Abstract
The TTAGGG motif is common to two seemingly unrelated dimensions of chromatin function—the vertebrate telomere repeat and the promoter regions of many Schizosaccharomyces pombe genes, including all of those encoding canonical histones. The essential S. pombe protein Teb1 contains two Myb-like DNA binding domains related to those found in telomere proteins and binds the human telomere repeat sequence TTAGGG. Here, we analyse Teb1 binding throughout the genome and the consequences of reduced Teb1 function. Chromatin immunoprecipitation (ChIP)-on-chip analysis reveals robust Teb1 binding at many promoters, notably including all of those controlling canonical histone gene expression. A hypomorphic allele, teb1-1, confers reduced binding and reduced levels of histone transcripts. Prompted by previously suggested connections between histone expression and centromere identity, we examined localization of the centromeric histone H3 variant Cnp1 and found reduced centromeric binding along with reduced centromeric silencing. These data identify Teb1 as a master regulator of histone levels and centromere identity.
Keywords: centromeres, fission yeast, histones, Teb1, transcription
Introduction
The regulation of histone levels is a multi-faceted phenomenon crucial to genomic integrity. While DNA is generally packaged into nucleosomes comprising two copies each of histones H2A, H2B, H3, and H4, histones stand as a double-edged sword for DNA metabolism. Although their extreme basicity greatly favours strong interaction with DNA, any non-specific binding can lead to dramatic outcomes that threaten cell survival (Gunjan et al, 2005). Therefore, a major challenge is reached when cells undergo chromosomal replication, when histone production and nucleosome assembly must be coordinated with replication fork progression (Schumperli, 1986). A delay between DNA synthesis and histone deposition triggers DNA damage, chromosomal rearrangements and loss of viability (Han et al, 1987; Kim et al, 1988). Conversely, an excess of histones is also ill tolerated, as even a slight or transient excess of histone synthesis threatens the integrity of chromosome structure and gene expression (Meeks-Wagner and Hartwell, 1986; Singh et al, 2010).
Regulation of histone levels also appears to play a key role in controlling the centromeric deposition of the histone H3 variant Centromere Protein A (CENP-A) (Castillo et al, 2007), which in turn nucleates the assembly of the kinetochores that connect chromosomes with spindle microtubules. While CENP-A and the kinetochore proteins are highly conserved, centromeric DNA sequences are poorly conserved and in many species, centromeres are assembled in a largely sequence-independent manner. Indeed, in mammals, flies, worms, and yeasts (Candida albicans and Schizosaccharomyces pombe), ‘neocentromeres’ can arise and fulfill proper kinetochore assembly in chromosomal regions devoid of normal centromere sequences (Williams et al, 1998; Ishii et al, 2008; Marshall et al, 2008; Ketel et al, 2009; Yuen et al, 2011). These observations suggest that epigenetic mechanisms control centromere identity. In all cases, perpetuation of the epigenetic mark depends on CENP-A binding, which is both necessary and sufficient for centromere inheritance (Mendiburo et al, 2011). The lack of sequence specificity also allows the spreading of CENP-A past its usual boundaries in cells overproducing CENP-A (Heun et al, 2006; Castillo et al, 2007). Conversely, overproduction of canonical histone H3 in fission yeast compromises centromere function by displacing CENP-A from the centromere central core (Castillo et al, 2007). The importance of the balance of core histone levels is further underscored by experiments in which the level of H3 relative to H4 is increased, leading to impaired CENP-A loading. In contrast, CENP-A loads normally if the H3/H4 ratio is decreased, suggesting that H3 and CENP-A compete for the available H4 histones.
Histone concentrations are regulated at multiple levels including that of transcription. In budding and fission yeast, histone genes are arranged in a tail-to-tail organization with a common intergenic sequence. Schizosaccharomyces pombe contains a single gene encoding H2Aβ (hta2+), the H2Aα-H2B-encoding gene pair (hta1+-htb1+), and three copies of the gene pair encoding H3 and H4 (hht1-hhf1+, hht2+-hhf2+, hht3+-hhf3+). The levels of all histone transcripts except hht2+ increase during S phase (Takayama and Takahashi, 2007). Repression of histone gene transcription outside S phase or in response to hydroxyurea treatment is accomplished by members of the HIRA histone chaperone complex (Blackwell et al, 2004; Takayama and Takahashi, 2007). During S phase, the GATA-like transcription factor Ams2 is required for histone upregulation (Takayama and Takahashi, 2007). Ams2 is thought to bind a 17-bp consensus sequence known as the AACCCT box, located in the common promoter of each divergent pair of histone genes. Ams2 protein levels oscillate through the cell cycle, being maximal in S phase and decreasing dramatically in G2 via ubiquitin-mediated degradation. This degradation is linked to passage through S phase by phosphorylation of Ams2, which is accomplished by the S-phase kinase DDK and is requisite for the interaction between Ams2 and the Pof3 subunit of the SCF ubiquitin ligase. These observations suggest a regulatory loop in which Ams2 is synthesized in G1 to favour histone transcription during S phase and degraded in a DDK-dependent manner (Takayama et al, 2010).
Ams2 is thought to play roles in chromatin assembly not only via modulation of histone levels, but also via assembly of CENP-A chromatin. Fission yeast centromeres comprise the pericentric heterochromatin region, which assembles at the so-called outer repeats (otr), and the central (cnt) and inner-most (imr) regions characterized by the presence of Cnp1CENP-A. Proper loading of Cnp1CENP-A at the central core requires both the establishment of heterochromatin at the otr and Ams2 function. Indeed, Ams2 was originally identified as a multicopy suppressor of the temperature-sensitive CENP-A mutant cnp1-1. A biphasic model for incorporation of Cnp1CENP-A into centromeric chromatin has been proposed in which Ams2 is required to favour Cnp1CENP-A loading during S phase while Cnp1CENP-A loading during G2 occurs through an Ams2-independent backup mechanism (Takayama et al, 2008).
Teb1, also known as SpX or Mug152, was initially reported as a potential telomeric factor as it harbours two helix-loop-helix dsDNA binding domains of the homeodomain subset of Myb domains (Figure 1A), which are recurrently found in telomeric proteins; later it was identified as the product of a meiotically upregulated gene (Vassetzky et al, 1999; Spink et al, 2000; Martin-Castellanos et al, 2005). Curiously, however, Teb1 was shown to have higher affinity for the vertebrate telomere repeat, TTAGGG, than to fission yeast telomere repeats in vitro (Vassetzky et al, 1999; Spink et al, 2000). Here, we investigate the function of this essential protein and show that Teb1 binds in vivo and regulates the activities of many promoters, including those controlling the expression of all four types of canonical histones. We also find that Teb1 is involved in the centromeric loading of Cnp1CENP-A and maintenance of centromere identity. Moreover, Teb1 regulates the expression of a protease capable of histone clipping. Hence, Teb1 is a newly recognized general transcription factor with prominent roles in controlling histone levels and stability.
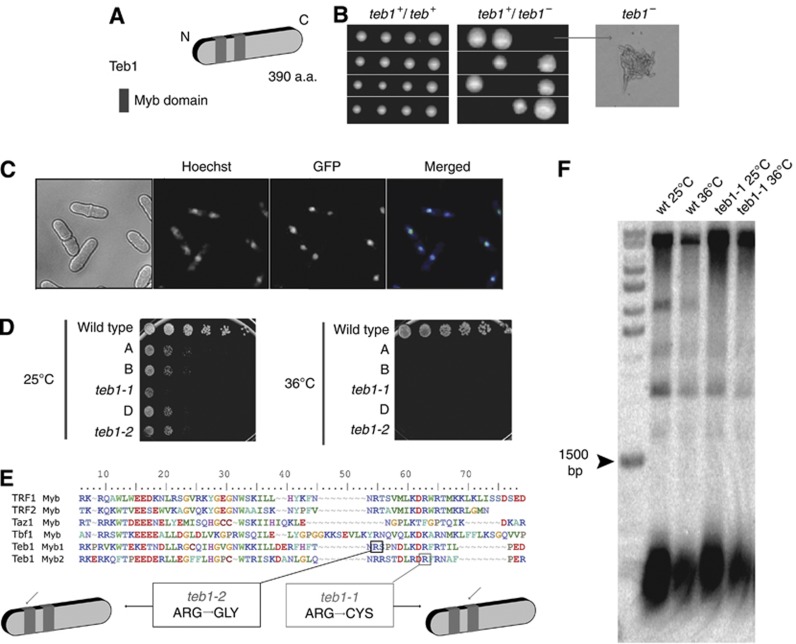
Figure 1.
Teb1 is essential and localizes to the nucleus. (A) Teb1 contains two Myb domains near its N-terminus. (B) Tetrad dissection of sporulated homozygous (teb1 +/+) and heterozygous (teb1+/−) diploids. While teb1+ spores are fully viable, teb1Δ spores germinate but die, forming microcolonies (arrow). (C) Endogenously GFP-tagged Teb1 localizes to the nucleus (counterstained with Hoechst 33342). (D) Serial dilution assay on complete media. teb1-1 and teb1-2 behave as hypomorphic alleles at 25°C and are inviable at 36°C. Rows A, B and D contain other teb1 TS mutants that were not further characterized. (E) Sequence alignment of myb domains from several proteins. Hypomorphic teb1-1 and teb1-2 display single point mutations in conserved regions of the first (teb1-1, Arg184Gly, black square) or second (teb1-2, Arg92Cys, grey square) Myb domains. (F) Southern blot analysis of telomere length. No size changes are conferred by the teb1-1 mutation. The same results were obtained for the other ts alleles (unpublished observations).
Results
Teb1 is an essential nuclear protein
To investigate the roles of Teb1, a loss-of-function approach was taken in which one copy of the teb1+ gene in a diploid strain was replaced by a G418 resistance marker (Table I). Sporulation of the resulting heterozygous teb1+/teb1Δ diploid revealed that teb1Δ haploids are inviable, forming microcolonies of elongated cells that divide only a few times before ceasing division (Figure 1B). Fusion of a GFP tag with the Teb1 C-terminus at its endogenous locus yields fully viable strains harbouring Teb1-GFP, which yields a diffuse nuclear localization pattern (Figure 1C).
Table 1. Strains used in this work.
| JCF number | Genotype | Mating type |
|---|---|---|
| 1 | Wt | h+ |
| 2 | Wt | h− |
| 24 | ade6-M210 leu1-32 ura4-D18 | h− |
| 1942 | ade6-M210 leu1-32 ura4-D18 teb1-1-3HA-KanMX6 | h− |
| 1944 | ade6-M210 leu1-32 ura4-D18 teb1-2-3HA-KanMX6 | h− |
| 1969 | ade6-M? leu1-32 ura4-D18 hht1-CFP-KanMX6 sid4-YFP-KanMx6 teb1-1-3HA-KanMX6 mis6-mcherry-natMX6 | h+ |
| 7401 | cnt:arg3 cnt3:ade6 otr2: ura4 tel1L:his3 ade6-210 arg3-D4 his3-D1 leu1-32 ura4-D18 teb1-3HA-KanMX6 | h− |
| 7404 | cnt:arg3 cnt3:ade6 otr2: ura4 tel1L:his3 ade6-210 arg3-D4 his3-D1 leu1-32 ura4-D18 teb1-1-3HA-KanMX6 | h− |
| 7419 | teb1-1-3HA-KanMX6 | h+ |
| 7429 | teb1-3HA-KanMX6 | h+ |
| 7433 | cnt:arg3 cnt3:ade6 otr2: ura4 tel1L:his3 ade6-210 arg3-D4 his3-D1 leu1-32 ura4-D18 teb1-3HA-Kan (gift from R Allshire) | h+ |
| 7450 | ade6-M210 leu1-32 ura4-D18 teb1-3HA-KanMX6 | h− |
| 7452 | cnt1:arg3 cnt3:ade6 otr2: ura4 tel1L:his3 ade6-210 arg3-D3 his3-D1 leu1-32 ura4-D18 rik1::LEU2+ (gift from R Allshire) | h? |
| 7453 | cnt1:arg3 cnt3:ade6 otr2: ura4 tel1L:his3 ade6-210 arg3-D3 his3-D1 leu1-32 ura4-D18 mis6-302 (gift from R Allshire) | h? |
| 7872 | teb1-1-3HA::KanMX Ams2-13MYC::Hyg | |
| VKS 201 | teb1-GFP-KanMX6 | h? |
To generate a tool for studying Teb1 function, we screened for conditional alleles. Random mutations were generated by PCR amplification of a cassette harbouring the teb1+ open reading frame and a kanR marker that confers resistance to G418. A wild-type (wt) strain was transformed with the PCR products and G418-resistant transformants were screened for temperature-sensitive growth. Several hypomorphic mutant alleles that display sickness at the permissive temperature (25°C) and lethality at the restrictive temperature (36°C) were identified (Figure 1D). Two of these conditional alleles, teb1-1 and teb1-2, harbour mutations of conserved arginine residues within one or the other Myb domain (Figure 1E). Backcrossing teb1-1 with a wt strain recapitulates sickness at 25°C and inviability at 36°C. When teb1-1 cells were transformed with a plasmid-borne library of overexpressed fission yeast genes and grown at 36°C, the only plasmid conferring viability at 36°C encoded the wt teb1+ sequence, confirming that the reduced viability of teb1-1 strains is due to loss of Teb1 function (unpublished observations).
To investigate whether Teb1 plays a role at telomeres, we examined the telomeres of teb1-1 cells grown at 25°C or following shift to the restrictive temperature of 36°C for 16 h. Southern blot analysis of terminal restriction fragments revealed that both teb1-1 and wt cells harbour telomeres of 300±50 bp at both temperatures (Figure 1F). Therefore, the teb1-1 mutation does not affect telomere length.
Teb1 binds to the promoters of many genes
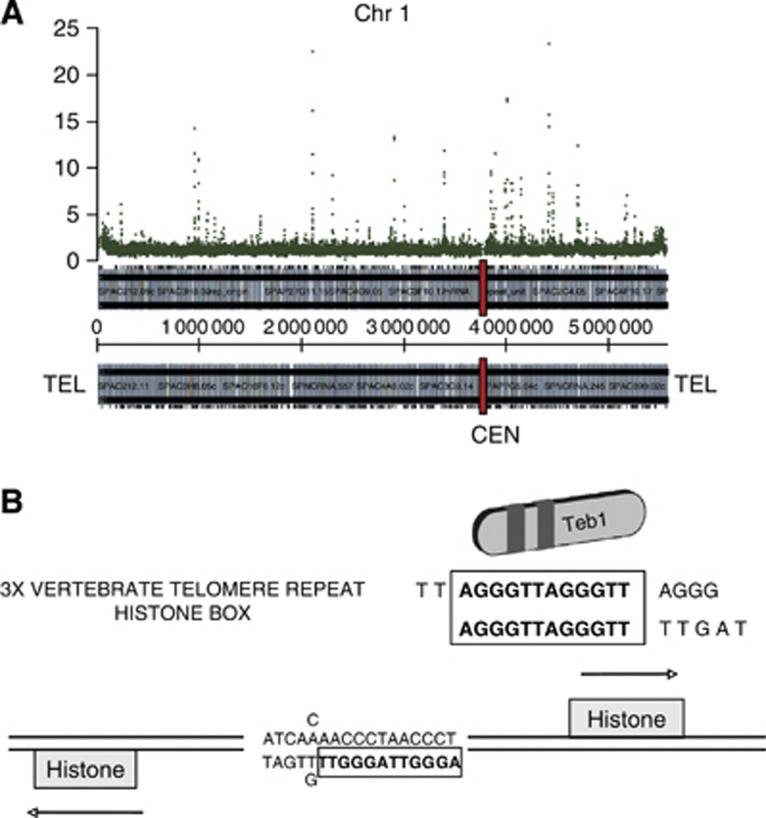
The in vitro binding specificity of Teb1 for the vertebrate telomeric repeat sequence (TTAGGG) (Vassetzky et al, 1999) along with the presence of tandem copies of this repeat in the promoters of several fission yeast genes suggested that Teb1 might bind the corresponding promoters. To investigate this, we immunoprecipitated (IP) endogenously haemaglutinin (HA)-tagged Teb1 and hybridized the IP with the oligonucleotide 4 × 44K Chromatin immunoprecipitation (ChIP)-on-chip whole genome DNA microarray platform (Agilent), which covers the majority of the fission yeast genome. The resulting ChIP-chip results show a distinct and reproducible pattern of Teb1 binding (Figure 2A; Supplementary Figure S1). As expected based on the in vitro binding data, the Teb1 binding sites often contain runs of TTAGGG repeats; when found in histone gene promoters, a slightly permuted 17-bp version of these repeats has been referred to as the AACCCT box (Matsumoto and Yanagida, 1985; Figure 2B). Accordingly, ChIP-chip analysis detected Teb1 binding to histone promoters. We found that using an enrichment value of two-fold as the criterion for Teb1 binding, Teb1 can be seen to bind all five promoters of the nine canonical histone genes (Figure 3; note that four pairs of histone genes share divergent promoters; see also Supplementary Table I). Hence, Teb1 binds in vivo to AACCCT-like boxes in all histone gene promoters.
Figure 2.
Teb1 binds to specific genomic loci. (A) Teb1 binding map of Chromosome 1, represented along the X axis. Teb1 enrichment values, based on ChIP-chip analysis of endogenously tagged functional Teb1, are represented on the Y axis. (B) The AACCCT box found in all canonical histone gene promoters is bound by Teb1.
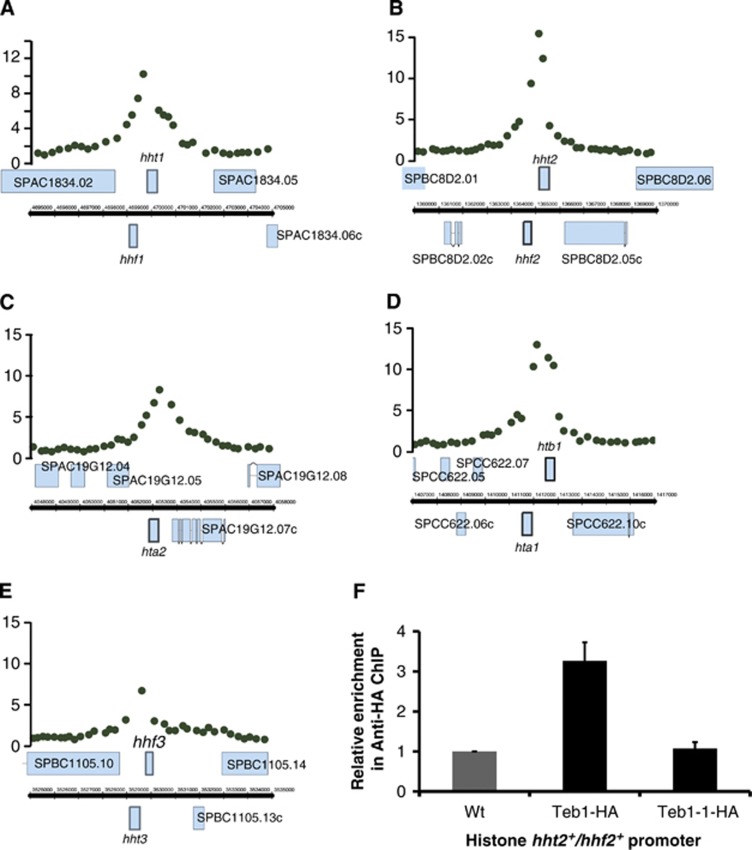
Figure 3.
Teb1 binds all histone promoters, and this binding is lost in teb1-1 cells. (A–E) Teb1 is specifically enriched at histone gene promoters. ChIP-chip data showing Teb1 binding at the indicated promoters. (F) ChIP of the indicated Teb1-HA variants at 25°C analysed by qPCR with primers for the histone hht2+/hhf2+ promoter region and the ade6+ control region. The values of ‘relative enrichment’ correspond to Teb1 binding at the hht2+/hhf2+ promoter normalized to the ade6+ promoter and to wt (non-tagged). The experiment was performed in triplicate and error bars correspond to the standard deviation. Unlike teb1+cells, teb1-1 cells do not harbour detectable levels of Teb1 binding to the hht2+/hhf2+ promoter.
Expression profiling reveals a role for Teb1 in transcriptional regulation
Given that Teb1 binds a number of gene promoters, we sought to determine whether the phenotypes of teb1-1 cells stem from altered transcription of these genes. We used microarray technology to assess the transcriptional profile of the teb1-1 strain and compare it to that of wt. RNA from wt and teb1-1 cells grown at 25°C was extracted and differentially labelled by reverse transcription with Cy3- and Cy5-labelled dCTP. The resulting labelled cDNA was hybridized to in-house-constructed glass slide microarrays containing probes for 99.3% of all known and predicted S. pombe genes (Lyne et al, 2003). This procedure was repeated for cells that had been subjected to growth for 1 h at the restrictive temperature of 36°C. The data were analysed with GenePix 6.0 software, with a two-fold difference in gene expression as the minimal change scored as upregulation or downregulation. A comprehensive list of the expression profile results can be found in Supplementary Table II.
Upon inhibition of Teb1 function, many genes become upregulated whereas only a few are downregulated (Supplementary Table III). Intriguingly, the downregulated genes can be classified into gene ontology groups. For example, one such group comprises four genes involved in iron homeostasis, all of which are downregulated by teb1-1 (Supplementary Table IV). In a random distribution of genes, one would expect only ∼0.2% of genes from this group in the list, as only around 0.2% of genes in the genome are involved in iron homeostasis. However, this group represents ∼5 and ∼10% of the genes downregulated by the teb1-1 mutation at 25 and 36°C, respectively. Nonetheless, it is unclear why these four genes (str1, str3, fip1 and frp1) are downregulated, as the only member of this group that shows detectable Teb1 promoter binding is str3 (Supplementary Table I). Hence, Teb1 may act indirectly on these genes, perhaps by regulating the expression of a transcription factor that controls the iron homeostasis genes. Alternatively, the levels of Teb1 binding to the str1, fip1 and frp1 promoters may be significant but below the detection limit used in our ChIP-chip analysis.
Another group that is overrepresented in the list of genes downregulated in a teb1-1 background at 25°C is the ribosomal protein (RP) genes (Supplementary Table II). Moreover, many ribosomal genes that fail to emerge as downregulated using a two-fold threshold show a reduction in expression of just under two-fold. As for the iron homeostasis group, no evidence of Teb1 binding to their promoters was found. Interestingly, the Candida albicans Myb-domain protein Tbf1 plays a role in controlling this same subset of ribosomal genes (Hogues et al, 2008).
Even more striking was the downregulation of expression of the canonical histone genes induced by Teb1 mutation (Supplementary Table V), considered below.
Teb1 is involved in histone transcriptional regulation
The correspondence between Teb1 promoter binding and significant downregulation by teb1-1 mutation for all nine canonical histone genes prompted us to examine this group in more detail. We confirmed the ChIP-chip data by quantitative real-time PCR using primers for the promoter driving hht2+ and hhf2+ (encoding histones H3.2 and histone H4.2), which showed significant enrichment (Figure 3F). To further probe the relationship between Teb1 promoter binding and transcriptional regulation, we investigated the ability of the mutant Teb1-1 protein to bind histone promoters in vivo. ChIP analysis shows a drastic reduction in Teb1-1 binding to the hht2 and hhf2 promoter at both permissive (Figure 3F) and restrictive (unpublished observations) temperatures, highlighting the critical importance of the single mutated arginine residue within the Myb domain for the DNA-binding ability of this protein. Moreover, the vastly reduced binding of Teb1-1 at 25°C provides a plausible explanation for why teb1-1 cells grow slowly, even at permissive temperature.
While the genes encoding all four types of canonical histones were among those downregulated by the teb1-1 mutation (Supplementary Table V), the level of downregulation was more modest for hta1+ and htb1+, which are regulated by the same divergent promoter, than for the other histones, being well under the two-fold cutoff used. Intriguingly, the slight downregulation of these two copies of histones is alleviated when cells are grown for 1 h at the restrictive temperature, 36°C. This suggests that Teb1 exerts only a minor effect on the regulation of hta1+/htb1+, and that additional factors promote transcription of these histones at high temperatures. Unlike the canonical histone genes, Teb1 does not appear to bind or regulate the expression of the H4 variant SPBC800.13, the H2 variant pht1 or the H3 variant cnp1 (Supplementary Figure S2; Supplementary Table II).
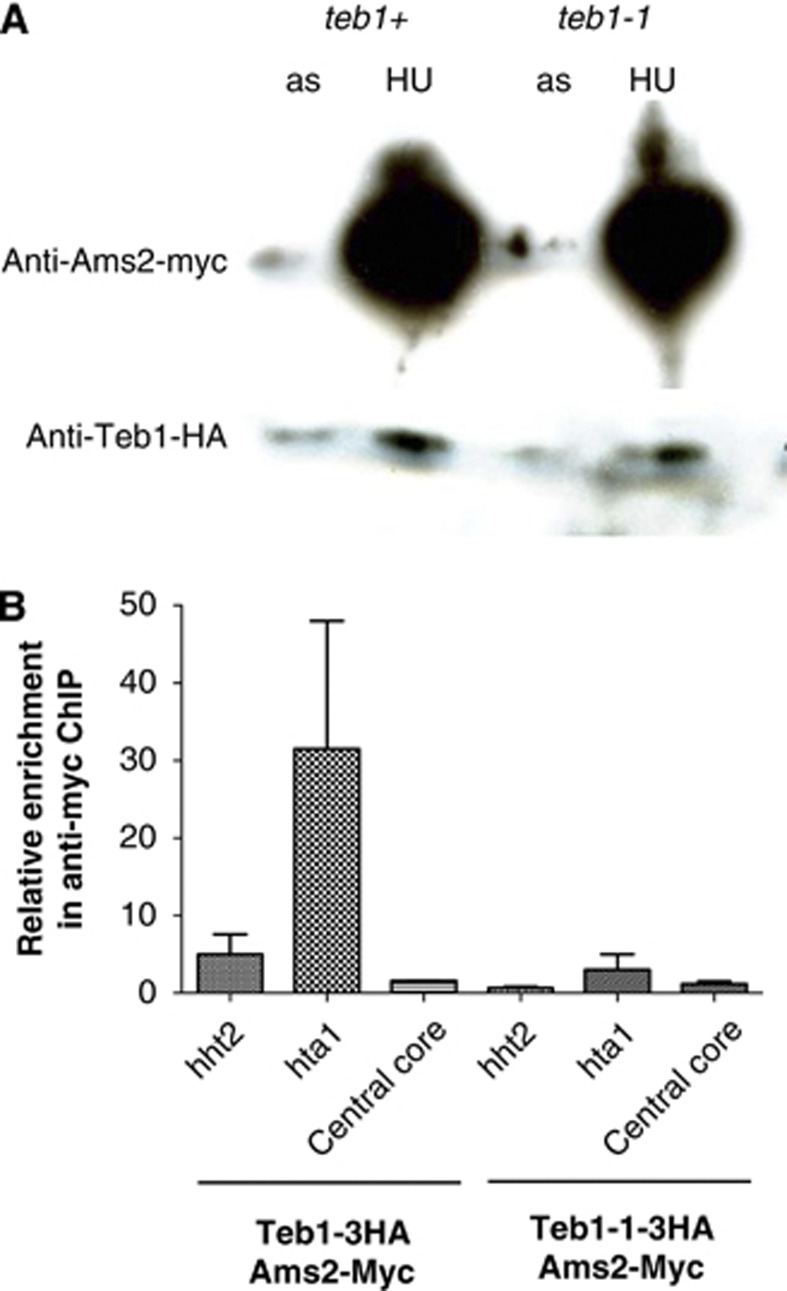
Transcriptional upregulation of the canonical histone genes in S phase is under the control of the GATA-like transcription factor Ams2. While it has been suggested that Ams2 binds histone promoters via the AACCCT motif, it remains unclear whether Ams2 binds directly or is a part of a complex, as there are no GATA sequences within the conserved motif. Our observation that Teb1 binds these motifs at sites known to be regulated by Ams2 suggests Teb1 as a mediator of Ams2 binding. To test this possibility, we fused a 13Myc tag with the C-terminus of Ams2 and HA tags at the C-termini of wt Teb1 and Teb1-1; all fusions were at the endogenous loci and yielded functional protein. As previously published, we observe higher levels of Ams2 in S phase; we also observe moderately elevated levels of Teb1 in S-phase cells, suggesting that this protein is also upregulated during the time window when canonical histones are transcribed (Figure 4A). Importantly, Ams2 levels are not altered by the teb1-1 mutation; our microarray data also failed to detect Teb1 binding to the ams2+ promoter and revealed no changes in ams2+ expression in the teb1-1 background. However, ChIP assays reveal that while Ams2 strongly binds the hht2+/hhf2+ and hta1/htb1+ promoters in a wt background, binding is significantly reduced by the teb1-1 mutation (Figure 4B). Hence, Ams2 binding to histone promoters requires the presence of wt Teb1.
Figure 4.
Binding of Ams2 to histone promoters is partially lost in teb1-1 cells. (A) Western blot showing the levels of Teb1-HA and Ams2-Myc in asynchronously growing and cultures synchronized in S phase by treatment with 15 mM HU for 4 h. Ponceau staining confirmed equal loading in each lane (data not shown). (B) ChIP analysis with an anti-Myc antibody and qPCR using primers for the histone hht2/hhf2 and hta1/htb1 promoter regions or the adh1 locus (which harbours no Teb1 binding; Supplementary Table I) in cells synchronized in S phase with 15 mM HU. Ams2 binding is lost in the teb1-1 background.
Teb1 is essential for maintenance of centromere identity
The regulation of histone levels has been implicated in the maintenance of centromere identity. For instance, the identification of Ams2 as a multicopy suppressor of the Cnp1CENP-A ts allele cnp1-1 (Chen et al, 2003) could be attributed either to direct binding of Ams2 to the centromere central core or to the altered histone levels imparted by Ams2 overexpression. Hence, we wished to explore the possibility that Teb1 plays a role in Cnp1CENP-A recruitment, a role that could comprise at least part of its essential function.
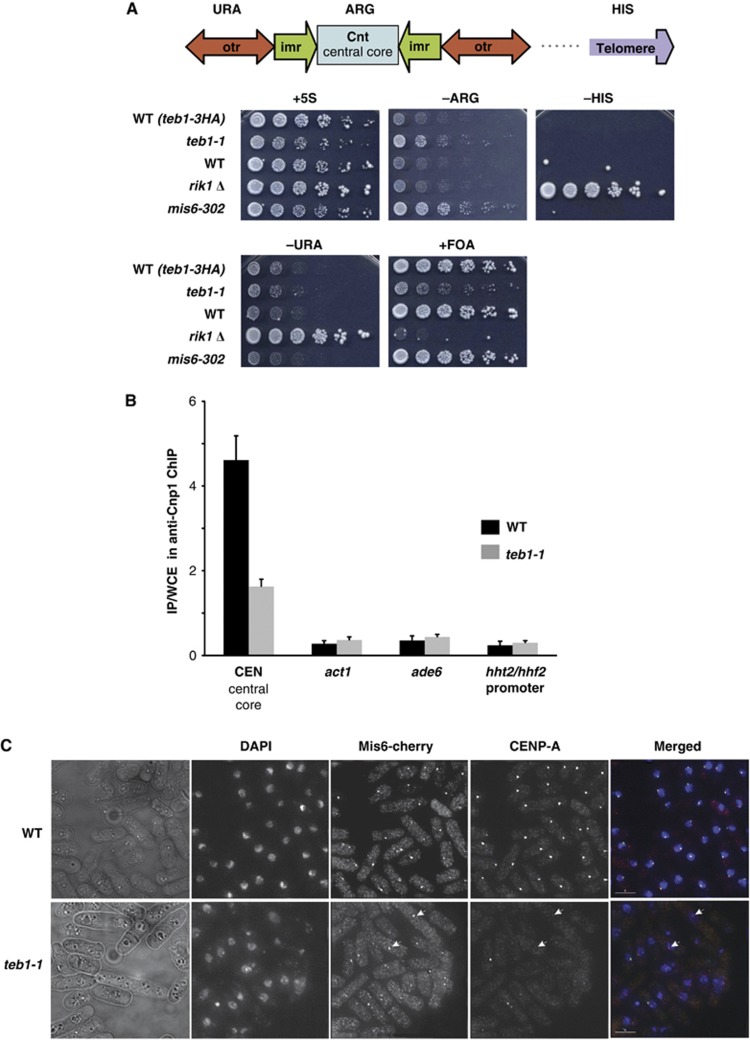
Centromeric Cnp1CENP-A confers local silencing of transcription; hence, expression of a marker gene inserted in the centromeric central core provides a readout for centromeric Cnp1CENP-A assembly (Pidoux et al, 2003). To assess the involvement of Teb1 in the Cnp1CENP-A deposition, we introduced the teb1-1 mutation into strains with an arg3+ marker integrated into the centromeric central core. To separate general effects on gene silencing from specific effects on centromere structure, we used a strain that also harbours markers in other silent regions of the genome, the pericentromeric outer repeats (otr-ura4+) and the telomere (telo-his3+). Two controls were also utilized, the rik1Δ mutation, which abolishes silencing at the centromeric outer repeats and the telomere, and the mis6-302 mutation, which abolishes silencing at the centromeric central core (Ekwall et al, 1996; Partridge et al, 2000). Efficient silencing of reporter genes in the outer repeats and telomere was observed in both wt and teb1-1 backgrounds. However, the reporter gene at the centromeric central core displayed a clear loss of silencing in teb1-1 cells at 25°C (Figure 5A).
Figure 5.
Teb1 is required for normal Cnp1 loading. (A) Silencing at the centromeric central core is specifically reduced in teb1-1 cells. Strains of the indicated genotypes harbouring a ura4+ marker in the otr region, an arg3+ marker in the central core and a his3+ marker in the subtelomere were analysed by serial dilution assay. (B) Cnp1 binding to the central core decreases in teb1-1 cells compared to wt. ChIP was performed with an anti-Cnp1 antibody. The experiment was performed in triplicate and error bars correspond to the standard deviation. (C) Indirect immunofluorescence shows that while Mis6 and Cnp1 colocalize at wt centromeres, Cnp1 foci fail to appear in many teb1-1 cells; Mis6 localization is not affected (white arrowheads).
The central core-specific silencing defect of teb1-1 cells suggests a problem with Cnp1CENP-A deposition and kinetochore formation. To address these latter parameters, we performed ChIP of Cnp1CENP-A in wt and teb1-1 cells and qPCR for central core sequences. Remarkably, levels of central core enrichment in Cnp1CENP-A ChIP were significantly reduced by the teb1-1 mutation (Figure 5B). Interestingly, Cnp1CENP-A levels are slightly higher at the control act1+ or ade6+ loci in teb1-1 cells (Figure 5B). Elevated levels of Cnp1CENP-A at non-centromeric sites may result from reduced loading at the centromere.
Two independent pathways of Cnp1CENP-A loading have been reported in fission yeast (Takayama et al, 2008), an S-phase pathway that depends on Ams2 and a G2 pathway dependent upon Mis6. To assess whether these pathways are differentially affected by the teb1-1 mutation, we carried out indirect immunofluorescence microscopy using antibodies against Cnp1CENP-A or Mis6 tagged with mCherry at its endogenous locus. Log-phase cultures, which comprise mainly G2 cells, were grown for 6 h at 36°C. All the wt cells displayed a single Mis6 or Cnp1CENP-A focus (Figure 5C). In teb1-1 cells, Mis6 localization was not altered. In contrast, most teb1-1 cells were either devoid of visible Cnp1 foci or displayed very weak signal intensity. Importantly, expression levels of Mis6, Ams2, Scm3 (Cnp1 chaperone) do not appear to be altered in the teb1-1 background (Supplementary Table II). These results confirm that Teb1 plays a role in Cnp1CENP-A deposition at the centromeric central core, and indicate that it acts in the Cnp1CENP-A loading pathway independently or downstream of Mis6.
Teb1 binds to subterminal regions of the chromosome where neocentromeres form
Recent studies have shown that excision of the centromere (cen1) on Chromosome I (Chr I) can be tolerated in cells that establish centromere function at an ectopic locus on Chr I referred to as a neocentromere (Ishii et al, 2008). Such neocentromeres assemble Cnp1CENP-A and other centromere components and are formed at one of the two ends of the chromosome in subtelomeric regions distinct from those generally bound by Swi6/HP-1 heterochromatin. Intriguingly, our ChIP-chip analysis reveals a reproducible broad peak of binding of Teb1 in this same region (Supplementary Figure S3), as well as at subtelomeric regions of Chr II; the corresponding region of Chr III comprises rDNA repeats and is not covered by the array. Taken together with the role of Teb1 in maintaining Cnp1 levels at the central core, this observation suggests that Teb1 might have a function in recruiting Cnp1CENP-A upon disruption of cen1, allowing neocentromeres to be formed.
Teb1 regulates a protease capable of clipping histone H3
Our observation that Teb1 status is crucial for histone transcription prompted us to examine the periodic variation in histone protein levels in wt versus teb1-1 settings. To synchronize cells, we arrested them in G1 by nitrogen starvation (Supplementary Figure S4). Western blot analysis of denatured cell extracts of G1-arrested cells using an antibody against histone H3 initially revealed specific degradation products (Supplementary Figure S4A); the Cdc2 protein used as a loading control also appeared vulnerable to G1 arrest-specific degradation (Supplementary Figure S4A). Following refeeding with a nitrogen source and consequent reentry into the cell cycle, full-length Cdc2 and histone H3 reappeared. This clipping depends on both Teb1 and the vacuolar serine protease Isp6, which has been implicated in the downregulation of protein levels during G1 arrest (Nakashima et al, 2006). As our genome-wide map reveals Teb1 binding at the isp6+promoter (Supplementary Table I), and the expression data indicate reduced isp6+ levels in teb1-1 cells (Supplementary Table II), we surmised that Teb1 regulates protein degradation in G1 cells via the control of isp6+ expression. This supposition initially appeared correct, as we found that isp6Δ cells failed to yield the G1-associated clipped protein products (Supplementary Figure S4B). However, western blot analysis of guanidinium-HCl extracted proteins revealed only full-length, unclipped histone H3 and Cdc2 (Supplementary Figure S4D), suggesting that the H3 and Cdc2 degradation products most likely occur in extracts of G1-arrested cells rather than in vivo. Hence, the Isp6 vacuolar protease is upregulated during G1 arrest in a Teb1-dependent manner, and this protease appears to be responsible for protein clipping in extracts of these cells. Further experiments should reveal whether this clipping is relevant to cell cycle arrest, for example, whether transport to the vacuolar compartment is enhanced in vivo.
Discussion
The function of Teb1 has been a matter of intrigue for investigators of Myb domain proteins for over a decade. While its domain organization made Teb1 a candidate telomere binding protein, its in vitro binding affinity for vertebrate, but not fission yeast, telomere repeats raised the possibility of a non-telomeric function. Moreover, Teb1 is essential, making a solely telomeric role unlikely since survivors harbouring circular chromosomes arise readily in the complete absence of telomeres in fission yeast (Naito et al, 1998; Nakamura et al, 1998). The localization pattern of Teb1-GFP reinforced the idea of a non-telomeric function as it shows a dispersed fluorescence throughout the nucleus rather than the clear foci displayed by telomeres; likewise, telomere sequences are not enriched in Teb1 ChIP samples. Rather, our data confirm earlier speculation that Teb1 is a transcriptional regulator that binds many promoter regions, often those containing stretches of TTAGGG repeats. Furthermore, numerous genes whose promoters bind Teb1 also depend on Teb1 for normal levels of transcription.
Among the groups of genes regulated by Teb1, perhaps the most consistent pattern is exhibited by the canonical histone genes, all of whose promoters bind Teb1 and mRNA levels are reduced in strains harbouring the teb1-1 mutation. These observations add a new player to the histone gene regulatory function previously attributed to Ams2, and suggest an explanation for how this GATA binding factor figures so prominently in regulating a set of genes whose promoters are generally devoid of GATA sequences. Indeed, only three of the nine histone-encoding genes have any GATA motif in their entire promoter region (Song et al, 2008). It was proposed that Ams2 might be able to bind GATA-like sequences present in the AACCCT box or that Ams2 might bind the AACCCT box indirectly through other proteins (Takayama and Takahashi, 2007). The binding profile and histone regulatory function of Teb1 make it an ideal candidate Ams2 recruiter, as does our finding that the enrichment of histone promoter sequences in Ams2 ChIP is drastically reduced by the teb1-1 mutation. Accordingly, levels of histone H3 and H4 transcripts drop by 50–70% in teb1-1 cells, a similar effect to that conferred by the loss of Ams2 in the ams2-shut-off strain (Takayama and Takahashi, 2007). Hence, our data support the hypothesis that Teb1 provides a platform for Ams2 binding.
The theme of Teb1 interacting with additional factors to regulate local gene transcription may be widespread, as some genes harbouring promoter-bound Teb1 show elevated expression in teb1-1 cells while others show reduced expression. In this regard, Teb1 function may be partially reminiscent of that of the budding yeast ‘repressor activator protein’ ScRap1, which not only binds telomeres and provides a platform for a complex of end protection factors, but also binds the promoter regions of the RP genes (Warner, 1999); stimulation of RP transcription by ScRap1 is thought to comprise its essential function (Moehle and Hinnebusch, 1991). A number of analogies between ScRap1 and S. pombe Teb1 can be drawn. Both harbour a pair of Myb domains and bind telomere-like sequences; both localize to promoters and appear to exert either repressive or activating effects depending on context. The suggested role of ScRap1 as a ‘placeholder’ that facilitates local assembly of either active or repressive chromatin regions may apply to Teb1 as well (Bhattacharya and Warner, 2008). Notably, both the S. pombe and mammalian Rap1 homologues harbour Myb domains but lack DNA binding activity and localize to telomeres via protein–protein interactions with additional Myb domain-containing telomere binding proteins (the mammalian TRFs or their fission yeast orthologue Taz1) (Li et al, 2000; Kanoh and Ishikawa, 2001). Moreover, expression of the fission yeast RP genes is significantly altered by the teb1-1 mutation (Supplementary Table II). A protein harbouring a single Myb domain, Tbf1, has also been implicated in RP transcription in Candida albicans (Hogues et al, 2008). Tbf1 has essential homologues in both S. cerevisiae and S. pombe as well; in both of the latter species, the essential function of Tbf1 remains to be delineated. Hence, further characterization of Teb1 function may shed light on the evolutionary relationships between this intriguing array of key transcriptional regulators and organizers of the specialized chromatin regions comprising telomeres and centromeres.
The range of genes whose expression is controlled by Teb1 also includes groups of genes whose promoters lack Teb1 binding sites. The effects of Teb1 on expression of genes lacking promoter binding may stem from Teb1-mediated regulation of expression of an additional transcription factor, or such effects may be secondary to downregulation of histone gene expression. For instance, a surfeit of histone-free regions in teb1-1 cells might lead to enhanced transcription of some genes. Alternatively, Teb1 may bind the relevant promoters but at levels that fail to emerge using the two-fold enrichment criterion employed in our genome-wide analysis.
In addition to regulating transcription levels, our results uncover a possible post-translational role for Teb1 in regulating the vacuolar protease Isp6, which appears to be upregulated during G1 arrest in a Teb1-dependent manner. However, the Isp6-mediated histone H3 and Cdc2 cleavage we have observed appears to occur during protein extraction rather than in vivo, since it is not apparent when cells are lysed in a guanidinium-HCl buffer that should more efficiently inhibit the activities of vacuolar proteases. Histone H3 cleavage under conditions of nutrient deprivation has been observed in other organisms including budding yeast, where an unidentified serine protease is thought to clip H3 at specific genomic sites, promoting local nucleosome eviction and transcriptional upregulation (Santos-Rosa et al, 2009); proteolytic clipping of histone H3 has also been reported to occur during differentiation of mouse embryonic stem cells (Duncan et al, 2008).
Teb1 function impacts chromosome structure not only by modulating levels of core histones, but also by influencing the assembly of Cnp1CENP-A chromatin at centromeres. Both centromeric Cnp1CENP-A levels and silencing at the centromeric central core are reduced by the teb1-1 mutation, although Mis6 localization appears normal. These data indicate that Teb1 acts independently or downstream of Mis6 to promote Cnp1CENP-A loading. ChIP experiments did not reveal Teb1 binding to centromeres, making it difficult to assess whether Teb1 controls Cnp1CENP-A loading directly (e.g., by promoting centromeric binding of Ams2) or indirectly through regulation of core histone levels. Moreover, contrary to what has been reported previously (Chen et al, 2003), we could not detect Ams2 enrichment at the central core, even in S phase-arrested wt cells (Figure 4B). Our results therefore suggest that the defect in Cnp1 loading has its origin in an alteration of the histone levels rather than a direct effect at the centromere. However, it is possible that the absence of detectable Ams2 levels at the centromere is an outcome of a less efficient ChIP at this locus, or that Teb1 and/or Ams2 bind the central core transiently during a specific window of early S phase that is not sampled in HU-arrested cells (Kim et al, 2003; Hayashi et al, 2007). Further studies are needed to address these possibilities. Proteins containing Myb-like domains have been implicated in centromere function in both C. elegans (KNL2) and human (Mis18BP1) (Maddox et al, 2007), suggesting a conserved role for Myb domain proteins in maintaining centromere identity. We also found that Teb1 binds the exact region where neo-centromeres form on Chr I upon cen1 excision, raising the possibility that Teb1 provides a signal or a seed for Cnp1CENP-A recruitment during neo-centromere formation. In this scenario, a model for direct interaction between Teb1 and CENP-A at the site of CENP-A loading would appear more relevant than a role for Teb1 in controlling CENP-A loading by modulating core histone levels. The possible validity of such a model awaits in-depth analysis of Teb1 binding at different cell-cycle stages and neocentromere formation in the teb1-1 setting.
Together, these findings not only reveal Teb1 as a general transcriptional regulator, but also provide insights into a remarkable range of roles of Teb1 in processes that involve histone regulation, from transcriptional regulation to the maintenance of centromere integrity.
Materials and methods
Strains
Strains constructed by gene replacement utilized the one-step gene replacement method with a kanMX6 cassette. The strains used are listed in Table I.
Media and growth conditions
Media and growth conditions were as described previously (Moreno et al, 1991). Cultures were grown at 32°C in rich (YES) or minimal media (EMM) unless otherwise indicated. Dilution assays were carried out by spotting five-fold serial dilutions of a starting concentration of 1 × 107 cells/ml. Plates were scanned after 3–5 days.
Construction of teb1 conditional mutants
For isolation of temperature-sensitive teb1 mutants, a cassette encoding C-terminally HA-tagged Teb1 and G418 resistance was randomly mutagenized by error-prone PCR using Vent DNA polymerase (New England Biolabs Ltd.) supplemented with 10 × deoxyguanosine triphosphate (dGTP). PCR fragments were purified and transformed into a wt strain. Approximately 4000 transformants were screened for temperature sensitivity at 36°C and dark red staining on phloxin media.
Live-cell analysis
Logarithmically growing cell cultures (at 32°C for non-ts strains and at 25 or 36°C for ts strains) stained with Hoescht 33342 were visualized on a Zeiss Axioplan 2 Microscope (Carl Zeiss Microimaging, Inc.) with an attached CCD camera (Hamamatsu); images were analysed using Volocity software (Improvision).
Chromatin immunoprecipitation
Cells were grown to log phase (OD=0.5–0.7) and crosslinked with formaldehyde (1% (v/v)) for 15 min at room temperature. Following resuspension in lysis buffer (50 mM HEPES pH 7.5, 140 mM NaCl, 1 mM EDTA, 1% IGEPAL CA-630, 0.1% Sodium deoxycholate, 1 mM PMSF, MG132 (SIGMA) and Inhibitor cocktail (CALBIOCHEM)), cells were lysed using a FastPrep (Q-Biogene), pelleted (30 min at 20 000 × g), resuspended in lysis buffer and sonicated with a Bioruptor (Diagenode). Magnetic beads (Dynabeads, Invitrogen) preincubated with HA antibody (Abcamab9110) or Cnp1CENP-A antibody (a generous gift from A Pidoux and R Allshire) were added to the sonicated DNA for 2 h at 4°C. Extensive washes were carried out in lysis buffer supplemented with 0.0025% SDS, high-salt buffer (50 mM HEPES, 1 M NaCl, 1 mM EDTA), LiCl buffer (20 mM Tris–HCl pH 7.5, 250 mM LiCl, 1 mM EDTA) and 20 mM Tris–HCl pH 7.5, 0.1 mM EDTA. IP complexes were eluted from the beads in 1% SDS at 65°C for 15 min before crosslink reversal for 12 h at 65°C. DNA was then purified using a PCR purification kit (Qiagen) before analysis by qPCR in presence of Sybr-Green (Invitrogen) on a MJ Research Chromo 4 machine. Each measurement was performed in triplicate; standard curves (serial dilution of WCE) and melting curves were performed for each PCR run to ensure amplification specificity. qPCR data were analysed with Opticon Monitor software and enrichment was normalized to WCE. Primers used for qPCR were the following: ade6 5′-AGG TAT AAC GAC AAC AAA CGT TGC-3′ and 5′-CAA GGC ATC AGT GTT AAT ATG CTC-3′ hsp90 5′-CCC TCT AGC ATC TTC TAG ATA CAC T-3′ and 5′-GGG TTT TCT TCT CAG CTA TAA CGT G-3′ hht2 5′-GCA CCA CCC TTT CCC AAT CC-3′ and 5′-GGT TCT TTC CAC GTC GGG TG-3′ act1 5′-GGA GGA AGA TTG AGC AGC AGT-3′ and 5′-GGA TTC CTA CGT TGG TGA TGA-3′ centromere central core 5′-AAC AAT AAA CAC GAA TGC CTC-3′ and 5′-ATA GTA CCA TGC GAT TGT CTG-3′.
Southern blot analysis
Southern blots were performed as described (Miller et al, 2006). Telomeres were detected using a synthetic telomeric fragment (Miller et al, 2006).
ChIP-chip
ChIP-chip experiments were carried out as previously described (Aligianni et al, 2009). Briefly, 6 × 106 cells/ml of JCF 7429 were fixed with formaldehyde (1% v/v) for 30 min, followed by addition of 2.5 M glycine. Cells were disrupted with glass beads (BioSpec) in lysis buffer. Cross-linked chromatin was sheared on a Bioruptor (Diagenode). Soluble protein DNA complexes were immunoprecipitated overnight at 4°C with anti-HA antibody (Abcam 9110) and protein A Sepharose beads (Amersham). The immunoprecipitated material was extensively washed before elution in TES, followed by Proteinase K treatment and crosslink reversal for 5 h at 65°C. DNA was treated with RNaseA for 1 h at 37°C, phenol extracted, ethanol precipitated and amplified by random PCR as previously described (Bernstein et al, 2004). Input and IP samples were labelled with Cy3 and Cy5, respectively, using a Bioprime labelling kit (Invitrogen). Hybridization and washes were carried out according to manufacturer’s instructions for the 4 × 44K ChIP-on-chip whole genome DNA microarray platform (Agilent). The Agilent arrays were scanned in a GenePix 4000B laser scanner at 5 μm resolution, and the acquired fluorescent signals subsequently processed for analysis with GenePix Pro 6.0 software (Axon instruments). The data were then imported to Bioconductor version 2.6.1, and systematic or array bias was removed by the variance stabilization algorithm (Huber et al, 2002). Briefly, each column (Cy3 and Cy5) on the array was calibrated by an affine transformation and then the data were transformed by a glog2 variance stabilizing transformation. After normalization, Cy5/Cy3 or Cy3/Cy5 (dye swap) ratios were obtained for each array element. To determine enrichment ratios at promoter regions, we calculated the mean intensity of all probes within 1000, bp upstream of each open reading frame. To assess statistically significant enrichment over a promoter or coding region, we applied SAM statistics (Significance Analysis of Microarrays) (Tusher et al, 2001). We compared four independent repeats at 36°C and three independent repeats at 25°C. We determined a conservative list of significantly enriched promoters using 0% FDR (false discovery rate). For analysis of Teb1 binding, we performed three independent biological ChIP-chip repeats for samples from cells collected at 25°C and four independent biological repeats for samples from cells grown 1 h at 36°C. The ChIP-chip data have been submitted to ArrayExpress database and assigned the identifier accession: E-MTAB-1253.
Expression microarrays
RNA was processed for microarray hybridization as described (Lyne et al, 2003). In all, 20 μg total RNA from sample and reference was directly labelled with Cy3 and Cy5 dCTP (GE Healthcare) using the Invitrogen Superscript direct cDNA labelling system. Four biological repeats were carried out with dye swap and averaged at analysis step. The resulting labelled cDNA was hybridized to glass slide microarrays containing probes for 99.3% of all known and predicted S. pombe genes. Microarrays were scanned using an Axon GenePix 4000B scanner and analysed with GenePix 6.0 software. Quality control and data normalization was carried out using a custom perl script as described (Lyne et al, 2003). Results were visualized with GeneSpring GX 7.3 (Agilent). The microarray data have been submitted to ArrayExpress database and assigned the identifier accession: E-MTAB-1251.
G1 arrest and release
Prototrophic strains were grown to log phase in EMM+NH4Cl, extensively washed and resuspended in EMM without NH4Cl, starved for 16–20 h at 25°C and shifted to 36°C for 1.5 h before being refed with NH4Cl (5 mg/l). Samples were taken for FACS analysis and western blot analysis at representative time points.
Protein extraction, SDS–PAGE and western blot analysis
TCA extractions were performed as follows: 10–15 ml of liquid cultures were grown to OD (600 nm)=0.4–0.6. Cells were resuspended in 20% TCA, washed in 1 M Tris-Base, and lysed using glass beads. Following centrifugation, the pellet was resuspended in loading buffer (NuPAGE LDS Sample Buffer, Invitrogen). For urea extraction, cell pellets were resuspended in lysis buffer (8 M urea, 200 mM NaCl, 100 mM Tris=7.5, 0.2% SDS, 1 mM PMSF, protease inhibitor cocktail) and disrupted with glass beads. Cleared lysates were supplemented with 1 mM DTT and loading buffer (Invitrogen). For guanidinium-HCl (Gu-HCl) protein extraction, cell pellets were resuspended in lysis buffer (6 M Gu-HCl, 200 mM NaCl, 100 mM Tris pH=7.5, 1 mM PMSF, protease inhibitors cocktail). Following clearing of the lysates, Gu-HCl was removed by TCA precipitation (20% vol/vol). Following cold acetone wash and drying, proteins were resuspended in loading buffer (Invitrogen) supplemented with 1 mM DTT. All protein samples were resolved by SDS–PAGE (NuPAGE Novex 4–12% Bis-Tris gel) and transferred onto an IMS-activated PVDF membrane. Membranes were blocked in PBS 0.1% Tween (PBST) 5% (w/v) milk prior to antibody incubation. Primary antibodies were incubated for 1 h at room temperature; membranes were washed 3 × 10 min in PBST, incubated with the secondary antibody for 45 min at room temp, washed again and detected by ECL (Amersham). Antibodies used: Rabbit anti-Cdc2 (p34 PSTAIRE, Santa Cruz Biotechnology)—1:1000; Rabbit anti-H3 (ab1791, Abcam)—1:3000; Anti-mouse IgG-HRP (NA931V, GE Healthcare)—1:4000; Anti-rabbit IgG-HRP (NA9340, Amersham Biosciences)—1:4000.
FACS analysis
Cells were fixed in cold 70% ethanol before resuspension in 50 mM NaCitrate for RNase A (0.1 mg/ml) treatment for 2 h at 37°C, addition of propidium iodide (16 μg/ml) to the suspension, sonication and processing in FACS (FACS scan).
Indirect immunofluorescence
Cells (OD=0.5) fixed with paraformaldehyde (1% v/v) for 15 min at room temperature, washed in PEM, resuspended in PEMS (100 mM PIPES, 1 mM EGTA, 1 mM MgSO4, pH 6.9 plus 1 M sorbitol) containing 1 mg/ml of Zymolase-100T (ImmunO) and incubated at 37°C for 90 min. After incubation in PEMS supplemented with 1% Triton X-100, cells were washed with PEMS, resuspended in PEMBAL (100 mM PIPES, 1 mM EGTA, 1 mM MgSO4, pH 6.9 plus 1% (w/v) BSA, 0.1% (w/v) NaN3 and 100 mM Lysine) and incubated for 30 min at room temperature. Following an overnight incubation with the primary antibody, cells were washed in PEMBAL and resuspended in 200–250 μl of PEMBAL containing a secondary antibody. Cells were mounted on poly-lysine slides using mounting media containing 1 μg/ml of DAPI solution and visualized on a Zeiss Axioplan 2 Microscope (Carl Zeiss MicroImaging, Inc.) with an attached CCD camera (Hamamatsu). Images were captured and analysed using Volocity software (Improvision). Antibodies used were Sheep anti-Cnp1CENP-A serum (kindly provided by A Pidoux and R Allshire) used at 1:3000 dilution, Rabbit anti-RFP (AB3216, Chemicon international) at 1:100 dilution, Anti-Sheep alexa 488 (A1101-5, Invitrogen) at 1:1000, and Anti-Rabbit Cy3 (C-2306, Sigma) at 1:200.
Supplementary Material
Acknowledgments
We thank Alison Pidoux, Ricardo Almeida, Yuko Takayama, Kohta Takahashi and our laboratory members for discussions; Robin Allshire and Takashi Toda for the generous gift of strains and reagents; and Samuel Marguerat for help with the microarray data submission. We are grateful to an anonymous reviewer for suggesting that we try Gu-HCl extraction to inhibit vacuolar protease activity during lysis. This work was funded by Cancer Research UK and Fundação para a Ciência e Tecnologia.
Author contributions: LPV and JPC conceived the project and wrote the paper. LPV performed the experiments in Figures 1, 2, 3 and 5 and Supplementary Figure S4A. P-MD performed the experiments in Supplementary Figure S4B–D, and provided crucial advice. MK performed the experiments in Figure 4A. Experiments for Figure 4B were done by MK and P-MD. SA and SW processed and analysed the ChIP-chip and expression microarray data, respectively, and JB provided both the resources and crucial support for the genome-wide analyses.
Footnotes
The authors declare that they have no conflict of interest.
References
- Aligianni S, Lackner DH, Klier S, Rustici G, Wilhelm BT, Marguerat S, Codlin S, Brazma A, de Bruin RAM, Bähler J (2009) The fission yeast homeodomain protein Yox1p binds to MBF and confines MBF-dependent cell-cycle transcription to G1-S via negative feedback. PLoS Genet 5: e1000626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Humphrey EL, Liu CL, Schreiber SL (2004) The use of chromatin immunoprecipitation assays in genome-wide analyses of histone modifications. Methods Enzymol 376: 349–360 [DOI] [PubMed] [Google Scholar]
- Bhattacharya A, Warner JR (2008) Tbf1 or Not Tbf1? Mol Cell 29: 537–538 [DOI] [PubMed] [Google Scholar]
- Blackwell C, Martin KA, Greenall A, Pidoux A, Allshire RC, Whitehall SK (2004) The Schizosaccharomyces pombe HIRA-like protein Hip1 is required for the periodic expression of histone genes and contributes to the function of complex centromeres. Mol Cell Biol 24: 4309–4320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo AG, Mellone BG, Partridge JF, Richardson W, Hamilton GL, Allshire RC, Pidoux AL (2007) Plasticity of fission yeast CENP-A chromatin driven by relative levels of histone H3 and H4. PLoS Genet 3: e121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ES, Saitoh S, Yanagida M, Takahashi K (2003) A cell cycle-regulated GATA factor promotes centromeric localization of CENP-A in fission yeast. Mol Cell 11: 175–187 [DOI] [PubMed] [Google Scholar]
- Duncan EM, Muratore-Schroeder TL, Cook RG, Garcia BA, Shabanowitz J, Hunt DF, Allis CD (2008) Cathepsin L proteolytically processes histone H3 during mouse embryonic stem cell differentiation. Cell 135: 284–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekwall K, Nimmo ER, Javerzat JP, Borgstrom B, Egel R, Cranston G, Allshire R (1996) Mutations in the fission yeast silencing factors clr4+ and rik1+ disrupt the localisation of the chromo domain protein Swi6p and impair centromere function. J Cell Sci 109: Pt 112637–2648 [DOI] [PubMed] [Google Scholar]
- Gunjan A, Paik J, Verreault A (2005) Regulation of histone synthesis and nucleosome assembly. Biochimie 87: 625–635 [DOI] [PubMed] [Google Scholar]
- Han M, Chang M, Kim UJ, Grunstein M (1987) Histone H2B repression causes cell-cycle-specific arrest in yeast: effects on chromosomal segregation, replication, and transcription. Cell 48: 589–597 [DOI] [PubMed] [Google Scholar]
- Hayashi M, Katou Y, Itoh T, Tazumi A, Tazumi M, Yamada Y, Takahashi T, Nakagawa T, Shirahige K, Masukata H (2007) Genome-wide localization of pre-RC sites and identification of replication origins in fission yeast. EMBO J 26: 1327–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heun P, Erhardt S, Blower MD, Weiss S, Skora AD, Karpen GH (2006) Mislocalization of the Drosophila centromere-specific histone CID promotes formation of functional ectopic kinetochores. Dev Cell 10: 303–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogues H, Lavoie H, Sellam A, Mangos M, Roemer T, Purisima E, Nantel A, Whiteway M (2008) Transcription factor substitution during the evolution of fungal ribosome regulation. Mol Cell 29: 552–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber W, von Heydebreck A, Sultmann H, Poustka A, Vingron M (2002) Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics (Oxford, England) 18: Suppl 1S96–S104 [DOI] [PubMed] [Google Scholar]
- Ishii K, Ogiyama Y, Chikashige Y, Soejima S, Masuda F, Kakuma T, Hiraoka Y, Takahashi K (2008) Heterochromatin integrity affects chromosome reorganization after centromere dysfunction. Science 321: 1088–1091 [DOI] [PubMed] [Google Scholar]
- Kanoh J, Ishikawa F (2001) spRap1 and spRif1, recruited to telomeres by Taz1, are essential for telomere function in fission yeast. Curr Biol 11: 1624–1630 [DOI] [PubMed] [Google Scholar]
- Ketel C, Wang HS, McClellan M, Bouchonville K, Selmecki A, Lahav T, Gerami-Nejad M, Berman J (2009) Neocentromeres form efficiently at multiple possible loci in Candida albicans. Plos Genet 5: e1000400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S-M, Dubey DD, Huberman JA (2003) Early-replicating heterochromatin. Genes Dev 17: 330–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim UJ, Han M, Kayne P, Grunstein M (1988) Effects of histone H4 depletion on the cell cycle and transcription of Saccharomyces cerevisiae. EMBO J 7: 2211–2219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Oestreich S, de Lange T (2000) Identification of human Rap1: implications for telomere evolution. Cell 101: 471–483 [DOI] [PubMed] [Google Scholar]
- Lyne R, Burns G, Mata J, Penkett CJ, Rustici G, Chen D, Langford C, Vetrie D, Bahler J (2003) Whole-genome microarrays of fission yeast: characteristics, accuracy, reproducibility, and processing of array data. BMC Genomics 4: 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddox PS, Hyndman F, Monen J, Oegema K, Desai A (2007) Functional genomics identifies a Myb domain-containing protein family required for assembly of CENP-A chromatin. J Cell Biol 176: 757–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall OJ, Chueh AC, Wong LH, Choo KH (2008) Neocentromeres: new insights into centromere structure, disease development, and karyotype evolution. Am J Hum Genet 82: 261–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Castellanos C, Blanco M, Rozalen AE, Perez-Hidalgo L, Garcia AI, Conde F, Mata J, Ellermeier C, Davis L, San-Segundo P, Smith GR, Moreno S (2005) A large-scale screen in S. pombe identifies seven novel genes required for critical meiotic events. Curr Biol 15: 2056–2062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto S, Yanagida M (1985) Histone gene organization of fission yeast: a common upstream sequence. EMBO J 4: 3531–3538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meeks-Wagner D, Hartwell LH (1986) Normal stoichiometry of histone dimer sets is necessary for high fidelity of mitotic chromosome transmission. Cell 44: 43–52 [DOI] [PubMed] [Google Scholar]
- Mendiburo MJ, Padeken J, Fülöp S, Schepers A, Heun P (2011) Drosophila CENH3 Is Sufficient for Centromere Formation. Science 334: 686–690 [DOI] [PubMed] [Google Scholar]
- Miller KM, Rog O, Cooper JP (2006) Semi-conservative DNA replication through telomeres requires Taz1. Nature 440: 824–828 [DOI] [PubMed] [Google Scholar]
- Moehle CM, Hinnebusch AG (1991) Association of RAP1 binding sites with stringent control of ribosomal protein gene transcription in Saccharomyces cerevisiae. Mol Cell Biol 11: 2723–2735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno S, Klar A, Nurse P (1991) Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol 194: 795–823 [DOI] [PubMed] [Google Scholar]
- Naito T, Matsuura A, Ishikawa F (1998) Circular chromosome formation in a fission yeast mutant defective in two ATM homologues. Nat Genet 20: 203–206 [DOI] [PubMed] [Google Scholar]
- Nakamura TM, Cooper JP, Cech TR (1998) Two modes of survival of fission yeast without telomerase. Science 282: 493–496 [DOI] [PubMed] [Google Scholar]
- Nakashima A, Hasegawa T, Mori S, Ueno M, Tanaka S, Ushimaru T, Sato S, Uritani M (2006) A starvation-specific serine protease gene, isp6+, is involved in both autophagy and sexual development in Schizosaccharomyces pombe. Curr Genet 49: 403–413 [DOI] [PubMed] [Google Scholar]
- Partridge JF, Borgstrøm B, Allshire RC (2000) Distinct protein interaction domains and protein spreading in a complex centromere. Genes Dev 14: 783–791 [PMC free article] [PubMed] [Google Scholar]
- Pidoux AL, Richardson W, Allshire RC (2003) Sim4: a novel fission yeast kinetochore protein required for centromeric silencing and chromosome segregation. J Cell Biol 161: 295–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Rosa H, Kirmizis A, Nelson C, Bartke T, Saksouk N, Cote J, Kouzarides T (2009) Histone H3 tail clipping regulates gene expression. Nat Struct Mol Biol 16: 17–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumperli D (1986) Cell-cycle regulation of histone gene expression. Cell 45: 471–472 [DOI] [PubMed] [Google Scholar]
- Singh RK, Liang D, Gajjalaiahvari UR, Kabbaj M-HM, Paik J, Gunjan A (2010) Excess histone levels mediate cytotoxicity via multiple mechanisms. Cell Cycle 9: 4236–4244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song JS, Liu X, Liu XS, He X (2008) A high-resolution map of nucleosome positioning on a fission yeast centromere. Genome Res 18: 1064–1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spink KG, Evans RJ, Chambers A (2000) Sequence-specific binding of Taz1p dimers to fission yeast telomeric DNA. Nucleic Acids Res 28: 527–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama Y, Mamnun YM, Trickey M, Dhut S, Masuda F, Yamano H, Toda T, Saitoh S (2010) Hsk1- and SCF(Pof3)-dependent proteolysis of S. pombe Ams2 ensures histone homeostasis and centromere function. Dev Cell 18: 385–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama Y, Sato H, Saitoh S, Ogiyama Y, Masuda F, Takahashi K (2008) Biphasic incorporation of centromeric histone CENP-A in fission yeast. Mol Biol Cell 19: 682–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama Y, Takahashi K (2007) Differential regulation of repeated histone genes during the fission yeast cell cycle. Nucleic Acids Res 35: 3223–3237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G (2001) Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA 98: 5116–5121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassetzky NS, Gaden F, Brun C, Gasser SM, Gilson E (1999) Taz1p and Teb1p, two telobox proteins in Schizosaccharomyces pombe, recognize different telomere-related DNA sequences. Nucleic Acids Res 27: 4687–4694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner JR (1999) The economics of ribosome biosynthesis in yeast. Trends Biochem Sci 24: 437–440 [DOI] [PubMed] [Google Scholar]
- Williams BC, Murphy TD, Goldberg ML, Karpen GH (1998) Neocentromere activity of structurally acentric mini-chromosomes in Drosophila. Nat Genet 18: 30–37 [DOI] [PubMed] [Google Scholar]
- Yuen KWY, Nabeshima K, Oegema K, Desai A (2011) Rapid de novo centromere formation occurs independently of heterochromatin protein 1 in C. elegans embryos. Curr Biol 21: 1800–1807 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.