Abstract
Development of a dense map of the horse genome is key to efforts aimed at identifying genes controlling health, reproduction, and performance. We herein report a high-resolution gene map of the horse (Equus caballus) X chromosome (ECAX) generated by developing and typing 116 gene-specific and 12 short tandem repeat markers on the 5,000-rad horse × hamster whole-genome radiation hybrid panel and mapping 29 gene loci by fluorescence in situ hybridization. The human X chromosome sequence was used as a template to select genes at 1-Mb intervals to develop equine orthologs. Coupled with our previous data, the new map comprises a total of 175 markers (139 genes and 36 short tandem repeats, of which 53 are fluorescence in situ hybridization mapped) distributed on average at ≈880-kb intervals along the chromosome. This is the densest and most uniformly distributed chromosomal map presently available in any mammalian species other than humans and rodents. Comparison of the horse and human X chromosome maps shows remarkable conservation of gene order along the entire span of the chromosomes, including the location of the centromere. An overview of the status of the horse map in relation to mouse, livestock, and companion animal species is also provided. The map will be instrumental for analysis of X linked health and fertility traits in horses by facilitating identification of targeted chromosomal regions for isolation of polymorphic markers, building bacterial artificial chromosome contigs, or sequencing.
Keywords: gene mapping, comparative map, mouse
Equine genome analysis has proceeded at an unprecedented pace during recent years. From the initial horse [Equus caballus (ECA)] gene map 6–7 years ago, organized international efforts have led to a 10-fold expansion in the map. The progress is evident from the recently published meiotic (1–3), cytogenetic (4), and radiation hybrid (RH) maps (5) that provide an array of polymorphic and gene-specific markers distributed over all equine autosomes and the X chromosome. Comparative information available through these maps is proving critical for accurate alignment of the horse genome with the sequenced genomes of human and mouse (4, 5) and with the gene maps of other livestock species. The maps, most of which are of low to medium density, have served as a starting point to initiate research aimed at identifying genes responsible for valuable traits associated with equine biology, health, and performance, including genes responsible for base coat color, overo lethal white, hyperkalemic periodic paralysis, and severe combined immunodeficiency (6–9). Further, marker-based studies are in progress to dissect molecular causes of various coat colors [e.g., gray (10–12) and appaloosa (13, 14)], genetic diseases [e.g., exertional rhabdomyolysis (15, 16); polysaccharide storage myopathy (17)], and other traits of interest. The major impediment associated with the latter group of studies is the lack of adequate resolution maps for individual equine chromosomes. Such maps could facilitate rapid targeted hunts for candidate genes associated with the traits, once they are mapped by genetic linkage analyses with highly polymorphic markers.
The X chromosome is the most conserved mammalian chromosome (18, 19). Extensive comparisons of structure, organization, and gene content of this chromosome in evolutionarily diverse mammals have revealed a remarkable degree of conservation (20–22). Until now, the chromosome has been best studied in humans and mice, where the focus of research has been the intriguing patterns of X inactivation and the involvement of various X specific genes in genetic diseases (23, 24), female and male fertility (25–27), and embryonic development (28). Relatively very little applied research has been conducted in livestock and companion/pet species, which is primarily attributable to the limited information available through the medium- to low-resolution X chromosome maps currently available in pigs (29, 30), cattle (31, 32), cats (21, 33), dogs (34–38), and sheep/goats/buffalo (31, 39, 40), which are insufficient to permit comprehensive studies aimed at dissecting traits of interest.
The current ECA X chromosome (ECAX) map is comparable to those of other livestock and companion species. Beginning with the physical assignment of three gene-specific loci, i.e., G6PD, HGPRT, and PGK, during the early 1970s (41), the ECAX map developed mainly through synteny (42, 43) and genetic linkage mapping approaches (3). During recent years, the map has benefited considerably from the availability of the RH panel (44). The current ECAX map comprises 26 gene and 17 short tandem repeat loci (5, 22), offering a basic RH and comparative map. However, the resolution of this map is inadequate to facilitate identification of economically important genes over the ≈153 Mbp of the chromosome (5).
Building on our recent success in developing high-resolution gene maps for ECA17 (45), ECA22 (A. L. Gustafson, T.R., M. L. Wagner, J.R.M., L.C.S., and B.P.C., unpublished work), and a targeted region of ECA26 (46), we undertook development of a high-resolution gene map of ECAX by stepwise selection of gene-specific markers from the human and mouse X chromosome sequence templates. This effort has resulted in a dense and comprehensive map second only to the detailed maps in humans and mice and will lay the foundation for identifying and analyzing X linked genes involved in equine reproduction and genetic disorders.
Materials and Methods
Marker Selection and Primer Design. The human genome sequence data available from National Center for Biotechnology Information build 34 of the human reference sequence (http://genome.ucsc.edu/cgi-bin/hgGateway) and ensembl l version (also from July 2003, www.ensembl.org/Homo_sapiens) were jointly interrogated for known genes from the human X chromosome. The genes were selected at ≈1-Mb intervals, beginning at 0 Mbp (distal tip of the short arm) and ending at 153 Mbp (distal end of the long arm). Whenever possible, human exonic sequences from selected genes were compared with orthologous sequences from other mammals by using blastn, nr, and est_others search engines (www.ncbi.nlm.nih.gov/BLAST). This was followed by multiple alignment of the sequences in clustalw (http://bioweb.pasteur.fr/seqanal/interfaces/clustalw-simple.html). The alignments were used to design heterologous primers for PCR amplification of horse DNA in a hamster DNA background, as described (45, 47, 48). Briefly, all primers were derived either from a single exon or from two adjacent exons, leaving an ≈500- to 700-bp intron between, and chosen for 100% sequence identity among human, cattle, pig, etc., orthologues but with one to three mismatches with the rodent (mouse, rat) sequences. Primers were designed by using primer3 software (www-genome.wi.mit.edu/cgi-bin/primer/primer3_www.cgi). A total of 88 gene-specific markers were developed by using this approach. Additionally, species-specific primers for all microsatellites (12) and some genes (28) were designed from the available equine genomic or EST sequences. Detailed information on all new markers (128) is presented in Table 1, which is published as supporting information on the PNAS web site.
Primer Optimization and Sequencing. Horse and hamster genomic DNAs were used to optimize the PCR conditions for individual primer pairs, such that only horse-specific DNA amplification was obtained, and all equine PCR amplification products were verified by sequencing. The identities of the sequences were confirmed through blast (www.ncbi.nlm.nih.gov/BLAST) and blat (http://genome.ucsc.edu/cgi-bin/hgBlat?command=start) searches, as described (45), and all comparisons were revalidated against the latest build of sequence data at University of California, Santa Cruz, and Ensembl. For RPS6KA3 (see Table 1), new primers were designed by using sequence data obtained from the first primer pair.
RH Typing Analysis, Bacterial Artificial Chromosome (BAC) Library Screening, and Fluorescence in Situ Hybridization (FISH) Mapping. PCR typing on the 5,000-rad horse × hamster RH panel and data analysis was performed as described (5). The TAMU and CHORI-241 equine genomic BAC libraries were screened by PCR to obtain clones specific for 29 ECAX genes. These clones ensured physical anchors for the RH map at regular intervals along the entire length of the chromosome. Individual BAC clones were labeled with biotin and/or digoxigenin by using BIO- and DIG-Nick Translation Mixes (Roche Molecular Biochemicals) and separately hybridized to horse metaphase chromosomes to confirm probe origin and determine precise physical locations. Additionally, nine closely positioned or overlapping markers in the RH map were cohybridized in differently labeled pairs or triplets on metaphase and interphase chromatin to refine their relative physical order. In situ hybridization, signal detection, microscopy, and image analysis were carried out as described (5).
Results
Generation of Gene-Specific Markers on ECAX. A total of 167 equine gene-specific primer pairs were designed by using the human X chromosome sequence map (Ensembl, www.ensembl.org; Human Genome Browser, http://genome.ucsc.edu/index.html?org=Human&db=hg16&hgsid=27764832). After the first round of optimization, 40 primer sets were excluded due to weak amplification of horse DNA, multiple PCR products, or amplification products of equal sizes for horse and hamster. Of the remaining 127 primer pairs, 28 were identified from equine EST or gene sequences, and the remaining 99 originated from multiple alignments of mammalian sequences (Table 1). Sequencing the PCR amplification products of individual primer pairs to verify the identity of the markers resulted in 11 primer pairs being discarded, because the sequences did not correspond to the expected genes. This yielded 116 equine orthologs for human X chromosome genes and represented a ≈70% success rate in developing horse-specific markers for use on the 5,000-rad RH panel.
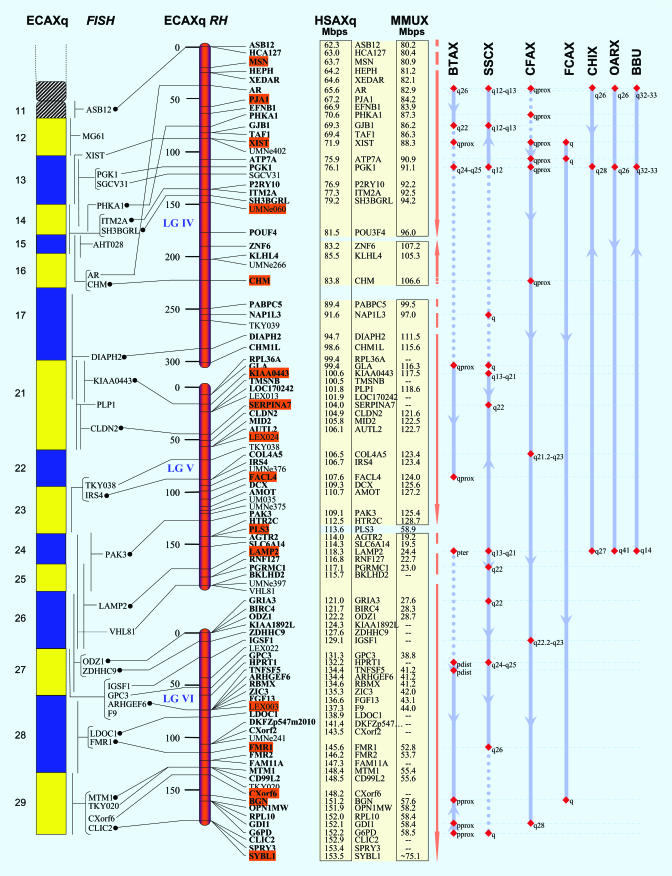
Generation of a Composite RH Map. A total of 128 new markers (116 genes and 12 microsatellites) were typed on the 5,000-rad horse × hamster whole genome RH panel (44). When integrated with 42 loci from the two previous RH maps (5, 22), the final map for the equine X chromosome comprised 169 markers (135 Type I and 34 Type II) uniformly distributed along the length of the chromosome (Fig. 1). With the estimated size of ECAX at ≈153 Mbp (5), the average marker density is 1 every 900 kbp, and the average gene marker density is 1 every ≈1 Mbp. The retention frequency of the 169 markers in the RH panel ranged from 5.4% (BRIA3, BIRC4, and ODZ1) to 31.5% (Adlican), with an average of ≈13% (Fig. 2, which is published as supporting information on the PNAS web site). This retention frequency is satisfactory considering the RH panel was made from a male horse. A relatively high retention of markers was observed toward the proximal and distal end of the short arm, whereas retention was relatively lower than average in the distal part of the long arm (Fig. 2).
Fig. 1.
High-resolution RH and comparative map of the long arm of horse X chromosome (ECAXq). A complete map and detailed figure legend are published as Fig. 3. From left to right are a schematic drawing of ECAXq, FISH localizations, RH map showing the distance and order of loci (ECAXqRH), and comparative location of orthologs in human (HSA) and mouse (MMU), followed by comparative map information on orthologs mapped in cattle (BTA), pig (SSC), dog (CFA), cat (FCA), goat (CHI), sheep (OAR), and river buffalo (BBU).
At logarithm of odds score 7 (2PT-RHMAP), the 169 markers clustered in six RH-linkage groups arranged tandemly from pter to qter on the chromosome (LGI–LGVI; Fig. 3, which is published as supporting information on the PNAS web site), each comprising 48, 11, 15, 30, 31, and 34 markers, respectively (Fig. 3). The proximal three linkage groups comprising 74 markers were located on the short arm, whereas the distal three linkage groups comprising 95 markers were located on the long arm. The order of markers within each RH linkage group was deduced by using the equal retention probability and stepwise locus ordering models (RHMAXLIK). Twenty-nine loci served as frameworks (odds of 1,000:1; shaded orange in Fig. 3). The comprehensive map thus generated spans of 1,344 centiRay (cR)5000, uniformly covering the entire length of ECAX (see Fig. 3 legend for details on map construction). The only two human X (HSAX) regions (each spanning ≈5–6 Mbp) not represented on ECAX were the centromeric region and the HSAXp21.1 region (spanning between 54–62 and 31–37 Mb positions, respectively, on the human sequence map). Otherwise, the representation/alignment of HSAX on ECAX was quite uniform, averaging one gene-specific marker every ≈1-Mbp interval.
Seven genes from the upper terminal part of the RH group 1 (a region corresponding to ECAXpter) produced PCR patterns characteristic to both the X and Y chromosomes. These genes gave the same-size PCR amplification products in males and females and also amplified in RH cell lines known to be Y specific (results not shown). Redesign of primers for most of these genes did not change the PCR typing pattern. However, the second set of primers generated by using sequence data from the first amplification products of STS produced an X specific pattern, as indicated with two adjacent map positions for this gene: STSX and STSXY (Fig. 3).
FISH Map. Equine BAC clones containing 29 selected genes FISH mapped to the expected chromosomal location (Table 1) based on the RH map and the previously FISH mapped loci (22). These localizations bring the total number of cytogenetically mapped markers on ECAX to 53 (46 gene-specific and 7 short tandem repeats; Fig. 3). The FISH markers are fairly uniformly distributed along the chromosome, from Xpter to Xqter (Fig. 3 and Fig. 4, which is published as supporting information on the PNAS web site), except in band Xp21. Two-color FISH on metaphase chromosomes provided the following physical order for overlapping loci:. Adlican-TMSB4 (pter⇒cen; Fig. 4h), CHM-DIAPH2 (cen⇒qter; Fig. 4f) and FMR1-MTM1 (cen⇒qter; Fig. 4h). Further, interphase FISH with combinations of differently labeled probes helped refine relative order of three loci (Adlican, NLGN4, and TMSB4) located in the terminal region of the short arm (Xpter). The interphase-FISH order for the three loci was pter⇒Adlican⇒NLGN4⇒TMSB4⇒cen (Fig. 4i). All FISH results were in agreement with the physical order of loci deduced by the RH map. However, there were minor exceptions: although double-color FISH showed that NUDT11 is distal to MAGEH1 on ECAXp13 (Fig. 4e), RH analysis gave a reversed order. Reexamination of typing results of these and adjacent loci did not show any genotyping/scoring errors. Last, our FISH assignment of LAMP2 is in agreement with our RH map but does not concur with earlier localization (4).
Comparative Mapping. The mapping of 137 equine orthologs of HSAX genes demonstrated complete synteny conservation between the X chromosomes of the two species (Fig. 3). Of these, 114 loci have also been mapped in mouse: 113 to MMUX and one (CLCN4) to MMU7 (49). The results reiterate conservation of the X chromosome across three evolutionarily distantly related mammals, with a remarkably higher degree of conservation between horse and human than between horse and mouse or human and mouse. As yet no mouse orthologs have been found for 23 of the horse/human X specific genes. Of these, nine genes are from the human pseudoautosomal regions (PAR), PAR1 and -2 (see Fig. 3).
To obtain a refined comparative map among horse, human, and mouse X chromosomes, we determined the precise sequence location of human and mouse orthologs for all 133 physically ordered equine genes from the available genomic draft sequences (see Materials and Methods). Four FISH-mapped genes (ANT3, MG61, PLP1, and F9) not mapped by RH analysis were also included in the comparison by placing them on the comparative map based on adjacent FISH markers, increasing the number of comparative loci to 137. As described earlier (5), if sequence locations of a group of human or mouse orthologs indicated conservation of gene order in relation to the derived order of equine genes, the data were clustered in boxes (see Fig. 3). A total of seven clusters were observed in human and 13 in mouse. These clusters, referred to as conserved linkages [maximally contiguous chromosomal region with identical gene content and order (50)], showed groups of genes with similar physical order in horse–human and horse–mouse. The clustering also highlighted smaller evolutionarily conserved segments among the three species that could be observed within the larger conserved linkages (Fig. 3, cream-shaded regions over human/mouse orthologs).
Discussion
We herein report a physical map of the horse X chromosome comprising 175 markers (139 genes and 36 microsatellites). The 128 markers mapped in this study by RH analysis, 90% of which are functional genes, expand the existing ECAX map (5, 22) by >4-fold. Further, a total of 53 FISH localizations substantially improve the number of physical anchor points between the chromosome and the RH map and concurrently provide excellent corroboration to the RH data. The map is the first high-resolution integrated gene map for a horse chromosome. With markers distributed on average at 880-kb intervals along the chromosome and gene-specific loci dispersed on average at ≈1-Mb intervals, this is the densest and most uniformly distributed map of the X chromosome presently available in any of the mammalian species other than humans and mice.
The currently available X chromosome maps of various livestock and pet/companion animals show a range of 12 (sheep/goat/buffalo) to 92 (pig) genes mapped using different approaches [cattle, ≈37 genes (31, 32, 39, 51); pig, ≈92 genes (29, 30, 52–57); dog, 22 genes (34, 35, 37); cat, 30 genes (21, 33); sheep/goat/buffalo, 12 genes (31, 39, 40)]. In most of these species except cat (33) and dog (36), either the different maps are not integrated and physically ordered into a single map or the distribution of the loci is not uniform. In contrast, the 137 gene human–horse comparative map presented in this study is 2- to 4-fold more informative for gene content and is conspicuously integrated into a single physically ordered map. This is evident also from the comparative status of the X chromosome map for all ECAX genes hitherto mapped in different species (see Fig. 3, right half of each sheet). Last, despite containing only ≈10% of the expected X homologs from human (1,695 genes http://genome.ucsc.edu/cgi-bin/hgGateway) or mouse (1,116 genes; draft sequence, www.ensembl.org/Mus_musculus), the map provides excellent alignment of ECAX with the X chromosome map of the two species.
Despite a total of 169 markers, the ECAX RH map was divided into six RH linkage groups, due to the threshold chosen (logarithm of odds, 7 RHMAP2PT) to develop the linkage groups. Assuming the linkage groups to be separated by at least 50 cR5000, the five gaps add ≈250 cR to the current map of 1,344 cR, thus increasing the effective map size to ≈1,600 cR. This implies that with the size of ECAX as 153 Mbp, the average span of the map for this chromosome in our 5,000-rad panel is ≈10 cR/Mbp. Further, of the 153 Mbp, ≈2–3 Mbp spans the centromeric region, whereas 5–6 Mbp spans the intercalary heterochromatic region on the long arm of the chromosome (Xq21). It is presumed that these regions are gene poor. These assumptions stem from the fact that gene density in the predominantly heterochromatic regions of HSA1, -9, and -16 is zero for stretches spanning 20, 16, and 9 Mbp of heterochromatin, respectively (http://bioinformatics.weizmann.ac.il/cards). A similar condition was also seen for satellite-bearing chromosome HSA22, where the proximal ≈14 Mbp are barren, and HSAY, where the distal ≈20 Mbp do not contain any genes (except for three genes in the PAR2 region).
XY Amplification and the Pseudoautosomal Region. Of significant interest in the ECAX map is the distal/terminal region of the short arm. Seven markers from this region (APXL, DXYS155E, ASMT, GYG2, Adlican, NLGN4, and STS) demonstrate both X and Y specific amplification with RH cell line DNA. In humans, all seven genes are known to have both X and Y homologs with sequence identities ranging from 60% to 96% [(58) GeneCards, http://bioinformatics.weizmann.ac.il/cards]. Presently 31 such genes are located on HSAY. These genes, referred to as X degenerates, are considered as surviving relics of the ancestral autosome from which X and Y chromosomes evolved (58). Four more X degenerate genes (TMSB4X, USP9X, KAL1, and SMCX; Fig. 3) were also mapped in this study, but primers for these genes show only X specific amplification. We suggest that, because the latter four loci are further away from the XY recombining region, sequences of these genes might have had more evolutionary time for X-Y divergence (59); however, this needs further investigation. Nevertheless, the identification of equine X-Y pattern genes in horse is an important starting point to analyze shared sequences between equine X and Y chromosomes, thus shedding light on their evolution.
Synaptonemal complex analysis in horses shows that the ECAXpter region is pseudoautosomal [PAR (60, 61)]. The three main characteristics of the PAR genes are that they (i) represent functional homologs of genes present on both X and Y chromosomes; (ii) escape X inactivation; and (iii) recombine during male meiosis (62–65). The human PAR1 spans ≈2.6 Mb and contains 13 genes (62, 64–66). Of these, two, DXYS155E and ASMT, are mapped to ECAX in this study. The human PAR2, located terminally on HSAXq, spans 320 kb and contains four genes [HSPRY3, SYBL1, IL9R, and CXYorf1 (65)]. Of these, two have been mapped in this study to the terminal end of ECAXq (Fig. 3). Analysis hitherto carried out in a range of mammalian species show significant species-specific differences in the PAR genes. A gene that is pseudoautosomal in one species may not be pseudoautosomal in the other. Of the 13 human PAR genes, eight do not have a known mouse homolog (64). Further, STS is a PAR gene in cattle (67), pig (54), dog, sheep, and mouse but not in human and primate (63, 64); KAL1 is pseudoautosomal in pig (54) and cattle (67) but not in human and primate. Surprisingly, this locus is not even found in mouse (64). The horse PAR has not yet been defined for gene content. The genes mapped in this study provide a foundation to begin investigating the Xpter region in horse for identification of prospective PAR genes. Subsequent expression/methylation studies of these genes can help validate their pseudoautosomal status and contribute to precise characterization and physical demarcation the equine PAR.
Comparative Map. The physically ordered ECAX map, comprising 139 equine genes, is in close agreement with our previous maps (5, 22) and provides a robust comparative map in relation to the observed order of these genes in humans and mice. The most striking feature of the horse–human comparison is that, barring minor exceptions involving four to five interruptions on the short arm, the relative order of loci in the two species is exceptionally conserved from Xpter to Xqter. This degree of gene order conservation has not yet been observed for any other chromosome between humans and other nonprimate mammals. The horse–mouse comparison, however, shows noticeably less conservation in gene order. The rearrangements are evident from the 13 conserved linkage blocks (boxes containing ordered loci) originating from different regions of MMUX, some with reversed centromere–telomere orientation in relation to horse and human (Fig. 3).
The comparative map of the equine X chromosome presented here helps to identify 13 blocks/clusters of loci demonstrating conserved gene order across horse, human, and mouse (Fig. 3, yellow shaded regions). These clusters potentially represent the most conserved X chromosome regions of the ancestor common to horse, human, and mouse. Additionally, the blocks provide a quick comparative overview of smaller conserved linkages and their relative organization in the three species. Although an exceptional degree of gene order conservation is observed between ECAX and HSAX, there are also minor rearrangements. The most noticeable of these is a small segment corresponding to ECAXp12–13 and the human sequence positions 46–54 Mb (Fig. 3, red bracket). Interestingly, the horse and mouse genomes are also relatively more rearranged in the same region. Based on studies focused on the evolution of mammalian X chromosome, it is proposed that the human X chromosome is composed of different evolutionary strata. These are roughly divided into the X conserved region XCR, corresponding to the proximal one-quarter of short arm and complete long arm of HSAX, and the recently added region XRA, corresponding to distal three-quarters of the short arm of HSAX (59, 68). Rearrangements observed by us in the horse, human, and mouse genomes are incidentally located close to the suggested ancestral fusion point of the conserved and the ancestral regions on HSAXp11.23 (68). A detailed study of this region aimed at identifying evolutionary breakpoints may also lead to discovery of signatures of these rearrangements in horse/mouse, which in turn will be useful for understanding the origin of gene order differences in this region across the three species.
Future Uses. The mammalian X chromosome contains a disproportionately high number of genes influencing development, female/male fertility, reproduction, and disease [Online Mendelian Inheritance in Man, www.ncbi.nlm.nih.gov/Sitemap/index.html#OMIM; Online Mendelian Inheritance in Animals, http://morgan.angis.su.oz.au/Databases/BIRX/omia (25, 26, 69, 70)] that are also of significance in horses. The human and pig X chromosomes carry an unexpectedly high number of genes specifically expressed in the skeletal muscle (55, 71, 72). Analysis of these genes could have implications for the performance of horses. Structural and numerical aberrations of the X chromosome are the most common documented chromosome abnormalities in the horse that invariably lead to reproductive failures, intersexuality, hermaphroditism, or sex reversal (73, 74). Additionally, a number of X linked conditions/diseases have been described in the horse, e.g., G6PD deficiency (75), X linked severe immunodeficiency (76), fragile X (77), agammaglobulinemia (78), and hydrocephalus (79). Presently very little information is available concerning the underlying molecular causes of these conditions. The high-resolution RH and comparative map of ECAX presented in this study, in conjunction with detailed map/sequence information on human and mouse X chromosomes (23, 26, 27, 70), will serve as a basis to rapidly converge on X specific genes significant in the horse. Efforts to isolate BAC clones for all mapped markers on this chromosome will considerably facilitate localized isolation of polymorphic markers, building BAC contigs in areas of interest and sequencing targeted chromosomal regions.
Supplementary Material
Acknowledgments
We thank Dee Honeycutt for excellent management of the horse RH panel and Glenda Goh and Kristi Hope for technical assistance. This project was funded by grants from the Texas Higher Education Board (ARP 010366-0162-2001; ATP 000517-0306-2003 BPC, to J.E.W.), National Research Initiative Competitive Grants Program (NRICGP)/U.S. Department of Agriculture (USDA) Grant 2003-03687 (to B.P.C.), NRICGP/USDA Grant 2000-03510 (to L.C.S.), Texas Equine Research Foundation (to B.P.C. and L.C.S.), Link Endowment (to B.P.C. and L.C.S.), American Quarter Horse Association, USDA–National Research Support Projects-8 Coordinators' Fund, and the Dorothy Russell Havemeyer Foundation.
Abbreviations: ECA, Equus caballus; ECAX, ECA X chromosome; RH, radiation hybrid; FISH, fluorescence in situ hybridization; BAC, bacterial artificial chromosome; cR, centiRay; PAR, pseudoautosomal region.
References
- 1.Guérin, G., Bailey, E., Bernoco, D., Anderson, I., Antczak, D. F., Bell, K., Binns, M. M., Bowling, A. T., Brandon, R., Cholewinski, G., et al. (1999) Anim. Genet. 30, 341–354. [DOI] [PubMed] [Google Scholar]
- 2.Guérin, G., Bailey, E., Bernoco, D., Anderson, I., Antczak, D. F., Bell, K., Biros, I., Bjornstad, G., Bowling, A. T., Brandon, R., et al. (2003) Anim. Genet. 34, 161–168. [DOI] [PubMed] [Google Scholar]
- 3.Swinburne, J., Gerstenberg, C., Breen, M., Aldridge, V., Lockhart, L., Marti, E., Antczak, D., Eggleston-Stott, M., Bailey, E., Mickelson, J., et al. (2000) Genomics 66, 123–134. [DOI] [PubMed] [Google Scholar]
- 4.Milenkovic, D., Oustry-Vaiman, A., Lear, T. L., Billault, A., Mariat, D., Piumi, F., Schibler, L., Cribiu, E. & Guerin, G. (2002) Mamm. Genome 13, 524–534. [DOI] [PubMed] [Google Scholar]
- 5.Chowdhary, B. P., Raudsepp, T., Kata, S. R., Goh, G., Millon, L. V., Allan, V., Piumi, F., Guerin, G., Swinburne, J., Binns, M., et al. (2003) Genome Res. 13, 742–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marklund, L., Moller, M. J., Sandberg, K. & Andersson, L. (1996) Mamm. Genome 7, 895–899. [DOI] [PubMed] [Google Scholar]
- 7.Santschi, E. M., Purdy, A. K., Valberg, S. J., Vrotsos, P. D., Kaese, H. & Mickelson, J. R. (1998) Mamm. Genome 9, 306–309. [DOI] [PubMed] [Google Scholar]
- 8.Rudolph, J. A., Spier, S. J., Byrns, G., Rojas, C. V., Bernoco, D. & Hoffman, E. P. (1992) Nat. Genet. 2, 144–147. [DOI] [PubMed] [Google Scholar]
- 9.Wiler, R., Leber, R., Moore, B. B., VanDyk, L. F., Perryman, L. E. & Meek, K. (1995) Proc. Natl. Acad. Sci. USA 92, 11485–11489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Swinburne, J. E., Hopkins, A. & Binns, M. M. (2002) Anim. Genet. 33, 338–342. [DOI] [PubMed] [Google Scholar]
- 11.Henner, J., Poncet, P. A., Guerin, G., Hagger, C., Stranzinger, G. & Rieder, S. (2002) Mamm. Genome 13, 535–537. [DOI] [PubMed] [Google Scholar]
- 12.Locke, M. M., Penedo, M. C., Bricker, S. J., Millon, L. V. & Murray, J. D. (2002) Anim. Genet. 33, 329–337. [DOI] [PubMed] [Google Scholar]
- 13.Terry, R. R., Bailey, E., Bernoco, D. & Cothran, E. G. (2001) Anim. Genet. 32, 98–101. [DOI] [PubMed] [Google Scholar]
- 14.Terry, R. B., Bailey, E., Lear, T. & Cothran, E. G. (2002) Anim. Genet. 33, 82–84. [DOI] [PubMed] [Google Scholar]
- 15.MacLeay, J. M., Valberg, S. J., Pagan, J. D., de laCorte, F., Roberts, J., Billstrom, J., McGinnity, J. & Kaese, H. (1999) Equine Vet. J. Suppl 30, 458–462. [DOI] [PubMed]
- 16.MacLeay, J. M., Sorum, S. A., Valberg, S. J., Marsh, W. E. & Sorum, M. D. (1999) Am. J. Vet. Res. 60, 1562–1566. [PubMed] [Google Scholar]
- 17.Valberg, S. J., Geyer, C., Sorum, S. A. & Cardinet, G. H., 3rd (1996) Am. J. Vet. Res. 57, 286–290. [PubMed] [Google Scholar]
- 18.Ohno, S. (1967) Sex Chromosomes and Sex-Linked Genes (Springer, Berlin).
- 19.Charlesworth, B. (1991) Science 251, 1030–1033. [DOI] [PubMed] [Google Scholar]
- 20.Graves, J. A., Gecz, J. & Hameister, H. (2002) Cytogenet. Genome Res. 99, 141–145. [DOI] [PubMed] [Google Scholar]
- 21.Murphy, W. J., Sun, S., Chen, Z. Q., Pecon-Slattery, J. & O'Brien, S. J. (1999) Genome Res. 9, 1223–1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Raudsepp, T., Kata, S. R., Piumi, F., Swinburne, J., Womack, J. E., Skow, L. C. & Chowdhary, B. P. (2002) Genomics 79, 451–457. [DOI] [PubMed] [Google Scholar]
- 23.Boyd, Y., Blair, H. J., Cunliffe, P., Masson, W. K. & Reed, V. (2000) Genome Res. 10, 277–292. [DOI] [PubMed] [Google Scholar]
- 24.Lyon, M. F. (2002) Acta Paediatr. Suppl 91, 107–112. [DOI] [PubMed]
- 25.Wang, Z. J., Jeffs, B., Ito, M., Achermann, J. C., Yu, R. N., Hales, D. B. & Jameson, J. L. (2001) Proc. Natl. Acad. Sci. USA 98, 7988–7993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vaiman, D. (2002) Cytogenet. Genome Res. 99, 224–228. [DOI] [PubMed] [Google Scholar]
- 27.Vialard, F., Cocquet, J., Christin-Maitre, S., Veitia, R. & Fellous, M. (2002) Cytogenet. Genome Res. 99, 218–223. [DOI] [PubMed] [Google Scholar]
- 28.Burgoyne, P. S., Ojarikre, O. A. & Turner, J. M. (2002) Cytogenet. Genome Res. 99, 252–256. [DOI] [PubMed] [Google Scholar]
- 29.McCoard, S. A., Fahrenkrug, S. C., Alexander, L. J., Freking, B. A., Rohrer, G. A., Wise, T. H. & Ford, J. J. (2002) Anim. Genet. 33, 178–185. [DOI] [PubMed] [Google Scholar]
- 30.Rink, A., Santschi, E. M., Eyer, K. M., Roelofs, B., Hess, M., Godfrey, M., Karajusuf, E. K., Yerle, M., Milan, D. & Beattie, C. W. (2002) Mamm. Genome 13, 578–587. [DOI] [PubMed] [Google Scholar]
- 31.Iannuzzi, L., Di Meo, G. P., Perucatti, A., Incarnato, D., Schibler, L. & Cribiu, E. P. (2000) Cytogenet. Cell Genet. 89, 171–176. [DOI] [PubMed] [Google Scholar]
- 32.Amaral, M. E., Kata, S. R. & Womack, J. E. (2002) Mamm. Genome 13, 268–271. [DOI] [PubMed] [Google Scholar]
- 33.Menotti-Raymond, M., David, V. A., Chen, Z. Q., Menotti, K. A., Sun, S., Schaffer, A. A., Agarwala, R., Tomlin, J. F., O'Brien, S. J. & Murphy, W. J. (2003) J. Hered. 94, 95–106. [DOI] [PubMed] [Google Scholar]
- 34.Breen, M., Jouquand, S., Renier, C., Mellersh, C. S., Hitte, C., Holmes, N. G., Cheron, A., Suter, N., Vignaux, F., Bristow, A. E., et al. (2001) Genome Res. 11, 1784–1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Everts, R. E., van Wolferen, M. E., Versteeg, S. A., Zijlstra, C., Engelen, J. J., Bosma, A. A., Rothuizen, J. & van Oost, B. A. (2002) Cytogenet. Genome Res. 98, 86–92. [DOI] [PubMed] [Google Scholar]
- 36.Guyon, R., Lorentzen, T. D., Hitte, C., Kim, L., Cadieu, E., Parker, H. G., Quignon, P., Lowe, J. K., Renier, C., Gelfenbeyn, B., et al. (2003) Proc. Natl. Acad. Sci. USA 100, 5296–5301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Spriggs, H. F., Holmes, N. G., Breen, M. G., Deloukas, P. G., Langford, C. F., Ross, M. T., Carter, N. P., Davis, M. E., Knights, C. E., Smith, A. E., et al. (2003) Mamm. Genome 14, 214–221. [DOI] [PubMed] [Google Scholar]
- 38.Kirkness, E. F., Bafna, V., Halpern, A. L., Levy, S., Remington, K., Rusch, D. B., Delcher, A. L., Pop, M., Wang, W., Fraser, C. M., et al. (2003) Science 301, 1898–1903. [DOI] [PubMed] [Google Scholar]
- 39.Piumi, F., Schibler, L., Vaiman, D., Oustry, A. & Cribiu, E. P. (1998) Cytogenet. Cell Genet. 81, 36–41. [DOI] [PubMed] [Google Scholar]
- 40.Schibler, L., Vaiman, D., Oustry, A., Giraud-Delville, C. & Cribiu, E. P. (1998) Genome Res. 8, 901–915. [DOI] [PubMed] [Google Scholar]
- 41.Deys, B. F. (1972) Ph.D. thesis (University of Leiden, Leiden, The Netherlands).
- 42.Caetano, A. R., Shiue, Y. L., Lyons, L. A., O'Brien, S. J., Laughlin, T. F., Bowling, A. T. & Murray, J. D. (1999) Genome Res. 9, 1239–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shiue, Y.-L., Millon, L. V., Skow, L. C., Honeycutt, D., Murray, J. D. & Bowling, A. T. (2000) Chromosome Res. 8, 45–55. [DOI] [PubMed] [Google Scholar]
- 44.Chowdhary, B. P., Raudsepp, T., Honeycutt, D., Owens, E. K., Piumi, F., Guerin, G., Matise, T. C., Kata, S. R., Womack, J. E. & Skow, L. C. (2002) Mamm. Genome 13, 89–94. [DOI] [PubMed] [Google Scholar]
- 45.Lee, E.-J., Raudsepp, T., Kata, S. R., Adelson, D., Womack, J. E., Skow, L. C. & Chowdhary, B. P. (2004) Genomics, 83, 203–215. [DOI] [PubMed] [Google Scholar]
- 46.Ward, T. L., Valberg, S. J., Lear, T. R., Guérin, G., Milenkovic, D., Swinburne, J. E., Binns, M., Raudsepp, T., Skow, L., Chowdhary, B. P., et al. (2003) Cytogenet. Genome Res., in press. [DOI] [PubMed]
- 47.Jiang, Z., He, H., Hamasima, N., Suzuki, H. & Verrinder, G. (2002) Genome 45, 147–156. [DOI] [PubMed] [Google Scholar]
- 48.Jiang, Z., Priat, C. & Galibert, F. (1998) Mamm. Genome 9, 577–587. [DOI] [PubMed] [Google Scholar]
- 49.Rugarli, E. I., Adler, D. A., Borsani, G., Tsuchiya, K., Franco, B., Hauge, X., Disteche, C., Chapman, V. & Ballabio, A. (1995) Nat. Genet. 10, 466–471. [DOI] [PubMed] [Google Scholar]
- 50.Nadeau, J. H. & Sankoff, D. (1998) Trends Genet. 14, 495–501. [DOI] [PubMed] [Google Scholar]
- 51.Band, M. R., Larson, J. H., Rebeiz, M., Green, C. A., Heyen, D. W., Donovan, J., Windish, R., Steining, C., Mahyuddin, P., Womack, J. E., et al. (2000) Genome Res. 10, 1359–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hu, Z., Rohrer, G. A., Murtaugh, M. P., Stone, R. T. & Beattie, C. W. (1997) Mamm. Genome 8, 608–610. [DOI] [PubMed] [Google Scholar]
- 53.Hawken, R. J., Murtaugh, J., Flickinger, G. H., Yerle, M., Robic, A., Milan, D., Gellin, J., Beattie, C. W., Schook, L. B. & Alexander, L. J. (1999) Mamm. Genome 10, 824–830. [DOI] [PubMed] [Google Scholar]
- 54.Quilter, C. R., Blott, S. C., Mileham, A. J., Affara, N. A., Sargent, C. A. & Griffin, D. K. (2002) Mamm. Genome 13, 588–594. [DOI] [PubMed] [Google Scholar]
- 55.Davoli, R., Fontanesi, L., Zambonelli, P., Bigi, D., Gellin, J., Yerle, M., Milc, J., Braglia, S., Cenci, V., Cagnazzo, M., et al. (2002) Anim. Genet. 33, 3–18. [DOI] [PubMed] [Google Scholar]
- 56.Cirera, S., Jorgensen, C. B., Sawera, M., Raudsepp, T., Chowdhary, B. P. & Fredholm, M. (2003) Mamm. Genome 14, 405–426. [DOI] [PubMed] [Google Scholar]
- 57.Lahbib-Mansais, Y., Tosser-Klopp, G., Leroux, S., Cabau, C., Karsenty, E., Milan, D., Barillot, E., Yerle, M., Hatey, F. & Gellin, J. (2003) Mamm. Genome 14, 275–288. [DOI] [PubMed] [Google Scholar]
- 58.Skaletsky, H., Kuroda-Kawaguchi, T., Minx, P. J., Cordum, H. S., Hillier, L., Brown, L. G., Repping, S., Pyntikova, T., Ali, J., Bieri, T., et al. (2003) Nature 423, 825–837. [DOI] [PubMed] [Google Scholar]
- 59.Lahn, B. T. & Page, D. C. (1999) Science 286, 964–967. [DOI] [PubMed] [Google Scholar]
- 60.Safronova, L. D. & Pimenova, T. I. (1988) Genetika 24, 708–714. [PubMed] [Google Scholar]
- 61.Power, M. M., Gustavsson, I., Switonski, M. & Ploen, L. (1992) Cytogenet. Cell Genet. 61, 202–207. [DOI] [PubMed] [Google Scholar]
- 62.Ellison, J. W., Li, X., Francke, U. & Shapiro, L. J. (1996) Mamm. Genome 7, 25–30. [DOI] [PubMed] [Google Scholar]
- 63.Blaschke, R. J. & Rappold, G. A. (1997) Genome Res. 7, 1114–1117. [DOI] [PubMed] [Google Scholar]
- 64.Gianfrancesco, F., Sanges, R., Esposito, T., Tempesta, S., Rao, E., Rappold, G., Archidiacono, N., Graves, J. A., Forabosco, A. & D'Urso, M. (2001) Genome Res. 11, 2095–2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Charchar, F. J., Svartman, M., El-Mogharbel, N., Ventura, M., Kirby, P., Matarazzo, M. R., Ciccodicola, A., Rocchi, M., D'Esposito, M. & Graves, J. A. (2003) Genome Res. 13, 281–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rappold, G. A. (1993) Hum. Genet. 92, 315–324. [DOI] [PubMed] [Google Scholar]
- 67.Moore, S. S., Byrne, K., Johnson, S. E., Kata, S. & Womack, J. E. (2001) Anim. Genet. 32, 102–104. [DOI] [PubMed] [Google Scholar]
- 68.Wilcox, S. A., Watson, J. M., Spencer, J. A. & Graves, J. A. (1996) Genomics 35, 66–70. [DOI] [PubMed] [Google Scholar]
- 69.Graves, J. A. & Delbridge, M. L. (2001) BioEssays 23, 1091–1094. [DOI] [PubMed] [Google Scholar]
- 70.Liao, D. J., Du, Q. Q., Yu, B. W., Grignon, D. & Sarkar, F. H. (2003) Cancer Invest. 21, 641–658. [DOI] [PubMed] [Google Scholar]
- 71.Pallavicini, A., Zimbello, R., Tiso, N., Muraro, T., Rampoldi, L., Bortoluzzi, S., Valle, G., Lanfranchi, G. & Danieli, G. A. (1997) Hum. Mol. Genet. 6, 1445–1450. [DOI] [PubMed] [Google Scholar]
- 72.Bortoluzzi, S., Rampoldi, L., Simionati, B., Zimbello, R., Barbon, A., d'Alessi, F., Tiso, N., Pallavicini, A., Toppo, S., Cannata, N., et al. (1998) Genome Res. 8, 817–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Makinen, A., Hasegawa, T., Makila, M. & Katila, T. (1999) Equine Vet. J. 31, 346–349. [DOI] [PubMed] [Google Scholar]
- 74.Makinen, A., Suojala, L., Niini, T., Katila, T., Tozaki, T., Miyake, Y. & Hasegawa, T. (2001) Equine Vet. J. 33, 527–530. [DOI] [PubMed] [Google Scholar]
- 75.Stockham, S. L., Harvey, J. W. & Kinden, D. A. (1994) Vet. Pathol. 31, 518–527. [DOI] [PubMed] [Google Scholar]
- 76.Felsburg, P. J., Somberg, R. L. & Perryman, L. E. (1992) Immunodefic. Rev. 3, 277–303. [PubMed] [Google Scholar]
- 77.Ronne, M. (1992) Hereditas 117, 127–136. [DOI] [PubMed] [Google Scholar]
- 78.Perryman, L. E., McGuire, T. C. & Banks, K. L. (1983) Am. J. Pathol. 111, 125–127. [PMC free article] [PubMed] [Google Scholar]
- 79.Ojala, M. & Ala-Huikku, J. (1992) Equine Vet. J. 24, 140–143. [DOI] [PubMed] [Google Scholar]
- 80.Bowling, A. T., Breen, M., Chowdhary, B. P., Hirota, K., Lear, T., Millon, L. V., Ponce de Leon, F. A., Raudsepp, T. & Stranzinger, G. (1997) Chromosome Res. 5, 433–443. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.