Abstract
FOF1-ATP synthases are ubiquitous proton- or ion-powered membrane enzymes providing ATP for all kinds of cellular processes. The mechanochemistry of catalysis is driven by two rotary nanomotors coupled within the enzyme. Their different step sizes have been observed by single-molecule microscopy including videomicroscopy of fluctuating nanobeads attached to single enzymes and single-molecule Förster resonance energy transfer. Here we review recent developments of approaches to monitor the step size of subunit rotation and the transient elastic energy storage mechanism in single FOF1-ATP synthases.
Keywords: FOF1-ATP synthase, rotary motor, subunit compliance, single-molecule FRET
1. Introduction
Adenosine triphosphate (ATP) is the universal energy currency of the cell. When it is hydrolysed to adenosine diphosphate (ADP) and inorganic phosphate (Pi), its biochemical standard free energy ΔG0’(ATP) of −30 kJ mol−1 [1–3] can be used for a variety of biochemical reactions. However, under physiological conditions, the free energy of ATP hydrolysis is even higher, about −50 to −60 kJ mol−1 [4]. As the amount of ATP is limited—for example, our human body contains about 50 g—it must constantly be recycled as we consume 1000-fold more ATP than we have [5,6]. Some 4.5 billion years ago, a class of enzymes named ATP synthases evolved to regenerate ATP from ADP and Pi. Depending on structural properties of subunits and the primary physiological function in different cells or cellular compartments, three major types of enzymes are discriminated [7,8]. FOF1-ATP synthases are found in the thylakoid membrane of chloroplasts, in the inner mitochondrial membrane and in the bacterial plasma membrane. Their function is the synthesis of ATP. By contrast, VOV1-ATPases are ATP-driven proton pumps [9,10]. Archaeal AOA1-ATP synthases are ATP-producing enzymes with structures similar to those of VOV1-ATPases.
ATP synthesis in FOF1-ATP synthases is powered by an electrochemical potential difference of protons (or Na+ in some cells), over the respective membranes [11]. About 50 years ago, the principal mechanism of the ATP synthase was postulated by Peter Mitchell in his chemiosmotic theory in 1961 [12]. He claimed that ATP is not synthesized by a phosphorylated intermediate in mitochondria, but by an enzyme integrated in the inner mitochondrial membrane, using the difference in proton concentrations (ΔpH) in the two compartments plus the electric potential (Δψ) across the separating membrane [12]. Both components comprise the proton motive force, PMF. The ΔpH is generated during photosynthesis in chloroplasts, or during aerobic phosphorylation in mitochondria or bacteria. Although it took more than a decade before the chemiosmotic theory was commonly accepted, this mechanism is now part of every biochemical textbook. However, the way that single protons drive the catalytic reaction cycle within the enzyme still had to be unravelled at the atomic level.
2. The components of the two rotary motors of FOF1-ATP synthase
Today, a wealth of structural information about the enzyme is available. FOF1-ATP synthase consists of two domains. In the bacterial enzyme, subunits α3β3γδɛ comprise the F1 domain (we will use the Escherichia coli nomenclature in the following) while subunits ab2cn form the membrane-embedded FO domain. The number of c subunits varies between species and seems to depend on the available proton (or Na+) motive force. The smallest number of c subunits is eight for the mitochondrial enzyme from bovine heart [13], and the largest known so far is 15 in enzymes from cyanobacteria [14]. The bacterial enzyme from E. coli has 10 c subunits [15].
The first pioneering crystal structure of the F1 portion from bovine heart mitochondria was described by John Walker and co-workers in 1994 [16]. The main features were the alternating arrangement of α3β3 subunits, which formed a hexagonal structure, and a central stalk formed by subunit γ. The catalytic nucleotide binding sites were located mainly on the β subunits, at the interface with α. Subunits named βTP and βDP contained AMP-PNP, a non-hydrolysable ATP analogue, or ADP, respectively, while the third β subunit (βE) was empty. It seemed that the β subunits changed their conformation depending on the bound nucleotide. Three additional nucleotide binding sites were located at the other interfaces of αβ. However, they were non-catalytic, and all contained AMP-PNP. Consequently, the crystal structure was interpreted as a still picture of the active enzyme. It was shown later that it resembled the catalytic dwell (see below) [17,18].
Another feature of subunit β was the amino acid sequence DELSEED, which apparently formed a lever that can act to open and close the associated nucleotide binding site. It seemed that parts of subunit γ can interact with this lever and, thereby, define the conformational state of each catalytic binding site, because γ sequentially changes its orientation with respect to the three β subunits in the active enzyme. Moreover, subunit γ consisted of a globular domain that, together with ε, faced the membrane-embedded FO portion, and two N- and C-terminal α-helices that formed a coiled coil and extended into the central cavity of the α3β3 hexagon.
Recently, the first F1 structure of the E. coli enzyme was published [19]. With a high overall similarity to the mitochondrial F1 structures, it was confirmed that the globular domain of γ is located at the membrane side of F1 and probably interacts with the c subunits of FO (which were not present in this structure). Subunit ε was attached to γ and is therefore also part of the central stalk of E. coli F1. Subunit δ was not present in this structure, but is located at the top of the enzyme.
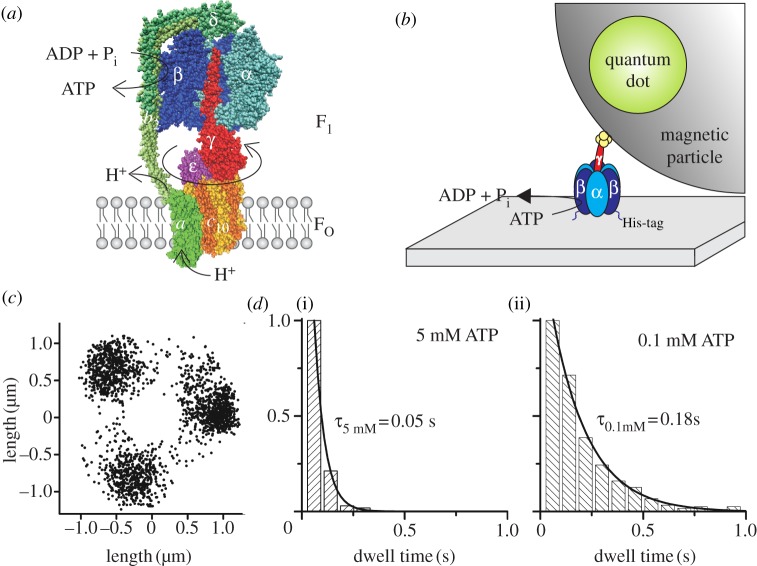
In the FO portion, a ring of c subunits is embedded in the membrane. The ring interacts with subunits γ and ε of F1 [20,21] and with subunits a [22,23] and b2 of FO [24,25]. Subunit a forms the two proton (or Na+ in some organisms) conducting half channels that end on either side of the membrane. The dimeric subunits b2 form a right-handed coiled coil with a small residue off-set [26,27] that encompasses the whole protein from the membrane to the top of the α3β3 hexagon. The b subunits are regarded as the eccentric stalk bound tightly to F1 [28,29]. This peripheral connection of ab2 to F1 combined with the three possible orientations of γ within F1 results in an asymmetric structure of the enzyme, with an orientation of the central axis that is stochastic with respect to the peripheral b2 subunits. The structural model of E. coli FOF1-ATP synthase is shown in figure 1a. Homology modelling based on the mitochondrial enzyme structures and assumptions about subunit localization based on cross-link data and single-molecule Förster resonance energy transfer (FRET) triangulation were used.
Figure 1.
(a) Structural model of E. coli FOF1-ATP synthase with one α (light blue) and one β (dark blue) subunit removed to expose subunit γ (red) in the centre of the α3β3-pseudohexagon. Together with ɛ (pink) and δ on top (dark green), they form the F1 portion. The membrane-embedded FO portion consists of 10 c subunits (yellow/orange), which form a ring structure within the membrane. Subunit a (light green) and two b subunits (green) form an eccentric stalk to hold both portions together. Subunits γ, ε and the c10-ring comprise the rotor of FOF1 (reddish colours), which rotates clockwise (if viewed from the membrane) during ATP synthesis with respect to the stator subunits α3β3δab2 (blue/green colours). ATP synthesis in β is driven by proton (or Na+) flow through FO owing to an electrochemical potential difference over the membrane. This homology model of FOF1 is a composite of several partial structures (PDB codes 1BMF [16], 2CK3 [30], 1H8E [31] and 1YCE [32] in the Protein Data Bank, http://www.pdb.org) and was drawn with VMD v. 1.9.1 [33]. Subunits a, b2 and δ have been modelled in roughly their correct size and manually attached using cross-link data. (b) Set-up of the rotation assay with a magnetic bead attached to the F1 portion. F1 (α3β3γ) is attached to a cover glass via histidine (His)-tags in each β subunit. The streptavidin-covered magnetic bead is coupled to a cysteine in subunit γ via biotin. The bead is doped with fluorescent biotinylated quantum dots to visualize ATP-driven rotation of the γ-bead complex. (c) Endpoint distribution of a rotating actin filament (1 µm) coupled to the c10-ring of E. coli FOF1 in the rotation assay showing apparent 120° stepping rotation during ATP hydrolysis at high [ATP] in the presence of detergent (adapted from [34]). (d) Dwell times of stepped rotation of E. coli FOF1 in the presence of (i) 5 mM or (ii) 0.1 mM ATP. The histograms show the averaged dwells of the stepping positions of one enzyme after 200 or 120 rotations, respectively.
The question still remains how proton (or Na+) translocation at a remote part in FO is coupled to the synthesis of ATP in the three nucleotide binding sites in the β subunits of F1. A concept for ATP synthesis was first proposed by Paul Boyer in 1981 before detailed structural information was available [35,36]. According to his ‘binding change mechanism’, two of the three nucleotide binding sites either bind ADP and Pi or generate ATP, and these reactions are synchronized by a rotating, central, asymmetric γ subunit. Proton-translocation through FO is the driving force for γ rotation. In this concept, FOF1-ATP synthase comprises a double motor, formed by the rotating c-ring in FO and γε in F1. The rotor is stabilized by the stator subunits ab2 in FO and α3β3δ in F1. In figure 1a, the rotary subunits γ, ε and c are depicted in red, pink and orange, respectively.
Since the first crystal structure was published supporting the rotary mechanism of catalysis, many research groups tried to reveal the molecular mechanisms of this motor enzyme, applying a variety of biochemical and spectroscopic techniques to study its function [37–39]. However, the asymmetric structure of the holoenzyme limited the results owing to ensemble averaging. FOF1-ATP synthase is a relatively robust enzyme that can easily be manipulated, making it suitable for single-molecule microscopy (which has evolved since 1989 from the first experiments with dyes in organic crystals at liquid helium temperature [40]). Observing sequential conformational changes of single enzymes as time trajectories, i.e. one enzyme at a time or separated in space, does not require a synchronization of asymmetric proteins for a well-defined starting position or conformation in the catalytic cycle. Here we will present some of the single-molecule experiments that unravelled the rotational mechanism of the two coupled rotary nanomotors within FOF1-ATP synthase.
3. Direct evidence: ATP-driven rotation assays resolve 120° steps and sub-steps in F1
In 1997, the ground-breaking experiment that unambiguously demonstrated the rotational mechanism in the F1 portion of FOF1-ATP synthase was reported by Noji et al. [41]. They attached the α3β3γ sub-complex of TF1 (i.e. F1 from the thermophilic Bacillus strain PS3) to a Ni-nitrilotriacetic acid covered glass coverslip via histidine-tags in each β subunit and coupled a fluorescent actin filament to a cysteine in the globular domain of subunit γ via a biotin–streptavidin–biotin linker. They used videomicroscopy to record single molecules of this F1–actin complex. After addition of ATP, hydrolysis reaction in F1 caused a unidirectional rotational movement of subunit γ that was visualized in real time by the actin filament. This rotation assay served as a starting point for a series of single-molecule experiments that eventually resulted in the decryption of the successive reaction steps in F1 during ATP hydrolysis. Besides fluorescent actin filaments, gold nanobeads [42], gold nanorods [43–45], polystyrene [46] or magnetic beads [47–50], or the fluorescence anisotropy of single fluorophores [51–53] served as reporters of subunit rotation in F1 as well as in FOF1. In the latter, the reporter was attached to the c-ring [53–55].
The principal set-up for the rotation assay is shown in figure 1b with a fluorescent bead as the marker of rotation. The bright bead could be located with nanometre precision in high-speed videomicroscopy. During γ subunit rotation, the centre of the bead position moved only a few nanometres in the x- and y-direction of the microscopic images. Alternatively, using a fluorescent actin filament as marker of rotation, the ATP-driven rotation was analysed by locating the ends of the actin filament in each frame of the recorded video. The changing end position was attributed to the free, moving end of the filament, whereas the other end that stayed at the same position in each image was associated with the streptavidin connection to the rotary γ subunit in F1 (or rotary c subunits in FOF1 from E. coli in the presence of detergent as shown here in figure 1c for convenience [34]). Plotting all changing end positions resulted in three major stopping positions, which were separated by micrometres in the x- and y-direction. Therefore, the related rotary angles were easily distinguishable. The dwell times for each assigned area of stopping positions were calculated and added to histograms (figure 1d, c-ring rotation data from E. coli FOF1 in detergent as an example [34]). Dwell time distributions depended on the ATP concentration ([ATP]). At high [ATP], i.e. in the millimolar range, the dwell times were significantly affected by the viscous drag of the actin filament slowing down rotation, but for low [ATP], less than 1 µM, dwell times corresponded to the biochemical turnover rates in solution [56].
Subsequent videomicroscopy experiments aimed to associate different reaction steps of ATP hydrolysis, i.e. ATP binding, ATP hydrolysis, ADP and Pi release, to angular-resolved sub-steps of γ subunit rotation [42]. Small 40-nm gold beads were used to lower the friction on the enzyme during rotation, and a high-speed camera with 8000 frames per second was required to observe a three-stepped rotation at 2 mM [ATP]. Each stop after a 120° rotational step corresponded to an ATP hydrolysis reaction in one of the three catalytic sites in the β subunits. By lowering [ATP] to 2 µM, each rotary transition was further resolved into two sub-steps of 80° and 40°. Substrate was limited at low [ATP] and, therefore, binding of an ATP molecule was slowed down. The duration of the dwell before the 80° rotational sub-step depended on [ATP], indicating that (i) the 80° sub-step is associated with the binding process of an ATP molecule to an empty nucleotide binding site, and (ii) the dwell before the 80° sub-step is the ‘ATP-waiting state’ of the enzyme. The dwell before the rotary 40° sub-step was independent of [ATP] and was associated with the ATP hydrolysis reaction and product release. Therefore, it was named the ‘catalytic dwell’.
Next, rotation of subunit γ and the binding event of a fluorescent ATP derivative to subunit β were monitored simultaneously. In this rotation assay, polystyrene or magnetic beads attached to TF1 served as the reporters to follow the rotation of γ by bright-field microscopy [57,58]. A total of 10 per cent of the ATP molecules were fluorescently labelled with the cyanine dye Cy3 (Cy3-ATP). By total internal reflection fluorescence microscopy (TIRFM), an evanescent wave was created above the coverslip to only observe Cy3-ATP molecules bound to TF1, but not the diffusing molecules in the bulk solution. The polarization of the evanescent wave was oscillated with a frequency of 1 Hz. Thereby, fluorescence of bound Cy3-ATP in any of the three β subunits was detected through angle-resolved fluorescence anisotropy imaging. Fluorescence and bright-field light were separated by a dichroic mirror and simultaneously recorded with a conventional video camera. At 0.6 µM [ATP + Cy3-ATP], subunit γ progressed in 120° steps and a binding event of Cy3-ATP immediately induced a rotation of the bead by 120°. This Cy3-ATP molecule (or its hydrolysed derivative Cy3-ADP) remained bound at least for a 240° rotation of γ. The nucleotide binding site in β then remained empty until binding of another ATP molecule occurred. These results excluded that each β subunit acts on its own, but confirmed that all three catalytic sites—together with subunit γ—hydrolyse ATP in a concerted action. When only Cy3-ATP without unlabelled ATP was used, the hydrolysis reaction was slowed down (see docking calculations and turnover rates in [59]), and the 80° and 40° sub-steps became visible.
From these and subsequent single-molecule rotation experiments with surface-attached F1 from the thermophilic Bacillus strain PS3, the following reaction scheme for ATP hydrolysis in the F1 motor emerged [17,60,61]. If the ATP-waiting dwell at a rotational angle of subunit γ is defined as 0°, ATP binding to the empty catalytic side induces an 80° rotational movement of γ. At the same time, the nucleotide binding site at +120° (i.e. in the forward direction of γ rotation) releases ADP, while the site at −120° contains ATP from the previous binding event. After the first 80° sub-step, the enzyme reaches the catalytic dwell. ATP that was bound during the previous step is hydrolysed into ADP and Pi. In a second reaction, the release of phosphate triggers the following 40° sub-step. ADP is released during the second 80° sub-step from the side that was previously at −120°. The rotational movement of γ in the separated F1 motor is [ATP]-dependent and occurs in three or six distinguished rotary steps. While the rotation of γ is counter clockwise when viewed from the membrane, the sequence of events in the β subunits is in the opposite direction, i.e. after binding of one ATP to an empty site in one β subunit, the ATP in the neighbouring β subunit in clockwise direction will be hydrolysed during the following catalytic dwell.
Most single-molecule studies on the mechanism of the F1-ATPase described above were carried out with the α3β3γ sub-complex of TF1, mainly by attaching actin filaments or beads to subunit γ. The mesophilic E. coli F1 enzyme, as studied by the group of Masamitsu Futai with 40–60-nm sized gold beads, rotated slightly faster at similar [ATP] and temperatures. Their protein preparation contained also the second rotary subunit ε, but not the static subunit δ of F1 in the single-molecule assay. At saturating [ATP], a rotational rate of γ in these single F1 molecules of 400 rps (or 1200 ATP s−1, respectively), was reported [62,63]. The single-molecule catalytic dwell times were 0.2 ms, i.e. about 10 times shorter than expected from the biochemical ensemble measurements of ATP hydrolysis, and the transition time for the 120° steps was 0.6 ms [64]. Thus, different stochastic pausing mechanisms (ADP inhibition and ε inhibition with distinct dwells) could be the causes for the apparent evidence that about 90 per cent of the F1 molecules in the ensemble measurements are inhibited. Compared with TF1, the E. coli enzyme has a shorter catalytic dwell but a slightly slower transition for the 120° step [64–66]. On the other hand, Wayne Frasch and colleagues observed the rotation of 75-nm long gold nanorods attached to γ of E. coli F1 under saturating [ATP] with a very high temporal resolution of 2.5 µs. They found catalytic dwell times of about 8 ms in single EF1, comparable to their bulk ATP hydrolysis measurements of 7.7 ms [43]. Because differences in rotational rates were also reported for ATP-driven c-ring rotation in FOF1 from E. coli (see below), any comparison of rotational rates of a single molecule has to be done carefully, and the sites of cysteine mutations for marker attachment, marker sizes and shapes, other mutations, completeness of subunits in the enzyme, or the presence of detergents or lipids for FOF1 have to be distinguished. In addition, the time resolution of the experimental approach has to match the rotational details to be resolved.
4. Mechanically driven ATP synthesis in single F1 using external magnetic force
The F1 complex is capable of only ATP hydrolysis because it lacks the FO motor. However, when γ in F1 was turned in the opposite direction by an external magnetic force acting on a magnetic bead attached to γ, it could also synthesize ATP, demonstrating that mechanical energy can be transformed into chemical energy [47,48]. The applied external magnetic field was used to drive the enzyme in either catalytic direction, that is, to enforce ATP hydrolysis or synthesis, or to stall the γ subunit at any given rotational angle. In sealed picolitre reaction chambers, it was possible to detect and quantify the few synthesized ATP molecules by the luciferin/luciferase luminescence reactions, or by monitoring ATP-driven rotation after release of the external magnetic force, respectively.
5. Proton-driven subunit rotation can be monitored by single-molecule FRET
A different experimental approach was developed to monitor rotational movements in the holoenzyme FOF1-ATP synthase during proton-driven ATP synthesis. In the group of Gräber, single-molecule Förster resonance energy transfer (FRET) was applied to observe the rotation of the central shaft in individual E. coli FOF1-ATP synthases. Two fluorescent dyes in close proximity, that is in the range of up to 10 nm, can transfer energy from one to the other by the non-radiative Förster mechanism, provided the emission spectrum of the donor fluorophore overlaps with the absorption spectrum of the acceptor, and the transition dipole moments of the dyes are not oriented orthogonal to each other [67]. Then the resonance energy transfer efficiency depends on the distance between the two dyes. Therefore, FRET can be used as a molecular ruler within single proteins for dye distances between 3 and 8 nm. Börsch and colleagues used two fluorophores attached to subunit γ or ε on the rotor and to both b subunits on the stator via genetically introduced cysteines [68–72]. Specific labelling of the respective cysteines was achieved by separate labelling of the F1 and FO portions, and subsequent reassembly into a fully functional double-labelled holoenzyme, which was reconstituted in liposomes. These proteoliposomes contained less than one enzyme on average and showed ATP hydrolysis and synthesis rates similar to the non-modified wild-type enzymes from E. coli.
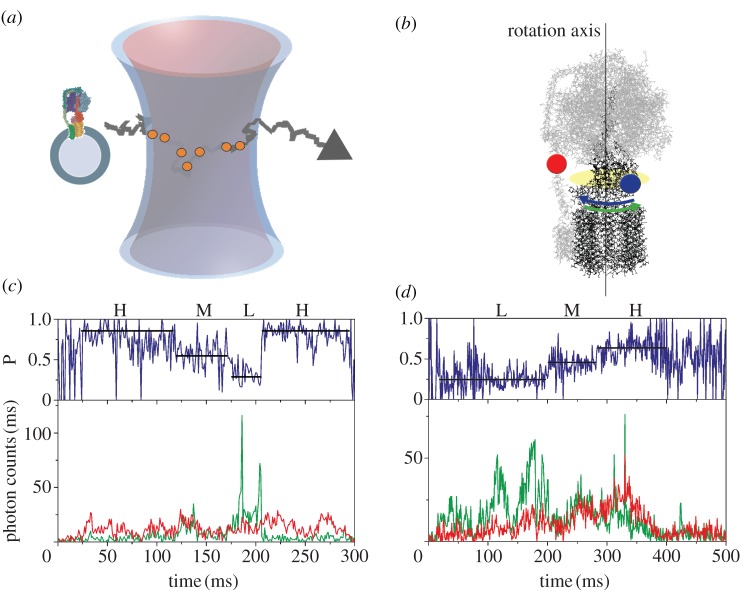
The measurement principle of confocal single-molecule FRET with freely diffusing proteoliposomes in solution is shown in figure 2a. The dye rhodamine 110 (Rh110) on the rotor subunit γ was excited by a laser at 488 nm and acted as the FRET donor, while the second dye cyanine 5 (Cy5) on the stator subunits b2 served as the FRET acceptor [73] (figure 2b). The fluorescence of both dyes was detected by sensitive avalanche photo-diodes and recorded using time-correlated single photon counting electronics with picosecond time resolution. Once a proteoliposome entered the femtolitre-sized confocal excitation/detection volume owing to Brownian motion, the FRET donor Rh110 on FOF1 was excited repeatedly and emitted photons in a burst. The fluorescence intensity depended on the actual position of the dye within the laser focus, which had an approximate three-dimensional Gaussian intensity distribution. The mean transit time for the proteoliposomes was about 30–50 ms, but a few proteoliposomes could be observed for up to several hundred milliseconds (figure 2c,d). To power ATP synthesis, the proteoliposomes with a single enzyme were energized by a ΔpH using buffer mixing in the presence of an electrical potential difference created by a K+ diffusion potential [11]. For rotation measurements during ATP hydrolysis, 1 mM ATP was added to the buffer in the presence of 2.5 mM Mg2+. The concentration of liposomes containing only one labelled FOF1-ATP synthase was adjusted to about 100 pM so that on time average, only one enzyme was detected in the focus of the confocal microscope. In the presence of AMP-PNP, each enzyme showed one of three different FRET efficiencies corresponding to three possible orientations of the γ subunit within the F1 portion. From the FRET efficiencies, the dye-to-dye distances of about 4.5, 6.5 and 8 nm were calculated for each γ subunit orientation. These distances were in agreement with the model of FOF1 from E. coli.
Figure 2.
(a) Monitoring subunit rotation in a single reconstituted FOF1-ATP synthase by confocal FRET microscopy. The proteoliposome arbitrarily traverses the confocal excitation/detection volume owing to Brownian motion. (b) Positions of FRET fluorophores in FOF1-ATP synthase. FRET donor Rh110 (blue dot) on subunit ɛ of the rotor shown in black was attached off the central axis of rotation and was expected to move according to the blue arrow during ATP hydrolysis. Rotation was opposite for ATP synthesis (green arrow). FRET acceptor Cy5 (red dot) was attached to the b subunit dimer of the stator shown in grey. (c) Photon burst of a FRET-labelled FOF1-ATP synthase during ATP hydrolysis. The lower panel shows fluorescence intensity trajectories of FRET donor Rh110 (green trace. ID) and acceptor Cy5 (red trace, IA). The upper panel shows the corresponding proximity factor P as a blue trace, with P = IA / (ID + IA). FRET efficiency levels were assigned manually and called ‘high FRET’ H, ‘medium FRET’ M, and ‘low FRET’ L. The FRET sequence was →H → M → L → H→ (adapted from [73]). (d) Photon burst of a FRET-labelled FOF1-ATP synthase during ATP synthesis. The lower panel shows fluorescence intensity trajectories and the upper panels shows the proximity factor trace. The FRET sequence was reversed →L → M → H → L→ (adapted from [73]).
During catalysis, also, three FRET efficiencies were found. However, they changed fast and stepwise within photon bursts of single enzymes. Their sequence was reversed, depending on the catalytic conditions as seen in figures 2c for ATP hydrolysis and 2d for ATP synthesis. The mean dwell times for the FRET levels were found in the range of 12–20 ms for ATP hydrolysis, or between 17 and 50 ms for ATP synthesis, respectively. These dwells corresponded to a single enzyme turnover of approximately 70 ATP s−1 for ATP hydrolysis and up to 50 ATP s−1 for ATP synthesis. Single-molecule rates were in good agreement with the biochemical ensemble rates for the same enzyme preparations reconstituted in liposomes. The progression of the three FRET levels was the same in about 80 per cent of all FRET levels, but reversed for the opposing biochemical conditions. For ATP hydrolysis and synthesis, the three FRET levels were similar, that is independent from the direction of rotation. This demonstrated not only that γ and ε are indeed part of the rotor in the holoenzyme FOF1-ATP synthase, but also that the catalytic events during ATP synthesis and ATP hydrolysis are in principle reversible. In the presence of the non-competitive inhibitor aurovertin B, differences in the rotational movement of γ during ATP synthesis and ATP hydrolysis could be unravelled, indicating that inhibitors not only serve as controls for biochemical activity measurements but can also be used to obtain additional mechanistic information. Single-molecule FRET measurements can discriminate between a general slowing down of rotation during ATP hydrolysis and partial blockade of rotation during ATP synthesis [73].
In a subsequent single-molecule FRET experiment, the step size of the rotating c-ring in FOF1-ATP synthase was resolved [74]. Enhanced green fluorescent protein (EGFP) was fused to the stator subunit a in FO and served as a FRET donor [75], while the acceptor fluorophore Alexa568 was covalently bound to a cysteine of one c subunit. Reconstituted single enzymes were driven by a proton motive force to synthesize ATP. Double-labelled, rotating enzymes were detected by pulsed alternating laser excitation with an optimized duty cycle [76,77]. Up to five different FRET levels were discriminated. However, the FRET efficiency changes were smaller and the dwell times shorter than before, as expected for a 10-stepped rotary movement with the same rotational rate for the full rotation as the three-step motor in F1. Small and large FRET changes were manually assigned in the dataset, but the progression of FRET levels was not always clear and unidirectional. The time resolution of the single-molecule FRET experiment was limited to 1 ms owing to a reduced brightness and photophysical properties of the fluorophores, so that some short dwells could be missed. Simulated single-molecule FRET time trajectories were generated by a Monte Carlo approach to facilitate the unravelling of the c-ring step size. Simulations of a 10-stepped c-ring with five different FRET distances for ring symmetry reasons were compared to a c-ring rotating with three 120° steps. Afterwards the experimental FRET results could be fitted best when assuming a 10-stepped rotation. Thus it was concluded that the rotary c-ring of the enzyme indeed proceeds in 36° steps during ATP synthesis. However, it could not be ruled out that the single-molecule FRET level distribution would also support a combination of 80° + 40° sub-steps, which were reported for the thermophilic TF1 during ATP hydrolysis.
The one-step-after-another rotation of the c-ring was supported for the ATP hydrolysis mode using nanorods [44] or nanobeads [78]. However, mutations in the proton-translocation pathway through subunit a or increased viscous drag were required to resolve the sub-steps in these experiments. As discussed for the case of γ subunit rotation in F1 above, it is important to compare the single-molecule rotational speed and step sizes with caution. Using closed liposomes in the FRET experiments in contrast to surface-supported lipid bilayer patches or lipid nanodiscs will affect and slow down ATP-driven subunit rotation owing to the build-up of a counteracting proton concentration difference by the coupled enzyme. In addition, the amino acid positions for cysteine mutations not only will allow attachment of the fluorophores or other markers, but might alter the catalytic rates of the mutant enzymes as well. Nevertheless, because there is a mismatch between the 10-step rotation in FO and the three-step rotation in F1, these results strengthened the model of two elastically coupled nanomotors that was developed by Junge and co-workers [4,79–82].
5. Different step sizes of the two coupled motors require internal elasticity
In FOF1-ATP synthase, two nanomotors work against each other. Depending on the proton motive force, the E. coli enzymes in proteoliposomes can switch from ATP synthesis to ATP hydrolysis [3]. The chemical potential of ATP synthesis or hydrolysis in F1 works against the electrochemical potential difference over the membrane that drives the motor in FO. The two motors run with different gears. F1 is a 3-stepper motor, while FO is a 10-stepper motor in E. coli (or 8–15 stepper, depending on the organism). The question arises how the energy is transmitted between theses two motors, and how the enzyme can overcome the energy barriers for a system where two motors with different gears are tightly coupled. It has been suggested that the motors are elastically coupled [4,79–82]. Instead of a rigid system, certain parts of this enzyme are elastically soft and can store energy transiently until needed for the chemical reaction. Thereby, the enzyme can run with a high rotational rate and kinetic efficiency.
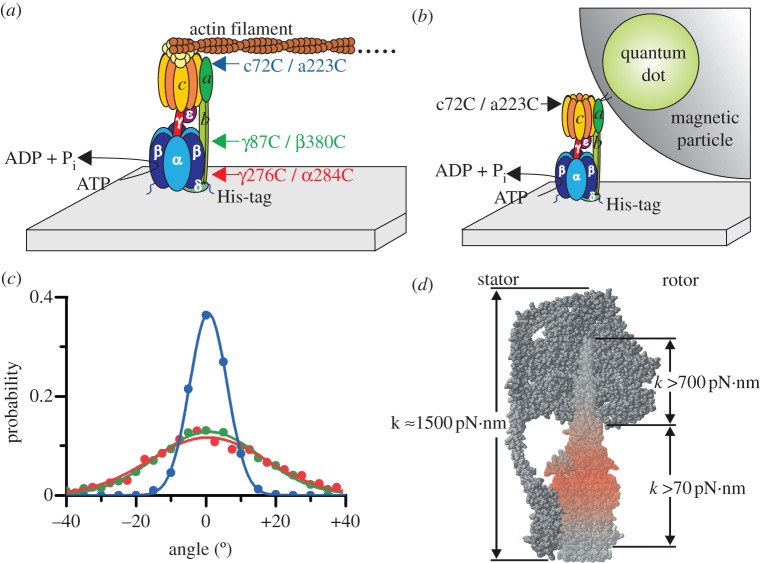
In a series of single-molecule experiments, Sielaff and co-workers have determined these elastic elements [49,50]. They used mutants of E. coli F1 or FOF1 that contained two engineered cysteines, one in the rotor (subunits γ or c) and the other in the stator (subunits β, α or a) opposing the former cysteine (figure 3a). The cysteine pairs, namely aI223C/cL72C (blue in figure 3a), βD380C/γA87C (green) and αE284C/γA276C (red), were placed over the length of the rotor stalk to scan for its different elasticities. In the rotation assay, the enzyme was attached to the glass surface via histidine-tags in each β subunit, while a short fluorescently labelled actin filament (approx. 0.5 µm) or a quantum dot (Q-dot) doped magnetic bead (1 µm) linked to the enzyme on the other side served as a reporter for videomicroscopy. The reporters were bound to either the c-ring in FOF1, or to γ in F1, respectively (figure 3a,b).
Figure 3.
(a) Rotation assay with E. coli FOF1 attached to a cover glass via histidine (His)-tags in subunit β, driven by ATP hydrolysis. A TMR-labelled fluorescent actin filament is coupled to strep-tags in each c subunit via a streptactin–biotin link. To monitor the internal elasticity of the rotor, three mutants each with two opposing cysteines on the rotor and the stator were engineered as indicated. After oxidation, they served to form a disulphide bridge and to stall the enzyme in a certain position. (b) A Q-dot doped magnetic particle instead of an actin filament was used to monitor the internal elasticity of the stator, i.e. subunits b2. The magnetic particle was covered with streptavidin or streptactin, to link it to b2, either directly via two cysteines/biotins in the membrane integral part of subunits b, or indirectly to the c-ring, that was cross-linked to subunit a via two cysteines as indicated, respectively. (c) Compliance of the enzyme–filament complex for the three double mutants shown in (a), determined by the thermal fluctuations of the cross-linked protein. The histograms were fitted with Gaussians and the torsional stiffness was derived from their inverse width resulting in 450, 59 and 47 pN·nm for the blue, green and red curve, respectively (adapted from [50]). (d) Model of FOF1 showing in red the site of the highest elasticity, namely the contact site of c-ring with γε. The compliance in units of pN·nm is given for different domains of the enzyme (figure provided by S. Engelbrecht and W. Junge).
After addition of ATP and under reducing conditions, the subunits started to rotate as expected. Upon oxidation, the two cysteines formed a disulphide bridge and the enzyme stopped to rotate. In this state, only the thermal fluctuations of the enzyme-reporter-system were visible. A histogram of these fluctuations showed Gaussian-like distributions (figure 3c). The width σ of the Gaussians was inversely proportional to the torsional stiffness κ (in pN·nm) by σ = kBT/κ, where kB is the Boltzmann constant, and T the absolute temperature. The coiled coil shaft of γ had a medium stiffness with κ = 320 pN·nm. The portion with the smallest torsional stiffness was located right between the respective sites of torque generation in F1 and FO—that is, at the interface of the globular portion of γ and the c-ring. With a torsional stiffness of less than 70 pN·nm, it can store up to 14 kJ mol−1 elastic energy to smooth the cooperation of the two motors when they operate against each other. In the unrestricted enzyme, the compliance was even lower (about 35 pN·nm as inferred from the dwell positions of the enzyme), caused by the flexible hinge motion of the lever in the β subunits. Moreover, a molecular dynamics simulation of the free and the cross-linked system revealed a striking agreement with the experimental data [83].
On the other hand, the stator was found be at least 10 times as stiff as the most compliant portion of the rotor. The elasticity of the wild-type b2 dimer was compared with mutated b subunits that either were elongated by 11 amino acid residues (‘long’) or destabilized by substituting three consecutive residues with glycines (‘Gly3-mutant’) [49]. In the rotational assay, a Q-dot doped magnetic bead coupled to the stator served as the reporter to monitor the motion of FOF1 (figure 3b). As the stator did not rotate by itself, the motion was artificially induced by applying an external rotating magnetic field, which drove the bead forwards or backwards. Two modes of operation were tested. (i) When the magnetic bead was coupled to the membrane integral end of the b2 dimer by a cysteine–biotin–streptavidin coupling, the dimer was twisted around its axis. (ii) The physiological bending motion of b2 was studied by coupling the magnetic bead to the c-ring that was cross-linked to subunit a. In all cases, the compliance was about 500 pN·nm, except for the bending of the Gly3-mutant. Here, the compliance was three times lower compared with the wild-type enzyme. The ATP-dependent H+-pumping activity of the mutants was halved compared with the wild-type FOF1-ATP synthase. Nonetheless, these mutants with a destabilized stator were active, because lowering its compliance three-fold (that is, still one magnitude larger than the rotor compliance) did not grossly affect the stator stability. The elastic properties are summarized in figure 3d. These data supported the theoretical model for the energy transduction in FOF1-ATP synthase by transient reversible deformations in the rotor.
6. Three-colour single-molecule FRET reveals rotor twisting up to 120°
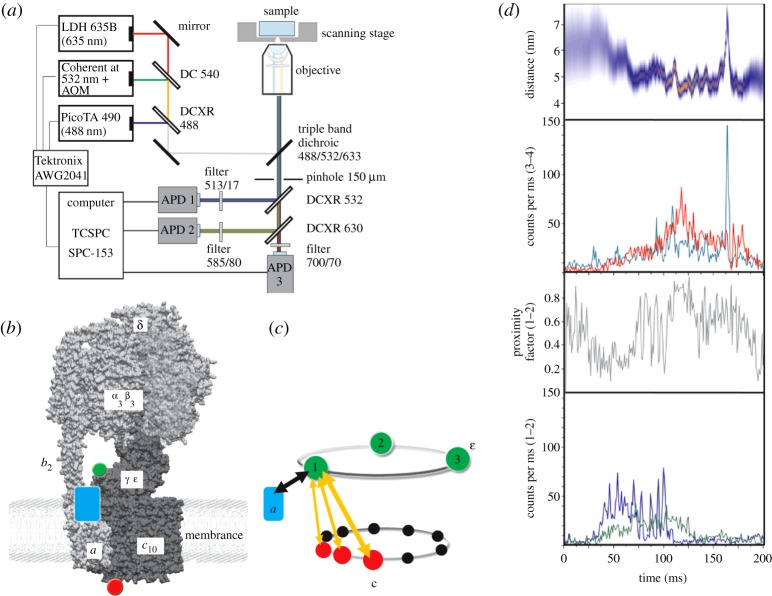
To identify elastic deformations of the rotor subunits without attachment of large beads or filaments and to observe these conformational fluctuations during ATP synthesis, the single-molecule FRET approach with freely diffusing single FOF1-ATP synthases in liposomes was extended recently to a three fluorophore experiment [77,84]. The multi-colour confocal microscope set-up is shown in figure 4a. A triple-mutant of E. coli FOF1-ATP synthase was used comprising the fluorescent protein EGFP fused to the C-terminus of the a subunit as a primary FRET donor, the ε subunit labelled with Alexa532 as secondary FRET donor at residue 56, as well as a Cy5 label as the FRET acceptor on a cysteine introduced at residue 2 on one of the ten c subunits (figure 4b). The aim of this spectroscopic set-up was to correlate the internal deformations of the rotor subunits with an overall rotation of the three subunits γεc10 as a proof of the catalytic activity of the enzyme. Thus, photophysical artefacts on the single-molecule fluorescence level like spectral fluctuations and blinking could be discriminated from a transient twisting of ε versus c subunits. Indications for relative movements between ε and c were found also by Gräber and co-workers [85].
Figure 4.
(a) Experimental set-up for confocal three-colour FRET measurements using three alternating lasers in optimized duty cycle (adapted from [77]). (b) Positions of the three fluorophores on a single FOF1-ATP synthase in a liposome. EGFP (blue rectangle) is the first FRET donor on the static a subunit, which is excited by the 488 nm laser. Alexa532 (green dot) is attached to the rotary subunit ε and acts as FRET acceptor for EGFP by 488 nm excitation, but subsequently becomes the secondary FRET donor when excited with the 532 nm laser pulse. Cy5 (red dot) on one of 10 rotary c subunits is the FRET acceptor for both EGFP and Alexa532. (c) Sketch of the alternating FRET distance measurements in a single enzyme between the static position of EGFP on a and three stopping positions of the rotary ε subunit (green dots with positions 1, 2 and 3). Twisting of the rotor between ε and c is due to different step sizes of both rotary subunits and results in FRET distance changes according to the orange arrows. (d) Photon burst of a single FOF1-ATP synthase comprising two simultaneous FRET measurements in one enzyme. Bottom fluorescence trajectories show FRET donor EGFP (blue) and combined FRET acceptors Alexa532 plus Cy5 (green). Relative intensities changed during observation time and yielded stepwise changes of the proximity factor (shown as grey trace above). The associated FRET trajectory for Alexa532 as FRET donor (cyan) and Cy5 as FRET acceptor (red) was recorded during 532 nm excitation. The distance trajectory (blue/orange) in the top panel indicated stepwise twisting between ε and c as well as elastic distance fluctuations within the second half of this photon burst (figure 4b,c and d adapted from [84]).
Briefly, the cysteine mutant εH56C in F1 was produced in E. coli strain RA1. F1 was prepared and labelled with Alexa532-maleimide. The mean labelling efficiency was determined to be 35 per cent. The double-mutant FOF1-ATP synthase with EGFP on a and a cysteine introduced at residue position 2 of c was purified separately. After substoichiometric labelling of the c-ring of about 7.4 per cent with Cy5-monomaleimide, the enzyme was reconstituted into preformed liposomes, and its unlabelled F1 was exchanged with the Alexa532-labelled F1. Success rates of the F1 exchange procedures were not quantified. Thus, three laser wavelengths (488, 532 and 635 nm) were overlaid in the same confocal excitation volume and a pulse sequence was applied for a duty cycle-optimized alternating laser excitation scheme (DCO-ALEX, figure 4a) to identify the few FOF1-ATP synthases with all three fluorophores present [77,84].
Two simultaneous FRET measurements on each single FOF1-ATP synthase were combined as shown in figure 4b,c. The first distance measurement between EGFP on the static subunit a (blue) and the two markers on the rotor subunits ε (green) and c (red) revealed an active enzyme when stepwise FRET fluctuations were detected as ɛ/c rotated from stop position 1 to stop positions 2 and 3. Subsequently, the second distance measurement across the rotor subunits ε and c, indicated by orange arrows in figure 4c, unravelled the angular twist between these two dyes on the same enzyme. One example of such a photon burst is shown in figure 4d. Pulsed excitation with the 488 nm laser probed the movement of the rotor subunits with respect to the stator. In the lowest panel, relative fluorescence intensities of FRET donor EGFP and the FRET acceptors Alexa532 plus Cy5 changed step wise from low-to-medium-to-high FRET efficiencies, as seen in the corresponding proximity factor trace above (grey trajectory). Exciting the fluorophore Alexa532 on ε with 532 nm now acting as the actual FRET donor showed FRET efficiency changes to the Cy5 acceptor on c by relative intensity changes within this photon burst. The corresponding FRET distance trajectory in the top panel of figure 4d (blue–orange) indicated a large distance change from 6 to 5 nm at the beginning of the photon burst, which was followed by distance fluctuations of about 0.5–1 nm. On the basis of these FRET experiments, we quantitatively analysed the relative movements of ε and c during both ATP synthesis and ATP hydrolysis [86]. We found an extensive twisting in this section of the rotor, encompassing the β-barrel domain of ε and the c-ring, significantly greater than 36° and possibly in the range of up to 120°.
These single-molecule FRET data and the bead-based angular fluctuation measurements confirmed the hypothesis that a substantial fraction of the total rotor torsional elastic energy is stored in the flexible rotor portion, while the stator is very rigid and does not contribute to the high compliance of the FOF1-ATP synthase. The softest portion is probably located at the interface of the two stepping nanomotors, i.e. where it is mechanically needed to operate the enzyme with high kinetic efficiency.
7. Outlook
What did we learn in 15 years from the single-molecule rotation experiments with F1 and FOF1-ATP synthases? First of all, experimental proofs of subunit rotation and discrimination of 120° steps at high [ATP] and sub-steps at low [ATP] in F1 allowed us to refine the model of rotary catalysis. The four chemical steps of ATP hydrolysis, i.e. from binding to bond cleavage to release of products, were attributed not only by their timing but also with respect to the three synchronized catalytic binding sites on the enzyme. Advantages of the direct visualization of rotating beads or filaments attached to surface-bound enzymes were (i) very long observation times of a single molecule with many turnovers and good statistics, and (ii) the possibility to change biochemical conditions and to monitor their effects on the rotary behaviour of the same enzyme. Single-molecule elasticity measurements required both benefits. The angular and the time resolutions for stepsizes of the rotary nanomotors are very high using non-bleaching bright nanoparticles as reporters. Attaching magnetic beads on the rotor ultimately allowed making and measuring ‘man-made ATP’ by forced rotation in small reaction chambers.
Other single-molecule manipulation and imaging methods such as atomic force microscopy [87] or very soft optical tweezers will increase the resolution of the detailed mechanochemical catalytic cycle in this ATPase. Experiments are under way that probe mechanical stimulation of different domains of F1 to correlate directional forces with induced γ subunit rotation. The extraordinary statistics of conformational changes based on a single surface-attached molecule enable advanced theoretical analyses such as fluctuation theorems [88,89].
Single-molecule FRET measurements appear to be complementary to this approach. Direction of rotation has been measured by FRET for both ATP hydrolysis and ATP synthesis. The small dye molecules did not contribute to viscous drag. However, the positions of the cysteine mutations in the FOF1-ATP synthase still had to be controlled to avoid reduced turnover rates. The function of the inhibitor dicyclohexylcarbodiimide (binding to the c subunit and blocking rotation in FO) could be discriminated from the inhibitor aurovertin B, which slowed down rotation during ATP hydrolysis but apparently blocked rotation during ATP synthesis. In contrast to single-molecule FRET measurements of the P-type ATPase KdpFABC, which is a bacterial K+ transporter with a single flexible nucleotide binding domain [90,91], or to the ATP-driven ABC transporter P-glycoprotein [92] with two ATP binding sites, the FOF1-ATP synthase showed a direct correlation of ATP binding and stepwise conformational changes. Because stepped rotation could be completely blocked by inhibitors, the discrimination of thermal fluctuations of the labelled protein domain from transitions between conformations during catalysis was possible. Similar rotary behaviour can be anticipated for the structurally related A-type (including the Thermus thermophilus enzyme [78,93]) and vacuolar V-type ATPases [94,95]. The time resolution for conformational transitions detected by single-molecule FRET with FOF1-ATP synthase reached a sub-millisecond range [72,96] but depended on the signal-to-noise ratio. Therefore, fast transitions are followed more accurately using light-scattering probes [43]. As the single-molecule FRET approach is always limited by the photophysics of the dyes, further improvements are expected by new fluorophores with higher photostability [97,98] or by fluorescent nanocrystals [99].
Monitoring freely diffusing proteoliposomes is limited to less than a second because of the transit time through the laser focus. New non-optical particle trapping devices such as the anti-Brownian electrokinetic trap [100–104] (ABEL trap) invented by Cohen and Moerner can be used to catch and actively push a single FOF1-ATP synthase to a target position for extended confocal FRET measurements [105]. Then, the different numbers of transported protons required to make one ATP molecule in the FOF1-ATP synthases of chloroplasts [3,106], bacteria [3] or mitochondria [107] (as calculated from ensemble measurements) can be compared to the step sizes of the rotary c-rings of the respective single FO motors.
Finally, single-molecule rotation measurements of FOF1-ATP synthases need to be established in living cells [108]. The present in vitro rotation experiments for ATP synthesis required artificially high ΔpH > 4 for the proton motive force. However, the regular ΔpH is much lower in vivo. Therefore, solutions have to be developed regarding how to get the fluorophores attached specifically to a single enzyme in a living cell. The signal-to-background ratio in the presence of cellular autofluorescence still has to be increased. Eventually, the regulation of the bacterial enzyme can be addressed by single-molecule FRET, i.e. how ATP hydrolysis is prevented in living cells under variable external conditions. Understanding the basic mechanochemical details during the catalytic processes of F1 and FOF1 has been achieved by monitoring subunit rotations and elastic deformations in single enzymes. Simultaneously, single-molecule microscopy instrumentation and data analysis tools have been improved, so that we can start studying this ‘splendid molecular machine’ [109] at work in its native environment.
Acknowledgements
The authors want to thank W. Junge, S. D. Dunn, G. D. Glick and P. Gräber for their ongoing support of our biophysical and biochemical work on single FOF1-ATP synthase, and in particular S. Ernst, M. G. Düser and N. Zarrabi for preparing, measuring and analysing the three-colour FRET data. We thank S. Engelbrecht and W. Junge for providing the E. coli homology model and for figure 3d. Parts of this work have been supported financially by the ‘Deutsche Forschungsgemeinschaft’ (grant nos BO 1891/8-1 and BO 1891/10-2 to M.B.).
References
- 1.Rosing J, Slater EC. 1972. The value of G degrees for the hydrolysis of ATP. Biochim. Biophys. Acta 267, 275–290 10.1016/0005-2728(72)90116-8 (doi:10.1016/0005-2728(72)90116-8) [DOI] [PubMed] [Google Scholar]
- 2.Alberty RA. 2003. Thermodynamics of the hydrolysis of adenosine triphosphate as a function of temperature, pH, pMg, and ionic strength. J. Phys. Chem. B 107, 12 324–12 330 10.1021/jp030576l (doi:10.1021/jp030576l) [DOI] [Google Scholar]
- 3.Steigmiller S, Turina P, Graber P. 2008. The thermodynamic H+/ATP ratios of the H+-ATP synthases from chloroplasts and Escherichia coli. Proc. Natl Acad. Sci. USA 105, 3745–3750 10.1073/pnas.0708356105 (doi:10.1073/pnas.0708356105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Junge W, Panke O, Cherepanov DA, Gumbiowski K, Muller M, Engelbrecht S. 2001. Inter-subunit rotation and elastic power transmission in F0F1-ATPase. FEBS Lett. 504, 152–160 10.1016/S0014-5793(01)02745-4 (doi:10.1016/S0014-5793(01)02745-4) [DOI] [PubMed] [Google Scholar]
- 5.Capald RA, Aggeler R. 2002. Mechanism of the F1F0-type ATP synthase, a biological rotary motor. Trends Biochem. Sci. 27, 154–160 10.1016/S0968-0004(01)02051-5 (doi:10.1016/S0968-0004(01)02051-5) [DOI] [PubMed] [Google Scholar]
- 6.von Ballmoos C, Cook GM, Dimroth P. 2008. Unique rotary ATP synthase and its biological diversity. Annu. Rev. Biophys. 37, 43–64 10.1146/annurev.biophys.37.032807.130018 (doi:10.1146/annurev.biophys.37.032807.130018) [DOI] [PubMed] [Google Scholar]
- 7.Gruber G, Wieczorek H, Harvey WR, Muller V. 2001. Structure–function relationships of A-, F- and V-ATPases. J. Exp. Biol. 204, 2597–2605 [DOI] [PubMed] [Google Scholar]
- 8.Muench SP, Trinick J, Harrison MA. 2011. Structural divergence of the rotary ATPases. Q. Rev. Biophys. 44, 311–356 10.1017/S0033583510000338 (doi:10.1017/S0033583510000338) [DOI] [PubMed] [Google Scholar]
- 9.Kane PM. 2006. The where, when, and how of organelle acidification by the yeast vacuolar H+-ATPase. Microbiol. Mol. Biol. Rev. 70, 177–191 10.1128/MMBR.70.1.177-191.2006 (doi:10.1128/MMBR.70.1.177-191.2006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Forgac M. 2007. Vacuolar ATPases: rotary proton pumps in physiology and pathophysiology. Nat. Rev. Mol. Cell. Biol. 8, 917–929 10.1038/nrm2272 (doi:10.1038/nrm2272) [DOI] [PubMed] [Google Scholar]
- 11.Fischer S, Graber P. 1999. Comparison of ΔpH- and Δψ-driven ATP synthesis catalyzed by the H+-ATPases from Escherichia coli or chloroplasts reconstituted into liposomes. FEBS. Lett. 457, 327–332 10.1016/S0014-5793(99)01060-1 (doi:10.1016/S0014-5793(99)01060-1) [DOI] [PubMed] [Google Scholar]
- 12.Mitchell P. 1961. Coupling of phosphorylation to electron and hydrogen transfer by a chemi-osmotic type of mechanism. Nature 191, 144–148 10.1038/191144a0 (doi:10.1038/191144a0) [DOI] [PubMed] [Google Scholar]
- 13.Watt IN, Montgomery MG, Runswick MJ, Leslie AG, Walker JE. 2010. Bioenergetic cost of making an adenosine triphosphate molecule in animal mitochondria. Proc. Natl Acad. Sci. USA 107, 16 823–16 827 10.1073/pnas.1011099107 (doi:10.1073/pnas.1011099107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pogoryelov D, Reichen C, Klyszejko AL, Brunisholz R, Muller DJ, Dimroth P, Meier T. 2007. The oligomeric state of c rings from cyanobacterial F-ATP synthases varies from 13 to 15. J. Bacteriol. 189, 5895–5902 10.1128/JB.00581-07 (doi:10.1128/JB.00581-07) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang W, Hermolin J, Fillingame RH. 2001. The preferred stoichiometry of c subunits in the rotary motor sector of Escherichia coli ATP synthase is 10. Proc. Natl Acad. Sci. USA 98, 4966–4971 10.1073/pnas.081424898 (doi:10.1073/pnas.081424898) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abrahams JP, Leslie AG, Lutter R, Walker JE. 1994. Structure at 2.8 Å resolution of F1-ATPase from bovine heart mitochondria. Nature 370, 621–628 10.1038/370621a0 (doi:10.1038/370621a0) [DOI] [PubMed] [Google Scholar]
- 17.Sielaff H, Rennekamp H, Engelbrecht S, Junge W. 2008. Functional halt positions of rotary FOF1-ATPase correlated with crystal structures. Biophys. J. 95, 4979–4987 10.1529/biophysj.108.139782 (doi:10.1529/biophysj.108.139782) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Okuno D, Fujisawa R, Iino R, Hirono-Hara Y, Imamura H, Noji H. 2008. Correlation between the conformational states of F1-ATPase as determined from its crystal structure and single-molecule rotation. Proc. Natl Acad. Sci. USA 105, 20 722–20 727 10.1073/pnas.0805828106 (doi:10.1073/pnas.0805828106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cingolani G, Duncan TM. 2011. Structure of the ATP synthase catalytic complex (F1) from Escherichia coli in an autoinhibited conformation. Nat. Struct. Mol. Biol. 18, 701–707 10.1038/nsmb.2058 (doi:10.1038/nsmb.2058) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stock D, Leslie AG, Walker JE. 1999. Molecular architecture of the rotary motor in ATP synthase. Science 286, 1700–1705 10.1126/science.286.5445.1700 (doi:10.1126/science.286.5445.1700) [DOI] [PubMed] [Google Scholar]
- 21.Tsunoda SP, Aggeler R, Yoshida M, Capaldi RA. 2001. Rotation of the c subunit oligomer in fully functional F1F0 ATP synthase. Proc. Natl Acad. Sci. USA 98, 898–902 10.1073/pnas.98.3.898 (doi:10.1073/pnas.98.3.898) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang W, Fillingame RH. 1998. Interacting helical faces of subunits a and c in the F1F0 ATP synthase of Escherichia coli defined by disulfide cross-linking. Proc. Natl Acad. Sci. USA 95, 6607–6612 10.1073/pnas.95.12.6607 (doi:10.1073/pnas.95.12.6607) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hakulinen JK, Klyszejko AL, Hoffmann J, Eckhardt-Strelau L, Brutschy B, Vonck J, Meier T. 2012. Structural study on the architecture of the bacterial ATP synthase Fo motor. Proc. Natl Acad. Sci. USA 109, E2050–E2056 10.1073/pnas.1203971109 (doi:10.1073/pnas.1203971109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jones PC, Hermolin J, Jiang W, Fillingame RH. 2000. Insights into the rotary catalytic mechanism of FOF1 ATP synthase from the cross-linking of subunits b and c in the Escherichia coli enzyme. J. Biol. Chem. 275, 31 340–31 346 10.1074/jbc.M003687200 (doi:10.1074/jbc.M003687200) [DOI] [PubMed] [Google Scholar]
- 25.Fillingame RH, Angevine CM, Dmitriev OY. 2002. Coupling proton movements to c-ring rotation in F(1)F(o) ATP synthase: aqueous access channels and helix rotations at the a–c interface. Biochim. Biophys. Acta 1555, 29–36 10.1016/S0005-2728(02)00250-5 (doi:10.1016/S0005-2728(02)00250-5) [DOI] [PubMed] [Google Scholar]
- 26.Del Rizzo PA, Bi Y, Dunn SD. 2006. ATP synthase b subunit dimerization domain: a right-handed coiled coil with offset helices. J. Mol. Biol. 364, 735–746 10.1016/j.jmb.2006.09.028 (doi:10.1016/j.jmb.2006.09.028) [DOI] [PubMed] [Google Scholar]
- 27.Steigmiller S, Borsch M, Graber P, Huber M. 2005. Distances between the b-subunits in the tether domain of F0F1-ATP synthase from E. coli. Biochim. Biophys. Acta 1708, 143–153 10.1016/j.bbabio.2005.03.013 (doi:10.1016/j.bbabio.2005.03.013) [DOI] [PubMed] [Google Scholar]
- 28.Krebstakies T, Zimmermann B, Graber P, Altendorf K, Borsch M, Greie JC. 2005. Both rotor and stator subunits are necessary for efficient binding of F1 to F0 in functionally assembled Escherichia coli ATP synthase. J. Biol. Chem. 280, 33 338–33 345 10.1074/jbc.M506251200 (doi:10.1074/jbc.M506251200) [DOI] [PubMed] [Google Scholar]
- 29.Bottcher B, Bertsche I, Reuter R, Graber P. 2000. Direct visualisation of conformational changes in EF0F1 by electron microscopy. J. Mol. Biol. 296, 449–457 10.1006/jmbi.1999.3435 (doi:10.1006/jmbi.1999.3435) [DOI] [PubMed] [Google Scholar]
- 30.Bowler MW, Montgomery MG, Leslie AG, Walker JE. 2007. Ground state structure of F1-ATPase from bovine heart mitochondria at 1.9 Å resolution. J. Biol. Chem. 282, 14 238–14 242 10.1074/jbc.M700203200 (doi:10.1074/jbc.M700203200) [DOI] [PubMed] [Google Scholar]
- 31.Menz RI, Walker JE, Leslie AG. 2001. Structure of bovine mitochondrial F1-ATPase with nucleotide bound to all three catalytic sites: implications for the mechanism of rotary catalysis. Cell 106, 331–341 10.1016/S0092-8674(01)00452-4 (doi:10.1016/S0092-8674(01)00452-4) [DOI] [PubMed] [Google Scholar]
- 32.Meier T, Polzer P, Diederichs K, Welte W, Dimroth P. 2005. Structure of the rotor ring of F-Type Na+-ATPase from Ilyobacter tartaricus. Science 308, 659–662 10.1126/science.1111199 (doi:10.1126/science.1111199) [DOI] [PubMed] [Google Scholar]
- 33.Humphrey W, Dalke A, Schult K. 1996. VMD: visual molecular dynamics. J. Mol. Graph. 14, 33–38 10.1016/0263-7855(96)00018-5 (doi:10.1016/0263-7855(96)00018-5) [DOI] [PubMed] [Google Scholar]
- 34.Sielaff H. 2007. Der Rotationsmechanismus und die elastische Kopplung der F-ATP-Synthase. PhD thesis, University of Osnabrück, Germany [Google Scholar]
- 35.Boyer PD, Kohlbrenner WE. 1981. The present status of the binding-change mechanism and its relation to the ATP formation by chloroplasts. In Energy coupling in photosynthesis (eds Selman B, Selman-Reiner S.), pp. 231–240 New York, NY: Elsevier [Google Scholar]
- 36.Boyer PD. 1993. The binding change mechanism for ATP synthase—some probabilities and possibilities. Biochim. Biophys. Acta 1140, 215–250 10.1016/0005-2728(93)90063-L (doi:10.1016/0005-2728(93)90063-L) [DOI] [PubMed] [Google Scholar]
- 37.Duncan TM, Bulygin VV, Zhou Y, Hutcheon ML, Cross RL. 1995. Rotation of subunits during catalysis by Escherichia coli F1-ATPase. Proc. Natl Acad. Sci. USA 92, 10 964–10 968 10.1073/pnas.92.24.10964 (doi:10.1073/pnas.92.24.10964) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou Y, Duncan TM, Cross RL. 1997. Subunit rotation in Escherichia coli FOF1-ATP synthase during oxidative phosphorylation. Proc. Natl Acad. Sci. USA 94, 10 583–10 587 10.1073/pnas.94.20.10583 (doi:10.1073/pnas.94.20.10583) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sabbert D, Engelbrecht S, Junge W. 1996. Intersubunit rotation in active F-ATPase. Nature 381, 623–625 10.1038/381623a0 (doi:10.1038/381623a0) [DOI] [PubMed] [Google Scholar]
- 40.Moerner WE, Kador L. 1989. Optical detection and spectroscopy of single molecules in a solid. Phys. Rev. Lett. 62, 2535–2538 10.1103/PhysRevLett.62.2535 (doi:10.1103/PhysRevLett.62.2535) [DOI] [PubMed] [Google Scholar]
- 41.Noji H, Yasuda R, Yoshida M, Kinosita K., Jr 1997. Direct observation of the rotation of F1-ATPase. Nature 386, 299–302 10.1038/386299a0 (doi:10.1038/386299a0) [DOI] [PubMed] [Google Scholar]
- 42.Yasuda R, Noji H, Yoshida M, Kinosita K, Jr, Itoh H. 2001. Resolution of distinct rotational substeps by submillisecond kinetic analysis of F1-ATPase. Nature 410, 898–904 10.1038/35073513 (doi:10.1038/35073513) [DOI] [PubMed] [Google Scholar]
- 43.Spetzler D, York J, Daniel D, Fromme R, Lowry D, Frasch W. 2006. Microsecond time scale rotation measurements of single F1-ATPase molecules. Biochemistry 45, 3117–3124 10.1021/bi052363n (doi:10.1021/bi052363n) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ishmukhametov R, Hornung T, Spetzler D, Frasch WD. 2010. Direct observation of stepped proteolipid ring rotation in E. coli FF-ATP synthase. Embo J. 29, 3911–3923 10.1038/emboj.2010.259 (doi:10.1038/emboj.2010.259) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Spetzler D, Ishmukhametov R, Hornung T, Day LJ, Martin J, Frasch WD. 2009. Single molecule measurements of F1-ATPase reveal an interdependence between the power stroke and the dwell duration. Biochemistry (Mosc). 48, 7979–7985 10.1021/bi9008215 (doi:10.1021/bi9008215) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hirono-Hara Y, Noji H, Nishiura M, Muneyuki E, Hara KY, Yasuda R, Kinosita K, Jr, Yoshida M. 2001. Pause and rotation of F1-ATPase during catalysis. Proc. Natl Acad. Sci. USA 98, 13 649–13 654 10.1073/pnas.241365698 (doi:10.1073/pnas.241365698) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Itoh H, Takahashi A, Adachi K, Noji H, Yasuda R, Yoshida M, Kinosita K. 2004. Mechanically driven ATP synthesis by F1-ATPase. Nature 427, 465–468 10.1038/nature02212 (doi:10.1038/nature02212) [DOI] [PubMed] [Google Scholar]
- 48.Rondelez Y, Tresset G, Nakashima T, Kato-Yamada Y, Fujita H, Takeuchi S, Noji H. 2005. Highly coupled ATP synthesis by F1-ATPase single molecules. Nature 433, 773–777 10.1038/nature03277 (doi:10.1038/nature03277) [DOI] [PubMed] [Google Scholar]
- 49.Wachter A, Bi Y, Dunn SD, Cain BD, Sielaff H, Wintermann F, Engelbrecht S, Junge W. 2011. Two rotary motors in F-ATP synthase are elastically coupled by a flexible rotor and a stiff stator stalk. Proc. Natl Acad. Sci. USA 108, 3924–3929 10.1073/pnas.1011581108 (doi:10.1073/pnas.1011581108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sielaff H, Rennekamp H, Wachter A, Xie H, Hilbers F, Feldbauer K, Dunn SD, Engelbrecht S, Junge W. 2008. Domain compliance and elastic power transmission in rotary F0F1-ATPase. Proc. Natl Acad. Sci. USA 105, 17 760–17 765 10.1073/pnas.0807683105 (doi:10.1073/pnas.0807683105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hasler K, Engelbrecht S, Junge W. 1998. Three-stepped rotation of subunits gamma and epsilon in single molecules of F-ATPase as revealed by polarized, confocal fluorometry. FEBS Lett. 426, 301–304 10.1016/S0014-5793(98)00358-5 (doi:10.1016/S0014-5793(98)00358-5) [DOI] [PubMed] [Google Scholar]
- 52.Adachi K, Yasuda R, Noji H, Itoh H, Harada Y, Yoshida M, Kinosita K., Jr 2000. Stepping rotation of F1-ATPase visualized through angle-resolved single-fluorophore imaging. Proc. Natl Acad. Sci. USA 97, 7243–7247 10.1073/pnas.120174297 (doi:10.1073/pnas.120174297) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kaim G, Prummer M, Sick B, Zumofen G, Renn A, Wild UP, Dimroth P. 2002. Coupled rotation within single FOF1 enzyme complexes during ATP synthesis or hydrolysis. FEBS Lett. 525, 156–163 10.1016/S0014-5793(02)03097-1 (doi:10.1016/S0014-5793(02)03097-1) [DOI] [PubMed] [Google Scholar]
- 54.Sambongi Y, Iko Y, Tanabe M, Omote H, Iwamoto-Kihara A, Ueda I, Yanagida T, Wada Y, Futai M. 1999. Mechanical rotation of the c subunit oligomer in ATP synthase (FOF1): direct observation. Science 286, 1722–1724 10.1126/science.286.5445.1722 (doi:10.1126/science.286.5445.1722) [DOI] [PubMed] [Google Scholar]
- 55.Panke O, Gumbiowski K, Junge W, Engelbrecht S. 2000. F-ATPase: specific observation of the rotating c subunit oligomer of EF0EF1. FEBS Lett. 472, 34–38 10.1016/S0014-5793(00)01436-8 (doi:10.1016/S0014-5793(00)01436-8) [DOI] [PubMed] [Google Scholar]
- 56.Yasuda R, Noji H, Kinosita K, Jr, Yoshida M. 1998. F1-ATPase is a highly efficient molecular motor that rotates with discrete 120 degree steps. Cell 93, 1117–1124 10.1016/S0092-8674(00)81456-7 (doi:10.1016/S0092-8674(00)81456-7) [DOI] [PubMed] [Google Scholar]
- 57.Nishizaka T, Oiwa K, Noji H, Kimura S, Muneyuki E, Yoshida M, Kinosita K., Jr 2004. Chemomechanical coupling in F1-ATPase revealed by simultaneous observation of nucleotide kinetics and rotation. Nat. Struct. Mol. Biol. 11, 142–148 10.1038/nsmb721 (doi:10.1038/nsmb721) [DOI] [PubMed] [Google Scholar]
- 58.Adachi K, Oiwa K, Nishizaka T, Furuike S, Noji H, Itoh H, Yoshida M, Kinosita K., Jr 2007. Coupling of rotation and catalysis in F1-ATPase revealed by single-molecule imaging and manipulation. Cell 130, 309–321 10.1016/j.cell.2007.05.020 (doi:10.1016/j.cell.2007.05.020) [DOI] [PubMed] [Google Scholar]
- 59.Steinbrecher T, Hucke O, Steigmiller S, Borsch M, Labahn A. 2002. Binding affinities and protein ligand complex geometries of nucleotides at the F1 part of the mitochondrial ATP synthase obtained by ligand docking calculations. FEBS Lett. 530, 99–103 10.1016/S0014-5793(02)03433-6 (doi:10.1016/S0014-5793(02)03433-6) [DOI] [PubMed] [Google Scholar]
- 60.Watanabe R, Iino R, Noji H. 2010. Phosphate release in F1-ATPase catalytic cycle follows ADP release. Nat. Chem. Biol. 6, 814–820 10.1038/nchembio.443 (doi:10.1038/nchembio.443) [DOI] [PubMed] [Google Scholar]
- 61.Okuno D, Iino R, Noji H. 2011. Rotation and structure of FOF1-ATP synthase. J. Biochem. 149, 655–664 10.1093/jb/mvr049 (doi:10.1093/jb/mvr049) [DOI] [PubMed] [Google Scholar]
- 62.Nakanishi-Matsui M, Kashiwagi S, Hosokawa H, Cipriano DJ, Dunn SD, Wada Y, Futai M. 2006. Stochastic high-speed rotation of Escherichia coli ATP synthase F1 sector: the epsilon subunit-sensitive rotation. J. Biol. Chem. 281, 4126–4131 10.1074/jbc.M510090200 (doi:10.1074/jbc.M510090200) [DOI] [PubMed] [Google Scholar]
- 63.Nakanishi-Matsui M, Futai M. 2008. Stochastic rotational catalysis of proton pumping F-ATPase. Phil. Trans. R. Soc. B 363, 2135–2142 10.1098/rstb.2008.2266 (doi:10.1098/rstb.2008.2266) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sekiya M, Nakamoto RK, Al-Shawi MK, Nakanishi-Matsui M, Futai M. 2009. Temperature dependence of single molecule rotation of the Escherichia coli ATP synthase F1 sector reveals the importance of gamma-beta subunit interactions in the catalytic dwell. J. Biol. Chem. 284, 22 401–22 410 10.1074/jbc.M109.009019 (doi:10.1074/jbc.M109.009019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Futai M, Nakanishi-Matsui M, Okamoto H, Sekiya M, Nakamoto RK. 2012. Rotational catalysis in proton pumping ATPases: from E. coli F-ATPase to mammalian V-ATPase. Biochim. Biophys. Acta 1817, 1711–1721 10.1016/j.bbabio.2012.03.015 (doi:10.1016/j.bbabio.2012.03.015) [DOI] [PubMed] [Google Scholar]
- 66.Sekiya M, Hosokawa H, Nakanishi-Matsui M, Al-Shawi MK, Nakamoto RK, Futai M. 2011. Single molecule behavior of inhibited and active states of Escherichia coli ATP synthase F1 rotation. J. Biol. Chem. 285, 42 058–42 067 10.1074/jbc.M110.176701 (doi:10.1074/jbc.M110.176701) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Förster T. 1946. Energiewanderung und Fluoreszenz. Naturwissenschaften 33, 166–175 10.1007/BF00585226 (doi:10.1007/BF00585226) [DOI] [Google Scholar]
- 68.Borsch M, Diez M, Zimmermann B, Reuter R, Graber P. 2002. Stepwise rotation of the gamma-subunit of EF0F1-ATP synthase observed by intramolecular single-molecule fluorescence resonance energy transfer. FEBS Lett. 527, 147–152 10.1016/S0014-5793(02)03198-8 (doi:10.1016/S0014-5793(02)03198-8) [DOI] [PubMed] [Google Scholar]
- 69.Borsch M, Diez M, Zimmermann B, Reuter R, Graber P. 2002. Monitoring gamma-subunit movement in reconstituted single EFOF1 ATP synthase by fluorescence resonance energy transfer. In Fluorescence spectroscopy, imaging and probes. New tools in chemical, physical and life sciences (eds Kraayenhof R, Visser AJW, Gerritsen HC.), pp. 197–207 Berlin, Germany: Springer [Google Scholar]
- 70.Diez M, et al. 2004. Proton-powered subunit rotation in single membrane-bound FOF1-ATP synthase. Nat. Struct. Mol. Biol. 11, 135–141 10.1038/nsmb718 (doi:10.1038/nsmb718) [DOI] [PubMed] [Google Scholar]
- 71.Zimmermann B, Diez M, Zarrabi N, Graber P, Borsch M. 2005. Movements of the epsilon-subunit during catalysis and activation in single membrane-bound H+-ATP synthase. Embo. J. 24, 2053–2063 10.1038/sj.emboj.7600682 (doi:10.1038/sj.emboj.7600682) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zimmermann B, Diez M, Borsch M, Graber P. 2006. Subunit movements in membrane-integrated EFOF1 during ATP synthesis detected by single-molecule spectroscopy. Biochim. Biophys. Acta 1757, 311–319 10.1016/j.bbabio.2006.03.020 (doi:10.1016/j.bbabio.2006.03.020) [DOI] [PubMed] [Google Scholar]
- 73.Johnson KM, Swenson L, Opipari AW, Jr, Reuter R, Zarrabi N, Fierke CA, Borsch M, Glick GD. 2009. Mechanistic basis for differential inhibition of the F1F0-ATPase by aurovertin. Biopolymers 91, 830–840 10.1002/bip.21262 (doi:10.1002/bip.21262) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Duser MG, Zarrabi N, Cipriano DJ, Ernst S, Glick GD, Dunn SD, Borsch M. 2009. 36° Step size of proton-driven c-ring rotation in FOF1-ATP synthase. Embo J. 28, 2689–2696 10.1038/emboj.2009.213 (doi:10.1038/emboj.2009.213) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Duser MG, Bi Y, Zarrabi N, Dunn SD, Borsch M. 2008. The proton-translocating a subunit of F0F1-ATP synthase is allocated asymmetrically to the peripheral stalk. J. Biol. Chem. 283, 33 602–33 610 10.1074/jbc.M805170200 (doi:10.1074/jbc.M805170200) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zarrabi N, Duser MG, Ernst S, Reuter R, Glick GD, Dunn SD, Wrachtrup J, Borsch M. 2007. Monitoring the rotary motors of single FOF1-ATP synthase by synchronized multi channel TCSPC. Proc. SPIE 6771, 67710F. 10.1117/12.734301 (doi:10.1117/12.734301) [DOI] [Google Scholar]
- 77.Zarrabi N, Ernst S, Duser MG, Golovina-Leiker A, Becker W, Erdmann R, Dunn SD, Borsch M. 2009. Simultaneous monitoring of the two coupled motors of a single FOF1-ATP synthase by three-color FRET using duty cycle-optimized triple-ALEX. Proc. SPIE 7185, 718 505. 10.1117/12.809610 (doi:10.1117/12.809610) [DOI] [Google Scholar]
- 78.Furuike S, Nakano M, Adachi K, Noji H, Kinosita K, Jr, Yokoyama K. 2011. Resolving stepping rotation in Thermus thermophilus H+-ATPase/synthase with an essentially drag-free probe. Nat. Commun. 2, 233. 10.1038/ncomms1215 (doi:10.1038/ncomms1215) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Junge W, Lill H, Engelbrecht S. 1997. ATP synthase: an electrochemical transducer with rotatory mechanics. Trends Biochem. Sci. 22, 420–423 10.1016/S0968-0004(97)01129-8 (doi:10.1016/S0968-0004(97)01129-8) [DOI] [PubMed] [Google Scholar]
- 80.Cherepanov DA, Mulkidjanian AY, Junge W. 1999. Transient accumulation of elastic energy in proton translocating ATP synthase. FEBS Lett. 449, 1–6 10.1016/S0014-5793(99)00386-5 (doi:10.1016/S0014-5793(99)00386-5) [DOI] [PubMed] [Google Scholar]
- 81.Panke O, Cherepanov DA, Gumbiowski K, Engelbrecht S, Junge W. 2001. Viscoelastic dynamics of actin filaments coupled to rotary F-ATPase: angular torque profile of the enzyme. Biophys. J. 81, 1220–1233 10.1016/S0006-3495(01)75780-3 (doi:10.1016/S0006-3495(01)75780-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Junge W, Sielaff H, Engelbrecht S. 2009. Torque generation and elastic power transmission in the rotary FOF1-ATPase. Nature 459, 364–370 10.1038/nature08145 (doi:10.1038/nature08145) [DOI] [PubMed] [Google Scholar]
- 83.Czub J, Grubmuller H. 2011. Torsional elasticity and energetics of F1-ATPase. Proc. Natl Acad. Sci. USA 108, 7408–7413 10.1073/pnas.1018686108 (doi:10.1073/pnas.1018686108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ernst S, Duser MG, Zarrabi N, Borsch M. 2012. Three-color Förster resonance energy transfer within single FOF1-ATP synthases: monitoring elastic deformations of the rotary double motor in real time. J. Biomed. Opt. 17, 011004. 10.1117/1.JBO.17.1.011004 (doi:10.1117/1.JBO.17.1.011004) [DOI] [PubMed] [Google Scholar]
- 85.Rombach-Riegraf V, Petersen J, Galvez E, Gräber P. 2008. Observation of rotation of subunit c in the membrane integrated EFOF1 by single molecule fluorescence. Biochim. Biophys. Acta– Bioenerg. 1777(Suppl.), S13. 10.1016/j.bbabio.2008.05.057 (doi:10.1016/j.bbabio.2008.05.057) [DOI] [Google Scholar]
- 86.Ernst S, Düser MG, Zarrabi N, Dunn SD, Börsch M. 2012. Elastic deformations of the rotary double motor of single c synthases detected in real time by Förster resonance energy transfer. Biochim. Biophys. Acta– Bioenerg. 1817, 1722–1731 10.1016/j.bbabio.2012.03.034 (doi:10.1016/j.bbabio.2012.03.034) [DOI] [PubMed] [Google Scholar]
- 87.Uchihashi T, Iino R, Ando T, Noji H. 2011. High-speed atomic force microscopy reveals rotary catalysis of rotorless F1-ATPase. Science 333, 755–758 10.1126/science.1205510 (doi:10.1126/science.1205510) [DOI] [PubMed] [Google Scholar]
- 88.Seifert U. 2005. Fluctuation theorem for a single enzyme or molecular motor. Europhys. Lett. 70, 36–41 10.1209/epl/i2005-10003-9 (doi:10.1209/epl/i2005-10003-9) [DOI] [Google Scholar]
- 89.Hayashi K, Ueno H, Iino R, Noji H. 2010. Fluctuation theorem applied to F1-ATPase. Phys. Rev. Lett. 104, 218 103. 10.1103/PhysRevLett.104.218103 (doi:10.1103/PhysRevLett.104.218103) [DOI] [PubMed] [Google Scholar]
- 90.Zarrabi N, Heitkamp T, Greie J-C, Borsch M. 2008. Monitoring the conformational dynamics of a single potassium transporter by ALEX-FRET. Proc. SPIE 6862, 68620M. 10.1117/12.768262 (doi:10.1117/12.768262) [DOI] [Google Scholar]
- 91.Heitkamp T, Kalinowski R, Bottcher B, Borsch M, Altendorf K, Greie JC. 2008. K+-translocating KdpFABC P-type ATPase from Escherichia coli acts as a functional and structural dimer. Biochemistry 47, 3564–3575 10.1021/bi702038e (doi:10.1021/bi702038e) [DOI] [PubMed] [Google Scholar]
- 92.Verhalen B, Ernst S, Borsch M, Wilkens S. 2012. Dynamic ligand induced conformational rearrangements in P-glycoprotein as probed by fluorescence resonance energy transfer spectroscopy. J. Biol. Chem. 287, 1112–1127 10.1074/jbc.M111.301192 (doi:10.1074/jbc.M111.301192) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Imamura H, Nakano M, Noji H, Muneyuki E, Ohkuma S, Yoshida M, Yokoyama K. 2003. Evidence for rotation of V1-ATPase. Proc. Natl Acad. Sci. USA 100, 2312–2315 10.1073/pnas.0436796100 (doi:10.1073/pnas.0436796100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hirata T, Iwamoto-Kihara A, Sun-Wada GH, Okajima T, Wada Y, Futai M. 2003. Subunit rotation of vacuolar-type proton pumping ATPase: relative rotation of the G and C subunits. J. Biol. Chem. 278, 23 714–23 719 10.1074/jbc.M302756200 (doi:10.1074/jbc.M302756200) [DOI] [PubMed] [Google Scholar]
- 95.Diepholz M, Borsch M, Bottcher B. 2008. Structural organization of the V-ATPase and its implications for regulatory assembly and disassembly. Biochem. Soc. Trans. 36, 1027–1031 10.1042/BST0361027 (doi:10.1042/BST0361027) [DOI] [PubMed] [Google Scholar]
- 96.Margittai M, et al. 2003. Single-molecule fluorescence resonance energy transfer reveals a dynamic equilibrium between closed and open conformations of syntaxin 1. Proc. Natl Acad. Sci. USA 100, 15 516–15 521 10.1073/pnas.2331232100 (doi:10.1073/pnas.2331232100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Peneva K, Mihov G, Nolde F, Rocha S, Hotta J, Braeckmans K, Hofkens J, Uji-i H, Herrmann A, Müllen K. 2008. Water-soluble monofunctional perylene and terrylene dyes: powerful labels for single-enzyme tracking. Angew. Chem. Int. Ed. Engl. 47, 3372–3375 10.1002/anie.200705409 (doi:10.1002/anie.200705409) [DOI] [PubMed] [Google Scholar]
- 98.Peneva K, Mihov G, Herrmann A, Zarrabi N, Borsch M, Duncan TM, Mullen K. 2008. Exploiting the nitrilotriacetic acid moiety for biolabeling with ultrastable perylene dyes. J. Am. Chem. Soc. 130, 5398–5399 10.1021/ja711322g (doi:10.1021/ja711322g) [DOI] [PubMed] [Google Scholar]
- 99.Borsch M, Wrachtrup J. 2011. Improving FRET-based monitoring of single chemomechanical rotary motors at work. Chemphyschem 12, 542–553 10.1002/cphc.201000702 (doi:10.1002/cphc.201000702) [DOI] [PubMed] [Google Scholar]
- 100.Cohen AE, Moerner WE. 2005. The anti-Brownian electrophoretic trap (ABEL trap): fabrication and software. Proc. SPIE 5699, 296–305 10.1117/12.598689 (doi:10.1117/12.598689) [DOI] [Google Scholar]
- 101.Cohen AE, Moerner WE. 2006. Suppressing Brownian motion of individual biomolecules in solution. Proc. Natl Acad. Sci. USA 103, 4362–4365 10.1073/pnas.0509976103 (doi:10.1073/pnas.0509976103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Cohen AE, Moerner WE. 2008. Controlling Brownian motion of single protein molecules and single fluorophores in aqueous buffer. Opt. Express. 16, 6941–6956 10.1364/OE.16.006941 (doi:10.1364/OE.16.006941) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Fields AP, Cohen AE. 2011. Electrokinetic trapping at the one nanometer limit. Proc. Natl Acad. Sci. USA 108, 8937–8942 10.1073/pnas.1103554108 (doi:10.1073/pnas.1103554108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Goldsmith RH, Moerner WE. 2010. Watching conformational- and photodynamics of single fluorescent proteins in solution. Nat. Chem. 2, 179–186 10.1038/nchem.545 (doi:10.1038/nchem.545) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Borsch M. 2011. Single-molecule fluorescence resonance energy transfer techniques on rotary ATP synthases. Biol. Chem. 392, 135–142 10.1515/BC.2011.001 (doi:10.1515/BC.2011.001) [DOI] [PubMed] [Google Scholar]
- 106.Turina P, Samoray D, Graber P. 2003. H+/ATP ratio of proton transport-coupled ATP synthesis and hydrolysis catalysed by CF0F1-liposomes. Embo. J. 22, 418–426 10.1093/emboj/cdg073 (doi:10.1093/emboj/cdg073) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Petersen J, Forster K, Turina P, Graber P. 2012. Comparison of the H+/ATP ratios of the H+-ATP synthases from yeast and from chloroplast. Proc. Natl Acad. Sci. USA 109, 11 150–11 155 10.1073/pnas.1202799109 (doi:10.1073/pnas.1202799109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Seyfert K, Oosaka T, Yaginuma H, Ernst S, Noji H, Iino R, Borsch M. 2011. Subunit rotation in a single FOF1-ATP synthase in a living bacterium monitored by FRET. Proc. SPIE 7905, 79050K. 10.1117/12.873066 (doi:10.1117/12.873066) [DOI] [Google Scholar]
- 109.Boyer PD. 1997. The ATP synthase—a splendid molecular machine. Annu. Rev. Biochem. 66, 717–749 10.1146/annurev.biochem.66.1.717 (doi:10.1146/annurev.biochem.66.1.717) [DOI] [PubMed] [Google Scholar]