Abstract
The classic tumor clonal evolution theory postulates that cancers change over time to produce unique molecular subclones within a parent neoplasm, presumably including regional differences in gene expression. More recently, however, this notion has been challenged by studies showing that tumors maintain a relatively stable transcript profile. To examine these competing hypotheses, we microdissected discrete subregions containing approximately 3000 to 8000 cells (500 to 1500 μm in diameter) from ex vivo esophageal squamous cell carcinoma (ESCC) specimens and analyzed transcriptomes throughout three-dimensional tumor space. Overall mRNA profiles were highly similar in all 59 intratumor comparisons, in distinct contrast to the markedly different global expression patterns observed in other dissected cell populations. For example, normal esophageal basal cells contained 1918 and 624 differentially expressed genes at a greater than twofold level (95% confidence level of <5% false positives), compared with normal differentiated esophageal cells and ESCC, respectively. In contrast, intratumor regions had only zero to four gene changes at a greater than twofold level, with most tumor comparisons showing none. The present data indicate that, when analyzed using a standard array-based method at this level of histological resolution, ESCC contains little regional mRNA heterogeneity.
Esophageal squamous cell carcinoma (ESCC) is the predominant histological subtype of esophageal cancer, ranking as the eighth most common neoplasm in the world, with the sixth highest mortality.1,2 Detection at a late stage and a paucity of effective chemotherapeutic and other clinical modalities make ESCC difficult to treat, and overall survival is poor. Today, only 19% of patients live for 5 years or longer after diagnosis.3 Better understanding of the molecular etiology of this type of cancer is needed, both to identify detection markers for early diagnosis and to identify new therapeutic targets for advanced disease. Moreover, given that ESCC shares several histopathological and molecular features with other SCCs (eg, head and neck, and lung), greater understanding of ESCC may improve our overall understanding of SCCs, the class that results in the largest number of cancer deaths worldwide.4–6
ESCC contains extensive genomic alterations.7–10 At present, however, little is known about the amount of transcriptomic heterogeneity that exists within this tumor type. One theory of cancer progression postulates that molecular heterogeneity is a dominant mechanism of tumor development, with unique subclones differentiating from the parent neoplasm in three-dimensional space over time, producing a heterogeneous collection of spatially distinct tumor subfields that differ in molecular profile.11 In this model, clonal evolution based on molecular status is an important process that underlies the progression of tumors into aggressive, faster-growing forms, ultimately conferring metastatic capability on certain subclones within the tumor that then disseminate via the bloodstream or lymphatic system.
An alternative view of mRNA profiles in tumor progression is that tumors arise de novo with their transcriptome more or less intact, including the key changes that mediate metastasis.12–17 In this model, tumor cells share essentially the same transcript profile throughout the three-dimensional space of a neoplasm, and the rise of heterogeneous regional subclones with altered transcript profiles is not a fundamental feature of cancer progression. To date, studies examining gene expression patterns in cancer have tended to support the latter view, that there is only a limited amount of intratumor transcript profile variation.12–17
To address the issue of mRNA heterogeneity in ESCC, we measured global gene expression levels in microdissected tumor cell populations from discrete three-dimensional microanatomical regions. Two separate analyses were performed: the central region of tumors versus the peripheral area, and a three-dimensional comparison of multiple subregions throughout tumors.
Materials and Methods
Tissue Specimens
All cases and samples were obtained from subjects residing in the Taihang Mountain region of north-central China, and the study was approved by the Institutional Review Boards of the collaborating institutions [Shanxi Cancer Hospital and Institute, Taiyuan, Shanxi Province, China (Single Project Assurance no. S-12118-01) and the National Cancer Institute, NIH, Bethesda, Maryland]. After informed consent was obtained, patients were interviewed to obtain information on demographics, cancer risk factors (smoking, alcohol drinking, and detailed family history of cancer), and clinical information (Supplemental Table S1). None of the patients had prior therapy, and none of the ESCC cases were from patients with a family history of the disease. Using accepted inclusion criteria,18 nine cases having sufficient tumor and matched normal epithelium were evaluated and selected by a pathologist (J.R.-C). Resected specimens were fresh frozen, optimal cutting temperature media embedded and stored in liquid nitrogen according to standard practices19 until assays could be performed.
Tissue Processing
Immediately before use, the paired normal and tumor samples were cut into sections (8 μm thick) using a cryostat (Leica Microsystems, Wetzlar, Germany), placed onto glass slides, and stored for <2 weeks at −80°C. Before dissection, each section was individually removed from storage and immediately stained and dehydrated using an H&E protocol designed for microdissection.20–22
Laser Capture Microdissection
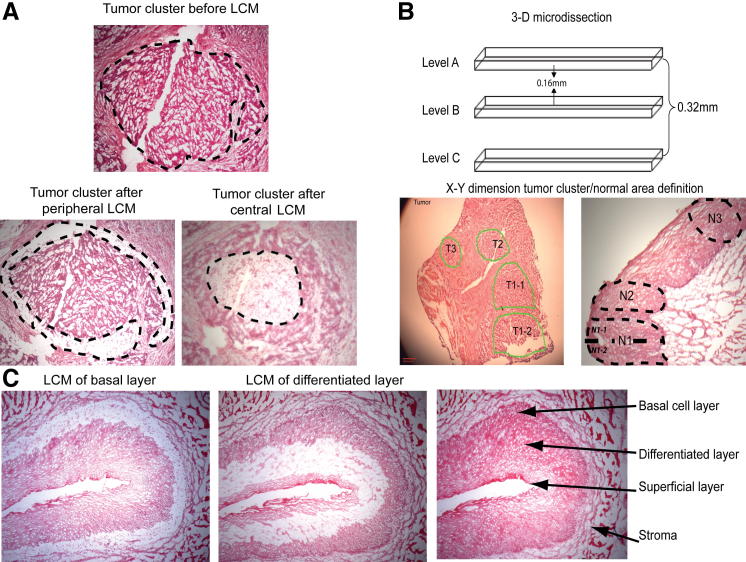
The peripheral and central tumor regions from each of the nine cases were morphologically selected and dissected by laser capture microdissection with a PixCell IIe system (Arcturus Engineering, Mountain View, CA) using 4000 medium-sized laser shots (laser spot size, 15 μm; approximately two cells per laser pulse), procuring approximately 8000 cells in total (Supplemental Table S2). Because molecular comparison between the regions was a critical aspect of the study, overlapping dissection of the two areas was avoided (Figure 1A). In addition, three-dimensional microdissections were performed on tumor regions from five of the same cases (Figure 1B). Approximately 3000 cells were procured from each dissection using large-sized laser shots (laser spot size, 30 μm; approximately three cells per laser pulse) and two to four serial recut slides per case (Supplemental Table S2).
Figure 1.
Microdissection design. A: H&E staining showing an ESCC focus before and after dissection of the tumor periphery and center. B: Three-dimensional microdissection design and definition of the tumor cluster and normal area. Dissected areas are outlined in green. C: Histology of normal esophageal epithelium compartment before and after microdissection.
RNA Isolation and Assessment
The time from slide removal from the freezer to completion of laser capture microdissection did not exceed 30 minutes. After laser capture microdissection, RNA was isolated using a PicoPure RNA isolation kit (Arcturus Engineering) and digested with DNase. The quantity and quality of the RNA was measured as described previously,20,22 using a Bioanalyzer 2100 (Agilent Technologies, Palo Alto, CA) and a NanoDrop ND-100 spectrophotometer (Thermo Scientific, Wilmington, DE), respectively.
RNA Amplification and Microarray Hybridization
Total RNA was used as the template for amplification, because of bias reduction compared with using mRNA alone.23 To ensure that all samples contained a similar overall representation of transcriptome, 50 ng of total RNA was used for each sample. Two rounds of linear amplification were performed by combining reagents supplied in the Ambion MessageAmp II biotin-enhanced, single-round a(ntisense)RNA amplification kit (AM1791; Life Technologies, Foster City, CA) and the Ambion MessageAmp II aRNA amplification kit (AM1751; Life Technologies), resulting in biotin-labeled antisense complimentary (c)RNA.24 For each sample 15 μg of biotin-labeled cRNA sample was fragmented and processed for hybridization to a GeneChip human genome U133A 2.0 array (Affymetrix, Santa Clara, CA) according to the expression kit user manual. Arrays were washed and stained using the Midi euk2v3 protocol (version 4) on a GeneChip Fluidics Station 450 (Affymetrix). The fluorescent intensity emitted by the labeled target was measured using a GeneChip 3000 7G scanner (Affymetrix). The microarray gene expression data discussed in this publication have been deposited in the NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo; accession number GSE33426).
For Affymetrix GeneChip studies, a single-round linear amplification is the gold standard for processing labeled aRNA.25 In earlier experiments, we compared various quality metrics after one round of amplification (using 1 μg total RNA) versus lower RNA concentrations using two and even three rounds of amplification. These data suggest that integrity and the detection percentage of genes were similar to one round of amplification, down to concentrations as low as 1 ng (Supplemental Table S3). The overall signal intensity (ie, scaling factors) was closely related, and the gene-present percentage varied by only approximately 8% to 9%. Moreover, scatter-plot analysis of the samples showed significant correlation in probe-set signaling (Supplemental Figure S1).
Data Analysis and Statistics
Microarray Data Quality Control, Preprocessing, and Gene Filtering
Quality assessment of each microarray was done using probe-level, model-based quality statistics: developed by Richard Simon and the BRB-ArrayTools Development Team; (NUSE) and relative log expression (RLE).26,27 Arrays with their respective median NUSE and RLE values exceeding the upper control limits were considered as poor quality and excluded from analysis. Robust multiarray analysis with quantile normalization was completed for the good-quality arrays, using the Bioconductor suite of array analysis tools (http://www.bioconductor.org) under R version 2.8.0 (http://www.r-project.org). Different probe-set filtering criteria were applied to the two analyses. For the comparison of gene expression between different cell types (central tumor, peripheral tumor, normal basal, and normal differentiated cells), probe sets showing minimal variation across the arrays were excluded from the analysis. Probe sets were selected if their expression differed by ≥1.5-fold from the median in ≥20% of the arrays. Overall, 10,725 probe sets were retained and used for the analysis. Because the goal was to assess the homogeneity in expression among the three-dimensional samples, no filtering criterion was applied for the three-dimensional comparison of tumor subregions.
Class Comparison
For the analysis of gene expression from different cell types, because a large number of differentially expressed genes between tumor and normal cells were expected, a stringent multiple testing procedure was used to identify differentially expressed genes while controlling for the proportion of false positives. Specifically, a multivariate permutation test28,29 was used to provide a false positive proportion of <5% with 95% confidence. The test statistics used were paired t-statistics for each probe set. Although paired t-statistics were used, the multivariate permutation test is nonparametric and does not require the assumption of normal distributions for gene expression measurements. For the analysis of comparing gene expression among tumor three-dimensional samples, because a much smaller number of differentially expressed genes were expected, and to allow for higher false positive rate, probe sets with expression difference at P < 0.001 based on the paired t-test were identified. The multivariate permutation test was performed using BRB-ArrayTools version 3.8.0 software (http://linus.nci.nih.gov/BRB-ArrayTools.html).
Multidimensional Scaling
Multidimensional scaling (MDS) was used to evaluate gene expression profiles of specimens in three dimensions. The data points were generated based on the principal coordinate analysis, using one minus the correlation coefficient (1 − r) as the distance metric.
Pathway Analysis
Genes identified by class comparison as described above were used for network and gene ontology analyses. Data were analyzed through the use of IPA (Ingenuity Systems, Redwood City, CA). Gene accession numbers were imported into the Ingenuity Pathways Analysis system, and networks were algorithmically generated based on their connectivity. Biochemical pathway analysis was performed using the ratio of the number of molecules in a given pathway to the total number of molecules that make up that pathway, to generate networks in which the differentially regulated genes are related to known associations between genes or proteins but are independent of established canonical pathways.
Intratumor and Intertumor Correlation Coefficients Analysis
Intratumor versus intertumor variability for microarray gene expression data can be assessed in two ways: one assesses the Pearson correlation coefficient across all genes on different arrays and the other assesses the intratumor heterogeneity on a gene-by-gene basis by one minus the intraclass correlation coefficient (1 − ICC). The ICC is calculated by fitting a variance component model for each gene. Two variance components were estimated: variance between cases (ie, intertumor variance, denoted by B) and variance within a case (ie, intratumor variance, denoted by W). The ICC is defined as B/(W + B), and intratumor heterogeneity is defined as 1 − ICC = W/(W + B). For the variance component analysis, gene expression data of tumor specimens in both experiments were filtered to remove genes that were invariant (<20% of the arrays with expression differing from median by >1.5-fold).
Using these analytic and statistical methods, we have previously demonstrated reproducible and valid gene expression array and quantitative RT-PCR measurements with microdissected specimens.20,22,24
Results
Technical Parameters
A total of 74 microdissected samples from nine patients were generated: 18 samples were procured from the tumor center (TC) or tumor periphery (TP) of a neoplastic focus (Figure 1A), 44 came from a set of dissections performed as three-dimensional tumor maps (Figure 1B), and 12 samples were from a control three-dimensional map of normal squamous epithelium (Figure 1B). The recovered RNA had a preamplification mean RNA integrity number of 6.1 (range, 3.6 to 8.6) (Supplemental Table S2). One of the 74 samples did not generate enough cRNA after two rounds of linear amplification, and two samples failed NUSE and RLE array quality testing (Supplemental Figure S2); however, the remaining 71 samples were of sufficient quality for analysis.
Tumor Heterogeneity
Central versus Peripheral Regions
We first analyzed the expression profiles of cells located in the TC versus those at the TP (Figure 1A and Supplemental Table S4). On average, the microdissected TC was 660 μm in diameter, whereas the TP was two to four cell layers thick (40 to 80 μm) and was in direct contact with the surrounding stroma.
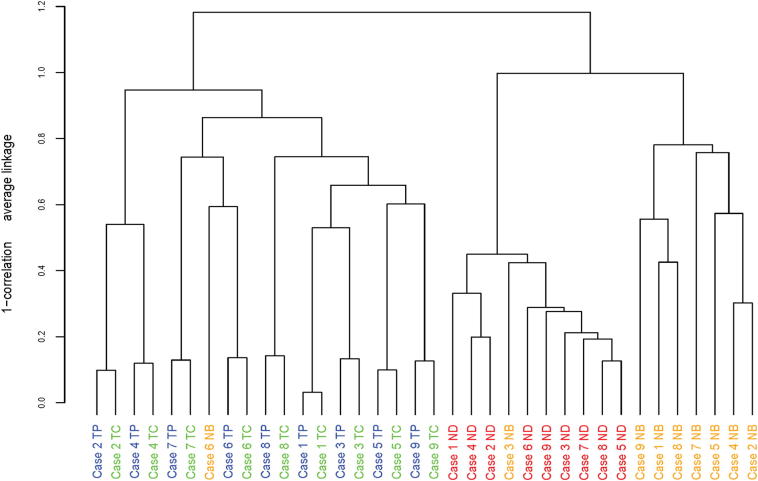
Striking similarity in global transcript levels was observed between the two tumor areas (Supplemental Table S4); thrombospondin 2 (THBS2) was the only differentially expressed gene, with a 2.4-fold increase in the TP. As a metric for determining relative transcriptome heterogeneity, we compared and contrasted the present TC and TP data with those from normal squamous epithelium and cancer from a previous study.30 That analysis showed that expression profiles of microdissected normal basal (NB) squamous epithelium, adjacent normal differentiated (ND) cells, and ESCC cells from the same patient were significantly different from each other, with a large number of differentially expressed genes. For example, comparison of dissected NB cells versus adjacent ND cells (Figure 1C) revealed 4994 gene alterations at false positive ≤ 5% with 95% confidence level, including 1918 with fold change ≥ 2 and 202 with fold change ≥ 4 (Supplemental Table S4). Similarly, comparison of both TP versus NB cells and TC versus ND cells showed large numbers of differentially expressed genes, in sharp contrast to the single differentially expressed gene in the TP versus TC analysis (Supplemental Table S4).
The data from the dissected cell populations are illustrated in both hierarchical (Figure 2) and graphical (Supplemental Figure S3) format. The dendrogram (Figure 2) illustrates the separation of the three different cell types (NB, ND, and ESCC) into distinct groups after unsupervised clustering. In contrast, the TC and TP samples were closely aligned, because of the similarity of their transcript profiles. This dendrogram also shows that the intrapatient TC and TP profiles were more closely aligned than the patient–patient comparisons, which were less than the distance between normal and tumor cells. This same pattern is seen with MDS (Supplemental Figure S3), in which the two sets of intratumor dissections are admixed but are distinct from the NB and ND cells.
Figure 2.
Differential gene expression comparisons of TP/TC (blue/green) versus NB/ND (orange/red) in nine cases by hierarchical cluster analysis. Each bar presents one case.
Three-Dimensional Mapping
We next analyzed tumor transcriptome heterogeneity by microdissecting multiple regions throughout the three-dimensional anatomical field of five neoplasms. In each tumor, four regions were dissected in the x–y plane and, depending on the thickness of the tissue block, in two or three levels in the z dimension (Figure 1B). For each x–y dissection, the tumor areas were selected according to the following criteria: T1-1 and T1-2 were located within a single tumor focus and were dissected as two separate but closely associated areas; T2 was an independent tumor region, separated from T1 by stroma; T3 was an independent cluster, located as far from the T-1 and T-2 regions as possible. The same three-dimensional microdissection procedure was performed on a specimen of normal esophageal epithelium from one case in the study, which served as a normal tissue comparator.
The number of differentially expressed genes varied among the several tumor subregions (Table 1). As with the TC versus TP measurements, three-dimensional transcriptome mapping identified relatively little heterogeneity in ESCC. At the P < 0.001 level, the number of genes was slightly under 20 per comparison (range, 2 to 54; false discovery proportion, 24.7% to 100%), whereas at the twofold level or greater there were only a few differentially expressed genes, with most comparisons showing none.
Table 1.
Class Comparisons of Three-Dimensional Microdissected Intratumor Subregions
| Comparison | Comparison definition∗ | Differentially expressed genes (No.) |
|
|---|---|---|---|
| P < 0.001 | Fold change ≥2 | ||
| X-Y dimension | |||
| A1 | T1-1 vs T1-2 at level A | 24 | 1 |
| A2 | (T1-1 + T1-2) vs T2 at level A | 10 | 0 |
| A3 | (T1-1 + T1-2) vs T3 at level A | 5 | 0 |
| A4 | T2 vs T3 at level A | 3 | 0 |
| C1 | T1-1 vs T1-2 at level C | 2 | 0 |
| C2 | (T1-1 + T1-2) vs T2 at level C | 30 | 0 |
| C3 | (T1-1 + T1-2) vs T3 at level C | 33 | 0 |
| C4 | T2 vs T3 at level C | 21 | 0 |
| AC1 | T1-1 vs T1-2 across level A and B | 7 | 0 |
| AC2 | (T1-1 + T1-2) vs T2 across level A and B | 19 | 0 |
| AC3 | (T1-1 + T1-2) vs T3 across level A and B | 24 | 0 |
| AC4 | T2 vs T3 across level A and B | 10 | 0 |
| Z dimension | |||
| V1.1 | T1-1 at level A vs T1-1 at level C | 32 | 2 |
| V1.2 | T1-2 at level A vs T1-2 at level C | 10 | 0 |
| V2 | T2 at level A vs T2 at level C | 54 | 4 |
| V3 | T3 at level A vs T3 at level C | 27 | 0 |
| Overall difference between level A and level C | 93 | 0 | |
Tumor subregions (T) and levels (A–C) are as shown in Figure 1B.
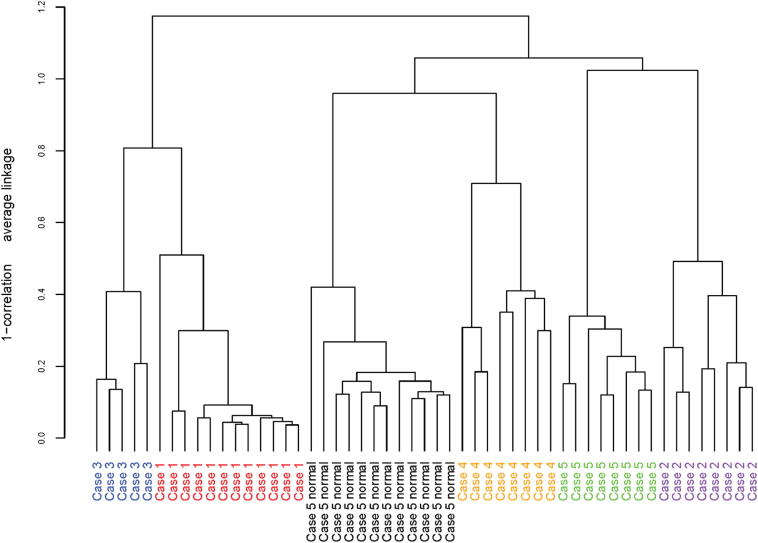
The expression profiles are illustrated using both hierarchical clustering (Figure 3) and MDS (Supplemental Figure S4). Again, as with the TC versus TP comparison, the case-to-case heterogeneity was greater than the intratumor heterogeneity, which showed little difference across the three-dimensional field. The normal case also exhibited minimal heterogeneity among the three-dimensional dissections of esophageal squamous epithelium (Supplemental Figure S4).
Figure 3.
Differential gene expression comparisons of three-dimensional-microdissected ESCC in five cases by hierarchical cluster analysis. Each bar represents one dissection (including normal epithelium for case 5). Microdissection was performed as shown in Figure 1B.
We next compared the three-dimensional ESCC data with microdissected normal and tumor prostate cells from a previous study,24 as a technical assessment of relative transcriptome heterogeneity. The 1 − r average linkage distance of prostate cells versus esophageal cells was approximately an order of magnitude greater than that of the largest inter-ESCC comparison (Supplemental Figure S5), indicating that the methodological approach used in the study was capable of detecting large differences present in transcriptome profiles.
Overall, the expression data from both the TC versus TP analysis and the three-dimensional mapping measurements showed that a relatively stable transcriptome is present in ESCC. We found little evidence of a significant departure from this profile with respect to the number of expression differences in any tumor subregion, in contrast to the relatively large number of differentially expressed mRNAs that characterized other dissected cell populations.
Intratumor and Intertumor Correlation Coefficients
The intratumor versus intertumor variability of microarray gene expression data were assessed in two different ways. We first determined the Pearson correlation coefficient across all genes on the arrays and found a higher correlation between intratumor specimens than intertumor specimens, implying low intratumor heterogeneity. The intratumor heterogeneity was less than the intertumor heterogeneity (Figures 2 and 3). All of the intratumor specimens clustered together, and the distance as measured by 1 − r was shorter between the intratumor specimens than between the intertumor specimens.
We then assessed intratumor heterogeneity on a gene-by-gene basis, using 1 − ICC. For one set of experiments (nine TP and nine TC specimens, taken from nine cases), 10,301 probe sets were included for the variance component analysis. For the second set of experiments (41 three-dimensional specimens taken from five cases), 9834 probe sets were included. For both analyses, the histogram of intratumor heterogeneity was plotted (Supplemental Figure S6). Intratumor heterogeneity was less than intertumor heterogeneity in 97% of the genes, with a median intratumor heterogeneity of 0.13 (ICC = 0.87). Thus, for almost all genes, there was a greater variation between tumors (cases) than within a tumor. In the second analysis, intratumor heterogeneity was less than intertumor heterogeneity in approximately two thirds of the genes (67.2%), with a median intratumor heterogeneity of 0.37 (ICC = 0.63). Overall, both analyses demonstrated that intratumor heterogeneity was less than intertumor heterogeneity.
Individual Genes and Pathways
To determine whether there were small but biologically consequential expression changes among the intratumor regions, we examined expression of individual genes and the status of specific biochemical pathways. Thrombospondin 2 (THBS2) was the only gene significantly differentially expressed in TP versus TC comparison. Transcripts that were altered in one or more of the three-dimensional mapping comparisons are listed in Table 2. Seven genes (CEP55, CDK5R1, RGS5, GGH, DICER1, SET, and MAP4K3) showed fold change ≥ 2 in 1 of the 16 subregion comparisons, and six genes (YYI, RYBP, LAX1, PGGT1B, CDC42EP4, and LARS2) were differentially expressed (fold change ≥ 2) in 2 of the 16 subregions (Table 2). However, no one individual mRNA was altered in more than two comparisons. Thus, the individual gene-level measurements mirrored the transcriptome overall, in that little intratumor heterogeneity was observed.
Table 2.
Individual Genes Differentially Expressed within the Three-Dimensional Intratumor Comparisons
| Gene symbol | UniGene ID | Probeset | Chromosomal location | Gene name | Comparison∗ |
|---|---|---|---|---|---|
| Fold change ≥ 2 in 1 of the 16 subregion comparisons | |||||
| CEP55 | Hs.14559 | 218542_at | 10q23.33 | Centrosomal protein 55kDa | A1 |
| CDK5R1 | Hs.93597 | 204995_at | 17q11.2 | Cyclin-dependent kinase 5, regulatory subunit 1 (p35) | V1.1 |
| RGS5 | Hs.24950 | 209071_s_at | 1q23.1 | Regulator of G-protein signaling 5 | V1.1 |
| GGH | Hs.78619 | 203560_at | 8q12.3 | Gamma-glutamyl hydrolase (conjugase, folylpolygammaglutamyl hydrolase) | V2 |
| DICER1 | Hs.87889 | 206061_s_at | 14q32.13 | Dicer 1, ribonuclease type III | V2 |
| SET | Hs.436687 | 213047_x_at | 9q34 | SET nuclear oncogene | V2 |
| MAP4K3 | Hs.655750 | 218311_at | 2p22.1 | Mitogen-activated protein kinase kinase kinase kinase 3 | V2 |
| Fold change ≥ 2 in 2 of the 16 subregion comparisons | |||||
| YY1 | Hs.388927 | 200047_s_at | 14q | YY1 transcription factor | A2, AC2 |
| RYBP | Hs.7910 | 201844_s_at | 3p13 | RING1 and YY1 binding protein | AC3, V2 |
| LAX1 | Hs.272794 | 207734_at | 1q32.1 | Lymphocyte transmembrane adaptor 1 | A2, AC2 |
| PGGT1B | Hs.254006 | 216288_at | 5q22.3 | Protein geranylgeranyltransferase type I, beta subunit | AC2, V1.2 |
| CDC42EP4 | Hs.3903 | 218063_s_at | 17q24∼q25 | CDC42 effector protein (Rho GTPase binding) 4 | AC3, C4 |
| LARS2 | Hs.526975 | 34764_at | 3p21.3 | Leucyl-tRNA synthetase 2, mitochondrial | V1.2, V3 |
Comparisons are as defined in Table 1.
The biochemical pathway analysis also showed minimal differences among the three-dimensional intratumor comparisons. Only two pathways, the mitochondrial dysfunction pathway and the valine, leucine, and isoleucine biosynthesis pathway, showed an alteration in any tumor subregion, and they were identified in only 1 or 2 of the 16 comparisons (Table 3). Moreover, the top networks from the 16 comparisons typically had a score of 2 (Supplemental Table S5). In contrast, the scores of the top networks in other dissected cell populations, including normal versus tumor, and NB versus ND, ranged from 10 to 28 (data not shown), much larger than the intratumor comparisons.
Table 3.
Pathway Analysis of Three-Dimensional Intratumor Comparisons
| Top canonical pathways | Comparison∗ | P value |
|---|---|---|
| Mitochondrial dysfunction | A1, C3 | 0.0151, 0.0337 |
| RAN signaling | A1 | 0.0285 |
| Wnt/β-catenin signaling | A1 | 0.0302 |
| cAMP-mediated signaling | A1 | 0.0463 |
| Cell cycle control of chromosomal replication | A2 | 0.0229 |
| Semaphorin signaling in neurons | A2 | 0.0395 |
| Glioma invasiveness signaling | A2 | 0.0445 |
| DNA methylation and transcriptional repression signaling | A3 | 0.00683 |
| Glycine, serine, and threonine metabolism | A3 | 0.0223 |
| Tyrosine metabolism | A3 | 0.0230 |
| Melatonin signaling | A3 | 0.0245 |
| Phospholipid degradation | A3 | 0.0249 |
| FXR/RXR activation | C2 | 0.0117 |
| Taurine and hypotaurine metabolism | C2 | 0.0187 |
| DNA double-strand break repair by homologous recombination | C2 | 0.0269 |
| Pyrimidine metabolism | C2 | 0.0269 |
| BMP signaling pathway | C3 | 0.0159 |
| TGF-β signaling | C3 | 0.0171 |
| Oxidative phosphorylation | C3 | 0.0354 |
| AMPK signaling | C3 | 0.0455 |
| T helper cell differentiation | C4 | 0.00477 |
| IL-10 signaling | C4 | 0.00556 |
| RhoA signaling | C4 | 0.0106 |
| mTOR signaling | C4 | 0.0201 |
| Corticotropin releasing hormone signaling | V1.1 | 0.00264 |
| CDK5 signaling | V1.1 | 0.0181 |
| Neuregulin signaling | V1.1 | 0.0208 |
| Rac signaling | V1.1 | 0.0272 |
| Valine, leucine, and isoleucine biosynthesis | V1.2, V3 | 0.00910, 0.0271 |
| Ascorbate and aldarate metabolism | V1.2 | 0.0129 |
| Aminoacyl-tRNA biosynthesis | V1.2 | 0.0248 |
| Histidine metabolism | V1.2 | 0.0323 |
| Regulation of actin-based motility by Rho | V2 | 0.0377 |
| Estrogen receptor signaling | V3 | 0.0303 |
| Aryl hydrocarbon receptor signaling | V3 | 0.0312 |
| Folate biosynthesis | V3 | 0.0337 |
| C21-steroid hormone metabolism | V3 | 0.0381 |
Comparisons are as defined in Table 1.
Discussion
ESCC was selected as a tumor type to evaluate transcriptome heterogeneity in patients, in part because of the high degree of genomic alterations that are present,7–10 potentially providing fertile ground for finding any associated mRNA heterogeneity. However, little evidence of regional intratumor heterogeneity at the mRNA level was observed in ESCC, at least at the dissection resolution used here (3000 to 8000 cells), suggesting that clonal evolution that significantly affects transcriptomes is not a predominant mechanism in this cancer type and that a relatively stable expression profile is maintained throughout three-dimensional tumor space.
To assess heterogeneity, we examined mRNA levels in several ways: tumor center versus tumor periphery, three-dimensional mapping, overall transcriptome profiles, individual gene differences, and biochemical pathways. None of the comparisons identified any pronounced intratumor expression differences (which is consistent with the findings from six of seven previous studies that have examined this question in cancer12–17,31), thus challenging the theory that cancer progression produces subclones with significantly different mRNA profiles11 and supporting the alternative view that ESCC arises with and then maintains an intact and relatively stable transcriptome.32
Analysis of heterogeneity of DNA mutations and gene expression in nonmicrodissected samples represents the majority of the intratumor heterogeneity literature to date.12–17,32–34 In contrast to the heterogeneity observed in DNA, multiple RNA samples from a single patient tumor have demonstrated low intratumor heterogeneity, irrespective of organ type, irrespective of whether the RNA was procured from core needle biopsies or samples as large as 1-cm3 tissue chunks, and irrespective of whether RNA amplification was performed before hybridization.12–17 To further investigate this apparent discrepancy of genomic variability but transcriptional stability, more powerful molecular discovery tools for analyzing small numbers of cells need to be used. For example, once next-generation deep-sequencing technologies are validated for use with small quantities of DNA and RNA from dissected cells, these technologies should allow for the combined in-depth analysis of genomic and transcriptomic intratumor heterogeneity necessary to examine this issue in more detail.
The 14 genes identified as differentially expressed in 1 or 2 of the 16 ESCC three-dimensional comparisons have been reported in previous gene expression profiling studies of ESCC and other tumors. Although none are known to be influenced by genomic instability,35 a new mechanism involving the SEI1/SET/NM23H1 pathway in ESCC has been identified. In this pathway, SEI1 is able to up-regulate SET expression and promote translocation of NM23H1 to the nucleus from the cytoplasm.36 In three-dimensional peripheral zone analysis of our ESCC samples, THBS2 was significantly up-regulated, which may indicate that cells in the tumor periphery were under the transcriptional influence of endogenous angiogenesis and tumor growth inhibitor signaling.37 In an analysis of two-dimensional peripheral and core specimens from leiomyosarcoma, minimal intratumor heterogeneity was observed, with only 13 genes differentially expressed between the two zones.16 Comparison of these genes with those altered in ESCC identified RGS5 as common to both tumor types. The RGS5 protein is a member of the regulator of G-protein signaling group, and its expression has been associated with clinical outcome in renal cell carcinoma38 and with chemoresistance in ovarian cancer.39 Recent global gene expression profiling of ESCC demonstrated that CEP55 was significantly up-regulated in tumor compared with normal tissue.40 CEP55 expression has also been shown to be related to tumor progression in head and neck SCC.41 Although we do not have human papillomavirus infection data on the ESCC cases used in the current study, it is interesting to note the differential expression of transcription factor YYI, which is known to bind to human papillomavirus in head and neck SCC42 and is highly correlated with human papillomavirus infection in cervical cancers.43
Zonal gene expression heterogeneity was observed in a microdissected xenograft pancreatic carcinoma study.31 However, other analyses of mRNA levels from clinical specimens demonstrating minimal heterogeneity12–17 have not used microdissection, and thus these measurements represent an aggregate profile from multiple different cell types, including tumor, stroma, and inflammatory cells.32,34,44 Our strategy for overcoming tissue heterogeneity and potential masking of tumor specific profiles was to use microdissection45,46 of discrete tumor areas. This is particularly important, because previous microdissection-based studies by our research group showed that tumor cells and adjacent tumor stroma have opposite changes in the directionality of gene expression, with tumor cells decreasing the number of differentially expressed genes and stroma increasing them; this characteristic of the tumor microenvironment is missed in bulk specimen analysis, which can mask important biological phenomena.24,30 Thus, to ensure that we were accurately and fully measuring tumor cell heterogeneity, we procured and analyzed carefully dissected samples.
Five caveats to the current study require brief mention. First, the question of what constitutes heterogeneity versus homogeneity is somewhat arbitrary. Indeed, the intratumor comparisons were not identical, but were judged to be relatively similar, compared with other dissected cell populations. Second, the microdissection process was performed blindly, in that the tumor cells were procured based only on spatial location and not on either a priori knowledge of molecular status or on phenotypic features of the targeted cells. Thus, it is possible that one or more hidden tumor regions with a significantly altered transcriptome were present but were missed during the dissection process; however, the marked similarity of more than 50 individual tumor areas analyzed to date does not provide evidence for this phenomenon. In future studies, it will be possible to address this question in more detail by using molecularly targeted laser dissection methods (eg, expression microdissection47 or immunoguided laser capture microdissection48) to prescreen for and then dissect tumor regions that are markedly different from each other based on expression of a target protein or transcript. Third, the study did not examine heterogeneity at the level of a single cell or a few cells; approximately 10,000 cells were procured in each regional dissection to provide a deep and robust transcriptome analysis. This issue also can be addressed in future studies, as the reproducibility of single-cell measurements improves. Fourth, we used standard bioinformatics tools in the present study, and it is possible these methods were incapable of detecting subtle but important mRNA-level heterogeneity. We hope that the publicly available data sets from our work will be analyzed in unique ways by other investigators to examine these results from various perspectives.
Finally, we did not independently validate the present results at the mRNA level, because we and others have previously shown that expression array technology provides robust and reliable mRNA measurements from microdissected tissue.24,30,40,49 To date, the vast majority of transcripts that we have evaluated by both array and quantitative RT-PCR have shown the same result, although the expression differences are often less on the array, given the complex nature of the hybridization reaction. We did attempt to validate four differentially expressed genes at the protein level, but we were not successful because of technical challenges. This is a frequent limitation of immunohistochemical staining of tissue, especially for proteins (and these are a majority) that are not highly abundant in a given cell type. The same difficulty has also been observed previously in a larger cohort of ESCC cases.40
The ESCC data have implications for molecular diagnostics, because tumor heterogeneity can have important clinical consequences. For example, DNA clonal diversity measures are known to be strong predictors of cancer progression in Barrett’s esophagus,50 and are used in guiding treatment decisions.33,51,52 Moreover, it has been demonstrated experimentally that clonally heterogeneous tumors can either inhibit or increase the therapeutic efficacy of cytotoxic drugs33,53 and can contribute to drug resistance.54 In the clinic, however, obtaining specimens from multiple regions of a patient’s tumor to assess heterogeneity would be difficult and in many cases impractical. Thus, the question of whether a small core needle biopsy is representative of an entire tumor is coming to the forefront of molecular diagnostics and personalized medicine, such as with the clinical trial Biomarker-Integrated Approaches of Targeted Therapy for Lung Cancer Elimination (BATTLE).55 The feasibility of making precise microarray-based predictions of clinical outcome or tumorigenesis from a single core has been demonstrated in breast cancer,13 but in cervical cancer the number of cores necessary for obtaining reliable gene expression estimates is dependent on the intratumor heterogeneity level for the probe sets or genes of interest.12 The predominantly homogeneous transcriptomic expression data from microdissected tumor epithelium in ESCC suggest that molecular diagnostic transcriptomic profiles obtained from clinical core needle biopsies will indeed be sufficient to accurately represent a patient’s entire tumor, which supports prior ESCC analysis of two-dimensional nonmicrodissected tumor core needle biopsies.56
In summary, global expression measurements of ESCC indicate that regional mRNA heterogeneity is not a prominent feature, and that a relatively stable transcriptome is maintained throughout three-dimensional space, at least at a resolution on the order of 500 to 1500 μm in diameter. This finding may extend to SCCs in other organs, which as a group produce significant cancer morbidity and mortality worldwide. Future studies that measure molecular profiles in smaller cell numbers and in other geographic regions, and that include DNA mutations and proteomic status, are needed to further assess what degree of molecular heterogeneity exists in this clinically aggressive histological class of tumors.
Footnotes
Supported in part by the Cohen-Reinauch BATTLE-2 Fund, the Novartis-IPCT BATTLE Program, and the NIH Intramural Research Program of the Center for Cancer Research and the Division of Cancer Epidemiology and Genetics, National Cancer Institute.
Disclosures: M.R.E.-B. is an inventor on all NIH patents covering laser capture microdissection and expression microdissection and is entitled to receive royalty-based payments through the NIH Technology Transfer Program.
Contributor Information
Michael R. Emmert-Buck, Email: buckm@mail.nih.gov.
Heidi S. Erickson, Email: hserickson@mdanderson.org.
Supplemental Data
Correlation analysis of gene expression data points performed using the one-round versus the two-round amplification method. Jurkat total RNA (1 μg) was amplified using the one-round method and hybridized to the Affymetrix U133A GeneChip according to the manufacturer’s recommendations. In a separate experiment, Jurkat total RNA (10 ng) was amplified using the two-round amplification method as described under Materials and Methods. Excluding probe-set signal intensities corresponding to Affymetrix control genes, probe-set signals for 1 μg were compared with those generated for the 10-ng sample using the GeneChip operating system scatter-plot analysis. Correlation between signals generated by the two methods was excellent (R2 = 0.91). Signals corresponding to genes or expressed sequence tags scored as present under both methods are shown in red, those scored marginal under both methods are shown in blue, and those scored absent under both methods are shown in orange.
Quality control assessment of each array using NUSE and RLE. Each bar represents one dissection. Case 1, (red); case 2, (purple); case 3, (blue); case 4, (orange); case 5, (green).
Differential gene expression comparisons of TP/TC (blue/green) versus NB/ND (orange/red) in nine cases through MDS analysis. Each spot represents one case.
Differential gene expression comparisons of three-dimensional-microdissected ESCC in five cases through MDS analysis. Each spot represents one dissection (including normal epithelium for case 5). Microdissection was performed as shown in Figure 1B.
Global differential gene expression comparisons on three-dimension microdissected ESCC tumor clusters. A: Principal component analysis (PCA) mapping on ESCC tumor clusters, ESCC matched normal epithelium fragments, and prostate cell populations. B: Hierarchical cluster analysis on ESCC tumor clusters, ESCC matched normal epithelium fragments, and prostate cell populations. For ESCC samples, different colors indicate different cases; for prostate samples, colors indicate different cell populations. Each spot (A) or bar (B) represents one cluster, fragment, or cell population. E, epithelium; N, normal; P, prostate; S, stroma; T, tumor.
Histogram of intratumor heterogeneity calculated as W/(W + B). A: The distribution of 10,301 probe sets as a function of W/(W + B) values for tumor periphery versus tumor center in nine cases. B: The distribution of 9834 probe sets as a function of W/(W + B) for 41 three-dimensional microdissected tumor subregions in five cases. Values of W/(W + B) > 0.5 correspond to intratumor heterogeneity being less than intertumor heterogeneity.
Supplemental Data
Supplemental material for this article can be found at http://dx.doi.org/10.1016/j.ajpath.2012.10.028.
References
- 1.Kamangar F., Dores G.M., Anderson W.F. Patterns of cancer incidence, mortality, and prevalence across five continents: defining priorities to reduce cancer disparities in different geographic regions of the world. J Clin Oncol. 2006;24:2137–2150. doi: 10.1200/JCO.2005.05.2308. [DOI] [PubMed] [Google Scholar]
- 2.Parkin D.M., Bray F., Ferlay J., Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 3.Jemal A., Siegel R., Xu J., Ward E. Cancer Statistics, 2010. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [Erratum appeared in CA Cancer J Clin 2011, 61:133–134] [DOI] [PubMed] [Google Scholar]
- 4.Pisani P., Bray F., Parkin D.M. Estimates of the world-wide prevalence of cancer for 25 sites in the adult population. Int J Cancer. 2002;97:72–81. doi: 10.1002/ijc.1571. [DOI] [PubMed] [Google Scholar]
- 5.Travis W.D. Pathology of lung cancer. Clin Chest Med. 2002;23:65–81. doi: 10.1016/s0272-5231(03)00061-3. viii. [DOI] [PubMed] [Google Scholar]
- 6.Yan W., Wistuba I.I., Emmert-Buck M.R., Erickson H.S. Squamous cell carcinoma - similarities and differences among anatomical sites. Am J Cancer Res. 2011;1:275–300. [PMC free article] [PubMed] [Google Scholar]
- 7.Hu N., Su H., Li W.J., Giffen C., Goldstein A.M., Hu Y., Wang C., Roth M.J., Li G., Dawsey S.M., Xu Y., Taylor P.R., Emmert-Buck M.R. Allelotyping of esophageal squamous-cell carcinoma on chromosome 13 defines deletions related to family history. Genes Chromosomes Cancer. 2005;44:271–278. doi: 10.1002/gcc.20242. [DOI] [PubMed] [Google Scholar]
- 8.Hu N., Wang C., Hu Y., Yang H.H., Kong L.H., Lu N., Su H., Wang Q.H., Goldstein A.M., Buetow K.H., Emmert-Buck M.R., Taylor P.R., Lee M.P. Genome-wide loss of heterozygosity and copy number alteration in esophageal squamous cell carcinoma using the Affymetrix GeneChip Mapping 10 K array. BMC Genomics. 2006;7:299. doi: 10.1186/1471-2164-7-299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu N., Wang C., Ng D., Clifford R., Yang H.H., Tang Z.Z., Wang Q.H., Han X.Y., Giffen C., Goldstein A.M., Taylor P.R., Lee M.P. Genomic characterization of esophageal squamous cell carcinoma from a high-risk population in China. Cancer Res. 2009;69:5908–5917. doi: 10.1158/0008-5472.CAN-08-4622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bandla S., Pennathur A., Luketich J.D., Beer D.G., Lin L., Bass A.J., Godfrey T.E., Litle V.R. Comparative genomics of esophageal adenocarcinoma and squamous cell carcinoma. Ann Thorac Surg. 2012;93:1101–1106. doi: 10.1016/j.athoracsur.2012.01.064. [Erratum appeared in Thorac Surg 2012, 94:1042] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shipitsin M., Campbell L.L., Argani P., Weremowicz S., Bloushtain-Qimron N., Yao J., Nikolskaya T., Serebryiskaya T., Beroukhim R., Hu M., Halushka M.K., Sukumar S., Parker L.M., Anderson K.S., Harris L.N., Garber J.E., Richardson A.L., Schnitt S.J., Nikolsky Y., Gelman R.S., Polyak K. Molecular definition of breast tumor heterogeneity. Cancer Cell. 2007;11:259–273. doi: 10.1016/j.ccr.2007.01.013. [DOI] [PubMed] [Google Scholar]
- 12.Bachtiary B., Boutros P.C., Pintilie M., Shi W., Bastianutto C., Li J.H., Schwock J., Zhang W., Penn L.Z., Jurisica I., Fyles A., Liu F.F. Gene expression profiling in cervical cancer: an exploration of intratumor heterogeneity. Clin Cancer Res. 2006;12:5632–5640. doi: 10.1158/1078-0432.CCR-06-0357. [DOI] [PubMed] [Google Scholar]
- 13.Barry W.T., Kernagis D.N., Dressman H.K., Griffis R.J., Hunter J.D., Olson J.A., Marks J.R., Ginsburg G.S., Marcom P.K., Nevins J.R., Geradts J., Datto M.B. Intratumor heterogeneity and precision of microarray-based predictors of breast cancer biology and clinical outcome. J Clin Oncol. 2010;28:2198–2206. doi: 10.1200/JCO.2009.26.7245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Francis P., Fernebro J., Edén P., Laurell A., Rydholm A., Domanski H.A., Breslin T., Hegardt C., Borg A., Nilbert M. Intratumor versus intertumor heterogeneity in gene expression profiles of soft-tissue sarcomas. Genes Chromosomes Cancer. 2005;43:302–308. doi: 10.1002/gcc.20191. [DOI] [PubMed] [Google Scholar]
- 15.Jochumsen K.M., Tan Q., Hølund B., Kruse T.A., Mogensen O. Gene expression in epithelial ovarian cancer: a study of intratumor heterogeneity. Int J Gynecol Cancer. 2007;17:979–985. doi: 10.1111/j.1525-1438.2007.00908.x. [DOI] [PubMed] [Google Scholar]
- 16.Shmulevich I., Hunt K., El-Naggar A., Taylor E., Ramdas L., Labordé P., Hess K.R., Pollock R., Zhang W. Tumor specific gene expression profiles in human leiomyosarcoma: an evaluation of intratumor heterogeneity. Cancer. 2002;94:2069–2075. doi: 10.1002/cncr.10425. [DOI] [PubMed] [Google Scholar]
- 17.Trautmann K., Steudel C., Grossmann D., Aust D., Ehninger G., Miehlke S., Thiede C. Expression profiling of gastric cancer samples by oligonucleotide microarray analysis reveals low degree of intra-tumor variability. World J Gastroenterol. 2005;11:5993–5996. doi: 10.3748/wjg.v11.i38.5993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Subramanian J., Simon R. Gene expression-based prognostic signatures in lung cancer: ready for clinical use? J Natl Cancer Inst. 2010;102:464–474. doi: 10.1093/jnci/djq025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tumor Analysis Best Practices Working Group Expression profiling–best practices for data generation and interpretation in clinical trials. Nat Rev Genet. 2004;5:229–237. doi: 10.1038/nrg1297. [DOI] [PubMed] [Google Scholar]
- 20.Erickson H.S., Albert P.S., Gillespie J.W., Wallis B.S., Rodriguez-Canales J., Linehan W.M., Gonzalez S., Velasco A., Chuaqui R.F., Emmert-Buck M.R. Assessment of normalization strategies for quantitative RT-PCR using microdissected tissue samples. Lab Invest. 2007;87:951–962. doi: 10.1038/labinvest.3700659. [DOI] [PubMed] [Google Scholar]
- 21.Erickson H.S., Gillespie J.W., Emmert-Buck M.R. Tissue microdissection. Methods Mol Biol. 2008;424:433–448. doi: 10.1007/978-1-60327-064-9_34. [DOI] [PubMed] [Google Scholar]
- 22.Erickson H.S., Albert P.S., Gillespie J.W., Rodriguez-Canales J., Marston Linehan W., Pinto P.A., Chuaqui R.F., Emmert-Buck M.R. Quantitative RT-PCR gene expression analysis of laser microdissected tissue samples. Nat Protoc. 2009;4:902–922. doi: 10.1038/nprot.2009.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao H., Hastie T., Whitfield M.L., Børresen-Dale A.L., Jeffrey S.S. Optimization and evaluation of T7 based RNA linear amplification protocols for cDNA microarray analysis. BMC Genomics. 2002;3:31. doi: 10.1186/1471-2164-3-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Richardson A.M., Woodson K., Wang Y., Rodriguez-Canales J., Erickson H.S., Tangrea M.A., Novakovic K., Gonzalez S., Velasco A., Kawasaki E.S., Emmert-Buck M.R., Chuaqui R.F., Player A. Global expression analysis of prostate cancer-associated stroma and epithelia. Diagn Mol Pathol. 2007;16:189–197. doi: 10.1097/PDM.0b013e3180de20ac. [DOI] [PubMed] [Google Scholar]
- 25.Kralj J.G., Player A., Sedrick H., Munson M.S., Petersen D., Forry S.P., Meltzer P., Kawasaki E., Locascio L.E. T7-based linear amplification of low concentration mRNA samples using beads and microfluidics for global gene expression measurements. Lab Chip. 2009;9:917–924. doi: 10.1039/b811714d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brettschneider J., Collin F., Bolstad B.M., Speed T.P. Quality assessment for short oligonucleotide microarray data. Technometrics. 2008;50:241–264. [Google Scholar]
- 27.Bolstad B.M., Collin F., Brettschneider J., Simpson K., Cope L., Irizarry R.A., Speed T.P. Quality assessment of Affymetrix GeneChip data. In: Gentleman R., Carey V., Huber W., Irizarry R., Dudoit S., editors. Bioinformatics and Computational Biology Solutions Using R and Bioconductor. Springer; 2005. pp. 33–47. [Google Scholar]
- 28.Korn E.L., Troendle J.F., McShane L.M., Simon R. Controlling the number of false discoveries: application to high-dimensional genomic data. J Stat Plan Inference. 2004;124:20. [Google Scholar]
- 29.Korn E.L., Li M.C., McShane L.M., Simon R. An investigation of two multivariate permutation methods for controlling the false discovery proportion. Stat Med. 2007;26:4428–4440. doi: 10.1002/sim.2865. [DOI] [PubMed] [Google Scholar]
- 30.Yan W., Shih J.H., Rodriguez-Canales J., Tangrea M.A., Ylaya K., Hipp J., Player A., Hu N., Goldstein A.M., Taylor P.R., Emmert-Buck M.R., Erickson H.S. Identification of unique expression signatures and therapeutic targets in esophageal squamous cell carcinoma. BMC Res Notes. 2012;5:73. doi: 10.1186/1756-0500-5-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakamura T., Kuwai T., Kitadai Y., Sasaki T., Fan D., Coombes K.R., Kim S.J., Fidler I.J. Zonal heterogeneity for gene expression in human pancreatic carcinoma. Cancer Res. 2007;67:7597–7604. doi: 10.1158/0008-5472.CAN-07-0874. [DOI] [PubMed] [Google Scholar]
- 32.Ramaswamy S., Ross K.N., Lander E.S., Golub T.R. A molecular signature of metastasis in primary solid tumors. Nat Genet. 2003;33:49–54. doi: 10.1038/ng1060. [DOI] [PubMed] [Google Scholar]
- 33.Marusyk A., Polyak K. Tumor heterogeneity: causes and consequences. Biochim Biophys Acta. 2010;1805:105–117. doi: 10.1016/j.bbcan.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Perou C.M., Sorlie T., Eisen M.B., van de Rijn M., Jeffrey S.S., Rees C.A., Pollack J.R., Ross D.T., Johnsen H., Akslen L.A., Fluge O., Pergamenschikov A., Williams C., Zhu S.X., Lonning P.E., Børresen-Dale A.L., Brown P.O., Botstein D. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 35.Hu N., Clifford R.J., Yang H.H., Wang C., Goldstein A.M., Ding T., Taylor P.R., Lee M.P. Genome wide analysis of DNA copy number neutral loss of heterozygosity (CNNLOH) and its relation to gene expression in esophageal squamous cell carcinoma. BMC Genomics. 2010;11:576. doi: 10.1186/1471-2164-11-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li Y., Nie C.J., Hu L., Qin Y., Liu H.B., Zeng T.T., Chen L., Fu L., Deng W., Chen S.P., Jia W.H., Zhang C., Xie D., Guan X.Y. Characterization of a novel mechanism of genomic instability involving the SEI1/SET/NM23H1 pathway in esophageal cancers. Cancer Res. 2010;70:5695–5705. doi: 10.1158/0008-5472.CAN-10-0392. [DOI] [PubMed] [Google Scholar]
- 37.Streit M., Riccardi L., Velasco P., Brown L.F., Hawighorst T., Bornstein P., Detmar M. Thrombospondin-2: a potent endogenous inhibitor of tumor growth and angiogenesis. Proc Natl Acad Sci USA. 1999;96:14888–14893. doi: 10.1073/pnas.96.26.14888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yao M., Huang Y., Shioi K., Hattori K., Murakami T., Sano F., Baba M., Kondo K., Nakaigawa N., Kishida T., Nagashima Y., Yamada-Okabe H., Kubota Y. A three-gene expression signature model to predict clinical outcome of clear cell renal carcinoma. Int J Cancer. 2008;123:1126–1132. doi: 10.1002/ijc.23641. [DOI] [PubMed] [Google Scholar]
- 39.Hooks S.B., Callihan P., Altman M.K., Hurst J.H., Ali M.W., Murph M.M. Regulators of G-Protein signaling RGS10 and RGS17 regulate chemoresistance in ovarian cancer cells. Mol Cancer. 2010;9:289. doi: 10.1186/1476-4598-9-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Su H., Hu N., Yang H.H., Wang C., Takikita M., Wang Q.H., Giffen C., Clifford R., Hewitt S.M., Shou J.Z., Goldstein A.M., Lee M.P., Taylor P.R. Global gene expression profiling and validation in esophageal squamous cell carcinoma and its association with clinical phenotypes. Clin Cancer Res. 2011;17:2955–2966. doi: 10.1158/1078-0432.CCR-10-2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Janus J.R., Laborde R.R., Greenberg A.J., Wang V.W., Wei W., Trier A., Olsen S.M., Moore E.J., Olsen K.D., Smith D.I. Linking expression of FOXM1, CEP55 and HELLS to tumorigenesis in oropharyngeal squamous cell carcinoma. Laryngoscope. 2011;121:2598–2603. doi: 10.1002/lary.22379. [DOI] [PubMed] [Google Scholar]
- 42.Ferris R.L., Martinez I., Sirianni N., Wang J., López-Albaitero A., Gollin S.M., Johnson J.T., Khan S. Human papillomavirus-16 associated squamous cell carcinoma of the head and neck (SCCHN): a natural disease model provides insights into viral carcinogenesis. Eur J Cancer. 2005;41:807–815. doi: 10.1016/j.ejca.2004.11.023. [DOI] [PubMed] [Google Scholar]
- 43.Baritaki S., Sifakis S., Huerta-Yepez S., Neonakis I.K., Soufla G., Bonavida B., Spandidos D.A. Overexpression of VEGF and TGF-beta1 mRNA in Pap smears correlates with progression of cervical intraepithelial neoplasia to cancer: implication of YY1 in cervical tumorigenesis and HPV infection. Int J Oncol. 2007;31:69–79. [PubMed] [Google Scholar]
- 44.Liotta L.A., Kohn E.C. Cancer’s deadly signature. Nat Genet. 2003;33:10–11. doi: 10.1038/ng0103-10. [DOI] [PubMed] [Google Scholar]
- 45.Emmert-Buck M.R., Bonner R.F., Smith P.D., Chuaqui R.F., Zhuang Z., Goldstein S.R., Weiss R.A., Liotta L.A. Laser capture microdissection. Science. 1996;274:998–1001. doi: 10.1126/science.274.5289.998. [DOI] [PubMed] [Google Scholar]
- 46.Harrell J.C., Dye W.W., Harvell D.M., Sartorius C.A., Horwitz K.B. Contaminating cells alter gene signatures in whole organ versus laser capture microdissected tumors: a comparison of experimental breast cancers and their lymph node metastases. Clin Exp Metastasis. 2008;25:81–88. doi: 10.1007/s10585-007-9105-7. [DOI] [PubMed] [Google Scholar]
- 47.Tangrea M.A., Chuaqui R.F., Gillespie J.W., Ahram M., Gannot G., Wallis B.S., Best C.J., Linehan W.M., Liotta L.A., Pohida T.J., Bonner R.F., Emmert-Buck M.R. Expression microdissection: operator-independent retrieval of cells for molecular profiling. Diagn Mol Pathol. 2004;13:207–212. doi: 10.1097/01.pdm.0000135964.31459.bb. [DOI] [PubMed] [Google Scholar]
- 48.Tangrea M.A., Hanson J.C., Bonner R.F., Pohida T.J., Rodriguez-Canales J., Emmert-Buck M.R. Immunoguided microdissection techniques. Methods Mol Biol. 2011;755:57–66. doi: 10.1007/978-1-61779-163-5_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lassmann S., Kreutz C., Schoepflin A., Hopt U., Timmer J., Werner M. A novel approach for reliable microarray analysis of microdissected tumor cells from formalin-fixed and paraffin-embedded colorectal cancer resection specimens. J Mol Med (Berl) 2009;87:211–224. doi: 10.1007/s00109-008-0419-y. [DOI] [PubMed] [Google Scholar]
- 50.Merlo L.M., Shah N.A., Li X., Blount P.L., Vaughan T.L., Reid B.J., Maley C.C. A comprehensive survey of clonal diversity measures in Barrett’s esophagus as biomarkers of progression to esophageal adenocarcinoma. Cancer Prev Res (Phila) 2010;3:1388–1397. doi: 10.1158/1940-6207.CAPR-10-0108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Marx A.H., Zielinski M., Kowitz C.M., Dancau A.M., Thieltges S., Simon R., Choschzick M., Yekebas E., Kaifi J.T., Mirlacher M., Atanackovic D., Brümmendorf T.H., Fiedler W., Bokemeyer C., Izbicki J.R., Sauter G. Homogeneous EGFR amplification defines a subset of aggressive Barrett’s adenocarcinomas with poor prognosis. Histopathology. 2010;57:418–426. doi: 10.1111/j.1365-2559.2010.03643.x. [DOI] [PubMed] [Google Scholar]
- 52.Bonavia R., Inda M.M., Cavenee W.K., Furnari F.B. Heterogeneity maintenance in glioblastoma: a social network. Cancer Res. 2011;71:4055–4060. doi: 10.1158/0008-5472.CAN-11-0153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Michor F., Polyak K. The origins and implications of intratumor heterogeneity. Cancer Prev Res (Phila) 2010;3:1361–1364. doi: 10.1158/1940-6207.CAPR-10-0234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hoey T. Drug resistance, epigenetics, and tumor cell heterogeneity. Sci Transl Med. 2010;2:28ps19. doi: 10.1126/scitranslmed.3001056. [DOI] [PubMed] [Google Scholar]
- 55.Kim E.S., Herbst R.S., Wistuba I.I., Lee J.J., Blumenschein G.R., Jr., Tsao A., Stewart D.J., Hicks M.E., Erasmus J., Jr., Gupta S., Alden C.M., Liu S., Tang X., Khuri F.R., Tran H.T., Johnson B.E., Heymach J.V., Mao L., Fossella F., Kies M.S., Papadimitrakopoulou V., Davis S.E., Lippman S.M., Hong W.K. The BATTLE trial: personalizing therapy for lung cancer. Cancer Discov. 2011;1:44–53. doi: 10.1158/2159-8274.CD-10-0010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Motoori M., Takemasa I., Yamasaki M., Komori T., Takeno A., Miyata H., Takiguchi S., Fujiwara Y., Yasuda T., Yano M., Matsuura N., Matsubara K., Monden M., Mori M., Doki Y. The feasibility of using biopsy samples from esophageal cancer for comprehensive gene expression profiling. Int J Oncol. 2009;35:265–271. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Correlation analysis of gene expression data points performed using the one-round versus the two-round amplification method. Jurkat total RNA (1 μg) was amplified using the one-round method and hybridized to the Affymetrix U133A GeneChip according to the manufacturer’s recommendations. In a separate experiment, Jurkat total RNA (10 ng) was amplified using the two-round amplification method as described under Materials and Methods. Excluding probe-set signal intensities corresponding to Affymetrix control genes, probe-set signals for 1 μg were compared with those generated for the 10-ng sample using the GeneChip operating system scatter-plot analysis. Correlation between signals generated by the two methods was excellent (R2 = 0.91). Signals corresponding to genes or expressed sequence tags scored as present under both methods are shown in red, those scored marginal under both methods are shown in blue, and those scored absent under both methods are shown in orange.
Quality control assessment of each array using NUSE and RLE. Each bar represents one dissection. Case 1, (red); case 2, (purple); case 3, (blue); case 4, (orange); case 5, (green).
Differential gene expression comparisons of TP/TC (blue/green) versus NB/ND (orange/red) in nine cases through MDS analysis. Each spot represents one case.
Differential gene expression comparisons of three-dimensional-microdissected ESCC in five cases through MDS analysis. Each spot represents one dissection (including normal epithelium for case 5). Microdissection was performed as shown in Figure 1B.
Global differential gene expression comparisons on three-dimension microdissected ESCC tumor clusters. A: Principal component analysis (PCA) mapping on ESCC tumor clusters, ESCC matched normal epithelium fragments, and prostate cell populations. B: Hierarchical cluster analysis on ESCC tumor clusters, ESCC matched normal epithelium fragments, and prostate cell populations. For ESCC samples, different colors indicate different cases; for prostate samples, colors indicate different cell populations. Each spot (A) or bar (B) represents one cluster, fragment, or cell population. E, epithelium; N, normal; P, prostate; S, stroma; T, tumor.
Histogram of intratumor heterogeneity calculated as W/(W + B). A: The distribution of 10,301 probe sets as a function of W/(W + B) values for tumor periphery versus tumor center in nine cases. B: The distribution of 9834 probe sets as a function of W/(W + B) for 41 three-dimensional microdissected tumor subregions in five cases. Values of W/(W + B) > 0.5 correspond to intratumor heterogeneity being less than intertumor heterogeneity.