Summary
Among trypanosomatid protozoa the mechanism of RNA interference (RNAi) has been investigated in Trypanosoma brucei and to a lesser extent in Leishmania braziliensis. Although these two parasitic organisms belong to the same family, they are evolutionarily distantly related raising questions about the conservation of the RNAi pathway. Here we carried out an in-depth analysis of small interfering RNAs (siRNAs) associated with L. braziliensis Argonaute1 (AGO1). In contrast to T. brucei, Leishmania siRNAs are sensitive to 3’-end oxidation, indicating the absence of blocking groups, and the Leishmania genome does not code for a HEN1 RNA 2’-O-methyltransferase, which modifies small RNA 3’ends. Consistent with this observation, ~20% of siRNA 3’ ends carry non-templated uridine. Thus siRNA biogenesis, and most-likely their metabolism, is different in these organisms. Similarly to T. brucei, putative mobile elements and repeats constitute the major Leishmania siRNA-producing loci and AGO1 ablation leads to accumulation of long transcripts derived from putative mobile elements. However, contrary to T. brucei, no siRNAs were detected from other genomic regions with the potential to form double-stranded RNA, namely sites of convergent transcription and inverted repeats. Thus, our results indicate that organism-specific diversification has occurred in the RNAi pathway during evolution of the trypanosomatid lineage.
Keywords: Leishmania, Argonaute, retrotransposon, tandem repeat, convergent transcription unit, siRNA
Introduction
Gene silencing by small regulatory RNAs has been one of the major discoveries of the last two decades and has profoundly changed our view how gene expression is controlled in eukaryotes, shifting the emphasis from transcriptional and epigenetic regulation to post-transcriptional mechanisms. This field has been built on four fundamental discoveries. First, in the early 90’s genetic analysis in Caenorhabditis elegans brought to light two small non-coding RNAs, which regulate larval development (Lee et al., 1993, Wightman et al., 1993) and are the founders of the micro RNA (miRNA) family. Second, in 1998 Fire and Mello (Fire et al., 1998) made the seminal discovery that long double-stranded RNA (dsRNA) triggers degradation of homologous transcripts, a phenomenon dubbed RNA interference (RNAi). Third, Baulcombe and colleagues (Hamilton & Baulcombe, 1999) correlated post-transcriptional gene silencing (PTGS) in plants with the accumulation of a ~25 nt antisense RNA species, which they proposed “may represent the specificity determinant of PTGS”. Fourth, these small RNAs, which became known as “small/short interfering RNA (siRNAs), were shown to be the mediators of RNAi (Elbashir et al., 2001).
After two decades of research it has become evident that small regulatory RNAs come in a variety of types, some of which are organism-specific, with the main known classes being siRNAs, miRNAs and Piwi-associated RNAs or piRNAs (Kim et al., 2009, Liu & Paroo, 2010) siRNAs are generated by cleavage of long dsRNA by Dicer, an RNase III-family endonuclease in a complex with a dsRNA binding protein. The processing of miRNAs from their primary transcripts is more elaborate and in general requires the assistance of the microprocessor in the nucleus followed by export of precursors to the cytoplasm and cleavage by Dicer. However, in certain instances miRNA maturation can occur independently of Dicer (Cifuentes et al., 2010, Cheloufi et al., 2010, Yang et al., 2010). So far miRNAs have been shown to be derived from long, primary transcripts coding for multiple miRNAs, from introns or from other cellular RNAs, including the small nucleolar RNAs (Ender et al., 2008, Saraiya & Wang, 2008). Additionally, tRNAs have been implicated as sources of miRNAs (Cole et al., 2009, Lee et al., 2009, Haussecker et al., 2010).
Invariably, small regulatory RNAs are found in a complex with a member of the Argonaute (AGO) protein family (Ender & Meister, 2010), which is characterized by conserved domains (MID, PAZ and Piwi). Certain AGO family members have been termed “slicers” because they are endowed with an RNase H-like cleavage activity, which resides in the Piwi domain. siRNAs are associated with AGO slicers, are usually perfectly complementary to the target transcript and, upon binding of the ribonucleoprotein complex, target transcript cleavage ensues. In contrast, most miRNAs are only partially complementary to their targets and elicit down-regulation of gene expression by accelerating transcript decay and/or inhibiting translation (Guo et al., 2010, Fabian et al., 2010, Djuranovic et al., 2012, Bazzini et al., 2012).
The endogenous RNAi pathway triggered by long dsRNA is by and large a genome defense mechanism to control spreading of potentially dangerous parasitic nucleic acids, such as mobile elements, repeats and viruses (Huang et al., 2011). However, in the last few years deep sequencing of endogenous siRNAs has revealed further complexities of the siRNA repertoire with the identification of loci that upon transcription give rise to structured RNAs (long inverted repeats) or sense and antisense transcripts with the potential to form dsRNA, as it can occur at sites of convergent transcription. However, the functional significance of siRNAs from long hairpins and convergent transcription is at present unclear. Furthermore, siRNAs derived from the pairing of gene and pseudogene transcripts have been implicated in regulating gene expression in certain organisms (Watanabe et al., 2008), including Trypanosoma brucei (Wen et al., 2011).
Members of the order Kinetoplastida, which includes many human protozoan parasites, are considered early divergent eukaryotes and thus can inform us about ancient traits of the RNAi pathway. T. brucei, the causative agent of African sleeping sickness in humans and nagana in cattle, was the first member of this group where RNAi was experimentally demonstrated (Ngo et al., 1998). More recently, RNAi has been shown to be functional in Leishmania Viannia braziliensis and other members of the Viannia subgenus (Lye et al., 2010), the cause of cutaneous and mucocutaneous leishmaniasis in South America. Additionally, Crithidia fasciculata, a non-parasitic ‘outgroup’, is endowed with RNAi genes and generates siRNA-size molecules upon expression of a green fluorescent protein (GFP) hairpin (Lye et al., 2010).
In T. brucei the RNAi pathway depends on five core components, which are also present in L. braziliensis (Lye et al., 2010, Barnes et al., 2012), namely a single Argonaute, AGO1 (Shi et al., 2004, Durand-Dubief & Bastin, 2003), two Dicer-like enzymes, the cytoplasmic DCL1 (Shi et al., 2006) and the nuclear DCL2 (Patrick et al., 2009), and two novel factors termed RIF4, an exonuclease that together with AGO1 is required for the maturation of siRNAs from duplex to single-stranded form, and RIF5, a DCL1 cofactor (Barnes et al., 2012). Trypanosome siRNAs are produced by both Dicers and are characterized by a 5’ phosphate and a blocked 3’ end (Patrick et al., 2009). Past (Djikeng et al., 2001) and recent (Tschudi et al., 2012) siRNA profiling in T. brucei procyclic forms indicates that the endogenous siRNA-producing loci have been conserved from trypanosomes to animals and include retroposons, satellite-like repeats, long inverted repeats, sites of convergent transcription and two pseudogenes.
Although T. brucei and L. braziliensis belong to the same family and have conserved mechanisms of gene expression, including polycistronic transcription of long gene arrays and trans-splicing of pre-mRNA, these organisms are evolutionarily distantly related (Fernandes et al., 1993), raising the question whether the structure of siRNAs and the scope of the RNAi pathway have been conserved through the evolution of the trypanosomatid lineage. Here, we have undertaken an in-depth analysis of siRNAs in L. braziliensis promastigotes. Structurally, Leishmania siRNAs are slightly shorter than those in T. brucei and, importantly, are not modified at the 3’ end. As in T. brucei (Djikeng et al., 2001, Tschudi et al., 2012) the major classes of Leishmania siRNAs are derived from putative mobile elements and repeats, namely the SLACS retroposon (Spliced Leader Associated Conserved Sequence; (Peacock et al., 2007) and the transposable element TATE (Telomere-Associated Transposable Element; (Peacock et al., 2007)), TAS-like sequences (L. braziliensis-specific Telomere Associated Sequence; (Fu & Barker, 1998, Fu et al., 1998) and a family of 74-nucleotide long tandem repeats, termed CIR74 (Chromosomal Internal Repeats, 74-nucleotide long; this paper). Differently from T. brucei we did not find evidence that transcripts from convergent transcription units (CTUs) are a source of siRNAs. Bioinformatics analysis revealed that approximately 20% of siRNAs are subject to tailing at the 3’ end, with uridine being the most common addition. Thus, the T. brucei and L. braziliensis RNAi pathways show organism-specific diversifications with implications for the production of dsRNA and siRNA metabolism.
Results
Structure of Leishmania braziliensis siRNAs
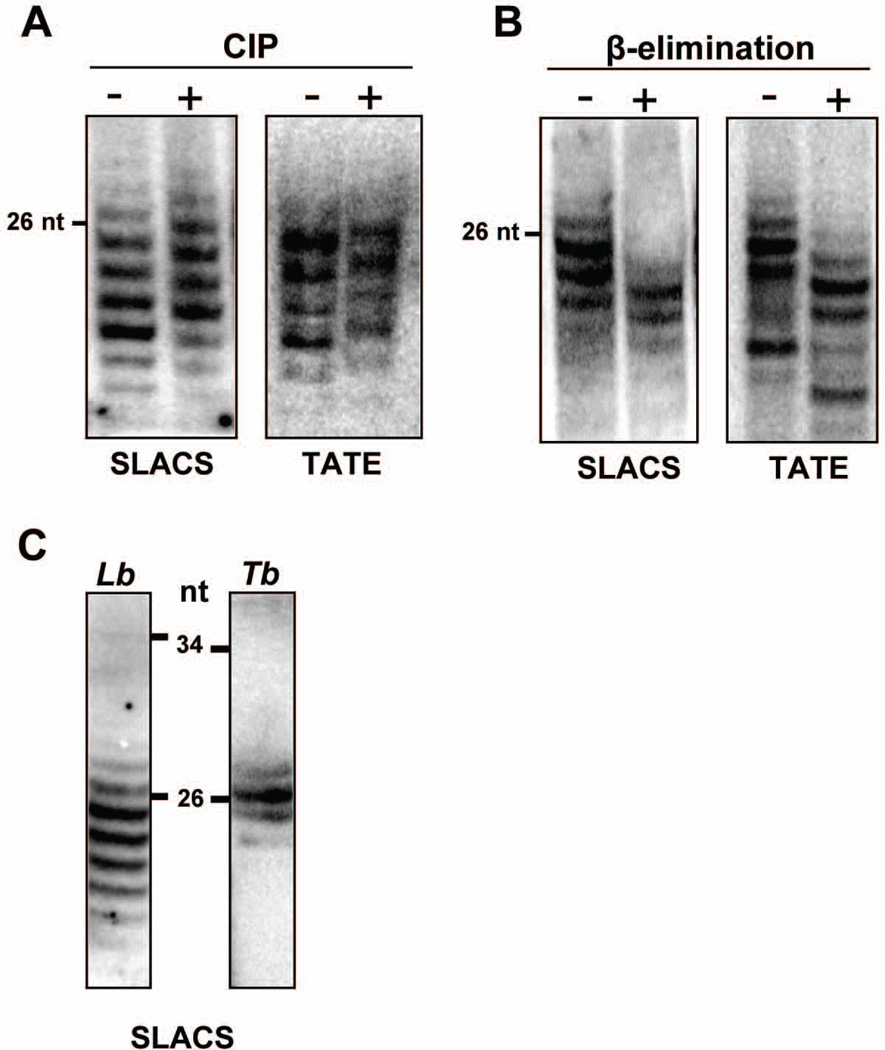
Previous work from our laboratories showed that L. braziliensis accumulates 20–25 nt small RNAs originating from the retroposon SLACS and a GFP hairpin transgene (Lye et al., 2010). To corroborate that these molecules are indeed siRNAs, we analyzed the 5’ and 3’ terminus of L. braziliensis small RNAs by enzymatic and chemical treatment, respectively. siRNAs generated by Dicer cleavage are expected to carry a 5’ phosphate group. Thus, we exposed total low molecular weight RNAs to calf intestine phosphatase (CIP), which removes the 5’ phosphate. The relative mobility of treated and untreated RNAs was then revealed by electrophoresis on a sequencing gel followed by Northern hybridization with probes for SLACS and TATE, the two major classes of known L. braziliensis putative transposable elements. After CIP treatment (Fig. 1A), SLACS- and TATE-derived small RNAs showed a slightly diminished electrophoretic mobility as compared to the untreated sample, consistent with the presence of a phosphate group at the 5’ end.
Fig. 1. Structure of L. braziliensis siRNAs.
A. Low molecular weight RNAs were treated with calf intestine phosphatase (CIP) or B. submitted to periodate oxidation/ β-elimination, fractionated on a 15% sequencing gel and analyzed by Northern blotting with SLACS or TATE oligonucleotide probes.
C. L. braziliensis or T. brucei low molecular weight RNAs were fractionated as described above and analyzed by Northern blotting with species-specific SLACS oligonucleotide probes. The samples were run on two separate gels and the lanes aligned using the positions of the molecular weight markers. The positions of relevant 5’ end-labeled pBR322/MspI fragments are indicated.
Cleavage of dsRNA by Dicer produces a 3’ OH, but in a number of organisms, including T. brucei (Patrick et al., 2009), the 2’-O position of the ribose of certain siRNAs and miRNAs is modified by the addition of a methyl group, a reaction carried out by a member of the HEN1 methyltransferase family (Ji & Chen, 2012). As a result, these small RNAs become resistant to periodate oxidation and β-elimination. Fig. 1B shows that this chemical treatment resulted in an increased mobility of SLACS and TATE small RNAs, which is expected if the siRNAs are unmodified. Thus, on the basis of the above observations and the association of small RNAs with LbrAGO1 (see below), we concluded that these small RNAs represent authentic siRNAs, and, in contrast to T. brucei (Patrick et al., 2009), they are not modified at the 3’ end. Lastly, by Northern blot analysis we compared the size distribution of SLACS siRNAs in T. brucei and L. braziliensis and found that Leishmania siRNAs were more heterogeneous in size and on average slightly shorter than the trypanosome ones (Fig. 1C).
Epitope-tagged LbrAGO1 complements LbrAGO1 deficiency and is in a complex with single-stranded siRNAs
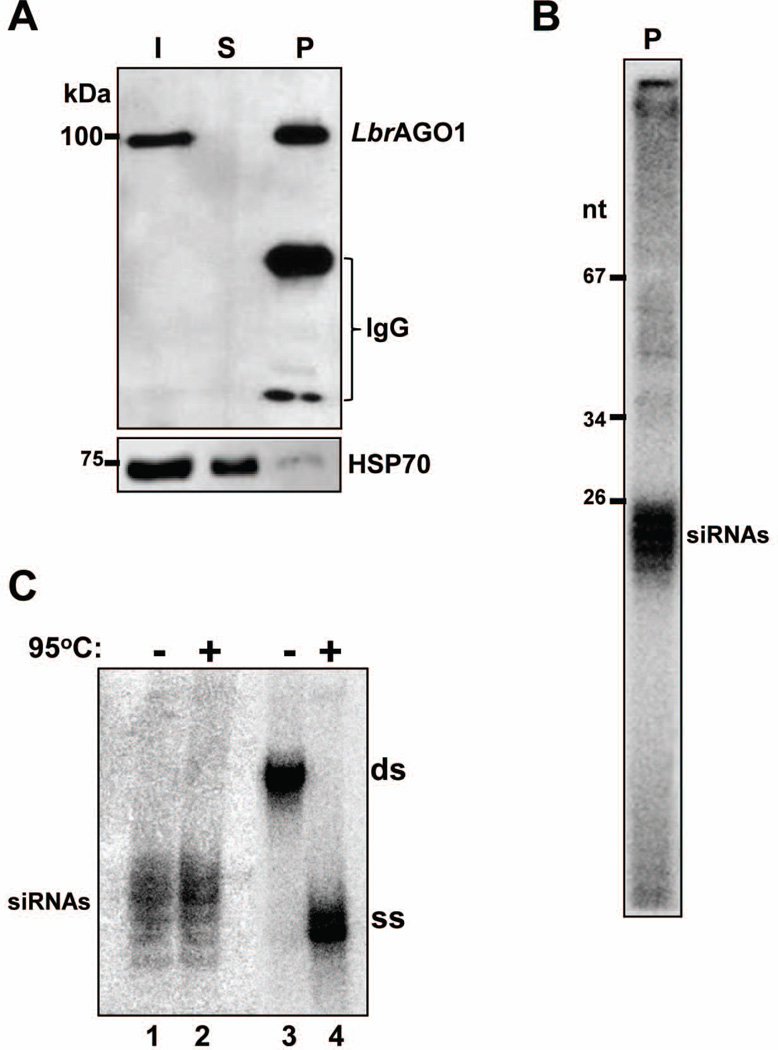
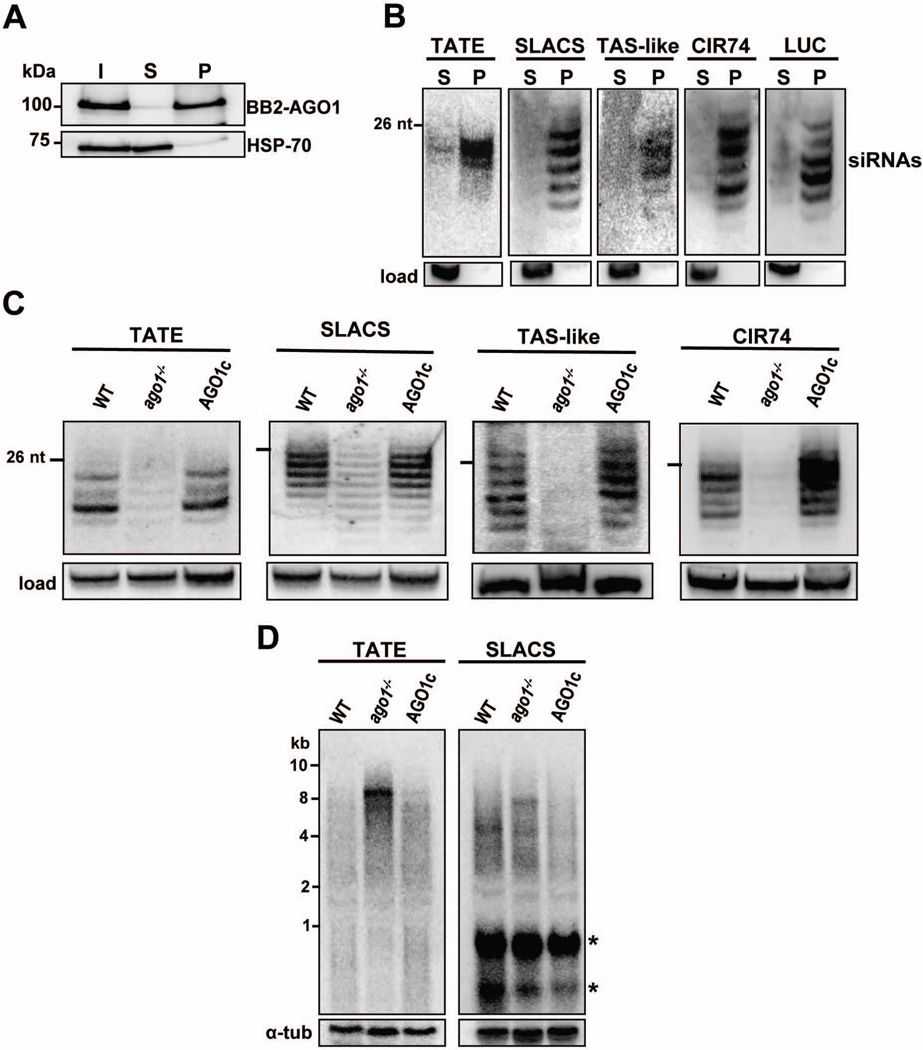
To analyze the repertoire of L. braziliensis siRNAs associated with AGO1, we modified a promastigote ago1−/− cell line by introducing a BB2-LbrAGO1 cassette carrying the BB2 epitope (Bastin et al., 1996) at the N-terminus of AGO1 (LbrAGO1c cell line; see Experimental Procedures). Additionally, the LbrAGO1c cell line expressed a firefly luciferase (LUC) gene and a homologous hairpin. In preliminary experiments we found that, similarly to T. brucei (Djikeng et al., 2003), most BB2-LbrAGO1 was soluble after centrifugation of a detergent extract for 1 hr at 100,000×g, with a small amount (<10%) co-sedimenting with the polyribosome pellet (data not shown). Thus, to purify BB2-LbrAGO1 a 100,000×g supernatant (S100) was subjected to affinity purification using the BB2 monoclonal antibody bound to Protein-G Sepharose beads. The efficiency and specificity of the selection was evaluated by Western blot analysis (Fig. 2A), and the presence of siRNAs in the immunoprecipitate was revealed by 5’ end labeling of the RNA followed by electrophoresis on a sequencing gel (Fig. 2B). Lastly, we verified that siRNAs associated with BB2-LbrAGO1 were in single-stranded form by examining their electrophoretic mobility on a native polyacrylamide gel (Fig. 2C). By this method duplex molecules display retarded mobility as compared to single-stranded forms, but acquire single-strand mobility upon denaturation of the sample before electrophoresis, as illustrated by the control synthetic 25-nt duplex oligonucleotide (lanes 3 and 4). By this analysis the majority of BB2-LbrAGO1-associated SLACS siRNAs were single-stranded (lanes 1 and 2).
Fig. 2. siRNAs co-purify with BB2-LbrAGO1 and are mostly single-stranded.
A. Pull-down of BB2-LbrAGO1 from LbrAGO1c soluble extract. Aliquots of the input (I), supernatant (S) or pellet (P), each representing 107 promastigote cells, were fractionated on a 10% SDS-PAGE and Western blotted with anti-BB2 or anti-HSP70 monoclonal antibody.
B. The immunoprecipitated BB2-LbrAGO1 material was deproteinized, radiolabeled at the 5’ end and fractionated on a 20% sequencing gel.
C. Purified RNA from the BB2-LbrAGO1 immunoprecipitate was electrophoresed on a 15% native polyacrylamide gel with (lane 1) or without (lane 2) heating at 95°C before loading. The siRNAs were revealed by hybridization to a SLACS oligonucleotide probe. As a control for single- and double-stranded migration, we used a 32P-labeled synthetic 25 nt double-stranded RNA (lanes 3 and 4). ss, single-stranded; ds, double-stranded.
The siRNA repertoire
To examine the repertoire of L. braziliensis siRNAs, we generated two independent libraries of BB2-LbrAGO1-associated siRNAs, using the established phosphate-dependent method, which preferentially identifies molecules with a 5’ phosphate group (see supporting information). The libraries were constructed by ligation of Illumina proprietary adaptors, followed by limited PCR amplification and sequencing of the resulting material on the Illumina Genome Analyzer IIx Platform. Since upon examining the various classes of siRNAs (see below) the two libraries gave essentially the same results (Fig. S1A), we will present the analysis of the second library because it had the highest number of reads (The sequence data from this study have been submitted to the NCBI Sequence Read Archive - SRA at http://www.ncbi.nlm.nih.gov/Traces/sra/sra.cgi - under accession no. SRA059332). We obtained 27,868,280 RNA-Seq reads, which were pre-processed by clipping the 3’ adapter (5’-CTGTAGGCACCATCAAT-3’) and then aligned to the 35 L. braziliensis chromosomes of clone M2904 (MHOM/BR/75M2904) using the Bowtie Short Read Aligner (Langmead et al., 2009). Reads with more than two mismatches in the first 22 nucleotides were discarded and reads reporting multiple alignments were retained and aligned pseudo-randomly (using Bowtie’s default options), resulting in 20,710,782 (74.3%) aligned reads. The reads not matching might arise from un-sequenced regions of the genome or from sequencing errors. The alignments were uploaded onto a customized local installation of the generic Genome Browser (Stein et al., 2002).
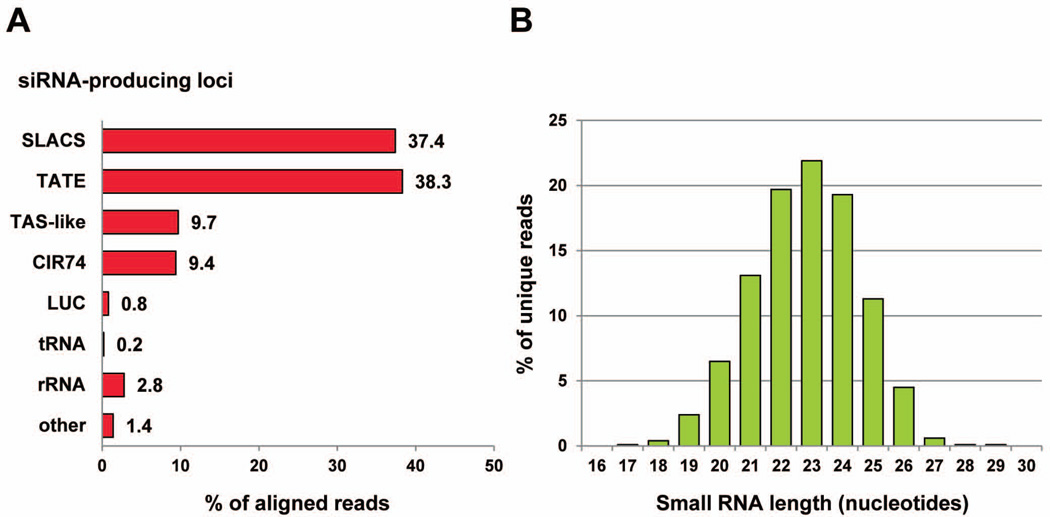
To categorize siRNA-producing loci we manually inspected each chromosome for reads that aligned to annotated features as well as to regions without annotation. In addition, we surveyed short tandem repeats, which were catalogued using the program tandem repeats finder (Benson, 1999) and regions in-between convergent transcription units (CTUs). We also searched individual chromosomes for the presence of long inverted repeats (longer than 100 nt) using BLAST and the program inverted repeat finder (Warburton et al., 2004) and identified numerous regions ranging in size between 100 and 1,800 nt. It is unknown whether or not hairpin RNA structures are generated from these inverted repeats. However, we did not find small RNA reads mapping to these regions of the genome suggesting that they do not form RNA hairpins recognized by the Lbr RNAi machinery. Our analysis identified four abundant categories of endogenous siRNA-producing loci (Fig. 3A), which included SLACS, TATE, TAS-like repeats related to the previously identified TAS family (Fu et al., 1998, Fu & Barker, 1998), and the CIR74 family, a novel class of tandem repeats that are found on chromosomes 2 and 27 in sub-telomeric and internal regions, respectively. In addition, as mentioned above LbrAGO1c cells stably express a LUC hairpin, and siRNAs derived from this transgene represented 0.8% of the reads. Together, the above four classes plus the LUC-derived small RNA reads accounted for 95.6% of the aligned reads. Similarly to other small RNA libraries, our collection also contained reads derived from tRNAs, rRNAs and a class termed “other” (Fig. 3A), consisting of reads from abundant small nuclear and nucleolar RNAs, mRNA coding regions and reads dispersed throughout the genome. These reads were likely to represent degradation products and were not analyzed further.
Fig. 3. The L. braziliensis siRNA repertoire.
A. RNA-Seq reads were aligned to the L. braziliensis genome version 2.0 allowing up to 2 mismatches and categorized into various classes as indicated in the diagram. “Other” includes snoRNAs, snRNAs, mRNAs and unidentified reads.
B. Size distribution of unique reads that aligned to the L. braziliensis genome after trimming the 3’ adapter and subtracting putative degradation products.
Next, to eliminate redundancy the reads were collapsed resulting in 1,879,394 unique sequences with approximately 87% of the reads ranging in size between 21 and 25 nt (Fig. 3B). A similar size distribution was observed for each of the four major siRNA classes listed in Fig. 3A and for the siRNAs derived from the LUC hairpin transgene (Fig. S1B).
Transposable elements
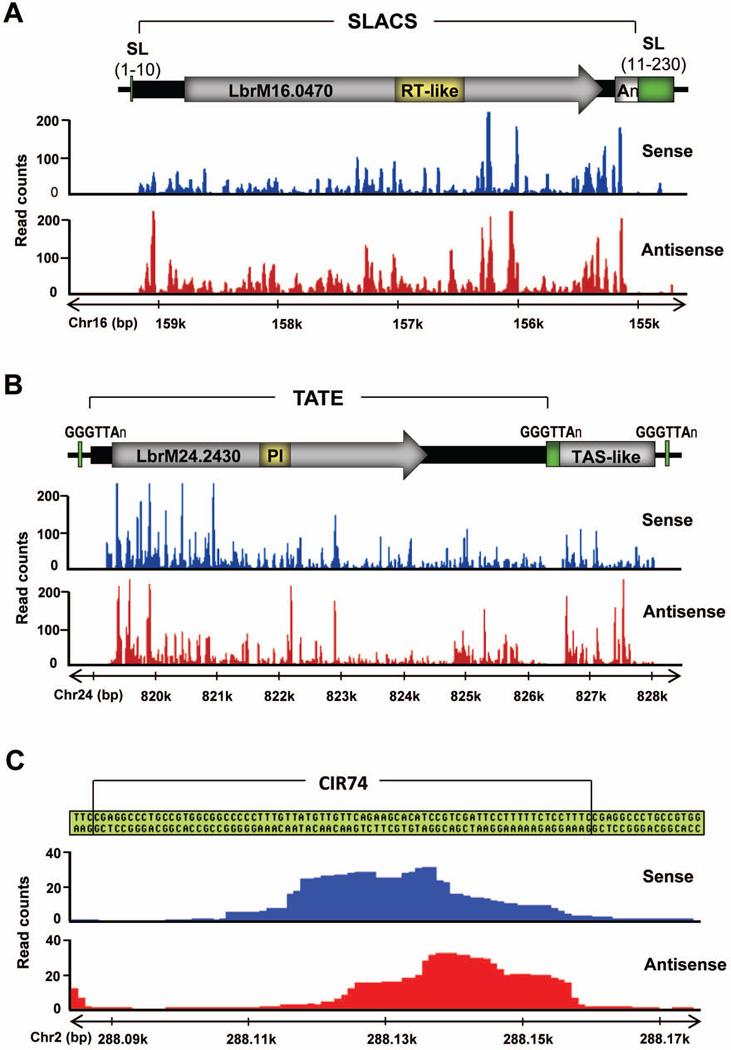
SLACS and TATE transposable elements constituted the majority of siRNAs bound to LbrAGO1, representing ~75% of the siRNA repertoire. Whereas SLACS are site-specific retroposons that interrupt tandem arrays of spliced leader RNA genes, (Aksoy et al., 1987, Peacock et al., 2007), TATE elements are mostly present at telomeres and are inserted in the telomeric hexameric sequence GGGTTA (Peacock et al., 2007). However, the precise number and organization of TATE and SLACS in the L. braziliensis genome are not known. Alignment of the reads along one of the SLACS (LbrM16.0470; Fig. 4A) or TATE (LbrM24.2430; Fig. 4B) complete sequences revealed that the small RNAs are quite evenly distributed over the entire elements and are derived from both the sense and antisense strands.
Fig. 4. Distribution of small RNA reads through SLACS, TATE, TAS-like and CIR74 elements.
The images show the schematic representation of each element with non-redundant small RNA reads aligned. Sense/plus or antisense/minus reads are colored in blue and red, respectively.
A. Schematic representation of a SLACS element (LbrM16.0470) from chromosome 16 (Chr16) inserted into a spliced leader (SL) RNA gene repeat (green boxes). The reverse transcriptase-like (RT-like) domain and the poly(A) tail are represented by the yellow and light gray boxes, respectively.
B. Schematic representation of a TATE element (LbrM24.2430) from chromosome 24 (Chr24) associated with a TAS-like element. Green boxes, telomeric repeats (GGGTTAn); yellow box, phage integrase (PI) domain.
C. Schematic representation of a CIR74 repeat unit from chromosome 2 (Chr2).
Repeats
Our analysis also revealed reads with similarity to the previously described L. braziliensis-specific 1.6 kb TAS family (Fig.4B), which is found on mini- and megabase chromosomes (clone telo10, AF031224; (Fu et al., 1998, Fu & Barker, 1998). Gauging from the distribution of the reads, TAS-like sequences are spread throughout the genome and have gained or lost sequences, resulting in elements ranging in size from 0.38 to 4.8 kb. Some TAS-like sequences contain the repeated boxes GTACAGT and GGAGAGGGTGT described for the prototypical TAS, but do not include telomeric repeats (Fu & Barker, 1998), which we did not find represented in the small RNA libraries. Most of the copies of TAS-like sequences were found at the 3’end of TATE elements (Fig. 4B), but a few appeared to be “solitary”.
CIR74 sequences form satellite-like DNA clusters, have no identifiable open reading frame and are present on chromosomes 2 and 27 (Table 1). On chromosome 2 we found a single CIR74 locus (nt 287,900–290,339) located at the end of a transcription unit immediately after a hypothetical protein (LbrM02.0680). On chromosome 27 there are three CIR74 loci, all located in intergenic regions. We found that small RNA reads were not uniformly distributed along a single CIR74 repeat (Fig. 4C), suggesting that CIR74-derived dsRNA is not uniformly processed and/or that certain siRNAs are preferentially loaded into LbrAGO1 or preferentially degraded.
Table 1.
Summary of features of siRNA-producing loci.
| Element | Chromosomes | Regions | Characteristics |
|---|---|---|---|
| SLACS | 2, 8, 16, 25, 31 | all |
|
| TATE | 1, 3–5, 7, 11–13, 18, 24–32, 34, 35 | telomeric, subtelomeric |
|
| TAS-like | 1–5, 7–13, 16–19, 23–25, 27–35 | telomeric, subtelomeric |
|
| CIR74 | 2 and 27 | internal, subtelomeric |
|
The LbrAGO1c cell line contains the LUC gene and a homologous hairpin integrated at one of the SSU RNA loci. Inspection of the distribution of siRNAs (120,077 reads) along the LUC coding region (Fig. S2) revealed that they were exclusively derived from the region of the gene used to generate the hairpin structure (nt 281–788 of the ORF).
From the data presented above it appeared that the landscape of siRNA-producing loci was very similar between L. braziliensis and T. brucei (Djikeng et al., 2001, Barnes et al., 2012), with most siRNAs derived from putative mobile elements and repeats. However, we also uncovered a significant difference, namely that in L. braziliensis we did not find evidence for reads mapping to convergent transcription units (CTUs; Fig. S3), which instead are well represented in the T. brucei siRNA repertoire (Tschudi et al., 2012). We surveyed 38 CTUs in the L. brazilienesis genome, excluding the ones containing transposable elements and/or repeats, and measured the read occurrence within 10 kb on either side (Table S1). We found on average 69 reads per 20 kb, which is comparable to a similar analysis of genomic regions with no siRNAs above background, namely 32 divergent transcription units, where on average 36 reads mapped.
Validation of the library and consequences of AGO1 ablation on accumulation of siRNAs and mobile element-derived transcripts
To validate the high-throughput sequencing results we first immunoprecipitated BB2-LbrAGO1 (Fig. 5A) and then performed Northern hybridizations with probes for SLACS, TATE, TAS-like and CIR74 elements, as well as for the LUC hairpin region (Fig. 5B). In all cases we found that the corresponding siRNAs were enriched in the immunoprecipitated material, thus confirming that siRNAs from the four major siRNA-producing loci and from the LUC hairpin were mostly associated with BB2-LbrAGO1. Hybridization with a serine or arginine tRNA probe did not show hybridization in the size range of siRNAs (data not shown), an indication that the tRNA reads in our libraries were indeed degradation products.
Fig. 5. LbrAGO1 controls the accumulation of siRNAs and transcripts derived from putative mobile elements.
A. Pull-down of BB2-LbrAGO1 from a soluble extract of LbrAGO1c cells. Aliquots of the input (I), supernatant (S) or the bead pellet (P), each representing 107 promastigote cells, were fractionated on a 10% SDS-PAGE and Western blotted with anti-BB2 or anti-HSP70 monoclonal antibody.
B. L. braziliensis siRNAs were extracted from immunoprecipitated BB2-LbAGO1, fractionated on a 15% sequencing gel, transferred to a nylon membrane and hybridized with the indicated probes. The membranes were stripped and re-hybridized with a 5S rRNA probe as a loading control (load). S, supernatant; P, pellet.
C. Low molecular weight RNAs from L. braziliensis wild-type (WT) promastigotes, LbrAGO1 null cells (Lbrago1−/−) or the latter complemented with the BB2-LbrAGO1 gene (LbrAGO1c) were fractionated on a 15% sequencing gel, transferred to a nylon membrane and hybridized with TATE, SLACS, TAS-like or CIR74 probes. A cross-reacting band was used as a loading control, except for the TAS-like panel where 5S RNA was the loading control.
D. Northern blot analysis of total RNA separated on a 1.2% agarose-formaldehyde gel and hybridized to a TATE or SLACS probe. The identity of the various cell lines is indicated above each lane. Hybridization to α-tubulin mRNA was used as a loading control. Asterisks denote bands cross-hybridizing with the SLACS probe.
Next, we asked whether LbrAGO1 controls the accumulation of siRNAs and of transcripts derived from mobile elements. We found that the levels of TATE, SLACS, TAS-like and CIR74 siRNAs were significantly down-regulated in the Lbrago1−/− as compared to wild-type (WT) and LbrAGO1c cells and that complementation with the BB2-LbrAGO1 transgene restored accumulation of siRNAs (Fig. 5C). However, a proportion of siRNAs were still detected after ablation of AGO1, most prominently in the case of SLACS elements. Residual endogenous siRNAs are also observed in T. brucei ago1−/− cells (Shi et al., 2004). Next, we surveyed the steady-state levels of SLACS and TATE transcripts by Northern blotting of total RNA separated by formaldehyde agarose gel electrophoresis (Fig. 5D). In the absence of AGO1 we detected significant accumulation of TATE and SLACS transcripts, and specifically a ~8.5 kb TATE and a ~8.0 kb SLACS transcript, which may be derived from transcription of full-length genomic elements. Taken together the above observations indicated that most siRNAs are associated with LbrAGO1 and that AGO1 controls the accumulation of all classes of siRNAs and of long transcripts derived from putative mobile elements.
Non-templated nucleotides at the 3’ end of small RNAs
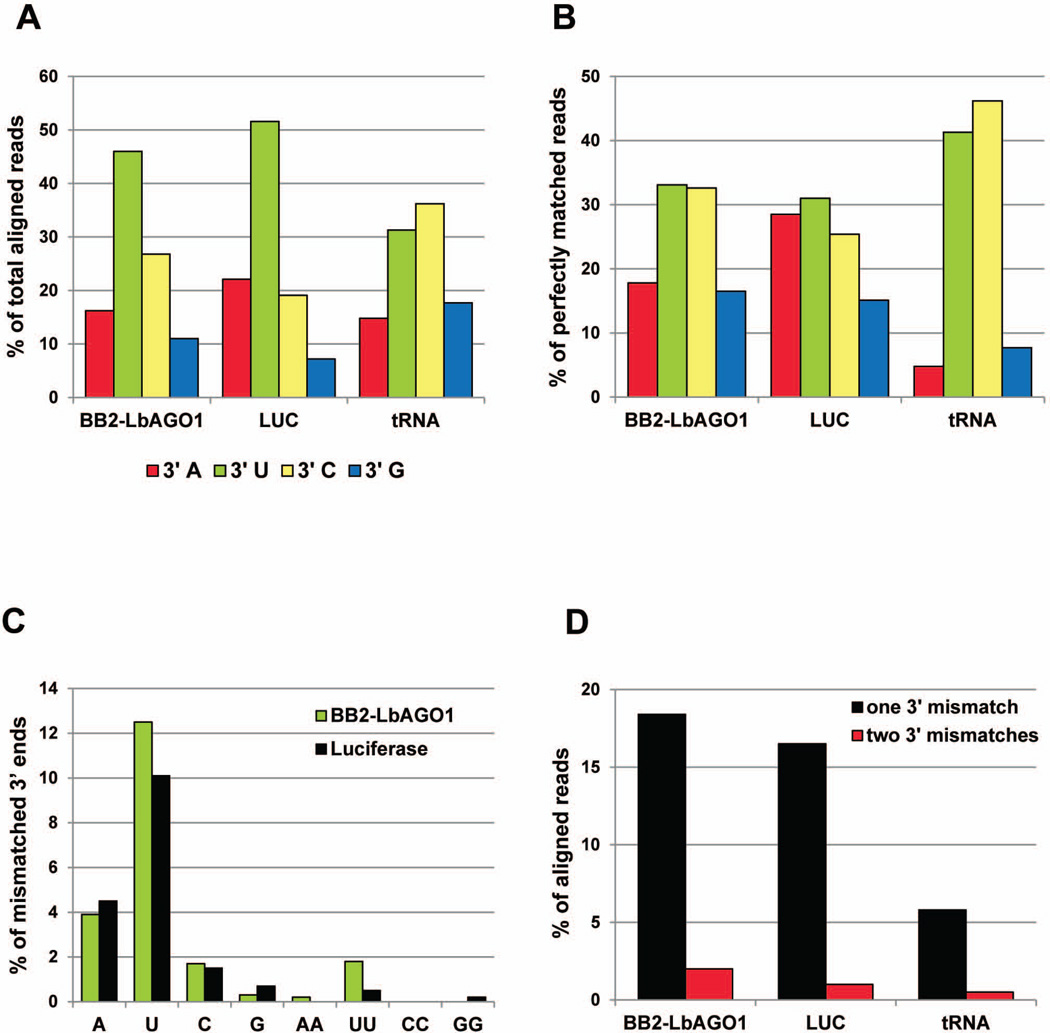
As the 3’ ribose of L. braziliensis siRNAs does not appear to be blocked (Fig. 1B), there is potential for the 3’ end to be subject to tailing activities. To address this possibility we examined the relative representation of 3’ nucleotides of reads aligned to the genome allowing up to two mismatches. This analysis revealed enrichment for uridine, both for the total library reads and for those derived from the LUC hairpin (Fig. 6A). In contrast, tRNA-derived reads showed no U bias at the 3’end. However, when we analyzed the 3’ends of perfectly matched reads we no longer observed a bias in the 3’ nucleotide distribution, suggesting that a proportion of the 3’ Us in the total reads was not templated in the genome (Fig. 6B). The majority of 3’-extended reads had a single nucleotide addition, with uridine having the highest representation and adenine being the second highest nucleotide, but 2- to 3-fold less abundant than uridine (Fig. 6C). By this analysis approximately 20% of the total reads and 18% of the LUC-derived reads contained one or two 3’ end mismatches (Fig. 6D), whereas for the tRNA-derived reads this value was approximately 6%. Lastly, we compared the size distribution of reads with perfect 3’ ends to that of reads having one and two nucleotide extensions (Fig. S4) and found that the latter sequences were on average longer than those with perfectly matched 3’ ends. Thus, collectively the above observations suggested that a significant proportion of siRNAs carry non-templated nucleotides at the 3’ terminus and that 3’ additions occurred on all size classes of siRNAs.
Fig. 6. Non-templated nucleotides are present at the 3’ end of small RNA-derived reads.
A. Analysis of 3’ nucleotide distribution in the BB2-LbrAGO1 library, the LUC- and tRNA-derived reads. The analysis was carried out on redundant reads.
B. Analysis of the 3’ nucleotides of reads aligned to the genome with 0 mismatches.
C. 3’ nucleotide prevalence of non-templated nucleotides expressed as percentage of total aligned reads.
D. Percentage of reads with one or two non-templated 3’ nucleotides.
Discussion
Here we have documented the main structural features and repertoire of endogenous siRNAs associated with AGO1 in L. braziliensis. Our results expose similarities and differences between L. braziliensis and T. brucei siRNAs and highlight diversifications of the RNAi pathway during the evolution of the trypanosomatid lineage.
Both Northern blotting and bioinformatics analysis indicated that the majority of Leishmania siRNAs are 20–25 nt long and thus shorter than those in T. brucei, which are 24–26 nt in size (Djikeng et al., 2001, Tschudi et al., 2012). The shorter size of Leishmania siRNAs may reflect differences in the cleavage specificity of the Dicer-like endonucleases in these two organisms. Additionally, the Leishmania siRNAs are more heterogeneous in size than those in T. brucei. Trimming by exonucleases as well as tailing by terminal nucleotidyl transferases (see below) may contribute to the observed size heterogeneity. Since siRNA size heterogeneity was observed in RNA isolated by the Trizol procedure, which minimizes RNA degradation, we believe that it is a feature of Leishmania siRNAs and most likely not an artifact introduced by experimental manipulation. It should be noted that in a previous publication from our laboratories (Lye et al., 2010) the L. braziliensis siRNAs did not show the same extent of size heterogeneity we report here. This apparent discrepancy is mostly due to the improvement of the Northern blotting methodology and the use of oligonucleotide probes as opposed to long riboprobes.
Leishmania siRNAs do not appear to be modified at the 3’ end, as they are susceptible to periodate oxidation and β-elimination. This observation contrasts with our previous findings in T. brucei, which showed that siRNAs are insensitive to this chemical treatment (Patrick et al., 2009) and predicted that they carry a modified 3’ terminus. Recently, we have gathered evidence that the T. brucei HEN1-like gene, which belongs to the family of HEN1 2’-O-methyltransferases, modifies the terminal ribose of siRNAs (unpublished observations). This modification was first discovered in Arabidopsis thaliana (Li et al., 2005), where both siRNAs and miRNAs are HEN1 substrates, and was later on described in a variety of non-coding RNAs, including the piRNAs (Saito et al., 2007, Kirino & Mourelatos, 2007), a class of small RNAs in Tetrahymena thermophila (Kurth & Mochizuki, 2009) and Drosophila siRNAs (Horwich et al., 2007, Pelisson et al., 2007). Additionally, C. reinhardtii small RNAs are modified at the 3’ end (Molnar et al., 2007). Consistent with the absence of a blocked siRNA 3’ terminus, the genome of L. braziliensis does not appear to code for a HEN1-like gene, as determined by using the BLAST search algorithm at low stringency (Expected value=10).
Methylation of the terminal ribose is thought to protect the small RNA 3’ end from the attack by exonucleolytic and tailing activities (Kim et al., 2010, Cerutti & Ibrahim, 2010). For instance, in A. thaliana in the absence of HEN1 uridine residues are added to the 3’ end of mi- and siRNAs (Li et al., 2005), and recently it has been shown that the nucleotidyl transferase AtHESO1 uridylates unmethylated small RNAs to trigger their degradation (Zhao et al., 2012). Similarly, in HEN11-deficient Drosophila AGO2-associated siRNAs, which normally carry a 2'-O-methyl group on the terminal ribose, are tailed and trimmed (Ameres et al., 2010). The important role that 3’ end remodeling by nucleotydyl transferases play in small RNA metabolism is also underscored by the findings that in C. elegans the absence of the nucleotidyl transferase CDE-1, which uridylates siRNAs bound to a specific AGO protein, results in increased levels of such siRNAs and in defects in chromosome segregation, and in C. reinhardtii the MUT68 nucleotidyl transferase, which adds uridines to a small proportion of unmodified and shorter than average small RNAs, has been shown to cooperate with the exosome subunit RRP6 to eliminate these RNAs (Ibrahim et al., 2010). It has been proposed that the MUT68/RRP6 is part of a quality control that eliminates damaged or “dysfunctional” small RNAs, although what constitutes a “dysfunctional” small RNA is not yet understood (Ibrahim et al., 2010). Thus, it was unsurprising to find that in L. braziliensis a proportion of siRNAs carried untemplated nucleotides at the 3’ end, with uridine being the most frequent addition. We estimated that about 20% of endogenous siRNAs are tailed, but this is probably an underestimate, considering that a single nucleotide addition may frequently match the genomic sequence. In contrast, deep sequencing of T. brucei AGO1-associated siRNAs, which carry a modified terminal ribose (Patrick et al., 2009), did not reveal any bias in the representation of 3’ nucleotides (unpublished observations). The presence of untemplated nucleotides on all size classes of siRNAs and the significant fraction of tailed molecules (~20%) suggest that tailing of Lbr siRNAs does not preferentially target shorter molecules and is a rather frequent occurrence. This phenomenon clearly indicates that the 3’ end of certain siRNAs is exposed to the action of nucleotydyl transfereases. Establishing whether tailing occurs before or after the loading of siRNAs into AGO1 and whether tailed siRNAs are eliminated will require further investigation.
By analogy to other systems (Ibrahim et al., 2010, van Wolfswinkel et al., 2009, Ren et al., 2012, Zhao et al., 2012) tailing of Lbr siRNAs is most likely carried out by one or more nucleotidyl transferases. The Leishmania homologs of T. brucei cytosolic terminal uridilyl transferase (TUTase) 3 (LbrM19.1680) and TUTase 4 (LbrM32.2690; (Aphasizhev et al., 2004, Stagno et al., 2007), are good candidates for adding uridine to siRNAs and we are currently exploring this possibility.
In L. braziliensis we found four major classes of siRNA-producing loci (Table 1), all derived from either putative mobile elements or repeats, mirroring in this respect the siRNA repertoire in T. brucei (Djikeng et al., 2001, Tschudi et al., 2012). Thus, similarly to trypanosomes and many other organisms one of the functions of the RNAi pathway in Leishmania is to down-regulate expression of putative mobile elements that may affect genome integrity. Indeed, genetic ablation of LbrAGO1 leads to the accumulation of SLACS and TATE transcripts. Of note, we found that both in T. brucei and Leishmania only one family of satellite-like repeats, namely the CIR147 family in T. brucei (Patrick et al., 2009, Tschudi et al., 2012) and the CIR74 family in Leishmania (this paper), are sources of abundant siRNAs. However, whether the repeat-derived sense and antisense transcripts and the corresponding siRNAs have any specific function is at present not known. On the other hand, in Leishmania we failed to identify siRNAs derived from regions of convergent transcription, which are instead well represented in the T. brucei siRNA repertoire (Tschudi et al., 2012). Thus, our data point to the possibility that in L. braziliensis putative CTU-derived sense and antisense transcripts, with the potential to form dsRNA, are either produced in much lower amounts than in T. brucei or they are not processed by the Dicer enzymes. Importantly, it has been recently shown that in L. major and L. tarentolae modified thymine residues, termed base J, are present at the ends of CTUs and appear to prevent read-through transcription (van Luenen et al., 2012). A similar mechanism is likely to operate in L. braziliensis to decrease formation of sense and antisense transcripts from CTUs.
In addition, we surveyed the reads for the presence of small RNAs derived from the SIDER family, which is found in T. brucei, T. cruzi and Leishmania ssp. (Bringaud et al., 2011). This family consists of short, truncated versions of ingi-related retroposons, has expanded in the genome of Leishmania ssp., where it has acquired an independent function in regulation of gene expression (Boucher et al., 2002, McNicoll et al., 2005, Muller et al., 2010, Bringaud et al., 2007). L. braziliensis harbors about 2,000 copies of SIDER (Smith et al., 2009), but we found no evidence of SIDER-derived reads, suggesting that these elements are not a significant source of dsRNA that is processed by the RNAi machinery.
A standing question in the trypanosomatid RNAi pathway is whether miRNAs exist. As a first step toward answering this question we manually inspected each chromosome for reads that did not map to the major siRNA-producing loci and had a common 5’ end, the latter being a feature anticipated for miRNA-derived reads. Unfortunately, we did not find evidence of such reads. Since our analysis was carried out on small RNAs associated with AGO1 in L. braziliensis promastigotes, we cannot exclude the possibility that miRNA-like molecules are produced in other life-cycle stages or that they may be not associated with AGO1, Thus, it would be important to determine the entire repertoire of small non-coding RNAs during the L. braziliensis life-cycle and irrespective of their association with AGO1.
Experimental Procedures
Cell lines
L. braziliensis M2903 strain (MHOM/BR/75/M2903) was obtained from Prof. Diane McMahon-Pratt (Yale School of Public Health). Promastigote forms were grown in freshly prepared Schneider’s Insect Medium (Sigma-Aldrich Cat. No. S9895) supplemented with 10% heat-inactivated fetal bovine serum (FBS), 2 mM L-glutamine, 500 units ml−1 penicillin and 50 μg ml−1 streptomycin (Gibco Cat. No. 5070). Prior to use, and following each genetic manipulation, parasites were inoculated into gamma-interferon KO BL6 mice and recovered 1 month later by needle aspiration.
Stable transfections of L. braziliensis M2903 strain (MHOM/BR/75/M2903) were performed using the high-voltage (1400 V) procedure. The parasites were grown to mid-log phase, pelleted at 1,300 × g, washed once with cytomix electroporation buffer (120 mM KCl, 0.15 mM CaCl2, 10 mM K2HPO4, 25 mM HEPES-KOH, pH 7.6, 2 mM EDTA, and 5 mM MgCl2) and resuspended in cytomix at a final concentration of 2 × 108 cells/ml. For transfections, 2 to 4 µg DNA was mixed with 500 µl of cells and electroporated twice in a 0.4-cm gap cuvette at 25 mF, 1400 V (3.75 kV/cm), waiting 10 s in-between zaps. Following electroporation, cells were incubated at 26°C for 24 h in drug-free media and then plated on semisolid media containing the appropriate drug to select clonal lines. For selections using blasticidin deaminase (BSD gene), hygromycin phosphotransferase (HYG gene), streptothricin acetyltransferase (SAT gene) and the bleomycin binding protein from Streptoalloteichus hindustanus (PHLEO gene) markers, parasites were plated on 10–20 µg/ml blasticidin, 30–80 µg/ml hygromycin B, 50−100 µg/ml nourseothricin and 0.2–2 µg/ml phleomycin, respectively (ranges reflect differences when using drugs singly or in combination). Colonies normally appeared after 14 days, at which point they were recovered, grown to stationary phase in 1 ml and passaged with the appropriate drugs. We routinely have plating efficiencies ranging from 60–95% for the un-transfected L. braziliensis M2903 strain; transfection efficiencies varied from 2–50 colonies per µg of SwaI-digested pIR1 vectors controls.
To generate ago1 knockout parasites, two replacement constructs were assembled by fusing 800 nucleotides immediately upstream of the L. braziliensis AGO1 start codon to the open reading frame for the selectable marker encoding blasticidin (BSD) or hygromycin B (HYG) resistance. The selectable marker genes were then combined at the 3’ end with 300 nucleotides immediately downstream of the AGO1 termination codon. Targeting fragments prepared from these constructs were used successively to replace both AGO1 alleles by the Leishmania transfection methodology described above. Following the final selection, PCR tests established the absence of the AGO1 gene in several clones, consistent with homozygous ∆AGO1::BSD/∆AGO1::HYG replacements. For this work we selected clone 16 (lane 3 in Fig. S5) and this cell line is hereafter referred to as ago1−/−. We then integrated into the SSU rRNA locus an RNAi reporter expressing both a luciferase (LUC) ORF and a 1 kb LUC stem-loop hairpin (SSU:IR2SAT-LUC-SR; construct B6386, (Lye et al., 2010). Lastly, a BB2 epitope tag was added to the 5’ end of the AGO1 gene and integrated into the ribosomal RNA SSU locus (SSU:IR1PHLEO-BB2-AGO1(b); construct B6263; (Lye et al., 2010), yielding ago1−/SSU:BB2-AGO1/SSU:LUCSR, hereafter termed LbrAGO1c; transfectant clone 603 was used in this work (see lane 4 in Fig. S5). All constructs were verified by DNA sequencing and all transfectants confirmed by multiple PCR tests as described (Lye et al., 2010).
RNA preparation and Northern blotting
Total RNA from L. braziliensis promastigotes was isolated using the Trizol reagent (Invitrogen), Low molecular weight RNAs were enriched by centrifugation through a Microcon-YM100 filter (Millipore) and used for Northern blot analyses as previously described (Patrick et al., 2009). DNA oligonucleotide probes (corresponding to the forward strand of each element) were end-labeled with T4 polynucleotide kinase (NEB) and [γ-32P] ATP and purified on a P6 column (BIO-RAD). Hybridizations were carried out in ExpressHyb solution (Clontech) overnight at 30°C. Northern blot analysis of total RNA separated on 1.2% agarose-formaldehyde gels was carried out as previously described (Shi et al., 2004).
Oligonucleotide probes
SLACS: 5’-TATAAAGACGCAAACAGGGGCCACAA-3’
TATE: 5’-ATCTTCATTATGAAGCACTGGATCCAGGGCT-3’
TAS-like: 5’-ATGCCACCGTC CTGTGTGATAGGGTG-3’
CIR74: 5’-TTTGTTATGTTGTTCAGAAGCACATCCG TCGATTC-3’
LUC: 5’-TTGAACGTGCAAAAAAAGCTCCCAAT-3’
Enzymatic treatments of small RNAs
Low molecular weight RNAs were treated with Calf intestinal alkaline phosphatase (CIP, Amershan) following the manufacturer’s instructions for 1 h at 37°C. Periodate oxidation/β-elimination was carried out as previously described (Patrick et al., 2009).
Preparation of S100 extracts and BB2-LbAGO1 immunoprecipitation
S100 extracts were prepared from 5×109 LbrAGO1c cells and BB2-LbrAGO1 was immunoprecipitated with anti-BB2 antibodies bound to protein G-Sepharose beads as described (Shi et al., 2004). Equivalent amounts of input, supernatant and beads were processed for Western blot analysis.
Preparation of small RNA libraries
RNA immunoprecipitated with BB2-LbrAGO1 was phenol extracted and ethanol precipitated. The detailed method for construction of the library is provided in the supplemental material. Sequencing was carried out at the Yale Genomic Center on an Illumina Genome Analyzer Platform IIx.
Bioinformatics
Small RNA reads obtained from the Illumina GA2 platform (36 nt in length) were first processed by clipping the 3’ adapter (5’-CTGTAGGCACCATCAAT-3’) and by collapsing the reads into unique reads, using the collection of command line tools for short reads pre-processing, the FASTX-Toolkit (hannonlab.cshl.edu/fastx_toolkit/). After pre-processing, the reads were aligned to the 35 chromosomes of the L. braziliensis Genome Version 2 (genedb.org) using Bowtie (Langmead et al., 2009). Alignments with more than two mismatches in the first 22 nucleotides were discarded. Reads reporting multiple alignments were retained and aligned pseudo-randomly (using Bowtie’s default options). Aligned RNA-Seq reads were loaded into a customized, local installation of the Generic Genome Browser (Stein et al., 2002), for interactive analysis of the reads distributed through the L. braziliensis genome. For the 3’ nucleotide analysis, small RNA reads were pre-processed to produce count statistics (reads without the adaptor or smaller than 16 nt were rejected). Processed reads were collapsed to produce a list of uniquely occurring reads that were aligned against the reference sequences or contaminants (i.e. rRNA) using Bowtie (default options, with the exception of allowing for up to three mismatches). Any read that aligned to contaminants was removed from further consideration. A custom Python script was then written to process reads aligned only to the reference sequences. The script computed per position and overall statistics for these reads, based on the Bowtie alignment reports.
Supplementary Material
Acknowledgements
Research in the author’s laboratory was supported by NIH grants AI28798 and AI56333 to E.U. and AI29646 to S.M.B. V. D.A. was supported by a James Hudson Brown-Alexander B. Coxe post-doctoral fellowship. We thank Prof. Jay Bangs (University of Madison, Wisconsin) for providing Hsp70 antibodies, the “Yale University Biomedical High Performance Computing Center” for supporting bioinformatics analyses and Erin Acino Brettman for assistance in confirming the correct structure for the Lbrago1− replacements.
References
- Aksoy S, Lalor TM, Martin J, Van der Ploeg LH, Richards FF. Multiple copies of a retroposon interrupt spliced leader RNA genes in the African trypanosome, Trypanosoma gambiense. EMBO J. 1987;6:3819–3826. doi: 10.1002/j.1460-2075.1987.tb02718.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ameres SL, Horwich MD, Hung JH, Xu J, Ghildiyal M, Weng Z, Zamore PD. Target RNA-directed trimming and tailing of small silencing RNAs. Science. 2010;328:1534–1539. doi: 10.1126/science.1187058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aphasizhev R, Aphasizheva I, Simpson L. Multiple terminal uridylyltransferases of trypanosomes. FEBS Lett. 2004;572:15–18. doi: 10.1016/j.febslet.2004.07.004. [DOI] [PubMed] [Google Scholar]
- Barnes RL, Shi H, Kolev NG, Tschudi C, Ullu E. Comparative genomics reveals two novel RNAi factors in Trypanosoma brucei and provides insight into the core machinery. PLoS Pathog. 2012;8(e1002678) doi: 10.1371/journal.ppat.1002678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastin P, Bagherzadeh Z, Matthews KR, Gull K. A novel epitope tag system to study protein targeting and organelle biogenesis in Trypanosoma brucei. Mol Biochem Parasitol. 1996;77:235–239. doi: 10.1016/0166-6851(96)02598-4. [DOI] [PubMed] [Google Scholar]
- Bazzini AA, Lee MT, Giraldez AJ. Ribosome profiling shows that miR-430 reduces translation before causing mRNA decay in zebrafish. Science. 2012;336:233–237. doi: 10.1126/science.1215704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999;27:573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucher N, Wu Y, Dumas C, Dube M, Sereno D, Breton M, Papadopoulou B. A common mechanism of stage-regulated gene expression in Leishmania mediated by a conserved 3'-untranslated region element. J Biol Chem. 2002;277:19511–19520. doi: 10.1074/jbc.M200500200. [DOI] [PubMed] [Google Scholar]
- Bringaud F, Berriman M, Hertz-Fowler C. TSIDER1, a short and non-autonomous Salivarian trypanosome-specific retroposon related to the ingi6 subclade. Mol Biochem Parasitol. 2011;179:30–36. doi: 10.1016/j.molbiopara.2011.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bringaud F, Muller M, Cerqueira GC, Smith M, Rochette A, El-Sayed NM, et al. Members of a large retroposon family are determinants of post-transcriptional gene expression in Leishmania. PLoS Pathog. 2007;3:1291–1307. doi: 10.1371/journal.ppat.0030136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerutti H, Ibrahim F. Turnover of mature miRNAs and siRNAs in plants and algae. Adv Exp Med Biol. 2010;700:124–139. [PubMed] [Google Scholar]
- Cheloufi S, Dos Santos CO, Chong MM, Hannon GJ. A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature. 2010;465:584–589. doi: 10.1038/nature09092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cifuentes D, Xue H, Taylor DW, Patnode H, Mishima Y, Cheloufi S, et al. A novel miRNA processing pathway independent of Dicer requires Argonaute2 catalytic activity. Science. 2010;328:1694–1698. doi: 10.1126/science.1190809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole C, Sobala A, Lu C, Thatcher SR, Bowman A, Brown JW, et al. Filtering of deep sequencing data reveals the existence of abundant Dicer-dependent small RNAs derived from tRNAs. RNA. 2009;15:2147–2160. doi: 10.1261/rna.1738409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djikeng A, Shi H, Tschudi C, Shen S, Ullu E. An siRNA ribonucleoprotein is found associated with polyribosomes in Trypanosoma brucei. RNA. 2003;9:802–808. doi: 10.1261/rna.5270203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djikeng A, Shi H, Tschudi C, Ullu E. RNA interference in Trypanosoma brucei: cloning of small interfering RNAs provides evidence for retroposon-derived 24-26-nucleotide RNAs. RNA. 2001;7:1522–1530. [PMC free article] [PubMed] [Google Scholar]
- Djuranovic S, Nahvi A, Green R. miRNA-mediated gene silencing by translational repression followed by mRNA deadenylation and decay. Science. 2012;336:237–240. doi: 10.1126/science.1215691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand-Dubief M, Bastin P. TbAGO1, an Argonaute protein required for RNA interference is involved in mitosis and chromosome segregation in Trypanosoma brucei. BMC Biol. 2003;1 doi: 10.1186/1741-7007-1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir SM, Lendeckel W, Tuschl T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 2001;15:188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ender C, Krek A, Friedlander MR, Beitzinger M, Weinmann L, Chen W, et al. A human snoRNA with microRNA-like functions. Mol Cell. 2008;32:519–528. doi: 10.1016/j.molcel.2008.10.017. [DOI] [PubMed] [Google Scholar]
- Ender C, Meister G. Argonaute proteins at a glance. J Cell Sci. 2010;123:1819–1823. doi: 10.1242/jcs.055210. [DOI] [PubMed] [Google Scholar]
- Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem. 2010;79:351–379. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- Fernandes AP, Nelson K, Beverley SM. Evolution of nuclear ribosomal RNAs in kinetoplastid protozoa: perspectives on the age and origins of parasitism. Proc Natl Acad Sci U S A. 1993;90:11608–11612. doi: 10.1073/pnas.90.24.11608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Fu G, Barker DC. Characterisation of Leishmania telomeres reveals unusual telomeric repeats and conserved telomere-associated sequence. Nucleic Acids Res. 1998;26:2161–2167. doi: 10.1093/nar/26.9.2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu G, Perona-Wright G, Barker DC. Leishmania braziliensis: characterisation of a complex specific subtelomeric repeat sequence and its use in the detection of parasites. Exp Parasitol. 1998;90:236–243. doi: 10.1006/expr.1998.4326. [DOI] [PubMed] [Google Scholar]
- Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton AJ, Baulcombe DC. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science. 1999;286:950–952. doi: 10.1126/science.286.5441.950. [DOI] [PubMed] [Google Scholar]
- Haussecker D, Huang Y, Lau A, Parameswaran P, Fire AZ, Kay MA. Human tRNA-derived small RNAs in the global regulation of RNA silencing. RNA. 2010;16:673–695. doi: 10.1261/rna.2000810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horwich MD, Li C, Matranga C, Vagin V, Farley G, Wang P, Zamore PD. The Drosophila RNA methyltransferase, DmHen1, modifies germline piRNAs and single-stranded siRNAs in RISC. Curr Biol. 2007;17:1265–1272. doi: 10.1016/j.cub.2007.06.030. [DOI] [PubMed] [Google Scholar]
- Huang HY, Houwing S, Kaaij LJ, Meppelink A, Redl S, Gauci S, et al. Tdrd1 acts as a molecular scaffold for Piwi proteins and piRNA targets in zebrafish. EMBO J. 2011;30:3298–3308. doi: 10.1038/emboj.2011.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim F, Rymarquis LA, Kim EJ, Becker J, Balassa E, Green PJ, Cerutti H. Uridylation of mature miRNAs and siRNAs by the MUT68 nucleotidyltransferase promotes their degradation in Chlamydomonas. Proc Natl Acad Sci U S A. 2010;107:3906–3911. doi: 10.1073/pnas.0912632107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji L, Chen X. Regulation of small RNA stability: methylation and beyond. Cell Res. 2012;22:624–636. doi: 10.1038/cr.2012.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- Kim YK, Heo I, Kim VN. Modifications of small RNAs and their associated proteins. Cell. 2010;143:703–709. doi: 10.1016/j.cell.2010.11.018. [DOI] [PubMed] [Google Scholar]
- Kirino Y, Mourelatos Z. The mouse homolog of HEN1 is a potential methylase for Piwi-interacting RNAs. RNA. 2007;13:1397–1401. doi: 10.1261/rna.659307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurth HM, Mochizuki K. 2'-O-methylation stabilizes Piwi-associated small RNAs and ensures DNA elimination in Tetrahymena. RNA. 2009;15:675–685. doi: 10.1261/rna.1455509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, Ambros V. The Celegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- Lee YS, Shibata Y, Malhotra A, Dutta A. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs) Genes Dev. 2009;23:2639–2649. doi: 10.1101/gad.1837609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Yang Z, Yu B, Liu J, Chen X. Methylation protects miRNAs and siRNAs from a 3'-end uridylation activity in Arabidopsis. Curr Biol. 2005;15:1501–1507. doi: 10.1016/j.cub.2005.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Paroo Z. Biochemical principles of small RNA pathways. Annu Rev Biochem. 2010;79:295–319. doi: 10.1146/annurev.biochem.052208.151733. [DOI] [PubMed] [Google Scholar]
- Lye LF, Owens K, Shi H, Murta SM, Vieira AC, Turco SJ, et al. Retention and loss of RNA interference pathways in trypanosomatid protozoans. PLoS Pathog. 2010;6(10):e1001161. doi: 10.1371/journal.ppat.1001161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNicoll F, Muller M, Cloutier S, Boilard N, Rochette A, Dube M, Papadopoulou B. Distinct 3'-untranslated region elements regulate stage-specific mRNA accumulation and translation in Leishmania. J Biol Chem. 2005;280:35238–35246. doi: 10.1074/jbc.M507511200. [DOI] [PubMed] [Google Scholar]
- Molnar A, Schwach F, Studholme DJ, Thuenemann EC, Baulcombe DC. miRNAs control gene expression in the single-cell alga Chlamydomonas reinhardtii. Nature. 2007;447:1126–1129. doi: 10.1038/nature05903. [DOI] [PubMed] [Google Scholar]
- Muller M, Padmanabhan PK, Rochette A, Mukherjee D, Smith M, Dumas C, Papadopoulou B. Rapid decay of unstable Leishmania mRNAs bearing a conserved retroposon signature 3'-UTR motif is initiated by a site-specific endonucleolytic cleavage without prior deadenylation. Nucleic Acids Res. 2010;38:5867–5883. doi: 10.1093/nar/gkq349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngo H, Tschudi C, Gull K, Ullu E. Double-stranded RNA induces mRNA degradation in Trypanosoma brucei. Proc. Natl. Acad. Sci. USA. 1998;95:14687–14692. doi: 10.1073/pnas.95.25.14687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrick KL, Shi H, Kolev NG, Ersfeld K, Tschudi C, Ullu E. Distinct and overlapping roles for two Dicer-like proteins in the RNA interference pathways of the ancient eukaryote Trypanosoma brucei. Proc Natl Acad Sci USA. 2009;106:17933–17938. doi: 10.1073/pnas.0907766106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peacock CS, Seeger K, Harris D, Murphy L, Ruiz JC, Quail MA, et al. Comparative genomic analysis of three Leishmania species that cause diverse human disease. Nat Genet. 2007;39:839–847. doi: 10.1038/ng2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelisson A, Sarot E, Payen-Groschene G, Bucheton A. A novel repeat-associated small interfering RNA-mediated silencing pathway downregulates complementary sense gypsy transcripts in somatic cells of the Drosophila ovary. J Virol. 2007;81:1951–1960. doi: 10.1128/JVI.01980-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren G, Chen X, Yu B. Uridylation of miRNAs by hen1 suppressor1 in Arabidopsis. Curr Biol. 2012;22:695–700. doi: 10.1016/j.cub.2012.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Sakaguchi Y, Suzuki T, Siomi H, Siomi MC. Pimet, the Drosophila homolog of HEN1, mediates 2'-O-methylation of Piwi- interacting RNAs at their 3' ends. Genes Dev. 2007;21:1603–1608. doi: 10.1101/gad.1563607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saraiya AA, Wang CC. snoRNA, a novel precursor of microRNA in Giardia lamblia. PLoS Pathog. 2008;4(11):e1000224. doi: 10.1371/journal.ppat.1000224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Djikeng A, Tschudi C, Ullu E. Argonaute protein in the early divergent eukaryote Trypanosoma brucei: control of small interfering RNA accumulation and retroposon transcript abundance. Mol Cell Biol. 2004;24:420–427. doi: 10.1128/MCB.24.1.420-427.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Tschudi C, Ullu E. An unusual Dicer-like1 protein fuels the RNA interference pathway in Trypanosoma brucei. RNA. 2006;12:2063–2072. doi: 10.1261/rna.246906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stagno J, Aphasizheva I, Aphasizhev R, Luecke H. Dual role of the RNA substrate in selectivity and catalysis by terminal uridylyl transferases. Proc Natl Acad Sci U S A. 2007;104:14634–14639. doi: 10.1073/pnas.0704259104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein LD, Mungall C, Shu S, Caudy M, Mangone M, Day A, et al. The generic genome browser: a building block for a model organism system database. Genome Res. 2002;12:1599–1610. doi: 10.1101/gr.403602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschudi C, Shi H, Franklin JB, Ullu E. Small interfering RNA-producing loci in the ancient parasitic eukaryote Trypanosoma brucei . BMC Genomics. 2012;13:427. doi: 10.1186/1471-2164-13-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Luenen HG, Farris C, Jan S, Genest PA, Tripathi P, Velds A, et al. Glucosylated hydroxymethyluracil, DNA base j, prevents transcriptional readthrough in Leishmania. Cell. 2012;150:909–921. doi: 10.1016/j.cell.2012.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wolfswinkel JC, Claycomb JM, Batista PJ, Mello CC, Berezikov E, Ketting RF. CDE-1 affects chromosome segregation through uridylation of CSR-1-bound siRNAs. Cell. 2009;139:135–148. doi: 10.1016/j.cell.2009.09.012. [DOI] [PubMed] [Google Scholar]
- Warburton PE, Giordano J, Cheung F, Gelfand Y, Benson G. Inverted repeat structure of the human genome: the X-chromosome contains a preponderance of large, highly homologous inverted repeats that contain testes genes. Genome Res. 2004;14:1861–1869. doi: 10.1101/gr.2542904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Totoki Y, Toyoda A, Kaneda M, Kuramochi-Miyagawa S, Obata Y, et al. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature. 2008;453:539–543. doi: 10.1038/nature06908. [DOI] [PubMed] [Google Scholar]
- Wen YZ, Zheng LL, Liao JY, Wang MH, Wei Y, Guo XM, et al. Pseudogene-derived small interference RNAs regulate gene expression in African Trypanosoma brucei. Proc Natl Acad Sci U S A. 2011;108:8345–8350. doi: 10.1073/pnas.1103894108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in Celegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- Yang JS, Maurin T, Robine N, Rasmussen KD, Jeffrey KL, Chandwani R, et al. Conserved vertebrate mir-451 provides a platform for Dicer-independent, Ago2-mediated microRNA biogenesis. Proc Natl Acad Sci U S A. 2010;107:15163–15168. doi: 10.1073/pnas.1006432107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Yu Y, Zhai J, Ramachandran V, Dinh TT, Meyers BC, et al. The Arabidopsis nucleotidyl transferase HESO1 uridylates unmethylated small RNAs to trigger their degradation. Curr Biol. 2012;22:689–694. doi: 10.1016/j.cub.2012.02.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.