Abstract
Pulmonary surfactant protein A (SP-A) plays a key role in innate lung host defense, in surfactant-related functions, and in parturition. In the course of evolution, the genetic complexity of SP-A has increased, particularly in the regulatory regions (i.e. promoter, untranslated regions). Although most species have a single SP-A gene, two genes encode SP-A in humans and primates (SFTPA1and SFTPA2). This may account for the multiple functions attributed to human SP-A, as well as the regulatory complexity of its expression by a relatively diverse set of protein and non-protein cellular factors. The interplay between enhancer cis-acting DNA sequences and trans-acting proteins that recognize these DNA elements is essential for gene regulation, primarily at the transcription initiation level. Furthermore, regulation at the mRNA level is essential to ensure proper physiological levels of SP-A under different conditions. To date, numerous studies have shown significant complexity of the regulation of SP-A expression at different levels, including transcription, splicing, mRNA decay, and translation. A number of trans-acting factors have also been described to play a role in the control of SP-A expression. The aim of this report is to describe the genetic complexity of the SFTPA1 and SFTPA2 genes, as well as to review regulatory mechanisms that control SP-A expression in humans and other animal species.
Keywords: surfactant proteins, innate immunity, genetic variants, transcriptional regulation, evolution
1. Introduction
Pulmonary surfactant is a lipoprotein complex essential for life, as it prevents alveolar collapse at low lung volumes. In addition to maintaining alveolar integrity, surfactant plays an important role in lung host defense, and the control of inflammation, primarily by specific functions of the protein component (i.e. surfactant proteins). The genetic variability of surfactant proteins has been extensively studied, and variants have been associated with acute and chronic lung disease throughout life in a variety of population studies. The roles of the hydrophilic surfactant proteins (SP-A and SP-D) as innate immunity molecules and inflammatory mediators have also been extensively reported. We have recently reviewed the current knowledge of SP-A and SP-D genetic variations in pulmonary disease and pathogenesis (Silveyra and Floros, 2012b). In the present review, we focus on the human SP-A genes, their variants, and regulatory regions. We present an overview of the known regulatory mechanisms involved in SP-A gene expression at the genetic and epigenetic level, as well as the experimental models used to study this regulation. Moreover, we discuss evolutionary aspects of this regulation by comparing SP-A regulatory regions in humans and other animal species.
2. Pulmonary Surfactant
In the lung alveolar epithelium, type II pneumocytes synthesize and secrete a lipoprotein complex essential for life known as pulmonary surfactant. The main function of this complex is to reduce surface tension at the alveolar air-liquid interface, and consequently prevent alveolar collapse at low lung volumes. The composition of surfactant is approximately 90–92% lipids, and 8–10% proteins (Postle et al., 2001; Perez-Gil and Weaver, 2010). The lipid fraction is primarily phospholipids, the key components of the surface tension lowering function. The protein component participates in surfactant functions, as well as in the modulation of the innate immune response, and the regulation of inflammatory processes (Crouch, 1998; Crouch et al., 2000; Phelps, 2001). Surfactant homeostasis is not only critical for breathing (and thus survival) in the prematurely born infant, but also for maintaining lung health, and normal lung function throughout life, as quantitative and/or qualitative derangement in surfactant composition and/or function are associated with respiratory diseases (Floros and Phelps, 1997; Floros and Kala, 1998; Floros and Wang, 2001; Whitsett et al., 2010).
3. Surfactant proteins
Approximately 8–10% of pulmonary surfactant is composed by proteins. Surfactant-associated proteins are classified into two groups, based on their hydrophobicity properties. The hydrophobic, surfactant proteins B (SP-B), and C (SP-C) are primarily involved in the prevention of alveolar collapse. SP-B expression is altered in a number of acute and chronic lung diseases and absence of SP-B is not compatible with life, whereas SP-C stabilizes surfactant at low lung volumes, and plays a role in innate immunity (Wang et al., 1996; deMello and Lin, 2001; Glasser et al., 2001; Augusto et al., 2003; Nogee, 2004; Whitsett et al., 2010). The remaining two proteins, SP-A and SP-D, are hydrophilic proteins that belong to the C-type lectin family (collectins), and are primarily host defense proteins (Crouch and Wright, 2001; Hawgood and Poulain, 2001; Phelps, 2001; Floros et al., 2009). Members of the collectin family are characterized by an N-terminal collagen-like domain and a C-terminal carbohydrate recognition domain (CRD) that allows binding to various types of macromolecules, pathogens and allergens (Crouch and Wright, 2001). SP-A and SP-D are found in large oligomeric structures that have the ability to bind and opsonize several pathogens and allergens (Kishore et al., 2005; Kishore et al., 2006). SP-A and SP-D also play a role in surfactant homeostasis (Botas et al., 1998; Ikegami et al., 2000; LeVine et al., 2000; Hawgood and Poulain, 2001), and alterations of the levels of these two proteins have been reported in several pulmonary pathologies (Silveyra and Floros, 2012b).
3.1. Surfactant protein A
3.1.1. Function and structure
SP-A, the most abundant protein of surfactant, is involved in both host defense and surfactant-related functions. The role of SP-A in innate immunity is primarily mediated by its ability to bind several pathogens, enhance phagocytosis and chemotaxis of alveolar macrophages, induce proliferation of immune cells, and stimulate proinflammatory cytokine production, as well as to modulate the generation of reactive oxygen species (reviewed in (Phelps, 2001; Wright, 2005; Wang and Reid, 2007)). Furthermore, SP-A participates in several other functions that involve secretion of surfactant, signaling the initiation of parturition, as well as maintaining the structure of the extracellular form of surfactant tubular myelin (Williams et al., 1991; Condon et al., 2004; Wang et al., 2010; Snegovskikh et al., 2011; Yadav et al., 2011). Moreover, the expression and function of SP-A is not restricted to alveolar epithelial cells. Several studies have reported SP-A expression in the lower and higher airway epithelial cells, as well as extrapulmonary tissues including the lacrimal system (Bräuer et al., 2007), parotid glands (Bräuer et al., 2009), gingiva (Bräuer et al., 2012), epithelial cells of small and large intestine (Rubio et al., 1995) and Eustachian tube epithelium (Paananen et al., 2001), and vagina (MacNeill et al., 2004).
The structure of SP-A consists of four domains: an N-terminal, a collagen-like domain, a neck region, and a carbohydrate recognition domain (CRD) (Floros, 2001; Floros et al., 2009). The most conserved domains are the neck and N-terminal. The CRD consists of 115 amino acids, including four cysteine residues that are conserved across species. These form two pairs of disulfide bonds within the CRD that play a critical role in the stability of the SP-A structure (Floros et al., 2009). A cysteine residue located near the N-terminal (position 6 of the mature protein) has been proposed to participate in the supratrimeric assembly of SP-A (Sánchez-Barbero et al., 2007). Furthermore, the human and baboon SP-A1 gene contain an extra cysteine (Cys85) that may generate additional intermolecular disulfide bonds that affect SP-A1 structure, function, and biochemical properties (Wang et al., 2007a).
3.1.2. Human SFTPA1 and SFTPA2
The majority of species studied to date have a single SP-A gene. However, in primates and humans there are two functional genes (SFTPA1 and SFTPA2). In humans, the mature SP-A monomer is a 35kDa protein composed by 248 amino acids, and the two gene products (SP-A1 and SP-A2) differ in four amino acids at the coding region (Floros and Hoover, 1998; DiAngelo et al., 1999). The amino acid differences that distinguish between SP-A1 and SP-A2 genes and among their corresponding variants are located in the collagen-like domain. These are Met66, Asp 73, Ile81 and Cys85 for SP-A1, and Thr66, Asn73, Val81, and Arg85 for SP-A2. One of these has shown to affect its structure and function (Wang et al., 2007a). Furthermore, nucleotide differences that do or do not change the encoded amino acid among SFTPA1 or SFTPA2 variants are located within the sequence for the signal peptide, collagen-like region, and CRD (Floros, 2005; Floros et al., 2009). Several studies have identified functional differences between SP-A1 and SP-A2 in a variety of innate immunity and surfactant related functions including cytokine production (Wang et al., 2000; Wang et al., 2002; Huang et al., 2004), modulation of surfactant secretion (Wang et al., 2004), phagocytosis by alveolar macrophages (Mikerov et al., 2005; Mikerov et al., 2007; Mikerov et al., 2008a), as well as other structural characteristics (Garcia-Verdugo et al., 2002; Oberley and Snyder, 2003; Wang et al., 2007a; Wang et al., 2007b). However, no differences were found between SP-A1 and SP-A2 in the inhibition of hemagglutination activity of influenza A virus (Mikerov et al., 2008a). Moreover, functional divergence between SP-A1 and SP-A2 has been shown for an extracellular form of surfactant, the tubular myelin (Wang et al., 2010). Whether further functional divergence exists among SP-A1 and SP-A2 variants remains to be determined. Differences in structure, and posttranslational modifications have been also described between SP-A1 and SP-A2 (Floros et al., 2009), and some of the structural differences between SP-A1 and SP-A2 may be responsible for functional differences (Wang et al., 2007a). Moreover, differences in the expression of SFTPA1 and SFTPA2, as assessed by the protein ratio of SP-A1 to total SP-A, have been observed in human bronchoalveolar lavage samples as a function of age and lung health status (e.g. healthy vs. cystic fibrosis, culture positive vs. negative; asthmatic vs. control) (Tagaram et al., 2007; Wang et al., 2011).
SP-A is found in a variety of oligomeric structures. A 630 kDa hexameric bouquet-like structure contains a total of eighteen SP-A1 and SP-A2 monomers composed of six trimeric structural subunits of 105kDa. Both hetero-oligomers (i.e. consisting of both SP-A1, and SP-A2 monomers), and homo-oligomers (i.e. consisting of SP-A1 or SP-A2 monomers) are functional, and both gene products are required for tubular myelin formation (Voss et al., 1991; Wang et al., 2002; Mikerov et al., 2005; Mikerov et al., 2007; Wang et al., 2007a; Mikerov et al., 2008a; Wang et al., 2010). Differences between SP-A1 and SP-A2 in oligomerization properties, aggregation, structural stability, sugar-binding capacity, and ability to form phospholipids monolayers and tubular myelin have also been observed (Wang et al., 2000; Garcia-Verdugo et al., 2002; Wang et al., 2002; Oberley and Snyder, 2003; Wang et al., 2004; Mikerov et al., 2007; Sánchez-Barbero et al., 2007; Wang et al., 2007a; Wang et al., 2007b; Wang et al., 2010).
3.1.3. SP-A variants
More than 30 variants or intragenic haplotypes have been identified and characterized for SFTPA1 and SFTPA2 based on various combinations of single nucleotide polymorphisms (SNPs) within the coding region that may or may not change the encoded amino acids (Karinch and Floros, 1995a; DiAngelo et al., 1999; Floros, 2001; Floros and Wang, 2001; Floros et al., 2009). Among SFTPA1 variants, SNPs are found in the codons of amino acids 19, 50, 62, 133, and 219. Two of these polymorphisms are silent (amino acids 62 and 133), and the rest result in amino acid substitutions. Similarly, among SFTPA2 variants, SNPs are found in four codons: three silent (amino acids 9, 91, and 223) and one synonymous (amino acid 140) (DiAngelo et al., 1999). Of the variants identified, four SFTPA1 (6A, 6A2, 6A3, 6A4), and six SFTPA2 (1A, 1A0, 1A1, 1A2, 1A3, 1A5) variants have been observed in higher frequency in the general population (DiAngelo et al., 1999; Floros, 2001; Silveyra and Floros, 2012b). Furthermore, the frequency of these variants has been found to be variable in the population (DiAngelo et al., 1999; Floros, 2001; Liu et al., 2003).
3.1.4. SP-A receptors
As a critical component of innate immunity, SP-A helps combat infections caused by bacteria, viruses, fungi, and other pathogens by mechanisms that involve binding, aggregation, agglutination, inhibition of growth, and promotion of phagocytosis by activation of alveolar macrophages, an important cellular component of the lung first line of defense (McNeely and Coonrod, 1994; Schagat et al., 2001; Wu et al., 2003; Ding et al., 2004; Mikerov et al., 2007; Mikerov et al., 2008b; Mikerov et al., 2008c). SP-A can either bind pathogens to promote their destruction (opsonization), and/or activate immune cells by direct binding (Haagsman, 1998; LeVine and Whitsett, 2001; Wright, 2005), and/or stimulate the production of pro- and anti-inflammatory cytokines (Wang and Reid, 2007). Soluble receptors, as well as membrane receptors in the surface of alveolar macrophages have been shown to interact with SP-A. These include: CD35 (CR1), C1qR (CD93), CD14, CD91/calreticulin, SIRPα, SP-R210, gp-340, P63, TLR-2 and TLR-4, and others (reviewed in (Kishore et al., 2006; Bates, 2010; Silveyra and Floros, 2012b)). Some of these have been identified in the cell surface of alveolar type II pneumocytes, but not in alveolar macrophages, indicating that they may play a role in surfactant function, whereas others have been found to be ubiquitous.
3.2. Human SP-A Gene structure
The genomic locus of the human SP-A genes is located in the long arm of chromosome 10 (Bruns et al., 1987; Floros and Hoover, 1998), and it consists of the two functional genes, SFTPA1 and SFTPA2, and a pseudogene (SFTPA3P) (Hoover and Floros, 1998). The two functional genes are located in opposite transcriptional orientation in 10q22.3 (GenBank, NC_000010.10) as shown in Figure 1, and are in linkage disequilibrium (Liu et al., 2003). According to GenBank, the SFTPA1 genomic sequence (Gene ID: 653509) is located between positions 81370695-81375199 of the positive strand, whereas the SFTPA2 (Gene ID: 729238) is located in the complement strand, positions 81315608-81320163. The SFTPA3P pseudogene (Gene ID: 100288405) is also located in the complement strand, between positions 81353703-81355415. Similarly, the Ensembl database locates SFTPA1 between positions 81370701 81375202 of the positive strand (ENSG00000225827), and the pseudogene between positions 81355416-81355050 of the complement strand (ENSG00000225827). The SFTPA2 gene location is the same for both GenBank and Ensembl (ENSG00000185303). While both human SFTPA1 and SFTPA2 genes are approximately 4.5kb in length, and share approximately 94% nucleotide identity, their protein products share approximately 96% amino acid sequence identity. The structure of the two human genes is similar and consists of four coding exons (I–IV), and several untranslated exons (A, B, B′, C, C′,D, D′) (Karinch and Floros, 1995a) at 5′UTR as illustrated in Figure 1 (reviewed in (Floros, 2001; Floros and Wang, 2001; Floros, 2005; Floros et al., 2009)).
Figure 1.
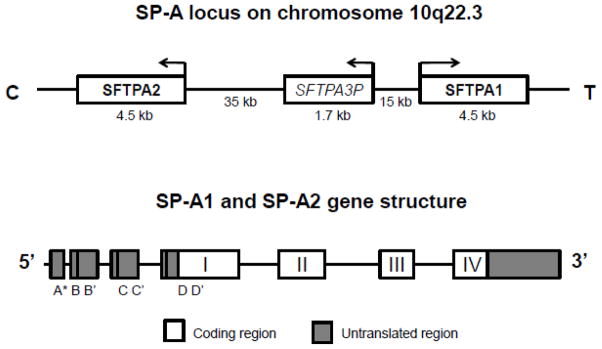
Upper panel Diagrammatic representation of the 10q22–23 locus of the human chromosome 10. The two human SP-A genes SFTPA1 and SFTPA2 are located in opposite transcriptional orientation, flanking a pseudogene (SFTPA3P). Lower panel. SP-A1 and SP-A2 common gene structure. Coding exons are indicated with numbers, and untranslated exons are indicated with letters. A* represents exons A, A′, and A″.
In general, the genetic complexity of SP-A has been shown to increase as one ascends the mammalian evolutionary ladder. Almost all mammalian species have been shown to contain a single SP-A gene, except from baboons and humans that have two. A gene duplication event occurred more than 26.5 million years ago (Gao et al., 1996), resulting in the generation of SFTPA1 and SFTPA2. Additional SP-A sequences have been reported in some species, e.g. three genes in opossum (SP-A1, SP-A2, and SP-A3) and two variants in chicken (SP-A and an SP-A-like gene) (Gao et al., 1996; Hogenkamp et al., 2006; Hughes, 2007). All SP-A genes of various species are about 5 Kb in length, with the basic exon-intron structure being conserved in all species studied so far. The sequence similarity among species is higher in exons than in introns. The human and baboon genes contain additional exons/introns at the 5′ untranslated region that are alternatively spliced generating mRNA splice variants (Karinch and Floros, 1995a; Gao et al., 1996). Furthermore, in humans, splice and sequence differences between the functional genes and/or their variants give rise to a number of variable transcripts, that have been shown to differentially affect SP-A gene expression (Karinch and Floros, 1995a; Karinch et al., 1998; Wang et al., 2003; Wang et al., 2005; Wang et al., 2009; Silveyra et al., 2010; Silveyra et al., 2011). This increase in genetic complexity, in particular at the regulatory regions, may reflect the functional importance of SP-A during evolution, and the necessity to control protein levels in specific physiological or pathological situations (Mendelson et al., 1998; Floros, 2005; Tagaram et al., 2007; Bruce et al., 2009; Vlachaki et al., 2010; Wang et al., 2011; Silveyra and Floros, 2012b).
3.3. SP-A promoter and control of transcription
To date, several regulatory regions have been identified within the 5′ flanking regions and the human SFTPA1 and SFTPA2 gene sequences and in particular in the first 300 base pairs of the proximal promoter, immediately upstream of the transcription start site. These include, cAMP response elements (CRE), and GT-boxes important for high basal and c-AMP mediated regulation of SFTPA2 in alveolar epithelial cells (Young and Mendelson, 1996; Young and Mendelson, 1997), as well as TTF-1 binding elements (TBE), important for hormonal regulation of lung development, as well as temporal and spatial control of SP-A gene expression (Yi et al., 2002; Liu et al., 2008). Some of these elements are conserved among species. A number of lung cell-specific transcription factors and small molecules have been shown to interact with regulatory sequences present at the human SP-A promoters, in particular during the late stages of development, where the expression of SP-A transitions from practically silent to dramatically upregulated (Gao et al., 2003; Alcorn et al., 2004; Liu et al., 2008). Some of the molecules shown to affect SP-A expression include: estrogen receptor alpha (Liu et al., 2006), dexamethasone (Hoover et al., 1999; Alcorn et al., 2004; Islam and Mendelson, 2008), NfkB (Islam and Mendelson, 2002; Wu et al., 2011), C/EBP (Rosenberg et al., 2002), phorbol esters (Hoover et al., 2000), c-AMP (Young and Mendelson, 1996; Young and Mendelson, 1997; Liu et al., 2009), TTF-1 (Li et al., 1998; Alcorn et al., 2004; Liu et al., 2008), and others (Karinch et al., 1998; Kumar and Snyder, 998). Several of these factors, in addition to modulating the expression of SP-A in humans, affect the expression of rabbit, murine, and baboon SP-A genes (Lacaze-Masmonteil et al., 1992; Alcorn et al., 1993; Kouretas et al., 1993; Bruno et al., 1995; Rosenberg et al., 1999; Bruno et al., 2000; Rosenberg et al., 2002).
In addition to these sequence-specific interacting factors at the SP-A promoters, another level of complexity appears to play a role in the control of expression of human SP-A genes, known as epigenetic modifications. Epigenetic modifications have been shown to affect the expression of SP-A1 and SP-A2 mRNAs in a variety of physiological and pathological conditions. Recent unpublished data from our group identified significant differences in the methylation status of specific cytosines of the SFTPA2 5′ flanking region between normal and adenocarcinoma human paired samples. The hypermethylated DNA in cancerous tissue correlated with decreased SP-A2 mRNA expression. These findings may hold promise for future use of SP-A2 as a biomarker for the diagnosis and/or therapies of lung cancer. Moreover, previous studies from our group reported associations of altered methylation patterns of CpG sites in the human SFTPA1 promoter with lung cancer (Lin et al., 2007). Furthermore, histone acetylation and methylation at regulatory regions of the SP-A gene promoters have been shown to affect SP-A expression in lung cells during development and during hypoxia (Islam and Mendelson, 2006; Lin et al., 2007; Islam and Mendelson, 2008; Vaid and Floros, 2009; Benlhabib and Mendelson, 2011; Silveyra and Floros, 2012a).
In summary, gene regulation is a dynamic and complex process. The promoter region of SP-A may serve as an enhanceosome (Benlhabib and Mendelson, 2011) through which SP-A expression is modulated via the interaction of several transcription factors with specific sequence elements, some of which are subject to epigenetic regulation, and may be modulated by pathological conditions (e.g. cancer) and environmental insults (Silveyra and Floros, 2012a). Given the functional differences between SFTPA1 and SFTPA2 and their genetic complexity, differences in the regulation of expression among SP-A variants may underlie individual susceptibility in response to different insults such as oxidative stress and disease (Mendelson et al., 1998; Tagaram et al., 2007; Floros et al., 2009; Wang et al., 2011).
3.4. SP-A mRNA
Transcription of SFTPA1 and SFTPA2 genes results in a variety of mRNA transcripts. The source of this variability involves differences in the transcription start sites, as well as alternative splicing of exons (A, B, B′, C, C′, D, D′) at the 5′UTR. The formation of SP-A 5′UTR splice variants is not a random process as there are major, minor, and rare splice-variants for SP-A1 and SP-A2 transcripts (Karinch and Floros, 1995a). Furthermore, alternative transcription start sites have been observed for SFTPA1 and SFTPA2 by primer extension in two independent studies (McCormick et al., 1994; Karinch and Floros, 1995a); these identified three transcription start sites for SFTPA1, and one common transcript start site for SFTPA2, although some minor discrepancies observed between the two studies were attributed to variations in the individual lung RNA samples, and/or the limited number of clones tested (Floros et al., 2009).
The predominant human SP-A transcript splice variants and the frequencies with which they are found differ between the two genes. Similarly, frequency differences may also exist among individuals for these mRNAs (Karinch and Floros, 1995b). For SP-A1, the major splice variant is AD′ (81%), followed by ACD′ and AB′D′ (7%), and for SP-A2 the major variants are ABD′ (49%), and ABD (44%) (Karinch and Floros, 1995a). Although more than 30 variants for human SFTPA1and SFTPA2 have been identified in the population, the sequence for those found in the population with higher frequencies is available in GenBank under the accession numbers indicated in Table 1.
Table 1.
Gen Bank sequences for SP-A1 and SP-A2 mRNA transcripts
| Gene | Variant id | 5′UTR splice | Coding | 3′JTR sequence | GenBank id |
|---|---|---|---|---|---|
| SP-A1 | AD′6A | AD′ | 6A | 6A | HQ021433 |
| SP-A1 | AD′6A2 | AD′ | 6A2 | 6A2 | HQ021434 |
| SP-A1 | AD′6A3 | AD′ | 6A3 | 6A3 | HQ021435 |
| SP-A1 | AD′6A4 | AD′ | 6A4 | 6A4 | HQ021436 |
| SP-A1 | AB′D′6A | AB′D′ | 6A | 6A | JX502764 |
| SP-A1 | AB′D′6A2 | AB′D′ | 6A2 | 6A2 | HQ021437 |
| SP-A1 | AB′D′6A3 | AB′D′ | 6A3 | 6A3 | HQ021435 |
| SP-A1 | AB′D′6A4 | AB′D′ | 6A4 | 6A4 | HQ021439 |
| SP-A1 | ACD′6A | ACD′ | 6A | 6A | JX502765 |
| SP-A1 | ACD′6A2 | ACD′ | 6A2 | 6A2 | HQ021440 |
| SP-A1 | ACD′6A3 | ACD′ | 6A3 | 6A3 | HQ021441 |
| SP-A1 | ACD′6A4 | ACD′ | 6A4 | 6A4 | HQ021442 |
| SP-A2 | ABD1A | ABD | 1A | 1A | HQ021432 |
| SP-A2 | ABD1A0 | ABD | 1A0 | 1A0 | HQ021421 |
| SP-A2 | ABD1A1 | ABD | 1A1 | 1A1 | HQ021422 |
| SP-A2 | ABD1A2 | ABD | 1A2 | 1A2 | HQ021423 |
| SP-A2 | ABD1A3 | ABD | 1A3 | 1A3 | HQ021424 |
| SP-A2 | ABD1A5 | ABD | 1A5 | 1A5 | HQ021425 |
| SP-A2 | ABD′1A | ABD′ | 1A | 1A | HQ021426 |
| SP-A2 | ABD′1A0 | ABD′ | 1A0 | 1A0 | HQ021427 |
| SP-A2 | ABD′1A1 | ABD′ | 1A1 | 1A1 | HQ021423 |
| SP-A2 | ABD′1A2 | ABD′ | 1A2 | 1A2 | HQ021429 |
| SP-A2 | ABD′1A3 | ABD′ | 1A3 | 1A3 | HQ021430 |
| SP-A2 | ABD′1A5 | ABD′ | 1A5 | 1A5 | HQ021431 |
| SP-A1 | variant 1 | AB′D′ | 6A3 | 6A3 | NM_005411.4 |
| SP-A1 | variant 2 | ACD′ | 6A3 | 6A3 | NM_001093770.2 |
| SP-A1 | variant 3 | ABD′ | 6A3 | 6A3 | NM_001164644.1 |
| SP-A1 | variant 4 | AD′ | 6A3 | 6A3 | NM_001164647.1 |
| SP-A1 | variant 5 | ACD′ | 6A3 (truncated)a | 6A3 | NM_001164645.1 |
| SP-A1 | variant 6 | AB′D′ | 6A3 (truncated)a | 6A3 | NM_001164646.1 |
| SP-A2 | ABD′ | 1A2 | 1A0 | NM_001098668.2 |
missing a fragment of 147 nt (positions 238–435 mRNA; positions 84–231 from start codon)
SP-A1 and SP-A2 mRNA variants also show sequence differences at the 3′UTRs (Krizkova et al., 1994; Hoover and Floros, 1999; Wang et al., 2003; Silveyra et al., 2010). These, together with several other mRNA elements, such as the 5′CAP, poly(A) tail, secondary structures, and other structural elements, may modulate mRNA translation and thus differentially control SP-A1 and SP-A2 protein levels.
3.4.1 Role of SP-A UTRs in the control of transcription and translation
Several studies from our laboratory have reported that SP-A 5′UTR splice variants affect gene expression with variable efficiency, and that variants containing the untranslated exon B have an increased rate of transcription, and translation in both in vitro (Silveyra et al., 2010) and in lung epithelial cell culture systems (Wang et al., 2005; Silveyra et al., 2010). When compared to SP-A1 5′UTR variants ACD′, AD′, and AB′D′, the SP-A2 5′UTR variant ABD displayed a significantly lower rate of mRNA decay and a higher translation efficiency (Wang et al., 2005), and postulated that alternative splicing at the SP-A 5′UTR is a major regulatory mechanism for differential SP-A1 and/or SP-A2 variant expression (Wang et al., 2005; Silveyra et al., 2010; Silveyra et al., 2011). In support of this, our preliminary studies in which lung cells were exposed to diesel exhaust particulate matter showed that the splicing pattern of SP-A 5′UTRs is altered (our unpublished data), and the efficiency of SP-A translation is increased in variants containing exon B (Wang et al., 2009). Furthermore, the SP-A2 5′UTRs that contain exon B (absent in SP-A1 variants) has been shown, via a specific double-loop secondary structure, to form an internal ribosomal entry site (IRES) and mediate cap-independent translation, a process that often occurs under cellular stress, hypoxia, and other insults (Wang et al., 2009; Komar and Hatzoglou, 2011). Moreover, infectious agents such as respiratory syncytial virus also affect the rate of SP-A translation in pulmonary epithelial cells, although the mechanisms involved in this inhibition are still unknown (Bruce et al., 2009).
A recent study identified potential regulatory sequence elements in the 30 nucleotide SP-A2 untranslated exon B that may modulate gene expression via interaction with trans-acting factors. The exon B DNA sequence was shown to exhibit characteristics of a transcriptional enhancer (Silveyra et al., 2011), and at the mRNA level, exon B enhanced translation when placed in its natural sequence environment (ABD) or within orthologous sequences (Silveyra et al., 2011). The enhancing effect of exon B in SP-A expression may involve sequence motifs and/or secondary structures that potentially interact with specific RNA-binding factors and modulate a variety of post-transcriptional events. Preliminary results from our group using RNA gel shift assays identified several exon B binding proteins, and mapped specific nucleotides of exon B important for these interactions (our unpublished data). Moreover, the splice variant ACD′, found in SP-A1 transcripts (but not in SP-A2 transcripts), contains two upstream AUG codons (uAUGs) at its unique untranslated exon C that may affect translational efficiency. One of these is in frame with the main start codon (Karinch and Floros, 1995b), and shown to be functional (our preliminary data).
Variant-specific sequences located in the SP-A 3′UTRs may differentially regulate SP-A expression at the translational level. A pyrimidine-rich domain of 37 nucleotides is present in the 3′UTRs of SP-A1 and SP-A2 variants, but 7 of the nucleotides in this region differ between SP-A1 and SP-A2. Moreover, the 3′-UTRs of SP-A2 variants contain two AU-rich elements (AUUUA), whereas the SP-A1 sequences contain only one. The published literature indicates that these elements may influence mRNA stability, as well as the regulation of SP-A1 and SP-A2 variants by glucocorticoids (Hoover and Floros, 1999; Wang et al., 2003). An 11-nt element, present in the 3′UTR of all the SP-A2 variants and the SP-A1 variant 6A2 has been shown to negatively affect translation efficiency, by mechanisms that involve formation of secondary structures with different stabilities, as well as differences in predicted miRNA binding to this element (Silveyra et al., 2010). Interestingly, the effects observed were more evident when the transcripts were polyadenylated, indicating a potential role of the poly(A) tail in the control of translation of SP-A transcripts (Silveyra et al., 2010; Silveyra et al., 2011). Given the nucleotide differences among SP-A variants at the 3′UTR, it is highly likely that differences observed in their translational regulation are mediated by miRNAs. Our preliminary studies in cell lines expressing different SP-A variants indicate a role of specific miRNAs in the control of SP-A translation (our unpublished data).
In summary, the UTRs of SP-A mRNA transcripts are implicated in the control of SP-A expression. Several gene-specific, and variant-specific elements have been described and studied in these regions. It is possible that the ability of the variants to bind regulatory factors at the UTRs differs, and that this may account for differential protein expression of SP-A1 and SP-A2 variants under normal or compromised conditions (i.e. lung disease).
4. Genetic associations of SP-A variants with lung disease
The influence of genetic variants in acute and chronic lung disease susceptibility have been studied within different biological contexts, and correlated with environmental factors, and/or other conditions including prematurity, concurrent diseases, or need for mechanical ventilation (Leikauf et al., 2002; Hallman and Haataja, 2003; Villar et al., 2003; Christie, 2004; Grigoryev et al., 2004; Kishore et al., 2005; Nonas et al., 2005; Meyer and Garcia, 2007; Lam and dos Santos, 2008; Reddy and Kleeberger, 2009; Meyers, 2010). Numerous associations between SFTPA1 and SFTPA2 genetic variants and acute and chronic lung disease have been identified in several populations and study groups including neonates, children, and adults. For example, the SFTPA2 1A3 variant has been associated with susceptibility to tuberculosis in both Mexican and Ethiopians (Floros et al., 2000; Malik et al., 2006). In neonates, a specific SFTPA1/SFTPA2 haplotype, 6A2/1A0, has been shown to associate with risk for respiratory distress syndrome (RDS) in Finnish and non-Finnish Caucasians (Rämet et al., 2000; Floros et al., 2001). RDS is a condition where insufficient amounts of surfactant proteins have been observed, as well as absence of the extracellular form of surfactant, the tubular myelin, where SP-A is an essential component. The SFTPA1 and SFTPA2 genetic associations have been reviewed elsewhere (Floros and Kala, 1998; Floros and Pavlovic, 2003; Floros and Thomas, 2009; Silveyra and Floros, 2012b).
5. Concluding remarks
Human SP-A expression is controlled developmentally and by tissue specificity at both transcriptional and translational levels. The SP-A 5′ and 3′UTRs contain several regulatory elements implicated in the differential regulation of SP-A1 and SP-A2 expression, as well as their genetic variants. Study of mechanisms involved in these processes may help explain the altered ratio of SP-A1 and SP-A2 protein content in certain lung diseases or conditions (Tagaram et al., 2007; Wang et al., 2011), as well as gain insight into the basis of individual disease susceptibility.
Although there is considerable evidence that the human SP-A complexity is associated with various pulmonary diseases, and that differences in the function and structure of SP-A1 and SP-A2 exist as well as differences in the relative levels of SP-A1 to total SP-A in several diseases, the underlying mechanisms are not yet entirely known. We postulate that given the functional differences between SP-A1 and SP-A2, the overall SP-A functional activity in the lung depends on the relative levels of SP-A1 and SP-A2 rather than the total SP-A content (i.e., without regard to the SP-A1 and SP-A2 proportions), and that an altered regulation of SP-A1 or SP-A2 gene expression could result in an unfavorable SP-A1 to SP-A2 ratio for normal host defense.
Highlights.
Structure of surfactant protein A genes
Characteristics of SP-A1 and SP-A2 variants
Regulatory aspects of SP-A1 and SP-A2 expression
Acknowledgments
This work was supported by NIH-HL34788.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alcorn JL, Gao E, Chen Q, Smith ME, Gerard RD, Mendelson CR. Genomic elements involved in transcriptional regulation of the rabbit surfactant protein-A gene. Mol Endocrinol. 1993;7:1072–85. doi: 10.1210/mend.7.8.8232306. [DOI] [PubMed] [Google Scholar]
- Alcorn JL, Islam KN, Young PP, Mendelson CR. Glucocorticoid inhibition of SP-A gene expression in lung type II cells is mediated via the TTF-1-binding element. Am J Physiol Lung Cell Mol Physiol. 2004;286:L767–76. doi: 10.1152/ajplung.00280.2003. [DOI] [PubMed] [Google Scholar]
- Augusto LA, Synguelakis M, Johansson J, Pedron T, Girard R, Chaby R. Interaction of pulmonary surfactant protein C with CD14 and lipopolysaccharide. Infect Immun. 2003;71:61–7. doi: 10.1128/IAI.71.1.61-67.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates SR. P63 (CKAP4) as an SP-A receptor: implications for surfactant turnover. Cell Physiol Biochem. 2010;25:41–54. doi: 10.1159/000272062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benlhabib H, Mendelson CR. Epigenetic regulation of surfactant protein A gene (SP-A) expression in fetal lung reveals a critical role for Suv39h methyltransferases during development and hypoxia. Mol Cell Biol. 2011;31:1949–58. doi: 10.1128/MCB.01063-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botas C, Poulain F, Akiyama J, Brown C, Allen L, Goerke J, Clements J, Carlson E, Gillespie AM, Epstein C, Hawgood S. Altered surfactant homeostasis and alveolar type II cell morphology in mice lacking surfactant protein D. Proc Natl Acad Sci U S A. 1998;95:11869–74. doi: 10.1073/pnas.95.20.11869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce SR, Atkins CL, Colasurdo GN, Alcorn JL. Respiratory syncytial virus infection alters surfactant protein A expression in human pulmonary epithelial cells by reducing translation efficiency. Am J Physiol Lung Cell Mol Physiol. 2009;297:L559–67. doi: 10.1152/ajplung.90507.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruno MD, Bohinski RJ, Huelsman KM, Whitsett JA, Korfhagen TR. Lung cell-specific expression of the murine surfactant protein A (SP-A) gene is mediated by interactions between the SP-A promoter and thyroid transcription factor-1. J Biol Chem. 1995;270:6531–6. doi: 10.1074/jbc.270.12.6531. [DOI] [PubMed] [Google Scholar]
- Bruno MD, Korfhagen TR, Liu C, Morrisey EE, Whitsett JA. GATA-6 activates transcription of surfactant protein A. J Biol Chem. 2000;275:1043–9. doi: 10.1074/jbc.275.2.1043. [DOI] [PubMed] [Google Scholar]
- Bruns G, Stroh H, Veldman GM, Latt SA, Floros J. The 35 kd pulmonary surfactant-associated protein is encoded on chromosome 10. Hum Genet. 1987;76:58–62. doi: 10.1007/BF00283051. [DOI] [PubMed] [Google Scholar]
- Bräuer L, Kindler C, Jäger K, Sel S, Nölle B, Pleyer U, Ochs M, Paulsen FP. Detection of surfactant proteins A and D in human tear fluid and the human lacrimal system. Invest Ophthalmol Vis Sci. 2007;48:3945–53. doi: 10.1167/iovs.07-0201. [DOI] [PubMed] [Google Scholar]
- Bräuer L, Möschter S, Beileke S, Jäger K, Garreis F, Paulsen FP. Human parotid and submandibular glands express and secrete surfactant proteins A, B, C and D. Histochem Cell Biol. 2009;132:331–8. doi: 10.1007/s00418-009-0609-x. [DOI] [PubMed] [Google Scholar]
- Bräuer L, Schicht M, Stengl C, Heinemann F, Götz W, Scholz M, Paulsen F. Detection of surfactant proteins A, B, C, and D in human gingiva and saliva. Biomed Tech (Berl) 2012;57:59–64. doi: 10.1515/bmt-2011-0031. [DOI] [PubMed] [Google Scholar]
- Christie JD. Genetic epidemiology of acute lung injury: choosing the right candidate genes is the first step. Crit Care. 2004;8:411–3. doi: 10.1186/cc2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condon J, Jeyasuria P, Faust J, Mendelson CR. Surfactant protein secreted by the maturing mouse fetal lung acts as a hormone that signals the initiation of parturition. Proc Natl Acad Sci U S A. 2004;101:4978–4983. doi: 10.1073/pnas.0401124101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crouch E, Hartshorn K, Ofek I. Collectins and pulmonary innate immunity. Immunol Rev. 2000;173:52–65. doi: 10.1034/j.1600-065x.2000.917311.x. [DOI] [PubMed] [Google Scholar]
- Crouch E, Wright JR. Surfactant proteins A and D and pulmonary host defense. Annu Rev Physiol. 2001;63:521–54. doi: 10.1146/annurev.physiol.63.1.521. [DOI] [PubMed] [Google Scholar]
- Crouch EC. Collectins and pulmonary host defense. Am J Respir Cell Mol Biol. 1998;19:177–201. doi: 10.1165/ajrcmb.19.2.140. [DOI] [PubMed] [Google Scholar]
- deMello DE, Lin Z. Pulmonary alveolar proteinosis: a review. Pediatr Pathol Mol Med. 2001;20:413–32. [PubMed] [Google Scholar]
- DiAngelo S, Lin Z, Wang G, Phillips S, Ramet M, Luo J, Floros J. Novel, non-radioactive, simple and multiplex PCR-cRFLP methods for genotyping human SP-A and SP-D marker alleles. Dis Markers. 1999;15:269–81. doi: 10.1155/1999/961430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding J, Umstead TM, Floros J, Phelps DS. Factors affecting SP-A-mediated phagocytosis in human monocytic cell lines. Respir Med. 2004;98:637–50. doi: 10.1016/j.rmed.2003.12.018. [DOI] [PubMed] [Google Scholar]
- Floros J. Human surfactant protein A (SP-A) variants: why so many, why such a complexity? Swiss Med Wkly. 2001;131:87–90. doi: 10.4414/smw.2001.06136. [DOI] [PubMed] [Google Scholar]
- Floros J, Fan R, Matthews A, DiAngelo S, Luo J, Nielsen H, Dunn M, Gewolb IH, Koppe J, van Sonderen L, Farri-Kostopoulos L, Tzaki M, Ramet M, Merrill J. Family-based transmission disequilibrium test (TDT) and case-control association studies reveal surfactant protein A (SP-A) susceptibility alleles for respiratory distress syndrome (RDS) and possible race differences. Clin Genet. 2001;60:178–87. doi: 10.1034/j.1399-0004.2001.600303.x. [DOI] [PubMed] [Google Scholar]
- Floros J, Hoover RR. Genetics of the hydrophilic surfactant proteins A and D. Biochim Biophys Acta. 1998;1408:312–22. doi: 10.1016/s0925-4439(98)00077-5. [DOI] [PubMed] [Google Scholar]
- Floros J, Kala P. Surfactant proteins: molecular genetics of neonatal pulmonary diseases. Annu Rev Physiol. 1998;60:365–84. doi: 10.1146/annurev.physiol.60.1.365. [DOI] [PubMed] [Google Scholar]
- Floros J, Lin H, García A, Salazar M, Guo X, DiAngelo S, Montano M, Luo J, Pardo A, Selman M. Surfactant protein genetic marker alleles identify a subgroup of tuberculosis in a Mexican population. J Infect Dis. 2000;182:1473–8. doi: 10.1086/315866. [DOI] [PubMed] [Google Scholar]
- Floros J, Pavlovic J. Genetics of acute respiratory distress syndrome: challenges, approaches, surfactant proteins as candidate genes. Semin Respir Crit Care Med. 2003;24:161–8. doi: 10.1055/s-2003-39015. [DOI] [PubMed] [Google Scholar]
- Floros J, Phelps DS. Pulmonary surfactant. In: Yaksh TL, et al., editors. Anesthesia: Biologic Foundations. Lippincott-Raven Publishers; Philadelphia: 1997. pp. 1259–1280. [Google Scholar]
- Floros J, Thomas N. Research Signpost, K., India. Genetic variations of surfactant proteins and lung injury. In: Nakos G, Papathanasiou A, editors. Surfactant Pathogenesis and Treatment of Lung Diasease. 2009. pp. 25–48. [Google Scholar]
- Floros J, Wang G. A point of view: quantitative and qualitative imbalance in disease pathogenesis; pulmonary surfactant protein A genetic variants as a model. Comp Biochem Physiol A Mol Integr Physiol. 2001;129:295–303. doi: 10.1016/s1095-6433(01)00325-7. [DOI] [PubMed] [Google Scholar]
- Floros J, Wang G, Mikerov AN. Genetic complexity of the human innate host defense molecules, surfactant protein A1 (SP-A1) and SP-A2--impact on function. Crit Rev Eukaryot Gene Expr. 2009;19:125–37. doi: 10.1615/critreveukargeneexpr.v19.i2.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floros J, Wang G, Lin Z. Genetic diversity of human SP-A, a molecule with innate host defense and surfactan-related function; characteristics, primary function, and significance. Current Pharmacogenomics. 2005;3:87–95. [Google Scholar]
- Gao E, Wang Y, Alcorn JL, Mendelson CR. Transcription factor USF2 is developmentally regulated in fetal lung and acts together with USF1 to induce SP-A gene expression. Am J Physiol Lung Cell Mol Physiol. 2003;284:L1027–36. doi: 10.1152/ajplung.00219.2002. [DOI] [PubMed] [Google Scholar]
- Gao E, Wang Y, McCormick SM, Li J, Seidner SR, Mendelson CR. Characterization of two baboon surfactant protein A genes. Am J Physiol. 1996;271:L617–30. doi: 10.1152/ajplung.1996.271.4.L617. [DOI] [PubMed] [Google Scholar]
- Garcia-Verdugo I, Wang G, Floros J, Casals C. Structural analysis and lipid-binding properties of recombinant human surfactant protein a derived from one or both genes. Biochemistry. 2002;41:14041–53. doi: 10.1021/bi026540l. [DOI] [PubMed] [Google Scholar]
- Glasser SW, Burhans MS, Korfhagen TR, Na CL, Sly PD, Ross GF, Ikegami M, Whitsett JA. Altered stability of pulmonary surfactant in SP-C-deficient mice. Proc Natl Acad Sci U S A. 2001;98:6366–71. doi: 10.1073/pnas.101500298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigoryev DN, Finigan JH, Hassoun P, Garcia JG. Science review: searching for gene candidates in acute lung injury. Crit Care. 2004;8:440–7. doi: 10.1186/cc2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haagsman HP. Interactions of surfactant protein A with pathogens. Biochim Biophys Acta. 1998;1408:264–77. doi: 10.1016/s0925-4439(98)00072-6. [DOI] [PubMed] [Google Scholar]
- Hallman M, Haataja R. Genetic influences and neonatal lung disease. Semin Neonatol. 2003;8:19–27. doi: 10.1016/s1084-2756(02)00196-3. [DOI] [PubMed] [Google Scholar]
- Hawgood S, Poulain FR. The pulmonary collectins and surfactant metabolism. Annu Rev Physiol. 2001;63:495–519. doi: 10.1146/annurev.physiol.63.1.495. [DOI] [PubMed] [Google Scholar]
- Hogenkamp A, van Eijk M, van Dijk A, van Asten AJ, Veldhuizen EJ, Haagsman HP. Characterization and expression sites of newly identified chicken collectins. Mol Immunol. 2006;43:1604–16. doi: 10.1016/j.molimm.2005.09.015. [DOI] [PubMed] [Google Scholar]
- Hoover RR, Floros J. Organization of the human SP-A and SP-D loci at 10q22-q23. Physical and radiation hybrid mapping reveal gene order and orientation. Am J Respir Cell Mol Biol. 1998;18:353–62. doi: 10.1165/ajrcmb.18.3.3035. [DOI] [PubMed] [Google Scholar]
- Hoover RR, Floros J. SP-A 3′-UTR is involved in the glucocorticoid inhibition of human SP-A gene expression. Am J Physiol. 1999;276:L917–24. doi: 10.1152/ajplung.1999.276.6.L917. [DOI] [PubMed] [Google Scholar]
- Hoover RR, Pavlovic J, Floros J. Induction of AP-1 binding to intron 1 of SP-A1 and SP-A2 is implicated in the phorbol ester inhibition of human SP-A promoter activity. Exp Lung Res. 2000;26:303–17. doi: 10.1080/019021400408272. [DOI] [PubMed] [Google Scholar]
- Hoover RR, Thomas KH, Floros J. Glucocorticoid inhibition of human SP-A1 promoter activity in NCI-H441 cells. Biochem J. 1999;340 ( Pt 1):69–76. [PMC free article] [PubMed] [Google Scholar]
- Huang W, Wang G, Phelps DS, Al-Mondhiry H, Floros J. Human SP-A genetic variants and bleomycin-induced cytokine production by THP-1 cells: effect of ozone-induced SP-A oxidation. Am J Physiol Lung Cell Mol Physiol. 2004;286:L546–53. doi: 10.1152/ajplung.00267.2003. [DOI] [PubMed] [Google Scholar]
- Hughes AL. Evolution of the lung surfactant proteins in birds and mammals. Immunogenetics. 2007;59:565–72. doi: 10.1007/s00251-007-0218-6. [DOI] [PubMed] [Google Scholar]
- Ikegami M, Whitsett JA, Jobe A, Ross G, Fisher J, Korfhagen T. Surfactant metabolism in SP-D gene-targeted mice. Am J Physiol Lung Cell Mol Physiol. 2000;279:L468–76. doi: 10.1152/ajplung.2000.279.3.L468. [DOI] [PubMed] [Google Scholar]
- Islam KN, Mendelson CR. Potential role of nuclear factor kappaB and reactive oxygen species in cAMP and cytokine regulation of surfactant protein-A gene expression in lung type II cells. Mol Endocrinol. 2002;16:1428–40. doi: 10.1210/mend.16.6.0856. [DOI] [PubMed] [Google Scholar]
- Islam KN, Mendelson CR. Permissive effects of oxygen on cyclic AMP and interleukin-1 stimulation of surfactant protein A gene expression are mediated by epigenetic mechanisms. Mol Cell Biol. 2006;26:2901–12. doi: 10.1128/MCB.26.8.2901-2912.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam KN, Mendelson CR. Glucocorticoid/glucocorticoid receptor inhibition of surfactant protein-A (SP-A) gene expression in lung type II cells is mediated by repressive changes in histone modification at the SP-A promoter. Mol Endocrinol. 2008;22:585–96. doi: 10.1210/me.2007-0412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karinch AM, Deiter G, Ballard PL, Floros J. Regulation of expression of human SP-A1 and SP-A2 genes in fetal lung explant culture. Biochim Biophys Acta. 1998;1398:192–202. doi: 10.1016/s0167-4781(98)00047-5. [DOI] [PubMed] [Google Scholar]
- Karinch AM, Floros J. 5′ splicing and allelic variants of the human pulmonary surfactant protein A genes. Am J Respir Cell Mol Biol. 1995a;12:77–88. doi: 10.1165/ajrcmb.12.1.7811473. [DOI] [PubMed] [Google Scholar]
- Karinch AM, Floros J. Translation in vivo of 5′ untranslated-region splice variants of human surfactant protein-A. Biochem J. 1995b;307 ( Pt 2):327–30. doi: 10.1042/bj3070327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishore U, Bernal AL, Kamran MF, Saxena S, Singh M, Sarma PU, Madan T, Chakraborty T. Surfactant proteins SP-A and SP-D in human health and disease. Arch Immunol Ther Exp (Warsz) 2005;53:399–417. [PubMed] [Google Scholar]
- Kishore U, Greenhough TJ, Waters P, Shrive AK, Ghai R, Kamran MF, Bernal AL, Reid KB, Madan T, Chakraborty T. Surfactant proteins SP-A and SP-D: structure, function and receptors. Mol Immunol. 2006;43:1293–315. doi: 10.1016/j.molimm.2005.08.004. [DOI] [PubMed] [Google Scholar]
- Komar AA, Hatzoglou M. Cellular IRES-mediated translation: the war of ITAFs in pathophysiological states. Cell Cycle. 2011;10:229–40. doi: 10.4161/cc.10.2.14472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouretas D, Karinch AM, Rishi A, Melchers K, Floros J. Conservation analysis of rat and human SP-A gene identifies 5′ flanking sequences of rat SP-A that bind rat lung nuclear proteins. Exp Lung Res. 1993;19:485–503. doi: 10.3109/01902149309064359. [DOI] [PubMed] [Google Scholar]
- Krizkova L, Sakthivel R, Olowe S, Rogan P, Floros J. Human SP-A: genotype and single-strand conformation polymorphism analysis. Am J Physiol. 1994;266:L519–27. doi: 10.1152/ajplung.1994.266.5.L519. [DOI] [PubMed] [Google Scholar]
- Kumar AR, Snyder JM. Differential regulation of SP-A1 and SP-A2 genes by cAMP, glucocorticoids, and insulin. Am J Physiol. 1998;274:L177–85. doi: 10.1152/ajplung.1998.274.2.L177. [DOI] [PubMed] [Google Scholar]
- Lacaze-Masmonteil T, Fraslon C, Bourbon J, Raymondjean M, Kahn A. Characterization of the rat pulmonary surfactant protein A promoter. Eur J Biochem. 1992;206:613–23. doi: 10.1111/j.1432-1033.1992.tb16966.x. [DOI] [PubMed] [Google Scholar]
- Lam E, dos Santos CC. Advances in molecular acute lung injury/acute respiratory distress syndrome and ventilator-induced lung injury: the role of genomics, proteomics, bioinformatics and translational biology. Curr Opin Crit Care. 2008;14:3–10. doi: 10.1097/MCC.0b013e3282f42211. [DOI] [PubMed] [Google Scholar]
- Leikauf GD, McDowell SA, Wesselkamper SC, Hardie WD, Leikauf JE, Korfhagen TR, Prows DR. Acute lung injury: functional genomics and genetic susceptibility. Chest. 2002;121:70S–75S. doi: 10.1378/chest.121.3_suppl.70s. [DOI] [PubMed] [Google Scholar]
- LeVine AM, Whitsett JA. Pulmonary collectins and innate host defense of the lung. Microbes Infect. 2001;3:161–6. doi: 10.1016/s1286-4579(00)01363-0. [DOI] [PubMed] [Google Scholar]
- LeVine AM, Whitsett JA, Gwozdz JA, Richardson TR, Fisher JH, Burhans MS, Korfhagen TR. Distinct effects of surfactant protein A or D deficiency during bacterial infection on the lung. J Immunol. 2000;165:3934–40. doi: 10.4049/jimmunol.165.7.3934. [DOI] [PubMed] [Google Scholar]
- Li J, Gao E, Mendelson CR. Cyclic AMP-responsive expression of the surfactant protein-A gene is mediated by increased DNA binding and transcriptional activity of thyroid transcription factor-1. J Biol Chem. 1998;273:4592–600. doi: 10.1074/jbc.273.8.4592. [DOI] [PubMed] [Google Scholar]
- Lin Z, Thomas NJ, Bibikova M, Seifart C, Wang Y, Guo X, Wang G, Vollmer E, Goldmann T, Garcia EW, Zhou L, Fan JB, Floros J. DNA methylation markers of surfactant proteins in lung cancer. Int J Oncol. 2007;31:181–91. [PubMed] [Google Scholar]
- Liu D, Benlhabib H, Mendelson CR. cAMP enhances estrogen-related receptor alpha (ERRalpha) transcriptional activity at the SP-A promoter by increasing its interaction with protein kinase A and steroid receptor coactivator 2 (SRC-2) Mol Endocrinol. 2009;23:772–83. doi: 10.1210/me.2008-0282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Hinshelwood MM, Giguère V, Mendelson CR. Estrogen related receptor-alpha enhances surfactant protein-A gene expression in fetal lung type II cells. Endocrinology. 2006;147:5187–95. doi: 10.1210/en.2006-0664. [DOI] [PubMed] [Google Scholar]
- Liu D, Yi M, Smith M, Mendelson CR. TTF-1 response element is critical for temporal and spatial regulation and necessary for hormonal regulation of human surfactant protein-A2 promoter activity. Am J Physiol Lung Cell Mol Physiol. 2008;295:L264–71. doi: 10.1152/ajplung.00069.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Bentley CM, Floros J. Study of human SP-A, SP-B and SP-D loci: allele frequencies, linkage disequilibrium and heterozygosity in different races and ethnic groups. BMC Genet. 2003;4:13. doi: 10.1186/1471-2156-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacNeill C, Umstead TM, Phelps DS, Lin Z, Floros J, Shearer DA, Weisz J. Surfactant protein A, an innate immune factor, is expressed in the vaginal mucosa and is present in vaginal lavage fluid. Immunology. 2004;111:91–9. doi: 10.1111/j.1365-2567.2003.01782.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik S, Greenwood CM, Eguale T, Kifle A, Beyene J, Habte A, Tadesse A, Gebrexabher H, Britton S, Schurr E. Variants of the SFTPA1 and SFTPA2 genes and susceptibility to tuberculosis in Ethiopia. Hum Genet. 2006;118:752–9. doi: 10.1007/s00439-005-0092-y. [DOI] [PubMed] [Google Scholar]
- McCormick SM, Boggaram V, Mendelson CR. Characterization of mRNA transcripts and organization of human SP-A1 and SP-A2 genes. Am J Physiol. 1994;266:L354–66. doi: 10.1152/ajplung.1994.266.4.L354. [DOI] [PubMed] [Google Scholar]
- McNeely TB, Coonrod JD. Aggregation and opsonization of type A but not type B Hemophilus influenzae by surfactant protein A. Am J Respir Cell Mol Biol. 1994;11:114–22. doi: 10.1165/ajrcmb.11.1.8018334. [DOI] [PubMed] [Google Scholar]
- Mendelson CR, Gao E, Li J, Young PP, Michael LF, Alcorn JL. Regulation of expression of surfactant protein-A. Biochim Biophys Acta. 1998;1408:132–49. doi: 10.1016/s0925-4439(98)00063-5. [DOI] [PubMed] [Google Scholar]
- Meyer NJ, Garcia JG. Wading into the genomic pool to unravel acute lung injury genetics. Proc Am Thorac Soc. 2007;4:69–76. doi: 10.1513/pats.200609-157JG. [DOI] [PubMed] [Google Scholar]
- Meyers DA. Genetics of asthma and allergy: what have we learned? J Allergy Clin Immunol. 2010;126:439–46. doi: 10.1016/j.jaci.2010.07.012. quiz 447–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikerov A, Umstead T, Gan X, Huang W, Guo X, Wang G, Phelps D, Floros J. Impact of ozone exposure on the phagocytic activity of human surfactant protein A (SP-A) and SP-A variants. Am J Physiol Lung Cell Mol Physiol. 2008a;294:L121–30. doi: 10.1152/ajplung.00288.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikerov A, Wang G, Umstead T, Zacharatos M, Thomas N, Phelps D, Floros J. Surfactant protein A2 (SP-A2) variants expressed in CHO cells stimulate phagocytosis of Pseudomonas aeruginosa more than do SP-A1 variants. Infect Immun. 2007;75:1403–12. doi: 10.1128/IAI.01341-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikerov A, White M, Hartshorn K, Wang G, Floros J. Inhibition of hemagglutination activity of influenza A viruses by SP-A1 and SP-A2 variants expressed in CHO cells. Med Microbiol Immunol. 2008b;197:9–12. doi: 10.1007/s00430-007-0051-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikerov AN, Haque R, Gan X, Guo X, Phelps DS, Floros J. Ablation of SPA has a negative impact on the susceptibility of mice to Klebsiella pneumoniae infection after ozone exposure: sex differences. Respir Res. 2008c;9:77. doi: 10.1186/1465-9921-9-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikerov AN, Umstead TM, Huang W, Liu W, Phelps D, Floros J. SP-A1 and SP-A2 variants differentially enhance association of Pseudomonas aeruginosa with rat alveolar macrophages. Am J Physiol Lung Cell Mol Physiol. 2005;288:L150–L158. doi: 10.1152/ajplung.00135.2004. [DOI] [PubMed] [Google Scholar]
- Nogee LM. Alterations in SP-B and SP-C expression in neonatal lung disease. Annu Rev Physiol. 2004;66:601–23. doi: 10.1146/annurev.physiol.66.032102.134711. [DOI] [PubMed] [Google Scholar]
- Nonas SA, Finigan JH, Gao L, Garcia JG. Functional genomic insights into acute lung injury: role of ventilators and mechanical stress. Proc Am Thorac Soc. 2005;2:188–94. doi: 10.1513/pats.200501-005AC. [DOI] [PubMed] [Google Scholar]
- Oberley RE, Snyder JM. Recombinant human SP-A1 and SP-A2 proteins have different carbohydrate-binding characteristics. Am J Physiol Lung Cell Mol Physiol. 2003;284:L871–81. doi: 10.1152/ajplung.00241.2002. [DOI] [PubMed] [Google Scholar]
- Paananen R, Glumoff V, Sormunen R, Voorhout W, Hallman M. Expression and localization of lung surfactant protein B in Eustachian tube epithelium. Am J Physiol Lung Cell Mol Physiol. 2001;280:L214–20. doi: 10.1152/ajplung.2001.280.2.L214. [DOI] [PubMed] [Google Scholar]
- Perez-Gil J, Weaver TE. Pulmonary surfactant pathophysiology: current models and open questions. Physiology (Bethesda) 2010;25:132–41. doi: 10.1152/physiol.00006.2010. [DOI] [PubMed] [Google Scholar]
- Phelps DS. Surfactant regulation of host defense function in the lung: a question of balance. Pediatr Pathol Mol Med. 2001;20:269–92. [PubMed] [Google Scholar]
- Postle AD, Heeley EL, Wilton DC. A comparison of the molecular species compositions of mammalian lung surfactant phospholipids. Comp Biochem Physiol A Mol Integr Physiol. 2001;129:65–73. doi: 10.1016/s1095-6433(01)00306-3. [DOI] [PubMed] [Google Scholar]
- Reddy AJ, Kleeberger SR. Genetic polymorphisms associated with acute lung injury. Pharmacogenomics. 2009;10:1527–39. doi: 10.2217/pgs.09.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg E, Li F, Reisher SR, Wang M, Gonzales LW, Ewing JR, Malek S, Ballard PL, Notarfrancesco K, Shuman H, Feinstein SI. Members of the C/EBP transcription factor family stimulate expression of the human and rat surfactant protein A (SP-A) genes. Biochim Biophys Acta. 2002;1575:82–90. doi: 10.1016/s0167-4781(02)00287-7. [DOI] [PubMed] [Google Scholar]
- Rosenberg E, Li F, Smith CI, Reisher SR, Feinstein SI. Transcriptional activation and protein binding by two regions of the rat surfactant protein A promoter. Am J Physiol. 1999;277:L134–41. doi: 10.1152/ajplung.1999.277.1.L134. [DOI] [PubMed] [Google Scholar]
- Rubio S, Lacaze-Masmonteil T, Chailley-Heu B, Kahn A, Bourbon JR, Ducroc R. Pulmonary surfactant protein A (SP-A) is expressed by epithelial cells of small and large intestine. J Biol Chem. 1995;270:12162–9. doi: 10.1074/jbc.270.20.12162. [DOI] [PubMed] [Google Scholar]
- Rämet M, Haataja R, Marttila R, Floros J, Hallman M. Association between the surfactant protein A (SP-A) gene locus and respiratory-distress syndrome in the Finnish population. Am J Hum Genet. 2000;66:1569–79. doi: 10.1086/302906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schagat TL, Wofford JA, Wright JR. Surfactant protein A enhances alveolar macrophage phagocytosis of apoptotic neutrophils. J Immunol. 2001;166:2727–33. doi: 10.4049/jimmunol.166.4.2727. [DOI] [PubMed] [Google Scholar]
- Silveyra P, Floros J. Air pollution and epigenetics: effects on SP-A and innate host defence in the lung. Swiss Med Wkly. 2012a;142:w13579. doi: 10.4414/smw.2012.13579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silveyra P, Floros J. Genetic variant associations of human SP-A and SP-D with acute and chronic lung injury. Front Biosci. 2012b;17:407–29. doi: 10.2741/3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silveyra P, Raval M, Simmons BP, Diangelo S, Wang G, Floros J. The Untranslated Exon B of Human Surfactant Protein A2 (SFTPA2) mRNAs is an Enhancer for Transcription and Translation. Am J Physiol Lung Cell Mol Physiol. 2011 doi: 10.1152/ajplung.00439.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silveyra P, Wang G, Floros J. Human SP-A1 (SFTPA1) variant-specific 3′ UTRs and poly(A) tail differentially affect the in vitro translation of a reporter gene. Am J Physiol Lung Cell Mol Physiol. 2010;299:L523–34. doi: 10.1152/ajplung.00113.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snegovskikh VV, Bhandari V, Wright JR, Tadesse S, Morgan T, Macneill C, Foyouzi N, Park JS, Wang Y, Norwitz ER. Surfactant protein-A (SP-A) selectively inhibits prostaglandin F2alpha (PGF2alpha) production in term decidua: implications for the onset of labor. J Clin Endocrinol Metab. 2011;96:E624–32. doi: 10.1210/jc.2010-1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Barbero F, Rivas G, Steinhilber W, Casals C. Structural and functional differences among human surfactant proteins SP-A1, SP-A2 and co-expressed SP-A1/SP-A2: role of supratrimeric oligomerization. Biochem J. 2007;406:479–89. doi: 10.1042/BJ20070275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagaram H, Wang G, Umstead T, Mikerov A, Thomas N, Graff G, Hess J, Thomassen M, Kavuru M, Phelps D, Floros J. Characterization of a human surfactant protein A1 (SP-A1) gene-specific antibody; SP-A1 content variation among individuals of varying age and pulmonary health. Am J Physiol Lung Cell Mol Physiol. 2007;292:L1052–63. doi: 10.1152/ajplung.00249.2006. [DOI] [PubMed] [Google Scholar]
- Vaid M, Floros J. Surfactant protein DNA methylation: a new entrant in the field of lung cancer diagnostics? (Review) Oncol Rep. 2009;21:3–11. [PMC free article] [PubMed] [Google Scholar]
- Villar J, Flores C, Mendez-Alvarez S. Genetic susceptibility to acute lung injury. Crit Care Med. 2003;31:S272–5. doi: 10.1097/01.CCM.0000057903.11528.6D. [DOI] [PubMed] [Google Scholar]
- Vlachaki EM, Koutsopoulos AV, Tzanakis N, Neofytou E, Siganaki M, Drositis I, Moniakis A, Schiza S, Siafakas NM, Tzortzaki EG. Altered surfactant protein-A expression in type II pneumocytes in COPD. Chest. 2010;137:37–45. doi: 10.1378/chest.09-1029. [DOI] [PubMed] [Google Scholar]
- Voss T, Melchers K, Scheirle G, Schäfer KP. Structural comparison of recombinant pulmonary surfactant protein SP-A derived from two human coding sequences: implications for the chain composition of natural human SP-A. Am J Respir Cell Mol Biol. 1991;4:88–94. doi: 10.1165/ajrcmb/4.1.88. [DOI] [PubMed] [Google Scholar]
- Wang G, Bates-Kenney SR, Tao JQ, Phelps DS, Floros J. Differences in biochemical properties and in biological function between human SP-A1 and SP-A2 variants, and the impact of ozone-induced oxidation. Biochemistry. 2004;43:4227–39. doi: 10.1021/bi036023i. [DOI] [PubMed] [Google Scholar]
- Wang G, Guo X, Diangelo S, Thomas N, Floros J. Humanized SFTPA1 and SFTPA2 transgenic mice reveal functional divergence of SP-A1 and SP-A2: Formation of tubular myelin in vivo requires both gene products. J Biol Chem. 2010 doi: 10.1074/jbc.M109.046243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Guo X, Floros J. Human SP-A 3′-UTR variants mediate differential gene expression in basal levels and in response to dexamethasone. Am J Physiol Lung Cell Mol Physiol. 2003;284:L738–48. doi: 10.1152/ajplung.00375.2002. [DOI] [PubMed] [Google Scholar]
- Wang G, Guo X, Floros J. Differences in the translation efficiency and mRNA stability mediated by 5′-UTR splice variants of human SP-A1 and SP-A2 genes. Am J Physiol Lung Cell Mol Physiol. 2005;289:L497–508. doi: 10.1152/ajplung.00100.2005. [DOI] [PubMed] [Google Scholar]
- Wang G, Guo X, Silveyra P, Kimball S, Floros J. Cap-independent translation of human SP-A 5′-UTR variants: a double-loop structure and cis-element contribution. Am J Physiol Lung Cell Mol Physiol. 2009;296:L635–47. doi: 10.1152/ajplung.90508.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Myers C, Mikerov A, Floros J. Effect of cysteine 85 on biochemical properties and biological function of human surfactant protein A variants. Biochemistry. 2007a;46:8425–35. doi: 10.1021/bi7004569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Phelps D, Umstead T, Floros J. Human SP-A protein variants derived from one or both genes stimulate TNF-alpha production in the THP-1 cell line. Am J Physiol Lung Cell Mol Physiol. 2000;278:L946–54. doi: 10.1152/ajplung.2000.278.5.L946. [DOI] [PubMed] [Google Scholar]
- Wang G, Taneva S, Keough K, Floros J. Differential effects of human SP-A1 and SP-A2 variants on phospholipid monolayers containing surfactant protein B. Biochim Biophys Acta. 2007b;1768:2060–9. doi: 10.1016/j.bbamem.2007.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Umstead T, Phelps D, Al-Mondhiry H, Floros J. The effect of ozone exposure on the ability of human surfactant protein a variants to stimulate cytokine production. Environ Health Perspect. 2002;110:79–84. doi: 10.1289/ehp.0211079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Reid K. The immunoregulatory roles of lung surfactant collectins SP-A, and SP-D, in allergen-induced airway inflammation. Immunobiology. 2007;212:417–25. doi: 10.1016/j.imbio.2007.01.002. [DOI] [PubMed] [Google Scholar]
- Wang Y, Voelker DR, Lugogo NL, Wang G, Floros J, Ingram JL, Chu HW, Church TD, Kandasamy P, Fertel D, Wright JR, Kraft M. Surfactant Protein-A is Defective in Abrogating Inflammation in Asthma. Am J Physiol Lung Cell Mol Physiol. 2011 doi: 10.1152/ajplung.00381.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Gurel O, Baatz JE, Notter RH. Differential activity and lack of synergy of lung surfactant proteins SP-B and SP-C in interactions with phospholipids. J Lipid Res. 1996;37:1749–60. [PubMed] [Google Scholar]
- Whitsett JA, Wert SE, Weaver TE. Alveolar surfactant homeostasis and the pathogenesis of pulmonary disease. Annu Rev Med. 2010;61:105–19. doi: 10.1146/annurev.med.60.041807.123500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams MC, Hawgood S, Hamilton RL. Changes in lipid structure produced by surfactant proteins SP-A, SP-B, and SP-C. Am J Respir Cell Mol Biol. 1991;5:41–50. doi: 10.1165/ajrcmb/5.1.41. [DOI] [PubMed] [Google Scholar]
- Wright JR. Immunoregulatory functions of surfactant proteins. Nat Rev Immunol. 2005;5:58–68. doi: 10.1038/nri1528. [DOI] [PubMed] [Google Scholar]
- Wu H, Kuzmenko A, Wan S, Schaffer L, Weiss A, Fisher JH, Kim KS, McCormack FX. Surfactant proteins A and D inhibit the growth of Gram-negative bacteria by increasing membrane permeability. J Clin Invest. 2003;111:1589–602. doi: 10.1172/JCI16889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu TT, Chen TL, Loon WS, Tai YT, Cherng YG, Chen RM. Lipopolysaccharide stimulates syntheses of toll-like receptor 2 and surfactant protein-A in human alveolar epithelial A549 cells through upregulating phosphorylation of MEK1 and ERK1/2 and sequential activation of NF-κB. Cytokine. 2011;55:40–7. doi: 10.1016/j.cyto.2011.03.005. [DOI] [PubMed] [Google Scholar]
- Yadav AK, Madan T, Bernal AL. Surfactant proteins A and D in pregnancy and parturition. Front Biosci (Elite Ed) 2011;3:291–300. doi: 10.2741/e244. [DOI] [PubMed] [Google Scholar]
- Yi M, Tong GX, Murry B, Mendelson CR. Role of CBP/p300 and SRC-1 in transcriptional regulation of the pulmonary surfactant protein-A (SP-A) gene by thyroid transcription factor-1 (TTF-1) J Biol Chem. 2002;277:2997–3005. doi: 10.1074/jbc.M109793200. [DOI] [PubMed] [Google Scholar]
- Young PP, Mendelson CR. A CRE-like element plays an essential role in cAMP regulation of human SP-A2 gene in alveolar type II cells. Am J Physiol. 1996;271:L287–99. doi: 10.1152/ajplung.1996.271.2.L287. [DOI] [PubMed] [Google Scholar]
- Young PP, Mendelson CR. A GT box element is essential for basal and cyclic adenosine 3′,5′-monophosphate regulation of the human surfactant protein A2 gene in alveolar type II cells: evidence for the binding of lung nuclear factors distinct from Sp1. Mol Endocrinol. 1997;11:1082–93. doi: 10.1210/mend.11.8.9950. [DOI] [PubMed] [Google Scholar]


