Abstract
Lactococcus lactis is an attractive vehicle for biotechnological production of proteins and clinical delivery of therapeutics. In many such applications using this host, it is desirable to maximize secretion of recombinant proteins into the extracellular space, which is typically achieved by using the native signal peptide from a major secreted lactococcal protein, Usp45. In order to further increase protein secretion from L. lactis, inherent limitations of the Usp45 signal peptide (Usp45sp) must be elucidated. Here, we performed extensive mutagenesis on Usp45sp to probe the effects of both the mRNA sequence (silent mutations) and the peptide sequence (amino acid substitutions) on secretion. We screened signal peptides based on their resulting secretion levels of Staphylococcus aureus nuclease and further evaluated them for secretion of Bacillus subtilis α-amylase. Silent mutations alone gave an increase of up to 16% in the secretion of α-amylase through a mechanism consistent with relaxed mRNA folding around the ribosome binding site and enhanced translation. Targeted amino acid mutagenesis in Usp45sp, combined with additional silent mutations from the best clone in the initial screen, yielded an increase of up to 51% in maximum secretion of α-amylase while maintaining secretion at lower induction levels. The best sequence from our screen preserves the tripartite structure of the native signal peptide but increases the positive charge of the n-region. Our study presents the first example of an engineered L. lactis signal peptide with a higher secretion yield than Usp45sp and, more generally, provides strategies for further enhancing protein secretion in bacterial hosts.
INTRODUCTION
Lactococcus lactis, a Gram-positive bacterium, has been widely used in the food fermentation industry for the production of cheese and buttermilk (1). Over the past two decades, there has been increasing interest in the use of this bacterium for the biotechnological production of heterologous proteins and clinical delivery of therapeutic molecules. A major advantage of using L. lactis or other Gram-positive bacteria rather than Gram-negative bacteria (e.g., Escherichia coli) for protein production is that proteins can be easily secreted into the medium, which streamlines the often expensive and inefficient downstream purification process (2). In contrast to many commonly used Gram-positive bacteria (e.g., Bacillus species), laboratory strains of L. lactis possess only one exported housekeeping protease, HtrA (3), and secrete only one major extracellular protein, Usp45 (4), thus minimizing protein degradation by the extracellular proteolytic system and further simplifying downstream purification. Given its simple and well-known metabolism, its completely sequenced genome (5, 6), and the availability of various genetic tools (7), L. lactis is an attractive host for many biotechnological applications. Furthermore, this bacterium has generally regarded as safe (GRAS) status in humans and has been used as a mucosal delivery vehicle because it can survive passage through the stomach acid and contact with bile (8, 9). L. lactis has been engineered to express and secrete a variety of therapeutic proteins, including interleukin-10 for treatment of inflammatory bowel disease (10), bovine β-lactoglobulin for immunity against this cow milk allergen (11), and single-chain insulin for treatment of diabetes (12). In such biomedical applications, it is often desirable to maximize secretion of the recombinant protein into the extracellular space in order to reduce the dose and administration frequency of the therapeutic bacteria.
In L. lactis, most proteins are secreted unfolded via the secretion (Sec) pathway (13). Proteins are synthesized as precursors containing the mature moiety of the protein with an N-terminal signal peptide. The signal peptide plays an important role in targeting the protein to the cytoplasmic membrane, where the protein precursor is subsequently translocated by the Sec machinery (14). Following cleavage of the signal peptide, the mature protein is released extracellularly (15). The lactococcal signal peptides follow the common tripartite structure, including a positively charged N terminus (n-region), a central hydrophobic core (h-region), and a more polar C terminus (c-region) containing the signal peptide cleavage site (16). The most widely used signal peptide for L. lactis secretion is that from Usp45 (Usp45sp), which has the best secretion efficiency (17). Other reported lactococcal signal peptides, both natural (e.g., SP310 [18], SPExp4 [19], and AL9 [20]) and engineered (e.g., SP310mut2 [21]), have secretion efficiencies only as good as, and often worse than, that of Usp45sp (2, 21, 22). Although improvements in secretion have been achieved by either introducing a synthetic propeptide (23) or complementing the lactococcal translocation machinery with SecDF (24), the inherent limitations of Usp45sp itself have not been addressed. Thus, optimization of this widely used signal peptide should not only directly lead to enhancements in the secretion efficiencies of a wide range of proteins but may also be combined with other improvements, such as those mentioned above, to fully exploit the secretion potential of L. lactis.
As the signal peptide of the only major secretory protein in L. lactis, it is unknown whether Usp45sp has been subjected to stringent evolutionary pressures that have maximized its secretory efficiency. Here, we screened libraries of silent and targeted mutations in Usp45sp and demonstrated that the secretion efficiency of this signal peptide can indeed be improved. We first showed that silent mutations of Usp45sp alone can give an increase of up to 16% in the activity of secreted α-amylase. We then used a simple mathematical model to provide a quantitative explanation for this observed effect based on the free energy of the mRNA secondary structure around the ribosome binding site (RBS) and its consequent effect on protein translation. We further introduced nonsilent mutations in Usp45sp using a targeted mutagenesis approach and achieved an increase of up to ∼50% in the activity of secreted α-amylase at the maximum induction level. Using the best clone from the targeted mutation library, we introduced silent mutations into this sequence and identified clones that also increase secretion levels at lower concentrations of inducer. Analyzing these silent mutation clones with our mathematical model, we show that the effect of optimizing the amino acid sequence is more dominant than that of relaxing mRNA folding in enhancing secretion levels. In summary, we have used two complementary directed-evolution approaches to achieve progressive improvements in L. lactis secretion efficiencies solely by engineering the signal peptide; this report represents the first successful effort in achieving secretion yields higher than that attainable with Usp45sp and also provides general strategies for enhancing protein secretion in bacterial hosts.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Bacterial strains and plasmids used in this work are listed in Table S2 in the supplemental material. E. coli was grown in TY medium (8 g tryptone, 5 g yeast extract, and 5 g sodium chloride per liter) at 37°C with shaking, and L. lactis was statically grown in M17 medium (Oxoid, Hampshire, United Kingdom) containing 0.5% glucose (GM17) at 30°C. Solid media were prepared by adding agar (15 g/liter) to the corresponding broths. For the nuclease plate assay, brain heart infusion (BHI) agar (Acumedia, Lansing, MI) was used to grow L. lactis. When needed, chloramphenicol (Cm) was used at a final concentration of 10 μg/ml.
DNA manipulations and plasmid construction.
Isolation of plasmid DNA from E. coli and L. lactis was performed as described previously (12). Phusion high-fidelity DNA polymerase, restriction enzymes, and T4 DNA ligase were purchased from NEB (Ipswich, MA) and were used as recommended by the manufacturer. Primers used for DNA amplification (Integrated DNA Technologies, Coralville, IA) are listed in Table S3 in the supplemental material. pNZ8048m:usp45sp-MazF was constructed by isolating the mazF gene from E. coli K-12 (GenBank accession no. NC000913) by colony PCR with primers 1 and 2, digesting with BspEI and BamHI, and ligating into similarly cut pNZ8048m:usp45sp-SCI57his (12) (Usp45sp from GenBank accession no. M60178 but with two silent mutations to introduce a BspEI site, as described previously [12]). To incorporate an NcoI site at the junction between the coding sequences for Usp45sp and mazF, pNZ8048m:usp45sp-MazF was PCR amplified with primers 3 and 2, digested with BspEI and BamHI, and ligated into similarly cut pNZ8048m:usp45sp-SCI57his, giving rise to pNZ8048m:usp45sp-MazF. pNZ8048m:usp45sp-Nuc (Fig. 1A) was constructed by isolating the nuclease gene from Staphylococcus aureus (GenBank accession no. AM990992) by colony PCR with primers 4 and 5, digesting with NcoI and BamHI, and ligating into similarly cut pNZ8048m:usp45sp-MazF. pNZ8048m:usp45sp-AmyE was constructed using the same method as described above after isolating Bacillus subtilis α-amylase (GenBank accession no. FJ643607) with primers 6 and 7, except that an internal EcoRI site was eliminated by a silent mutation to facilitate subsequent library construction. This was done by PCR amplification of the α-amylase with primers 6 and 9 and primers 8 and 7, fusion of the two fragments by assembly PCR, digestion of the full-length product with NcoI and BamHI, and ligation into NcoI- and BamHI-digested pNZ8048m:usp45sp-MazF. To replace Usp45sp with the SP310 signal peptide, SP310 was constructed by extension PCR with primers 10 and 11 followed by digestion with EcoRI and NcoI and ligation into similarly cut pNZ8048m:usp45sp-Nuc and pNZ8048m:usp45sp-AmyE to yield pNZ8048m:SP310-Nuc and pNZ8048m:SP310-AmyE, respectively. All ligation mixtures were transformed into chemically competent E. coli EC1000, sequenced, and electroporated into electrocompetent L. lactis NZ9000 as described previously (12).
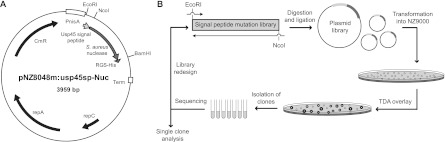
Fig 1.
Schematics of the expression constructs and screening process. (A) Plasmid map of pNZ8048m:usp45sp-Nuc, showing promoter PnisA, usp45 signal peptide sequence, S. aureus nuclease gene, and RGS-His tag in the modified pNZ8048 backbone (12). (B) Flow diagram of the mutagenesis and screening process. The signal peptide mutations were encoded by degenerate PCR primers. Nucleotide libraries were constructed, digested, and ligated into the pNZ8048m vector backbone via the EcoRI and NcoI sites. After plasmid transformation into L. lactis NZ9000, the resulting library was plated and screened by carefully overlaying a toluidine blue-DNA agar (TDA) mixture with inducer (10 ng/ml nisin) onto the agar plates. Since the size of the pink halo around each colony corresponds to the amount of nuclease secreted, clones could be quickly screened by eye and subjected to further analyses (sequencing, amylase secretion assays, Western blotting, etc.). Subsequent libraries were designed based on data collected in the initial screen.
Library construction and selection procedures.
The selection process is outlined in Fig. 1B. To construct the mutation libraries for Usp45sp silent mutations, Usp45sp targeted mutations, and Usp45TM8 silent mutations, three separate extension PCRs were performed with forward primer 12 and degenerate reverse primers 14, 15, and 16, respectively. The constructs were digested with EcoRI and NcoI and ligated with a 1:1 molar ratio into >1 μg similarly digested pNZ8048m:SP310-Nuc. The ligation products were electroporated directly into L. lactis NZ9000 and plated onto 10 150-mm-diameter petri dishes with BHI agar containing Cm. Colonies were allowed to grow overnight at 30°C.
Toluidine blue-DNA agar (TDA) was made with 36 mg toluidine blue O (Sigma-Aldrich, St. Louis, MO), 4 g NaCl, 4 g agar, 0.12 g salmon DNA (EMD Chemicals, Billerica, MA), and 0.4 ml of 0.01 M CaCl2 dissolved in 400 ml of 0.05 M Tris (pH 9) (25). Molten TDA (13 ml) containing 10 ng/ml nisin (Sigma-Aldrich, St. Louis, MO) was carefully overlaid on each BHI agar plate and allowed to solidify. After incubation at 30°C for 1 h, colonies were evaluated by the size of their pink halos. Single colonies were picked, inoculated into 5 ml GM17–Cm, and grown overnight at 30°C. Plasmids in selected clones were isolated and analyzed by DNA sequencing. To clone the selected signal peptide upstream of α-amylase, signal peptides were PCR amplified with primers 12 and 13, digested with EcoRI and NcoI, and ligated into similarly digested pNZ8048m:usp45sp-AmyE.
Nuclease plate assay.
To evaluate the secretion efficiency and activity of nuclease secreted by individual clones, 10 ml molten TDA was spread into each 100-mm-diameter petri dish and allowed to solidify. Wells were created in the TDA with P1000 pipet tips (USA Scientific, Ocala, FL). Overnight cultures of L. lactis NZ9000 containing pNZ8048m:usp45sp-Nuc (or similar constructs with other signal peptides) were diluted 1:25 into fresh GM17–Cm. Cultures were grown for 1.5 h and induced with the appropriate amount of nisin for another 2 h. The optical density at 600 nm (OD600) was measured on a plate reader (Infinite M200; Tecan, Männedorf, Switzerland) to ensure that different cultures had similar growth rates. Cells were removed from the supernatant by a 5-min centrifugation at 5,000 × g, 1 μl supernatant was added to each TDA well, and the plates were incubated at 30°C for 1 h. The size of the pink halo around each clone was measured.
α-Amylase activity assay and Western blotting.
To evaluate the secretion efficiency and activity of α-amylase secreted by individual clones, starch azure assays were carried out as previously described (17) but with modifications. Cultures of L. lactis were grown, induced with nisin, and centrifuged as mentioned above. The supernatant (100 μl) was incubated with 20 mg starch azure (Sigma-Aldrich) and 900 μl α-amylase buffer (50 mM Tris-HCl [pH 7.5], 50 mM NaCl, 5 mM CaCl2) at 37°C for 1 h with shaking. Centrifugation was carried out at 15,000 × g for 5 min, and supernatant absorbance at 595 nm was measured on a plate reader. α-Amylase activity was calculated from a standard curve (see Fig. S1 in the supplemental material). The standard curve was obtained by incubating 0.0038 to 3.8 units of purified B. subtilis α-amylase (Sigma-Aldrich) with starch azure under the conditions described above.
For Western blotting, the supernatant was passed through a 0.22-μm-pore-size filter (Millipore, Billerica, MA) to remove any cells, and 13 μl supernatant (adjusted with phosphate-buffered saline as needed to achieve equivalent cell densities) was mixed with 2 μl 500 mM dithiothreitol and 5 μl lithium dodecyl sulfate sample buffer for analysis by SDS-PAGE in a 4% to 12% NuPAGE Bis-Tris gel (Invitrogen, Carlsbad, CA). Proteins were transferred and blotted as described previously (12).
mRNA structure analysis and mathematical modeling.
The mRNA sequence from the transcription start site to 6 nucleotides after the end of the signal peptide-coding region (135 bases total) was analyzed using the mfold web server (26). The structure with the lowest (most negative) folding energy was used to calculate the free energy (ΔG) of the ribosome binding site (RBS). ΔGRBS was calculated by adding up the ΔG contributions of the 6 bases in AGGAGG and an initiation ΔG of 3.4 kcal/mol (27).
To relate mRNA folding to translation and secretion efficiencies, a mathematical model was developed using modifications of a previous model (28). The details of this thermodynamic model, including parameter values, are provided in the supplemental material. Briefly, the model predicts that translational efficiency correlates with ribosome binding to accessible RBSs, which depends on the fraction of RBSs in the unfolded state, which, in turn, is determined by the ΔG of the RBS for a given mRNA sequence. Secretion efficiency is determined as a fraction of the total amount of translated protein and is specific to a particular amino acid sequence.
RESULTS
Signal peptide screening with nuclease and α-amylase reporters.
To allow efficient screening of a large library of signal peptides, the Staphylococcus aureus nuclease was used as the reporter protein (29). Secretion of S. aureus nuclease from L. lactis colonies can be easily detected by overlaying the library transformants with toluidine blue-DNA agar (TDA) (25); the size of the pink halo indicates nuclease activity, which corresponds directly to secretion efficiency. For better quantification of secretion efficiency, selected signal peptides were fused upstream of Bacillus subtilis α-amylase. Activity of secreted α-amylase in liquid culture can be easily quantified by incubation with starch azure (17) (see Fig. S1 in the supplemental material). Furthermore, the use of two different heterologous proteins to evaluate secretion efficiencies can elucidate any protein-specific dependence on secretion and increase the likelihood that the best signal peptide selected would improve secretion of other useful proteins. These two proteins, S. aureus nuclease and B. subtilis α-amylase, were evaluated for their abilities to report secretion efficiencies of two known signal peptides, Usp45sp (17) and SP310 (18). A TDA plate test, carried out on L. lactis cultures secreting nuclease fused to either signal peptide, showed a larger pink halo with Usp45sp, indicating secretion levels qualitatively higher than those of SP310 (see Fig. S2A in the supplemental material). A more quantitative starch azure activity test on cultures secreting α-amylase also demonstrated that secretion with SP310 was only ∼62% of that achieved with Usp45sp (see Fig. S2B in the supplemental material), in agreement with previous results (21). We therefore used Usp45sp as the starting sequence for directed evolution to probe any inherent sequence limitations in using this signal peptide and to identify and alleviate bottlenecks to improve protein secretion in L. lactis. The signal peptide mutagenesis and screening process was performed as shown in Fig. 1B. To construct silent mutation libraries, we used primers with appropriate degeneracies in the third nucleotide of each codon, giving a maximum theoretical diversity of ∼1012. For the targeted mutation library, degenerate codons (NNK) were used to fully randomize specific amino acid residues, giving a maximum diversity of ∼1019. Our actual library sizes were limited by the transformation efficiency in L. lactis (∼106 CFU/μg DNA). A TDA-nisin overlay allowed us to rapidly screen a large number of nuclease-secreting colonies by visualizing the sizes of the pink halos and selecting individual clones for more quantitative testing with α-amylase. The maximum subinhibitory concentration of nisin (10 ng/ml) was used for all screens (30).
Silent mutations in Usp45sp modestly increase secretion.
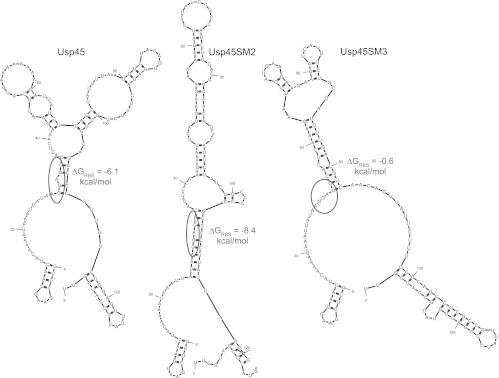
First, a Usp45sp silent mutation library was constructed to examine if mRNA secondary structure, independent of amino acid sequence, has an effect on secretion and if secretion can be improved by relaxing mRNA structure, assuming that translation and secretion are directly correlated. It is known that silent mutations in the gene itself can alter mRNA folding and translation initiation, thus influencing gene expression (31). Wild-type Usp45sp is predicted to have a considerable amount of mRNA folding and secondary structure, especially around the RBS (Fig. 2), and preliminary studies on signal peptides with very highly folded mRNAs revealed that secretion was completely abolished (results not shown). We therefore created a library to allow all possible silent mutations of Usp45sp and screened for (i) clones with representative low, medium, and high secretion levels to further probe the correlation between secondary structure and secretion rate and (ii) clones with secretion levels significantly higher than that of wild-type Usp45sp. Individual silent mutation clones were assayed for secretion of nuclease or α-amylase and were ordered by their ΔG values around the RBS (Table 1), which we hypothesized to be a major determinant of translation initiation and, thus, protein expression and secretion (28). The majority of both nuclease and α-amylase secretion levels follows the ΔG trend: signal peptides with ΔGRBS values that are more negative generally exhibited worse secretion than Usp45sp, and those with ΔGRBS values that were less negative generally yielded better secretion than Usp45sp. To visualize putative mRNA folding, secondary structure predictions for Usp45sp, Usp45SM2, and Usp45SM3—representing the wild-type sequence, a worse secreting sequence, and a better secreting sequence, respectively—were computed using mfold (26) (Fig. 2). As shown, Usp45SM2 has greater secondary structure around the RBS, which would be expected to hinder ribosome binding and lead to lower expression and secretion. On the other hand, Usp45SM3 shows a more relaxed mRNA conformation around the RBS, which would more readily allow translation initiation and, thus, result in higher secretion. Three of the best clones, Usp45SM10, Usp45SM5, and Usp45SM3, showed a small (∼15%) but significant increase in secretion of α-amylase.
Fig 2.
Representative mRNA structures, showing the differences in secondary structure and ΔG around the ribosome binding site (RBS) for wild-type Usp45 signal peptide, Usp45SM2, and Usp45SM3. mRNAs from the transcript start site to 6 nucleotides after the end of the signal peptide sequence were entered into mfold to obtain the secondary structures (26). Structures with the lowest ΔG values are shown, with the RBS (AGGAGG) circled.
Table 1.
mRNA folding energies of the AGGAGG ribosome binding site and activities of secreted nuclease and α-amylase of selected clones from the Usp45sp silent mutation librarya
| Usp45sp silent mutation clone | ΔG of ribosome binding site (AGGAGG) | Secreted nuclease activity (compared to Usp45sp)b | Secreted α-amylase activity (% of Usp45sp)c |
|---|---|---|---|
| Usp45SM2 | −8.4 | − | 79 ± 5†† |
| Usp45SM14 | −8.4 | −− | 83 ± 13 |
| Usp45SM11 | −6.1 | −− | 85 ± 3†† |
| Usp45sp | −6.1 | * | 100 |
| Usp45SM16 | −5.9 | − | 91 ± 12 |
| Usp45SM19 | −5.9 | + | 106 ± 16 |
| Usp45SM4 | −5.7 | + | 107 ± 6 |
| Usp45SM10 | −3.8 | + | 116 ± 5† |
| Usp45SM20 | −3.7 | ++ | 103 ± 15 |
| Usp45SM5 | −3.7 | + | 113 ± 5† |
| Usp45SM8 | −3.3 | + | 92 ± 5 |
| Usp45SM15 | −3.3 | + | 100 ± 6 |
| Usp45SM9 | −3.3 | + | 114 ± 7 |
| Usp45SM7 | −3.2 | + | 105 ± 14 |
| Usp45SM6 | −2.5 | ++ | 99 ± 12 |
| Usp45SM12 | −1.6 | ++ | 96 ± 12 |
| Usp45SM3 | −0.6 | * | 114 ± 3†† |
Nuclease and α-amylase secretion levels are reported for cultures induced with 10 ng/ml nisin for 2 h at mid-log phase. ΔG, mRNA folding energy. Nucleotide sequences are given in Table S4 in the supplemental material.
Data represent levels of nuclease secretion. *, secretion similar to Usp45sp level; −, secretion slightly (<10%) lower than Usp45sp level; −−, secretion much (>10%) lower than Usp45sp level; +, secretion slightly (<10%) higher than Usp45sp level; ++, secretion much (>10%) higher than Usp45sp level.
α-Amylase secretion data represent the means ± standard errors of the means (SEM) of the results of three independent experiments. †, P < 0.05; ††, P < 0.01 (for statistical comparison of α-amylase activities of the respective clone and Usp45sp using one-tailed Student's t test).
Mathematical model captures secretion levels based on ΔG around the RBS.
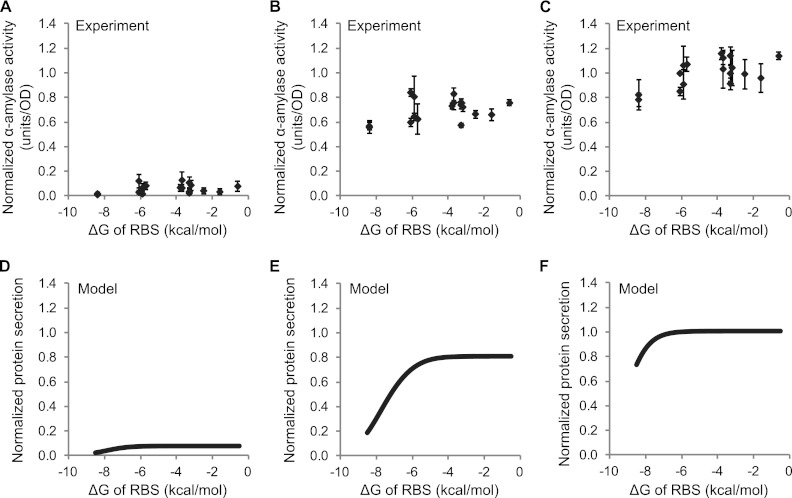
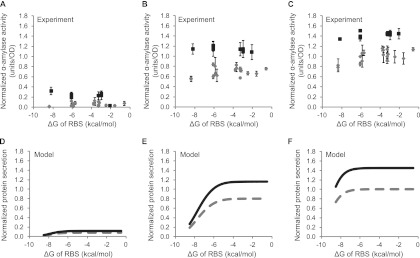
To more quantitatively link predicted ΔGRBS values and experimentally measured secretion levels, we developed a mathematical model based on modifications of previous work (28). The model considers translation to be initiated once the ribosome binds to the RBS, which is assumed to occur only when the initiation region of the mRNA is unfolded (32, 33) and is therefore determined by ΔGRBS. In the model, translation is proportional to the amount of ribosome-bound mRNA and secretion is a fraction of the total translated protein (see Methods in the supplemental material for detailed model and parameter values). We plotted α-amylase secretion versus ΔGRBS for three different nisin induction levels (0.1, 1, and 10 ng/ml) (Fig. 3A to C) and compared these experimental findings to model simulations with three analogously different mRNA transcript levels (all 10-fold apart) (Fig. 3D to F). Model predictions of protein secretion for all three transcript amounts show good agreement with experimental results. At all induction levels, tightly folded mRNA around the RBS yields low secretion. Secretion gradually increases as the mRNA structure around RBS is relaxed, but it eventually plateaus at high ΔGRBS when either the mRNA transcripts or ribosomes become limiting. The agreement between experiment and model further supports the notion that the mRNA conformation around the RBS can explain differences in secretion among the Usp45sp silent mutation clones. The relaxation of the mRNA structure alone in this context can augment secretion by an ∼15% increase at the maximum induction level. This modest improvement is consistent with the model, since the value determined for Usp45sp (−6.1 kcal/mol) is close to the plateau region of the curve (Fig. 3F).
Fig 3.
Secretion efficiencies of clones selected from the Usp45sp silent mutation library and comparison to results from a mathematical model. α-Amylase activity was plotted against the folding energy (ΔG) of the ribosome binding site (RBS) for induction at (A) 0.1 ng/ml, (B) 1 ng/ml, and (C) 10 ng/ml nisin. α-Amylase activity was normalized to the amount secreted by Usp45sp culture induced with 10 ng/ml nisin. ΔGRBS was calculated for the AGGAGG RBS sequence from the mRNA structure with the lowest overall ΔG. The data represent the means ± standard errors of the means (SEM) of the results of three independent experiments. Simulation results show protein secretion levels plotted against ΔG for (D) low (3.2 μM), (E) medium (32 μM), and (F) high (320 μM) mRNA transcript levels. Parameter values used in the model are given in Table S1 in the supplemental material.
Amino acid mutations in Usp45sp more significantly augment secretion of both nuclease and α-amylase.
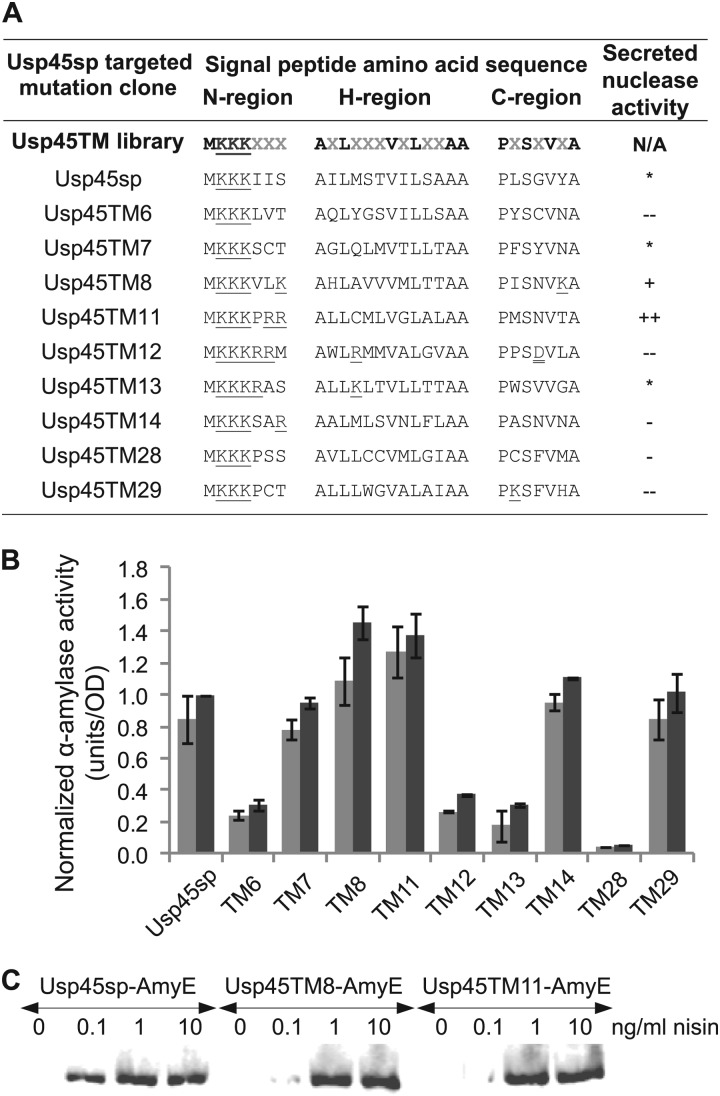
Since our experimental and modeling results suggest that the mRNA structure of Usp45sp is not a major bottleneck in achieving enhanced protein secretion, we constructed another signal peptide library consisting of targeted amino acid substitutions in Usp45sp. The length and tripartite structure of wild-type Usp45sp were kept: 27 amino acids in total, with 7, 13, and 7 residues in the n-, h-, and c-regions, respectively. Residues that were considered significant in maintaining the identities of the three regions were kept as follows: (i) charged lysines in the n-region, (ii) hydrophobic residues in the h-region, and (iii) residues important for structure and cleavage of the signal peptide in the c-region. The resulting library had 13 possible positions at which amino acid mutations could occur (Fig. 4A). After screening for library members with nuclease secretion, little to no conservation was observed in the 13 randomized positions (Fig. 4A). Five selected clones (Usp45TM8, -TM11, -TM12, -TM13, and -TM14) had increased positive charge in the n-region (from +3 to +4 or +5), but only two of these clones (Usp45TM8 and -TM11) exhibited significantly higher secretion for both nuclease and α-amylase (Fig. 4A and B). The poorer performances of Usp45TM12 and -TM13 may be due to the fact that these sequences newly introduced a positive charge in the stretch of hydrophobic residues in the h-region. Comparing nuclease and α-amylase secretion across all clones, there was clearly a protein-specific effect on secretion for signal peptides Usp45TM13, -TM14, and -TM29 (Fig. 4A and B), but the two best clones from the nuclease screen, Usp45TM8 and -TM11, also exhibited enhanced secretion of α-amylase at both 1 and 10 ng/ml nisin induction (Fig. 4B). Western blotting showed no differences among wild-type Usp45sp, Usp45TM8, and Usp45TM11 signal peptides in the size of the secreted α-amylase, indicating no change in the cleavage pattern when the engineered signal peptides were used (Fig. 4C). The best clone, Usp45TM8, gave an ∼45% increase in α-amylase secretion, 30% higher than what was achieved by silent mutations alone.
Fig 4.
Secretion efficiencies of clones selected from the targeted mutagenesis library of the Usp45 signal peptide. (A) Amino acid sequences and secreted nuclease activity for cultures induced at 10 ng/ml nisin. Amino acid mutation positions, indicated by gray Xs in the Usp45TM library sequence, were encoded by degenerate NNK nucleotides. *, −, −−, +, and ++ indicate secretion levels similar to and <10% lower, >10% lower, <10% higher, and >10% higher than Usp45sp levels, respectively. Single and double underlines indicate positively and negatively charged residues, respectively. Nucleotide sequences are given in Table S5 in the supplemental material. (B) α-Amylase activity of different clones normalized to the amount secreted by Usp45sp culture induced with 10 ng/ml nisin. Light gray bars represent induction at 1 ng/ml nisin, and dark gray bars represent induction at 10 ng/ml nisin. The data represent the means ± SEM of the results of two independent experiments. (C) Western blot showing secreted α-amylase in the supernatant detected by anti-RGS-His antibody. The different targeted mutations did not alter the size of the secreted protein, indicating that the signal peptide cleavage site had been maintained.
Silent mutations in Usp45TM8 further increase secretion.
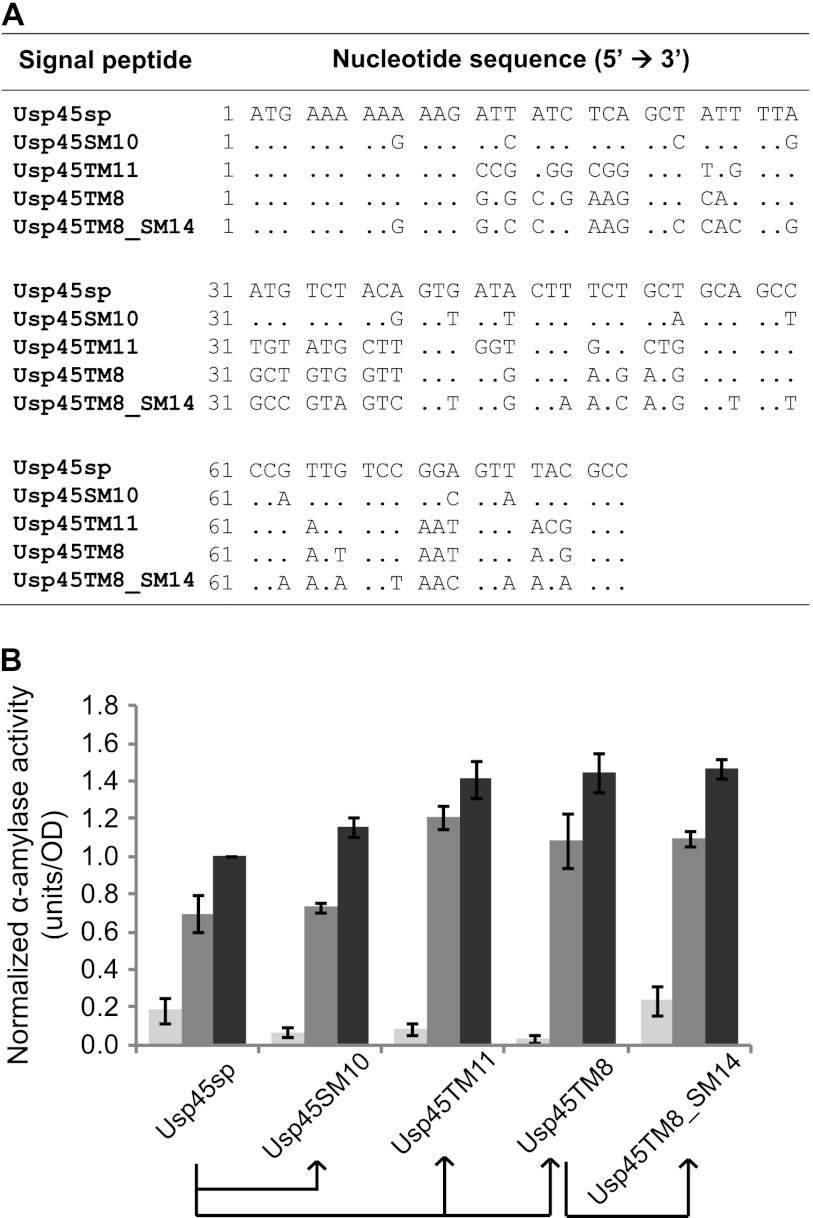
Finally, another round of silent mutation was performed on the best targeted mutation clone to combine the benefits of the two mutation strategies. We constructed a library with silent mutations in Usp45TM8 in order to determine if relaxing the mRNA secondary structure encoding this new signal peptide would have any consequent effect on secretion efficiency. In addition, comparison of Usp45TM8 and Usp45sp silent mutation clones with highly folded mRNA structures could reveal if any secretion enhancement achieved by amino acid substitution could be eliminated by tighter mRNA folding. We again screened for representative low-, medium-, and high-secretion clones and analyzed them individually for α-amylase secretion. Table 2 shows Usp45TM8 silent mutation clones ordered by ΔGRBS. All analyzed clones had an increase in secretion of at least 30% compared to wild-type Usp45sp, even though the lowest ΔGRBS (Usp45TM8_SM15) was as low as those for the most poorly secreting Usp45sp silent mutation clones (Usp45SM2 and -SM14). This shows that the secretion enhancement brought about by the amino acid sequence of Usp45TM8 is more dominant than any inhibition of translation initiation due to mRNA folding (at least in this range of ΔGRBS). As in the Usp45sp silent mutation library, α-amylase secretion generally increased (up to ∼50%) when ΔGRBS increased, which is consistent with our hypothesis that tight mRNA folding around the RBS inhibits translation initiation and subsequent secretion. We then plotted α-amylase secretion against ΔGRBS for both the Usp45TM8 and Usp45sp silent mutation clones for comparison (Fig. 5A to C). At all induction levels, clones from the two silent mutation libraries exhibited the same trend, with low secretion at more-negative ΔGRBS and higher saturated secretion at less-negative ΔGRBS. Usp45TM8 silent mutation clones generally exhibited higher secretion than Usp45sp silent mutation clones at comparable ΔGRBS values, with more pronounced differences observed at higher levels of induction. We also simulated secretion using the same mathematical model as described above, except we used a higher secretion constant for the Usp45TM8 amino acid sequence (see Discussion and the supplemental material), and there is good agreement between our modeling and experimental results (Fig. 5). Our best clones, Usp45TM8_SM14 and -TM8_SM25, yielded an ∼50% increase in α-amylase secretion compared to Usp45sp.
Table 2.
mRNA folding energies of the AGGAGG ribosome binding site and activities of secreted α-amylase of selected clones from the Usp45TM8 silent mutation librarya
| Usp45TM8 silent mutation clone | ΔG of ribosome binding site (AGGAGG) | Secreted α-amylase activity (% of Usp45sp)b |
|---|---|---|
| Usp45TM8_SM15 | −8.2 | 135 ± 2 |
| Usp45TM8_SM7 | −6.1 | 137 ± 6 |
| Usp45TM8_SM22 | −6.1 | 140 ± 2 |
| Usp45TM8_SM25 | −6.1 | 151 ± 3 |
| Usp45TM8_SM9 | −3.3 | 145 ± 5 |
| Usp45TM8_SM10 | −3.0 | 143 ± 9 |
| Usp45TM8_SM14 | −3.0 | 147 ± 5 |
| Usp45TM8 | −2.1 | 145 ± 10 |
α-Amylase secretion levels are reported for cultures induced with 10 ng/ml nisin for 2 h at mid-log phase. ΔG, mRNA folding energy. Nucleotide sequences are given in Table S6 in the supplemental material.
α-Amylase secretion data represent the means ± SEM of the results of three independent experiments. For each value in this column, P < 0.01 (for statistical comparison of α-amylase activities of the respective clone and Usp45sp using one-tailed Student's t test).
Fig 5.
Secretion efficiencies of clones selected from Usp45TM8 silent mutation library compared to clones from Usp45sp silent mutation library. α-Amylase activity (normalized to the amount secreted by wild-type Usp45sp at 10 ng/ml nisin) was plotted against ΔG of the RBS for induction at (A) 0.1 ng/ml, (B) 1 ng/ml, and (C) 10 ng/ml nisin for Usp45TM8 silent mutation clones (black squares; Usp45sp silent mutation clones as described for Fig. 3 are shown as gray diamonds for comparison). The data represent the means ± SEM of the results of three independent experiments. Simulation results show protein expression levels against folding energy for (D) low (3.2 μM), (E) medium (32 μM), and (F) high (32 μM) mRNA transcript levels. Solid black lines represent Usp45TM8 silent mutation clones, and dashed gray lines represent Usp45sp silent mutation clones (data from Fig. 3).
DISCUSSION
Using directed evolution, we have systematically explored the mRNA and amino acid sequence space of Usp45sp to achieve enhanced protein secretion in L. lactis (Fig. 6). A minor limitation of wild-type Usp45sp lies in its significant mRNA secondary structure, which may lead to less-efficient translation initiation. By relaxing mRNA folding around the RBS (−6.1 kcal/mol for Usp45sp and −3.8 kcal/mol for Usp45SM10), we could slightly improve secretion (up to 16%). The modest nature of this effect could be quantitatively captured by a mathematical model that relates the free energy of folding around the RBS to protein secretion. A more significant limitation lies in the actual amino acid sequence of Usp45sp, which likely impacts the efficacy of the physical process of protein secretion. By mutating nonconserved positions in the tripartite signal peptide, we significantly improved secretion yields (by 41% and 45% for Usp45TM11 and Usp45TM8, respectively). As quantitatively predicted by our mathematical model and confirmed by additional directed-evolution experiments, introducing silent mutations into the best amino acid sequence (Usp45TM8) minimally improved protein secretion at maximum induction (6% additional enhancement for Usp45TM8_SM14 compared to Usp45TM8).
Fig 6.
Progressive improvements in L. lactis secretion achieved by directed evolution. (A) Nucleotide sequence alignment of selected clones (only differences from Usp45sp are shown). (B) α-Amylase activity of selected clones at 0.1 ng/ml (light gray), 1 ng/ml (medium gray), and 10 ng/ml (dark gray) nisin induction. The data represent the means ± SEM of the results of three independent experiments. Arrows below the graph show the evolutionary relationships between the different selected clones.
Intriguingly, the best silent mutation clones for Usp45TM8 were able to enhance secretion at lower induction levels. The Western blot in Fig. 4C and the starch azure assay in Fig. 6B show that secretion of α-amylase by Usp45TM8 and -TM11 at 0.1 ng/ml nisin is lower than that by wild-type Usp45sp despite enhancing secretion at higher nisin concentrations (1 and 10 ng/ml). Silent mutations in Usp45TM8 were able to restore wild-type secretion levels at 0.1 ng/ml nisin, and Usp45TM8_SM14 is shown as an illustrative example (Fig. 6B). Although the mechanism by which silent mutations enhance submaximal secretion is not entirely clear, combining amino acid and silent mutation strategies has enabled us to augment the maximum secretion enhancement of 51% without jeopardizing secretion at lower induction level.
Our mathematical model is most successful in explaining the relationship between mRNA folding and protein secretion at a high nisin induction level (e.g., 10 ng/ml nisin) (Fig. 3 and 5). In this regime, the availability of ribosomes is limiting and translation rate is at its maximum. At lower nisin concentrations (1 or 0.1 ng/ml nisin), the number of mRNA transcripts becomes limiting and other factors not considered by the model (e.g., degradation of mRNA [34]) might become more important. Thus, it is possible that Usp45TM8, while as efficient as Usp45TM8_SM14 in translation and secretion, is subjected to a higher rate of mRNA degradation. This effect may be negligible at the high induction levels but becomes more pronounced when the mRNA transcript level becomes limiting. The importance of such secondary effects at lower induction levels (0.1 and 1 ng/ml nisin) may account for the disparity between the experiments and model simulations at more-negative ΔGRBS. Clones with stronger folding around the RBS also tend to have a higher level of secondary structure overall, which might reduce mRNA degradation. By neglecting the counterbalancing effect of reduced mRNA degradation in the model, the simulations should underestimate secretion at low ΔGRBS and this is indeed what is seen (Fig. 3 and 5). Another assumption in the model is that the mRNA transcript level varies linearly with nisin concentration for the range of inducer used in our experiments.
In analyzing our experimental results, we initially attempted to use ΔG of the entire translation initiation region to predict expression, as has been done in other work (31, 35), but we did not see any correlations even when different mRNA windows were used. In our study, small windows could not capture the contributions of the entire signal peptide region (81 nucleotides from the start codon), while large windows included significant, but distal, mRNA secondary structures that did not affect translation initiation. It was also not possible to use ΔG of the untranslated region around the RBS (28) because our silent mutations are contained in the coding region. Therefore, we simulated folding of the mRNA sequence from the transcription start site to six nucleotides after the signal peptide to capture all silent mutation contributions, but the relevant ΔG was calculated only from base pairing involving the RBS. These values were computed using ΔG data from mfold (26), but since ΔG contributions of individual base pairs were parsed from the overall mRNA structure, an initiation term of 3.4 kcal/mol was added (27). The ΔG for unfolded mRNA binding to the 30S ribosomal subunit was −14 kcal/mol (28). This value is consistent with experimentally measured values (36) with the additional consideration of translational standby sites (37).
With a limited number of targeted mutation clones, it is difficult to present a comprehensive picture of the molecular determinants in Usp45sp that govern protein secretion. Nevertheless, two of the best clones in our study, Usp45TM8 and -TM11, appeared to increase the overall positive charge in the n-region without introducing destabilizing charges in the hydrophobic h-region (Fig. 4). It has been suggested that positive charges in the n-region are responsible for interacting with the Sec translocation machinery and the negatively charged phospholipids in the membrane during translocation (38). Increasing the positive charge in this region has been shown to improve secretion efficiency in E. coli and B. brevis (21, 39, 40). In the hydrophobic h-region, Usp45TM8 has an increased number of polar residues (four) and Usp45TM11 has a decreased number (two) compared to wild-type Usp45sp (three). Previous studies investigating the effect of introducing hydrophobic residues such as leucines in this region have given contradictory results; secretion has been shown to increase in E. coli and B. brevis but has been shown to decrease in L. lactis (21, 39, 40). In the light of these findings and our own results, it is possible that the specific amino acid identities and their actual positions within the h-region are important factors in regulating secretion. For example, in Usp45TM11, a glycine is found in position 8 of the h-region, and this helix-breaking residue is commonly present in the middle of the hydrophobic core to allow the signal peptide to form a hairpin-like structure to facilitate membrane insertion (38). Given our library design, mutations in the c-region should have relatively minor influences on the overall secretion efficiency since most of the important residues (proline and serine favoring β-turns; −3 and −1 residues for signal peptidase cleavage) (41) were held constant (Fig. 4A). Overall, the best signal peptides from our screens preserve the tripartite structure of the signal peptide but increase the positive charge of the n-region.
Many other factors, such as the overall charge balance, hydrophobicity profile, and interactions among the n-, h-, and c-regions, can influence secretion efficiency, so further design of signal peptides may still necessitate extensive trial-and-error. The lack of a blueprint for signal peptide engineering motivated us to use a directed evolution approach to discover sequences that enhance secretion. In designing our targeted mutation library, we attempted to retain features that appear to be generally conserved in signal peptides while allowing for full randomization in the remaining positions. This library was far from exhaustive, but nevertheless, our simple screen, requiring only a transformation step and a plate assay, was successfully used to identify signal peptide mutants with an increase of up to 51% in secretion relative to the most efficient natural signal peptide in L. lactis.
Additional studies are needed to determine whether even greater improvements in secretion can be achieved solely by engineering the signal peptide, since other steps in the secretion process such as translocation efficiency (24), modification by chaperones (15), and transportation across the cell wall (42) might become limiting. Indeed, protein secretion in L. lactis has been improved by introduction of a synthetic propeptide (23) or by complementation with the SecDF machinery (24). Thus, other natural or engineered bacterial species with higher levels of protein secretion may serve as better fundamental models for studying inherent limitations of signal peptides. Nonetheless, this study produced signal peptides that can replace wild-type Usp45sp in many biotechnological and clinical applications of L. lactis and it also highlights the potential of directed evolution as a general approach for signal peptide engineering in bacteria.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by a CAREER Award from the National Science Foundation (CBET-1055231) to C.A.S.
We are grateful to Mark Goulian for providing the S. aureus and B. subtilis strains.
Footnotes
Published ahead of print 2 November 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02667-12.
REFERENCES
- 1. Leroy F, De Vuyst L. 2004. Lactic acid bacteria as functional starter cultures for the food fermentation industry. Trends Food Sci. Technol. 15:67–78 [Google Scholar]
- 2. Morello E, Bermúdez-Humarán LG, Llull D, SolÉ V, Miraglio N, Langella P, Poquet I. 2008. Lactococcus lactis, an efficient cell factory for recombinant protein production and secretion. J. Mol. Microbiol. Biotechnol. 14:48–58 [DOI] [PubMed] [Google Scholar]
- 3. Poquet I, Saint V, Seznec E, Simoes N, Bolotin A, Gruss A. 2000. HtrA is the unique surface housekeeping protease in Lactococcus lactis and is required for natural protein processing. Mol. Microbiol. 35:1042–1051 [DOI] [PubMed] [Google Scholar]
- 4. van Asseldonk MV, Rutten G, Oteman M, Siezen RJ, de Vos WM, Simons G. 1990. Cloning of usp45, a gene encoding a secreted protein from Lactococcus lactis subsp. lactis MG1363. Gene 95:155–160 [DOI] [PubMed] [Google Scholar]
- 5. Bolotin A, Wincker P, Mauger S, Jaillon O, Malarme K, Weissenbach J, Ehrlich SD, Sorokin A. 2001. The complete genome sequence of the lactic acid bacterium Lactococcus lactis ssp. lactis IL1403. Genome Res. 11:731–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wegmann U, O'Connell-Motherway M, Zomer A, Buist G, Shearman C, Canchaya C, Ventura M, Goesmann A, Gasson MJ, Kuipers OP, van Sinderen D, Kok J. 2007. Complete genome sequence of the prototype lactic acid bacterium Lactococcus lactis subsp. cremoris MG1363. J. Bacteriol. 189:3256–3270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mierau I, Kleerebezem M. 2005. 10 years of the nisin-controlled gene expression system (NICE) in Lactococcus lactis. Appl. Microbiol. Biotechnol. 68:705–717 [DOI] [PubMed] [Google Scholar]
- 8. Klijn N, Weerkamp AH, de Vos WM. 1995. Genetic marking of Lactococcus lactis shows its survival in the human gastrointestinal tract. Appl. Environ. Microbiol. 61:2771–2774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wells JM, Mercenier A. 2008. Mucosal delivery of therapeutic and prophylactic molecules using lactic acid bacteria. Nat. Rev. Microbiol. 6:349–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Steidler L, Hans W, Schotte L, Neirynck S, Obermeier F, Falk W, Fiers W, Remaut E. 2000. Treatment of murine colitis by Lactococcus lactis secreting interleukin-10. Science 289:1352–1355 [DOI] [PubMed] [Google Scholar]
- 11. Chatel J, Langella P, Adel-Patient K, Commissaire J, Wal J, Corthier G. 2001. Induction of mucosal immune response after intranasal or oral inoculation of mice with Lactococcus lactis producing bovine beta-lactoglobulin. Clin. Diagn. Lab. Immunol. 8:545–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ng DTW, Sarkar CA. 2011. Nisin-inducible secretion of a biologically active single-chain insulin analog by Lactococcus lactis NZ9000. Biotechnol. Bioeng. 108:1987–1996 [DOI] [PubMed] [Google Scholar]
- 13. Nouaille S, Ribeiro LA, Miyoshi A, Pontes D, Le Loir Y, Oliveira SC, Langella P, Azevedo V. 2003. Heterologous protein production and delivery systems for Lactococcus lactis. Genet. Mol. Res. 2:102–111 [PubMed] [Google Scholar]
- 14. Desvaux M, Hébraud M, Talon R, Henderson IR. 2009. Secretion and subcellular localizations of bacterial proteins: a semantic awareness issue. Trends Microbiol. 17:139–145 [DOI] [PubMed] [Google Scholar]
- 15. Le Loir Y, Azevedo V, Oliveira SC, Freitas DA, Miyoshi A, Bermúdez-Humarán LG, Nouaille S, Ribeiro LA, Leclercq S, Gabriel JE, Guimaraes VD, Oliveira MN, Charlier C, Gautier M, Langella P. 2005. Protein secretion in Lactococcus lactis: an efficient way to increase the overall heterologous protein production. Microb. Cell Fact. 4:2 doi:10.1186/1475-2859-4-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. von Heijne G. 1990. The signal peptide. J. Membr. Biol. 115:195–201 [DOI] [PubMed] [Google Scholar]
- 17. van Asseldonk M, de Vos WM, Simons G. 1993. Functional analysis of the Lactococcus lactis usp45 secretion signal in the secretion of a homologous proteinase and a heterologous α-amylase. Mol. Gen. Genet. 240:428–434 [DOI] [PubMed] [Google Scholar]
- 18. Ravn P, Arnau J, Madsen SM, Vrang A, Israelsen H. 2000. The development of TnNuc and its use for the isolation of novel secretion signals in Lactococcus lactis. Gene 242:347–356 [DOI] [PubMed] [Google Scholar]
- 19. Poquet I, Ehrlich SD, Gruss A. 1998. An export-specific reporter designed for gram-positive bacteria: application to Lactococcus lactis. J. Bacteriol. 180:1904–1912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Perez-Martinez G, Kok J, Venema G, van Dijl JM, Smith H, Bron S. 1992. Protein export elements from Lactococcus lactis. Mol. Gen. Genet. 234:401–411 [DOI] [PubMed] [Google Scholar]
- 21. Ravn P, Arnau J, Madsen SM, Vrang A, Israelsen H. 2003. Optimization of signal peptide SP310 for heterologous protein production in Lactococcus lactis. Microbiology 149:2193–2201 [DOI] [PubMed] [Google Scholar]
- 22. Mercenier A, Hols P, Roussel Y, Perez-Martinez G, Buesa J, Wilks M, Pozzi G, Remaut E, Morelli L, Grangette C, Monedero V, Palumbo E, Foligne B, Steidler L, Nutten S. 2004. Screening and construction of probiotic strains with enhanced protective properties against intestinal disorders. Microb. Ecol. Health Dis. 16:86–95 [Google Scholar]
- 23. Le Loir Y, Gruss A, Ehrlich S, Langella P. 1998. A nine-residue synthetic propeptide enhances secretion efficiency of heterologous proteins in Lactococcus lactis. J. Bacteriol. 180:1895–1903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nouaille S, Morello E, Cortez-Peres N, Le Loir Y, Commissaire J, Gratadoux JJ, Poumerol E, Gruss A, Langella P. 2006. Complementation of the Lactococcus lactis secretion machinery with Bacillus subtilis SecDF improves secretion of staphylococcal nuclease. Appl. Environ. Microbiol. 72:2272–2279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lachica RVF, Genigeorgis C, Hoeprich PD. 1971. Metachromatic agar-diffusion methods for detecting staphylococcal nuclease activity. Appl. Microbiol. 21:585–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zuker M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Freier SM, Kierzek R, Jaeger JA, Sugimoto N, Caruthers MH, Neilson T, Turner DH. 1986. Improved free-energy parameters for predictions of RNA duplex stability. Proc. Natl. Acad. Sci. U. S. A. 83:9373–9377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. de Smit MH, van Duin J. 1990. Secondary structure of the ribosome binding site determines translational efficiency: a quantitative analysis. Proc. Natl. Acad. Sci. U. S. A. 87:7668–7672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Le Loir Y, Gruss A, Ehrlich SD, Langella P. 1994. Direct screening of recombinants in Gram-positive bacteria using the secreted staphylococcal nuclease as a reporter. J. Bacteriol. 176:5135–5139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pontes DS, de Azevedo MSP, Chatel J, Langella P, Azevedo V, Miyoshi A. 2011. Lactococcus lactis as a live vector: heterologous protein production and DNA delivery systems. Protein Expr. Purif. 79:165–175 [DOI] [PubMed] [Google Scholar]
- 31. Kudla G, Murray AW, Tollervey D, Plotkin JB. 2009. Coding-sequence determinants of gene expression in Escherichia coli. Science 324:255–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. de Smit MH, van Duin J. 1994. Control of translation by mRNA secondary structure in Escherichia coli: a quantitative analysis of literature data. J. Mol. Biol. 244:144–150 [DOI] [PubMed] [Google Scholar]
- 33. Kaminishi T, Wilson DN, Takemoto C, Harms JM, Kawazoe M, Schluenzen F, Hanawa-Suetsugu K, Shirouzu M, Fucini P, Yokoyama S. 2007. A snapshot of the 30S ribosomal subunit capturing mRNA via the Shine-Dalgarno interaction. Structure 15:289–297 [DOI] [PubMed] [Google Scholar]
- 34. Schlax PJ, Worhunsky DJ. 2003. Translational repression mechanisms in prokaryotes. Mol. Microbiol. 48:1157–1169 [DOI] [PubMed] [Google Scholar]
- 35. Na D, Lee S, Lee D. 2010. Mathematical modeling of translation initiation for the estimation of its efficiency to computationally design mRNA sequences with desired expression levels in prokaryotes. BMC Syst. Biol. 4:71 doi:10.1186/1752-0509-4-71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Calogero RA, Pon CL, Canonaco MA, Gualerzi CO. 1988. Selection of the mRNA translation initiation region by Escherichia coli ribosomes. Proc. Natl. Acad. Sci. U. S. A. 85:6427–6431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. de Smit MH, van Duin J. 2003. Translational standby sites: how ribosomes may deal with the rapid folding kinetics of mRNA. J. Mol. Biol. 331:737–743 [DOI] [PubMed] [Google Scholar]
- 38. Tjalsma H, Bolhuis A, Jongbloed JDH, Bron S, van Dijl JM. 2000. Signal peptide-dependent protein transport in Bacillus subtilis: a genome-based survey of the secretome. Microbiol. Mol. Biol. Rev. 64:515–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Izard JW, Doughty MB, Kendall DA. 1995. Physical and conformational properties of synthetic idealized signal sequences parallel their biological function. Biochemistry 34:9904–9912 [DOI] [PubMed] [Google Scholar]
- 40. Takimura Y, Kato M, Ohta T, Yamagata H, Udaka S. 1997. Secretion of human interleukin-2 in biologically active form by Bacillus brevis directly into culture medium. Biosci. Biotechnol. Biochem. 61:1858–1861 [DOI] [PubMed] [Google Scholar]
- 41. von Heijne G. 1983. Patterns of amino acids near signal-sequence cleavage sites. Eur. J. Biochem. 133:17–21 [DOI] [PubMed] [Google Scholar]
- 42. Forster BM, Marquis H. 2012. Protein transport across the cell wall of monoderm Gram-positive bacteria. Mol. Microbiol. 84:405–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.