Background: Snail plays an important role in chemoresistance, but the mechanism is still unclear.
Results: Up-regulation of microRNA-125b through Wnt signaling by snail enriches cancer stem cells and increases chemoresistance.
Conclusion: MicroRNA-125b is a key mediator for Snail-induced stem cell propagation and chemoresistance.
Significance: We reveal a novel mechanism for Snail-induced stem cell maintenance and chemoresistance.
Keywords: Cancer Stem Cells, Cancer Therapy, Cell Death, Drug Resistance, MicroRNA, Chemoresistance, Snail, miR-125b
Abstract
Chemoresistance is a major obstacle in cancer treatment. Our previous studies have shown that miR-125b plays an important role in chemoresistance. Here we report a novel mechanism that up-regulation of miR-125b through Wnt signaling by Snail enriches cancer stem cells. Overexpression of Snail dramatically increases the expression of miR-125b through the Snail-activated Wnt/β-catenin/TCF4 axis. Snail confers chemoresistance by repressing Bak1 through up-regulation of miR-125b. Restoring the expression of Bak1 or depleting miR-125b re-sensitizes Snail-expressing cancer cells to Taxol, indicating that miR-125b is critical in Snail-induced chemoresistance. Moreover, overexpression of miR-125b significantly increases the cancer stem cell population (CD24-CD44+), while depletion of miR-125b or rescue of the expression of Bak1 increases the non-stem cell population (CD24+CD44+) in Snail-overexpressing cells. These findings strongly support that miR-125b functions as a key mediator in Snail-induced cancer stem cell enrichment and chemoresistance. This novel mechanism for Snail-induced stem cell propagation and chemoresistance may have important implications in the development of strategies for overcoming cancer cell resistance to chemotherapy.
Introduction
The chemotherapeutic resistance of cancer is one of the main barriers for cancer treatment. Resistance to chemotherapy can occur prior to drug treatment or may develop over time following drug exposure (1). Paclitaxel (Taxol) is a mitotic inhibitor, widely used in breast cancer chemotherapy (2–6). Taxol has been found to kill non-stem cells more efficiently than cancer stem cells and led to enrichment of the cancer stem cell population (7). The resistance of cancer cells to Taxol results in cancer recurrence and metastasis (8). The mechanism of cancer cell resistance to Taxol may involve dysregulation of P-glycoprotein, non-P-glycoprotein, and tubulin structure (9–10). However, the molecular mechanisms conferring Taxol resistance are still not fully understood.
Snail, a zinc-finger transcription factor, plays a major role in epithelial-mesenchymal transition (EMT).2 Overexpression of Snail has been reported to be responsible for tumor metastasis and recurrence (11–12). Snail has also been shown to induce EMT along with cancer stem cell properties in a breast cancer model (13). However, the mechanism of Snail-induced cancer stem cells is not fully understood.
MicroRNAs (miRNAs) are short ribonucleic acid (RNA) molecules with only 22 nucleotides and are found in all eukaryotic cells. Deregulation of miRNA expression has been discovered in a variety of tumors (14–16). In our previous studies, we found that miR-125b confers breast cancer cell resistance to Taxol (19). Our finding was supported by a recent study that showed a higher expression of circulating miR-125b is associated with poor clinical response of primary breast cancer to chemotherapy and is significantly correlated with tumor grade and lymph node metastasis of the patients (20). However, it is unknown how miR-125b is regulated in cancer cells.
In this study, we report that Snail is up-regulated in Taxol-resistant cancer cells. Moreover, overexpression of Snail dramatically increases the expression of miR-125b in both immortalized breast epithelial cells and in breast cancer cells. Snail regulates the expression of miR-125b through activation of the miR-125b promoter, confers resistance of cancer cells to Taxol, and enriches the cancer stem cell pool size by repressing Bak1 through miR-125b. Rescued expression of Bak1 or depletion of miR-125b decreased the Snail-induced cancer stem cell population and reduced Snail-induced drug resistance. These findings reveal a novel mechanism for Snail-induced stem cell maintenance and chemoresistance, and identify miR-125b as a key mediator for Snail-induced stem cell propagation.
MATERIALS AND METHODS
Cells and Cell Culture
Human breast cancer cell lines MDA-MB-435 (MDA-435), MDA-MB-231 (MDA-231), MCF-7, SKBr3, MDA-MB-468 (MDA-468), and BT474 were purchased from American Type Culture Collection (ATCC). Taxol-resistant cell lines 435TRP, SKBR3TRP, and HMLETRP were developed from parental MDA-MB-435, SKBR3, and HMLE cell lines, respectively, as we previously described (19). MDA-MB-435, MDA-MB-231, MCF-7, and SKBR3 were cultured in DMEM/F-12 (Mediatech Inc.) supplemented with 10% FBS and penicillin/streptomycin. The HMLETRP, HMLE, and HMLE-Snail (kindly provided by Dr. R. A. Weinberg) cell lines were cultured in 1:1 Dulbecco's Modified Eagle's Medium (DMEM)/Ham's F-12 medium (Mediatech Inc) supplemented with 5% FBS (Clontech), 100 units/ml penicillin-streptomycin (Invitrogen), 2 mm l-glutamine (Invitrogen), 10 ng/ml human epidermal growth factor (EGF) (Invitrogen), 0.5 μg/ml hydrocortisone (Sigma), and 10 μg/ml insulin (Sigma).
Mammosphere Formation Assay
Cells were plated in ultra-low attachment 96-well plates (Corning) at different densities per well and grown in DMEM/F12 medium (serum-free) with 20 ng/ml EGF and 20 ng/ml bFGF. 50 μl of fresh media was added every 3 days. Mammospheres were counted at 3–5 days after seeding and pictures were taken. Mammosphere (diameter >75 μm) number was counted.
Flow Cytometry Analysis
Cells (1 × 106) were incubated with CD44-APC and CD24-PE (BD Biosciences) conjugated antibodies and placed on ice 45 min, then washed with blocking buffer (PBS with 0.1% Na2N3, 1%FBS). CD44/CD24 markers were analyzed using a FACSCanto Flow Cytometer (BD Biosciences).
Cell Sorting
Parental HMLE cells (1 × 107) were in incubated with CD44-APC and CD24-PE (BD Biosciences)-conjugated antibodies and placed on ice 45 min then washed with blocking buffer (PBS with 1% FBS) twice. HMLE CD24+CD44+ and CD24-CD44+ cells were sorted from parental HMLE by FACSAria sorter (BD Biosciences) for further experiments. After sorting, the purity of CD24+CD44+ and CD24-CD44+ population was analyzed by Flow Cytometry (99 and 95%, respectively).
Vector Construction and Establishment of Stable Cell Lines
Full-length human precursor miR-125b together with 150 bp of flanking sequence was amplified from human genomic DNA and cloned into the pMSCVpuro expression vector (Clontech). Primers used for the genomic amplification of miR-125b were: CGCAGATCTGCTTAAACGGAATCTCAATT; CTAGAATTCAACAGAAATCCAGAGCTGCC. Snail was amplified from MDA231 cDNA with primers: Snail XhoI F: TGACTCGAGATGCCGCGCTCTTTCCTCGT; Snail EcoRI R: CTAGAATTCTCAGCGGGGACATCCTGAGC, and cloned into pMSCVpuro vector with XhoI and EcoRI and named pMSCV-Snail. eGFP was amplified from pire2EGFP with primers: TCAAGATCTCACGATGATAATATGGCCAC; TGACTCGAGCTGATTATGATCTAGAGTCG; and cloned into pMSCVpuro with BglII and XhoI and named pMSCV-GFP. miR-125b spong and control were designed as previously described (21–22). Spong was cloned into pMSCV-GFP. miR-125b spong1 had 8 miR-125b binding sites, and spong3 had 4 miR-125b binding sites. All cloned fragments were verified by sequencing. Infectious and replication-incompetent retroviral particles were produced according to the manufacturer's protocol. MCF-7, BT474, and HMLE cells were infected with retroviral particles expressing miR-125b or the puromycin resistance gene only (vector control). T47D and MDA468 were infected with retroviral particles expressing Snail or the puromycin resistance gene only (vector control). MDA231 and MDA435TRP were infected with retroviral particles expressing miR-125b spong1, spong3, or scramble. After retroviral infection and primary puromycin selection at 3 μg/ml, cell pools of MCF-7–125b, MCF-7-vector, BT474–125b, BT474-vector, HMLE-125b, HMLE-vector, T47D-vector, T47D-Snail, MDA468-vector, and MDA468-Snail were maintained for multiple culture passages under lower puromycin concentration (1 μg/ml). Bak1 was digested from pIRES2-Bak1 with NheI and EcoRI, and subcloned into pIRESPuro3 (Clontech) and named Puro3-Bak1. Plasmids were transfected with Lipofactamine 2000 (Invitrogen) according to the manufacturer's protocol.
Quantitative Real-time PCR (qRT-PCR)
Total RNA was isolated from cultured cells using TRIzol reagent (Invitrogen). For miRNA expression analysis, qRT-PCR was performed using qRT-PCR miRNA Detection Kit and mirVana qRT-PCR Primer Sets (Applied Biosystems) according to the manufacturer's protocols. Human U6 served as an internal control. Primers for pri-miR125b-1 and miR-125b-2 were devised as reported in the literature (23). Precursor miR-125b-1 RT primer: GTCGTATCCAGTGC-AGGGTCCGAGGTATTCGCACTGGATACGACAGCACG; Precursor miR-125b-2 RT primer: GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACTCCCCT. For quantitative PCR, cDNA was mixed with 2× SYBR Green PCR Master Mix (Applied Biosystems) and various sets of gene-specific primers and then subjected to RT-PCR quantification using the iQ5 Real-Time PCR system (Bio-Rad). All reactions were performed in triplicate. The relative amounts of mRNA were calculated by using the comparative CT method. The results are presented as fold-change of each miRNA.
siRNA Experiments
siRNA oligonucleotides for TCF4, β-catenin, and Snail were purchased from Sigma, with a scrambled siRNA (Sigma) serving as a control. Transfection was performed using Lipofectamine 2000 Transfection Reagent (Invitrogen) according to the manufacturer's protocol. 48 h after transfection, the cell lysates were prepared for further analysis by Western blotting.
Pre-miRNA and anti-miRNA Transfection
miRNA precursors (pre-miRNAs) and antisense miRNAs (anti-miRNAs) were purchased from Applied Biosystems. Pre-miR-Negative and anti-miR-Negative were used as negative controls. Lipofectamine 2000 (Invitrogen) was used for the transfection of pre-miRNAs or anti-miRNAs. 48 h after transfection, the expression of Bak1, a target of miR-125b, was tested by Western blotting.
Luciferase Reporter Assay
The miR-125b promoter pGL3 basic luciferase vector was constructed according to the literature and TSSG prediction. pGL3-miR-125b promoter (Wt) with forward primer: ACTGGTACCAGAAGAACAAGAAGAAGAAAG; pGL3-miR-125b promoter with deletion of one TCF4 binding site named (pGL3-miR-125b del-1) with forward primer: ACTGGTACCAAAGGGTCATCTTCCC-ATCTG; Reverse primer was: ACTAGATCTTAT-TTCTTCTCTAAGCTCCTT. The above two different forward primers with reverse primer were amplified from human genomic DNA and cloned into pGL3 basic luciferase with BglII and KpnI. pGL3-miR-125b promoter with deletion of two TCF4 binding sites named (pGL3-miR-125b del-2) was amplified from pGL3-miR-125b del-1 with forward primer and reverse primer: TTCCCAC-TTCGTGTCTACACAGCCTGGTGCTCGCTC, GAGCGAGCACCAGGCTGTGTAGACACGAAGTGGGAA. We also generated pGL3-miR-125b del-3, pGL3-miR-125b del-4, and pGL3-miR-125b del-5 (pGL3-miR-125b promoter with deletion of three, four, or five TCF4 binding sites, respectively) by the same mutation method as above. HMLEvector and HMLE-Snail were transfected with PGL3-miR-125 or pGL3-miR-125b promoters with different deletions of TCF4, using Lipofectamine 2000 reagent. 48 h later, cells were harvested and lysed with passive lysis buffer (Promega). Luciferase activity was measured by using a dual luciferase reporter assay (Promega). The pRL-SV40 vector (Promega) was used as an internal control. The results were expressed as relative luciferase activity (Firefly Luc/Renilla Luc).
Western Blotting
Cells were harvested and lysed in NETN (20 mm Tris-HCl, pH 8.0, 100 mm NaCl, 1 mm EDTA, 0.5% Nonidet P-40) for 10 min on ice. Lysates were cleared by centrifugation at 13,200 rpm at 4 °C for 10 min. Supernatants were collected and protein concentrations were determined by the Bradford assay (Bio-Rad). The proteins were then separated with a SDS/polyacrylamide gel and transferred to a Nitrocellulose membrane (Bio-Rad). After blocking in TBS with 5% BSA (Sigma) for 1 h, the membranes were incubated overnight at 4–8 °C with the primary antibodies in TBST containing 1% BSA. The following antibodies were utilized: Bak1, Snail, and TCF4 antibodies were purchased from Cell Signaling, the β-actin antibody was purchased from Sigma, and the tubulin antibody was from Santa Cruz Biotechnology. Membranes were extensively washed with TBST and incubated with horseradish peroxidase-conjugated secondary anti-mouse antibody or anti-rabbit antibody (dilution 1:2,500, Bio-Rad). After additional washes with TBST, antigen-antibody complexes were visualized with the enhanced chemiluminescence kit (Pierce).
Cell Viability Assay
A total of 5 × 103∼1 × 104 cells/well were seeded in 96-well plates. 24 h later, the medium was replaced with fresh medium with or without Taxol and then incubated for 48 h. Cell viability was determined using CellTiter 96 AQueous One Solution Cell Proliferation Assay kit (Promega).
Statistical Analysis
Statistical evaluation for data analysis was determined by Unpaired Student's t test. All data are shown as the means ± S.E. p < 0.05 was considered statistically significant.
RESULTS
miR-125b Is Transcriptionally Activated by Snail through Wnt/β-catenin/TCF4
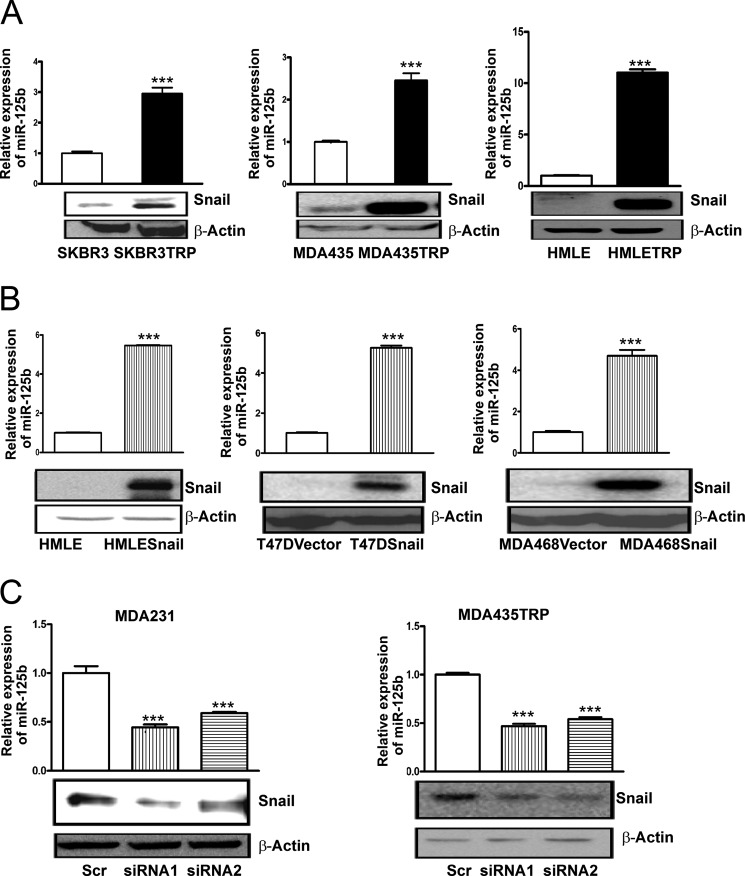
miR-125b plays a critical role in breast cancer resistance to Taxol (19). However, the mechanism of miR-125b regulation in cancer cells is unknown. Snail has been reported to confer drug resistance in cancer cells (24–27), but how Snail induces chemoresistance is not fully understood. To examine whether Snail is overexpressed in Taxol-resistant cancer cells, we compared the Snail protein level between parental SKBR3 and Taxol-resistant SKBR3TRP, parental MDA435, and Taxol-resistant MDA435TRP cells, as well as between parental HMLE and Taxol-resistant HMLETRP cells (supplemental Fig. S1), three pairs of Taxol-sensitive and -resistant cell lines established in our laboratory (19), by immunoblotting. Compared with their parental cells, Taxol-resistant cells showed much higher expression levels of Snail. Meanwhile, higher expression of miR-125b was also found in Taxol-resistant cells compared with their parental cells (Fig. 1A). To test whether miR-125b is regulated by Snail, we forced ectopic expression of Snail in the Snail low-expressing breast cell lines HMLE, T47D, and MDA468 (Fig. 1B). Compared with cells transfected with vector alone, Snail overexpressing cells showed a dramatically higher expression of miR-125b. To confirm this finding, we knocked down the expression of Snail in natural Snail-overexpressing cell lines MDA-MB-231 and MDA435TRP by siRNAs (Fig. 1C). Silencing of Snail with two independent different siRNA sequences markedly decreased the expression of miR-125b in both cell lines. Taken together, these results show that Snail regulates the expression of miR-125b.
FIGURE 1.
miR-125b is activated by Snail in breast cancer cell lines. A, qRT-PCR was performed to examine the expression of miR-125b in Taxol-resistant SKBR3TRP, 435TRP, HMLETRP, and their parental SKBR3, MDA-435, and HMLE cell lines. RNU6B was used as an internal control and for normalization of the data. Cell lysates were prepared for Western blotting with an antibody against Snail, and β-actin was used as a loading control. B, HMLE, HMLE-Snail, T47D-vector, T47D-Snail, MDA468-vector, and MDA468-Snail stable cell lines were collected. Samples underwent RNA extraction or were prepared for cell lysates. qRT-PCR was performed to examine the expression of miR-125b in breast cells. Cell lysates were prepared for Western blotting with an antibody against Snail, and β-actin was used as a loading control. C, MDA231 or MDA435TRP cells were transfected with scramble siRNA or Snail siRNA in duplicate wells. 48 h after transfection, one panel of samples was prepared for lysates, and Western blotting was carried out with antibody against Snail. β-Actin was used as a loading control. Additional samples from the same cell line were used for extracting RNA. qRT-PCR was performed to validate the expression of miR-125b in MDA231 and MDA435TRP with depletion of Snail by siRNA. RNU6B was used as an internal control and for normalization of the data. Columns represent the mean of three independent experiments; bars represent S.E. ***, p < 0.001.
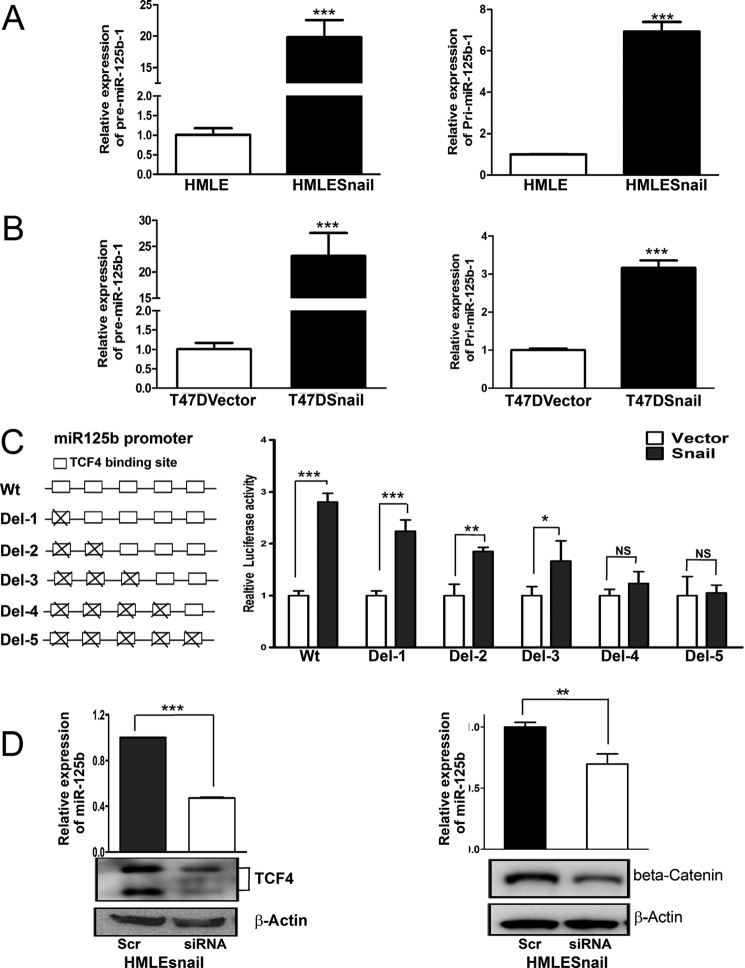
Mature miR-125b is processed from two different precursor miRNAs (pre-miRNAs) and two primary miRNA (pri-miRNAs). miR-125b-1 is located on chromosome 11 while miR-125b-2 is located on chromosome 21 (23). To determine which miR-125b is activated by Snail, we examined the expression of precursor miR-125b-1 (pre-miR-125b-1) and precursor miR-125b-2 (pre-miR-125b-2), as well as primary miR-125b-1 (pri-miR-125b-1) and primary miR-125b-2 (pri-miR-125b-2) in isogenic Snail low- and high-expressing HMLE and T47D cells. Compared with HMLE and T47D vector, there was a dramatic increase in the expression of precursor miR-125b-1 and pri-miR-125b-1 in HMLE-Snail and T47D Snail cells (Fig. 2, A and B). However, expression of precursor miR-125b-2 was almost not detectable in HMLE, HMLESnail, T47D vector, and T47DSnail cells (data not shown). These results indicate that Snail regulates the transcription of miR-125b-1; however, we cannot exclude the possibility that Snail may regulate miR-125b-2.
FIGURE 2.
Snail up-regulates miR-125b through transcriptional activation of the miR-125b promoter. A and B, pre-(Precursor) and pri-miRNA of miR-125b1 were measured by qRT-PCR in HMLE, HMLE-Snail, T47D-vector, and T47D-Snail cells. Columns represent the mean of three independent experiments; bars represent S.E. ***, p < 0.001. C, the full-length of the miR-125b promoter (WT) and miR-125b1 promoters with different TCF4 sites deletion were co-transfected with pRL-SV40 into HMLE and HMLE-Snail cells. 48 h later, cells were harvested and lysed with passive lysis buffer. Luciferase activity was measured by using a dual luciferase reporter assay. The pRL-40 vector was used as an internal control. The results were expressed as relative luciferase activity. Columns represent the mean of three independent experiments; bars represent S.E. NS, no statistical significance; *, p < 0.05; **, p < 0.01; ***, p < 0.001. D, HMLE-Snail cells were transfected with scrambled siRNA, TCF4, or β-catenin siRNA in duplicate wells. 48 h after siRNA transfection, the expression of miR-125b was measured by qRT-PCR. Columns represent the mean of three independent experiments; bars represent S.E. ***, p < 0.001. 48 h after transfection, cell lysates were prepared, and immunoblot analyses were carried out with antibodies against TCF4, β-catenin, or β-actin.
It has been reported that Snail activates Wnt signaling by interacting with β-catenin and TCF4 to activate the expression of target genes (28–29). We found that there are five putative TCF4 binding sites (CANNTG) within the miR-125b-1 promoter. To investigate whether Snail activates the miR-125b-1 promoter through TCF4, we cloned the full-length promoter, and promoters containing deletions of one, two, three, four, and five TCF4 binding sites of pri-125b-1, into the pGL3-basic vector and named them miR-125b1 promoter, miR-125b1-TCF4 del1, miR-125b1-TCF4 del2, miR-125b1-TCF4 del3, miR-125b1-TCF4 del4, or miR-125b1-TCF4 del5, respectively. We co-transfected the miR-125b-1 promoter and miR-125b-1-TCF4 del promoters with internal control vector pRL-SV40 into Snail-low expressing HMLE vector cells and Snail-high expressing HMLESnail cells (Fig. 2C). Compared with HMLE vector cells, the full-length miR-125b-1 promoter in HMLE-Snail showed much higher activity, indicating that the promoter of miR-125b is activated by Snail. Compared with the full length promoter (miR-125b-1 promoter), deletion of TCF4 binding sites decreased the promoter activity of miR-125b-1 in Snail-expressing cells in a dose-dependent manner (Fig. 2C). These results indicate that TCF4 is required for Snail to fully activate the promoter of miR-125b. To further confirm this result, we knocked down the expression of TCF4 and β-catenin (Fig. 2D) in HMLE-Snail cells with TCF4 siRNA and β-catenin siRNA. Knockdown of both TCF4 and β-catenin expression decreased the expression of miR-125b in Snail expressing cells, which further confirms that Snail regulates miR-125b through TCF4 and β-catenin.
Snail Regulates the Expression of Bak1
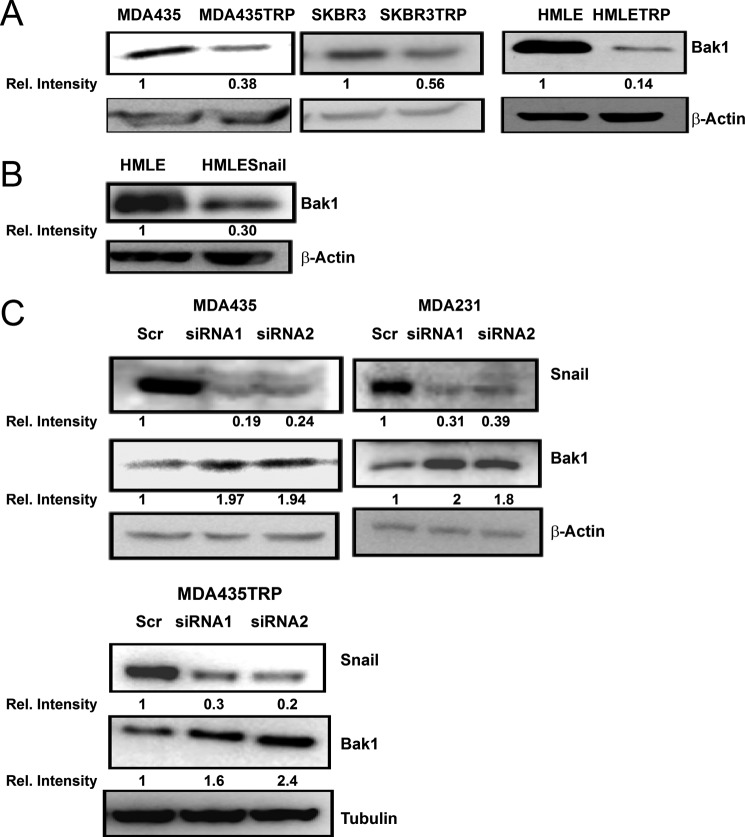
Bak1 is a member of the Bcl-2 gene family, which is involved in initiating apoptosis. Deregulation of Bak1 is important for cancer development and drug resistance (19). Because overexpression of Snail confers chemotherapeutic resistance (25), and Snail was up-regulated in Taxol-resistant cells (Fig. 1A), we next examined whether Snail induces drug resistance through Bak1. To explore the potential link between Snail and Bak1, we compared the expression of Bak1 in Taxol-resistant cells to their parental cells by immunoblotting (Fig. 3A). In comparison with parental cell lines, there were low expression levels of Bak1 in Taxol-resistant SKBR3TRP, MDA435TRP, and HMLETRP cells, which have high expression of Snail. To verify whether Snail regulates the expression of Bak1, we enforced ectopic expression of Snail in HMLE cells. We found that expression of Snail represses Bak1 expression in HMLE cells (Fig. 3B). To confirm this, we examined the expression of Bak1 after knockdown of endogenous Snail by siRNAs in three Snail high expressing cell lines MDA231, MDA435, and MDA435TRP (Fig. 3C). Silencing Snail resulted in increased expression of Bak1 in all three cell lines, indicating that endogenous Snail regulates Bak1.
FIGURE 3.
Bak1 is inversely correlated with Snail in breast cancer cells. A, SKBR3TRP, MDA-435TRP, HMLETRP, and their parental SKBR3, MDA-435, and HMLE cells were harvested. Cell lysates were prepared for Western blotting with an antibody against Bak1. β-Actin was used as a loading control. B, Snail ectopic expressing HMLE and parental HMLE cells were harvested. Cell lysates were prepared for Western blotting with an antibody against Bak1, and β-actin was used as a loading control. C, MDA231, MDA435, and MDA435TRP were transfected with scrambled siRNA or Snail siRNA. 48 h after transfection, cell lysates were prepared for Western blotting with an antibody against Snail, Bak1, or β-actin.
Snail Regulates Bak1 through miR-125b
Bak1 is a confirmed target of miR-125b (19, 30). Since overexpression of Snail increases the expression of miR-125b while repressing the expression of Bak1 in cancer cells, we explored whether Snail regulates Bak1 through miR-125b. To this end, we knocked down Snail in MDA231 (Fig. 4A) and MDA435TRP (Fig. 4B) with siRNA in negative control miRNA or miR-125b-transfected cells. Depletion of Snail in MDA231 and MDA435TRP cells increased the expression of Bak1 in comparison with scrambled siRNA. Moreover, as shown in Fig. 4, A and B, re-expression of miR-125b in the Snail-depleted MDA-231 and MDA435TRP cells decreased the expression of Bak1 to normal expression levels in the control cells. To verify this result, we transfected HMLESnail cells with control (anti-neg) and miR-125b inhibitor (anti-miR-125b) (Fig. 4C). Inhibition of miR-125b in Snail-expressing cells restored the expression of Bak1 to the level in the parental HMLE cells. These results further support that Snail regulates Bak1 through miR-125b.
FIGURE 4.
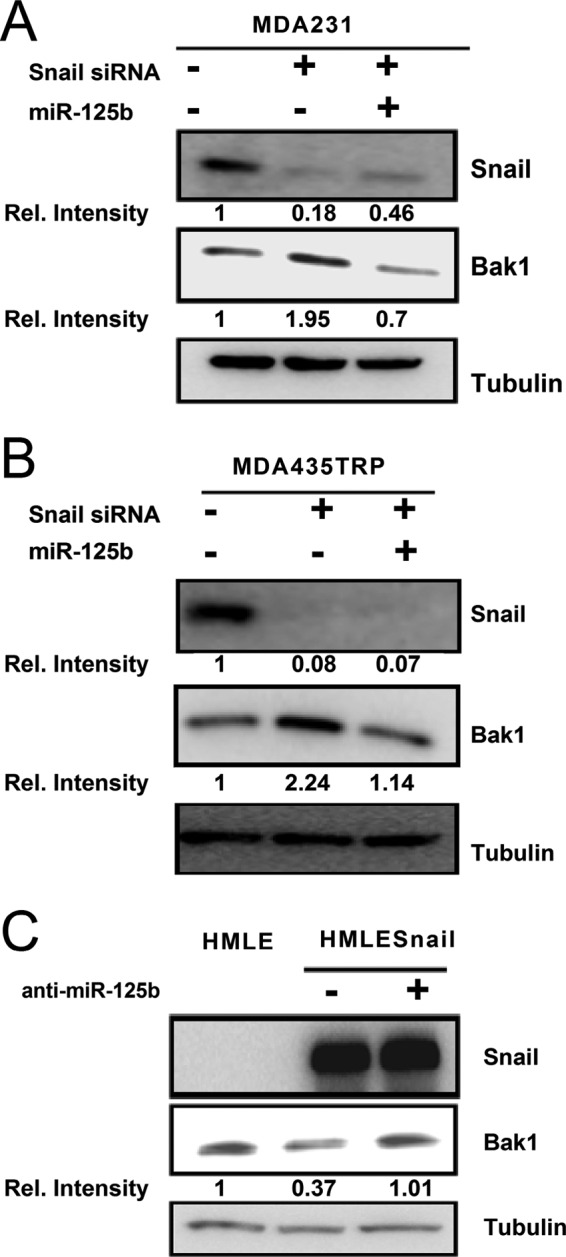
Bak1 is repressed by Snail through miR-125b in breast cancer cells. A and B, MDA-231 or MDA435TRP cells were transfected with 100 nm scrambled siRNA or with 100 nm Snail siRNA alone or with 100 nm Snail siRNA in combination with 100 nm miR-125b. Western blotting was performed to detect the expression of Snail and Bak1. α-Tubulin was used as a loading control. C, Snail ectopic expressing HMLE cells were transfected with 100 nm anti-neg-miRNA or anti-miR-125b. Western blotting was performed to detect the expression of Snail and Bak1. α-Tubulin was used as a loading control.
Snail Increases Chemoresistance by Repressing Bak1 through miR-125b
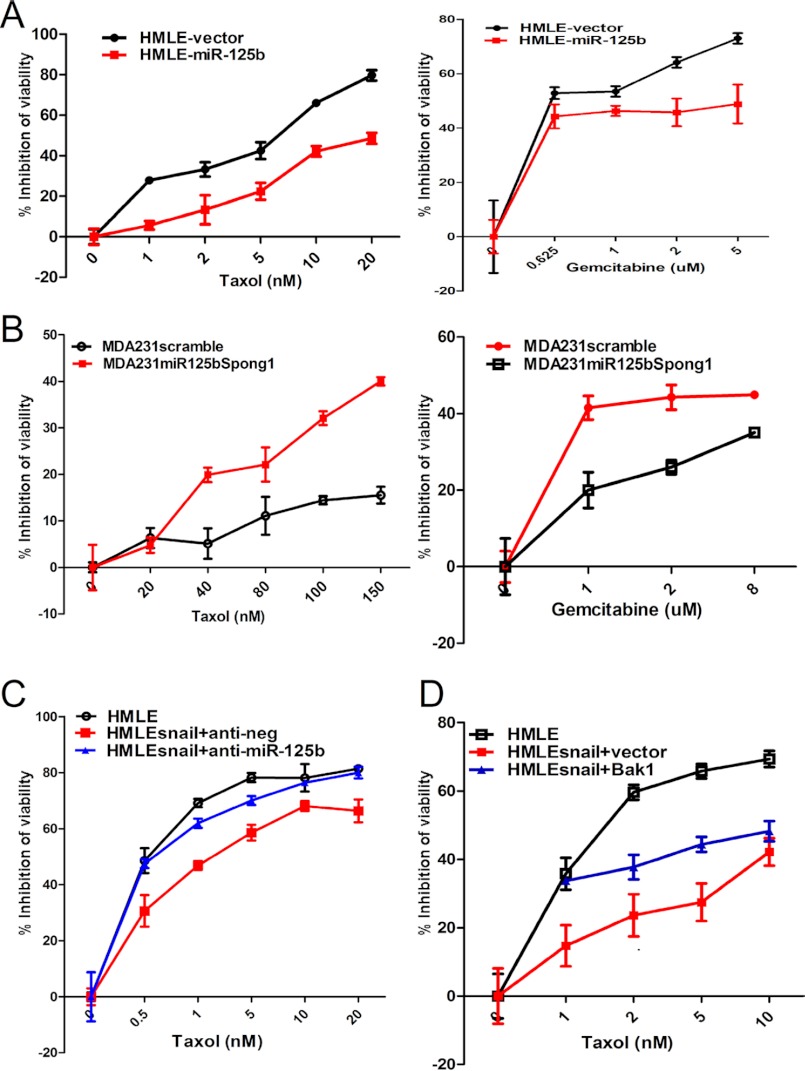
We next sought to determine if Snail increases chemoresistance by repressing Bak1 through miR-125b. First, we treated HMLE and HMLE-Snail cells with different concentrations of Taxol, Gemcitabine, and doxorubicin HCl (supplemental Fig. S2). Overexpression of Snail dramatically increased the resistance of HMLE cells to these chemotherapeutic agents, indicating that overexpression of Snail confers cancer cell drug resistance. We next examined the role of miR-125b in Snail-induced multi-drug resistance. We overexpressed miR-125b in HMLE cells, and then we treated HMLE-vector and HMLE-miR-125b cells with different concentrations of Taxol or Gemcitabine (Fig. 5A). We found that miR-125b increased HMLE cells' resistance to Taxol and Gemcitabine. Then, we inhibited the expression of miR-125b by transfecting miR-125b high expressing MDA231 cells with a miRNA inhibitor (miR-125b spong1). miRNA spong is a decoy target of miRNA that has a specific miRNA seed region and allow them to block the specific miRNAs (21–22). Compared with the scrambled miRNA, miR-125b spong1 re-sensitized MDA231 cells to Taxol and Gemcitabine (Fig. 5B), indicating that miR-125b confers multi-drug resistance in these cells. Next, we determined whether Snail induces multi-drug resistance through miR-125b. We transfected HMLE-Snail cells with anti-neg (Control) or anti-miR-125b (miR-125b inhibitor). Inhibition of miR-125b with anti-miR-125b significantly re-sensitized HMLE-Snail cells to Taxol (Fig. 5C) and slightly re-sensitized HMLE-Snail cells to Gemcitabine (supplemental Fig. S3, left). Moreover, restoration of miR-125b target gene Bak1 expression re-sensitized HMLE-Snail cells to Taxol and Gemcitabine (Fig. 5D and supplemental Fig. S3, right). These results indicate that down-regulation of Bak1 through miR-125b plays a pivotal role in Snail-induced multi-drug resistance.
FIGURE 5.
Snail increases drug resistance by repressing Bak1 through miR-125b. A and B, HMLE-vector, HMLE-miR-125b, MDA231 scramble, and MDA231-miR-125bspong1 cells were seeded into 96-well plates at a density of 8 × 103 cells per well and then treated with different concentrations of Taxol or Gemcitabine for 48 h. The inhibition of cell viability was detected. Points represent the mean of three independent experiments; Bars represent S.E. C, HMLE-Snail was transfected with 100 nm anti-neg (Ctr) or anti-miR-125b. Cells were seeded 24 h after transfection into 96-well plates at a density of 5 × 103 cells per well, incubated for 8 h, and then treated with different concentrations of Taxol. After 48 h of Taxol treatment, inhibition of viability was measured. D, HMLE-Snail cells were transfected with 4 μg of puro3vector and puro3-Bak1. 24 h after transfection, cells were seeded into 96-well plates at the density of 5 × 103 cells per well, incubated for 8 h, and then treated with different concentrations of Taxol. 48 h after Taxol treatment, inhibition of viability was assessed.
miR-125b Increases the Cancer Stem Cell Pool Size
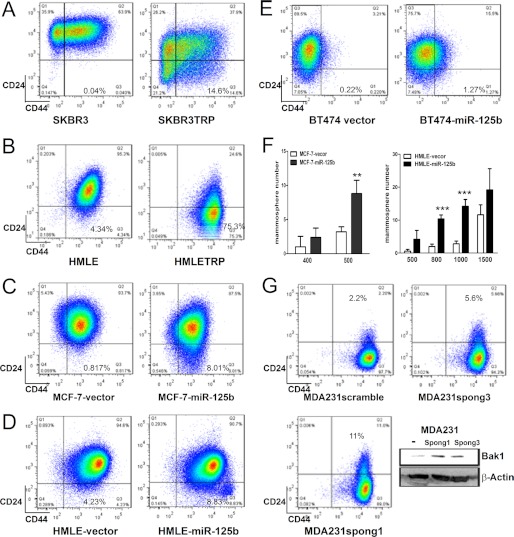
Emerging evidence supports the theory that chemoresistance arises from cancer stem cells. To test this hypothesis, CD24+CD44+ and CD24-CD44+ cells were sorted from parental HMLE, and then treated with Taxol or Gemcitabine. Compared with CD24+CD44+ cells, CD24-CD44+ cells were more resistant to Taxol and Gemcitabine (supplemental Fig. S4A and S5). In addition, it has been reported that miR-125b, in human lymphocytes, blocks cell differentiation and maintains CD4+ T cells in their naïve state (31). This implies that miR-125b may play a role in maintaining cancer stem cells. To further determine whether miR-125b confers cancer cell to chemoresistance through increasing cancer stem cell population, two Taxol-resistant cell lines SKBR3TRP and HMLETRP, which express high level of miR-125b, were analyzed for CD24 and CD44 (Fig. 6, A and B). Compared with the parental SKBR3 and HMLE cells, SKBR3TRP and HMLETRP cells have a dramatically increased CD24-CD44+ cell population (0.04% versus 14.6%, 4.34% versus 75.3%). Furthermore, higher expression of miR-125b was also found in CD24-CD44+ cells compared with CD24+CD44+ cells (supplemental Fig. S4B). We next examined whether overexpression of miR-125b leads to the enrichment of CD24-CD44+ cancer stem cells. MCF-7, HMLE, and BT474 cells were transfected with a control pMSCV-vector and a miR-125b-expressing pMSCV-miR-125b vector (Fig. 6, C–E). As anticipated, the CD24-CD44+ population in MCF-7-miR-125b cells, HMLE-miR-125b cells, and BT474-miR-125b cells all dramatically increased compared with the cells transfected with vector alone. These results indicate that miR-125b increases the cancer stem cell population in breast cancer cells. Previous literature reported that CD24-CD44+ population of immortalized human breast epithelial cells (HMLE) form mammospheres efficiently (13). To determine whether miR-125b enhances mammosphere formation in breast cells, we seeded different cell numbers of MCF-7-vector, MCF-7-miR-125b, as well as HMLE-vector and HMLE-miR-125b in 96-well plates (Fig. 6F). Compared with vector transfection alone, overexpression of miR-125b dramatically increased mammosphere formation in MCF-7-miR-125b and HMLE-miR-125b cells. These results further support that miR-125b plays a role in maintaining breast cancer stem cells. To further confirm these results, we next determined whether depletion of miR-125 increases the non-stem cell population (CD24+/CD44+). We blocked the expression of miR-125b in miR-125b high expressing MDA231 and MDA435TRP cells with miRNA competitive inhibitors (miR-125b spong1 and miR-125b spong3) (Fig. 6G and supplemental Fig. S4). The expression of Bak1 was examined as an indicator of the depletion efficiency of miR-125b. We found that both miR-125b spong1 and miR-125b spong3 increased the expression of Bak1, but miR-125b spong1 (with 8 competitive binding sites) inhibits the expression of miR-125 more efficiently than miR-125b spong3, which has only 4 binding sites (Fig. 6G). Subsequently, depletion of miR-125b in MDA-231 cells increased the non-stem cell population (CD24+CD44+) in a dosage-dependent manner in terms of inhibition of miR-125b (11% versus 5.6% versus 2.2%). Similar results were also obtained in MDA-435TRP cells (supplemental Fig. S4). These result further support the role of miR-125b in breast cancer stem cells.
FIGURE 6.
miR-125b increases the cancer stem cell pool size. A–E, 1 × 106 SKBR3, SKBR3TRP, HMLE, HMLETRP, MCF-7-vector, MCF-7-miR-125b, HMLE-vector, HMLE-miR-125b, BT474-vector, and BT474-miR-125b stable cell lines were incubated with CD24 and CD44 antibodies for 45 min, washed, and then analyzed by flow cytometry. F, MCF-7-vector, MCF-7-mir-125b, HMLE-vector, and HMLE-miR-125b were seeded in ultralow attachment 96-well plates at different cell numbers with conditioned medium. After 3–5 days incubation, the mammosphere (diameter >75 μm) number was counted. X-axis represents the number of seeded cells per well. Columns represent the mean of three independent experiments; Bars represent S.E. **, p < 0.01; ***, p < 0.001. G, MDA231 scramble-spong, MDA231-miR-125b-spong3, and MDA231-miR-125b-spong1 cells were incubated with CD24 and CD44 antibodies for 45min, washed, and then analyzed by flow cytometry. Cell lysates of MDA231 scramble-spong, MDA231-miR-125b-spong3, MDA231-miR-125b-spong1 were prepared for Western blotting. Antibodies against Bak1 and β-actin were used.
miR-125b Is a Key Mediator for Snail-induced Stem Cell Propagation
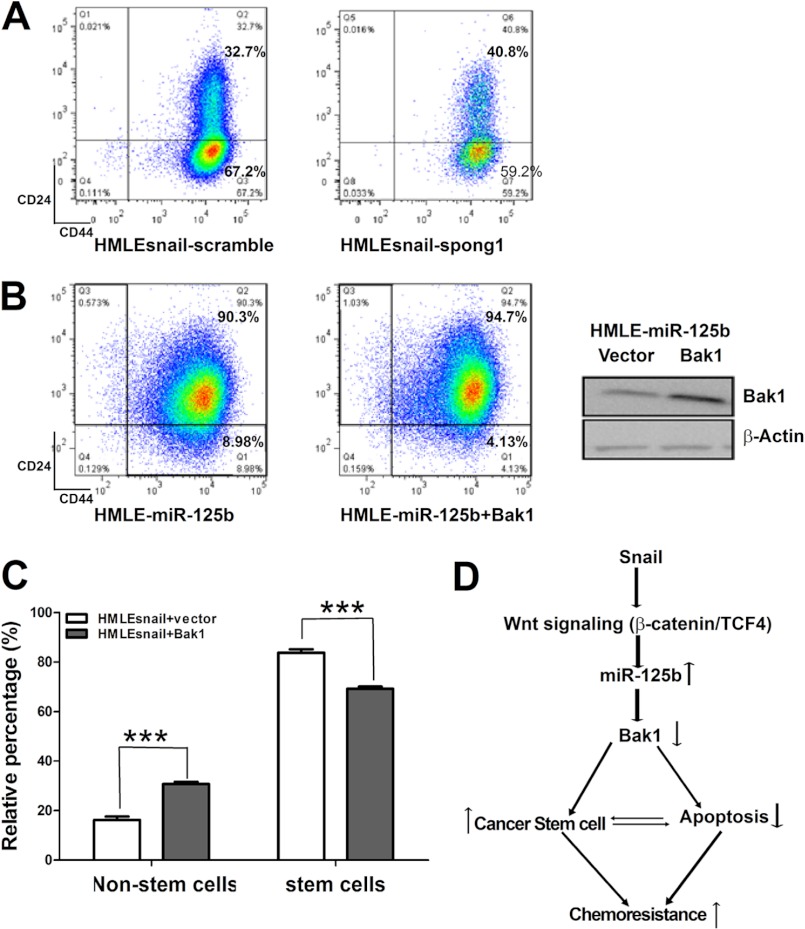
We next determined whether miR-125b is required for maintaining Snail-induced cancer stem cells. HMLE-Snail cells were transfected with control and miR125b inhibitor miR-125b-spong1 (Fig. 7A). Depletion of miR-125b increased the non-stem cell population while decreasing the stem cell population in HMLE-Snail cells, indicating that miR-125b plays an important role in the Snail-increased breast cancer stem cell population.
FIGURE 7.
miR-125b and Bak1 are required for Snail-induced breast cancer stem cell enrichment. A, HMLE-Snail-scramble and HMLE-Snail-mir-125b-spong1 cells were incubated with CD24 and CD44 antibodies for 45 min, washed, and then analyzed by flow cytometry. B, HMLE-miR-125b cells were transfected with puro-vector or puro-Bak1. 24 h after transfection, cells were harvested and incubated with CD24 and CD44 for 45 min, washed, and then analyzed by flow cytometry. Cell lysates were prepared for Western blotting. Antibodies against Bak1 and β-actin were used. C, HMLE-Snail cells were transfected with puro-vector or puro-bak1. 24 h after transfection, cells were harvested and were incubated with CD24 and CD44 antibodies for 45 min, washed, and then analyzed by flow cytometry. Non-stem cells represent non-stem cell population. Stem cells represent enriched stem cell population. Columns represent the mean of three independent experiments; bars represent S.E. ***, p < 0.001.
It is known that Bak1 plays an important role in maintaining hematopoietic stem cell number (32). Since Snail represses the expression of Bak1 through miR-125b, we evaluated the role of Bak1 in miR125b-induced cancer stem cells. First, we performed a Bak1 rescue experiment in which miR-125b expressing HMLE-miR-125b cells were transduced with either a vector alone or a Bak1-expressing vector. Re-introduction of Bak1 in miR-125b-expressing cells reduced stem cell population (Fig. 7B). Furthermore, we determined whether Bak1 affects the pool size of Snail-induced cancer stem cells. We restored the expression of Bak1 in HMLE-Snail cells with either puro3-vector or puro3-Bak1 (Fig. 7C). Restoration of Bak1 expression in Snail-expressing cells also increased the non-stem cell population while decreasing the stem cell population in HMLE-Snail cells. Moreover, compared with non-stem cell population, CD24-CD44+ cells show decreased Bak1 expression and increased Snail expression (supplemental Fig. S4B), which further support our results. Taken together, these data demonstrate that Snail enriches the stem cell population through miR-125b and its target gene Bak1, indicating that miR-125b is a key mediator for Snail-induced stem cell propagation.
DISCUSSION
Cancer stem cells are thought to play a major role in chemoresistance. Conventional chemotherapies only kill differentiated cancer cells, but do not completely destroy cancer stem cells. The residual cancer cells after conventional therapy may be enriched for cells with tumor initiating and mesenchymal features (33). Growing evidence supports that cancer stem cells are critical for tumor growth, metastasis, and recurrence (34). Snail is a transcription factor and a master regulator of EMT in breast cancer cells (13). Overexpression of Snail correlates with tumor recurrence and reduced patient survival (35). Snail is reported to mediate radioresistance and chemoresistance through induction of cancer stem cell-like phenotypes (25, 27). However, little is known about the mechanisms of how Snail enriches for cancer stem cells and promotes chemoresistance.
miR-125b is highly expressed in hematopoietic stem cells (36). It was also reported that miR-125b blocks cell differentiation (37–38). The deregulation of miR-125b in hematopoietic cells results in leukemia (39–40), and overexpression of miR-125b was shown to stimulate the growth of prostate cancer (41). A recent report shows that malignant myoepithelioma breast cancer has higher expression levels of miR-125b than the other breast cancer subtypes. However, the precise function of miR-125b remains elusive in breast cancer (42), and how miR-125b is regulated is unknown.
This is the first report to show that Snail activates the expression of miR-125b through transcriptional regulation via Wnt signaling. We found that Snail is also up-regulated in Taxol-resistant cells. The expression of miR-125b is correlated with Snail. Further studies showed that expression of Snail in cancer cells dramatically up-regulated miR-125b. Snail transcriptionally increases the expression of miR-125b through Wnt signaling (β-catenin and TCF4). Depletion of TCF4 binding sites in the promoter of miR-125b-1 inhibited the luciferase activity of the miR-125b-1 promoter activated by Snail in a dose-dependent manner. Meanwhile, down-regulation of TCF4 or β-catenin by siRNA also blocked the expression of miR-125b by Snail.
We previously reported that miR-125b confers breast cancer cells resistant to Taxol by targeting Bak1 (19). Here, we also found that Bak1 is inversely correlated with Snail in cancer cells. Expression of Snail represses Bak1 expression, while depletion of Snail in Snail-overexpressing cells increases the expression of Bak1. Moreover, restoration of miR-125b expression in Snail-depleted cells decreased the expression of Bak1 to basal levels. Furthermore, the expression of Bak1 in HMLE-Snail cells was increased after down-regulation of miR-125b in Snail-overexpressing cells. These data showed that Snail represses the expression of Bak1 through miR-125b. Depletion of miR-125b or restoration of Bak1 expression reversed Snail-induced Taxol resistance, indicating that the miR-125b-Bak1 axis is required for Snail-induced drug resistance.
This is the first report to show that miR-125b is a key mediator for Snail-induced stem cell propagation. Recently, miRNAs have been reported to play important roles in stem cell homeostasis (17–18, 43). miR-93 and miR-106b greatly enhanced iPSC induction and modulated mesenchymal-to-epithelial transition in the initiation stage of reprogramming while inhibition of these miRNAs significantly decreased the reprogramming efficiency (44). Here we found that overexpression of miR-125b in breast cancer cells increased the cancer stem cell pool size, while depletion of miR-125b increased the non-stem cell population size in Snail expressing breast cancer cells. Our data, for the first time, demonstrate that the miR-125b-Bak1 pathway contributes to the Snail-induced enrichment of cancer stem cells. It was reported that p53 represses EMT and cancer stem cells (45). p53 is one of the target genes of miR-125b (46). In our model, restoration of p53 does not decrease the miR-125b-induced enrichment of cancer stem cells (data now shown), which may due to different cell lines and experimental models. Snail may also increase the cancer stem cell pool through other pathways such as E-caderin (7).
In summary, here we report that Snail activates miR-125b expression through β-catenin/TCF-4. The miR-125b-Bak1 axis plays a key role in Snail-induced enrichment of cancer stem cells and chemoresistance (Fig. 7D). These novel findings reveal a new mechanism whereby miR-125b is required for Snail-induced cancer cells to be resistant to Taxol. Thus, the development of miR-125b targeted therapeutics or therapy that activates Bak1 may overcome cancer stem cell-mediated drug resistance.
Supplementary Material
Acknowledgment
We thank Dr. Robert Weinberg for providing HMLE and HMLE-Snail cell lines.
This work was supported, in whole or in part, by National Institutes of Health/NCI Grant R01CA149646 (to M. T.), the Vincent F. Kilborn, Jr. Cancer Research Foundation (to M. T.), and The Norwegian Radiumhospitalet Legater (Project 334003, to M. T. and O. F.).

This article contains supplemental Figs. S1—S5.
- EMT
- epithelial-mesenchymal transition
- miRNA
- microRNA
- qRT-PCR
- quantitative real-time PCR.
REFERENCES
- 1. Giaccone G., Pinedo H. M. (1996) Drug Resistance. Oncologist 1, 82–87 [PubMed] [Google Scholar]
- 2. Arbuck S. G., Dorr A., Friedman M. A. (1994) Paclitaxel (Taxol) in breast cancer. Hematol. Oncol. Clin. North Am. 8, 121–140 [PubMed] [Google Scholar]
- 3. Hortobagyi G. N., Holmes F. A., Theriault R. L., Buzdar A. U. (1994) Use of Taxol (paclitaxel) in breast cancer. Oncology 51, 29–32 [DOI] [PubMed] [Google Scholar]
- 4. Meden H., Rath W., Kuhn W. (1994) Taxol–a new cytostatic drug for therapy of ovarian and breast cancer. Geburtshilfe Frauenheilkd 54, 187–193 [DOI] [PubMed] [Google Scholar]
- 5. Sledge G. W., Jr., Robert N., Sparano J. A., Cobeligh M., Goldstein L. J., Neuberg D., Rowinsky E., Baughman C., McCaskill-Stevens W. (1994) Paclitaxel (Taxol)/doxorubicin combinations in advanced breast cancer: the Eastern Cooperative Oncology Group experience. Semin. Oncol. 21, 15–18 [PubMed] [Google Scholar]
- 6. Bishop J. F., Dewar J., Toner G. C., Tattersall M. H., Olver I. N., Ackland S., Kennedy I., Goldstein D., Gurney H., Walpole E., Levi J., Stephenson J. (1997) Paclitaxel as first-line treatment for metastatic breast cancer. The Taxol Investigational Trials Group, Australia and New Zealand. Oncology 11, 19–23 [PubMed] [Google Scholar]
- 7. Gupta P. B., Onder T. T., Jiang G., Tao K., Kuperwasser C., Weinberg R. A., Lander E. S. (2009) Identification of selective inhibitors of cancer stem cells by high-throughput screening. Cell 138, 645–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Donnenberg V. S., Donnenberg A. D. (2005) Multiple drug resistance in cancer revisited: the cancer stem cell hypothesis. J. Clin. Pharmacol. 45, 872–877 [DOI] [PubMed] [Google Scholar]
- 9. Parekh H., Wiesen K., Simpkins H. (1997) Acquisition of taxol resistance via P-glycoprotein- and non-P-glycoprotein-mediated mechanisms in human ovarian carcinoma cells. Biochem. Pharmacol. 53, 461–470 [DOI] [PubMed] [Google Scholar]
- 10. Orr G. A., Verdier-Pinard P., McDaid H., Horwitz S. B. (2003) Mechanisms of Taxol resistance related to microtubules. Oncogene 22, 7280–7295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Blanco M. J., Moreno-Bueno G., Sarrio D., Locascio A., Cano A., Palacios J., Nieto M. A. (2002) Correlation of Snail expression with histological grade and lymph node status in breast carcinomas. Oncogene 21, 3241–3246 [DOI] [PubMed] [Google Scholar]
- 12. Côme C., Magnino F., Bibeau F., De Santa Barbara P., Becker K. F., Theillet C., Savagner P. (2006) Snail and slug play distinct roles during breast carcinoma progression. Clin. Cancer Res. 12, 5395–5402 [DOI] [PubMed] [Google Scholar]
- 13. Mani S. A., Guo W., Liao M. J., Eaton E. N., Ayyanan A., Zhou A. Y., Brooks M., Reinhard F., Zhang C. C., Shipitsin M., Campbell L. L., Polyak K., Brisken C., Yang J., Weinberg R. A. (2008) The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell 133, 704–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Iorio M. V., Ferracin M., Liu C. G., Veronese A., Spizzo R., Sabbioni S., Magri E., Pedriali M., Fabbri M., Campiglio M., Ménard S., Palazzo J. P., Rosenberg A., Musiani P., Volinia S., Nenci I., Calin G. A., Querzoli P., Negrini M., Croce C. M. (2005) MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 65, 7065–7070 [DOI] [PubMed] [Google Scholar]
- 15. Ozen M., Creighton C. J., Ozdemir M., Ittmann M. (2008) Widespread deregulation of microRNA expression in human prostate cancer. Oncogene 27, 1788–1793 [DOI] [PubMed] [Google Scholar]
- 16. Zhang L., Volinia S., Bonome T., Calin G. A., Greshock J., Yang N., Liu C. G., Giannakakis A., Alexiou P., Hasegawa K., Johnstone C. N., Megraw M. S., Adams S., Lassus H., Huang J., Kaur S., Liang S., Sethupathy P., Leminen A., Simossis V. A., Sandaltzopoulos R., Naomoto Y., Katsaros D., Gimotty P. A., DeMichele A., Huang Q., Bützow R., Rustgi A. K., Weber B. L., Birrer M. J., Hatzigeorgiou A. G., Croce C. M., Coukos G. (2008) Genomic and epigenetic alterations deregulate microRNA expression in human epithelial ovarian cancer. Proc. Natl. Acad. Sci. U.S.A. 105, 7004–7009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yu F., Yao H., Zhu P., Zhang X., Pan Q., Gong C., Huang Y., Hu X., Su F., Lieberman J., Song E. (2007) let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell 131, 1109–1123 [DOI] [PubMed] [Google Scholar]
- 18. Shimono Y., Zabala M., Cho R. W., Lobo N., Dalerba P., Qian D., Diehn M., Liu H., Panula S. P., Chiao E., Dirbas F. M., Somlo G., Pera R. A., Lao K., Clarke M. F. (2009) Downregulation of miRNA-200c links breast cancer stem cells with normal stem cells. Cell 138, 592–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhou M., Liu Z., Zhao Y., Ding Y., Liu H., Xi Y., Xiong W., Li G., Lu J., Fodstad O., Riker A. I., Tan M. (2010) MicroRNA-125b confers the resistance of breast cancer cells to paclitaxel through suppression of pro-apoptotic Bcl-2 antagonist killer 1 (Bak1) expression. J. Biol. Chem. 285, 21496–21507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang H., Tan G., Dong L., Cheng L., Li K., Wang Z., Luo H. (2012) Circulating MiR-125b as a marker predicting chemoresistance in breast cancer. PLoS One 7, e34210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ebert M. S., Neilson J. R., Sharp P. A. (2007) MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat. Methods 4, 721–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Edbauer D., Neilson J. R., Foster K. A., Wang C. F., Seeburg D. P., Batterton M. N., Tada T., Dolan B. M., Sharp P. A., Sheng M. (2010) Regulation of synaptic structure and function by FMRP-associated microRNAs miR-125b and miR-132. Neuron 65, 373–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhou R., Hu G., Liu J., Gong A. Y., Drescher K. M., Chen X. M. (2009) NF-kappaB p65-dependent transactivation of miRNA genes following Cryptosporidium parvum infection stimulates epithelial cell immune responses. PLoS Pathog. 5, e1000681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yin T., Wang C., Liu T., Zhao G., Zha Y., Yang M. (2007) Expression of snail in pancreatic cancer promotes metastasis and chemoresistance. J. Surg. Res. 141, 196–203 [DOI] [PubMed] [Google Scholar]
- 25. Kurrey N. K., Jalgaonkar S. P., Joglekar A. V., Ghanate A. D., Chaskar P. D., Doiphode R. Y., Bapat S. A. (2009) Snail and slug mediate radioresistance and chemoresistance by antagonizing p53-mediated apoptosis and acquiring a stem-like phenotype in ovarian cancer cells. Stem Cells 27, 2059–2068 [DOI] [PubMed] [Google Scholar]
- 26. Wang Z., Li Y., Kong D., Banerjee S., Ahmad A., Azmi A. S., Ali S., Abbruzzese J. L., Gallick G. E., Sarkar F. H. (2009) Acquisition of epithelial-mesenchymal transition phenotype of gemcitabine-resistant pancreatic cancer cells is linked with activation of the notch signaling pathway. Cancer Res. 69, 2400–2407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chen W. J., Wang H., Tang Y., Liu C. L., Li H. L., Li W. T. (2010) Multidrug resistance in breast cancer cells during epithelial-mesenchymal transition is modulated by breast cancer resistant protein. Chin. J. Cancer 29, 151–157 [DOI] [PubMed] [Google Scholar]
- 28. Stemmer V., de Craene B., Berx G., Behrens J. (2008) Snail promotes Wnt target gene expression and interacts with β-catenin. Oncogene 27, 5075–5080 [DOI] [PubMed] [Google Scholar]
- 29. Medici D., Hay E. D., Olsen B. R. (2008) Snail and Slug promote epithelial-mesenchymal transition through β-catenin-T-cell factor-4-dependent expression of transforming growth factor-β3. Mol. Biol. Cell 19, 4875–4887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shi X. B., Xue L., Yang J., Ma A. H., Zhao J., Xu M., Tepper C. G., Evans C. P., Kung H. J., deVere White R. W. (2007) An androgen-regulated miRNA suppresses Bak1 expression and induces androgen-independent growth of prostate cancer cells. Proc. Natl. Acad. Sci. U.S.A. 104, 19983–19988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rossi R. L., Rossetti G., Wenandy L., Curti S., Ripamonti A., Bonnal R. J., Birolo R. S., Moro M., Crosti M. C., Gruarin P., Maglie S., Marabita F., Mascheroni D., Parente V., Comelli M., Trabucchi E., De Francesco R., Geginat J., Abrignani S., Pagani M. (2011) Distinct microRNA signatures in human lymphocyte subsets and enforcement of the naive state in CD4+ T cells by the microRNA miR-125b. Nat. Immunol. 12, 796–803 [DOI] [PubMed] [Google Scholar]
- 32. Guo S., Lu J., Schlanger R., Zhang H., Wang J. Y., Fox M. C., Purton L. E., Fleming H. H., Cobb B., Merkenschlager M., Golub T. R., Scadden D. T. (2010) MicroRNA miR-125a controls hematopoietic stem cell number. Proc. Natl. Acad. Sci. U.S.A. 107, 14229–14234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Creighton C. J., Li X., Landis M., Dixon J. M., Neumeister V. M., Sjolund A., Rimm D. L., Wong H., Rodriguez A., Herschkowitz J. I., Fan C., Zhang X., He X., Pavlick A., Gutierrez M. C., Renshaw L., Larionov A. A., Faratian D., Hilsenbeck S. G., Perou C. M., Lewis M. T., Rosen J. M., Chang J. C. (2009) Residual breast cancers after conventional therapy display mesenchymal as well as tumor-initiating features. Proc. Natl. Acad. Sci. U.S.A. 106, 13820–13825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Dean M., Fojo T., Bates S. (2005) Tumour stem cells and drug resistance. Nat. Rev. Cancer 5, 275–284 [DOI] [PubMed] [Google Scholar]
- 35. Moody S. E., Perez D., Pan T. C., Sarkisian C. J., Portocarrero C. P., Sterner C. J., Notorfrancesco K. L., Cardiff R. D., Chodosh L. A. (2005) The transcriptional repressor Snail promotes mammary tumor recurrence. Cancer Cell 8, 197–209 [DOI] [PubMed] [Google Scholar]
- 36. O'Connell R. M., Chaudhuri A. A., Rao D. S., Gibson W. S., Balazs A. B., Baltimore D. (2010) MicroRNAs enriched in hematopoietic stem cells differentially regulate long-term hematopoietic output. Proc. Natl. Acad. Sci. U.S.A. 107, 14235–14240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mizuno Y., Yagi K., Tokuzawa Y., Kanesaki-Yatsuka Y., Suda T., Katagiri T., Fukuda T., Maruyama M., Okuda A., Amemiya T., Kondoh Y., Tashiro H., Okazaki Y. (2008) miR-125b inhibits osteoblastic differentiation by down-regulation of cell proliferation. Biochem. Biophys. Res. Commun. 368, 267–272 [DOI] [PubMed] [Google Scholar]
- 38. Gururajan M., Haga C. L., Das S., Leu C. M., Hodson D., Josson S., Turner M., Cooper M. D. (2010) MicroRNA 125b inhibition of B cell differentiation in germinal centers. Int. Immunol. 22, 583–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bousquet M., Quelen C., Rosati R., Mansat-De Mas V., La Starza R., Bastard C., Lippert E., Talmant P., Lafage-Pochitaloff M., Leroux D., Gervais C., Viguié F., Lai J. L., Terre C., Beverlo B., Sambani C., Hagemeijer A., Marynen P., Delsol G., Dastugue N., Mecucci C., Brousset P. (2008) Myeloid cell differentiation arrest by miR-125b-1 in myelodysplastic syndrome and acute myeloid leukemia with the t(2;11)(p21;q23) translocation. J. Exp. Med. 205, 2499–2506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bousquet M., Harris M. H., Zhou B., Lodish H. F. (2010) MicroRNA miR-125b causes leukemia. Proc. Natl. Acad. Sci. U.S.A. 107, 21558–21563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Shi X. B., Xue L., Ma A. H., Tepper C. G., Kung H. J., White R. W. (2011) miR-125b promotes growth of prostate cancer xenograft tumor through targeting pro-apoptotic genes. Prostate 71, 538–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bockmeyer C. L., Christgen M., Muller M., Fischer S., Ahrens P., Langer F., Kreipe H., Lehmann U. (2011) MicroRNA profiles of healthy basal and luminal mammary epithelial cells are distinct and reflected in different breast cancer subtypes. Breast Cancer Res. Treat. 130, 735–745 [DOI] [PubMed] [Google Scholar]
- 43. Li X., Zhang J., Gao L., McClellan S., Finan M. A., Butler T. W., Owen L. B., Piazza G. A., Xi Y. (2011) MiR-181 mediates cell differentiation by interrupting the Lin28 and let-7 feedback circuit. Cell Death Differ. 19, 378–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Li Z., Yang C. S., Nakashima K., Rana T. M. (2011) Small RNA-mediated regulation of iPS cell generation. EMBO J. 30, 823–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Chang C. J., Chao C. H., Xia W., Yang J. Y., Xiong Y., Li C. W., Yu W. H., Rehman S. K., Hsu J. L., Lee H. H., Liu M., Chen C. T., Yu D., Hung M. C. (2011) p53 regulates epithelial-mesenchymal transition and stem cell properties through modulating miRNAs. Nat. Cell Biol. 13, 317–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Le M. T., Teh C., Shyh-Chang N., Xie H., Zhou B., Korzh V., Lodish H. F., Lim B. (2009) MicroRNA-125b is a novel negative regulator of p53. Genes Dev. 23, 862–876 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.