Abstract
Sexual reproduction in fungi is regulated by the mating-type (MAT) locus where recombination is suppressed. We investigated the evolution of MAT loci in eight fungal species belonging to Grosmannia and Ophiostoma (Sordariomycetes, Ascomycota) that include conifer pathogens and beetle symbionts. The MAT1-2 idiomorph/allele was identified from the assembled and annotated Grosmannia clavigera genome, and the MAT locus is flanked by genes coding for cytoskeleton protein (SLA) and DNA lyase. The synteny of these genes is conserved and consistent with other members in Ascomycota. Using sequences from SLA and flanking regions, we characterized the MAT1-1 idiomorph from other isolates of G. clavigera and performed dotplot analysis between the two idiomorphs. Unexpectedly, the MAT1-2 idiomorph contains a truncated MAT1-1-1 gene upstream of the MAT1-2-1 gene that bears the high-mobility-group domain. The nucleotide and amino acid sequence of the truncated MAT1-1-1 gene is similar to its homologous copy in the MAT1-1 idiomorph in the opposite mating-type isolate, except that positive selection is acting on the truncated gene and the alpha(α)-box that encodes the transcription factor has been deleted. The MAT idiomorphs sharing identical gene organization were present in seven additional species in the Ophiostomatales, suggesting that the presence of truncated MAT1-1-1 gene is a general pattern in this order. We propose that an ancient unequal recombination event resulted in the ancestral MAT1-1-1 gene integrated into the MAT1-2 idiomorph and surviving as the truncated MAT1-1-1 genes. The α-box domain of MAT1-1-1 gene, located at the same MAT locus adjacent to the MAT1-2-1 gene, could have been removed by deletion after recombination due to mating signal interference. Our data confirmed a 1:1 MAT/sex ratio in two pathogen populations, and showed that all members of the Ophiostomatales studied here including those that were previously deemed asexual have the potential to reproduce sexually. This ability can potentially increase genetic variability and can enhance fitness in new, ecological niches.
Keywords: heterothallism, homothallism, mating system evolution, outcrossing, selfing
Sexual reproduction in fungi is controlled by the mating-type (MAT) loci, in which the genes determine mating incompatibility and regulate key mating processes (Coppin et al. 1997; Debuchy et al. 2010). Most haploid ascomycete fungi have a single MAT locus termed MAT1, which has two alleles called MAT1-1 and MAT1-2 (Turgeon and Yoder 2000). Like large portions of the human X and Y chromosomes, the DNA and amino acid sequences of MAT1-1 and MAT1-2 are no more similar than expected by chance; thus, they have been termed idiomorphs (Metzenberg and Glass 1990). The idiomorphs encode transcription factors with conserved DNA binding domains involved in the regulation of mating identity and sexual development (Coppin et al. 1997). The two idiomorphs are distinguished by the presence of either an alpha(α)-box domain in MAT1-1 or a high-mobility-group (HMG) domain in MAT1-2 (Coppin et al. 1997). Heterothallic (outcrossing) ascomycete fungi carry either one of the two idiomorphs at the MAT locus, and two isolates bearing complementary MAT1 idiomorphs are required to mate. In contrast, homothallic ascomycetes can undergo haploid selfing (completion of the sexual cycle with a clonemate) because MAT genes with α-box and HMG domains are both present in the mating partners (Debuchy et al. 2010; Billiard et al. 2011). This gene arrangement may have evolved under selection for universal compatibility (Billiard et al. 2011, 2012). The MAT1-1 idiomorph usually comprises a gene, MAT1-1-1, whereas the MAT1-2 idiomorph has a different gene, MAT1-2-1. However, additional genes such as MAT1-1-2 and MAT1-1-3 have been reported in the MAT1-1 idiomorphs in various sordariomycetous ascomycetes (Turgeon and Yoder 2000; Debuchy et al. 2010). The questions of which mode of sexual reproduction, heterothallism or homothallism, is ancestral and which genetic/evolutionary mechanism mediates the change from one to the other have received much attention from fungal biologists (Debuchy et al. 2010). Therefore, understanding the MAT locus organization can provide insight into the genetics and evolution of mating systems and life history in ascomycetes.
The fungi in Ophiostomatales (Sordariomycetes, Ascomycota) are diverse, with more than 100 species, including many important or aggressive tree pathogens responsible for wilt diseases, blue stain in commercial timber; in addition, they play an important role as insect symbionts and associates, as well as saprophytes (Zipfel et al. 2006). Most species belong to sexual genera Grosmannia, Ophiostoma, and the asexual genus Leptographium. In North America, Grosmannia clavigera is an important conifer pathogen that has a symbiotic relationship with its vector the mountain pine beetle (Dendroctonus ponderosae). Grosmannia clavigera and other fungal pathogens and symbionts, such as Leptographium longiclavatum and Ophiostoma montium, are carried in the mycangia and on the exoskeleton of mountain pine beetles because these fungi produce abundant slimy spores that attach themselves to the insect’s body. These fungi can grow into the sapwood and damage the host tree’s water transport system (Yamaoka et al. 1995; Solheim and Krokene 1998). The mountain pine beetles and its fungal symbionts have destroyed more than 17.5 million ha of lodgepole pine forests in western Canada in the last decade (http://www.for.gov.bc.ca/hfp/mountain_pine_beetle/facts.htm), and the magnitude of devastation is the largest in recorded history in Canada (Kurz et al. 2008; Safranyik et al. 2010).
G. clavigera is very aggressive and it may be capable of detoxifying terpenoids that are an important class of defense compounds in pines (DiGuistini et al. 2011). Because G. clavigera plays an important role in the pine−beetle−fungus dynamics and epidemics, it is essential to understand its biology, genetics, and population structure. Populations of G. clavigera are polymorphic and form distinct genetic clusters yet with some gene flow and admixture among clusters (Lee et al. 2007; Tsui et al. 2012). Evidence of random mating and linkage equilibrium were also revealed, indicating sexual reproduction occurred in G. clavigera populations (Tsui et al. 2012). However, the sexual fruiting bodies of G. clavigera are rarely observed (Lee et al. 2005), even though G. clavigera is reported to have a life cycle comprising both the asexual and sexual stages. It is important to understand the ability of a fungal pathogen to perform sexual reproduction because such information could provide clues to the population biology of these fungi, which could be useful to further our understanding of the recent unprecedented epidemic.
Grosmannia clavigera lineage Gs sensu (Alamouti et al. 2011) is heterothallic because a gene homologous to MAT1-2-1 is characterized at the MAT locus in the isolate for which the genome was sequenced but no gene coding for the α-box domain was reported (DiGuistini et al. 2011). Although fungi in Ophiostomatales are diverse in breeding strategies with worldwide distribution, the MAT locus has been characterized in only a limited number of species, such as Ophiostoma ulmi, O. novo-ulmi, and O. himal-ulmi, which are responsible for the Dutch elm disease epidemic in Europe and North America (Paoletti et al. 2005a; Jacobi et al. 2010). The genes corresponding to opposite mating-types recently were reported in the MAT locus of Ophiostoma quercus, which causes significant sapstain in hardwood (Wilken et al. 2012). Because the MAT1-1 idiomorph has not been characterized in G. clavigera and the MAT locus organization has not been well investigated in other fungi of Ophiostomatales, we used genomics and primer walking approaches to study the organization and evolution of the MAT locus in G. clavigera and eight related fungi. To address the question of whether heterothallism or homothallism is a derived character state, we also compared the MAT locus organization with other ascomycete species in the Sordariomycetes. The aim of the present investigation was (1) to characterize the mating-type locus organization of G. clavigera bearing a MAT1-1 idiomorph; (2) to investigate the evolution of MAT genes and mating systems in fungi belonging to Ophiostomatales with respect to other ascomycetes; and (3) to determine the mating-type ratio in the populations of G. clavigera.
Materials and Methods
Fungal materials and culture collection
Thirty-four fungal isolates belonging to nine species of Ophiostomatales were studied: G. clavigera (Gc), L. longiclavatum (Llo), Leptographium terebrantis (Lt), Grosmannia aurea (Ga), Leptographium wingfieldii (Lw), Grosmannia robusta (Gr), Grosmannia huntii (Gh), Leptographium lundbergii (Llun), and Ophiostoma montium (Table 1). The identities of many isolates were previously characterized and confirmed using the DNA sequences of rRNA genes and additional protein coding genes (Lim et al. 2004). They were cultured and maintained in malt extract agar (MEA) (Tsui et al. 2012). Twenty isolates have MAT1-1 idiomorphs, and 14 isolates have MAT1-2 idiomorphs.
Table 1. Taxa and isolates used to characterize the mating-type (MAT) loci.
| Species | Isolate Code | Geographic Origin | Substrate | Year of Isolation | Idiomorph | GenBank Accession No. |
|---|---|---|---|---|---|---|
| Grosmannia clavigera (Gc) | SL-KW1407/UAMH11150 (genome isolate) | Kamloops, Canada | Pinus contorta | 2001 | MAT1-2 | ACXQ02000000 (locus GL629756) |
| B13 | Banff, Canada | Pinus contorta | 2003 | MAT1-2 | JX402933 | |
| SS274 | Fairview, Canada | Pinus contorta × banksiana hybrid | 2007 | MAT1-2 | JX402934 | |
| B101 | Banff, Canada | Pinus contorta | 2003 | MAT1-1 | JX402947 | |
| ATCC18086 (holotype) | Cache Creek, Canada | Pinus ponderosa | 1965 | MAT1-1 | JX402948 | |
| SS278 | Canmore, Canada | Pinus contorta | 2007 | MAT1-1 | JX402945 | |
| M6 | Manning Park, Canada | Pinus contorta | 2003 | MAT1-1 | JX402943 | |
| M11 | Manning Park, Canada | Pinus contorta | 2003 | MAT1-1 | JX402944 | |
| BW28 | Banff, Canada | Pinus contorta | 2003 | MAT1-1 | JX402946 | |
| Leptographium longiclavatum (Llo) | SS86 | Kakwa, Canada | Pinus contora | 2007 | MAT1-2 | JX402931 |
| HV7 | Hidden Valley, USA | Dendroctonus ponderosae | 2003 | MAT1-2 | JX402932 | |
| SS88 | Kakwa, Canada | Pinus contora | 2007 | MAT1-1 | JX402953 | |
| SL-KW1436 (holotype) | Williams Lake, Canada | Pinus contorta | 2004 | MAT1-1 | JX402954 | |
| HV18 | Hidden Valley, USA | Dendroctonus ponderosae | 2003 | MAT1-1 | JX402955 | |
| Leptographium terebrantis (Lt) | T26 (LPKRLT-3) | BC, Canada | Pinus contorta | 2003 | MAT1-2 | JX402936 |
| T27 (CBS337.7) | Louisiana, USA | Pinus taeda | 1966 | MAT1-2 | JX402937 | |
| SS394 | Fox Creek, Canada | Pinus contorta × banksiana hybrid | 2007 | MAT1-2 | JX402935 | |
| SS403 | Crowsnest Pass, Canada | Pinus contorta | 2007 | MAT1-1 | JX402956 | |
| Grosmannia aurea (Ga) | SS419 | Grande Prairie, Canada | Pinus contorta × banksiana hybrid | 2007 | MAT1-2 | JX402938 |
| SS471 | Fox Creek, Canada | Pinus contorta × banksiana hybrid | 2007 | MAT1-2 | JX402939 | |
| CBS438.69 (OA18-A27) (holotype) | Invermore, Canada | Pinus contorta var. latifolia | 1969 | MAT1-1 | JX402951 | |
| AU98-Pr2-169 | Princeton, Canada | Pinus contorta | NA | MAT1-1 | JX402952 | |
| Leptographium wingfieldii (Lw) | CMW2095 | NA | Pinus strobus | 2004 | MAT1-1 | JX402950 |
| CMW2096 | NA | Pinus sylvestris | NA | MAT1-1 | JX402949 | |
| Leptographium lundbergii (Llun) | UAMH9584 | Skutskar, Uppland, Sweden | Pinus sylvestris | NA | MAT1-2 | JX402940 |
| UM1434 | NA | NA | 2004 | MAT1-1 | JX402941 | |
| DAOM64706 | Ontario, Canada | P. strobus | 1961 | MAT1-1 | JX402958 | |
| Grosmannia huntii (Gh) | CBS398.77 | NY | P. monticola | 1963 | MAT1-1 | JX402942 |
| CMW185 | South Africa | NA | 2001 | MAT1-2 | JX402930 | |
| Grosmannia robusta (Gr) | CMW668 | South Africa | Picea abies | 2001 | MAT1-1 | JX402957 |
| Ophiostoma montium (Om) | UAMH 1363 | British Columbia, Canada | Pinus contorta | 1959 | MAT1-1 | JX402993 |
| UAMH 4875 | Alberta, Canada | Pinus contorta | 1983 | MAT1-1 | JX402995 | |
| UAMH 11095 | Fox Creek, Canada | Pinus contorta x banksiana hybrid | 2007 | MAT1-2 | JX402994 | |
| UAMH 4838 | Alberta, Canada | Pinus contorta | 1986 | MAT1-2 | JX402996 |
DNA extraction, polymerase chain reaction (PCR) amplification, sequencing
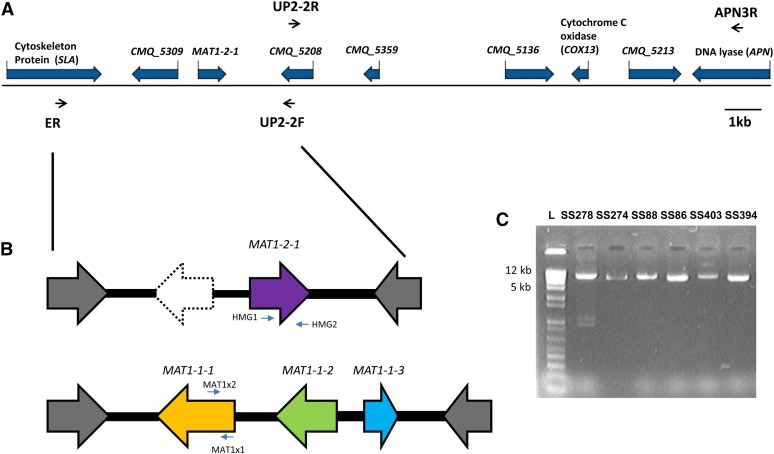
DNA was extracted from all isolates using the procedures described in Roe et al. (2011) and Tsui et al. (2012). The genome sequencing, assembly, and annotation of G. clavigera has been previously described in DiGuistini et al. (2011). Based on the genome sequence of G. clavigera (GenBank accession number: ACXQ02000000), genes homologous to cytoskeleton assembly protein (SLA), HMG-domain of MAT, and DNA lyase (APN), as well as a few hypothetical proteins without known functions, were identified and characterized (Figure 1) (DiGuistini et al. 2011). To characterize the MAT1-1 idiomorph, we used a long-range PCR amplification approach coupled with primer walking sequencing. Primers ER and UP2-2F targeting the SLA, and a hypothetical protein-encoding gene (CMQ_5208) located downstream from the MAT locus were designed for long-range PCR reaction (Figure 1A) (Table 2). Primers UP2-2R and APN3R, which targeting CMQ_5208 and APN, were also designed to amplify a 15 kb fragment of a putative MAT1-1 isolate (Figure 1A).
Figure 1 .
(A) Sequence arrangement and annotation of the MAT idiomorph and adjacent genes from annotated genome of Grosmannia clavigera SL-KW1407. (B) The MAT genes of opposite mating-type isolates as determined from long-range PCR and primer walking using primers ER and UP2-2F. Genes are indicated by different colors, and the arrows indicate the predicted directions of gene translation. (C) Amplicons of MAT fragments after long-range PCR were run in an agarose gel to indicate the size variation between MAT1-2 and MAT1-1 isolates.
Table 2. Major primers used in this investigation.
| Target Gene | Primer Name | Sequence (5′-3′) |
|---|---|---|
| Regular PCR | ||
| SLA of G. clavigera | ER | GCCACGTCGTTCAACAACTA |
| Hypothetical protein (CMQ_5309 of | UP2-2F | AGATGGTCATCTCCCGTGAC |
| G. clavigera) | ||
| UP2-2R | AGATGGTCATCTCCCGTGAC | |
| HMG domain in G. clavigera | HMG1 | CCGCGCCCACCCAATGCGTACAT |
| HMG2 | CGAGGGTTGTATCTGTAGTCAGG | |
| alpha-box domain in G. clavigera | MAT1x1 | CGTCCACTGAATGCCTTCATG |
| MAT1x2 | GTGGGCAATCATAGCCAAAGT | |
| Cytochrome C oxidase of G. clavigera | COX13A | GCTTGACGCAACTATCTCTGC |
| COX13B | TGCATCCCCTACTCGATACAC | |
| DNA lyase (APN) of G. clavigera | cAPNR | GATTCCTTTTACAGCTTTCCCCAC |
| APN2R | GACGAGGAGCTGCATCAGG | |
| APN3F | GACAGGATCACGAACACAACC | |
| APN3R | TCTTCGATTGGCTCTTTAGGG | |
| SLA of O. montium | OM7R | CAACACGCTCATTGAGAC |
| HMG domain in O. montium | OM-HMG1 | CGCCCCCCCAATGCCTACATTC |
| OM-HMG2 | CGGGGATTGTACTTGTAGTGCGG | |
| alpha-box domain in O. montium | OM-A1 | GAATGCCTTCATGGCCTTCC |
| OM-A2 | ACCTTTGCCATCAACGTCCATTT | |
| Real-time PCR | ||
| truncated MAT1-1-1 | pMF2 | GATCAGATGGGCAAGCTCAG |
| pMR2 | AAGGCTTGGAAGGACGTGTT |
PCR, polymerase chain reaction.
Long-range PCR amplifications of DNA were carried out in 50 μL using a PTC-100 thermocycler (MJ Research Inc., Watertown, MA). Reaction mixtures contained 100 ng of DNA, 1× PCR buffer, 200 μM each dNTP, 0.6 μM of each primer (Eurofins MWG Operon, Huntsville, AL), 1.5 μL of dimethyl sulfoxide and 2 U of Phusion DNA polymerase (Finnzymes; New England BioLabs, Ipswich, MA). The PCR amplifications were performed for 30 sec at 98°, followed by 35 cycles of 10 sec at 98°, 30 sec at 60−62° and 4 min at 72°, and final extension at 72° for 10 min. Sequencing reactions with primer walking were performed at the Centre de recherche du CHUQ, Québec, Canada. Primers for sequencing are listed in (Supporting Information, Table S1).
After MAT idiomorph characterization, mating-type specific PCR assay was performed by designing primers HMG1 and HMG2 targeting the HMG domain, as well as primers MAT1x1 and MAT1x2 targeting the α-box domain (Figure 1B, Table 2). Fragments of cytochrome c oxidase subunit gene VIa (COX13) and APN also were amplified for selected species and representatives (Table 2). PCR amplifications were carried out in 25 μL using a PTC-100 thermocycler (MJ Research Inc.). Reaction mixtures contained 20−40 ng DNA, 1× PCR buffer, 200 μM each dNTP, 1.5 μM of each primer (Eurofins MWG Operon), and 1 U of Paq polymerase (Stratagene, Integrated Sciences). The PCR amplifications were performed for 3 min at 94°, followed by 30 cycles of 35 sec at 94°, 35 sec at 52−55°, and 35 sec at 72°, and final extension at 72° for 7 min.
Gene annotation and analyses
The genome and transcriptomes of G. clavigera were assembled and annotated as described in (DiGuistini et al. 2011). Gene models were predicted using GLEAN, and the putative gene function assignments were generated from searches of the NCBInr and Swiss-Prot databases using BLAST with PFAM domain assignments (DiGuistini et al. 2011).
Sequence reads after primer walking were assembled using the Staden package (Staden 1996) and Geneious Pro (http://www.geneious.com/), and they were compared with genes present in GenBank using BLASTx and BLASTn. The assembled sequences were submitted to FGENESH+ within Softberry (http://www.softberry.ru/berry.phtml?topic=fgenes_plus&group=programs&subgroup=gfs) for gene prediction and to determine the location of the coding/non coding regions, and manually annotated with Artemis (Rutherford et al. 2000).
The nucleotide sequences of both MAT idiomorphs were compared using dotplot (matrix) analyses implemented in Geneious. The sequences of the MAT loci were also compared across different species using the online Artemis Comparison Tool (http://www.webact.org/WebACT/home) with BLASTn algorithm.
Evolutionary analyses of MAT genes
The amino acid sequences encoded by the MAT1-1 α-box and the MAT1-2 HMG domain were aligned to sequences of other ascomycetes from GenBank with Clustal W (Thompson et al. 1997) implemented in Geneious. Phylogenetic analysis was carried out with the Neighbor-joining (NJ) and maximum likelihood algorithm in MEGA 5 (Tamura et al. 2011), PhyML(Dereeper et al. 2008) implemented in (http://www.phylogeny.fr/version2_cgi/index.cgi), as well as PRODIST and PROML in PHYLIP 3.69 (Felsenstein 2005).
The nucleotide sequences were aligned in Geneious and manually adjusted with Se-Al v2 (Rambaut 1999) and analyzed with PAUP v4.b10 (Swofford 2003) and MEGA 5 (Tamura et al. 2011). Nucleotide data of different genes were subjected to parsimony analysis implemented in PAUP. Bootstrap support for the branches was based on 1000 replicates with TBR branch swapping algorithms and simple sequence addition. The individual data sets were also subjected to NJ implemented in PAUP using GTR model corrections with the proportion of variable sites and gamma shape estimated from Modeltest 3.7 (Posada 2006). Bootstrap values were estimated based on 1000 replicates. Sequences were deposited in GenBank (accession numbers of MAT idiomorphs: JX402930-JX402958, JX402993-JX402996; of other genes: JX402959-JX402992).
Signatures of purifying or positive selection acting on the MAT genes were tested at the codon level. A maximum likelihood analysis was used to fit codon substitution models to the data using the CODEML program within PAML (Yang 2007). Four random site models were used to describe the variation of ω (= dN/dS) among codon sites in an alignment containing the full length MAT1-1-1 and truncated MAT1-1-1 genes of six species. Random site models M1A (neutral), M2A (selection), M7 (beta), and M8 (beta and selection) were used to describe the variation of ω among codon sites within each MAT1 gene. M1a assumes two site classes in proportions p0 and p1 = 1 − p0 with 0 < ω0< 1(purifying selection) and ω1 = 1 (neutral). M2a adds an additional class of site with ω2 as a free parameter, allowing for sites with ω2 > 1 (positive selection) with proportion p2. M7 is a flexible null model in which a v ratio for each codon is randomly selected from a beta distribution between 0 and 1. M8 adds one additional site class to M7 allowing for positive selection. A test for positive selection was implemented using likelihood ratio tests that compare models pair M1a/M2a (Yang 2007). Three different starting ω values (0.2, 1.0, and 2.0) were implemented for each model fitting, as described in (Joly et al. 2010). Codon sites under positive selection were then identified using the empirical Bayes method to calculate the posterior probability that a particular amino acid belongs to a given selection class (neutral, deleterious, or advantageous) (Yang 2007).
Analysis of mating-type distribution in populations of G. clavigera and L. longiclavatum
We used the aforementioned mating-type primers (HMG 1, HMG2, MAT1x1, MAT1x2) to identify mating-types in populations from Canada and the USA (Table 2). We performed PCR on DNA extracted from 335 isolates of G. clavigera characterized in a previous study (Tsui et al. 2012), as well as more than 100 isolates of L. longiclavatum currently being investigated (Farfan et al. 2011). Mating-type distributions were tested for deviation from the expected ratios of 1:1 using chi-square goodness-of-fit tests.
Reverse transcriptase (RT) quantitative PCR amplification
RNA was extracted and RT was performed from G. clavigera isolate SL-KW1407 cultured on 1% MEA overlaid with cellophane for 4 d (DiGuistini et al. 2011) to determine the level of expression from hypothetical protein encoding gene CMQ_5309 (truncated MAT1-1-1 gene) and MAT1-2-1. RT-quantitative PCR was also carried out with SsoFast EvaGreen Supermix (Bio-Rad) in a volume of 20 µL in a Bio-Rad CFX384 system. Primer pairs were internal to regions of MAT1-2-1 and truncated MAT1-1-1 to yield amplicons of 100−120 bp (Table 2). PCRs were described as follows: 96° for 45 sec, followed by 35 cycles of 95° for 15 sec, and 57.5° for 30 s, followed by a melt-curve analysis. The Cq value for amplification of the β-tubulin gene was used as a reference.
Results
Organization of the mating-type locus in G. clavigera
The MAT1-2 idiomorph and flanking genes of G. clavigera isolate SL-KW1407 were identified from the genome (Figure 1A), and the amplification of the MAT1-2 idiomorph from isolate G. clavigera SS274 demonstrated greater than 99.9% sequence similarity to the sequence of the reference genome. The gene order and orientation near the MAT locus was syntenic with other Sordariomycetes. Two ORFs were predicted in the MAT1-2 idiomorph. One of the translated proteins bearing the HMG domain (285 amino acids; EFX05114) was homologous to the MAT1-2-1 of other ascomycetes and shared 65% similarity to that of Ophiostoma himal-ulmi. In contrast, CMQ_5309, encoding a hypothetical protein (EFX04946.1), had no significant similarity to any genes in the NCBI sequence database. SLA2, which encodes the cytoskeleton assembly control protein (EFX04935.1), was located upstream from the MAT locus, whereas cytochrome c oxidase subunit (EFX05020.1) gene COX13 and DNA lyase (EFX04950.1) gene APN2 were located downstream from the MAT (Figure 1A). The deduced amino acids of SLA and APN had 68% and 62% identities, respectively, to those of Neurospora crassa, a relative in the Sordariomycetes. However, the intergenic distance between MAT and APN loci in G. clavigera was large, spanning greater than 10 kb with several putative proteins coding genes of unknown functions identified (Figure 1A). One of the putative proteins (CMQ_5213; EFX04951) had a protease-associated domain and was homologous (35% similarity) to a RING-9 protein in Verticillium albo-atrum (XP_003007854). The functions of these additional genes to mating activity are not yet established. tBLAST searches using the α-box domain containing genes from different ascomycetes as queries did not return any significant matches in the SL-KW1407 genome, confirming the absence of MAT1-1-1 gene in the reference isolate.
Using the primer walking approach, we obtained the full sequence (22,910 bp) of the MAT1-1 idiomorph of G. clavigera isolate B101. The gene arrangement was syntenic to the assembled genome of G. clavigera isolate SL-KW1407. The genes of SLA, MAT, COX13, and APN were located in the identical order and orientation in both MAT1-1 and MAT1-2 idiomorphs. At the nucleotide level, the sequence of the >15-kb fragment spanning from hypothetical protein coding gene CMQ_5208 to APN was 99.9% identical in isolates B101 and SL-KW1407.
Three open reading frames (ORFs) were predicted in the MAT1-1 idiomorph of isolate B101, encoding proteins of 619, 340, and 171 amino acids, respectively (Figure 1B). The first protein had α-box domain and was homologous (52% similarity) to the MAT1-1-1 (ACZ53927.1) of Ophiostoma novo-ulmi subsp. novo-ulmi. The second and third proteins were also 24% and 52% similar to the MAT1-1-2 (ACZ53926.1) and MAT1-1-3 (ACZ53925.1) of O. novo-ulmi subsp. novo-ulmi, respectively.
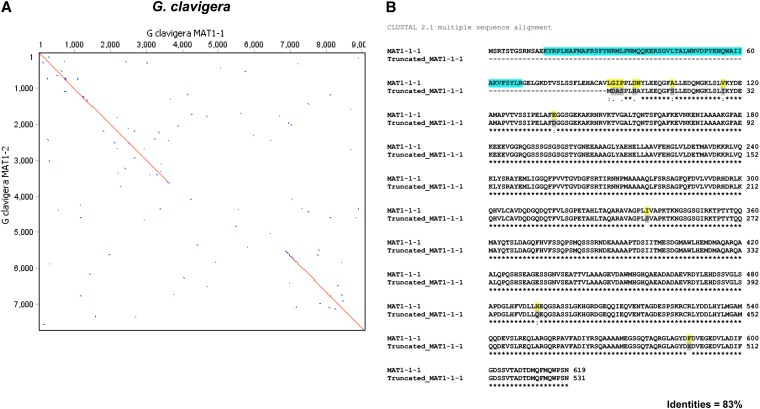
Dotplot comparison of the MAT1-1 and MAT1-2 idiomorphs revealed nucleotide sequence similarity (99%) in the SLA encoding gene, regions upstream from the MAT locus, as well as the gene encoding protein CMQ_5208 (Figure 2A). Surprisingly the gene encoding putative protein (CMQ_5309; EFX05047.1) located upstream from the MAT1-2-1 gene in isolate SL-KW1407 was homologous (>80% similarity in amino acids) to the MAT1-1-1 gene in the MAT1-1 idiomorph but shorter in length and without introns. Sequence comparison revealed α-box domain truncation/deletion (89 amino acids) at the N-terminus of the putative protein. The start codon was, however, present in the putative protein coding sequence (now called truncated MAT1-1-1) followed by five codons that are not homologous to the MAT1-1-1 on the MAT1-1 idiomorph (Figure 2B). This 15bp 5′ end sequence was used to search the G. clavigera genome as well as the NCBI database but no significant match was returned.
Figure 2 .
Comparison of MAT loci in Grosmannia clavigera. (A) Dotplot comparison/pairwise alignment of DNA sequence data for MAT1-1 and MAT1-2 idiomorphs of G. clavigera. Sequence lengths are given along the axes. (B) The amino acid alignment of MAT1-1-1 in MAT1-2 idiomorph to the truncated MAT1-1-1 in MAT1-2 idiomorph of G. clavigera by Clustal W. The comparison indicates the deletion of α-box domain (in square) in truncated MAT1-1-1.
The MAT loci of additional G. clavigera isolates from different geographic locations were also sequenced using the identical long-range PCR approach to investigate whether the truncated MAT1-1-1 gene could be unique solely to isolate SL-KW1407 (Table 1). Sequencing data confirmed that the isolates bearing the same mating-type have identical idiomorph size (data not shown). The presence of this truncated gene in multiple G. clavigera isolates confirms that it is not a spurious result or a unique feature of the reference isolate.
Organization of the MAT idiomorphs in fungal species related to G. clavigera
The presence of the truncated/incomplete gene suggested that the ancestor of G. clavigera may have had both MAT1-1 and MAT1-2 copies and was homothallic. To understand the evolutionary history leading to the presence of the truncated MAT1-1-1 gene, we used the same primer pair ER and UP2-2F (targeting SLA and hypothetical protein-coding gene CMQ_5208) to characterize the MAT loci of several fungi related to G. clavigera (Lim et al. 2004) (Table 1). If the truncated gene has been inherited from a common ancestor, it may be preserved in other related species since it may confer an evolutionary or ecological advantage.
Sequences containing orthologous genes of the MAT locus and flanking regions were characterized from 21 isolates belonging to seven species within the Ophiostomatales: Llo, Lw, Lt, Llun, Ga, Gr, and Gh (Table 1, Figure 1C). Interspecific variations in fragment size ranged from 5787 to 8011 bp in MAT1-1 isolates, and from 5209 to 7727 bp in MAT1-2 isolates among different species. The variations in fragment size are also partly due to incomplete sequencing of the flanking sequences of SLA and the putative protein-coding gene CMQ_5208. The 3′end of mating-type loci in Llun and Gh were incomplete because the primers targeting the CMQ_5208 failed to amplify, and a new primer GCM3 was designed to target the conserved regions in the MAT1-1-3 for MAT1-1 idiomorph characterization. Conversely, primer HMG2 targeting MAT1-2-1 gene was used to amplify the partial MAT1-2 idiomorph fragment. Only the MAT1-1 idiomorphs of Lw and Gr were obtained because MAT1-2 isolates were not available (Table 1). The MAT1-1 idiomorph of L. lundbergii isolate DAOM64706 was shorter and incomplete when compared with other taxa, so its sequence was not included in the analyses.
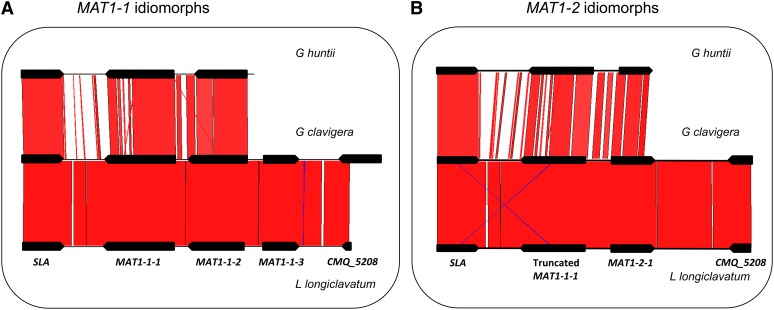
Dotplot comparison of the MAT1-1 and MAT1-2 idiomorphs in each of the species also revealed nucleotide sequence similarity (95–99%) in the SLA protein-coding regions, regions upstream from the MAT locus, as well as the CMQ_5208 (Figure S1). The organization of the idiomorphs among these seven species was identical, with the presence of three predicted ORFs in the MAT1-1 idiomorph and two ORFs in the MAT1-2 idiomorph, in addition to the truncated MAT1-1-1 gene (the comparison among Gh, Gc, and Llo is illustrated in Figure 3). The nucleotide sequence of the MAT idiomorphs was similar among these seven species within Grosmannia: it ranged from 72% between G. clavigera and G. huntii to 97.5% between G. clavigera and L. longiclavatum (Table S2 and Figure 3, A and B). Sequence similarity was much lower when compared to other ascomycetes.
Figure 3 .
Homology among the MAT locus. (A) MAT1-1 idiomorph; (B) MAT1-2 idiomorph, of G. huntii, G. clavigera, and L. longiclavatum. The diagram was prepared from the output of Artemis Comparison Tool. Regions of strong homology are shaded and connected by lines. The intensity of shading indicates the strength of homology. Genes are represented by box arrows. The MAT1-2 sequence of G. clavigera SL-KW1407 was obtained from the genome sequence, while the MAT1-1 sequence of G. clavigera and opposite MAT isolates of other fungi were obtained in this investigation.
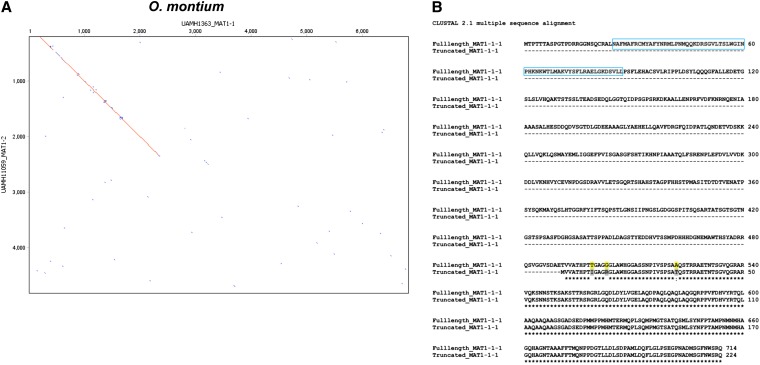
Ophiostoma is a sister genus to Grosmannia (Zipfel et al. 2006). A genomic DNA library of O. montium populations (R. C. Hamelin, unpublished data) was searched to design primers anchoring the SLA and the flanking regions of the MAT locus in both mating-types. Using a long-range PCR amplification and primer walking approach, we also amplified the MAT locus from four isolates of O. montium including both mating-types (Tables 1 and 2). We found fragments of ca. 6 kb and 5 kb in size that corresponded to both MAT1-1 and MAT1-2 idiomorphs. Dotplot comparison of the MAT1-1 and MAT1-2 idiomorphs were similar to the orthologs from Grosmannia species and revealed nucleotide sequence similarity in the SLA protein and intergenic regions upstream from the MAT locus (Figure 4A). The MAT1-1 idiomorph had three putative ORFs that corresponded to MAT1-1-1, MAT1-1-2, and MAT1-1-3 genes, and they were similar (53–76% at amino acids level) to those of O. novo-ulmi subsp. novo-ulmi (Figure S2). The MAT1-2 idiomorph contained a MAT1-2-1 gene encoding a protein (263 aa) with HMG domain (75% similar to that of O. novo-ulmi subsp. novo-ulmi), as well as a truncated MAT1-1-1 gene without the α-box domain located upstream from the MAT1-2-1 gene. The truncated MAT1-1-1 (224 amino acids) was short when compared with the original MAT1-1-1 (714 aa) in the MAT1-2 idiomorph (Figure 4B) and the extent of deletion/truncation was greater than that in Grosmannia species.
Figure 4 .
Comparison of MAT loci in Ophiostoma montium. (A) Dotplot comparison/pairwise alignment of DNA sequence data for MAT1-1 and MAT1-2 idiomorphs of O. montium. Sequence lengths are given along the axes. (B) The amino acid alignment of MAT1-1-1 in MAT1-2 idiomorph to the truncated MAT1-1-1 in MAT1-2 idiomorph of O. montium by Clustal W. The comparison indicates the truncated MAT1-1-1 was highly eroded and the absence of α-box domain (in square in full length MAT1-1-1).
Evolutionary analyses of the MAT loci
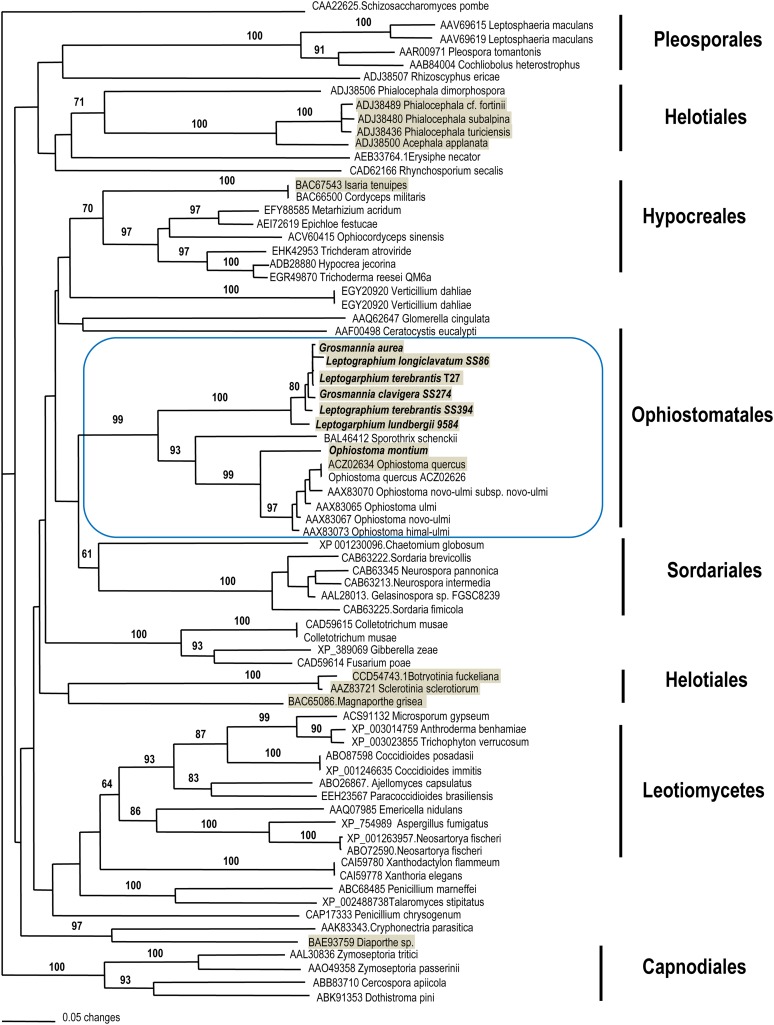
Phylogenetic analysis inferred from 129 amino acid characters from HMG-domain of 74 ascomycetes supported the monophyletic origin of fungi in Ophiostomatales, and Grosmannia and Ophiostoma formed a sister relationship with strong statistical support (Figure 5). Representatives of these two genera also clustered with the members of Neurospora and Sordaria in Sordariomycetes, with strong likelihood support. Within the cluster of Ophiostomatales, G. clavigera, L. longiclavatum, and L. terebrantis together with either G. aurea or L. wingfieldii formed a monophyletic clade with >90% bootstrap support. O. montium formed a sister relationship to a cluster containing O. novo-ulmi and its relatives. Sporothrix schenckii, a human pathogen, also nested within the same cluster with strong support (93%). Sequence analysis of α-box domain from 39 ascomycetes species also supported the same sister relationship between Grosmannia and Ophiostoma but the relationships among the major ascomycete families were not well resolved (Figure S3).
Figure 5 .
NJ tree generated from MEGA showing the phylogenetic relationships among ascomycetes inferred from the HMG domain (129 amino acid characters) of the MAT1-2-1. Number on branches indicated bootstrap support (1000 pseudoreplicates) more than 60% from NJ and PhyML (from left to right). The taxa shaded in gray indicate the presence of truncated MAT genes in opposite MAT isolates.
MAT genes have been useful for evaluating the phylogenetic relationships among different fungal species (Debuchy and Turgeon 2006; Devier et al. 2009). Gene genealogies of MAT1-1-2, MAT1-1-3, and MAT1-2-1 (Figure S4) were concordant with previous species relationships established based on rRNA and other protein coding genes (Lim et al. 2004). Also, phylogenetic analyses of SLA, intergenic regions upstream of full-length MAT1-1-1 and truncated MAT1-1-1, COX13, and APN, demonstrated that sequences of opposite mating-types corroborate species phylogenies rather than showing trans-specific polymorphism (Figure S5). There were no strong conflicts in tree topologies inferred from genes at and flanking the MAT locus. Also there was no conflict in topologies among various tree-building algorithms.
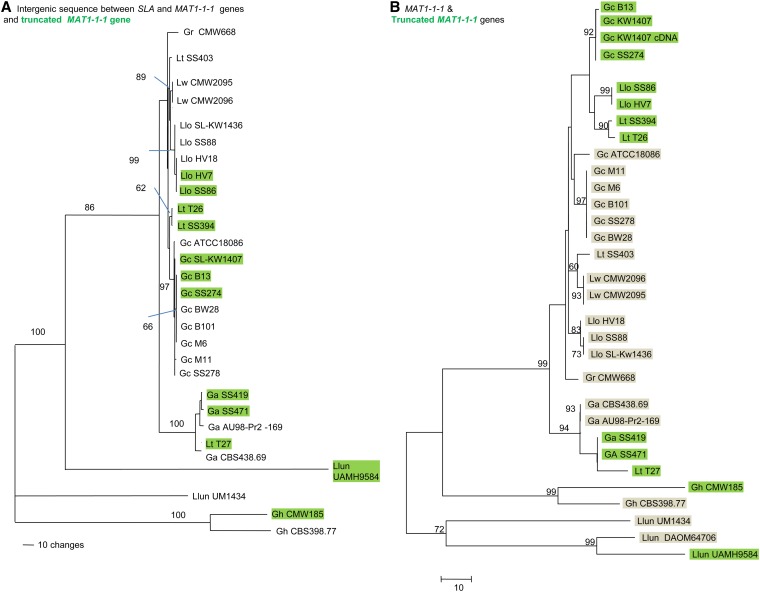
Phylogenetic analyses of the nucleotide sequences inferred that recombination occurred in the flanking sequences upstream from MAT1-1-1 and truncated MAT1-1-1 genes (Figure 6, A and B). The tree topology of flanking sequence did not conflict with that of SLA coding sequences (Figure S5) because representatives of both mating-types clustered within a species (Figure 6A). Without recombination, the flanking sequence on MAT1-1 idiomorph would be expected to cluster separately to the flanking sequence on the MAT1-2 idiomorph in the gene genealogy. The nucleotide sequence of truncated MAT1-1-1 genes diverged from the full-length MAT1-1-1 genes as a result of independent accumulation of mutations (Figure 6B). Sequences of the opposite mating-type within a species did not cluster together as for the SLA gene, but were divergent even though they had high similarity (83%) at the amino acid level.
Figure 6 .
Gene genealogies showing the phylogenetic relationships between G. clavigera and relatives. (A) Intergenic sequences after 3′ end of SLA to the 5′ end of the truncated MAT1-1-1 and MAT1-1-1 genes (1349 characters), and (B) the full-length and truncated MAT1-1-1 genes. MAT1-2 isolates are indicated in green while MAT1-1 isolates are highlighted in gray (1968 characters). Number on branches indicated bootstrap support (500 pseudoreplicates) greater than 60%.
Based on the tests of selection using likelihood ratio tests, diversifying (positive) selection was detected in the truncated MAT1-1-1 from the full-length MAT1-1-1 at the intraspecific level (Tables 3, A and B). The truncated MAT1-1-1 genes in the MAT1-2 idiomorph are under positive selection from the full-length MAT1-1-1 genes on the MAT1-1 idiomorph in G. clavigera, L. longiclavatum and G. aurea, but not L. terebrantis (Tables 3, A and B). The truncated/incomplete MAT1-1-1 gene may go through neutral evolution due to loss of function or adaptive evolution. In contrast, tests of selection on MAT genes (MAT1-2-1, MAT1-1-1, and truncated MAT1-1-1 individually) at the interspecific level revealed purifying selection (Table S3C), indicating that these genes are preserved for proper function of the sexual cycle.
Table 3. Parameter estimates and likelihood values of the various models of codon evolution using CODEML in PAML.
| Model | Model Parameters | -lnL | Models Comparison | 2ΔL | Pr. | Sites Under Positive Selection (Bayes Empirical Bayes, Pr. ω>1) | |
|---|---|---|---|---|---|---|---|
| A. Test on the truncated and full-length MAT1-1-1 genes within a species (datasets with positive selection detected) | |||||||
| G. clavigera (9 isolates) | M1a (neutral) | P0 = 0.26, P1 = 0,74 | 2172.75 | M1a vs. M2a | 17.70 | <0.001 | 2 G (0.99**), 3 I (0.99**), 8 N (0.99**) |
| M2a (selection) | P0 = 0.99, P1 = 0.00, P2 = 0.004, ω2 = 227.05 | 2163.90 | |||||
| M7 (beta) | P = 2.30, q = 0.005 | 2172.75 | M7 vs. M8 | 17.41 | <0.001 | 2 G (0.99**), 3 I (0.99**), 8 N (0.99**) | |
| M8 (beta + ω) | P0 = 0.98, P = 0.005, q = 3.27, P1 = 0.02, ω = 81.42 | 2164.05 | |||||
| G. aurea (four isolates) | M1a (neutral) | P0 = 0.77, P1 = 0.23 | 2102.86 | M1a vs. M2a | 21.18 | <0.001 | 3 A (0.94), 8 A (0.94) |
| M2a (selection) | P0 = 0.99, P1 = 0.00, P2 = 0.003, ω2 = 282.78 | 2092.27 | |||||
| M7 (beta) | P = 0.005, q = 0,012 | 2102.96 | M7 vs. M8 | 21.37 | <0.001 | 3 A (0.97*), 8 A (0.97*) | |
| M8 (beta + ω) | P0 = 0.99, P = 0.005, q = 15.47, P1 = 0.003, ω = 282.82 | 2092.27 | |||||
| L. longiclavatum (five isolates) | M1a (neutral) | P0 = 0.72, P1 = 0.28 | 2183.24 | M1a vs. M2a | 18.88 | <0.001 | 3 A (0.93), 8 A (0,93) |
| M2a (selection) | P0 = 0.99, P1 = 0.00, P2 = 0.002, ω2= 209.51 | 2173.80 | |||||
| M7 (beta) | P = 0.005, q= 0.011 | 2183.25 | M7 vs. M8 | 18.90 | <0.001 | 3 A (0.97*), 8 A (0.97*) | |
| M8 (beta + ω) | P0 = 0.99, P = 0.51, q = 1.49, P1 = 0.001, ω = 209.58 | 2173.80 | |||||
| B. Test on the truncated and full-length MAT1-1-1 genes within a species (dataset without positive selection detected) | |||||||
| L. terebrantis (4 isolates) | M1a (neutral) | 2462.37 | M1a vs. M2a | 1.30 | ns | ||
| M2a (selection) | 2461.72 | ||||||
| M7 (beta) | 2462.54 | M7 vs. M8 | 1.65 | ns | |||
| M8 (beta + ω) | 2461.72 | ||||||
Parameter estimates and likelihood values of the various models of codon evolution using CODEML in PAML. Notes for G. clavigera: Models assuming positive selection (M2a and M8) fit better the data than neutral models (M1a and M7) according to likelihood ratio tests. Model M2 assumes that 0.4% of the sites have dN/dS value = 227.05. Three sites under strong positive selection (P > 0.99) are identified by this model with Bayes empirical Bayes methods. Model M8 showed that approximately 98% of sites have dN/dS from a U-shaped beta distribution (hence, data fit strongly this model) and approximately 2% of site are under strong positive selection with dN/dS = 81.4. Both models M2 and M8 identified the same positive selection sites with Bayes empirical Bayes methods (even if model M8 would assume more sites under positive selection).
Transcript analysis of MAT genes
The MAT1-1-1 gene with α-box domain is a master regulator of sexual reproduction and is involved in gamete fertilization and the formation of ascogenous hyphae; the deletion of MAT1-1-1 gene can lead to incomplete development of perithecia in some ascomycetes (Debuchy et al. 2010). To determine whether the truncated MAT1-1-1 gene is transcribed, PCR and RT-qPCR experiments were performed on the cDNA. The MAT1-2-1 gene (Cq = 28) and truncated MAT1-1-1 gene (Cq = 32.9) were expressed during the vegetative stage in mycelia (4d culture with cellophane overlaid on MEA plates), even though the expression level was low compared with the reference β-tubulin gene (Cq = 25.5). The data were also consistent with the low level of sex gene expression reported in transcriptomic data in various terpenoid compound treatments (DiGuistini et al. 2011) (Table S4).
Determination of MAT type ratio in populations of G. clavigera and L. longiclavatum
Using specific primers targeting the α-domain and HMG domain, we detected both the MAT1-1 and MAT1-2 idiomorphs in G. clavigera. We tested the null hypothesis of balanced numbers of the two MAT idiomorphs within the epidemic population samples in Tsui et al. (2012). Both mating-types were present in the populations at all of the spatial scales. MAT1-1 and MAT1-2 frequencies did not significantly deviate from a 1:1 ratio in any of the 19 populations, the four genetic clusters inferred from Bayesian estimation during population genetic studies, nor the entire population (P < 0.05) (Table 4A). Similarly both MAT1-1 and MAT1-2 also were detected in populations of L. longiclavatum, and its mating-type ratio did not deviate significantly from 1:1 at small spatial scale (Table 4B). However the MAT ratio was significantly different at larger landscape level, where MAT1-1 isolates appeared more frequent in the Rocky Mountain (Cluster Rocky) whereas MAT1-2 isolates were predominant in the new epidemics area (Cluster North; northern British Columbia and Alberta) (Table 4B).
Table 4. MAT ratio tests on populations of Grosmannia clavigera and Leptographium longiclavatum.
| Population (Location, Province, or State) | Number Total | Clone Corrected (293 Isolates) |
||
|---|---|---|---|---|
| MAT1-1 | MAT1-2 | χ2 (P Value) | ||
| A. MAT ratio of Grosmannia clavigera population, including location and sample size (after clone correction) (*p< 0.05) | ||||
| Houston, BC | 16 | 5 | 11 | 2.25, P = 0.133 |
| Fort St. James, BC | 23 | 13 | 10 | 0.39, P = 0.532 |
| Tumbler Ridge, BC | 13 | 6 | 7 | 0.0769, P = 0.7815 |
| Fairview, BC | 11 | 4 | 7 | 0.8182, P = 0.3657 |
| Grande Prairie, AB | 20 | 8 | 12 | 0.8, 0.3722 |
| Fox Creek, AB | 12 | 5 | 7 | 0.333, 0.5637 |
| Kakwa, AB | 17 | 5 | 12 | 2.8824, 0.08956 |
| Valemount, BC | 8 | 4 | 4 | 0,1 |
| Williams Lake, BC | 15 | 4 | 11 | 3.2667, 0.0707 |
| Manning Park, BC | 19 | 10 | 9 | 0.0526, 0.8185 |
| Golden, BC | 8 | 4 | 4 | 0,1 |
| Yoho, BC | 7 | 3 | 4 | 0.1429, 0.7055 |
| Banff, AB | 20 | 11 | 9 | 0.2, 0.6547 |
| Canmore, AB | 39 | 14 | 25 | 3.1026, 0.07817 |
| Cypress Hills, AB | 5 | 2 | 3 | 0.2, 0.6547 |
| Sparwood, BC | 7 | 3 | 4 | 0.1429,0.7055 |
| Crowsnest Pass, AB | 9 | 5 | 4 | 0.1111,0.7389 |
| Hidden Valley, MT, USA | 20 | 11 | 9 | 0.2, 0.6547 |
| Hell Roaring, ID, USA | 24 | 15 | 9 | 1.5, 0.2207 |
| Total | 293 | 132 | 161 | 2.8703, 0.09023 |
| Genetic clusters inferred from Tsui et al. (2012) | ||||
| NBC | 39 | 18 | 21 | 0.231,0.63 |
| NORTH | 81 | 32 | 49 | 3.57, 0.059 |
| SBC | 34 | 14 | 20 | 1.059, 0.303 |
| ROCKY | 139 | 68 | 71 | 0.065, 0799 |
| B. MAT ratio of Leptogaphium longiclavatum populations, including locations and sample size (after clone correction) (*p<0.05) | ||||
| Canmore | 15 | 4 | 11 | 3.27, 0.07 |
| Crownsnest Pass | 6 | 2 | 4 | 0.667, 0.414 |
| Cypress Hills | 2 | 1 | 1 | 0, 1 |
| Golden | 8 | 3 | 5 | 0.5, 0.48 |
| Sparwood | 7 | 3 | 4 | 0.143, 0.705 |
| Yoho | 5 | 2 | 3 | 0.2, 0.655 |
| Cluster Rocky | 43 | 15 | 28 | 3.93, 0.047* |
| Fairview | 15 | 9 | 6 | 0.6, 0.439 |
| Fox Creek | 21 | 12 | 9 | 0.429, 0.512 |
| Grande Prairies | 29 | 19 | 10 | 2.793, 0.09 |
| Kakwa | 22 | 11 | 11 | 0, 1 |
| Tumbler Ridge | 26 | 16 | 10 | 1.385, 0.239 |
| Valemount | 7 | 4 | 3 | 0.143, 0.705 |
| Cluster North | 120 | 71 | 49 | 4.033, 0.045* |
| Total | 163 | 86 | 77 | 0.497, 0.48 |
MAT, mating type.
Discussion
The MAT locus organization indicates heterothallism
All fungal species in this study are heterothallic because they have a locus with one of the two alternative single-copy idiomorphs, MAT1-1 or MAT1-2. The organization of opposite MAT loci was highly conserved among L. longiclavatum, L. terebrantis, G. aurea, G. huntii, L. lundbergii, L. wingfieldii, and O. montium, thus supporting a common origin in Ophiostomatales (Butler 2007). Also the general SLA-MAT-APN pattern, as well as the synteny and orientation of MAT1-1-1, MAT1-1-2, and MAT1-1-3 genes on MAT1-1 idiomorph are consistent with other representatives within Sordariomycetes, such as Neurospora, and Podospora (Debuchy and Turgeon 2006). This suggests a common evolutionary origin of the MAT organization/structure for members of the Ophiostomatales and even the Sordariomycetes.
Whether heterothallism is the ancestral state in ascomycetes has been a major biological question in fungal evolution (Coppin et al. 1997; Butler 2007). Evolution from heterothallism (outcrossing) to homothallism (haploid selfing) could be the most likely scenario based on population genetics models (Nauta and Hoekstra 1992; Billiard et al. 2011). Recent analyses of mating-type evolution have ascertained Neurospora to be heterothallic in ancestry (Strandberg et al. 2010; Nygren et al. 2011; Gioti et al. 2012). We speculate that the representative of Grosmannia and Ophiostoma may have diverged from the same common heterothallic ancestor as Neurospora. However, it is important to determine the mating-type organization from additional species in Ophiostomatales because their mating systems and breeding strategies (Wilken et al. 2012) may be different from other Sordariomycetes.
Another special feature in the MAT locus is that COX13 is located between MAT and APN genes in G. clavigera, and this is similar to the organization found in the human pathogen Histoplasma capsulatum (Fraser et al. 2007). Although the MAT and APN loci were at least 15 kb apart, there was no evidence to suggest the presence of transposable elements, which have been reported in MAT locus expansion in the obligate biotroph Blumeria graminis (Spanu et al. 2010), H. capsulatum (Fraser et al. 2007) and the endophyte Phialocephala fortiniii (Zaffarano et al. 2010).
Unequal recombination defined the evolution of the truncated MAT1-1-1 genes in Grosmannia and Ophiostoma
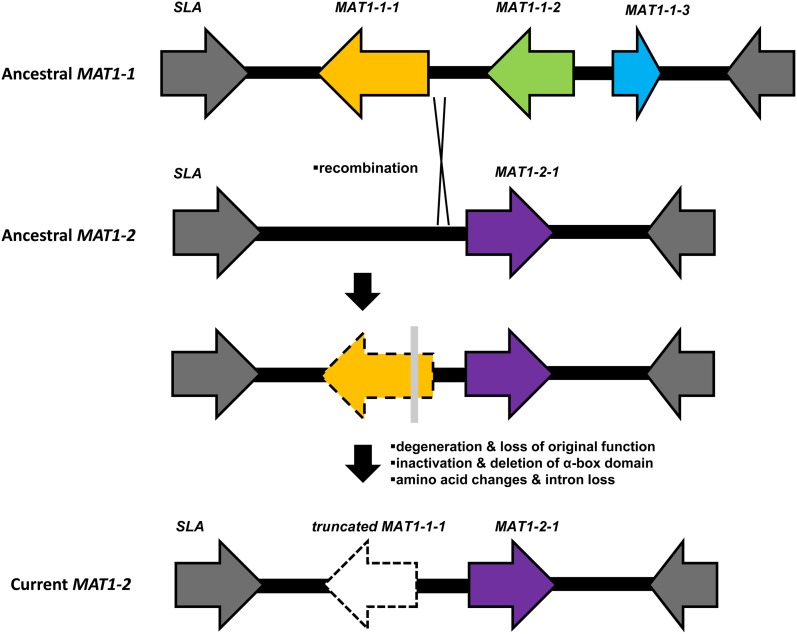
Most fungi in this investigation carried the truncated MAT1-1-1 in the MAT1-2 idiomorphs. The polymorphism between full-length and truncated MAT1-1-1 indicated that the event leading to the current MAT loci organization was ancient—before the radiation of G. clavigera from other species. A number of competing scenarios may account for the evolutionary origin of this unique organization.
Unequal recombination/crossover at the MAT locus between opposite mating-type idiomorphs during the pairing of chromosomes should be the most favorable mechanism to account for the truncated MAT1-1-1 (Gioti et al. 2012) (Figure 7). The ancestral MAT1-2 idiomorph of Grosmannia and Ophiostoma members contained one ORF corresponding to the MAT1-2-1 gene bearing the HMG domain. A fragment of the MAT1-1 idiomorph, for instance the MAT1-1-1 gene and the 5′end flanking sequence, could have become integrated into the ancestral MAT1-2 idiomorph during the crossover in sexual reproduction (Figure 7). Afterward the α-box domain had been deleted over evolutionary time (Figure 7).
Figure 7 .
Proposed model for the evolution of MAT locus in the common ancestor of Grosmannia and Ophiostoma (Ophiostomatales).
Unequal recombination at the MAT locus is not unique to members of Grosmannia and Ophiostoma. This event has clearly happened many times, and its footprints have been demonstrated in other ascomycete species (Yokoyama et al. 2003; Paoletti et al. 2005b; Seidl et al. 2009; Amselem et al. 2011; Wilken et al. 2012) (Figure 5). Fragments of the MAT1-1-1 and MAT1-1-3 genes were reported in 10 isolates of O. quercus that contained the full, complete MAT1-2-1 gene in their MAT1-2 idiomorphs (Wilken et al. 2012). Similarly, truncated MAT1-1-1 genes were identified in the MAT1-2 idiomorphs of at least five Phialocephala species (Zaffarano et al. 2010) and the one in Hypocrea jecorina (Seidl et al. 2009). In contrast, a partial MAT1-2-1 sequence was found in the MAT1-1 idiomorph of Aspergillus fumigatus (Paoletti et al. 2005b). Also fragments of homologous MAT1-2-1 and MAT1-1-1 genes were detected bordering the mating-type idiomorphs in Botrytis cinerea isolates (Amselem et al. 2011).
Recombination is supposed to be suppressed and rare at the MAT locus in ascomycetes (Idnurm 2011), but recombination or crossover events have been reported. Homologs of MAT1-1-2 and MAT1-1-3 also were reported in the MAT1-2 idiomorph of Diaporthe W- and G-type species, possibly due to recombination of idiomorphs (Kanematsu et al. 2007). The MAT1-2-2 gene in the MAT1-2 idiomorph of Magnaporthe oryzae was partially homologous to the MAT1-1-3 gene in opposite mating-types (Kanamori et al. 2007). Recombination breakpoints and unequal crossover events have been revealed in the MAT loci of Neurospora, Cochliobolus, Stemphylium, Ascochyta and Phoma during the comparison of MAT idiomorphs between heterothallic to homothallic representatives (Yun et al. 1999; Inderbitzin et al. 2005; Gioti et al. 2012; Woudenberg et al. 2012).
Acquisition of MAT genes by transposition has been independently reported in several ascomycetes (Rydholm et al. 2007; Poggeler et al. 2011; Gioti et al. 2012). Therefore, the intronless MAT1-1-1 in MAT1-2 idiomorph may have arisen by retrotransposition from the MAT1-1-1 cDNA from the ancestral MAT1-1 idiomorph, but the MAT loci do not contain transposon-related and repetitive sequences. Interspecific introgression of MAT1-1-1 gene and vegetative incompatibility genes from Ophiostoma ulmi into O. novo-ulmi has been proposed (Paoletti et al. 2006). Introgression or non-random acquisition of MAT genes was also demonstrated in the evolutionary histories among multiple Neurospora species (Menkis et al. 2010; Strandberg et al. 2010). However, the molecular data do not support introgression to account for the presence of the truncated MAT1-1-1 gene because the gene genealogies of MAT and flanking genes do not contradict the species phylogeny of Grosmannia and Ophiostoma known from the housekeeping genes (Lim et al. 2004), suggesting all these genes shared the same evolutionary history.
The occurrence of truncated mating-type genes in opposite MAT idiomorphs may suggest a common homothallic ancestor carrying a complete set of MAT genes, for instance all MAT1-1-1, MAT1-1-2, MAT1-1-3, and MAT1-2-1 located at a single locus in the same order as Sordaria macrospora [Figure 15.2 in (Debuchy and Turgeon 2006)]. The MAT1-1 and MAT1-2 idiomorphs could have arisen from the loss of HMG and α-box domain sequences as a result of multiple translocation breaks and segregations (Paoletti et al. 2005b). This scenario had been proposed to explain the MAT locus evolution in Cordyceps takamontana (Yokoyama et al. 2003), members of Botrytis and Sclerotinum (Amselem et al. 2011), as well as the Aspergilli (Galagan et al. 2005; Paoletti et al. 2005b). However, this model of evolution is very unlikely based on the criterion of parsimony. Also G. clavigera and related fungi have syntenic order of MAT genes and may share a common heterothallic ancestor within the Sordariomycetes as discussed above.
Deletion of the α-box domain due to evolutionary degeneration of MAT1-1-1 genes
The deletion/removal of the α-box domain in the truncated MAT1-1-1 gene may have been selected to avoid universal compatibility or haploid selfing (self-fertility) that involves no sexual recombination in reproduction (Billiard et al. 2011, 2012). In fact, the expression of additional α-box domain may interfere or compete with the signal from the “resident/ original” HMG domain at the same locus (Coppin et al. 1997), therefore the “additional” MAT1-1-1 gene may have been “inactivated” for functions in mating (Coppin et al. 1997) after the integration into the MAT locus as a result of unequal recombination. Under laboratory conditions, artificial association of both mating-type loci in the same nucleus of a heterothallic Neurospora crassa isolate led to inhibition of growth and the formation of barren perithecia in crosses with a tester (Perkins 1972). Transgenic dual maters (carrying genes of both mating-types) were also unable to produce progeny in isolates of N. crassa (Glass et al. 1990), Podospora anserina (Coppin and Debuchy 2000) and Cochliobolus heterostrophus (Turgeon et al. 1993).
In the absence of purifying selection, genes that have lost their original functions accumulate mutations and degenerate to become pseudogenes. Our data illustrated that the truncated MAT1-1-1 gene in O. montium is highly eroded (31% in length compared with its ‘full length’ homolog) and could be a pseudogene. Similarly, the truncated MAT1-1-1 gene reported in Cordyceps takamontana (known as Isaria tenuipes) was a pseudogene with accumulated mutations and stop codons (Yokoyama et al. 2005). The 3′-end of the MAT1-1-1 gene in the MAT1-2 idiomorph of H. jecorina was also considered to be nonfunctional because the flanking region contained multiple stop codons with no translational start point detected (Seidl et al. 2009). Also, the duplicated homeodomain transcription factors at A mating-type locus in Coprinopsis cinerea (Basidiomycota) were deleted and inactivated progressively (Kues et al. 2012). The MAT locus in yeasts was considered a “deletion hotspot” with a continued process of evolutionary deletions, gene truncation and transpositions on chromosomal genes located beside the MAT locus after the mating-type switching event (Gordon et al. 2011).
The truncated MAT1-1-1 genes in G. clavigera and relatives could have lost their original function in sexual reproduction but the degree of degeneration is not the same as the homolog in O. montium. However, these genes do not contain any stop codons and introns. The CODMEL tests indicated purifying selection at the inter-specific level due to accelerated rate of amino acid changes. The deletion of the α-box domain in the truncated MAT1-1-1 genes also did not prevent its expression as it has already been demonstrated in Magnaporthe orzyae (Kanamori et al. 2007). It is possible that the truncated genes have evolved new functions through adaptive evolution because the organization has been maintained in the entire phylogenetic clade. In contrast, the 3′-terminal truncated SXI1α gene at the MAT locus of Cryptococcus neoformans serotype-AD hybrid (Basidiomycota) is still functional in sexual reproduction and may even promote cell fusion (Lin et al. 2007). Additional in-depth molecular genetics studies and experiments are necessary to elucidate the possible biological functions of the truncated MAT1-1-1 gene.
Implications for the mating strategies and breeding systems in fungi
Fungi within the Ophiostomatales have complex mating behavior that range from strict outcrossing (heterothallism) to haploid-selfing (homothallism) (Gorton and Webber 2000; Carlier et al. 2006). Also, Ophiostoma ulmi had been thought to perform ‘pseudoselfing’ by a process involved in mutation at the MAT locus or introgressed MAT genes that led to a mating-type switch (Brasier and Gibbs 1975). Our results reflected that most members in Grosmannia are heterothallic in genetic makeup and they require a partner (outcrossing) to produce perithecia in life histories. These molecular data are largely congruent to the classical data from pairing-cultures, except that G. robusta was reported to produce perithecia readily in culture without pairing cultures (Jacobs and Wingfield 2001). Also evidence of incongruence between molecular data and classical data were reported in O. quercus, as MAT1-2-1 genes appeared to be present in both mating partners that are able to cross (Wilken et al. 2012). Previous phylogenetic data demonstrated that homothallism has evolved multiple times independently from within heterothallic ascomycetes (Yun et al. 1999; Strandberg et al. 2010; Billiard et al. 2011). Further characterization of the MAT loci from selfing species in Ophiostomatales based on classical mating studies could verify if homothallic members have been derived from a heterothallic ancestor, infer the phylogenetic relationships between the MAT locus organizations and reveal the mechanisms underlying the lifestyle changes.
Heterothallic (outcrossing) fungi gain benefits from recombination by increased genetic diversity and repaired mutation (Heitman et al. 2007). The presence of both mating-type isolates in G. clavigera at different spatial scales is consistent with high levels of genotypic diversity found in this fungus (Tsui et al. 2012). The finding of linkage equilibrium among microsatellite markers further indicated that sexual reproduction is a major factor influencing the population genetic structure and epidemiology of G. clavigera (Tsui et al. 2012). If G. clavigera undergoes a sexual cycle regularly, the ascospores and ascomata should be discovered without difficulty. It was proposed that the inability to find the ascomata in nature is due to the inappropriate sampling methodology, or collecting plant material at the wrong stage in the life cycle (Sommerhalder et al. 2006). Unfortunately, several attempts to cross complement isolates in vitro proved unsuccessful. Other possible explanations may be mutations in genes regulating the sexual development or environmental factors that reduce ascomata formation (Bennett et al. 2003).
Finally, our results revealed the presence of homologous MAT genes in Llo, Lt, Lw, and Llun, which have long been considered to be asexual (Jacobs and Wingfield 2001). We also found 1:1 mating-type ratio in L. longiclavatum populations. Evidence of purifying selection on the MAT genes at the inter-specific level indicated that sexual reproduction is important in nature or occurs regularly (López-Villavicencio et al. 2010). Our data showed that these fungi previously deemed asexual have the potential to reproduce sexually, as was demonstrated for other asexual fungi such as Penicillium, Aspergillus, and Fusarium (Butler 2007; Ropars et al. 2012). This ability potentially increases genetic variability and can enhance fitness of fungal pathogens in new, ecological niches (Coppin et al. 1997).
Supplementary Material
Acknowledgments
We are grateful to the associate editor and the anonymous reviewers for their comments on the submitted manuscript. We acknowledge Paul Dyer and Carly Eagle (University of Nottingham, UK) for technical advice on the long-range PCR experiment; Tatiana Giraud, Evelyne Coppin, and Robert Debuchy (Université Paris-Sud, France); Colette Breuil, Jim Kronstad, and Mary Berbee (University of British Columbia, Canada) for advice and discussion on experimental design and data analyses. We are also grateful to Amanda Roe, Adrianne Rice, Felix Sperling, and Janice Cooke (University of Alberta, Canada) for providing additional cultures and DNA samples. We also thank Sepideh Alamouti, H.-J. Chen, Juan Valle, Ben Lai (UBC, Canada), and Forest Health Officers from the Alberta Sustainable Resource Development Fund for technical assistance, as well as Marc-André Rodrigue (Centre de recherche du CHUQ, Québec) for assistance in primer-walking sequencing. Financial assistance for this research was provided by Genome Canada, Genome British Columbia, Genome Alberta, and the Government of Alberta (AAET/AFRI-859-G07) in support of the Tria I and Tria II Projects <http://www.thetriaproject.ca>, of which J.B. and R.C.H. are Principal Investigators. Funding was also provided by the Genomic Research and Development Initiative of Natural Resources Canada.
Footnotes
Communicating editor: J. H. McCusker
Literature Cited
- Alamouti S. M., Wang V., DiGuistini S., Six D., Bohlmann J., et al. , 2011. Gene genealogies reveal cryptic species and host preferences for the pine fungal pathogen Grosmannia clavigera. Mol. Ecol. 12: 2581–2602 [DOI] [PubMed] [Google Scholar]
- Amselem J., Cuomo C. A., van Kan J. A., Viaud M., Benito E. P., et al. , 2011. Genomic analysis of the necrotrophic fungal pathogens Sclerotinia sclerotiorum and Botrytis cinerea. PLoS Genet. 7: e1002230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett R. S., Yun S., Lee T. Y., Turgeon B. G., Arseniuk E., et al. , 2003. Identity and conservation of mating type genes in geographically diverse isolates of Phaeosphaeria nodorum. Fungal Genet. Biol. 40: 25–37 [DOI] [PubMed] [Google Scholar]
- Billiard S., López-Villavicencio M., Devier B., Hood M. E., Fairhead C., et al. , 2011. Having sex, yes, but with whom? Inferences from fungi on the evolution of anisogamy and mating types. Biol. Rev. Camb. Philos. Soc. 86: 421–442 [DOI] [PubMed] [Google Scholar]
- Billiard S., Lopez-Villavicencio M., Hood M. E., Giraud T., 2012. Sex, outcrossing and mating types: unsolved questions in fungi and beyond. J. Evol. Biol. 25: 1020–1038 [DOI] [PubMed] [Google Scholar]
- Brasier C. M., Gibbs J. N., 1975. Highly fertile form of the aggressive strain of Ceratocystis ulmi. Nature 257: 128–131 [Google Scholar]
- Butler G., 2007. The evolution of MAT: the ascomycetes, pp. 3–18 in Sex in Fungi: Molecular Determination and Evolutionary Implications, edited by Heitman J., Kronstad J. W., Taylor J. W., Casselton L. A. ASM Press, Washington, DC [Google Scholar]
- Carlier F.-X., Decock C., Jacobs K., Maraite H., 2006. Ophiostoma arduennense sp. nov. (Ophiostomatales, Ascomycota) from Fagus sylvatica in southern Belgium. Mycol. Res. 110: 801–810 [DOI] [PubMed] [Google Scholar]
- Coppin E., Debuchy R., Arnaise S., Picard M., 1997. Mating types and sexual development in filamentous ascomycetes. Microbiol. Mol. Biol. Rev. 61: 411–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppin E., Debuchy R., 2000. Co-expression of the mating-type genes involved in internuclear recognition is lethal in Podospora anserina. Genetics 155: 657–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debuchy R., Turgeon B. G., 2006. Mating-type structure, function, and evolution in Euascomycetes, pp. 293–323 in The Mycota I Growth, Differentiation, and Sexuality, edited by Kües U., Fischer R. Springer-Verlag, Berlin, Heidelberg [Google Scholar]
- Debuchy R., Berteaux-Lecellier V., Silar P., 2010. Mating systems and sexual morphogenesis in ascomycetes, pp. 501–535 in Cellular and Molecular Biology of Filamentous Fungi, edited by Borkovich K. A., Ebbole D. J. American Society for Microbiology Press, Washington, DC [Google Scholar]
- Dereeper A., Guignon V., Blanc G., Audic S., Buffet S., et al. , 2008. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 36: W465–W469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devier B., Aguileta G., Hood M. E., Giraud T., 2009. Ancient trans-specific polymorphism at pheromone receptor genes in Basidiomycetes. Genetics 181: 209–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiGuistini S., Wang Y., Liao N. Y., Taylor G., Tanguay P., et al. , 2011. Genome and transcriptome analyses of the mountain pine beetle-fungal symbiont Grosmannia clavigera. Proc. Natl. Acad. Sci. USA 108: 2504–2509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farfan L., Tsui C. K. M., El-Kassaby Y. A., Hamelin R. C., 2011. Population genetic analysis of Leptographium longiclavatum a pathogen associate with the mountain pine beetle Dendroctonus ponderosae. Phytopathology 101: S51 [Google Scholar]
- Felsenstein J., 2005. PHYLIP (Phylogeny Inference Package), version 3.6a3. Department of Genome Science. University of Washington, Seattle, WA [Google Scholar]
- Fraser J. A., Stajich J. E., Tarcha E. J., Cole G. T., Inglis D. O., et al. , 2007. Evolution of the mating type locus: Insights gained from the dimorphic primary fungal pathogens Histoplasma capsulatum, Coccidioides immitis, and Coccidioides posadasii. Eukaryot. Cell 6: 622–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galagan J. E., Calvo S. E., Cuomo C., Ma L. J., Wortman J. R., et al. , 2005. Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae. Nature 438: 1105–1115 [DOI] [PubMed] [Google Scholar]
- Gioti A., Mushegian A. A., Strandberg R., Stajich J. E., Johannesson H., 2012. Unidirectional evolutionary transitions in fungal mating systems and the role of transposable elements. Mol. Biol. Evol. 29: 3215–3226 [DOI] [PubMed] [Google Scholar]
- Glass N. L., Grotelueschen J., Metzenberg R. L., 1990. Neurospora crassa A mating-type region. Proc. Natl. Acad. Sci. USA 87: 4912–4916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon J. L., Armisén D., Proux-Wéra E., Óhéigeartaigh S. S., Byrne K. P., et al. , 2011. Evolutionary erosion of yeast sex chromosomes by mating-type switching accidents. Proc. Natl. Acad. Sci. USA 108: 20024–20029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorton C., Webber J. F., 2000. Reevaluation of the status of the bluestain fungus and bark beetle associate Ophiostoma minus. Mycologia 92: 1071–1079 [Google Scholar]
- J. Heitman, Kronstad J. W., Taylor J. W., Casselton L. A. (Editors), 2007. Sex in Fungi: Molecular Determination and Evolutionary Implications. ASM Press, Washington, DC [Google Scholar]
- Idnurm A., 2011. Sex and speciation: The paradox that non-recombining DNA promotes recombination. Fungal Biol. Rev. 25: 121–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inderbitzin P., Harkness J., Turgeon B. G., Berbee M. L., 2005. Lateral transfer of mating system in Stemphylium. Proc. Natl. Acad. Sci. USA 102: 11390–11395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobi V., Dufour J., Bouvet G. F., Aoun M., Bernier L., 2010. Identification of transcripts up-regulated in asexual and sexual fruiting bodies of the Dutch elm disease pathogen Ophiostoma novo-ulmi. Can. J. Microbiol. 58: 697–705 [DOI] [PubMed] [Google Scholar]
- Jacobs K., Wingfield M. J., 2001. Leptographium Species: Tree Pathogens, Insect Associates, and Agents of Blue-Stain. American Phytopathological Society Press, St. Paul, MN [Google Scholar]
- Joly D. L., Feau N., Tanguay P., Hamelin R. C., 2010. Comparative analysis of secreted protein evolution using expressed sequence tags from four poplar leaf rusts (Melampsora spp. ). BMC Genomics 11: 422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanamori M., Kato H., Yasuda N., Koizumi S., Peever T. L., et al. , 2007. Novel mating type-dependent transcripts at the mating type locus in Magnaporthe oryzae. Gene 403: 6–17 [DOI] [PubMed] [Google Scholar]
- Kanematsu S., Adachi Y., Ito T., 2007. Mating-type loci of heterothallic Diaporthe spp.: homologous genes are present in opposite mating-types. Curr. Genet. 52: 11–22 [DOI] [PubMed] [Google Scholar]
- Kües U., Yu Y.-D., Navarro-Gonzalez M., 2012. A mating loci in Coprinopsis cinerea differ in numbers of homeodomain transcription factor genes, p. 37 in Abstract Book of 11th European Conference on Fungal Genetics, Marburg, Germany [Google Scholar]
- Kurz W. A., Dymond C. C., Stinson G., Rampley G. J., Neilson E. T., et al. , 2008. Mountain pine beetle and forest carbon feedback to climate change. Nature 452: 987–990 [DOI] [PubMed] [Google Scholar]
- Lee S., Kim J., Breuil C., 2005. Leptographium longiclavatum sp. nov., a new species associated with the mountain pine beetle, Dendroctonus ponderosae. Mycol. Res. 109: 1162–1170 [DOI] [PubMed] [Google Scholar]
- Lee S., Hamelin R. C., Six D. L., Breuil C., 2007. Genetic diversity and the presence of two distinct groups in Ophiostoma clavigerum associated with Dendroctonus ponderosae in British Columbia and the Northern Rocky Mountains. Phytopathology 97: 1177–1185 [DOI] [PubMed] [Google Scholar]
- Lim Y. W., Alamouti S. M., Kim J., Lee S., Breuil C., 2004. Multigene phylogenies of Ophiostoma clavigerum and closely related species from bark beetle-attacked Pinus in North America. FEMS Microbiol. Lett. 237: 89–96 [DOI] [PubMed] [Google Scholar]
- Lin X., Litvintseva A. P., Nielsen K., Patel S., Floyd A., et al. , 2007. alpha AD alpha hybrids of Cryptococcus neoformans: evidence of same-sex mating in nature and hybrid fitness. PLoS Genet. 3: 1975–1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Villavicencio M., Aguileta G., Giraud T., de Vienne D. M., Lacoste S., et al. , 2010. Sex in Penicillium: Combined phylogenetic and experimental approaches. Fungal Genet. Biol. 47: 693–706 [DOI] [PubMed] [Google Scholar]
- Menkis A., Whittle C. A., Johannesson H., 2010. Gene genealogies indicate abundant gene conversions and independent evolutionary histories of the mating-type chromosomes in the evolutionary history of Neurospora tetrasperma. BMC Evol. Biol. 10: 234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzenberg R. L., Glass N. L., 1990. Mating type and mating strategies in Neurospora. Bioessays 12: 53–59 [DOI] [PubMed] [Google Scholar]
- Nauta M. J., Hoekstra H. F., 1992. Evolution of reproductive systems in filamentous ascomycetes. I. Evolution of mating types. Evolution 68: 405–410 [DOI] [PubMed] [Google Scholar]
- Nygren K., Strandberg R., Wallberg A., Nabholz B., Gustafsson T., et al. , 2011. A comprehensive phylogeny of Neurospora reveals a link between reproductive mode and molecular evolution in fungi. Mol. Phylogenet. Evol. 59: 649–663 [DOI] [PubMed] [Google Scholar]
- Paoletti M., Buck K. W., Brasier C. M., 2005a Cloning and sequence analysis of the MAT-B (MAT-2) genes from the three Dutch elm disease pathogens, Ophiostoma ulmi. Mycol. Res. 109: 983–991 [DOI] [PubMed] [Google Scholar]
- Paoletti M., Rydholm C., Schwier E. U., Anderson M. J., Szakacs G., et al. , 2005b Evidence for sexuality in the opportunistic fungal pathogen Aspergillus fumigatus. Curr. Biol. 15: 1242–1248 [DOI] [PubMed] [Google Scholar]
- Paoletti M., Buck K. W., Brasier C. M., 2006. Selective acquisition of novel mating type and vegetative incompatibility genes via interspecies gene transfer in the globally invading eukaryote Ophiostoma novo-ulmi. Mol. Ecol. 15: 249–262 [DOI] [PubMed] [Google Scholar]
- Perkins D. D., 1972. An insertional translocation in Neurospora that generates duplications heterozygous for mating type. Genetics 71: 25–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poggeler S., O’Gorman C. M., Hoff B., Kuck U., 2011. Molecular organization of the mating-type loci in the homothallic Ascomycete Eupenicillium crustaceum. Fungal Biol. 115: 615–624 [DOI] [PubMed] [Google Scholar]
- Posada D., 2006. ModelTest Server: a web-based tool for the statistical selection of models of nucleotide substitution online. Nucleic Acids Res. 34: W700–S703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut, A., 1999 Se-Al: Sequence Alignment Editor. Available at: <http://evolve.zoo.ox.ac.uk/>. Accessed: January 15, 2013.
- Roe A., Rice S. V., Coltman D. W., Cooke J. E. K., Sperling F. A. H., 2011. Comparative phylogeography, genetic differentiation and contrasting reproductive modes in three fungal symbionts of a multipartite bark beetle symbiosis. Mol. Ecol. 20: 584–600 [DOI] [PubMed] [Google Scholar]
- Ropars J., Dupont J., Fontanillas E., Rodríguez de la Vega R. C., Malagnac F., et al. , 2012. Sex in cheese: evidence for sexuality in the fungus Penicillium roqueforti. PLoS ONE 7: e49665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford K., Parkhil J., Crook J., Horsnell T., Rice P., 2000. Artemis: sequence visualization and annotation. Bioinformatics 16: 944–945 [DOI] [PubMed] [Google Scholar]
- Rydholm C., Dyer P. S., Lutzoni F., 2007. DNA sequence characterization and molecular evolution of MAT1 and MAT2 mating-type loci of the self-compatible ascomycete mold Neosartorya fischeri. Eukaryot. Cell 6: 868–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safranyik L., Carroll A. L., Régnière J., Langor D. W., Riel W. G., et al. , 2010. Potential for range expansion of mountain pine beetle into the boreal forest of North America. Canadian Entomologist 142: 415–442 [Google Scholar]
- Seidl V., Seibel C., Kubicek C. P., Schmoll M., 2009. Sexual development in the industrial workhorse Trichoderma reesei. Proc. Natl. Acad. Sci. USA 106: 13909–13914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solheim H., Krokene P., 1998. Growth and virulence of mountain pine beetle associated blue-stain fungi, Ophiostoma clavigerum and Ophiostoma montium. Can. J. Bot. 76: 561–566 [Google Scholar]
- Sommerhalder R. J., Mcdonald B. A., Zhan J., 2006. The frequencies and spatial distribution of mating types in Stagonospora nodorum are consistent with recurring sexual reproduction. Phytopathology 96: 234–239 [DOI] [PubMed] [Google Scholar]
- Spanu P. D., Abbott J. C., Amselem J., Burgis T. A., Soanes D. M., et al. , 2010. Genome expansion and gene loss in powdery mildew fungi reveal tradeoffs in extreme parasitism. Science 330: 1543–1546 [DOI] [PubMed] [Google Scholar]
- Staden R., 1996. The Staden sequence analysis package. Mol. Biotechnol. 5: 233–241 [DOI] [PubMed] [Google Scholar]
- Strandberg R., Nygren K., Menkis A., James T. Y., Wik L., Stajich J. E., et al. , 2010. Conflict between reproductive gene trees and species phylogeny among heterothallic and pseudohomothallic members of the filamentous ascomycete genus Neurospora. Fungal Genet. Biol. 47: 869–878 [DOI] [PubMed] [Google Scholar]
- Swofford D. L., 2003. PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods). Sinauer Associates, Sunderland, MA [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., et al. , 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28: 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J. D., Gibson T. J., Plewniak F., Jeanmougin F., Higgins D. G., 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25: 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsui C. K. M., Roe A. D., El-Kassaby Y. A., Rice A. V., Alamounti S. M., et al. , 2012. Population structure and migration pattern of a conifer pathogen, Grosmannia clavigera, as influenced by its symbiont, the mountain pine beetle. Mol. Ecol. 21: 71–86 [DOI] [PubMed] [Google Scholar]
- Turgeon B. G., Yoder O. C., 2000. Proposed nomenclature for Mating type genes of filamentous ascomycetes. Fungal Genet. Biol. 31: 1–5 [DOI] [PubMed] [Google Scholar]
- Turgeon B. G., Bohlmann H., Ciuffetti L. M., Christiansen S. K., Yang G., et al. , 1993. Cloning and analysis of the mating type genes from Cochliobolus heterostrophus. Mol. Gen. Genet. 238: 270–284 [DOI] [PubMed] [Google Scholar]
- Wilken P. M., Steenkamp E. T., Hall T. A., de Beer Z. W., Wingfield M. J., et al. , 2012. Both mating types in the heterothallic fungus Ophiostoma quercus contain MAT1–1 and MAT1–2 genes. Fungal Biol. 116: 427–437 [DOI] [PubMed] [Google Scholar]
- Woudenberg J. H. C., de Gruyter J., Crous P. W., Zwiers L. H., 2012. Analysis of the mating-type loci of co-occurring and phylogenetically related species of Ascochyta and Phoma. Mol. Plant Pathol. 13: 350–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaoka Y., Hiratsuka Y., Maruyama P. J., 1995. The ability of Ophiostoma clavigerum to kill mature lodgepole-pine trees. Eur. J. Forest Pathol. 25: 401–404 [Google Scholar]
- Yang Z., 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24: 1586–1591 [DOI] [PubMed] [Google Scholar]
- Yokoyama E., Yamagishi K., Hara A., Yokoyama E., Yamagishi K., et al. , 2003. Structures of the mating-type loci of Cordyceps takaomontana. Appl. Environ. Microbiol. 69: 5019–5022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama E., Yamagishi K., Hara A., 2005. Heterothallism in Cordyceps takaomontana. FEMS Microbiol. Lett. 250: 145–150 [DOI] [PubMed] [Google Scholar]
- Yun S. H., Berbee M. L., Yoder O. C., Turgeon B. G., 1999. Evolution of the fungal self-fertile reproductive life style from self-sterile ancestors. Proc. Natl. Acad. Sci. USA 96: 5592–5597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaffarano P. L., Duò A., Grünig C. R., 2010. Characterization of the mating-type (MAT) locus in the Phialocephala fortinii s. l. – Acephala applanata species complex. Fungal Genet. Biol. 47: 761–772 [DOI] [PubMed] [Google Scholar]
- Zipfel R. D., de Beer Z. W., Jacobs K., Wingfield B. D., Wingfield M. J., 2006. Multi-gene phylogenis define Ceratocystiopsis and Grosmannia distinct from Ophiostoma. Stud. Mycol. 55: 75–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.