Abstract
The accessory Sec system of Streptococcus gordonii is comprised of SecY2, SecA2, and five proteins (Asp1 through -5) that are required for the export of a serine-rich glycoprotein, GspB. We have previously shown that a number of the Asps interact with GspB, SecA2, or each other. To further define the roles of these Asps in export, we examined their subcellular localization in S. gordonii and in Escherichia coli expressing the streptococcal accessory Sec system. In particular, we assessed how the locations of these accessory Sec proteins were altered by the presence of other components. Using fluorescence microscopy, we found in E. coli that SecA2 localized within multiple foci at the cell membrane, regardless of whether other accessory Sec proteins were expressed. Asp2 alone localized to the cell poles but formed a similar punctate pattern at the membrane when SecA2 was present. Asp1 and Asp3 localized diffusely in the cytosol when expressed alone or with SecA2. However, these proteins redistributed to the membrane in a punctate arrangement when all of the accessory Sec components were present. Cell fractionation studies with S. gordonii further corroborated these microscopy results. Collectively, these findings indicate that Asp1 to -3 are not integral membrane proteins that form structural parts of the translocation channel. Instead, SecA2 serves as a docking site for Asp2, which in turn attracts a complex of Asp1 and Asp3 to the membrane. These protein interactions may be important for the trafficking of GspB to the cell membrane and its subsequent translocation.
INTRODUCTION
The accessory Sec (SecA2-SecY2) system is a specialized export pathway conserved among Gram-positive bacteria, including a number of the streptococcal and staphylococcal species (1–5). The system mediates the transport of large serine-rich repeat, cell wall-anchored glycoproteins to the bacterial cell surface. The accessory Sec substrates are adhesins that contribute to a variety of diseases, including infective endocarditis (6, 7), pneumonia (3), meningitis (8, 9), and sepsis (9, 10). In Streptococcus gordonii M99, the substrate of the accessory Sec system is GspB, which mediates binding of the bacterium to the GPIbα receptor on platelets (11, 12). This interaction appears to have a key role in the pathogenesis of infective endocarditis, as measured by animal models of infection (7, 11, 13).
The accessory Sec system is encoded within an operon on the chromosome (1). The gene organization of the accessory Sec locus tends to be conserved among species expressing this system (14, 15). In addition to the export substrate, the locus contains genes that encode proteins mediating its export and glycosylation (Fig. 1A). Prior to export, GspB is glycosylated in the cytoplasm of S. gordonii by four glycosyltransferases (GtfA, GtfB, Gly, and Nss [16]). GspB export requires five highly conserved proteins: SecA2, SecY2, Asp1, Asp2, and Asp3 (17). Two additional components, Asp4 and Asp5, are present in only some species (14, 15). However, GspB export requires both Asp4 and Asp5 (18).
Fig 1.
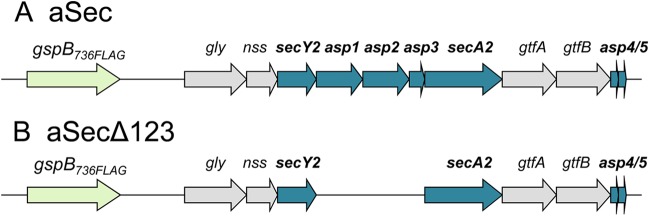
Genetic map of the S. gordonii accessory Sec operon. (A) The accessory Sec operon (aSec), with replacement of native gspB with gspB736FLAG. GspB736FLAG is identical to GspB, except for a shorter C-terminal serine-rich-repeat region and a 3×FLAG tag, and has the same export requirements as the full-length export substrate (43). (B) The accessory Sec operon with deleted asp1, asp2, and asp3 (aSecΔ123), encoding gspB736FLAG (green), the four glycosyltransferases (gray), SecA2, and the components of the accessory Sec translocon (blue).
SecA2 and SecY2 are paralogs of canonical SecA and SecY and are likely to have similar transport functions, with SecA2 serving as a motor protein for translocation (19) and SecY2 functioning as a transmembrane channel for the passage of the substrate (1, 20). Asp4 and Asp5 may function as SecG and SecE homologs, respectively, based on the similarity of their sizes and their predicted transmembrane topologies (18). Together, SecY2, Asp4, and Asp5 presumably form the membrane translocation complex (translocon) of the accessory Sec system, with associated SecA2 working as an ATPase to power substrate transport. It is uncertain whether SecA2 also functions as a receptor for the export substrate, as has been described for Escherichia coli SecA. In E. coli, SecA receives a subset of preproteins from chaperone SecB and then relays these preproteins to the SecYEG channel for translocation (21).
Asp1, Asp2, and Asp3 do not have homologs beyond the accessory Sec system, and their functions in transport are largely unknown. In S. gordonii, these proteins are present in the cytoplasm and are essential for GspB export (17). Asp2 and Asp3 can bind the unstructured serine-rich-repeat domains of GspB before the regions are glycosylated (22). These findings suggest that the two Asps may function in part as chaperones to transport the substrate to the translocon. Our most recent studies showed that Asp1 to -3 also facilitate a specific interaction between the AST domain of GspB and SecA2 that occurs during transport and may directly alter the conformation of either GspB or SecA2 (23). In support of the latter possibility, Asp3 can also bind SecA2 (24) and thus could have a role in modulating SecA2 activity. Furthermore, Asp3 can bind Asp1 in vitro and in vivo, and this interaction is required for GspB export (24). Together, these findings suggest that these Asps have multiple functions in GspB transport.
Our previous findings provided evidence for interactions among the accessory Sec components and suggested possible roles for these proteins in GspB transport. However, no information was available regarding the subcellular locations of the proteins that could reveal how the accessory Sec components interact to facilitate the transport of GspB from the cytosol to and through the membrane translocon. To address these questions, we examined the subcellular localization of these accessory Sec proteins and assessed how their trafficking is affected by the presence or absence of other components of the system. Our findings indicate that some of these accessory Sec proteins are cytosolic proteins, while others are membrane associated. The interaction of the Asps with one another and with SecA2 can result in redistribution of the proteins between these compartments. Because two of the Asps can also interact with the preprotein (22), trafficking of the Asps to the membrane may be important for mediating substrate transport to the accessory Sec translocon.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The plasmids used in this study are presented in Table 1. Streptococcal and E. coli strains used in this study are listed in Tables 2 and 3, respectively. Streptococci were grown in Todd-Hewitt broth (THB) in 5% CO2 at 37°C, whereas E. coli was grown in Luria-Bertani (LB) medium with aeration. For microscopy and cross-linking studies, E. coli was grown for 3 h at 30°C (until the optical density at 600 nm [OD600] reached ∼0.5). The expression of the recombinant proteins fused with the enhanced green fluorescent protein (EGFP) from pBAD/His A (Life Technologies) was then induced for 4 h at 25°C with l-arabinose (Life Technologies). Based on empirical testing, we used concentrations of the sugar that produced minimally detectable expression levels of the EGFP-fused proteins by fluorescence microscopy: 0.0002% for EGFP, 0.002% for untagged Asp3 (negative control), 0.0012% for EGFPSecA2, 0.0008% for EGFPAsp1, 0.0008% for EGFPAsp2, and 0.002% for EGFPAsp3. Untagged accessory Sec components coexpressed with the EGFP fusion proteins were expressed constitutively from pVA891 (25), pACYCDuet (Novagen), or pColaDuet (Novagen) without induction. For H6SecA2 and HAAsp2 copurification studies, overnight cultures of E. coli were diluted 1:50 in fresh LB medium and grown at 30°C for 3 h and then at 25°C for 4 h without induction for protein expression.
Table 1.
Plasmids used in this study
| Plasmid | Characteristicsa | Description | Source |
|---|---|---|---|
| pVA891 | Cmr | 25 | |
| pSwaR | pVA891 + M99 accessory Sec operon (Fig. 1A) | Expressing GspB736FLAG and the components of the accessory Sec operon | 23 |
| pSwaRΔKpn | pVA891 + M99 accessory Sec operon with deleted asp1, asp2, and asp3 (Fig. 1B) | Expressing GspB736FLAG, the glycosyltransferases, SecA2, and the components of the accessory Sec translocon | 23 |
| pBAD/His A | Ampr | Vector with l-arabinose-inducible promoter for dose-dependent protein expression in E. coli | Life Technologies |
| pBAD.egfp | pBAD + egfp | Expressing EGFP | This study |
| pBAD.asp3 | pBAD + asp3 | Expressing untagged Asp3 | This study |
| pBAD.EGFPsecA2 | pBAD + EGFPsecA2 | Expressing EGFPSecA2 | This study |
| pBAD.EGFPasp1 | pBAD + EGFPasp1 | Expressing EGFPAsp1 | This study |
| pBAD.EGFPasp2 | pBAD + EGFPasp2 | Expressing EGFPAsp2 | This study |
| pBAD.EGFPasp3 | pBAD + EGFPasp3 | Expressing EGFPAsp3 | This study |
| pACYCDuet | Cmr | Expression vector with T7 promoter | Novagen |
| pACYC.secA2 | pACYCDuet + secA2 | Expressing untagged SecA2 | This study |
| pCOLADuet | Kanr | Expression vector with T7 promoter | Novagen |
| pCola.secA2 | pColaDuet + secA2 | Expressing untagged SecA2 | This study |
| pCola.asp1-asp3 | pCOLADuet + asp1 + asp3 | Coexpressing untagged Asp1 and Asp3 | This study |
| pCola.asp1-asp2 | pCOLADuet + asp1 + asp2 | Coexpressing untagged Asp1 and Asp2 | This study |
| pCola.asp3-asp2 | pCOLADuet + asp3 + asp2 | Coexpressing untagged Asp2 and Asp3 | This study |
| pACYC.H6secA2 | pACYCDuet + H6secA2 | Expressing H6SecA2 | This study |
| pETDuet.HAasp2 | pETDuet + HAasp2 | Expressing HAAsp2 | 22 |
| pVA.secA2FLAG | pVA891 + 3′ of secA2FLAG | For FLAG tagging of chromosomal secA2 in S. gordonii | This study |
| pVA.asp1FLAG | pVA891 + 3′ of asp1FLAG | For FLAG tagging of chromosomal asp1 in S. gordonii | This study |
| pVA.asp2FLAG | pVA891 + 3′ of asp2FLAG | For FLAG tagging of chromosomal asp2 in S. gordonii | This study |
| pVA.asp3FLAG | pVA891 + 3′ of asp3inf>FLAG | For FLAG tagging of chromosomal asp3 in S. gordonii | This study |
| pMSP3535 and pMSP3545 | Ermr | Cloning vector containing a replication origin for Gram-positive bacteria and nisin-inducible promoter | 27 |
| pMSP.asp1 | pMSP3545 + asp1 | Expressing untagged Asp1 | 17 |
| pMSP.asp2 | pMSP3545 + asp2 | Expressing untagged Asp2 | 17 |
| pMSP.asp3 | pMSP3545 + asp3 | Expressing untagged Asp3 | 17 |
| pMSP.secA2 | pMSP3535 + secA2 | Expressing untagged SecA2 | 19 |
| pMSP.EGFPasp1 | pMSP3545 + asp1 | Expressing EGFPAsp1 | This study |
| pMSP.EGFPasp2 | pMSP3545 + asp2 | Expressing EGFPAsp2 | This study |
| pMSP.EGFPasp3 | pMSP3545 + asp3 | Expressing EGFPAsp3 | This study |
| pMSP.EGFPsecA2 | pMSP3545 + secA2 | Expressing EGFPSecA2 | This study |
Cmr, chloramphenicol resistance; Ampr, ampicillin resistance; Kanr, kanamycin resistance; Ermr, erythromycin resistance.
Table 2.
Streptococcal strains used in this study
| S. gordonii strain | Characteristicsa | Description | Source |
|---|---|---|---|
| M99 | Parental strain | 42 | |
| PS846 | M99 ΔgspB::pEVP Cmr | M99 with deleted gspB | 20 |
| PS1225 | M99 gspB736FLAG::pEVP Cmr | M99 expressing GspB736FLAG | This study |
| PS1574 | PS1225 ΔsecY2FLAG::spc Spcr | M99 expressing GspB736FLAG but not SecY2 | This study |
| PS2190 | PS1574 secY2FLAG | M99 expressing SecY2FLAG and GspB736FLAG | This study |
| PS2962 | PS2190 secA2FLAG::pVA Ermr | M99 expressing SecY2FLAG, GspB736FLAG, and SecA2FLAG | This study |
| PS2963 | PS2190 asp1FLAG::pVA Ermr | M99 expressing SecY2FLAG, GspB736FLAG, and Asp1FLAG | This study |
| PS2964 | PS2190 asp2FLAG::pVA Ermr | M99 expressing SecY2FLAG, GspB736FLAG, and Asp2FLAG | This study |
| PS2965 | PS2190 asp3FLAG::pVA Ermr | M99 expressing SecY2FLAG, GspB736FLAG, and Asp3FLAG | This study |
| PS1242 | PS1225 Δasp1::spc Spcr | M99 with deleted asp1 and expressing GspB736FLAG | This study |
| PS1243 | PS1225 Δasp2::spc Spcr | M99 with deleted asp2 and expressing GspB736FLAG | This study |
| PS1244 | PS1225 Δasp3::spc Spcr | M99 with deleted asp3 and expressing GspB736FLAG | 24 |
| PS1226 | PS516 gspB::pB736flagC Cmr | M99 with deleted secA2 and expressing GspB736FLAG | 19 |
| PS2988 | PS1242/pMSP.asp1 Ermr | M99 expressing GspB736FLAG from the chromosome and Asp1 from pMSP3545 | This study |
| PS2989 | PS1243/pMSP.asp2 Ermr | M99 expressing GspB736FLAG from the chromosome and Asp2 from pMSP3545 | This study |
| PS1345 | PS1244/pMSP.asp3 Ermr | M99 expressing GspB736FLAG from the chromosome and Asp3 from pMSP3545 | 24 |
| PS1270 | PS1226/pMSP.secA2 Ermr | M99 expressing GspB736FLAG from the chromosome and SecA2 from pMSP3535 | 19 |
| PS2990 | PS1242/pMSP.EGFPasp1 Ermr | M99 expressing GspB736FLAG from the chromosome and EGFPAsp1 from pMSP3545 | This study |
| PS2991 | PS1243/pMSP.EGFPasp2 Ermr | M99 expressing GspB736FLAG from the chromosome and EGFPAsp2 from pMSP3545 | This study |
| PS2992 | PS1244/pMSP.EGFPasp3 Ermr | M99 expressing GspB736FLAG from the chromosome and EGFPAsp3 from pMSP3545 | This study |
| PS2993 | PS1226/pMSP.EGFPsecA2 Ermr | M99 expressing GspB736FLAG from the chromosome and EGFPSecA2 from pMSP3535 | This study |
Cmr, chloramphenicol resistance; Ermr, erythromycin resistance; Spcr, spectinomycin resistance.
Table 3.
Combinations of plasmids used in E. coli Top10a
| Figure | Group or lane | Plasmid | Expression |
|---|---|---|---|
| 2A | 1 | pBAD.EGFPsecA2/pSwaRΔKpn | EGFPSecA2, SecA2, SecY2, Asp4, Asp5, GspB, and glycosyltransferases |
| 2A | 2 | pBAD.EGFPsecA2 | EGFPSecA2 |
| 2A | 3 | pBAD.asp3 | Asp3 |
| 2A | 4 | pBAD.egfp | EGFP |
| 3A | 1 | pSwaR | Entire accessory Sec components |
| 3A | 2 | pVA891 | |
| 4A | 1 | pBAD.EGFPasp2 | EGFPAsp2 |
| 4A | 2 | pBAD.EGFPasp2/sparΔKpn/pCola.secA2 | EGFPAsp2, SecA2, SecY2, Asp4, Asp5, GspB, and glycosyltransferases |
| 4A | 3 | pBAD.EGFPasp2/pACYC.secA2/pCola.secA2 | EGFPAsp2 and SecA2 |
| 4A | 4 | pBAD.EGFPasp2/pACYC.secA2/pCola.asp1-asp3 | EGFPAsp2, SecA2, Asp1, and Asp3 |
| 4A | 5 | pBAD.EGFPasp2/pCola.asp1-asp3 | EGFPAsp2, Asp1, and Asp3 |
| 5A | 1 | pBAD.EGFPasp2/pACYC.secA2/pCola.secA2 | EGFPAsp2 and SecA2 |
| 5A | 2 | pACYC.secA2/pCola.secA2 | SecA2 |
| 5B | 1 | pACYC.H6secA2/pETDuet.HAasp2 | H6SecA2 and HAAsp2 |
| 5B | 2 | pACYCDuet/pETDuet.HAasp2 | HAAsp2 |
| 6A | 1 | pBAD.EGFPasp1 | EGFPAsp1 |
| 6A | 2 | pBAD.EGFPasp3 | EGFPAsp3 |
| 6B | 1 | pBAD.EGFPasp1/pSwaRΔKpn/pCola.secA2 | EGFPAsp1, SecA2, SecY2, Asp4, Asp5, GspB, and glycosyltransferases |
| 6B | 2 | pBAD.EGFPasp1/pACYC.secA2/pCola.secA2 | EGFPAsp1, SecA2 |
| 6B | 3 | pBAD.EGFPasp3/pSwaRΔKpn/pCola.secA2 | EGFPAsp3, SecA2, SecY2, Asp4, Asp5, GspB, and glycosyltransferases |
| 6B | 4 | pBAD.EGFPasp3/pACYC.secA2/pCola.secA2 | EGFPAsp3, SecA2 |
| 7A | 1 | pBAD.EGFPasp3/pSwaRΔKpn/pCola.asp1-asp2 | EGFPAsp3, SecA2, SecY2, Asp4, Asp5, Asp1, Asp2, GspB, and glycosyltransferases |
| 7A | 2 | pBAD.EGFPasp3/pSwaRΔKpn/pCola.asp1 | EGFPAsp3, SecA2, SecY2, Asp4, Asp5, Asp1, GspB, and glycosyltransferases |
| 7A | 3 | pBAD.EGFPasp3/pSwaRΔKpn/pCola.asp2 | EGFPAsp3, SecA2, SecY2, Asp4, Asp5, Asp2, GspB, and glycosyltransferases |
| 8A | 1a and 1b | pBAD.EGFPasp1/pSwaRΔKpn/pCola.asp3-asp2 | EGFPAsp1, SecA2, SecY2, Asp4, Asp5, Asp2, Asp3, GspB, and glycosyltransferases |
| 8A | 2 | pBAD.EGFPasp1/pSwaRΔKpn | EGFPAsp1, SecA2, SecY2, Asp4, Asp5, GspB, and glycosyltransferases |
From Life Technologies.
Plasmid construction.
The plasmids used in this study were constructed with the primers listed in Table 4. To express EGFP fusion proteins from pBAD, egfp was amplified by PCR from pEGFP-C1 (Clontech) using primers NcoI-EGFP and EGFP-XhoI. The genes encoding SecA2, Asp1, Asp2, and Asp3 were amplified from the M99 chromosome or pETDuet-HAasp2 (with silent mutations at the NcoI and NdeI sites of asp2 [22]), using EcoRI- and HindIII-linked primers for the asp genes and PstI- and KpnI-linked primers for secA2. The PCR products were ligated into pBAD to create constructs expressing an N-terminal EGFP. C-terminal EGFP fusions were found to be unstable and so were not used in this study. The construction of pVA891 plasmids expressing either the entire accessory Sec operon (aSec) or the operon with asp1, asp2, and asp3 genes deleted (aSecΔ123), resulting in pSwaR or pSwaRΔKpn, respectively, has been described elsewhere (23). To express untagged Asps in E. coli, genes were amplified from the M99 chromosome or pETDuet.HAasp2 using NcoI- and NotI-linked primers and NdeI- and XhoI-linked primers and cloned into the first and second multiple-cloning sites within the Duet vectors (Novagen), respectively. To express SecA2 from the Duet vectors, primers BamHI-SecA2 and SecA2-AscI were used for secA2 insertion into the first multiple-cloning site of the plasmids. The construction of pETDuet.HAAsp2 has been described previously (22).
Table 4.
Primers used in this study
| Primer | Sequence (5′–3′)a |
|---|---|
| NcoI-EGFP | CCGGTCGCGACCATGGTGAGC |
| EGFP-XhoI | CATGGACGAGCTGTACAAGCTCGAGCT |
| EcoRI-Asp1 | ATTAGAATTCTTATGTATTATTTTATTCCTTCATGGAGTGGAAG |
| Asp1-HindIII | TAATAAGCTTTTATTTTTCATCTATAGCCTCCTTGAGC |
| EcoRI-Asp2 | ATTAGAATTCTTATGAAAAATAAGCTGAAGATCTTACAGATTGG |
| Asp2-HindIII | AATAAGCTTTCATTTCTTTCTTCCAAATTCTTTTTCCAATATGATG |
| EcoRI-Asp3 | ATTAGAATTCTTATGAAGATTCAAAAACATAAGGAAATTTACTG |
| Asp3-HindIII | TAATAAGCTTTTAACCATTTGACTCCTCTAAAATTTCTTC |
| PstI-SecA2 | ATTACTGCAGCATGGTTAAAAATTTTTTTCATATTCGGCGC |
| SecA2-KpnI(s) | TAATGGTACCCTATGGGAAGTACATTACGACC |
| NcoI-Asp1 (orf1C4) | GGTACCATGGATTATTTTATTCCTTCAT (17) |
| Asp1-NotI | TTGCGGCCGCTTATTTTTCATCTATAGCCTCC |
| NcoI-Asp3 | ATTACCATGGCAAAGATTCAAAAACATAAGGAAATTTACTGG |
| Asp3-NotI | TTGCGGCCGCTTAACCATTTGACTCCTCTAA |
| NdeI-Asp2 | ATTACATATGAAAAATAAGCTGAAGATCTTACAGATTGG |
| Asp2-XhoI | AAACGTTCTCGAGCTATTTCTTTCTTCCAAATTCTTT |
| NdeI-Asp3 | GCATATGAAGATTCAAAAACATAAGGAA |
| Asp3-XhoI | TAATCTCGAGACCATTTGACTCCTCTAAAATTTCTTC |
| BamHI-SecA2 | ACTCGGGATCCAATGGTTAAAAATTTTTTTCATATTCGGCG |
| SecA2-AscI | TATGGCGCGCCAACTATGGGAAGTACATTACGA |
| Asp1-XbaI | GCTCTAGAGTTATTTCTTTCTTCCAAATTCTTT |
| Asp2-XbaI | GCTCTAGAGTTATTTTTCATCTATAGCCTCC |
| Asp3-SpeI (orf3C2) | TATGACTAGTATTTTTAACCATTTGACTCC |
| SecA2-SpeI | TAATGGTACCAAACTAGTCTATGGGAAGTACATTACGACCTC |
| BamHI-pVAf.SecA2 | AAATAGGATCCTGAGATGTCTGAAATCTATTCTTCTC |
| pVAf. SecA2-SpeI | TATTTACTAGTTGGGAAGTACATTACGACCTCTC |
| BamHI-pVAf.Asp1 | AAATAGGATCCGAAAACCCTTTGCCAGAAAAC |
| pVAf.Asp1-SpeI | TATTTACTAGTTTTTTCATCTATAGCCTCCTTGAGC |
| BamHI-pVAf.Asp2 | AAATAGGATCCGCTCAACGTGGCCGTCTTAG |
| pVAf.Asp2-SpeI | TATTTACTAGTTTTCTTTCTTCCAAATTCTTTTTCCAATATGATGCG |
| BamHI-pVAf.Asp3 | AAATAGGATCCTATTTTGAAAACAAATTAATTGCTTCTGGTC |
| pVAf.Asp3-SpeI | TATTTACTAGTACCATTTGACTCCTCTAAAATTTCTTCAATTC |
Restriction sites are underlined.
The following plasmids were used in the construction of streptococcal strains. The plasmid pB736flagC (19) was used for the transformation of S. gordonii M99, resulting in strain PS1225, which expresses GspB736FLAG from the chromosome. Plasmids based on pS326 were used to delete asp1, asp2, asp3, or secA2 in strain PS1225, and the pMSP-based plasmids were used to complement these deletion strains with native Asp/SecA2, as reported previously (17, 19). To express EGFP fusion proteins from pMSP3545 (27) for complementation studies, egfp fusions from the pBAD vector were amplified with NcoI-EGFP and the XbaI-linked primers for asp1 and asp2, or the SpeI-linked primers for asp3 and secA2, before insertion into the vector. To construct strain PS2190, which has a markerless replacement of secY2 with secY2FLAG, we used the same strategies described for strains expressing other SecY2 variants (20). In brief, wild-type secY2 was replaced with a spectinomycin resistance cassette by transformation with pY2KO, generating strain PS1574. Next, pY2FLAG was used to replace the spectinomycin resistance cassette. In the cloning of pVA.secA2FLAG or pVA.aspFLAG for the FLAG tagging of chromosomal secA2 and the asp genes in S. gordonii, ∼350 nucleotides in the 3′ end of each gene were amplified by PCR with BamHI- and SpeI-linked primers. The amplicon replaced the gspB truncated variant in pVA.gspB736FLAG (22) by restriction digestion and ligation. The resulting pVA.secA2FLAG or pVA.aspFLAG was used to transform S. gordonii. These plasmids were integrated into the chromosome by homologous recombination. Thus, the 3×FLAG-tagged fusion replaced the native gene copy on the chromosome.
Fluorescence and immunofluorescence microscopy.
E. coli cells expressing EGFP fusion proteins were collected after induction and suspended in Dulbecco's phosphate-buffered saline (PBS) (Sigma), pH. 7.4. The cells were mounted on slides with 0.5% agarose and covered with coverslips. Bacteria that adhered to the coverslips in a uniform layer were imaged with an Applied Precision Deltavision Spectris DV4 (inverted) microscope with 5 s UV excitation at 490 nm. For immunofluorescence microscopy, E. coli cells expressing aSec or the empty cloning vector were harvested and fixed for 1 h at room temperature in a PBS buffer containing 2.5% paraformaldehyde and 0.03% glutaraldehyde. Glycine-PBS (250 mM) was then added to the suspension to quench excess formaldehyde. The cells were washed once with PBS and permeabilized with 0.1% Triton X-100 in PBS for 1 h, followed by washing and treatment with 2 mg ml−1 of lysozyme for 1 h. After three more washes with PBS, the cells were suspended in PBS, mounted on poly-l-lysine slides, and incubated with anti-SecA antibody, followed by incubation with anti-rabbit IgG conjugated to Alexa Fluor 568 (Life Technologies). Eight washes were performed with PBS containing 0.05% Tween 20 after each antibody incubation. After an additional wash with only PBS, the cells were protected with coverslips and imaged with a Deltavision Spectris DV4 microscope with 2 s UV excitation at 555 nm.
A chi-square test was performed to assess the significance of differences in the numbers of cells with punctate versus nonpunctate phenotypes. When assessing the difference in these values between two samples (strains), a two-by-two contingency table was used in the test.
Protein cross-linking.
E. coli cells coexpressing pBAD.EGFPasp2, pACYC.secA2, and pCola.secA2 were used for in vivo cross-linking. After induction, the cells were collected by centrifugation, washed once with PBS, and suspended in PBS containing 2 mM dithiobissuccinimidylpropionate (DSP), a membrane-permeable, amine-reactive, and cleavable cross-linking agent (Thermo Scientific). After incubation in DSP for 1 h on ice, cross-linking was terminated by the addition of 20 mM Tris, pH 7.3. The cells were then collected and stored at −80°C. To purify EGFPAsp2, cell pellets were thawed and suspended in Bugbuster Protein Extraction Reagent (EMD) supplemented with Complete Protease Inhibitors (Roche). The lysates were sonicated and clarified by centrifugation at 11,000 × g for 20 min at 4°C. The clarified lysates (CL) were then mixed with anti-green fluorescent protein (GFP)-conjugated agarose (MBL International) and incubated for 3 h at 4°C. The agarose was recovered and washed with 1% Bugbuster reagent in PBS four times. Before resolving proteins bound to the agarose by SDS-PAGE, the agarose was boiled in a lithium dodecyl sulfate (LDS) sample buffer (Life Technologies) supplemented with 5% β-mercaptoethanol, which cleaves the disulfide bonds of cross-linked proteins. This reducing agent was used to improve the resolution of cross-linked proteins, which otherwise migrated as broad streaks on SDS-PAGE. Western blotting with anti-SecA2 antibody was carried out to detect SecA2 that had been cross-linked to and copurified with EGFPAsp2.
Protein copurification.
E. coli cells coexpressing pACYC.H6sec2 and pETDuet.HAasp2, and the negative control (expressing pACYCDuet and pETDuet.HAasp2), were grown without induction for protein overexpression. H6SecA2 was affinity purified with Ni-nitrilotriacetic acid (NTA) agarose (Qiagen) as described previously (22). The agarose was recovered and boiled in LDS sample buffer supplemented with 5% β-mercaptoethanol prior to resolving recovered proteins by SDS-PAGE. Detection of H6Sec2 and HAAsp2 was carried out by Western blotting with anti-H6 (GE Healthcare) or anti-hemagglutinin (HA) (Sigma) antibody, respectively.
Cell fractionation of S. gordonii.
Overnight cultures of M99 variants expressing 3×FLAG-tagged accessory Sec components were collected by centrifugation, and the cells were treated with PBS containing 100 U ml−1 mutanolysin (Sigma), 5 mg ml−1 lysozyme, 0.02% (wt/vol) MgCl2, and 2.6% (wt/vol) raffinose. The spheroplasts were then collected by centrifugation at 4,500 × g and frozen overnight at −80°C. The spheroplasts were thawed and suspended in 20 mM Tris, pH. 7.3, containing 1 mM EDTA and Complete Protease Inhibitors. The suspension was passed through a French pressure cell three times at 2,000 lb/in2 (gauge reading), and the whole-cell lysates (WCL) were clarified by centrifugation at 11,000 × g for 20 min at 4°C. The soluble and insoluble fractions of the CL were separated by centrifugation at 100,000 × g for 2 h at 4°C. The supernatants and pellets were collected, and the pellets were solubilized in Bugbuster Protein Extraction Reagent prior to resolution by SDS-PAGE.
RESULTS
Visualization of the streptococcal accessory Sec components in E. coli.
We chose to examine the localization and trafficking of the accessory Sec components by microscopy, as this approach allows the assessment of protein distribution in intact cells (28–30). Our initial attempts to detect the accessory Sec components in S. gordonii by fluorescence microscopy were unsuccessful, regardless of whether we used immunofluorescence or GFP fusions (data not shown). Our inability to detect these proteins was most likely due to the extremely low expression levels of the components in the organism (data not shown). In view of these technical limitations, we decided to examine the localization of the accessory Sec components when expressed as EGFP fusion proteins in E. coli. We have recently shown that the reconstituted streptococcal accessory Sec system functions in E. coli and can export glycosylated GspB736FLAG to the periplasm of the organism (23). When tested in S. gordonii, the EGFP-fused accessory Sec proteins retained their physiological functions, being capable of mediating the export of GspB736FLAG (see Fig. S1 in the supplemental material). These findings indicate that the mechanisms for GspB transport mediated by the streptococcal accessory Sec proteins are functionally conserved when the system is expressed in E. coli and that the subcellular localization of the fusion proteins is likely to reflect those in S. gordonii. Thus, the system appears to be a valid model for studying the interaction and localization of the accessory Sec components.
SecA2 localizes to multiple foci along the cell membrane.
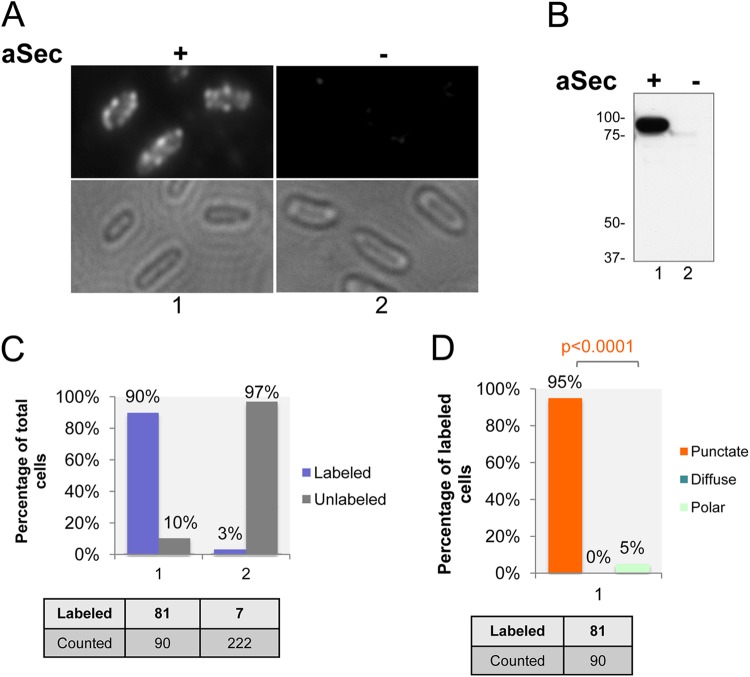
When fused with EGFP, SecA2 formed multiple foci along the cell membrane in more than 70% of fluorescent cells (Fig. 2A and B, groups 1 and 2), whereas EGFP alone was diffusely distributed in the cytosol of all of the fluorescent cells (group 4). The punctate pattern of EGFPSecA2 membrane localization was seen when the protein was expressed alone (Fig. 2A, group 2) or when coexpressed with the accessory Sec translocon but without Asp1 to -3 (group 1; for simplicity, the group of proteins expressed in the latter case is referred to as aSecΔ123 [Fig. 1B]). In parallel studies, the localization of SecA2 expressed in E. coli from a plasmid encoding an intact streptococcal accessory Sec operon was assessed by immunofluorescence microscopy with anti-SecA2 antibody. Under these conditions, native SecA2 also localized to the membrane in a punctate pattern in 95% of the labeled cells (Fig. 3A and D). Thus, these findings indicate that SecA2 can localize to the cell membrane independently of other accessory Sec components.
Fig 2.
Punctate distribution of EGFPSecA2 at the cell membrane. (A) Micrographs of EGFPSecA2 localization in E. coli. The fusion protein was expressed with aSecΔ123 (group 1) or alone (group 2). Detection of EGFP fusions was performed by fluorescence microscopy with live cells. Untagged Asp3 (group 3) and EGFP (group 4) served as negative controls. The orange label denotes proteins expressed constitutively from the pVA891 vector, whereas the black label represents proteins expressed with l-arabinose induction from the pBAD vector. (B) Quantification of strains shown in panel A, presented in the same labeling order. Cells that showed fluorescent signals were further quantified for punctate (red), diffuse (blue), and polar (green) localization phenotypes. Chi-square analysis was performed to assess differences in phenotype frequencies observed between two strains (EGFPSecA2 and EGFP; P < 0.0001) or between the punctate and nonpunctate (diffused and polar) phenotypes of a strain (EGFPSecA2/aSecΔ123, P < 0.0001; EGFPSecA2, P < 0.0002). (C) Western blot of the WCL of constructs shown in panel A, presented in the same labeling order, with anti-GFP detecting expression of EGFPSecA2 or EGFP. Values at left are expressed in kilodaltons.
Fig 3.
Punctate distribution of native SecA2 at the cell membrane. (A) Immunofluorescence micrographs of SecA2. E. coli expressing the entire accessory Sec operon (aSec +; group 1) or the empty vector (aSec −; group 2) were fixed, permeabilized, and probed with anti-SecA2 polyclonal antibody and then fluorophore-conjugated secondary antibody. (B) Western blot of the WCL of constructs shown in panel A, presented in the same labeling order. The blot was probed with anti-SecA2 antibody. Values at left are expressed in kilodaltons. (C) Quantification of strains shown in panel A, presented in the same labeling order. Fluorescence-labeled and unlabeled cells were further differentiated and quantified. (D) Quantification of punctate, diffuse, and polar phenotypes shown by fluorescence-labeled cells of the construct in group 1 of panel A. Chi-square analysis was performed to assess differences in frequencies observed between the punctate and nonpunctate phenotypes of fluorescence-labeled cells.
The localization of Asp2 is differentially affected by SecA2, Asp1, and Asp3.
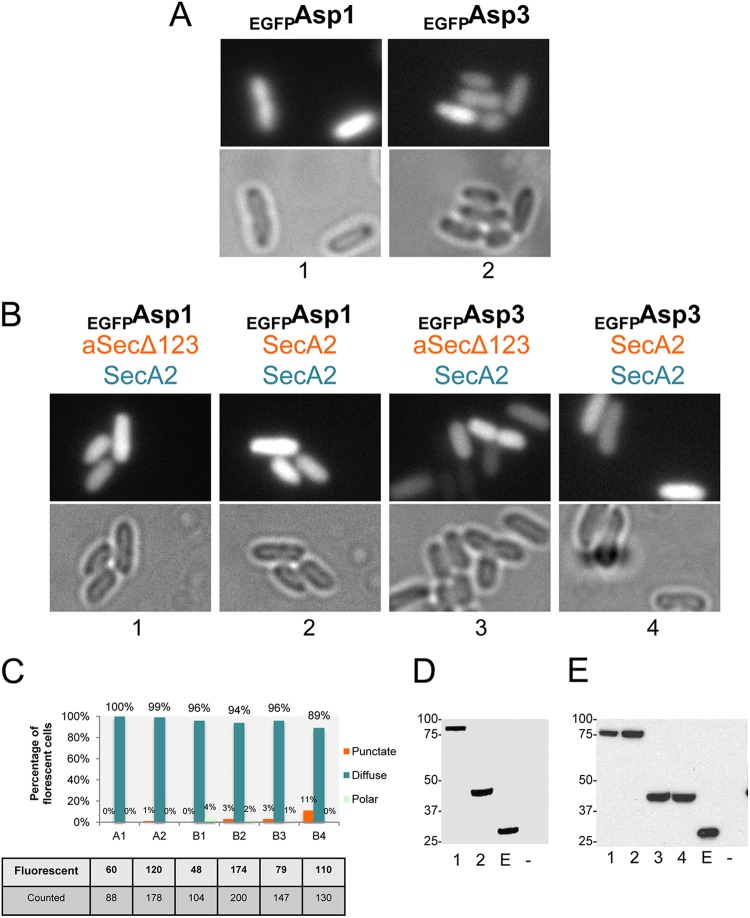
We next analyzed the localization of Asp2. EGFPAsp2 expressed alone predominantly localized to one or both poles at the membrane (Fig. 4A and B, group 1). However, when EGFPAsp2 was coexpressed with SecA2 and aSecΔ123, EGFPAsp2 formed multiple foci along the cell membrane in 80% of the fluorescent cells (Fig. 4A and B, group 2). A similar increase in the fraction of cells (64%) having a punctate pattern of EGFPAsp2 localization was also seen when the protein was expressed with SecA2 alone (Fig. 4A and B, compare group 3 to group 1). Thus, the targeting of Asp2 to multiple foci at the cell membrane is dependent on SecA2.
Fig 4.
Asp2 requires SecA2 for its punctate distribution along the cell membrane and Asp1 and Asp3 to traffic to the cytosol. (A) Micrographs of EGFPAsp2 localization in E. coli. EGFPAsp2 was expressed alone (group 1); with SecA2 and aSecΔ123 (group 2); with SecA2 (group 3); with SecA2, Asp1, and Asp3 (group 4); or with only Asp1 and Asp3 (group 5). The black labels represent proteins expressed with l-arabinose induction from the pBAD vector. The orange labels represent proteins expressed constitutively from the pVA891 (aSecΔ123) or pACYCDuet (SecA2) vector. The blue labels represent proteins expressed constitutively from the pColaDuet vector. (B) Quantification of strains shown in panel A, presented in the same labeling order. Cells that showed fluorescent signals were further quantified for punctate, diffuse, and polar phenotypes. Chi-square analysis was performed to assess differences in phenotype frequencies observed between two strains (EGFPAsp2/SecA2/SecA2 and EGFPAsp2/−/Asp1 + Asp3; P < 0.0001) or between the punctate and nonpunctate phenotypes of each indicated strain. (C) Western blot with anti-GFP antibody assessing expression of EGFPAsp2 from the constructs shown in panel A, presented in the same labeling order. Lane E, EGFP; lane −, untagged Asp3. Values at left are expressed in kilodaltons.
We then assessed the impacts of Asp1 and Asp3 on the distribution of Asp2. When EGFPAsp2 was coexpressed with SecA2, Asp1, and Asp3, it still localized in a punctate pattern in 45% of cells. In the remainder of the population, however, EGFPAsp2 was distributed in the cytosol (Fig. 4A and B, group 4). In contrast, when expressed only with Asp1 and Asp3, EGFPAsp2 distributed in the cytosol in a predominantly diffuse pattern (Fig. 4A and B, group 5). Thus, the localization of EGFPAsp2 at the cell membrane depends on the presence of SecA2, whereas its distribution to the cytosol requires Asp1 and Asp3.
Asp2 and SecA2 form a complex.
Because the localization of Asp2 to multiple foci at the cell membrane requires SecA2, it is possible that the two proteins interact directly. To assess this possibility, EGFPAsp2 and SecA2 were coexpressed in E. coli and cross-linked using DSP. EGFPAsp2 was then recovered by GFP affinity purification, and the material was probed by Western blotting for the presence of SecA2. In control studies, no SecA2 was recovered from E. coli expressing SecA2 alone (Fig. 5A, lane 4), indicating that the protein did not adhere nonspecifically to the anti-GFP agarose. In contrast, when the two proteins were coexpressed, SecA2 and EGFPAsp2 were recovered as a cross-linked complex (Fig. 5A, lane 3). As a complementary study, H6SecA2 and HAAsp2 were coexpressed constitutively from E. coli, and H6SecA2 was then affinity purified with Ni-NTA agarose. We found that HAAsp2 copurified with H6SecA2 (Fig. 5B, lane 3). Moreover, no HAAsp2 was found in the material recovered from the negative-control strain expressing only HAAsp2 (lane 4). These results indicate that Asp2 and SecA2 form a complex and provide further evidence that Asp2 requires SecA2 for its localization at the cell membrane.
Fig 5.

Asp2 and SecA2 form a complex. (A) Cross-linking and copurification of Asp2 and SecA2. E. coli cells coexpressing EGFPAsp2 and SecA2 were treated with the membrane-permeable cross-linker DSP. EGFPAsp2 was then recovered from the clarified CL by affinity purification. Proteins in the recovered material were resolved by SDS-PAGE under reducing conditions and probed for SecA2 by Western blotting. Lane 1, CL from E. coli expressing EGFPAsp2 and SecA2; lane 2, CL from E. coli expressing SecA2 but not EGFPAsp2; lane 3, proteins recovered by anti-GFP (αGFP) purification from material in lane 1; lane 4, proteins recovered by anti-GFP purification from material in lane 2. (B) Copurification of Asp2 and SecA2. H6SecA2 and HAAsp2 were coexpressed constitutively in E. coli, and H6SecA2 was recovered from the CL by affinity purification. Proteins in the recovered material were resolved by SDS-PAGE and probed for HAAsp2 with anti-HA antibody by Western blotting. Lane 1, CL from E. coli expressing H6SecA2 and HAAsp2; lane 2, CL from E. coli expressing HAAsp2 but not H6SecA2; lane 3, proteins recovered by Ni-NTA purification from material in lane 1; lane 4, proteins recovered by Ni-NTA purification from material in lane 2.
Asp1 and Asp3 are cytosolic proteins but can traffic to the membrane in the presence of the entire accessory Sec system.
When expressed individually, EGFPAsp1 and EGFPAsp3 both localized to the cytosol of E. coli in a diffuse distribution (Fig. 6A). Because our previous findings indicated that Asp3 could individually interact with Asp1, Asp2, and SecA2 (24), we asked whether Asp1 and Asp3 could also traffic to the membrane in the presence of other accessory Sec components and, if so, whether SecA2 played a direct role in retaining these proteins at the membrane, as was seen with Asp2. When EGFPAsp1 or EGFPAsp3 was expressed with either SecA2 alone or aSecΔ123, EGFPAsp1 or EGFPAsp3 remained diffusely distributed in the cytosol in more than 89% of the fluorescent cells (Fig. 6B and C). Thus, SecA2, alone or with the translocon, is insufficient for the trafficking of Asp1 or Asp3 to the cell membrane.
Fig 6.
SecA2, alone or with the translocon, is insufficient for influencing the localization of Asp1 or Asp3. (A) Micrographs of E. coli expressing EGFPAsp1 (group 1) or EGFPAsp3 (group 2). (B) Micrographs of E. coli expressing each fusion protein in conjunction with SecA2 and aSecΔ123 (groups 1 and 3) or with SecA2 alone (groups 2 and 4). The black, orange, and blue labels represent proteins expressed from the pBAD, pVA891/pACYCDuet, or pColaDuet vector, respectively. (C) Quantification of strains shown in panels A and B, presented in the same labeling order. Cells that showed fluorescent signals were further quantified for punctate, diffuse, and polar phenotypes. (D and E), Western blotting with anti-GFP examining the expression of the fusion proteins. WCL from constructs in panels A (D) and B (E) are shown in the same respective labeling order. Lanes E, EGFP; lanes −, untagged Asp3. Values at left are expressed in kilodaltons.
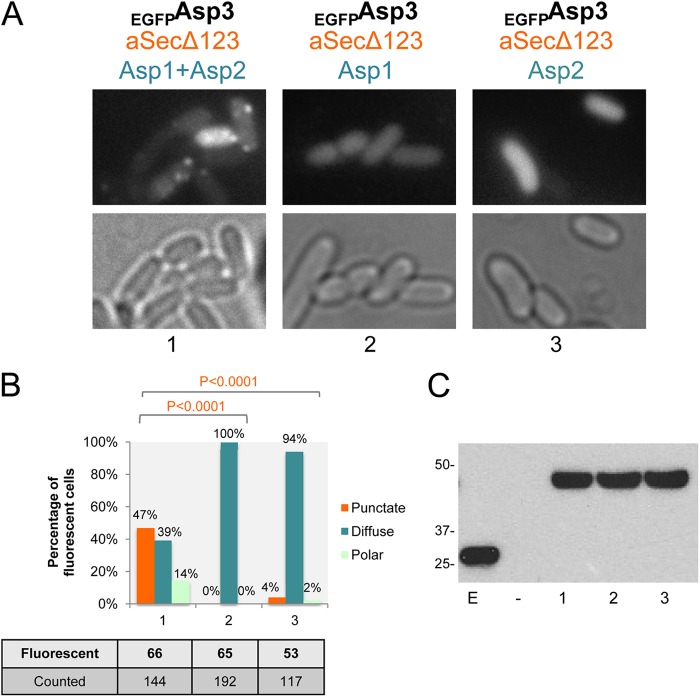
In contrast, when EGFPAsp3 was expressed with the entire accessory Sec system, it localized to the membrane as multiple foci in 47% of the cells with fluorescent signals (Fig. 7A and B, group 1). When EGFPAsp3 was expressed without either Asp2 (group 2) or Asp1 (group 3), but with the rest of the accessory Sec components still present, EGFPAsp3 redistributed to the cytosol in 100% or 94% of the fluorescent cells, respectively (Fig. 7A and B, compare groups 2 and 3 to group 1). These observations indicate that Asp3 can traffic to the cell membrane in the presence of the entire accessory Sec system, but SecA2 (alone or in combination with the translocon) is insufficient to enable the process. Instead, both Asp1 and Asp2 are also required for Asp3 trafficking to the membrane.
Fig 7.
Trafficking of Asp3 to the membrane requires both Asp1 and Asp2. (A) Micrographs of E. coli expressing EGFPAsp3 with aSecΔ123, in conjunction with both Asp1 and Asp2 (group 1), only Asp1 (group 2), or only Asp2 (group 3). The black, orange, and blue labels represent proteins expressed from the pBAD, pVA891, or pColaDuet vector, respectively. (B) Quantification of strains shown in panelA, presented in the same labeling order. Cells that showed fluorescent signals were further quantified for different phenotypes. Chi-square analysis was performed to assess differences in phenotype frequencies observed between two strains: EGFPAsp3/aSecΔ123/Asp1 + Asp3 and EGFPAsp3/aSecΔ123/Asp1 (P < 0.0001) or EGFPAsp3/aSecΔ123/Asp1 + Asp3 and EGFPAsp3/aSecΔ123/Asp3 (P < 0.0001). (C) Western blotting with anti-GFP detecting the expression of EGFP fusions from the WCL of constructs shown in panel A, presented in the same labeling order. Lane E, EGFP; lane −, untagged Asp3. Values at left are expressed in kilodaltons.
We then examined which cofactors influenced the localization of Asp1. Even in the presence of the entire accessory Sec system, EGFPAsp1 was predominantly distributed diffusely in the cytosol (Fig. 8A, group 1a). However, in 10% of fluorescent cells, EGFPAsp1 localized to the cell membrane and formed weakly detectable focal arrangements (Fig. 8A, group 1b). When EGFPAsp1 was expressed without Asp2 and Asp3, the number of fluorescent cells exhibiting a punctate phenotype decreased to 1% (P < 0.0025) (Fig. 8A and B, compare group 2 to group 1). These findings suggest that Asp1 localizes transiently at the cell membrane, and only in the presence of the entire accessory Sec system.
Fig 8.
Trafficking of Asp1 to the membrane requires both Asp2 and Asp3. (A) Micrographs of E. coli expressing EGFPAsp1. In groups 1a and 1b, the fusion protein was coexpressed with aSecΔ123, Asp2, and Asp3. In group 2, EGFPAsp1 was coexpressed with aSecΔ123. The black, orange, and blue labels represent proteins expressed from the pBAD, pVA891, or pColaDuet vector, respectively. The arrowheads point to signals from EGFPAsp1 forming multiple foci. (B) Quantification of strains shown in panel A, presented in the same labeling order. Cells that showed fluorescent signals were further quantified for different phenotypes. Chi-square analysis was performed to assess differences in phenotype frequencies observed between the two strains. (C) Western blotting with anti-GFP probing the expression of EGFPAsp1 from WCL of constructs shown in panel A, presented in the same labeling order. Lane E, EGFP; lane −, untagged Asp3. Values at left are expressed in kilodaltons.
The punctate localization of the Asps is not an artifact of protein overexpression.
A potential technical concern with expressing the accessory Sec components at the levels needed for detection by microscopy is that it may result in artifacts, such as inclusion bodies. To avoid this possibility, we expressed the fusion proteins at minimally detectable levels in all of the above-described experiments. To address the issue more directly, we deliberately induced high levels of Asp expression and examined the localization patterns of these Asps under such conditions. We found that when overexpressed at high levels, EGFPAsp2 and EGFPAsp3 localized predominantly to the cell poles, suggesting inclusion body formation (see Fig. S3, groups 2a and 3a, in the supplemental material). This polar localization pattern was very different in appearance from the punctate formation seen when the proteins were expressed at low levels (Fig. 4A, group 2, and 7A, group 1). These findings thus demonstrate that the punctate phenotype we observed was not an artifact of protein expression levels.
The accessory Sec components partition to different subcellular compartments of S. gordonii.
To corroborate our findings in E. coli, we assessed by cell fractionation the subcellular localization of the accessory Sec components in S. gordonii M99. FLAG-tagged Asp1, Asp2, Asp3, or SecA2 was expressed from the chromosome of S. gordonii, in conjunction with SecY2FLAG and GspB736FLAG. SecY2FLAG served as a membrane control for cell fractionation, whereas GspB736FLAG was used to monitor substrate export. S. gordonii strains expressing the tagged forms of the accessory Sec components were able to secrete GspB736FLAG (see Fig. S2 in the supplemental material), indicating that the proteins had retained their physiological functions. To examine the localization of these components, the clarified CL of bacterial spheroplasts were fractionated into the soluble (cytosolic) and the insoluble (membrane) fractions and analyzed by Western blotting (Fig. 9). As would be expected of a cytosolic protein, the β-subunit of RNA polymerase (RNAPβ) was detected only in the soluble fraction and served as a control for efficient fractionation. SecY2FLAG was found exclusively in the insoluble fraction, consistent with its predicted structure and function as an integral membrane channel. Asp3FLAG and nonexported GspB736FLAG were present only in the soluble fraction, indicating that they are cytosolic proteins. SecA2FLAG, Asp1FLAG, and Asp2FLAG could be found in both the soluble and the insoluble fractions. The partitioning of these proteins into both fractions of disrupted cells indicates that they are not integral membrane proteins but instead may be membrane associated or can redistribute between the membrane and cytosol compartments. These results further validate our observations of protein localization made in E. coli and indicate that the Asps traffic from the cytosol to the cell membrane to form a protein complex mediating substrate transport.
Fig 9.
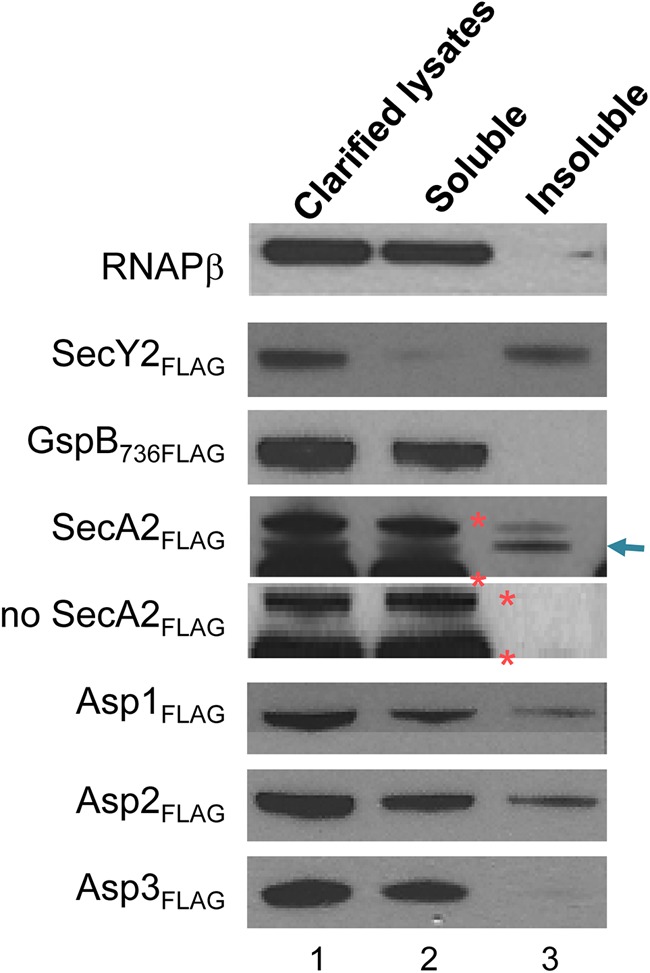
Cellular fractionation of S. gordonii. The clarified lysates (lane 1) of the M99 variants expressing 3×FLAG-tagged accessory components were separated by ultracentrifugation into the soluble (lane 2) and the insoluble (lane 3) fractions. Each fraction contained proteins from an equal volume of cell culture. The presence of the FLAG-tagged accessory Sec component in each fraction was detected by Western blotting with anti-FLAG antibody. The RNAPβ in each fraction was probed with anti-RNAPβ antibody (22, 44). The arrow indicates the position of SecA2FLAG. Two streptococcal proteins of 75 and 100 kDa that cross-reacted with anti-FLAG antibody are marked with orange asterisks.
DISCUSSION
SecA2, SecY2, Asp1, Asp2, and Asp3 are five core proteins in the accessory Sec system of Gram-positive bacteria, and each component is conserved and indispensable for the export of the glycoprotein substrate (1, 2, 17). While SecA2 is likely to associate with SecY2 to form a functioning translocation channel, the roles of Asp1, Asp2, and Asp3 in substrate transport are less clear. Recent studies have shown that the three Asps facilitate a specific interaction between GspB and SecA2 and that this interaction may be required for initiating the translocation of the preprotein through the translocon (23). Because Asp2 and Asp3 can bind GspB (22) and Asp3 also can bind Asp1, Asp2, and SecA2 (24), both Asp2 and Asp3 may also function as chaperones to transport GspB to the translocon through interactions among the Asps and with SecA2. To better understand the roles of the accessory Sec components in GspB transport, we examined the subcellular distribution of these proteins. Because we were unable to detect the proteins expressed at native levels in S. gordonii by fluorescence microscopy, we opted to reconstitute the accessory Sec system in E. coli. As noted above, the system functions comparably in this background and has all of the key export requirements found in the native organism. In conjunction with the fractionation studies performed in S. gordonii, our findings not only reveal the subcellular locations of the accessory Sec components but, further, show the differential trafficking patterns of these proteins between the two cellular compartments.
Using these combined approaches, we found that SecA2 partitioned to both the soluble and insoluble fractions of S. gordonii and thus could be present in the cytosol or at the cell membrane. Microscopy studies in E. coli showed that at the membrane, SecA2 forms multiple foci. These results indicate that SecA2 can be associated with the membrane and that it forms a membrane localization pattern similar to that of canonical SecA of Streptococcus pneumoniae (31), Bacillus subtilis (28), and E. coli (29) but is different from the single “exportal” localization seen in Streptococcus pyogenes (32). Regardless of the number of SecA2 foci, these observations suggest that the accessory Sec system forms specialized translocation centers at the cell membrane for the export of GspB.
We also found that SecA2 localized to the membrane independently of the accessory Sec translocon (SecY2, Asp4, and Asp5) or other proteins in the accessory Sec system (GspB, Asp1, Asp2, Asp3, and the glycosyltransferases). This is consistent with a previous report indicating that Streptococcus parasanguinis SecA2 can associate with the cell membrane without SecY2 (33). Although the mechanism for the punctate distribution of SecA2 is presently unknown, it has been suggested that membrane anionic phospholipids are targets for SecA binding in B. subtilis (28), E. coli (29), and S. pneumoniae (31). Of note, anionic phospholipids in the bacterial cell membrane are also distributed in a similar punctate pattern (34, 35). This distribution might represent lipid domains within the cell membrane to which proteins are anchored. Presumably, binding to the lipids serves as the first step of SecA activation, as membrane-associated SecA exhibits a higher ATPase activity than the soluble form (36). SecA binds SecYEG with an even higher affinity than its association with the lipids (21). This interaction of SecA to SecYEG is likely to be important for substrate export, since it changes the conformation of SecA in an ATP-dependent fashion (37). Whether SecA2 interacts with the cell membrane or the translocon in a similar manner remains to be investigated.
Unlike SecA2, the subcellular distribution of Asp2 is dependent on a number of other accessory Sec components. When EGFPAsp2 was expressed alone, it mainly localized to the cell poles, suggesting that it formed inclusion bodies. When coexpressed with Asp1 and Asp3, EGFPAsp2 became diffusely distributed in the cytosol. In the presence of SecA2, EGFPAsp2 was targeted to the cell membrane, where it formed a punctate arrangement similar to what was observed for SecA2. Further studies with cross-linking and copurification of Asp2 and SecA2 demonstrated that these proteins form a complex. These combined findings thus indicate that Asp2 can partition between the two cellular compartments due to its interaction with SecA2 or Asp1/Asp3 and suggest that membrane-associated SecA2 serves as a receptor for Asp2, whereas Asp1 and Asp3 facilitate Asp2 trafficking to the cytosol. Surprisingly, we did not see a difference between the localization patterns of SecA2 or Asp2 in the presence of GspB (Fig. 2A, group 1, and 4A, group 2) and in its absence (Fig. 2A, group 2, and 4A, group 3). Thus, the localization and trafficking of SecA2 and Asp2 might be dependent on the interactions with anionic phospholipids or other accessory Sec proteins, respectively, rather than the movement of the preprotein.
Consistent with the finding that Asp1 and Asp3 can influence EGFPAsp2 to redistribute to the cytosol, we found that both Asp1 and Asp3 are very soluble and that, by themselves, these proteins partitioned mainly to the cytosol of S. gordonii and E. coli. However, in the presence of the entire accessory Sec system, EGFPAsp3 displayed a punctate distribution along the cell membrane in half of the observed cells. This localization pattern was lost in the absence of either Asp1 or Asp2, indicating that Asp1 and Asp2 can mediate Asp3 targeting to the membrane. Asp1 was found at times at the cell membrane in the presence of both Asp2 and Asp3 and the rest of the accessory Sec components. Thus, Asp2 and Asp3 are also likely to mediate trafficking of Asp1 to the membrane. Collectively, these results suggest that Asp1, Asp2, and Asp3 traffic as a complex from the cytosol to the cell membrane. This is consistent with our previous studies showing that Asp3 can independently bind Asp2 and Asp1 (24).
These results are also consistent with studies in S. parasanguinis, where Gap3 (a homolog of Asp3) has been shown to bind the Asp1 homolog Gap1 (38). This interaction appears to be needed for the stability of Gap3 (39). In contrast, we have found Asp3 to be quite stable in both S. gordonii and E. coli, regardless of whether Asp1 is present. In addition, while Gap1 and Gap3 can individually bind SecA2 independently of Gap2 (40), we found that Asp1 and Asp3 require Asp2 to form a membrane complex with SecA2. We also did not see a direct interaction between Asp1 and SecA2, unlike what was reported for S. parasanguinis. These contrasting findings indicate that, while many of the key features of accessory Sec export are shared by S. gordonii and S. parasanguinis, there may be significant differences in the mechanistic details of transport.
With respect to GspB transport, our combined data suggest a model for the roles of the Asps in GspB transport (Fig. 10). We propose that Asp2 and Asp3 (in complex with Asp1) bind nascent GspB in the cytosol. This binding occurs prior to or concomitant with glycosylation of the preprotein (22). The GspB-Asp complex traffics to the cell membrane and docks at SecA2 through the subsequent interaction of Asp2 and SecA2. At the membrane, the three Asps relay GspB to SecA2 by facilitating the binding of SecA2 to the AST domain of GspB (23). The latter interaction might also be important for initiating preprotein translocation through the translocon. Upon successful preprotein translocation, the Asp complex returns to the cytosol for the next cycle of substrate transport. This recycling probably relies on conformational changes of the Asps that occur after the release of bound GspB. It is possible that upon the transfer of GspB to SecA2, both Asp2 and Asp3 assume different conformations that weaken the binding of the Asps with SecA2 but strengthen the interaction between Asp2 and Asp3, thereby promoting the return of these proteins to the cytosol. The model thus describes a transport mechanism consistent with our fractionation and localization findings and is similar to the way in which a preprotein is chaperoned and targeted by SecB to SecA in the canonical Sec system of E. coli. However, it is still unclear why the transport of a glycosylated substrate by the accessory Sec system should require more than one chaperone. Perhaps a higher degree of complexity within the system is necessary to differentiate, recognize, and transport a single substrate by this specialized pathway. Alternatively, multiple proteins might be needed to coordinate the glycosylation and transport processes of an accessory Sec substrate. Indeed, our latest findings suggest that Asp2 might also play a role in modulating carbohydrate deposition on GspB (41). These multiple functions of the Asps are among the novel aspects of accessory Sec transport and are currently under active investigation.
Fig 10.
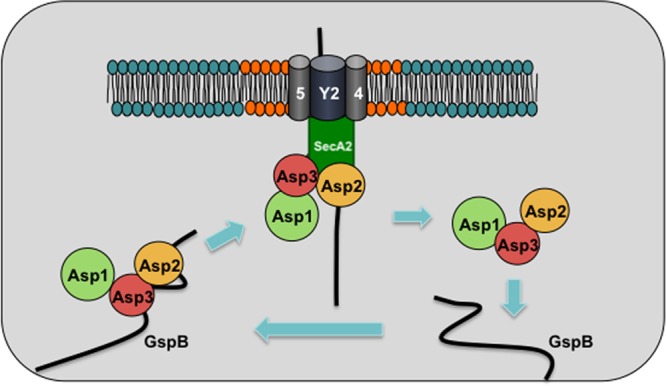
Model for trafficking and localization of the accessory Sec components. SecY2, Asp4, and Asp5 form the membrane translocon, which may be associated with anionic phospholipids (shown in orange). SecA2 may also be attracted to anionic phospholipids at the membrane, and this association is likely to be further strengthened by the interaction between SecA2 and the components of the translocon. Asp2 and Asp3 (in complex with Asp1) bind GspB before it becomes fully glycosylated (22). Through the interaction of Asp2 and SecA2, the Asp complex traffics from the cytosol to the cell membrane, thereby transporting GspB to SecA2 docked at SecY2. At the translocon, the Asps also facilitate a specific interaction between SecA2 and the AST domain of GspB (23). In turn, SecA2 transfers the preprotein to the translocon for export. After the initiation of GspB translocation, the Asps dissociate from SecA2 and return to the cytosol for the next cycle of GspB transport.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Department of Veterans Affairs, the VA Merit Review program, the Northern California Institute for Research and Education, grants 10POST3010018 and 12POST11040013 (to Y.T.Y.) from the American Heart Association, grants R01 AI41513 and R01 AI057433 (to P.M.S.) from the National Institutes of Health, and grant MCB-0343566 (to P.C.Z.) from the National Science Foundation.
Footnotes
Published ahead of print 30 November 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.01742-12.
REFERENCES
- 1. Bensing BA, Sullam PM. 2002. An accessory sec locus of Streptococcus gordonii is required for export of the surface protein GspB and for normal levels of binding to human platelets. Mol. Microbiol. 44:1081–1094 [DOI] [PubMed] [Google Scholar]
- 2. Siboo IR, Chaffin DO, Rubens CE, Sullam PM. 2008. Characterization of the accessory Sec system of Staphylococcus aureus. J. Bacteriol. 190:6188–6196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Obert C, Sublett J, Kaushal D, Hinojosa E, Barton T, Tuomanen EI, Orihuela CJ. 2006. Identification of a candidate Streptococcus pneumoniae core genome and regions of diversity correlated with invasive pneumococcal disease. Infect. Immun. 74:4766–4777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wu H, Fives-Taylor PM. 1999. Identification of dipeptide repeats and a cell wall sorting signal in the fimbriae-associated adhesin, Fap1, of Streptococcus parasanguis. Mol. Microbiol. 34:1070–1081 [DOI] [PubMed] [Google Scholar]
- 5. Mistou MY, Dramsi S, Brega S, Poyart C, Trieu-Cuot P. 2009. Molecular dissection of the secA2 locus of group B Streptococcus reveals that glycosylation of the Srr1 LPXTG protein is required for full virulence. J. Bacteriol. 191:4195–4206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Siboo IR, Chambers HF, Sullam PM. 2005. Role of SraP, a serine-rich surface protein of Staphylococcus aureus, in binding to human platelets. Infect. Immun. 73:2273–2280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Xiong YQ, Bensing BA, Bayer AS, Chambers HF, Sullam PM. 2008. Role of the serine-rich surface glycoprotein GspB of Streptococcus gordonii in the pathogenesis of infective endocarditis. Microb. Pathog. 45:297–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. van Sorge NM, Quach D, Gurney MA, Sullam PM, Nizet V, Doran KS. 2009. The group B streptococcal serine-rich repeat 1 glycoprotein mediates penetration of the blood-brain barrier. J. Infect. Dis. 199:1479–1487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Seo HS, Mu R, Kim BJ, Doran KS, Sullam PM. 2012. Binding of glycoprotein Srr1 of Streptococcus agalactiae to fibrinogen promotes attachment to brain endothelium and the development of meningitis. PLoS Pathog. 8:e1002947 doi:10.1371/journal.ppat.1002947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Seifert KN, Adderson EE, Whiting AA, Bohnsack JF, Crowley PJ, Brady LJ. 2006. A unique serine-rich repeat protein (Srr-2) and novel surface antigen (epsilon) associated with a virulent lineage of serotype III Streptococcus agalactiae. Microbiology 152:1029–1040 [DOI] [PubMed] [Google Scholar]
- 11. Pyburn TM, Bensing BA, Xiong YQ, Melancon BJ, Tomasiak TM, Ward NJ, Yankovskaya V, Oliver KM, Cecchini G, Sulikowski GA, Tyska MJ, Sullam PM, Iverson TM. 2011. A structural model for binding of the serine-rich repeat adhesin GspB to host carbohydrate receptors. PLoS Pathog. 7:e1002112 doi:10.1371/journal.ppat.1002112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bensing BA, Lopez JA, Sullam PM. 2004. The Streptococcus gordonii surface proteins GspB and Hsa mediate binding to sialylated carbohydrate epitopes on the platelet membrane glycoprotein Ibalpha. Infect. Immun. 72:6528–6537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sullam PM, Frank U, Yeaman MR, Tauber MG, Bayer AS, Chambers HF. 1993. Effect of thrombocytopenia on the early course of streptococcal endocarditis. J. Infect. Dis. 168:910–914 [DOI] [PubMed] [Google Scholar]
- 14. Rigel NW, Braunstein M. 2008. A new twist on an old pathway—accessory Sec [corrected] systems. Mol. Microbiol. 69:291–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhou M, Wu H. 2009. Glycosylation and biogenesis of a family of serine-rich bacterial adhesins. Microbiology 155:317–327 [DOI] [PubMed] [Google Scholar]
- 16. Takamatsu D, Bensing BA, Sullam PM. 2004. Four proteins encoded in the gspB-secY2A2 operon of Streptococcus gordonii mediate the intracellular glycosylation of the platelet-binding protein GspB. J. Bacteriol. 186:7100–7111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Takamatsu D, Bensing BA, Sullam PM. 2004. Genes in the accessory sec locus of Streptococcus gordonii have three functionally distinct effects on the expression of the platelet-binding protein GspB. Mol. Microbiol. 52:189–203 [DOI] [PubMed] [Google Scholar]
- 18. Takamatsu D, Bensing BA, Sullam PM. 2005. Two additional components of the accessory sec system mediating export of the Streptococcus gordonii platelet-binding protein GspB. J. Bacteriol. 187:3878–3883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bensing BA, Sullam PM. 2009. Characterization of Streptococcus gordonii SecA2 as a paralogue of SecA. J. Bacteriol. 191:3482–3491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bensing BA, Sullam PM. 2010. Transport of preproteins by the accessory Sec system requires a specific domain adjacent to the signal peptide. J. Bacteriol. 192:4223–4232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hartl FU, Lecker S, Schiebel E, Hendrick JP, Wickner W. 1990. The binding cascade of SecB to SecA to SecY/E mediates preprotein targeting to the E. coli plasma membrane. Cell 63:269–279 [DOI] [PubMed] [Google Scholar]
- 22. Yen YT, Seepersaud R, Bensing BA, Sullam PM. 2011. Asp2 and Asp3 interact directly with GspB, the export substrate of the Streptococcus gordonii accessory Sec system. J. Bacteriol. 193:3165–3174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bensing BA, Yen YT, Seepersaud R, Sullam PM. 2012. A specific interaction between SecA2 and a region of the preprotein adjacent to the signal peptide occurs during transport via the accessory Sec system. J. Biol. Chem. 287:24438–24447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Seepersaud R, Bensing BA, Yen YT, Sullam PM. 2010. Asp3 mediates multiple protein-protein interactions within the accessory Sec system of Streptococcus gordonii. Mol. Microbiol. 78:490–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Macrina FL, Evans RP, Tobian JA, Hartley DL, Clewell DB, Jones KR. 1983. Novel shuttle plasmid vehicles for Escherichia-Streptococcus transgeneric cloning. Gene 25:145–150 [DOI] [PubMed] [Google Scholar]
- 26. Li Y, Chen Y, Huang X, Zhou M, Wu R, Dong S, Pritchard DG, Fives-Taylor P, Wu H. 2008. A conserved domain of previously unknown function in Gap1 mediates protein-protein interaction and is required for biogenesis of a serine-rich streptococcal adhesin. Mol. Microbiol. 70:1094–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bryan EM, Bae T, Kleerebezem M, Dunny GM. 2000. Improved vectors for nisin-controlled expression in gram-positive bacteria. Plasmid 44:183–190 [DOI] [PubMed] [Google Scholar]
- 28. Campo N, Tjalsma H, Buist G, Stepniak D, Meijer M, Veenhuis M, Westermann M, Muller JP, Bron S, Kok J, Kuipers OP, Jongbloed JD. 2004. Subcellular sites for bacterial protein export. Mol. Microbiol. 53:1583–1599 [DOI] [PubMed] [Google Scholar]
- 29. Shiomi D, Yoshimoto M, Homma M, Kawagishi I. 2006. Helical distribution of the bacterial chemoreceptor via colocalization with the Sec protein translocation machinery. Mol. Microbiol. 60:894–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Brandon LD, Goehring N, Janakiraman A, Yan AW, Wu T, Beckwith J, Goldberg MB. 2003. IcsA, a polarly localized autotransporter with an atypical signal peptide, uses the Sec apparatus for secretion, although the Sec apparatus is circumferentially distributed. Mol. Microbiol. 50:45–60 [DOI] [PubMed] [Google Scholar]
- 31. Tsui HC, Keen SK, Sham LT, Wayne KJ, Winkler ME. 2011. Dynamic distribution of the SecA and SecY translocase subunits and septal localization of the HtrA surface chaperone/protease during Streptococcus pneumoniae D39 cell division. MBio 2 pii: e00202-11 doi:10.1128/mBio.00202-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rosch JW, Caparon MG. 2005. The ExPortal: an organelle dedicated to the biogenesis of secreted proteins in Streptococcus pyogenes. Mol. Microbiol. 58:959–968 [DOI] [PubMed] [Google Scholar]
- 33. Chen Q, Wu H, Kumar R, Peng Z, Fives-Taylor PM. 2006. SecA2 is distinct from SecA in immunogenic specificity, subcellular distribution and requirement for membrane anchoring in Streptococcus parasanguis. FEMS Microbiol. Lett. 264:174–181 [DOI] [PubMed] [Google Scholar]
- 34. Mileykovskaya E, Dowhan W. 2000. Visualization of phospholipid domains in Escherichia coli by using the cardiolipin-specific fluorescent dye 10-N-nonyl acridine orange. J. Bacteriol. 182:1172–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kawai F, Shoda M, Harashima R, Sadaie Y, Hara H, Matsumoto K. 2004. Cardiolipin domains in Bacillus subtilis Marburg membranes. J. Bacteriol. 186:1475–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lill R, Dowhan W, Wickner W. 1990. The ATPase activity of SecA is regulated by acidic phospholipids, SecY, and the leader and mature domains of precursor proteins. Cell 60:271–280 [DOI] [PubMed] [Google Scholar]
- 37. Natale P, den Blaauwen T, van der Does C, Driessen AJ. 2005. Conformational state of the SecYEG-bound SecA probed by single tryptophan fluorescence spectroscopy. Biochemistry 44:6424–6432 [DOI] [PubMed] [Google Scholar]
- 38. Zhou M, Peng Z, Fives-Taylor P, Wu H. 2008. A conserved C-terminal 13-amino-acid motif of Gap1 is required for Gap1 function and necessary for the biogenesis of a serine-rich glycoprotein of Streptococcus parasanguinis. Infect. Immun. 76:5624–5631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhou M, Zhu F, Li Y, Zhang H, Wu H. 2012. Gap1 functions as a molecular chaperone to stabilize its interactive partner Gap3 during biogenesis of serine-rich repeat bacterial adhesin. Mol. Microbiol. 83:866–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhou M, Zhang H, Zhu F, Wu H. 2011. Canonical SecA associates with an accessory secretory protein complex involved in biogenesis of a streptococcal serine-rich repeat glycoprotein. J. Bacteriol. 193:6560–6566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Seepersaud R, Bensing BA, Yen YT, Sullam PM. 2012. The accessory Sec protein Asp2 modulates GlcNAc deposition onto the serine-rich repeat glycoprotein GspB. J. Bacteriol. 194:5564–5575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sullam PM, Valone FH, Mills J. 1987. Mechanisms of platelet aggregation by viridans group streptococci. Infect. Immun. 55:1743–1750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bensing BA, Takamatsu D, Sullam PM. 2005. Determinants of the streptococcal surface glycoprotein GspB that facilitate export by the accessory Sec system. Mol. Microbiol. 58:1468–1481 [DOI] [PubMed] [Google Scholar]
- 44. Seepersaud R, Needham RH, Kim CS, Jones AL. 2006. Abundance of the delta subunit of RNA polymerase is linked to the virulence of Streptococcus agalactiae. J. Bacteriol. 188:2096–2105 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.