Abstract
Rex1/Zfp42 is a nuclear protein that is highly conserved in mammals, and widely used as an embryonic stem (ES) cell marker. Although Rex1 expression is associated with enhanced pluripotency, loss-of-function models recently described do not exhibit major phenotypes, and both preimplantation development and ES cell derivation appear normal in the absence of Rex1. To better understand the functional role of Rex1, we examined the expression and localization of Rex1 during preimplantation development. Our studies indicated that REX1 is expressed at all stages during mouse preimplantation development, with a mixed pattern of nuclear, perinuclear, and cytoplasmic localization. Chromatin association seemed to be altered in 8-cell embryos, and in the blastocyst, we found REX1 localized almost exclusively in the nucleus. A functional role for Rex1 in vivo was assessed by gain- and loss-of-function approaches. Embryos with attenuated levels of Rex1 after injection of zygotes with siRNAs did not exhibit defects in preimplantation development in vitro. In contrast, overexpression of Rex1 interfered with cleavage divisions and with proper blastocyst development, although we failed to detect alterations in the expression of lineage and pluripotency markers. Rex1 gain- and loss-of-function did alter the expression levels of Zscan4, an important regulator of preimplantation development and pluripotency. Our results suggest that Rex1 plays a role during preimplantation development. They are compatible with a role for Rex1 during acquisition of pluripotency in the blastocyst.
Introduction
Preimplantation development starts at fertilization and lasts until implantation. Upon fertilization, arrested oocytes resume development and finish the second meiotic division. Sperm chromatin is decondensed and converted into a functional pronucleus. After pronuclear fusion, zygotic genome activation (ZGA) initiates, although up to the mid 2-cell stage in mice, development is mainly governed by maternally stored RNAs and proteins [1–3]. The transition to the embryonic genetic program governed by de novo transcription occurs in 2 major transient waves of de novo transcription [4]. The first wave during the 1- to 2-cell stage corresponds to ZGA. The second wave occurs during the 4- to 8-cell stage, and is known as mid-preimplantation gene activation (MGA). The novel expression landscape resulting from combined ZGA and MGA specifies the totipotent state of blastomeres during the cleavage stage of embryogenesis. Proper progression of cleavage and subsequent compaction and blastocoel formation are a prerequisite for future cell differentiation, lineage separation, and commitment. In the first lineage separation step, the preimplantation blastocyst is divided in the inner cell mass (ICM), from which embryonic stem (ES) cells can be derived [5,6], as well as the trophectoderm (TE).
ES cells can be maintained in culture for an apparently unlimited number of cell divisions (self-renewal) and maintain the defining property of pluripotency or the ability to differentiate into cell lineages of all 3 primary layers of the embryo. Oct4, Sox2, and Nanog participate in a transcriptional network with essential functions in the formation and/or maintenance of both the ICM during mouse preimplantation development and murine ES cells in culture [7–10]. Additional transcription factors have been implicated in stem cell biology and pluripotency, based on specific expression patterns [11–13], loss-of-function studies [14,15], or epigenetic contributions [16]. Among those genes activated during ZGA and restrictively expressed in preimplantation embryos is Zscan4 [17], which is expressed in 2-cell stage embryos [18] and a subset of ES cells [19]. Zscan4 is essential for long-term proliferation of ES cells, and contributes to telomere elongation and enhanced genomic stability [20].
Another example is Rex1 (for reduced expression-1, also known as Zfp42), which encodes a protein containing 4 Cys-His-type zinc-fingers. Rex1 was generated by duplication from Yy1 by retrotransposition, displays significant similarity to the Yy1 transcription factor in the zinc-finger domains, and is present exclusively in eutherian mammals [21]. Rex1 was first discovered as a result of its specific expression in pluripotent F9 embryonal carcinoma cells [22]. Rex1 was subsequently shown to be expressed in other pluripotent cell types, specifically undifferentiated ES cells [23], multipotent adult progenitor cells [24], and amniotic fluid cells [25]. During mouse development, mRNA encoding Rex1 was detected during mouse development in the ICM of the blastocyst and in trophoblast-derived tissues [23]. Rex1 mRNA is also expressed in spermatocytes actively undergoing meiosis [23], and Rex1 has been detected in dividing cells of the human testes and ovaries [26].
Rex1 is widely used as a pluripotency marker, as Rex1 expression has been positively linked to increased pluripotency in both murine ES cells [11,27,28] and human ES and induced pluripotent stem (iPS) cells [29,30]. In contrast, conflicting results have been reported regarding the functional role of Rex1. Gene silencing by RNA interference results in loss of self-renewal in ES cells [14], and overexpression of Rex1 negatively affects self-renewal (D. Guallar, M. Sánchez and J. Schoorlemmer, unpublished data). However, Rex1 does not have to be provided for efficient reprogramming of differentiated cells toward iPS [11,27], and Rex1 is dispensable for the maintenance of self-renewing pluripotent ES cells [31], and ES cell lines can efficiently be derived from Rex1-deficient blastocysts [32]. Gene expression studies have indicated subtle differentiation defects in Rex1−/− murine ES lines [31,32], in line with both the potential association of Rex1 to polycomb-regulated gene expression [33] and a functional connection of Rex1 to genomic imprinting [34].
The in vivo roles of Rex1 have been analyzed in different mouse lines in which the transcription unit was interrupted. Similar to the results in ES cells, and contrary to initial expectations, mutant mice lacking Rex1 were viable and fertile [32]. Breeding experiments showed, however, that litter sizes correlated well with the combined gene dosage of Rex1 in the parents, and that both homozygous (Rex1−/−) and heterozygous (Rex1−/+) animals were present in numbers below Mendelian ratios [32,34]. Embryos were shown to die during the late-gestation and neonatal stages [32], in line with a functional connection of Rex1 to genomic imprinting [34], although Rex1 expression is restricted to the early stages of development [4,23]. Rex1 dosage is more critical during spermatogenesis than during oogenesis [34]. This result is consistent with the fact that Rex1 expression is mainly detected in spermatogenesis [23,35].
Although Rex1 is expressed during the early stages of development, the actual presence, levels, and contributions at different stages of preimplantation have been poorly defined. Considering the unique presence of Rex1 in eutherian mammals and its invariable association with pluripotency in ES cells, the absence of phenotypes in Rex1-deficient ES cells and mice is very surprising. We reasoned that understanding the signaling, transcriptional, and epigenetic mechanisms underlying preimplantation development may unravel molecular mechanisms that govern pluripotency in ES cells and operate during the reprogramming of somatic cells toward an induced pluripotent state (iPS).
We therefore studied the presence and function of Rex1 during preimplantation mouse development. We report a detailed expression analysis of REX1, which appears to transition into the nucleus at both the 8-cell and at the blastocyst stage. No phenotype was apparent in Rex1 loss-of-function studies using siRNA technology. By contrast, REX1 overexpression caused developmental delay during cleavage stages and interfered with blastocyst development. We show that Rex1 regulates expression of Zscan4, providing a potential alternative mechanism for future developmental defects.
Materials and Methods
Immunofluorescence and confocal microscopy
The rabbit α-REX1 serum described previously [33] was further affinity-purified over REXΔ-GST protein [36]. Monoclonal α-HA (clone HA-7) was obtained as an unpurified ascites fraction (Sigma H9658). Mouse embryos (CD-1; Charles River) for immunostaining were obtained from natural matings using standard methods. To stain embryos after injection with siRNAs, superovulated B6D2F1/J mice were used. Immunostaining and confocal microscopy were routinely performed as described previously [33], with a few modifications. The application of alternative protocols [37] (M. Torres-Padilla, personal communication) did not improve nonspecific staining (M. Climent, data not shown). After storage for at least a week in 0.1% Triton X-100 in phosphate-buffered saline (PBS), embryos were permeabilized with 0.5% Triton X-100 in PBS for 20′. Nuclei were counterstained with DAPI; the purified αREX1 was diluted 1:2,400. To extract soluble protein, embryos transferred to KSOM (Millipore) were incubated for 10′ at 4°C in 0.5% Triton X-100 in PBS, washed 3 times in PBS, and subsequently fixed in 2.5% paraformaldehyde and processed as usual. Images were captured on a Leica TCS SP2 AOBS confocal microscope at the Centro de Investigaciones Biológicas (C.I.B. del CSIC), or with Olympus FV10i at the IACS (Aragon Health Sciences Institute).
REX1 staining intensities in confocal data were quantified in maximum projections over the Z-axis [8 sections (1 μm) per embryo] using ImageJ 1.45 g software. Five regions were drawn in each embryo to measure the fluorescence levels. Triplicate areas of identical size (without fluorescent objects) were used for background subtraction. The net-corrected total cell fluorescence was calculated for each condition.
Plasmid construction and preparation
For overexpression of REX1 or eGFP, cDNAs were inserted into the chicken β-actin promoter-driven expression vector pCAGIP [38]. To generate pCAG-REX1-IRESeGFP, Rex1/Zfp42 cDNA [33] was transferred as an EcoR1 fragment into pCAG and fused to an IRES-eGFP derived from pIRES2-eGFP (Clontech). Further details of the plasmids used are available on request. All constructs were verified by DNA sequencing. The plasmids for microinjection were purified on PureLink™ kits and columns (Invitrogen), linearized with ScaI, phenol-extracted twice, ethanol precipitated, resuspended in TE, purified on GeneClean Turbo cartridges (MP Biomedicals), and eluted in Tris/EDTA (10/0.1 mM). Concentration was measured using Nanodrop.
siRNA and dsRNA used for microinjection
Stealth siRNA duplexes against mRex1 and control siRNAs (Ref 12935-112) were purchased from Invitrogen. Two different siRNAs were used to target Rex1, corresponding to positions 658–682 and 742–766 of mRex1 (NM_9556.3), respectively. The siRNAs were annealed according to the manufacturer's instructions, and buffer-changed repeatedly by dilution in Tris 10 mM/EDTA 0.1 mM and subsequent concentration on YM-10 columns (Millipore).
Oocyte and embryo collection, culture, and microinjection
All procedures were carried out under the Project License PI29/08 approved by the in-house Ethics Committee for Animal Experiments from the University of Zaragoza. Animals were taken care of and used according to the Spanish Policy for Animal Protection RD1201/05, which meets the European Union Directive 86/609 on the protection of animals used for experimental and other scientific purposes.
Oocyte and embryo collection, culture, and microinjection were performed according to standard procedures [39]. Embryos were obtained from B6D2F1/J mice mated to B6D2F1/J males (Charles River). Four- to 6-week-old females were superovulated by an intraperitoneal (i.p.) injection of PMSG (5 IU), followed 48 h later by an i.p. injection of human chorionic gonadotropin (hCG; 7.5 IU). After mating, fertilized eggs were harvested at 20 h post-hCG. After removing cumulus cells with hyaluronidase, zygotes were thoroughly washed and selected for good morphology and collected. Fertilized eggs (1-cell embryos) were cultured in KSOM (Embriomax®; Millipore MR-020P) at 37°C in an atmosphere of 5% CO2, 90% relative humidity.
One-cell embryos were microinjected (defined as day 1) in an M-2 medium (Sigma M7167) using either siRNAs at 100 μM or a linearized plasmid at 2 ng/μL and 0.4 mg/mL Dextran-coupled Texas Red (D1829; Invitrogen). Plasmid vectors expressing REX1, eGFP, or REX1-IRESeGFP in pPyCAGIP [9] were microinjected into the male pronucleus, while transient RNA interference experiments were carried out by microinjecting siRNA duplexes into the cytoplasm of zygotes.
Successful injection was monitored in Texas Red; dead embryos were eliminated immediately after injection, and 18 h after. At each time indicated, the embryos were scored for the number of cells or, following compaction, for development to the morula or blastocyst stage. Images were captured using a Leica DFC360Fx camera adapted to a Leica M165FC stereomicroscope using Leica Application Suite 3.2.0. To compare Rex1-overexpressing embryos with injected controls, embryos were injected on day 1, selected for development to at least the 2-cell stage on day 2, and to the 5–8-cell stage on day 3. Embryos injected with pCAG-REX1-IRESeGFP were separated on day 3 into three groups representing low (absent), intermediate, and high eGFP expression. Only the first and latter groups were used in experiments and are identified as low and high expressers, respectively. Embryos were harvested in TRIzol and stored frozen at −80°C.
Gene expression in preimplantation embryos
For the analysis of expression in mouse preimplantation embryos, embryos for reverse transcription (RT)–polymerase chain reaction (PCR) experiments were selected for morphology at the appropriate stages (20, 30, 43, 55, 66, 80, and 102 h post-hCG for 1-cell, early 2-cell, late 2-cell, 4-cell, 8-cell, morulae, and blastocyst embryos, respectively) and homogenized in 100 μL TRIzol® reagent (Invitrogen). An identical number of experimental and control embryos was processed from the same experiment (typically 8–10 embryos). Total RNA was isolated from TRIzol by chloroform extraction, precipitated with ethanol in the presence of glycogen (Roche) as a carrier, pelleted by centrifugation, resuspended in diethylpyrocarbonate (DEPC)-treated water, treated with RQ1 RNase-free DNase (Promega), phenol extracted, reprecipitated with ethanol, and resuspended in DEPC-treated water. RNA was reverse-transcribed with the ThermoScript® RT-PCR System (Invitrogen) using random hexamer primers. After reverse transcription, 1 to 2 oocytes or embryo equivalents were used as a template for each PCR; products were separated on 2% agarose gels, visualized using ethidium bromide, and photographed on a Gel Doc transilluminator (BioRad). Data were quantified using Quantity One software (BioRad), and expression levels were recalculated using H2afz as a reference gene. All primers used in this study are shown in Table 1, and were used at 200 μM. PCRs were carried out in the linear range of amplification for 35 cycles of denaturation at 94°C for 30 s, followed by 30 s of annealing, 1 min extension at 72°C, and final extension for 5 min.
Table 1.
Primers Used in Reverse Transcription–Polymerase Chain Reaction Assays
| Target | Forward (5′→3′) | Reverse (5′→3′) |
|---|---|---|
| H2afz | CGTATCACCCCTCGTCACTT | AAGCCTCCAACTTGCTCAAA |
| Rex1 (Exon IV) | AAGCCGTATCAGTGCACGTTCGAAGGCT | ATGCGTGTATCCCCAGTGCCTCTGTCAT |
| Rex1 (Exon III-IV) | TCACTGTGCTGCCTCCAAGT | CCCTTTCTGGCCACTTGTCT |
| Oct4 | GGCGTTCTCTTTGGAAAGGTGTTC | CTCGAACCACATCCTTCTCT |
| Cdx2 | GCAGTCCCTAGGAAGCCAAGTGA | CTCTCGGAGAGCCCAAGTGTG |
| Stella | AGGCTCGAAGGAAATGAGTTTG | TCCTAATTCTTCCCGATTTTCG |
| Nanog | CACCCACCCATGCTAGTCTT | ACCCTCAAACTCCTGGTCCT |
| Zscan4 | 5′GAGGTCGAATTCTCTAGATTCAAGTGTGAAGAATGTTCTAG3′ | CTCGACGAATTCTCAGTCAGATCTGTGGTAATTC |
| Zscan4 | GAGATTCATGGAGAGTCTGACTGATGAGTG | GCTGTTGTTTCAAAAGCTTGATGACTTC |
| Tcstv3 | ACCAGCTGAAACATCCATCC | CCATGGATCCCTGAAGGTAA |
Transfection and western blot to test siRNAs
Human embryonic kidney 293T cells were transiently cotransfected on poly-d-Lysine-coated dishes using Lipofectamine™ reagent (Invitrogen) with a mixture of plasmids that direct the expression of HA-tagged REX1 and the siRNAs indicated. The day after transfection, cells were rinsed once with 1×ice-cold PBS, and cells were scraped in 40 mM HEPES, pH 7.6, 200 mM NaCl, 0.1% NP40, and Complete™ protease inhibitors without EDTA (Roche). Cells were lysed by standard sonication, and lysates were spun at 4°C for 10 min in an Eppendorf centrifuge at 12,000 rpm. Aliquots of the lysate were supplemented with Laemmli sample buffer, separated by SDS-PAGE, and analyzed by standard western blot.
Statistical analysis
Differences between groups of embryos were statistically evaluated using the Pearson's Chi-square test using a 95% confidence interval. Differences with a P value of<0.05 were considered statistically significant.
Results
Expression of Rex1 mRNA and protein during preimplantation development
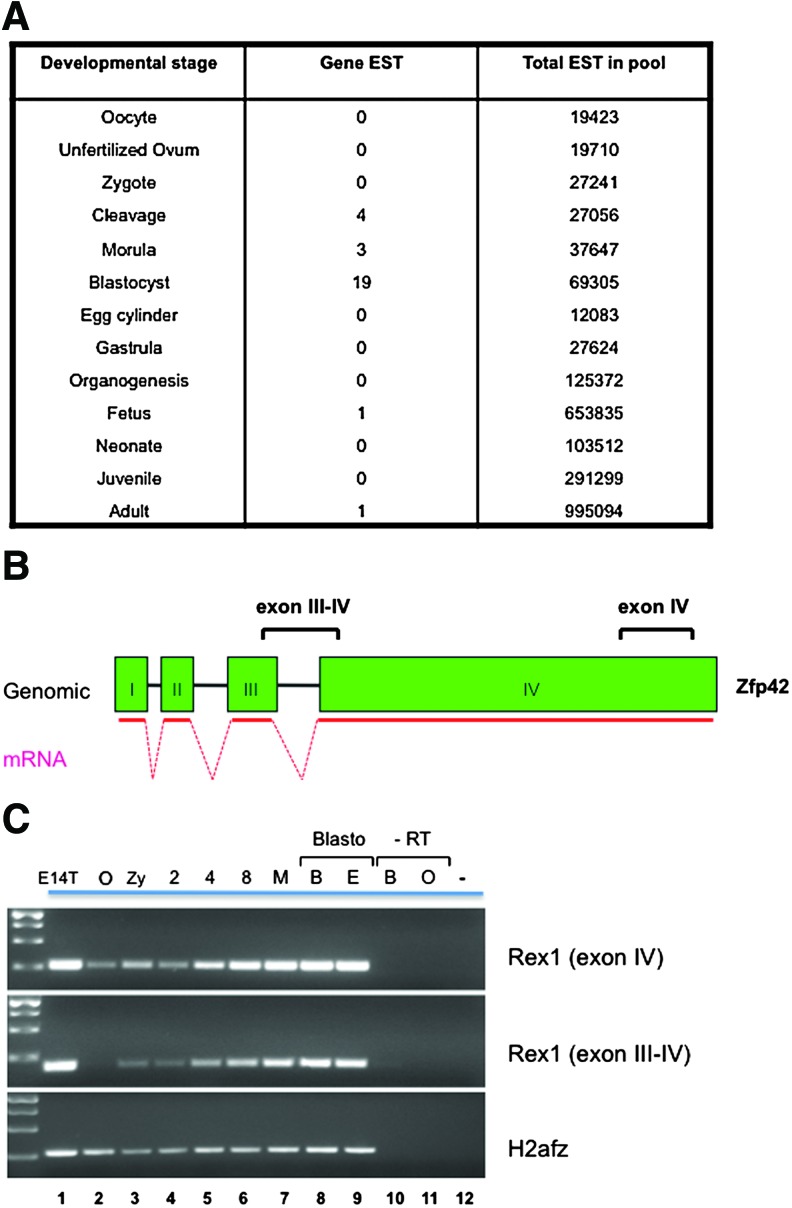
In silico analysis indicated that Rex1 is expressed in cleavage-stage embryos and in the blastocyst. The transcript levels are probably upregulated early during ZGA (1- to 2-cell stages) based on profiling data [40]. Using publicly available databases of expressed sequence tags, among 31 cDNA clones described most originated from blastocyst and ES cells (8 and 10, respectively) or 2- to 8-cell stage libraries (Unigene database) (Fig. 1A).
FIG. 1.
(A) Expression sequence tag (EST) frequencies in Unigene cDNA libraries. Out of 4.7 million mouse ESTs, 28 Rex1 sequences were restrictedly detected at the cleavage stages, morula and blastocyst. (B) Representation of the genomic structure of the Rex1 locus. Exons I–IV are indicated as well as the resulting mRNA. Fragments amplified by reverse transcription (RT)–polymerase chain reaction (PCR) are depicted as exon IV (primers 153–154, 260nt) and exon III–IV (primers 223–224, 250nt). (C) Detection by RT-PCR of mRNA encoding Rex1 in preimplantation mouse embryos. RNA extracted from 25 oocytes or embryos at each stage was reverse-transcribed and subjected to PCR using primers indicated in (B) or H2afz as a control. RNA extracted from E14T embryonic stem (ES) cells was included as a positive control. O, oocyte at metaphase II; Zy, zygote; 2, 2-cell; 4, 4-cell; 8, 8-cell embryos; M, morula; B, early blastocyst; E, expanded blastocyst E4.5. The lower band of the molecular-weight markers in the left-most lane corresponds to a band of 250 base pairs. O-RT and B-RT are oocyte-RT, and blastocyst-RT controls without reverse transcription, a PCR without input (−) is shown to the right. Color images available online at www.liebertpub.com/scd
We experimentally confirmed the expression pattern of Rex1 mRNA during preimplantation stages suggested by earlier studies [4,23] and in silico analysis. Significant expression of Rex1 was detected in ES cells used as a positive control (Fig. 1C). RT-PCR analysis for preimplantation embryos indicated some expression of Rex1 in the zygote, with a slight decrease in Rex1 expression in the 2-cell embryo. From the 4-cell stage onward, Rex1 mRNA levels gradually increased during the 8-cell-to-blastocyst transition. Although a different set of primers spanning the exon III-IV boundary produced a slightly lower yield of product as compared to exon IV primers, the overall pattern was no different. These results suggest that Rex1 is already present (likely derived from maternal stores) before the major burst of ZGA, and increases expression once zygotic transcription is initiated.
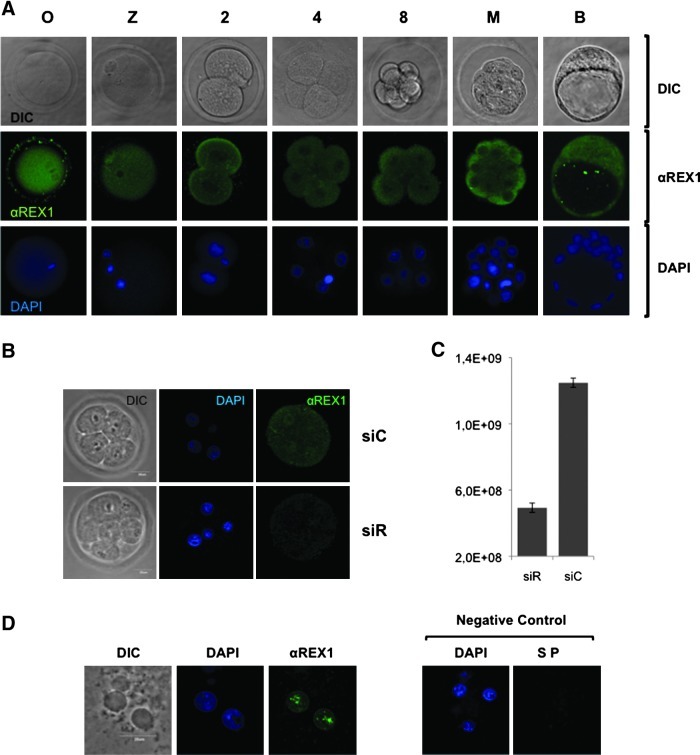
To study the temporal and spatial expression pattern of the REX1 protein, we used an affinity-purified polyclonal serum raised against the aminoterminus of REX1. Using this anti-REX1 antibody, we detected a 41-kDa protein uniquely present in mouse ES cells by western blot analysis of extracts from mouse ES cells (in addition to some additional bands that have not been characterized) [36]. The serum had been tested previously by preincubation with the REX1 fragment used for immunization [33]. In accordance with the presence of Rex1 transcripts throughout preimplantation development (Fig. 1C), immunostaining revealed REX1 expression at all stages tested, starting at the 1-cell stage until the blastocyst stage. We detected anti-REX1 immunoreactivity in all cells, although αREX1 immunoreactivity showed a dynamic subcellular localization pattern in a mixed pattern of nuclear, perinuclear, and cytoplasmic localization from the 2-cell stage onward. Although a nuclear protein in mouse ES cells [33], REX1 staining was evenly distributed in the MII oocyte and also in the cytoplasm of embryos up to the 4-cell stage (Fig. 2A). At the 8-cell stage, staining was more concentrated in the nuclei, followed by a return to a more even distribution between the cytoplasm and the nucleus at the 16-cell stage.
FIG. 2.
(A) REX1 is present throughout mouse preimplantation development. Indirect immunofluorescent detection of REX1 in confocal sections of representative mouse embryos depicted as differential interference contrast microscopy images. DAPI staining indicates chromatin (blue), while REX1 (A488, green) appears in a mixed nuclear, perinuclear, and cytoplasmatic pattern in most embryos. Negative control embryos incubated without primary antibody processed in parallel are depicted in Supplementary Fig. S1. O, oocyte at metaphase II; Z, zygote; 2, 4, 8: 2-, 4-, and 8-cell embryos; M, morula, B, blastocyst. (B, C) REX1 in 8-cell embryos. Indirect immunofluorescent detection of REX1 in embryos injected with Rex1-specific siRNAs (siR) or control siRNAs (siC), scale bar=20 μm. (B) Confocal sections as in (A). (C) Quantification of staining intensities using maximum projections over the Z-axis for 4 embryos per condition. Data show the corrected total cell fluorescence; error bars represent the standard deviation. (D) REX1 is chromatin associated. Confocal sections as in (A) show a representative example of anti-REX1 immunoreactivity in 8-cell embryos pretreated with Tx100 before fixation to extract nonchromatin-bound protein.
Localization of REX1 in cleavage-stage embryos
To further confirm the observed patterns, we stained 8-cell embryos injected with siRNAs (see below) to attenuate Rex1 levels. Anti-REX1 immunoreactivity was dependent on the presence of REX1, as staining was several-fold stronger in embryos injected with control siRNAs (Fig. 2B, top panels; siC) as opposed to embryos injected with siRNAs directed against Rex1 (Fig. 2B, lower panels; siR). Quantification of intensities confirmed this difference (Fig. 2C). Staining of the control embryos also confirmed a more nuclear localization in 8-cell embryos (Fig. 2B, top panels; siC), as opposed to more even distribution between the cytoplasm and the nucleus at both 4- and 16-cell stage embryos (Fig. 2A). To better evaluate this phenomenon, we tested for the presence of chromatin-associated REX1 after Triton extraction of soluble protein. Under these conditions, we observed REX1 localized toward the nuclear periphery in 8-cell embryos (Fig. 2D), as opposed to the more even distribution throughout the nucleus in 2- and 4-cell embryos (Supplementary Fig. S2A; Supplementary Data are available online at www.liebertpub.com/scd). In control experiments, staining was dependent on the primary antibody (see SP in Fig. 2D) and the presence of the nucleus in the confocal plane (Supplementary Fig. S2B). The distinct pattern of REX1 localization observed in 8-cell embryos is compatible with gene regulatory functions at this stage.
Nuclear localization of REX1 in the blastocyst
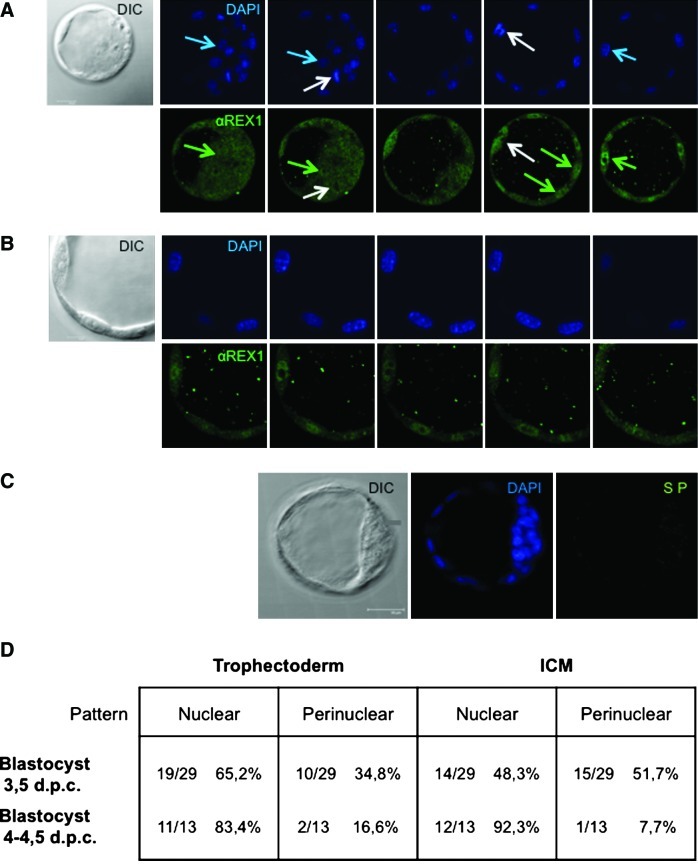
As reported previously [33], REX1 protein was detected in cells corresponding to both the ICM and the TE in E3.5 blastocysts (Fig. 2A), localizing in the cytoplasmic, nuclear, and perinuclear compartments. REX1 protein is not associated with chromatin in mitotic cells (data not shown and Fig. 3A) and is excluded from the nucleoli at all times. Starting at the early blastocyst stage (E3.5, Fig. 3A), staining turned progressively more nuclear in a variable number of cells, and a predominantly nuclear localization was obtained in late blastocysts (Fig. 3B and Supplementary Fig. S3), especially in the TE. To quantify this shift, staining in individual embryos was arbitrarily designated Nuclear (primarily nuclear with a cytoplasmic component) or Perinuclear, assigning separate values for the ICM and TE. Data obtained from 29 pre-expansion (E3.5) blastocysts are shown in Fig. 3D. In the TE, 34.8% of embryos displayed perinuclear staining, as opposed to predominantly nuclear staining in 65.2%. In the ICM by contrast, about equal numbers of embryos displayed perinuclear-versus-predominantly nuclear staining (Fig. 3D). Upon expansion, E4.5 blastocysts lost perinuclear staining, and staining turned nuclear (with a cytoplasmatic component) in most embryos stained. The majority of embryos display uniquely nuclear staining in the TE (11 out of 13) and ICM (12/13). Differences observed between E3.5 and E4.5 blastocysts were statistically significant both in the TE (P=0.026) and in the ICM (P=0.005). Hence, we propose that during blastocyst development in vitro, REX1 localization shifts toward a predominantly nuclear localization throughout the embryo.
FIG. 3.
REX1 intracellular localization in blastocyst. Detection of anti-REX1 immunoreactivity in virtual confocal sections as in Fig. 2. (A) Early blastocyst (E3.5). DAPI staining and the corresponding αREX1 immunoreactivity are shown in selected confocal sections across the embryo. Scale bar=20μm. White arrows point toward mitotic cells. Green arrows highlight nuclei that stained positive for REX1 in green, corresponding. Blue arrows locate the DAPI staining of the same nuclei. (B) Detail of the trophectoderm (TE) of a representative E3.5 embryo to highlight predominant nuclear staining. DAPI staining and the corresponding αREX1 immunoreactivity are shown in every fifth section. Scale bar=10μm. (C) Control embryo without primary antibody (SP). Scale bar=30μm. (D) Localization of REX1 in either the TE or the inner cell mass (ICM) in early (E3.5) and late (E4.5) blastocysts. The table shows the number of embryos displaying predominantly nuclear staining (with a cytoplasmatic component) or perinuclear staining, in relation to the total number of embryos analyzed. Differences between E3.5 and E4.5 are statistically significant [χ2 test; 95% confidence index (P≤0.05)] in the predominance at E4.5 of nuclear localization both in the TE (P=0.026) and in the ICM (P=0.005). Color images available online at www.liebertpub.com/scd
In vitro development of Rex1-depleted embryos
We designed siRNAs to attenuate Rex1 expression, which were obtained as regular siRNAs or as Stealth™ RNAs (Materials and Methods section). All siRNAs were tested by cotransfection with HA-REX1 in 293T cells. The results show that expression of HA-REX1 (Fig. 4A, control) was abolished by cotransfection of S2 Stealth RNAs (and S1 and S3 to a lesser extent), but not by other siRNAs tested or control siRNAs (Fig. 4A).
FIG. 4.
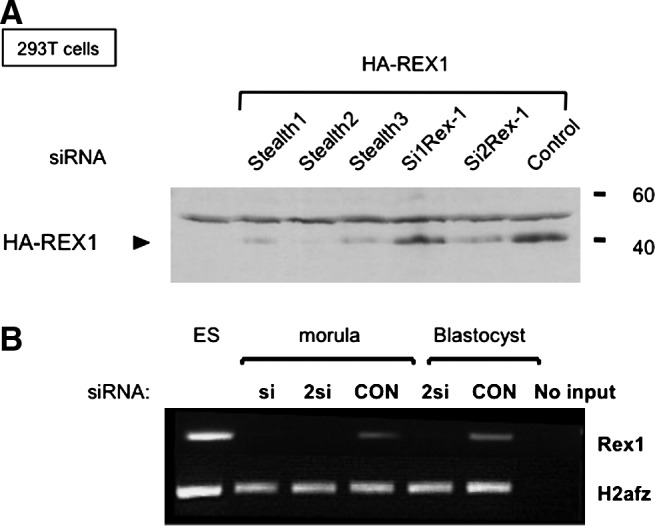
Attenuation of Rex1 levels using siRNA technology. (A) Western blot to detect HA-tagged REX1 in cell lysates from transiently transfected 293T cells using αHA. Figure shows HA-REX cotransfected with Stealth™ RNAs (Stealth1–3) or with regular siRNAs (si1Rex1 and si2Rex1). Nontransfected 293T cells were included as a negative control. The band corresponding to HA-REX is indicated with an arrow. The antibody also generates a nonspecific band in all lanes. (B) Rex1 levels in injected embryos. Transcript levels of Rex1 were assessed by semiquantitative RT-PCR analysis in embryos injected with control siRNA (Con), Rex1 Stealth-2 siRNA (si), or a combination of Stealth-1 and Stealth-2 siRNAs (2si). Expression levels were normalized using H2afz as a reference gene. PCRs without input did not yield any products (no input), and ES cell RNA was used as a positive control for RT and amplification.
To examine whether transient depletion of Rex1 affected early embryonic development, 1-cell embryos were collected and injected with control siRNAs or RNAs directed against Rex1, and developmental progress was monitored daily. Similar numbers of embryos were injected with either Rex1 or control siRNAs (nonsense oligonucleotides) in each experiment, and successful injection was visualized using coinjected Texas-Red (see Materials and Methods section; Supplementary Fig. S4). Approximately 80% of each group cleaved to the 2-cell stage during overnight culture (data not shown). The 2-cell embryos were selected for further culture, and developmental progress was examined daily. After injection of embryos on 5 separate occasions and monitoring development of over 200 embryos, we were unable to observe differences in developmental progress between embryos injected with control siRNAs or siRNAs directed against Rex1 (Supplementary Table S1). Control- and siRex-injected embryos developed into morulae (76.7% and 78.1%, respectively) and blastocyst (51.9% and 54.3%, respectively) with equal efficiency. No statistically significant differences were found between the 2 groups at the stages analyzed (Supplementary Table S1).
Rex1 mRNA levels were monitored in injected embryos on days 3 and 4, corresponding to morula and early blastocyst stages, respectively. Expression was severely suppressed in the siRex-injected embryos, and was significantly lower than those in control embryos (Fig. 4B).
Preimplantation development of embryos overexpressing Rex1
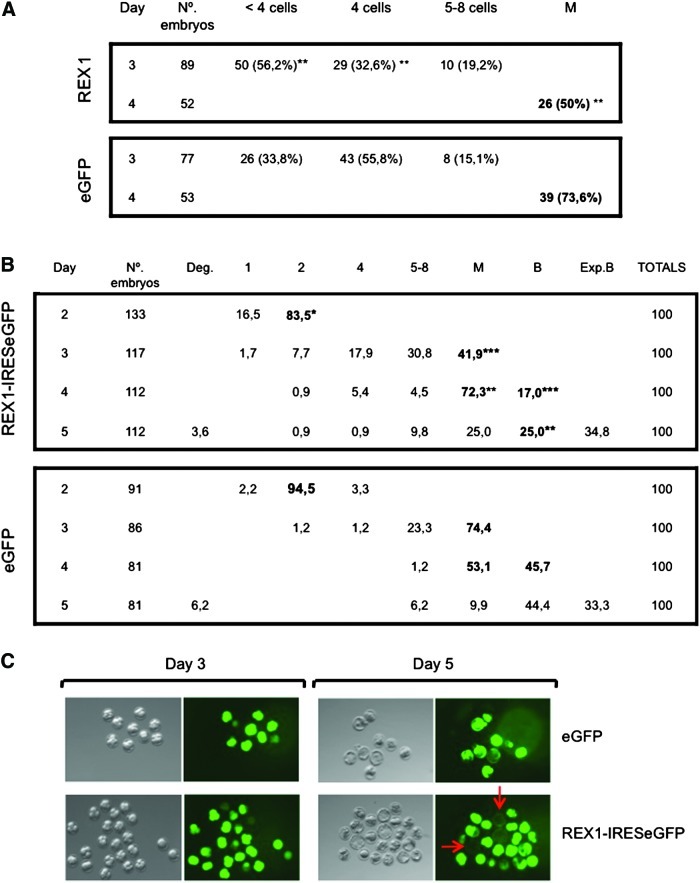
In the absence of a clear preimplantation phenotype in embryos with attenuated levels of Rex1, we sought to examine the effects of overexpression. One-cell embryos were injected with a vector driving ectopic expression of REX1 or eGFP as a control, and subsequent development in vitro was monitored in parallel on days 3 and 4 (42–44 and 66–68 h after injection, respectively). We observed that as a group, the Rex1-overexpressing embryos consistently developed more slowly than the control-injected embryos. By 42 h postinjection, 56.2% of Rex1-injected embryos had not reached the 4-cell stage, compared with 33.8% of the control-injected embryos (Fig. 5A). This delay in development was maintained at the early morula stage (66 h, 50% vs. 73.6% morula). These results establish that overexpression of Rex1 was associated with delayed progression through the early cleavage divisions up to compaction.
FIG. 5.
REX1 overexpression in preimplantation embryos. (A) Development of embryos injected on day 1 with vectors that direct overexpression of REX1 or eGFP as a control. Data represent the total of 4 independent experiments. At days 3 and 4, the embryos were scored for the number of cells and for the initiation of compaction (morula). The table shows the number and fraction (in brackets) of embryos present at the stages listed. Development of REX1 and eGFP embryos differs significantly at each time point (χ2 test; **P<0.01). (B) Preimplantation development of embryos injected at the 1-cell stage with vectors directing expression of REX1-IRESeGFP or eGFP. Cumulative results of 4 independent experiments are expressed as a percentage of embryos that have reached a defined stage on a given day. Stages are indicated as follows: 1, 2, 4 indicate 1-,2-, 4-cell embryos; 5–8 indicates 5–8-cell embryos; M, morula, B, blastocyst. Some dead embryos observed on day 5 are listed as degenerate (deg.). Significant statistical differences observed between the 2 groups are indicated as follows (χ2 test; *P<0.05;**P<0.01; ***P<0.001). (C) Preimplantation development of embryos injected at the 1-cell stage with vectors directing expression of REX1-IRESeGFP or eGFP. The progress is shown in a typical experiment (out of many). One-cell embryos were injected (day 1), and after overnight culture, those that cleaved to the 2-cell stage were selected for further culture. Well-expressing embryos were selected on day 3 (left panel), and development was assessed on day 5 (right panel). The red arrows point out the well-developed blastocysts in the REX1-overexpressing group, which seem to have lost high ectopic expression. Color images available online at www.liebertpub.com/scd
In a next set of experiments, we followed development over a slightly longer period to detect potential effects on postcleavage development and used vectors driving expression of REX1 linked to IRESeGFP or eGFP as a control. Similar numbers of 1-cell embryos were injected (day 1) with each construct in each experiment, and subsequent development in vitro was monitored in parallel every day. Cumulative results of several experiments are shown in Fig. 5B. Again, the Rex1-overexpressing embryos consistently developed more slowly than the control-injected embryos, in statistically significant numbers. By 18 h postinjection (day 2), only 83.5% of Rex1-injected embryos had reached the 2-cell stage, compared to 94.5% of the control-injected embryos (Fig. 5B). This delay in development was maintained during the following days. By 42 h (day 3), the delay was even more pronounced, as the number of Rex1-injected embryos at the morula stage was only 41.9% as opposed to 74.4% in the control embryos. This difference was maintained at days 4 and 5, as the fraction of embryo reaching the blastocyst stage was diminished in the overexpressing embryos (17% vs. 45.7% and 25% vs. 44.4% on days 4 and 5, respectively).
We wished to relate more directly Rex1 overexpression in individual embryos with the phenotype observed. We injected embryos with either a vector driving expression of eGFP as a control, or expression of a REX1-IRESeGFP cassette. The majority of embryos in each group cleaved to the 2-cell stage during overnight culture (data not shown). After further culture in vitro, development to early (compacted) morula was scored on day 3 or to blastocyst on day 5, and compared to eGFP expression. Embryos in both groups that expressed high levels of eGFP and developed into morulae on day 3 were further cultured in vitro and analyzed again on day 5 (Fig. 5C). In both groups, blastocysts were observed, but much less in the REX1-IRESeGFP group (27.3% vs. 100%, data not shown). Overexpressing embryos displayed retarded development (Fig. 5C), as expected from previous experiments (Fig. 5A). Interestingly, embryos that developed into blastocysts in the REX1-IRESeGFP group (indicated with arrows, Fig. 5C) seemed to have lost transgene expression, which underscores that Rex1 overexpression interferes with blastocyst development. In addition, these results confirm that eGFP overexpression does not interfere with blastocyst development in our experimental setting.
Rex1 overexpression interferes with blastocyst development
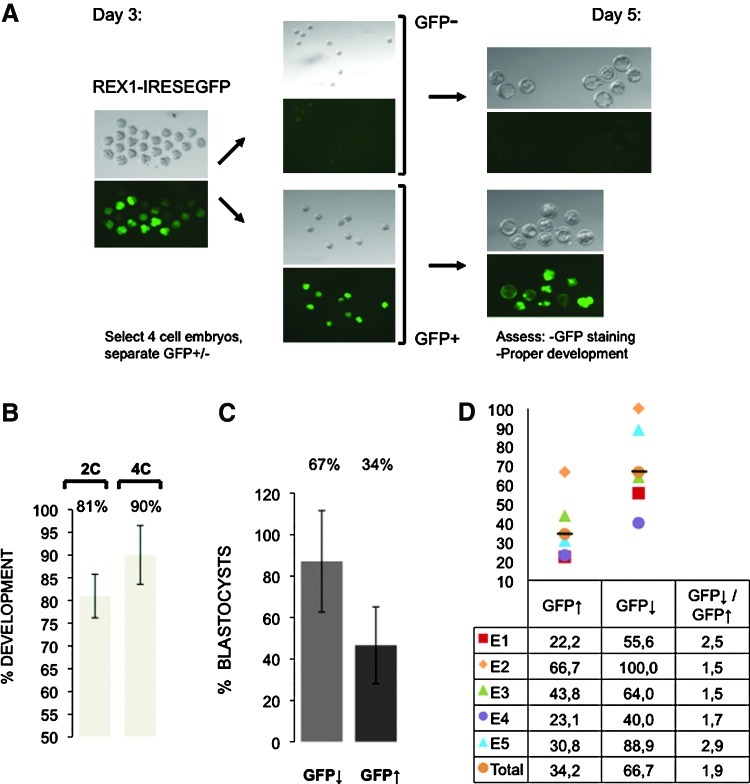
To assess the effect of Rex1 overexpression in individual embryos, we used GFP expression to separate embryos as a function of expression levels of GFP, and limited the assay to embryos with normal cleavage-stage development. We injected zygotes with either a vector driving expression of eGFP as a control, or expression of a REX1-IRESeGFP cassette. Injected embryos were selected for development into the 2-cell stage the following morning (81%, day 2, Fig. 6B), and beyond the 4-cell stage on day 3 (90%, Fig. 6B). On day 3, embryos were selected for low (absent) or high eGFP expression (Fig. 6A), cultured further in vitro up to day 5, and analyzed and processed separately.
FIG. 6.
Blastocyst development in REX1-IRESeGFP-injected embryos. (A) Zygotes were injected with REX1-IRESeGFP-expressing plasmids and cultured in vitro. The following days those that progressed to the 2-cell stage on day 2, and cleaved to >4-cell stages early on day 3, were selected for further culture. On day 3, embryos were separated in pools that express high levels (GFP↑) or low levels (GFP↓) of REX1 (as judged by epifluorescence). Further development to blastocyst and gene expression levels were compared between the 2 groups. (B) Development of injected embryos. The fraction of injected embryos in 3 independent experiments that develop into the 2- or 4-cell stage as indicated. (C) Development after GFP selection. The fraction of >4-cell embryos that develop into blastocyst in 5 independent experiments is represented for overexpressing (GFP↑) versus nonexpressing embryos (GFP↓). (D) Data from separate experiments (Supplementary Table S2) were recalculated as the fold blastocyst development in nonexpressing embryos (GFP↓) versus overexpressing (GFP↑) in each experiment. The differences observed between the 2 groups are statistically significant (χ2 test; P<0.001). Color images available online at www.liebertpub.com/scd
Using this direct comparison between control embryos (low or absent eGFP levels) and Rex1-overexpressing embryos (high eGFP levels), we show an almost 2-fold difference (Fig. 6D, 1.95) in blastocyst development between the 2 groups. While 66.7% of control embryos reached the blastocyst stage, only 34.2% of Rex1-overexpressing embryos did (Fig. 6C). Importantly, using the protocol developed (outlined in detail in the Materials and Methods section), this particular difference in blastocyst development could be observed in every experiment analyzed (n=5; Fig. 6D and Supplementary Table S2), although fold difference varied between 1.5 and 2.9 (mean 1.95; Fig. 6D). We conclude that Rex1 overexpression interferes with blastocyst development.
Marker gene expression associated with Rex1 gain- and loss-of-function
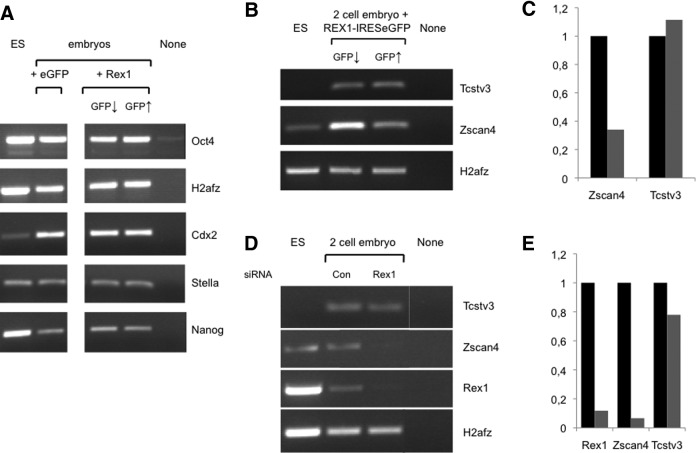
As Rex1 overexpression interfered with proper blastocyst development, we assessed the expression of a variety of lineage and pluripotency markers, that is, Oct4/Pou5f1, Stella, Nanog, and Cdx2, in control embryos (low or absent eGFP levels) and Rex1-overexpressing embryos (high eGFP levels). No marked differences were observed between control and Rex1-overexpressing blastocysts in expression levels of any of the preimplantation markers tested (Fig. 7A). These results suggest that although Rex1 overexpression interferes with proper blastocyst development, TE and ICM lineages are initially established, although not necessarily properly maintained.
FIG. 7.
Gene expression in Rex1 gain- and loss-of-function mouse embryos. (A) One-cell embryos were microinjected (day 1) with plasmids that direct expression of REX1-IRESeGFP or eGFP as a control, and embryos were separated in (GFP↑) or (GFP↓) groups as explained in the legend to Fig. 6A. RNA levels were analyzed by semiquantitative RT-PCR in embryos taken on day 5. RT-cDNA from equivalent amounts of embryos was amplified with primers specific for the genes indicated; H2afz was used as a control for input. The figure shows an ethidium bromide-stained gel scan from a representative experiment. RNA extracted from E14T ES cells was processed alongside as a positive control. The following templates were used for amplification: E14T ES cells (ES), embryos overexpressing eGFP as a control (eGFP), embryos with low or neglectable Rex1 overexpression levels (GFP↓), embryos with high Rex1 overexpression (GFP↑), or Milli-Q water as a negative control (none). (B–E) Deregulation of Zscan4. (B) Embryos were injected on day 1, separated in (GFP↑) and (GFP↓) groups on day 2, and RNA levels were analyzed by semiquantitative RT-PCR in embryos taken on day 2 as described in the legend to Fig. 7A. The figure shows an ethidium bromide-stained gel from 1 representative experiment out of 2 performed. None stands for no cDNA input. (C) Quantification of data shown in (B). Expression levels are represented relative to H2afz levels (gray and black bars for GFP↑and GFP↓, respectively). (B) Embryos were injected with siRNAs and analyzed as described in the legend to Fig. 4B (si). (E) Quantification of data shown in (D). Expression levels are represented relative to H2afz levels (gray and black bars for Rex1 and Control, respectively).
As Rex1 overexpression also slowed down cleavage divisions, we assessed the expression levels of several genes with distinct expression in 2–4-cell-stage embryos, including Zscan4 and Tcstv3 [17,18]. We injected zygotes with the REX1-IRESeGFP overexpression vectors (Fig. 6). Embryos with either high (GFP↑) or low (GFP↓) expression of GFP were selected in the 2-cell stage and assayed by RT-PCR. We observed expression in 2-cell embryos of all markers analyzed (Fig. 7B). Overexpression of Rex1 in these embryos (Fig. 7B) suppressed expression of Zscan4 ∼3-fold (Fig. 7B, C). As a control, Tcstv3 levels were hardly affected by REX1-IRESeGFP overexpression (Fig. 7B, C).
To confirm a potential role for Rex1 in control of Zscan4 expression, we assessed expression levels in Rex1 loss-of-function embryos generated by microinjection of zygotes with siRNAs directed against Rex1 (Fig. 4). Under these conditions, Rex1 mRNA levels were about 9-fold attenuated in 2-cell embryos (Fig. 7D, E). As opposed to Tcstv3, expression of Zscan was downregulated about 10-fold by the injection of siRNAs directed against Rex1 (Fig. 7D, E). We conclude that both attenuation of Rex1 levels and overexpression interfere with Zscan4 expression. These results demonstrate that Rex1 controls expression of Zscan4, a crucial regulator of preimplantation development in the mouse [20].
Discussion
Although Rex1 is best known as a highly specific stem cell marker [27,28,30], previous reports [4,23] have suggested its presence in early-stage embryos. We confirm the presence of Rex1 both at the mRNA and protein level throughout mouse preimplantation development, suggesting a functional role in developmental control at early stages. Our data show that αREX1 immunoreactivity is present in all cells of murine preimplantation embryos at all stages. The extranuclear localization of REX1 we observe (Fig. 2) is not unprecedented. Cytoplasmic/perinuclear staining has been described previously for several chromatin regulators, that is, CBX [41], TBP [42], and most interestingly YY1 [43]. Transcription factors with a strict nuclear distribution in postimplantation embryos may be localized in the cytoplasm earlier, as reported for HP1γ in 8-cell embryos [41] and YY1 in both the ICM and trophoblast of an E3.5 blastocyst [43]. TBP protein also shows nuclear as well as cytoplasmic expression in both the ICM and TE at a similar stage [42].
We observe that αREX1 immunoreactivity displays a combination of cytoplasmic, perinuclear, and nuclear localization, with dynamic changes at various stages. Relocalization of REX1 is observed in 8-cell embryos (Fig. 2), and during blastocyst development in vitro REX1 localization shifts toward a predominantly nuclear localization throughout the embryo (Fig. 3). These changes might coincide with a change in regulatory contributions. In 8-cell embryos, the shift may coincide with downregulation of muERV-L [36], or may be related to the initiation of Tsix expression [44]. On the other hand, in the late blastocyst, nuclear REX1 may contribute to epigenetic modulation of differentiation-specific genes [31] and to regulation of genes coupled to degenerated endogenous retroviral elements (ERV)-derived elements, which we have identified recently (J. Schoorlemmer et al., manuscript in preparation). An intriguing possibility is that in the late-blastocyst Rex1 may contribute to biallelic expression of Nanog [45] in the ICM.
Our results showing absence of major phenotypes in embryos with reduced levels of Rex1 are compatible with previous reports on genetic loss-of-function models. In several mouse models, homozygous Rex1-deficient offspring are viable and fertile, although weak phenotypes are observed. Rex1 phenotypes comprise the birth of a reduced number of Rex1-deficient individuals [32], reduced litter size proportional to the extent of Rex1 deficiency [34], and testicular germ cell defects [35]. Important functions of Rex1 during preimplantation development may be compensated for by Yy1 and Yy2, genes whose zinc fingers are related to Rex1. All 3 Yy1 family members are present during preimplantation development [23,43]. Alternatively, changes at the molecular level caused by Rex1 deficiency may be resolved or compensated without doing harm, in such a way that embryos that manage to escape such a bottleneck may resume development and are no further affected.
In contrast to Rex1 loss-of-function, we show that overexpression of Rex1 interferes with proper preimplantation development. Rex1-overexpressing embryos show developmental delay maybe as early as day 2, delayed entry into the 4-cell stage, and a reduced percentage of embryos progressing to the blastocyst stage (Fig. 5B). Moreover, we were able to directly relate overexpression with aborted blastocyst development (Figs. 5C and 6). As impaired blastocyst development was observed in embryos without previous defects (Fig. 6A, B), Rex1 might exert 2 different and independent functions during cleavage stages and in the blastocyst. We present the earliest molecular phenotype that can be attributed to Rex1 loss- and gain-of-function, which is deregulation of Zscan4 expression. We propose that Rex1 regulates Zscan4 expression by direct binding to so-far unidentified regulatory elements and recruiting LSD1/KDM1A (see below). Although attenuation of Rex1 interferes with Zscan4 expression, (Fig. 7D), phenotypes of Zscan4 loss-of-function [18] are much more severe than those of Rex1 loss-of-function (Supplementary Fig. S4 and Supplementary Table S1). Partial loss of Rex1 might allow for sufficient Zscan4 expression to alleviate the preimplantation phenotype.
In contrast to recovery after cleavage-stage delays, Rex1 overexpression interferes with development into the blastocyst stage, and disintegration occurs later on. Surprisingly, levels of preimplantation (Oct4/Pou5f1 and Stella) or lineage markers (Nanog and Cdx2) appear normal in embryos deformed by Rex1 overexpression (Fig. 7A), indicating that lineage separation is properly initiated. It will be important to determine the amount of Nanog-positive cells in GFP+/GFP− cells. Preliminary results show that in the blastocyst mitosis might be affected under our experimental conditions, similar to results obtained in ES cells (D. Guallar, M. Climent et al., unpublished data).
In the absence of hard evidence, we can only speculate regarding the physiological relevance of the gene regulation defects caused by Rex1 overexpression that we have observed. We do note that, however, both in the case of muERV-L [36] and Zscan (Fig. 7), overexpression and attenuation of Rex1 levels affect the same target genes. In accordance with the presence of Rex1 throughout preimplantation development, we demonstrate changes in gene expression caused by overexpression of Rex1 at different time points: in 2-cell embryos (Zscan4, Fig. 7B) and in 8-cell embryos (muERV-L, [36]). Separately, we show that Rex1 controls the expression of a group of genes regulated through cis-acting degenerated ERV-derived elements during blastocyst development (J. Schoorlemmer et al., manuscript in preparation). It is presently unknown whether the molecular phenotypes we observe at different stages are the result of a cascade of instructive events that necessarily follow each other. Alternatively, they might represent independent regulation events controlled by localization or by stage-dependent processes such as the presence of cofactors and target access.
Furthermore, it remains an open question whether any of these deregulated genes may directly interfere with blastocyst development. Considering the interplay between Rex1 and LSD1 in the regulation of muERV-L expression [36], we have initiated studies to determine lineage determination and fixation defects in Rex1-overexpressing embryos similar to those caused by LSD1 deficiency [46]. We have recently shown that REX1 can interact with LSD1 [36], raising the intriguing possibility that REX1 titrates LSD1 levels. The observation that the Rex1 target Zscan4 is transiently induced during ZGA raises the possibility that Rex1, its close relatives Yy1 and Yy2, as well as interacting proteins Trim28 (TIF1/Kap1) and LSD1 may play roles in ZGA. Although suggested previously [47], the importance of gene regulation through degenerated ERV-derived elements during these stages may be even more pervasive than appreciated so far. Irrespective of the exact molecular events underlying the Rex1 overexpression phenotype, we believe that our results reinforce the notion that Rex1 function is relevant during preimplantation development.
Supplementary Material
Acknowledgments
We thank Ignacio García-Tuñon for initial experiments with REX1-GFP. We acknowledge technical assistance at several core facilities at I+CS: Ana Benítez and David García-Domingo (UCC) for tissue culture, María Royo (UMIG) for additional confocal microscopy, and Javier Godino (USC) for citometry. Jon Schoorlemmer wishes to express his gratitude toward the Departamento de Anatomía, Embriología y Genética Animal, Facultad de Veterinaria, Universidad de Zaragoza, for its hospitality. Work in the authors' laboratory has been supported by the grant PI07119 from FIS/CarlosIII, Spanish Ministry of Health; grant PI110/09 from the Dept. de Ciencia, Tecnología y Universidad, DGA, Spain; and PAMER grants, Aragon Health Sciences Institute, Spain.
Author Disclosure Statement
The authors declare that no competing financial interests exist.
References
- 1.Edwards RG. Aspects of the molecular regulation of early mammalian development. Reprod Biomed Online. 2003;6:97–113. doi: 10.1016/s1472-6483(10)62061-5. [DOI] [PubMed] [Google Scholar]
- 2.Rossant J. Stem cells from the mammalian blastocyst. Stem Cells. 2001;19:477–482. doi: 10.1634/stemcells.19-6-477. [DOI] [PubMed] [Google Scholar]
- 3.Johnson MH. McConnell JM. Lineage allocation and cell polarity during mouse embryogenesis. Semin Cell Dev Biol. 2004;15:583–597. doi: 10.1016/j.semcdb.2004.04.002. [DOI] [PubMed] [Google Scholar]
- 4.Hamatani T. Carter MG. Sharov AA. Ko MS. Dynamics of global gene expression changes during mouse preimplantation development. Dev Cell. 2004;6:117–131. doi: 10.1016/s1534-5807(03)00373-3. [DOI] [PubMed] [Google Scholar]
- 5.Evans MJ. Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- 6.Martin GR. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc Natl Acad Sci U S A. 1981;78:7634–7638. doi: 10.1073/pnas.78.12.7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nichols J. Zevnik B. Anastassiadis K. Niwa H. Klewe-Nebenius D. Chambers I. Schöler H. Smith A. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell. 1998;95:379–391. doi: 10.1016/s0092-8674(00)81769-9. [DOI] [PubMed] [Google Scholar]
- 8.Avilion A. Nicolis S. Pevny L. Perez L. Vivian N. Lovell-Badge R. Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev. 2003;17:126–140. doi: 10.1101/gad.224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chambers I. Colby D. Robertson M. Nichols J. Lee S. Tweedie S. Smith A. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell. 2003;113:643–655. doi: 10.1016/s0092-8674(03)00392-1. [DOI] [PubMed] [Google Scholar]
- 10.Mitsui K. Tokuzawa Y. Itoh H. Segawa K. Murakami M. Takahashi K. Maruyama M. Maeda M. Yamanaka S. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell. 2003;113:631–642. doi: 10.1016/s0092-8674(03)00393-3. [DOI] [PubMed] [Google Scholar]
- 11.Takahashi K. Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 12.Chen X. Xu H. Yuan P. Fang F. Huss M. Vega VB. Wong E. Orlov YL. Zhang W, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- 13.Yoshikawa T. Piao Y. Zhong J. Matoba R. Carter MG. Wang Y. Goldberg I. Ko MS. High-throughput screen for genes predominantly expressed in the ICM of mouse blastocysts by whole mount in situ hybridization. Gene Expr Patterns. 2006;6:213–224. doi: 10.1016/j.modgep.2005.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang J. Gao W. Yang H. Zhang B. Zhu Z. Xue Y. Screening for genes essential for mouse embryonic stem cell self-renewal using a subtractive RNA interference library. Stem Cells. 2006;24:2661–2668. doi: 10.1634/stemcells.2006-0017. [DOI] [PubMed] [Google Scholar]
- 15.Hu G. Kim J. Xu Q. Leng Y. Orkin SH. Elledge SJ. A genome-wide RNAi screen identifies a new transcriptional module required for self-renewal. Genes Dev. 2009;23:837–848. doi: 10.1101/gad.1769609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hemberger M. Dean W. Reik W. Epigenetic dynamics of stem cells and cell lineage commitment: digging Waddington's canal. Nat Rev Mol Cell Biol. 2009;10:526–537. doi: 10.1038/nrm2727. [DOI] [PubMed] [Google Scholar]
- 17.Zhang W. Walker E. Tamplin OJ. Rossant J. Stanford WL. Hughes TR. Zfp206 regulates ES cell gene expression and differentiation. Nucleic Acids Res. 2006;34:4780–4790. doi: 10.1093/nar/gkl631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Falco G. Lee SL. Stanghellini I. Bassey UC. Hamatani T. Ko MS. Zscan4: a novel gene expressed exclusively in late 2-cell embryos and embryonic stem cells. Dev Biol. 2007;307:539–550. doi: 10.1016/j.ydbio.2007.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carter MG. Stagg CA. Falco G. Yoshikawa T. Bassey UC. Aiba K. Sharova LV. Shaik N. Ko MS. An in situ hybridization-based screen for heterogeneously expressed genes in mouse ES cells. Gene Expr Patterns. 2008;8:181–198. doi: 10.1016/j.gep.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zalzman M. Falco G. Sharova LV. Nishiyama A. Thomas M. Lee SL. Stagg CA. Hoang HG. Yang HT, et al. Zscan4 regulates telomere elongation and genomic stability in ES cells. Nature. 2010;464:858–863. doi: 10.1038/nature08882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim J. Faulk C. Kim J. Retroposition and evolution of the DNA-binding motifs of YY1, YY2 and REX1. Nucleic Acids Res. 2007;35:3442–3452. doi: 10.1093/nar/gkm235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hosler B. LaRosa G. Grippo J. Gudas L. Expression of REX-1, a gene containing zinc finger motifs, is rapidly reduced by retinoic acid in F9 teratocarcinoma cells. Mol Cell Biol. 1989;9:5623–5629. doi: 10.1128/mcb.9.12.5623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rogers M. Hosler B. Gudas L. Specific expression of a retinoic acid-regulated, zinc-finger gene, Rex-1, in preimplantation embryos, trophoblast and spermatocytes. Development. 1991;113:815–824. doi: 10.1242/dev.113.3.815. [DOI] [PubMed] [Google Scholar]
- 24.Jiang Y. Jahagirdar B. Reinhardt R. Schwartz R. Keene C. Ortiz-Gonzalez X. Reyes M. Lenvik T. Lund T, et al. Pluripotency of mesenchymal stem cells derived from adult marrow. Nature. 2002;418:41–49. doi: 10.1038/nature00870. [DOI] [PubMed] [Google Scholar]
- 25.Karlmark K. Freilinger A. Marton E. Rosner M. Lubec G. Hengstschläger M. Activation of ectopic Oct-4 and Rex-1 promoters in human amniotic fluid cells. Int J Mol Med. 2005;16:987–992. [PubMed] [Google Scholar]
- 26.Kristensen DM. Nielsen JE. Skakkebaek NE. Graem N. Jacobsen GK. Rajpert-De Meyts E. Leffers H. Presumed pluripotency markers UTF-1 and REX-1 are expressed in human adult testes and germ cell neoplasms. Hum Reprod. 2008;23:775–782. doi: 10.1093/humrep/den010. [DOI] [PubMed] [Google Scholar]
- 27.Okita K. Ichisaka T. Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313–317. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- 28.Toyooka Y. Shimosato D. Murakami K. Takahashi K. Niwa H. Identification and characterization of subpopulations in undifferentiated ES cell culture. Development. 2008;135:909–918. doi: 10.1242/dev.017400. [DOI] [PubMed] [Google Scholar]
- 29.Brivanlou A. Gage F. Jaenisch R. Jessell T. Melton D. Rossant J. Stem cells. Setting standards for human embryonic stem cells. Science. 2003;300:913–916. doi: 10.1126/science.1082940. [DOI] [PubMed] [Google Scholar]
- 30.Chan E. Ratanasirintrawoot S. Park I. Manos P. Loh Y. Huo H. Miller J. Hartung O. Rho J, et al. Live cell imaging distinguishes bona fide human iPS cells from partially reprogrammed cells. Nat Biotechnol. 2009;27:1033–1037. doi: 10.1038/nbt.1580. [DOI] [PubMed] [Google Scholar]
- 31.Scotland K. Chen S. Sylvester R. Gudas L. Analysis of Rex1 (zfp42) function in embryonic stem cell differentiation. Dev Dyn. 2009;238:1863–1877. doi: 10.1002/dvdy.22037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Masui S. Ohtsuka S. Yagi R. Takahashi K. Ko M. Niwa H. Rex1/Zfp42 is dispensable for pluripotency in mouse ES cells. BMC Dev Biol. 2008;8:45. doi: 10.1186/1471-213X-8-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Garcia-Tuñon I. Guallar D. Alonso-Martin S. Benito AA. Benítez-Lázaro A. Pérez-Palacios R. Muniesa P. Climent M. Sánchez M. Vidal M. Schoorlemmer J. Association of Rex-1 to target genes supports its interaction with polycomb function. Stem Cell Res. 2011;7:1–16. doi: 10.1016/j.scr.2011.02.005. [DOI] [PubMed] [Google Scholar]
- 34.Kim JD. Kim H. Ekram MB. Yu S. Faulk C. Kim J. Rex1/Zfp42 as an epigenetic regulator for genomic imprinting. Hum Mol Genet. 2011;20:1353–1362. doi: 10.1093/hmg/ddr017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rezende NC. Lee MY. Monette S. Mark W. Lu A. Gudas LJ. Rex1 (Zfp42) null mice show impaired testicular function, abnormal testis morphology, and aberrant gene expression. Dev Biol. 2011;356:370–382. doi: 10.1016/j.ydbio.2011.05.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guallar D. Pérez-Palacios R. Climent M. Martínez-Abadía I. Larraga A. Fernández-Juan M. Vallejo C. Muniesa P. Schoorlemmer J. Expression of endogenous retroviruses is negatively regulated by the pluripotency marker Rex1/Zfp42. Nucleic Acids Res. 20122012 doi: 10.1093/nar/gks686. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Torres-Padilla M. Bannister A. Hurd P. Kouzarides T. Zernicka-Goetz M. Dynamic distribution of the replacement histone variant H3.3 in the mouse oocyte and preimplantation embryos. Int J Dev Biol. 2006;50:455–461. doi: 10.1387/ijdb.052073mt. [DOI] [PubMed] [Google Scholar]
- 38.Niwa H. Burdon T. Chambers I. Smith A. Self-renewal of pluripotent embryonic stem cells is mediated via activation of STAT3. Genes Dev. 1998;12:2048–2060. doi: 10.1101/gad.12.13.2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nagy A. Gertsenstein M. Vintersten K. Behringer R. Manipulating the Mouse Embryo: A Laboratory Manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2003. [Google Scholar]
- 40.Zeng F. Baldwin DA. Schultz RM. Transcript profiling during preimplantation mouse development. Dev Biol. 2004;272:483–496. doi: 10.1016/j.ydbio.2004.05.018. [DOI] [PubMed] [Google Scholar]
- 41.Ruddock-D'Cruz N. Prashadkumar S. Wilson K. Heffernan C. Cooney M. French A. Jans D. Verma P. Holland M. Dynamic changes in localization of Chromobox (Cbx) family members during the maternal to embryonic transition. Mol Reprod Dev. 2008;75:477–488. doi: 10.1002/mrd.20752. [DOI] [PubMed] [Google Scholar]
- 42.Worrad DM. Ram PT. Schultz RM. Regulation of gene expression in the mouse oocyte and early preimplantation embryo: developmental changes in Sp1 and TATA box-binding protein, TBP. Development. 1994;120:2347–2357. doi: 10.1242/dev.120.8.2347. [DOI] [PubMed] [Google Scholar]
- 43.Donohoe M. Zhang X. McGinnis L. Biggers J. Li E. Shi Y. Targeted disruption of mouse Yin Yang 1 transcription factor results in peri-implantation lethality. Mol Cell Biol. 1999;19:7237–7244. doi: 10.1128/mcb.19.10.7237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Navarro P. Oldfield A. Legoupi J. Festuccia N. Dubois A. Attia M. Schoorlemmer J. Rougeulle C. Chambers I. Avner P. Molecular coupling of Tsix regulation and pluripotency. Nature. 2010;468:457–460. doi: 10.1038/nature09496. [DOI] [PubMed] [Google Scholar]
- 45.Miyanari Y. Torres-Padilla ME. Control of ground-state pluripotency by allelic regulation of Nanog. Nature. 2012;483:470–473. doi: 10.1038/nature10807. [DOI] [PubMed] [Google Scholar]
- 46.Macfarlan TS. Gifford WD. Agarwal S. Driscoll S. Lettieri K. Wang J. Andrews SE. Franco L. Rosenfeld MG. Ren B. Pfaff SL. Endogenous retroviruses and neighboring genes are coordinately repressed by LSD1/KDM1A. Genes Dev. 2011;25:594–607. doi: 10.1101/gad.2008511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Peaston AE. Evsikov AV. Graber JH. de Vries WN. Holbrook AE. Solter D. Knowles BB. Retrotransposons regulate host genes in mouse oocytes and preimplantation embryos. Dev Cell. 2004;7:597–606. doi: 10.1016/j.devcel.2004.09.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.