maize bZIP transcription factor family
In this paper, 170 proteins were confirmed as maize bZIP transcription factors which were encoded by 125 bZIP genes. Each maize bZIP gene was assigned a unique identifier from ZmbZIP1 to 125 as proposed by Jakoby et al.7 The nomenclature was based on the exact position of these genes on maize chromosomes 1–10 and from top to bottom. For the distinct transcripts encoded by the same gene locus share the same gene number with an additional decimal part, such as point 1 or 2 and so forth. These orthologous bZIP proteins in Arabidopsis and rice are identified by the BLASTP search with an E-value of <1E−10. All of the related information on ZmbZIP transcription factors are listed in the Supplementary Table S1.
3.2. Phylogenetic analysis and classification of the ZmbZIP transcription factor family
To analyse the phylogenetic relationship of bZIP transcription factors among maize, rice and Arabidopsis, 170 sequences from maize, 89 sequences from rice and 74 sequences from Arabidopsis, 333 sequences in total, were analysed (Supplementary Fig. S1). It was notable to see that the phylogenetic tree can be subdivided into 11 clades except some ambiguous branches, and the grouping in maize complies with the classification of Arabidopsis, whereas in rice, they are categorized into 11 groups I–XI according to the DNA-binding specificity. Although the phylogenetic tree of the three plant species has some divergences against the classification of OsbZIP proteins because certain members of groups III, IV and VI were separated from their clusters which has also been observed by Nijhawan et al.8 The interspecies clustering indicates a parallel evolution of bZIP transcription factors in three plants and the homologous bZIP proteins which develop similar functions can be observed.33 Here, we integrate the features of both phylogenetic relationship among three plants and the basic and hinge regions to classify 170 ZmbZIP proteins into 11 groups and name the groups following Arabidopsis. The two proteins with the nearest evolutionary distance to the two unclassified AtbZIP62 and AtbZIP72 and OsbZIP80 in group XI are divided into group U. This classification can be further rationalized by the later analyses of the gene structures and additional conserved motifs outside the bZIP domain.
Considering the distinct functions the basic–hinge region and the Leu zipper may have, we separate them from the bZIP domains and make comparisons. Three phylograms are generated based on bZIP domains, basic and hinge regions and Leu zippers defined from the first leucine, respectively (Supplementary Fig. S2). The phylogram of the basic and hinge regions presents a nice consistency with the tree built by the bZIP domain, which strongly supports our classification, while the phylogram of the Leu zipper region displays some differences. Members of group A are divided into three clusters; at the same time, ZmbZIP2 in group G inserts into a separated A cluster. This divergence between the basic–hinge region and the Leu zipper region signifies that the ZmbZIP transcription factors having the same DNA-binding sites might be required for different dimerization properties, and will be able to control a wide range of transcriptional responses as a result.
3.3. Gene structure of ZmbZIP genes
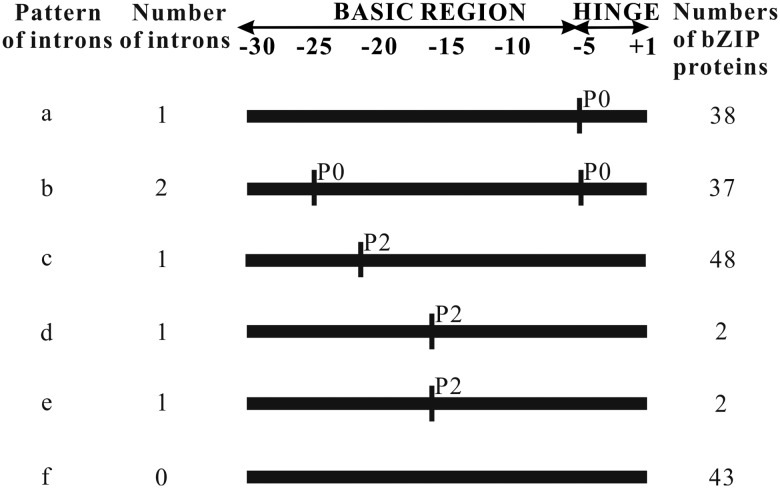
As a kind of evolutionary relic, the intron–exon arrangement carries the imprint of the evolution of a gene family. It is interesting to find a similar gene structure in each group (Supplementary Fig. S3). There are 26 (15.3%) of the total ZmbZIP genes having no intron. This phenomenon occurs exclusively in groups S and F, accounting for 84.6 and 15.4%, respectively. Among those having introns, the number of introns within the open reading frame (ORF) ranges from 1 to 14, showing a great difference in the ZmbZIP family. A greater degree of variation in the number of introns exists in groups D and G, varying from 5 to 12 and 3 to 14, respectively, while the number of introns in the rest groups changes in a smaller range of numbers, mostly from 1 to 3. The intron positions within the ORF are diverse and the phases of the splicing sites differ from each other, but the positions and phases of introns in the basic and hinge regions of the bZIP domain are highly conserved. Six intron patterns of the ZmbZIP genes, from a to f, are identified based on the intron position, number and splicing phase within the basic and hinge regions (Fig. 1, Supplementary Fig. S4 and Table S1). Our result was supported by Nijhawan et al.8 Patterns a, b and c are in the majority. Pattern a has just one intron in phase 0 (P0) within the hinge region at the −5 position, which inserts between the amino acids Gln and Ala. Pattern b has two introns both in P0: one in the basic region at the −25 position and the other in the hinge region at the −5 position interrupting Lys and Ala. Pattern b only emerges in group D. Pattern c has a single intron at the −20 position in P2, inserting into the codon which encodes Arg. Patterns d and e are exclusive in group E and each is found in two ZmbZIP genes. Both of these patterns insert in the same position and same splicing P2. The only difference is that pattern d interrupts Arg, whereas pattern e interrupts Gln instead. Pattern f lacks any intron in the basic region and in the hinge region. Among 43 genes in pattern f, 26 members are intronless, while the remaining 17 have introns outside the basic and hinge regions. Pattern c occurs in most of the groups and consists of large members.
Figure 1.
Intron pattern within the basic and hinge regions of the bZIP domains of ZmbZIP proteins. P0 represent the intron splicing site between codons, P2 means the intron splicing site locating between the second nucleotide and the third nucleotide in one codon.
3.4. The identification of additional conserved motifs in ZmbZIP transcription factors
One hundred and seventy ZmbZIP protein sequences were loaded into the MEME analysis tool and a total of 30 additional conserved motifs outside the bZIP domain have been identified (Supplementary Fig. S5 and Table S2). It can be observed that some motifs are shared by several groups, such as motifs 4, 10 and 15 present in three groups and motifs 6, 13, 14, 21, 22 and 25 present in two groups, respectively, while most of the conserved motifs appear in specific groups, which preliminarily proves our hypothesis that the group-specific motifs aid in determining specific functions for members in each group.
Evidence suggests that members in group A have roles in abscisic acid response or stress signalling. Fifteen, 15 and 12 ZmbZIP proteins in group A contain motifs 14, 15 and 18, respectively. A part of these three motifs represent potential casein kinase II phosphorylation sites (S/TxxD/E), presented as T[DA]E[EA], [TG][LM]GE and T[VAS][DE]E in motifs 14, 15 and 18, respectively. These phosphorylation sites have been reported in many ABF (ABRE-binding factor) and AREB (ABA-responsive element-binding protein) in Arabidopsis.34–37 Motif 18 also contains a phosphorylation site for the Ca2+-dependent protein kinase (R/KxxS/T) presented as [KR][ED][IF][SVT]. ZmbZIP23 (ZmbZIP72 in a previous report) is one of the proteins in group A which has been experimentally verified to function as an ABA-dependent transcription factor in positive modulation of abiotic stress tolerance.38 Motif 15 can also be found in four members of group C and eight members of group D. Motifs 11 and 29 characterized by a part of the proline-rich domain are observed in group G exclusively, which have been shown to have transcriptional activation potential. Most of the members in group D share motifs 1, 2 and 5, which have unclear function. Besides, this largest group in the ZmbZIP protein family possesses the most unique motifs, for their dual-function of defence against pathogen and developmental regulation.39 Interestingly, there are some common motifs that co-exist in maize and rice.8 For example, motifs 1, 2 and 5 in group D of maize are in common with motifs 18, 20 and 19 in rice, respectively; motif 9 in group I of maize is the same as motif 9 in rice.
3.5. The features of the three-dimensional structure of ZmbZIP proteins
The 170 ZmbZIP protein sequences were aligned and the Chimera software was used to generate a conservation map (Fig. 2A). ZmbZIP14 is used as a model to display the conservation of the whole family. The red sphere represents the highly conserved regions and the labelled amino acids N280, R288, L298, L305 and L312 in red are the most conserved sites shared by all of the aligned ZmbZIP protein sequences. The conserved asparagine and arginine are located in the basic region and responsible for DNA binding, while the three conserved leucines are in the Leu zipper and help to form dimers.
Further, the homology modelling was performed and the results show four models to be the best hits to our proteins, including PDB accession number 1T2K (H. sapiens), 1DH3 (M. musculus), 1YSA (S. cerevisiae) and 2OQQ (A. thaliana). Two of these models form homodimers (1DH3 and 2OQQ), whereas the other two form heterodimers (1T2K and 1YSA). The three-dimensional superposition map of four dimers is displayed (Fig. 2B). The overall structure of these dimers has been modelled as a Y-shaped complex, called ‘scissors-grip’ model.40 Also, the potential distribution of the bZIP domain shows that the enrichment region of positive charges is located at the top of Y, the basic region (Fig. 2C). The negative charges distribute across the stem region of Y, the leucine zipper region. As for dimerization in this region, situations must be more complicated due to the participation of more active forces.
3.6. The prediction of DNA-binding site specificity of ZmbZIP transcription factors
Experiments of mutant proteins demonstrated that binding specificity is determined by the core basic region and the hinge region independently, and the two regions have an additive effect on DNA-binding specificity.41,42 The amino acid sequence alignment of the basic and hinge regions reveals some highly conserved amino acid residues within each group (Supplementary Fig. S6). The first leucine of the leucine zipper was regarded as +1, and asparagine and arginine are numbered −18 and −10, respectively. Experiments have shown that these two invariant sites can be functionally replaced by other amino acids, but their replacement will lead to new DNA-binding specificities.43 In the maize bZIP family, these replacements are infrequent and just occur in two groups. ZmbZIP51 and ZmbZIP118 in group U have a hydrophobic Ile residue at position −10 instead of Arg/Lys, indicating that they might not be able to bind DNA or might possess a unique DNA-binding specificity. Four members of group E have a conserved Lys substitution at position −18 instead of Arg, implying a different requirement for dimerization. Further, we can predict the DNA-binding specificity in a group manner, as described in Supplementary Table S3.
3.7. Analysis of the 170 maize Leu zippers and prediction of dimerization property
The procedure of sequence-specific DNA binding requires dimerization. To define the basis of the dimerization stability and predict the dimerization specificity of 170 ZmbZIP proteins, we follow the standard nomenclature for the seven unique amino acid positions (a, b, c, d, e, f and g) to arrange the Leu zipper44 (Supplementary Fig. S7). The first heptad is from four amino acids before the occurrence of the first leucine in the bZIP domain and from g to f. The boundaries of both N-terminal and C-terminal of the ZmbZIP Leu zippers have been demarcated following the criteria once used for the bZIP proteins of Arabidopsis, rice and humans.45
Because of their special locations near the Leu zipper interface, the amino acids at the a, d, e and g positions play key roles in regulating leucine zipper oligomerization, dimerization stability and dimerization specificity. Thus, we focus on the characteristics of the amino acids at these positions to perform our prediction. Amino acids at the a and d positions are typically hydrophobic and on the surface of the α-helix.46 These features are perfect for the interaction between two monomers because they create a hydrophobic core which is essential for dimer stability.47 The positions of e and g abound in charged amino acids, including acidic amino acids E and D, and basic amino acids R and K. The electrostatic interactions between amino acids at position g and oppositely charged amino acids at position e’ (the prime means a residue on the opposite helix of the leucine zipper) can form interhelical salt bridges which mediate the dimerization and determine the dimerization specificity as well as stability.48 Experiments of the replacements of charged amino acids into alanine at position g or e lead to the formation of tetramers instead of dimers,49 thus charged amino acids at the g and e positions prevent formation of higher-order oligomers. At the same time, the asparagine at the a position can form a polar pocket in the hydrophobic interface that limits oligomerization when interacting interhelically.
In ZmbZIP proteins, hydrophobic amino acids are predominant at the a and d positions as they account for more than 84% of the total (Fig. 2D). About 22% of amino acids present at the a position are asparagines with nearly equivalent amount of rice (23%). Specific to each heptad (Fig. 2E), it displays the high frequency of Asn at the a position both in the second and fifth heptads, accounting for 47.6 and 49.4%, respectively. It differs from rice and Arabidopsis, which have the highest frequency of Asn at the a position in the second heptad and followed by the fifth heptad. The fourth and eighth heptads also have the appearances of Asn with the frequency of 8 and 12.4%, respectively. The high frequency of Asn at the a position implies that it will form a good number of homodimerizing Leu zippers among the bZIP family, because asparagines produce more stable N–N interactions at the a↔a′ position than other a position amino acids.50 There are also charged amino acids found at the a position which will drive heterodimer formation. Strikingly, the frequency of stabilizing leucine at the d position in ZmbZIPs is found to be 70%, which is much the same as rice (71%) but is significantly greater than in Arabidopsis (56%). These similarities might not be a coincidence; it implies a homologous relationship between these two monocotyledons. The charged amino acids occupy almost half of the e and g positions, with frequencies of 42 and 59%, respectively.
To analyse the contribution of charged residues at the e and g positions in governing dimerization properties of ZmbZIP proteins, we calculate the presence of attractive and repulsive g↔e′ pairs in each heptad of maize leucine zippers (Supplementary Fig. S7), and the histogram of their frequency is presented in Fig. 2F. If both the g and the following e position amino acids are charged, they refer to the complete g↔e′ pairs and the successive six amino acids from g to e are colour coded. If only the g or e position is charged, it refers to the incomplete g↔e′ pairs and only the charged amino acids are colour coded. The complete g↔e′ pairs are classified into four groups according to the electrostatic charges at the g and e positions. A blue box indicates that the g position is acidic and the following e position is basic (±attractive; blue). An orange box indicates that the g position is basic and the following e position is acidic (±attractive; orange). The pink and green boxes indicate that the g and the following e positions have a similar charge, either both basic (basic repulsive; pink) or both acidic (acidic repulsive; green). It can be found that the maximum frequency of the complete g↔e′ pairs appears in the first heptads, among which, the pink boxes are in the majority. The frequencies of the complete g↔e′ pairs in the next three heptads decrease dramatically. In the fifth heptads, the frequency of attractive g↔e′ pairs increases. Few complete g↔e′ pairs are observed in the eighth heptads except two repulsive g↔e′ pairs. Moreover, only blue boxes are present in the ninth heptads. When two helices with blue or orange boxes dimerize, the acidic and basic residues on opposite helices interact to create two self-complementary salt bridges. It is all possible that the helices are identical or not; thus, the interaction between two attractive g↔e′ pairs would not prefer homodimerization or heterodimerization. However, the helices with pink and green boxes prefer to interact with each other rather than themselves; thus, they favour the formation of heterodimers. Biophysical measurements also show that repulsive g↔e′ pairs are more important than attractive g↔e′ pairs in driving dimerization specificity. There are 32% of the g↔e′ interactions containing single-charged amino acids. These leucine zippers with incomplete g↔e′ pairs will have more promiscuous dimerization activity and contribute little to the stability of the homodimer. However, in a heterodimer, they can form complete attractive g↔e′ interactions and contribute to stability through complementation.
According to the analyses above, we categorize 170 ZmbZIP proteins into 26 subfamilies (BZ1–BZ26) on the basis of the defining properties of dimerization specificity, which was proposed by Vinson et al. We can conclude that four subfamilies (BZ1–BZ4) favour homodimerization because of attractive g↔e′ pairs in the first heptad and lacking of any repulsive interactions. Most of the subfamilies have both homo- and heterodimerization properties (BZ5–BZ23) for three reasons: (i) all have more than one asparagines at the a position, which favour homodimerizing strongly; (ii) the existence of repulsive g↔e′ pairs in these subfamilies can inhibit homodimerization; (iii) the incomplete g↔e′ pairs can promote promiscuous heterodimerization. Only three subfamilies (BZ24–BZ26) can be considered as having apparent heterodimerizing specificity because they contain only repulsive interhelical interactions. All these go to prove the complexity and diversity of the dimerization patterns in the Leu zipper of the ZmbZIP protein family, with the potential to homodimerize with themselves or with members in the same subfamily as well as heterodimerize with other subfamily members.
Based on the criteria to define the boundaries and natural C terminus, we can observe that the length of the Leu zipper in the ZmbZIP transcription factor family is variable, ranging from three to nine heptads. Of the ZmbZIP proteins, 27% have only three short zippers (BZ1 and BZ26), and more than 17% have no α-helix breakers for 10 or more heptads (BZ18–BZ23). These longer Leu zippers have been used to define multiple families of homodimerizing bZIP proteins in Arabidopsis.
3.8. Chromosomal distribution of the bZIP gene family and genetic analysis
The results of the chromosomal distribution map (Supplementary Fig. S8) along with the histogram of the number of ZmbZIP genes on each chromosome (Supplementary Fig. S9) show that the 125 ZmbZIP genes dispersed non-uniformly across the 10 chromosomes, varying from 9 to 17. Chromosome 1 distributes most of the bZIP genes with the number 17, accounting for 13.6% of the total, whereas chromosomes 9 and 10 have the least genes with the number of 9, accounting for 7.2% of the total. Genes in different groups also distribute unevenly on chromosomes. Members of group D distribute more evenly than other groups across all chromosomes on a minor scale. No group of genes displays a preferential distribution on a specific chromosome.
Gene duplication and positive selection play an extremely important part in gene family expansion and protein functional diversification.51 To explore the contribution of duplication events to this family, we analyse the occurrences of tandem duplication and large-scale segmental duplication during the evolution of this gene family. Among them, the gene cluster is one of the results of gene duplication. In our analysis, six ZmbZIP gene clusters are composed of 12 bZIP genes existing in the maize genome (Supplementary Table S4) and define clusters where the linked genes in the same gene family were not interrupted by more than 8 other ORFs as explained in Richly et al.52 The majority of ZmbZIP genes are singletons. Within the gene clusters, a pair of genes can be identified as a tandem duplication cluster, since they meet the three conditions according to Shiu et al.53 Thus, we can make an inference that the expansion of the ZmbZIP transcription factor family might not rely on the independent duplication of individual sequences and might be the consequence of segmental chromosomal duplication and rearrangement events. Similar analyses from rice, Arabidopsis and sorghum also support this inference.
Since the segmentally duplicated chromosome blocks in maize have been identified at a genome-wide level, we can use the SyMap to compute and view these syntenic blocks.54 The syntenic blocks on each chromosome and the distribution map of bZIP genes on each chromosome are vertically arranged together to view if the genes are located on the syntenic duplicated segmental regions (Supplementary Fig. S10). Fifty-eight ZmbZIP genes located on the duplicated segmental regions of maize chromosomes at the minimum identity score for collinear gene pairs of 0.4 are identified via the GenoPix 2D Plotter software (Supplementary Table S5). Interestingly, all these collinear ZmbZIP gene pairs located on the syntenic regions belong to the same groups. Among them, eighteen members of group D constitute the most collinear gene pairs (49 of 75), accounting for 65%, and more than half the members of group I (12 of 20) are observed to be present on the duplicated segments of maize chromosomes. Thirty-three of these genes segmentally duplicated once and the rest duplicated more than once. Surprisingly, all but four members of group D located in syntenic regions segmentally duplicate at a high frequency, with the maximum of 11 times. On the contrary, no member of group S is found to be located in the syntenic duplicated region, implying that this group might evolve after the emergence of large-scale segmental duplication events. All these results suggest that the expansion of ZmbZIP genes is largely a consequence of segmental chromosomal duplication events.
3.9. Expression analysis of ZmbZIP transcription factors in global transcriptome at different developmental stages in specific organs
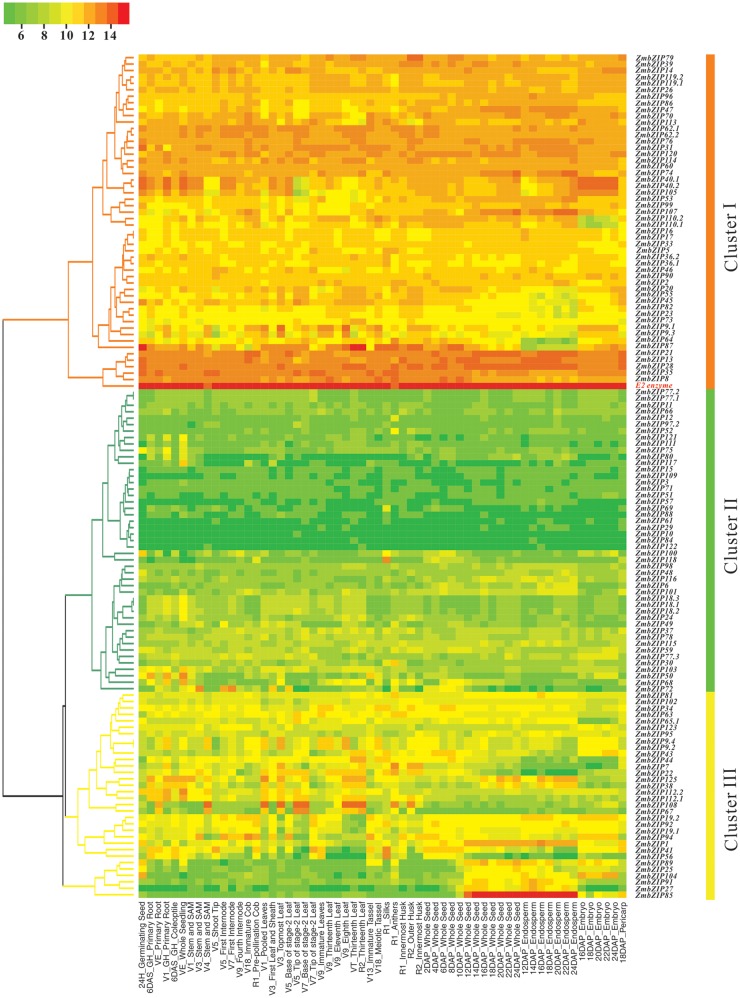
A total number of 154 probes detected in R represent 116 ZmbZIP genes and can be assigned to 154 corresponding transcripts (Supplementary Table S6). To understand the temporal and spatial transcription patterns of the ZmbZIP genes in the maize life cycle, a hierarchy cluster was performed to visualize a global transcription profile of the ZmbZIP genes across the 11 organs at different developmental stages. As illustrated in Fig. 3, the heatmap can be apparently divided into three clusters. Cluster I has 51 members (excluding the E2 enzyme as the reference). Cluster II has 47 members and 32 genes belong to cluster III. Genes in cluster I obviously have relatively high expression levels, with the mean of the log-signal values of each gene ranging from 10 to 14. On the contrary, cluster II contains the genes with relatively low expression levels and the mean of the log-signal values of each gene is within the range of four to eight. In cluster III, genes are expressed in a mid-level with the mean of log-signal values from 6 to 10. To identify putative differentially expressed genes in specific organs or stages, we calculate the coefficient of variation (CV value; CV = SD/mean, where SD represents the standard deviation and mean describes the mean expression level of a gene across all the tissues) of each gene in the three clusters to compare the degree of variation of each gene among distinct organs55 (Supplementary Table S7). The results show huge variation among all the genes with the CV values ranging from 1 to 44%. The gene with the least CV value is the reference gene, E2 enzyme. It suggests that E2 enzyme is a housekeeping gene in maize, which is expressed uniformly in all tissues. And the gene with the highest CV value is ZmbZIP85 in cluster III. Interestingly, ZmbZIP85 is one of the few well-documented bZIP genes in maize, O2, which regulate the synthesis of the main storage proteins in seeds. As shown in the figure, ZmbZIP85 specifically accumulates in the developing endosperm from the 12 DAP (days after pollination) to 24 DAP. Our data confirm the known fact that O2 is a pivotal positive regulator of seed development.
Figure 3.
Clustering of ZmbZIP genes according to their expression profiles of 130 detected transcripts 60 tissues at different stages in the 11 organs in maize. A hierarchical clustering in the left showing three kinds of expression patterns of ZmbZIP genes in maize development. The colour scale representing average signal values is shown at the top.
Cluster I has the least expression variability from 1 to 18%, indicating a most stable expression pattern relative to other ZmbZIP genes. The most changeable genes in this cluster are ZmbZIP87 and 64 with CV values 14 and 18%, respectively. It is notable that these two genes seem to have an opposite effect compared with ZmbZIP85. Their expression levels significantly decrease during endosperm development, which, in turn, implies that they might act as negative regulators in seed development. It can be further divided into two subclusters in cluster II. Genes in the first subcluster have lower signal values and more stable expression pattern than those in the second subcluster. There are seven genes with CV values more than 15% in cluster II, among which three genes have CV values more than 20%. These three genes are ZmbZIP50, 68 and 72. It should be pointed out that the transcriptional activation of bZIP transcription factors are required for dimerization. Therefore, it is reasonable to observe a synergistic effect formed in many bZIP proteins in specific stages, such as ZmbZIP50 and 68 in the V1 (vegetative 1, with first leaf fully extended) primary root stage and the VE (vegetative emergence) whole seedling stage. Obviously, ZmbZIP118 might play a role in the development of pistil, deducing from the significantly up-regulated expression level in silk. Genes in cluster III show an apparent fluctuation, with the CV value ranging from 6 to 44%. In this cluster, 56.3% (18 genes) are with a CV value more than 15%, including 12 genes with a CV value greater than 20%. Among them, ZmbZIP41 and 56 from group E increase their expression levels in silk and the early developmental stages of seeds. ZmbZIP108 and 67 from group H have a similar expression pattern and increase their expression levels in the early and late developmental stages of the leaf. Besides ZmbZIP85, many genes show the up-regulation pattern in the development of the seed and the endosperm, such as ZmbZIP74, −107, −125, −38, −1, −89, −25, −104, −91 and −27. In addition, some genes exhibit novel and differential expression during the development of the embryo. For instance, ZmbZIP110.1, 110.2 and 65.1 are with down-regulated expression in the developing embryo, whereas ZmbZIP40.1, −40.2, −105, −19.2, −94, −104 and −85 are with up-regulated expression.
3.10. Expression of ZmbZIP genes under drought stress and fungal infections
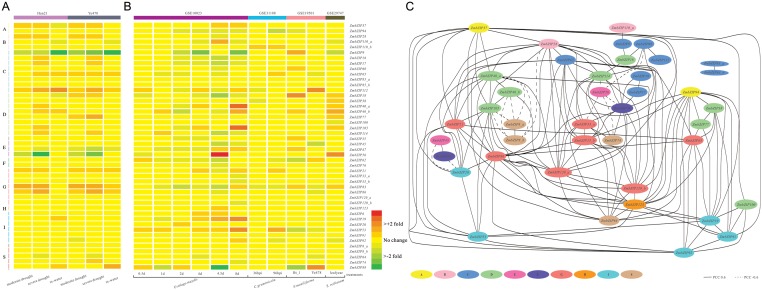
As a kind of ubiquitous transcription factors, bZIP proteins regulate the expression of a wide spectrum of stress-related genes. The log2 (treated/control) ratio values are illustrated by a heatmap (Fig. 4A and B), showing the fold change of each ZmbZIP gene compared with the control. A total of 46 probe sets discovered on the maize18k GeneChip could be assigned to 40 ZmbZIP genes with matches of more than eight (Supplementary Table S8). The probe sets that represent the same gene are named _a to _c and ordered by group.
Figure 4.
Expression profiles of 46 ZmbZIP genes in drought stress and four kinds of fungal infections, and a network showing the co-regulatory pattern of ZmbZIP genes in different fungal infections. (A) Normalized signal intensities of 46 probe sets representing 40 ZmbZIP genes displayed for the microarrary experiments under different degrees of drought stress. Log2 (treated/control) ratio values (shown by a green–red gradient) represented the fold change of the gene expression in the fungal infections. ZmbZIP genes are ordered by group. Gene expression data were extracted from GEO. (B) Normalized signal intensities of 46 probe sets representing 40 ZmbZIP genes displayed for four microarrary experiments, which were treated with U. maydis, C. graminicola, F. moniliforme and S. reiliana, respectively. (C) The co-regulatory network of ZmbZIP genes. For each pair of ZmbZIP genes, the Pearson correlation coefficient (PCC value) was calculated to measure the correlation of expression levels in these four fungal infections, based on 62 maize microarray experiments. Each pair of correlated ZmbZIP genes is shown in the figure with an edge connecting them. PCC values higher than 0.6 are represented by solid lines and values lower than −0.6 are represented by dashed lines.
To explore the roles of ZmbZIP genes in drought stress, the microarrary data of two different inbred lines, a drought-tolerant line, Han21 line, and a drought-sensitive line, Ye478 line, under three treatments have been used (Fig. 4A). Interestingly, most of the genes which show a response to drought stress show a similar expression pattern in two inbred lines, whereas although still some distinct differences can be carefully observed. Many bZIP transcription factors in plants have been reported to be closely related to drought stress. In rice, two proteins that have close phylogenetic relationship with members of group A in maize, OsbZIP23 and OsbZIP72, play a role in ABA response and drought tolerance.56,57 Their orthologous gene ZmbZIP37 also increases its expression level under drought stress, implying a possible positive regulatory role. Another protein, OsbZIP52/RISBZ5, an orthologous gene of ZmbZIP112, can significantly down-regulate the drought-related genes by overexpression in rice.58 Contrary to OsbZIP52, ZmbZIP112 is highly expressed under a drought stress condition; AtbZIP1 is reported as a positive regulator in Arabidopsis which shows tolerance to salt, osmotic and drought stresses.59 Its ortholog, ZmbZIP74, has a divergence in two lines. In the Han21 line, ZmbZIP74 has no change in the expression level, while in the drought-sensitive line, Ye478, it is up-regulated under severe drought condition. This phenomenon can be modestly explained as a more complicated regulatory mechanism being involved in drought stress in the Han21 line irrelevant to the bZIP protein, compared with the bZIP-dependent drought stress regulatory mechanism active in the Ye478 line.
To discover the ZmbZIP genes involved in maize pathogen response, we detect differentially expressed genes with microarray data. Data sets of four experiments under the treatments of Ustilago maydis, Colletotrichum graminicola, Fusarium moniliforme and Sphacelotheca reiliana infection have been analysed. The first two series of experiments focus on the time course, and the last two series focus on the comparisons between the resistant line and the susceptible line or between treatment and control, respectively.
As shown in Fig. 4B, after the infection of U. maydis, four members in group D increase their expression levels and accumulate mostly on the 8th day. And five members from group I also show up-regulated expression levels over time, especially of ZmbZIP19 and ZmbZIP53. It can be concluded that the gene expression profiles are more variable with time under U. maydis infection. A similar situation can be found under an infection of C. graminicola. The gene expression levels at 96 h post-infection are more changeable than those at 36 h post-infection. Among them, ZmbZIP65, 21 and 53 show up-regulated expression, while the gene expression levels of ZmbZIP94, 62 and 92 are significantly down-regulated when compared with the control. It is very interesting to observe the differences between the resistant line Bt-1 and the susceptible line Ye478 under an infection of F. moniliforme. More genes are up-regulated in the Bt-1 line (seven genes) than those in the Ye478 line (three genes). Strikingly, two genes, ZmbZIP62 and 88, up-regulated in the Ye478 line are apparently down-regulated in the Bt-1 line, whereas the reverse situation occurs in ZmbZIP83, implying the counter effects these three genes might have on these two lines. Compared with mock-infected controls, seven genes in group D are up-regulated when infected with S. reiliana, suggesting that many genes in this group might participate in the pathogen response. This result as with the situation under U. maydis infection is consistent with the fact that group D proteins might be involved in integrating different systemic signals (SA and ethylene) and response to pathogen attack in Arabidopsis.
To test whether ZmbZIP genes are co-expressed in maize pathogen responses especially for their dimerization character, we calculate the Pearson correlation coefficient (PCC) among pairwise genes60 based on the transcriptome data from 62 microarray experiments as mentioned above (Supplementary Table S9). A network is made to show the strongly correlated gene pairs which have a PCC value of 0.6 or higher (in bold edge) and −0.6 or lower (in dashed edge), where the negative sign here means negative correlation (Fig. 4C).
In the network, calculations have been made and strong correlations exist between 156 pairs of genes. Among them, 13 pairs of genes show strong negative correlations with a PCC value of ≤0.6, and this study also demonstrates strongly positive correlations between the rest of the 143 gene pairs with the PCC value of ≥0.6. Interestingly, connections from ZmbZIP8_a, ZmbZIP8_b and ZmbZIP56 are all dashed lines, which signify strongly negative correlations of these two genes with other genes. In addition, most of the negative correlations are related to these two genes, accounting for 84.6% of the total. Other negative correlations are presented by two gene pairs, ZmbZIP20–45 and ZmbZIP20–62. As for the gene pairs with positive correlation, we concentrate on the six gene pairs with a PCC value of >0.9. These gene pairs all belong to the same groups, indicating that they might be paralogous genes sharing the nearest phylogenetic relationship with each other. For example, the ZmbZIP40_a–105 gene pair from group D has a complete positive correlation with a PCC value of 1 and shows similar expression patterns in the fungal infections. In addition, the point to be noted is that high correlation may give two clues: co-functional genes or duplication of promoters. Since some of the genes within a group may have similar promoter cis-elements, they would have similar patterns of expression involved in potential roles in dimerization.
Supplementary data
Supplementary data are available at www.dnaresearch.oxfordjournals.org.
Funding
The project was supported by the Natural Science Foundation of Fujian Province, China (Grant No. B0810040).
Supplementary Material
Acknowledgements
We are grateful to the providers who submitted the microarray data to the public expression databases which can be applied freely.
Footnotes
Edited by Kazuo Shinozaki
References
- 1.Hurst H.C. Transcription factors. 1: bZIP proteins. Protein Profile. 1994;1:123–68. [PubMed] [Google Scholar]
- 2.Izawa T., Foster R., Chua N.H. Plant bZIP protein DNA binding specificity. J. Mol. Biol. 1993;230:1131–44. doi: 10.1006/jmbi.1993.1230. doi:10.1006/jmbi.1993.1230. [DOI] [PubMed] [Google Scholar]
- 3.Landschulz W.H., Johnson P.F., McKnight S.L. The leucine zipper: a hypothetical structure common to a new class of DNA binding proteins. Science. 1988;240:1759–64. doi: 10.1126/science.3289117. doi:10.1126/science.3289117. [DOI] [PubMed] [Google Scholar]
- 4.Ellenberger T.E., Brandl C.J., Struhl K., Harrison S.C. The GCN4 basic region leucine zipper binds DNA as a dimer of uninterrupted alpha helices: crystal structure of the protein-DNA complex. Cell. 1992;71:1223–37. doi: 10.1016/s0092-8674(05)80070-4. doi:10.1016/S0092-8674(05)80070-4. [DOI] [PubMed] [Google Scholar]
- 5.Fassler J., Landsman D., Acharya A., Moll J.R., Bonovich M., Vinson C. B-ZIP proteins encoded by the Drosophila genome: evaluation of potential dimerization partners. Genome Res. 2002;12:1190–200. doi: 10.1101/gr.67902. doi:10.1101/gr.67902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vinson C., Myakishev M., Acharya A., Mir A.A., Moll J.R., Bonovich M. Classification of human B-ZIP proteins based on dimerization properties. Mol. Cell. Biol. 2002;22:6321–35. doi: 10.1128/MCB.22.18.6321-6335.2002. doi:10.1128/MCB.22.18.6321-6335.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jakoby M., Weisshaar B., Droge-Laser W., et al. bZIP transcription factors in Arabidopsis. Trends Plant Sci. 2002;7:106–11. doi: 10.1016/s1360-1385(01)02223-3. doi:10.1016/S1360-1385(01)02223-3. [DOI] [PubMed] [Google Scholar]
- 8.Nijhawan A., Jain M., Tyagi A.K., Khurana J.P. Genomic survey and gene expression analysis of the basic leucine zipper transcription factor family in rice. Plant Physiol. 2008;146:333–50. doi: 10.1104/pp.107.112821. doi:10.1104/pp.107.112821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang J., Zhou J., Zhang B., Vanitha J., Ramachandran S., Jiang S.Y. Genome-wide expansion and expression divergence of the basic leucine zipper transcription factors in higher plants with an emphasis on sorghum. J. Integr. Plant Biol. 2011;53:212–31. doi: 10.1111/j.1744-7909.2010.01017.x. doi:10.1111/j.1744-7909.2010.01017.x. [DOI] [PubMed] [Google Scholar]
- 10.Liao Y., Zou H.F., Wei W., et al. Soybean GmbZIP44, GmbZIP62 and GmbZIP78 genes function as negative regulator of ABA signaling and confer salt and freezing tolerance in transgenic Arabidopsis. Planta. 2008;228:225–40. doi: 10.1007/s00425-008-0731-3. doi:10.1007/s00425-008-0731-3. [DOI] [PubMed] [Google Scholar]
- 11.Gaut B.S., Le Thierry d'Ennequin M., Peek A.S., Sawkins M.C. Maize as a model for the evolution of plant nuclear genomes. Proc. Natl Acad. Sci. USA. 2000;97:7008–15. doi: 10.1073/pnas.97.13.7008. doi:10.1073/pnas.97.13.7008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wei F., Zhang J., Zhou S., et al. The physical and genetic framework of the maize B73 genome. PLoS Genet. 2009;5:e1000715. doi: 10.1371/journal.pgen.1000715. doi:10.1371/journal.pgen.1000715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schnable P.S., Ware D., Fulton R.S., et al. The B73 maize genome: complexity, diversity, and dynamics. Science. 2009;326:1112–5. doi: 10.1126/science.1178534. doi:10.1126/science.1178534. [DOI] [PubMed] [Google Scholar]
- 14.Schmidt R.J., Burr F.A., Aukerman M.J., Burr B. Maize regulatory gene opaque-2 encodes a protein with a ‘leucine-zipper’ motif that binds to zein DNA. Proc. Natl Acad. Sci. USA. 1990;87:46–50. doi: 10.1073/pnas.87.1.46. doi:10.1073/pnas.87.1.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brochetto-Braga M.R., Leite A., Arruda P. Partial purification and characterization of lysine-ketoglutarate reductase in normal and opaque-2 maize endosperms. Plant Physiol. 1992;98:1139–47. doi: 10.1104/pp.98.3.1139. doi:10.1104/pp.98.3.1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lohmer S., Maddaloni M., Motto M., et al. The maize regulatory locus Opaque-2 encodes a DNA-binding protein which activates the transcription of the b-32 gene. EMBO J. 1991;10:617–24. doi: 10.1002/j.1460-2075.1991.tb07989.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yunes J.A., Cord Neto G., Leite A., Ottoboni L.M., Arruda P. The role of the Opaque2 transcriptional factor in the regulation of protein accumulation and amino acid metabolism in maize seeds. An. Acad. Bras. Cienc. 1994;66 Su 1(Pt 2):227–37. [PubMed] [Google Scholar]
- 18.Schmidt R.J., Ketudat M., Aukerman M.J., Hoschek G. Opaque-2 is a transcriptional activator that recognizes a specific target site in 22-kD zein genes. Plant Cell. 1992;4:689–700. doi: 10.1105/tpc.4.6.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carlini L.E., Ketudat M., Parsons R.L., Prabhakar S., Schmidt R.J., Guiltinan M.J. The maize EmBP-1 orthologue differentially regulates opaque2-dependent gene expression in yeast and cultured maize endosperm cells. Plant Mol. Biol. 1999;41:339–49. doi: 10.1023/a:1006338727053. doi:10.1023/A:1006338727053. [DOI] [PubMed] [Google Scholar]
- 20.Pysh L.D., Aukerman M.J., Schmidt R.J. OHP1: a maize basic domain/leucine zipper protein that interacts with opaque2. Plant Cell. 1993;5:227–36. doi: 10.1105/tpc.5.2.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Muszynski M.G., Dam T., Li B., et al. Delayed flowering1 encodes a basic leucine zipper protein that mediates floral inductive signals at the shoot apex in maize. Plant Physiol. 2006;142:1523–36. doi: 10.1104/pp.106.088815. doi:10.1104/pp.106.088815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Walsh J., Freeling M. The liguleless2 gene of maize functions during the transition from the vegetative to the reproductive shoot apex. Plant J. 1999;19:489–95. doi: 10.1046/j.1365-313x.1999.00541.x. doi:10.1046/j.1365-313X.1999.00541.x. [DOI] [PubMed] [Google Scholar]
- 23.Walsh J., Waters C.A., Freeling M. The maize gene liguleless2 encodes a basic leucine zipper protein involved in the establishment of the leaf blade-sheath boundary. Genes Dev. 1998;12:208–18. doi: 10.1101/gad.12.2.208. doi:10.1101/gad.12.2.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harper L., Freeling M. Interactions of liguleless1 and liguleless2 function during ligule induction in maize. Genetics. 1996;144:1871–82. doi: 10.1093/genetics/144.4.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kusano T., Berberich T., Harada M., Suzuki N., Sugawara K. A maize DNA-binding factor with a bZIP motif is induced by low temperature. Mol. Gen. Genet. 1995;248:507–17. doi: 10.1007/BF02423445. doi:10.1007/BF02423445. [DOI] [PubMed] [Google Scholar]
- 26.Singh K., Dennis E.S., Ellis J.G., et al. OCSBF-1, a maize ocs enhancer binding factor: isolation and expression during development. Plant Cell. 1990;2:891–903. doi: 10.1105/tpc.2.9.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.De Vetten N.C., Ferl R.J. Characterization of a maize G-box binding factor that is induced by hypoxia. Plant J. 1995;7:589–601. doi: 10.1046/j.1365-313x.1995.7040589.x. doi:10.1046/j.1365-313X.1995.7040589.x. [DOI] [PubMed] [Google Scholar]
- 28.Foley R.C., Grossman C., Ellis J.G., et al. Isolation of a maize bZIP protein subfamily: candidates for the ocs-element transcription factor. Plant J. 1993;3:669–79. doi:10.1111/j.1365-313X.1993.00669.x. [PubMed] [Google Scholar]
- 29.Guindon S., Dufayard J.F., Lefort V., Anisimova M., Hordijk W., Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 2010;59:307–21. doi: 10.1093/sysbio/syq010. doi:10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 30.Darriba D., Taboada G.L., Doallo R., Posada D. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics. 2011;27:1164–5. doi: 10.1093/bioinformatics/btr088. doi:10.1093/bioinformatics/btr088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Letunic I., Bork P. Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics. 2007;23:127–8. doi: 10.1093/bioinformatics/btl529. doi:10.1093/bioinformatics/btl529. [DOI] [PubMed] [Google Scholar]
- 32.Pettersen E.F., Goddard T.D., Huang C.C., et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–12. doi: 10.1002/jcc.20084. doi:10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 33.Wei K.F., Wu L.J., Chen J., Chen Y.F., Xie D.X. Structural evolution and functional diversification analyses of argonaute protein. J. Cell. Biochem. 2012;113:2576–85. doi: 10.1002/jcb.24133. doi:10.1002/jcb.24133. [DOI] [PubMed] [Google Scholar]
- 34.Choi H., Hong J., Ha J., Kang J., Kim S.Y. ABFs, a family of ABA-responsive element binding factors. J. Biol. Chem. 2000;275:1723–30. doi: 10.1074/jbc.275.3.1723. doi:10.1074/jbc.275.3.1723. [DOI] [PubMed] [Google Scholar]
- 35.Finkelstein R.R., Lynch T.J. The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell. 2000;12:599–609. doi: 10.1105/tpc.12.4.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lopez-Molina L., Mongrand S., Chua N.H. A postgermination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis. Proc. Natl Acad. Sci. USA. 2001;98:4782–7. doi: 10.1073/pnas.081594298. doi:10.1073/pnas.081594298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Uno Y., Furihata T., Abe H., Yoshida R., Shinozaki K., Yamaguchi-Shinozaki K. Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proc. Natl Acad. Sci. USA. 2000;97:11632–7. doi: 10.1073/pnas.190309197. doi:10.1073/pnas.190309197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ying S., Zhang D.F., Fu J., et al. Cloning and characterization of a maize bZIP transcription factor, ZmbZIP72, confers drought and salt tolerance in transgenic Arabidopsis. Planta. 2012;235:253–66. doi: 10.1007/s00425-011-1496-7. doi:10.1007/s00425-011-1496-7. [DOI] [PubMed] [Google Scholar]
- 39.Xiang C., Miao Z., Lam E. DNA-binding properties, genomic organization and expression pattern of TGA6, a new member of the TGA family of bZIP transcription factors in Arabidopsis thaliana. Plant Mol. Biol. 1997;34:403–15. doi: 10.1023/a:1005873500238. doi:10.1023/A:1005873500238. [DOI] [PubMed] [Google Scholar]
- 40.Vinson C.R., Sigler P.B., McKnight S.L. Scissors-grip model for DNA recognition by a family of leucine zipper proteins. Science. 1989;246:911–6. doi: 10.1126/science.2683088. doi:10.1126/science.2683088. [DOI] [PubMed] [Google Scholar]
- 41.Suckow M., von Wilcken-Bergmann B., Muller-Hill B. Identification of three residues in the basic regions of the bZIP proteins GCN4, C/EBP and TAF-1 that are involved in specific DNA binding. EMBO J. 1993;12:1193–200. doi: 10.1002/j.1460-2075.1993.tb05760.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Niu X., Renshaw-Gegg L., Miller L., Guiltinan M.J. Bipartite determinants of DNA-binding specificity of plant basic leucine zipper proteins. Plant Mol. Biol. 1999;41:1–13. doi: 10.1023/a:1006206011502. doi:10.1023/A:1006206011502. [DOI] [PubMed] [Google Scholar]
- 43.Suckow M., Schwamborn K., Kisters-Woike B., von Wilcken-Bergmann B., Muller-Hill B. Replacement of invariant bZip residues within the basic region of the yeast transcriptional activator GCN4 can change its DNA binding specificity. Nucleic Acids Res. 1994;22:4395–404. doi: 10.1093/nar/22.21.4395. doi:10.1093/nar/22.21.4395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McLachlan A.D., Stewart M. Tropomyosin coiled-coil interactions: evidence for an unstaggered structure. J. Mol. Biol. 1975;98:293–304. doi: 10.1016/s0022-2836(75)80119-7. doi:10.1016/S0022-2836(75)80119-7. [DOI] [PubMed] [Google Scholar]
- 45.Deppmann C.D., Acharya A., Rishi V., et al. Dimerization specificity of all 67 B-ZIP motifs in Arabidopsis thaliana: a comparison to Homo sapiens B-ZIP motifs. Nucleic Acids Res. 2004;32:3435–45. doi: 10.1093/nar/gkh653. doi:10.1093/nar/gkh653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moitra J., Szilak L., Krylov D., Vinson C. Leucine is the most stabilizing aliphatic amino acid in the d position of a dimeric leucine zipper coiled coil. Biochemistry. 1997;36:12567–73. doi: 10.1021/bi971424h. doi:10.1021/bi971424h. [DOI] [PubMed] [Google Scholar]
- 47.Thompson K.S., Vinson C.R., Freire E. Thermodynamic characterization of the structural stability of the coiled-coil region of the bZIP transcription factor GCN4. Biochemistry. 1993;32:5491–6. doi: 10.1021/bi00072a001. doi:10.1021/bi00072a001. [DOI] [PubMed] [Google Scholar]
- 48.Vinson C.R., Hai T., Boyd S.M. Dimerization specificity of the leucine zipper-containing bZIP motif on DNA binding: prediction and rational design. Genes Dev. 1993;7:1047–58. doi: 10.1101/gad.7.6.1047. doi:10.1101/gad.7.6.1047. [DOI] [PubMed] [Google Scholar]
- 49.Krylov D., Mikhailenko I., Vinson C. A thermodynamic scale for leucine zipper stability and dimerization specificity: e and g interhelical interactions. EMBO J. 1994;13:2849–61. doi: 10.1002/j.1460-2075.1994.tb06579.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Acharya A., Ruvinov S.B., Gal J., Moll J.R., Vinson C. A heterodimerizing leucine zipper coiled coil system for examining the specificity of a position interactions: amino acids I, V, L, N, A, and K. Biochemistry. 2002;41:14122–31. doi: 10.1021/bi020486r. doi:10.1021/bi020486r. [DOI] [PubMed] [Google Scholar]
- 51.Wei K.F., Chen Y.F., Chen J., Wu L.J., Xie D.X. Evolution and adaptation of hemagglutinin gene of human H5N1 influenza virus. Virus Genes. 2012;44:450–8. doi: 10.1007/s11262-012-0717-x. doi:10.1007/s11262-012-0717-x. [DOI] [PubMed] [Google Scholar]
- 52.Richly E., Kurth J., Leister D. Mode of amplification and reorganization of resistance genes during recent Arabidopsis thaliana evolution. Mol. Biol. Evol. 2002;19:76–84. doi: 10.1093/oxfordjournals.molbev.a003984. doi:10.1093/oxfordjournals.molbev.a003984. [DOI] [PubMed] [Google Scholar]
- 53.Shiu S.H., Karlowski W.M., Pan R., Tzeng Y.H., Mayer K.F., Li W.H. Comparative analysis of the receptor-like kinase family in Arabidopsis and rice. Plant Cell. 2004;16:1220–34. doi: 10.1105/tpc.020834. doi:10.1105/tpc.020834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wei F., Coe E., Nelson W., et al. Physical and genetic structure of the maize genome reflects its complex evolutionary history. PLoS Genet. 2007;3:e123. doi: 10.1371/journal.pgen.0030123. doi:10.1371/journal.pgen.0030123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wei K.F., Chen J., Chen Y.F., Wu L.J., Xie D.X. Molecular phylogenetic and expression analysis of the complete WRKY transcription factor family in maize. DNA Res. 2012;19:153–64. doi: 10.1093/dnares/dsr048. doi:10.1093/dnares/dsr048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lu G., Gao C., Zheng X., Han B. Identification of OsbZIP72 as a positive regulator of ABA response and drought tolerance in rice. Planta. 2009;229:605–15. doi: 10.1007/s00425-008-0857-3. doi:10.1007/s00425-008-0857-3. [DOI] [PubMed] [Google Scholar]
- 57.Xiang Y., Tang N., Du H., Ye H., Xiong L. Characterization of OsbZIP23 as a key player of the basic leucine zipper transcription factor family for conferring abscisic acid sensitivity and salinity and drought tolerance in rice. Plant Physiol. 2008;148:1938–52. doi: 10.1104/pp.108.128199. doi:10.1104/pp.108.128199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu C., Wu Y., Wang X. bZIP transcription factor OsbZIP52/RISBZ5: a potential negative regulator of cold and drought stress response in rice. Planta. 2012;235:1157–69. doi: 10.1007/s00425-011-1564-z. doi:10.1007/s00425-011-1564-z. [DOI] [PubMed] [Google Scholar]
- 59.Sun X., Li Y., Cai H., et al. The Arabidopsis AtbZIP1 transcription factor is a positive regulator of plant tolerance to salt, osmotic and drought stresses. J. Plant Res. 2012;125:429–38. doi: 10.1007/s10265-011-0448-4. doi:10.1007/s10265-011-0448-4. [DOI] [PubMed] [Google Scholar]
- 60.Wei K.F., Chen J., Chen Y.F., Wu L.J., Xie D.X. Multiple-strategy analyses of ZmWRKY subgroups and functional exploration of ZmWRKY genes in pathogen responses. Mol. Biosyst. 2012;8:1940–9. doi: 10.1039/c2mb05483c. doi:10.1039/c2mb05483c. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.