Abstract
The structural features that enable replicative DNA polymerases to synthesize DNA rapidly and accurately also limit their ability to copy damaged DNA. Direct replication of DNA damage is termed translesion synthesis (TLS), a mechanism conserved from bacteria to mammals and executed by an array of specialized DNA polymerases. This chapter examines how these translesion polymerases replicate damaged DNA and how they are regulated to balance their ability to replicate DNA lesions with the risk of undesirable mutagenesis. It also discusses how TLS is co-opted to increase the diversity of the immunoglobulin gene hypermutation and the contribution it makes to the mutations that sculpt the genome of cancer cells.
Cells can use specialized translesion polymerases to replicate through damaged DNA. These polymerases can also be co-opted to generate mutations in the immunoglobulin genes during the vertebrate immune response.
Genome replication is normally an extremely accurate process with an error rate as low as one in 1010 bases in mammalian cells. This extraordinary fidelity is the result of a combination of factors: the intrinsic ability of the replicative polymerases to select the correct nucleotide, their ability to recognize and excise misincorporated nucleotides, and the activity of postreplicative mismatch repair (Loeb and Monnat 2008; Arana and Kunkel 2010). However, the features that give the replicative polymerases their high intrinsic fidelity also mean that they are easily stalled by DNA damage. Since the DNA at the fork is unwound, any attempt to excize the damage will result in DNA breaks and replication fork collapse. To avoid this, cells replicate through the damage, restoring it to duplex DNA before attempting excision repair. This is known as damage tolerance. Damage tolerance can be achieved by recombinational mechanisms or directly by translesion synthesis (TLS).
TLS is conserved throughout evolution. It relies on specialized DNA polymerases that have active sites capable of accommodating damaged or distorted templates. However, use of these polymerases carries an increased risk of mutagenesis because damaged bases are frequently non- or misinstructional and because the structural adaptations that allow lesion replication also result in reduced fidelity when copying undamaged DNA. The principal DNA polymerases responsible for TLS during replication belong to the Y-family (Sale et al. 2012), along with the B-family enzyme Pol ζ (Gan et al. 2008). In addition, DNA polymerases of the X and A family are capable of TLS and can contribute to mutagenesis during DNA-repair reactions such as base excision repair (BER) and nonhomologous end joining (NHEJ). Whereas TLS plays an important role in creating mutations, the TLS polymerases possess structural features that often help them extract the correct code from diversely damaged templates. Coupled with control mechanisms that ensure their access to DNA templates is strictly regulated, this helps limit the opportunities for these polymerases to cause undesirable mutagenesis.
THE MECHANISMS OF TRANSLESION SYNTHESIS BY THE EUKARYOTIC Y-FAMILY POLYMERASES AND Pol ζ
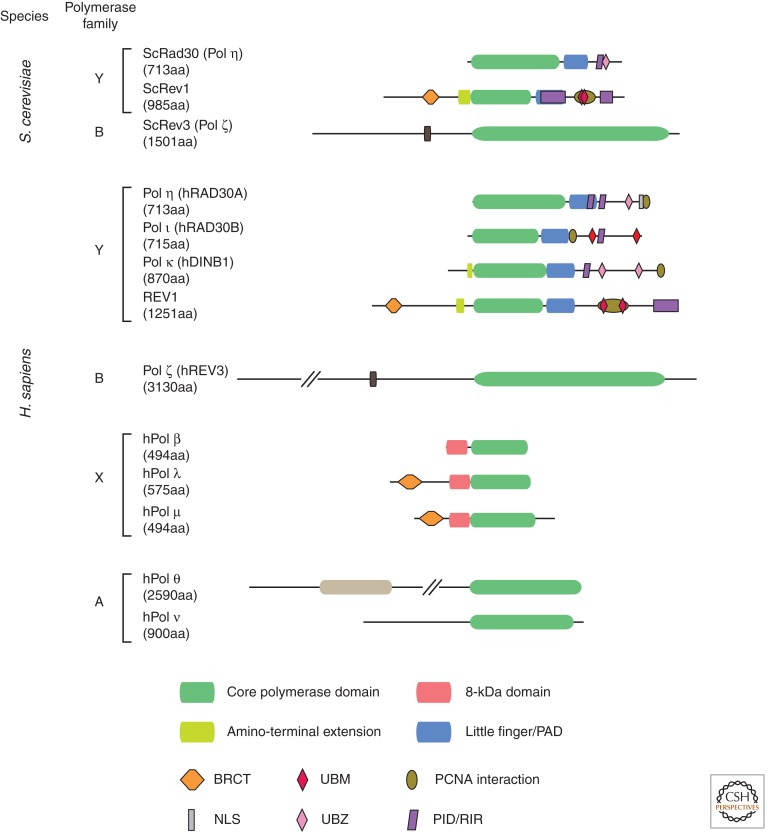
The budding yeast Saccharomyces cerevisiae has three DNA polymerases specialized for TLS: RAD30 (Pol η) and Rev1, both from the Y-family, and the B-family enzyme Pol ζ. Vertebrate cells possess two further Y-family polymerases Pol κ and Pol ι (Fig. 1).
Figure 1.
Domain structure of TLS polymerases in S. cerevisiae and Homo sapiens. The diagrams are to scale except that hREV3 and hPol θ are truncated, as indicated by parallel diagonal lines. The REV7 subunit of Pol ζ is not shown. PAD, polymerase-associated domain (also known as the little finger); BRCT, a domain with homology to the BRCA1 carboxyl terminus; UBM and UBZ, ubiquitin-binding domains; PID, polymerase-interacting domain (of REV1); RIR, REV1-interacting region (of other Y-family polymerases).
General Features of the Y-Family Polymerases
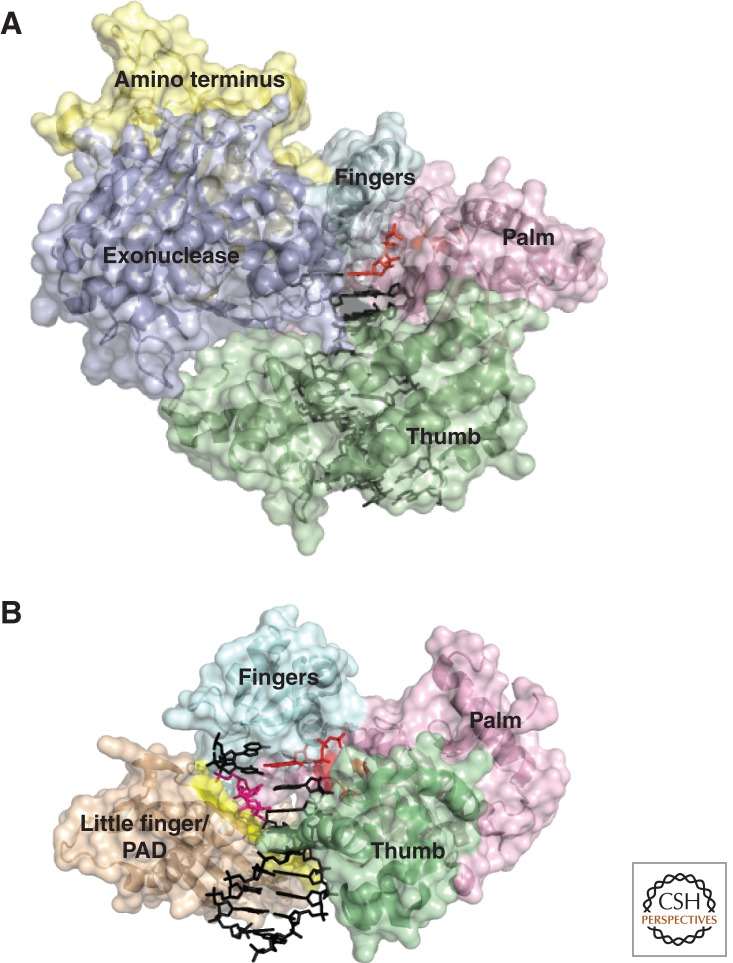
The Y-family polymerases all adopt the same overall right-handed topology as the replicative polymerases, despite little conservation of primary sequence between them (Yang and Woodgate 2007) (Fig. 2). As in replicative polymerases, the key carboxylate residues necessary for catalysis reside in the “palm” domain, but they are housed within a much larger active site pocket that can accommodate bulky or distorted templates. The fingers and thumb domains, which in the replicative polymerases are required to detect the fit of a correctly paired incoming nucleotide, are generally shorter and make fewer contacts with both the template and incoming nucleotide. This reduces the enzymes’ ability to discriminate the correct incoming base and thus contributes to their diminished fidelity and processivity. An additional little-finger domain, also known as the polymerase-associated domain (PAD), helps stabilize the Y-family polymerases on DNA. Critically, the Y-family TLS polymerases also lack a 3′-5′ proofreading exonuclease domain. Together, these features result in genuinely error-prone enzymes with incorporation error rates of up to one in 10 (McCulloch and Kunkel 2008).
Figure 2.
Comparative anatomy of a replicative polymerase and a TLS polymerase. (A) S. cerevisiae polymerase δ, PDB 3IAY (Swan et al. 2009), a replicative B-family polymerase. The domains are shaded: palm, pink; thumb, green; fingers, cyan; exonuclease, purple. The DNA is in black, and the active site in the palm and incoming nucleotide triphosphate is in red. (B) H. sapiens polymerase η, PDB 3SI8 (Biertumpfel et al. 2010), a Y-family TLS polymerase with a T-T CPD in the +2 position. The domains in common with Pol δ are shaded the same. The little-finger domain/PAD is shaded in light brown. The DNA is in black, except the CPD, which is pink. The active site and incoming nucleotide triphosphate pairing with the first base after the CPD are in red. The β-strand splint in the little finger/PAD domain that constrains the CPD is highlighted in yellow.
Pol η
Pol η, also known as RAD30 in yeast (McDonald et al. 1997), plays a key role in the accurate replication of the principal form of UV-induced DNA damage, the cyclobutane pyrimidine dimer (CPD) (Johnson et al. 1999b). On undamaged DNA, however, Pol η is significantly error prone (Washington et al. 1999; Johnson et al. 2000c; Matsuda et al. 2000; Matsuda et al. 2001). Mutations in Pol η are linked to the clinical syndrome xeroderma pigmentosum variant (XPV) (Johnson et al. 1999a; Masutani et al. 1999), characterized by hypersensitivity to UV-induced skin-pigmentation changes and an increased incidence of skin cancers (Cleaver 1972). Pol η-deficient cells are hypermutable following UV exposure (Stary et al. 2003; Choi and Pfeifer 2005; Busuttil et al. 2008), which results from inaccurate bypass of CPDs by Pol κ or Pol ι working in cooperation with Pol ζ (Wang et al. 2007; Gueranger et al. 2008; Ziv et al. 2009) and from more frequent DNA breaks and chromosome aberrations (Cleaver et al. 1999; Limoli et al. 2002a,b).
Structurally, the ability of Pol η to replicate CPDs accurately is due to its large active site, which is able to accommodate both bases of a CPD in such a manner that the linked Ts can pair correctly with incoming dA (Biertumpfel et al. 2010; Silverstein et al. 2010) (Fig. 2B). The structural distortion of the newly synthesized duplex caused by the T-T dimer is controlled by a positively charged β-strand in the little-finger domain that acts as a splint (yellow patch in Fig. 2B) that ensures the reading frame is maintained. Further DNA synthesis beyond the CPD is limited to three bases by steric clashes that likely result in the enzyme being displaced from the DNA, thereby limiting the extent of low-fidelity synthesis (Biertumpfel et al. 2010). Pol η, however, is not able to completely replicate the highly helix-distorting (6-4) pyrimidine-pyrimidone photoproduct, the second most abundant UV lesion, although Pol η can incorporate dG opposite the 3′ base of this lesion in vitro (Masutani et al. 2000; Johnson et al. 2001). In vivo, however, Pol η-deficient cells do not exhibit a significant defect in the replication of (6-4) photoproducts, suggesting that other polymerases, possibly Pol ζ, are normally responsible for accurate replication of this lesion (Gibbs et al. 2005; Hendel et al. 2008; Szüts et al. 2008; Yoon et al. 2010).
The ability of Pol η to accommodate a dinucleotide lesion in its active site also contributes to its ability to replicate intrastrand cross-links at G-G dinucleotides caused by cisplatin, a commonly used chemotherapeutic agent. The active site stabilizes the lesion, allowing the 3′ G to Watson–Crick pair with incoming dCTP, while still providing the rigidly inclined 5′ G space within the active site (Alt et al. 2007; Ummat et al. 2012; Zhao et al. 2012). Replication of the 5′ G is much less efficient and accurate, and, in vivo, this step is catalyzed by Pol ζ (Shachar et al. 2009). Pol η also contributes to the replication of single-base lesions, notably 7,8-dihydro-8-oxoguanine (8-oxoG), the most common oxidative lesion in DNA, opposite which it incorporates the correct dC (de Padula et al. 2004). Again, however, extension requires Pol ζ (Haracska et al. 2003).
Pol ι
Pol ι evolved as a gene duplication of RAD30 (McDonald et al. 1999) and is present in most vertebrates and in some lower eukaryotes, but notably not birds or S. cerevisiae (Waters et al. 2009). It is remarkably error prone, exhibiting markedly different fidelities depending on the template base (Tissier et al. 2000b). This property is explained by its unusual active site, which restricts the conformation of template purines to syn, promoting Hoogsteen pairing with the incoming base (Nair et al. 2004). In the case of 8-oxoG, the syn conformation would normally result in pairing with incorrect dA. However, the narrow active site of Pol ι prevents this, favoring instead pairing with the smaller, and correct, dC (Kirouac and Ling 2011).
The in vivo roles of Pol ι have been more difficult to establish firmly. After some debate about the role of Pol ι in bypassing CPDs (Johnson et al. 2000b; Tissier et al. 2000a; Choi et al. 2006; Vaisman et al. 2006), several studies have now established that Pol ι can indeed contribute to replication of UV lesions in vivo. When both Pol ι and Pol η are disrupted, the formation of UV-induced skin tumors in mice is accelerated (Dumstorf et al. 2006; Ohkumo et al. 2006). Further, in a human Burkitt lymphoma cell line lacking both Pol ι and Pol η, mutational hotspots present in a Pol η single mutant were lost (Gueranger et al. 2008).
Pol ι is also implicated in BER. It possesses dRP lyase activity, interacts with XRCC1, and can substitute for Pol β in an in vitro short-patch BER reaction (Bebenek et al. 2001; Petta et al. 2008). However, the extent to which use of Pol ι in this context contributes to mutagenesis is not known.
Pol κ
Pol κ is the most widely conserved of the Y-family polymerases with homologs in bacteria, archaea, and eukaryotes. Interestingly, however, it is not found in S. cerevisiae (Ohmori et al. 2001). It is the most accurate of the Y-family polymerases on undamaged DNA (Johnson et al. 2000a) and has a less marked tendency for –1 frameshifts than its bacterial homolog dinB (Ohashi et al. 2000). Pol κ can bypass many lesions, albeit with relatively low efficiency, but is blocked by dinucleotide lesions (Bavoux et al. 2005a) as its catalytic site can only accommodate a single Watson–Crick base pair (Lone et al. 2007). It can, however, replicate some N2-dG adducts such as N2-furfuryl-dG (Jarosz et al. 2006); N2-(1-carboxyethyl)-2′-dG, a byproduct of glycolysis (Yuan et al. 2008); and N2-benzo[a]pyrene-diol-epoxide-dG (Zhang et al. 2000, 2002b; Rechkoblit et al. 2002) efficiently and accurately. However, N2-dG adducts formed by the important lipid-oxidation products acrolein and 4-hydroxynonenal (γ-hydroxy-1,N2-propano-2′-dG and trans-4-hydroxy-2-nonenal-dG respectively) block Pol κ. In these cases, Pol κ can operate in a two-step TLS reaction by extending from dC inserted by Pol ι (Washington et al. 2004b; Wolfle et al. 2006). Indeed, Pol κ is adept at extending a diverse range of mismatched termini (Washington et al. 2002), and this is likely to be related to the stability it gains from the manner in which it encircles DNA, largely a function of a unique domain in its amino terminus termed the N-clasp (Lone et al. 2007). However, in vivo evidence suggests that the role of extender polymerase (Fig. 3) is dominated by Pol ζ (Shachar et al. 2009), even for lesions such as N2-benzo[a]pyrene-dG, that in vitro can be efficiently bypassed by Pol κ alone (Zhang et al. 2000; Avkin et al. 2004).
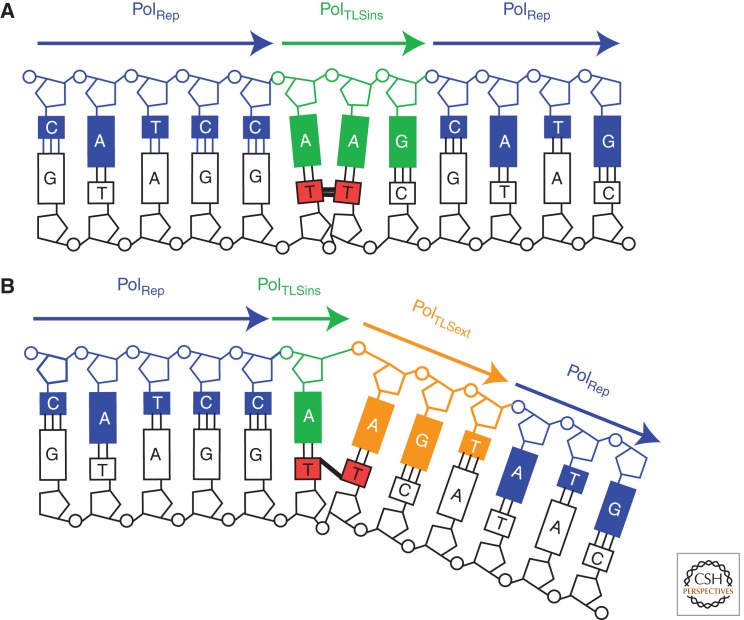
Figure 3.
One-step versus two-step TLS. (A) The tracts of DNA synthesized during one-step bypass of a T-T CPD. PolRep, the replicative polymerases Pol δ and Pol ε. In the case illustrated, bypass of a CPD, the inserter and extender polymerase (PolTLSins/ext) is Pol η, which can replicate this lesion on its own, aided by its large active site pocket and molecular split (see text). The extent of its synthesis patch is limited either by its dissociation from the DNA induced by a steric clash between the lesion and the little-finger domain as the chain elongates or by the primer at the end of a single-stranded gap. (B) The tracts of DNA synthesized during two-step bypass of a T-T (6-4) photoproduct. Like many lesions, the highly distorting (6-4) photoproduct is not bypassed in a single step. Instead, an inserter polymerase (PolTLSins) incorporates a base opposite the 3′T of the lesion, but cannot extend opposite the 5′T. Instead, this is carried out by an extender polymerase (PolTLSext), most commonly Pol ζ, which can deal with the highly misaligned terminus.
Pol κ also cooperates with Pol δ in the gap-filling step of nucleotide excision repair (NER), being recruited by ubiquitinated proliferating-cell-nuclear-antigen (PCNA) and the scaffold protein XRCC1 (Ogi and Lehmann 2006). The recruitment of Pol κ may facilitate repair-patch synthesis in conditions of low dNTPs, as pertains to G1, because of the relatively low Km of the enzyme. However, it would also be expected to contribute to mutagenesis at the rate of about one mutation per 30 repair patches (each of 30 nucleotides) (Ogi et al. 2010).
The Catalytic Activity of REV1
REV1 can insert dC opposite an abasic site (Nelson et al. 1996a) a limited number of adducts, notably N2-adducts of guanine (Zhang et al. 2002c; Washington et al. 2004a; Nair et al. 2008), and template dG (Nelson et al. 1996a; Haracska et al. 2002). However, it does this in a most unusual way. It does not directly pair the incoming dC with the DNA template. Instead, it swings the template dG out of the helix and temporarily coordinates it with a specialized loop within the little finger. In place of the template dG, REV1 places its own Arg324, which hydrogen bonds with incoming dC (Nair et al. 2005). This mechanism thus allows the bypass of bulky dG adducts (Nair et al. 2008) and suggests how REV1 can act as a template-independent dC transferase. The catalytic activity of REV1 is employed during the bypass of abasic sites in the immunoglobulin (Ig) loci of vertebrate B cells (Jansen et al. 2006; Ross and Sale 2006; Masuda et al. 2009) (see below) and during abasic site bypass in yeast (Kim et al. 2011). REV1 has also been implicated in the replication of sequences capable of forming DNA secondary structures, notably triplet repeats (Collins et al. 2007) and G-quadruplexes (Sarkies et al. 2010). However, inactivation of the dC transferase of REV1 has no measurable effect on the sensitivity of chicken DT40 cells to a range of DNA-damaging agents (Ross et al. 2005) or on murine development (Masuda et al. 2009), but it is required for budding yeast to survive exposure to the G-adducting agent 4-nitroquinoline-1-oxide (Wiltrout and Walker 2011). Nonetheless, complete loss of REV1 results in defective TLS through lesions that are not substrates for its catalytic activity (Gibbs et al. 2000; Simpson and Sale 2003; Ross et al. 2005; Jansen et al. 2009). This is attributable to loss of a second function of REV1 in the coordination of other TLS polymerases, which is discussed further below.
Pol ζ
Pol ζ is a B-family enzyme related to the replicative polymerases Pols δ, ε, and α. It is essential for practically all DNA-damage-induced and a significant proportion of spontaneous mutagenesis in yeast (Lemontt 1971; Cassier et al. 1980; Quah et al. 1980; Morrison et al. 1989; Roche et al. 1994; Harfe and Jinks-Robertson 2000), despite the relatively modest sensitivity of pol ζ mutants to DNA-damaging agents (Lawrence and Christensen 1976). The core of Pol ζ is made up of two subunits: Rev3, the catalytic subunit, and Rev7, an accessory subunit that stimulates the activity of Rev3 (Nelson et al. 1996b) and links Pol ζ to Rev1 (Acharya et al. 2005). Like the Pol δ catalytic subunit (Pol3), Rev3 associates with the accessory proteins Pol31 and Pol32 via its carboxyl terminus (Acharya et al. 2009; Johnson et al. 2012), which harbors an iron–sulphur cluster (Netz et al. 2012). This suggests a common element to the recruitment and regulation of the two polymerases.
Pol ζ lacks proofreading exonuclease activity, and the accuracy of the yeast enzyme is lower than Pol δ and Pol ε at about 10–4, similar to Pol α (Johnson et al. 2000b). It is not significantly modified in vitro by the presence of the accessory proteins RPA, RFC, and PCNA (Zhong et al. 2006). In vitro, Pol ζ is generally inefficient at inserting bases opposite lesions but is markedly adept at extending from mismatched primer termini and those containing abnormal structures (Johnson et al. 2000b; Haracska et al. 2003; Washington et al. 2004a). It has been proposed that this is its principal in vivo role (Prakash et al. 2005). However, genetic evidence argues that in vivo Pol ζ is also capable of significant single-step bypass of lesions such as (6-4) photoproducts (Gibbs et al. 2005; Szüts et al. 2008; Yoon et al. 2010).
Pol ζ has been notoriously hard to study in vertebrate cells. Vertebrate Pol ζ also comprises REV3 and REV7 subunits, but REV3 is considerably larger than its yeast counterpart at some 350 kDa, mostly because of the insertion of a large exon of unknown function toward its amino terminus (Gan et al. 2008). Currently, the only structural information on the complex concerns the REV7-interacting peptide from human REV3 in complex with REV7 showing how the interaction could produce an interface for the binding of REV1 (Hara et al. 2010). Despite the report of an antibody that can detect endogenous REV3 (Wang et al. 2011), the size of Pol ζ, along with its apparently very low levels of expression in cells (Ogawara et al. 2010), has significantly hampered biochemical and cell biological studies. Further, unlike its yeast counterpart, deletion of Pol ζ in mice results in embryonic lethality (Bemark et al. 2000; Esposito et al. 2000; Wittschieben et al. 2000; Kajiwara et al. 2001; O-Wang et al. 2002; Van Sloun et al. 2002) and senescence of primary fibroblasts (Lange et al. 2012). However, transformed mouse and chicken cells are able to proliferate in the absence of REV3 (Sonoda et al. 2003; Zander and Bemark 2004).
Pol ζ also plays a role in double-strand break repair. The DNA synthesis associated with recombination in S. cerevisiae is error prone compared with that of replication, and this mutagenesis is dependent on Pol ζ (Strathern et al. 1995; Holbeck and Strathern 1997). Further, immunoprecipitation shows that yeast Pol ζ and REV1 are associated with HO-endonuclease-induced double-strand breaks (Hirano and Sugimoto 2006). Vertebrate Pol ζ and REV1 also appear to be involved in double-strand break repair, although their precise role in this process remains poorly understood (Sonoda et al. 2003; Sharma et al. 2011).
COOPERATION BETWEEN TLS POLYMERASES IN BYPASSING LESIONS
It is evident from the examples discussed above that, in some cases, lesion bypass can be effected in a single step by a single DNA polymerase, for example, the bypass of CPDs by Pol η (Fig. 3A). However, in general, this type of TLS appears to be the exception rather than the rule. Many in vitro and structural studies have highlighted constraints that polymerases face in solving both problems of incorporation opposite a lesion and extension from the resulting mismatched terminus. Certain polymerases seem to be particularly good at the insertion step, for instance, Pol ι and REV1, whereas others are more proficient at extension, notably Pol ζ and, to a lesser extent, Pol κ. These findings led to the proposal that lesion bypass is often a two-step process (Johnson et al. 2000b) (Fig. 3B). Recent work has confirmed that two-step bypass is the norm for several important lesions and that Pol ζ plays a crucial role as an extender in vivo, even in cases where bypass can be carried out by a single polymerase in vitro (Shachar et al. 2009).
CONTROL OF TRANSLESION SYNTHESIS
The potential mutagenicity of TLS polymerases means that they must be carefully controlled. Control of TLS has been most extensively studied in relation to the Y-family polymerases and Pol ζ (Gan et al. 2008; Sale et al. 2012). The regulation of the mutagenic X- and A-family polymerases, discussed below, is much less well understood but is likely to depend on the DNA-repair mechanisms in which they participate.
Unlike in Escherichia coli, changes in the overall cellular levels of the translesion polymerases do not appear to play a major role in the regulation of eukaryotic TLS. The exception is Rev1 in S. cerevisiae, which is under profound cell-cycle control with markedly elevated levels in G2 (Waters and Walker 2006). Rather, the preferred method seems to be to locally concentrate the enzymes in replication factories (Kannouche et al. 2001; Sabbioneda et al. 2008; Sale et al. 2012). Formation of these factories is driven by a number of factors, including PCNA ubiquitination (discussed below), although importantly, focus formation is not strictly necessary for polymerase function (Ross et al. 2005; Gueranger et al. 2008). Nonetheless, local concentration of the Y-family polymerases in the vicinity of distressed forks may be a first step to increase the likelihood of their recruitment to stalled termini (Sabbioneda et al. 2008).
Ubiquitination of Proliferating Cell Nuclear Antigen
Following replication arrest, eukaryotic PCNA is monoubiquitinated at lysine 164 in a reaction mediated by the E3 ubiquitin ligase RAD18 working in conjunction with the E2 ubiquitin-conjugating enzyme RAD6 (Hoege et al. 2002; Stelter and Ulrich 2003; Haracska et al. 2004; Kannouche et al. 2004). In vertebrates, there are additional ubiquitin ligases capable of introducing this modification (Simpson et al. 2006; Zhang et al. 2008; Terai et al. 2010). The activity of RAD18 is stimulated by its association with RPA-coated single-stranded DNA (Davies et al. 2008), and PCNA monoubiquitation serves to recruit the Y-family polymerases to sites of replication arrest by enhancing their direct association with PCNA through an interaction between ubiquitin-PCNA and ubiquitin-binding motif (UBM) or ubiquitin-binding zinc (UBZ) domains in the carboxyl terminus of the polymerases (Bienko et al. 2005).
PCNA can also be polyubiquitinated with K63-linked chains built on the initial ubiquitin conjugated to K164 of PCNA (Hoege et al. 2002; Chiu et al. 2006). In yeast, at least, PCNA can also be SUMOylated (Papouli et al. 2005; Pfander et al. 2005). These modifications are principally linked to recombinational modes of damage bypass. Nonetheless, it is clear that there is complex cross-talk between the mechanisms that control recombination and TLS (Sale 2012). For example, the E3 ligase responsible for PCNA polyubiquitination is also required for efficient TLS in both S. cerevisiae (Pages et al. 2008) and Schizosaccharomyces pombe (Coulon et al. 2010).
S. cerevisiae lacking ubiquitinatable PCNA (pol30-119, which carries a K164R substitution) have a generalized defect in UV-induced mutagenesis that is indistinguishable from either a rad6 or a rev3 mutant (Stelter and Ulrich 2003; Haracska et al. 2004). However, both mouse and chicken cells with a pcnaK164R mutation retain significant TLS activity (Arakawa et al. 2006; Langerak et al. 2007; Szüts et al. 2008; Hendel et al. 2011; Krijger et al. 2011). At least in DT40 cells, the majority of this PCNA ubiquitination-independent TLS is dependent on a noncatalytic function of REV1 (Arakawa et al. 2006; Edmunds et al. 2008; Szüts et al. 2008).
The Noncatalytic Role of REV1 in Translesion Synthesis
In addition to its catalytic function, REV1 plays an important part in the coordination of TLS through protein–protein interactions with other TLS polymerases, PCNA, and ubiquitin. The extreme carboxyl terminus of vertebrate REV1 interacts with the other Y-family polymerases (Guo et al. 2003; Ohashi et al. 2004; Tissier et al. 2004) and with Pol ζ via its REV7 subunit (Murakumo et al. 2001). Deletion of this domain results in the same damage-tolerance defect seen in a complete rev1 mutant (Ross et al. 2005; Edmunds et al. 2008). The carboxyl terminus of REV1 in S. cerevisiae also binds to Rad30 (Pol η) and to Rev7 (Pol ζ), although the way in which it interacts with Pol η appears to be distinct from the vertebrate protein (Acharya et al. 2007; D’Souza et al. 2008; Kosarek et al. 2008). Structural studies of the mammalian REV1 carboxyl terminus have revealed an atypical four-helix bundle with distinct interaction surfaces for REV7 and the REV1-interacting regions (RIRs) of Pol η and κ (Pozhidaeva et al. 2012; Wojtaszek et al. 2012). Adjacent to this carboxy-terminal domain are two UBM domains that may facilitate the binding of REV1 to ubiquitinated PCNA (Garg and Burgers 2005; Guo et al. 2006b) or other ubiquitinated proteins in the vicinity of stalled replication forks (Edmunds et al. 2008). The region in which the UBMs are embedded also mediates a direct interaction with PCNA (Ross et al. 2005; Wood et al. 2007). This arrangement suggests that REV1 could act as an adaptor to coordinate the interaction between PCNA, the inserter Y-family polymerases, and the extender polymerase Pol ζ during TLS.
This noncatalytic function of REV1 is required for normal replication-fork progression following DNA damage in mouse and chicken DT40 cells (Edmunds et al. 2008; Jansen et al. 2009), suggesting that REV1 plays a role at, or close to, the replication fork. In DT40 cells, this early role appears to be independent of PCNA ubiquitination (Edmunds et al. 2008). REV1 also plays a later role in filling postreplicative gaps, left when replication reprimes downstream of an arrested fork. This role is very prominent in mouse cells (Jansen et al. 2009) but much less obvious in DT40 (Edmunds et al. 2008), probably because it is masked by the very active recombinational pathways in the latter system.
The role of the amino-terminal BRCT domain of REV1 remains incompletely understood. The rev1-1 mutant of S. cerevisiae carries a G193R substitution within the BRCT domain and exhibits defective mutagenic tolerance of (6-4) photoproducts despite retaining almost full catalytic activity (Nelson et al. 2000). Mouse cells lacking the BRCT domain of REV1 exhibit defective DNA damage tolerance, an increase in UVC-induced chromosome aberrations, and spontaneous intragenic deletions in the HPRT locus (Jansen et al. 2005). Further, there is strong evidence that the BRCT domain is required for the early role of REV1 close to replication forks (Jansen et al. 2009). The reported binding of the BRCT domain to PCNA (Guo et al. 2006a) has been challenged (de Groote et al. 2011). Instead, it is proposed to preferentially bind recessed 5′ phosphorylated primer-template junctions (de Groote et al. 2011). This raises the interesting possibility that REV1, like the 9-1-1 checkpoint complex, is recruited to postreplicative gaps by binding to the 5′ end of the gap.
TLS Polymerase Phosphorylation and the 9-1-1 Clamp
The exposure of RPA-coated single-stranded DNA at stalled replication forks not only triggers PCNA ubiquitination, but also ATR-dependent checkpoint activation (Byun et al. 2005). Both REV1 and Pol η are phosphorylated by ATR, although how this regulates TLS is still unclear (Sabbioneda et al. 2007; Pages et al. 2009; Gohler et al. 2011). Significantly, full activation of the ATR-dependent replication checkpoint requires downstream repriming and recruitment of the 9-1-1 checkpoint clamp to the 5′ phosphorylated end of the resulting RPA-coated gap (Zou and Elledge 2003; Majka et al. 2006). As well as stimulating ATR-dependent phosphorylation, this PCNA-like clamp also directly interacts with Pol κ and Pol ζ/Rev1 (Kai and Wang 2003; Sabbioneda et al. 2005). This has led to the suggestion that the apparatus needed for TLS of highly distorting lesions is actually recruited to the 5′ end of a postreplicative gap, subsequently translocating back to the arrested 3′ terminus. This mechanism would intrinsically restrict access of TLS to forks at which stalling is sufficiently persistent to give rise to downstream repriming (Jansen et al. 2007).
Ubiquitination of the TLS Polymerases
An emerging level of polymerase regulation is posttranslational modification of the polymerases themselves by ubiquitination (Bienko et al. 2005; Guo et al. 2006b; Wimmer et al. 2008; Bienko et al. 2010; Jung et al. 2010). The significance of polymerase ubiquitination is still under investigation, the best understood case being Pol η. Pol η is monoubiquitinated on its carboxyl terminus, probably by PIRH2 (Jung et al. 2010), and this inhibits its interaction with PCNA, suggesting that deubiquitination of Pol η is needed for its recruitment to stalled forks (Bienko et al. 2010).
The Fanconi Anemia Pathway
As has already been noted, TLS plays an important role in the cellular response to DNA interstrand cross-links. The replication-dependent repair of cross-links is coordinated by the Fanconi anemia (FA) complex, which is instrumental in ensuring the correct recruitment of both homologous recombination and TLS (Niedzwiedz et al. 2004; Raschle et al. 2008; Knipscheer et al. 2009; Long et al. 2011). However, evidence is emerging that the FA pathway may also play a role in the coordination of TLS that is independent of cross-link repair, the FA pathway appearing to be particularly important for REV1-dependent TLS (Mirchandani et al. 2008; Kee and D’Andrea 2010; Kim et al. 2012).
The Role of the Lesion Itself
Despite the emergence of these refined mechanisms for polymerase recruitment, it remains largely unclear what determines polymerase selection at a particular lesion. It seems likely, based on some of the parameters discussed above, that the lesion itself plays a role in determining which polymerase is ultimately employed, even if this is simply a question of which enzyme can perform the reaction in the most straightforward manner, seen for example in the replication of CPDs by Pol η. It may also be that the context in which the lesion is found limits the options for its bypass (Sale 2012).
OTHER POTENTIALLY MUTAGENIC POLYMERASES IN VERTEBRATES
Higher eukaryotes, and particularly vertebrates, have an expanded repertoire of potentially error-prone DNA polymerases of the X and A families. These enzymes are capable of TLS and are likely to contribute to mutagenesis in vivo through the specialized support they provide to specific DNA-repair pathways.
Translesion Synthesis and Mutagenesis by Eukaryotic X-Family Polymerases
S. cerevisiae has a single X-family DNA polymerase, Pol 4, which plays a role in NHEJ (Wilson and Lieber 1999; Tseng and Tomkinson 2002) and possibly BER (Bebenek et al. 2005). It can fill in small gaps and extend primer termini that have limited homology but has a high error rate, partly because of lack of proofreading exonuclease activity (Bebenek et al. 2005). Thus, even though the patches it synthesizes are short, its deployment is likely to be mutagenic.
Mammals have four X-family polymerases: Pol β, terminal deoxynucleotidyl transferase (TdT), Pol λ, and Pol μ, of which Pols β, λ, and μ are also capable of lesion bypass. Pol β can incorporate A or C opposite 8-oxoG with roughly equal efficiency because the templating 8-oxoG can adopt both syn and anti conformations in the active site of the enzyme (Batra et al. 2012). Pol μ is closely related to TdT and, like TdT (Desiderio et al. 1984), plays a role in V(D)J recombination of Ig genes, increasing junctional diversity (Bertocci et al. 2006). However, it is more widely expressed and participates more broadly in NHEJ (Chayot et al. 2012). Pol λ is the closest relative of S. cerevisiae Pol 4. It possesses dRP lyase activity and can participate in BER in vitro (Garcia-Diaz et al. 2001). However, its major in vivo role also appears to be in NHEJ, both during V(D)J recombination and more generally. Pol μ and Pol λ are both capable of TLS of a range of lesions in vitro (Duvauchelle et al. 2002; Maga et al. 2002; Zhang et al. 2002a) but have a propensity for primer misalignment that leads to –1 frameshifts (Bebenek et al. 2003; Covo et al. 2004; Bebenek et al. 2008). However, Pol λ is also capable of high-fidelity bypass, including of 8-oxoG, especially in the presence of the auxiliary factors replication protein A (RPA) and PCNA (Maga et al. 2007), helped by its novel proofreading mechanism in which misincorporated dA can be removed by pyrophosphorolysis (Crespan et al. 2012). The in vivo significance of this TLS capability for repair and for mutagenesis remains to be fully explored, but it is likely to facilitate the processing of complex breaks and gaps that also contain damaged bases (Picher and Blanco 2007; Zhou et al. 2008; Skosareva et al. 2012).
Translesion Synthesis and Mutagenesis by the Vertebrate A-Family Polymerases, Pol θ and Pol ν
Pol θ (Mus308) is an unusual enzyme in that it contains both helicase and polymerase domains (Harris et al. 1996; Sharief et al. 1999; Seki et al. 2003), of which there are additional paralogs in vertebrates: Pol ν (Marini et al. 2003), which lacks the helicase domain, and Hel308, which lacks DNA polymerase domains (Marini and Wood 2002). Unlike most A-family polymerases, such as E. coli Pol I, Pol θ, and Pol ν are significantly error prone (Seki et al. 2004; Takata et al. 2006). Pol θ exhibits a similar base substitution frequency to Pol κ but also has a high rate of indel mutagenesis (Arana et al. 2008, 2011), and Pol ν is unusual in incorporating T opposite template G with a frequency approaching 0.5 (Takata et al. 2006). Both enzymes are also capable of TLS. Pol θ can bypass abasic sites in a single step, incorporating A almost as readily as opposite template T (Seki et al. 2004), whereas both can bypass thymine glycol, again incorporating A. Neither Pol θ nor Pol ν can replicate the dinucleotide CPD and (6-4) photoproduct lesions completely (Seki et al. 2004; Takata et al. 2006). However, Pol θ is able to extend from mismatched primer termini including from bases incorporated opposite these UV-induced lesions (Seki and Wood 2008). Recently, Pol ν has also been shown to be capable of bypassing certain extremely large major groove adducts, including interstrand cross-links (Yamanaka et al. 2010), which is facilitated by its ability to catalyze strand-displacement DNA synthesis (Takata et al. 2006).
Evidence points to Pol θ and Pol ν being involved in a number of repair pathways, although firm clues as to their regulation are sparse. Both enzymes are processive and, given their error rates, it seems likely that their access to DNA is controlled. Mus308 was initially identified through its involvement in tolerance of interstrand cross-linking agents (Harris et al. 1996), and the role Pol θ and Pol ν play in DNA interstrand cross-link repair may reflect their ability to perform TLS or participate in certain forms of homologous recombination (Zietlow et al. 2009; Kohzaki et al. 2010; Moldovan et al. 2010; Yamanaka et al. 2010). Importantly, Pol ν both physically and functionally interacts with the FA pathway (Moldovan et al. 2010). Additionally, Drosophila Mus308 is involved in microhomology-mediated NHEJ (Chan et al. 2010; Yu and McVey 2010) and Pol θ in BER (Yoshimura et al. 2006; Prasad et al. 2009), which is likely facilitated by its terminal-transferase (Hogg et al. 2012) and dRP-lyase (Prasad et al. 2009) activities. As with the X-family polymerases, it seems likely that these roles are important in contexts involving complex or clustered damage.
THE ROLE OF TRANSLESION SYNTHESIS IN SOMATIC MUTATION OF IMMUNOGLOBULIN GENES
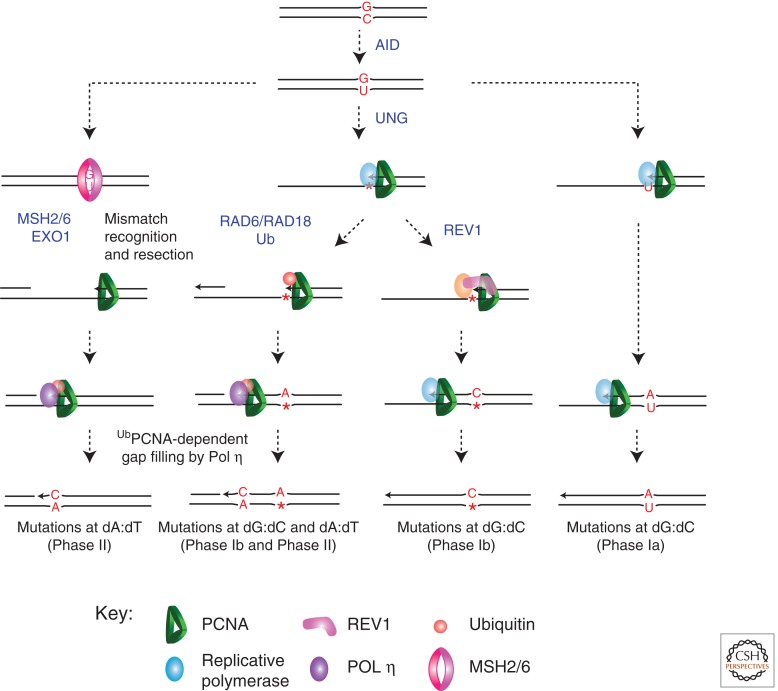
During the vertebrate immune response, TLS is co-opted to help generate an extraordinarily high level of mutagenesis in the Ig genes. Ig somatic hypermutation is driven by targeted deamination of C by activation-induced deaminase (AID) (Di Noia and Neuberger 2007). Although AID can only deaminate dC, its action is responsible for mutagenesis at all four bases. This mutagenesis can be divided into two genetically distinguishable phases, one targeted to C/G base pairs and a second targeted to A/T (Rada et al. 1998) (Fig. 4). These phases reflect the two broad mechanisms by which the translesion polymerases generate mutations; many mutations at C/G reflect error-prone replication of abasic sites, whereas mutations at A/T result from intrinsically error-prone DNA synthesis, principally by Pol η.
Figure 4.
The role of TLS in Ig gene somatic hypermutation. Ig gene hypermutation is initiated by AID, which deaminates dC in the Ig variable-region genes to produce uracil. This triggers a number of mutagenic consequences. In the right column, U is replicated by Pol δ or Pol ε, resulting in C/G to T/A transitions (Phase Ia [Neuberger et al. 2003]). Alternatively, U is excised to generate an abasic site (central columns). This can be replicated by a polymerase capable of TLS to generate all possible mutations at dC (Phase Ib). REV1 is illustrated and is responsible for C/G to G/C transversions. Alternatively, replication of the abasic site stimulates PCNA ubiquitination, possibly by gap formation following replication arrest, and this recruits Pol η. This can result in mutations at the site of dC deamination to generate Phase Ib mutation and could also lead to Phase II mutations at A/T base pairs generated by misincorporation on undamaged DNA by Pol η. Finally, the initial U/G mismatch can be recognized as a mismatch by MSH2/6, resulting in resection by EXO1 and formation of a gap. This, in turn, stimulates PCNA ubiquitination, recruitment of Pol η, and Phase II mutagenesis at A/T base pairs.
Mutagenesis at C/G base pairs is conceptually simple. It can result from direct replication of U generated by AID to give C/G to T/A transitions (Petersen-Mahrt et al. 2002), which account for about 60% of mutations at C/G. Alternatively, U can be excised by uracil DNA glycosylase (UNG) (Di Noia and Neuberger 2002; Rada et al. 2002; Imai et al. 2003) forming an abasic site, from which TLS can generate all possible substitutions. There is strong evidence that the deoxycytidyl transferase activity of REV1 contributes to generating C to G transversion mutations (Jansen et al. 2006; Ross and Sale 2006; Masuda et al. 2009), but it is less clear which other polymerases are involved. A role for Pol θ has been proposed (Masuda et al. 2005; Zan et al. 2005; Masuda et al. 2006), but is controversial (Martomo et al. 2008). The role played by Pol ζ is also enigmatic. Early experiments showed that lowering Pol ζ levels reduced hypermutation (Diaz et al. 2001; Zan et al. 2001), but it has been argued that this may simply be attributable to fewer cell divisions (Schenten et al. 2009). However, careful analysis of the pattern of hypermutation in Pol η−/− Pol ζ+/− animals revealed a decrease in tandem base substitutions compared with Pol η−/− Pol ζ+/+ animals, suggesting that Pol ζ does indeed contribute (Saribasak et al. 2012). Further, genetic analysis in DT40 cells suggests that Pol ζ contributes to the extension of at least some bypass events in the Ig locus (Hirota et al. 2010).
The origin of mutations at A/T base pairs is more complex. While still dependent on AID, A/T mutagenesis requires Pol η (Zeng et al. 2001; Delbos et al. 2005, 2007) and is likely to be the result of intrinsically error-prone synthesis by the enzyme (Pavlov et al. 2002). How is this linked to deamination of dC? A/T mutagenesis is only partially dependent on uracil excision by UNG. A second pathway, involving recognition and processing of the U/G mismatch by MSH2/6 and EXO1 is actually quantitatively more prominent (Rada et al. 2004). It seems likely that both uracil excision and mismatch recognition generate a common substrate, possibly a single-stranded gap (Sale et al. 2009), that promotes ubiquitination of PCNA and recruitment of Pol η (Langerak et al. 2007; Roa et al. 2008; Krijger et al. 2011). Under normal circumstances Pol η is the only polymerase responsible for A/T mutagenesis (Delbos et al. 2007). Pol ι does not appear to contribute to Ig diversification in mice (McDonald et al. 2003; Delbos et al. 2005; Martomo et al. 2006), whereas Pol κ can partially substitute for Pol η, resulting in an alteration in the spectrum of mutagenesis to A to C and T to G transversions (Delbos et al. 2005; Faili et al. 2009). It is thus far unclear how and why Pol η is preferentially selected.
MUTAGENESIS AND THE CANCER GENOME
Cancer is driven by the stepwise accumulation of mutations in key genes that control normal cellular behavior. The advent of massive sequencing technologies has revealed the extraordinary extent of mutagenesis in many tumors, with more than 100,000 individual point mutations per genome in some cases (Stratton 2011). The pattern of somatic mutations in cancer has been likened to an archaeological record that contains the history of all the mutagenic processes operational during the evolution of the tumor (Stratton 2011). Deciphering these patterns is going to be extremely complex and will require a detailed knowledge of how different agents damage DNA and how this damage is converted into mutations through the lesion-replication mechanisms discussed here. In some cases, it is already possible to distinguish patterns that make sense. For instance, tobacco smoke-associated lung-cancer genomes exhibit high levels of G:C to T:A transversions, characteristic of oxidative and adduct damage at G (Pleasance et al. 2010b). Likewise, the melanoma genome exhibits a high number of C to T transitions at dipyrimidine sequences characteristic of error-prone bypass of UV-induced damage (Pleasance et al. 2010a).
However, in many cases the patterns are more complex, and recent approaches have applied machine-learning algorithms to deconvolve individual mutational patterns (Nik-Zainal et al. 2012). A particularly striking example is a class of mutations detected in breast-cancer genomes, often in highly clustered mutational showers or kataegis (Nik-Zainal et al. 2012). These mutations are C to T and C to G substitutions at TpCpX trinucleotides, a signature consistent with the action of some members of the AID/APOBEC family (Beale et al. 2004). Since these enzymes act on single-stranded DNA, the clusters of mutation may reflect areas where single-stranded DNA has been exposed, for instance by replication arrest or resection during homology-directed repair (Robert et al. 2012). By analogy with Ig-gene somatic hypermutation the C to T transitions may reflect direct replication of uracil, whereas the C to G transversions may be the result of REV1-dependent abasic site replication.
TRANSLESION SYNTHESIS AND CANCER: A CAUSE AND POTENTIAL TARGET?
Although an inherited deficiency in Pol η results in a marked predisposition to skin cancer, spontaneously arising mutation of the TLS polymerases has not emerged as a common event in human cancer (Lange et al. 2011). Nonetheless, polymorphisms in the TLS polymerases (Wang et al. 2004; Sakiyama et al. 2005; He et al. 2008) and dysregulation of their expression (O-Wang et al. 2001; Yang et al. 2004; Albertella et al. 2005b; Bavoux et al. 2005b; Lee and Matsushita 2005) have been linked to cancer.
In addition to TLS potentially contributing to the development of some cancers, there is considerable interest in modulating TLS as an adjunct to conventional cancer chemotherapy. There are two potentially important ways in which this could improve current treatments. First, downregulation of TLS sensitizes cells to commonly used chemotherapeutics, notably the widely used platinum-based DNA cross-linking agents such as cisplatin and oxaliplatin (Simpson and Sale 2003; Sonoda et al. 2003; Wu et al. 2004; Albertella et al. 2005a; Sharma et al. 2012). Proof of the potential of this approach has been provided by experiments in mice in which suppression of REV3 resulted in sensitization of an otherwise chemoresistant lung tumor to cisplatin (Doles et al. 2010). Second, many current cancer chemotherapeutics act by damaging DNA, and TLS could contribute to chemotherapy-induced mutagenesis that leads to the acquisition of resistance and, potentially, secondary tumor formation. The role of TLS in the acquisition of resistance to chemotherapeutics has been demonstrated both in cell lines (Wu et al. 2004; Okuda et al. 2005) and in mice (Xie et al. 2010). Thus, modulation of TLS holds significant promise as an adjunct to conventional chemotherapy, both as a means of increasing its effectiveness and reducing the emergence of resistance and secondary tumors. More work will be needed, however, to understand the balance between the beneficial effects of TLS inhibition and the hazardous consequences of inducing potentially more damaging genomic instability.
Footnotes
Editors: Errol C. Friedberg, Stephen J. Elledge, Alan R. Lehmann, Tomas Lindahl, and Marco Muzi-Falconi
Additional Perspectives on DNA Repair, Mutagenesis, and Other Responses to DNA Damage available at www.cshperspectives.org
REFERENCES
- Acharya N, Haracska L, Johnson RE, Unk I, Prakash S, Prakash L 2005. Complex formation of yeast Rev1 and Rev7 proteins: A novel role for the polymerase-associated domain. Mol Cell Biol 25: 9734–9740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acharya N, Haracska L, Prakash S, Prakash L 2007. Complex formation of yeast Rev1 with DNA polymerase η. Mol Cell Biol 27: 8401–8408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acharya N, Johnson R, Pages V, Prakash L, Prakash S 2009. Yeast Rev1 protein promotes complex formation of DNA polymerase ζ with Pol32 subunit of DNA polymerase δ. Proc Natl Acad Sci 106: 9631–9636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albertella MR, Green CM, Lehmann AR, O’Connor MJ 2005a. A role for polymerase η in the cellular tolerance to cisplatin-induced damage. Cancer Res 65: 9799–9806 [DOI] [PubMed] [Google Scholar]
- Albertella MR, Lau A, O’Connor MJ 2005b. The overexpression of specialized DNA polymerases in cancer. DNA Repair (Amst) 4: 583–593 [DOI] [PubMed] [Google Scholar]
- Alt A, Lammens K, Chiocchini C, Lammens A, Pieck JC, Kuch D, Hopfner KP, Carell T 2007. Bypass of DNA lesions generated during anticancer treatment with cisplatin by DNA polymerase η. Science 318: 967–970 [DOI] [PubMed] [Google Scholar]
- Arakawa H, Moldovan GL, Saribasak H, Saribasak NN, Jentsch S, Buerstedde JM 2006. A role for PCNA ubiquitination in immunoglobulin hypermutation. PLoS Biol 4: e366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arana ME, Kunkel TA 2010. Mutator phenotypes due to DNA replication infidelity. Semin Cancer Biol 20: 304–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arana ME, Seki M, Wood RD, Rogozin IB, Kunkel TA 2008. Low-fidelity DNA synthesis by human DNA polymerase θ. Nucleic Acids Res 36: 3847–3856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arana ME, Potapova O, Kunkel TA, Joyce CM 2011. Kinetic analysis of the unique error signature of human DNA polymerase ν. Biochemistry 50: 10126–10135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avkin S, Goldsmith M, Velasco-Miguel S, Geacintov N, Friedberg EC, Livneh Z 2004. Quantitative analysis of translesion DNA synthesis across a benzo[a]pyrene-guanine adduct in mammalian cells: The role of DNA polymerase κ. J Biol Chem 279: 53298–53305 [DOI] [PubMed] [Google Scholar]
- Batra VK, Shock DD, Beard WA, McKenna CE, Wilson SH 2012. Binary complex crystal structure of DNA polymerase β reveals multiple conformations of the templating 8-oxoguanine lesion. Proc Natl Acad Sci 109: 113–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bavoux C, Hoffmann JS, Cazaux C 2005a. Adaptation to DNA damage and stimulation of genetic instability: The double-edged sword mammalian DNA polymerase κ. Biochimie 87: 637–646 [DOI] [PubMed] [Google Scholar]
- Bavoux C, Leopoldino AM, Bergoglio VJOW, Ogi T, Bieth A, Judde JG, Pena SD, Poupon MF, Helleday T, et al. 2005b. Up-regulation of the error-prone DNA polymerase κ promotes pleiotropic genetic alterations and tumorigenesis. Cancer Res 65: 325–330 [PubMed] [Google Scholar]
- Beale RC, Petersen-Mahrt SK, Watt IN, Harris RS, Rada C, Neuberger MS 2004. Comparison of the differential context-dependence of DNA deamination by APOBEC enzymes: Correlation with mutation spectra in vivo. J Mol Biol 337: 585–596 [DOI] [PubMed] [Google Scholar]
- Bebenek K, Tissier A, Frank EG, McDonald JP, Prasad R, Wilson SH, Woodgate R, Kunkel TA 2001. 5′-Deoxyribose phosphate lyase activity of human DNA polymerase ι in vitro. Science 291: 2156–2159 [DOI] [PubMed] [Google Scholar]
- Bebenek K, Garcia-Diaz M, Blanco L, Kunkel TA 2003. The frameshift infidelity of human DNA polymerase λ: Implications for function. J Biol Chem 278: 34685–34690 [DOI] [PubMed] [Google Scholar]
- Bebenek K, Garcia-Diaz M, Patishall SR, Kunkel TA 2005. Biochemical properties of Saccharomyces cerevisiae DNA polymerase IV. J Biol Chem 280: 20051–20058 [DOI] [PubMed] [Google Scholar]
- Bebenek K, Garcia-Diaz M, Foley MC, Pedersen LC, Schlick T, Kunkel TA 2008. Substrate-induced DNA strand misalignment during catalytic cycling by DNA polymerase λ. EMBO Rep 9: 459–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bemark M, Khamlichi AA, Davies SL, Neuberger MS 2000. Disruption of mouse polymerase ζ (Rev3) leads to embryonic lethality and impairs blastocyst development in vitro. Curr Biol 10: 1213–1216 [DOI] [PubMed] [Google Scholar]
- Bertocci B, De Smet A, Weill J, Reynaud C 2006. Nonoverlapping functions of DNA polymerases μ, λ, and terminal deoxynucleotidyltransferase during immunoglobulin V(D)J recombination in vivo. Immunity 25: 31–41 [DOI] [PubMed] [Google Scholar]
- Bienko M, Green C, Crosetto N, Rudolf F, Zapart G, Coull B, Kannouche P, Wider G, Peter M, Lehmann A, et al. 2005. Ubiquitin-binding domains in Y-family polymerases regulate translesion synthesis. Science 310: 1821–1824 [DOI] [PubMed] [Google Scholar]
- Bienko M, Green CM, Sabbioneda S, Crosetto N, Matic I, Hibbert RG, Begovic T, Niimi A, Mann M, Lehmann AR, et al. 2010. Regulation of translesion synthesis DNA polymerase η by monoubiquitination. Mol Cell 37: 396–407 [DOI] [PubMed] [Google Scholar]
- Biertumpfel C, Zhao Y, Kondo Y, Ramon-Maiques S, Gregory M, Lee JY, Masutani C, Lehmann AR, Hanaoka F, Yang W 2010. Structure and mechanism of human DNA polymerase η. Nature 465: 1044–1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busuttil RA, Lin Q, Stambrook PJ, Kucherlapati R, Vijg J 2008. Mutation frequencies and spectra in DNA polymerase η-deficient mice. Cancer Res 68: 2081–2084 [DOI] [PubMed] [Google Scholar]
- Byun TS, Pacek M, Yee MC, Walter JC, Cimprich KA 2005. Functional uncoupling of MCM helicase and DNA polymerase activities activates the ATR-dependent checkpoint. Genes Dev 19: 1040–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassier C, Chanet R, Henriques JA, Moustacchi E 1980. The effects of three PSO genes on induced mutagenesis: A novel class of mutationally defective yeast. Genetics 96: 841–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SH, Yu AM, McVey M 2010. Dual roles for DNA polymerase θ in alternative end-joining repair of double-strand breaks in Drosophila. PLoS Genet 6: e1001005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chayot R, Montagne B, Ricchetti M 2012. DNA polymerase μ is a global player in the repair of non-homologous end-joining substrates. DNA Repair 11: 22–34 [DOI] [PubMed] [Google Scholar]
- Chiu RK, Brun J, Ramaekers C, Theys J, Weng L, Lambin P, Gray DA, Wouters BG 2006. Lysine 63-polyubiquitination guards against translesion synthesis-induced mutations. PLoS Genet 2: e116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi JH, Pfeifer GP 2005. The role of DNA polymerase η in UV mutational spectra. DNA Repair 4: 211–220 [DOI] [PubMed] [Google Scholar]
- Choi JH, Besaratinia A, Lee DH, Lee CS, Pfeifer GP 2006. The role of DNA polymerase ι in UV mutational spectra. Mutat Res 599: 58–65 [DOI] [PubMed] [Google Scholar]
- Cleaver JE 1972. Xeroderma pigmentosum: Variants with normal DNA repair and normal sensitivity to ultraviolet light. J Invest Dermatol 58: 124–128 [DOI] [PubMed] [Google Scholar]
- Cleaver JE, Afzal V, Feeney L, McDowell M, Sadinski W, Volpe JP, Busch DB, Coleman DM, Ziffer DW, Yu Y, et al. 1999. Increased ultraviolet sensitivity and chromosomal instability related to P53 function in the xeroderma pigmentosum variant. Cancer Res 59: 1102–1108 [PubMed] [Google Scholar]
- Collins NS, Bhattacharyya S, Lahue RS 2007. Rev1 enhances CAG.CTG repeat stability in Saccharomyces cerevisiae. DNA Repair (Amst) 6: 38–44 [DOI] [PubMed] [Google Scholar]
- Coulon S, Ramasubramanyan S, Alies C, Philippin G, Lehmann A, Fuchs RP 2010. Rad8Rad5/Mms2-Ubc13 ubiquitin ligase complex controls translesion synthesis in fission yeast. EMBO J 29: 2048–2058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covo S, Blanco L, Livneh Z 2004. Lesion bypass by human DNA polymerase μ reveals a template-dependent, sequence-independent nucleotidyl transferase activity. J Biol Chem 279: 859–865 [DOI] [PubMed] [Google Scholar]
- Crespan E, Maga G, Hubscher U 2012. A new proofreading mechanism for lesion bypass by DNA polymerase λ. EMBO Rep 13: 68–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies AA, Huttner D, Daigaku Y, Chen S, Ulrich HD 2008. Activation of ubiquitin-dependent DNA damage bypass is mediated by replication protein A. Mol Cell 29: 625–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Groote FH, Jansen JG, Masuda Y, Shah DM, Kamiya K, de Wind N, Siegal G 2011. The Rev1 translesion synthesis polymerase has multiple distinct DNA binding modes. DNA Repair 10: 915–925 [DOI] [PubMed] [Google Scholar]
- Delbos F, De Smet A, Faili A, Aoufouchi S, Weill JC, Reynaud CA 2005. Contribution of DNA polymerase η to immunoglobulin gene hypermutation in the mouse. J Exp Med 201: 1191–1196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delbos F, Aoufouchi S, Faili A, Weill JC, Reynaud CA 2007. DNA polymerase η is the sole contributor of A/T modifications during immunoglobulin gene hypermutation in the mouse. J Exp Med 204: 17–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Padula M, Slezak G, Auffret van Der Kemp P, Boiteux S 2004. The post-replication repair RAD18 and RAD6 genes are involved in the prevention of spontaneous mutations caused by 7,8-dihydro-8-oxoguanine in Saccharomyces cerevisiae. Nucleic Acids Res 32: 5003–5010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desiderio SV, Yancopoulos GD, Paskind M, Thomas E, Boss MA, Landau N, Alt FW, Baltimore D 1984. Insertion of N regions into heavy-chain genes is correlated with expression of terminal deoxytransferase in B cells. Nature 311: 752–755 [DOI] [PubMed] [Google Scholar]
- Diaz M, Verkoczy LK, Flajnik MF, Klinman NR 2001. Decreased frequency of somatic hypermutation and impaired affinity maturation but intact germinal center formation in mice expressing antisense RNA to DNA polymerase ζ. J Immunol 167: 327–335 [DOI] [PubMed] [Google Scholar]
- Di Noia J, Neuberger MS 2002. Altering the pathway of immunoglobulin hypermutation by inhibiting uracil-DNA glycosylase. Nature 419: 43–48 [DOI] [PubMed] [Google Scholar]
- Di Noia JM, Neuberger MS 2007. Molecular mechanisms of antibody somatic hypermutation. Annu Rev Biochem 76: 1–22 [DOI] [PubMed] [Google Scholar]
- Doles J, Oliver TG, Cameron ER, Hsu G, Jacks T, Walker GC, Hemann MT 2010. Suppression of Rev3, the catalytic subunit of Polζ, sensitizes drug-resistant lung tumors to chemotherapy. Proc Natl Acad Sci 107: 20786–20791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Souza S, Waters L, Walker G 2008. Novel conserved motifs in Rev1 C-terminus are required for mutagenic DNA damage tolerance. DNA Repair (Amst) 7: 1455–1470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumstorf CA, Clark AB, Lin Q, Kissling GE, Yuan T, Kucherlapati R, McGregor WG, Kunkel TA 2006. Participation of mouse DNA polymerase ι in strand-biased mutagenic bypass of UV photoproducts and suppression of skin cancer. Proc Natl Acad Sci 103: 18083–18088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvauchelle JB, Blanco L, Fuchs RP, Cordonnier AM 2002. Human DNA polymerase μ (Pol mu) exhibits an unusual replication slippage ability at AAF lesion. Nucleic Acids Res 30: 2061–2067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmunds CE, Simpson LJ, Sale JE 2008. PCNA ubiquitination and REV1 dDefine temporally distinct mechanisms for controlling translesion synthesis in the avian cell line DT40. Mol Cell 30: 519–529 [DOI] [PubMed] [Google Scholar]
- Esposito G, Godindagger I, Klein U, Yaspo ML, Cumano A, Rajewsky K 2000. Disruption of the Rev3l-encoded catalytic subunit of polymerase ζ in mice results in early embryonic lethality. Curr Biol 10: 1221–1224 [DOI] [PubMed] [Google Scholar]
- Faili A, Stary A, Delbos F, Weller S, Aoufouchi S, Sarasin A, Weill JC, Reynaud CA 2009. A backup role of DNA polymerase κ in Ig gene hypermutation only takes place in the complete absence of DNA polymerase η. J Immunol 182: 6353–6359 [DOI] [PubMed] [Google Scholar]
- Gan GN, Wittschieben JP, Wittschieben BO, Wood RD 2008. DNA polymerase ζ (pol zeta) in higher eukaryotes. Cell Res 18: 174–183 [DOI] [PubMed] [Google Scholar]
- Garcia-Diaz M, Bebenek K, Kunkel TA, Blanco L 2001. Identification of an intrinsic 5′-deoxyribose-5-phosphate lyase activity in human DNA polymerase λ: A possible role in base excision repair. J Biol Chem 276: 34659–34663 [DOI] [PubMed] [Google Scholar]
- Garg P, Burgers PM 2005. Ubiquitinated proliferating cell nuclear antigen activates translesion DNA polymerases η and REV1. Proc Natl Acad Sci 102: 18361–18366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs PE, Wang XD, Li Z, McManus TP, McGregor WG, Lawrence CW, Maher VM 2000. The function of the human homolog of Saccharomyces cerevisiae REV1 is required for mutagenesis induced by UV light. Proc Natl Acad Sci 97: 4186–4191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs PE, McDonald J, Woodgate R, Lawrence CW 2005. The relative roles in vivo of Saccharomyces cerevisiae Pol η, Pol ζ, Rev1 protein and Pol32 in the bypass and mutation induction of an abasic site, T-T (6–4) photoadduct and T-T cis-syn cyclobutane dimer. Genetics 169: 575–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gohler T, Sabbioneda S, Green CM, Lehmann AR 2011. ATR-mediated phosphorylation of DNA polymerase η is needed for efficient recovery from UV damage. J Cell Biol 192: 219–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gueranger Q, Stary A, Aoufouchi S, Faili A, Sarasin A, Reynaud CA, Weill JC 2008. Role of DNA polymerases η, ι and ζ in UV resistance and UV-induced mutagenesis in a human cell line. DNA Repair (Amst) 7: 1551–1562 [DOI] [PubMed] [Google Scholar]
- Guo C, Fischhaber PL, Luk-Paszyc MJ, Masuda Y, Zhou J, Kamiya K, Kisker C, Friedberg EC 2003. Mouse Rev1 protein interacts with multiple DNA polymerases involved in translesion DNA synthesis. EMBO J 22: 6621–6630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo C, Sonoda E, Tang TS, Parker JL, Bielen AB, Takeda S, Ulrich HD, Friedberg EC 2006a. REV1 protein interacts with PCNA: Significance of the REV1 BRCT domain in vitro and in vivo. Mol Cell 23: 265–271 [DOI] [PubMed] [Google Scholar]
- Guo C, Tang TS, Bienko M, Parker JL, Bielen AB, Sonoda E, Takeda S, Ulrich HD, Dikic I, Friedberg EC 2006b. Ubiquitin-binding motifs in REV1 protein are required for its role in the tolerance of DNA damage. Mol Cell Biol 26: 8892–8900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara K, Hashimoto H, Murakumo Y, Kobayashi S, Kogame T, Unzai S, Akashi S, Takeda S, Shimizu T, Sato M 2010. Crystal structure of human REV7 in complex with a Human REV3 fragment and structural implication of the interaction between DNA polymerase ζ and REV1. J Biol Chem 285: 12299–12307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haracska L, Prakash S, Prakash L 2002. Yeast Rev1 protein is a G template-specific DNA polymerase. J Biol Chem 277: 15546–15551 [DOI] [PubMed] [Google Scholar]
- Haracska L, Prakash S, Prakash L 2003. Yeast DNA polymerase ζ is an efficient extender of primer ends opposite from 7,8-dihydro-8-Oxoguanine and O6-methylguanine. Mol Cell Biol 23: 1453–1459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haracska L, Torres-Ramos C, Johnson R, Prakash S, Prakash L 2004. Opposing effects of ubiquitin conjugation and SUMO modification of PCNA on replicational bypass of DNA lesions in Saccharomyces cerevisiae. Mol Cell Biol 24: 4267–4274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harfe BD, Jinks-Robertson S 2000. DNA polymerase ζ introduces multiple mutations when bypassing spontaneous DNA damage in Saccharomyces cerevisiae. Mol Cell 6: 1491–1499 [DOI] [PubMed] [Google Scholar]
- Harris PV, Mazina OM, Leonhardt EA, Case RB, Boyd JB, Burtis KC 1996. Molecular cloning of Drosophila mus308, a gene involved in DNA cross-link repair with homology to prokaryotic DNA polymerase I genes. Mol Cell Biol 16: 5764–5771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, Ye F, Zhang J, Cheng Q, Shen J, Chen H 2008. REV1 genetic variants associated with the risk of cervical carcinoma. Eur J Epidemiol 23: 403–409 [DOI] [PubMed] [Google Scholar]
- Hendel A, Ziv O, Gueranger Q, Geacintov N, Livneh Z 2008. Reduced efficiency and increased mutagenicity of translesion DNA synthesis across a TT cyclobutane pyrimidine dimer, but not a TT 6–4 photoproduct, in human cells lacking DNA polymerase η. DNA Repair (Amst) 7: 1636–1646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendel A, Krijger PH, Diamant N, Goren Z, Langerak P, Kim J, Reissner T, Lee KY, Geacintov NE, Carell T, et al. 2011. PCNA Ubiquitination is important, but not essential for translesion DNA synthesis in mammalian cells. PLoS Genet 7: e1002262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano Y, Sugimoto K 2006. ATR homolog Mec1 controls association of DNA polymerase ζ-Rev1 complex with regions near a double-strand break. Curr Biol 16: 586–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota K, Sonoda E, Kawamoto T, Motegi A, Masutani C, Hanaoka F, Szuts D, Iwai S, Sale JE, Lehmann A, et al. 2010. Simultaneous disruption of two DNA polymerases, Polη and Polζ, in Avian DT40 cells unmasks the role of Polη in cellular response to various DNA lesions. PLoS Genet 6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S 2002. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 419: 135–141 [DOI] [PubMed] [Google Scholar]
- Hogg M, Sauer-Eriksson AE, Johansson E 2012. Promiscuous DNA synthesis by human DNA polymerase θ. Nucleic Acids Res 40: 2611–2622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holbeck SL, Strathern JN 1997. A role for REV3 in mutagenesis during double-strand break repair in Saccharomyces cerevisiae. Genetics 147: 1017–1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai K, Slupphaug G, Lee WI, Revy P, Nonoyama S, Catalan N, Yel L, Forveille M, Kavli B, Krokan HE, et al. 2003. Human uracil-DNA glycosylase deficiency associated with profoundly impaired immunoglobulin class-switch recombination. Nat Immunol 4: 1023–1028 [DOI] [PubMed] [Google Scholar]
- Jansen JG, Tsaalbi-Shtylik A, Langerak P, Calleja F, Meijers CM, Jacobs H, de Wind N 2005. The BRCT domain of mammalian Rev1 is involved in regulating DNA translesion synthesis. Nucleic Acids Res 33: 356–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen JG, Langerak P, Tsaalbi-Shtylik A, van den Berk P, Jacobs H, de Wind N 2006. Strand-biased defect in C/G transversions in hypermutating immunoglobulin genes in Rev1-deficient mice. J Exp Med 203: 319–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen J, Fousteri M, de Wind N 2007. Send in the clamps: Control of DNA translesion synthesis in eukaryotes. Mol Cell 28: 522–529 [DOI] [PubMed] [Google Scholar]
- Jansen J, Tsaalbi-Shtylik A, Hendriks G, Gali H, Hendel A, Johansson F, Erixon K, Livneh Z, Mullenders L, Haracska L, et al. 2009. Separate domains of Rev1 mediate two modes of DNA damage bypass in mammalian cells. Mol Cell Biol 29: 3113–3123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarosz DF, Godoy VG, Delaney JC, Essigmann JM, Walker GC 2006. A single amino acid governs enhanced activity of DinB DNA polymerases on damaged templates. Nature 439: 225–228 [DOI] [PubMed] [Google Scholar]
- Johnson RE, Kondratick CM, Prakash S, Prakash L 1999a. hRAD30 mutations in the variant form of xeroderma pigmentosum. Science 285: 263–265 [DOI] [PubMed] [Google Scholar]
- Johnson RE, Prakash S, Prakash L 1999b. Efficient bypass of a thymine-thymine dimer by yeast DNA polymerase, Polη. Science 283: 1001–1004 [DOI] [PubMed] [Google Scholar]
- Johnson RE, Prakash S, Prakash L 2000a. The human DINB1 gene encodes the DNA polymerase Polθ. Proc Natl Acad Sci 97: 3838–3843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson RE, Washington MT, Haracska L, Prakash S, Prakash L 2000b. Eukaryotic polymerases ι and ζ act sequentially to bypass DNA lesions. Nature 406: 1015–1019 [DOI] [PubMed] [Google Scholar]
- Johnson RE, Washington MT, Prakash S, Prakash L 2000c. Fidelity of human DNA polymerase eta. J Biol Chem 275: 7447–7450 [DOI] [PubMed] [Google Scholar]
- Johnson RE, Haracska L, Prakash S, Prakash L 2001. Role of DNA polymerase η in the bypass of a (6–4) TT photoproduct. Mol Cell Biol 21: 3558–3563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson RE, Prakash L, Prakash S 2012. Pol31 and Pol32 subunits of yeast DNA polymerase delta are also essential subunits of DNA polymerase ζ. Proc Natl Acad Sci 109: 12455–12460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung YS, Liu G, Chen X 2010. Pirh2 E3 ubiquitin ligase targets DNA polymerase η for 20S proteasomal degradation. Mol Cell Biol 30: 1041–1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kai M, Wang TS 2003. Checkpoint activation regulates mutagenic translesion synthesis. Genes Dev 17: 64–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajiwara K, O-Wang J, Sakurai T, Yamashita S, Tanaka M, Sato M, Tagawa M, Sugaya E, Nakamura K, Nakao K, et al. 2001. Sez4 gene encoding an elongation subunit of DNA polymerase ζ is required for normal embryogenesis. Genes Cells 6: 99–106 [DOI] [PubMed] [Google Scholar]
- Kannouche P, Broughton BC, Volker M, Hanaoka F, Mullenders LH, Lehmann AR 2001. Domain structure, localization, and function of DNA polymerase η, defective in xeroderma pigmentosum variant cells. Genes Dev 15: 158–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannouche PL, Wing J, Lehmann AR 2004. Interaction of human DNA polymerase η with monoubiquitinated PCNA: A possible mechanism for the polymerase switch in response to DNA damage. Mol Cell 14: 491–500 [DOI] [PubMed] [Google Scholar]
- Kee Y, D’Andrea AD 2010. Expanded roles of the Fanconi anemia pathway in preserving genomic stability. Genes Dev 24: 1680–1694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim N, Mudrak SV, Jinks-Robertson S 2011. The dCMP transferase activity of yeast Rev1 is biologically relevant during the bypass of endogenously generated AP sites. DNA Repair 10: 1262–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Yang K, Dejsuphong D, D’Andrea AD 2012. Regulation of Rev1 by the Fanconi anemia core complex. Nat Struct Mol Biol 19: 164–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirouac KN, Ling H 2011. Unique active site promotes error-free replication opposite an 8-oxo-guanine lesion by human DNA polymerase ι. Proc Natl Acad Sci 108: 3210–3215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knipscheer P, Raschle M, Smogorzewska A, Enoiu M, Ho TV, Scharer OD, Elledge SJ, Walter JC 2009. The Fanconi anemia pathway promotes replication-dependent DNA interstrand cross-link repair. Science 326: 1698–1701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohzaki M, Nishihara K, Hirota K, Sonoda E, Yoshimura M, Ekino S, Butler JE, Watanabe M, Halazonetis TD, Takeda S 2010. DNA polymerases ν and θ are required for efficient immunoglobulin V gene diversification in chicken. J Cell Biol 189: 1117–1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosarek JN, Woodruff RV, Rivera-Begeman A, Guo C, D’Souza S, Koonin EV, Walker GC, Friedberg EC 2008. Comparative analysis of in vivo interactions between Rev1 protein and other Y-family DNA polymerases in animals and yeasts. DNA Repair (Amst) 7: 439–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krijger PH, van den Berk PC, Wit N, Langerak P, Jansen JG, Reynaud CA, de Wind N, Jacobs H 2011. PCNA ubiquitination-independent activation of polymerase η during somatic hypermutation and DNA damage tolerance. DNA Repair 10: 1051–1059 [DOI] [PubMed] [Google Scholar]
- Lange SS, Takata K, Wood RD 2011. DNA polymerases and cancer. Nat Rev Cancer 11: 96–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange SS, Wittschieben JP, Wood RD 2012. DNA polymerase ζ is required for proliferation of normal mammalian cells. Nucleic Acids Res 40: 4473–4482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langerak P, Nygren AO, Krijger PH, van den Berk PC, Jacobs H 2007. A/T mutagenesis in hypermutated immunoglobulin genes strongly depends on PCNAK164 modification. J Exp Med 204: 1989–1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence CW, Christensen R 1976. UV mutagenesis in radiation-sensitive strains of yeast. Genetics 82: 207–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee GH, Matsushita H 2005. Genetic linkage between Pol ι deficiency and increased susceptibility to lung tumors in mice. Cancer Sci 96: 256–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemontt JF 1971. Mutants of yeast defective in mutation induced by ultraviolet light. Genetics 68: 21–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limoli CL, Giedzinski E, Bonner WM, Cleaver JE 2002a. UV-induced replication arrest in the xeroderma pigmentosum variant leads to DNA double-strand breaks, γ-2AX formation, and Mre11 relocalization. Proc Natl Acad Sci 99: 233–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limoli CL, Laposa R, Cleaver JE 2002b. DNA replication arrest in XP variant cells after UV exposure is diverted into an Mre11-dependent recombination pathway by the kinase inhibitor wortmannin. Mutat Res 510: 121–129 [DOI] [PubMed] [Google Scholar]
- Loeb LA, Monnat RJ Jr. 2008. DNA polymerases and human disease. Nat Rev Genet 9: 594–604 [DOI] [PubMed] [Google Scholar]
- Lone S, Townson SA, Uljon SN, Johnson RE, Brahma A, Nair DT, Prakash S, Prakash L, Aggarwal AK 2007. Human DNA polymerase κ encircles DNA: Implications for mismatch extension and lesion bypass. Mol Cell 25: 601–614 [DOI] [PubMed] [Google Scholar]
- Long DT, Raschle M, Joukov V, Walter JC 2011. Mechanism of RAD51-dependent DNA interstrand cross-link repair. Science 333: 84–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maga G, Villani G, Ramadan K, Shevelev I, Tanguy Le Gac N, Blanco L, Blanca G, Spadari S, Hubscher U 2002. Human DNA polymerase λ functionally and physically interacts with proliferating cell nuclear antigen in normal and translesion DNA synthesis. J Biol Chem 277: 48434–48440 [DOI] [PubMed] [Google Scholar]
- Maga G, Villani G, Crespan E, Wimmer U, Ferrari E, Bertocci B, Hubscher U 2007. 8-oxo-guanine bypass by human DNA polymerases in the presence of auxiliary proteins. Nature 447: 606–608 [DOI] [PubMed] [Google Scholar]
- Majka J, Binz SK, Wold MS, Burgers PM 2006. Replication protein A directs loading of the DNA damage checkpoint clamp to 5′-DNA junctions. J Biol Chem 281: 27855–27861 [DOI] [PubMed] [Google Scholar]
- Marini F, Wood RD 2002. A human DNA helicase homologous to the DNA cross-link sensitivity protein Mus308. J Biol Chem 277: 8716–8723 [DOI] [PubMed] [Google Scholar]
- Marini F, Kim N, Schuffert A, Wood RD 2003. POLN, a nuclear PolA family DNA polymerase homologous to the DNA cross-link sensitivity protein Mus308. J Biol Chem 278: 32014–32019 [DOI] [PubMed] [Google Scholar]
- Martomo SA, Yang WW, Vaisman A, Maas A, Yokoi M, Hoeijmakers JH, Hanaoka F, Woodgate R, Gearhart PJ 2006. Normal hypermutation in antibody genes from congenic mice defective for DNA polymerase ι. DNA Repair 5: 392–398 [DOI] [PubMed] [Google Scholar]
- Martomo SA, Saribasak H, Yokoi M, Hanaoka F, Gearhart PJ 2008. Reevaluation of the role of DNA polymerase θ in somatic hypermutation of immunoglobulin genes. DNA Repair 7: 1603–1608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda K, Ouchida R, Takeuchi A, Saito T, Koseki H, Kawamura K, Tagawa M, Tokuhisa T, Azuma T, O-Wang J 2005. DNA polymerase θ contributes to the generation of C/G mutations during somatic hypermutation of Ig genes. Proc Natl Acad Sci 102: 13986–13991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda K, Ouchida R, Hikida M, Nakayama M, Ohara O, Kurosaki T, O-Wang J 2006. Absence of DNA polymerase θ results in decreased somatic hypermutation frequency and altered mutation patterns in Ig genes. DNA Repair 5: 1384–1391 [DOI] [PubMed] [Google Scholar]
- Masuda K, Ouchida R, Li Y, Gao X, Mori H, Wang JY 2009. A critical role for REV1 in regulating the induction of C:G transitions and A:T mutations during Ig gene hypermutation. J Immunol 183: 1846–1850 [DOI] [PubMed] [Google Scholar]
- Masutani C, Kusumoto R, Yamada A, Dohmae N, Yokoi M, Yuasa M, Araki M, Iwai S, Takio K, Hanaoka F 1999. The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase η. Nature 399: 700–704 [DOI] [PubMed] [Google Scholar]
- Masutani C, Kusumoto R, Iwai S, Hanaoka F 2000. Mechanisms of accurate translesion synthesis by human DNA polymerase η. EMBO J 19: 3100–3109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda T, Bebenek K, Masutani C, Hanaoka F, Kunkel TA 2000. Low fidelity DNA synthesis by human DNA polymerase η. Nature 404: 1011–1013 [DOI] [PubMed] [Google Scholar]
- Matsuda T, Bebenek K, Masutani C, Rogozin IB, Hanaoka F, Kunkel TA 2001. Error rate and specificity of human and murine DNA polymerase η. J Mol Biol 312: 335–346 [DOI] [PubMed] [Google Scholar]
- McCulloch SD, Kunkel TA 2008. The fidelity of DNA synthesis by eukaryotic replicative and translesion synthesis polymerases. Cell Res 18: 148–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald JP, Levine AS, Woodgate R 1997. The Saccharomyces cerevisiae RAD30 gene, a homologue of Escherichia coli dinB and umuC, is DNA damage inducible and functions in a novel error-free postreplication repair mechanism. Genetics 147: 1557–1568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald JP, Rapic-Otrin V, Epstein JA, Broughton BC, Wang X, Lehmann AR, Wolgemuth DJ, Woodgate R 1999. Novel human and mouse homologs of Saccharomyces cerevisiae DNA polymerase η. Genomics 60: 20–30 [DOI] [PubMed] [Google Scholar]
- McDonald JP, Frank EG, Plosky BS, Rogozin IB, Masutani C, Hanaoka F, Woodgate R, Gearhart PJ 2003. 129-derived strains of mice are deficient in DNA polymerase ι and have normal immunoglobulin hypermutation. J Exp Med 198: 635–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirchandani KD, McCaffrey RM, D’Andrea AD 2008. The Fanconi anemia core complex is required for efficient point mutagenesis and Rev1 foci assembly. DNA Repair (Amst) 7: 902–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moldovan GL, Madhavan MV, Mirchandani KD, McCaffrey RM, Vinciguerra P, D’Andrea AD 2010. DNA polymerase POLN participates in cross-link repair and homologous recombination. Mol Cell Biol 30: 1088–1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison A, Christensen RB, Alley J, Beck AK, Bernstine EG, Lemontt JF, Lawrence CW 1989. REV3, a Saccharomyces cerevisiae gene whose function is required for induced mutagenesis, is predicted to encode a nonessential DNA polymerase. J Bacteriol 171: 5659–5667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakumo Y, Ogura Y, Ishii H, Numata S, Ichihara M, Croce CM, Fishel R, Takahashi M 2001. Interactions in the error-prone postreplication repair proteins hREV1, hREV3, and hREV7. J Biol Chem 276: 35644–35651 [DOI] [PubMed] [Google Scholar]
- Nair DT, Johnson RE, Prakash S, Prakash L, Aggarwal AK 2004. Replication by human DNA polymerase ι occurs by Hoogsteen base-pairing. Nature 430: 377–380 [DOI] [PubMed] [Google Scholar]
- Nair DT, Johnson RE, Prakash L, Prakash S, Aggarwal AK 2005. Rev1 employs a novel mechanism of DNA synthesis using a protein template. Science 309: 2219–2222 [DOI] [PubMed] [Google Scholar]
- Nair DT, Johnson RE, Prakash L, Prakash S, Aggarwal AK 2008. Protein-template-directed synthesis across an acrolein-derived DNA adduct by yeast Rev1 DNA polymerase. Structure 16: 239–245 [DOI] [PubMed] [Google Scholar]
- Nelson JR, Lawrence CW, Hinkle DC 1996a. Deoxycytidyl transferase activity of yeast REV1 protein. Nature 382: 729–731 [DOI] [PubMed] [Google Scholar]
- Nelson JR, Lawrence CW, Hinkle DC 1996b. Thymine-thymine dimer bypass by yeast DNA polymerase ζ. Science 272: 1646–1649 [DOI] [PubMed] [Google Scholar]
- Nelson JR, Gibbs PE, Nowicka AM, Hinkle DC, Lawrence CW 2000. Evidence for a second function for Saccharomyces cerevisiae Rev1p. Mol Microbiol 37: 549–554 [DOI] [PubMed] [Google Scholar]
- Netz DJ, Stith CM, Stumpfig M, Kopf G, Vogel D, Genau HM, Stodola JL, Lill R, Burgers PM, Pierik AJ 2012. Eukaryotic DNA polymerases require an iron-sulfur cluster for the formation of active complexes. Nat Chem Biol 8: 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuberger MS, Harris RS, Di Noia J, Petersen-Mahrt SK 2003. Immunity through DNA deamination. Trends Biochem Sci 28: 305–312 [DOI] [PubMed] [Google Scholar]
- Niedzwiedz W, Mosedale G, Johnson M, Ong CY, Pace P, Patel KJ 2004. The Fanconi anaemia gene FANCC promotes homologous recombination and error-prone DNA repair. Mol Cell 15: 607–620 [DOI] [PubMed] [Google Scholar]
- Nik-Zainal S, Alexandrov LB, Wedge DC, Van Loo P, Greenman CD, Raine K, Jones D, Hinton J, Marshall J, Stebbings LA, et al. 2012. Mutational processes molding the genomes of 21 breast cancers. Cell 149: 979–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawara D, Muroya T, Yamauchi K, Iwamoto TA, Yagi Y, Yamashita Y, Waga S, Akiyama M, Maki H 2010. Near-full-length REV3 L appears to be a scarce maternal factor in Xenopus laevis eggs that changes qualitatively in early embryonic development. DNA Repair 9: 90–95 [DOI] [PubMed] [Google Scholar]
- Ogi T, Lehmann AR 2006. The Y-family DNA polymerase κ (pol kappa) functions in mammalian nucleotide-excision repair. Nat Cell Biol 8: 640–642 [DOI] [PubMed] [Google Scholar]
- Ogi T, Limsirichaikul S, Overmeer RM, Volker M, Takenaka K, Cloney R, Nakazawa Y, Niimi A, Miki Y, Jaspers NG, et al. 2010. Three DNA polymerases, recruited by different mechanisms, carry out NER repair synthesis in human cells. Mol Cell 37: 714–727 [DOI] [PubMed] [Google Scholar]
- Ohashi E, Bebenek K, Matsuda T, Feaver WJ, Gerlach VL, Friedberg EC, Ohmori H, Kunkel TA 2000. Fidelity and processivity of DNA synthesis by DNA polymerase κ, the product of the human DINB1 gene. J Biol Chem 275: 39678–39684 [DOI] [PubMed] [Google Scholar]
- Ohashi E, Murakumo Y, Kanjo N, Akagi J, Masutani C, Hanaoka F, Ohmori H 2004. Interaction of hREV1 with three human Y-family DNA polymerases. Genes Cells 9: 523–531 [DOI] [PubMed] [Google Scholar]
- Ohkumo T, Kondo Y, Yokoi M, Tsukamoto T, Yamada A, Sugimoto T, Kanao R, Higashi Y, Kondoh H, Tatematsu M, et al. 2006. UV-B radiation induces epithelial tumors in mice lacking DNA polymerase η and mesenchymal tumors in mice deficient for DNA polymerase ι. Mol Cell Biol 26: 7696–7706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohmori H, Friedberg EC, Fuchs RP, Goodman MF, Hanaoka F, Hinkle D, Kunkel TA, Lawrence CW, Livneh Z, Nohmi T, et al. 2001. The Y-family of DNA polymerases. Mol Cell 8: 7–8 [DOI] [PubMed] [Google Scholar]
- Okuda T, Lin X, Trang J, Howell SB 2005. Suppression of hREV1 expression reduces the rate at which human ovarian carcinoma cells acquire resistance to cisplatin. Mol Pharmacol 67: 1852–1860 [DOI] [PubMed] [Google Scholar]
- O-Wang J, Kawamura K, Tada Y, Ohmori H, Kimura H, Sakiyama S, Tagawa M 2001. DNA polymerase κ, implicated in spontaneous and DNA damage-induced mutagenesis, is overexpressed in lung cancer. Cancer Res 61: 5366–5369 [PubMed] [Google Scholar]
- O-Wang J, Kajiwara K, Kawamura K, Kimura M, Miyagishima H, Koseki H, Tagawa M 2002. An essential role for REV3 in mammalian cell survival: Absence of REV3 induces p53–independent embryonic death. Biochem Biophys Res Commun 293: 1132–1137 [DOI] [PubMed] [Google Scholar]
- Pages V, Bresson A, Acharya N, Prakash S, Fuchs R, Prakash L 2008. Requirement of Rad5 for DNA polymerase ζ-dependent translesion synthesis in Saccharomyces cerevisiae. Genetics 180: 73–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pages V, Santa Maria SR, Prakash L, Prakash S 2009. Role of DNA damage-induced replication checkpoint in promoting lesion bypass by translesion synthesis in yeast. Genes Dev 23: 1438–1449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papouli E, Chen S, Davies AA, Huttner D, Krejci L, Sung P, Ulrich HD 2005. Crosstalk between SUMO and ubiquitin on PCNA is mediated by recruitment of the helicase Srs2p. Mol Cell 19: 123–133 [DOI] [PubMed] [Google Scholar]
- Pavlov YI, Rogozin IB, Galkin AP, Aksenova AY, Hanaoka F, Rada C, Kunkel TA 2002. Correlation of somatic hypermutation specificity and A-T base pair substitution errors by DNA polymerase η during copying of a mouse immunoglobulin κ light chain transgene. Proc Natl Acad Sci 99: 9954–9959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen-Mahrt SK, Harris RS, Neuberger MS 2002. AID mutates E. coli suggesting a DNA deamination mechanism for antibody diversification. Nature 418: 99–103 [DOI] [PubMed] [Google Scholar]
- Petta TB, Nakajima S, Zlatanou A, Despras E, Couve-Privat S, Ishchenko A, Sarasin A, Yasui A, Kannouche P 2008. Human DNA polymerase ι protects cells against oxidative stress. EMBO J 27: 2883–2895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfander B, Moldovan GL, Sacher M, Hoege C, Jentsch S 2005. SUMO-modified PCNA recruits Srs2 to prevent recombination during S phase. Nature 436: 428–433 [DOI] [PubMed] [Google Scholar]
- Picher AJ, Blanco L 2007. Human DNA polymerase λ is a proficient extender of primer ends paired to 7,8-dihydro-8-oxoguanine. DNA Repair 6: 1749–1756 [DOI] [PubMed] [Google Scholar]
- Pleasance ED, Cheetham RK, Stephens PJ, McBride DJ, Humphray SJ, Greenman CD, Varela I, Lin ML, Ordonez GR, Bignell GR, et al. 2010a. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature 463: 191–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleasance ED, Stephens PJ, O’Meara S, McBride DJ, Meynert A, Jones D, Lin ML, Beare D, Lau KW, Greenman C, et al. 2010b. A small-cell lung cancer genome with complex signatures of tobacco exposure. Nature 463: 184–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pozhidaeva A, Pustovalova Y, D’Souza S, Bezsonova I, Walker GC, Korzhnev DM 2012. NMR structure and dynamics of the C-terminal domain from human Rev1 and its complex with Rev1 interacting region of DNA polymerase η. Biochem 51: 5506–5520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash S, Johnson RE, Prakash L 2005. Eukaryotic translesion synthesis DNA polymerases: Specificity of structure and function. Annu Rev Biochem 74: 317–353 [DOI] [PubMed] [Google Scholar]
- Prasad R, Longley MJ, Sharief FS, Hou EW, Copeland WC, Wilson SH 2009. Human DNA polymerase θ possesses 5′-dRP lyase activity and functions in single-nucleotide base excision repair in vitro. Nucleic Acids Res 37: 1868–1877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quah SK, von Borstel RC, Hastings PJ 1980. The origin of spontaneous mutation in Saccharomyces cerevisiae. Genetics 96: 819–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada C, Ehrenstein MR, Neuberger MS, Milstein C 1998. Hot spot focusing of somatic hypermutation in MSH2-deficient mice suggests two stages of mutational targeting. Immunity 9: 135–141 [DOI] [PubMed] [Google Scholar]
- Rada C, Williams GT, Nilsen H, Barnes DE, Lindahl T, Neuberger MS 2002. Immunoglobulin isotype switching is inhibited and somatic hypermutation perturbed in UNG-deficient mice. Curr Biol 12: 1748–1755 [DOI] [PubMed] [Google Scholar]
- Rada C, Di Noia JM, Neuberger MS 2004. Mismatch recognition and uracil excision provide complementary paths to both Ig switching and the A/T-focused phase of somatic mutation. Mol Cell 16: 163–171 [DOI] [PubMed] [Google Scholar]
- Raschle M, Knipsheer P, Enoiu M, Angelov T, Sun J, Griffith J, Ellenberger T, Scharer O, Walter J 2008. Mechanism of replication-coupled DNA interstrand crosslink repair. Cell 134: 969–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rechkoblit O, Zhang Y, Guo D, Wang Z, Amin S, Krzeminsky J, Louneva N, Geacintov NE 2002. trans-lesion synthesis past bulky benzo[a]pyrene diol epoxide N2-dG and N6-dA lesions catalyzed by DNA bypass polymerases. J Biol Chem 277: 30488–30494 [DOI] [PubMed] [Google Scholar]
- Roa S, Avdievich E, Peled JU, Maccarthy T, Werling U, Kuang FL, Kan R, Zhao C, Bergman A, Cohen PE, et al. 2008. Ubiquitylated PCNA plays a role in somatic hypermutation and class-switch recombination and is required for meiotic progression. Proc Natl Acad Sci 105: 16248–16253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts SA, Sterling J, Thompson C, Harris S, Mav D, Shah R, Klimczak LJ, Kryukov GV, Malc E, Mieczkowski PA, et al. 2012. Clustered mutations in yeast and in human cancers can arise from damaged long single-strand DNA regions. Mol Cell 46: 424–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roche H, Gietz RD, Kunz BA 1994. Specificity of the yeast rev3Δ antimutator and REV3 dependency of the mutator resulting from a defect (rad1Δ) in nucleotide excision repair. Genetics 137: 637–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross AL, Sale JE 2006. The catalytic activity of REV1 is employed during immunoglobulin gene diversification in DT40. Mol Immunol 43: 1587–1594 [DOI] [PubMed] [Google Scholar]
- Ross AL, Simpson LJ, Sale JE 2005. Vertebrate DNA damage tolerance requires the C-terminus but not BRCT or transferase domains of REV1. Nucleic Acids Res 33: 1280–1289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabbioneda S, Minesinger BK, Giannattasio M, Plevani P, Muzi-Falconi M, Jinks-Robertson S 2005. The 9-1-1 checkpoint clamp physically interacts with polζ and is partially required for spontaneous polζ-dependent mutagenesis in Saccharomyces cerevisiae. J Biol Chem 280: 38657–38665 [DOI] [PubMed] [Google Scholar]
- Sabbioneda S, Bortolomai I, Giannattasio M, Plevani P, Muzi-Falconi M 2007. Yeast Rev1 is cell cycle regulated, phosphorylated in response to DNA damage and its binding to chromosomes is dependent upon MEC1. DNA Repair (Amst) 6: 121–127 [DOI] [PubMed] [Google Scholar]
- Sabbioneda S, Gourdin AM, Green CM, Zotter A, Giglia-Mari G, Houtsmuller A, Vermeulen W, Lehmann AR 2008. Effect of proliferating cell nuclear antigen ubiquitination and chromatin structure on the dynamic properties of the Y-family DNA polymerases. Mol Biol Cell 19: 5193–5202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakiyama T, Kohno T, Mimaki S, Ohta T, Yanagitani N, Sobue T, Kunitoh H, Saito R, Shimizu K, Hirama C, et al. 2005. Association of amino acid substitution polymorphisms in DNA repair genes TP53, POLI, REV1 and LIG4 with lung cancer risk. Int J Cancer 114: 730–737 [DOI] [PubMed] [Google Scholar]
- Sale JE 2012. Competition, collaboration and coordination-determining how cells bypass DNA damage. J Cell Sci 125: 1633–1643 [DOI] [PubMed] [Google Scholar]
- Sale JE, Batters C, Edmunds CE, Phillips LG, Simpson LJ, Szuts D 2009. Timing matters: Error-prone gap filling and translesion synthesis in immunoglobulin gene hypermutation. Philos Trans R Soc Lond B Biol Sci 364: 595–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sale JE, Lehmann AR, Woodgate R 2012. Y-family DNA polymerases and their role in tolerance of cellular DNA damage. Nat Rev Mol Cell Biol 13: 141–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saribasak H, Maul RW, Cao Z, Yang WW, Schenten D, Kracker S, Gearhart PJ 2012. DNA polymerase ζ generates tandem mutations in immunoglobulin variable regions. J Exp Med 209: 1075–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkies P, Reams C, Simpson LJ, Sale JE 2010. Epigenetic instability due to defective replication of structured DNA. Mol Cell 40: 703–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenten D, Kracker S, Esposito G, Franco S, Klein U, Murphy M, Alt F, Rajewsky K 2009. Pol ζ ablation in B cells impairs the germinal center reaction, class switch recombination, DNA break repair, and genome stability. J Exp Med 206: 477–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki M, Wood RD 2008. DNA polymerase θ (POLQ) can extend from mismatches and from bases opposite a (6–4) photoproduct. DNA Repair (Amst) 7: 119–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki M, Marini F, Wood R 2003. POLQ (Pol θ), a DNA polymerase and DNA-dependent ATPase in human cells. Nucleic Acids Res 31: 6117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki M, Masutani C, Yang LW, Schuffert A, Iwai S, Bahar I, Wood RD 2004. High-efficiency bypass of DNA damage by human DNA polymerase Q. EMBO J 23: 4484–4494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shachar S, Ziv O, Avkin S, Adar S, Wittschieben J, Reissner T, Chaney S, Friedberg E, Wang Z, Carell T, et al. 2009. Two-polymerase mechanisms dictate error-free and error-prone translesion DNA synthesis in mammals. EMBO J 28: 383–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharief FS, Vojta PJ, Ropp PA, Copeland WC 1999. Cloning and chromosomal mapping of the human DNA polymerase θ (POLQ), the eighth human DNA polymerase. Genomics 59: 90–96 [DOI] [PubMed] [Google Scholar]
- Sharma S, Hicks JK, Chute CL, Brennan JR, Ahn JY, Glover TW, Canman CE 2011. REV1 and polymerase ζ facilitate homologous recombination repair. Nucleic Acids Res 40: 682–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Shah NA, Joiner AM, Roberts KH, Canman CE 2012. DNA polymerase ζ is a major determinant of resistance to platinum-based chemotherapeutic agents. Mol Pharmacol 81: 778–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstein TD, Johnson RE, Jain R, Prakash L, Prakash S, Aggarwal AK 2010. Structural basis for the suppression of skin cancers by DNA polymerase η. Nature 465: 1039–1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson LJ, Sale JE 2003. Rev1 is essential for DNA damage tolerance and non-templated immunoglobulin gene mutation in a vertebrate cell line. EMBO J 22: 1654–1664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson LJ, Ross AL, Szuts D, Alviani CA, Oestergaard VH, Patel KJ, Sale JE 2006. RAD18-independent ubiquitination of proliferating-cell nuclear antigen in the avian cell line DT40. EMBO Rep 7: 927–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skosareva LV, Lebedeva NA, Rechkunova NI, Kolbanovskiy A, Geacintov NE, Lavrik OI 2012. Human DNA polymerase λ catalyzes lesion bypass across benzo[a]pyrene-derived DNA adduct during base excision repair. DNA Repair 11: 367–373 [DOI] [PubMed] [Google Scholar]
- Sonoda E, Okada T, Zhao GY, Tateishi S, Araki K, Yamaizumi M, Yagi T, Verkaik NS, van Gent DC, Takata M, et al. 2003. Multiple roles of Rev3, the catalytic subunit of polζ in maintaining genome stability in vertebrates. EMBO J 22: 3188–3197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stary A, Kannouche P, Lehmann AR, Sarasin A 2003. Role of DNA polymerase η in the UV mutation spectrum in human cells. J Biol Chem 278: 18767–18775 [DOI] [PubMed] [Google Scholar]
- Stelter P, Ulrich HD 2003. Control of spontaneous and damage-induced mutagenesis by SUMO and ubiquitin conjugation. Nature 425: 188–191 [DOI] [PubMed] [Google Scholar]
- Strathern JN, Shafer BK, McGill CB 1995. DNA synthesis errors associated with double-strand-break repair. Genetics 140: 965–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stratton MR 2011. Exploring the genomes of cancer cells: Progress and promise. Science 331: 1553–1558 [DOI] [PubMed] [Google Scholar]
- Swan MK, Johnson RE, Prakash L, Prakash S, Aggarwal AK 2009. Structural basis of high-fidelity DNA synthesis by yeast DNA polymerase δ. Nat Struct Mol Biol 16: 979–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szüts D, Marcus AP, Himoto M, Iwai S, Sale JE 2008. REV1 restrains DNA polymerase ζ to ensure frame fidelity during translesion synthesis of UV photoproducts in vivo. Nucleic Acids Res 36: 6767–6780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takata K, Shimizu T, Iwai S, Wood RD 2006. Human DNA polymerase N (POLN) is a low fidelity enzyme capable of error-free bypass of 5S-thymine glycol. J Biol Chem 281: 23445–23455 [DOI] [PubMed] [Google Scholar]
- Terai K, Abbas T, Jazaeri AA, Dutta A 2010. CRL4(Cdt2) E3 ubiquitin ligase monoubiquitinates PCNA to promote translesion DNA synthesis. Mol Cell 37: 143–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tissier A, Frank EG, McDonald JP, Iwai S, Hanaoka F, Woodgate R 2000a. Misinsertion and bypass of thymine-thymine dimers by human DNA polymerase ι. EMBO J 19: 5259–5266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tissier A, McDonald JP, Frank EG, Woodgate R 2000b. Polι, a remarkably error-prone human DNA polymerase. Genes Dev 14: 1642–1650 [PMC free article] [PubMed] [Google Scholar]
- Tissier A, Kannouche P, Reck MP, Lehmann AR, Fuchs RP, Cordonnier A 2004. Co-localization in replication foci and interaction of human Y-family members, DNA polymerase polη and REVl protein. DNA Repair (Amst) 3: 1503–1514 [DOI] [PubMed] [Google Scholar]
- Tseng HM, Tomkinson AE 2002. A physical and functional interaction between yeast Pol4 and Dnl4-Lif1 links DNA synthesis and ligation in nonhomologous end joining. J Biol Chem 277: 45630–45637 [DOI] [PubMed] [Google Scholar]
- Ummat A, Rechkoblit O, Jain R, Roy Choudhury J, Johnson RE, Silverstein TD, Buku A, Lone S, Prakash L, Prakash S, et al. 2012. Structural basis for cisplatin DNA damage tolerance by human polymerase η during cancer chemotherapy. Nat Struct Mol Biol 19: 628–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaisman A, Takasawa K, Iwai S, Woodgate R 2006. DNA polymerase ι-dependent translesion replication of uracil containing cyclobutane pyrimidine dimers. DNA Repair 5: 210–218 [DOI] [PubMed] [Google Scholar]
- Van Sloun PP, Varlet I, Sonneveld E, Boei JJ, Romeijn RJ, Eeken JC, De Wind N 2002. Involvement of mouse Rev3 in tolerance of endogenous and exogenous DNA damage. Mol Cell Biol 22: 2159–2169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Devereux TR, Vikis HG, McCulloch SD, Holliday W, Anna C, Wang Y, Bebenek K, Kunkel TA, Guan K, et al. 2004. Pol ι is a candidate for the mouse pulmonary adenoma resistance 2 locus, a major modifier of chemically induced lung neoplasia. Cancer Res 64: 1924–1931 [DOI] [PubMed] [Google Scholar]
- Wang Y, Woodgate R, McManus TP, Mead S, McCormick JJ, Maher VM 2007. Evidence that in xeroderma pigmentosum variant cells, which lack DNA polymerase η, DNA polymerase ι causes the very high frequency and unique spectrum of UV-induced mutations. Cancer Res 67: 3018–3026 [DOI] [PubMed] [Google Scholar]
- Wang S, Wen R, Shi X, Lambrecht A, Wang H, Xiao W 2011. RAD5a and REV3 function in two alternative pathways of DNA-damage tolerance in Arabidopsis. DNA Repair 10: 620–628 [DOI] [PubMed] [Google Scholar]
- Washington MT, Johnson RE, Prakash S, Prakash L 1999. Fidelity and processivity of Saccharomyces cerevisiae DNA polymerase η. J Biol Chem 274: 36835–36838 [DOI] [PubMed] [Google Scholar]
- Washington MT, Johnson RE, Prakash L, Prakash S 2002. Human DINB1-encoded DNA polymerase κ is a promiscuous extender of mispaired primer termini. Proc Natl Acad Sci 99: 1910–1914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washington MT, Minko IG, Johnson RE, Haracska L, Harris TM, Lloyd RS, Prakash S, Prakash L 2004a. Efficient and error-free replication past a minor-groove N2-guanine adduct by the sequential action of yeast Rev1 and DNA polymerase ζ. Mol Cell Biol 24: 6900–6906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washington MT, Minko IG, Johnson RE, Wolfle WT, Harris TM, Lloyd RS, Prakash S, Prakash L 2004b. Efficient and error-free replication past a minor-groove DNA adduct by the sequential action of human DNA polymerases ι and κ. Mol Cell Biol 24: 5687–5693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters LS, Walker GC 2006. The critical mutagenic translesion DNA polymerase Rev1 is highly expressed during G2/M phase rather than S phase. Proc Natl Acad Sci 103: 8971–8976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters L, Minesinger B, Wiltrout M, D’Souza S, Woodruff R, Walker G 2009. Eukaryotic translesion polymerases and their roles and regulation in DNA damage tolerance. Microbiol Mol Biol Rev 73: 134–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson TE, Lieber MR 1999. Efficient processing of DNA ends during yeast nonhomologous end joining: Evidence for a DNA polymerase β (Pol4)-dependent pathway. J Biol Chem 274: 23599–23609 [DOI] [PubMed] [Google Scholar]
- Wiltrout ME, Walker GC 2011. The DNA polymerase activity of Saccharomyces cerevisiae Rev1 is biologically significant. Genetics 187: 21–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimmer U, Ferrari E, Hunziker P, Hubscher U 2008. Control of DNA polymerase λ stability by phosphorylation and ubiquitination during the cell cycle. EMBO Rep 9: 1027–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittschieben J, Shivji MK, Lalani E, Jacobs MA, Marini F, Gearhart PJ, Rosewell I, Stamp G, Wood RD 2000. Disruption of the developmentally regulated Rev3l gene causes embryonic lethality. Curr Biol 10: 1217–1220 [DOI] [PubMed] [Google Scholar]
- Wojtaszek J, Liu J, D’Souza S, Wang S, Xue Y, Walker GC, Zhou P 2012. Multifaceted recognition of vertebrate Rev1 by translesion polymerases ζ and κ. J Biol Chem 287: 26400–26408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfle WT, Johnson RE, Minko IG, Lloyd RS, Prakash S, Prakash L 2006. Replication past a trans-4-hydroxynonenal minor-groove adduct by the sequential action of human DNA polymerases ι and κ. Mol Cell Biol 26: 381–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood A, Garg P, Burgers PM 2007. A Ubiquitin-binding motif in the translesion DNA polymerase rev1 mediates its essential functional interaction with ubiquitinated PCNA in response to DNA damage. J Biol Chem 282: 20256–20263 [DOI] [PubMed] [Google Scholar]
- Wu F, Lin X, Okuda T, Howell SB 2004. DNA polymerase ζ regulates cisplatin cytotoxicity, mutagenicity, and the rate of development of cisplatin resistance. Cancer Res 64: 8029–8035 [DOI] [PubMed] [Google Scholar]
- Xie K, Doles J, Hemann MT, Walker GC 2010. Error-prone translesion synthesis mediates acquired chemoresistance. Proc Natl Acad Sci 107: 20792–20797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka K, Minko IG, Takata K, Kolbanovskiy A, Kozekov ID, Wood RD, Rizzo CJ, Lloyd RS 2010. Novel enzymatic function of DNA polymerase ν in translesion DNA synthesis past major groove DNA-peptide and DNA–DNA cross-links. Chem Res Toxicol 23: 689–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Woodgate R 2007. What a difference a decade makes: Insights into translesion DNA synthesis. Proc Natl Acad Sci 104: 15591–15598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Chen Z, Liu Y, Hickey RJ, Malkas LH 2004. Altered DNA polymerase ι expression in breast cancer cells leads to a reduction in DNA replication fidelity and a higher rate of mutagenesis. Cancer Res 64: 5597–5607 [DOI] [PubMed] [Google Scholar]
- Yoon JH, Prakash L, Prakash S 2010. Error-free replicative bypass of (6–4) photoproducts by DNA polymerase ζ in mouse and human cells. Genes Dev 24: 123–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura M, Kohzaki M, Nakamura J, Asagoshi K, Sonoda E, Hou E, Prasad R, Wilson SH, Tano K, Yasui A, et al. 2006. Vertebrate POLQ and POLβ cooperate in base excision repair of oxidative DNA damage. Mol Cell 24: 115–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu AM, McVey M 2010. Synthesis-dependent microhomology-mediated end joining accounts for multiple types of repair junctions. Nucleic Acids Res 38: 5706–5717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan B, Cao H, Jiang Y, Hong H, Wang Y 2008. Efficient and accurate bypass of N2-(1-carboxyethyl)-2′-deoxyguanosine by DinB DNA polymerase in vitro and in vivo. Proc Natl Acad Sci 105: 8679–8684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zan H, Komori A, Li Z, Cerutti A, Schaffer A, Flajnik MF, Diaz M, Casali P 2001. The translesion DNA polymerase ζ plays a major role in Ig and bcl-6 somatic hypermutation. Immunity 14: 643–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zan H, Shima N, Xu Z, Al-Qahtani A, Evinger Iii AJ, Zhong Y, Schimenti JC, Casali P 2005. The translesion DNA polymerase θ plays a dominant role in immunoglobulin gene somatic hypermutation. EMBO J 24: 3757–3769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zander L, Bemark M 2004. Immortalized mouse cell lines that lack a functional Rev3 gene are hypersensitive to UV irradiation and cisplatin treatment. DNA Repair (Amst) 3: 743–752 [DOI] [PubMed] [Google Scholar]
- Zeng X, Winter D, Kasmer C, Kraemer K, Lehmann A, Gearhart P 2001. DNA polymerase eta is an A-T mutator in somatic hypermutation of immunoglobulin variable genes. Nat Immunol 2: 537–541 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Yuan F, Wu X, Wang M, Rechkoblit O, Taylor JS, Geacintov NE, Wang Z 2000. Error-free and error-prone lesion bypass by human DNA polymerase kappa in vitro. Nucleic Acids Res 28: 4138–4146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wu X, Guo D, Rechkoblit O, Taylor JS, Geacintov NE, Wang Z 2002a. Lesion bypass activities of human DNA polymerase μ. J Biol Chem 277: 44582–44587 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Wu X, Guo D, Rechkoblit O, Wang Z 2002b. Activities of human DNA polymerase κ in response to the major benzo[a]pyrene DNA adduct: Error-free lesion bypass and extension synthesis from opposite the lesion. DNA Repair 1: 559–569 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Wu X, Rechkoblit O, Geacintov NE, Taylor JS, Wang Z 2002c. Response of human REV1 to different DNA damage: Preferential dCMP insertion opposite the lesion. Nucleic Acids Res 30: 1630–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Chea J, Meng X, Zhou Y, Lee E, Lee M 2008. PCNA is ubiquitinated by RNF8. Cell Cycle 7: 3399–3404 [DOI] [PubMed] [Google Scholar]
- Zhao Y, Biertumpfel C, Gregory MT, Hua YJ, Hanaoka F, Yang W 2012. Structural basis of human DNA polymerase η-mediated chemoresistance to cisplatin. Proc Natl Acad Sci 109: 7269–7274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong X, Garg P, Stith C, Nick McElhinny S, Kissling G, Burgers P, Kunkel T 2006. The fidelity of DNA synthesis by yeast DNA polymerase ζ alone and with accessory proteins. Nucleic Acids Res 34: 4731–4742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou RZ, Blanco L, Garcia-Diaz M, Bebenek K, Kunkel TA, Povirk LF 2008. Tolerance for 8-oxoguanine but not thymine glycol in alignment-based gap filling of partially complementary double-strand break ends by DNA polymerase λ in human nuclear extracts. Nucleic Acids Res 36: 2895–2905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zietlow L, Smith LA, Bessho M, Bessho T 2009. Evidence for the involvement of human DNA polymerase N in the repair of DNA interstrand cross-links. Biochemistry 48: 11817–11824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziv O, Geacintov N, Nakajima S, Yasui A, Livneh Z 2009. DNA polymerase ζ cooperates with polymerases κ and ι in translesion DNA synthesis across pyrimidine photodimers in cells from XPV patients. Proc Natl Acad Sci 106: 11552–11557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L, Elledge SJ 2003. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 300: 1542–1548 [DOI] [PubMed] [Google Scholar]