Abstract
DNA glycosylases safeguard the genome by locating and excising a diverse array of aberrant nucleobases created from oxidation, alkylation, and deamination of DNA. Since the discovery 28 years ago that these enzymes employ a base flipping mechanism to trap their substrates, six different protein architectures have been identified to perform the same basic task. Work over the past several years has unraveled details for how the various DNA glycosylases survey DNA, detect damage within the duplex, select for the correct modification, and catalyze base excision. Here, we provide a broad overview of these latest advances in glycosylase mechanisms gleaned from structural enzymology, highlighting features common to all glycosylases as well as key differences that define their particular substrate specificities.
1. Introduction
The integrity of the chemical structure of DNA and its interactions with replication and transcription machinery is important for the faithful transmission and interpretation of genetic information. Oxidation, alkylation, and deamination of the nucleobases by a number of endogenous and exogenous agents create aberrant nucleobases (Figure 1) that alter normal cell progression, cause mutations and genomic instability, and can lead to a number of diseases including cancer [reviewed in 1]. Many of these lesions are removed by the base excision repair (BER) pathway [2], which is initiated by a DNA glycosylase specialized for a particular type of chemical damage. Upon locating a particular lesion within the DNA, glycosylases catalyze the excision of the nucleobase from the phosphoribose backbone by cleaving the N-glycosidic bond, generating an apurinic/apyrimidinic (AP) site (Figure 2). Monofunctional glycosylases catalyze only base excision, whereas bifunctional glycosylases also contain a lyase activity that cleaves the backbone immediately 3′ to the AP site. The resulting single-stranded and nicked AP sites are processed by AP endonuclease 1 (APE1), which hydrolyzes the phosphodiester bond 5′ to the AP site. This generates a 3′ hydroxyl substrate for replacement synthesis by DNA polymerase β, followed by sealing of the resulting nick by DNA ligase.
Figure 1.
Common DNA lesions referenced in this review. (A) Oxidized nucleobases. 8-OHG, 7,8-dihydro-8-hydroxyguanine; 8oxoG, 7,8-dihydro-8oxoGuanine; FapyG, 2,6-diamino-4-hydroxy-5-formamidopyrimidine; mFapyG, N7-methylFapyG; Tg, thymine glycol; Sp, spiroiminodihydantoin; Gh, guanidinohydantoin; Ia, iminoallantion; 5-OHU, 5-hydroxyuracil; DHU, dihydrouracil; 5-OHC, 5-hydroxycytosine; DHT, dihydrothymine. (B) Alkylated nucleobases. εA, 1,N6-ethenoadenine; εC, 3,N4-ethenocytosine; 3mA, N3-methyladenine; 3mG, N3-methylguanine; 7mG, N7-methylguanine; Hx, hypoxanthine. (C) Nucleobases repaired by the UDG/TDG family of DNA glycosylases. U, uracil; T, thymine; 5mC, 5-methylcytosine; 5hmC, 5-hydroxymethylcytosine; 5fC, 5-formylcytosine; 5caC, 5-carboxylcytosine.
Figure 2.
Chemical reaction catalyzed by DNA glycosylases. (A,B) Monofunctional glycosylases cleave the N-glycosidic bond to liberate free nucleobase (N) from the phosphoribose backbone through either associative (A) or dissociative (B) mechanisms. (C) Bifunctional mechanism, in which both the N-glycosidic bond and the DNA backbone are cleaved.
Since the glycosylases are the first line of defense against a vast array of DNA damage, they have been the subject of a large body of work to understand their mechanisms of action and cellular roles [3–12]. The first crystal structures of DNA glycosylases were reported in 1992 for bacteriophage T4 Endonuclease V (EndoV) and Escherichia coli (E. coli) Endonuclease III (EndoIII), which remove pyrimidine dimers and oxidized pyrimidines, respectively [13, 14]. Soon thereafter, DNA or inhibitor-bound structures of EndoV and uracil DNA glycosylase (UDG) established that these enzymes use a base-flipping mechanism to gain access to modified nucleobases in DNA [15–19]. Subsequent studies established that glycosylases fall into one of six structural superfamilies (Figure 3). Despite their divergent architectures, these proteins, with the exception of the ALK family (see section 3.3) [12], have evolved the base-flipping strategy to correctly identify and orient their substrates for catalysis. Recognition of the target modification likely proceeds in several steps, in which the protein probes the stability of the base pairs through processive interrogation of the DNA duplex, followed by extrusion of the aberrant nucleobase into a specific active site pocket on the enzyme [9, 20]. The enzyme-substrate complex is stabilized by nucleobase contacts within the active site and a pair of side chains that plug the gap in the DNA left by the extrahelical nucleotide and wedge into the DNA base stack on the opposite strand [3–12].
Figure 3.
DNA glycosylase structural superfamilies. Representative crystal structures from each class shown are: EndoV, T4 pyrimidine dimer DNA glycosylase EndoV (PDB ID 1VAS); UDG, human uracil-DNA glycosylase UDG (1EMH); Helix-hairpin-Helix (HhH), human 8oxoGuanine DNA glycosylase OGG1 (1YQK); Helix-two turn-helix (H2TH), Bacillus stearothermophilus 8oxoGuanine DNA glycosylase MutM (1L1T); AAG, human alkyladenine DNA glycosylase AAG/MPG (1EWN); ALK, Bacillus cereus alkylpurine DNA glycosylase AlkD (3JXZ). Proteins are colored according to secondary structure with the HhH and H2TH domains magenta. DNA is shown as gray sticks.
In this review, we focus on the most recent advances toward understanding the mechanisms by which each class of DNA glycosylase locates, selects, and removes its target lesions. A growing number of structures and mechanistic studies of glycosylases specific for oxidized nucleobases (Section 2), alkylation damage (Section 3), and cytosine deamination products (Section 4) have elucidated many of the structural determinants of substrate specificity and have provided new insights into catalysis of N-glycosidic bond cleavage. Some aspects of substrate selection and excision are common across different structural classes or substrate specificities, while others are specific to a given enzyme. Our goal in this review, therefore, is to provide a broad overview of the structural mechanisms for the entire repertoire of DNA glycosylases in order to highlight key similarities and differences between each structural class. We note that the roles of DNA glycosylases in the cell and in the context of BER have been the subject of recent reviews, and thus we focus our discussion on the structural enzymology.
2. Oxidative damage
DNA bases undergo oxidative damage from chemical oxidants, free radicals and reactive oxygen species (ROS) produced from cellular respiration, inflammatory responses, and ionizing radiation [21–23]. Oxidized bases are often used as biomarkers for oxidative stress and cancer [22, 24]. Guanines are especially susceptible to oxidation, leading to a number of lesions that are substrates for BER (Figure 1A) [25]. Attack of a hydroxyl radical at the C8 position of guanine produces 7,8-dihydro-8-hydroxyguanine (8-OHG), which tautomerizes to 8-oxo-7,8-dihydroguanine (8oxoG), or the ring-opened 2,6-diamino-5-formamido-4-hydroxy-pyrimidine (FapyG), two of the most abundant oxidative DNA adducts [26, 27]. 8oxoG is a particularly insidious lesion because of its dual coding potential by replicative polymerases, leading to G→T transversion mutations likely as a result of its ability to form both 8oxoG(syn)•A(anti) and 8oxoG(anti)•C(anti) base pairs [22, 23, 28–30]. Oxidation of guanine and 8oxoG also produces a variety of ring-opened purines in addition to FapyG, including hydantoin lesions, spiroiminodihydantoin (Sp), guanidinohydantoin (Gh), and its isomer iminoallantoin (Ia) (Figure 1A) [31–33]. Fapy lesions inhibit DNA polymerases and are potentially mutagenic [34]. Hydantoin lesions have been suggested to lead to an increase in G→T and G→C transversions and stall the replication machinery [31, 32, 35, 36]. In addition to purines, reaction of hydroxyl radicals at positions 5 or 6 of thymine produces 5,6-dihydroxy-5,6-dihydrothymine (thymine glycol, Tg), a cytotoxic lesion that distorts the DNA duplex and can inhibit replication [26, 37]. Other potentially harmful pyrimidines include dihydrothymine (DHT), dihydrouracil (DHU), 5-hydroxyuracil (5-OHU), 5-hydroxycytosine (5-OHC), 5-hydroxymethyluracil (5hmU), and 5-formyluracil (5fU) [38–43].
DNA glycosylases that remove oxidative DNA damage can be categorized on the basis of their preferences for purine or pyrimidine lesions and their structural folds (Table 1). Oxidized purines, including 8oxoG and FapyG, are removed from DNA by 8oxoG DNA glycosylase (OGG1) in eukaryotes and MutM (also known as FapyG DNA glycosylase, Fpg) in bacteria [recently reviewed in 23 ]. Oxidized pyrimidines are removed by endonuclease III (EndoIII, or Nth) and endonuclease VIII (Endo VIII, or Nei), and their eukaryotic orthologs, NTH1 and NEIL1 (Nei-like1), respectively. Despite their different substrates, OGG1 and EndoIII/Nth adopt a common architecture characteristic of the Helix-hairpin-Helix (HhH) superfamily of DNA glycosylases [44]. MutM/Fpg and EndoVIII/Nei are also structurally similar, with helix-two turn-helix (H2TH) and antiparallel β-hairpin zinc finger motifs, and they share a common bifunctional catalytic mechanism involving both base excision and AP lyase activities [45–49].
Table 1.
DNA glycosylases specific for oxidized, alkylated, mismatched, uracil, and 5-methylcytosine bases
| Eukaryotes | Archaea | Prokaryotes | Protein Fold | Substrates | PDB entries | |
|---|---|---|---|---|---|---|
| Oxidation | ||||||
| OGG1 | OGG | Ogg | HhH | 8oxoG•C, FapyG, FapyA |
1KO9 (hOGG1) 1EBM (K249Q/8oxoG-DNA) 1FN7 (THF-DNA) 1HU0, 1LWV, 1LWW, 1LWY (NaBH4-trapped DNA complex) 1M3H (D268E/nicked-DNA) 1M3Q (D268E/abasic-DNA/8-aminoG) 1N39 (D268E/THF-DNA) 1N3A (D268Q/THF-DNA) 1N3C (D268N/THF-DNA) 1YQK (N149C/G•C-DNA XL) 1YQL (N149C, K249Q/7-deaza-8-azaguanine-DNA XL) 1YQM (N149C, K249Q/7-deazaguanine-DNA XL) 1YQR (N149C, K249Q/8oxoG•C-DNA XL) 2I5W (N149C/8oxoG•G-DNA XL) 2NOB (N149C, K249Q, H270A/8oxoG•C-DNA XL) 2NOE (G42A, K249Q/8oxoG•C-DNA) 2NOF (N149C, Q315F/8oxoG•C/DNA XL) 2NOH (K249Q, Q315A/8oxoG•C-DNA) 2NOI (G42A, N149C, K249Q/8oxoG•C-DNA XL) 2NOL (K249Q, S292C/8oxoG•C-DNA) 2NOZ (S292C, Q315F/8oxoG•C-DNA) 2XHI (K249C, C253K, D268N/8oxoG•C-DNA) 3KTU (2′F-8oxoG-DNA) |
|
| OGG2 | HhH | 8oxoG (paired with any base) |
3FHF (MjOGG) 3KNT (MjOGG K129G/8oxoG•C-DNA) 3FHG (SsOGG) |
|||
| AGOG | HhH | 8oxoG (ssDNA, dsDNA) | 1XQO 1XQP (free 8oxoG) |
|||
| MutM/Fpg | H2TH | 8oxoG, FaPy, 7mFapyG, Sp, Gh, Tg, Ug, DHT, DHU, 5- OHU, 5-OHC, FU, urea, oxazolone, oxaluric acid, oxidized εA derivatives, sulfur mustard guanine N7- adduct, ring-opened oxidized aminofluorene guanine C8-adduct, 5- hydroxy-5- methylhydantoin, 3- [(aminocarbonyl)amino ]-(2R)-hydroxy-2- methylpropanoic acid | 1EES TtMutM 2F5Q, 2F5S (E3Q GsMutM/8oxoG•C-DNA XL) 1R2Y (E3Q GsMutM/8oxoG•C-DNA) 1R2Z (E3Q GsMutM/DHU-DNA) 1LIT (GsMutM/reduced abasic site-DNA) 1LIZ (GsMutM/NaBH4-trapped DNA complex) 1L2B (GsMutM/nicked DNA complex) 1L2C (GsMutM/HPD•T-DNA) 1L2D (GsMutM/HPD•G-DNA) 2F5N, 2F5P (Q166C GsMutM/A•T-DNA XL) 2F5O (Q166C GsMutM/G•C-DNA XL) 2F5Q, 2F5S (E3Q, Q166C GsMutM/8oxoG•C-DNA XL) 3JR4, 3JR5 (N174C GsMutM/G•C-DNA XL) 3GP1 (Q166C, V222P GsMutM/8oxoG•C-DNA XL) 3GPP (Q166C, T224P GsMutM/8oxoG•C-DNA XL) 3GPU, 3GQ3, 3GO8 (Δ220-235, Q166C GsMutM/8oxoG•C-DNA XL) 3GPX (Δ220-235, Q166C GsMutM/G•C-DNA XL) 3GPY (Q166C GsMutM/8oxoG•C-DNA XL) 3GQ4 (GsMutM/8oxoG•C-DNA XL) 3GQ5 (Q166C, T224P GsMutM/G•C-DNA XL) 3SAR, 3SAU (Q166C GsMutM/A•T-DNA XL) 3SAS, 3SAT (Q166C, R112A GsMutM/G•C-DNA XL) 3SAV (A149S, Q166C, GsMutM/G•C-DNA XL) 3SAW (GsMutM/G•C-DNA XL) 3SBJ (Q166C, V222P GsMutM/A•T-DNA XL) 1XC8, 1TDZ (LlMutM/FapyG•C-DNA) 1PJI, 1NNJ (LlMutM/PDI•C-DNA) 1PJJ, 1PM5 (LlMutM/THF•C-DNA) 1KFV(P1G LlMutM/PDI•C-DNA) 3C58 (LlMutM/7bFapyG•C-DNA) 2XZF (LlMutM-HC-DNA) 2XZU (LlMutM-HC-DNA XL) |
|||
| NTH1 | EndoIII | Nth/EndoIII | HhH/FeS2 | Tg, Ug, DHU, 5-OHU, 5-OHC, urea | 2ABK (EcEndoIII) 1ORN, 1ORP (GsEndoIII/NaBH4-trapped DNA complexes) 1P59 (GsEndoIII/THF-DNA) |
|
| NEIL1 | Nei/Endo VIII | H2TH | Tg, DHT, DHU, 5- OHU, 5-OHC, 5fU, 5hmU, FapyG, FapyA, urea, 8oxoA, Gh, Sp, Ia; (Nei only: Ug, 8oxoG, 7mFapyG, 5,6dhC, 5- OHT) | 1Q39 (EcEndoVIII) 1Q3B, 1Q3C (EcEndoVIII R252 and E2A mutants) 1K3W, 1K3X (EcEndoVIII/NaBH4-trapped DNA complexes) 2EA0, 2OPF, 2OQ4 (EcEndoVIII/PED-DNA) 1TDH (NEIL1) 3A45 (MvNei1) 3A46 (MvNei1/THF-DNA) 3VK8, 3VK7 (MvNei1/Tg-DNA, MvNei1/5-OHU-DNA) |
||
| NEIL2 | H2TH | Gh/Ia, 5-OHU, FapyG | [Ref. 115, 124] | |||
| NEIL3 | H2TH | Sp, Gh, FapyG, FapyA | [Ref. 115, 129] | |||
| Alkylation | ||||||
| AAG | AAG | 3mA, 7mG, εA, Hx, A, G | 1EWN (εA-DNA) 1BNK (pyrrolidine-DNA) 3QI5 (εC-DNA) |
|||
| MAG, Mag1 | HhH | 3mA, 3mG, 7mG, 7- CEG, 7-HEG, εA, Hx, G | 3S6I (SpMag1/THF-DNA) | |||
| AfAlkA | HhH | 3mA, 7mG, εA, 1mA, 3mC | 2JHN (AfAlkA) | |||
| MpgII | HhH | 3mA, 7mG | [Ref. 182] | |||
| AlkA | HhH | 3mA, 3mG, 7mG, 7- CEG, 7-HEG, 7-EG, O2-mT, O2-mC, εA, Hx, A, G, T, C, Xa | 1MPG (EcAlkA) 1DIZ (EcAlkA/1-azaribose-DNA) 1PVS (EcAlkA/Hx base) 3OGD, 3OH6, 3OH9 (EcAlkA/undamaged DNA XL) 2H56 (BhAlkA) 2YG9 (DrAlkA) |
|||
| MagIII | HhH | 3mA, mispaired 7mG | 1PU6 1PU7 (MagIII/3,9-dimethylA) 1PU8 (MagIII/εA) |
|||
| TAG | HhH1 | 3mA, 3mG | 2OFK (StTAG) 2OFI (StTAG/THF-DNA/3mA) 1NKU, 1LMZ (EcTAG NMR) 1P7M (EcTAG/3mA NMR) 4AIA (SaTAG) |
|||
| AlkC, AlkD | ALK/HEAT | 3mA, 3mG, 7mG, 7- POB-G, O2-POB-C | 3BVS (AlkD) 3JX7 (AlkD/3d3mA-DNA) 3JXY (AlkD/G•T-DNA) 3JXZ (AlkD/THF•T-DNA) 3JY1 (AlkD/THF•C-DNA) |
|||
| Adenine | ||||||
| MUTYH | MutY | MutY | HhH/FeS2 | A•8oxoG, A•G, | 1MUY, 1KG2, 1KG3 (EcMutY CD) 1RRQ (GsMutY/A•8oxoG-DNA XL) 1RRS, IVRL (GsMutY/HPD•8oxoG-DNA XL/adenine) 3N5N (HsMUTYH) 1WEF (K20A EcMutY CD) 1WEG, 1KG4 (K142A EcMutY CD) 1WEI (K20A EcMutY CD/adenine) 1KG5 (K142Q EcMutY CD) 1KG6 (K142R EcMutY CD) 1KG7 (E161A EcMutY CD) 1KQJ (C199H EcMutY CD) 1MUD (D138N EcMutY CD/adenine) 1MUN (D138N EcMutY CD) |
|
| U/T/5mC | ||||||
| UDG | Ung | UDG-1 | U•G | 1AKZ (HsUDG) 1SSP (HsUDG/U-DNA) 1LAU, 1UDG (HSV1 UDG) 1UDH (HSV1 UDG/uracil) 1EUG, 2EUG, 3EUG, 5EUG (EcUng/U, EcUng/glycerol) |
||
| SMUG | UDG-3 | U (ssDNA), U•G, U•A, 5hmU, 5-OHU, 5fU | 1OE4 (XlSMUG/THF-DNA) 1OE5 (XlSMUG/THF-DNA/U) 1OE6 (XlSMUG/THF-DNA/5hmU) |
|||
| TDG | MUG | UDG-2 | T•G, U•G, U•A, 5fC, 5caC, 5FU•G, 5FU•A, 5BrU•G, 5BrU•A, 5hmU•G 5-OHU•G, Tg•G, εC•G, εC•A, Hx•G, 8hmεC, εG Xa | 2D07 (HsTDG/SUMO3) 1WYW (HsTDG/SUMO1) 2RBA (HsTDG/THF-DNA) 3UO7 (HsTDG/5caC-DNA) 3UFJ (HsTDG/dU analog) 1MWJ (EcMUG/U-DNA) 1MTL, 1MWI (EcMUG/AP-DNA) 1MUG (EcMUG) |
||
| UDG | UDG-4 | U (ssDNA), U•G | 1UI0 (TtUDG) | |||
| MBD4 | HhH | T•G, U•G, 5FU•G, εC, 5mC | 1NGN (MmMBD4 CD) 3IHO (HsMBD4 CD) 4DK9 (HsMBD4/THF-DNA) 4EVV (MmMBD4/T•G-DNA) 4EW0 (MmMBD4/5hmU•G-DNA) 4EW4 (MmMBD4/AP-DNA) |
|||
| MIG | HhH/FeS2 | T•G | 1KEA | |||
| DME, ROS1, DML2, DML3 | HhH/FeS2 | 5mC, T•G | Plants only; [Ref. 308, 311, 315 323] |
TAG adopts the HhH architecture, but lacks the conserved catalytic aspartate and lysine residues present in mono- and bifunctional HhH glycosylases
EndoIII, MutY, MIG, and DME/ROS incorporate Fe4S4-type iron sulfur clusters (FeS) into their HhH architecture
Abbreviations: AP, abasic site; THF, tetrahydrofuran; HPD, 1-hydroxypentane-3,4-diol; PDI, 3-hydroxypropyl; PED, pentane-3,4-diol; HC; hydantoin carbanucleoside; 8oxoG, 8-oxo-7,8-dihydroguanine; FapyG, 2,6-diamino-4-hydroxy-5-formamidopyrimidine; FapyA, 4,6-diamino-5-formamidopyrimidine; 7mFapyG, N7-methylFapyG; 7bFapyG; N7-benzylFapyG;Tg, thymine glycol; DHT, dihydrothymine; DHU, dihydrouracil; 5-OHC, 5-hydroxycytosine; 5-OHT, 5-hydroxythymine; 5,6dhC, 5,6-dihydroxycytosine; 5-OHU, 5-hydroxyuracil; 5mC, 5-methylcytosine; 5hmC, 5-hydroxymethylcytosine; 5fC, 5-formylcytosine; 5caC, 5-carboxylcytosine; 5hmU, 5-hydroxymethyluracil; 5fU, 5-formyluracil; 5FU, 5-fluorouracil; 5BrU, 5-bromouracil; Gh, guanidinohydantoin; Ia, iminoallantion; Sp, spiroiminodihydantoin; 3mA, N3-methyladenine; 3mG, N3-methylguanine, 7mG, N7-methylguanine; 7-CEG, 7-(2-chloroethyl)guanine; 7-HEG, 7-(2-hydroxyethyl)guanine; 7-POB-G, N7- pyridyloxobutylguanine; O2-POB-C, O2-pyridyloxobutylcytosine; εA, 1,N6-ethenoadenine; εG, 1,N2-ethenoguanine; εC, 3,N4-ethenocytosine; 8hmεC, 8-(hydroxymethyl)-3,N4-ethenocytosine; 3d3mA, 3-deaza-N3-methyladenine; Hx, hypoxanthine; Xa, xanthine; ssDNA, single-stranded DNA; XL, covalent cross-linked protein-DNA; CD, catalytic domain; O2-mT, O2-methylthymine;O2-mC, O2-methylcytosine
Organisms: Hs, Homo Sapeiens; Mm, Mus musculus; Xl, Xenopus laevis; Mv, mimivirus; HSV1, Herpes simplex virus 1; Sc, Saccharomyces cerevisiae; Sp, Schizosaccharomyces pombe; Mt, Methanothermobacter thermautotrophicus; Af, Archaeoglobus fulgidus; Ss, Sulfolobus solfataricus; Tt, Thermus thermophilus; Gs, Geobacillus stearothermophilus; Mj, Methanocaldococcus jannaschii; Ec, Escherichia coli; Ll, Lactobacillus lacti; St, Salmonella typhi; Sa, Staphylococcus aureus; Bh, Bacillus halodurans; Dr, Deinococcus radiodurans
2.1. 8oxoG Repair
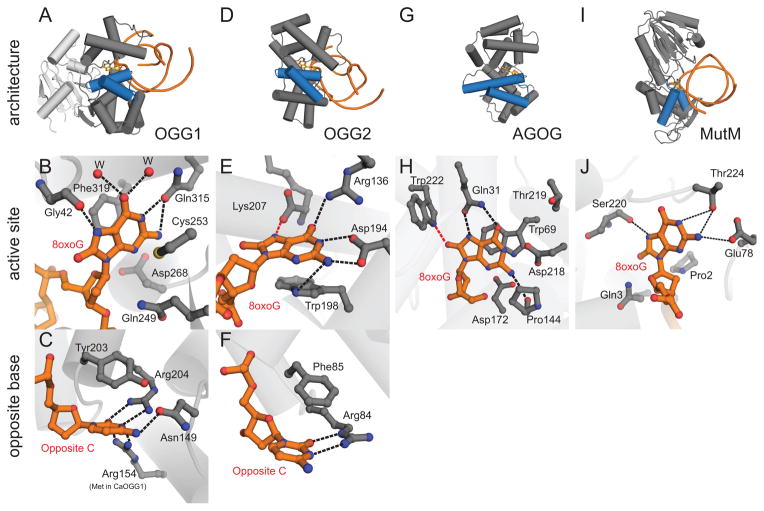
Eukaryotic OGG1 and bacterial MutM/Fpg preferentially catalyze removal of 8oxoG paired with C [50, 51]. Both enzymes are bifunctional in that they contain both base excision and AP lyase activities, although a recent report suggests that human OGG1 (hOGG1) may function as a monofunctional glycosylase under physiological conditions (see section 2.1.1) [44, 52, 53]. The OGG enzymes can be subdivided into three structural families (Figure 4): (1) OGG1, including human OGG1 and the recently discovered Clostridium acetobutylicum (CaOGG) enzyme (Figure 4A–C) [54–63], (2) archaeal OGG2 (Figure 4D–F) [64, 65], and (3) archaeal 8oxoG glycosylase (AGOG), represented by the Pyrobaculum aerophilum enzyme (Figure 4G–H) [66]. Structural studies of the various OGG orthologs [67] and of MutM have elucidated the molecular details required for 8oxoG recognition and excision from two distinct protein architectures, and in recent years have advanced our understanding how DNA glycosylases in general scan unmodified DNA in search of damage [for an excellent review, see ref. 4].
Figure 4.
Oxidative DNA glycosylases. (A–C) OGG1, represented by human OGG1 (PDB ID 1EBM), (D–F) OGG2, represented by MjOGG (PDB ID 3KNT), (G–H) Pyrobaculum aerophilum AGOG (PDB ID 1XQP) and (I–J) Geobacillus stearothermophilus MutM. The overall folds of each enzyme are shown on the top row (blue HhH motif), active sites on the second row, and opposing base on the bottom row. In the close-up views, the protein side-chains are grey and the DNA orange. Water molecules are represented by red spheres and hydrogen bonds are shown as dashed lines. (B) The human OGG1 8oxoG recognition pocket. The only 8oxoG specific contact is the hydrogen bond from the carbonyl group of Gly42 to the protonated N7 of 8oxoG. (C) The high specificity of hOGG1 for 8oxoG•C base pairs can be rationalized by the 5 hydrogen bonds between the opposite cytosine and 3 side chains. (E) In MjOGG, the 8oxoG N7 donates a hydrogen bond (red dashed line) to the C-terminal Lys207 carboxylate. (F) The opposite cytosine in MjOGG is contacted by only one side chain. (H) 8oxoG nucleoside bound inside the AGOG active site, with a unique 8oxoG-specific contact to Trp222 (red dashed line). (J) Active site of MutM (PDB ID 1R2Y) shows multiple contacts to 8oxoG but lacks the aromatic residues seen in the OGG1, OGG2, and AGOG enzymes.
2.1.1. OGG1
A battery of recent structures of hOGG1 in complex with DNA containing an 8oxoG•C base pair (Lesion Recognition Complex, LRC) or a normal G•C base pair (Interrogation Complex, IC) from the Verdine group has been invaluable in understanding how DNA glycosylases recognize and discriminate their substrates from normal DNA [52, 68–70] (the Km values of murine OGG1 (mOGG1) are 42.7 ± 14.6 nM for 8oxoG•C and 694 ± 145 nM for G•C [71]). The original hOGG1 LRC structure was obtained from a catalytically inactive Lys249Gln mutant bound to DNA containing an 8oxoG•C base pair [52], which revealed how hOGG1 utilizes the HhH architecture to kink the DNA duplex, disrupt the 8oxoG•C base pair, and extrude the 8oxoG out of the helix and into a base binding pocket [52]. Of the multiple contacts to the extrahelical 8oxoG, only one—between the carbonyl oxygen of Gly42 and the N7 hydrogen of 8oxoG—is specific to 8oxoG (Figure 4B) and was thus proposed to account for OGG1’s ability to distinguish 8oxoG from G. However, the position of the backbone and the integrity of the 8oxoG-specific hydrogen bond are not dependent on glycine in this position, as a Gly42Ala substitution did not alter the protein backbone conformation, disrupt the hydrogen bond, or affect the Kd (~15 nM) of the interaction with 8oxoG-DNA [70].
In the hOGG1 IC structure, which used a disulfide crosslinking strategy to trap the enzyme bound to a G•C base pair, the extrahelical guanine was situated in a pocket adjacent to the active site that the authors termed the ‘exo’ site [68]. In a subsequent IC structure, in which the enzyme was forcibly presented with a G•C base pair adjacent to 8oxoG, the extrahelical guanine was not observed in the active or exo sites, likely as a result of steric and electrostatic clashes imposed by the 8oxoG [69]. In both of these ICs, the protein (Asn149Cys) was crosslinked to the cytosine opposite the extrahelical G. In a more recent structure of a catalytically active hOGG1/G•C-DNA complex that was crosslinked at a more remote location from the lesion (Ser292Cys), the target guanine was fully engaged inside the active site in a virtually identical position as 8oxoG in the LRC. In the IC, however, the guanine remained uncleaved, presumably because it lacks the N7 hydrogen present in 8oxoG that forms a specific hydrogen bond with the carbonyl of Gly42 [72]. The alignment of active site residues other than Gly42 are also important for catalysis, as observed in a phototrapped, uncleaved hOGG1/8oxoG-DNA complex that showed an intact 8oxoG-Gly42 interaction amidst a collection of side chain conformers that differed from their position in the LRC [73]. Taken together, these data demonstrated that hOGG1 recognition of 8oxoG within DNA occurs in multiple steps, and that 8oxoG excision relies on precise chemical compatibility within the base binding pocket.
hOGG1 has been regarded as a bifunctional DNA glycosylase involving two key catalytic residues, Asp268 and Lys249 [52, 74–76]. The proposed catalytic mechanism involves Asp268-dependent deprotonation of the Lys249 ε-amino group, which forms a Schiff base with ribose C1′ of the 8oxoG nucleotide, resulting in β-elimination. However, various groups have reported monofunctional glycosylase activity for hOGG1 in vivo [71, 77–80]. Recently, Dalhus and colleagues used structural and mutational analysis to show that the weak AP lyase activity in hOGG1 is an artifact of the proximity of Lys249 to the C1′ and may not reflect a physiological role [53]. A double Lys↔Cys swap mutant (Lys249Cys/Cys253Lys) abrogated AP lyase activity while maintaining 8oxoG excision activity, and a Lys249Cys/Cys253Lys/Asp268Asn triple mutant also eliminated the base excision activity. A crystal structure of the triple mutant revealed that Lys253 was too far (4.7 Å) away from the incoming C1′ to form the Schiff base, whereas Asn268 was in the same position as Asp268 in the wild-type enzyme. These results provided additional evidence for hOGG1 acting as monofunctional enzyme, in which Asp268 stabilizes an oxocarbenium intermediate during base hydrolysis [76, 81] and Lys249 helps to position 8oxoG in the active site.
In addition to discrimination of 8oxoG from G, OGG1 shows a preference for the nucleobase opposite the lesion [56]. Km values of mOGG1 are 42.7+/−14.6 nM (8oxoG•C), 114+/−28 nM (8oxoG•T), 233 +/− 9.5 nM (8oxoG•G), and 2164 +/− 502 nM (8oxoG•A) [71]. Specificity of hOGG1 for 8oxoG•C base pairs likely results from the five hydrogen bonds between the enzyme (Arg204, Asn149 and Arg154) and the opposing C, and substitution of Arg154 with histidine eliminates the specificity [52] (Figure 4C). Structures of an OGG ortholog from the bacterium Clostridium acetobutylicum CaOGG provided additional insight into specificity for the opposing base [61–63]. Whereas OGG1 has high preference for 8oxoG opposite C [56, 71], CaOGG can excise 8oxoG opposite any base [61]. Structures of CaOGG in complex with DNA containing 8oxoG•C and 8oxoG•A showed that the bacterial protein maintains the fold and general DNA interactions as hOGG1, but lacks two of the five hydrogen bonds with the opposing nucleobase as a result of Met132 residing in place of the Arg154 in hOGG1 (Figure 4C) [52, 62]. In addition, the Asn149-cytosine hydrogen bond in hOGG1 is stabilized by Asn149’s interaction with the hydroxyl group of Tyr203, which is missing in CaOGG (Phe179 at this position). A CaOGG Phe179Tyr mutant was 14-fold less efficient than the wild-type enzyme at excising 8oxoG•A, but did not affect 8oxoG•C excision. Moreover, the double mutant Phe179Tyr/Met132Arg, which mimics two of the critical interactions in hOGG1, was 50-fold less efficient at excising 8oxoG•A compared to the wild-type protein [61]. Thus, the fewer number of stabilizing contacts with and around the opposite base in CaOGG creates an environment that can accommodate other nucleobases at this position [52, 62, 63].
2.1.2. OGG2
The OGG2 family of DNA glycosylases consists of enzymes from various archaeal species that were predicted to be structurally similar to the OGG1 catalytic domain [64, 82, 83]. Despite very low sequence identity with hOGG1, structures of OGG2 from Methanocaldococcus janischii (MjOGG) and Sulfolobus solfataricus (SsOGG) confirmed that these enzymes adopt the HhH fold and contain the catalytic lysine and aspartate residues present in OGG1, but lack the N-terminal β-sheet domain [65] (Figure 4D). The structure of MjOGG in complex with 8oxoG-DNA illustrated that the OGG2 family of enzymes utilize a distinct mechanism for identification of 8oxoG in the active site, in which the C-terminal carboxylate group of Lys207, as opposed to the Gly42 backbone carbonyl interaction in hOGG1, interacts with the N7 of 8oxoG [84] (Figure 4E). Deletion of the three C-terminal residues abolished 8oxoG excision activity in MgOGG, but did not significantly affect enzyme integrity since the truncation only slightly diminished lyase activity [65]. Similar to CaOGG1, the OGG2 enzymes do not significantly discriminate against the base opposite 8oxoG [64, 82, 83], and this lack of specificity in OGG2 can be explained by the fewer contacts to the orphaned base relative to hOGG1; OGG2 forms two hydrogen bonds from a single residue, Arg84 (Figure 4F), compared to the hydrogen bond pentad observed in hOGG1 [52] (Figure 4C).
2.1.3. AGOG
AGOG is a recently discovered 8oxoG-specific DNA glycosylase from the aerobic hyperthermophillic archaeon, Pyrobaculum aerophilum, that removes 8oxoG from both ssDNA and dsDNA [66, 85]. Like OGG2, AGOG has similar overall HhH fold and active site composition as that of hOGG1, but the specific residues contacting 8oxoG are not conserved in the two enzymes [86] (Figure 4G). An 8oxoG base soaked into the crystal shows that the 8oxoG-specific hydrogen bond from N7 of the nucleobase (to Gly42 main chain carbonyl in OGG1) is mediated by the Gln31 side chain in AGOG. Substitution of Gln31 to serine caused a 180-fold reduction in catalytic activity (Gln31Ser kcat = 0.011 ± 0.0004 min−1) [87]. Unlike other 8oxoG glycosylases, AGOG also forms a direct hydrogen bond to the 8oxo moiety via the indole nitrogen of Trp69 (Figure 4H), although this interaction may be dispensable since a Trp69Phe mutant did not significantly reduce activity [86, 87]. Mutational analysis confirmed the roles of residues Trp222, Gln31 and Lys147 in substrate recognition and Asp172 and Lys140 in catalysis [87]. Like CaOGG and OGG2, AGOG shows no significant preference for the nucleobase paired with 8oxoG, with single turnover rates of 8oxoG excision of 3.15 ± 0.03 (8oxoG•C), 3.12 ± 0.06 min−1 (8oxoG•A), and 6.8 ± 0.6 min−1 (8oxoG•G) [87]. The basis for this cannot be determined from the current structure, although the robust activity for 8oxoG in ssDNA, which occurs at a rate of 5.4 ± 0.4 min−1 (nearly two-fold faster than 8oxoG•C), argues that the enzyme primarily contacts only the lesion-containing strand within the duplex [87].
2.1.4. MutM/Fpg
MutM/Fpg excises a number of oxidized nucleobases in addition to 8oxoG, including FapyG, hydantoins, Tg, DHU, and 5-OHU [49, 88–91]. The crystal structure of Thermus thermophilus MutM/Fpg defined the structural architecture as distinct N- and C- terminal domains separated by a flexible hinge [47] (Figure 4I). The N-terminal domain is comprised of a two layer β-sandwich flanked by α-helices on either side and contains the catalytically important N-terminal proline and glutamate residues. The predominantly α-helical C-terminal domain contains the hallmark H2TH motif essential for DNA binding [49]. DNA-bound structures of MutM/Fpg from Lactococcus lactis [92] and Geobacillus stearothermophilus [93] revealed that the DNA was severely kinked by ~75° with the lesion flipped into the active site similar to other DNA glycosylases (Figure 4J). Subsequent structures detailed the interactions of the enzyme with various substrates and abasic analogs, including 8oxoG, FapyG, DHU, tetrahydrofuran (THF), 1,3-propanediol (Pr), hydroxy propanediol and hydantoin carbanucleoside [94–97]. These structures illustrated that even though specific amino acids contacting the base in the active site may differ, the orientation of the backbone deoxyribose remains relatively unchanged, suggesting that catalysis proceeds by properly positioning the deoxyribose ring [97]. In addition, the MutM/abasic-DNA complexes suggested that β-elimination occurs concurrently with depurination, as opposed to sequential depurination-β-elimination reactions proposed previously for hOGG1, based on the fact that the enzyme sterically clashes with the cyclic, but not the ring-opened form of the deoxyribose [97, 98].
More recently, a series of crystal structures of Geobacillus stearothermophilus MutM/Fpg from the Verdine laboratory provided detailed snapshots along the reaction pathway, illustrating how the enzyme actively interrogates the DNA duplex to differentiate between 8oxoG and guanine in the context of duplex DNA [4, 20, 93, 95]. MutM ICs crosslinked with DNA containing normal A•T or G•C base pairs showed Phe114 probing the minor groove, with the interrogated base pairs severely buckled but remaining intrahelical [93, 99]. In the LRC structure of MutM crosslinked to 8oxoG-DNA, the Phe114 residue fully penetrates the base stack and helps to induce a severe kink in the DNA that allows the target 8oxoG to become extrahelical [20]. In E. coli MutM, mutation of this phenylalanine to alanine (Phe111Ala) resulted in significantly reduced activity for 8oxoG excision and altered diffusion along DNA in single molecule studies [99]. The side chains of Met77 and Arg112 fill the space vacated by the flipped 8oxoG, with the Arg112 guanidinium moiety interacting with the Watson-Crick face of the estranged cytosine [20]. A third set of so-called encounter complexes (ECs) with 8oxoG-DNA or Gua-DNA were determined using a variant form of G. stearothermophilus MutM that has an altered or absent 8oxoG capping loop, which normally interacts with 8oxoG in the active site [100, 101]. These complexes showed that MutM can detect the presence of intrahelical 8oxoG in the duplex based on local steric effects that influence the surrounding phosphate backbone. Recent data from the E. coli enzyme showed that the interaction with the 8oxoG capping loop is specific for 8oxoG, since an EcMutM/Fpg variant lacking the tip of the capping loop can efficiently excise mFapyG, DHU, Sp, and Gh but not 8oxoG [102]. Furthermore, a recent study showed that hydrophobic isosteres of 8oxoG are good, and in some cases better, substrates for Fpg, demonstrating that hydrogen bonding to the base is not important for efficient excision by Fpg [103].
2.2. Repair of oxidized pyrimidines and hydantoins
2.2.1. EndoIII/Nth/NTH1
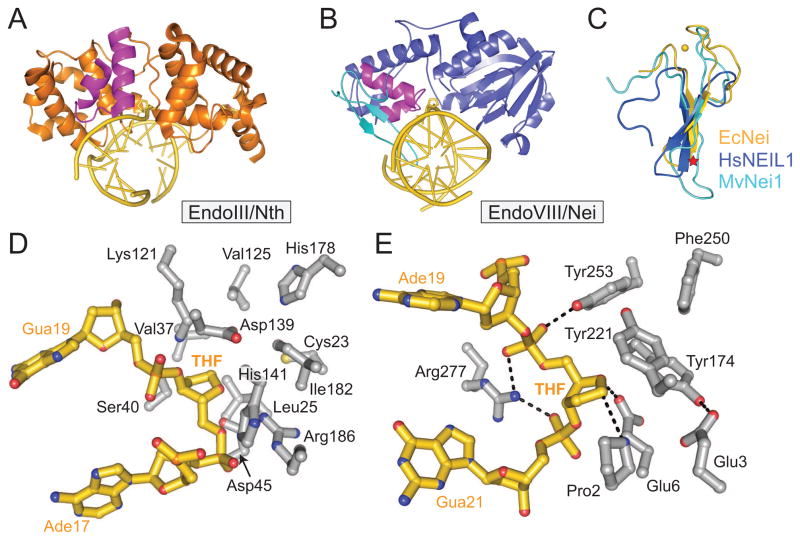
Bacterial EndoIII/Nth and human NTH1 are bifunctional DNA glycosylases that use an aspartate/lysine catalytic pair to excise a variety of oxidized pyrimidine lesions (Table 1) and nick the backbone at the resulting AP site [104]. Tg, the preferred substrate with a Km of 10 nM, is excised three- to four-fold faster than DHT, which is preferred over 5-OHC and 5-OHU [91, 105, 106]. The structure of the E. coli enzyme was the first to describe the HhH architecture for any glycosylase and the inclusion of a [4Fe-4S]-type iron-sulfur cluster in any DNA binding protein [14]. More recently, the crystal structure of Geobacillus stearothermophilus EndoIII covalently tethered to DNA, along with subsequent modeling experiments, suggested that the broad substrate specificity is a consequence of the highly polar nature of the active site, and that the enzyme may recognize its substrates on the basis of glycosidic bond stability [107]. EndoIII binds DNA in the minor groove, bends the DNA at the site of the lesion, and extrudes the modified nucleobase into an active site pocket (Figure 5A). A unique feature of this particular HhH enzyme is the extensive contacts made to the DNA backbone of the strand opposite the lesion. A glutamine side chain plugs the DNA gap and an intercalated leucine stabilizes the estranged base [107] (Figure 5D). There are currently no known structures of human NTH1, although sequence and substrate similarities [108] suggest the current bacterial and archaeal EndoIII/Nth structures are accurate representations of the human ortholog. An outstanding question remains regarding the significance of the iron-sulfur cluster in these and other DNA repair enzymes, although studies point to a possible role in DNA damage detection based on the iron-sulfur redox potential [109–111].
Figure 5.
EndoIII/Nth and EndoVIII/Nei. (A) Bacillus stearothermophilus Endonuclease III (BsEndoIII) (PDB ID 1P59) bound to THF-DNA (gold). The THF moiety and iron-sulfur cluster are shown as sticks in the center and right side of the figure, respectively. The HhH DNA binding motif is magenta. (B) Mimivirus NEIL1 ortholog MvNei1 (3A46) bound to THF-DNA (gold). The H2TH motif is colored magenta and the zincless finger is cyan. (C) Overlay of the zinc finger from Escherichia coli Endonuclease VIII (EcNei) (1K3W) (gold) with zincless finger motifs from human NEIL1 (1TDH, blue) and MvNei1 (cyan). The zinc ion in EcNei is depicted as a gold sphere. A red star denotes the general location of arginine residues that contact the DNA. (D) Active site of BsEndoIII (grey) bound to THF-DNA (gold). (E) Active site of MvNei1 bound to THF-DNA.
2.2.2. EndoVIII/Nei
Like Nth, bacterial Nei (EndoVIII) is a bifunctional DNA glycosylase specific for oxidized pyrimidines [112]. Whereas Nth is a member of the HhH superfamily, Nei is structurally similar to MutM/Fpg and contains tandem H2TH/antiparallel β-hairpin zinc finger motifs that bind and stabilize the kinked DNA substrate, and a catalytic N-terminal proline [48]. The disparate substrate specificities of Nei and MutM/Fpg are reflected in the fact that residues involved in substrate recognition differ between these enzymes [48]. The structure of a covalently trapped DNA complex of E. coli Nei revealed that the protein undergoes a significant interdomain conformational change upon DNA binding [113]. This conformational switch between free (open) and DNA bound (closed) states, similar to that observed in other DNA binding proteins (e.g., lac repressor bound to target site [114]), has not been observed in other DNA glycosylases, although it has been proposed that the MutM/Fpg proteins may have some degree of conformational flexibility in solution [47, 48, 113]. For a more in depth review of EndoVIII/Nei, see reference [112].
2.2.3. NEIL1
Three eukaryotic Nei-like orthologs, NEIL1-3, have been discovered in humans [115–119] and have been characterized to some extent both functionally and structurally [recently reviewed in 120]. Like Nei, the NEIL orthologs are bifunctional H2TH glycosylase/AP lyases that excise a broad spectrum of oxidized pyrimidines and ring-opened purines (Table 1). Specifically, NEIL1 has a preference for Gh, Sp, Tg, 5-OHU, DHU, and Fapy, but also has been shown to have activity toward DHT, 5fU, 5hmU, 5-OHC, urea, and even abasic sites within a variety of structural contexts, including ssDNA, dsDNA, bulges, and bubbles [106, 116–118, 121–128]. NEIL2 primarily cleaves 5-OHU, but has not been shown to excise Tg or 8oxoG [115]. NEIL3 has a preference for various oxidized purines and pyrimidines in ssDNA and bubble structures [129] and has been reported to remove Gh and Sp hydantoins from both ss- and dsDNA [129]. In contrast, NEIL2 removes Gh and Ia from ss- and dsDNA, but Sp from ssDNA only [130]. NEIL1 has weak activity for 8oxoG in dsDNA, but unlike NTH1 and OGG1, NEIL1 can excise 8oxoG located near the 3′ end of single-strand breaks, suggesting that NEIL1 is not simply a back-up glycosylase for NTH1 and OGG1 but instead reinforces its unique substrate specificity [131, 132]. The ability of NEIL enzymes to remove ssDNA lesions and their interactions with several replication proteins implicates them in DNA repair during S-phase [116, 121, 127]. Interestingly, NEIL1 was shown to remove psoralen-induced monoadducts and interstrand crosslinks (ICLs) in dsDNA, implicating it in nucleotide excision repair (NER) [133].
The NEIL enzymes are proposed to operate by a mechanism similar to Nei despite a few differences among them. Most notably, NEIL1 lacks the zinc finger motif present in Nei, NEIL2 and NEIL3 (Figure 5C). This so-called zincless finger retains many aspects of the antiparallel β-hairpin zinc finger motif, but the loops that coordinate a zinc ion are missing [134]. The crystal structure of NEIL1 also revealed the position of a conserved arginine in the zincless finger that was confirmed by mutagenesis to be critical for glycosylase activity [134]. Structures of a viral NEIL1 ortholog (MvNei1) bound to THF-DNA illustrated how this longer β-hairpin loop of the zincless finger interacts with the strand opposite the lesion [135] (Figure 5B). Like in other glycosylases, the DNA bound to MvNei1 is kinked at the site of the lesion and the THF moiety is flipped out of the duplex [135]. Both DNA-bound and free MvNei1 structures superimpose on the closed conformation of Nei, demonstrating that the large-scale domain movements notable in EcNei are not observed in MvNei1, although small-scale movements in the catalytic proline and the zincless finger to accommodate the DNA are evident upon DNA binding [48, 113, 135]. A tyrosine residue (Tyr221) in the proposed lesion recognition loop stacks against the abasic site (Figure 5E) and most likely is in an alternate conformation in the presence of substrate, although the side chain could not be discerned in structures of MvNei1 bound to Tg- and 5-OHU-DNA [135, 136]. Recognition of the pyrimidine ring takes place through a hydrogen bond interaction between the main chain amide of Tyr221 and the O4 of Tg and 5-OHU [136]. Two other residues (Glu6 and Tyr253) are within hydrogen-bonding distance, and mutation of these residues decreased the rate of Tg excision 7-fold (Tyr253) and 4-fold (Glu6), but had no significant effect on 5-OHU activity [136].
The plasticity of the active site to accommodate different oxidative lesions while discriminating against 8oxoG has been illustrated by homology modeling and molecular dynamics simulations [137]. Interestingly, A to I editing by adenosine deamination on dsRNA leads to NEIL1 variants that contain either an Arg and Lys at position 242 in the lesion recognition loop and have different substrate specificities, implying that the substrate specificity of NEIL1 changes in response to cellular conditions and may even be modulated by protein binding partners [138–141]. For example, an interaction between the C-terminal domain of NEIL1 and flap endonuclease 1 (FEN-1) (Kd=0.2 μM) stimulates 5-OHU excision activity by 5-fold [138]. NEIL1 also interacts with BER enzymes pol β and DNA ligase IIIα through the C-terminal region of NEIL1 [142].
2.3. Repair of A•8oxoG mismatches by MutY/MUTYH
Failure of MutM/OGG1 to excise 8oxoG prior to replication results in 8oxoG•A mispairs, the adenine of which is the substrate for MutY/MUTYH glycosylase [143, 144]. BER of the resulting AP site restores the 8oxoG•C pair, providing another chance for MutM/OGG1 to eliminate the 8oxoG from the DNA [reviewed in 145]. Structures of the catalytic domain of E. coli (Ec) MutY bound to adenine base revealed a HhH-FeS architecture similar to EndoIII and provided details of the active site and a proposed catalytic mechanism for adenine excision [146, 147]. Transition state analysis from kinetic isotope effect measurements confirmed a stepwise, dissociative (SN1) reaction mechanism whereby Glu43 acts as a general acid to protonate adenine N7, creating a positive charge on the nucleobase that facilitates cleavage of the N-glycosidic bond. The resulting oxocarbenium ion in the DNA is likely stabilized by nearby Asp144 and converted to the product AP site upon nucleophilic attack by water [148]. A high-resolution crystal structure of EcMutY bound to adenine provided evidence that MutY-catalyzed β-elimination, involving Lys142, Lys20 and possibly Glu161, is an activity secondary to and separable from the depurination reaction, similar to that observed in hOGG1 (see section 2.1.1) [147].
A similar disulfide crosslinking strategy employed in the OGG1 and MutM structures was used to obtain structures of the full-length B. stearothermophilus homolog (BsMutY) anchored to 8oxoG•A-DNA [149, 150] (Figure 6A). In this structure, the adenine is flipped into the glycosylase active site but remains uncleaved as a result of mutation of the catalytic aspartate (Asp144Asn) [149]. Surprisingly, no direct hydrogen bonds were observed between the catalytic domain and the extrahelical adenine substrate. A subsequent structure of a catalytically proficient (Asp144) BsMutY crosslinked to DNA containing a non-hydrolyzable 2′-fluorinated deoxyadenosine showed adenine deeper into the active site and directly hydrogen bonded to Gln43, Tyr126, Arg31, Glu188, and Trp30 [150] (Figure 6B). Mutation of the Glu188 residue in EcMutY (Gln182) decreased binding and activity for 8oxoG•A and G•A mismatches but increased binding affinity toward 8oxoG•T and G•T mismatches, which are not normal substrates for MutY [151]. Cellular repair assays on the E. coli enzyme confirmed the importance of Asp138 (BsMutY144) and Glu37 (BsMutY Glu43) for the excision of adenine opposite 8oxoG [152].
Figure 6.

Crystal structure of Bacillus stearothermophilus MutY. (A) Overall structure of BsMutY (PDB ID 1RRQ) colored by domain (green, iron-sulfur cluster domain; cyan, catalytic domain; blue, C-terminal (8oxoG recognition) domain. The DNA is colored gold with adenine substrate in purple and opposite 8oxoG in magenta. (B) Active site details of the MutY fluorinated lesion recognition complex (FLRC) bound to adenine (3G0Q). Protein (silver) and nucleic acid (gold) atoms are shown as sticks, water molecules are shown as red spheres. Hydrogen bonds are shown as dashed lines.
The C-terminal domain contributes specific contacts to the stacked 8oxoG lesion that are functionally important for lesion recognition and enzyme activity. Tyr88 intercalates the duplex and stacks against the 8oxoG nucleobase, and Gly260 contacts the phosphate 5′ to 8oxoG [149]. Inherited mutations at these positions in MUTYH (Tyr165Cys and Gly382Asp) have been implicated in the development of colorectal cancer [153]. Substitution of the analogous residues in EcMutY (Tyr82Cys and Gly253Asp) reduce the DNA binding and base excision activities relative to the wild-type enzyme and the glycine has been implicated in discrimination of 8oxoG from G [154, 155]. Furthermore, enzymatic studies with modified substrates in vivo demonstrated that MutY cannot effectively process adenine paired with guanine or modified forms of 8oxoG, whereas changes made to the target adenine are tolerated [156], implying that recognition of the 8oxoG by the C-terminal domain is necessary for locating the misincorporated adenine.
A crystal structure of a human MUTYH consisting of the catalytic domain and the interdomain connector (IDC) that tethers the catalytic and C-terminal domains was recently determined [157]. The human IDC sequence, which is not conserved in prokaryotic MutY, has been reported to recruit the Rad9, Rad1, Hus1 (9-1-1) complex involved in genome maintenance in eukaryotes [158–160]. Mutations in the IDC disrupted the MUTYH-9-1-1 interaction and decreased DNA repair of oxidative lesions in vivo, suggesting that structural studies of the human enzyme will reveal insights into its broader role in maintaining genome integrity [157, 160].
3. Alkylation damage
A diverse array of alkylated DNA adducts are produced by environmental mutagens, cellular metabolites, and chemotherapeutic agents (Figure 1B) [1, 161–163]. The major and minor groove-exposed N7 and N3 positions of purines make them susceptible to reaction with electrophiles, with guanine N7 being the most nucleophilic [164]. Whereas N7-methylguanine (7mG) is relatively innocuous compared to larger N7-alkyl substituents, the positive charge generated from N7-substitution destabilizes the base and leads to spontaneous depurination and ring decomposition to produce, for example, 5-N-methyl-2,6-diamino-4-hydroxyformamidopyrimidine (mFapyG). The glycosidic linkage of N3-methyladenine (3mA) is especially unstable, with a half-life for 3mA depurination as short as 24 h at 37°C [165]. Reactive aldehydes and epoxides generated from lipid peroxidation produce a number of ethenoadducts with A, G, and C, including 1,N6-ethenoadenine (εA), 1,N2- and N2,3-ethenoguanine, and 3,N4-ethenocytosine (εC) [1, 166, 167]. In general, these lesions cause genomic instability through mutations and strand breaks [1]. 3mA is cytotoxic, likely as a result of inhibition of DNA synthesis caused by disruption of the contacts between DNA polymerase and the adenine N3 position in the minor groove [168–171].
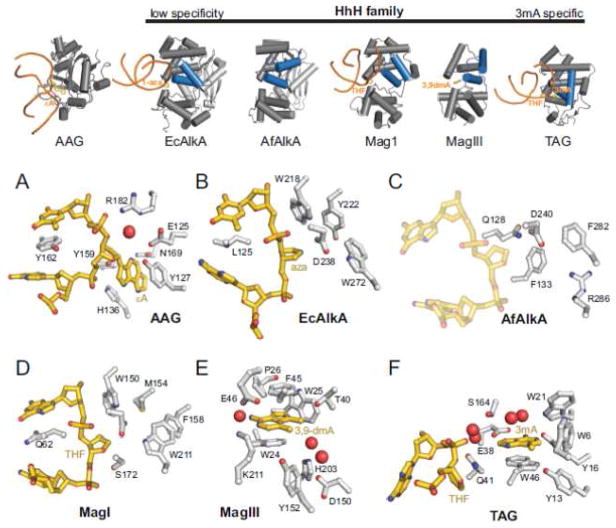
DNA glycosylases specific for alkylation damage have been characterized from eukaryotes, archaea, and bacteria. These include human AAG/MPG/ANPG [172, 173], Saccharomyces cerevisiae MAG and Schizosaccharomyces pombe Mag1 [174–176], E. coli 3mA DNA glycosylase I (TAG) and II (AlkA) [177, 178], Archaeoglobus fulgidus AlkA (AfAlkA) [179, 180], Deinococcus radiodurans AlkA (DrAlkA) [181], Thermotoga maritima MpgII [182], Helicobacter pylori MagIII [183], and Bacillus cereus AlkC and AlkD [184]. AAG, AlkA, and MAG/Mag1 excise a broad range of alkylated and deaminated bases [179, 185–191]. Interestingly, AfAlkA has robust activity toward N1-methyladenine (1mA) and N3-methylcytosine (3mC), which are normally repaired by oxidative demethylation [179, 180, 188]. In contrast, TAG is highly specific for N3-methylpurines 3mA and 3mG [192], and MagIII, MpgII and AlkC/D are selective for positively charged lesions (e.g., 3mA and 7mG) [182–184]. The alkylpurine DNA glycosylases can be grouped into three structural classes: 1) AAG, defined by the human enzyme, 2) ALK, including AlkC and AlkD, and 3) HhH, comprising all others (Figure 3) [193]. Despite their different architectures, AAG and the HhH enzymes have similar active sites that contain aromatic, electron-rich side chains that stack against the extrahelical alkylpurine substrate (Figure 7) [193–196], whereas the ALK family is distinct structurally and mechanistically from the canonical base-flipping enzymes [12].
Figure 7.
Alkylpurine DNA glycosylases. Overall architectures are shown on the top row, with HhH enzymes arranged in order of increasing specificity for 3mA. (A–F) Active sites. Protein and nucleic acid atoms are shaded grey and gold, respectively, and waters are shown as red spheres. (A) Human AAG/εA-DNA substrate complex (1EWN). (B) E. coli AlkA bound to 1-azaribose-DNA (1DIZ). (C) A. fulgidus AlkA (2JHJ) with THF-DNA modeled from the S. pombe MagI/DNA complex (3S6I). (D) S. pombe Mag1/THF-DNA (3S6I). (E) H. pylori MagIII/3,9-dimethyladenine (1PU7). (F) E. coli TAG/THF-DNA/3mA product complex (2OFI).
3.1 AAG
Human AAG, also known as MPG and ANPG, excises a variety of alkylated purines, including 3mA, 7mG, and εA, as well as hypoxanthine (Hx), the oxidative deamination product of adenine (Figure 1B) [197, 198]. The exceptional rate enhancement of Hx excision relative to alkylated substrates suggests that Hx is the predominant biological substrate [199]. AAG has also been shown to excise N1-methylguanine and 1,N2-εG [200, 201]. Crystal structures of a catalytic fragment of AAG bound to oligonucleotides containing either a pyrrolidine transition-state analog or an εA nucleobase showed that AAG is a single domain protein with a mixed α/β structure and a positively charged DNA binding surface [195, 202]. The flipped εA base is stacked between two tyrosine residues (Tyr127 and Tyr159) and His136 inside the active site cavity, while Tyr162 on the tip of a β-hairpin plugs the gap in the DNA left by the flipped nucleotide (Figure 7A). These structures provided a framework for a number of recent kinetic and thermodynamic studies aimed at dissecting AAG’s mechanism, substrate specificity, and collaboration with other BER enzymes. These studies are described in detail and referenced in the following sections.
3.1.1. Mechanism of base flipping and substrate discrimination
A series of careful biochemical examinations of substrate binding, flipping, and excision by AAG has recently been reported. As is typically true for other glycosylases, substrates that decrease the stability of the DNA increase the efficiency of excision by AAG, with bulged nucleotides excised more efficiently than mismatched base pairs [199, 203]. Interestingly, the strength of AAG binding to bulges correlates with increased spontaneous frameshift mutations upon overexpression of the enzyme, which may be a result of AAG shielding bulged bases from mismatch repair [204]. Discrimination of nucleobases on the basis of their stability within the DNA duplex can be rationalized by the barrier to base flipping. Kinetic analysis using intrinsic εA fluorescence revealed that εA flipping by AAG is highly favorable, which helps to explain discrimination of this lesion from undamaged bases [205]. These experiments also generated a two-step binding regime in which distinct DNA-bound and base flipped complexes form on the millisecond to second time scale, whereas N-glycosidic bond cleavage takes place on the minute time scale [205, 206]. Thus, destabilized base pairing allows AAG to selectively excise DNA lesions. More stringent selection takes place inside the active site, in which side chains create steric clashes with unmodified A and G bases [199, 202, 207].
Excision of neutral substrates by AAG has been shown by pH-activity profiles to employ both a general acid and general base [208]. The general acid acts to protonate the nucleobase, facilitating its dissociation, and the general base would deprotonate a catalytic water molecule to attack C1′ [208]. Consistent with such a mechanism, excision of positively charged lesions (e.g., 7mG) does not require the general acid [208]. Although the identity of the general acid has not been determined, the necessity to protonate the base explains the specificity of AAG for purines versus pyrimidines [208]. Quantum mechanical modeling studies indicate that base excision by AAG is facilitated by π-π interactions between the enzyme and its substrate DNA, consistent with the structures, and suggest that the nucleobase is not fully protonated but rather hydrogen bond donation by a protein-bound water molecule lowers the catalytic barrier [209].
3.1.2. Structural basis of AAG inhibition by εC
In addition to εA, AAG has a modest activity toward 1,N2-εG [200]. Although AAG binds εC with a 2-fold greater affinity than εA [210], AAG is incapable of excising εC, which is normally removed by the uracil/thymine DNA glycosylase family of enzymes (see section 4) [211–213]. A recent structure of AAG in complex with εC-DNA showed εC to reside in the active site in a virtually identical position as εA [210] (Figure 8). The hydrogen bond between His136 and εA (N6) is preserved to εC (N4), and as a consequence the εC nucleotide is pulled slightly farther into the binding pocket. The enhanced binding to εC may be explained by one additional hydrogen bond between the protein (Asn169) and O2 of εC, which is not present in εA. Regarding inhibition, protonation of substrate purines likely occurs at the N7 nitrogen [208], and crystal structures suggest that a protonated N7 would be stabilized by a hydrogen bond to the backbone oxygen of Ala134 (Figure 8). The AAG/εC-DNA structure proposes that inhibition by εC is due to the inability of AAG to protonate εC, which lacks a nitrogen at the position corresponding to N7 of εA. In addition, this structure also showed an octahedral coordinate Mn2+ ion bound to the guanine opposite the εC that perturbed the guanine sugar pucker. This was the first observation of bound divalent ion to AAG and suggested that inhibition of the enzyme by divalent ions might be a consequence of impaired base flipping or duplex opening to expose the substrate base [210]. AAG has also been trapped onto εC-DNA in a non-specific orientation, providing a structural basis for the enzyme’s ability to bind single-base bulges [204, 214].
Figure 8.
Binding of εA and εC to AAG. The εA complex (PDB ID 1EWN) is colored blue (protein) and salmon (DNA), and the εC complex (PDB ID 3QI5) is silver and gold. Hydrogen bonds are depicted as dashed lines. A water molecule (red sphere) is in position to protonate N7 of εA, and protonated N7 would donate a hydrogen bond to Ala134 (green dashed line). εC does not have an ionizable group at this position.
3.1.3. Product release and diffusion along DNA
Single- and multiple-turnover kinetic experiments have shown that the rate-limiting step of hypoxanthine hydrolysis by AAG is the release of the abasic DNA product [215]. In fact, the tight binding of AAG to product DNA enables AAG to catalyze the reverse reaction to re-form the N-glycosidic bond [216]. Product release is promoted by APE1, the next enzyme in the BER pathway [217]. Displacement of the glycosylase by APE1 has also been observed for TDG and OGG1 [218–220]. The nonspecific binding of both AAG and APE1 to DNA suggests that these enzymes may bind DNA simultaneously and facilitate a handoff of the abasic site from AAG to APE1. Baldwin & O’Brien propose that APE1 displaces AAG from the AP site without a direct protein-protein interaction, and that AAG remains bound to the DNA upon AP dissociation [215]. The processivity of AAG along DNA is dependent on ionic strength, indicating a reliance on electrostatic interactions with the DNA backbone. Furthermore, the amino terminal 80 amino acids, which are not necessary for catalysis by AAG, contribute to the enzyme’s ability to diffuse along DNA [221].
3.2. HhH superfamily
The majority of yeast, archaeal, and bacterial alkylpurine DNA glycosylases adopt the HhH protein fold, with the exception of AlkC/AlkD and bacterial orthologs of human AAG [222, 223]. The HhH glycosylases contain two α-helical domains with the active site cleft located at their interface. The domain containing the HhH motif and DNA intercalating residues is formed from an internal region of the primary structure and has a relatively conserved tertiary structure. The HhH anchors the protein to the DNA through a series of hydrogen bonds between main-chain atoms of the hairpin and the phosphoribose backbone downstream of the lesion. At the damage site, bulky side chains from neighboring loops fill the void left by the extrahelical nucleobase target and wedge into the base stack opposite the flipped out nucleotide. Both plug and wedge residues are important for stabilizing the bent conformation of the DNA and have been implicated in probing the DNA helix during the search process [224]. The second domain, formed from the N- and C-termini, is more structurally divergent and often contains additional structural elements, such as a zinc ion (TAG), iron-sulfur cluster (MpgII), or carbamylated lysine (MagIII) [193].
Comparative analysis of the HhH alkylpurine glycosylases has been instrumental in deciphering the physical and chemical determinants of substrate recognition [225]. On one hand, we have learned that the HhH scaffold accommodates a diverse array of nucleobase binding pockets that discriminate between lesions on the basis of shape complementarity. For example, AlkA’s nucleobase binding surface is a shallow cleft that can accommodate a variety of alkylpurines, whereas the active sites of TAG and MagIII are more constrained and perfectly shaped for 3mA. On the other hand, this steric selection is not the only determinant of specificity since some active sites can accommodate nucleobases for which they do not excise (e.g., Mag1) [191]. In addition, the catalytic requirements for excision of cationic lesions 3mA and 7mG differ from the uncharged alkylpurines (e.g., εA) by virtue of their weaker N-glycosidic bonds [9]. Hence, the inherent instability of these lesions render their excision highly dissociative, and recent reports suggest that cationic lesions may be removed and even detected within DNA differently than neutral lesions [191, 193, 226].
3.2.1. E. coli AlkA
Crystal structures of unliganded AlkA identified the enzyme as a member of the HhH superfamily and revealed a shallow nucleobase binding surface that can accommodate a variety of alkylpurines, a feature that helped to explain its broad specificity [194, 227] (Figure 7B). In addition to the two-domain HhH architecture, AlkA contains an amino-terminal β-sheet domain of unknown function that is also present in OGG1 (Figures 4 and 7). A structure of AlkA bound to DNA containing 1-azaribose, which mimics the oxocarbenium reaction intermediate, has contributed greatly to our understanding of these enzymes [196, 228]. The HhH anchors the protein to the DNA and does not directly participate in lesion recognition. The DNA is kinked by ~60° around the 1-azaribose, which is rotated 180° around the phosphoribose backbone and stabilized by the Leu125 plug in the gap left behind (Figure 7B). Rotation of the 1-azaribose into the active site places the N1′ nitrogen directly adjacent to the carboxylate group of the catalytic Asp238, which is in a prime location to stabilize the oxocarbenium intermediate [196]. In addition to this lesion-specific binding mode, AlkA has the ability to bind to DNA ends [229], which may explain why a structure of AlkA bound to a substrate DNA has not been determined. Nonetheless, this feature was exploited to develop a host-guest crystallization strategy to determine structures of various lesions in DNA [230].
High resolution structures of AlkA cross-linked to undamaged DNA bases provided insight into how the enzyme detects damage within the context of unmodified DNA [224]. Not surprisingly, the most notable differences between these undamaged DNA complexes (UDCs) and the 1-azaribose lesion recognition complex (LRC) are centered around the lesion. The UDCs do not exhibit the kink present in the LRC DNA. The domain containing most of the catalytically important residues, including Asp238, is shifted 2.4 Å toward the lesion strand in the LRC compared to the UDCs. This movement, combined with a modest 1-Å shift of the Leu125 plug residue toward the lesion strand, clamps the lesion between the two domains and creates additional protein contacts that stabilize the LRC. In contrast, the HhH motif makes the same DNA contacts in LRC and UDC structures, providing additional evidence that the HhH motif is a non-specific DNA binding element and is not involved in distorting the DNA for catalysis. Leu125 in the UDCs does not interact with the DNA, although it is still present in the minor groove. The phosphate backbone in the LRC is significantly (~9 Å) closer to the protein, which allows the Leu125 side-chain to intercalate into the DNA base stack in that structure. A 3mA base modeled in place of a centrally located cytosine indicated that Leu125 likely makes van der Waals contacts to the N3-methyl group of the 3mA [224]. These observations suggest that AlkA employs a passive scanning mechanism along the minor groove and uses the Leu125 side chain to detect abnormal bases and flip them into the active site.
3.2.2. Archaeal AlkA
An AlkA ortholog from the archaeon Archaeoglobus fulgidus (AfAlkA), has been shown to excise 1mA and 3mC in addition to 3mA, 7mG, εA and Hx from DNA [179, 180, 188]. The crystal structure of this ortholog shows that the nucleobase binding pockets of AfAlkA and E. coli AlkA are strikingly different despite the similarity in their overall fold [180] (Figure 7B,C). Mutation of the catalytic Asp240 (Asp238 in EcAlkA) completely eliminates base excision activity in AfAlkA. The substrate nucleobase is predicted to stack between Phe133 and Phe282, similar to stabilization of 3mA by MagIII (section 3.2.4, Figure 7E). In support of this, substitution of Phe133 or Phe282 with alanine diminishes εA and 1mA base excision, and the double mutant abrogates activity. Arg286 is predicted to orient εA in the active site through hydrogen bonding, but would potentially repel the protonated amine groups of 1mA and 3mC [180]. Thus, the AfAlkA structure is a nice example of how the versatility of the HhH scaffold allows for inclusion of various active sites that dramatically alters the enzyme-substrate specificity.
3.2.3. Yeast MAG/Mag1
S. cerevesiae MAG and S. pombe Mag1 are 42% and 47% similar in sequence to E. coli AlkA, respectively, but have a more restricted substrate specificity (Table 1) [225]. MAG excises 3mA, 7mG, εA, Hx, and guanine, but not oxidized substrates (e.g., O2-methylthymine) from DNA, while Mag1 is more restricted to 3mA, 3mG, and 7mG and has only a modest activity toward εA [185, 189–191, 231–233]. These differences suggest that these proteins have different roles in protecting cells against alkylation damage [234, 235]. For example, MAG deletion strains are more sensitive to alkylation agents than are S. pombe mag1, and MAG expression is induced to higher levels than Mag1 upon exposure to alkylation agents [235, 236].
Our laboratory recently determined crystal structures of Mag1 bound to DNA containing a THF abasic analog [191] and of free MAG (unpublished results) (Figure 9). Neither MAG nor Mag1 contain the mixed α/β domain present at the N-terminus of the AlkA orthologs (Figure 9A). Nevertheless, Mag1 engages the THF-DNA similarly to AlkA, with the DNA bent by ~60° and the THF moiety rotated around the phosphate backbone toward the nucleobase binding pocket. Inside the active site, there are only two notable differences between MAG and Mag1. Mag1 residues Phe158 and Ser159 at the back of the binding cleft are occupied by Ser197 and Gly198 in MAG (Figures 7D and 9B). Swapping these residues (Mag1 FS→SG and MAG SG→FS double mutants) did not affect their relative εA activities, providing evidence that the bulky Phe residue in the binding pocket is not responsible for the lower εA excision activity of Mag1 [191]. Interestingly, substitution of the catalytic aspartate residues had dramatically different effects. MAG Asp209Asn completely abrogated εA and 7mG excision activities similar to that observed for AlkA Asp238 [194], while Mag1 Asp170Asn had a more modest effect, implying that this residue in MAG plays a more significant role in catalysis, possibly explaining the broader substrate preference of this enzyme [191].
Figure 9.
Yeast 3-methyladenine DNA glycosylases MAG and Mag1. In all panels, the unbound Saccharomyces cerevisiae MAG (grey) free enzyme is superimposed onto the Schizosaccharomyces pombe Mag1/THF-DNA complex (blue/gold, PDB ID 3S6I). (A) Overall structures. (B) Active sites. Mag1 residues Phe158 and Ser159 are the only two active site residues that differ between the two enzymes. (C) Close-up of Mag1-DNA contacts at the lesion. Swapping Mag1 His64 and MAG Ser97 between the two enzymes effectively swaps their respective abilities to remove εA (see text for details).
Outside of the active site, there is a notable difference between MAG and Mag1 at the point of contact with the DNA minor groove flanking the damage site (Figure 9C). In addition to the plug and wedge residues, His64 in Mag1 is in position to hydrogen bond with either the N3 of the adenine immediately 5′ to the lesion or to the exocyclic N2 of the guanine on the opposite strand [191]. MAG and AlkA orthologs, including those from Bacillus halodurans and Deinococcus radiodurans, which have broad substrate preferences and for which crystal structures are available, contain a serine residue at this position (Figure 9C) [181, 191, 225]. Surprisingly, swapping histidine and serine between Mag1 and MAG led to dramatic increase in εA excision rate in Mag1 and a decrease in εA excision in MAG, whereas the 7mG excision rates in both enzymes remained the same [191]. Thus, contacts to the minor groove may be important for damage detection and/or stabilizing a specific enzyme-substrate complex for catalysis. These results also suggest that cationic and uncharged lesions may be detected or stabilized differently, although more work is required to test this hypothesis.
3.2.4. H. pylori MagIII and T. maritima MpgII
MagIII and MpgII are related alkylpurine glycosylases identified by their sequence similarity to EndoIII [182, 183]. MagIII is highly specific for 3mA but can excise mispaired 7mG, whereas MpgII can excise both 3mA and 7mG [182, 183]. The crystal structure of MagIII showed a unique feature in the N/C-terminal domain, which contains a carbamylated lysine (Lys205) that neutralizes an otherwise highly positively charged region of the protein [237]. MagIII’s preference for 3mA can be explained by the snug fit of 3mA inside the active site, which partially excludes N7-substituted purines. Structures of MagIII bound to positively charged 3,9-dimethyladenine (3,9-dmA) and uncharged εA bases showed the nucleobases stacked between Phe45 and Trp24 and bounded on three sides by Trp25, Pro26, and Lys211 (Figure 7E). Other than these van der Waals and π-stacking interactions, there are no specific hydrogen bonding or polar contacts to the adenine ring like those observed in TAG (see section 3.2.5). Similar to Mag1, mutation of the putative catalytic aspartate Asp150 in MagIII does not completely abrogate base excision activity, again suggesting that the catalytic power of this residue determines the ability of the HhH enzymes to remove more stable, neutral nucleobases from DNA, and that little catalytic assistance is required for hydrolysis of the labile 3mA glycosidic bond [9, 237].
Unlike MagIII, MpgII contains an iron-sulfur cluster and shows robust activity toward 7mG, which is intriguing given the sequence similarity between MagIII and MpgII [182, 225]. Although there is no structure for MpgII, sequence comparison predicts that only two residues differ within the active site: MpgII Trp52 and Lys53 are occupied by Phe45 and Glu46 in MagIII, respectively. The MagIII active site is constrained by a salt bridge between Glu46-Lys211. Substitution of Glu46 with the corresponding lysine residue (Lys53) in MpgII should relieve this constraint from electrostatic repulsion. Indeed, a MagIII Glu46Lys mutant resulted in an 8-fold increase in 7mG•T activity, suggesting that steric exclusion of 7mG partially accounts for MagIII’s low activity toward methylguanine bases [237].
3.2.5. E. coli TAG
TAG substrate preference is strictly limited to N3-substituted purines 3mA and 3mG [192]. NMR studies of E. coli TAG showed it to be a structurally divergent member of the HhH family, containing a zinc ion in the N/C-terminal domain and lacking the catalytic aspartate residue present in other 3mA DNA glycosylases [238–240]. Similar to MagIII, TAG’s specificity can be partially attributed to the fact that the 3mA binding pocket would sterically exclude all other nucleobases (Figure 7F). Binding studies and NMR investigation of 3mA in the active site led to the suggestion that TAG enhances the rate of 3mA depurination by binding tightly to the nucleobase, thereby destabilizing the ground state of the enzyme-substrate complex [240]. This idea was illustrated by crystal structures of a TAG/abasic-DNA/3mA product complex using the Salmonella typhi ortholog, which is 82% identical and 92% conserved overall with E. coli TAG [226]. In that structure, the bound DNA is more B-form when compared to the highly distorted 1-azaribose DNA bound to AlkA, and there was a large (7 Å) separation between the THF, which is not fully engaged inside the active site, and 3mA, which is buried deep inside the cleft. These observations indicated that the DNA undergoes significant relaxation upon breakage of the N-glycosidic bond, suggesting that steric strain may contribute to bond cleavage [226]. A recent structure of Staphylococcus aureus TAG recapitulates the structural features observed in the E. coli and S. typhi structures, and the authors suggested that tautomerization of 3mA contributes to its recognition by TAG [241].
3.3. AlkC and AlkD
Recently, AlkC and AlkD were identified in Bacillus cereus as two related alkylpurine glycosylases to be highly specific for 3mA and 7mG [184], and were predicted to represent a new structural superfamily of DNA glycosylases on the basis of their sequence similarity to an unpublished entry in the Protein DataBank (2B6C) [242]. The crystal structure of Bacillus cereus AlkD confirmed this prediction [222]. AlkD is composed exclusively of HEAT repeats (Figure 3)—tandem pairs of short α-helices that generate extended, non-enzymatic scaffolds that typically mediate protein but not nucleic acid interactions. To our knowledge, AlkD is the first HEAT repeat protein identified to interact with nucleic acids or to contain enzymatic activity [12]. AlkD’s positively-charged, concave surface is perfectly suited to bind a DNA duplex, and is lined with highly conserved residues that are important for 7mG excision and DNA binding activities and for protection against bacterial sensitivity to alkylating agents [193, 222, 242].
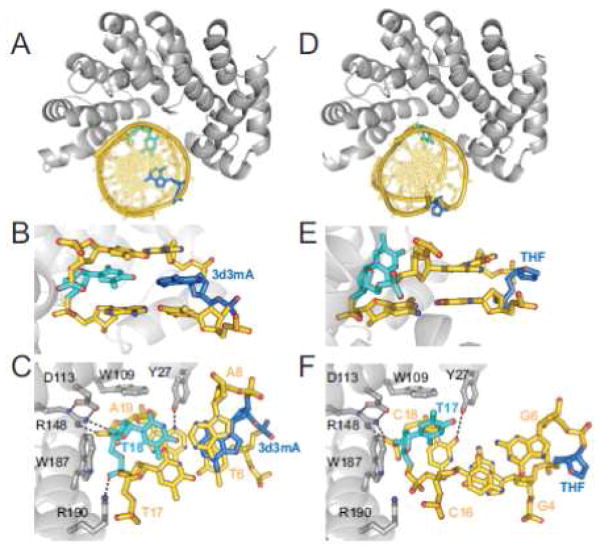
High resolution crystal structures of AlkD in complex with DNAs resembling the substrate (3-deaza-3-methyladenine, 3d3mA) and product (THF) of 3mA excision confirmed that the DNA duplex is positioned via electrostatic interactions along AlkD’s concave surface and revealed a novel lesion capture mechanism distinct from other glycosylases [193]. The 3d3mA and THF moieties are positioned on the side of the DNA facing away from the protein with no contact to the protein whatsoever (Figure 10A,D) [193]. In the substrate structure, the 3d3mA•T base pair is sheared as a result of movement of the thymine into the minor groove toward the protein (Figure 10 B,C). In the product structures, both the abasic site and its opposing nucleobase are rotated out of the helix to create a single-base bulge with base stacking maintained by the flanking base pairs (Figure 10E,F). The THF is flipped 180° around the phosphoribose backbone into a solvent exposed orientation, while the opposing base is tipped up and sandwiched between the minor groove and the protein.
Figure 10.
Crystal structures of AlkD in complex with 3d3mA-DNA (PDB ID 3JX7) (A–C) and THF-DNA (3JXZ) (D–F). The protein is colored silver, DNA is gold, 3d3mA and THF lesions are blue and opposite thymines are cyan. (A,D) Overall structures showing DNA bound to the concave surface of the protein. (B,E) Side views of the base pairs flanking the lesions. (C,F) View rotated 90° with respect to panels A/D and B/E. Hydrogen bonds are shown as dashed lines.
Several distinguishing structural and biochemical features of AlkD indicate that it utilizes a unique mechanism to liberate positively charged bases from DNA [193]. Unlike the base-flipping glycosylases, AlkD lacks the plug residue universally used by DNA glycosylases to prevent the flipped substrate base from re-entering the DNA base stack. Second, AlkD is not inhibited by high concentrations of free nucleobase. Third, AlkD does not discriminate against the base opposite the lesion, and activity is dramatically reduced by a bulky pyrene opposite the lesion, counter to that found for the case of base flipping by UDG [243]. Fourth, AlkD liberates bulky, positively-charged pyridyloxobutyl (POB)-bases from DNA [193]. Thus, AlkD does not employ a specific nucleobase binding pocket to recognize or remove its substrates, suggesting that depurination of N3- or N7-alkylpurines can be facilitated without direct contact to the protein.
The 3d3mA and THF structures suggested that AlkD has the ability to detect and trap destabilized base pairs but would only excise modified nucleobases that contained weak N-glycosidic bonds. We therefore trapped AlkD in complex with a G•T mismatch in order to evaluate how the enzyme restructures DNA by comparing G•T-DNA in the free and AlkD-bound states. AlkD significantly resculpts the non-Watson-Crick base pair from the canonical wobble G•T structure in order to create an optimized protein-DNA binding surface by maximizing contacts between the phosphoribose backbone of the thymine strand and the concave cleft [193]. These specific protein-DNA contacts are identical to the 3d3mA•T structure (Figure 10C), and substitution of the participating side chains either abolish or severely impair 7mG excision, indicating that the specific DNA capture mechanism is a prerequisite for catalysis.
The specific structure of the DNA trapped in the AlkD complexes provides a rationale for the enzyme’s specificity toward bases with a low threshold for depurination. As a result of the collapsed duplex, the phosphoribose backbone is highly kinked, which places the flipped THF in close proximity to a phosphate immediately 5′ to the lesion [193]. We have recently found that chemical perturbation of this phosphate to a methylphosphonate abolishes 7mG excision activity by AlkD (unpublished results), indicating that this phosphate participates in catalysis either directly, by stabilizing the oxocarbenium ion intermediate, or indirectly, by maintaining a specific kink in the duplex that weakens the N-glycosidic bond. Interestingly, a direct role of the DNA in catalysis of base excision has also been observed in uracil DNA glycosylase [244–246]. More work will be necessary to verify the structure of a bound 7mG-DNA that is activated for hydrolysis.
4. Uracil/Thymine/5mC
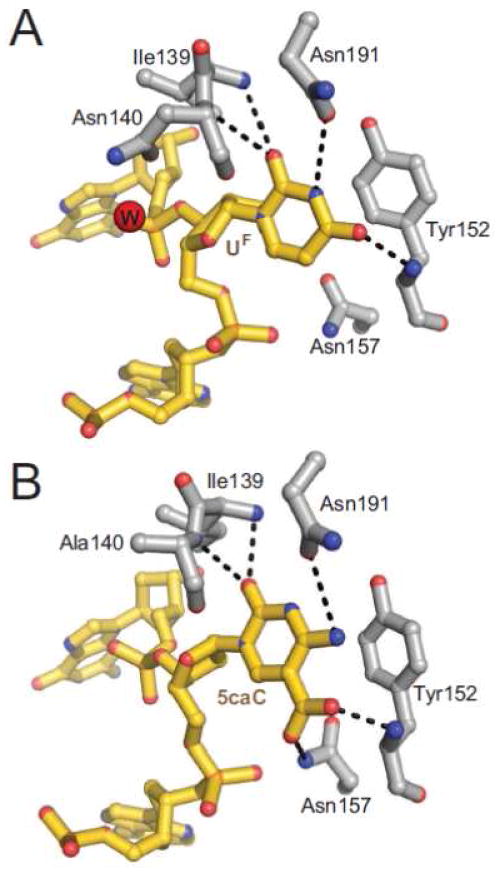
G•U and G•T mismatches arise from deamination of cytosine and 5-methylcytosine (5mC), respectively, and lead to A•T transition mutations [247, 248]. Uracil is excised in eukaryotes by uracil DNA glycosylase (UDG, also known as UNG), single-stranded monofunctional uracil glycosylase (SMUG), and to a lesser extent by thymine DNA glycosylase (TDG). In bacteria, uracil is removed by the UDG ortholog, Ung, and mispaired uracil glycosylase (MUG) [249–252]. Thymine is removed from G•T mismatches by TDG and methyl binding domain 4 (MBD4) in eukaryotes and by archaeal mismatch specific glycosylase (MIG) [253–255]. With the exception of MBD4 and MIG, which belong to the HhH superfamily, the UDG/TDG glycosylases adopt a highly conserved α/β fold (Figure 3) and can be divided into 4 subfamilies on the basis of sequence similarity and substrate specificity [16, 256, 257] (Table 1). UDG family 1 contains UDG/UNG and is defined by the landmark structures of the human and viral enzymes in various states, which revealed mechanistic details about substrate recognition and catalysis common to the entire superfamily [16–19]. Family 2 is composed of thymine-specific TDG and MUG, which are homologous to UDG in structure but not sequence [258–261]. The third family is defined by SMUG, and the fourth by Thermus thermophilus TDG. The common α/β fold of the UDG superfamily contains a positively-charged groove approximately the width of a DNA duplex that is ideal for binding double-stranded DNA [16].
UDG has served as a model for understanding the structural and biochemical functions of DNA glycosylases in general, and recent work has focused on the mechanism by which the enzyme locates uracil amidst undamaged DNA. This collective body of work on UDG has been the subject of several recent reviews [5–7, 10, 262–264], and thus will not be discussed here. We instead focus on recent structural results for TDG in light of new evidence implicating this enzyme in active 5mC demethylation [265, 266].
4.1. A possible role of BER in DNA demethylation
In addition to repair of thymine mismatches, the biological functions of TDG and MBD4 may extend beyond DNA repair as a defense against mutation to a potential role in regulating gene expression and DNA demethylation [265, 267]. 5mC is an important marker for gene expression, X-chromosome inactivation, and transposon silencing among other developmental processes [268–270]. Whereas DNA methylation mechanisms are relatively well understood [271], the demethylation pathways are not. Demethylation can occur passively after replicative synthesis of unmethylated daughter strands or actively by demethylase enzymes. In plants, active demethylation takes place by the DME/ROS1 family of 5mC DNA glycosylases (see section 4.4 below), but an analogous 5mC glycosylase has not been discovered in mammals. TDG and MBD4 have been implicated in active demethylation on the basis of their abilities to excise mispaired thymine produced from AID- or APOBEC-dependent 5mC deamination, and recent studies have shown TDG to be necessary for maintenance of epigenetic stability [265, 272, 273]. In addition, the recent discoveries that the ten-eleven translocation (TET) proteins oxidize 5mC to 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC), and 5-carboxylcytosine (5caC) (Figure 1C), and that 5hmC is found at transcriptional start sites and within actively transcribed genes raises the distinct possibility that these 5mC derivatives and their deamination products are intermediates in a BER-dependent active demethylation pathway [274–283]. Indeed, TDG is capable of excising 5mC oxidation products 5fC and 5caC, with single-turnover rate constants (2.6 ± 0.1 min−1, 5fC•G; 0.5 ± 0.01 min−1, 5caC•G) comparable to T•G (kcat = 1.8 ± 0.04 min−1) [284, 285], further implicating the thymine glycosylases in active demethylation.
4.2. Structural insight into TDG function
Structures of the catalytic domain of TDG (residues 111-308) bound to substrate and product DNA and conjugated by the regulatory protein SUMO have provided a basis to understand TDG’s sequence specificity and its mechanisms of base excision and product release [261, 285–287]. In addition, we review an NMR study of the N-terminal regulatory domain of TDG (residues 1-111) that supports models for allosteric control of TDG activity [288, 289].
4.2.1. TDG-DNA complexes
Structures of TDG bound to duplex DNA containing a THF product mimic provided the general features of DNA binding and the first glimpse into thymine recognition [261]. In this structure, two TDG molecules are bound to a single 22-nucleotide DNA duplex, with one protein anchored at the abasic site and the other positioned at an undamaged site, although biochemical analysis indicated that only one protein per lesion is required for catalysis [261, 290]. The TDG complex is very similar to DNA-bound structures of UDG and E. coli MUG, with notable exceptions. Both TDG and UDG impose a ~43° bend in the substrate DNA at the abasic site, although MUG does not significantly bend the DNA [9, 258, 260, 291]. TDG utilizes an arginine (Arg275) to plug the gap created by the flipped nucleotide, whereas UDG and MUG have leucine plugs. Substitution of Arg275 with either alanine or leucine results in a significant decrease in both the rate of thymine excision and substrate binding [292]. Another unique aspect of the TDG family are lysine residues Lys246 and Lys248, which make contacts to the DNA backbone of the non-damaged strand, 8 and 9 nucleotides away from the damage site, providing an explanation for TDG’s requirement for 9–10 bases 5′ to the lesion [261, 293].
Two TDG-substrate-DNA complexes were recently determined that provided a snapshot of an uncleaved nucleobase in the active site [285, 287]. One structure contained the wild-type enzyme bound to DNA containing a non-hydrolyzable dU mimetic (2′-deoxy-2′-fluoroarabinouridine, UF) (Figure 11A) and the other trapped 5caC in the active site by utilizing a variant TDG containing an Asn140Ala substitution (Figure 11B), which had previously been shown to decrease the rate of thymine excision while having only marginal effect on substrate binding [261, 292]. The overall structures of UF and 5caC substrate complexes are similar to the THF product complex, and the active sites reveal common modes of recognition of the two substrates. A hydrogen bond was observed from Asn191 to N4 in the 5caC structure and to pyrimidine N3 in the UF structure, and this contact is conserved in UNG and SMUG1 but not MUG. In UNG and SMUG1, this asparagine side chain forms an additional hydrogen bond to the pyrimidine O4 [260, 291, 294]. Maiti et al suggest the differential orientation of this residue in UNG and SMUG1 prevents these enzymes from excising cytosine analogs such as 5fC and 5caC [287]. In addition, 5caC and likely thymine participate in van der Waals interactions with Ala145 and both UF and 5caC form hydrogen bonds with main chain atoms of Tyr152 at either the O4 of uracil (and thymine) or the carboxyl group of 5caC. The 5caC forms an additional interaction with Asn157. Even though modeling a thymine base into the active site of the UF structure shows a steric clash with Ala145, this residue is able to accommodate the 5-carboxyl group in the 5caC structure. Nevertheless, Ala145Gly and His151Ala mutants both increase TDG’s thymine excision activity by 13-fold over the wild-type. Maiti et al propose that His151 slows the cleavage reaction by destabilizing the partial negative charge that develops during the reaction [295]. The mutations showed an even greater increase in activity for thymine from normal A•T base pairs, suggesting that these highly conserved residues are needed to limit aberrant action on undamaged DNA [295]. Zhang, et al. found that TDG binds DNA containing 5caC with higher affinity than 5fC, U, and T, which they attribute to the hydrogen bonds to the electronegative 5caC carboxyl group from Asn157, His151, and Tyr152 [285]. The inability of other members of the UDG family, including SMUG1 and UDG, to bind 5caC and 5fC may result from the presence of residues that interfere with these interactions [285].
Figure 11.
Protein-DNA contacts within the TDG active site. (A) TDG bound to DNA containing 2′-deoxy-2′-fluoroarabinouridine (UF) (PDB ID 3UFJ). Hydrogen bonds are shown as dashed lines and the putative catalytic water molecule is a red sphere. (B) TDG in complex with 5-carboxylcytosine (5caC)-DNA.
Regarding catalysis, stabilization of Asn140 and the β2-α4 catalytic loop, which encircles the active site, by Thr197 was found to be important for TDG function, as a Thr197Ala substitution resulted in a 32-fold reduction in base excision activity [295]. Interestingly, the UF structure suggested that a putative water nucleophile, which is absent from other TDG structures and the enzyme-substrate complex for related MUG enzymes, is positioned by the side chain and main chain of Asn140 and Thr197, respectively. The low resolution and extensive merohedral twinning of the crystallographic data precludes an unambiguous assignment of such a water molecule. However, the presence of a water at this position in TDG is consistent with those observed in the high-resolution structure of free MUG [258] and with the putative water nucleophiles located in high-resolution structures of UNG [291, 296].
TDG makes several contacts to the guanine opposite the lesion that offer an explanation for the enzyme’s specificity for thymine in certain sequence contexts. Specificity for G•T mispairs [297] is dictated by Ala274 and Pro280, which make guanine-specific hydrogen bonds from their backbone oxygen atoms to N1 and N2 of the G opposite the abasic site (Figure 12A). The Ala274 contact is conserved by UDG enzymes, whereas the Pro280 interaction is unique to TDG. In addition, TDG has the greatest excision activity for thymine that is immediately 5′ to a guanine (i.e., in a TpG/CpG dinucleotide step) [298, 299]. Interestingly, TDG does not contact the 5′ cytosine on the non-lesion strand. The specificity for TpG/CpG is likely explained by contacts to the TpG guanine from the conserved Gln278 side chain and Ala277 main chain, as well as the Ala274/Pro280 contacts to the G•T guanine described above (Figure 12A) [261, 297, 299].
Figure 12.
Comparison of TDG (A) and MBD4 (B) contacts to the strand opposite the lesion. The guanine base opposite the THF abasic site is marked with an asterisk. (A) TDG/THF-DNA complex (PDB ID 2RBA). (B) MBD4/THF-DNA complex (4DK9).
4.2.2. TDG-SUMO interaction
Posttranslational modification of TDG by Small Ubiquitin-like Modifier (SUMO) proteins occurs at the C-terminal end of the catalytic domain (Lys330). TDG sumoylation facilitates dissociation of TDG from AP-DNA and modulates enzymatic activity through a mechanism involving conformation changes of the N-terminal regulatory domain [289, 300, 301]. In addition, sumoylation of TDG is essential for activation of CREB-binding protein (CBP)-dependent transcription and localization to promyelocytic leukemia protein oncogenic domains (PODs) [302]. The crystal structure of human TDG conjugated with SUMO1 was determined in 2005 and provided insight into how SUMO1 modulates TDG-DNA binding [286]. C-terminal residues of TDG (307-330) form a crossover β-strand that extends a β-sheet with SUMO1. In addition, TDG and SUMO1 interact through a series of main chain hydrogen bonds and side chain hydrophobic and polar interactions at its SUMO-interacting site (SIM). A mutational analysis revealed that these noncovalent bonds are necessary for SUMO1-induced disruption of DNA binding by TDG, confirming an earlier study [286, 300]. The covalent tethering of SUMO1 to the C-terminus of TDG places helix α7 in an outwardly extended conformation from the rest of the protein. Superposition of sumoylated and DNA-bound forms of TDG predict that helix α7 in this orientation collides with the DNA strand immediately opposite the lesion (Figure 13). The nature of the conformational change in the C-terminal end of the TDG glycosylase domain is uncertain since helix α7 was not present in the DNA-bound structures. Nevertheless, the SUMO1-TDG structure suggests that sumoylation of TDG locks helix α7 in an orientation that promotes DNA release. A structure of SUMO3 conjugated to TDG shows very similar binding between the two proteins as with SUMO1 [303].
Figure 13.

SUMO1 modified TDG creates steric clash with DNA. The SUMO1-modified TDG structure (blue TDG, green SUMO1, PDB ID 1WYW) is superimposed onto the TDG/THF-DNA complex (silver/gold, 2RBA). SUMO1 modification holds helix α7 in a position that would presumably clash with the DNA.
4.2.3. TDG regulatory domain
TDG contains at its N-terminus a lysine-rich regulatory domain (RD) that interacts with DNA and a number of proteins involved in genome maintenance. The RD binds to DNA methyltransferase DNMT3a [304] and is a target for acetylation by transcriptional co-activators CBP/p300, which aids in recruitment of APE1 [305]. In addition, the RD is important for TDG specificity for G•T mismatches [252]. This regulatory domain is highly flexible and contains a non-specific DNA binding function that is modulated by sumoylation of the catalytic domain, thereby affecting its enzymatic activity [289]. A recent NMR study of this domain revealed residues 1-50 to be unstructured even in the context of the full protein. Residues 51-111, on the other hand, showed a modest degree of structure and an extended conformation that contacts the catalytic domain in the context of the full-length protein [288]. The authors proposed that an electrostatic interaction between the regulatory and catalytic domains modulate rates of thymine and uracil excision, supporting previous models for allosteric control of G•T specificity [288, 289].
4.3. MBD4
Mismatch specific thymine glycosylase MBD4 contains a methyl-CpG-binding domain (MBD) and a C-terminal glycosylase domain that preferentially excises T mispaired with G [254]. MBD4 excises thymine from G•T and G•U mispairs at rates (kcat) of 0.012 s−1 and 0.05 s−1, respectively [306]. Crystal structures of the glycosylase domains of mouse and human MBD4 showed that the enzyme belongs to the helix-hairpin-helix structural superfamily [256, 307]. A very recent structure of MBD4’s glycosylase domain bound to THF-DNA showed the overall DNA binding regime characteristic of HhH glycosylases, including a 57° bend in the DNA, an extrahelical THF moiety, the opposite base (guanine) stacked in the duplex, and plug (Leu506) and wedge (Arg468) residues that stabilize the flipped nucleotide and distorted duplex [295]. Like TDG, MBD4 makes several guanine-specific contacts to the base opposite the thymine, and thus the structure provides a rationale for discrimination of G•T over A•T base pairs [306] (Figure 12B). Specifically, Leu506 and Arg468 mainchain oxygens participate in hydrogen bonds or polar interactions with the N1 and exocyclic N2 of guanine, contacts which cannot be made to adenine [261, 295] (Figure 12B). Finally, although MBD4 has been proposed to process G•T mispairs created by active demethylation [273], MBD4 is not able to excise 5fC or 5caC [283, 295]. Manvilla et al attribute this lack of activity to incompatibility between the MBD4 active site and the negative charges that develop on 5fC and 5caC upon dissociation [295].
4.4. DME/ROS1
Plants contain a family of 5mC glycosylases, represented by the Arabidopsis proteins DEMETER (DME), Repressor of Silencing 1 (ROS1), DME like 2 (DML2) and DML3, that are responsible for active demethylation via BER. DME is responsible for endosperm gene imprinting and is necessary for seed viability [308]. The DML enzymes, including ROS1, likely function as genome wide demethylases, particularly at sites 5′ and 3′ to genes to regulate transcription, protecting plants from erroneous gene silencing [309–312]. DME, ROS1, and DML3 excise 5mC in CpG, CpNpG, and CpNpN contexts, and DML2 was shown to have 5mC excision activity in a CpG context [313–315].
The DME/ROS1 family enzymes utilize a bifunctional glycosylase-lyase mechanism and, although there are no crystal structures available, sequence analysis predicts an iron-sulfur-containing HhH glycosylase domain similar to EndoIII and MutY [308, 313, 315, 316] (Figure 14A). A homology model of ROS1 using BsEndoIII as a template helped to identify several residues conserved among the HhH superfamily that are important for ROS1 function [317]. Tyr606 and Asp611 are predicted to reside near the base recognition pocket and are necessary for the excision of both 5mC and thymine from G•T mismatches. A Tyr606Leu mutant slightly reduced the DNA binding activity, whereas Asp611Val mutation increased DNA binding with respect to wild-type ROS1 [317]. The homology model also revealed that two aromatic residues (Phe589 and Tyr1028) conserved in the DME family are positioned to interact with the lesion. Interestingly, Phe589Ala and Tyr1028Ser mutations changed the preference of ROS1 from 5mC to T•G mismatches, indicating that they are important for substrate recognition [317]. A similar homology model for the glycosylase domain of DME predicts analogous residues that may be necessary for catalysis (S. Brooks, B.F. Eichman, unpublished) (Figure 14).
Figure 14.
DME domain structure and distribution of critical residues. (A) Schematic of A. thaliana DEMETER (DME), showing the location of the glycosylase domain (green) and domains A (blue) and B (gold) of unknown function. IDR1, inter-domain region 1; IDR2, inter-domain region 2. Red bars mark the locations of residues identified by random mutagenesis to be critical for DME function. Putative residues important for DNA binding and catalysis and conserved in other HhH enzymes are marked by symbols below the schematic (Asn778 DNA plug, magenta triangle; Met1238 DNA wedge, blue triangle; catalytic Lys1286, yellow star). (B) Homology model of the C-terminal half of domain A and the glycosylase domain with DNA superimposed from the structure of Bacillus stearothermophilus EndoIII bound to abasic DNA (PDB ID 1P59). The putative catalytic Lys1286 (cyan) Met1238 wedge (slate) residues are contributed by the glycosylase domain (green), the putative Asn778 plug residue (magenta) is from domain A.
Unlike other glycosylases, DME/ROS1 proteins contain two additional domains flanking the glycosylase domain and that are essential for 5mC excision (Figure 14A) [314, 318, 319]. The C-terminal domain lacks any identifiable sequence homology to proteins outside of the DME/ROS1 family, and thus additional structural and functional studies will be necessary to ascertain its function. The N-terminal domain, on the other hand, contains a conserved lysine-rich region required for ROS1 binding to non-methylated DNA and enhances ROS1 specificity for 5mC over T. Deletion of this domain caused ROS1 to process long DNA substrates less effectively [319]. On its own, the N-terminal region binds DNA with a high affinity. Deletion of the N-terminal domain reduces the ability of ROS1 to bind DNA and excise 5mC, but does not affect the ability of DME to bind methylated and non-methylated DNA [318, 319]. The N-terminal and glycosylase domains are separated by ~240 and 400 residues in ROS1 and DME, respectively. Deletion of this interdomain region (IDR1) in DME does not affect 5mC excision activity [318]. The ROS1-EndoIII homology model predicted that this linker region is an inserted sequence within the glycosylase domain and that the lesion-intercalating plug residue is N-terminal to the IDR1 insertion [317]. Interestingly, although ROS1 Asn608 aligns with the Gln42 plug in BsEndoIII, elimination of the Asn608 side chain did not affect base excision, whereas Gln607 was shown to be essential for both 5mC and T excision and binding of both methylated and non-methylated DNA [317]. In contrast, a DME homology model (Figure 14B), predicts the plug residue to be Asn778, which was shown by a random mutagenesis study to be critical for 5mC excision, whereas Gln777 was not identified as an important residue in that assay [318]. A better understanding of these 5mC glycosylases awaits structural information for DME and ROS1.
5. Summary and Perspectives
Recent structural and enzymological studies on previously known and newly discovered DNA glycosylases that process a wide variety of oxidized, alkylated, and deaminated nucleobases have dramatically advanced our understanding of the inner workings of these amazing DNA repair machines. One of the most significant questions has focused on how each glycosylase imparts specificity for a particular type of damage. On the most basic level, the specificity of each glycosylase can be considered to be a function of the chemical complementarity of the modified nucleobase within the active site. That is, glycosylases are a collection of specifically evolved active sites situated among a variety of scaffolds. On the other hand, recent work on several enzymes within the oxidation (e.g., OGG1, MutM/Fpg, EndoVIII/Nei) and alkylation (e.g., AlkA, Mag1) classes indicates that some aspect of substrate recognition takes place within the DNA duplex, before the modified nucleobase has been extruded into the active site [68, 100, 136, 191, 224]. A series of structures of OGG1 and MutM crosslinked to unmodified DNA have detailed this search process for the oxidative enzymes, and together with a crosslinked AlkA-DNA structure highlight the role of the intercalating plug residue in probing the minor groove. What follows from this notion of substrate discrimination prior to base flipping is that the intrinsic structure of the DNA lesion itself certainly plays a large role in enzymatic recognition and excision. Related to this, work on UDG and alkylation damage specific glycosylases (e.g., TAG, AlkD) has provided several recent examples for how the DNA conformation and the intrinsic stability of the lesion contributes to substrate-assisted catalysis of base excision [193, 226, 244–246, 320].
Most of the work to date has focused on their catalytic domains, but many glycosylases contain extra domains, interact with other proteins, or undergo post-translational modification, all of which may serve to regulate base excision activity or localize the protein to specific locations on the chromosome. The structure of the full-length MutY protein in complex with an 8oxoG•A mispair is an excellent example of how an extra domain serves to provide enhanced specificity for the base opposite the lesion [149], and work has begun to address the interaction between human MutY and the 9-1-1 complex involved in genome maintenance [157–159]. Sumoylation of TDG and its modulation of DNA affinity through conformational changes involving the N-terminal, regulatory domain is a more complex example involving both covalent modification and a non-catalytic domain [286, 289, 300, 301]. The 5mC glycosylase, DME, contains N- and C-terminal domains flanking the HhH glycosylase domain, but in that case, the extra domains seem to be part of the catalytic core as their mutation or deletion renders the protein inactive [S. Brooks and B.F. Eichman, unpublished results and ref. 318]. Finally, it is becoming increasingly evident that glycosylases do not perform their duties in isolation, but are part of larger complexes that exist to efficiently shuttle DNA damage through the BER pathway by molecular handoff of intermediates, or even to shunt damage into alternative repair pathways. For example, physical interactions of the BER scaffolding protein XRCC1 with AAG and hNTH1 and hNEIL2 have been shown to stimulate their base excision repair activity [321, 322], and APE1 has been shown to stimulate the activities of AAG and TDG, effectively coordinating the first 2 steps of BER [217, 219]. In order to fully understand the role of DNA glycosylases in the context of BER, work in the future will need to more closely explore how these protein interactions and modifications modulate glycosylase behavior.
Highlights.
Recent structural and mechanistic studies of DNA glycosylases are reviewed
DNA glycosylases excise oxidized, alkylated, or deaminated nucleobases
Five structural folds use base flipping to recognize and remove damage
A sixth architecture traps destabilized base pairs without a base binding pocket
Acknowledgments
The authors thank Sylvie Doublié, Sheila David, and Alex Drohat for critique of the manuscript. Work on DNA glycosylases in the author’s laboratory is supported by grants from the National Institutes of Health (R01 ES019625) and the National Science Foundation (MCB-1122098). S.C.B. is supported by a National Science Foundation Graduate Research Fellowship under Grant No. 0909667. E.H.R. was supported in part by the Vanderbilt Training Program in Environmental Toxicology (T32 ES07028).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Friedberg EC, Aguilera A, Gellert M, Hanawalt PC, Hays JB, Lehmann AR, Lindahl T, Lowndes N, Sarasin A, Wood RD. DNA repair: from molecular mechanism to human disease. DNA Repair (Amst) 2006;5:986–996. doi: 10.1016/j.dnarep.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 2.Lindahl T. Suppression of spontaneous mutagenesis in human cells by DNA base excision-repair. Mutation research. 2000;462:129–135. doi: 10.1016/s1383-5742(00)00024-7. [DOI] [PubMed] [Google Scholar]
- 3.Dalhus B, Laerdahl JK, Backe PH, Bjørås M. DNA base repair--recognition and initiation of catalysis. FEMS Microbiol Rev. 2009;33:1044–1078. doi: 10.1111/j.1574-6976.2009.00188.x. [DOI] [PubMed] [Google Scholar]
- 4.David SS, O’Shea VL, Kundu S. Base-excision repair of oxidative DNA damage. Nature. 2007;447:941–950. doi: 10.1038/nature05978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Friedman JI, Stivers JT. Detection of damaged DNA bases by DNA glycosylase enzymes. Biochemistry. 2010;49:4957–4967. doi: 10.1021/bi100593a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fromme JC, Banerjee A, Verdine GL. DNA glycosylase recognition and catalysis. Curr Opin Struct Biol. 2004;14:43–49. doi: 10.1016/j.sbi.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 7.Huffman JL, Sundheim O, Tainer JA. DNA base damage recognition and removal: new twists and grooves. Mutat Res. 2005;577:55–76. doi: 10.1016/j.mrfmmm.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 8.Li GM. Novel molecular insights into the mechanism of GO removal by MutM. Cell Res. 2010;20:116–118. doi: 10.1038/cr.2010.15. [DOI] [PubMed] [Google Scholar]
- 9.Stivers JT, Jiang YL. A mechanistic perspective on the chemistry of DNA repair glycosylases. Chem Rev. 2003;103:2729–2759. doi: 10.1021/cr010219b. [DOI] [PubMed] [Google Scholar]
- 10.Zharkov DO, Mechetin GV, Nevinsky GA. Uracil-DNA glycosylase: Structural, thermodynamic and kinetic aspects of lesion search and recognition. Mutat Res. 2010;685:11–20. doi: 10.1016/j.mrfmmm.2009.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scharer OD, Jiricny J. Recent progress in the biology, chemistry and structural biology of DNA glycosylases. Bioessays. 2001;23:270–281. doi: 10.1002/1521-1878(200103)23:3<270::AID-BIES1037>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 12.Rubinson EH, Eichman BF. Nucleic acid recognition by tandem helical repeats. Curr Opin Struct Biol. 2012;22:101–109. doi: 10.1016/j.sbi.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morikawa K, Matsumoto O, Tsujimoto M, Katayanagi K, Ariyoshi M, Doi T, Ikehara M, Inaoka T, Ohtsuka E. X-ray structure of T4 endonuclease V: an excision repair enzyme specific for a pyrimidine dimer. Science. 1992;256:523–526. doi: 10.1126/science.1575827. [DOI] [PubMed] [Google Scholar]
- 14.Kuo CF, McRee DE, Fisher CL, O’Handley SF, Cunningham RP, Tainer JA. Atomic structure of the DNA repair [4Fe-4S] enzyme endonuclease III. Science. 1992;258:434–440. doi: 10.1126/science.1411536. [DOI] [PubMed] [Google Scholar]
- 15.Vassylyev DG, Kashiwagi T, Mikami Y, Ariyoshi M, Iwai S, Ohtsuka E, Morikawa K. Atomic model of a pyrimidine dimer excision repair enzyme complexed with a DNA substrate: structural basis for damaged DNA recognition. Cell. 1995;83:773–782. doi: 10.1016/0092-8674(95)90190-6. [DOI] [PubMed] [Google Scholar]
- 16.Mol CD, Arvai AS, Slupphaug G, Kavli B, Alseth I, Krokan HE, Tainer JA. Crystal structure and mutational analysis of human uracil-DNA glycosylase: structural basis for specificity and catalysis. Cell. 1995;80:869–878. doi: 10.1016/0092-8674(95)90290-2. [DOI] [PubMed] [Google Scholar]
- 17.Mol CD, Kuo CF, Thayer MM, Cunningham RP, Tainer JA. Structure and function of the multifunctional DNA-repair enzyme exonuclease III. Nature. 1995;374:381–386. doi: 10.1038/374381a0. [DOI] [PubMed] [Google Scholar]
- 18.Savva R, McAuley-Hecht K, Brown T, Pearl L. The structural basis of specific base-excision repair by uracil-DNA glycosylase. Nature. 1995;373:487–493. doi: 10.1038/373487a0. [DOI] [PubMed] [Google Scholar]
- 19.Slupphaug G, Mol CD, Kavli B, Arvai AS, Krokan HE, Tainer JA. A nucleotide-flipping mechanism from the structure of human uracil-DNA glycosylase bound to DNA. Nature. 1996;384:87–92. doi: 10.1038/384087a0. [DOI] [PubMed] [Google Scholar]
- 20.Banerjee A, Santos WL, Verdine GL. Structure of a DNA glycosylase searching for lesions. Science. 2006;311:1153–1157. doi: 10.1126/science.1120288. [DOI] [PubMed] [Google Scholar]
- 21.Valko M, Rhodes CJ, Moncol J, Izakovic M, Mazur M. Free radicals, metals and antioxidants in oxidative stress-induced cancer. Chem Biol Interact. 2006;160:1–40. doi: 10.1016/j.cbi.2005.12.009. [DOI] [PubMed] [Google Scholar]
- 22.Klaunig JE, Kamendulis LM. The role of oxidative stress in carcinogenesis. Annu Rev Pharmacol Toxicol. 2004;44:239–267. doi: 10.1146/annurev.pharmtox.44.101802.121851. [DOI] [PubMed] [Google Scholar]
- 23.van Loon B, Markkanen E, Hubscher U. Oxygen as a friend and enemy: How to combat the mutational potential of 8-oxo-guanine. DNA repair. 2010;9:604–616. doi: 10.1016/j.dnarep.2010.03.004. [DOI] [PubMed] [Google Scholar]
- 24.Kryston TB, Georgiev AB, Pissis P, Georgakilas AG. Role of oxidative stress and DNA damage in human carcinogenesis. Mutation research. 2011;711:193–201. doi: 10.1016/j.mrfmmm.2010.12.016. [DOI] [PubMed] [Google Scholar]
- 25.Neeley WL, Essigmann JM. Mechanisms of formation, genotoxicity, and mutation of guanine oxidation products. Chem Res Toxicol. 2006;19:491–505. doi: 10.1021/tx0600043. [DOI] [PubMed] [Google Scholar]
- 26.Evans MD, Dizdaroglu M, Cooke MS. Oxidative DNA damage and disease: induction, repair and significance. Mutat Res. 2004;567:1–61. doi: 10.1016/j.mrrev.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 27.Burrows CJ, Muller JG. Oxidative Nucleobase Modifications Leading to Strand Scission. Chem Rev. 1998;98:1109–1152. doi: 10.1021/cr960421s. [DOI] [PubMed] [Google Scholar]
- 28.Cheng KC, Cahill DS, Kasai H, Nishimura S, Loeb LA. 8-Hydroxyguanine, an abundant form of oxidative DNA damage, causes G----T and A----C substitutions. The Journal of biological chemistry. 1992;267:166–172. [PubMed] [Google Scholar]
- 29.Brieba LG, Eichman BF, Kokoska RJ, Doublié S, Kunkel TA, Ellenberger T. Structural basis for the dual coding potential of 8-oxoguanosine by a high-fidelity DNA polymerase. EMBO J. 2004;23:3452–3461. doi: 10.1038/sj.emboj.7600354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hsu GW, Ober M, Carell T, Beese LS. Error-prone replication of oxidatively damaged DNA by a high-fidelity DNA polymerase. Nature. 2004;431:217–221. doi: 10.1038/nature02908. [DOI] [PubMed] [Google Scholar]
- 31.Burrows CJ, Muller JG, Kornyushyna O, Luo W, Duarte V, Leipold MD, David SS. Structure and potential mutagenicity of new hydantoin products from guanosine and 8-oxo-7,8-dihydroguanine oxidation by transition metals. Environ Health Perspect. 2002;110(Suppl 5):713–717. doi: 10.1289/ehp.02110s5713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Henderson PT, Delaney JC, Muller JG, Neeley WL, Tannenbaum SR, Burrows CJ, Essigmann JM. The hydantoin lesions formed from oxidation of 7,8-dihydro-8-oxoguanine are potent sources of replication errors in vivo. Biochemistry. 2003;42:9257–9262. doi: 10.1021/bi0347252. [DOI] [PubMed] [Google Scholar]
- 33.Luo W, Muller JG, Rachlin EM, Burrows CJ. Characterization of hydantoin products from one-electron oxidation of 8-oxo-7,8-dihydroguanosine in a nucleoside model. Chem Res Toxicol. 2001;14:927–938. doi: 10.1021/tx010072j. [DOI] [PubMed] [Google Scholar]
- 34.Tudek B. Imidazole ring-opened DNA purines and their biological significance. J Biochem Mol Biol. 2003;36:12–19. doi: 10.5483/bmbrep.2003.36.1.012. [DOI] [PubMed] [Google Scholar]
- 35.Leipold MD, Muller JG, Burrows CJ, David SS. Removal of hydantoin products of 8-oxoguanine oxidation by the Escherichia coli DNA repair enzyme, FPG. Biochemistry. 2000;39:14984–14992. doi: 10.1021/bi0017982. [DOI] [PubMed] [Google Scholar]
- 36.Delaney S, Neeley WL, Delaney JC, Essigmann JM. The substrate specificity of MutY for hyperoxidized guanine lesions in vivo. Biochemistry. 2007;46:1448–1455. doi: 10.1021/bi061174h. [DOI] [PubMed] [Google Scholar]
- 37.Aller P, Rould MA, Hogg M, Wallace SS, Doublié S. A structural rationale for stalling of a replicative DNA polymerase at the most common oxidative thymine lesion, thymine glycol. Proc Natl Acad Sci U S A. 2007;104:814–818. doi: 10.1073/pnas.0606648104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kreutzer DA, Essigmann JM. Oxidized, deaminated cytosines are a source of C --> T transitions in vivo. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:3578–3582. doi: 10.1073/pnas.95.7.3578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shikazono N, Pearson C, O’Neill P, Thacker J. The roles of specific glycosylases in determining the mutagenic consequences of clustered DNA base damage. Nucleic acids research. 2006;34:3722–3730. doi: 10.1093/nar/gkl503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu J, Doetsch PW. Escherichia coli RNA and DNA polymerase bypass of dihydrouracil: mutagenic potential via transcription and replication. Nucleic acids research. 1998;26:1707–1712. doi: 10.1093/nar/26.7.1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Purmal AA, Kow YW, Wallace SS. Major oxidative products of cytosine, 5-hydroxycytosine and 5-hydroxyuracil, exhibit sequence context-dependent mispairing in vitro. Nucleic acids research. 1994;22:72–78. doi: 10.1093/nar/22.1.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boorstein RJ, Teebor GW. Mutagenicity of 5-hydroxymethyl-2′-deoxyuridine to Chinese hamster cells. Cancer Res. 1988;48:5466–5470. [PubMed] [Google Scholar]
- 43.Yoshida M, Makino K, Morita H, Terato H, Ohyama Y, Ide H. Substrate and mispairing properties of 5-formyl-2′-deoxyuridine 5′-triphosphate assessed by in vitro DNA polymerase reactions. Nucleic acids research. 1997;25:1570–1577. doi: 10.1093/nar/25.8.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nash HM, Bruner SD, Scharer OD, Kawate T, Addona TA, Spooner E, Lane WS, Verdine GL. Cloning of a yeast 8-oxoguanine DNA glycosylase reveals the existence of a base-excision DNA-repair protein superfamily. Curr Biol. 1996;6:968–980. doi: 10.1016/s0960-9822(02)00641-3. [DOI] [PubMed] [Google Scholar]
- 45.Bailly V, Verly WG, O’Connor T, Laval J. Mechanism of DNA strand nicking at apurinic/apyrimidinic sites by Escherichia coli [formamidopyrimidine]DNA glycosylase. Biochem J. 1989;262:581–589. doi: 10.1042/bj2620581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jiang D, Hatahet Z, Melamede RJ, Kow YW, Wallace SS. Characterization of Escherichia coli endonuclease VIII. J Biol Chem. 1997;272:32230–32239. doi: 10.1074/jbc.272.51.32230. [DOI] [PubMed] [Google Scholar]
- 47.Sugahara M, Mikawa T, Kumasaka T, Yamamoto M, Kato R, Fukuyama K, Inoue Y, Kuramitsu S. Crystal structure of a repair enzyme of oxidatively damaged DNA, MutM (Fpg), from an extreme thermophile, Thermus thermophilus HB8. Embo J. 2000;19:3857–3869. doi: 10.1093/emboj/19.15.3857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zharkov DO, Golan G, Gilboa R, Fernandes AS, Gerchman SE, Kycia JH, Rieger RA, Grollman AP, Shoham G. Structural analysis of an Escherichia coli endonuclease VIII covalent reaction intermediate. EMBO J. 2002;21:789–800. doi: 10.1093/emboj/21.4.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zharkov DO, Shoham G, Grollman AP. Structural characterization of the Fpg family of DNA glycosylases. DNA Repair (Amst) 2003;2:839–862. doi: 10.1016/s1568-7864(03)00084-3. [DOI] [PubMed] [Google Scholar]
- 50.van der Kemp PA, Thomas D, Barbey R, de Oliveira R, Boiteux S. Cloning and expression in Escherichia coli of the OGG1 gene of Saccharomyces cerevisiae, which codes for a DNA glycosylase that excises 7,8-dihydro-8-oxoguanine and 2,6-diamino-4-hydroxy-5-N-methylformamidopyrimidine. Proc Natl Acad Sci U S A. 1996;93:5197–5202. doi: 10.1073/pnas.93.11.5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Castaing B, Geiger A, Seliger H, Nehls P, Laval J, Zelwer C, Boiteux S. Cleavage and binding of a DNA fragment containing a single 8-oxoguanine by wild type and mutant FPG proteins. Nucleic Acids Res. 1993;21:2899–2905. doi: 10.1093/nar/21.12.2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bruner SD, Norman DP, Verdine GL. Structural basis for recognition and repair of the endogenous mutagen 8-oxoguanine in DNA. Nature. 2000;403:859–866. doi: 10.1038/35002510. [DOI] [PubMed] [Google Scholar]
- 53.Dalhus B, Forsbring M, Helle IH, Vik ES, Forstrom RJ, Backe PH, Alseth I, Bjørås M. Separation-of-function mutants unravel the dual-reaction mode of human 8-oxoguanine DNA glycosylase. Structure. 2011;19:117–127. doi: 10.1016/j.str.2010.09.023. [DOI] [PubMed] [Google Scholar]
- 54.Aburatani H, Hippo Y, Ishida T, Takashima R, Matsuba C, Kodama T, Takao M, Yasui A, Yamamoto K, Asano M. Cloning and characterization of mammalian 8-hydroxyguanine-specific DNA glycosylase/apurinic, apyrimidinic lyase, a functional mutM homologue. Cancer Res. 1997;57:2151–2156. [PubMed] [Google Scholar]
- 55.Arai K, Morishita K, Shinmura K, Kohno T, Kim SR, Nohmi T, Taniwaki M, Ohwada S, Yokota J. Cloning of a human homolog of the yeast OGG1 gene that is involved in the repair of oxidative DNA damage. Oncogene. 1997;14:2857–2861. doi: 10.1038/sj.onc.1201139. [DOI] [PubMed] [Google Scholar]
- 56.Bjørås M, Luna L, Johnsen B, Hoff E, Haug T, Rognes T, Seeberg E. Opposite base-dependent reactions of a human base excision repair enzyme on DNA containing 7,8-dihydro-8-oxoguanine and abasic sites. The EMBO journal. 1997;16:6314–6322. doi: 10.1093/emboj/16.20.6314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nagashima M, Sasaki A, Morishita K, Takenoshita S, Nagamachi Y, Kasai H, Yokota J. Presence of human cellular protein(s) that specifically binds and cleaves 8-hydroxyguanine containing DNA. Mutation research. 1997;383:49–59. doi: 10.1016/s0921-8777(96)00045-6. [DOI] [PubMed] [Google Scholar]
- 58.Roldan-Arjona T, Wei YF, Carter KC, Klungland A, Anselmino C, Wang RP, Augustus M, Lindahl T. Molecular cloning and functional expression of a human cDNA encoding the antimutator enzyme 8-hydroxyguanine-DNA glycosylase. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:8016–8020. doi: 10.1073/pnas.94.15.8016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Radicella JP, Dherin C, Desmaze C, Fox MS, Boiteux S. Cloning and characterization of hOGG1, a human homolog of the OGG1 gene of Saccharomyces cerevisiae. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:8010–8015. doi: 10.1073/pnas.94.15.8010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rosenquist TA, Zharkov DO, Grollman AP. Cloning and characterization of a mammalian 8-oxoguanine DNA glycosylase. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:7429–7434. doi: 10.1073/pnas.94.14.7429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Robey-Bond SM, Barrantes-Reynolds R, Bond JP, Wallace SS, Bandaru V. Clostridium acetobutylicum 8-oxoguanine DNA glycosylase (Ogg) differs from eukaryotic Oggs with respect to opposite base discrimination. Biochemistry. 2008;47:7626–7636. doi: 10.1021/bi800162e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Faucher F, Wallace SS, Doublié S. Structural basis for the lack of opposite base specificity of Clostridium acetobutylicum 8-oxoguanine DNA glycosylase. DNA Repair (Amst) 2009;8:1283–1289. doi: 10.1016/j.dnarep.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Faucher F, Robey-Bond SM, Wallace SS, Doublié S. Structural characterization of Clostridium acetobutylicum 8-oxoguanine DNA glycosylase in its apo form and in complex with 8-oxodeoxyguanosine. J Mol Biol. 2009;387:669–679. doi: 10.1016/j.jmb.2009.01.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gogos A, Clarke ND. Characterization of an 8-oxoguanine DNA glycosylase from Methanococcus jannaschii. The Journal of biological chemistry. 1999;274:30447–30450. doi: 10.1074/jbc.274.43.30447. [DOI] [PubMed] [Google Scholar]
- 65.Faucher F, Duclos S, Bandaru V, Wallace SS, Doublié S. Crystal structures of two archaeal 8-oxoguanine DNA glycosylases provide structural insight into guanine/8-oxoguanine distinction. Structure. 2009;17:703–712. doi: 10.1016/j.str.2009.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sartori AA, Lingaraju GM, Hunziker P, Winkler FK, Jiricny J. Pa-AGOG, the founding member of a new family of archaeal 8-oxoguanine DNA-glycosylases. Nucleic acids research. 2004;32:6531–6539. doi: 10.1093/nar/gkh995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Faucher F, Doublié S, Jia Z. 8-Oxoguanine DNA Glycosylases: One Lesion, Three Subfamilies. International Journal of Molecular Sciences. 2012;13:6711–6729. doi: 10.3390/ijms13066711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Banerjee A, Yang W, Karplus M, Verdine GL. Structure of a repair enzyme interrogating undamaged DNA elucidates recognition of damaged DNA. Nature. 2005;434:612–618. doi: 10.1038/nature03458. [DOI] [PubMed] [Google Scholar]
- 69.Banerjee A, Verdine GL. A nucleobase lesion remodels the interaction of its normal neighbor in a DNA glycosylase complex. Proc Natl Acad Sci U S A. 2006;103:15020–15025. doi: 10.1073/pnas.0603644103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Radom CT, Banerjee A, Verdine GL. Structural characterization of human 8-oxoguanine DNA glycosylase variants bearing active site mutations. J Biol Chem. 2007;282:9182–9194. doi: 10.1074/jbc.M608989200. [DOI] [PubMed] [Google Scholar]
- 71.Zharkov DO, Rosenquist TA, Gerchman SE, Grollman AP. Substrate specificity and reaction mechanism of murine 8-oxoguanine-DNA glycosylase. J Biol Chem. 2000;275:28607–28617. doi: 10.1074/jbc.M002441200. [DOI] [PubMed] [Google Scholar]
- 72.Crenshaw CM, Nam K, Oo K, Kutchukian PS, Bowman BR, Karplus M, Verdine GL. Enforced presentation of an extrahelical guanine to the lesion-recognition pocket of the human 8-oxoguanine DNA glycosylase, hOGG1. J Biol Chem. 2012 doi: 10.1074/jbc.M111.316497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lee S, Radom CT, Verdine GL. Trapping and structural elucidation of a very advanced intermediate in the lesion-extrusion pathway of hOGG1. J Am Chem Soc. 2008;130:7784–7785. doi: 10.1021/ja800821t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nash HM, Lu R, Lane WS, Verdine GL. The critical active-site amine of the human 8-oxoguanine DNA glycosylase, hOgg1: direct identification, ablation and chemical reconstitution. Chem Biol. 1997;4:693–702. doi: 10.1016/s1074-5521(97)90225-8. [DOI] [PubMed] [Google Scholar]
- 75.Bjørås M, Seeberg E, Luna L, Pearl LH, Barrett TE. Reciprocal “flipping” underlies substrate recognition and catalytic activation by the human 8-oxo-guanine DNA glycosylase. J Mol Biol. 2002;317:171–177. doi: 10.1006/jmbi.2002.5400. [DOI] [PubMed] [Google Scholar]
- 76.Norman DP, Chung SJ, Verdine GL. Structural and biochemical exploration of a critical amino acid in human 8-oxoguanine glycosylase. Biochemistry. 2003;42:1564–1572. doi: 10.1021/bi026823d. [DOI] [PubMed] [Google Scholar]
- 77.Hill JW, Hazra TK, Izumi T, Mitra S. Stimulation of human 8-oxoguanine-DNA glycosylase by AP-endonuclease: potential coordination of the initial steps in base excision repair. Nucleic Acids Res. 2001;29:430–438. doi: 10.1093/nar/29.2.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kuznetsov NA, Koval VV, Zharkov DO, Nevinsky GA, Douglas KT, Fedorova OS. Kinetics of substrate recognition and cleavage by human 8-oxoguanine-DNA glycosylase. Nucleic Acids Res. 2005;33:3919–3931. doi: 10.1093/nar/gki694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Morland I, Luna L, Gustad E, Seeberg E, Bjørås M. Product inhibition and magnesium modulate the dual reaction mode of hOgg1. DNA Repair (Amst) 2005;4:381–387. doi: 10.1016/j.dnarep.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 80.Vidal AE, Hickson ID, Boiteux S, Radicella JP. Mechanism of stimulation of the DNA glycosylase activity of hOGG1 by the major human AP endonuclease: bypass of the AP lyase activity step. Nucleic Acids Res. 2001;29:1285–1292. doi: 10.1093/nar/29.6.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Norman DP, Bruner SD, Verdine GL. Coupling of substrate recognition and catalysis by a human base-excision DNA repair protein. J Am Chem Soc. 2001;123:359–360. doi: 10.1021/ja003144m. [DOI] [PubMed] [Google Scholar]
- 82.Chung JH, Suh MJ, Park YI, Tainer JA, Han YS. Repair activities of 8-oxoguanine DNA glycosylase from Archaeoglobus fulgidus, a hyperthermophilic archaeon. Mutation research. 2001;486:99–111. doi: 10.1016/s0921-8777(01)00081-7. [DOI] [PubMed] [Google Scholar]
- 83.Im EK, Hong CH, Back JH, Han YS, Chung JH. Functional identification of an 8-oxoguanine specific endonuclease from Thermotoga maritima. Journal of biochemistry and molecular biology. 2005;38:676–682. doi: 10.5483/bmbrep.2005.38.6.676. [DOI] [PubMed] [Google Scholar]
- 84.Faucher F, Wallace SS, Doublié S. The C-terminal lysine of Ogg2 DNA glycosylases is a major molecular determinant for guanine/8-oxoguanine distinction. J Mol Biol. 2010;397:46–56. doi: 10.1016/j.jmb.2010.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Volkl P, Huber R, Drobner E, Rachel R, Burggraf S, Trincone A, Stetter KO. Pyrobaculum aerophilum sp. nov., a novel nitrate-reducing hyperthermophilic archaeum. Appl Environ Microbiol. 1993;59:2918–2926. doi: 10.1128/aem.59.9.2918-2926.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lingaraju GM, Sartori AA, Kostrewa D, Prota AE, Jiricny J, Winkler FK. A DNA glycosylase from Pyrobaculum aerophilum with an 8-oxoguanine binding mode and a noncanonical helix-hairpin-helix structure. Structure. 2005;13:87–98. doi: 10.1016/j.str.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 87.Lingaraju GM, Prota AE, Winkler FK. Mutational studies of Pa-AGOG DNA glycosylase from the hyperthermophilic crenarchaeon Pyrobaculum aerophilum. DNA Repair (Amst) 2009;8:857–864. doi: 10.1016/j.dnarep.2009.03.009. [DOI] [PubMed] [Google Scholar]
- 88.Wiederholt CJ, Delaney MO, Pope MA, David SS, Greenberg MM. Repair of DNA containing Fapy.dG and its beta-C-nucleoside analogue by formamidopyrimidine DNA glycosylase and MutY. Biochemistry. 2003;42:9755–9760. doi: 10.1021/bi034844h. [DOI] [PubMed] [Google Scholar]
- 89.Tchou J, Kasai H, Shibutani S, Chung MH, Laval J, Grollman AP, Nishimura S. 8-oxoguanine (8-hydroxyguanine) DNA glycosylase and its substrate specificity. Proc Natl Acad Sci U S A. 1991;88:4690–4694. doi: 10.1073/pnas.88.11.4690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gasparutto D, Muller E, Boiteux S, Cadet J. Excision of the oxidatively formed 5-hydroxyhydantoin and 5-hydroxy-5-methylhydantoin pyrimidine lesions by Escherichia coli and Saccharomyces cerevisiae DNA N-glycosylases. Biochim Biophys Acta. 2009;1790:16–24. doi: 10.1016/j.bbagen.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 91.Hatahet Z, Kow YW, Purmal AA, Cunningham RP, Wallace SS. New substrates for old enzymes. 5-Hydroxy-2′-deoxycytidine and 5-hydroxy-2′-deoxyuridine are substrates for Escherichia coli endonuclease III and formamidopyrimidine DNA N-glycosylase, while 5-hydroxy-2′-deoxyuridine is a substrate for uracil DNA N-glycosylase. The Journal of biological chemistry. 1994;269:18814–18820. [PubMed] [Google Scholar]
- 92.Serre L, Pereira de Jesus K, Boiteux S, Zelwer C, Castaing B. Crystal structure of the Lactococcus lactis formamidopyrimidine-DNA glycosylase bound to an abasic site analogue-containing DNA. EMBO J. 2002;21:2854–2865. doi: 10.1093/emboj/cdf304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Fromme JC, Verdine GL. Structural insights into lesion recognition and repair by the bacterial 8-oxoguanine DNA glycosylase MutM. Nat Struct Biol. 2002;9:544–552. doi: 10.1038/nsb809. [DOI] [PubMed] [Google Scholar]
- 94.Gilboa R, Zharkov DO, Golan G, Fernandes AS, Gerchman SE, Matz E, Kycia JH, Grollman AP, Shoham G. Structure of formamidopyrimidine-DNA glycosylase covalently complexed to DNA. J Biol Chem. 2002;277:19811–19816. doi: 10.1074/jbc.M202058200. [DOI] [PubMed] [Google Scholar]
- 95.Fromme JC, Verdine GL. DNA lesion recognition by the bacterial repair enzyme MutM. J Biol Chem. 2003;278:51543–51548. doi: 10.1074/jbc.M307768200. [DOI] [PubMed] [Google Scholar]
- 96.Coste F, Ober M, Carell T, Boiteux S, Zelwer C, Castaing B. Structural basis for the recognition of the FapydG lesion (2,6-diamino-4-hydroxy-5-formamidopyrimidine) by formamidopyrimidine-DNA glycosylase. J Biol Chem. 2004;279:44074–44083. doi: 10.1074/jbc.M405928200. [DOI] [PubMed] [Google Scholar]
- 97.Pereira de Jesus K, Serre L, Zelwer C, Castaing B. Structural insights into abasic site for Fpg specific binding and catalysis: comparative high-resolution crystallographic studies of Fpg bound to various models of abasic site analogues-containing DNA. Nucleic Acids Res. 2005;33:5936–5944. doi: 10.1093/nar/gki879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Fromme JC, Bruner SD, Yang W, Karplus M, Verdine GL. Product-assisted catalysis in base-excision DNA repair. Nat Struct Biol. 2003;10:204–211. doi: 10.1038/nsb902. [DOI] [PubMed] [Google Scholar]
- 99.Dunn AR, Kad NM, Nelson SR, Warshaw DM, Wallace SS. Single Qdot-labeled glycosylase molecules use a wedge amino acid to probe for lesions while scanning along DNA. Nucleic acids research. 2011;39:7487–7498. doi: 10.1093/nar/gkr459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Qi Y, Spong MC, Nam K, Banerjee A, Jiralerspong S, Karplus M, Verdine GL. Encounter and extrusion of an intrahelical lesion by a DNA repair enzyme. Nature. 2009;462:762–766. doi: 10.1038/nature08561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Qi Y, Spong MC, Nam K, Karplus M, Verdine GL. Entrapment and structure of an extrahelical guanine attempting to enter the active site of a bacterial DNA glycosylase, MutM. The Journal of biological chemistry. 2010;285:1468–1478. doi: 10.1074/jbc.M109.069799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Duclos S, Aller P, Jaruga P, Dizdaroglu M, Wallace S, Doublié S. Structural and biochemical studies of a plant formamidopyrimidine-DNA glycosylase reveal why eukaryotic Fpg glycosylases do not excise 8-oxoguanine. DNA Repair (Amst) 2012 doi: 10.1016/j.dnarep.2012.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.McKibbin PL, Kobori A, Taniguchi Y, Kool ET, David SS. Surprising repair activities of nonpolar analogs of 8-oxoG expose features of recognition and catalysis by base excision repair glycosylases. Journal of the American Chemical Society. 2012;134:1653–1661. doi: 10.1021/ja208510m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ikeda S, Biswas T, Roy R, Izumi T, Boldogh I, Kurosky A, Sarker AH, Seki S, Mitra S. Purification and characterization of human NTH1, a homolog of Escherichia coli endonuclease III. Direct identification of Lys-212 as the active nucleophilic residue. The Journal of biological chemistry. 1998;273:21585–21593. doi: 10.1074/jbc.273.34.21585. [DOI] [PubMed] [Google Scholar]
- 105.Higgins SA, Frenkel K, Cummings A, Teebor GW. Definitive characterization of human thymine glycol N-glycosylase activity. Biochemistry. 1987;26:1683–1688. doi: 10.1021/bi00380a029. [DOI] [PubMed] [Google Scholar]
- 106.Miller H, Fernandes AS, Zaika E, McTigue MM, Torres MC, Wente M, Iden CR, Grollman AP. Stereoselective excision of thymine glycol from oxidatively damaged DNA. Nucleic Acids Res. 2004;32:338–345. doi: 10.1093/nar/gkh190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Fromme JC, Verdine GL. Structure of a trapped endonuclease III-DNA covalent intermediate. Embo J. 2003;22:3461–3471. doi: 10.1093/emboj/cdg311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Aspinwall R, Rothwell DG, Roldan-Arjona T, Anselmino C, Ward CJ, Cheadle JP, Sampson JR, Lindahl T, Harris PC, Hickson ID. Cloning and characterization of a functional human homolog of Escherichia coli endonuclease III. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:109–114. doi: 10.1073/pnas.94.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Boon EM, Livingston AL, Chmiel NH, David SS, Barton JK. DNA-mediated charge transport for DNA repair. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:12543–12547. doi: 10.1073/pnas.2035257100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lukianova OA, David SS. A role for iron-sulfur clusters in DNA repair. Curr Opin Chem Biol. 2005;9:145–151. doi: 10.1016/j.cbpa.2005.02.006. [DOI] [PubMed] [Google Scholar]
- 111.Yavin E, Boal AK, Stemp ED, Boon EM, Livingston AL, O’Shea VL, David SS, Barton JK. Protein-DNA charge transport: redox activation of a DNA repair protein by guanine radical. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:3546–3551. doi: 10.1073/pnas.0409410102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wallace SS, Bandaru V, Kathe SD, Bond JP. The enigma of endonuclease VIII. DNA Repair (Amst) 2003;2:441–453. doi: 10.1016/s1568-7864(02)00182-9. [DOI] [PubMed] [Google Scholar]
- 113.Golan G, Zharkov DO, Feinberg H, Fernandes AS, Zaika EI, Kycia JH, Grollman AP, Shoham G. Structure of the uncomplexed DNA repair enzyme endonuclease VIII indicates significant interdomain flexibility. Nucleic acids research. 2005;33:5006–5016. doi: 10.1093/nar/gki796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kalodimos CG, Biris N, Bonvin AM, Levandoski MM, Guennuegues M, Boelens R, Kaptein R. Structure and flexibility adaptation in nonspecific and specific protein-DNA complexes. Science. 2004;305:386–389. doi: 10.1126/science.1097064. [DOI] [PubMed] [Google Scholar]
- 115.Hazra TK, Kow YW, Hatahet Z, Imhoff B, Boldogh I, Mokkapati SK, Mitra S, Izumi T. Identification and characterization of a novel human DNA glycosylase for repair of cytosine-derived lesions. J Biol Chem. 2002;277:30417–30420. doi: 10.1074/jbc.C200355200. [DOI] [PubMed] [Google Scholar]
- 116.Hazra TK, Izumi T, Boldogh I, Imhoff B, Kow YW, Jaruga P, Dizdaroglu M, Mitra S. Identification and characterization of a human DNA glycosylase for repair of modified bases in oxidatively damaged DNA. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:3523–3528. doi: 10.1073/pnas.062053799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Bandaru V, Sunkara S, Wallace SS, Bond JP. A novel human DNA glycosylase that removes oxidative DNA damage and is homologous to Escherichia coli endonuclease VIII. DNA Repair (Amst) 2002;1:517–529. doi: 10.1016/s1568-7864(02)00036-8. [DOI] [PubMed] [Google Scholar]
- 118.Takao M, Kanno S, Kobayashi K, Zhang QM, Yonei S, van der Horst GT, Yasui A. A back-up glycosylase in Nth1 knock-out mice is a functional Nei (endonuclease VIII) homologue. J Biol Chem. 2002;277:42205–42213. doi: 10.1074/jbc.M206884200. [DOI] [PubMed] [Google Scholar]
- 119.Morland I, Rolseth V, Luna L, Rognes T, Bjørås M, Seeberg E. Human DNA glycosylases of the bacterial Fpg/MutM superfamily: an alternative pathway for the repair of 8-oxoguanine and other oxidation products in DNA. Nucleic Acids Res. 2002;30:4926–4936. doi: 10.1093/nar/gkf618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Grin IR, Zharkov DO. Eukaryotic endonuclease VIII-like proteins: new components of the base excision DNA repair system. Biochemistry (Mosc) 2011;76:80–93. doi: 10.1134/s000629791101010x. [DOI] [PubMed] [Google Scholar]
- 121.Dou H, Mitra S, Hazra TK. Repair of oxidized bases in DNA bubble structures by human DNA glycosylases NEIL1 and NEIL2. J Biol Chem. 2003;278:49679–49684. doi: 10.1074/jbc.M308658200. [DOI] [PubMed] [Google Scholar]
- 122.Hu J, de Souza-Pinto NC, Haraguchi K, Hogue BA, Jaruga P, Greenberg MM, Dizdaroglu M, Bohr VA. Repair of formamidopyrimidines in DNA involves different glycosylases: role of the OGG1, NTH1, and NEIL1 enzymes. J Biol Chem. 2005;280:40544–40551. doi: 10.1074/jbc.M508772200. [DOI] [PubMed] [Google Scholar]
- 123.Rosenquist TA, Zaika E, Fernandes AS, Zharkov DO, Miller H, Grollman AP. The novel DNA glycosylase, NEIL1, protects mammalian cells from radiation-mediated cell death. DNA repair. 2003;2:581–591. doi: 10.1016/s1568-7864(03)00025-9. [DOI] [PubMed] [Google Scholar]
- 124.Katafuchi A, Nakano T, Masaoka A, Terato H, Iwai S, Hanaoka F, Ide H. Differential specificity of human and Escherichia coli endonuclease III and VIII homologues for oxidative base lesions. J Biol Chem. 2004;279:14464–14471. doi: 10.1074/jbc.M400393200. [DOI] [PubMed] [Google Scholar]
- 125.Ocampo-Hafalla MT, Altamirano A, Basu AK, Chan MK, Ocampo JE, Cummings A, Jr, Boorstein RJ, Cunningham RP, Teebor GW. Repair of thymine glycol by hNth1 and hNeil1 is modulated by base pairing and cis-trans epimerization. DNA Repair (Amst) 2006;5:444–454. doi: 10.1016/j.dnarep.2005.12.004. [DOI] [PubMed] [Google Scholar]
- 126.Zhang QM, Yonekura S, Takao M, Yasui A, Sugiyama H, Yonei S. DNA glycosylase activities for thymine residues oxidized in the methyl group are functions of the hNEIL1 and hNTH1 enzymes in human cells. DNA Repair (Amst) 2005;4:71–79. doi: 10.1016/j.dnarep.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 127.Zhao X, Krishnamurthy N, Burrows CJ, David SS. Mutation versus repair: NEIL1 removal of hydantoin lesions in single-stranded, bulge, bubble, and duplex DNA contexts. Biochemistry. 2010;49:1658–1666. doi: 10.1021/bi901852q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Krishnamurthy N, Zhao X, Burrows CJ, David SS. Superior removal of hydantoin lesions relative to other oxidized bases by the human DNA glycosylase hNEIL1. Biochemistry. 2008;47:7137–7146. doi: 10.1021/bi800160s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Liu M, Bandaru V, Bond JP, Jaruga P, Zhao X, Christov PP, Burrows CJ, Rizzo CJ, Dizdaroglu M, Wallace SS. The mouse ortholog of NEIL3 is a functional DNA glycosylase in vitro and in vivo. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:4925–4930. doi: 10.1073/pnas.0908307107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Hailer MK, Slade PG, Martin BD, Rosenquist TA, Sugden KD. Recognition of the oxidized lesions spiroiminodihydantoin and guanidinohydantoin in DNA by the mammalian base excision repair glycosylases NEIL1 and NEIL2. DNA Repair (Amst) 2005;4:41–50. doi: 10.1016/j.dnarep.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 131.Parsons JL, Zharkov DO, Dianov GL. NEIL1 excises 3′ end proximal oxidative DNA lesions resistant to cleavage by NTH1 and OGG1. Nucleic Acids Res. 2005;33:4849–4856. doi: 10.1093/nar/gki816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Parsons JL, Kavli B, Slupphaug G, Dianov GL. NEIL1 is the major DNA glycosylase that processes 5-hydroxyuracil in the proximity of a DNA single-strand break. Biochemistry. 2007;46:4158–4163. doi: 10.1021/bi0622569. [DOI] [PubMed] [Google Scholar]
- 133.Couve S, Mace-Aime G, Rosselli F, Saparbaev MK. The human oxidative DNA glycosylase NEIL1 excises psoralen-induced interstrand DNA cross-links in a three-stranded DNA structure. J Biol Chem. 2009;284:11963–11970. doi: 10.1074/jbc.M900746200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Doublié S, Bandaru V, Bond JP, Wallace SS. The crystal structure of human endonuclease VIII-like 1 (NEIL1) reveals a zincless finger motif required for glycosylase activity. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:10284–10289. doi: 10.1073/pnas.0402051101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Imamura K, Wallace SS, Doublié S. Structural characterization of a viral NEIL1 ortholog unliganded and bound to abasic site-containing DNA. J Biol Chem. 2009;284:26174–26183. doi: 10.1074/jbc.M109.021907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Imamura K, Averill A, Wallace SS, Doublie S. Structural characterization of viral ortholog of human DNA glycosylase NEIL1 bound to thymine glycol or 5-hydroxyuracil-containing DNA. The Journal of biological chemistry. 2012;287:4288–4298. doi: 10.1074/jbc.M111.315309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Jia L, Shafirovich V, Geacintov NE, Broyde S. Lesion specificity in the base excision repair enzyme hNeil1: modeling and dynamics studies. Biochemistry. 2007;46:5305–5314. doi: 10.1021/bi062269m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Hegde ML, Theriot CA, Das A, Hegde PM, Guo Z, Gary RK, Hazra TK, Shen B, Mitra S. Physical and functional interaction between human oxidized base-specific DNA glycosylase NEIL1 and flap endonuclease 1. J Biol Chem. 2008;283:27028–27037. doi: 10.1074/jbc.M802712200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Dou H, Theriot CA, Das A, Hegde ML, Matsumoto Y, Boldogh I, Hazra TK, Bhakat KK, Mitra S. Interaction of the human DNA glycosylase NEIL1 with proliferating cell nuclear antigen. The potential for replication-associated repair of oxidized bases in mammalian genomes. J Biol Chem. 2008;283:3130–3140. doi: 10.1074/jbc.M709186200. [DOI] [PubMed] [Google Scholar]
- 140.Muftuoglu M, de Souza-Pinto NC, Dogan A, Aamann M, Stevnsner T, Rybanska I, Kirkali G, Dizdaroglu M, Bohr VA. Cockayne syndrome group B protein stimulates repair of formamidopyrimidines by NEIL1 DNA glycosylase. J Biol Chem. 2009;284:9270–9279. doi: 10.1074/jbc.M807006200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Yeo J, Goodman RA, Schirle NT, David SS, Beal PA. RNA editing changes the lesion specificity for the DNA repair enzyme NEIL1. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:20715–20719. doi: 10.1073/pnas.1009231107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Wiederhold L, Leppard JB, Kedar P, Karimi-Busheri F, Rasouli-Nia A, Weinfeld M, Tomkinson AE, Izumi T, Prasad R, Wilson SH, Mitra S, Hazra TK. AP endonuclease-independent DNA base excision repair in human cells. Mol Cell. 2004;15:209–220. doi: 10.1016/j.molcel.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 143.Nghiem Y, Cabrera M, Cupples CG, Miller JH. The mutY gene: a mutator locus in Escherichia coli that generates G.C----T.A transversions. Proceedings of the National Academy of Sciences of the United States of America. 1988;85:2709–2713. doi: 10.1073/pnas.85.8.2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Tominaga Y, Ushijima Y, Tsuchimoto D, Mishima M, Shirakawa M, Hirano S, Sakumi K, Nakabeppu Y. MUTYH prevents OGG1 or APEX1 from inappropriately processing its substrate or reaction product with its C-terminal domain. Nucleic Acids Res. 2004;32:3198–3211. doi: 10.1093/nar/gkh642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Michaels ML, Miller JH. The GO system protects organisms from the mutagenic effect of the spontaneous lesion 8-hydroxyguanine (7,8-dihydro-8-oxoguanine) J Bacteriol. 1992;174:6321–6325. doi: 10.1128/jb.174.20.6321-6325.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Guan Y, Manuel RC, Arvai AS, Parikh SS, Mol CD, Miller JH, Lloyd S, Tainer JA. MutY catalytic core, mutant and bound adenine structures define specificity for DNA repair enzyme superfamily. Nat Struct Biol. 1998;5:1058–1064. doi: 10.1038/4168. [DOI] [PubMed] [Google Scholar]
- 147.Manuel RC, Hitomi K, Arvai AS, House PG, Kurtz AJ, Dodson ML, McCullough AK, Tainer JA, Lloyd RS. Reaction intermediates in the catalytic mechanism of Escherichia coli MutY DNA glycosylase. J Biol Chem. 2004;279:46930–46939. doi: 10.1074/jbc.M403944200. [DOI] [PubMed] [Google Scholar]
- 148.McCann JA, Berti PJ. Transition-state analysis of the DNA repair enzyme MutY. Journal of the American Chemical Society. 2008;130:5789–5797. doi: 10.1021/ja711363s. [DOI] [PubMed] [Google Scholar]
- 149.Fromme JC, Banerjee A, Huang SJ, Verdine GL. Structural basis for removal of adenine mispaired with 8-oxoguanine by MutY adenine DNA glycosylase. Nature. 2004;427:652–656. doi: 10.1038/nature02306. [DOI] [PubMed] [Google Scholar]
- 150.Lee S, Verdine GL. Atomic substitution reveals the structural basis for substrate adenine recognition and removal by adenine DNA glycosylase. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:18497–18502. doi: 10.1073/pnas.0902908106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Chang PW, Madabushi A, Lu AL. Insights into the role of Val45 and Gln182 of Escherichia coli MutY in DNA substrate binding and specificity. BMC Biochem. 2009;10:19. doi: 10.1186/1471-2091-10-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Brinkmeyer MK, Pope MA, David SS. Catalytic contributions of key residues in the adenine glycosylase MutY revealed by pH-dependent kinetics and cellular repair assays. Chemistry & biology. 2012;19:276–286. doi: 10.1016/j.chembiol.2011.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Al-Tassan N, Chmiel NH, Maynard J, Fleming N, Livingston AL, Williams GT, Hodges AK, Davies DR, David SS, Sampson JR, Cheadle JP. Inherited variants of MYH associated with somatic G:C-->T:A mutations in colorectal tumors. Nat Genet. 2002;30:227–232. doi: 10.1038/ng828. [DOI] [PubMed] [Google Scholar]
- 154.Chmiel NH, Livingston AL, David SS. Insight into the functional consequences of inherited variants of the hMYH adenine glycosylase associated with colorectal cancer: complementation assays with hMYH variants and pre-steady-state kinetics of the corresponding mutated E.coli enzymes. J Mol Biol. 2003;327:431–443. doi: 10.1016/s0022-2836(03)00124-4. [DOI] [PubMed] [Google Scholar]
- 155.Livingston AL, Kundu S, Henderson Pozzi M, Anderson DW, David SS. Insight into the roles of tyrosine 82 and glycine 253 in the Escherichia coli adenine glycosylase MutY. Biochemistry. 2005;44:14179–14190. doi: 10.1021/bi050976u. [DOI] [PubMed] [Google Scholar]
- 156.Livingston AL, O’Shea VL, Kim T, Kool ET, David SS. Unnatural substrates reveal the importance of 8-oxoguanine for in vivo mismatch repair by MutY. Nat Chem Biol. 2008;4:51–58. doi: 10.1038/nchembio.2007.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Luncsford PJ, Chang DY, Shi G, Bernstein J, Madabushi A, Patterson DN, Lu AL, Toth EA. A structural hinge in eukaryotic MutY homologues mediates catalytic activity and Rad9-Rad1-Hus1 checkpoint complex interactions. J Mol Biol. 2010;403:351–370. doi: 10.1016/j.jmb.2010.08.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Lu AL, Bai H, Shi G, Chang DY. MutY and MutY homologs (MYH) in genome maintenance. Frontiers in bioscience: a journal and virtual library. 2006;11:3062–3080. doi: 10.2741/2033. [DOI] [PubMed] [Google Scholar]
- 159.Chang DY, Lu AL. Interaction of checkpoint proteins Hus1/Rad1/Rad9 with DNA base excision repair enzyme MutY homolog in fission yeast, Schizosaccharomyces pombe. J Biol Chem. 2005;280:408–417. doi: 10.1074/jbc.M406800200. [DOI] [PubMed] [Google Scholar]
- 160.Shi G, Chang DY, Cheng CC, Guan X, Venclovas C, Lu AL. Physical and functional interactions between MutY glycosylase homologue (MYH) and checkpoint proteins Rad9-Rad1-Hus1. Biochem J. 2006;400:53–62. doi: 10.1042/BJ20060774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Lawley PD, Brookes P. The Action of Alkylating Agents on Deoxyribonucleic Acid in Relation to Biological Effects of the Alkylating Agents. Exp Cell Res. 1963;24(SUPPL9):512–520. doi: 10.1016/0014-4827(63)90291-x. [DOI] [PubMed] [Google Scholar]
- 162.Gates KS. An overview of chemical processes that damage cellular DNA: spontaneous hydrolysis, alkylation, and reactions with radicals. Chem Res Toxicol. 2009;22:1747–1760. doi: 10.1021/tx900242k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Sedgwick B. Repairing DNA-methylation damage. Nat Rev Mol Cell Biol. 2004;5:148–157. doi: 10.1038/nrm1312. [DOI] [PubMed] [Google Scholar]
- 164.Gates KS, Nooner T, Dutta S. Biologically relevant chemical reactions of N7-alkylguanine residues in DNA. Chem Res Toxicol. 2004;17:839–856. doi: 10.1021/tx049965c. [DOI] [PubMed] [Google Scholar]
- 165.Margison GP, O’Connor PJ. Biological implications of the instability of the N-glycosidic bone of 3-methyldeoxyadenosine in DNA. Biochim Biophys Acta. 1973;331:349–356. doi: 10.1016/0005-2787(73)90021-x. [DOI] [PubMed] [Google Scholar]
- 166.Barbin A. Etheno-adduct-forming chemicals: from mutagenicity testing to tumor mutation spectra. Mutation research. 2000;462:55–69. doi: 10.1016/s1383-5742(00)00014-4. [DOI] [PubMed] [Google Scholar]
- 167.Nair J, Barbin A, Velic I, Bartsch H. Etheno DNA-base adducts from endogenous reactive species. Mutation research. 1999;424:59–69. doi: 10.1016/s0027-5107(99)00008-1. [DOI] [PubMed] [Google Scholar]
- 168.Boiteux S, Huisman O, Laval J. 3-Methyladenine residues in DNA induce the SOS function sfiA in Escherichia coli. Embo J. 1984;3:2569–2573. doi: 10.1002/j.1460-2075.1984.tb02175.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Larson K, Sahm J, Shenkar R, Strauss B. Methylation-induced blocks to in vitro DNA replication. Mutat Res. 1985;150:77–84. doi: 10.1016/0027-5107(85)90103-4. [DOI] [PubMed] [Google Scholar]
- 170.Plosky BS, Frank EG, Berry DA, Vennall GP, McDonald JP, Woodgate R. Eukaryotic Y-family polymerases bypass a 3-methyl-2′-deoxyadenosine analog in vitro and methyl methanesulfonate-induced DNA damage in vivo. Nucleic Acids Res. 2008;36:2152–2162. doi: 10.1093/nar/gkn058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Fronza G, Gold B. The biological effects of N3-methyladenine. J Cell Biochem. 2004;91:250–257. doi: 10.1002/jcb.10698. [DOI] [PubMed] [Google Scholar]
- 172.Karran P, Lindahl T. Hypoxanthine in deoxyribonucleic acid: generation by heat-induced hydrolysis of adenine residues and release in free form by a deoxyribonucleic acid glycosylase from calf thymus. Biochemistry. 1980;19:6005–6011. doi: 10.1021/bi00567a010. [DOI] [PubMed] [Google Scholar]
- 173.Brent TP. Partial purification and characterization of a human 3-methyladenine-DNA glycosylase. Biochemistry. 1979;18:911–916. doi: 10.1021/bi00572a028. [DOI] [PubMed] [Google Scholar]
- 174.Chen J, Derfler B, Samson L. Saccharomyces cerevisiae 3-methyladenine DNA glycosylase has homology to the AlkA glycosylase of E. coli and is induced in response to DNA alkylation damage. Embo J. 1990;9:4569–4575. doi: 10.1002/j.1460-2075.1990.tb07910.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Berdal KG, Bjørås M, Bjelland S, Seeberg E. Cloning and expression in Escherichia coli of a gene for an alkylbase DNA glycosylase from Saccharomyces cerevisiae; a homologue to the bacterial alkA gene. Embo J. 1990;9:4563–4568. doi: 10.1002/j.1460-2075.1990.tb07909.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Memisoglu A, Samson L. Cloning and characterization of a cDNA encoding a 3-methyladenine DNA glycosylase from the fission yeast Schizosaccharomyces pombe. Gene. 1996;177:229–235. doi: 10.1016/0378-1119(96)00308-3. [DOI] [PubMed] [Google Scholar]
- 177.Riazuddin S, Lindahl T. Properties of 3-methyladenine-DNA glycosylase from Escherichia coli. Biochemistry. 1978;17:2110–2118. doi: 10.1021/bi00604a014. [DOI] [PubMed] [Google Scholar]
- 178.Thomas L, Yang CH, Goldthwait DA. Two DNA glycosylases in Escherichia coli which release primarily 3-methyladenine. Biochemistry. 1982;21:1162–1169. doi: 10.1021/bi00535a009. [DOI] [PubMed] [Google Scholar]
- 179.Mansfield C, Kerins SM, McCarthy TV. Characterisation of Archaeglobus fulgidus AlkA hypoxanthine DNA glycosylase activity. FEBS Lett. 2003;540:171–175. doi: 10.1016/s0014-5793(03)00257-6. [DOI] [PubMed] [Google Scholar]
- 180.Leiros I, Nabong MP, Grosvik K, Ringvoll J, Haugland GT, Uldal L, Reite K, Olsbu IK, Knaevelsrud I, Moe E, Andersen OA, Birkeland NK, Ruoff P, Klungland A, Bjelland S. Structural basis for enzymatic excision of N1-methyladenine and N3-methylcytosine from DNA. Embo J. 2007;26:2206–2217. doi: 10.1038/sj.emboj.7601662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Moe E, Hall DR, Leiros I, Monsen VT, Timmins J, McSweeney S. Structure-function studies of an unusual 3-methyladenine DNA glycosylase II (AlkA) from Deinococcus radiodurans. Acta Crystallogr D Biol Crystallogr. 2012;68:703–712. doi: 10.1107/S090744491200947X. [DOI] [PubMed] [Google Scholar]
- 182.Begley TJ, Haas BJ, Noel J, Shekhtman A, Williams WA, Cunningham RP. A new member of the endonuclease III family of DNA repair enzymes that removes methylated purines from DNA. Curr Biol. 1999;9:653–656. doi: 10.1016/s0960-9822(99)80288-7. [DOI] [PubMed] [Google Scholar]
- 183.O’Rourke EJ, Chevalier C, Boiteux S, Labigne A, Ielpi L, Radicella JP. A novel 3-methyladenine DNA glycosylase from helicobacter pylori defines a new class within the endonuclease III family of base excision repair glycosylases. J Biol Chem. 2000;275:20077–20083. doi: 10.1074/jbc.M001071200. [DOI] [PubMed] [Google Scholar]
- 184.Alseth I, Rognes T, Lindback T, Solberg I, Robertsen K, Kristiansen KI, Mainieri D, Lillehagen L, Kolsto AB, Bjørås M. A new protein superfamily includes two novel 3-methyladenine DNA glycosylases from Bacillus cereus, AlkC and AlkD. Mol Microbiol. 2006;59:1602–1609. doi: 10.1111/j.1365-2958.2006.05044.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Saparbaev M, Kleibl K, Laval J. Escherichia coli, Saccharomyces cerevisiae, rat and human 3-methyladenine DNA glycosylases repair 1,N6-ethenoadenine when present in DNA. Nucleic Acids Res. 1995;23:3750–3755. doi: 10.1093/nar/23.18.3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.McCarthy TV, Karran P, Lindahl T. Inducible repair of O-alkylated DNA pyrimidines in Escherichia coli. Embo J. 1984;3:545–550. doi: 10.1002/j.1460-2075.1984.tb01844.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Bjelland S, Birkeland NK, Benneche T, Volden G, Seeberg E. DNA glycosylase activities for thymine residues oxidized in the methyl group are functions of the AlkA enzyme in Escherichia coli. J Biol Chem. 1994;269:30489–30495. [PubMed] [Google Scholar]
- 188.Birkeland NK, Anensen H, Knaevelsrud I, Kristoffersen W, Bjørås M, Robb FT, Klungland A, Bjelland S. Methylpurine DNA glycosylase of the hyperthermophilic archaeon Archaeoglobus fulgidus. Biochemistry. 2002;41:12697–12705. doi: 10.1021/bi020334w. [DOI] [PubMed] [Google Scholar]
- 189.Lingaraju GM, Kartalou M, Meira LB, Samson LD. Substrate specificity and sequence-dependent activity of the Saccharomyces cerevisiae 3-methyladenine DNA glycosylase (Mag) DNA repair. 2008;7:970–982. doi: 10.1016/j.dnarep.2008.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Alseth I, Osman F, Korvald H, Tsaneva I, Whitby MC, Seeberg E, Bjørås M. Biochemical characterization and DNA repair pathway interactions of Mag1-mediated base excision repair in Schizosaccharomyces pombe. Nucleic Acids Res. 2005;33:1123–1131. doi: 10.1093/nar/gki259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Adhikary S, Eichman BF. Analysis of substrate specificity of Schizosaccharomyces pombe Mag1 alkylpurine DNA glycosylase. EMBO Rep. 2011;12:1286–1292. doi: 10.1038/embor.2011.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Bjelland S, Bjørås M, Seeberg E. Excision of 3-methylguanine from alkylated DNA by 3-methyladenine DNA glycosylase I of Escherichia coli. Nucleic Acids Res. 1993;21:2045–2049. doi: 10.1093/nar/21.9.2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Rubinson EH, Gowda AS, Spratt TE, Gold B, Eichman BF. An unprecedented nucleic acid capture mechanism for excision of DNA damage. Nature. 2010;468:406–411. doi: 10.1038/nature09428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Labahn J, Scharer OD, Long A, Ezaz-Nikpay K, Verdine GL, Ellenberger TE. Structural basis for the excision repair of alkylation-damaged DNA. Cell. 1996;86:321–329. doi: 10.1016/s0092-8674(00)80103-8. [DOI] [PubMed] [Google Scholar]
- 195.Lau AY, Scharer OD, Samson L, Verdine GL, Ellenberger T. Crystal structure of a human alkylbase-DNA repair enzyme complexed to DNA: mechanisms for nucleotide flipping and base excision. Cell. 1998;95:249–258. doi: 10.1016/s0092-8674(00)81755-9. [DOI] [PubMed] [Google Scholar]
- 196.Hollis T, Ichikawa Y, Ellenberger T. DNA bending and a flip-out mechanism for base excision by the helix-hairpin-helix DNA glycosylase, Escherichia coli AlkA. Embo J. 2000;19:758–766. doi: 10.1093/emboj/19.4.758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Wyatt MD, Allan JM, Lau AY, Ellenberger TE, Samson LD. 3-methyladenine DNA glycosylases: structure, function, and biological importance. Bioessays. 1999;21:668–676. doi: 10.1002/(SICI)1521-1878(199908)21:8<668::AID-BIES6>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 198.McCullough AK, Dodson ML, Lloyd RS. Initiation of base excision repair: glycosylase mechanisms and structures. Annual review of biochemistry. 1999;68:255–285. doi: 10.1146/annurev.biochem.68.1.255. [DOI] [PubMed] [Google Scholar]
- 199.O’Brien PJ, Ellenberger T. Dissecting the broad substrate specificity of human 3-methyladenine-DNA glycosylase. J Biol Chem. 2004;279:9750–9757. doi: 10.1074/jbc.M312232200. [DOI] [PubMed] [Google Scholar]
- 200.Saparbaev M, Langouet S, Privezentzev CV, Guengerich FP, Cai H, Elder RH, Laval J. 1,N(2)-ethenoguanine, a mutagenic DNA adduct, is a primary substrate of Escherichia coli mismatch-specific uracil-DNA glycosylase and human alkylpurine-DNA-N-glycosylase. The Journal of biological chemistry. 2002;277:26987–26993. doi: 10.1074/jbc.M111100200. [DOI] [PubMed] [Google Scholar]
- 201.Lee CY, Delaney JC, Kartalou M, Lingaraju GM, Maor-Shoshani A, Essigmann JM, Samson LD. Recognition and processing of a new repertoire of DNA substrates by human 3-methyladenine DNA glycosylase (AAG) Biochemistry. 2009;48:1850–1861. doi: 10.1021/bi8018898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Lau AY, Wyatt MD, Glassner BJ, Samson LD, Ellenberger T. Molecular basis for discriminating between normal and damaged bases by the human alkyladenine glycosylase, AAG. Proc Natl Acad Sci U S A. 2000;97:13573–13578. doi: 10.1073/pnas.97.25.13573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Lyons DM, O’Brien PJ. Efficient recognition of an unpaired lesion by a DNA repair glycosylase. J Am Chem Soc. 2009;131:17742–17743. doi: 10.1021/ja908378y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Klapacz J, Lingaraju GM, Guo HH, Shah D, Moar-Shoshani A, Loeb LA, Samson LD. Frameshift mutagenesis and microsatellite instability induced by human alkyladenine DNA glycosylase. Mol Cell. 2010;37:843–853. doi: 10.1016/j.molcel.2010.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Wolfe AE, O’Brien PJ. Kinetic mechanism for the flipping and excision of 1,N(6)-ethenoadenine by human alkyladenine DNA glycosylase. Biochemistry. 2009;48:11357–11369. doi: 10.1021/bi9015082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Hendershot JM, Wolfe AE, O’Brien PJ. Substitution of active site tyrosines with tryptophan alters the free energy for nucleotide flipping by human alkyladenine DNA glycosylase. Biochemistry. 2011;50:1864–1874. doi: 10.1021/bi101856a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Connor EE, Wyatt MD. Active-site clashes prevent the human 3-methyladenine DNA glycosylase from improperly removing bases. Chem Biol. 2002;9:1033–1041. doi: 10.1016/s1074-5521(02)00215-6. [DOI] [PubMed] [Google Scholar]
- 208.O’Brien PJ, Ellenberger T. Human alkyladenine DNA glycosylase uses acid-base catalysis for selective excision of damaged purines. Biochemistry. 2003;42:12418–12429. doi: 10.1021/bi035177v. [DOI] [PubMed] [Google Scholar]
- 209.Rutledge LR, Wetmore SD. Modeling the chemical step utilized by human alkyladenine DNA glycosylase: a concerted mechanism AIDS in selectively excising damaged purines. Journal of the American Chemical Society. 2011;133:16258–16269. doi: 10.1021/ja207181c. [DOI] [PubMed] [Google Scholar]
- 210.Lingaraju GM, Davis CA, Setser JW, Samson LD, Drennan CL. Structural basis for the inhibition of human alkyladenine DNA glycosylase (AAG) by 3,N4-ethenocytosine-containing DNA. The Journal of biological chemistry. 2011;286:13205–13213. doi: 10.1074/jbc.M110.192435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Saparbaev M, Laval J. 3,N4-ethenocytosine, a highly mutagenic adduct, is a primary substrate for Escherichia coli double-stranded uracil-DNA glycosylase and human mismatch-specific thymine-DNA glycosylase. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:8508–8513. doi: 10.1073/pnas.95.15.8508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Gros L, Ishchenko AA, Saparbaev M. Enzymology of repair of etheno-adducts. Mutation research. 2003;531:219–229. doi: 10.1016/j.mrfmmm.2003.07.008. [DOI] [PubMed] [Google Scholar]
- 213.Gros L, Maksimenko AV, Privezentzev CV, Laval J, Saparbaev MK. Hijacking of the human alkyl-N-purine-DNA glycosylase by 3,N4-ethenocytosine, a lipid peroxidation-induced DNA adduct. The Journal of biological chemistry. 2004;279:17723–17730. doi: 10.1074/jbc.M314010200. [DOI] [PubMed] [Google Scholar]
- 214.Setser J, Lingaraju G, Davis C, Samson L, Drennan C. Searching for DNA lesions: Structural evidence for lower and higher-affinity DNA binding conformations of human alkyladenine DNA glycosylase (AAG) Biochemistry. 2011 doi: 10.1021/bi201484k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Baldwin MR, O’Brien PJ. Nonspecific DNA binding and coordination of the first two steps of base excision repair. Biochemistry. 2010;49:7879–7891. doi: 10.1021/bi100889r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Admiraal SJ, O’Brien PJ. N-glycosyl bond formation catalyzed by human alkyladenine DNA glycosylase. Biochemistry. 2010;49:9024–9026. doi: 10.1021/bi101380d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Baldwin MR, O’Brien PJ. Human AP endonuclease 1 stimulates multiple-turnover base excision by alkyladenine DNA glycosylase. Biochemistry. 2009;48:6022–6033. doi: 10.1021/bi900517y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Waters TR, Gallinari P, Jiricny J, Swann PF. Human thymine DNA glycosylase binds to apurinic sites in DNA but is displaced by human apurinic endonuclease 1. J Biol Chem. 1999;274:67–74. doi: 10.1074/jbc.274.1.67. [DOI] [PubMed] [Google Scholar]
- 219.Fitzgerald ME, Drohat AC. Coordinating the initial steps of base excision repair. Apurinic/apyrimidinic endonuclease 1 actively stimulates thymine DNA glycosylase by disrupting the product complex. The Journal of biological chemistry. 2008;283:32680–32690. doi: 10.1074/jbc.M805504200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Sidorenko VS, Nevinsky GA, Zharkov DO. Mechanism of interaction between human 8-oxoguanine-DNA glycosylase and AP endonuclease. DNA Repair (Amst) 2007;6:317–328. doi: 10.1016/j.dnarep.2006.10.022. [DOI] [PubMed] [Google Scholar]
- 221.Hedglin M, O’Brien PJ. Human alkyladenine DNA glycosylase employs a processive search for DNA damage. Biochemistry. 2008;47:11434–11445. doi: 10.1021/bi801046y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Rubinson EH, Metz AH, O’Quin J, Eichman BF. A new protein architecture for processing alkylation damaged DNA: the crystal structure of DNA glycosylase AlkD. J Mol Biol. 2008;381:13–23. doi: 10.1016/j.jmb.2008.05.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Aamodt RM, Falnes PO, Johansen RF, Seeberg E, Bjoras M. The Bacillus subtilis counterpart of the mammalian 3-methyladenine DNA glycosylase has hypoxanthine and 1,N6-ethenoadenine as preferred substrates. The Journal of biological chemistry. 2004;279:13601–13606. doi: 10.1074/jbc.M314277200. [DOI] [PubMed] [Google Scholar]
- 224.Bowman BR, Lee S, Wang S, Verdine GL. Structure of Escherichia coli AlkA in complex with undamaged DNA. J Biol Chem. 2010;285:35783–35791. doi: 10.1074/jbc.M110.155663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Rubinson EH, Adhikary S, Eichman BF. Structural Studies of Alkylpurine DNA Glycosylases. In: Stone MP, editor. ACS Symposium Series: Structural Biology of DNA Damage and Repair. Vol. 1041. American Chemical Society; Washingon, D.C: 2010. pp. 29–45. [Google Scholar]
- 226.Metz AH, Hollis T, Eichman BF. DNA damage recognition and repair by 3-methyladenine DNA glycosylase I (TAG) Embo J. 2007;26:2411–2420. doi: 10.1038/sj.emboj.7601649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Yamagata Y, Kato M, Odawara K, Tokuno Y, Nakashima Y, Matsushima N, Yasumura K, Tomita K, Ihara K, Fujii Y, Nakabeppu Y, Sekiguchi M, Fujii S. Three-dimensional structure of a DNA repair enzyme, 3-methyladenine DNA glycosylase II, from Escherichia coli. Cell. 1996;86:311–319. doi: 10.1016/s0092-8674(00)80102-6. [DOI] [PubMed] [Google Scholar]
- 228.Hollis T, Lau A, Ellenberger T. Structural studies of human alkyladenine glycosylase and E. coli 3-methyladenine glycosylase. Mutat Res. 2000;460:201–210. doi: 10.1016/s0921-8777(00)00027-6. [DOI] [PubMed] [Google Scholar]
- 229.Zhao B, O’Brien PJ. Kinetic mechanism for the excision of hypoxanthine by Escherichia coli AlkA and evidence for binding to DNA ends. Biochemistry. 2011;50:4350–4359. doi: 10.1021/bi200232c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Bowman BR, Lee S, Wang S, Verdine GL. Structure of the E. coli DNA glycosylase AlkA bound to the ends of duplex DNA: a system for the structure determination of lesion-containing DNA. Structure. 2008;16:1166–1174. doi: 10.1016/j.str.2008.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Saparbaev M, Laval J. Excision of hypoxanthine from DNA containing dIMP residues by the Escherichia coli, yeast, rat, and human alkylpurine DNA glycosylases. Proc Natl Acad Sci U S A. 1994;91:5873–5877. doi: 10.1073/pnas.91.13.5873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Bjørås M, Klungland A, Johansen RF, Seeberg E. Purification and properties of the alkylation repair DNA glycosylase encoded the MAG gene from Saccharomyces cerevisiae. Biochemistry. 1995;34:4577–4582. doi: 10.1021/bi00014a010. [DOI] [PubMed] [Google Scholar]
- 233.Berdal KG, Johansen RF, Seeberg E. Release of normal bases from intact DNA by a native DNA repair enzyme. Embo J. 1998;17:363–367. doi: 10.1093/emboj/17.2.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Memisoglu A, Samson L. Base excision repair in yeast and mammals. Mutat Res. 2000;451:39–51. doi: 10.1016/s0027-5107(00)00039-7. [DOI] [PubMed] [Google Scholar]
- 235.Memisoglu A, Samson L. Contribution of base excision repair, nucleotide excision repair, and DNA recombination to alkylation resistance of the fission yeast Schizosaccharomyces pombe. J Bacteriol. 2000;182:2104–2112. doi: 10.1128/jb.182.8.2104-2112.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Chen J, Samson L. Induction of S.cerevisiae MAG 3-methyladenine DNA glycosylase transcript levels in response to DNA damage. Nucleic acids research. 1991;19:6427–6432. doi: 10.1093/nar/19.23.6427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Eichman BF, O’Rourke EJ, Radicella JP, Ellenberger T. Crystal structures of 3-methyladenine DNA glycosylase MagIII and the recognition of alkylated bases. Embo J. 2003;22:4898–4909. doi: 10.1093/emboj/cdg505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Drohat AC, Kwon K, Krosky DJ, Stivers JT. 3-Methyladenine DNA glycosylase I is an unexpected helix-hairpin-helix superfamily member. Nat Struct Biol. 2002;9:659–664. doi: 10.1038/nsb829. [DOI] [PubMed] [Google Scholar]
- 239.Kwon K, Cao C, Stivers JT. A novel zinc snap motif conveys structural stability to 3-methyladenine DNA glycosylase I. J Biol Chem. 2003;278:19442–19446. doi: 10.1074/jbc.M300934200. [DOI] [PubMed] [Google Scholar]
- 240.Cao C, Kwon K, Jiang YL, Drohat AC, Stivers JT. Solution structure and base perturbation studies reveal a novel mode of alkylated base recognition by 3-methyladenine DNA glycosylase I. J Biol Chem. 2003;278:48012–48020. doi: 10.1074/jbc.M307500200. [DOI] [PubMed] [Google Scholar]
- 241.Zhu X, Yan X, Carter LG, Liu H, Graham S, Coote PJ, Naismith J. A model for 3-methyladenine recognition by 3-methyladenine DNA glycosylase I (TAG) from Staphylococcus aureus. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2012;68:610–615. doi: 10.1107/S1744309112016363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Dalhus B, Helle IH, Backe PH, Alseth I, Rognes T, Bjørås M, Laerdahl JK. Structural insight into repair of alkylated DNA by a new superfamily of DNA glycosylases comprising HEAT-like repeats. Nucleic Acids Res. 2007;35:2451–2459. doi: 10.1093/nar/gkm039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Jiang YL, Kwon K, Stivers JT. Turning On uracil-DNA glycosylase using a pyrene nucleotide switch. J Biol Chem. 2001;276:42347–42354. doi: 10.1074/jbc.M106594200. [DOI] [PubMed] [Google Scholar]
- 244.Dinner AR, Blackburn GM, Karplus M. Uracil-DNA glycosylase acts by substrate autocatalysis. Nature. 2001;413:752–755. doi: 10.1038/35099587. [DOI] [PubMed] [Google Scholar]
- 245.Jiang YL, Ichikawa Y, Song F, Stivers JT. Powering DNA repair through substrate electrostatic interactions. Biochemistry. 2003;42:1922–1929. doi: 10.1021/bi027014x. [DOI] [PubMed] [Google Scholar]
- 246.Parker JB, Stivers JT. Uracil DNA glycosylase: revisiting substrate-assisted catalysis by DNA phosphate anions. Biochemistry. 2008;47:8614–8622. doi: 10.1021/bi800854g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Coulondre C, Miller JH, Farabaugh PJ, Gilbert W. Molecular basis of base substitution hotspots in Escherichia coli. Nature. 1978;274:775–780. doi: 10.1038/274775a0. [DOI] [PubMed] [Google Scholar]
- 248.Duncan BK, Miller JH. Mutagenic deamination of cytosine residues in DNA. Nature. 1980;287:560–561. doi: 10.1038/287560a0. [DOI] [PubMed] [Google Scholar]
- 249.Lindahl T. An N-glycosidase from Escherichia coli that releases free uracil from DNA containing deaminated cytosine residues. Proc Natl Acad Sci U S A. 1974;71:3649–3653. doi: 10.1073/pnas.71.9.3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Kavli B, Sundheim O, Akbari M, Otterlei M, Nilsen H, Skorpen F, Aas PA, Hagen L, Krokan HE, Slupphaug G. hUNG2 is the major repair enzyme for removal of uracil from U:A matches, U:G mismatches, and U in single-stranded DNA, with hSMUG1 as a broad specificity backup. J Biol Chem. 2002;277:39926–39936. doi: 10.1074/jbc.M207107200. [DOI] [PubMed] [Google Scholar]
- 251.Haushalter KA, Todd Stukenberg MW, Kirschner MW, Verdine GL. Identification of a new uracil-DNA glycosylase family by expression cloning using synthetic inhibitors. Curr Biol. 1999;9:174–185. doi: 10.1016/s0960-9822(99)80087-6. [DOI] [PubMed] [Google Scholar]
- 252.Gallinari P, Jiricny J. A new class of uracil-DNA glycosylases related to human thymine-DNA glycosylase. Nature. 1996;383:735–738. doi: 10.1038/383735a0. [DOI] [PubMed] [Google Scholar]
- 253.Brown TC, Jiricny J. A specific mismatch repair event protects mammalian cells from loss of 5-methylcytosine. Cell. 1987;50:945–950. doi: 10.1016/0092-8674(87)90521-6. [DOI] [PubMed] [Google Scholar]
- 254.Hendrich B, Hardeland U, Ng HH, Jiricny J, Bird A. The thymine glycosylase MBD4 can bind to the product of deamination at methylated CpG sites. Nature. 1999;401:301–304. doi: 10.1038/45843. [DOI] [PubMed] [Google Scholar]
- 255.Begley TJ, Cunningham RP. Methanobacterium thermoformicicum thymine DNA mismatch glycosylase: conversion of an N-glycosylase to an AP lyase. Protein Eng. 1999;12:333–340. doi: 10.1093/protein/12.4.333. [DOI] [PubMed] [Google Scholar]
- 256.Wu P, Qiu C, Sohail A, Zhang X, Bhagwat AS, Cheng X. Mismatch repair in methylated DNA. Structure and activity of the mismatch-specific thymine glycosylase domain of methyl-CpG-binding protein MBD4. The Journal of biological chemistry. 2003;278:5285–5291. doi: 10.1074/jbc.M210884200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Mol CD, Arvai AS, Begley TJ, Cunningham RP, Tainer JA. Structure and activity of a thermostable thymine-DNA glycosylase: evidence for base twisting to remove mismatched normal DNA bases. J Mol Biol. 2002;315:373–384. doi: 10.1006/jmbi.2001.5264. [DOI] [PubMed] [Google Scholar]
- 258.Barrett TE, Savva R, Panayotou G, Barlow T, Brown T, Jiricny J, Pearl LH. Crystal structure of a G:T/U mismatch-specific DNA glycosylase: mismatch recognition by complementary-strand interactions. Cell. 1998;92:117–129. doi: 10.1016/s0092-8674(00)80904-6. [DOI] [PubMed] [Google Scholar]
- 259.Barrett TE, Savva R, Barlow T, Brown T, Jiricny J, Pearl LH. Structure of a DNA base-excision product resembling a cisplatin inter-strand adduct. Nature structural biology. 1998;5:697–701. doi: 10.1038/1394. [DOI] [PubMed] [Google Scholar]
- 260.Barrett TE, Scharer OD, Savva R, Brown T, Jiricny J, Verdine GL, Pearl LH. Crystal structure of a thwarted mismatch glycosylase DNA repair complex. Embo J. 1999;18:6599–6609. doi: 10.1093/emboj/18.23.6599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Maiti A, Morgan MT, Pozharski E, Drohat AC. Crystal structure of human thymine DNA glycosylase bound to DNA elucidates sequence-specific mismatch recognition. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:8890–8895. doi: 10.1073/pnas.0711061105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Stivers JT. Extrahelical damaged base recognition by DNA glycosylase enzymes. Chemistry. 2008;14:786–793. doi: 10.1002/chem.200701501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.O’Brien PJ. Catalytic promiscuity and the divergent evolution of DNA repair enzymes. Chem Rev. 2006;106:720–752. doi: 10.1021/cr040481v. [DOI] [PubMed] [Google Scholar]
- 264.Krokan HE, Drablos F, Slupphaug G. Uracil in DNA--occurrence, consequences and repair. Oncogene. 2002;21:8935–8948. doi: 10.1038/sj.onc.1205996. [DOI] [PubMed] [Google Scholar]
- 265.Cortellino S, Xu J, Sannai M, Moore R, Caretti E, Cigliano A, Le Coz M, Devarajan K, Wessels A, Soprano D, Abramowitz LK, Bartolomei MS, Rambow F, Bassi MR, Bruno T, Fanciulli M, Renner C, Klein-Szanto AJ, Matsumoto Y, Kobi D, Davidson I, Alberti C, Larue L, Bellacosa A. Thymine DNA glycosylase is essential for active DNA demethylation by linked deamination-base excision repair. Cell. 2011;146:67–79. doi: 10.1016/j.cell.2011.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Jacobs AL, Schar P. DNA glycosylases: in DNA repair and beyond. Chromosoma. 2011 doi: 10.1007/s00412-011-0347-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Cortazar D, Kunz C, Saito Y, Steinacher R, Schar P. The enigmatic thymine DNA glycosylase. DNA repair. 2007;6:489–504. doi: 10.1016/j.dnarep.2006.10.013. [DOI] [PubMed] [Google Scholar]
- 268.Chan SW, Henderson IR, Jacobsen SE. Gardening the genome: DNA methylation in Arabidopsis thaliana. Nature reviews. Genetics. 2005;6:351–360. doi: 10.1038/nrg1601. [DOI] [PubMed] [Google Scholar]
- 269.Huh JH, Bauer MJ, Hsieh TF, Fischer RL. Cellular programming of plant gene imprinting. Cell. 2008;132:735–744. doi: 10.1016/j.cell.2008.02.018. [DOI] [PubMed] [Google Scholar]
- 270.Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nature reviews. Genetics. 2010;11:204–220. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Goll MG, Bestor TH. Eukaryotic cytosine methyltransferases. Annual review of biochemistry. 2005;74:481–514. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- 272.Cortazar D, Kunz C, Selfridge J, Lettieri T, Saito Y, MacDougall E, Wirz A, Schuermann D, Jacobs AL, Siegrist F, Steinacher R, Jiricny J, Bird A, Schar P. Embryonic lethal phenotype reveals a function of TDG in maintaining epigenetic stability. Nature. 2011;470:419–423. doi: 10.1038/nature09672. [DOI] [PubMed] [Google Scholar]
- 273.Rai K, Huggins IJ, James SR, Karpf AR, Jones DA, Cairns BR. DNA demethylation in zebrafish involves the coupling of a deaminase, a glycosylase, and gadd45. Cell. 2008;135:1201–1212. doi: 10.1016/j.cell.2008.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, Rao A. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Ito S, D’Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C, Zhang Y. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Wu H, Zhang Y. Mechanisms and functions of Tet protein-mediated 5-methylcytosine oxidation. Genes Dev. 2011;25:2436–2452. doi: 10.1101/gad.179184.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Pastor WA, Pape UJ, Huang Y, Henderson HR, Lister R, Ko M, McLoughlin EM, Brudno Y, Mahapatra S, Kapranov P, Tahiliani M, Daley GQ, Liu XS, Ecker JR, Milos PM, Agarwal S, Rao A. Genome-wide mapping of 5-hydroxymethylcytosine in embryonic stem cells. Nature. 2011;473:394–397. doi: 10.1038/nature10102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Ficz G, Branco MR, Seisenberger S, Santos F, Krueger F, Hore TA, Marques CJ, Andrews S, Reik W. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature. 2011;473:398–402. doi: 10.1038/nature10008. [DOI] [PubMed] [Google Scholar]
- 280.Veron N, Peters AH. Epigenetics: Tet proteins in the limelight. Nature. 2011;473:293–294. doi: 10.1038/473293a. [DOI] [PubMed] [Google Scholar]
- 281.Guo JU, Su Y, Zhong C, Ming GL, Song H. Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell. 2011;145:423–434. doi: 10.1016/j.cell.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Guo JU, Su Y, Zhong C, Ming GL, Song H. Emerging roles of TET proteins and 5-hydroxymethylcytosines in active DNA demethylation and beyond. Cell Cycle. 2011;10:2662–2668. doi: 10.4161/cc.10.16.17093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.He YF, Li BZ, Li Z, Liu P, Wang Y, Tang Q, Ding J, Jia Y, Chen Z, Li L, Sun Y, Li X, Dai Q, Song CX, Zhang K, He C, Xu GL. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333:1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Maiti A, Drohat AC. Thymine DNA glycosylase can rapidly excise 5-formylcytosine and 5-carboxylcytosine: potential implications for active demethylation of CpG sites. The Journal of biological chemistry. 2011;286:35334–35338. doi: 10.1074/jbc.C111.284620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Zhang L, Lu X, Lu J, Liang H, Dai Q, Xu GL, Luo C, Jiang H, He C. Thymine DNA glycosylase specifically recognizes 5-carboxylcytosine-modified DNA. Nat Chem Biol. 2012;8:328–330. doi: 10.1038/nchembio.914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Baba D, Maita N, Jee JG, Uchimura Y, Saitoh H, Sugasawa K, Hanaoka F, Tochio H, Hiroaki H, Shirakawa M. Crystal structure of thymine DNA glycosylase conjugated to SUMO-1. Nature. 2005;435:979–982. doi: 10.1038/nature03634. [DOI] [PubMed] [Google Scholar]
- 287.Maiti A, Noon MS, Mackerell AD, Jr, Pozharski E, Drohat AC. Lesion processing by a repair enzyme is severely curtailed by residues needed to prevent aberrant activity on undamaged DNA. Proceedings of the National Academy of Sciences of the United States of America. 2012 doi: 10.1073/pnas.1201010109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Smet-Nocca C, Wieruszeski JM, Chaar V, Leroy A, Benecke A. The thymine-DNA glycosylase regulatory domain: residual structure and DNA binding. Biochemistry. 2008;47:6519–6530. doi: 10.1021/bi7022283. [DOI] [PubMed] [Google Scholar]
- 289.Steinacher R, Schar P. Functionality of human thymine DNA glycosylase requires SUMO-regulated changes in protein conformation. Curr Biol. 2005;15:616–623. doi: 10.1016/j.cub.2005.02.054. [DOI] [PubMed] [Google Scholar]
- 290.Morgan MT, Maiti A, Fitzgerald ME, Drohat AC. Stoichiometry and affinity for thymine DNA glycosylase binding to specific and nonspecific DNA. Nucleic acids research. 2011;39:2319–2329. doi: 10.1093/nar/gkq1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Parikh SS, Mol CD, Slupphaug G, Bharati S, Krokan HE, Tainer JA. Base excision repair initiation revealed by crystal structures and binding kinetics of human uracil-DNA glycosylase with DNA. Embo J. 1998;17:5214–5226. doi: 10.1093/emboj/17.17.5214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Maiti A, Morgan MT, Drohat AC. Role of two strictly conserved residues in nucleotide flipping and N-glycosylic bond cleavage by human thymine DNA glycosylase. J Biol Chem. 2009;284:36680–36688. doi: 10.1074/jbc.M109.062356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Scharer OD, Kawate T, Gallinari P, Jiricny J, Verdine GL. Investigation of the mechanisms of DNA binding of the human G/T glycosylase using designed inhibitors. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:4878–4883. doi: 10.1073/pnas.94.10.4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Wibley JE, Waters TR, Haushalter K, Verdine GL, Pearl LH. Structure and specificity of the vertebrate anti-mutator uracil-DNA glycosylase SMUG1. Mol Cell. 2003;11:1647–1659. doi: 10.1016/s1097-2765(03)00235-1. [DOI] [PubMed] [Google Scholar]
- 295.Manvilla BA, Maiti A, Begley MC, Toth EA, Drohat AC. Crystal Structure of Human Methyl-Binding Domain IV Glycosylase Bound to Abasic DNA. J Mol Biol. 2012 doi: 10.1016/j.jmb.2012.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Xiao G, Tordova M, Jagadeesh J, Drohat AC, Stivers JT, Gilliland GL. Crystal structure of Escherichia coli uracil DNA glycosylase and its complexes with uracil and glycerol: structure and glycosylase mechanism revisited. Proteins. 1999;35:13–24. [PubMed] [Google Scholar]
- 297.Waters TR, Swann PF. Kinetics of the action of thymine DNA glycosylase. J Biol Chem. 1998;273:20007–20014. doi: 10.1074/jbc.273.32.20007. [DOI] [PubMed] [Google Scholar]
- 298.Sibghat U, Gallinari P, Xu YZ, Goodman MF, Bloom LB, Jiricny J, Day RS., 3rd Base analog and neighboring base effects on substrate specificity of recombinant human G:T mismatch-specific thymine DNA-glycosylase. Biochemistry. 1996;35:12926–12932. doi: 10.1021/bi961022u. [DOI] [PubMed] [Google Scholar]
- 299.Morgan MT, Bennett MT, Drohat AC. Excision of 5-halogenated uracils by human thymine DNA glycosylase. Robust activity for DNA contexts other than CpG. J Biol Chem. 2007;282:27578–27586. doi: 10.1074/jbc.M704253200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Hardeland U, Steinacher R, Jiricny J, Schar P. Modification of the human thymine-DNA glycosylase by ubiquitin-like proteins facilitates enzymatic turnover. Embo J. 2002;21:1456–1464. doi: 10.1093/emboj/21.6.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Smet-Nocca C, Wieruszeski JM, Leger H, Eilebrecht S, Benecke A. SUMO-1 regulates the conformational dynamics of thymine-DNA Glycosylase regulatory domain and competes with its DNA binding activity. BMC Biochem. 2011;12:4. doi: 10.1186/1471-2091-12-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Mohan RD, Rao A, Gagliardi J, Tini M. SUMO-1-dependent allosteric regulation of thymine DNA glycosylase alters subnuclear localization and CBP/p300 recruitment. Mol Cell Biol. 2007;27:229–243. doi: 10.1128/MCB.00323-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Baba D, Maita N, Jee JG, Uchimura Y, Saitoh H, Sugasawa K, Hanaoka F, Tochio H, Hiroaki H, Shirakawa M. Crystal structure of SUMO-3-modified thymine-DNA glycosylase. Journal of molecular biology. 2006;359:137–147. doi: 10.1016/j.jmb.2006.03.036. [DOI] [PubMed] [Google Scholar]
- 304.Li YQ, Zhou PZ, Zheng XD, Walsh CP, Xu GL. Association of Dnmt3a and thymine DNA glycosylase links DNA methylation with base-excision repair. Nucleic Acids Res. 2007;35:390–400. doi: 10.1093/nar/gkl1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Tini M, Benecke A, Um SJ, Torchia J, Evans RM, Chambon P. Association of CBP/p300 acetylase and thymine DNA glycosylase links DNA repair and transcription. Mol Cell. 2002;9:265–277. doi: 10.1016/s1097-2765(02)00453-7. [DOI] [PubMed] [Google Scholar]
- 306.Petronzelli F, Riccio A, Markham GD, Seeholzer SH, Stoerker J, Genuardi M, Yeung AT, Matsumoto Y, Bellacosa A. Biphasic kinetics of the human DNA repair protein MED1 (MBD4), a mismatch-specific DNA N-glycosylase. The Journal of biological chemistry. 2000;275:32422–32429. doi: 10.1074/jbc.M004535200. [DOI] [PubMed] [Google Scholar]
- 307.Zhang W, Liu Z, Crombet L, Amaya MF, Liu Y, Zhang X, Kuang W, Ma P, Niu L, Qi C. Crystal structure of the mismatch-specific thymine glycosylase domain of human methyl-CpG-binding protein MBD4. Biochem Biophys Res Commun. 2011;412:425–428. doi: 10.1016/j.bbrc.2011.07.091. [DOI] [PubMed] [Google Scholar]
- 308.Choi Y, Gehring M, Johnson L, Hannon M, Harada JJ, Goldberg RB, Jacobsen SE, Fischer RL. DEMETER, a DNA glycosylase domain protein, is required for endosperm gene imprinting and seed viability in arabidopsis. Cell. 2002;110:33–42. doi: 10.1016/s0092-8674(02)00807-3. [DOI] [PubMed] [Google Scholar]
- 309.Zilberman D, Gehring M, Tran RK, Ballinger T, Henikoff S. Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nat Genet. 2007;39:61–69. doi: 10.1038/ng1929. [DOI] [PubMed] [Google Scholar]
- 310.Zhang X, Yazaki J, Sundaresan A, Cokus S, Chan SW, Chen H, Henderson IR, Shinn P, Pellegrini M, Jacobsen SE, Ecker JR. Genome-wide high-resolution mapping and functional analysis of DNA methylation in arabidopsis. Cell. 2006;126:1189–1201. doi: 10.1016/j.cell.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 311.Gong Z, Morales-Ruiz T, Ariza RR, Roldan-Arjona T, David L, Zhu JK. ROS1, a repressor of transcriptional gene silencing in Arabidopsis, encodes a DNA glycosylase/lyase. Cell. 2002;111:803–814. doi: 10.1016/s0092-8674(02)01133-9. [DOI] [PubMed] [Google Scholar]
- 312.Zhu J, Kapoor A, Sridhar VV, Agius F, Zhu JK. The DNA glycosylase/lyase ROS1 functions in pruning DNA methylation patterns in Arabidopsis. Curr Biol. 2007;17:54–59. doi: 10.1016/j.cub.2006.10.059. [DOI] [PubMed] [Google Scholar]
- 313.Gehring M, Huh JH, Hsieh TF, Penterman J, Choi Y, Harada JJ, Goldberg RB, Fischer RL. DEMETER DNA glycosylase establishes MEDEA polycomb gene self-imprinting by allele-specific demethylation. Cell. 2006;124:495–506. doi: 10.1016/j.cell.2005.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Morales-Ruiz T, Ortega-Galisteo AP, Ponferrada-Marin MI, Martinez-Macias MI, Ariza RR, Roldan-Arjona T. DEMETER and REPRESSOR OF SILENCING 1 encode 5-methylcytosine DNA glycosylases. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:6853–6858. doi: 10.1073/pnas.0601109103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Penterman J, Zilberman D, Huh JH, Ballinger T, Henikoff S, Fischer RL. DNA demethylation in the Arabidopsis genome. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:6752–6757. doi: 10.1073/pnas.0701861104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Ponferrada-Marin MI, Roldan-Arjona T, Ariza RR. ROS1 5-methylcytosine DNA glycosylase is a slow-turnover catalyst that initiates DNA demethylation in a distributive fashion. Nucleic Acids Res. 2009;37:4264–4274. doi: 10.1093/nar/gkp390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Ponferrada-Marin MI, Parrilla-Doblas JT, Roldan-Arjona T, Ariza RR. A discontinuous DNA glycosylase domain in a family of enzymes that excise 5-methylcytosine. Nucleic Acids Res. 2011;39:1473–1484. doi: 10.1093/nar/gkq982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Mok YG, Uzawa R, Lee J, Weiner GM, Eichman BF, Fischer RL, Huh JH. Domain structure of the DEMETER 5-methylcytosine DNA glycosylase. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:19225–19230. doi: 10.1073/pnas.1014348107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Ponferrada-Marin MI, Martinez-Macias MI, Morales-Ruiz T, Roldan-Arjona T, Ariza RR. Methylation-independent DNA binding modulates specificity of Repressor of Silencing 1 (ROS1) and facilitates demethylation in long substrates. The Journal of biological chemistry. 2010;285:23032–23039. doi: 10.1074/jbc.M110.124578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Parker JB, Bianchet MA, Krosky DJ, Friedman JI, Amzel LM, Stivers JT. Enzymatic capture of an extrahelical thymine in the search for uracil in DNA. Nature. 2007;449:433–437. doi: 10.1038/nature06131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Campalans A, Marsin S, Nakabeppu Y, O’Connor RT, Boiteux S, Radicella JP. XRCC1 interactions with multiple DNA glycosylases: a model for its recruitment to base excision repair. DNA Repair (Amst) 2005;4:826–835. doi: 10.1016/j.dnarep.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 322.Mutamba JT, Svilar D, Prasongtanakij S, Wang XH, Lin YC, Dedon PC, Sobol RW, Engelward BP. XRCC1 and base excision repair balance in response to nitric oxide. DNA Repair (Amst) 2011;10:1282–1293. doi: 10.1016/j.dnarep.2011.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Ortega-Galisteo AP, Morales-Ruiz T, Ariza RR, Roldan-Arjona T. Arabidopsis DEMETER-LIKE proteins DML2 and DML3 are required for appropriate distribution of DNA methylation marks. Plant Mol Biol. 2008;67:671–681. doi: 10.1007/s11103-008-9346-0. [DOI] [PubMed] [Google Scholar]