Abstract
Clinical isolates of Helicobacter pylori show marked diversity, which may derive from genomic changes that occur during the often lifelong association of the bacterium with its human host. We used the rhesus macaque model, together with DNA microarrays, to examine genomic changes in H. pylori that occur early during experimental infection. Microarray analysis showed that H. pylori recovered from challenged macaques had deleted babA, a member of a large family of paralogous outer membrane proteins (OMPs) that mediates attachment of H. pylori to the Lewis B blood group antigen on gastric epithelium. In some cases the babA gene was replaced by babB, an uncharacterized OMP that is closely related to babA. In other cases the babA gene was present but was not expressed because of alteration in dinucleotide CT repeats in the 5′ coding region. In either case, strains lacking babA did not adhere to Lewis B, which is expressed on macaque gastric epithelium. Absence of babA and duplication of babB was also seen in H. pylori isolates derived from human clinical samples, suggesting that this gene conversion event is not unique to experimentally infected rhesus monkeys. These results demonstrate in real time with a relevant animal model that H. pylori regulates OMP expression in vivo by using both antigenic variation and phase variation. We suggest that changes in babA and babB after experimental infection of macaques represent a dynamic response in the H. pylori outer membrane that facilitates adherence to the gastric epithelium and promotes chronic infection.
The gastric pathogen Helicobacter pylori shows marked genetic diversity that exceeds that seen with other bacterial species (1, 2). Comparison of two complete H. pylori genome sequences (3, 4) revealed that although most genes are highly conserved between the two strains (26695 and J99), often differing only in synonymous substitutions, ≈6-7% of the genes are present in one strain but absent in the other. Subsequent analysis of 15 strains by whole-genome DNA microarray analysis showed that fully 22% of genes are strain specific (5). H. pylori alleles at independent loci are rarely coinherited for long periods of time (linkage equilibrium), which is consistent with a panmictic population structure (2, 6). Like other panmictic bacterial species such as Neisseria gonorrhoeae and Bacillus subtilis, H. pylori is naturally competent for DNA transformation. These observations have led to the hypothesis that during chronic infection of an individual host H. pylori can diversify by mutation, excision, and acquisition of genetic material from other H. pylori strains, or even other species that transiently colonize the gastric environment (5).
This hypothesis has been supported by occasional opportunities in which H. pylori strains have been isolated from the same host several years apart (7). Recently, microarray analysis was used to examine paired isolates of H. pylori strain J99 obtained from the same host after a 6-year interval (8). Considerable diversity (3%) was found among loci from multiple J99 isolates, although much less so than among unrelated isolates (22%) studied in the same fashion (5). These studies are important because they confirm by example that H. pylori can undergo genetic flux within an individual host over time. However, whether the acquired loci were originally present in a subpopulation of J99 or, alternatively, were acquired by horizontal gene transfer, could not be determined. Furthermore, the timing of acquisition of genetic diversity could not be determined. Although it has generally been assumed that diversity develops incrementally over years during chronic infection of the host, the possibility of acute genetic changes as an organism adapts to a new host has not been studied.
The rhesus monkey (Macaca mulatta) model offers the opportunity to study experimentally the development of H. pylori genomic diversity in a relevant animal host. Rhesus macaques are commonly infected with H. pylori that is nearly identical to human isolates by 16S rRNA sequence analysis (9). Experimental inoculation of macaques with H. pylori results in persistent infection and a histologic gastritis that mimics that seen in humans (10, 11). Here, we describe the use of the rhesus model, together with DNA microarray analysis, to examine genomic changes in H. pylori that occur during experimental infection. The results show that transmission of H. pylori from one host to another selects changes in outer membrane protein (OMP) expression, which may represent a dynamic response in the H. pylori outer membrane that is designed to adhere maximally to gastric epithelium and promote chronic infection.
Methods
Bacterial Strains and Culture. We recently confirmed the observation (11) that H. pylori J166 is adapted to rhesus macaques, because in each of three animals it blocked by competition colonization with two other strains (10). Isolates demonstrated to be J166 by repetitive palindromic PCR were recovered from each monkey at 2, 4, 8, and 17 wk postinoculation (PI). Comparisons between the input J166 and these output isolates from each monkey served as the primary basis for this article. All strains were routinely cultivated on Brucella agar (Difco) containing 5% bovine calf serum (GIBCO/BRL) supplemented with TVPA (trimethoprim, 5 mg/liter; vancomycin, 10 mg/liter; polymyxin B, 2.5 units/liter; amphotericin B, 4 mg/liter, Sigma) and incubated at 37°C in 5% CO2. For isolation of RNA, bacteria were cultivated in Brucella broth containing 5% bovine calf serum with TVPA and incubated as above with rotation at 60 rpm.
Molecular Biology Procedures. Chromosomal DNA was prepared from plate-grown bacteria. PCRs were performed by using standard conditions, and products were visualized by agarose gel electrophoresis. Primers (Table 1, which is published as supporting information on the PNAS web site) were derived from published sequences of H. pylori J99 (3) or sequence analysis of H. pylori J166. DNA sequencing was performed on both strands of purified PCR fragments (QIAquick PCR Purification Kit, Qiagen, Valencia, CA) by using dye terminator sequencing chemistry. High-stringency Southern blots were performed by blotting 500 ng of genomic DNA restricted with HindIII or SspI (New England Biolabs) and probing with ≈400-bp fragments of babA (primers AF2, AR5) or babB (primers BF2, BR3) amplified from the input J166. Probes were labeled with horseradish peroxidase (ECL Direct DNA Labeling System, Amersham Pharmacia Biotech), and chemiluminescence was detected with ECL reagents (Amersham Pharmacia Biotech).
Whole-Genome DNA Microarray. Microarray design and hybridization conditions were as described (5). The array consists of 1,660 unique PCR products that represent the superset of ORFs from both published H. pylori genomes. Data points were excluded because of low signal or slide abnormalities, and only those genes for which three measurements were obtained are reported. The assay was found to be 96% sensitive and 98% specific for detecting the presence of a gene (5).
Lewisb (Leb) Adhesion Assay. Attachment of H. pylori strains to the Leb blood group antigen was measured in duplicate by using an ELISA as described (12). Briefly, Leb conjugated to human serum albumin (Isosep, Tullinge, Sweden) was immobilized on polystyrene 96-well microtiter plates (Nalge Nunc) and incubated with digoxigenin-labeled (Roche Molecular Biochemicals) H. pylori from 48-h plates (13). Bound bacteria were detected by using anti-digoxigenin antibody conjugated to horseradish peroxidase and ABTS solution (Roche Molecular Biochemicals). Extinction was quantified in a microplate reader (BioRad) by using dual wavelength (405/490 nm) and normalized to uncoated control wells.
Real-Time Quantitative RT-PCR. Total bacterial RNA was prepared from midlog phase liquid cultures by using Trizol (GIBCO/BRL). RNA was treated with DNase I (Roche Molecular Biochemicals), purified with RNeasy (Qiagen), and suspended in molecular biology grade water (BioWhittaker) at 20 ng/μl. Gene-specific oligonucleotide primers were designed for quantitative detection of mRNA from babA (AF1, AR3) and babB (BF1, BR2) by using OLIGO 6.0 software (Molecular Biology Insights, Cascade, CO) and known DNA sequence from H. pylori J166. All primer pairs had a calculated melting temperature of 68-70°C and amplified products of ≈250 bp.
Reverse transcriptase and amplification of cDNA from each gene were performed essentially as described (14), except that ×0.4 SYBR green (Molecular Probes) was used as the fluorophore and 5% DMSO (Sigma) was added to each reaction. To eliminate PCR carryover effects, dUTP was incorporated during PCR, and each reaction included 0.4 unit uracil-N-glycosylase (New England Biolabs), which is active during an initial 3 min at 50°C and hydrolyzes any UTP-containing PCR products. During the second phase of the reaction, RNA was reverse-transcribed to cDNA at 60°C for 30 min, which severely reduces uracil-DNA glycosylase activity. Two-step amplification was then performed in a BioRad iCycler for 45 cycles (95°C for 20 sec, 59.5°C for 1 min). Cycle threshold was defined as the crossover point of an arbitrary fluorescence level at 490 nm that was at least 10 SD above a baseline determined from cycles 2 to 10. The appropriate size of each PCR product was confirmed for each primer pair by agarose gel electrophoresis. Absence of contaminating DNA was examined for each sample by performing RT-PCRs and replacing Mn(OAc)2 with 2.4 mM MgCl2, in which Tth has DNA polymerase but not reverse-transcriptase activity. Copy number of mRNA for babA and babB was calculated based on standard curves by using serial dilutions of cloned babA and babB.
Real-Time Quantitative PCR. A modification of the real-time RT-PCR method was used for quantitation of DNA copy number of babA and babB. Because there was no reverse-transcriptase step, Mn(OAc)2 was replaced with MgCl2 in the reaction mixture. Copy number of babA and babB were calculated based on standard curves from cloned target DNA.
Immunodetection of Leb. Gastric biopsy sections (10 μm) were deparaffinized and treated with 3% hydrogen peroxide for 10 min, followed by antigen retrieval in citrate buffer, pH 6.0 (Lab Vision, Fremont, CA) at 97°C for 15 min. Slides were cooled to room temperature, washed in PBS containing 0.01% Triton X-100, and incubated with anti-Leb IgG mAb (LWB01, Lab Vision, undiluted) for 1 h at room temperature. Slides were washed again and incubated with biotinylated goat anti-mouse IgG (1:1,000, Vector Laboratories) overnight at 4°C. After washing, slides were incubated with avidin-biotin complex-peroxidase (ABC, Vector Laboratories) for 90 min. Diaminobenzidine (Vector Laboratories) was applied as a substrate. Slides were counterstained with hematoxylin before dehydration and mounting.
Results
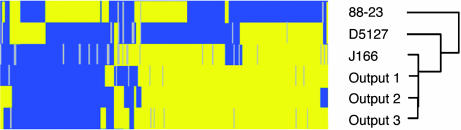
DNA Microarray Analysis. We previously cultured gastric biopsies at 2, 4, 8, and 17 wk PI from three monkeys inoculated with a mixture of H. pylori strains D5127, 88-23, and J166. Molecular fingerprinting with repetitive palindromic PCR showed that the predominant strain isolated from all three animals at each time point was J166 (10). To more closely compare the inoculated (J166input) and recovered (J166output) isolates, we used a whole-genome DNA microarray. The three input strains and three (presumptively) J166 output strains recovered from each animal 17 wk PI were studied. Cluster analysis showed that the three output strains were most closely related to J166, which confirmed the results obtained by repetitive palindromic PCR (Fig. 1). Interestingly, the gene for one of the H. pylori OMPs, babA, was present in J166input but absent in each of the J166output strains. Because babA is thought to be important in H. pylori pathogenesis, we examined this observation further.
Fig. 1.
Presence (blue) and absence (yellow) of genes in each of the H. pylori input strains (88-23, D5127, J166) and output strains obtained from each monkey (output 1, 2, 3) 17 wk PI. Data are shown only for the 193 genes that were either not present or not absent in all strains. Genes for which data were insufficient to make a determination of present or absent are shown in gray. Data were simplified into a binary score (present = 1, absent = 0), analyzed with xcluster software (http://genome-www.stanford.edu) and displayed with treeview (28). Each of the input strains was babA+ and babB+, but all J166output strains were babA-.
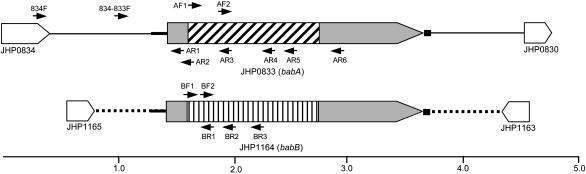
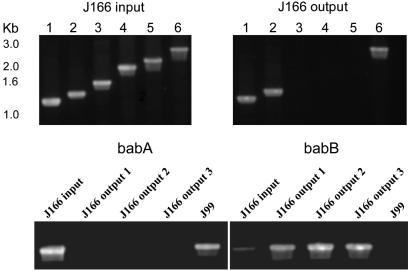
Confirmation of Microarray Data by PCR and Southern Blot. H. pylori has a large family of ≈30 paralogous OMPs that show significant amino acid homology in the NH2-terminal and COOH-terminal domains (4). BabA, which is the best studied of these OMPs, mediates adherence to the fucosylated Leb histo-blood group antigen present on gastric epithelium (15). The presence of transcriptionally active babA is clinically relevant because it is associated with peptic ulcer disease and gastric adenocarcinoma (12, 16). We next used PCR to determine whether the entire babA gene or only some portion of it was deleted in J166output strains recovered 17 wk PI. Preliminary experiments (data not shown) demonstrated that the babA gene (JHP0833) in H. pylori J166, like that in J99, is flanked by JHP0834 upstream (Fig. 2). Downstream of babA in J166 there is an 811-bp UTR followed by JHP0830. We amplified genomic DNA from J166input and the three J166output strains studied by microarray, using a primer in JHP0834 (834F) together with one of several downstream primers (AR1-AR6) that walked progressively down the babA gene. Appropriate-sized fragments were obtained for each PCR when chromosomal DNA from J166input was the template, which was expected because the microarray suggested that babA was present (Fig. 3 Upper Left). However, in each J166output strain, PCR products were obtained only when the babA primer was in the 5′ or 3′ region of the gene (AR1, AR2, AR6), but not when it was in the midregion (AR3-AR5) of babA (Fig. 3 Upper Right). The fragment size amplified with the 3′-most primer in babA was identical for J166input and each J166output strain, suggesting that babA in the J166output strains was in fact not deleted, but rather that the midportion of the gene had been replaced by a divergent sequence.
Fig. 2.
Schematic diagram of the organization of babA (JHP0833) and babB (JHP1164) in H. pylori J166 and J99. ORFs are shown by rectangles with direction of transcription indicated. Regions of homology are shown by shading, and unique regions are shown by angled or vertical cross hatch. Intergenic regions are shown with solid (babA) or dotted (babB) lines, which are broad in regions of homology. Direction (5′ to 3′) and approximate position of primers listed in Table 1 are shown as arrows.
Fig. 3.
Ethidium bromide-stained agarose gels of PCR products. (Upper) Amplification of babA from J166input (Left) and J166output (Right). Forward and reverse primers were 834F and AR1-6, respectively (lanes 1-6). Absence of bands with primers AR3-5 in J166output suggested the midportion of babA was replaced by a divergent sequence. (Lower) Amplification of DNA from J166input and J166output using a forward primer upstream of babA (834F) and a reverse primer specific for the unique region of babA (AR3) or babB (BR1). (Left) Amplification with the babA-specific primer showed the expected 1.5-kb band for J166input and the control strain J99 but no product for each J166output strain. (Right) Amplification with the babB-specific primer showed the expected fragment for the output strains (which had replaced babA with babB) and no band for J99, which has babA downstream of JHP0834. Surprisingly, J166input showed a faint band when amplified with the babB-specific primer, suggesting that both babA and babB can occupy the locus downstream of JHP0834.
BabA is closely related to another H. pylori OMP of unknown function called BabB. BabA and BabB are nearly identical in the NH2-terminal domain and completely identical in the COOH-terminal domains (≈300 aa), but they are divergent in the middle region. This finding suggested the possibility that in the J166output strains, babA had been replaced by a second copy of babB. To examine this we performed a Southern blot on J166input and J166output strains (17-wk time point) with a probe amplified from the unique region of babA or babB. As expected, J166input had single copies of babA and babB, whereas in J166output babA was absent and there was both the original and a second copy (babB2) of babB (Fig. 7, which is published as supporting information on the PNAS web site).
To confirm these results by PCR, we amplified genomic DNA from J166output with a primer upstream of J166 babA in JHP0834 (834F) and a downstream primer specific for the middle region of babA (AR3) or babB (BR1). Amplification with the babA-specific primer pair yielded the expected 1.5-kb product from J166input and J99, in which babA is also downstream of JHP0834 (Fig. 3 Lower Left). No product was obtained from each of the three J166output strains isolated at 17 wk. Amplification with the babB-specific primer pair yielded the expected product with each of the three J166output strains but not with J99, which has babA in this position (Fig. 3 Lower Right). Surprisingly, amplification with the babB-specific primer pair also gave a faint band for J166input, and DNA sequence of this fragment confirmed that it was babB. This finding suggested that in J166input babB was downstream of JHP0834, which contradicted the results from amplification with babA-specific primers (Fig. 3 Lower Left) and Southern blot (Fig. 7). The possibility that DNA from J166input or PCR reagents were contaminated with small amounts of DNA from J166output was excluded by control experiments, which included preparing fresh DNA from single colonies of all H. pylori strains in parallel with Escherichia coli DNA that was used as a negative control.
Quantitative PCR of babA and babB. An alternative explanation for the apparent presence of both babA and babB downstream of JHP0834 in J166input is that the bacterial population is heterogenous and that a minority of the population may have deleted babA and replaced it with babB2. To examine this idea further we used real-time PCR to quantitate the fraction of J166input that contains babA or babB downstream of JHP0834. A primer in the noncoding region between HP0834 and HP0833 (834-833F) was paired with a primer in either babA (AR3) or babB (BR2) and used in a quantitative PCR. We constructed standard curves (Fig. 8, which is published as supporting information on the PNAS web site) using single copies of babA or babB together with the upstream gene (JHP0834) cloned into pBR322. Using the standard curves and adjusting for starting template quantity, we estimate that ≈1 in 104 cells of J166input contains babB2 downstream of JHP0834. Similar results were obtained after up to 13 in vitro passages of J166input. Therefore, whereas passage in vivo selects for babB2 downstream of JHP0834, passage in vitro does not. However, passage in vitro shows that populations of J166input must be continuously giving rise to variants with babB2 at the babA locus.
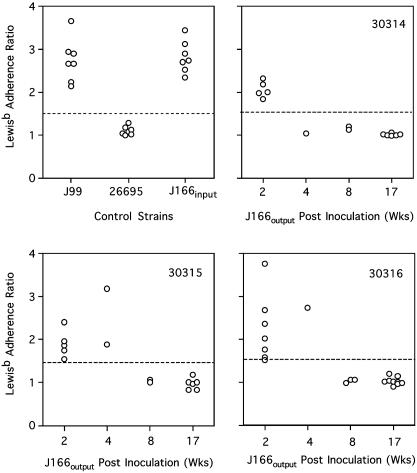
Leb Adhesion Assays. To functionally evaluate the babA deletion we assayed Leb adhesion of J166input and J166output colonies collected up to 17 wk PI from each monkey (Fig. 4). Adherence to Leb was demonstrated for J166input and J166output isolated up to 4 wk PI, but not in any J166output strains isolated 8 wk or more after inoculation. To confirm the relationship between Leb adhesion and the presence of babA, we amplified genomic DNA from each of 49 J166output colonies isolated from the three monkeys, using primers in HP0834 (834F) and the unique region of babA (AR3). All colonies that showed adhesion to Leb also had the babA gene by PCR. However, nine colonies that did not adhere to Leb were positive for babA by PCR. DNA sequence from these discordant clones showed that there was one more (seven colonies) or one fewer (two colonies) CT repeat in the 5′ coding region of babA, resulting in a frameshift mutation. These results confirm that the loss of functional babA, either by gene conversion or frameshift mutation in a 5′ poly(CT) tract, is accompanied by loss of adherence to Leb. Furthermore, these data suggest that, although strains expressing babA can initially colonize rhesus monkeys, they are not maintained during persistent infection.
Fig. 4.
Leb adherence ratio (Leb/control) for replicates of H. pylori control strains (Upper Left) and individual colonies isolated from each monkey (30314, 30315, 30316) at 2-17 wk PI. Values >1.5 (dotted line) are considered positive.
DNA Sequence Analysis. The simplest explanation for the insertion of babB2 at the babA locus (with loss of babA) is by a DNA homology-dependent recombination event via the 5′ and 3′ regions of shared sequence between babA and babB. To gain a better understanding of this we sequenced babA/babB/babB2 from J166input and babB/babB2 from one J166output strain obtained 17 wk PI. The two copies of babB in J166output differed at only a single nucleotide, which is consistent with the hypothesis that the second copy arose by an intrastrain recombination event. Conserved sequence upstream of the ATG (Fig. 9, which is published as supporting information on the PNAS web site) contained a consensus E. coli -10 hexamer (TATAAT), an extended -10 E. coli promoter sequence (TnTGn), and sequences matching 4 or 5 bp in the E. coli cognate -35 hexamer (TTGACA). Interestingly, the input babB2 has a promoter element that is a mosaic of babA (upstream of the -10 hexamer) and babB (downstream of the -10 hexamer), whereas the output babB2 promoter is identical to that from babB except for the length of a poly(A) tract. These results suggest that, at least in some cases, generation of the input and output babB2 may be independent events, rather than simple selection of a preexisting subclone present in the population. Differences in the poly(A) tract between J166input and J166output may reflect reciprocal changes of gene conversion or replication errors that are common in the population.
Southern Blot Analysis of babA and babB in H. pylori Isolates from Naturally Infected Rhesus Monkeys and Humans. Captive rhesus monkeys are commonly infected with H. pylori that is indistinguishable from that which infects humans (9). Because infection in macaques appears to select against H. pylori with babA, we hypothesized that natural rhesus strains of H. pylori would have the babA gene replaced by a second copy of babB. Southern blot analysis of 10 rhesus-derived H. pylori strains probed with a PCR-amplified fragment of the midregion of babA or babB showed in all 10 strains that babA was absent (data not shown) and two copies of babB were present (Fig. 10, which is published as supporting information on the PNAS web site). Similar analysis of 20 low-passage human clinical isolates of H. pylori showed that in four strains babA was absent and two copies of babB were identified (data not shown).
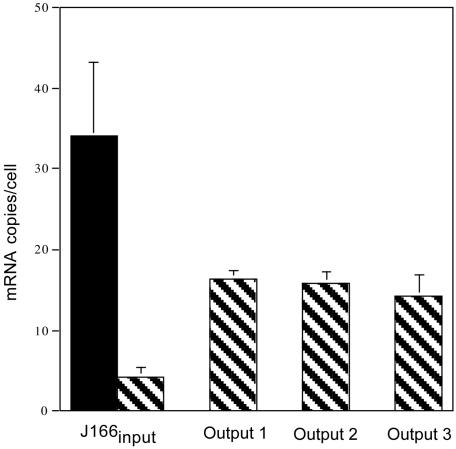
Real-Time RT-PCR of babA and babB mRNA. One explanation for replacement of babA with babB might be that babB is normally silent and that transfer into the babA locus switches on babB expression. To address this hypothesis, we used quantitative real-time RT-PCR to measure mRNA levels of babA and babB in J166input and J166output strains cultured from each monkey 17 wk PI. Standard curves were constructed from cloned babA and babB by using primer pairs AF1/AR3 and BF1/BR2, respectively (Fig. 11, which is published as supporting information on the PNAS web site). In J166input expression of babA was >8-fold greater than expression of babB (Fig. 5). In J166output babB expression was increased ≈4-fold compared with J166input and no babA expression was detected. Although babB is apparently not silent in J166input, increased expression in J166output is consistent with duplication of babB.
Fig. 5.
Mean (SD) mRNA copies per cell for babA (filled bar) and babB (hatched bars) in J166input and one J166output obtained from each of the three monkeys (30314, 30315, 30316) 17 wk PI. Data were calculated from standard curves (Fig. 8) of the relationship between Ct and log10 copies of babA and babB mRNA. No babA message was detected in any of the output strains.
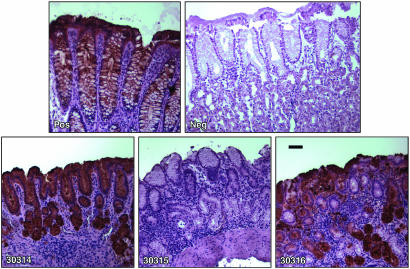
Detection of Leb in Gastric Mucosa of Rhesus Macaques. To better understand the relevance of the Leb adhesin (BabA) in the macaque model, we examined rhesus gastric tissue by immunohistochemistry using mAb to human Leb. Two of three monkeys showed evidence of Leb expression (Fig. 6). We also examined gastric tissue from four other arbitrarily selected rhesus monkeys, three of which were positive for Leb expression (data not shown). These results indicate that, like in humans (17), expression of Leb in gastric mucosa is common among rhesus monkeys. Presuming that BabA binds rhesus Leb, these results indicate that the loss of expression of the Leb adhesin (BabA) after experimental infection does not reflect its biological irrelevance in macaques.
Fig. 6.
Immunohistochemistry of gastric biopsies from each of the three macaques (30314, 30315, 30316) probed with anti-Leb mAb. Positive and negative controls (Upper) are human gastric tissue provided by the antibody supplier (Lab Vision) and courtesy of Ben Appelmelk (Vrije Universiteit Medical Center, Amsterdam), respectively. (Bar = 50 μm.)
Discussion
The remarkable genomic diversity of H. pylori has been exploited to understand numerous aspects of its pathogenesis, most notably the association of clinical disease with presence of the Cag pathogenicity island (18). H. pylori diversity has also been used to advantage in studies that seek to determine routes of transmission, for example within families (19), and even to serve as an archive of human migration over the millennia (20). Approximately 4% of the genome from both sequenced strains of H. pylori, significantly more than that of any other known bacterial genome, is composed of five paralogous gene families that are predicted to encode OMPs (21). All members of these gene families have one domain of similarity at the amino-terminal end and seven domains of similarity at the carboxyl-terminal end. This finding has led to the suggestion that recombination events might lead to a mosaic organization of OMPs that could be the basis for antigenic variation to avoid host immunity (4). This suggestion is supported by the observation that the two H. pylori strains whose genomes are sequenced have babA and babB in complementary loci (3, 4).
In this article, we use the rhesus macaque model and whole-genome DNA microarrays to examine strains recovered after a mixed inoculation with three unique H. pylori isolates. Analysis of the microarray data confirmed at high resolution our previous repetitive palindromic PCR results (10) and demonstrated that strain J166 preferentially colonizes rhesus macaques. Of particular interest was the observation that strains of J166 recovered from each of three monkeys lost expression of the babA gene, which codes for the Leb blood group binding adhesin. Loss of babA expression and Leb adherence occurred by one of two mechanisms in different isolates. In some cases a gene conversion event occurred, in which babA was replaced with the closely related babB whose function is unknown, yielding strains that have deleted babA and duplicated babB. RecA-dependent recombination has previously been proposed to explain the concerted evolution between the 3′ conserved segments of babA and babB (22). In other cases, a change in the number of dinucleotide CT repeats in the 5′ coding region of babA resulted in a frameshift and loss of Leb adherence. Although babA has not previously been reported to have CT repeats in the 5′ coding region, H. pylori strains 26695 and J99 each have five OMPs with 5′ CT repeats, which have been postulated to regulate their expression by slipped strand mispairing (3, 4). Our results demonstrate in real time with a relevant animal model that H. pylori regulates OMP expression in vivo by using both antigenic variation and phase variation. Both of these mechanisms are also operative in other bacteria. For example, in N. gonorrhea, frameshifts in a poly(C) tract produce reversible on-off switching of pilin expression, and movement of silent pilS genes into an expressed pilE locus produces variant pilin (23). Interestingly, like H. pylori, the N. gonorrhea genome contains few (five) putative two-component regulatory sequences (David W. Dyer, personal communication), which is approximately one-third that found in E. coli (4). Frequent use of phase and antigenic variation, together with natural transformation, may be a common mechanism of gene regulation for bacteria that occupy a restricted host niche and are therefore not exposed to multiple environmental conditions.
The promoter elements of babA and babB2 in J166input are identical, but they differ in the putative -35 hexamer and -10 to -35 spacing from J166output babB and babB2, which are themselves identical (Fig. 9). This may explain why babB in J166output is not expressed at a higher level. We expected that because J166output has two copies of babB, one of which is in the babA locus, that the babB message in J166output would be equivalent to that found for babA plus that found for babB in J166input. However, this was not the case (Fig. 5). The lower than expected level of message for babB in J166output may reflect differences in the -10 to -35 spacing, the -35 hexamer, or both. The presence of poly(A) or poly(T) tracts in the 5′ intergenic region has been found in other H. pylori OMPs and may be a common mechanism for regulation of OMP gene expression (4).
Why, then, is babA expression selected against when H. pylori is passaged through rhesus monkeys? First of all, this appears to be a phenomenon that is not unique to the rhesus monkey. In the United States ≈15-30% of H. pylori strains do not express babA (16), which is consistent with our finding using Southern blot that babA was absent in 4 of 20 human strains. Like in strains isolated from experimentally or naturally infected rhesus monkeys, in each of these four strains there were two copies of babB (data not shown). Therefore, whatever conditions favor deletion of babA and duplication of babB in macaques may sometimes be present in humans as well. Second, we have observed loss of babA and duplication of babB in each of five monkeys inoculated with J166 alone, so this phenomenon does not depend on an initial mixed inoculum as was done in these studies. Third, selection against expression of babA in macaques is unlikely to be explained by supposing that the gene is irrelevant, because the Leb antigen is expressed on rhesus gastric mucosa. Future studies will have to determine whether BabA binds rhesus Leb, but this seems likely based on cross reactivity of anti-human Leb antibody.
It is instructive to ask whether passage through the rhesus macaque is selecting for deletion of babA or duplication and therefore overexpression of babB. One possibility is that BabA (but not BabB) is immunogenic in rhesus macaques and that loss of babA reflects antigenic variation that the bacterium uses to avoid the host immune response. To examine this, we purified partial fragments of recombinant BabA and BabB as described (24). By ELISA we were unable to demonstrate seroreactivity to either BabA or BabB up to 5 months after inoculation in the monkeys from which we isolated J166output, although as expected they did demonstrate increased seroreactivity to H. pylori whole-cell antigen (Fig. 12, which is published as supporting information on the PNAS web site). These results are consistent with other reports that have failed to identify BabA or BabB as immunodominant antigens in humans (25, 26). Taken together, the findings that rhesus-derived strains of H. pylori and most J166output strains have two copies of babB suggest to us that BabB may function as an adhesin and that overexpression of BabB is advantageous. Perhaps the inflammatory response to H. pylori induces expression of a receptor for BabB, much like was demonstrated recently for the sialyl-dimeric-Lewis x glycosphingolipid that binds H. pylori SabA (27). It would be of interest to inoculate macaques with a strain containing a frameshift in babA. In this case, duplication of babB in output strains would suggest an advantage to overexpression of babB, because babA would already be functionally deleted. For the moment, our working hypothesis is that change in babA and babB after experimental infection of macaques represents a dynamic response in the H. pylori outer membrane that is designed to adhere maximally to gastric epithelium and promote chronic infection.
Supplementary Material
Acknowledgments
We thank Stanley Falkow for critical reading of the manuscript and Stephan Odenbreit and Rainer Haas for providing clones expressing babA and babB. This work was supported in part by Public Health Service Grants AI42081, AI43274, and RR14298 from the National Institutes of Health.
Abbreviations: OMP, outer membrane protein; PI, postinoculation; Leb, Lewisb.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AY428590 -AY428593).
References
- 1.Falush, D., Kraft, C., Taylor, N. S., Correa, P., Fox, J. G., Achtman, M. & Suerbaum, S. (2001) Proc. Natl. Acad. Sci. USA 98, 15056-15061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Suerbaum, S., Smith, J. M., Bapumia, K., Morelli, G., Smith, N. H., Kunstmann, E., Dyrek, I. & Achtman, M. (1998) Proc. Natl. Acad. Sci. USA 95, 12619-12624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alm, R. A., Ling, L. L., Moir, D. T., King, B. L., Brown, E. D., Doig, P. C., Smith, D. R., Noonan, B., Guild, B. C., deJonge, B. L., et al. (1999) Nature 397, 176-180. [DOI] [PubMed] [Google Scholar]
- 4.Tomb, J. F., White, O., Kerlavage, A. R., Clayton, R. A., Sutton, G. G., Fleischmann, R. D., Ketchum, K. A., Klenk, H. P., Gill, S., Dougherty, B. A., et al. (1997) Nature 388, 539-547. [DOI] [PubMed] [Google Scholar]
- 5.Salama, N., Guillemin, K., McDaniel, T. K., Sherlock, G., Tompkins, L. S. & Falkow, S. (2000) Proc. Natl. Acad. Sci. USA 97, 14668-14673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Achtman, M., Azuma, T., Berg, D. E., Ito, Y., Morelli, G., Pan, Z. J., Suerbaum, S., Thompson, S. A., van der Ende, A. & van Doorn, L. J. (1999) Mol. Microbiol. 32, 459-470. [DOI] [PubMed] [Google Scholar]
- 7.Kuipers, E. J., Israel, D. A., Kusters, J. G., Gerrits, M. M., Weel, J., van Der Ende, A., van Der Hulst, R. W., Wirth, H. P., Hook-Nikanne, J., Thompson, S. A. & Blaser, M. J. (2000) J. Infect. Dis. 181, 273-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Israel, D. A., Salama, N., Krishna, U., Rieger, U. M., Atherton, J. C., Falkow, S. & Peek, R. M. (2001) Proc. Natl. Acad. Sci. USA 98, 14625-14630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Drazek, E. S., Dubois, A. & Holmes, R. K. (1994) J. Clin. Microbiol. 32, 1799-1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Solnick, J. V., Hansen, L. M., Canfield, D. R. & Parsonnet, J. (2001) Infect. Immun. 69, 6887-6892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dubois, A., Berg, D. E., Incecik, E. T., Fiala, N., Heman-Ackah, L. M., Perez-Perez, G. I. & Blaser, M. J. (1996) Infect. Immun. 64, 2885-2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gerhard, M., Lehn, N., Neumayer, N., Borén, T., Rad, R., Schepp, W., Miehlke, S., Classen, M. & Prinz, C. (1999) Proc. Natl. Acad. Sci. USA 96, 12778-12783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Falk, P., Boren, T., Haslam, D. & Caparon, M. (1994) Methods Cell Biol. 45, 165-192. [DOI] [PubMed] [Google Scholar]
- 14.Kang, J. J., Watson, R. M., Fisher, M. E., Higuchi, R., Gelfand, D. H. & Holland, M. J. (2000) Nucleic Acids Res. 28, e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ilver, D., Arnqvist, A., Ogren, J., Frick, I. M., Kersulyte, D., Incecik, E. T., Berg, D. E., Covacci, A., Engstrand, L. & Borén, T. (1998) Science 279, 373-377. [DOI] [PubMed] [Google Scholar]
- 16.Yamaoka, Y., Souchek, J., Odenbreit, S., Haas, R., Arnqvist, A., Boren, T., Kodama, T., Osato, M. S., Gutierrez, O., Kim, J. G. & Graham, D. Y. (2002) J. Clin. Microbiol. 40, 2244-2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Taylor, D. E., Rasko, D. A., Sherburne, R., Ho, C. & Jewell, L. D. (1998) Gastroenterology 115, 1113-1122. [DOI] [PubMed] [Google Scholar]
- 18.Censini, S., Lange, C., Xiang, Z., Crabtree, J. E., Ghiara, P., Borodovsky, M., Rappuoli, R. & Covacci, A. (1996) Proc. Natl. Acad. Sci. USA 93, 14648-14653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang, J. T., Sheu, J. C., Lin, J. T., Wang, T. H. & Wu, M. S. (1993) J. Infect. Dis. 168, 1544-1548. [DOI] [PubMed] [Google Scholar]
- 20.Falush, D., Wirth, T., Linz, B., Pritchard, J. K., Stephens, M., Kidd, M., Blaser, M. J., Graham, D. Y., Vacher, S., Perez-Perez, G. I., et al. (2003) Science 299, 1582-1585. [DOI] [PubMed] [Google Scholar]
- 21.Doig, P., de Jonge, B. L., Alm, R. A., Brown, E. D., Uria-Nickelsen, M., Noonan, B., Mills, S. D., Tummino, P., Carmel, G., Guild, B. C., et al. (1999) Microbiol. Mol. Biol. Rev. 63, 675-707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pride, D. T. & Blaser, M. J. (2002) J. Mol. Biol. 316, 629-642. [DOI] [PubMed] [Google Scholar]
- 23.Seifert, H. S. (1996) Mol. Microbiol. 21, 433-440. [DOI] [PubMed] [Google Scholar]
- 24.Odenbreit, S., Kavermann, H., Puls, J. & Haas, R. (2002) Int. J. Med. Microbiol. 292, 257-266. [DOI] [PubMed] [Google Scholar]
- 25.Kimmel, B., Bosserhoff, A., Frank, R., Gross, R., Goebel, W. & Beier, D. (2000) Infect. Immun. 68, 915-920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haas, G., Karaali, G., Ebermayer, K., Metzger, W. G., Lamer, S., Zimny-Arndt, U., Diescher, S., Goebel, U. B., Vogt, K., Roznowski, A. B., et al. (2002) Proteomics 2, 313-324. [DOI] [PubMed] [Google Scholar]
- 27.Mahdavi, J., Sonden, B., Hurtig, M., Olfat, F. O., Forsberg, L., Roche, N., Angstrom, J., Larsson, T., Teneberg, S., Karlsson, K. A., et al. (2002) Science 297, 573-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eisen, M. B., Spellman, P. T., Brown, P. O. & Botstein, D. (1998) Proc. Natl. Acad. Sci. USA 95, 14863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.