Using a combination of genetic and expression analyses, this study reveals a reproductive function of the Armadillo repeat gene ZAK IXIK in promoting early embryo and endosperm development through a distinctive gametophytic maternal effect involving mechanisms for maternal allele-specific expression that are not implicated in the well-established pathways.
Abstract
The proper balance of parental genomic contributions to the fertilized embryo and endosperm is essential for their normal growth and development. The characterization of many gametophytic maternal effect (GME) mutants affecting seed development indicates that there are certain classes of genes with a predominant maternal contribution. We present a detailed analysis of the GME mutant zak ixik (zix), which displays delayed and arrested growth at the earliest stages of embryo and endosperm development. ZIX encodes an Armadillo repeat (Arm) protein highly conserved across eukaryotes. Expression studies revealed that ZIX manifests a GME through preferential maternal expression in the early embryo and endosperm. This parent-of-origin–dependent expression is regulated by neither the histone and DNA methylation nor the DNA demethylation pathways known to regulate some other GME mutants. The ZIX protein is localized in the cytoplasm and nucleus of cells in reproductive tissues and actively dividing root zones. The maternal ZIX allele is required for the maternal expression of MINISEED3. Collectively, our results reveal a reproductive function of plant Arm proteins in promoting early seed growth, which is achieved through a distinct GME of ZIX that involves mechanisms for maternal allele-specific expression that are independent of the well-established pathways.
INTRODUCTION
The embryo and the endosperm of sexually reproducing flowering plants are derived from male and female gametes and thus carry genomes of different parental origin. The gametes are formed by the haploid gametophytes, which develop inside the diploid sporophytic reproductive organs of the flowers. During double fertilization, the male gametes (two haploid sperm cells) unite with the female gametes (the haploid egg cell and the diploid central cell) to form the embryo and endosperm, respectively. The two female gametes are harbored within the female gametophyte or embryo sac. The two male gametes are contained inside the male gametophyte or pollen grain. Since developmental progression of the fertilization products occurs within female tissues of both gametophytic and sporophytic origin, the extent of maternal contributions to embryo and endosperm growth and development has been a research subject of intense interest. In particular, several large-scale screens for Ds transposon or T-DNA insertion mutants defective in Arabidopsis thaliana embryo sac development and other reproductive functions have been performed to identify gametophytic maternal effect (GME) mutants (Christensen et al., 1998; Grossniklaus and Schneitz, 1998; Howden et al., 1998; Moore, 2002; reviewed in Brukhin et al., 2005; Pagnussat et al., 2005).
The genetic principle of these screens is based on the distorted inheritance of mutant alleles that affect gametophytic functions, which does not follow the classical Mendelian rules of segregation typical of mutations affecting sporophytic functions (Moore et al., 1997; Howden et al., 1998). In these screens, three classes of gametophytic mutants were categorized according to the transmission efficiency of mutant alleles and their effects on developmental stages. (1) Female- or male-specific gametophytic mutants that affect the development or function of the gametophytes: If they are fully penetrant, such mutant alleles can only be transmitted via the other, nonaffected gametophyte. Thus, these mutants can only be maintained as heterozygotes. (2) General gametophytic mutants affecting both female and male gametophytes, often to a different degree: These will not be transmitted if fully penetrant but can be maintained as either heterozygotes and/or homozygotes if they are only partially penetrant. (3) Mutants affecting the fertilization products, the embryo and/or endosperm, exhibit maternal or paternal effects on seed development and hence are classified as gametophytic maternal/paternal effect mutants (Grossniklaus et al., 1998; Bayer et al., 2009). Mutants of this third class may also affect gametophyte development and either seed development does not initiate or seeds carrying the mutant alleles abort if the affected gene products are essential for seed development. Consequently, mutants of all three classes have higher rates of unfertilized ovules and/or seed abortion than wild-type plants, even when these mutant plants are grown under optimal, healthy conditions (Feldmann et al., 1997; Moore et al., 1997).
Over 220 Arabidopsis and maize (Zea mays) mutants in the GME class have been identified to date. The most intensively characterized GME mutants belong to the fertilization independent seed (fis) class; disruption of any of the four genes, MEDEA (MEA), FIS2, FERTILIZATION INDEPENDENT ENDOSPERM (FIE), or MULTICOPY SUPPRESSOR OF IRA1 (MSI1), leads to autonomous development of the endosperm (and embryo in the case of msi1) in the absence of fertilization as well as overproliferation of the fertilization products if fertilization occurs (Grossniklaus et al., 1998; Luo et al., 1999; Ohad et al., 1999; Köhler et al., 2003). All four genes encode core subunits of the FIS Polycomb Repressive Complex 2 (FIS-PRC2); in addition, MSI1 is also a part of the Chromatin Assembly Factor 1 complex (Hennig et al., 2003). MEA and FIS2 are regulated by genomic imprinting, such that only transcripts derived from the maternal alleles can be detected after fertilization (Kinoshita et al., 1999; Vielle-Calzada et al., 1999; Jullien et al., 2006a). Activation of the maternal MEA allele involves the DNA glycosylase DEMETER (DME), disruption of which leads to a GME seed abortion phenotype (Choi et al., 2002; Jullien et al., 2006b). Paternal silencing of MEA, FIS2, and FIE is directly or indirectly regulated by DNA methylation mediated by DNA METHYLTRANSFERASE1 (MET1) or DECREASED DNA METHYLATION1 (DDM1) (Vielle-Calzada et al., 1999; Vinkenoog et al., 2000; Yadegari et al., 2000; Jullien et al., 2006b; Wöhrmann et al., 2012) and trimethylation of histone H3 Lys-27 (H3K27me3), which is deposited by FIS-PRC2 (Baroux et al., 2006; Gehring et al., 2006; Jullien et al., 2006b).
Besides the fis class mutants, a handful of other Arabidopsis and maize GME mutants have been described and display a variety of phenotypes affecting developmental progression and patterning of the seed. In the three Arabidopsis mutants prolifera (prl) (Springer et al., 1995, 2000), maternally expressed pab c-terminal (mpc) (Tiwari et al., 2008), and expo1a;expo1b (xpo1) (Blanvillain et al., 2008), primarily the female gametophyte development and fertilization, and less frequently the development of both fertilization products, are arrested at various stages. Moreover, three GME mutants that have a normal embryo sac and only affect the young embryo and/or endosperm have been described. The maize stunter1 mutant produces miniature kernels containing an undersized embryo and endosperm (Phillips and Evans, 2011). The Arabidopsis mutants glauce (Ngo et al., 2007; Leshem et al., 2012) and capulet2 (Grini et al., 2002) both support normal embryo development to the globular or transition stage, but the endosperm either does not develop due to an unfertilized central cell or is severely retarded, respectively. Other mutants affecting later embryonic and/or endosperm developmental stages in addition to patterning are the maize mutants maternal effect lethal1 (Evans and Kermicle, 2001) and baseless1 (Gutiérrez-Marcos et al., 2006) and the Arabidopsis mutants Arabidopsis formin homolog5 (fh5) (Ingouff et al., 2005; Fitz Gerald et al., 2009). Disrupted at both early and late stages with both gametophytic and zygotic embryo lethal effects is the Arabidopsis ligase1 (lig1) mutant (Andreuzza et al., 2010). Two of the genes underlying these various mutants, MPC (Tiwari et al., 2008) and At-FH5 (Fitz Gerald et al., 2009), are maternally expressed imprinted genes like MEA and FIS2, and their silenced paternal alleles are under the control of MET1 and FIE, respectively.
The largest GME mutant class in plants, maternal effect embryo arrest (mee), comprises 56 mee mutants (Pagnussat et al., 2005) and the capulet1 mutant (Grini et al., 2002), sharing the phenotype of a very early developmental arrest during seed development but apparently normal embryo sac prior to fertilization. None of the genes underlying these phenotypes have been studied in detail. Here, we report the characterization of a selected mutant from this class and the affected gene, originally named MEE50 (Pagnussat et al., 2005). Delay and arrest of growth and development are observed at the zygotic or one-cell embryo stage in mee50 seeds. We show that MEE50 encodes an Armadillo repeat (Arm) protein. Hence, we renamed this mutant zak ixik (zix) after the ancient Mayan goddess of fertility and childbirth. In Mayan glyphs, Zak Ixik, the White Woman, is often depicted together with an armadillo or other animals (Thompson, 1972). ZIX is predominantly maternally expressed in the zygote, early embryo, and endosperm, and this parent-of-origin–dependent expression is regulated neither by the DNA and histone methylation nor the DNA demethylation pathways that regulate other well-characterized GME mutants. In addition, we demonstrate that ZIX is localized in the cytoplasm and nucleus of cells in reproductive tissues and actively dividing root zones. Disruption of ZIX does not interfere with the expression of several cell cycle and transcription factor genes expressed in seeds, except for the maternal allele of MINISEED3 (MINI3). Collectively, our study reveals that the multitude of plant Arm proteins encompasses the reproductive function of ZIX, which promotes early seed development through mechanisms that are distinct from that of other GME mutants and do not involve certain types of DNA or histone methylation or FIS-PRC2 repression.
RESULTS
The zix GME Mutant Is Defective in Early Embryo and Endosperm Development
The original zix allele was isolated through the distorted segregation ratio of a Ds-carrying mutant allele in the Landsberg erecta (Ler) background, in which the Ds element had inserted in the first exon of At4g00231 (Pagnussat et al., 2005). From the same mutant screen, we subsequently identified two additional alleles with Ds insertions at two other locations in the first exon of the same gene (Figure 1). Seed abortion frequencies in mature siliques of heterozygous zix/ZIX plants reached between 35 and 37% compared with 6% in wild-type plants (Table 1). Transmission of all mutant alleles, which carried the kanamycin (Kan) resistance gene, assessed by the ratio of Kan-resistant to Kan-sensitive seedlings, deviated from the expected Mendelian ratios of 3:1 and 1:1 for self-fertilized and outcrossed heterozygotes, respectively (Table 2). This segregation ratio distortion, with the transmission of the female alleles being more severely reduced than that of the male alleles, indicates that these are gametophytic mutants. Moreover, we could not obtain 100% Kan-resistant progeny populations from any mutant heterozygote despite reasonable transmission of both male and female mutant alleles (Table 2), which implies that additional zygotic embryo lethality underlies the genetics of these mutant alleles.
Figure 1.
Ds Insertion Locations in zix Alleles and Protein Domains in ZIX.
Unnamed shaded box, exon; open box, untranslated region; line, intron; named shaded box, protein domain; and ARM, Armadillo repeat. Bar = 100 nt.
Table 1. Seed Abortion Phenotype of zix Mutant and Complemented Alleles.
| Genotype | Aborted | n |
|---|---|---|
| Ler wild type | 6.1% | 459 |
| zix-1/ZIX | 36.8% | 446 |
| zix-2/ZIX | 35% | 446 |
| zix-3/ZIX | 37.3% | 434 |
| zix-1/ZIX;Lg/- (1) | 12.7% | 433 |
| zix-1/ZIX;Lg/- (2) | 12.6% | 405 |
| zix-1/ZIX;Lg/- (3) | 10.1% | 431 |
| zix-2/ZIX;Lg/- (1) | 12.6% | 507 |
| zix-2/ZIX;Lg/- (2) | 10.3% | 507 |
| zix-2/ZIX;Lg/- (3) | 8.6% | 500 |
| zix-3/ZIX;Lg/- (1) | 5.1% | 430 |
| zix-3/ZIX;Lg/- (2) | 12.6% | 446 |
| zix-3/ZIX;Lg/- (3) | 6.7% | 433 |
For the wild type and three original zix alleles, eight mature siliques were taken randomly from three plants. For complemented zix plants, three plants for each allele numbered (1), (2), and (3) were selected, and eight mature siliques were taken randomly from each of these three plants. Lg, genomic At4g00231 transgene; n, numbers of ovules and seeds.
Table 2. Transmission Rates of Mutant and Complemented zix Alleles.
| Genotype | Kanr/Kans | n |
|---|---|---|
| Mutant alleles | ||
| zix-1/ZIX selfed | (229/166) 1.38 | 395 |
| zix-2/ZIX selfed | (483/273) 1.77 | 756 |
| zix-3/ZIX selfed | (588/353) 1.66 | 941 |
| zix-1/ZIX × ZIX/ZIX | (94/251) 0.38 | 345 |
| ZIX/ZIX × zix-1/ZIX | (193/278) 0.7 | 471 |
| zix-2/ZIX × ZIX/ZIX | (98/242) 0.37 | 340 |
| ZIX/ZIX × zix-2/ZIX | (308/518) 0.6 | 826 |
| zix-3/ZIX × ZIX/ZIX | (85/233) 0.37 | 317 |
| ZIX/ZIX × zix-3/ZIX | (276/343) 0.8 | 619 |
| Complemented alleles with genomic ZIX | ||
| zix-1/ZIX;Lg/- (1) selfed | (540/212) 2.55 | 754 |
| zix-1/ZIX;Lg/- (2) selfed | (603/227) 2.66 | 830 |
| zix-1/ZIX;Lg/- (3) selfed | (954/280) 3.41 | 1234 |
| zix-2/ZIX;Lg/- (1) selfed | (931/354) 2.63 | 1285 |
| zix-2/ZIX;Lg/- (2) selfed | (897/383) 2.34 | 1280 |
| zix-2/ZIX;Lg/- (3) selfed | (949/390) 2.43 | 1289 |
| zix-3/ZIX;Lg/- (1) selfed | (935/195) 4.8 | 1130 |
| zix-3/ZIX;Lg/- (2) selfed | (648/254) 2.55 | 902 |
| zix-3/ZIX;Lg/- (3) selfed | (798/196) 4.7 | 994 |
| Complemented alleles with ProZIX:ZIX-GFP | ||
| zix-3/ZIX;ZG/- (1) selfed | (345/147) 2.35 | 492 |
| zix-3/ZIX;ZG/- (2) selfed | (195/104) 1.88 | 299 |
| zix-3/ZIX;ZG/- (3) selfed | (179/81) 2.2 | 260 |
| zix-3/ZIX;ZG/- (4) selfed | (279/128) 2.18 | 407 |
| zix-3/ZIX;ZG/- (5) selfed | (228/81) 2.8 | 309 |
A detailed phenotypic characterization of the female gametophytes in ovules of unpollinated zix/ZIX pistils revealed a slight delay in embryo sac maturation: 2 d after emasculation, stage 12 flowers of zix/ZIX plants had more ovules with unfused polar nuclei (14%, n = 501) than those of wild-type plants (4%, n = 490) (Figures 2A and 2B). The most prominent mutant phenotype, however, was observed during early seed development after fertilization when we followed the developmental progression of seeds derived from zix mutant embryo sacs (hereafter referred to as zix seeds).
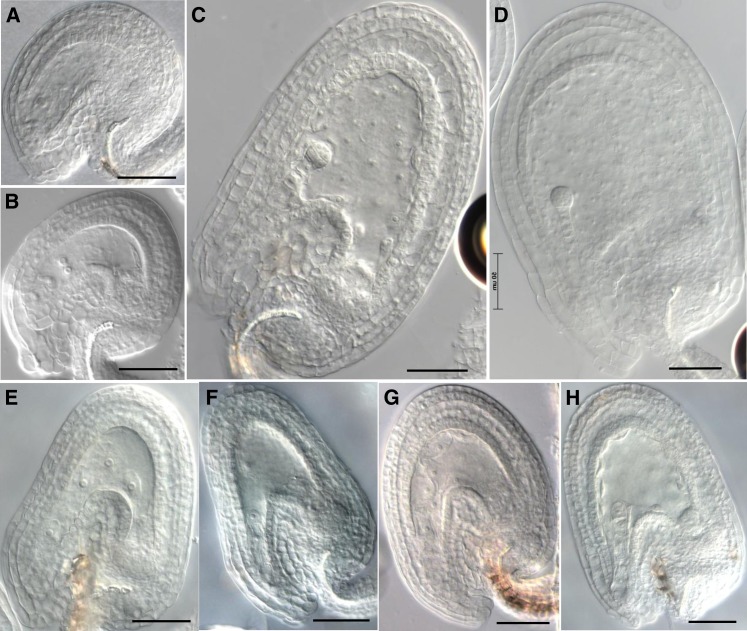
Figure 2.
Ovule and Seed Phenotypes of zix before Fertilization and 3 DAP.
(A) Wild-type mature embryo sac 2 d after emasculation.
(B) zix embryo sac 2 d after emasculation.
(C) and (D) Wild-type seeds at the eight-cell embryo and early globular stages 3 DAP.
(E) and (F) zix seeds at the elongated zygote and one-cell embryo stages 3 DAP.
(G) and (H) Wild-type seeds at the elongated zygote and one-cell embryo stages 1.5 DAP.
Bars = 50 µm.
[See online article for color version of this figure.]
At 1.5 d after pollination (DAP), zix zygotes and early endosperm grew more slowly than those in wild-type embryo sacs: In 59% of wild-type seeds, the zygote had elongated and the endosperm had undergone three to four rounds of nuclear division; on the other hand, only 19 to 35% of zix seeds had reached a comparable stage of zygote and endosperm development (Table 3). In 24% of wild-type seeds, the zygotes were short and symmetric and the endosperm had undergone only one or two division cycles (Table 3), while up to 57% of zix seeds still remained at this stage (Table 3). This slow growth phenotype resulted regardless of whether the pollen were derived from wild-type or heterozygous zix/ZIX flowers, with the zix-2 allele displaying the most severe delay (Table 3), probably due to the Ds insertion being closest to the start codon (Figure 1).
Table 3. Phenotypic Classes of zix Alleles 1.5 DAP.
| Genotype | zy+(1-4end) | zy+(6-8end) | ez+(2-4end) | ez+(6-12end) | 1c+(8-16end) | 2c | unf | pn | n |
|---|---|---|---|---|---|---|---|---|---|
| Ler wild type s | 24.2% | 3.6% | 4.6% | 59.4% | 3.1% | 4.1% | 0.5% | 0.5% | 322 |
| zix-1/ZIX s | 41.6% | 3.1% | 6.2% | 34.2% | 2.5% | 3.1% | 7.5% | 1.8% | 322 |
| zix-1/ZIX c | 38.7% | 8.5% | 8.3% | 27.1% | 2.6% | 3.9% | 9.7% | 1.8% | 362 |
| zix-2/ZIX s | 56.8% | 6.7% | 5% | 19.4% | 0.6% | 0 | 11% | 0.4% | 315 |
| zix-2/ZIX c | 50.5% | 6.9% | 4.5% | 30% | 0.7% | 0 | 8.2% | 0 | 291 |
| zix-3/ZIX s | 41.5% | 9% | 5.4% | 35.5% | 1% | 1.6% | 5.4% | 0.6% | 299 |
| zix-3/ZIX c | 44.6% | 9.8% | 4.2% | 32.2% | 0 | 0.3% | 7.4% | 1.4% | 285 |
Two days after emasculation, stage 12 zix/ZIX flowers were manually pollinated with self or wild-type pollen and the pistils were dissected 1.5 d later. Whole-mount ovules and seeds were cleared with chloral hydrate and scored under a DIC microscope. s, selfed; c, crossed with the wild type; :n, numbers of ovules from five to six siliques; zy, early symmetric zygote; end, endosperm nuclei; ez, elongated zygote; 1c, one-cell embryo; 2c, two-cell embryo; unf, unfertilized ovules; pn, unfused polar nuclei.
At 3 DAP, in wild-type self-fertilized siliques, ∼70% of the seeds were past the four-cell embryo stage, with most being at the early globular stage, while 25% lagged behind and were between the one- and four-cell embryo stages with well-developed endosperm typical of wild-type seeds at these stages (Table 4, Figures 2C, 2D, 2G, and 2H). By contrast, in zix/ZIX siliques crossed with either self or wild-type pollen, up to 29% of the seeds were arrested at the zygotic or one-cell embryo stage with less than eight endosperm nuclei, and the suspensors of these one-cell embryos did not divide any further in most cases (Figures 2E and 2F compared with wild-type seeds in Figures 2G and 2H, Table 4). In addition, 1.9 to 6.6% of the seeds were arrested at the two- to eight-cell stage, but the endosperm underwent only two or three rounds of division, a twofold reduction compared with the more than five divisions of wild-type seeds at this stage (Table 4). In 1.9 to 9% of the seeds, the embryo sac resembled a mature wild-type prefertilization embryo sac with no sign of either embryo or endosperm development and thus appeared unfertilized (Table 4). Beyond 3 DAP, most mutant seeds collapsed.
Table 4. Phenotypic Classes of zix Alleles at 3 DAP.
| Genotype | glob | ea.gl/8c | 4c/2c/1c | unf | 8c.m/4c.m/2c.m | 1c.m/zy.m | col | n |
| Ler wild type s | 23 | 96/38 | 27/25/5 | 1 | 0/0/0 | 0/0 | 14 | 229 |
| 10% | 58.5% | 25% | 0.5% | 0% | 0% | 6% | 100% | |
| zix-1/ZIX s | 0 | 151/19 | 10/4/1 | 10 | 3/5/5 | 46/21 | 22 | 297 |
| 0% | 57.2% | 5% | 3.3% | 4.4% | 22.7% | 7.4% | 100% | |
| zix-1/ZIX c | 7 | 105/37 | 27/3/1 | 5 | 6/5/4 | 30/11 | 19 | 260 |
| 2.7% | 54.6% | 11.9% | 1.9% | 5.8% | 15.8% | 7.3% | 100% | |
| zix-2/ZIX s | 0 | 68/52 | 29/16/5 | 20 | 1/2/11 | 49/26 | 21 | 300 |
| 0% | 40% | 16.6% | 6.7% | 4.7% | 25% | 7% | 100% | |
| zix-2/ZIX c | 1 | 81/58 | 18/20/3 | 8 | 0/5/10 | 32/5 | 21 | 262 |
| 0.4% | 53% | 15.7% | 3.1% | 5.8% | 14% | 8% | 100% | |
| zix-3/ZIX s | 2 | 64/37 | 18/14/4 | 24 | 0/1/4 | 50/27 | 20 | 265 |
| 0.8% | 38.1% | 13.6% | 9% | 1.9% | 29% | 7.6% | 100% | |
| zix-3/ZIX c | 0 | 77/52 | 16/11/5 | 13 | 0/7/11 | 57/7 | 16 | 272 |
| 0% | 47.4% | 11.8% | 4.8% | 6.6% | 23.5% | 5.9% | 100% |
Two days after emasculation, stage 12 zix/ZIX flowers were manually pollinated with self or wild-type pollen and the pistils were dissected 3 d later. Whole-mount ovules and seeds were cleared with chloral hydrate and scored under a DIC microscope. The percentage of each class or underlined group is given underneath the class or group. s, selfed; c, crossed with the wild type; n, numbers of ovules from five siliques; glob, globular stage; ea.gl, early globular stage; 8c, eight-cell stage; 4c, four-cell stage; 2c, two-cell stage; 1c, one-cell stage; unf, unfertilized ovules; 8c.m, eight-cell mutant; 4c.m, four-cell mutant; 2c.m, two-cell mutant; 1c.m, one-cell mutant; zy.m, zygotic mutant; col, collapsed ovules. The 1c.m mutant class had a one-cell embryo with nondividing suspensor and ≤8 endosperm nuclei. 2c.m, 4c.m, and 8c.m mutant classes had greater than or equal to twofold reduction in endosperm nuclei compared to wild-type seeds at the same stage.
The slight reduction in the male transmission efficiency of zix alleles (Table 2) prompted us to question whether the zix male allele contributes to the delayed growth and early embryo and endosperm postfertilization arrest. However, neither aberrant seed phenotypes were observed when heterozygous zix/ZIX flowers were used as pollen donors on wild-type flowers nor was any obvious abnormality observed in zix/ZIX pollen. Introgressing a DNA fragment encompassing the genomic At4g00231 locus into plants having any of the three zix alleles rescued the mutant phenotypes: In transgenic zix/ZIX plants carrying additional At4g00231 copy/copies, the seed abortion rates were significantly reduced and the transmission of mutant alleles significantly increased in comparison to those of nontransformed zix/ZIX plants (P values < 0.0001, at 95% confidence interval) (Tables 1 and 2).
Based on these results, we conclude that the disruption of At4g00231 causes the loss-of-function GME phenotype in zix/ZIX plants and that maternal ZIX promotes early embryo and endosperm development in wild-type seeds.
ZIX Encodes an Arm Protein Highly Conserved across Eukaryotic Kingdoms
At4g00231 is a single-copy gene in the Arabidopsis genome (http://www.Arabidopsis.org). Searching the nonredundant public databases by BLASTp (http://blast.ncbi.nlm.nih.gov) with the ZIX virtual translation sequence uncovered similar proteins across eukaryotic kingdoms, all sharing a sequence of ∼100 amino acids at the C terminus of unknown function (pfam09759), annotated as “Ataxin-10 related” (Figure 1; see Supplemental Figure 1 online). The rat homolog of the human protein Ataxin-10, which is associated with a cerebellar dysfunction disorder (Matsuura et al., 2000), is predicted to belong to the Arm repeat protein family (März et al., 2004), the secondary structure of which is arranged into three α-helical turns interspersed by coils (Hatzfeld, 1999). Using the HHpred algorithm (Söding, 2005), we found that the ZIX secondary structure contains multiple α-helices connected by coil domains (see Supplemental Figure 2 online). Although only two putative Arm repeat motifs similar to the archetypal Arm repeat (pfam00514 and smart00185) (Bateman et al., 2004; Letunic et al., 2004) were identified by this algorithm at the N and C termini of ZIX (Figure 1; see Supplemental Figure 2 online), upon scrutiny of the sequence and the helix alignments, we recognized a third Arm repeat motif at the ZIX N terminus (Figure 1; see Supplemental Figure 3 online). The two best-fitting three dimensional models for ZIX predicted by I-TASSER (Zhang, 2008) and PHYRE (Kelley and Sternberg, 2009), using the presumed ZIX secondary structure and the Protein Data Bank (Bourne et al., 2004), were importin-α and β-catenin (see Supplemental Figure 4 online). These are the two Arm repeat proteins with a known crystal structure, the latter being the vertebrate equivalent of the Drosophila melanogaster armadillo protein (Riggleman et al., 1989). The finding that the predicted three-dimensional structure of ZIX is similar to these two Arm proteins indicates that there could be additional Arm domains in the ZIX protein that were not recognized by the algorithms used because of lower primary sequence homology.
The Maternal Effect of ZIX Is Mediated by Predominant Maternal Allele Expression in Early Seeds
Based on Arabidopsis Genevestigator expression data (Hruz et al., 2008), ZIX transcripts are present in many different tissues throughout the plant life cycle, with the highest expression levels in seeds, especially the endosperm (see Supplemental Figures 5A and 5B online). AtProteom (Baerenfaller et al., 2008) detected ZIX peptides in young roots and young siliques (see Supplemental Figure 5C online). These results potentially point to diverse roles of ZIX in various Arabidopsis developmental processes, in addition to early embryo and endosperm development. We investigated ZIX expression specifically in Arabidopsis gametophytes and seeds 1 to 2 DAP by RNA in situ hybridization. ZIX transcripts were present in pollen and all four cell types of the mature embryo sac (central cell, egg cell, and synergids), as well as in the early embryo and endosperm, with the strongest expression in the prefertilization central cell (Figure 3). Thus, the expression of ZIX is consistent with its functional relevance for early seed development as revealed by the mutant phenotypes.
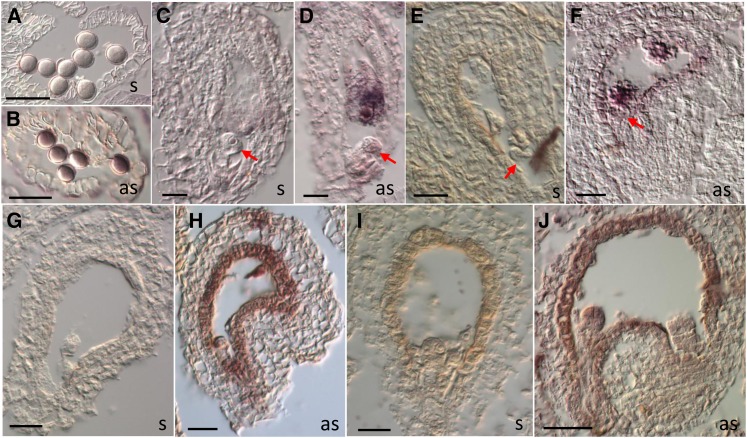
Figure 3.
ZIX Expression in Mature Gametophytes and Early Seeds.
In situ hybridization of ZIX RNAs in pollen ([A] and [B]), mature embryo sac ([C] and [D]), fertilized seeds at the zygotic stage ([E] and [F]), one-cell embryo stage ([G] and [H]), and eight-cell embryo stage ([I] and [J]). as, antisense probe; s, sense probe. Arrows point to the egg cell ([C] and [D]) or the zygote ([E] and [F]). Bars = 40 µm.
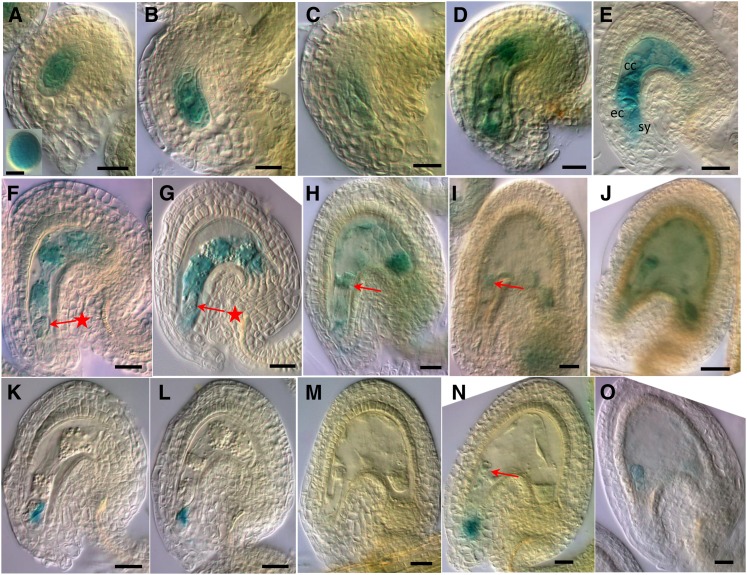
To better understand ZIX’s GME on early embryo and endosperm development, we focused our study on ZIX expression in the female gametophyte and early seeds from 1 to 3 DAP, when the zix phenotypes are manifest. In the ZIX:GUS (for β-glucuronidase) reporter lines, GUS expression was prominent in pollen and throughout the development of the female gametophyte, from the functional megaspore to the mature embryo sac (Figures 4A to 4E). In fertilized seeds of reciprocal crosses between the reporter lines and the wild type up to 3 DAP, activities of the maternal ZIX promoter were primarily detected in the early embryo and endosperm from the elongated zygote until at least the globular stage (Figures 4F to 4J; see Supplemental Figure 6 online). Notably, there was a pause in maternal ZIX promoter activity at the early zygote stage before elongation (Figures 4F and 4G), which was preceded by a low level of expression in the unfertilized egg cell (Figures 3D, 4D, and 4E). The paternal ZIX promoter, on the other hand, only became active in both the embryo and endosperm from the one-cell embryo stage onwards (Figures 4K to 4O; see Supplemental Figure 6 online). In some seeds, residual GUS activity of the ruptured pollen tube in the degenerating synergid space could be observed at the micropyle (Figures 4K, 4L, and 4N). Furthermore, for both parental alleles, GUS expression in the suspensor was detected at a later stage than in the embryo proper (Figures 4I, 4J, 4N, and 4O). These results suggest postfertilization de novo expression of both parental ZIX alleles in the embryo and of the paternal allele in the endosperm. Whether postfertilization maternal ZIX expression in the endosperm was due to carryover of prefertilization transcripts from the central cell and/or involved de novo expression could not be determined due to the continuous presence of GUS activity. This parent-of-origin–dependent expression pattern was observed in both the Ler and Columbia (Col) accessions harboring the ZIX reporter construct.
Figure 4.
ZIX Promoter Activities in Gametophytes and Early Seeds.
Prefertilization ovules or seeds at 1 to 3 DAP of ZIX:GUS lines from reciprocal crosses with wild-type plants were stained with GUS solution and imaged with DIC microscopy. cc, central cell; ec, egg cell; sy, synergids. Arrows with a star point to the early zygote ([F] and [G]); normal arrows point to the elongated zygote (H) or one-cell embryo ([I] and [N]). Bars = 4 µm in the inset in (A) and 20 µm in all others.
(A) Functional megaspore; inset: pollen.
(B) Two-nucleate embryo sac.
(C) Four-nucleate embryo sac.
(D) Eight-nucleate/seven-cell embryo sac.
(E) Four-cell embryo sac.
(F) to (J) Seeds from ZIX:GUS × wild-type crosses.
(K) to (O) Seeds from wild-type × ZIX:GUS crosses.
(F), (G), (K), and (L) Early zygote.
(H) and (M) Elongated zygote.
(I) and (N) One-cell embryo.
(J) and (O) Two-cell embryo.
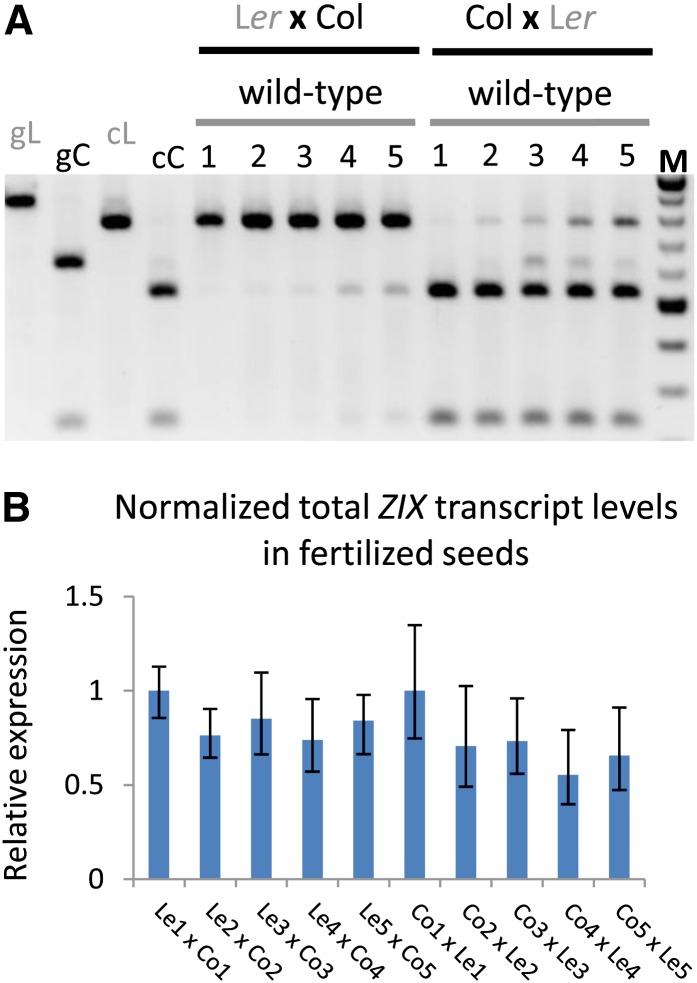
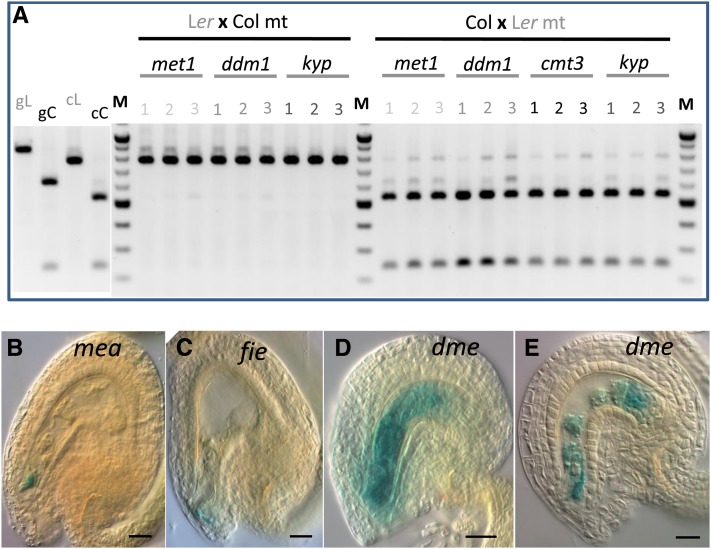
Using a single nucleotide polymorphism of a DraI restriction site present in the second exon of ZIX in the Col accession but not in the Ler accession (Jander et al., 2002; Ilic et al., 2004), we further investigated the parental origin of the endogenous ZIX transcripts in developing wild-type seeds of reciprocal crosses between these two accessions at 1 to 5 DAP. The results of RT-PCR followed by DraI restriction digestion of ZIX PCR products not only confirmed the preferential maternal postfertilization expression of ZIX but also showed an apparent increase of paternal ZIX transcript levels during early seed development, independent of the two genetic backgrounds studied (Figure 5A). Although we were not yet able to define the experimental parameters for allele-specific quantitative PCR (qPCR) to distinguish these two alleles, which differ by only a single nucleotide, qPCR for total ZIX expression levels in fertilized seeds 1 to 5 DAP showed that ZIX expression normalized to the expression of the reference gene ACTIN11 (ACT11) did not change significantly during this developmental window (Figure 5B). This result further underscores that the de novo paternal ZIX expression, despite increasing gradually, was still substantially lower than maternal ZIX expression as total ZIX transcript levels did not increase significantly.
Figure 5.
Allele-Specific ZIX Transcript Levels in Fertilized Seeds.
(A) Endogenous ZIX transcripts were amplified by RT-PCR and the PCR products digested with DraI and separated by agarose gel electrophoresis. The Col allele was smaller than the Ler allele after DraI digestion. Numbers refer to days after pollination. Genomic Ler and Col ZIX alleles were included in the assay as controls for genomic DNA contamination in the cDNA pool. cC, cDNA Col allele; cL, cDNA Ler allele; gC, genomic Col allele; gL, genomic Ler allele; M, DNA ladder.
(B) Relative ZIX expression levels in fertilized seeds of reciprocal crosses shown in (A) measured by qPCR and normalized by ACT11 expression. Means of expression levels were calculated from three technical replicates and two biological replicates. Error bars indicate se. Numbers refer to days after pollination. The primers used in these qPCRs generate products that span an exon-intron junction and hybridize specifically to ZIX cDNA (see Methods).
[See online article for color version of this figure.]
To investigate allele-specific ZIX expression in the developing young embryo, we examined the transcriptome RNA-sequencing data of isolated embryos from previous reports (Autran et al., 2011; Nodine and Bartel, 2012). Although the two Arabidopsis accessions used in Autran et al. (2011) are Ler and Col as in this study, maternal and paternal ZIX transcripts could not be distinguished in this data set. In the report by Nodine and Bartel (2012), the normalized ZIX maternal-to-paternal expression ratios for one-cell/two-cell, eight-cell, and 32-cell embryos of the Col × Cape Verde Islands crosses are 4.4, 1.76, and 1.57, respectively, and of the Cape Verde Islands × Col crosses are 0.7, 1.7, and 0.85, respectively. Since these ratios are inconsistent in reciprocal crosses, and since different Arabidopsis accessions were studied, we cannot draw any clear conclusion regarding allele-specific expression of ZIX in the embryo from these data.
In conclusion, the parent-of-origin–dependent expression patterns of ZIX revealed by both promoter reporter and allele-specific RT-PCR analyses provide the basis of the GME of the zix mutant during early embryo and endosperm development.
Regulation of Parent-of-Origin–Dependent ZIX Expression Is Distinct from That of Other Known GME Mutants
The differential expression of maternal and paternal ZIX alleles during early seed development raises the question of how parent-of-origin–dependent ZIX expression is regulated. We tested several trans-acting factors that were previously shown to repress silenced paternal alleles or to activate expressed maternal alleles of known genes with a GME on embryo and/or endosperm development (Vielle-Calzada et al., 1999; Yadegari et al., 2000; Jullien et al., 2006b). As repression is commonly associated with DNA and histone methylation, we tested whether the following genes are regulators of parent-of-origin–dependent ZIX expression: MET1, DDM1, and CHROMOMETHYLASE3 (CMT3) genes, which maintain CG and non-CG DNA methylation (Vongs et al., 1993; Lindroth et al., 2001; Kankel et al., 2003); KRYPTONITE (KYP), which establishes H3K9me2 and DNA methylation at CHG sites (Jackson et al., 2002); and the FIS-PRC2 genes MEA and FIE, which partially repress the paternal MEA allele by H3K27me3 (Baroux et al., 2006; Gehring et al., 2006; Jullien et al., 2006b; Wöhrmann et al., 2012).
To test whether a paternal hypomethylation background could alleviate paternal ZIX allele silencing, we evaluated the endogenous paternal ZIX transcript levels by RT-PCR in reciprocal crosses using wild-type plants and the DNA/histone hypomethylated mutants met1, ddm1, cmt3, and kyp as pollen donors in either the Ler or Col background. We could not detect any change of paternal ZIX expression levels in fertilized seeds up to 3 DAP, regardless of the mutant and accession used (Figure 6A). Moreover, hypomethylated pollen of these mutants rescued neither the early seed developmental arrest nor the seed abortion phenotype of zix. To examine whether the maternal mea or fie allele could derepress paternal ZIX, we monitored paternal ZIX promoter activities in mea and fie seeds fertilized by ZIX:GUS pollen. As in wild-type seeds, no paternal ZIX reactivation was observed (Figures 6B and 6C compared with Figures 4K to 4M). Collectively, these results indicate that early paternal ZIX repression in the zygote, young embryo, and early endosperm is not regulated by the DNA and histone methylation pathways mediated by these genes.
Figure 6.
Effects of Various Parental Mutant Backgrounds on Parent-of-Origin–Dependent ZIX Expression in Early Seeds.
(A) Endogenous ZIX transcript levels, as determined by RT-PCR, in early seeds fertilized by pollen from hypomethylated mutants (met1, ddm1, cmt3, and kyp). Numbers refer to days after pollination. cC, cDNA Col allele; cL, cDNA Ler allele; gC, genomic Col allele; gL, genomic Ler allele; M, DNA ladder; mt, mutant pollen.
(B) to (E) Promoter activities of paternal ZIX:GUS alleles in the mea (B) and fie (C) maternal background and of maternal ZIX:GUS alleles in the dme ([D] and [E]) maternal background. Bars = 20 µm.
Regarding the regulation of expression of the maternal ZIX allele, we tested whether the DNA glycosylase DME, which demethylates DNA, is required for activation, depending on the context, as is the case for both MEA and FIS2 (Baroux et al., 2006; Gehring et al., 2006; Jullien et al., 2006b; Wöhrmann et al., 2012). We crossed a ZIX:GUS line in the Col background with the dme-7 mutant and examined maternal ZIX promoter activity in unfertilized ovules or fertilized seeds of dme-7/DME;ZIX:GUS/- F1 plants. Fifty percent of the ovules or seeds in pistils of these plants carried the ZIX:GUS allele, and half of these also contained the dme-7 allele, while the other half harbored the wild-type DME allele. We observed that 53% (n = 246) of the unfertilized ovules and 51.6% (n = 184) of the fertilized seeds at 1.5 DAP showed maternal GUS expression similar to that in the wild type (Figures 6D and 6E compared with Figures 4E to 4G), indicating that maternal DME is not necessary for maternal ZIX expression.
Collectively, our results on the zix reproductive phenotype, the spatial and temporal expression of ZIX, and the regulation of its parent-of-origin–dependent expression pattern establish that ZIX represents a GME gene in plants that is not regulated by the pathways known to be involved in the regulation of other GME loci.
We further searched for potential regulators of ZIX expression by surveying public Arabidopsis epigenome databases for the presence of epigenetic marks at the ZIX locus. These database compilations integrate genome-wide data on DNA cytosine methylation (Vaughn et al., 2007; Chodavarapu et al., 2010; Cokus et al., 2008; Lister et al., 2008), histone methylation (Zhang et al., 2007; Bernatavichute et al., 2008; Oh et al., 2008; Zhang et al., 2009; Bouyer et al., 2011; Roudier et al., 2011), histone acetylation and ubiquitylation (Roudier et al., 2011), and the presence of small RNAs (Lister et al., 2008) of young seedlings or immature floral tissues. This analysis revealed a low level of DNA cytosine methylation at the 3′ end of ZIX only in the CG context, while most histone modification marks commonly associated with highly and broadly expressed genes, H3K4me1/2/3, H3K36me3, H2Bub, and H3K56Ac, were present in the ZIX gene body and, in some cases, the promoter (see Supplemental Figure 7 online), consistent with the broad expression pattern of ZIX (see Supplemental Figure 5 online). Histone modifications associated with genes having a low or tissue-specific expression (H3K27me3) or with transposons (H2K9me2), as well as small RNAs, were absent from the ZIX locus (see Supplemental Figure 7 online). Whether these epigenetic marks display a similar distribution at the ZIX locus in the gametophytes, embryo, and endosperm is not known. In the future, determining the epigenetic landscape in these reproductive tissues will bring us a step closer to understanding the regulation of parent-of-origin–dependent expression of ZIX and other genes with a GME.
The ZIX Protein Is Localized to Both Cytoplasm and Nuclei of Cells in Reproductive Tissues and Roots
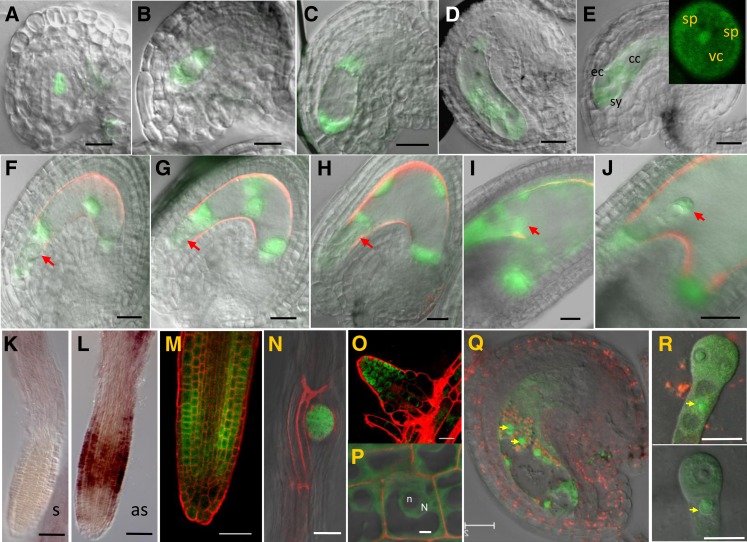
To study the subcellular localization of the ZIX protein, we introduced a ZIX-Green Fluorescent Protein (GFP) translational fusion construct driven by the native ZIX promoter (ProZIX:ZIX-GFP) into wild-type Ler plants. This fusion protein was functional, as the construct significantly increased zix allele transmission in zix/ZIX plants (Table 2), indicating successful complementation of the mutant phenotype. Corroborating the expression analysis of the ZIX:GUS reporter lines, in addition to expression throughout female gametophyte development (Figures 7A to 7D), ZIX-GFP was also detected in the vegetative cell but not the sperm cells of pollen (Figure 7E). On the other hand, ZIX-GFP was observed in accessory (synergids and antipodals) as well as gametic cells of the mature embryo sac (Figure 7E). In seeds, ZIX-GFP was detected from the one-cell embryo stage and two-nuclear endosperm stage to at least the early heart stage (Figures 7F to 7J; see Supplemental Figure 8 online). The absence of ZIX-GFP in the zygote (Figures 7F and 7G) was in agreement with the absence of ZIX promoter activity at this stage (Figures 4F, 4G, and 4K to 4M). Furthermore, as reported by the AtProteom database (see Supplemental Figure 5C online), ZIX transcripts (Figures 7K and 7L) and ZIX-GFP proteins (Figures 7M to 7P) were also observed in roots, in particular at the zones with active cellular proliferation (i.e., the root cell division zone [Figures 7L and 7M] and the lateral root initiation zone [Figures 7N and 7O]). In all cases, ZIX-GFP was localized in both the cytoplasm and the nucleus (Figures 7E and 7M to 7R), implicating ZIX in various cellular processes in which Arm proteins are known to play a role (Shapiro, 2001).
Figure 7.
ZIX-GFP Protein Localization in Gametophytes, Early Seeds, and Roots.
(A) to (J) Overlays of DIC and GFP pseudo-colored wide-field microscopy images. Red is background autofluorescence. Red arrows point to the early zygote ([F] and [G]), elongated zygote (H), or one-cell embryo (I).
(A) Functional megaspore.
(B) Two-nucleate embryo sac.
(C) Four-nucleate embryo sac.
(D) Eight-nucleate/seven-cell embryo sac.
(E) Four-cell embryo sac. cc, central cell; ec, egg cell; sy, synergids. Inset in (E): pollen grain. sp, sperm; vc, vegetative cell.
(F) and (G) Early zygote.
(H) Elongated zygote.
(I) One-cell embryo.
(J) Two-cell embryo.
(K) and (L) Whole-mount RNA in situ hybridization images of ZIX transcripts in roots. as, antisense probe; s, sense probe.
(M) to (R) Overlays of DIC, FM-64 (red), and GFP pseudo-colored confocal laser scanning microscopy images.
(M) Primary roots.
(N) Lateral root initiation zone.
(O) Emerging lateral roots.
(P) Close-up of root cells. N, nucleus; n, nucleolus.
(Q) Early seed with four-endosperm nuclei.
(R) Isolated two-cell (top panel) and one-cell embryos (bottom panel). Yellow arrows point to GFP-ZIX in the nuclei.
Bars = 20 µm in ovule, seeds, and embryo images, 40 µm in root images, and 5 µm in (P).
ZIX Regulates the Transcription Factor Gene MINI3 during Early Seed Development
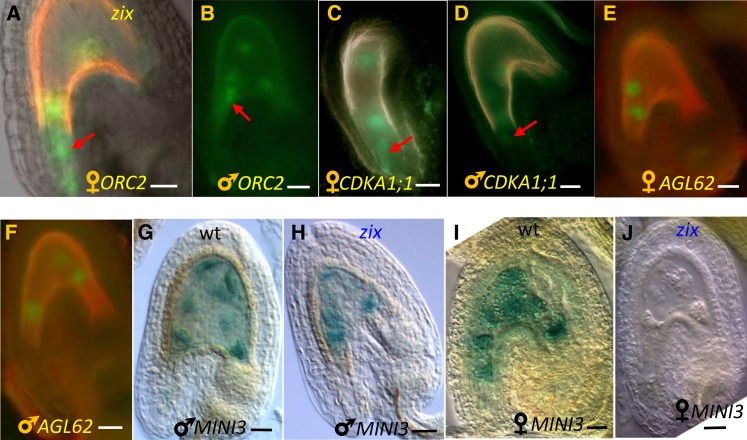
The presence of ZIX in actively dividing root tissues and the slow growth and early arrest of zix seeds suggest a link between ZIX function and cell cycle regulation. Thus, we examined whether the maternal zix mutant allele affects the expression of two Arabidopsis cell cycle genes, ORIGIN OF REPLICATION2 (ORC2) and CYCLIN DEPENDENT KINASE A1;1 (CDKA1;1). As both genes are biparentally active soon after fertilization, they could be studied in zix mutants prior to the early zix developmental arrest. ORC2 is a subunit of the Arabidopsis Origin of Replication Complex, and disruption of the gene leads to zygotic lethality due to embryo and endosperm arrest after a couple of cell cycles (Collinge et al., 2004). Mutated cdka1;1, a cyclin-dependent kinase, results in a delay of the second mitotic division of the generative cell, such that the sperm cells are only produced during pollen tube growth; consequently, only the sperm fertilizing the egg cell contributes its genome to the developing fertilization products, while the second sperm fertilizing the central cell does not (Nowack et al., 2006; Aw et al., 2010).
To examine the paternal allele expression of these genes in zix seeds, we monitored their promoter activities of the promoter:gene-GFP or -GUS lines used as pollen donors in crosses with zix/ZIX flowers. To study the maternal allele expression of these genes in zix seeds, we examined reporter gene expression in a zix embryo sac fertilized by wild-type sperm cells. No aberrant expression of either parental allele of both ORC2 and CDKA1:;1 reporters was detected in the zygote, one-cell embryo, and early endosperm of zix ovules at 2 DAP (Figures 8A to 8D), indicating that these genes are not regulated by ZIX. Overexpression of ZIX driven by the 35S promoter in wild-type plants did not confer any overgrowth phenotype of any tissues and organs. Therefore, an association of ZIX with cell cycle regulation cannot be established from these results. Whether other cell cycle genes functioning in early embryo and endosperm are affected by a lack of maternal ZIX expression remains an open question. Alternatively, ZIX might not be associated with cell cycle regulation, but rather with the competency to divide.
Figure 8.
Effects of Maternal zix Allele on the Expression of Other Genes in Early Seeds 2 DAP.
(A) Overlay of DIC and GFP pseudo-colored image.
(B) to (F) GFP pseudo-colored images (red, yellow, and pink are background autofluorescence).
(G) to (J) DIC images. wt, the wild type.
(A) and (B) Maternal and paternal ORC2:ORC2-GFP alleles in zix seeds.
(C) and (D) Maternal and paternal CDKA1;1:CDKA1;1-YFP alleles in zix seeds.
(E) and (F) Maternal and paternal AGL62:AGL62-GFP alleles in zix seeds.
(G) and (H) Paternal MINI3:GUS allele in wild-type and zix seeds.
(I) and (J) Maternal MINI3:GUS allele in wild-type and zix seeds.
Arrows point to the GFP expression in the zygote or one-cell embryo. Bars = 20 µm.
A well-characterized function of Arm proteins is their role as coactivators in conjunction with other transcription factors that initiate gene expression of cellular proliferation and differentiation pathways (Willert and Jones, 2006). Therefore, we evaluated whether the early biallelic expression of two transcription factors expressed early in the endosperm and/or embryo, AGAMOUS-LIKE62 (AGL62) (Kang et al., 2008) and MINI3 (Luo et al., 2005), was affected in zix seeds. Mutants disrupting either AGL62 (Kang et al., 2008) or MINI3 (Luo et al., 2005) show precocious endosperm cellularization. We examined the parental promoter activities of reporter lines for these genes (Luo et al., 2005; Bemer et al., 2010) in zix mutants as described above for ORC2 and CDKA1;1. At 2 DAP, AGL62 did not show any difference in parental expression in zix compared with wild-type seeds of a similar developmental stage (Figures 8E and 8F). On the other hand, although the paternal MINI3 expression in zix seeds (Figure 8H) was indistinguishable from that of wild-type seeds of a similar stage (Figures 8G), maternal MINI3 expression was lost (83%, n = 156 for zix-1; 94.5%, n = 182 for zix-3) (Figure 8J compared with Figure 8I) or strongly reduced in zix seeds (17%, n = 156 for zix-1; 5.5%, n = 182 for zix-3) (see Supplemental Figure 9 online), suggesting that MINI3 acts downstream of ZIX at least during early embryo and endosperm development. However, this loss of maternal expression of MINI3 alone is unlikely to be responsible for the GME of ZIX as MINI3 does not display a maternal effect on its own (Luo et al., 2005). Yet, when combined with the misregulation of other genes, MINI3 could be a contributing factor. Later effects of ZIX on MINI3-dependent endosperm differentiation are likely but cannot be evaluated due to the early GME seed abortion phenotype of zix.
DISCUSSION
ZIX Reveals a Novel Female Gametophytic Function of Arm Repeat Proteins in Plants
In animals and other nonplant eukaryotes, Arm proteins are involved in a variety of cellular processes vital for growth and development, such as cell–cell adhesion, signal transduction, nuclear translocation, and transcriptional regulation (reviewed in Hatzfeld, 1999; Coates, 2003; Mudgil et al., 2004). Likewise, plant Arm proteins exist in numerous forms and functions. More than 100 Arabidopsis proteins have been predicted to contain Arm repeats (Coates, 2003; Mudgil et al., 2004; Samuel et al., 2006); yet, the low primary sequence conservation of the repeats renders these predictions nonexhaustive. Several of the 20 plant Arm-encoding genes characterized and described to date, including ZIX, are not in the predicted list or come from other plant species (Brassica spp, tobacco [Nicotiana tabacum], tomato [Solanum lycopersicum], and potato [Solanum tuberosum]) (Rook et al., 2006; reviewed in Coates, 2007; Gebert et al., 2008; Masuda et al., 2008; Wang et al., 2009). The wide-ranging biological roles of plant Arm proteins encompass housekeeping and transcriptional regulation activities (Rook et al., 2006; Masuda et al., 2008; Wang et al., 2009), regulation of growth and development mediated by hormone signaling or photoperiod (Amador et al., 2001; Downes et al., 2003; El Refy et al., 2003; Kim et al., 2004; Coates et al., 2006), plant cell architecture (Oh et al., 2005; G. Yang et al., 2007; Gebert et al., 2008; Sakai et al., 2008), and programmed cell death in self-incompatibility reactions (Gu et al., 1998; Stone et al., 1999, 2003; Liu et al., 2007) or the hypersensitive response of innate immunity (Zeng et al., 2004; Palma et al., 2005; González-Lamothe et al., 2006; Mosher et al., 2006; Yang et al., 2006; W. Yang et al., 2007). While most of these plant Arm repeat–encoding genes play a role during vegetative development, only two are involved in reproductive development: TWO-IN-ONE is essential for cytokinesis of sporophytic growth as well as male and female gametophyte development (Oh et al., 2005), and ARMADILLO REPEAT ONLY1 is required during pollen tube growth (Gebert et al., 2008). To date, ZIX is the only reported Arm protein in plants with the function of promoting early embryo and endosperm growth and development, achieved through a GME that acts on the earliest stages of sporophyte development.
The delayed and arrested growth phenotype of zix as well as the presence of ZIX in actively dividing root zones suggest that ZIX functions during nuclear/cellular proliferation. Although the molecular mechanisms through which ZIX accomplishes its developmental role remain unknown, the ZIX gene structure and protein localization provide some hints for speculation on its acting mechanism. At the tertiary sequence level, ZIX resembles importin-α and β-catenin. Since ZIX does not contain the importin β-binding domain characteristic of importin-α, ZIX is more likely to be functionally similar to β-catenin. The cytoplasmic and nuclear localization of ZIX is also similar to that of β-catenin. In human cells, β-catenin remains in the cytoplasm as an adaptor of E-cadherin for proper cellular adhesion (Shapiro, 2001). Triggered by a Wnt signaling cascade, β-catenin translocates into the nucleus to form a coactivator complex with other transcription factors and chromatin remodeling factors to activate gene expression of the cellular proliferation and/or differentiation pathways (Shapiro, 2001; Willert and Jones, 2006). Along this line, we can postulate that during Arabidopsis early seed development or in the mitotically active zones of the root, cellular proliferation is stimulated by a developmental cue. Upon triggering by this signal, ZIX shuttles from the cytoplasm to the nucleus to effect the transcriptional activation of genes essential for nuclear/cellular proliferation or differentiation, such as the transcription factor MINI3, which regulates the timing of endosperm differentiation (Luo et al., 2005). Additional biochemical and cytological studies will be needed to test this hypothesis.
Of particular interest is the Ataxin-10 domain at the C terminus of ZIX, which is conserved across eukaryotic kingdoms and thus likely confers conserved functions for all ZIX homologs. Despite being annotated as “Ataxin-10 related,” no mutations in this domain for any eukaryote have been reported to be linked to the ataxia phenotype, a human neurological disorder (Matsuura et al., 2000). Moreover, the ataxia phenotype is associated with the gross expansion of the pentanucleotide ATTCT repeat in intron 9 of human Ataxin-10 (Matsuura et al., 2000), and this expansion does not interfere with the full-length Ataxin-10 transcript processing in Ataxin-10 patients (Wakamiya et al., 2006). These results do not substantiate the relevance of this “Ataxin-10 related” domain to the ataxia phenotype, especially for Arabidopsis where the only ZIX intron contains no such pentanucleotide repeat. Thus, the biological function of this conserved domain awaits further investigation.
The sporophytic effects of zix could not be assessed in this study due to its gametophytic lethality and the inability to recover zix homozygotes. In mice, the Ataxin-10 null homozygotes die at early postimplantation (Wakamiya et al., 2006). On the other hand, mouse Ataxin-10 does not exert a maternal effect on embryo development because litters of intercrossed Ataxin-10 null heterozygous mice segregate two heterozygotes:one wild type, typical for zygotic embryonic lethal mutants (Wakamiya et al., 2006). Nevertheless, one can still surmise that ZIX homologs in both animals and plants play certain common biological roles during early embryonic development despite the unknown specific cellular processes.
zix Represents a Distinct Class of GME Mutants Affecting Seed Development
Previously, we used our genetic analysis of the loss-of-function mutant glauce, the molecular identity of which has recently been discovered (Leshem et al., 2012), to demonstrate that early seed development requires both maternal growth promoters and growth repressors (Ngo et al., 2007). In this study of the loss-of-function zix mutant, we present molecular evidence for a growth-promoting maternal effect. Maternally acting negative regulators of embryo and endosperm proliferation are represented by the extensively characterized fis class of mutants, which includes MEA (Grossniklaus et al., 1998; Kiyosue et al., 1999; Luo et al., 1999), FIS2 (Luo et al., 1999), FIE (Ohad et al., 1999), and MSI1 (Köhler et al., 2003; Guitton et al., 2004). These Arabidopsis FIS-PRC2 genes repress embryo and endosperm growth, in a fertilization-dependent or -independent manner (Vielle-Calzada et al., 1999; Luo et al., 2000; Yadegari et al., 2000; Spillane et al., 2000; Köhler et al., 2003; Leroy et al., 2007). ZIX parent-of-origin–dependent expression patterns are spatially and temporally different from those of the FIS-PRC2 genes as well as all other genes with a known GME on seed development, such as PRL (Springer et al., 2000), DME (Choi et al., 2002), XPO1 (Blanvillain et al., 2008), MPC (Tiwari et al., 2008), At-FH5 (Fitz Gerald et al., 2009), and At-LIG1 (Andreuzza et al., 2010). These other genes are expressed prior to fertilization in the embryo sac (with the exception of At-FH5) and either maternally in the fertilized endosperm or biparentally in both the embryo and the endosperm from the zygote stage onwards. ZIX, on the other hand, exhibits a biphasic expression pattern correlating with phenotypes at two distinct developmental stages. Before fertilization, ZIX is expressed throughout the embryo sac, and mutant female gametophyte maturation is delayed. Soon after fertilization, an ephemeral “off” state of the maternal ZIX allele in the early zygote precedes the “on” state primarily from the late zygote/one-cell embryo onwards, and this “off-on” switch differs from the continuous maternal expression in the endosperm. Such a difference between embryo and endosperm has also been reported for certain epigenetic marks and the requirement for polymerase II (Pillot et al., 2010).
Expression of ZIX is not observed in the sperm cells, and paternal ZIX expression after fertilization starts at the one-cell embryo stage concomitantly in the embryo and endosperm. Therefore, ZIX products are missing in young zix mutant seeds due to a silent paternal allele and a mutated maternal allele, likely causing the very early growth delay and growth arrest seen in zix seeds. These biphasic expression patterns and developmental phenotypes of ZIX have two implications. First, in wild-type seeds, the absence of ZIX activity in the zygote in comparison to the early endosperm might reflect the distinct chromatin states in the two fertilization products as reported at the global scale of the nucleus (Pillot et al., 2010). The chromatin state of ZIX in the early zygote might have been specifically reset from being active to inactive, rather than the active state being inherited from the egg cell. Second, although paternal ZIX expression in the embryo and endosperm is clearly de novo, as is maternal ZIX in the embryo, maternal ZIX transcript levels in the endosperm could be contributed by both the prefertilization transcripts deposited in the central cell and the de novo expression. Although the source of maternal dominance of ZIX postfertilization expression as revealed by the RT-PCR assay could not be pinpointed to the embryo or the endosperm, ZIX might be imprinted in either or both tissues.
If the postfertilization seed phenotype is not entirely due to the ZIX gametophytic expression and deposition, but rather also depends on zygotic transcription of the maternal ZIX allele, this would argue for the existence of imprinted ZIX expression in the embryo. Current data cannot provide a definitive answer as to whether ZIX is imprinted in the embryo or not. Yet, this is an important question for future investigation for two reasons. First, the existence of genomic imprinting in the plant embryo is still debated (see discussion in Raissig et al., 2011); second, it would shed light on the parental conflict theory for the evolution of imprinting. The parental conflict theory predicts that maternally expressed imprinted genes suppress offspring growth, while paternally expressed imprinted genes have the opposite effect (Haig and Westoby, 1989; Haig and Westoby, 1991). At present, the parental conflict theory is consistent with all developmental phenotypes that are caused by disruption of imprinted genes in plants, although this is based on an extremely small number of imprinted genes for which the function is known (Raissig et al., 2011). However, although predominantly maternally expressed, ZIX promotes growth. Thus, should ZIX indeed be regulated by genomic imprinting, this would constitute a first exception.
Another feature distinguishing the zix mutant from other GME mutants is that during early seed development, paternal ZIX repression and maternal ZIX activation are not regulated by several well-characterized DNA and histone methylation pathways or by the DNA demethylation pathway, respectively, which regulate other known GME loci. Neither a release of paternal ZIX allele early silencing in mutants affecting these pathways nor rescue of seed abortion by hypomethylated pollen, as reported for the mea, fis2, fie, and athfh5 mutants (Luo et al., 1999; Vielle-Calzada et al., 1999; Vinkenoog et al., 2000; Yadegari et al., 2000; Baroux et al., 2006; Gehring et al., 2006; Jullien et al., 2006b; Fitz Gerald et al., 2009), was observed for zix. Likewise, DNA demethylation by the DNA glycosylase DME, which is involved in the activation of the maternal MEA and FIS2 alleles (Gehring et al., 2006; Jullien et al., 2006b), is not involved in the regulation of the maternal ZIX allele. However, the list of genes implicated in silencing or activation by DNA methylation and/or histone modification in this study is not exhaustive. Public epigenome databases do not reveal the presence of small RNAs at the ZIX locus in young seedlings or inflorescences. However, the small RNA landscape of the ZIX locus in the embryo and endosperm could be different. Therefore, we cannot exclude that the maternal polymerase IV–dependent small RNA pathway, recently discovered to regulate many paternally repressed genes during early embryo development (Autran et al., 2011), may also regulate the paternal ZIX allele. An extended search for paternal ZIX regulators should thus include mutants affecting small RNA pathways in addition to mutants affecting various types of histone modifications and higher order chromatin organization (Shahbazian and Grunstein, 2007). On the maternal side, whether the deposition of other transcriptionally permissive marks by histone modifications (Shahbazian and Grunstein, 2007) can activate the maternal ZIX allele during early seed development remains to be determined.
In addition to 55 other uncharacterized mee mutants (Pagnussat et al., 2005), a GME mutant with a very early seed developmental arrest phenotype similar to that of zix was described for capulet1 (Grini et al., 2002). It is not known whether the genes disrupted in these mutants are maternal growth promoters like zix and whether there is a common mechanism underlying their GME. Further research on this mutant class would reveal whether the GME manifest by ZIX imposes a more widespread early growth promotion effect than currently documented.
METHODS
Plant Materials
zix-1, zix-2, and zix-3 are Ds insertion lines in the Ler background (Pagnussat et al., 2005). The met1 allele in the Col background was generated by six backcross generations of the original met1 allele in the Ler background (Eric Richards, personal communication). ddm1-1, cmt3-7 (CS6365), kyp-2 (CS6367), mea-1, and fie-2 are in the Ler background. ddm1-2, kyp-4 (SALK_044606), and dme-7 (SALK_075424) are in the Col background. All T-DNA insertion lines were obtained from the ABRC stock center (www.Arabidopsis.org). Homozygous met1, ddm1, and cmt3 mutants were selected from a segregating population of heterozygous parents. Plant selection on antibiotic plates and plant growth conditions were as previously described (Ngo et al., 2007).
Mutant Allele Identification and Confirmation
Genomic DNA extraction and Thermal Asymmetric Interlaced-PCR to identify the insertion sites of the Ds elements in the three zix alleles were performed as previously reported (Ngo et al., 2007). The insertion 3′ and 5′ ends were confirmed by PCR with Ds primers and gene-specific primers (see Supplemental Table 1 online). met1, ddm1, and cmt3 mutant alleles were identified by the presence of cleaved-amplified polymorphic sequence markers that distinguish the mutant allele from the wild-type allele (primers and PCR conditions in Supplemental Table 1 online). kyp, mea, and fie mutant alleles were identified as described elsewhere (Baroux et al., 2006; Autran et al., 2011). Genotyping of the T-DNA insertion in dme-7 allele was performed with the LBb3.1 primer (5′-ATTTTGCCGATTTCGGAAC-3′) and dme-7 LP and dme-7 RP primers (see Supplemental Table 1 online).
RNA Expression Studies
RNA in situ hybridization for ZIX transcripts on sectioned reproductive tissues was performed as reported elsewhere (Wuest et al., 2010). ZIX sense and antisense RNA probes were in vitro synthesized from a pDrive vector (Qiagen) harboring a partial ZIX cDNA amplified by the two primers 5′-ATGGAAGCTTCTCTACCGGAA-3′ and 5′-GCCTTGACAGCTACAAAGC-3′. One-hundred ng of the respective probes were used per slide of sectioned pistils. Development of signals from digoxigenin-conjugated probes was run overnight. Whole amount RNA in situ hybridization on roots was performed on 10-d-old seedlings as described elsewhere (Drea et al., 2009), with a modified Proteinase K treatment of 1.4 µg/mL after dehydration but before the acetylation reaction. Hybridization was performed with 50 ng of probes per sample at 55°C and the signal development was stopped after 3 h.
For RT-PCR analysis of endogenous ZIX transcripts, Arabidopsis thaliana ovules and seeds freshly dissected from 15 to 50 pistils before and after manual pollination were immediately frozen in liquid nitrogen. Total RNA extraction from the ground tissues was performed with 2× CTAB extraction buffer (2% CTAB, 2 M NaCl, 100 mM Tris-HCl, and 25 mM EDTA both at pH8, and 2% mercaptoethanol freshly added before extraction) at 65°C for 30 min, RNase-free DNaseI treated at 37°C for 15 min, extracted with an equal volume of chloroform, precipitated with 3 M LiCl (final concentration) after 2 h of incubation at −80°C, washed with 70% ethanol, dissolved in 20 μL of Diethylpyrocarbonate-treated water, and quantified with the Nanodrop ND6000. All RNA samples displayed the A260/A280 ratio of 2.0 to 2.3. Two micrograms of extracted RNA from each sample was then reverse transcribed with SuperScriptII (Invitrogen) in the presence of RNaseOut (Invitrogen) in a total volume of 20 µL. Two microliters of cDNAs from the reverse transcription reactions was used for allele-specific PCR (primers and PCR parameters in Supplemental Table 1 online). Five microliters of PCR products was digested with DraI (New England Biolabs) overnight at 37°C, and the digests were imaged by UV light after electrophoretic separation on Tris-acetate-EDTA gels.
For real-time qPCR of total ZIX transcripts in 1 to 5 DAP seeds of reciprocal crosses between the Ler and Col accessions, two biological replicates and three technical replicates were performed in 20-μL reactions. One microgram of total RNA from each biological replicate was reversed transcribed into cDNAs in a 20-μL reaction, and 1 μL of the 1:5 dilution of the cDNAs was used for subsequent PCR. ACTIN11, a gene stably expressed in the early embryo and endosperm, was used as reference for transcript level normalization. qPCR primers for ZIX (forward, 5′-TGTCAAGGGCTCACCATCATCG-3′, and reverse, 5′-AATAATCCTCCACAGACCCAACGG-3′) and for ACT11 (forward, 5′-GTGGTCGTACTACTGGTATTGTGTTG-3′, and reverse, 5′-CGCAGAATAGCATGTGGAAGAGC-3′) were designed such that the PCR product spans an exon-intron junction of each gene so that only ZIX or ACT11 cDNA was amplified with the incorporated SYBR Green I dye in an Applied Biosystems 7500 Fast Real-time PCR machine. For the amplification phase, the parameters were as followed: 50°C/2 min, 95°C/10 min, 40 cycles of 95°C/15 s and 67°C/1 min. For the melting curve phase, the parameters included 95°C/15 s, 60°C/1 min, 95°C/15 s, and 60°C/30 s. All samples displayed only one melting curve peak. No amplification was detected in any of the non-DNA and genomic DNA controls. The relative quantification of ZIX expression normalized by ACT11 expression was analyzed by the ΔΔCT (cycle threshold) method (Livak and Schmittgen, 2001).
Plasmid Construction and Plant Transformation
For mutant complementation, a 4-kb fragment of At4g00231, including 2 kb upstream of the ATG start codon, the gene body of two exons and the middle intron, and 0.4 kb of the 3′ untranslated region was amplified from the genomic DNA of wild-type Ler leaves by PCR with primers containing the adaptors and BP sites for Gateway cloning (primers in Supplemental Table 1 online). The PCR products were cloned into pDONR221 (Invitrogen) by the BP reaction, and the LR reaction was performed between the pDONR221 containing the genomic At4g00231 fragment and the destination binary vector pMDC123 (Curtis and Grossniklaus, 2003). The final selected pMDC123 clone containing the genomic At4g00231 fragment was sequence verified before being introduced into the Agrobacterium tumefaciens strain GV3101 to shuttle this binary vector into zix/ZIX plants by the floral dip method (Clough and Bent, 1998). T1 transgenic mutant plants carrying the T-DNA harboring the genomic At4g00231 fragment and the Basta resistance gene were selected on Murashige and Skoog (MS) plates containing 50 mg/L Kan and 10 mg/L glufosinate. Complementation assays were performed with T2 transgenic mutant plants selected on MS plates containing only 50 mg/L Kan.
For ZIX:GUS promoter reporter lines and ZIX:ZIX-GFP fusion lines, the genomic fragment covering the 2 kb upstream region of the At4g00231 start codon and the genomic fragment used for the complementation assay excluding the stop codon and the 3′ untranslated region, respectively (primers in Supplemental Table 1 online), were PCR amplified from genomic DNA of Ler leaves and cloned into pDONR207 by the BP reactions (Invitrogen). LR reactions were performed for the BP products with the pMDC164 and pMDC111 plasmids, respectively (Curtis and Grossniklaus, 2003). For the ORC2:ORC2-GFP line, the At-ORC2 (At2g37560) genomic sequence was PCR amplified with the primers Orc2gsPac (5′-CGTTAATTAAACGGGAGAACAACTGATGGG-3′) and Orc2gaAsc (5′-TGCGCGCCACTGATTGAGATCAAGCAAAAGCTG-3′) and digested with PacI and AscI, and the digested PCR product was ligated into the vector pMDC110 (Curtis and Grossniklaus, 2003), the Gateway cassette of which had been excised between these two sites. Destination vectors carrying the transgenes ZIX:GUS, ZIX:ZIX-GFP, or ORC2:ORC2-GFP were sequence verified prior to Agrobacterium transformation of wild-type and zix/ZIX plants. T1 transgenic plants with the fusion constructs in the wild-type background were selected on MS plates containing 20 mg/L hygromycin and in the zix/ZIX background on MS plates containing 50 mg/L Kan and 20 mg/L hygromycin.
Microscopy Analysis
The clearing and GUS staining procedures for mutant phenotypic characterization and GUS expression analysis of ovules, seeds, and pollen were performed as previously described (Ngo et al., 2007). For GFP examination in ovules and seeds, the pistils were dissected freshly in water to expose the ovules and seeds, which were embedded in water during observation with a Leica DM6000 microscope. Differential interference contrast (DIC) and GFP images were taken sequentially, processed, and superimposed by Adobe Photoshop or the GNU Image Manipulation Program v2.6 (http://gimpshop.com/). The GFP signals of isolated embryos, endosperm, and roots were examined by a Leica SP2 confocal laser scanning microscope as described elsewhere (Pillot et al., 2010).
Bioinformatics and Sequence Analysis
Expression data of ZIX were extracted from the public database links at The Arabidopsis Information Resource website (http://www.Arabidopsis.org). BLASTp of the ZIX virtual translated sequence was done with default parameters (http://blast.ncbi.nlm.nih.gov/Blast.cgi). The secondary structure and putative Arm repeat motifs of ZIX were predicted by HHpred (http://toolkit.lmb.uni-muenchen.de/hhpred) with the following selected HHM databases: pdb70_16Jan10, interpro_16.2, pfamA_23.0, smart_17Jun09, and pfam_17Jun09. Based on these results, ZIX putative Arm repeats were manually aligned with the consensus Arm repeat sequences of pfam00514, smart00185, and the known Arm repeats of the yeast importin-α (Karyopherin) and the mouse β-catenin extracted from the protein database SMART (http://smart.embl-heidelberg.de/). ZIX three-dimensional models were predicted by I-TASSER (http://zhanglab.ccmb.med.umich.edu/I-TASSER) and PHYRE (http://www.sbg.bio.ic.ac.uk/phyre/). Chromatin mark and small RNA distributions at the ZIX locus were extracted from the Arabidopsis epigenome public databases available at http://neomorph.salk.edu/epigenome/epigenome.html, http://epigara.biologie.ens.fr/cgi-bin/gbrowse/a2e/, https://www.mcdb.ucla.edu/Research/Jacobsen/LabWebSite/P_EpigenomicsData.shtml, and http://chromatin.cshl.edu/cgi-bin/gbrowse/epivariation/. The data from all databases were compared, and the common data were selected for presentation in the supplemental data.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative databases under the following The Arabidopsis Information Resource accession numbers: ZIX (AT4G00231), 505006409; MET1 (AT5G49160), 2155959; DDM1 (AT5g66750), 2173644; CMT3 (AT1G69770), 2205015; KYP (AT5G13960), 2159133; MEA (AT1G02580), 2196110; FIE (AT3G20740), 2091876; DME (AT5G04560), 2184432; CDKA1;1 (AT3G48750), 2099478; ORC2 (AT2G37560), 2040706; AGL62 (AT5G60440), 2175188; MINI3 (AT1g55600), 2020467; and ACT11 (AT3G12110), 2099302.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Alignment of the ZIX C Terminus with the Ataxin-10–Related Domain pfam09759 of Other Eukaryotes.
Supplemental Figure 2. Predicted Secondary Structure of ZIX by HHpred Based on Sequence Alignment with Proteins of Known Structures.
Supplemental Figure 3. Alignment of Putative ZIX Arm Repeat Motifs with Known Arm Repeats.
Supplemental Figure 4. Two Top 3D Thread Models of ZIX Predicted by I-TASSER Using Proteins of Known Crystallized Structures.
Supplemental Figure 5. Expression Data of ZIX Transcripts and Proteins from Public Databases.
Supplemental Figure 6. ZIX Promoter Activities in Early Seeds.
Supplemental Figure 7. Chromatin Mark and Small RNA Distributions at ZIX Locus from Public Databases.
Supplemental Figure 8. ZIX-GFP Protein Localization in Early Seeds.
Supplemental Figure 9. Maternal MINI3:GUS Expression in 1 DAP Wild-Type Seed and 2 DAP zix Seeds.
Supplemental Table 1. Primers and PCR Parameters.
Supplementary Material
Acknowledgments
The met1/MET1 and ddm1/DDM1 seeds were gifts from Eric Richards (Cornell University, Ithaca, NY), fie-2/+ seeds from Abed Chaudhury (Commonwealth Scientific and Industrial Research Organization, Plant Industry, Australia), CDKA1;1:CDKA-YFP seeds from Moritz Nowack (University of Cologne, Germany), AGL62:GFP-GUS seeds from Marian Bemer (Radboud University, Nijmegen, The Netherlands), and MINI3:MINI3-GUS seeds from Ming Luo (Commonwealth Scientific and Industrial Research Organization, Plant Industry). We thank Jacqueline Gheyselinck (University of Zurich) for instructions on in situ hybridization, Johan Jaenisch and Sharme Thirugnanarajah (University of Zurich) for genotyping the dme-7 plants, Anja Schmidt (University of Zurich) for helpful discussions on seed RNA isolation, Marian Bemer and Bruno Müller (University of Zurich) for valuable tips on qPCR, Pat Hogan (University of California, Davis, CA), Arturo Bolanos, Christof Eichenberger, and Peter Kopf (University of Zurich) for general lab support, Valeria Gagliardini (University of Zurich) for assistance with sequencing the genomic complementation construct, and Wei-Cai Yang (Institute of Genetics and Developmental Biology, Beijing, China) for sharing unpublished information confirming the phenotype of zix. We thank Sandra Noble (Foundation for the Advancement of Mesoamerican Studies, Crystal River, FL) for information on the goddess Zak Ixik and two anonymous reviewers for their insightful comments and constructive criticism on the article. This work was supported by the University of Zurich, a grant from the Swiss National Science Foundation to U.G., National Science Foundation Grants IOS-0313501 and IOS-0745167 to V.S., and, in part, through the National Science Foundation International Research Fellowship Program Award 0754305 to Q.A.N.
AUTHOR CONTRIBUTIONS
C.B. performed confocal laser scanning microscopy for ZIX-GFP. D.G. performed whole-mount in situ hybridization of ZIX RNA on roots. P.M. and M.A.C. generated the ORC2:ORC2-GFP lines. Q.A.N. conceived and designed the study, performed all other experiments, analyzed the data, and wrote the first draft of the article. V.S. and U.G. provided the infrastructure, reagents and materials, and helped in data analysis. C.B., V.S., and U.G. contributed to writing the article. All authors critically read and commented on the article and approved of its final version for submission.
Glossary
- GME
gametophytic maternal effect
- FIS-PRC2
FIS Polycomb Repressive Complex 2
- H3K27me3
histone H3 Lys-27
- Ler
Landsberg erecta
- Kan
kanamycin
- DAP
days after pollination
- GUS
β-glucuronidase
- Col
Columbia
- qPCR
quantitative PCR
- MS
Murashige and Skoog
- GFP
green fluorescent protein
- DIC
differential interference contrast
References
- Amador V., Monte E., García-Martínez J.L., Prat S. (2001). Gibberellins signal nuclear import of PHOR1, a photoperiod-responsive protein with homology to Drosophila armadillo. Cell 106: 343–354 [DOI] [PubMed] [Google Scholar]
- Andreuzza S., Li J., Guitton A.E., Faure J.E., Casanova S., Park J.S., Choi Y., Chen Z., Berger F. (2010). DNA LIGASE I exerts a maternal effect on seed development in Arabidopsis thaliana. Development 137: 73–81 [DOI] [PubMed] [Google Scholar]
- Autran D., et al. (2011). Maternal epigenetic pathways control parental contributions to Arabidopsis early embryogenesis. Cell 145: 707–719 [DOI] [PubMed] [Google Scholar]
- Aw S.J., Hamamura Y., Chen Z., Schnittger A., Berger F. (2010). Sperm entry is sufficient to trigger division of the central cell but the paternal genome is required for endosperm development in Arabidopsis. Development 137: 2683–2690 [DOI] [PubMed] [Google Scholar]
- Baerenfaller K., Grossmann J., Grobei M.A., Hull R., Hirsch-Hoffmann M., Yalovsky S., Zimmermann P., Grossniklaus U., Gruissem W., Baginsky S. (2008). Genome-scale proteomics reveals Arabidopsis thaliana gene models and proteome dynamics. Science 320: 938–941 [DOI] [PubMed] [Google Scholar]
- Baroux C., Gagliardini V., Page D.R., Grossniklaus U. (2006). Dynamic regulatory interactions of Polycomb group genes: MEDEA autoregulation is required for imprinted gene expression in Arabidopsis. Genes Dev. 20: 1081–1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman A., et al. (2004). The Pfam protein families database. Nucleic Acids Res. 32(Database issue): D138–D141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayer M., Nawy T., Giglione C., Galli M., Meinnel T., Lukowitz W. (2009). Paternal control of embryonic patterning in Arabidopsis thaliana. Science 323: 1485–1488 [DOI] [PubMed] [Google Scholar]
- Bemer M., Heijmans K., Airoldi C., Davies B., Angenent G.C. (2010). An atlas of type I MADS box gene expression during female gametophyte and seed development in Arabidopsis. Plant Physiol. 154: 287–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernatavichute Y.V., Zhang X., Cokus S., Pellegrini M., Jacobsen S.E. (2008). Genome-wide association of histone H3 lysine nine methylation with CHG DNA methylation in Arabidopsis thaliana. PLoS ONE 3: e3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanvillain R., Boavida L.C., McCormick S., Ow D.W. (2008). Exportin1 genes are essential for development and function of the gametophytes in Arabidopsis thaliana. Genetics 180: 1493–1500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourne P.E., et al. (2004). The distribution and query systems of the RCSB Protein Data Bank. Nucleic Acids Res. 32(Database issue): D223–D225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouyer D., Roudier F., Heese M., Andersen E.D., Gey D., Nowack M.K., Goodrich J., Renou J.P., Grini P.E., Colot V., Schnittger A. (2011). Polycomb Repressive Complex 2 controls the embryo-to-seedling phase transition. PLoS Genet. 7: e1002014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brukhin V., Curtis M., Grossniklaus U. (2005). The female gametophyte: no longer the forgotten generation. Curr. Sci. 89: 1844–1852 [Google Scholar]
- Chodavarapu R.K., et al. (2010). Relationship between nucleosome positioning and DNA methylation. Nature 466: 388–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y., Gehring M., Johnson L., Hannon M., Harada J.J., Goldberg R.B., Jacobsen S.E., Fischer R.L. (2002). DEMETER, a DNA glycosylase domain protein, is required for endosperm gene imprinting and seed viability in Arabidopsis. Cell 110: 33–42 [DOI] [PubMed] [Google Scholar]
- Christensen C.A., Subramanian S., Drews G.N. (1998). Identification of gametophytic mutations affecting female gametophyte development in Arabidopsis. Dev. Biol. 202: 136–151 [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Coates J.C. (2003). Armadillo repeat proteins: Beyond the animal kingdom. Trends Cell Biol. 13: 463–471 [DOI] [PubMed] [Google Scholar]
- Coates J.C. (2007). Armadillo repeat proteins: Versatile regulators of plant development and signalling. In Plant Growth Signaling, Vol. 10, L. Bӧgre and G. Beemster, eds (Berlin, Heidelberg: Springer-Verlag), pp. 299–314.
- Coates J.C., Laplaze L., Haseloff J. (2006). Armadillo-related proteins promote lateral root development in Arabidopsis. Proc. Natl. Acad. Sci. USA 103: 1621–1626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cokus S.J., Feng S., Zhang X., Chen Z., Merriman B., Haudenschild C.D., Pradhan S., Nelson S.F., Pellegrini M., Jacobsen S.E. (2008). Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature 452: 215–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collinge M.A., Spillane C., Köhler C., Gheyselinck J., Grossniklaus U. (2004). Genetic interaction of an origin recognition complex subunit and the Polycomb group gene MEDEA during seed development. Plant Cell 16: 1035–1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis M.D., Grossniklaus U. (2003). A Gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downes B.P., Stupar R.M., Gingerich D.J., Vierstra R.D. (2003). The HECT ubiquitin-protein ligase (UPL) family in Arabidopsis: UPL3 has a specific role in trichome development. Plant J. 35: 729–742 [DOI] [PubMed] [Google Scholar]
- Drea S., Derbyshire P., Koumproglou R., Dolan L., Doonan J.-H., Shaw P. (2009). In situ analysis of gene expression in plants. Methods Mol. Biol. 513: 229–242 [DOI] [PubMed] [Google Scholar]
- El Refy A., Perazza D., Zekraoui L., Valay J.G., Bechtold N., Brown S., Hülskamp M., Herzog M., Bonneville J.M. (2003). The Arabidopsis KAKTUS gene encodes a HECT protein and controls the number of endoreduplication cycles. Mol. Genet. Genomics 270: 403–414 [DOI] [PubMed] [Google Scholar]
- Evans M.M., Kermicle J.L. (2001). Interaction between maternal effect and zygotic effect mutations during maize seed development. Genetics 159: 303–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldmann K.A., Coury D.A., Christianson M.L. (1997). Exceptional segregation of a selectable marker (KanR) in Arabidopsis identifies genes important for gametophytic growth and development. Genetics 147: 1411–1422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitz Gerald J.N., Hui P.S., Berger F. (2009). Polycomb group-dependent imprinting of the actin regulator AtFH5 regulates morphogenesis in Arabidopsis thaliana. Development 136: 3399–3404 [DOI] [PubMed] [Google Scholar]
- Gebert M., Dresselhaus T., Sprunck S. (2008). F-actin organization and pollen tube tip growth in Arabidopsis are dependent on the gametophyte-specific Armadillo repeat protein ARO1. Plant Cell 20: 2798–2814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring M., Huh J.H., Hsieh T.F., Penterman J., Choi Y., Harada J.J., Goldberg R.B., Fischer R.L. (2006). DEMETER DNA glycosylase establishes MEDEA polycomb gene self-imprinting by allele-specific demethylation. Cell 124: 495–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Lamothe R., Tsitsigiannis D.I., Ludwig A.A., Panicot M., Shirasu K., Jones J.D. (2006). The U-box protein CMPG1 is required for efficient activation of defense mechanisms triggered by multiple resistance genes in tobacco and tomato. Plant Cell 18: 1067–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grini P.E., Jürgens G., Hülskamp M. (2002). Embryo and endosperm development is disrupted in the female gametophytic capulet mutants of Arabidopsis. Genetics 162: 1911–1925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossniklaus U., Schneitz K. (1998). The molecular and genetic basis of ovule and megagametophyte development. Semin. Cell Dev. Biol. 9: 227–238 [DOI] [PubMed] [Google Scholar]
- Grossniklaus U., Vielle-Calzada J.P., Hoeppner M.A., Gagliano W.B. (1998). Maternal control of embryogenesis by MEDEA, a polycomb group gene in Arabidopsis. Science 280: 446–450 [DOI] [PubMed] [Google Scholar]
- Gu T., Mazzurco M., Sulaman W., Matias D.D., Goring D.R. (1998). Binding of an arm repeat protein to the kinase domain of the S-locus receptor kinase. Proc. Natl. Acad. Sci. USA 95: 382–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guitton A.E., Page D.R., Chambrier P., Lionnet C., Faure J.E., Grossniklaus U., Berger F. (2004). Identification of new members of FERTILISATION INDEPENDENT SEED Polycomb group pathway involved in the control of seed development in Arabidopsis thaliana. Development 131: 2971–2981 [DOI] [PubMed] [Google Scholar]
- Gutiérrez-Marcos J.F., Costa L.M., Evans M.M. (2006). Maternal gametophytic baseless1 is required for development of the central cell and early endosperm patterning in maize (Zea mays). Genetics 174: 317–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haig D., Westoby M. (1989). Parent specific gene expression and the triploid endosperm. Am. Nat. 134: 147–155 [Google Scholar]
- Haig D., Westoby M. (1991). Genomic imprinting in the endosperm: Its effect on seed development in crosses between species, and between different ploidies of the same species, and its implications for the evolution of apomixis. Philos. Trans. R. Soc. Lond. B Biol. Sci. 333: 1–13 [Google Scholar]
- Hatzfeld M. (1999). The Armadillo family at structural proteins. Int. Rev. Cytol. 186: 179–224 [DOI] [PubMed] [Google Scholar]
- Hennig L., Taranto P., Walser M., Schönrock N., Gruissem W. (2003). Arabidopsis MSI1 is required for epigenetic maintenance of reproductive development. Development 130: 2555–2565 [DOI] [PubMed] [Google Scholar]
- Howden R., Park S.K., Moore J.M., Orme J., Grossniklaus U., Twell D. (1998). Selection of T-DNA-tagged male and female gametophytic mutants by segregation distortion in Arabidopsis. Genetics 149: 621–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hruz T., Laule O., Szabo G., Wessendorp F., Bleuler S., Oertle L., Widmayer P., Gruissem W., Zimmermann P. (2008). Genevestigator v3: A reference expression database for the meta-analysis of transcriptomes. Adv. Bioinforma. 2008: 420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilic K., Berleth T., Provart N.J. (2004). BlastDigester—A web-based program for efficient CAPS marker design. Trends Genet. 20: 280–283 [DOI] [PubMed] [Google Scholar]
- Ingouff M., Fitz Gerald J.N., Guérin C., Robert H., Sørensen M.B., Van Damme D., Geelen D., Blanchoin L., Berger F. (2005). Plant formin AtFH5 is an evolutionarily conserved actin nucleator involved in cytokinesis. Nat. Cell Biol. 7: 374–380 [DOI] [PubMed] [Google Scholar]
- Jackson J.P., Lindroth A.M., Cao X., Jacobsen S.E. (2002). Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature 416: 556–560 [DOI] [PubMed] [Google Scholar]
- Jander G., Norris S.R., Rounsley S.D., Bush D.F., Levin I.M., Last R.L. (2002). Arabidopsis map-based cloning in the post-genome era. Plant Physiol. 129: 440–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jullien P.E., Katz A., Oliva M., Ohad N., Berger F. (2006a). Polycomb group complexes self-regulate imprinting of the Polycomb group gene MEDEA in Arabidopsis. Curr. Biol. 16: 486–492 [DOI] [PubMed] [Google Scholar]
- Jullien P.E., Kinoshita T., Ohad N., Berger F. (2006b). Maintenance of DNA methylation during the Arabidopsis life cycle is essential for parental imprinting. Plant Cell 18: 1360–1372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang I.H., Steffen J.G., Portereiko M.F., Lloyd A., Drews G.N. (2008). The AGL62 MADS domain protein regulates cellularization during endosperm development in Arabidopsis. Plant Cell 20: 635–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kankel M.W., Ramsey D.E., Stokes T.L., Flowers S.K., Haag J.R., Jeddeloh J.A., Riddle N.C., Verbsky M.L., Richards E.J. (2003). Arabidopsis MET1 cytosine methyltransferase mutants. Genetics 163: 1109–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley L.A., Sternberg M.J. (2009). Protein structure prediction on the Web: A case study using the Phyre server. Nat. Protoc. 4: 363–371 [DOI] [PubMed] [Google Scholar]
- Kim S., Choi H.I., Ryu H.J., Park J.H., Kim M.D., Kim S.Y. (2004). ARIA, an Arabidopsis arm repeat protein interacting with a transcriptional regulator of abscisic acid-responsive gene expression, is a novel abscisic acid signaling component. Plant Physiol. 136: 3639–3648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita T., Yadegari R., Harada J.J., Goldberg R.B., Fischer R.L. (1999). Imprinting of the MEDEA Polycomb gene in the Arabidopsis endosperm. Plant Cell 11: 1945–1952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyosue T., Ohad N., Yadegari R., Hannon M., Dinneny J., Wells D., Katz A., Margossian L., Harada J.J., Goldberg R.B., Fischer R.L. (1999). Control of fertilization-independent endosperm development by the MEDEA Polycomb gene in Arabidopsis. Proc. Natl. Acad. Sci. USA 96: 4186–4191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler C., Hennig L., Bouveret R., Gheyselinck J., Grossniklaus U., Gruissem W. (2003). Arabidopsis MSI1 is a component of the MEA/FIE Polycomb group complex and required for seed development. EMBO J. 22: 4804–4814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leroy O., Hennig L., Breuninger H., Laux T., Köhler C. (2007). Polycomb group proteins function in the female gametophyte to determine seed development in plants. Development 134: 3639–3648 [DOI] [PubMed] [Google Scholar]
- Leshem Y., Johnson C., Wuest S.E., Song X., Ngo Q.A., Grossniklaus U., Sundaresan V. (2012). Molecular characterization of the glauce mutant: a central cell-specific function is required for double fertilization in Arabidopsis. Plant Cell 24: 3264–3277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I., Copley R.R., Schmidt S., Ciccarelli F.D., Doerks T., Schultz J., Ponting C.P., Bork P. (2004). SMART 4.0: Towards genomic data integration. Nucleic Acids Res. 32(Database issue): D142–D144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindroth A.M., Cao X., Jackson J.P., Zilberman D., McCallum C.M., Henikoff S., Jacobsen S.E. (2001). Requirement of CHROMOMETHYLASE3 for maintenance of CpXpG methylation. Science 292: 2077–2080 [DOI] [PubMed] [Google Scholar]
- Lister R., O’Malley R.C., Tonti-Filippini J., Gregory B.D., Berry C.C., Millar A.H., Ecker J.R. (2008). Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell 133: 523–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P., Sherman-Broyles S., Nasrallah M.E., Nasrallah J.B. (2007). A cryptic modifier causing transient self-incompatibility in Arabidopsis thaliana. Curr. Biol. 17: 734–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Luo M., Bilodeau P., Dennis E.S., Peacock W.J., Chaudhury A. (2000). Expression and parent-of-origin effects for FIS2, MEA, and FIE in the endosperm and embryo of developing Arabidopsis seeds. Proc. Natl. Acad. Sci. USA 97: 10637–10642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M., Bilodeau P., Koltunow A., Dennis E.S., Peacock W.J., Chaudhury A.M. (1999). Genes controlling fertilization-independent seed development in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 96: 296–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M., Dennis E.S., Berger F., Peacock W.J., Chaudhury A. (2005). MINISEED3 (MINI3), a WRKY family gene, and HAIKU2 (IKU2), a leucine-rich repeat (LRR) KINASE gene, are regulators of seed size in Arabidopsis. Proc. Natl. Acad. Sci. USA 102: 17531–17536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- März P., Probst A., Lang S., Schwager M., Rose-John S., Otten U., Ozbek S. (2004). Ataxin-10, the spinocerebellar ataxia type 10 neurodegenerative disorder protein, is essential for survival of cerebellar neurons. J. Biol. Chem. 279: 35542–35550 [DOI] [PubMed] [Google Scholar]
- Masuda H.P., Cabral L.M., De Veylder L., Tanurdzic M., de Almeida Engler J., Geelen D., Inzé D., Martienssen R.A., Ferreira P.C., Hemerly A.S. (2008). ABAP1 is a novel plant Armadillo BTB protein involved in DNA replication and transcription. EMBO J. 27: 2746–2756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura T., et al. (2000). Large expansion of the ATTCT pentanucleotide repeat in spinocerebellar ataxia type 10. Nat. Genet. 26: 191–194 [DOI] [PubMed] [Google Scholar]
- Mudgil Y., Shiu S.H., Stone S.L., Salt J.N., Goring D.R. (2004). A large complement of the predicted Arabidopsis ARM repeat proteins are members of the U-box E3 ubiquitin ligase family. Plant Physiol. 134: 59–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore J.M. (2002). Isolation and Characterization of Gametophytic Mutants in Arabidopsis thaliana PhD dissertation (Stony Brook, NY: State University of New York)
- Moore J.M., Calzada J.P., Gagliano W., Grossniklaus U. (1997). Genetic characterization of hadad, a mutant disrupting female gametogenesis in Arabidopsis thaliana. Cold Spring Harb. Symp. Quant. Biol. 62: 35–47 [PubMed] [Google Scholar]
- Mosher R.A., Durrant W.E., Wang D., Song J., Dong X. (2006). A comprehensive structure-function analysis of Arabidopsis SNI1 defines essential regions and transcriptional repressor activity. Plant Cell 18: 1750–1765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngo Q.A., Moore J.M., Baskar R., Grossniklaus U., Sundaresan V. (2007). Arabidopsis GLAUCE promotes fertilization-independent endosperm development and expression of paternally inherited alleles. Development 134: 4107–4117 [DOI] [PubMed] [Google Scholar]
- Nodine M.D., Bartel D.P. (2012). Maternal and paternal genomes contribute equally to the transcriptome of early plant embryos. Nature 482: 94–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowack M.K., Grini P.E., Jakoby M.J., Lafos M., Koncz C., Schnittger A. (2006). A positive signal from the fertilization of the egg cell sets off endosperm proliferation in angiosperm embryogenesis. Nat. Genet. 38: 63–67 [DOI] [PubMed] [Google Scholar]
- Oh S., Park S., van Nocker S. (2008). Genic and global functions for Paf1C in chromatin modification and gene expression in Arabidopsis. PLoS Genet. 4: e1000077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh S.A., Johnson A., Smertenko A., Rahman D., Park S.K., Hussey P.J., Twell D. (2005). A divergent cellular role for the FUSED kinase family in the plant-specific cytokinetic phragmoplast. Curr. Biol. 15: 2107–2111 [DOI] [PubMed] [Google Scholar]
- Ohad N., Yadegari R., Margossian L., Hannon M., Michaeli D., Harada J.J., Goldberg R.B., Fischer R.L. (1999). Mutations in FIE, a WD polycomb group gene, allow endosperm development without fertilization. Plant Cell 11: 407–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagnussat G.C., Yu H.J., Ngo Q.A., Rajani S., Mayalagu S., Johnson C.S., Capron A., Xie L.F., Ye D., Sundaresan V. (2005). Genetic and molecular identification of genes required for female gametophyte development and function in Arabidopsis. Development 132: 603–614 [DOI] [PubMed] [Google Scholar]
- Palma K., Zhang Y., Li X. (2005). An importin alpha homolog, MOS6, plays an important role in plant innate immunity. Curr. Biol. 15: 1129–1135 [DOI] [PubMed] [Google Scholar]
- Phillips A.R., Evans M.M. (2011). Analysis of stunter1, a maize mutant with reduced gametophyte size and maternal effects on seed development. Genetics 187: 1085–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillot M., Baroux C., Vazquez M.A., Autran D., Leblanc O., Vielle-Calzada J.P., Grossniklaus U., Grimanelli D. (2010). Embryo and endosperm inherit distinct chromatin and transcriptional states from the female gametes in Arabidopsis. Plant Cell 22: 307–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raissig M.T., Baroux C., Grossniklaus U. (2011). Regulation and flexibility of genomic imprinting during seed development. Plant Cell 23: 16–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riggleman B., Wieschaus E., Schedl P. (1989). Molecular analysis of the armadillo locus: uniformly distributed transcripts and a protein with novel internal repeats are associated with a Drosophila segment polarity gene. Genes Dev. 3: 96–113 [DOI] [PubMed] [Google Scholar]
- Rook F., Corke F., Baier M., Holman R., May A.G., Bevan M.W. (2006). IMPAIRED SUCROSE INDUCTION1 encodes a conserved plant-specific protein that couples carbohydrate availability to gene expression and plant growth. Plant J. 46: 1045–1058 [DOI] [PubMed] [Google Scholar]
- Roudier F., et al. (2011). Integrative epigenomic mapping defines four main chromatin states in Arabidopsis. EMBO J. 30: 1928–1938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai T., et al. (2008). Armadillo repeat-containing kinesins and a NIMA-related kinase are required for epidermal-cell morphogenesis in Arabidopsis. Plant J. 53: 157–171 [DOI] [PubMed] [Google Scholar]
- Samuel M.A., Salt J.N., Shiu S.H., Goring D.R. (2006). Multifunctional arm repeat domains in plants. Int. Rev. Cytol. 253: 1–26 [DOI] [PubMed] [Google Scholar]
- Shahbazian M.D., Grunstein M. (2007). Functions of site-specific histone acetylation and deacetylation. Annu. Rev. Biochem. 76: 75–100 [DOI] [PubMed] [Google Scholar]
- Shapiro L. (2001). Beta-catenin and its multiple partners: Promiscuity explained. Nat. Struct. Biol. 8: 484–487 [DOI] [PubMed] [Google Scholar]
- Söding J. (2005). Protein homology detection by HMM-HMM comparison. Bioinformatics 21: 951–960 [DOI] [PubMed] [Google Scholar]
- Spillane C., MacDougall C., Stock C., Köhler C., Vielle-Calzada J.P., Nunes S.M., Grossniklaus U., Goodrich J. (2000). Interaction of the Arabidopsis polycomb group proteins FIE and MEA mediates their common phenotypes. Curr. Biol. 10: 1535–1538 [DOI] [PubMed] [Google Scholar]
- Springer P.S., Holding D.R., Groover A., Yordan C., Martienssen R.A. (2000). The essential Mcm7 protein PROLIFERA is localized to the nucleus of dividing cells during the G(1) phase and is required maternally for early Arabidopsis development. Development 127: 1815–1822 [DOI] [PubMed] [Google Scholar]
- Springer P.S., McCombie W.R., Sundaresan V., Martienssen R.A. (1995). Gene trap tagging of PROLIFERA, an essential MCM2-3-5-like gene in Arabidopsis. Science 268: 877–880 [DOI] [PubMed] [Google Scholar]
- Stone S.L., Anderson E.M., Mullen R.T., Goring D.R. (2003). ARC1 is an E3 ubiquitin ligase and promotes the ubiquitination of proteins during the rejection of self-incompatible Brassica pollen. Plant Cell 15: 885–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone S.L., Arnoldo M., Goring D.R. (1999). A breakdown of Brassica self-incompatibility in ARC1 antisense transgenic plants. Science 286: 1729–1731 [DOI] [PubMed] [Google Scholar]
- Thompson J.E.S. (1972). A Commentary on the Dresden Codex. (Philadelphia: American Philosophical Society)
- Tiwari S., Schulz R., Ikeda Y., Dytham L., Bravo J., Mathers L., Spielman M., Guzmán P., Oakey R.J., Kinoshita T., Scott R.J. (2008). MATERNALLY EXPRESSED PAB C-TERMINAL, a novel imprinted gene in Arabidopsis, encodes the conserved C-terminal domain of polyadenylate binding proteins. Plant Cell 20: 2387–2398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughn M.W., et al. (2007). Epigenetic natural variation in Arabidopsis thaliana. PLoS Biol. 5: e174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vielle-Calzada J.P., Thomas J., Spillane C., Coluccio A., Hoeppner M.A., Grossniklaus U. (1999). Maintenance of genomic imprinting at the Arabidopsis MEDEA locus requires zygotic DDM1 activity. Genes Dev. 13: 2971–2982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinkenoog R., Spielman M., Adams S., Fischer R.L., Dickinson H.G., Scott R.J. (2000). Hypomethylation promotes autonomous endosperm development and rescues postfertilization lethality in fie mutants. Plant Cell 12: 2271–2282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vongs A., Kakutani T., Martienssen R.A., Richards E.J. (1993). Arabidopsis thaliana DNA methylation mutants. Science 260: 1926–1928 [DOI] [PubMed] [Google Scholar]
- Wakamiya M., Matsuura T., Liu Y., Schuster G.C., Gao R., Xu W., Sarkar P.S., Lin X., Ashizawa T. (2006). The role of ataxin 10 in the pathogenesis of spinocerebellar ataxia type 10. Neurology 67: 607–613 [DOI] [PubMed] [Google Scholar]
- Wang Z., Yuan T., Yuan C., Niu Y., Sun D., Cui S. (2009). LFR, which encodes a novel nuclear-localized Armadillo-repeat protein, affects multiple developmental processes in the aerial organs in Arabidopsis. Plant Mol. Biol. 69: 121–131 [DOI] [PubMed] [Google Scholar]
- Willert K., Jones K.A. (2006). Wnt signaling: Is the party in the nucleus? Genes Dev. 20: 1394–1404 [DOI] [PubMed] [Google Scholar]
- Wöhrmann H.J.P., Gagliardini V., Raissig M.T., Wehrle W., Arand J., Schmidt A., Tierling S., Page D.R., Schöb H., Walter J., Grossniklaus U. (2012). Identification of a DNA methylation-independent imprinting control region at the Arabidopsis MEDEA locus. Genes Dev. 26: 1837–1850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuest S.E., Vijverberg K., Schmidt A., Weiss M., Gheyselinck J., Lohr M., Wellmer F., Rahnenführer J., von Mering C., Grossniklaus U. (2010). Arabidopsis female gametophyte gene expression map reveals similarities between plant and animal gametes. Curr. Biol. 20: 506–512 [DOI] [PubMed] [Google Scholar]
- Yadegari R., Kinoshita T., Lotan O., Cohen G., Katz A., Choi Y., Katz A., Nakashima K., Harada J.J., Goldberg R.B., Fischer R.L., Ohad N. (2000). Mutations in the FIE and MEA genes that encode interacting Polycomb proteins cause parent-of-origin effects on seed development by distinct mechanisms. Plant Cell 12: 2367–2382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C.W., González-Lamothe R., Ewan R.A., Rowland O., Yoshioka H., Shenton M., Ye H., O’Donnell E., Jones J.D., Sadanandom A. (2006). The E3 ubiquitin ligase activity of Arabidopsis PLANT U-BOX17 and its functional tobacco homolog ACRE276 are required for cell death and defense. Plant Cell 18: 1084–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang G., Gao P., Zhang H., Huang S., Zheng Z.L. (2007). A mutation in MRH2 kinesin enhances the root hair tip growth defect caused by constitutively activated ROP2 small GTPase in Arabidopsis. PLoS ONE 2: e1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W., Devaiah S.P., Pan X., Isaac G., Welti R., Wang X. (2007). AtPLAI is an acyl hydrolase involved in basal jasmonic acid production and Arabidopsis resistance to Botrytis cinerea. J. Biol. Chem. 282: 18116–18128 [DOI] [PubMed] [Google Scholar]
- Zeng L.R., Qu S., Bordeos A., Yang C., Baraoidan M., Yan H., Xie Q., Nahm B.H., Leung H., Wang G.L. (2004). Spotted leaf11, a negative regulator of plant cell death and defense, encodes a U-box/armadillo repeat protein endowed with E3 ubiquitin ligase activity. Plant Cell 16: 2795–2808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Bernatavichute Y.V., Cokus S., Pellegrini M., Jacobsen S.E. (2009). Genome-wide analysis of mono-, di- and trimethylation of histone H3 lysine 4 in Arabidopsis thaliana. Genome Biol. 10: R62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Clarenz O., Cokus S., Bernatavichute Y.V., Pellegrini M., Goodrich J., Jacobsen S.E. (2007). Whole-genome analysis of histone H3 lysine 27 trimethylation in Arabidopsis. PLoS Biol. 5: e129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. (2008). I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 9: 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.