Abstract
Mutations in superoxide dismutase 1 (SOD1) are found in approximately 20% of patients with familial amyotrophic lateral sclerosis. The propensity of mutant SOD1 to form aggregates in pathologically affected cells (i.e. motor neurons) has implicated these poorly soluble protein aggregates and/or their misfolded soluble precursors as being instrumental to the disease process. We investigated the relative solubility and toxicity of four different mutant SOD1 proteins in a cell-based model system and demonstrate that the mutant, misfolded SOD1 proteins that are the most soluble are also the most toxic. This toxicity was ameliorated by upregulating heat-shock protein chaperones in order to refold the soluble, misfolded protein, regardless of the presence of poorly soluble SOD1. We further demonstrate that increasing the solubility of a SOD1 mutant protein that is both poorly soluble and non-toxic, as compared to other mutant proteins, resulted in remarkably increased toxicity of the mutant SOD1. Again, this increased toxicity was attenuated by upregulating heat-shock protein chaperones in order to refold the soluble, misfolded proteins. These findings implicate easily soluble, misfolded SOD1 as being toxic to the cell and support the hypothesis that reducing solubility of mutant SOD1 proteins through aggregation may occur as a self-protective response in the cell.
Keywords: SOD1, amyotrophic lateral sclerosis, aggregation, solubility, misfold
Introduction
Amyotrophic lateral sclerosis (ALS1) is an invariably fatal neurodegenerative disorder that primarily targets the motor system, affecting 1–2 people per every 100,000 persons worldwide (Logroscino, Traynor, Hardiman, et al., 2007, Worms, 2001). Approximately 10% of ALS cases are familial, and about 20% of these families harbor a disease causing mutation in the gene superoxide dismutase 1 (SOD1) (Chio, Traynor, Lombardo, et al., 2008, Gaudette, Hirano and Siddique, 2000, Rosen, Siddique, Patterson, et al., 1993). SOD1 is a ubiquitously expressed, 153 amino acid homodimeric protein whose primary function is detoxification of oxygen free radicals in the cell (Fridovich, 1986, Mccord and Fridovic.I, 1969). However, mutant SOD1 proteins do not lose their enzymatic function and their role in ALS pathogenesis is related to an, as of yet, undefined toxic gain of function (Borchelt, Lee, Slunt, et al., 1994, Gurney, Pu, Chiu, et al., 1994, Wong, Pardo, Borchelt, et al., 1995). Rodents expressing the mutated human SOD1 transgene reliably mimic disease pathogenesis and are commonly used to model ALS (Gurney, Pu, Chiu, et al., 1994, Wong, Pardo, Borchelt, et al., 1995).
Protein aggregation is a pathological hallmark of many neurodegenerative diseases, including ALS (Ross and Poirier, 2004). SOD1 is a primary component of protein aggregates found in patients carrying SOD1 mutations and in mutant SOD1 rodent disease models. Antibodies specific for misfolded forms of SOD1 have shown that granular SOD1 inclusions can be found in autopsy tissues from sporadic ALS patients, providing a possible mechanistic link between the sporadic and familial forms of disease (Bosco, Morfini, Karabacak, et al., 2010, Forsberg, Andersen, Marklund, et al., 2011, Forsberg, Jonsson, Andersen, et al., 2010). The role of these aggregates in disease pathogenesis is unclear. Past research has focused heavily on aggregates being cytotoxic, but recent evidence implies that the aggregates may actually be innocuous, or even cytoprotective (Bodner, Outeiro, Altmann, et al., 2006, Finkbeiner, Arrasate, Mitra, et al., 2004, Walsh, Klyubin, Fadeeva, et al., 2002). Mutant SOD1 is known to misfold into a disordered tertiary structure, and it may be this soluble, misfolded form of SOD1 that is cytotoxic (Banci, Bertini, Durazo, et al., 2007, Durazo, Shaw, Chattopadhyay, et al., 2009, Shaw, Durazo, Nersissian, et al., 2006). In this paradigm, aggregates form as a compensatory response to sequester away toxic improperly folded soluble protein. Supporting this theory are data from transgenic mutant SOD1 rodents demonstrating that soluble, misfolded SOD1 is enriched in pathologically affected tissues prior to disease onset, whereas SOD1 aggregates increase most markedly after symptom onset (Johnston, Dalton, Gurney, et al., 2000, Jonsson, Ernhill, Andersen, et al., 2004, Wang, Xu and Borchelt, 2002, Zetterstrom, Stewart, Bergemalm, et al., 2007). The relative dearth of SOD1 aggregates pre-symptomatically implies that SOD1 aggregates may not be the primary driver of disease initiation. In contrast, because improperly folded soluble SOD1 is present prior to symptom onset and accumulates in pathologically affected tissue leading up to symptom onset, the culpable, toxic, SOD1 species may be a soluble, non-aggregated form (Wang, Farr, Zeiss, et al., 2009, Zetterstrom, Stewart, Bergemalm, et al., 2007).
We examined four disease-causing human SOD1 (hSOD1) mutants – A4V, G37R, G93A, and G93C- and the wild-type hSOD1 protein, and compared their relative solubility and cytotoxicity. Our data demonstrate that the degree of cellular toxicity consistently correlates with the amount of easily soluble, but misfolded mutant SOD1 protein. Indeed, for each of the mutants, refolding of misfolded soluble mutant protein reduced toxicity, and increasing the level of soluble mutant protein increased toxicity. These data support the hypothesis that a readily soluble, non-aggregated form of mutant SOD1 may be an important mediator of mutant SOD1-mediated toxicity.
Materials and Methods
Cell Culture
Chinese Hamster Ovary (CHO) cells were maintained in Gibco F-12 Hams media supplemented with fetal bovine serum, L-glutamine and penicillin/streptomycin. Cells were transiently transfected at 95% confluency with Lipofectamine 2000 according to manufacturer protocols. In co-transfections, the ratio of wild-type: mutant hSOD1 DNA was 1:1, and the total amount of hSOD1 DNA transfected in co-transfection or mutant only conditions was constant.
Pharmacological administration
Cells were treated with 2.5 μM MG-132 (Calbiochem), 10 μM geldanamcyin (Alomone Labs), or DMSO vehicle 24 hours after transfection. Drug treatment protocols were as follows: 24 hour treatment with MG-132, geldanamycin, or vehicle, 48 hour treatment with MG-132 alone, 48 hour treatment with MG-132 with geldanamycin also being present for the last 24 of those 48 hours, or 48 hour treatment with DMSO vehicle.
Cytotoxicity
Cytotoxicity was evaluated using Invitrogen's Live/Dead Viability/Cytotoxicity Kit for Mammalian Cells (L-3224) according to manufacturer protocols. All viability experiments were repeated three times and a minimum of 100 cells were counted per condition during each trial. Cell counts were obtained using the automated cell counting software Image-Pro Plus 5.1 and were verified by human observation.
Solubility Fractionation
Cells were homogenized through 3 cycles of freeze/thaw on dry ice in phosphate buffered saline in the presence of protease inhibitors (Roche, Mini Complete cocktail tablets) and then centrifuged at 16,000 × g for 30 minutes at 4°C. The supernatant was removed as the “readily soluble” fraction, and the pellet was washed 3 times in PBS prior to suspension in a HEPES-based buffer containing 1.5% NP-40, 2% sodium dodecyl sulfate (SDS), and 0.25% deoxycholic acid and protease inhibitors. The homogenate was sonicated 3 × 5 seconds; the resulting suspension contained the “detergent-soluble” protein fraction. Uniform amounts of total protein were separated through SDS-PAGE on a 4–20% gradient gel and transferred to PVDF membrane through semi-dry transfer. Membranes were blocked for 1 hour in 5% filtered non-fat milk prior to overnight incubation with primary antibody at 4°C (Calbiochem pan-SOD1 sheep polyclonal 1:1000, Sigma-Aldrich β-actin mouse monoclonal, 1:5,000, Millipore ubiquitin mouse monocolonal 1:1,000, or EnzoLife Science Hsp70/Hsp72 mouse monoclonal, 1:100). The next morning, membranes were washed in PBS/tween before 1 hour incubation in near-infrared (IR) dye conjugated secondary antibody (Rockland) at room temperature. The membrane was washed extensively and visualized on an Odyssey (Li-Cor Biosciences) scanner. Optical densitometry (OD) values for hSOD1 bands were obtained with NIH ImageJ software. The percent “readily soluble” protein was defined as: readily soluble hSOD1 OD/ (readily soluble + detergent-soluble OD) *100 for each mutant. All solubility experiments were performed and quantified 3 times.
Proteasome Activity Assay
CHO cells were treated for 24 hours with 2.5 νM MG-132 or DMSO vehicle and proteasome activity was evaluated with Millipore Proteasome Activity Assay (Cat No APT280) according to manufacturer protocols.
Heat Shock
CHO cells were placed in a 42°C incubator for 1 hour (or maintained at 37°C) before being returned to a 37°C incubator overnight for recovery. Cells that were both treated with MG-132 and heat-shocked were treated with 2.5 μM MG-132 directly before heat shock.
Statistical Analysis
For each experimental paradigm, the statistical analysis (GraphPad InStat 3 software) was based on at least 3 independent experiments. For comparisons of wild-type and mutant protein solubility and toxicity, we performed one-way ANOVA with a post-hoc Tukey's test (figures 1B and 1C). The same statistical analysis was used for the pharmacological treatments for each mutant in figures 3, 5, and SI figures 4 and 5. Unpaired t-tests were used to determine differences between drug and vehicle treatments in figures 2 and 4D, between mut-mut and WTFLAG-mut for figures 4B, 4C, and SI figure 7, between heat shock conditions in SI figure 6, and for proteasome activity in SI fig 2.
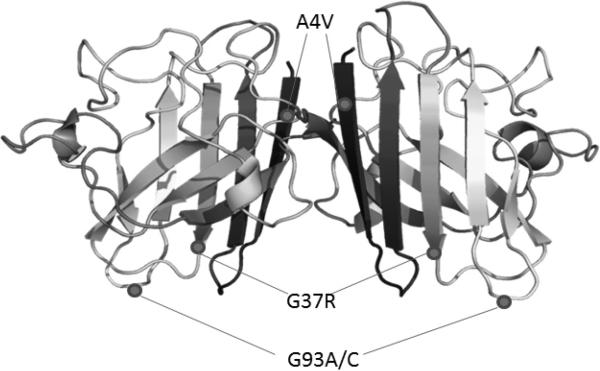
FIGURE 1. Localization of SOD1 mutants in metallated homodimeric structure.
A4V, G37R, G93A, and G93C are identified in the hSOD1 homodimer. Structure modified from Protein Data Bank, PDB 2C9V (Strange, Antonyuk, Hough, et al., 2006).
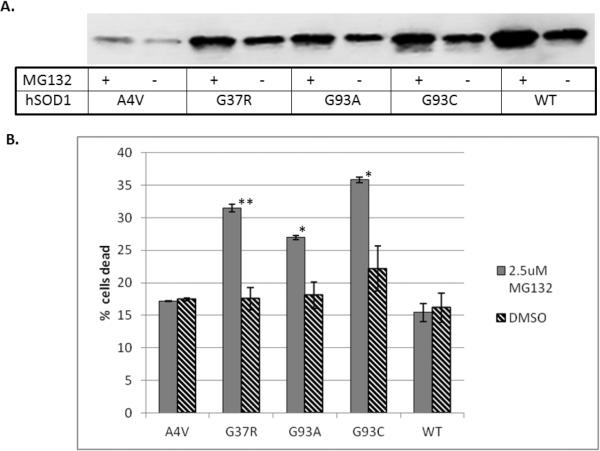
FIGURE 3. SOD1-related toxicity correlates directly with relative solubility.
A) 24 hours after SOD1 transfection, CHO cells were exposed to the proteasome inhibitor MG-132 (2.5 μM) for 24 hours. The homogenates were separated into readily soluble and detergent-soluble fractions and then detected through immunoblot using a pan-SOD1 antibody. The immunoblots demonstrate that the levels of readily soluble protein increase in the presence of MG-132. B) Proteasome inhibition differentially affects SOD1-related toxicity in cells expressing the different hSOD1 mutants. Values represent the means of at least 3 independent experiments, ± SEM. Significance is based on comparison of means between MG132 and DMSO treatments (* = p<0.05, ** = p<0.005).
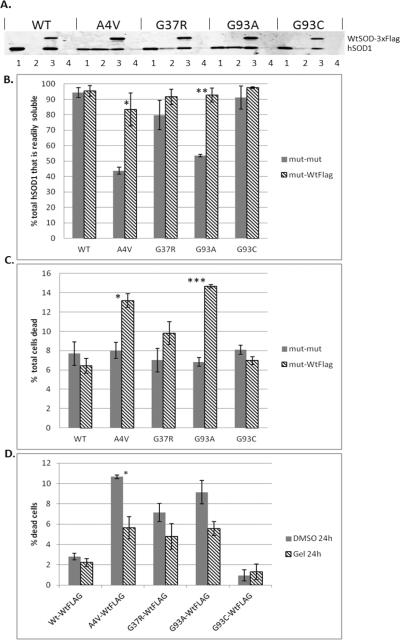
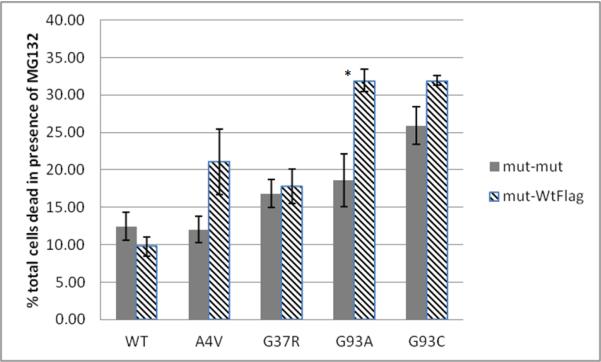
FIGURE 5. Wild-type hSOD1 increases both mutant hSOD1 solubility and toxicity; thus increased toxicity can be rescued by upregulating Hsp70.
A) CHO cells were transiently transfected with equal total amounts of mutant hSOD1 (mut-mut) or mutant and wild-type-3xFLAG (mut-WtFLAG) hSOD1 DNA; 48 hours later protein was separated into readily soluble and detergent-soluble fractions, and detected on immunoblot with a pan-SOD1 antibody. Lane (1) = readily soluble mut-mut, (2) = detergent-soluble mut-mut, (3) = readily soluble mut-WtFLAG, (4) = detergent-soluble mut-WtFLAG; the Wt-3xFLAG protein has a slightly higher molecular weight. B) Bands were quantified using optical densitometry, confirming that co-expression with WT hSOD1 increases the proportion of readily soluble mutant protein for A4V and G93A. C) The increased proportion of readily soluble mutant SOD1 protein corresponds with toxicity. Wild-type hSOD1 induced toxicity in A4V and G93A expressing cells relative to the mut-mut condition. D) Induction of heat shock proteins with geldanamycin (10 μM for 24 hours) protects against toxicity created by co-expressing A4V with WT hSOD1 protein. For B and C, statistical comparisons are mut-WtFLAG vs. the mut-mut condition. For D statistical comparisons are with the DMSO condition for each hSOD1 mutant. Values represent the means of at least 3 independent experiments ± SEM (* = p<0.05, ** = p< 0.001, *** = p<0.0001).
FIGURE 2. Solubility, but not toxicity, differs across non-stressed hSOD1 mutants.
A) CHO cells transiently expressing wild-type (WT), A4V, G37R, G93A, or G93C hSOD1 for 48 hours were separated into readily and detergent-soluble fractions and detected through immunoblot using a pan-SOD1 antibody. β-actin levels demonstrate equal protein loading. NT indicates non-transfected cells. B) Levels of readily and detergent-soluble hSOD1 were quantified through optical densitometry; the percent readily soluble hSOD1 was expressed over the total hSOD1 detected for each mutant. Wild-type and G93C protein were the most soluble, A4V was the least soluble. C) 48 hours following transient transfection, viability was assessed through membrane integrity. Toxicity was expressed as the percentage of dead cells over the total number of cells. The mutants did not differ from each other in terms of toxicity, however all mutants exhibited a weak toxicity in comparison to wild-type hSOD1. Values represent the means ± SEM and significance is relative to the wild-type hSOD1 condition (* = p<0.05, ** = p<0.001).
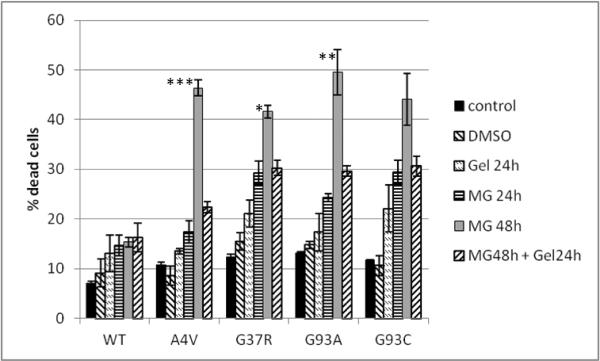
FIGURE 4. Upregulating Hsp70 ameliorates toxicity induced by proteasome inhibition.
Transiently transfected CHO cells were treated at 24 hours post transfection with: DMSO (veh); 2.5 μM MG-132 for 24 hours;10 μM geldanamycin (Gel) for 24 hours; 2.5 μM MG-132 for 48 hours; or 2.5 μM MG-132 for 48 hours with the cells simultaneously being exposed to 10 μM geldanamycin for the last 24 hours. Values represent the means of 3 independent experiments, ± SEM. Statistical analysis was based on comparisons of cell death in the MG48 condition vs. the MG48 + Gel24h condition for each mutant (*=p<0.05, **=p<0.01, ***=p<0.001).
Results
In this study, we present data correlating toxicity and solubility of four different hSOD1 mutants in Chinese Hamster Ovary (CHO) cells. We chose CHO cells both because they are strongly adherent and because they do not endogenously express human SOD1. It is important to use a model system that does not endogenously express hSOD1 in order to ensure dimers between the endogenous and transiently expressed SOD1 do not form. Additionally, rodent and human SOD1 run at slightly different molecular weights and so in this model, endogenous and transiently transfected SOD1 can be differentiated in electrophoretic gels. Past studies indicate that mutant SOD1 pathology is conserved in CHO cells (Prudencio and Borchelt, 2011, Stieber, Gonatas, Moore, et al., 2004). The mutants chosen were selected because they are well-characterized and are scattered throughout the three-dimensional structure of the SOD1 homodimer (figure 1). A4V, the most common SOD1 mutation among ALS patients in North America, is associated with a rapidly progressive form of disease (Rosen, Bowling, Patterson, et al., 1994) and is located at the dimer interface. G37R and G93A hSOD1 mice well-characterized rodent models of disease; G37R is located within the SOD1 β-barrel and G93A is located in the solvent exposed β-barrel plug. The G93C mutation represents a variation on G93A.
Solubility differs among hSOD1 mutants
The mutant and wild-type hSOD1 proteins did not demonstrate uniform solubility. hSOD1 transgenes were transiently expressed in CHO cells for 48 hours prior to harvesting. Similar levels of protein expression were demonstrated for each of the mutant hSOD1 proteins (SI figure 1). Cellular homogenates were separated into two components: 1) a “readily soluble,” fraction harvested without the addition of exogenous solubilizing agents, and 2) a “detergent-soluble” fraction (1.5% NP-40, 2% SDS, and 0.25% deoxycholic acid) that contained the majority of remaining cellular proteins. This strategy was used to isolate the SOD1 protein that is inherently soluble within the cytosol from the SOD1 protein that is less soluble, and potentially contained within some aggregated form. The solubility fractions were subjected to immunoblot analysis, probed with a pan-SOD1 antibody, and the relative amounts of SOD1 in each fraction were quantified through densitometry. For wild-type (WT) and G93C hSOD1, the majority of SOD1 protein was found in the readily soluble fraction, whereas a significant proportion of SOD1 protein was found in the detergent-soluble fraction for A4V, G37R, and G93A (figure 2A, B). The “least soluble” mutant, that is, the mutant that had the greatest proportion of SOD1 in the detergent-soluble fraction, was A4V.
The effect of the expression of each of the mutant proteins on cell viability was evaluated using Invitrogen's cell viability assay, as described under Methods. In this assay, live cells are identified by intracellular esterase activity, and dead cells are detected through loss of plasma membrane integrity. All of the cells expressing mutant SOD1 proteins showed weak toxicity when compared to cells expressing the wild-type hSOD1 protein; however, there were no differences in cellular toxicity among the four mutants (figure 2C).
Increased levels of readily soluble mutant hSOD1 correlates with toxicity
In order to investigate the relative toxicities of mutant hSOD1 proteins in our model, we needed to stress the cells to induce significant levels of cell death. We chose proteasome inhibition as the stressor because the primary function of the proteasome is protein turnover, and misfolded and/or damaged proteins are degraded through the ubiquitin-proteasome pathway (Hershko and Ciechanover, 1982, Hershko and Ciechanover, 1986, Hershko and Ciechanover, 1998, Hershko, Heller, Elias, et al., 1983). Therefore, inhibition of the proteasome is expected to increase levels of both soluble misfolded, and aggregated proteins. Additionally, proteasome activity in the lumbar spinal cord of G93A mice is decreased throughout disease, making this a relevant form of cellular stress (Kabashi, Agar, Taylor, et al., 2004, Kabashi and Durham, 2006). When hSOD1-expressing cells were exposed to the proteasome inhibitor MG-132 for 24 hours, we found a corresponding increase in levels of hSOD1 in both readily soluble (figure 3A) and detergent-soluble fractions. Notably, whereas the amount of hSOD1 present in each of these fractions increased in the presence of MG-132, the relative proportions of hSOD1 found in the different fractions did not differ from the control (DMSO treated) condition. Proteasome inhibition was verified using both a proteasome activity assay and estimation of relative levels of polyubiquitinated proteins (SI figure 2).
As predicted, proteasome inhibition produced significant toxicity in cells expressing the mutant hSOD1 proteins. Importantly, no changes in toxicity were detected in cells expressing wild-type hSOD1. The degree of cellular toxicity varied among the cells expressing the four different mutant proteins (figure 3B), and relative toxicity correlated directly with the relative solubility of each mutant protein (figure 2B). Specifically, the cells expressing G93C, which showed the highest proportion of readily soluble protein, exhibited the greatest increase in toxicity when exposed to MG-132 for 24 hours, whereas cells expressing A4V, the least soluble among the mutant proteins, showed very little increase in toxicity with proteasome inhibition. Cells expressing G37R and G93A showed intermediate toxicities relative to A4V and G93C. The strong correlation between solubility and toxicity across the hSOD1 mutants suggests that the toxic hSOD1 species is found in the readily soluble fraction. These data also support the idea that the soluble, toxic SOD1 protein is likely misfolded, since the proteasome is responsible for turnover and degradation of misfolded proteins and proteasome inhibition increased toxicity in cells expressing mutant, but not wild-type, hSOD1 protein. However, because proteasome inhibition increased levels of both overexpressed wild-type and mutant hSOD1, it was necessary to further verify that the accumulating mutant protein was indeed misfolded.
To test the hypothesis that the toxic, soluble SOD1 proteins were misfolded, we sought to promote their refolding, predicting that this would reduce toxicity. To refold the soluble, misfolded SOD1 protein we induced transcription of heat shock chaperones, including heat shock protein 70 (Hsp70). Hsp70 is instrumental to protein quality control and is known to act in concert with other heat shock co-chaperones to assist in nascent protein folding, as well as refolding of soluble misfolded proteins, and targeting of proteins for degradation (Batulan, Shinder, Minotti, et al., 2003, Batulan, Taylor, Aarons, et al., 2006, Beckmann, Mizzen and Welch, 1990, Bukau, Weissman and Horwich, 2006, Meacham, Patterson, Zhang, et al., 2001, Rudiger, Buchberger and Bukau, 1997). Previous studies of aggregate-related toxicity could not distinguish between the effects of SOD1 aggregates and their misfolded, readily soluble precursors because aggregates are formed from these precursors and so reducing levels of misfolded, soluble protein would necessarily also reduce the presence of aggregates. To circumvent this issue, we designed an experiment where we could separate the toxic effects of readily soluble versus detergent-soluble SOD1 protein. We used proteasome inhibition to increase levels of both pools and then selectively refolded the soluble, misfolded fraction by upregulating heat shock chaperones. Because aggregate formation mediated by proteasome inhibition is a reversible process (Puttaparthi, Wojcik, Rajendran, et al., 2003), we maintained proteasome inhibition throughout the experiment. This paradigm allowed us to observe the toxic effects of the detergent-soluble SOD1 in the absence of the misfolded, readily soluble precursors.
Heat shock chaperones were induced pharmacologically with the Hsp90 inhibitor geldanamycin, and increased Hsp70 levels were confirmed by immunoblot (SI figure 3). In the absence of cell stress, Hsp90 complexes with heat shock transcription factor 1 (Hsf-1) and holds it in an inactive state, thereby preventing induction of Hsp70 and other co-chaperones. Inhibiting Hsp90 with geldanamycin releases Hsf-1 from this complex, allowing Hsf-1 to induce transcription of heat shock chaperones, including Hsp70 (Shen, He, Wang, et al., 2005). So, in this paradigm, cells are first stressed by exposure to the proteasome inhibitor MG-132 for 24 hours to induce accumulation of misfolded proteins, and then are exposed to both MG-132 and geldanamycin for the next 24 hours in order to up-regulate heat shock chaperones to specifically target refolding of the misfolded, readily soluble protein, while maintaining levels of the detergent-soluble protein. There are reports that the heat shock chaperone Hsp70 may play a role in disaggregating already formed inclusions (Liberek, Lewandowska and Zietkiewicz, 2008). However, we found that up-regulating Hsp70 in the presence of an inhibited proteasome did not decrease levels of already accumulated “less soluble” protein in our system (SI figure 4).
Using this experimental paradigm, we found that up-regulating heat shock chaperones with geldanamycin reduced the cellular toxicity initiated by proteasome inhibition (figure 4). As with our previous experiments using proteasome inhibition, the mutants exhibited different levels of toxicity in the presence of MG-132 at 24 hours, and this toxicity increased with continued exposure to MG-132 for the next 24 hours. However, when cells first were exposed to MG-132 for 24 hours and then subsequently exposed to both MG-132 and geldanamcyin for the next 24 hours, the increased cellular toxicity of MG-132 at 48 hours was ameliorated. Similarly, when cells were exposed to both MG-132 and geldanamycin from the outset, geldanamycin prevented the MG-132-mediated toxicity (SI figure 5). In order to confirm the protective effect of heat shock chaperones in this model, we also induced expression of heat shock proteins using heat shock (42°C × 1 hour). This paradigm yielded the same results as treatment with geldanamycin (SI figure 6). The convergence of geldanamycin treatment and heat shock in both up-regulating heat shock chaperones and ameliorating cell death supports the hypothesis that these induced chaperones are responsible for attenuating MG-132-induced toxicity.
The protective effect of up-regulating heat shock chaperones was not due to a decrease in mutant SOD1. Solubility profiles showed that the relative levels of readily soluble and detergent-soluble proteins were conserved with pharmacologic manipulation (SI figure 4). This suggests that the attenuation of toxicity with geldanamycin was not due to an overall reduction in levels of detergent-soluble protein or a transfer of the detergent-soluble protein to the readily soluble fraction. Indeed, inducing heat shock chaperones in mutant hSOD1-expressing cells attenuated MG-132-induced toxicity regardless of the presence of detergent-soluble SOD1, supporting the hypothesis that the toxic SOD1 species is not only misfolded, but is also very soluble. These data may seem to contradict the hypothesis that levels of soluble SOD1 correlate with toxicity. However, we interpret the data as showing that it is the SOD1 conformer within the soluble fraction that determines toxicity, and not simply levels of soluble SOD1. That is, the toxic SOD1 protein species is both readily soluble and misfolded. Refolding of the toxic misfolded protein with heat shock chaperones reduced toxicity, even though the total level of soluble SOD1 did not change.
Increasing mutant SOD1 solubility increases cellular toxicity
In the experiments up to this point, we had increased total levels of both readily soluble and detergent-soluble misfolded SOD1, showing a corresponding increase in toxicity in the mutants demonstrating the highest proportion of readily soluble protein. Targeted refolding of readily soluble misfolded protein by inducing heat shock chaperones ameliorated this toxicity, suggesting that the toxic form of the mutant SOD1 protein is both easily soluble and misfolded. To more directly address the question of how mutant SOD1 solubility impacts toxicity, we needed to increase the inherent solubility of the individual SOD1 mutants to show that specifically increasing the proportion of mutant hSOD1 in the readily soluble fraction would also increase its toxicity. Past studies demonstrated that co-expressing mutant and wild-type hSOD1 increases mutant SOD1 solubility (Witan, Gorlovoy, Kaya, et al., 2009, Witan, Kern, Koziollek-Drechsler, et al., 2008). Therefore, we transfected either equal amounts of wild-type and mutant hSOD1 DNA, or transfected double the amount of mutant hSOD1 DNA. In this manner, the total amount of hSOD1 DNA transfected was uniform regardless of whether the cells were expressing mutant only, or mutant plus wild-type protein. To differentiate the co-expressed wild-type and mutant hSOD1 proteins, an N-terminal 3X-FLAG tag was added to the wild-type hSOD1.
As predicted, the co-expression of wild-type hSOD1-FLAG and mutant hSOD1 increased the relative solubility of the mutant protein (figure 5A, B). The relative solubility of mutant hSOD1 was evaluated independently for the mut-mut and mut-WTFLAG conditions. The greatest increase in mutant SOD1 solubility in the presence of wild-type hSOD1, as compared to the mutant-mutant condition, was found in cells expressing A4V and G93A proteins. These were also the mutants that were the least soluble in the absence of wild-type protein (figure 2) and so had the greatest potential for increased solubility. We next evaluated the effect of co-expressing wild-type hSOD1 on toxicity; the presence of wild-type protein increased toxicity in cells expressing A4V and G93A hSOD1 (figure 5C). The greatest increase in toxicity as compared to the mutant-mutant condition was found in A4V and G93A hSOD1-expressing cells. Notably, these are the same mutants that showed the greatest increase in solubility in the presence of the wild-type protein.
The increase in toxicity caused by co-expression of wild-type SOD1 was rescued by treatment with geldanamycin (figure 5D), again suggesting that the increased soluble fraction of mutSOD1 contains misfolded, toxic protein. Heat-shocking the cells at 42°C for one hour similarly rescued viability, supporting the hypothesis that geldanamycin mediates viability via the heat shock pathway (SI figure 7). Additionally, increased solubility due to co-expression of wild-type and mutant hSOD1 also increased the SOD1-related toxicity in cells exposed to proteasome inhibition (figure 6). Again, A4V and G93A, the SOD1 mutants that exhibited the greatest increase in solubility when co-expressed with the wild-type protein, showed the greatest sensitivity to MG-132 when co-expressed with wild-type hSOD1. Notably, A4V-expressing cells were resistant to MG-132-induced toxicity in the absence of wild-type protein (figure 3), and the co-expression of wild-type hSOD1 with A4V transformed a comparatively non-toxic mutant into a toxic one, presumably by increasing its solubility.
FIGURE 6. Presence of wild-type hSOD1 increases mutant hSOD1 vulnerability to proteasome inhibition.
CHO cells were transiently transfected with equal total levels of mutant (mut-mut) hSOD1 or mutant and wild-type-3×FLAG (mut-WtFLAG) hSOD1 DNA and allowed to express for 24 hours before being exposed to 2.5 μM MG-132 for 24 hours. Viability was assessed via membrane integrity. Values represent the means of at least 3 independent experiments ± SEM. Statistical comparisons are mut-WtFLAG vs. the mut-mut condition (* = p< 0.05).
Discussion
We present data correlating the toxicity of mutant hSOD1 proteins, under a number of different conditions, with their relative solubilities. We demonstrate that under conditions of stress induced by proteasome inhibition, the most easily solubilized mutant proteins are also the most toxic, and that targeted re-folding of improperly folded, soluble mutant protein ameliorates this toxicity, even in the presence of less soluble SOD1 species. Furthermore, we demonstrate that increasing mutant hSOD1 solubility by co-expressing wild-type hSOD1 increases cell toxicity, as well as sensitivity to the effects of proteasome inhibition. The mutants demonstrating the greatest increase in solubility when co-expressed with wild-type hSOD1 also demonstrated the greatest increase in toxicity. This toxicity was rescued by inducing molecular heat shock chaperones, suggesting that the toxicity of mutant hSOD1 is related to levels of readily soluble misfolded hSOD1. It may be that the less soluble hSOD1 serves a protective function by sequestering away the more soluble toxic species (figure 7). This less soluble fraction of SOD1 may include protein aggregates, though our experimental data do not address this issue. However, there is precedent for the hypothesis of protective sequestering of toxic proteins in other neurodegenerative diseases. Examples include models of Huntington's and Parkinson's disease, where pharmacologically enhancing aggregate size (and correspondingly decreasing levels of diffuse, soluble protein) increases the viability of mutant huntingtin- and synuclein-expressing cells (Bodner, Outeiro, Altmann, et al., 2006, Finkbeiner, Arrasate, Mitra, et al., 2004). Conformation-specific SOD1 antibodies are available, however we chose not to use them because a) many of them are not thoroughly characterized, and b) separate mutants may not exhibit uniform misfolded conformations, decreasing the usefulness of such an approach.
FIGURE 7. Misfolded and soluble SOD1 is more toxic than aggregated SOD1.
Mutant SOD1 is initially present as soluble, natively folded (circles) and soluble, misfolded (hexagons) protein. Proteasome inhibition slows protein turnover, increasing levels of soluble, misfolded and poorly soluble, potentially aggregated SOD1 (hexagon conglomerates) and is associated with increased cell toxicity. Up-regulating heat shock protein chaperones while maintaining proteasome inhibition targets soluble, misfolded protein for re-folding but does not decrease levels of poorly soluble, aggregated SOD1 and is associated with low levels of cell toxicity. The presence of poorly soluble, aggregated SOD1 alone is not enough to initiate toxicity; in contrast, misfolded, soluble SOD1 must be present for cell toxicity.
Our findings are consistent with previous data suggesting that the toxic hSOD1 species is localized in the soluble protein fraction (Wang, Farr, Zeiss, et al., 2009, Zetterstrom, Stewart, Bergemalm, et al., 2007). There is debate over when protein aggregates first appear in rodent models of disease, however there is a general consensus that their presence drastically increases after the animals are already symptomatic (Johnston, Dalton, Gurney, et al., 2000, Wang, Xu and Borchelt, 2002). As detectable symptom onset is actually preceded by tissue pathology (e.g. denervation of the neuromuscular junction), factors other than protein aggregation are likely instrumental to disease initiation (Fischer, Culver, Tennant, et al., 2004). Indeed, both hydrophobic, soluble misfolded mutant hSOD1, and soluble mutant hSOD1 oligomers are present in pathologically affected tissue prior to symptom onset (Wang, Farr, Zeiss, et al., 2009, Zetterstrom, Stewart, Bergemalm, et al., 2007). Furthermore, these protein species are selectively increased in pathologically affected tissue leading up to disease onset, supporting a role for soluble misfolded protein in disease initiation (Wang, Farr, Zeiss, et al., 2009, Zetterstrom, Stewart, Bergemalm, et al., 2007).
A study on mutant SOD1 aggregation and toxicity utilizing live-cell imaging in PC-12 cells showed that those cells expressing mutant SOD1 aggregates were more likely to die than cells exhibiting a diffuse form of the protein (Matsumoto, Stojanovic, Holmberg, et al., 2005). However, in this study toxicity was also observed in cells lacking aggregates, suggesting that aggregation is not necessary for cell death. A separate study using relative fluorescence of GFP-tagged SOD1 as a proxy for proper SOD1 folding showed that those mutants with the most disrupted folding were also the most sensitive to oxidative stress (Zhang and Zhu, 2006). There is also evidence that increasing the presence of molecular chaperones ameliorates mutant SOD1 toxicity both in vitro and in vivo (Batulan, Taylor, Aarons, et al., 2006, Bruening, Roy, Giasson, et al., 1999, Kalmar, Novoselov, Gray, et al., 2008, Kieran, Kalmar, Dick, et al., 2004). However, these previous experiments could not differentiate the effects on viability of aggregates versus soluble aggregate precursors. Our experimental paradigm provides data showing the differential effects of separate SOD1 solubility states, providing further insight into the nature of the mutant SOD1 protein that is responsible for cytotoxicity.
The observed effect of wild-type hSOD1 on both mutant hSOD1 solubility and toxicity is consistent with that reported in other studies (Borchelt, Prudencio, Durazo, et al., 2009, Prudencio and Borchelt, 2009, Prudencio, Durazo, Whitelegge, et al., 2010, Witan, Gorlovoy, Kaya, et al., 2009, Witan, Kern, Koziollek-Drechsler, et al., 2008). Both solubility and toxicity of mutant hSOD1 was enhanced by the presence of wild-type protein. Ours is the first study showing that the increased solubility generated by co-expression of wild-type hSOD1 correlates with the relative increase in toxicity and that the enhanced toxicity can be attenuated by refolding the soluble misfolded protein through up-regulating heat shock chaperones. It may be that the wild-type protein somehow stabilizes soluble mutant protein, thereby increasing its toxic effect on the cell, and that inducing heat shock chaperones rescues this effect by stimulating re-folding of the stabilized, misfolded mutant protein. The mechanism through which wild-type hSOD1 interacts with the mutant protein is still under debate; separation of tagged SOD1 species through electrophoretic gels has suggested that co-expressed wild-type and mutant hSOD1 form a heterodimer (Witan, Gorlovoy, Kaya, et al., 2009, Witan, Kern, Koziollek-Drechsler, et al., 2008), yet immunocytochemical studies showed that these species do not intimately interact (Prudencio, Durazo, Whitelegge, et al., 2010). Even so, the presence of wild-type hSOD1 exacerbates mutant hSOD1-induced toxicity both in vitro and in rodent models, and has even been shown to initiate disease in otherwise unaffected A4V-expressing mice (Deng, Shi, Furukawa, et al., 2006, Prudencio, Durazo, Whitelegge, et al., 2010, Wang, Deng, Grisotti, et al., 2009). Our findings showing enhanced solubility and toxicity of A4V in the presence of wild-type hSOD1 may provide insight into the A4V paradox. It is well-established that humans possessing the A4V mutation have one of the most rapidly progressing forms of ALS, whereas mice expressing A4V do not develop disease unless it is expressed in the presence of wild-type hSOD1. Similarly in our studies, A4V-expressing cells were the least toxic in the context of proteasome inhibition, yet when co-expressed with wild-type hSOD1, A4V itself became toxic to the cell and the toxicity was further increased in the presence of a proteasome inhibitor. It thus appears that the presence of wild-type hSOD1 enhances mutant hSOD1 toxicity, potentially by stabilizing a soluble, misfolded form of the protein and/or inhibiting sequestration of toxic protein into aggregates.
In conclusion, these data add to the growing literature implicating soluble, misfolded protein as the most cytotoxic protein species in various neurodegenerative diseases. We demonstrate that the toxicity and solubility of different SOD1 mutants are correlated in vitro, with the most soluble mutants also being the most toxic, and that this toxicity is attenuated by targeting improperly folded soluble protein for refolding, independent of the presence of less soluble SOD1 species. We further demonstrate that increasing mutant SOD1 solubility both induces toxicity and increases sensitivity to proteasome inhibition in vitro, and that this can be tempered through up-regulating molecular heat shock chaperones. These results support the hypothesis that toxic mutant hSOD1 is localized to the soluble fraction, and that a reduction in solubility may function as a protective, compensatory mechanism to sequester away this protein species. Future studies focusing on improperly folded soluble hSOD1 may shed light on the pathogenesis of ALS and other protein misfolding disorders.
Supplementary Material
Highlights
Four separate SOD1 mutant proteins are evaluated for correlation between solubility and toxicity
The most soluble SOD1 mutants examined were also the most toxic with proteasome inhibition
Stimulating refolding of soluble misfolded proteins with protein chaperones decreased toxicity
The presence of aggregated proteins was not toxic in the absence of soluble misfolded precursors
Increasing solubility of mutant, misfolded SOD1 increased toxicity
Acknowledgements
We would like to thank Dr. David R Borchelt (Department of Neuroscience and McKnight Brain Institute, University of Florida) for providing the hSOD1 plasmids and Dr. Lary Walker (Department of Neurology, Emory University) and Dr. Rick Kahn (Department of Biochemistry, Emory University) for helpful feedback and discussion. We are grateful to the patients, families, and staff of the Emory ALS Center for their continued efforts to make a difference in ALS care and research.
This work was supported by Emory Neuroscience NINDS Core Facilities (P30 NS055077), and the Packard Center for ALS Research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
There are no real or perceived conflicts of interest in this work.
The abbreviations used are: ALS, amyotrophic lateral sclerosis; hSOD, human superoxide dismutase 1; Hsp70, heat shock protein 70; CHO cells: Chinese Hamster Ovary cells
References
- Banci L, Bertini I, Durazo A, Girotto S, Gralla EB, Martinelli M, et al. Metal-free superoxide dismutase forms soluble oligomers under physiological conditions: A possible general mechanism for familial ALS. P Natl Acad Sci USA. 2007;104:11263–11267. doi: 10.1073/pnas.0704307104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batulan Z, Shinder GA, Minotti S, He BP, Doroudchi M, Nalbantoglu J, et al. High threshold for induction of the stress response in motor neurons is associated with failure to activate HSF1. J Neurosci. 2003;23:5789–5798. doi: 10.1523/JNEUROSCI.23-13-05789.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batulan Z, Taylor DM, Aarons RJ, Minotti S, Doroudchi MM, Nalbantoglu J, et al. Induction of multiple heat shock proteins and neuroprotection in a primary culture model of familial amyotrophic lateral sclerosis. Neurobiol Dis. 2006;24:213–225. doi: 10.1016/j.nbd.2006.06.017. [DOI] [PubMed] [Google Scholar]
- Beckmann RP, Mizzen LA, Welch WJ. Interaction of Hsp-70 with Newly Synthesized Proteins - Implications for Protein Folding and Assembly. Science. 1990;248:850–854. doi: 10.1126/science.2188360. [DOI] [PubMed] [Google Scholar]
- Bodner RA, Outeiro TF, Altmann S, Maxwell MM, Cho SH, Hyman BT, et al. Pharmacological promotion of inclusion formation: A therapeutic approach for Huntington's and Parkinson's diseases. P Natl Acad Sci USA. 2006;103:4246–4251. doi: 10.1073/pnas.0511256103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borchelt DR, Lee MK, Slunt HS, Guarnieri M, Xu ZS, Wong PC, et al. Superoxide-Dismutase-1 with Mutations Linked to Familial Amyotrophic-Lateral-Sclerosis Possesses Significant Activity. P Natl Acad Sci USA. 1994;91:8292–8296. doi: 10.1073/pnas.91.17.8292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borchelt DR, Prudencio M, Durazo A, Whitelegge JP. Modulation of mutant superoxide dismutase 1 aggregation by co-expression of wild-type enzyme. J Neurochem. 2009;108:1009–1018. doi: 10.1111/j.1471-4159.2008.05839.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosco DA, Morfini G, Karabacak NM, Song YY, Gros-Louis F, Pasinelli P, et al. Wild-type and mutant SOD1 share an aberrant conformation and a common pathogenic pathway in ALS. Nat Neurosci. 2010;13:1396–U1133. doi: 10.1038/nn.2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruening W, Roy J, Giasson B, Figlewicz DA, Mushynski WE, Durham HD. Up-regulation of protein chaperones preserves viability of cells expressing toxic Cu/Zn-superoxide dismutase mutants associated with amyotrophic lateral sclerosis. J Neurochem. 1999;72:693–699. doi: 10.1046/j.1471-4159.1999.0720693.x. [DOI] [PubMed] [Google Scholar]
- Bukau B, Weissman J, Horwich A. Molecular chaperones and protein quality control. Cell. 2006;125:443–451. doi: 10.1016/j.cell.2006.04.014. [DOI] [PubMed] [Google Scholar]
- Chio A, Traynor BJ, Lombardo F, Fimognari M, Calvo A, Ghiglione P, et al. Prevalence of SOD1 mutations in the Italian ALS population. Neurology. 2008;70:533–537. doi: 10.1212/01.wnl.0000299187.90432.3f. [DOI] [PubMed] [Google Scholar]
- Deng HX, Shi Y, Furukawa Y, Zhai H, Fu RG, Liu ED, et al. Conversion to the amyotrophic lateral sclerosis phenotype is associated with intermolecular linked insoluble aggregates of SOD1 in mitochondria. P Natl Acad Sci USA. 2006;103:7142–7147. doi: 10.1073/pnas.0602046103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durazo A, Shaw BF, Chattopadhyay M, Faull KF, Nersissian AM, Valentine JS, et al. Metal-free Superoxide Dismutase-1 and Three Different Amyotrophic Lateral Sclerosis Variants Share a Similar Partially Unfolded beta-Barrel at Physiological Temperature. J Biol Chem. 2009;284:34382–34389. doi: 10.1074/jbc.M109.052076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkbeiner S, Arrasate M, Mitra S, Schweitzer ES, Segal MR. Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature. 2004;431:805–810. doi: 10.1038/nature02998. [DOI] [PubMed] [Google Scholar]
- Fischer LR, Culver DG, Tennant P, Davis AA, Wang MS, Castellano-Sanchez A, et al. Amyotrophic lateral sclerosis is a distal axonopathy: evidence in mice and man. Exp Neurol. 2004;185:232–240. doi: 10.1016/j.expneurol.2003.10.004. [DOI] [PubMed] [Google Scholar]
- Forsberg K, Andersen PM, Marklund SL, Brannstrom T. Glial nuclear aggregates of superoxide dismutase-1 are regularly present in patients with amyotrophic lateral sclerosis. Acta Neuropathol. 2011;121:623–634. doi: 10.1007/s00401-011-0805-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsberg K, Jonsson PA, Andersen PM, Bergemalm D, Graffmo KS, Hultdin M, et al. Novel Antibodies Reveal Inclusions Containing Non-Native SOD1 in Sporadic ALS Patients. Plos One. 2010;5 doi: 10.1371/journal.pone.0011552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridovich I. Superoxide Dismutases. Adv Enzymol Ramb. 1986;58:61–97. doi: 10.1002/9780470123041.ch2. [DOI] [PubMed] [Google Scholar]
- Gaudette M, Hirano M, Siddique T. Current status of SOD1 mutations in familial amyotrophic lateral sclerosis. Amyotroph Lateral Scler Other Motor Neuron Disord. 2000;1:83–89. doi: 10.1080/14660820050515377. [DOI] [PubMed] [Google Scholar]
- Gurney ME, Pu HF, Chiu AY, Dalcanto MC, Polchow CY, Alexander DD, et al. Motor-Neuron Degeneration in Mice That Express a Human Cu,Zn Superoxide-Dismutase Mutation. Science. 1994;264:1772–1775. doi: 10.1126/science.8209258. [DOI] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A. Mechanisms of Intracellular Protein Breakdown. Annu Rev Biochem. 1982;51:335–364. doi: 10.1146/annurev.bi.51.070182.002003. [DOI] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A. The Ubiquitin Pathway for the Degradation of Intracellular Proteins. Prog Nucleic Acid Re. 1986;33:19–56. doi: 10.1016/s0079-6603(08)60019-7. [DOI] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- Hershko A, Heller H, Elias S, Ciechanover A. Components of Ubiquitin-Protein Ligase System - Resolution, Affinity Purification, and Role in Protein Breakdown. J Biol Chem. 1983;258:8206–8214. [PubMed] [Google Scholar]
- Johnston JA, Dalton MJ, Gurney ME, Kopito RR. Formation of high molecular weight complexes of mutant Cu,Zn-superoxide dismutase in a mouse model for familial amyotrophic lateral sclerosis. P Natl Acad Sci USA. 2000;97:12571–12576. doi: 10.1073/pnas.220417997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonsson PA, Ernhill K, Andersen PM, Bergemalm D, Brannstrom T, Gredal O, et al. Minute quantities of misfolded mutant superoxide dismutase-1 cause amyotrophic lateral sclerosis. Brain. 2004;127:73–88. doi: 10.1093/brain/awh005. [DOI] [PubMed] [Google Scholar]
- Kabashi E, Agar JN, Taylor DM, Minotti S, Durham HD. Focal dysfunction of the proteasome: a pathogenic factor in a mouse model of amyotrophic lateral sclerosis. Journal of Neurochemistry. 2004;89:1325–1335. doi: 10.1111/j.1471-4159.2004.02453.x. [DOI] [PubMed] [Google Scholar]
- Kabashi E, Durham HD. Failure of protein quality control in amyotrophic lateral sclerosis. Biochim Biophys Acta. 2006;1762:1038–1050. doi: 10.1016/j.bbadis.2006.06.006. [DOI] [PubMed] [Google Scholar]
- Kalmar B, Novoselov S, Gray A, Cheetham ME, Margulis B, Greensmith L. Late stage treatment with arimoclomol delays disease progression and prevents protein aggregation in the SOD1(G93A) mouse model of ALS. J Neurochem. 2008;107:339–350. doi: 10.1111/j.1471-4159.2008.05595.x. [DOI] [PubMed] [Google Scholar]
- Kieran D, Kalmar B, Dick JR, Riddoch-Contreras J, Burnstock G, Greensmith L. Treatment with arimoclomol, a coinducer of heat shock proteins, delays disease progression in ALS mice. Nat Med. 2004;10:402–405. doi: 10.1038/nm1021. [DOI] [PubMed] [Google Scholar]
- Liberek K, Lewandowska A, Zietkiewicz S. Chaperones in control of protein disaggregation. Embo J. 2008;27:328–335. doi: 10.1038/sj.emboj.7601970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logroscino G, Traynor B, Hardiman O, Chio A, Mitchell DJ, Swingler RJ, et al. Incidence of amyotrophic lateral sclerosis in Europe: A collaborative study of the population-based cohorts. Neurology. 2007;68:A90–A91. [Google Scholar]
- Matsumoto G, Stojanovic A, Holmberg CI, Kim S, Morimoto RI. Structural properties and neuronal toxicity of amyotrophic lateral sclerosis-associated Cu/Zn superoxide dismutase 1 aggregates. J Cell Biol. 2005;171:75–85. doi: 10.1083/jcb.200504050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mccord JM, Fridovic I. Superoxide Dismutase an Enzymic Function for Erythrocuprein (Hemocuprein) J Biol Chem. 1969;244:6049. [PubMed] [Google Scholar]
- Meacham GC, Patterson C, Zhang WY, Younger JM, Cyr DM. The Hsc70 co-chaperone CHIP targets immature CFTR for proteasomal degradation. Nat Cell Biol. 2001;3:100–105. doi: 10.1038/35050509. [DOI] [PubMed] [Google Scholar]
- Prudencio M, Borchelt DR. Expression of Human Wt Sod1 Slows Aggregation of Mutant Sod1, but Eventually Co-Aggregates with the Mutant Protein. J Neurochem. 2009;108:146–146. [Google Scholar]
- Prudencio M, Borchelt DR. Superoxide dismutase 1 encoding mutations linked to ALS adopts a spectrum of misfolded states. Mol Neurodegener. 2011;6 doi: 10.1186/1750-1326-6-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prudencio M, Durazo A, Whitelegge JP, Borchelt D. An examination of wild-type SOD1 in modulating the toxicity and aggregation of ALS-associated mutant SOD1. Hum Mol Genet. 2010;19:4774–4789. doi: 10.1093/hmg/ddq408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puttaparthi K, Wojcik C, Rajendran B, DeMartino GN, Elliott JL. Aggregate formation in the spinal cord of mutant SOD1 transgenic mice is reversible and mediated by proteasomes. J Neurochem. 2003;87:851–860. doi: 10.1046/j.1471-4159.2003.02028.x. [DOI] [PubMed] [Google Scholar]
- Rosen DR, Bowling AC, Patterson D, Usdin TB, Sapp P, Mezey E, et al. A Frequent Ala-4 to Val Superoxide Dismutase-1 Mutation Is Associated with a Rapidly Progressive Familial Amyotrophic-Lateral-Sclerosis. Hum Mol Genet. 1994;3:981–987. doi: 10.1093/hmg/3.6.981. [DOI] [PubMed] [Google Scholar]
- Rosen DR, Siddique T, Patterson D, Figlewicz DA, Sapp P, Hentati A, et al. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature. 1993;362:59–62. doi: 10.1038/362059a0. [DOI] [PubMed] [Google Scholar]
- Ross CA, Poirier MA. Protein aggregation and neurodegenerative disease. Nat Med. 2004;10(Suppl):S10–17. doi: 10.1038/nm1066. [DOI] [PubMed] [Google Scholar]
- Rudiger S, Buchberger A, Bukau B. Interaction of Hsp70 chaperones with substrates. Nat Struct Biol. 1997;4:342–349. doi: 10.1038/nsb0597-342. [DOI] [PubMed] [Google Scholar]
- Shaw BF, Durazo A, Nersissian AM, Whitelegge JP, Faull KF, Valentine JS. Local unfolding in a destabilized, pathogenic variant of superoxide dismutase 1 observed with H/D exchange and mass spectrometry. J Biol Chem. 2006;281:18167–18176. doi: 10.1074/jbc.M600623200. [DOI] [PubMed] [Google Scholar]
- Shen HY, He JC, Wang Y, Huang QY, Chen JF. Geldanamycin induces heat shock protein 70 and protects against MPTP-induced dopaminergic neurotoxicity in mice. J Biol Chem. 2005;280:39962–39969. doi: 10.1074/jbc.M505524200. [DOI] [PubMed] [Google Scholar]
- Stieber A, Gonatas JO, Moore JS, Bantly A, Yim HS, Yim MB, et al. Disruption of the structure of the Golgi apparatus and the function of the secretory pathway by mutants G93A and G85R of Cu, Zn superoxide dismutase (SOD1) of familial amyotrophic lateral sclerosis. J Neurol Sci. 2004;219:45–53. doi: 10.1016/j.jns.2003.12.004. [DOI] [PubMed] [Google Scholar]
- Strange RW, Antonyuk SV, Hough MA, Doucette PA, Valentine JS, Hasnain SS. Variable metallation of human superoxide dismutase: Atomic resolution crystal structures of Cu-Zn, Zn-Zn and as-isolated wild-type enzymes. Journal of Molecular Biology. 2006;356:1152–1162. doi: 10.1016/j.jmb.2005.11.081. [DOI] [PubMed] [Google Scholar]
- Walsh DM, Klyubin I, Fadeeva JV, Rowan MJ, Selkoe DJ. Amyloid-beta oligomers: their production, toxicity and therapeutic inhibition. Biochem Soc Trans. 2002;30:552–557. doi: 10.1042/bst0300552. [DOI] [PubMed] [Google Scholar]
- Wang J, Farr GW, Zeiss CJ, Rodriguez-Gil DJ, Wilson JH, Furtak K, et al. Progressive aggregation despite chaperone associations of a mutant SOD1-YFP in transgenic mice that develop ALS. P Natl Acad Sci USA. 2009;106:1392–1397. doi: 10.1073/pnas.0813045106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Xu G, Borchelt DR. High molecular weight complexes of mutant superoxide dismutase 1: Age-dependent and tissue-specific accumulation. Neurobiol Dis. 2002;9:139–148. doi: 10.1006/nbdi.2001.0471. [DOI] [PubMed] [Google Scholar]
- Wang LJ, Deng HX, Grisotti G, Zhai H, Siddique T, Roos RP. Wild-type SOD1 overexpression accelerates disease onset of a G85R SOD1 mouse. Hum Mol Genet. 2009;18:1642–1651. doi: 10.1093/hmg/ddp085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witan H, Gorlovoy P, Kaya AM, Koziollek-Drechsler I, Neumann H, Behl C, et al. Wild-type Cu/Zn superoxide dismutase (SOD1) does not facilitate, but impedes the formation of protein aggregates of amyotrophic lateral sclerosis causing mutant SOD1. Neurobiol Dis. 2009;36:331–342. doi: 10.1016/j.nbd.2009.07.024. [DOI] [PubMed] [Google Scholar]
- Witan H, Kern A, Koziollek-Drechsler I, Wade R, Behl C, Clement AM. Heterodimer formation of wild-type and amyotrophic lateral sclerosis-causing mutant Cu/Zn-superoxide dismutase induces toxicity independent of protein aggregation. Hum Mol Genet. 2008;17:1373–1385. doi: 10.1093/hmg/ddn025. [DOI] [PubMed] [Google Scholar]
- Wong PC, Pardo CA, Borchelt DR, Lee MK, Copeland NG, Jenkins NA, et al. An Adverse Property of a Familial Als-Linked Sod1 Mutation Causes Motor-Neuron Disease Characterized by Vacuolar Degeneration of Mitochondria. Neuron. 1995;14:1105–1116. doi: 10.1016/0896-6273(95)90259-7. [DOI] [PubMed] [Google Scholar]
- Worms PA. The epidemiology of motor neuron diseases: a review of recent studies. J Neurol Sci. 2001;191:3–9. doi: 10.1016/s0022-510x(01)00630-x. [DOI] [PubMed] [Google Scholar]
- Zetterstrom P, Stewart HG, Bergemalm D, Jonsson PA, Graffmo KS, Andersen PM, et al. Soluble misfolded subfractions of mutant superoxide dismutase-1s are enriched in spinal cords throughout life in murine ALS models. P Natl Acad Sci USA. 2007;104:14157–14162. doi: 10.1073/pnas.0700477104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang FJ, Zhu HN. Intracellular conformational alterations of mutant SOD1 and the implications for fALS-associated SOD1 mutant induced motor neuron cell death. Bba-Gen Subjects. 2006;1760:404–414. doi: 10.1016/j.bbagen.2005.11.024. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.