Abstract
The anaerobic oxidation of methane (AOM) is carried out by a globally distributed group of uncultivated Euryarchaeota, the anaerobic methanotrophic arachaea (ANME). In this work, we used G+C analysis of 16S rRNA genes to identify a putatively thermophilic ANME group and applied newly designed primers to study its distribution in low-temperature diffuse vent fluids from deep-sea hydrothermal vents. We found that the G+C content of the 16S rRNA genes (PGC) is significantly higher in the ANME-1GBa group than in other ANME groups. Based on the positive correlation between the PGC and optimal growth temperatures (Topt) of archaea, we hypothesize that the ANME-1GBa group is adapted to thrive at high temperatures. We designed specific 16S rRNA gene-targeted primers for the ANME-1 cluster to detect all phylogenetic groups within this cluster, including the deeply branching ANME-1GBa group. The primers were successfully tested both in silico and in experiments with sediment samples where ANME-1 phylotypes had previously been detected. The primers were further used to screen for the ANME-1 microorganisms in diffuse vent fluid samples from deep-sea hydrothermal vents in the Pacific Ocean, and sequences belonging to the ANME-1 cluster were detected in four individual vents. Phylotypes belonging to the ANME-1GBa group dominated in clone libraries from three of these vents. Our findings provide evidence of existence of a putatively extremely thermophilic group of methanotrophic archaea that occur in geographically and geologically distinct marine hydrothermal habitats.
INTRODUCTION
Over the past decade, many studies on the anaerobic oxidation of methane (AOM) have been published (for a review, see reference 1), emphasizing the global distribution of this process in marine sediments and its importance as a factor that reduces the level of methane emissions to the atmosphere. The oxidation of methane by archaea is assumed to be a reverse methanogenesis coupled to the reduction of sulfate by sulfate-reducing bacteria (2). This process occurs in marine habitats where sulfate from the seawater and methane of biological and geochemical origin from deeper layers meet (1). Based on the 16S rRNA phylogeny, all anaerobic methanotrophic archaea (ANME) are grouped into three distinct clusters of Euryarchaeota, namely, ANME-1, ANME-2, and ANME-3 (3, 4). Microorganisms of the ANME-2 and ANME-3 clusters belong to Methanosarcinales, whereas the ANME-1 cluster is a deep phylogenetic branch of Euryarchaeota only distantly related to the orders Methanomicrobiales and Methanosarcinales (5). Despite the great interest in these microorganisms, all three clusters remain uncultured. It is assumed that one of the major obstacles to the isolation of AOM-mediating microorganisms is their slow growth, the main reason for which is presumably bioenergetic limitations caused by the very low energy yield of AOM (6). According to theoretical calculations, the free-energy yield (ΔG) of sulfate-dependent oxidation of methane increases with increasing temperature (7), suggesting that high-temperature AOM should occur. There are several lines of evidence that anaerobic methanotrophs can thrive at high temperatures. Sequences of the 16S rRNA genes of all three clusters of methane-oxidizing archaea (ANME-1, ANME-2, and ANME-3) as well as ANME-specific core lipids have been found in deep-sea hydrothermal vents and warm sediments (8–14). Moreover, several researchers managed to determine in vitro methane oxidation rates at different temperatures by using hydrothermal sediment samples from Guaymas Basin and Middle Valley (7, 15, 16). These studies have shown maximum AOM activity between 45 and 60°C. Here, we provide one more line of evidence of thermophilic AOM by analysis of the G+C content of the 16S rRNA genes (hereafter PGC) of ANME phylotypes and application of newly designed specific primers.
The positive correlation between the G+C content of the rRNA and optimal growth temperatures of prokaryotes (hereafter Topt) was noted by the founders of 16S rRNA-based phylogeny (see, e.g., reference 17). The relevant data were systematized by Galtier and Lobry (18) and further formalized by Kimura et al. (19), who obtained the correlation equation PGC = 0.17 Topt + 49.0 (R2 = 0.828) but mentioned the occurrence of deviations from the general trend. These deviations are especially notable in the domain Bacteria. In the present work, we show that, in Archaea, the correlation is stringent enough to allow confident predictions of the thermophilic adaptation of uncultured microorganisms with PGC higher than a particular value. We identify a particular phylogenetic group of methane-oxidizing archaea with a high PGC as putatively thermophilic. We then designed, tested, and applied a specific primer set to assess distribution of putatively thermophilic anaerobic methanotrophs in geographically and geologically distinct deep-sea hydrothermal vent fluids.
MATERIALS AND METHODS
Analysis of the correlation between the PGC of archaea and their Topt.
The list of the currently recognized archaeal species (a total of 374) and their type strains was taken from J. P. Euzéby's LPSN site (http://www.bacterio.cict.fr/) (20); for the final check, the update of 13 April 2012 was used. The 16S rRNA gene sequences were downloaded from RDP release 10 (http://rdp.cme.msu.edu/) or from GenBank following links at the Euzéby's site. The G+C content was determined using BioEdit 7.0.9.0 program (21). The Topt values were taken from relevant original publications. For 19 type strains, the sequences were not available, were too short (<1,200 nucleotides), or contained introns or the Topt could not be found. Thus, the correlation was analyzed for a total of 355 archaeal type strains.
The PGC was also determined for all archaeal isolates with 16S rRNA gene sequences longer than 1,200 nt available from RDP release 10. For the final check, update 28, 12 January 2012 was used, comprising 2,022 sequences. Then, we examined the information available on those of the isolates that were not affiliated with known thermophilic genera but exhibited a PGC higher than a particular value (see Results and Discussion).
Analysis of the PGC of ANME phylotypes.
A total of 945 sequences of 16S rRNA genes affiliated with five ANME groups (ANME-1a, ANME-1b, ANME-2ab, ANME-2c, and ANME-3) were downloaded from the ARB-SILVA database (http://www.arb-silva.de) (22). Eight sequences affiliated with the ANME-1AT and ANME-1GBa groups were downloaded from the database developed by German Jurgens (5). Five more sequences affiliated with the ANME-1GBa group were taken from GenBank according to accession numbers given in reference 14. The G+C content of ANME phylotypes was determined using the BioEdit 7.0.9.0 program.
Design and specificity verification of primers targeting the ANME-1 cluster.
For the design of the ANME-1-specific primers, we retrieved 275 sequences affiliated with the ANME-1a and ANME-1b groups from the ARB-SILVA database and 8 sequences affiliated with the ANME-1AT and ANME-1GBa groups from the database developed by German Jurgens (5). The primers were designed using homemade software briefly described in reference 23 with weighting mismatches as described in reference 24. At the stage of primer design, a restricted outgroup sampling was used. However, the specificity of the primers was further confirmed by using the RDP Probe Match tool and the OligoReport program (K. E. Ashelford, unpublished data) with the RDP database for individual primers and by using the OligoCheck program (Ashelford, unpublished) with the ARB-SILVA database for primer pairs. The newly designed primers, as well as the previously published primers and probes targeting phylotypes of the ANME-1 cluster, are listed in Table 1.
Table 1.
In silico specificity test of primers and probes targeting 16S rRNA genes of ANME-1 group and its subgroups
| Oligonucleotide name (orientation) | Sequence (5′→3′) | Reference |
In silico specificity test result (%)a |
||||
|---|---|---|---|---|---|---|---|
| ANME-1a | ANME-1b | ANME-1AT | ANME-1GBa | Negative controls | |||
| ANME1-305(F) | AGCCCGGAGATGGGTTCT | 25 | 96 | 75 | 80 | 0 | 0, 12, 26 |
| ANME1-350(R) | AGTTTTCGCGCCTGATGC | 98 | 98 | 80 | 0 | 0, 20, 90 | |
| ANME1-632(F) | TCAGGGAATACTGCTTGG | 50 | 75 | 0 | 0 | 0, 0, 0 | |
| ANME1-830(R) | TCGCAGTAATGCCAACAC | 84 | 88 | 0 | 0 | 0, 0 0 | |
| Mix of primers ANME1-395(F) | AACTCTGAGTGCCTCCTA | 26 | 80 | 92 | 80 | 0 | 0, 0, 0 |
| AACTCTGAGTGCCTCCAA | |||||||
| AACTCTGAGTGCCCCCTA | |||||||
| ANME1-1417(R) | CCTCACCTAAAYCCCACT | 63 | 94 | 0 | 0 | 0, 0, 6 | |
| ANME1GBHS-183(F) | ATACCTGGAATGGGCGGA | 0 | 0 | 0 | 100 | 0, 0, 0 | |
| ANME1GBHS-841(R) | AACACCGGCACCACTCGT | 0 | 0 | 0 | 100 | 0, 0, 0 | |
| ANME-1-337(F) | AGGTCCTACGGGACGCAT | 27 | 94 | 96 | 80 | 0 | 0, 0, 0 |
| ANME-1-724(R) | GGTCAGACGCCTTCGCT | 92 | 85 | 80 | 0 | 0, 0, 11 | |
| ANME-1-25(F) | GAGGCYACTGCYATCAGMGT | This study | 95 | 97 | 100 | 100 | 0, 0, 11 |
| ANME-1-1118(F) | CYCRCAGTTGCCAGCATCTC | 68 | 96 | 100 | 83 | 0, 0, 4 | |
| ANME-1-1406(R) | AYCYCACTCGGYTGGCTTGA | 100 | 100 | 100 | 100 | 0, 0, 13 | |
| ANME-1-GI812(R) | CTGGCCCACATCGTTTAC | 7 | 16 | 0 | 0 | 0 | 0, 0, 11 |
| cANME-1-GI812(R) | CTAGCCCGCATCGTTTAC | 56 | 15 | 80 | 0 | 10, 23, 50 | |
| ANME-1-GII186(R) | GGACATCCTGCATTCCAG | 0 | 0 | 60 | 0 | 0, 0, 0 | |
| ANME1-628(F) | GCTTTCAGGGAATACTGC | 28 | 38 | 74 | 0 | 0 | 0, 0, 2 |
| ARCH-915(R) | GTGCTCCCCCGCCAATTCCT | 29 | 80 | 96 | 100 | 100 | 91, 97, 98 |
The results of in silico specificity tests are presented as percentages of perfectly matching sequences and, for negative controls, as a series of three numbers, the first of which shows the percentage of perfectly matching sequences, and the second and third of which show the percentages of sequences exhibiting one and two mismatches, respectively. As negative controls, we used the 16S rRNA gene sequences of the type strains of archaeal species available from the RDP database (RDP release 10, update 28, 12 January 2012), representing 342 out of the 374 currently recognized archaeal species.
Field sites and sampling.
Hydrothermally heated sediments characterize the Guaymas Basin hydrothermal vent site in the Gulf of California. In 2010, during the research cruise BIG, a core sample of sediments from Guaymas Basin was obtained (location BIG 1). Sediments in the sampling area were covered with a white microbial mat. For DNA extraction, the layer 4 to 10 cm below the sediment surface was used. The temperature in this layer ranged from 50°C to 70°C.
Mississippi Canyon Block 118 (MC118) in the Gulf of Mexico is characterized by methane hydrate deposits and thermogenic hydrocarbon-rich fluids (30). It is located offshore of Louisiana at a water depth of ∼890 m. Samples of sediments covered with a white microbial mat were taken in 2006 using the Johnson-Sea-Link submersible. The temperature of the bottom water was 5.5°C. A detailed description of sediments of the MC118 site is provided in references 30 and 31.
From 2006 through 2009, vent fluids were collected from seven hydrothermal sites in the Pacific Ocean: Axial Seamount and the Endeavor Segment (32), both on the Juan de Fuca Ridge, and five volcanoes along the Mariana Arc (33) (see Fig. S1 and Table S1 in the supplemental material). All fluid samples were collected from low-temperature vents using the hydrothermal fluid and particle sampler (HFPS) (34) mounted on the deep-sea research submersibles Jason 2 and Alvin. While on the seafloor, fluids were passed through a Sterivex-GP (0.22-μm pore size). These filters were immediately frozen at −80°C upon vehicle recovery.
Characteristics of all samples that produced positive PCR results with the newly designed primer sets are shown in Table 2.
Table 2.
Description of samples that yielded positive results of ANME-1 16S rRNA gene amplification
| Sample | Geographical location, site, and vent | Coordinates (depth, m) | Temp (°C) | Total cell concn (cells ml−1)a | PCR result with indicated primer setb (no. of OTUs) |
No. of clones in clonal library | Detection of group (% of clone library): |
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | ANME-1AT | ANME-1a | ANME-1b | ANME-1GBa | ||||||
| MC118 | Gulf of Mexico, Mississippi Canyon, Block 118 | 28°51.47′N, 88°29.52′W (880) | 5 | NDc | + (2) | + (1) | + (2) | 24 | − | + (83)d | + (17)d | − |
| FS611 | JFR,e Axial Seamount, Bag City | 45°54.974′N, 129°59.354′W (1,410) | 11.2 | 1.7 × 105 | ND | ND | + (1) | 96 | − | − | − | + (100) |
| FS625 | JFR, Endeavor Segment, Easter Island | 47°56.88′N, 129°5.967′W, (2,150) | 17.8 | 1.3 × 105 | ND | ND | + (2) | 96 | − | − | − | + (100) |
| FS725 | JFR, Endeavor Segment, Boardwalk | 47°58.11′N, 129°5.24′W (2,150) | 16.4 | 7.1 × 104 | ND | ND | + (2) | 96 | + (2) | − | − | + (98) |
| FS448 | Mariana Arc, NW Rota-1, Fault Shrimp | 14°36.06′N, 144°46.62′E (520) | 25 | 3.49 × 105 | ND | ND | + (1) | 96 | − | + (100) | − | − |
| BG410 | Gulf of California, Guaymas Basin, BIG 1 | 27°00.37′N, 111°24.56′W (2,000) | 50–70 | ND | + (4) | ND | ND | 28 | + (7) | − | − | + (93) |
Primer sets: 1, ANME-1-25(F)–ANME-1-1406(R); 2, ANME-1-1118(F)–ANME-1-1406(R); 3, ANME-1-25(F)–ARCH-915(R).
ND, not determined.
Results for the clone library constructed by using the ANME-1-25(F)–ANME-1-1406(R) primer pair.
JFR, Juan de Fuca Ridge.
Nucleic acid extraction.
DNA from vent fluid filters was extracted as previously described (35). DNA from a sediment core of a Guaymas Basin hydrothermal field was extracted as previously described (36). DNA of the sediments of the MC118 site was kindly provided by the Andreas Teske Laboratory (University of North Carolina at Chapel Hill).
PCR amplification of 16S rRNA gene fragments.
PCR amplification was performed in a 50-μl reaction mixture containing 1 μl of template DNA, 1× GoTaq Green reaction buffer (Promega, San Luis Obispo, CA), 1 μl of 10 mM 4 deoxynucleoside triphosphate (dNTP) mix (200 μM final concentration for each dNTP) (Promega) and 1 U of GoTaq DNA polymerase (Promega). Concentrations of the primers were calculated using the PCR Optimization program (http://molbiol.ru/eng/scripts/01_14.html). PCR programs were adjusted experimentally based on the standard PCR protocol (37) and on the calculations of the annealing temperatures of primers made using the OligoAnalyzer program (http://eu.idtdna.com/analyzer/Applications/OligoAnalyzer/). For the primer set ANME-1-25(F)–ARCH-915(R), the annealing temperature was 59°C, and the concentration of each primer was 0.45 μM. For the primer set ANME-1-25(F)–ANME-1-1406(R) the annealing temperature was 58°C and the concentration of each primer was 0.3 μM. For the primer set ANME-1-1118(F)–ANME-1-1406(R) the annealing temperature was 61°C and the concentration of each primer was 1.25 μM. As negative controls, reactions without added DNA, reactions with the addition of DNA isolated from the surrounding seawater, and reactions with the addition of genomic DNA of Methanocaldococcus jannaschii or Archaeoglobus profundus were used. As positive controls, reactions with the addition of plasmid DNA harboring a cloned ANME-1 16S rRNA gene fragment (kindly provided by the A. Teske laboratory, University of North Carolina at Chapel Hill) were used. PCRs were performed using a Mastercycler gradient (Eppendorf, Hamburg, Germany). Amplicons were visualized with ethidium bromide on 1% agarose gels in 1× Tris-acetate-EDTA (TAE) buffer.
Cloning and sequencing of PCR-amplified 16S rRNA gene fragments.
PCR products were purified and concentrated using the MinElute PCR purification kit (Qiagen) according to the manufacturer's instructions. Product quality was assessed on 0.8% agarose gels stained with ethidium bromide. Bands were excised and DNA was extracted using the MinElute gel extraction kit (Qiagen). This purified product was ligated into pCR4-TOPO vector for 5 min at room temperature and transformed into electrocompetent cells according to the manufacturer's instructions (Invitrogen). For each library, 24 to 96 clones were randomly selected and grown in SuperBroth with 50 mg ml−1 kanamycin in 96 deep-well blocks overnight at 37°C with vigorous shaking. Cells were collected by centrifugation, and plasmid DNA was isolated using a standard alkaline-lysis procedure (38). Plasmids were sequenced bidirectionally with primers T3 (5′-ATTAACCCTCACTAAAGGGA) and T7 (5′-TAATACGACTCACTATAGGG) using the BigDye Terminator v.3.1 kit on an ABI 3730 sequencer (Applied Biosystems). If necessary, the intermediate primer ARCH-915(R) (29) was used.
Sequence analysis.
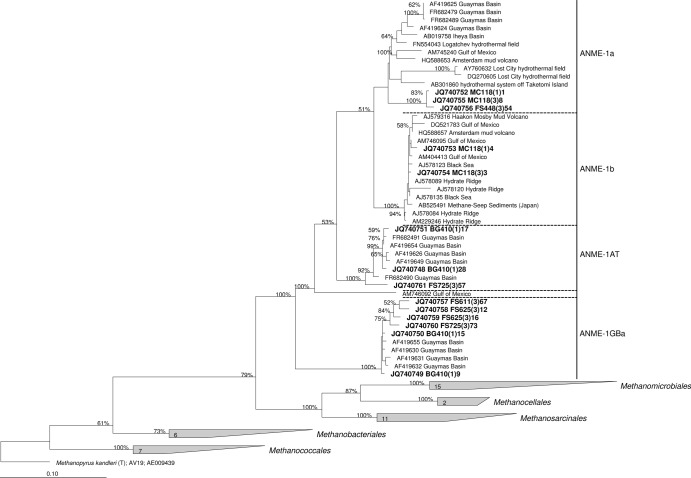
Sequences were analyzed and edited in the Chromas Lite 2.01 program (http://www.technelysium.com.au). Forward and reverse reads were assembled into contigs using the BioEdit 7.0.9.0 program. Sequences were aligned in the ClustalW program (39). All sequences were analyzed by the Pintail program (40) in order to detect chimeric 16S rRNA gene sequences. Sequences originating from the same samples were grouped into 98% similar operational taxonomic units (OTUs) using the cd-hit program (41). The phylogenetic tree (see Fig. 3) was constructed with the ARB software package (42) using the maximum likelihood (PHYML) algorithm and nonparametric bootstrap analysis (100 replicates).
Fig 3.
Dendrogram showing the phylogenetic position of 16S rRNA gene sequences retrieved from the samples analyzed in this study among selected reference sequences of the domain Archaea. Sequences from this study are grouped into 14 98% similar OTUs and printed in boldface type. The tree was constructed in the ARB software package using the maximum likelihood (PHYML) algorithm and nonparametric bootstrap analysis. The percent bootstrap values are based on 100 replicates and are indicated at the nodes with ≥50% bootstrap support. Bar, 10% estimated sequence divergence.
Nucleotide sequence accession numbers.
The sequences obtained in this study have been deposited in the NCBI GenBank database under accession no. JQ740748 to JQ740762.
RESULTS AND DISCUSSION
Correlation between the PGC of archaea and their Topt.
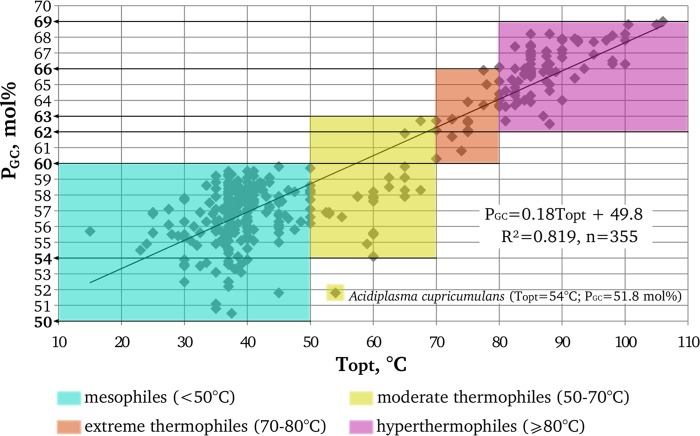
We plotted the G+C contents of 355 16S rRNA genes of archaeal type strains against their optimal growth temperatures. Our data can be approximated by the linear correlation equation PGC = 0.18 Topt + 49.8 (R2 = 0.819) (Fig. 1), which is close to the equation of Kimura et al. (19), obtained for a less strictly defined organism sampling. Since G·C pairs are more stable than A·U pairs due to an additional hydrogen bond (43), we interpret the correlation as indicative of the necessity for the 16S rRNA molecule to have a definite rigidity level of its secondary structure.
Fig 1.
Correlation between the optimal growth temperatures (Topt) of archaeal type strains and the G+C contents of their 16S rRNA genes (PGC).
The distribution of the dots in the plot in Fig. 1 shows that there exist particular PGC ranges characteristic of mesophilic (Topt < 50°C), moderately thermophilic (50°C ≤ Topt < 70°C), extremely thermophilic (70°C ≤ Topt < 80°C), and hyperthermophilic (Topt ≥ 80°C) groups of archaeal type strains. For the present work, the most significant conclusion from Fig. 1 is that all archaeal type strains with PGC above 60 mol% are thermophiles and those with PGC above 63 mol% are extreme thermophiles or hyperthermophiles. Moreover, our analysis of all sequences of the 16S rRNA genes of archaeal isolates available from the RDP database with checking the Topt of the corresponding microorganisms in relevant publications showed that no archaeal isolates with a PGC above 60 mol% have been reported to be mesophilic. The information on the four isolates that had a PGC above 60 mol% and were not affiliated with known thermophilic genera is presented in Table S2 in the supplemental material. These are haloarchaea with unreported temperature optima. It follows that a PGC value exceeding 60 mol% may be accepted as sound evidence of the thermophilic nature of the analyzed archaeal phylotypes.
Analysis of PGC of ANME phylotypes.
We have analyzed the G+C content of over 900 sequences of 16S rRNA genes of seven ANME groups: ANME-1a (8), ANME-1b (4), ANME-1AT (5), ANME-1GBa (5), ANME-2ab (3), ANME-2c (3), and ANME-3 (4) (Fig. 2). We found that for six ANME groups (ANME-1a, ANME-1b, ANME-1AT, ANME-2ab, ANME-2c, and ANME-3), PGC does not exceed 59 mol%. Meanwhile, the minimum value of this parameter for the group ANME-1GBa is 62.5 mol%, and the average value is 63.5 ± 0.7 mol%.
Fig 2.
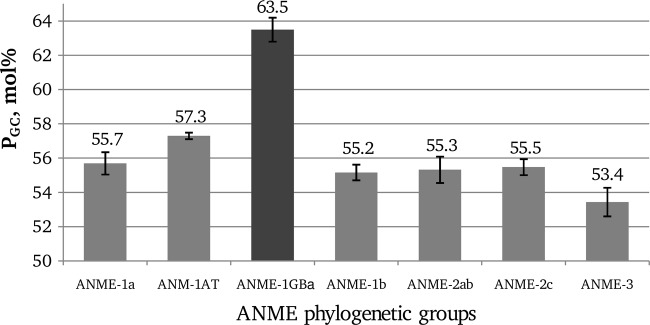
PGC values of different groups of ANME archaea. Standard deviations are indicated by error bars.
We also made a separate examination of the G+C contents of 16S rRNA sequences representing ANME microorganisms that were detected in or enriched from different hydrothermal-associated environments as some of such sequences were not in Silva database or do not have a defined phylogenetic affiliation there. For this analysis, 76 16S rRNA gene sequences representing all known ANME clusters and subgroups were taken from relevant publications (see Table S3 in the supplemental material, showing results of our analysis of the PGC values for these sequences in conjunction with the corresponding in situ or incubation temperatures). All of these ANME sequences except those that belong to the ANME-1GBa group have a PGC lower than 59 mol%. Based on the obtained correlation between the PGC of archaea and their Topt, we can predict that these sequences may represent moderate thermophilic or mesophilic microorganisms, which is consistent with in situ or incubation temperatures reported in corresponding publications.
Based on our 60 mol% cutoff value, only microorganisms of the ANME-1GBa group can confidently be predicted to be adapted to thrive at a high temperature. In addition, considering the PGC of other ANME groups and the absence of extreme thermophiles or hyperthermophiles with PGC below 60 mol% in our plot (Fig. 1), we may state that, among the currently recognized methanotrophic archaea (Fig. 2), microorganisms of the ANME-1GBa group are the only candidates for extreme thermophily or hyperthermophily (Topt, ≥70°C). Previously, phylotypes of the ANME-1GBa group were found only in hydrothermal sediments of the Guaymas Basin (8, 14). The ANME-1GBa group is the deepest branch within the ANME-1 cluster and is phylogenetically distant from other members of this cluster (5). We failed to find published ANME-1-targeted primers that would be able to amplify all groups within this cluster, including the ANME-1GBa group (Table 1). Therefore, we developed a new primer system with the goal of screening for ANME-1GBa phylotypes and other phylotypes of the ANME-1 cluster in low-temperature fluids from deep-sea hydrothermal vents. We rejected the idea of developing ANME-1GBa-specific primers because such primers would have to be based on a small number of highly similar sequences, and thus there was a risk of overspecificity to particular sequences.
In silico verification of the specificity of the ANME-1-targeted primers.
Three ANME-1-targeted primers that we designed perfectly matched most of the sequences of all four phylogenetic groups within the ANME-1 cluster (Table 1): ANME-1-25(F) (5′-GAGGCYACTGCYATCAGMGT-3′), ANME-1-1118(F) (5′-CYCRCAGTTGCCAGCATCTC-3′), and ANME-1-1406(R) (5′-AYCYCACTCGGYTGGCTTGA-3′). Table 1 presents the results of our analysis of the specificity of the newly designed primers, as well as of previously published primers and probes. In contrast to our primers, none of the previously published primers and probes covered the ANME-1 cluster entirely, as expected since these primers were designed to target specific subsets of ANME-1 clusters.
All previously published and newly designed primers and probes were also checked in silico by using 16S rRNA gene sequences of archaeal type strains as negative controls (Table 1). Moreover, the specificity of the newly designed systems of primers was confirmed by using all archaeal 16S rRNA gene sequences (of cultured and uncultured archaea) from the ARB-SILVA database (more than 25,000 sequences). This analysis showed that the primer pairs ANME-1-25F–ANME-1-1406(R) and ANME-1-1118F–ANME-1-1406(R) did not match any sequences except for those of the ANME-1 group, even if one mismatch per primer was allowed. All three new primers did not match any bacterial 16S rRNA gene sequences.
In silico analysis showed that the designed systems of primers do not form homo- or heterodimers with significant ΔG0. The pair of primers ANME-1-25(F)–ANME-1-1406(R) allows amplification of nearly the entire 16S rRNA gene, while the ANME-1-1118(F)–ANME-1-1406(R) primer pair will be suitable for applications that require a smaller amplicon (∼300 bp).
Verification of the specificity of the newly designed primers in experiments with environmental samples.
The specificity of the designed primers was also confirmed in experiments with sediment samples where ANME-1 phylotypes had previously been detected: those from the hydrocarbon seeps in the Gulf of Mexico (MC118 site) (31) and the hydrothermal sediments of the Guaymas Basin (8, 14).
Three sets of primers were tested with the sample from the Gulf of Mexico: ANME-1-25(F)–ANME-1-1406(R), ANME-1-1118(F)–ANME-1-1406(R), and ANME-1-25(F)–ARCH-915(R) (Table 2 and Fig. 3). Using two different combinations of primers [ANME-1-25(F)–ANME-1-1406(R) and ANME-1-25(F)–ARCH-915(R)], sequences related to ANME-1a and ANME-1b were detected. In terms of the diversity of sequences and their ratio in clone libraries, the results of using these two systems of primers were nearly identical. However, only the ANME-1b group was detected by using a third primer set, ANME-1-1118(F)–ANME-1-1406(R) (not shown in Fig. 2), in accordance with the reduced in silico specificity of the ANME-1-1118(F) primer to the ANME-1a group (Table 1). Previous work detected many different uncultured archaeal lineages in the MC118 site (31), but only the 16S rRNA genes of the ANME-1 cluster were detected with our new primers. The primer set ANME-1-25(F)–ARCH-915(R), in which the reverse primer is universal for archaea, also showed high specificity to the ANME-1 cluster.
The ANME-1-25(F)–ANME-1-1406(R) primer set was also verified by testing it with hydrothermal sediments from Guaymas Basin (site BIG 1). A broad diversity of ANME groups was previously detected in Guaymas Basin sediments, including two distinct offshoots of the ANME-1 cluster: ANME-1AT and ANME-1GBa (8). Both of these groups were detected using the ANME-1-25(F)–ANME-1-1406(R) primer set with a sediment sample from site BIG 1 (Table 1 and Fig. 3).
Detection of phylotypes of the ANME-1 cluster in diffuse vent fluids.
In contrast to focused fluids that circulate through the oceanic crust at high temperatures (generally 250° to 350°C), diffuse flows provide large stable habitats beneath the seafloor with a gradient of both temperature and redox pairs suitable for the development of microbial communities (44–46). In order to study the distribution of microorganisms of the ANME-1GBa group as well as of other groups of the ANME-1 cluster in diffuse hydrothermal ecosystems, we screened 50 hydrothermal vent fluid samples representing 42 different diffuse flow hydrothermal vents from the Axial Seamount, the Endeavor Segment, and from the volcanoes of the Mariana Arc for the 16S genes of the ANME-1 cluster using the ANME-1-25(F)–ARCH-915(R) primer set. A positive PCR result was obtained for four samples. Amplified gene fragments were cloned and sequenced (Table 2).
Axial Volcano is an active submarine volcano located on the Juan de Fuca Ridge (47). Here we detected anaerobic methanotrophs of the ANME-1 cluster at Bag City vent, a stable, basalt-hosted diffuse vent located in lobate lavas (35). All ANME-1 microorganisms detected in this site were classified as members of the ANME-1GBa group (Table 2 and Fig. 3).
The 16S rRNA genes of microorganisms of the ANME-1 cluster were also amplified from two vent fluid samples of Endeavor Segment—Easter Island and Boardwalk (32). In both cases, samples were taken in an area of primarily flat basalt with diffuse venting out of cracks, near high-temperature black smokers. All sequences obtained from Easter Island and the majority of sequences obtained from Boardwalk were also related to the ANME-1GBa group (Table 2 and Fig. 3).
Finally, microorganisms of the ANME-1 group were detected at Fault Shrimp, a diffuse vent located at NW Rota-1, an actively erupting volcano on the Mariana Arc (48). Fault Shrimp has venting out of cracks in basaltic andesite with microbial mat and shrimp present. All ANME-1 microorganisms detected in this site were classified as members of the ANME-1a group (Table 2 and Fig. 3).
Like the sequences reported in previous works (8, 14), the ANME-1GBa OTUs that we detected at deep-sea hydrothermal vents had a significantly higher G+C content (62.1 to 64.5 mol%) than the OTUs of other ANME-1 groups. Taking into account the sequences obtained in our study, the PGC of the ANME-1GBa group is 63.6 ± 0.8 mol%. Considering the obtained correlation (Fig. 1) and the fact that there are no mesophilic or moderately thermophilic archaeal type strains with a PGC above 63 mol%, we predict that the optimal growth temperatures of microorganisms of ANME-1GBa group is above 70°C. The putatively thermophilic methanotrophic archaea of the ANME-1GBa group were previously detected only in hydrothermal sediments in the Guaymas Basin (8, 14). Our findings provide evidence for wider distribution of this group in geographically and geologically distinct hydrothermal habitats. Moreover, the ANME-1GBa group appears to be the most frequent subgroup of the ANME-1 cluster in diffuse hydrothermal vent fluids. It should be noted that phylotypes of the ANME-1GBa group have only been found in environments associated with hydrothermal activity (hydrothermal sediments of Guaymas Basin and diffuse hydrothermal ecosystems). The same is true for mcrA genes that may be assigned to the ANME-1GBa group (see below).
Microorganisms of ANME-1 cluster were detected in only 4 out of the 42 unsedimented basalt-hosted deep-sea hydrothermal vents studied. This suggests only sporadic occurrence of these microorganisms in such environments. Previous works indicate that nonsedimented environments are not typical habitats for methanotrophic archaea. This can be explained by the absence of large sources of methane in such habitats. However, while there are no visible sediments at the surface of the studied sites of hydrothermal venting, chemical data suggest that there are large deposits of buried organic-rich sediments beneath the ridge crest at the Endeavor Segment (49–51). Here, due to the putatively buried sediments, methane levels in hydrothermal fluids are extremely high (up to 3 mM), suggesting a large source of methane for methanotrophy to occur in the subseafloor. There is no evidence for such sediments at Axial or NW Rota-1, but at each site, subsets of diffuse vents do have micromolar concentrations of methane, likely due to methanogenesis (32, 33). Taking into account (i) evidence of thermophilic nature of the ANME-1GBa group, (ii) the low temperature of the studied fluids at the surface, (iii) the absence of sediments at the surface of the studied sites, and (iv) evidence of the presence of organic-rich sediments beneath the surface of the Easter Island and Boardwalk sites (Endeavor Segment), it can be assumed that microorganisms of this group are a component of the subseafloor ecosystem and are advected to the surface from zones with higher temperature. From points i and ii above, it follows that ANME-1GBa organisms are most probably inactive in the surface zone.
To date, there is no direct evidence of the involvement of microorganisms of ANME-1GBa group in the process of anaerobic methane oxidation, although such evidence does exist for representatives of other groups of the ANME-1 cluster (7, 27). However, in another study of ours (32), we detected two closely related mcrA phylotypes (GenBank accession no. HQ635702 and HQ635704) in the FS625 sample studied also in this work (Easter Island vent). The mcrA gene encodes the α subunit of methyl-coenzyme M reductase (MCR), which catalyzes the methane-forming step in methanogenic archaea (53) and is also involved in oxidation of methane catalyzed by microorganisms of ANME clusters (54). Amino acid sequences deduced from the mcrA genes that we detected in the FS625 sample exhibited 91 to 92% identity to crystallized and well-described MCR of methanotrophic archaea (GenBank accession no. CBH39484) (54, 55), and our analysis placed the mcrA genes from sample FS625 together with mcrA phylotypes from Guaymas Basin (14) in the root of the ANME-1 cluster (see Fig. S2 in the supplemental material). In the present work, we also detected in the same sample just two closely related 16S rRNA genes affiliated with the ANME-1GBa group and no other ANME-1 phylotypes. Thus, there are grounds to assign the previously detected mcrA sequences to the ANME-1GBa group. Considering the similarity of these mcrA sequences to those of methanotrophic archaea, we regard these data as substantial evidence of the methanotrophic nature of the ANME-1GBa microorganisms. Close relatives of mcrA phylotypes that we assign to the ANME-1GBa group were previously detected only in Guaymas Basin hydrothermal sediments (mcrA-Guaymas) (14).
In recent studies, Holler et al. (7) and Wankel et al. (16) determined high rates of AOM between 42 and 65°C in incubation experiments. To quantify the abundance of anaerobic methanotrophs, molecular techniques such as quantitative PCR (qPCR) and catalyzed reporter deposition-fluorescent in situ hybridization (CARD-FISH) were used. However, the primers and probes that were used in these studies excluded ANME-1GBa microorganisms. In future studies, ANME-1GBa-targeted primers such as ANME1GBHS-183(F)–ANME1GBHS-841(R) (26) (Table 1) can be applied to both detection and quantification of the ANME-1GBa microorganisms. Further manipulation of vent samples or samples of Guaymas Basin sediments in incubation experiments over a broad range of temperatures coupled with such molecular tools may provide direct evidence of moderate or extreme thermophily of ANME-1GBa microorganisms.
Supplementary Material
ACKNOWLEDGMENTS
We thank the research staff of Andreas Teske Laboratory (University of North Carolina at Chapel Hill) for providing DNA of the MC118 sample, A. Godfroy, T. Sokolova, and the scientific team of the BIG 2010 cruise for providing the BG410 sample, and D. Tokar, S. Murdock, J. Holden, and D. Butterfield for sample collection and field support.
This work was supported by the Molecular and Cell Biology Program of the Russian Academy of Sciences, National Science Foundation grant OCE-0929167, National Aeronautics and Space Administration Astrobiology Institute Cooperative Agreement NNA04CC04A Director's Discretionary Fund, and a L'Oreal USA Fellowship (to J.A.H.). A.Y.M. was partially supported by the National Aeronautics and Space Administration Planetary Biology Internship Program.
Footnotes
Published ahead of print 26 November 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03034-12.
REFERENCES
- 1. Knittel K, Boetius A. 2009. Anaerobic oxidation of methane: progress with an unknown process. Annu. Rev. Microbiol. 63:311–334 [DOI] [PubMed] [Google Scholar]
- 2. Thauer RK. 2011. Anaerobic oxidation of methane with sulfate: on the reversibility of the reactions that are catalyzed by enzymes also involved in methanogenesis from CO2. Curr. Opin. Microbiol. 14:292–299 [DOI] [PubMed] [Google Scholar]
- 3. Orphan VJ, Hinrichs KU, Ussler W, III, Paull CK, Taylor LT, Sylva SP, Hayes JM, Delong EF. 2001. Comparative analysis of methane-oxidizing archaea and sulfate-reducing bacteria in anoxic marine sediments. Appl. Environ. Microbiol. 67:1922–1934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Knittel K, Lösekann T, Boetius A, Kort R, Amann R. 2005. Diversity and distribution of methanotrophic archaea at cold seeps. Appl. Environ. Microbiol. 71:467–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Schleper C, Jurgens G, Jonuscheit M. 2005. Genomic studies of uncultivated archaea. Nat. Rev. Microbiol. 3:479–488 [DOI] [PubMed] [Google Scholar]
- 6. Nauhaus K, Albrecht M, Elvert M, Boetius A, Widdel F. 2007. In vitro cell growth of marine archaeal-bacterial consortia during anaerobic oxidation of methane with sulfate. Environ. Microbiol. 9:187–196 [DOI] [PubMed] [Google Scholar]
- 7. Holler T, Widdel F, Knittel K, Amann R, Kellermann MY, Hinrichs KU, Teske A, Boetius A, Wegener G. 2011. Thermophilic anaerobic oxidation of methane by marine microbial consortia. ISME J. 5:1946–1956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Teske A, Hinrichs KU, Edgcomb V, de Vera Gomez A, Kysela D, Sylva SP, Sogin ML, Jannasch HW. 2002. Microbial diversity of hydrothermal sediments in the Guaymas Basin: evidence for anaerobic methanotrophic communities. Appl. Environ. Microbiol. 68:1994–2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Schouten S, Wakeham SG, Hopmans EC, Sinninghe Damsté JS. 2003. Biogeochemical evidence that thermophilic archaea mediate the anaerobic oxidation of methane. Appl. Environ. Microbiol. 69:1680–1686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kelley DS, Karson JA, Früh-Green GL, Yoerger DR, Shank TM, Butterfield DA, Hayes JM, Schrenk MO, Olson EJ, Proskurowski G, Jakuba M, Bradley A, Larson B, Ludwig K, Glickson D, Buckman K, Bradley AS, Brazelton WJ, Roe K, Elend MJ, Delacour A, Bernasconi SM, Lilley MD, Baross JA, Summons RE, Sylva SP. 2005. A serpentinite-hosted ecosystem: the Lost City hydrothermal field. Science 307:1428–1434 [DOI] [PubMed] [Google Scholar]
- 11. Brazelton WJ, Schrenk MO, Kelley DS, Baross JA. 2006. Methane- and sulfur-metabolizing microbial communities dominate the Lost City hydrothermal field ecosystem. Appl. Environ. Microbiol. 72:6257–6270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Inagaki F, Kuypers MM, Tsunogai U, Ishibashi J, Nakamura K, Treude T, Ohkubo S, Nakaseama M, Gena K, Chiba H, Hirayama H, Nunoura T, Takai K, Jørgensen Horikoshi BBK, Boetius A. 2006. Microbial community in a sediment-hosted CO2 lake of the southern Okinawa Trough hydrothermal system. Proc. Natl. Acad. Sci. U. S. A. 103:14164–14169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Roussel EG, Bonavita MA, Querellou J, Cragg BA, Webster G, Prieur D, Parkes RJ. 2008. Extending the sub-sea-floor biosphere. Science 320:1046 doi:10.1126/science.1154545 [DOI] [PubMed] [Google Scholar]
- 14. Biddle JF, Cardman Z, Mendlovitz H, Albert DB, Lloyd KG, Boetius A, Teske A. 2012. Anaerobic oxidation of methane at different temperature regimes in Guaymas Basin hydrothermal sediments. ISME J. 6:1018–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kallmeyer J, Boetius A. 2004. Effects of temperature and pressure on sulfate reduction and anaerobic oxidation of methane in hydrothermal sediments of Guaymas Basin. Appl. Environ. Microbiol. 70:1231–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wankel SD, Adams MM, Johnston DT, Hansel CM, Joye SB, Girguis PR. 25 July 2012. Anaerobic methane oxidation in metalliferous hydrothermal sediments: influence on carbon flux and decoupling from sulfate reduction. Environ. Microbiol. [Epub ahead of print.] doi:10.1111/j.1462-2920.2012.02825.x [DOI] [PubMed] [Google Scholar]
- 17. Woese CR, Achenbach L, Rouviere P, Mandelco L. 1991. Archaeal phylogeny: reexamination of the phylogenetic position of Archaeoglobus fulgidus in light of certain composition-induced artifacts. Syst. Appl. Microbiol. 14:364–371 [DOI] [PubMed] [Google Scholar]
- 18. Galtier N, Lobry JR. 1997. Relationships between genomic G+C content, RNA secondary structures, and optimal growth temperature in prokaryotes. J. Mol. Evol. 44:632–636 [DOI] [PubMed] [Google Scholar]
- 19. Kimura H, Sugihara M, Kato K, Hanada S. 2006. Selective phylogenetic analysis targeted at 16S rRNA genes of thermophiles and hyperthermophiles in deep-subsurface geothermal environments. Appl. Environ. Microbiol. 72:21–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Euzéby JP. 1997. List of Bacterial Names with Standing in Nomenclature: a folder available on the Internet. Int. J. Syst. Bacteriol. 47:590–592 [DOI] [PubMed] [Google Scholar]
- 21. Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95–98 [Google Scholar]
- 22. Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, Glöckner FO. 2007. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 35:7188–7196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Subbotina IV, Chernyh NA, Sokolova TG, Kublanov IV, Bonch-Osmolovskaya EA, Lebedinsky AV. 2003. Oligonucleotide probes for the detection of representatives of the genus Thermoanaerobacter. Microbiology 72:331–339 [Google Scholar]
- 24. Slobodkina GB, Chernyh NA, Slobodkin AI, Subbotina IV, Bonch-Osmolovskaya EA, Lebedinsky AV. 2004. PCR-based identification of hyperthermophilic archaea of the family Thermococcaceae. Appl. Environ. Microbiol. 70:5701–5703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Boetius A, Ravenschlag K, Schubert CJ, Rickert D, Widdel F, Gieseke A, Amann R, Jørgensen Witte BBU, Pfannkuche O. 2000. A marine microbial consortium apparently mediating anaerobic oxidation of methane. Nature 407:623–626 [DOI] [PubMed] [Google Scholar]
- 26. Miyashita A, Mochimaru H, Kazama H, Ohashi A, Yamaguchi T, Nunoura T, Horikoshi K, Takai K, Imachi H. 2009. Development of 16S rRNA gene-targeted primers for detection of archaeal anaerobic methanotrophs (ANMEs). FEMS Microbiol. Lett. 297:31–37 [DOI] [PubMed] [Google Scholar]
- 27. Girguis PR, Cozen AE, DeLong EF. 2005. Growth and population dynamics of anaerobic methane-oxidizing archaea and sulfate-reducing bacteria in a continuous-flow bioreactor. Appl. Environ. Microbiol. 71:3725–3733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lloyd KG, Alperin MJ, Teske A. 2011. Environmental evidence for net methane production and oxidation in putative ANaerobic MEthanotrophic (ANME) archaea. Environ. Microbiol. 13:2548–2564 [DOI] [PubMed] [Google Scholar]
- 29. Stahl DA, Amann R. 1991. Development and application of nucleic acid probes in bacterial systematic, p 205–248 In Stackebrandt E, Goodfellow M. (ed), Nucleic acid techniques in bacterial systematics. John Wiley and Sons, Ltd, Chichester, United Kingdom [Google Scholar]
- 30. Lapham LL, Chanton JP, Martens CS, Higley PD, Jannasch HW, Woolsey JR. 2008. Measuring temporal variability in pore-fluid chemistry to assess gas hydrate stability: development of a continuous pore-fluid array. Environ. Sci. Technol. 42:7368–7373 [DOI] [PubMed] [Google Scholar]
- 31. Lloyd KG, Albert DB, Biddle JF, Chanton JP, Pizarro O, Teske A. 2010. Spatial structure and activity of sedimentary microbial communities underlying a Beggiatoa spp. mat in a Gulf of Mexico hydrocarbon seep. PLoS One 5:e8738 doi:10.1371/journal.pone.0008738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ver Eecke HC, Butterfield DA, Huber JA, Lilley MD, Olson EJ, Roe KK, Evanse LJ, Merkel AY, Cantin HV, Holden JF. 2012. Hydrogen-limited growth of hyperthermophilic methanogens at deep-sea hydrothermal vents. Proc. Natl. Acad. Sci. U. S. A. 109:13674–13679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Huber JA, Cantin HV, Huse SM, Welch DB, Sogin ML, Butterfield DA. 2010. Isolated communities of Epsilonproteobacteria in hydrothermal vent fluids of the Mariana Arc seamounts. FEMS Microbiol. Ecol. 73:538–549 [DOI] [PubMed] [Google Scholar]
- 34. Butterfield DA, Lilley MD, Huber JA, Roe KK, Embley RW, Baross JA, Embley RW, Massoth GJ. 2004. Mixing, reaction, and microbial activity in the sub-seafloor revealed by temporal and spatial variation in diffuse flow vents at Axial Volcano, p 269–289 In Wilcock WSD, DeLong EF, Kelley DS, Baross JA, Cary SC. (ed), The subseafloor biosphere at mid-ocean ridges. American Geophysical Union, Washington, DC [Google Scholar]
- 35. Huber JA, Mark Welch DB, Morrison HG, Huse SM, Neal PR, Butterfield DA, Sogin ML. 2007. Microbial population structures in the deep marine biosphere. Science 318:97–100 [DOI] [PubMed] [Google Scholar]
- 36. Tsai YL, Olson BH. 1991. Rapid method for direct extraction of DNA from soil and sediments. Appl. Environ. Microbiol. 57:1070–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Saiki R, Gelfand D, Stoffel S, Scharf S, Higuchi R, Horn G, Mullis K, Erlich H. 1988. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science 239:487–491 [DOI] [PubMed] [Google Scholar]
- 38. Gibson TJ, Sulston JE. 1987. Preparation of large numbers of plasmid DNA samples in microtiter plates by the alkaline lysis method. Gene Anal. Tech. 4:41–44 [DOI] [PubMed] [Google Scholar]
- 39. Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ashelford KE, Chuzhanova NA, Fry JC, Jones AJ, Weightman AJ. 2005. At least 1 in 20 16S rRNA sequence records currently held in public repositories is estimated to contain substantial anomalies. Appl. Environ. Microbiol. 71:7724–7736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Li W, Jaroszewski L, Godzik A. 2001. Clustering of highly homologous sequences to reduce the size of large protein database. Bioinformatics 17:282–283 [DOI] [PubMed] [Google Scholar]
- 42. Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar Buchner A, Lai T, Steppi S, Jobb G, Förster W, Brettske I, Gerber S, Ginhart AW, Gross O, Grumann S, Hermann S, Jost R, König A, Liss T, Lüssmann R, May M, Nonhoff B, Reichel B, Strehlow R, Stamatakis A, Stuckmann N, Vilbig A, Lenke M, Ludwig T, Bode A, Schleifer KH. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363–1371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wada A, Suyama A. 1986. Local stability of DNA and RNA secondary structure and its relation to biological functions. Prog. Biophys. Mol. Biol. 47:113–157 [DOI] [PubMed] [Google Scholar]
- 44. Huber JA, Holden JF. 2008. Modeling the impact of diffuse vent microorganisms along mid-ocean ridges and flanks, p 215–231 In Lowell RP, Perfit MR, Seewald J, Metaxas A. (ed), Magma to microbe: modeling hydrothermal processes at oceanic spreading ridges. Geophysical Monograph Series, vol 178 American Geophysical Union, Washington, DC [Google Scholar]
- 45. Wankel SD, Germanovich LN, Lilley MD, Genc G, DiPerna CJ, Bradley AS, Olson EJ, Girguis PR. 2011. Influence of subsurface biosphere on geochemical fluxes from diffuse hydrothermal fluids. Nat. Geosci. 4:461–468 [Google Scholar]
- 46. Bemis K, Lowell RP, Farough A. 2012. Diffuse flow on and around hydrothermal vents at mid-ocean ridges. Oceanography 25:182–191 [Google Scholar]
- 47. Johnson HP, Embley RW. 1990. Axial Seamount: an active ridge axis volcano on the central Juan de Fuca Ridge. J. Geophys. Res. 95:12689–12696 [Google Scholar]
- 48. Embley RW, Baker ET, Butterfield DA, Chadwick WW, Jr, Lupton JE, Resing JA, de Ronde CEJ, Nakamura Tunnicliffe K-IV, Dower JF, Merle SG. 2007. Exploring the submarine ring of fire: Mariana Arc—Western Pacific. Oceanography 20:68–79 [Google Scholar]
- 49. Lilley MD, Butterfield DA, Olson EJ, Lupton JE, Macko SA, McDuff RE. 1993. Anomalous CH4 and NH4+ concentrations at an unsedimented mid-ocean-ridge hydrothermal system. Nature 364:45–47 [Google Scholar]
- 50. Butterfield DA, McDuff RE, Mottl MJ, Lilley MD, Lupton JE, Massoth GJ. 1994. Gradients in the composition of hydrothermal fluids from the Endeavour Segment vent field: phase separation and brine loss. J. Geophys. Res. 99:9561–9583 [Google Scholar]
- 51. Cruse AM, Seewald JS. 2010. Low-molecular weight hydrocarbons in vent fluids from the Main Endeavour Field, northern Juan de Fuca Ridge. Geochim. Cosmochim. Acta 74:6126–6140 [Google Scholar]
- 52. Reference deleted.
- 53. Friedrich MW. 2005. Methyl-coenzyme M reductase genes: unique functional markers for methanogenic and anaerobic methane-oxidizing Archaea. Methods Enzymol. 397:428–442 [DOI] [PubMed] [Google Scholar]
- 54. Shima S, Krueger M, Weinert T, Demmer U, Kahnt J, Thauer RK, Ermler U. 2012. Structure of a methyl-coenzyme M reductase from Black Sea mats that oxidize methane anaerobically. Nature 481:98–101 [DOI] [PubMed] [Google Scholar]
- 55. Meyerdierks A, Kube M, Kostadinov I, Teeling H, Glöckner FO, Reinhardt R, Amann R. 2010. Metagenome and mRNA expression analyses of anaerobic methanotrophic archaea of the ANME-1 group. Environ. Microbiol. 12:422–439 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.