Abstract
Protein turnover is an effective way of maintaining a functional proteome, as old and potentially damaged polypeptides are destroyed and replaced by newly synthesized copies. An increasing number of intracellular proteins, however, have been identified that evade this turnover process and instead are maintained over a cell’s lifetime. This diverse group of long-lived proteins might be particularly prone to accumulation of damage and thus play a critical role in the functional deterioration of key regulatory processes during ageing.
Ageing is a universal phenomenon that challenges all biological systems at multiple levels and ultimately results in their functional decline. Age-dependent changes can be seen in a wide range of organisms, from the decrease in replicative potential observed in unicellular yeast1, to the reduced performance of vital organs in more complex organisms such as humans. The rate by which cells and organisms age varies widely and genetic and environmental factors have been shown to be involved in the age-dependent decline of cell and tissue function. Although ageing is a complex phenomenon, it is becoming clear that a cell’s failure to maintain proper protein homeostasis plays a major role in ageing and age-related disease2. Constant protein turnover is one of the major strategies utilized in maintaining this homeostasis, and has been the focus of much work. Recent ageing studies have now placed a new emphasis on literally old culprits: long-lived proteins, which evade turnover3, 4. In this Opinion article, we discuss the different contexts in which long-lived proteins have been characterized and the possible functional consequences of their persistence. We argue that these long-lived proteins play a larger role in organismal ageing than previously appreciated.
Exceptions to the rule
Proteins are constantly being degraded and subsequently replaced with newly synthesized copies. This turnover process ensures a constant supply of new and functional proteins, allowing non-functional, damaged, or even toxic species to be destroyed. The rate of turnover, however, can vary widely from protein to protein, with half-lives spanning orders of magnitude within the same cell. Studies in budding yeast (~1.5 hour cell cycle) have found the median and mean protein half-life under normal growth conditions to be ~43 minutes5. This figure increases to 0.5–35 hours in dividing mammalian cells (~24 hour cell cycle) and ~43 hours in non-dividing cells6, 7. Turnover studies in mice (lifespan ~1.5 years) found the average half-lives of proteins in the brain, liver, and blood to be between 3 and 9 days8. Although half-lives for different proteins in the cell may range from minutes to days, protein turnover rates often correlate with their function or subcellular localization. For example, proteins within the mitochondria and endoplasmic reticulum on average have longer half-lives than other proteins8. Large complexes, such as ribosomes and proteasomes, also have highly similar rates of turnover for each of their components6, 8.
Of all the studies on protein turnover to date, most have concentrated on turnover during relatively short timescales (i.e. significantly shorter than cellular and organismal life span), leaving few studies that focus on proteins with long half-lives. Although long-lived proteins have recently received more attention, the existence of a long-lived protein has been a well-established fact for several decades. As early as 1966, radioisotope pulse labeling (Box 1) was used to identify histones as having long half-lives9. Later studies from the 1970s also used radioisotopes to identify myelin and myelin proteolipid protein as long-lived10, 11. Also in the 1970s, an alternative technique, L- D-aspartic acid racemization (Box 1), was used to monitor protein turnover, identifying several collagens, elastin, eye lens crystallins, tooth enamel, and tooth dentine to have half-lives on the order of years (table 1)12–17.
Box 1. Methods for identifying long-lived proteins.
L/D aspartic acid racemization. All living organisms exclusively incorporate the L-enantiomers of amino acids into their proteins. Through a process that is time and temperature dependent, however, these amino acids very slowly racemize to a mixture of L- and D-enantiomers. Therefore, with careful calibration, ratios of the abundance of L- to D-enantiomers can provide the age of a protein. Although the slow rate of racemization is more suited for determining the age of proteins which are thousands of years old, racemization of aspartic acid is fastest among the amino acids and has been used to determine the half-life of particularly long-lived proteins12, 13, 15–17. Radio isotope pulse-labeling. Incorporation, and subsequent persistence, of radio isotopes can be used to identify long-lived proteins. Here, a radio-labeled precursor, often leucine for proteins, is injected into an animal’s tissue of interest, or added to the media in cell culture. Injected animals are sacrificed, or cells harvested, at various timepoints, specific proteins extracted or immunoprecipitated, and radioactivity measured. Persistent radioactivity indicates a stable, or long-lived protein, and by taking multiple measurements post-injection, a half-life of the protein of interest can be determined3, 9–11, 18, 72–74, 77, 78.
Stable isotope pulse-chase mass spectrometry. Incorporation, and subsequent persistence, of the stable 15N isotope can also be used for long-lived protein identification. Here, 2 generations of rats are fed a diet where the sole nitrogen source is from 15N algae, resulting in a uniformly 15N-labeled animal19. The diet is then switched to a normal 14N diet for the chase period, and animals sacrificed at appropriate time points4. Tissues can then be harvested and digested followed by MS. The MS not only identifies each detectable protein, but can also determine relative ratios of the 15N versions to their 14N equivalents79. Any identified proteins with significant 15N after a long chase period would be considered to be long-lived.
Table 1.
Known long-lived proteins and molecules. Listed are all known long-lived proteins or molecules. Half-lives are listed when determined. Otherwise, the listed age is how old at least a subset of the molecule was confirmed to be (lifetime). Methods used to determine longevity are: L/D-aspartic acid racemization (L/D Race), radio isotope pulse labeling (RI-PL), and stable isotope pulse/chase labeling analyzed by MS (SI-PC-MS).
| Molecule | Age | Measure | Organism | Method | Reference |
|---|---|---|---|---|---|
| Eye lens crystallin | >70 years | Lifetime | Human | L/D Race | 15 |
|
| |||||
| Collagen | 117 years | Halflife | Human | L/D Race | 12 |
|
| |||||
| Elastin | >78 years | Lifetime | Human | L/D Race | 14 |
|
| |||||
| Enamel/Dentine | >70 years | Lifetime | Human | L/D Race | 16, 17 |
|
| |||||
| Histones | 223 days | Halflife | Mouse | RI-PL | 77 |
| 117 days | Halflife | Mouse | RI-PL | 9 | |
| 218 days | Halflife | Rat | RI-PL | 78 | |
|
| |||||
| Nuclear Pore proteins | >1 month | Lifetime | Worms | RI-PL | 3 |
| > 1 year | Lifetime | Rat | SI-PC-MS | 4 | |
|
| |||||
| Myelin | 95 days | Halflife | Rat | RI-PL | 11 |
| >100 days | Halflife | Mouse | RI-PL | 10 | |
|
| |||||
| Myelin Proteolipid protein | >100 days | Halflife | Mouse | RI-PL | 10 |
|
| |||||
| Rec8 cohesion | >weeks | Lifetime | Mouse | RI-PL | 18 |
|
| |||||
| mRNA | indefinite? | Lifetime | Plant Seed | N/D | 71 |
| >2 years | Halflife | Frog Oocytes | RI-PL | 72 | |
|
| |||||
| Cholesterol | >18 months | Lifetime | Rabbit | RI-PL | 74 |
|
| |||||
| Phospholipids | >192 days | Lifetime | Rabbit | RI-PL | 73 |
Recent advancements in pulse-chase labeling provide strong evidence that more long-lived proteins remain to be discovered. In two recent studies, it was found that nuclear pore complex (NPC) proteins and a major component of the DNA cohesin complex were long-lived3, 18. While previous studies that identified these long-lived proteins necessarily relied on a priori knowledge that they may indeed be long-lived, recent advances in high-resolution mass spectrometry (MS) coupled with stable isotopic pulse-chase labeling of whole animals (Box 1)19, has allowed the non-biased identification of long-lived proteins. These studies confirmed the longevity of the myelin and histone proteins, and extended the known lifespan of NPC proteins to over 1 year4. The surprising message from all these studies is that a small but critical part of the cellular proteome is as old as the host cells, and they may play a major role in the age-dependent functional decline of their respective tissues.
Long-lived proteins and ageing
To exemplify how a lack of protein turnover can contribute to the ageing process, we will use an analogy of a modern car. The cars we drive today are complex machines made up of many components. Parts of the car that are under constant use wear down over time. This often is not a problem, as these parts such as tires and filters are replaced, extending the life of the machine. The engine and chassis, however, are large and expensive, and typically are not replaced for the life of the car. They are long-lived components, and as they lose their function, so does the car.
Although biological systems such as cells are not mechanical devices but rather do operate on principles of statistical thermodynamics, the idea of continuous replacement of individual parts in order to maintain the entire system can be applied. We have already mentioned the process of protein turnover, in which polypeptides are continuously being degraded and replaced with newly synthesized copies, eliminating damage or toxic protein species. However, if a protein cannot be turned-over, its persistence may lead to damage that is not easily repaired, which in turn might adversely affect cell function (Fig 1.). To illustrate the potential impact that a long lifetime has on protein function, we will first discuss crystallin, a well-studied long-lived protein that is confined to eye lens cells and experiences well-characterized age-dependent changes and modifications but has physiological consequences that are isolated to the eye lens, and the extra-cellular matrix (ECM) proteins collagen and elastin. Second, we will discuss the NPC, a multiprotein assembly containing long-lived components that are not solely structural in nature, but also play an active role in the transport of molecules into and out of all nuclei. These are just a few examples of long-lived proteins with important functions; as these long-lived proteins age their functions decline, which may play a major role in the ageing of their respective cells and tissues.
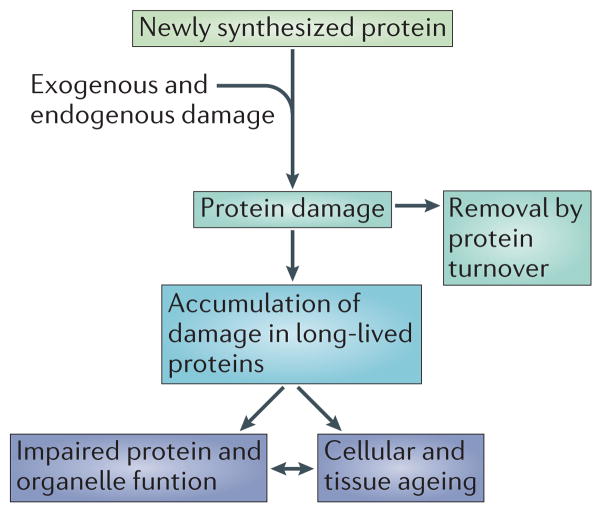
Figure 1.
Long-lived proteins and the accumulation of damage. Most proteins participate in a constant cycle of synthesis and degradation. Some proteins, however, evade degradation and are long-lived. Due to their longevity, these long-lived proteins are more prone to the accumulation of damage, which can lead to impaired protein function and cellular ageing.
Crystallin and lens structure
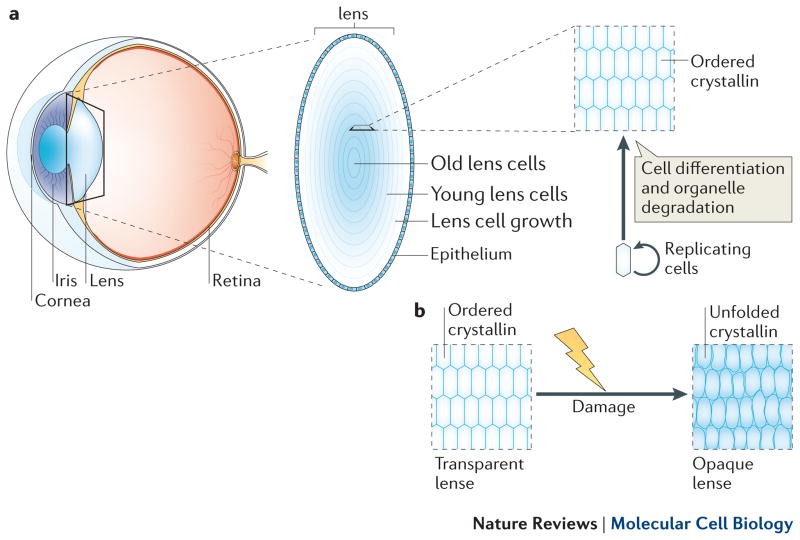
Crystallins from the eye lens were among the first long-lived proteins discovered, and represent perhaps the most complete picture, to date, linking protein longevity and adverse consequences15. The crystallin proteins, which are expressed as three different isoforms (alpha, beta and gamma), comprise over 90% of the total protein content in eye lens fibre cells, and greater than 35% of their wet weight20, 21. Their longevity is derived from the unique developmental programme that the lens fibre cells undergo during embryogenesis and adult life. The bulk of the lens is composed of non-dividing lens fibre cells that vary with age, with the oldest fibre cells in the centre ‘nucleus’, and cells of decreasing age with each layer outward (Fig. 2a)22. The outer most layer is composed of a replicating population of cells that gives rise to the underlying fibre cells. Upon differentiation into non-dividing fibre lens cells, their organelles, including the nucleus, are all degraded through regulated processes that utilize proteases and nucleases, leaving behind membrane-enclosed bags of crystallin protein23. This organelle evacuation is necessary to ensure the transparent quality of the lens, as organelles scatter light whereas ordered proteins (crystallins) do not24. This differentiation process occurs throughout adult life, producing rings of new fibre cells analogous to the rings of a tree. As the fibre cells have no organelles, protein synthesis is minimal or non-existent as is protein degradation, resulting in crystallin proteins, and perhaps others, that are synthesized at the birth of the cell persisting throughout the life of the organism.
Figure 2.
Eye lens structure. a) The position of the eye lens is depicted in a cross-section of the eye, as well as the general organization of lens structure (inset). The eye lens experiences radial growth, with the older lens fiber cells residing in the center and the younger cells on the outer rings. b) Lens cells through ageing. As the eye lens ages, long-lived crystallin molecules accumulate damage, leading to their aggregation into larger ordered structures which disrupts their transparency properties.
As lens fibre lack the ability to maintain protein homeostasis through protein turnover, other mechanisms must be in place to help preserve the proper folded state of the crystallins. For one, alpha-crystallin functions as a molecular chaperone, binding and maintaining the folded state of the beta and gamma crystallins,25, 26 the absence or mutations of which accelerates aggregation of lens proteins26, 27. A second mechanism is the use of reactive oxygen species scavengers in fibre cells. Compounds such as reduced glutathione (GSH) circulate from the outer lens epithelium throughout the lens interior, clearing oxygen radicals and other oxidizing compounds from fibre cells28, 29. These systems help prevent the misfolding, aggregation, and accumulation of damage on long-lived crystallins. Nonetheless, their activity does decrease with damage and age30–33.
These quality control mechanisms for crystallin proteins are limited, and over time cannot meet the demand of the increasingly stressed lens-cell proteome. Lens crystallins must cope with unique stresses such as ultraviolet radiation as well as common ones such as oxidative stresses. These insults result in several modifications of the lens proteins, including deamidation, glycation, mixed disulphides, and truncation34–39 (Box 2). In response to damage, the crystallin proteins unfold and begin to aggregate into large molecular weight species40, 41. These large aggregates are less transparent than the natively folded protein, resulting in opaque lenses and impaired vision, also known as cataracts (Fig 2b.). Although biological repair mechanisms become exhausted, modern medicine has devised a way to revive these ageing lenses using surgery to replace cataract lenses. Other long-lived proteins, however, may not have so elegant a solution.
Box 2. Protein Damage.
Proteins can accumulate diverse forms of damage and modifications, far too many to list here. Below are the major forms of damage mentioned in this review.
Deamidation: Deamidation is the nonenzymatic loss of amide groups on the side chains of asparagine and glutamine residues. This process is context dependent, modulated by surrounding amino acids as well as structure, and results in a negative charge in the side chain, which may affect the degree to which proteins are hydrolyzed80. Advanced Glycation End (AGE) products: A diverse group of modifications, AGEs are the conjugation of carbohydrates or carbohydrate fragments onto proteins. This modification can result in crosslinking of different peptides, and include glyoxal, methylglyoxal, and carboxymethyl-lysine among many others81. Levels of AGE can be increased in aged tissues such as the aged myocardium82.
Mixed disulfides: As the burden of oxidative stress increases, aberrant disulfide crosslinks can be made between protein cysteins and GSH, or to other cysteins forming intra- or intermolecular crosslinks83, 84. These crosslinks can result in large protein species often found in aggregates84.
Truncation: Proteins can experience truncations, where amino acids are lost from the Nor C- termini85, or cleaved at internal residues86. Truncation may be causes or products of other types of damage.
Protein aggregation: Protein aggregation is formation of non-native high molecular structures. These aggregates typically incapacitate the aggregated protein, and may be the result of other modifications such as deamidation, glycation and mixed disulfides. Aggregates are also a hallmark of a number of neurodegenerative diseases, such as Huntington’s and Alzheimer’s diseases87.
Carbonylation: One of the most studied oxidative damage modifications, carbonylation is an irreversible addition of a carbonyl group, typically on lysine, argenine, or proline residues. This metal-catalized oxidation (MCO) modification is the result of reactive oxygen species (ROS) produced within the cell88.
Collagen and Elastin
Studies on the longevity of collagen and elastin have followed a similar trajectory as lens crystallins, and analogous to the inactive environment of lens fiber cells, collagens and elastin typically reside in extracellular regions, which are isolated from the cellular milieu. Collagens and elastins are aggregates of proteins that are synthesized by the cell as pre-pro-proteins, and subsequently cleaved and secreted into the ECM where they crosslink and form higher order structures. Although equipped with a robust network of proteases used for remodeling of the ECM, the ECM isn’t under the constant flux of protein turnover that intracellular proteins are subjected to42.
Like lens crystallins, collagen and elastin are also particularly prone to damage, presumably due to their longevity. These chemical modifications include glycation, crosslinking, and fragmentation of their large protein structures13, 43–45. As this damage accumulates, concomitant changes in the physical properties of collagen and elastin has been observed, such as increased rigidity, resistance to denaturation13, 44, 45, and impaired performance of the underlying tissue45–48. Although collagen and elastin were determined to be long-lived, unlike crystallins there is considerable evidence for their continued expression and degradation throughout the life of the host organism13. Thus, the exact contributions of the long-lived versus newly synthesized collagens and elastins to the ageing process is yet to be fully understood.
Long-lived proteins in metabolically active cells
The discovery that the NPC contained long-lived proteins was a surprise because, unlike the crystallins, it is an intracellular structure and is ubiquitously expressed. The NPC is a ~90MDa protein structure of eightfold symmetry that is composed of ~30 nucleoporins (Nups) in multiple copies49, 50. This complex is embedded in the nuclear envelope (NE) at sites where the inner nuclear membrane (INM) and outer nuclear membrane (ONM) are joined51. NPCs act as exclusive transport channels that mediate all nuclear trafficking and thus maintain proper nuclear/cytoplasmic compartmentalization52. Indeed, the NPC represents one of the most active transport channels as each NPC can mediate cargo transport up to 1,000 times per second53. With thousands of nuclear pores per nucleus54, the NPCs of a given cell are responsible for correctly allowing passage of millions of transport cargos while excluding all other complexes. Breakdown of this essential gate would result in loss of proper nuclear/cytoplasmic compartmentalization, and consequently, all cellular regulation that relies on this sequestration.
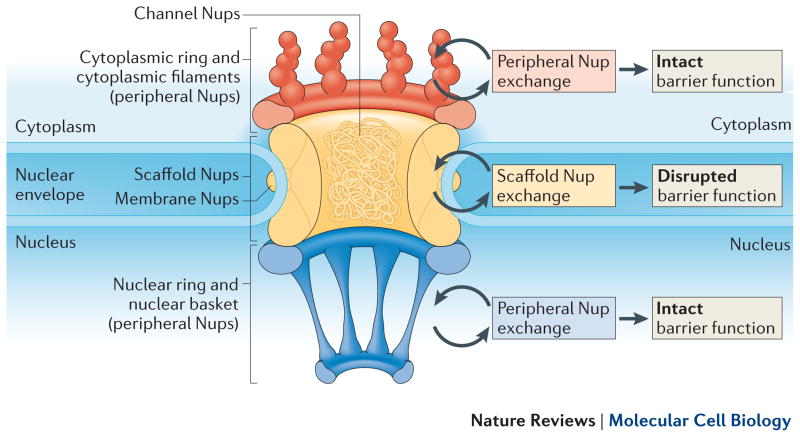
Similarly to other large protein assemblies, the NPC is composed of several subcomplexes, each of which contribute specific functions and biophysical properties to the pore (Fig. 3). First are at least three transmembrane Nups which are thought to anchor the NPC to the membrane55–57. Lining the inside of the pore are the scaffold Nups, made up of the multi-component Nup107/160 and Nup205 complexes58, 59. These scaffold complexes are thought to provide a structural foundation to which several other Nup complexes are bound60. Attached to the scaffold in the center of the pore are the FG-Nups, which contain phenylalanine/glycine rich domains61 and have been shown to establish the nuclear permeability barrier, allowing signal-dependent the passage of cargo62, 63. Finally, are the peripheral Nups, which extend into the cytoplasmic and nuclear space and form the cytoplasmic filaments and nuclear basket, respectively. These peripheral complexes aid in transport and transport directionality, providing docking sites for cargo and other transport molecules49, 52.
Figure 3.
Schematic of the nuclear pore complex. The NPC is embedded in a double membrane structure separating the nucleus and cytoplasm. It is composed of several subcomplexes, including the peripheral, membrane, channel FG, and scaffold Nups. Whereas the peripheral, channel, and membrane Nups turnover continuously, the scaffold Nups are stable. We propose this stability is necessary and that rapid exchange or loss of these components would result in a disrupted permeability barrier. Thus, maintenance or turnover of these scaffold complexes is necessarily slow, if at all.
The first evidence that NPC proteins are long-lived came from a study of C. elegans NPCs, where it was discovered that expression of scaffold Nups, but not peripheral Nups, was almost absent by adulthood, although the respective proteins were still present3. This suggested that whereas the peripheral Nups are continuously turned over, the scaffold of the NPC is built during embryogenesis and lasts the lifetime of the worm, which is on the order of weeks. These results complemented other work demonstrating that scaffold NPC proteins could persist on the nuclear pore without exchange for hours64. This was in contrast to peripheral Nups, which resided on the pore for mere seconds, representing a 4-orders-of-magnitude difference in residence time of different components of the same complex.
A more recent study used stable isotopic pulse-chase labeling analyzed by MS (Box 1) to determine NPC longevity in the rat, which has a more complex physiology and longer lifespan, on the order of 2 years4. In this study, rats were sacrificed at 6 and 12 months of chase, and nuclei from liver and brain tissue were analyzed by MS. Data for every NPC protein was obtained from the nuclei of brain and liver tissues, and indeed found that only scaffold NPC proteins were still labeled with the pulse isotope after 6 and 12 months, specifically in the brain and not the liver. This indicated that these scaffold Nups were in excess of 1 year old, far exceeding the normal life-span of a protein and on par with some of the longest-lived proteins known to date.
Both the worm and rat studies similarly found that only the scaffold Nup107/160 and Nup205 complexes, and not the peripheral, membrane, or FG Nups, were long-lived3, 4. Thus, unlike large protein complexes such as the ribosome or proteasome, it appears that the NPC does not turnover as a single unit, but instead individual sub-complexes have different rates of turnover, and some exhibit no or very little turnover.
The persistence of these long-lived NPC components does not seem to occur without consequences, and we now know some of the ramifications of their longevity. Nuclei isolated from older worms and rats were both found to have reduced levels of scaffold Nups as well as an increase in oxidative damage compared with younger nuclei3. In both cases, this resulted in a compromised nuclear permeability barrier and the surprising discovery of tubulin aggregates inside nuclei from old rat brains. The presence of nuclear aggregates in old brain nuclei is significant, as these types of aggregates have been linked to several neurodegenerative diseases65.
Why might a dynamic protein complex contain long-lived components, particularly when there are known ramifications? Perhaps this due to the NPC being a membrane-embedded multicomplex structure that needs to maintain a selective permeability barrier at all times. Many proteins function as part of large multi-component complexes, which need to be disassembled first before being turned over. This disassembly results in the loss of function of that complex, which is often of no consequence as replacement complexes are already present. However, what would happen if loss of function of just a few complexes had major consequences? This may be the case for NPCs, as loss of a few nuclear pores can result in a loss of cytoplasmic/nuclear compartmentalization66. Thus, the turnover of an NPC would strike a delicate balance between removing and replacing components, while maintaining enough nuclear pore integrity to maintain the permeability barrier. As peripheral Nups come on and off the NPC at high rates and may be present in excess, they can be replaced with little difficulty64. The same cannot be said of the scaffold Nups, which are responsible for the structural integrity of the NPC, and act as points to which peripheral and central channel Nups are bound. Thus, removing large portions of the scaffold might not only destabilize the NPC, but would also necessarily displace any bound peripheral and channel Nups. Loss of scaffold Nups such as Nup93 results in a loss of permeability barrier, so continuous exchange of these core Nups while also ensuring pore integrity and proper transport functions may not be possible3 (Fig 3). This could be thought of as similar to changing parts of a car’s engine while the car is running.
For dividing cells, there are times when the NPC is ‘turned off’ and therefore a window of opportunity for NPC turnover exists. During mitosis in dividing cells, the NE breaks down (NEBD) resulting in NPC disassembly and dispersion into the cytosol and ER67. This might present the only time for scaffold Nups to be replaced with newly synthesized copies as cytoplasmic/nuclear compartmentalization is not present or needed. Once mitosis has completed and Nups have had an opportunity to turnover, the NE then reassembles around chromatin along with NPCs, and the permeability barrier is soon restored68. If a cell has exited the cell cycle and no longer divides (for example, neurons), or if there is no NEBD in mitosis (for example, in budding yeast), there may be no easy way to replace potentially damaged NPCs. Thus, other novel ways of turnover or maintenance must be used to combat the accumulation of damage in Nups, but ultimately this may have negative consequences for ageing NPCs.
How might the negative consequences of NPC ageing in mammalian non-dividing cells be attenuated? Careful inspection of the rat NPC turnover data might yield some clues. In this study, there were long-lived 15N-labeled scaffold Nups even after a 1 year chase, but over 50% of peptides from the respective proteins were 14N4. One option might be through limited cell growth or tissue turnover in the chase period, as these 15N peptides were found in the brain which contain a complex mixture of glial and neuronal cells. Another possibility would be insertion of new NPCs that would dilute out the old pulse labeled peptides. Although in interphase NPCs can indeed be inserted into an intact NE69, such a process would have to be coupled to whole NPC removal in post-mitotic cells to avoid the uncontrolled increase of nuclear pores. Finally, the most likely scenario is that there is minimal replacement of individual subcomplexes, while largely maintaining overall NPC structure and function. Although residence times are long for scaffold Nups, they are not infinite64, and small and infrequent temporary loss of some scaffold Nups may be tolerated. This slow exchange process would therefore help prevent accumulation of damaged scaffold Nups through a normal, although slow, protein turnover mechanism. Irrespective of which scenario will turn out to be correct, there is still a significant population of NPC components that are >1 year old. This makes NPC components some of the oldest polypeptides that are part of an active protein machine. The presence of these long-lived proteins in non-dividing cells such as neurons might present a functional problem as unlike eye lenses, neurons are almost irreplaceable, and degeneration of their function may result in their permanent loss.
Other long-lived molecules
Long-lived molecules are not limited to proteins; they appear in several other biological processes. The cell’s genome is replicated in a semi-conservative manner before mitosis, and as a result, is never replaced once the cell stops dividing. Thus, DNA in neurons is as old as the organism itself, which has been confirmed through 14C carbon dating70. Other nucleic acids can be long-lived; studies in plant seeds found pools of mRNAs that lay dormant, for perhaps years, until germination is initiated71, and RNA pools in Xenopus laevis oocytes were found to have half-lives of over 2 years72. Finally, early isotopic studies also found that cholesterol and lipids associated with the myelin sheath were long-lived73, 74. This is consistent with the observation that two other components of the sheath, myelin basic protein and proteolipid protein, are also long-lived. The longevity of these non-protein molecules emphasizes that a lack of turnover may not be a problem limited to the aging proteome, but may impact the age-dependent accumulation of damage on other components of the cell.
Concluding thoughts
Our appreciation that a subset of nuclear proteins is long-lived and therefore evades protein turnover has important implications for cellular and organismal aging as was discussed for eye lens crystallin and nuclear pore proteins. Might long-lived histones be responsible for the observed loss of youthful gene expression programs in post-mitotic tissues75? Does Rec8 longevity contribute to age-related increases in meiosis errors as was speculated by the authors18? The perhaps most important question at this point is, whether additional long-lived proteins exist that remain to be discovered in the brain, or other tissues with limited turnover such as the heart and ovaries76? It is important to address these questions, since with perhaps limited means to repair age-associated damage, long-lived proteins may prove to be the Achilles heel of the ageing proteome.
Acknowledgments
The authors thank members of the Hetzer laboratory and E.Q. Toyama for helpful suggestions and critical reading of the manuscript. B.H.T is supported by the Hewitt Foundation; M.W.H by the Ellison Medical Foundation, by the US National Institutes of Health (NIH) (R01GM098749) and the National Cancer Institute (award number P30CA014195).
References
- 1.Steinkraus KA, Kaeberlein M, Kennedy BK. Replicative aging in yeast: the means to the end. Annu Rev Cell Dev Biol. 2008;24:29–54. doi: 10.1146/annurev.cellbio.23.090506.123509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Taylor RC, Dillin A. Aging as an event of proteostasis collapse. Cold Spring Harb Perspect Biol. 2011;3 doi: 10.1101/cshperspect.a004440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.D’Angelo MA, Raices M, Panowski SH, Hetzer MW. Age-dependent deterioration of nuclear pore complexes causes a loss of nuclear integrity in postmitotic cells. Cell. 2009;136:284–95. doi: 10.1016/j.cell.2008.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Savas JN, Toyama BH, Xu T, Yates JR, 3rd, Hetzer MW. Extremely long-lived nuclear pore proteins in the rat brain. Science. 2012;335:942. doi: 10.1126/science.1217421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Belle A, Tanay A, Bitincka L, Shamir R, O’Shea EK. Quantification of protein half-lives in the budding yeast proteome. Proc Natl Acad Sci U S A. 2006;103:13004–9. doi: 10.1073/pnas.0605420103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cambridge SB, et al. Systems-wide proteomic analysis in mammalian cells reveals conserved, functional protein turnover. J Proteome Res. 2011;10:5275–84. doi: 10.1021/pr101183k. [DOI] [PubMed] [Google Scholar]
- 7.Yen HC, Xu Q, Chou DM, Zhao Z, Elledge SJ. Global protein stability profiling in mammalian cells. Science. 2008;322:918–23. doi: 10.1126/science.1160489. [DOI] [PubMed] [Google Scholar]
- 8.Price JC, Guan S, Burlingame A, Prusiner SB, Ghaemmaghami S. Analysis of proteome dynamics in the mouse brain. Proc Natl Acad Sci U S A. 2010;107:14508–13. doi: 10.1073/pnas.1006551107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Piha RS, Cuenod M, Waelsch H. Metabolism of histones of brain and liver. J Biol Chem. 1966;241:2397–404. [PubMed] [Google Scholar]
- 10.Fischer CA, Morell P. Turnover of proteins in myelin and myelin-like material of mouse brain. Brain Res. 1974;74:51–65. doi: 10.1016/0006-8993(74)90111-5. [DOI] [PubMed] [Google Scholar]
- 11.Rodrijguez de Lores A, Alberici de Canal M, De Robertis E. Turnover of proteins in subcellular fractions of rat cerebral cortex. Brain Res. 1971;31:179–84. doi: 10.1016/0006-8993(71)90642-1. [DOI] [PubMed] [Google Scholar]
- 12.Verzijl N, et al. Effect of collagen turnover on the accumulation of advanced glycation end products. J Biol Chem. 2000;275:39027–31. doi: 10.1074/jbc.M006700200. [DOI] [PubMed] [Google Scholar]
- 13.Sell D, Monnier V. Comprehensive Physiology. 235–305. American Physiological Society; 2011. [Google Scholar]
- 14.Shapiro SD, Endicott SK, Province MA, Pierce JA, Campbell EJ. Marked longevity of human lung parenchymal elastic fibers deduced from prevalence of D-aspartate and nuclear weapons-related radiocarbon. J Clin Invest. 1991;87:1828–34. doi: 10.1172/JCI115204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Masters PM, Bada JL, Zigler JS., Jr Aspartic acid racemisation in the human lens during ageing and in cataract formation. Nature. 1977;268:71–3. doi: 10.1038/268071a0. [DOI] [PubMed] [Google Scholar]
- 16.Helfman PM, Bada JL. Aspartic acid racemization in tooth enamel from living humans. Proc Natl Acad Sci U S A. 1975;72:2891–4. doi: 10.1073/pnas.72.8.2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Helfman PM, Bada JL. Aspartic acid racemisation in dentine as a measure of ageing. Nature. 1976;262:279–81. doi: 10.1038/262279b0. [DOI] [PubMed] [Google Scholar]
- 18.Tachibana-Konwalski K, et al. Rec8-containing cohesin maintains bivalents without turnover during the growing phase of mouse oocytes. Genes Dev. 2010;24:2505–16. doi: 10.1101/gad.605910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McClatchy DB, Dong MQ, Wu CC, Venable JD, Yates JR., 3rd 15N metabolic labeling of mammalian tissue with slow protein turnover. J Proteome Res. 2007;6:2005–10. doi: 10.1021/pr060599n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bloemendal H. The vertebrate eye lens. Science. 1977;197:127–38. doi: 10.1126/science.877544. [DOI] [PubMed] [Google Scholar]
- 21.Graw J. Genetics of crystallins: cataract and beyond. Exp Eye Res. 2009;88:173–89. doi: 10.1016/j.exer.2008.10.011. [DOI] [PubMed] [Google Scholar]
- 22.Wride MA. Lens fibre cell differentiation and organelle loss: many paths lead to clarity. Philos Trans R Soc Lond B Biol Sci. 2011;366:1219–33. doi: 10.1098/rstb.2010.0324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bassnett S. Lens organelle degradation. Exp Eye Res. 2002;74:1–6. doi: 10.1006/exer.2001.1111. [DOI] [PubMed] [Google Scholar]
- 24.Bassnett S, Shi Y, Vrensen GF. Biological glass: structural determinants of eye lens transparency. Philos Trans R Soc Lond B Biol Sci. 2011;366:1250–64. doi: 10.1098/rstb.2010.0302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Horwitz J. Alpha-crystallin can function as a molecular chaperone. Proc Natl Acad Sci U S A. 1992;89:10449–53. doi: 10.1073/pnas.89.21.10449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rao PV, Huang QL, Horwitz J, Zigler JS., Jr Evidence that alpha-crystallin prevents non-specific protein aggregation in the intact eye lens. Biochim Biophys Acta. 1995;1245:439–47. doi: 10.1016/0304-4165(95)00125-5. [DOI] [PubMed] [Google Scholar]
- 27.Xi JH, et al. Mechanism of small heat shock protein function in vivo: a knock-in mouse model demonstrates that the R49C mutation in alpha A-crystallin enhances protein insolubility and cell death. J Biol Chem. 2008;283:5801–14. doi: 10.1074/jbc.M708704200. [DOI] [PubMed] [Google Scholar]
- 28.Giblin FJ. Glutathione: a vital lens antioxidant. J Ocul Pharmacol Ther. 2000;16:121–35. doi: 10.1089/jop.2000.16.121. [DOI] [PubMed] [Google Scholar]
- 29.Lou MF. Redox regulation in the lens. Prog Retin Eye Res. 2003;22:657–82. doi: 10.1016/s1350-9462(03)00050-8. [DOI] [PubMed] [Google Scholar]
- 30.Xing KY, Lou MF. Effect of age on the thioltransferase (glutaredoxin) and thioredoxin systems in the human lens. Invest Ophthalmol Vis Sci. 2010;51:6598–604. doi: 10.1167/iovs.10-5672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sharma KK, Ortwerth BJ. Effect of cross-linking on the chaperone-like function of alpha crystallin. Exp Eye Res. 1995;61:413–21. doi: 10.1016/s0014-4835(05)80136-8. [DOI] [PubMed] [Google Scholar]
- 32.Gupta R, Srivastava OP. Deamidation affects structural and functional properties of human alphaA-crystallin and its oligomerization with alphaB-crystallin. J Biol Chem. 2004;279:44258–69. doi: 10.1074/jbc.M405648200. [DOI] [PubMed] [Google Scholar]
- 33.Harding JJ. Free and protein-bound glutathione in normal and cataractous human lenses. Biochem J. 1970;117:957–60. doi: 10.1042/bj1170957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sharma KK, Santhoshkumar P. Lens aging: effects of crystallins. Biochim Biophys Acta. 2009;1790:1095–108. doi: 10.1016/j.bbagen.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hanson SR, Hasan A, Smith DL, Smith JB. The major in vivo modifications of the human water-insoluble lens crystallins are disulfide bonds, deamidation, methionine oxidation and backbone cleavage. Exp Eye Res. 2000;71:195–207. doi: 10.1006/exer.2000.0868. [DOI] [PubMed] [Google Scholar]
- 36.Ortwerth BJ, Olesen PR. Studies on the solubilization of the water-insoluble fraction from human lens and cataract. Exp Eye Res. 1992;55:777–83. doi: 10.1016/0014-4835(92)90004-c. [DOI] [PubMed] [Google Scholar]
- 37.Ahmed N, et al. Methylglyoxal-derived hydroimidazolone advanced glycation end-products of human lens proteins. Invest Ophthalmol Vis Sci. 2003;44:5287–92. doi: 10.1167/iovs.03-0573. [DOI] [PubMed] [Google Scholar]
- 38.Takemoto LJ. Quantitation of asparagine-101 deamidation from alpha-A crystallin during aging of the human lens. Curr Eye Res. 1998;17:247–50. doi: 10.1076/ceyr.17.3.247.5218. [DOI] [PubMed] [Google Scholar]
- 39.Wilmarth PA, et al. Age-related changes in human crystallins determined from comparative analysis of post-translational modifications in young and aged lens: does deamidation contribute to crystallin insolubility? J Proteome Res. 2006;5:2554–66. doi: 10.1021/pr050473a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bloemendal H, et al. Ageing and vision: structure, stability and function of lens crystallins. Prog Biophys Mol Biol. 2004;86:407–85. doi: 10.1016/j.pbiomolbio.2003.11.012. [DOI] [PubMed] [Google Scholar]
- 41.Roy D, Spector A. Absence of low-molecular-weight alpha crystallin in nuclear region of old human lenses. Proc Natl Acad Sci U S A. 1976;73:3484–7. doi: 10.1073/pnas.73.10.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kerrigan JJ, Mansell JP, Sandy JR. Matrix turnover. J Orthod. 2000;27:227–33. doi: 10.1179/ortho.27.3.227. [DOI] [PubMed] [Google Scholar]
- 43.Braverman IM, Fonferko E. Studies in cutaneous aging: I. The elastic fiber network. J Invest Dermatol. 1982;78:434–43. doi: 10.1111/1523-1747.ep12507866. [DOI] [PubMed] [Google Scholar]
- 44.Kragstrup TW, Kjaer M, Mackey AL. Structural, biochemical, cellular, and functional changes in skeletal muscle extracellular matrix with aging. Scand J Med Sci Sports. 2011;21:749–57. doi: 10.1111/j.1600-0838.2011.01377.x. [DOI] [PubMed] [Google Scholar]
- 45.Sell DR, Monnier VM. Molecular basis of arterial stiffening: role of glycation - a mini-review. Gerontology. 2012;58:227–37. doi: 10.1159/000334668. [DOI] [PubMed] [Google Scholar]
- 46.Fonck E, et al. Effect of aging on elastin functionality in human cerebral arteries. Stroke. 2009;40:2552–6. doi: 10.1161/STROKEAHA.108.528091. [DOI] [PubMed] [Google Scholar]
- 47.Gosselin LE, Adams C, Cotter TA, McCormick RJ, Thomas DP. Effect of exercise training on passive stiffness in locomotor skeletal muscle: role of extracellular matrix. J Appl Physiol. 1998;85:1011–6. doi: 10.1152/jappl.1998.85.3.1011. [DOI] [PubMed] [Google Scholar]
- 48.Haus JM, Carrithers JA, Trappe SW, Trappe TA. Collagen, cross-linking, and advanced glycation end products in aging human skeletal muscle. J Appl Physiol. 2007;103:2068–76. doi: 10.1152/japplphysiol.00670.2007. [DOI] [PubMed] [Google Scholar]
- 49.Hoelz A, Debler EW, Blobel G. The structure of the nuclear pore complex. Annu Rev Biochem. 2011;80:613–43. doi: 10.1146/annurev-biochem-060109-151030. [DOI] [PubMed] [Google Scholar]
- 50.Cronshaw JM, Krutchinsky AN, Zhang W, Chait BT, Matunis MJ. Proteomic analysis of the mammalian nuclear pore complex. J Cell Biol. 2002;158:915–27. doi: 10.1083/jcb.200206106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hetzer MW, Wente SR. Border control at the nucleus: biogenesis and organization of the nuclear membrane and pore complexes. Dev Cell. 2009;17:606–16. doi: 10.1016/j.devcel.2009.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wente SR, Rout MP. The nuclear pore complex and nuclear transport. Cold Spring Harb Perspect Biol. 2010;2:a000562. doi: 10.1101/cshperspect.a000562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ribbeck K, Gorlich D. Kinetic analysis of translocation through nuclear pore complexes. EMBO J. 2001;20:1320–30. doi: 10.1093/emboj/20.6.1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Garcia-Segura LM, Lafarga M, Berciano MT, Hernandez P, Andres MA. Distribution of nuclear pores and chromatin organization in neurons and glial cells of the rat cerebellar cortex. J Comp Neurol. 1989;290:440–50. doi: 10.1002/cne.902900311. [DOI] [PubMed] [Google Scholar]
- 55.Stavru F, et al. NDC1: a crucial membrane-integral nucleoporin of metazoan nuclear pore complexes. J Cell Biol. 2006;173:509–19. doi: 10.1083/jcb.200601001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hallberg E, Wozniak RW, Blobel G. An integral membrane protein of the pore membrane domain of the nuclear envelope contains a nucleoporin-like region. J Cell Biol. 1993;122:513–21. doi: 10.1083/jcb.122.3.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Greber UF, Senior A, Gerace L. A major glycoprotein of the nuclear pore complex is a membrane-spanning polypeptide with a large lumenal domain and a small cytoplasmic tail. EMBO J. 1990;9:1495–502. doi: 10.1002/j.1460-2075.1990.tb08267.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Siniossoglou S, et al. A novel complex of nucleoporins, which includes Sec13p and a Sec13p homolog, is essential for normal nuclear pores. Cell. 1996;84:265–75. doi: 10.1016/s0092-8674(00)80981-2. [DOI] [PubMed] [Google Scholar]
- 59.Grandi P, et al. Nup93, a vertebrate homologue of yeast Nic96p, forms a complex with a novel 205-kDa protein and is required for correct nuclear pore assembly. Mol Biol Cell. 1997;8:2017–38. doi: 10.1091/mbc.8.10.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Alber F, et al. The molecular architecture of the nuclear pore complex. Nature. 2007;450:695–701. doi: 10.1038/nature06405. [DOI] [PubMed] [Google Scholar]
- 61.Hu T, Guan T, Gerace L. Molecular and functional characterization of the p62 complex, an assembly of nuclear pore complex glycoproteins. J Cell Biol. 1996;134:589–601. doi: 10.1083/jcb.134.3.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Frey S, Richter RP, Gorlich D. FG-rich repeats of nuclear pore proteins form a three-dimensional meshwork with hydrogel-like properties. Science. 2006;314:815–7. doi: 10.1126/science.1132516. [DOI] [PubMed] [Google Scholar]
- 63.Hulsmann BB, Labokha AA, Gorlich D. The permeability of reconstituted nuclear pores provides direct evidence for the selective phase model. Cell. 2012;150:738–51. doi: 10.1016/j.cell.2012.07.019. [DOI] [PubMed] [Google Scholar]
- 64.Rabut G, Doye V, Ellenberg J. Mapping the dynamic organization of the nuclear pore complex inside single living cells. Nat Cell Biol. 2004;6:1114–21. doi: 10.1038/ncb1184. [DOI] [PubMed] [Google Scholar]
- 65.Woulfe JM. Abnormalities of the nucleus and nuclear inclusions in neurodegenerative disease: a work in progress. Neuropathol Appl Neurobiol. 2007;33:2–42. doi: 10.1111/j.1365-2990.2006.00819.x. [DOI] [PubMed] [Google Scholar]
- 66.Lenart P, et al. Nuclear envelope breakdown in starfish oocytes proceeds by partial NPC disassembly followed by a rapidly spreading fenestration of nuclear membranes. J Cell Biol. 2003;160:1055–68. doi: 10.1083/jcb.200211076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Burke B, Ellenberg J. Remodelling the walls of the nucleus. Nat Rev Mol Cell Biol. 2002;3:487–97. doi: 10.1038/nrm860. [DOI] [PubMed] [Google Scholar]
- 68.Anderson DJ, Hetzer MW. Shaping the endoplasmic reticulum into the nuclear envelope. J Cell Sci. 2008;121:137–42. doi: 10.1242/jcs.005777. [DOI] [PubMed] [Google Scholar]
- 69.Doucet CM, Talamas JA, Hetzer MW. Cell cycle-dependent differences in nuclear pore complex assembly in metazoa. Cell. 2010;141:1030–41. doi: 10.1016/j.cell.2010.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Spalding KL, Bhardwaj RD, Buchholz BA, Druid H, Frisen J. Retrospective birth dating of cells in humans. Cell. 2005;122:133–43. doi: 10.1016/j.cell.2005.04.028. [DOI] [PubMed] [Google Scholar]
- 71.Dure L, Waters L. Long-Lived Messenger Rna: Evidence from Cotton Seed Germination. Science. 1965;147:410–2. doi: 10.1126/science.147.3656.410. [DOI] [PubMed] [Google Scholar]
- 72.Ford PJ, Mathieson T, Rosbash M. Very long-lived messenger RNA in ovaries of Xenopus laevis. Dev Biol. 1977;57:417–26. doi: 10.1016/0012-1606(77)90226-3. [DOI] [PubMed] [Google Scholar]
- 73.Davison AN, Morgan RS, Wajda M, Wright GP. Metabolism of myelin lipids: incorporation of [3-14c]serine in brain lipids of the developing rabbit and their persistence in the central nervous system. J Neurochem. 1959;4:360–365. [Google Scholar]
- 74.Davison AN, Wajda M. Metabolism of myelin lipids: estimation and separation of brain lipids in the developing rabbit. J Neurochem. 1959;4:353–9. doi: 10.1111/j.1471-4159.1959.tb13217.x. [DOI] [PubMed] [Google Scholar]
- 75.Feser J, Tyler J. Chromatin structure as a mediator of aging. FEBS Lett. 2011;585:2041–8. doi: 10.1016/j.febslet.2010.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bergmann O, et al. Evidence for cardiomyocyte renewal in humans. Science. 2009;324:98–102. doi: 10.1126/science.1164680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Commerford SL, Carsten AL, Cronkite EP. Histone turnover within nonproliferating cells. Proc Natl Acad Sci U S A. 1982;79:1163–5. doi: 10.1073/pnas.79.4.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Duerre JA, Lee CT. In vivo methylation and turnover of rat brain histones. J Neurochem. 1974;23:541–7. doi: 10.1111/j.1471-4159.1974.tb06057.x. [DOI] [PubMed] [Google Scholar]
- 79.Park SK, Venable JD, Xu T, Yates JR., 3rd A quantitative analysis software tool for mass spectrometry-based proteomics. Nat Methods. 2008;5:319–22. doi: 10.1038/nmeth.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Robinson NE, Robinson AB. Deamidation of human proteins. Proc Natl Acad Sci U S A. 2001;98:12409–13. doi: 10.1073/pnas.221463198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Semba RD, Nicklett EJ, Ferrucci L. Does accumulation of advanced glycation end products contribute to the aging phenotype? J Gerontol A Biol Sci Med Sci. 2010;65:963–75. doi: 10.1093/gerona/glq074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Schleicher ED, Wagner E, Nerlich AG. Increased accumulation of the glycoxidation product N(epsilon)-(carboxymethyl)lysine in human tissues in diabetes and aging. J Clin Invest. 1997;99:457–68. doi: 10.1172/JCI119180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Liang JN, Pelletier MR. Destabilization of lens protein conformation by glutathione mixed disulfide. Exp Eye Res. 1988;47:17–25. doi: 10.1016/0014-4835(88)90020-6. [DOI] [PubMed] [Google Scholar]
- 84.Spector A, Roy D. Disulfide-linked high molecular weight protein associated with human cataract. Proc Natl Acad Sci U S A. 1978;75:3244–8. doi: 10.1073/pnas.75.7.3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Miesbauer LR, et al. Post-translational modifications of water-soluble human lens crystallins from young adults. J Biol Chem. 1994;269:12494–502. [PubMed] [Google Scholar]
- 86.Lund AL, Smith JB, Smith DL. Modifications of the water-insoluble human lens alpha-crystallins. Exp Eye Res. 1996;63:661–72. doi: 10.1006/exer.1996.0160. [DOI] [PubMed] [Google Scholar]
- 87.Morimoto RI. The heat shock response: systems biology of proteotoxic stress in aging and disease. Cold Spring Harb Symp Quant Biol. 2011;76:91–9. doi: 10.1101/sqb.2012.76.010637. [DOI] [PubMed] [Google Scholar]
- 88.Moller IM, Rogowska-Wrzesinska A, Rao RS. Protein carbonylation and metal-catalyzed protein oxidation in a cellular perspective. J Proteomics. 2011;74:2228–42. doi: 10.1016/j.jprot.2011.05.004. [DOI] [PubMed] [Google Scholar]