Abstract
To understand the diversity of transcripts in yeast (Saccharomyces cerevisiae) we analyzed the transcriptional landscapes for cells grown under 18 different environmental conditions. Each sample was analyzed using RNA-sequencing, and a total of 670,446,084 uniquely mapped reads and 377,263 poly-adenylated end tags were produced. Consistent with previous studies, we find that the majority of yeast genes are expressed under one or more different conditions. By directly comparing the 5′ and 3′ ends of the transcribed regions, we find extensive differences in transcript ends across many conditions, especially those of stationary phase, growth in grape juice, and salt stimulation, suggesting differential choice of transcription start and stop sites is pervasive in yeast. Relative to the exponential growth condition (i.e., YPAD), transcripts differing at the 5′ ends and 3′ ends are predicted to differ in their annotated start codon in 21 genes and their annotated stop codon in 63 genes. Many (431) upstream open reading frames (uORFs) are found in alternate 5′ ends and are significantly enriched in transcripts produced during the salt response. Mutational analysis of five genes with uORFs revealed that two sets of uORFs increase the expression of a reporter construct, indicating a role in activation which had not been reported previously, whereas two other uORFs decreased expression. In addition, RNA binding protein motifs are statistically enriched for alternate ends under many conditions. Overall, these results demonstrate enormous diversity of transcript ends, and that this heterogeneity is regulated under different environmental conditions. Moreover, transcript end diversity has important biological implications for the regulation of gene expression. In addition, our data also serve as a valuable resource for the scientific community.
Keywords: yeast, RNA-sequencing, environmental conditions, UTRs
Regulation of gene expression occurs on many levels, both transcriptionally and posttranscriptionally. Measuring RNA levels as a proxy for protein levels and protein function is inadequate. In yeast, RNA levels and protein levels do not perfectly correlate, instead having an r2 value of 0.77 (Lee et al. 2011). Thereby, there must be features of messenger RNAs (mRNAs) or the RNA-handling machinery that affects mRNA levels, translational efficiency, and/or protein turnover.
It has been known for some time that untranslated regions (UTRs) play a significant role in the regulation of gene expression (e.g., van der Velden and Thomas 1999). Sequences in UTRs may control translation, subcellular localization, and transcript stability through a variety of mechanisms (e.g., Mignone et al. 2002). The UTR of a transcript often changes through the use of differential transcription start sites [e.g., (Law et al. 2005). These differential transcript ends—referred to as differential ends in this article—may thus have a role in posttranscriptional regulation of gene expression.
The yeast Saccharomyces cerevisiae provides an ideal organism in which to study this phenomenon. A wealth of molecular, genetic, and genomic information exists for S. cerevisiae, much of it assembled and conveniently available at the Saccharomyces Genome Database (Cherry et al. 1998). Although the complete genome sequence has been known since 1996 (Goffeau et al. 1996), information on the exact length of mRNAs has historically been scarce. Several investigators have attempted to provide this information by using cDNA sequencing (Miura et al. 2006), microarrays (David et al. 2006), Illumina-based RNA sequencing (Nagalakshmi et al. 2008), Illumina-based 3′ RNA-Seq (Yoon 2010), and Helicos-based RNA sequencing for 3′ ends (Ozsolak et al. 2010). Furthermore, previous studies in which authors examined alternative UTRs in yeast have only considered cells growing in one condition, with the exception of the studies by Miura et al. and Yoon et al., whereby analysis was performed under two or three different conditions, respectively (Miura et al. 2006; Yoon 2010). Miura et al. found that most genes had multiple 5′ and 3′ ends, but they did not examine how these were regulated under different incubation conditions in the same strain. A large-scale systematic analysis of differential transcript ends under varying environmental conditions and stressors has thus not previously been performed. Our study examined not only which and how many transcripts have alternate forms but also how differential ends vary under a large set of different conditions and how these differential ends affect gene expression.
There has long been evidence that alternative 5′ UTR sequences in mRNAs can have biological relevance by affecting protein synthesis or protein function. Previous authors have observed that transcripts in yeast may vary in length and result in (1) a relative increase in protein level (Law et al. 2005), (2) a decrease in protein level (Law et al. 2005), (3) secretion of a particular protein (Carlson and Botstein 1982), and (4) an entirely different protein from an overlapping out-of-frame open reading fame (ORF) (Guittaut et al. 2001; Coelho et al. 2002; Kumar et al. 2002). Existing knowledge on alternative UTRs is compiled in several reviews, such as (van der Velden and Thomas 1999; Mignone et al. 2002; Landry et al. 2003; Kochetov 2006).
RNA sequencing (RNA-Seq) provides a powerful tool to study alternative UTRs. Previous studies (Nagalakshmi et al. 2008; Yoon 2010; Ozsolak et al. 2010) have used RNA-Seq to study the yeast transcriptome in a few conditions and identified many genes with multiple 3′ transcript boundaries. RNA-Seq provides much better resolution than microarrays (Wang et al. 2009), and differential transcript ends have been identified in RNA-Seq data by examining expression drop-offs (Nagalakshmi et al. 2008); in addition, poly-adenylation in eukaryotic transcripts can be identified in RNA-Seq data as “end tags,” runs of adenines that do not map back to the genome that lie as the ends of “mappable sequences” (see e.g., Wang et al. 2009) for an explanation).
This study generated deep RNA-Seq data from yeast incubated under many different environmental conditions and stresses, selected to activate a broad variety of gene expression patterns and regulatory networks. We focused on differential transcript ends and found that alternative UTRs are pervasive in yeast. Importantly, we also found that the uORFs introduced in at least two genes with longer 5′ ends enhance gene expression.
Materials and Methods
The BY4741 strain used in (Nagalakshmi et al. 2008) was used in this study. RNA-Seq was performed using the protocol developed in (Nagalakshmi et al. 2008), further described in (Nagalakshmi et al. 2010) and (Waern et al. 2011), and using the modifications developed by (Parkhomchuk et al. 2009) to generate strand-specific reads.
Analysis was performed on custom software developed in-house using BowTie (Langmead et al. 2009) to map reads to the S288C reference genome available on SGD, downloaded on May 17, 2010. Python, NumPy, SciPy, and matplotlib were used to further process the data. The software’s source code is available (Saccharomyces Genome Database). Of note, to maximize the information gleaned, unmappable reads were trimmed by four bases from the 3′ end and remapping was attempted; this was done iteratively until only 28 bp remained, at which point the read was considered unmappable. This end trimming typically doubled or more the number of mappable reads.
Expression levels were calculated by adding, for each base pair, the number of times that base pair was encountered in the RNA-Seq data. For example, if 14 reads overlapped a particular base pair, that base pair would have an expression level of 14. The expression level of a gene was calculated as the mean expression level across the annotated ORF; these values were quantile normalized for comparisons between multiple conditions.
The mappability of genes was calculated because if a gene is in or near an area of the genome with high homology to another genomic area, it becomes impossible to assign the genomic origin of an RNA-Seq read stemming from one of the homologous areas. Mappability of genes was determined by creating, for the plus and minus strands, simulated 76-mer and 28-mer reads starting at each base pair in the annotated genome and processing these reads in the pipeline. (Note from above that reads may have been 28, 32, 36, Δ, 76-mers.) Perfectly mappable genes would thus have an expression level of 104, as all 76 of the 76-mer reads and all 28 of the 28-mer reads would have intersected every base pair of the gene. Genes were considered mappable if the mean expression level across the gene was 90% or more of that (94 reads).
Calling 3′ end tags was done as in Nagalakshmi et al. 2008, which is further described in Wang et al. 2009; three or more nongenomic A bases were required for each tag.
For a full explanation of the end-calling algorithm, refer to the software source code. In summary, for each annotated ORF the log2 expression levels for YPAD exponential growth and the other condition were retrieved and median normalized. The standard deviation of the expression level difference over the annotated ORF was calculated as the measure of signal noise, here called n, since the median-normalized expression of the annotated ORF should have been the same in both conditions. Ends were called as the first region—determined by both a 10-bp and 80-bp sliding window—which was expressed at a level of 3.5 times n or more away from (i.e., above or below) the expression level in YPAD exponential growth, although not less than a fourfold difference in expression. This cutoff was chosen by manual inspection and represents a conservative cutoff level. Tests were then applied to make sure that the called end was sufficiently expressed, was not an annotated intron, and was in a mappable region of the yeast genome. Each differential end also had to be at least 40 bp long.
RNA-binding protein (RBP) motifs from (Riordan et al. 2011) were called by using the consensus motifs in Supporting Information, Table S1 as a standard text search algorithm, for example, their motif AAACACAW could be matched to either AAACACAA or AAACACAU; no probabilistic weighting of nucleotide combinations was performed.
Differential expression was determined using the DESeq software package (Anders and Huber 2010). Subsequent Gene Ontology (GO) (Ashburner et al. 2000) analyses were conducted using the GO::TermFinder software (Boyle et al. 2004).
3′ RACE on the CDC19 gene was carried out using the RLM-RACE kit from Ambion, following all instructions therein. The 3′ RACE outer primer had sequence CACCGAAACCGTCGCTGCCT and the 3′ RACE inner primer had sequence TTTTCGAACAAAAGGCCAAG.
Luciferase assays were performed using firefly luciferase with Renilla luciferase as a control, as described previously (McNabb and Reed 2005). Instead of integrating the luciferase constructs, however, they were used on a plasmid, as described in (Chu et al. 2011). Plasmid inserts were produced by DNA 2.0, and sequences are provided in Table S8.
Results
Mapping expressed regions of the yeast genome in 18 different conditions
In this study, we used strand-specific RNA-Seq (Parkhomchuk et al. 2009) to analyze the transcriptional landscapes of yeast under 18 different environmental conditions, detailed in Table 1. Each condition was analyzed using two biological replicates, and one additional technical replicate was performed for exponential growth in YPAD medium. In total, 850,570,328 reads of 76-bp length were sequenced on an Illumina GA IIx [see (Shendure and Ji 2008) for platform details], of which 670,446,084 reads and 377,263 poly-adenylated end tags (a total of 41,663,340,382 bases) were uniquely mapped to the yeast genome using Bowtie (Langmead et al. 2009).
Table 1. The 18 conditions under which RNA-Seq was performed.
| Condition | Description | No. of Sequenced Reads |
|---|---|---|
| Exponential growth | YPAD medium | 52,497,608 |
| Salt | 1 M NaCl for 45 min | 29,363,375 |
| DNA damage | 1 mM MMS for 1 hr | 29,427,647 |
| Alpha factor | 2.5 mM for 45 min; add another 50 μL to 25 mL yeast for another 30 min | 54,107,345 |
| Sorbitol | 1 M sorbitol for 45 min | 55,210,660 |
| Oxidative stress | 0.4 M H2O2 for 45 min | 34,262,447 |
| Heat shock | in 37° shaker for 1 hr | 36,362,297 |
| Stationary phase | 18 d in 30° incubator | 32,982,185 |
| SC media | Synthetic complete medium | 30,783,099 |
| SC glycerol media | 4% glycerol instead of glucose in SC medium | 35,241,114 |
| High calcium | 10 mM calcium chloride medium | 37,389,705 |
| Low nitrogen | 1/5 the normal amount of Yeast Nitrogen Base in YPAD | 27,864,332 |
| Low phosphate | see File S1 | 30,373,844 |
| Calcofluor | 0.1% for 1 hr | 31,921,767 |
| Hydroxyurea | 0.075M for 1 hr | 27,825,606 |
| Grape juice | Walgreen’s brand grape juice, filtered | 50,208,736 |
| Benomyl | 5 μg/mL for 1 hr | 40,974,093 |
| Congo red | 30 μg/mL of Congo red for 1 hr | 33,650,224 |
Unless otherwise noted, the conditions consist of YPAD medium with the indicated reagents added for the indicated incubation time. SC, synthetic complete.
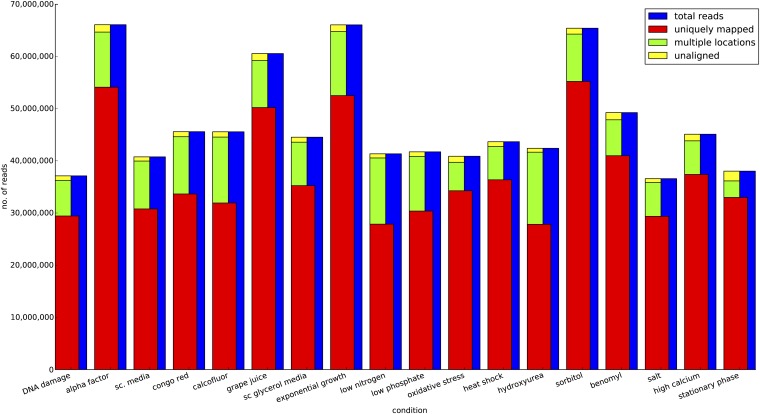
The number of uniquely mapped reads per condition ranged from 28 to 55 million (hydroxyurea and sorbitol, respectively), and the biological replicates showed high correlation. The Pearson r values of gene expression levels, calculated as described in the Methods section, ranged from 0.981 to 0.999 (alpha factor and high calcium, respectively). Figure 1 shows the number of reads sequenced for each condition, and Table S1 shows the Pearson correlations (r values) of the biological replicates for a given condition.
Figure 1 .
Sequencing done for each condition. Shown is the number of sequence tags generated for each condition (i.e., two biological replicates, pooled). Of the reads labeled as “unaligned,” some still provide useful information as poly-A tags.
Approximately 93% of the yeast genome can be uniquely mapped with 76-bp reads. Most of the yeast genome is expressed, consistent with previous studies (e.g., David et al. 2006; Nagalakshmi et al. 2008), and the data in this study demonstrate that approximately 80% (range 77–87%) of the base pairs in the yeast genome are expressed from one or both strands (i.e., as a percentage of the 12-Mb genome) at a depth of five sequencing reads of coverage or more per base (Figure S1). At the same level of coverage, approximately 45% (range 40–51%) of the base pairs are expressed as a percentage of the 24-Mb genome, i.e., separating out the Watson and Crick strands. Although we attempted to minimize potential artifacts from second-strand synthesis from reverse transcriptase, it is still possible that some of the observed transcription might be due to reverse transcription. The RNA-Seq method used in this study did not yield lower levels of antisense transcription when actinomycin D was used (Parkhomchuk et al. 2009), unlike that found in previous studies (Perocchi et al. 2007; Levin et al. 2010). Examination of annotated genes, however, revealed that transcription from one strand was accompanied by only a fraction of that amount of transcription occurring on the opposite strand. The median gene’s ratio of sense to anti-sense transcription was 5.1 to 6.4 log-two power (Figure S3). These results indicate that most of the yeast genome is truly expressed under different conditions and that some true antisense transcription is present, as expected (Xu et al. 2009).
Most yeast genes are differentially expressed
We analyzed gene expression across the 18 conditions. Of 6058 mappable genes (i.e., not in repetitive regions), we found that 5958 genes were expressed in at least one condition (with a threshold of each ORF base detected five or more times on average); 100 genes fell below this threshold, and 85 of these were classified as “dubious.” Interestingly, 611 dubious genes are expressed at or above our threshold.
Analysis of differential expression for the 5362 genes which are both mappable and nondubious was done using DESeq (Anders and Huber 2010). Using a binomial test (P ≤ 0.01), the majority (5245; 97.8%) was differentially expressed relative to exponential growth in YPAD media in at least one condition. Adding a criterion of 1.5-fold enrichment reduced this number to 4800 (89.5%), and this figure further reduces to 3421 (63.8%) when a 2-fold threshold is used. Interestingly, only 117 genes never differ in their overall RNA levels when we use the 1.5-efold enrichment threshold, listed in Table S9. These are enriched for the GO categories “biological process,” “intron homing,” “cellular component,” “molecular function,” and “endonuclease activity” (Ashburner et al. 2000) at a P-value of 0.05 (corrected) against a background set of mappable genes; the GO::TermFinder software suite (Boyle et al. 2004) was used to calculate enrichment.
GO-enrichment analysis of the genes that are differentially expressed using the binomial test revealed largely expected results, e.g., alpha factor−stimulated cells had as their top five categories “conjugation,” “conjugation with cellular fusion,” “sexual reproduction,” “interaction between organisms,” and “response to pheromone.” Similarly, the top five categories for Congo red stimulation, which damages cell walls, were “cell wall organization and biogenesis,” “external encapsulating structure organization and biogenesis,” “cell morphogenesis,” “morphogenesis,” and “anatomical structure development”; one of the top categories for heat shocked cells was “protein folding” (see Table S2 for complete results).
Interestingly, cells grown in commercial grape juice had as their top GO category “vitamin metabolism,” and the 41 genes in this category contained many vitamin biosynthetic genes related to thiamine, nicotinamide adenine dinucleotide/nicotinamide adenine dinucleotide phosphate, flavin, and carnitine, and are aimed at rapidly processing sugar. Two genes among these, PYC1 and PYC2, are associated with lactic acidosis when their human homolog is mutated [gene annotations taken from the Saccharomyces Genome Database (Cherry et al. 1998)].
Extensive differences in 5′ ends in stress conditions
We were particularly interested in examining differential transcript end diversity in yeast. A custom software pipeline was developed for the identification of differentially expressed transcript ends. This pipeline compared signal tracks for the exponential growth condition to every other condition, one at a time, and called differential ends when the difference in signal between the two conditions exceeded a stringent threshold calculated for each gene based on internal signal noise (see Materials and Methods; Figure S4). As transcriptional landscapes were directly compared, the term “topology comparisons” was used to describe this approach. Table S3 contains the complete list of alternate ends.
Extensive differences were observed in 5′ and 3′ ends, and Figure 2 summarizes the number of genes with differential ends. Relative to cells grown in rich medium, we find 613 cases of longer 5′ ends and 598 instances of shorter 5′ ends across the conditions, affecting 471 and 364 unique ORFs, respectively. For some conditions (e.g., alpha factor) cells exhibit minimal use of alternative 5′ ends, and, of those that do, the differential 5′ ends most often are shorter relative to cells grown in rich medium, that is, the transcription start site is closer to the start codon for translation for the nonrich medium condition.
Figure 2 .
Number of differential ends. Shown is the number of genes with differential longer and shorter ends on either the 5′ or 3′ side. All comparisons were with the exponential growth condition.
Two conditions, salt stimulation and stationary phase, show an abundance of transcripts with increased 5′ UTR size, with more than 200 genes each expressing a longer variant of its transcript relative to growth in rich medium. Of 15 transcripts with an alternative 5′ UTR longer than 300 base pairs, seven were found in stationary phase and four under salt stimulation. The longest alternative 5′ UTR, for the gene CDC8, was 853 base pairs longer in cells in stationary phase.
One interesting effect is when a differential transcript is shortened to the point that the annotated start codon is not transcribed. This is observed in 21 instances and 18 unique genes across 10 conditions; the affected genes are AAC1, ATG16, BRL1, CCM1, COS6, COX23, EMI2, FYV10, GEP7, GLR1, LAP2, MAD1, MRN1, MSN4, PRP39, RDH54, SUC2, and UBP7 (Table S1). Of these, only SUC2 has been characterized previously as a gene that encodes two transcript forms that produce proteins with differential amino termini (Carlson and Botstein 1982).
Upstream ORFs lie in many differential 5′ ends
Examining differential 5′ ends identified across all conditions revealed a total of 614 instances of uORFs in the differential transcribed region (see Materials and Methods). These affect 446 transcripts and a total of 543 unique uORFs (47 of which consist of solely a start and stop codon, with no intervening codon). These results are presented in Table S4.
We next compared the number of uORFs found in differential ends to the number expected by chance in differential ends of the same sizes, using random sampling of promoter sequences within 1000 bp of annotated start codons in the yeast genome. We found that for cells incubated in the presence of high salt, transcripts were statistically enriched for uORFs in the differential 5′ ends (z-value > 3). Interestingly, there was no statistical enrichment of uORFs in shortened 5′ UTRs in salt, but only in longer 5′ UTRs. Of the 222 genes with a longer alternative 5′ UTR in the salt response, 104 had one or more uORFs whereas a total of 148 uORFs were introduced in the longer-form UTR, where only 107 (std. dev.: 9.7) would have been expected by chance alone (Table S5). Overall, these results indicate that the differential 5′ ends, particularly long ones, are found under cell growth in extreme conditions such as high salt.
Diverse roles of differential uORFs in salt response
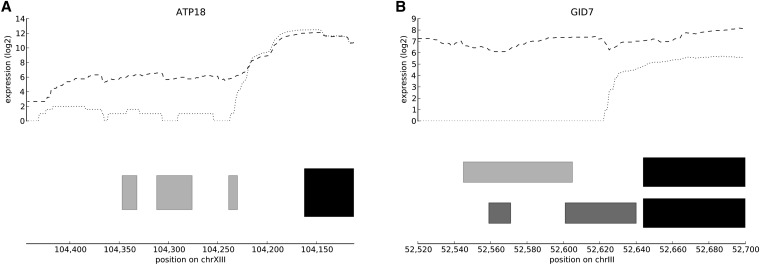
To further analyze the role of differential uORFs in gene expression, we selected five genes with uORFs for further study, and fused their upstream sequences from the gene’s start codon to the nearest neighboring gene upstream of a firefly luciferase reporter. A Renilla luciferase gene fused to a constitutive promoter was used as a loading control for these same cells (McNabb and Reed 2005; plasmid pTH650 described in Chu et al. 2011). A version of the upstream sequence in which the “in register uORFs” had been removed also was produced based on sequential AUGs, where the AUG start codon was changed to UUG (see Figure 3 for an explanation). In each case, the longest uORFs and the first accessible uORFs were removed. Results of the luciferase assays are presented in Figure 4.
Figure 3 .
uORFs knockouts. Illustration of the manner in which uORFs were mutated. The upper track shows expression, the dotted line under exponential growth in YPAD, and the dashed line under salt stimulation conditions. The lower track shows the main ORF in solid black and the knocked-out uORFs in light gray. Out-of-register ORFs, which were not knocked out, are shown in dark gray. (A) ATP18; (B) GID7. In the case of GID7, the shorter of those two out-of-register ORFs is conserved across Saccharomyces paradoxus, Saccharomyces mikatae, and Saccharomyces bayanus (see sequence data from Cliften et al. 2003). Please also note that the exact insert placed into the plasmid ran from the far left of each panel to the beginning of the main ORF; the latter was replaced with luciferase.
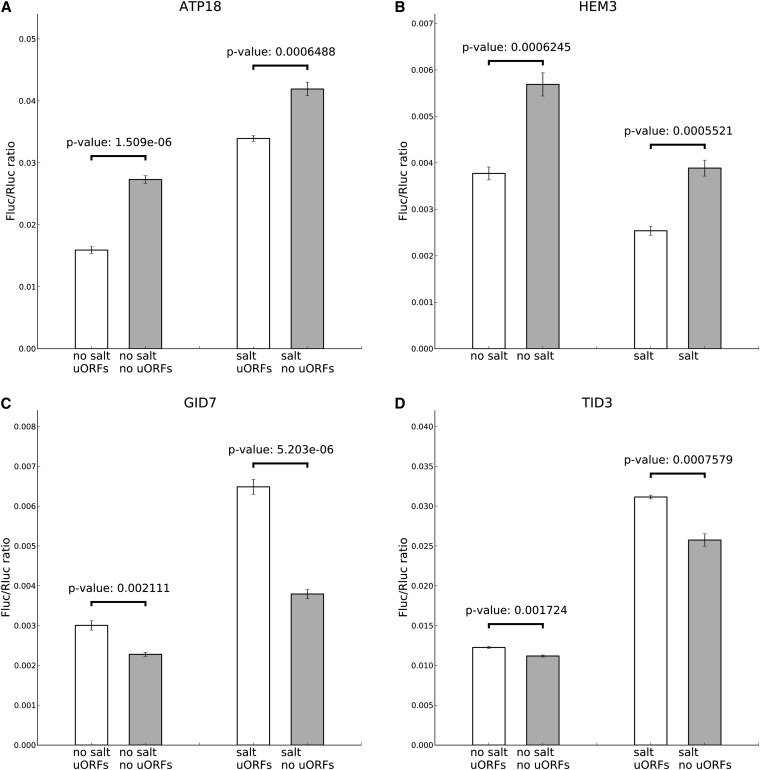
Figure 4 .
Luciferase analysis of uORFs. Luciferase assays confirming the relevance of uORFs in four genes with longer alternative 5′ UTRs in the salt stress condition. (A) ATP18; (B) HEM3; (C) GID7; and (D) TID3. The left two bars show results in SC–ura media and the right two bars show results in SC–ura media with 1 M salt for 1 hr. White bars are the results using the leader sequences, and gray bars have had uORF start codons mutated.
Of note, HEM3 and ATP18 were expressed less in salt relative to YPAD according to the RNA-Seq data (0.6× and 0.5×, respectively), and the uORFs were found to play a repressive role, i.e., the mutated, uORF-less construct was expressed greater. GID7 and TID3, however, were expressed greater in salt (2.5× more in both cases), and the plasmids with uORF knockouts express less reporter indicating that the uORFs for these genes played an activating role in protein expressed. RPH1 also was examined, although mutations analyses revealed no change indicating that its uORFs did not appear to play a significant role controlling gene expression.
As a further control that the uORFs themselves were responsible for the translation changes and not a condition-specific splicing event (e.g., Cheah et al. 2007). TopHat (Trapnell et al. 2009) was used to analyze the transcripts for splicing events. No evidence of alternative splicing was found. Thus, we conclude that most uORFs affect gene expression; for some genes they activate gene expression and in other instances they repress gene expression
Extensive differences in mRNA 3′ ends
Analysis of 3′ ends revealed a number of genes with differential 3′ ends in each of the various conditions as determined by topology comparisons (Figure 2). Relative to exponential growth in YPAD, there were 451 instances of longer 3′ ends, and 1991 instances of shorter 3′ ends across all conditions, representing 328 and 993 unique ORFs, respectively. Table S3 contains the complete list of altered 3′ lengths. Yeast grown in grape juice was clearly distinct, with 764 genes having a shorter transcript. Interestingly, one gene, REE1, was shortened in every condition relative to rich medium.
In addition to topology comparisons, analysis of 377,263 poly-adenylated end tags in our data set can be used to analyze 3′ ends. These are sequencing reads with a nongenomic run of A bases at the ends of mappable sequences are presumed to be a poly-adenylation event and thus mark the end of a transcript. These reads are recovered from the population of reads that did not initially map to the yeast genome, as the poly-adenylated tail will prevent alignment. Determining differential ends using this approach from our sequencing method is sensitive to thresholding and vulnerable to noise from low sampling levels; with 377,263 such reads across 18 conditions and a 6000 gene organism, only 3.5 reads per gene would be expected in each condition, assuming a uniform distribution of gene expression levels. In addition, poly-adenylated tags may be hampered by artifacts due to oligo-dT primer ligation to A-rich genomic sequences. Topology comparisons are thus relied upon as the main approach in this study.
Ozsolak et al. (Ozsolak et al. 2010) used poly-adenylation to call transcript ends because their protocol only sequenced poly-adenylated regions of the transcriptome and does not suffer from undersampling. They found 91,891 poly-adenylation sites (File S1), and reported that 72.1% of yeast genes have at least two poly-adenylation sites further than 50 bp apart.
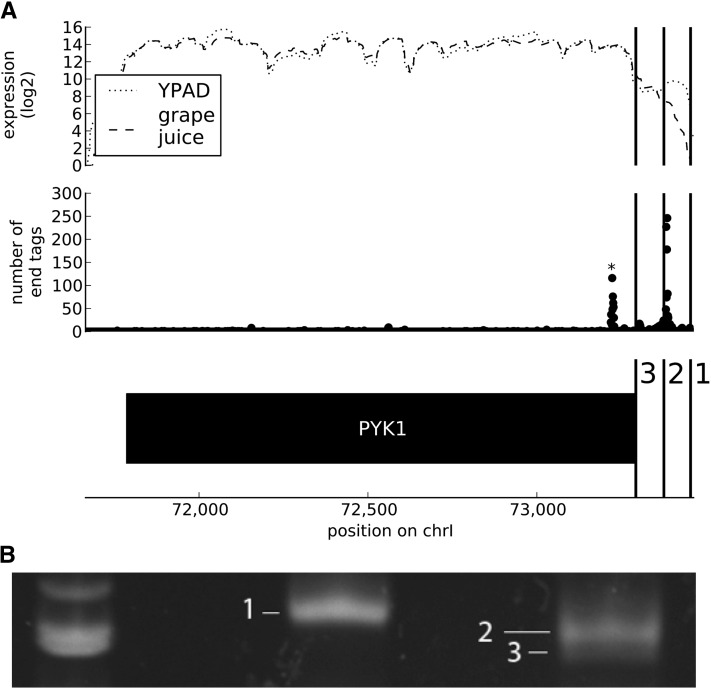
Pooling the results for all conditions in our RNA-Seq data, we found that 171 genes had more than 10 end tags. Of those, 128 (74.9%) had two or more discrete poly-adenylation start sites, indicating variability in 3′ end structure. Among our interesting results, we found one gene, CDC19/PYK1 (pyruvate kinase), which has an alternative 3′ end (Figure 5). Rapid amplification of cDNA ends polymerase chain reaction (RACE PCR) confirmed the existence of three distinct ends, one of which is visible only as an expression drop, another only from poly-adenylation tags, and a third visible from both. In a study of 3′ UTRs in Caenorhabditis elegans using polyA capture, 3′ RACE, cDNA sequencing, and RNA-Seq, 27% of called ends were only called using one technique (Mangone et al. 2010); thus, the use of multiple methods for calling ends is generally useful for 3′ end mapping.
Figure 5 .
Pyruvate kinase differential end validation. (A) The top track shows the expression profile for CDC19/PYK1 (pyruvate kinase) under exponential growth in YPAD and grape juice conditions. The middle track shows the number of sequenced end tags from the study as a whole. The bottom track shows the annotated ORF. The numbers 1, 2, and 3 show Sanger sequencing results from the 3′ RACE bands in part B. (B) Gel showing the results of 3′ RACE PCR for CDC19. The ladder shows the 600- and 500-bp markers. The center lane is the result for RNA from the exponential growth condition, and the right lane is from grape juice. The bands labeled 1, 2, and 3 were sequenced and correspond to the marked locations in part A.
Although the effects were often less dramatic than those seen at the 5′ end, expression at the 3′ end can decrease to levels where the main stop codon is not part of the dominant form of the transcript, or not present at all in the data. Figure S2B shows the 3′ end of the VMA5 gene, which does not appear to have a stop codon at all in synthetic complete media. Under exponential growth, there are 32 poly-adenylated end tags near VMA5 between genomic co-ordinates 285,831 and 285,837 on chromosome XI, although there is clearly some expression past that point, which would expose the annotated stop codon in a minority of transcripts.
In another example, PPZ2, which encodes a phosphatase involved in regulating cell cycle progression and shown in Figure S2A, the transcript level for the gene as a whole increases by 2.2-fold in the shift from exponential growth to stationary phase, but the level of the differential end, called algorithmically from genomic co-ordinates 1,336,923 to 1,336,988 on chromosome IV, increases by 15.8-fold (after we adjusted for sequencing library size). A total of 97 other instances of a shortened 3′ end were found using topology comparisons, represented in 63 unique ORFs in examples from all 17 non-exponential growth conditions. They are listed in the Table S6. Thus, many yeast transcripts may lack a translation termination codon.
The differential ends called by topology comparisons were examined for RBP motifs using the motifs published in (Riordan et al. 2011). Most conditions were statistically enriched for RBP motifs among the 3′ sequences that were shortened relative to rich YPD medium, with the exception of the oxidative stress and sorbitol conditions (Table S7). However, only five conditions were enriched for RBPs among the lengthened 3′ sequences (grape juice, oxidative stress, heat shock, sorbitol, and stationary phase). Statistical enrichment was calculated, similarly to that of uORFs, based on random sampling from the yeast genome of sequences of equal length to the differential ends, and a z-value cutoff greater than 3. Thus, transcripts with differential ends are likely to affect differences in RNA binding proteins and presumably gene regulation.
Discussion
Prematurely terminated transcripts
One particularly surprising finding of our study is that 63 ORFs were found which, under at least one condition, showed a 3′ end that likely terminated before the annotated stop codon. These transcripts would be left without a stop codon, potentially triggering a nonstop decay of the mRNA (Frischmeyer et al. 2002) and/or production of a polylysine tail on the protein that is produced. PPZ2, whose transcript exhibits this property, is a serine/threonine phosphatase involved in regulating cell-cycle progression, so its appearance in the stationary phase condition is of interest. The phenomenon of premature poly-adenylation has been noted before and described as an attenuating regulator of human mobile elements (Perepelitsa-Belancio and Deininger 2003), and the nonstop decay pathway has been described in yeast (Frischmeyer et al. 2002). Although the pathway is understood, there is little understanding of how prevalent it is (Garneau et al. 2007). This study indicates the particular genes that may be controlled in this fashion and that the phenomenon made be more widespread that previously appreciated.
5′ ORF truncations
Of the 18 genes found to have 5′ ORF truncations, i.e., the dominant form of the transcript did not contain the annotated start codon, one, SUC2, has been extensively characterized previously. For SUC2 the long form contains the signal sequence that causes the sucrose invertase to be secreted (Carlson and Botstein 1982). Carlson and Botstein found that glucose-repressed cells produce a longer form of SUC2, which is consistent with the results of our study, in which the RNA-Seq data show a longer transcript present in media using glycerol as the carbon source. In addition to SUC2, 17 other genes produce shorter proteins. Only SUC2, however, is predicted to encode a protein that contains a signal sequence that is missing in the shorter transcript [performed using SignalP 4.0 (Petersen et al. 2011)]. The alternative amino terminal sequence in these 17 instances may affect protein function in other ways.
RBP motifs
In 2442 differential 3′ ends (1991 shortened and 451 lengthened) across 17 non-YPAD exponential growth conditions, 6420 RBP motifs were present (motifs from Riordan et al. 2011). RBP motifs are, to date, quite degenerate in the method they are defined, and this likely leads to many false positives. Nevertheless, statistical enrichment was more often seen in shortened 3′ ends, suggesting that cells may remove the layer of control offered by RBPs under stress conditions.
uORFs
Upstream ORFs are a known regulatory feature. The availability of genomic sequences for many organisms, yeast since 1996 (Goffeau et al. 1996), has allowed for a theoretical search for uORFs, and a large abundance has been found. Whether they have meaning as a method for translational control depends first and foremost on whether they are transcribed; these data have been harder to produce. Using a random lacZ insertion method, Burns et al. and Ross-Macdonald et al. revealed extensive translation of short and out of frame ORF expression through the yeast genome (Burns et al. 1994; Ross-Macdonald et al. 1999). In a recent RNA-Seq study, Nagalakshmi et al. (2008) concluded that 6% of genes had uORFs. Estimates vary and range up to 20% (Hood et al. 2009). Our data indicate that many of these lie in differential UTRs.
Two broad mechanisms of uORF action are known (see Vilela and MCcarthy 2003): control by uORFs modulating posttermination behavior of ribosomes and control modulated by the actual peptide produced by a uORF. The canonical example of the first is GCN4, characterized in (Mueller and Hinnebusch 1986), and of the second the arginine attenuator peptide found before the CPA1 gene (Gaba et al. 2001).
Even if transcribed, whether uORFs have regulatory function further depends on their interaction with the translational machinery. Genes such as GCN4 (Mueller and Hinnebusch 1986), CPA1 (Gaba et al. 2001), and YAP1/YAP2 (Vilela et al. 1998) are well characterized, and further examples do exist. Some, however, show no effect [see (Zhang and Dietrich 2005) where not all validate], and some show an effect dependent on RNA processing [see (Zhang and Dietrich 2005), for an example involving riboswitches in N. crassa]. Most studies to date have used bioinformatic approaches solely and analyze sequence context and/or conservation, which does not offer proof of uORF relevance [see e.g., (Cvijović et al. 2007; Lawless et al. 2009; Selpi et al. 2009), all in S. cerevisiae]. In our study, in which we performed luciferase validation experiments, we examine five leader sequences with uORFs, only four of which had a direct impact on luciferase levels (Zhang and Dietrich 2005). In another study, authors analyzed the ribosome footprint to mRNA ratios in meiosis and concluded that uORFs in differential ends could alter translational efficiency of mRNA. They also reported that in the case of uORFs with an AUG start codon, the translational efficiency and ribosome occupancy showed a “strong negative correlation” (Brar et al. 2012). A subsequent microarray-based study of meiotic yeast confirmed the presence of abundant transcript architecture changes in meiotic and sporulating yeast (Kim Guisbert et al. 2012).
In this study, there was only a statistical enrichment for uORFs relative to promoter sequences in longer 5′ ends in the salt condition. Under no conditions was a statistically significant enrichment of uORFs present in shortened 5′ UTRs, implying that normally used uORFs are seldom removed by alternative promoter choices.
In each of the four cases of a uORF affecting translation reported in (Zhang and Dietrich 2005), removing the uORFs led to increased luciferase activity. In this study, a new phenomenon is reported: uORFs which increase luciferase activity (GID7 and TID3; see Figure 4). That uORFs might increase translational efficiency has been hypothesized (Neafsey and Galagan 2007; see also Hood et al. 2009), but no previous examples have been found or directed demonstrated using mutational analyses. One possibly is that these uORFs have RNA binding sites or ribosome entry sites that attracting ribosomes to those mRNAs.
Overall, this study demonstrates pervasive differential transcript ends occur in yeast and that in a least several cases these differences likely affect gene expression. The mechanisms by which differential ends are selected remains to be determined, and such information will be important to obtain a details understanding of how eukaryotic gene expression is controlled.
Supplementary Material
Acknowledgments
Jennifer Li-Pook-Than and Jennifer E.G. Gallagher provided comments on the manuscript. This research was supported by National Institute of Health grants NIH P50 HG 002357 and NIH/CA077808.
Footnotes
Communicating editor: B. J. Andrews
Literature Cited
- Anders S., Huber W., 2010. Differential expression analysis for sequence count data. Genome Biol. 11: R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M., Ball C. A., Blake J., Botstein D., Butler H., et al. , 2000. Gene Ontology: tool for the unification of biology. Nat. Genet. 25: 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle E., Weng S., Gollub J., Jin H., Botstein D., et al. , 2004. GO:TermFinder—open source software for accessing Gene Ontology information and finding significantly enriched Gene Ontology terms associated with a list of genes. Bioinformatics 20: 3710–3715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brar G. A., Yassour M, Friedman N, Regev A, Ingolia N. T., et al. , 2012. High-resolution view of the yeast meiotic program revealed by ribosome profiling. Science 335: 552–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns N., Grimwade B., Ross-Macdonald P. B., Choi E. Y., Finberg K., et al. , 1994. Large-scale analysis of gene expression, protein localization, and gene disruption in Saccharomyces cerevisiae. Genes Dev. 8: 1087–1105 [DOI] [PubMed] [Google Scholar]
- Carlson M., Botstein D., 1982. Two differentially regulated mRNAs with different 5′ ends encode secreted with intracellular forms of yeast invertase. Cell 28: 145–154 [DOI] [PubMed] [Google Scholar]
- Cheah M. T., Wachter A., Sudarsan N., Breaker R. R., 2007. Control of alternative RNA splicing and gene expression by eukaryotic riboswitches. Nature 447: 497–500 [DOI] [PubMed] [Google Scholar]
- Cherry J. M., Adler C., Ball C., Chervitz S. A., Dwight S. S., et al. , 1998. SGD: Saccharomyces Genome Database. Nucleic Acids Res. 26: 73–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu D., Barnes D. J., von der Haar T., 2011. The role of tRNA and ribosome competition in coupling the expression of different mRNAs in Saccharomyces cerevisiae. Nucleic Acids Res. 39: 6705–6714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cliften P, Sudarsanam P., Desikan A., Fulton L., Fulton B., et al. , 2003. Finding functional features in Saccharomyces genomes by phylogenetic footprinting. Science 301: 71–76 [DOI] [PubMed] [Google Scholar]
- Coelho P. S. R., Bryan A. C., Kumar A., Shadel G. S., Snyder M., 2002. A novel mitochondrial protein, Tar1p, is encoded on the antisense strand of the nuclear 25S rDNA. Genes Dev. 16: 656–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cvijović M., Dalevi D., Bilsland E., Kemp G. J. L., Sunnerhagen P., 2007. Identification of putative regulatory upstream ORFs in the yeast genome using heuristics and evolutionary conservation. BMC Bioinformatics 8: 295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David L., Huber W., Granovskaia M., Toedling J., Palm C. J., et al. , 2006. A high-resolution map of transcription in the yeast genome. Nat. Biotechnol. 103: 5320–5325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frischmeyer P. A., van Hoof A., O’Donnell K., Guerrerio A. L., Parker R., et al. , 2002. An mRNA surveillance mechanism that eliminates transcripts lacking termination codons. Science 295: 2258–2261 [DOI] [PubMed] [Google Scholar]
- Gaba A., Wang Z., Krishnamoorthy T., Hinnebusch A. G., Sachs M. S., 2001. Physical evidence for distinct mechanisms of translational control by upstream open reading frames. EMBO J. 20: 6453–6463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garneau N. L., Wilusz J., Wilusz C. J., 2007. The highways and byways of mRNA decay. Nat. Rev. Mol. Cell Biol. 8: 113–126 [DOI] [PubMed] [Google Scholar]
- Goffeau A., Barrell B., Bussey H., Davis R., 1996. Life with 6000 genes. Science 274: 546–567 [DOI] [PubMed] [Google Scholar]
- Guittaut M., Charpentier S., Normand T., Dubois M., Raimond J., et al. , 2001. Identification of an internal gene to the human Galectin-3 gene with two different overlapping reading frames that do not encode Galectin-3. J. Biol. Chem. 276: 2652–2657 [DOI] [PubMed] [Google Scholar]
- Hood H. M., Neafsey D. E., Galagan J., Sachs Matthew S., 2009. Evolutionary roles of upstream open reading frames in mediating gene regulation in fungi. Annu. Rev. Microbiol. 63: 385–409 [DOI] [PubMed] [Google Scholar]
- Kim Guisbert K. S., Zhang Y., Flatow J., Hurtado S., Staley J. P., et al. , 2012. Meiosis-induced alterations in transcript architecture and noncoding RNA expression in S. cerevisiae. RNA 18: 1142–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochetov V., 2006. Alternative translation start sites and their significance for eukaryotic proteomes. Mol. Biol. 40: 705–712 [PubMed] [Google Scholar]
- Kumar A., Harrison P. M., Cheung K. H., Lan N., Echols N., et al. , 2002. Serial analysis of gene expression. Nat. Biotechnol. 20: 58–63 [DOI] [PubMed] [Google Scholar]
- Landry J.-R., Mager D. L., Wilhelm B. T., 2003. Complex controls: the role of alternative promoters in mammalian genomes. Trends Genet. 19: 640–648 [DOI] [PubMed] [Google Scholar]
- Langmead B., Trapnell C., Pop M., Salzberg S. L., 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law G. L., Bickel K. S., MacKay V. L., Morris D. R., 2005. The undertranslated transcriptome reveals widespread translational silencing by alternative 5′ transcript leaders. Genome Biol. 6: R111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawless C., Pearson R. D., Selley J. N., Smirnova J. B., Grant C. M., et al. , 2009. Upstream sequence elements direct post-transcriptional regulation of gene expression under stress conditions in yeast. BMC Genomics 10: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M. V., Topper S. E., Hubler S. L., Hose J., Wenger C. D., et al. , 2011. A dynamic model of proteome changes reveals new roles for transcript alteration in yeast. Mol. Syst. Biol. 7: 514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin J. Z., Yassour M., Adiconis X., Nusbaum C., Thompson D. A., et al. , 2010. Comprehensive comparative analysis of strand-specific RNA sequencing methods. Nat. Methods 7: 709–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangone M., Manoharan A. P., Thierry-Mieg D., Thierry-Mieg J., Han T., et al. , 2010. The landscape of C. elegans 3′UTRs. Science 329: 432–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNabb D., Reed R., 2005. Dual luciferase assay system for rapid assessment of gene expression in Saccharomyces cerevisiae. Eukaryot. Cell 4: 1539–1549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mignone F, Gissi C., Liuni S., Pesole G., 2002. Untranslated regions of mRNAs. Genome Biol 3: REVIEWS0004.1–0004.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura F., Kawaguchi N., Sese J., Toyoda A., Hattori M., et al. , 2006. A large-scale full-length cDNA analysis to explore the budding yeast transcriptome. Proc. Natl. Acad. Sci. USA 103: 17846–17851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller P. P., Hinnebusch A. G., 1986. Multiple upstream AUG codons mediate translational control of GCN4. Cell 45: 201–207 [DOI] [PubMed] [Google Scholar]
- Nagalakshmi U., Wang Z., Waern K., Shou C., Raha D., et al. , 2008. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science 320: 1344–1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagalakshmi U., Karl W., Michael S., 2010. RNA-Seq: a method for comprehensive transcriptome analysis. Curr. Protoc. Mol. Biol. 89: 4.11.1–4.11.13 [DOI] [PubMed] [Google Scholar]
- Neafsey D. E., Galagan J. E., 2007. Dual modes of natural selection on upstream open reading frames. Mol. Biol. Evol. 24: 1744–1751 [DOI] [PubMed] [Google Scholar]
- Ozsolak F, Kapranov P., Foissac S., Kim S. W., Fishilevich E., et al. , 2010. Comprehensive polyadenylation site maps in yeast and human reveal pervasive alternative polyadenylation. Cell 143: 1018–1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkhomchuk D., Borodina T., Amstislavskiy V., Banaru M., Hallen L., et al. , 2009. Transcriptome analysis by strand-specific sequencing of complementary DNA. Nucleic Acids Res. 37: e123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perepelitsa-Belancio V., Deininger P., 2003. RNA truncation by premature polyadenylation attenuates human mobile element activity. Nat. Genet. 35: 363–366 [DOI] [PubMed] [Google Scholar]
- Perocchi F., Xu Z., Clauder-Münster S., Steinmetz Lars M., 2007. Antisense artifacts in transcriptome microarray experiments are resolved by actinomycin D. Nucleic Acids Res. 35: e128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen T. N., Brunak S., von Heijne G., Nielsen H., 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8: 785–786 [DOI] [PubMed] [Google Scholar]
- Riordan D. P., Herschlag D., Brown P. O., 2011. Identification of RNA recognition elements in the Saccharomyces cerevisiae transcriptome. Nucleic Acids Res. 39: 1501–1509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross-Macdonald P., Coelho P. S., Roemer T., Agarwal S., Kumar A., et al. , 1999. Large-scale analysis of the yeast genome by transposon tagging and gene disruption. Nature 402: 413–418 [DOI] [PubMed] [Google Scholar]
- Selpi S., Bryant C. H., Kemp G. J., Sarv J., Kristiansson E., et al. , 2009. Predicting functional upstream open reading frames in Saccharomyces cerevisiae. BMC Bioinformatics 10: 451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shendure J., Ji H., 2008. Next-generation DNA sequencing. Nat. Biotechnol. 26: 1135–1145 [DOI] [PubMed] [Google Scholar]
- Trapnell C., Pachter L., Salzberg S. L., 2009. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Velden A. W., Thomas A. A., 1999. The role of the 5′ untranslated region of an mRNA in translation regulation during development. Int J Biochem Cell Biol 31: 87–106 [DOI] [PubMed] [Google Scholar]
- Vilela C., McCarthy J., 2003. Regulation of fungal gene expression via short open reading frames in the mRNA 5′untranslated region. Mol. Microbiol. 49: 859–867 [DOI] [PubMed] [Google Scholar]
- Vilela C., Linz B., Rodrigues-Pousada C., McCarthy J., 1998. The yeast transcription factor genes YAP1 and YAP2 are subject to differential control at the levels of both translation and mRNA stability. Nucleic Acids Res. 26: 1150–1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waern K., Nagalakshmi U., Snyder M., 2011. RNA sequencing. Methods Mol. Biol. 759: 125–132 [DOI] [PubMed] [Google Scholar]
- Wang Z., Gerstein M., Snyder M., 2009. RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 10: 57–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z., Wei W., Gagneur J., Perocchi F., Clauder-Münster S., et al. , 2009. Bidirectional promoters generate pervasive transcription in yeast. Nature 457: 1033–1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon O. K., Brem R. B., 2010. Noncanonical transcript forms in yeast and their regulation during environmental stress. RNA 16: 1256–1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Dietrich F., 2005. Identification and characterization of upstream open reading frames (uORF) in the 5′ untranslated regions (UTR) of genes in Saccharomyces cerevisiae. Curr. Op. Genet. 48: 77–87 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.