Abstract
Nucleoside analogs are frequently used to label newly synthesized DNA. These analogs are toxic in many cells, with the exception of the budding yeast. We show that Schizosaccharomyces pombe behaves similarly to metazoans in response to analogs 5-bromo-2′-deoxyuridine (BrdU) and 5-ethynyl-2′-deoxyuridine (EdU). Incorporation causes DNA damage that activates the damage checkpoint kinase Chk1 and sensitizes cells to UV light and other DNA-damaging drugs. Replication checkpoint mutant cds1Δ shows increased DNA damage response after exposure. Finally, we demonstrate that the response to BrdU is influenced by the ribonucleotide reductase inhibitor, Spd1, suggesting that BrdU causes dNTP pool imbalance in fission yeast, as in metazoans. Consistent with this, we show that excess thymidine induces G1 arrest in wild-type fission yeast expressing thymidine kinase. Thus, fission yeast responds to nucleoside analogs similarly to mammalian cells, which has implications for their use in replication and damage research, as well as for dNTP metabolism.
Keywords: dNTP metabolism, DNA damage response, replication checkpoint, thymidine arrest, thymidine salvage pathway
UNDERSTANDING mechanisms that maintain DNA replication fidelity is key to understanding cancer (Barlogie et al. 1976; Johnson et al. 1981; Christov and Vassilev 1988). Replication must be controlled to avoid mutations, promote DNA repair, and restrain rereplication. Predictably, abnormally replicating and proliferating cells are a hallmark of tumorigenesis (Barlogie et al. 1976; Johnson et al. 1981; Christov and Vassilev 1988). Thus, accurate analysis of replication states is essential to understanding the response of a cell population under study.
Studies of replication dynamics rely on manipulations of DNA nucleotide metabolism. For example, the drug hydroxyurea (HU) is commonly used to inhibit nucleotide synthesis. This causes replication fork stalling and checkpoint activation until dNTP levels are reestablished (Santocanale and Diffley 1998; Kim and Huberman 2001; Lopes et al. 2001; Alvino et al. 2007; Poli et al. 2012). Dysregulation of nucleotide levels is associated with disruptions in cell-cycle and checkpoint control (Kumar et al. 2010, 2011; Davidson et al. 2012; Poli et al. 2012). The regulatory response to nucleotide levels is different between the yeasts Saccharomyces cerevisiae and Schizosaccharomyces pombe. Budding yeast S. cerevisiae is highly resistant to HU, and its checkpoint proteins directly regulate nucleotide biosynthesis during the normal cell cycle (Chabes et al. 2003; Poli et al. 2012). In contrast, fission yeast S. pombe is sensitive to much lower levels of HU and uses checkpoint-independent mechanisms to control nucleotide levels during normal cell growth (Hakansson et al. 2006; Nestoras et al. 2010).
Exposure to exogenous nucleosides alters metazoan dNTP metabolism and cell-cycle dynamics (Meuth and Green 1974a,b), which is a potential problem in assays that rely on incorporation of modified or antigenic nucleoside analogs. By measuring incorporation of analog, the replicative capacity of a culture is inferred (Bohmer and Ellwart 1981; Crissman and Steinkamp 1987; Frum and Deb 2003), providing a direct metric of DNA synthesis. Thymidine analogs 5-bromo-2′-deoxyuridine (BrdU) and 5-ethynyl-2′-deoxyuridine (EdU) (Diermeier-Daucher et al. 2009) are commonly used nucleosides. BrdU is detected using antibodies (Hodson et al. 2003), while EdU is covalently labeled by bio-orthogonal, copper-based chemistry (Buck et al. 2008). In yeasts, which lack a thymidine salvage pathway, analog incorporation requires an exogenous thymidine kinase (e.g., herpes simplex virus, hsv-tk+) (Sclafani and Fangman 1986; Hodson et al. 2003; Sivakumar et al. 2004; Viggiani and Aparicio 2006).
A disadvantage to this method is the inherent toxicity of nucleoside analogs such as BrdU, FdU, and IdU (Sivakumar et al. 2004). Observations in bacteria and mammals indicate that BrdU is both a mutagen and a teratogen (Davidson and Kaufman 1978; Kaufman and Davidson 1978; Lasken and Goodman 1984), able to cause T-C transition mutations (Goodman et al. 1985). However, up to 400 µg/ml BrdU may be added to a culture of S. cerevisiae before toxic effects are observed (Lengronne et al. 2001). In fission yeast, both EdU and BrdU analogs show toxicity at much lower doses and may activate a Rad3 (ATR/Mec1)-dependent damage response pathway (Hodson et al. 2003; Sivakumar et al. 2004; Hua and Kearsey 2011). Differences in dosage sensitivity may reflect the differences in nucleotide metabolism between these different yeast species.
We examine the range of toxicity and the induction of DNA damage in S. pombe cells that incorporate either BrdU or EdU. Checkpoint mutants rad3Δ, chk1Δ, and cds1Δ show hypersensitivity to BrdU and EdU, which is exacerbated in minimal media. Cells exposed to low doses of BrdU are sensitized to additional DNA-damaging agents and show increased mutation rates. Even in low-dose analog, we observe induction of damage markers including phosphorylated histone H2A and Rad52 foci. Chk1 and Cdc2 phosphorylation indicates the DNA damage checkpoint is activated, showing that analog incorporation generates DNA damage.
Consistent with this, we observe that completion of S phase and entry to the next cell cycle is delayed with increased analog dose. This effect is not limited to analogs, however, because fission yeast cells exposed to thymidine undergo G1 arrest, similar to thymidine synchronization in human cells. Finally, the ribonucleotide reductase (RNR) inhibitor Spd1 modulates cell response to BrdU and EdU. While spd1Δ cells are less sensitive to chronic exposure, acute viability is decreased and surviving spd1Δ cells frequently carry mutations. Thus, the power of using analog incorporation to detect new DNA synthesis must be tempered by appropriate cautions to maximize relevant conclusions and minimize disruptions to normal nucleotide metabolism.
Materials and Methods
Yeast strains, analog addition, growth, and mutagenesis
Fission yeast strains are described in Table 1. Cells were grown as described in Sabatinos and Forsburg (2010), and BrdU (5 mg/ml in water) or EdU (10 mM stock; Invitrogen, Carlsbad, CA) was added as required to liquid cultures or plates. Media were yeast extract with supplements (YE5S, hereafter “YES”), Edinburgh minimal medium (EMM–ammonium chloride nitrogen source), or S. pombe minimal glutamate medium (PMG– sodium glutamate nitrogen source) (Sabatinos and Forsburg 2010). Physiology experiments including Chk1 protein, Rad52 foci, mutagenesis, and flow cytometry were all performed in liquid EMM cultures. Protein extracts were prepared from 0.3 M NaOH-treated cells and lysed in 2× SDS–PAGE sample buffer by boiling for 5 min, as described in Sabatinos and Forsburg (2010). The choice of HA monoclonal produced a different nonspecific background band location on Chk1-HA blots (Roche 12CA5 or Covance 16B12).
Table 1 . Fission yeast strains used in this study.
| Strain no. | Genotype | Source |
|---|---|---|
| FY2317 | h+ leu1-32::hENT1-leu1+(pJAH29) his7-366::hsv-tk-his7+(pJAH31) ura4-D18 ade6-M210 | Hodson et al. (2003) |
| FY3179 | h+ mrc1Δ::ura4+ leu1-32::hENT1-leu1+(pJAH29) his7-366::hsv-tk-his7+(pJAH31) ura4-D18 ade6-M210 | This study |
| FY3454 | h+ ura4-D18 ade6-M210 | This study |
| FY5148 | h+ cds1Δ::ura4 leu1-32::hENT1-leu1+(pJAH29) his7-366::hsv-tk-his7+(pJAH31) ura4-D18 ade6-M210 | This study |
| FY5149 | h+ chk1Δ::ura4+ leu1-32::hENT1-leu1+(pJAH29) his7-366::hsv-tk-his7+(pJAH31) ura4-D18 ade-704 | This study |
| FY5150 | h+ rad3Δ::ura4+ leu1-32::hENT1-leu1+(pJAH29) his7-366::hsv-tk-his7+(pJAH31) ura4-D18 ade6-M210 | This study |
| FY5155 | h− cds1Δ::ura4+ pola-FLAG::ura4+ rad11-myc::KanMX6 rad22-YFP::natMX leu1-32::[hENT-leu1+] his7-366::[hsv-tk his7+] ura4-D18 ade6-M210 | This study |
| FY5159 | h− pola-FLAG::ura4+ rad11-myc::KanMX6 rad22-YFP::natMX leu1-32::[hENT-leu1+] his7-366::[hsv-tk his7+] ura4-D18 ade6-M210 | This study |
| FY5030 | h− cds1-13myc::KanMX leu1-32::[hENT leu1+] his7-366::[his7+] ade6-M210 ura4-D18 | This study |
| FY5031 | h+ cds1-13myc::KanMX chk1HA leu1-32::[hENT leu1+] his7-366::[hsv-tk his7+] ade6-M216 ura4-D18 | This study |
| FY6247 | h+ Δspd1::ura4+ ura4-D18 leu1-32::hENT1-leu1+(pJAH29) his7-366::hsv-tk-his7+(pJAH31) ade6-? | This study |
Mutation rate was determined by splitting cultures (±BrdU), treating them with 32.6 µM BrdU for 2 hr, and then plating them on YES and PMG + canavanine. Canavanine plates (70 µg/ml in PMG+ supplements) were scored after 8 days growth (32°) and numbers of Can1− colonies were compared to the concentration derived from titer plates (YES). Grouped experiments for Table 2 and Figure 7 were performed independently and rates calculated using FALCOR (http://www.keshavsingh.org/protocols/FALCOR.html) and the Ma–Sandri–Sarkar Maximum-Likelihood Estimator (MSS-MLE) or the Lea–Coulson method of the median. MSS-MLE results were analyzed by a two-tailed t-test, and Mann–Whitney U-tests were used for Lea–Coulson significance testing (http://vassarstats.net/utest.html). Frequency of hsv-tk+ loss was scored as the number of FUdR-resistant or sectored colonies per total, and the proportions were assessed with Z-tests (http://vassarstats.net/propdiff_ind.html).
Table 2 . Mutation rates for spontaneous and BrdU-induced canavanine forward mutation analysis.
| Strain | Treatmenta | No. mutations (m)b | can1− rate per 107 generationsb | 95% C.I.b | t-testc |
|---|---|---|---|---|---|
| Wild type, non-inc (FY3454) | Untreated (n = 8) | 4.669 | 5.59 | 3.02/8.70 | * |
| +BrdU (n = 8) | 4.77 | 6.82 | 3.70/10.60 | ||
| Wild type (FY2317) | Untreated (n = 8) | 5.601 | 9.56 | 5.37/14.59 | P < 0.001 |
| +BrdU (n = 7) | 9.079 | 14.19 | 8.41/21.00 | ||
| cds1Δ (FY5148) | Untreated (n = 8) | 5.617 | 9.84 | 5.53/15.01 | P < 0.025 |
| +BrdU (n = 8) | 7.350 | 14.21 | 8.43/21.05 | ||
| chk1Δ (FY5149) | Untreated (n = 8) | 8.079 | 15.82 | 9.55/23.19 | * |
| +BrdU (n = 8) | 8.714 | 16.80 | 10.28/24.45 | ||
| rad3Δ (FY5150) | Untreated (n = 8) | 4.272 | 9.68 | 5.12/15.24 | P < 0.025 |
| +BrdU (n = 8) | 5.868 | 9.56 | 5.43/14.52 | ||
| mrc1Δ (FY3179) | Untreated (n = 8) | 6.525 | 11.36 | 6.59/17.04 | P < 0.025 |
| +BrdU (n = 8) | 8.393 | 9.23 | 5.61/13.48 |
Experiments were plated in duplicate and results summed for analysis. Total number of biological replicate experiments is indicated (“n”) for each sample.
Number of mutations (m), mutation rate, and 95% confidence interval (C.I.) calculated using the Ma–Sandri–Sarkar maximum-likelihood estimator (MSS-MLE) method FALCOR calculator (http://www.keshavsingh.org/protocols/FALCOR.html). Data are from eight independent assays (Hall et al. 2009).
Pairwise two-tailed t-test calculated within genotypes with or without BrdU from MSS-MLE results, using mutation number (m) and calculated variance, with (nwithBrdU + nwithoutBrdU − 2) d.f. *Not significant, P > 0.05.
Figure 7 .
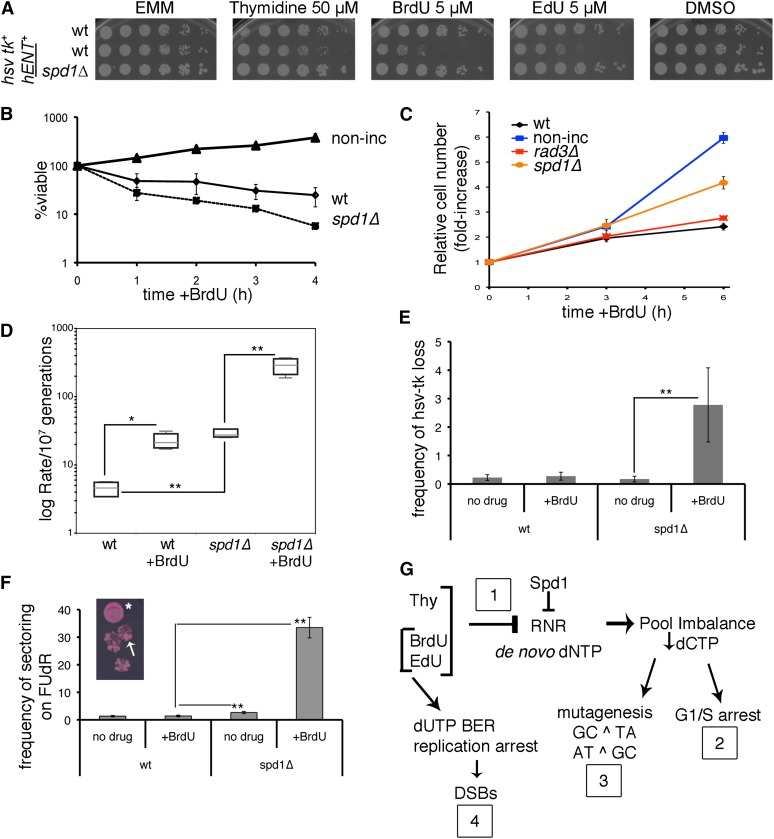
Spd1 protects cells from division and mutation during dNTP imbalance. (A) Comparison between wild-type and spd1Δ hsv-tk+ hENT+ strains by spot test on EMM plates containing BrdU, EdU, or thymidine. DMSO is a vehicle control for EdU. Shown is the minimal dose where wild-type cells began to show sensitivity to analogs. Strains FY2317, -3454, and -6247 are shown. (B) Cultures were treated with 32.6 µM BrdU and plated to calculate viability relative to 0 hr. Wild-type (wt) and spd1Δ strains express hsv-tk+ hENT+ (FY2317 and -6247), while the nonincorporating control (non-inc, FY3454) does not. Shown are mean viability values from three independent experiments ± SEM. (C) Proliferation was monitored by counting cell concentration during BrdU treatment for cultures as in A, in addition to rad3Δ hsv-tk+ hENT+ (FY5150). Shown are mean viability values (n = 3 experiments) ± SEM. (D) Canavanine mutation was scored for incorporating wild-type and spd1Δ strains (FY2317 and -6247), with or without 32.6 µM BrdU treatment (2 hr, 32°). Lea and Coulson fluctuation analysis was used to calculate the rate of can1− forward mutation (per 107 generations) in independent cultures over three experiments (wt, n = 12; spd1Δ, n = 15). Shown are median mutation rates with quartile bounding boxes and 95% C.I. error whiskers. Significance was assessed by two-tailed pairwise Mann–Whitney U-tests, *P = 0.0001, **P < 0.0001. (E) Colonies of wild-type or spd1Δ cells, untreated (no drug) or following 2 hr in 32.6 µM BrdU, were grown on YES and then replicated onto medium with FUdR to score for hsv-tk+ loss. Significance was assessed by two-tailed Z-test (**P < 0.0002). (F) Enhanced sectoring of colonies on FUdR was noted for spd1Δ cells either untreated or following BrdU exposure as in E (two-tailed Z-test, ** P < 0.0002), compared to BrdU-treated wild-type cells. Inset, example of spd1Δ colony with hsv-tk+ loss (*) or sectored area (arrow). Frequencies were calculated from independent experiments, presented with 95% C.I. (G) Model for the effect of exogenous thymidine (Thy) and nucleoside analogs in fission yeast cells expressing a reconstituted thymidine salvage pathway (hsv-tk+). Details are in Discussion.
Microscopy
Cells were fixed in cold 70% ethanol for cell-cycle analysis or microscopy. Cells were rehydrated in water and incubated for 10 min in 1 mg/ml of aniline blue [Sigma (St. Louis) M6900]. Cells in mount (50% glycerol, 1 µg/ml DAPI, and 1 µg/ml p-phenylenediamine) were photographed on a Leica DMR wide-field epifluorescent microscope, using a 63× objective lens (NA 1.32 Plan Apo), a 100-W Hg arc lamp for excitation, and a 12-bit Hamamatsu ORCA-100 CCD camera. OpenLab v3.1.7 (Improvision, Lexington, MA) software was used at acquisition and ImageJ (Schneider et al. 2012) for analysis. DAPI counterstaining did not significantly affect BrdU or EdU signal intensity.
Live-cell Rad52-YFP foci were imaged on a DeltaVision Core microscope (with softWoRx v4.1; Applied Precision, Issaquah, WA), using a 60× (NA1.4 Plan Apo) lens, a solid-state illuminator, and a 12-bit Roper CoolSnap HQII CCD camera. The system’s x-y pixel size was 0.1092 µm. YFP fluorescence for single time points was acquired as eighteen 0.3-µm z-sections. Long-term time-lapse movies used nine z-steps of 0.5 µm. Rad52-YFP images were deconvolved and maximum-intensity projected (softWoRx). Movies were performed in CellAsics (Hayward, CA) microfluidics plates (Y04C series), with supplemented EMM medium. Transmitted light images were fused with DAPI or Rad52-projected images. Images were contrast adjusted using a histogram stretch and equivalent scale in all samples. A threshold of 2× over the average nuclear YFP signal was used for focus discrimination. Rad52 foci are presented as the proportion of nuclei with Rad52 foci ±95% confidence interval (C.I.). Significance was assessed with chi-square tests.
Flow cytometry
Whole-cell SytoxGreen flow cytometry (FACS) was performed as described in Sabatinos and Forsburg (2009). Whole-cell FACS for EdU was performed using Click-iT (Invitrogen) with AlexaFluor 488 on rehydrated cells. FACS for BrdU was performed on “ghosts” (Carlson et al. 1997), prepared by spheroplasting cells in 0.1 M KCl with 5 mg/ml Zymolyase 20T and then 1% Triton X-100 before sonicating to release nuclei. Nuclei were blocked (10% FCS, 1% BSA) for 30 min and then incubated with mouse anti-BrdU (Becton Dickinson; B44). Secondary antibody was Alexa-488–conjugated anti-mouse (Invitrogen), and nuclei were counterstained with propidium iodide for total DNA content.
Results
Cell signal and viability vary with analog dose
Acute nucleoside analog toxicity is proportional to dose in human cells (Popescu 1999). To compare this with fission yeast we used BrdU and EdU concentrations typically used in replication studies. Our strains contained hsv-tk+ and the human equilibrative nucleoside transporter (hENT), which allowed us to use lower analog doses and achieve effective transport into cells (Hodson et al. 2003; Sivakumar et al. 2004). Since BrdU dose is frequently presented in micrograms per milliliter, we began with a starting BrdU dose of 100 µg/ml (326 µM), half the dose used in fission yeast genome labeling work without hENT (e.g., Hayano et al. 2011) and less than commonly used in budding yeast (e.g., Lengronne et al. 2001). However, to compare between BrdU (typically milligrams per milliliter) and EdU (typically micromolar), we report all doses in molarity. The starting dose for each (326 µM BrdU and 10 µM EdU) was determined by previous protocols.
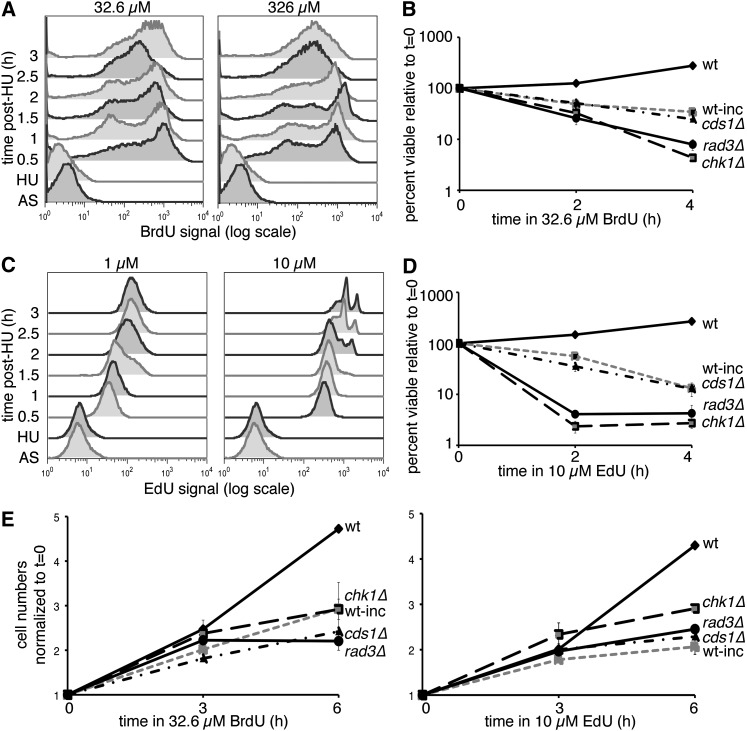
We compared BrdU signals, using flow cytometry on HU-blocked cells released into BrdU (32.6 or 326 µM) or EdU (1 or 10 µM), to determine whether there was a corresponding decrease in the resulting signal (Figure 1). Low-dose BrdU (32.5 µM) produced a similar signal to the full dose of 326 µM, but with better viability (Figure 1A). EdU toxicity appeared at a dose of 10 µM (Figure 1D), consistent with results in Hua and Kearsey (2011), yet a 10-fold decrease in EdU dose to 1 µM significantly decreased EdU signal intensity (Figure 1C). Thus, minimum doses of 32.6 µM BrdU and 10 µM EdU are required for optimal short-term labeling in hsv-tk+ hENT+ strains. Higher doses of nucleoside analog may be required to detect small incorporation differences during short incubations, but enhanced signal comes at a cost to cell viability.
Figure 1 .
BrdU and EdU doses affect signal, viability, and cell division. (A) Time course of incorporation at 32.6 or 326 µM BrdU in HU-synchronized cells after release. Asynchronous (AS) cultures were blocked for 4 hr in HU (HU time point) before release at 32° in medium with BrdU at the indicated concentration. BrdU signal was detected in isolated nuclei. (B) Relative viability during 32.6 µM BrdU incubation, comparing nonincorporating (wt) or hsv-tk+ hENT+ cells, both wild type (wt-inc) and checkpoint mutants. Means ± SEM of three experiments are shown. (C) As in A, time course of EdU incorporation at 1 and 10 µM doses in HU-synchronized cells postrelease. Whole cells were treated with ClickIt reaction before flow cytometry. (D) Relative viability in 10 µM EdU treatment over time for nonincorporating (wt) or incorporating wild-type (wt-inc) or checkpoint-mutant incorporating cells. Means ± SEM of three experiments are shown. (E) Cells were counted during BrdU or EdU treatment to determine proliferation in nonincorporating (wt) or hsv-tk+ hENT+ wild-type (wt-inc) and checkpoint-mutant cells. Cell concentrations were normalized to the 0-hr sample for each cell line/condition and are shown as means ± SEM (n = 3).
Disruptions in DNA replication and repair activate checkpoint pathways (reviewed in Sabatinos and Forsburg 2012). The master regulator in fission yeast is the Rad3ATR kinase, which activates Chk1 kinase at DNA double-strand breaks (DSBs). During replication fork stalling, Cds1 kinase is activated by Rad3 via the fork processivity factor Mrc1. Mutations in these checkpoint proteins cause sensitivity to drugs that activate the replication checkpoint. Previous work indicated that Rad3 is required for viability in EdU, but did not investigate the downstream pathways required (Hua and Kearsey 2011).
We examined cell viability in replication checkpoint mutant cds1Δ and DNA damage checkpoint mutant chk1Δ, compared to wild-type and rad3Δ incorporating strains (Figure 1, B and D). We found chk1Δ and rad3Δ are strikingly sensitive to BrdU and EdU treatment. While cds1Δ behaved similarly to wild-type cells during the first 4 hr of incubation, prolonged BrdU exposure (8 hr) caused decreased cds1Δ viability (similar results for mrc1Δ, data not shown). Lower BrdU and EdU doses improved viability in all genotypes (Supporting Information, Figure S1), yet rad3Δ and chk1Δ cells were still the most EdU sensitive.
Wild-type cells without the incorporation cassette continued to divide normally during exposure (Figure 1E). Wild-type hsv-tk+ hENT+ cells had reduced division during analog exposure, as did cds1Δ, rad3Δ, and mrc1Δ (data not shown). In contrast, the chk1Δ incorporating strain continued to divide in BrdU and EdU, suggesting that Chk1 activation is required to inhibit cell division during exposure.
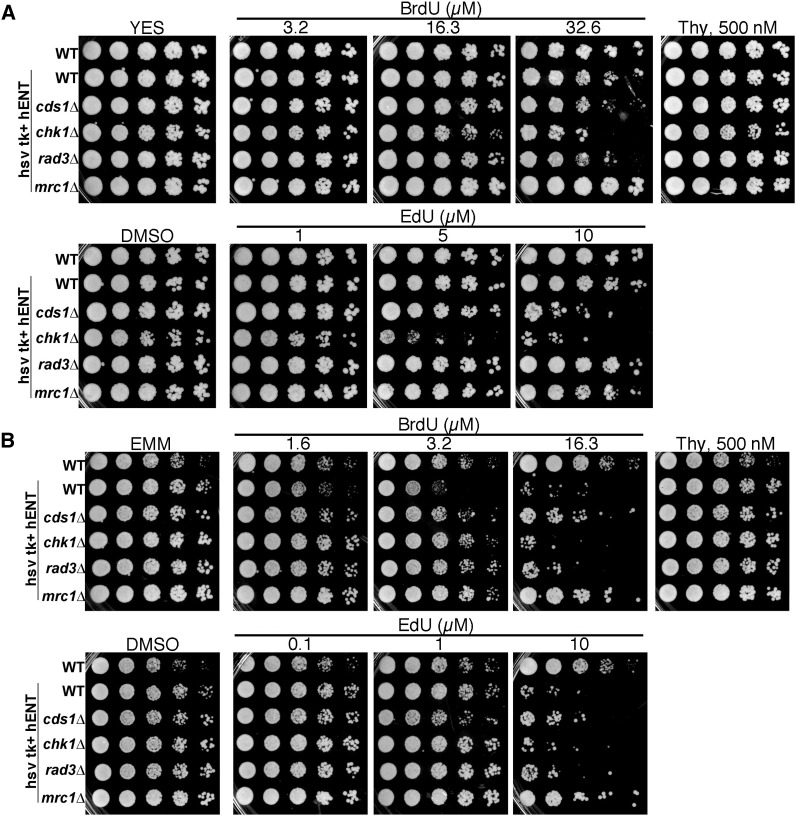
Checkpoint and cell-cycle phenotypes can be sensitive to media composition. We examined sensitivity to BrdU and EdU in spot tests on three media types: rich YES, defined EMM, and lower-nitrogen PMG (Figure 2 and Figure S1C). On YES, chk1Δ incorporating cells show modest sensitivity at 16.3 µM, while growth inhibition in rad3Δ and cds1Δ began to show at 32.6 µM (Figure 2A). EdU sensitivity was highest in chk1Δ on YES (5 µM) followed by cds1Δ (10 µM). Unexpectedly, rad3Δ sensitivity was similar to wild type at 10 µM EdU in YES (Figure 2A).
Figure 2 .
Media formulation alters BrdU and EdU sensitivity. (A) Serial dilution assay in YES medium of nonincorporating wild-type (WT) and hsv-tk+ hENT+ WT and checkpoint-mutant strains cds1Δ, chk1Δ, rad3Δ, and mrc1Δ. Plates containing BrdU, thymidine (Thy) control, or EdU were compared using 1/5 dilutions of cells, grown at 32° for 3 days. (B) As in A, spot tests on defined nitrogen-rich EMM medium. Refer to Figure S1 for PMG media effects.
On EMM, which contains high-level nitrogen, we found that wild-type, chk1Δ, and rad3Δ hsv-tk+ hENT+ cells were all BrdU sensitive (Figure 2B). Yet, mrc1Δ and cds1Δ were less sensitive. Wild-type, chk1Δ, and rad3Δ hsv-tk+ hENT+ cells were most sensitive at 10 µM EdU in EMM, followed by cds1Δ and mrc1Δ. Nonincorporating cells were not sensitive to EdU or BrdU on any media.
PMG medium contains low levels of nitrogen. All incorporator strains were sensitive to 16.3 µM BrdU in PMG (Figure S1C), above which there was little growth. Similarly, EdU sensitivity was higher in PMG and chk1Δ cells were most sensitive to EdU in PMG, followed by cds1Δ. Thus, cells are less sensitive to analogs on rich medium than on minimal medium, while chk1Δ restricts growth in all cases.
S-phase progression is slowed by analog incorporation
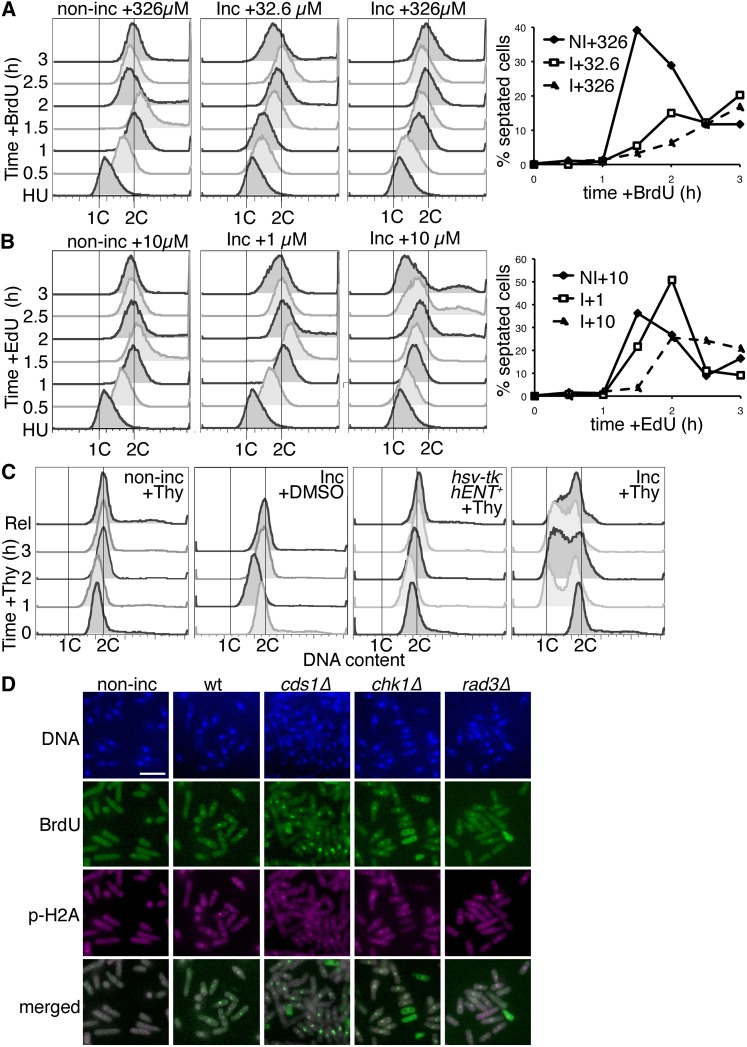
We next tested whether analog incorporation has an effect on DNA synthesis. We blocked cells in early S phase with HU and released them into medium with low- or high-dose BrdU or EdU to monitor replication. Non-incorporating cells completed S phase by 1 hr postrelease, with the appearance of a 2C DNA peak. A second S phase was detected 1.5–2 hr postrelease by 4C DNA content and a septation index (Figure 3, A and B, and Figure S2A). We detected a shift in forward scatter (FSC) to small cells (Figure S2B), correlating with cell division at 2 hr postrelease.
Figure 3 .
BrdU and EdU cause prolonged DNA synthesis, cell-cycle slowing, and DNA damage. (A) DNA synthesis profiles of wild-type nonincorporating (non-inc) and incorporating (Inc) cells, out of hydroxyurea arrest (HU), released into medium with 32.6 or 326 µM BrdU to detect DNA replication. Left, whole-cell DNA content (SytoxGreen) FACS profiles. Right, septation index for non-inc (NI) and Inc (I) cells stained with aniline blue and DAPI, at different BrdU doses (µM). (B) As in A, cells released from HU were released into medium with 1 or 10 µM EdU. Left, FACS profiles of whole-cell DNA content. Right, septation index at different EdU doses (µM). (C) Asynchronous (AS) cells were treated with 2 mM thymidine (+Thy) or DMSO (vehicle control) for 3 hr (32°) and then released for 0.75 hr. DNA content (SytoxGreen FACS) for each time point, analyzed by FACS, is shown at each point. DMSO control was to test response to DMSO only and was not released. Similar results were seen with aqueous thymidine solution (not shown). (D) Cells were exposed to 32.6 µM BrdU for 2 hr (32°) and then processed for BrdU and phospho-histone H2A (p-H2A) immunofluorescence. DNA was counterstained with DAPI. Merged image is BrdU and p-H2A signals. Bar, 10 µm.
BrdU exposure at both doses delayed S phase completion in incorporating strains. The DNA peak moved slowly to a 2C position, completing by 2 hr postrelease. Septation was highest at 3 hr, coincident with a small 4C peak observed by FACS (Figure 3A and Figure S2A). FSC confirmed the BrdU-incorporated cells were elongated (Figure S2B), consistent with incomplete septation.
Cells treated with 1 µM EdU completed the first S phase by 1 hr and entered the second S phase at 2 hr postrelease with increased septa and 4C DNA (Figure 3B and Figure S2A). Cells in 10 µM EdU completed a first S phase within 2 hr but had much slower transit through the second S phase, which started at 2 hr and was not resolved by 3 hr (Figure S2A). Thus, EdU causes a dose-dependent inhibition of cell-cycle progression in incorporating cells.
Thymidine is commonly used in human cell cultures to arrest cells in G1 (Harper 2005), but does not affect yeast since S. pombe does not have a thymidine salvage pathway (nonincorporating cells). We asked whether thymidine changes cell cycle in hENT+ hsv-tk+ strains (Figure 3C). We treated cells with 2 mM thymidine (Harper 2005) and saw no effect in nonincorporating cells or in cells with hENT+ but no hsv-tk. However, an incorporating strain treated with 2 mM thymidine arrested with a 1C DNA content. After thymidine removal, cells shifted toward 2C DNA content. Thus, thymidine reversibly arrests hENT+ hsv-tk+ S. pombe cells with a 1C DNA content, as in mammals.
Nucleoside analogs induce DNA damage response
Reduced viability of checkpoint mutants and delayed S-phase progression during analog incorporation suggested that DNA damage was generated. We examined molecular markers of DNA damage after BrdU treatment. First, we detected histone H2A phosphorylated at S129 (p-H2A) and BrdU. Rad3 and Tel1 kinases phosphorylate p-H2A in response to DNA DSBs and replication stress (Nakamura et al. 2004; Bailis et al. 2008). Nonincorporating cells had few p-H2A foci (Figure 3D), while BrdU-labeled cells frequently costained with p-H2A. These results suggest that BrdU incorporation activates a DNA damage response, which is enhanced in cds1Δ cells that cannot properly respond to S-phase stress.
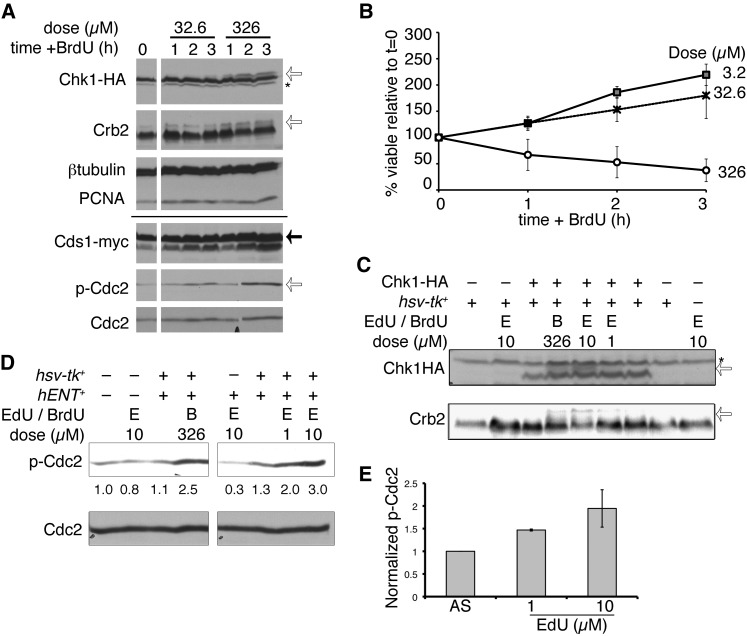
Next, we looked for activation of checkpoint components. We examined HA-tagged Chk1 protein (Figure 4A), the G2-M kinase of the S. pombe DNA damage response (DDR). We observed that Chk1HA shows a characteristic phospho-shift (Capasso et al. 2002) after 1 hr exposure to 326 µM BrdU in hENT+ hsv-tk+ cells (Figure 4A) or after 3 hr in 32.6 µM BrdU. Thus, BrdU dose influences G2-DDR activation. However, Cds1-myc kinase, required for response to replication stress, was not significantly modified relative to overall protein level in BrdU (Figure 4A).
Figure 4 .
BrdU exposure triggers the DNA damage response. (A) Cells were treated with the indicated dose of BrdU and harvested for protein extraction hourly. Chk1-HA was detected with anti-HA (16B12); the asterisk indicates nonspecific background signal and the open arrow indicates phospho-Chk1HA. Crb2 modification is also indicated by an open arrow. Cds1-myc was detected with anti-myc (solid arrow). Phospho-Cdc2 (p-Cdc2, open arrow) and Cdc2 were also detected. PCNA and β-tubulin are loading controls. The solid line indicates a split between two independent gels, with identical lysates. (B) Incorporating strain viability (FY5031) proportional to BrdU dose is shown as means of three independent experiments ± SEM. (C) Chk-HA and Crb2 phosphorylation after 3 hr EdU (µM), in wild-type hsv-tk+ or nonincorporating cells. BrdU (326 µM) is included as a control. Chk1-HA and Crb2 band shifts are indicated with open arrows. The asterisk indicates nonspecific background band (above Chk1HA) detected using a different antibody from A (αHA, 12CA5). (D) Phospho-Cdc2 (p-Cdc2) after 3 hr BrdU or EdU exposure (doses µM). Below bands are the quantified band intensities of p-Cdc2, normalized to total Cdc2 (below). (E) Quantification of p-Cdc2, relative to total Cdc2 levels, from three independent experiments. Mean values are ± SEM.
We observed a phospho-shift in the upstream G2-DDR checkpoint mediator Crb2 in BrdU (Figure 4A), again consistent with G2 checkpoint activation. Activated Chk1 leads to Cdc2 phosphorylation at tyrosine 15 to maintain G2 checkpoint arrest (O’Connell et al. 1997), and we saw phospho-Cdc2 accumulated in BrdU-treated incorporating cells (Figure 4A). A dose of 326 µM BrdU was lethal to most cells during 3 hr exposure (Figure 4B), suggesting the damage is catastrophic.
Similar results were seen in EdU, causing Chk1HA and Crb2 phospho-shifts (Figure 4C) and increased phospho-Cdc2 (Figure 4D), which was proportional to EdU dose (Figure 4E). These data indicate that the DNA damage checkpoint is fully activated during analog exposure.
Loss of the replication checkpoint influences damage response
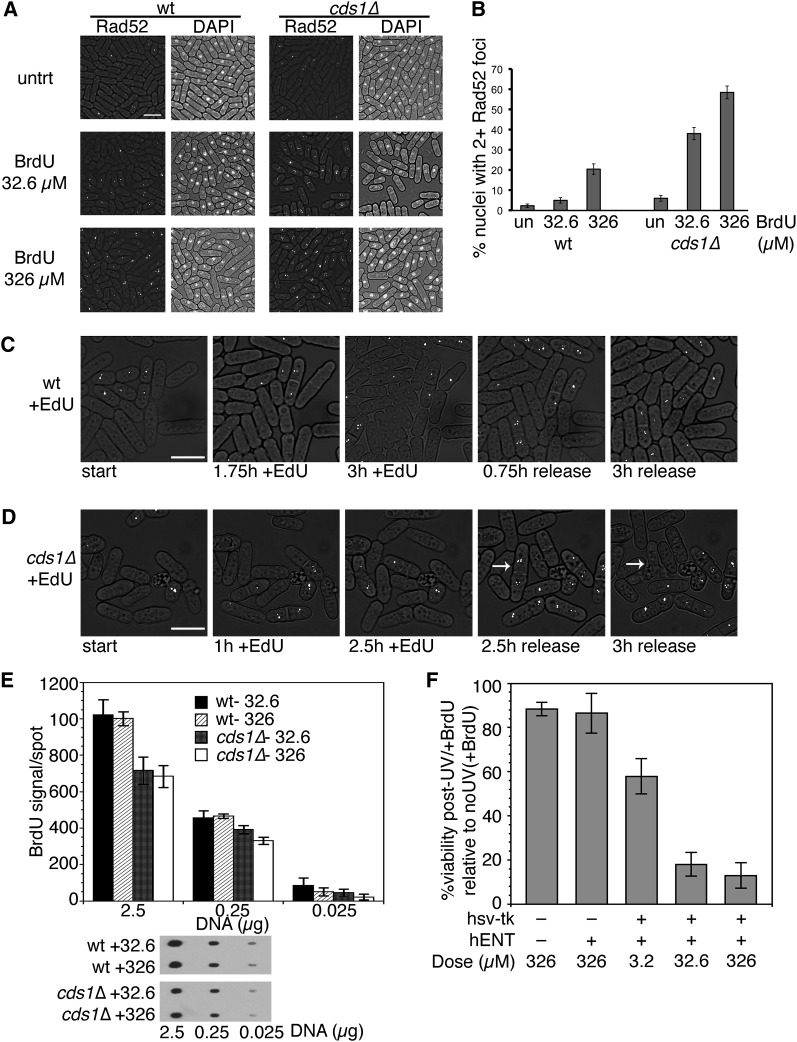
Our data suggest that the Chk1-DNA damage checkpoint pathway is a primary response to analogs. However, the modest sensitivity caused by cds1Δ indicated a role for the replication checkpoint during treatment. Cds1 is particularly important for replication fork stalling and restart (e.g., Lindsay et al. 1998; Kim and Huberman 2001; Miyabe et al. 2009). The homologous recombination protein Rad52 forms nuclear foci in response to a variety of lesions, including double-strand breaks and collapsed or restarting replication forks (Meister et al. 2005; Bailis et al. 2008). We monitored Rad52-YFP focus formation in wild-type and cds1Δ incorporating strains after 3 hr in 32.6 or 326 µM BrdU (Figure 5A). Incorporating cells formed more Rad52 foci than without BrdU treatment, consistent with increased damage (Figure 5B). This effect was dose dependent, and more foci were detected at 326 µM BrdU (wild type) and at all BrdU doses (cds1Δ).
Figure 5 .
BrdU and EdU induce a DNA damage response. (A) Rad52-YFP foci were monitored in untreated cells (untrt) or after 3 hr BrdU at 32°. Rad52 foci (left) or DAPI-stained nuclei (right) are shown on a transmitted light background. Bar, 10 µm. (B) Quantification of three independent experiments in A. Shown are proportions of nuclei with two or more Rad52-YFP foci after 3 hr BrdU ± 95% confidence interval (C.I.). (C and D) Time points selected from movies of wild-type (C; and File S3) or cds1Δ (D; and File S4) cells treated with 10 µM EdU. Arrow (in D) indicates cell that forms foci and lyses. Bar, 10 µm. (E) BrdU incorporated is similar at 32.6 and 326 µM doses. Shown is the mean BrdU signal per dot (±SEM, three independent experiments) at 2.5, 0.25, or 0.025 µg of heat-denatured total DNA, blotted and detected with BrdU antibody. Below, example of BrdU detection on DNA spots. (F) Wild-type nonincorporating or hsv-tk+ hENT+ cells were treated BrdU doses for 3 hr, plated on YES, and then irradiated with 100 J/m2 UV light. Comparison plates were not treated with BrdU, to calculate percentage of viability after BrdU+UV treatment. Shown is the mean viability after BrdU+UV relative to BrdU only, for three independent experiments ± SEM.
Live cell analysis in a microfluidics chamber allowed us to track individual cells during and after analog exposure (File S1, File S2, File S3, and File S4). We monitored Rad52 foci during 3 hr exposure to 32.6 µM BrdU (File S1 and File S2) or 10 µM EdU (File S3 and File S4) in wild-type and cds1Δ hsv-tk+ hENT+ cells. We used a dose of 32.6 µM BrdU because this was the lowest dose that produced a difference in Rad52 foci (Figure 5B) and was close to the 10 µM EdU dose required to effectively label cells for replication studies. BrdU movies recapitulated our static time-point data (Figure 5, A and B), confirming that cds1Δ cells generated more Rad52 foci during analog exposure. We also monitored recovery during 3 hr after analog was removed, observing that multiple Rad52 foci (two or more) frequently resolved into one focus. Further, cells that septated and presumably entered the next cell cycle during BrdU (File S1 and File S2) or EdU exposure (File S3 and File S4) promptly formed Rad52 foci, suggesting an immediate response is mounted during S phase. However, cds1Δ cells that entered S phase during analog exposure frequently lysed during recovery (Figure 5, C and D).
We next asked whether differences in Rad52 foci levels in different BrdU doses could be attributed to different levels of BrdU incorporation in DNA. We extracted total DNA from cells after 3 hr exposure to 32.6 or 326 µM BrdU and blotted several amounts of heat-denatured DNA to probe with BrdU antibody (Figure 5E). Surprisingly, we found that the amount of BrdU incorporated at either 32.6 or 326 µM did not change. Thus, BrdU incorporation is saturated at 32.6 µM, consistent with our earlier FACS results (Figure 1A). However, wild-type cell viability was higher in 32.6 µM BrdU than in 326 µM BrdU (Figure 4B). This suggests that additional stress occurs at the higher dose of BrdU that does not involve BrdU-base substitution. Interestingly, we detected less BrdU incorporation in cds1Δ compared to wild type in the samples with the higher levels of DNA. Thus, enhanced analog sensitivity in cds1Δ is accompanied by less efficient BrdU incorporation. This could reflect replisome disruption in cds1Δ or Cds1-mediated interactions with dNTP metabolism.
Enhanced sensitivity to “second-hit” damage after BrdU exposure
Since hsv-tk+ hENT+ cells acquired DNA damage and G2 arrest signals, we investigated whether BrdU incorporation changes cell sensitivity to other DNA damage, as reported for human cells (e.g., Ackland et al. 1988; Cecchini et al. 2005). UV treatment on BrdU-substituted human cells induces DSBs, demonstrating that BrdU-DNA is sensitized to additional lesions. To test this in S. pombe, we treated cells with 3 hr of low-dose (32.6 µM) BrdU for minimal toxicity with saturated incorporation and examined viability with UV treatment (Figure 5F). Nonincorporating cells experienced a slight viability decrease, to 88 ± 3%, after UV dose of 100 J/m2. However, BrdU incorporation significantly decreased UV survival and further, was dose dependent: cells treated with 3.2 µM BrdU were slightly more UV sensitive (58% viability), while saturating doses of 32.6 or 326 µM had the same effect on UV survival to <20% viability.
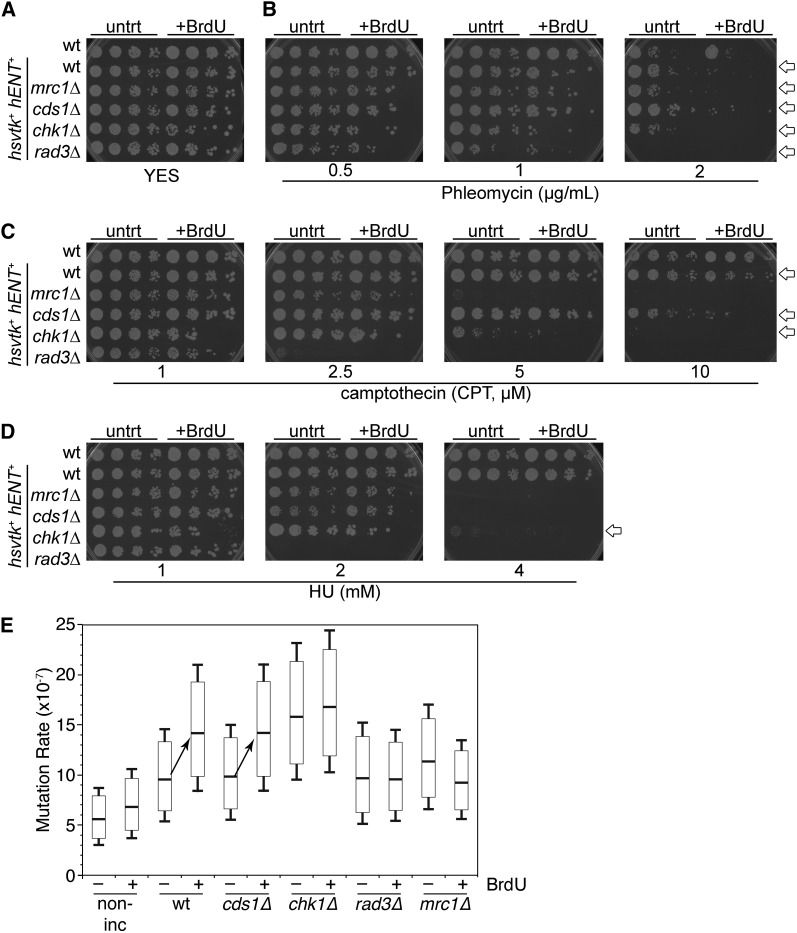
Next, we incubated cells with 32.6 µM BrdU for 2 hr and then examined their sensitivity to a panel of DNA-damaging drugs by a spot test (Figure 6A). First, we examined sensitivity to phleomycin, a radiomimetic (Figure 6B). All cells showed increased sensitivity to phleomycin after BrdU pretreatment. DDR pathway mutants rad3Δ and chk1Δ were most severely affected at lower doses, followed by mrc1Δ and cds1Δ and then wild-type hsv-tk+ hENT+ cells.
Figure 6 .
BrdU pretreatment changes sensitivity to DNA-damaging drugs. (A–D) Cells were either untreated (untrt) or pretreated (+BrdU) with 32.6 µM BrdU for 2 hr at 32° and then spotted onto drug plates in a 1/5 serial dilution. All plates are YES medium. Arrows, far right, indicate strains that were more sensitive to drug following BrdU pretreatment. Strains FY3454, -2317, -3179, -5148, -5149, and -5150 are shown. Also refer to Figure S3. (A) YES control for plating efficiency. (B) Sensitivity to phleomycin. (C) Camptothecin (CPT) sensitivity. (D) Hydroxyurea (HU) sensitivity. (E) Forward mutation analysis for loss of Can1 wild-type status. Cells were incubated with 32.6 µM BrdU for 2 hr (32°) and then plated to assess colony number on titer dishes. Remaining culture was plated on PMG + canavanine and incubated 7 days at 32°. Mutation rate, per 107 generations, was calculated comparing can1− mutants that grew on canavanine plates to the total number plated. Refer to Table 2 for significance results.
BrdU substitution also enhanced sensitivity to camptothecin (CPT) in chk1Δ (5 µM) and cds1Δ (10 µM) (Figure 6C). CPT causes single-strand DNA breaks by covalently linking topoisomerase I to DNA, damage that is later converted to a DSB (Hsiang et al. 1989). Although rad3Δ and mrc1Δ are highly sensitive to CPT, we detected no increased CPT sensitivity following BrdU exposure in rad3Δ and mrc1Δ cells.
The alkylating agent methanemethyl sulfonate (MMS) methylates DNA bases and stalls replication forks. The rad3Δ cells were already highly sensitive to MMS regardless of BrdU pretreatment (Figure S3B). However, the remaining hsv-tk+ hENT+ strains were all more MMS sensitive following BrdU incorporation, with the highest sensitivity in chk1Δ (Figure S3B). HU stalls replication forks, but acts through dNTP depletion as opposed to DNA base damage (Figure 6D). BrdU pretreatment enhanced HU sensitivity only in the chk1Δ hsv-tk+ hENT+ strain. UV lesions stall replication and transcription, so we looked at different doses of UV post-BrdU. All strains except rad3Δ showed increased UV sensitivity after BrdU, with chk1Δ being the most affected (Figure S3A).
We then determined spontaneous mutation frequencies after 32.6 µM BrdU treatment. We used the can1+ gene, which encodes an arginine transporter that imports the toxic precursor canavanine; can1− mutants are resistant to canavanine (Fantes and Creanor 1984). Increased can1 mutation occurred after BrdU exposure in wild-type and cds1Δ hsv-tk+ hENT+ cells (Figure 6E and Table 2), but not in chk1Δ and nonincorporating cells. Intriguingly, rad3Δ and mrc1Δ hsv-tk+ hENT+ cells had a significantly lower mutation rate after BrdU treatment.
We amplified the can1 locus from can1+ colonies in all genotypes (with or without BrdU) to determine whether can1 mutation occurred by gross chromosomal rearrangement. Instead, we saw a 3.8-kb can1 band in 91% of BrdU-treated hsv-tk+ hENT+ can1− isolates (Figure S3C). We checked for smaller deletions by restriction fragment length polymorphism (RFLP) analysis and did not see differences between fragments. Thus, we infer that BrdU-induced mutation in wild-type or cds1Δ cells at can1 is largely due to point mutations, as in metazoans (Goodman et al. 1985).
dNTP pools influence BrdU toxicity and mutagenesis
We next asked whether BrdU affects dNTP pools, which would cause toxicity or mutation. Fission yeast Spd1 inhibits RNR to regulate RNR activity throughout the cell cycle and in response to DNA damage and repair. Previous work has shown that spd1Δ cells have higher endogenous dNTP pools (Holmberg et al. 2005). We reasoned that increased pools of normal dNTPs might dampen the sensitivity to exogenous nucleotides. Consistent with this prediction, we found that spd1Δ hENT hsv-tk+ cells were less sensitive to thymidine, BrdU, or EdU than wild type (Figure 7A and Figure S4). spd1Δ cells were minimally sensitive to BrdU or EdU in YES (Figure S4A). In EMM with BrdU or EdU, we observed two colony sizes in spd1Δ strains: large colonies, similar to other incorporating cells, amid a high background population of small colonies (Figure S4B). We found that spd1Δ cells incorporate BrdU similarly to wild type and produce p-H2A foci in BrdU-labeled nuclei (Figure S4C).
Because spd1Δ hsv-tk+ hENT+ cells were less sensitive to analogs than wild-type incorporating cells in chronic exposure, we were surprised to find that spd1Δ viability was lower than wild type during acute BrdU treatment in liquid culture (Figure 7B). However, spd1Δ hsv-tk+ hENT+ cells continued to divide in 32.6 µM BrdU, while wild-type and rad3Δ cells delayed division (Figure 7C). We examined can1 reversion, in a direct comparison between wild type and spd1Δ after 32.6 µM BrdU for 2 hr at 32° (Figure 7D). Consistent with Figure 6E, wild-type hsv-tk+ hENT+ cells experienced an increase in can1 mutation with 32.6 μM BrdU treatment. Notably, spd1Δ cells already had a higher rate of mutation before BrdU, which increased a further 10-fold after BrdU incorporation.
We reasoned that enhanced spd1Δ survival on plates (despite poor viability and enhanced mutagenesis in liquid) might be attributed to genetic selection combined with the increased rate of mutation, leading to loss of the hsv-tk+ cassette. If this were to happen, a subpopulation of cells would accumulate that is insensitive to further analog incorporation, which could explain the background small cells on streaked plates (Figure S4B). Cells expressing hsv-tk+ are sensitive to FudR, so loss of the cassette can be scored by assessing the frequency of FudR-resistant colonies or sectors (Kiely et al. 2000). We observed that untreated spd1Δ cells give rise to FudR-resistant colonies infrequently, at a rate similar to wild type (Figure 7E and Table 3). However, BrdU treatment caused a significant increase in FudR resistance in spd1Δ compared to wild-type cells. Significantly, higher colony sectoring (Figure 7F) is also consistent with an increase in mutation frequency and suggests that spd1Δ has higher genomic plasticity even within single colonies.
Table 3 . Frequency of hsv-tk+ loss or sectoring in incorporating wild-type and spd1Δ cultures, with or without BrdU treatment.
| Strain | Treatment | FUdR resistance (%)a | Sectored colonies (%)a |
|---|---|---|---|
| Wild type (FY2317) | Untreated (n = 8422) | 0.226 (±0.101) | 1.35 (±0.25) |
| +BrdU (n = 5474) | 0.274 (±0.138) | 1.41 (±0.31) | |
| spd1Δ (FY6247) | Untreated (n = 6968) | 0.172 (±0.097) | 2.65 (±0.38) |
| +BrdU (n = 612) | 2.78 (±1.30) | 33.50 (±3.74) |
FUdR resistance and sectoring frequencies are presented with 95% confidence intervals (C.I.). Data from two (wild type) or three (spd1Δ) independent experiments are shown.
Discussion
Previous studies in budding yeast indicated that BrdU incorporation does not perturb yeast cell growth (Lengronne et al. 2001), although specific mutants may be more sensitive to BrdU during replication (Hodgson et al. 2007). We propose that BrdU, EdU, and thymidine all skew dNTP pools in hsv-tk+ hENT+ fission yeast. Our data show that S. cerevisiae and S. pombe are different in their response to nucleotide analogs and to dNTP imbalance. This is consistent with a 10-fold difference in HU sensitivities between the yeasts; HU also acts by depleting dNTPs.
Studies in metazoans found that BrdU treatment inhibits RNR and causes a “dCTP-less” state (Meuth and Green 1974b) (Figure 7G, part 1). BrdU-induced mutagenesis is attributed to low levels of dCTP (Hopkins and Goodman 1980; Meuth 1989), which is suppressed by coculture with exogenous cytidine (Meuth and Green 1974b; Davidson and Kaufman 1978). Thymidine arrest in mammalian cells (Harper 2005) is also caused by decreased dCTP levels, which are restored once excess thymidine is removed from the medium (Meuth 1989; Kunz and Kohalmi 1991). Unlike BrdU, thymidine is minimally toxic over short time periods (Lockshin et al. 1985), although high doses (>2 mM) induce mutation (Phear and Meuth 1989), polyploidy, and chromosomal aberrations (Potter 1971). Interestingly, lower BrdU concentrations (e.g., 32.6 µM) skew dNTP pools similarly to much higher thymidine doses (2 mM) (Meuth and Green 1974a; Meuth et al. 1976).
We hypothesize that exposure to exogenous nucleosides creates dNTP pool imbalance in S. pombe. During thymidine incubation, hsv-tk+ hENT+ cells arrest with 1C DNA content and release into S phase within 45 min. Previous work by Mitchison and Creanor (1971) determined that thymidine had no effect on S. pombe, consistent with the absence of a thymidine salvage pathway in wild-type cells. However, our incorporating strains have a reconstituted thymidine salvage pathway and are able to convert exogenous thymidine, BrdU, or EdU into nucleotides.
Mitchison and Creanor (1971) also demonstrated that deoxyadenosine treatment induces a short-term G1 arrest in wild-type cells (Mitchison and Creanor 1971). This is consistent with our data and suggests that dNTP pools are skewed by exogenous nucleosides in S. pombe if they are converted into useable nucleotides, to create a transient G1 arrest. We propose that Br- and ethynyl-dUTP (BrdU and EdU, respectively) or thymidine, in a hsv-tk+ strain, cause an apparent increase in dTTP. Excess thymidine or analog may also inhibit RNR as in human cells (Meuth and Green 1974a,b), depleting the dCTP pool (Figure 7G, parts 1 and 2). If so, BrdU and cytidine cotreatment will suppress toxicity and mutagenesis as in metazoans (Davidson and Kaufman 1978; Popescu 1999).
Future work will determine how dNTP pools are altered with these nucleosides, but we predict that thymidine, BrdU, and EdU all cause a dCTP-less state as in human cells. Interestingly, the fission yeast dCMP deaminase mutant (dcd1Δ) has an opposite effect: levels of CTP pools become high while that of dTTP is low. The effects on cell fitness are similar to what we observed. Consistent with our model, dcd1Δ cells expressing hsv-tk+ increase dTTP pools, relieving growth defects and dcd1Δ sensitivities to UV, HU, and bleomycin (Sanchez et al. 2012).
A second observation supports dNTP pool imbalance as a cause of observed phenotypes in S. pombe hsv-tk+ hENT+ cells: while BrdU >32.6 µM does not increase BrdU substitution in DNA, cell viability strikingly decreases above this dose. We observe no evidence for additional BrdU substitution, which excludes further T^C point mutations as the source of high-dose sensitivity. Instead, we propose that this dosage sensitivity reflects increased changes in cellular dNTP pools, perhaps via negative allosteric inhibition of RNR by analogs/thymidine (Meuth and Green 1974b). To confirm this hypothesis, measurements of cellular dNTPs and RNR activity during BrdU and EdU are required.
Changes in dNTP pools cause point mutations in metazoans (Meuth 1989; Phear and Meuth 1989), by substituting the nucleotide in excess during replication (Phear and Meuth 1989). We find that BrdU treatment increases can1 forward mutations in wild-type, spd1Δ, and cds1Δ hsv-tk+ hENT+ cells. However, the rate of mutagenesis is not significantly changed in chk1Δ, while rad3Δ and mrc1Δ experience decreased mutation. We suggest that the replication checkpoint does not actively prevent BrdU-dependent mutation, while an active G2/M checkpoint (chk1+) promotes mutagenesis. This may reflect the increased survival of checkpoint-intact cells or checkpoint effects on Spd1 and nucleotide metabolism (see below). We assume that can1− mutations are A•T ^G•C transitions caused by Br-dUTP substitution for the lowered dCTP analog or G•C ^A•T transitions via enhanced Br-dUTP pairing with guanine in template DNA (Hopkins and Goodman 1980; Lasken and Goodman 1984; Goodman et al. 1985) (Figure 7G, part 3). This is confirmed by our RFLP analysis of can1 mutants, which did not show any gross structural changes.
Fission yeast dNTP metabolism is controlled by RNR, which is inhibited by Spd1 (Holmberg et al. 2005; Hakansson et al. 2006; Nestoras et al. 2010). Spd1 is ubiquitinated during stress so that the dNTP pool is expanded, which ties nucleotide metabolism to the cell cycle and DDR (Moss et al. 2010; Nestoras et al. 2010). We found that spd1Δ cells are less sensitive to chronic nucleoside exposure, but their relative viability in acute BrdU is worse. Intriguingly, spd1Δ cells showed a 10-fold increase in mutations of can1+ or hsv-tk+ and frequent sectoring, indicating that spd1Δ cells are unstable and prone to mutation. Previous work showed spd1Δ suppresses mutation in ddb1Δ cells that are incapable of activating RNR (Holmberg et al. 2005). However, we show that spd1Δ cells are intrinsic mutators, an effect worsened after BrdU treatment.
Increased basal dNTP pools in spd1Δ (Holmberg et al. 2005) could promote this mutagenesis by facilitating replication during exogenous thymidine/dUTP treatment and allowing additional Br-dUTP incorporation opposite G (Lasken and Goodman 1984; 1985). Alternatively, transient changes in one dNTP during spd1Δ replication may not prompt arrest. Our data are consistent with models linking dNTP regulation to genome stability in S. pombe (Holmberg et al. 2005; Moss et al. 2010; Nestoras et al. 2010). While chk1Δ hsv-tk+ hENT+ cells do not show increased mutation post-BrdU, we hypothesize that a spd1Δ chk1Δ double mutant may have a very high mutation rate due to extra damage and larger dNTP pools.
Rad3 is required to survive BrdU as reported in Hua and Kearsey (2011). We show this occurs through the Chk1 G2-DDR path downstream of Rad3, resulting in Cdc2 phosphorylation and Rad52 focus formation. BrdU also increases phospho-histone H2A, a signal of DSBs and/or replication fork stress (e.g., Bailis et al. 2008; Rozenzhak et al. 2010). Our model suggests that halogenated dUTP causes DNA damage (Figure 7G, part 4), perhaps via removal of substituted bases by base excision repair (BER) (Krych et al. 1979; Szyszko et al. 1983; Morgan et al. 2007). BER under dNTP depletion could additionally cause single-strand DNA breaks (SSBs) that stall replication forks and/or convert to DSBs during S phase. This would promote cell accumulation in S phase and a modest requirement for the Cds1 pathway.
Alternatively, topoisomerase I has RNaseH activity on a double-strand DNA template with only one dUTP substitution (Sekiguchi and Shuman 1997) and could contribute to SSB and DSB accumulation. While BrdU-induced DNA damage in human cells is documented, the cause is not known (Dillehay et al. 1984; Ackland et al. 1988). The role of postreplication repair in surviving BrdU may prove essential.
As in mammals, we observe that a second DNA-damaging agent is more dangerous after BrdU substitution. Sensitivity of chk1Δ cells to other drugs following BrdU incorporation suggests that BrdU substitution increases the DNA damage “load” in treated cells. In mammalian cells, UV exposure following BrdU substitution causes DSBs and interstrand cross-links (Murray and Martin 1989; Cecchini et al. 2005) and enhanced sensitivity to bleomycin (Ackland et al. 1988) and cisplatin (Russo et al. 1986). Budding yeast is also UV sensitive after BrdU exposure (Sclafani and Fangman 1986). Some part of this sensitivity may result from S-phase slowing in BrdU, a more vulnerable time for DNA damage. We show increased sensitivity in cds1Δ strains, implying that replication fork stability is diminished, perhaps from imperfect BrdU base pairing (Lasken and Goodman 1985). We find that rad3Δ response to CPT, UV, MMS, or HU is unchanged with BrdU pretreatment, probably because of catastrophic failure of rad3Δ in these drugs.
Our results point to challenges in the use of nucleoside analogs to analyze DNA replication. While we do not directly compare between the two analogs, our results indicate that fission yeast is extremely sensitive to nucleoside analogs BrdU and EdU, when treated at doses similar to those used in human cells. Consistent with replication stability problems, the increased toxicity of EdU, and cell-cycle effects at lower doses, may reflect a larger ethynyl side group and thus greater steric interference during replication. The long-term effects associated with BrdU and EdU exposure mean that these analogs are most useful for analysis in a single cell cycle. Further, thymidine may be a potential reversible blocking agent in S. pombe, yet its effects must be more clearly described. In all cases, appropriate care must be taken to mitigate analog effects, lest disruption of nucleotide levels interfere with the very process under study.
Supplementary Material
Acknowledgments
We thank Antony Carr for the spd1Δ strain, Myron Goodman for helpful discussions, Oscar Aparicio for flow cytometer access, members of the Forsburg laboratory for comments, and anonymous reviewers for suggestions. This work is supported by National Institutes of Health (NIH) grant R01 GM59321 and NIH grant R01 GM081418 (to S.L.F.).
Footnotes
Communicating editor: N. M. Hollingsworth
Literature Cited
- Ackland S. P., Schilsky R. L., Beckett M. A., Weichselbaum R. R., 1988. Synergistic cytotoxicity and DNA strand break formation by bromodeoxyuridine and bleomycin in human tumor cells. Cancer Res. 48: 4244–4249 [PubMed] [Google Scholar]
- Alvino G. M., Collingwood D., Murphy J. M., Delrow J., Brewer B. J., et al. , 2007. Replication in hydroxyurea: it’s a matter of time. Mol. Cell. Biol. 27: 6396–6406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailis J. M., Luche D. D., Hunter T., Forsburg S. L., 2008. Minichromosome maintenance proteins interact with checkpoint and recombination proteins to promote s-phase genome stability. Mol. Cell. Biol. 28: 1724–1738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlogie B., Spitzer G., Hart J. S., Johnston D. A., Buchner T., et al. , 1976. DNA histogram analysis of human hemopoietic cells. Blood 48: 245–258 [PubMed] [Google Scholar]
- Bohmer R. M., Ellwart J., 1981. Combination of BUdR-quenched Hoechst fluorescence with DNA-specific ethidium bromide fluorescence for cell cycle analysis with a two-parameter flow cytometer. Cell Tissue Kinet. 14: 653–658 [DOI] [PubMed] [Google Scholar]
- Buck S. B., Bradford J., Gee K. R., Agnew B. J., Clarke S. T., et al. , 2008. Detection of S-phase cell cycle progression using 5-ethynyl-2′-deoxyuridine incorporation with click chemistry, an alternative to using 5-bromo-2′-deoxyuridine antibodies. Biotechniques 44: 927–929 [DOI] [PubMed] [Google Scholar]
- Capasso H., Palermo C., Wan S., Rao H., John U. P., et al. , 2002. Phosphorylation activates Chk1 and is required for checkpoint-mediated cell cycle arrest. J. Cell Sci. 115: 4555–4564 [DOI] [PubMed] [Google Scholar]
- Carlson C. R., Grallert B., Bernander R., Stokke T., Boye E., 1997. Measurement of nuclear DNA content in fission yeast by flow cytometry. Yeast 13: 1329–1335 [DOI] [PubMed] [Google Scholar]
- Cecchini S., Masson C., La Madeleine C., Huels M. A., Sanche L., et al. , 2005. Interstrand cross-link induction by UV radiation in bromodeoxyuridine-substituted DNA: dependence on DNA conformation. Biochemistry 44: 16957–16966 [DOI] [PubMed] [Google Scholar]
- Chabes A., Georgieva B., Domkin V., Zhao X., Rothstein R., et al. , 2003. Survival of DNA damage in yeast directly depends on increased dNTP levels allowed by relaxed feedback inhibition of ribonucleotide reductase. Cell 112: 391–401 [DOI] [PubMed] [Google Scholar]
- Christov K., Vassilev N., 1988. Flow cytometric analysis of DNA and cell proliferation in ovarian tumors. Cancer 61: 121–125 [DOI] [PubMed] [Google Scholar]
- Crissman H. A., Steinkamp J. A., 1987. A new method for rapid and sensitive detection of bromodeoxyuridine in DNA-replicating cells. Exp. Cell Res. 173: 256–261 [DOI] [PubMed] [Google Scholar]
- Davidson M. B., Katou Y., Keszthelyi A., Sing T. L., Xia T., et al. , 2012. Endogenous DNA replication stress results in expansion of dNTP pools and a mutator phenotype. EMBO J. 31: 895–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson R. L., Kaufman E. R., 1978. Bromodeoxyuridine mutagenesis in mammalian cells is stimulated by thymidine and suppressed by deoxycytidine. Nature 276: 722–723 [DOI] [PubMed] [Google Scholar]
- Diermeier-Daucher S., Clarke S. T., Hill D., Vollmann-Zwerenz A., Bradford J. A., et al. , 2009. Cell type specific applicability of 5-ethynyl-2′-deoxyuridine (EdU) for dynamic proliferation assessment in flow cytometry. Cytometry. Part A. J. Int. Soc. Anal. Cytol. 75: 535–546 [DOI] [PubMed] [Google Scholar]
- Dillehay L. E., Thompson L. H., Carrano A. V., 1984. DNA-strand breaks associated with halogenated pyrimidine incorporation. Mutat. Res. 131: 129–136 [DOI] [PubMed] [Google Scholar]
- Fantes P. A., Creanor J., 1984. Canavanine resistance and the mechanism of arginine uptake in the fission yeast Schizosaccharomyces pombe. J. Gen. Microbiol. 130: 3265–3273 [DOI] [PubMed] [Google Scholar]
- Frum R., Deb S. P., 2003. Flow cytometric analysis of MDM2-mediated growth arrest. Methods Mol. Biol. 234: 257–267 [DOI] [PubMed] [Google Scholar]
- Goodman M. F., Hopkins R. L., Lasken R., Mhaskar D. N., 1985. The biochemical basis of 5-bromouracil- and 2-aminopurine-induced mutagenesis. Basic Life Sci. 31: 409–423 [DOI] [PubMed] [Google Scholar]
- Hakansson P., Dahl L., Chilkova O., Domkin V., Thelander L., 2006. The Schizosaccharomyces pombe replication inhibitor Spd1 regulates ribonucleotide reductase activity and dNTPs by binding to the large Cdc22 subunit. J. Biol. Chem. 281: 1778–1783 [DOI] [PubMed] [Google Scholar]
- Hall B.M., Ma C., Liang P., Singh K. K., 2009. Fluctuation AnaLysis CalculatOR (FALCOR): a web tool for the determination of mutation rate using Luria-Delbruck fluctuation analysis. Bioinformatics 25: 1564–1565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper J. V., 2005. Synchronization of cell populations in G1/S and G2/M phases of the cell cycle. Methods Mol. Biol. 296: 157–166 [DOI] [PubMed] [Google Scholar]
- Hayano M., Kanoh Y., Matsumoto S., Masai H., 2011. Mrc1 marks early-firing origins and coordinates timing and efficiency of initiation in fission yeast. Mol. Cell. Biol. 31: 2380–2391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgson B., Calzada A., Labib K., 2007. Mrc1 and Tof1 regulate DNA replication forks in different ways during normal S phase. Mol. Biol. Cell 18: 3894–3902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodson J. A., Bailis J. M., Forsburg S. L., 2003. Efficient labeling of fission yeast Schizosaccharomyces pombe with thymidine and BUdR. Nucleic Acids Res. 31: e134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmberg C., Fleck O., Hansen H. A., Liu C., Slaaby R., et al. , 2005. Ddb1 controls genome stability and meiosis in fission yeast. Genes Dev. 19: 853–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins R. L., Goodman M. F., 1980. Deoxyribonucleotide pools, base pairing, and sequence configuration affecting bromodeoxyuridine- and 2-aminopurine-induced mutagenesis. Proc. Natl. Acad. Sci. USA 77: 1801–1805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiang Y. H., Lihou M. G., Liu L. F., 1989. Arrest of replication forks by drug-stabilized topoisomerase I-DNA cleavable complexes as a mechanism of cell killing by camptothecin. Cancer Res. 49: 5077–5082 [PubMed] [Google Scholar]
- Hua H., Kearsey S. E., 2011. Monitoring DNA replication in fission yeast by incorporation of 5-ethynyl-2′-deoxyuridine. Nucleic Acids Res. 39: e60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson T. S., Raju M. R., Giltinan R. K., Gillette E. L., 1981. Ploidy and DNA distribution analysis of spontaneous dog tumors by flow cytometry. Cancer Res. 41: 3005–3009 [PubMed] [Google Scholar]
- Kaufman E. R., Davidson R. L., 1978. Bromodeoxyuridine mutagenesis in mammalian cells: mutagenesis is independent of the amount of bromouracil in DNA. Proc. Natl. Acad. Sci. USA 75: 4982–4986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiely J., Haase S. B., Russell P., Leatherwood J., 2000. Functions of fission yeast orp2 in DNA replication and checkpoint control. Genetics 154: 599–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. M., Huberman J. A., 2001. Regulation of replication timing in fission yeast. EMBO J. 20: 6115–6126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krych M., Pietrzykowska I., Szyszko J., Shugar D., 1979. Genetic evidence for the nature, and excision repair, of DNA lesions resulting from incorporation of 5-bromouracil. Mol. Gen. Genet. 171: 135–143 [DOI] [PubMed] [Google Scholar]
- Kumar D., Viberg J., Nilsson A. K., Chabes A., 2010. Highly mutagenic and severely imbalanced dNTP pools can escape detection by the S-phase checkpoint. Nucleic Acids Res. 38: 3975–3983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar D., Abdulovic A. L., Viberg J., Nilsson A. K., Kunkel T. A., et al. , 2011. Mechanisms of mutagenesis in vivo due to imbalanced dNTP pools. Nucleic Acids Res. 39: 1360–1371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunz B. A., Kohalmi S. E., 1991. Modulation of mutagenesis by deoxyribonucleotide levels. Annu. Rev. Genet. 25: 339–359 [DOI] [PubMed] [Google Scholar]
- Lasken R. S., Goodman M. F., 1984. The biochemical basis of 5-bromouracil-induced mutagenesis. Heteroduplex base mispairs involving bromouracil in G x C—-A x T and A x T—-G x C mutational pathways. J. Biol. Chem. 259: 11491–11495 [PubMed] [Google Scholar]
- Lasken R. S., Goodman M. F., 1985. A fidelity assay using “dideoxy” DNA sequencing: a measurement of sequence dependence and frequency of forming 5-bromouracil X guanine base mispairs. Proc. Natl. Acad. Sci. USA 82: 1301–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengronne A., Pasero P., Bensimon A., Schwob E., 2001. Monitoring S phase progression globally and locally using BrdU incorporation in TK(+) yeast strains. Nucleic Acids Res. 29: 1433–1442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsay H. D., Griffiths D. J., Edwards R. J., Christensen P. U., Murray J. M., et al. , 1998. S-phase-specific activation of Cds1 kinase defines a subpathway of the checkpoint response in Schizosaccharomyces pombe. Genes Dev. 12: 382–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockshin A., Giovanella B. C., Stehlin J. S., Jr, Kozielski T., Quian C., et al. , 1985. Antitumor activity and minimal toxicity of concentrated thymidine infused in nude mice. Cancer Res. 45: 1797–1802 [PubMed] [Google Scholar]
- Lopes M., Cotta-Ramusino C., Pellicioli A., Liberi G., Plevani P., et al. , 2001. The DNA replication checkpoint response stabilizes stalled replication forks. Nature 412: 557–561 [DOI] [PubMed] [Google Scholar]
- Meister P., Taddei A., Vernis L., Poidevin M., Gasser S. M., et al. , 2005. Temporal separation of replication and recombination requires the intra-S checkpoint. J. Cell Biol. 168: 537–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meuth M., 1989. The molecular basis of mutations induced by deoxyribonucleoside triphosphate pool imbalances in mammalian cells. Exp. Cell Res. 181: 305–316 [DOI] [PubMed] [Google Scholar]
- Meuth M., Green H., 1974a. Alterations leading to increased ribonucleotide reductase in cells selected for resistance to deoxynucleosides. Cell 3: 367–374 [DOI] [PubMed] [Google Scholar]
- Meuth M., Green H., 1974b. Induction of a deoxycytidineless state in cultured mammalian cells by bromodeoxyuridine. Cell 2: 109–112 [DOI] [PubMed] [Google Scholar]
- Meuth M., Aufreiter E., Reichard P., 1976. Deoxyribonucleotide pools in mouse-fibroblast cell lines with altered ribonucleotide reductase. Eur. J. Biochem. 71: 39–43 [DOI] [PubMed] [Google Scholar]
- Mitchison J. M., Creanor J., 1971. Induction synchrony in the fission yeast. Schizosaccharomyces pombe. Exp. Cell Res. 67: 368–374 [DOI] [PubMed] [Google Scholar]
- Miyabe I., Morishita T., Shinagawa H., Carr A. M., 2009. Schizosaccharomyces pombe Cds1Chk2 regulates homologous recombination at stalled replication forks through the phosphorylation of recombination protein Rad60. J. Cell Sci. 122: 3638–3643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan M. T., Bennett M. T., Drohat A. C., 2007. Excision of 5-halogenated uracils by human thymine DNA glycosylase. Robust activity for DNA contexts other than CpG. J. Biol. Chem. 282: 27578–27586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss J., Tinline-Purvis H., Walker C. A., Folkes L. K., Stratford M. R., et al. , 2010. Break-induced ATR and Ddb1-Cul4(Cdt)(2) ubiquitin ligase-dependent nucleotide synthesis promotes homologous recombination repair in fission yeast. Genes Dev. 24: 2705–2716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray V., Martin R. F., 1989. The degree of ultraviolet light damage to DNA containing iododeoxyuridine or bromodeoxyuridine is dependent on the DNA sequence. Nucleic Acids Res. 17: 2675–2691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T. M., Du L. L., Redon C., Russell P., 2004. Histone H2A phosphorylation controls Crb2 recruitment at DNA breaks, maintains checkpoint arrest, and influences DNA repair in fission yeast. Mol. Cell. Biol. 24: 6215–6230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nestoras K., Mohammed A. H., Schreurs A. S., Fleck O., Watson A. T., et al. , 2010. Regulation of ribonucleotide reductase by Spd1 involves multiple mechanisms. Genes Dev. 24: 1145–1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell M. J., Raleigh J. M., Verkade H. M., Nurse P., 1997. Chk1 is a wee1 kinase in the G2 DNA damage checkpoint inhibiting cdc2 by Y15 phosphorylation. EMBO J. 16: 545–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phear G., Meuth M., 1989. The genetic consequences of DNA precursor pool imbalance: sequence analysis of mutations induced by excess thymidine at the hamster aprt locus. Mutat. Res. 214: 201–206 [DOI] [PubMed] [Google Scholar]
- Poli J., Tsaponina O., Crabbe L., Keszthelyi A., Pantesco V., et al. , 2012. dNTP pools determine fork progression and origin usage under replication stress. EMBO J. 31: 883–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popescu N. C., 1999. Sister chromatid exchange formation in mammalian cells is modulated by deoxyribonucleotide pool imbalance. Somat. Cell Mol. Genet. 25: 101–108 [DOI] [PubMed] [Google Scholar]
- Potter C. G., 1971. Induction of polyploidy by concentrated thymidine. Exp. Cell Res. 68: 442–448 [DOI] [PubMed] [Google Scholar]
- Rozenzhak S., Mejia-Ramirez E., Williams J. S., Schaffer L., Hammond J. A., et al. , 2010. Rad3 decorates critical chromosomal domains with gammaH2A to protect genome integrity during S-Phase in fission yeast. PLoS Genet. 6: e1001032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo A., DeGraff W., Kinsella T. J., Gamson J., Glatstein E., et al. , 1986. Potentiation of chemotherapy cytotoxicity following iododeoxyuridine incorporation in Chinese hamster cells. Int. J. Radiat. Oncol. Biol. Phys. 12: 1371–1374 [DOI] [PubMed] [Google Scholar]
- Sabatinos S. A., Forsburg S. L., 2009. Measuring DNA content by flow cytometry in fission yeast. Methods Mol. Biol. 521: 449–461 [DOI] [PubMed] [Google Scholar]
- Sabatinos S. A., Forsburg S. L., 2010. Molecular genetics of Schizosaccharomyces pombe. Methods Enzymol. 470: 759–795 [DOI] [PubMed] [Google Scholar]
- Sabatinos S. A., Forsburg S. L., 2012. The replication fork collapse point: a new view of arrest and recovery, in The Mechanisms of DNA Replication, edited by D. Stuart. InTech, Rijeka, Croatia (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez A., Sharma S., Rozenzhak S., Roguev A., Krogan N. J., et al. , 2012. Replication fork collapse and genome instability in dCMP deaminase mutant. Mol. Cell. Biol. 32: 4445–4454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santocanale C., Diffley J. F., 1998. A Mec1- and Rad53-dependent checkpoint controls late-firing origins of DNA replication. Nature 395: 615–618 [DOI] [PubMed] [Google Scholar]
- Schneider C. A., Rasband W. S., Eliceiri K. W., 2012. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9: 671–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sclafani R. A., Fangman W. L., 1986. Thymidine utilization by tut mutants and facile cloning of mutant alleles by plasmid conversion in S. cerevisiae. Genetics 114: 753–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiguchi J., Shuman S., 1997. Site-specific ribonuclease activity of eukaryotic DNA topoisomerase I. Mol. Cell 1: 89–97 [DOI] [PubMed] [Google Scholar]
- Sivakumar S., Porter-Goff M., Patel P. K., Benoit K., Rhind N., 2004. In vivo labeling of fission yeast DNA with thymidine and thymidine analogs. Methods 33: 213–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szyszko J., Pietrzykowska I., Twardowski T., Shugar D., 1983. Identification of uracil as a major lesion in E. coli DNA following the incorporation of 5-bromouracil, and some of the accompanying effects. Mutat. Res. 108: 13–27 [DOI] [PubMed] [Google Scholar]
- Viggiani C. J., Aparicio O. M., 2006. New vectors for simplified construction of BrdU-incorporating strains of Saccharomyces cerevisiae. Yeast 23: 1045–1051 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.