Myo1 becomes immobile and acts as a scaffold for proteins involved in septum formation and coordination with the actomyosin ring during yeast cytokinesis.
Abstract
Core components of cytokinesis are conserved from yeast to human, but how these components are assembled into a robust machine that drives cytokinesis remains poorly understood. In this paper, we show by fluorescence recovery after photobleaching analysis that Myo1, the sole myosin-II in budding yeast, was mobile at the division site before anaphase and became immobilized shortly before cytokinesis. This immobility was independent of actin filaments or the motor domain of Myo1 but required a small region in the Myo1 tail that is thought to be involved in higher-order assembly. As expected, proteins involved in actin ring assembly (tropomyosin and formin) and membrane trafficking (myosin-V and exocyst) were dynamic during cytokinesis. Strikingly, proteins involved in septum formation (the chitin synthase Chs2) and/or its coordination with the actomyosin ring (essential light chain, IQGAP, F-BAR, etc.) displayed Myo1-dependent immobility during cytokinesis, suggesting that Myo1 plays a scaffolding role in the assembly of a cytokinesis machine.
Introduction
Cytokinesis in animal and fungal cells requires concerted functions of an actomyosin ring (AMR), membrane trafficking, and localized ECM remodeling at the division site (Balasubramanian et al., 2004; Strickland and Burgess, 2004; Eggert et al., 2006; Barr and Gruneberg, 2007). The AMR, which consists of myosin-II and actin filaments, is thought to generate a contractile force that powers the ingression of the plasma membrane (PM). The AMR may also guide membrane deposition and ECM remodeling during cytokinesis (Vallen et al., 2000; Fang et al., 2010). Targeted membrane deposition is thought to increase surface area at the division site (Strickland and Burgess, 2004; Barr and Gruneberg, 2007) and may also deliver enzymatic cargoes for chitinous primary septum (PS) formation in the budding yeast Saccharomyces cerevisiae (Chuang and Schekman, 1996; VerPlank and Li, 2005) or localized ECM remodeling in animal cells. Importantly, defects in PS formation cause cytokinesis block in budding yeast (Bi, 2001; Schmidt et al., 2002; VerPlank and Li, 2005; Nishihama et al., 2009), and defects in ECM remodeling cause embryonic lethality with cells arrested in cytokinesis in Caenorhabditis elegans (Mizuguchi et al., 2003) and mice (Izumikawa et al., 2010). Thus, there are complex and interdependent relationships among the cellular events involved in cytokinesis.
It is generally assumed that the AMR consists of several sarcomere-like structures, in which myosin-II bipolar filaments slide on actin filaments to generate force for contraction (Schroeder, 1972; Sanger and Sanger, 1980; Satterwhite and Pollard, 1992). In addition, AMR contraction must be coupled with disassembly, as the volume of the ring decreases during furrow ingression (Schroeder, 1972; Bi, 2010; Mendes Pinto et al., 2012). This is different from muscle contraction, during which the number of sarcomeres (or contractile units) remains unchanged (Huxley, 1969). Experimental evidence in support of the “sarcomere disassembly” hypothesis is still lacking. Direct EM examination of the cleavage furrow has revealed that actin filaments are organized into parallel arrays of opposing polarity (Sanger and Sanger, 1980; Kamasaki et al., 2007) or as bundles of actin filaments (Maupin and Pollard, 1986) but failed to unambiguously identify myosin filaments. Thus, it is not clear how myosin-II is organized at the division site. FRAP analysis indicates that myosin-II is largely immobile at the cleavage furrow during cytokinesis in C. elegans (Carvalho et al., 2009) and Drosophila melanogaster (Goldbach et al., 2010; Uehara et al., 2010) but undergoes rapid turnover in the fission yeast Schizosaccharomyces pombe (Pelham and Chang, 2002), Dictyostelium discoideum (Yumura, 2001; Zhou et al., 2010), and mammalian cells (Kondo et al., 2011). In S. cerevisiae, myosin-II is reported to be dynamic in one study (Lister et al., 2006) but undergoes a dynamic to immobile transition from G2/M to cytokinesis in another study (Dobbelaere and Barral, 2004). These observations suggest that there are at least two modes of myosin-II organization at the division site, but their functional significance and regulatory mechanisms remain unknown. In this study, we show that Myo1, the sole myosin-II heavy chain in S. cerevisiae (Bi et al., 1998; Lippincott and Li, 1998a), displays cell cycle–regulated changes in mobility at the division site and that Myo1 immobility during cytokinesis is regulated through a putative assembly domain in its tail.
Core components of cytokinesis are conserved from yeast to human, but how these components are assembled into a robust machine that drives cell cleavage is not well understood. In this study, we performed FRAP analysis on 13 cytokinesis proteins in budding yeast to compare their individual and collective behaviors. We also performed similar analysis on these proteins in different mutants to determine their dependency relationships. We found that different proteins display distinct dynamics during the cell cycle and that the immobility of all other proteins during cytokinesis depends on Myo1, but not vice versa, suggesting that Myo1 plays a scaffolding role in the assembly of a cytokinesis machine.
Results
Myosin-II is mobile at the division site during the early part of the cell cycle and becomes progressively immobilized from anaphase to the onset of cytokinesis
To determine the dynamics of the AMR components, we first analyzed the dynamics of Myo1, the sole myosin-II heavy chain in budding yeast, during the cell cycle. When the entire Myo1-GFP ring at the bud neck was photobleached, the mean of the maximal fluorescence recovery was <8% regardless of cell cycle stages (Fig. S1, A–C; Video 1; and the entire FRAP data with individual curves and quantitative analyses were also shown in Figs. S3 and S4), which are marked by bud size (see Materials and methods for details) and septin–hourglass splitting (see associated videos; not depicted in figures), a cellular event that coincides with the onset of cytokinesis (Lippincott et al., 2001). The recovery was noticeably higher in small-budded cells (7.2 ± 1.1%) than in cells undergoing cytokinesis (1.1 ± 1.3%). These FRAP data suggest that there is a limited exchange of Myo1 between the bud neck and the cytosol throughout the cell cycle, which could be caused by a slow rate of exchange and/or a small pool of Myo1 in the cytosol.
As a complementary approach to FRAP, we also used fluorescence loss in photobleaching (FLIP) to probe the dynamics of Myo1 at the bud neck. A cytosolic region of a mother cell with a small bud was photobleached sequentially four times (Fig. S1 D). The fluorescence intensity of Myo1-GFP at the bud neck decreased only 30% in 23 min after the initial bleaching in the cytosol, in comparison to a 20% fluorescence loss in an unbleached control cell in the same imaging field during the same period (Fig. S1 E). In contrast, when a similar experiment was performed on a cell carrying Tpm2-GFP (tropomyosin), which is highly dynamic (see Fig. 5 A), the fluorescence intensity of Tpm2 at the bud neck decreased 50% in 50 s even after one bleach in the cytosol, whereas little or no change was observed for the control cell in the same field during the same period (Fig. S1, F–H). These FLIP data suggest that Myo1 cycles between the bud neck and the cytosol slowly. We also monitored the kinetics of Myo1 localization during the cell cycle, and found that Myo1 signal at the bud neck reached its peak from bud emergence to the small-budded stage (in <30 min; unpublished data). From this point on, Myo1 remained fairly constant at the bud neck (Fig. S1, I and J). Approximately 20 min before septin–hourglass splitting, Myo1 intensity was briefly increased by ∼20% and then decreased in a linear fashion during AMR constriction (Fig. S1, I and J; Tully et al., 2009). Direct measurement of Myo1-GFP (the sole source of Myo1 expressed from its native promoter at its physiological locus) showed that nearly all of the Myo1 molecules were localized to the bud neck around the small-budded stage. Because of the very dim signal of Myo1-GFP in the cytosol, it was rather difficult to obtain an accurate and meaningful measurement of Myo1 in this pool. However, based on fluorescence recovery after full-ring bleaching (Fig. S1 A) and the measurement of Myo1 intensity at the bud neck throughout the cell cycle (Fig. S1, I and J), we estimate that ≥80% of the total cellular Myo1 is localized to the bud neck before cytokinesis. Together, these data indicate that the majority of Myo1 are localized to the division site early in the cell cycle and maintained there with little flux between the ring and the cytosol.
Figure 5.
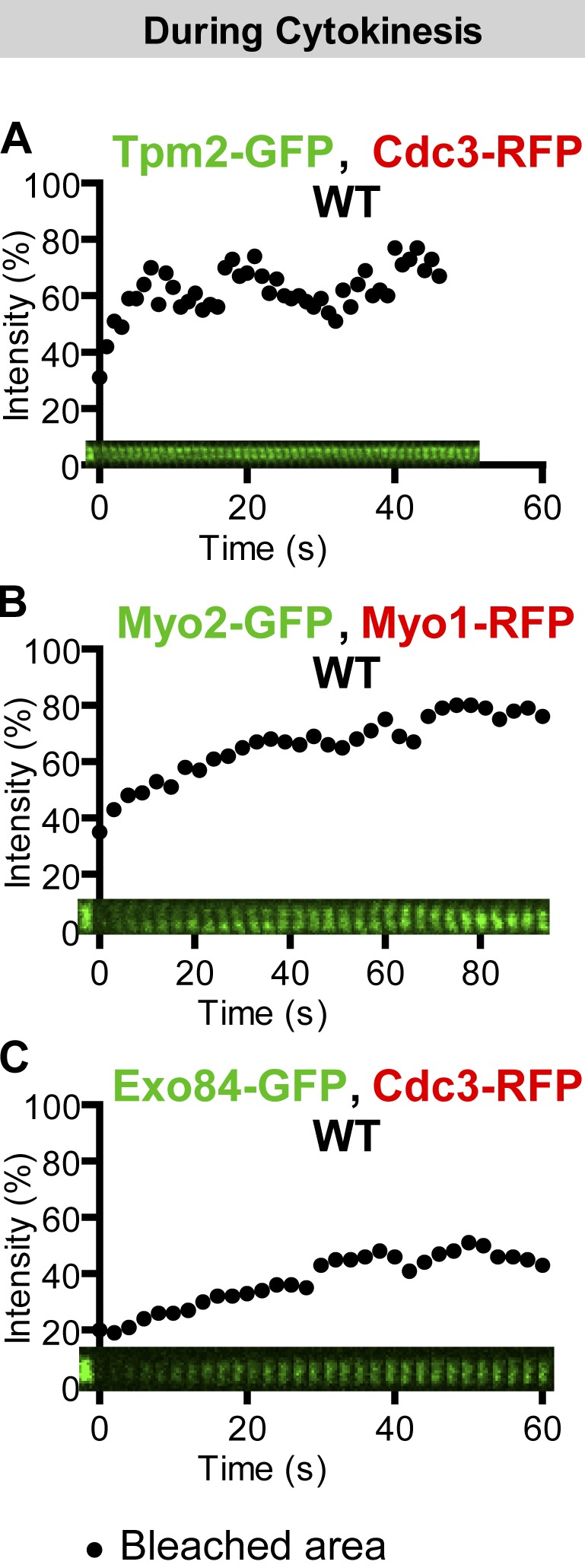
Actin ring–associated proteins and membrane trafficking components are dynamic during cytokinesis. (A) Tropomyosin is highly dynamic during cytokinesis. The full ring of Tpm2-GFP from a cell of the strain YEF6197 (TPM2-GFP CDC3-RFP) undergoing cytokinesis was analyzed by FRAP. (B and C) Membrane trafficking components are dynamic during cytokinesis. The full rings of Myo2-GFP and Exo84-GFP from cells of the strains YEF6001 (MYO2-GFP, pRS316-MYO1-mCherry) and YEF5862 (EXO84-GFP CDC3-RFP) during cytokinesis were analyzed by FRAP. WT, wild type.
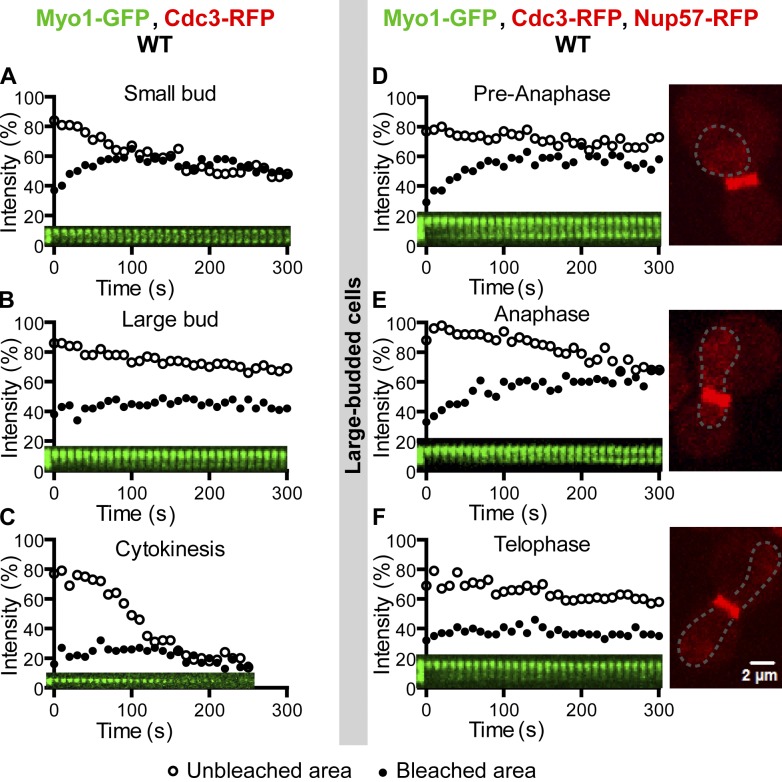
To determine Myo1 dynamics within the ring structure at the bud neck, we bleached a half of the ring in cells at different stages of the cell cycle. In small-budded cells, Myo1-GFP signal in the bleached region recovered quickly with a recovery rate (t1/2) of 20.4 ± 1.3 s and a maximal recovery of 19.3 ± 0.8% (Fig. 1 A, solid circles; and Video 2, left), whereas the GFP signal in the unbleached region decreased correspondingly (Fig. 1 A, open circles), suggesting that Myo1 moves laterally from the unbleached region to the bleached region. The magnitude of recovery is significant, considering that the maximally possible recovery would be 50% if all the recovery in the bleached half were attributed to lateral movement within the ring structure. This is a reasonable assumption given that Myo1 displays little flux between the ring and the cytosol (see the preceding paragraphs). In large-budded cells, the maximal recovery was limited to 7.1 ± 1.1% (Fig. 1 B and Video 2, middle). In cells undergoing cytokinesis, virtually no recovery was observed (Fig. 1 C and Video 2, right). Thus, Myo1 is mobile within the ring structure during the early part of the cell cycle and becomes immobilized toward the late part of the cell cycle.
Figure 1.
Myo1 displays cell cycle–regulated dynamics at the division site. (A–C) Myo1 is mobile at the division site in small-budded cells (A), becomes less mobile in large-budded cells (B), and is immobilized during cytokinesis (C), indicated by the presence of a double septin ring at the division site, which is not shown in any figure but can be seen in the figure-associated video. A half of the Myo1-GFP ring from cells at different stages of the cell cycle (indicated by bud size) of the strain XDY286 (MYO1-GFP CDC3-RFP) was photobleached, and fluorescence recovery in the bleached and unbleached regions was followed over time. Except where noted, FRAP analysis was performed in a similar way throughout the study. Quantitative analysis and the corresponding kymograph for a single representative cell are shown in each plot of the main figures. Individual recovery curves for all cells examined for a given protein at a specific cell cycle stage and their associated quantitative analyses are shown in Figs. S3 and S4. (D–F) Myo1 becomes immobilized in late anaphase or telophase. Large-budded cells (n = 12) of the strain YEF6036 (MYO1-GFP CDC3-RFP NUP57-RFP) at different phases of the cell cycle (indicated by the nuclear pore protein Nup57-RFP) were subjected to FRAP analysis to determine the precise timing of Myo1 immobilization during the cell cycle. WT, wild type.
To determine the transition point in Myo1 mobility within the ring structure more precisely, we performed similar FRAP analysis on cells carrying mCherry-labeled Nup57, a component of the nuclear pore complex (Alber et al., 2007) that marks the nuclear position in the cell. In wild-type cells, the penetration of the nucleus from the mother into the daughter cell compartment is correlated with the onset of anaphase (Yang et al., 1997). We found that Myo1 became increasingly immobile from middle or late anaphase to the onset of cytokinesis (Fig. 1, D–F). In addition, Myo1 remained highly mobile in mutants arrested at the onset of anaphase (cdc16 and cdc23) and was nearly immobile in mutants arrested at the mitotic exit (cdc15-2 and dbf2-1 dbf20Δ; unpublished data). Together, these data indicate that Myo1 is mobile within the ring structure before the onset of anaphase, increasingly immobilized from anaphase to telophase, and becomes completely immobile during cytokinesis.
We then probed the dynamics of the regulatory light chain (RLC) and the essential light chain (ELC) for Myo1. Mlc2, the RLC for Myo1, displays a localization pattern identical to Myo1 throughout the cell cycle, and its localization to the bud neck completely depends on its binding to Myo1 (Luo et al., 2004). Mlc2-GFP showed nearly identical dynamics as Myo1-GFP during the cell cycle when either the entire ring (not depicted) or a half of the ring (Fig. S2, A–C) was bleached.
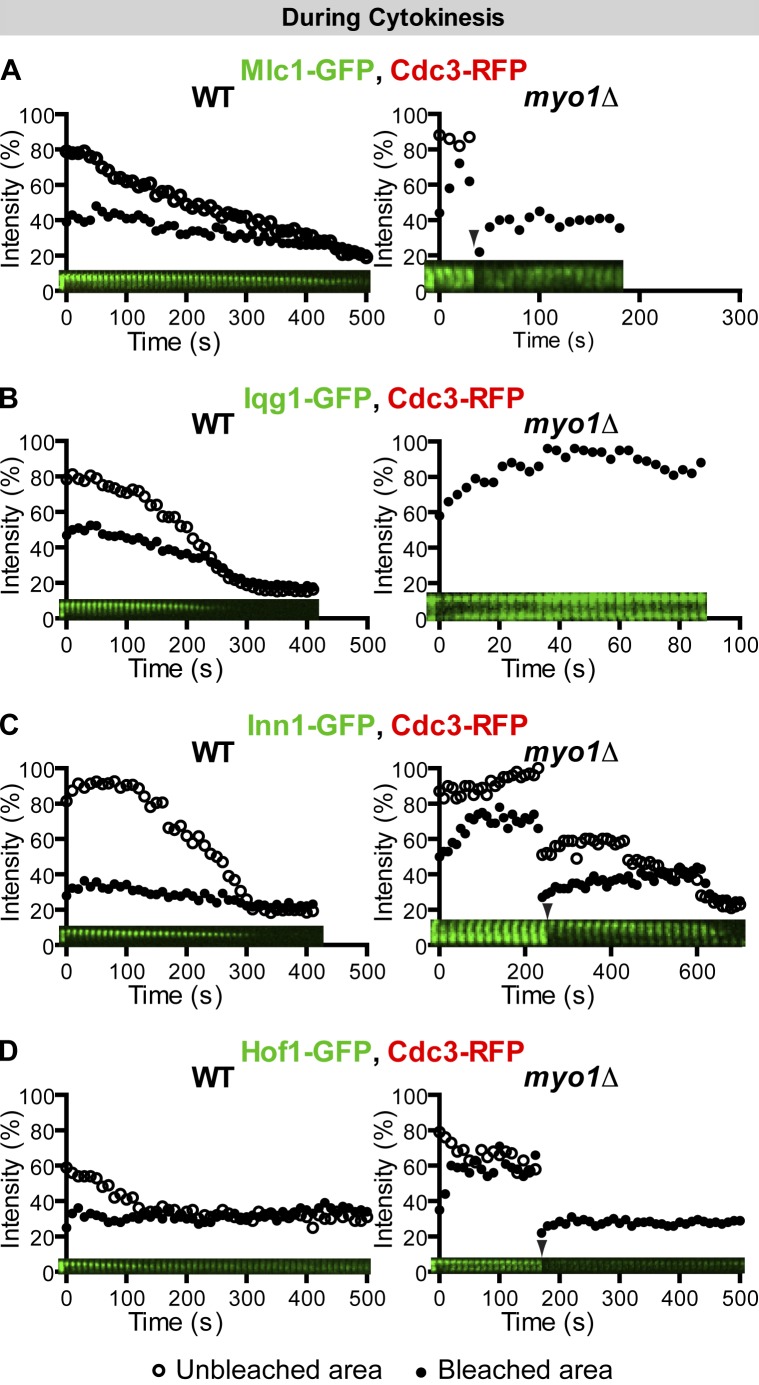
Mlc1, the ELC for Myo1 (Luo et al., 2004), also binds to Myo2 (a myosin-V in budding yeast) and Iqg1 (the sole IQGAP in budding yeast) via their respective IQ motifs (Stevens and Davis, 1998; Boyne et al., 2000; Shannon and Li, 2000). In small-budded cells, Mlc1 localizes to the bud cortex as puncta, reflecting its association with myosin-V and secretory vesicles (Wagner et al., 2002; Luo et al., 2004). These puncta were highly dynamic (Fig. S2 D). Mlc1 localizes to the bud neck only in large-budded cells and cells undergoing cytokinesis (Shannon and Li, 2000; Wagner et al., 2002; Luo et al., 2004). In these cells, Mlc1 displayed immobility from its initial localization to the completion of cytokinesis (see Fig. 7 A and Video 8). Together, these data indicate that myosin-II undergoes cell cycle–regulated changes in dynamics, being mobile within the ring structure early in the cell cycle and becoming progressively immobilized from anaphase to the onset of cytokinesis.
Figure 7.
Proteins involved in septum formation or its coordination with the AMR display Myo1-dependent immobility during cytokinesis. (A–D) The immobility of Mlc1, Iqg1, Inn1, and Hof1 during cytokinesis depends on Myo1. FRAP analysis was performed on the indicated proteins in cells undergoing cytokinesis of the following strains: (A) YEF6065 (CDC3-RFP, pUG34-MLC1; left) and YEF6351 (myo1Δ CDC3-RFP, pUG34-MLC1; right); (B) YEF6140 (CDC3-RFP, pUG35-IQG1; left) and YEF6356 (myo1Δ CDC3-RFP, pUG35-IQG1; right, full-ring bleaching); (C) YEF6138 (INN1-GFP CDC3-RFP; left) and YEF6357 (myo1Δ INN1-GFP CDC3-RFP; n = 5; right); and (D) YEF6131 (HOF1-GFP CDC3-RFP; left) and YEF6358 (myo1Δ HOF1-GFP CDC3-RFP; right). Arrowheads denote additional half-ring or full-ring bleaching. WT, wild type.
The change in Myo1 dynamics is also reflected by the dynamics of its binding partners during the cell cycle. Bni5, a septin-binding protein (Lee et al., 2002), mediates Myo1 targeting to the division site from late G1 to the onset of telophase, whereas Mlc1 and Iqg1 mediate Myo1 targeting from the onset of anaphase to the end of cytokinesis (Fang et al., 2010). Not surprisingly, Bni5 was mobile at the division site during its entire stay in the presence or absence of Myo1 (Fig. S2, E–H; and Video 3, left); in contrast, Mlc1 (see Fig. 7 A and Video 8) and Iqg1 (see Fig. 7 B) displayed immobility throughout their localization at the division site.
Actin filaments, motor domain, and light chain binding sites are not required for Myo1 immobility during cytokinesis
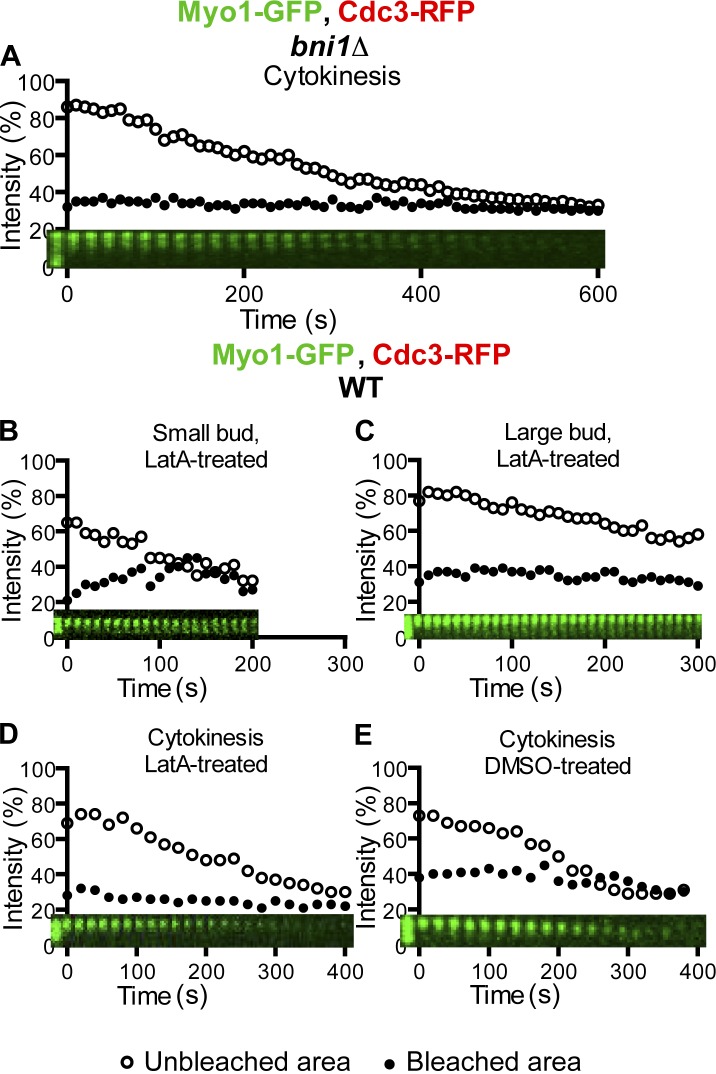
The timing of Myo1 mobility changes during the cell cycle correlates with the timing of actin ring assembly and AMR constriction (Epp and Chant, 1997; Bi et al., 1998; Lippincott and Li, 1998a). To determine the possible role of actin ring assembly in regulating Myo1 immobility, we examined Myo1 dynamics in bni1Δ cells during the cell cycle. Bni1 and Bnr1 are two formins in budding yeast that share an essential role in nucleating actin cable assembly to mediate polarized cell growth during budding (Pruyne et al., 2002; Sagot et al., 2002). During cytokinesis, Bni1 is the only formin localized at the division site (Buttery et al., 2007) and plays an important role in actin ring assembly and cytokinesis (Vallen et al., 2000; Tolliday et al., 2002). We found that deletion of BNI1 did not affect Myo1 dynamics during the cell cycle, including its immobility during cytokinesis (a cell in which Myo1-GFP failed to constrict presumably because of the absence of the actin ring; Fig. 2 A and Video 3, right). Because about one third of bni1Δ cells are still able to form a faint actin ring, we disrupted all actin filaments using latrunculin A (LatA; Ayscough et al., 1997) and then examined Myo1 dynamics. The LatA treatment did not affect Myo1 behavior during the cell cycle (Fig. 2, B–E; and Video 3, right). Myo1 was mobile at the division site in small-budded cells (t1/2, 27.8 ± 7.5; maximal recovery, 18.6 ± 3.7%; n = 6), less mobile in large-budded cells (maximal recovery, 6.9 ± 1.2%; n = 5), and became immobile during cytokinesis (n = 6). As expected, Myo1-GFP failed to constrict in LatA-treated cells (Fig. 2 D and Video 3, right), in contrast to the DMSO-treated control cell (Fig. 2 E). These data indicate that the actin ring and thus AMR constriction are not required for Myo1 immobility during cytokinesis.
Figure 2.
Actin filaments are not required for Myo1 immobility during cytokinesis. (A) Myo1 remains immobile during cytokinesis in bni1Δ cells. Myo1-GFP ring in a cell of the strain YEF6116 (bni1Δ MYO1-GFP CDC3-RFP) undergoing cytokinesis was analyzed by FRAP. (B–E) Myo1-GFP displays similar dynamics in LatA- and DMSO-treated cells. Cells of the strain XDY286 (MYO1-GFP CDC3-RFP) were subjected to FRAP analysis in the presence LatA (B–D) or DMSO (E, control) during the indicated stages of the cell cycle. WT, wild type.
The tail of Myo1 is sufficient for directing the assembly of a “headless” AMR (Fang et al., 2010), which largely fulfills its role in cytokinesis (Lord et al., 2005; Fang et al., 2010). However, this headless AMR constricts with ∼70–80% of the constriction rate of a normal AMR (Lord et al., 2005; Fang et al., 2010). To determine whether the head domain of Myo1, which includes its motor domain, a putative actin binding site, and the binding sites for both ELC and RLC, plays any role in regulating its immobility during cytokinesis, we performed FRAP analysis on yeast cells in which the chromosomal copy of MYO1 was precisely replaced with the Myo1 tail (residues 856–1,928)–coding sequence (Fang et al., 2010). Surprisingly, the dynamic behavior of the Myo1 tail was very similar to that of the full-length protein, being mobile at the division site in small-budded cells (t1/2, 22.7 ± 3.2; maximal recovery, 27.3 ± 2.8%; n = 12; Fig. 3 A and Video 4, left) and less mobile in large-budded cells (maximal recovery, 12.4 ± 1.6%; n = 7; Fig. 3 B and Video 4, middle) but becoming immobilized during cytokinesis (n = 7; Fig. 3 C and Video 4, right). Thus, the regulation of Myo1 dynamics during the cell cycle is largely mediated by its tail not its head domain.
Figure 3.
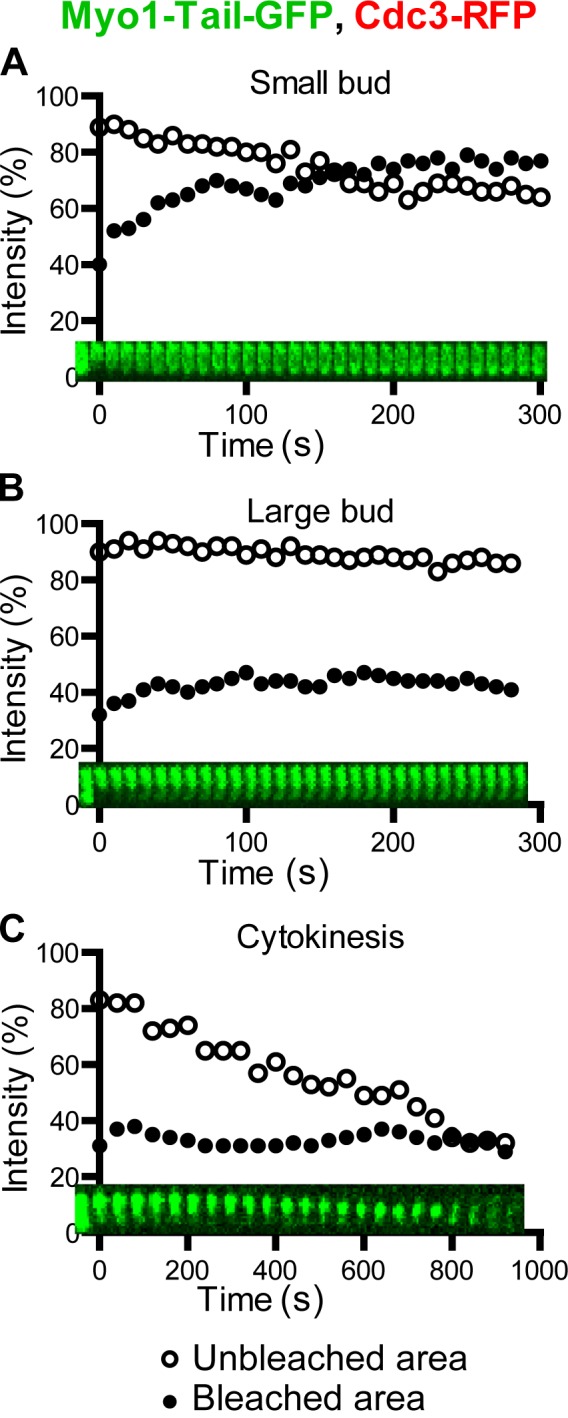
The tail of Myo1 confers its dynamic property during the cell cycle. (A–C) Myo1-Tail-GFP in a small-budded cell (A), a large-budded cell (B), and a cell undergoing cytokinesis (C) of the strain XDY288 (myo1-Tail-GFP CDC3-RFP) was analyzed by FRAP.
A small region near the C terminus of Myo1 is required for its immobility during cytokinesis
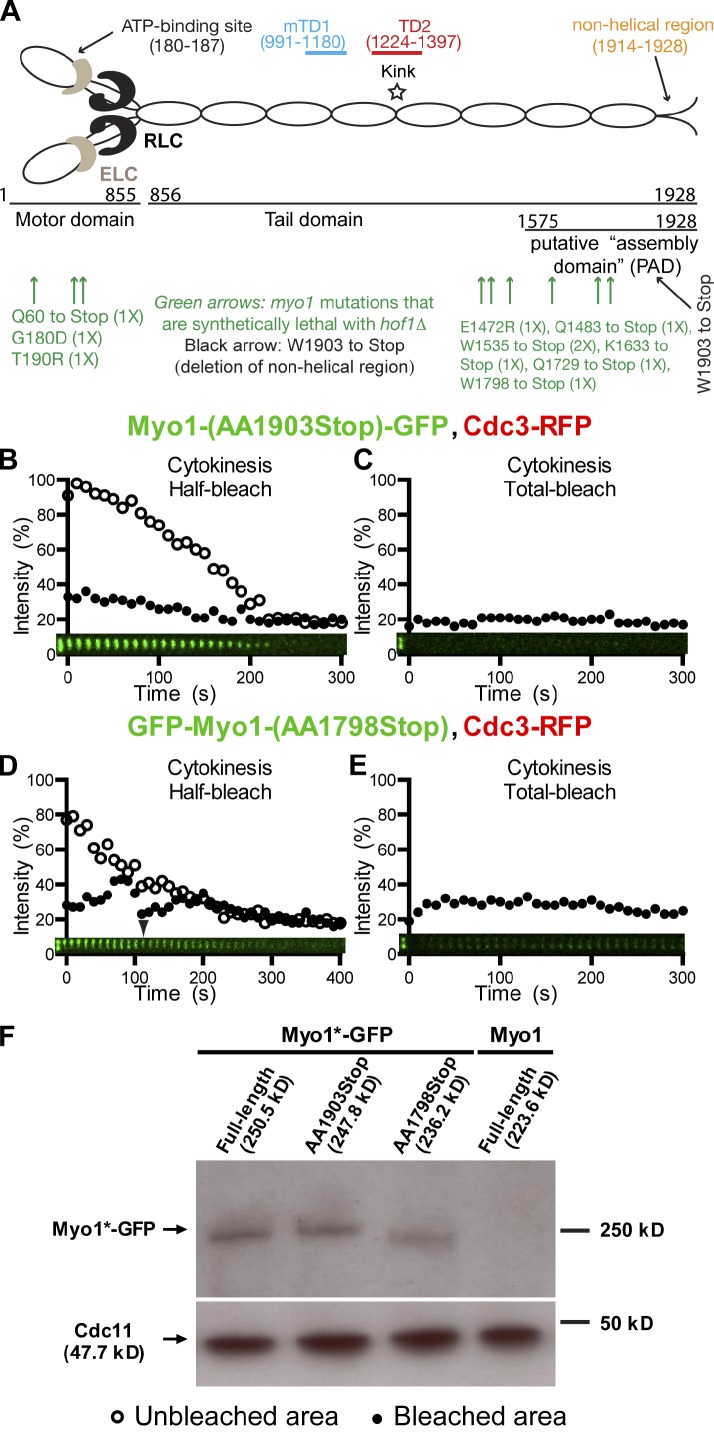
All myosin-IIs from animal cells can assemble into bipolar filaments in vitro, and this assembly invariably depends on a region near their C termini, called the assembly domain, which is required for antiparallel interaction of myosin-II molecules (Trybus, 1991; Tan et al., 1992). Myo1 tail also contains a putative assembly domain near its C terminus, which can only localize to the division site if it coexists with a Myo1 molecule harboring the targeting domains (mTD1 and TD2) near the middle of its tail (Fig. 4 A; Fang et al., 2010). Strikingly, seven of the 10 point mutations in MYO1 that are synthetically lethal with the deletion of HOF1 (Nishihama et al., 2009), which encodes an F-BAR protein involved in cytokinesis (Kamei et al., 1998; Lippincott and Li, 1998b; Vallen et al., 2000), are clustered within or near the putative assembly domain, highlighting the importance of this region in Myo1 function and cytokinesis (Fig. 4 A). Six of the seven are stop codon mutations, which define five distinct truncation alleles of MYO1 (Fig. 4 A). Based on these observations, we hypothesized that Myo1 may undergo cell cycle–triggered higher-order assembly, forming bipolar filaments during cytokinesis that account for its immobility.
Figure 4.
A small C-terminal region of Myo1 is required for its immobility during cytokinesis. (A) Myo1 motifs and the positions of myo1 mutations that are synthetically with hof1Δ. (B and C) Myo1 dynamics are not affected by the deletion of its putative nonhelical region. Half (B) and full (C) rings of Myo1-(AA1903Stop)-GFP in cells of the strain YEF6617 (myo1-(AA1903Stop)-GFP CDC3-RFP) were photobleached during cytokinesis. (C and D) The putative assembly domain of Myo1 is required for its immobility during cytokinesis. Half (C) and full (D) rings of GFP-Myo1-(AA1798Stop) in cells of the strain YEF6616 (GFP-myo1-(AA1798Stop) CDC3-RFP) were photobleached during cytokinesis. Except where noted, the first bleaching always corresponds to time 0, and additional bleaching (either half bleach or total bleach) is indicated by the arrowhead. (F) Expression level of the full-length and truncation alleles of MYO1. Cell lysates of strains YEF6618 (GFP-MYO1), YEF6617, YEF6616, and YEF473 (MYO1, negative control) were probed for the expression levels of Myo1 variants by Western blotting using a GFP antibody. As a loading control, the levels of the septin Cdc11 from the same cell lysates were probed using an anti-Cdc11 antibody.
To test this hypothesis, we performed FRAP analysis on cells carrying one of four truncation alleles of MYO1 (stopped at residue 1,483, 1,535, 1,633, or 1,798) isolated from the hof1 synthetic lethal screen as well as a truncation allele deleted for the coding sequence of a smaller C-terminal region (stopped at residue 1,903), including the predicted nonhelical tailpiece (Fig. 4 A). Similar nonhelical regions have been implicated in filament assembly for smooth muscle and nonmuscle myosin-IIs (Trybus, 1991). Like the wild-type protein, Myo1 lacking the putative nonhelical region was immobile during cytokinesis, as indicated by the half-ring bleaching (Fig. 4 B and Video 5, left), and virtually no recovery was observed when the entire ring was bleached (Fig. 4 C). Thus, the putative nonhelical region of Myo1 is not required for its immobility during cytokinesis. In contrast, Myo1-(AA1–1798) was mobile at the division site during cytokinesis (t1/2, 12.7 ± 3.4; maximal recovery, 18.3 ± 2.1%; n = 8; Fig. 4 D and Video 5, right), even though its expression level was similar to that of the full-length protein or Myo1 lacking the putative nonhelical region (Fig. 4 F). In addition, when the entire ring was bleached during cytokinesis, the recovery was noticeably higher than that of the full-length protein (Fig. 4 E compare with Fig. S1 C). The other three Myo1 variants with larger truncations also displayed mobility, although their overall signal at the bud neck was dimmer than the full-length or Myo1-(AA1–1798) proteins during cytokinesis (unpublished data). Together, these data demonstrate that a 105-aa fragment (residues 1,798–1,903) near the C terminus of Myo1 is essential for the establishment and/or maintenance of its immobility during cytokinesis.
Actin ring–associated proteins and membrane trafficking components are dynamic during cytokinesis
To gain insight into the construction of the cytokinesis machinery, we compared the dynamics of Myo1 and other cytokinesis proteins by performing FRAP analysis on those proteins involved in actin ring assembly, membrane trafficking, and septum formation. Because GFP-tagged actin is not functional and does not label actin cables or the actin ring (Doyle and Botstein, 1996), we probed the dynamics of actin ring–associated proteins (formins and tropomyosin) instead of actin itself. Both formin and tropomyosin are universally required for nucleating and stabilizing the actin filaments in the AMR (Balasubramanian et al., 2004; Moseley and Goode, 2006; Pollard, 2008). The formin Bni1 localizes to the sites of polarized growth during the cell cycle, including the bud neck during cytokinesis, whereas the formin Bnr1 localizes to the mother side of the bud neck from bud emergence to the onset of cytokinesis (Pruyne et al., 2004; Buttery et al., 2007). Thus, Bni1 is the only formin associated with the actin ring during cytokinesis. FRAP analysis indicates that Bni1-3GFP, expressed from its native promoter, is dynamic at the bud cortex as well as at the bud neck (Buttery et al., 2007). In contrast, the neck-localized Bnr1-GFP, expressed from its native promoter, is relatively immobile (Buttery et al., 2007). These observations are essentially confirmed in our study. In our experiments, GFP-tagged Bni1, expressed from its native promoter and carried on a high-copy plasmid (a single GFP-tagged Bni1 did not produce a signal strong enough for our study), localized to the division site at the onset of cytokinesis and was highly dynamic (Fig. S2 J). After full-ring bleaching, Bni1 displayed a recovery rate (t1/2) of 14.6 ± 1.3 s and a maximal recovery of 31.1 ± 1.9%. As expected, GFP-tagged Bnr1, expressed from a methionine promoter carried on a plasmid, was largely immobile throughout the cell cycle (Fig. S2 I).
GFP-tagged tropomyosins have been used to label the actin ring in live cells of the budding and fission yeasts (Pelham and Chang, 2002; Yoshida et al., 2006). We found that GFP-tagged Tpm2 localized to the division site 1–2 min before the onset of cytokinesis and was highly dynamic, with a recovery rate (t1/2) of 2.0 ± 0.5 s and a maximal recovery of 18.6 ± 2.9% (Fig. 5 A and Video 6, left). Together, these data indicate that actin ring–associated proteins, in contrast to myosin-II, are dynamic at the bud neck throughout cytokinesis.
To determine the dynamics of membrane trafficking components during cytokinesis, we performed FRAP analysis on cells carrying GFP-tagged Myo2 (myosin-V) and Exo84 (a subunit of the exocyst). Myo2 is required for the transport of post-Golgi vesicles along actin cables to the sites of polarized growth, including the bud neck during cytokinesis (Bretscher, 2003). Exo84 is a vesicle-associated subunit of the exocyst that is required for the tethering of post-Golgi vesicles to the PM during polarized cell growth and cytokinesis (Guo et al., 2000; Boyd et al., 2004). Both Myo2 and Exo84 arrive at the division site around the onset of cytokinesis (Fang et al., 2010; Wloka et al., 2011). Upon full-ring bleaching, Myo2-GFP recovered with a fast rate (t1/2) of 11.0 ± 1.3 s and a maximal recovery of 47.1 ± 5.0% (Fig. 5 B and Video 6, middle) throughout cytokinesis (AMR constriction was marked by RFP-tagged Myo1 in the same cells). Similarly, Exo84 recovered with a fast rate (t1/2) of 12.2 ± 0.6 s and a maximal recovery of 35.9 ± 2.0% (Fig. 5 C and Video 6, right; Boyd et al., 2004). The dynamic properties of Myo2 and Exo84 are consistent with their role in membrane trafficking during cytokinesis.
Proteins involved in septum formation or its coordination with the AMR display Myo1-dependent immobility during cytokinesis
Chs2, the chitin synthase II, is delivered to the bud neck at the onset of cytokinesis by the exocytic machinery to execute its essential role in PS formation (Sburlati and Cabib, 1986; Chuang and Schekman, 1996; VerPlank and Li, 2005). Bleaching of the full (unpublished data)- or half-ring of Chs2-GFP within the first 3–4 min after its localization to the bud neck led to a full recovery (Fig. 6 A and Video 7), which presumably reflects its timed and continuous delivery to the division site by the exocytic machinery. After this period, Chs2 became immobile (Fig. 6 A and Video 7). Together, these results suggest that (a) Chs2 delivery to the division site is complete within a few minutes of its initial localization; (b) once delivered, Chs2 may be closely associated with Myo1; and (c) Chs2 is not recycled back to the furrow membrane after its endocytic removal during the late stage of cytokinesis. Thus, Chs2 displays biphasic dynamics during cytokinesis.
Figure 6.
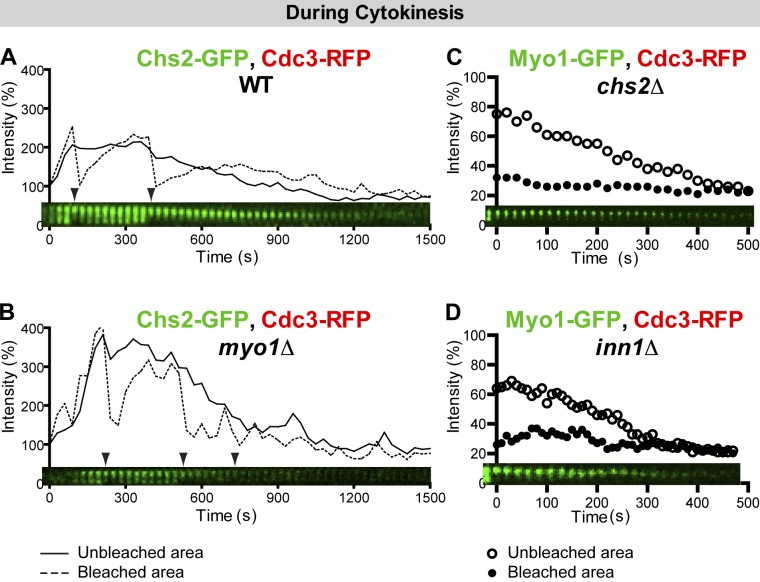
Chs2 displays biphasic dynamics during cytokinesis, and its immobility depends on Myo1. (A) Chs2 is dynamic and then immobile during cytokinesis. Chs2-GFP in the strain YEF5874 (CHS2-GFP CDC3-RFP) was bleached sequentially during cytokinesis. Time 0 corresponds to the initial localization of Chs2 to the bud neck. Arrowheads denote bleaching events. Solid and dashed lines indicate the intensity measurements for the unbleached and bleached areas, respectively. (B) Chs2 immobility depends on Myo1. Chs2-GFP in the strain YEF6336 (myo1Δ CHS2-GFP CDC3-RFP) was subjected to FRAP analysis. Note that the duration of Chs2 at the bud neck was shorter, and the Chs2 signal was weaker in the myo1Δ strain than in the wild-type (WT) strain. (C and D) Myo1 immobility during cytokinesis does not depend on Chs2 and Inn1. Myo1-GFP in strains YEF6273 (chs2Δ MYO1-GFP CDC3-RFP) and YEF6230 (inn1Δ MYO1-GFP CDC3-RFP) was analyzed by FRAP.
To determine whether the immobility of Chs2 during cytokinesis depends on Myo1, we examined Chs2 dynamics in myo1Δ cells. Strikingly, Chs2 became mobile throughout cytokinesis and, as expected, failed to constrict (Fig. 6 B and Video 7), suggesting that Chs2 is immobilized in a Myo1-dependent manner.
To determine whether Myo1 and other major components of the division machinery act in unison during cytokinesis such that deletion of one component will change the organization and/or the dynamics of other components, we examined Myo1 dynamics during cytokinesis in several cytokinesis mutants. In chs2Δ or inn1Δ cells, in which PS formation is completely blocked and cytokinesis is more defective than in myo1Δ cells (Bi, 2001; Schmidt et al., 2002; VerPlank and Li, 2005; Sanchez-Diaz et al., 2008; Nishihama et al., 2009; Meitinger et al., 2010), Myo1 remained immobile (Fig. 6, C and D; and Video 7). The Myo1 immobility was also observed in mlc2Δ, bni1Δ, bnr1Δ, hof1Δ, and cyk3Δ cells (unpublished data). Myo1 dynamics during cytokinesis could not be probed in mlc1Δ and iqg1Δ cells, as both Mlc1 and Iqg1 are required for Myo1 targeting to the division site during cytokinesis (Fang et al., 2010), and mlc1Δ and iqg1Δ are lethal in most strain backgrounds under normal growth conditions (Epp and Chant, 1997; Stevens and Davis, 1998). Together, these data indicate that the immobility of Chs2 during cytokinesis, but not its delivery to the division site, depends on Myo1; in contrast, Myo1 immobility does not depend on any other aforementioned cytokinesis proteins.
Besides Chs2, several other proteins (Mlc1, Iqg1, Inn1, Hof1, and Cyk3) have been implicated in PS formation or its coordination with the AMR (Korinek et al., 2000; Wagner et al., 2002; Nishihama et al., 2009; Meitinger et al., 2010). To determine their dynamics and explore their relationships with Myo1 during cytokinesis, we performed FRAP analysis of these proteins in wild-type and myo1Δ cells. As described earlier, in a wild-type strain, Mlc1, the ELC for Myo1, was immobile upon its localization to the bud neck in large-budded cells and remained immobile during cytokinesis (Fig. 7 A, left; and Video 8). However, in myo1Δ cells, Mlc1 became dynamic (Fig. 7 A, right; and Video 8). Both half-ring and full-ring (Fig. 7 A, arrowhead) bleaching indicates that the recovery is largely caused by cytosol–neck exchange. Like Mlc1, Iqg1 was immobile during cytokinesis (Fig. 7 B, left; and Video 8) but became dynamic in myo1Δ cells (Fig. 7 B, right; and Video 8). These data suggest that even though Mlc1 and Iqg1 are recruited to the neck in a Myo1-independent fashion and are actually required for Myo1 localization at the division site during cytokinesis (Fang et al., 2010). Myo1, in turn, is required for their organization.
Inn1 plays an essential role in PS formation and interacts with the SH3 domain of the F-BAR protein Hof1 and of the transglutaminase domain–containing protein Cyk3 through distinct PXXP motifs in its C terminus (Nishihama et al., 2009). Both Hof1 and Cyk3 are also involved in PS formation (Korinek et al., 2000; Vallen et al., 2000; Meitinger et al., 2010). Inn1 localized to the bud neck at the onset of cytokinesis (Sanchez-Diaz et al., 2008; Nishihama et al., 2009) and remained immobile throughout the division process (Fig. 7 C, left; and Video 9). However, in myo1Δ cells, Inn1 became more dynamic (Fig. 7 C, right; and Video 9). Hof1 was relatively dynamic until the onset of cytokinesis when it quickly became immobile and remained so during cytokinesis (Fig. 7 D, left; Video 9; and not depicted). Hof1 became much more dynamic in myo1Δ cells (Fig. 7 D, right; and Video 9). Thus, the immobility of Inn1 and Hof1 depends on Myo1. Interestingly, Cyk3 was dynamic throughout cytokinesis (t1/2 of 7.4 ± 0.9 s and maximal recovery of 31.7 ± 1.9% after full-ring bleaching; Fig. S2 K). These results suggest that Inn1 and Hof1 are closely associated with Myo1 during cytokinesis, whereas Cyk3 is fluxing between the bud neck and the cytosol. Together, our data indicate that proteins involved in PS formation, with the exception of Cyk3, display Myo1-dependent immobility during cytokinesis.
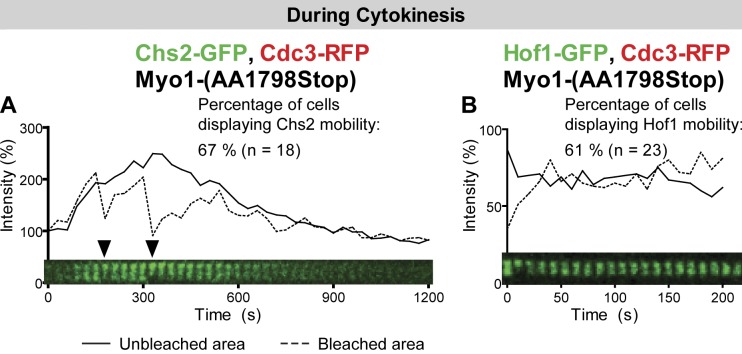
Because Myo1 immobility depends on a small region near its C-terminal end, we examined Chs2 and Hof1 dynamics in cells carrying the myo1-(AA1798Stop) allele (compare with Fig. 4, A and D). Both Chs2 (Fig. 8 A and Video 10, left) and Hof1 (Fig. 8 B and Video 10, right) became mobile at the division site during cytokinesis in the majority of the cells examined. We also probed the dynamics of Chs2 and Hof1 in cells carrying the myo1-(AA1535Stop) allele (compare with Fig. 4 A). In this case, Chs2 and Hof1 were mobile in all cells examined (unpublished data). Thus, the immobility of Chs2 and Hof1 depends on the immobility, not just the presence, of Myo1 at the division site.
Figure 8.
The immobility of Chs2 and Hof1 depends on a small region near the C-terminal end of Myo1. (A and B) Chs2 and Hof1 become mobile at the division site during cytokinesis in myo1-(AA1798Stop) cells. Chs2-GFP in strain YEF6771 (myo1-(AA1798Stop) CHS2-GFP CDC3-RFP; A) and Hof1-GFP in strain YEF6769 (myo1-(AA1798Stop) HOF1-GFP CDC3-RFP; B) were analyzed by FRAP during cytokinesis. Arrowheads denote bleaching events. Solid and dashed lines indicate the intensity measurements for the unbleached and bleached areas, respectively.
Discussion
Myo1 tail dictates its dynamics during the cell cycle
In this study, we show that Myo1 undergoes only limited exchange between the bud neck and the cytosol. However, Myo1 organization at the bud neck is clearly cell cycle regulated. Myo1 moves laterally at the bud neck before anaphase and then becomes immobilized from late anaphase to telophase and remains immobile during cytokinesis. This progressive immobilization of myosin-II from anaphase to cytokinesis also occurs in Drosophila (Goldbach et al., 2010; Uehara et al., 2010). Similarly, myosin-II does not undergo rapid exchange between the AMR and the cytosol during cytokinesis in C. elegans (Carvalho et al., 2009). In contrast, myosin-II exchanges rapidly between the AMR and the cytosol in D. discoideum (Yumura, 2001; Yumura et al., 2008; Zhou et al., 2010) yet displays no lateral movement within the AMR during cytokinesis (Yumura, 2001). On the other hand, myosin-II in the fission yeast S. pombe and mammalian cells is clearly mobile at the division site during cytokinesis (Pelham and Chang, 2002; Clifford et al., 2008; Kondo et al., 2011). However, in both cases, only one of the myosin-II isoforms (Myo2 in fission yeast and myosin-IIA in mammalian cells) has been analyzed. Our preliminary study indicates that Myp2, the other myosin-II in fission yeast, is immobile during cytokinesis (unpublished data). In mammalian cells, isoform-specific myosin-II dynamics have been observed in the context of cell migration (Sandquist and Means, 2008). It is possible that different isoforms of myosin-II are optimized for different functions during cytokinesis. For example, the immobile myosin-IIs, such as Myo1 in budding yeast or Myp2 in fission yeast, may have evolved to efficiently coordinate AMR constriction with PS formation (or localized ECM remodeling in animal cells) during cytokinesis, whereas the fast turnover myosin-IIs, such as Myo2 in fission yeast, may have evolved to mainly drive PM ingression through force production. Thus, it will be very informative to investigate whether and why different isoforms of myosin-II in a given organism display distinct dynamics during cytokinesis.
Regulation of myosin-II dynamics during cytokinesis remains poorly understood. In this study, we show that actin filaments, motor domain, and light chain (ELC and RLC) binding sites in the head domain are largely dispensable for the establishment of the Myo1 immobility during cytokinesis. In contrast, Myo1 tail, especially a 105-aa region within the putative assembly domain (Fang et al., 2010), is required for this process. Thus, we speculate that Myo1 might form bipolar filaments during cytokinesis, which accounts for its immobility as well as maximal maintenance at the division site. Indeed, cell cycle–regulated filament assembly is associated with myosin-II immobility during cytokinesis in Drosophila (Uehara et al., 2010). In this case, when the RLC is locked in a state promoting myosin filament assembly, myosin-II becomes much more immobile even in metaphase, when it is usually highly dynamic (Uehara et al., 2010). Unfortunately, the role of the assembly domain in regulating myosin-II immobility during cytokinesis could not be examined in Drosophila, as myosin-II lacking this domain fails to accumulate at the division site (Uehara et al., 2010). However, it has been demonstrated that the assembly domain dictates isoform-specific myosin-II dynamics in migrating mammalian cells (Sandquist and Means, 2008). Collectively, it is likely that cell cycle–regulated Myo1 filament assembly accounts for its immobility during cytokinesis.
Myo1 plays a scaffolding role during cytokinesis
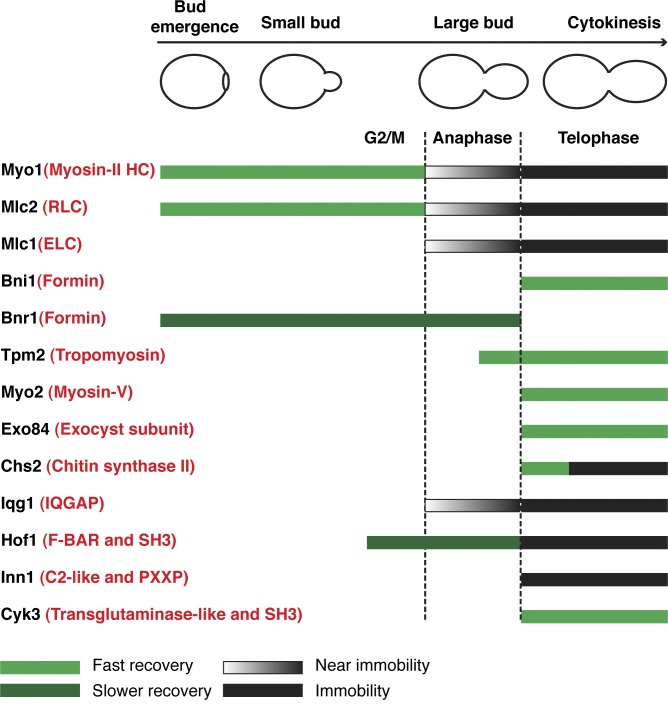
How does the cell build a division machine? To address this question, we analyzed the dynamics of 13 cytokinesis proteins in budding yeast during the cell cycle under the same experimental conditions (Fig. 9). Different cytokinesis proteins display distinct and functionally relevant dynamics, ranging from rapid turnover to complete immobility. The AMR displays a dichotomy in dynamics, with myosin-II (myosin heavy chain, RLC, and ELC) being immobile and actin ring–associated components (formin and tropomyosin) being dynamic. Actin and actin-associated proteins are also dynamic during cytokinesis in fission yeast (Pelham and Chang, 2002) and cultured mammalian cells (Guha et al., 2005; Murthy and Wadsworth, 2005). As expected, components of the membrane trafficking machine, such as myosin-V and the exocyst, are dynamic during cytokinesis. Surprisingly, the secretory cargo Chs2 displays a unique biphasic behavior during cytokinesis, showing myosin-V–like dynamics during its initial localization to the division site, followed by myosin-II–like dynamics during the rest of cytokinesis, suggesting that Chs2, after its full delivery to the division site, is immobilized onto myosin-II for PS formation. This strongly supports the hypothesis that the AMR guides PS formation in yeast (or localized ECM remodeling in animal cells) during cytokinesis (Fang et al., 2010). Like Myo1, proteins involved in PS formation or its coordination with the AMR, such as ELC, IQGAP, F-BAR, and Inn1, except Cyk3, are immobile during cytokinesis regardless of their dynamic state early in the cell cycle.
Figure 9.
Localization and dynamics of cytokinesis proteins during the yeast cell cycle. Straight bars indicate the duration of the cytokinesis proteins at the mother–bud neck during the cell cycle. Green bars indicate rapid recovery caused by cytosol–neck exchange or lateral movement within the ring structure. Dark green bars (for Bnr1 and Hof1 before cytokinesis) indicate less recovery. Black gradient bars indicate near-immobile behaviors. Black bars indicate immobility. Vertical dashed lines indicate the onset of anaphase and telophase, respectively. The generic names of yeast proteins are highlighted in red in parentheses.
Taking the advantage of yeast genetics and combining it with FRAP analysis, we found that the immobility of all other cytokinesis proteins depends on Myo1, but Myo1 immobility does not depend on other proteins. These findings and the previous observation that Myo1 is required for actin ring assembly (Bi et al., 1998) suggest that Myo1 plays a scaffolding role during cytokinesis. This novel role of Myo1 may define a general principle for the assembly of a division machine in other systems, as the core components and the mechanisms of cytokinesis are conserved from yeast to humans.
Materials and methods
Strains and growth conditions
Yeast strains used in this study are listed in Table 1. Standard culture media and genetic methods were used throughout this study (Guthrie and Fink, 1991). For LatA experiments, cells were grown in synthetic complete (SC) media at 23°C to exponential phase and then treated with 200 µM LatA (Wako Chemicals USA) or DMSO (solvent in which LatA was dissolved) for 10 min before being subjected to time-lapse and FRAP analysis.
Table 1.
Yeast strains used in this study
| Strain | Genotype | Reference or source |
| YEF473 | a/α his3/his3 leu2/leu2 lys2/lys2 trp1/trp1 ura3/ura3 | Bi and Pringle, 1996 |
| YEF473A | a his3 leu2 lys2 trp1 ura3 | Bi and Pringle, 1996 |
| YEF473B | α his3 leu2 lys2 trp1 ura3 | Bi and Pringle, 1996 |
| YEF5804 | As YEF473A, except CDC3-mCherry:LEU2 | This study |
| YEF5862 | As YEF5804, except EXO84-GFP:URA3 | This study |
| YEF5874 | As YEF5804, except CHS2-GFP:HIS3 | This study |
| YEF5986 | As YEF5804, except MYO2-ARG-GFP:HIS3 | This study |
| XDY286 | As YEF473A, except MYO1-GFP CDC3-mCherry:LEU2 | Fang et al., 2010 |
| XDY288 | As YEF473A, except myo1-Tail-GFP CDC3-mCherry:LEU2 | Fang et al., 2010 |
| YEF6001 | As YEF5986, except (pRS316-MYO1-mCherry) | This study |
| YEF6036 | As XDY286, except NUP57-mCherry:His3MX6 | This study |
| YEF6065 | a ade2-1 ura3-52 his3 leu2-3, 112, trp1-1 can1-100 CDC3-mCherry:LEU2 (pUG34-MLC1) | This study |
| YEF6069 | As YEF473A, except MLC2-GFP:KanMX6 CDC3-mCherry:LEU2 | This study |
| YEF6116 | As YEF473, except bni1Δ::HIS3/bni1Δ::HIS3 MYO1-GFP:KanMX6/MYO1-GFP:KanMX6 CDC3-mCherry:LEU2/CDC3 | This study |
| YEF6130 | As YEF473, except CYK3-GFP:KanMX6/CYK3-GFP:KanMX6 CDC3-mCherry:LEU2/CDC3 | This study |
| YEF6131 | As YEF473, except HOF1-GFP:KanMX6/HOF1-GFP:KanMX6 CDC3-mCherry:LEU2/CDC3 | This study |
| YEF6134 | As YEF473, except CDC3-mCherry:TRP1/CDC3 (YEp13-BNI1-GFP) | This study |
| YEF6135 | As YEF473, except CDC3-mCherry:LEU2/CDC3 (pUG23-BNR1) | This study |
| YEF6138 | As YEF473, except INN1-GFP:KanMX6/INN1-GFP:KanMX6 CDC3-mCherry:LEU2/CDC3 | This study |
| YEF6140 | As YEF473, except CDC3-mCherry:LEU2/CDC3 (pUG35-IQG1) | This study |
| YEF6197 | As YEF473A, except TPM2-GFP:TRP1 CDC3-mCherry:LEU2 | This study |
| YEF6230 | As YEF473A, except inn1Δ::KanMX6 MYO1-GFP:His3MX6 CDC3- mCherry:LEU2 | This study |
| YEF6273 | As YEF473A, except chs2Δ::His3MX6 MYO1-GFP:KanMX6 CDC3- mCherry:TRP1 | This study |
| YEF6336 | As YEF473A, except myo1Δ::KanMX6 CHS2-GFP:KanMX6 CDC3-mCherry:LEU2 | This study |
| YEF6349 | As YEF473A, except myo1Δ::His3MX6 CDC3-mCherry:LEU2 | This study |
| YEF6351 | As YEF473A, except myo1Δ::KanMX6 CDC3-mCherry:LEU2 (pUG34-MLC1) | This study |
| YEF6356 | As YEF6349, except (pUG35-IQG1) | This study |
| YEF6357 | As YEF6349, except INN1-GFP:KanMX6 | This study |
| YEF6358 | As YEF6349, except HOF1-GFP:KanMX6 | This study |
| YEF6616 | As YEF473, except GFP-myo1-(AA1798Stop):KanMX6/GFP-myo1-(AA1798Stop):KanMX6 CDC3-mCherry:LEU2/CDC3 | This study |
| YEF6617 | As YEF473, except myo1-(AA1903Stop)-GFP/myo1-(AA1903Stop)-GFP CDC3-mCherry:LEU2/CDC3 | This study |
| YEF6618 | As YEF473, except GFP-MYO1/GFP-MYO1 CDC3-mCherry:LEU2/CDC3 | This study |
| YEF6769 | As YEF473B, except myo1-(AA1798Stop):His3MX6 HOF1-GFP:KanMX6 CDC3-mCherry:TRP1 | This study |
| YEF6771 | As YEF473B, except myo1-(AA1798Stop):His3MX6 CHS2-GFP:KanMX6 CDC3-mCherry:TRP1 | This study |
| YEF6899 | As YEF473A, except CDC3-mCherry:LEU2 BNI5-GFP:KanMX6 | This study |
| YEF6904 | As YEF473A, except myo1Δ::His3MX6 CDC3-mCherry:LEU2 BNI5-GFP:KanMX6 | This study |
Construction of plasmids and yeast strains
All primers were purchased from Integrated DNA Technologies. Sequencing of constructs was performed at the DNA Sequencing Facility, University of Pennsylvania. Plasmid YIp128-CDC3-mCherry (integrative, LEU2) carries N-terminally mCherry-tagged CDC3 under the control of its own promoter (Gao et al., 2007). Plasmid YIp204-CDC3-mCherry (integrative, TRP1) was constructed by subcloning a 5.3-kb SalI–EcoRI fragment carrying mCherry-CDC3 from YIp128-CDC3-mCherry into YIpalc204 (Gietz and Sugino, 1988). Plasmid pRS316-MYO1-mCherry (CEN, URA3) carries C-terminally mCherry-tagged MYO1 under the control of its own promoter (Fang et al., 2010). Plasmids pUG34-MLC1 (CEN, HIS3) and pUG35-IQG1 (CEN, URA3) carry N-terminally GFP-tagged MLC1 and C-terminally GFP-tagged IQG1 under the control of the MET25 promoter, respectively (provided by A. Ragnini-Wilson, Tor Vergata University of Rome, Rome, Italy; Wagner et al., 2002). Plasmid pUG23-BNR1 (CEN, HIS3) was constructed by gap repairing the BNR1 ORF into EcoRI-digested pUG23 (supplied by J.H. Hegemann, Heinrich-Heine-Universität Düsseldorf, Düsseldorf, Germany), resulting in C-terminally GFP-tagged BNR1 under the control of the MET25 promoter. Plasmid YEp13-BNI1-GFP (2μ, LEU2), carrying C-terminally GFP-tagged BNI1 under the control of its own promoter, was constructed by a PCR-based method (Longtine et al., 1998). The PCR products were generated using the plasmid pFA6a-GFP-KanMX6 (Longtine et al., 1998) as the template DNA and a pair of primers composed of sequences flanking the stop codon of BNI1 and then transformed into a yeast strain carrying the plasmid YEp13-BNI1, which was isolated from a YEp13-based genomic library. Plasmids NRB884 (integrative, URA3) and pG1331 (integrative, TRP1) carry the 3′ region of EXO84 and TPM2 with a GFP inserted in frame after their last codon, respectively (W. Guo, University of Pennsylvania, Philadelphia, PA). These plasmids were digested with BglII and integrated at the EXO84 and TPM2 loci, respectively.
Diploid strains carrying a specific truncation allele of MYO1 were constructed as follows. Plasmid pRS316-N-MYO1-GFP, which carries an N-terminally GFP-tagged MYO1 (GFP inserted immediately after the start codon; Caviston et al., 2003), was digested with ClaI and SalI and then transformed into Masa1243 (a myo1Δ::URA3-KanMX6 [pUG23-MYO1]; Fang et al., 2010). After a few selective steps (Fang et al., 2010), a haploid strain (Masa1284) carrying the GFP-tagged MYO1 allele in precise replacement of the endogenous MYO1 was generated. PCR products that had been amplified from pFA6a-KanMX6 using pairs of F3 and R1 primers (Longtine et al., 1998) were transformed into Masa1284 to generate haploid strains carrying truncated alleles of MYO1 that were marked with KanMX6. F3 primers consist of sequences upstream of the desired truncation site (codons 1,483, 1,535, 1,633, 1,729, 1,798, or 1,903) in MYO1, including a stop codon. The haploid strains were crossed with a wild-type strain of opposite mating type to yield diploid strains, which were then sporulated, and tetrads were dissected to select for haploid strains of opposite mating types carrying a specific truncation allele of MYO1 (GFP-myo1*:KanMX6). These haploid strains were then crossed to generate diploid strains homozygous for a specific truncation allele. Except for strain YEF6771 in which CHS2-GFP:KanMX6 was PCR-amplified from the template plasmid pFA6a-GFP-KanMX6 (Longtine et al., 1998), all other yeast strains were made by transferring gene deletions or tagged alleles of genes from one strain to another using PCR-based approach coupled with standard yeast genetics. All strains carrying CDC3-RFP were constructed by digesting plasmid YIp128-CDC3-mCherry or YIp204-CDC3-mCherry with BglII and integrating it at the CDC3 locus of the respective strains.
Live-cell imaging and quantitative analysis
For all the FRAP and FLIP experiments, cells were grown at 23°C to exponential phase in SC media with selection for the presence of specific plasmids carried in the yeast strain. Cells were concentrated by centrifugation, spotted on SC-dropout media containing 2% agarose, and sealed with nail polish. Images were acquired at 23°C on a spinning-disk confocal microscope equipped with a scanhead (CSU10; Yokogawa Corporation of America) combined with a microscope (IX71; Olympus) and a 100× objective (1.4 NA, Plan S-Apochromat oil immersion; Olympus). Acquisition and hardware were controlled by MetaMorph version 7.7 (Molecular Devices). An electron multiplying charge-coupled device camera (model C9100-13; ImagEM; Hamamatsu Photonics) was used for capture. Diode lasers for excitation (488 nm for GFP and 561 nm for mCherry/RFP) were housed in a launch constructed by Spectral Applied Research. FRAP was performed using a computer-controlled ablation system (MicroPoint; Photonic Instruments) consisting of a nitrogen-pumped dye laser (wavelength of 435 nm) controlled by MetaMorph. Images were taken every 1 or 3 s (no z stack) or 10–30 s (with a z stack consisting of 11 × 0.3–µm steps). Whenever feasible, a maximum projection (as the best adaptation to cell movement) was created with MetaMorph version 7.7, and quantification was performed with ImageJ (National Institutes of Health), drawing a respective polygon on the region of interest to yield the integrated density for the region. This integrated density was used in Prism Version 5 (GraphPad Software) to create plots and, whenever feasible, to calculate the half-time (t1/2) (equal to recovery rate) and a maximum recovery using the one-phase association function Y = Y0 + (plateau − Y0) × (1 − exp (−K × x)) in Prism, in which K is the rate constant, x is the time (in seconds), Y0 is the intensity (in percent) right after the bleach (time = 0), and plateau is the intensity after which no more recovery occurred. t1/2 was calculated as ln(2)/K, similar to that described before (Lister et al., 2006), and the maximal recovery was calculated as plateau − Y0. It is worth pointing out that all the t1/2 values presented in Figs. S3 and S4 were calculated based on individual recovery curves, and then, the mean and the SEM were determined.
Unless noted otherwise, the following description applies to the entire study. First, all proteins were GFP-tagged at their C termini and expressed from their native promoters at their endogenous loci and were the only copy of the gene present in the cell. Second, for clear and accurate description of the FRAP data, both the quantification and the kymograph of the same cell are presented in a single plot. The number of cells examined and the range of their behaviors are described in the text and/or summarized in Figs. 9, S3, and S4. Finally, bud size, which increases with nuclear cycle progression, and septin–hourglass splitting (septin structures were marked by mCherry-labeled Cdc3), which is controlled by the mitotic exit network and coincides with the onset of cytokinesis (Cid et al., 2001; Lippincott et al., 2001), were used as crude and precise cell cycle markers, respectively. Using the combination of these markers, three cell cycle stages were defined: small bud refers to cells with a daughter volume ∼20% of its mother; large bud refers to cells with a daughter volume ∼40–60% of its mother without septin–hourglass splitting during the entire period of image acquisition; and cytokinesis refers to cells with split septin rings at the point of bleaching.
Detection of truncated Myo1-GFP proteins by Western blotting
Overnight cultures of yeast strains were diluted into 50 ml YM-P rich medium (Lillie and Pringle, 1980) and grown to an OD600 of ∼1.00 at 23°C. Cells were pelleted by centrifugation and then resuspended in PBS containing 0.1% NP-40 and a cocktail of protease inhibitors (Complete; Roche). Cells were lysed by using acid-washed glass beads in a bead beater for six cycles of 30-s beating and 1-min chilling on ice. Cell lysates were centrifuged, and 200-µl supernatants were mixed with 50 µl of 6× sample buffer and boiled for 10 min. After centrifugation, the supernatants were subjected to SDS-PAGE. After transfer to a polyvinylidene difluoride membrane (Immobilon-P; EMD Millipore), the membrane was cut in the middle for separate incubation with anti-GFP and -Cdc11 antibodies. The primary antibodies used for Western blotting were a mouse monoclonal antibody against GFP (catalogue no. MMS-118P and lot no. 14812801; used at 1:7,500 dilution; Covance) and a rabbit polyclonal antibody against yeast Cdc11 (y-415; catalogue no. sc-7170 and lot no. C0804; used at 1:30,000 dilution; Santa Cruz Biotechnology, Inc.). The secondary antibodies were goat anti–mouse IgG (used at 1:7,500 dilution) and goat anti–rabbit IgG (used at 1:20,000 dilution) conjugated to HRP (Jackson ImmunoResearch Laboratories, Inc.). Proteins were detected by using chemiluminescent HRP substrate obtained from EMD Millipore (Immobilon).
Online supplemental material
Fig. S1 shows the full-ring bleaching of Myo1-GFP at the division site during the cell cycle, FLIP analysis of Myo1-GFP and the control Tpm2-GFP, and Myo1-GFP intensity at the bud neck during the cell cycle. Fig. S2 shows the FRAP analyses of Mlc2-GFP, Mlc1-GFP (at the bud cortex), Bni5-GFP, Bnr1-GFP, Bni1-GFP, and Cyk3-GFP during the cell cycle. Fig. S3 shows individual recovery curves and quantitative analyses for the indicated proteins. Fig. S4 shows individual curves and quantitative analyses for the indicated proteins. Videos 1 and 2 show the full-ring and half-ring bleaching of Myo1-GFP at different phases of the cell cycle, respectively. Video 3 shows the half-ring and full-ring bleaching of Bni5-GFP in wild-type and myo1Δ cells before the onset of cytokinesis and also the half-ring bleaching of Myo1-GFP in bni1Δ and LatA-treated cells. Video 4 shows the half-ring bleaching of Myo1-Tail-GFP during the cell cycle. Video 5 shows the half-ring bleaching of Myo1-(AA1903Stop) and Myo1-(AA1798Stop) during cytokinesis. Video 6 shows the full-ring bleaching of Tpm2-GFP, Myo2-GFP, and Exo84-GFP during cytokinesis. Video 7 shows the half-ring bleaching of Chs2-GFP in wild-type and myo1Δ cells and of Myo1-GFP in chs2Δ and inn1Δ cells. Video 8 shows the half-ring and/or full-ring bleaching of Mlc1-GFP and Iqg1-GFP in wild-type and myo1Δ cells during cytokinesis. Video 9 shows the half-ring and full-ring bleaching of Inn1-GFP and Hof1-GFP in wild-type and myo1Δ cells during cytokinesis. Video 10 shows the half-ring bleaching of Chs2-GFP and Hof1-GFP in cells carrying the myo1-(AA1798Stop) allele. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201208030/DC1.
Supplementary Material
Acknowledgments
We thank C. Palmer, K. Bass, S. Tcheperegine, E. Falls, J. Schreiter, and E. Chen for constructing plasmids and yeast strains; Drs. W. Guo, A. Ragnini-Wilson, J. Hegemann, J.R. Pringle, and K. Tanaka for plasmids and yeast strains; Dr. A. Stout and J. Hanna for technical assistance; Dr. S. Okada for valuable discussion; and Drs. R. Nishihama, P. Tran, and M. Kraub for critically reading the manuscript.
This work was supported by the National Institutes of Health grants GM59216 and GM87365 to E. Bi and R15GM065883 to E. Vallen, a Howard Hughes Medical Institute grant to Swarthmore College, and a Ph.D. fellowship from the Boehringer Ingelheim Fonds to C. Wloka.
Footnotes
Abbreviations used in this paper:
- AMR
- actomyosin ring
- ELC
- essential light chain
- FLIP
- fluorescence loss in photobleaching
- LatA
- latrunculin A
- PM
- plasma membrane
- PS
- primary septum
- RLC
- regulatory light chain
- SC
- synthetic complete
References
- Alber F., Dokudovskaya S., Veenhoff L.M., Zhang W., Kipper J., Devos D., Suprapto A., Karni-Schmidt O., Williams R., Chait B.T., et al. 2007. The molecular architecture of the nuclear pore complex. Nature. 450:695–701 10.1038/nature06405 [DOI] [PubMed] [Google Scholar]
- Ayscough K.R., Stryker J., Pokala N., Sanders M., Crews P., Drubin D.G. 1997. High rates of actin filament turnover in budding yeast and roles for actin in establishment and maintenance of cell polarity revealed using the actin inhibitor latrunculin-A. J. Cell Biol. 137:399–416 10.1083/jcb.137.2.399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian M.K., Bi E., Glotzer M. 2004. Comparative analysis of cytokinesis in budding yeast, fission yeast and animal cells. Curr. Biol. 14:R806–R818 10.1016/j.cub.2004.09.022 [DOI] [PubMed] [Google Scholar]
- Barr F.A., Gruneberg U. 2007. Cytokinesis: placing and making the final cut. Cell. 131:847–860 10.1016/j.cell.2007.11.011 [DOI] [PubMed] [Google Scholar]
- Bi E. 2001. Cytokinesis in budding yeast: the relationship between actomyosin ring function and septum formation. Cell Struct. Funct. 26:529–537 10.1247/csf.26.529 [DOI] [PubMed] [Google Scholar]
- Bi E. 2010. Lord of the ring. Nat. Rev. Mol. Cell Biol. 11:606 10.1038/nrm2953 [DOI] [PubMed] [Google Scholar]
- Bi E., Pringle J.R. 1996. ZDS1 and ZDS2, genes whose products may regulate Cdc42p in Saccharomyces cerevisiae. Mol. Cell. Biol. 16:5264–5275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi E., Maddox P., Lew D.J., Salmon E.D., McMillan J.N., Yeh E., Pringle J.R. 1998. Involvement of an actomyosin contractile ring in Saccharomyces cerevisiae cytokinesis. J. Cell Biol. 142:1301–1312 10.1083/jcb.142.5.1301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd C., Hughes T., Pypaert M., Novick P. 2004. Vesicles carry most exocyst subunits to exocytic sites marked by the remaining two subunits, Sec3p and Exo70p. J. Cell Biol. 167:889–901 10.1083/jcb.200408124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyne J.R., Yosuf H.M., Bieganowski P., Brenner C., Price C. 2000. Yeast myosin light chain, Mlc1p, interacts with both IQGAP and class II myosin to effect cytokinesis. J. Cell Sci. 113:4533–4543 [DOI] [PubMed] [Google Scholar]
- Bretscher A. 2003. Polarized growth and organelle segregation in yeast: the tracks, motors, and receptors. J. Cell Biol. 160:811–816 10.1083/jcb.200301035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buttery S.M., Yoshida S., Pellman D. 2007. Yeast formins Bni1 and Bnr1 utilize different modes of cortical interaction during the assembly of actin cables. Mol. Biol. Cell. 18:1826–1838 10.1091/mbc.E06-09-0820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho A., Desai A., Oegema K. 2009. Structural memory in the contractile ring makes the duration of cytokinesis independent of cell size. Cell. 137:926–937 10.1016/j.cell.2009.03.021 [DOI] [PubMed] [Google Scholar]
- Caviston J.P., Longtine M., Pringle J.R., Bi E. 2003. The role of Cdc42p GTPase-activating proteins in assembly of the septin ring in yeast. Mol. Biol. Cell. 14:4051–4066 10.1091/mbc.E03-04-0247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang J.S., Schekman R.W. 1996. Differential trafficking and timed localization of two chitin synthase proteins, Chs2p and Chs3p. J. Cell Biol. 135:597–610 10.1083/jcb.135.3.597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cid V.J., Adamiková L., Sánchez M., Molina M., Nombela C. 2001. Cell cycle control of septin ring dynamics in the budding yeast. Microbiology. 147:1437–1450 [DOI] [PubMed] [Google Scholar]
- Clifford D.M., Wolfe B.A., Roberts-Galbraith R.H., McDonald W.H., Yates J.R., III, Gould K.L. 2008. The Clp1/Cdc14 phosphatase contributes to the robustness of cytokinesis by association with anillin-related Mid1. J. Cell Biol. 181:79–88 10.1083/jcb.200709060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobbelaere J., Barral Y. 2004. Spatial coordination of cytokinetic events by compartmentalization of the cell cortex. Science. 305:393–396 10.1126/science.1099892 [DOI] [PubMed] [Google Scholar]
- Doyle T., Botstein D. 1996. Movement of yeast cortical actin cytoskeleton visualized in vivo. Proc. Natl. Acad. Sci. USA. 93:3886–3891 10.1073/pnas.93.9.3886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggert U.S., Mitchison T.J., Field C.M. 2006. Animal cytokinesis: from parts list to mechanisms. Annu. Rev. Biochem. 75:543–566 10.1146/annurev.biochem.74.082803.133425 [DOI] [PubMed] [Google Scholar]
- Epp J.A., Chant J. 1997. An IQGAP-related protein controls actin-ring formation and cytokinesis in yeast. Curr. Biol. 7:921–929 10.1016/S0960-9822(06)00411-8 [DOI] [PubMed] [Google Scholar]
- Fang X., Luo J., Nishihama R., Wloka C., Dravis C., Travaglia M., Iwase M., Vallen E.A., Bi E. 2010. Biphasic targeting and cleavage furrow ingression directed by the tail of a myosin II. J. Cell Biol. 191:1333–1350 10.1083/jcb.201005134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X.D., Sperber L.M., Kane S.A., Tong Z., Tong A.H., Boone C., Bi E. 2007. Sequential and distinct roles of the cadherin domain-containing protein Axl2p in cell polarization in yeast cell cycle. Mol. Biol. Cell. 18:2542–2560 10.1091/mbc.E06-09-0822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz R.D., Sugino A. 1988. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 74:527–534 10.1016/0378-1119(88)90185-0 [DOI] [PubMed] [Google Scholar]
- Goldbach P., Wong R., Beise N., Sarpal R., Trimble W.S., Brill J.A. 2010. Stabilization of the actomyosin ring enables spermatocyte cytokinesis in Drosophila. Mol. Biol. Cell. 21:1482–1493 10.1091/mbc.E09-08-0714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guha M., Zhou M., Wang Y.L. 2005. Cortical actin turnover during cytokinesis requires myosin II. Curr. Biol. 15:732–736 10.1016/j.cub.2005.03.042 [DOI] [PubMed] [Google Scholar]
- Guo W., Sacher M., Barrowman J., Ferro-Novick S., Novick P. 2000. Protein complexes in transport vesicle targeting. Trends Cell Biol. 10:251–255 10.1016/S0962-8924(00)01754-2 [DOI] [PubMed] [Google Scholar]
- Guthrie C., Fink G.R., 1991. Guide to yeast genetics and molecular biology. Methods in Enzymology, Vol. 194 San Diego: Academic Press [Google Scholar]
- Huxley H.E. 1969. The mechanism of muscular contraction. Science. 164:1356–1365 10.1126/science.164.3886.1356 [DOI] [PubMed] [Google Scholar]
- Izumikawa T., Kanagawa N., Watamoto Y., Okada M., Saeki M., Sakano M., Sugahara K., Sugihara K., Asano M., Kitagawa H. 2010. Impairment of embryonic cell division and glycosaminoglycan biosynthesis in glucuronyltransferase-I-deficient mice. J. Biol. Chem. 285:12190–12196 10.1074/jbc.M110.100941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamasaki T., Osumi M., Mabuchi I. 2007. Three-dimensional arrangement of F-actin in the contractile ring of fission yeast. J. Cell Biol. 178:765–771 10.1083/jcb.200612018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamei T., Tanaka K., Hihara T., Umikawa M., Imamura H., Kikyo M., Ozaki K., Takai Y. 1998. Interaction of Bnr1p with a novel Src homology 3 domain-containing Hof1p. Implication in cytokinesis in Saccharomyces cerevisiae. J. Biol. Chem. 273:28341–28345 10.1074/jbc.273.43.28341 [DOI] [PubMed] [Google Scholar]
- Kondo T., Hamao K., Kamijo K., Kimura H., Morita M., Takahashi M., Hosoya H. 2011. Enhancement of myosin II/actin turnover at the contractile ring induces slower furrowing in dividing HeLa cells. Biochem. J. 435:569–576 10.1042/BJ20100837 [DOI] [PubMed] [Google Scholar]
- Korinek W.S., Bi E., Epp J.A., Wang L., Ho J., Chant J. 2000. Cyk3, a novel SH3-domain protein, affects cytokinesis in yeast. Curr. Biol. 10:947–950 10.1016/S0960-9822(00)00626-6 [DOI] [PubMed] [Google Scholar]
- Lee P.R., Song S., Ro H.S., Park C.J., Lippincott J., Li R., Pringle J.R., De Virgilio C., Longtine M.S., Lee K.S. 2002. Bni5p, a septin-interacting protein, is required for normal septin function and cytokinesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 22:6906–6920 10.1128/MCB.22.19.6906-6920.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lillie S.H., Pringle J.R. 1980. Reserve carbohydrate metabolism in Saccharomyces cerevisiae: responses to nutrient limitation. J. Bacteriol. 143:1384–1394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippincott J., Li R. 1998a. Sequential assembly of myosin II, an IQGAP-like protein, and filamentous actin to a ring structure involved in budding yeast cytokinesis. J. Cell Biol. 140:355–366 10.1083/jcb.140.2.355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippincott J., Li R. 1998b. Dual function of Cyk2, a cdc15/PSTPIP family protein, in regulating actomyosin ring dynamics and septin distribution. J. Cell Biol. 143:1947–1960 10.1083/jcb.143.7.1947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippincott J., Shannon K.B., Shou W., Deshaies R.J., Li R. 2001. The Tem1 small GTPase controls actomyosin and septin dynamics during cytokinesis. J. Cell Sci. 114:1379–1386 [DOI] [PubMed] [Google Scholar]
- Lister I.M., Tolliday N.J., Li R. 2006. Characterization of the minimum domain required for targeting budding yeast myosin II to the site of cell division. BMC Biol. 4:19 10.1186/1741-7007-4-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M.S., McKenzie A., III, Demarini D.J., Shah N.G., Wach A., Brachat A., Philippsen P., Pringle J.R. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 14:953–961 [DOI] [PubMed] [Google Scholar]
- Lord M., Laves E., Pollard T.D. 2005. Cytokinesis depends on the motor domains of myosin-II in fission yeast but not in budding yeast. Mol. Biol. Cell. 16:5346–5355 10.1091/mbc.E05-07-0601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J., Vallen E.A., Dravis C., Tcheperegine S.E., Drees B.L., Bi E. 2004. Identification and functional analysis of the essential and regulatory light chains of the only type II myosin Myo1p in Saccharomyces cerevisiae. J. Cell Biol. 165:843–855 10.1083/jcb.200401040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maupin P., Pollard T.D. 1986. Arrangement of actin filaments and myosin-like filaments in the contractile ring and of actin-like filaments in the mitotic spindle of dividing HeLa cells. J. Ultrastruct. Mol. Struct. Res. 94:92–103 10.1016/0889-1605(86)90055-8 [DOI] [PubMed] [Google Scholar]
- Meitinger F., Petrova B., Lombardi I.M., Bertazzi D.T., Hub B., Zentgraf H., Pereira G. 2010. Targeted localization of Inn1, Cyk3 and Chs2 by the mitotic-exit network regulates cytokinesis in budding yeast. J. Cell Sci. 123:1851–1861 10.1242/jcs.063891 [DOI] [PubMed] [Google Scholar]
- Mendes Pinto I., Rubinstein B., Kucharavy A., Unruh J.R., Li R. 2012. Actin depolymerization drives actomyosin ring contraction during budding yeast cytokinesis. Dev. Cell. 22:1247–1260 10.1016/j.devcel.2012.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuguchi S., Uyama T., Kitagawa H., Nomura K.H., Dejima K., Gengyo-Ando K., Mitani S., Sugahara K., Nomura K. 2003. Chondroitin proteoglycans are involved in cell division of Caenorhabditis elegans. Nature. 423:443–448 10.1038/nature01635 [DOI] [PubMed] [Google Scholar]
- Moseley J.B., Goode B.L. 2006. The yeast actin cytoskeleton: from cellular function to biochemical mechanism. Microbiol. Mol. Biol. Rev. 70:605–645 10.1128/MMBR.00013-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murthy K., Wadsworth P. 2005. Myosin-II-dependent localization and dynamics of F-actin during cytokinesis. Curr. Biol. 15:724–731 10.1016/j.cub.2005.02.055 [DOI] [PubMed] [Google Scholar]
- Nishihama R., Schreiter J.H., Onishi M., Vallen E.A., Hanna J., Moravcevic K., Lippincott M.F., Han H., Lemmon M.A., Pringle J.R., Bi E. 2009. Role of Inn1 and its interactions with Hof1 and Cyk3 in promoting cleavage furrow and septum formation in S. cerevisiae. J. Cell Biol. 185:995–1012 10.1083/jcb.200903125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelham R.J., Chang F. 2002. Actin dynamics in the contractile ring during cytokinesis in fission yeast. Nature. 419:82–86 10.1038/nature00999 [DOI] [PubMed] [Google Scholar]
- Pollard T.D. 2008. Progress towards understanding the mechanism of cytokinesis in fission yeast. Biochem. Soc. Trans. 36:425–430 10.1042/BST0360425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruyne D., Evangelista M., Yang C., Bi E., Zigmond S., Bretscher A., Boone C. 2002. Role of formins in actin assembly: nucleation and barbed-end association. Science. 297:612–615 10.1126/science.1072309 [DOI] [PubMed] [Google Scholar]
- Pruyne D., Gao L., Bi E., Bretscher A. 2004. Stable and dynamic axes of polarity use distinct formin isoforms in budding yeast. Mol. Biol. Cell. 15:4971–4989 10.1091/mbc.E04-04-0296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagot I., Rodal A.A., Moseley J., Goode B.L., Pellman D. 2002. An actin nucleation mechanism mediated by Bni1 and profilin. Nat. Cell Biol. 4:626–631 [DOI] [PubMed] [Google Scholar]
- Sanchez-Diaz A., Marchesi V., Murray S., Jones R., Pereira G., Edmondson R., Allen T., Labib K. 2008. Inn1 couples contraction of the actomyosin ring to membrane ingression during cytokinesis in budding yeast. Nat. Cell Biol. 10:395–406 10.1038/ncb1701 [DOI] [PubMed] [Google Scholar]
- Sandquist J.C., Means A.R. 2008. The C-terminal tail region of nonmuscle myosin II directs isoform-specific distribution in migrating cells. Mol. Biol. Cell. 19:5156–5167 10.1091/mbc.E08-05-0533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger J.M., Sanger J.W. 1980. Banding and polarity of actin filaments in interphase and cleaving cells. J. Cell Biol. 86:568–575 10.1083/jcb.86.2.568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satterwhite L.L., Pollard T.D. 1992. Cytokinesis. Curr. Opin. Cell Biol. 4:43–52 10.1016/0955-0674(92)90057-J [DOI] [PubMed] [Google Scholar]
- Sburlati A., Cabib E. 1986. Chitin synthetase 2, a presumptive participant in septum formation in Saccharomyces cerevisiae. J. Biol. Chem. 261:15147–15152 [PubMed] [Google Scholar]
- Schmidt M., Bowers B., Varma A., Roh D.-H., Cabib E. 2002. In budding yeast, contraction of the actomyosin ring and formation of the primary septum at cytokinesis depend on each other. J. Cell Sci. 115:293–302 [DOI] [PubMed] [Google Scholar]
- Schroeder T.E. 1972. The contractile ring. II. Determining its brief existence, volumetric changes, and vital role in cleaving Arbacia eggs. J. Cell Biol. 53:419–434 10.1083/jcb.53.2.419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon K.B., Li R. 2000. A myosin light chain mediates the localization of the budding yeast IQGAP-like protein during contractile ring formation. Curr. Biol. 10:727–730 10.1016/S0960-9822(00)00539-X [DOI] [PubMed] [Google Scholar]
- Stevens R.C., Davis T.N. 1998. Mlc1p is a light chain for the unconventional myosin Myo2p in Saccharomyces cerevisiae. J. Cell Biol. 142:711–722 10.1083/jcb.142.3.711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strickland L.I., Burgess D.R. 2004. Pathways for membrane trafficking during cytokinesis. Trends Cell Biol. 14:115–118 10.1016/j.tcb.2004.01.006 [DOI] [PubMed] [Google Scholar]
- Tan J.L., Ravid S., Spudich J.A. 1992. Control of nonmuscle myosins by phosphorylation. Annu. Rev. Biochem. 61:721–759 10.1146/annurev.bi.61.070192.003445 [DOI] [PubMed] [Google Scholar]
- Tolliday N., VerPlank L., Li R. 2002. Rho1 directs formin-mediated actin ring assembly during budding yeast cytokinesis. Curr. Biol. 12:1864–1870 10.1016/S0960-9822(02)01238-1 [DOI] [PubMed] [Google Scholar]
- Trybus K.M. 1991. Assembly of cytoplasmic and smooth muscle myosins. Curr. Opin. Cell Biol. 3:105–111 10.1016/0955-0674(91)90172-U [DOI] [PubMed] [Google Scholar]
- Tully G.H., Nishihama R., Pringle J.R., Morgan D.O. 2009. The anaphase-promoting complex promotes actomyosin-ring disassembly during cytokinesis in yeast. Mol. Biol. Cell. 20:1201–1212 10.1091/mbc.E08-08-0822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehara R., Goshima G., Mabuchi I., Vale R.D., Spudich J.A., Griffis E.R. 2010. Determinants of myosin II cortical localization during cytokinesis. Curr. Biol. 20:1080–1085 10.1016/j.cub.2010.04.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallen E.A., Caviston J., Bi E. 2000. Roles of Hof1p, Bni1p, Bnr1p, and myo1p in cytokinesis in Saccharomyces cerevisiae. Mol. Biol. Cell. 11:593–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- VerPlank L., Li R. 2005. Cell cycle-regulated trafficking of Chs2 controls actomyosin ring stability during cytokinesis. Mol. Biol. Cell. 16:2529–2543 10.1091/mbc.E04-12-1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner W., Bielli P., Wacha S., Ragnini-Wilson A. 2002. Mlc1p promotes septum closure during cytokinesis via the IQ motifs of the vesicle motor Myo2p. EMBO J. 21:6397–6408 10.1093/emboj/cdf650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wloka C., Nishihama R., Onishi M., Oh Y., Hanna J., Pringle J.R., Krauss M., Bi E. 2011. Evidence that a septin diffusion barrier is dispensable for cytokinesis in budding yeast. Biol. Chem. 392:813–829 10.1515/BC.2011.083 [DOI] [PubMed] [Google Scholar]
- Yang S.S., Yeh E., Salmon E.D., Bloom K. 1997. Identification of a mid-anaphase checkpoint in budding yeast. J. Cell Biol. 136:345–354 10.1083/jcb.136.2.345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida S., Kono K., Lowery D.M., Bartolini S., Yaffe M.B., Ohya Y., Pellman D. 2006. Polo-like kinase Cdc5 controls the local activation of Rho1 to promote cytokinesis. Science. 313:108–111 10.1126/science.1126747 [DOI] [PubMed] [Google Scholar]
- Yumura S. 2001. Myosin II dynamics and cortical flow during contractile ring formation in Dictyostelium cells. J. Cell Biol. 154:137–146 10.1083/jcb.200011013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yumura S., Ueda M., Sako Y., Kitanishi-Yumura T., Yanagida T. 2008. Multiple mechanisms for accumulation of myosin II filaments at the equator during cytokinesis. Traffic. 9:2089–2099 10.1111/j.1600-0854.2008.00837.x [DOI] [PubMed] [Google Scholar]
- Zhou Q., Kee Y.S., Poirier C.C., Jelinek C., Osborne J., Divi S., Surcel A., Will M.E., Eggert U.S., Müller-Taubenberger A., et al. 2010. 14-3-3 coordinates microtubules, Rac, and myosin II to control cell mechanics and cytokinesis. Curr. Biol. 20:1881–1889 10.1016/j.cub.2010.09.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.