Abstract
V(D)J recombination is the process by which antibody and T-cell receptor diversity is attained. During this process, antigen receptor gene segments are cleaved and rejoined by non-homologous DNA end joining for the generation of combinatorial diversity. The major players of the initial process of cleavage are the proteins known as RAG1 (recombination activating gene 1) and RAG2. In this review, we discuss the physiological function of RAGs as a sequence-specific nuclease and its pathological role as a structure-specific nuclease. The first part of the review discusses the basic mechanism of V(D)J recombination, and the last part focuses on how the RAG complex functions as a sequence-specific and structure-specific nuclease. It also deals with the off-target cleavage of RAGs and its implications in genomic instability.
Keywords: altered DNA structures, chromosomal translocation, class switch recombination, DNA damage, double-strand break, genomic instability, non-homologous DNA end joining, recombination activating gene
Introduction
Lymphocytes detect the antigens in the environment by means of antibodies on the surface of B cells and T-cell receptors (TCR) on the T cells. With the diverse and expanding array of antigens, the generation of antibody/TCR diversity using limited genetic resources remained a question that baffled scientists for decades. An almost limitless number of antigens exist in the environment and recent research suggests that among the millions of lymphocytes, each one expresses a structurally different antigen receptor to combat this plethora of antigens. How is the genetic information for all of these antigen receptors encoded in the DNA? Do cells carry enough DNA to encode all the antibody specificities? Or is it that random mutations generate this enormously diverse repertoire of antibodies? Two theories arose initially to answer these questions. Somatic mutation/variation theory suggested that a few inherited genes, with time, encountered mutations or recombinations to encode each antibody.1 In contrast, germline theory proposed that the genome contains a large repertoire of antigen receptor genes and each of them encodes for separate, specific antibody.2 Arguments supporting and opposing these theories were put forward and remained unresolved for several years. In this review, we summarize the basic principles that presently govern the generation of diversity of antibody and TCR with special emphasis on V(D)J recombination. We also discuss the role of recombination activating genes (RAGs) in the generation of antibody diversity and chromosomal translocations.
The discovery of RAG genes
In the early 1990s, it was shown that two tightly linked genes, RAG1 and RAG2, which were unique to vertebrates, were responsible for the generation of antigen receptor diversity.3,4 An elegant series of experiments involving genomic DNA transfections into mouse 3T3 fibroblasts lacking V(D)J recombination activity, showed that the transfer of a single genomic locus could make these cells proficient for V(D)J recombination.5 Following this, using the technique of ‘genome walking’, the RAG1 gene was discovered. Comparative sequence analysis of RAG1 genes from various species indicated that they were evolutionarily conserved.3 Further studies demonstrated that the locus contained two closely linked genes, RAG1 and RAG2 on chromosome 11p in humans and chromosome 2p in mice.4,6 The coding and 3′ untranslated sequences of RAG1 and RAG2 were contained in a single exon.6 The proteins encoded by the RAG genes play a crucial role in the generation of antigen receptor diversity as discussed below.
Antigen receptors of lymphoid system
There are two major antigen receptors for the lymphoid system, antibodies and TCR in B and T cells, respectively. Antibodies or immunoglobulins are glycoproteins that are either secreted out from B cells or remain bound to their membrane. Each antibody is composed of four polypeptides – two identical heavy chains (H) and two identical light (L) chains held together by disulphide bonds.7,8 Amino acid sequence at the N-terminus of both chains varies greatly among different antibodies, whereas the C-terminal sequence remains strikingly similar.9 These two regions are referred to as the variable (V) and constant (C) regions, respectively. The V region composed of 110–130 amino acids, gives the antibody its specificity for binding to antigen. The exon encoding the variable region is assembled from two (or three) individual gene segments,2,10 which are classified into variable (V),11 diversity (D) (present only in immunoglobulin heavy chains, not in the light chains)12–14 and joining (J)15,16 regions (Fig. 1). To obtain a functional variable region, recombination between D and J occurs to give a DJ segment, followed by another recombinational event involving V to yield the final V(D)J fragment. The germline consists of multitudes of V, D and J gene segments and random recombination among these results in the generation of approximately 106 different combinations, accounting for the dramatic expansion in the variability of the sequence (Fig. 1).
Figure 1.
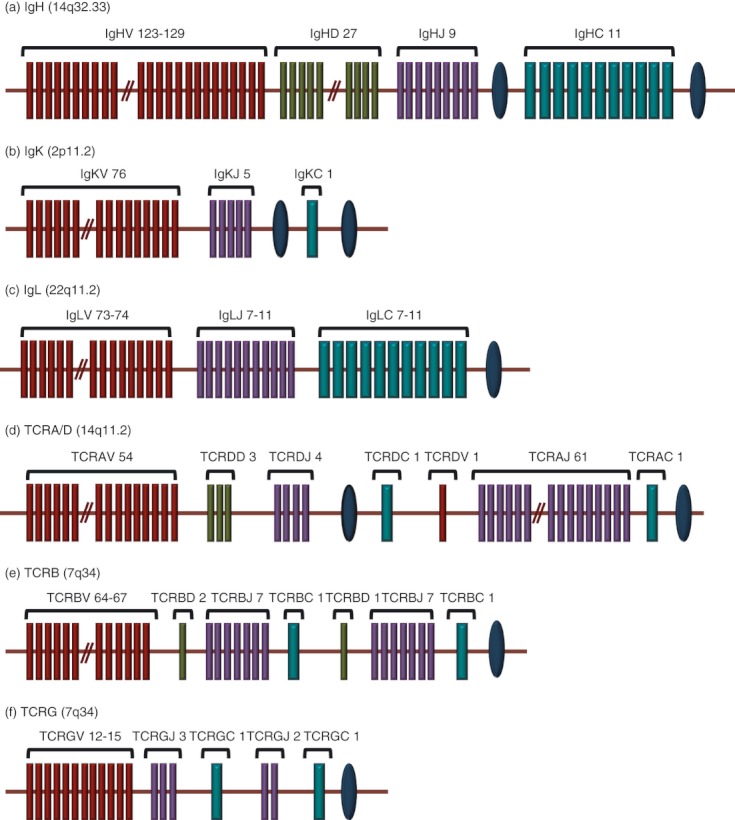
Genomic organization of the antigen receptor loci in human beings. The maroon, green, purple and blue rectangles represent V, D, J and C segments, respectively and the blue ovals indicate enhancers. (a) The human IgH locus, located on chromosome 14 at band 14q32.33. It has 123 to 129 IgHV genes depending on haplotypes, 27 IgHD segments, and nine IgHJ segments and in the most frequent haplotype, 11 IgHC genes. (b) The human IgK locus located on the chromosome 2, at band 2p11.2. It consists of 76 IgKV genes, five IgKJ segments, and a unique IgKC gene. (c) The human IgL locus located on chromosome 22 at band 22q11.2. It consists of 73 to 74 IgLV genes, 7 to 11 IgLJ and 7 to 11 IgLC genes depending on the haplotypes. (d) The human TCRA locus located on chromosome 14 at band 14q11.2. It consists of 116 genes: 54 TCRAV, 61 TCRAJ and a unique TCRAC gene. TCRD locus is embedded in the TCRA locus, between the TCRAV and TCRAJ genes. Though represented in the figure as ‘TCRAV 54’, among the 54 V genes, five can be rearranged to either TCRAJ or TCRDD genes and therefore can be used in the synthesis of α or δ chains. (e) The human TCRB locus on chromosome 7 at 7q34, consisting of 64–67 TCRBV, 2 TCRBD, 14 TCRBJ and two TCRBC genes. (f) The human TCRG locus is on chromosome 7 at 7p14 and consists of 12–15 TCRGV, five TCRGJ and two TCRGC1 genes. The data presented are in accordance with IMGT (http://www.imgt.org).
The TCR is structurally similar to the antigen-binding fragment [F(ab)] of the antibody. Similar to the antibodies, it has two glycoprotein subunits and each is encoded by a somatically rearranged gene. The TCRs are composed of either an αβ or a γδ pair of subunits. The structure of TCR is further stabilized by interchain disulphide bonds. At the 5′ end of each of the TCR loci there is a cluster of V segments followed by J segments (Fig. 1). In the TCR-β and TCR-δ chain loci, these segments are interrupted by a series of D segments similar to that of the immunoglobulin heavy chain (Fig. 1).
Somatic recombination occurs in a strict regimen, with D to J recombination preceding V to DJ on the heavy chain and the heavy chain recombination in turn occurring before that of the light chains.17 Similarly, the TCR-β rearrangement always precedes that of TCR-α. Besides, the TCR rearrangement is restricted to early stages of the T-cell development and immunoglobulin rearrangement to early B cells. Adherence to this chronological order relies on the cell lineage and cell cycle restricted expression of participating enzymes as well as on chromosomal accessibility of the recombining loci.18
A mature B lymphocyte expresses a single species of antibody possessing a unique specificity in spite of having multiple allelic loci for different antibody chains. This specificity is acquired by a process termed allelic exclusion.19 Initially, two models were put forward to explain this process. In the case of the ‘regulated model’, gene assembly proceeds on one chromosome at a time and the protein products suppress further rearrangements by feedback inhibition.20 The ‘stochastic model’ suggests that inefficient V(D)J rearrangement results in allelic exclusion.21 Later studies demonstrated that the process was regulated by several factors including nuclear localization, non-coding RNA transcription, alteration in chromatin structure by histone modifications, epigenetic alterations at the DNA level, and feedback signalling from expressed alleles.19
Antigen receptor loci
By 1998, immunoglobulin and TCR genes were fully identified and sequenced. There are seven major loci, which undergo somatic recombination in developing B and T cells during the formation of antigen receptors. These are immunoglobulin heavy chain (IgH), light chain κ (IgK) and light chain λ (IgL) in B cells and TCR-α (TCRA), TCR-β (TCRB), TCR-γ (TCRG) and TCR-δ (TCRD) in T cells. Each of these is further divided into subexons, which undergo the recombination (Fig. 1).
Mechanism of V(D)J recombination
Recombination signal sequence and 12/23 rule
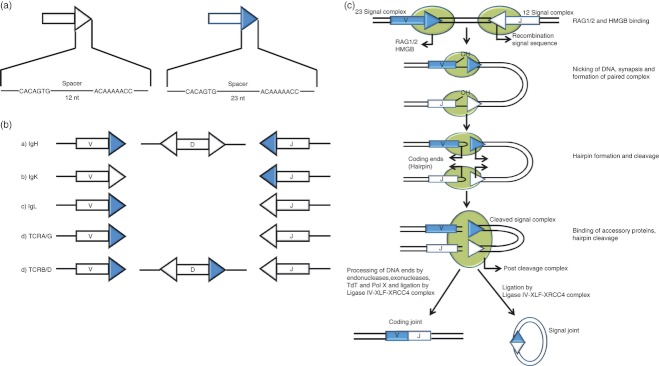
A fairly conserved DNA sequence known as recombination signal sequence (RSS) resides adjacent to each subexon and consists of a palindromic heptamer (CACAGTG) and an A/T-rich nonamer (ACAAAAACC)14,22–24 (Fig. 2a,b). The first three nucleotides of the heptamer are crucial for the recombination activity.25,26 Though the nonamer binding domain of RAG1 is well characterized, the region of the RAG complex that recognizes the heptamer is yet to be deciphered.27,28 The heptamer and nonamer are separated by a spacer DNA sequence of either 12 bp (12RSS) or 23 bp (23RSS) (Fig. 2a). Although the length of the spacer is conserved, its sequence is not of much importance.12,24 Generally, a 12RSS recombines only with a 23RSS and vice versa, a restriction termed as the ‘12/23 rule’ (Fig. 2b), which prevents non-productive rearrangements. The coupled cleavage of a 12RSS and 23RSS requires Mg2+, whereas Mn2+ supports RAG-mediated nicking of a single RSS.29
Figure 2.
Mechanism of the V(D)J recombination. (a). Recombination Signal Sequence (RSS). In 12RSS (represented by open triangle), the heptamer is separated from nonamer by 12 bp and in 23RSS (represented by closed triangle) by 23 bp. (b). Distribution of RSS at the seven antigen receptor loci in human beings and the 12/23 rule. The V, D and J regions of the coding segments are depicted as rectangles; the flanking 12RSS is represented by an open triangle and 23RSS by a closed triangle. As per the ‘12/23 rule’, V(D)J recombination is restricted to gene segments that are flanked by signals of different spacer lengths. (c). Mechanism of V(D)J recombination. Schematic model of the protein–DNA complexes in V(D)J recombination. The 12RSS and 23RSS are represented as white and blue triangles, respectively. Coding segments are shown as rectangles and proteins as shaded ovals. ‘V’ stands for variable and ‘J’ for joining gene segments. Recombination activating genes (RAGs; shaded ovals) bind the RSS and introduce a single-strand nick in the DNA precisely at the border between the heptamer of RSS and the coding segment. This nick generates a free 3′-OH group on the coding end which is then covalently linked to the opposing phosphodiester bond, leaving a covalently closed hairpin on the coding end and a blunt 5′ phosphorylated signal end. The ends remain associated with RAGs; constituting a transitory structure referred to as ‘post-cleavage complex’. Following this, the coding ends are processed and joined by NHEJ to create the exon, which forms the antigen-binding region of antigen receptors. Signal end remains bound to RAGs constituting a ‘signal end complex’ that forms the signal joint.
Recently, the ‘beyond 12/23’ rule has been proposed to explain the exclusion of direct TCRBV to TCRBJ joining in the TCR-β region, in spite of the incidence of appropriately oriented pairs of 12RSS and 23RSS.30 The exclusion was enforced during the DNA cleavage step of the V(D)J recombination and was attributed to several factors, like relatively slow nicking of the TCRB substrates and poor synapsis of the TCRBV and the TCRBJ.31 Extrachromosomal V(D)J recombination assays could recapitulate the ‘beyond 12/23 rule’ in the TCRBV, implying that it is solely the RAG proteins and RSSs, which play a role in establishing this restriction.32 In contrast, with respect to TCRDV locus, the involvement of other factors was also suggested.33
The perfect cleavage
RAG1 and RAG2 initiate recombination by introducing a single-strand nick in DNA precisely at the border between the heptamer of RSS and the coding segment.34 The 3′-OH group of the nick at the coding end then becomes covalently linked to the opposing phosphodiester bond of antiparallel strand by a transesterification reaction resulting in hairpin structure at the coding end and blunt signal end.35 The signal ends remain associated with RAG proteins resulting in a transitory structure referred to as a ‘post-cleavage complex’.36,37 In the presence of Mg2+, nicking can take place, and with Mn2+, both nicking and hairpin formation are observed.37 However, this was demonstrated only in vitro in a non-physiological concentration of MnCl2 using isolated RSS substrates and not in the physiological 12RSS and 23RSS pair.37 The in vivo scenario is still unclear though it is commonly thought that Mg2+ is the physiological divalent metal ion involved in RAG-mediated cleavage. RAG1 and RAG2 are assisted by high mobility group proteins of the HMG-box family (HMGB1 and HMGB2) for bringing two signal ends together. The HMG proteins interact with the nonamer binding domain of RAG1 in the absence of DNA and enhance its intrinsic DNA bending activity.38 Following resolution of the hairpin structure, the coding ends are joined to create the exon, which forms the antigen-binding region of the antigen receptors (Fig. 2c). The signal ends remain bound to RAGs, which in turn protect the ends from further nuclease digestion 36,39 (Fig. 2c).
The imperfect joining
The blunt-ended signal ends can be directly ligated without any modification, while the coding ends undergo further processing (Fig. 2c).34,35 The hairpin at the coding end is opened and joined together by non-homologous end joining (NHEJ), the DNA double-strand break repair pathway.40,41 Artemis, in conjunction with DNA-PKcs, acts as an endonuclease and resolves the hairpins formed during V(D)J recombination.42 Ku heterodimer, consisting of Ku70 and Ku80, binds to the broken DNA ends and forms a complex with DNA-PKcs.43 Artemis has an inherent 5′-3′ exonuclease activity, whereas in association with DNA-PKcs it acts as a 5′-3′ endonuclease.42 The ends are filled in by the Pol X family of polymerases namely Pol μ and Pol λ. Mice deficient in Pol μ are shown to have shorter immunoglobulin light chain V to J junctions,44 while those lacking Pol λ have shorter immunoglobulin heavy-chain D to J and V to DJ junctions.45 In addition, Pol μ plays a role in the processing of 3′ ends while Pol λ processes the 5′ ends.45 This would suggest that Pol μ is involved in V(D)J recombination, but not Pol λ.46 The ligase IV/XRCC4 complex ligates the processed ends47,48 with the help of XLF.49,50 Ku70, Ku80, XRCC4 and Ligase IV are considered to be the ‘core’ NHEJ factors as these proteins were conserved during evolution and are required for all known NHEJ reactions.51,52 These are also inevitable for the joining of both coding and signal ends. On the other hand, DNA-PKcs and Artemis are believed to have evolved more recently and are needed only for the joining of coding ends.52
Diversity at the junction
At the time of joining of V, D and J subexons, several modifications like insertions and deletions can occur at the junctions resulting in further increase in the antigen receptor diversity. Asymmetric hairpin opening at the coding ends due to nicking a few bases away from the terminus results in one DNA strand longer than the other. The shorter strand is extended by addition of nucleotides complementary to the longer strand resulting in the insertion of palindromic (P) nucleotides at the coding joints.53 Terminal deoxynucleotidyl transferase (TdT) and DNA Pol μ further diversify these junctional sequences by catalysing the addition of non-templated nucleotides (N-nucleotides) to the coding ends.54 The junctional diversification can expand the diversity upto 1011 from the earlier 106 through the combinatorial diversification.
Non-standard recombination products
Alternative outcomes of V(D)J recombination reported were ‘hybrid joint’ and ‘open-shut joint’. During the formation of ‘hybrid joint’, the coding end of one subexon is joined to the signal end of another following the initial cleavage step of V(D)J recombination. In certain cases, the original pair of coding and signal ends, which was separated during the RAG cleavage phase, rejoins leading to formation of an ‘open-shut joint’.55 When there are no modifications at the joints, ‘open-shut joints’ are hard to detect. The released signal ends may non-specifically attack double-stranded DNA leading to transposition.55
Other modes of generation of the diversity
The antigen receptors are further modified by two processes, namely class switch recombination (CSR) and somatic hypermutation (SHM). The CSR refers to the rearrangements of the constant regions of antigen receptors upon encountering an antigen. This further expands the variability in the constant region following rearrangement at the variable region. The CSR replaces the expression from Cμ to Cγ, Cε or Cα, resulting in the switching of immunoglobulin isotype from IgM to IgG, IgE, or IgA without changing the antigen specificity (Fig. 3).56 The immunoglobulin CH locus comprises an array of CH genes, flanked by a switch (S) region at its 5′ region. The CSR takes place between two S regions, resulting in the loop-out deletion of the intervening DNA segments as circular DNA 57 (Fig. 3). The SHM refers to the random genetic mutations that occur in the B cells (and not T cells) at certain hotspots in the antigen-binding regions following an antigen encounter and results in the increased affinity of the receptor to the antigen.58 As a result of this, a fraction of the antibodies possessing low-affinity receptors to the defined antigen, further increase their affinity and undergo expansion. The SHM takes place in the V region of both H and L chain genes (VL/H), introducing a million times more point mutations than the genome-wide background leading to the generation of high-affinity antibodies. Hence, CSR and SHM act on entirely different targets, i.e. CH and VL/H, respectively. Therefore, it was believed that these two processes were regulated differently. However, recently, it has been shown that the same enzyme, the activation-induced cytidine deaminase initiates both CSR and SHM in mice and humans.57,59,60
Figure 3.
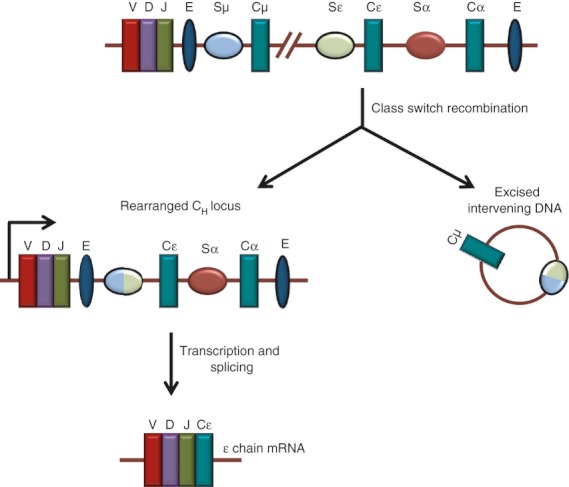
Mechanism of generation of the antigen receptor diversity by class switch recombination (CSR). CSR determines the type of constant region of antigen receptor by exchanging the gene encoding the immunoglobulin heavy chain constant region (CH) with one of the downstream CH genes. The figure depicts CSR between Sμ and Sα in the human immunoglobulin heavy chain locus. V, D and J segments of antigen receptor loci are represented as maroon, purple and green rectangles. The ovals represent the switch region and enhancers are marked by ‘E’. During the process, DNA double-strand breaks are introduced at the switch (S) regions followed by their rejoining. This leads to the deletion of the intervening sequence between S regions and juxtaposition of rearranged VDJ segment with the downstream CH fragment.
Mechanism of action of RAG proteins
RAG1
Murine RAG1 comprises 1040 amino acids. The ‘core region’ of RAG1 (cRAG1) consisting of amino acids 384–1008, is essential for all activities in vivo and in vitro.61,62 RAG1 exists as a homodimer in solution.63 It harbours all the elements that are indispensible for activity including the active site, DNA-binding domain and RAG2-interacting domain. The core regions acted as focal points of subsequent research, mainly because they were more soluble than their full-length counterparts. Using surface plasmon resonance and in vivo one-hybrid experiments, it was shown that the N-terminus of cRAG1 (amino acids 384–460) harbours the nonamer binding region.28 The heptamer recognition region of RAGs still remains obscure. The DDE motif (a triad of three acidic amino acids: D600, D708 and E962) of RAG1 forms the catalytic centre of the RAG1/RAG2 complex,64–66 which plays a role in chelating the two divalent metal ions essential for catalysis.67 The N-terminal non-core region (amino acids 1–383) contains a RING domain fold, which exhibits ubiquitin ligase activity.68
Studies by Rodgers's group63 using limited proteolysis showed that murine cRAG1 is composed of topologically independent domains that can function individually. These include the N-terminal, the central and the C-terminal domains. The central domain has the heptamer binding site, RAG2 binding site and zinc finger motif. The C-terminal domain has the dimerization region and binds DNA co-operatively. Murine cRAG1 was successfully expressed in Escherichia coli as a fusion protein with Maltose binding protein (MBP) tag with high yield and solubility and was active when combined with cRAG2 expressed in human embryonic kidney cell line.69 However, there is no report of successful bacterial expression of RAG2.
RAG2
Murine ‘core RAG2’ consists of amino acids 1–383 out of the total 527. The molecular function of core RAG2 remains elusive. RAG2 consists of an N-terminal 6-bladed beta-propeller domain and a C-terminal plant homeo domain (PHD).70,71 The PHD is a motif characteristic of chromatin remodelling proteins.72 It has been predicted to facilitate the ordered rearrangement of IgH chains and the binding of core histone proteins.72–74 The C-terminus of RAG2 contains a threonine residue (T490) that acts as a target of Chk2 kinase.75 Phosphorylation of this amino acid regulates the proteosomal degradation of RAG2 at the G1/S transition of the cell cycle.76 This regulatory mechanism ensures that RAG2 is degraded in a cell-cycle-dependent manner preventing RAG-induced DNA breaks during replication. Biochemical analysis of recombinant RAG2 has identified several basic residue mutants defective in catalysis. Accordingly, Schatz's group77 has proposed a model for the interaction of RAG2 with DNA in which the amino acids K119 and K283 directly contact DNA. It was shown that the PHD finger specifically recognizes histone 3 trimethylated at lysine 4 (H3K4me3).78 The H3K4me3 increases the catalytic turnover number (Kcat) of RAGs as well as tethering it to DNA.79 Recently, it was reported that mice harbouring RAG2 with a deleted C-terminal end rapidly developed thymic lymphomas with chromosomal translocations, amplifications and deletions involving TCRA/D and IgH loci implying its role in genomic stability.80
Interaction between RAG1 and RAG2
Protein–DNA cross-linking studies have suggested that cRAG1/cRAG2 may form specific contacts with the heptamer of RSS.81,82 In addition, binding assays have shown that cRAG1, even in the absence of RAG2, could bind to RSS in a manner that was specific to both heptamer and nonamer.38,82 RAG1 and RAG2 can interact and exist as a complex.83 Modification interference and direct footprinting studies indicated that RAG1 alone could make extensive contacts with the nonamer, whereas RAG2 is needed for the heptamer occupancy.81 Direct interaction between RAG2 and DNA in the RAG1/RAG2/RSS complex has also been reported.77 Fluorescence anisotropy and gel mobility shift assays indicate that RAG1 exhibits sequence specific binding character. It has been suggested that this property is masked by its non-specific DNA interactions and addition of RAG2 effectively rescues RAG1 from this non-specific binding.84
Using chromatin immunoprecipitation, it was shown that in vivo RAG binding was focused at the ‘recombination centres’ involving a small region encompassing J (and where present, J-proximal D) subexons in the IgH, IgK, TCRB and TCRA loci.85,86 The study showed that RAG2 binds throughout the genome at sites with substantial levels of H3K4me3. RAG1 binding was shown to be restricted to highly active chromatin in the presence of arrays of RSSs.85,86
Role of RAGs in receptor editing
Expression of RAG in early B cells occurs in two waves, the former is responsible for V(D)J rearrangement of IgH genes in pro-B and pre-B-I cells and the latter for VJ recombination of the IgL genes in small pre-B-II cells.87 There are also reports of a third wave of RAG expression, induced in activated mature B cells in vitro and in vivo.88 Spleen B cells from mice activated with lipopolysaccharide or interleukin-4 showed expression of RAGs.88 Mice immunized with the antigen trinitrophenyl-keyhole limpet haemocyanin also showed expression of RAG1 and RAG2 in the inguinal and popliteal cells of draining lymph nodes.88 In addition, immature B cells carrying self-reactive receptors exhibited up-regulated RAG expression. Following B-cell antigen receptor cross-linking, an increased level of RAG mRNA was reported in cultures of developing B cells.89,90 The sustained RAG expression allowed secondary V(D)J recombination that involves the replacement of self-reactive antibody V region genes by the V(D)J recombination. During this process, B cells with a non-self-reactive receptor are allowed to exit from prolonged V(D)J recombination, whereas the self-reactive cells are retained for further editing, until they produce a non-self-reactive receptor.91 ‘Receptor editing’ refers to the secondary rearrangement that occurs in immature B and T cells, which plays a role in mediating tolerance.92–95 ‘Receptor revision’ refers to the secondary rearrangement in mature B and T cells which alters their receptors in response to antigenic stimulation.96 In the case of immunoglobulin light chain and TCRA that lacks the D gene segments, the secondary rearrangement occurs between unrearranged V gene segments upstream and J segments downstream with deletion of the original rearranged VJ segment. These rearrangements do not violate the 12/23 rule. However, in the case of IgH and TCRB, rearranged gene contains a D segment and all other unused D segments are lost during DJ and VDJ rearrangements leaving behind only non-compatible RSSs. This obstacle is overcome by the presence of a 3′ sequence of the V segment, which plays the role of a surrogate RSS, thereby replacing the previously rearranged V, while retaining the already rearranged DJ.96,97
RAG mediated transposition
RAGs have been shown to exhibit transposition activity by integrating excised RSS-flanked signal ends into a target DNA molecule, in vitro. Integration can be intermolecular wherein the target DNA is a plasmid or intramolecular in which the target can be the intervening sequence stretching between RSSs.98–100 Integration was not sequence-specific but was targeted to altered DNA structures like hairpins.101 Several lines of studies compared RAGs with bacterial transposons and revealed striking similarities.102 Isolated studies have shown that RAG transposition can occur in vivo.103,104 The first among these demonstrated interchromosomal transpositions, wherein TCR-α signal ends from chromosome 14 inserted into the X-linked hypoxanthine-guanine phosphoribosyl transferase locus, resulted in gene inactivation.103 It was also shown that RAG expression in yeast could lead to transposition.104 The transib transposase from the insect Helicoverpa zea was shown to be active in vitro and its breakage and joining activities mimicked that of RAG, providing strong evidence that RAGs and transib transposases were derived from a common progenitor.105 However, there is no evidence that RAG-mediated transposition can occur in the mammalian genome. This can be the result of the stringent regulation of the process in the mammalian system.106
Structure-specific nuclease activity of RAGs
In contrast to the standard function of being a recombinase, later studies pointed out that the RAG complex can also act as a structure-specific nuclease and this property has several implications in the pathological roles of the RAG complex (Fig. 4). Studies suggested that RAGs possess a structure-specific 3′ flap endonuclease activity that can remove single-strand (ss) extensions from branched DNA structures.107 RAGs also showed hairpin opening activity in the presence of MnCl2.108,109 The fragility of the BCL2 major breakpoint region was attributed to its acquiring a stable non-B DNA structure in the genome, which was prone to RAG cleavage.110 Further, it was shown that RAGs could cleave symmetric bubbles, heterologous loops and potential G-quadruplex structures at the physiological concentrations of MgCl2111,112 (Fig. 4). RAG nicking efficiency was dependent on size of heteroduplex DNA and the proximity to RSS sequence did not affect nicking efficiency.113 It was also observed that structure specificity of RAGs could be attributed to the sequence at the single-stranded region. Cytosines were the most preferred, followed by thymines while purines were not cleaved at all. A consensus sequence of ‘C(d)C(s)C(s)’ (d, double-stranded; s, single-stranded) was also proposed for the generation of breaks at single-strand/double-strand transitions.114 The nonamer binding region of RAG1 was not important for RAG cleavage at non-B DNA structures, in contrast to that at RSS.115 The study showed low cleavage kinetics and a lack of cleavage complex formation at heteroduplex DNA, as the two mechanisms that ensured the control of the pathological activity of RAGs.115
Figure 4.
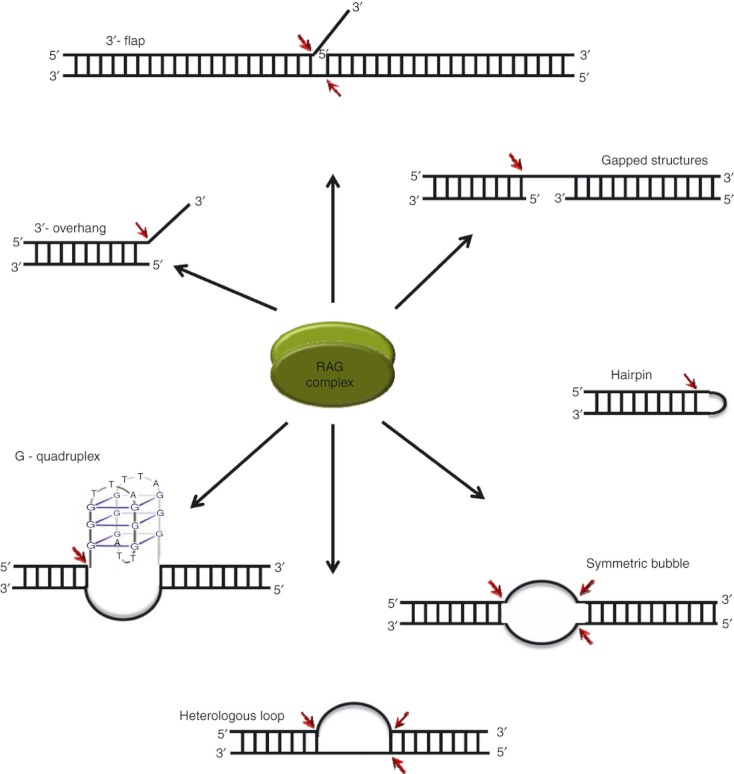
Structure-specific nuclease action of recombination activating genes (RAGs). The cleavage targets of the RAG complex when it acts as a structure-specific nuclease. The shaded ovals represent the RAG complex and the cleavage sites are indicated by red arrows.
Off-target cleavage by RAGs and its pathological relevance
In an ideal scenario, RAGs target RSS within the immunoglobulin/TCR loci. However, a large number of RSS-like sequences (cryptic RSS) exist throughout the genome and this would lead to the non-specific targeting of RAGs leading to DNA double-strand breaks outside the immunoglobulin/TCR loci, resulting in genomic rearrangements. If the rearrangement juxtaposes the immunoglobulin/TCR regulatory sequences like promoters or enhancers to proto-oncogenes, it could lead to over-expression of the oncogenes culminating in lymphoid malignancies. RAGs are known to generate breaks at sequences resembling heptamer or nonamer because of misrecognition in several leukaemias and lymphomas, which include translocations like MTS1, LMO2, TTG-1, SIL and SCL.116–119 The discovery that RAGs can detect and cleave non B-DNA structures further increased the spectrum of non-specific cleavage by RAGs.110 In case of t(14;18) translocation at follicular lymphoma wherein nearly 75% of the breakpoints are dispersed over a 150-bp region called major breakpoint region of BCL2,120 it has been shown that a non-B structure can form, which is specifically targeted and cleaved by RAGs.110,111 Later, the nature of this structure was identified as a G-quadruplex.112,121 It has also been shown that RAGs can cleave at an eight-nucleotide motif ‘CCACCTCT’ in the minor breakpoint cluster of the BCL2 in a nonamer-independent manner.122
Conclusions
To generate a functional antibody or TCR, several of the genomic segments propagating in the embryo have to select each other, merge in various combinations and further modify themselves. Though the main players in the process have been identified, the mechanism by which each of the individual proteins acts and broadly how the chronological order is regulated are not known. The structure of RAG proteins still remains elusive. Several questions regarding the structure specificity of RAGs are unclear. Biochemical and biophysical studies on the domains within the core and non-core regions of these proteins, studies on the full length proteins in vivo, and detection of their interacting partners are being pursued.
Acknowledgments
We would like to thank Dr Mridula Nambiar, Ms Mrinal Srivastava, Dr Abani Naik, Ms Rupa Kumari and Ms Deepthi R for critical reading of the manuscript. This work was supported by grant (SR/SO/BB/0037/2011) from DST, India. NM is supported by a Senior Research Fellowship from CSIR, India.
References
- 1.Lederberg J. Genes and antibodies. Science. 1959;129:1649–53. doi: 10.1126/science.129.3364.1649. [DOI] [PubMed] [Google Scholar]
- 2.Dreyer WJ, Bennett JC. The molecular basis of antibody formation: a paradox. Proc Natl Acad Sci U S A. 1965;54:864–9. doi: 10.1073/pnas.54.3.864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schatz DG, Oettinger MA, Baltimore D. The V(D)J recombination activating gene, RAG-1. Cell. 1989;59:1035–48. doi: 10.1016/0092-8674(89)90760-5. [DOI] [PubMed] [Google Scholar]
- 4.Oettinger MA, Schatz DG, Gorka C, Baltimore D. RAG-1 and RAG-2, adjacent genes that synergistically activate V(D)J recombination. Science. 1990;248:1517–23. doi: 10.1126/science.2360047. [DOI] [PubMed] [Google Scholar]
- 5.Schatz DG, Baltimore D. Stable expression of immunoglobulin gene V(D)J recombinase activity by gene transfer into 3T3 fibroblasts. Cell. 1988;53:107–15. doi: 10.1016/0092-8674(88)90492-8. [DOI] [PubMed] [Google Scholar]
- 6.Oettinger MA, Stanger B, Schatz DG, Glaser T, Call K, Housman D, Baltimore D. The recombination activating genes, RAG 1 and RAG 2, are on chromosome 11p in humans and chromosome 2p in mice. Immunogenetics. 1992;35:97–101. doi: 10.1007/BF00189518. [DOI] [PubMed] [Google Scholar]
- 7.Porter RR. Structural studies of immunoglobulins. Science. 1973;180:713–6. doi: 10.1126/science.180.4087.713. [DOI] [PubMed] [Google Scholar]
- 8.Edelman GM. Antibody structure and molecular immunology. Science. 1973;180:830–40. doi: 10.1126/science.180.4088.830. [DOI] [PubMed] [Google Scholar]
- 9.Hilschmann N, Craig LC. Amino acid sequence studies with Bence-Jones proteins. Proc Natl Acad Sci U S A. 1965;53:1403–9. doi: 10.1073/pnas.53.6.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tonegawa S. Somatic generation of antibody diversity. Nature. 1983;302:575–81. doi: 10.1038/302575a0. [DOI] [PubMed] [Google Scholar]
- 11.Weigert MG, Cesari IM, Yonkovich SJ, Cohn M. Variability in the lambda light chain sequences of mouse antibody. Nature. 1970;228:1045–7. doi: 10.1038/2281045a0. [DOI] [PubMed] [Google Scholar]
- 12.Sakano H, Kurosawa Y, Weigert M, Tonegawa S. Identification and nucleotide sequence of a diversity DNA segment (D) of immunoglobulin heavy-chain genes. Nature. 1981;290:562–5. doi: 10.1038/290562a0. [DOI] [PubMed] [Google Scholar]
- 13.Sakano H, Maki R, Kurosawa Y, Roeder W, Tonegawa S. Two types of somatic recombination are necessary for the generation of complete immunoglobulin heavy-chain genes. Nature. 1980;286:676–83. doi: 10.1038/286676a0. [DOI] [PubMed] [Google Scholar]
- 14.Kurosawa Y, von Boehmer H, Haas W, Sakano H, Trauneker A, Tonegawa S. Identification of D segments of immunoglobulin heavy-chain genes and their rearrangement in T lymphocytes. Nature. 1981;290:565–70. doi: 10.1038/290565a0. [DOI] [PubMed] [Google Scholar]
- 15.Brack C, Hirama M, Lenhard-Schuller R, Tonegawa S. A complete immunoglobulin gene is created by somatic recombination. Cell. 1978;15:1–14. doi: 10.1016/0092-8674(78)90078-8. [DOI] [PubMed] [Google Scholar]
- 16.Bernard O, Hozumi N, Tonegawa S. Sequences of mouse immunoglobulin light chain genes before and after somatic changes. Cell. 1978;15:1133–44. doi: 10.1016/0092-8674(78)90041-7. [DOI] [PubMed] [Google Scholar]
- 17.Alt FW, Yancopoulos GD, Blackwell TK, et al. Ordered rearrangement of immunoglobulin heavy chain variable region segments. EMBO J. 1984;3:1209–19. doi: 10.1002/j.1460-2075.1984.tb01955.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bassing CH, Swat W, Alt FW. The mechanism and regulation of chromosomal V(D)J recombination. Cell. 2002;109(Suppl):S45–55. doi: 10.1016/s0092-8674(02)00675-x. [DOI] [PubMed] [Google Scholar]
- 19.Corcoran AE. Immunoglobulin locus silencing and allelic exclusion. Semin Immunol. 2005;17:141–54. doi: 10.1016/j.smim.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 20.Alt FW, Enea V, Bothwell AL, Baltimore D. Activity of multiple light chain genes in murine myeloma cells producing a single, functional light chain. Cell. 1980;21:1–12. doi: 10.1016/0092-8674(80)90109-9. [DOI] [PubMed] [Google Scholar]
- 21.Coleclough C. Chance, necessity and antibody gene dynamics. Nature. 1983;303:23–6. doi: 10.1038/303023a0. [DOI] [PubMed] [Google Scholar]
- 22.Sakano H, Huppi K, Heinrich G, Tonegawa S. Sequences at the somatic recombination sites of immunoglobulin light-chain genes. Nature. 1979;280:288–94. doi: 10.1038/280288a0. [DOI] [PubMed] [Google Scholar]
- 23.Max EE, Seidman JG, Leder P. Sequences of five potential recombination sites encoded close to an immunoglobulin kappa constant region gene. Proc Natl Acad Sci U S A. 1979;76:3450–4. doi: 10.1073/pnas.76.7.3450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Early P, Huang H, Davis M, Calame K, Hood L. An immunoglobulin heavy chain variable region gene is generated from three segments of DNA: VH D and JH. Cell. 1980;19:981–92. doi: 10.1016/0092-8674(80)90089-6. [DOI] [PubMed] [Google Scholar]
- 25.Hesse JE, Lieber MR, Mizuuchi K, Gellert M. V(D)J recombination: a functional definition of the joining signals. Genes Dev. 1989;3:1053–61. doi: 10.1101/gad.3.7.1053. [DOI] [PubMed] [Google Scholar]
- 26.Ramsden DA, McBlane JF, van Gent DC, Gellert M. Distinct DNA sequence and structure requirements for the two steps of V(D)J recombination signal cleavage. EMBO J. 1996;15:3197–206. [PMC free article] [PubMed] [Google Scholar]
- 27.Difilippantonio MJ, McMahan CJ, Eastman QM, Spanopoulou E, Schatz DG. RAG1 mediates signal sequence recognition and recruitment of RAG2 in V(D)J recombination. Cell. 1996;87:253–62. doi: 10.1016/s0092-8674(00)81343-4. [DOI] [PubMed] [Google Scholar]
- 28.Spanopoulou E, Zaitseva F, Wang FH, Santagata S, Baltimore D, Panayotou G. The homeodomain region of Rag-1 reveals the parallel mechanisms of bacterial and V(D)J recombination. Cell. 1996;87:263–76. doi: 10.1016/s0092-8674(00)81344-6. [DOI] [PubMed] [Google Scholar]
- 29.van Gent DC, Ramsden DA, Gellert M. The RAG1 and RAG2 proteins establish the 12/23 rule in V(D)J recombination. Cell. 1996;85:107–13. doi: 10.1016/s0092-8674(00)81086-7. [DOI] [PubMed] [Google Scholar]
- 30.Bassing CH, Alt FW, Hughes MM, et al. Recombination signal sequences restrict chromosomal V(D)J recombination beyond the 12/23 rule. Nature. 2000;405:583–6. doi: 10.1038/35014635. [DOI] [PubMed] [Google Scholar]
- 31.Drejer-Teel AH, Fugmann SD, Schatz DG. The beyond 12/23 restriction is imposed at the nicking and pairing steps of DNA cleavage during V(D)J recombination. Mol Cell Biol. 2007;27:6288–99. doi: 10.1128/MCB.00835-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jung D, Bassing CH, Fugmann SD, Cheng HL, Schatz DG, Alt FW. Extrachromosomal recombination substrates recapitulate beyond 12/23 restricted VDJ recombination in nonlymphoid cells. Immunity. 2003;18:65–74. doi: 10.1016/s1074-7613(02)00507-1. [DOI] [PubMed] [Google Scholar]
- 33.Olaru A, Petrie HT, Livak F. Beyond the 12/23 rule of VDJ recombination independent of the Rag proteins. J Immunol. 2005;174:6220–6. doi: 10.4049/jimmunol.174.10.6220. [DOI] [PubMed] [Google Scholar]
- 34.Schlissel M, Constantinescu A, Morrow T, Baxter M, Peng A. Double-strand signal sequence breaks in V(D)J recombination are blunt, 5′-phosphorylated, RAG-dependent, and cell cycle regulated. Genes Dev. 1993;7:2520–32. doi: 10.1101/gad.7.12b.2520. [DOI] [PubMed] [Google Scholar]
- 35.Roth DB, Zhu C, Gellert M. Characterization of broken DNA molecules associated with V(D)J recombination. Proc Natl Acad Sci U S A. 1993;90:10788–92. doi: 10.1073/pnas.90.22.10788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Agrawal A, Schatz DG. RAG1 and RAG2 form a stable postcleavage synaptic complex with DNA containing signal ends in V(D)J recombination. Cell. 1997;89:43–53. doi: 10.1016/s0092-8674(00)80181-6. [DOI] [PubMed] [Google Scholar]
- 37.Hiom K, Gellert M. A stable RAG1-RAG2-DNA complex that is active in V(D)J cleavage. Cell. 1997;88:65–72. doi: 10.1016/s0092-8674(00)81859-0. [DOI] [PubMed] [Google Scholar]
- 38.Aidinis V, Bonaldi T, Beltrame M, Santagata S, Bianchi ME, Spanopoulou E. The RAG1 homeodomain recruits HMG1 and HMG2 to facilitate recombination signal sequence binding and to enhance the intrinsic DNA-bending activity of RAG1-RAG2. Mol Cell Biol. 1999;19:6532–42. doi: 10.1128/mcb.19.10.6532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hiom K, Gellert M. Assembly of a 12/23 paired signal complex: a critical control point in V(D)J recombination. Mol Cell. 1998;1:1011–9. doi: 10.1016/s1097-2765(00)80101-x. [DOI] [PubMed] [Google Scholar]
- 40.Ramsden DA, Gellert M. Formation and resolution of double-strand break intermediates in V(D)J rearrangement. Genes Dev. 1995;9:2409–20. doi: 10.1101/gad.9.19.2409. [DOI] [PubMed] [Google Scholar]
- 41.Taccioli GE, Rathbun G, Oltz E, Stamato T, Jeggo PA, Alt FW. Impairment of V(D)J recombination in double-strand break repair mutants. Science. 1993;260:207–10. doi: 10.1126/science.8469973. [DOI] [PubMed] [Google Scholar]
- 42.Ma Y, Pannicke U, Schwarz K, Lieber MR. Hairpin opening and overhang processing by an Artemis/DNA-dependent protein kinase complex in nonhomologous end joining and V(D)J recombination. Cell. 2002;108:781–94. doi: 10.1016/s0092-8674(02)00671-2. [DOI] [PubMed] [Google Scholar]
- 43.Rooney S, Sekiguchi J, Zhu C, et al. Leaky Scid phenotype associated with defective V(D)J coding end processing in Artemis-deficient mice. Mol Cell. 2002;10:1379–90. doi: 10.1016/s1097-2765(02)00755-4. [DOI] [PubMed] [Google Scholar]
- 44.Bertocci B, De Smet A, Berek C, Weill JC, Reynaud CA. Immunoglobulin kappa light chain gene rearrangement is impaired in mice deficient for DNA polymerase mu. Immunity. 2003;19:203–11. doi: 10.1016/s1074-7613(03)00203-6. [DOI] [PubMed] [Google Scholar]
- 45.Bertocci B, De Smet A, Weill JC, Reynaud CA. Nonoverlapping functions of DNA polymerases mu, lambda, and terminal deoxynucleotidyltransferase during immunoglobulin V(D)J recombination in vivo. Immunity. 2006;25:31–41. doi: 10.1016/j.immuni.2006.04.013. [DOI] [PubMed] [Google Scholar]
- 46.Nick McElhinny SA, Havener JM, Garcia-Diaz M, et al. A gradient of template dependence defines distinct biological roles for family X polymerases in nonhomologous end joining. Mol Cell. 2005;19:357–66. doi: 10.1016/j.molcel.2005.06.012. [DOI] [PubMed] [Google Scholar]
- 47.Rathmell WK, Chu G. A DNA end-binding factor involved in double-strand break repair and V(D)J recombination. Mol Cell Biol. 1994;14:4741–8. doi: 10.1128/mcb.14.7.4741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Taccioli GE, Gottlieb TM, Blunt T, et al. Ku80: product of the XRCC5 gene and its role in DNA repair and V(D)J recombination. Science. 1994;265:1442–5. doi: 10.1126/science.8073286. [DOI] [PubMed] [Google Scholar]
- 49.Buck D, Malivert L, de Chasseval R, et al. Cernunnos, a novel nonhomologous end-joining factor, is mutated in human immunodeficiency with microcephaly. Cell. 2006;124:287–99. doi: 10.1016/j.cell.2005.12.030. [DOI] [PubMed] [Google Scholar]
- 50.Ahnesorg P, Smith P, Jackson SP. XLF interacts with the XRCC4-DNA ligase IV complex to promote DNA nonhomologous end-joining. Cell. 2006;124:301–13. doi: 10.1016/j.cell.2005.12.031. [DOI] [PubMed] [Google Scholar]
- 51.Lieber MR. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem. 2010;79:181–211. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sharma S, Raghavan SC. Nonhomologous DNA end joining in cell-free extracts. J Nucleic Acids. 2012 doi: 10.4061/2010/389129. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lafaille JJ, DeCloux A, Bonneville M, Takagaki Y, Tonegawa S. Junctional sequences of T cell receptor gamma delta genes: implications for gamma delta T cell lineages and for a novel intermediate of V-(D)-J joining. Cell. 1989;59:859–70. doi: 10.1016/0092-8674(89)90609-0. [DOI] [PubMed] [Google Scholar]
- 54.Desiderio SV, Yancopoulos GD, Paskind M, Thomas E, Boss MA, Landau N, Alt FW, Baltimore D. Insertion of N regions into heavy-chain genes is correlated with expression of terminal deoxytransferase in B cells. Nature. 1984;311:752–5. doi: 10.1038/311752a0. [DOI] [PubMed] [Google Scholar]
- 55.Gellert M. Molecular analysis of V(D)J recombination. Annu Rev Genet. 1992;26:425–46. doi: 10.1146/annurev.ge.26.120192.002233. [DOI] [PubMed] [Google Scholar]
- 56.Stavnezer J, Guikema JE, Schrader CE. Mechanism and regulation of class switch recombination. Annu Rev Immunol. 2008;26:261–92. doi: 10.1146/annurev.immunol.26.021607.090248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Honjo T, Kinoshita K, Muramatsu M. Molecular mechanism of class switch recombination: linkage with somatic hypermutation. Annu Rev Immunol. 2002;20:165–96. doi: 10.1146/annurev.immunol.20.090501.112049. [DOI] [PubMed] [Google Scholar]
- 58.Neuberger MS, Milstein C. Somatic hypermutation. Curr Opin Immunol. 1995;7:248–54. doi: 10.1016/0952-7915(95)80010-7. [DOI] [PubMed] [Google Scholar]
- 59.Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, Honjo T. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–63. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- 60.Revy P, Muto T, Levy Y, et al. Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the Hyper-IgM syndrome (HIGM2) Cell. 2000;102:565–75. doi: 10.1016/s0092-8674(00)00079-9. [DOI] [PubMed] [Google Scholar]
- 61.Sadofsky MJ, Hesse JE, McBlane JF, Gellert M. Expression and V(D)J recombination activity of mutated RAG-1 proteins. Nucleic Acids Res. 1993;21:5644–50. doi: 10.1093/nar/21.24.5644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Silver DP, Spanopoulou E, Mulligan RC, Baltimore D. Dispensable sequence motifs in the RAG-1 and RAG-2 genes for plasmid V(D)J recombination. Proc Natl Acad Sci U S A. 1993;90:6100–4. doi: 10.1073/pnas.90.13.6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Arbuckle JL, Fauss LA, Simpson R, Ptaszek LM, Rodgers KK. Identification of two topologically independent domains in RAG1 and their role in macromolecular interactions relevant to V(D)J recombination. J Biol Chem. 2001;276:37093–101. doi: 10.1074/jbc.M105988200. [DOI] [PubMed] [Google Scholar]
- 64.Fugmann SD, Villey IJ, Ptaszek LM, Schatz DG. Identification of two catalytic residues in RAG1 that define a single active site within the RAG1/RAG2 protein complex. Mol Cell. 2000;5:97–107. doi: 10.1016/s1097-2765(00)80406-2. [DOI] [PubMed] [Google Scholar]
- 65.Landree MA, Wibbenmeyer JA, Roth DB. Mutational analysis of RAG1 and RAG2 identifies three catalytic amino acids in RAG1 critical for both cleavage steps of V(D)J recombination. Genes Dev. 1999;13:3059–69. doi: 10.1101/gad.13.23.3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kim DR, Dai Y, Mundy CL, Yang W, Oettinger MA. Mutations of acidic residues in RAG1 define the active site of the V(D)J recombinase. Genes Dev. 1999;13:3070–80. doi: 10.1101/gad.13.23.3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Steitz TA, Steitz JA. A general two-metal-ion mechanism for catalytic RNA. Proc Natl Acad Sci U S A. 1993;90:6498–502. doi: 10.1073/pnas.90.14.6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yurchenko V, Xue Z, Sadofsky M. The RAG1 N-terminal domain is an E3 ubiquitin ligase. Genes Dev. 2003;17:581–5. doi: 10.1101/gad.1058103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rodgers KK, Villey IJ, Ptaszek L, Corbett E, Schatz DG, Coleman JE. A dimer of the lymphoid protein RAG1 recognizes the recombination signal sequence and the complex stably incorporates the high mobility group protein HMG2. Nucleic Acids Res. 1999;27:2938–46. doi: 10.1093/nar/27.14.2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Corneo B, Moshous D, Callebaut I, de Chasseval R, Fischer A, de Villartay JP. Three-dimensional clustering of human RAG2 gene mutations in severe combined immune deficiency. J Biol Chem. 2000;275:12672–5. doi: 10.1074/jbc.275.17.12672. [DOI] [PubMed] [Google Scholar]
- 71.Gomez CA, Ptaszek LM, Villa A, et al. Mutations in conserved regions of the predicted RAG2 kelch repeats block initiation of V(D)J recombination and result in primary immunodeficiencies. Mol Cell Biol. 2000;20:5653–64. doi: 10.1128/mcb.20.15.5653-5664.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Callebaut I, Mornon JP. The V(D)J recombination activating protein RAG2 consists of a six-bladed propeller and a PHD fingerlike domain, as revealed by sequence analysis. Cell Mol Life Sci. 1998;54:880–91. doi: 10.1007/s000180050216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Elkin SK, Ivanov D, Ewalt M, et al. A PHD finger motif in the C terminus of RAG2 modulates recombination activity. J Biol Chem. 2005;280:28701–10. doi: 10.1074/jbc.M504731200. [DOI] [PubMed] [Google Scholar]
- 74.Liu Y, Subrahmanyam R, Chakraborty T, Sen R, Desiderio S. A plant homeodomain in RAG-2 that binds Hypermethylated lysine 4 of histone H3 is necessary for efficient antigen-receptor-gene rearrangement. Immunity. 2007;27:561–71. doi: 10.1016/j.immuni.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lin WC, Desiderio S. Regulation of V(D)J recombination activator protein RAG-2 by phosphorylation. Science. 1993;260:953–9. doi: 10.1126/science.8493533. [DOI] [PubMed] [Google Scholar]
- 76.Li Z, Dordai DI, Lee J, Desiderio S. A conserved degradation signal regulates RAG-2 accumulation during cell division and links V(D)J recombination to the cell cycle. Immunity. 1996;5:575–89. doi: 10.1016/s1074-7613(00)80272-1. [DOI] [PubMed] [Google Scholar]
- 77.Fugmann SD, Schatz DG. Identification of basic residues in RAG2 critical for DNA binding by the RAG1-RAG2 complex. Mol Cell. 2001;8:899–910. doi: 10.1016/s1097-2765(01)00352-5. [DOI] [PubMed] [Google Scholar]
- 78.Matthews AG, Kuo AJ, Ramon-Maiques S, et al. RAG2 PHD finger couples histone H3 lysine 4 trimethylation with V(D)J recombination. Nature. 2007;450:1106–10. doi: 10.1038/nature06431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Shimazaki N, Tsai AG, Lieber MR. H3K4me3 stimulates the V(D)J RAG complex for both nicking and hairpinning in trans in addition to tethering in cis: implications for translocations. Mol Cell. 2009;34:535–44. doi: 10.1016/j.molcel.2009.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Deriano L, Chaumeil J, Coussens M, et al. The RAG2 C terminus suppresses genomic instability and lymphomagenesis. Nature. 2011;471:119–23. doi: 10.1038/nature09755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Swanson PC, Desiderio S. V(D)J recombination signal recognition: distinct, overlapping DNA-protein contacts in complexes containing RAG1 with and without RAG2. Immunity. 1998;9:115–25. doi: 10.1016/s1074-7613(00)80593-2. [DOI] [PubMed] [Google Scholar]
- 82.Eastman QM, Villey IJ, Schatz DG. Detection of RAG protein-V(D)J recombination signal interactions near the site of DNA cleavage by UV cross-linking. Mol Cell Biol. 1999;19:3788–97. doi: 10.1128/mcb.19.5.3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Santagata S, Aidinis V, Spanopoulou E. The effect of Me2+ cofactors at the initial stages of V(D)J recombination. J Biol Chem. 1998;273:16325–31. doi: 10.1074/jbc.273.26.16325. [DOI] [PubMed] [Google Scholar]
- 84.Zhao S, Gwyn LM, De P, Rodgers KK. A non-sequence-specific DNA binding mode of RAG1 is inhibited by RAG2. J Mol Biol. 2009;387:744–58. doi: 10.1016/j.jmb.2009.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Schatz DG, Ji Y. Recombination centres and the orchestration of V(D)J recombination. Nat Rev Immunol. 2011;11:251–63. doi: 10.1038/nri2941. [DOI] [PubMed] [Google Scholar]
- 86.Ji Y, Resch W, Corbett E, Yamane A, Casellas R, Schatz DG. The in vivo pattern of binding of RAG1 and RAG2 to antigen receptor loci. Cell. 2010;141:419–31. doi: 10.1016/j.cell.2010.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Grawunder U, Leu TM, Schatz DG, Werner A, Rolink AG, Melchers F, Winkler TH. Down-regulation of RAG1 and RAG2 gene expression in preB cells after functional immunoglobulin heavy chain rearrangement. Immunity. 1995;3:601–8. doi: 10.1016/1074-7613(95)90131-0. [DOI] [PubMed] [Google Scholar]
- 88.Hikida M, Mori M, Takai T, Tomochika K, Hamatani K, Ohmori H. Reexpression of RAG-1 and RAG-2 genes in activated mature mouse B cells. Science. 1996;274:2092–4. doi: 10.1126/science.274.5295.2092. [DOI] [PubMed] [Google Scholar]
- 89.Hertz M, Nemazee D. BCR ligation induces receptor editing in IgM+ IgD– bone marrow B cells in vitro. Immunity. 1997;6:429–36. doi: 10.1016/s1074-7613(00)80286-1. [DOI] [PubMed] [Google Scholar]
- 90.Melamed D, Nemazee D. Self-antigen does not accelerate immature B cell apoptosis, but stimulates receptor editing as a consequence of developmental arrest. Proc Natl Acad Sci U S A. 1997;94:9267–72. doi: 10.1073/pnas.94.17.9267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Jankovic M, Casellas R, Yannoutsos N, Wardemann H, Nussenzweig MC. RAGs and regulation of autoantibodies. Annu Rev Immunol. 2004;22:485–501. doi: 10.1146/annurev.immunol.22.012703.104707. [DOI] [PubMed] [Google Scholar]
- 92.Papavasiliou F, Casellas R, Suh H, et al. V(D)J recombination in mature B cells: a mechanism for altering antibody responses. Science. 1997;278:298–301. doi: 10.1126/science.278.5336.298. [DOI] [PubMed] [Google Scholar]
- 93.Han S, Dillon SR, Zheng B, Shimoda M, Schlissel MS, Kelsoe G. V(D)J recombinase activity in a subset of germinal center B lymphocytes. Science. 1997;278:301–5. doi: 10.1126/science.278.5336.301. [DOI] [PubMed] [Google Scholar]
- 94.Gay D, Saunders T, Camper S, Weigert M. Receptor editing: an approach by autoreactive B cells to escape tolerance. J Exp Med. 1993;177:999–1008. doi: 10.1084/jem.177.4.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Tiegs SL, Russell DM, Nemazee D. Receptor editing in self-reactive bone marrow B cells. J Exp Med. 1993;177:1009–20. doi: 10.1084/jem.177.4.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Nemazee D, Hogquist KA. Antigen receptor selection by editing or downregulation of V(D)J recombination. Curr Opin Immunol. 2003;15:182–9. doi: 10.1016/s0952-7915(03)00008-6. [DOI] [PubMed] [Google Scholar]
- 97.Chen C, Nagy Z, Prak EL, Weigert M. Immunoglobulin heavy chain gene replacement: a mechanism of receptor editing. Immunity. 1995;3:747–55. doi: 10.1016/1074-7613(95)90064-0. [DOI] [PubMed] [Google Scholar]
- 98.Agrawal A, Eastman QM, Schatz DG. Transposition mediated by RAG1 and RAG2 and its implications for the evolution of the immune system. Nature. 1998;394:744–51. doi: 10.1038/29457. [DOI] [PubMed] [Google Scholar]
- 99.Hiom K, Melek M, Gellert M. DNA transposition by the RAG1 and RAG2 proteins: a possible source of oncogenic translocations. Cell. 1998;94:463–70. doi: 10.1016/s0092-8674(00)81587-1. [DOI] [PubMed] [Google Scholar]
- 100.Fugmann SD. RAG1 and RAG2 in V(D)J recombination and transposition. Immunol Res. 2001;23:23–39. doi: 10.1385/IR:23:1:23. [DOI] [PubMed] [Google Scholar]
- 101.Lee GS, Neiditch MB, Sinden RR, Roth DB. Targeted transposition by the V(D)J recombinase. Mol Cell Biol. 2002;22:2068–77. doi: 10.1128/MCB.22.7.2068-2077.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Fugmann SD, Lee AI, Shockett PE, Villey IJ, Schatz DG. The RAG proteins and V(D)J recombination: complexes, ends, and transposition. Annu Rev Immunol. 2000;18:495–527. doi: 10.1146/annurev.immunol.18.1.495. [DOI] [PubMed] [Google Scholar]
- 103.Messier TL, O'Neill JP, Hou SM, Nicklas JA, Finette BA. In vivo transposition mediated by V(D)J recombinase in human T lymphocytes. EMBO J. 2003;22:1381–8. doi: 10.1093/emboj/cdg137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Clatworthy AE, Valencia MA, Haber JE, Oettinger MA. V(D)J recombination and RAG-mediated transposition in yeast. Mol Cell. 2003;12:489–99. doi: 10.1016/s1097-2765(03)00305-8. [DOI] [PubMed] [Google Scholar]
- 105.Hencken CG, Li X, Craig NL. Functional characterization of an active Rag-like transposase. Nat Struct Mol Biol. 2012;19:834–6. doi: 10.1038/nsmb.2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Matthews AG, Oettinger MA. Regulation of RAG transposition. Adv Exp Med Biol. 2009;650:16–31. doi: 10.1007/978-1-4419-0296-2_2. [DOI] [PubMed] [Google Scholar]
- 107.Santagata S, Besmer E, Villa A, et al. The RAG1/RAG2 complex constitutes a 3′ flap endonuclease: implications for junctional diversity in V(D)J and transpositional recombination. Mol Cell. 1999;4:935–47. doi: 10.1016/s1097-2765(00)80223-3. [DOI] [PubMed] [Google Scholar]
- 108.Besmer E, Mansilla-Soto J, Cassard S, et al. Hairpin coding end opening is mediated by RAG1 and RAG2 proteins. Mol Cell. 1998;2:817–28. doi: 10.1016/s1097-2765(00)80296-8. [DOI] [PubMed] [Google Scholar]
- 109.Shockett PE, Schatz DG. DNA hairpin opening mediated by the RAG1 and RAG2 proteins. Mol Cell Biol. 1999;19:4159–66. doi: 10.1128/mcb.19.6.4159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Raghavan SC, Swanson PC, Wu X, Hsieh CL, Lieber MR. A non-B-DNA structure at the Bcl-2 major breakpoint region is cleaved by the RAG complex. Nature. 2004;428:88–93. doi: 10.1038/nature02355. [DOI] [PubMed] [Google Scholar]
- 111.Raghavan SC, Swanson PC, Ma Y, Lieber MR. Double-strand break formation by the RAG complex at the bcl-2 major breakpoint region and at other non-B DNA structures in vitro. Mol Cell Biol. 2005;25:5904–19. doi: 10.1128/MCB.25.14.5904-5919.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Nambiar M, Goldsmith G, Moorthy BT, Lieber MR, Joshi VM, Choudhary B, Hosur RV, Raghavan SC. Formation of a G-quadruplex at the BCL2 major breakpoint region of the t(14;18) translocation in follicular lymphoma. Nucleic Acids Res. 2011;39:936–48. doi: 10.1093/nar/gkq824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Raghavan SC, Gu J, Swanson PC, Lieber MR. The structure-specific nicking of small heteroduplexes by the RAG complex: implications for lymphoid chromosomal translocations. DNA Repair. 2007;6:751–9. doi: 10.1016/j.dnarep.2006.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Naik AK, Lieber MR, Raghavan SC. Cytosines, but not purines, determine recombination activating gene (RAG)-induced Breaks on heteroduplex DNA structures: implications for genomic instability. J Biol Chem. 2010;285:7587–97. doi: 10.1074/jbc.M109.089631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Naik AK, Raghavan SC. Differential reaction kinetics, cleavage complex formation, and nonamer binding domain dependence dictate the structure-specific and sequence-specific nuclease activity of RAGs. J Mol Biol. 2012;415:475–88. doi: 10.1016/j.jmb.2011.11.002. [DOI] [PubMed] [Google Scholar]
- 116.Raghavan SC, Kirsch IR, Lieber MR. Analysis of the V(D)J recombination efficiency at lymphoid chromosomal translocation breakpoints. J Biol Chem. 2001;276:29126–33. doi: 10.1074/jbc.M103797200. [DOI] [PubMed] [Google Scholar]
- 117.Lewis SM, Agard E, Suh S, Czyzyk L. Cryptic signals and the fidelity of V(D)J joining. Mol Cell Biol. 1997;17:3125–36. doi: 10.1128/mcb.17.6.3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Marculescu R, Le T, Simon P, Jaeger U, Nadel B. V(D)J-mediated translocations in lymphoid neoplasms: a functional assessment of genomic instability by cryptic sites. J Exp Med. 2002;195:85–98. doi: 10.1084/jem.20011578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zhang M, Swanson PC. V(D)J recombinase binding and cleavage of cryptic recombination signal sequences identified from lymphoid malignancies. J Biol Chem. 2008;283:6717–27. doi: 10.1074/jbc.M710301200. [DOI] [PubMed] [Google Scholar]
- 120.Jager U, Bocskor S, Le T, et al. Follicular lymphomas' BCL-2/IgH junctions contain templated nucleotide insertions: novel insights into the mechanism of t(14;18) translocation. Blood. 2000;95:3520–9. [PubMed] [Google Scholar]
- 121.Nambiar M, Raghavan SC. How does DNA break during chromosomal translocations? Nucleic Acids Res. 2011;39:5813–25. doi: 10.1093/nar/gkr223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Nambiar M, Raghavan SC. Mechanism of fragility at BCL2 gene minor breakpoint cluster region during t(14;18) chromosomal translocation. J Biol Chem. 2012;287:8688–701. doi: 10.1074/jbc.M111.307363. [DOI] [PMC free article] [PubMed] [Google Scholar]