Abstract
Notch signaling is highly conserved in all metazoan animals and plays critical roles in cell fate specification, cell proliferation, apoptosis, and stem cell maintenance. Although core components of the Notch signaling cascade have been identified, many gaps in the understanding of the Notch signaling pathway remain to be filled. One form of posttranslational regulation, which is controlled by the ubiquitin-proteasome system, is known to modulate Notch signaling. The ubiquitination pathway is a highly coordinated process in which the ubiquitin moiety is either conjugated to or removed from target proteins by opposing E3 ubiquitin ligases and deubiquitinases (DUBs). Several E3 ubiquitin ligases have been implicated in ubiquitin conjugation to the receptors and the ligands of the Notch signaling cascade. In contrast, little is known about a direct role of DUBs in Notch signaling in vivo. Here, we report an in vivo RNA interference screen in Drosophila melanogaster targeting all 45 DUBs that we annotated in the fly genome. We show that at least four DUBs function specifically in the formation of the fly wing margin and/or the specification of the scutellar sensory organ precursors, two processes that are strictly dependent on the balanced Notch signaling activity. Furthermore, we provide genetic evidence suggesting that these DUBs are necessary to positively modulate Notch signaling activity. Our study reveals a conserved molecular mechanism by which protein deubiquitination process contributes to the complex posttranslational regulation of Notch signaling in vivo.
Keywords: deubiquitinase, Drosophila melanogaster, Notch signaling, ubiquitination
Notch signaling is an evolutionarily conserved developmental pathway that is strictly required to direct the specification of almost every cell type (Artavanis-Tsakonas et al. 1999; Lai 2004). Furthermore, Notch signaling functions to regulate stem cell maintenance and adult tissue homeostasis (Liu et al. 2010). Given the wide-ranging importance of Notch signaling in development, it is not surprising that dysregulation of Notch signaling in humans results in birth defects as well as tumor formation in different organs (Bolós et al. 2007; Ranganathan et al. 2011; Louvi and Artavanis-Tsakonas 2012; Penton et al. 2012; South et al. 2012).
First identified in Drosophila, the Notch gene encodes a type I transmembrane receptor protein with two known ligands, Delta and Serrate, which themselves are type I transmembrane proteins. Binding of the Notch receptor with its ligands expressed by the neighboring cell initiates the signaling cascade, leading to a sequence of proteolytic events that releases the Notch intracellular domain (NICD). The NICD subsequently translocates into the nucleus, where it associates with Suppressor of Hairless [Su(H)] and Mastermind (Mam) proteins to assemble an active transcription complex that selectively turns on the expression of downstream target genes (Fortini 2009; Kopan and Ilagan 2009).
Notch signaling is tightly controlled both in time and space, and such complex regulation can occur at multiple levels. For example, the regulation of the amount of Notch receptors and respective ligands, proteolytic processing to generate active NICD, the formation of transcriptional repressive or active complexes on the chromatin, as well as trafficking of both receptors and ligands have all been shown as crucial steps for modulating Notch signaling output (Schweisguth 2004; Bray 2006; Fortini 2009; Andersson et al. 2011).
Protein ubiquitination represents a major form of posttranslational regulatory mechanism that modulates protein quality control, stability and trafficking (Grabbe et al. 2011). The ubiquitination process is catalyzed by three distinct enzyme complexes, which conjugate small modifier protein ubiquitin (Ub) to specific substrates in a step-wise fashion. A single E1 (Ub-activating enzyme) and a single E2 (Ub-conjugating enzyme) complex are responsible for activating and conjugating Ub moiety, respectively. In contrast to a limited number of E1 and E2 enzymes, distinct E3 complexes serve as the Ub protein ligases to transfer Ub moiety from the E2 enzyme onto specific substrates. A fourth type of enzyme, the deubiquitinating enzyme (also known as deubiquitinase or DUB), counteracts the ubiquitination process by removing Ub from substrate proteins (Ciechanover 2005; Grabbe et al. 2011).
During the past decade, genetic screens performed in Drosophila and other model organisms, combined with rigorous biochemistry, have begun to unveil the importance of ubiquitination in the regulation of Notch activity. Several E3 ubiquitin ligases have been identified to regulate ubiquitination of the Notch receptor (Lai 2002; LeBras et al. 2011; Weinmaster and Fischer 2011). Suppressor of deltex [Su(dx)] was first discovered in Drosophila as a negative regulator of Notch signaling, acting in an antagonist manner to another E3 ubiquitin ligase Deltex identified in later studies (Matsuno et al. 1995; Cornell et al. 1999; Qiu et al. 2000; Matsuno et al. 2002). Along with the other two E3 ligases, Nedd4 and c-Cbl, Su(dx) plays important roles in sorting and lysosomal degradation of unactivated Notch receptor (Jehn et al. 2002; Sakata et al. 2004; Wilkin et al. 2004; Wang et al. 2010). An evolutionarily conserved F-box protein Fbw7 (also known as Sel-10) has been shown to ubiquitinate NICD, resulting in proteasomal degradation of NICD in Caenorhabditis elegans and mammals, although a direct demonstration of its role in Drosophila is missing (Hubbard et al. 1997; Oberg et al. 2001; Wu et al. 2001; Tetzlaff et al. 2004; Tsunematsu et al. 2004; Matsumoto et al. 2011; Nicholson et al. 2011). In addition, work from several laboratories indicated that two distinct E3 ligases, Neuralized (Neur) and Mind bomb (Mib1), directly promote mono-ubiquitination of the ligand proteins Delta and Serrate to facilitate their endocytosis (Deblandre et al. 2001; Lai et al. 2001; Pavlopoulos et al. 2001; Yeh et al. 2001; Itoh et al. 2003; Chen and Corliss 2004; Lai et al. 2005; Le Borgne et al. 2005). Furthermore, clonal analysis in Drosophila suggested that neur and mib1 function in the signal-sending cell and are required for most Notch-mediated processes (Le Borgne and Schweisguth 2003; Li and Baker 2004; Pitsouli and Delidakis 2005; Wang and Struhl 2005).
On the contrary, very little is known about the role of DUBs in the regulation of Notch signaling. To date, the only DUB enzyme that has been characterized in Notch signaling in Drosophila is Fat facets (Faf), a ubiquitin carboxyl-terminal hydrolase domain-containing protein that shares homology with vertebrate USP9 (Chen and Fischer 2000). Studies in the fly eye development revealed that Faf enhances Delta endocytosis to promote Notch signaling to ensure a correct recruitment of photoreceptor precursors (Cadavid et al. 2000; Overstreet et al. 2004). In this context, Faf deubiquitinates Liquid facets (Lqf), the Drosophila Epsin homolog, to increase the activity of Lqf, which in turn enhances the efficiency of Delta internalization (Chen et al. 2002). Very recently, the eukaryotic translation initiation factor 3 complex subunit F (eIF3F) was identified as a DUB to positively regulate Notch signaling in cultured mammalian cells (Moretti et al. 2010), but whether it functions in the same manner in vivo is unknown. In addition, three DUBs have been implicated as potential regulators of Notch signaling in a genome-wide screen for zebrafish development (Tse et al. 2009). However, whether they function directly on the Notch signaling pathway is undetermined.
The patterning of the adult fly wing blade represents a simple but efficient in vivo system for studying Notch signaling (Blair 2007). Here, we describe an in vivo RNA interference (RNAi) screen targeting all 45 DUBs that we annotated in the fly genome (Supporting Information, Table S1) to identify DUBs as novel modulators of Notch signaling. By monitoring the effects of reduced expression of individual DUBs on the formation of the wing marginal vein and the specification of scutellar macrochaetae (bristles), we identified at least four DUBs as potential Notch signaling modulators. Our genetic analyses provided evidence demonstrating that CG8445 (calypso), CG9124 (eIF-3p40), and CG9769, which encode the Drosophila orthologs of vertebrate BAP1 and the eIF3 complex subunits H and F, respectively, play a conserved role in regulating Notch signaling in vivo. In addition, we characterized CG32479, the Drosophila ortholog of vertebrate USP10, whose activity is necessary and sufficient to positively regulate Notch signaling.
Materials and Methods
Fly genetics
Act5C-Gal4, Act5C>yw > Gal4, C96-Gal4, dpp-Gal4, GMR-Gal4, ptc-Gal4, tub-Gal4, Gbe+Su(H)m8-lacZ [Su(H)-lacZ], and neuralizedA101-lacZ (neur-lacZ) were described previously (Huang et al. 1991; Furriols and Bray 2001; Ahmed et al. 2003; Kim et al. 2006; Du et al. 2011; Su et al. 2011). Transgenic RNAi flies targeting all 45 DUB genes predicted in the fly genome were obtained from the Vienna Drosophila RNAi Center [VDRC, Vienna, Austria (Dietzl et al. 2007)], and the Fly Stocks of National Institute of Genetics (NIG-Fly; National Institute of Genetics, Shizuoka, Japan; Table S2). Our primary RNAi screens were conducted at 29° by crossing individual RNAi lines with the dpp-Gal4 and c96-Gal4 drivers, respectively, for defects on patterning the adult wing margin and scutellar bristles. Crosses were shifted to lower temperatures (18° or 21°) for RNAi lines whose overexpression resulted in embryonic or larval lethality at 29° (Du et al. 2011).
We took three approaches to ensure the specificity and effectiveness of DUB RNAi transgenes used in our targeted screens. (1) In most cases, we obtained at least two RNAi lines targeting different regions of a specific DUB, thus avoiding potential off-target effects associated with a particular dsRNA construct. (2) For those DUBs with only a single RNAi line available, we obtained transgenic GS (Gene Search) trap fly lines (Toba et al. 1999) from the Drosophila Genetic Resource Center (Kyoto, Japan) to test effects of overexpressed DUB. One example using this approach was CG32479, in which overexpression or downregulation of CG32479 in the scutellum resulted in opposite phenotypes on the differentiation of scutellar bristles (Figure 8). (3) For those DUBs with a single RNAi line whose RNAi knockdown did not cause any obvious defects in the wing or the scutellum, we searched databases of existing genome-wide RNAi screens for the effectiveness of these RNAi lines when overexpressed by other Gal4 drivers in other cellular processes (Cronin et al. 2009; Mummery-Widmer et al. 2009; Pospisilik et al. 2010; Neely et al. 2010a,b; Saj et al. 2010; Neumüller et al. 2011; Valakh et al. 2012). For example, a single RNAi line for CG8830 had no effect in our screen (Table S2). However, the same RNAi line, when overexpressed by the pnr-Gal4 driver, resulted in lethality during pupal development (Mummery-Widmer et al. 2009).
Figure 8 .
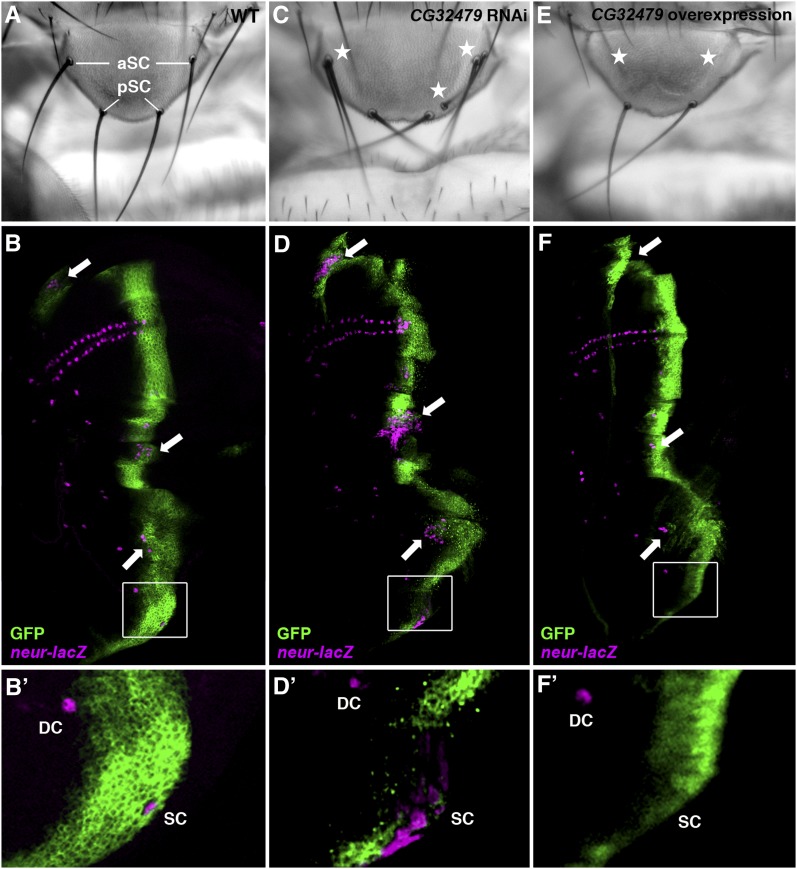
CG32479 is both necessary and sufficient to control the formation of scutellar bristles. The adult scutellar bristles (A) are differentiated from macrochaete SOP cells in early third-instar wing imaginal discs, which can be marked by the neur-lacZ reporter (magenta; B-F′). Knocking down the expression of CG32479 by the ptc-Gal4 driver (green) led to increased numbers of scutellar bristles in the adult notum (stars; C): 64% of flies with 5−6 scutellar bristles and 36% of flies with more than 6 scutellar bristles (n = 83). This fully penetrant scutellar bristle phenotype was a consequence of expanded SOP fate observed in wing discs along the ptc-Gal4 expression pattern (arrows; D). In contrast, overexpressing CG32479 resulted in loss of anterior scutellar bristles (100% penetrant, n = 25; stars; E) and reduction of SOP cells (arrows; F). Enlarged boxed areas shown in B′, D′, and F′ highlight distinct neur-lacZ staining in the scutellar (SC) and dorsal-central (DC) SOPs resulted from altered expression of CG32479. Note that the expression pattern of the ptc-Gal4 driver (marked by GFP; B-F′) overlaps the SC but not the DC SOPs.
In a secondary screen to establish a direct involvement of candidate DUBs in the regulation of Notch signaling, expression patterns of Notch signaling targets [Cut, Wingless (Wg) and/or Su(H)-lacZ] as well as different forms of the Notch receptor protein [Notch extracellular domain (NECD) and Notch intracellular domain (NICD)] were examined in early third-instar larval wing discs in which DUB RNAi was induced by the dpp-Gal4 or ptc-Gal4. In addition, cell-autonomous effects of CG9769 or CG32479 on Notch signaling were examined in FLIPout clones overexpressing respective RNAi using the Act5C>yw > Gal4 recombined with UAS-gfp, which allow the distinction between RNAi-overexpressing (positively marked by GFP) and wild-type control cells (GFP-negative) in the developing wing (Ito et al. 1997). The conditions to induce FLIPout clones in the wing disc are listed as follows: 2-day-old larval progenies from the cross of hs-flp;; Act5C > yw>Gal4 and UAS-CG9769 RNAi (V101465; VDRC); UAS-P35 or UAS-CG32479 RNAi (V37858 or V37859) were heat-shocked at 37° for 30 min (Su et al. 2011). Heat-shocked larvae were then cultured either at 25° (for CG9769 RNAi) or at 18° (for CG32479 RNAi) until the third-instar larval stage.
For RNAi lines displaying scutellar bristle defects, ptc-Gal4; neur-lacZ flies were used to examine the effect of DUBs in the specification of scutellar sensory organ precursor (SOP) cells in the notal region of the wing disc. In some experiments, Notch RNAi lines (V27228 and V27229; VDRC), gfp RNAi lines (9330 and 9331; Bloomington Drosophila Stock Center) and GS lines inserted in cis at the CG32479 locus [202182 and 204397; Drosophila Genetic Resource Center at Kyoto, (DGRC), Japan] were used.
Immunofluorescence of wing imaginal discs and image acquisition of adult fly structures
Wing discs dissected from third-instar larvae were fixed in 4% paraformaldehyde and labeled with the following primary antibodies: mouse anti-Cut [1;100; 2B10; Developmental Studies Hybridoma Bank (DSHB)], mouse anti-Hindsight (Hnt; 1:50; 1G9; DSHB), mouse anti-Wg (1:200; 4D4; DSHB), mouse anti-NECD (1:200; C458.2H; DSHB), mouse anti-NICD (1:200; C17.9C6; DSHB), and rabbit anti-β-Galactosidase (1:4000; Cappel). Alexa fluor-conjugated secondary antibodies (1:400; Invitrogen) were used. In some experiments, DAPI (0.05 μg/mL; Sigma-Aldrich) was used to visualize nuclei. The fluorescence images were acquired with a Zeiss Axio Imager2 microscope equipped with an ApoTome.
Adult eyes, wings, and nota were dissected and mounted as described previously (Zhu et al. 2003). The images of these adult structures were acquired with a Leica DMIL inverted microscope (wings) or a Leica MZ16F stereomicroscope equipped with a QImaging QICAM Fast 1394 digital camera (eyes and nota). The figures were assembled in Adobe Photoshop CS5. Minor image adjustments (brightness and/or contrast) were done in AxioVision 4.8.1 or Adobe Photoshop.
Results
The genomic inventory of DUBs encoded in Drosophila melanogaster
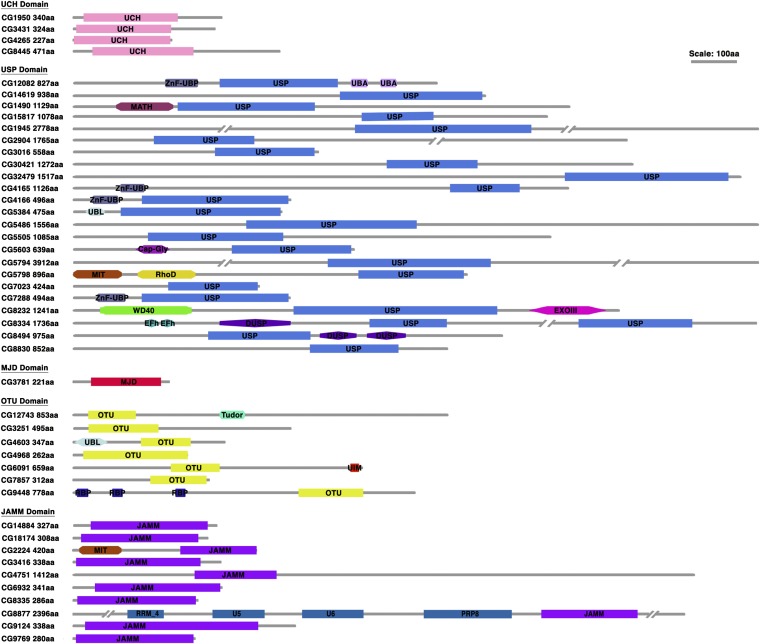
DUBs are highly conserved proteases that catalyze the cleavage of the isopeptide bond between the C-terminal glycine of ubiquitin or ubiquitin-like protein and a unique lysine of a target protein. Recent bioinformatic studies in yeast and vertebrates (Buus et al. 2009; Sowa et al. 2009; Tse et al. 2009; Kouranti et al. 2010) further classified DUBs into five subfamilies on the basis of their distinct signature DUB catalytic domains: ubiquitin C-terminal hydrolases (UCHs), ubiquitin-specific proteases (USPs), Machado-Joseph disease domain proteases (MJDs), otubain proteases (OTUs) and JAB1/MPN/Mov34 domain proteases (JAMMs). Among them, the UCHs, USPs, MJDs, and OTUs are cysteine proteases whereas the JAMMs are metalloproteases (Nijman et al. 2005).
A handful of DUBs have been studied, and their function has been linked to various cellular processes, including ubiquitin processing, histone modification, cell-cycle regulation, and developmental signaling (Nijman et al. 2005; Reyes-Turcu et al. 2009; Sowa et al. 2009). More importantly, mutations in several DUBs in humans have been identified in diseases, such as cancer, neurodegenerative diseases, and inflammatory diseases (Nijman et al. 2005; Singhal et al. 2008; Hussain et al. 2009; Huang et al. 2011), suggesting that the activity of DUBs must be tightly regulated in order to maintain the necessary levels of ubiquitination. Despite the apparent importance of DUBs in many physiological and pathological processes, molecular functions of the majority of DUBs remain largely unknown.
The Drosophila melanogaster represents an ideal model system for systematically studying in vivo functions of DUBs in many cellular processes, due to its genomic and functional conservation with vertebrates and a large collection of already existing genetic resources. However, the annotation of the fly DUBs is incomplete (Chen and Fischer 2000). We used several bioinformatic tools [Interpro Protein Sequence Analysis (http://www.ebi.ac.uk/interpro/), NCBI Conserved Domain database (http://www.ncbi.nlm.nih.gov/cdd/), and Pfam 26.0 (http://pfam.sanger.ac.uk/)] and found that the fly genome encodes a set of proteins with distinct signature DUB catalytic domains (Figure 1). A total of 45 putative DUBs belonging to five typical DUB sub-families were identified. These included four UCHs, 23 USPs, one MJD proteases, seven OTU proteases, and 10 JAMM proteases (Table S1). Among the 45 fly DUBs identified, 43 were found to have direct orthologs in vertebrates as revealed by NCBI HomoloGene (http://www.ncbi.nlm.nih.gov/homologene) or OrthoDB (http://cegg.unige.ch/orthodb5). The only exceptions are two Otu domain-containing proteases, including the founding member of the Otu sub-family, Ovarian cancer [Otu; CG12743 (Makarova et al. 2000)].
Figure 1 .
Inventory and domain architectures of annotated DUBs in Drosophila. Drosophila DUBs are characterized into five subfamilies on the basis of their signature DUB catalytic domains. The UCH, USP, MJD, OUT, and JAMM domain-containing proteases are shown. Apart from signature DUB domains, we retrieved domain architectures for each DUB by using the Pfam and the NCBI Conserved Domain database. The abbreviations for additional domains are listed as follows: Cap-Gly, cytoskeleton-associated proteins, glycine-rich domain; DUSP, domain in ubiquitin-specific proteases; EFh, EF-hand, calcium binding motif; EXOIII, exonuclease, RNase T/DNA polymerase III; MATH, meprin and TRAF-homology; MIT, microtubule interacting and trafficking molecule domain; PRP8, pre-mRNA processing splicing factor 8; RBP, zinc finger, RanBP2-type; RhoD, rhodanese homology domain; RPT, internal repeats; RRM_4, RNA recognition motif of the spliceosomal PrP8; Tudor, Tudor domain; U5 and U6, U5 and U6 snRNA binding domains; UBA, ubiquitin-associated; UBL, ubiquitin-like; UIM, ubiquitin interaction motif; WD40, WD40-repeat-containing domain; and ZnF-UBP, zinc finger ubiquitin binding domain. Proteins and domains are plotted on an approximate scale.
DUBs have been demonstrated to play essential roles in multiple cellular processes other than ubiquitin processing. Additional architecture domains that are present in the primary amino acid sequence of these enzymes may provide hints to distinct functions of DUBs. Therefore, we performed domain search by using the Pfam and the NCBI Conserved Domain database, which revealed a wide range of functionally important domains in the fly DUBs (Figure 1). One example is the ZnF-UBP domain (zinc-finger ubiquitin binding domain), which is present at the N-terminal to the USP domain in several fly DUBs, including CG12082, CG4165, CG4166 (Nonstop, Not), and CG7288. CG4166 and its orthologs (yeast Ubp8 and vertebrate USP22; Figure S1) function as an essential component of the SAGA complex to deubiquitinate histone H2B (Henry et al. 2003; Weake et al. 2008; Zhang et al. 2008; Zhao et al. 2008). Crystal structure analyses revealed that the Ubp8 ZnF-UBP domain does not form the ubiquitin tail-binding pocket as found in USP5 (vertebrate ortholog of CG12082) and USP16. Instead, it functions as the scaffold to facilitate the assembly of the SAGA complex, thus providing structural insight as to why CG4166 and its orthologs alone do not possess the DUB activity (Reyes-Turcu et al. 2006; Allen and Bycroft 2007; Pai et al. 2007; Köhler et al. 2010; Samara et al. 2010). We believe that the conservation of functionally important architecture domains present in the fly DUBs, such as the ZnF-UBP domain, established guidelines for detailed structure/function analyses of specific roles of the fly DUBs in a variety of cellular processes.
A targeted in vivo RNAi screen to identify novel DUB genes in the regulation of Notch signaling
Most of the components of the Notch signaling pathway were identified through classical forward genetic screens conducted in Drosophila (Artavanis-Tsakonas et al. 1999; Bray 2006; Andersson et al. 2011). However, these screens failed to uncover a role of DUBs in the regulation of Notch signaling, probably attributable to the fact that many DUBs exhibit pleiotropic effects in early development. The only exception is faf, which encodes a DUB that regulates Notch signaling specifically in the developing eye (Fischer-Vize et al. 1992). Recently, the availability of two collections of transgenic RNAi libraries housed in the VDRC (Dietzl et al. 2007) and the NIG-Fly Stock Center made it possible to conduct reverse genetic screens to identify DUBs that regulate Notch signaling at later stages of Drosophila development. Notch signaling is critical for patterning the wing margin and scutellar bristle (de Celis et al. 1991; Artavanis-Tsakonas et al. 1999; Brennan et al. 1999). We therefore examined the adult wing margin morphology and counted the number of scutellar bristles in the scutellum as simple readouts in our primary in vivo RNAi screens to identify DUB genes that function in Notch signaling.
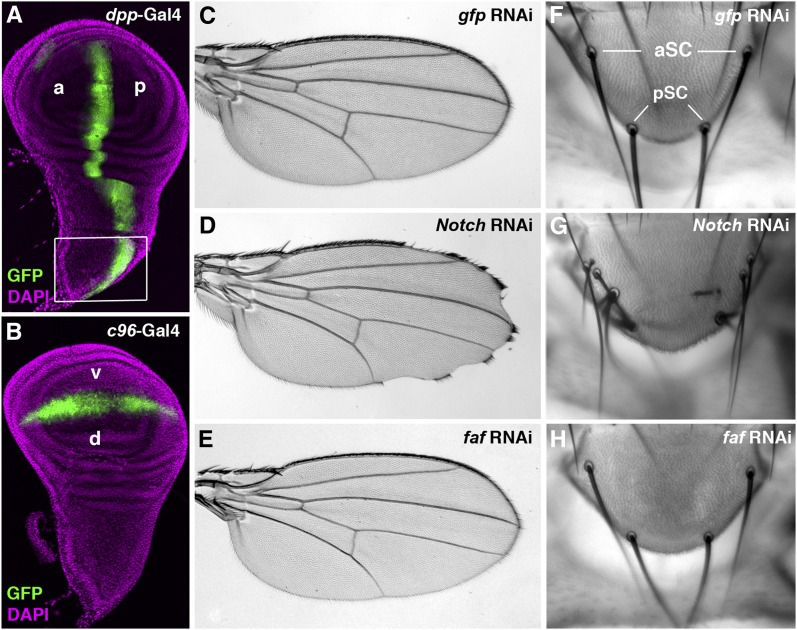
We first reduced the expression of individual DUB genes in wing discs by RNAi using the dpp-Gal4 and C96-Gal4 drivers, respectively, and then examined the resulting phenotypes in the adult fly. The dpp-Gal4 driver confers the RNAi knockdown in the region between longitudinal veins L3 and L4 in the adult wing as well as in the notum (Figure 2A), potentially affecting both the wing margin formation and scutellar bristle differentiation. However, strong dpp-Gal4 activity is also observed in embryonic segmentation (Staehling-Hampton et al. 1994), a stage that is essential for embryogenesis. In contrast, the C96-Gal4 driver dictates the RNAi expression along the wing margin (Figure 2B), thus minimizing a potential effect of embryonic lethality resulted from RNAi overexpression by the dpp-Gal4 driver. Note that the C96-Gal4 driver also confers a restricted expression in late embryos (Kim et al. 2006).
Figure 2 .
The adult wing margin and scutellar bristle phenotypes are simple but effective readouts for altered Notch signaling. The larval wing imaginal disc is the primordial tissue of the adult wing blade and notum. The expression patterns of the dpp-Gal4 (A) and the C96-Gal4 drivers (B) in wing discs were marked by the UAS-gfp transgene (green). The dpp-Gal4 drives transgene expression abutting the anterior-posterior (a-p) boundary (A), whereas the C96-Gal4 confers gfp expression along the dorsal-ventral (d-v) boundary, i.e., presumptive wing margin in the wing pouch (B). Note that the dpp-Gal4 is also expressed in regions where the adult scutellar structures are derived (box; A). DAPI staining was used to mark nuclei in wing discs (magenta; A and B). As expected, ectopic expression of gfp RNAi by the C96-Gal4 did not produce any effect in the adult wing margin (C). In contrast, reduced expression of Notch receptor gene by RNAi resulted in serrations along the wing margin (D). However, knockdown of the expression of faf, which encodes a DUB that specifically regulates Notch signaling in the developing Drosophila eye, had no effect on patterning of the wing margin (E). Note that wings dissected from female flies are shown in all figures. Two pairs of scutellar bristles, anterior SC and posterior SC (aSC and pSC), are present in the scutellum (F). When the expression of Notch was knocked down by RNAi using the dpp-Gal4, an increased number of scutellar bristles was observed (G). In contrast, overexpressing RNAi targeting either gfp (F) or faf (H) had no effect on the specification of scutellar bristles. Phenotypes shown in panels D (n > 20) and G (n = 10) are fully penetrant.
To test whether the aforementioned strategy was feasible for our screens, we examined the effect of RNAi specifically targeting two known regulators of Notch signaling on the wing margin formation and scutellar bristle differentiation. The Notch receptor gene is ubiquitously expressed. Flies with reduced Notch expression often exhibit scalloped wing margin morphology and defects in the notum (de Celis et al. 1991; Artavanis-Tsakonas et al. 1999; Brennan et al. 1999). Similarly, knocking down Notch expression by RNAi using the C96-Gal4 driver led to frequent serrations along the wing margin (Figure 2D). Furthermore, we found that Notch RNAi, when overexpressed in the notum by the dpp-Gal4 driver, led to formation of more scutellular bristles (Figure 2G). faf, on the other hand, is an eye-specific regulator of Notch signaling; overexpressed faf RNAi by the tub-Gal4 driver led to a rough eye appearance (Figure S4B). However, no obvious morphologic defect was observed in the wing (Figure 2E) or scutellum (Figure 2H) when faf RNAi was misexpressed in the wing disc by the C96-Gal4 or dpp-Gal4, which is consistent with the observation that faf mutant flies do not display any abnormalities in the wing or the notum (Overstreet et al. 2004). These proof-of-principle experiments therefore demonstrated that specific adult phenotypes in the wing and notum associated with defective Notch signaling could be efficiently scored in targeted RNAi screens for all DUBs in the Notch signaling cascade.
We obtained 99 RNAi lines targeting all 45 DUB genes that we annotated in the fly genome (Table S2). We found that reduced expression of six DUB genes by either the C96-Gal4 or dpp-Gal4 altered adult wing margin morphology and/or the number of scutellar bristles (Figures 3 and 4; Table S2). As several developmental signaling systems, including Notch, Hedgehog, Wnt and growth factor signaling, collaborate to control the wing development (Blair 2007), a secondary screen was performed in the third-instar larval wing imaginal disc, a primordial tissue of the adult wing, to identify DUBs specifically functioning on Notch signaling. The distribution patterns of Notch signaling targets, Cut and Wg (Figure 5, A−D), along the presumptive wing margin, i.e. the dorsal-ventral boundary of the wing disc, were examined when candidate DUB RNAi was respectively overexpressed by either the ptc-Gal4 or the dpp-Gal4 driver (Table S3). Note that both Gal4 drivers confer RNAi expression along the anterior-posterior border, thus allowing us to compare directly expression patterns of Notch target genes in RNAi expressing cells (i.e. anteroposterior border cells intersecting the future wing margin) with those in wild-type margin cells in the same disc (see an example of the effect of Notch RNAi on the activation of Cut and Wg in Figure 5, E−H). Only those candidate DUB genes, exhibiting significantly altered target expression when knocked down by RNAi, were chosen as potential Notch regulators.
Figure 3 .
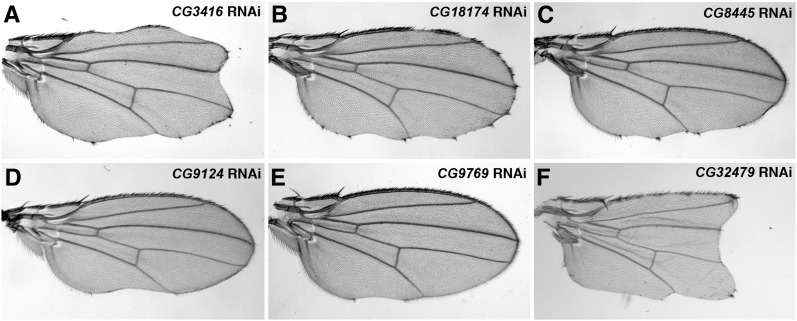
Reduced expression of several DUBs leads to defects on the formation of the margin vein in the adult fly wing. Reduced expression of CG3416 (A), CG18174 (B), CG8445 (C), CG9124 (D), CG9769 (E), or CG32479 (F) in the wing imaginal disc by respective RNAi transgenes driven by the c96-Gal4 driver led to defective wing margin formation, with different degrees of severity. Note that the wing margin defects in panels A and F are stronger than those caused by Notch RNAi (cf. Figure 2D), suggesting that CG3416, which encodes an essential component of the 19S proteasome, and probably CG32479, play additional roles other than those in Notch signaling. Phenotypes shown in panels A (n = 19), B (n = 9), C (n = 15), D (n = 11), E (n = 6), and F (n = 23) are fully penetrant.
Figure 4 .
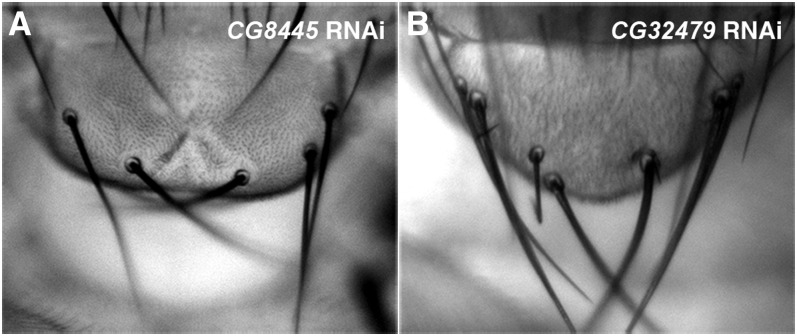
Two candidate DUBs regulate the specification of scutellar bristles in the adult fly notum. Reduced expression of CG8445 (A) or CG32479 (B) in the wing imaginal disc by respective RNAi driven by the dpp-Gal4 driver resulted in an increased number of scutellar bristles, with a penetrance of 18% (n = 11) for CG8445 RNAi and 100% (n = 20) for CG32479 RNAi.
Figure 5 .
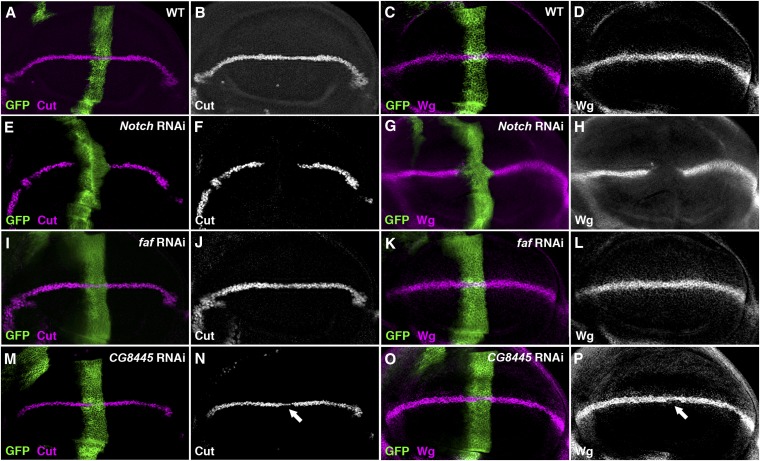
The Effect of CG8445 RNAi on the expression of Notch signaling targets in the wing imaginal disc. The expression of Wg (magenta; A) and Cut (magenta; C) along the dorsal-ventral boundary (i.e., presumptive wing margin) is activated by Notch signaling in a wild-type (WT) wing disc. Knocking down the expression of Notch receptor by the ptc-Gal4 driver along the anterior-posterior boundary (marked by GFP) led to a complete loss of downstream targets, Cut (E, F) and Wg (G, H), on the presumptive wing margin cells intersecting the dorsal-ventral boundary. In contrast, the expression of both Cut (J) and Wg (L) was unaffected in wing discs ectopically expressing faf RNAi, consistent with previous studies indicating that faf mutant flies did not exhibit any defect in the wing. Knockdown of the expression of CG8445 by RNAi in the wing disc resulted in modulate reduction of Cut (arrow; N) and Wg (arrow; P), suggesting that CG8445 is necessary to positively regulate Notch signaling. All phenotypes are fully penetrant (n > 20 discs).
One of these candidate DUBs, CG3416 (Mov34), encodes the Drosophila ortholog of vertebrate Rpn8, an essential regulatory subunit of the 19S proteasome (Glickman et al. 1998). As proteasome function is important in many cellular processes, it was not surprising that reduced CG3416 expression by RNAi with either the ptc-Gal4 or dpp-Gal4 resulted in early larval lethality, precluding a study of its function in Notch signaling in the developing wing disc (Table S3). In addition, the expression pattern of Cut or Wg was not significantly affected in wing discs overexpressing RNAi specific for CG18174, another candidate DUB (Table S3), suggesting that the wing margin defects associated with reduced CG18174 expression may not be a direct consequence of defective Notch signaling.
In contrast, reducing the expression of each of the remaining four candidate DUBs all led to strong reduction in the expression of Wg and Cut along the future wing margin (Table S3; Figures 5−7), very similar to the effects resulted from reduced Notch expression (cf. Figure 5, E−H). One of these candidate DUBs, CG8445 (calypso), encodes a Drosophila ortholog of vertebrate BRCA1 associated protein 1 (BAP1). Interestingly, a recent study demonstrated that zebrafish BAP1, when knocked down by specific morpholinos, expanded the number of hindbrain neurons in the developing embryo, a neurogenic process dependent on appropriate levels of Notch signaling (Tse et al. 2009). Consistent with the function of vertebrate BAP1, we observed increased number of scutellar bristles in the adult fly notum (Figure 4A) and reduced Cut and Wg expression in the wing disc (Figure 5, M−P) when CG8445 RNAi was overexpressed. These data strongly suggested that CG8455 plays a conserved role like BAP1 to positively regulate Notch signaling.
Figure 7 .
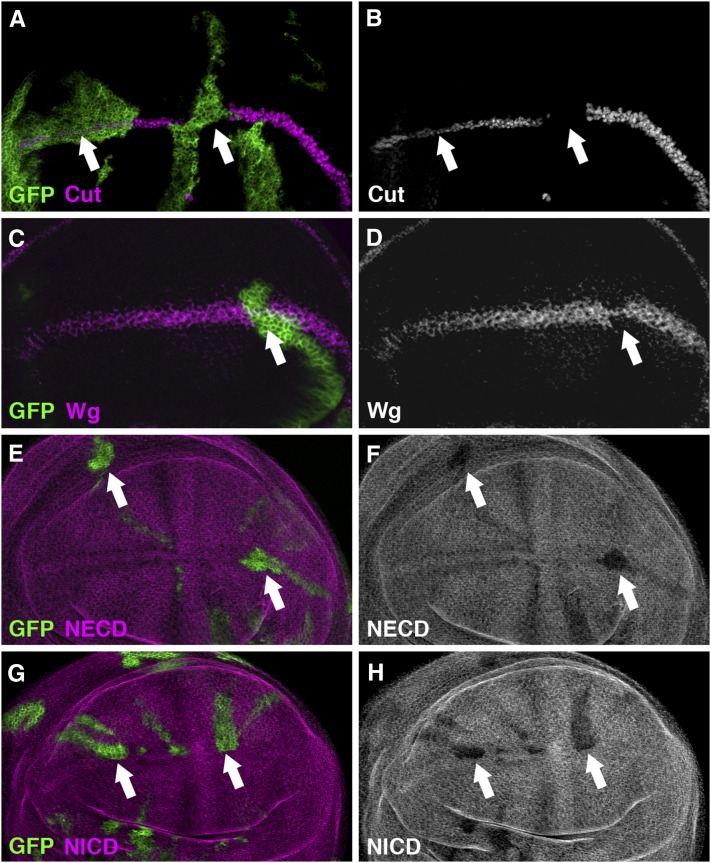
CG32479 positively regulates Notch signaling in the wing imaginal disc. Random clones overexpressing CG32479 RNAi were generated by the FLIPout technique. Knocking down the expression of CG32479 by RNAi in these clones (positively marked by GFP, green) cell-autonomously reduced the expression of Cut (arrows; B) and Wg (arrow; D). Consistently, the production of Notch protein was cell-autonomously downregulated in random clones overexpressing CG32479 RNAi (positively marked by GFP), as demonstrated by immunostaining with antibodies specific for the NECD (arrows; F) and NICD (arrows; H), respectively. All phenotypes are fully penetrant (n > 20 discs).
To our knowledge, the other three DUB genes, CG9124 (eIF-3p40) CG9769, and CG32479, have not yet been functionally studied in vivo. Therefore, we conducted genetic studies to unveil the functions of these genes in the regulation of Notch signaling in Drosophila.
Components of the eIF3 complex function as potential DUBs for the Notch receptor In Vivo
Both CG9124 (eIF-3p40) and CG9769 encode the MJD domain-containing DUB enzymes that are highly conserved from insects to humans (Figure 1). The vertebrate orthologs of protein products of these two DUBs are components of the eukaryotic translation initiation factor 3 (eIF3) complex. CG9769 is the fly ortholog of the eIF3 subunit F (eIF3F) with 45.4% homology, whereas CG9124 shares 50% conservation with the subunit H (eIF3H) (Lasko 2000).
Reduced expression of either CG9124 or CG9769 by RNAi in the wing disc driven by the dpp-Gal4 or ptc-Gal4 resulted in lethality at the pupal stage. However, when the respective RNAi was overexpressed using the C96-Gal4 driver, we observed a weak loss-of-function Notch phenotype along the wing margin (Figure 3, D and E). To confirm the requirement of the fly eIF3 proteins in Notch signaling, we investigated the effect of reduced expression of CG9124 or CG9769 on the activation of Cut and Wg. As expected, reduced expression of CG9124 or CG9769 by RNAi in the wing disc led to abolished Cut and Wg expression along the dorsal-ventral boundary (Figure 6, A−D; Figure S2, A−D), providing in vivo evidence to support a role of the eIF3 complex in modulating Notch signaling.
Figure 6 .
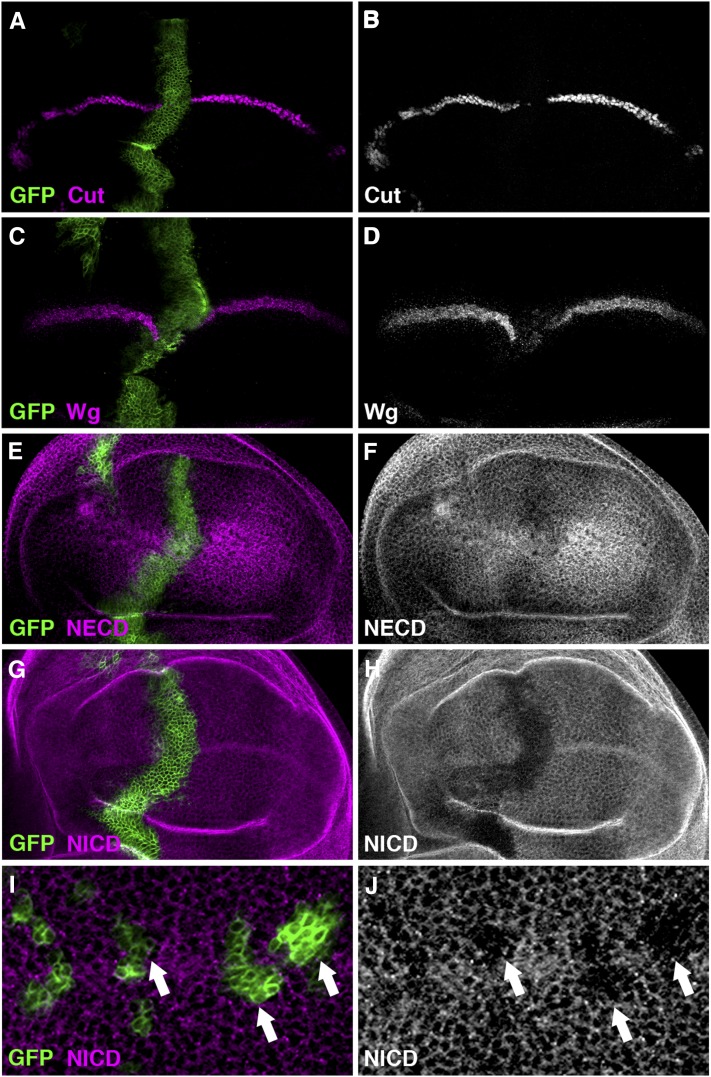
CG9769 positively regulates Notch signaling in the wing disc. When the expression of CG9769 was knocked down by RNAi at the anteroposterior boundary of the wing disc by the dpp-Gal4 driver (marked by GFP; A and C), Notch signaling activity was largely compromised as indicated by decreased expression of Cut (B) and Wg (D). The abundance of Notch protein, as examined by specific antibodies raised against either the extracellular domain (NECD; E and F) or the intracellular domain (NICD; G and H) of the Notch receptor, also was reduced. The effect of CG9769 on the production of Notch protein was cell-autonomous (arrows; J), as demonstrated in FLIPout clones (Ito et al. 1997) overexpressing CG9769 RNAi (positively marked by GFP; I). All phenotypes are fully penetrant (n > 20 discs).
Having confirmed their role in Notch signaling, the next question we asked was at which step along the Notch signaling cascade these two components of the fly eIF3 complex function in the wing disc. The clue to this question came from a recent study in cultured mammalian cells, in which eIF3F functions at the level of activated Notch protein (i.e., NICD) to assure its proper entry to the nucleus (Moretti et al. 2010). To test whether this mode of regulation is conserved in vivo, we examined the expression pattern of Notch protein in the wing disc when either component was knocked down by RNAi. Because the full-length Notch protein is subject to proteolytic cleavages, two Notch antibodies, C458.2H (Diederich et al. 1994) and C17.9C6 (Fehon et al. 1990), were used to specifically recognize the resulting extracellular (NECD) and intracellular domains (NICD) of the Notch receptor, respectively. We found that the levels of both NECD and NICD were significantly reduced along the anteroposterior boundary when CG9124 or CG9769 RNAi was overexpressed by the dpp-Gal4 or ptc-Gal4 (Figure 6, E−H; Figure S2, E−F). Further analyses of clones overexpressing CG9769 RNAi, which were positively marked by GFP (Figure 6I), confirmed that CG9769 acted cell-autonomously to regulate the abundance of the Notch receptor (Figure 6J). Taken together, our study uncovered an in vivo role of components of the eIF3 complex in the positive regulation of the Notch signal transduction, most likely acting at the level of the Notch receptor.
Identification of CG32479 as a novel DUB that regulates Notch signaling
The last candidate DUB gene CG32479 encodes the fly ortholog of vertebrate USP10, which contains a carboxy-terminal USP signature domain (Figure 1). Reduced expression of CG32479 by RNAi resulted in a typical loss-of-function Notch phenotype in the adult fly wing and notum: big serrations along the wing margin (Figure 3F) and increased number of scutellar bristles (Figure 4B). Consistent with the adult wing margin phenotype, we found that the expression of Cut (Figure 7B), Wg (Figure 7D) and Su(H)-lacZ (Figure S3B) was all downregulated in the wing disc cells overexpressing CG32479 RNAi. These results, together with the observation that CG32479 RNAi led to reduced production of Notch proteins (Figure 7, E−H), suggested that CG32479 is cell-autonomously required in Notch signaling to specify the wing margin.
To gain further insight on the role of CG32479 in Notch signaling, we investigated whether CG32479 activity was sufficient for activating Notch signaling. As the wing margin formation is not a reliable readout for activated Notch signaling, we investigate consequences of manipulated CG32479 expression on scutellar bristle differentiation. The number of scutellar bristles in the fly notum is determined through a lateral inhibition process in which the fate of sensory organ precursors (SOPs) is specified from the scutellar proneural cluster. It is known that an appropriate Notch signaling activity is required for correct SOP specification. Reduced Notch signaling leads to increased SOP specification to generate extra scutellar bristles in the adult fly notum, whereas heightened Notch activity often results in reduced number of scutellar bristles (de Celis et al. 1991; Brennan et al. 1999). The development of the adult notum is prepatterned in the wing disc where SOP cells can be visualized by specific markers, including the neur-lacZ enhancer trap used in our study [Figure 8B (Huang et al. 1991)]. We manipulated the expression of CG32479 in SOP cells using the ptc-Gal4 driver; scutellar SOPs (Figure 8B′) as well as additional SOPs in the wing pouch and other regions of the notum (arrows; Figure 8B) overlap the stripe of ptc expression (marked by GFP) in the developing wing disc (Brennan et al. 1999). As shown in Figure 8D′, overexpressing CG32479 RNAi by the ptc-Gal4 significantly expanded the number of scutellar (SC) SOP cells as marked by an increased neur-lacZ labeling. In addition, we identified two GS (Gene Search) trap fly lines (Toba et al. 1999) that can be used to overexpress CG32479. Heightened CG32479 activity conferred by either GS transgene reduced the number of bristles in the scutellum (Figure 8E), a typical phenotype associated with activated Notch signaling. Consistently, we demonstrated that this gain-of-function Notch phenotype was a direct consequence of failed scutellar SOP specification (Figure 8F′); the expression of neuralized-lacZ was diminished specifically in SC SOPs but not in dorsal-central (DC) SOPs where CG32479 was not overexpressed. Apart from its effect on scutellar SOPs, altered expression of CG32479 was sufficient to control SOP specification in other areas of the wing pouch and notum in the wing disc where the ptc-Gal4 was expressed (arrows; Figure 8, D and F). Taken together, our results indicated that CG32479 is not only necessary but also sufficient to positively regulate Notch signaling in vivo.
Discussion
The ubiquitination state of a protein substrate is tightly coordinated by opposing actions of ubiquitination and deubiquitination. Although the ubiquitination process has been extensively studied in the last two decades, the complex roles of deubiquitination have only recently been revealed through the study of a small number of DUBs. Protein deubiquitination is likely to represent an important regulatory mechanism in processes ranging from proteasomal regulation, cell-cycle control, histone modification, membrane trafficking, to stem cell maintenance (Reyes-Turcu et al. 2009; Huang et al. 2011). Despite progress in uncovering a role for DUBs in several cellular processes, their substrate specificity and modes of regulation in an in vivo system are largely unknown. Drosophila melanogaster has been widely recognized as an ideal in vivo system for understanding complex biological processes due to its genomic conservation with vertebrates and highly amenable genetics. However, only a handful of fly DUBs, including CG1945 (Faf), CG4166 (Not), CG5798 (Ubpy), and CG8445 (Calypso), have been functionally studied in detail (Chen et al. 2002; Zhao et al. 2008; Mukai et al. 2010; Scheuermann et al. 2010).
In the current study, we identified and annotated 45 DUBs encoded in the Drosophila genome. Like their vertebrate counterparts, the fly DUBs can be categorized into five subfamilies, each with a distinct DUB signature domain. Previous forward genetic screens have yielded several genes that play critical roles in the regulation of the Notch signaling transduction. However, only a single DUB (Faf) was identified in these screens, probably due to the fact that most DUBs are either maternally provided or are pleiotropically required in processes essential for cell survival or differentiation. In contrast, our RNAi-based screen, designed to examine the role of DUBs specifically on the formation of the adult wing and notum, led us to successfully uncover at least four candidate DUBs that potentially regulate Notch signaling. It should be noted that these DUBs were not identified in previous RNAi screens for novel regulators of Notch signaling (Mummery-Widmer et al. 2009; Saj et al. 2010). One of these screens was based on the usage of the pnr-Gal4 driver, which strongly expresses individual RNAi transgenes in the proximal-most part of the presumptive notum in the wing disc (Mummery-Widmer et al. 2009). Most of the RNAi transgenes tested in this screen resulted in early lethality or general growth defects in the fly nota, thus making it difficult to identify specific Notch signaling regulators. In our screen, however, we took advantage of the c96-Gal4 driver, which confers a much weaker and restricted expression of the RNAi transgenes along the presumptive wing margin. This unique property of the c96-Gal4 allowed us to uncover a specific role of DUBs in a complex biological pathway, such as Notch signaling, from their pleiotropic effects in other cellular and developmental processes in Drosophila. Our in vivo RNAi screen for DUBs in Notch signaling, coupled with morphological and molecular analyses, provided a framework in which specific roles of individual DUBs can be studied in fly development.
At least four potential DUB regulators of Notch signaling were identified in this study. Knocking down either of these DUBs by RNAi not only significantly altered the adult wing margin formation and/or notal bristle formation but also reduced the expression of Notch signaling targets, thus suggesting a positive role of candidate DUBs in Notch signaling. Previous studies suggested that orthologs of these fly DUBs function at the level of transcription or posttranscription in a variety of cellular processes (Bomberger et al. 2010; Moretti et al. 2010; Scheuermann et al. 2010; Yuan et al. 2010). It is highly plausible that the fly DUBs confer a multi-level regulation along the Notch signaling cascade.
One of candidate DUBs, CG8445 (Calypso), has been reported to function in the Polycomb repressive deubiquitinase complex (PR-DUB) to remove mono-ubiquitination from histone H2A in nucleosomes. Reduced expression of CG8445 disrupts the PR-DUB complex, resulting in elevated levels of mono-ubiquitinated H2A to globally de-repress target gene transcription in Drosophila (Scheuermann et al. 2010). Accordingly, the effect of CG8445 RNAi observed in our study could be a consequence of epigenetic effects of the PR-DUB on a negative regulator of Notch signaling; increased transcription of this negative regulator downregulates Notch signaling. Alternatively, CG8445 could act independently of the PR-DUB, thereby directly deubiquitinating a key component of the Notch signaling pathway. This notion is supported by the fact that a genome-wide chromatin immunoprecipitation assay failed to detect any significant binding of the PR-DUB on genomic loci of known Notch signaling regulator genes (Scheuermann et al. 2010).
Our work on CG9769 and CG9124 (eIF-3p40), which encode the fly orthologs of eIF3F and eIF3H, respectively (Lasko 2000), reveals an in vivo function of the eIF3 complex members in Notch signaling. The eIF3 complex serves as a scaffold to facilitate interactions among several other eIF complexes to participate in the different reactions involved in translation (Hinnebusch 2006). eIF3F and eIF3H are two indispensable components among the 13 eIF3 components identified to date (Masutani et al. 2007; Zhou et al. 2008). Furthermore, reduced expression of fly orthologs of other eIF3 proteins, including eIF3I (CG8882, Trip1) and eIF3L (CG5642), did not result in any adult phenotype associated with Notch signaling (Mummery-Widmer et al. 2009; Saj et al. 2010). Thus, we believe that the function of CG9124 and CG9769 on Notch signaling may be independent of their role on translation initiation. Indeed, a recent study suggests that eIF3F could act potentially as a JAMM-containing DUB to regulate the translocation of NICD to the nucleus in cultured mammalian cells (Moretti et al. 2010). The JAMM domain in eIF3F and eIF3H is largely conserved across different species. CG9124 and CG9769 may function in a similar manner as their vertebrate counterparts on Notch signaling. It is interesting to note that CG9769 RNAi was able to downregulate both NICD and NECD, raising an intriguing possibility that the fly eIF3F may function at the level of full-length Notch receptor.
The last candidate DUB identified in our screen is CG32479, which is both necessary and sufficient to positively regulate Notch signaling. Our results further suggested that CG32479 might function at the same level as or upstream of Notch. Additional genetic and biochemical studies will be needed to identify its direct substrate protein. The vertebrate ortholog of CG32479 is USP10, which is known to regulate membrane protein trafficking and deubiquitination of tumor suppressor p53 (Faus et al. 2005; Bomberger et al. 2009, 2010; Yuan et al. 2010; Draker et al. 2011; Liu et al. 2011). As the regulatory functions of two other DUBs identified in our screen on Notch signaling are conserved, it would be interesting to investigate whether USP10 also participates in the regulation of Notch signaling activity in mammals.
Several key components of the Notch signaling cascade are subject to ubiquitination (Lai 2002; LeBras et al. 2011; Weinmaster and Fischer 2011). It is believed that the nature of the ubiquitination pattern (mono- v.s. poly-ubiquitination) or specific lysine residues that are modified by ubiquitin on a single substrate protein may result in distinct outcomes in its subcellular localization, stability or signaling activity (Ciechanover 2005; Mukhopadhyay and Riezman 2007; Clague and Urbé 2010; Ikeda et al. 2010; Grabbe et al. 2011). Thus, multiple DUBs could be employed to target the same substrate protein, such as the Notch receptor, to confer its different activities in the Notch signaling transduction. Moreover, the requirement of Notch signaling and its regulation are heavily context dependent in vivo. For example, Faf, the only known DUB for Notch signaling, functions specifically in the developing eye (Fischer-Vize et al. 1992; Cadavid et al. 2000; Overstreet et al. 2004). Similarly, although all four candidate DUBs play a role in the formation of the wing margin and the patterning of the eye (Figure S4, C−F) in our study, only CG32479 and CG8445 are required for the specification of scutellar bristles as knocking down the expression of CG9124 and CG9769 had no effect on SOP differentiation (Figure S4, I and J).
Finally, it should be pointed out that we could not rule out the possibility that additional DUBs, other than those identified in our screen, may regulate Notch signaling during the development of other organs/tissues. We surveyed the Flybase (http://flybase.org) for potential alleles of the individual DUB genes, and found molecularly defined P-element insertions at the loci of 17 fly DUBs. Additional P-element insertions also are present less than 1.5 kbp away from the loci of another 27 DUBs; CG1950 is the only DUB that does not contain ready-to-use genetic resources at or near its locus. Nevertheless, all these genetic resources are potentially useful for generating loss-of-function DUB alleles, which will help obtain a complete profile of specific DUBs in Notch signaling as well as identify their bona fide in vivo targets in development.
Supplementary Material
Acknowledgments
We thank Gabrielle Boulianne and Sarah Bray, the Bloomington Drosophila Stock Center, the Drosophila Genetic Resource Center at Kyoto (DGRC), the Developmental Studies Hybridoma Bank (DSHB), Fly Stocks of National Institute of Genetics (NIG-Fly), and the Vienna Drosophila RNAi Center (VDRC) for fly stocks and antibodies. This work was supported by a National Institutes of Health/National Institute of General Medical Sciences grant (R01GM085175) and a March of Dimes Basil O’Connor Starter Scholar Award (5-FY07-41) to A.J.Z., American Heart Association Postdoctoral Fellowship Awards (10POST4110011 and 0825591D) to J.Z. and Y.S., and an American Heart Association Scientist Development Grant Award (12SDG8870002) to Y.S.
Footnotes
Communicating editor: H. D. Lipshitz
Literature Cited
- Ahmed A., Chandra S., Magarinos M., Vaessin H., 2003. Echinoid mutants exhibit neurogenic phenotypes and show synergistic interactions with the Notch signaling pathway. Development 130: 6295–6304 [DOI] [PubMed] [Google Scholar]
- Allen M. D., Bycroft M., 2007. The solution structure of the ZnF UBP domain of USP33/VDU1. Protein Sci. 16: 2072–2075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson E. R., Sandberg R., Lendahl U., 2011. Notch signaling: simplicity in design, versatility in function. Development 138: 3593–3612 [DOI] [PubMed] [Google Scholar]
- Artavanis-Tsakonas S., Rand M. D., Lake R. J., 1999. Notch signaling: cell fate control and signal integration in development. Science 284: 770–776 [DOI] [PubMed] [Google Scholar]
- Blair S. S., 2007. Wing vein patterning in Drosophila and the analysis of intercellular signaling. Annu. Rev. Cell Dev. Biol. 23: 293–319 [DOI] [PubMed] [Google Scholar]
- Bolós V., Grego-Bessa J., de la Pompa J. L., 2007. Notch signaling in development and cancer. Endocr. Rev. 28: 339–363 [DOI] [PubMed] [Google Scholar]
- Bomberger J. M., Barnaby R. L., Stanton B. A., 2009. The deubiquitinating enzyme USP10 regulates the post-endocytic sorting of cystic fibrosis transmembrane conductance regulator in airway epithelial cells. J. Biol. Chem. 284: 18778–18789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bomberger J. M., Barnaby R. L., Stanton B. A., 2010. The deubiquitinating enzyme USP10 regulates the endocytic recycling of CFTR in airway epithelial cells. Channels 4: 150–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray S. J., 2006. Notch signalling: a simple pathway becomes complex. Nat. Rev. Mol. Cell Biol. 7: 678–689 [DOI] [PubMed] [Google Scholar]
- Brennan K., Tateson R., Lieber T., Couso J. P., Zecchini V., et al. , 1999. The Abruptex mutations of Notch disrupt the establishment of proneural clusters in Drosophila. Dev. Biol. 216: 230–242 [DOI] [PubMed] [Google Scholar]
- Buus R., Faronato M., Hammond D. E., Urbé S., Clague M. J., 2009. Deubiquitinase activities required for hepatocyte growth factor-induced scattering of epithelial cells. Curr. Biol. 19: 1463–1466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadavid A. L., Ginzel A., Fischer J. A., 2000. The function of the Drosophila fat facets deubiquitinating enzyme in limiting photoreceptor cell number is intimately associated with endocytosis. Development 127: 1727–1736 [DOI] [PubMed] [Google Scholar]
- Chen W., Corliss D. C., 2004. Three modules of zebrafish Mind bomb work cooperatively to promote Delta ubiquitination and endocytosis. Dev. Biol. 267: 361–373 [DOI] [PubMed] [Google Scholar]
- Chen X., Fischer J. A., 2000. In vivo structure/function analysis of the Drosophila fat facets deubiquitinating enzyme gene. Genetics 156: 1829–1836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Zhang B., Fischer J. A., 2002. A specific protein substrate for a deubiquiting enzyme: liquid facets is the substrate of Fat facet. Genes Dev. 16: 289–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciechanover A., 2005. Proteolysis: from the lysosome to ubiquitin and the proteasome. Nat. Rev. Mol. Cell Biol. 6: 79–86 [DOI] [PubMed] [Google Scholar]
- Clague M. J., Urbé S., 2010. Ubiquitin: same molecule, different degradation pathways. Cell 143: 682–685 [DOI] [PubMed] [Google Scholar]
- Cornell M., Evans D., Mann R., Fostier M., Flasza M., et al. , 1999. The Drosophila melanogaster Suppressor of deltex gene, a regulator of the Notch receptor signaling pathway, is an E3 class ubiquitin ligase. Genetics 152: 567–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronin S. J., Nehme N. T., Limmer S., Liegeois S., Pospisilik J. A., et al. , 2009. Genome-wide RNAi screen identifies genes involved in intestinal pathogenic bacterial infection. Science 325: 340–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deblandre G. A., Lai E. C., Kintner C., 2001. Xenopus neuralized is a ubiquitin ligase that interacts with XDelta1 and regulates Notch signaling. Dev. Cell 1: 795–806 [DOI] [PubMed] [Google Scholar]
- de Celis J. F., Marí-Beffa M., García-Bellido A., 1991. Cell-autonomous role of Notch, an epidermal growth factor homologue, in sensory organ differentiation in Drosophila. Proc. Natl. Acad. Sci. USA 88: 632–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diederich R. J., Matsuno K., Hing H., Artavanis-Tsakonas S., 1994. Cytosolic interaction between deltex and Notch ankyrin repeats implicates deltex in the Notch signaling pathway. Development 120: 473–481 [DOI] [PubMed] [Google Scholar]
- Dietzl G., Chen D., Schnorrer F., Su K. C., Barinova Y., et al. , 2007. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature 448: 151–156 [DOI] [PubMed] [Google Scholar]
- Draker R., Sarcinella E., Cheung P., 2011. USP10 deubiquitylates the histone variant H2A.Z and both are required for androgen receptor-mediated gene activation. Nucleic Acids Res. 39: 3529–3542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J., Zhang J., Su Y., Liu M., Ospina J. K., et al. , 2011. In vivo RNAi screen reveals neddylation genes as novel regulators of Hedgehog signaling. PLoS ONE 6: e24168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faus H., Meyer H. A., Huber M., Bahr I., Haendler B., 2005. The ubiquitin-specific protease USP10 modulates androgen receptor function. Mol. Cell. Endocrinol. 245: 138–146 [DOI] [PubMed] [Google Scholar]
- Fehon R. G., Kooh P. J., Rebay I., Regan C. L., Xu T., et al. , 1990. Molecular interactions between the protein products of the neurogenic loci Notch and Delta, two EGF-homologous genes in Drosophila. Cell 61: 523–534 [DOI] [PubMed] [Google Scholar]
- Fischer-Vize J. A., Rubin G. M., Lehmann R., 1992. The fat facets gene is required for Drosophila eye and embryo development. Development 116: 985–1000 [DOI] [PubMed] [Google Scholar]
- Fortini M. E., 2009. Notch signaling: the core pathway and its posttranslational regulation. Dev. Cell 16: 633–647 [DOI] [PubMed] [Google Scholar]
- Furriols M., Bray S., 2001. A model Notch response element detects Suppressor of Hairless-dependent molecular switch. Curr. Biol. 11: 60–64 [DOI] [PubMed] [Google Scholar]
- Glickman M. H., Rubin D. M., Coux O., Wefes I., Pfeifer G., et al. , 1998. A subcomplex of the proteasome regulatory particle required for ubiquitin-conjugate degradation and related to the COP9-signalosome and elF3. Cell 94: 615–623 [DOI] [PubMed] [Google Scholar]
- Grabbe C., Husnjak K., Dikic I., 2011. The spatial and temporal organization of ubiquitin networks. Nat. Rev. Mol. Cell Biol. 12: 295–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry K. W., Wyce A., Lo W. S., Duggan L. J., Emre N. C., et al. , 2003. Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8. Genes Dev. 17: 2648–2663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch A. H., 2006. eIF3: a versatile scaffold for translation initiation complexes. Trends Biochem. Sci. 31: 553–562 [DOI] [PubMed] [Google Scholar]
- Huang F., Dambly-Chaudiere C., Ghysen A., 1991. The emergence of sense organs in the wing disc of Drosophila. Development 111: 1087–1095 [DOI] [PubMed] [Google Scholar]
- Huang Z., Wu Q., Guryanova O. A., Cheng L., Shou W., et al. , 2011. Deubiquitylase HAUSP stabilizes REST and promotes maintenance of neural progenitor cells. Nat. Cell Biol. 13: 142–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard E. J., Wu G., Kitajewski J., Greenwald I., 1997. sel-10, a negative regulator of lin-12 activity in Caenorhabditis elegans, encodes a member of the CDC4 family of proteins. Genes Dev. 11: 3182–3193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain S., Zhang Y., Galardy P. J., 2009. DUBs and cancer: the role of deubiquitinating enzymes as oncogenes, non-oncogenes and tumor suppressors. Cell Cycle 8: 1688–1697 [DOI] [PubMed] [Google Scholar]
- Ikeda F., Crosetto N., Dikic I., 2010. What determines the specificity and outcomes of ubiquitin signaling? Cell 143: 677–681 [DOI] [PubMed] [Google Scholar]
- Ito K., Awano W., Suzuki K., Hiromi Y., Yamamoto D., 1997. The Drosophila mushroom body is a quadruple structure of clonal units each of which contains a virtually identical set of neurones and glial cells. Development 124: 761–771 [DOI] [PubMed] [Google Scholar]
- Itoh M., Kim C. H., Palardy G., Oda T., Jiang Y. J., et al. , 2003. Mind bomb is a ubiquitin ligase that is essential for efficient activation of Notch signaling by Delta. Dev. Cell 4: 67–82 [DOI] [PubMed] [Google Scholar]
- Jehn B. M., Dittert I., Beyer S., Mark K., Bielke W., 2002. c-Cbl binding and ubiquitin-dependent lysosomal degradation of membrane associated Notch1. J. Biol. Chem. 277: 8033–8040 [DOI] [PubMed] [Google Scholar]
- Kim S. Y., Renihan M. K., Boulianne G. L., 2006. Characterization of big bang, a novel gene encoding for PDZ domain-containing proteins that are dynamically expressed throughout Drosophila development. Gene Expr. Patterns 6: 504–518 [DOI] [PubMed] [Google Scholar]
- Köhler A., Zimmerman E., Schneider M., Hurt E., Zheng N., 2010. Structural basis for assembly and activation of the heterotetrameric SAGA histone H2B deubiquitinase module. Cell 141: 606–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopan R., Ilagan M. X., 2009. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell 137: 216–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouranti I., McLean J. R., Feoktistova A., Liang P., Johnson A. E., et al. , 2010. A global census of fission yeast deubiquitinating enzyme localization and interaction networks reveals distinct compartmentalization profiles and overlapping functions in endocytosis and polarity. PLoS Biol. 8: e1000471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai E. C., 2002. Protein degradation: four E3s for the Notch pathway. Curr. Biol. 12: R74–R78 [DOI] [PubMed] [Google Scholar]
- Lai E. C., 2004. Notch signaling: control of cell communication and cell fate. Development 131: 965–973 [DOI] [PubMed] [Google Scholar]
- Lai E. C., Deblandre G. A., Kintner C., Rubin G. M., 2001. Drosophila Neuralized is a ubiquitin ligase that promotes the internalization and degradation of Delta. Dev. Cell 1: 783–794 [DOI] [PubMed] [Google Scholar]
- Lai E. C., Roegiers F., Qin X., Jan Y. N., Rubin G. M., 2005. The ubiquitin ligase Drosophila Mind bomb promotes Notch signaling by regulating the localization and activity of Serrate and Delta. Development 132: 2319–2332 [DOI] [PubMed] [Google Scholar]
- Lasko P., 2000. The Drosophila melanogaster genome: translation factors and RNA binding proteins. J. Cell Biol. 150: F51–F56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Borgne R., Schweisguth F., 2003. Unequal segregation of Neuralized biases Notch activation during asymmetric cell division. Dev. Cell 5: 139–148 [DOI] [PubMed] [Google Scholar]
- Le Borgne R., Remaud S., Hamel S., Schweisguth F., 2005. Two distinct E3 ubiquitin ligases have complementary functions in the regulation of Delta and Serrate signaling in Drosophila. PLoS Biol. 3: e96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBras S., Loyer N., Le Borgne R., 2011. The multiple facets of ubiquitination in the regulation of notch signaling pathway. Traffic 12: 149–161 [DOI] [PubMed] [Google Scholar]
- Li Y., Baker N. E., 2004. The roles of cis-inactivation by Notch ligands and of neuralized during eye and bristle patterning in Drosophila. BMC Dev. Biol. 4: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Sato C., Cerletti M., Wagers A., 2010. Notch signaling in the regulation of stem cell self-renewal and differentiation. Curr. Top. Dev. Biol. 92: 367–409 [DOI] [PubMed] [Google Scholar]
- Liu J., Xia H., Kim M., Xu L., Li Y., et al. , 2011. Beclin1 controls the levels of p53 by regulating the deubiquitination activity of USP10 and USP13. Cell 147: 223–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louvi A., Artavanis-Tsakonas S., 2012. Notch and disease: a growing field. Semin. Cell Dev. Biol. 23: 473–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova K. S., Aravind L., Koonin E. V., 2000. A novel superfamily of predicted cysteine proteases from eukaryotes, viruses and Chlamydia pneumoniae. Trends Biochem. Sci. 25: 50–52 [DOI] [PubMed] [Google Scholar]
- Masutani M., Sonenberg N., Yokoyama S., Imataka H., 2007. Reconstitution reveals the functional core of mammalian eIF3. EMBO J. 26: 3373–3383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto A., Onoyama I., Sunabori T., Kageyama R., Okano H., et al. , 2011. Fbxw7-dependent degradation of Notch is required for control of “stemness” and neuronal-glial differentiation in neural stem cells. J. Biol. Chem. 286: 13754–13764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuno K., Diederich R., Go M., Blaumueller C., Artavanis-Tsakonas S., 1995. Deltex acts as a positive regulator of Notch signaling through interactions with the Notch ankyrin repeats. Development 121: 2633–2644 [DOI] [PubMed] [Google Scholar]
- Matsuno K., Ito M., Hori K., Miyashita F., Suzuki S., et al. , 2002. Involvement of a proline-rich motif and RING-H2 finger of Deltex in the regulation of Notch signaling. Development 129: 1049–1059 [DOI] [PubMed] [Google Scholar]
- Moretti J., Chastagner P., Gastaldello S., Heuss S. F., Dirac A. M., et al. , 2010. The translation initiation factor 3f (eIF3f) exhibits a deubiquitinase activity regulating Notch activation. PLoS Biol. 8: e1000545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukai A., Yamamoto-Hino M., Awano W., Watanabe W., Komada M., et al. , 2010. Balanced ubquitinylation and deubiquitylation of Frizzled regulate cellular responsiveness to Wg/Wnt. EMBO J. 29: 2114–2125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay D., Riezman H., 2007. Proteasome-independent functions of ubiquitin in endocytosis and signaling. Science 315: 201–205 [DOI] [PubMed] [Google Scholar]
- Mummery-Widmer J. L., Yamazaki M., Stoerger T., Novatchkova M., Bhalerao S., et al. , 2009. Genome-wide analysis of Notch signaling in Drosophila by transgenic RNAi. Nature 458: 987–992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neely G. G., Kuba K., Cammarato A., Isobe K., Amann S., et al. , 2010a. A global in vivo Drosophila RNAi screen identifies NOT3 as a conserved regulator of heart function. Cell 141: 142–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neely G. G., Hess A., Costigan M., Keene A. C., Goulas S., et al. , 2010b. A genome-wide Drosophila screen for heat nociception identifies α2δ3 as an evolutionarily conserved pain gene. Cell 143: 628–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumüller R. A., Richter C., Fischer A., Novatchkova M., Neumüller K. G., et al. , 2011. Genome-wide analysis of self-renewal in Drosophila neural stem cells by transgenic RNAi. Cell Stem Cell 8: 580–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson S. C., Nicolay B. N., Frolov M. V., Moberg K. H., 2011. Notch-dependent expression of the archipelago ubiquitin ligase subunit in the Drosophila eye. Development 138: 251–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijman S. M., Luna-Vargas M. P., Velds A., Brummelkamp T. R., Dirac A. M., et al. , 2005. A genomic and functional inventory of deubiquitinating enzymes. Cell 123: 773–786 [DOI] [PubMed] [Google Scholar]
- Oberg C., Li J., Pauley A., Wolf E., Gurney M., et al. , 2001. The Notch intracellular domain is ubiquitinated and negatively regulated by the mammalian Sel-10 homolog. J. Biol. Chem. 276: 35847–35853 [DOI] [PubMed] [Google Scholar]
- Overstreet E., Fitch E., Fischer J. A., 2004. Fat facets and Liquid facets promote Delta endocytosis and Delta signaling in the signaling cells. Development 131: 5355–5366 [DOI] [PubMed] [Google Scholar]
- Pai M. T., Tzeng S. R., Kovacs J. J., Keaton M. A., Li S. S., et al. , 2007. Solution structure of the Ubp-M BUZ domain, a highly specific protein module that recognizes the C-terminal tail of free ubiquitin. J. Mol. Biol. 370: 290–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlopoulos E., Pitsouli C., Klueg K. M., Muskavitch M. A., Moschonas N. K., et al. , 2001. neuralized encodes a peripheral membrane protein involved in Delta signaling and endocytosis. Dev. Cell 1: 807–816 [DOI] [PubMed] [Google Scholar]
- Penton A. L., Lenoard L. D., Spinner N. B., 2012. Notch signaling in human development and disease. Semin. Cell Dev. Biol. 23: 450–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitsouli C., Delidakis C., 2005. The interplay between DSL proteins and ubiquitin ligases in Notch signaling. Development 132: 4041–4050 [DOI] [PubMed] [Google Scholar]
- Pospisilik J. A., Schramek D., Schnidar H., Cronin S. J., Nehme N. T., et al. , 2010. Drosophila genome-wide obesity screen reveals hedgehog as a determinant of brown vs. white adipose cell fate. Cell 140: 148–160 [DOI] [PubMed] [Google Scholar]
- Qiu L., Joazeiro C., Fang N. H. W., Elly C., Altman Y., et al. , 2000. Recognition and ubiquitination of Notch by Itch, a HECT-type E3 ubiquitin ligase. J. Biol. Chem. 275: 35734–35737 [DOI] [PubMed] [Google Scholar]
- Ranganathan P., Weaver K. L., Capobianco A. J., 2011. Notch signalling in solid tumours: a little bit of everything but not all the time. Nat. Rev. Cancer 11: 338–351 [DOI] [PubMed] [Google Scholar]
- Reyes-Turcu F. E., Horton J. R., Mullally J. E., Heroux A., Cheng X., et al. , 2006. The ubiquitin binding domain ZnF UBP recognizes the C-terminal diglycine motif of unanchored ubiquitin. Cell 124: 1197–1208 [DOI] [PubMed] [Google Scholar]
- Reyes-Turcu F. E., Ventii K. H., Wilkinson K. D., 2009. Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu. Rev. Biochem. 78: 363–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saj A., Arziman Z., Stempfle D., van Belle W., Sauder U., et al. , 2010. A combined ex vivo and in vivo RNAi screen for Notch regulators in Drosophila reveals an extensive Notch interaction network. Dev. Cell 18: 862–876 [DOI] [PubMed] [Google Scholar]
- Sakata T., Sakaguchi H., Tsuda L., Higashitani A., Aigaki T., et al. , 2004. Drosophila Nedd4 regulates endocytosis of Notch and suppresses its ligand-independent activation. Curr. Biol. 14: 2228–2236 [DOI] [PubMed] [Google Scholar]
- Samara N. L., Datta A. B., Berndsen C. E., Zhang X., Yao T., et al. , 2010. Structural insights into the assembly and function of the SAGA deubiquitinating module. Science 328: 1025–1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheuermann J. C., de Ayala Alonso A. G., Oktaba K., Ly-Hartig N., McGinty R. K., et al. , 2010. Histone H2A deubiquitinase activity of the Polycomb repressive complex PR-DUB. Nature 465: 243–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweisguth F., 2004. Regulation of notch signaling activity. Curr. Biol. 14: R129–R138 [PubMed] [Google Scholar]
- Singhal S., Taylor M. C., Baker R. T., 2008. Deubiquitylating enzymes and disease. BMC Biochem. 9: S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- South A. P., Cho R. J., Aster J. C., 2012. The double-edged sword of Notch signaling in cancer. Semin. Cell Dev. Biol. 23: 458–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowa M. E., Bennett E. J., Gygi S. P., Harper J. W., 2009. Defining the human deubiquitinating enzyme interaction landscape. Cell 138: 389–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staehling-Hampton K., Jackson P. D., Clark M. J., Brand A. H., Hoffmann F. M., 1994. Specificity of bone morphogenetic protein-related factors: cell fate and gene expression changes in Drosophila embryos induced by decapentaplegic but not 60A. Cell Growth Differ. 5: 585–593 [PubMed] [Google Scholar]
- Su Y., Ospina J. K., Zhang J., Michelson A. P., Schoen A. M., et al. , 2011. Sequential phosphorylation of Smoothened transduces graded Hedgehog signaling. Sci. Signal. 4: ra43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tetzlaff M. T., Yu W., Li M., Zhang P., Finegold M., et al. , 2004. Defective cardiovascular development and elevated cyclin E and Notch proteins in mice lacking the Fbw7 F-box protein. Proc. Natl. Acad. Sci. USA 101: 3338–3345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toba G., Ohsako T., Miyata N., Ohtsuka T., Seong K. H., et al. , 1999. The gene search system. A method for efficient detection and rapid molecular identification of gnes in Drosophila melanogaster. Genetics 151: 725–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tse W. K., Eisenhaber B., Ho S. H., Ng Q., Eisenhaber F., et al. , 2009. Genome-wide loss-of-function analysis of deubiquitylating enzymes for zebrafish development. BMC Genomics 10: 637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsunematsu R., Nakayama K., Oike Y., Nishiyama M., Ishida N., et al. , 2004. Mouse Fbw7/Sel-10/Cdc4 is required for Notch degradation during vascular development. J. Biol. Chem. 279: 9417–9423 [DOI] [PubMed] [Google Scholar]
- Valakh V., Naylor S. A., Berns D. S., DiAntonio A., 2012. A large-scale RNAi screen identifies functional classes of genes shaping synaptic development and maintenance. Dev. Biol. 366: 163–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Struhl G., 2005. Distinct roles for Mind bomb, Neuralized and Epsin in mediating DSL endocytosis and signaling in Drosophila. Development 132: 2883–2894 [DOI] [PubMed] [Google Scholar]
- Wang Y., Chen Z., Bergmann A., 2010. Regulation of EGFR and Notch signaling by distinct isoforms of D-cbl during Drosophila development. Dev. Biol. 342: 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weake V. M., Lee K. K., Guelman G., Lin C. H., Seidel C., et al. , 2008. SAGA-mediated H2B deubiquitination controls the development of neuronal connectivity in the Drosophila visual system. EMBO J. 27: 394–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinmaster G., Fischer J. A., 2011. Notch ligand ubiquitylation: what is it good for? Dev. Cell 21: 134–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkin M. B., Carbery A. M., Fostier M., Aslam H., Mazaleyrat S. L., et al. , 2004. Regulation of Notch endosomal sorting and signaling by Drosophila Nedd4 family proteins. Curr. Biol. 14: 2237–2244 [DOI] [PubMed] [Google Scholar]
- Wu G., Lyapina S., Das I., Li J., Gurney M., et al. , 2001. SEL-10 is an inhibitor of notch signaling that targets Notch for ubiquitin-mediated protein degradation. Mol. Cell. Biol. 21: 7403–7415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh E., Dermer M., Commisso C., Zhou L., McGlade C. J., et al. , 2001. Neuralized functions as an E3 ubiquitin ligase during Drosophila development. Curr. Biol. 11: 1675–1679 [DOI] [PubMed] [Google Scholar]
- Yuan J., Luo K., Zhang L., Cheville J. C., Lou Z., 2010. USP10 regulates p53 localization and stability by deubiquitinating p53. Cell 140: 384–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X. Y., Varthi M., Sykes S. M., Phillips C., Warzecha C., et al. , 2008. The putative cancer stem cell marker USP22 is a subunit of the human SAGA complex required for activated transcription and cell-cycle progression. Mol. Cell 29: 102–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Lang G., Ito S., Bonnet J., Metzger E., et al. , 2008. A TFTC/STAGA module mediates histone H2A and H2B deubiquitination, coactivates nuclear receptors, and counteracts heterochromatin silencing. Mol. Cell 29: 92–101 [DOI] [PubMed] [Google Scholar]
- Zhou M., Sandercock A. M., Fraser C. S., Ridlova G., Stephens E., et al. , 2008. Mass spectrometry reveals modularity and a complete subunit interaction map of the eukaryotic translation factor eIF3. Proc. Natl. Acad. Sci. USA 105: 18139–18144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu A. J., Zheng L., Suyama K., Scott M. P., 2003. Altered localization of Drosophila Smoothened protein activates Hedgehog signal transduction. Genes Dev. 17: 1240–1252 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.