Abstract
In the arterial circulation, regions of disturbed flow (DF), which are characterized by flow separation and transient vortices, are susceptible to atherogenesis, whereas regions of undisturbed laminar flow (UF) appear protected. Coordinated regulation of gene expression by endothelial cells (EC) may result in differing regional phenotypes that either favor or inhibit atherogenesis. Linearly amplified RNA from freshly isolated EC of DF (inner aortic arch) and UF (descending thoracic aorta) regions of normal adult pigs was used to profile differential gene expression reflecting the steady state in vivo. By using human cDNA arrays, ≈2,000 putatively differentially expressed genes were identified through false-discovery-rate statistical methods. A sampling of these genes was validated by quantitative real-time PCR and/or immunostaining en face. Biological pathway analysis revealed that in DF there was up-regulation of several broad-acting inflammatory cytokines and receptors, in addition to elements of the NF-κB system, which is consistent with a proinflammatory phenotype. However, the NF-κB complex was predominantly cytoplasmic (inactive) in both regions, and no significant differences were observed in the expression of key adhesion molecules for inflammatory cells associated with early atherogenesis. Furthermore, there was no histological evidence of inflammation. Protective profiles were observed in DF regions, notably an enhanced antioxidative gene expression. This study provides a public database of regional EC gene expression in a normal animal, implicates hemodynamics as a contributory mechanism to athero-susceptibility, and reveals the coexistence of pro- and antiatherosclerotic transcript profiles in susceptible regions. The introduction of additional risk factors may shift this balance to favor lesion development.
There is a strong correlation between flow characteristics and the focal and regional nature of atherogenesis. Sites of disturbed flow (DF) (e.g., the inner wall of curved vessels and the wall opposite the flow divider at branches and bifurcations) are susceptible to lesion development, whereas regions of undisturbed laminar flow (UF) are relatively protected (1–3). Throughout the initiation and development of atherosclerotic lesions, the endothelium is retained as the interface between blood and arterial tissues (4, 5), where its function both before and during lesion formation is critical to the pathological outcome. It has been hypothesized that coordinated regulation of endothelial gene expression in response to local biomechanical forces results in differing regional phenotypes that promote athero-protection or athero-susceptibility (6). This hypothesis has been addressed in principle by gene expression studies of cultured endothelium (7–20). However, it has only been addressed in vivo for a limited number of candidate genes and proteins (21–25).
Studies of candidate gene expression by endothelial cells (EC) in culture (12, 18, 23) and in knock-out animals (22, 24, 25) suggest that transcriptional activity is linked to flow disturbances. In EC cultures, unidirectional laminar shear stress (LSS) was correlated with protective profiles of gene expression (e.g., antioxidative, antiinflammatory, and/or antiproliferative) when compared to the absence of flow (7–11, 13, 14, 17, 19) and selected examples of the protective genes have been shown to be expressed in endothelium in vivo (14, 19). A small number of microarray studies comparing EC gene expression between DF and UF conditions in vitro have been conducted (15, 16); however, transcriptional profiling in vivo has been limited by the small sample size available within hemodynamic regions of interest. The in vivo correlation between local gene expression and athero-susceptibility has therefore been generated primarily by extrapolation of in vitro findings. Using RNA amplification to overcome sampling limitations, we have profiled regional EC gene expression in the normal adult pig aorta to investigate athero-susceptibility as a function of differential hemodynamics in vivo.
Materials and Methods
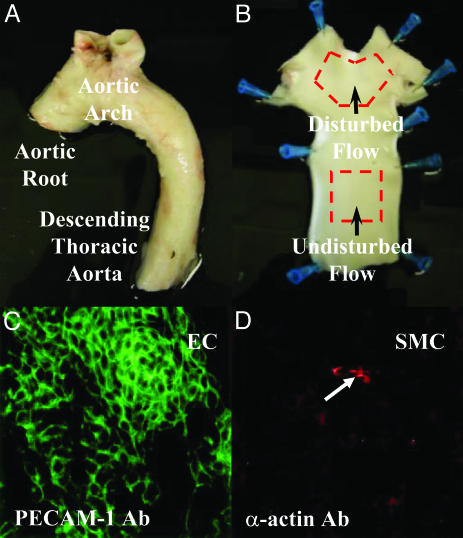
Sample Collection and Characterization. Endothelial cells were freshly harvested from eight adult pig aortas (Landrace X Yorkshire males, 230–250 pounds; Hatfield Industries, Hatfield, PA). The ascending aorta, aortic arch, and descending thoracic aorta were dissected from the surrounding tissue (Fig. 1A), flushed with cold sterile PBS, and incised lengthwise. EC were gently scraped from a 1-cm2 region located at the inner-curve and lateral walls of the aortic arch (DF) and separately from the dorsal descending thoracic aorta (UF) as illustrated in Fig. 1B, with each sample representing ≈10,000 cells. The cells were transferred directly to a lysis buffer containing RNase inhibitors. Representative cell scrapes were periodically transferred to glass microscope slides to monitor cell purity by immunostaining with EC-specific antiplatelet-endothelial cell adhesion molecule 1 and smooth muscle cell (SMC)-specific anti-α-actin antibodies. Additional tissue samples were fixed in 4% paraformaldehyde for en face immunostaining, regional EC nuclear staining (Hoechst 33258), and cross-sectional vessel histology.
Fig. 1.
(A and B) Illustration of the endothelial cell isolation procedure. About 10,000 cells were scraped from precisely defined regions (≈1 cm2) of the aortic arch (DF) and the descending thoracic aorta (UF). (C and D) A representative field of EC isolated by mechanical scraping and stained with anti-platelet-endothelial cell adhesion molecule 1 (PECAM-1) and anti-α-actin antibodies. EC purity (green) was routinely >99% with only occasional contamination by isolated SMC (red; arrow).
Microarray Procedures. These procedures have been described (26) and are provided in full in Supporting Materials and Methods, which is published as supporting information on the PNAS web site. Total RNA (100 ng; ≈1 ng of mRNA) was linearly amplified (27), and 2 μg of amplified antisense RNA was used to synthesize 33P-labeled DNA probes. These probes were hybridized to nylon microarray filters, custom designed, and printed by AstraZeneca Pharmaceuticals (Alderley Park, UK), which contained 13,824 3′-biased, sequence-verified human cDNA clones (1.5–2.0 kb) spotted in duplicate. Matched DF and UF arrays for the same animal (n = 8) were processed simultaneously.
Data Analysis and Bioinformatics. Image files were quantified with arrayvision 6.3 (Imaging Research, St. Catherine's, ON, Canada) as described (26). A putative set of differentially expressed genes was identified by taking the union of predictions made through the methods of arraystat 1.2 (Imaging Research) (28) in “unpaired” mode and sam 1.15 (Significance Analysis of Microarrays) (29) in “one class” mode. Both methods used an expected false-discovery rate (30) of 5%. The putative set of differentially expressed genes was imported into genespring (Silicon Genetics, Redwood City, CA) for annotation by using information available in public databases and hierarchical classification according to a simple gene ontology construction. A detailed description of the data analysis is provided in Supporting Materials and Methods. Results obtained by other analytical approaches can be accessed at www.cbil.upenn.edu/RAD/normal_pig_study. In accordance with proposed standards of the Microarray Gene Expression Data Society (www.mged.org), the complete annotated study is publicly available in a miame compliant framework through the RNA Abundance Database (www.cbil.upenn.edu/RAD) (31).
Validation Studies. Immunostaining on fixed tissue sections was performed according to standard procedures for selected proteins. Quantitative real-time PCR (QRT-PCR) was performed on selected genes as described (26). Expression was measured in paired DF and UF samples from six animals, and a ratio was calculated for each animal based on a minimum of three replicate observations. If the PCR ratios for at least four of six animals were >1.1 or <0.9, then the PCR result was designated “up-regulation” or “down-regulation,” respectively. If the PCR ratios for at least four of six animals were between 0.9 and 1.1, then the designation was “unchanged.” In all other cases, no decision was made based on the PCR results. Where the PCR ratios were >1.1 in three animals and <0.9 in the remaining animals, a note was made about this interanimal discrepancy.
Results
Regional Characterization of EC. The presence of flow reversal in the aortic arch of adult boars similar to that measured in humans (32) was confirmed by ultrasound (data not shown). EC isolated from DF and UF regions (Fig. 1 A and B) displayed differences in cell shape and alignment that reflected the local hemodynamic conditions (Fig. 4 A and B, which is published as supporting information on the PNAS web site). In situ immunostaining with EC-specific platelet-endothelial cell adhesion molecule 1 and SMC-specific α-actin antibodies confirmed the presence of an intact endothelium (Fig. 4 C and D). There was no histological evidence of inflammation at either site (Fig. 5, which is published as supporting information on the PNAS web site). To monitor the specificity of the isolation procedure for EC, cell scrapes were transferred to glass microscope slides where cell-specific staining confirmed EC purity of >99% (Fig. 1 C and D). Thus near-pure isolates of EC showing morphologies consistent with differential hemodynamic environments in vivo were rapidly obtained for transcript profiling.
Differential Expression. Putative sets of differentially expressed EC genes (DF compared with UF) identified in the normal pig aorta through the predictions of arraystat and/or sam are summarized in Table 1, which also shows their annotation by biological classifications of known relevance to the initiation and progression of atherosclerosis. sam analysis identified a larger number of genes (1,823) than did arraystat (1,048) at the same false-discovery rate of 5%, while capturing ≈75% of the arraystat set. An expanded version of Table 1 with links to fully annotated gene lists is available online (www.cbil.upenn.edu/RAD/normal_pig_study).
Table 1. Genes identified as differentially expressed (DF vs. UF) in the normal pig aorta by biological classification.
| Biological classification | No. represented on array | No. differentially expressed |
|---|---|---|
| All genes | 13824 | 2091 |
| Adhesion | 298 | 44 |
| Apoptosis | 244 | 47 |
| Coagulation | 63 | 8 |
| Complement | 71 | 19 |
| Extracellular matrix | 270 | 47 |
| Growth factors | 238 | 50 |
| Immune response | 136 | 28 |
| Inflammation | 150 | 28 |
| Lipid/cholesterol metabolism | 167 | 20 |
| NF-κB | 51 | 10 |
| Oxidative mechanisms | 166 | 27 |
| Proliferation | 391 | 73 |
| Signal transduction | 2430 | 412 |
| TNF-α | 69 | 8 |
| Transcription factors | 557 | 94 |
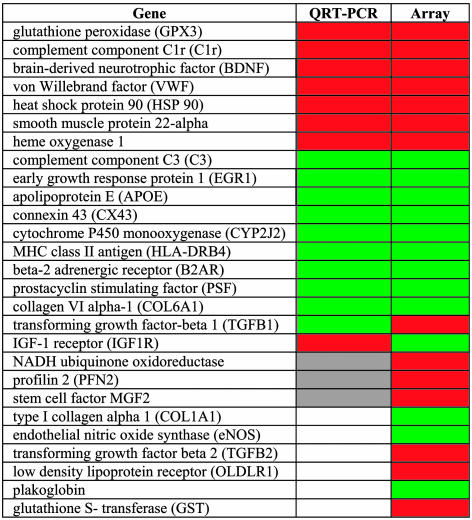
QRT-PCR. Twenty-seven genes identified as significantly differentially expressed were selected for validation by QRT-PCR. Fig. 2 summarizes the concordance of differential expression determined by QRT-PCR with that predicted by array analysis. The array prediction was validated for 60% (16/27) of the genes sampled, and there was disagreement with the array prediction for 18% (5/27). For 22% (6/27) of the gene sample, high interanimal variability prevented a definitive conclusion (reported as “undetermined” in Fig. 2) despite highly reproducible measurements for each animal (n ≥ 3). The basis of the interanimal variability for these genes is unclear, but it may be due to confounding factors other than the influence of regional hemodynamics, and it may be genetic in origin. The validation of about two of three genes after linear amplification is consistent with our previous report, for which we used a model system (26).
Fig. 2.
Comparison of QRT-PCR results with microarray predictions for selected genes: red, up-regulated in DF; green, down-regulated in DF; gray, unchanged; white, undetermined (high interanimal variability).
Mining of Biological Pathways. Table 2 shows selected differentially expressed genes (DF vs. UF) in the biological classifications of inflammation, cell adhesion, and oxidative mechanisms. Also shown in Table 2 are the arraystat ratio (mean, n = 8) and the putative positive or negative impact, deduced from the literature, on mechanisms contributing to atherogenesis (references are provided as Supporting References to Table 2).
Table 2. Selected genes identified as differentially expressed (DF vs. UF) in the context of a putative role in atherosclerosis.
| Up-regulated
|
Down-regulated
|
||||||
|---|---|---|---|---|---|---|---|
| Gene | GenBank ID | Ratio | Putative effect* | Gene | GenBank ID | Ratio | Putative effect* |
| Inflammatory/immune response | |||||||
| IL-1A | X02851 | 2.01 | + | IL-10 | M57627 | 0.67 | + |
| IL-1R1 | M27492 | 1.46 | + | IKBKAP | AF153419 | 0.77 | + |
| IL-6 | X04430 | 1.68 | + | CEZANNE | AJ293573 | 0.66 | + |
| MCP-1 | M37719 | 1.57 | + | IL-8 | M28130 | 0.62 | – |
| RAGE | AB036432 | 4.02 | + | CXCR4 | AF005058 | 0.29 | – |
| IL-8RB | M99412 | 1.65 | + | FOS | BC004490 | 0.43 | – |
| NFKB1 | M58603 | 1.34 | + | PAFR | D10202 | 0.66 | – |
| NFKB2 | S76638 | 1.35 | + | C3 | J04763 | 0.27 | – |
| NFKBIA | BC004983 | 2.76 | + | HLADRB4 | M19556 | 0.55 | – |
| EPCR | L35545 | 1.90 | – | TRAF6 | H12612 | 0.62 | – |
| PTGS2 | D28235 | 1.55 | – | IGFBP1 | R81994 | 0.66 | – |
| IL-14 | L15344 | 0.67 | ? | ||||
| Oxidative mechanisms | |||||||
| GPX3 | D00632 | 5.64 | – | TXNIP | S73591 | 0.44 | – |
| SOD3 | U10116 | 1.39 | – | COX6B | BC001015 | 0.61 | – |
| NQO1 | BC000474 | 1.50 | – | COX7A2 | AY007643 | 0.74 | – |
| POR | AF258341 | 1.36 | – | NDUFC1 | AK023115 | 0.69 | – |
| HMOX1 | X06985 | 1.66 | – | NDUFS4 | AF020351 | 0.64 | – |
| GSTT1 | AB057594 | 1.55 | – | NDUFS6 | AF044959 | 0.56 | – |
| MGST2 | U77604 | 2.83 | – | NDUFS3 | AL135819 | 0.52 | – |
| NDUFA7 | Y16007 | 1.40 | + | NDUFA3 | AF044955 | 0.42 | – |
| NDUFB7 | AF112200 | 1.35 | + | NDUFV2 | M22538 | 0.75 | – |
| NDUFB10 | AF088991 | 1.35 | + | NDUFA6 | BC002772 | 0.76 | – |
| NDUFS8 | U65579 | 0.70 | – | ||||
| CYBB | X04011 | 0.46 | – | ||||
| NOS3 | M93718 | 0.69 | + | ||||
| SOD1 | K00065 | 0.77 | + | ||||
| Cell adhesion† | |||||||
| VWF | X04385 | 5.04 | + | CTNND1 | AF062343 | 0.59 | ? |
| CD44 | AJ251595 | 1.47 | + | CDH1 | Z13009 | 0.50 | ? |
| CDH3 | X63629 | 1.43 | – | CDH13 | L34058 | 0.65 | ? |
| FN1 | AJ276395 | 2.51 | ? | ITGA6 | X53586 | 0.43 | ? |
| ITGB4 | X51841 | 1.43 | ? | ITGA5 | BC008786 | 0.69 | ? |
| MO1A | J03270 | 1.34 | ? | JUP | Z68228 | 0.19 | ? |
IL-1A, IL-1α; IL-1R1, IL-1 receptor 1; IL-6, IL-6 (interferon β2); MCP-1, monocyte chemotactic protein 1; RAGE, advanced glycosylation end product-specific receptor; IL-8RB (CXCR2), IL-8 receptor β; NFKB1, nuclear factor of κ light polypeptide gene enhancer in B cells 1 (p105); NFKB2, nuclear factor of κ light polypeptide gene enhancer in B cells 2 (p49/p100); NFKBIA, NF-κB inhibitor α; EPCR, endothelial protein C receptor; PTGS2, prostaglandin endoperoxide synthase-2 (COX-2); IKBKAP, inhibitor of κ light polypeptide gene enhancer in B-cells, kinase complex-associated protein; CEZANNE, cellular zinc finger anti-NF-κB; CXCR4, chemokine (C-X-C motif) receptor 4; FOS, v-fos FBJ murine osteosarcoma viral oncogene; PAFR, platelet-activating factor receptor; C3, complement component C3; HLADRB4, MHC class II HLA-DRw53-beta (DR4,w4); TRAF6, tumor necrosis factor receptor-associated factor 6; IGFBP1, insulin-like growth factor binding protein 1; GPX3, glutathione peroxidase 3; SOD3, superoxide dismutase 3; NQO1, NADH quinone oxidoreductase; POR, cytochrome P450 oxidoreductase; HMOX1, heme oxygenase (decycling) 1; GSTT1, GST θ1; MGST2, microsomal GST 2; NDUFA7, NADH ubiquinone oxidoreductase; NDUFB7, NADH dehydrogenase (ubiquinone) component B7; NDUFB10, NADH dehydrogenase (ubiquinone) component B10; TXNIP, thioredoxin interacting protein (VDUP1); COX6B, cytochrome c oxidase subunit VIb; COX7A2, subunit VIIa; NDUFC1, NADH dehydrogenase (ubiquinone) component C1; NDUFS4, component S4; NDUFS6, component S6; NDUFS3, component S3; NDUFA3, component A3; NDUFV2, component V2; NDUFA6, component A6; NDUFS8, component S8; CYBB, cytochrome b-245 beta (NADPH oxidase component p91 phox); NOS3, endothelial nitric oxide synthase; SOD1, superoxide dismutase 1; VWF, von Willebrand factor; CD44, cell adhesion molecule (CD44); CDH3, cadherin 3 (P-cadherin); FN1, fibronectin 1; ITGB4, integrin β4; MO1A, leukocyte adhesion glycoprotein Mo1α; CTNND1, catenin δ1; CDH1, cadherin 1 (E-cadherin); CDH13, cadherin 13 (H-cadherin); ITGA6, integrin α6; ITGA5, integrin α5 (fibronectin receptor); JUP, junction plakoglobin.
+, pro-atherosclerotic; –, anti-atherosclerotic; ?, unknown.
References to the putative effect provided as Supporting References to Table 2, which is published as supporting information on the PNAS web site
Vascular cell adhesion molecule 1 (VCAM-1), intercellular adhesion molecule 1, P-selectin, E-selectin were not significantly different
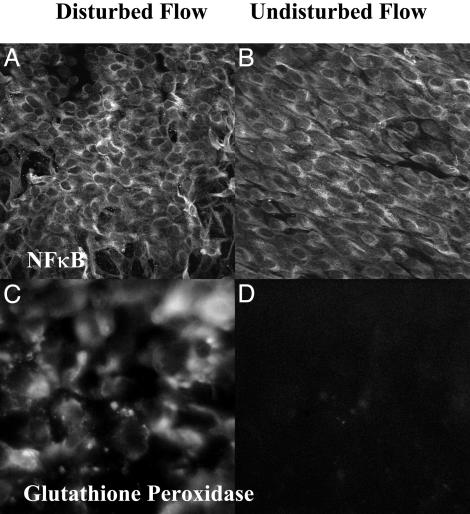
Proinflammatory gene expression in DF. The differential regulation of inflammatory and immune-related genes revealed an endothelium primed for inflammation in DF. Consistent with a proinflammatory response, there was up-regulation of several broad-acting cytokines, chemokines, and receptors (interleukin 1α, interleukin 1 receptor 1, interleukin 6, interleukin 8 receptor β, advanced glycosylation end-product-specific receptor, monocyte chemotactic protein 1) as well as down-regulation of the antiinflammatory interleukin 10 in DF when compared with UF. Furthermore, up-regulation of elements of the NF-κB system (nuclear factor of kappa light polypeptide gene enhancer in B cells 1 and 2 and NF-κB inhibitor α) and simultaneous down-regulation of transcription for the IκB kinase-complex-associated protein involved in assembling an active kinase complex (33), and the cellular zinc finger anti-NF-κB protein (Cezanne) which down-regulates NF-κB (34) were noted (Table 2). However, NF-κB was inactive in both DF and UF regions as shown by cytoplasmic localization and nuclear exclusion of the NF-κB complex (p65 antibody) (Fig. 3A and B). These observations are consistent with an NF-κB system primed but inactive in DF as described by Hajra et al. (22). Differentially expressed genes that may mitigate an inflammatory tendency included the up-regulation of endothelial protein C receptor and the down-regulation of platelet-activating factor receptor, chemokine receptor 4, complement component C3, several cathepsins, and the MHC class II antigen HLA-DRB4.
Fig. 3.
En face immunostaining in DF and UF regions. (A and B) Subcellular localization of NF-κB complex (p65 antibody) was visualized by 2D projection of confocal imaging through the EC layer. The staining was predominantly cytoplasmic with nuclear exclusion in both aortic regions. Also clearly illustrated are the EC morphologies characteristic of DF (A) and UF (B). (C and D) Epifluorescence microscopy for the prominent differentially expressed antioxidative enzyme GPX3 revealed strong expression in DF (C) in contrast to low expression in UF (D), consistent with microarray predictions.
Absence of differences in adhesion molecule expression. Several adhesion-related molecules were differentially regulated by flow type, including von Willebrand factor, CD44, fibronectin 1, several integrins, cadherins, and junction plakoglobin. The up-regulation of von Willebrand factor (24) and CD44 (25) in particular may have implications for atherogenesis. However, VCAM-1, intercellular adhesion molecule 1, E-selectin, and P-selectin, all associated with early NF-κB-mediated inflammatory responses and the onset of atherogenesis, were not detected as differentially expressed despite the increased transcriptional levels of proinflammatory cytokines and NF-κB pathway components noted above. Because the statistical methods aimed at detecting putative differential expression do not allow us to make any definitive statement regarding unchanged genes, we also measured differential expression of these genes by QRT-PCR (data not shown). There was no clear pattern of differential expression, with the results indicating slight upregulation in some animals, but slight down-regulation or no change in others. Immunostaining for VCAM-1 was weak and indistinguishable between the two regions (data not shown). Collectively, these observations in the context of normal histology suggest that, although the endothelium in DF regions may be primed for an inflammatory response, the process is held in check, possibly at the level of NF-κB.
Protective antioxidative expression profile in DF. Reactive oxygen species (ROS) and prooxidative pathways are implicated in the initiation and progression of atherosclerosis (35). Furthermore, ROS is a potent activator of NF-κB (36, 37). Data mining revealed an antioxidative profile in endothelium located in regions of DF that may protect against NF-κB-mediated inflammation. Significant suppression of a component of superoxide-generating NADH oxidase was noted. An antioxidative state was supported by concomitant up-regulation of key antioxidative enzymes, including GPX3, extracellular superoxide dismutase, NADH quinone oxidoreductase, heme oxygenase, and two forms of GST (GST-θ1 and microsomal MGST2). Several of these genes have been reported to be up-regulated by laminar shear stress in vitro in a response mediated by a shear-responsive antioxidant response element (38). Down-regulation of components of the cytochrome c oxidase and NADH dehydrogenase complexes (intramitochondrial sources of ROS through their involvement in electron transport), and of endothelial nitric oxide synthase 3 were noted. Also protective was the down-regulation of thioredoxin-interacting protein [i.e., vitamin D3-up-regulated protein 1 (VDUP-1)], an inhibitor of thioredoxin that, in turn, is an important regulator of cell redox balance (39). Berk and colleagues have shown that shear stress influences VDUP-1 gene and protein expression (B. Berk; personal communication). Down-regulation of VDUP-1 leads to increased thioredoxin activity and, ultimately, down-regulation of VCAM-1 expression through the mitogen-activated protein kinase/Jun kinase pathway. Few prooxidative genes were differentially expressed, and the balance was decisively shifted in the direction of a protective antioxidative state in DF regions in these healthy animals. The expression of antioxidative enzymes GPX3 and heme oxygenase were validated by QRT-PCR (Fig. 2). The prominent up-regulation of GPX3 in DF was confirmed by immunostaining (Fig. 3 C and D). The antioxidative expression profile observed here may be critical to maintaining a delicate balance in gene expression in vulnerable regions of normal animals.
Gene expression in other biological classifications. Although it is not within the scope of this paper to exhaustively analyze the entire database of differentially expressed genes, this study resulted in a very rich dataset, which is now provided as a public resource. Reference to the online database also revealed a balance of gene expression related to coagulation mechanisms with a shift toward protective anticoagulant and profibrinolytic mechanisms in the DF region. Notably, all components of the plasminogen-activator system identified by our analyses were regulated in a manner consistent with antithrombotic mechanisms in DF. Lipid/cholesterol metabolism-related gene expression reflected a balance in DF regions between the expression of genes, which might promote cholesterol deposition and reverse cholesterol transport. Numerous growth factors and receptors were shown to be differentially regulated. The differential expression of numerous transcription factors may provide new insight into the modulation of the endothelial response to hemodynamics through the identification of common regulatory binding elements.
Discussion
Although the localization of atherosclerotic lesions to predictable regions in mammalian arteries has been recognized for over a century, compelling evidence implicating the local hemodynamics is more recent (6). We have exploited RNA amplification to demonstrate the heterogeneity of endothelial phenotypes from hemodynamically distinct regions in vivo, demonstrating that a delicate balance of pro- and antiatherosclerotic mechanisms may exist simultaneously in endothelium of lesion-prone sites of the adult porcine aorta to create a setting of vulnerability to atherogenesis. Atherosclerosis has been described as a chronic inflammatory disease involving many cell types and complex cross-talk between them (40). Many interrelated physiological mechanisms are recruited very early in the process, including cell adhesion, oxidative metabolism, lipid metabolism, apoptosis, innate and adaptive immunity, and coagulation (recently reviewed in refs. 40–46). The predisposition for expression of key molecules in multiple pathways is therefore likely to be important in determining the susceptibility of a site to lesion initiation when additional risk factors are prevalent, thereby influencing the threshold or timing for atherogenesis.
NF-κB is a transcription factor proposed to play a central role as a mediator/integrator of atherogenesis (36). Its association with inhibitory IκBs masks the nuclear localization sequence, retaining the complex in the cytoplasm. Activation of IκB kinase results in phosphorylation of IκBs and their targeting for degradation by the proteosome, releasing NF-κB dimers for translocation to the nucleus, where they activate transcription in target genes. These genes include cytokines, chemokines, adhesion molecules, and genes involved in cell proliferation and cell survival. NF-κB is controlled by the redox state of the cell, which may be a common signaling pathway leading to NF-κB activation through ROS (37). Activation of NF-κB and NF-κB-regulated genes has been observed in atherosclerotic plaques (47), and NF-κB-activated by shear stress has been observed in cultured cells (48, 49) and regions of DF (22). Collins et al. (36) have suggested that the survival/protective genes induced by NF-κB limit the inflammatory response at low levels, although a strong challenge results in the expression of adhesion molecules/cytokines.
In one of the few in vivo regional arterial studies, Hajra et al. (22) evaluated NF-κB regulation in regions of high and low probability for atherosclerosis in mouse aorta by using en face immunostaining and confocal microscopy. They demonstrated preferential activation of NF-κB (translocation to the nucleus) and induction of NF-κB-responsive gene expression in high probability regions of the aortas of lipopolysaccharide-treated or hypercholesterolemic LDLR-/- mice. In normal mice, however, although p65 and IκBs were elevated in such regions, NF-κB activation remained low, suggesting that the pathway was primed but not activated. Our findings are consistent with and greatly extend such an interpretation.
Several recent reports have used microarrays to assess gene expression in EC exposed to unidirectional steady flow in vitro (9, 13, 20) and compared expression profiles to those of control cells in no-flow conditions. The results, however, are not easily compared with our study, which captures the profile of two distinct regions long-adapted to differential hemodynamic environments in vivo. More relevant are two in vitro microarray studies that compared flow conditions analogous to DF and UF.
In the first, Garcia-Cardena et al. (16) compared differential expression in cultured human umbilical vein endothelial cells between turbulent shear stress (TSS) and LSS, analogous to DF vs. UF regions in vivo in this study. One hundred genes were identified as differentially expressed (TSS vs. LSS) after 24 h (68 up-regulated and 32 down-regulated). The authors identified a set of genes that was down-regulated in LSS but up-regulated in TSS as potentially the most pathologically relevant. They considered highly regulated genes of known or putative function in signaling, response to injury, or atherogenesis and focused on linking the protective effects of LSS to matrix biology and cell cycle. Brooks et al. (15) compared gene expression in cultured human aortic EC under pulsatile DF conditions with LSS by using microarrays (588 genes) and subtraction cloning. They identified >100 genes as differentially expressed at 24 h, many of which were up-regulated genes associated with mechanisms known to be proatherosclerotic, particularly inflammatory molecules, adhesion factors, and oxidation-related molecules.
Few genes identified as differentially expressed by these in vitro studies were common to those identified by our in vivo study. Furthermore, there was little agreement between the in vitro studies. The reasons may be the different nature of the DF in each study, different cell origins, sensitivities of microarray analysis, or a myriad of technical considerations. Despite significantly different outcomes, however, both of these in vitro studies demonstrated that conditions of DF resulted in significant differential endothelial gene expression compared with conditions of unidirectional steady flow.
The methods used here have refined endothelial transcriptional profiling to regions small enough to represent exposure to discrete flow characteristics in vivo, but these methods also provide sufficient cells to amplify RNA and profile gene expression with reasonable confidence. We have previously evaluated the linear amplification protocol by using similar amounts of human matched probe/target and found it to identify differential expression with fidelity and enhanced sensitivity (26). In agreement with this previous study, the QRT-PCR results reported in the current study corroborate approximately two of three genes tested. When considering differential expression with cross-species hybridization, differences in sequence homology are expected to lead to fewer genes hybridizing than for the equivalent same-species hybridization, thus contributing to false negatives. However, both samples should hybridize in the same way so that false positives will not be affected. The linear amplification protocol minimizes bias possibly introduced by other amplification methods, but it is clear that certain transcripts may be reproducibly misrepresented. Although we verified EC purity at >99%, we cannot rule out the possibility that a small number of contaminating SMC could lead to some false positives with the use of amplification. Given the increased sensitivity of amplification, it is possible that strongly differentially expressed genes in a small number of contaminating cells could be detected.
Inflammatory responses leading to atherogenesis in DF regions may share common signaling pathways as responses mediated by cytokines, such as tumor necrosis factor (TNF)-α. In particular, the two may share common mechanisms mediated by NF-κB. We have previously profiled EC gene expression in response to TNF-α in vitro (26), and we find the overlap with the present in vivo study to be small. Up-regulated genes common to both studies included monocyte chemotactic protein 1, interleukin 1α, NF-κB1, NF-κB2, NF-κB inhibitor α, TNF-α-induced protein 6, and cycloxygenase-2, whereas common down-regulated genes included TNF receptor-associated factor 6 and the chemokine receptor CXCR4. An important distinction between these studies is that differential expression of VCAM-1, intercellular adhesion molecule 1, and E-selectin was induced by TNF-α, but not by differential hemodynamics in the normal animal.
Because this is the first in vivo study profiling regional endothelial gene expression on a large scale, we have attempted to bridge the findings to published candidate gene studies in vivo (21–24) and experiments of flow disturbance in vitro (8, 15, 16, 19, 23). Furthermore, we have presented specific observations relating to some pathways that are central to atherogenesis. Further mining of these data with emphasis on different biological pathways (such as those highlighted in Table 1) will provide additional insights into the regional susceptibility to atherosclerosis.
In summary, a baseline is established for gene expression profiling by using endothelium from DF and UF regions in the normal adult pig aorta, which defines in vivo the existing relationships between many classes of molecules responsible for vascular homeostasis and implicated in early atherogenesis. This study revealed distinctly different patterns of gene expression than reported by in vitro studies to date. The hemodynamic characteristics of DF may prime the endothelium toward inflammation (and by inference, atherogenesis) but protective profiles in gene expression (e.g., a net antioxidative profile) prevent disease initiation in the absence of additional risk factors.
Supplementary Material
Acknowledgments
We thank Drs. Garret FitzGerald, Aron Fisher, and Paul Janmey of the University of Pennsylvania for critical reading of the manuscript, Rebecca Riley for expert technical assistance, and Dr. Richard Magid for help with NF-κB imaging. This work was supported by National Institutes of Health Grants HL62250, HL70128, K25-HG-02296, and K25-HG-00052; National Space Biomedical Research Institute (National Aeronautics and Space Administration) Grant NSBRI-01-102; and a Sponsored Research Award from AstraZeneca Pharmaceuticals.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: DF, disturbed flow; UF, undisturbed flow; EC, endothelial cells; LSS, laminar shear stress; SMC, smooth muscle cell; QRT-PCR, quantitative real-time PCR; VCAM-1, vascular cell adhesion molecule 1; ROS, reactive oxygen species; GPX3, glutathione peroxidase 3; TNF, tumor necrosis factor.
References
- 1.Cornhill, J. F. & Roach, M. R. (1976) Atherosclerosis 23, 489-501. [DOI] [PubMed] [Google Scholar]
- 2.Glagov, S., Zarins, C., Giddens, D. P. & Ku, D. N. (1988) Arch. Pathol. Lab. Med. 112, 1018-1031. [PubMed] [Google Scholar]
- 3.Wissler, R. W. (1994) Atherosclerosis 108, S3-S20. [DOI] [PubMed] [Google Scholar]
- 4.Davies, P. F., Reidy, M. A., Goode, T. B. & Bowyer, D. E. (1976) Atherosclerosis 25, 125-130. [DOI] [PubMed] [Google Scholar]
- 5.Faggiotto, A., Ross, R. & Harker, L. (1984) Arteriosclerosis 4, 323-340. [DOI] [PubMed] [Google Scholar]
- 6.Gimbrone, M. A., Jr., Anderson, K. R., Topper, J. N., Langille, B. L., Clowes, A. W., Bercel, S., Davies, M. G., Stenmark, K. R., Frid, M. G., Weiser-Evans, M. C., et al. (1999) J. Vasc. Surg. 29, 1104-1151. [DOI] [PubMed] [Google Scholar]
- 7.Nagel, T., Resnick, N., Atkinson, W. J., Dewey, C. F., Jr., & Gimbrone, M. A., Jr. (1994) J. Clin. Invest. 94, 885-891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Topper, J. N., Cai, J., Falb, D. & Gimbrone, M. A., Jr. (1996) Proc. Natl. Acad. Sci. USA 93, 10417-10422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, B. P., Li, Y. S., Zhao, Y., Chen, K. D., Li, S., Lao, J., Yuan, S., Shyy, J. Y. & Chien, S. (2001) Physiol. Genomics 7, 55-63. [DOI] [PubMed] [Google Scholar]
- 10.De Keulenaer, G. W., Chappell, D. C., Ishizaka, N., Nerem, R. M., Alexander, R. W. & Griendling, K. K. (1998) Circ. Res. 82, 1094-1101. [DOI] [PubMed] [Google Scholar]
- 11.Bongrazio, M., Baumann, C., Zakrzewicz, A., Pries, A. R. & Gaehtgens, P. (2000) Cardiovasc. Res. 47, 384-393. [DOI] [PubMed] [Google Scholar]
- 12.Passerini, A. G., Milsted, A. & Rittgers, S. E. (2003) J. Vasc. Surg. 37, 182-190. [DOI] [PubMed] [Google Scholar]
- 13.McCormick, S. M., Eskin, S. G., McIntire, L. V., Teng, C. L., Lu, C. M., Russell, C. G. & Chittur, K. K. (2001) Proc. Natl. Acad. Sci. USA 98, 8955-8960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wasserman, S. M., Mehraban, F., Komuves, L. G., Yang, R. B., Tomlinson, J. E., Zhang, Y., Spriggs, F. & Topper, J. N. (2002) Physiol. Genomics 12, 13-23. [DOI] [PubMed] [Google Scholar]
- 15.Brooks, A. R., Lelkes, P. I. & Rubanyi, G. M. (2002) Physiol. Genomics 9, 27-41. [DOI] [PubMed] [Google Scholar]
- 16.Garcia-Cardena, G., Comander, J., Anderson, K. R., Blackman, B. R. & Gimbrone, M. A., Jr. (2001) Proc. Natl. Acad. Sci. USA 98, 4478-4485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin, K., Hsu, P. P., Chen, B. P., Yuan, S., Usami, S., Shyy, J. Y., Li, Y. S. & Chien, S. (2000) Proc. Natl. Acad. Sci. USA 97, 9385-9389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.DePaola, N., Davies, P. F., Pritchard, W. F., Jr., Florez, L., Harbeck, N. & Polacek, D. C. (1999) Proc. Natl. Acad. Sci. USA 96, 3154-3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Topper, J. N., Cai, J., Qiu, Y., Anderson, K. R., Xu, Y. Y., Deeds, J. D., Feeley, R., Gimeno, C. J., Woolf, E. A., Tayber, O., et al. (1997) Proc. Natl. Acad. Sci. USA 94, 9314-9319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peters, D. G., Zhang, X. C., Benos, P. V., Heidrich-O'Hare, E. & Ferrell, R. E. (2002) Physiol. Genomics 12, 25-33. [DOI] [PubMed] [Google Scholar]
- 21.Iiyama, K., Hajra, L., Iiyama, M., Li, H., DiChiara, M., Medoff, B. D. & Cybulsky, M. I. (1999) Circ. Res. 85, 199-207. [DOI] [PubMed] [Google Scholar]
- 22.Hajra, L., Evans, A. I., Chen, M., Hyduk, S. J., Collins, T. & Cybulsky, M. I. (2000) Proc. Natl. Acad. Sci. USA 97, 9052-9057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.De Nigris, F., Lerman, L. O., Ignarro, S. W., Sica, G., Lerman, A., Palinski, W., Ignarro, L. J. & Napoli, C. (2003) Proc. Natl. Acad. Sci. USA 100, 1420-1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Methia, N., Andre, P., Denis, C. V., Economopoulos, M. & Wagner, D. D. (2001) Blood 98, 1424-1428. [DOI] [PubMed] [Google Scholar]
- 25.Cuff, C. A., Kothapalli, D., Azonobi, I., Chun, S., Zhang, Y., Belkin, R., Yeh, C., Secreto, A., Assoian, R. K., Rader, D. J. & Pure, E. (2001) J. Clin. Invest. 108, 1031-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Polacek, D. C., Passerini, A. G., Shi, C., Francesco, N. M., Manduchi, E., Grant, G. R., Powell, S., Bischof, H., Winkler, H., Stoeckert, C. J., Jr., & Davies, P. F. (2003) Physiol. Genomics 13, 147-156. [DOI] [PubMed] [Google Scholar]
- 27.Van Gelder, R. N., von Zastrow, M. E., Yool, A., Dement, W. C., Barchas, J. D. & Eberwine, J. H. (1990) Proc. Natl. Acad. Sci. USA 87, 1663-1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nadon, R. & Shoemaker, J. (2002) Trends Genet. 18, 265-271. [DOI] [PubMed] [Google Scholar]
- 29.Tusher, V. G., Tibshirani, R. & Chu, G. (2001) Proc. Natl. Acad. Sci. USA 98, 5116-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Benjamini, Y. & Hockberg, Y. (1995) J. R. Stat. Soc. B 57, 289-300. [Google Scholar]
- 31.Manduchi, E., Grant, G. R., He, H., Liu, J., Mailman, M. D., Pizarro, A. D., Whetzel, P. L. & Stoeckert, C. J., Jr. (2004) Bioinformatics, in press. [DOI] [PubMed]
- 32.Kilner, P. J., Yang, G. Z., Mohiaddin, R. H., Firmin, D. N. & Longmore, D. B. (1993) Circulation 88, 2235-2247. [DOI] [PubMed] [Google Scholar]
- 33.Cohen, L., Henzel, W. J. & Baeuerle, P. A. (1998) Nature 395, 292-296. [DOI] [PubMed] [Google Scholar]
- 34.Evans, P. C., Taylor, E. R., Coadwell, J., Heyninck, K., Beyaert, R. & Kilshaw, P. J. (2001) Biochem. J. 357, 617-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Griendling, K. K., Sorescu, D. & Ushio-Fukai, M. (2000) Circ. Res. 86, 494-501. [DOI] [PubMed] [Google Scholar]
- 36.Collins, T. & Cybulsky, M. I. (2001) J. Clin. Invest. 107, 255-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li, N. & Karin, M. (1999) FASEB J. 13, 1137-1143. [PubMed] [Google Scholar]
- 38.Chen, X. L., Varner, S. E., Rao, A. S., Grey, J. Y., Thomas, S., Cook, C. K., Wasserman, M. A., Medford, R. M., Jaiswal, A. K. & Kunsch, C. (2003) J. Biol. Chem. 278, 703-711. [DOI] [PubMed] [Google Scholar]
- 39.Schulze, P. C., De Keulenaer, G. W., Yoshioka, J., Kassik, K. A. & Lee, R. T. (2002) Circ. Res. 91, 689-695. [DOI] [PubMed] [Google Scholar]
- 40.Steinberg, D. (2002) Nat. Med. 8, 1211-1217. [DOI] [PubMed] [Google Scholar]
- 41.Binder, C. J., Chang, M. K., Shaw, P. X., Miller, Y. I., Hartvigsen, K., Dewan, A. & Witztum, J. L. (2002) Nat. Med. 8, 1218-1226. [DOI] [PubMed] [Google Scholar]
- 42.Ruggeri, Z. M. (2002) Nat. Med. 8, 1227-1234. [DOI] [PubMed] [Google Scholar]
- 43.Li, A. C. & Glass, C. K. (2002) Nat. Med. 8, 1235-1242. [DOI] [PubMed] [Google Scholar]
- 44.Repa, J. J. & Mangelsdorf, D. J. (2002) Nat. Med. 8, 1243-1248. [DOI] [PubMed] [Google Scholar]
- 45.Dzau, V. J., Braun-Dullaeus, R. C. & Sedding, D. G. (2002) Nat. Med. 8, 1249-1256. [DOI] [PubMed] [Google Scholar]
- 46.Libby, P. & Aikawa, M. (2002) Nat. Med. 8, 1257-1262. [DOI] [PubMed] [Google Scholar]
- 47.Brand, K., Page, S., Rogler, G., Bartsch, A., Brandl, R., Knuechel, R., Page, M., Kaltschmidt, C., Baeuerle, P. A. & Neumeier, D. (1996) J. Clin. Invest. 97, 1715-1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lan, Q., Mercurius, K. O. & Davies, P. F. (1994) Biochem. Biophys. Res. Commun. 201, 950-956. [DOI] [PubMed] [Google Scholar]
- 49.Khachigian, L. M., Resnick, N., Gimbrone, M. A., Jr., & Collins, T. (1995) J. Clin. Invest. 96, 1169-1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.