Abstract
Two 4-chloro-2-methylphenoxyacetic acid (MCPA)-degrading enrichment cultures selected from an aquifer on low (0.1 mg liter−1) or high (25 mg liter−1) MCPA concentrations were compared in terms of metabolic activity, community composition, population growth, and single cell physiology. Different community compositions and major shifts in community structure following exposure to different MCPA concentrations were observed using both 16S rRNA gene denaturing gradient gel electrophoresis fingerprinting and pyrosequencing. The communities also differed in their MCPA-mineralizing activities. The enrichments selected on low concentrations mineralized MCPA with shorter lag phases than those selected on high concentrations. Flow cytometry measurements revealed that mineralization led to cell growth. The presence of low-nucleic acid-content bacteria (LNA bacteria) was correlated with mineralization activity in cultures selected on low herbicide concentrations. This suggests that LNA bacteria may play a role in degradation of low herbicide concentrations in aquifers impacted by agriculture. This study shows that subpopulations of herbicide-degrading bacteria that are adapted to different pesticide concentrations can coexist in the same environment and that using a low herbicide concentration enables enrichment of apparently oligotrophic subpopulations.
INTRODUCTION
Herbicides are used worldwide for controlling weeds during agricultural practice. Extensive use of herbicides has led to detection of residues in surface waters and groundwaters serving as public drinking water resources. These residues are typically present in groundwater in the concentration range of traces to micrograms per liter (1). Such concentrations may still exceed the European drinking water limits of 0.1 μg liter−1 for individual pesticides and be a threat to drinking water resources (2). One of the major groups of herbicides detected in drinking water resources are the phenoxy acid (PA) herbicides used for controlling broadleaf weeds in cereal crops; 4-chloro-2-methylphenoxyacetic acid (MCPA) is a PA that has been among the five top-selling herbicides during the last decades in Denmark (2). Even though it has been reported to be readily biodegradable in top soil (3), its broad application and weak retention in soil result in frequent occurrence of MCPA in surface waters and groundwaters (2).
To investigate the degradation of such pesticide concentrations, enrichment culturing has long been the method of choice for selecting and isolating catabolic microorganisms (4, 5), and it has been used successfully to obtain bacteria capable of degrading xenobiotic compounds such as PA herbicides (6, 7). The medium composition, including pollutant concentrations, used in the enrichment procedure is likely to be the main determining factor for the selection of organisms with specific characteristics and catabolic pathways (8, 9). Since high substrate concentrations are commonly used for isolation purposes (10–13), the obtained degraders may not be very efficient to degrade low pollutant concentrations (14–16) and subsequently not very appropriate for use in bioaugmentation studies. Thus, populations existing in the same microniches which are adapted to metabolize substrates at low concentrations may have been overlooked although they could be more efficient in remediating low levels of organic pollutants in contaminated water resources (5). This raises two important questions: first, whether specific populations adapted to metabolize low herbicide concentrations exist in oligotrophic aquifers impacted by agriculture and, second, whether relevant populations able to degrade low pollutant concentrations are selected using high substrate concentrations. Thus, comparing enrichments on low and high substrate concentrations could facilitate the development of new and more efficient approaches for remediation of pollutants at low concentrations.
Microorganisms have adapted and developed strategies to survive in oligotrophic environments by adjusting their cellular composition with respect to both structure and metabolic function. They are typically small in cell size and have relatively small genomes (17). Flow cytometry (FCM) studies in combination with nucleic acid stains revealed that bacterial cells tend to cluster into two major fractions: high-nucleic acid-content bacteria (HNA bacteria) and low-nucleic acid-content bacteria (LNA bacteria) based on their distinctly different fluorescence intensities (related to the nucleic acid content) and side scatter signals (related to cellular size) (18, 19). LNA bacteria are better adapted to oligotrophic conditions, and their dominance decreases with nutrient input in aquatic ecosystems (20). However, to our knowledge, there are no reports investigating links between the growth of LNA bacteria and the metabolic activity of these bacteria at low substrate concentrations.
The objective of this study was to investigate the differences in the dynamics of the metabolism, the bimodal distribution of LNA and HNA bacteria during metabolic activity, and the phylogenetic diversity of two MCPA-mineralizing microbial communities selected from an aquifer on low (0.1 mg liter−1) or high (25 mg liter−1) MCPA concentrations. We hypothesized that communities enriched with low substrate concentrations will provide more efficient communities with regard to metabolic functionality and community structure. The MCPA mineralization by the bacterial cultures was quantified by radiorespirometry using 14C-ring-labeled MCPA. The physiology and the nucleic acid content of single cells and the growth of the cultures were analyzed by flow cytometry. The structure and composition of the cultures at appropriate time points of extensive mineralization activity were analyzed by 16S rRNA gene denaturing gradient gel electrophoresis (DGGE) fingerprinting and subsequently more in depth by pyrosequencing.
MATERIALS AND METHODS
Cultivation and metabolic activity measurements.
Saturated sediment was sampled from an aquifer in Fladerne Creek, Denmark. The samples were collected from 2.5 m below the surface (mbs) to 3.25 mbs. Details and characterization of the site have been published previously (21). Samples were passed through a sieve (mesh size, 2 mm) and homogenized thoroughly using a sterile spatula prior to setting up the laboratory experiments. Bacterial cultures were enriched by inoculating 5 g (wet weight) of sediment into sterilized 100-ml glass flasks containing 25 ml of autoclaved mineral salt solution (MSN) (pH 7.1) (22) and MCPA as the sole carbon source at concentrations of 0.1 mg liter−1 or 25 mg liter−1. Each enrichment was conducted in triplicate. Cultures enriched with 0.1 mg liter−1 MCPA were called low-enrichment cultures (LECs) and those with 25 mg liter−1 were called high-enrichment culture (HECs). LECs were subcultured five times by transferring aliquots of 500 μl, 200 μl, 100 μl, 100 μl, and 50 μl at each step to fresh MCPA-containing medium when maximum MCPA mineralization was reached. HECs were similarly subcultured three times by transferring aliquots of 500 μl, 200 μl, and 100 μl (see Fig. S1 in the supplemental material). After these transfers over 2 to 3 months, the sediment-free cultures from the last round of the enrichments were stored in 40% (vol/vol) glycerol stock solutions at −80°C and revived by two washes in MSN before inoculation into sterilized 100-ml glass flasks in triplicates. LECs were transferred to flasks amended with either 0.1 mg liter−1 MCPA (LEC-L), or 25 mg liter−1 MCPA (LEC-H). Similarly, HECs were transferred to flasks amended with 25 mg liter−1 MCPA (HEC-H) and 0.1 mg liter−1 MCPA (HEC-L). For the mineralization experiments, U-14C-ring-labeled MCPA (specific activity of 5,908,900 Bq/mg, radiochemical purity of >95%; Izotop, Budapest, Hungary) was added to equal 187 Bq per flask. Unlabeled MCPA (97.5% purity; Ehrenstorfer GmbH, Augsburg, Germany) was added from stock solutions in high-pressure liquid chromatography-grade methanol (Sigma-Aldrich, MI) to the flasks to reach the desired concentrations. The solvent was evaporated before the addition of the MSN. Control experiments without herbicide additions were prepared with methanol that was similarly evaporated. Following inoculation, the flasks were equipped with a 10-ml glass tube containing 2 ml of 0.5 M NaOH to trap 14CO2 generated from [14C]MCPA mineralization and sealed with airtight glass stoppers before incubation in the dark at 20°C. NaOH was replaced at regular intervals, mixed with 10 ml of Wallac OptiPhase HiSafe 3 scintillation cocktail (Turku, Finland), and counts were determined in a Wallac 1409 liquid scintillation counter. At each sampling point 50 μl of medium was collected from each flask for the flow cytometric measurements. During the mineralization, 1.8-ml samples were collected at appropriate intervals—at time zero (t0), early exponential phase (∼5% mineralization) (t1), maximum mineralization level (t2), and after mineralization (t3)—and immediately frozen in liquid nitrogen and subsequently stored at −80°C until DNA extraction.
Flow cytometric measurements and staining procedure.
Two fluorescent dyes, SYBR Green I and propidium iodide (PI), were used to visualize the difference between cells with intact and damaged cytoplasmic membranes (23). The staining procedure and flow cytometry were performed as described by De Roy et al. (24). Samples were diluted in cell-free water before measurements to obtain an even rate of <1,000 events s−1. The cell populations were gated on a two-parameter dot plot of green fluorescence, as indicative of apparent nucleic acid content (18), and red fluorescence and counted separately. This allowed us to distinguish two different groups of bacteria, namely, low-nucleic acid- and high-nucleic acid-content bacteria (LNA and HNA bacteria, respectively) (17, 19). All samples were measured three times.
Bacterial community analysis.
Total genomic DNA was extracted from 1.8 ml of suspension of each sample using an UltraClean microbial DNA isolation kit (Mobio Laboratories, Carlsbad, CA) following the manufacturer's recommendations, with the following modifications: addition of proteinase K (Qiagen, Valencia, CA) prior to lysis followed by incubation at 56°C for 30 min; increases in the initial centrifugation time to 10 min and the speed to 13,000 × g. The 16S rRNA gene fragments were amplified by PCR using a Taq polymerase kit (Fermentas, Germany) with the universal bacterial primers PRBA338fGC and P518r and a GC-clamp of 40 bp on the forward primer (25). PCR products were analyzed by DGGE using the PhorU system (Ingeny, Leiden, Netherlands) running 8% (wt/vol) polyacrylamide gels with a denaturing gradient ranging from 45% to 60% (26). The DGGE patterns were processed using BioNumerics software, version 2.0 (Applied Maths, Belgium). Clustering was done with the Pearson correlation and the unweighted-pair group method using average linkages (UPGMA). The matrix of similarities for the densitometric curves of the band patterns was calculated based on the Pearson product-moment correlation coefficients and was used to perform cluster analysis. To calculate the approximate changes within the community structure between different mineralization time points, moving-window analysis was performed based on the similarity matrix (27). Range-weighted richness (Rr) (approximate environmental carrying capacity for microbial diversity) values of the communities at different time intervals were calculated as described previously (28). In order to graphically represent the evenness of the bacterial communities, Lorenz distribution curves were set up based on the DGGE profiles as previously described (28). Community organization (Co) values (the species relative abundance distribution in the microbial community) were defined as 100 times the Gini coefficient (29).
454 pyrosequencing.
Amplicon pyrosequencing was performed on total DNA extracted from cultures before (t0) and after (t3) MCPA mineralization. V4, V5, and V6 regions of the 16S rRNA gene were targeted using the primers 530F (30) and 1061R (31), extended as amplicon fusion primers with the respective primer L adaptor, a key sequence, and multiplex identifiers (MID) on the forward primer. The PCR mix of 50 μl contained 1× PCR buffer, 1.44 mM MgCl2, 0.1 mM concentrations of the deoxynucleoside triphosphates (dNTPs), 1% dimethyl sulfoxide, 1.25 U of Taq polymerase, 0.32 mg ml−1 bovine serum albumin (BSA), 0.3 mM each primer, and 1 μl of 5 ng μl−1 template DNA. PCR was performed as described by Larsen et al. (32). Amplicons were purified with a High Pure PCR Product Purification Kit (Roche) and as specified by the manufacturer. The quality of the PCR products was verified on a 1% agarose gel. Amplicon concentrations were measured with PicoGreen (Invitrogen, Ltd., United Kingdom) according to the manufacturer's instructions, and all amplicons were pooled together in equal molar amounts. Emulsion PCR, emulsion breaking, and sequencing were performed by using a 454 GS FLX Titanium Pyrosequencer (454 Life Sciences, Branford, CT) as recommended by the manufacturer. For this study, six amplicons in a pool of 17 mixed samples were sequenced on a quarter of an FLX PicoTiterPlate. Quality filtering of the pyrosequencing reads was performed using the automatic amplicon pipeline of the GS Run Processor (Roche), with a modification of the valley filter (vfScanAll using flow false instead of TiOnly) to extract sequences. Afterwards, reads were quality trimmed using the TRIM function of Greengenes (33) with the following settings: good-quality score of 20, window size of 40 bp, and window threshold at 90%. Subsequently, reads were batched per sample based on MID identifiers with BioEdit (34), and reads with inferior read lengths (<250 bp) were excluded from further analysis. Sequences were analyzed in the software package QIIME (35) as described by Kostka et al. (36). Bacterial diversity was determined by means of sampling-based analysis of operational taxonomic units (OTUs) and visualized as rarefaction curves (see Fig. S2 in the supplemental material).
Sequencing data accession number.
The sequences have been deposited in the NCBI Sequence Read Archive ([SRA] http://www.ncbi.nlm.nih.gov/Traces/sra) under study accession number SRR585238.
RESULTS
MCPA mineralization.
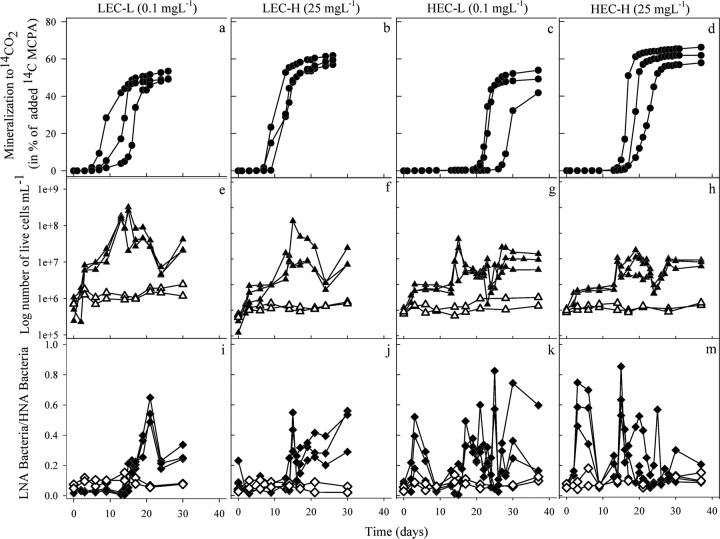
The MCPA mineralization by the cultures enriched at low (LEC-L and LEC-H) and high (HEC-L and HEC-H) MCPA concentrations were investigated. All experiments were done in triplicates and repeated once, giving highly reproducible results. The shape of all mineralization curves was sigmoidal, with a distinct initial lag period with zero or very low mineralization rate, followed by rapid 14CO2 production and eventually a plateau in the mineralization (Fig. 1). LEC-L cultures mineralized 50.7% ± 2.4% and LEC-H cultures mineralized 59.5% ± 2.4% of the added [14C]MCPA to 14CO2 within 26 days (Fig. 1a and b). In comparison, 11 additional days were required by HEC-L and HEC-H cultures to mineralize 48.4% ± 6.1% and 62.0% ± 4.2% of the added MCPA, respectively (Fig. 1c and d). The lag phase of LECs was approximately 10 days at both MCPA concentrations. In contrast, longer lag phases of 16 and 21 days were measured with HEC-H and HEC-L cultures, respectively, before rapid mineralization. Once MCPA mineralization was initiated, the degradation rates were similar among all cultures (data not shown).
Fig 1.
Mineralization of 14C-labeled MCPA to 14CO2 (filled circles), number of living cells (filled triangles), and the ratio of LNA to HNA bacteria (filled diamonds) in the enrichment cultures during MCPA mineralization. The open symbols indicate the control microcosms without pesticide addition. These data are shown individually for triplicate samples.
Total cell counts.
The growth patterns of the studied microbial communities revealed a clear response to the presence of MCPA (Fig. 1e to h). No growth was detected in inoculated control experiments without MCPA. The growth coincided with the phases of extensive MCPA mineralization in LEC-L and LEC-H cultures (Fig. 1a and b). The initial cell density was 6.2 × 105 ± 4.4 × 105 cells ml−1 for LEC-L and 3.7 × 105 ± 2.3 × 105 cells ml−1 for LEC-H cultures. The number of cells increased to 2.0 × 108 ± 1.6 × 108 cells ml−1 for LEC-L and 3.3 × 108 ± 1.9 × 108 cells ml−1 for LEC-H cultures during MCPA mineralization (Fig. 1e and f). Similar patterns between growth and mineralization were observed with HEC-L and HEC-H cultures (Fig. 1c and d). The initial cell density was 1.0 × 106 ± 0.2 × 106 cells ml−1 for HEC-L and 1.1 × 106 ± 0.1 × 106 for HEC-H cultures. The number of cells reached 1.2 × 109 ± 0.9 × 109 cells ml−1 for HEC-L and 1.7 × 109 ± 1.6 × 109 cells ml−1 for HEC-H cultures during MCPA mineralization.
Single cell analysis.
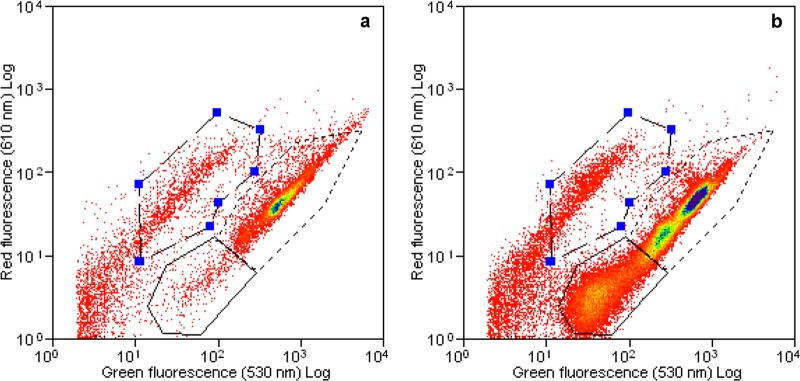
The LNA and HNA bacteria were clearly distinguishable on FCM dot plots of green fluorescence against red fluorescence (Fig. 2). The average ratio of LNA bacteria to HNA bacteria was initially 0.08 ± 0.05. This ratio continuously fluctuated in HECs (Fig. 1k and m). However, in LECs LNA bacteria proliferated in the phase where extensive MCPA mineralization occurred (Fig. 1i and j). With LEC-L cultures, the growth of LNA bacteria started after a lag period of 14 days, and exponential growth occurred in parallel to the MCPA mineralization, with a maximum average LNA/HNA ratio of 0.56 ± 0.15 at day 21 (Fig. 1i). The trend was similar in LEC-H cultures (Fig. 1j), but here the correlation between the LNA/HNA ratio and the metabolic activity was not as clear as for LEC-L cultures. The LNA/HNA ratio was not changed in the control experiments without MCPA throughout the experiment (Fig. 1). Furthermore, enrichment of LNA bacteria corresponded with the change in the community structure in LECs (Fig. 1a and b and 3a).
Fig 2.
Flow cytometric dot plot of one replicate of low-enrichment culture (LEC-L), stained with SYBR green I and propidium iodide, before (a) and after (b) MCPA mineralization. Solid lines indicate LNA bacteria, dotted lines indicate HNA bacteria, and dashed lines indicate dead cells.
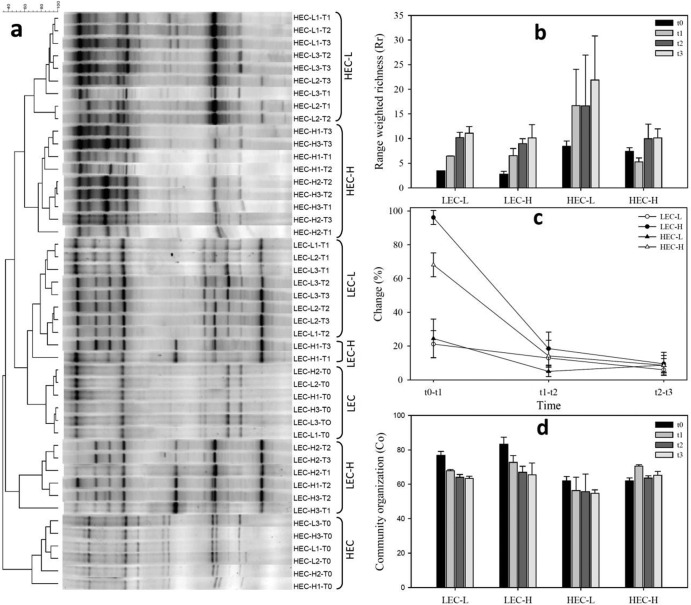
Fig 3.
DGGE profiles and UPGMA (unweighted pair-group method with arithmetic averages) tree (a), range-weighted richness (Rr) (b), moving-window analysis (dynamics) (c), and community organization (Co) (d) of 16S rRNA gene fingerprints of the enrichment cultures. Error bars represent SDs of means of triplicate samples.
DGGE fingerprint analysis of the enrichment cultures.
The changes in the communities during MCPA mineralization were analyzed by cluster analysis of the DGGE banding patterns with focus on four phases in the mineralization, namely, time zero (t0), early exponential mineralization (t1), maximum mineralization level (t2), and after mineralization (t3). Six main clusters were observed, demonstrating four major shifts from the original two communities (LECs and HECs) in relation to metabolism of the different MCPA concentrations (Fig. 3a). The calculated similarity value within the clusters was 81.7% ± 7.0%. The clusters for LECs and HECs were distinct from each other, with a similarity of only 16.5% ± 9.5% at t0. Clusters for LEC-L and LEC-H cultures had, after growth at 0.1 mg liter-1 and 25 mg liter-1, a similarity of 59.0% ± 20.7%. When these two clusters were compared to the cluster from the original LEC enrichment, the similarity values were 67.1% ± 12.0% for cluster LEC-L and 24.9% ± 19.4% for cluster LEC-H. The LEC-H cluster was subdivided into two subclusters. The reason for that subclustering was most probably the appearance of a dominant band that was closely related to cluster LEC-L and HEC-L. This subclustering was neglected throughout the calculations. HEC-H and HEC-L clusters had a similarity of 60.0% ± 12.8%, and the HEC-H cluster had a similarity to the HEC cluster of 31.9% ± 11.5%, whereas the HEC-L cluster had a similarity to the HEC cluster of 58.0% ± 10.0%.
The number of bands was 9 ± 2 (mean ± standard deviation [SD]) for LECs and 12 ± 3 for HECs spreading over a denaturing gradient range of ∼8% in all samples (Fig. 3a). This corresponds with range-weighted richness (Rr) of 3.1 ± 0.5 for LECs and 7.9 ± 0.7 for HECs at t0 (Fig. 3b). Richness of the enrichment cultures increased gradually over time, reaching an average Rr value of 10.6 ± 0.7 for LEC-L and LEC-H cultures, 21.9 ± 8.9 for HEC-L cultures, and 10.1 ± 1.8 for HEC-H cultures. According to the classification of Marzorati et al. (28), all the communities had low Rr values except HEC-L, which reached a medium Rr during the MCPA mineralization. Figure 3c shows the results of the moving-window analysis. Here, the correlation between different time points (i.e., t0 − t1) was plotted. The Δt values were calculated as the average and SD of the respective percent change values. The Δt values of samples with a low MCPA amendment were 13.3% ± 7.6% for LEC-L and 12.7% ± 10.3% for HEC-L cultures. Due to the high change in community structure from t0 to t1 in samples amended with high MCPA concentration, separate Δt values were calculated for t0 − t1 and t1 − t3. During the initial phase (t0 − t1), the enrichment cultures had very high Δt values of 96.1% ± 4.1% for LEC-H and 68.1% ± 7.1% for HEC-H. The community structure changed at lower rates between t1 and t3 with Δt values of 14.0% ± 6.4% for LEC-H and 11.4% ± 3.9% for HEC-H cultures. Figure 3d shows the community organization values (Co), representing a numerical value for the species evenness. A low value represents an even community, while a community with a high value has one or more dominant species. Prior to mineralization, LECs showed a less evenly distributed community structure, with a Co value of 80.0 ± 4.6, compared to HECs that had a Co value of 61.9 ± 0.1. In LECs, the Co values decreased gradually over time, resulting in a more evenly distributed community, with an average Co value of 64.4 ± 1.5. No big change was observed in the evenness of HECs over time.
454 pyrosequencing results.
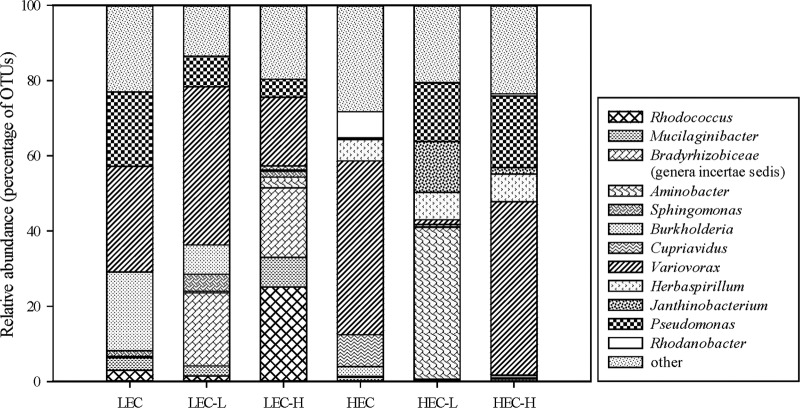
The diversity and community structure of the cultures before (t0) and after MCPA mineralization (t3) were analyzed by 454 pyrosequencing (Fig. 4). One representative sample was chosen from each treatment group based on the highly similar clustering (>95%) of these groups in DGGE analysis. Most sequences were identified as Bacteria by the Ribosomal Database Project (RDP) Classifier (37), and only 31 out of 72,175 sequences remained unclassified. A total of 52 genera were identified. Prior to MCPA mineralization, species of Variovorax were the most abundant group in both original enrichments, comprising 28% of the communities for LECs and 46% for HECs. Following MCPA mineralization, the abundance of Variovorax species increased slightly in LEC-L cultures (47%) but did not show any significant change in HEC-H cultures, i.e., when the incubation conditions remained the same as previously. However, when the substrate concentration was altered substantially, the abundance of Variovorax spp. decreased in both LEC-H (22%) and HEC-L (2%) cultures. The second most dominant genus in the data set was Pseudomonas. In LECs, 20% of the whole community belonged to this genus. However, after MCPA mineralization (t3), the abundance of Pseudomonas bacteria decreased to 9% in LEC-L and 6% in LEC-H cultures. In contrast, the proportion of Pseudomonas species was very low (<1%) in HECs prior to mineralization, but increased during mineralization to 19% in HEC-L and 23% in HEC-H cultures. The greatest increase in abundance was detected in HEC-L cultures, where 40% of the community belonged to the genus Aminobacter. Another interesting observation was the increase in Rhodococcus spp. to an abundance of 25% in LEC-H cultures during MCPA mineralization.
Fig 4.
Relative abundance of most frequently found bacterial genera in the enrichment cultures before and after MCPA mineralization. Bradyrhizobiaceae (genera incertae sedis) are poorly described genera belonging to the family Bradyrhizobiaceae. Other genera represent all the identified genera not included in the eight most abundant ones. LEC, low-enrichment culture at t0; LEC-L, low-enrichment culture amended with 0.1 mg liter−1 MCPA at t3; LEC-H, low-enrichment culture amended with 25 mg liter−1 MCPA at t3; HEC, high-enrichment culture at t0; HEC-L, high-enrichment culture amended with 0.1 mg liter−1 MCPA at t3; HEC-H, high-enrichment culture amended with 25 mg liter−1 MCPA at t3.
DISCUSSION
Metabolic functionality of the enrichments.
By employing a conventional enrichment approach based on different MCPA concentrations, two different bacterial cultures were obtained from an aquifer. They represented different metabolic efficiencies, community compositions, physiologies, and nucleic acid contents at the single cell level. This could reflect different affinities and catabolic enzyme systems, where populations adapted to handle different substrate concentrations might be present in the same microenvironments. We have shown that populations selected on low MCPA concentrations (LECs) have shorter lag phases in mineralizing both low and high MCPA concentrations than those enriched with high MCPA concentrations (HECs) (Fig. 1). These findings may be explained by the presence of catabolic enzyme systems with different affinities and different abilities to produce enzymes performing the catabolic reactions. These types of enzyme systems have been reported for single species possessing systems with different affinities for the same substrate (38–41) or for different species within mixed populations with different affinities for the substrate (42, 43). In these studies, the systems active at low concentrations can be categorized as high-affinity, low-capacity systems, and vice versa for the ones operating at high concentrations. Our results suggest that the obtained enrichment cultures may have similar enzyme systems with different affinities for MCPA. The possible high-affinity enzyme systems of LEC-L cultures may allow greater utilization of MCPA with shorter lag phases than HEC-L cultures (Fig. 1a and c). Shorter lag phases observed in LEC-H as opposed to the long adaptation times required by HEC-L cultures (Fig. 1b and c) might be related to the ability of communities enriched with low MCPA concentrations to switch quickly between high- and low-affinity enzyme systems. These findings suggest that cultures obtained by using excessive amounts of substrates are not necessarily better in terms of their degradative capacities and their adaptation to changes in substrate levels than those obtained with low concentrations that are closer to environmentally relevant concentrations.
Presence of LNA bacteria.
A promising application of FCM is the fractioning of bacterial cells into distinct fractions based on differences in the individual cell fluorescence intensities (related to the nucleic acid content) and in the side and forward light scatter signals, with two major fractions: high-nucleic acid-content bacteria and low-nucleic acid-content bacteria (HNA and LNA bacteria, respectively) (18, 19). We applied this technique to investigate the relative abundance and dynamics of these two fractions in our samples following the protocol by Wang et al. (17). Previous studies have suggested that the LNA bacteria represent the dead or inactive part of the community, whereas HNA bacteria represent the active part of the community (18, 20, 44, 45). However, we found an enrichment of LNA bacteria in the phase where extensive MCPA mineralization occurred in communities selected with low MCPA concentrations. This is in line with recent research in which this group of bacteria was shown to be active and equally abundant with equal or even higher growth rates than the HNA bacteria (17, 46–48). The proliferation of LNA bacteria in connection with MCPA utilization in LEC-L and LEC-H cultures strongly suggests that this group could play a role in degradation of low herbicide concentrations in our samples. These results are contradictory to the observations of Servais et al. (45), who suggested that nucleic acid content is positively correlated with the rates of substrate utilization. Proliferation of LNA bacteria in our experiments also coincided with major shifts in community compositions. (Fig. 3a and b and Fig. 4). This observation is in parallel with the observations of Zubkov et al. (49), suggesting that the LNA population may be a different group of bacteria rather than a different physiological state of HNA cells. However, the observed clustering of LNA and HNA cells in the shape and dispersion of their respective cytometric clouds (Fig. 2) supports a different scenario, suggested by Bouvier et al. (50). These authors explained this fractioning as a result of a complex process that involves both the passage of cells from one fraction to the other as well as bacterial groups that are characteristic for either the HNA or LNA fraction. Overall, our results suggest that LNA bacteria are active populations which could play a role in degradation of low pollutant concentrations, and they might be overlooked in cultivation studies which typically use high levels of substrate. Our findings were supported by a recent study where we tested cytometric characteristics of five different MCPA-degrading bacterial strains isolated from the enrichment cultures used here. The isolates obtained from LECs were dominated by LNA cells, and they were more efficient in mineralizing MCPA at low concentrations (1 μg liter−1) with higher mineralization capacities than the ones obtained from cultures enriched with high concentrations (HECs). Furthermore, these cells shifted to the HNA fraction with the increasing MCPA concentration. On the other hand, the isolates obtained from HECs did not harbor LNA cells (E. Gözdereliler, K. De Roy, N. Boon, and S. R. Sørensen, unpublished data).
Changes in community composition and structure during metabolic activity.
Apart from the catabolic enzyme systems and cellular nucleic acid contents, the bacterial cultures may also respond to shifts in substrate concentration as a change in the community composition. Therefore, these changes were monitored by 16S rRNA gene DGGE. The distinct clustering of HECs and LECs at time zero (t0) was a clear indication of the selective pressures of substrate concentration since these communities were under high selective pressure during the initial enrichment process. Ghosh et al. (51) reported similar results, showing that substrate concentration is an important parameter determining the community structure in atrazine-degrading microbial communities. More interestingly, we have observed four different clusters significantly different from both the original enrichments and each other after MCPA mineralization. To overcome the lack of direct phylogenetic information from DGGE profiles, a pragmatic analysis was applied to the DGGE patterns (28, 52). The derived ecological descriptors provided a useful tool to deliver additional information on community structure, functioning, and dynamics. We found low Rr values (<10) in both communities before mineralization initiated (t0). Marzorati et al. (28) described communities having low Rr as “particularly adverse or restricted to colonization.” This attribution agrees with our findings, considering the selective pressure during the enrichment procedure. The Rr values gradually increased during MCPA mineralization, suggesting an increasing diversity in the cultures. Higher species richness may increase ecosystem functionality and its functional stability (53, 54). Nonetheless, community diversity alone does not determine ecosystem stability. Species evenness and functional redundancy may have an equal or even a greater importance for understanding the stability and functionality of microbial communities and for the functions they perform in the ecosystem (29, 55, 56). Therefore, community organization values (Co) were calculated, representing a numerical value for the species evenness (52). All the studied cultures had similar Co values (∼60), indicating a medium community organization according to the range reported in Marzorati et al. (28). This community structuring suggests that few groups of MCPA-degrading species were numerically dominant in the enrichment cultures. Thus, a pool of MCPA degrader species is capable of replacing the dominant degrader species and performing the necessary function when required, according to the concept of functional redundancy (55). The rate of change of the bacterial communities during MCPA mineralization was visualized by moving-window analysis based on the DGGE profiles (27). The high Δt values between t0 and t1 in LEC-H and HEC-H cultures clearly indicate that high MCPA concentrations had a strong diverging effect on the community structure of the enrichment cultures. Such a high rate of change indicates a highly dynamic community (28). This suggests that use of low, rather than high, substrate concentrations provides less dynamic communities. Similar results were reported in a study by Udiković-Kolić et al. (57), where high atrazine input induced a shift in the taxonomical and functional composition of atrazine-degrading cultures adapted to low atrazine concentrations.
In order to elucidate the changes in microbial diversity before and after mineralization, 454 pyrosequencing was employed to analyze samples from each cluster identified by DGGE (Fig. 4). The most interesting finding from this analysis was the changes within the enrichments shifted to different MCPA concentrations. Variovorax spp., having members known to be PA herbicide degraders (58), were the most abundant group in the enrichment cultures prior to mineralization. The dominance of Variovorax bacteria did not change in cultures that were amended with the same MCPA concentration as the original enrichments, while it decreased significantly in the cultures shifted to a different MCPA concentration. The decline of Variovorax species in LEC-H and HEC-L cultures suggests that those bacteria are highly specialized in regard to their optimal MCPA concentrations and can be outcompeted when the initial MCPA concentration is drastically changed. In LEC-H cultures, the dominance of Variovorax spp. was taken over by Rhodococcus spp. Few studies reported the ability of Rhodococcus spp. to degrade PA herbicides, and to our knowledge only one Rhodococcus sp. was isolated when 2,4-dichlorophenoxyacetic acid was the sole carbon source (59). In contrast to findings in LEC-H cultures, the dominance of Variovorax spp. was taken over by Aminobacter spp. in HEC-L cultures. These bacteria have previously been shown to degrade pesticide residues such as atrazine (60) and 2,6-dichlorobenzamide (61). Sørensen et al. (62) reported that Aminobacter spp. are highly efficient at mineralizing low concentrations of 2,6-dichlorobenzamide in the range of micrograms per liter. These findings could explain the proliferation of Aminobacter bacteria in cultures shifted to a low MCPA concentration. To explain this phenomenon, one should first understand the survival of Aminobacter bacteria in HECs despite the high selective pressure and competition through the enrichments. According to Monod (63), the substrate affinity of a system shifts from low to high as a natural result of substrate depletion in batch systems. This depletion results in threshold concentrations below which fast-growing bacteria are not able to utilize the substrate. Aminobacter spp., most probably having high-affinity enzyme systems, can degrade low MCPA concentrations more rapidly than bacteria having low-affinity enzyme systems, thereby surviving in these highly selective environments. In the current study, this hypothesis is supported by the proliferation of Aminobacter bacteria in populations shifted from high to low MCPA concentrations. Once the affinity for the substrate increases, Aminobacter species that seem to be more efficient at low MCPA concentrations take over the dominance in the population. One could then question the absence of Aminobacter spp. in LECs. One possible explanation for this could be that other members of LECs have even better enzymatic systems for handling low substrate concentrations than Aminobacter spp., which consequently are outcompeted.
Conclusions.
In summary, our results demonstrate that different populations of herbicide-degrading bacteria adapted to metabolizing substrates at different concentrations can exist in a community and can be selected using conventional enrichment techniques. We have shown that using environmentally relevant substrate concentrations, in contrast to the classical high concentrations, would provide bacterial communities that are more efficient in terms of metabolic functionality and stability. Furthermore, the proliferation of LNA bacteria linked to metabolic activity and major changes in community composition suggest that this group of bacteria could play a role in degradation of low herbicide concentrations in aquifers impacted by agricultural practice. To our knowledge, this is the first report showing a link between the growth of LNA bacteria and specific metabolic activity.
Supplementary Material
ACKNOWLEDGMENTS
This study was funded by the Seventh Framework Programme (2007 to 2013) of the European Commission within the Goodwater Marie Curie Initial Training Network (grant number 212683) and by the EU Biotreat project (contract number 266039, call FP7-KBBE-2010.3.5.01). S.R.S. was supported by the Center for Environmental and Agricultural Microbiology (CREAM) funded by the Villum Kann Rasmussen Foundation. K.D.R. was supported by the Flemish Fund for Scientific Research (FWO-Vlaanderen, 3G070010).
We thank Pascal Boeckx and Samuel Bodé for their help during radiorespirometry measurements, Chris Callewaert for his assistance during DGGE analysis, and Jacob Bælum and Carsten Suhr Jacobsen for their help during pyrosequencing analysis. We thank Marek Stibal, Arnaud Dechesne, and Gamze Gulez for critical reading of the manuscript.
Footnotes
Published ahead of print 2 November 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02536-12.
REFERENCES
- 1. Thorling L, Hansen B, Langtofte C, Brüsch W, Møller RR, Mielby S, Højberg AL. 2010. Grundvand. Status og udvikling 1989–2009. Teknisk rapport, GEUS 2010. Geological Survey of Denmark and Greenland, Copenhagen, Denmark [Google Scholar]
- 2. Scheidleder A, Grath J, Winkler G, Stark U, Koreimann C, Gmeiner C. 1999. Groundwater quality and quantity in Europe. European Environment Agency, Copenhagen, Denmark [Google Scholar]
- 3. Sørensen SR, Schultz A, Jacobsen OS, Aamand J. 2006. Sorption, desorption and mineralisation of the herbicides glyphosate and MCPA in samples from two Danish soil and subsurface profiles. Environ. Pollut. 141:184–194 [DOI] [PubMed] [Google Scholar]
- 4. Caldwell DE, Wolfaardt GM, Korber DR, Lawrence JR. 1997. Cultivation of microbial consortia and communities, p 79–90 In Hurst JH, Knudsen GR, McInerney MJ, Stetzenbach LD, Walter MV. (ed), Manual of environmental microbiology. American Society for Microbiology, Washington, DC [Google Scholar]
- 5. Dunbar J, White S, Forney L. 1997. Genetic diversity through the looking glass: effect of enrichment bias. Appl. Environ. Microbiol. 63:1326–1331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Itoh K, Kanda R, Momoda Y, Sumita Y, Kamagata Y, Suyama K, Yamamoto H. 2000. Presence of 2,4-D catabolizing bacteria in a Japanese arable soil that belong to BANA (Bradyrhizobium-Agromonas-Nitrobacter-Afipia) cluster in α-Proteobacteria. Microbes Environ. 15:113–117 [Google Scholar]
- 7. Pemberton JM, Corney B, Don RH. 1979. Evolution and spread of pesticide degrading ability among soil microorganisms, p 287–299 In Timmis KN, Puhler A. (ed), Plasmids of medical environmental and commercial importance. Elsevier, Amsterdam, The Netherlands [Google Scholar]
- 8. Harder W, Dijkhuizen L. 1982. Strategies of mixed substrate utilization in microorganisms. Philos. Trans. R. Soc. Lond. B Biol. Sci. 297:459–480 [DOI] [PubMed] [Google Scholar]
- 9. Veldkamp H, Kuenen JG. 1973. The chemostat as a model system for ecological studies. Bull. Ecol. Res. Comm. 17:347–355 [Google Scholar]
- 10. Breugelmans P, D'Huys PJ, DeMot R, Springael D. 2007. Characterization of novel linuron mineralizing bacterial consortia enriched from long-term linuron-treated agricultural soils. FEMS Microbiol. Ecol. 62:374–385 [DOI] [PubMed] [Google Scholar]
- 11. Hussain S, Sørensen SR, Devers-Lamrani M, El-Sebai T, Martin-Laurent F. 2009. Characterization of an isoproturon mineralizing bacterial culture enriched from a French agricultural soil. Chemosphere 77:1052–1059 [DOI] [PubMed] [Google Scholar]
- 12. Topp E, Mulbry WM, Zhu H, Nour SM, Cuppels D. 2000. Characterization of S-triazine herbicide metabolism by a Nocardioides sp. isolated from agricultural soils. Appl. Environ. Microbiol. 66:3134–3141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zakaria D, Lappin-Scott H, Burton S, Whitby C. 2007. Bacterial diversity in soil enrichment cultures amended with 2 (2-methyl-4-chlorophenoxy) propionic acid (mecoprop). Environ. Microbiol. 9:2575–2587 [DOI] [PubMed] [Google Scholar]
- 14. Goldstein RM, Mallory LM, Alexander M. 1985. Reasons for possible failure of inoculation to enhance biodegradation. Appl. Environ. Microbiol. 50:977–983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lehtomaki M, Niemela S. 1975. Improving microbial degradation of oil in soil. Ambio 4:126–129 [Google Scholar]
- 16. Pham H, Boon N, Marzorati M, Verstraete W. 2009. Enhanced removal of 1,2-dichloroethane by anodophilic microbial consortia. Water Res. 43:2936–2946 [DOI] [PubMed] [Google Scholar]
- 17. Wang Y, Hammes F, Boon N, Chami M, Egli T. 2009. Isolation and characterization of low nucleic acid (LNA)-content bacteria. ISME J. 3:889–902 [DOI] [PubMed] [Google Scholar]
- 18. Gasol JM, Zweifel UL, Peters F, Fuhrman JA, Hagström A. 1999. Significance of size and nucleic acid content heterogeneity as measured by flow cytometry in natural planktonic bacteria. Appl. Environ. Microbiol. 65:4475–4483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lebaron P, Servais P, Agogue H, Courties C, Joux F. 2001. Does the high nucleic acid content of individual bacterial cells allow us to discriminate between active cells and inactive cells in aquatic systems? Appl. Environ. Microbiol. 67:1775–1782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li WKW, Jellett JF, Dickie PM. 1995. DNA distributions in planktonic bacteria stained with TOTO or TO-PRO. Limnol. Oceanogr. 40:1485–1495 [Google Scholar]
- 21. Larsen L, Sørensen SR, Aamand J. 2000. Mecoprop, isoproturon, and atrazine in and above a sandy aquifer: vertical distribution of mineralization potential. Environ. Sci. Technol. 34:2426–2430 [Google Scholar]
- 22. Sørensen SR, Aamand J. 2003. Rapid mineralization of the herbicide isoproturon in soil from a previously treated Danish agricultural field. Pest Manag. Sci. 59:1118–1124 [DOI] [PubMed] [Google Scholar]
- 23. Berney M, Hammes F, Bosshard F, Weilenmann HU, Egli T. 2007. Assessment and interpretation of bacterial viability by using the Live/Dead BacLight kit in combination with flow cytometry. Appl. Environ. Microbiol. 73:3283–3290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. De Roy K, Clement L, Thas O, Wang Y, Boon N. 2012. Flow cytometry for fast microbial community fingerprinting. Water Res. 46:907–919 [DOI] [PubMed] [Google Scholar]
- 25. Muyzer G, Dewaal EC, Uitterlinden AG. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Boon N, Gelder LD, Lievens H, Siciliano SD, Top E, Verstraete W. 2002. Bioaugmenting bioreactors for the continuous removal of 3-chloroaniline by a slow release approach. Environ. Sci. Technol. 36:4698–4704 [DOI] [PubMed] [Google Scholar]
- 27. Boon N, Pycke BF, Marzorati M, Hammes F. 2011. Nutrient gradients in a granular activated carbon biofilter drives bacterial community organization and dynamics. Water Res. 45:6355–6361 [DOI] [PubMed] [Google Scholar]
- 28. Marzorati M, Wittebolle L, Boon N, Daffonchio D, Verstraete W. 2008. How to get more out of molecular fingerprints: practical tools for microbial ecology. Environ. Microbiol. 10:1571–1581 [DOI] [PubMed] [Google Scholar]
- 29. Wittebolle L, Marzorati M, Clement L, Balloi A, Daffonchio D, Heylen K, Verstraete W, Boon N. 2009. Initial community evenness favours functionality under selective stress. Nature 458:623–626 [DOI] [PubMed] [Google Scholar]
- 30. Dowd SE, Sun Y, Secor PR, Rhoads DD, Wolcott BM, James GA, Wolcott RD. 2008. Survey of bacterial diversity in chronic wounds using pyrosequencing, DGGE, and full ribosome shotgun sequencing. BMC Microbiol. 8:43 doi:10.1186/1471-2180-8-43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Andersson AF, Lindberg M, Jakobsson H, Backhed F, Nyren P, Engstrand L. 2008. Comparative analysis of human gut microbiota by barcoded pyrosequencing. PLoS One 3:e2836 doi:10.1371/journal.pone.0002836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Larsen N, Vogensen FK, van den Berg FWJ, Nielsen DS, Andreasen AS, Pedersen BK, Al-Soud WA, Sørensen SJ, Hansen LH, Jakobsen M. 2010. Gut microbiota in human adults with type 2 diabetes differs from non-diabetic adults. PLoS One 5:e9085 doi:10.1371/journal.pone.0009085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. 2006. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72:5069–5072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hall T. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95–98 [Google Scholar]
- 35. Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pe AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knoghts D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knoght R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7:335–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kostka JE, Prakash O, Overholt WA, Green SJ, Freyer G, Canion A, Delgardio J, Norton N, Hazen TC, Huettel M. 2011. Hydrocarbon-degrading bacteria and the bacterial community response in Gulf of Mexico beach sands impacted by the deepwater horizon oil spill. Appl. Environ. Microbiol. 77:7962–7974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wang Q, Garrity GM, Tiedje JM, Cole JR. 2007. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73:5261–5267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ishida Y, Imai I, Kadota H. 1982. Growth and uptake kinetics of a facultatively oligotrophic bacterium at low nutrient concentrations. Microb. Ecol. 8:23–32 [DOI] [PubMed] [Google Scholar]
- 39. Lewis DL, Hodson RE, Freeman LF. 1985. Multiphasic kinetics for transformation of methyl parathion by Flavobacterium species. Appl. Environ. Microbiol. 50:553–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rapp P. 2001. Multiphasic kinetics of transformation of 1,2,4-trichlorobenzene at nano- and micromolar concentrations by Burkholderia sp. strain PS14. Appl. Environ. Microbiol. 67:3496–3500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tros ME, Schraa G, Zehnder AJB. 1996. Transformation of low concentrations of 3-chlorobenzoate by Pseudomonas sp. strain B13: kinetics and residual concentrations. Appl. Environ. Microbiol. 62:437–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Murray RE, Parson LL, Smith MH. 1989. Kinetics of nitrate utilization by mixed populations of denitrifying bacteria. Appl. Environ. Microbiol. 55:717–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Novák L, Larrea L, Wanner J. 1994. Estimation of maximum specific growth rate of heterotrophic and autotrophic biomass: a combined technique of mathematical modelling and batch cultivations. Water Sci. Technol. 30:171–180 [Google Scholar]
- 44. Lebaron P, Servais P, Baudoux AC, Bourrain M, Courties C, Parthuisot N. 2002. Variations of bacterial specific activity with cell size and nucleic acid content assessed by flow cytometry. Aquat. Microb. Ecol. 28:131–140 [Google Scholar]
- 45. Servais P, Casamayor EO, Courties C, Catala P, Parthuisot N, Lebaron P. 2003. Activity and diversity of bacterial cells with high and low nucleic acid content. Aquat. Microb. Ecol. 33:41–51 [Google Scholar]
- 46. Jochem FJ, Lavrentyev PJ, First MR. 2004. Growth and grazing rates of bacteria groups with different apparent DNA content in the Gulf of Mexico. Mar. Biol. 145:1213–1225 [Google Scholar]
- 47. Longnecker K, Sherr BF, Sherr EB. 2005. Activity and phylogenetic diversity of bacterial cells with high and low nucleic acid content and electron transport system activity in an upwelling ecosystem. Appl. Environ. Microbiol. 71:7737–7749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Mary I, Heywood JL, Fuchs BM, Amann R, Tarran GA, Burkill PH, Zubkov MV. 2006. SAR11 dominance among metabolically active low nucleic acid bacterioplankton in surface waters along an Atlantic meridional transect. Aquat. Microb. Ecol. 45:107–113 [Google Scholar]
- 49. Zubkov MV, Fuchs BM, Tarran GA, Burkill PH, Amann R. 2002. Mesoscale distribution of dominant bacterioplankton groups in the northern North Sea in early summer. Aquat. Microb. Ecol. 29:135–144 [Google Scholar]
- 50. Bouvier T, del Giorgio PA, Gasol JM. 2007. A comparative study of the cytometric characteristics of high and low nucleic-acid bacterioplankton cells from different aquatic ecosystems. Environ. Microbiol. 9:2050–2066 [DOI] [PubMed] [Google Scholar]
- 51. Ghosh D, Roy K, Srinivasan V, Mueller T, Tuovinen OH, Sublette K, Peacock A, Radosevich M. 2009. In-situ enrichment and analysis of atrazine-degrading microbial communities using atrazine-containing porous beads. Soil Biol. Biochem. 41:1131–1334 [Google Scholar]
- 52. Read S, Marzorati M, Guimarães BC, Boon N. 2011. Microbial resource management revisited: successful parameters and new concepts. Appl. Microbiol. Biotechnol. 90:861–871 [DOI] [PubMed] [Google Scholar]
- 53. Bell T, Newman JA, Silverman BW, Turner SL, Lilley AK. 2005. The contribution of species richness and composition to bacterial services. Nature 436:1157–1160 [DOI] [PubMed] [Google Scholar]
- 54. Tilman D. 1996. Biodiversity: population versus ecosystem stability. Ecology 77:350–363 [Google Scholar]
- 55. Fernandez A, Huang SY, Seston S, Xing J, Hickey R, Criddle C, Tiedje J. 1999. How stable is stable? Function versus community composition. Appl. Environ. Microbiol. 65:3697–3704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wu T, Chellemi DO, Graham JH, Martin KJ, Rosskopf EN. 2008. Comparison of soil bacterial communities under diverse agricultural land management and crop production practices. Microb. Ecol. 55:293–310 [DOI] [PubMed] [Google Scholar]
- 57. Udiković-Kolić N, Devers-Lamrani M, Petrić I, Hršak D, Martin-Laurent F. 2011. Evidence for taxonomic and functional drift of an atrazine-degrading culture in response to high atrazine input. Appl. Microbiol. Biotechnol. 90:1547–1554 [DOI] [PubMed] [Google Scholar]
- 58. Vallaeys T, Albino1 Soulas LG, Wright AD, Weightman AJ. 1998. Isolation and characterization of a stable 2,4-dichlorophenoxyacetic acid degrading bacterium, Variovorax paradoxus, using chemostat culture. Biotechnol. Lett. 20:1073–1076 [Google Scholar]
- 59. McGhee I, Burns RG. 1995. Biodegradation of 2,4-dichlorophenoxyacetic acid (2,4-D) and 2-methyl-4-chlorophenoxyacetic acid (MCPA) in contaminated soil Appl. Soil. Ecol. 2:143–154 [Google Scholar]
- 60. Rousseaux S, Hartmann A, Soulas G. 2001. Isolation and characterisation of new Gram-negative and Gram-positive atrazine degrading bacteria from different French soils. FEMS Microbiol. Ecol. 36:211–222 [DOI] [PubMed] [Google Scholar]
- 61. Simonsen A, Holtze MS, Sørensen SR, Sørensen SJ, Aamand J. 2006. Mineralisation of 2,6-dichlorobenzamide (BAM) in dichlobenil exposed soils and isolation of a BAM-mineralising Aminobacter sp. Environ. Pollut. 144:289–295 [DOI] [PubMed] [Google Scholar]
- 62. Sørensen SR, Holtze MS, Simonsen A, Aamand J. 2007. Degradation and mineralization of nanomolar concentrations of the herbicide dichlobenil and its persistent metabolite 2,6-dichlorobenzamide by Aminobacter spp. isolated from dichlobenil-treated soils. Appl. Environ. Microbiol. 73:399–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Monod J. 1949. The growth of bacterial cultures. Annu. Rev. Microbiol. 3:371–394 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.