Abstract
Ldb1 and Ldb2 are coregulators that mediate Lin11-Isl1-Mec3 (LIM)–homeodomain (HD) and LIM-only transcription factor–driven gene regulation. Although both Ldb1 and Ldb2 mRNA were produced in the developing and adult pancreas, immunohistochemical analysis illustrated a broad Ldb1 protein expression pattern during early pancreatogenesis, which subsequently became enriched in islet and ductal cells perinatally. The islet-enriched pattern of Ldb1 was similar to pan-endocrine cell–expressed Islet-1 (Isl1), which was demonstrated in this study to be the primary LIM-HD transcription factor in developing and adult islet cells. Endocrine cell–specific removal of Ldb1 during mouse development resulted in a severe reduction of hormone+ cell numbers (i.e., α, β, and δ) and overt postnatal hyperglycemia, reminiscent of the phenotype described for the Isl1 conditional mutant. In contrast, neither endocrine cell development nor function was affected in the pancreas of Ldb2−/− mice. Gene expression and chromatin immunoprecipitation (ChIP) analyses demonstrated that many important Isl1-activated genes were coregulated by Ldb1, including MafA, Arx, insulin, and Glp1r. However, some genes (i.e., Hb9 and Glut2) only appeared to be impacted by Ldb1 during development. These findings establish Ldb1 as a critical transcriptional coregulator during islet α-, β-, and δ-cell development through Isl1-dependent and potentially Isl1-independent control.
The vertebrate pancreas is composed of acinar, endocrine, and ductal cells critical for maintaining nutritional homeostasis. Although acinar and ductal cells secrete and transport enzymes important for food digestion, endocrine-derived α, β, δ, and pancreatic polypeptide (PP) cells of the islets of Langerhans produce hormones essential for regulating glucose homeostasis. For example, insulin secreted from β-cells is critical for glucose uptake in peripheral tissues, whereas glucagon released from α-cells acts in a counterregulatory manner to promote gluconeogenesis and glycogenolysis (1).
Pancreatic organogenesis begins at embryonic day (E)9.5 when dorsal and ventral buds form from a regionalized domain of the foregut endoderm. This process is influenced by both extrinsic signals from the adjacent mesenchyme and intrinsic lineage-specific transcription factors like Ptf1a and Pdx1 (2–5). Several days later, endocrine cells arise from neurogenin 3 (Ngn3) expressing endocrine progenitors that reside within the ductal epithelium (6–9). These cells delaminate and aggregate into hormone+ endocrine clusters, which then proliferate between E13 and 18.5 (2). Differentiation of endocrine lineages occurs in part through cell type–specific expression of several islet-enriched transcription factors, including Arx, Isl1, MafB, Nkx6.1, Nkx2.2, Pax4, and Pax6 (10). The levels of Pdx1, MafA, and Nkx6.1 (11–13) are enhanced in mature islet β-cells, whereas Arx and MafB are restricted to α-cells (14–17).
Interestingly, Isl1 is produced in both the developing pancreatic epithelium and surrounding lateral and dorsal mesenchyme at ∼E9.5, with expression then becoming restricted to all postnatal islet cells (18,19). Although Isl1−/− mice die embryonically from severe heart defects (i.e., ∼E10.5), impaired formation of the dorsal pancreatic epithelium and mesenchyme was observed (19). A conditional deletion strategy was subsequently used to study the function of Isl1 in endocrine progenitors by crossing floxed (F) Isl1 mice with transgenic Pdx1Late-Cre mice (20,21), which catalyzed recombination in pancreatic epithelial cells by E13.5. Isl1-deficient mice became overtly hyperglycemic due to greatly reduced numbers of α-, β-, and δ-cells, a phenotype caused by effects on proliferation, apoptosis, and maturation due (in part) to actions on MafA and Arx transcription (22,23).
LIM domain [derived from Lin11-Isl1-Mec3 (24)] factors like Isl1 (LIM-homeodomain [HD]) and related LIM-only (Lmo) proteins act through binding with the LIM-domain–binding coregulators Ldb1 (also called CLIM2, Nuclear LIM Interactor, and Chip) and/or Ldb2 (25–27). Strikingly, there are ∼250 known coregulators (www.nursa.org), yet only a few have been associated with pancreatic development or adult islet cell function [e.g., cAMP-responsive element–binding protein (CBP)/p300, p300/CBP–associated factor, Pdx-1 COOH terminus–interacting factor 1, Set7/9, and Bridge-1 (28–34)]. In this study, we analyzed how Ldb1 and/or Ldb2 influence pancreatic endocrine cell development. Ldb1 and Ldb2 mRNA was expressed in developing pancreatic and adult islet cells, with Ldb1 more abundant. In addition, Ldb1 protein was widely distributed in the early pancreatic epithelium and surrounding mesenchyme, eventually becoming enriched in endocrine and ductal cells. Ldb1 removal in developing mouse Pax6+ endocrine cells reduced insulin+ (i.e., β), glucagon+ (α), and somatostatin+ (δ) cell formation in a manner similar to Pdx1Late-Cre;Isl1F/F mice, perhaps not surprisingly considering the relative abundance of Isl1 mRNA levels to other pancreatic LIM-HD–expressed genes. In contrast, endocrine cell development in Ldb2−/− mutants was unaffected. Gene expression and chromatin immunoprecipitation (ChIP) analyses showed that Ldb1 control was primarily linked to Isl1 activation (22,23). However, distinct and novel Ldb1 regulatory actions were also found during development, suggesting essential contributions of other LIM-HD and/or Lmo factors in islet cell formation and function.
RESEARCH DESIGN AND METHODS
Animals.
Ldb1F/F (35), Ldb2−/− [Mouse Genome Informatics, Lexicon Genetics (36)], Pdx1Late-Cre;Isl1F/F (22), and Pax6-Cre (also called Le-Cre) (37) mice have been described previously; the lines were maintained on a mixed (B6) background. Cre was first visualized at ∼E11.5 in Pdx1Late-Cre endocrine cells (21), at least 24 h after other transgenic ∼4.5-kb promoter-driven Pdx1-Cre lines (38). Pax6-Cre;Ldb1F/F and control littermate mice (Ldb1F/+, Ldb1F/F, and Pax6-Cre;Ldb1F/+ genotypes) were generated by mating Pax6-Cre;Ldb1F/+ to Ldb1F/F mice. The morning of vaginal plug discovery was considered E0.5. The Vanderbilt University and Children’s Hospital of Philadelphia Institutional Use and Care Committees approved all of the animal experiments.
Fasting blood glucose measurements.
Postnatal (P) day 10 to P26 mice were fasted for 6 h, and blood glucose was measured from the tail vein using a BD-Logic glucometer (Nova Biomedical, Waltham, MA) and Nova Max test strips (Nova Diabetes Care). Some Pax6-Cre;Ldb1F/F animals exceeded the 600 mg/dL limit of the meter, but were still referred to as 600 mg/dL. All numerical data are presented ± SEM. Significance was determined after performing an unpaired t test, for which P < 0.05.
RNA isolation, cDNA synthesis, and quantitative real-time PCR.
Control, Ldb1-mutant, and Isl1-mutant E15.5–18.5 pancreata were quickly excised and stored at −20°C in RNA-Later Ice (#AM7030; Ambion/Life Technologies, Carlsbad, CA). RNA was extracted using the RNeasy Mini kit (#74104; Qiagen, Valencia, CA) after homogenizing the tissue with a T8 Ultra-Turrax disperser (IKA Works, Wilmington, NC). cDNA was prepared from this RNA using oligo(dT) and the iScript cDNA synthesis kit (#170-8891; Bio-Rad, Hercules, CA). Quantitative real-time PCR (qPCR) reactions were performed in triplicate with reference gene normalization using the SYBR Green PCR master mix (Roche, Indianapolis, IN) in a LightCycler 480 II (Roche). See Supplementary Table 1 for primer sequences.
Immunohistochemical and in situ RNA hybridization analyses.
Staged embryonic and postnatal littermate-matched control, Pax6-Cre;Ldb1F/F, and Pdx1Late-Cre;Isl1F/F pancreatic tissues were fixed in 4% paraformaldehyde and embedded in paraffin or Optimal Cutting Temperature (Tissue-Tek). Sections were cut to 6–12 μm and blocked with 5% normal donkey serum in 1% BSA/1× PBS and then incubated with primary antibodies overnight at 4°C (see Supplementary Table 2 for antibody type, dilution, and specific staining conditions). Cy2-, Cy3-, or Cy5-conjugated donkey anti–guinea pig, anti-mouse, anti-goat, or anti-rabbit IgG secondary antibodies (1:500; Jackson ImmunoResearch Laboratories, West Grove, PA) were used for detection. Lmo4, Glp1r, some insulin, and Ldb1 primary signals were visualized using a Vectastain Elite ABC kit (Vector Laboratories, Burlingame, CA). Slides were imaged by confocal microscopy using a Zeiss LSM510 or by fluorescent/brightfield microscopy using a Zeiss Axioimager M2 (Zeiss) and the images processed by LSM (Zeiss) or ImageJ (National Institutes of Health) software.
In situ hybridization (ISH) analysis was performed on E15.5 embryos fixed in 4% paraformaldehyde followed by overnight saturation in 30% sucrose solution. Embryos were embedded in Optimal Cutting Temperature (Tissue-Tek) and sectioned to 10 μm. Prehybridization was performed in a solution of 50% formamide, 5× SSC (pH 4.5), 50 μg/mL yeast tRNA (Sigma-Aldrich), 1% SDS, and 50 μg/mL heparin (Sigma-Aldrich) at 55°C for 1 to 2 h, and hybridization was at 70°C overnight using Ldb1 (600 base pairs) and Ldb2 (578 base pairs) coding region probes at 300–400 ng/mL. Slides were washed in a solution of 2% blocking reagent (Roche), 10% heat-inactivated sheep serum, 0.1% Tween-20, and 1× maleic acid buffer for 1 h at room temperature. Anti-digoxigenin antibody (1:2,000; Roche) was diluted in blocking solution and incubated overnight at 4°C. Slides were washed first in 1× maleic acid buffer/0.1% Tween and then with 0.1% Tween before BM Purple (Roche) was added. After the signal developed, slides were rinsed in PBS and mounted with Fluoroshield (Axell).
ChIP.
βTC-3 and αTC-6 monolayer cells (∼4 × 106 cells) were cross-linked with 1% formaldehyde and chromatin fragmentation performed as described previously (22). Chromatin was precleared with protein G-Sepharose (#101242; Invitrogen/Life Technologies, Carlsbad, CA) and then incubated with anti-Ldb1 (sc-11198X; Santa Cruz Biotechnology), anti-Isl1 (DSHB, 39.4D5), species-matched preimmune IgG (Santa Cruz Biotechnology), or without antibody. Bound complexes were precipitated with BSA- and salmon sperm DNA-blocked protein G-Sepharose. The eluted and immunoprecipitated DNA (1:20) was used in a PCR reaction with Taq polymerase Hotstart Mastermix (5 Prime, Gaithersburg, MD) and 12.5 pmol of each primer (see Supplementary Table 1). PCR parameters were: 95°C for 2 min (1 cycle), 95°C for 30 s, 59 to 60°C for 30 s, and 72°C for 30 s (29 to 30 cycles). Reaction products were separated on 1.5–2.0% agarose gels using 1× Tris–acetate–ethylenediaminetetraacetic acid buffer and visualized with ethidium bromide. Experiments were performed with at least three independently isolated chromatin preparations.
Coimmunoprecipitation and immunoblotting.
βTC-3 nuclear extract was prepared in the presence of protease inhibitor cocktail (Sigma-Aldrich) as described previously (39). Covalent antibody-bound anti-Ldb1–, anti-Isl1–, or control IgG Dynabeads (Invitrogen) were incubated with 400 μg of extract protein for 3 h at 4°C. The beads were then washed five times with PBS and bound proteins eluted with RIPA buffer (50 mmol/L Tris [pH 7.4], 150 mmol/L NaCl, 1% Nonidet P-40, 0.5% deoxycholic acid, 0.1% SDS) at 37°C. The eluted material was separated by 10% SDS-PAGE (NuPAGE; Invitrogen) and electrophoretically transferred to polyvinylidene difluoride membrane. The membrane was blocked in PBS/Tween supplemented with 5% nonfat dry milk followed by incubation overnight with anti-Ldb1 (1:2,000; see Supplementary Table 2 for additional antibody sources), anti-Isl1 (1:2,000), anti-Pdx1 (1:20,000), anti-Pax6 (1:1,000), anti-NeuroD1 (1:3,000; 3181-1; Epitomics), anti-Hnf1α (1:2,000, sc-6548X; Santa Cruz Biotechnology), and anti-MafA (1:2,000, A300 BL-1225; Bethyl Laboratories). The washed membrane was incubated with horseradish peroxidase-conjugated secondary antibody followed by detection using Western-Lightning Plus-ECL (PerkinElmer, Waltham, MA).
Transient transfection and reporter gene assays.
The wild-type pFox-mouse MafA region 3-luciferase plasmid (40) was cotransfected using Lipofectamine reagent (Invitrogen) into βTC-3 cells with cytomegalovirus (CMV) enhancer-driven Ldb1 dominant-negative acting Ldb1ΔN and the Renilla phRL-TK internal control. Ldb1ΔN spans amino acids 200–375 and contains the LIM interaction domain, but not the dimerization domain (41). Lysates were prepared 48 h posttransfection and analyzed using the Dual Luciferase assay according to the manufacturer’s protocol (Promega, Madison, WI). Each transfection was performed in triplicate on at least three independent occasions; firefly luciferase activity levels were normalized to Renilla.
RESULTS
Ldb1 is broadly expressed in the early pancreatic epithelium and surrounding mesenchyme, then becomes enriched in islet and ductal cells perinatally.
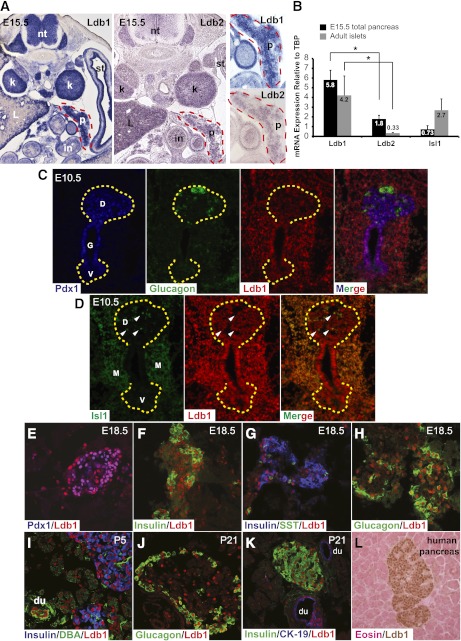
Ldb1 and Ldb2 mRNA expression was observed in the pancreas, neural tube, and kidney by ISH (Fig. 1A). Notably, Ldb1 levels were ∼3.2-fold higher than Ldb2 in E15.5 pancreata by qPCR analysis and 12.7-fold in adult islets (Fig. 1B). Ldb1 mRNA was also much more abundant than Isl1 in the E15.5 pancreas (Fig. 1B), presumably reflecting a broader cell distribution than endocrine cell–specific Isl1 (18,19).
FIG. 1.
Ldb1 is enriched in islet and ductal cells. A: Ldb1 (left) and Ldb2 (middle) mRNA expression was visualized by RNA ISH under identical conditions in E15.5 tissue. A higher-magnification view of pancreatic Ldb1 and Ldb2 expression is shown on the right. B: qPCR was performed to measure Ldb1, Ldb2, and Isl1 mRNA levels in E15.5 total pancreas (black bars) and 3-month-old isolated islets (gray bars). Expression levels are displayed relative to TATA-binding protein (TBP), which is set as onefold. Error bars represent ± SEM (n = 5). Ldb1 mRNA is significantly more abundant than Ldb2 in E15.5 and adult samples. C–K: Ldb1, Pdx1, Isl1, hormone (insulin, glucagon, and somatostatin), and ductal (DBA, CK-19) markers were visualized at E10.5, E18.5, P5, and P21 by coimmunofluorescence. Yellow dashed lines mark dorsal and ventral pancreas domains in C and D. Notably, only a few of the pancreatic Ldb1+ cells in D are copositive for Isl1 at this stage (some marked by white arrowheads). L: Immunohistochemical analysis illustrates enriched Ldb1 protein (brown) expression in adult human islet cells; the sample is eosin (pink) counterstained. *P < 0.05. D, dorsal pancreas; du, duct; G, gut tube; in, intestine; k, kidney; L, liver; M, mesenchyme; nt, neural tube; P, pancreas (outlined with red dashed line); st, stomach; V, ventral pancreas. (A high-quality digital representation of this figure is available in the online issue.)
Immunostaining analysis was next performed to characterize temporal and spatial Ldb1 protein expression in the developing and adult pancreas. At E10.5, Ldb1 was widely produced in early Pdx1+ pancreas-specified endoderm, mesenchyme, and early glucagon+ cell population (Fig. 1C). Isl1 expression was also detected in Ldb1+ cells in lateral and dorsal mesenchyme (Fig. 1D) and in a small subset of Ldb1+ cells in the pancreatic epithelium (Fig. 1D) (19). Later in pancreatic development, Ldb1 expression became more enriched in insulin+/Pdx1+, glucagon+, somatostatin+, and pancreatic polypeptide+ cells than the surrounding acinar cells (Fig. 1E–H and data not shown). Islet cell–enriched Ldb1 expression persisted postnatally and was found in all hormone+ subtypes, overlapping with Isl1 expression (Fig. 1I–K) (18). In addition, Ldb1 was present in cytokeratin-19+ (CK-19) and Dolichos biflorus agglutinin (DBA)+ ductal cells, which lack Isl1 (Fig. 1I and K) (18). The Ldb1 protein was also enriched in human islet cells (Fig. 1L), consistent with recent findings examining both Ldb1 and Isl1 mRNA expression in this context (42).
Isl1 is the most abundantly expressed LIM-HD family member in the developing and adult pancreas.
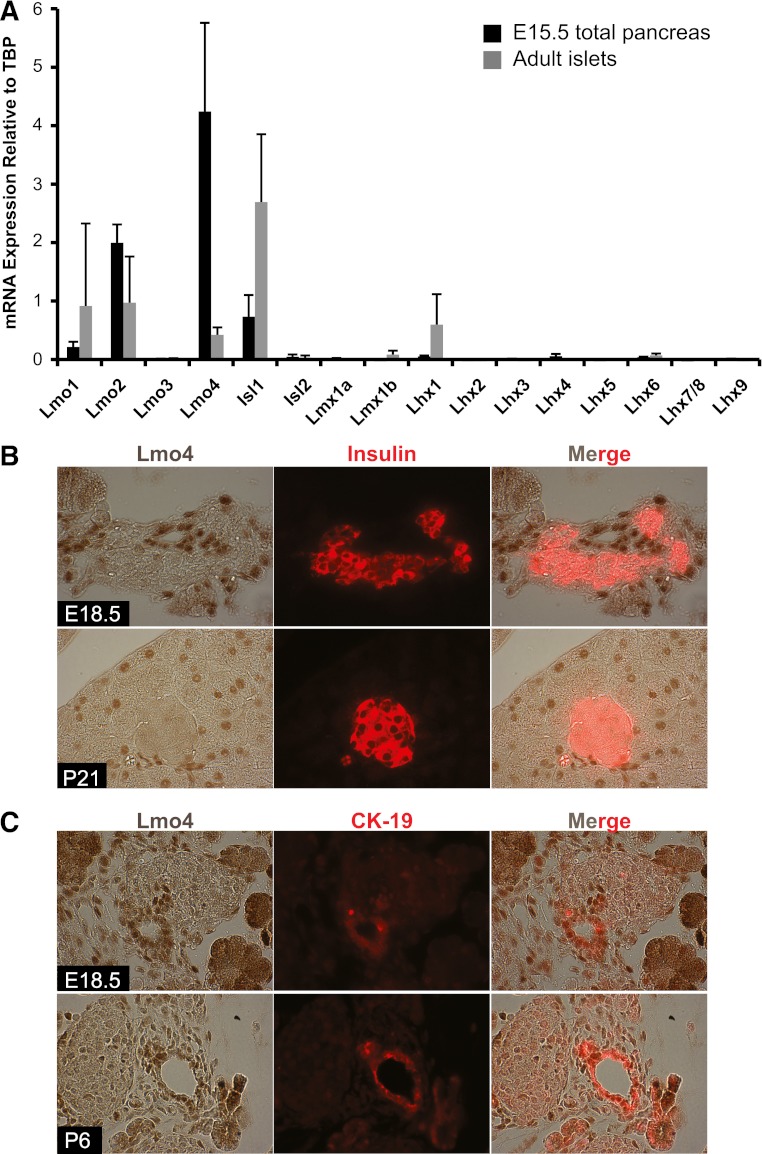
Ldb1 and Ldb2 interact with the LIM domains of LIM-HD and Lmo proteins (43). Significantly, these obligate coregulators cannot function in the absence of an associated LIM-factor because they lack the capacity to independently bind cis-element DNA, transactivate, or remodel chromatin (43). Expression of all mouse LIM-HD and known Ldb1-interacting Lmo genes was measured in E15.5 pancreata and isolated 3 month-old islets by qPCR to evaluate their abundance. Lmo2 and Lmo4 were highly expressed relative to Isl1 at E15.5, but were reduced in adult islets (Fig. 2A). Immunohistochemical analysis showed that Lmo4 was present in E18.5 and adult acinar and ductal cell nuclei, but not endocrine cells (Fig. 2B and C). As a consequence, the relatively small islet Lmo4 mRNA signal presumably represents acinar and ductal cell sample contamination. Notably, this distribution pattern was quite distinct from Isl1 (18,19), suggesting a role for Ldb1/2 in Lmo4 gene regulation in the exocrine pancreas. Isl1 was the principal LIM-HD mRNA detected in E15.5 and adult islet pancreatic samples, with much lower levels of Lhx1 in adult (Fig. 2A). These observations paralleled human mRNA data demonstrating acinar cell enrichment of Lmo4 and Isl1 in islets (42). Taken together, these data suggest an important transcriptional relationship among Isl1, Ldb1, and/or Ldb2 in regulating endocrine cell development in the mouse pancreas.
FIG. 2.
Analysis of LIM-HD and Lmo levels in the developing and adult pancreas. A: LIM-HD and Lmo mRNA expression was measured by qPCR in the E15.5 pancreas (black bars) and 3-month-old islets (gray bars). Values are relative to TATA-binding protein (TBP), set at onefold. Error bars represent ± SEM (n = 5). B: Lmo4 (brown) and insulin (red) staining was performed on E18.5 and P21 pancreas tissue. Lmo4 was not found in hormone+ cells. C: Lmo4 (brown) colocalized with the ductal CK-19 (red) marker in E18.5 and P6 pancreata. (A high-quality digital representation of this figure is available in the online issue.)
Endocrine cell deletion of Ldb1, and not Ldb2, led to reduced hormone production, postnatal islet cell loss, and hyperglycemia.
Our studies next focused on determining the significance of Ldb1 and Ldb2 to endocrine hormone+ cell development in vivo. Notably, Ldb1−/− mice fail to develop past ∼E8.5 due to defects in heart formation, foregut indentation, and anterior–posterior axis patterning (44), whereas Ldb2−/− animals appear overtly normal [Mouse Genome Informatics, Lexicon Genetics (36)]. To circumvent embryonic lethality, Pax6-Cre mice (37) were crossed to Ldb1F/F mice (35) to specifically remove this coregulator from developing endocrine cells. Immunostaining analyses showed that a majority of Ldb1 protein was selectively removed from hormone+ cells of Pax6-Cre;Ldb1F/F animals by E15.5 and absent from E18.5 endocrine cells, with expression remaining in surrounding exocrine cells (Supplementary Fig. 1).
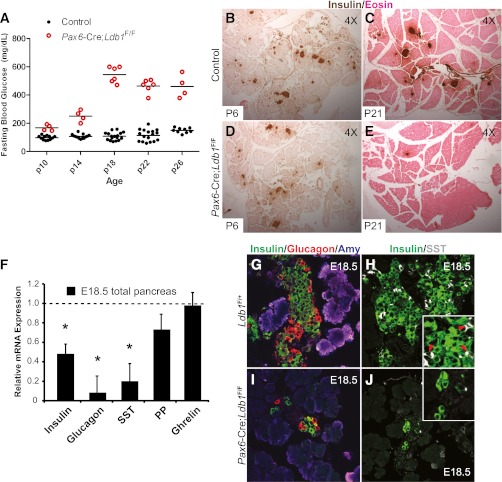
Pax6-Cre;Ldb1F/F mice were born at the predicted Mendelian ratio, with no overt change in body length or weight (data not shown). Both male and female Ldb1-deficient mice had elevated fasting blood glucose levels by P10 that worsened with age (Fig. 3A). Notably, a detectible loss of insulin+ islet cells was visible in P6 Pax6-Cre;Ldb1F/F mice despite their mild hyperglycemic phenotype, with virtually no islets remaining in the 3–5-week-old pancreata (Fig. 3 and data not shown). All mutant mice were killed by P35 because of the severe hyperglycemia.
FIG. 3.
Deletion of Ldb1 in endocrine hormone+ cells causes reduced pancreatic hormone production, postnatal islet loss, and hyperglycemia in vivo. A: Six-hour fasting blood glucose levels in littermate control and Pax6-Cre;Ldb1F/F pups. Horizontal bars indicate mean blood glucose values within each genotype, and values were significantly different between control and mutants at all ages (P < 0.0001). Insulin (brown) and eosin (pink) staining at P6 and P21 in the control (B and C) and Pax6-Cre;Ldb1F/F (D and E) pancreas. F: Islet hormone insulin, glucagon, and somatostatin (SST) mRNA levels are significantly reduced in E18.5 Ldb1 mutant pancreata (n = 4–6). Data are presented relative to littermate controls, which are set at onefold and marked by the dashed line. Error bars represent ± SEM. The number of pancreatic insulin+ (green), glucagon+ (red), and somatostatin+ (white) cells is greatly reduced between E18.5 control (G and H) and Pax6-Cre;Ldb1F/F (I and J) tissue. The insets in H and J show a magnified view of the cell clusters. *P < 0.05. (A high-quality digital representation of this figure is available in the online issue.)
A significant reduction in insulin, glucagon, and somatostatin mRNA levels was observed in E18.5 Ldb1 mutant pancreata, whereas there was little to no effect on PP or ghrelin expression (Fig. 3F). Immunohistochemical analysis of hormone protein expression revealed a similar trend, as insulin+, glucagon+, and somatostatin+ cell numbers were compromised and amylase+, pancreatic polypeptide+, and ghrelin+ cells unchanged in Pax6-Cre;Ldb1F/F pancreata (Fig. 3I and J and data not shown). Notably, these cell types were impacted in an analogous manner in Pdx1Late-Cre;Isl1F/F mice, which results in postnatal endocrine cell apoptosis in both circumstances (22 and data not shown). In contrast, there was no effect on hormone+ cell development or glucose homeostasis in Ldb2−/− mice (Supplementary Fig. 2 and data not shown), despite a broad and overlapping expression pattern with Ldb1 in the developing pancreas (Fig. 1A). Collectively, these data demonstrated that Ldb1 was the principal coregulator of LIM-HD and Lmo transcription factor activity in the pancreas. Moreover, as the impact of Ldb1 deficiency on endocrine cell formation and function was comparable to that observed in Pdx1Late-Cre;Isl1F/F mice (22), these data strongly supported a functional link between Ldb1 and Isl1 in islet α-, β-, and δ-cell development.
Ldb1:Isl1 activate MafA, Arx, and Glp1r transcription.
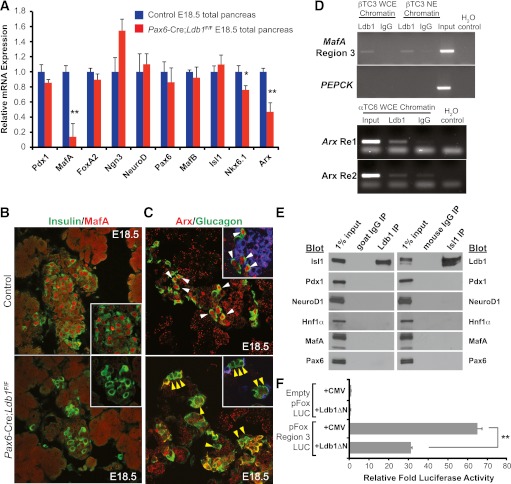
To gain mechanistic insight into the linkage between Ldb1:Isl1 control in the developing pancreas, mRNA levels of many key islet-enriched transcriptional regulators were evaluated in E18.5 Pax6-Cre;Ldb1F/F pancreata (Fig. 4A). The expression of two known Isl1-controlled genes, MafA and Arx (22,23), were significantly reduced in the Pax6-Cre;Ldb1F/F pancreata (Fig. 4A–C). Isl1 regulates by binding to the MafA 5′-flanking region 3, Arx intronic Re1, and Arx 3′-flanking Re2 domains (15,22,23,45). Importantly, Ldb1 bound to these same transcriptional control sequences in ChIP experiments performed with βTC-3 (MafA+) and αTC-6 (Arx+) cells (Fig. 4D), providing compelling support for direct Ldb1:Isl1 complex activation. Endogenous Ldb1 and Isl1 were also found to interact in coimmunoprecipitation assays, whereas Ldb1 and Isl1 did not bind to the islet cell Pdx1, Pax6, NeuroD1, MafA, or Hnf1α transcriptional regulators (Fig. 4E). Moreover, dominant-negative acting Ldb1ΔN, which only produces the COOH-terminal LIM-interaction domain of Ldb1 (41), significantly reduced the activity of the Isl1-responsive MafA region 3-driven luciferase reporter in transfected βTC-3 cells (Fig. 4F). Collectively, these results demonstrate that specific interactions between Ldb1 and Isl1 are essential to expression of key endocrine genes like MafA and Arx.
FIG. 4.
Isl1-regulated MafA and Arx expression is greatly reduced in the E18.5 Ldb1 mutant pancreas. A: mRNA levels of islet-enriched transcription factors in E18.5 littermate control (blue bars) and Pax6-Cre;Ldb1F/F (red bars) pancreas (n = 4–6). Littermate control mRNA level was set at onefold ± SEM. Immunostaining levels of β-cell MafA (red) (B) and α-cell Arx (red) (C) were greatly reduced in E18.5 Pax6-Cre;Ldb1F/F pancreata. Arrowheads in C mark Arx+ glucagon+ (white, top) or Arx− glucagon+ cells (yellow, bottom), with some magnified hormone+ cell clusters shown. D: ChIP analysis of Ldb1 binding to MafA Region 3 (top) as well as Arx Re1 and Re2 (bottom). The PEPCK promoter served as the negative background control. Dilute input as well as Ldb1- and IgG-enriched DNA were analyzed by PCR using βTC-3 and αTC-6 chromatin isolated from whole-cell extract (WCE) and/or nuclear extract (NE). H2O control serves as a negative control for the PCR. E: Binding between endogenous Ldb1 and Isl1 were found in coimmunoprecipitation experiments using βTC-3 nuclear extracts, whereas Ldb1 and Isl1 did not bind to Pdx1, NeuroD1, Hnf1α, MafA, or Pax6. Diluted βTC-3 nuclear extract served as input positive control (1%), and immunoprecipitation (IP) results were compared with species-matched IgG treatments. F: Dominant-negative acting Ldb1ΔN significantly reduced MafA region 3-driven reporter expression in βTC-3 cells. Data are presented as mean fold reporter activity, with the empty pFox-Luc + CMV cotransfection set at onefold ± SEM; n = 3. *P < 0.05, **P < 0.01. Blot, immunoblot antibody probe. (A high-quality digital representation of this figure is available in the online issue.)
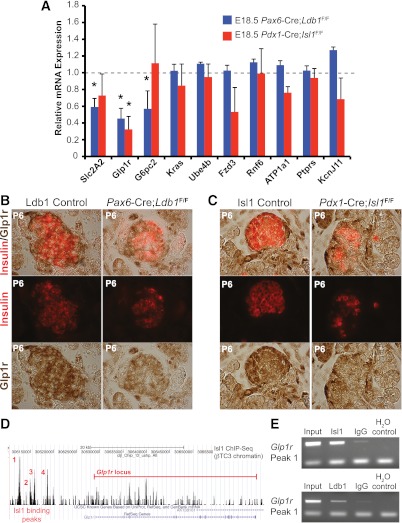
To broaden our understanding of Isl1:Ldb1 regulation, we next analyzed in Isl1- and Ldb1-deficient pancreata the mRNA expression level of various Isl1 ChIP-sequencing (ChIP-Seq)–identified genes predicted to contribute to the deficiencies in islet cell formation and function in Pdx1late-Cre;Isl1F/F mice (22). These candidates were selected from Isl1 ChIP-Seq data generated in βTC-3 cells (B.E., C.S.H., J. Liu, A. Du, E. Walp, R.S., C.L.M., unpublished observations). Intriguingly, only glucagon-like peptide 1 receptor (Glp1r) mRNA levels were significantly reduced in both the Isl1 and Ldb1 mutants (Fig. 5A). However, Slc2A2 (encoding the Glut2 glucose transporter) and glucose-6-phosphatase 2 expression were exclusively decreased in the Ldb1 mutant (Fig. 5A). Described below are immunohistochemical and ChIP data illustrating that Glp1r represents a new Ldb1:Isl1 activated gene (Fig. 5), whereas Slc2A2/Glut2 appears to represent a class of genes dependent upon Ldb1 and not Isl1 (see section “Ldb1, and not Isl1, stimulates Glut2 and Hb9 expression during islet cell development”).
FIG. 5.
Glp1r is a novel Ldb1- and Isl1-activated target gene. A: qPCR quantification of Isl1 ChIP-Seq candidates from Ldb1- (blue bars) and Isl1-deficient (red bars) E18.5 pancreata (n = 4–6). Data are presented as fold of the littermate control, which was set at 1 (marked by the dashed line), ± SEM. B and C: Immunostaining of Glp1r (brown) and insulin (red) at P6 illustrates reduced Glp1r protein levels in insulin+ cells lacking Ldb1 or Isl1. D: βTC-3 ChIP-Seq pictograph demonstrating the four distal 5′ peaks of Isl1 occupancy near Glp1r. The red line denotes the Glp1r locus. E: ChIP enrichment of peak 1 Glp1r 5′ DNA in Isl1 (top panel) and Ldb1 βTC-3 immunoprecipitates (bottom panel) as compared with IgG control-treated DNA. H2O serves as a negative control for the PCR. *P < 0.05. (A high-quality digital representation of this figure is available in the online issue.)
Glp1r is an important signaling hub for incretins like Glp1 and exendin in pancreatic β-cells and other extrapancreatic tissues (reviewed in Ref. 46). Glp1r and insulin protein staining was decreased in the β-cells of Pax6-Cre;Ldb1F/F and Pdx1Late-Cre;Isl1F/F mice at P6 (Fig. 5B and C), paralleling their mRNA changes (Figs. 3F and 5A). There were four Isl1 ChIP-Seq binding peaks clustered between 16- and 10-kb pairs of the Glp1r start site (Fig. 5D). At least peak 1 (base pair −16,250 to −16,050) bound both Isl1 and Ldb1 (Fig. 5E), suggesting that these factors also act together to regulate Glp1r expression in developing β-cells.
Ldb1, and not Isl1, stimulates Glut2 and Hb9 expression during islet cell development.
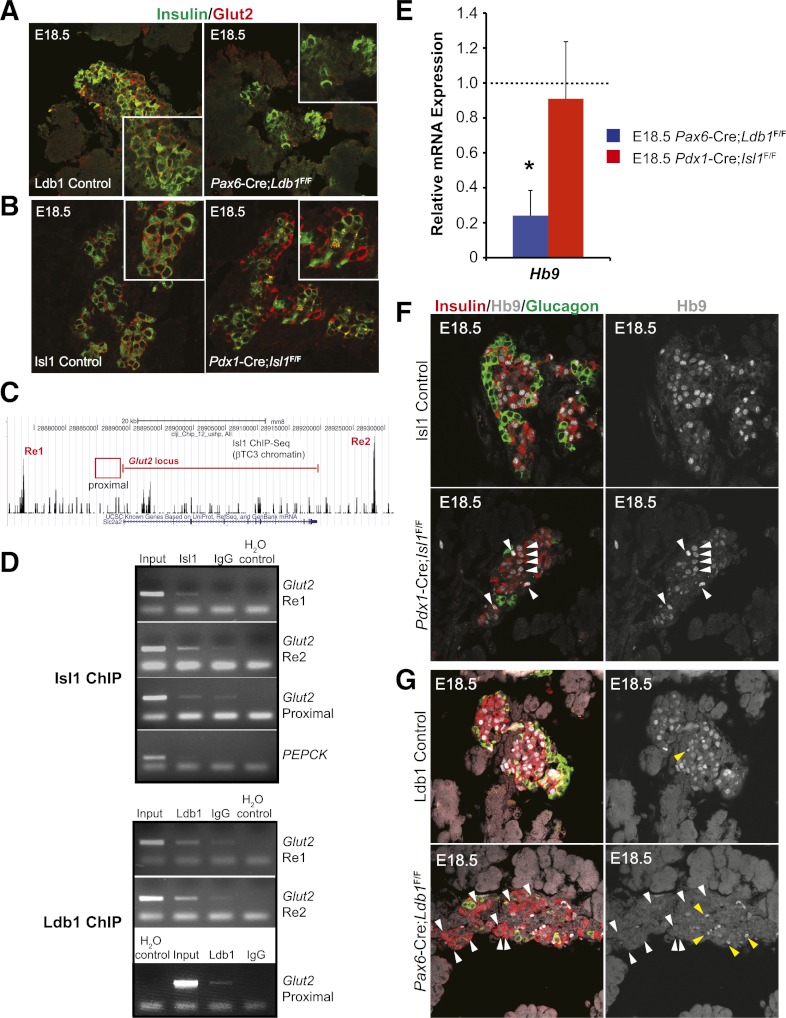
Glut2 is the major glucose transporter of islet β-cells (47). Strikingly, although the Re1 and Re2 control regions of Slc2A2/Glut2 were Isl1-bound in βTC-3 cells (Fig. 6C), mRNA expression was only significantly reduced in the E18.5 pancreata of the Ldb1 mutant (Fig. 5A). As expected, immunostaining revealed a discernible difference of this membrane protein from the control in the Ldb1 mutant, but not the Isl1 mutant (Fig. 6A and B). ChIP analysis was next performed over Isl1-bound Re1 and Re2 and the well-characterized proximal transcriptional control domain in βTC-3 cells (48,49). Interestingly, Ldb1 alone bound the proximal control domain, whereas both Isl1 and Ldb1 binding was detected within Re1 and Re2 (Fig. 6D). Collectively, these data suggest that Ldb1 and an unidentified LIM regulator(s) stimulate E18.5 Glut2 transcription.
FIG. 6.
E18.5 Glut2 and Hb9 mRNA and protein expression is only compromised in Ldb1 mutant mice. Immunofluorescence analysis of Glut2 (red) and insulin (green) in the E18.5 control, mutant Ldb1 (A), and mutant Isl1 (B) pancreas. Insets show magnified insulin+ cell clusters. C: ChIP-Seq pictograph demonstrating Isl1 occupancy at distal Glut2 Re1 (5′) and Re2 (3′) domains in βTC-3 cells. The red line denotes the Glut2-coding region, whereas ChIP-tested proximal 5′ promoter region is represented by the red box. D: βTC-3 ChIP analysis of Isl1 (top panel) and Ldb1 (bottom panel) occupancy of Glut2 Re1, Re2, and the proximal domain compared with the PEPCK control (from top to bottom, respectively). H2O serves as a negative PCR control. Results recapitulate observed Isl1 ChIP-Seq occupation of Glut2 Re1 and Re2, whereas Ldb1 also binds to the proximal domain. E: qPCR analysis of E18.5 Hb9 mRNA levels in pancreata from Ldb1- (blue bar) and Isl1-deficient (red bar) pancreata. Littermate control mRNA level was set at onefold (dashed line) ± SEM. F: E18.5 immunostaining analysis demonstrates that Hb9 protein (white) is maintained in the insulin+ (red) nuclei of Pdx1-Cre;Isl1F/F pancreata as compared with littermate controls, as denoted by the white arrowheads. G: However, Hb9 is lost from most remaining insulin+ cells in the Pax6-Cre;Ldb1F/F pancreata seen by comparing white arrowhead–labeled Hb9+ cells of control and mutant in F and G. The nuclear Hb9 signals are shown in the right panels. Yellow arrowheads in G illustrate autofluorescence from erythrocytes. *P < 0.05. (A high-quality digital representation of this figure is available in the online issue.)
A complex composed of Ldb1, Isl1, and Lhx3 is essential to Hb9 (also known as Mnx1 and Hlxb9) expression in motor neurons (50), a transcription factor critical to both early pancreatic development (51) and motor neuron identity in the developing spinal cord (52). Because Isl1 binding was not found within or around the Hb9 gene by ChIP-Seq analysis in βTC-3 cells (B.E., C.S.H., J. Liu, A. Du, E. Walp, R.S., C.L.M., unpublished observations), it was not surprising that Pdx1Late-Cre;Isl1F/F pancreas mRNA and immunohistochemical staining levels at E18.5 were indistinguishable from wild-type (Fig. 6E and F). Nonetheless, Hb9 expression was significantly compromised in the Ldb1 mutant (Fig. 6E and G). Because Ldb1 binding in β-cells could not be linked to Hb9 control regions involved in either motor neuron (50) or islet β-cell transcription (53) by ChIP, it remains unclear if Ldb1 directly or indirectly stimulates expression.
DISCUSSION
Regulation of gene transcription involves the recruitment of coregulators by DNA-bound transcriptional activators and repressors. These protein–protein interactions ultimately influence the activity of the RNA polymerase II transcriptional machinery. We conclude from the analysis of pancreatic endocrine cell knockout mice in this study that the LIM-HD and Lmo coregulator Ldb1 is specifically required in α-, β-, and δ-cell production from islet progenitors during development. The inability of these cell types to properly form leads to overt hyperglycemia soon after birth. These findings were very similar to those observed upon removal of Isl1 from Pdx1Late-Cre;Isl1F/F mice pancreata, the predominant LIM-HD transcription factor in the developing and adult pancreas. However, Ldb1 also likely mediates activation by other LIM-HD and Lmo transcription factors, as suggested both by its much broader expression than Isl1 in very early pancreatic Pdx1+ dorsal and ventral lobe cell populations and unique impact in Hb9 and Glut2 transcription in later hormone+ cell formation. Clearly, the mechanisms by which Ldb1 controls pancreatic cell development and function warrant further investigation.
The Ldb1 and Ldb2 coregulators lack DNA-binding, transactivation, and enzymatic capacity. In contrast, the few other characterized coregulators associated with islet-enriched transcription factors possess enzymatic capabilities. For example, Pdx1 recruits coregulators capable of catalyzing modifications influencing protein stability [PDX-1 COOH terminus–interacting factor 1 (29)] and epigenetic control [CBP/p300 (32–34) and Set7/9 (30,48)]. Ldb1 and Ldb2 act by binding to LIM domain proteins through their COOH-terminal LIM interaction domain to regulate the stoichiometry, positioning, and abundance of Ldb-LIM complexes on target gene promoters (50,54,55). Notably, Isl1-dependent MafA region 3-driven activation was inhibited by a dominant-negative Ldb1 mutant spanning this COOH-terminal interaction surface (Fig. 4F). Although Ldb1 is important in developing heart (44), spinal cord (50), pituitary gland (26), and limb (36,56), the studies in this paper were the first to examine Ldb1 and Ldb2 expression and function in the developing pancreas.
Ldb1 was widely produced in the developing pancreas by E10.5, including throughout the Pdx1+ pancreatic epithelium and surrounding mesenchyme (Fig. 1). Later in development, Ldb1 became enriched in islet and ductal cells, with much lower expression in the exocrine pancreas. Ldb2 mRNA was also expressed in the pancreas and many surrounding tissues at E15.5, although the cellular protein distribution is unclear due to lack of immunostaining reagents. Notably, our analysis of Ldb1 (Fig. 3) and Ldb2 (Supplementary Fig. 2) mutant mice demonstrated that only Ldb1 was impactful to endocrine hormone+ cell production, specifically insulin-, glucagon-, and somatostatin-expressing cells. Because Ldb2−/− animals appear overtly normal [Mouse Genome Informatics, Lexicon Genetics (36)], in stark contrast to Ldb1−/− (44), it was not surprising to find that Ldb2 was of lesser (if any) relevance. The low abundance of Ldb2 mRNA compared with Ldb1 further suggests that Ldb2 has a limited (if any) functional role in adult islet cells (Fig. 1).
Pax6-Cre specifically eliminated Ldb1 expression from the majority of hormone+ cells in Ldb1F/F mice by E15.5 and compromised all but islet PP and ε (i.e., ghrelin+) cell formation, glycemic control, and viability (Fig. 3, data not shown). The widespread impact was analogous to removal of Isl1 from the developing pancreas with Pdx1Late-Cre (22). Thus, both Ldb1 and Isl1 activated critical effectors of islet α-cell [Arx (23)] and β-cell (MafA and Glp1r) formation and/or function (Fig. 4) (22). For example, Ldb1:Isl1 regulation of Glp1r was established by ChIP-Seq and mRNA expression analysis in E18.5 Pdx1Late-Cre;Isl1F/F and Pax6-Cre;Ldb1F/F pancreata (Fig. 5), with loss of Glp1r synthesis likely also contributing to compromised β-cell mass and function in the Isl1 and Ldb1 mutants (57).
Whereas Isl1 was the major LIM-HD factor expressed in the pancreas, Lmo1, Lmo2, and Lmo4 mRNA were produced in E15.5 pancreas and adult islets (Fig. 2A). Lmo1 and Lmo2 mRNA levels are also islet-enriched in humans (42). Binding of Lmo2 factors to Gata1/2, Tal1/Scl (basic helix-loop-helix), and Ldb1 is essential to α- and β-globin transcription during erythropoiesis (54,58). Existing antibody reagents limited our analysis only to Lmo4 in the pancreas, which was found enriched in ductal and acinar cells (Fig. 2B and C) (42). This distribution pattern is quite distinct from endocrine cell–enriched Isl1, suggesting that interactions among Ldb1, Lmo4, and exocrine-enriched Gata4 (59) could be critical to ductal and acinar cell formation. Moreover, regulatory complexes containing Ldb1, endocrine cell–enriched Gata6 (59,60), and Lmo1/2 could potentially contribute to Hb9, Nkx6.1, and Glut2 transcription during development, which represent genes unaffected in the E18.5 Isl1-deficient pancreata (Figs. 4 and 6). However, the impactful nature of Isl1 loss in the pancreas of the Pdx1Late-Cre;Isl1F/F mice on α-, β-, and δ-cell formation implies that such complexes are of reduced regulatory importance. In contrast, the relative abundance of Ldb1 to Isl1 in the Pdx1+ cells of the E10.5 dorsal and ventral pancreatic buds strongly indicates that distinct LIM-HD and Lmo proteins will be essential to Ldb1-mediated production of these pancreatic progenitor cells (Fig. 1).
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Institutes of Health (NIH) (grants DK-078606 to R.S. and C.L.M., DK-090570-02 to R.S., F32-DK-083160 to C.S.H., and T32-GM-07229 to B.E.) and the Juvenile Diabetes Research Foundation (Grant 2-2007-730 to C.L.M.). Partial support was also provided by the Vanderbilt University Diabetes Research and Training Center (Public Health Service Grant P60-DK-20593). Confocal microscopy was performed in the NIH-supported Vanderbilt University Medical Center Cell Imaging Shared Resource (NIH grants CA-68485, DK-20593, DK-58404, HD-15052, DK-59637, and EY-08126).
No potential conflicts of interest relevant to this article were reported.
C.S.H. researched data and wrote the manuscript. S.D., T.C., B.E., C.W., and M.F. researched data. H.W. reviewed and edited manuscript. R.S. and C.L.M. supervised research and wrote the manuscript. R.S. and C.L.M. are the guarantors of this work and, as such, had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
The authors thank Dr. Jingxuan Liu (Children’s Hospital of Philadelphia) for conducting the Isl1 ChIP-Seq and Dr. Jonathan Schug (University of Pennsylvania) as well as members of the Functional Genomics Core of the Penn Diabetes Center (Diabetes Research Center: P30-DK-19525) for performing sequencing and data analysis. The authors also thank Drs. Paul Love (National Institutes of Health), Jane Visvader (Walter and Eliza Hall Institute of Medical Research, Australia), Patrick Collombat (INSERM Unité Mixte de Recherche 636), Chris Wright (Vanderbilt University), and Maureen Gannon (Vanderbilt University) for providing Ldb1, Lmo4, Arx, Pdx1, and DBA antibodies, respectively. Dr. Stephen Brandt (Vanderbilt University) generously provided the Ldb1ΔN and control CMV plasmids.
Footnotes
This article contains Supplementary Data online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db12-0952/-/DC1.
REFERENCES
- 1.Slack JM. Developmental biology of the pancreas. Development 1995;121:1569–1580 [DOI] [PubMed] [Google Scholar]
- 2.Pan FC, Wright C. Pancreas organogenesis: from bud to plexus to gland. Dev Dyn 2011;240:530–565 [DOI] [PubMed] [Google Scholar]
- 3.Offield MF, Jetton TL, Labosky PA, et al. PDX-1 is required for pancreatic outgrowth and differentiation of the rostral duodenum. Development 1996;122:983–995 [DOI] [PubMed] [Google Scholar]
- 4.Kawaguchi Y, Cooper B, Gannon M, Ray M, MacDonald RJ, Wright CV. The role of the transcriptional regulator Ptf1a in converting intestinal to pancreatic progenitors. Nat Genet 2002;32:128–134 [DOI] [PubMed] [Google Scholar]
- 5.Krapp A, Knöfler M, Ledermann B, et al. The bHLH protein PTF1-p48 is essential for the formation of the exocrine and the correct spatial organization of the endocrine pancreas. Genes Dev 1998;12:3752–3763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gradwohl G, Dierich A, LeMeur M, Guillemot F. neurogenin3 is required for the development of the four endocrine cell lineages of the pancreas. Proc Natl Acad Sci USA 2000;97:1607–1611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sander M, Neubüser A, Kalamaras J, Ee HC, Martin GR, German MS. Genetic analysis reveals that PAX6 is required for normal transcription of pancreatic hormone genes and islet development. Genes Dev 1997;11:1662–1673 [DOI] [PubMed] [Google Scholar]
- 8.Gu G, Dubauskaite J, Melton DA. Direct evidence for the pancreatic lineage: NGN3+ cells are islet progenitors and are distinct from duct progenitors. Development 2002;129:2447–2457 [DOI] [PubMed] [Google Scholar]
- 9.St-Onge L, Sosa-Pineda B, Chowdhury K, Mansouri A, Gruss P. Pax6 is required for differentiation of glucagon-producing alpha-cells in mouse pancreas. Nature 1997;387:406–409 [DOI] [PubMed] [Google Scholar]
- 10.Oliver-Krasinski JM, Stoffers DA. On the origin of the beta cell. Genes Dev 2008;22:1998–2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gannon M, Ables ET, Crawford L, et al. pdx-1 function is specifically required in embryonic beta cells to generate appropriate numbers of endocrine cell types and maintain glucose homeostasis. Dev Biol 2008;314:406–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matsuoka TA, Artner I, Henderson E, Means A, Sander M, Stein R. The MafA transcription factor appears to be responsible for tissue-specific expression of insulin. Proc Natl Acad Sci USA 2004;101:2930–2933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sander M, Sussel L, Conners J, et al. Homeobox gene Nkx6.1 lies downstream of Nkx2.2 in the major pathway of beta-cell formation in the pancreas. Development 2000;127:5533–5540 [DOI] [PubMed] [Google Scholar]
- 14.Collombat P, Mansouri A, Hecksher-Sorensen J, et al. Opposing actions of Arx and Pax4 in endocrine pancreas development. Genes Dev 2003;17:2591–2603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Collombat P, Hecksher-Sørensen J, Broccoli V, et al. The simultaneous loss of Arx and Pax4 genes promotes a somatostatin-producing cell fate specification at the expense of the alpha- and beta-cell lineages in the mouse endocrine pancreas. Development 2005;132:2969–2980 [DOI] [PubMed] [Google Scholar]
- 16.Heller RS, Stoffers DA, Liu A, et al. The role of Brn4/Pou3f4 and Pax6 in forming the pancreatic glucagon cell identity. Dev Biol 2004;268:123–134 [DOI] [PubMed] [Google Scholar]
- 17.Artner I, Le Lay J, Hang Y, et al. MafB: an activator of the glucagon gene expressed in developing islet alpha- and beta-cells. Diabetes 2006;55:297–304 [DOI] [PubMed] [Google Scholar]
- 18.Thor S, Ericson J, Brännström T, Edlund T. The homeodomain LIM protein Isl-1 is expressed in subsets of neurons and endocrine cells in the adult rat. Neuron 1991;7:881–889 [DOI] [PubMed] [Google Scholar]
- 19.Ahlgren U, Pfaff SL, Jessell TM, Edlund T, Edlund H. Independent requirement for ISL1 in formation of pancreatic mesenchyme and islet cells. Nature 1997;385:257–260 [DOI] [PubMed] [Google Scholar]
- 20.Herrera PL. Adult insulin- and glucagon-producing cells differentiate from two independent cell lineages. Development 2000;127:2317–2322 [DOI] [PubMed] [Google Scholar]
- 21.Heiser PW, Lau J, Taketo MM, Herrera PL, Hebrok M. Stabilization of beta-catenin impacts pancreas growth. Development 2006;133:2023–2032 [DOI] [PubMed] [Google Scholar]
- 22.Du A, Hunter CS, Murray J, et al. Islet-1 is required for the maturation, proliferation, and survival of the endocrine pancreas. Diabetes 2009;58:2059–2069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu J, Hunter CS, Du A, et al. Islet-1 regulates Arx transcription during pancreatic islet alpha-cell development. J Biol Chem 2011;286:15352–15360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hunter CS, Rhodes SJ. LIM-homeodomain genes in mammalian development and human disease. Mol Biol Rep 2005;32:67–77 [DOI] [PubMed] [Google Scholar]
- 25.Agulnick AD, Taira M, Breen JJ, Tanaka T, Dawid IB, Westphal H. Interactions of the LIM-domain-binding factor Ldb1 with LIM homeodomain proteins. Nature 1996;384:270–272 [DOI] [PubMed] [Google Scholar]
- 26.Bach I, Carrière C, Ostendorff HP, Andersen B, Rosenfeld MG. A family of LIM domain-associated cofactors confer transcriptional synergism between LIM and Otx homeodomain proteins. Genes Dev 1997;11:1370–1380 [DOI] [PubMed] [Google Scholar]
- 27.Jurata LW, Kenny DA, Gill GN. Nuclear LIM interactor, a rhombotin and LIM homeodomain interacting protein, is expressed early in neuronal development. Proc Natl Acad Sci USA 1996;93:11693–11698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thomas MK, Yao KM, Tenser MS, Wong GG, Habener JF. Bridge-1, a novel PDZ-domain coactivator of E2A-mediated regulation of insulin gene transcription. Mol Cell Biol 1999;19:8492–8504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu A, Desai BM, Stoffers DA. Identification of PCIF1, a POZ domain protein that inhibits PDX-1 (MODY4) transcriptional activity. Mol Cell Biol 2004;24:4372–4383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chakrabarti SK, Francis J, Ziesmann SM, Garmey JC, Mirmira RG. Covalent histone modifications underlie the developmental regulation of insulin gene transcription in pancreatic beta cells. J Biol Chem 2003;278:23617–23623 [DOI] [PubMed] [Google Scholar]
- 31.Rocques N, Abou Zeid N, Sii-Felice K, et al. GSK-3-mediated phosphorylation enhances Maf-transforming activity. Mol Cell 2007;28:584–597 [DOI] [PubMed] [Google Scholar]
- 32.Mosley AL, Corbett JA, Ozcan S. Glucose regulation of insulin gene expression requires the recruitment of p300 by the beta-cell-specific transcription factor Pdx-1. Mol Endocrinol 2004;18:2279–2290 [DOI] [PubMed] [Google Scholar]
- 33.Qiu Y, Guo M, Huang S, Stein R. Insulin gene transcription is mediated by interactions between the p300 coactivator and PDX-1, BETA2, and E47. Mol Cell Biol 2002;22:412–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qiu Y, Sharma A, Stein R. p300 mediates transcriptional stimulation by the basic helix-loop-helix activators of the insulin gene. Mol Cell Biol 1998;18:2957–2964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao Y, Kwan KM, Mailloux CM, et al. LIM-homeodomain proteins Lhx1 and Lhx5, and their cofactor Ldb1, control Purkinje cell differentiation in the developing cerebellum. Proc Natl Acad Sci USA 2007;104:13182–13186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Narkis G, Tzchori I, Cohen T, Holtz A, Wier E, Westphal H. Isl1 and Ldb co-regulators of transcription are essential early determinants of mouse limb development. Dev Dyn 2012;241:787–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ashery-Padan R, Zhou X, Marquardt T, et al. Conditional inactivation of Pax6 in the pancreas causes early onset of diabetes. Dev Biol 2004;269:479–488 [DOI] [PubMed] [Google Scholar]
- 38.Gannon M, Herrera PL, Wright CV. Mosaic Cre-mediated recombination in pancreas using the pdx-1 enhancer/promoter. Genesis 2000;26:143–144 [DOI] [PubMed] [Google Scholar]
- 39.Schreiber E, Matthias P, Müller MM, Schaffner W. Rapid detection of octamer binding proteins with ‘mini-extracts’, prepared from a small number of cells. Nucleic Acids Res 1989;17:6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hunter CS, Maestro MA, Raum JC, et al. Hnf1α (MODY3) regulates β-cell-enriched MafA transcription factor expression. Mol Endocrinol 2011;25:339–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu Z, Huang S, Chang LS, Agulnick AD, Brandt SJ. Identification of a TAL1 target gene reveals a positive role for the LIM domain-binding protein Ldb1 in erythroid gene expression and differentiation. Mol Cell Biol 2003;23:7585–7599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dorrell C, Schug J, Lin CF, et al. Transcriptomes of the major human pancreatic cell types. Diabetologia 2011;54:2832–2844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Matthews JM, Visvader JE. LIM-domain-binding protein 1: a multifunctional cofactor that interacts with diverse proteins. EMBO Rep 2003;4:1132–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mukhopadhyay M, Teufel A, Yamashita T, et al. Functional ablation of the mouse Ldb1 gene results in severe patterning defects during gastrulation. Development 2003;130:495–505 [DOI] [PubMed] [Google Scholar]
- 45.Raum JC, Gerrish K, Artner I, et al. FoxA2, Nkx2.2, and PDX-1 regulate islet beta-cell-specific mafA expression through conserved sequences located between base pairs -8118 and -7750 upstream from the transcription start site. Mol Cell Biol 2006;26:5735–5743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Drucker DJ. The biology of incretin hormones. Cell Metab 2006;3:153–165 [DOI] [PubMed] [Google Scholar]
- 47.Thorens B, Sarkar HK, Kaback HR, Lodish HF. Cloning and functional expression in bacteria of a novel glucose transporter present in liver, intestine, kidney, and beta-pancreatic islet cells. Cell 1988;55:281–290 [DOI] [PubMed] [Google Scholar]
- 48.Deering TG, Ogihara T, Trace AP, Maier B, Mirmira RG. Methyltransferase Set7/9 maintains transcription and euchromatin structure at islet-enriched genes. Diabetes 2009;58:185–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bonny C, Thompson N, Nicod P, Waeber G. Pancreatic-specific expression of the glucose transporter type 2 gene: identification of cis-elements and islet-specific trans-acting factors. Mol Endocrinol 1995;9:1413–1426 [DOI] [PubMed] [Google Scholar]
- 50.Lee SK, Pfaff SL. Synchronization of neurogenesis and motor neuron specification by direct coupling of bHLH and homeodomain transcription factors. Neuron 2003;38:731–745 [DOI] [PubMed] [Google Scholar]
- 51.Harrison KA, Thaler J, Pfaff SL, Gu H, Kehrl JH. Pancreas dorsal lobe agenesis and abnormal islets of Langerhans in Hlxb9-deficient mice. Nat Genet 1999;23:71–75 [DOI] [PubMed] [Google Scholar]
- 52.Thaler J, Harrison K, Sharma K, Lettieri K, Kehrl J, Pfaff SL. Active suppression of interneuron programs within developing motor neurons revealed by analysis of homeodomain factor HB9. Neuron 1999;23:675–687 [DOI] [PubMed] [Google Scholar]
- 53.Thompson N, Gésina E, Scheinert P, Bucher P, Grapin-Botton A. RNA profiling and chromatin immunoprecipitation-sequencing reveal that PTF1a stabilizes pancreas progenitor identity via the control of MNX1/HLXB9 and a network of other transcription factors. Mol Cell Biol 2012;32:1189–1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Visvader JE, Mao X, Fujiwara Y, Hahm K, Orkin SH. The LIM-domain binding protein Ldb1 and its partner LMO2 act as negative regulators of erythroid differentiation. Proc Natl Acad Sci USA 1997;94:13707–13712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thaler JP, Lee SK, Jurata LW, Gill GN, Pfaff SL. LIM factor Lhx3 contributes to the specification of motor neuron and interneuron identity through cell-type-specific protein-protein interactions. Cell 2002;110:237–249 [DOI] [PubMed] [Google Scholar]
- 56.Tzchori I, Day TF, Carolan PJ, et al. LIM homeobox transcription factors integrate signaling events that control three-dimensional limb patterning and growth. Development 2009;136:1375–1385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lamont BJ, Li Y, Kwan E, Brown TJ, Gaisano H, Drucker DJ. Pancreatic GLP-1 receptor activation is sufficient for incretin control of glucose metabolism in mice. J Clin Invest 2012;122:388–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wadman IA, Osada H, Grütz GG, et al. The LIM-only protein Lmo2 is a bridging molecule assembling an erythroid, DNA-binding complex which includes the TAL1, E47, GATA-1 and Ldb1/NLI proteins. EMBO J 1997;16:3145–3157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ketola I, Otonkoski T, Pulkkinen MA, et al. Transcription factor GATA-6 is expressed in the endocrine and GATA-4 in the exocrine pancreas. Mol Cell Endocrinol 2004;226:51–57 [DOI] [PubMed] [Google Scholar]
- 60.Decker K, Goldman DC, Grasch CL, Sussel L. Gata6 is an important regulator of mouse pancreas development. Dev Biol 2006;298:415–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.