Abstract
Physiological responses, developmental programs, and cellular functions rely on complex networks of interactions at different levels and scales. Systems biology brings together high-throughput biochemical, genetic, and molecular approaches to generate omics data that can be analyzed and used in mathematical and computational models toward uncovering these networks on a global scale. Various approaches, including transcriptomics, proteomics, interactomics, and metabolomics, have been employed to obtain these data on the cellular, tissue, organ, and whole-plant level. We summarize progress on gene regulatory, cofunction, protein interaction, and metabolic networks. We also illustrate the main approaches that have been used to obtain these networks, with specific examples from Arabidopsis thaliana, and describe the pros and cons of each approach.
INTRODUCTION
Physiological responses, developmental programs, and cellular functions rely on diverse interactions, such as those between DNA and proteins, between proteins, or in a sequence of enzymatic activities, that form complex networks to control biological processes (Vidal et al., 2011). Minor perturbations at one part of the system could cause ripples, or even waves, of effects across the entire network. Alternatively, regulation within a network could render the system robust to any minor perturbation. Understanding the nature of these complex phenomena requires in-depth and integrated knowledge of the molecular, biochemical, and physiological aspects of a biological process.
Characterizing mutant phenotypes has gradually increased our understanding of specific functions and modes of action of individual proteins, resulting in the description of small gene regulatory networks (GRNs) that underlie specific responses and developmental or biochemical steps. Examples of this include stem cell fate in the shoot apical meristem (Dodsworth, 2009), circadian clocks (Pokhilko et al., 2010, 2012), auxin signaling in the embryo (Lau et al., 2011), and light signal transduction (Rausenberger et al., 2011). While such a focused approach is informative, a holistic systems biology view allows us to gain comprehensive insight and to connect the dots of the molecular networks underlying plant growth and development. A full understanding of a process on a systems level also requires knowledge of the dynamic behavior of these components and their interactions (Albert, 2007).
Uncovering the complex set of transcriptional and protein interactions that regulate development and response to the environment requires the identification of networks on a genome and proteome scale. Interactions can be either physical (e.g., association between two proteins or protein and DNA) or functional (in a biochemical or signaling pathway), and these interactions often can be inferred from available data (Yellaboina et al., 2007). Systems-level network analysis approaches can be quantitative or qualitative. The former aims at precise network structure and kinetics data to simulate the dynamics of a biological process, while the latter relies on collective topological features (Albert, 2007). Here, we summarize the progress made in uncovering genome-wide gene cofunction, gene regulatory, protein interaction, and metabolic networks. Network science identifies individual entities (nodes) and their links (edges) (Vidal et al., 2011). We will illustrate some of the main experimental and computational methods with specific examples from Arabidopsis thaliana and describe the pros and cons of different approaches.
GENOME-WIDE COFUNCTIONAL NETWORK INFERENCE AND ANALYSIS
Advances in high-throughput technologies have led to the generation of extensive gene expression data sets on a genome-wide scale. The observation that genes sharing a common function can be transcriptionally coordinated (Hughes et al., 2000; Kim et al., 2001; Wu et al., 2002; Stuart et al., 2003) has enabled these data sets to be analyzed such that putative functional associations between genes can be inferred. Coexpression approaches use statistical metrics to establish correlations between gene expression profiles across many samples based on the principle of guilt-by-association (Figure 1A) (Usadel et al., 2009). Given the abundance of existing gene expression data, the generation of coexpression networks is a feasible top-down approach to generate genome-wide cofunctional network models in plants.
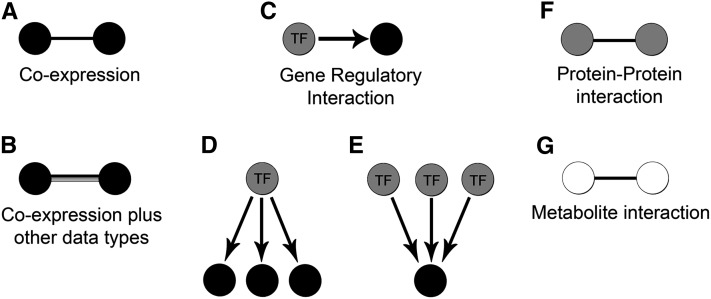
Figure 1.
Schematic Representation of Different Types of Networks.
(A) and (B) Cofunctional networks.
(A) A coexpression association where two genes are linked based on a common expression pattern.
(B) A cofunctional association where two genes are linked based on coexpression and/or other shared properties.
(C) A gene regulatory interaction where a TF directly binds the promoter to regulate the expression of a target gene.
(D) and (E) Two different approaches for uncovering GRNs using either a TF-centered approach (D) or a target-centered approach (E).
(F) Protein–protein interactions reflecting an experimentally determined physical interaction between two proteins.
(G) Metabolite interaction where two metabolites are linked through a common edge that represents a biochemical reaction converting one metabolite into the other.
Genes are colored in black, proteins are colored in gray, and metabolites are colored white.
Coexpression networks most commonly use the Pearson correlation coefficient to establish linear pairwise correlations between gene pairs in an adjacency matrix. Another associative metric that can be used is the Spearman correlation coefficient which enables nonlinear correlations between genes to be uncovered (Usadel et al., 2009). A modified graphical Gaussian model that takes into account partial correlations between genes after removing the effects of other adjacent genes has also been used (Ma et al., 2007). Following the establishment of gene associations, a cutoff threshold is then set, and pairwise interaction values exceeding this selected threshold are kept. The strength of the correlations between gene pairs can be considered as edge weights indicating the strength of coregulation between gene pairs. The end result of such an approach is a network consisting of nodes representing genes connected by edges representing a significant similarity in their common expression pattern. It is important to note that both positively and negatively acting components of a biological process can be coexpressed (Bassel et al., 2011b; Lee et al., 2011), so it is not possible to predict the function of putative coregulated gene pairs using this approach.
Functional modules consisting of subsets of highly interconnected nodes can be identified within networks. These cofunctional modules comprise strongly connected nodes, based on their collective interaction strength, with only weak interactions to other nodes in the network. In the case of coexpression networks, the modules represent groups of coexpressed genes or modular units of gene expression that may act together within a biological process with which they are associated. A variety of network clustering algorithms have been developed to identify modules in networks, including molecular complex detection (Bader and Hogue, 2003), Markov clustering algorithm (Enright et al., 2002), MCLUST (Fraley and Raftery, 2003), biclustering (Prelić et al., 2006), and the heuristic cluster chiseling algorithm (Mutwil et al., 2010). Some of these methods place genes into mutually exclusive clusters, which may not be realistic in a biological context where individual genes are capable of acting in multiple pathways, while other methods do not take into account interaction strength. A recently developed clustering algorithm named ClusterONE allows for overlap in module detection while considering edge weights (Nepusz et al., 2012). These algorithms produce clusters of varying gene members and sizes based on the subjective modification of their respective thresholds. Variability in module detection can also be compounded by the generation of different networks from the same data when using different methods (Aoki et al., 2007). Another source of variation is in the decision for placing the significance cutoff in the correlation coefficient affecting overall network size and topology. Too high a cutoff would not enable weakly coordinated processes to be captured, while too low a cutoff leads to large gene networks and a greater false positive rate. The use of known network topology to guide threshold cutoffs when generating cofunctional networks is also ambiguous, as conflicting reports have shown biological networks either to be scale free (follow a power law distribution; Barabasi and Albert, 1999) or not scale free (Khanin and Wit, 2006; Daudin et al., 2008).
Once a coexpression network has been generated, identifying modules by clustering can help extract biological meaning from the network. A module that contains genes that are annotated to overrepresented Gene Ontology (GO) categories (Ashburner et al., 2000) suggests this unit of genes may be involved in a given biological process. Uncharacterized genes within such a module can be candidates for participating in the same process. Similarly, genes directly connected to (or coexpressed with) known central regulators of a developmental process are candidates within this cofunctional framework. Identifying statistically overrepresented cis-elements in the promoters of the genes within a coexpression module may help infer regulatory interactions between transcription factors (TFs) and their targets. Given that members of a module in a coexpression network are highly transcriptionally coordinated across multiple samples, common upstream regulatory factors likely act through shared promoter elements. Shared cis-elements may be significantly overrepresented within a given module, providing clustered targets for TFs known to bind these sites. This information can be used to infer GRNs between TFs and their targets (see below).
Another means of extracting biologically relevant information from coexpression networks is to focus on genes with the greatest number of edges, so-called hub nodes, which show the greatest amount of transcriptional coordination with other genes. The centrality hypothesis states that if these connections are functionally relevant, removal of this central hub would severely perturb the network, possibly leading to an altered phenotype. The exploration of hub nodes often results in pleiotropic phenotypes due to the genes of interest being involved in multiple processes and hence can be enriched in false positives (Han et al., 2004; Yu et al., 2008a).
Coexpression networks created using various combinations of these approaches have led to the identification of previously uncharacterized genes in Arabidopsis involved in cell wall synthesis (Persson et al., 2005), flower development (Usadel et al., 2009), seed germination (Bassel et al., 2011b), and plant growth (Mutwil et al., 2010). The conservation of coexpression networks has been investigated across diverse plant species within a consensus network named PlaNet (Mutwil et al., 2011). This work demonstrated that the net transcriptional output of plant cells is at least partially conserved across model and crop species.
Integrating multiple data types can enhance the confidence by which cofunctional relationships are inferred (Figure 1B). A drawback of cofunction networks utilizing coexpression alone is that not all biological processes are transcriptionally coordinated. In addition, the guilt-by-association principle has been reported to have limited applicability in the inference of gene function (Gillis and Pavlidis, 2012). This conclusion was reached based on the observation that functional information is not encoded throughout coexpression networks, but rather within a subset of key interactions. These observations limit the general applicability of predicting cofunctional relationships based solely on coexpression analysis.
A genome-wide cofunction network of Arabidopsis called AraNet has been constructed using a number of omics data sets from Arabidopsis in addition to functional linkage data from homologous gene pairs in other model organisms (Lee et al., 2010). These data were vetted against experimentally characterized genes in Arabidopsis through standardized GO annotations available from The Arabidopsis Information Resource (Lamesch et al., 2010, 2012) using a Bayesian probabilistic model. Several previously uncharacterized genes have been assigned a function based on the functions of their neighboring genes in AraNet and subsequent experimental validation. These genes include genes associated with lateral root formation (Lee et al., 2010) and drought sensing (Lee et al., 2010).
Another cofunction network named RiceNet has been generated by combining gene coexpression data from this species with diverse proteomic data sets (Lee et al., 2011). A subset of predicted cofunctional interactions was found to interact physically at the protein level, and three novel regulators were reported to function in planta as components of the biotic stress response in this key crop species. These examples show that combining multiple data types can enhance predictive capacity of cofunctional relationships. Web-based tools to generate cofunctional networks of genes of interest using multiple data types are described later in this review.
Many studies predicting cofunctional relationships have focused on using the greatest quantity of gene expression data available, generally coming from diverse plant tissues and cell types. While more data increase the statistical power of the analysis, a lack of context specificity within data sets will only capture processes that are common in the samples used. Examples of this are the identification of photosynthesis and flavonoid synthesis pathways conserved across PlaNet (Mutwil et al., 2011). However, different tissues are defined by distinct regulatory networks, as are the discrete cell types that comprise them. A condition-dependent approach using data from a single tissue (Bassel et al., 2011b) or from single cell types (Birnbaum et al., 2003; Brady et al., 2007; Dinneny et al., 2008) can increase the context specificity by which gene cofunctional interactions can be inferred. An example of this is SeedNet, which was inferred using data generated exclusively from whole Arabidopsis seeds. A core module of seed dormancy–related coexpressed genes was identified within a dormancy subdomain of the network. Characterizing this module led to the identification of 10 novel regulators controlling seed dormancy (Bassel et al., 2011b). These predicted gene interactions were absent in other condition-independent cofunctional networks, while the overall network topology was found to be conserved in seeds of the monocot crop species wheat (Triticum aestivum) when equivalent data sets were compared.
When using condition-independent data sets, strategies have been developed to uncover biologically significant cofunctional interactions. One approach to enrich for causal interactions is to remove from the network all edges that are conditionally dependent (i.e., those that cannot be explained by other variables). Such a selection of conditionally independent edges has been used successfully for small metabolic and developmental networks by Gaussian Graphical Modeling (Wille et al., 2004). However, computational complexity prevents genome-wide applications at this time.
The spatial and temporal modulation of gene networks during cellular development may be limited by the availability of a sufficient number of data sets capturing cell type–specific processes over time. AraNet is capable of detecting genes preferentially coexpressed within specific root cell types with greater accuracy than randomly generated networks, despite using diverse whole tissue data sets as input. This predictive capacity may be increased through the generation of condition-dependent cofunctional networks.
A novel computational approach to construct a cofunctional network using gene expression data has recently been presented (Bassel et al., 2011a), wherein a rule-based machine learning algorithm called BioHEL was used to associate genes functionally. This algorithm generates rules that predict the developmental fate of a biological system through the automated exploration of subsets of annotated gene expression data. This cofunctional association metric, termed “coprediction,” relies on the hypothesis that groups of genes that collectively are able to predict a developmental output have a greater probability of being functionally related. This approach was used to generate a seed coprediction network (SCoPNet), which was functionally validated with the identification of four novel regulators of seed germination. A key feature of coprediction using gene expression data is that it does not rely on establishing cofunctional relationships using common gene expression patterns. Additional computational approaches beyond correlation-based methods may therefore be used to associate genes functionally using large-scale datasets.
LARGE-SCALE TRANSCRIPTIONAL REGULATORY NETWORKS
Mapping and systems analysis of large scale regulatory networks allow for the discovery of emergent properties (Long et al., 2008). A GRN describes the interaction of regulatory factors and their targets; here, we focus on TFs and their regulation of target gene expression (Figure 1C). There are two complementary ways to map GRNs: TF centered and target centered (Figures 1D and 1E).
Chromatin immunoprecipitation (ChIP) coupled with microarrays (ChIP-chip) or next generation sequencing (ChIP-seq) is a TF-centered approach to map GRNs. This technique involves immunoprecipitation of a TF and its bound target DNA with subsequent identification of downstream targets. The ChIP approach results in a network that describes the in vivo binding targets of a single TF (Table 1). Yeast one-hybrid (Y1H) assays are a target-centered approach that identifies upstream TFs binding to a promoter or motif of interest. This assay requires the initial development of a TF collection or cDNA library of prey TFs, which are assayed against bait promoters of interest in yeast (Deplancke et al., 2004).
Table 1. List of ChIP-Chip and ChIP-Seq Studies in Plants: The General Topic, Gene(s) of Focus, Tissue Isolated, and Experimental Validation Technique to Generate GRNs and Test Their in Vivo Relevance.
| Targets of Listed TFs | Technique | Tissue | Topic | Method for Experimental Validation | No. of Interactions Experimentally Confirmed | Reference |
|---|---|---|---|---|---|---|
| AT1G24260 (SEPALLATA3) | ChIP-Seq and ChIP-chip | Inflorescence | Floral development | Induction quantitative RT-PCR | 16 | Kaufmann et al. (2009) |
| Mutant analysis | 1 | |||||
| At1g69120 (APETALA1) | ChIP-Seq and ChIP-chip | Inflorescence | Floral development | Binding motif complimentation | 1 | Kaufmann et al. (2010) |
| Induction quantitative RT-PCR | 9 | |||||
| AT5G41315 (GLABRA3), | ChIP-chip | Seedlings | Trichome development | Semiquantitive RT-PCR | 12 | Morohashi and Grotewold (2009) |
| AT3G27950 (GLABRA1) | ||||||
| Inducible GL3 and GL1 lines | 14 | |||||
| Mutant analysis | 1 | |||||
| AT1G14350 (FOUR LIPS), | ChIP-chip | Seedlings | Stomatal development | ChIP-PCR | 1 | Xie et al. (2010) |
| AT2G02820 (MYB88) | ||||||
| EMSA | 1 | |||||
| ChIP-qPCR | 7 | |||||
| AT5G60690 (REVOLUTA) | ChIP-Seq | Seedlings | Leaf development | Induction quantitative RT-PCR | 9 | Brandt et al. (2012) |
| Mutant and overexpression analysis | 6 | |||||
| Binding motif analysis | n/a | |||||
| ChIP-qPCR | 5 | |||||
| AT5G13790 (AGAMOUS-LIKE15) | ChIP-chip | Embryonic culture | Embryogenesis | Quantitative RT-PCR | 3 | Zheng et al. (2009) |
| Binding motif analysis | n/a | |||||
| AT2G20180 (PHYTOCHROME INTERACTING FACTOR3-LIKE5) | ChIP-chip | Seeds | Seed germination | Quantitative RT-PCR | 21 | Oh et al. (2009) |
| Binding motif analysis | n/a | |||||
| ChIP-qPCR | 3 | |||||
| AT1G75080 (BRASSINAZOLE-RESISTANT1) | ChIP-chip | Seedlings | Brassinosteroid signaling | Semiquantitative RT-PCR | 1 | Sun et al. (2010) |
| EMSA | 2 | |||||
| Binding motif analysis | n/a | |||||
| Quantitative RT-PCR | 22 | |||||
| AT1G19350 (BRI1 EMS SUPRESSOR1) | ChIP-chip | Seedlings | Brassinosteroid signaling | EMSA | 1 | Yu et al. (2011) |
| Mutant analysis | 4 | |||||
| AT3G47640 (POPEYE) | ChIP-chip | Roots | Iron deficiency response | Quantitative RT-PCR | 3 | Long et al. (2010) |
| AT2G32780 (UPBEAT1) | ChIP-chip | Roots | Reactive oxygen species | Mutant analysis | 1 | Tsukagoshi et al. (2010) |
| AT4G37650 (SHORTROOT), | ChIP-chip | Root ground tissue | Root development | Quantitative RT-PCR | 14 | Sozzani et al. (2010) |
| AT3G54220 (SCARECROW) | ||||||
| ChIP-qPCR | 1 | |||||
| Semiquantitative RT-PCR | 11 | |||||
| AT5G11260 (ELONGATED HYPOCOTYL5) | ChIP-chip | Seedlings | Light regulation | ChIP-qPCR | 4 | Lee et al. (2007) |
| Binding motif analysis | n/a | |||||
| ChIP-qPCR | 8 | Huang et al. (2012) | ||||
| AT5G61380 (TIMING OF CAB EXPRESSION1) | ChIP-Seq | Seedlings | Circadian clock | Induction quantitative RT-PCR | 2 | |
| Mutant and overexpression analysis | 4 | |||||
| Binding motif analysis | n/a | |||||
| Quantitative RT-PCR | 1 | |||||
| Mutant analysis | 1 | |||||
| AT3G22170 (FAR-RED ELONGATED HYPOCOTYL3) | ChIP-Seq and ChIP-chip | Seedlings | Phytochrome signaling | Mutated promoter (Y1H) | 1 | Ouyang et al. (2011) |
| Inducible FHY3 line | 1 | |||||
| Binding motif analysis | n/a | |||||
| AT5G06950 (TGA2) | ChIP-chip | Leaves | Salicylic acid response | ChIP-qPCR | 3 | Thibaud-Nissen et al. (2006) |
| Binding motif analysis | n/a | |||||
| AT3G54610 (GENERAL CONTROL NON-REPRESSIBLE5) | ChIP-chip | Seedlings | Histone acetyltransferase | Semiquantitative RT-PCR | 7 | Benhamed et al. (2008) |
| GRMZM2G017087 (Zm KNOTTED1) | ChIP-Seq | Ears, tassels, and leaves | Meristem development | ChIP-qPCR | 34 | Bolduc et al. (2012) |
| EMSA | 4 | |||||
| Binding motif analysis | n/a | |||||
| GRMZM2G084799 (Zm PERICARP COLOR1) | ChIP-Seq | Pericarps and silks | ChIP-qPCR | 19 | Morohashi et al. (2012) | |
| Transient expression assay | 4 | |||||
| EMSA | 2 | |||||
| Binding motif analysis | n/a |
EMSA, Electrophoretic Mobility Shift Assay; n/a, not applicable.
ChIP studies have focused on TFs that are well characterized and typically abundant. In plants, ChIP studies have been performed primarily in Arabidopsis, but occasionally also in rice (Oryza sativa) and maize (Zea mays) (Fornalé et al., 2010; Moreno-Risueno et al., 2010) (Table 1). A protein expressed in few cell types or developmental stages can be difficult to precipitate due to lack of abundance. Traditionally this has been overcome using overexpression and conditional induction of the gene of interest by fusion to the glucocorticoid receptor or to a hormone inducible promoter, such as the estrogen-inducible promoter. Although this provides the researcher with potential binding targets, it obscures the developmental or tissue/cell type–specific context of the transcriptional regulation identified. Additionally, high-quality antibodies against the TFs are needed to achieve specific and sufficient immunoprecipitation. Using epitope-tagged TFs or fusion with a protein to which high-quality antibodies have been developed (i.e., green fluorescent protein) has been a way to circumvent the development of protein-specific antibodies. However, the tagged protein must be verified to ensure that its expression reflects the wild type and that it can functionally complement a loss-of-function phenotype.
ChIP studies provide evidence for in vivo DNA binding but do not show the regulatory effect of the TF on its targets. Similarly, Y1H does not show the regulatory effect of the binding. Additionally, Y1H is limited by the heterologous nature of the assay and does not provide direct in planta evidence for TF-DNA binding. For example, a plant TF may not be able to form obligate heterodimers or higher order protein complexes to bind to promoter sequences. This will produce false negatives that cannot be detected in the yeast system. Therefore, this approach requires biological validation to be confident of the in vivo physical interactions through independent validation techniques, including directed ChIP experiments. However, Y1H can be adapted to high-throughput techniques to screen the promoters of multiple genes of interest simultaneously, leading to a typically larger and more comprehensive network (Brady et al., 2011; Gaudinier et al., 2011). The assay can be tailored to screen specific biologically relevant promoter sequences of interest, focusing the experimental approach and enriching for the identification of relevant targets when mapping GRNs.
Although previously only 1 to 10% of interactions identified by ChIP have been determined to be regulatory in nature (Farnham, 2009; Moreno-Risueno et al., 2010), more recently biological validation rates have been increasing (Kaufmann et al., 2011). Using Y1H assays to determine TF-DNA interactions, two of three interactions tested were reported to be regulatory in planta by ChIP and genetic analysis. Thirty-two of 66 additional interactions were confirmed through genetic approaches with the supposition that the same proportion above (two out of three) would bind in planta (Brady et al., 2011). Comprehensive biological validation of GRNs predicted by either of the above methods is a challenge. The ChIP approach provides in planta binding data and the Y1H assays show binding, at least in yeast, but neither method is sufficient to probe the regulatory nature of the interactions. Perturbing the network using loss-of-function and/or conditional induction lines of the TF (such as using glucocorticoid receptor fusions or estrogen-inducible promoters) and analyzing expression of the TF’s targets relative to the control will show if the TF acts as an activator, repressor, or has no effect. In vivo confirmation can also be performed using a transient expression system in protoplasts in which a TF is induced to see its effect on target gene expression (Pruneda-Paz et al., 2009; Lau et al., 2011), although the developmental and tissue-specific context of the interactions can be missed using this approach.
Both ChIP and Y1H led to genome-wide understanding of transcriptional regulatory networks. For example, ChIP-chip and ChIP-seq technologies have been used to identify binding targets of the general floral regulator SEPALLATA3 (SEP3) (Kaufmann et al., 2009) and of the A-function floral identity regulator APETALA1 (AP1). As many as 2298 genes were found as AP1 targets, with 10.8% of these genes as high-confidence targets that show binding and a moderate change in gene regulation by AP1 as determined by induction of AP1 and ChIP-seq analysis. AP1 bound to the SEP3 promoter and activated SEP3 expression. A comparison of AP1 and SEP3 targets (Kaufmann et al., 2010) showed overlap, suggesting that these proteins may function as a heterodimer to activate genes involved in early flower organ development. Using a combination of ChIP and network predictions, an opposing relationship between two proteins involved in the response to iron deprivation, POPEYE (PYE), a basic helix-loop-helix TF, and BRUTUS (BTS), a putative E3 ligase, has been described (Long et al., 2010). Using ChIP-chip of PYE, 70 targets were identified. Using the ATTED-II coexpression network tool with PYE as bait, BTS, which is involved in iron homeostasis, was identified. The pye-1 mutant plants are small and chlorotic when grown on iron-deficient media, while BTS plays an antagonistic role in negatively regulating the iron deprivation response as bts-1 plants are green and have an increased tolerance to iron deficiency. Y1H assays have been used at various scales to identify upstream transcriptional regulators. A directed Y1H screen, with a library of 186 Arabidopsis TFs that are circadian regulated, against the CCA1 promoter was used to identify CCA1 HIKING EXPEDITION (CHE), a TCP TF. Using tiled promoter fragments, the binding region was narrowed down to 171 bp (Pruneda-Paz et al., 2009). A transient expression assay in protoplasts determined that CHE acts as a repressor of CCA1, and in planta binding was confirmed by ChIP–quantitative PCR (qPCR). Yeast two-hybrid (Y2H) and coimmunopurification experiments additionally demonstrated a protein–protein interaction between CHE and TOC1 as well as between TOC1 and CCA1, indicating further putative combinatorial action between these TFs (Pruneda-Paz et al., 2009).
Finally, combining approaches will lead to more comprehensive networks. For example, the combination of Y1H and Y2H assays between 167 TFs and 93 TF and microRNA promoters as well as computationally and experimentally derived microRNA–mRNA interactions resulted in a more holistic Arabidopsis root stele–enriched GRN (Brady et al., 2011). A network of 103 interactions was mapped and interactions were tested in planta with a variety of biological validation techniques, including ChIP, mutant and conditional induction analyses to confirm 59% of the interactions. Only 16% of the GRN TF mutants screened showed a root phenotype but 65% showed a molecular phenotype. This demonstrates the robustness of the stele-enriched network as TFs regulating the same gene can compensate for a mutation in one of the regulators. Increasing this TF collection from 24% to 92% of stele-expressed TFs (Gaudinier et al., 2011) provides the ability to generate a more comprehensive network of transcriptional regulation across the root stele.
LARGE-SCALE PROTEIN–PROTEIN INTERACTIONS: THE INTERACTOME
To understand fully genotype-to-phenotype relationships at the systems level, comprehensive knowledge about the complex and dynamic protein–protein interactions that exist within an organism is required. Identifying binary protein–protein interactions and components of protein complexes is a crucial step toward elucidating associated biological activities (Figure 1F). Several assays have been developed to identify and predict protein interactions in yeast, in cell suspension, in planta, and in silico, and various public databases have compiled these interactions (Table 2).
Table 2. Freely Accessible Databases and Tools for Network Analyses.
| Application | Tools |
|---|---|
| Databases and warehouses | |
| Gene expression | BAR, Genevestigator, ATTED, AT-TAX, TileViz |
| Protein expression | Pep2pro |
| Protein–protein interactions | ANAP, BAR, PAIR, IntAct, AtPIN, AtPID, AthPPI, Associomics, PRIN (rice), BioGRID |
| Protein localization | SUBA, PREDOTAR, TargetP |
| Gene set analysis | BAR, AMIGO, DAVID, FuncAssociate, BiNGO, ATCOECIS, MapMan, AraNet, AgriGO |
| Comparative genomics | Plaza, OrthologID, DoOP |
| Gene regulation and promoter architecture | |
| Motif detection | BAR, ATCOECIS, TAIR patmatch, DoOP, STAMP, Weeder |
| Binding site analysis | PLACE, AthaMap, AGRIS |
| Data integration and network inference | LeMoNe, ENIGMA, CORNET, Ondex, AraNet, GeneMania |
| Generic computation and modeling environments | R, MatLab, Systems Biology Workbench |
| Data visualization | AraCyc, PlantCyc, Cytoscape, MapMan, REACTOME |
As there is still a long way to full coverage of in planta protein–protein interaction networks, computational approaches, including coexpression, have been used to infer protein–interactions. Protein interactions can be inferred by homology to known interactions in other organisms (interaction ortholog or interlog) (Geisler-Lee et al., 2007), using indirect evidence or literature (Cui et al., 2008) or integrated methods (Xu et al., 2010). Interlogs can be filtered using functional association data to improve prediction reliability (De Bodt et al., 2009). Several databases of inferred protein interactions have been established (Geisler-Lee et al., 2007; Cui et al., 2008; Brandão et al., 2009; De Bodt et al., 2009; Lin et al., 2009, 2011a, 2011b) (Table 2). The predicted Arabidopsis interactome resource provides an interactome inferred from multiple indirect lines of evidence, including coexpression, colocalization, coevolution, annotation similarity, domain interaction, and homologous interactions in other species. The BAR Arabidopsis Interactions Viewer also provides similar functionality (Geisler-Lee et al., 2007). These computational approaches can guide the identification of potential in vivo and in planta protein–protein interactions. In this respect, coexpression is relevant because proteins generally should be expressed in the same cell at the same time for in vivo interaction. Care must be used when inferring protein expression from transcriptome data as transcript and protein abundance do not always correlate (Boruc et al., 2010; Van Leene et al., 2010; Arabidopsis Interactome Mapping Consortium, 2011; Petricka et al., 2012).
While inferring interactions using computational approaches can be useful, validation in a biological system is required to draw more meaningful conclusions. This is limited by only a few available in planta high-throughput assays. To gain insight into protein–protein interactions and to obtain interactome networks, one can identify binary interactions using Y2H, bimolecular fluorescence complementation (BiFC), split ubiquitin or split luciferase, or alternatively isolate components of a protein complex in planta using coimmunopurification or tandem affinity purification (TAP) (Van Leene et al., 2011). Uses of these different approaches are described below.
On a small, protein family scale, a Y2H approach was used to identify binary protein–protein interactions, such as AUXIN/INDOLE-3-ACETIC ACID (AUX/IAA)–AUXIN RESPONSE FACTOR (ARF) interactions (De Rybel et al., 2010; Vernoux et al., 2011), to map the dimerization network of MADS domain TFs (de Folter et al., 2005) and, in combination with BiFC assays, to reveal functional modules in core cell cycle binary protein–protein interactions (Boruc et al., 2010). To obtain a more global interactome network, a high-throughput Y2H approach has been used to map binary interactions between soluble proteins in plants (Arabidopsis Interactome Mapping Consortium, 2011). This represents the first large-scale experimentally validated protein interactome in plants and has also been used to determine a plant-pathogen immune network (Mukhtar et al., 2011).
However, Y2H is mainly suitable for soluble proteins that can traffic to the nucleus, ignoring membrane proteins, which often play essential roles in fundamental biological processes, including signaling, homeostasis, nutrient acquisition, and metabolism. Despite their importance, we know little about the functions of most membrane proteins. To systematically elucidate a map of membrane protein interactions, a mating-based split ubiquitin system (mbSUS) was developed (Obrdlik et al., 2004; Miller et al., 2005). The split ubiquitin system is similar to the classical Y2H as it uses yeast as a heterologous system and has a similar readout, but it specifically allows the detection of interactions of full-length membrane proteins. The concept of mbSUS relies on the release of a TF from a membrane protein if it interacts with another membrane (or soluble) protein. Similar to other detection systems based on the reconstitution of two halves of a protein, mbSUS uses an ubiquitin molecule split into two halves. The use of a mutated N-terminal domain of ubiquitin (Nub) and C-terminal half (Cub) fused to a TF to test for physical interaction between two fused proteins is the basis of mbSUS, whereby a functional ubiquitin is reconstituted only when brought into vicinity via interaction between two fusion partners (Johnsson and Varshavsky, 1994). Cleaving the reunited ubiquitin molecule by an endogenous ubiquitin-specific protease releases the TF to move into the nucleus and activate reporter genes. The mbSUS was used to test for interactions of the translocon complex at the outer chloroplast membrane (Rahim et al., 2009) and to screen for potential interactions among 490 Arabidopsis membrane and signaling proteins (Lalonde et al., 2010). However, the currently available membrane interaction networks for Arabidopsis cover only a small portion of the membrane-bound proteome. A recent study used a computational classification system to predict putative membrane protein interactions and decrease the sample space of interactions to be proved (Chen et al., 2012). These predictions were validated through the verification of 541 interactions between 239 membrane proteins, enriched in transporters.
The above-described yeast-based systems are informative, though high-throughput proteome-wide in planta interaction data are more biologically relevant. An alternative approach is TAP, which allows investigation of protein interactions through in situ purification of protein complexes. In a purification experiment, complexes that are associated with a protein of interest at the time of extraction can be isolated. The TAP approach is based on the expression of a bait protein fused to a double affinity tag and a two-step purification process followed by mass spectrometry of protein complex members (Van Leene et al., 2008). A targeted mapping of the cell cycle interactome, using TAP, provided a first draft of the basic cell cycle complex machinery in Arabidopsis (Van Leene et al., 2010). The TAP approach has also been employed to isolate the core jasmonate signaling module and to find new interactors of JASMONATE ZIM-DOMAIN repressor proteins (Pauwels et al., 2010; Fernández-Calvo et al., 2011) to determine whether PROHIBITINS are present within a multimeric complex (Van Aken et al., 2007), to purify 14-3-3 complexes (Chang et al., 2009), and to identify components of complexes, including the COP9 signalosome 3 complex (Rubio et al., 2005), novel WPP2-interacting proteins (Zhao et al., 2008), the BRASSINAZOLE-RESISTANT1–dephosphorylating phosphatase (Tang et al., 2011), and E2F TARGET GENE1–associated proteins (Takahashi et al., 2008). However, current studies using TAP rely on the use of cell suspension cultures. Implementing this technology in planta within the context of multicellular tissues will provide further biological insights. Furthermore, improving the methodology to identify membrane-associated complexes awaits development.
For the cell cycle interactome study, interaction between 17 protein pairs was validated using split luciferase assays. Here, the firefly luciferase (LUC) protein is split into two halves and fused to two different proteins. LUC activity is only reconstituted when the N- and C-terminal LUC moieties are brought together by the two interacting proteins. The validation rate was 41%, which is a great improvement over a similar type of split protein reconstitution performed following TAP tagging in yeast (Yu et al., 2008a; Van Leene et al., 2010). The actual false positive rate is expected to be much smaller because in a TAP tagging experiment proteins may interact with each other indirectly and therefore may not be confirmed when validated by other assays. Furthermore, these interactions likely occur in specific cell types that may require additional temporal cues not present in transient systems. The LUC-based approach was also used to detect SNARE–SNARE interactions in Arabidopsis protoplasts (Kato et al., 2010). BiFC, which uses a similar strategy of bringing together two parts of a protein, in this case yellow fluorescent protein, was also recently used to validate 78 G-protein interactions in planta that were previously identified by Y2H analysis. Here, the validation rate was a striking 95%, which is the highest validation rate reported for any organism (Klopffleisch et al., 2011). However, this bait set of proteins was biased in their original selection by their common participation in a biological process, which may partially explain the high rate of validation relative to other studies. In addition, the high sensitivity of the stable reconstituted fluorescent protein can lead to unspecific reconstitution, especially upon overexpression of the fusion proteins of interest (Lalonde et al., 2008).
Another approach to probe protein interactions over a large scale is to use protein microarrays (Popescu et al., 2007, 2009). This involves the generation of recombinant proteins in planta through transient expression and spotting the purified proteins onto microchips. These chips may then be hybridized with fluorescently tagged proteins of interest or subjected to enzymatic modification with a labeled substrate. Imaging of the arrays following incubation can identify protein–protein interactions and substrates of posttranslational modification. Using this approach, large-scale calmodulin interaction (Popescu et al., 2007) and mitogen-activated protein kinase phosphorylation networks have been indentified (Popescu et al., 2009).
In addition to these approaches, direct in planta immunoprecipitation of tagged proteins of interest, followed by mass spectrometry of coimmunoprecipitated proteins has been successful in identifying protein complex members for several individual proteins in Arabidopsis, such as SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASE1 (Karlova et al., 2006), CELLULOSE SYNTHASE CATALYTIC SUBUNITs (Desprez et al., 2007), MADS domain TFs (Smaczniak et al., 2012), and the ESCRT-III component VPS2.2 (Ibl et al., 2012). While this is a powerful approach, uncovering rare, dynamic complexes in specific plant cells and tissues remains challenging.
A combination of the above approaches led to the description of a number of interactomes in Arabidopsis, not only for the cell cycle (Boruc et al., 2010; Van Leene et al., 2010, 2011), but also for the chloroplast (Yu et al., 2008b) and for ESCRT, TOPLESS (TPL), TPL-related (TPR), and G-proteins (Klopffleisch et al., 2011; Shahriari et al., 2011; Causier et al., 2012; Ibl et al., 2012). For example, the TPL and TPR interactomes revealed that TPL proteins have been co-opted multiple times in evolution to cause transcriptional repression. In any systems-level analysis of protein–protein interactions, in planta validation of the entire spectrum of interactions that were observed or predicted is unlikely, but the tools above can be used to confirm a subset of these interactions. In addition, the cell cycle interactome showed that various techniques may be complementary as the overlap was rather small between Y2H and BiFC (Boruc et al., 2010).
Genetic analysis is an additional method of in planta validation. In an experiment mapping transcriptional regulatory interactions in the Arabidopsis stele, physically interacting TFs were identified using Y2H. TFs that interact with each other previously have been shown to influence each other’s expression, either directly or indirectly (Cui et al., 2007). For each TF of an interacting pair, expression of the interactor was measured in each respective TF loss-of-function background. The majority of interacting TFs was found to have a regulatory influence on each other’s expression (Brady et al., 2011). In the Y2H interactome study to determine a plant-pathogen immune network, interaction between Pseudomonas syringae and Hyaloperonspora arabidopsidis effectors with Arabidopsis proteins was determined (Mukhtar et al., 2011). A prediction was that Arabidopsis proteins that were targeted by effectors from both pathogens could display pathogen resistance or susceptibility. Fifteen of the 17 proteins targeted by effectors from both pathogens showed mutant phenotypes consistent with immune system functions (Mukhtar et al., 2011).
METABOLIC NETWORKS
Genome-scale metabolic network models have been built and applied to study a wide variety of topics, including adaptive evolution, network properties, and metabolic engineering (Figure 1G) (Feist and Palsson, 2008). Genome-scale metabolic models in metabolic engineering have been used to couple growth rate with overproduction of lactate (Fong et al., 2005) and l-Val (Park et al., 2011) in Escherichia coli and prioritization of alternative engineering strategies for overproducing ethanol in yeast (Bro et al., 2006). Although most of the work in metabolic network reconstruction and application has been performed in microbes, several advances in plant metabolic network reconstruction and modeling have been made recently, paving the way for rational plant metabolic engineering.
Two types of plant metabolic networks have been developed: descriptive and predictive. Genome-wide descriptive metabolic networks have been predicted from metabolic pathway databases that have been curated from experimental data in the literature (Caspi et al., 2012; Kanehisa et al., 2012). These genome-wide metabolic network representations are now available for several plant species, such as Arabidopsis (Mueller et al., 2003; Zhang et al., 2010), Populus trichocarpa (Zhang et al., 2010), Chlamydomonas reinhardtii (May et al., 2009), Medicago truncatula (Urbanczyk-Wochniak and Sumner, 2007), grasses (Youens-Clark et al., 2011), and shade plants (Bombarely et al., 2011). Recently, the plant metabolic network group published genome-wide metabolic networks for maize, soybean (Glycine max), cassava (Manihot esculenta), and wine grape (Vitis vinifera) online (www.plantcyc.org; Zhang et al., 2010). Most of these networks (except for Arabidopsis) are not associated with extensive validation and curation with experimental data from the literature, and caution should be taken when interpreting the data. These networks have been used for a variety of studies, including identification of novel genes in glucosinolate metabolism (Chan et al., 2011), the role of duplicated enzymes (Hanada et al., 2011), and metabolomic responses to UV-B (Kusano et al., 2011). While the descriptive, qualitative networks have been useful in many studies, they are not predictive and are therefore limited in facilitating metabolic engineering.
Predictive metabolic modeling approaches can be broadly grouped into kinetic and stoichiometric modeling (Sweetlove et al., 2008). Kinetic modeling uses enzyme kinetics numerically to simulate and test metabolic fluxes and can explain mechanism of flux changes. However, the difficulty of determining in vivo enzyme kinetics has limited this modeling to a small number of pathways. The most widely adapted modeling approach used for genome-wide metabolism is constraints-based modeling that includes stoichiometric, thermodynamic, and flux capacity constraints to model the fluxes of metabolites (Thiele and Palsson, 2010). This approach has been used to build predictive models of metabolism for Arabidopsis (Poolman et al., 2009; de Oliveira Dal’Molin et al., 2010), maize (Saha et al., 2011), and C. reinhardtii (Chang et al., 2011). Most of these models have not been validated extensively using flux measurements, though advances in metabolic flux analysis using 13C-labeling and metabolomics approaches hold promise (Schwender, 2008; Sweetlove et al., 2008; Allen et al., 2009).These predictive models have been applied in studies in many fields, such as metabolic engineering, drug discovery, drug target discovery, identification of novel gene function, evolutionary processes, network behaviors, and interpretations of mutant phenotypes (Feist and Palsson, 2008). The most common algorithm used in such studies has been flux balance analysis (FBA), which attempts to balance the stoichiometry of the metabolites within the metabolic network system with a goal (objective function) of maximal growth or maximal biomass accumulation. While prediction of fluxes using FBA matches well with experimental data (Burgard and Maranas, 2003), its assumptions may not always hold true, especially for engineered mutant lines. Several algorithms that have the goal of minimizing the change in the metabolic network upon perturbation have been developed and appear to perform better than FBA in explaining fluxes of mutants (Segrè et al., 2002; Shlomi et al., 2005; Herrgård et al., 2006).
The field of metabolic network analysis in plants is still young, and there are several challenges ahead. First, the process of generating high-quality networks is still time consuming and requires much manual input. More advances in automating these steps, such as the Model SEED system, are needed (Henry et al., 2010). Second, compartmentalization at the subcellular and tissue level must be considered for more accurate modeling of plant metabolic networks. Recent advances, such as the large-scale C4 photosynthesis model that incorporates mesophyll and bundle sheath–specific reactions (Dal’Molin et al., 2010) and Arabidopsis tissue-specific network models (Mintz-Oron et al., 2012), are early examples of the next-generation metabolic network models. Third, metabolic network models need to be integrated with regulatory and signaling models to allow more accurate and comprehensive understanding of plant metabolism. For example, large-scale experimental validations showed that the metabolic network in E. coli can explain fluxes in ∼50% of the conditions (Covert et al., 2004). Adding transcriptional regulatory constraints increased predictability to ∼70%. Transcriptional regulatory constraints have been incorporated into FBA analysis using the steady state regulatory FBA approach (Shlomi et al., 2007). More recently, an attempt has been made to develop a system to integrate signaling, metabolic, and regulatory networks into a common framework (Lee et al., 2008). Finally, metabolomics (comprehensive measurement of small molecules) in plants promises to be a useful source for validating the structure of, identifying missing links in, and understanding the dynamics and evolution of, metabolic networks (Fiehn et al., 2000; Kliebenstein, 2009; Nakabayashi et al., 2010; Tohge et al., 2011). However, several issues remain before these data can be integrated into metabolic network analysis, the biggest of which may be the high portion of unnamed metabolites in these studies. In addition, the mechanistic relationships among the levels of metabolites, transcripts, proteins, and enzyme activities are as yet unclear. An elegant study examining diurnal cycles of Arabidopsis suggests that metabolites regulate transcripts and not vice versa (Gibon et al., 2006). Despite these challenges, the advances in the omics and visualization technologies will help move this field forward in both expected and unexpected ways.
TOOLS AND TOYS FOR NETWORK ANALYSES
Any systems biology study depends on suitable software and data access, and Table 2 provides a selection of some useful tools. Much can be learned from available information, and online databases and data warehouses provide access to this body of prior knowledge. Repository databases focus on data storage and retrieval, while data warehouses contain extensively curated data and often include analysis tools. Many of the popular data warehouses in plant biology provide access to transcriptome data. Tools such as Genevestigator, the Bio-Array Resource for Plant Biology, and ATTED II are useful to search for genes or proteins with particular expression profiles or coexpression with bait genes (Toufighi et al., 2005; Hruz et al., 2008; Obayashi et al., 2011). Functional modules in biological networks are often composed of coexpressed and colocalized proteins that are highly connected in protein interaction graphs. A large set of databases is dedicated to protein–protein interaction data, some of which include information on coexpression. Several initiatives, such as IntAct (Kerrien et al., 2006), the Molecular Interaction database (Chatr-aryamontri et al., 2007), the Database of Interacting Proteins (Salwinski et al., 2004), the Biomolecular Interaction Network Database (Bader et al., 2001, 2003; Alfarano et al., 2005), and BioGRID (Stark et al., 2006), have been established to systematically collect and organize the interaction data reported by proteome-scale high throughput experiments and low-throughput studies focusing on individual proteins and pathways. In this context, the Arabidopsis network analysis pipeline is a useful tool because it integrates 11 Arabidopsis protein interaction databases (Wang et al., 2012). One of the most comprehensive resources for protein localization is the subcellular location database for Arabidopsis proteins, which collects both experimental and predicted subcellular protein localizations in Arabidopsis (Heazlewood et al., 2007).
Gene set analysis helps characterizing user-defined gene lists. Gene set analysis searches for distinguishing properties of gene lists, for instance, the overrepresentation of certain GO categories or functional bins for metabolic and regulatory processes, such as those defined in the MapMan software (Usadel et al., 2005). Gene set analysis should be supported by statistical evidence that frequencies of called categories are different from what is expected by chance. The hypergeometric test is an appropriate statistical framework to evaluate overrepresentation in small gene lists. Note that testing of multiple categories requires control of type I (false positive) error rates (e.g., the Bonferroni correction) (Dudoit and VanderLaan, 2008). The reference population used to construct the test’s null hypothesis must be chosen carefully to include only elements from the sampled space and not from the entire genome. Gene set analysis together with cofunction networks can be used to infer gene function. Gene associations solely based on coexpression are not always informative, but predictive power increases when associations include other type of data, such as cocitation or protein interactions. AraNet, GeneMania, and STRING are three such tools that are user friendly (Warde-Farley et al., 2010; Hwang et al., 2011; Szklarczyk et al., 2011).
Traditionally, systems biology often focuses on single-model species. Nevertheless, much can be learned by exploiting comparative genomics tools, which can help assign functions to multigene family members by defining ortholog sets, identify relevant gene regulatory motifs, or study how evolution or breeding shaped biological systems. The Plaza database, for instance, makes genomic data from 25 plant species available (Van Bel et al., 2012). Deciphering transcriptional regulation involves identification of both cis-regulatory elements, for instance, by motif samplers, and of regulatory trans-acting TFs. Several algorithms and tools are available for such analyses (Ladunga, 2010), but the currently fragmented knowledge about plant TF binding sites makes reverse engineering of promoter functions a nontrivial task.
Data integration and network inference are two major needs in systems biology. LeMoNe and ENIGMA, for instance, are software packages to extract module networks from gene expression data (Michoel et al., 2007; Maere et al., 2008). Coexpression-based correlation networks can also be constructed with TMEV (Saeed et al., 2006). Similarly, CORNET constructs coexpression networks but also provides workflows to integrate such networks with protein–protein interaction and localization data (De Bodt et al., 2010) (Figure 2). Network inference is an active field in machine learning and statistics communities, and powerful new algorithms are continuously being developed. To unleash the full potential of available algorithms, software environments with more open access are needed. Here, the general computation environments R (www.R-project.org/) and MatLab (www.mathworks.co.uk/products/matlab/index.html), with their large package libraries for specialized tasks, and the Systems Biology Workbench, with its rich model library (Sauro et al., 2003), are powerful alternatives. These environments lack much of the convenient graphical user interfaces characteristic of most of the previously mentioned tools, but their inherent scripting design standardizes reproducing and transferring analysis workflows within or between laboratories. In particular, the Systems Biology Workbench offers great modeling options for biological systems with minimal scripting needs and is well suited to analyze biochemical reaction models (Sauro et al., 2003).
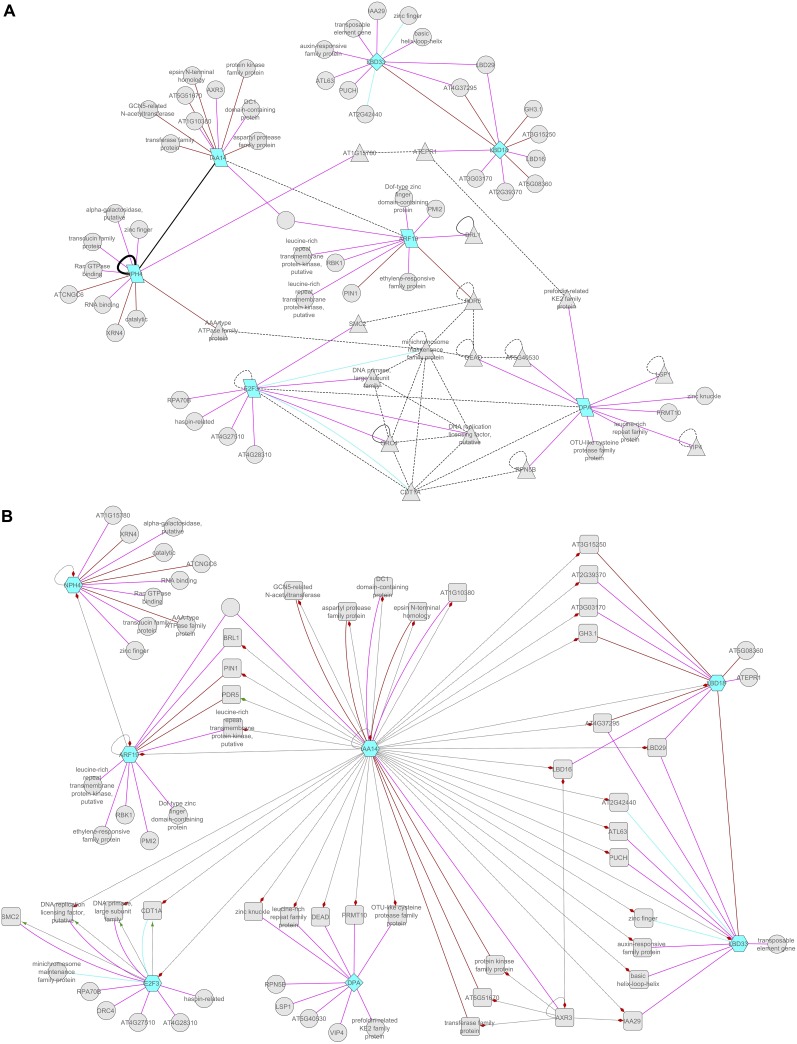
Figure 2.
Network Examples for Protein–Protein and Regulatory Transcriptional Interactions.
Examples of networks starting from a few lateral root initiation components with proven protein–DNA and protein–protein interactions. We selected ARF7/NPH4 (AT5G20730), ARF19 (AT1G19220), SLR (AT4G14550), LBD18 (AT2G45420), LBD33 (AT5G06080), E2FA (AT2G36010), and DPA (AT5G02470) to illustrate the complexity of the networks at various levels. A coexpression network was tested for protein–protein interactions (A) and for regulatory interactions (B). We used standard settings (0.6 to 1.0) for the Pearson’s correlation coefficient, and all the available data sets in the CORNET database (https://cornet.psb.ugent.be/) (with at least meeting the conditions in three data sets) were used.
For identification of protein–DNA interactions, ChIP-chip and ChIP-seq data analysis is a multistep procedure. In the case of ChIP-seq, reads first need to be aligned to a reference genome with a read-aligner, such as bowtie (Langmead et al., 2009), to generate genome coverage data. Then, peaks must be detected from coverage data or microarray signals. Finally, results should be visualized as an important validation step to test whether parameters and program settings were adequate. The experienced user will perform many of these tasks in R using packages such as CSAR (Muiño et al., 2011b). Others can choose from a number of software suites and analysis pipelines, such as the stand-alone program CisGenome (Ji et al., 2008), the Web tool PRI-CAT (Muiño et al., 2011a), or the commercial CLC Genomics Workbench (www.clcbio.com). More thorough descriptions of procedures to analyze ChIP-chip and ChIP-seq data can be found elsewhere (Pepke et al., 2009; Wilbanks and Facciotti, 2010). For visualization, various genome browsers are available. We found the integrated genome browser (Nicol et al., 2009) user friendly and robust. Like other such browsers, it allows sharing of results via network access. Researchers need only to upload their results to a server and make the URL publicly available to allow the community to browse through their findings.
CONCLUSION AND FUTURE PERSPECTIVES
Network science provides crucial information and the ability both to develop hypotheses and to address specific biological questions. Nevertheless, the above-described interactions, either metabolic, protein–protein interaction, or protein-DNA, need to be integrated and fine-tuned to provide a realistic view of the biological process(es). Networks can be generated through many complementary platforms. The integration of various approaches and data sets adds depth (and, therefore, confidence) and breadth to a network and leads to the identification of general network properties, such as the location of hubs and their roles at particular times and under varying conditions.
A major challenge will be to validate the interactions generated by the various screens and predictions and incorporate these interactions and their functional consequences in a spatiotemporal manner. Protein–protein interaction screens are performed mostly in systems that do not provide the spatiotemporal context of complex formation and with protein levels exceeding the native levels. Insights into these networks are needed at the cellular, tissue, and whole-plant levels. It is also important to decipher how these networks are interwoven to generate an organism that can develop, grow, and reproduce. Cell- and tissue-specific transcript profiling and ChIP-chip studies have been applied successfully to explore gene regulation in developmental processes and cell-type/tissue functions to understand transcriptional dynamics (Birnbaum et al., 2003; Brady et al., 2007; Dinneny et al., 2008; Deal and Henikoff, 2010; Weinhofer et al., 2010). By contrast, protein–protein interactions have yet to be explored at this resolution. While we are indeed making progress in understanding regulatory networks and protein–protein interactions, it will be important to implement structural features and posttranslational modifications. For example, reversible phosphorylation mediated by kinases and phosphatases has hardly been addressed to date (Kline-Jonakin et al., 2011), and high-throughput cell- and tissue-specific research is limited and challenging.
Discovery and inference of molecular networks has made great progress, and the time is ripe to combine networks with phenotype data. Although mathematical modeling can help by prescreening predictions in silico and thus limiting the extent of experimentation, high-throughput approaches to assay the impact of network perturbations on phenotypes (phenomics) must now be developed and employed.
Acknowledgments
This work was supported by a Biotechnology and Biological Science Research Council David Phillips Fellowship (BB_BB/H022457/1) and a Marie Curie European Reintegration grant (PERG06-GA-2009-256354) to I.D.S. I.D.S. and S.M.B. are involved in a Biotechnology and Biological Science Research Council–funded U.S. Partnering Award (BB/J020117/1). G.W.B. was funded by a Birmingham Fellowship and a Marie Curie International Incoming Fellowship S.M.B. is funded by National Science Foundation- Integrative Organismal Systems 1052395. S.Y.R. is funded by grants from the National Science Foundation (IOS-1026003, MCB-1052348, MCB-820823, and DBI-640769). L.H. is funded by grants from the Swiss National Science Foundation and the Swedish Research Council.
AUTHOR CONTRIBUTIONS
All authors contributed to writing the article.
References
- Albert R. (2007). Network inference, analysis, and modeling in systems biology. Plant Cell 19: 3327–3338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfarano C., et al. (2005). The Biomolecular Interaction Network Database and related tools 2005 update. Nucleic Acids Res. 33: D418–D424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen D.K., Libourel I.G., Shachar-Hill Y. (2009). Metabolic flux analysis in plants: coping with complexity. Plant Cell Environ. 32: 1241–1257 [DOI] [PubMed] [Google Scholar]
- Aoki K., Ogata Y., Shibata D. (2007). Approaches for extracting practical information from gene co-expression networks in plant biology. Plant Cell Physiol. 48: 381–390 [DOI] [PubMed] [Google Scholar]
- Arabidopsis Interactome Mapping Consortium (2011). Evidence for network evolution in an Arabidopsis interactome map. Science 333: 601–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M., et al. The Gene Ontology Consortium (2000). Gene ontology: Tool for the unification of biology. Nat. Genet. 25: 25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bader G.D., Donaldson I., Wolting C., Ouellette B.F., Pawson T., Hogue C.W. (2001). BIND-The biomolecular interaction network database. Nucleic Acids Res. 29: 242–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bader G.D., Hogue C.W. (2003). An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics 4: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bader G.D., Betel D., Hogue C.W. (2003). BIND: the Biomolecular Interaction Network Database. Nucleic Acids Res. 31: 248–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barabasi A.L., Albert R. (1999). Emergence of scaling in random networks. Science 286: 509–512 [DOI] [PubMed] [Google Scholar]
- Bassel G.W., Glaab E., Marquez J., Holdsworth M.J., Bacardit J. (2011a). Functional network construction in Arabidopsis using rule-based machine learning on large-scale data sets. Plant Cell 23: 3101–3116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassel G.W., Lan H., Glaab E., Gibbs D.J., Gerjets T., Krasnogor N., Bonner A.J., Holdsworth M.J., Provart N.J. (2011b). Genome-wide network model capturing seed germination reveals coordinated regulation of plant cellular phase transitions. Proc. Natl. Acad. Sci. USA 108: 9709–9714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benhamed M., et al. (2008). Genome-scale Arabidopsis promoter array identifies targets of the histone acetyltransferase GCN5. Plant J. 56: 493–504 [DOI] [PubMed] [Google Scholar]
- Birnbaum K., Shasha D.E., Wang J.Y., Jung J.W., Lambert G.M., Galbraith D.W., Benfey P.N. (2003). A gene expression map of the Arabidopsis root. Science 302: 1956–1960 [DOI] [PubMed] [Google Scholar]
- Bolduc N., Yilmaz A., Mejia-Guerra M.K., Morohashi K., O’Connor D., Grotewold E., Hake S. (2012). Unraveling the KNOTTED1 regulatory network in maize meristems. Genes Dev. 26: 1685–1690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bombarely A., Menda N., Tecle I.Y., Buels R.M., Strickler S., Fischer-York T., Pujar A., Leto J., Gosselin J., Mueller L.A. (2011). The Sol Genomics Network (solgenomics.net): Growing tomatoes using Perl. Nucleic Acids Res. 39(Database issue): D1149–D1155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boruc J., Van den Daele H., Hollunder J., Rombauts S., Mylle E., Hilson P., Inzé D., De Veylder L., Russinova E. (2010). Functional modules in the Arabidopsis core cell cycle binary protein-protein interaction network. Plant Cell 22: 1264–1280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady S.M., Orlando D.A., Lee J.Y., Wang J.Y., Koch J., Dinneny J.R., Mace D., Ohler U., Benfey P.N. (2007). A high-resolution root spatiotemporal map reveals dominant expression patterns. Science 318: 801–806 [DOI] [PubMed] [Google Scholar]
- Brady S.M., et al. (2011). A stele-enriched gene regulatory network in the Arabidopsis root. Mol. Syst. Biol. 7: 459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandão M.M., Dantas L.L., Silva-Filho M.C. (2009). AtPIN: Arabidopsis thaliana protein interaction network. BMC Bioinformatics 10: 454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt R., et al. (2012). Genome-wide binding-site analysis of REVOLUTA reveals a link between leaf patterning and light-mediated growth responses. Plant J. 72: 31–42 [DOI] [PubMed] [Google Scholar]
- Bro C., Regenberg B., Förster J., Nielsen J. (2006). In silico aided metabolic engineering of Saccharomyces cerevisiae for improved bioethanol production. Metab. Eng. 8: 102–111 [DOI] [PubMed] [Google Scholar]
- Burgard A.P., Maranas C.D. (2003). Optimization-based framework for inferring and testing hypothesized metabolic objective functions. Biotechnol. Bioeng. 82: 670–677 [DOI] [PubMed] [Google Scholar]
- Caspi R., et al. (2012). The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res. 40(Database issue): D742–D753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Causier B., Ashworth M., Guo W., Davies B. (2012). The TOPLESS interactome: A framework for gene repression in Arabidopsis. Plant Physiol. 158: 423–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan E.K., Rowe H.C., Corwin J.A., Joseph B., Kliebenstein D.J. (2011). Combining genome-wide association mapping and transcriptional networks to identify novel genes controlling glucosinolates in Arabidopsis thaliana. PLoS Biol. 9: e1001125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang I.F., Curran A., Woolsey R., Quilici D., Cushman J.C., Mittler R., Harmon A., Harper J.F. (2009). Proteomic profiling of tandem affinity purified 14-3-3 protein complexes in Arabidopsis thaliana. Proteomics 9: 2967–2985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang R.L., Ghamsari L., Manichaikul A., Hom E.F., Balaji S., Fu W., Shen Y., Hao T., Palsson B.O., Salehi-Ashtiani K., Papin J.A. (2011). Metabolic network reconstruction of Chlamydomonas offers insight into light-driven algal metabolism. Mol. Syst. Biol. 7: 518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatr-aryamontri A., Ceol A., Palazzi L.M., Nardelli G., Schneider M.V., Castagnoli L., Cesareni G. (2007). MINT: the Molecular INTeraction database. Nucleic Acids Res. 35: D572–D574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Lalonde S., Obrdlik P., Noorani Vatani A., Parsa S.A., Vilarino C., Revuelta J.L., Frommer W.B., Rhee S.Y. (2012). Uncovering Arabidopsis membrane protein interactome enriched in transporters using mating-based split ubiquitin assays and classification models. Front. Plant Sci. 3: 124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covert M.W., Knight E.M., Reed J.L., Herrgard M.J., Palsson B.O. (2004). Integrating high-throughput and computational data elucidates bacterial networks. Nature 429: 92–96 [DOI] [PubMed] [Google Scholar]
- Cui H., Levesque M.P., Vernoux T., Jung J.W., Paquette A.J., Gallagher K.L., Wang J.Y., Blilou I., Scheres B., Benfey P.N. (2007). An evolutionarily conserved mechanism delimiting SHR movement defines a single layer of endodermis in plants. Science 316: 421–425 [DOI] [PubMed] [Google Scholar]
- Cui J., Li P., Li G., Xu F., Zhao C., Li Y.H., Yang Z.N., Wang G., Yu Q.B., Li Y.X., Shi T.L. (2008). AtPID: Arabidopsis thaliana protein interactome database—An integrative platform for plant systems biology. Nucleic Acids Res. 36(Database issue): D999–D1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dal’Molin C.G., Quek L.E., Palfreyman R.W., Brumbley S.M., Nielsen L.K. (2010). C4GEM, a genome-scale metabolic model to study C4 plant metabolism. Plant Physiol. 154: 1871–1885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daudin J.J., Picard F., Robin S. (2008). A mixture model for random graphs. Stat. Comput. 18: 173–183 [Google Scholar]
- Deal R.B., Henikoff S. (2010). A simple method for gene expression and chromatin profiling of individual cell types within a tissue. Dev. Cell 18: 1030–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bodt S., Carvajal D., Hollunder J., Van den Cruyce J., Movahedi S., Inzé D. (2010). CORNET: A user-friendly tool for data mining and integration. Plant Physiol. 152: 1167–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bodt S., Proost S., Vandepoele K., Rouzé P., Van de Peer Y. (2009). Predicting protein-protein interactions in Arabidopsis thaliana through integration of orthology, gene ontology and co-expression. BMC Genomics 10: 288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Folter S., Immink R.G., Kieffer M., Parenicová L., Henz S.R., Weigel D., Busscher M., Kooiker M., Colombo L., Kater M.M., Davies B., Angenent G.C. (2005). Comprehensive interaction map of the Arabidopsis MADS Box transcription factors. Plant Cell 17: 1424–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Oliveira Dal’Molin C.G., Quek L.E., Palfreyman R.W., Brumbley S.M., Nielsen L.K. (2010). AraGEM, a genome-scale reconstruction of the primary metabolic network in Arabidopsis. Plant Physiol. 152: 579–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deplancke B., Dupuy D., Vidal M., Walhout A.J. (2004). A gateway-compatible yeast one-hybrid system. Genome Res. 14(10B): 2093–2101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rybel B., et al. (2010). A novel aux/IAA28 signaling cascade activates GATA23-dependent specification of lateral root founder cell identity. Curr. Biol. 20: 1697–1706 [DOI] [PubMed] [Google Scholar]
- Desprez T., Juraniec M., Crowell E.F., Jouy H., Pochylova Z., Parcy F., Höfte H., Gonneau M., Vernhettes S. (2007). Organization of cellulose synthase complexes involved in primary cell wall synthesis in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 104: 15572–15577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinneny J.R., Long T.A., Wang J.Y., Jung J.W., Mace D., Pointer S., Barron C., Brady S.M., Schiefelbein J., Benfey P.N. (2008). Cell identity mediates the response of Arabidopsis roots to abiotic stress. Science 320: 942–945 [DOI] [PubMed] [Google Scholar]
- Dodsworth S. (2009). A diverse and intricate signalling network regulates stem cell fate in the shoot apical meristem. Dev. Biol. 336: 1–9 [DOI] [PubMed] [Google Scholar]
- Dudoit S., VanderLaan M.J. (2008). Multiple testing procedures with applications to genomics. In Springer Series in Statistics. (Berlin, Germany: Springer) [Google Scholar]
- Enright A.J., Van Dongen S., Ouzounis C.A. (2002). An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 30: 1575–1584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farnham P.J. (2009). Insights from genomic profiling of transcription factors. Nat. Rev. Genet. 10: 605–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feist A.M., Palsson B.O. (2008). The growing scope of applications of genome-scale metabolic reconstructions using Escherichia coli. Nat. Biotechnol. 26: 659–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Calvo P., et al. (2011). The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell 23: 701–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiehn O., Kopka J., Dörmann P., Altmann T., Trethewey R.N., Willmitzer L. (2000). Metabolite profiling for plant functional genomics. Nat. Biotechnol. 18: 1157–1161 [DOI] [PubMed] [Google Scholar]
- Fong S.S., Burgard A.P., Herring C.D., Knight E.M., Blattner F.R., Maranas C.D., Palsson B.O. (2005). In silico design and adaptive evolution of Escherichia coli for production of lactic acid. Biotechnol. Bioeng. 91: 643–648 [DOI] [PubMed] [Google Scholar]
- Fornalé S., et al. (2010). ZmMYB31 directly represses maize lignin genes and redirects the phenylpropanoid metabolic flux. Plant J. 64: 633–644 [DOI] [PubMed] [Google Scholar]
- Fraley C., Raftery A.E. (2003). Enhanced model-based clustering, density estimation, and discriminant analysis software: MCLUST. J. Classif. 20: 263–286 [Google Scholar]
- Gaudinier A., et al. (2011). Enhanced Y1H assays for Arabidopsis. Nat. Methods 8: 1053–1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler-Lee J., O’Toole N., Ammar R., Provart N.J., Millar A.H., Geisler M. (2007). A predicted interactome for Arabidopsis. Plant Physiol. 145: 317–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibon Y., Usadel B., Blaesing O.E., Kamlage B., Hoehne M., Trethewey R., Stitt M. (2006). Integration of metabolite with transcript and enzyme activity profiling during diurnal cycles in Arabidopsis rosettes. Genome Biol. 7: R76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillis J., Pavlidis P. (2012). “Guilt by association” is the exception rather than the rule in gene networks. PLoS Comput. Biol. 8: e1002444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J.D., Bertin N., Hao T., Goldberg D.S., Berriz G.F., Zhang L.V., Dupuy D., Walhout A.J., Cusick M.E., Roth F.P., Vidal M. (2004). Evidence for dynamically organized modularity in the yeast protein-protein interaction network. Nature 430: 88–93 [DOI] [PubMed] [Google Scholar]
- Hanada K., Sawada Y., Kuromori T., Klausnitzer R., Saito K., Toyoda T., Shinozaki K., Li W.H., Hirai M.Y. (2011). Functional compensation of primary and secondary metabolites by duplicate genes in Arabidopsis thaliana. Mol. Biol. Evol. 28: 377–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heazlewood J.L., Verboom R.E., Tonti-Filippini J., Small I., Millar A.H. (2007). SUBA: The Arabidopsis Subcellular Database. Nucleic Acids Res. 35(Database issue): D213–D218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry C.S., DeJongh M., Best A.A., Frybarger P.M., Linsay B., Stevens R.L. (2010). High-throughput generation, optimization and analysis of genome-scale metabolic models. Nat. Biotechnol. 28: 977–982 [DOI] [PubMed] [Google Scholar]
- Herrgård M.J., Fong S.S., Palsson B.O. (2006). Identification of genome-scale metabolic network models using experimentally measured flux profiles. PLoS Comput. Biol. 2: e72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hruz T., Laule O., Szabo G., Wessendorp F., Bleuler S., Oertle L., Widmayer P., Gruissem W., Zimmermann P. (2008). Genevestigator v3: A reference expression database for the meta-analysis of transcriptomes. Adv. Bioinforma. 2008: 420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W., Pérez-García P., Pokhilko A., Millar A.J., Antoshechkin I., Riechmann J.L., Mas P. (2012). Mapping the core of the Arabidopsis circadian clock defines the network structure of the oscillator. Science 336: 75–79 [DOI] [PubMed] [Google Scholar]
- Hughes T.R., et al. (2000). Functional discovery via a compendium of expression profiles. Cell 102: 109–126 [DOI] [PubMed] [Google Scholar]
- Hwang S., Rhee S.Y., Marcotte E.M., Lee I. (2011). Systematic prediction of gene function in Arabidopsis thaliana using a probabilistic functional gene network. Nat. Protoc. 6: 1429–1442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibl V., Csaszar E., Schlager N., Neubert S., Spitzer C., Hauser M.T. (2012). Interactome of the plant-specific ESCRT-III component AtVPS2.2 in Arabidopsis thaliana. J. Proteome Res. 11: 397–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji H., Jiang H., Ma W., Johnson D.S., Myers R.M., Wong W.H. (2008). An integrated software system for analyzing ChIP-chip and ChIP-seq data. Nat. Biotechnol. 26: 1293–1300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnsson N., Varshavsky A. (1994). Split ubiquitin as a sensor of protein interactions in vivo. Proc. Natl. Acad. Sci. USA 91: 10340–10344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M., Goto S., Sato Y., Furumichi M., Tanabe M. (2012). KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 40(Database issue): D109–D114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlova R., Boeren S., Russinova E., Aker J., Vervoort J., de Vries S. (2006). The Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASE1 protein complex includes BRASSINOSTEROID-INSENSITIVE1. Plant Cell 18: 626–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato N., Fujikawa Y., Fuselier T., Adamou-Dodo R., Nishitani A., Sato M.H. (2010). Luminescence detection of SNARE-SNARE interaction in Arabidopsis protoplasts. Plant Mol. Biol. 72: 433–444 [DOI] [PubMed] [Google Scholar]
- Kaufmann K., Muiño J.M., Jauregui R., Airoldi C.A., Smaczniak C., Krajewski P., Angenent G.C. (2009). Target genes of the MADS transcription factor SEPALLATA3: Integration of developmental and hormonal pathways in the Arabidopsis flower. PLoS Biol. 7: e1000090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann K., Nagasaki M., Jáuregui R. (2011). Modelling the molecular interactions in the flower developmental network of Arabidopsis thaliana. Stud. Health Technol. Inform. 162: 279–297 [PubMed] [Google Scholar]
- Kaufmann K., Wellmer F., Muiño J.M., Ferrier T., Wuest S.E., Kumar V., Serrano-Mislata A., Madueño F., Krajewski P., Meyerowitz E.M., Angenent G.C., Riechmann J.L. (2010). Orchestration of floral initiation by APETALA1. Science 328: 85–89 [DOI] [PubMed] [Google Scholar]
- Kerrien S., et al. (2007). IntAct - open source resource for molecular interaction data. Nucleic Acids Res. 35: D561–D565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanin R., Wit E. (2006). How scale-free are biological networks. J. Comput. Biol. 13: 810–818 [DOI] [PubMed] [Google Scholar]
- Kim S.K., Lund J., Kiraly M., Duke K., Jiang M., Stuart J.M., Eizinger A., Wylie B.N., Davidson G.S. (2001). A gene expression map for Caenorhabditis elegans. Science 293: 2087–2092 [DOI] [PubMed] [Google Scholar]
- Kliebenstein D. (2009). Advancing genetic theory and application by metabolic quantitative trait loci analysis. Plant Cell 21: 1637–1646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kline-Jonakin K.G., Barrett-Wilt G.A., Sussman M.R. (2011). Quantitative plant phosphoproteomics. Curr. Opin. Plant Biol. 14: 507–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klopffleisch K., et al. (2011). Arabidopsis G-protein interactome reveals connections to cell wall carbohydrates and morphogenesis. Mol. Syst. Biol. 7: 532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusano M., et al. (2011). Metabolomics reveals comprehensive reprogramming involving two independent metabolic responses of Arabidopsis to UV-B light. Plant J. 67: 354–369 [DOI] [PubMed] [Google Scholar]
- Ladunga I. (2010). An overview of the computational analyses and discovery of transcription factor binding sites. Methods Mol. Biol. 674: 1–22 [DOI] [PubMed] [Google Scholar]
- Lalonde S., Ehrhardt D.W., Loqué D., Chen J., Rhee S.Y., Frommer W.B. (2008). Molecular and cellular approaches for the detection of protein-protein interactions: Latest techniques and current limitations. Plant J. 53: 610–635 [DOI] [PubMed] [Google Scholar]
- Lalonde S., et al. (2010). A membrane protein/signaling protein interaction network for Arabidopsis version AMPv2. Front. Physiol. 1: 24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamesch P., et al. (2012). The Arabidopsis Information Resource (TAIR): Improved gene annotation and new tools. Nucleic Acids Res. 40(Database issue): D1202–D1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamesch P., Dreher K., Swarbreck D., Sasidharan R., Reiser L., Huala E. (2010). Using the Arabidopsis information resource (TAIR) to find information about Arabidopsis genes. Curr. Protoc. Bioinformatics 1: 11 [DOI] [PubMed] [Google Scholar]
- Langmead B., Trapnell C., Pop M., Salzberg S.L. (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau S., De Smet I., Kolb M., Meinhardt H., Jürgens G. (2011). Auxin triggers a genetic switch. Nat. Cell Biol. 13: 611–615 [DOI] [PubMed] [Google Scholar]
- Lee I., Ambaru B., Thakkar P., Marcotte E.M., Rhee S.Y. (2010). Rational association of genes with traits using a genome-scale gene network for Arabidopsis thaliana. Nat. Biotechnol. 28: 149–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I., Seo Y.S., Coltrane D., Hwang S., Oh T., Marcotte E.M., Ronald P.C. (2011). Genetic dissection of the biotic stress response using a genome-scale gene network for rice. Proc. Natl. Acad. Sci. USA 108: 18548–18553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., He K., Stolc V., Lee H., Figueroa P., Gao Y., Tongprasit W., Zhao H., Lee I., Deng X.W. (2007). Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell 19: 731–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.M., Gianchandani E.P., Eddy J.A., Papin J.A. (2008). Dynamic analysis of integrated signaling, metabolic, and regulatory networks. PLoS Comput. Biol. 4: e1000086 Erratum. PLoS Comput. Biol. 4: 10.1371/annotation/5594348b-de00-446a-bdd0-ec56e70b3553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin M., Hu B., Chen L., Sun P., Fan Y., Wu P., Chen X. (2009). Computational identification of potential molecular interactions in Arabidopsis. Plant Physiol. 151: 34–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin M., Shen X., Chen X. (2011a). PAIR: The predicted Arabidopsis interactome resource. Nucleic Acids Res. 39(Database issue): D1134–D1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin M., Zhou X., Shen X., Mao C., Chen X. (2011b). The predicted Arabidopsis interactome resource and network topology-based systems biology analyses. Plant Cell 23: 911–922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long T.A., Brady S.M., Benfey P.N. (2008). Systems approaches to identifying gene regulatory networks in plants. Annu. Rev. Cell Dev. Biol. 24: 81–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long T.A., Tsukagoshi H., Busch W., Lahner B., Salt D.E., Benfey P.N. (2010). The bHLH transcription factor POPEYE regulates response to iron deficiency in Arabidopsis roots. Plant Cell 22: 2219–2236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma S.S., Gong Q.Q., Bohnert H.J. (2007). An Arabidopsis gene network based on the graphical Gaussian model. Genome Res. 17: 1614–1625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maere S., Van Dijck P., Kuiper M. (2008). Extracting expression modules from perturbational gene expression compendia. BMC Syst. Biol. 2: 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May P., Christian J.O., Kempa S., Walther D. (2009). ChlamyCyc: An integrative systems biology database and web-portal for Chlamydomonas reinhardtii. BMC Genomics 10: 209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michoel T., Maere S., Bonnet E., Joshi A., Saeys Y., Van den Bulcke T., Van Leemput K., van Remortel P., Kuiper M., Marchal K., Van de Peer Y. (2007). Validating module network learning algorithms using simulated data. BMC Bioinformatics 8(suppl. 2): S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J.P., Lo R.S., Ben-Hur A., Desmarais C., Stagljar I., Noble W.S., Fields S. (2005). Large-scale identification of yeast integral membrane protein interactions. Proc. Natl. Acad. Sci. USA 102: 12123–12128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mintz-Oron S., Meir S., Malitsky S., Ruppin E., Aharoni A., Shlomi T. (2012). Reconstruction of Arabidopsis metabolic network models accounting for subcellular compartmentalization and tissue-specificity. Proc. Natl. Acad. Sci. USA 109: 339–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Risueno M.A., Busch W., Benfey P.N. (2010). Omics meet networks - Using systems approaches to infer regulatory networks in plants. Curr. Opin. Plant Biol. 13: 126–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morohashi K., et al. (2012). A genome-wide regulatory framework identifies maize pericarp color1 controlled genes. Plant Cell 24: 2745–2764
- Morohashi K., Grotewold E. (2009). A systems approach reveals regulatory circuitry for Arabidopsis trichome initiation by the GL3 and GL1 selectors. PLoS Genet. 5: e1000396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller L.A., Zhang P., Rhee S.Y. (2003). AraCyc: A biochemical pathway database for Arabidopsis. Plant Physiol. 132: 453–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muiño J.M., Hoogstraat M., van Ham R.C., van Dijk A.D. (2011a). PRI-CAT: A web-tool for the analysis, storage and visualization of plant ChIP-seq experiments. Nucleic Acids Res. 39(Web Server issue): W524–W527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muiño J.M., Kaufmann K., van Ham R.C., Angenent G.C., Krajewski P. (2011b). ChIP-seq Analysis in R (CSAR): An R package for the statistical detection of protein-bound genomic regions. Plant Methods 7: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhtar M.S., et al. European Union Effectoromics Consortium (2011). Independently evolved virulence effectors converge onto hubs in a plant immune system network. Science 333: 596–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutwil M., Klie S., Tohge T., Giorgi F.M., Wilkins O., Campbell M.M., Fernie A.R., Usadel B., Nikoloski Z., Persson S. (2011). PlaNet: Combined sequence and expression comparisons across plant networks derived from seven species. Plant Cell 23: 895–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutwil M., Usadel B., Schütte M., Loraine A., Ebenhöh O., Persson S. (2010). Assembly of an interactive correlation network for the Arabidopsis genome using a novel heuristic clustering algorithm. Plant Physiol. 152: 29–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakabayashi R., Yamazaki M., Saito K. (2010). A polyhedral approach for understanding flavonoid biosynthesis in Arabidopsis. New Biotechnol. 27: 829–836 [DOI] [PubMed] [Google Scholar]
- Nepusz T., Yu H.Y., Paccanaro A. (2012). Detecting overlapping protein complexes in protein-protein interaction networks. Nat. Methods 9: 471–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicol J.W., Helt G.A., Blanchard S.G., Jr, Raja A., Loraine A.E. (2009). The Integrated Genome Browser: Free software for distribution and exploration of genome-scale datasets. Bioinformatics 25: 2730–2731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obayashi T., Nishida K., Kasahara K., Kinoshita K. (2011). ATTED-II updates: Condition-specific gene coexpression to extend coexpression analyses and applications to a broad range of flowering plants. Plant Cell Physiol. 52: 213–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obrdlik P., et al. (2004). K+ channel interactions detected by a genetic system optimized for systematic studies of membrane protein interactions. Proc. Natl. Acad. Sci. USA 101: 12242–12247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E., Kang H., Yamaguchi S., Park J., Lee D., Kamiya Y., Choi G. (2009). Genome-wide analysis of genes targeted by PHYTOCHROME INTERACTING FACTOR 3-LIKE5 during seed germination in Arabidopsis. Plant Cell 21: 403–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang X., et al. (2011). Genome-wide binding site analysis of FAR-RED ELONGATED HYPOCOTYL3 reveals its novel function in Arabidopsis development. Plant Cell 23: 2514–2535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J.H., Kim T.Y., Lee K.H., Lee S.Y. (2011). Fed-batch culture of Escherichia coli for L-valine production based on in silico flux response analysis. Biotechnol. Bioeng. 108: 934–946 [DOI] [PubMed] [Google Scholar]
- Pauwels L., et al. (2010). NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature 464: 788–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepke S., Wold B., Mortazavi A. (2009). Computation for ChIP-seq and RNA-seq studies. Nat. Methods 6(11 suppl.): S22–S32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson S., Wei H.R., Milne J., Page G.P., Somerville C.R. (2005). Identification of genes required for cellulose synthesis by regression analysis of public microarray data sets. Proc. Natl. Acad. Sci. USA 102: 8633–8638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petricka J.J., Schauer M.A., Megraw M., Breakfield N.W., Thompson J.W., Georgiev S., Soderblom E.J., Ohler U., Moseley M.A., Grossniklaus U., Benfey P.N. (2012). The protein expression landscape of the Arabidopsis root. Proc. Natl. Acad. Sci. USA 109: 6811–6818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokhilko A., Hodge S.K., Stratford K., Knox K., Edwards K.D., Thomson A.W., Mizuno T., Millar A.J. (2010). Data assimilation constrains new connections and components in a complex, eukaryotic circadian clock model. Mol. Syst. Biol. 6: 416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokhilko A., Fernández A.P., Edwards K.D., Southern M.M., Halliday K.J., Millar A.J. (2012). The clock gene circuit in Arabidopsis includes a repressilator with additional feedback loops. Mol. Syst. Biol. 8: 574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poolman M.G., Miguet L., Sweetlove L.J., Fell D.A. (2009). A genome-scale metabolic model of Arabidopsis and some of its properties. Plant Physiol. 151: 1570–1581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popescu S.C., Popescu G.V., Bachan S., Zhang Z.M., Gerstein M., Snyder M., Dinesh-Kumar S.P. (2009). MAPK target networks in Arabidopsis thaliana revealed using functional protein microarrays. Genes Dev. 23: 80–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popescu S.C., Popescu G.V., Bachan S., Zhang Z.M., Seay M., Gerstein M., Snyder M., Dinesh-Kumar S.P. (2007). Differential binding of calmodulin-related proteins to their targets revealed through high-density Arabidopsis protein microarrays. Proc. Natl. Acad. Sci. USA 104: 4730–4735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prelić A., Bleuler S., Zimmermann P., Wille A., Bühlmann P., Gruissem W., Hennig L., Thiele L., Zitzler E. (2006). A systematic comparison and evaluation of biclustering methods for gene expression data. Bioinformatics 22: 1122–1129 [DOI] [PubMed] [Google Scholar]
- Pruneda-Paz J.L., Breton G., Para A., Kay S.A. (2009). A functional genomics approach reveals CHE as a component of the Arabidopsis circadian clock. Science 323: 1481–1485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahim G., Bischof S., Kessler F., Agne B. (2009). In vivo interaction between atToc33 and atToc159 GTP-binding domains demonstrated in a plant split-ubiquitin system. J. Exp. Bot. 60: 257–267 [DOI] [PubMed] [Google Scholar]
- Rausenberger J., Tscheuschler A., Nordmeier W., Wüst F., Timmer J., Schäfer E., Fleck C., Hiltbrunner A. (2011). Photoconversion and nuclear trafficking cycles determine phytochrome A’s response profile to far-red light. Cell 146: 813–825 [DOI] [PubMed] [Google Scholar]
- Rubio V., Shen Y., Saijo Y., Liu Y., Gusmaroli G., Dinesh-Kumar S.P., Deng X.W. (2005). An alternative tandem affinity purification strategy applied to Arabidopsis protein complex isolation. Plant J. 41: 767–778 [DOI] [PubMed] [Google Scholar]
- Saeed A.I., Bhagabati N.K., Braisted J.C., Liang W., Sharov V., Howe E.A., Li J., Thiagarajan M., White J.A., Quackenbush J. (2006). TM4 microarray software suite. Methods Enzymol. 411: 134–193 [DOI] [PubMed] [Google Scholar]
- Saha R., Suthers P.F., Maranas C.D. (2011). Zea mays iRS1563: A comprehensive genome-scale metabolic reconstruction of maize metabolism. PLoS ONE 6: e21784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salwinski L., Miller C.S., Smith A.J., Pettit F.K., Bowie J.U., Eisenberg D. (2004). The Database of Interacting Proteins: 2004 update. Nucleic Acids Res. 32: D449–D451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauro H.M., Hucka M., Finney A., Wellock C., Bolouri H., Doyle J., Kitano H. (2003). Next generation simulation tools: The Systems Biology Workbench and BioSPICE integration. OMICS 7: 355–372 [DOI] [PubMed] [Google Scholar]
- Schwender J. (2008). Metabolic flux analysis as a tool in metabolic engineering of plants. Curr. Opin. Biotechnol. 19: 131–137 [DOI] [PubMed] [Google Scholar]
- Segrè D., Vitkup D., Church G.M. (2002). Analysis of optimality in natural and perturbed metabolic networks. Proc. Natl. Acad. Sci. USA 99: 15112–15117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahriari M., Richter K., Keshavaiah C., Sabovljevic A., Huelskamp M., Schellmann S. (2011). The Arabidopsis ESCRT protein-protein interaction network. Plant Mol. Biol. 76: 85–96 [DOI] [PubMed] [Google Scholar]
- Shlomi T., Berkman O., Ruppin E. (2005). Regulatory on/off minimization of metabolic flux changes after genetic perturbations. Proc. Natl. Acad. Sci. USA 102: 7695–7700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlomi T., Eisenberg Y., Sharan R., Ruppin E. (2007). A genome-scale computational study of the interplay between transcriptional regulation and metabolism. Mol. Syst. Biol. 3: 101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smaczniak C., et al. (2012). Characterization of MADS-domain transcription factor complexes in Arabidopsis flower development. Proc. Natl. Acad. Sci. USA 109: 1560–1565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sozzani R., Cui H., Moreno-Risueno M.A., Busch W., Van Norman J.M., Vernoux T., Brady S.M., Dewitte W., Murray J.A., Benfey P.N. (2010). Spatiotemporal regulation of cell-cycle genes by SHORTROOT links patterning and growth. Nature 466: 128–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark C., Breitkreutz B.J., Reguly T., Boucher L., Breitkreutz A., Tyers M. (2006). BioGRID: a general repository for interaction datasets. Nucleic Acids Res. 34: D535–D539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart J.M., Segal E., Koller D., Kim S.K. (2003). A gene-coexpression network for global discovery of conserved genetic modules. Science 302: 249–255 [DOI] [PubMed] [Google Scholar]
- Sun Y., et al. (2010). Integration of brassinosteroid signal transduction with the transcription network for plant growth regulation in Arabidopsis. Dev. Cell 19: 765–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweetlove L.J., Fell D., Fernie A.R. (2008). Getting to grips with the plant metabolic network. Biochem. J. 409: 27–41 [DOI] [PubMed] [Google Scholar]
- Szklarczyk D., Franceschini A., Kuhn M., Simonovic M., Roth A., Minguez P., Doerks T., Stark M., Muller J., Bork P., Jensen L.J., von Mering C. (2011). The STRING database in 2011: Functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 39(Database issue): D561–D568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi N., Lammens T., Boudolf V., Maes S., Yoshizumi T., De Jaeger G., Witters E., Inzé D., De Veylder L. (2008). The DNA replication checkpoint aids survival of plants deficient in the novel replisome factor ETG1. EMBO J. 27: 1840–1851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W., et al. (2011). PP2A activates brassinosteroid-responsive gene expression and plant growth by dephosphorylating BZR1. Nat. Cell Biol. 13: 124–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibaud-Nissen F., Wu H., Richmond T., Redman J.C., Johnson C., Green R., Arias J., Town C.D. (2006). Development of Arabidopsis whole-genome microarrays and their application to the discovery of binding sites for the TGA2 transcription factor in salicylic acid-treated plants. Plant J. 47: 152–162 [DOI] [PubMed] [Google Scholar]
- Thiele I., Palsson B.O. (2010). A protocol for generating a high-quality genome-scale metabolic reconstruction. Nat. Protoc. 5: 93–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tohge T., Ramos M.S., Nunes-Nesi A., Mutwil M., Giavalisco P., Steinhauser D., Schellenberg M., Willmitzer L., Persson S., Martinoia E., Fernie A.R. (2011). Toward the storage metabolome: Profiling the barley vacuole. Plant Physiol. 157: 1469–1482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toufighi K., Brady S.M., Austin R., Ly E., Provart N.J. (2005). The Botany array resource: e-Northerns, expression angling, and promoter analyses. Plant J. 43: 153–163 [DOI] [PubMed] [Google Scholar]
- Tsukagoshi H., Busch W., Benfey P.N. (2010). Transcriptional regulation of ROS controls transition from proliferation to differentiation in the root. Cell 143: 606–616 [DOI] [PubMed] [Google Scholar]
- Urbanczyk-Wochniak E., Sumner L.W. (2007). MedicCyc: A biochemical pathway database for Medicago truncatula. Bioinformatics 23: 1418–1423 [DOI] [PubMed] [Google Scholar]
- Usadel B., et al. (2005). Extension of the visualization tool MapMan to allow statistical analysis of arrays, display of corresponding genes, and comparison with known responses. Plant Physiol. 138: 1195–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usadel B., Obayashi T., Mutwil M., Giorgi F.M., Bassel G.W., Tanimoto M., Chow A., Steinhauser D., Persson S., Provart N.J. (2009). Co-expression tools for plant biology: opportunities for hypothesis generation and caveats. Plant Cell Environ. 32: 1633–1651 [DOI] [PubMed] [Google Scholar]
- Van Aken O., Pecenková T., van de Cotte B., De Rycke R., Eeckhout D., Fromm H., De Jaeger G., Witters E., Beemster G.T., Inzé D., Van Breusegem F. (2007). Mitochondrial type-I prohibitins of Arabidopsis thaliana are required for supporting proficient meristem development. Plant J. 52: 850–864 [DOI] [PubMed] [Google Scholar]
- Van Bel M., Proost S., Wischnitzki E., Movahedi S., Scheerlinck C., Van de Peer Y., Vandepoele K. (2012). Dissecting plant genomes with the PLAZA comparative genomics platform. Plant Physiol. 158: 590–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Leene J., Boruc J., De Jaeger G., Russinova E., De Veylder L. (2011). A kaleidoscopic view of the Arabidopsis core cell cycle interactome. Trends Plant Sci. 16: 141–150 [DOI] [PubMed] [Google Scholar]
- Van Leene J., et al. (2010). Targeted interactomics reveals a complex core cell cycle machinery in Arabidopsis thaliana. Mol. Syst. Biol. 6: 397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Leene J., Witters E., Inzé D., De Jaeger G. (2008). Boosting tandem affinity purification of plant protein complexes. Trends Plant Sci. 13: 517–520 [DOI] [PubMed] [Google Scholar]
- Vernoux T., et al. (2011). The auxin signalling network translates dynamic input into robust patterning at the shoot apex. Mol. Syst. Biol. 7: 508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal M., Cusick M.E., Barabási A.L. (2011). Interactome networks and human disease. Cell 144: 986–998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Marshall A., Zhang D., Wilson Z.A. (2012). ANAP: An integrated knowledge base for Arabidopsis protein interaction network analysis. Plant Physiol. 158: 1523–1533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warde-Farley D., et al. (2010). The GeneMANIA prediction server: Biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 38(Web Server issue): W214–W220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinhofer I., Hehenberger E., Roszak P., Hennig L., Köhler C. (2010). H3K27me3 profiling of the endosperm implies exclusion of polycomb group protein targeting by DNA methylation. PLoS Genet. 6: e1001152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilbanks E.G., Facciotti M.T. (2010). Evaluation of algorithm performance in ChIP-seq peak detection. PLoS ONE 5: e11471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wille A., et al. (2004). Sparse graphical Gaussian modeling of the isoprenoid gene network in Arabidopsis thaliana. Genome Biol. 5: R92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L.F., Hughes T.R., Davierwala A.P., Robinson M.D., Stoughton R., Altschuler S.J. (2002). Large-scale prediction of Saccharomyces cerevisiae gene function using overlapping transcriptional clusters. Nat. Genet. 31: 255–265 [DOI] [PubMed] [Google Scholar]
- Xie Z., Li D., Wang L., Sack F.D., Grotewold E. (2010). Role of the stomatal development regulators FLP/MYB88 in abiotic stress responses. Plant J. 64: 731–739 [DOI] [PubMed] [Google Scholar]
- Xu F., Li G.A., Zhao C., Li Y.H., Li P., Cui J.A., Deng Y.P., Shi T.L. (2010). Global protein interactome exploration through mining genome-scale data in Arabidopsis thaliana. BMC Genomics 11(suppl. 2): S2 doi:10.1186/1471-2164-11-S2-S2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yellaboina S., Goyal K., Mande S.C. (2007). Inferring genome-wide functional linkages in E. coli by combining improved genome context methods: Comparison with high-throughput experimental data. Genome Res. 17: 527–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youens-Clark K., et al. (2011). Gramene database in 2010: Updates and extensions. Nucleic Acids Res. 39(Database issue): D1085–D1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H., et al. (2008a). High-quality binary protein interaction map of the yeast interactome network. Science 322: 104–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Q.B., et al. (2008b). Construction of a chloroplast protein interaction network and functional mining of photosynthetic proteins in Arabidopsis thaliana. Cell Res. 18: 1007–1019 [DOI] [PubMed] [Google Scholar]
- Yu X., Li L., Zola J., Aluru M., Ye H., Foudree A., Guo H., Anderson S., Aluru S., Liu P., Rodermel S., Yin Y. (2011). A brassinosteroid transcriptional network revealed by genome-wide identification of BESI target genes in Arabidopsis thaliana. Plant J. 65: 634–646 [DOI] [PubMed] [Google Scholar]
- Zhang P., et al. (2010). Creation of a genome-wide metabolic pathway database for Populus trichocarpa using a new approach for reconstruction and curation of metabolic pathways for plants. Plant Physiol. 153: 1479–1491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q., Brkljacic J., Meier I. (2008). Two distinct interacting classes of nuclear envelope-associated coiled-coil proteins are required for the tissue-specific nuclear envelope targeting of Arabidopsis RanGAP. Plant Cell 20: 1639–1651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y., Ren N., Wang H., Stromberg A.J., Perry S.E. (2009). Global identification of targets of the Arabidopsis MADS domain protein AGAMOUS-Like15. Plant Cell 21: 2563–2577 [DOI] [PMC free article] [PubMed] [Google Scholar]