Abstract
We have developed dual recombinase-mediated cassette exchange (dRMCE) to efficiently re-engineer the thousands of available conditional alleles in mouse embryonic stem cells. dRMCE takes advantage of the wild-type loxP and FRT sites present in these conditional alleles and in many gene-trap lines. dRMCE is a scalable, flexible tool to introduce tags, reporters and mutant coding regions into an endogenous locus of interest in an easy and highly efficient manner.
Gene targeting by homologous recombination in mouse embryonic stem cells is a powerful tool for tailored manipulation of the mouse genome. However, the efficiency of this technology is limited by the great variation in targeting frequencies for different loci1. In particular, targeting frequencies for a substantial number of loci are very low (<1%), which renders repeated genetic manipulation tedious, especially as the screening strategy usually involves Southern blotting or long-range PCR. In light of the increasing need to re-engineer more subtle mutations at the same locus, recombinase-mediated cassette exchange (RMCE; reviewed in ref. 2) was developed to facilitate this process. RMCE takes advantage of pairs of heterotypic, noninteracting recombination sites for a particular site-specific recombinase, but these sites first need to be introduced into the locus of interest through conventional homologous recombination. Subsequently, co-transfection of such modified mouse embryonic stem cells with the custom-designed RMCE exchange vector and the appropriate recombinase expression plasmid will result in replacement at the endogenous locus2.
It was previously established that coexpression of both Cre and Flp recombinases catalyzes the exchange of sequences flanked by single loxP and FRT sites (their respective standard target sites) integrated into the genome at a random location3. However, this study did not explore whether such an approach could be used to modify conditional mouse alleles carrying single or multiple loxP and FRT sites. Therefore, we decided to develop dual RMCE (dRMCE) as a re-engineering tool applicable to the vast numbers of mouse conditional alleles that harbor wild-type loxP and FRT sites and therefore are not compatible with conventional RMCE. In particular, dRMCE will expand the long-term value of the rapidly increasing collection of targeted alleles produced by the International Knockout Mouse Consortium (IKMC; http://www.knockoutmouse.org/). This large-scale program is generating ‘knockout-first’ conditional alleles for most protein-coding genes in mouse embryonic stem cells, and several thousand alleles are already available4.
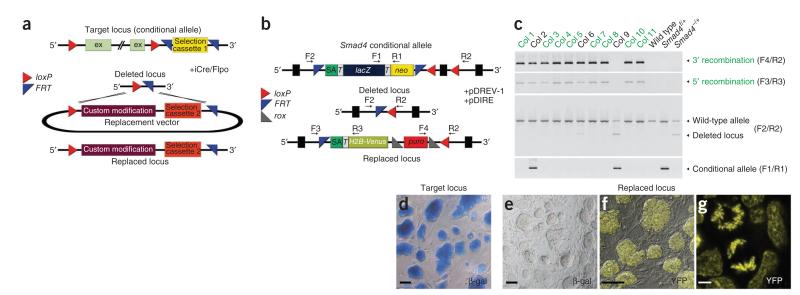
The general dRMCE strategy takes advantage of the fact that most conditional alleles encode a selection cassette flanked by FRT sites, in addition to loxP sites that flank functionally relevant exons (‘floxed’ exons; Fig. 1a). The FRT-flanked selection cassette is in general placed outside the loxP-flanked region, which renders these alleles directly compatible with dRMCE. Simultaneous expression of Cre and Flp recombinases should rapidly induce cis recombination and formation of the deleted allele, which would then serve as a ‘docking site’ at which to insert the replacement vector by trans recombination (Fig. 1a). The correctly replaced locus would encode the custom modification and a different drug-selection cassette flanked by single loxP and FRT sites.
Figure 1.
The principle of dRMCE to re-engineer mouse conditional alleles. (a) Schematic of the target locus shows the configuration of a conditional mouse allele with a genomic region flanked by two loxP sites and an outside selection cassette flanked by two FRT sites. Upon transfection, the combination of iCre- and Flpo-mediated recombination in cis results in a deleted allele flanked by single loxP and FRT sites, which serves as a ‘docking site’ for insertion of the replacement vector. ex, exon. (b) Schematic representation of replacement in the Smad4 locus by dRMCE. The target locus is a Smad4 conditional allele (Smad4f) with a promoterless selection cassette. Co-transfection of the pDIRE and pDREV-1 plasmids induces replacement, probably through production of the Smad4− deleted allele as intermediate. Correct trans insertion of the replacement vector results in the Smad4YFP allele. F1–F4 and R1–R3 denote primers used for PCR screening of colonies (see supplementary table 2 for sequences). H2B-Venus, YFP fusion protein with histone 2B; lacZ, β-galactosidase coding region; neo, neomycin resistance coding region; puro, puromycin resistance cassette; rox, Dre recombinase target sites11; SA, splice acceptor; T, autocleavable T2A peptide coding region14. (c) PCR screening reveals a large number of clones with correct 3′ and 5′ replacement (69%). Col, colony; 3′ recombination, 5′ recombination, correct replacement at the 3′ and 5′ ends, respectively. (d) The parental Smad4f cells are β-galactosidase positive. (e,f) Clones with correct replacement (Smad4YFP) lack β-galactosidase activity but show YFP fluorescence. (g) Micrograph shows single cells expressing the H2B-Venus fusion protein engineered by dRMCE. Scale bars: 100 μm (d–f), 5 μm (g).
In developing dRMCE, we used optimized versions of the Cre (iCre)5 and Flp (Flpo)6 recombinases to assemble the pDIRE expression vector (see Supplementary Fig. 1a and Supplementary Data for complete vector sequences). We also developed the pDREV vector series (Supplementary Fig. 1b and Supplementary Data) that allows insertion and efficient expression of reporters and/or coding regions of choice in any IKMC knockout-first allele. We tested the feasibility of dRMCE-mediated replacement using loci representing these readily available knock-out-first alleles as well as standard conditional loss-of-function mutations generated by individual researchers.
The floxed Smad4 (Smad4f) knockout-first allele generated by the IKMC contains a promoterless gene-trap selection cassette (lacZ-T2A-neo) that is flanked by FRT sites and followed by loxP sites flanking a critical Smad4 coding exon (Fig. 1b). This results in expression of a lacZ reporter and the neomycin resistance (neo) genes under control of the endogenous Smad4 promoter, which is active in embryonic stem cells. Heterozygous Smad4f mouse embryonic stem cells were co-transfected with the pDIRE and pDREV-1 vectors, the latter of which encodes an H2B-Venus YFP reporter (Fig. 1b). Puromycin-resistant colonies were screened by short-range PCR at the 3′ (loxP) and the 5′ (FRT) junctions for correct replacement events, which resulted in the YFP-tagged Smad4 allele (Smad4YFP; Fig. 1c). Most of the colonies were correctly replaced and of clonal origin (69%: 33 out of 48 clones; Supplementary Table 1). In contrast to the β-galactosidase–positive Smad4f cells (Fig. 1d), Smad4YFP colonies have lost the lacZ reporter and have become YFP positive (Fig. 1e,f). The H2B-Venus fusion protein appeared to be functional, as it was nuclear and bound to chromosomes during mitosis (Fig. 1g)7.
A small number of colonies were mixed, as judged by PCR analysis (Fig. 1c and Supplementary Table 1). These mixed colonies were easily recognized because they were composed of cells positive for either β-galactosidase activity (Smad4f allele) or YFP fluorescence (Smad4YFP allele) but not both (Supplementary Fig. 2). Some other mixed colonies contained cells carrying the Smad4 loss-of-function allele (Smad4−; Fig. 1c and Supplementary Fig. 2). This analysis indicated that mixed colonies originate as a consequence of incomplete recombination. Therefore, dRMCE-mediated replacement must always be validated by confirming the absence of both the floxed and the deleted allele.
The floxed Zp503 (Zfp503f) knockout-first allele from the IKMC encodes a promoter-driven selection cassette and three loxP and two FRT sites (Supplementary Fig. 3a). We co-transfected recipient heterozygous Zfp503f mouse embryonic stem cells with the pDIRE and pDREV-0 plasmids and selected colonies on the basis of their resistance to puromycin. Generation of the correctly replaced YFP-tagged Zfp503 (Zfp503YFP) allele was again highly efficient (52%; Supplementary Fig. 3b and Supplementary Table 1).
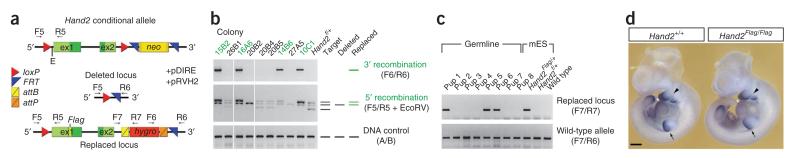
To establish dRMCE as a universally applicable method for re-engineering of conditional alleles, we also targeted two other loci. We first selected the Hand2 locus, as it is very difficult to target by homologous recombination (0.17%)8, and chose to use dRMCE to introduce a Flag epitope tag into the endogenous Hand2 coding region (Hand2Flag). To this end, we co-transfected mouse embryonic stem cells heterozygous for the floxed Hand2 (Hand2f) allele with the pDIRE and a pDRAV-type Hand2Flag replacement vectors (Fig. 2a and Supplementary Fig. 1). Molecular analysis revealed that 13% of all drug-resistant colonies had undergone correct replacement (Fig. 2b and Supplementary Table 1). Second, we carried out dRMCE-mediated replacement of a Gli3 conditional allele (J.L.-R. and R.Z., unpublished data) with the Hand2Flag vector, resulting in 33% correct replacement (Supplementary Table 1 and data not shown). These results indicate that dRMCE permits highly efficient re-engineering of conditional alleles with wild-type loxP and FRT sites in one step, irrespective of the difficulty of targeting the parental locus by homologous recombination.
figure 2.
dRMCE for efficient modification of difficult-to-target loci. (a) Schematic shows the conditional Hand2 allele (Hand2f) used as a target locus for insertion of a Flag epitope tag into the Hand2 coding region. After co-transfection of the replacement vector (pRVH2) and pDIRE plasmid into heterozygous Hand2f mouse embryonic stem cells, dRMCE-mediated replacement results in the Hand2Flag allele. The PGK-hygro (hygromycin resistance gene) selection cassette is flanked by the attB and attP target sites for excision by the ɸC31 recombinase. F5–F7 and R5–R7 denote primers used for PCR screening and genotyping. E, EcoRV site required to detect correct 5′ replacement by combining PCR amplification with an EcoRV restriction digestion. (b) PCR screening at both ends of the locus identified Hand2 colonies with correct replacement (13%). Scheme at right shows PCR fragment patterns indicative of particular genomic configurations. 3′ recombination, 5′ recombination, correct replacement at the 3′ and 5′ ends, respectively. A, B, primers to amplify a region serving as positive control (see supplementary table 2 for sequences). (c) Gels show germline transmission of the Hand2Flag allele (lanes 1, 5, 6). Above, PCR analysis to detect Hand2Flag allele. Below, PCR detection of wild-type allele. (d) In situ detection of Hand2 transcripts in a wild-type (Hand2+/+; left) and Hand2Flag/Flag (right) mouse embryo at embryonic day 10.5. Note expression in the posterior limb bud mesenchyme (arrow), branchial arches (black arrowhead) and heart (white arrowhead). Scale bar, 500 μm.
To assess the potential effects of dRMCE on germline transmission, we injected two correctly engineered Hand2Flag mouse embryonic stem cell clones (Supplementary Fig. 4) separately into mouse blastocysts, which resulted in several highly chimeric offspring in each case (80–100% as judged by coat color). These chimeric males transmitted the Hand2Flag allele to their F1 progeny within the first two litters (Fig. 2c). These results indicated that dRMCE neither compromises germline transmission potential nor causes frequent chromosomal abnormalities, as judged by karyotyping of all clones9 before injection (data not shown). Furthermore, the mouse embryonic stem cells used by the IKMC to generate their conditional alleles have been proven to remain highly germline competent even after multiple rounds of gene targeting and exposure to site-specific recombinases10.
Most notably, dRMCE-mediated engineering did not alter the temporal and spatial expression of Hand2 transcripts in mouse embryos homozygous for the Hand2Flag allele (Fig. 2d and Supplementary Fig. 5). The Hand2Flag allele was fully functional, as homozygous embryos and mice were phenotypically wild type, which contrasts with the lethality of Hand2-deficient mouse embryos8. In summary, dRMCE allowed re-engineering of conventional targeted alleles with frequencies of 10–70% correct replacement (Table 1). Minimally, this represented a 5- to 65-fold increase in efficiency in comparison to homologous recombination, which is particularly beneficial for difficult to target loci. Even at the lowest efficiency (13% for Hand2, Table 1), very few colonies needed to be analyzed to identify correctly replaced clones (for example, one 48-well plate of colonies).
Table 1.
dRMCE allows re-engineering of different loci at frequencies much higher than homologous recombination
| Gene locus | dRMCE | Homologous recombination | Fold increase |
|---|---|---|---|
| Smad4 | 69% | 6%15 | 12× |
| Zfp503 | 52% | 11%a | 5× |
| Hand2 | 13% | 0.2%8 | 65× |
| Gli3 | 33% | 3%b | 11× |
IKMC.
Unpublished.
To facilitate the generation of dRMCE targeting vectors for the large number of alleles containing both loxP and FRT sites (Fig. 1a), we prepared a tool kit consisting of the pDIRE expression vector and various dRMCE vectors (Supplementary Fig. 1 and Supplementary Data). In all cases, the selection cassette can be removed either in cells or directly in mice by breeding them to Dre11 or ɸC31o6 deleter mouse strains. The vector backbones allow the use of validated vector-specific primers for PCR screening in combination with locus-specific primers (Supplementary Table 2).
Taken together, our data indicate that dRMCE is a highly efficient and universal tool for re-engineering of conditional mouse alleles. dRMCE also provides a one-step alternative to the Floxin strategy12 recently developed for gene trap lines available from the International Gene Trap Consortium (IGTC; http://www.genetrap.org/)13. dRMCE is conceptually straightforward and allows a wide range of precise genomic modifications to be engineered. For example, dRMCE can be used to express mutant gene products and introduce epitope or fluorescent tags or heterologous genes into the endogenous locus of choice. Because of its efficiency, the dRMCE technology is well suited for high-throughput approaches such as functional screening of disease-causing mutations in pathways or genes of interest directly in mouse embryonic stem cells (for example, upon induced differentiation into specific cell types) or in mice generated from engineered clones. Finally, dRMCE technology provides nonspecialists with access to advanced mouse genetic engineering and enhances the long-term value of the existing large collections of genetically altered mouse embryonic stem cell lines.
METHODS
Methods and any associated references are available in the online version of the paper at http://www.nature.com/naturemethods/.
ONLINE METHODS
dRMCE protocols and plasmids
The step-by-step protocols used for dRMCE are available online through the Nature Protocols Network (http://www.natureprotocols.com/). All dRMCE plasmids are available from the Addgene repository (http://www.addgene.org/).
Construction of the pDIRE expression vector
The iCre coding sequence was amplified by PCR from the pBOB-CAG-iCre-SD plasmid (Addgene) using primers with specific restriction sites. After digestion with SalI and NotI, the iCre fragment was cloned into pBluescript IIKS (Stratagene). The human EF1A promoter was later inserted 5′ as a HindIII-BamHI fragment derived from the pBS513 EF1alpha-Cre plasmid (Addgene). The SV40 polyadenylation (SV40 pA) site was inserted as a SpeI-SpeI fragment after PCR amplification from the pEGFP-N1 plasmid. These cloning steps resulted in the pEF1α-iCre cassette, which was completely sequenced. This iCre expression unit was isolated as an EcoRV-EcoRV fragment and inserted into the PsiI site of the pPGKFlpobpA plasmid (Addgene) to generate the pDIRE expression vector (Supplementary Fig. 1a and Supplementary Data). To prevent potential promoter competition, the pDIRE vector was designed such that iCre is expressed under the control of the EF1A promoter, while the expression of Flpo is controlled by the PGK promoter.
Construction of the pDREV replacement vector series
The H2B-Venus fusion protein was selected as a versatile and sensitive reporter because Venus is one of the brightest and best validated fluorescent proteins and the fusion of Venus with H2B7 allows tracking of individual H2B-expressing cells in vivo. A 1.75-kb DNA fragment encoding H2B-Venus downstream of the autocleavable T2A peptide14 and upstream of the SV40 pA site and the rox, XhoI, rox and loxP sites was synthesized by GeneArt and cloned as a BglII-HindIII fragment into the pL1L2_GT vector series (B.R. and W.C.S., unpublished data) engineered in all three reading frames (Supplementary Fig. 1b). The PGK-puro selection cassette was excised as a SalI restriction fragment from the pPGKpuro plasmid (Addgene) and inserted into the XhoI site of the L1L2-gt-H2B-Venus plasmid series. This resulted in the definitive pDREV replacement vector collection (pDREV-0, pDREV-1 and pDREV-2), which is compatible with all three open reading frames (Supplementary Fig. 1b and Supplementary Data).
Construction of the pDRAV replacement backbone vectors
The pBluescript IIKS plasmid was modified by inserting linkers to produce all possible orientations of the loxP and FRT sites together with a lox2272 site that permits subsequent use of conventional RMCE. The attB-pGK-Hygro-attP resistance cassette was cloned into the BamHI and SalI sites. The multiple cloning sites of all pDRAV plasmids consist of unique NotI-NsiI-HpaI-PacI-BamHI restriction sites that can be used to insert the sequences of interest.
Construction of the Hand2-Flag replacement vector (pRVH2)
Linkers were inserted into pBluescript IIKS plasmid to produce the following restriction/recombinase site configuration: SacI-loxP-NarI-NotI-BamHI-SalI-ClaI-FRT-HindIII-KpnI. A NarI-NotI fragment of the Hand2 5′ untranslated region and a NotI-BamHI fragment corresponding to the remainder of the Hand2 transcription unit (with a Flag-epitope tag inserted into coding exon 1) were sequentially inserted into the pBluescript IIKS backbone. A DNA fragment encoding the attB-pGK-hygro-attP resistance cassette with HindIII and PacI sites (to enable Southern blot screening) was cloned into the BamHI and SalI sites of the pBluescript IIKS backbone to produce the final replacement vector (Fig. 2a).
Mouse embryonic stem cell transfection and selection
50 μg of the appropriate replacement vector were coelectroporated with 50 μg of pDIRE plasmid into mouse embryonic stem cells (1.5 × 107 cells per cuvette; 240 kV, 475 μF). IKMC mouse embryonic stem cells10 were grown in Knockout DMEM (4.5 g l−1 glucose) containing 10% FBS, 2 mM d-glutamine, 1× penicillin-streptomycin, 0.1 mM β-mercaptoethanol and 103 U ml−1 LIF/ESGRO (Chemicon; all other reagents from Gibco-Invitrogen). R1 embryonic stem cells were grown in DMEM (4.5 g l−1 glucose) containing 15% FCS (HyClone), 2 mM d-glutamine, 1× non-essential amino acids, 2 mM sodium pyruvate, 1× penicillin-streptomycin, 0.1 mM β-mercaptoethanol and 103 U ml−1 LIF/ESGRO. The culture medium was changed daily and from the second day onward; resistant colonies were selected in the presence of 175 μg ml−1 hygromycin or 0.5 μg ml−1 puromycin (Sigma). After 8 d in selection medium, drug-resistant colonies were picked and analyzed by PCR. Clones with correct replacement were expanded, frozen in several aliquots and the correct replacement verified by Southern blot analysis. General protocols for handling, culture and manipulation of mouse embryonic stem cells are available as a standard textbook9. Detailed protocols for the culture of IKMC targeted mouse embryonic stem cells can be downloaded from the EUCOMM website (http://www.eucomm.org/information/protocols/).
Immunodetection of YFP and β-galactosidase
Embryonic stem cell colonies were grown on eight-well micro-Slides (Ibidi), fixed with cold acetone and incubated with rabbit anti-GFP–Alexa Fluor-488 (1:1,000; Invitrogen) and mouse anti–β-galactosidase (1:50; Developmental Studies Hybridoma Bank, Department of Biology, University of Iowa) primary antibodies, followed by an incubation step with a goat anti-mouse–Alexa Fluor-594 (1:1,000; Invitrogen). Fluorescent images were acquired using a Leica SP5 confocal microscope and software.
Whole-mount in situ hybridization
Mouse embryos were processed for whole-mount in situ hybridization using digoxigenin-labeled Hand2 antisense riboprobes8. Images were acquired using a Leica MZ16A stereomicroscope and software.
Image processing
All images were processed and composites made using the Adobe Photoshop CS4 software package. Only contrast and brightness of the original images were adjusted minimally and within the linear range when necessary.
Mice
All animal experiments were performed in accordance with Swiss law and have been approved by the veterinary authorities of Basel.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to D. Klewe-Nebenius and T. Hennek (University of Basel) for generating the chimeric mice. We thank I. Verma (Salk Institute, San Diego, California, USA), B. Sauer (Stowers Institute, Kansas City, Missouri, USA), R. Jaenisch (Whitehead Institute, Cambridge, Massachusetts, USA) and P. Soriano (Mount Sinai School of Medicine, New York, New York, USA) for plasmids obtained via Addgene and A.-K. Hadjantonakis (Sloan-Kettering Institute, New York, New York, USA) for suggesting the use of the H2B-Venus fusion protein. We thank A. Schauerte and P. Lorentz for expert technical assistance and are indebted to P. Bovolenta, G. Nusspaumer, A. Zuniga and members of our research groups for helpful input and discussions. Our research is supported by the Swiss National Science Foundation (to R.Z.), by a Marie Curie Intra-European Fellowship and a European Reintegration Grant (to J.L.-R.) and by both cantons of Basel, Switzerland.
Footnotes
Note: Supplementary information is available on the Nature Methods website.
COMPETING FINANCIAL INTERESTS The authors declare competing financial interests: details accompany the full-text HTML version of the paper at http://www.nature.com/naturemethods/.
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions/.
References
- 1.Glaser S, Anastassiadis K, Stewart AF. Nat. Genet. 2005;37:1187–1193. doi: 10.1038/ng1668. [DOI] [PubMed] [Google Scholar]
- 2.Branda CS, Dymecki SM. Dev. Cell. 2004;6:7–28. doi: 10.1016/s1534-5807(03)00399-x. [DOI] [PubMed] [Google Scholar]
- 3.Lauth M, Spreafico F, Dethleffsen K, Meyer M. Nucleic Acids Res. 2002;30:e115. doi: 10.1093/nar/gnf114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Collins FS, Rossant J, Wurst W. Cell. 2007;128:9–13. doi: 10.1016/j.cell.2006.12.018. [DOI] [PubMed] [Google Scholar]
- 5.Shimshek DR, et al. Genesis. 2002;32:19–26. doi: 10.1002/gene.10023. [DOI] [PubMed] [Google Scholar]
- 6.Raymond CS, Soriano P. PLoS ONE. 2007;2:e162. doi: 10.1371/journal.pone.0000162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kanda T, Sullivan KF, Wahl GM. Curr. Biol. 1998;8:377–385. doi: 10.1016/s0960-9822(98)70156-3. [DOI] [PubMed] [Google Scholar]
- 8.Galli A, et al. PLoS Genet. 2010;6:e1000901. doi: 10.1371/journal.pgen.1000901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nagy A. Manipulating the Mouse Embryo: A Laboratory Manual. 3rd edn Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York, USA: 2003. [Google Scholar]
- 10.Pettitt SJ, et al. Nat. Methods. 2009;6:493–495. doi: 10.1038/nmeth.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anastassiadis K, et al. Dis. Model, Mech. 2009;2:508–515. doi: 10.1242/dmm.003087. [DOI] [PubMed] [Google Scholar]
- 12.Singla V, et al. Nat. Methods. 2010;7:50–52. doi: 10.1038/nmeth.1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Skarnes WC, et al. Nat. Genet. 2004;36:543–544. doi: 10.1038/ng0604-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Szymczak AL, et al. Nat. Biotechnol. 2004;22:589–594. doi: 10.1038/nbt957. [DOI] [PubMed] [Google Scholar]
- 15.Chu GC, Dunn NR, Anderson DC, Oxburgh L, Robertson EJ. Development. 2004;131:3501–3512. doi: 10.1242/dev.01248. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.