Abstract
The epithelial Na+ channel (ENaC) is a heteromeric protein complex playing a fundamental role in Na+ homeostasis and blood pressure regulation. Specific mutations inactivating PY motifs in ENaC C termini cause Liddle's syndrome, an inherited form of hypertension. Previously we showed that these PY motifs serve as binding sites for the E3 enzyme Nedd4-2, implying ubiquitination as a regulatory mechanism of ENaC. Ubiquitination involves the sequential action of E1, E2, and E3 enzymes. Here we identify the E2 enzyme UBE2E3, which acts in concert with Nedd4-2, and show by coimmunoprecipitation that UBE2E3 and Nedd4-2 interact together. In Xenopus laevis oocytes, UBE2E3 reduces ENaC activity marginally, consistent with Nedd4-2 being the rate-limiting factor in this process, whereas a catalytically inactive mutant of UBE2E3 (UBE2E3-CS) causes elevated ENaC activity by increasing cell surface expression. No additive effect is observed when UBE2E3-CS is coexpressed with an inactive Nedd4-2 mutant, and the stimulatory role of UBE2E3-CS depends on the integrity of the PY motifs (Nedd4-2 binding sites) and the ubiquitination sites on ENaC. In renal mpkCCDcl4 cells, displaying ENaC-dependent transepithelial Na+ transport, Nedd4-2 and UBE2E3 can be coimmunoprecipitated and overexpression of UBE2E3 affects Na+ transport, corroborating the concept of a concerted action of UBE2E3 and Nedd4-2 in ENaC regulation.
Covalent attachment of ubiquitin to proteins is a posttranslational modification that targets membrane proteins for internalization and/or degradation (17). Ubiquitination involves the successive action of a ubiquitin-activating enzyme (E1), a ubiquitin-conjugating or ubiquitin carrier enzyme (E2), and a ubiquitin-protein ligase (E3) (26). Possible involvement of E4 enzymes may lead to the formation of polyubiquitin chains (19, 24, 41). The unique E1 enzyme activates free ubiquitin in an ATP-dependent manner by forming a thioester bond with its catalytic cysteine and then transfers it to the cysteine of an E2 enzyme. Approximately 30 E2 enzymes are encoded in the human genome, and they can be separated into four classes on the basis of their primary structure. Class I E2 enzymes consist of approximately 150 residues which bear a highly conserved catalytic domain named UBC domain (ubiquitin conjugating domain, including a catalytic cysteine). Class II and III enzymes possess additional C- and N-terminal extensions, respectively (46). The class IV E2s have both N- and C-terminal elongations (4, 33). E2s usually form a complex with an E3 enzyme, which recognizes the target protein and catalyzes the attachment of ubiquitin via the formation of an isopeptide bond on lysine ɛ-NH2 groups. There are probably hundreds of E3 enzymes comprising different structures. They can be divided into several main groups on the basis of the presence of one of the following domains: (i) RING fingers; (ii) U-boxes; (iii) PHD domains; (iv) HECT (homologous to E6-AP-carboxy-terminal) domains.
The epithelial Na+ channel (ENaC) is located at the apical membrane of Na+-transporting epithelia of the kidney, colon, lung, and other tissues and plays a crucial role in the control of Na+ balance, blood volume, and pressure (40). It is composed of three homologous subunits (α, β, and γ), each containing two transmembrane domains, a large extracellular loop, and short cytosolic N and C termini. Each subunit also contains a PY motif (xPPxYxxL) in the C-terminal region. Interestingly, the PY motifs of β- or γ-ENaC are either deleted or mutated in most forms of Liddle's syndrome, an inherited form of human hypertension, which is characterized by severe salt-sensitive hypertension, hypokalemia, and metabolic alkalosis (20, 30, 43, 64, 71).
Using the Xenopus laevis oocyte system, which has been proven to be a particularly powerful tool to study the properties and certain regulatory aspects of ENaC, it has been shown that channels containing Liddle mutations display increased amiloride-sensitive Na+ currents (a measure of ENaC activity) (59, 67). These augmented currents can be explained by increased channel number at the plasma membrane, elevated open probability (15), and reduced Na+ feedback regulation (39). We and others have shown that the PY motifs interact with WW domains of a HECT (homologous to E6-AP-carboxy terminal [29]) domain-containing subfamily of E3 enzymes, the Nedd4/Nedd4-like family of ubiquitin-protein ligases (58). Specifically, binding to the WW domains of Nedd4-2 (6, 11, 21, 25, 34), Nedd4-1 (5, 12, 14, 16, 22, 25, 36, 37, 44, 62, 65, 69), WWP1 (54), and WWP2 (48, 54) was demonstrated by various approaches, suggesting that ENaC is regulated by ubiquitination. Indeed, it could be confirmed that ENaC subunits become ubiquitinated (70) and exhibit rapid turnover (47, 70), a hallmark of ubiquitinated proteins. This rapid turnover could be slowed by inhibition with either lysosomal or proteasomal inhibitors (45, 70). It was also found that Nedd4-2 regulates ENaC activity in a ubiquitination-dependent manner when coexpressed either in X. laevis oocytes (2, 11, 16, 21, 25, 34, 35, 42, 66) or in epithelial cells (66), although regulation by other members of the Nedd4/Nedd4-like family of E3 enzymes was also demonstrated, namely, Nedd4-1 (12, 14, 18, 22, 35, 65) and WWP2 (48). The relative efficiencies of human Nedd4-1 and Nedd4-2 with respect to ENaC regulation in X. laevis oocytes have been compared, and it has been shown that human Nedd4-2 is more efficient than Nedd4-1 (35). In support of a regulatory role of Nedd4-2 is the recent finding that Sgk1 (serum- and glucocorticoid-induced kinase 1), an aldosterone-induced kinase in cells of the cortical collecting duct, phosphorylates Nedd4-2, which perturbs ENaC-Nedd4-2 interaction, suggesting a link between aldosterone-dependent regulation of ENaC and Nedd4-2 (11, 66).
To date, it is not known which E2 enzyme(s) is involved in Nedd4-2-dependent regulation of ENaC. A number of E2s have been shown to be able to transfer ubiquitin onto the HECT domain of either Rsp5 (the Saccharomyces cerevisiae orthologue of Nedd4) or Nedd4-1, including Ubc4/Ubc5 (S. cerevisiae), UbcH5B, UbcH5C, UbcH6, and UbcH7 (all human) (3, 23, 51). Moreover, it has been demonstrated that E2 enzymes interact via their UBC domain with a HECT domain (23, 28, 50). Accordingly, we performed a two-hybrid screen using the C-terminal region of Nedd4-2 (including the HECT domain) as bait and isolated the E2 enzyme UBE2E3 from X. laevis. We demonstrate that UBE2E3 binds to Nedd4-2, transfers ubiquitin to Nedd4-2, and acts in concert with this enzyme in the ubiquitination of bacterial proteins in vitro. Additionally, we show in X. laevis oocytes that UBE2E3 can influence ENaC cell surface expression in a Nedd4-2-dependent manner. Moreover, UBE2E3 affects transepithelial Na+ transport in mpkCCDcl4 cells. These data suggest that UBE2E3 is the E2 enzyme involved in Nedd4-2-dependent regulation of ENaC and for the first time, demonstrate the involvement of a ubiquitin-conjugating enzyme in ion channel regulation.
MATERIALS AND METHOD
Yeast two-hybrid screen
The C-terminal part of Xenopus Nedd4-2 (residues 581 to 971) was amplified by PCR (nucleotides 1841 to 3019) and cloned into the LexA-based bait plasmid pBTM116 (72) (pBTM116-Nedd4-2-CT) using EcoRI/SalI linkers. For the two-hybrid screen, the yeast strain L40 was cotransformed with pBTM116-Nedd4-2-CT and a X. laevis oocyte cDNA library (Matchmaker; Clontech) by standard yeast techniques. Positive clones were selected on medium lacking leucine, tryptophan, and histidine and checked with a filter assay for β-galactosidase activity. Prey plasmids were then isolated and retransformed into yeast with pBTM116-Nedd4-2-CT or with pBTM116-βrENaC-N terminus as a negative control to confirm the specificity of the interaction. Sequencing of the positive clones revealed the presence of a ubiquitin-conjugating enzyme which, in view of its high homology, likely represents the X. laevis orthologue of mammalian UBE2E3. Quantitative β-galactosidase assays were performed as described previously (69).
cDNA constructs
DNA fragment encoding the full-length Xenopus UBE2E3 (GenBank accession no. AY281323) (nucleotides 91 to 741) was subcloned into pSDeasySB (57), pGEX-4T-1 (Amersham), and pET-30a (Novagen) vectors using EcoRI/XhoI linkers. To create the catalytically inactive mutants (UBE2E3-CS), Cys145 was mutated to Ser in the pSDeasySB, pGEX-4T-1, and pET-30a constructs by a PCR-based method (49). The mutant UBE2E3 F122N CS was generated by mutating Phe122 into Asn from the pSDeasySB-UBE2E3-CS construct. Mouse UBE2E3 (also known as UbcM2; GenBank accession no. AF003346), was subcloned into pSDEasySB (nucleotides 4 to 627) from an expressed sequence tag clone (GenBank accession no. BE335264) using HindIII/XhoI linkers. Mouse UBE2E3 was labeled with an S-tag (KETAAAKFERNHMDS; Novagen) at its C terminus. For retroviral expression in mpkCCDcl4 cells, mouse UBE2E3 enzymes (wild type, catalytically inactive [UBE2E3-CS], or labeled with an S-tag) were subcloned into the Epstein-Barr virus-based retroviral vector (LZRS) (53). Mouse Ubc4 (mUbc4) (GenBank accession no. U62483) was subcloned into pGEX-4T-1 (nucleotides 377 to 832) from an expressed sequence tag clone (GenBank accession no. AA276008) using EcoRI/NotI linkers and into pSDeasyBS (nucleotides 377 to 1572) using EcoRI/StuI linkers. The mutant mUbc4-CS was obtained by mutating Cys85 into Ser. Xenopus Nedd4-2 (nucleotides 103 to 3019) was subcloned into pET-30a from the pSDeasy construct described previously (2) using BamHI/SalI linkers. The following rat ENaC (rENaC) constructs were used: wild-type α, β, and γ rENaC subunits or rENaC subunits lacking a functional PY motif (ENaCΔPY). These mutants were generated by mutating Tyr673 to Ala in αrENaC, Tyr618 to His in βrENaC, and Tyr628 to Ala in γrENaC (as described previously [60]). α- and γ-rENaC with conserved lysine residues at the N terminus mutated to arginine (ENaCΔK) to generate a ubiquitination-deficient ENaC channel were described before (70). The rat ENaC subunits labeled with a FLAG epitope were described previously by Firsov et al. (15). For the filter and liquid β-galactosidase activity assays, the N-terminal part of UBE2E3 (residues 1 to 60) or UBE2E3 lacking its N-terminal part (residues 61 to 207) were cloned into the pACT2 vector (Clontech), using PCR-based methods and subcloning into the EcoRI/blunt or EcoRI/XhoI sites, respectively.
Expression in Xenopus oocytes, electrophysiological measurements, and cell surface binding assay
pSDeasy plasmids encoding ENaC, wild-type UBE2E3, UBE2E3-CS, UBE2E3 F122N CS, wild-type mUbc4, mUbc4-CS, and X. laevis Nedd4-2-CS (2) proteins were linearized and transcribed as described previously (60), the cRNA was injected into Xenopus oocytes, and after 16 to 24 h, amiloride-sensitive Na+ currents were measured by the two-electrode voltage-clamp method (60). The following quantities of cRNA were injected: wild-type rENaC, 3 ng of each subunit; rENaCΔPY or rENaCΔK, 1.5 or 0.15 ng (1/10) of each subunit; FLAG-tagged rENaC, 1.5 ng of each subunit; X. laevis Nedd4-2-CS, 10 ng; wild-typeUBE2E3 or UBE2E3-CS, 10 ng. For the dose-response analysis, increasing amounts of UBE2E3-CS were used (0, 2, 5, 10, and 20 ng). For the experiment with brefeldin A (BFA), the oocytes were incubated in modified Barth's solution (MBS) (60) containing 10 μg of BFA per ml for the indicated time. The binding assay using anti-FLAG antibodies was performed in MBS as described previously (15). Statistical analysis was performed with the two-tailed Student t test analysis for unpaired data.
In vitro ubiquitination assay
X. laevis Nedd4-2 (xNedd4-2) was translated and radiolabeled with [35S]methionine (Redivue l-[35S]methionine; Amersham) using the TNT Quick coupled transcription/translation system (Promega). The ubiquitination assay was performed in ubiquitination buffer (25 mM Tris-HCl, 125 mM NaCl, 2 mM MgCl2, 50 μM dithiothreitol, 2 mM ATP). Reaction mixtures contained 0.5 μg of E1 enzyme (rabbit; Calbiochem), 0.8 μg of the respective E2 enzyme (glutathione S-transferase [GST]-UBE2E3, GST-UBE2E3-CS, or GST-mUbc4), 3 μg of GST-ubiquitin, and 2 μl of the radiolabeled xNedd4-2. After 1 h at 30°C with shaking, reactions were stopped by boiling the mixtures for 5 min at 95°C in sodium dodecyl sulfate (SDS) sample buffer (25 mM sucrose, 5 mM EDTA, 30 mM Tris-HCl [pH 8.8], 0.25% bromophenol blue, 10% SDS). Samples were then analyzed by SDS-PAGE (8% polyacrylamide) and autoradiography. For the ubiquitination of bacterial proteins, 5 μl of crude lysate from Escherichia coli was mixed in ubiquitination buffer with 1 μg of ubiquitin (Sigma), 0.5 μg of E1 (Calbiochem), 1 μg of His6-tagged wild-type UBE2E3 or His6-tagged UBE2E3-CS, and 0.5 μg of His6-tagged xNedd4-2, and incubated for 2 h at 30°C with moderate shaking. Reactions were stopped by boiling for 5 min at 95°C in SDS sample buffer. Samples were separated by SDS-PAGE (8% polyacrylamide), transferred to nitrocellulose membranes, and analyzed by Western blotting using FK2 mouse monoclonal antibody directed against ubiquitin (Affiniti Research).
In vitro binding assay and in vivo coimmunoprecipitation
For the binding assay, 2 μl of [35S]methionine-labeled xNedd4-2 translated in vitro was mixed with 2 μg of the respective purified GST fusion protein (GST, GST-UBE2E3, or GST-mUbc4) and 20 μl of glutathione agarose beads in pull-down buffer (100 mM NaCl, 20 mM Tris-HCl [pH 7.4], 1% Triton X-100). Reaction mixtures were incubated for 4 h at 4°C on a wheel and washed four times with 1 ml of pull-down buffer. Samples were then boiled in SDS sample buffer for 5 min at 95°C and analyzed by SDS-PAGE (8% polyacrylamide) followed by autoradiography.
For the coimmunoprecipitation experiment, X. laevis oocytes were injected with 5 ng of mouse Nedd4-2 and 5 ng of S-tagged or nontagged mouse UBE2E3. After overnight incubation in MBS, oocytes were lysed in Triton X-100 homogenization buffer (20 mM Tris-HCl [pH 7.4], 100 mM NaCl, 1% Triton X-100, 1 mM phenylmethylsulfonyl fluoride, 10 μg of leupeptin per ml, 10 μg of pepstatin A per ml, 10 μg of aprotinin per ml) (25 μl per oocyte). For the coimmunoprecipitation in mpkCCDcl4 cells, cells grown on plastic dishes were lysed in lysis buffer (50 mM HEPES [pH 7.5], 150 mM NaCl, 1.5 mM MgCl2, 1 mM EGTA, 10% glycerol, 1% Triton X-100, 1 mM phenylmethylsulfonyl fluoride, 10 μg of leupeptin per ml, 10 μg of pepstatin A per ml, 10 μg of aprotinin per ml). After centrifugation at 4°C for 15 min at 20,000 × g, the supernatant was recovered, and immunoprecipitation was performed with anti-mouse Nedd4-2 antibodies and protein A Sepharose. The immunoprecipitated material was analyzed by SDS-PAGE (10% polyacrylamide). Separated proteins were transferred to nitrocellulose membranes and then affinity blotted using the S-protein horseradish peroxidase (HRP) conjugate (Novagen) at a 1:10,000 dilution.
Expression of UBE2E3 and electrophysiological measurements in mpkCCDcl4 cells
Virus packaging cells (Phoenix cells) were transfected with retroviral vector (LZRS) containing wild-type UBE2E3, UBE2E3-CS, or S-tagged UBE2E3 to produce recombinant viral particles. The recombinant viruses harvested were immediately used for transduction of the mpkCCDcl4 cells as described previously (7). The expression of UBE2E3 was checked by Western blotting using an anti-UBE2E3 antibody or by affinity blotting using S-protein HRP conjugate. Electrophysiological measurements (amiloride-sensitive short-circuit currents [ISC]) were performed on confluent cell monolayers grown on collagen-coated filters (4.7-cm2 Transwell; Corning Costar, Cambridge, Mass.) as described by Auberson et al. (7).
Northern blot analysis
Total RNA and poly(A+) mRNA were isolated from mpkCCDcl4 cells grown on collagen-coated filters, separated on a 1% agarose gel, and transferred to nitrocellulose membranes. The blot was then hybridized using a radiolabeled probe specific for mouse UBE2E3 (GenBank accession no. AF003346; nucleotides 531 to 1011) and analyzed by autoradiography.
Production of fusion proteins
pGEX-4T-1 constructs were transformed into E. coli K-12, and protein expression was induced with 0.1 mM isopropyl β-thiogalactoside (IPTG) for 3 h at 30°C. Proteins were affinity purified with glutathione-agarose beads. pET-30a constructs were transformed into E. coli BL-21, and protein expression was induced with 1 mM IPTG for 3 h at 30°C. Proteins were then affinity purified on Ni2+-agarose beads, eluted, and dialyzed overnight against phosphate-buffered saline at 4°C.
Antibodies
The anti-UBE2E3 antibody was raised against a GST fusion protein containing amino acids 1 to 60 of Xenopus UBE2E3 and produced in a rabbit by Cocalico Biologicals, Inc. (Reamstown, United Kingdom). For the competition experiment with the antigen (see Fig. 8B), the anti-UBE2E3 antibody was incubated for 30 min at 4°C with 0.5 μg of purified GST-UBE2E3 diluted in phosphate-buffered saline prior to Western blotting. The anti-Nedd4-2 antibody has been described previously (35).
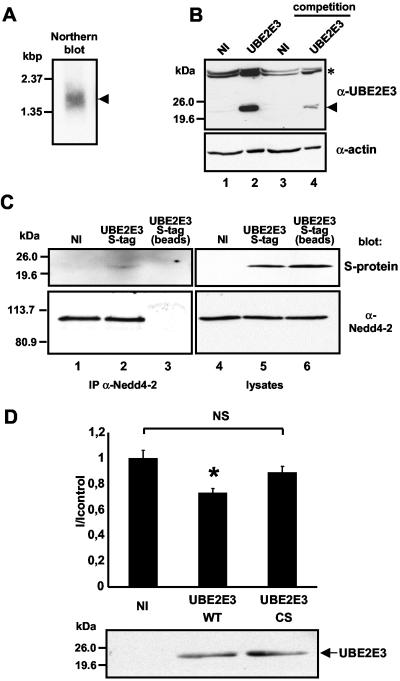
FIG. 8.
UBE2E3 is expressed in renal epithelial cells and does influence transepithelial Na+ transport in mpkCCDcl4 cells. (A) Northern blot analysis of poly(A+) RNA isolated from mpkCCDcl4 cells using a specific probe for mouse UBE2E3 (arrowhead). (B) Western blot analysis on lysates from mpkCCDcl4 cells either not infected (NI) or overexpressing UBE2E3 using an anti-UBE2E3 antibody (α-UBE2E3). Endogenous and exogenous UBE2E3 are detected at the expected position (lanes 1 and 2, arrowhead). For the competition (lanes 3 and 4), the anti-UBE2E3 antibody was incubated with 0.5 μg of purified GST-UBE2E3 prior to Western blotting. An antiactin antibody (α-actin) was used as a loading control. Cross-reacting bands of higher molecular size (*) are indicated. (C) Lysates from wild-type mpkCCDcl4 cells (not infected [NI]) or mpkCCDcl4 cells overexpressing S-tag UBE2E3 (S-tag UBE2E3) were subjected to immunoprecipitation (IP) with an anti-Nedd4-2 antibody (α-Nedd4-2). Immunoprecipitated material was analyzed by SDS-PAGE and affinity blotted with the S-protein-HRP conjugate (top blot, lanes 1 to 3). The same nitrocellulose membrane was stripped and Western blotted using an anti-Nedd4-2 antibody (bottom blot, lanes 1 to 3). For a control, immunoprecipitation with only the beads (protein A-Sepharose) was performed (lane 3). Protein expression in the lysates was checked using either the S-protein-HRP conjugate (top blot, lanes 4 to 6) or anti-Nedd4-2 antibody (bottom blot, lanes 4 to 6). (D) Transepithelial Na+ current measurements on mpkCCDcl4 cells, either not infected (NI) or overexpressing wild-type UBE2E3 (UBE2E3 WT) or catalytically inactive UBE2E3 (UBE2E3 CS). Values were normalized against wild-type mpkCCDcl4. The mean current of control was 6.36 ± 0.55 μA/cm2. Nine oocytes were used. Values that were significantly different (P < 0.05) from the values for noninfected cells or cells expressing UBE2E3-CS (*). The values for noninfected cells or cells expressing UBE2E3-CS were not significantly different from each other (NS). Protein expression was checked in lysates from the same cells using an anti-UBE2E3 antibody.
RESULT
Identification of UBE2E3 as an E2 enzyme interacting with Nedd4-2
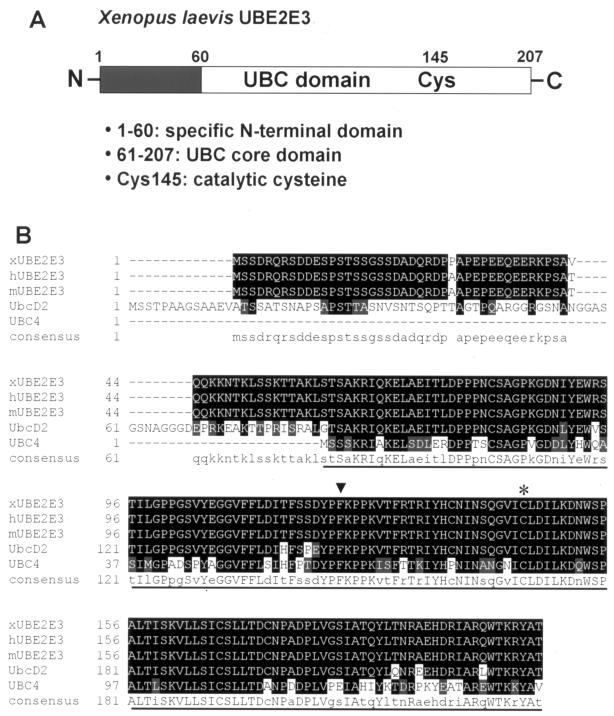
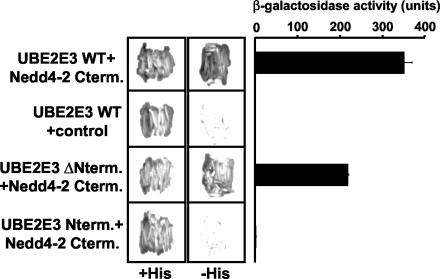
To identify the ubiquitin-conjugating enzyme (E2) involved in Nedd4-2-dependent regulation of ENaC in X. laevis oocytes, we screened a X. laevis oocyte cDNA library (3.5 × 106 independent clones) by the yeast two-hybrid system, using the C-terminal region (including the HECT domain) of X. laevis Nedd4-2 (xNedd4-2) as bait. This bait was chosen because of previous reports suggesting that the HECT domain is involved in the interaction with E2 enzymes (23, 28). The screening led to the isolation of 34 positive clones, 15 of which encode the X. laevis orthologue of mammalian UBE2E3, a ubiquitin-conjugating enzyme, also known as UbcM2, UbcH9, or UbcD2 (32, 46) (Fig. 1). None of the other clones encoded an E2 enzyme. As shown in Fig. 1, X. laevis UBE2E3 is a 207-residue protein with a predicted molecular mass of 23 kDa. It is highly conserved, with only two amino acid changes between X. laevis and the mouse or human orthologue. Moreover, there are homologues of UBE2E3 in Saccharomyces cerevisiae (Ubc4/5) and Drosophila melanogaster (UbcD2). UBE2E3 contains a UBC core domain, which represents the catalytic domain (Fig. 1B). It contains a specific N-terminal extension of unknown function (46), which makes it a class III E2 enzyme. There is an essential cysteine in position 145 in the UBC domain that is important. Further analysis with the two-hybrid system revealed that it is primarily the UBC domain which mediates the interaction with Nedd4-2 (Fig. 2), as coexpression of a UBE2E3 construct containing only the UBC domain with Nedd4-2 displayed a positive β-galactosidase signal and was able to grow on medium lacking histidine, whereas the N terminus of UBE2E3 did not interact with Nedd4-2, despite proper expression in the yeast cells (not shown).
FIG. 1.
X. laevis UBE2E3. (A) Linear view of UBE2E3 showing the N-terminal extension (black box) and the UBC core domain (white) with the catalytic cysteine (Cys145). The numbers are amino acid positions. (B) Amino acid sequence alignment of the primary sequences of X. laevis UBE2E3 (xUBE2E3) and its orthologues in human (hUBE2E3), mouse (mUBE2E3), D. melanogaster (UbcD2), and yeast (UBC4). The highly conserved UBC core domain is underlined. The catalytic cysteine at position 145 (asterisk) and the phenylalanine at position 122 (arrowhead), the key residue for the interaction with HECT E3s, are indicated. Conserved amino acids in the different species are indicated by white letters on black background and are shown as capital letters in the consensus sequence. Conservative changes in the different species are indicated by white letters on gray shaded background and are shown as lowercase letters in the consensus sequence. Gaps introduced to maximize sequence alignment are indicated by dashes.
FIG. 2.
Identification of UBE2E3 as a binding partner of Nedd4-2. The C-terminal part of X. laevis Nedd4-2 (Nedd4-2 Cterm.) cloned into pBTM116 was transformed into L40 yeast strain together with pACT2 vector containing wild-type UBE2E3 (UBE2E3 WT), UBE2E3 without N-terminal extension (UBE2E3 ΔNterm.), or UBE2E3 N-terminal extension alone (UBE2E3 Nterm.). For a control, pBTM116 containing the N-terminal part of rat βENaC was transformed into the L40 yeast strain in combination with wild-type UBE2E3 (UBE2E3 WT + control). Double transformants selected on Trp− Leu− plates were restreaked on plates containing histidine or on plates not containing histidine to check for interaction. Quantitative analysis of the interaction was performed using a liquid β-galactosidase assay.
UBE2E3 binds Nedd4-2 in vitro and in Xenopus oocytes
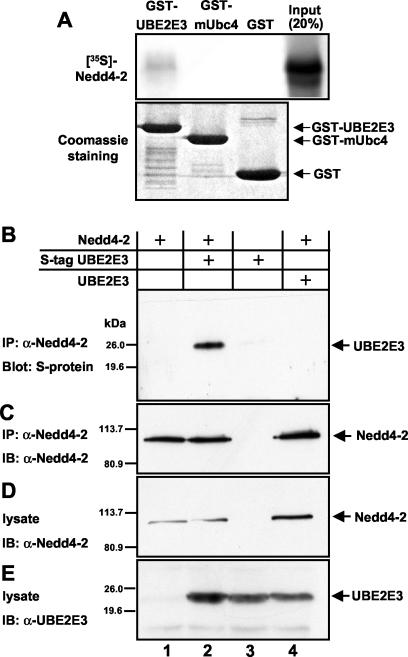
Having identified UBE2E3 as a potential binding partner of Nedd4-2, we wished to confirm the two-hybrid data by biochemical means. We purified UBE2E3 as a GST fusion protein and performed pull-down experiments with Nedd4-2, expressed as a 35S-labeled in vitro translation product. Immobilized GST fusion protein containing UBE2E3 was able to weakly bind Nedd4-2 (Fig. 3A), whereas GST alone was not. The weak binding between UBE2E3 and Nedd4-2 may be due to low concentrations of the radiolabeled Nedd4-2. In contrast, the mUbc4 protein, which is identical to human UbcH5B and is known to interact with Nedd4-1 (3), did not bind Nedd4-2.
FIG. 3.
UBE2E3 binds Nedd4-2 in vitro and in Xenopus oocytes. (A) Equal amounts of GST-UBE2E3 and GST-mUbc4 fusion proteins or GST alone (Coomassie blue staining) (10% of input in the bottom gel) immobilized on glutathione-Sepharose beads were incubated with in vitro-translated 35S-labeled Nedd4-2. After extensive washing, bound material was analyzed by SDS-PAGE and autoradiography (top gel). (B to E) Lysates from oocytes expressing Nedd4-2 alone, S-tagged UBE2E3 plus Nedd4-2, S-tagged UBE2E3 alone, or nontagged UBE2E3 plus Nedd4-2 were subjected to immunoprecipitation (IP) with an anti-Nedd4-2 antibody (α-Nedd4-2). (B) Immunoprecipitated material was analyzed by SDS-PAGE and affinity blotted with the S-protein-HRP conjugate. IB, immunoblotting. (C) The same nitrocellulose membrane was stripped and Western blotted using an anti-Nedd4-2 antibody. Protein expression in the lysate was checked using either an anti-Nedd4-2 antibody (D) or an anti-UBE2E3 antibody (E).
To demonstrate Nedd4-2-UBE2E3 interaction in oocytes, we expressed UBE2E3 as an C-terminally S-tagged protein in X. laevis oocytes, together with Nedd4-2. After overnight incubation, oocytes were lysed, and immunoprecipitation was performed with an anti-Nedd4-2 antibody (35). The precipitated proteins were analyzed by SDS-PAGE and Western blotting using either an anti-Nedd4-2 antibody or a HRP-conjugated S-protein recognizing the N-terminal S-tag on UBE2E3 (Fig. 3B). As can be seen in Fig. 3, UBE2E3 immunoprecipitates with Nedd4-2, when the two proteins were coexpressed (Fig. 3B and C, lane 2). No UBE2E3 was detected when either protein was expressed alone (Fig. 3B and C, lanes 1 and 3), or if nontagged UBE2E3 was coexpressed with Nedd4-2 (lane 4).
UBE2E3 participates in a Nedd4-2-dependent ubiquitination cascade
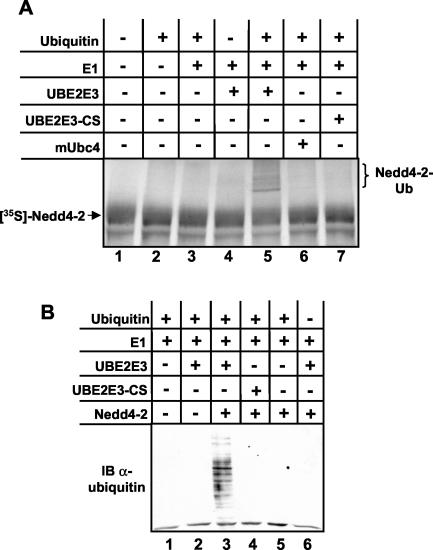
An important intermediate step in the Nedd4-2-dependent ubiquitination is the formation of a thioester complex between Nedd4-2 and ubiquitin, which is expected to depend on E1 and the E2-interacting protein UBE2E3. It is also known that several HECT-type ubiquitin-protein ligases become autoubiquitinated in the presence of E1 and E2 enzymes in vitro (29, 52), referred to as intrinsic ubiquitin-protein ligase activity (19). Therefore, we investigated whether UBE2E3 was able to transfer ubiquitin to Nedd4-2 in vitro. We translated Nedd4-2 in vitro in the presence of [35S]methionine using a reticulocyte lysate and mixed the translation product with purified E1, different E2s, and GST-ubiquitin. Indeed, the ubiquitin-activating enzyme E1, together with wild-type UBE2E3, transfers GST-ubiquitin to Nedd4-2, as suggested by the presence of high-molecular-weight products (Fig. 4A, lane 5, brackets). When any of the components was omitted or when the catalytically inactive UBE2E3, in which cysteine 145 was mutated to serine (UBE2E3-CS) or mUbc4 was used, no such bands were visible. Moreover, when a catalytically inactive Nedd4-2 mutant (Nedd4-2-CS) was used, the high-molecular-weight bands were not seen (not shown), indicating that the bands do indeed represent autoubiquitinated Nedd4-2 species. We also determined that UBE2E3 was able to ubiquitinate cellular proteins in concert with Nedd4-2. A bacterial lysate from E. coli (which does not contain ubiquitinated proteins) was incubated with ubiquitin, purified E1, His-tagged UBE2E3, and Nedd4-2. We then separated these bacterial proteins by SDS-PAGE and performed Western blot analysis with an antiubiquitin antibody. As illustrated by Fig. 4B, E1, UBE2E3 and Nedd4-2 had the capacity to conjugate ubiquitin on many bacterial proteins. Again, when any of the components was excluded or when UBE2E3-CS was used, ubiquitination was not detected.
FIG. 4.
UBE2E3 participates in the Nedd4-2-dependent ubiquitination cascade. (A) In vitro-translated 35S-labeled Nedd4-2 was incubated (+) for 1 h at 30°C with ubiquitin, E1, and different E2s. Samples were then analyzed by SDS-PAGE and autoradiography. The position of autoubiquitinated Nedd4-2 species (Nedd4-2-Ub) in lane 5 are indicated by brackets to the right of the gel. (B) Equal amounts of crude lysate from E. coli were incubated (+) for 2 h at 30°C with ubiquitin, E1, wild-type UBE2E3, or UBE2E3-CS and Nedd4-2 and then analyzed by SDS-PAGE and Western blotting using a monoclonal antibody directed against ubiquitin (α-ubiquitin). IB, immunoblotting.
UBE2E3 regulates ENaC in concert with Nedd4-2
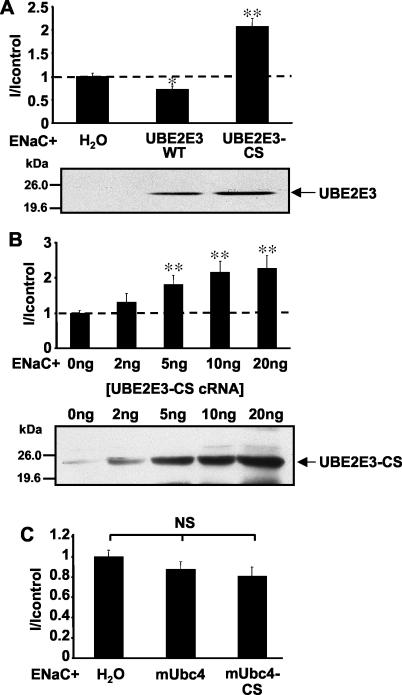
Having shown that UBE2E3 binds Nedd4-2 and acts in concert with this ubiquitin-protein ligase in a ubiquitination cascade, we wished to know whether UBE2E3 plays a role in ENaC regulation. To address this question, we took advantage of the X. laevis oocyte system, in which we had previously shown that Nedd4-2 was able to efficiently downregulate ENaC activity (>90%) and that a catalytically inactive Nedd4-2 mutant increased channel activity most probably by competing with endogenous Nedd4-2 (2, 34). These experimental results suggest that Nedd4-2 is a limiting factor in this inhibitory pathway. Similarly, we expressed ENaC alone or together with either wild-type UBE2E3 or the catalytically inactive mutant of UBE2E3 (UBE2E3-CS) and measured amiloride-sensitive Na+ currents (INa+, a measurement of ENaC activity) using the two-electrode voltage-clamp technique. We found that coexpression of wild-type UBE2E3 only slightly decreased ENaC activity compared to control oocytes expressing ENaC alone (H2O) (Fig. 5A), consistent with the idea that Nedd4-2, but not UBE2E3, is the rate-limiting element. On the other hand, the catalytically inactive UBE2E3-CS mutant significantly increased INa+ about twofold (Fig. 5A) in a dose-dependent manner (Fig. 5B), probably by competing against endogenous UBE2E3 protein, which was detected in lysates of Xenopus oocytes (Fig. 5B, top panel, 0 ng of ENaC, or on the overexposed fluorogram of Fig. 5A, ENaC + H2O). Because the amount of endogenous UBE2E3 was close to the detection limit, it was not always observed on our blots. In contrast, mUbc4 displayed no effect on ENaC activity, either in its wild-type form, or as a catalytically inactive mutant (Fig. 5C).
FIG. 5.
UBE2E3 affects ENaC activity. (A) Oocytes were injected either with ENaC cRNA alone or together with cRNA encoding wild-type UBE2E3 (UBE2E3 WT) or a catalytically inactive mutant of UBE2E3 (UBE2E3-CS). Amiloride-sensitive Na+ currents (I) were measured after 16 to 24 h. Values were normalized to control oocytes (H2O) expressing ENaC alone (mean current of control, 1.56 ± 0.16 μA). After measurements, the oocytes were lysed, and the expression of UBE2E3 was checked by SDS-PAGE and Western blotting using an anti-UBE2E3 antibody. (B) Same as panel A, but increasing amounts (0, 2, 5, 10, and 20 ng) of UBE2E3-CS cRNA were injected with cRNA encoding ENaC (mean current of control, 1.59 ± 0.05 μA). (C) Oocytes were injected with ENaC cRNA either alone or in combination with mUbc4 cRNA or its inactive form mUbc4-CS (mean current of control, 1.53 ± 0.13 μA). All experiments show the results for 15 to 18 oocytes from three different animals per condition. Values that were significantly different from the control value (H2O) are indicated as follows: *, P < 0.05; **, P < 0.01. Values that were not significantly different (NS) from each other are indicated.
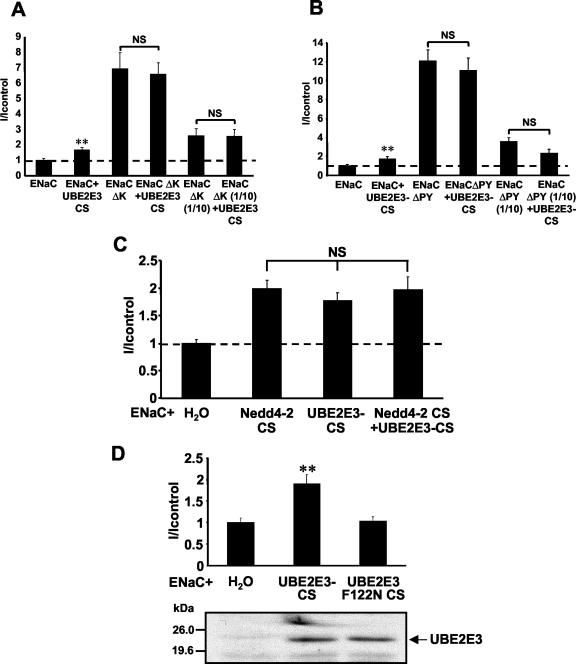
As mentioned previously, ENaC has been shown to be directly ubiquitinated on the N termini of its α and γ subunits, and this ubiquitination has been proposed to control the cell surface density of ENaC (70). If UBE2E3 affected the ubiquitination of ENaC, one would predict that UBE2E3-CS would not influence the activity of a mutant ENaC channel in which all the N-terminal lysines of the α and γ subunits, the ubiquitin conjugation sites, are mutated to arginine. Indeed this prediction could be confirmed. When we expressed such an ubiquitination-deficient ENaC mutant, we observed increased ENaC activity as reported before (70). Coexpression of UBE2E3-CS did not affect this activity, even if 10 times less cRNA encoding ENaCΔK was injected to assure that ENaC function was not saturated (Fig. 6A).
FIG. 6.
UBE2E3-CS regulates ENaC in concert with Nedd4-2. (A) Currents (I) of oocytes expressing either wild-type ENaC or mutant ENaC lacking ubiquitination sites (conserved lysine residues in the N termini of α- and γ-ENaC mutated to Arg; ENaCΔK) with or without UBE2E3-CS. Ten times less cRNA encoding mutant ENaC [ENaCΔK (1/10)] was injected for lower Na+ currents (mean current of control, 1.92 ± 0.27 μA) (B) Currents of oocytes expressing either wild-type ENaC or mutant ENaC lacking all PY motifs (ENaCΔPY) with or without UBE2E3-CS. Ten times less cRNA encoding mutant ENaC [ENaCΔPY (1/10)] was injected for lower Na+ currents. Values were normalized to control oocytes (ENaC) (mean current of control, 1.81 ± 0.44 μA). (C) Oocytes were injected with ENaC cRNA either alone (H2O) or with Nedd4-2-CS cRNA, UBE2E3-CS, cRNA or both. Amiloride-sensitive Na+ currents were measured, and values were normalized to control oocytes (H2O) (mean current of control, 3.86 ± 0.25 μA). (D) Same as panel C, but ENaC cRNA and either UBE2E3-CS cRNA or UBE2E3-F122N-CS cRNA were injected. After measurements, the oocytes were lysed, and the expression was checked by SDS-PAGE and Western blotting using an anti-UBE2E3 antibody (mean current of control, 2.98 ± 0.54 μA). All experiments show the results for 15 to 18 oocytes from three different animals per condition. Values that were significantly different (P < 0.01) from the control values (ENaC alone) (**) and values that were not significantly different (NS) from each other are indicated.
To acquire further evidence that UBE2E3-CS was interfering with the Nedd4-2-dependent pathway, we investigated whether its expression affected channels in which all the PY motifs (i.e., the binding sites for Nedd4-2) had been mutated. We found that amiloride-sensitive Na+ currents mediated by such channels were approximately 12-fold higher than those of the wild-type channel (Fig. 6B, compare ENaC with ENaCΔPY), explained by the loss of the binding sites for the negative regulator Nedd4-2. As shown before, UBE2E3-CS increased the wild-type channel activity by twofold (ENaC versus ENaC + UBE2E3-CS), whereas UBE2E3-CS did not increase the activity of the PY mutated channel (ENaCΔPY). This was also true when limiting quantities of ENaCΔPY cRNA were injected to exclude the possibility that the system was saturated [ENaCΔPY(1/10) versus ENaCΔPY(1/10) + UBE2E3-CS].
To further confirm that UBE2E3 and Nedd4-2 act in concert, we expressed Nedd4-2-CS, the catalytically inactive form of Nedd4-2, and UBE2E3-CS either individually or together, and found that both inactive mutant proteins increased ENaC activity to approximately the same level. Consistent with a model in which UBE2E3 and Nedd4-2 act in the same pathway, we observed no additive effect when they were expressed together (Fig. 6C). In addition, it has been shown that there is a conserved phenylalanine in the HECT-specific E2 subfamily (50), which is also present in UBE2E3 (Fig. 1B, F122, arrowhead), that is essential for the interaction with the HECT domain (28). Mutating this residue to Asn disrupts the interaction between UBE2E3 and Nedd4-2 in the two-hybrid system (data not shown). It could therefore be expected that a double mutant (UBE2E3 F122N CS) in which Phe122 is mutated to Asn and Cys145 is mutated to Ser, will not compete with endogenous UBE2E3 and therefore will not affect ENaC activity. This was indeed the case (Fig. 6D), thus providing further evidence that UBE2E3 and Nedd4-2 act in concert.
UBE2E3 is involved in the control of ENaC cell surface expression
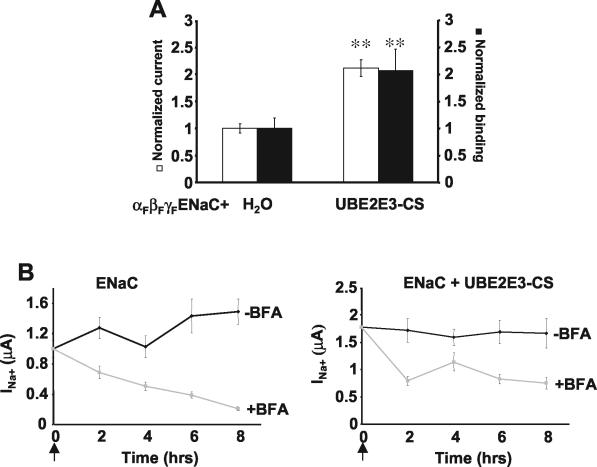
Previously, we have shown that Nedd4-2 controls cell surface density of ENaC (2, 11). To investigate whether UBE2E3 has similar effects, oocytes were injected with cRNA encoding α-, β-, and γ-ENaC (each containing a FLAG epitope at the extracellular loop) with or without cRNA encoding UBE2E3-CS. Oocytes expressing these proteins were incubated with 125I-labeled FLAG antibodies, which allowed simultaneous quantitation of FLAG-binding sites at the cell surface (representing the level of ENaC cell surface expression), and measurement of the amiloride-sensitive Na+ currents (15). We observed that UBE2E3-CS proportionally increased the amount of bound antibodies and currents, suggesting that UBE2E3-CS affects channel number at the cell surface without modifying intrinsic channel properties (such as open probability and/or single-channel conductance) (Fig. 7A).
FIG. 7.
UBE2E3-CS is involved in the control of ENaC cell surface expression and acts at an endocytosis step. (A) Oocytes were injected with cRNA encoding FLAG-tagged ENaC alone (H2O) or with cRNA encoding UBE2E3-CS. Amiloride-sensitive Na+ currents (white bars) and binding of iodinated anti-FLAG antibodies (black bars) were measured in the same oocytes to quantitate the number of channels at the cell surface as described previously (12). Values were normalized to control (H2O) values (mean current of control, 0.82 ± 0.13 μA). Eighteen oocytes from three animals were used. Values that were significantly different (P < 0.01) from the control values are indicated (**). (B) Oocytes injected with ENaC cRNA alone (ENaC) or with UBE2E3-CS cRNA (ENaC + UBE2E3-CS) were treated (+BFA) or not treated (−BFA) with 10 μg of BFA per ml at time zero (arrow). Amiloride-sensitive Na+ currents were measured at time zero and after 2, 4, 6, and 8 h of treatment. Values were normalized to the control value (ENaC) at time zero. Fifteen oocytes from three animals were used.
We wished to determine whether the increased number of ENaC channels at the plasma membrane in the presence of UBE2E3-CS was caused by increased delivery of ENaC to the cell surface, decreased internalization of cell surface-associated channels, or both. To address this point, we tested the effect of BFA on ENaC activity in Xenopus oocytes. BFA inhibits the endoplasmic reticulum-to-Golgi transport of newly synthesized membrane proteins; we and others have already shown that it affects ENaC activity at the plasma membrane (63, 70). We injected ENaC cRNA with or without UBE2E3-CS cRNA into Xenopus oocytes. After overnight incubation, we treated the oocytes with 10 μg of BFA per ml for up to 8 h and measured amiloride-sensitive Na+ currents. We found that the ENaC currents decreased by more than 75% after 8 h of treatment, whereas coexpression of UBE2E3-CS led to the appearance of a BFA-resistant pool (Fig. 7B). These findings are comparable to previous data, which showed that ENaC channels devoid of the ubiquitination sites also displayed a BFA-resistant pool at the cell surface (70) and suggest that ubiquitination (likely via UBE2E3 and Nedd4-2) plays a role in controlling ENaC density at the cell surface. However, the existence of a BFA-sensitive pool in the presence of UBE2E3 suggests that other ubiquitination-independent mechanisms are involved in the control of ENaC turnover in Xenopus oocytes, as proposed previously (63, 67).
UBE2E3 is expressed in renal epithelial cells and does influence transepithelial Na+ transport in mpkCCDcl4 cells
To play a role in Nedd4-2-dependent regulation of ENaC in renal cells, UBE2E3 must be expressed in such cells. We performed Northern blot analysis on RNA isolated from mpkCCDcl4 cells, a well-characterized murine cell model to study transepithelial Na+ transport of the cortical collecting duct (CCD), and known to express both ENaC and Nedd4-2 (8, 34). Using an UBE2E3-specific probe, we detected a mRNA species of approximately 1.7 kb, corresponding to the reported size of murine UBE2E3 mRNA (46) (Fig. 8A).
We also performed Western blot analysis on mpkCCDcl4 lysates either transfected or not transfected with UBE2E3, using our anti-UBE2E3 antibody. As can be seen in Fig. 8B, lane 1, a faint band at approximately 22 kDa was observed in noninfected mpkCCDcl4 cells, migrating to the same position as UBE2E3 ectopically expressed in mpkCCDcl4 cells (Fig. 8B, lane 2). Both bands could be reduced by competition of the antibody with its antigen (Fig. 8B, lanes 3 and 4). This demonstrates that UBE2E3 is expressed at low levels in mpkCCDcl4 cells. We then wanted to know whether UBE2E3 can interact with Nedd4-2 in mpkCCDcl4 cells. We expressed S-tagged UBE2E3, taking advantage of a retroviral expression system, and performed immunoprecipitation with an anti-Nedd4-2 antibody, thereby precipitating endogenous Nedd4-2 (Fig. 8C). When the immunoprecipitated material was affinity blotted with S-protein, coimmunoprecipitated S-tagged UBE2E3 was detected (Fig. 8C, lane 2), whereas no protein was visible in noninfected cells or when the immunoprecipitating antibody was omitted (lanes 1 and 3).
Next, we wanted to determine whether UBE2E3 plays a role in the regulation of transepithelial Na+ transport. We therefore expressed either wild-type or mutant UBE2E3 in mpkCCDcl4 cells, cultured the cells on collagen-coated permeable filters, and measured the amiloride-sensitive Na+ transport by the ISC method. Because the transepithelial Na+ transport in such cells is controlled by a number of different pathways, one may expect relatively small effects by UBE2E3. Indeed, we found that overexpression of UBE2E3 inhibited ISC to about 73% of control currents, whereas the catalytically inactive UBE2E3 had no significant effect compared to noninfected mpkCCDcl4 cells. This demonstrates that UBE2E3 is able to regulate the endogenous, amiloride-sensitive transepithelial Na+ transport in cells derived from the CCD.
DISCUSSION
Ubiquitination of target proteins requires the sequential action of an enzymatic cascade including the ubiquitin-activating enzyme E1, ubiquitin-conjugating E2 enzymes, and ubiquitin-protein ligases E3. It has been shown that the cell surface expression of ENaC is regulated via ubiquitination involving the E3 enzyme Nedd4-2. Here we provide information about the nature of another member of the Nedd4-2-dependent cascade. Specifically, our data suggest that the ubiquitin-conjugating enzyme UBE2E3 participates in the regulation of ENaC density in the plasma membranes of X. laevis oocytes. This is based on the following observations. (i) Nedd4-2 and UBE2E3 interact both in vitro and in cells, as suggested by the results of two-hybrid analysis, pull-down assays, and coimmunoprecipitation experiments. (ii) UBE2E3 mediates, together with E1, autoubiquitination of Nedd4-2, and the three enzymes in concert are able to ubiquitinate bacterial proteins. (iii) Catalytically inactive UBE2E3-CS (but not mUbc4-CS, an unrelated E2 enzyme from mice) increases ENaC activity, by elevating ENaC channel density at the cell surface. (iv) The effect of UBE2E3-CS depends on the presence of the PY motifs and ubiquitination sites on ENaC, and no additive effect on ENaC activity is observed when it is coexpressed with the catalytically inactive Nedd4-2-CS, corroborating the notion that UBE2E3 and Nedd4-2 act on the same pathway.
It is not yet known how ubiquitination via UBE2E3 and Nedd4-2 controls the density of ENaC channels. Three hypotheses or any combination of the three hypotheses can be envisioned. (i) Ubiquitination may direct proteins from the trans-Golgi level to the lysosome (24, 68). (ii) Ubiquitination may promote internalization of ENaC at the cell surface (27). (iii) Ubiquitination may facilitate translocation of internalized ENaC channels into the lysosome (38). The present data with BFA and UBE2E3-CS (Fig. 7B), which are similar to previous findings using ubiquitination-deficient ENaC mutants (70), show the appearance of a BFA-resistant pool of ENaC at the plasma membrane. These data make it unlikely that ubiquitination and UBE2E3-CS acted at the trans-Golgi level but suggest that UBE2E3-CS interferes with an internalization step or with the translocation into the late endosomal or lysosomal system, impairment of which may lead to increased recycling of channels back to the cell surface. The existence of a BFA-sensitive pool of ENaC (in the presence of UBE2E3-CS), which was also found with ubiquitination-deficient ENaC mutants (70), points to the existence of additional, ubiquitination-independent regulatory mechanisms in the X. laevis oocytes, as proposed previously (63, 67).
Our data show that it is primarily the UBC domain of UBE2E3 that interacts with Nedd4-2. This finding is consistent with the crystal structure data of the complex between the HECT domain of E6AP and the ubiquitin-conjugating enzyme UbcH7, which contains little more than the UBC domain (28). This crystal structure also demonstrates that a phenylalanine (Phe63 in UbcH7) is the most critical contact point, as it binds in the deepest part in the center of the HECT groove and establishes van der Waals contacts with six hydrophobic and aromatic E6AP-HECT side chains. This phenylalanine has been found to be conserved in all E2 enzymes interacting with HECT ubiquitin-protein ligases (50) and is also present in UBE2E3 (Fig. 1B, F122, arrowhead). Our data confirm the importance of F122 for the interaction with Nedd4-2, as a UBE2E3 double mutant (F122N C145S) fails to stimulate ENaC activity (Fig. 6D), consistent with the idea that this mutant cannot bind Nedd4-2 and therefore cannot compete with endogenous UBE2E3. Moreover, mutation of F122 in the UBE2E3 construct abolishes interaction with Nedd4-2 in the two-hybrid system (not shown). However, the presence of F122 is not sufficient for binding to a HECT domain, as the results of our binding and functional studies with mUbc4 (containing also the conserved phenylalanine) (Fig. 3A and 5C) suggest, indicating that other residues in the UBC domain and/or the N-terminal extension may play a role in the specificity of the interaction.
The tissue distribution and cellular localization of UBE2E3 have been studied. Ito et al. cloned human UBE2E3 and analyzed the tissue distribution of the corresponding mRNA in various organs (32). Strong expression was seen in skeletal muscle, but low levels of UBE2E3 mRNA were expressed in all the other tissues analyzed. We detected UBE2E3 in mpkCCDcl4 cells, both at the mRNA and protein levels (Fig. 8A and B). Moreover, we demonstrated the presence of UBE2E3 cDNA in a cDNA library made from dissected CCD, and in an independent serial analysis of gene expression study, tags corresponding to UBE2E3 were found in the library of human dissected CCD (10). This indicates that UBE2E3 is expressed in the same cells as ENaC and Nedd4-2 and that it may be a potential endogenous regulator of ENaC. Indeed, this is further supported by the observation that ectopically expressed UBE2E3 (i) coimmunoprecipitates in mpkCCDcl4 cells with endogenous Nedd4-2 and (ii) negatively regulates amiloride-sensitive transepithelial Na+ current in these cells, whereas the catalytically inactive UBE2E3-CS has only minor effects on this activity. Interestingly, the orthologues of UBE2E3 in yeast, Ubc4 and Ubc5 (46), are involved in the control of internalization of Ste2 (27), in concert with Rsp5, the orthologue of Nedd4 (13), showing that such mechanism of regulation is conserved in eukaryotes. However, the wide tissue distribution of UBE2E3 suggests that it has other functions in addition to the regulation of ENaC via Nedd4-2. This is indeed confirmed by several reports, which show that UBE2E3 may play a role in prostate cancer (9) or interact with RING-finger ubiquitin-protein ligases (31) or with nuclear import receptors (55, 56).
In conclusion, we have identified a ubiquitin-conjugating enzyme that is involved in Nedd4-2-dependent regulation of ENaC in X. laevis oocytes and in renal cells, providing further information on this important regulatory pathway of ENaC. To our knowledge, this is the first demonstration of the regulation of an ion channel involving an E2 enzyme. Moreover, in view that other ion channels have been found to be regulated by the Nedd4/Nedd4-like family of ubiquitin-protein ligases, such as the cardiac voltage-gated Na+ channel (1) or the ClC-5 channel (61), UBE2E3 may also be important for the control of these channels.
Acknowledgments
We thank Bernard Rossier, Laurent Schild, Jean-Daniel Horisberger, Dmitri Firsov, Hugues Abriel, Dario Diviani, Laura Stanasila, Phil Shaw, Miguel VanBemmelen, and the members of our laboratory for critically reading the manuscript. We are grateful to Elena Kamynina for helpful suggestions on the use of S-tag.
This work was supported in part by grants from the Swiss National Science Foundation (grant 31-64052.00) and the Leenaards Foundation in Lausanne, Switzerland.
REFERENCE
- 1.Abriel, H., E. Kamynina, J.-D. Horisberger, and O. Staub. 2000. Regulation of the cardiac voltage-gated Na+ channel (rH1) by the ubiquitin-protein ligase Nedd4. FEBS Lett. 466:377-380. [DOI] [PubMed] [Google Scholar]
- 2.Abriel, H., J. Loffing, J. F. Rebhun, J. H. Pratt, J.-D. Horisberger, D. Rotin, and O. Staub. 1999. Defective regulation of the epithelial Na+ channel (ENaC) by Nedd4 in Liddle's syndrome. J. Clin. Investig. 103:667-673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anan, T., Y. Nagata, H. Koga, Y. Honda, N. Yabuki, C. Miyamoto, A. Kuwano, I. Matusuda, F. Endo, H. Saya, and M. Nakao. 1999. Human ubiquitin-protein ligase Nedd4: expression, subcellular localization, and selective interaction with ubiquitin-conjugating enzymes. Genes Cells 3:751-763. [DOI] [PubMed] [Google Scholar]
- 4.Aristarkhov, A., E. Eytan, A. Moghe, A. Admon, A. Hershko, and J. V. Ruderman. 1996. E2-C, a cyclin-selective ubiquitin carrier protein required for the destruction of mitotic cyclins. Proc. Natl. Acad. Sci. USA 93:4294-4299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Asher, C., A. Chigaev, and H. Garty. 2001. Characterization of interactions between Nedd4 and β and γENaC using surface plasmon resonance. Biochem. Biophys. Res. Commun. 286:1228-1231. [DOI] [PubMed] [Google Scholar]
- 6.Asher, C., I. Sinha, and H. Garty. 2003. Characterization of the interactions between Nedd4-2, ENaC, and sgk-1 using surface plasmon resonance. Biochim. Biophys. Acta 1612:59-64. [DOI] [PubMed] [Google Scholar]
- 7.Auberson, M., N. Hoffmann-Pochon, A. Vandewalle, S. Kellenberger, and L. Schild. 2003. Epithelial Na+ channel mutants causing Liddle's syndrome retain ability to respond to aldosterone and vasopressin. Am. J. Physiol. 285:F459-F471. [DOI] [PubMed] [Google Scholar]
- 8.Bens, M., V. Vallet, F. Cluzeaud, L. Pascual-Letallec, A. Kahn, M. E. Rafestin-Oblin, B. C. Rossier, and A. Vandewalle. 1999. Corticosteroid-dependent sodium transport in a novel immortalized mouse collecting duct principal cell line. J. Am. Soc. Nephrol. 10:923-934. [DOI] [PubMed] [Google Scholar]
- 9.Bull, J. H., G. Ellison, A. Patel, G. Muir, M. Walker, M. Underwood, F. Khan, and L. Paskins. 2001. Identification of potential diagnostic markers of prostate cancer and prostatic intraepithelial neoplasia using cDNA microarray. Br. J. Cancer 84:1512-1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chabardès-Garonne, D., A. Méjean, J.-C. Aude, L. Chevel, A. Di Stefano, M.-C. Gaillard, M. Imbert-Teboul, M. Wittner, C. Balian, V. Anthouard, C. Robert, B. Ségurens, P. Wincker, J. Weissenbach, A. Doucet, and J. M. Elalouf. 2003. A panoramic view of gene expression in the human kidney. Proc. Natl. Acad. Sci. USA 100:13710-13715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Debonneville, C., S. Y. Flores, E. Kamynina, P. J. Plant, C. Tauxe, M. A. Thomas, C. Munster, A. Chraibi, J. H. Pratt, J. D. Horisberger, D. Pearce, J. Loffing, and O. Staub. 2001. Phosphorylation of Nedd4-2 by Sgk1 regulates epithelial Na+ channel cell surface expression. EMBO J. 20:7052-7059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dinudom, A., B. J. Harvey, P. Komwatana, J. A. Young, S. Kumar, and D. I. Cook. 1998. Nedd4 mediates control of an epithelial Na+ channel in salivary duct cells by cytosolic Na+. Proc. Natl. Acad. Sci. USA 95:7169-7173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dunn, R., and L. Hicke. 2001. Domains of the Rsp5 ubiquitin-protein ligase required for receptor-mediated and fluid-phase endocytosis. Mol. Biol. Cell 12:421-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Farr, T. J., S. J. Coddington-Lawson, P. M. Snyder, and F. J. McDonald. 2000. Human Nedd4 interacts with the human epithelial Na+ channel: WW3 but not WW1 binds to Na+-channel subunits. Biochem. J. 345:503-509. [PMC free article] [PubMed] [Google Scholar]
- 15.Firsov, D., L. Schild, I. Gautschi, A.-M. Mérillat, E. Schneeberger, and B. C. Rossier. 1996. Cell surface expression of the epithelial Na+ channel and a mutant causing Liddle syndrome: a quantitative approach. Proc. Natl. Acad. Sci. USA 93:15370-15375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fotia, A. B., A. Dinudom, K. E. Shearwin, J. P. Koch, C. Korbmacher, D. I. Cook, and S. Kumar. 2003. The role of individual Nedd4-2 (KIAA0439) WW domains in binding and regulating epithelial sodium channels. FASEB J. 17:70-72. [DOI] [PubMed] [Google Scholar]
- 17.Glickman, M. H., and A. Ciechanover. 2002. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol. Rev. 82:373-428. [DOI] [PubMed] [Google Scholar]
- 18.Goulet, C. C., K. A. Volk, C. M. Adams, L. S. Prince, J. B. Stokes, and P. M. Snyder. 1998. Inhibition of the epithelial Na+ channel by interaction of Nedd4 with a PY motif deleted in Liddle's syndrome. J. Biol. Chem. 273:30012-30017. [DOI] [PubMed] [Google Scholar]
- 19.Grossman, S. R., M. E. Deato, C. Brignone, H. M. Chan, A. L. Kung, H. Tagami, Y. Nakatani, and D. M. Livingston. 2003. Polyubiquitination of p53 by a ubiquitin ligase activity of p300. Science 300:342-344. [DOI] [PubMed] [Google Scholar]
- 20.Hansson, J. H., C. Nelson-Williams, H. Suzuki, L. Schild, R. A. Shimkets, Y. Lu, C. M. Canessa, T. Iwasaki, B. C. Rossier, and R. P. Lifton. 1995. Hypertension caused by a truncated epithelial sodium channel gamma subunit: genetic heterogeneity of Liddle syndrome. Nat. Genet. 11:76-82. [DOI] [PubMed] [Google Scholar]
- 21.Harvey, K. F., A. Dinudom, D. I. Cook, and S. Kumar. 2001. The Nedd4-like protein KIAA0439 is a potential regulator of the epithelial sodium channel. J. Biol. Chem. 276:8597-8601. [DOI] [PubMed] [Google Scholar]
- 22.Harvey, K. F., A. Dinudom, P. Komwatana, C. N. Jolliffe, M. L. Day, G. Parasivam, D. I. Cook, and S. Kumar. 1999. All three WW domains of murine Nedd4 are involved in the regulation of epithelial sodium channels by intracellular Na+. J. Biol. Chem. 274:12525-12530. [DOI] [PubMed] [Google Scholar]
- 23.Hatakeyama, S., J. P. Jensen, and A. M. Weissman. 1997. Subcellular localization and ubiquitin-conjugating enzyme (E2) interactions of mammalian HECT family ubiquitin protein ligases. J. Biol. Chem. 272:15085-15092. [DOI] [PubMed] [Google Scholar]
- 24.Helliwell, S. B., S. Losko, and C. A. Kaiser. 2001. Components of a ubiquitin ligase complex specify polyubiquitination and intracellular trafficking of the general amino acid permease. J. Cell Biol. 153:649-662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Henry, P. C., V. Kanelis, C. M. O'Brien, B. Kim, I. Gautschi, J. Forman-Kay, L. Schild, and D. Rotin. 2003. Affinity and specificity of interactions between Nedd4 isoforms and the epithelial Na+ channel. J. Biol. Chem. 278:20019-20028. [DOI] [PubMed] [Google Scholar]
- 26.Hershko, A., and A. Ciechanover. 1998. The ubiquitin system. Annu. Rev. Biochem. 67:425-479. [DOI] [PubMed] [Google Scholar]
- 27.Hicke, L., and H. Riezman. 1996. Ubiquitination of a yeast plasma membrane receptor signals its ligand-stimulated endocytosis. Cell 84:277-287. [DOI] [PubMed] [Google Scholar]
- 28.Huang, L., E. Kinnucan, G. Wang, S. Beaudenon, P. M. Howley, J. M. Huibregtse, and N. P. Pavletich. 1999. Structure of an E6AP-UbcH7 complex: insights into ubiquitination by the E2-E3 enzyme cascade. Science 286:1321-1326. [DOI] [PubMed] [Google Scholar]
- 29.Huibregtse, J. M., M. Scheffner, S. Beaudenon, and P. M. Howley. 1995. A family of proteins structurally and functionally related to the E6-AP ubiquitin-protein ligase. Proc. Natl. Acad. Sci. USA 92:2563-2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Inoue, J., T. Iwaoka, H. Tokunaga, K. Takamune, S. Naomi, M. Araki, K. Takahama, K. Yamaguchi, and K. Tomita. 1998. A family with Liddle's syndrome caused by a new missense mutation in the β subunit of the epithelial sodium channel. J. Clin. Endocrinol. Metab. 83:2210-2213. [DOI] [PubMed] [Google Scholar]
- 31.Ito, K., S. Adachi, R. Iwakami, H. Yasuda, Y. Muto, N. Seki, and Y. Okano. 2001. N-terminally extended human ubiquitin-conjugating enzymes (E2s) mediate the ubiquitination of RING-finger proteins, ARA54 and RNF8. Eur. J. Biochem. 268:2725-2732. [DOI] [PubMed] [Google Scholar]
- 32.Ito, K., S. Kato, Y. Matsuda, M. Kimura, and Y. Okano. 1999. cDNA cloning, characterization, and chromosome mapping of UBE2E3 (alias UbcH9), encoding an N-terminally extended human ubiquitin-conjugating enzyme. Cytogenet. Cell. Genet. 84:99-104. [DOI] [PubMed] [Google Scholar]
- 33.Kaiser, P., W. Seufert, L. Höfferer, B. Kofler, C. Sachsenmaier, H. Herzog, S. Jentsch, M. Schweiger, and R. Schneider. 1994. A human ubiquitin-conjugating enzyme homologous to yeast UBC8. J. Biol. Chem. 269:8797-8802. [PubMed] [Google Scholar]
- 34.Kamynina, E., C. Debonneville, M. Bens, A. Vandewalle, and O. Staub. 2001. A novel mouse Nedd4 protein suppresses the activity of the epithelial Na+ channel. FASEB J. 15:204-214. [DOI] [PubMed] [Google Scholar]
- 35.Kamynina, E., C. Tauxe, and O. Staub. 2001. Differential characteristics of two human Nedd4 proteins with respect to epithelial Na+ channel regulation. Am. J. Physiol. 281:F469-F477. [DOI] [PubMed] [Google Scholar]
- 36.Kanelis, V., N. A. Farrow, L. E. Kay, D. Rotin, and J. D. Forman-Kay. 1998. NMR studies of tandem WW domains of Nedd4 in complex with a PY motif-containing region of the epithelial sodium channel. Biochem. Cell Biol. 76:341-350. [DOI] [PubMed] [Google Scholar]
- 37.Kanelis, V., D. Rotin, and J. D. Forman-Kay. 2001. Solution structure of a Nedd4 WW domain-ENaC peptide complex. Nat. Struct. Biol. 8:1-6. [DOI] [PubMed] [Google Scholar]
- 38.Katzmann, D. J., M. Babst, and S. D. Emr. 2001. Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell 106:145-155. [DOI] [PubMed] [Google Scholar]
- 39.Kellenberger, S., I. Gautschi, B. C. Rossier, and L. Schild. 1998. Mutations causing Liddle syndrome reduce sodium-dependent downregulation of the epithelial sodium channel in the xenopus oocyte expression system. J. Clin. Investig. 101:2741-2750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kellenberger, S., and L. Schild. 2002. Epithelial sodium channel/degenerin family of ion channels: a variety of functions for a shared structure. Physiol. Rev. 82:735-767. [DOI] [PubMed] [Google Scholar]
- 41.Koegl, M., T. Hoppe, S. Schlenker, H. D. Ulrich, T. U. Mayer, and S. Jentsch. 1999. A novel ubiquitination factor, E4, is involved in multiubiquitin chain assembly. Cell 96:635-644. [DOI] [PubMed] [Google Scholar]
- 42.Konstas, A. A., L. M. Shearwin-Whyatt, A. B. Fotia, B. Degger, D. Riccardi, D. I. Cook, C. Korbmacher, and S. Kumar. 2002. Regulation of the epithelial sodium channel by N4WBP5A, a novel Nedd4/Nedd4-2-interacting protein. J. Biol. Chem. 277:29406-29416. [DOI] [PubMed] [Google Scholar]
- 43.Liddle, G. W., T. Bledsoe, and W. S. Coppage, Jr. 1963. A familial renal disorder simulating primary aldosteronism but with negligible aldosterone secretion. Trans. Assoc. Am. Physicians 76:199-213. [Google Scholar]
- 44.Lott, J. S., S. J. Coddington-Lawson, P. H. Teesdale-Spittle, and F. J. McDonald. 2002. A single WW domain is the predominant mediator of the interaction between the human ubiquitin-protein ligase Nedd4 and the human epithelial sodium channel. Biochem. J. 361:481-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Malik, B., L. Schlanger, O. Al-Khalili, H. F. Bao, G. Yue, S. R. Price, W. E. Mitch, and D. C. Eaton. 2001. Enac degradation in A6 cells by the ubiquitin-proteosome proteolytic pathway. J. Biol. Chem. 276:12903-12910. [DOI] [PubMed] [Google Scholar]
- 46.Matuschewski, K., H.-P. Hauser, M. Treier, and S. Jentsch. 1996. Identification of a novel family of ubiquitin-conjugating enzymes with distinct amino-terminal extensions. J. Biol. Chem. 271:2789-2794. [DOI] [PubMed] [Google Scholar]
- 47.May, A., A. Puoti, H.-P. Gaeggeler, J.-D. Horisberger, and B. C. Rossier. 1997. Early effect of aldosterone on the rate of synthesis of the epithelial sodium channel α subunit in A6 renal cells. J. Am. Soc. Nephrol. 8:1813-1822. [DOI] [PubMed] [Google Scholar]
- 48.McDonald, F. J., A. H. Western, J. D. McNeil, B. C. Thomas, D. R. Olson, and P. M. Snyder. 2002. Ubiquitin-protein ligase WWP2 binds to and downregulates the epithelial Na+ channel. Am. J. Physiol. 283:F431-F436. [DOI] [PubMed] [Google Scholar]
- 49.Nelson, R. M., and G. L. Long. 1989. A general method of site-specific mutagenesis using a modification of the Thermus aquaticus polymerase chain reaction. Anal. Biochem. 180:147-151. [DOI] [PubMed] [Google Scholar]
- 50.Nuber, U., and M. Scheffner. 1999. Identification of determinants in E2 ubiquitin-conjugating enzymes required for hect E3 ubiquitin-protein ligase interaction. J. Biol. Chem. 274:7576-7582. [DOI] [PubMed] [Google Scholar]
- 51.Nuber, U., S. Schwarz, P. Kaiser, R. Schneider, and M. Scheffner. 1996. Cloning of human ubiquitin-conjugating enzymes UbcH6 and UbcH7 (E2-F1) and characterization of their interaction with E6-AP and Rsp5. J. Biol. Chem. 271:2795-2800. [DOI] [PubMed] [Google Scholar]
- 52.Nuber, U., S. E. Schwarz, and M. Scheffner. 1998. The ubiquitin-protein ligase E6-associated protein (E6-AP) serves as its own substrate. Eur. J. Biochem. 234:643-649. [DOI] [PubMed] [Google Scholar]
- 53.Pear, W. S., G. P. Nolan, M. L. Scott, and D. Baltimore. 1993. Production of high-titer helper-free retroviruses by transient transfection. Proc. Natl. Acad. Sci. USA 90:8392-8396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pirozzi, G., S. J. McConnell, A. J. Uveges, J. M. Carter, A. B. Sparks, B. K. Kay, and D. M. Fowlkes. 1997. Identification of novel human WW domain-containing proteins by cloning of ligand targets. J. Biol. Chem. 272:14611-14616. [DOI] [PubMed] [Google Scholar]
- 55.Plafker, S. M., and I. G. Macara. 2000. Importin-11, a nuclear import receptor for the ubiquitin-conjugating enzyme, UbcM2. EMBO J. 19:5502-5513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Plafker, S. M., and I. G. Macara. 2002. Ribosomal protein L12 uses a distinct nuclear import pathway mediated by importin 11. Mol. Biol. Cell 22:1266-1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Puoti, A., A. May, B. C. Rossier, and J.-D. Horisberger. 1998. Novel isoforms of the α and γ subunits of the Xenopus epithelial Na+ channel provide information about the amiloride binding site and extracellular sodium sensing. Proc. Natl. Acad. Sci. USA 94:5949-5954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rotin, D., O. Staub, and R. Haguenauer-Tsapis. 2000. Ubiquitination and endocytosis of plasma membrane proteins: role of Nedd4/Rsp5 family of ubiquitin-protein ligases. J. Membr. Biol. 176:1-17. [DOI] [PubMed] [Google Scholar]
- 59.Schild, L., C. M. Canessa, R. A. Shimkets, D. G. Warnock, R. P. Lifton, and B. C. Rossier. 1995. A mutation in the epithelial sodium channel causing Liddle's disease increases channel activity in the Xenopus laevis oocyte expression system. Proc. Natl. Acad. Sci. USA 92:5699-5703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schild, L., Y. Lu, I. Gautschi, E. Schneeberger, R. P. Lifton, and B. C. Rossier. 1996. Identification of a PY motif in the epithelial Na+ channel subunits as a target sequence for mutations causing channel activation found in Liddle syndrome. EMBO J. 15:2381-2387. [PMC free article] [PubMed] [Google Scholar]
- 61.Schwake, M., T. Friedrich, and T. J. Jentsch. 2001. An internalization signal in ClC-5, an endosomal Cl-channel mutated in Dent's disease. J. Biol. Chem. 276:12049-12054. [DOI] [PubMed] [Google Scholar]
- 62.Shi, H., C. Asher, A. Chigaev, Y. Yung, E. Reuveny, R. Seger, and H. Garty. 2002. Interactions of beta and gamma ENaC with Nedd4 can be facilitated by an EKR-mediated phosphorylation. J. Biol. Chem. 277:13539-13547. [DOI] [PubMed] [Google Scholar]
- 63.Shimkets, R. A., R. P. Lifton, and C. M. Canessa. 1997. The activity of the epithelial sodium channel is regulated by clathrin-mediated endocytosis. J. Biol. Chem. 272:25537-25541. [DOI] [PubMed] [Google Scholar]
- 64.Shimkets, R. A., D. G. Warnock, C. M. Bositis, C. Nelson-Williams, J. H. Hansson, M. Schambelan, J. R. Gill, S. Ulick, R. V. Milora, J. W. Findling, C. M. Canessa, B. C. Rossier, and R. P. Lifton. 1994. Liddle's syndrome: heritable human hypertension caused by mutations in the β subunit of the epithelial sodium channel. Cell 79:407-414. [DOI] [PubMed] [Google Scholar]
- 65.Snyder, P. M., D. R. Olson, F. J. McDonald, and D. B. Bucher. 2001. Multiple WW domains, but not the C2 domain, are required for the inhibition of ENaC by human Nedd4. J. Biol. Chem. 276:28321-28326. [DOI] [PubMed] [Google Scholar]
- 66.Snyder, P. M., D. R. Olson, and B. C. Thomas. 2002. Serum and glucocorticoid-regulated kinase modulates Nedd4-2-mediated inhibition of the epithelial Na+ channel. J. Biol. Chem. 277:5-8. [DOI] [PubMed] [Google Scholar]
- 67.Snyder, P. M., M. P. Price, F. J. McDonald, C. M. Adams, K. A. Volk, B. G. Zeiher, J. B. Stokes, and M. J. Welsh. 1995. Mechanism by which Liddle's syndrome mutations increase activity of a human epithelial Na+ channel. Cell 83:969-978. [DOI] [PubMed] [Google Scholar]
- 68.Soetens, O., J. O. De Craene, and B. Andre. 2001. Ubiquitin is required for sorting to the vacuole of the yeast general amino acid permease, Gap1. J. Biol. Chem. 276:43949-43957. [DOI] [PubMed] [Google Scholar]
- 69.Staub, O., S. Dho, P. C. Henry, J. Correa, T. Ishikawa, J. McGlade, and D. Rotin. 1996. WW domains of Nedd4 bind to the proline-rich PY motifs in the epithelial Na+ channel deleted in Liddle's syndrome. EMBO J. 15:2371-2380. [PMC free article] [PubMed] [Google Scholar]
- 70.Staub, O., I. Gautschi, T. Ishikawa, K. Breitschopf, A. Ciechanover, L. Schild, and D. Rotin. 1997. Regulation of stability and function of the epithelial Na+ channel (ENaC) by ubiquitination. EMBO J. 16:6325-6336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tamura, H., L. Schild, N. Enomoto, N. Matsui, F. Marumo, B. C. Rossier, and S. Sasaki. 1996. Liddle disease caused by a missense mutation of beta subunit of the epithelial sodium channel gene. J. Clin. Investig. 97:1780-1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vojtek, A. B., S. M. Hollenberg, and J. A. Cooper. 1993. Mammalian Ras interacts directly with the serine/threonine kinase Raf. Cell 74:205-214. [DOI] [PubMed] [Google Scholar]