Abstract
Eukaryotic RNA polymerases (Pol) I, II, III and archaeal Pol use a related set of general transcription factors to recognize promoter sequences, recruit Pol to promoters and to function at key points in the transcription initiation mechanism. The TFIIB-like general transcription factors (GTFs) function during several important and conserved steps in the initiation pathway for Pol II, III, and archaeal Pol. Until recently, the mechanism of Pol I initiation seemed unique, since it appeared to lack a GTF paralogous to the TFIIB-like proteins. The surprising recent discovery of TFIIB-related Pol I general factors in yeast and humans highlights the evolutionary conservation of transcription initiation mechanisms for all eukaryotic and archaeal Pols. These findings reveal new roles for the function of the Pol I GTFs and insight into the function of TFIIB-related factors. Models for Pol I transcription initiation are reexamined in light of these recent findings.
Keywords: Rrn7, TAF1B, Pol I, TFIIB, Brf, rDNA Transcription
1. Introduction
RNA polymerase (Pol) I is the most specialized Pol as it exclusively transcribes the ribosomal DNA (rDNA) gene unit into a single rRNA transcript [1, 2]. The large rRNA transcript or rRNA precursor is processed into the 18S, 28S, and 5.8S rRNAs that are key components of ribosomes [1, 2]. Pol I transcription is robust and accounts for the majority of total RNA in cells [3], and its upregulation in human cells is a hallmark of cancer [2, 4–6]. rDNA transcription takes place in a unique nuclear compartment called the nucleolus [7], where the majority of ribosome biogenesis occurs, producing over a million ribosomes per cell [3, 8]. In most eukaryotes, the nucleolus contains hundreds of tandem ribosomal genes, located at one or more chromosomal loci, but only a subset of these genes are actively transcribed at any one time [1, 2].
2. Pol I subunit composition
To synthesize rRNA, eukaryotic cells use a 14-subunit Pol I enzyme [9–11]. All three eukaryotic Pols (Pol I, II, III) contain 12 subunits that are either shared between the Pols, or are functionally and structurally related Pol-specific paralogs [9–11] (Table 1). Each Pol contains a core set of five shared subunits: Rpb5, 6, 8, 10, and 12 [9–12]. The Pol subunits AC40 and AC19 are shared between Pol I and III, and are paralogs to the Pol II subunits Rpb3 and Rpb11 [13–16]. Seven subunits are unique to Pol I, which include the two largest subunits that form the catalytic site (Rpa190 and Rpa135), the Rpb9/TFIIS/Rpc11-related paralog A12.2 [17], and the two Pol I stalk subunits A14 and A43 that are paralogous to Rpb4/7 and Rpc17/Rpc25 subunits from Pol II and III respectively [18–20].
Table 1.
Yeast RNA polymerase I and paralogous counterparts
| Pol I | Pol III | Pol II | Archaea | Bacteria | |
|---|---|---|---|---|---|
| Core1 | Rpa190 | Rpc160 | Rpb1 | Rpo1 | β' |
| Rpa135 | Rpc128 | Rpb2 | Rpo2 | β | |
| AC40 | AC40 | Rpb3 | Rpo3 | α | |
| AC19 | AC19 | Rpb11 | Rpo11 | α | |
| Rpb6 | Rpb6 | Rpb6 | Rpo6 | ω | |
| Rpb5 | Rpb5 | Rpb5 | Rpo5 | ||
| Rpb8 | Rpb8 | Rpb8 | Rpo8 | ||
| Rpb10 | Rpb10 | Rpb10 | Rpo10 | ||
| Rpb12 | Rpb12 | Rpb12 | Rpo12 | ||
| Rpa12 | Rpc11 | Rpb9 | TFS | ||
| Stalk | Rpa14 | Rpc17 | Rpb4 | Rpo4 | |
| Rpa43 | Rpc25 | Rpb7 | Rpo7 | ||
| GTFs | A49-NT2 | Rpc53 | TFIIFα4 | ||
| A34.5 | Rpc37 | TFIIFβ5 | |||
| A49-CT3 | Rpc34 | TFIIEβ6 | TFE | ||
Shared Pol I subunits are outlined;
NT, N-terminal dimerization domain;
CT, C-terminal tWH domain;
TFIIFα, Tfg1;
TFIIFβ, Tfg2;
TFIIEβ, Tfa1
The Pol I subunits A34.5 and A49 comprise a heterodimeric protein that contain domains structurally and functionally related to the Pol II general transcription factors (GTFs) TFIIF- and TFIIE [21–24]. A34.5 and the N-terminal domain of A49 dimerize with a triple barrel fold that is structurally related to the dimerization domain of the TFIIF subunits, Tfg1 and 2, and the Pol III TFIIF-like subunits, Rpc37 and Rpc53 [22]. A large unstructured linker connects the A49 dimerization domain to a tandem wing helix (tWH) domain that is structurally related to the tWH domains of the TFIIE subunit Tfa2 and the Pol III TFIIE-like subunit Rpc34. In terms of functional conservation, genetic evidence suggests that A34.5 and A49 perform TFIIF- and TFIIE-like functions in Pol I transcription elongation and PIC formation [22, 23, 25, 26]. Thus, both Pol I and III evolved to stably incorporate TFIIE- and TFIIF-like GTFs, unlike the more loosely associated Pol II GTFs, TFIIE and TFIIF [21, 27].
Given the sequence and structural similarity of each Pol, it is not surprising that the TFIIE and TFIIF subunits and their Pol I and III paralogs localize to evolutionary conserved positions on Pol. For Pol II and III, the dimerization domain of TFIIF and Rpc37/Rpc53 are positioned on the lobe domains of their respective Pols [28–30]. Likewise, previous EM studies and recent combined chemical crosslinking and mass spectrometry studies of Pol I show a similar position for the A49/A34.5 dimer on the Pol I lobe domain [31–33]. The crosslinking studies also showed that the A49 TFIIE-like tWH domain spans the Pol I active site cleft, bridging the clamp and protrusion domains [33], analogous to the positions observed for the TFIIE and Rpc34 tWH domains on Pol II and III [12, 34, 35]. Together, these findings suggest that each Pol transcription system follows a common structural, functional, and evolutionary framework.
3. Pol I transcription initiation factors
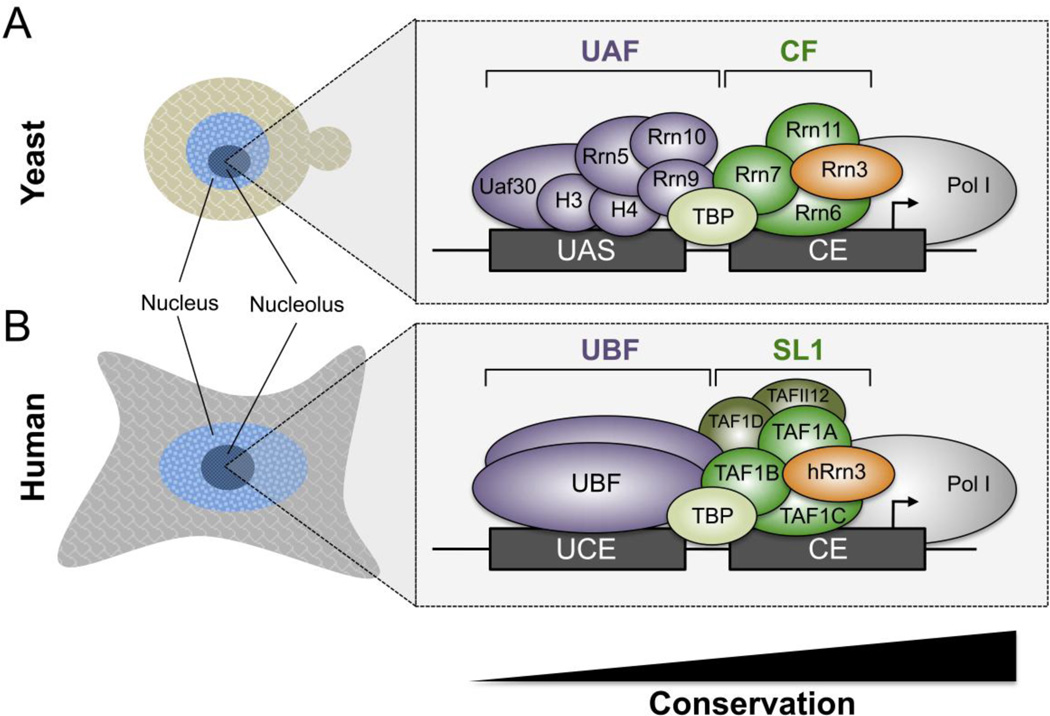
Transcription from rDNA repeats is controlled by a bipartite promoter with two essential cis-regulatory elements (Fig. 1). In yeast, Pol I promoters contain a Core Element (CE) that overlaps the transcription start site (TSS) and an Upstream Activating Sequence (UAS) that is centered ~100 bp upstream of the TSS [1, 2]. Yeast Pol I is recruited to the promoter via four different initiation factors including the UAS-binding Upstream Activating Factor (UAF), TATA-binding protein (TBP), the CE-binding protein Core Factor (CF), and Rrn3. UAF consists of six subunits: UAF30, Rrn9, Rrn10, Rrn5, and histones H3 and H4 [36, 37]. CF is composed of three subunits that include Rrn6, Rrn7, and Rrn11 [38, 39].
Figure 1. Comparison of yeast and human Pol I preinitiation complexes.
Schematic of Pol I factors that assemble on (A) yeast and (B) humans rDNA promoters. Yeast rDNA promoters contain two elements called the Upstream Activation Sequence (UAS) and Core Element (CE). Human rDNA promoters contain an Upstream Control Element (UCE) and Core Element (CE). Orthologous and analogous yeast and human factors are colored identically. Human Selectivity Factor 1 (SL1) and orthologous yeast Core Factor (CF) subunits are colored in identical shades of green, and unique SL1 subunits are colored in a darker shade of green. The relative evolutionary conservation of Pol I, CF/SL1, UAF/UBF is also indicated below the PIC models.
Pol I preinitiation complex (PIC) formation in yeast begins with UAF binding to the UAS followed by TBP recruitment, which next facilitates recruitment of CF and Rrn3-bound Pol I to the promoter [40]. In vitro, UAF remains stably bound to the promoter of immobilized templates after initiation and operates as a reinitiation scaffold for subsequent rounds of transcription while CF, TBP, and Rrn3 are released [40]. In vivo, chromatin immunoprecipitation (ChIP) studies show that Pol I crosslinks to the promoter and rDNA coding regions as expected, whereas UAF, CF, and Rrn3 are exclusively promoter associated [41]. In the absence of Pol I and Rrn3, UAF, CF, and TBP remain stably bound to the promoter, suggesting that CF and TBP build a reinitiation scaffold in vivo [42].
Pol I GTFs are only weakly conserved between yeast and humans, but their promoters retain a similar bipartite structure. In humans, Pol I promoters contain an Upstream Control Element positioned ~100 bp upstream of a TSS overlapping CE [1, 2]. The human CE is targeted by a CF ortholog called Selectivity Factor 1 [1, 2]. Three subunits of SL1 that include TAF1A, B, and C, that are orthologs of yeast Rrn6, 7, and 11, respectively [1, 43, 44]. Orthologous CF and SL1 components share ≤16% protein sequence identity. SL1 also contains three additional subunits that include TBP, the human-specific TAF1D subunit [45], and TAF12, a component of the conserved Pol II coactivators SAGA and TFIID [46, 47]. The mechanistic roles of TAF1D and TAF12 subunits remain unclear.
Most components of yeast UAF beside the histone components also lack clear orthologs to the human Pol I factors. The UCE of human rDNA promoters is targeted by a dimer called the Upstream Binding Factor (UBF) [1, 48]. UBF contains a series of tandem High Mobility Group (HMG) boxes [49] that bind and bend DNA, forming a loop that bridges the UCE and CE [50, 51]. None of the UAF subunits share homology with UBF, but a UBF-related HMG box protein termed Hmo1 binds throughout the yeast rDNA locus [52, 53], but it is still not clear whether UAF functions are replaced by UBF in humans or vice versa. A summary of yeast and human Pol I initiation factor orthology and homology is listed in Table 2.
Table 2.
Yeast Pol I GTFs and their ortholog, paralogs, and structural homologs
| Complex | Yeast | Human | Genome1 and PDB2 Homologs |
|---|---|---|---|
| yRrn3 | hRrn3 | HEAT repeats | |
| yCF/hSL1 | Rrn11 | TAF1A | Tetratricopeptide repeats |
| Rrn7 | TAF1B | TFIIB, Brf1, Brf2, TFB, Cyclin Fold | |
| Rrn11 | TAF1C | WD-40 repeats, HEAT repeats | |
| N.A. | TAF1D | Josephin domain | |
| N.A. | TAF12 | Histone fold | |
| yUAF/hUBF | Rrn5 | N.A. | SANT domain, Histone fold |
| Rrn9 | N.A. | TFIIA Toa1 subunit3 | |
| Rrn10 | N.A. | No significant matches | |
| Uaf30 | N.A. | Swi/Snf SWIB/MDM2 domain | |
| Histone H3 | N.A. | Histone fold | |
| Histone H4 | N.A. | Histone fold | |
| Hmo1 | UBF | HMG Homeobox repeats | |
HHpred search results against S. cerevisiae and H. sapien genome databases;
HHpred search results against PDB database;
Transcription relevant matches are listed and matches represent tentative sequence relationships that require further study;
Rrn9 matches small region of TFIIA Toa1 subunit with low probability but high protein identity; N.A. Not applicable due to absence of homology between yeast and human Pol I factors.
Current PIC assembly models for mammalian rDNA promoters suggest that UBF binds to the promoters first and then facilitates SL1 recruitment, while other models suggest that UBF and SL1 bind together, or that SL1 facilitates UBF recruitment [2, 54, 55]. UBF and SL1 promoter binding is immediately followed by recruitment of Rrn3 bound Pol I [2, 54, 55]. After human Pol I initiation and promoter clearance, UBF and SL1 remain stably bound to the promoter, creating a reinitiation scaffold like those found in yeast for continued cycles of rDNA transcription [2, 54, 55].
4. Discovery of a Pol I TFIIB-related factor
Soon after the discovery of TFIIB [56–58], a TFIIB-related factor (BRF1) was discovered among the Pol III GTFs [59–61]. Brf1 is a subunit of the Pol III GTF TFIIIB that also contains subunits Bpd1 and TBP. Previous searches for a Pol I TFIIB-related factor focused on components of CF and SL1 since both interact with TBP and Pol I. However, no detectable TFIIB homology could be detected among the Pol I factors by traditional protein homology search methods. Given the conservation of the core Pol enzymes and GTFs, Pol I seemed unique in lacking a protein with homology to TFIIB. For almost two decades, it became accepted that either Pol I uses a TFIIB-like factor that has diverged beyond recognition by sequence homology, or that the Pol I machinery evolved a unique initiation mechanism distinct from Pol II and III [62–64].
Over the past decade, more sensitive sequence homology detection methods have been developed, such as HHpred [65], that are designed specifically to detect homology between distantly related proteins. Two groups independently reexamined the question of whether the Pol I transcription system required a TFIIB-related factor. Using HHpred, both groups discovered sequence and predicted structural homology between the yeast CF subunit Rrn7 and its human ortholog TAF1B with known TFIIB-like proteins in archaea and eukaryotes [63, 64]. It was found that the N-terminus of Rrn7 and TAF1B share homology with TFIIB, TFB, and the N-terminus of Brf [63, 64]. Remarkably, the sequence identity between Rrn7 and other TFIIB-like proteins was very low, ranging from 8–13% [63, 64]. Even more surprising, the protein sequence identity between Rrn7 and TAF1B was only 16% [63, 64]. However, functional studies described below showed that these proteins share conserved function and structure. Together, these new findings uncovered the missing link between the Pol I transcription system and its Pol II and III counterparts, and shed new light on the Pol I initiation factor function.
5. Comparison of TFIIB family proteins
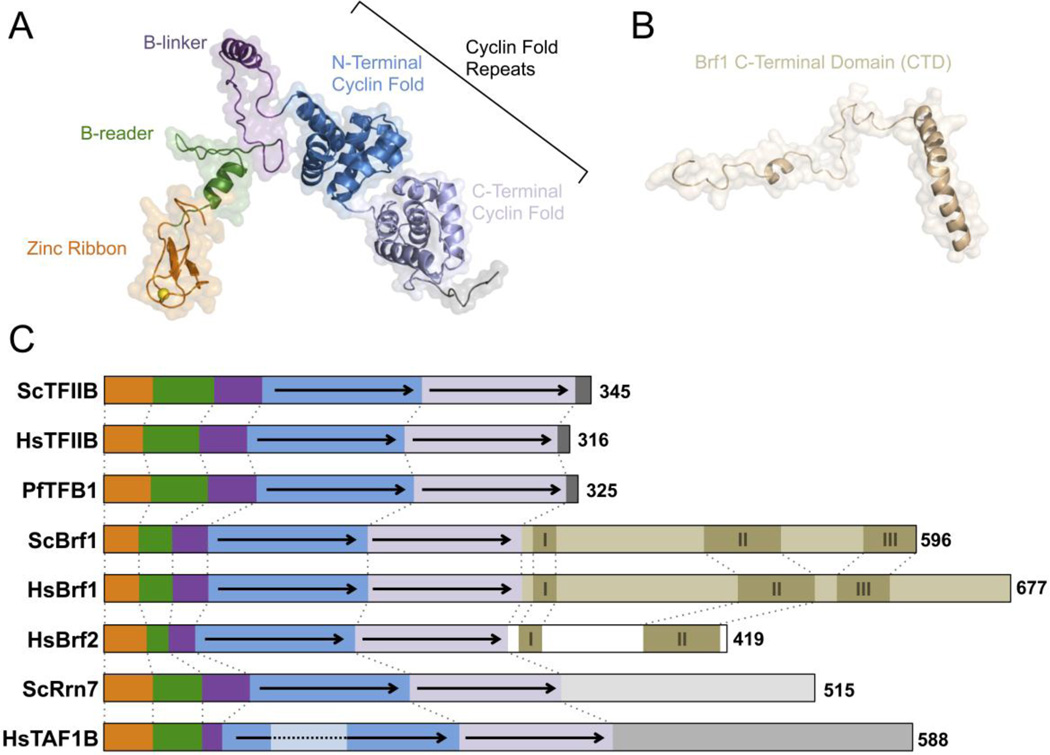
The overall domain architectures of TFIIB, TFB and Brf proteins are quite similar, but with several intriguing differences. All three TFIIB-related family members contain a zinc ribbon domain followed by a linker region that connects to tandem cyclin fold repeats [66, 67] (Fig. 2A). Brf1 contains an extended C-terminal domain (CTD) immediately after the second cyclin repeat [66, 68]. This unique Brf1 CTD folds into an elongated structure with three homology regions (I–III) where the second region tightly binds TBP [69, 70] (Fig. 2B). Human Brf2, a GTF that is specific to Pol III type 3 promoters such as the U6 snRNA gene, has a shortened CTD that contains segments with weak homology to Brf1 homology regions I and II [66]. In contrast, TFIIB and TFB lack a CTD and their TBP binding is mediated by the tandem cyclin repeat domains [67, 71].
Figure 2. TFIIB protein family domain architectures.
Transparent surface and ribbon representation of the (A) TFIIB structure (PDB, 3K1F) [74], and (B) the Brf1 C-terminal domain (PDB, 1NGM) [70]. The yellow sphere within the zinc ribbon structure represents the Zn2+ atom. (C) Schematic of selected TFIIB family proteins from the following organisms: Saccharomyces cerevisiae, Sc; Homo sapiens, Hs; Pyrococcus furious, Pf. Homologous domains and regions are identically colored as follows: Zinc Ribbon, orange; B-reader, green; B-linker, purple; N-terminal Cyclin Fold; blue; C-terminal Cyclin Fold, light purple; Brf1 CTD; tan; Brf2 CTD, white; Rrn7 CTD, light grey; TAF1B CTD, grey; and TAF1B-specific insertion, light blue. Conserved helical segments (I, II, III) are labeled and shaded in brown. HHpred searches of Brf family proteins were used to assign CTD homology regions in HsBrf1 and 2. Dotted lines connect homologous regions and domains.
The Brf1 and Brf2 CTDs are both essential and mediate interaction between Pol III and the TFIIIB subunits Bdp1 and TBP [66, 68]. Rrn7 and TAF1B also contain an extended CTD but with no sequence similarity to the Brf CTDs [63, 64]. There is no obvious homology between the CTD of Rrn7 and human TAF1B, other than they are predicted to fold into a series of tandem helices [63, 64]. Although the lack of similarity between the Rrn7 and TAF1B CTD is surprising, the lack of similarity with the Pol III factors is expected since they each interact with GTF subunits specific to their respective Pols.
The general domain organization of Rrn7 and TAF1B are nearly identical in that they contain the same three TFIIB-like regions: the N-terminal zinc ribbon domain, linker region, and tandem cyclin repeats [63, 64] (Fig. 2C). Despite the low protein sequence identity between Rrn7, TAF1B and TFIIB family proteins, the secondary structure predictions are quite similar [63, 64]. HHpred alignments were used to assign Rrn7 and TAF1B domain boundaries [63, 64]. The zinc ribbon domain of Rrn7 has an atypical consensus signature (Cys-X4-Cys-X13-Cys-X3-His) that contains additional residues within the zinc motifs and a shortened spacer between the motifs compared to the TFIIB/Brf consensus signature (Cys-X2-Cys-X15–17-Cys-X2-Cys/His) [72, 73]. Interestingly, the TAF1B zinc ribbon consensus signature (Cys-X2-Cys-X14-Cys-X2-Cys-His) is more typical, although the residues between the second and third cysteine are more similar to Rrn7 than TFIIB/BRF.
The TFIIB linker region can be divided into two important functional segments called the B-reader and B-linker that are noted for their roles in transcription start site (TSS) selection and promoter opening respectively [74]. The linker regions are very divergent between Rrn7, TAF1B, and other TFIIB family proteins [63, 64]. Linker sequence variability may be tied to the weak sequence conservation of Pol I initiation factors when compared to the more closely related Pol II and III paralogs [63, 64]. Pol I subunits and promoter sequences also vary considerably between species [75, 76], yet Pol I promoters retain a similar bipartite structure [77, 78]. Given these precedents, it is not surprising to observe significant linker sequence variation, since Pol I factors and promoters likely evolved to complement each other.
The Rrn7 linker contains two patches of conserved charged residues in the B-reader segment. In fungi, the size of the linker region varies (Fig. 3A) and S. cerevisiae and other sensu stricto strains contain a large linker [63, 64]. The linker regions of more divergent fungi and higher eukaryotes are missing a large portion of the B-linker [63, 64]. TAF1B also contains a large serine and tyrosine rich insertion between the first and second helices of the N-terminal cyclin fold that lies in close proximity to the B-linker domain [63]. Most yeast species lack this insertion, but some species like S. pombe contain a TAF1B-like serine patch (Fig. 3B). Likewise, the B-linker domain of S. pombe is also shortened (Fig. 3A). Though speculative, it will be interesting to see if a B-linker function is imparted by the insertion, and/or whether it can act as point of regulation by posttranslational modifications such as phosphorylation.
Figure 3. Protein sequence alignment of Rrn7 and TAF1B family protein domains.
Protein sequence alignment of Rrn7 and TAF1B (A) zinc ribbon and linker domains, and (B) Serine/Threonine/Tyrosine-rich TAF1B insertion. Amino acid residues are colored according to their conservation. Residues conserved in Rrn7 and TAF1B family protein are highlighted in orange. Residues conserved in either Rrn7 or TAF1B family proteins are colored in dark green, and less conserved residues are colored in lighter shades of green. In panel A, residues important for TAF1B and Rrn7 transcriptional activity are marked by by black boxes and triangles, respectively. In panel B, the Ser/Thr/Tyr-rich insertion is highlighted in light blue where Ser, Thr, and Tyr residues within the insertion are highlighted in purple with white lettering. The consensus secondary structure is also shown below the alignment. Pink rectangles and grey lines correspond to alpha helical and coiled secondary structure predictions respectively. Rrn7/TAF1B protein domains and regions are denoted by brackets above the alignments. Rrn7 and TAF1B protein sequences were used from the following organisms: Sc, Saccharomyces cerevisiae; Sk, Saccharomyces kudriavzevii; Mg, Meyerozyma guilliermondii; Dh, Debaryomyces hansenii; So, Schizosaccharomyces pombe; Bt, Bos taurus; Gg, Gallus gallus; Mm, Mus musculus; Hs, Homo sapiens.
6. General functions of TFIIB and Brf1
TFIIB, Brf1, and TFB play critical and essential roles during the transcription initiation process. By far the most attention has been placed on the founding family member TFIIB. During the transcription cycle, TFIIB is directly involved in numerous roles including facilitating Pol II recruitment to the promoter, TSS recognition, open-complex formation, abortive initiation, promoter clearance, and roles in termination and gene looping [47, 71, 79, 80]. A subset of these transcriptional functions have also been described for TFB and Brf1 [11, 66–68].
A major function of TFIIB is to facilitate Pol II recruitment to the promoter. To do this, the TFIIB zinc ribbon domain binds a cavity between the Rpb1 dock domain and the Rpb2 wall domain, while the N-terminal cyclin fold domain contacts the Rpb2 wall [81–83]. The TFIIB zinc ribbon is necessary for the interaction with Pol II and mutations in this domain are lethal. The binding of the two structured domains to distinct sites on Pol II positions the TFIIB linker domain to traverse the active site cleft [74, 84]. In this location, the B-reader segment lies near the TSS in the Pol II active site and the B-linker segment is positioned near the site of DNA unwinding in the open complex [74].
Although the Brf1 zinc ribbon is essential for yeast viability [72], it is not essential for Pol III recruitment, but rather plays an important post-recruitment role. The Brf ribbon-Pol III interaction is likely redundant with Pol III interactions made by other TFIIIB subunits. For example, the Brf1 CTD coordinates interactions between Bdp1 and TBP, and Brf1 and Bdp1 directly interact with Pol III subunits Rpc34 and Rpc17 [66, 68]. Therefore, the ability to interact with Pol III and TFIIIB complex integrity is retained in ribbon domain mutants. In contrast, Brf1 zinc ribbon mutants are defective in open complex formation [85], as promoter opening assays using potassium permanganate and Brf1 zinc ribbon mutants showed reduced sensitivity to DNA modification in a region surrounding the TSS [72]. Consistent with this finding, preopening of the promoter with a heteroduplex bubble, bypasses the requirement for the Brf ribbon domain [86].
7. Post-recruitment role of Pol I TFIIB-related factors
Biochemical characterization of Rrn7 and TAF1B indicated that they function very similarly to TFIIB and Brf1 in transcription initiation. First, mutation of TAF1B zinc ribbon cysteine residues and mutations in the linker region abolish transcription activity in vitro [64] and similar mutations are lethal in yeast Rrn7 [63] (Fig. 3A). However, the TAF1B zinc ribbon and linker mutants still assemble into SL1, interact with Pol I, and form Pol I PICs on an immobilized rDNA template [64]. These observations are identical to the aforementioned biochemical studies of yeast Brf1 lacking the zinc ribbon domain where the mutant factor can assemble into TFIIIB and form PICs but cannot initiate transcription [60, 72].
Unlike with Brf1, preopening the Pol I promoter template at the TSS could not restore transcription activity when using a TAF1B zinc ribbon mutant [64]. This may indicate that TAF1B has additional post-recruitment roles or that the size and position of the heteroduplex bubbles tested so far may need further optimization. For example, the rDNA heterduplex bubble encompassed only three unpaired nucleotides at positions −1 to +2 relative to the TSS at +1 [64], whereas those used in the Brf1 studies used five unpaired nucleotides [86]. The Brf1 studies also highlighted that the position of the bubble is important for bypassing the transcriptional defect of the zinc ribbon mutant. Recovery of wild-type levels of transcription activity was only observed with a Pol III bubble positioned between +2 and +6 [86], which is centered a few nucleotides downstream from the position used in the TAF1B studies. These studies are further complicated since the Pol I promoter TSS overlaps the essential CE. More detailed biochemical studies will be necessary to ascertain the post-recruitment roles for the TAF1B and Rrn7 zinc ribbon and linker.
The TFIIIB subunit Bdp1 also has a post-recruitment role. Like Brf1, activity of transcriptionally inactive Bdp1 mutants can be restored with preopened heteroduplexed templates [86]. By analogy, it is possible that other CF and SL1 subunits besides Rrn7 and TAF1B may contribute to post Pol I-recruitment steps. One current model for the post-recruitment role of Brf1 and Bdp1 suggests that they are not directly involved in DNA opening, but instead alter DNA interaction of Pol III subunits either close to or at the TSS and DNA bubble [87]. In contrast, the TFIIB zinc ribbon and linker play either a direct role in DNA opening or in stabilization of the open state since mutations in both of these regions are bypassed by preopening the DNA [74, 88]. Clearly, more work is needed to delineate whether CF and SL1 directly or indirectly participate in DNA opening and/or play a role in Pol I open complex stabilization.
8. Domains involved in the interaction between Rrn7 and Pol I
As described above, both TFIIB and Brf1 directly interact with Pol II and Pol III, respectively. Protein interaction assays also showed that yeast Rrn7 and Pol I directly interact. Full-length Rrn7 binds immobilized Pol I, and there are two separate Pol I binding domains in Rrn7; both the N-terminal half of Rrn7 and the CTD individually bind Pol I [63]. Within the N-terminal half of Rrn7, the zinc ribbon domain was sufficient to bind Pol I and this binding was dependent on the zinc coordinating residues [63]. The Rpa190 dock domain was essential for viability and predicted to have similar secondary and tertiary structure as the Pol II and III dock domains [63]. Rpa190 mutations in residues analogous to those in Rpb1 that were important for TFIIB zinc ribbon binding disrupted the interaction between Pol I and the Rrn7 zinc ribbon [63]. This indicated that the Rrn7 zinc ribbon binds to the Pol I dock domain and likely positions the Rrn7 linker within the Pol I active site cleft, similar to the Pol II-TFIIB linker [63, 74]. Therefore, it is likely that zinc ribbon mutants of Brf1, Rrn7, and TAF1B have a common defect in that they cannot properly position their respective linkers in the active site cleft. The TFIIB linker and possibly those of Brf1, Rrn7, and TAF1B are flexible and dynamic [35, 74, 84], so without the zinc ribbon anchor, the linker is misplaced and cannot function.
It is unclear whether Rrn7/TAF1B participate directly in Pol I recruitment to the rDNA promoter. For example, Pol I recruitment to the rDNA promoter seems independent of the TFIIB-related factor Rrn7/TAF1B. In yeast, CF recruits Rrn3-bound Pol I to rDNA promoters through interactions between the CF subunit Rrn6 and Rrn3 [89]. In humans, Rrn3 interacts with the SL1 subunit TAF1A as well as TAF1B [90]. Interestingly, CF and SL1 can also recruit Pol I to rDNA promoters in the absence of Rrn3, but these PICs are inactive [40, 91] Therefore, it is likely that additional protein contacts between CF/SL1 and Pol I, together with Rrn3, help facilitate Pol I recruitment, PIC formation, and initiation, and one of these additional protein contacts is provided by Rrn7/TAF1B.
9. Functional conservation of TFIIB family protein domains
The results of domain swapping between Rrn7, TFIIB, and Brf1 are also consistent with a common transcription initiation mechanism between these factors and also showed that Rrn7 is most closely related to Brf1. Systematic domain swaps between the TFIIB family proteins showed that the Rrn7 and Brf1 zinc ribbons are functionally interchangeable [63]. A surprising finding was that Brf1, TFIIB, and Rrn7 have interchangeable B-linker but not B-reader segments [63]. The TFB and TFIIB B-linkers are important for DNA opening, since B-linker mutations cannot support transcription from closed templates unless preopened [74, 88]. In yeast, the lethal TFIIB linker mutation L110P is also lethal in the context of the Rrn7-TFIIB B-linker chimera [63]. This further supports the conclusion that each TFIIB-related factor uses a similar mechanism for DNA opening. As mentioned previously, the Rrn7 and TAF1B B-linker regions are different in size [63], suggesting that Rrn7 can accommodate much smaller B-linker segments. A functional Rrn7 chimera containing the Brf1 B-linker segment also supports this conclusion since it is almost half the size of the Rrn7 linkers [63]. This indicates that the Rrn7 B-linker is flexible in terms of size and sequence, and fits well with sequence comparisons of yeast and metazoan B-linkers that vary considerably in size (Fig. 3A).
Even though yeast Rrn7 and human TAF1B share only 16% identity in their N-terminal regions, they perform compatible functions in transcription. Domain swaps of the Rrn7 zinc ribbon with the TAF1B ribbon are functional, although at reduced levels, while exchanging their entire N-terminal domains supports near wild-type growth [63]. However, full-length TAF1B is unable to support yeast growth, likely due to their divergent CTDs [63]. Consistent with this, domain swaps shows that the Rrn7 and TAF1B CTDs are not functionally compatible [63], indicating that they make species-specific interactions. For instance, the TAF1B CTD may be unable to assemble with yeast CF subunits or interact with yeast Pol I. It is likely that the TAF1B CTD makes unique contacts with SL1-specific subunits such as TAF1D and TAF12, or interacts differently with TBP. In total, these findings suggest that yeast CF and human SL1 perform similar transcriptional functions that are evolutionarily conserved from humans to yeast, yet there are important differences between CF and SL1 that require further attention.
10. Structural organization of a minimal Pol I PIC
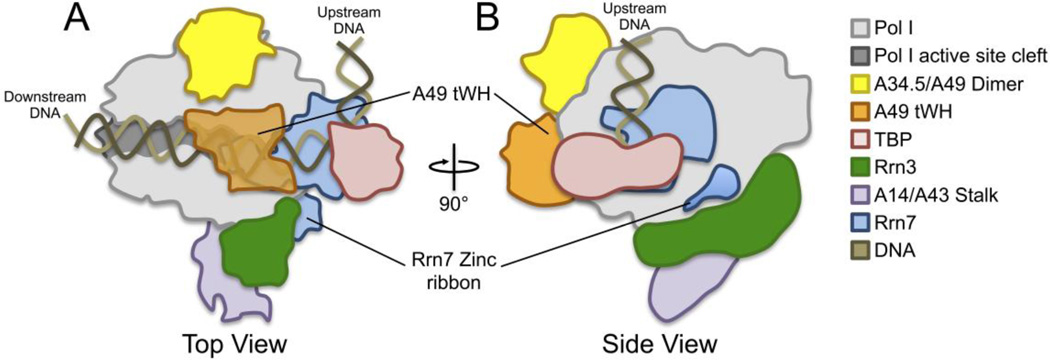
Recent modeling of the Pol I PIC suggested that there may be a direct functional link between Rrn7 and Rrn3 [92]. The structures of A34.5/A49 and Rrn3 have been solved by X-ray crystallography and their positions on Pol I have been mapped by combined chemical crosslinking and mass spectrometry. Modeling the position of Rrn7 in the PIC, based on the position of TFIIB in the Pol II PIC, predicts that the Rrn7 zinc ribbon domain and Rrn3 are in close proximity [92] (Fig. 4). Rrn3 is composed of an array of tandem HEAT repeats that average ~ 40 residues in length, encompassing the entire length of Rrn3 [92]. These repeats fold into an elongated and bent conformation that wraps itself across the side of Rpa19o and the Pol I stalk (Fig. 4), passing close to the Rpa190 dock domain [92]. Previous in vitro binding studies show that TAF1B can interact directly with human Rrn3 [90], but the TAF1B domains necessary for this interaction are not known. Given the close proximity of Rrn7 and Rrn3, an attractive model is that the Rrn7 zinc ribbon domain directly interacts with Rrn3 and that mutations of the ribbon domain could simultaneously disrupt interaction with Pol I and Rrn3 [92], although there is not yet any experimental evidence that Rrn7 interacts with Rrn3.
Figure 4. Model for the structural organization of a minimal Pol I PIC.
Surface cartoon representation of a minimal Pol I PIC modeled after the Pol II PIC structure (PDB,3K1F) [74]. Mapped positions of recently solved X-ray structures of Rrn3 (PDB,3TJ1) [92], A34.5/A49 dimerization domain (PDB,3NFG) [22], and A49 tWH domain (PDB,3NFI) [22] are shown. Rrn7 was modeled and placed into a similar position as TFIIB in the closed Pol II PIC. Pol I PIC components are colored according to the chart to the right. Top view (A) and rotated side view (B) are shown and the Rrn7 zinc ribbon and A49 tWH domains are labeled. The A49 tWH is transparent in panel A to show close proximity to hypothetical position of Rrn7 linker region. Upstream and downstream ends of DNA are labeled.
Since Rrn7 is also predicted to be close to A34.5/A49 (Fig. 4A), there may be a functional link between these factors within the Pol I PIC. Based on homology modeling, the Rrn7 linker domain is likely to be positioned near the A49 C-terminal tWH domain. This prediction is similar to the findings in the Pol II system where the TFIIB linker region is adjacent to the tWH domains of TFIIE [35]. A49 also has a large flexible linker between its dimerization and tWH domain that crosslinks to the Rpa135 protrusion and lobe domains and the Rpa190 clamp and active site cleft [33]. This is similar to the essential TFIIF linker domain of the Tfg2 subunit that lies close to the Rpb2 protrusion domain and likely participates in the initiation mechanism [28, 29, 33, 35]. Future protein-protein interaction studies will help differentiate between these intriguing models and reveal additional insights into the Pol I factor protein interaction network.
11. TBP interactions with CF and SL1
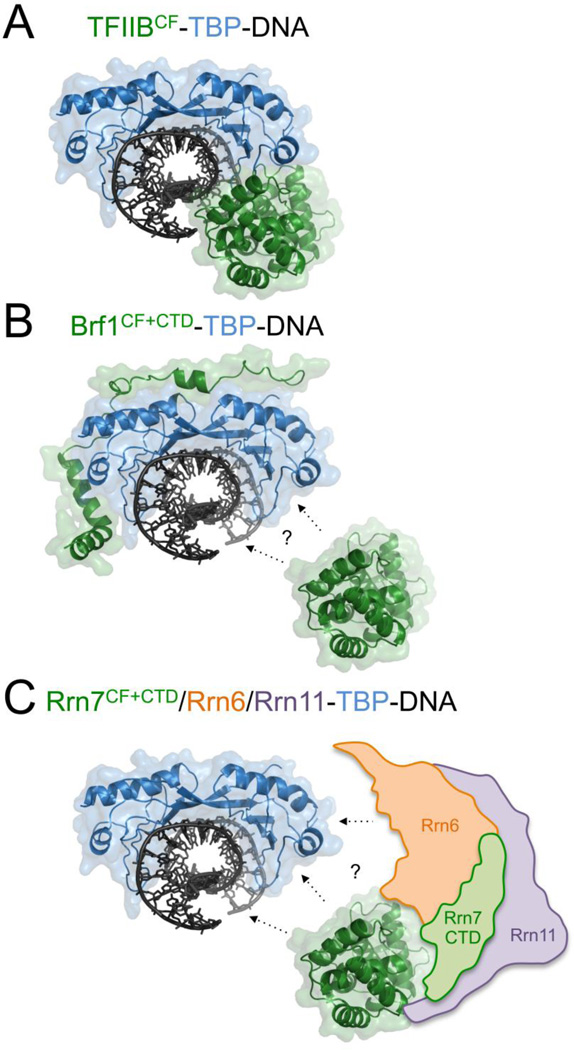
TFIIB and Brf1 interact directly with TBP. The C-terminal TBP stirrup interacts strongly with the TFIIB cyclin fold domain [93–95] (Fig. 5A), while Brf1 interacts strongly with the convex surface of TBP through its elongated CTD domain [70] (Fig. 5B). If the Brf1 and Brf2 cyclin fold domains interact with TBP, this interaction is much weaker than observed for TFIIB, and its contribution to TFIIIB function and stability is not yet clear [96–98].
Figure 5. TBP interaction models for TFIIB family proteins.
Ternary complex structure models of (A) TFIIBCF-TBP-DNA, (B) Brf1CF+CTD-TBP-DNA, and (C) Rrn7CF+CTD/Rrn6/Rrn11-TBP-DNA. CF, Cyclin Folds; CTD, C-terminal domain. The models are based on the 3D structure of the TFIIB-TBP-DNA ternary complex (PDB,1VOL) [95]. TBP and DNA are colored in blue and dark grey, and the TFIIB, Brf1, and Rrn7 CF and CTD domains are colored in green. Panel B and C use the same TBP-DNA model as in panel A, but the Brf1 and Rrn7 CF domain are moved away from TBP and DNA, since their positions and orientation in Pol II and III PICs are still under investigation. In panel B, the Brf1 CTD structure (PDB,1NGM) [70] is shown. Panel C includes the hypothetical positions of Core Factor subunits Rrn6 (orange), Rrn11 (purple), and the Rrn7-CTD (green). Proposed interactions are denoted by dotted arrows.
The role of TBP in Pol I transcription and the TBP interaction mechanism of CF and SL1 remains unresolved. Initial studies in vivo showed an essential role for TBP in Pol I transcription [99–102], but reconstitution studies in vitro showed that TBP is not required for basal transcription [103]. More recent in vitro studies using recombinant CF convincingly show that CF, Rrn3, and Pol I are sufficient for low level basal Pol I transcription, and addition of UAF and TBP support the highest levels of transcription [104]. These findings are the most consistent with a model where TBP plays a coactivator role to help facilitate interactions between UAF and CF, and possibly UBF and SL1.
It appears that Rrn7 has also evolved a distinct mechanism to interact with TBP. Rrn7 as well as Rrn11 interact weakly with TBP, while the CF subunit Rrn6 interacts strongly with TBP [38, 39, 105], suggesting that Rrn7 has delegated most of its TBP binding activity to the CF subunit Rrn6 (Fig. 5C). Given the weak affinity of TBP for Rrn7, it is possible that TBP interacts weakly with the Rrn7 cyclin folds, but the functional consequence of this potential interaction is unknown. Previous studies have also shown the Rrn7 interacts with the UAF subunit Rrn9 that also interacts with TBP, providing an indirect protein contact between CF and TBP [102]. Rrn6 may interact with the convex surface of TBP, analogous to the Brf1 CTD, but additional studies will be necessary to test this hypothesis. Similar TBP binding properties have also been observed for SL1 [106, 107], suggesting that the mechanism of Rrn7/TAF1B-TBP interaction is quite divergent from its Pol II and III counterparts.
12. A function for Rrn7 in Pol II transcription?
Recent studies in S. pombe showed an unexpected role for Rrn7 in Pol II-dependent transcription. Mobility shift assays and ChIP studies found that core promoters of Pol II-dependent TATA-less ribosomal protein genes (RPG) in S. pombe contain a HomoID box sequence that is specifically bound by Rrn7 [108]. In addition to the Pol II transcription machinery, Rrn7 is also required for RPG transcription, since immunodepletion of Rrn7 inhibited in vitro transcription activity [108]. ChIP studies showed promoter binding of Rrn7 in the absence of the CF subunit Rrn11 and likely Rrn6 [108]. This is quite surprising since CF subunits stably associate with each other and are expressed at relatively low and equal levels in S. cerevisiae [41, 104]. These results suggest that S. pombe may express an excess of Rrn7 that possibly assembles into a unique Pol II-dependent regulatory complex that targets RPG core promoters [109]. The HomoID box sequence is also found in S. pombe rDNA promoters, suggesting that Rrn7 may target a specific regulatory sequence that is found in both rDNA and RPG promoters, providing an additional level of ribosome biogenesis coordination [108]. It is not clear if this mechanism is specific to S. pombe or conserved in other eukaryotes, but this represents a unique Rrn7 function that requires further exploration into its functional relevance.
13. Plant-specific Pol I TFIIB-related factors
Rrn7 and TAF1B were not the first TFIIB-related factors proposed to play a role in Pol I transcription. A plant-specific TFIIB-related protein termed Brp1 was suggested to play a role in Pol I transcription in the primitive red algae Cyanidioschyzon merolae and the model flowering plant Arabidopsis thaliana [73]. C. merolae studies showed that CmBrp1 coprecipitates with CmRpa190, colocalizes with CmRpa190 in the nucleolus, crosslinks to rDNA promoter in vivo, and CmBrp1 antibodies inhibits Pol I dependent transcription in vitro [73]. Comparative studies in A. thaliana showed that AtBrp1 crosslinked to rDNA promoters in vivo and forms a stable ternary complex with AtTBP2 and rDNA promoter DNA in vitro [73]. Together, these studies suggest that Brp1 performs a general role in plant Pol I transcription.
In contrast to these findings, phylogenetic analysis of eukaryotic TFIIB family proteins in humans, yeast, and plants showed that Brp1 is more similar to TFIIB, while another plant TFIIB-related factor, AtMEE12, is part of a distinct clade containing its yeast and human orthologs Rrn7 and TAF1B [63, 64]. Less is known about the transcriptional function of AtMEE12. Although AtBrp1 is generally expressed in plant cells, the protein localizes to the cytosolic face of the outer plastid envelope of wild-type plant cells rather than the expected nucleolar localization expected for a Pol I transcription factor [110]. When plants cells are treated with proteasome inhibitors, AtBRP1 shuttles from the cytosol to the nucleus [110], suggesting that the function of AtBRP1 is highly specialized, whereas AtMEE12 localizes both in the nucleus and nucleolus [111]. Together, it seems that AtBrp1 is more specialized yet plays a redundant role in Pol I transcription, while AtMEE12 plays a general role in Pol I transcription as described for Rrn7 and TAF1B.
Conclusions and Perspectives
The discovery of TFIIB-related factors in Pol I transcription and conservation of their biochemical functions with their Pol II and III counterparts further highlights the structural and evolutionary conservation of eukaryotic and archaeal transcription initiation mechanisms. More generally, the findings described here unify the initiation mechanisms of eukaryotic and archaea transcription systems. Although many of the functions of Rrn7 and TAF1B are conserved with TFIIB and Brf1, there are several functional differences that garner closer attention such as how Pol I initiation factors integrate within the Pol I PIC. Over the past few years, a much clearer yet incomplete picture of the minimal Pol I PIC has emerged. Over the next decade, the architecture of the complete Pol I PIC along with its Pol II and III counterparts will likely be resolved. There is now a clear need for low- and high-resolution structural studies to determine the architecture of the unique Pol I initiation factors and structural organization of a complete Pol I PIC. Significant steps toward these goals are currently underway and recent advances in recombinant CF expression and the development and use of more sophisticated protein modeling and protein crosslinking technologies make these goals achievable.
Research Highlights.
A Pol I TFIIB-related factor has recently been discovered.
Pol I TFIIB-related factors share many basic functions with TFIIB and Brf1.
TAF1B and possibly Rrn7 function at a step post-polymerase recruitment.
Eukaryotic Pol I, II, and III systems share a common evolutionary origin.
Acknowledgements
We are grateful to Hahn lab members for insightful discussions and helpful comments on the manuscript. This work was supported by grant RO1 GM053451 to S.H.
Abbreviations
- Pol
RNA polymerase
- GTF
general transcription factor
- rRNA
ribosomal RNA
- rDNA
ribosomal DNA
- CE
core element
- TSS
transcription start site
- UAS
upstream activating sequence
- UCE
upstream control element
- UAF
upstream activating factor
- UBF
upstream binding factor
- TBP
TATA binding protein
- CF
core factor
- SL1
Selectivity factor 1
- tWH
tandem Winged Helix
- HMG
High Mobility Group protein
- ChIP
Chromatin immunoprecipitation
- PIC
preinitiation complex
- CTD
C-terminal domain
- Brf
TFIIB-related factor
- RPG
ribosomal protein genes
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Schneider DA. RNA polymerase I activity is regulated at multiple steps in the transcription cycle: recent insights into factors that influence transcription elongation. Gene. 2012;493:176–184. doi: 10.1016/j.gene.2011.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Drygin D, Rice WG, Grummt I. The RNA polymerase I transcription machinery: an emerging target for the treatment of cancer. Annu Rev Pharmacol Toxicol. 2010;50:131–156. doi: 10.1146/annurev.pharmtox.010909.105844. [DOI] [PubMed] [Google Scholar]
- 3.Warner JR. The economics of ribosome biosynthesis in yeast. Trends Biochem Sci. 1999;24:437–440. doi: 10.1016/s0968-0004(99)01460-7. [DOI] [PubMed] [Google Scholar]
- 4.Montanaro L, Trere D, Derenzini M. Nucleolus, ribosomes, and cancer. Am J Pathol. 2008;173:301–310. doi: 10.2353/ajpath.2008.070752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.White RJ. RNA polymerases I and III, growth control and cancer. Nat Rev Mol Cell Biol. 2005;6:69–78. doi: 10.1038/nrm1551. [DOI] [PubMed] [Google Scholar]
- 6.White RJ. RNA polymerases I and III, non-coding RNAs and cancer. Trends Genet. 2008;24:622–629. doi: 10.1016/j.tig.2008.10.003. [DOI] [PubMed] [Google Scholar]
- 7.Shaw P, Brown J. Nucleoli: composition, function, and dynamics. Plant Physiol. 2012;158:44–51. doi: 10.1104/pp.111.188052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cmarko D, Smigova J, Minichova L, Popov A. Nucleolus: the ribosome factory. Histol Histopathol. 2008;23:1291–1298. doi: 10.14670/HH-23.1291. [DOI] [PubMed] [Google Scholar]
- 9.Cramer P, Armache KJ, Baumli S, Benkert S, Brueckner F, Buchen C, Damsma GE, Dengl S, Geiger SR, Jasiak AJ, Jawhari A, Jennebach S, Kamenski T, Kettenberger H, Kuhn CD, Lehmann E, Leike K, Sydow JF, Vannini A. Structure of eukaryotic RNA polymerases. Annu Rev Biophys. 2008;37:337–352. doi: 10.1146/annurev.biophys.37.032807.130008. [DOI] [PubMed] [Google Scholar]
- 10.Werner M, Thuriaux P, Soutourina J. Structure-function analysis of RNA polymerases I and III. Curr Opin Struct Biol. 2009;19:740–745. doi: 10.1016/j.sbi.2009.10.005. [DOI] [PubMed] [Google Scholar]
- 11.Werner F, Grohmann D. Evolution of multisubunit RNA polymerases in the three domains of life. Nat Rev Microbiol. 2011;9:85–98. doi: 10.1038/nrmicro2507. [DOI] [PubMed] [Google Scholar]
- 12.Vannini A, Cramer P. Conservation between the RNA polymerase I, II, and III transcription initiation machineries. Mol Cell. 2012;45:439–446. doi: 10.1016/j.molcel.2012.01.023. [DOI] [PubMed] [Google Scholar]
- 13.Dequard-Chablat M, Riva M, Carles C, Sentenac A. RPC19, the gene for a subunit common to yeast RNA polymerases A (I) and C (III) The Journal of biological chemistry. 1991;266:15300–15307. [PubMed] [Google Scholar]
- 14.Mann C, Buhler JM, Treich I, Sentenac A. RPC40, a unique gene for a subunit shared between yeast RNA polymerases A and C. Cell. 1987;48:627–637. doi: 10.1016/0092-8674(87)90241-8. [DOI] [PubMed] [Google Scholar]
- 15.Woychik NA, McKune K, Lane WS, Young RA. Yeast RNA polymerase II subunit RPB11 is related to a subunit shared by RNA polymerase I and III. Gene expression. 1993;3:77–82. [PMC free article] [PubMed] [Google Scholar]
- 16.Kolodziej P, Young RA. RNA polymerase II subunit RPB3 is an essential component of the mRNA transcription apparatus. Molecular and cellular biology. 1989;9:5387–5394. doi: 10.1128/mcb.9.12.5387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ruan W, Lehmann E, Thomm M, Kostrewa D, Cramer P. Evolution of two modes of intrinsic RNA polymerase transcript cleavage. The Journal of biological chemistry. 2011;286:18701–18707. doi: 10.1074/jbc.M111.222273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Siaut M, Zaros C, Levivier E, Ferri ML, Court M, Werner M, Callebaut I, Thuriaux P, Sentenac A, Conesa C. An Rpb4/Rpb7-like complex in yeast RNA polymerase III contains the orthologue of mammalian CGRP-RCP. Molecular and cellular biology. 2003;23:195–205. doi: 10.1128/MCB.23.1.195-205.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sadhale PP, Woychik NA. C25, an essential RNA polymerase III subunit related to the RNA polymerase II subunit RPB7. Molecular and cellular biology. 1994;14:6164–6170. doi: 10.1128/mcb.14.9.6164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jasiak AJ, Armache KJ, Martens B, Jansen RP, Cramer P. Structural biology of RNA polymerase III: subcomplex C17/25 X-ray structure and 11 subunit enzyme model. Mol Cell. 2006;23:71–81. doi: 10.1016/j.molcel.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 21.Carter R, Drouin G. The increase in the number of subunits in eukaryotic RNA polymerase III relative to RNA polymerase II is due to the permanent recruitment of general transcription factors. Mol Biol Evol. 2010;27:1035–1043. doi: 10.1093/molbev/msp316. [DOI] [PubMed] [Google Scholar]
- 22.Geiger SR, Lorenzen K, Schreieck A, Hanecker P, Kostrewa D, Heck AJ, Cramer P. RNA polymerase I contains a TFIIF-related DNA-binding subcomplex. Mol Cell. 2010;39:583–594. doi: 10.1016/j.molcel.2010.07.028. [DOI] [PubMed] [Google Scholar]
- 23.Kuhn CD, Geiger SR, Baumli S, Gartmann M, Gerber J, Jennebach S, Mielke T, Tschochner H, Beckmann R, Cramer P. Functional architecture of RNA polymerase I. Cell. 2007;131:1260–1272. doi: 10.1016/j.cell.2007.10.051. [DOI] [PubMed] [Google Scholar]
- 24.Penrod Y, Rothblum K, Rothblum LI. Characterization of the Interactions of Mammalian RNA Polymerase I Associated Proteins PAF53 and PAF49. Biochemistry. 2012 doi: 10.1021/bi300408q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Desmoucelles C, Pinson B, Saint-Marc C, Daignan-Fornier B. Screening the yeast "disruptome" for mutants affecting resistance to the immunosuppressive drug, mycophenolic acid. The Journal of biological chemistry. 2002;277:27036–27044. doi: 10.1074/jbc.M111433200. [DOI] [PubMed] [Google Scholar]
- 26.Beckouet F, Labarre-Mariotte S, Albert B, Imazawa Y, Werner M, Gadal O, Nogi Y, Thuriaux P. Two RNA polymerase I subunits control the binding and release of Rrn3 during transcription. Molecular and cellular biology. 2008;28:1596–1605. doi: 10.1128/MCB.01464-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lane LA, Fernandez-Tornero C, Zhou M, Morgner N, Ptchelkine D, Steuerwald U, Politis A, Lindner D, Gvozdenovic J, Gavin AC, Muller CW, Robinson CV. Mass spectrometry reveals stable modules in holo and apo RNA polymerases I and III. Structure. 2011;19:90–100. doi: 10.1016/j.str.2010.11.009. [DOI] [PubMed] [Google Scholar]
- 28.Chen ZA, Jawhari A, Fischer L, Buchen C, Tahir S, Kamenski T, Rasmussen M, Lariviere L, Bukowski-Wills JC, Nilges M, Cramer P, Rappsilber J. Architecture of the RNA polymerase II-TFIIF complex revealed by cross-linking and mass spectrometry. The EMBO journal. 2010;29:717–726. doi: 10.1038/emboj.2009.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eichner J, Chen HT, Warfield L, Hahn S. Position of the general transcription factor TFIIF within the RNA polymerase II transcription preinitiation complex. The EMBO journal. 2010;29:706–716. doi: 10.1038/emboj.2009.386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu CC, Lin YC, Chen HT. The TFIIF-like Rpc37/53 dimer lies at the center of a protein network to connect TFIIIC, Bdp1, and the RNA polymerase III active center. Molecular and cellular biology. 2011;31:2715–2728. doi: 10.1128/MCB.05151-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bischler N, Brino L, Carles C, Riva M, Tschochner H, Mallouh V, Schultz P. Localization of the yeast RNA polymerase I-specific subunits. The EMBO journal. 2002;21:4136–4144. doi: 10.1093/emboj/cdf392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.De Carlo S, Carles C, Riva M, Schultz P. Cryo-negative staining reveals conformational flexibility within yeast RNA polymerase I. J Mol Biol. 2003;329:891–902. doi: 10.1016/s0022-2836(03)00510-2. [DOI] [PubMed] [Google Scholar]
- 33.Jennebach S, Herzog F, Aebersold R, Cramer P. Crosslinking-MS analysis reveals RNA polymerase I domain architecture and basis of rRNA cleavage. Nucleic Acids Res. 2012;40:5591–5601. doi: 10.1093/nar/gks220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vannini A, Ringel R, Kusser AG, Berninghausen O, Kassavetis GA, Cramer P. Molecular basis of RNA polymerase III transcription repression by Maf1. Cell. 2010;143:59–70. doi: 10.1016/j.cell.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 35.Grunberg S, Warfield L, Hahn S. Architecture of the RNA polymerase II preinitiation complex and mechanism of ATP-dependent promoter opening. Nat Struct Mol Biol. 2012 doi: 10.1038/nsmb.2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Keener J, Dodd JA, Lalo D, Nomura M. Histones H3 and H4 are components of upstream activation factor required for the high-level transcription of yeast rDNA by RNA polymerase I. Proc Natl Acad Sci U S A. 1997;94:13458–13462. doi: 10.1073/pnas.94.25.13458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Keys DA, Lee BS, Dodd JA, Nguyen TT, Vu L, Fantino E, Burson LM, Nogi Y, Nomura M. Multiprotein transcription factor UAF interacts with the upstream element of the yeast RNA polymerase I promoter and forms a stable preinitiation complex. Genes Dev. 1996;10:887–903. doi: 10.1101/gad.10.7.887. [DOI] [PubMed] [Google Scholar]
- 38.Lalo D, Steffan JS, Dodd JA, Nomura M. RRN11 encodes the third subunit of the complex containing Rrn6p and Rrn7p that is essential for the initiation of rDNA transcription by yeast RNA polymerase I. The Journal of biological chemistry. 1996;271:21062–21067. doi: 10.1074/jbc.271.35.21062. [DOI] [PubMed] [Google Scholar]
- 39.Lin CW, Moorefield B, Payne J, Aprikian P, Mitomo K, Reeder RH. A novel 66-kilodalton protein complexes with Rrn6, Rrn7, and TATA-binding protein to promote polymerase I transcription initiation in Saccharomyces cerevisiae. Molecular and cellular biology. 1996;16:6436–6443. doi: 10.1128/mcb.16.11.6436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aprikian P, Moorefield B, Reeder RH. New model for the yeast RNA polymerase I transcription cycle. Molecular and cellular biology. 2001;21:4847–4855. doi: 10.1128/MCB.21.15.4847-4855.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bier M, Fath S, Tschochner H. The composition of the RNA polymerase I transcription machinery switches from initiation to elongation mode. FEBS letters. 2004;564:41–46. doi: 10.1016/S0014-5793(04)00311-4. [DOI] [PubMed] [Google Scholar]
- 42.Goetze H, Wittner M, Hamperl S, Hondele M, Merz K, Stoeckl U, Griesenbeck J. Alternative chromatin structures of the 35S rRNA genes in Saccharomyces cerevisiae provide a molecular basis for the selective recruitment of RNA polymerases I and II. Molecular and cellular biology. 2010;30:2028–2045. doi: 10.1128/MCB.01512-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Boukhgalter B, Liu M, Guo A, Tripp M, Tran K, Huynh C, Pape L. Characterization of a fission yeast subunit of an RNA polymerase I essential transcription initiation factor, SpRrn7h/TAF(I)68, that bridges yeast and mammals: association with SpRrn11h and the core ribosomal RNA gene promoter. Gene. 2002;291:187–201. doi: 10.1016/s0378-1119(02)00597-8. [DOI] [PubMed] [Google Scholar]
- 44.Russell J, Zomerdijk JC. The RNA polymerase I transcription machinery. Biochem Soc Symp. 2006:203–216. doi: 10.1042/bss0730203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gorski JJ, Pathak S, Panov K, Kasciukovic T, Panova T, Russell J, Zomerdijk JC. A novel TBP-associated factor of SL1 functions in RNA polymerase I transcription. The EMBO journal. 2007;26:1560–1568. doi: 10.1038/sj.emboj.7601601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Denissov S, van Driel M, Voit R, Hekkelman M, Hulsen T, Hernandez N, Grummt I, Wehrens R, Stunnenberg H. Identification of novel functional TBP-binding sites and general factor repertoires. The EMBO journal. 2007;26:944–954. doi: 10.1038/sj.emboj.7601550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thomas MC, Chiang CM. The general transcription machinery and general cofactors. Crit Rev Biochem Mol Biol. 2006;41:105–178. doi: 10.1080/10409230600648736. [DOI] [PubMed] [Google Scholar]
- 48.McStay B, Frazier MW, Reeder RH. xUBF contains a novel dimerization domain essential for RNA polymerase I transcription. Genes Dev. 1991;5:1957–1968. doi: 10.1101/gad.5.11.1957. [DOI] [PubMed] [Google Scholar]
- 49.Jantzen HM, Admon A, Bell SP, Tjian R. Nucleolar transcription factor hUBF contains a DNA-binding motif with homology to HMG proteins. Nature. 1990;344:830–836. doi: 10.1038/344830a0. [DOI] [PubMed] [Google Scholar]
- 50.Stefanovsky VY, Pelletier G, Bazett-Jones DP, Crane-Robinson C, Moss T. DNA looping in the RNA polymerase I enhancesome is the result of non-cooperative in-phase bending by two UBF molecules. Nucleic Acids Res. 2001;29:3241–3247. doi: 10.1093/nar/29.15.3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bazett-Jones DP, Leblanc B, Herfort M, Moss T. Short-range DNA looping by the Xenopus HMG-box transcription factor, xUBF. Science. 1994;264:1134–1137. doi: 10.1126/science.8178172. [DOI] [PubMed] [Google Scholar]
- 52.Hall DB, Wade JT, Struhl K. An HMG protein, Hmo1, associates with promoters of many ribosomal protein genes and throughout the rRNA gene locus in Saccharomyces cerevisiae. Molecular and cellular biology. 2006;26:3672–3679. doi: 10.1128/MCB.26.9.3672-3679.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Berger AB, Decourty L, Badis G, Nehrbass U, Jacquier A, Gadal O. Hmo1 is required for TOR-dependent regulation of ribosomal protein gene transcription. Molecular and cellular biology. 2007;27:8015–8026. doi: 10.1128/MCB.01102-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Albert B, Perez-Fernandez J, Leger-Silvestre I, Gadal O. Regulation of ribosomal RNA production by RNA polymerase I: does elongation come first? Genet Res Int. 2012;2012:276948. doi: 10.1155/2012/276948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Birch JL, Zomerdijk JC. Structure and function of ribosomal RNA gene chromatin. Biochem Soc Trans. 2008;36:619–624. doi: 10.1042/BST0360619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ha I, Lane WS, Reinberg D. Cloning of a human gene encoding the general transcription initiation factor IIB. Nature. 1991;352:689–695. doi: 10.1038/352689a0. [DOI] [PubMed] [Google Scholar]
- 57.Pinto I, Ware DE, Hampsey M. The yeast SUA7 gene encodes a homolog of human transcription factor TFIIB and is required for normal start site selection in vivo. Cell. 1992;68:977–988. doi: 10.1016/0092-8674(92)90040-j. [DOI] [PubMed] [Google Scholar]
- 58.Reinberg D, Roeder RG. Factors involved in specific transcription by mammalian RNA polymerase II. Purification and functional analysis of initiation factors IIB and IIE. The Journal of biological chemistry. 1987;262:3310–3321. [PubMed] [Google Scholar]
- 59.Buratowski S, Zhou H. A suppressor of TBP mutations encodes an RNA polymerase III transcription factor with homology to TFIIB. Cell. 1992;71:221–230. doi: 10.1016/0092-8674(92)90351-c. [DOI] [PubMed] [Google Scholar]
- 60.Colbert T, Hahn S. A yeast TFIIB-related factor involved in RNA polymerase III transcription. Genes Dev. 1992;6:1940–1949. doi: 10.1101/gad.6.10.1940. [DOI] [PubMed] [Google Scholar]
- 61.Lopez-De-Leon A, Librizzi M, Puglia K, Willis IM. PCF4 encodes an RNA polymerase III transcription factor with homology to TFIIB. Cell. 1992;71:211–220. doi: 10.1016/0092-8674(92)90350-l. [DOI] [PubMed] [Google Scholar]
- 62.Hahn S. Structural biology: New beginnings for transcription. Nature. 2009;462:292–293. doi: 10.1038/462292a. [DOI] [PubMed] [Google Scholar]
- 63.Knutson BA, Hahn S. Yeast Rrn7 and human TAF1B are TFIIB-related RNA polymerase I general transcription factors. Science. 2011;333:1637–1640. doi: 10.1126/science.1207699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Naidu S, Friedrich JK, Russell J, Zomerdijk JC. TAF1B is a TFIIB-like component of the basal transcription machinery for RNA polymerase I. Science. 2011;333:1640–1642. doi: 10.1126/science.1207656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Soding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33:W244–W248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schramm L, Hernandez N. Recruitment of RNA polymerase III to its target promoters. Genes Dev. 2002;16:2593–2620. doi: 10.1101/gad.1018902. [DOI] [PubMed] [Google Scholar]
- 67.Bartlett MS. Determinants of transcription initiation by archaeal RNA polymerase. Curr Opin Microbiol. 2005;8:677–684. doi: 10.1016/j.mib.2005.10.016. [DOI] [PubMed] [Google Scholar]
- 68.Kassavetis GA, Geiduschek EP. Transcription factor TFIIIB and transcription by RNA polymerase III. Biochem Soc Trans. 2006;34:1082–1087. doi: 10.1042/BST0341082. [DOI] [PubMed] [Google Scholar]
- 69.Khoo B, Brophy B, Jackson SP. Conserved functional domains of the RNA polymerase III general transcription factor BRF. Genes Dev. 1994;8:2879–2890. doi: 10.1101/gad.8.23.2879. [DOI] [PubMed] [Google Scholar]
- 70.Juo ZS, Kassavetis GA, Wang J, Geiduschek EP, Sigler PB. Crystal structure of a transcription factor IIIB core interface ternary complex. Nature. 2003;422:534–539. doi: 10.1038/nature01534. [DOI] [PubMed] [Google Scholar]
- 71.Hahn S, Young ET. Transcriptional regulation in Saccharomyces cerevisiae: transcription factor regulation and function, mechanisms of initiation, and roles of activators and coactivators. Genetics. 2011;189:705–736. doi: 10.1534/genetics.111.127019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hahn S, Roberts S. The zinc ribbon domains of the general transcription factors TFIIB and Brf: conserved functional surfaces but different roles in transcription initiation. Genes Dev. 2000;14:719–730. [PMC free article] [PubMed] [Google Scholar]
- 73.Imamura S, Hanaoka M, Tanaka K. The plant-specific TFIIB-related protein, pBrp, is a general transcription factor for RNA polymerase I. The EMBO journal. 2008;27:2317–2327. doi: 10.1038/emboj.2008.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kostrewa D, Zeller ME, Armache KJ, Seizl M, Leike K, Thomm M, Cramer P. RNA polymerase II-TFIIB structure and mechanism of transcription initiation. Nature. 2009;462:323–330. doi: 10.1038/nature08548. [DOI] [PubMed] [Google Scholar]
- 75.Memet S, Saurin W, Sentenac A. RNA polymerases B and C are more closely related to each other than to RNA polymerase A. The Journal of biological chemistry. 1988;263:10048–10051. [PubMed] [Google Scholar]
- 76.Carter R, Drouin G. The evolutionary rates of eukaryotic RNA polymerases and of their transcription factors are affected by the level of concerted evolution of the genes they transcribe. Mol Biol Evol. 2009;26:2515–2520. doi: 10.1093/molbev/msp164. [DOI] [PubMed] [Google Scholar]
- 77.Heix J, Grummt I. Species specificity of transcription by RNA polymerase I. Curr Opin Genet Dev. 1995;5:652–656. doi: 10.1016/0959-437x(95)80035-2. [DOI] [PubMed] [Google Scholar]
- 78.Moss T, Stefanovsky VY. Promotion and regulation of ribosomal transcription in eukaryotes by RNA polymerase I. Prog Nucleic Acid Res Mol Biol. 1995;50:25–66. doi: 10.1016/s0079-6603(08)60810-7. [DOI] [PubMed] [Google Scholar]
- 79.Wang Y, Roberts SG. New insights into the role of TFIIB in transcription initiation. Transcription. 2010;1:126–129. doi: 10.4161/trns.1.3.12900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Singh BN, Hampsey M. A transcription-independent role for TFIIB in gene looping. Mol Cell. 2007;27:806–816. doi: 10.1016/j.molcel.2007.07.013. [DOI] [PubMed] [Google Scholar]
- 81.Chen HT, Hahn S. Binding of TFIIB to RNA polymerase II: Mapping the binding site for the TFIIB zinc ribbon domain within the preinitiation complex. Mol Cell. 2003;12:437–447. doi: 10.1016/s1097-2765(03)00306-x. [DOI] [PubMed] [Google Scholar]
- 82.Chen HT, Hahn S. Mapping the location of TFIIB within the RNA polymerase II transcription preinitiation complex: a model for the structure of the PIC. Cell. 2004;119:169–180. doi: 10.1016/j.cell.2004.09.028. [DOI] [PubMed] [Google Scholar]
- 83.Bushnell DA, Westover KD, Davis RE, Kornberg RD. Structural basis of transcription: an RNA polymerase II-TFIIB cocrystal at 4.5 Angstroms. Science. 2004;303:983–988. doi: 10.1126/science.1090838. [DOI] [PubMed] [Google Scholar]
- 84.Liu X, Bushnell DA, Wang D, Calero G, Kornberg RD. Structure of an RNA polymerase II-TFIIB complex and the transcription initiation mechanism. Science. 2010;327:206–209. doi: 10.1126/science.1182015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kassavetis GA, Kumar A, Letts GA, Geiduschek EP. A post-recruitment function for the RNA polymerase III transcription-initiation factor IIIB. Proc Natl Acad Sci U S A. 1998;95:9196–9201. doi: 10.1073/pnas.95.16.9196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kassavetis GA, Letts GA, Geiduschek EP. The RNA polymerase III transcription initiation factor TFIIIB participates in two steps of promoter opening. The EMBO journal. 2001;20:2823–2834. doi: 10.1093/emboj/20.11.2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kassavetis GA, Han S, Naji S, Geiduschek EP. The role of transcription initiation factor IIIB subunits in promoter opening probed by photochemical crosslinking. The Journal of biological chemistry. 2003;278:17912–17917. doi: 10.1074/jbc.M300743200. [DOI] [PubMed] [Google Scholar]
- 88.Fishburn J, Hahn S. Architecture of the yeast RNA polymerase II open complex and regulation of activity by TFIIF. Molecular and cellular biology. 2012;32:12–25. doi: 10.1128/MCB.06242-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Peyroche G, Milkereit P, Bischler N, Tschochner H, Schultz P, Sentenac A, Carles C, Riva M. The recruitment of RNA polymerase I on rDNA is mediated by the interaction of the A43 subunit with Rrn3. The EMBO journal. 2000;19:5473–5482. doi: 10.1093/emboj/19.20.5473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Miller G, Panov KI, Friedrich JK, Trinkle-Mulcahy L, Lamond AI, Zomerdijk JC. hRRN3 is essential in the SL1-mediated recruitment of RNA Polymerase I to rRNA gene promoters. The EMBO journal. 2001;20:1373–1382. doi: 10.1093/emboj/20.6.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cavanaugh AH, Evans A, Rothblum LI. Mammalian Rrn3 is required for the formation of a transcription competent preinitiation complex containing RNA polymerase I. Gene expression. 2008;14:131–147. [PMC free article] [PubMed] [Google Scholar]
- 92.Blattner C, Jennebach S, Herzog F, Mayer A, Cheung AC, Witte G, Lorenzen K, Hopfner KP, Heck AJ, Aebersold R, Cramer P. Molecular basis of Rrn3-regulated RNA polymerase I initiation and cell growth. Genes Dev. 2011;25:2093–2105. doi: 10.1101/gad.17363311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Littlefield O, Korkhin Y, Sigler PB. The structural basis for the oriented assembly of a TBP/TFB/promoter complex. Proc Natl Acad Sci U S A. 1999;96:13668–13673. doi: 10.1073/pnas.96.24.13668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bagby S, Kim S, Maldonado E, Tong KI, Reinberg D, Ikura M. Solution structure of the C-terminal core domain of human TFIIB: similarity to cyclin A and interaction with TATA-binding protein. Cell. 1995;82:857–867. doi: 10.1016/0092-8674(95)90483-2. [DOI] [PubMed] [Google Scholar]
- 95.Nikolov DB, Chen H, Halay ED, Usheva AA, Hisatake K, Lee DK, Roeder RG, Burley SK. Crystal structure of a TFIIB-TBP-TATA-element ternary complex. Nature. 1995;377:119–128. doi: 10.1038/377119a0. [DOI] [PubMed] [Google Scholar]
- 96.Schroder O, Bryant GO, Geiduschek EP, Berk AJ, Kassavetis GA. A common site on TBP for transcription by RNA polymerases II and III. The EMBO journal. 2003;22:5115–5124. doi: 10.1093/emboj/cdg476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Saxena A, Ma B, Schramm L, Hernandez N. Structure-function analysis of the human TFIIB-related factor II protein reveals an essential role for the C-terminal domain in RNA polymerase III transcription. Molecular and cellular biology. 2005;25:9406–9418. doi: 10.1128/MCB.25.21.9406-9418.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Cabart P, Murphy S. BRFU, a TFIIB-like factor, is directly recruited to the TATA-box of polymerase III small nuclear RNA gene promoters through its interaction with TATA-binding protein. The Journal of biological chemistry. 2001;276:43056–43064. doi: 10.1074/jbc.M108515200. [DOI] [PubMed] [Google Scholar]
- 99.Schultz MC, Reeder RH, Hahn S. Variants of the TATA-binding protein can distinguish subsets of RNA polymerase I, II, and III promoters. Cell. 1992;69:697–702. doi: 10.1016/0092-8674(92)90233-3. [DOI] [PubMed] [Google Scholar]
- 100.Cormack BP, Struhl K. The TATA-binding protein is required for transcription by all three nuclear RNA polymerases in yeast cells. Cell. 1992;69:685–696. doi: 10.1016/0092-8674(92)90232-2. [DOI] [PubMed] [Google Scholar]
- 101.Aprikian P, Moorefield B, Reeder RH. TATA binding protein can stimulate core-directed transcription by yeast RNA polymerase I. Molecular and cellular biology. 2000;20:5269–5275. doi: 10.1128/mcb.20.14.5269-5275.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Steffan JS, Keys DA, Vu L, Nomura M. Interaction of TATA-binding protein with upstream activation factor is required for activated transcription of ribosomal DNA by RNA polymerase I in Saccharomyces cerevisiae in vivo. Molecular and cellular biology. 1998;18:3752–3761. doi: 10.1128/mcb.18.7.3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Keener J, Josaitis CA, Dodd JA, Nomura M. Reconstitution of yeast RNA polymerase I transcription in vitro from purified components. TATA-binding protein is not required for basal transcription. The Journal of biological chemistry. 1998;273:33795–33802. doi: 10.1074/jbc.273.50.33795. [DOI] [PubMed] [Google Scholar]
- 104.Bedwell GJ, Appling FD, Anderson SJ, Schneider DA. Efficient transcription by RNA polymerase I using recombinant core factor. Gene. 2012;492:94–99. doi: 10.1016/j.gene.2011.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Steffan JS, Keys DA, Dodd JA, Nomura M. The role of TBP in rDNA transcription by RNA polymerase I in Saccharomyces cerevisiae: TBP is required for upstream activation factor-dependent recruitment of core factor. Genes Dev. 1996;10:2551–2563. doi: 10.1101/gad.10.20.2551. [DOI] [PubMed] [Google Scholar]
- 106.Comai L, Zomerdijk JC, Beckmann H, Zhou S, Admon A, Tjian R. Reconstitution of transcription factor SL1: exclusive binding of TBP by SL1 or TFIID subunits. Science. 1994;266:1966–1972. doi: 10.1126/science.7801123. [DOI] [PubMed] [Google Scholar]
- 107.Zomerdijk JC, Beckmann H, Comai L, Tjian R. Assembly of transcriptionally active RNA polymerase I initiation factor SL1 from recombinant subunits. Science. 1994;266:2015–2018. doi: 10.1126/science.7801130. [DOI] [PubMed] [Google Scholar]
- 108.Rojas DA, Moreira-Ramos S, Zock-Emmenthal S, Urbina F, Contreras-Levicoy J, Kaufer NF, Maldonado E. Rrn7 protein, an RNA polymerase I transcription factor, is required for RNA polymerase II-dependent transcription directed by core promoters with a HomolD box sequence. The Journal of biological chemistry. 2011;286:26480–26486. doi: 10.1074/jbc.M111.224337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Contreras-Levicoy J, Moreira-Ramos S, Rojas DA, Urbina F, Maldonado E. Transcription directed by human core promoters with a HomolD box sequence requires DDB1, RECQL and RNA polymerase II machinery. Gene. 2012 doi: 10.1016/j.gene.2012.05.059. [DOI] [PubMed] [Google Scholar]
- 110.Lagrange T, Hakimi MA, Pontier D, Courtois F, Alcaraz JP, Grunwald D, Lam E, Lerbs-Mache S. Transcription factor IIB (TFIIB)-related protein (pBrp), a plant-specific member of the TFIIB-related protein family. Molecular and cellular biology. 2003;23:3274–3286. doi: 10.1128/MCB.23.9.3274-3286.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Chen YH, Li HJ, Shi DQ, Yuan L, Liu J, Sreenivasan R, Baskar R, Grossniklaus U, Yang WC. The central cell plays a critical role in pollen tube guidance in Arabidopsis. Plant Cell. 2007;19:3563–3577. doi: 10.1105/tpc.107.053967. [DOI] [PMC free article] [PubMed] [Google Scholar]