Abstract
Elven, E., Bachmann, L. & Gusarov V. I. (2012) Molecular phylogeny of the Athetini–Lomechusini–Ecitocharini clade of aleocharine rove beetles (Insecta). —Zoologica Scripta, 41, 617–636.
It has previously been shown that the Aleocharinae tribes Athetini and Lomechusini form a well-supported clade, which also includes the small Neotropical tribe Ecitocharini. However, neither Athetini nor Lomechusini were recovered as monophyletic. In this study, we addressed the basal phylogenetic relationships among the three tribes using sequence data from (i) a mitochondrial fragment covering the COI, Leu2 and COII genes; (ii) a mitochondrial fragment covering part of the 16S gene, the Leu1 gene and part of the NADH 1 gene; and (iii) a part of the nuclear 18S gene, for 68 Athetini, 33 Lomechusini and 2 Ecitocharini species, plus representatives from 10 other tribes. The athetine subtribe Geostibina was recovered as sister group to the ‘true Lomechusini’, which included the type genus Lomechusa. The two clades formed a sister group to the main Athetini clade, which also included Ecitocharini and the ‘false Lomechusini’, a group of New World genera normally placed in Lomechusini. The following changes in classification are proposed: (i) Geostibina Seevers, 1978 is raised to tribal rank, and 13 Athetini genera are placed in Geostibini; (ii) Ecitodonia Seevers, 1965; Ecitopora Wasmann, 1887, and Tetradonia Wasmann, 1894 are moved from Lomechusini to Athetini; (iii) Ecitocharini Seevers, 1965 is placed in synonymy with Athetini; (iv) Discerota Mulsant & Rey, 1874 is tentatively included in Oxypodini; (v) Actocharina Bernhauer, 1907 is placed in synonymy with Hydrosmecta Thomson, 1858.
Introduction
The rove beetles (Staphylinidae) are one of the two largest families of Coleoptera, comprising (with Scydmaeninae) 32 extant subfamilies and more than 55 000 species (O’Keefe 2005; Thayer 2005; Grebennikov & Newton 2009; Bouchard et al. 2011). The Aleocharinae [ca. 1200 genera and 13 000 species (Thayer 2005)] are the largest subfamily within Staphylinidae and also one of the most challenging because of the large number of small and morphologically very similar species. Until now, relatively few phylogenetic studies of aleocharines have been published, whether morphology-based (e.g. Kistner & Jacobson 1990; Steidle & Dettner 1993; Muona 1997; Ahn 2001; Ahn & Ashe 2004; Ashe 2005; Paśnik 2010), molecular (Maus et al. 2001; Thomas 2009; Elven et al. 2010) or both (Ahn et al. 2010). The current classification of the subfamily (e.g. Smetana 2004; Ashe 2007; Bouchard et al. 2011) is based mainly on intuitive assessments of morphological characters, and even today, new aleocharine tribes are being erected without phylogenetic justification (e.g. Klimaszewski et al. 2010).
In a recent study, Elven et al. (2010) addressed the molecular phylogeny of Athetini, the largest of the aleocharine tribes. Their most important discovery was the Athetini–Lomechusini–Ecitocharini clade (further referred to as the ALE clade), a well-supported monophyletic group consisting of the genera traditionally included in three different tribes. Within the ALE clade, the tribes Lomechusini and Ecitocharini were found nested within Athetini. Furthermore, the Lomechusini were not recovered as monophyletic, but formed two separate clades within the ALE clade. These unexpected but statistically well-supported results indicated the need for a tribe-level revision of the whole ALE clade. However, Elven et al. (2010) could not do so as their taxon sampling was heavily biased towards their primary focus, the phylogeny of Athetini. The Lomechusini were represented by just three genera and four species, and Ecitocharini were represented by only a single species. A more comprehensive sampling of Lomechusini and Ecitocharini was needed before phylogeny-based changes in the tribe-level classification could be seriously considered. In this study, we specifically address this issue using a broader taxon sample for all three tribes.
The tribe Athetini Casey, 1910 is distributed worldwide and includes more than 170 genera and thousands of species (Newton et al. 2000). The genus Atheta alone includes 1700 species (A. F. Newton, unpublished data). The majority of athetines are free living, while a few are associated with ants or termites. The tribe is traditionally diagnosed based on a combination of several characters, for example, galea and lacinia of moderate length, tarsal formula 4-5-5, mesocoxae narrowly or moderately separated, mesoventral process narrow, athetine bridge of aedeagus present. However, none of these characters is unique to Athetini. An older available family group name, Callicerini Jakobson, 1908 (non Rondani, 1845), exists for this tribe, but Athetini Casey, 1910 is the name currently in prevailing use (Newton & Thayer 1992). An application has been submitted to the International Commission on Zoological Nomenclature to conserve the name Athetini and suppress Callicerini (Gusarov 2011). The tribe Lomechusini Fleming, 1821 is distributed worldwide, but is most diverse in tropical regions. It includes more than 200 genera and over 2200 species (Hlaváč et al. 2011), most of which are associated with ants or termites. The tribe is poorly defined and is traditionally diagnosed by a combination of characters, for example, galea and lacinia significantly elongate, tarsal formula 4-5-5, mesocoxae broadly separated, mesoventral process short and broad, athetine bridge of aedeagus present (Newton et al. 2000). Not surprisingly, some genera have repeatedly been moved in and out of Lomechusini (e.g. Meronera: cf. Newton et al. 2000 and Navarrete-Heredia et al. 2002). The Neotropical tribe Ecitocharini Seevers, 1965 includes only 10 genera of derived myrmecophiles associated with army ants of the genus Eciton (Kistner & Jacobson 1990). The tribe is diagnosed based on a combination of characters (galea and lacinia of moderate length, tarsal formula 4-5-5, particular glands and gland reservoirs present, mesoventral process narrow, body with distinct polygonal microsculpture) (Kistner & Jacobson 1990).
Phylogenetic relationships involving members of the ALE clade have been studied in several publications. The conclusions and limitations of the studies most relevant for the current study are briefly reviewed here in chronological order.
Kistner & Jacobson (1990) revised and redefined the tribe Ecitocharini and conducted a cladistic analysis of the tribe based on 22 morphological characters. The ingroup included all 10 genera of Ecitocharini, but the outgroup included only a single albeit very large and morphologically diverse genus, Zyras. The species used to code the character states for Zyras was/were not mentioned. Thus, this study was not designed to test the monophyly of Ecitocharini or to rigorously infer relationships among the ingroup taxa.
Steidle & Dettner (1993) studied the morphology and chemistry of the abdominal tergal gland of 22 aleocharine species from nine tribes (adjusted to current classification). Adding further data from published descriptions of the tergal gland and its products, they constructed a matrix of nine morphological and chemical characters for 27 species from 10 tribes, including six Lomechusini and eight Athetini species. Ecitocharini were not included. In their phylogenetic tree, Athetini, Lomechusini and Aleocharini formed a clade supported by just a single apomorphy (gland reservoir large). Together with Oxypodini (excluding the subtribe Dinardina), they formed a larger clade supported by the presence of two groups of products in the gland secretion. Species belonging to the same tribe/subtribe were lumped into a single terminal taxon, and the study was thus not designed to test the monophyly of the included tribes.
Muona (1997) used 87 binary morphological characters in an analysis of 41 genera from 12 aleocharine tribes aimed at testing the monophyly of Athetini. Ecitocharini were not represented, but the study included the dorylophilous tribe Mimanommatini (listed as Dorylomimini). Unfortunately, the character matrix, phylogenetic trees and other details have not been published, and the main results were only summarized in an abstract for the 15th Meeting of the Willi Hennig Society (Muona 1997). Athetini were recovered as paraphyletic, while Lomechusini were polyphyletic. Six tribes (Myllaenini, Lomechusini, Hoplandriini, Termitohospitini, Termitodiscini and Mimanommatini) formed a clade within Athetini. It is noteworthy that three morphologically highly derived tribes, the myrmecophilous Mimanommatini and the termitophilous Termitohospitini and Termitodiscini, were nested within Athetini.
Ashe (2005) used 27 larval and 133 adult morphological characters to infer a phylogeny of the tachyporine-group subfamilies of Staphylinidae. For the Aleocharinae, 29 genera from 13 tribes were included with the aim to resolve the basal phylogenetic relationships of the subfamily. Athetini were represented by the genera Atheta, Geostiba and Pontomalota, and Lomechusini by Zyras and Drusilla. Ecitocharini were not included. Lomechusini were recovered as monophyletic, while relationships among the three Athetini genera or between Athetini, Lomechusini and the other tribes of ‘higher’ Aleocharinae (sensu Ashe 2005) remained unresolved.
Thomas (2009) published the first molecular phylogeny of Aleocharinae, based on nucleotide sequences of the mitochondrial 12S and 16S RNA genes. The study included eight tribes, with Athetini being represented by three genera. Lomechusini and Ecitocharini were not included. In the resulting trees, the relationships among the athetine genera, or between these and the other included tribes, were not resolved.
Paśnik (2010) published a comprehensive morphological phylogeny of the tribe Tachyusini, usually considered a subtribe of Oxypodini (e.g. Seevers 1978; Smetana 2004). The study was based on 159 adult morphological characters and included 84 species from 14 aleocharine tribes. Athetini sensu Paśnik were represented by four genera: Aloconota, Atheta, Dinaraea and Liogluta. Lomechusini were represented by four genera: Amaurodera, Drusilla, Trachyota and Zyras. Also included was Meronera, which has been alternatively placed in Lomechusini (e.g. Newton et al. 2000), Oxypodini (Seevers 1978), Tachyusini (Ashe 1985) and Falagriini (Pace 2008). The central result of Paśnik’s study was the recovery of a strongly supported clade, Tachyusini sensu Paśnik. Many of the recovered relationships were surprising, and an investigation into the underlying character matrix revealed serious issues with the interpretation and weighting of many important tribal characters, a fact that strongly undermines the validity of Paśnik’s results. One example with a direct bearing on the current study is the athetine bridge in the median lobe of the aedeagus, an important character shared by the Athetini and Lomechusini, which Paśnik erroneously interpreted as missing in Lomechusini, the athetine genus Thamiaraea, and in Meronera.
Elven et al. (2010) presented the first comprehensive molecular phylogeny of Athetini. The study included 80 aleocharine species from 11 tribes. Athetini were represented by 27 genera and 58 species, Lomechusini by three genera and four species, and Ecitocharini by a single species. Also included was the genus Meronera (see above). They discovered the ALE clade, consisting of the three tribes Athetini, Lomechusini and Ecitocharini. Within the ALE clade, the athetine genera Geostiba and Earota formed a sister group to the lomechusine genera Pella and Drusilla. This clade in turn formed a sister group to the remaining Athetini (referred to as the ‘main Athetini clade’), which also included the tribe Ecitocharini and the lomechusine genus Myrmedonota. The (Geostiba, Earota) clade was an unexpected discovery, but there is at least one tentative morphological synapomorphy for this clade: sensillum a of the epipharynx being reduced (Yosii & Sawada 1976: fig. 47B; Gusarov 2002b: fig. 2). This character state is thus a potential synapomorphy of the subtribe Geostibina. The lomechusine genus Myrmedonota formed a weakly supported clade with Meronera within the main Athetini clade. Although the type genus of Lomechusini was not included in Elven et al. (2010), they argued that based on morphology, it seemed more closely related to Pella and Drusilla than to Meronera and Myrmedonota. In this study, the name ‘true Lomechusini’ will refer to the clade that includes the type genus Lomechusa, while the other clade will be referred to as the ‘false Lomechusini’. Ecitophya, the only genus of Ecitocharini included in Elven et al. (2010), formed a well-supported clade with the New World genus Stethusa, a generalized non-myrmecophile athetine.
The main goal of this study is to firmly resolve the phylogeny of the major lineages of the ALE clade discovered by Elven et al. (2010), to revise the tribe-level classification. With taxon sampling expanded in all three ALE tribes, the study aims to test the following hypotheses: (i) Geostibina are a sister clade to the ‘true Lomechusini’, the clade that includes Lomechusa; (ii) Geostibina and the ‘true Lomechusini’ form a sister group to the main Athetini clade; (iii) All athetine genera that have sensillum a of the epipharynx reduced belong to Geostibina; (iv) Several genera traditionally placed in Lomechusini are not members of the ‘true Lomechusini’ clade, but form a subclade (the ‘false Lomechusini’) within the main Athetini clade; and (v) Ecitocharini are monophyletic, nested within the main Athetini clade, and have Stethusa as their sister group.
Material and methods
Taxon sampling
Taxa used in this study are listed in Table 1. About half of the sequences were produced for this study, the remaining were taken from Elven et al. (2010). The taxa were chosen specifically to address the phylogenetic relationships between Athetini, Lomechusini and Ecitocharini, and to this end, we included a broad representation of the first two tribes. The study includes six athetine genera with reduced sensillum a of the epipharynx: Alevonota, Aloconota, Callicerus, Earota, Geostiba and Pelioptera. These genera were hypothesized to belong to a monophyletic Geostibina. We included the type species of the type genera of Athetini, Lomechusini and Geostibina, and the type species of many other genera including Alevonota, Aloconota, Callicerus, Drusilla, Earota and Meronera (indicated in Table 1). The large lomechusine genus Zyras is represented by at least three subgenera including the nominotypical subgenus, but not the type species. The tribe Ecitocharini is represented by two genera, but not the type genus.
Table 1.
List of specimens used in this study
| Species name | Tribe | Depository1 | ZMUN Barcode | Country of origin | GenBank accession numbers | ||
|---|---|---|---|---|---|---|---|
| COI–Leu2–COII | 16S–Leu1–NADH1 | 18S | |||||
| Subfamily Tachyporinae | |||||||
| Tachinus proximus Kraatz, 1855 | Tachyporini | ZMUN | 10002542 | Norway | GQ980859 | GQ980968 | GQ981067 |
| Subfamily Aleocharinae (except the ALE clade tribes) | |||||||
| Aleochara moerens Gyllenhal, 1827 #1 | Aleocharini | ZMUN | 10002579 | Norway | GQ980861 | GQ980970 | GQ981069 |
| Aleochara moerens Gyllenhal, 1827 #2 | Aleocharini | ZMUN | 10002570 | Norway | GQ980862 | GQ980971 | GQ981070 |
| Tetrasticta sp. 1 | Aleocharini | ZMUC | 10029285 | Laos | JN581929 | JN581761 | JN581846 |
| Tetrasticta sp. 2 | Aleocharini | ZMUC | 10029284 | Laos | JN581930 | JN581762 | JN581847 |
| •Cordalia obscura (Gravenhorst, 1802) | Falagriini | ZMUN | 10002651 | Greece | GQ980864 | GQ980973 | GQ981071 |
| Myrmecopora uvida (Erichson, 1840) #1 | Falagriini | ZMUN | 10030945 | Greece | JN581919 | JN581750 | JN581834 |
| Myrmecopora uvida (Erichson, 1840) #2 | Falagriini | ZMUN | 10029111 | Greece | JN581920 | JN581751 | JN581835 |
| Gymnusa variegata Kiesenwetter, 1845 | Gymnusini | ZMUN | 10002641 | Romania | GQ980860 | GQ980969 | GQ981068 |
| •Bolitochara pulchra (Gravenhorst, 1806) #1 | Homalotini | ZMUN | 10002596 | Norway | GQ980866 | GQ980974 | GQ981073 |
| •Bolitochara pulchra (Gravenhorst, 1806) #2 | Homalotini | ZMUN | 10002591 | Norway | GQ980865 | – | GQ981072 |
| Gyrophaena congrua Erichson, 1837 | Homalotini | ZMUN | 10002584 | Norway | GQ980867 | GQ980975 | GQ981074 |
| Gyrophaena fasciata (Marsham, 1802) #1 | Homalotini | ZMUN | 10002585 | Norway | GQ980868 | GQ980976 | GQ981075 |
| Gyrophaena fasciata (Marsham, 1802) #2 | Homalotini | ZMUN | 10002572 | Norway | GQ980869 | GQ980977 | GQ981076 |
| •Silusida marginella (Casey, 1893) #1 | Homalotini | ZMUN | 10002625 | USA | GQ980870 | GQ980978 | GQ981077 |
| •Silusida marginella (Casey, 1893) #2 | Homalotini | ZMUN | 10002624 | USA | GQ980871 | GQ980979 | GQ981078 |
| ••Hoplandria lateralis (Melsheimer, 1846) | Hoplandriini | ZMUN | 10002550 | USA | GQ980872 | GQ980980 | GQ981079 |
| Myllaena audax Casey, 1911 #1 | Myllaenini | ZMUN | 10030903 | USA | JN581918 | JN581749 | JN581833 |
| Myllaena audax Casey, 1911 #2 | Myllaenini | ZMUN | 10002598 | USA | GQ980881 | – | GQ981088 |
| Halobrecta cf. halensis Mulsant & Rey, 1873 | Oxypodini | ZMUN | 10002647 | Greece | GQ980966 | GQ981065 | GQ981172 |
| Oxypoda praecox Erichson, 1839 | Oxypodini | ZMUN | 10002637 | Germany | GQ980882 | GQ980989 | GQ981089 |
| Thendelecrotona sp. | Oxypodinini2 | ZMUN | 10002612 | South Africa | GQ980967 | GQ981066 | GQ981173 |
| Placusa sp. prope tachyporoides (Waltl, 1838) | Placusini | ZMUN | 10002541 | USA | GQ980883 | GQ980990 | GQ981090 |
| Tribe Athetini | |||||||
| Acrotona sp. prope assecla (Casey, 1910) | Athetini | ZMUN | 10002544 | USA | GQ980884 | GQ980991 | GQ981091 |
| Acrotona sp. prope austiniana (Casey, 1910) #1 | Athetini | ZMUN | 10002543 | USA | GQ980885 | GQ980992 | GQ981092 |
| Acrotona sp. prope austiniana (Casey, 1910) #2 | Athetini | ZMUN | 10002547 | USA | GQ980886 | GQ980993 | GQ981093 |
| •Actocharina leptotyphloides (Bernhauer, 1907) #1 | Athetini | ZMUN | 10002656 | Austria | JN581857 | JN581688 | JN581774 |
| •Actocharina leptotyphloides (Bernhauer, 1907) #2 | Athetini | ZMUN | 10002657 | Austria | JN581858 | JN581689 | JN581775 |
| Alevonota egregia (Rye, 1876) #1 | Athetini | ZMUN | 10030889 | France | JN581859 | JN581690 | – |
| Alevonota egregia (Rye, 1876) #2 | Athetini | ZMUN | 10030891 | France | JN581860 | JN581691 | – |
| •Alevonota rufotestacea (Kraatz, 1856) | Athetini | ZMUN | 10030810 | France | JN581861 | JN581692 | – |
| Aloconota cambrica (Wollaston, 1855) #1 | Athetini | ZMUN | 10030823 | Austria | JN581862 | JN581693 | JN581776 |
| Aloconota cambrica (Wollaston, 1855) #2 | Athetini | ZMUN | 10029295 | Austria | JN581863 | JN581694 | JN581777 |
| Aloconota currax (Kraatz, 1856) #1 | Athetini | ZMUN | 10029300 | Austria | JN581864 | JN581695 | JN581778 |
| Aloconota currax (Kraatz, 1856) #2 | Athetini | ZMUN | 10029301 | Austria | JN581865 | JN581696 | JN581779 |
| Aloconota gregaria (Erichson, 1839) | Athetini | ZMUN | 10029294 | Norway | JN581866 | JN581697 | JN581780 |
| Aloconota sp. #1 | Athetini | ZMUN | 10030840 | Uganda | JN581867 | JN581698 | JN581781 |
| Aloconota sp. #2 | Athetini | ZMUN | 10030746 | Uganda | JN581868 | – | JN581782 |
| Alpinia sp. prope alpicola (Miller, 1859) #1 | Athetini | ZMUN | 10029305 | Romania | JN581869 | JN581699 | JN581783 |
| Alpinia sp. prope alpicola (Miller, 1859) #2 | Athetini | ZMUN | 10002644 | Romania | GQ980897 | – | GQ981104 |
| •Amidobia talpa (Heer, 1841) | Athetini | ZMUN | 10002646 | Norway | GQ980898 | GQ981002 | GQ981105 |
| •Amischa analis (Gravenhorst, 1802) #1 | Athetini | ZMUN | 10029292 | Norway | JN581871 | JN581702 | JN581786 |
| •Amischa analis (Gravenhorst, 1802) #2 | Athetini | ZMUN | 10002623 | Norway | GQ980895 | – | GQ981102 |
| Amischa nigrofusca (Stephens, 1832) #1 | Athetini | ZMUN | 10029304 | Norway | JN581872 | JN581703 | JN581787 |
| Amischa nigrofusca (Stephens, 1832) #2 | Athetini | ZMUN | 10002622 | Norway | GQ980896 | – | GQ981103 |
| Atheta (Alaobia) gagatina (Baudi di Selve, 1848) #1 | Athetini | ZMUN | 10002578 | Norway | GQ980901 | GQ981005 | GQ981108 |
| Atheta (Alaobia) gagatina (Baudi di Selve, 1848) #2 | Athetini | ZMUN | 10002580 | Norway | GQ980902 | GQ981006 | GQ981109 |
| Atheta (Alaobia) membranata G. Benick, 1974 | Athetini | ZMUN | 10002653 | France | GQ980903 | GQ981007 | – |
| Atheta (Alaobia) scapularis (C.R.Sahlberg, 1831) #1 | Athetini | ZMUN | 10030796 | France | JN581873 | JN581704 | – |
| Atheta (Alaobia) scapularis (C.R.Sahlberg, 1831) #2 | Athetini | ZMUN | 10030807 | France | JN581874 | JN581705 | – |
| Atheta (crassicornis-gr.) crassicornis (Fabricius, 1793) | Athetini | ZMUN | 10002640 | Hungary | GQ980907 | GQ981011 | GQ981113 |
| Atheta (crassicornis-gr.) modesta (Melsheimer, 1844) #1 | Athetini | ZMUN | 10002621 | USA | GQ980908 | GQ981012 | GQ981114 |
| Atheta (crassicornis-gr.) modesta (Melsheimer, 1844) #2 | Athetini | ZMUN | 10002620 | USA | GQ980909 | GQ981013 | GQ981115 |
| Atheta (Datomicra) celata (Erichson, 1837) #1 | Athetini | ZMUN | 10002560 | Norway | GQ980910 | GQ981014 | GQ981116 |
| Atheta (Datomicra) celata (Erichson, 1837) #2 | Athetini | ZMUN | 10002556 | Norway | GQ980911 | GQ981015 | GQ981117 |
| Atheta (Datomicra) dadopora (Thomson, 1867) | Athetini | ZMUN | 10002554 | USA | GQ980912 | GQ981016 | GQ981118 |
| Atheta (Dimetrota) aeneipennis (Thomson, 1856) | Athetini | ZMUN | 10002583 | Norway | GQ980913 | GQ981017 | GQ981119 |
| Atheta (Dimetrota) cinnamoptera (Thomson, 1856) | Athetini | ZMUN | 10002582 | Norway | GQ980914 | GQ981018 | GQ981120 |
| Atheta (Dimetrota) setigera (Sharp, 1869) | Athetini | ZMUN | 10002639 | Romania | GQ980917 | GQ981021 | GQ981123 |
| Atheta (Dralica) vilis (Erichson, 1837) #1 | Athetini | ZMUN | 10029114 | Belarus | JN581875 | JN581706 | JN581788 |
| Atheta (Dralica) vilis (Erichson, 1837) #2 | Athetini | ZMUN | 10002666 | Belarus | JN581876 | JN581707 | JN581789 |
| Atheta (Mycetota) laticollis (Stephens, 1832) | Athetini | ZMUN | 10002606 | Norway | GQ980920 | GQ981024 | GQ981126 |
| Atheta (Mycetota) pasadenae Bernhauer, 1906 | Athetini | ZMUN | 10002642 | France | GQ980921 | GQ981025 | GQ981127 |
| Atheta (Oreostiba) bosnica Ganglbauer, 1895 | Athetini | ZMUN | 10002638 | Romania | GQ980922 | GQ981026 | GQ981128 |
| Atheta (Oxypodera) kenyamontis Pace, 1986 | Athetini | ZMUN | 10002586 | Kenya | GQ980923 | GQ981027 | GQ981129 |
| Atheta (Parameotica) laticeps (Thomson, 1856) #1 | Athetini | ZMUN | 10029115 | Belarus | JN581925 | JN581755 | JN581840 |
| Atheta (Parameotica) laticeps (Thomson, 1856) #2 | Athetini | ZMUN | 10029116 | Belarus | JN581926 | JN581756 | JN581841 |
| Atheta (ravilla-gr.) ravilla (Erichson, 1839) #1 | Athetini | ZMUN | 10002548 | Norway | GQ980924 | GQ981028 | GQ981130 |
| Atheta (ravilla-gr.) ravilla (Erichson, 1839) #2 | Athetini | ZMUN | 10002557 | Norway | GQ980925 | GQ981029 | GQ981131 |
| Atheta (s. str.) contristata (Kraatz, 1856) | Athetini | ZMUN | 10002635 | Romania | GQ980926 | GQ981030 | GQ981132 |
| ••Atheta (s. str.) graminicola (Gravenhorst, 1806) #1 | Athetini | ZMUN | 10002561 | Norway | GQ980927 | GQ981031 | GQ981133 |
| ••Atheta (s. str.) graminicola (Gravenhorst, 1806) #2 | Athetini | ZMUN | 10002562 | Norway | GQ980928 | GQ981032 | GQ981134 |
| Atheta (Thinobaena) vestita (Gravenhorst, 1806) | Athetini | ZMUN | 10002613 | Norway | GQ980929 | GQ981033 | GQ981135 |
| Atheta (vaga-gr.) vaga (Heer, 1839) | Athetini | ZMUN | 10002655 | France | GQ980930 | GQ981034 | – |
| Atheta sp. ex gr. lippa | Athetini | ZMUN | 10002564 | USA | GQ980918 | GQ981022 | GQ981124 |
| Boreophilia hyperborea (Brundin, 1940) | Athetini | ZMUN | 10002634 | Russia | GQ980933 | GQ981037 | GQ981138 |
| Boreostiba sp. | Athetini | ZMUN | 10002633 | Russia | GQ980934 | GQ981038 | GQ981139 |
| •Brundinia meridionalis (Mulsant & Rey, 1853) | Athetini | ZMUN | 10002667 | Ukraine | JN581877 | JN581708 | JN581790 |
| •Callicerus obscurus Gravenhorst, 1802 #1 | Athetini | ZMUN | 10030905 | Denmark | JN581878 | JN581709 | JN581791 |
| •Callicerus obscurus Gravenhorst, 1802 #2 | Athetini | ZMUN | 10030800 | Denmark | JN581879 | JN581710 | JN581792 |
| •Dadobia immersa (Erichson, 1837) | Athetini | ZMUN | 10002630 | Norway | GQ980953 | GQ981055 | GQ981159 |
| •Dalotia coriaria (Kraatz, 1856) | Athetini | ZMUN | 10002643 | France | – | GQ981039 | GQ981140 |
| •Discerota torrentum (Kiesenwetter, 1850) #1 | Athetini | ZMUN | 10029112 | France | JN581880 | JN581711 | JN581793 |
| •Discerota torrentum (Kiesenwetter, 1850) #2 | Athetini | ZMUN | 10029113 | France | JN581881 | JN581712 | JN581794 |
| Earota dentata (Bernhauer, 1906) | Athetini | ZMUN | 10002539 | USA | GQ980965 | GQ981064 | GQ981171 |
| •Earota reyi (Kiesenwetter, 1850) #1 | Athetini | ZMUN | 10029306 | France | JN581888 | JN581718 | JN581799 |
| •Earota reyi (Kiesenwetter, 1850) #2 | Athetini | ZMUN | 10029307 | France | JN581889 | JN581719 | JN581800 |
| •Geostiba (s. str.) circellaris (Gravenhorst, 1806) | Athetini | ZMUN | 10002587 | Norway | GQ980954 | GQ981056 | GQ981160 |
| Geostiba (Sibiota) bicarinata Lohse & Smetana, 1988 #1 | Athetini | ZMUN | 10030875 | USA | JN581895 | JN581725 | JN581807 |
| Geostiba (Sibiota) bicarinata Lohse & Smetana, 1988 #2 | Athetini | ZMUN | 10030948 | USA | JN581896 | JN581726 | JN581808 |
| Geostiba (Sibiota) nubigena Lohse & Smetana, 1988 #1 | Athetini | ZMUN | 10030888 | USA | JN581897 | JN581727 | JN581809 |
| Geostiba (Sibiota) nubigena Lohse & Smetana, 1988 #2 | Athetini | ZMUN | 10030736 | USA | JN581898 | JN581728 | JN581810 |
| Hydrosmecta eximia (Sharp, 1869) #1 | Athetini | ZMUN | 10002661 | Austria | JN581899 | JN581729 | JN581811 |
| Hydrosmecta eximia (Sharp, 1869) #2 | Athetini | ZMUN | 10002659 | Austria | JN581900 | JN581730 | JN581812 |
| Hydrosmecta gracilicornis (Erichson, 1839) | Athetini | ZMUN | 10002658 | Austria | JN581901 | JN581731 | JN581813 |
| Hydrosmecta valdieriana (Scheerpeltz, 1944) #1 | Athetini | ZMUN | 10002662 | Austria | JN581903 | JN581733 | JN581815 |
| Hydrosmecta valdieriana (Scheerpeltz, 1944) #2 | Athetini | ZMUN | 10002663 | Austria | JN581904 | JN581734 | JN581816 |
| Hydrosmecta sp. 1 | Athetini | ZMUN | 10002650 | USA | GQ980955 | GQ981057 | GQ981161 |
| Hydrosmecta sp. 2 | Athetini | ZMUN | 10002660 | Austria | JN581902 | JN581732 | JN581814 |
| Liogluta microptera Thomson, 1867 #1 | Athetini | ZMUN | 10002600 | Czech Republic | GQ980937 | GQ981041 | GQ981143 |
| Liogluta microptera Thomson, 1867 #2 | Athetini | ZMUN | 10002602 | Czech Republic | GQ980936 | – | GQ981142 |
| Liogluta nigropolita (Bernhauer, 1907) | Athetini | ZMUN | 10002636 | Russia | GQ980938 | – | GQ981144 |
| Lypoglossa lateralis (Mannerheim, 1830) | Athetini | ZMUN | 10002632 | Russia | GQ980887 | GQ980994 | GQ981094 |
| •Lyprocorrhe anceps (Erichson, 1837) | Athetini | ZMUN | 10002649 | Norway | GQ980939 | GQ981042 | GQ981145 |
| •Meronera venustula (Erichson, 1839) | Athetini3 | ZMUN | 10002576 | USA | GQ980875 | GQ980983 | GQ981082 |
| •Mocyta fungi (Gravenhorst, 1806) #1 | Athetini | ZMUN | 10002588 | Germany | GQ980888 | GQ980995 | GQ981095 |
| •Mocyta fungi (Gravenhorst, 1806) #2 | Athetini | ZMUN | 10002589 | Germany | GQ980889 | GQ980996 | GQ981096 |
| Mocyta scopula (Casey, 1893) #1 | Athetini | ZMUN | 10002540 | USA | GQ980890 | GQ980997 | GQ981097 |
| Mocyta scopula (Casey, 1893) #2 | Athetini | ZMUN | 10002559 | USA | GQ980891 | GQ980998 | GQ981098 |
| Pelioptera sp. prope micans (Kraatz, 1857) | Athetini | ZMUC | 10030959 | Laos | – | JN581758 | JN581843 |
| Philhygra debilis (Erichson, 1837) #1 | Athetini | ZMUN | 10002607 | Norway | GQ980941 | GQ981044 | GQ981147 |
| Philhygra debilis (Erichson, 1837) #2 | Athetini | ZMUN | 10002608 | Norway | GQ980942 | GQ981045 | GQ981148 |
| Philhygra fallaciosa (Sharp, 1869) #1 | Athetini | ZMUN | 10002610 | Czech Republic | GQ980943 | GQ981046 | GQ981149 |
| Philhygra fallaciosa (Sharp, 1869) #2 | Athetini | ZMUN | 10002609 | Czech Republic | GQ980944 | GQ981047 | GQ981150 |
| Philhygra iterans (Casey, 1910) #1 | Athetini | ZMUN | 10002595 | USA | GQ980945 | GQ981048 | GQ981151 |
| Philhygra iterans (Casey, 1910) #2 | Athetini | ZMUN | 10002594 | USA | GQ980946 | GQ981049 | GQ981152 |
| •Pontomalota opaca (LeConte, 1863) #1 | Athetini | ZMUN | 10002577 | USA | GQ980956 | GQ981058 | GQ981162 |
| •Pontomalota opaca (LeConte, 1863) #2 | Athetini | ZMUN | 10002574 | USA | GQ980957 | GQ981059 | GQ981163 |
| •Stethusa dichroa (Gravenhorst, 1802) #1 | Athetini | ZMUN | 10002567 | USA | GQ980948 | GQ981051 | GQ981154 |
| •Stethusa dichroa (Gravenhorst, 1802) #2 | Athetini | ZMUN | 10002568 | USA | GQ980949 | GQ981052 | GQ981155 |
| Stethusa spuriella (Casey, 1910) #1 | Athetini | ZMUN | 10002599 | USA | GQ980950 | GQ981053 | GQ981156 |
| Stethusa spuriella (Casey, 1910) #2 | Athetini | ZMUN | 10002628 | USA | GQ980951 | GQ981054 | GQ981157 |
| •Strigota ambigua (Erichson, 1839) #1 | Athetini | ZMUN | 10002571 | USA | GQ980893 | GQ981000 | GQ981100 |
| •Strigota ambigua (Erichson, 1839) #2 | Athetini | ZMUN | 10002575 | USA | GQ980894 | GQ981001 | GQ981101 |
| •Tarphiota fucicola (Mäklin in Mannerheim, 1852) | Athetini | ZMUN | 10002593 | USA | GQ980958 | GQ981060 | GQ981164 |
| Tribe Ecitocharini | |||||||
| Ecitomorpha sp. | Ecitocharini | ZMUN | 10002689 | Peru | JN581892 | JN581722 | JN581803 |
| Ecitophya gracillima Mann, 1925 #1 | Ecitocharini | ZMUN | 10002592 | Peru | GQ980863 | GQ980972 | JN581804 |
| Ecitophya gracillima Mann, 1925 #2 | Ecitocharini | ZMUN | 10029164 | Peru | JN581893 | JN581723 | JN581805 |
| Tribe Lomechusini | |||||||
| Amaurodera yaoana Pace, 1992 #1 | Lomechusini | ZMUC | 10030878 | Laos | JN581870 | JN581700 | JN581784 |
| Amaurodera yaoana Pace, 1992 #2 | Lomechusini | ZMUC | 10030812 | Laos | – | JN581701 | JN581785 |
| •Drusilla canaliculata (Fabricius, 1787) #1 | Lomechusini | ZMUN | 10002604 | Norway | GQ980873 | GQ980981 | GQ981080 |
| •Drusilla canaliculata (Fabricius, 1787) #2 | Lomechusini | ZMUN | 10002601 | Norway | GQ980874 | GQ980982 | GQ981081 |
| Drusilla sp. 1 #1 | Lomechusini | ZMUN | 10051252 | Thailand | JN581882 | JN581713 | – |
| Drusilla sp. 1 #2 | Lomechusini | ZMUN | 10051251 | Thailand | JN581883 | JN581714 | – |
| Drusilla sp. 2 #1 | Lomechusini | ZMUC | 10051193 | Laos | JN581884 | JN581715 | JN581795 |
| Drusilla sp. 2 #2 | Lomechusini | ZMUC | 10051194 | Laos | JN581885 | JN581716 | JN581796 |
| Drusilla sp. prope khamhengi Pace, 1984 #1 | Lomechusini | ZMUC | 10051166 | Laos | JN581886 | JN581717 | JN581797 |
| Drusilla sp. prope khamhengi Pace, 1984 #2 | Lomechusini | ZMUC | 10051165 | Laos | JN581887 | – | JN581798 |
| Ecitodonia sp. #1 | Lomechusini | ZMUN | 10029249 | Ecuador | JN581890 | JN581720 | JN581801 |
| Ecitodonia sp. #2 | Lomechusini | ZMUN | 10029248 | Ecuador | JN581891 | JN581721 | JN581802 |
| Ecitopora sp. | Lomechusini | ZMUN | 10029251 | Ecuador | JN581894 | JN581724 | JN581806 |
| ••Lomechusa emarginata (Paykull, 1789) #1 | Lomechusini | ZMUN | 10030941 | Norway | JN581905 | JN581735 | JN581817 |
| ••Lomechusa emarginata (Paykull, 1789) #2 | Lomechusini | ZMUN | 10030947 | Norway | JN581906 | JN581736 | JN581818 |
| Lomechusa pubicollis Brisout de Barneville, 1860 | Lomechusini | ZMUN | 10030917 | Germany | JN581907 | JN581737 | JN581819 |
| Lomechusini genus 1 | Lomechusini | ZMUN | 10029159 | Ecuador | JN581908 | JN581738 | JN581820 |
| Lomechusini genus 2 #1 | Lomechusini | ZMUN | 10002685 | Ecuador | JN581909 | JN581739 | JN581821 |
| Lomechusini genus 2 #2 | Lomechusini | ZMUN | 10029161 | Ecuador | JN581910 | – | JN581822 |
| Lomechusini genus 3 #1 | Lomechusini | ZMUN | 10029252 | Ecuador | – | JN581740 | JN581823 |
| Lomechusini genus 3 #2 | Lomechusini | ZMUN | 10029253 | Ecuador | – | JN581741 | JN581824 |
| Lomechusini genus 4 #1 | Lomechusini | ZMUN | 10029256 | Ecuador | JN581911 | JN581742 | JN581825 |
| Lomechusini genus 4 #2 | Lomechusini | ZMUN | 10029254 | Ecuador | JN581912 | – | JN581826 |
| Lomechusini genus 5 #1 | Lomechusini | ZMUN | 10029258 | Ecuador | JN581913 | JN581743 | JN581827 |
| Lomechusini genus 5 #2 | Lomechusini | ZMUN | 10029257 | Ecuador | JN581914 | JN581744 | JN581828 |
| Lomechusini genus 6 | Lomechusini | ZMUN | 10029279 | Ecuador | JN581915 | JN581745 | JN581829 |
| Lomechusini genus 7 #1 | Lomechusini | ZMUN | 10029280 | Ecuador | JN581916 | JN581746 | JN581830 |
| Lomechusini genus 7 #2 | Lomechusini | ZMUN | 10029282 | Ecuador | – | JN581747 | JN581831 |
| Lomechusoides amurensis (Wasmann, 1897) | Lomechusini | ZMUN | 10030868 | Russia | JN581917 | JN581748 | JN581832 |
| Myrmedonota sp. #1 | Lomechusini | ZMUN | 10002615 | USA | GQ980876 | GQ980984 | GQ981083 |
| Myrmedonota sp. #2 | Lomechusini | ZMUN | 10002614 | USA | GQ980877 | GQ980985 | GQ981084 |
| Orphnebius (Mesocephalobius) sp. 1 | Lomechusini | ZMUC | 10051237 | Laos | JN581921 | JN581752 | JN581836 |
| Orphnebius (Mesocephalobius) sp. 2 | Lomechusini | ZMUC | 10051236 | Laos | JN581922 | JN581753 | JN581837 |
| Orphnebius (Mesocephalobius) sp. 3 | Lomechusini | ZMUN | 10051239 | Thailand | JN581923 | – | JN581838 |
| Orphnebius (Mesocephalobius) sp. 4 | Lomechusini | ZMUN | 10051238 | Thailand | JN581924 | JN581754 | JN581839 |
| Pedinopleurus sp. | Lomechusini | ZMUN | 10051235 | Thailand | JN581927 | JN581757 | JN581842 |
| Pella caliginosa (Casey, 1893) #1 | Lomechusini | ZMUN | 10002617 | USA | GQ980878 | GQ980986 | GQ981085 |
| Pella caliginosa (Casey, 1893) #2 | Lomechusini | ZMUN | 10002616 | USA | GQ980879 | GQ980987 | GQ981086 |
| Pella humeralis (Gravenhorst, 1802) | Lomechusini | ZMUN | 10002569 | Norway | GQ980880 | GQ980988 | GQ981087 |
| Tetradonia sp. 1 | Lomechusini | ZMUN | 10029163 | Ecuador | – | JN581759 | JN581844 |
| Tetradonia sp. 2 | Lomechusini | ZMUN | 10029160 | Ecuador | JN581928 | JN581760 | JN581845 |
| Zyras (Glossacantha) perdecoratus Pace, 2005 #1 | Lomechusini | ZMUN | 10051274 | Thailand | JN581932 | JN581764 | JN581849 |
| Zyras (Glossacantha) perdecoratus Pace, 2005 #2 | Lomechusini | ZMUN | 10051273 | Thailand | JN581933 | JN581765 | JN581850 |
| Zyras (Glossacantha) sp. 3 #1 | Lomechusini | ZMUN | 10051280 | Thailand | JN581938 | JN581770 | JN581853 |
| Zyras (Glossacantha) sp. 3 #2 | Lomechusini | ZMUN | 10051279 | Thailand | JN581939 | JN581771 | JN581854 |
| Zyras (Glossacantha) sp. prope perdecoratus Pace, 2005 #1 | Lomechusini | ZMUN | 10051276 | Thailand | JN581940 | JN581772 | JN581855 |
| Zyras (Glossacantha) sp. prope perdecoratus Pace, 2005 #2 | Lomechusini | ZMUN | 10051275 | Thailand | JN581941 | JN581773 | JN581856 |
| Zyras (Rhynchodonia) sp. 2 #1 | Lomechusini | ZMUC | 10051233 | Laos | JN581936 | JN581768 | JN581851 |
| Zyras (Rhynchodonia) sp. 2 #2 | Lomechusini | ZMUC | 10051234 | Laos | JN581937 | JN581769 | JN581852 |
| Zyras (s. str.) collaris (Paykull, 1800) | Lomechusini | ZMUN | 10002669 | Abkhasia | JN581931 | JN581763 | JN581848 |
| Zyras sp. 1 #1 | Lomechusini | ZMUN | 10030963 | Thailand | JN581934 | JN581766 | – |
| Zyras sp. 1 #2 | Lomechusini | ZMUN | 10030758 | Thailand | JN581935 | JN581767 | – |
Type species are marked with a bullet (•), or two bullets (••) if the respective genus is the type of its tribe. Additional label information is provided in Table S1.
1) The specimens listed as deposited at ZMUC will be divided between ZMUC and ZMUN.
2) Thendelecrotona was moved from Athetini to Aleocharinae incertae sedis in Elven et al. (2010), but based on its similarity to the Malagasy genus Oxypodinus, we now treat it as a member of Oxypodinini.
3) Meronera is listed under Athetini where it was placed by Elven et al. (2010). The remaining members of the ‘false Lomechusini’ are listed under Lomechusini.
The tribes Lomechusini and Athetini both have global distributions. The Lomechusini in our data set have a good geographic coverage and include species from the Palaearctic, Nearctic, Neotropical and Oriental regions. The Athetini are mainly represented by Palaearctic and Nearctic species but also include one Oriental and two African species.
Whenever possible, two specimens of each species were sequenced as additional control for misidentifications. The total data set included 180 samples representing 120 species, of which 68 belong to Athetini, 33 to Lomechusini, 2 to Ecitocharini and 16 to nine other aleocharine tribes used as outgroup. Tachinus proximus from the subfamily Tachyporinae was included as a more distant outgroup taxon. There was one taxonomic change for the sequences taken from Elven et al. (2010): the genus Thendelecrotona is here recognized to be a member of the tribe Oxypodinini, based on its similarity to the Malagasy genus Oxypodinus in both external characters and the male genitalia.
For most of the Palaearctic and Nearctic specimens, identification to species level was straightforward. For samples from the tropical regions, identification to species level was often impossible and many samples were identified only to genus level. Seven Neotropical species (12 specimens) could not even be assigned to genera, and their initial tribal placement in Lomechusini is based on an assessment of tribal characters, including tarsal formula 4-5-5, mesocoxae relatively broadly separated and mesoventral process short and broad.
Most specimens were collected directly into >96% ethanol and stored at −20 °C prior to processing, but some trap material that had been exposed to high (+20 to +40 °C) temperatures and/or dilution by rain was also included. All specimens used for DNA extraction are labelled as vouchers and deposited at the Natural History Museum, University of Oslo (ZMUN) or the Zoological Museum, Natural History Museum of Denmark (ZMUC). Label information is provided in Table S1.
Molecular markers
Nucleotide sequences from one nuclear and two mitochondrial regions were targeted. The first mitochondrial region covered most of the cytochrome oxidase 1 and 2 (COI and COII) and the tRNA-Leucine 2 (Leu2) genes. The second mitochondrial region covered the 3′-end of the large ribosomal subunit (16S), the tRNA-Leucine 1 (Leu1) and a small part of the NADH dehydrogenase subunit 1 (NADH1) genes. The nuclear region covered an internal part of the small ribosomal subunit (18S) gene. These markers were used by Elven et al. (2010) and have proved suitable for the study of athetine phylogeny.
DNA extraction, amplification and sequencing
DNA was extracted from the head and prothorax using the Qiagen DNeasy Blood and Tissue Kit (Qiagen, Hilden, Germany) following the manufacturer’s protocol for animal tissue. For the very small specimens of Actocharina, the whole body was used. DNA extraction was performed on vacuum-dried samples without prior homogenization. Samples were incubated in lysis buffer for 20–30 h. After extraction, the exoskeletons were retrieved for dry mounting with the rest of the voucher.
The targeted regions were amplified using the primers listed in Table S2. The mitochondrial COI–Leu2–COII region was amplified in three overlapping fragments, while the mitochondrial 16S–Leu1–NADH1 region and the nuclear 18S region were each amplified in single fragments. PCRs were set-up in a 25-μL reaction volume containing 2.5 mm MgCl2 (Applied Biosystems, Foster City, CA, USA), 1 × ABI GeneAmp PCR buffer (Applied Biosystems), 0.8 mm GeneAmp dNTPs (Applied Biosystems), 0.5 μm of each primer (MWG-Biotech AG, Ebersberg, Germany), 1 U ABI AmpliTaq DNA Polymerase (Applied Biosystems) and 3 μL template DNA extract. Most reactions also included 1.1 mg dimethyl sulfoxide (Merck, Darmstadt, Germany) to improve PCR performance. When amplifying the 16S–Leu1–NADH1 region, the PCR set-up was adjusted to 2 mm MgCl2, 0.96 × PCR buffer, 0.64 mm dNTPs and 0.4 μm of each primer. The amplification profile consisted of an initial denaturation step of 94 °C for 30 s, followed by 30 cycles of 94 °C for 1 min, annealing temperature Ta for 30 s and 72 °C for 2 min, and finally a 10 min extension step at 72 °C. The annealing temperatures are listed in Table S2. For some difficult samples, PCR performance was improved by replacing dimethyl sulfoxide in the reaction mix with 0.4 μg bovine serum albumin (Sigma-Aldrich, Steinheim, Germany), by using alternative primers (listed in Table S2), by lowering the annealing temperature or by using an alternative PCR protocol employing HotStar Taq DNA Polymerase (Qiagen).
PCR products were purified using ExoSAP-IT (USB Corporation, Cleveland, Ohio, USA). If secondary products were detected on a standard agarose gel, the PCR product of appropriate size was cut out from 1% agarose gel and purified using the MN NucleoSpin Extract II gel extraction kit (Macherey-Nagel, Düren, Germany). Purified PCR products were sequenced in both directions using the terminal primers with the ABI BigDye Terminator Cycle Sequencing Kit v3.1 (Applied Biosystems) and analysed on the ABI 3730 DNA Analyzer (Applied Biosystems). All DNA sequencing was outsourced to the ABI-lab, Departments of Biology and of Molecular Biosciences, University of Oslo.
Sequence alignment and model selection
Alignment of the protein-coding genes was straightforward, as there were virtually no indels. For the RNA-coding genes, published secondary structures from other insect groups were used as a guide for manual alignment in MEGA 4 (Tamura et al. 2007). Secondary structures for Apis mellifera (Gillespie et al. 2006) were used to aid alignment of 16S and 18S, while secondary structures for Xenos vesparum (Carapelli et al. 2006) were used to aid alignment of Leu1 and Leu2. Loop regions that could not be aligned unambiguously were excluded from the subsequent analyses.
The concatenated alignment was partitioned by codon positions, stems vs. loops, and genomic origin (mitochondrial vs. nuclear) to produce a total of seven partitions (for details, see Elven et al. 2010). MrModelTest 2.3. (Nylander 2004) was used to determine a suitable evolutionary model for each partition under the Akaike information criterion.
Phylogenetic analyses
The partitioned data set was analysed under maximum likelihood (ML) and Bayesian inference. Two analyses were performed with each method; one with all sequences included and one where sequences with more than 20% missing information were excluded (i.e. those lacking data for one of the three targeted regions).
ML analyses were performed in RAxML 7.0.3 (Stamatakis 2006) using the Rapid Bootstrap algorithm (Stamatakis et al. 2008) followed by ML optimization. Each analysis was performed with 1000 bootstrap replicates, and every fifth bootstrap tree was used as a starting point for subsequent ML optimization on the original data set. The partitioned data set was analysed using the GTRMIX option (CAT approximation for the bootstrap, followed by final ML optimization under the GTR+Г model). ML optimization was also performed on 100 randomized parsimony trees to test whether this would yield a higher final likelihood score. The tree with the overall highest score was selected as the best tree, and bootstrap values were drawn on this tree.
Prior to the analysis, the rearrangement settings and number of rate categories were optimized following the author’s recommendations (RAxML 7.0.4 Manual). Two alternative rearrangement settings (i = auto vs. i = 10) were compared by performing ML optimization on 10 randomized parsimony trees with each setting, and choosing the setting resulting in the highest likelihood score for the optimized trees. Four alternative numbers of rate categories (10, 25, 40 and 55) were then compared in the same way using the best rearrangement setting from above. The same 10 starting trees were used for all comparisons. Based on the results of these comparisons, the analysis that included incomplete sequences was performed using i = auto, while the analysis using only complete sequences was performed using i = 10. Both analyses were performed using 25 rate categories.
Bayesian analyses were performed in MrBayes v3.1.2 (Ronquist & Huelsenbeck 2003) using the GTR+Г model for 3rd codon positions and GTR+I+Г for the other partitions. MrModelTest suggested GTR+I+Г for all partitions, but under this modelling regime, the runs did not converge. Closer inspection of the model parameters showed that several 3rd codon position parameters stabilized at different values in different runs and that the difference was most pronounced for the pinvar (I) and gamma (Г) parameters. When gamma alone was used to account for rate heterogeneity in this partition, the runs converged. The 3rd codon positions contained very few (2.1%) invariable sites, and there may have been insufficient data for estimating the pinvar parameter properly. All model parameters were unlinked across partitions and were allowed to evolve during the run starting from flat priors. All analyses were performed with four independent runs, each with three heated and one cold chain. Preliminary runs revealed poor mixing for the rate multiplier parameter under the default tuning parameter value of 500, and the value was therefore increased to 8000 (less bold proposals). The analyses were run for 100 million generations with sampling every 10 000 generations. Convergence was assessed by examining the average standard deviation of split frequencies between the four runs, the potential scale reduction factor for each model parameter and the mixing behaviour of the model parameters. The average standard deviation of split frequencies was calculated using a custom C++ program, SplitFreqs, which is available from the first author upon request. Model parameters and likelihood values were inspected in Tracer (Rambaut & Drummond 2007). The posterior tree distribution was summarized in a majority-rule consensus tree after discarding the first 25% of the samples as burn-in. The analyses were run at the Bioportal computer facility (http://www.bioportal.uio.no) at the University of Oslo, Norway.
Results
Sequence alignment
The concatenated sequence alignment of 180 samples included 3786 positions after trimming. Complete sequence information was obtained for 150 samples, while 30 lacked sequence data for one of the three target regions (Table 1). Because of alignment ambiguity, 284 positions were excluded from all phylogenetic analyses. Table S3 lists the number of parsimony informative, uninformative, invariant and excluded sites for each target gene. The alignment and partition definitions are included as a nexus file in the Supporting information (Data S1).
Phylogenetic analyses
The majority-rule consensus trees from the Bayesian analyses are shown in Figs 1 (complete sequences only) and 2 (incomplete sequences included). The trees with the highest likelihood score from the ML analyses are shown in Figs S1 (complete sequences only) and S2 (incomplete sequences included). In both ML analyses, the highest-scoring trees were found by using bootstrap trees rather than randomized parsimony trees as the starting points for ML optimization.
Figure 1.
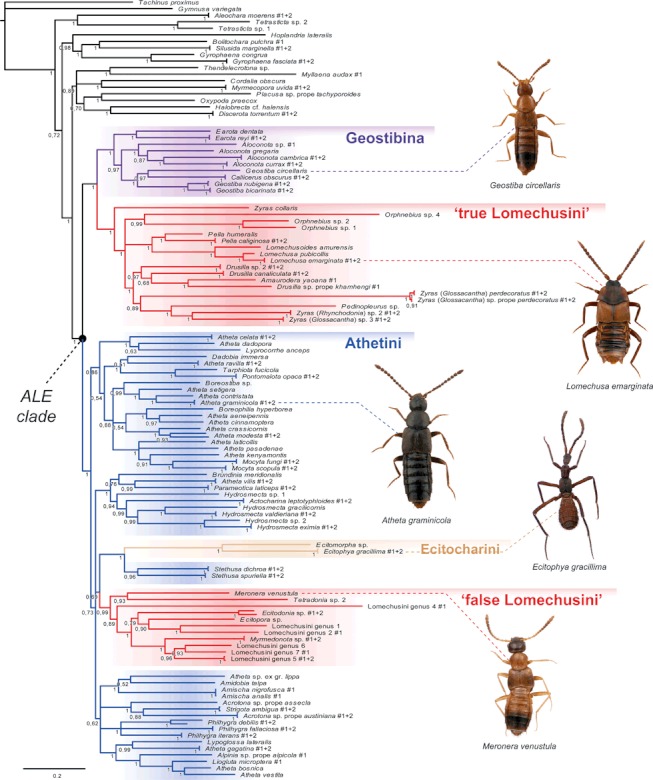
Majority-rule consensus tree from the Bayesian analysis with incomplete sequences excluded. Posterior probabilities are indicated under the branches. The labels of conspecific specimens have been combined to save space.
Figure 2.
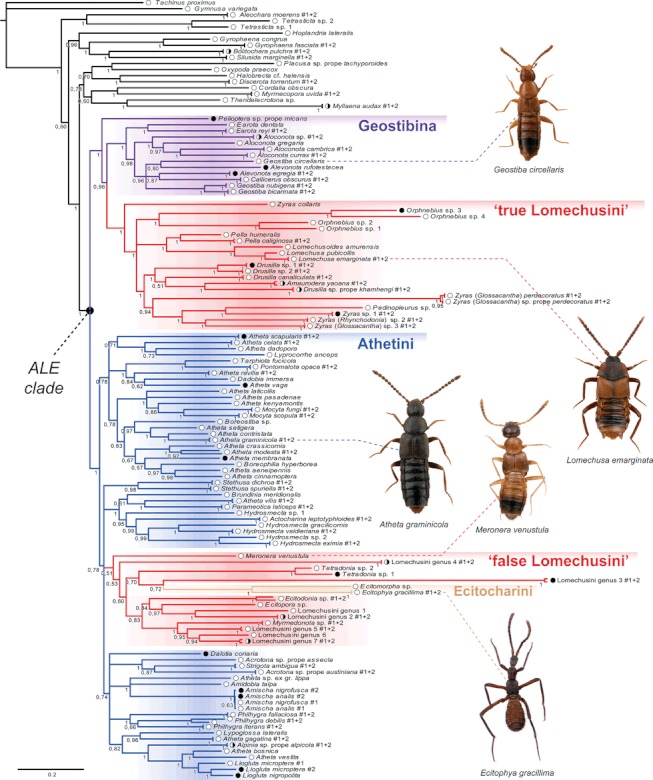
Majority-rule consensus tree from the Bayesian analysis with incomplete sequences included. Posterior probabilities are indicated under the branches. The labels of conspecific specimens have been combined to save space, except where the specimens did not group together. Complete sequences are indicated with open circles, incomplete with solid circles. Half-solid circles indicate pairs of conspecific specimens with one having incomplete sequence.
In both Bayesian analyses, the average standard deviation of split frequencies stabilized between 0.01 and 0.05 after about 20 million generations, indicating that stationarity was reached after about 5 million generations. The final standard deviation values after 100 million generations were 0.011 when incomplete sequences were excluded and 0.032 when they were included. The potential scale reduction factor approached 1 for all model parameters, never exceeding 1.009. Most model parameters showed good mixing, but the substitution rates and base frequencies for unpaired mitochondrial sites showed bimodal sampling when incomplete sequences were excluded. The chains swapped frequently between the two distinct optima, and both were thoroughly sampled during the run. The rate multiplier parameters showed alternating periods of good and poor mixing when the proposals tuning parameter value was adjusted to 8000, in contrast to uniformly poor mixing under the default value of 500.
The Bayesian and ML analyses produced largely congruent phylogenies (Figs 1, 2, S1 and S2) with very similar relative branch lengths, although the total inferred tree length was somewhat higher (up to 23%) under Bayesian inference. Many nodes were recovered with high statistical support both under ML and Bayesian inference. All trees contained some exceptionally long branches, particularly among the Lomechusini.
The ALE clade was recovered with high statistical support in all analyses (Bayesian posterior probability (PP) = 1.00, ML bootstrap support (BS) = 87%). When incomplete sequences were excluded (Figs 1 and S1), the Geostibina clade, represented by Aloconota, Callicerus, Earota and Geostiba, formed a sister group to the ‘true Lomechusini’ clade, which included Amaurodera, Drusilla, Lomechusa, Lomechusoides, Orphnebius, Pedinopleurus, Pella and Zyras. The Geostibina clade, the ‘true Lomechusini’ clade and the sister group relationship between them all received high statistical support (PP = 1.00, BS > 95%). In turn, the (Geostibina, ‘true Lomechusini’) clade formed a sister group to the main Athetini clade (PP = 1.00, BS = 83%), which included Ecitocharini and the ‘false Lomechusini’. The ‘false Lomechusini’ clade, consisting of Ecitodonia, Ecitopora, Meronera, Myrmedonota, Tetradonia and several not yet identified Neotropical taxa, was well supported in the Bayesian analysis (PP = 0.99) but not in the ML analysis (BS < 50%). The Ecitocharini, represented by two genera, were recovered as monophyletic (PP = 1.00, BS = 100%) and had Stethusa as its sister group (PP = 1.00, BS = 93%).
The inclusion of 30 incomplete sequences had little impact on the overall tree topology (Figs 2 and S2). Of the well-supported relationships, only the (Stethusa, Ecitocharini) clade was not recovered when incomplete sequences were included; Stethusa and Ecitocharini instead formed separate clades with poorly supported or unresolved relationships to other athetine clades. Also, the ‘false Lomechusini’ were not recovered as monophyletic when incomplete sequences were included.
All five genera included in the Geostibina clade have sensillum a of the epipharynx reduced. Among the other genera included in our study, only Pelioptera shares this character state. When examining the microscope slides of the entomological collection of the Natural History Museum, Oslo, as well as published illustrations of athetine genera, we identified additional genera (not included in our phylogenies) that have sensillum a of the epipharynx reduced. Table 2 lists taxa in which the presence of reduced sensillum a has been confirmed. In total, 13 genera were confirmed to share this character state. For nine of these, the type species was examined. Six other athetine genera that resemble Geostiba in having the ligula fully bilobed were found to have sensillum a fully developed (i.e. much longer than wide): Boreophilia (slide preparation examined), Liogluta (see Yosii & Sawada 1976: fig. 40B), Madeirostiba (see Assing & Wunderle 1995: fig. 5), Ousipalia (slide preparation examined), Saphocallus (see Assing 2001: fig. 16a), Schistoglossa (slide preparation examined) and Tomoglossa (see Sawada 1977: fig. 9c).
Table 2.
Genera and species confirmed to belong to the tribe Geostibini based on the presence of reduced sensillum a of the epipharynx. Type species are indicated in bold
| Genus | Species | Examined material/publications |
|---|---|---|
| Alevonota Gravenhorst, 1802 | A. gracilenta (Erichson, 1839) | ZMUN slide collection |
| Aloconota Thomson, 1858 | A. brunneipes (Casey, 1906) | ZMUN slide collection |
| A. bulbosa Sawada, 1989 | Sawada (1989a): fig. 12b | |
| A. cuspidata (Sawada, 1971) | Sawada (1971): fig. 2c (as Tomoglossa) | |
| A. gregaria (Erichson, 1839) | Yosii & Sawada (1976): fig. 43b | |
| A. insecta (Thomson, 1856) | Yosii & Sawada (1976): fig. 44b | |
| A. languida (Erichson, 1839) | Sawada (1984): fig. 3b (as Disopora) | |
| A. pfefferi (Roubal, 1929) | ZMUN slide collection | |
| A. punctifoveata (Sawada, 1970) | Sawada (1970): fig. 8c (as Tomoglossa) | |
| A. sulcifrons (Stephens, 1832) | ZMUN slide collection | |
| Callicerus Gravenhorst, 1802 | C. obscurus Gravenhorst, 1802 | Yosii & Sawada (1976): fig. 46b Assing (2001): fig. 2b |
| C. rigidicornis (Erichson, 1839) | Assing (2001): fig. 14a | |
| Chinecallicerus Assing, 2004 | C. laevigatus Assing, 2006 | Assing (2006): fig. 4 |
| Earota Mulsant & Rey, 1874 | E. dentata (Bernhauer, 1906) | Gusarov (2002b): fig. 2 |
| Enalodroma Thomson, 1859 | E. hepatica (Erichson, 1839) | Sawada (1984): fig. 4b (as Aloconota) |
| Geostiba Thomson, 1858 | G. alticola Lohse & Smetana, 1988 | ZMUN slide collection |
| G. appalachigena Gusarov, 2002 | ZMUN slide collection | |
| G. balsamensis Pace, 1997 | ZMUN slide collection | |
| G. bicarinata Lohse & Smetana, 1988 | ZMUN slide collection | |
| G. carteriensis Pace, 1997 | Gusarov (2002a): fig. 4 | |
| G. circellaris (Gravenhorst, 1806) | Yosii & Sawada (1976): fig. 47b Gusarov (2002a): fig. 2 | |
| G. crepusculigena Gusarov, 2002 | ZMUN slide collection | |
| G. daisetsuana Sawada, 1989 | Sawada (1989b): fig. 3b | |
| G. flava (Kraatz, 1856) | ZMUN slide collection | |
| G. graveyardensis Pace, 1997 | ZMUN slide collection | |
| G. infirma (Weise, 1878) | ZMUN slide collection | |
| G. nebuligena Gusarov, 2002 | ZMUN slide collection | |
| G. pluvigena Gusarov, 2002 | ZMUN slide collection | |
| G. sakhalinensis Pace, 1997 | ZMUN slide collection | |
| G. winkleri (Bernhauer, 1915) | ZMUN slide collection | |
| Homoiocalea Bernhauer, 1943 | H. toroenensis (Bernhauer, 1943) | Sawada (1984): fig. 1b (as Callicerus) |
| Micrearota Casey, 1910 | M. prolongata (Casey, 1910) | ZMUN slide collection |
| Pelioptera Kraatz, 1857 | P. acuticollis (Kraatz, 1859) | Sawada (1982): fig. 10b |
| P. babai Sawada, 1989 | Sawada (1989a): fig. 13b | |
| P. exasperata (Kraatz, 1859) | Sawada (1982): fig. 11b | |
| P. flavonitescens (Bernhauer, 1938) | Sawada (1977): fig. 17k (as Geostiba) | |
| P. luzonica (Bernhauer, 1916) | Sawada (1980): fig. 16b | |
| P. micans (Kraatz, 1857) | Sawada (1980): fig. 9b | |
| P. monticola Cameron, 1933 | Sawada (1980): fig. 15b | |
| P. nilgiriensis (Fauvel, 1904) | Sawada (1980): fig. 11b | |
| P. opaca (Kraatz, 1857) | Sawada (1980): fig. 10b | |
| P. ocyamensis (Bernhauer, 1914) | Sawada (1977): fig. 17b (as Geostiba) | |
| P. peguana (Bernhauer, 1915) | Sawada (1980): fig. 14b | |
| P. purpurascens (Cameron, 1920) | Sawada (1987): fig. 5b | |
| P. testaceipennis (Motschulsky, 1858) | Sawada (1977): fig. 18b (as Geostiba luchuensis (Cameron, 1933) Sawada (1980): fig. 13b | |
| P. vacillator (Cameron, 1933) | Sawada (1977): fig. 19b (as Geostiba) | |
| P. xylophila (Cameron, 1920) | Sawada (1980): fig. 12b | |
| Pseudosemiris Machulka, 1935 | P. kaufmanni (Eppelsheim, 1887) | Assing (2001): fig. 19a |
| Seeversiella Ashe, 1986 | S. globicollis (Bernhauer, 1907) | Gusarov (2003): fig. 2 |
| Tropimenelytron Pace, 1983 | T. americanum Gusarov, 2002 | ZMUN slide collection |
| T. tuberiventre (Eppelsheim in Leder, 1879) | Gusarov (2002c): fig. 2 | |
| T. unicum (Bernhauer, 1907) | Sawada (1977): fig. 15b (as Aloconota) |
Discussion
The molecular phylogeny of the Athetini–Lomechusini–Ecitocharini (ALE) clade of aleocharine rove beetles presented in this study is in line with the main finding of Elven et al. (2010). The monophyly of the ALE clade as well as the three major clades within it, the Geostibina, the ‘true Lomechusini’, and the main Athetini clade, was confirmed with high statistical support. All non-geostibine Athetini except Discerota torrentum were recovered as part of the main Athetini clade, which also included Ecitocharini and the ‘false Lomechusini’. Of the five initial hypotheses to be tested in this study, none was rejected. There was a strong statistical support for all hypothesized relationships except the monophyly of the ‘false Lomechusini’ (hypothesis 4), for which there was only moderate support. The five hypotheses are reviewed below.
H1: Geostibina are a sister clade to the ‘true Lomechusini’.
The Geostibina clade and the ‘true Lomechusini’ clade were both strongly supported, and the sister group relationship between them strongly corroborated.
Within the Geostibina clade, the monophyly of Earota (two species) and Aloconota (four species) was confirmed, but the monophyly of Geostiba (three species) and Alevonota (two species) was not. The inclusion of Callicerus in the Geostibina clade creates a nomenclatural problem. The rarely used family group name Callicerina Jakobson, 1908 has priority over the currently more widely used Geostibina Seevers, 1978; but the first is also a junior homonym of Callicerini Rondani, 1845, which is in use for a tribe of hoverflies (Diptera: Syrphidae). To solve the issue, an application proposing to suppress the name Callicerina Jakobson, 1908 and to keep the more widely used Geostibina Seevers, 1978 has been submitted to the International Commission on Zoological Nomenclature (Gusarov 2011). Pending the Commission’s ruling, we maintain current usage and treat the name Geostibina as valid.
The ‘true Lomechusini’ clade included Drusilla, Lomechusa, Pella and five other genera represented here by species from the Old World. Within this clade, the genera Lomechusa (two species), Orphnebius (four species) and Pella (two species) were recovered as monophyletic with high support. The genus Drusilla (four species) was recovered as paraphyletic with respect to the morphologically distinct Oriental genus Amaurodera (one species). As currently accepted, Drusilla is distributed worldwide and includes almost 200 species (Hlaváč et al. 2011). Within Drusilla, there is a fairly distinct group of Palaearctic species related to the type species of the genus, Drusilla canaliculata (revised by Assing 2005). However, Drusilla in the broad sense can only be defined by the lack of unusual characters (e.g. the distinct shape of pronotum in Amaurodera). A worldwide revision of Drusilla is needed to divide the genus into diagnosable monophyletic genera. The genus Zyras (represented in our analyses by six species) was recovered as polyphyletic. The nominotypical subgenus (represented by Z. collaris) was recovered as the sister group to all other ‘true Lomechusini’. Two closely related species of Zyras (Glossacantha) from Thailand were recovered in a separate clade with a very long branch. A third clade included yet another species of Zyras (Glossacantha), one species of Zyras (Rhynchodonia), one Zyras species not yet identified to subgenus and the morphologically very distinct genus Pedinopleurus. Accordingly, even the subgenus Glossacantha is not monophyletic. In the current classification, Zyras is divided into 54 subgenera and includes more than 800 species (Hlaváč et al. 2011). Some groups formerly treated as subgenera of Zyras have been raised to genus rank (e.g. Pella: Kistner 1972; Maruyama 2006) and new subgenera are being described regularly (e.g. Pace 1999). Like Drusilla, Zyras is a group in need of a worldwide revision. Our results confirm the opinion of Kistner (1972) and Maruyama (2006) that Pella is not related to Zyras and should be treated as a separate genus.
H2: Geostibina and the ‘true Lomechusini’ form a sister group to the main Athetini clade.
The main Athetini clade was recovered with strong support in a strongly supported sister group relationship with Geostibina and the ‘true Lomechusini‘. The basal nodes of the main Athetini clade were not resolved with good support.
H3: All athetine genera with sensillum a of the epipharynx reduced belong to Geostibina.
All five genera forming the Geostibina clade (Alevonota, Aloconota, Callicerus, Earota and Geostiba) have sensillum a of the epipharynx reduced. Among the other genera included in this study, only Pelioptera has this character state. In our analyses, Pelioptera formed a sister group to the (Geostibina, ‘true Lomechusini’) clade. The support for this placement was not strong, however, and is further weakened by the fact that the entire COI–COII region was missing from the Pelioptera sequence. Given that Pelioptera shares additional character states (see below) with the five genera of Geostibina, we hypothesize that Pelioptera is also a member of Geostibina and that the reduced sensillum a is a synapomorphy for this group. In addition to the six genera included in the molecular analyses, seven further athetine genera were confirmed to possess the reduced sensillum a of the epipharynx (Table 2). All 13 genera also share the shape of the ligula: broad at the base and divided into two separate lobes. We hypothesize that they all belong to Geostibina. Within Athetini, there are additional genera with a fully bilobed ligula. Two of these, Liogluta and Boreophilia, were included in our study and were not recovered as members of the Geostibina clade. Based on our examination, they also have a normally developed sensillum a. Five more genera with a bilobed ligula not included in this study (Madeirostiba, Ousipalia, Saphocallus, Schistoglossa and Tomoglossa) were also confirmed to have a normally developed sensillum a. Based on this evidence, we predict that they do not belong to the Geostibina clade.
H4: The ‘false Lomechusini’ are nested within the main Athetini clade.
The ‘false Lomechusini’ clade included Meronera, Myrmedonota and 10 other genera (11 species) from the New World. Seven of the included genera were unidentified. The clade was well supported only in the Bayesian analysis (Fig. 1), and when incomplete sequences were included, the ‘false Lomechusini’ were no longer recovered as monophyletic. The position of the ‘false Lomechusini’ within the main Athetini clade was not resolved. However, the ‘false Lomechusini’ are confirmed to belong to the main Athetini clade and thus not to be monophyletic with the ‘true Lomechusini’.
Ten of the included genera were collected in Ecuador and two in the USA. Of the five identified genera, two (Ecitopora and Ecitodonia) are exclusively Neotropical. Two genera (Tetradonia and Meronera) are distributed mostly in the Neotropical region, with a few species also in the Nearctic. The fifth genus, Myrmedonota, is known from both the Nearctic and the Oriental regions (but see below). The geographic distribution of the ‘false Lomechusini’ suggests that the clade may have originated in South America and dispersed into North America. More extensive taxon sampling in both Old and New World tropics is needed to test this hypothesis.
The placement of Meronera and Myrmedonota in the ‘false Lomechusini’ clade is in line with Elven et al. (2010). The genus Myrmedonota was represented by a species from the Eastern USA, but was originally described from Singapore (Cameron 1920) and is furthermore known from Malaysia, Indonesia and Papua New Guinea. It was only recently reported from North America by Maruyama et al. (2008), who provided a new diagnosis of the genus based on the type species M. cingulata, two new species from the Eastern United States and published descriptions of two species from New Guinea. The geographic distribution of Myrmedonota suggests that the Nearctic species may not be related to and congeneric with the Oriental. We do not propose to remove Myrmedonota from Lomechusini until additional species, in particular the type, have been examined in more detail.
H5: Ecitocharini form a monophyletic sister group to Stethusa within the main Athetini clade.
The Ecitocharini were recovered as a strongly supported monophyletic group within the main Athetini clade. When only complete sequences were used, Ecitocharini formed a well-supported sister group to the New World athetine genus Stethusa (Figs 1 and S1). However, when taxa with incomplete sequences were included in the analysis, Ecitocharini grouped (with weak support) with the longest branch of the ‘false Lomechusini’ formed by the unidentified genus 3 (Figs 2 and S2). This genus lacked sequence data for the COI–Leu2–COII region, and the 18S sequences of both genus 3 and the Ecitocharini were unusually divergent. We consider the weakly supported sister group relationship between Ecitocharini and genus 3 to be an artefact and treat the well-supported sister group relationship between Ecitocharini and Stethusa as phylogenetically correct.
The type genus of Ecitocharini, Ecitochara, was unfortunately not available for this study, which instead included the genera Ecitophya and Ecitomorpha as representatives of the tribe. However, the members of Ecitocharini share several derived morphological character states (Kistner & Jacobson 1990) and are furthermore connected by life style (i.e. association with army ants of the genus Eciton) and geographic distribution. It seems reasonable to assume that the tribe is monophyletic.
As the Ecitocharini are nested within the main Athetini clade, their inclusion in Athetini should be uncontroversial. Seevers (1965) rationale for erecting the tribe Ecitocharini was to ‘emphasize their evolutionary and ecological divergence from the Athetini’, but he believed that the former were derived from the latter, which is congruent with this study. We therefore place the name Ecitocharini Seevers, 1965 in synonymy with Athetini Casey, 1910. Ecitocharina may still be used as a valid name at the rank of subtribe, but lack of resolution at the base of the main Athetini clade does not currently allow us to divide the entire tribe into subtribes.
Tribe-level classification of the ALE clade
The aim of this study was to resolve the phylogeny of the major lineages of the ALE clade in order to revise the classification of the tribes involved. With strong support for all three subclades within the ALE clade, the ‘true Lomechusini’, the subtribe Geostibina and the main Athetini clade, we can address the issue of classification. A revised tribe-level classification needs to meet the following criteria: (i) all formally recognized taxa should be monophyletic; (ii) the classification should reflect the three main subclades and be compatible with their phylogenetic relationships: [(Geostibina, Lomechusini) (the main Athetini clade including the ‘false Lomechusini’)]; (iii) the principle of priority should be satisfied; (iv) the recognized family group taxa should be diagnosable using morphological characters, preferably apomorphic states; and (v) the choice of classification should promote stability of scientific names (ICZN, 1999: Preamble). We here discuss three possible alternative classifications, all of which satisfy criteria 1–3 and all of which meet criterion 4 equally well by recognizing the three main subclades of the ALE clade in one way or another. The main difference between the three alternatives is in the ranks of some taxa. Therefore, we will focus on how the changes in classification will affect the stability of names (criterion 5).
Alternative 1: raising the rank of Geostibina to tribe. Three tribes are recognized in the ALE clade: Athetini, Geostibini and Lomechusini. Most of the genera and species currently in Athetini and Lomechusini stay in their respective tribes. The members of subtribe Geostibina (most of which belong to the genus Geostiba) are removed from Athetini by raising the rank of the subtribe to tribe. The genera of the ‘false Lomechusini’ are moved from Lomechusini to Athetini. Further subdivision of the three tribes into subtribes is still possible.
Alternative 2: moving subtribe Geostibina to Lomechusini. Two tribes are recognized in the ALE clade: Athetini and Lomechusini (including subtribe Geostibina). Most of the genera and species currently in Athetini and Lomechusini stay in their respective tribes. The subtribe Geostibina is moved from Athetini to Lomechusini, while the genera of ‘false Lomechusini’ are moved from Lomechusini to Athetini. Subdivision of the two tribes into subtribes is still possible.
Alternative 3: expanding the Lomechusini to include all Athetini. Only one tribe is recognized in the ALE clade: Lomechusini. This tribe is further subdivided into three subtribes: Athetina, Geostibina and Lomechusina. This solution is similar to some earlier classifications (e.g. by Bernhauer & Scheerpeltz 1926) where Athetini and Lomechusini were treated as subtribes of the tribe Myrmedoniini Thomson, 1867. The members of ‘false Lomechusini’ stay in the tribe Lomechusini, but are moved to the subtribe Athetina. If this solution is implemented, hundreds of genera and thousands of species currently in Athetini will need to be moved to Lomechusini, and the largest aleocharine tribe will be abandoned. This solution does not promote stability of names. Furthermore, as the ranks of what are currently treated as tribes Athetini and Lomechusini are lowered to subtribes, it becomes impossible to recognize the taxa currently treated as subtribes of Athetini and Lomechusini, except by inserting a rank of infratribe.
The main difference between the three alternatives is in how the stability of classification is promoted. Alternative 1 seems to achieve that goal best and at the same time allows further subdivision of all three tribes into subtribes. We favour this alternative and, consequently, raise the rank of the subtribe Geostibina to tribe and move the genera of ‘false Lomechusini’ to Athetini.
The redefined tribes Athetini, Lomechusini and Geostibini
Most of the Athetini can be diagnosed by a combination of the following characters: sensillum a of the epipharynx fully developed, galea of moderate length, mesocoxae narrowly or moderately separated, and mesoventral process not broad. Most of the Lomechusini can be diagnosed by a combination of the following characters: sensillum a of the epipharynx fully developed, galea significantly elongate, mesocoxae broadly separated, and mesoventral process broad and short. Both in Athetini and Lomechusini, there are exceptions that do not fit the above diagnoses, and a detailed morphological study will be needed to improve the diagnoses of the two tribes.
The main diagnostic character and putative synapomorphy for the tribe Geostibini is sensillum a of the epipharynx reduced (e.g. as in Yosii & Sawada 1976: fig. 47B). Geostibini share with Athetini and Lomechusini the tarsal formula 4-5-5 and the presence of the athetine bridge of the aedeagus. Like most Athetini, but unlike Lomechusini, Geostibini have the galea moderately long, mesocoxae narrowly or moderately separated, and mesoventral process not broad. Like Lomechusini and some Athetini, Geostibini have the ligula broad at the base and divided into two separate lobes. Table 2 lists all genera in which reduction in sensillum a of the epipharynx has been confirmed. We place all these genera in the tribe Geostibini. There is no doubt that additional genera will need to be transferred from Athetini to Geostibini. Unfortunately, the main diagnostic character of Geostibini, sensillum a of the epipharynx, is rarely described or illustrated in published papers. Thus, for most athetine genera, direct examination of slide mounted specimens will be needed to assess their tribal placement.
Genus Discerota
The athetine genus Discerota (represented by D. torrentum) formed a well-supported clade with Halobrecta outside the ALE clade. In Elven et al. (2010), Halobrecta was removed from Athetini and tentatively placed in Oxypodini. Like Halobrecta, Discerota lacks the athetine bridge of the aedeagus and has both male and female genitalia similar to some Oxypodini. For these reasons, we move Discerota from Athetini and place it tentatively in Oxypodini as well.
The riparian clade of Athetini
Compared to Elven et al. (2010), we expanded the taxon sampling of Athetini by adding several species associated with riparian habitats. In addition to providing a more rigorous test of the monophyly of the main Athetini clade, this allowed us to test whether the riparian athetines are related to each other or if different lineages have colonized the riparian habitats independently.
Remarkably, many riparian taxa were recovered as members of a strongly supported monophyletic group. This riparian clade included Actocharina, Atheta (Dralica) vilis, Brundinia, Parameotica and Hydrosmecta (represented by five species).
The monotypic genus Actocharina, originally a subgenus of Atheta, is distributed in Austria and northern Italy where it inhabits sandy river banks in the Kalkalpen. The beetles are minuscule, only up to 1.4 mm long, have reduced eyes, wings and pigmentation and presumably move in the interstitial space between sand particles like many Hydrosmecta. When describing Atheta (Actocharina) leptotyphloides, Bernhauer (1907) mentioned its close relationship to two small species of Hydrosmecta, H. subtilissima and H. tenuissima. With respect to external morphology, Actocharina is indeed similar to the smallest species of Hydrosmecta, for example, H. delicatula or H. tenuissima (cf. figs 162:1 and 164:13, 17 in Benick & Lohse 1974). The main difference between the two genera is Actocharina being more derived in characters related to cryptic interstitial life style. In our analyses, Actocharina formed a well-supported clade with four of the five included species of Hydrosmecta, and the fifth Hydrosmecta species was sister to this clade. We, therefore, consider Actocharina a morphologically derived member of the genus Hydrosmecta and place the name Actocharina Bernhauer, 1907 in synonymy with Hydrosmecta Thomson, 1858.
Conclusions
Elven et al. (2010) demonstrated that the tribes Athetini and Lomechusini are not monophyletic and that the tribe Ecitocharini may belong to Athetini. In this study, we thoroughly assessed the basal relationships among the three tribes to propose a phylogenetically robust tribe-level classification. The athetine subtribe Geostibina was shown to be a sister group to the ‘true Lomechusini’. Five athetine genera are included in this clade, and eight more can be referred to it based on morphology. Geostibina and the ‘true Lomechusini’ together form a sister group to the main Athetini clade, which comprises all non-geostibine athetines in addition to the tribe Ecitocharini and the ‘false Lomechusini’. The resolution within the main Athetini clade was poor, but support for the clade itself was strong. The monophyly of the ‘false Lomechusini’ was not strongly supported; nevertheless, there is no doubt about their inclusion in Athetini and their separation from the ‘true Lomechusini’. We propose raising the subtribe Geostibina to the rank of tribe. Doing so will best promote stability of nomenclature, while complying with the criterion of monophyly. We furthermore propose including the ‘false Lomechusini’ and the Ecitocharini in Athetini.
It is likely that future revisional work on Athetini and Lomechusini and the inclusion of further genera in large-scale phylogenetic analyses will further change the definition of these tribes. The ‘false Lomechusini’ in particular raise many further questions, such as whether the group is actually monophyletic, which other New World genera belong to it, whether the group is also represented outside the New World, and which are its closest relatives within Athetini. Further studies are also needed on phylogeny of the ‘true Lomechusini’. The molecular markers used in this study may prove suitable for resolving relationships within Zyras and Drusilla, the two most challenging genera within the Lomechusini.
Proposed changes in classification
The following changes in classification of Aleocharinae are proposed (see Table 3 for details): (i) Geostibina, formerly a subtribe of Athetini, is raised to tribe rank as Geostibini Seevers, 1978, stat. nov. Thirteen genera are moved from Athetini to Geostibini. (ii) Three genera are moved from Lomechusini to Athetini. (iii) The family group name Ecitocharini Seevers, 1965 is placed in synonymy with the name Athetini Casey, 1910. All 10 genera formerly treated as members of the tribe Ecitocharini are moved to Athetini. (iv) The genus Discerota Mulsant & Rey, 1874 is removed from Athetini and tentatively included in Oxypodini. (v) The genus name Actocharina Bernhauer, 1907 is placed in synonymy with Hydrosmecta Thomson, 1858. The new combination Hydrosmecta leptotyphloides (Bernhauer, 1907) is established for the species originally described as Atheta (Actocharina) leptotyphloides Bernhauer, 1907.
Table 3.
Proposed changes in classification of Aleocharinae
| Name | Previous status/placement | New status/placement |
|---|---|---|
| Geostibina Seevers, 1978 | Valid subtribe of Athetini | Tribe Geostibini Seevers, 1978, stat. nov. |
| Alevonota Thomson, 1858 | Athetini | Geostibini |
| Aloconota Thomson, 1858 | Athetini | Geostibini |
| Callicerus Gravenhorst, 1802 | Athetini | Geostibini |
| Chinecallicerus Assing, 2004 | Athetini | Geostibini |
| Earota Mulsant & Rey, 1874 | Athetini | Geostibini |
| Enalodroma Thomson, 1859 | Athetini | Geostibini |
| Geostiba Thomson, 1858 | Athetini | Geostibini |
| Homoiocalea Bernhauer, 1943 | Athetini | Geostibini |
| Micrearota Casey, 1910 | Athetini | Geostibini |
| Pelioptera Kraatz, 1857 | Athetini | Geostibini |
| Pseudosemiris Machulka, 1835 | Athetini | Geostibini |
| Seeversiella Ashe, 1986 | Athetini | Geostibini |
| Tropimenelytron Pace, 1983 | Athetini | Geostibini |
| Ecitodonia Seevers, 1965 | Lomechusini | Athetini |
| Ecitopora Wasmann, 1887 | Lomechusini | Athetini |
| Tetradonia Wasmann, 1894 | Lomechusini | Athetini |
| Ecitocharini Seevers, 1965 | Valid tribe | New synonym of Athetini Casey, 1910 |
| Campbellia Kistner & Jacobson, 1990 | Ecitocharini | Athetini |
| Ecitochara Wasmann, 1887 | Ecitocharini | Athetini |
| Ecitodaemon Reichensperger, 1939 | Ecitocharini | Athetini |
| Ecitomorpha Wasmann, 1889 | Ecitocharini | Athetini |
| Ecitophya Wasmann, 1900 | Ecitocharini | Athetini |
| Ecitoschneirla Kistner & Jacobson, 1990 | Ecitocharini | Athetini |
| Ecitosymbia Bruch, 1923 | Ecitocharini | Athetini |
| Ecitoxenia Wasmann, 1900 | Ecitocharini | Athetini |
| Retteneciton Kistner & Jacobson, 1990 | Ecitocharini | Athetini |
| Seeverseciton Kistner & Jacobson, 1990 | Ecitocharini | Athetini |
| Discerota Mulsant & Rey, 1874 | Athetini | Tentatively Oxypodini |
| Actocharina Bernhauer, 1907 | Valid genus | New synonym of Hydrosmecta Thomson, 1858 |
| Actocharina leptotyphloides (Bernhauer, 1907) | Actocharina | Hydrosmecta leptotyphloides (Bernhauer, 1907), new combination |
Acknowledgments
We are grateful to Sinan Anlaş, Andrey Babenko, Igor Belousov, Alexander Derunkov, Erhard Lipkow, Martin Fikáček, Andrey Gontarenko, Ilya Kabak, Derek Lott, György Makranczy, Yuri Marusik, Jan Pedersen, Alexander Ryabukhin, Michael Sharkey, Andrea Schomann, Alexey Shavrin, Alexey Solodovnikov, Alexey Tishechkin, Marc Tronquet, Larry Watrous and Nikolay Yunakov for providing specimens for DNA analysis. Volker Assing gave valuable advice on collecting methods and habitats of some rare species needed for our study. We acknowledge the help of Heinrich Terlutter who shared his experience in collecting riparian aleocharines. We thank Galina Gussarova for helpful advice on the analyses and for constructive discussion of the results. The project was supported in part by the Norwegian Research Council grant 180681/D15 to V. Gusarov.
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Best tree from the maximum likelihood analysis with incomplete sequences excluded. Bootstrap values ≥ 50% are indicated under the branches. The labels of conspecific specimens have been combined to save space.
Best tree from the maximum likelihoodanalysis with incomplete sequences included. Bootstrap values ≥ 50% are indicated under the branches. The labels of conspecific specimens have been combined to save space, except where the specimens did not group together. Complete sequences are indicated with open circles, incomplete with solid circles. Half-solid circles indicate pairs of conspecific specimens with one having incomplete sequence.
Label information for the specimens included in this study. The specimens marked with asterisk (*) are on loan from the Natural History Museum of Denmark (ZMUC) and will be divided between ZMUC and the Natural History Museum, University of Oslo (ZMUN) upon completion of the project. The remaining specimens are deposited at ZMUN.
Primers used for amplification andsequencing. Alternative primers used to amplify certain difficultsamples are indicated with ‘alt’. Primers usedexclusively as internal sequencing primers are indicated with‘int’.
Alignment statistics on the molecular markers used in the study. The four genes COI, COII, 16S and 18S account for 94% of the total alignment, or 3568 sites.
ALE_alignment.nex. Sequence alignment in nexus format. Concatenated alignment of two mitochondrial and one nuclear region, covering 3568 bases for 180 samples.
References
- Ahn K-J. Phylogenetic relationships of the intertidal genus Halorhadinus Sawada and key to the genera of the Liparocephalini (Coleoptera: Staphylinidae: Aleocharinae) Insect Systematics and Evolution. 2001;32:123–132. [Google Scholar]
- Ahn K-J, Ashe JS. Phylogeny of the Myllaenini and related taxa (Coleoptera: Staphylinidae: Aleocharinae) Cladistics. 2004;20:123–138. doi: 10.1111/j.1096-0031.2004.00012.x. [DOI] [PubMed] [Google Scholar]
- Ahn K-J, Jeon M-J, Branham MA. Phylogeny, biogeography and the stepwise evolutionary colonization of intertidal habitat in the Liparocephalini based on morphological and molecular characters (Coleoptera: Staphylinidae: Aleocharinae) Cladistics. 2010;26:344–358. doi: 10.1111/j.1096-0031.2009.00290.x. [DOI] [PubMed] [Google Scholar]
- Ashe JS. Fecundity, development and natural history of Meronera venustula (Erichson) (Coleoptera: Staphylinidae: Aleocharinae) Psyche. 1985;92:181–204. [Google Scholar]
- Ashe JS. Phylogeny of the tachyporine group of subfamilies and ‘basal’ lineages of the Aleocharinae (Coleoptera: Staphylinidae) based on larval and adult characteristics. Systematic Entomology. 2005;30:3–37. [Google Scholar]
- Ashe JS. In The Tree of Life Web Project. 2007. http://tolweb.org/. Available via http://tolweb.org/Aleocharinae/9777/2007.04.25 (downloaded 2011, October 13) April 25 Aleocharinae. Version 25 April 2007.
- Assing V. A revision of Callicerus Gravenhorst, 1802, Pseudosemiris Machulka 1935, and Saphocallus Sharp, 1888 (Coleoptera: Staphylinidae, Aleocharinae, Athetini) Beiträge zur Entomologie. 2001;51:247–334. [Google Scholar]
- Assing V. On the western Palaearctic species of Drusilla Leach, with special reference to the species of the eastern Mediterranean (Coleoptera: Staphylinidae, Aleocharinae) Koleopterologische Rundschau. 2005;75:111–149. [Google Scholar]
- Assing V. A new genus and new species of Aleocharinae from China (Insects: Coleoptera: Staphylinidae) Entomological Problems. 2006;36:21–30. [Google Scholar]
- Assing V, Wunderle P. A new genus of Aleocharinae from Madeira (Insecta: Coleoptera: Staphylinidae) Reichenbachia. 1995;31:31–36. [Google Scholar]
- Benick G, Lohse GA. 14. Tribus: Callicerini (Athetae) In: Freude H, Harde KW, Lohse GA, editors. Die Käfer Mitteleuropas. Band 5. Staphylinidae II (Hypocyphtinae und Aleocharinae). Pselaphidae. Krefeld: Goecke & Evers; 1974. pp. 72–220. [Google Scholar]
- Bernhauer M. 1907;57:185–186. Atheta (nov. subg. Actocharina leptotyphloides Bernh. nov. spec Verhandlungen der Kaiserlich-Königlichen Zoologisch-Botanischen Gesellschaft in Wien. [Google Scholar]
- Bernhauer M, Scheerpeltz O. Staphylinidae VI. Coleopterorum Catalogus. 1926;82:499–988. [Google Scholar]
- Bouchard P, Bousquet Y, Davies AE, Alonso-Zarazaga MA, Lawrence JF, Lyal CHC, Newton AF, Reid CAM, Schmitt M, Ślipiński SA, Smith ABT. Family-group names in Coleoptera (Insecta) ZooKeys. 2011;88:1–972. doi: 10.3897/zookeys.88.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron M. New species of Staphylinidae from Singapore. Part III. The Transactions of the Entomological Society of London. 1920;1920:212–284. [Google Scholar]
- Carapelli A, Vannini L, Nardi F, Boore JL, Beani L, Dallai R, Frati F. The mitochondrial genome of the entomophagous endoparasite Xenos vesparum (Insecta: Strepsiptera) Gene. 2006;376:248–259. doi: 10.1016/j.gene.2006.04.005. [DOI] [PubMed] [Google Scholar]
- Elven H, Bachmann L, Gusarov VI. Phylogeny of the tribe Athetini (Coleoptera: Staphylinidae) inferred from mitochondrial and nuclear sequence data. Molecular Phylogenetics and Evolution. 2010;57:84–100. doi: 10.1016/j.ympev.2010.05.023. [DOI] [PubMed] [Google Scholar]
- Gillespie JJ, Johnston JS, Cannone JJ, Gutell RR. Characteristics of the nuclear (18S, 5.8S, 28S and 5S) and mitochondrial (12S and 16S) rRNA genes of Apis mellifera (Insecta: Hymenoptera): structure, organization, and retrotransposable elements. Insect Molecular Biology. 2006;15:657–686. doi: 10.1111/j.1365-2583.2006.00689.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grebennikov VV, Newton AF. Good-bye Scydmaenidae, or why the ant-like stone beetles should become megadiverse Staphylinidae sensu latissimo (Coleoptera) European Journal of Entomology. 2009;106:275–301. [Google Scholar]
- Gusarov VI. A revision of Nearctic species of the genus Geostiba Thomson, 1858 (Coleoptera: Staphylinidae: Aleocharinae) Zootaxa. 2002a;81:1–88. [Google Scholar]
- Gusarov VI. A revision of Nearctic species of the genus Earota Mulsant & Rey, 1874 (Coleoptera: Staphylinidae: Aleocharinae) Zootaxa. 2002b;92:1–16. [Google Scholar]
- Gusarov VI. A revision of Nearctic species of the genus Tropimenelytron Pace, 1983 (Coleoptera: Staphylinidae: Aleocharinae), a new genus for North America. Zootaxa. 2002c;114:1–24. [Google Scholar]
- Gusarov VI. A revision of the genus Seeversiella Ashe, 1986 (Coleoptera: Staphylinidae: Aleocharinae) Zootaxa. 2003;142:1–102. [Google Scholar]
- Gusarov VI. Athetini Casey, 1910 and Geostibina Seevers, 1978 (Insecta, Coleoptera, Staphylinidae, Aleocharinae): proposed conservation. Bulletin of Zoological Nomenclature. 2011;68:54–60. [Google Scholar]
- Hlaváč P, Newton AF, Maruyama M. World catalogue of the species of the tribe Lomechusini (Staphylinidae: Aleocharinae) Zootaxa. 2011;3075:1–151. [Google Scholar]
- ICZN. International Code of Zoological Nomenclature. 4th edn. London: The International Trust for Zoological Nomenclature; 1999. [Google Scholar]
- Kistner DH. Studies of Japanese myrmecophiles. Part I. The genera Pella and Falagria (Coleoptera, Staphylinidae) Entomological Essays to Commemorate Retirement of Prof. K. Yasumatsu in 1971. 1972. pp. 141–165.
- Kistner D, Jacobson H. Cladistic analysis and taxonomic revision of the ecitophilous tribe Ecitocharini with studies of their behavior and evolution (Coleoptera, Staphylinidae, Aleocharinae) Sociobiology. 1990;17:333–480. [Google Scholar]
- Klimaszewski J, Pace R, Center TD, Couture J. A remarkable new species of Himalusa Pace from Thailand (Coleoptera, Staphylinidae, Aleocharinae): phytophagous aleocharine beetle with potential for bio-control of skunkvine-related weeds in the United States. ZooKeys. 2010;35:1–12. [Google Scholar]
- Maruyama M. Revision of the Palearctic species of the myrmecophilous genus Pella (Coleoptera, Staphylinidae, Aleocharinae) National Science Museum Monographs. 2006;32:1–207. [Google Scholar]
- Maruyama M, Patrick LB, Klimaszewski J. First record of the genus Myrmedonota Cameron (Coleoptera, Staphylinidae) from North America, with descriptions of two new species. Zootaxa. 2008;1716:35–43. [Google Scholar]
- Maus C, Peschke K, Dobler S. Phylogeny of the genus Aleochara inferred from mitochondrial cytochrome oxidase sequences (Coleoptera: Staphylinidae) Molecular Phylogenetics and Evolution. 2001;18:202–216. doi: 10.1006/mpev.2000.0874. [DOI] [PubMed] [Google Scholar]
- Muona J. Aleocharinae cladistics (Coleoptera, Staphylinidae) In A Celebration of Systematics. Abstracts of the 15th Meeting of the Willi Hennig Society, 15–20 December 1996, University of Cape Town, South Africa Cladistics. 1997;13:161–186. [Google Scholar]
- Navarrete-Heredia JL, Newton AF, Thayer MK, Ashe JS, Chandler DS. Guía ilustrada para los géneros de Staphylinidae (Coleoptera) de México. México: Universidad de Guadalajara, CONABIO; 2002. [Google Scholar]
- Newton AF, Thayer MK. Current classification and family-group names in Staphyliniformia (Coleoptera) Fieldiana Zoology. 1992;67:i–iv. New Series 1–92. [Google Scholar]
- Newton AF, Thayer MK, Ashe JS, Chandler DS. 22. Staphylinidae Latreille, 1802. In: Arnett RH, Thomas MC, editors. American Beetles, vol. 1, Archostemata, Myxophaga, Adephaga, Polyphaga: Staphyliniformia. Boca Raton, FL: CRC Press, LLC; 2000. pp. 272–418. [Google Scholar]
- Nylander JAA. MrModeltest v2. [Computer software and manual] Uppsala: Evolutionary Biology Centre, Uppsala University; 2004. Program distributed by the author. [Google Scholar]
- O’Keefe STO. Scydmaenidae Leach, 1815. In: Beutel RG, Leschen RAB, editors. Coleoptera, Beetles Volume 1: Morphology and Systematics (Archostemata, Adephaga, Myxophaga, Polyphaga partim). Handbook of Zoology, Vol. IV. Part 38. Berlin: Walter de Gruyter; 2005. pp. 280–288. [Google Scholar]
- Pace R. Aleocharinae di Hong Kong (Coleoptera, Staphylinidae) Revue suisse de Zoologie. 1999;106:663–689. [Google Scholar]
- Pace R. New records of Aleocharinae from Ecuador and Peru, with the description of new species, new subgenera and new genera (Coleoptera, Staphylinidae) In: Giachino PM, editor. Biodiversity of South America I. Memoirs on Biodiversity Volume I. Verona: World Biodiversity Association Onlus; 2008. pp. 225–398. [Google Scholar]
- Paśnik G. Phylogeny and Generic Classification of Tachyusini (Coleoptera, Staphylinidae: Aleocharinae) Kraków: Institute of Systematics and Evolution of Animals, Polish Academy of Sciences; 2010. p. 127p. [Google Scholar]
- Rambaut A, Drummond AJ. 2007. Tracer v1.4. Available via http://beast.bio.ed.ac.uk/
- Ronquist F, Huelsenbeck JP. MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Sawada K. Aleocharinae (Staphylinidae, Coleoptera) of the IBP-Station in the Shiga Heights, Central Japan, II. Contributions from the Biological Laboratory. Kyoto University. 1970;23:33–60. [Google Scholar]
- Sawada K. Aleocharinae (Staphylinidae, Coleoptera) from the campus of the Seto Marine Biological Laboratory. Publications of the Seto Marine Biological Laboratory. 1971;18:291–315. [Google Scholar]
- Sawada K. Studies on the genus Atheta Thomson and its allies (Coleoptera, Staphylinidae). III. Japanese species described by the previous authors. Contributions from the Biological Laboratory, Kyoto University. 1977;25:171–222. [Google Scholar]
- Sawada K. Atheta and its allies of Southeast Asia (Coleoptera; Staphylinidae). II. Reexamination of the species mainly from Borneo. Contributions from the Biological Laboratory, Kyoto University. 1980;26:23–66. [Google Scholar]
- Sawada K. von Motschulsky V, Kraatz G. Atheta and its allies of Southeast Asia (Coleoptera; Staphylinidae). III. Oriental species. Contributions from the Biological Laboratory, Kyoto University. 1982. pp. 141–187.
- Sawada K. Studies on the genus Atheta Thomson and its allies (Coleoptera, Staphylinidae). IV. Systematic studies on Liogluta series with notes of taxa established in C. G. Thomson, 1858 and G. Kraatz, 1859. Contributions from the Biological Laboratory, Kyoto University. 1984;26:429–452. [Google Scholar]
- Sawada K. Atheta and its allies of Southeast Asia (Coleoptera; Staphylinidae). V. Singaporean species, described in Cameron, 1920. Contributions from the Biological Laboratory, Kyoto University. 1987;27:137–150. [Google Scholar]
- Sawada K. New species of Aleocharinae from Japan, I (Coleoptera; Staphylinidae) Contributions from the Biological Laboratory, Kyoto University. 1989a;27:273–307. [Google Scholar]
- Sawada K. Five new species of Atheta and its allies from Hokkaido, Japan (Coleoptera: Staphylinidae) Bulletin of the Sounkyo Museum of Natural History. 1989b;9:1–15. [Google Scholar]
- Seevers CH. The Systematics, evolution and zoogeography of Staphylinid beetles associated with army ants (Coleoptera, Staphylinidae) Fieldiana Zoology. 1965;47:137–351. [Google Scholar]
- Seevers CH. A generic and tribal revision of the North American Aleocharinae (Coleoptera: Staphylinidae) Fieldiana Zoology. 1978;71:1–289. [Google Scholar]
- Smetana A. Subfamily Aleocharinae Fleming, 1821. In: Löbl I, Smetana A, editors. Catalogue of Palaearctic Coleoptera, vol. 2. Hydrophyloidea–Histeroidea–Staphylinoidea. Stenstrup: Apollo Books; 2004. pp. 353–494. [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based Phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Stamatakis A, Hoover P, Rougemont J. A Rapid bootstrap Algorithm for the RAxML web servers. Systematic Biology. 2008;57:758–771. doi: 10.1080/10635150802429642. [DOI] [PubMed] [Google Scholar]
- Steidle JLM, Dettner K. Chemistry and morphology of the tergal gland of freeliving adult Aleocharinae (Coleoptera: Staphylinidae) and its phylogenetic significance. Systematic Entomology. 1993;18:149–168. [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Molecular Biology and Evolution. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Thayer MK. Staphylinidae Latreille, 1802. In: Beutel RG, Leschen RAB, editors. Coleoptera, Beetles Volume 1: Morphology and Systematics (Archostemata, Adephaga, Myxophaga, Polyphaga partim). Handbook of Zoology, Vol. IV. Part 38. Berlin: Walter de Gruyter; 2005. pp. 296–344. [Google Scholar]
- Thomas JC. A preliminary molecular investigation of aleocharine phylogeny (Coleoptera: Staphylinidae) Annals of the Entomological Society of America. 2009;102:189–195. [Google Scholar]
- Yosii R, Sawada K. Studies on the genus Atheta Thomson and its allies (Coleoptera, Staphylinidae). II. Diagnostic characters of genera and subgenera with descriptions of representative species. Contributions from the Biological Laboratory, Kyoto University. 1976;25:11–140. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Best tree from the maximum likelihood analysis with incomplete sequences excluded. Bootstrap values ≥ 50% are indicated under the branches. The labels of conspecific specimens have been combined to save space.
Best tree from the maximum likelihoodanalysis with incomplete sequences included. Bootstrap values ≥ 50% are indicated under the branches. The labels of conspecific specimens have been combined to save space, except where the specimens did not group together. Complete sequences are indicated with open circles, incomplete with solid circles. Half-solid circles indicate pairs of conspecific specimens with one having incomplete sequence.
Label information for the specimens included in this study. The specimens marked with asterisk (*) are on loan from the Natural History Museum of Denmark (ZMUC) and will be divided between ZMUC and the Natural History Museum, University of Oslo (ZMUN) upon completion of the project. The remaining specimens are deposited at ZMUN.
Primers used for amplification andsequencing. Alternative primers used to amplify certain difficultsamples are indicated with ‘alt’. Primers usedexclusively as internal sequencing primers are indicated with‘int’.
Alignment statistics on the molecular markers used in the study. The four genes COI, COII, 16S and 18S account for 94% of the total alignment, or 3568 sites.
ALE_alignment.nex. Sequence alignment in nexus format. Concatenated alignment of two mitochondrial and one nuclear region, covering 3568 bases for 180 samples.


