Abstract
Tef (Eragrostis tef) is a major cereal crop in Ethiopia. Lodging is the primary constraint to increasing productivity in this allotetraploid species, accounting for losses of ∼15–45% in yield each year. As a first step toward identifying semi-dwarf varieties that might have improved lodging resistance, an ∼6× fosmid library was constructed and used to identify both homeologues of the dw3 semi-dwarfing gene of Sorghum bicolor. An EMS mutagenized population, consisting of ∼21,210 tef plants, was planted and leaf materials were collected into 23 superpools. Two dwarfing candidate genes, homeologues of dw3 of sorghum and rht1 of wheat, were sequenced directly from each superpool with 454 technology, and 120 candidate mutations were identified. Out of 10 candidates tested, six independent mutations were validated by Sanger sequencing, including two predicted detrimental mutations in both dw3 homeologues with a potential to improve lodging resistance in tef through further breeding. This study demonstrates that high-throughput sequencing can identify potentially valuable mutations in under-studied plant species like tef and has provided mutant lines that can now be combined and tested in breeding programs for improved lodging resistance.
Keywords: brachytic2, dw3, ethyl methanesulfonate (EMS) mutagenesis, Eragrostis tef, green revolution genes
TEF [Eragrostis tef (Zucc.) Trotter] is an allotetraploid (2n = 4X = 40) species that is native to Ethiopia. Tef belongs to the grass subfamily Chloridoideae, a lineage that has received very little research attention. Tef has an estimated genome size of 713–733 Mbp (Ayele et al. 1996), which is ∼60% larger than the rice genome.
As a staple food in Ethiopia, covering more acreage than any other crop in that nation, tef possesses several advantageous characteristics, including excellent storage properties and a very nutritious seed with excellent protein and mineral composition (Stallknecht et al. 1993). In addition, tef plants grow well under extreme environmental conditions such as drought and water-logging. However, compared to other crops, the average yield of tef is quite low, at an average of ∼700 kg ha−1 (Central Statistics Authority 2005). Tef has a tall and tender stem that is susceptible to lodging caused by wind and rain. As a consequence, the yield of tef is reduced ∼15–45% each year, depending on the weather and the variety. Furthermore, an increased incidence of lodging is associated with fertilizer application. Using QTL to improve lodging resistance indicated a positive correlation between lodging and yield, such that the most lodging-resistant progeny also had the lowest yield (Yu et al. 2007). This is probably because such traits as larger seed size and higher seed number create a heavier panicle that leads to a greater likelihood of lodging.
In other cereal crops (Peng et al. 1999; Sasaki et al. 2002; Multani et al. 2003), semi-dwarf varieties have been found to provide both lodging resistance and higher yields. As shown in the earliest studies (Quinby and Karper 1954; Jennings 1964; Walcott and Laing 1976; Foster and Rutger 1978), semi-dwarf varieties have both strong stalks and a greater carbon partitioning into seed rather than vegetative material. Comparison of genetic-improvement-associated trait changes in tef and wheat revealed that tef harvest index and lodging susceptibility remain unaltered even though plant height and total biomass yield increased in some varieties (Assefa et al. 2011), further suggesting the importance of developing tef varieties with short stature. However, tef is a tetraploid species with most genes represented by two homeologous copies, thus raising doubts as to the likely effectiveness of the facile use of plant height screens for the outcome of single-gene knockouts.
Numerous studies have been carried out on plant dwarfing genes (Itoh et al. 2001; Monna et al. 2002; Sasaki et al. 2002; Hong et al. 2003; Multani et al. 2003; Muangprom 2005; Sakamoto et al. 2005; Tanabe et al. 2005; Zou et al. 2006; Asano et al. 2009), and mutant alleles of these genes have been seen to have negative effects on plant performance, often by decreasing fertility. However, mutant alleles of a few dwarfing genes have been found to be useful for crop improvement. Among them, two of the most successful applications have been to create the semi-dwarf wheat and rice varieties that led to the Green Revolution in the 1960s. The key genes employed were the reduced height-1 (Rht-B1b and Rht-D1b) genes in wheat (Peng et al. 1999) and the semi-dwarf (sd1) gene in rice (Sasaki et al. 2002; Spielmeyer 2002; Muangprom 2005). Cloning of these genes revealed that both are involved in the gibberellin (GA) response. The wild-type alleles of Rht-B1b and Rht-D1b encode DELLA proteins that are important components of the GA signal transduction pathway. Peng et al. (1999) found that a point mutation in each of the Rht-B1b and Rht-D1b alleles introduced a stop codon into the N-terminal coding region. The rice sd1 gene encodes GA20 oxidase, a key enzyme in the GA biosynthesis pathway. The studied sd1 allele has a 383-bp deletion that results in a frameshift mutation that conditions a greatly lowered GA20 oxidase level (Sasaki et al. 2002).
In sorghum, the gene dwarf3 (dw3) (Multani et al. 2003) was used for lodging resistance and higher yields in this crop decades before the wheat or rice Green Revolutions were conceptualized (Karper 1932; Quinby and Karper 1954). The maize ortholog of dw3, br2, has not been used extensively for maize improvement, perhaps because maize lodging is not so much a plant anatomy issue as it is associated with insect and fungal damage to stalks (Munkvold and Hellmich 2000; Multani et al. 2003). Multani and his colleagues (Multani et al. 2003) discovered that corn br2 and sorghum dw3 encode a p-glycoprotein whose loss leads to dwarf corn and sorghum plants by interfering with the movement of auxin, thus causing the compact lower stalk internodes characteristic of this mutation.
Mutagenesis has been used to identify the existence and involvement of genes in numerous biological processes (Auerbach and Robson 1946; Rapoport 1946; Ehrenberg and Gustafsson 1957; Natarajan and Ramanna 1966). Chemical mutagens, such as ethyl methanesulfonate (EMS) and N-methyl-N-nitrosourea (MNU), are applied to induce single nucleotide substitutions and create mutation libraries at the genome level. For instance, the original sd1 rice mutant was generated by mutagenesis (IRRI annual report for 1966). Recently, mutagenesis has taken a significant step forward by the utilization of reverse genetic approaches that allow the directed investigation of the mutational status of specific genes. TILLING (targeting induced local lesions in genomes) and EcoTILLING (Comai et al. 2004) have been applied to a variety of plants, resulting in identification of mutations in specific target genes in Arabidopsis (Greene et al. 2003; Till et al. 2003; Martin et al. 2009), rice (Till et al. 2007; Suzuki et al. 2008), wheat (Slade et al. 2005), maize (Till et al. 2004), sorghum (Xin et al. 2008), pea (Dalmais et al. 2008), soybean (Cooper et al. 2008), tomato (Minoia et al. 2010), and other plant species. However, TILLING is a time-consuming, expensive, and labor-intensive approach when applied to either large populations or large genomes and might be particularly challenging in a recent tetraploid like tef (Smith et al. 2012) where homeologues have not yet differentiated greatly in sequence.
Recent advances in high-throughput sequencing technologies have provided avenues for greatly improved detection of genome-wide variation. For instance, using 454 amplicon sequencing, over 1000 SNPs were detected from a sugarcane mapping population and 93% were found to be valid (Bundock et al. 2009). A 454 GS FLX platform based on KeyPoint technology has been used to screen for induced and naturally occurring sequence variation in more than 3000 M2 tomato families and identified two mutants and six haplotypes in the elF4E gene (Rigola et al. 2009). To enhance this approach, enrichment of a specific gene can be performed before high-throughput sequencing. Array capture and PCR amplification of targets are commonly used (Mamanova et al. 2010; Ng et al. 2010a). Array-enriched whole-exome sequencing has been widely used to identify candidate causative genes for human disease. For example, SNPs in genes DHODH and MLL2 were respectively identified as causal agents for Miller syndrome and Kabuki syndrome by this approach (Ng et al. 2010a,b).
Recently, Smith et al. (2012) cloned and sequenced homologs of the sd1 and rht1 genes from tef. Two rht1 homeologues, apparent orthologs of the wheat genes, were sequenced across 31 tef accessions, as were three tef sd1 homologs of unknown paralogous/orthologous relationship. This study indicated that all accessions contained genes that appeared to be fully functional and that there were both very few haplotypes per gene (one to five) and no clear history of recombination between these haplotypes. Hence, a search for naturally inactivated semi-dwarf genes in the tef germplasm would be an arduous and perhaps fruitless undertaking.
In this study, two dwarfing candidate genes, orthologs of dw3, were isolated from a fosmid genomic library containing tef DNA inserts. These two candidate genes, and two tef rht1 homeologues, were enriched by gene-specific PCR amplification from genomic DNA pools derived from an EMS-mutagenized tef population containing ∼21,210 individuals. Candidate mutations were identified, and several were further validated by Sanger sequencing, thereby demonstrating that high-throughput sequence analysis can be an efficient approach for mutant discovery in a tetraploid orphan crop. Although semi-dwarf phenotypes were not seen in heterozygous or homozygous mutant plants, future crosses to bring these two mutations into the same genetic background may yield progeny with a semi-dwarf phenotype that yields improved lodging resistance and yield.
Materials and Methods
Fosmid genomic library construction
Genomic DNA from tef USDA accession PI524434 was used for fosmid library construction. The genomic DNA was randomly sheared with 40 repetitions using a GeneMachines Hydroshear, and DNA fragments from 35–45 kb were selected. The sheared DNA was end-repaired and purified and then was ligated into the pCC1FOS vector (Epicentre Technologies, WI). For packaging of fosmid clones, MaxPlax Lambda Packaging Extract (Epicentre Technologies) was mixed with 10 μl of ligation reaction and processed according to the manufacturer’s instructions. Plates containing ∼100 fosmid clones were pooled and stored in deep-well plates. These pools were further combined into superpools for the initial PCR screening to identify clones containing genes of interest.
Fosmid library screening, sequencing, and data analysis
The tef genomic fosmid library was screened for dwarfing candidate gene dw3/br2 by a PCR-based strategy. This library was assigned to 25 superpools, with 48 pools in each superpool (Supporting Information, Table S1). All 25 superpools were screened by primers specific to putative dwarfing genes. One positive superpool identified in the previous step was selected and all of the 48 pools were screened. Then, one positive pool was selected and 96 clones from this pool were randomly picked and screened. During the entire screening process, tef genomic DNA was used as a positive control for PCR amplification. Finally, the identified positive clones were further verified by complete sequencing to identify a full-length copy of the target gene. The complete clone sequences were submitted to GenBank (accession nos. JN672669–JN672670).
The fosmid sequences were initially annotated by FGENESH (http://www.softberry.com) and GENESCAN (Burge and Karlin 1997). Putative proteins were identified using TBLASN and BLASTP (Altschul et al. 1997). Divergence times of the two homeologous dw3 homologs in the tetraploid tef were estimated by two approaches. One was a molecular clock approach. The divergence time was calculated based on the formula T = Ks/2r, where r corresponds to the absolute rate of substitutions/site/year and Ks is the estimated numbers of substitutions per site between homologous sequences. Synonymous substitutions were calculated by the method of Nei–Gojobori (Nei and Gojobori 1986) with the Jukes–Cantor correction. The other dating approach was based on comparative genomics. The number of synonymous mutation (Ds) was calculated for the orthologous dw3/br2 genes from several grass species, including maize and sorghum. Thus the divergence time of the two tef homeologues was dated using Ds(tef)/Ttef = Ds(AB)/T(AB), where Ds is a measure of the number of synonymous differences when comparing the two tef homeologues; Ds(AB) and T(AB) represent the level of synonymous mutation and the divergence time between maize and sorghum, respectively.
Tef mutagenesis and pooling
Tef seed used in the mutagenesis were from cultivar DZ-Cr-37, a released variety that provides early maturing plants that are drought tolerant (Teferra et al. 2000). Seed from DZ-Cr-37 were treated with EMS at concentrations of 0, 0.5, 1, and 1.5%. The seeds were soaked in water for 1 hr, then in the EMS solution for 5 hr, and were subsequently washed with water for 3 hr. Mutagenized seeds were planted in the greenhouse in a soilless growing mix. Plant heights measured 3 weeks after planting were used as surrogate scores for the efficacy of different concentrations of EMS. That is, only seeds with good germination efficiency but also showing some obvious detrimental effects of the mutagen were considered good candidates for a high level of mutation. EMS concentrations of 1–1.5% were found to be a good working dose for DZ-Cr-37, so plants grown from these seeds constituted the tef M1 generation, a total of 2,121 M1 plants. M1 plants were self-fertilized, and 10 M2 generation seeds were planted in the greenhouse from each M1 panicle. By definition, an M2 family is derived from a single M1 seed, because it is the self-pollinated seed on the panicle of an M1 plant. Genomic DNAs were isolated from a pool of 10 M2 leaves (one equivalent-size leaf per plant) from each M2 family, using the standard CTAB procedure (Murray and Thompson 1980). A total of 2121 pools were generated, which represented 2121 M2 families. Among them, 934 pools were from the 1.0% EMS treatment and 1187 pools were from the 1.5% EMS treatment. A total of 90–100 pools were combined into each superpool, generating 23 superpools for mutation screening by PCR and 454 sequencing. If a mutation of interest was identified in the superpool, all pools in this superpool were screened by Sanger sequencing. Once the target mutation was identified, 10–16 seeds from this pool were sown to collect M3 single plants for further validation. The seeds from mutationally confirmed M3 plants have been saved for future use.
Phenotypic analysis
The SIFT (sorting intolerant from tolerant; Ng 2003) program was used to predict the severity of the effects of each identified mutation on protein function. The mutagenized tef plants containing damaging mutation predicted by SIFT were selected and mutation phenotypes were further evaluated in the M3 generation. Each M3 plant height was carefully inspected at least three times during the growing season. Plants were grown both under standard greenhouse conditions (with high fertilizer inputs like those routinely used for maize) and under conditions where no fertilizer was added after the initial planting.
454 library preparation
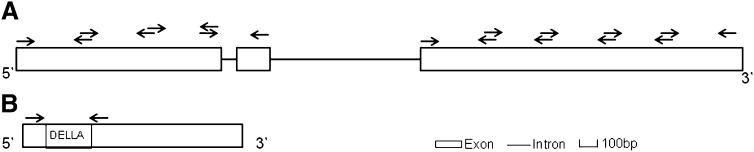
Exonic segments of four dwarfing candidate genes, dw3a, dw3b, rht1a, and rht1b (Smith et al. 2012) were chosen for 454 sequencing. Seven pairs of primers for the dw3/br2 genes and 1 pair of primers for the rht1 homeologues were designed to cover the appropriate coding regions (Figure 1) for initial PCR and sequencing. The primer sequences are listed in the supporting information (Table S2). For each primer, an 8- to 10-bp TI-MID tag was added at the 5′ end to differentiate the superpools. These tags differed by at least four nucleotides.
Figure 1.
Gene structure for tef dwarfing candidate genes of (A) dw3/br2 and (B) rht. The arrows indicate the primers used to screen the mutagenized populations in the 454 sequencing.
All PCR amplifications were performed in 1× Phusion HF Buffer, 1.5 mM MgCl2, 200 μM of each dNTP, 0.5 μM primer, 3% DMSO, and 2.5 units Phusion high-fidelity DNA Polymerase (New England Biolabs, MA), with 100 ng of genomic DNA in a 50 μl total volume. Amplification was carried out in a PTC-gradient cycler (MJ Research, MA) as follows: 30 sec at 98° followed by 30 cycles of 10 sec at 98°; 20 sec at 72°; and a final extension of 10 min at 72°. PCR products were isolated from 1.5% agarose gels and purified using a Quick gel extraction kit (QIAGEN, Hilden, Germany).
454 titanium sequencing, data processing, and candidate mutation validation
Sequencing was performed at the University of Georgia sequencing facility according to the manufacturer’s instructions (Roche Applied Science, IN). Base-called reads were trimmed, filtered for quality, and converted into FASTA format. During preprocessing, the origin of the reads was identified based on the target-specific primer sequences and the 8- to 10-bp TI-MID tags. Single-nucleotide mutations were then identified by mapping the 454 superpool data onto the reference sequences generated by the fosmid sequence analysis. A mutation was considered to be a candidate when it fulfilled the following criteria: (1) it was present in only one of the superpools, and with a frequency of three or more reads in that superpool, and (2) its quality score was >15. The mutation identification was implemented using custom PERL scripts. Candidate mutations identified in the superpools were validated by Sanger sequencing of the individual 10-plant pools, all from the M1 panicle. So the mutation was expected in 50% of the pooled DNA, yielding overlapping reads of ∼50% wild type and ∼50% mutant at the identified mutant nucleotide. These candidates were further investigated by Sanger sequencing of DNA from single M2 seedlings from the appropriate M1 panicle, indicating M2 progeny that segregated 1:2:1 for the mutant nucleotide. PCR products obtained from the M2 families and individuals were sequenced using the BigDye Terminator v. 3.1 kit on the 3730 DNA analyzer (Applied Biosystems, CA). Once a M2 plant was validated, the M2 seeds were planted again in the greenhouse to collect M3 seed.
Results
Fosmid sequencing and identification of dwarfing candidate genes
A total of 102,191 tef fosmid clones were isolated and transferred to 96-well microtiter plates, corresponding to ∼6× genome coverage. The probability of recovering a particular gene in this library is >99.5%, calculated by the formula N = ln(1 − P)/ln(1 − f), where P is the calculated probability, f is the proportion of the genome contained in a single clone, and N is the number of fosmid clones. This library was organized into 25 superpools and screened for dw3/br2 homologs. Two of the positive superpools were chosen to screen all pools and fosmids. Two fosmids containing dw3 target genes were sequenced to identify the full-length dw3 sequences. We designated these two genes as dw3a and dw3b in the allotetraploid tef genome. Annotation of these two fosmids resulted in a total of eight genes. The gene dw3a was located in the middle of one fosmid and dw3b at the end of the other fosmid (Table 1). Sequence comparison revealed 94.8% and 94.1% identities between dw3a and dw3b at the nucleotide and protein levels in coding regions, respectively. However, the nucleotide identity dropped to <70% in noncoding regions (Table S3). There were seven PCR positives for dw3 in the superpool screening of this ∼6× library, thus suggesting that these two genes represent the two homeologous copies from the two diploid ancestors of this allotetraploid. With a molecular clock method, these two dw3 homeologues were calculated to have shared a common ancestor approximately 9 million years ago (MYA). Similarly, with a comparative genomics approach, this divergence was dated at approximately 10 MYA, when one employs maize and sorghum genome divergences as 12 MYA (Swigonova et al. 2004). The gene lengths of the tef dw3 homeologues were similar to their maize, sorghum, and rice orthologs. However, the tef genes are missing one intron compared to maize and sorghum, and two introns compared to rice.
Table 1. Two fosmids containing tef dw3 homologs.
| Size of fosmid insert (bp) | No. of genes annotated | Target gene | Length of target gene (bp) | Target gene position in the fosmid (bp) |
|---|---|---|---|---|
| 39,729 | 8 | dw3a | 5,239 | 10,232–15,470 |
| 43,434 | 8 | dw3b | 5,181 | 37,617–42,797 |
Generation of EMS mutagenized tef populations and 454 sequencing
EMS is a chemical mutagen that predominantly induces C to T and G to A transitions across the genome (Krieg 1963). In this study, 2121 M2 families were grown for DNA isolation and screening. A total of 934 families were from seed treated with 1% EMS and 1187 families were from seed treated with 1.5% EMS. The M2 seed germination rates for both treatments were >95%, even after 9 years of storage at room temperature. Approximately equal amounts of leaf material were harvested from each of ∼10 individual M2 plants that were derived from a single M1 seed. As any mutation occurring in the M1 seed would segregate in the M2 offspring, pooling of leaf material from 10 individual M2 plants meant that the chance of recovery of this mutation was >99%.
A total of 327,696 quality-filtered reads, with a median read length of 393 bp (ranging from 347 to 413 bp), were obtained from half a plate of a 454 Titanium run, producing a total of 123 Mb of raw data (Table S4). This provided 9.7× sequence coverage, on average, for each M2 pool. Of the total reads, 312,786 (95%) could be assigned into one of the 23 superpools and one of the 184 amplicons (23 superpools × 8 primer pairs) to cover most of the coding portion of the dw3 homeologues and one key region (Peng et al. 1999) of the rht1 homeologues based on the target-specific primer sequences and the 8–10 bp TI-MID tags. For each amplicon, the number of reads from each superpool was fairly consistent (Figure S1). 454 sequencing would be expected to produce more reads for short fragments, and this was observed with our shortest amplicons. The remaining 5% of reads contained one or more deviations in the sample TI-MID tag sequences or were shorter than 100 bp, and thus were excluded from further analysis.
Mutation patterns
The mutagenized population was from cultivar DZ-Cr-37, while the cloned sequences were from accession PI524434, so it was expected that the clone sequence would differ from that of the cultivar that was mutagenized. This observation was confirmed by the amplicon sequencing, which indicated 0.3% nucleotide differences (1/ 3109 bp) in the DZ-Cr-37 amplicons (all from protein-encoding sequence) compared to PI524434. This single-nucleotide change does not cause an amino acid change. Compared to the wild-type sequence of the sequenced exonic DNA of the dw3 and rht1 homeologue amplicons from DZ-Cr-37, a total of 120 candidate mutations were identified in the four homeologous genes in the mutagenized population. Many additional sequence variations were observed, but these were low-frequency sequence changes that were likely to have been an outcome of sequencing errors. Of the 120 sequence changes represented three or more times in the amplicon sequence data set, 86 were missense/nonsense mutations (Table 2). The majority (78%) of the mutations detected in the superpools were derived from the 1.5% EMS treatment (Table S5). The mutation frequency, determined by screening 2121 M2 families, was ∼8.7 mutations/Mb for dw3 homeologues and 2.7 mutations/Mb for rht1 homeologues. Of these candidate mutations, the most abundant mutation types are G/C to A/T transitions, accounting for 69.2% and 84.1% of total mutations in the dw3a and dw3b genes, respectively. Transversions accounted for <20% of the candidate mutations. However, more detailed analysis of 10 candidate mutations from the set of 120 indicated that only 60% were truly mutant (and the rest were sequencing errors, see below), so the actual rates of sequence change of each type cannot be known unless validation of the entire set of 120 is performed.
Table 2. Candidate dwarfing gene mutation detected by 454 sequencing in a mutagenized tef population.
| Gene | Mutationa | Effectb | Frequencyc | Distribution | Functiond | SIFTd | Validatione |
|---|---|---|---|---|---|---|---|
| dw3a | A89G | E30G | 5 | sp2 | Damaging | 0 | |
| C104T | P35L | 7 | sp19 | Damaging | 0 | ||
| A200G | N67S | 3 | sp11 | Damaging | 0 | ||
| A238G | S80G | 17 | sp6 | Tolerated | 0.36 | ||
| C503T | A168V | 19 | sp19 | Tolerated | 1 | Yes | |
| A628T | T210S | 6 | sp9 | Damaging | 0.03 | ||
| T677A | V226D | 3 | sp17 | Damaging | 0 | ||
| G730A | A244T | 3 | sp2 | Tolerated | 0.15 | ||
| G742A | A248T | 10 | sp19 | Damaging | 0.02 | Yes | |
| G838A | A280T | 3 | sp3 | Tolerated | 0.2 | ||
| G863A | R288Q | 9 | sp11 | Tolerated | 0.11 | ||
| C1105T | R369C | 6 | sp14 | Tolerated | 0.06 | Yes | |
| G1124A | G375D | 11 | sp7 | Damaging | 0 | ||
| C1139T | T380I | 4 | sp7 | Tolerated | 0.07 | Yes | |
| C1177T | Q393* | 3 | sp14 | Stop Codon | 0 | No | |
| A1189T | S397C | 13 | sp1 | Tolerated | 0.23 | ||
| A1228G | K410E | 3 | sp19 | Damaging | 0.02 | ||
| G1270A | D424N | 4 | sp16 | Tolerated | 0.46 | ||
| A1294G | T432A | 3 | sp22 | Tolerated | 0.77 | ||
| C1295T | T432M | 8 | sp14 | Tolerated | 0.1 | ||
| G1315A | G439S | 5 | sp22 | Tolerated | 0.08 | ||
| T1526A | L509H | 10 | sp17 | Damaging | 0 | ||
| G1606A | D536N | 4 | sp15 | Tolerated | 0.17 | ||
| G1642A | A548T | 7 | sp23 | Damaging | 0.01 | ||
| G1811A | S604N | 8 | sp9 | Damaging | 0 | ||
| C1814A | A605E | 3 | sp15 | Damaging | 0 | ||
| G1840A | V614M | 11 | sp16 | Damaging | 0 | ||
| G1849A | A617T | 3 | sp2 | Damaging | 0 | ||
| A1960T | I654F | 3 | sp7 | Tolerated | 0.6 | ||
| G1964A | G655E | 4 | sp23 | Damaging | 0 | ||
| G1999A | G667S | 3 | sp5 | Tolerated | 0.09 | ||
| G2288A | R763K | 3 | sp17 | Tolerated | 0.21 | ||
| G2593A | A865T | 6 | sp14 | Damaging | 0.01 | ||
| C2855T | A952V | 5 | sp14 | Damaging | 0.01 | ||
| C2863T | R955C | 3 | sp6 | Damaging | 0 | ||
| C2867T | A956V | 5 | sp23 | Damaging | 0.01 | ||
| dw3b | C76T | P26S | 4 | sp18 | Damaging | 0 | |
| A95T | H32L | 6 | sp15 | Damaging | 0 | ||
| C106T | P36S | 4 | sp20 | Damaging | 0 | Yes | |
| C118T | Q40* | 3 | sp19 | Stop Codon | 0 | No | |
| G127A | G43R | 6 | sp21 | Tolerated | 0.22 | ||
| C230T | S77F | 6 | sp18 | Tolerated | 0.05 | ||
| A266G | Q89R | 4 | sp9 | Damaging | 0 | ||
| C377T | A126V | 9 | sp11 | Damaging | 0 | ||
| C427T | L143F | 4 | sp7 | Damaging | 0.01 | No | |
| T499G | Y167D | 3 | sp2 | Tolerated | 0.88 | ||
| G517A | A173T | 4 | sp20 | Tolerated | 0.08 | No | |
| C530T | A177V | 3 | sp16 | Tolerated | 1 | ||
| C689T | A230V | 19 | sp20 | Tolerated | 0.08 | ||
| C746T | A249V | 8 | sp22 | Tolerated | 0.2 | ||
| T764A | F255Y | 4 | sp21 | Tolerated | 0.09 | ||
| G829A | A277T | 4 | sp22 | Damaging | 0.04 | ||
| G850A | A284T | 8 | sp19 | Tolerated | 0.29 | ||
| A920T | Q307L | 8 | sp23 | Damaging | 0 | ||
| C929T | A310V | 6 | sp1 | Tolerated | 0.12 | ||
| G1051A | G351S | 4 | sp21 | Damaging | 0 | ||
| C1163T | S388F | 3 | sp14 | Damaging | 0.02 | ||
| C1273T | L425F | 6 | sp20 | Tolerated | 0.06 | ||
| G1318A | V440M | 3 | sp10 | Damaging | 0 | ||
| C1322T | T441M | 3 | sp17 | Tolerated | 0.1 | ||
| G1354A | A452T | 3 | sp21 | Tolerated | 0.17 | ||
| C1460T | T487M | 7 | sp19 | Damaging | 0 | ||
| G1481A | R494K | 11 | sp17 | Damaging | 0 | ||
| G1604A | S535N | 3 | sp8 | Damaging | 0.01 | ||
| G1738T | E580* | 6 | sp14 | Stop Codon | 0 | ||
| A1739G | E580G | 5 | sp14 | Damaging | 0 | ||
| G1760A | G587D | 4 | sp18 | Damaging | 0 | ||
| G1831A | A611T | 5 | sp18 | Damaging | 0 | ||
| C1841T | A614V | 5 | sp11 | Damaging | 0 | ||
| G1867A | V623M | 8 | sp2 | Damaging | 0 | ||
| G1984A | E662K | 6 | sp18 | Damaging | 0 | ||
| G2077A | A693T | 3 | sp19 | Tolerated | 0.66 | ||
| T2210C | F737S | 3 | sp3 | Tolerated | 0.29 | ||
| A2257T | M753L | 10 | sp2 | Tolerated | 0.72 | ||
| G2284A | A762T | 3 | sp17 | Tolerated | 0.57 | ||
| G2287A | G763R | 5 | sp3 | Tolerated | 0.57 | ||
| C2387G | A796G | 3 | sp1 | Damaging | 0.02 | ||
| C2440T | P814S | 7 | sp5 | Tolerated | 0.46 | ||
| G2797A | V933M | 3 | sp4 | Damaging | 0.03 | ||
| C2803T | P935S | 5 | sp23 | Damaging | 0 | ||
| G3035A | G1012E | 5 | sp20 | Damaging | 0 | ||
| C3059T | A1020V | 3 | sp6 | Damaging | 0.04 | ||
| C3133T | R1045C | 4 | sp21 | Damaging | 0 | ||
| rht1a | C358T | P120S | 4 | sp18 | Damaging | 0.04 | Yes |
| rht1b | G160A | A54T | 3 | sp13 | Tolerated | 0.05 | |
| C184T | Q62* | 3 | sp23 | Stop Codon | 0 |
Mutation is named as X#Y, where X is the wild-type nucleotide, # is the position of the substitution, and Y is the mutated nucleotide.
Effect is designated as X#Y, where X is the wild-type amino acid, # is the position of the substitution, and Y is the mutated amino acid.
The mutation density in the total data set.
Function and SIFT represent predicted protein function effect and the scores, respectively. A score value <0.05 is usually predicted as damaging effect.
PCR validation of selected mutations. Yes indicates that a mutation was validated by Sanger sequencing. No indicates that a mutation was found to be a false positive by Sanger sequencing.
For dw3 homeologues, candidate mutations that were predicted to be silent, missense and nonsense accounted for 31 (27.2%), 80 (70.2%), and 3 (2.6%) of the cases, respectively. This mutation distribution pattern was significantly (χ2 = 16.5, d.f. = 2, P < 0.001) different from the predicted pattern by CODDLE (choose codons to optimize the detection of deleterious lesions, http://www.proweb.org/coddle/) program. CODDLE predicted ∼50% missense, 47% silent, and 3% nonsense mutations (Table S6). For rht1 genes, only six mutations, including two missense, three silent, and one nonsense mutations, were predicted. In addition, using the SIFT program (Ng 2003), the putative impact of the missense mutations on the protein function of the target genes could be predicted. Fifty (58%) of the candidate missense mutations, 48 from the dw3 genes, and 2 from the rht1 genes were predicted as deleterious by SIFT scores <0.05 (Table 2).
Mutant validation and phenotypic characterization
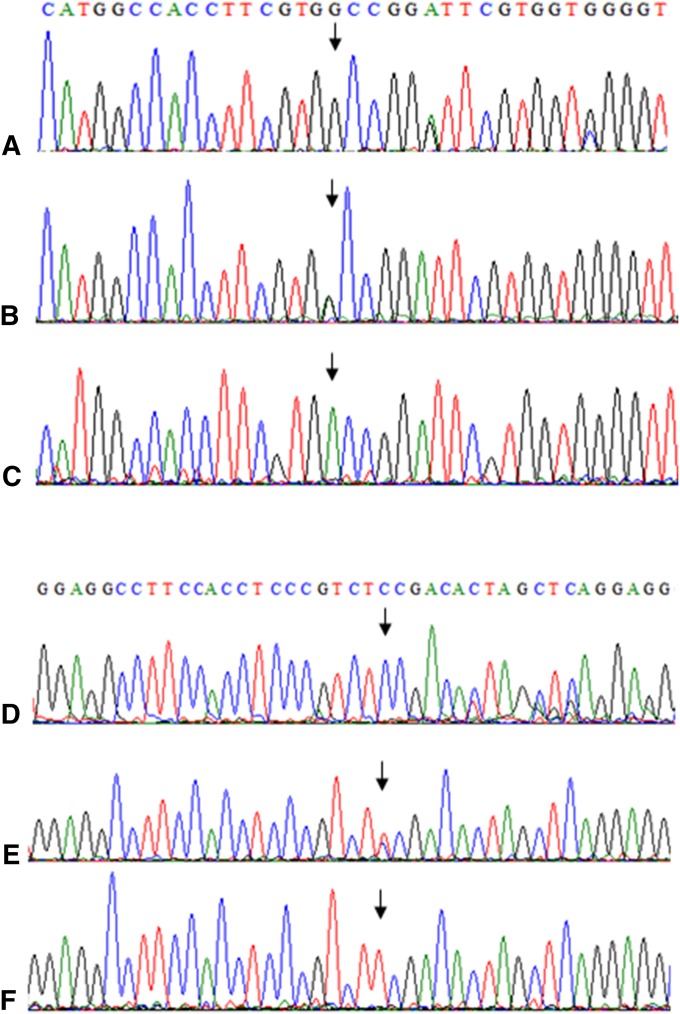
To validate these mutations, 10 candidate mutations were selected and DNA was analyzed from both 10-plant pools and single plants by Sanger sequencing. Six independent mutations, five at dw3 homeologues, and one at an rht1 homeologue, were validated in both pools and single plants (Table 2 and Figure 2). The other four, all with abundances less than or equal to four reads in the amplicon data set, were not found in any Sanger sequence, suggesting that they were low-frequency 454 sequencing errors. The mutation density of these validated mutations range from 4 to 19. We confirmed a G to A mutation at the 742-nt position (G742A) from the dw3a gene (Figure 2, i–iii) and a C to T mutation at the 106-nt position (C106T) from the dw3b gene (Figure 2, iv–vi) in the M2 tef lines p893 and p932, respectively. These two mutations encode alanine (nonpolar) to threonine (polar) and proline (nonpolar) to serine (polar) amino acid changes and were predicted to severely affect protein function by SIFT scores of 0.02 and 0.00, respectively, probably by alteration of the folding of the enzyme. Both nucleotides were located in a highly conserved domain of the predicted dw3 protein. These two amino acids are fully conserved across maize, sorghum, and tef. Sequencing the key region of the dw3 homeologue in individual M3 tef lines indicated that both G742A and C106T mutants were heterozygous at the target site in the M2 generation. (Figure S2). In an rht1 homeologue, a C to T mutation at the 358-nt position is also predicted as deleterious (Table 2). This amino acid (proline) is conserved across the grass family except in wheat, where it is a glutamine, while the predicted change in the tef mutation is to a serine.
Figure 2.
Mutation validation by Sanger sequence analysis. G742A from dw3a (A–C) and C106T from dw3b (D–F) have been validated in single plants. Mutation position is marked by an arrow. A nucleotide G (wild type) is detected at base 742 from superpool sp19 (A). It is identified as a G/A heterozygote in pool p892 (B) and A in the single plant p893-2 (C). Similarly, A nucleotide C (wild type) is detected as the 106th base in superpool sp20 (D). It is identified as a C/T heterozygote in pool p932 (E) and is confirmed as a T in the single plant p932-4 (F).
Possible phenotypic effects of the G742A and C106T mutations were examined in the M3 generations of tef lines p893 and p932. Regardless of high-input or low-input fertilizer regimens, homozygous wild-type plants were not distinguishable from either the heterozygous or homozygous mutations for either gene (Figure S3).
Discussion
The cloning and sequencing of two dw3/br2 homeologues from tef
A fosmid library representing approximately 6× coverage of the Eragrostis tef genome was constructed. The quality of this library was confirmed by PCR screening for dw3 homeologues. This screening identified seven positive clones in the superpools, suggesting that dw3 homeologues are present as single-copy genes (that is, one copy per diploid genome) in the tetraploid tef genome. This is consistent with previous studies on the diploids maize and sorghum, where br2 and dw3, respectively, are single-copy genes (Multani et al. 2003).
From sequence variation between the two homeologous genes, dw3a and dw3b, an ancestral divergence date of ∼9–10 MYA was calculated for the diploid genomes in tetraploid tef. For the two apparent rht1 homeologues used in this study, Smith and colleagues (Smith et al. 2012) calculated a divergence time of ∼4–5 MYA. Given the significant variation that exists in the rates of divergence for different plant nuclear genes (Zhang et al. 2002), it is clear that many more homeologous pairs need to have their divergence date ascertained in tef, so a relatively robust number can be derived.
Mutagenized tef populations and mutation discovery
For many decades, mutagenesis has been widely used for genetic analysis in plants and animals. Alkylating agents, including EMS, are particularly well suited for higher plants. One of the most challenging problems is to determine the ideal dosage of the mutagen. The toxicity and mutagenicity of EMS vary tremendously from one species to the next. A low-concentration treatment allows greater survival and less subsequent issues with segregation of desired mutations from undesired events, but requires more screening to identify the targeted mutants. In most crops, the commonly used EMS concentration is 0.25–1.5%. For instance, 1% EMS-treated tomato plants yield ∼1.8-fold more mutations per genome than a 0.7% EMS treatment, but the fertility rate was 41% lower than the 0.7% EMS (Minoia et al. 2010). In this tef study, seed treatments of up to 1.5% were well tolerated and provided significantly more mutations than 1% treatment. For the genes targeted in this study, the apparent mutation rate was 2.8× higher in the 1.5% treatment than in the 1% treatment, under the conditions employed.
The size of the mutagenized population is also important to ensure that a sufficient number of targeted mutations will be found. For example, in Arabidopsis, 18,000 EMS mutagenized lines were needed to find a nonsense mutation in any targeted 1.0-kb coding region with 95% confidence (Greene et al. 2003; Parry et al. 2009). Based on the apparent mutation frequency at the dw3 homeologues of tef from prediction, ∼2800 mutagenized lines would be required to identify a nonsense mutation with 95% confidence, given a coding region of 4 kb (dw3) and nonsense mutations that account for ∼3% of the events. In this study, ∼3 kb of the coding region was analyzed by amplicon sequencing for the 2,121 mutagenized pools, so the probability of finding a nonsense mutation was estimated to be 83%, assuming a binomial distribution. However, no nonsense mutation was confirmed.
A total of 120 candidate mutations were identified from four candidate genes, two of each of the dw3/br2 and rth1 homeologues, in 21,210 mutagenized tef lines. This number (120) was arrived at after low-frequency mutations (less than three reads in the amplicon data set) and low-quality reads were removed as probable false positives. Of the 10 candidate genes that were individually investigated by follow-up Sanger sequencing, the 4 mutations with >4 supporting reads (5–19) in the superpool data were all confirmed as true mutations, while the 2 mutations with only 3 supporting reads were found to be false positives. Of the 4 mutations with 4 supporting reads in the superpool data set, 2 were confirmed and 2 found to be false positives. Hence, if we extrapolate to the entire candidate pool, this suggests that the 61 candidate mutations with at least 5 sequences and half of the sequences with 4 reads in the superpool data (18/2 = 9) are likely to be real. This predicted level of confirmation, 70/120, also suggests that the EMS mutation rate is about ∼1.7× lower than our initial calculation. Some mutations may have been missed in this project if they were somewhat underrepresented by mere chance in the bulk sequencing data. A larger redundancy of amplicon reads could have helped solve this problem. Because each pool was derived from a single M1 seed, a mutation in a particular diploid seed embryo would be chimeric in the developing plant and would variably segregate in the M2 generation (due to both heterozygosity and the degree to which the mutated embryonic cell was represented by a clone of cells that made it into the gametophytic lineage). As the sequencing coverage for each pool was estimated at approximately 10×, we should observe 5 reads that harbor this mutation on average. The chance for a bona fide mutation to be observed with frequency <3 is 0.0547 (calculated by binomial distribution). Hence, mutations with a frequency <3 were not further considered. On the other hand, previous studies indicated that EMS mutations are predominantly G/C to A/T transitions, given the frequent alkylation of guanine residues by EMS (Krieg 1963). In Arabidopsis, maize and wheat, >99% of EMS-induced mutations were G/C to A/T transition (Greene et al. 2003; Till et al. 2004; Slade et al. 2005). In the mutagenized tef populations generated for this study, 69% of 1% EMS mutants and 82% of 1.5% EMS were found to be G/C to A/T transitions. A few transversions and A/T to G/C transitions were also observed, similar to the mutation spectrum reported in rice (Till et al. 2007), barley (Caldwell et al. 2004), and tomato (Minoia et al. 2010). The transversion mutations might result from EMS treatment or natural mutations in the population. In addition, PCR/sequencing errors may be responsible for some false positives even though we used high-fidelity Taq polymerase (error rate 4.4 × 10−7). Taq polymerase errors are heavily biased toward A/T to G/C changes (Keohavong and Thilly 1989).
The observed mutation frequency (8.7 mutations/Mbp) for dw3 genes was found to be higher than the rht1 genes (2.7 mutations/Mbp). These frequencies are comparable to previous studies on rice, sorghum, tomato, and Arabidopsis (2–6 mutations/Mbp) (Greene et al. 2003; Xin et al. 2008; Minoia et al. 2010). These values are notably higher than the 1 mutation/Mb that was found in diploid rice and barley as well as tetraploid peanut (Caldwell et al. 2004; Knoll et al. 2011), but much lower than the 24–42 mutations/Mbp documented in tetraploid and hexaploid wheat (Slade et al. 2005), the only species with a mutation rate >1 mutation/50 kb that has been reported so far. A higher mutation rate will, of course, reduce the population size required for effective screening, but requires tolerance of this mutational rate by the mutagenized lines. The mutagenized tef in this project showed excellent vigor and fertility, so it is likely that an even-higher level of mutagenesis would be tolerated.
Phenotypic analysis
We investigated two predicted deleterious mutations, one in each tef dw3 homeologue, in the corresponding M3 generations and validated both by Sanger sequence analysis. Plants segregating for these mutations exhibited no obvious height or other morphological differences at either high-fertilizer input conditions like those usually seen in greenhouse studies or under very low fertilizer inputs like those seen in standard Ethiopian tef agriculture. This suggests that each gene can complement mutations at the other locus.
Next steps
As a tetraploid species, it is challenging to discover mutation phenotypes from the tef genome because homeologous alleles are expected to be present even for a single-copy gene. Unless genes have been lost, subfunctionalized or neofunctionalized, it is likely that mutation in a single homeologous copy, even when homozygous, would not yield a phenotype. Fortunately, two mutations that were predicted to be quite deleterious, G742A and C106T, from each of the homeologous dw3 genes were found in this analysis. These lines will now be crossed to bring these two mutations together and then self or sibling pollinated to generate segregants that are homozygous for mutations in these two dw3 orthologs. It is hoped that some of the resulting lines will show a semi-dwarf character, lodging resistance and improved grain yield. If so, these dw3 alleles can be introgressed into a wide variety of locally adapted tef cultivars, thereby removing any undesired mutations in the mutagenized stock and allowing rigorous field experimentation regarding the value of this approach to tef improvement.
The results demonstrate that next-generation sequencing offers a nontransgenic alternative for the rapid identification of targeted genetic variation. In tef, semi-dwarfing mutations were found that might dramatically improve crop yield, once brought together into the same genetic background. This approach is universally applicable and may provide exceptional value to orphan crop species with a very limited research toolkit and/or research community and to agricultural environments where transgenic improvement might not be tolerated.
Supplementary Material
Acknowledgments
We thank Matt Estep and Michael Payne for laboratory assistance. This research was supported by the Umbarger Professorship at Purdue University and the Giles Professorship at the University of Georgia.
Footnotes
Communicating editor: M. Johnston
Sequence data from this article have been deposited with the GenBank Data Libraries under accession nos. JN672669–JN672670.
Literature Cited
- Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., et al. , 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano K., Hirano K., Ueguchi-Tanaka M., Angeles-Shim R. B., Komura T., et al. , 2009. Isolation and characterization of dominant dwarf mutants, Slr1-d, in rice. Mol. Gen. Genet. 281: 223–231. [DOI] [PubMed] [Google Scholar]
- Assefa K., Yu J. K., Zeid M., Belay G., Tefera H., et al. , 2011. Breeding tef [Eragrostis tef (Zucc.) trotter]: conventional and molecular approaches. Plant Breed. 130: 1–9. [Google Scholar]
- Auerbach C., Robson J. M., 1946. Chemical production of mutations. Nature 157: 302. [DOI] [PubMed] [Google Scholar]
- Ayele M., Doleže J., Duren M., Brunner H., Zapata-Arias F., 1996. Flow cytometric analysis of nuclear genome of the Ethiopian cereal Tef. [Eragrostis tef (Zucc.) Trotter] Genetica 98: 211–215. [Google Scholar]
- Bundock P. C., Eliott F. G., Ablett G., Benson A. D., Casu R. E., et al. , 2009. Targeted single nucleotide polymorphism (SNP) discovery in a highly polyploid plant species using 454 sequencing. Plant Biotechnol. J. 7: 347–354. [DOI] [PubMed] [Google Scholar]
- Burge C., Karlin S., 1997. Prediction of complete gene structures in human genomic DNA1. J. Mol. Biol. 268: 78–94. [DOI] [PubMed] [Google Scholar]
- Caldwell D. G., McCallum N., Shaw P., Muehlbauer G. J., Marshall D. F., et al. , 2004. A structured mutant population for forward and reverse genetics in Barley (Hordeum vulgare L.). Plant J. 40: 143–150. [DOI] [PubMed] [Google Scholar]
- Central Statistics Authority, 2005 Agricultural Sample Survey 2004/2005 Report on area and production for major crops, vol. 1. CSA, Addis Ababa, Ethiopia. [Google Scholar]
- Comai L., Young K., Till B. J., Reynolds S. H., Greene E. A., et al. , 2004. Efficient discovery of DNA polymorphisms in natural populations by Ecotilling. Plant J. 37: 778–786. [DOI] [PubMed] [Google Scholar]
- Cooper J. L., Till B. J., Laport R. G., Darlow M. C., Kleffner J. M., et al. , 2008. TILLING to detect induced mutations in soybean. BMC Plant Biol. 8: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalmais M., Schmidt J., Le Signor C., Moussy F., Burstin J., et al. , 2008. UTILLdb, a Pisum sativum in silico forward and reverse genetics tool. Genome Biol. 9: R43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrenberg L., Gustafsson A., 1957. On the mutagenic action of ethylene oxide and di-epoxybutane in barley. Hereditas 43: 595–602. [Google Scholar]
- Foster K. W., Rutger J. N., 1978. Inheritance of semidwarfism in rice, Oryza sativa L. Genetics 88: 559–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene E. A., Codomo C. A., Taylor N. E., Henikoff J. G., Till B. J., et al. , 2003. Spectrum of chemically induced mutations from a large-scale reverse-genetic screen in Arabidopsis. Genetics 164: 731–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Z., Ueguchi-Tanaka M., Umemura K., Uozu S., Fujioka S., et al. , 2003. A rice brassinosteroid-deficient mutant, ebisu dwarf (d2), is caused by a loss of function of a new member of cytochrome P450. Plant Cell 15: 2900–2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh H., Ueguchi-Tanaka M., Sentoku N., Kitano H., Matsuoka M., et al. , 2001. Cloning and functional analysis of two gibberellin 3-hydroxylase genes that are differently expressed during the growth of rice. Proc. Natl. Acad. Sci. USA 98: 8909–8914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings P. R., 1964. Plant type as a rice breeding objective. Crop Sci. 4: 13–15. [Google Scholar]
- Karper R. E., 1932. A dominant mutation of frequent recurrence in sorghum. Am. Nat. 66: 511–529. [Google Scholar]
- Keohavong P., Thilly W. G., 1989. Fidelity of DNA polymerases in DNA amplification. Proc. Natl. Acad. Sci. USA 86: 9253–9257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoll J. E., Ramos M. L., Zeng Y., Holbrook C. C., Chow M., et al. , 2011. TILLING for allergen reduction and improvement of quality traits in peanut (Arachis hypogaea L.). BMC Plant Biol. 11: 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieg D. R., 1963. Ethyl methanesulfonate-induced reversion of bacteriophage T4rII mutants. Genetics 48: 561–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamanova L., Coffey A. J., Scott C. E., Kozarewa I., Turner E. H., et al. , 2010. Target-enrichment strategies for next-generation sequencing. Nat. Methods 7: 111–118. [DOI] [PubMed] [Google Scholar]
- Martín B., Ramiro M., Martínez-Zapater J. M., Alonso-Blanco C., 2009. A high-density collection of EMS-induced mutations for TILLING in Landsberg erecta genetic background of Arabidopsis. BMC Plant Biol. 9: 147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minoia S., Petrozza A., D’Onofrio O., Piron F., Mosca G., et al. , 2010. A new mutant genetic resource for tomato crop improvement by TILLING technology. BMC Res. Notes 3: 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monna L., Kitazawa N., Yoshino R., Suzuki J., Masuda H., et al. , 2002. Positional cloning of rice semidwarfing gene, sd-1: rice “green revolution gene” encodes a mutant enzyme involved in gibberellin synthesis. DNA Res. 9: 11–17. [DOI] [PubMed] [Google Scholar]
- Muangprom A., 2005. A novel dwarfing mutation in a green revolution gene from Brassica rapa. Plant Physiol. 137: 931–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Multani D. S., Briggs S. P., Chamberlin M. A., Blakeslee J. J., Murphy A. S., et al. , 2003. Loss of an MDR transporter in compact stalks of maize br2 and sorghum dw3 mutants. Science 302: 81–84. [DOI] [PubMed] [Google Scholar]
- Munkvold G. P., Hellmich R. L., 2000. Genetically modified, insect resistant maize: implications for management of ear and stalk diseases. Plant Health Prog., http://www.planthealthprogress.org/current/reviews/maize/ 10.1094/PHP-2000-0912-01-RV. [DOI] [Google Scholar]
- Murray M. G., Thompson W., 1980. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 8: 4321–4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natarajan A. T., Ramanna M. S., 1966. Modification of relative mutagenic efficiency in barley of mesyloxy esters by different treatments. Nature 211: 1099–1100.5970109 [Google Scholar]
- Nei M., Gojobori T., 1986. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 3: 418–426. [DOI] [PubMed] [Google Scholar]
- Ng P. C., 2003. SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Res. 31: 3812–3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng S. B., Buckingham K. J., Lee C., Bigham A. W., Tabor H. K., et al. , 2010a Exome sequencing identifies the cause of a mendelian disorder. Nat. Genet. 42: 30–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng S. B., Bigham A. W., Buckingham K. J., Hannibal M. C., McMillin M. J., et al. , 2010b Exome sequencing identifies MLL2 mutations as a cause of Kabuki syndrome. Nat. Genet. 42: 790–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry M. A. J., Madgwick P. J., Bayon C., Tearall K., Hernandez-Lopez A., et al. , 2009. Mutation discovery for crop improvement. J. Exp. Bot. 60: 2817–2825. [DOI] [PubMed] [Google Scholar]
- Peng J., Richards D. E., Hartley N. M., Murphy G. P., Devos K. M., et al. , 1999. ‘Green revolution’ genes encode mutant gibberellin response modulators. Science 400: 256–261. [DOI] [PubMed] [Google Scholar]
- Quinby J., Karper R., 1954. Inheritance of height in sorghum. Agron. J. 46: 211–216. [Google Scholar]
- Rapoport I. A., 1946. Carbonyl compounds and the chemical mechanism of mutations. Compt. Rend. Acad. Sci. 54: 65–67. [Google Scholar]
- Rigola D., Van Oeveren J., Janssen A., Bonné A., Schneiders H., et al. , 2009. High-throughput detection of induced mutations and natural variation using KeyPoint technology. PLoS ONE 4: e4761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto T., Morinaka Y., Ohnishi T., Sunohara H., Fujioka S., et al. , 2005. Erect leaves caused by brassinosteroid deficiency increase biomass production and grain yield in rice. Nat. Biotechnol. 24: 105–109. [DOI] [PubMed] [Google Scholar]
- Sasaki A., Ashikari M., Ueguchi-Tanaka M., Itoh H., Nishimura A., et al. , 2002. A mutant gibberellin-synthesis gene in rice. Nature 416: 701–702. [DOI] [PubMed] [Google Scholar]
- Slade A. J., Fuerstenberg S. I., Loeffler D., Steine M. N., Facciotti D., 2005. A reverse genetic, nontransgenic approach to wheat crop improvement by TILLING. Nat. Biotechnol. 23: 75–81. [DOI] [PubMed] [Google Scholar]
- Smith S.M., Yuan Y., Doust A. N., Bennetzen J. L., 2012. Haplotype analysis and linkage disequilibrium at five loci in Eragrostis tef. G3: Genes Genomes Genetics 2: 407–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spielmeyer W., 2002. Semidwarf (sd-1), “green revolution” rice, contains a defective gibberellin 20-oxidase gene. Proc. Natl. Acad. Sci. USA 99: 9043–9048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stallknecht, G. F., K. M. Gilbertson, and J. Eckhoff, 1993 Teff: food crop for humans and animals, pp. 604–615 in New Crops, edited by J. Janick and J. E. Simon. Wiley, New York. [Google Scholar]
- Suzuki T., Eiguchi M., Kumamaru T., Satoh H., Matsusaka H., et al. , 2008. MNU-induced mutant pools and high performance TILLING enable finding of any gene mutation in rice. Mol. Gen. Genet. 279: 213–223. [DOI] [PubMed] [Google Scholar]
- Swigonova Z., Lai J., Ma J., Ramakrishna W., Llaca V., et al. , 2004. On the tetraploid origin of the maize genome. Comp. Funct. Genomics 5: 281–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanabe S., Ashikari M., Fujioka S., Takatsuto S., Yoshida S., et al. , 2005. A novel cytochrome P450 is implicated in brassinosteroid biosynthesis via the characterization of a rice dwarf mutant, dwarf11, with reduced seed length. Plant Cell 17: 776–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teferra T., Tefera H., Simane B., Tuinstra M., 2000. The influence of drought stress on yield of tef (Eragrostis tef). Trop. Sci. 40: 40–45. [Google Scholar]
- Till B. J., Reynolds S. H., Greene E. A., Codomo C. A., Enns L. C., et al. , 2003. Large-scale discovery of induced point mutations with high-throughput TILLING. Genome Res. 13: 524–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Till B. J., Reynolds S. H., Weil C., Springer N., Burtner C., et al. , 2004. Discovery of induced point mutations in maize genes by TILLING. BMC Plant Biol. 4: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Till B. J., Cooper J., Tai T. H., Colowit P., Greene E. A., et al. , 2007. Discovery of chemically induced mutations in rice by TILLING. BMC Plant Biol. 7: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walcott J. J., Laing D. R., 1976. Some physiological aspects of growth and yield in wheat crops: a comparison of a semidwarf and standard height cultivar. Aust. J. Exp. Agric. Anim. Husb. 16: 578–587. [Google Scholar]
- Xin Z., Wang L., Barkley N. A., Burow G., Franks C., et al. , 2008. Applying genotyping (TILLING) and phenotyping analyses to elucidate gene function in a chemically induced sorghum mutant population. BMC Plant Biol. 8: 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J.-K., Graznak E., Breseghello F., Tefera H., Sorrells M. E., 2007. QTL mapping of agronomic traits in tef. [Eragrostis tef (Zucc) Trotter] BMC Plant Biol. 7: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Vision T. J., Gaut B. S., 2002. Patterns of nucleotide substitution among simultaneously duplicated gene pairs in Arabidopsis thaliana. Mol. Biol. Evol. 19: 1464. [DOI] [PubMed] [Google Scholar]
- Zou J., Zhang S., Zhang W., Li G., Chen Z., et al. , 2006. The rice HIGH-TILLERING DWARF1 encoding an ortholog of Arabidopsis MAX3 is required for negative regulation of the outgrowth of axillary buds. Plant J. 48: 687–698. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.