Abstract
The cereal cyst nematodes belonging to Heterodera avenae group is a complex species consisting of 12 valid species and overlapping morphological characters make them difficult to be distinguished from one another. The non coding internal transcribed spacer sequences, ITS1 and ITS2 including 5.8S region of ribosomal DNA (rDNA) has been very useful for the accurate identification of the species and characterization of molecular genetic variation within the species of plant parasitic nematodes. In the present study, sequencing and PCR-RFLP of rDNA has been used to confirm the species identity. Further it was used to determine the genetic homogeneity of an Indian population used for whole genome and transcriptome sequencing. The Sequence of ITS1 and ITS2 including 5.8S showed approximately 99% similarity with the existing sequences of H. avenae from different countries and confirmed the species identity. Secondary structure of ITS2 region shows that the isolates from India, China, Israel and Australian possess more stable conformational energy than the German strain. Further to characterize the genetic variation within the population, about 200 individual cysts or females were analyzed separately by PCR-RFLP of rDNA with five restriction enzymes that could distinguish H. avenae from other closely related species within the group. This analysis did not reveal any variation within the population indicating it is genetically homogeneous and suitable for next generation sequencing using Illumina platform.
Keywords: Cereal cyst nematode, Heterodera avenae, PCR-RFLP, genetic homogeneity, ITS-rDNA
Background
Cereal cyst nematode (CCN), Heterodera avenae Wollenweber, 1924, has become an endemic problem in different agro-climatic zones from the Northern Hill regions to Central India since its first report in India. Yield loss of 41.6% has been recorded in wheat in the state of Rajasthan [1]. Based on comparison of yields from nematicide-treated and untreated fields, the average avoidable losses ranged from 32 – 44 % in barley and 24 – 35% in wheat [2]. Besides barley and wheat, in India, CCN is reported on oats, triticale, rye, maize and other graminaceous plants in India. Efforts are also underway to develop resistant cultivars in wheat. Several accessions from Australia and ICARDA (International Center for Agricultural Research in the Dry Areas) as well as species of Aegilops are being used in breeding for resistance to CCN [3]. Large variation in the disease reaction of these accessions and wild Triticum as well as their crosses with native cultivars has resulted in varying reports under multi-location trials. Although a part of this variation can be attributed to different experimental conditions, most of it could be due to the genetic variability of the nematode. Earlier work in India to determine variability in H. avenae using international differentials including clones of a few grasses, revealed the occurrence of 5 biotypes in the state of Rajasthan alone [4]. Later, five Indian CCN populations were screened against differential varieties listed under the International Testing Program in addition to some local wheat and barley varieties and grasses [5]. Based on the host responses, the authors grouped the populations into two biotypes. Biotype I comprised populations from the state of Rajasthan (Jaipur and Udaipur) and Haryana (Narnaul), the other hand, biotype II included populations from Punjab (Hoshiarpur and Ludhiana). Similar screening tests in subsequent years revealed that populations from the states of Delhi and Uttar Pradesh belonged to biotype I and those of Himachal Pradesh to biotype II [6]. However, these studies lack information on the genetic basis of the biotype differentiation. The H. avenae is a species complex presently containing eleven valid species and several undescribed species [7]. Though some of the species can be easily identified based on morphology, others are less distinct. The ‘Gotland strain’ of H. filipjevi is morphologically indistinguishable from true H. avenae. Differences in protein patterns and in rDNA between different populations of H. avenae and the ‘Gotland strain’ or H. filipjevi have been reported by several authors [8]. In India, H. filipjevi has been reported from Punjab, Ambala and Himachal Pradesh [9]. Considerable genetic variation was reported among seven Indian populations of H. avenae that could be responsible for differential host reactions in multi location trials [10].
The present management options to reduce the yield losses caused by H. avenae are not adequate as most of the nematicides are either not available or discontinued [11]. In view of this there is an urgent need to design some alternate modern management options for which nematode genomics will be very useful. Population homogeneity is essential for maintaining the quality control in data generation using Next Generation Sequencing approaches. In order to sequence the transcriptome and whole genome of the cereal cyst nematode, H. avenae from India, characterization of genetic uniformity of the population to be used for sequencing is a pre-requisite. As already mentioned, this species is more complex and morphological traits alone are not sufficient, molecular characterisation using coding and non coding regions of rDNA is of immense value to distinguish the species complex of H. avenea that consists of 12 sub species [12]. PCR-RFLP of rDNA is also a very useful approach for determining the population homogeneity within a given species [9, 12, 13]. Therefore, in the present study, sequencing and PCR-RFLP of rDNA was used to confirm the species identity and to determine the genetic homogeneity of an Indian population of H. avenae. That will be used in whole genome and transcriptome sequencing by next generation sequencing platform.
Methodology
Collection of biological material:
Soil and root samples were collected from H. avenae infested wheat fields from Indian Agricultural Research Institute (IARI), New Delhi. The samples were processed using Cobb's sieving technique and 25 and 60 mesh sieves were used for washing the soil samples. The cysts were collected using a stereobinocular microscope (ZEISS stemi 2000-C) and used for DNA extraction.
Sample preparation:
DNA was extracted from single cysts according to the method described by Subbotin [8]. DNA was extracted from about 200 cysts separately to study variation within the population (Figure 1). A single cyst was transferred into 10 µl of double distilled water in an Eppendorf tube and crushed with a microhomogenizer. Eight µl of nematode lysis buffer (125 mM KCl, 25 mM Tris-HCl pH 8.3, 3.75 mM MgCl2, 2.5 DTT, 1.125 % Tween 20, 0.025% gelatin) and 2 µl of proteinase K (600 (g/ml) were added. The tubes were incubated at 650 C (1h) and 950C (10 min) consecutively.
Figure 1.
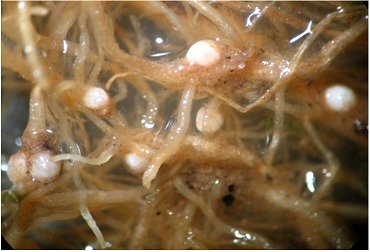
White females of Heterodera avenae from IARI fields
PCR amplification and PCR-RFLP:
After centrifugation (1 min: 13000 rpm) 10 µl of the DNA suspension was added to the PCR reaction mixture containing 0.2 mM dNTP's, 0.2 µM of each of the primer (reverse and forward) and 2.5 µl 10× buffer (10 mM Tris-HCl pH 8.3, 50 mM KCl, 1.5 mM MgCl2), 1 unit of Taq polymerase (Fermentas), and double distilled water to a final volume of 25 µl. Primers (5') TTGATTACGTCCCTGCCCTTT (3') and (5') TTCACTCGCCGTTACTAAGG (3') as described by Joyce et al., 1994 were used in the PCR reaction. Amplification reactions were performed in MJ Research DNA thermal cycler. Each cycle has denaturation at 95° C for 30 seconds, annealing at 55°C for 30 seconds, extension at 72°C for 90 seconds and the cycle was repeated 44 times with a final extension of 72°C for 5 min. Five µl aliquots of amplification products were loaded on 1.2 % agarose gels prepared in 1X TAE (tris-acetate-EDTA) buffer (pH 8.0), separated by electrophoresis (5 V/cm for 2.5 h) and stained with ethidium bromide. The gels were visualized using a gel documentation system (Alpha Image Analyzer, USA). Five to seven microliters of each PCR product was digested with one of the following 5 enzymes: Alu1, Pst1, Hinf1, BsuR1, and Msp1 (MBI Fermentas, Germany) in the buffer stipulated by the manufacturer. The digested DNA was loaded on a 1.5% agarose prepared in TAE buffer, separated by electrophoresis, stained with ethidium bromide and visualized using gel documentation system. Procedures for obtaining PCR amplified products and endonuclease digestion of these products was repeated twice to verify the results.
Cloning, sequencing and analysis:
Fresh PCR products were purified with a PCR purification kit (Qiagen, Germany) as per manufacturer's protocols, and 2 µl of purified product was used for TA cloning using pGEM T vector (Promega, Madison, USA). Recombinant colonies were identified using an ampicillin and blue white colony selection method, and were confirmed by PCR using the same primers used for PCR amplification. Plasmids were prepared and the inserts were sequenced and analyzed. Multiple sequence alignment (MSA) was done by MUSCLE, alignment curation by Gblock (www.phylogeny.fr). Secondary structure analysis was done by Mfold [14]. Restriction map was generated by NEBcutter [15].
Discussion
The wheat roots collected from fields were infested by H. avenae and white females attached to the roots were handpicked under the microscope and used for DNA extraction. PCR amplification of the rDNA ITS 1 and 2 regions of 200 individual cysts generated one fragment of approximately 1050 bp (Figure 2A)- ITS amplification is shown for only 96 individual cysts). Initial analysis by restriction digestion of the PCR product with all the five restriction enzymes AluI, PstI, HinfI, BsuRI, and MspI did not indicate any variation in the restriction profile of the ITS region among the different individuals (Figure 2 B-F - Restriction profile is shown only for 56 cysts for each restriction enzyme). Pst1 and Hinf1 can accurately differentiate H. avenae from H. filipjevi [9, 12]. The restriction profile generated by these two enzymes provided the primary confirmation of the species indicating that all the analyzed individual cysts were H. avenae. Therefore the PCR amplified ITS fragment of five individuals randomly selected were cloned separately and sequenced. Removal of primer sequences in the flanking 18S and 28S gave 566 and 215 bp ITS 1 and 2 sequences respectively.
Figure 2.
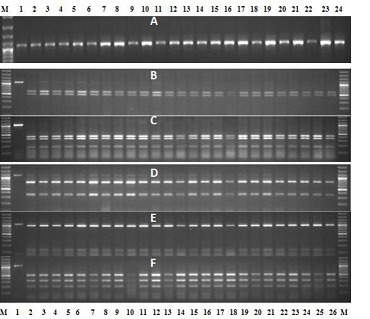
A) PCR-amplified 1050bp rDNA fragment from 24 individual cysts / females of Heterodera avenae; B-F) PCR-RFLP profile of rDNA of 26 individual females /cysts of H. avenae from IARI, New Delhi. M- 100bp ladder. B - Alu1: C-Pst1: D - Hinf1: E - BsuR1: F- Msp1.
After sequencing it was deposited in GenBank (accession numbers: KC152906). MSA of five geographical isolates taken for this study (India - KC152906, China - GU377252, Israel - AY148363, Germany - AY148387 and Australia - HM560737) and pairwise comparison showed differences in ITS1 ranging from 0.3–1.4%. Even though some sequence differences were seen in the ITS1 region, they were not significant enough in generating differences in the restriction profiles. Single base deletions were more common in the ITS1 region of the Australian, China and German isolates may be due to the geographical isolation (Figure 3). All isolates of H. avenae used in this study are given in Table 1 (see supplementary material). Subbotin et al [9] reported that absence of AluI and RsaI sites in the European population could be used to differentiate the European and Indian populations as these two restriction sites were present in the Indian population. However, the sequence analysis and comparison in the present study revealed that these two restriction sites were present in all the populations used in the analysis. Further, position of the restriction sites for MspI, AluI and RsaI remained identical in both Indian and the Chinese population and there was a slight shift in the German population. The length of ITS 2 was 215 bp and there was no difference in the ITS2 sequences of H. avenae isolates from different geographical locations except for the German population suggesting ITS2 region is more conserved than ITS1. Conformational analysis of the ITS2 - rDNA genes of H. avenae isolates resulted in secondary structure that predicted most energy efficient shape for each isolate. The delta G (ΔG) values were -81 for the isolates from India and Israel. Whereas 84.6 for Australian and -78.3 kcal/mol for the German population. This shows that the isolates with lower delta values are thermodynamically more stable so that they can possibly adapt to specific conditions prevalent in their respective habitat [16].
Figure 3.
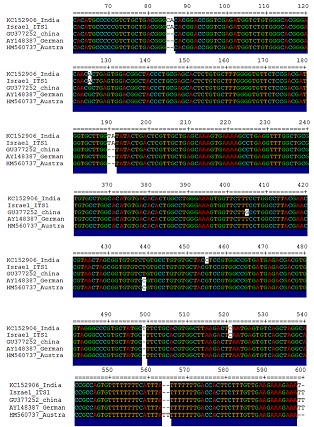
Multiple sequence alignment of ITS1 region of five geographical isolates of Heterodera avenae
Present study revealed that Indian isolate of cereal cyst nematode, H. avenae is quite homogeneous and suitable for generating unbiased sequence information by using next generation sequencing approach. Further, we observed no significant difference among the five geographical populations originating from five countries. However about 10 resistant genes have been reported globally to confer either total or partial resistance to different CCN pathotypes of H. avenae [17]. In view of this it will be informative to sequence one or more pathotypes so for better understanding of the biology of this important nematode pest on wheat for its management.
Competing interests
The authors declare that they have no competing interests.
Supplementary material
Acknowledgments
We would like to acknowledge and extend our gratitude to Department of Biotechnology, Government of India for financial support under the Indo Australia Strategic Research fund (PR12678) and IARI, ICAR, New Delhi, India for infrastructure facilities.
Footnotes
Citation:Rao et al, Bioinformation 9(2): 067-071 (2013)
References
- 1.Handa DK, et al. Plant Pathology. 1980;8:73. [Google Scholar]
- 2.Handa DK, et al. Current Nematolology. 1991;2:99. [Google Scholar]
- 3.Williams KJ, et al. Theoretical and Applied Genetics. 2006;112:1480. doi: 10.1007/s00122-006-0251-0. [DOI] [PubMed] [Google Scholar]
- 4.Mathur BN, et al. Nematologica. 1974;20:19. [Google Scholar]
- 5.Swarup G, et al. Int J Nematol. 1979;9:164. [Google Scholar]
- 6.Swarup G, et al. Current Science. 1982;51:896. [Google Scholar]
- 7.Robinson AJ, et al. Fundamental and Applied Nematology. 1996;19:109. [Google Scholar]
- 8.Subbotin SA, et al. Nematology. 2000;2:153. [Google Scholar]
- 9.Subbotin SA, et al. Nematology. 1999;1:195. [Google Scholar]
- 10.Umarao, et al. Int J Nematol. 2008;18:118. [Google Scholar]
- 11.Thakur, et al. Bioinformation. 2012;8:617. doi: 10.6026/97320630008617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Umarao, et al. Int J Nematol. 2007;17:29. [Google Scholar]
- 13.Gavas R, et al. Int J Nematol. 2009;19:189. [Google Scholar]
- 14.Zuker M. Nucleic Acids Res. 2003;31:3406. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vincze T, et al. Nucleic Acids Res. 2003;31:3688. doi: 10.1093/nar/gkg526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krüger D, A Gargas. Mycol Res. 2008;112:316. doi: 10.1016/j.mycres.2007.10.019. [DOI] [PubMed] [Google Scholar]
- 17.Barloy D, et al. Molecular Breeding. 2007;20:31. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


